Abstract
Background
Phosphatase and tensin homolog (PTEN) loss has long been associated with adverse findings in early prostate cancer. Studies to date have yet to employ quantitative methods (qPTEN) for measuring of prognostically relevant amounts of PTEN loss in postsurgical settings and demonstrate its clinical application.
Methods
PTEN protein levels were measured by immunohistochemistry in radical prostatectomy samples from training (n = 410) and validation (n = 272) cohorts. PTEN loss was quantified per cancer cell and per tissue microarray core. Thresholds for identifying clinically relevant PTEN loss were determined using log-rank statistics in the training cohort. Univariate (Kaplan-Meier) and multivariate (Cox proportional hazards) analyses on various subpopulations were performed to assess biochemical recurrence-free survival (BRFS) and were independently validated. All statistical tests were two-sided.
Results
PTEN loss in more than 65% cancer cells was most clinically relevant and had statistically significant association with reduced BRFS in training (hazard ratio [HR] = 2.48, 95% confidence interval [CI] = 1.59 to 3.87; P < .001) and validation cohorts (HR = 4.22, 95% CI = 2.01 to 8.83; P < .001). The qPTEN scoring method identified patients who recurred within 5.4 years after surgery (P < .001). In men with favorable risk of biochemical recurrence (Cancer of the Prostate Risk Assessment – Postsurgical scores <5 and no adverse pathological features), qPTEN identified a subset of patients with shorter BRFS (HR = 5.52, 95% CI = 2.36 to 12.90; P < .001) who may be considered for intensified monitoring and/or adjuvant therapy.
Conclusions
Compared with previous qualitative approaches, qPTEN improves risk stratification of postradical prostatectomy patients and may be considered as a complementary tool to guide disease management after surgery.
Among single gene biomarkers, loss of phosphatase and tensin homolog (PTEN) in prostate cancer is the most consistently associated with adverse events, including risk of upgrading at prostatectomy, disease recurrence after surgery, and progression to metastasis and death (1–3). Along with its prognostic significance, well-established assays for PTEN assessment make it an attractive candidate biomarker for risk stratification to guide disease management (2).
PTEN gene deletion is usually a subclonal event that produces molecular heterogeneity in primary prostate cancers, making it critical to assess with single-cell resolution (4–6). Because PTEN protein loss is highly concordant with gene deletion, PTEN status can be readily obtained by immunohistochemistry (IHC), which accurately reflects genomic loss and has been extensively validated and adapted to Clinical Laboratory Improvement Amendments laboratory standards (5,7,8). PTEN loss is very heterogeneous varying from partial (1%–99% cancer cells) to complete loss (100% cancer cells) (5,7). The prognostic significance of this variability has not been identified or validated, nor have specific clinical applications been identified for PTEN as a biomarker.
Because PTEN loss is strongly associated with prostate cancer recurrence after surgery, its assessment might help guide decisions around adjuvant radiation therapy (RT), which can improve survival for patients at higher risk for relapse (9,10). The intensity of postoperative monitoring and the optimal timing of RT are controversial (11,12). For many patients, risk may be unclear, and the choice between early (adjuvant) and late (salvage) RT can be difficult (9–12). In select populations, biomarkers of recurrence may improve decision-making in this context.
In the current study, we determine and independently validate a quantitative scoring method, qPTEN, analyze the prognostic significance of qPTEN thresholds, and investigate a potential clinical application of qPTEN in postoperative treatment decisions.
Methods
Cohort Description
We assembled 3 retrospective radical prostatectomy (RP) cohorts from separate institutions. University of Sao-Paulo (Brazil) (n = 125; RP years, 2006–2015) and the Centre Hospitalier d’University of Montreal (CHUM) (n = 285; RP years, 1993–2006) were combined into a single training (n = 410) cohort (Supplementary Table 1, available online). Archival tissues were used to construct tissue microarrays (TMAs), which contained three 1.0 mm cancer cores on average per case. A total of 272 patients and corresponding RP specimens from Kingston Health Services Center (KHSC) (RP year, 2000–2012) (Supplementary Table 1, available online) were used as a validation cohort, with six 0.6 mm cancer cores per case on TMAs.
RP slides for all cohorts were re-reviewed for stage, surgical margin status, and the World Health Organization grade group (13) by 2 urologic pathologists (TJ and DMB). Median follow-up was 5.7 years for training cohort and 4.9 years for validation cohort. See Supplementary Table 1 (available online) for clinical and pathological summaries. For all patients, biochemical recurrence was defined as 2 prostate-specific antigen values of no less than 0.2 ng/ml after prostatectomy and/or documentation of biochemical recurrence. Biochemical recurrence-free survival (BRFS) was calculated from date of surgery and used as the clinical endpoint. This retrospective study was approved by local Ethics Review Boards at Queen’s University, CHUM, Canada, and University of Sao Paulo, Brazil, which waived the requirement for informed consent.
