Abstract
Fluorescence live-cell and super-resolution microscopy methods have considerably advanced our understanding of the dynamics and mesoscale organization of macro-molecular complexes that drive cellular functions. However, different imaging techniques can provide quite disparate information about protein motion and organization, owing to their respective experimental ranges and limitations. To address these issues, we present here a robust computer program, called FluoSim, which is an interactive simulator of membrane protein dynamics for live-cell imaging methods including SPT, FRAP, PAF, and FCS, and super-resolution imaging techniques such as PALM, dSTORM, and uPAINT. FluoSim integrates diffusion coefficients, binding rates, and fluorophore photo-physics to calculate in real time the localization and intensity of thousands of independent molecules in 2D cellular geometries, providing simulated data directly comparable to actual experiments. FluoSim was thoroughly validated against experimental data obtained on the canonical neurexin-neuroligin adhesion complex at cell–cell contacts. This unified software allows one to model and predict membrane protein dynamics and localization at the ensemble and single molecule level, so as to reconcile imaging paradigms and quantitatively characterize protein behavior in complex cellular environments.
Subject terms: Biophysics, Computational biology and bioinformatics, Neuroscience
Introduction
Critical cellular functions such as membrane adhesion, receptor-mediated signaling, or synaptic transmission, involve the diffusional trapping of specific molecules in sub-cellular compartments1,2. To quantitatively describe such molecular dynamics in living cells, several fluorescence imaging techniques are currently available3,4: (1) single particle tracking (SPT) which resolves the motion of individual proteins at camera frame rate; (2) photo-activation and photo-bleaching methods, namely PhotoActivation of Fluorescence (PAF) and Fluorescence Recovery After Photobleaching (FRAP) which infer protein turnover at the population level; and (3) Fluorescence Correlation Spectroscopy (FCS), which analyzes molecular dynamics by correlating intensity fluctuations. More recent approaches based on single molecule localization such as PhotoActivated Localization Microscopy (PALM)5,6, direct Stochastic Optical Reconstruction Microscopy (dSTORM)7, and Point Accumulation In Nanoscopic Topography (PAINT)8, yield images of protein distribution at improved resolution (below 50 nm), giving unprecedented information about the nanoscale organization of biological structures9.
Despite such progress in imaging power, many experimental parameters remain difficult to estimate or control, including protein expression levels, probe labeling density, potential fixation artifacts, spatial and temporal sampling of the recordings, and protein motion below the system resolution, such that results from different experimental paradigms are often difficult to reconcile10. Thus, there is a pressing need for computer simulators that could unify those different imaging modes in a unique framework, estimate their respective biases, and serve as a predictive tool for experimenters, with the aim to quantitatively decipher protein organization and dynamics in living cells. Several particle-based packages relying on Monte Carlo simulations already exist to predict random motion and multi-state reactions of biological molecules, but either they do not integrate fluorescence properties or are limited to a specific type of imaging mode, and are usually not performing real-time visualization11–18.
In this context, we provide here fast, robust, and user-friendly software (FluoSim) that allows real time simulation of membrane protein dynamics in live-cell imaging (SPT, FRAP, PAF, and FCS) and super-resolution (PALM, dSTORM, uPAINT) modalities. We also show that FluoSim can be further used to produce large virtual data sets for training deep neural networks for image reconstruction19. This software should thus be of great interest to a wide community specialized in imaging methods applied to cell biology and neuroscience, with the common aim to better understand membrane dynamics and organization in cells.
Results
General principle of FluoSim
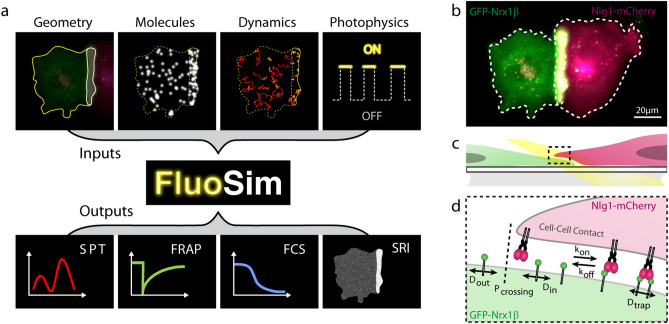
The FluoSim interface looks like performing a real experiment: the user imports a 2D cellular geometry from a microscopy image, and populates it with a realistic number of molecules (a parameter which depends on protein expression level, cell surface area, and labeling density) (Fig. 1a, Supplemental software and accompanying user manual). Kinetic parameters characterizing the diffusion and trapping of molecules in cellular regions of interest (ROI), are entered as inputs (Table 1). At each time increment (typically ~ 1–100 ms, adjusted to sensor acquisition rates), the algorithm updates the instantaneous positions of all molecules based on random number generation. Photo-switching rates of organic fluorophores or fluorescent proteins attached to the proteins of interest, further determine the fluorescence intensity associated to each molecule over time. The algorithm is optimized to visualize in real time the cellular system and provide post-processing information including molecule trajectories, image stacks, and output graphs (i.e. histograms of diffusion coefficients, FRAP, PAF, and FCS curves). Examples of simulated data sets for a realistic range of parameter values are given in Fig. S1. Moreover, since the positions of molecules are known with near-infinite accuracy, the program can generate super-resolved images comparable to those obtained with PALM or dSTORM, after introducing additional parameters describing protein labeling density, fluorophore duty cycle, and localization precision of the system. Finally, various levels of noise were introduced in the simulator, including a Poisson shot noise characterizing intensity fluctuations of individual fluorescent molecules, and a Gaussian readout noise applied around the camera offset20,21. This approach led to the simulation of very realistic images that closely matched those obtained by Total Internal Reflection Fluorescence (TIRF) imaging of single GFP-tagged molecules immobilized on a glass substrate (Fig. S2). A complete view of the parameters used in each experimental paradigm is available from the individual examples provided in the software menu.
Figure 1.
Schematics of the simulator and experimental system. (a) General principle of FluoSim. (b) Contact between two COS cells, one expressing GFP-Nrx1β (green) and the other expressing Nlg1-mCherry (magenta), resulting in molecule accumulation through adhesive interactions (yellow zone). (c) Diagram showing a zoomed section of the cell–cell interface at the coverslip; the yellow beam represents oblique laser illumination. (d) Cartoon of Nrx1β diffusional trapping by Nlg1 in a cell–cell contact.
Table 1.
Simulator parameters.
| Category | Parameter | Notation | Unit/format |
|---|---|---|---|
| Geometry | Reference image | .TIFF, PNG, JPG, GIF | |
| Image pixel size | µm/pixel | ||
| Region file | .rgn or .roi | ||
| FRAP laser spot | 2.8 µm | ||
| FCS laser spot width | σFCS | 0.25 µm | |
| Molecules | Numbera | 1–150,000 | |
| Initial position | Uniform or estimated | ||
| Times | Simulation durationa |
1000–100,000 frames (40–1600 s) |
|
| Time step | Δt | 2–100 ms | |
| Diffusion coefficients | Outside contact | Dout | 0.3 µm2/s |
| Inside contact | Din | 0.3 µm2/s | |
| Trapped | Dtrap | 0.04 µm2/s | |
| Crossing probability | Pcrossing | 25–70% | |
| Kinetics | Binding rate | kon | 0.15 s−1 |
| Unbinding rate | koff | 0.015 s−1 | |
| Photophysics | Switch-on ratea | konFluo | 0.005–5 s−1 |
| Switch-off ratea | koffFluo | 0–10 s−1 | |
| Bleaching ratea | koffBleach | 0.4–4 s−1 | |
| Activation ratea | koffActiv | 1–2 s−1 | |
| Export files | SPT or SRI images | .TIFF or multi-TIFF | |
| SPT trajectories | .trc | ||
| SPT histogram | .txt | ||
| FRAP curves | .txt | ||
| FCS fluctuations and autocorrelation | .txt |
aSee the methods above for the specific molecule numbers and photo-physical parameters used in the various imaging modes (SPT, STORM, FRAP, PAF, FCS).
