Abstract
Sialylation is an important carbohydrate modification of glycoconjugates that has been shown to modulate many cellular/molecular interactions in vertebrates. In Drosophila melanogaster (Dm), using sequence homology, several enzymes of the sialylation pathway have been cloned and their function tested in expression systems. Here we investigated whether sialic acid incorporation in cultured Dm central nervous system (CNS) neurons required endogenously expressed Dm sialic acid synthase (DmSAS). We compared neurons derived from wild type Dm larvae with those containing a DmSAS mutation (148bp deletion). The ability of these cells to produce Sia5NAz (sialic acid form) from Ac4ManNAz (azide-derivatized N-acetylmannosamine) and incorporate it into their glycoconjugates was measured by tagging the azide group of Sia5NAz with fluorescent agents via Click-iT chemistry. We found that most of the wild type Dm CNS neurons incorporated Sia5NAz into their glycoconjugates. Sialic acid incorporation was higher at the soma than at the neurite and could also be detected at perinuclear regions and the plasma membrane. In contrast, neurons from the DmSAS mutant did not incorporate Sia5NAz unless DmSAS was reintroduced (rescue mutant). Most of the neurons expressed α2,6-sialyltransferase. These results confirm that the mutation was a null mutation and that no redundant sialic acid biosynthetic activity exists in Dm cells, i.e., there is only one DmSAS. They also provide the strongest proof to date that DmSAS is a key enzyme in the biosynthesis of sialic acids in Dm CNS neurons, and the observed subcellular distribution of the newly synthesized sialic acids offers insights into their biological function.
Keywords: sialic acid synthase, Drosophila melanogaster, peracetylated-N-azido-mannosamine, peracetylated-N-acetyl-mannosamine, sialic acids, Click-iT chemistry, neurons
INTRODUCTION
The incorporation of sialic acids into macromolecules modulates numerous cellular and molecular interactions, including many that are involved in nervous system development and function(1). It was recently shown that Drosophila melanogaster (Dm) have N-linked glycans containing sialic acids(2, 3). Based on sequence homology with their counterparts in vertebrates, various enzymes in the sialylation pathway have been cloned from Dm. The sialic acid synthase (DmSAS), was found to generate phosphorylated forms of sialic acids when expressed in bacteria and in insect Sf9 cells (derived from Spodoptera frugiperda)(4). The CMP-sialic acid synthase (DmCSAS) was found to generate CMP-sialic acids when expressed in mammalian cells(5). The α2–6-sialyltransferase (DmSialT), however, is the only Dm sialylation pathway enzyme shown to be functional in live Dm S2 cells(6). These studies indicate that the sialylation pathway of Dm appears to be intact and similar to that of vertebrates(7). Recently it was shown that a mutation in the DmSialT, that resulted in the loss of DmSialT activity in vitro, affected various Dm functions in vivo, indicating that sialylation is important for normal function in Dm(8). Similar findings have been observed with a mutation in the DmSAS (Akan and Palter, unpublished). These reports indicate that the function of these various enzymes is important in Dm. Moreover, based on their characterization in expression systems these enzymes are likely involved in the sialylation pathway.
The investigation of sialylation in insect cells has been limited by a lack of facile labeling tools. Lectins, for example, have not proved adequate for the detection of the low levels of sialic acids found in Drosophila due to the generally low binding affinity and poor specificity of these probes. This project exploits bio-orthogonal labeling techniques developed by the metabolic glycoengineering field(9) to stably label newly-synthesized sialo-glycoconjugates with covalently-attached fluorophores. By using this approach with azido-modified N-acetymannosamine (ManNAc) analogs that serve as selective metabolic precursors for sialic acid biosynthesis(10), we used a commercial adaptation of “Click-iT chemistry” to label and detect sialic acids in cultured CNS neurons. We found that CNS neurons isolated from the wild type Dm, but not those isolated from the DmSAS mutant, incorporate sialic acids. The results support the view that endogenous DmSAS is functional in Dm neurons, and that its function is essential for sialic acid incorporation.
RESULTS AND DISCUSSION
Neuronal cultures
The CNS cultures from Dm third instar larvae contained the previously described cell types(11) (Supplemental Figure S1). To assess the proportion of neurons in these cultures, cells were stained with anti-horseradish peroxidase (anti-HRP) a neuronal marker in Drosophila(12). Most of the cells were HRP positive and displayed neurites (Figure 1). There was a small population of HRP negative cells that that did not have neurites and could be found individually and within clumps (Figure 1D).
Figure 1. Dissociated CNS cells displayed neuritic outgrowth and were horseradish peroxidase-(HRP)-positive.
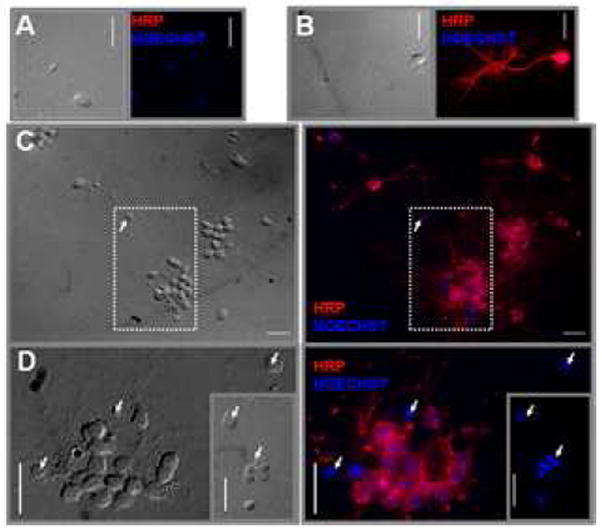
Dissociated central nervous system (CNS) cultures (2CNS/corverslip) were incubated for 1 day, immunostained with anti-HRP, and the nuclei stained with Hoechst. Left panels show the corresponding light picture. (A) Negative control for anti-HRP (exposed only to the secondary antibody: Alexa-Fluor-546-Goat-Anti-Rabbit). (B-D) Positive labeling with anti-HRP. The main panel in (D) corresponds to the inset area in (C) that was rotated by 90°. The inset in (D) corresponds to another field that shows a group of HRP-negative cells. The arrows indicate some of the HRP-negative cells. (63x objective, bar=10μm).
