Abstract
Nucleic acid sensing is a critical mechanism by which the immune system monitors for pathogen invasion. A set of germline-encoded innate immune receptors detect microbial DNA in various compartments of the cell, such as endosomes, the cytosol, and the nucleus. Sensing of microbial DNA through these receptors stimulates, in most cases, interferon regulatory factor-dependent type I IFN synthesis followed by JAK/STAT-dependent interferon-stimulated gene expression. In contrast, the detection of DNA in the cytosol by AIM2 assembles a macromolecular complex called the inflammasome, which unleashes the proteolytic activity of a cysteine protease caspase-1. Caspase-1 cleaves and activates the proinflammatory cytokines such as IL-1β and IL18 and a pore-forming protein, gasdermin D, which triggers pyroptosis, an inflammatory form of cell death. Research over the past decade has revealed that AIM2 plays essential roles not only in host defense against pathogens but also in inflammatory diseases, autoimmunity, and cancer in inflammasome-dependent as well as -independent manners. This review discusses the latest advancements in our understanding of AIM2 biology and its functions in health and disease.
Keywords: Inflammasome, cell death, Toll-like receptors/Pattern recognition receptors, cytokines, viral, bacterial
1. Introduction: AIM2 inflammasome as a cytosolic DNA sensor
The innate immune system is the first line of defense against infections and is critical for detecting and clearing pathogens. A broad repertoire of germline-encoded pattern recognition receptors (PRRs) enables the host to sense microbial infection by recognizing pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) generated during infection.(1) Intracellular signaling triggered by these receptors converge on common transcription factors, such as nuclear factor kappa B (NF-κB) and interferon regulatory factors (IRFs), resulting in the production of proinflammatory cytokines, chemokines, and type I interferons (IFNs) that control the infection. The strategic subcellular distribution of PRRs ensures surveillance for both extracellular and intracellular infections. Transmembrane PRRs on the surface of cells detect extracellular PAMPs and DAMPs and include Toll-like receptors (TLRs) and C-type lectin receptors (CLRs).(2, 3) Cytosolic PRRs detect intracellular PAMPs and DAMPs and include the retinoic acid-inducible gene- (RIG)-I- like receptors (RLRs), nucleotide-binding domain and leucine-rich-repeat-containing [NLR] receptors (NLRs), and absent in melanoma 2 (AIM2)-like receptor (ALRs).(4–6)
Inflammasomes are multiprotein complexes that detect a broad range of PAMPs and DAMPs, including nucleic acids, bacterial toxins, flagellin, microbial cell wall components, ATP, alum, and uric acid.(7) Canonical inflammasomes consist of a sensor protein that recognizes PAMPs, the adaptor molecule, apoptosis-associated speck-like protein containing CARD (ASC), and the effector protein caspase-1.(8) The receptor in the inflammasome complex belongs to the NLR and ALR families in most cases. Assembly of the inflammasome complex results in the activation of caspase-1 through an autoproteolysis process. Active caspase-1 then cleaves the precursor cytokines IL-1β and IL-18 into their mature active forms.(9) Additionally, active caspase-1 can induce an inflammatory form of cell death called pyroptosis by cleaving gasdermin D (GSDMD).(10) Cleavage of GSDMD results in the liberation of its N-terminal domain that forms pores in the plasma membrane, leading to osmotic imbalances, cell swelling, loss of membrane integrity, and terminal cell rupture.(11) In addition to the canonical inflammasomes, there is a noncanonical inflammasome pathway, in which caspase-11 and caspase-4 directly sense cytosolic lipopolysaccharide (LPS) from Gram-negative bacteria and their outer membrane vesicles.(12–15) Active caspase-11 and caspase-4, like caspase-1, cleave GSDMD to induce pyroptosis. During pyroptosis, IL-1β and IL-18 cytokines are released from the cell into the extracellular space, and they elicit a strong inflammatory response by activating the IL-1R/IL-18R-MyD88-NFκB axis. The DAMPs and alarmins, such as HMGB1 and IL-1α, released during pyroptosis also function to amplify the inflammatory response.(16, 17)
The AIM2 inflammasome is responsible for sensing DNA in the cytosol and is a critical host defense mechanism against bacterial and viral infections.(18) AIM2 sensing of cytosolic DNA is an important innate immune strategy since the presence of DNA in the cytosol can be indicative of pathogen invasion.(19) AIM2 was first identified as a gene that was lacking in melanoma cell lines using subtractive cDNA hybridization.(20) The function of AIM2 remained unknown for over a decade until independent laboratories discovered it as a cytosolic receptor for double-stranded DNA (dsDNA).(18, 21–23) AIM2 became the first member of the ALR family proteins to be characterized in terms of innate immune sensing. AIM2 is expressed in the cytosol, where it can directly bind to microbial as well as self-dsDNA in a sequence-independent manner.(18, 21–23). The binding of DNA to AIM2 the assembly of the AIM2 inflammasome complex (24); this results in ASC oligomerization and the subsequent recruitment of pro-caspase-1 to the inflammasome complex leading to the processing of caspase-1, pro-IL-1β, pro-IL-18, and GSDMD. Thus, the activation of this DNA sensing pathway during infection results in a strong innate immune response mediated by the release of IL-1β and IL-18 cytokines in combination with pyroptotic cell death. The AIM2-ASC platform has also been shown to activate caspase-8 leading to apoptosis.(25–27) While the ability of AIM2 to recognize DNA from multiple species of pathogens is beneficial for the host, the lack of sequence-specificity does not allow for the distinction between microbial DNA and self-DNA. Therefore, activation of AIM2 in response to self-DNA has important implications in autoimmunity. In this review, we summarize the recent advances in understanding the activation, functions, and regulations of the AIM2 inflammasome and its impact on health and disease.
2. Biochemical and structural basis of DNA sensing and inflammasome assembly by AIM2
Although AIM2 sensing of dsDNA is sequence-independent, the ability of dsDNA to effectively activate the AIM2 inflammasome is dependent on its length. Biochemical studies have shown that 70-base pairs (bp) of dsDNA is the minimum length necessary to activate the AIM2 inflammasome, but 200-bp of dsDNA allows for optimal AIM2 activation.(28) It has recently been reported that the AIM2 inflammasome assembly occurs more rapidly in the presence of longer dsDNA molecules; 300-bp dsDNA induced AIM2 polymerization into filaments significantly faster than 72-bp dsDNA.(29) Therefore, the length of dsDNA in the cytosol govern the activation of the AIM2 inflammasome and its kinetics and magnitude.
The structure of the AIM2 inflammasome has been extensively studied and is well characterized. AIM2 is a member of the ALR family of proteins, characterized by an N-terminal PYD (pyrin) domain and a C-terminal HIN (hematopoietic, interferon-inducible, and nuclear localization) domain.(30) The HIN domain is directly responsible for binding to dsDNA in the cytosol. Structural studies have revealed that the HIN domain is comprised of two OB (oligonucleotide/oligosaccharide binding) folds, which have been shown to bind to both strands of DNA.(28, 31) This explains why the AIM2 inflammasome is able to recognize dsDNA, rather than ssDNA.(22, 23, 28) Mechanistically, DNA binds to the HIN domain primarily through electrostatic interactions.(23) The OB folds contain positively charged arginine and lysine residues that associate with the negatively charged phosphates on the ribose backbone of DNA molecules.(30, 32) Additionally, minimal polar and hydrophobic interactions between the HIN domain and DNA have also been shown to contribute to AIM2 binding of DNA. Interestingly, structural analysis of the HIN domain and DNA interaction have revealed negligible contact of the HIN domain with the bases of DNA, potentially explaining sequence independent-sensing of DNA by AIM2.(30) Furthermore, multiple AIM2 molecules can bind to the same molecule of dsDNA allowing for the clustering of AIM2 molecules to initiate inflammasome formation.(33)
The PYD domain of AIM2 is responsible for recruiting the adapter protein ASC during inflammasome assembly. PYD belongs to the DD (death domain) superfamily and is distinct from CARD (caspase recruitment domain) subfamily.(34) Structural studies have revealed that the PYD domain contains a six-helix bundle and has a high affinity to associate with other PYD domains.(30, 32, 35) It was found that the AIM2 PYD domain contains a loop connecting α2 and α3 that is critical for self-association.(36) The PYD domain of AIM2 directly binds to the PYD domain of ASC. The interaction between the PYD domains of these two proteins results in the assembly of helical filaments, allowing for oligomerization of the AIM2 inflammasome.(30) In the ASC filaments, AIM2 PYD exists at only one end, while ASC PYD polymerizes to make up the remaining filament structure.(37) It is considered that AIM2 PYD functions as a nucleating platform for ASC PYD molecules to assemble and polymerize into filaments, similar to how actin nucleation factors drive actin polymerization. Currently, the exact mechanism of how DNA binding to the HIN domain leads to the recruitment of ASC via the PYD domain is unclear. According to one model, AIM2 exists in an autoinhibitory conformation during homeostatic conditions.(30) In this model, the HIN domain and PYD domain interact with each other to form an inhibitory state, in which the PYD domain is not available to recruit ASC. Once dsDNA binds to the HIN domain, a conformational change is thought to occur as a result of which the PYD domain gets displaced from the HIN domain and becomes accessible to the PYD domain of ASC.(28) A second model suggests an AIM2 concentration-dependent and oligomerization-driven mechanism for the AIM2-ASC assembly.(38)
ASC functions as a vital adaptor molecule to recruit the effector protein, caspase-1, to the inflammasome complex. In addition to the PYD domain, ASC contains a CARD domain that directly interacts with the CARD domain of caspase-1.(30) Electron micrographs have revealed that the AIM2 inflammasome forms a star-shaped ternary complex, in which ASC molecules were found to localize at the center, and caspase-1 CARD molecules were found along the arms of the stars.(32, 37) The formation of the AIM2 inflammasome allows caspase-1 molecules to be in close proximity to each other, resulting in the autocatalytic generation of active caspase-1 molecules.