Experimental system to validate FluoSim
To thoroughly validate FluoSim, we performed SPT, dSTORM, FRAP, and FCS experiments essentially on the canonical neurexin-neuroligin complex that mediates trans-synaptic adhesion in neurons22. We used COS-7 cells as a model expression system because they form large and flat lamellipodia that can be approximated as 2D environments for membrane diffusion. Cells were separately electroporated with recombinant GFP-tagged neurexin-1β (GFP-Nrx1β) and mCherry-tagged neuroligin-1 (Nlg1-mCherry), then cultured together for 24 h (Fig. 1b–d). Both molecules reached the cell membrane and accumulated at cell–cell contacts (GFP-Nrx1β enrichment = 3.9 ± 0.5, n = 20 cells), revealing adhesive interactions.
Simulations of SPT experiments
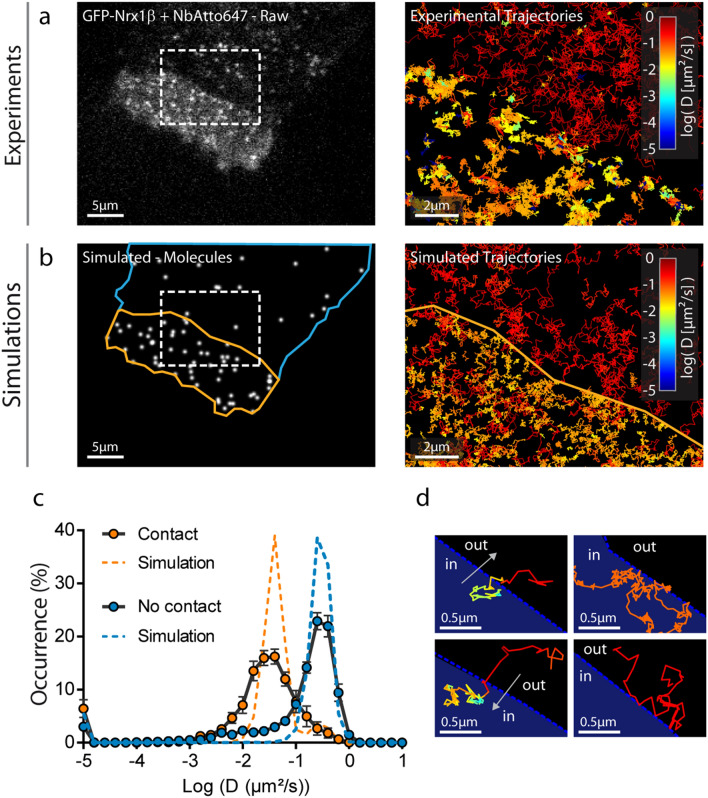
First, we experimentally tracked single GFP-Nrx1β molecules labeled with Atto647N-conjugated anti-GFP nanobody at 50 Hz using uPAINT8,23,24. Nrx1β exhibited fast free diffusion outside the adhesive contact (Dout = 0.3 µm2/s), and was slowed down by ~ tenfold in the contact, reflecting the formation of Nrx1β-Nlg1 bonds between apposed membranes (Dtrap = 0.04 µm2/s) (Fig. 2a,c). Nrx1β molecules often bounced at the contact border, revealing steric hindrance to penetrate the narrow cell–cell junction25, an effect which was described by a crossing probability (Pcrossing < 1) (Fig. 2d). Next, we simulated Nrx1β diffusion using FluoSim (Fig. 2b). We used the diffusion coefficients obtained experimentally and defined a sparse number of molecules (250) corresponding to the number of experimental detections per frame, together with two kinetic rates describing the Nrx1β-Nlg1 interaction taken from the literature (kon = 0.15 s−1 and koff = 0.015 s−1)26,27. We also chose realistic photophysics parameters (konFluo = 1 s−1 and koffFluo = 3 s−1) to match experimental trajectory number and duration, and single molecule rendering parameters for Atto647N (σ = 0.22 µm, FWHM = 0.53 µm). Using these parameters, FluoSim generated diffusion coefficient distribution curves inside and outside the contact that aligned well on experimental data (Fig. 2c). Experimental distributions were somewhat more spread than theoretical ones, most likely because of local membrane heterogeneities which are not accounted for in the model.
Figure 2.
Fitting SPT experiments. (a) Raw image of a COS-7 cell expressing GFP-Nrx1β sparsely labeled with Atto647N-conjugated GFP nanobody (white signal). (b) Image of simulated molecules in the same geometry. Rectangles highlight the interface between the cellular region with freely diffusing GFP-Nrx1β molecules (blue outline), and the contact region with a cell expressing Nlg1-mcherry (yellow outline). On the right, a zoom on this ROI shows single molecule trajectories for both experiments and simulations. The diffusion coefficient expressed in log scale is color coded. (c) Distributions of GFP-Nrx1β diffusion coefficients for experiments (circles, average ± sem of 3 cells, 4145 trajectories in contact and 4867 outside contacts) and simulations (dashed lines, 5 repetitions, 8757 trajectories in contact and 2645 trajectories outside contacts) on a semi-log plot. The Spearman correlation coefficient comparing experiment and simulation was r = 0.85 (P < 0.001, n = 36 bins) for contact regions, and r = 0.81 (P < 0.001, n = 36 bins) for outside regions. (d) Representative examples of single molecule trajectories at the interface: (left) a molecule escapes the contact and diffuses out freely, or enters the contact and gets trapped; (right) a molecule stays stuck in the contact with low diffusion, or bounces on the contact without entering it.
Simulations of dSTORM experiments
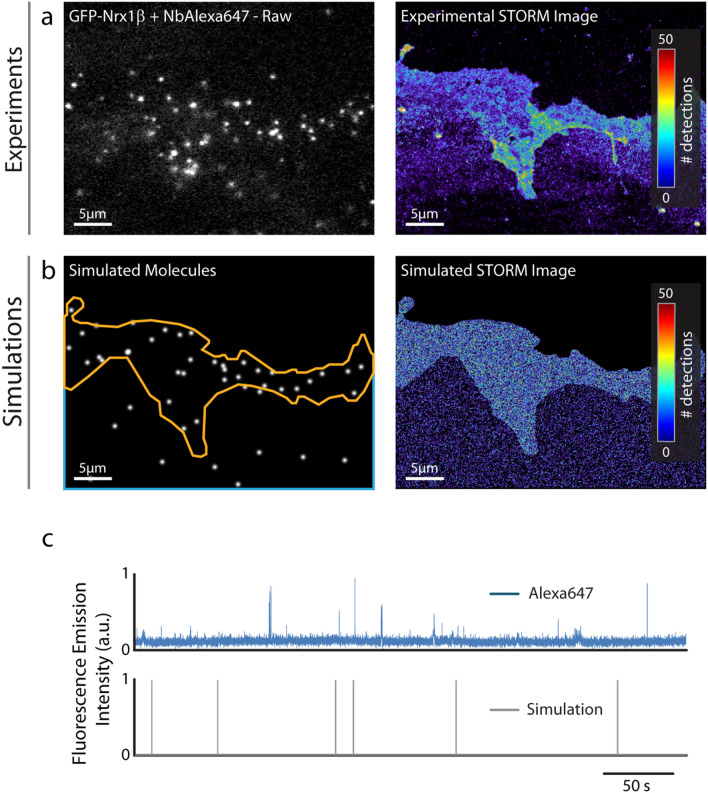
To simulate dSTORM experiments that were experimentally performed on GFP-Nrx1β using saturating labeling with Alexa647-conjugated GFP nanobody (Fig. 3a), we introduced a relatively large number of molecules in the imported geometry (70,000, corresponding to the sum of experimental detections per frame integrated over the cell surface area), and set all diffusion coefficients to zero to mimic cell fixation. To simulate stochastic fluorescence emission of Alexa64728, we calculated the switch-on (konFluo = 0.006 s−1) and switch-off (koffFluo = 9.3 s−1) rates of Alexa647 fluorophores from isolated Alexa647-conjugated nanobodies in dSTORM imaging conditions (Fig. 3c). We then simulated the accumulation of single molecule localizations for 80,000 frames, including a realistic localization precision (σ = 25 nm, FWHM = 58 nm), to mimic the experimental super-resolved maps of Nrx1β distribution (Fig. 3b). Simulated images had intensity values very similar to experimental ones, but as expected from an idealistic model, could not capture specific membrane features (e.g. ruffles) seen experimentally (also see Fig. S4 for a more homogeneous cell–cell contact).
