Abstract
Russula subsection Amoeninae is morphologically defined by a dry velvety pileus surface, a complete absence of cystidia with heteromorphous contents in all tissues, and spores without amyloid suprahilar spot. Thirty-four species within subsection Amoeninae have been published worldwide. Although most Russula species in South Korea have been assigned European or North American names, recent molecular studies have shown that Russula species from different continents are not conspecific. Therefore, the present study aims to: 1) define which species of Russula subsection Amoeninae occur on each continent using molecular phylogenetic analyses; 2) revise the taxonomy of Korean Amoeninae. The phylogenetic analyses using the internal transcribed spacer (ITS) and multilocus sequences showed that subsection Amoeninae is monophyletic within subgenus Heterophyllidiae section Heterophyllae. A total of 21 Russula subsection Amoeninae species were confirmed from Asia, Australia, Europe, North America, and Central America, and species from different continents formed separate clades. Three species were recognized from South Korea and were clearly separated from the European and North American species. These species are R. bella, also reported from Japan, a new species described herein, Russula orientipurpurea, and a new species undescribed due to insufficient material.
Keywords: Amoeninae , Heterophyllae , multilocus phylogeny, Russula orientipurpurea, species delimitation
Introduction
Russula Pers. is the largest genus in the family Russulaceae, with at least 2,000 described species worldwide (Adamčík et al. 2019). Compared to most other genera of Basidiomycetes, Russula has complex morphological and chemical features (Buyck et al. 2018). The initial period of diversification of the genus has been inferred as occurring in temperate regions of the Northern hemisphere, though there is some debate as to its origin given its high diversity in tropical areas (Looney et al. 2016; Buyck et al. 2018). Recent molecular studies have recognized eight subgenera within the genus: Russula subg. Archaeae Buyck & V. Hofst., R. subg. Brevipedum Buyck & V. Hofst., R. subg. Compactae (Fr.) Bon, R. subg. Crassotunicatae Buyck & V. Hofst., R. subg. Glutinosae Buyck & X.H. Wang, R. subg. Heterophyllidiae Romagnesi, R. subg. Malodorae Buyck & V. Hofst., and R. subg. Russula (Buyck et al. 2018; Buyck et al. 2020).
In the field, specimens of Russula subsection Amoeninae Buyck are identified by their velvety pileus surface and stipe, white spore print, mild taste, and stipe flushed with pink, red, or purple. Microscopically, species of Amoeninae display subglobose spores, typically with prominent amyloid and reticulate ornamentation but without amyloid suprahilar spot. Moreover, the basidiomes completely lack gloeocystidia, and both pileipellis and lamellar edges have large subulate hyphal terminations often arising from short unbranched cells (Buyck 1988, 1994). Because of the complete absence of gloeocystidia, Sarnari (1998) altered the rank of this subsection to subgenus, proposing Russula subg. Amoenula Sarnari. Recent multilocus phylogenetic studies, however, have shown that Amoeninae is in fact a small part of R. subg. Heterophyllidiae section Heterophyllae (see position of R. violeipes Quél. and R. mariae Peck in Buyck et al. 2018).
To date, 34 species have been published worldwide within Russula subsection Amoeninae (Suppl. material 1: Table S1), some of which were not originally placed in this group (e.g. R. diversicolor Pegler, R. epitheliosa Singer, and R. variegata Romagn.). However, their microscopic features are very similar to those of other Amoeninae species. Three species have been reported from Europe: R. amoena Quél., R. amoenicolor Romagn., and R. violeipes Quél. (Sarnari 1998). Although no relevant molecular studies have been performed that include species from North America, previous studies using morphological data have listed 14 species for this area (Suppl. material 1: Table S1). Regarding the tropics, R. diversicolor and R. epitheliosa were reported from neotropical areas of Latin America (Buyck 1988), as were six species from tropical areas of Africa (Buyck 1994; Buyck and Sharp 2007) and Madagascar (Heim 1938), and two species from arid areas of Australia (Lebel and Tonkin 2007, Hyde et al. 2016). Finally, six species native to Asia have been described, two from East Asia (Chiu 1945; Hongo 1968), and four from India (Das et al. 2005, 2017; Crous et al. 2016; Hyde et al. 2016).
Historically, taxonomic studies of Russula in Asia have been influenced by European or North American literature. That is why Russula species from Asia were often assigned names of morphologically similar counterparts from Europe or North America. However, recent molecular studies have revealed that many Asian Russula species are in fact not conspecific with European or North American species (Park et al. 2014; Lee et al. 2017; Paloi et al. 2018; Song et al. 2018). Although molecular data can provide additional information that may result in more robust phylogenies (Hebert et al. 2003; Savolainen et al. 2005; Hibbett et al. 2016; Adamčík et al. 2019), the use of this type of data may be hampered by misidentifications when there are limited reference databases or when low-resolution markers are used (Hofstetter et al. 2019).
The present study aims to: 1) clearly distinguish species of Russula subsection Amoeninae from different continents through a phylogenetic analysis using updated sequence data; and 2) revise the taxonomy of Korean Amoeninae based on materials obtained from recent collections from different habitats and areas of the Korean peninsula. Four Amoeninae species were reported in South Korea: R. amoena, R. bella, R. mariae, and R. violeipes (Park et al. 2013; Lee et al. 2015). Russula amoena and R. violeipes were described from Europe, R. bella from Japan, and R. mariae from North America. With the increase in available Russula sequence data, taxonomists can investigate more precisely the boundaries and distribution of Korean species. Therefore, the present study also aims to verify whether species are conspecific between continents using internal transcribed spacer (ITS) sequences from GenBank and generated for this study in a first analysis and, in a second analysis, using a concatenated dataset of other molecular markers including the second largest subunit of RNA polymerase II (rpb2), mitochondrial small subunit ribosomal DNA region (mtSSU), and the translation elongation factor 1-alpha (tef1α).
Methods
Sampling
A total of 15 collections from the Korean peninsula were included in this study. All Korean specimens were deposited in the Seoul National University Fungus Collection (SFC) and The Herbarium Conservation Center of the National Academy of Agricultural Science (HCCN). Because of a paucity of available sequence data from other continents, eight additional specimens from USA, four from Europe, and one from India were sequenced; all non-Asian samples are from the Herbarium of Plant Science and Biodiversity Centre of the Slovak Academy of Sciences (SAV) (Table 1 and Suppl. material 2: Table S2).
Table 1.
Specimens used for the multi-locus analyses in this study. Sequences produced in this study are presented in boldface.
| Taxon | Herbarium no. | Locality | GenBank accession no. | ||
|---|---|---|---|---|---|
| rpb2 | mtSSU | tef1α | |||
| Outgroup | |||||
| R. aff. delica | 1119/BB 12.086 | Italy | KU237879 | KU237442 | KU238020 |
| R. chloroides | 572/BB 07.209 | Slovakia | KU237845 | KU237407 | KU237990 |
| R. herrerae | 239/BB 06.532 | Mexico | KU237772 | KU237330 | KU237915 |
| Other subgenera | |||||
| R. aff. griseobrunnea | 741/BB 09.344 | New Caledonia | KU237877 | KU237440 | KU238018 |
| R. cfr. liberiensis | 91/BB 06.184 | Madagascar | KU237760 | KU237318 | KU237905 |
| R. compacta | 228/B 06.295 | USA | KU237766 | KU237324 | – |
| R. edulis | 579/BB 08.167 | Madagascar | KU237850 | KU237412 | KU237993 |
| Russula sp. | 569/BB 06.066 | Madagascar | KU237842 | KU237404 | KU237987 |
| Russula sp. | 570/BB 08.178 | Madagascar | KU237843 | KU237405 | KU237988 |
| Closely related groups in subg. Heterophyllidiae | |||||
| R. aff. crustosa | 31/BB 06.616 | Canada | KU237747 | KU237305 | KU237896 |
| R. aff. madagassensis | 93/BB 06.255 | Madagascar | KU237761 | KU237319 | KU237906 |
| R. aff. virescens | 721/BB 09.021 | New Caledonia | KU237868 | KU237430 | KU238009 |
| R. amoenolens | 577/ BB 08.675 | Italy | KU237410 | KU237848 | – |
| R. amoenolens cfr. annulata | 75/BB 06.048 | Madagascar | KU237756 | KU237314 | KU237902 |
| R. amoenolens cfr. illota | 36/ BB 06.380 | Mexico | KU237750 | KU237308 | KU237898 |
| R. amoenolens cfr. pseudocarmesina | 6/BB 06.030 | Madagascar | KU237739 | KU237296 | – |
| R. amoenolens cfr. roseoalba | 82/BB 06.105 | Madagascar | KU237758 | KU237316 | – |
| R. amoenolens cfr. vesca | 45/BB 06.525 | Mexico | KU237751 | KU237309 | KU237899 |
| R. flavobrunnea var. violaceotincta | 71/ BB 06.050 | Madagascar | KU237754 | KU237312 | KU237901 |
| R. grisea | 449/BB 07.184 | Slovakia | KU237795 | KU237355 | KU237939 |
| R. ionochlora | 448/BB 07.338 | Slovakia | KU237794 | KU237354 | KU237938 |
| R. langei | 450/ BB 07.792 | France | KU237796 | KU237356 | KU237940 |
| R. madagassensis | 21/BB 06.146 | Madagascar | KU237742 | KU237300 | KU237891 |
| R. maguanensis | XHW4765 | China | MH939989 | – | MH939983 |
| R. medullata | 555/BB 07.252 | Slovakia | KU237832 | KU237392 | KU237976 |
| R. mustelina | 1176/SA 09.88 | Slovakia | KU237881 | KU237444 | KU238022 |
| R. oleifera | 254/BB 98.024 | Tanzania | KU237776 | KU237334 | KU237919 |
| R. ornaticeps | 46/BB 06.530 | Mexico | KU237752 | KU237310 | – |
| R. prolifica | 18/BB 06.161 | Madagascar | KU237741 | KU237299 | KU237890 |
| R. pulverulenta | 578/ BB 05.160 | USA | KU237849 | KU237411 | – |
| Russula sp. | 545/BB 08.061 | Madagascar | KU237823 | KU237383 | KU237967 |