3. AIM2 sensing of microbial DNA and its role in host defense
Anti-bacterial host defense
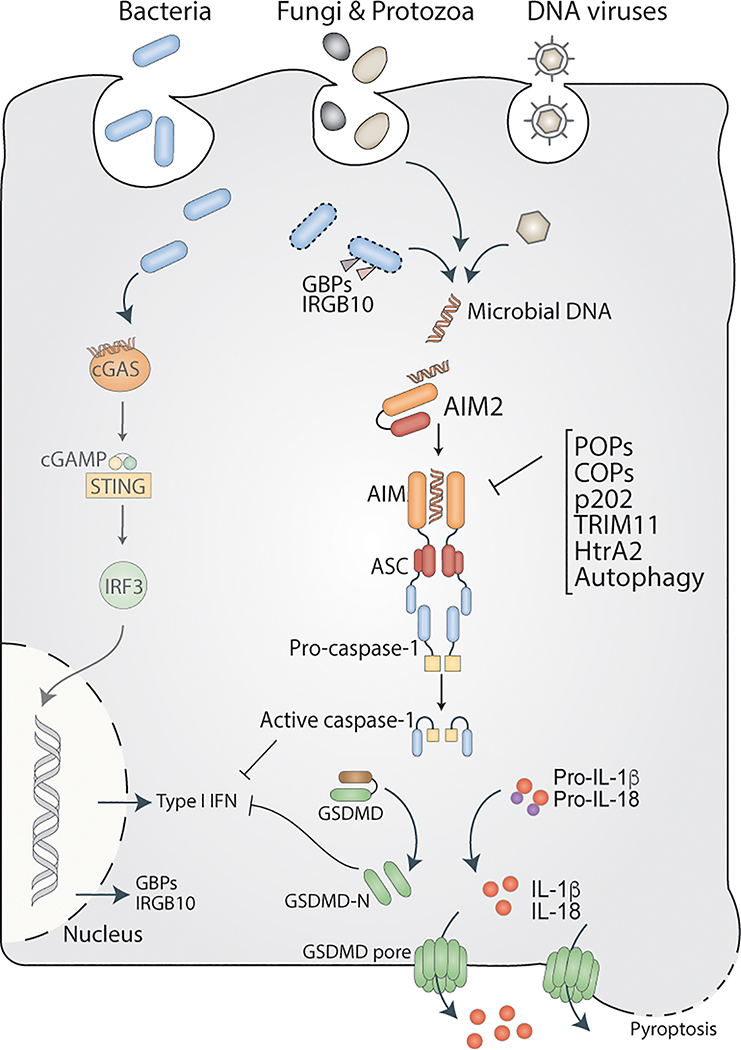
The AIM2 inflammasome sensing of microbial DNA alarms the innate immune system against pathogen invasion, and the antimicrobial host defense is a key function of the AIM2 inflammasome. AIM2 plays a protective role against various intracellular as well as extracellular bacterial infections. Listeria and Francisella are intracellular bacteria that successfully replicate in the cytosol of macrophages and dendritic cells (DCs), and AIM2 senses their DNAs released into the cytosol upon bacterial lysis (Figure 1). Correspondingly, mice deficient in Aim2 fail to mount inflammasome responses, clear bacteria, and survive F. novicida infection. However, despite detecting Listeria DNA in the cytosol, AIM2 inflammasome is only partially involved in inducing inflammasome response against this cytosolic bacterium.(39) The AIM2 inflammasome also plays a role in Mycobacterial infection; Aim2−/− mice were more susceptible to Mycobacteria tuberculosis (Mtb) infection compared to WT mice.(40) Interestingly, Mtb inhibits the inflammasome activation by M. smegmatis in DCs suggesting a dual effect of Mtb on the AIM2 inflammasome6. Along with the NLRP3 inflammasome, the AIM2 inflammasome plays a role in the control of bacterial replication during Brucella abortus infection.(41, 42) However, B. abortus-induced liver granuloma formation was not affected by the absence of ASC-inflammasome pathways.(42) AIM2 also mediates inflammasome activation by M. bovis, Porphyromonas gingivalis, Legionella pneumophila ΔSdhA, Chlamydia muridarum, and C. trachomatis in macrophages in vitro.(43–45). Furthermore, the AIM2 inflammasome prevents dysbiosis and intestinal inflammation through the regulation of the IL-18/IL-22BP/IL-22 and STAT3 pathway and the expression of antimicrobial peptides Reg3β and Reg3γ15.(46, 47)
Figure 1. AIM2 sensing of microbial DNA.
During infection with bacterial, viral, fungal, and protozoan pathogens, microbial DNA gains access to the cytosol, where AIM2 senses it. Interferon inducible proteins such as GBPs and IRGB10 target and permeabilize the bacterial membrane leading to the release of DNA into the cytosol. Upon sensing DNA in the cytosol, AIM2 assembles an ASC-caspase-1-inflammasome, which activates IL-1, IL18, as well as GSDMD. The N terminal fragment of GSDMD (GSDMD-N) forms pores on the plasma membrane, causing a lytic form of cell death. cGAS also detects viral and bacterial DNA in the cytosol leading to the activation of the STING-IRF3 signaling to stimulate type I IFN synthesis. Caspase-1 and GSDMD activated by the AIM2 inflammasome inhibit cGAS, thereby suppress type I IFN responses to cytosolic DNA (For the sake of simplicity, the lines depicting inhibition are drawn from caspase-1 and GSDMD-N to type I IFN instead of cGAS). AIM2 inflammasome activation is negatively regulated by several mechanisms involving POPs, COPs, and autophagy.
Besides sensing intracellular bacteria, AIM2 also senses infections with certain extracellular bacteria such as Staphylococcus aureus and Streptococcus pneumoniae.(48–50) AIM2 protects mice against acute central nervous system (CNS) infection with S. aureus; Aim2−/− mice have decreased levels of IL‐1β and other key inflammatory mediators, including IL‐6, CXCL1, CXCL10, and CCL2 in the CNS and were more susceptible to S. aureus infection.(50) This phenotype was similar to that of Asc−/− mice. Unlike Aim2−/− and Asc−/− mice, Nlrp3−/− mice were comparable to WT mice in their resistance to S. aureus CNS infection. During S. pneumoniae infection, AIM2 inflammasome is necessary for caspase-1 and IL-1β maturation in macrophages.(49) Similarly, the secretion of IL-1β, but not IL-12, TNF, and IL-6 into broncho-alveolar lavage (BAL) fluid was reduced in Aim2−/− mice infected intranasally with S. pneumoniae. In correspondence with this, S. pneumoniae load, pulmonary pathology, and lethality were higher in Aim2−/− mice. As S. pneumoniae and S. aureus are primarily extracellular bacteria, it is not yet clear how their DNA accesses the cytosol.
Anti-fungal and -protozoan host defense
AIM2 cooperates with NLRP3 to protect the host against infection with Aspergillus fumigatus. Whereas Aim2−/− Nlrp3−/−, Asc−/−, and Casp1−/− mice were significantly more susceptible to A. fumigatus infection-mediated tissue damage and -lethality, Aim2−/− and Nlrp3−/− were comparatively less susceptible, and their mortality rates were similar to WT mice.(51) Though the cooperative function of AIM2 and NLRP3 inflammasomes during A. fumigatus infection is clear, the underlying mechanisms remain to be elucidated. A similar dual requirement for AIM2 and NLRP3 has been observed for the inflammasome activation by Plasmodium berghei. It has been shown that the stimulation of macrophages with P. berghei-infected RBCs induces the activation of AIM2 and NLRP3 inflammasomes in macrophages.(52) Particularly, Plasmodium DNA-hemazoin (Hz) complexes were immunostimulatory for AIM2 and NLRP3 inflammasomes in macrophages. Hz-mediated phagolysosomal destabilization is considered to activate the Nlrp3 inflammasome as well as facilitate the release of P. berghei DNA into the cytosol for AIM2 sensing. Aim2 and Nlrp3 inflammasomes have been shown to impair anti-malarial host defense as mice deficient in Aim2, Nlrp3, and caspase-1 have lower parasitemia and lethality during P. yoelii infection.(53) Increased expression of AIM2 has been reported in localized cutaneous leishmaniasis (LCL) biopsies. In a murine model of leishmaniasis, Aim2 has been shown to suppress pathogen control and promote tissue inflammation.(54) Toxoplasma gondii has also been reported to activate the AIM2 inflammasome, interestingly the outcome of which is an atypical caspase-8-dependent apoptosis rather than caspase-1-dependent pyroptosis.(27)
Antiviral host defense
AIM2 inflammasome plays a vital role during DNA viral infections. Murine cytomegalovirus (MCMV), a herpesvirus of the subfamily beta-Herpesviridae, is sensed by AIM2 in macrophages and DCs leading to caspase-1-dependent IL-1 responses (Figure 1). Aim2−/− mice infected with MCMV contain reduced amounts of IL-18 in serum compared to WT mice, and consistent with that, IL-18-dependent NK cell activation in the spleen was also reduced in Aim2−/− mice. Importantly, Aim2−/− mice were more susceptible to MCMV infection and failed to control viral replication.(39) Similarly, UV-inactivated human cytomegalovirus (HCMV) infection induces AIM2- and caspase-1-dependent IL-1β secretion in THP1 cells.(55) However, UV-inactivated HCMV activation of the AIM2 inflammasome is suppressed by live HCMV infection. (55) Furthermore, AIM2 senses vaccinia virus, a double-stranded DNA virus, and AIM2 deficiency abrogates inflammasome-mediated response to vaccinia viral infection in macrophages and DCs in vitro.(39) Hepatitis B virus (HBV) is an enveloped virus but contains a smaller genome (~3 kb). Detailed profiling of AIM2 expression and inflammasome responses in patients with acute and chronic HBV infections revealed that AIM2 expression was higher in acute HBV patients and that AIM2 expression positively correlated with serum level of IL-1β and IL-18, but negatively correlated with serum HBV titer.(56) Similarly, higher AIM2 expression was observed in the kidney of patients with HBV-induced chronic glomerulonephritis (HBV-GN) than with HBV negative chronic glomerulonephritis. Here also, AIM2 expression positively correlated with caspase-1 and IL-1β levels and negatively correlated with the viral load.(57) Although these studies implicate a role for AIM2 in HBV infection, whether AIM2 senses HBV DNA and its precise role during HBV infection are not yet known. Human Papilloma Virus (HPV) is a dsDNA virus with a small genome (~8 kb). HPV16 and 18 are the high-risk type strains that commonly cause genital, anal, or skin cancers. HPV-positive skin cancer lesions contain high amounts of cytosolic AIM2 as well as HPV16 DNA. Further immunohistochemical analysis revealed the presence of cleaved caspase-1 and IL-1β in HPV-infected lesions, but not in healthy control skin.(58) The transfection of HPV16 DNA induced the secretion of IL-1β from keratinocytes in an AIM2 inflammasome-dependent manner.(58) However, whether HPV directly activates the AIM2 inflammasome and its functional relevance during HPV infection in the skin are unclear. AIM2 has also been shown to mediate inflammasome activation by Epstein‐Barr virus and modified vaccinia virus Ankara in primary human monocytes and keratinocytes, respectively.(59, 60) Interestingly, AIM2 has been documented to play roles during infections with RNA viruses, such as influenza, Enterovirus 71 (EV 71), Chikungunya virus, and West Nile virus.(61, 62) How AIM2 is engaged during RNA viral infections and whether self-DNA is the ligand for AIM2 in these scenarios are not clear.