Figure 3.
Fitting SRI experiments. (a) Representative single frame image of a STORM sequence performed on GFP-Nrx1β labeled with Alexa647-conjugated GFP nanobody, and corresponding super-resolved image generated from 3.57 × 106 single molecule localizations (pixel size 32 nm, total acquisition time 1600 s). The number of molecules detected per pixel is color coded. (b) Simulated image showing single molecule fluorescence emission in the same cell geometry, and corresponding super-resolved map with a localization precision of 58 nm (FWHM). The total number of detections is 3.61 × 106. (c) Fluorescence emission over time from an isolated Alexa647-conjugated GFP nanobody bound to the glass coverslip in a STORM sequence, and simulated emission of fluorescence of an immobile molecule obtained with switch-on and -off rates of 0.006 s−1 and 9.3 s−1, respectively.
Training of deep neural networks for image reconstruction
Deep learning is becoming increasingly popular for image reconstruction in fluorescence microscopy19,29,30. Convolutional Neural Networks (CNNs) are especially relevant for image treatment and have to be trained using large exemplary data sets obtained either experimentally, or from simulations. In this context, we tested the ability of FluoSim to train deep CNNs based on simulated data. We generated large image sets of randomly distributed single molecules represented as Gaussian intensity profiles plus Poisson noise, together with their localization maps as ground truth, and trained the previously described CNN called Deep-STORM19. FluoSim-trained Deep-STORM network was able to faithfully reconstruct super-resolved maps of both simulated and experimentally-observed microtubules, from single molecule localization image stacks (Figs. S3-S4). Strikingly, the CNN worked well at both low and high single molecule density, thus offering a considerable gain in acquisition time (× 20) for an equivalent resolution29. FluoSim-trained Deep-STORM network also allowed the reconstruction of images of Nrx1β-Nlg1 contacts that were comparable to images obtained with PALM-Tracer31, or to images directly simulated by FluoSim (Fig. S5). FluoSim can therefore be used to train deep CNNs.
Simulations of FRAP experiments
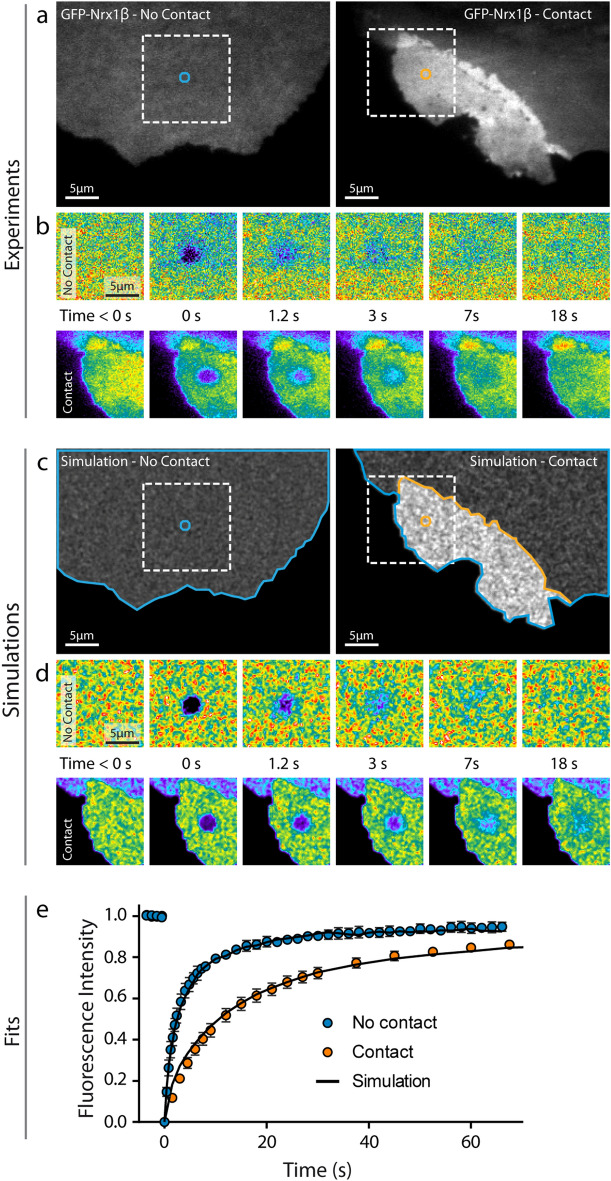
To challenge the simulator against ensemble measurements, we performed FRAP experiments on GFP-Nrx1β expressed in COS-7 cells. GFP-Nrx1β accumulates at cell–cell contacts when the opposite cell expresses its molecular partner Nlg1 (Fig. 4a) and shows slower recovery in the adhesive contact as compared to membrane regions not in contact with other cells (Fig. 4b,e). To mimic these experiments, we introduced a large number of molecules in the simulator (up to 150,000) and generated fluorescence-like images by defining a Gaussian intensity profile for each GFP-tagged molecule (σ = 0.17 µm, FWHM = 0.47 µm). Taking kinetic parameters from the literature (kon = 0.15 s−1 and koff = 0.015 s−1) and an intermediate porosity (Pcrossing = 0.3), the simulated images at steady state predicted Nrx1β enrichment in the contact area that matched experimental values (Fig. 4c,d). To induce local photo-bleaching, we chose a bleaching rate (koffBleach = 4.25 s−1) reproducing the initial drop of fluorescence observed experimentally (~ 75% in 400 ms). Using those coefficients plus the diffusion parameters determined from SPT, FRAP simulations accurately reproduced experimental data (Fig. 4e).
Figure 4.
Fitting FRAP experiments. (a) Representative images of COS-7 cells expressing GFP-Nrx1β either not forming contact (left), or forming contact with another cell expressing Nlg1-mCherry (right). (b) Corresponding FRAP sequences on the zoomed square areas (bleached circle diameter = 2.8 µm). Intensity is color coded. (c,d) Simulated images of the same cell geometries filled with 150,000 fluorescent molecules, and corresponding FRAP sequences. (e) Normalized FRAP curves obtained by experiment (closed circles) outside (mean ± sem of 6 cells, 44 bleached regions) or inside contact regions (mean ± sem of 18 cells, 144 bleached regions) and corresponding simulations (solid curves, average of 30 repetitions each, sem < 1% mean, not shown). The Spearman correlation coefficient comparing experiment and simulation was r = 1.0 (P < 0.001, n = 21 time points) for contact regions, and r = 0.98 (P < 0.001, n = 43 time points) for outside regions.