| R. substriata | XHW4785 | China | MH939994 | – | MH939988 |
| Subsect. Amoeninae | |||||
| R. aff. mariae | SAV F–4484 | USA, New York State | – | MT417192 | – |
| R. aff. mariae | SAV F–4493 | USA, New York State | – | MT417193 | MT417213 |
| R. aff. mariae | SAV F–4564 | USA, New York State | – | MT417194 | MT417214 |
| R. alachuana | SAV 1252 | USA, Florida | MT417198 | MT417186 | MT417204 |
| R. alachuana | SAV F–20108 | USA, Florida | MT417199 | MT417187 | MT417206 |
| R. amoena | SAV F–1352 | Slovakia | MT417200 | MT417185 | – |
| R. amoena | SAV F–3147 | Slovakia | MT417202 | MT417190 | MT417211 |
| R. cf. amoenicolor | SAV F–20302 | Greece | MT417196 | MT417188 | MT417209 |
| R. cf. amoenicolor | SAV F–20324 | Greece | MT417197 | MT417189 | MT417210 |
| R. bella | SFC20120722–03 | South Korea | MT199642 | MT196930 | – |
| R. bella | SFC20170819–05 | South Korea | MT199643 | MT196931 | MT199655 |
| R. bella | SFC20170819–10 | South Korea | MT199644 | MT196932 | MT199656 |
| R. bella | HCCN16818 | South Korea | KF361734 | MT196933 | – |
| R. bella | HCCN15410 | South Korea | – | MT196934 | – |
| R. bella | HCCN21655 | South Korea | KF361736 | MT196935 | MT199657 |
| R. bella | SFC20170731–02 | South Korea | MT199645 | MT196936 | MT199658 |
| R. mariae | 546/BB 07.038 | USA | KU237824 | KU237384 | KU237968 |
| R. orientipurpurea sp. nov. | HCCN19111 | South Korea | KF361712 | MT196923 | MT199648 |
| R. orientipurpurea sp. nov. | HCCN18725 | South Korea | KF361710 | MT196924 | MT199649 |
| R. orientipurpurea sp. nov. | HCCN21685 | South Korea | KF361714 | MT196925 | MT199650 |
| R. orientipurpurea sp. nov. | SFC20170819–08 | South Korea | MT199638 | MT196926 | MT199651 |
| R. orientipurpurea sp. nov. | SFC20170725–37 | South Korea | MT199639 | MT196927 | MT199652 |
| R. orientipurpurea sp. nov. | SFC20170821–22b | South Korea | MT199640 | MT196928 | MT199653 |
| R. orientipurpurea sp. nov. | SFC20170726–47 | South Korea | MT199641 | MT196929 | MT199654 |
| R. pseudoamoenicolor | India | MT199646 | MT196937 | MT199659 | |
| R. violeipes | 542/BB 07.273 | Slovakia | KU237820 | KU237380 | KU237964 |
| Russula sp. | SFC20160726–13 | South Korea | MT199647 | MT196938 | MT199660 |
| Russula sp. | SAV F–20134 | USA, Florida | – | – | MT417205 |
| Russula sp. | SAV F–20117 | USA, Florida | MT417195 | – | MT417208 |
| Russula sp. | SAV F–4063 | USA, Tennessee | MT417203 | MT417191 | MT417212 |
Morphological study
Macromorphological characters were described from fresh specimens using the terminology of Vellinga (1988). The color standard codes in Kornerup and Wanscher (1978) were followed for describing the colour of the basidiomes. All microscopic characters were measured from dried herbarium samples using an Eclipse 80i light microscope (Nikon, Japan) with immersion lenses at the magnification of 1000× and using the software NIS ELEMENT BR v3.2 (Nikon, Japan). The description templates and terminology of Adamčík et al. (2019) were used for the observations of microscopic structures. The exception is that the sterile elements in hymenium have no distinct heteromorphous contents unlike hymenial cystidia of majority of Russula members, i.e. gloeocystidia. Because it is not certain if they correspond to “true gloeocystidia”, we refer to them as hymenial cystidia when observed on lamellae sides and marginal cells in case of lamellar edges. Spore ornamentation was observed using a light microscope, and a scanning electron microscope (SEM, SUPRA 55VP, Carl Zeiss, Germany) at 5,000× and 10,000× magnification. For each collection, statistics of the measurements of microscopic characters were based on 20 measurements per character. Spore measurements excluded ornamentation. We followed the protocols of chemical tests for micro-morphological observation in Adamčik et al. (2019). Statistics of microscopic characters are expressed as the mean ± standard deviation with extreme values in parenthesis. The mean values are indicated by underline. When multiple samples were available, individual measurements of all microscopic characters of a species were obtained from at least three samples and diagnostic characters of species were further used to compare with the remaining samples.
Molecular studies
DNA was extracted from fresh or dried basidiomes using a modified CTAB extraction method (Roger and Bendich 1994). Four molecular markers were used for species-level identification and to infer evolutionary relationships among species. The following primer pairs were used in the amplifications: NSI1 and NLB4 for the ITS region (Martin and Rygiewicz 2005), bRBP2-6F1 and RPB2-7R for the partial rpb2 locus (Matheny et al. 2007), MS1 and MS2 for part of the mtSSU region (White et al. 1990), and EF1-983F and EF1-2218R for the partial tef1α locus (Matheny et al. 2007). The PCR conditions were: initial denaturation of 5 min at 95 °C, 35 cycles that varied for each marker (60 s at 95 °C, 40 s at 50 °C, and 60 s at 72 °C for ITS; 40 s at 95 °C, 40 s at 58 °C, and 60 s at 72 °C for rpb2; 30 s at 94 °C, 30 s at 55 °C, and 60 s at 72 °C for mtSSU; 30 s at 94 °C, 30 s at 56 °C, and 60 s at 72 °C for tef1α), and final incubation of 7 min at 72 °C. All PCR products were checked on 1% agarose gel stained with EcoDye DNA staining solution (SolGent Co., Daejeon, South Korea) and purified with the Expin PCR purification kit (GeneAll Biotechnology, Seoul, South Korea) following the manufacturer’s instructions. DNA sequencing was conducted using an ABI3730 automated DNA Sequencer by Macrogen (Seoul, South Korea). The obtained sequences were checked and manually edited using the software FINCHTV v1.4 (Geospiza, Inc.), and then assembled manually using MEGA 7 (Kumar et al. 2016).
Phylogenetic analysis
For species delimitation of the Korean samples, ITS sequences of R. subsect. Amoeninae were downloaded from GenBank and aligned with the newly generated ITS sequences using MAFFT v7, with the E-INS-I strategy (Katoh and Standley 2013). Russula grisea and R. virescens were used as outgroup based on the results of previous studies (Buyck et al. 2008; Park et al. 2013). Maximum Likelihood (ML) analyses were conducted using RAxML 8.2.10 (Stamatakis 2014) and the GTR + G model with 1000 rapid bootstrap replicates. For rpb2, mtSSU, and tef1α regions, sequences of each locus were separately aligned and analyzed after introns were excluded. Seven partitions were assigned; mtSSU, rpb2pos1, rpb2pos2, rpb2pos3, tef1αpos1, tef1αpos2, and tef1αpos3. Substitution models of all partitions were tested using ModelTest-NG (Darriba et al. 2020). The best substitution models for the different loci under BIC were GTR+I+G for mtSSU, K80+I+G for rpb2 partitions, and TrN+I+G for tef1α partitions. Bayesian inference (BI) analysis was performed with MRBAYES v. 3.2.6 (Ronquist and Huelsenbeck 2003), with four independent runs of four chains each. The TrN substitution model for tef1α was replaced by the GTR model for this analysis. The analysis was run for 20 million generations, with sampling every 1,000th generation. At the end of the run, the average standard deviation of split frequency of runs was 0.001412. The convergence and burn-in values of runs were then checked in Tracer 1.6 (Rambaut et al. 2014). We considered clades with the bootstrap values and posterior probabilities exceeding 70% and 0.95 as well-supported. The ITS dataset and the combined dataset (rpb2-mtSSU-tef1α) are available in TreeBase (http://treebase.org/treebaseweb/; submission ID 26896 and 22640, respectively). All phylogenetic analyses were executed on the CIPRES Science Gateway (Miller et al. 2010). Three species of R. subg. Malodorae were chosen as outgroup.
Results
Phylogenetic analysis
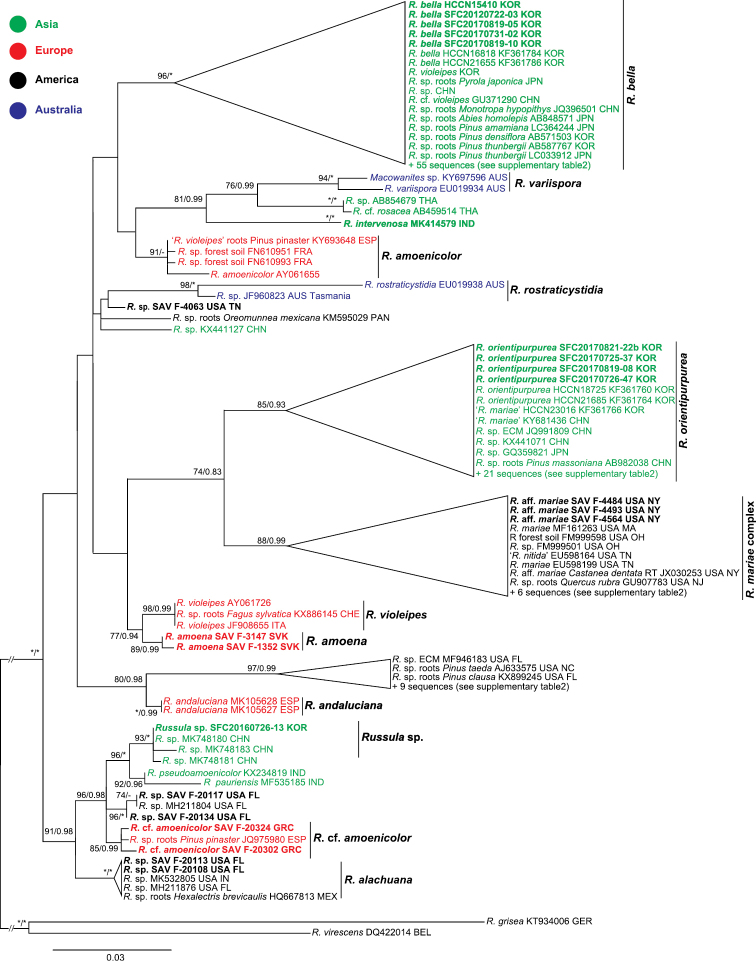
The ITS region was amplified and sequenced from 22 specimens for this study. A total of 152 ITS sequences belonging to Amoeninae were downloaded from GenBank and used in the analysis (Suppl. material 2: Table S2). The phylogenetic analysis of the ITS sequences indicated the existence of more than 21 Russula species-level clades: eight Asian species (two names undetermined), five European (one undetermined), five North American (three undetermined), two Australian, and one Central American (undetermined) (Fig. 1). However, none of the African or Malagasy species were included in this analysis as no ITS sequences were available. The Korean samples represented three phylogenetic species, and they were grouped with Asian samples and clearly separated from specimens of Australia, Europe, and North America.
Figure 1.
Maximum Likelihood (ML) tree based on internal transcribed spacer (ITS) sequences of most Russula subsect. Amoeninae and closely related species. Species in boldface are described in this study. ML bootstrap values >70 and Bayesian Inference posterior probability >0.90 are shown. Stars indicate clades with 100 ML bootstrap values and 1.0 Bayesian posterior probabilities.
A total of 72 ITS sequences were confirmed as R. bella: 5 from this study and 67 from GenBank. All of these sequences are from specimens in East Asia, i.e. from South Korea, China, and Japan. Of the ITS sequences in the R. bella clade, 31 were initially misidentified as the European R. violeipes, 35 ambiguously labelled as “Russula sp.”, and one labelled as Russula cf. violeipes. A total of 33 specimens for which ITS sequences were newly generated or retrieved from GenBank belonged to a new species clade, R. orientipurpurea. The twenty nine ITS sequences from GenBank originated from South Korea, China, and Japan. Of these, 22 were mislabelled as the North American R. mariae and seven were labelled as “Russula sp.” (Suppl. material 1: Table S1). The R. orientipurpurea clade was closely related to R. mariae, but they were clearly separated (Figs 1, 2). One specimen (SFC20160726-13) formed a unique clade with three Chinese specimens and we define it as an undetermined species. Two Indian species were positioned as a sister clade of the Russula sp. clade.