4. AIM2 sensing of self-DNA and its functional implications
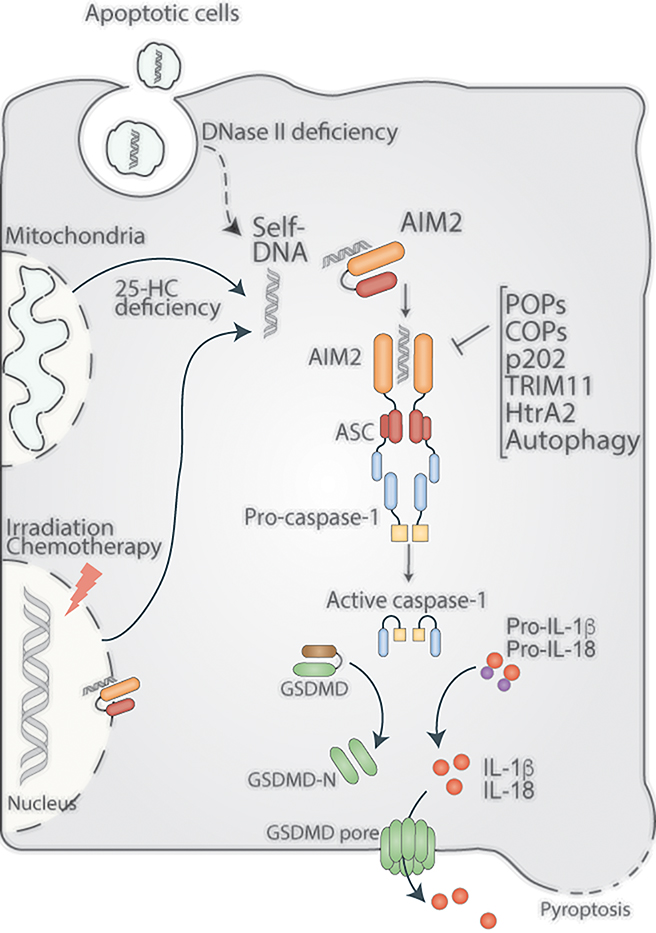
The lack of sequence specificity for AIM2 binding of DNA makes self-DNA a potential ligand for AIM2. Though the spatial separation of AIM2 in the cytosol and self-DNA in the nucleus and mitochondria prevents AIM2 sensing of self-DNA, there are several scenarios where self-DNA mislocalizes in the cytosol and is sensed by AIM2 with pathological consequences (Figure 2). The nuclear envelope (NE) serves as a physical barrier shielding the nuclear DNA from the innate immune surveillance pathways in the cytosol. Thus, NE plays a vital role in preventing self-DNA mediated aberrant innate immune activation. Impaired maturation of NE proteins could compromise NE integrity and thereby facilitate DNA leakage into the cytosol. Nelfinavir, an anti-HIV drug that limits viral replication by inhibiting aspartyl protease, prevents maturation of lamin A and facilitates the disintegration of NE. Following NE disintegration, genomic DNA leaks into the cytosol and is recognized by AIM2, leading to the activation of caspase-1 and IL-1β in THP1 cells and BMDMs.(63) Similarly, the impairment of mitochondrial structure and function results in the spilling of mitochondrial DNA into the cytosol; for example, the excessive accumulation of cholesterol owing to the inadequate cellular levels of 25-hydroxycholesterol leads to the impaired mitochondrial respiration and membrane polarization. As a result of which, mitochondrial DNA is released into the cytosol leading to AIM2 inflammasome activation.(64) Importantly, self-DNA serves as an AIM2 agonist in neurons during development and injury.(65) Previous studies indicated that AIM2 deficiency altered neuronal morphology and influenced behavior in mice.(66) A recent study shows the activation of AIM2 inflammasome in the developing brain in response to double-stranded DNA damage. AIM2 inflammasome activation in response to genotoxic stress during neurodevelopment contributes to normal brain development by purging unfit neurons via GSDMD-mediated pyroptosis. As a result, AIM2 deficiency leads to the retention of more cells (damaged as well as normal) in the brain and the development of anxiety-related behaviors in mice.(67)
Figure 2. AIM2 sensing of self-DNA.
The compartmentalization of cell-intrinsic DNA in the nucleus and mitochondria and cell-extrinsic DNA (derived from apoptotic cells) in lysosomes prevents cytosolic AIM2 from sensing self-DNA. Nonetheless, irradiation- and chemotherapy-induced nuclear DNA damage leads to the sensing of the nuclear DNA of AIM2. DNase II and 25-hydroxycholesterol (25-HC) deficiencies enable cytosolic mislocalization of self-DNA from lysosomes and mitochondria, respectively, triggering AIM2 activation. AIM2 sensing of self-DNA and the resulting inflammasome responses play critical roles in autoimmunity, chemotherapy-induced toxicity, as well as in neurodevelopment.
In addition to cell-intrinsic DNA, cell-extrinsic DNA can also become mislocalized in the cytosol, thus becoming a ligand for AIM2. Phagocytic cells, such as macrophages, internalize cell-free DNA released upon cellular damage, self-DNA-immune complexes, and apoptotic bodies containing DNA. This DNA cargo is routed to lysosomes, where it undergoes degradation by DNase II. However, defects in lysosome maturation and Dnase II deficiency lead to defective clearance of apoptotic bodies and accumulation of self-DNA in the lysosomes, which eventually leaks into the cytosol.(68) Aberrant AIM2 activation resulting from defective clearance of DNA-containing cargoes has been implicated in autoimmune disorders such as systemic lupus erythematosus and arthritis, as discussed later. Cellular damage caused by flu infection has been reported to trigger the release of DNA in the lungs, which is sensed by AIM2.(69) The self-DNA sensing by AIM2 plays critical roles in cytokine responses, immune cell recruitment, and mortality during flu infection depending upon the dose of the viral infection.(70)
Surprisingly, self-DNA is recognized by the AIM2 inflammasome in the nucleus also (71, 72). AIM2 has been shown to sense DNA in the nucleus upon radiation or chemotherapy induced-double-strand DNA breaks. The resulting inflammasome activation triggers pyroptosis in intestinal epithelial cells and bone marrow cells and contributes to the radiation-induced intestinal toxicity as well as hematopoietic failure.(71) Additionally, the pharmacological blockade of the translocation and assembly of AIM2 inflammasome in the nucleus by andrographolide alleviates pyroptosis and organ damage during radiotherapy.(72) Furthermore, chemotherapy induces the release of massive amounts of DNA from the damaged gastrointestinal lining through exosomes. These DNA-containing exosomes are taken up by immune cells and activate the AIM2 inflammasome leading to the release of IL-1β and IL-18.(73) During the early phase of chemotherapy, these proinflammatory cytokines could trigger the anti-tumor immunity by inducing the infiltration of immune cells into the tumor. However, uncontrolled inflammasome activation leads to gastrointestinal toxicity and diarrhea. The genetic deficiency of AIM2 and the pharmacological inhibition of AIM2 by thalidomide significantly reduced chemotherapy-induced diarrhea in mice.(73)
Autoimmune and Sterile inflammatory diseases.