Immunohistochemistry
IHC staining was performed on automated staining platforms: Benchmark XT (Ventana Medical System, Inc, Tucson, AZ) for CHUM cohort and Discovery XT (Ventana Medical System, Inc, Tucson, AZ) for KHSC and Brazil cohorts. Briefly, paraffin-embedded formalin-fixed high-density TMA blocks were sectioned at 5 μm. After antigen retrieval (CC1, Ventana), 2 different but equivalent rabbit monoclonal anti-PTEN antibodies were used: Clone-D4.3 XP, dilution-1:100, Cell Signaling Technologies for KHSC and Brazil; Clone-SP218, dilution-1:50, Spring Biosciences for CHUM (5,7,14). Bound primary antibody was visualized using antirabbit IgG conjugated-HRP (DISCOVERY OmniMap, Ventana). Negative and positive cell line controls included cell blocks containing PC3 and LNCaP for PTEN loss and RWPE-1 for intact PTEN. Expression of the ETS transcription factor ERG was scored using dichotomous (+ve and -ve grouping) scoring as previously described (15,16) (Supplementary Figure 1, available online). Staining conditions are further detailed in Supplementary Table 2 (available online).
Qualitative and Quantitative Evaluation of PTEN Loss (qPTEN)
To minimize variation, identical approaches and algorithms were used for both training and validation cohorts. Stained tissue microarrays were scanned at 20× magnification. Using a validated rubric (5,7), protein expression was independently visually scored by 2 urologic pathologists (TJ and DMB), blinded to patient outcome. When PTEN scoring was discrepant, a third pathologist (TLL) served as a tiebreaker. As previously described, intact PTEN was defined as cytoplasmic and/or nuclear staining above background, and subsequently, PTEN loss was defined as any loss of cytoplasmic and/or nuclear staining (Supplementary Figure 1, available online) (1,5,7).
Digital scoring was a semiautomated process. It was used to count cells and determine the extent of PTEN loss. The intensity of staining was not evaluated or quantified digitally. Because digital scoring software did not reliably differentiate cancer cells from benign, we visually identified and annotated areas containing 1) all cancer cells and 2) all cancer cells exhibiting PTEN loss. Cells in each region were counted using the automated Cytonuclear v1.4 algorithm, HALO software, v1.94 (Indica Labs, Inc, Albuquerque, NM).
Percentage of cells with PTEN loss was calculated as (number of cancer cells with PTEN loss/total cancer cells) x 100. Likewise, percentage of cores with PTEN loss was calculated as the (number of cores with any PTEN loss /total cores containing cancer) x 100. The range varied from 1% to 100%. Intact PTEN status was represented as 0%.
Statistical Analysis
Fisher exact and χ2 tests were performed to test association of PTEN loss with available clinical and pathological variables using SPSS v24. Kaplan-Meier estimates of BRFS were plotted.
Cox proportional hazards models were generated for univariate and multivariate analysis, and multiple pathological variables and molecular markers were adjusted for potential confounding effects (Tables 1 and 2). For each variable, the proportional hazards assumption was tested by checking statistical significance of its interaction with BRFS using the Survival Package in R (version 3.4.4) (17). All cases exhibiting PTEN loss in the training cohort were selected, and log-rank statistics were used to determine ideal thresholds for both percentage of PTEN deleted cores and cells to better identify patients at risk of experiencing shorter BRFS, using the Maxstat package in R (18).
Table 1.
