Abstract
The symbiotic microbial communities, or “microbiomes,” that reside on animals are dynamic, and can be affected by the behavior and physiology of the host. These communities provide many critical beneficial functions for their hosts, but they can also include potential pathogens. In birds, bacteria residing in the cloaca form a complex community, including both gut and sexually-transmitted bacteria. Transmission of cloacal bacteria among individuals is likely during the breeding season, when there is direct cloacal contact between individuals. In addition, the major energetic investment in reproduction can draw resources away from immune responses that might otherwise prevent the successful establishment of microbes. We assessed dynamic variation in the cloacal microbiome of free-living rufous-collared sparrows (Zonotrichia capensis) through sequential breeding and non-breeding seasons. We found that the cloacal bacterial communities differed between the sexes when they were in breeding condition. Further, in males, but not in females, the bacterial community became more diverse with the onset of reproduction, and then decreased in diversity as males transitioned to non-breeding condition. Individuals sampled across sequential breeding seasons did not accumulate more bacterial taxa over seasons, but bacterial community composition did change. Our results suggest that the cloacal microbiome in birds is dynamic and, especially in males, responsive to breeding condition.
Introduction
The microbiome of animals is dynamic and can change through time (Turnbaugh et al. 2007; McFall-Ngai et al. 2013). The relationship of microbes with their animal hosts likely has major implications for the animal’s evolution, behavior, and physiology, as the microbiome contributes many critical functions (Archie and Theis 2011; Ezenwa et al. 2012; Clay 2014). These microbial symbionts come from many sources. For example, microbes colonize the vertebrate gut early in life from a mixture of environmental and, for some species, maternal sources (van Dongen et al. 2013; Smith and Mueller 2015). Microbes are abundant in the vertebrate gut, but they also line most other mucosal surfaces, including the nasal passages, lungs, and urogenital tract. However, some microbial symbionts are picked up almost entirely from direct contact with other individuals.
The urogenital tract is an interesting case because pathogenic microbes can invade the normal, healthy community of commensal microbes through sexual contact, and can thus represent a potential cost of sexual reproduction if the microbes are pathogenic (Sheldon 1993; Lockhart et al. 1996). The basic natural history of animals and the microbes that reside in the urogenital tract is not well understood. For example, male and female animals vary in physiology and reproductive behavior, and yet it is not clear how these differences relate to the community composition of the urogenital microbiome. Further, as vertebrate reproductive activity often varies seasonally, the urogenital microbiome can be expected to vary seasonally as well. The widespread presence of sexually transmitted infections (STIs) in animals, and their potential impact on host health, underlines the importance of understanding the factors that contribute to variation in the urogenital microbiome.
The cloaca of birds, reptiles, and amphibians is a combined opening for the digestive, urinary, and reproductive systems. Thus, the cloacal microbiome is a mixture of bacteria coming from these sources, as well as from contacts during copulation, resulting in a particularly complex bacterial community. Establishment success of new members of the bacterial community depends on the host’s immune system, the physical environment of the cloaca (e.g., pH), and the current bacterial community structure, which can alter colonization success due to microbial interactions (Poiani 2010).
Not all bacteria that colonize hosts are pathogenic, and not all potential pathogen exposure results in disease. As most bacteria transferred during cloacal contact are likely not pathogenic, we refer to them as STIs, and not as sexually transmitted pathogens (Lombardo et al. 1999; Smith and Mueller 2015). In fact, some colonizing bacteria may be beneficial, providing crucial physiological functions for their host. For example, microbes may help to extract energy and nutrients from food, resulting in increased host survival (Ley et al. 2008), and microbes can also have longer-term benefits, as individuals with beneficial microbes could be preferred as mates (Lombardo et al. 1999; Smith and Mueller 2015). However, there are also several sexually transmitted bacteria that are pathogens, such as Chlamydia, and these may ultimately impact the evolution of polygamous mating systems (Sheldon 1993; Lombardo 1998; Poiani 2010).
From an epidemiological perspective, the spread of STIs through the population is largely influenced by individuals who have a large number of sexual partners (Smith and Dobson 1992; Ashby and Gupta 2013). For instance, promiscuity can increase cloacal bacterial diversity in polygamous lizards (White et al. 2011), and vaginal bacterial diversity of mice (MacManes 2011) and primates (Yildirim et al. 2014). Thus, multiple mating may lead to a greater sampling of the bacterial community within the broader host population, making the evolution of promiscuity dependent on the costs and benefits of STIs.
In vertebrates, life-cycle events are often partitioned into discrete stages, such as growth, reproduction, and offspring rearing. Coupled to those stages are specific behaviors and physiological changes (e.g., in the endocrine and immune systems) that can also influence symbiotic bacterial communities. In particular, there are at least three features of the reproductive life stage that can affect bacterial communities living on individual animals. First, there is an increase in physical contacts, and consequently, more exposure to infectious microorganisms (Anderson 1979). In female kittiwakes (Rissa tridactyla) the cloacal microbiome changed after cloacal contacts were experimentally blocked by a contraceptive device, suggesting that the cloacal microbiome is dynamic and can be affected by copulatory activity (White et al. 2010). Furthermore, it has been hypothesized that there could be a sex bias in the sexual transmission of bacteria from males to females due to the transfer of bacteria in seminal fluid. Kulkarni and Heeb (2007) experimentally infected zebra finches (Taeniopygia guttata) with Bacillus licheniformis and demonstrated that this bacterium was transmitted through sexual contact at a higher rate from males to females. Second, male testosterone levels are usually elevated during reproduction, and testosterone has often been investigated as a mediator of life-history trade-offs between immediate reproduction and self-maintenance/survival (Hau 2007). Testosterone could influence symbiotic bacterial communities by promoting behaviors that increase contact rates, such as extra pair copulations (Wingfield 1984; Raouf et al. 1997; Grear et al. 2009), or by suppressing immune function (Folstad and Karter 1992). Immune suppression by testosterone has been hypothesized (Immuno-competence Handicap Hypothesis) as a cost of testosterone-dependent sexual signals, but this hypothesis has only mixed support (Roberts et al. 2004; Roberts and Peters 2009). Along these lines, we previously found a positive relationship between testosterone and cloacal bacterial phylogenetic diversity in wild male rufous-collared sparrows (Zonotrichia capensis) sampled during the breeding season from a different population than the current study (Escallón et al. 2017). Stress hormones can also have an impact on the cloacal microbiome, as was seen in wild yellow-legged gulls (Larus michahellis), where corticosterone-implanted birds lost several microbial taxa (Noguera et al. 2018). Third, reproduction is an energetically challenging life-history stage in which animals often invest more in reproduction than in self-maintenance (Stearns 1992). This could result in changes in the cloacal environment (i.e., changes in pH, mucus secretion, and tissue bactericidal capacity) and facilitate colonization by opportunistic bacteria or by bacterial species dependent on the bird’s breeding cycles, such as STIs (Martinez-Bakker and Helm 2015).
In this study, we investigated whether reproductive condition is associated with changes in the cloacal bacterial communities in free-living rufous-collared sparrows by tracking the individual cloacal bacterial communities of males and females through sequential breeding and non-breeding seasons. This bird species is socially monogamous, but has a high level of extra pair paternity, with an average of 48% of the nests containing nestlings that do not belong to the social father (Eikenaar et al. 2013). Thus, cross infection of cloacal bacteria among individuals is likely. Rufous-collared sparrows are year-round residents in tropical regions, where there is constant food availability, and in some populations they are able to breed several times a year (Miller 1962). Male rufous-collared sparrows increase their testosterone levels during the breeding season, and decrease testosterone and territoriality during the non-breeding season (Moore et al. 2005; Addis et al. 2010). We tested two hypotheses that could explain temporal variation in bacterial cloacal communities in rufous-collared sparrows. With the bacterial clearance hypothesis, we predicted that bacteria are cyclically added and lost each breeding season, such that cloacal bacterial diversity of adults would be higher during the breeding season than in the non-breeding season. With the bacterial accumulation hypothesis, we predicted that cloacal bacterial diversity would increase through sequential breeding seasons, as each season presents additional opportunities for bacterial transmission through copulations and accumulation of diverse bacteria over time. Additionally, males and females differ in many aspects of their physiology and behavior, which can make the risk of infection sex-dependent, so we tested for the effect of sex on each of the hypotheses. As a comparison, we also sampled the cloacas of juveniles, who should have low bacterial diversity levels, because they have not yet copulated and should thus primarily have bacteria associated with the digestive tract and the physical environment. Finally, we investigated the relationship between blood plasma testosterone concentration and cloacal bacterial communities in males during the breeding and non-breeding seasons to see if there was any relationship between an individual’s hormonal environment and the cloacal bacterial community.