The differential expression of AIM2 has been observed in several autoimmune diseases such as arthritis, psoriasis, atopic dermatitis, venous ulcer, contact dermatitis, colitis, Sjogren’s syndrome (SS), and systemic lupus erythematosus (SLE) in humans.(74, 75) Accumulation of extranuclear DNA, high expression of AIM2, and ASC specks have been observed in the ductal epithelia in SS patients.(76) Additionally, damaged genomic DNA co-localized with AIM2 in the specimens of SS patients but not controls. The low Dnase I expression in the salivary gland epithelial cell-line (SGEC) lines and the ductal tissues of SS patients implied that the defective cytosolic DNA degradation underlies AIM2 activation in SS. Similarly, increased expression of AIM2 and the cytosolic presence of DNA were observed in keratinocytes in psoriatic lesions but not in healthy skin. Cytosolic DNA is capable of activating the AIM2 inflammasome in keratinocytes. The antimicrobial peptide LL37, which is highly expressed in the psoriatic lesions, sequesters DNA and restricts AIM2 activation in keratinocytes. It can be inferred from these observations that AIM2 senses self-DNA during psoriasis, and that this surveillance process is suppressed by LL-37(77). Dnase II deficiency leads to lethal accumulation of self-DNA during embryonic stages and type I IFN-driven embryonic lethality.(78) The deletion of type I IFN receptor in Dnase II−/− mice rescues embryonic lethality, but Ifnar−/− Dnase II−/− adult mice develop polyarthritis.(78–80) AIM2 inflammasome appears to play a critical role in the induction of polyarthritis. Increased accumulation of cleaved caspase-1 and IL-1β were observed in the joints of Ifnar−/− Dnase II−/− animals.(79) Importantly, AIM2 deficiency ameliorated arthritis characterized by reduced macrophage infiltration, joint inflammation, bone and cartilage destruction, and enhanced proliferation of synovial cells.(79) Furthermore, high expression of AIM2 is noticed in the kidney biopsy samples of diabetic and non-diabetic chronic kidney disease (CKD) patients. Proinflammatory actions of inflammasome-activated macrophages in kidney tubules cause local inflammation and tissue damage.(81) Similarly, Aim2 deficiency ameliorated renal injury in the unilateral ureteral obstruction model.(81) The AIM2 inflammasome also plays detrimental roles in several types of brain injury such as ischemic brain injury, cerebral ischemic reperfusion injury, subarachnoid hemorrhagia, and traumatic brain injury.(82–85)
Additional inflammatory roles of AIM2 have been reported in vascular diseases such as atherosclerosis and cardiomyopathy.(86) Atherosclerosis-prone ApoE−/− mice fed a high fat diet showed enhanced expression of AIM2 in vascular smooth muscle cells (VSMCs), which correlated with high ICAM-1 expression in these cells. AIM2 expression in VSMCs gradually increases with the increasing concentration of oxidized-LDL. Increased extracellular accumulation of dsDNA was observed during the advanced stages of atherosclerosis.(87) AIM2 silencing reduces cell death, macrophage accumulation in atherosclerotic plaques, and the size of atherosclerotic lesions in high fat diet-fed ApoE−/− mice.(86) AIM2 colocalizes with DNA in macrophages in the atherosclerotic plaques, and the atherosclerotic plaques in Aim2−/−ApoE−/− have decreased IL-1β and IL-18.(87) The deletion of Aim2 or inhibition of Aim2 with synthetic oligonucleotide A151 in high fat diet-fed ApoE−/− mice improved atherosclerotic lesion stability.(87) Similarly, the silencing of Aim2 with siRNA in a diabetic rat model ameliorated diabetes associated cardiomyopathy.(88) The AIM2 inflammasome has also been suggested to play a role in post-operative ileus.(89)
Cancer
AIM2 plays critical roles in oncogenesis in inflammasome-dependent as well as inflammasome-independent manners. The inflammasome-independent function of AIM2 in colorectal carcinoma is described elsewhere in the review. Depending upon the type of cancer, AIM2 plays pro- or anti-cancerous roles; for example, AIM2 plays a pro-oncogenic role in skin carcinomas, however, anti-cancerous role in colonic sarcomas. High expression of AIM2 is observed in non-small cell lung cancer (NSCLC), and AIM2 plays both inflammasome-dependent and independent roles in NSCLC.(90–92) In an in vitro study with lung cancer epithelial cell-lines A549 and H460, depletion of AIM2 and ASC with shRNA and siRNA, respectively, as well as inhibition of caspase-1 with VX765, inhibited cell-growth and metastasis. Furthermore, the depletion of AIM2 and ASC and the inhibition of caspase-1 promoted cell-cycle arrest at G2/M phase, which paralleled with cyclin B1 reduction in these cell lines.(90) Similarly, luteolin (a natural flavonoid) treatment reduced the expression of AIM2, and the activation of caspase-1 and IL-1β in A549 and H460 cell lines as well as in their xenografts in mice. Furthermore, luteolin depleted cyclin B1 and promoted growth arrest at the G2/M phase in these cell-lines.(92) These observations suggest a likely involvement of the AIM2 inflammasome in NSCLC. In HBV-infected hepatocellular carcinoma (HCC) patients, low AIM2 expression was associated with a shorter overall and disease‐free survival.(93) Diminished expression of AIM2 was observed in liver cancer tissue, but not in normal tissue of HCC patients and AIM2 expression negatively correlated with tumor progression in HCC patients.(93, 94) AIM2 deficiency enhanced Fibronectin-1 expression and epithelial‐mesenchymal transition (EMT), which may have implications for HCC metastasis.(93) In HCC cells, the AIM2 inflammasome suppresses the HCC growth by inhibiting the mTOR/S6K1 pathway.(94) The high expression of AIM2 and its inflammasome components correlated with improved patient survival in EBV induced nasopharyngeal carcinoma (NPC).(95) The AIM2 function in NPC likely involves IL-1β and the recruitment of immunostimulatory neutrophils into tumor mass that could mediate the anti-tumor activities. Although, differential expression of AIM2 and AIM2-inflammasome components are seen in additional cancers such as glioblastoma multiforme and prostate cancer in comparison to healthy tissue, its physiological relevance is not yet known (96, 97).
5. Crosstalk between the AIM2 inflammasome and other DNA sensing pathways
Additional to AIM2, cyclic GMP–AMP synthase (cGAS) senses intracellular DNA. In contrast to AIM2, cGAS is a nucleotidyl transferase that synthesizes the second messenger, cyclic GMP–AMP (cGAMP), which triggers the STING-TBK1 signaling cascade to activate type I IFN expression. A surprising observation that emerged from the earlier loss-of-function studies on AIM2 was that while the DNA-induced secretion of IL-1β was diminished, the production of IFN-β was enhanced.(18, 39, 98–102) The negative effect of AIM2 on cGAS pathway was inflammasome-dependent, and recently, GSDMD activated by the AIM2 inflammasome has been shown to suppress the cGAS-STING pathway by altering the intracellular ionic milieu optimal for cGAS activation (Figure 1).(103) This is reminiscent of GSDMD-activation of the NLRP3 inflammasome during noncanonical inflammasome signaling elicited by cytosolic LPS.(104) It has been shown by previous studies that type I IFNs play a detrimental role during infections with intracellular bacteria including F. novicida. By suppressing cGAS-mediated IFN response to F. novicida, the AIM2 inflammasome limits the detrimental activation of apoptotic caspases and enhances the host survival. Thus, the suppression of IFN-β via GSDMD is integral to the role of AIM2 in anti-bacterial host defense.
Furthermore, caspase-1 activated by the AIM2 inflammasome has also been shown to cleave and inactivate cGAS leading to the downregulation of type I IFN expression during DNA virus infections (Figure 1).(102) Similarly, inflammasome-activated GSDMD inhibits cGAS-dependent IFN response during Rickettsia infection as well.(105) However, as ISGs are important for the restriction of Rickettsia and viruses, the inflammasome antagonism of IFN is beneficial for the pathogen rather than the host.(102, 105) The inflammasome activation during Plasmodium infection also negatively regulates type I interferon signaling via IL-1β, which impairs parasite control and host survival. (53) The cross-talk between inflammasome and cGAS occurs not only during infections but sterile inflammation as well; GSDMD deficiency augments cGAS-dependent type I IFN response and worsens intestinal inflammation in a DSS colitis model.(106) Overall, these studies clearly establish a crosstalk between the inflammasome and type I interferon pathways and its critical role in a broad range of host-pathogen interactions as well as sterile inflammation.
Regulation of AIM2 inflammasome
Positive regulation by interferon inducible factors
Type I IFNs are key cytokines induced upon cytosolic sensing of DNA by cGAS. Interferons play critical roles in AIM2 inflammasome signaling via two mechanisms. First, type I IFN signaling upregulates AIM2 expression; AIM2 is expressed at basal levels during homeostatic conditions particularly in human cells, but can be upregulated by type I IFN signaling.(6) Secondly, type I IFN signaling has been shown to promote bacterial lysis and ligand availability during bacterial infections.(39, 99) For AIM2 inflammasome activation during bacterial infections, it is imperative that bacterial DNA becomes accessible to AIM2, and published reports indicate that this process is coordinated by a number of interferon-inducible host proteins including guanylate-binding proteins (GBP).(107, 108) GBPs induced by type I IFN signaling and IRF1 are targeted to cytosolic F. novicida and cause bacterial lysis leading to the release of bacterial DNA into the cytosol and AIM2 activation. Similarly, interferon response gene B10 (IRGB10) was found to associate with GBP2 and GBP5 and disrupt membrane integrity of cytosolic F. novicida and mediate liberation of bacterial DNA for AIM2 recognition.(109) Correspondingly, mice lacking GBPs or IRGB10 fail to mount adequate AIM2-dependent inflammasome responses to F. novicida and succumb to the infection. Thus, interferon inducible factors such as GBPs and IRGB10 enable AIM2 detection of bacterial DNA by enhancing ligand accessibility (Figure 1). Surprisingly, type I IFN signaling is dispensable for the activation of AIM2 inflammasome by viruses, such as MCMV,(39) perhaps due to DNA being more readily accessible than during bacterial infections.