Simulations of PAF experiments
PAF, which can be viewed as the mirror of a FRAP experiment, is also very popular to estimate membrane molecule dynamics in specific cellular compartments32. We thus confronted FluoSim with actual PAF experiments performed on COS-7 cells expressing Nrx1β fused to photoactivatable GFP (PAGFP-Nrx1β). Because PAGFP is non-fluorescent in the absence of photo-stimulation, cells were co-electroporated with BFP for detection. Characteristic adhesive contacts were formed between cells expressing PAGFP-Nrx1β, and co-cultured cells expressing Nlg1-mcherry (Fig. 5a,c). PAGFP-Nrx1β was turned on with a brief pulse of 405 nm laser and the fluorescence decay due to protein diffusion and turnover was recorded for 2 min (Fig. 5b,d). As expected from SPT and FRAP data, the fluorescence decay was slower for PAGFP-Nrx1β making contact with Nlg1-mcherry, compared to freely diffusing PAGFP-Nrx1β outside contact regions (Fig. 5e). In FluoSim, we introduced similar numbers of molecules, diffusion coefficients and kinetic rates as for FRAP. To match the sudden increase in PAGFP-Nrx1β fluorescence intensity at time zero, the photo-activation rate was raised from 0 to 1 s−1 for 300 ms. Given this set of parameters, the PAF simulations fit very well the experimental data (Fig. 5e).
Figure 5.
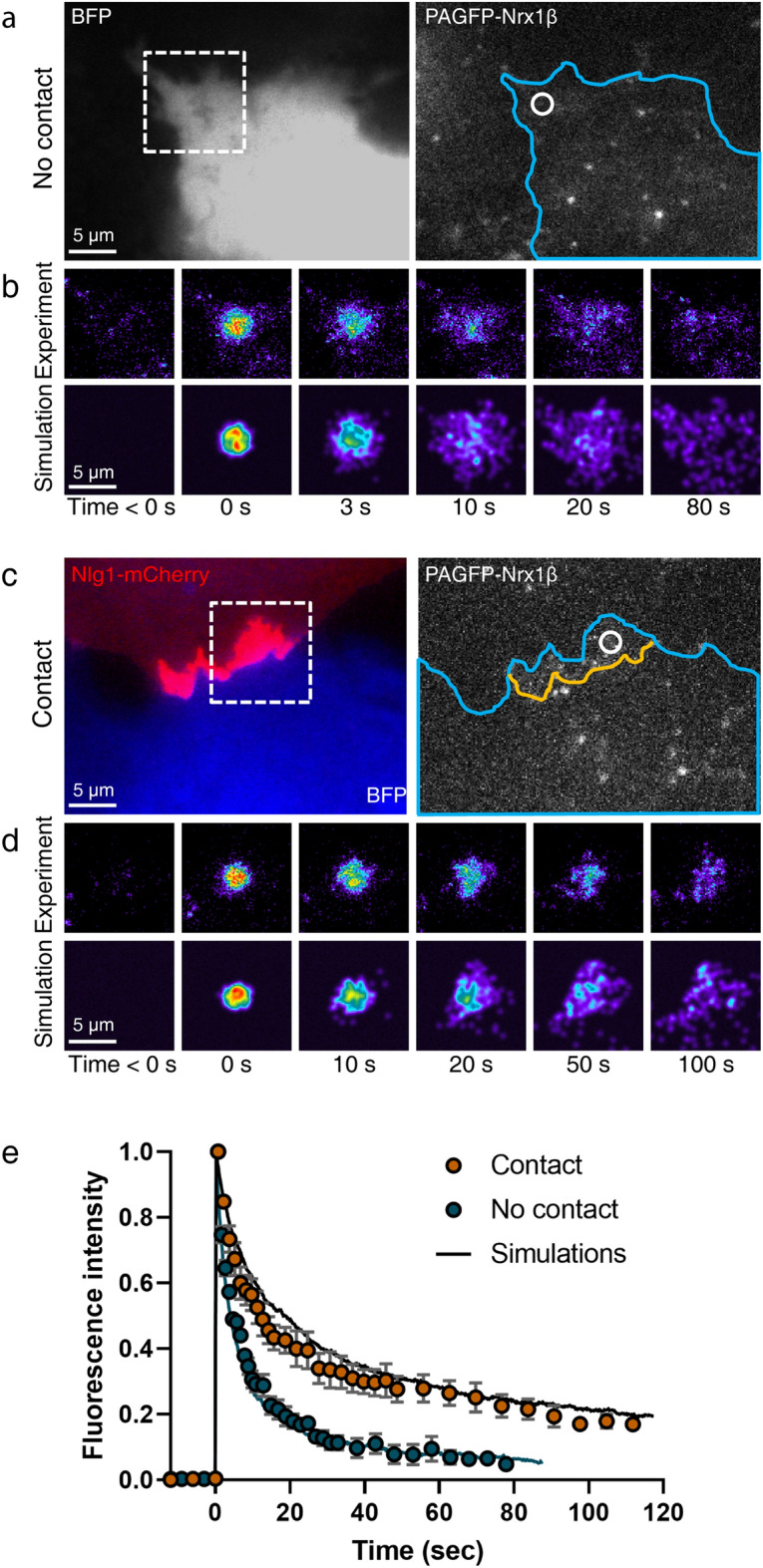
Fitting PAF experiments. (a,c) Representative images of COS-7 cells co-expressing BFP (blue) plus PAGFP-Nrx1β, and either not forming contact, or forming contact with another cell expressing Nlg1-mCherry (red). The cell geometry (blue contour) is entered in the simulator, together with the contact region (yellow line). (b,d) Corresponding PAF sequences on zoomed areas (dashed squares), where the photoactivated spot is indicated in white (circle of diameter = 2.8 µm). The experimental and simulated images are shown on top of each other for the indicated time points. Fluorescence intensity is color coded. (e) Graph showing the fluorescence intensity normalized between 0 and 1 for PAF experiments performed outside (blue circles) or inside (orange circles) contact regions (mean ± sem of 5 cells/1 PAF region per cell, and 3 cells/2 PAF regions per cell, respectively). The corresponding simulations (solid curves) represent the average of 20 repetitions (sem < 2% mean, not shown). The Spearman correlation coefficients comparing experiment and simulation was r = 0.998 for contact regions, and r = 0.995 for outside regions (P < 0.0001, n = 32 time points in both conditions).
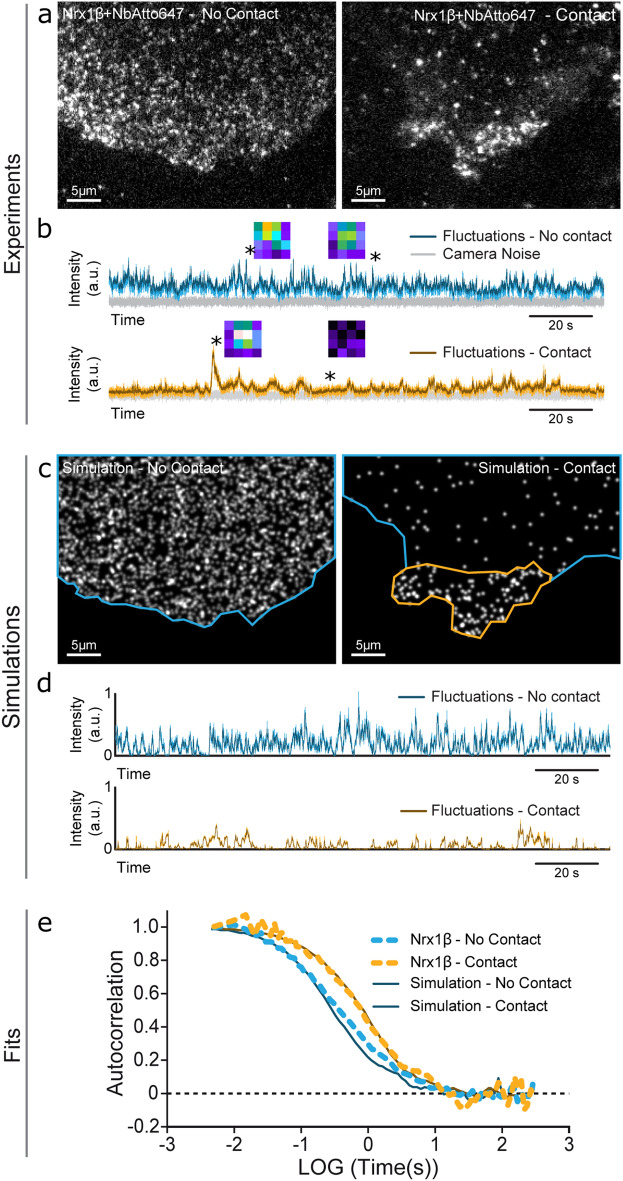
Simulations of FCS experiments
To further validate the simulator in conditions of intermediate molecular density, we performed FCS experiments33 by recording intensity fluctuations over time for Atto647N-nanobody bound to GFP-Nrx1β, in a diffraction-limited laser spot (Fig. 6a,b). As expected from slower diffusion, the autocorrelation function obtained for Nrx1β in the adhesive contact was shifted to the right compared to the free region, with a small photo-bleaching bias due to longer residence times (Fig. 6e). Again, the simulations performed with an intermediary number of molecules introduced in FluoSim, matched experimental FCS curves with the same set of kinetic and photophysical parameters used for SPT, FRAP, and PAF (Fig. 6c–e).