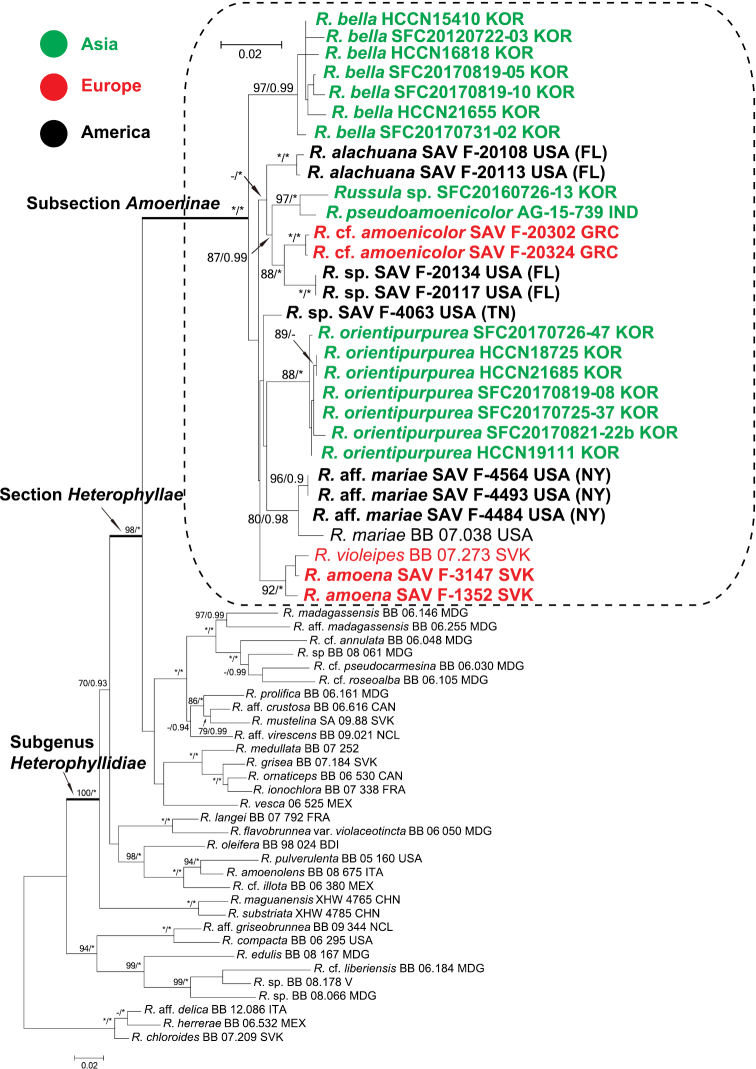
Figure 2.
Maximum Likelihood (ML) tree based on the second largest subunit of RNA polymerase II (rpb2), mitochondrial small subunit ribosomal DNA region (mtSSU), and the translation elongation factor 1-alpha (tef1α) sequences of Russula subsect. Amoeninae species and representatives from closely related subgenera. Species in boldface are described in this study. ML bootstrap values >70 and Bayesian Inference posterior probability >0.90 are shown. Stars indicate clades with 100 ML bootstrap values and 1.0 Bayesian posterior probabilities.
Russula amoena, R. amoenicolor, R. andaluciana, and R. violeipes were monophyletic (Fig. 1). The European Mediterranean samples have similar morphology with R. amoenicolor. However, phylogenetic analysis showed that they are likely not conspecific. Therefore, we named them as “R. cf. amoenicolor” in this study. The North American samples formed five clades. Of these, only two clades are labelled with species names (R. mariae and R. alachuana). The four Australian samples formed two well-supported clades that do not overlap with samples from other continents.
Sequences of three loci (rpb2, mtSSU, and tef1α) were obtained for 28 samples (Table 1) and combined with 102 sequences of 34 samples obtained from GenBank. Two short introns were detected only in tef1α and they were excluded in the phylogenetic analysis. The results of this multilocus phylogenetic analysis were similar to those of the ITS analysis. Subsection Amoeninae formed a well-supported monophyletic group (Fig. 2). Three species were found for South Korea, and the East Asian Russula species were clearly separated from European and North American species.
Morphological analysis
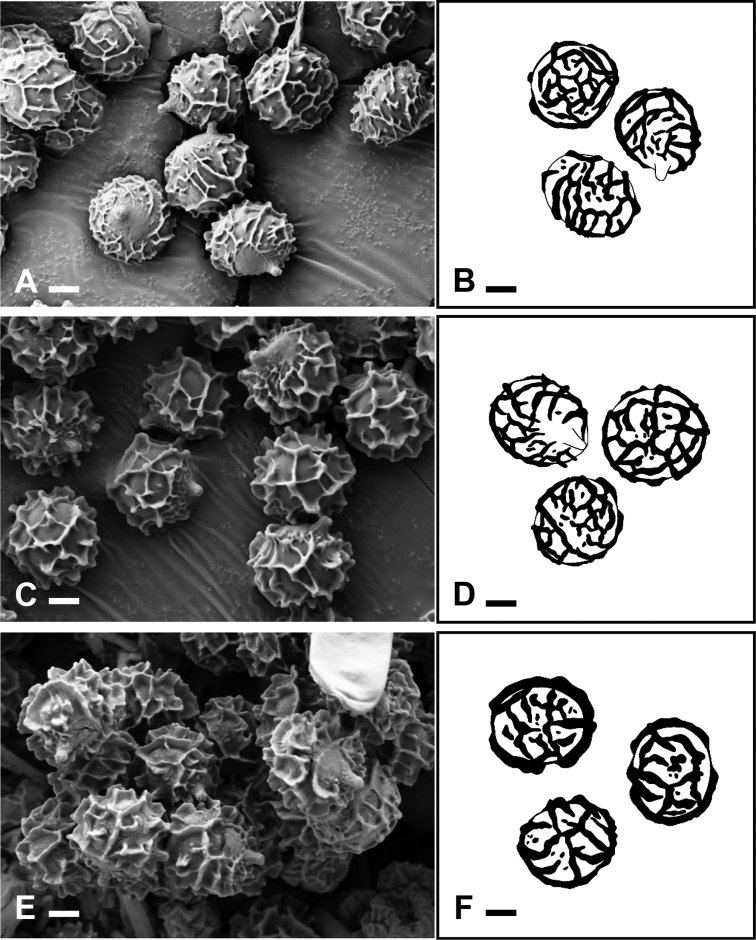
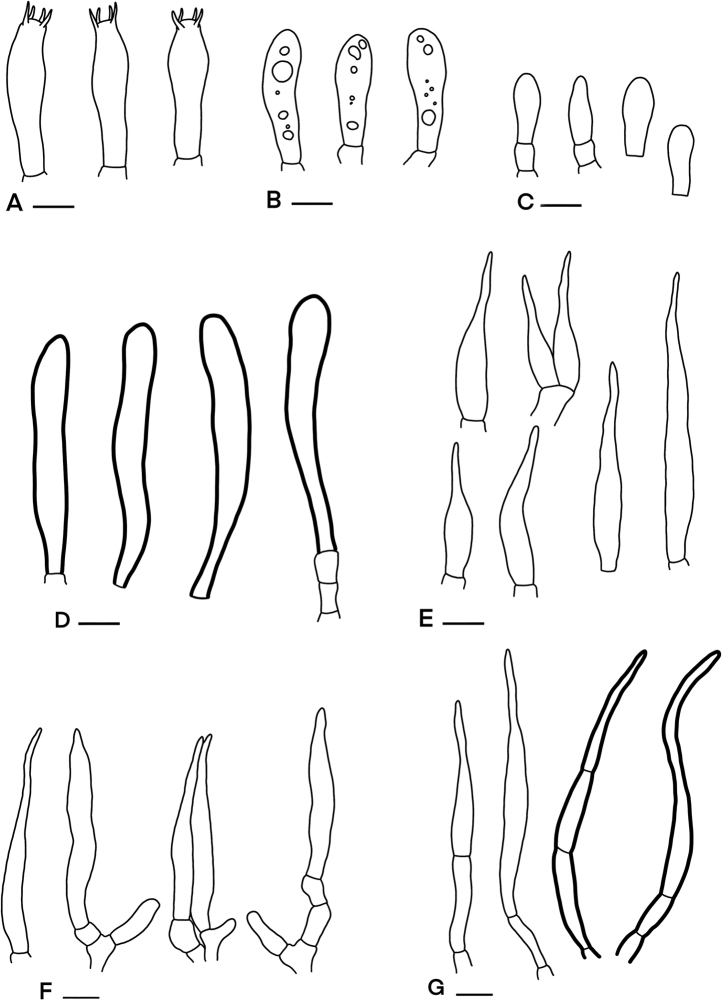
The three species found in South Korea have the typical morphological characters of subsection Amoeninae: pruinose dry pileus surface, mild taste, and stipe flushed with pink, red, or purple. In fact, they have almost completely white stipes, usually with only a faint pink color (Fig. 3). Microscopically, these species display moderately large spores with crested and subreticulate to reticulate ornamentation (Fig. 4), an absence of cystidia with well-defined contents reactant with sulfovanillin, and a pileipellis comprising mainly attenuated terminal cells of hyphae usually subtended by one or two shorter ellipsoid or subglobose cells. The pileipellis of all three species have dimorphous hyphal terminations, some with long subulate or lageniform terminal cells and others with shorter, cylindrical, or ellipsoid terminal cells (Figs 5–7). The three Korean species were easily distinguished in the field. The morphological features of the Korean R. bella are consistent with the original description of the species (Table 2). The other two Korean species, however, do not entirely agree with the description of any previously described Russula species.
Figure 3.
Basidiomes of Korean Amoeninae from this study. A, BRussula bella (A-SFC20170802-03, B-SFC20170819-05) C–ER. orientipurpurea (SFC20170819-08) F, GRussula sp. (SFC20160726-13). Scale bars: 20 mm.
Figure 4.