Negative Regulatory mechanisms controlling the AIM2 inflammasome
AIM2 is not a sequence specific DNA sensor and in fact, AIM2 detection of self-DNA has been reported in several autoimmune disorders.(74) Therefore, strict regulatory mechanisms are required to avoid inadvertent activation of the AIM2 inflammasome by the self-DNA as well as to avoid its excessive activation by microbial DNA, which could also be harmful to the host. Cytoplasmic localization of AIM2 versus the nuclear and mitochondrial localization of self-DNA ensures the spatial separation of the sensor from the endogenous ligand. Further, in resting cells, AIM2 is believed to exist in an auto-inhibitory conformation with intramolecular interactions between the PYD and HIN domains preventing auto-oligomerization.(28, 110) The presence of dsDNA possibly reduces the threshold required for AIM2 oligomerization and liberates the PYD domain to interact with ASC and initiate inflammasome activation.
Negative regulation of the AIM2 inflammasome by POP, COP, and other ALR proteins.
A new mechanism of inflammasome regulation involving decoy proteins encoding PYRIN only domains has been elucidated.(111, 112) Pyrin only proteins (POPs) are believed to act as decoys and sequester key inflammasome components like NLRs, ALRs, and ASC from undergoing productive inflammasome assembly. Interestingly, their expression is undetectable basally and induced by IL-1β and IFN-β suggesting a feedback mechanism that likely functions to attenuate excessive inflammasome activation. Three cellular POPs have been identified in humans including POP1, POP2, and POP3.(113–115) Although the mouse genome is predicted to encode two POPs, Pydc3, and Pydc4, POPs are poorly conserved in mammals and share weak homology with each other. POP1 and POP2 share a high degree of similarity with the PYD domain of ASC and are believed to generally inhibit inflammasome activation through their ability to sequester ASC from associating with NLRs and ALRs.(116, 117) Interestingly, POP2 regulates inflammasome pathway in a dual way, directly as well as indirectly via the NF-κB pathway to interfere with the priming phase of inflammasome activation.(116, 118) Transgenic mouse models expressing these POP proteins validate their function as inflammasome inhibitors; POP1 and POP2 transgenic mice have reduced systemic IL-1β and IL-18, inflammatory cell recruitment, and lethality during endotoxin shock.
Unlike POP1 and POP2, POP3 is reported to be a specific regulator for AIM2 inflammasome.(115, 119) POP3 is located on the same IFN-β inducible genomic cluster as AIM2. Moreover, it shares 61% identity with AIM2 and is believed to have arisen from the duplication of the gene segment encoding the PYD domain of AIM2. POP3 co-localizes with AIM2 upon viral infection and is considered to abrogate AIM2-ASC interaction by binding to the PYD of AIM2. Consistently, silencing of POP3 in human macrophages enhanced IL-1β and IL-18 secretion upon AIM2 activation. Macrophages from transgenic mice expressing human POP3 display impaired AIM2 activation in response to dsDNA and DNA viral infection. The AIM2 inflammasome is a critical mediator of the protection against MCMV infection. In accordance with this, POP3-transgenic mice displayed heightened susceptibility to MCMV infection and harbored higher viral load in the spleen, further corroborating POP3 as AIM2 inflammasome inhibitor.(115)
In addition to POPs, there is another set of decoy proteins expressing only the CARD domain called COPs. Three COPs have been identified so far, COP (pseudo-ICE), INCA, and ICEBERG, which are all homologs of caspase-1 and predicted to have arisen from caspase-1 gene duplication.(120–123) Like POPs, the expression of COPs is upregulated by proinflammatory stimuli like IFN-γ and TNF, and COPs are widely expressed in most tissues like placenta, spleen, bone marrow, and lymph nodes. As in the case of POPs, the mouse genome does not encode for any orthologues of COPs. The function of COPs has been explored only in over-expression systems; COPs interact with the CARD of pro-caspase-1 to block its interaction with upstream inflammasome components and activation. As a general inhibitor of caspase-1 activation, COPs are likely to negatively regulate AIM2-dependent inflammasome responses to DNA.
The mouse genome harbors additional PYHIN genes, of which, IFI202/p202 acts as a suppressor of AIM2 inflammasome signaling. It was initially identified as a disease locus for lupus susceptibility.(124) p202 encodes two tandem HIN domains homologous to those of IFI16, but no PYD domain. The HIN A domain of p202 binds to dsDNA with high affinity, thus sequestering DNA, and its HIN B domain binds to AIM2 HIN domain, making it unavailable for DNA binding.(125, 126) Recently, Wang et al., identified IFI16-β, an isoform of IFI16, which has a similar domain structure as mouse p202.(127) IFI16-β is shown to similarly sequester cytoplasmic dsDNA and inhibit AIM2 inflammasome activation.
Additional mechanisms.
Autophagy is a highly conserved basic cellular process that recycles organelles, proteins, and macromolecules during starvation to maintain homeostasis and cell survival. Apart from its role during nutrient deprivation, emerging evidence shows that autophagy makes critical contributions to immune responses, such as antigen presentation, clearance of intracellular pathogens, and cytokine release.(128) Autophagy has been shown to be a key negative regulator of inflammasome signaling via several means including the removal of the inflammasome complex and recycling of damaged organelles, such as mitochondria, that otherwise activate inflammasome signaling.(129) In the case of the AIM2 inflammasome, E3 ubiquitin ligase tripartite motif 11 (TRIM11) has been reported to be a specific negative regulator.(130) Upon infection with DNA viruses, TRIM11 binds to AIM2 and undergoes auto-polyubiquitination, which promotes association between TRIM11 and the autophagic cargo receptor p62 to mediate AIM2 degradation via selective autophagy. Interestingly, several TRIM proteins have been implicated in the regulation of other inflammasomes via selective autophagy.(131) Recently a mitochondrial protease HtrA2 has been identified as an NLRP3 and AIM2 inflammasome regulator.(132) HtrA2 has been shown to regulate autophagic recycling of inflammasome adaptor ASC in a protease activity-dependent manner to modulate inflammasome signaling.
Microbial antagonism of AIM2 inflammasome.
In addition to the host mechanisms, pathogens have also evolved diverse strategies to control AIM2 inflammasome activation. F. tularensis, L. monocytogenes, and L. pneumophila restrict ligand availability to prevent AIM2 activation(133); Legionella Dot/Icm type IVB secretion system effector, SdhA, regulates trafficking of Legionella containing vacuoles (LCV) and augments its integrity to prevent release of bacterial DNA into the cytosol and AIM2 activation.(45) Similarly, several proteins expressed by F. tularensis, such as mviN, FTL_0325 and ripA, prevent AIM2 activation either by maintaining bacterial membrane integrity and limiting DNA release or by suppressing the priming signals (134–137). Mutation in L. monocytogenes lmo2473 enhances bacterial lysis in the cytosol thus leading to increased AIM2 activation.(138) Mtb has been reported to inhibit AIM2 inflammasome activation via its type VII secretion system ESX-1-mediated secretion of virulence factors.(139) This inhibition does not involve changes in AIM2 mRNA or protein expression, but may involve inhibition of upstream IFN-β production and signaling. However, the exact mechanism underlying ESX-1-dependent inhibition of AIM2 by Mtb is yet to be established. DNA viruses have also been shown to interfere with AIM2 inflammasome activation. Herpes simplex virus-1 (HSV-1) tegument protein VP22 directly interacts with AIM2 HIN domain and inhibits its oligomerization, an initial step in AIM2 inflammasome activation.(140) Consistently, the VP22 mutant of HSV-1 robustly activates the AIM2 inflammasome in vitro and is efficiently cleared in vivo. Likewise, human HCMV tegument protein pUL83 interacts with human AIM2 and disrupts the activation of the AIM2 inflammasome.(141) In contrast, HBV has been shown to modulate AIM2 expression transcriptionally to evade detection.(142) Mechanistically, HBV surface antigen HBs reduces the stability of upstream transcription factor IRF7 mRNA to decrease AIM2 transcription in Kupffer cells. Overall, the microbes have several mechanisms to evade the immune surveillance by the AIM2 inflammasome.
6. Inflammasome-independent functions of AIM2
In addition to its function as an innate immune sensor for DNA, AIM2 has inflammasome-independent functions. Interestingly, AIM2 was identified as a tumor suppressor gene in melanoma cancer cells.(20) Lack of AIM2 expression has also been detected in many other types of cancers e.g. breast cancer, prostate cancer and colorectal cancer.(97, 143, 144) Mutation in the AIM2 gene has been correlated with tumorigenesis and poor prognosis in colorectal cancer.(143) Two independent studies investigating the role of AIM2 in colorectal cancer show that AIM2 acts in an inflammasome-independent manner to regulate tumor development.(145, 146) AIM2 has been shown to interact with and inhibit the activation of DNA-dependent protein kinase DNA-PK (a phosphoinositide-3 kinase-related family kinase).(145) Inhibition of DNA-PK by AIM2 attenuates Akt phosphorylation, and as a result, the proliferation of tumor cells. Furthermore, AIM2 has been shown to restrict the proliferation of intestinal stem cells and gut microbiota dysbiosis to regulate tumor onset and progression.(146) AIM2 has also been reported to participate in the preservation of epithelial barrier integrity in an inflammasome-independent manner; AIM2-mediated AKT activation regulates expression of tight junction proteins such as claudin 3 and occludins in the intestinal epithelium, thereby promoting barrier integrity and averting mucosal invasion by enteric pathogens such as Salmonella.(147). Unlike in colorectal cancers, AIM2 has been found to function as an oncogene that promotes growth and proliferation in an inflammasome-dependent manner in lung carcinoma cell lines such as A549 and H460.(90) In line with this, high expression of AIM2 has been observed in NSCLC tissue and cell lines.(148) Furthermore, it has been recently reported that AIM2 expression positively correlates with the growth and proliferation of NSCLC cells in vitro and in vivo and that AIM2 expression in NSCLC tumors is associated with poor prognosis in patient.(91) AIM2 promotion of cell proliferation was independent of its inflammasome function; AIM2 has been found to colocalize with mitochondria in NSCLC cells and modulate mitochondrial fission and fusion dynamics to facilitate growth and proliferation of cancer cells. Overall, the inflammasome-independent function of AIM2 is critical in tumor development.