DmSAS and sialic acid incorporation
To assess whether Dm CNS neurons incorporate sialic acids into glycoconjugates, cultures were incubated with 30-50 μM of the sialic acid precursor peracetylated- N-azido-mannosamine (Ac4ManNAz or NAZ). The cellular biosynthetic utilization of NAZ was detected using the Click-iT assay to label Sia5NAz (a sialic acid form) that had been incorporated into cellular glycoconjugates, as described under methods and schematically represented in Figure 2A-B. As controls, cultures were incubated with 30-50 μM of the non-azide, natural sialic acid precursor peracetylated-N-acetyl-mannosamine (Ac4ManNAc or NAC). Figure 2 shows pictures of neurons from the wild type (w1118) (Figure 2C-D) and from the DmSAS mutant (2d/2d) (Figure 2E) stained with anti-HRP and labeled for Sia5NAz incorporation. Only the somas displaying fluorescence levels that were above that displayed by the somas in the NAC group were considered to have detectable levels of Sia5NAz. It was found that an average of 22% of the w1118 neurons displayed detectable levels of Sia5NAz incorporation; while none of the neurons from the DmSAS mutant (2d/2d) displayed detectable levels of Sia5NAz incorporation (Figure 2F). Moreover, reintroduction of DmSAS to 2d/2d, that is the 4-4 rescue mutant, allowed Sia5NAz incorporation of an average of 31% of the neurons (Figure 2F). The DmSAS gene was reintroduced using a heat shock promoter, hence it is expressed in all cells. These results indicate that the DmSAS mutation was responsible for blocking incorporation of sialic acids.
Figure 2. Cultured CNS neurons from w1118 and 4-4, but not those from 2d/2d, incorporate sialic acids.
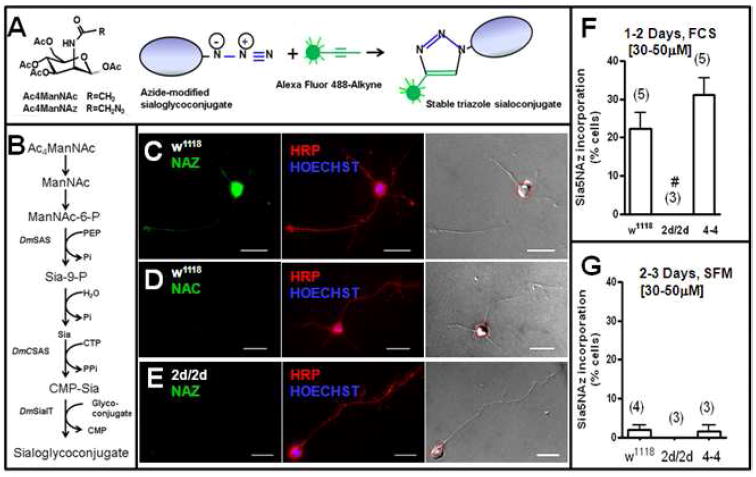
(A) Molecular structure of Ac4ManNAc (NAC) and Ac4ManNAz (NAZ); and a schematic representation of the Click-iT reaction between the azide-modified sialoglycoconjugates and alkyne group on the fluorescent agent. (B) General pathway for sialic acid incorporation into glycoconjugates in Dm. Once in the cytoplasm non-specific esterases remove the acetyl groups in NAC and NAZ converting them into ManNAc and ManNAz. ManNAc (or ManNAz) is processed to become CMP-Sia (cytidine monophosphate-sialic acid) (or CMP-Sia5NAz) which is the sialic acid donor molecule. DmSAS: Dm sialic acid synthetase; DmCSAS: Dm CMP sialic acid synthetase; DmSialT: Dm sialyltransferase; PEP: phosphoenolpyruvate; Sia-9-P: sialic acid 9-phosphate; Pi: phosphate; CTP: cytidine triphosphate; PPi: pyrophosphate. (C-G) Dissociated CNS cultures (1-3CNS/coverslip) were cultured in either serum containing medium (C-F) or in SFM (G) for one to three days in the presence of NAZ or NAC (30-50μM). (C-E) Show pictures of neurons that were immunostained with anti-HRP and the nuclei stained with Hoechst (middle panels). Staining with the Alexa-Fluor-488-Alkyne (left panels). The red dotted lines outline the soma (light pictures, right panels). Incorporation of Sia5NAz (fluorescent intensity labeling) was measured in individual neuronal somas. (63x objective, bar=10μm). (F) The mean percentage of neurons that incorporated Sia5NAz when cultured in serum containing medium; n=3-5 separate experiments. One-way analysis of variance (P<0.001); post test Bonferroni, #P<0.05 between the 2d/2d group and the w1118 group. No difference was found between the 4-4 group and the w1118 group. Sia5NAz incorporation was measured in 25-100 neurons per condition in one experiment; and in 102-150 neurons per condition in four experiments. (G) The mean percentage of neurons that incorporated Sia5NAz when cultured in SFM; n=3-4 separate experiments. No significant difference was found between any of the groups (One-way analysis of variance). Sia5NAz incorporation was measured in 103-146 neurons per condition in the four experiments.
Serum helps sialic acid incorporation
The number of neurons labeled for Sia5NAz incorporation was highly reduced when the cells were cultured in serum-free medium (SFM) (Figure 2G). On average only ~2% of the w1118 and 4-4 neurons cultured in SFM in the presence of 30-50 μM NAZ, displayed detectable levels of Sia5NAz incorporation (Figure 2G). This percentage was significantly lower compared to that observed when neurons were cultured with the same level of NAZ in serum-containing medium (Figure 2F). By heat inactivating the serum, we sought to determine if the serum contained an active protein that assisted in either NAZ uptake or processing. We found that cultures containing heat-inactivated serum and either 30μM (not shown) or 250μM of NAZ showed no reduction in Sia5NAz incorporation as compared to cultures containing non-heat-inactivated serum (e.g. Figure 3). We have not yet identified the serum component(s) that facilitate the incorporation of sialic acids. However, this effect does not appear to involve an active protein since it was observed even when CNS neurons were cultured in medium containing heat-inactivated serum. Although we have not rigorously ruled out the possibility, the serum effect also does not appear to involve attachment factors since the attachment of CNS neurons appear to be comparable in both SFM and serum-containing media. Under SFM, however, increasing the levels of NAZ from 30-50 μM to 250 μM increased the percentage neurons incorporating Sia5NAz from 2% to ~31% in 2-3 days cultures. Hence, the presence of serum is not required, but it facilitates the incorporation of Sia5NAz.
Figure 3. Incorporation of Sia5NAz into the soma and neurites of CNS cells.