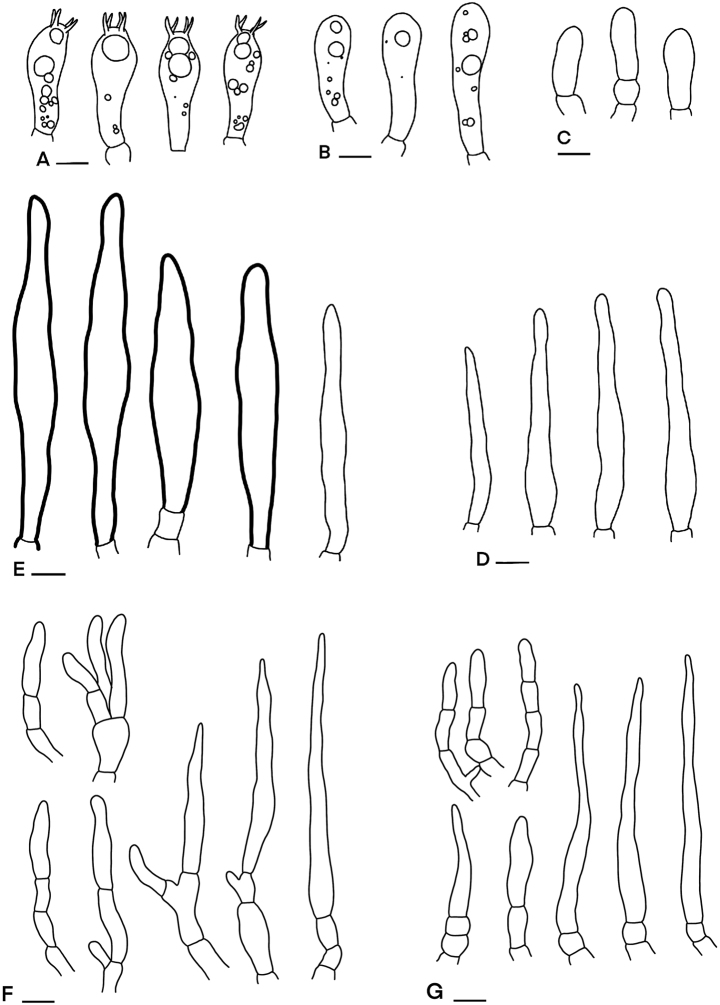
Basidiospores with scanning electron microscopy images (A, C, E) and drawing images (B, D, F) A, BRussula bella (SFC20170819-05) C, DR. orientipurpurea; (SFC20170821-22b) E, FRussula sp. (SFC20160726-13). Scale bars: 2 µm.
Figure 5.
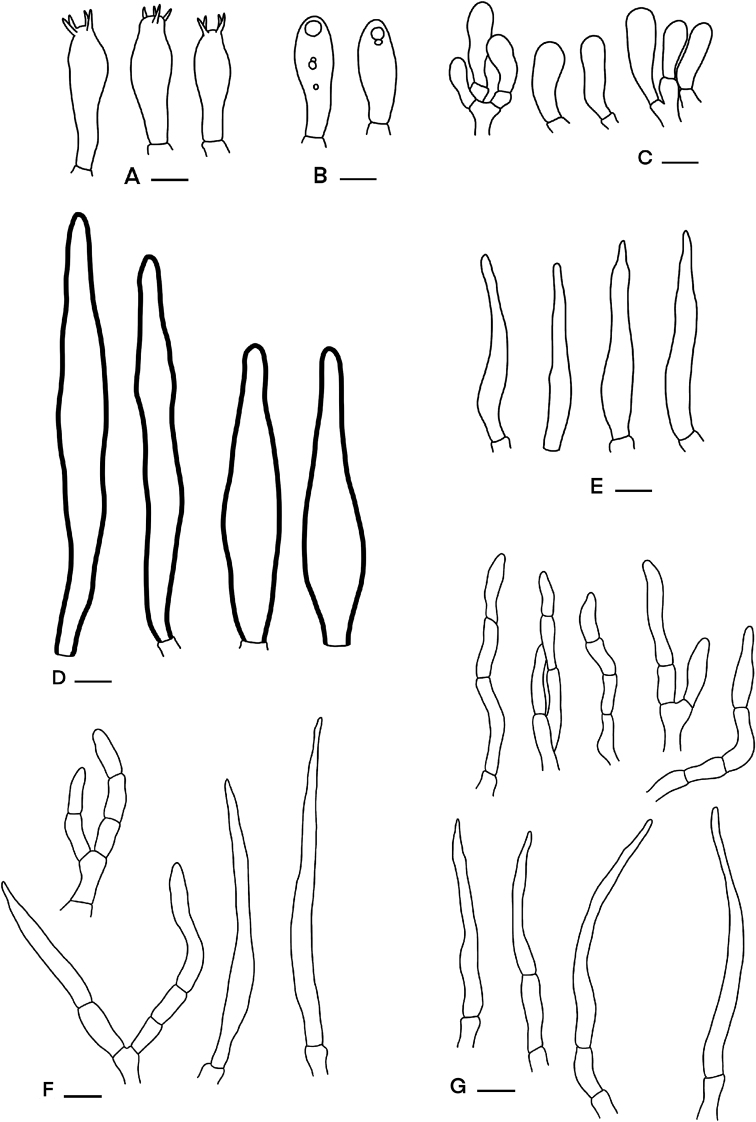
Microscopic features of Russula bella (SFC20170819-05) A basidia B basidiola C clavate marginal cells D hymenial cystidia on lamellae sides E subulate marginal cells F hyphal termination at pileus margin G hyphal termination at pileus centre. Scale bars: 10 µm.
Figure 7.
Microscopic features of Russula sp. (SFC20160726-13) A basidia B basidiola C clavate marginal cells D hymenial cystidia on lamellae sides E subulate marginal cells F hyphal termination at pileus margin G hyphal termination at pileus centre. Scale bars: 10 µm.
Table 2.
Morphological characteristics of Asian R. subsect. Amoeninae species.
| R. bella a | R. bella | R. intervenosa b | R. mukteshwarica c | R. orientipurpurea | R. pauriensis d | R. pseudoamoenicolor e | R. punicea f | Russula sp. | ||
| Pileus size (mm) | 20–45 | 20–50 | 26–49 | 65–130 | 52–60 | 53–63 | 50–100 | 25–60 | 60 | |
| Pileus colour | bright red | √ | √ | √ | √ | |||||
| pink | √ | √ | √ | √ | ||||||
| grey | √ | √ | √ | |||||||
| brown | ||||||||||
| purple | √ | √ | √ | |||||||
| violet | √ | √ | √ | √ | ||||||
| green | √ | √ | ||||||||
| bright yellow | √ | √ | ||||||||
| cream or pale yellow | √ | √ | ||||||||
| Stipe colour | almost white | √ | √ | √ | √ | |||||
| partly pink | √ | √ | √ | √ | √ | |||||
| partly purple | √ | √ | √ | √ | ||||||
| partly violet | √ | |||||||||
| Spore size | length (µm) | 6.5–7.5 | 6.5–7.7 | 7–8 | 7.6–9.3 | 6.9–7.8 | 6–8 | 6–9.5 | 6.5–7 | 6.5–7 |
| width (µm) | 5.5–6 | 5.3–6.0 | 6.5–7 | 7.3–8.2 | 6–6.9 | 5.5–7 | 5–8 | 5–6 | 5.6–6.2 | |
| mean (length × width) | 7.1 × 5.7 | 7.5 × 6.7 | 7.3 × 6.4 | 6.9–6.3 | 7.3–6.3 | 6.8 × 5.9 | ||||
| Q value | 1.17–1.33 | 1.07–1.19 | 1.00–1.15 | 1.09–1.19 | 1.00–1.17 | 1.03–1.33 | 1.10–1.19 | |||
| Spore ornamentation | subreticulate | √ | √ | √ | √ | √ | √ | √ | √ | √ |
| reticulate | √ | √ | ||||||||
| height (µm) | 0.4–1.0 | 0.6–0.9 | 0.75 | 0.6–1 | -2 | -2 | 0.7–1.2 | |||
| Cystidia on lamellae sides | length (µm) | 44–55 | 52–75 | 29–34 | 80–110 | 74.5–101 | 55–135 | 90–117 | 45–60 | 66.5–91.5 |
| width (µm) | 5.5–7 | 7.5–10.5 | 10–12.5 | 11–17 | 10.5–15 | 12–22 | 10–21 | 9–12 | 12.5–16.5 | |
| cylindrical or clavate | √ | √ | √ | |||||||
| subulate or lageniform | √ | |||||||||
| fusiform | √ | √ | √ | √ | √ | √ | √ | |||
| obtuse | √ | √ | √ | √ | √ | |||||
| acute | √ | √ | √ | √ | ||||||
| Cystidia on lamellae edges | length (µm) | 37–65 | 38.5–63 | 32–39 | 70–100 | 48–88 | 36–68 | 30–85 | 42.5–56.5 | |
| width (µm) | 5.5–7 | 5.5–7.5 | 5.5–7 | 11–17 | 5.5–10.5 | 8–15 | 7–10 | 5.5–7 | ||
| different from sides | √ | √ | √ | |||||||
| TC (margin) | length (µm) | 44–80 | 47–76 | 39–47 | 55.5–89 | 9–64 | 11–65 | 60–85 | ||
| width (µm) | 5.3–8 | 5–7 | 2.5–4.5 | 5–11 | 5–7 | 4–10 | 4–10 | 6–9 | 4.5–6 | |
| Short cells | number | 1–2 | 2–4 | 0–2 | 2–4 | 1–2 | ||||
| subterminal width (µm) | -12 | -14 | ||||||||
| TC centre | width (µm) | 4.5–7.5 | 4.5–7.5 | 3–4 | ||||||
References: a-Hongo 1968, b-Crous et al. 2016, c-Das et al. 2005, d-Das et al. 2017, e-Hyde et al. 2016, f-Chiu 1945, Size unit of pileus is mm and the other microscopic characters are µm. Species in boldface are described in this study.
Taxonomy
Russula bella
Hongo, Memoirs of Shiga University 18:50 (1968)
01ADF8C7-693F-504F-91A2-3BAB8ABB498B
Diagnosis.
Pileus medium-sized, 20–50 mm diam., plano-convex to convex when young, applanate with depressed center to infundibuliform when mature, often lobate, margin with short striation, sometimes cracking in age; cuticle smooth, pruinose, viscid and shiny when wet, cuticle peeling approximately to half of the pileus radius or sometimes almost to the center, color variable, typically darker at the disk, greyish red (11D5), brownish violet (11D6) to bluish red (12A6-A8, 12B7), few with yellowish brown spots, margin pink (11A5) to greyish rose (11B5). Lamellae 2–3 mm deep, brittle, adnate, approximately 11–19 per cm near the pileus margin, moderately distant to crowded, yellowish white to pale cream; lamellulae absent; edge entire and concolorous. Stipe 17–34 × 5–9 mm, centrally attached or eccentric, cylindrical to slightly tapered towards the base, surface dry, longitudinally striate, whitish with a pinkish flush; solid when young, becoming hollow in age. Context 1–3 mm thick near the stipe, white, unchanging after cutting, turning greenish with FeSO4, turning quickly yellow with KOH, pale violet with PDAB; taste mild; odor slightly fruity. Spore print pale cream to white.