Figure 6.
Fitting FCS experiments. (a) Images of Atto647N-conjugated GFP nanobody bound to COS cells expressing GFP-Nrx1β not forming contact (left), or forming contact with a cell expressing Nlg1-mCherry (right). A 642 nm focused laser beam of 0.6 µm FWHM was parked in contact or no-contact regions, and square images of the illuminated area (1 µm × 1 µm) were collected at 200 or 500 Hz, respectively. (b) Intensity fluctuations over time in the two regions (color), above the camera noise (grey). Insets represent images acquired at the indicated times (stars). Intensity is color coded. (c,d) Simulated images using the same cell geometries populated with 2500 and 200 molecules, respectively, and corresponding intensity fluctuations. (e) Normalized autocorrelation functions for FCS experiments (dashed curves) performed outside (2 cells/10 recordings) and inside contact regions (1 cell/3 recordings), and corresponding simulations (solid curves, average of 10 repetitions each).
Discussion
In summary, FluoSim allows a prediction and comparison of membrane protein dynamics in a wide range of fluorescence cell imaging modes, with a precise control of the relevant kinetic, photo-physical, and acquisition parameters determining measurement outputs. The program is intended to help biologists adjust and interpret their experiments on a variety of cellular systems, and serve as a teaching resource in bio-imaging programs.
FluoSim reproduced a wide range of experimental results (SPT, FRAP, PAF, FCS, SRI) on the Nrx1β-Nlg1 membrane complex using a unique set of parameters extracted from published in-vitro studies and/or taken from our own measurements26,27, thereby giving strong credit to the correlative approach. The program is very fast and robust, and should be applicable to model a wide range of 2D-like dynamic molecular systems experiencing membrane diffusion and transient confinement, for example integrins at focal contacts in fibroblasts34,35, cadherins at cell–cell contacts36–38, neuronal adhesion proteins and neurotransmitter receptors at synapses24,39,40, and trapping of membrane molecules by lipid rafts or cytoskeletal interactions41–43.
Independently of changes in the fluorescence parameters, FluoSim can also be used to predict changes in the sub-cellular distribution of membrane proteins over time, induced by the modification of diffusion coefficients (Dout, Din, Dtrap) or kinetic rates (kon, koff). For example, the effect of cross-linking surface receptors with antibodies can be simulated by a decrease in diffusion coefficients44,45. Moreover, changes in the parameters kon and koff in FluoSim can mimic the action of pharmacological agents, competing peptides, and chemo- or opto-genetic stimulations that modulate specific protein–protein interaction rates. For instance, cell-permeant peptides competing with glutamate receptor anchorage to scaffolding proteins rapidly decrease receptor density at synapses46, a process that is mimicked by changing the binding rates of receptors to their post-synaptic scaffold45. Similarly, long term potentiation (LTP) and long term depression (LTD) can be simulated by modulating the AMPA receptor/scaffold affinity, providing insight on the molecular mechanisms underlying synaptic plasticity45,47.
We believe that this quantitative simulation approach will be of great help to optimize experimental design, especially regarding the choice of various parameters such as fluorophores, control of laser powers, acquisition frame rate, and overall timing of the experiment with respect to the internal dynamics of the molecular system, thus replacing lengthy experimental adjustments and saving research time. The software can also be used to test the robustness and predictions of single molecule tracking algorithms that have been implemented those past years, e.g. SR-Tesseler and InferenceMap48–50. Compared to existing software such as PyFRAP, SuReSim, FERNET, or MCell13–15, FluoSim integrates many fluorescence modalities into a single program and achieves real-time display (Supplementary Table 1). In its present version, FluoSim is limited to Brownian motion and first order molecular reactions, but sub- or super-diffusive behaviors as well as more complex multi-state molecular reactions might be implemented on a case-by-case basis, depending on user needs (Table 1).
Finally, with the advent of single-molecule based super-resolution microscopy such as PALM and STORM, many new questions arise regarding the degree of labeling needed, and the number of single molecule localizations to accumulate in order to reconstruct a realistic image of biological structures, with the risk of finding artificial properties in under-sampling conditions10. By varying the number of molecules, type of fluorophore attached to them, localization precision, and length of the acquisition sequence, the software is able to determine the conditions for faithful detection of the biological sample. The capacity of FluoSim to mimic molecule dynamics might also be used in the near future to train the next generation of CNNs for the analysis of live imaging experiments51.
Methods
DNA plasmids
GFP-Nrx1β was a gift from M. Missler (Münster University, Germany). PA-GFP-Nrx1β was generated by amplifying PA-GFP32 (a gift of J. Lippincott-Schwartz, Janelia Research Campus, USA) with the two primers PAGFP-3F (5′-cgGCTAGCGGAGCAGGAatggtgagcaagggcgagg-3′) and PAGFP-4R (5′-cgGCTAGCTCCTGCTCCCTTGTACAGCTCGTCCATGC-3′) and inserting it into GFP-Nrx1β in place of GFP at NheI/NheI sites. Proper insert orientation was verified by restriction and sequencing. HA-tagged Nlg1 obtained from P. Scheiffele (Biozentrum, Basel) was used as a backbone to construct Nlg1-mCherry, by inserting mCherry intracellularly at position -21aa before the C-terminus. The HA-Nlg1 sequence was moved from the pNice vector into a pcDNA vector at the HindIII/ NotI sites. Two PCRs were performed on Nlg1: the first one from the KpnI site (located inside the Nlg1 sequence) to the insert position of mCherry (AgeI site added), and the other one from the mCherry site insertion (Nhe added) to the end of Nlg1 (NotI). The mCherry gene (AgeI/NheI) was obtained by PCR on pmCherry-N1 (Clontech). A four-fragment ligation was then done to obtain the final construct (HindIII-HA-Nlg1-AgeI-mCherry-NheI-Nlg1Cter-NotI). The plasmid for bacterial expression of the anti-GFP nanobody52 was a kind gift from A. Gautreau (Gif-sur-Yvette, France). The bacterial production of anti-GFP nanobody, purification, and conjugation to organic dyes (Atto647N or Alexa647), was described previously24.