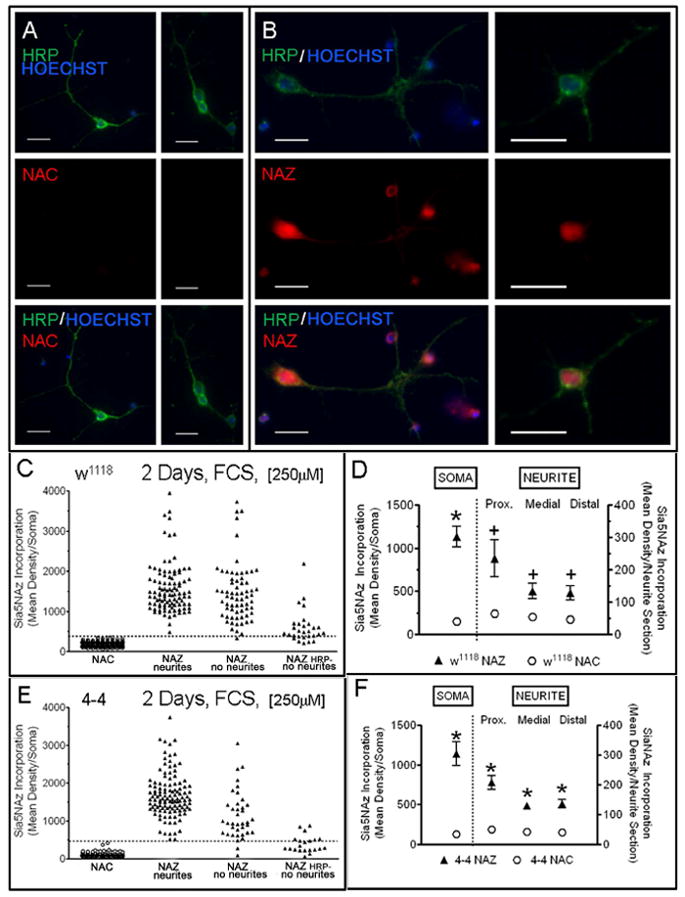
Cells were cultured for 2 days in medium containing heat-inactivated serum and 250μM of NAC or NAZ (5CNS/coverslip). Dissociated neurons from w1118 (A, B) and 4-4 (not shown), immunostained with anti-HRP; and stained with the Alexa-Fluor 546 Alkyne, and Hoechst for nuclei. (63x objective, bar=10μm). (C, E) Show fluorescent intensity labeling for all the cells. For the NAZ group the cells were separated into 3 groups: cells with neurites and that were HRP-positive (“with neurites”); cells that have no neurites and were HRP-positive (“no neurites”); and cells that have no neurites and were HRP-negative (“no neurites HRP-”). The total number of cells for w1118 were 226(NAC), 101(NAZ, “with neurites”), 67(NAZ, “no neurites”) and 29(NAZ, “no neurites HRP-”). For 4-4 were 279(NAC), 118(NAZ, “with neurites”), 39(NAZ, “no neurites”) and 21(NAZ, “no neurites HRP-”). (D, F) Show fluorescent intensity labeling in the somata and their corresponding neurites. Fluorescent intensity labeling in the neurites was measured in 20 randomly selected neurons in the NAZ and NAC groups from w1118 (D) and 4-4 (F) cultures. We selected neurons that had neurites that were longer than 5 times their diameter and in which we could identify the location of the neurite terminal. If the neuron had more than one neurite we selected the predominant one. Measurements were done in three regions (10μm in length) Proximal (Prox.): close to the soma; Distal: close to the neurite terminal but prior to any terminal branches. Unpaired t-test, two-tailed; *P<0.0001;+ P<0.01, group with respect to the NAC group.
Sialic acid incorporation in neurites
To determine if neurites incorporated sialic acids, cells were incubated with a high level of NAZ (250 μM) in the presence of serum. Neurites were visualized through HRP labeling (Figure 3A-B). Under these conditions all of the w1118 (Figure 3A-D) and 4-4 (Figure 3E-F) neurons (cells with neurites) incorporated Sia5NAz in the soma and, to a lower extent, in their neurites. Within the neurites, the level of Sia5NAz incorporation was higher in the Proximal than in the Medial and Distal regions (Figure 3D,F). Many of the cells without neurites (round cells) were HRP-positive and also incorporated Sia5NAz (Figure 3A). Interestingly, some of the round HRP-negative cells also incorporated Sia5NAz, but to a lower extent than the HRP-positive cells (Figure 3C,E). Under the same conditions, none of the cells in 2d/2d cultures displayed Sia5NAz incorporation (not shown). Since neuronal precursors have been shown to be HRP-positive(13), then some of the round cells could be neuronal precursors or neurons that failed to extend neurite outgrowth.
Giant neurons incorporate sialic acids
Giant neurons are part of the giant fiber system that in many invertebrates mediate the “escape” or “startle” response(14). Transcripts of SialT have been reported in a subset of CNS neurons in third instar larvae, particularly where giant neurons are located(7). However, expression of SialT was shown to be present in a larger number of neurons, but not in glia, throughout the third-instar larval CNS(8). Expression of SialT was highest in the region where giant neurons are located(8). Hence we decided to also measure sialic acid incorporation in the giant neurons. We first used Dm in which their giant neurons were labeled with GFP (membrane-targeted green fluorescent protein) to determine whether we could identify giant neurons in the dissociated CNS cultures based solely on their morphology/size. We confirmed that GFP labeling was detected within the CNS region where giant neurons are located (Figure 4A) and found that the giant neurons could easily be identified from the rest of the cultured cells based solely on their size (diameter ≥20 μm) (Figure 4B). Hence we identified giant neurons in CNS cultures of our experimental flies based solely on their size. Most of the giant neurons did not survive in culture. Two days after dissociation, some of them attached and their diameter were more than double that of non-attached giant neurons (Figure 4C-F). Although the culture conditions are not optimal for giant neurons, (indicated by the low level of HRP-staining and their short survival time of 1-2 days), the giant neurons incorporated Sia5NAz to a higher extent than the smaller CNS Neurons (Figure 4G). This finding is consistent with previous observations where SialT expression was reported to be highest in third instar larvae brain regions where giant neurons are located(7, 8)
Figure 4. Cultured giant neurons incorporate sialic acids.