Univariate Cox proportional hazards models for BRFS
| Training cohort |
Validation cohort |
|||
|---|---|---|---|---|
| Variable | HR (95% CI) | P * | HR (95% CI) | P * |
| Age | ||||
| 1-unit increase (continuous) | 1 (0.97 to 1.02) | .92 | 1.05 (1.00 to 1.10) | .07 |
| PSA | ||||
| 1-unit increase (continuous) | 1.041 (1.02 to 1.06) | <.001 | 1.023 (0.97 to 1.08) | .43 |
| ERG status | ||||
| Fusion vs no fusion | 0.89 (0.63 to 1.25) | .49 | 1.20 (0.66 to 2.19) | .55 |
| Grade groups (GG) | ||||
| GG2 vs GG1 | 1.56 (1.01 to 2.42) | .046 | 3.29 (1.39 to 7.80) | .007 |
| ≥GG3 vs GG1 | 2.63 (1.73 to 3.99) | <.001 | 4.73 (1.72 to 13.03) | .003 |
| pStage | ||||
| T3a vs T2 | 2.38(1.57 to 3.62) | <.001 | 2.97 (1.65 to 5.34) | <.001 |
| T3b vs T2 | 5.04 (3.38 to 7.51) | <.001 | 6.23 (2.58 to 15.04) | <.001 |
| Extraprostatic extension | ||||
| Yes vs no | 3.20 (2.30 to 4.47) | <.001 | 3.42 (1.99 to 5.86) | <.001 |
| Seminal vesicle invasion | ||||
| Yes vs no | 4.38 (2.79 to 6.88) | <.001 | 4.66 (1.99 to 10.94) | <.001 |
| Surgical margin | ||||
| Yes vs no | 2.95 (2.11 to 4.12) | <.001 | 3.17 (1.72 to 5.86) | <.001 |
| PTEN status | ||||
| Loss vs intact | 1.92 (1.35 to 2.73) | <.001 | 2.67 (1.55 to 4.62) | <.001 |
| PTEN loss (% cores) | ||||
| Low (1%–50%) vs intact (0%) | 1.61 (1.00 to 2.60) | .05 | 1.53 (0.64 to 4.67) | .34 |
| High (>50%) vs intact (0%) | 2.24 (1.46 to 3.44) | <.001 | 3.71 (2.02 to 6.79) | <.001 |
| PTEN loss (% cells) | ||||
| Low (1%–65%) vs intact (0%) | 1.47 (0.89 to 2.41) | .13 | 1.82 (0.91 to 3.64) | .09 |
| High (>65%) vs intact (0%) | 2.39 (1.57 to 3.62) | <.001 | 4.95 (2.48 to 9.90) | <.001 |
Two-sided log-rank test. BRFS = biochemical recurrence-free survival; CI = confidence interval; HR = hazard ratio; PSA = prostate-specific antigen; pStage = pathological stage; PTEN = phosphatase and tensin homolog.
Table 2.
Multivariate Cox proportional hazards model for BRFS in association with percent of cells and cores with PTEN loss using threshold
| Variable | BRFS in association with % of cells with PTEN loss |
BRFS in association with % of cores with PTEN loss |
||||||
|---|---|---|---|---|---|---|---|---|
| Training cohort |
Validation cohort |
Training cohort |
Validation cohort |
|||||
| HR (95% CI) | P * | HR (95% CI) | P * | HR (95% CI) | P * | HR (95% CI) | P * | |
| Age, 1-unit increase, y | 0.97 (0.94 to 0.99) | .04 | 1.05 (0.99 to 1.11) | .08 | 0.97 (0.94 to 1.00) | .046 | 1.04 (0.98 to 1.10) | .16 |
| PSA, 1-unit increase | 1.02 (1.00 to 1.05) | .05 | 1.00 (0.94 to 1.07) | .94 | 1.03 (1.00 to 1.05) | .04 | 0.99 (0.93 to 1.06) | .81 |
| ERG, fusion vs no fusion | 0.86 (0.59 to 1.24) | .41 | 0.74 (0.39 to 1.41) | .36 | 0.84 (0.58 to 1.23) | .38 | 0.82 (0.43 to 1.57) | .55 |
| Grade groups (GG) | ||||||||
| GG2 vs GG1 | 1.03 (0.64 to 1.66) | .91 | 1.67 (0.62 to 4.50) | .31 | 1.05 (0.65 to 1.69) | .85 | 1.96 (0.73 to 5.27) | .18 |
| ≥GG3 vs GG1 | 1.75 (1.09 to 2.82) | .02 | 2.19 (0.68 to 7.10) | .19 | 1.75 (1.09 to 2.82) | .02 | 2.48 (0.77 to 7.97) | .13 |
| Extraprostatic extension, yes vs no | 1.52 (1.01 to 2.29) | .04 | 1.69 (0.82 to 3.46) | .15 | 1.53 (1.01 to 2.29) | .04 | 1.66 (0.81 to 3.38) | .16 |
| Seminal vesicle invasion, yes vs no | 4.19 (2.40 to 7.33) | <.001 | 2.62 (0.98 to 7.00) | .05 | 4.21 (2.41 to 7.36) | <.001 | 2.21 (0.82 to 5.97) | .12 |
| Surgical margin (SM), Yes vs no | 2.40 (1.63 to 3.53) | <.001 | 2.14 (1.06 to 4.34) | .03 | 2.39 (1.63 to 3.52) | <.001 | 2.1 (1.05 to 4.20) | .03 |
| PTEN loss (% cells) | ||||||||
| Low (1%–65%) vs intact (0%) | 1.61 (0.95 to 2.75) | .08 | 1.45 (0.70 to 3.01) | .32 | 1.96 (1.18 to 3.26) | .009 | 1.25 (0.48 to 3.28) | .64 |
| High (>65%) vs intact (0%) | 2.48 (1.59 to 3.87) | <.001 | 4.22 (2.01 to 8.83) | <.001 | 2.15 (1.36 to 3.42) | .001 | 2.75 (1.46 to 5.17) | .002 |
Two-sided log-rank test. BRFS = biochemical recurrence-free survival; CI = confidence interval; HR = hazard ratio; PSA = prostate-specific antigen; PTEN = phosphatase and tensin homolog.