Basidiospores (n = 60, 3, 3) (6–)6.5–7.1–7.7(–8.8) × (5–)5.3–5.7–6(–6.6) µm, broadly ellipsoid, Q = (1.11–)1.17–1.25–1.33(–1.49), ornamentation thin, 0.4–1 µm high ridges forming an incomplete reticulum (2–7 in a 3 µm circle) with some dispersed isolated warts (0–2 warts in a 3 µm circle), suprahilar spot not amyloid, smooth. Basidia (25.1–)30–35.3–40.5(–48) × (7–)8.5–9.4–10.5(–11.5) µm, 4-spored, narrowly clavate, with guttate or granular contents. Basidiola (27.3–)28.4–32–35.5(–41.7) × (8.1–)8.7–9.4–10.3(–10.6) µm, narrowly clavate, with guttate or granular contents. Hymenial cystidia on lamellae sides inconspicuous, widely dispersed, up to 100 per mm2, (37.5–)52–63.5–75(–90) × (5.5–)7.5–9–10.5(–11.5) µm, clavate to subcylindrical, originating from subhymenium, apically obtuse, thick-walled with walls up to 0.8 µm; contents optically empty, negative in sulfovanillin. Lamellar edge with dispersed basidia, true gloeocystidia (with differentiated contents) absent; marginal cells very abundant, resembling terminal cells in the pileus, typically narrowly lageniform or subulate (24–)38.5–50.8–63(–83) × (4–)5.5–6.8–7.5(–9.5) µm; shorter clavate to subcylindrical with obtuse apex present, (9–)12.5–15.7–18.5(–23.5) × (3.5–)4.5–5.7–7(–7.5) µm. Pileipellis orthochromatic in Cresyl blue, trichoderm, sharply delimited from the underlying context, 280‒400 µm thick, with a well-defined, gelatinized, 150‒200 µm thick suprapellis of ascending or erect hyphal terminations forming a trichoderm, subpellis 130–200 µm thick, dense, horizontally oriented, sldender and gelatinized hyphae; acid-resistant incrustrations absent. Hyphal terminations near the pileus margin unbranched, often slightly flexuous, either long and attenuated or subcylindrical and obtuse, the attenuated ones more abundant, with terminal cells (20.5–)47–61.4–76(–85) × (3.5–)5–5.9–7(–8) µm, subulate or narrowly lageniform, rarely fusiform, apically acute to subacute, thin-walled, usually followed by 1–2 shorter and often more inflated cells before the branching; subcylindrical ones shorter, with terminal cells (7.5–)33.5–51.2–69(–107.5) × (2–)3.5–4.7–6(–7) µm, frequently originate from branched cells, sometimes with one shorter unbranched subterminal cell. Hyphal termination near the pileus center also dimorphous, the attenuated ones with terminal cells (10.5–)18.5–50.5–82.5(–104.5) × (3.5–)4.5–6.0–7.5(–10) µm, mainly subulate, occasionally narrowly fusiform, apically acute and sometimes acute-pointed, often with thickened walls (up to 0.8 µm), shorter cylindrical hyphae with terminal cells (9–)12.5–18.6–24.5(–32) × (2.5–)3–4–5(–6) µm; followed by 1–3 unbranched shorter cells, subterminal cells not usually not distinctly wider. Pileocystidia absent. Cystidioid or oleipherous hyphae in subpellis or context absent.
Ecology.
gregarious to scattered on soil in mixed forest with Quercus aliena, Pinus densiflora, and Populus sp.
Studied materials.
South Korea. Jeollanam-do, Haenam-gun, Mt. Duryun, 614 m elev., 34°27'24"N, 126°37'07"E, Yang Seop Kim, 5 September 1985, HCCN1457A (HCCN!); Chungcheongbuk-do, Danyang-gun, Mt. Sobaek, 790 m elev., 36°57'29"N, 128°26'35"E, Soon Ja Seok, 13 July 2007, HCCN15410 (HCCN!); Gyeonggi-do, Suwon-si, Seonggyungwan University, 58 m elev., 37°17'42"N, 126°58'22"E, Soon Ja Seok, 1 August 2008, HCCN16735 (HCCN!); Gangwon-do, Wonju-si, 275 m elev., 37°19'59"N, 127°54'35"E, Soon Ja Seok, 6 August 2008, HCCN16818 (HCCN!); Gyeonggi-do, Suwon-si, Seonggyungwan University, 48 m elev., 37°17'28"N, 126°58'24"E, Hye Yeong Choi, 5 August 2011, HCCN21655 (HCCN!); Daejeon-si, Yuseong-gu, 105 m elev., 36°23'48"N, 127°20'13"E, Myung Soo Park, 31 July 2012, SFC20120731-02 (SFC!); Gyeongsangbuk-do, Ulleung-gun, Mt. Seonginbong, 420 m elev., 37°30'50"N, 130°52'10"E, Seung-Yoon Oh, Won Ju Kim, Young Woon Lim, 14 August 2012, SFC20120814-23 (SFC!); Chungcheongnam-do, Seosan-si, Yonghyeon Natural Recreation Forest, 151 m elev., 36°45'53"N, 126°36'10"E, Young Ju Min, Won Ju Kim, Hyun Lee, 10 October 2012, SFC20121010-06 (SFC!); Seoul, Gwanak-gu, Seoul National University, 103 m elev., 37°27'26"N, 126°56'59"E, Komsit Wisitrassameewong, 31 July 2017, SFC20170731-02 (SFC!); ibid., 19 August 2017, SFC20170819-05 (SFC!).
Comments.
Russula bella is morphological similar to R. pseudoamoenicolor, R. punicea, and R. violeipes. Russula pseudoamoenicolor has a more vividly colored pileus and a larger basidiome and hymenial cystidia than those of R. bella (Hyde et al. 2016). Russula punicea differs from R. bella in the shape of hymenial cystidia; the former has acute cystidia, whereas the latter has obtuse cystidia (Chiu 1945; Hongo 1968). Russula violeipes differs from R. bella in the yellowish to greenish smeared violet color of the pileus (Kränzlin 2005). Moreover, R violeipes has larger basidia (45–65 × 11–14 µm) and pleurocystidia (80–115 × 12–15 µm) than those of R. bella (Kränzlin 2005).
Russula orientipurpurea
Wisitr., H. Lee & Y.W. Lim sp. nov.
47144CFE-DB12-566F-978B-B64361E338E2
MycoBank No: 835272
Figure 6.
Microscopic features of Russula orientipurpurea (SFC20170725-37) A basidia B basidiola C clavate marginal cells D hymenial cystidia on lamellae sides E subulate marginal cells F hyphal termination at pileus margin G hyphal termination at pileus centre. Scale bars: 10 µm.
Material examined.
Holotype. South Korea. Jullanam-do, Yeosu-si, Dolsando islands, 202 m elev., 34°35'24"N, 127°47'57"E, Komsit Wisitrassameewong, Jae Young Park, 25 July 2017, SFC20170725-37 (Holotype, SFC!).
Etymology.
‘orientipurpurea’ refers to the origin of the species, East Asia, and its typical purple color of pileus.
Diagnosis.
Pileus surface with pale cream with flushed pale purple to purple stains; spores with almost complete to complete reticulum; subfusiform to fusiform hymenial cystidia.
Pileus medium-sized, 52–60 mm diam., plano-convex to applanate with the deeply depressed center, margin inconspicuously striate up to 2 mm, acute, even; surface smooth, pruinose, slightly waxy, matt, slightly viscid when wet, cuticle peeling 1/2 to 3/4 of the pileus radius, color pale cream to cream, with darker shade of cream towards the center, typically flushed with pale or darker purple stains, sometimes with radial stripes of greyish ruby (12E5). Lamellae 4–5 mm deep, adnate, moderately distant, approximately 11–18 per cm near the pileus margin, white to pale yellow (3A3), furcations sometimes present near the stipe, lamellulae occasional, edge even. Stipe 40–50 × 11–13 mm, centrally attached, cylindrical, surface smooth, longitudinally striate, color white and sometimes with a greyish red (11D4-D5) flush; hollow. Context 2–3 mm thick at half of the pileus radius, white, rather firm but fragile in stipe when mature, turning slowly greenish with FeSO4 and pale orange to orange white with KOH; taste mild; odor slightly fruity. Spore print cream white to white.
Basidiospores (n = 80, 4, 4) (6.3–)6.9–7.3–7.8(–8.6) × (5.4–)6–6.4–6.9(–7.6) µm, subglobose to broadly ellipsoid, Q = (1.06–)1.09–1.14–1.19(–1.28), ornamentation of thin to moderately thick, 0.6–1.4 µm high ridges forming an incomplete or complete reticulum (3–7 in a 3 µm circle), in a 3 µm circle isolated warts rare (0–1 in a 3 µm circle), suprahilar spot not amyloid, small, surrounded by fine and less prominent reticulation. Basidia (28–)32–36.2–40(–45) × (7.5–)9.5–10.9–12.5(–14.5) µm, 4-spored, clavate. Basidiola (27.8–)33.4–37.7–42.1(–43.4) × (9–)9.7–10.5–11.4(–12.2) µm, narrowly clavate, with guttate or granular contents. Hymenial cystidia on lamellae sides widely dispersed to dispersed, 200–700 per mm2, (64–)74.5–87.9–101(–131) × (8.5–)10.5–12.8–15(–18.5) µm, mostly fusiform or narrowly fusiform, occasionally clavate, originating from subhymenium, emergent or not beyond basidia, apically constricted but obtuse, usually with thickened walls (up to 0.8 µm), contents optically empty, negative in sulfovanillin. Lamellar edge with dispersed basidia, true gloeocystidia (with differentiated contents) absent; marginal cells very abundant, resembling terminal cells in the pileus, usually narrowly lagenifom or subfusiform, apically narrowed but obtuse (28.5–)48–67.8–88(–121) × (3–)5.5–8.2–11(–14.5) µm; shorter narrowly clavate to clavate, (10.5–)13.5–18–22.5(–27.5) × (3.5–)5–6.5–8(–10.5) µm. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 170–240 µm thick, with a well-defined, gelatinized, 30–60 µm thick suprapellis of ascending or erect hyphal terminations forming a trichoderm, subpellis 180–250 µm thick, dense, horizontally oriented, gelatinized and branched cylindrical to inflated hyphae; acid-resistant incrustrations absent. Hyphal terminations near the pileus margin unbranched, apically often flexuous, either long and attenuated or subcylindrical, short and obtuse, the attenuated ones (39.0–)55.5–72.3–89.0(–112.0) × (2.5–)5.0–6.1–7.2(–8.5) µm, subulate or narrowly fusiform, sometimes slightly moniliform, apically subacute, thin-walled, subterminal cells shorter, cylindrical ones with terminal cells (10–)16.5–25.3–34(–57) × (3–)4–5.2–6(–7.5) µm, apically sometimes slightly constricted, apically obtuse; followed by 0–2 unbranched short, equally wide cells, sometimes originate from branched cells. Hyphal termination near the pileus center also dimorphous, the attenuated ones prevailing with terminal cells (10.5–)18.5–50.5–82.5(–104.5) × (3.5–)4.5–6.0–7.5(–10) µm, subulate, thin-walled, apically subacute, cylindrical ones with terminal cells (13.5–)19–42.9–66.5(–85.5) × (3.5–)4.5–5.6–6.5(–9.0) µm. Pileocystidia absent. Cystidioid or oleipherous hyphae in subpellis or context absent.
Ecology.
solitary to scattered on soil in mixed forest with Quercus and Pinus trees.
Studied materials.