Methods
Sampling of cloacal bacteria
We studied a wild breeding population of rufous-collared sparrows (Z.capensis) in the rural town of Guatavita, in the Colombian Andes (4.978158°; −73.791961°; 2940 m a.s.l.). At this site, there is a bimodal pattern of rainfall with high variability throughout the year, including a small average decrease in rainfall from December to February and from May to September. In addition, temperature is nearly constant (∼13°C) throughout the year (Instituto de Hidrología, Meteorología y Estudios Ambientales, Colombia). Tropical rufous-collared sparrows have an extended breeding season, and there can be some asynchrony in the timing of the breeding season among individuals (Moore et al. 2005; Class et al. 2011). In our study population, the largest proportion of individuals were in breeding condition between May and September (Males, 77%, N = 181/235; Females, 61%, N = 89/142), whereas from December to January the proportion of breeding individuals was lower (Males, 18.4%, N = 14/76; Females, 17.6%, N = 9/51).
We sampled rufous-collared sparrows by capturing them passively in mist nets only during the 2 h immediately after dawn. Upon capture, a cloacal swab sample was taken from each bird with a PurFlock® micro-ultrafine nylon-tipped sterile swab (Puritan, USA), as in Escallón et al. (2017). Swabs were placed in RNALater (Ambion, USA), frozen, and transported to Virginia Tech for DNA extraction. For males, we determined breeding condition based on the height of the cloacal protuberance, with a height >5 mm corresponding to fully grown testes, indicative of breeding condition (Moore et al. 2005). For females, we defined breeding condition as the presence of a brood patch. Juveniles and adults can be differentiated based on plumage coloration (Miller 1961). Juveniles have regressed gonads and are unable to reproduce until they molt into adult plumage (Miller and Miller 1968).
The sampling for cloacal bacteria in this population was done approximately every 6 months for three consecutive years (2011–2013). All captured birds were banded with a numbered metal leg band, which allowed individual identification on subsequent recaptures. To analyze the changes in cloacal bacterial diversity between breeding and non-breeding seasons, we selected the birds that had been sampled in either breeding or non-breeding condition and then resampled 5–6 months later in the other breeding condition (N = 20 males, 11 females). Similarly, to analyze changes in bacterial diversity between breeding seasons, we selected individuals that had been sampled in breeding condition and then were sampled again 10–12 months later in breeding condition (N = 16 males, 13 females). This left us with paired samples across breeding stages, with a total of 122 cloacal swabs collected from 27 males, 22 females, and 9 juveniles during that time.
Bacterial sample preparation and sequencing
To remove the bacteria from the collection swab, we vortexed the tube and then centrifuged it at 12,000 g for 5 min to form a pellet. The RNALater was pipetted out and the remaining pellet was used to extract bacterial DNA using the DNeasy blood and tissue kit (Qiagen Inc., Valencia, CA, USA). We then amplified the V4 region of the 16S rRNA gene using primers 515F and 806R, which contained unique barcode to label each sample (Caporaso et al. 2011). Samples were run in triplicate, and each PCR contained 1 μL template DNA, 0.5 μL of each primer at 10 μM concentration, 12 μL DNA-free PCR water (MO-BIO, Carlsbad, CA, USA), 10 μL 2.5× HotMasterMix (5 PRIME, Gaithersburg, Maryland), and 1 μL of 20 mg/mL bovine serum albumin (Fisher Scientific, Pittsburgh, PA, USA). A control without DNA template was run for each sample. Extracted DNA samples with PCR inhibitors were diluted 1–10 in PCR water. Thermocycler conditions were: an initial cycle for 3 min at 94°C followed by 35 cycles of 34 s at 94°C, 60 s at 50°C, and 90 s at 72°C, and a final cycle for 10 min at 72°C. The triplicate reactions for each sample were then pooled together, visualized on a 1% agarose gel, and quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). Sequencing of an equimolar mixture of the samples was done on an Illumina MiSeq with a 250-bp paired-end strategy at the Dana-Farber Cancer Institute of Harvard University, as in Caporaso et al. (2012). 10% PhiX was added to compensate for the low base diversity. Base calling and quality scoring were done with MiSeq Real-Time Analysis software (Illumina V. 1.18.54).
Sequence data processing
The reverse reads were of low quality, therefore we only used the forward reads in our analysis. Using the Quantitative Insights Into Microbial Ecology pipeline (QIIME v. 1.9.1; Caporaso et al. 2010), sequences were de-multiplexed and quality-filtered by discarding sequences if there were any ambiguous base calls, errors in the barcode, less than 50% of the read length had consecutive base calls with a phred quality score greater than 20, or there were more than 10 consecutive low-quality base calls. After quality filtering, the number of reads retained per sample ranged from 2,408 to 177,819 (Average 36,116). We imported the demultiplexed sequences into Geneious (Biomatters, Ltd) and filtered out reads assembled to PhiX. We exported the remaining 250 bp sequences (5,145,224 reads). In QIIME, we then clustered the quality-filtered sequences into operational taxonomic units (OTUs) at a sequence similarity threshold of 97%, as in Escallón et al. (2017). In the resulting OTU table, we filtered out mitochondria and chloroplast sequences and OTUs with fewer than 0.001% of the total reads (Bokulich et al. 2013), and rarefied all samples to 2,400 reads prior to analysis. Rarefaction resulted in the loss of two samples with low reads (<2,400), so the final dataset consisted of 120 samples and 7101 unique OTUs.
Hormone analysis
Testosterone was measured in males during the non-breeding and breeding seasons. Briefly, we took blood samples (c. 250 µL) from the brachial vein within 10 min of capture. Blood samples were kept on ice (<4 h) until centrifugation, when plasma was separated and frozen for later analysis. Plasma volumes ranged from 16 to 100 µL (mean: 48 µL). Total plasma testosterone concentration was measured by direct radioimmunoassay following the procedures of Moore et al. (2002). We measured total androgens (e.g., testosterone, 5α-dihydrotestosterone, and others) in our direct assay; however, testosterone is the major androgen in birds, so we will refer to our measured androgen levels as testosterone levels. The samples were run in a single assay, with a mean extraction efficiency of 60% and an intra-assay variation of 18.8%. The limit of detection for the assay was c. 0.16 ng/mL. The testosterone antibody used was T-3003s (Fitzgerald, Concord, MA, USA: Catalog #20R-TR018W).
Statistical analysis
To test the bacterial clearance hypothesis, we compared individuals between breeding and non-breeding seasons. We assessed differences in bacterial phylogenetic diversity and the number of observed OTUs (richness) between breeding and non-breeding seasons using paired t-tests of individual birds. We also calculated, for each individual’s paired sample, the net change in diversity from breeding to non-breeding season, by subtracting the initial diversity value from the final value and compared the breeding transitions with a t-test. Analyzing the net change in diversity gave us information on the magnitude of the change, as well as whether there were differences between groups.
We assessed whether the bacterial community composition differed between breeding and non-breeding conditions, and based on sex and age by calculating pairwise weighted (OTU relative abundance) and unweighted (presence/absence of OTUs) UniFrac distances with the phyloseq package in R (McMurdie and Holmes 2013). UniFrac uses branch length overlap to calculate phylogenetic distance between pairs of communities, and avoids some of the disadvantages associated with comparing communities at only a single level of taxonomic resolution (Hamady and Knight 2009). We used principal coordinate analysis (PCoA) of weighted and unweighted UniFrac distances to visualize bacterial community composition. Differences between groups were evaluated for weighted and unweighted UniFrac distances with a PERMANOVA using the adonis function from the vegan package with 1000 permutations (Oksanen et al. 2013). As PERMANOVA can confound mean tendency and dispersion differences among groups, we also tested for differences in dispersion (∼multivariate variance) among groups, using the betadisper function. To test for shifts in the community composition between breeding conditions, we performed t-tests with the first axis of the PCoA.