P values less than .05 were considered statistically significant. All statistical tests were two-sided.
Results
Quantitative Assessment of PTEN Loss (qPTEN)
We quantified PTEN loss in percentage of cancer cells and percentage of cancer-containing cores. Thresholds for both PTEN loss measures were trained using IHC data on 410 patients from CHUM and Brazil. The 2 measures were highly correlated (Spearman r = 0.773; P < .001).
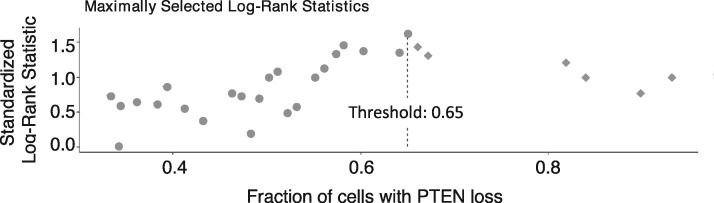
By maximizing log-rank statistics, we identified a threshold greater than 65% of cancer cells with PTEN loss that best identified patients with shorter BRFS (Figure 1; Supplementary Figure 2, available online). In both univariate (training, HR = 2.39, 95% confidence interval [CI] = 1.57 to 3.62; P < .001; validation, HR = 4.95, 95% CI = 2.48 to 9.90; P < .001) (Table 1) and multivariate analysis (training, HR = 2.48, 95% CI = 1.59 to 3.87; P < .001; validation, HR = 4.22, 95% CI = 2.01 to 8.83; P < .001) (Table 2) cases, above threshold had statistically significant association with shorter BRFS. Similarly, a threshold of more than 50% cancer cores with PTEN loss was associated with shorter BRFS in training and validation cohorts (see Tables 1 and 2; Supplementary Figure 2, available online). No statistical significance was found between the high PTEN loss (>65%) and low PTEN loss (1%–65%) groups in univariate (P = .10) and multivariate (P = .16) analysis in the training cohort (Supplementary Figure 2, available online). In the validation cohort, however, patients with high PTEN loss (>65%) cancer cells had statistically significantly shorter BRFS compared with patients with low PTEN loss (1%–65%), both in univariate (P = .02) and multivariate (P = .02) analysis (Supplementary Figure 2, available online).
Figure 1.
Threshold selection for percent cancer cells with qPTEN loss using maximally log-rank statistics. All 98 cases with PTEN loss from the training cohort were included in the analysis. Each data point in this figure represents between 1 and 3 patients because several cases showed overlapping values of % PTEN loss in cancer cells. Standardized log-rank statistics values were computed for each distinct threshold value of percent cancer cells with PTEN loss in the training cohort. The highest log-rank statistics value corresponded to a threshold of 65% and was chosen as an optimal threshold for identifying patients with shorter biochemical recurrence-free survival. Patients with low PTEN loss (1%–65% cancer cells) are shown as ● and high PTEN loss (>65% cancer cells) is represented as ♦. The thresholds for both percent cells and cores were selected using the same approach. PTEN = phosphatase and tensin homolog.
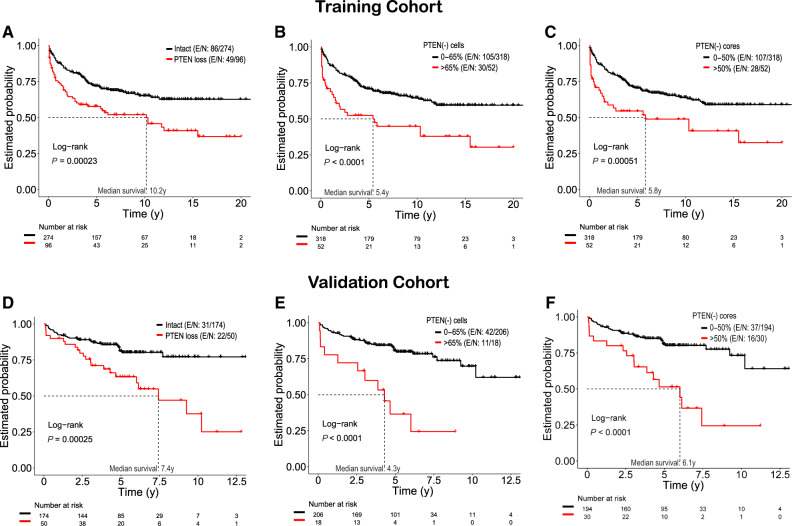
To further assess the clinical importance of patients with higher level PTEN loss (>65% for cells or >50% for cores), the patients with intact and lower PTEN loss were merged into one group (0%–65% for cells and 0%–50% for cores) in Kaplan-Meier analysis (Figure 2). Using an identical approach in both cohorts, we found that patients with high level PTEN loss above threshold (>65%) of cancer cells had statistically significantly decreased BRFS times in both training (10.2 years vs 5.4 years; P < .001; Figure 2A vs 2B) and validation cohorts (7.4 years vs 4.3 years; P < .001; Figure 2D vs 2E). In addition, 45.8% (44 of 96; Figure 2A vs 2B) of patients PTEN loss below threshold was reclassified as not aggressive and experienced excellent (>10 years median) survival rates (Figure 2A vs 2B).