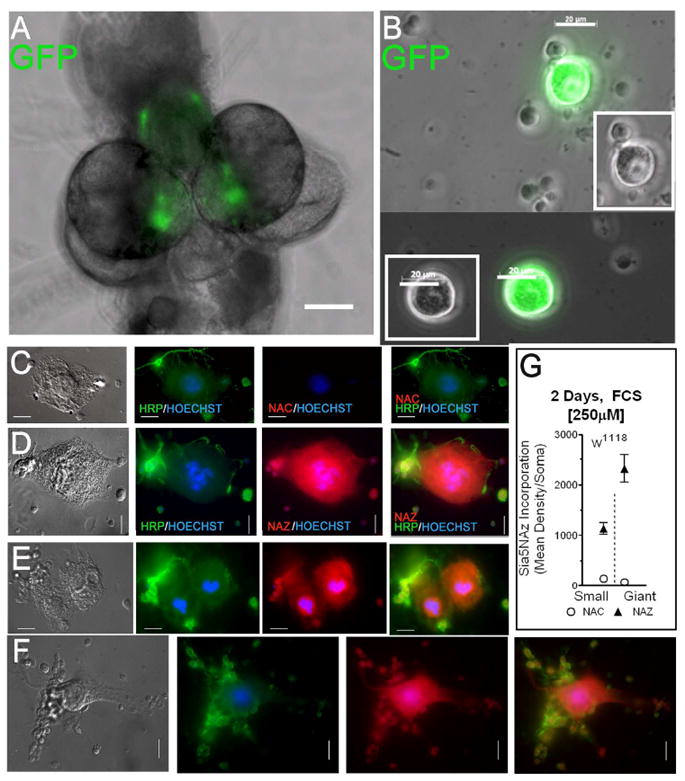
(A,B) Identification of giant neurons in third instar larvae in whole CNS (10x objective, bar=100μm) (A) and dissociated CNS (40x objective, bar=20μm) (B). (A,B) The giant neurons were labeled with membrane-targeted green fluorescent protein (GFP) by using UAS-mc8-GFP driven by OK307-Gal4. (C-F) Show pictures of giant and smaller CNS neurons derived from w1118 Dm. Cells (5 CNS/coverslip) were cultured for 2 days in medium containing heat-inactivated serum and 250μM of either NAC (C) or NAZ (D-F). Cells were immunostained with anti-HRP; and stained with the Alexa-Fluor-546-Alkyne and Hoechst for nuclei. (63x objective, bar=10μm). (G) Shows the quantification of Sia5NAz incorporation in giant neurons (n=7) as compared to that of close by smaller neurons (n=34) (mean ± sem).
Sialic acids in the plasma membrane
Visual inspection shows, as expected, strong HRP-labeling at the plasma membrane, whereas Sia5NAz labeling occurs throughout the cell (Figure 3B). In some neurons, Sia5NAz labeling appears to be highest at the perinuclear region (Figure 3B). To confirm that sialo-glycoconjugates are also located at the plasma membrane we carried out Sia5NAz labeling in live cells. In live cells, labeling of Sia5NAz was limited to the cell surface and displayed high co-localization with HRP labeling (Figure 5A). Detectable levels of Sia5NAz incorporation in the plasma membrane were observed in an average of 30% of the w1118 neurons, in none of the DmSAS mutant (2d/2d) neurons, and in 33% in the 4-4 rescue mutant neurons (Figure 5C). These results indicate that sialo- glycoconjugates are present in the plasma membrane of Dm CNS neurons.
Figure 5. Cultured CNS neurons from w1118 and 4-4, but not those from 2d/2d, incorporate sialo-glycoconjugates in the plasma membrane that were sensitive to sialidase treatment.
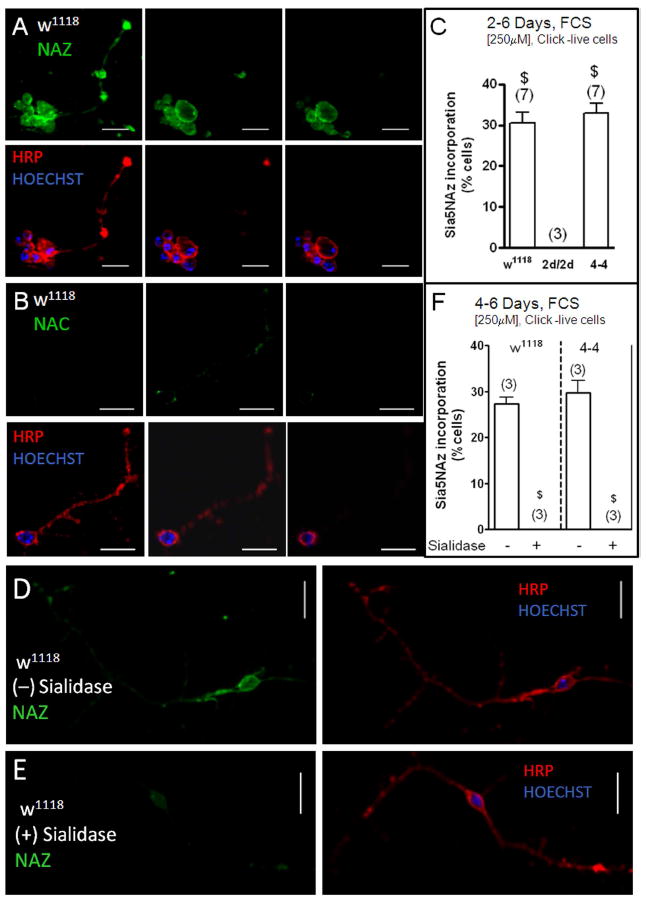
Cells were cultured for 2-6 days in serum-containing medium and 250μM NAZ (or NAC) (1-3 CNS/coverslip). The Click-iT reaction with Alexa-Fluor-488-Alkyne was carried out in live cells; while the immunostaining with anti-HRP and nuclear labeling with Hoechst were done following fixation. (A-B) Pictures of neurons taken using the Apotome Zeiss system, (63x objective, bar=10μm), panels going from left to right show pictures of 1μm thickness taken from the bottom towards the top. (C) Mean percentage of neurons that incorporated Sia5NAz in the plasma membrane. Sia5NAz incorporation was measured in 62-121 neurons per condition in each experiment. (n=# experiments). One-way analysis of variance (P<0.0001); post test Bonferroni, $P<0.001 between the group and the 2d/2d group. No significance difference was found between the w1118 and the 4-4 groups. (D-E) Pictures taken with the Apotome Zeiss system, (1μm thickness, 63x objective, bar=10μm) of untreated and sialidase treated cells. (F) Mean percentage of w1118 and 4-4 neurons that incorporated Sia5NAz in the plasma membrane in untreated and sialidase treated cultures. Sia5NAz incorporation was measured in 78-121 neurons per condition in each experiment. n=3 separate experiments. One-way analysis of variance (P<0.0001); post test Bonferroni, $P<0.001 between the sialidase treated and untreated group. No significance difference was found between the untreated w1118 and the 4- 4 groups.