To identify which OTUs were driving the differences between breeding condition and sex of the birds, we used indicator species analysis with the multipatt function in the indicspecies package (Dufrene and Legendre 1997; De Cáceres et al. 2012). Indicator species analysis calculates an indicator value for each OTU in the dataset by computing the product of the relative abundance and frequency of each OTU in our predefined groups (non-breeding male, breeding male, non-breeding female, breeding female, and juvenile). Significance of the relationship between OTUs and breeding status was based on permutation tests using 999 random permutations to estimate P-values. OTUs with an indicator value >0.5 and P < 0.05 were considered as indicator species. All multiple comparisons were corrected with the false discovery rate procedure (Benjamini and Hochberg 1995). A heatmap of the relative abundance of each OTU in each predefined group was used to visualize the differences.
Changes in the core bacterial community could be indicative of differences in abundant bacteria between breeding conditions. To identify differences between the relative abundance of shared OTUs in the bird population, we calculated the core bacterial community, defined as OTUs that were present in at least 90% of the birds using the rarefied data set. We then compared the change in relative abundance of the core OTUs between breeding conditions for males and females using a paired t-test with log-transformed data and plotted the results on a heat map.
To test the bacterial accumulation hypothesis, we assessed differences across sequential breeding seasons (10–12 months apart). We analyzed differences in phylogenetic diversity and OTU richness using paired t-tests. Note that we were comparing the same bird across two consecutive breeding seasons and no bird was compared across three consecutive breeding seasons. We assessed whether the bacterial community composition differed between sequential breeding seasons by calculating pairwise weighted and unweighted UniFrac distances with the phyloseq package in R (McMurdie and Holmes 2013) and then used the first axis of the PCoA to test for shifts in the community composition with a t-test.
We examined the relationship between circulating testosterone concentrations and both phylogenetic diversity and OTU richness using linear mixed models, assigning males as a random factor and fitting a separate intercept for each breeding condition. To normalize circulating testosterone concentrations, we log-transformed this variable. All statistical analyses were carried out in the R environment (version 3.5.1) (R Development Core Team 2018).
Results
Bacterial clearance hypothesis: breeding and non-breeding seasons
Male birds transitioning from non-breeding to breeding condition increased their cloacal phylogenetic diversity by 38% from 19.8 to 27.3 (t = −4.3, P = 0.001, Fig. 1a) and the number of observed OTUs by 58% from 222.8 to 351.7 (t = −4.1, P = 0.001, Supplementary Fig. S1a). When males transitioned from breeding to non-breeding, there was a strong trend toward a decrease for phylogenetic diversity (−23% from 26.0 to 20.1, t = 2.3, P = 0.058, Fig. 1b), but no change in the number of observed OTUs (t = 1.6, P = 0.15, Supplementary Fig. S1b). For females, the phylogenetic diversity and number of observed OTUs did not change during either reproductive transition (non-breeding to breeding: phylogenetic diversity t = −1.2, P = 0.28, Fig. 1c and the number of observed OTUs t =−0.4, P = 0.67, Supplementary Fig. S1c; breeding to non-breeding: phylogenetic diversity t = 1.7, P = 0.17, Fig. 1d and the number of observed OTUs t = 1.6, P = 0.19, Supplementary Fig. S1d).
Fig. 1.
Cloacal bacterial diversity in males changes between breeding conditions. Individual shifts in phylogenetic diversity from non-breeding to breeding condition (A and C) and breeding to non-breeding condition (B and D). Time between sampling was five to six months. Lines connect data from the same individual across breeding conditions. Males are the top row (A and B) and females are the bottom row (C and D).
Analysis of the net diversity change within individuals from breeding to non-breeding season revealed a similar pattern, with a significant difference between male reproductive states in phylogenetic diversity (P = 0.0003, Supplementary Fig. S2a) and in the number of observed OTUs (P = 0.001, Supplementary Fig. S2b). For females there was a negative trend in phylogenetic diversity (P = 0.07, Supplementary Fig. S2a), but no change in the number of observed OTUs (P = 0.2, Supplementary Fig. S2b).
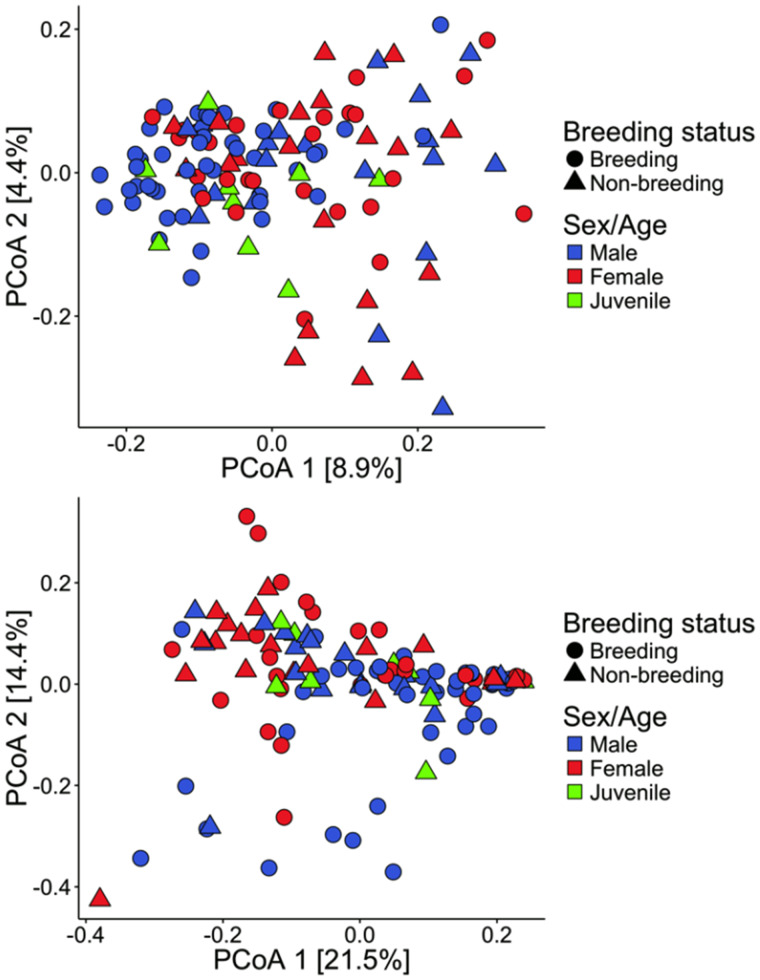
When comparing the cloacal community structure between sex and breeding condition groups, the breeding males had different cloacal community composition than the breeding females both when considering only the presence/absence of OTUs (Unweighted UniFrac, Adonis, pseudo =2.1, P = 0.001, R2 =0.03, Fig. 2a) and when considering OTU relative abundance (Weighted UniFrac, Adonis, pseudoF =3.5, P = 0.001, R2 =0.05, Fig. 2b), but the cloacal community composition was not different between non-breeding males and non-breeding females (Unweighted UniFrac, Adonis, pseudoF =1.0, P = 0.4, R2 =0.03, Fig. 2a; Weighted UniFrac, Adonis, pseudoF = 1.2, P = 0.2, R2 =0.03, Fig. 2b). Juveniles had a cloacal community composition that was not different from adult males (Unweighted UniFrac, Adonis, pseudoF = 1.3, P = 0.07, R2 =0.02, Fig. 2a; Weighted UniFrac, Adonis, pseudoF = 1.3, P = 0.19, R2 = 0.02, Fig. 2b), but did differ significantly from that of females when considering unweighted distance (Unweighted UniFrac, Adonis, pseudoF = 1.3, P = 0.03, R2 = 0.02, Fig. 2a), but not when considering weighted distance, despite some overlap in the PCoA ordination (Weighted UniFrac, Adonis, pseudoF = 0.8, P = 0.57, R2 =0.01, Fig. 2b). Significant multivariate dispersion among groups (Unweighted UniFrac, Beta dispersion, F = 3.5, P = 0.03) could be driving the significant Adonis results, as significant differences between females and juveniles may be caused by different within-group dispersion rather than different mean tendency across groups. In general, cloacal communities of breeding males and juveniles clustered together and formed a distinct group from non-breeding males and females in both breeding conditions.