Figure 2.
Quantitative analysis of PTEN loss (qPTEN) better stratifies risk of recurrence after surgery. Graphs show Kaplan-Meier curves for biochemical recurrence-free survival stratified by any PTEN loss vs qPTEN loss. Training cohort (A, B, C): Both models (A vs B and C) were statistically significant (any PTEN loss, P < .001 vs % of cells/cores, P < .001). Patients with PTEN loss above thresholds had statistically significant decreased median biochemical recurrence-free survival time from 10.2 years (any PTEN loss) to 5.4 years (>65% cells) (P < .001) and to 5.8 years (>50% cores) (P < .001). Using the thresholds, 45.8% (44 of 96) of patients reclassified as clinically insignificant loss (A vs B), whereas the remaining 54.2% (52 of 96) of cases showed shorter time to recurrence (A vs B). Corresponding results for the validation cohort (D, E, F) were similar to the training cohort. Note: In Kaplan-Meier curves, patients with intact PTEN are included in groups 0%–65% (cells) and 0%–50% (cores). Two-sided log-rank test was used to assess Kaplan-Meier estimates. E/N = event/total number of patients; PTEN = phosphatase and tensin homolog.
Focusing on percent cores with PTEN loss, patients in the training cohort with loss in more than 50% of cancer-containing cores experienced shorter BRFS rates compared with patients with intact PTEN (0%) or PTEN loss below 50% (10.2 years vs 5.8 years; P < .001; Figure 2A vs 2 C). However, in the validation cohort, only a modest survival difference was seen (7.4 years vs 6.1 years; P < .001; Figure 2D vs 2 F). ERG status did not have a statistically significant effect on BRFS (Tables 1 and 2) and was not associated with any clinical or pathological variables (Supplementary Table 3, available online).
Potential Clinical Applications for qPTEN Assessment After Radical Prostatectomy
We assessed whether qPTEN might add prognostic information to Cancer of the Prostate Risk Assessment Postsurgical (CAPRA-S), a widely accepted risk calculator for postsurgical recurrence (19,20). Patients with higher CAPRA-S scores (>5) or with adverse pathological features such as positive surgical margins, seminal vesicle invasion, and lymph node metastasis are known to be at high risk for reduced BRFS9. In contrast, patients with low CAPRA-S scores (<5) and without adverse pathological features are considered to have favorable outcomes, although a subset of these patients experience biochemical recurrence (BCR) and shorter BRFS. A biomarker for recurrence in this patient subset might be useful in guiding postoperative management. In these men, multivariate analysis showed that PTEN loss in more than 65% cancer cells identified an increased risk of developing BCR in both training (HR = 2.36, 95% CI = 1.06 to 5.26; P = .04) and validation cohorts (HR = 5.52, 95% CI = 2.36 to 12.9; P < .001) (Table 3). Addition of PTEN variable in a Cox model containing CAPRA-S increased the c-index from 0.62 to 0.69 in the validation cohort (Table 3). These findings indicate that in selected patient subsets, qPTEN assessment can add clinically significant value to standard risk assessment tools.
Table 3.