South Korea. Chungcheongnam-do, Gongju-si, Mt. Museong, 341 m elev., 36°35'52"N, 127°01'59"E, Hyun Lee, Seung-Yoon Oh, 26 July 2012, SFC20120726-37 (Paratype SFC!); Incheon-si, Gangwha-gun, Mt. Goryeo, 228 m elev., 37°44'54"N, 126°26'01"E, Young Woon Lim, 4 August 2012, SFC20120804-09 (Paratype, SFC!); Seoul, Gwanak-gu, Mt. Gwanak, 154 m elev., 37°27'06"N, 126°56'34"E, Hyun Lee, Won Ju Kim, 25 August 2012, SFC20120825-02 (Paratype SFC!); ibid, 202 m elev., 37°27'34"N, 126°56'19"E, Hyun Lee, Myung Soo Park, 31 August 2012, SFC20120831-04 (Paratype SFC!); ibid, 238 m elev., 37°26'53"N, 126°54'11"E, Hyun Lee, Komsit Wisitrassameewong, 19 August 2017, SFC20170819-08 (Paratype SFC!); Gyeongsangnam-do, Hapcheon-gun, Mt. Gaya, 631 m elev., 35°47'59"N, 128°05'46"E, Jae Young Park, Komsit Wisitrassameewong, Ki Hyeong Park, 26 July 2017, SFC20170726-47 (Paratype SFC!); Gyeongsangbuk-do, Ulleung-gun, Nari basin, 395 m elev., 37°31'03"N, 130°52'11"E, Jae Young Park, Nam Kyu Kim, 21 August 2017, SFC20170821-22b (Paratype SFC!).
Comments.
Russula orientipurpurea is common in mixed forests in South Korea. This species was misidentified as the North American R. mariae (Park et al. 2013). The spores in R. orientipurpurea have more complete ridge connections (3–7 lines in the 3 µm circle) and a smaller number of warts (0–1 warts in the 3 µm circle) than those in R. mariae (lines 1–4, warts 4–6). Russula orientipurpurea is morphologically similar to R. intervenosa. However, the dark red pileus centere, broader hymenial cystidia (up to 18 µm), and the thicker pileipellis (250–600 µm) of R. intervenosa (Crous et al. 2016) distinguishes this species from R. orientipurpurea (see Table 2).
Russula
sp.
9701A921-8289-5DF9-8505-D2CE73E4F70C
Diagnosis.
Pileus medium-sized, 60 mm diam., applanate with deeply depressed center, margin distinctly striate, crenulate, radially cracking; cuticle dry, viscid when moist, pruinose, peeling to 1/2 of the pileus radius, color greyish violet (18E5-E6), with dark violet (18F4-F6) patches, towards margin violet grey (18D2) to dull violet (18D3). Lamellae 3 mm deep, adnate, dense, pale cream; lamellulae rare, forked near the stipe; edge smooth and concolorous. Stipe 55 × 8–10 mm, centrally attached, tapering downwards base, surface dry, longitudinally-striated, white and flushed with purple. Context 2 mm thick at half of the pileus radius, white, unchanging; taste and odor not recorded. Spore print cream white.
Basidiospores (n = 20, 1, 1) (6.4–)6.5–6.8–7(–7.2) × (5.5–)5.6–5.9–6.2(–6.5) µm, Q = (1.08–)1.1–1.14–1.19(–1.24), subglobose to broadly ellipsoid, ornamentation of thin to moderately thick, 0.8–1.4 µm high ridges forming an incomplete reticulum (2–6 in a 3 µm circle), isolated warts rare (0–2 in a 3 µm circle), suprahilar spot not amyloid, small. Basidia (32–)34–36.2–38.5(–40) × (9–)9.5–10.5–11(–11.5) µm, 4-spored, clavate. Basidiola (24–)25.8–31–35(–42.5) × (8.3–)8.9–9.9–10.8(–11.4) µm, narrowly clavate, with guttate or granular contents. Hymenial cystidia on lamellae sides widely dispersed to dispersed, 100–500 per mm2, (63)66.5–79–91.5(–109) × (10.5–)12.5–14.5–16.5(–18) µm, narrowly fusiform or lageniform, originating from subhymenium and emergent beyond basidia, apically acute or obtuse but narrowed, thick-walled (walls up to 0.9 µm), contents optically empty, negative in sulfovanillin. Lamellar edge with dispersed basidia, true gloeocystidia (with differentiated contents) absent; marginal cells very abundant, resembling terminal cells in the pileus, narrowly lageniform or subulate (36.5–)42.5–49.6–56.5(–70.1) × (5–)5.5–6.3–7(–7.5) µm, narrowly lageniform or subulate, apically acute; narrowly clavate to clavate with optuse apex, (11.5–)14.5–17.8–21.2(–23.2) × (4–)5.2–6.2–7(–7.5) µm. Pileipellis orthochromatic in Cresyl blue, sharply delimited from the underlying context, 250–400 µm thick, with gelatinous matter, cystidoid hyphae prevailing, shorter cylindrical terminal hyphae present, parallel or repent scattered, pileocystidia absent, with a well-defined, gelatinized, 70–120 µm thick suprapellis of ascending or erect hyphal terminations forming a trichoderm, subpellis 230–350 µm thick, dense, horizontally oriented, gelatinized and branched cylindrical hyphae; acid-resistant incrustrations absent. Hyphal terminations near the pileus margin unbranched, usually only slightly flexuous, either long and attenuated or subcylindrical, short and obtuse; the attenuated ones more frequent, with terminal cells (45.0–)60.0–72.6–85.0(–96.5) × (3.5–)4.5–5.1–6.0(–6.5) µm, subulate, apically acute, thin-walled, shorter ones with terminal cells (18–)20.5–28.7–37.0(–48.0) × (3.0–)3.5–4.4–5.0(–6.0) µm, cylindrical or subcylindrical, apically obtuse but often constricted, thin walled; followed by (0–)1–2(–3) unbranched shorter and equally wide cells. Hyphal termination near the pileus center similar but shorter and narrower, attenuated longer ones with terminal cells (38.5–)43.5–53.4–63.0(–73.0) × (2.5–)3–3.5–4.0(–4.5) µm; shorter cylindrical ones with terminal cells (11.5–)13.5–16.4–19.0(–22.0) × (3.0–)3.5–4.1–4.5(–5.5) µm. Pileocystidia absent. Cystidioid or oleipherous hyphae in subpellis or context absent.
Ecology.
solitary on soil in deciduous forest, near Quercus mongolica.
Material examined.
South Korea. Incheon-si, Ongjin-gun, Jangbongdo islands, 72 m elev., 37°31'55"N, 126°21'10"E, Jae Young Park, Nam Kyu Kim, Suldbold Jargalmaa, 26 July 2016, SFC20160726-13 (Holotype, SFC!).
Comments.
This species is morphologically similar to R. mukteshwarica, but phylogenetically close to R. pseudoamoenicolor and R. pauriensis. Russula mukteshwarica and R. pauriensis differ from Russula sp. in the greenish yellow region at the pileus center, which is present in the first two species but absent in the latter (Das et al. 2005, 2017). Russula pseudoamoenicolor has a discolorous lamella edge (Hyde et al. 2016), whereas Russula sp. has a concolorous lamellae edge. Russula sp. has a purple pileus similar to that of R. violeipes; however, spore ornamentation height (up to 0.7 µm) and pleurocystidia size (80–115 × 12–15 µm) distinguishes Russula sp. from R. violeipes (Kränzlin 2005).
Key characters to Asian species in R. subsect. Amoeninae
| 1 | Basidiome with pinkish pileus | 2 |
| – | Basidiome with violet or purple pileus | 4 |
| 2 | Obtuse hymenial cystidia, subreticulate spore ornamentation, in mixed Pinus and Quercus forests | R. bella |
| – | Acute hymenial cystidia, subreticulate to reticulate spore ornamentation | 3 |
| 3 | Reticulate spore ornamentation, large hymenial cystidia (46‒60 × 9‒12 µm), broad hyphal terminations (6‒9 µm wide) in the pileipellis | R. punicea |
| – | Subreticulate spore ornamentation, small hymenial cystidia (29‒34 × 10‒12.5 µm), narrow hyphal termination (2.5‒4.5 µm wide) in the pileipellis | R. intervenosa |
| 4 | Reticulate spore ornamentation, pileus with pale cream to yellowish grey color and purplish tinges | R. orientipurpurea |
| – | Subreticulate spore ornamentation, pileus with vivid color | 5 |
| 5 | Pileus with violet/greenish/yellowish shades, stipe with purplish flush, large and broad hymenial cystidia | 6 |
| – | Pileus without greenish or yellowish shades | 7 |
| 6 | Small spores with high ornamentation (up to 2 µm), lamellae yellowish, lamellulae absent | R. pauriensis |
| – | Large spores with low ornamentation (up to 0.75 µm), lamellae yellowish to greenish, lamellulae present | R. mukteshwarica |
| 7 | Lamellar edge discolorous (pastel violet), spore ornamentation up to 2 µm high | R. pseudoamoenicolor |
| – | Lamellar edge concolorous, spore ornamentation up to 1.2 µm high | Russula sp. (SFC20160726-13) |
Discussion
The phylogenetic analyses showed that subsection Amoeninae forms a well-supported monophyletic group. Moreover, the species of Asian, Australian, European, and North American origin form separate clades. Similar results have been reported for other species groups in the various ectomycorrhizal genera of Russulaceae, which are often endemic to specific geographical regions or a single continent (Cho et al. 2018; De Crop et al. 2018; Wang et al. 2018; Lee et al. 2019; Looney et al 2020). Their host plants may have acted as bridges for species dispersal and diversification (Looney et al. 2018), and geographic distance and climate disjunctions may have caused species divergence (Caboň et al. 2019; Lee et al. 2019). The present study included recently obtained sequences of well-known species from different continents except Africa, which will provide useful information for understanding the diversity in this section.
Four Amoeninae species had been previously reported from South Korea based on morphological characteristics: two European, one North American, and one Japanese species (Lee et al. 2015). Based on ITS, large ribosomal subunit (LSU), and rpb2 data, Park et al. (2013) found that the Korean Amoeninae species were R. mariae (North American) and R. violeipes (European). The differences between those results and the present study may be traced back to the limited amount of sequences available at the time. The dramatic increase in the number of sequences available in public databases and the data obtained in the present study allow us to conclude that the species previously identified as R. mariae and R. violeipes in South Korea are in fact R. orientipurpurea and R. bella, respectively. We also showed that R. orientipurpurea forms a distinct clade that is quite distantly related to North American species, R. mariae. Moreover, the color of the pileus and geographical disjunction distinguishes R. orientipurpurea from R. mariae.
Phylogenetic analysis of LSU sequence data showed a close relationship among R. bella, R. mariae, and R. violeipes (Shimono et al. 2004). Although there are no ITS sequences available of R. bella from Japan, one LSU sequence of R. bella was identical to that of Korean samples (Park et al. 2013). The European R. violeipes is clearly distinguished from R. bella (Figs 1, 2). Therefore, considering the available morphological and molecular information, we conclude that all Korean samples previously designated as R. violeipes are in fact R. bella.
Russula sp. (SFC20160726-13) was confirmed to be identical to the three unidentified Chinese samples. The Chinese samples are from Taishan of Shandong Province, which is geographically close to South Korea. They formed a distinct clade and might be a new species. However, there are limited specimens to describe it as a new species. It would be better to introduce this new species after observing more specimens.