Fig. 2.
Cloacal community composition of males in breeding condition clustered with juveniles, whereas communities of males in non-breeding condition clustered with females. Principal coordinates illustrating dissimilarities between the cloacal microbiome of males, females, and juveniles. PCoA with Unweighted UniFrac pairwise distances (a), and PCoA with Weighted UniFrac pairwise distances (b). Each point represents a cloacal sample from an individual. Blue points represent males, red points females, and green points juveniles. Breeding status is represented by different shapes. Increasing distance between points indicates increasing dissimilarity in cloacal community composition.
When comparing the cloacal community composition between breeding seasons, males in breeding condition clustered separately from males in non-breeding condition when analyzing the presence/absence of OTUs (Unweighted UniFrac, Adonis, pseudoF = 2.8, P = 0.001, R2 =0.04, Supplementary Fig. S3a) and OTU relative abundance (Weighted UniFrac, Adonis, pseudoF = 2.9, P = 0.003, R2 =0.04, Supplementary Fig. S3b). In fact, with only one exception, the community composition had a consistent direction of change, as revealed by the change in the first multivariate axis of the unweighted analysis (non-breeding to breeding, t =−4.5, P = 0.001, Supplementary Fig. S4a; breeding to non-breeding, t = 2.7, P = 0.03, Supplementary Fig. S4b), and a similar strong trend in the weighted analysis for the non-breeding to breeding (t =−2.1, P = 0.058, Supplementary Fig. S5a), but not for breeding to non-breeding (t = 0.1, P = 0.91, Supplementary Fig. S5b). In contrast, females’ cloacal community composition was not significantly different between breeding and non-breeding stages (Unweighted UniFrac, Adonis, pseudoF = 1.1, P = 0.19, R2 = 0.02, Supplementary Fig. S3c; Weighted UniFrac, Adonis, pseudoF = 1.3, P = 0.2, R2 =0.03, Supplementary Fig. S3d). However, there was a consistent direction of change, similar to that observed in males, for the first multivariate axis of the unweighted analysis (non-breeding to breeding, t =−3.1, P = 0.03, Supplementary Fig. S4c; breeding to non-breeding, t = 3.1, P = 0.04, Supplementary Fig. S4d). For the weighted analysis, there was no consistent change from non-breeding to breeding (t =−1.8, P = 0.14, Supplementary Fig. S5c) but a strong trend toward a consistent direction from breeding to non-breeding (t = 2.6, P = 0.06, Supplementary Fig. S5d). In general, male cloacal bacterial communities changed consistently in one direction along the first axis of the PCoA ordination between breeding conditions, but the female cloacal bacterial communities did not change as consistently.
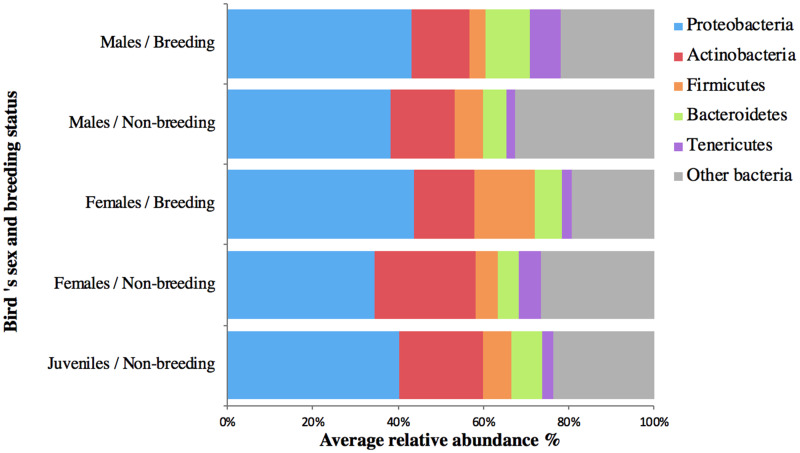
The most dominant bacterial phyla (mean relative abundance >1%) in the cloacal bacterial communities of the population of rufous-collared sparrows were as follows: Proteobacteria (41% of total relative abundance), Actinobacteria (16%), Bacteroidetes (7%), Firmicutes (7%), Tenericutes (5%), and other bacteria (24%) (Fig. 3). These five phyla were present in the cloacas of all of the birds, although there was some variation among sex and breeding status in the relative abundance of each phylum. The indicator species analysis revealed a diverse array of 32 OTUs that contributed predominantly to the dissimilarity between sexes and breeding status (Fig. 4). The cloacal community composition of breeding males was different from the other groups; in the indicator species analysis it is characterized by 10 OTUs belonging to the classes Alphaproteobacteria, Betaproteobacteria, Chitinophagales, Sphingobacteriia, Cytophagia, and Acidobacteriia. Juveniles had a large proportion (50%) of unique species based on the indicator species analysis, whereas breeding females were not characterized by any unique taxa. When comparing the number of shared OTUs in the whole microbiome, juveniles shared 35% of the OTUs (1,193 out of 3,368) with males and 42% of the OTUs (1005 out of 2,380) with females.
Fig. 3.
Average relative abundance of bacterial phyla in the cloaca of rufous-collared sparrows (n = 121). Five dominant phyla represented 76% of the cloacal microbiome (Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Tenericutes). Birds are organized by sex and breeding status.
Fig. 4.
Heat map of the relative abundances of indicator OTUs from indicator species analysis associated with the cloaca of rufous-collared sparrows in different breeding conditions. Rows indicate unique OTUs and columns indicate individual birds. The lowest taxonomic resolution that could be defined for OTU identification is listed; bacterial phylum for each core OTU is listed in parentheses (Pro = Proteobacteria, Act = Actinobacteria, Fir = Firmicutes, Bac = Bacteroidetes, and Cya = Cyanobacteria).
The core microbiome (bacteria present on >90% of birds) was composed of eight OTUs belonging to the phyla Proteobacteria and Actinobacteria. When comparing the relative abundance of only the core OTUs between breeding condition, there were no significant differences for the males (t =−1.4, P = 0.22, Table 1) or for the females (t = 0.8, P = 0.44, Table 1). Juveniles had in high abundance many of the core OTUs of breeding males. Juveniles had a larger core microbiome than adults with 25 OTUs (Table 2) and shared 40% of the OTUs (10 out of 25) with the core microbiome of males and 32% of the OTUs (8 out of 25) with the core microbiome of females.
Table 1.
Heat map of the core OTUs for males and females showing the change in OTU relative abundance between breeding conditions, and the OTU relative abundance in Juveniles.
| Males |
Females |
|||||
|---|---|---|---|---|---|---|
| Consensus lineage | Greengenes OTU No. | Non-breeding | Breeding | Non-breeding | Breeding | Juveniles |
| (Pro) Sphingomonas | 4449609 | 3.09 | 5.92 | 2.41 | 3.36 | 4.65 |
| (Act) Frigoribacterium | 589376 | 1.30 | 1.61 | 2.76 | 2.64 | 4.32 |
| (Act) Curtobacterium | 842941 | 1.56 | 0.67 | 7.41 | 1.02 | 1.33 |
| (Pro) Methylobacterium | 4303249 | 1.22 | 2.13 | 1.40 | 1.45 | 1.85 |
| (Pro) Pseudomonas | 4386920 | 2.17 | 2.42 | 1.28 | 0.44 | 0.30 |
| (Pro) Methylobacterium | 979344 | 0.89 | 1.15 | 0.75 | 0.80 | 1.04 |
| (Act) Rhodococcus fascians | 4460853 | 0.51 | 0.77 | 0.70 | 1.04 | 1.20 |
| (Pro) Methylobacterium | 4396717 | 0.57 | 0.89 | 0.51 | 0.72 | 0.94 |
Notes: The number on each cell and color scale represent the average percentage relative abundance of the core OTUs found on all birds from the population. Blue represents higher abundance and white lower abundance. The lowest taxonomic resolution that could be defined for OTU identification is listed; bacterial phylum for each core OTU is listed in parentheses (Pro, Proteobacteria; Act, Actinobacteria). Core OTUs are sorted in descending order of the sum of mean relative abundances across all groups.