Multivariate Cox proportional hazards model for BRFS in association with % of cells in patients with CAPRA-S scores less than 5 and without adverse pathological features (positive surgical margin, seminal vesicle invasion, or lymph node involvement)
| Variable | Model without PTEN loss (% cells) |
Model with PTEN loss (% cells) |
||||||
|---|---|---|---|---|---|---|---|---|
| Training cohort |
Validation cohort |
Training cohort |
Validation cohort |
|||||
| HR (95% CI) | P * | HR (95% CI) | P * | HR (95% CI) | P * | HR (95% CI) | P * | |
| ERG fusion (Yes vs No) | 0.78 (0.39 to 1.56) | .49 | 1.38 (0.62 to 3.10) | .43 | 0.77 (0.39 to 1.54) | .46 | 1.32 (0.58 to 2.97) | .51 |
| CAPRA-S (1-unit increase) | 1.49 (1.13 to 1.96) | .005 | 1.46 (1.06 to 2.02) | .02 | 1.49 (1.13 to 1.97) | .005 | 1.42 (1.02 to 1.99) | .04 |
| PTEN loss (% cells) | ||||||||
| >65% vs intact (0%–65%)† | — | — | — | — | 2.36 (1.06 to 5.26) | .036 | 5.52 (2.36 to 12.90) | <.001 |
| C-index (SE) of the model | 0.66 (0.04) | 0.62 (0.05) | 0.68 (0.04) | 0.69 (0.05) | ||||
Two-sided log-rank test. BRFS = biochemical recurrence-free survival; CAPR-S = Cancer of the Prostate Risk Assessment Postsurgical; CI = confidence interval; HR = hazard ratio; PSA = prostate specific antigen; PTEN = phosphatase and tensin homolog.
65% group includes PTEN intact cases.
Discussion
This work is the first to report an association between BRFS and the extent of PTEN loss as assessed using a quantitative approach. Digital scoring of PTEN loss per cancer cell and visual scoring of percent cancer cores with PTEN loss were compared with a well-established visual scoring method that reports any PTEN loss. Given conflicting previous results regarding the prognostic distinction between different extents of PTEN loss (5,21), we took a more systematic approach to define the extent of PTEN loss that is most prognostically relevant. When we tested various thresholds, we found that more than 65% cancer cells and more than 50% cores with PTEN loss were stronger indicators of poor prognosis than previously reported scoring methods (5,7). Cell-based qPTEN more accurately identified cases with poor BRFS, increasing the hazard ratios associated with PTEN loss. Thus, if PTEN status is considered in guiding treatment, the more stringent quantitative threshold of more than 65% cancer cells proposed here could potentially decrease overtreatment. In support of this finding, Trotman et al. (22) demonstrated dose-dependent effects of PTEN inactivation on murine prostate cancer progression. Representing only half of patients with detectable PTEN loss, patients with levels above thresholds experienced biochemical recurrence twice as fast as patients below these thresholds.
CAPRA-S is a well-validated risk assessment tool used for prognostication of prostate cancer patients. Patients with a CAPRA-S score above 5 are considered to be at high risk of experiencing biochemical recurrence (19). Although numerous studies have demonstrated prognostic utility of CAPRA-S risk assessment tool in prostate cancer, current treatment guidelines do not include CAPRA-S, and postoperative treatment decisions are made based on the presence of either adverse pathological features (surgical margin, pathological stage, and extraprostatic extension) or rising prostate-specific antigen without adverse pathology (9,10,12). Timing, dosage, or combination of various postoperative treatment types still remain unclear and are currently assessed in ongoing clinical trials (11,12). We hypothesized that within a subgroup of patients with a CAPRA-S score less than 5 and without any adverse pathological features, qPTEN loss could help identify patients at risk of experiencing shorter BRFS who might benefit for early postoperative management. With CAPRA-S included in multivariate Cox model, PTEN variable was found statistically significant in training (HR = 2.36, 95% CI = 1.06 to 5.26; P = .036) and validation (HR = 5.52, 95% CI = 2.36 to 12.9; P < .001) cohorts. Addition of PTEN variable in Cox models containing CAPRA-S increased the c-index in both training and validation cohorts (Table 3).
We considered using PTEN loss as a continuous variable to assess risk of recurrence, an approach that could be explored further. Instead, we chose a dichotomous single threshold–based approach, which is often favored because it is more reproducible and more easily adopted in clinical practice guidelines (23). Thus, the main purpose for the qPTEN threshold was to accurately identify a group of patients who experience shorter BRFS. Graphing P values for BRFS (Figure 1) indicated a peak effect at a threshold of 65% loss. As shown in Supplementary Figure 2 (available online), a threshold of more than 65% clearly identified a group of patients who experienced statistically significant shorter BRFS with median survival of less than 6 years compared with the other groups (intact and low PTEN loss 1%–65%), whose median survival exceeded 10 years in both the training and the validation cohorts.
This is the first study to show prognostic significance for quantitative assessment of PTEN loss for personalized disease management and could be used as a proof of principle for future biomarker(s) quantification. Although the statistical significance of the threshold in multivariate analysis will depend on competing risk factors such as surgical margins, stage, and grade, the differences in BRFS above and below threshold are clearly similar in both the training and validation cohorts, which differ in patient composition (see below).
When quantifying PTEN loss, we observed a statistically significant improvement in risk stratification for percent cells over percent TMA cores. Reporting percent of cells with PTEN loss is feasible, whether by digital quantitation or visual estimation. Visual estimation may not be much different for pathologists than reporting percent high-grade cancer in biopsy specimens, which is currently being incorporated into a number of practice guidelines (24), although future studies should address whether PTEN loss is best assessed on a per-cell or per-sample basis.