7. Conclusions and future directions
The AIM2 inflammasome plays a critical role in alarming the innate immune system during pathogen invasion and cellular perturbations. AIM2-mediated maturation of IL-1β and IL-18 and/or pyroptotic cell death contribute to host defense, autoimmunity, and neurodevelopment. While the inflammasome-mediated effects of AIM2 are relatively well understood, the inflammasome-independent arm of AIM2 remains poorly understood. The cell type-specific functions of AIM2, both inflammasome-dependent and inflammasome-independent, in various disease contexts is also largely unknown. Genotoxic stress and cell cycle dysregulation trigger the formation of micronuclei, the secondary nuclei. Emerging work in the past few years have demonstrated that cGAS senses DNA contained within the micronuclei to stimulate type I IFN expression.(149, 150) It would be of interest to determine if AIM2 gains access to the micronuclei and senses the micronuclear DNA to trigger inflammasome responses. Recent studies show the existence of a parallel DNA-PK-dependent, but cGAS-STING-independent DNA sensing pathway in humans but not in mice.(151) Whether the AIM2 inflammasome has any regulatory effect on this new DNA sensing pathway is not known. Although the differential expression of AIM2 is associated with various sterile inflammatory diseases and cancers, the roles of AIM2 in these disease conditions remain largely unknown. Furthermore, the biological relevance of AIM2 inflammasome activation in cancers associated with infections with oncogenic DNA viruses, such as HPV and HCMV, should be explored. Considering that AIM2 has been shown to negatively regulate the synthesis of IFN-β and that type I IFN promotes anti-tumor immunity,(152, 153) whether the IFN-suppressive role of AIM2 impacts anti-tumor immunity should be determined. A comprehensive understanding of inflammasome-dependent and -independent functions of AIM2 will provide novel insights into the pathogenesis of infectious and inflammatory diseases and cancer.
Acknowledgements:
The authors apologize to those investigators whose original papers could not be cited because of the space limitation. Research in the Rathinam laboratory is supported by the US National Institutes of Health (R01AI119015 and R21AI 135528).
Footnotes
Conflict of interest: The authors have no conflict of interest.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2(8):675–80. [DOI] [PubMed] [Google Scholar]
- 3.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9(7):465–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27(4):549–59. [DOI] [PubMed] [Google Scholar]
- 6.Brunette RL, Young JM, Whitley DG, Brodsky IE, Malik HS, Stetson DB. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J Exp Med. 2012;209(11):1969–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Russo AJ, Behl B, Banerjee I, Rathinam VAK. Emerging Insights into Noncanonical Inflammasome Recognition of Microbes. J Mol Biol. 2018;430(2):207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16(7):407–20. [DOI] [PubMed] [Google Scholar]
- 9.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–26. [DOI] [PubMed] [Google Scholar]
- 10.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–5. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535(7610):153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514(7521):187–92. [DOI] [PubMed] [Google Scholar]
- 13.Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD, et al. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell. 2016;165(5):1106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kayagaki N, Wong MT, Stowe IB, Ramani SR, Gonzalez LC, Akashi-Takamura S, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341(6151):1246–9. [DOI] [PubMed] [Google Scholar]
- 15.Hagar JA, Powell DA, Aachoui Y, Ernst RK, Miao EA. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341(6151):1250–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamkanfi M, Sarkar A, Vande Walle L, Vitari AC, Amer AO, Wewers MD, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185(7):4385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang, Han Z, Oppenheim JJ. Alarmins and immunity. Immunol Rev. 2017;280(1):41–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu W, Zu X, Liu S, Zhang H. The absent in melanoma 2 (AIM2) inflammasome in microbial infection. Clin Chim Acta. 2019;495:100–8. [DOI] [PubMed] [Google Scholar]
- 20.DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, et al. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15(4):453–7. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–72. [DOI] [PubMed] [Google Scholar]
- 23.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–60. [DOI] [PubMed] [Google Scholar]
- 24.Lugrin J, Martinon F. The AIM2 inflammasome: Sensor of pathogens and cellular perturbations. Immunol Rev. 2018;281(1):99–114. [DOI] [PubMed] [Google Scholar]
- 25.Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013;20(9):1149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pierini R, Juruj C, Perret M, Jones CL, Mangeot P, Weiss DS, et al. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 2012;19(10):1709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisch D, Bando H, Clough B, Hornung V, Yamamoto M, Shenoy AR, et al. Human GBP1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 2019;38(13):e100926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, et al. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36(4):561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matyszewski M, Morrone SR, Sohn J. Digital signaling network drives the assembly of the AIM2-ASC inflammasome. Proc Natl Acad Sci U S A. 2018;115(9):E1963–E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B, Tian Y, Yin Q. AIM2 Inflammasome Assembly and Signaling. Adv Exp Med Biol. 2019;1172:143–55. [DOI] [PubMed] [Google Scholar]
- 31.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B, Yin Q. AIM2 inflammasome activation and regulation: A structural perspective. J Struct Biol. 2017;200(3):279–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu A, Li Y, Yin Q, Ruan J, Yu X, Egelman E, et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 2015;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin T, Perry A, Smith P, Jiang J, Xiao TS. Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J Biol Chem. 2013;288(19):13225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu A, Kabaleeswaran V, Fu T, Magupalli VG, Wu H. Crystal structure of the F27G AIM2 PYD mutant and similarities of its self-association to DED/DED interactions. J Mol Biol. 2014;426(7):1420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 2014;156(6):1193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrone SR, Matyszewski M, Yu X, Delannoy M, Egelman EH, Sohn J. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat Commun. 2015;6:7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11(5):395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, et al. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24(10):637–44. [DOI] [PubMed] [Google Scholar]
- 41.Costa Franco MMS, Marim FM, Alves-Silva J, Cerqueira D, Rungue M, Tavares IP, et al. AIM2 senses Brucella abortus DNA in dendritic cells to induce IL-1beta secretion, pyroptosis and resistance to bacterial infection in mice. Microbes Infect. 2019;21(2):85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gomes MT, Campos PC, Oliveira FS, Corsetti PP, Bortoluci KR, Cunha LD, et al. Critical role of ASC inflammasomes and bacterial type IV secretion system in caspase-1 activation and host innate resistance to Brucella abortus infection. J Immunol. 2013;190(7):3629–38. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y, Zhou X, Kouadir M, Shi F, Ding T, Liu C, et al. the AIM2 inflammasome is involved in macrophage activation during infection with virulent Mycobacterium bovis strain. J Infect Dis. 2013;208(11):1849–58. [DOI] [PubMed] [Google Scholar]
- 44.Park E, Na HS, Song YR, Shin SY, Kim YM, Chung J. Activation of NLRP3 and AIM2 inflammasomes by Porphyromonas gingivalis infection. Infect Immun. 2014;82(1):112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge J, Gong YN, Xu Y, Shao F. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc Natl Acad Sci U S A. 2012;109(16):6193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ratsimandresy RA, Indramohan M, Dorfleutner A, Stehlik C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol. 2017;14(1):127–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu S, Peng L, Kwak YT, Tekippe EM, Pasare C, Malter JS, et al. The DNA Sensor AIM2 Maintains Intestinal Homeostasis via Regulation of Epithelial Antimicrobial Host Defense. Cell Rep. 2015;13(9):1922–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng S, Chen T, Lei G, Hou F, Jiang J, Huang Q, et al. Absent in melanoma 2 inflammasome is required for host defence against Streptococcus pneumoniae infection. Innate Immun. 2019;25(7):412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang R, Tsuchiya K, Kawamura I, Shen Y, Hara H, Sakai S, et al. Critical roles of ASC inflammasomes in caspase-1 activation and host innate resistance to Streptococcus pneumoniae infection. J Immunol. 2011;187(9):4890–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanamsagar R, Aldrich A, Kielian T. Critical role for the AIM2 inflammasome during acute CNS bacterial infection. J Neurochem. 2014;129(4):704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karki R, Man SM, Malireddi RKS, Gurung P, Vogel P, Lamkanfi M, et al. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 2015;17(3):357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalantari P, DeOliveira RB, Chan J, Corbett Y, Rathinam V, Stutz A, et al. Dual engagement of the NLRP3 and AIM2 inflammasomes by plasmodium-derived hemozoin and DNA during malaria. Cell Rep. 2014;6(1):196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu X, Du Y, Cai C, Cai B, Zhu M, Xing C, et al. Inflammasome activation negatively regulates MyD88-IRF7 type I IFN signaling and anti-malaria immunity. Nat Commun. 2018;9(1):4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupta G, Santana AKM, Gomes CM, Turatti A, Milanezi CM, Bueno Filho R, et al. Inflammasome gene expression is associated with immunopathology in human localized cutaneous leishmaniasis. Cell Immunol. 2019;341:103920. [DOI] [PubMed] [Google Scholar]