Table 2.
List of core OTUs (>90% prevalence in each age/sex group) from cloacas of rufous-collared sparrows, and the mean relative abundances of those OTUs.
| Males | Females | Juveniles | ||
|---|---|---|---|---|
| N | 27 | 22 | 9 | |
| Number of OTUs | 3,368 | 2,380 | 1,353 | |
| Number of core OTUs | 10 | 8 | 25 | |
| Consensus lineage | Greengenes OTU No. | Mean relative abundances (%) of core OTUs | ||
| (Pro) Sphingomonas | 4449609 | 5.1 | 3.0 | 4.7 |
| (Act) Frigoribacterium | 589376 | 1.5 | 2.8 | 4.3 |
| (Act) Curtobacterium | 842941 | 0.9 | 4.0 | 1.3 |
| (Pro) Methylobacterium | 4303249 | 1.9 | 1.4 | 1.8 |
| (Pro) Pseudomonas | 4386920 | 2.3 | 0.8 | 0.3 |
| (Pro) Methylobacterium | 979344 | 1.1 | 0.8 | 1.0 |
| (Act) Rhodococcus fascians | 4460853 | 0.7 | 0.8 | 1.2 |
| (Pro) Methylobacterium | 4396717 | 0.8 | 0.6 | 0.9 |
| Unassigned | denovo13229 | 0.9 | 0.2 | |
| (Act) Rhodococcus | 73846 | 0.4 | 0.5 | |
| (Pro) Agrobacterium | 4333206 | 3.1 | ||
| (Pro) Agrobacterium | 220269 | 1.8 | ||
| (Act) Rathayibacter caricis | 1821516 | 0.8 | ||
| (Pro) Rhizobiaceae | 5364 | 0.7 | ||
| (Pro) Aurantimonadaceae | 662915 | 0.7 | ||
| (Pro) Erwinia | 1123414 | 0.7 | ||
| (Pro) Methylobacterium adhaesivum | 591699 | 0.7 | ||
| (Act) Aeromicrobium | 140401 | 0.7 | ||
| (Pro) Methylobacteriaceae | denovo16236 | 0.6 | ||
| (Pro) Aurantimonadaceae | 982912 | 0.5 | ||
| (Pro) Sphingomonas echinoides | 4453466 | 0.3 | ||
| (Pro) Enterobacteriaceae | 274754 | 0.3 | ||
| (Pro) Methylobacterium | 1109067 | 0.2 | ||
| (Act) Microbacteriaceae | 808021 | 0.1 | ||
| (Act) Rhodococcus | denovo26817 | 0.1 | ||
Notes: The lowest taxonomic resolution that could be defined for OTU identification is listed. Bacterial phylum for each core OTU is listed in parentheses (Pro, Proteobacteria; Act, Actinobacteria). Core OTUs are sorted in descending order of the sum of mean relative abundances across all groups.
Bacterial accumulation hypothesis: sequential breeding seasons
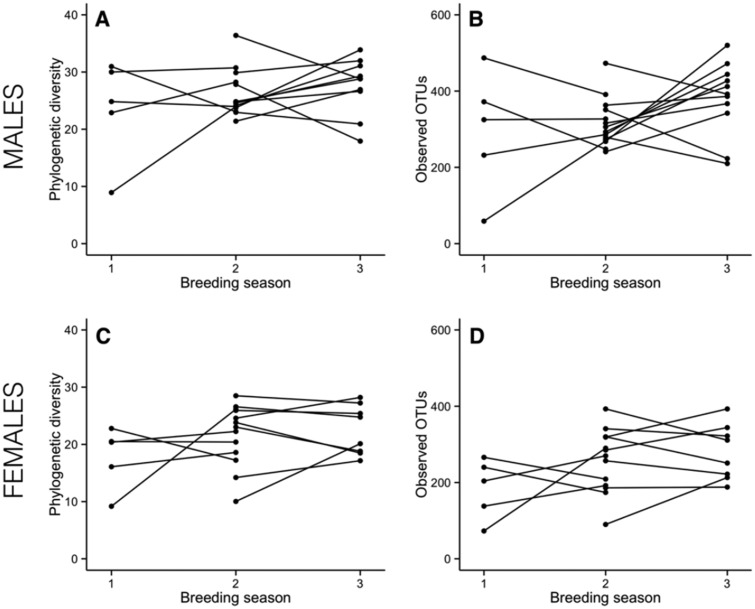
There was significant variation across the three sequential breeding seasons separated by 10–12 months, but the bacterial diversity did not consistently change in the birds’ cloacas from one breeding season to the next. The cloacal bacterial phylogenetic diversity and number of observed OTUs in males and females did not significantly change over the breeding seasons (Table 3 and Fig. 5). Similarly, for both males and females the cloacal bacterial community composition did not shift across sequential breeding seasons, as the first multivariate axis of the unweighted PCoA remained unchanged (Table 3, Supplementary Fig. S6). However, for females, there were strong trends in community composition change, as revealed by the weighted distance in the first axis of the PCoA (Table 3, Supplementary Fig. S6).
Table 3.
Results for the paired t-test between sequential breeding seasons, diversity metrics, and community composition.
| Breeding season |
||||||||
|---|---|---|---|---|---|---|---|---|
| Sex | Metric | 1 | 2 Mean (min−max) | 3 | Mean change | SE | T | P-value |
| Males | Phylogenetic diversity | 23.5 (8.9 − 31.0) | 26.0 (21.4 − 36.4) | 27.9 (17.9 − 33.9) | 1.87 | 1.76 | −1.1 | 0.31 |
| Observed OTUs | 295 (59 − 487) | 312 (241 − 473) | 381 (210 − 520) | 40.33 | 31.56 | −1.3 | 0.22 | |
| PCoA1 (Unweighted UniFrac) | −1.8 | 0.08 | ||||||
| PCoA1 (Weighted UniFrac) | −1.8 | 0.09 | ||||||
| Females | Phylogenetic diversity | 17.8 (9.2 − 22.8) | 21.6 (10.0 − 28.5) | 22.5 (17.1 − 28.2) | 1.48 | 1.75 | −0.8 | 0.41 |
| Observed OTUs | 184 (73 − 266) | 256 (90 − 393) | 280 (188 − 393) | 20.54 | 24.50 | −0.8 | 0.42 | |
| PCoA1 (Unweighted UniFrac) | −1.6 | 0.14 | ||||||
| PCoA1 (Weighted UniFrac) | −2.3 | 0.06 | ||||||
Fig. 5.
Cloacal bacterial diversity in males and females did not change across breeding season. Individual shifts in phylogenetic diversity (a and c) and number of observed OTUs (b and d) across sequential breeding seasons. Time between sampling was 10–12 months. Lines connect data from the same individual across breeding seasons. Only one male was sampled across the three seasons, all other birds were sampled in just two seasons. Males are the top row (a and b) and females are the bottom row (c and d).
Testosterone and cloacal bacterial diversity
Males in breeding condition (cloacal protuberance >5 mm) had testosterone levels that varied from 0.06 to 9.9 ng/mL (average: 1.08 ng/mL), whereas males in non-breeding condition had testosterone levels that varied from 0.05 to 0.5 ng/mL (average: 0.13 ng/mL). In breeding males, bacterial phylogenetic diversity was positively correlated with testosterone concentration (LMM, t = 2.4, d.f. =34, P = 0.03, Fig. 6a), but there was no correlation present in non-breeding males (LMM, t =−0.5, d.f. =34, P = 0.6, Fig. 6a). For males in breeding conditions there was a trend for a positive relationship between OTU richness and testosterone concentration (LMM, t = 1.9, d.f. =34, P = 0.07), but none during the non-breeding period (LMM, t =−0.8, d.f. =34, P = 0.4, Fig. 6b). In general, males with higher testosterone had a more taxonomically diverse cloacal microbiome, but not higher OTU richness.
Fig. 6.
Males in breeding condition with higher testosterone levels had a more taxonomically diverse cloacal bacterial community. Relationship between circulating testosterone levels with (a) cloacal bacterial phylogenetic diversity and (b) bacterial richness. Black points represent males in breeding condition and gray points males in non-breeding condition.