The current findings may also have relevance in preoperative settings. If the current observations are confirmed in biopsies (which may require different thresholds), qPTEN assessment at diagnosis may assist with decisions to choose active surveillance or definitive treatment. Based on the findings reported here, only PTEN loss above threshold levels should be considered a reliable indicator of aggressive cancer.
We note that qPTEN was far more prognostic in the validation cohort than the training cohort. This finding was surprising, given higher BCR event rates in the training cohort (Supplementary Table 1, available online). The composition of the training and validation cohorts is different as shown in Supplementary Table 1 (available online). The validation cohort is clinically less aggressive with BCR rate of 19.9%, whereas the training cohort exhibits BCR rate of 34.9% (Supplementary Table 1, available online). Therefore, it is not surprising to find variability in hazard ratios between 2 cohorts as seen in Tables 1–3. Adjustment of prognostic power between variables is common in multivariate Cox proportional hazards analysis. In Table 2, some variables in multivariate analysis of validation cohort become statistically nonsignificant because of the presence of other powerful pathological and molecular variables such as seminal vesicle invasion and positive surgical margins in the model. Regardless of the differences in cohort compositions and tissue sampling (see below), PTEN loss (% cell) remained statistically significant with a hazard ratio greater than 2 in both cohorts compared with other clinicopathological variables (Tables 2 and 3), demonstrating generalizability of the proposed models to wider populations, which can be seen as a strength of this study (25).
We note that the training cohort sampled each patient’s cancer with 3 TMA cores, whereas the validation cohort sampled 6 cores. As expected, the training cohort sampled less than half as many cancer cells per case than the validation cohort (median 2500 cells vs 6250 cells, respectively; Supplementary Figure 3, available online). We speculate that optimal performance of qPTEN may require assessment of a certain minimum number of samples and/or cancer cells. Future studies in more clinically relevant biopsy and surgical samples are needed to explore this possibility. In addition, the subjects in this study were typical surgical candidates with relatively low risk of progression after surgery. Therefore, a larger number of events may need to be studied to further refine PTEN loss dosage effects and thresholds.
These results indicate that PTEN is a powerful prognostic biomarker for low- and intermediate-risk prostate cancer after surgery. PTEN loss is highly prognostic for BRFS regardless of ERG status. In assessing risk of BCR, the extent of PTEN loss is particularly important. Rather than combining all cases with PTEN loss into a single unfavorable category, evaluating the extent of PTEN loss with specific thresholds should allow better separation of favorable and unfavorable prognosis, especially in low- and intermediate-risk cancers where clinical risk assessment tools sometimes fail. In particular, PTEN IHC might be useful in counseling patients regarding postoperative monitoring and/or therapy. qPTEN loss on surgical and diagnostic tissues has the potential to more accurately identify clinically significant prostate cancers while reducing the rate of overtreatment.
Funding
Work by TJ, PP, and DMB was awarded by Prostate Cancer Canada and is proudly funded by the Movember Foundation (grant no. T2014-01). TJ was supported by a Transformative Pathology Fellowship funded by the Ontario Institute for Cancer Research through funding provided by the government of Ontario. PP was supported by Terry Fox Transdisciplinary Fellowship.
VO, A-MM-M, and FS are researchers of the Centre de recherche du Centre hospitalier de l’Universitéde Montréal, which receives support from the Fonds de Recherche Québec - Santé. Biobanking was done in collaboration with the Réseau de Recherche sur le cancer of the Fonds de Recherche Québec - Santé that is affiliated with the Canadian Tumor Repository Network. TMA construction was supported by the Terry Fox Research Institute. FS holds the Montreal University Research Chair in Prostate Cancer.
JAS and TV are supported by FAPESP grant no. 2015/09111–5; JAS by CNPq Bolsa Produtividade em Pesquisa - Nível: PQ-1B grant no. 306864/2014–2. MK is supported by funding from Prostate Cancer Canada, Terry Fox Research Institute-Canadian Prostate Cancer Biomarker Network, and Canadian Institutes for Health Research.
Notes
The authors have no conflicts of interest to disclose.
Study funders played no role in the design, execution, analysis, interpretation, or reporting of this work.
We would like to thank Veronique Barrès and Lee R Boudreau for tissue microarray construction and Shakeel Virk for assistance in digital scoring software. Also, we thank Queen’s Laboratory for Molecular Pathology for TMA construction, slides scanning, and digital imaging software (HALO, Indica Labs, Inc) and Drs Leszek Kotula and Vladimir Kuznetsov for critical reading of the manuscript.