The occurrence of a previously reported species from South Korea (Lee et al. 2015), R. amoena, was not confirmed in this study. This species was originally described from Europe (Sarnari 1998). Two European specimens of R. amoena were included in our phylogenetic analyses, but none of the Korean samples match with these European collections. Thus, so far, R. amoena has not been confirmed in South Korea.
Most Korean specimens of R. bella and R. orientipurpurea were collected from mixed forests with pine and oak trees, which are very common in South Korea. Previous studies have reported that R. bella is commonly found as ectomycorrhizal root tips of the conifer species Abies homolepsis (Miyamoto et al. 2014), Pinus amamiana (Sugiyama et al. 2019), P. densiflora (direct GenBank submission), P. thenderbergii (Obase et al. 2011; Nakashima et al. 2016), and P. yunnanensis (Xie et al. 2010). Some sequences of R. orientipurpurea were also obtained from the roots of other Pinus spp. (Wen et al. 2015). This indicates that these two species are associated with conifers, but symbiotic relationships with deciduous trees cannot be excluded as the information available in the literature is scarce. Russula sp. (SFC20160726-13) seems rare and was collected in a forest dominated by Quercus mongolica. Since we have limited sample and data, additional specimens are needed to complete a morphological and ecological characterization of the species. Ecological information can be useful for the identification of morphologically similar ectomycorrhizal fungi species (Nuytinck and Verbeken 2003; Lee et al. 2019). Further investigations focused on ecological information are necessary to obtain a better understanding of the three Korean Russula species.
In conclusion, the East Asian Russula species in subsection Amoeninae are distinct from the European and North American species. Three species were identified from South Korea based on molecular and morphological data. However, the molecular data available in GenBank are still limited and comprise only some Russula species in subsection Amoeninae. The amount of ITS data for this group has continuously increased, but protein-coding gene sequences are still insufficient. An overall increase in sequence information would allow for a better understanding of the phylogenetic relationships and global diversity of this group.
Supplementary Material
Acknowledgements
This study was supported by the National Institute of Biological Resources (NIBR 201801105), Korea National Arboretum (KNA1-1-25, 19-2), and the Korean government (NRF-2015M3A9B8029237). The research of Miroslav Caboň and Slavomír Adamčík was funded by Slovak Grant APVV 15-0210.
Citation
Wisitrassameewong K, Park MS, Lee H, Ghosh A, Das K, Buyck B, Looney BP, Caboň M, Adamčík S, Kim C, Kim CS, Lim YW (2020) Taxonomic revision of Russula subsection Amoeninae from South Korea. MycoKeys 75: 1–29. https://doi.org/10.3897/mycokeys.75.53673
Supplementary materials
Table S1. List of validly published names classified in R. subsect. Amoeninae and allied species around the world
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Komsit Wisitrassameewong, Myung Soo Park, Hyun Lee, Aniket Ghosh, Kanad Das, Bart Buyck, Brian P. Looney, Miroslav Caboň, Slavomír Adamčík, Changmu Kim, Chang Sun Kim, Young Woon Lim
Data type
species list
Explanation note
List of validly published names classified in R. subsect. Amoeninae and allied species around the world.
Table S2. Sequences used for the ITS analyses in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Komsit Wisitrassameewong, Myung Soo Park, Hyun Lee, Aniket Ghosh, Kanad Das, Bart Buyck, Brian P. Looney, Miroslav Caboň, Slavomír Adamčík, Changmu Kim, Chang Sun Kim, Young Woon Lim
Data type
ITS sequences
Explanation note
Sequences used for the ITS analyses in this study. ITS sequences generated in this study are presented in boldface. (T) indicates the type specimen. Species names in bracket are the original species epithet in GenBank or Park et al. (2013).
References
- Adamčik S, Jančovičová BB. (2018) The Russulas described by Charles Horton Peck. Cryptogamie Mycologie 39(1): 3–110. 10.7872/crym/v39.iss1.2018.3 [DOI] [Google Scholar]
- Adamčík S, Looney B, Caboň M, Jančovičová S, Adamčíková K, Avis PG, Barajas M, Bhatt RP, Corrales A, Das K, Hampe F, Ghosh A, Gates G, Kälviäinen V, Khalid AN, Kiran M, De Lange R, Lee H, Lim YW, Kong A, Manz C, Ovrebo C, Saba M, Taipale T, Verbeken A, Wisitrassameewong K, Buyck B. (2019) The quest for a globally comprehensible Russula language. Fungal Diversity 99: 369–449. 10.1007/s13225-019-00437-2. [DOI] [Google Scholar]
- Beeli M. (1936) Contribution a l’étude la flore mycologique de Congo. XI. Fungi Geossensiani XII. Bulletin du Jardin Botanique de l’État à Bruxelles 14: 83–91. 10.2307/3666668 [DOI] [Google Scholar]
- Bills GF, Miller OK. (1984) Southern Appalachian Russulas. I. Mycologia 76: 975–1002. 10.1080/00275514.1984.12023944 [DOI] [Google Scholar]
- Buyck B. (1987) Trois Russules nouvelles du Zaïre. Bulletin du Jardin Botanique National de Belgique 57(3–4): 385–387. 10.2307/3668111 [DOI] [Google Scholar]
- Buyck B. (1988) Etude microscopique des specimens-types de Russules tropicales de la sous-section Diversicolores. Mycotaxon 33: 57–70. [Google Scholar]
- Buyck B. (1992) Checklist of tropical Russulae and their type specimens. Russulales News special issue 1, 99 pp.
- Buyck B. (1994) Flore illustrée des champignons d’Afrique Centrale. Fascicule 16. Russula II (Russulaceae). Ministère de l’Agriculture, Jardin Botanique National de Belgique, 411–542.
- Buyck B. (1995) Russula subsection Amoeninae in tropical African miombo woodland. Documents Mycologiques 98–100: 103–112.
- Buyck B, Sharp C. (2007) Two new species and first records for 13 other Russula (Russulales) from Zimbabwe. Cryptogamie Mycologie 28(1): 13–27. [Google Scholar]
- Buyck B, Hofstetter V, Eberhardt U, Verbeken A, Kauff F. (2008) Walking the thin line between Russula and Lactarius: the dilemma of Russula subsect. Ochricompactae. Fungal Diversity 28: 15–40. [Google Scholar]
- Buyck B, Zoller S, Hofstetter V. (2018) Walking the thin line… ten years later: the dilemma of above- versus below-ground features to support phylogenies in the Russulaceae. Fungal Diversity 89(1): 267–292. 10.1007/s13225-018-0397-5 [DOI] [Google Scholar]
- Buyck B, Wang XH, Adamčíková K, Caboň M, Jančovičová S, Hofstetter V, Adamčík S. (2020) One step closer to unravelling the origin of Russula: subgenus Glotinosae subg. Nov. Mycosphere 11(1): 285‒304. 10.5943/mycosphere/11/1/6 [DOI]
- Caboň M, Li G-J, Saba M, Kolařík M, Jančovičová S, Khalid AN, Moreau P-A, Wen H-A, Pfister DH, Adamčík S. (2019) Phylogenetic study documents different speciation mechanisms within the Russula globispora lineage in boreal and arctic environments of the Northern Hemisphere. IMA fungus 10: e5. 10.1186/s43008-019-0003-9 [DOI] [PMC free article] [PubMed]
- Chiu WF. (1945) The Russulaceae of Yunnan. Lloydia 8(1): 31‒59.
- Cho HJ, Park MS, Lee H, Oh S-Y, Wilson AW, Mueller GM, Lim YW. (2018) A systematic revision of the ectomycorrhizal genus Laccaria from Korea. Mycologia 110(5): 948–961. 10.1080/00275514.2018.1507542 [DOI] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Le Roux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernández-Restrepo M, Kolařík M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, Čmoková A, Dimitrov RA, Dyakov MYu, Dueñas M, Dutta AK, Esteve-Raventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GEStJ, Held BW, Jurjević Ž, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-Ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin HD, Silva BDB, Silva GA, Smith MTh, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ. (2016) Fungal Planet description sheets 400‒468: Russula intervenosa S. Paloi, A.K. Dutta & K. Acharya sp. nov. Persoonia: Molecular Phylogeny and Evolution of Fungi 36: 316‒458. 10.3767/003158516X692185 [DOI] [PMC free article] [PubMed]
- Das K, Ghosh A, Chakraborty D, Li J, Qiu L, Baghela A, Halama M, Hembrom ME, Mehmood T, Parihar A, Pencakowski B, Bielecka M, Recyńska K, Sasiela D, Singh U, Song Y, Świerkoz K, Szczęśniak K, Uniyal P, Zhang J, Buyck B. (2017) Fungal biodiversity profile 31–40. Cryptogamie Mycologie 38(3): 353–406. 10.7872/crym/v38.iss3.2017.353 [DOI] [Google Scholar]
- Das K, Miller SL, Sharma JR, Sharma P, Bhatt RP. (2005) Russula in Himalaya 1: A new species of subgenus Amoenula. Mycotaxon 94: 85–88. [Google Scholar]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. (2020) ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37(1): 291‒294. 10.1093/molbev/msz189 [DOI] [PMC free article] [PubMed]
- De Crop E, Hampe F, Wisitrassameewong K, Stubbe D, Nuytinck J, Verbeken A. (2018) Novel diversity in Lactifluus section Gerardii from Asia: five new species with pleurotoid or small agaricoid basidiocarps. Mycologia 110: 962–984. 10.1080/00275514.2018.1508979 [DOI] [PubMed] [Google Scholar]
- Härkönen M, Buyck B, Saarimäki T, Mwasumbi L. (1993) Tanzanian mushrooms and their uses 1. Russula. Karstenia 33: 11–50. 10.29203/ka.1993.297 [DOI] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, deWaard JR. (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London. Series B: Biological Sciences 270: 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R. (1938) Diagnoses latines d’espèces et variétés nouvelles de Lactario-Russulés du domaine oriental de madagascar. Candollea (7): 375–393.