Cell culture and electroporation
COS-7 cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM; GIBCO/BRL) supplemented with 10% fetal bovine serum (FBS), 100 units mL−1 penicillin, and 100 µg mL−1 streptomycin, in a 37 °C-5% CO2 atmosphere. One day before the experiments, cells were rinsed twice in warm PBS, trypsinized for 5 min, mixed in culture medium, and centrifuged for 5 min at 1000 rpm. The cell pellet was suspended in 100 µL electroporation medium and electroporated for either GFP-Nrx1β or NLG1-mCherry plasmids with the Amaxa Nucleofector system (Lonza), using 500,000 cells per cuvette and 3 µg DNA. Electroporated cells were mixed in culture medium, seeded on 18 mm glass coverslips at a concentration of 50,000–80,000 cells per coverslip, cultured in 12-well plates, and imaged 24–48 h after electroporation.
Single molecule pull-down
Single molecule pull-down of GFP-Nrx1β molecules was performed essentially as described53,54. Briefly, cleaned glass coverslips (VWR) were treated with N-(2-aminoethyl)-3-aminopropyltrimethoxysilane (United Chemical Technologies, #A0700), then incubated with mPEG-succinimidyl valerate (SVA) containing a 1:100 fraction of biotin PEG-SVA (both from Laysan Bio MW 5000 Da). Coverslips were dried and stored at − 20 °C. Just before the experiment, coverslips were incubated with 1 µM NeutrAvidin for 5 min (Invitrogen, A2666), followed by 10 nM biotinylated anti-GFP antibody for 15 min (ABCAM, 6658), both diluted in T50 buffer (10 mM Tris-HCl, 50 mM NaCl, pH = 7.5). Two days before the experiment, HEK cells were transfected with GFP-Nrx1β using Lipofectamine (Invitrogen) in optiMEM buffer. Two hours before the experiment, cells were detached in Ca-free PBS (30 min, 37 °C), then centrifuged at 3000 rpm for 2 min. The cell pellet was dissolved in 100 μL lysis buffer containing 150 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% Igepal, and protease inhibitor cocktail (Pierce). Samples were rotated for 2 h at 4 °C to solubilize proteins, then centrifuged at 14,000 rpm for 15 min to remove aggregates. The supernatant was kept on ice and diluted 1:200 in observation solution (135 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, 4 mM Trolox, 40 mM d-glucose, 0.03% Igepal, pH = 7.4). 100 µL of this mix was added to glass substrates for several minutes, then rinsed out with fresh solution when the molecular density reached about 100 per field of view (65 × 65 µm). Single GFP-Nrx1β molecules were observed with a 488 nm laser under TIRF illumination on an inverted Olympus microscope equipped with a 63 ×/1.45 NA objective and an EMCCD camera (Andor Ixon). GFP-Nrx1β molecules did not stick to a control coverslip containing no anti-GFP antibody, revealing binding specificity to the GFP tag. Acquisition sequences of 500 frames were recorded at 30 Hz, resulting in the photobleaching of nearly all immobilized GFP-Nrx1β molecules. Images were analyzed using Metamorph by measuring the average fluorescence intensity in small regions drawn around individual molecules.
Single molecule tracking (uPAINT)
Universal point accumulation in nanoscale topography (uPAINT) experiments were carried out as reported8. Cells were mounted in Tyrode solution (15 mM d-glucose, 108 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2 and 25 mM HEPES, pH 7.4) containing 1% globulin-free BSA (Sigma A7638) in an open Inox observation chamber (Life Imaging Services, Basel, Switzerland). The chamber was placed on a fully motorized inverted microscope (Nikon Ti-E Eclipse) equipped with perfect focus system, a thermostatic box (Life Imaging Services) providing air at 37 °C, and an APO TIRF 100 ×/1.49 NA oil immersion objective. GFP-and mCherry expressing cells were detected using a mercury lamp (Nikon Xcite) and the following filter sets (SemROCK): EGFP (Excitation: FF01-472/30; Dichroic: FF-495Di02; Emission: FF01-525/30) and mCherry (Excitation: FF01-543/22; Dichroic: FF-562Di02; Emission: FF01-593/40). Cells expressing GFP-Nrx1β were labeled using a low concentration of Atto647N-conjugated GFP nanobody (1 nM). A four-colour laser bench (405/488/561 nm lines, 100 mW each; Roper Scientific, and 1 W 642 nm line, MPB Communications Inc., Canada) is connected through an optical fiber to the Total Internal Reflection Fluorescence (TIRF) illumination arm of the microscope. Laser power was controlled through an acousto-optical tunable filter (AOTF) driven by the Metamorph software (Molecular Devices). Atto647N was excited with the 642 nm laser line (~ 2 mW at the objective front lens), through a four-band beam splitter (BS R405/488/561/635, SemRock). Samples were imaged by oblique laser illumination, allowing the excitation of individual Atto-conjugated ligands bound to the cell surface, without illuminating ligands in solution. Fluorescence was collected on an EMCCD camera with 16 µm pixel size (Evolve, Roper Scientific, Evry, France), using a FF01-676/29 nm emission filter (SemRock). Stacks of 4000 consecutive frames were obtained from each cell with an integration time of 20 ms, using the Nikon perfect focus system to avoid axial drift. Images were analyzed using PALM-Tracer, a program running on Metamorph and based on wavelet segmentation for molecule localization and simulated annealing algorithms for tracking (generously provided by JB Sibarita, Bordeaux)55. This program allows the tracking of localized molecules through successive images. Trajectories longer than 20 frames (400 ms) were selected. The diffusion coefficient, D, was calculated for each trajectory, from linear fits of the first 4 points of the mean square displacement (MSD) function versus time. Trajectories with displacement inferior to the pointing accuracy (~ 50 nm in uPAINT conditions) whose MSD function cannot be fitted are arbitrarily taken as D = 10−5 µm2 s−1.
dSTORM experiments
COS-7 cells expressing GFP-Nrx1β were surface-labeled with a high concentration (100 nM) of Alexa647-conjugated GFP Nanobody in Tyrode solution containing 1% globulin-free BSA (Sigma A7638) for 10 min, rinsed and fixed with 4% PFA-0.2% glutaraldehyde in PBS for 10 min at room temperature, and stored in PBS at 4 °C until imaging. For microtubule staining, COS-7 cells were rinsed twice in PBS, fixed using 4% PFA-20% sucrose for 15 min at room temperature, washed 3 times in PBS, and incubated with 50 mM NH4Cl in PBS for 10 min. After fixation, cells were washed 3 times in PBS, permeabilized using 0.2% Triton-X 100 for 10 min, washed in PBS, blocked in 1% BSA-PBS for 30 min, and incubated with anti-α-tubulin (Thermofisher MA1-80017, 1:500) overnight at 4 °C. The next day, cells were washed 3 times in PBS, and incubated 45 min with secondary goat-anti-rat-Alexa647 antibody (ThermoFisher A21247, 1:800) and kept in PBS before dSTORM imaging.
Cells were imaged in Tris‐HCl buffer (pH 7.5) containing 10% glycerol, 10% glucose, 0.5 mg/mL glucose oxidase (Sigma), 40 mg/mL catalase (Sigma C100-0.1% w/v), and 50 mM β-mercaptoethylamine (MEA) (Sigma M6500)56. The same microscope described for uPAINT was used. Pumping of Alexa647 dyes into their triplet state was performed for several seconds using ~ 60 mW of the 642 nm laser at the objective front lens. Then, a lower power (~ 30 mW) was applied to detect the stochastic emission of single-molecule fluorescence, which was collected using the same optics and detector as described above for uPAINT. 10–20 streams of 3000–4000 frames each were acquired at 50 Hz. To generate images intended to test deep CNN algorithms, a higher density of fluorescent molecules was generated by turning on the 405 nm laser power to 5 mW during acquisition. Multi-color 100-nm fluorescent beads (Tetraspeck, Invitrogen) were used to register long-term acquisitions and correct for lateral drift. The localization precision of our imaging system in STORM conditions is around 60 nm (FWHM). Stacks were analyzed using the PALM-Tracer program, allowing the reconstruction of a unique super-resolved image of 32 nm pixel size (zoom 5 compared to the original images) by summing the intensities of all localized single molecules (1 detection per frame is coded by an intensity value of 1).