Discussion
Our results are consistent with the bacterial clearance hypothesis and not with the bacterial accumulation hypothesis. That is, while the core microbiome stayed relatively stable, the phylogenetic diversity, richness, and community composition of the overall microbiome tended to increase with active reproductive condition of the birds. Thus, it is likely that birds are cyclically acquiring new bacteria during the breeding season, and then losing them during the non-breeding season. In particular, the cloacal microbiome of males changed more drastically than that of females, increasing in phylogenetic diversity and OTU richness during the breeding season and then decreasing in diversity with the transition to a non-breeding condition. Similarly, the bacterial community composition of males differed between breeding conditions. These differences were not driven by changes between breeding conditions in the relative abundance of the eight core OTUs, but mostly driven by 10 OTUs directly linked to males in breeding condition, as shown in the indicator species analysis. When analyzing the cloacal microbiome across sequential breeding seasons, the bacterial diversity did not change, indicating that birds do not accumulate more bacteria with additional reproductive seasons, although in some cases, community composition may change.
The primary changes we saw between breeding and non-breeding conditions were in males, whereas the female cloacal microbiome did not change consistently. These findings were contrary to our initial predictions because males and females copulate in stable social pair bonds, and thus both sexes should change their cloacal community composition correspondingly. Further, we predicted that females would exhibit greater changes in their cloacal microbiome, as they receive a higher proportion of bacteria during copulations and normally experience higher physiological costs of reproduction compared with males (Kulkarni and Heeb 2007; Cox and Calsbeek 2010; Cox et al. 2010). We determined females’ breeding status based on the presence of a brood patch, which starts before egg laying and continues during incubation. Use of this definition might have masked changes in the cloacal microbiome over the breeding cycle. On the other hand, the changes in the cloacal microbiome of the males could be related to an intrinsic characteristic of that sex.
Testosterone is a hormone that mediates many aspects of male reproduction, yet maintaining high levels of this hormone may have detrimental physiological consequences (Folstad and Karter 1992). Here, we found that testosterone levels were positively related to the phylogenetic diversity of bacteria in the cloacas of male rufous-collared sparrows. This confirms what we have previously found; testosterone likely influences cloacal bacterial community structure (Escallón et al. 2017). This hormone could be directly affecting immune defenses, and thus facilitating the invasion of new bacterial taxa. Alternatively, it could indirectly be affecting male reproductive behavior by making them more promiscuous, and thus increasing the opportunity for receiving STIs, thereby increasing their cloacal bacterial diversity. Future experimental studies are needed to distinguish between these two hypotheses. It is also possible that the stress hormone, corticosterone, could be playing an important role in lowering the cloacal microbial diversity of females in reproductive condition, but we did not measure that hormone in this study. During reproduction, corticosterone levels rise moderately to mobilize energy stores (Wada et al. 2006). High corticosterone levels have been associated with lower gut bacterial diversity in red squirrels (Tamiasciurus hudsonicus) (Stothart et al. 2016) and yellow-legged gulls (L.michahellis) (Noguera et al. 2018).
Reproduction is a physiologically demanding activity wherein organisms temporarily change their behavior and self-maintenance priorities to maximize their fitness (Stearns 1992). During breeding, there are increases in metabolic rate (Harshman and Zera 2007), oxidative stress (Monaghan et al. 2009), and potentially even increased susceptibility to infection by parasites (Oppliger et al. 1996; Norris and Evans 2000). This increased susceptibility to infection is consistent with what we report here for males, as the richness and diversity of cloacal bacteria increased, and community composition changed, when birds were in breeding condition. It is possible that lost mating opportunities due to sickness may be more costly for males, suggesting they may be using a strategy of tolerating higher bacterial diversity during the breeding season (Lozano et al. 2013). Alternatively, birds may benefit from acquiring a more diverse microbiome, as some of these bacteria could be beneficial symbionts that support host’s physiological functions or even enhance sexually-selected signals (Smith and Mueller 2015).
Four other, non-exclusive processes could also alter the cloacal microbiome of males. First, during reproduction there is an increase in physical contacts that could facilitate the sexual transmission of bacteria between individuals (Kulkarni and Heeb 2007). For example, when cloacal contacts during copulations were experimentally blocked in kittiwake (R.tridactyla) pairs, the bacterial diversity in the female’s cloaca started to decrease, presumably because it stopped receiving bacteria from the male (White et al. 2010). Similarly, in barn swallows (Hirundo rustica erythrogaster) females with more contacts with males had higher cloacal microbial diversity (Levin et al. 2016). In this same species, the cloacal microbiome in male–female pairs was more similar to each other than to that of other individuals in the population (Kreisinger et al. 2015; but see Pearce et al. 2017). Thus, social interactions in a reproductive context can promote transfer of bacteria. Reliably observing copulatory activity in most wild birds is challenging, as these events last mere seconds. However, one can measure rates of extra-pair paternity as a proxy for copulatory activity. Rufous-collared sparrows have high levels of extra pair paternity, with approximately 48% of the nests sired by a father who is not the social partner (Eikenaar et al. 2013). Future studies quantifying paternity could provide information on the intensity of copulatory activity of each individual and determine how it affects inter-individual cloacal microbiome variability. Second, when birds are in reproductive condition the internal environment of the cloaca may change. Such alterations can be directly related to the presence or absence of certain key species that secrete acidic fermentation products to create an acidic environment that restricts the growth of other bacteria (Witkin et al. 2007), or changes in the level of mucous secretion that affects the capacity of bacteria to colonize. Third, at the same time, these changes in the cloacal environment can affect competition and cooperation between the hundreds of microorganisms living there, and create niches for additional bacteria (Faust et al. 2012). Fourth, seasonal shifts in environmental bacterial communities (i.e., in soil, water, or food) could be reflected in the cloacal microbiome. The cloacal microbiome of migratory birds can vary between spring and fall migrants in Swainson’s thrushes (Catharus ustulatus) and gray catbirds (Dumetella carolinensis) (Lewis et al. 2016). Similarly, the gut microbiome in red squirrels (T.hudsonicus) changes across seasons, mostly driven by differences in diet availability (Ren et al. 2017). Our study population is subject to minimal ecological variation across seasons, but bacterial communities could still be sensitive to these small environmental changes. There is no obvious seasonal change in the diet of these omnivorous birds. However, if environmental factors had such an influence on the cloacal microbiome, then one would expect to see the same changes in the cloacal microbiome in both sexes, and it was only in males that we saw significant changes, suggesting that sex-specific effects are playing an important role.
Only during the breeding season were there differences in the cloacal community composition between the sexes, which suggests that sex-specific physiological factors could be playing an important role. The differences in cloacal community composition might be driven by male testosterone levels, as was described previously and confirmed here (Escallón et al. 2017). Interestingly, we found that the cloacal community composition of juveniles was different from females and non-breeding males, but not from breeding males. This was a somewhat unexpected result, as we predicted that juveniles would have a naïve microbiome that did not resemble that of adults. For example, in chinstrap penguins (Pygoscelis antarctica), there are clear differences in the cloacal microbiome between adults and nestlings (Barbosa et al. 2016). As revealed by the core microbiome analysis, the juveniles we sampled did share a portion of their microbiome with adults. Presumably, the juveniles could be transitioning to an adult microbiome as they had already left the nest when captured and were experiencing the same environment as adults. However, as juveniles they had regressed gonads and were not reproductively active. As such, the gastrointestinal portion of the microbiome may be driving the differences between adults and juveniles as described in barn swallows (H.rustica) (Kreisinger et al. 2017). If this is the case, the sexually transmitted portion of the cloacal bacterial community is just a small proportion of the total community and it will later become differentiated depending on the sex of the bird. Knowing the sex of the juveniles in future studies could uncover potential sex-specific effects on the microbiome during early development.