Sialidase treatment
To confirm that NAZ was converted to Sia5NAz and eventually incorporated into glycoconjugates, we treated live CNS neurons with sialidase prior to carrying out Sia5NAz labeling. Figure 5 shows images of w1118 CNS neurons that were cultured with NAZ for 6 days and then either not treated (Figure 5D) or sialidase treated (Figure 5E). Sialidase treatment completely eliminated the Sia5NAz labeling in w1118 and 4-4 CNS neurons (Figure 5F). These results support the view that the labeling observed in NAZ-treated cells represents the actual incorporation of Sia5NAz into sialylated glycoconjugates.
DmSialT expression in cultured neurons
In western blots the anti-DmSialT detected a strong band at the molecular weight expected for DmSialT only in HEK293 (Human Embryonic Kidney) cells that were transfected with the DmSialT (Figure 6A). Moreover, immunostaining with anti-DmSialT gave a positive signal only in HEK293 cells that were transfected with the DmSialT (Figure 6B). For immunostaining of CNS neurons we used three negative controls, in which the primary antibody was replaced with nothing, pre-immune serum, or purified rabbit IgG. The background level was comparable for the three negative controls (Figure 6C-G). Although the level of expression differed, essentially all of the neurons from w1118 (Figure 6H-J) and 2d/2d (Figure 6K-M) expressed SialT. These data also indicate that the expression of SialT was not modified in the 2d/2d mutant.
Figure 6. Most of the CNS neurons express SialT.
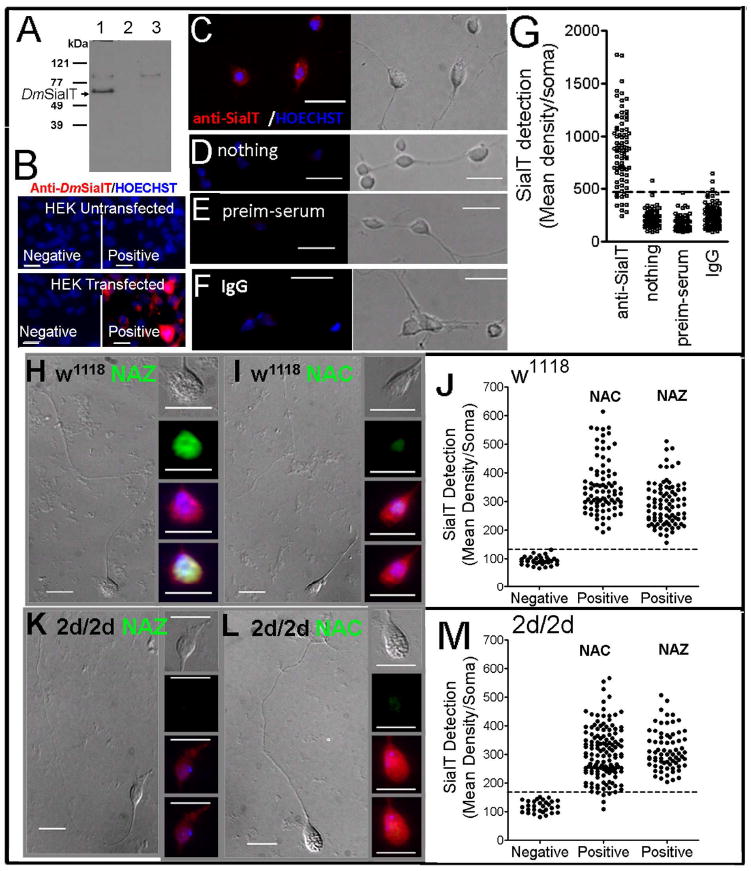
(A) Western blot for the DmSialT (Dm α2–6-sialyltransferase). The arrow indicates the location of DmSialT. The locations of the molecular weights ladder are indicated. Lane 1: HEK293 (Human Embryonic Kidney) cells transfected with DmSialT; Lane 2: loading buffer; lane 3: untransfected HEK293 cells. For each lane: 50 μg protein was mixed with 5XSDS loading buffer, heated up to 80°C for 20 minutes, then loaded onto 8%(w/v) SDS-PAGE. The blot was probed overnight at 4°C with anti-DmSialT (1:1000 dilution). (B) Untransfected (top) and transfected (bottom) HEK293 cells with DmSialT. Cells were immunostained with anti-DmSialT (1:10,000 dilution) and the nuclei stained with Hoechst (63x objective, bar=20μm). (A,B) HEK293 cells were transfected with 1 μg pCDNA3.1 DmSialT plasmid by using lipofactamine (Invitrogen). (C-G) Dissociated DmCNS cells immunostaining with anti-DmSialT. Cells were cultured for one day in serum containing medium (2-3 CNS/coverslip). (C) Cells were first incubated with the anti-DmSialT (1:10,000 dilution, 0.1μg ml-1) and then with the secondary antibody Alexa-Fluor-546-Goat-Anti-Rabbit. (D-F) Three negative controls were done, in which the primary antibody was replaced by either (D) nothing; (E) preimmune serum (0.05 μl preimmune serum per ml~ 0.1μg IgG ml-1); or (F) with purify rabbit IgG (0.1μg ml-1). The secondary antibody Alexa-Fluor-546-Goat-Anti-Rabbit was added in all cases. The nuclei were stained with Hoechst. The corresponding light pictures are shown, (63x objective, bar=10μm). (G) Quantification for SialT intensity labeling was done by measuring the fluorescent intensity labeling per each soma (each dot represent a soma). The background level was given by the three negative controls (dashed line). Most of the neurons displayed positive labeling for SialT. Pictures of dissociated neurons from w1118 (H,I) and 2d/2d (K,L) were immunostained with anti-DmSialT; and stained with the Alexa-Fluor-488-Alkyne, and Hoechst for nuclei. (63x objective, bar=10μm). Cells were cultured for 7 days in SFM. Most of the neurons from w1118 (J)and from 2d/2d (M) were positive for anti-DmSialT.