- 55.Botto S, Abraham J, Mizuno N, Pryke K, Gall B, Landais I, et al. Human Cytomegalovirus Immediate Early 86-kDa Protein Blocks Transcription and Induces Degradation of the Immature Interleukin-1beta Protein during Virion-Mediated Activation of the AIM2 Inflammasome. mBio. 2019;10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu DL, Xu GH, Lu SM, Ma BL, Miao NZ, Liu XB, et al. Correlation of AIM2 expression in peripheral blood mononuclear cells from humans with acute and chronic hepatitis B. Hum Immunol. 2013;74(5):514–21. [DOI] [PubMed] [Google Scholar]
- 57.Du W, Zhen J, Zheng Z, Ma S, Chen S. Expression of AIM2 is high and correlated with inflammation in hepatitis B virus associated glomerulonephritis. J Inflamm (Lond). 2013;10(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinholz M, Kawakami Y, Salzer S, Kreuter A, Dombrowski Y, Koglin S, et al. HPV16 activates the AIM2 inflammasome in keratinocytes. Arch Dermatol Res. 2013;305(8):723–32. [DOI] [PubMed] [Google Scholar]
- 59.Torii Y, Kawada JI, Murata T, Yoshiyama H, Kimura H, Ito Y. Epstein-Barr virus infection-induced inflammasome activation in human monocytes. PLoS One. 2017;12(4):e0175053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strittmatter GE, Sand J, Sauter M, Seyffert M, Steigerwald R, Fraefel C, et al. IFN-gamma Primes Keratinocytes for HSV-1-Induced Inflammasome Activation. J Invest Dermatol. 2016;136(3):610–20. [DOI] [PubMed] [Google Scholar]
- 61.Yogarajah T, Ong KC, Perera D, Wong KT. AIM2 Inflammasome-Mediated Pyroptosis in Enterovirus A71-Infected Neuronal Cells Restricts Viral Replication. Sci Rep. 2017;7(1):5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ekchariyawat P, Hamel R, Bernard E, Wichit S, Surasombatpattana P, Talignani L, et al. Inflammasome signaling pathways exert antiviral effect against Chikungunya virus in human dermal fibroblasts. Infect Genet Evol. 2015;32:401–8. [DOI] [PubMed] [Google Scholar]
- 63.Di Micco A, Frera G, Lugrin J, Jamilloux Y, Hsu ET, Tardivel A, et al. AIM2 inflammasome is activated by pharmacological disruption of nuclear envelope integrity. Proc Natl Acad Sci U S A. 2016;113(32):E4671–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dang EV, McDonald JG, Russell DW, Cyster JG. Oxysterol Restraint of Cholesterol Synthesis Prevents AIM2 Inflammasome Activation. Cell. 2017;171(5):1057–71 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adamczak SE, de Rivero Vaccari JP, Dale G, Brand FJ 3rd, Nonner D, Bullock MR, et al. Pyroptotic neuronal cell death mediated by the AIM2 inflammasome. J Cereb Blood Flow Metab. 2014;34(4):621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu PJ, Liu HY, Huang TN, Hsueh YP. AIM 2 inflammasomes regulate neuronal morphology and influence anxiety and memory in mice. Sci Rep. 2016;6:32405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lammert CR, Frost EL, Bellinger CE, Bolte AC, McKee CA, Hurt ME, et al. AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature. 2020;580(7805):647–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monteith AJ, Kang S, Scott E, Hillman K, Rajfur Z, Jacobson K, et al. Defects in lysosomal maturation facilitate the activation of innate sensors in systemic lupus erythematosus. Proc Natl Acad Sci U S A. 2016;113(15):E2142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H, Luo J, Alcorn JF, Chen K, Fan S, Pilewski J, et al. AIM2 Inflammasome Is Critical for Influenza-Induced Lung Injury and Mortality. J Immunol. 2017;198(11):4383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schattgen SA, Gao G, Kurt-Jones EA, Fitzgerald KA. Cutting Edge: DNA in the Lung Microenvironment during Influenza Virus Infection Tempers Inflammation by Engaging the DNA Sensor AIM2. J Immunol. 2016;196(1):29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu B, Jin C, Li HB, Tong J, Ouyang X, Cetinbas NM, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354(6313):765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gao J, Peng S, Shan X, Deng G, Shen L, Sun J, et al. Inhibition of AIM2 inflammasome-mediated pyroptosis by Andrographolide contributes to amelioration of radiation-induced lung inflammation and fibrosis. Cell Death Dis. 2019;10(12):957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lian Q, Xu J, Yan S, Huang M, Ding H, Sun X, et al. Chemotherapy-induced intestinal inflammatory responses are mediated by exosome secretion of double-strand DNA via AIM2 inflammasome activation. Cell Res. 2017;27(6):784–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46(2):269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma BR, Karki R, Kanneganti TD. Role of AIM2 inflammasome in inflammatory diseases, cancer and infection. Eur J Immunol. 2019;49(11):1998–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vakrakou AG, Svolaki IP, Evangelou K, Gorgoulis VG, Manoussakis MN. Cell-autonomous epithelial activation of AIM2 (absent in melanoma-2) inflammasome by cytoplasmic DNA accumulations in primary Sjogren’s syndrome. J Autoimmun. 2020;108:102381. [DOI] [PubMed] [Google Scholar]
- 77.Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Goss C, Anz D, et al. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3(82):82ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6(1):49–56. [DOI] [PubMed] [Google Scholar]
- 79.Jakobs C, Perner S, Hornung V. AIM2 Drives Joint Inflammation in a Self-DNA Triggered Model of Chronic Polyarthritis. PLoS One. 2015;10(6):e0131702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kawane K, Ohtani M, Miwa K, Kizawa T, Kanbara Y, Yoshioka Y, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443(7114):998–1002. [DOI] [PubMed] [Google Scholar]
- 81.Komada T, Chung H, Lau A, Platnich JM, Beck PL, Benediktsson H, et al. Macrophage Uptake of Necrotic Cell DNA Activates the AIM2 Inflammasome to Regulate a Proinflammatory Phenotype in CKD. J Am Soc Nephrol. 2018;29(4):1165–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yuan B, Zhou XM, You ZQ, Xu WD, Fan JM, Chen SJ, et al. Inhibition of AIM2 inflammasome activation alleviates GSDMD-induced pyroptosis in early brain injury after subarachnoid haemorrhage. Cell Death Dis. 2020;11(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang MJ, Zhao QC, Xia MX, Chen J, Chen YT, Cao X, et al. The HDAC3 inhibitor RGFP966 ameliorated ischemic brain damage by downregulating the AIM2 inflammasome. FASEB J. 2020;34(1):648–62. [DOI] [PubMed] [Google Scholar]
- 84.Liang J, Wang Q, Li JQ, Guo T, Yu D. Long non-coding RNA MEG3 promotes cerebral ischemia-reperfusion injury through increasing pyroptosis by targeting miR-485/AIM2 axis. Exp Neurol. 2020;325:113139. [DOI] [PubMed] [Google Scholar]
- 85.Ge X, Li W, Huang S, Yin Z, Xu X, Chen F, et al. The pathological role of NLRs and AIM2 inflammasome-mediated pyroptosis in damaged blood-brain barrier after traumatic brain injury. Brain Res. 2018;1697:10–20. [DOI] [PubMed] [Google Scholar]
- 86.Pan J, Han L, Guo J, Wang X, Liu D, Tian J, et al. AIM2 accelerates the atherosclerotic plaque progressions in ApoE−/− mice. Biochem Biophys Res Commun. 2018;498(3):487–94. [DOI] [PubMed] [Google Scholar]
- 87.Paulin N, Viola JR, Maas SL, de Jong R, Fernandes-Alnemri T, Weber C, et al. Double-Strand DNA Sensing Aim2 Inflammasome Regulates Atherosclerotic Plaque Vulnerability. Circulation. 2018;138(3):321–3. [DOI] [PubMed] [Google Scholar]
- 88.Wang X, Pan J, Liu H, Zhang M, Liu D, Lu L, et al. AIM2 gene silencing attenuates diabetic cardiomyopathy in type 2 diabetic rat model. Life Sci. 2019;221:249–58. [DOI] [PubMed] [Google Scholar]
- 89.Hupa KJ, Stein K, Schneider R, Lysson M, Schneiker B, Hornung V, et al. AIM2 inflammasome-derived IL-1beta induces postoperative ileus in mice. Sci Rep. 2019;9(1):10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang M, Jin C, Yang Y, Wang K, Zhou Y, Zhou Y, et al. AIM2 promotes non-small-cell lung cancer cell growth through inflammasome-dependent pathway. J Cell Physiol. 2019;234(11):20161–73. [DOI] [PubMed] [Google Scholar]
- 91.Qi M, Dai D, Liu J, Li Z, Liang P, Wang Y, et al. AIM2 promotes the development of non-small cell lung cancer by modulating mitochondrial dynamics. Oncogene. 2020;39(13):2707–23. [DOI] [PubMed] [Google Scholar]
- 92.Yu Q, Zhang M, Ying Q, Xie X, Yue S, Tong B, et al. Decrease of AIM2 mediated by luteolin contributes to non-small cell lung cancer treatment. Cell Death Dis. 2019;10(3):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen SL, Liu LL, Lu SX, Luo RZ, Wang CH, Wang H, et al. HBx-mediated decrease of AIM2 contributes to hepatocellular carcinoma metastasis. Mol Oncol. 2017;11(9):1225–40. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 94.Ma X, Guo P, Qiu Y, Mu K, Zhu L, Zhao W, et al. Loss of AIM2 expression promotes hepatocarcinoma progression through activation of mTOR-S6K1 pathway. Oncotarget. 2016;7(24):36185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen LC, Wang LJ, Tsang NM, Ojcius DM, Chen CC, Ouyang CN, et al. Tumour inflammasome-derived IL-1beta recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol Med. 2012;4(12):1276–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen PA, Shrivastava G, Balcom EF, McKenzie BA, Fernandes J, Branton WG, et al. Absent in melanoma 2 regulates tumor cell proliferation in glioblastoma multiforme. J Neurooncol. 2019;144(2):265–73. [DOI] [PubMed] [Google Scholar]
- 97.Ponomareva L, Liu H, Duan X, Dickerson E, Shen H, Panchanathan R, et al. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol Cancer Res. 2013;11(10):1193–202. [DOI] [PubMed] [Google Scholar]