FRAP experiments and analysis
COS-7 cells expressing GFP-Nrx1β in co-culture with cells expressing Nlg1-mCherry were mounted in Tyrode solution, and observed under the same set-up used for uPAINT and STORM. The laser bench has a second optical fiber output connected to an illumination device containing two x/y galvanometric scanning mirrors (ILAS, Roper Instrument) steered by MetaMorph. It allows precise spatial and temporal control of the focused laser beam at any user-selected region of interest within the sample for targeted photo-bleaching. Switching between the two fibers for alternating between imaging and bleaching is performed in the ms time range using an AOTF. Oblique illumination was performed using the 491 nm beam at low power (0.3 mW at the front of the objective) to image GFP-Nrx1β molecules in the plasma membrane close to the substrate plane. After acquiring a 10 s baseline at 1 Hz frame rate, rapid selective photo-bleaching of 3–9 circular areas of diameter 2.8 µm was achieved at higher laser power (3 mW at the objective front lens), during 400 ms. Fluorescence recovery was then recorded immediately after the bleach sequence for 80 s. The recording period included three phases with decreasing frame rate ranging from 2 to 0.1 Hz. Observational photo-bleaching was kept very low, as assessed by observing control unbleached areas nearby. FRAP curves were obtained by computing the average intensity in the photobleached area, after background subtraction, and normalized between 1 (baseline) and 0 (time zero after photo-bleaching).
PAF experiments and analysis
COS-7 cells co-expressing BFP and PAGFP-Nrx1β in co-culture with cells expressing Nlg1-mCherry were mounted in Tyrode solution, and observed under the microscope as described for FRAP experiments. Oblique illumination was carried out using the 491 nm beam at low power (0.3 mW at the front of the objective) to image membrane PAGFP-Nrx1β molecules close to the substrate. After acquiring a 12 s baseline at 0.3 Hz frame rate, photo-activation of 1–2 circular areas of diameter 2.8 µm was achieved with the focused 405 nm laser beam (1.5 mW at the objective front lens), during 300 ms. Fluorescence decay was then recorded for 80–100 s, with decreasing frame rate ranging from 2 to 0.1 Hz. PAF curves were normalized between 0 and 1, by dividing the intensity in the photoactivated area over time by the maximal intensity recorded immediately after the photo-activation pulse, after subtracting the baseline from both values.
FCS experiments and analysis
COS-7 cells expressing GFP-Nrx1β were surface-labeled at an intermediate concentration (10 nM) of Atto647N-conjugated GFP nanobody for 5 min at 37 °C, then mounted in Tyrode solution on the same microscope. The 642 nm laser beam was parked on a region of interest, either on the free cell membrane, or in the cell–cell contact area. The laser was kept at low power (3% of 100 mW, i.e. ~ 0.2 mW at the objective front lens) to avoid photo-bleaching the organic dye. Using glass coverslips coated with polylysine and higher concentrations of Atto647N-conjugated nanobody (100 nM) to provide a uniform density, we independently measured the Gaussian intensity profile of the laser beam, which gave a standard deviation σFCS = 0.26 µm (FWHM = 0.6 µm). Intensity fluctuations due to Nrx1β molecules entering and leaving the laser spot by membrane diffusion, were detected using a second camera (Hamamatsu Orca Flash 4.0) on the opposite port of the microscope. A square area of 16 × 16 pixels (1 µm × 1 µm) centered on the laser spot was chosen, and streams of 60,000 images at binning 4 were acquired through the Hokawo software (Hamamatsu) at 200 Hz or 500 Hz, for the study of cell–cell contacts or free regions, respectively. Control stacks with the laser off were acquired to record the camera readout noise. The integrated intensity of each image was read from the multi-TIFF stacks, from which the average camera noise was subtracted, and the autocorrelation function was computed using custom-made routines written in C++, and normalized by its first value.
FluoSim simulations
A general description of the FluoSim algorithm and the list of simulations performed to match the experiments, together with the precise set of parameters used in each imaging mode, is available in the Supplementary material.
Supplementary information
Acknowledgements
We thank M. Missler, P. Scheiffele, and J. Lippincott-Schwartz for the generous gift of plasmids; B. Tessier, Z. Karatas, and S. Benquet for molecular biology; R. Sterling and the cell culture facility of the Institute; J.B. Sibarita, C. Butler, and A. Kechkar for the kind gift of the PALM-Tracer program, M. Sainlos for the gift of dye-conjugated GFP nanobody, R. Vafabakhsh (Northwestern University, Evanston, USA) for help with single molecule pull-down, E. Isacoff (University of California, Berkeley, USA) for providing generous access to laboratory resources, L. Barusseau (Explora Nova) and F. Levet for programming tips, A. Disher (Hamamatsu Photonics) for help on camera acquisition parameters for FCS, E. Nehme (Technion, Haifa, ISR) for helpful advice on Deep-STORM, O. Rossier and G. Giannone for scientific insight, A. Mehidi and T. Orré for interesting feedback, and A. Florea for help with the FluoSim movie. This work received funding from the Fondation pour la Recherche Médicale (“Equipe FRM” DEQ20160334916 to O.T.), the French Ministry of Research (fellowship to M.L.), the Agence Nationale de la Recherche (Grants «SynAdh» ANR-13-BSV4-0005-01 and «Synthesyn» ANR-17-CE16-0028-01 to O.T.), the national infrastructure France BioImaging (Grant ANR-10INBS-04-01), the French-American Commission (Fulbright grant to O.T.), and the Conseil Régional Aquitaine («SiMoDyn» to O.T.).
Author contributions
M.L. developed FluoSim, performed the SPT and FRAP experiments, carried out the simulations, and made most figures. M.L. and O.T. designed FluoSim, performed the FCS experiments, and wrote the user manual. I.C. and M.L. did proof-of-concept SPT and FRAP experiments. I.C. performed dSTORM acquisition and analysis, and made two figures. E.B. and M.N. installed and run the CNN Deep-STORM. E.B. and M.L. adapted the Deep-STORM MATLAB and Python scripts to train the CNN with FluoSim simulation outputs. O.T. came up with the original idea, supervised the work, performed the PAF experiments, and wrote the manuscript. All authors reviewed the manuscript.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. The software FluoSim (v1.0) is released under a GNU GPL v3 license as supplementary material accompanying this manuscript, and as an archive that can be downloaded at https://www.iins.u-bordeaux.fr/SOFTWARE. The FluoSim (v1.0) release and its source code are available at https://github.com/mlagardere/FluoSim under the same license.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-75814-y.