Our data show that in culture a large proportion of the Dm CNS neurons express SialT and are capable of incorporating Sia5NAz, a somewhat surprising finding considering that previously SialT transcript was detected only in a small number of CNS neurons(6-8). However, another study found that the proportion of CNS neurons in the third instar larvae that express SialT is much larger although not 100%(8). One explanation for the nearly 100% of cells with neurites found to stain for SialT is that since insect cells release SialT(6), the released SialT was possibly taken up by neighboring CNS neurons. Another possibility is that the neuronal populations in vivo are not equally represented in culture. Supporting this view is the observation that after one day in culture most of the giant neurons and some other cells are lost; whereas neuroblast-like cells continuously undergo asymmetrical division, giving rise to new neurons. Another possibility is that the SialT expression in Dm CNS neurons is enhanced in culture as a result of environmental changes; as it has been shown in cultured mammalian cells(15). Finally, it is also possible that the proportion of neurons that incorporate sialic acids in vivo may actually be higher than previously described, and that in whole tissue only the fraction of cells expressing the highest enzymatic levels can be easily detected. Our data show that Giant neurons appear to incorporate significantly larger amounts of Sia5NAz than other smaller CNS neurons (Figure 4). This observation is consistent with previous findings that SialT is predominantly, but not solely, detected in third instar larvae CNS regions where the giant neurons are located(7, 8).
Implications
Our finding that DmSAS activity is required for sialic acid incorporation in CNS neurons suggests that the lack of sialylation in CNS neurons contributes to the observed behavioral changes of the DmSAS mutant (Akan and Palter, unpublished). Recently, it was reported that the DmSialT mutant also displays behavioral changes; this mutant has a lower number of muscle synaptic branches and boutons and displays smaller evoked excitatory junction potentials and sodium (Na+) currents with lower tetrodotoxin (TTX) sensitivity(8). The correlation between the loss of SialT activity of the mutated DmSialT in vitro and the functional changes in vivo strongly supports the notion that sialylation is important for normal function in Dm(8). Changes in TTX sensitivity have been shown to be due to changes in the amino acid sequence of Na+ channels(16); hence the change in TTX-sensitivity of Na+ currents in the SialT mutant could reflect changes in the expression or trafficking of different Na+ channels. In fact, the trafficking of Na+ channels(17) and of some K+ channels(18, 19) have been shown to be affected by glycosylation.
In vertebrates, sialylation has been shown to affect the function of some channels including Na+(20-22) and some K+(18, 23) channels. When expressed in mammalian cells, sialylation also affects the function of Dm shaker-K+ channels(24). In addition, sialylation of membrane lipids can also modify the function of Na+ channels(25). Hence, alterations of electrophysiological measurements in SialT mutant can also involve functional changes in both Na+ and K+ channels. Our demonstration that some of the sialo-glycoconjugates are located at the plasma membrane indicates that these molecules have the correct spatial localization to be Na+ and K+ channels and thus suggests that changes in sialylation in Dm neurons could affect their excitability. In summary, this is the first study that shows that an endogenously expressed Dm enzyme is functional in the sialylation pathway. We further found that DmSAS is functional in most, if not all, cultured CNS neurons from third instar larvae and that DmSAS is required for the incorporation of sialic acids in CNS neurons.
METHODS
Neuronal cultures
The third-instar larva is one of the stages in the developmental cycle of Dm. The eggs hatch and give rise to first-instar larvae. The larvae grow while molting twice into second- and third-instar larvae. Then the larvae encapsulate in the puparium and undergo metamorphosis after which the adult flies emerge(26). CNS neuronal cultures from third-instar larvae were prepared by modifying a previously reported protocol(11). Third-instar larvae were placed into a well (staining glass plate) containing ~300μl of 70%(v/v) ethanol for 2 minutes. Larvae were rinsed by transferring them consecutively through three wells containing phosphate buffered saline (PBS). While holding the body with a forceps placed at about 1/3 of the body length from the mouth, the mouth part was held with a second forceps and pulled away from the body. The mouth parts with the attached CNS (brain hemispheres and ventral nerve cord) were moved into a new well containing PBS, and the CNS was isolated under a dissecting microscope. Dissociation was done by placing the pooled CNS into a 1.5ml conical eppendorf tube containing 0.025%(w/v) of trypsin and incubating for 15 minutes at room temperature (RT). Afterwards, the tube was placed in a hybridization incubator rotating shaker (Robbins Scientific, model 1000) at full speed (20rpm) for 15 minutes at 37°C. Then, 500μl of the culture media with or without 10%(v/v) fetal calf serum (FCS) was added. The culture media consisted of Scheneider’s Drosophila Medium (Gibco) supplemented with 2%(v/v) B-27 Serum-Free Supplement (GIBCO), 50μg ml-1 insulin, 100μg ml-1 transferrin; penicillin (100U ml-1) and streptomycin (100μg ml-1) (Sigma). The tissue was dissociated by first triturating 40 times with a 1ml micropipette; and then 20 times with a fired polished glass Pasteur pipette. Clumps were allowed to settle down and the supernatant removed and collected in a 15ml conical tube. More culture media (~1 ml) was added to the clumps and the tissue was triturated again 20 times. The last two steps were repeated three to five times. The pooled supernatants were spun for 5 minutes in a tissue culture centrifuge (Beckman JS 4.2: 2510g). The pellet was resuspended by adding fresh medium (~20μl per two-five brains; ~7200cells/CNS). Then 20μl of cell suspension was placed in the middle of each Poly-D-lysine precoated (12μg ml-1 for 5 minutes) round coverslip (12mm diameter) that were placed in 4 well culture dishes. Cells were allowed to attach for 20 minutes while in the incubator (humidified air, 25°C). Following cell attachment, 800μl of culture medium was added to each coverslip. The medium was changed every other day.
Click-iT reaction
Sialo-glycoconjugates that have incorporated Sia5NAz can be detected by the “Click-iT” commercial adaptation (Invitrogen Molecular Probes; for fixed cells Catalog #C10269; for live cells Catalog #C10405) of fluorescent labeling strategies reported by Wong’s laboratory that exploit the Click-iT reaction between azide and alkyne groups(27). Control cells were grown in the absence of NAZ or in the presence of NAC(28). The NAC analog reproduces the increased flux through the sialic acid pathway supported by NAZ and is used to control for potential dose limiting toxicity of these analogs(28, 29).