- 98.Corrales L, Woo SR, Williams JB, McWhirter SM, Dubensky TW Jr., Gajewski TF. Antagonism of the STING Pathway via Activation of the AIM2 Inflammasome by Intracellular DNA. J Immunol. 2016;196(7):3191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11(5):385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gray EE, Winship D, Snyder JM, Child SJ, Geballe AP, Stetson DB. The AIM2-like Receptors Are Dispensable for the Interferon Response to Intracellular DNA. Immunity. 2016;45(2):255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107(21):9771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Ning X, Gao P, Wu S, Sha M, Lv M, et al. Inflammasome Activation Triggers Caspase-1-Mediated Cleavage of cGAS to Regulate Responses to DNA Virus Infection. Immunity. 2017;46(3):393–404. [DOI] [PubMed] [Google Scholar]
- 103.Banerjee I, Behl B, Mendonca M, Shrivastava G, Russo AJ, Menoret A, et al. Gasdermin D Restrains Type I Interferon Response to Cytosolic DNA by Disrupting Ionic Homeostasis. Immunity. 2018;49(3):413–26 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–71. [DOI] [PubMed] [Google Scholar]
- 105.Burke TP, Engstrom P, Chavez RA, Fonbuena JA, Vance RE, Welch MD. Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat Microbiol. 2020;5(5):688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma C, Yang D, Wang B, Wu C, Wu Y, Li S, et al. Gasdermin D in macrophages restrains colitis by controlling cGAS-mediated inflammation. Science Advances. 2020;6(21):eaaz6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, et al. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015;16(5):467–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015;16(5):476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Man SM, Karki R, Sasai M, Place DE, Kesavardhana S, Temirov J, et al. IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell. 2016;167(2):382–96 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A. 2014;111(1):E62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dorfleutner A, Chu L, Stehlik C. Inhibiting the inflammasome: one domain at a time. Immunol Rev. 2015;265(1):205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Indramohan M, Stehlik C, Dorfleutner A. COPs and POPs Patrol Inflammasome Activation. J Mol Biol. 2018;430(2):153–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dorfleutner A, Bryan NB, Talbott SJ, Funya KN, Rellick SL, Reed JC, et al. Cellular pyrin domain-only protein 2 is a candidate regulator of inflammasome activation. Infect Immun. 2007;75(3):1484–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stehlik C, Krajewska M, Welsh K, Krajewski S, Godzik A, Reed JC. The PAAD/PYRIN-only protein POP1/ASC2 is a modulator of ASC-mediated nuclear-factor-kappa B and pro-caspase-1 regulation. Biochem J. 2003;373(Pt 1):101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, et al. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat Immunol. 2014;15(4):343–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ratsimandresy RA, Chu LH, Khare S, de Almeida L, Gangopadhyay A, Indramohan M, et al. The PYRIN domain-only protein POP2 inhibits inflammasome priming and activation. Nat Commun. 2017;8:15556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Almeida L, Khare S, Misharin AV, Patel R, Ratsimandresy RA, Wallin MC, et al. The PYRIN Domain-only Protein POP1 Inhibits Inflammasome Assembly and Ameliorates Inflammatory Disease. Immunity. 2015;43(2):264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bedoya F, Sandler LL, Harton JA. Pyrin-only protein 2 modulates NF-kappaB and disrupts ASC:CLR interactions. J Immunol. 2007;178(6):3837–45. [DOI] [PubMed] [Google Scholar]
- 119.Krishnaswamy JK, Liu D, Eisenbarth SC. POP goes the inflammasome. Nat Immunol. 2014;15(4):311–3. [DOI] [PubMed] [Google Scholar]
- 120.Lamkanfi M, Denecker G, Kalai M, D’Hondt K, Meeus A, Declercq W, et al. INCA, a novel human caspase recruitment domain protein that inhibits interleukin-1beta generation. J Biol Chem. 2004;279(50):51729–38. [DOI] [PubMed] [Google Scholar]
- 121.Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000;103(1):99–111. [DOI] [PubMed] [Google Scholar]
- 122.Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8(6):649–57. [DOI] [PubMed] [Google Scholar]
- 123.Lee SH, Stehlik C, Reed JC. Cop, a caspase recruitment domain-containing protein and inhibitor of caspase-1 activation processing. J Biol Chem. 2001;276(37):34495–500. [DOI] [PubMed] [Google Scholar]
- 124.Panchanathan R, Duan X, Shen H, Rathinam VA, Erickson LD, Fitzgerald KA, et al. Aim2 deficiency stimulates the expression of IFN-inducible Ifi202, a lupus susceptibility murine gene within the Nba2 autoimmune susceptibility locus. J Immunol. 2010;185(12):7385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, et al. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Rep. 2013;4(2):327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ru H, Ni X, Zhao L, Crowley C, Ding W, Hung LW, et al. Structural basis for termination of AIM2-mediated signaling by p202. Cell Res. 2013;23(6):855–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang PH, Ye ZW, Deng JJ, Siu KL, Gao WW, Chaudhary V, et al. Inhibition of AIM2 inflammasome activation by a novel transcript isoform of IFI16. EMBO Rep. 2018;19(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harris J, Lang T, Thomas JPW, Sukkar MB, Nabar NR, Kehrl JH. Autophagy and inflammasomes. Mol Immunol. 2017;86:10–5. [DOI] [PubMed] [Google Scholar]
- 130.Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, et al. TRIM11 Suppresses AIM2 Inflammasome by Degrading AIM2 via p62-Dependent Selective Autophagy. Cell Rep. 2016;16(7):1988–2002. [DOI] [PubMed] [Google Scholar]
- 131.Kimura T, Jain A, Choi SW, Mandell MA, Johansen T, Deretic V. TRIM-directed selective autophagy regulates immune activation. Autophagy. 2017;13(5):989–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rodrigue-Gervais IG, Doiron K, Champagne C, Mayes L, Leiva-Torres GA, Vanie P Jr., et al. The mitochondrial protease HtrA2 restricts the NLRP3 and AIM2 inflammasomes. Sci Rep. 2018;8(1):8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ulland TK, Ferguson PJ, Sutterwala FS. Evasion of inflammasome activation by microbial pathogens. J Clin Invest. 2015;125(2):469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ulland TK, Buchan BW, Ketterer MR, Fernandes-Alnemri T, Meyerholz DK, Apicella MA, et al. Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J Immunol. 2010;185(5):2670–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dotson RJ, Rabadi SM, Westcott EL, Bradley S, Catlett SV, Banik S, et al. Repression of inflammasome by Francisella tularensis during early stages of infection. J Biol Chem. 2013;288(33):23844–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peng K, Broz P, Jones J, Joubert LM, Monack D. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell Microbiol. 2011;13(10):1586–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang MT, Mortensen BL, Taxman DJ, Craven RR, Taft-Benz S, Kijek TM, et al. Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J Immunol. 2010;185(9):5476–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7(5):412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shah S, Bohsali A, Ahlbrand SE, Srinivasan L, Rathinam VA, Vogel SN, et al. Cutting edge: Mycobacterium tuberculosis but not nonvirulent mycobacteria inhibits IFN-beta and AIM2 inflammasome-dependent IL-1beta production via its ESX-1 secretion system. J Immunol. 2013;191(7):3514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Maruzuru Y, Ichinohe T, Sato R, Miyake K, Okano T, Suzuki T, et al. Herpes Simplex Virus 1 VP22 Inhibits AIM2-Dependent Inflammasome Activation to Enable Efficient Viral Replication. Cell Host Microbe. 2018;23(2):254–65 e7. [DOI] [PubMed] [Google Scholar]
- 141.Han Y, Chen Z, Hou R, Yan D, Liu C, Chen S, et al. Expression of AIM2 is correlated with increased inflammation in chronic hepatitis B patients. Virol J. 2015;12:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zannetti C, Roblot G, Charrier E, Ainouze M, Tout I, Briat F, et al. Characterization of the Inflammasome in Human Kupffer Cells in Response to Synthetic Agonists and Pathogens. J Immunol. 2016;197(1):356–67. [DOI] [PubMed] [Google Scholar]
- 143.Dihlmann S, Tao S, Echterdiek F, Herpel E, Jansen L, Chang-Claude J, et al. Lack of Absent in Melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int J Cancer. 2014;135(10):2387–96. [DOI] [PubMed] [Google Scholar]
- 144.Chen IF, Ou-Yang F, Hung JY, Liu JC, Wang H, Wang SC, et al. AIM2 suppresses human breast cancer cell proliferation in vitro and mammary tumor growth in a mouse model. Mol Cancer Ther. 2006;5(1):1–7. [DOI] [PubMed] [Google Scholar]
- 145.Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21(8):906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015;162(1):45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hu GQ, Song PX, Li N, Chen W, Lei QQ, Yu SX, et al. AIM2 contributes to the maintenance of intestinal integrity via Akt and protects against Salmonella mucosal infection. Mucosal Immunol. 2016;9(5):1330–9. [DOI] [PubMed] [Google Scholar]
- 148.Kong H, Wang Y, Zeng X, Wang Z, Wang H, Xie W. Differential expression of inflammasomes in lung cancer cell lines and tissues. Tumour Biol. 2015;36(10):7501–13. [DOI] [PubMed] [Google Scholar]
- 149.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548(7668):461–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Harding SM, Benci JL, Irianto J, Discher DE, Minn AJ, Greenberg RA. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature. 2017;548(7668):466–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Burleigh K, Maltbaek JH, Cambier S, Green R, Gale M Jr., James RC, et al. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci Immunol. 2020;5(43). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kumari P, Saha I, Narayanan A, Narayanan S, Takaoka A, Kumar NS, et al. Essential role of HCMV deubiquitinase in promoting oncogenesis by targeting anti-viral innate immune signaling pathways. Cell Death Dis. 2017;8(10):e3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nat Rev Immunol. 2015;15(7):405–14. [DOI] [PubMed] [Google Scholar]