References
- 1.Choquet D, Triller A. The dynamic synapse. Neuron. 2013;80:691–703. doi: 10.1016/j.neuron.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Dustin ML, Choudhuri K. Signaling and polarized communication across the T cell immunological synapse. Annu. Rev. Cell Dev. Biol. 2016;32:303–325. doi: 10.1146/annurev-cellbio-100814-125330. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol. Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Lippincott-Schwartz J, Snapp EL, Phair RD. The development and enhancement of FRAP as a key tool for investigating protein dynamics. Biophys. J. 2018;115:1146–1155. doi: 10.1016/j.bpj.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 6.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 7.Dani A, Huang B, Bergan J, Dulac C, Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannone G, et al. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys. J. 2010;99:1303–1310. doi: 10.1016/j.bpj.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: Super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deschout H, et al. Precisely and accurately localizing single emitters in fluorescence microscopy. Nat. Methods. 2014;11:253–266. doi: 10.1038/nmeth.2843. [DOI] [PubMed] [Google Scholar]
- 11.Stefan, M. I., Bartol, T. M., Sejnowski, T. J. & Kennedy, M. B. Multi-state Modeling of Biomolecules. PLoS Comput. Biol.10, (2014). [DOI] [PMC free article] [PubMed]
- 12.Kerr RA, et al. Fast Monte Carlo simulation methods for biological reaction-diffusion systems in solution and on surfaces. SIAM J. Sci. Comput. 2008;30:3126. doi: 10.1137/070692017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bläßle A, et al. Quantitative diffusion measurements using the open-source software PyFRAP. Nat. Commun. 2018;9:1–14. doi: 10.1038/s41467-018-03975-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkataramani V, Herrmannsdörfer F, Heilemann M, Kuner T. SuReSim: simulating localization microscopy experiments from ground truth models. Nat. Methods. 2016;13:319–321. doi: 10.1038/nmeth.3775. [DOI] [PubMed] [Google Scholar]
- 15.Angiolini J, Plachta N, Mocskos E, Levi V. Exploring the dynamics of cell processes through simulations of fluorescence microscopy experiments. Biophys. J. 2015;108:2613–2618. doi: 10.1016/j.bpj.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim B, et al. Spatial rule-based modeling: a method and its application to the human mitotic kinetochore. Cells. 2013;2:506–544. doi: 10.3390/cells2030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tapia J-J, et al. MCell-R: a particle-resolution network-free spatial modeling framework. Methods Mol. Biol. 2019;1945:203–229. doi: 10.1007/978-1-4939-9102-0_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolle DP, Le Novère N. Meredys, a multi-compartment reaction–diffusion simulator using multistate realistic molecular complexes. BMC Syst. Biol. 2010;4:24. doi: 10.1186/1752-0509-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nehme E, Weiss L, Michaeli T, Shechtman Y. Deep-STORM: super-resolution single-molecule microscopy by deep learning. Optica. 2018;18:2334–2536. [Google Scholar]
- 20.Mandracchia B, et al. Fast and accurate sCMOS noise correction for fluorescence microscopy. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-019-13841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foi A, Trimeche M, Katkovnik V, Egiazarian K. Practical Poissonian–Gaussian noise modeling and fitting for single-image raw-data. IEEE Trans. Image Process. 2007;17:1–18. doi: 10.1109/TIP.2008.2001399. [DOI] [PubMed] [Google Scholar]
- 22.Südhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171:745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ries J, Kaplan C, Platonova E, Eghlidi H, Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 24.Chamma I, et al. Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin. Nat. Commun. 2016;7:10773. doi: 10.1038/ncomms10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka H, et al. Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin. Cell Rep. 2012;2:101–110. doi: 10.1016/j.celrep.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Comoletti D, et al. Characterization of the Interaction of a recombinant soluble neuroligin-1 with neurexin-1β. J. Biol. Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- 27.Saint-Michel E, Giannone G, Choquet D, Thoumine O. Neurexin/neuroligin interaction kinetics characterized by counting single cell-surface attached quantum dots. Biophys. J. 2009;97:480–489. doi: 10.1016/j.bpj.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1040. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang W, Aristov A, Lelek M, Hao X, Zimmer C. Deep learning massively accelerates super-resolution localization microscopy. Nat. Biotechnol. 2018;36:460–468. doi: 10.1038/nbt.4106. [DOI] [PubMed] [Google Scholar]
- 30.Weigert M, et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods. 2018;15:1090–1097. doi: 10.1038/s41592-018-0216-7. [DOI] [PubMed] [Google Scholar]
- 31.Kechkar A, Nair D, Heilemann M, Choquet D, Sibarita J-B. Real-time analysis and visualization for single-molecule based super-resolution microscopy. PLoS ONE. 2013;8:e62918. doi: 10.1371/journal.pone.0062918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 33.Haustein E, Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu. Rev. Biophys. Biomol. Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- 34.Rossier O, et al. Integrins β 1 and β 3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat. Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- 35.Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 2001;155:1319–1332. doi: 10.1083/jcb.200107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J. Biol. Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia M, et al. Two-tiered coupling between flowing actin and immobilized N-cadherin/catenin complexes in neuronal growth cones. Proc. Natl. Acad. Sci. 2015;112:6997–7002. doi: 10.1073/pnas.1423455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chazeau A, et al. Mechanical coupling between transsynaptic N-cadherin adhesions and actin flow stabilizes dendritic spines. Mol. Biol. Cell. 2015;26:859–873. doi: 10.1091/mbc.E14-06-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair D, et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J. Neurosci. 2013;33:13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renner M, Wang L, Levi S, Hennekinne L, Triller A. A simple and powerful analysis of lateral subdiffusion using single particle tracking. Biophys. J. 2017;113:2452–2463. doi: 10.1016/j.bpj.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenworthy AK, et al. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenne PF, et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heinemann F, Vogel SK, Schwille P. Lateral membrane diffusion modulated by a minimal actin cortex. Biophys. J. 2013;104:1465–1475. doi: 10.1016/j.bpj.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heine M, et al. Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science. 2008;320:201–205. doi: 10.1126/science.1152089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Czöndör K, et al. Unified quantitative model of AMPA receptor trafficking at synapses. Proc. Natl. Acad. Sci. USA. 2012;109:3522–3527. doi: 10.1073/pnas.1109818109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sainlos M, et al. Biomimetic divalent ligands for the acute disruption of synaptic AMPAR stabilization. Nat. Chem. Biol. 2011;7:81–91. doi: 10.1038/nchembio.498. [DOI] [PubMed] [Google Scholar]
- 47.Letellier M, Lagardère M, Tessier B, Janovjak H, Thoumine O. Optogenetic control of excitatory post-synaptic differentiation through neuroligin-1 tyrosine phosphorylation. Elife. 2020;9:e52027. doi: 10.7554/eLife.52027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beheiry ME, Dahan M, Masson JB. InferenceMAP: mapping of single-molecule dynamics with Bayesian inference. Nat. Methods. 2015;12:594–595. doi: 10.1038/nmeth.3441. [DOI] [PubMed] [Google Scholar]
- 49.Levet F, et al. SR-Tesseler: a method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods. 2015;12:1065–1071. doi: 10.1038/nmeth.3579. [DOI] [PubMed] [Google Scholar]
- 50.Briane V, Salomon A, Vimond M, Kervrann C. A computational approach for detecting micro-domains and confinement domains in cells: a simulation study. Phys. Biol. 2020;17:ab5e1d. doi: 10.1088/1478-3975/ab5e1d. [DOI] [PubMed] [Google Scholar]
- 51.Granik N, et al. Single-particle diffusion characterization by deep learning. Biophys. J. 2019;117:185–192. doi: 10.1016/j.bpj.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rothbauer U, et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods. 2006;3:887–889. doi: 10.1038/nmeth953. [DOI] [PubMed] [Google Scholar]
- 53.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vafabakhsh R, Levitz J, Isacoff EY. Conformational dynamics of a class C G-protein-coupled receptor. Nature. 2015;524:497–501. doi: 10.1038/nature14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izeddin I, et al. Wavelet analysis for single molecule localization microscopy. Opt. Exp. 2012;20:2081–2095. doi: 10.1364/OE.20.002081. [DOI] [PubMed] [Google Scholar]
- 56.Heilemann M, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. The software FluoSim (v1.0) is released under a GNU GPL v3 license as supplementary material accompanying this manuscript, and as an archive that can be downloaded at https://www.iins.u-bordeaux.fr/SOFTWARE. The FluoSim (v1.0) release and its source code are available at https://github.com/mlagardere/FluoSim under the same license.