NAC and NAZ were synthesized as previously described(30, 31) and the stock solutions of (25mM) were prepared in 95%(v/v) ethanol. The Click-iT reaction was carried out following the manufacture instructions. For fixed cells, the attached cells were washed once with PBS, then fixed with 4%(w/v) paraformaldehyde (pH=7.4; 15 minutes, RT). Coverslips were rinsed three times with PBS at RT. The cells were permeabilized by incubating with 0.5%(v/v) Triton X-100 in PBS (15 minutes, RT). Coverslips were rinsed three times with PBS at RT, and left in 1%(w/v) BSA (or 10%(v/v) goat serum when immunostaining will follow) for 1 hour. Under dark conditions the reaction-cocktail containing the Alexa-fluor-488-Alkyne or the Alexa-fluor-546-Alkyne (2.4μM) was added. The samples were rinsed three times with PBS. The nuclei were counter stained with Hoechst 33342 (20μg ml-1, 10 minutes, RT), and coverslips were mounted into slides using 1 drop of Aqua Poly/Mount (Polysciences, Inc.). In the initial experiments the cells were rinsed with PBS containing 1%(w/v) BSA.
Drosophila stocks
A deletion of DmSAS was created by imprecise excision of a P element; in the selected line (2d) the P element was deleted as well as 480 bp of flanking DNA including 148bp of a highly conserved coding region of DmSAS (Akan and Palter, unpublished). The 4-4 rescue line carries a cDNA for DmSAS on both second chromosomes (Akan and Palter, unpublished). The w1118 was used as the control fly strain as it is nearly isogenic with 2d/2d and is wild type for the sialic acid pathway enzymes. Third instar larvae heterozygous for the SAS deletion 2d/TM6Tb, produced a shortened body shape, tubby. Third instar larvae homozygous for the SAS deletion 2d/2d have normal body shape. To confirm the state of our fly stocks we used single larva PCR. We also used PCR (on the pooled bodies parts left after removing the CNS) to confirm that the larvae from which the CNS tissue was isolated from were all homozygous for the DmSAS mutation. PCR was done with the Extract-N-Amp™ Tissue PCR Kit (Sigma) following the manufacturer’s instructions. DNA was extracted by first adding 10μL of Extraction Solution and then 2.5μL of Tissue Preparation Solution per fly. The mixture was incubated for 10 minutes at RT and then for 3 minutes at 95°C. Finally 10μL of Neutralization Solution B was added per fly and vortexed. A quick spin was done to pull the debris down. We used 4μl of extract for the PCR reaction.
Drosophila melanogaster stocks UAS-mc8-GFP(3rd) and OK307-Gal4 were kindly provided by Dr. Greg Suh (NYU Langone Medical Center). Labeling (with membrane-targeted green fluorescent protein (GFP)) of giant neurons was done by using UAS-mc8-GFP(32, 33) driven by OK307-Gal4(34, 35).
Immunocytochemistry
Cells attached to coverslips were rinsed 1 time with PBS, fixed (4%(w/v) paraformaldehyde, pH 7.4; 14 minutes), and rinsed 3 times with PBS at RT. Cells were permeabilized (0.5%(v/v) Triton X-100 in PBS, 15 minutes), rinsed 3 times with PBS, and incubated with a blocking solution (10%(v/v) Goat serum in PBS, 1hour, RT). Then coverslips were incubated overnight (4°C) with the primary antibody, rabbit anti sialyltransferase (anti-DmSialT; 1:10,000; final 0.10μg ml-1) or rabbit anti horseradish peroxidase (anti-HRP; 1:20,000 Sigma-Aldrich: #P7899) in 2%(v/v) goat serum. After incubation with the primary antibody, coverslips were rinsed 3 times with PBS. Then coverslips were incubated (90 minutes, RT) with the secondary antibody Alexa-Fluor-546-Goat-Anti-Rabbit or Alexa-Fluor-488-Goat-Anti-Rabbit (1:1,500), and rinsed 3 times with PBS at RT. Anti-DmSialT was raised using the peptide sequence: CRFNHAPTQGHEVDVGSK-NH2 (243-260aa). The peptide was conjugated to KLH and BSA and was produced by CPC Scientific, Inc. The Rabbit IgG was from Jackson InmunoResearch. When the Click-iT reaction was combined with immunocytochemistry the Click-iT reaction was done first.
Sialidase treatment
The protocol previously used for cultured live mammalian cells(36) was adapted for cultured live drosophila CNS neurons. Cells were cultured in the presence of NAZ (250μM) for 2- 6 days in serum-containing medium. The serum-containing medium was removed, and the cells were washed once for 5 minutes with Scheneider’s media. Then the Scheneider’s media was replaced with fresh Scheneider’s media with or without sialidase (0.005U ml-1; neuraminidase from Clostridium perfringens C. welchii; Sigma) and incubated for 15 minutes at RT. The enzyme was removed by rinsing 3 times with PBS containing 0.1%FCS(v/v) (5 minutes/rinse, RT). Following the sialidase treatment the Click-iT reaction was carried out in live cells.
Image acquisition
Images were captured (and analyzed) with a Zeiss Axiovert 200 (Germany) inverted microscope equipped with fluorescence and Nomarski optics, ApoTome (for optical sections), using an AxioCam camera and AxioVision Software version 4.6.3 (Zeiss). The filter set for detecting Alexa 488(green): Excitation filter BP450-490; Beam Splitter FT 510; Emission BP 515-565. The filter set for detecting Alexa 546(red): Excitation filter BP546/12; Bean Splitter FT 580; Emission filter LP 590. The filter set for detecting Hoechst(blue): Excitation filter G365; Bean Splitter FT 395 and Emission filter LP 420 or BP445/50. Images were taken from 20-30 random fields per coverslip. In each experiment they were two coverslips per treatment. The neuronal soma was outlined by hand.