The bacterial diversity within individual birds did not change from one breeding season to the next (i.e., when sampling 10–12 months apart). We thought that independent of the bird’s age, additional opportunities for copulation could result in an accumulation of different bacterial species in their cloacas. Instead, we found that in both sexes the levels of diversity and richness remained unchanged from one breeding season to the next. This result is consistent with what was found in blue tits (Cyanistes caeruleus), where cloacal bacterial richness was constant across breeding seasons (Benskin et al. 2015). This could occur because birds are essentially resetting their cloacal microbiome during the non-breeding season and gradually losing many of the new bacteria that they accumulated during the previous breeding season. White et al. (2011) found that older female common lizards (Zootoca vivipara) had lower levels of cloacal bacterial diversity and hypothesized that older individuals could have communities that are more resistant to colonization by foreign bacteria. In the present study, we were not able to determine the birds’ adult age, but doing so would have allowed for further comparison of the bacterial community composition across age groups. Long term studies following the microbiome of individual birds would be necessary to disentangle the effect of age and multiple reproductive opportunities.
Unlike most studies that analyze the microbiome composition at a single time-point, in this study we incorporated important information about the temporal and inter-individual variability of the microbiome, showing that the cloacal microbiome in birds is dynamic and responsive to breeding condition and sex of the host. We found support for the bacterial clearance hypothesis, where bacteria are cyclically added and lost in each breeding season. Even though the environment plays an important role in shaping the bird’s microbiome (Hird et al. 2015; Waite and Taylor 2015), here we show that specific behavioral and physiological traits of individuals should not be overlooked, as these could alter susceptibility to STIs. Further studies should analyze the costs and benefits of acquiring a more diverse microbiome and consider the factors responsible for the observed variability across individuals. Birds and their cloacal bacterial communities are involved in complex reciprocal interactions, with potential fitness costs to the host (Jacob et al. 2015) and repercussions for the population (Lockhart et al. 1996), thus likely driving the evolution of life history strategies in ways that have yet to be revealed (Thrall et al. 2000; Kokko et al. 2002; Smith and Mueller 2015; Evans et al. 2017).
Data accessibility
Raw 16S rRNA amplicon sequences from Illumina sequencing have been deposited in the NCBI SRA (accession number SRP151469).
Ethics statement
This investigation adhered to animal-care protocols approved by the Institutional Animal Care and Use Committee of Virginia Tech.
Author contributions
C.E., L.K.B., and I.T.M. contributed to the design of the study. C.E. performed research and C.E. and L.K.B. processed and analyzed data. C.E. wrote the manuscript with input from L.K.B. and I.T.M. All authors contributed to revisions and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We are grateful to the landowners of Rodamonte for field accommodations and giving us access to work in their land. We thank N. Guerrero, D. Rozo, V. Gómez, M. A. Camargo, and B Mogollón for help in the field. We thank J. Molina for help with the permits. We are thankful for the insightful comments and careful review of the manuscript provided by W. Hopkins, J. Walters, D. Hawley, and J. McGlothlin as well as technical assistance provided by M. Hughey and D. Medina. Permission to study Z.capensis was provided by the Colombian environmental authority.
Funding
This research was funded by the Fralin Life Sciences Institute, Organismal Biology and Ecology group at Virginia Tech, and the National Science Foundation [DEB-1136640 to L.K.B. as well as IOS-0545735 and IOS-1353093 to I.T.M.].
Supplementary data
Supplementary data are available at IOB online.
References
- Addis EA, Busch DS, Clark AD, Wingfield JC.. 2010. Seasonal and social modulation of testosterone in Costa Rican rufous-collared sparrows (Zonotrichia capensis costaricensis). Gen Comp Endocrinol 166:581–9. [DOI] [PubMed] [Google Scholar]
- Anderson RM. 1979. Population biology of infectious diseases: Part I. Nature 280:361–7. [DOI] [PubMed] [Google Scholar]
- Archie EA, Theis KR.. 2011. Animal behaviour meets microbial ecology. Anim Behav 82:425–36. [Google Scholar]
- Ashby B, Gupta S.. 2013. Sexually transmitted infections in polygamous mating systems. Philos Trans R Soc Lond B Biol Sci 368:20120048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A, Balagué V, Valera F, Martínez A, Benzal J, Motas M, Diaz JI, Mira A, Pedrós-Alió C.. 2016. Age-related differences in the gastrointestinal microbiota of chinstrap penguins (Pygoscelis antarctica) (ed. H-U Peter). PLoS One 11:e0153215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 57:289–300. [Google Scholar]
- Benskin CMH, Rhodes G, Pickup RW, Mainwaring MC, Wilson K, Hartley IR.. 2015. Life history correlates of fecal bacterial species richness in a wild population of the blue tit Cyanistes caeruleus. Ecol Evol 5:821–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG.. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 10:57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer NN, Peña AG, Goodrich JK, Gordon JI, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer NN, Owens SM, Betley J, Fraser L, Bauer M, et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer NN, Knight R.. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Class AM, Wada H, Lynn SE, Moore IT.. 2011. The timing of life-history stages across latitudes in Zonotrichia Sparrows. Condor 113:438–48. [Google Scholar]
- Clay K. 2014. Defensive symbiosis: a microbial perspective (ed. C. Fox). Funct Ecol 28:293–8. [Google Scholar]
- Cox RM, Calsbeek R.. 2010. Severe costs of reproduction persist in Anolis lizards despite the evolution of a single-egg clutch. Evolution 64:1321–30. [DOI] [PubMed] [Google Scholar]
- Cox RM, Parker EU, Cheney DM, Liebl AL, Martin LB, Calsbeek R.. 2010. Experimental evidence for physiological costs underlying the trade-off between reproduction and survival. Funct Ecol 24:1262–9. [Google Scholar]
- De Cáceres M, Legendre P, Wiser SK, Brotons L.. 2012. Using species combinations in indicator value analyses (ed. R.B. O’Hara). Methods Ecol Evol 3:973–82. [Google Scholar]
- Dufrene M, Legendre P.. 1997. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345. [Google Scholar]
- Eikenaar C, Bonier F, Martin PR, Moore IT.. 2013. High rates of extra-pair paternity in two equatorial populations of rufous-collared sparrow, Zonotrichia capensis. J Avian Biol 44:600–2. [Google Scholar]
- Escallón C, Becker MH, Walke JB, Jensen RV, Cormier G, Belden LK, Moore IT.. 2017. Testosterone levels are positively correlated with cloacal bacterial diversity and the relative abundance of Chlamydiae in breeding male rufous‐collared sparrows (eds S.W. Nicolson, G.A. Wright, and J. Grindstaff). Funct Ecol 31:192–203. [Google Scholar]
- Evans JK, Buchanan KL, Griffith SC, Klasing KC, Addison B.. 2017. Ecoimmunology and microbial ecology: Contributions to avian behavior, physiology, and life history. Horm Behav. 88:112–121. [DOI] [PubMed] [Google Scholar]
- Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB.. 2012. Animal behavior and the microbiome. Science 338:198–199. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C.. 2012. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8:e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstad I, Karter AJ.. 1992. Parasites, bright males, and the immunocompetence handicap. Am Nat 139:603–22. [Google Scholar]
- Grear DA, Perkins SE, Hudson PJ.. 2009. Does elevated testosterone result in increased exposure and transmission of parasites? Ecol Lett 12:528–37. [DOI] [PubMed] [Google Scholar]
- Hamady M, Knight R.. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman LG, Zera AJ.. 2007. The cost of reproduction: the devil in the details. Trends Ecol Evol 22:80–6. [DOI] [PubMed] [Google Scholar]
- Hau M. 2007. Regulation of male traits by testosterone: implications for the evolution of vertebrate life histories. BioEssays 29:133–44. [DOI] [PubMed] [Google Scholar]