- Hibbett D, Abarenkov K, Kõljalg U, Öpik M, Chai B, Cole J, Wang Q, Crous P, Robert V, Helgason T, Herr JR, Kirk P, Lueschow S, O’Donnell K, Nilsson RH, Oono R, Schoch C, Smyth C, Walker DM, Porras-Alfaro A, Taylor JW, Geiser DM. (2016) Sequence-based classification and identification of Fungi. Mycologia 108(6): 1049–1068. [DOI] [PubMed] [Google Scholar]
- Hofstetter V, Buyck B, Eyssartier G, Schnee S, Gindro K. (2019) The unbearable lightness of sequenced-based identification. Fungal Diversity 96(1): 243–284. 10.1007/s13225-019-00428-3 [DOI] [Google Scholar]
- Hongo T. (1968) Notulae Mycologicae (7). Memoir of Shiga University 18: 47–52. 10.2307/1293963 [DOI] [Google Scholar]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui B-K, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang S-K, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li W-J, Lin C-G, Liu J-K, Lu Y-Z, Luo Z-L, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de Santiago ALCMA, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu H-X, Yang J, Zeng X-Y, Zhang H, Zhang J-F, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai Y-C, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang J-C, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu Z-Y, Liu X-Z, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su H-Y, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen T-C, Xu J-C, Zhang Z-K, Zhao Y-C, Zhou J-L, Zhu L. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contribution to fungal taxa. Fungal Diversity 80(1): 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvement in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibby G, Fatto R. (1990) Keys to the species of Russula in Northeastern North America, 3rd edition. Kibby-Fatto Enterprises, Somerville, 61 pp. [Google Scholar]
- Kornerup A, Wanscher JH. (1978) Metheun handbook of colour, 3rd edn. Eyre Metheun Ltd., London, 252 pp. [Google Scholar]
- Kränzlin F. (2005) Fungi of Switzerland, vol 6, Russulaceae: Lactarius, Russula. Verlag Mykologia, Luzern, 320 pp. [Google Scholar]
- Kumar S, Stecher G, Tamura K. (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870‒1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed]
- Lebel T, Tonkin J. (2007) Australian species of Macowanites are sequestrate species of Russula (Russulaceae, Basidiomycota). Australian Systematic Botany 20: 355–381. 10.1071/SB07007 [DOI] [Google Scholar]
- Lee YS, Lim YW, Kim JJ, Yun HY, Kim C, Park JY. (2015) National list of species of Korea: Basidiomycota. National Institute of Biological Resources, South Korea, 364 pp. [Google Scholar]
- Lee H, Park MS, Jung PE, Eimes JA, Seok SJ, Lim YW. (2017) Re-evaluation of the taxonomy and diversity of Russula section Foetentinae (Russulales, Basidiomycota) in Korea. Mycoscience 58(5): 351–360. 10.1016/j.myc.2017.04.006 [DOI] [Google Scholar]
- Lee H, Wissitrassameewong K, Park MS, Verbeken A, Eimes J, Lim YW. (2019) Taxonomic revision of the genus Lactarius (Russulales, Basidiomycota) in Korea. Fungal Diversity 95(1): 275–335. 10.1007/s13225-019-00425-6 [DOI] [Google Scholar]
- Looney BP. (2015) Molecular annotation of type specimens of Russula species described by W.A. Murrill from the southeast United States. Mycotaxon 129(2): 255–268. 10.5248/129.255 [DOI] [Google Scholar]
- Looney BP, Ryberg M, Hampe F, Sánchez-García M, Matheny PB. (2016) Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Molecular Ecology 25: 630–647. 10.1111/mec.13506 [DOI] [PubMed] [Google Scholar]
- Looney BP, Meidl P, Piatek MJ, Miettinen O, Martin FM, Matheny PB, Labbé JL. (2018) Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytologist 218(1): 54–65. 10.1111/nph.15001 [DOI] [PubMed] [Google Scholar]
- Looney BP, Adamčík S, Matheny PB. (2020) Coalescent-based delimitation and species-tree estimations reveal Appalachian origin and Neogene diversification in Russula subsection Roseinae Molecular Phylogenetics and Evolution 147: e106787. 10.1016/j.ympev.2020.106787 [DOI] [PubMed]
- Maire R. (1910) Les bases de la classification dan le genre Russula. Bulletin de la Société Mycologique de France 26: 49–125. [Google Scholar]
- Martin KJ, Rygiewicz PT. (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiology 5(28): 1–11. 10.1186/1471-2180-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Frøslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS. (2007) Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 43: 430–451. 10.1016/j.ympev.2006.08.024 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceeding of the Gateway Computing Environments Workshop (GCE): 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Miyamoto Y, Nakano T, Hattori M, Nara K. (2014) The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. The ISME Journal 8(8): 1739–1746. 10.1038/ismej.2014.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrill WA. (1940) Additon to Florda fungi: 1. Bulletin of the Terrey Botanical Club 67(1): 57–66. 10.2307/2485361 [DOI] [Google Scholar]
- Murrill WA. (1941) More Florida novelties. Mycologia 33: 434–448. 10.1080/00275514.1941.12020837 [DOI] [Google Scholar]
- Murrill WA. (1943) More new fungi from Florida. Lloydia 6: 207–228. 10.1056/NEJM194302112280612 [DOI] [Google Scholar]
- Nakashima H, Eguchi N, Uesugi T, Yamashita N, Matsuda Y. (2016) Effect of ectomycorrhizal composition on survival and growth of Pinus thunbergii seedlings varying in resistance to the pine wilt nematode. Mycorrhiza 30(2): 475–481. 10.1007/s00468-015-1217-0 [DOI] [Google Scholar]
- Nuytinck J, Verbeken A. (2003) Lactarius sanguifluus versus Lactarius vinosus–molecular and morphological analyses. Mycological Progress 2(3): 227–234. 10.1007/s11557-006-0060-5 [DOI] [Google Scholar]
- Obase K, Lee JK, Lee SY, Chun KW. (2011) Diversity and community structure of ectomycorrhizal fungi in Pinus thunbergii coastal forests in the eastern region of Korea. Mycoscience 52: 383–391. 10.1007/S10267-011-0123-6 [DOI] [Google Scholar]
- Paloi S, Das K, Acharya K. (2018) Russula darjeelingensis, a new species from Eastern Himalaya, India. Phytotaxa 358(1): 83–90. 10.11646/phytotaxa.358.1.6 [DOI] [Google Scholar]
- Park MS, Fong JJ, Lee H, Oh SY, Jung PE, Min YJ, Seok SJ, Lim YW. (2013) Delimitation of Russula subgenus Amoenula in Korea using three molecular markers. Mycobiology 41(4): 191–201. 10.5941/MYCO.2013.41.4.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, Lee H, Oh SY, Jung PE, Seok SJ, Fong JJ, Lim YW. (2014) Species delimitation of three species within the Russula subgenus Compacta in Korea: R. eccentrica, R. nigricans, and R. subnigricans. Journal of Microbiology 52(8): 631–638. 10.1007/s12275-014-4168-z [DOI] [PubMed] [Google Scholar]
- Peck CH. (1872) Report of the botanist (1870). Annual report on the New York state museum of natural history 24: 41–108. [Google Scholar]
- Posada D. (2008) jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25(7): 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Quélet L. (1880) Quelques especes critique ou nouvelles de la Flore Mycologique de France. Comptes Rendus de l’Association Française pour l’Avancement des Sciences 9: 661–675. [Google Scholar]
- Quélet L. (1898) Quelques espèces critiques ou nouvelles pour la Flore mycologique de France. Comptes Rendus de l’Association Française pour l’Avancement des Sciences 26(2): 446–452. [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer 1.6. http://beast.bio.ed.ac.uk
- Roger SO, Bendich AJ. (1994) Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperport RA. (Eds) Plant molecular biology manual.Kluwer Academic Publisher, Boston, 183–190. 10.1007/978-94-011-0511-8_12 [DOI]
- Romagnesi H. (1962) Taxa nova ex genere Russula. Bulletin Mensuel de la Société Linnéenne de Lyon 31(1): 172–177. 10.3406/linly.1962.7058 [DOI] [Google Scholar]
- Romagnesi H. (1985) Russula amoena Quél. Var. acystidiata n.f. Bulletin de la Société Mycologique de France 101: 2–5. [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sarnari M. (1998) Monographia Illustrata del Genere Russula in Europa tomo primo. Associazioni Micologica Bresadola, Trento, 800 pp. [Google Scholar]
- Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. (2005) Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philosophical Transactions of the Royal Society B – Biological Sciences 360: 1805–1811. 10.1098/rstb.2005.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Kato M, Takamatsu S. (2004) Molecular phylogeny of Russulaceae (Basidiomycetes; Russulales) inferred from the nucleotide sequences of nuclear large subunit rDNA. Mycoscience 45: 303–316. 10.1007/S10267-004-0189-5 [DOI] [Google Scholar]
- Singer R. (1932) Monographie der Gattung Russula. Beihefte zum Botanischen Centralblatt 49(2): 205–380. [Google Scholar]
- Singer R. (1942a) Type studies on Basidiomycetes. I. Mycologia 34: 64–93. 10.1080/00275514.1942.12020874 [DOI] [Google Scholar]
- Singer R. (1942b) Das system der Agaricales. II. Annales Mycologici 40: 1–108. [Google Scholar]
- Singer R. (1958) New and interesting species of Basidiomycetes. V. Sydowia 11: 141–374. 10.2307/3756045 [DOI] [Google Scholar]
- Singer R. (1975) The Agaricales in modern taxonomy. 3rd edn. J. Cramer, Vaduz, 912 pp. [Google Scholar]
- Song Y, Li J, Buyck B, Zheng J, Qiu L. (2018) Russula verrucospora sp. nov. and R. xanthovirens sp. nov., two novel species of Russula (Russulaceae) from southern China. Cryptogamie Mycologie 39(1): 129–142. 10.7872/crym/v39.iss1.2018.129 [DOI] [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama Y, Murata M, Kanetani S, Nara K. (2019) Towards the conservation of ectomycorrhizal fungi on endangered trees: native fungal species on Pinus amamiana are rarely conserved in trees planted ex situ. Mycorrhiza 29(3): 195–205. 10.1007/s00572-019-00887-1 [DOI] [PubMed] [Google Scholar]
- Vellinga EC. (1988) Glossary. In: Bas C, Kuyper TW, Noordeloos ME, Vellinga EC. (Eds) Flora Agaricana Neerlandica, vol.1 A.A. Balkema, Rotterdam, 1–182.
- Wang X-H, Halling RE, Hofstetter V, Lebel T, Buyck B. (2018) Phylogeny, biogeography and taxonomic re-assessment of Multifurca (Russulaceae, Russulales) using three-locus data. PLoS ONE 13(11): e0205840. 10.1371/journal.pone.0205840 [DOI] [PMC free article] [PubMed]
- Wen Z, Murata M, Xu Z, Chen Y, Nara K. (2015) Ectomycorrhizal fungal communities on the endangered Chinese Douglas-fir (Pseudotsuga sinensis) indicating regional fungal sharing overrides host conservatism across geographical regions. Plant and Soil 387: 189–199. 10.1007/s11104-014-2278-3 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Xie X D, Liu PG, Yu FQ. (2010) Species diversity of russuloid mycorrhizae-forming fungi on Pinus yunnanensis seedlings and the mycorrhizal morphology. Acta Botanica Yunnanica 32: 211–220. 10.3724/SP.J.1143.2010.10001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of validly published names classified in R. subsect. Amoeninae and allied species around the world
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Komsit Wisitrassameewong, Myung Soo Park, Hyun Lee, Aniket Ghosh, Kanad Das, Bart Buyck, Brian P. Looney, Miroslav Caboň, Slavomír Adamčík, Changmu Kim, Chang Sun Kim, Young Woon Lim
Data type
species list
Explanation note
List of validly published names classified in R. subsect. Amoeninae and allied species around the world.
Table S2. Sequences used for the ITS analyses in this study
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
Komsit Wisitrassameewong, Myung Soo Park, Hyun Lee, Aniket Ghosh, Kanad Das, Bart Buyck, Brian P. Looney, Miroslav Caboň, Slavomír Adamčík, Changmu Kim, Chang Sun Kim, Young Woon Lim
Data type
ITS sequences
Explanation note
Sequences used for the ITS analyses in this study. ITS sequences generated in this study are presented in boldface. (T) indicates the type specimen. Species names in bracket are the original species epithet in GenBank or Park et al. (2013).