Supplementary Material
References
- 1. Lotan TL, Carvalho FLF, Peskoe SB, et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol. 2015;28(1):128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jamaspishvili T, Berman DM, Ross AE, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018;15(4):222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Troyer DA, Jamaspishvili T, Wei W, et al. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. Prostate. 2015;75(11):1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Krohn A, Freudenthaler F, Harasimowicz S, et al. Heterogeneity and chronology of PTEN deletion and ERG fusion in prostate cancer. Mod Pathol. 2014;27(12):1612–1620. [DOI] [PubMed] [Google Scholar]
- 5. Lotan TL, Wei W, Ludkovski O, et al. Analytic validation of a clinical-grade PTEN immunohistochemistry assay in prostate cancer by comparison with PTEN FISH. Mod Pathol. 2016;29(8):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yun JW, Lee S, Ryu D, et al. Biomarkers associated with tumor heterogeneity in prostate cancer. Transl Oncol. 2019;12(1):43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lotan TL, Gurel B, Sutcliffe S, et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res. 2011;17(20):6563–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Picanço-Albuquerque CG, Morais CL, Carvalho FLF, et al. In prostate cancer needle biopsies, detections of PTEN loss by fluorescence in situ hybridization (FISH) and by immunohistochemistry (IHC) are concordant and show consistent association with upgrading. Virchows Arch. 2016;468(5):607–617. [DOI] [PubMed] [Google Scholar]
- 9. Dess RT, Morgan TM, Nguyen PL, et al. Adjuvant versus early salvage radiation therapy following radical prostatectomy for men with localized prostate cancer. Curr Urol Rep. 2017;18(7):55. [DOI] [PubMed] [Google Scholar]
- 10. Hwang WL, Tendulkar RD, Niemierko A, et al. Comparison between adjuvant and early-salvage postprostatectomy radiotherapy for prostate cancer with adverse pathological features. JAMA Oncol. 2018;4(5):e175230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morgan TM, Hawken SR, Ghani KR, et al. for the Michigan Urological Surgery Improvement Collaborative. Variation in the use of postoperative radiotherapy among high-risk patients following radical prostatectomy. Prostate Cancer Prostatic Dis. 2016;19(2):216–221. [DOI] [PubMed] [Google Scholar]
- 12. Dal Pra A, Abramowitz MC, Stoyanova R, Pollack A. Contemporary role of postoperative radiotherapy for prostate cancer. Transl Androl Urol. 2018;7(3):399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gordetsky J, Epstein J. Grading of prostatic adenocarcinoma: current state and prognostic implications. Diagn Pathol. 2016;11(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mingo J, Luna S, Gaafar A, et al. Precise definition of PTEN C-terminal epitopes and its implications in clinical oncology. NPJ Precis Oncol. 2019;3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chaux A, Albadine R, Toubaji A, et al. Immunohistochemistry for ERG expression as a surrogate for TMPRSS2-ERG fusion detection in prostatic adenocarcinomas. Am J Surg Pathol. 2011;35(7):1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park K, Tomlins SA, Mudaliar KM, et al. Antibody-based detection of ERG rearrangement-positive prostate cancer. Neoplasia. 2010;12(7):590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Statistics for Biology and Health. New York, NY: Springer; 2000:39–77.
- 18. Hothorn T, Lausen B. On maximally selected rank statistics. R News. 2002;2(1):3–5. [Google Scholar]
- 19. Cooperberg MR, Hilton JF, Carroll PR. The CAPRA-S score. Cancer. 2011;117(22):5039–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Punnen S, Freedland SJ, Presti JC, et al. Multi-institutional validation of the CAPRA-S score to predict disease recurrence and mortality after radical prostatectomy. Eur Urol. 2014;65(6):1171–1177. [DOI] [PubMed] [Google Scholar]
- 21. Ahearn TU, Pettersson A, Ebot EM, et al. A prospective investigation of PTEN Loss and ERG expression in lethal prostate cancer. J Natl Cancer Inst. 2015;108(2):djv346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Trotman LC, Niki M, Dotan ZA, et al. PTEN dose dictates cancer progression in the prostate. PLoS Biol. 2003;1(3):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huber F, Montani M, Sulser T, et al. Comprehensive validation of published immunohistochemical prognostic biomarkers of prostate cancer—what has gone wrong? A blueprint for the way forward in biomarker studies. Br J Cancer. 2015;112(1):140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grignon DJ. Prostate cancer reporting and staging: needle biopsy and radical prostatectomy specimens. Mod Pathol. 2018;31(S1):96–109. [DOI] [PubMed] [Google Scholar]
- 25. Taylor JMG, Ankerst DP, Andridge RR. Validation of biomarker-based risk prediction models. Clin Cancer Res. 2008;14(19):5977–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.