- Hird SM, Sánchez C, Carstens BC, Brumfield RT.. 2015. Comparative gut microbiota of 59 neotropical bird species. Front Microbiol 6:1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S, Parthuisot N, Vallat A, Ramon-Portugal F, Helfenstein F, Heeb P.. 2015. Microbiome affects egg carotenoid investment, nestling development and adult oxidative costs of reproduction in Great tits (ed. L. Martin). Funct Ecol 29:1048–58. [Google Scholar]
- Kokko H, Ranta E, Ruxton G., Lundberg P.. 2002. Sexually transmitted disease and the evolution of mating systems. Evolution 56:1091–1100. [DOI] [PubMed] [Google Scholar]
- Kreisinger J, Čížková D, Kropáčková L, Albrecht T.. 2015. Cloacal microbiome structure in a long-distance migratory bird assessed using deep 16sRNA pyrosequencing. PLoS One 10:e0137401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreisinger J, Kropáčková L, Petrželková A, Adámková M, Tomášek O, Martin J-F, Michálková R, Albrecht T.. 2017. . Temporal stability and the effect of transgenerational transfer on fecal microbiota structure in a long distance migratory bird. Front Microbiol 8:2838–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Heeb P.. 2007. Social and sexual behaviours aid transmission of bacteria in birds. Behav Process 74:88–92. [DOI] [PubMed] [Google Scholar]
- Levin II, Zonana DM, Fosdick BK, Song SJ, Knight R, Safran RJ.. 2016. Stress response, gut microbial diversity and sexual signals correlate with social interactions. Biol Lett 12:20160352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis WB, Moore FR, Wang S.. 2016. Characterization of the gut microbiota of migratory passerines during stopover along the northern coast of the Gulf of Mexico. J Avian Biol 47:659–68. [Google Scholar]
- Ley RER, Lozupone CA, Hamady M, Knight R, Gordon JI.. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart AB, Thrall PH, Antonovics J.. 1996. Sexually transmitted diseases in animals: ecological and evolutionary implications. Biol Rev Camb Philos Soc 71:415–71. [DOI] [PubMed] [Google Scholar]
- Lombardo MP. 1998. On the evolution of sexually transmitted diseases in birds. J Avian Biol 29:314. [Google Scholar]
- Lombardo MP, Thorpe PA, Power HW.. 1999. The beneficial sexually transmitted microbe hypothesis of avian copulation. Behav Ecol 10:333–7. [Google Scholar]
- Lozano GA, Lank DB, Addison B.. 2013. Immune and oxidative stress trade-offs in four classes of Ruffs (Philomachus pugnax) with different reproductive strategies. Can J Zool 91:212–8. [Google Scholar]
- MacManes MD. 2011. Promiscuity in mice is associated with increased vaginal bacterial diversity. Naturwissenschaften 98:951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Bakker M, Helm B.. 2015. The influence of biological rhythms on host–parasite interactions. Trends Ecol Evol 30:314–26. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S.. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data (ed. M. Watson). PLoS One 8:e61217–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AH. 1961. Molt cycles in equatorial Andean sparrows. Condor 63:143–61. [Google Scholar]
- Miller AH. 1962. Bimodal occurrence of breeding in an equatorial sparrow. Proc Natl Acad Sci U S A 48:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AH, Miller VD.. 1968. The behavioral ecology and breeding biology of the Andean sparrow, Zonotrichia capensis. Caldasia 10:83–154. [Google Scholar]
- Monaghan P, Metcalfe NB, Torres R.. 2009. Oxidative stress as a mediator of life history trade-offs: mechanisms, measurements and interpretation. Ecol Lett 12:75–92. [DOI] [PubMed] [Google Scholar]
- Moore IT, Bonier F, Wingfield JC.. 2005. Reproductive asynchrony and population divergence between two tropical bird populations. Behav Ecol 16:755–62. [Google Scholar]
- Moore IT, Perfito N, Wada H, Sperry TS, Wingfield JC.. 2002. Latitudinal variation in plasma testosterone levels in birds of the genus Zonotrichia. Gen Comp Endocrinol. 129:13–19. [DOI] [PubMed] [Google Scholar]
- Noguera JC, Aira M, Pérez-Losada M, Domínguez J, Velando A.. 2018. Glucocorticoids modulate gastrointestinal microbiome in a wild bird. R Soc Open Sci 5:171743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris K, Evans MR.. 2000. Ecological immunology: life history trade-offs and immune defense in birds. Behav Ecol 11:19–26. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H, Oksanen MJ. 2013. Package ‘vegan’. Community ecology package, version, 2(9).
- Oppliger A, Christe P, Richner H.. 1996. Clutch size and malaria resistance. Nature 381:565. [DOI] [PubMed] [Google Scholar]
- Pearce DS, Hoover BA, Jennings S, Nevitt GA, Docherty KM.. 2017. Morphological and genetic factors shape the microbiome of a seabird species (Oceanodroma leucorhoa) more than environmental and social factors. Microbiome 5:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiani A. 2010. Do cloacal pathogenic microbes behave as sexually transmitted parasites in birds? Open Ornithol J 3:72–85. [Google Scholar]
- R Development Core Team. 2018. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Raouf SA, Parker PG, Ketterson ED, Nolan V Jr, Ziegenfus C.. 1997. Testosterone affects reproductive success by influencing extra-pair fertilizations in male dark-eyed juncos (Aves: Junco hyemalis). Proc R Soc Lond B Biol Sci 264:1599–603. [Google Scholar]
- Ren T, Boutin S, Humphries MM, Dantzer B, Gorrell JC, Coltman DW, McAdam AG, Wu M.. 2017. Seasonal, spatial, and maternal effects on gut microbiome in wild red squirrels. Microbiome 5:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ML, Peters A.. 2009. Is testosterone immunosuppressive in a condition-dependent manner? An experimental test in blue tits. J Exp Biol 212:1811–8. [DOI] [PubMed] [Google Scholar]
- Roberts ML, Buchanan KL, Evans MR.. 2004. Testing the immunocompetence handicap hypothesis: a review of the evidence. Anim Behav 68:227–39. [Google Scholar]
- Sheldon BC. 1993. Sexually transmitted disease in birds: occurrence and evolutionary significance. Philos Trans R Soc Lond B Biol Sci 339:491–7. [DOI] [PubMed] [Google Scholar]
- Smith CC, Mueller UG.. 2015. Sexual transmission of beneficial microbes. Trends Ecol Evol 30:438–40. [DOI] [PubMed] [Google Scholar]
- Smith G, Dobson AP.. 1992. Sexually transmitted diseases in animals. Parasitol Today 8:159–66. [DOI] [PubMed] [Google Scholar]
- Stearns SC. 1992. The evolution of life histories. Oxford: Oxford University Press. [Google Scholar]
- Stothart MR, Bobbie CB, Schulte-Hostedde AI, Boonstra R, Palme R, Mykytczuk NCS, Newman AEM.. 2016. Stress and the microbiome: linking glucocorticoids to bacterial community dynamics in wild red squirrels. Biol Lett 12:20150875–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrall PH, Antonovics J, Dobson AP.. 2000. Sexually transmitted diseases in polygynous mating systems: prevalence and impact on reproductive success. Proc Biol Sci. 267:1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RER, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI.. 2007. The human microbiome project. Nature 449:804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen WF, White J, Brandl HB, Moodley Y, Merkling T, Leclaire S, Blanchard P, Danchin T, Hatch SA, Wagner RH.. 2013. Age-related differences in the cloacal microbiota of a wild bird species. BMC Ecol 13:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Moore IT, Breuner CW, Wingfield JC.. 2006. Stress responses in tropical sparrows: comparing tropical and temperate Zonotrichia. Physiol Biochem Zool 79:784–92. [DOI] [PubMed] [Google Scholar]
- Waite DW, Taylor MW.. 2015. Exploring the avian gut microbiota: current trends and future directions. Front Microbiol 6:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Mirleau P, Danchin E, Mulard H, Hatch SA, Heeb P, Wagner RH.. 2010. Sexually transmitted bacteria affect female cloacal assemblages in a wild bird. Ecol Lett 13:1515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Richard M, Massot M, Meylan S.. 2011. Cloacal bacterial diversity increases with multiple mates: evidence of sexual transmission in female common lizards. PLoS One 6:e22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield JC. 1984. Androgens and mating systems: testosterone-induced polygyny in normally monogamous birds. Auk 101:665–71. [Google Scholar]
- Witkin SS, Linhares IM, Giraldo P.. 2007. Bacterial flora of the female genital tract: function and immune regulation. Best Pract Res Clin Obstet Gynaecol 21:347–54. [DOI] [PubMed] [Google Scholar]
- Yildirim S, Yeoman CJ, Janga SC, Thomas SM, Ho M, Leigh SR Primate Microbiome Consortium White BA, Wilson BA, Stumpf RM.. 2014. Primate vaginal microbiomes exhibit species specificity without universal Lactobacillus dominance. ISME J 8:2431–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA amplicon sequences from Illumina sequencing have been deposited in the NCBI SRA (accession number SRP151469).