Supplementary Material
Acknowledgments
This research was supported by the NSF Project Grant EFRI-073600 (M. Betenbaugh., K. Palter, E. Recio-Pinto) and the NIH/NIBIB Grant (EB005692-04) (K. Yarema).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 2.Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic developmental elaboration of N-linked glycan complexity in the Drosophila melanogaster embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- 3.Koles K, Lim JM, Aoki K, Porterfield M, Tiemeyer M, Wells L, Panin V. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology. 2007;17:1388–1403. doi: 10.1093/glycob/cwm097. [DOI] [PubMed] [Google Scholar]
- 4.Kim K, Lawrence SM, Park J, Pitts L, Vann WF, Betenbaugh MJ, Palter KB. Expression of a functional Drosophila melanogaster N-acetylneuraminic acid (Neu5Ac) phosphate synthase gene: evidence for endogenous sialic acid biosynthetic ability in insects. Glycobiology. 2002;12:73–83. doi: 10.1093/glycob/12.2.73. [DOI] [PubMed] [Google Scholar]
- 5.Viswanathan K, Tomiya N, Park J, Singh S, Lee YC, Palter K, Betenbaugh MJ. Expression of a functional Drosophila melanogaster CMP-sialic acid synthetase. Differential localization of the Drosophila and human enzymes. J Biol Chem. 2006;281:15929–15940. doi: 10.1074/jbc.M512186200. [DOI] [PubMed] [Google Scholar]
- 6.Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. J Biol Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 7.Koles K, Repnikova E, Pavlova G, Korochkin LI, Panin VM. Sialylation in protostomes: a perspective from Drosophila genetics and biochemistry. Glycoconj J. 2009;26:313–324. doi: 10.1007/s10719-008-9154-4. [DOI] [PubMed] [Google Scholar]
- 8.Repnikova E, Koles K, Nakamura M, Pitts J, Li H, Ambavane A, Zoran MJ, Panin VM. Sialyltransferase regulates nervous system function in Drosophila. J Neurosci. 2010;30:6466–6476. doi: 10.1523/JNEUROSCI.5253-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VD, Yarema KJ. Metabolic glycoengineering: Sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 11.Wu CF, Suzuki N, Poo MM. Dissociated neurons from normal and mutant Drosophila larval central nervous system in cell culture. J Neurosci. 1983;3:1888–1899. doi: 10.1523/JNEUROSCI.03-09-01888.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jan LY, Jan YN. Antibodies to horseradish peroxidase as specific neuronal markers in Drosophila and in grasshopper embryos. Proc Natl Acad Sci U S A. 1982;79:2700–2704. doi: 10.1073/pnas.79.8.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salvaterra PM, Bournias-Vardiabasis N, Nair T, Hou G, Lieu C. In vitro neuronal differentiation of Drosophila embryo cells. J Neurosci. 1987;7:10–22. doi: 10.1523/JNEUROSCI.07-01-00010.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Augustin H, Allen MJ, Partridge L. Electrophysiological recordings from the giant fiber pathway of D. melanogaster. J Vis Exp. doi: 10.3791/2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolinska J, Ivanov S, Chelibonova-Lorer H. Effect of hydrocortisone on sialyltransferase activity in the rat small intestine during maturation. Changes along the villuscrypt axis and in fetal organ culture. FEBS Lett. 1988;242:57–60. doi: 10.1016/0014-5793(88)80984-0. [DOI] [PubMed] [Google Scholar]
- 16.Fozzard HA, Lipkind GM. The tetrodotoxin binding site is within the outer vestibule of the sodium channel. Mar Drugs. 8:219–234. doi: 10.3390/md8020219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt JW, Catterall WA. Palmitylation, sulfation, and glycosylation of the alpha subunit of the sodium channel. Role of post-translational modifications in channel assembly. J Biol Chem. 1987;262:13713–13723. [PubMed] [Google Scholar]
- 18.Watanabe I, Zhu J, Sutachan JJ, Gottschalk A, Recio-Pinto E, Thornhill WB. The glycosylation state of Kv1.2 potassium channels affects trafficking, gating, and simulated action potentials. Brain Res. 2007;1144:1–18. doi: 10.1016/j.brainres.2007.01.092. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe I, Zhu J, Recio-Pinto E, Thornhill WB. Glycosylation affects the protein stability and cell surface expression of Kv1.4 but Not Kv1.1 potassium channels. A pore region determinant dictates the effect of glycosylation on trafficking. J Biol Chem. 2004;279:8879–8885. doi: 10.1074/jbc.M309802200. [DOI] [PubMed] [Google Scholar]
- 20.Recio-Pinto E, Thornhill WB, Duch DS, Levinson SR, Urban BW. Neuraminidase treatment modifies the function of electroplax sodium channels in planar lipid bilayers. Neuron. 1990;5:675–684. doi: 10.1016/0896-6273(90)90221-z. [DOI] [PubMed] [Google Scholar]
- 21.Bennett E, Urcan MS, Tinkle SS, Koszowski AG, Levinson SR. Contribution of sialic acid to the voltage dependence of sodium channel gating. A possible electrostatic mechanism. J Gen Physiol. 1997;109:327–343. doi: 10.1085/jgp.109.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Hartmann HA, Satin J. Glycosylation influences voltage-dependent gating of cardiac and skeletal muscle sodium channels. J Membr Biol. 1999;171:195–207. doi: 10.1007/s002329900571. [DOI] [PubMed] [Google Scholar]
- 23.Sutachan JJ, Watanabe I, Zhu J, Gottschalk A, Recio-Pinto E, Thornhill WB. Effects of Kv1.1 channel glycosylation on C-type inactivation and simulated action potentials. Brain Res. 2005;1058:30–43. doi: 10.1016/j.brainres.2005.07.050. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D, Bennett ES. Gating of the shaker potassium channel is modulated differentially by N-glycosylation and sialic acids. Pflugers Arch. 2008;456:393–405. doi: 10.1007/s00424-007-0378-0. [DOI] [PubMed] [Google Scholar]
- 25.Salazar BC, Castano S, Sanchez JC, Romero M, Recio-Pinto E. Ganglioside GD1a increases the excitability of voltage-dependent sodium channels. Brain Res. 2004;1021:151–158. doi: 10.1016/j.brainres.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M, Golic KG, Hawley RS. Drosophila: A laboratory Handbook. Cold Spring: Harbor Laboratory Press; 2005. [Google Scholar]
- 27.Hsu TL, Hanson SR, Kishikawa K, Wang SK, Sawa M, Wong CH. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc Natl Acad Sci U S A. 2007;104:2614–2619. doi: 10.1073/pnas.0611307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswanathan K, Lawrence S, Hinderlich S, Yarema KJ, Lee YC, Betenbaugh M. Engineering sialic acid synthetic ability into insect cells: Identifying metabolic bottlenecks and devising strategies to overcome them. Biochemistry. 2003;42:15215–15225. doi: 10.1021/bi034994s. [DOI] [PubMed] [Google Scholar]
- 29.Kim EJ, Sampathkumar S-G, Jones MB, Rhee JK, Baskaran G, Yarema KJ. Characterization of the metabolic flux and apoptotic effects of O-hydroxyl- and N-acetylmannosamine (ManNAc) analogs in Jurkat (human T-lymphoma-derived) cells. J Biol Chem. 2004;279:18342–18352. doi: 10.1074/jbc.M400205200. [DOI] [PubMed] [Google Scholar]
- 30.Kim EJ, Sampathkumar SG, Jones MB, Rhee JK, Baskaran G, Goon S, Yarema KJ. Characterization of the metabolic flux and apoptotic effects of O-hydroxyl- and N-acylmodified N-acetylmannosamine analogs in Jurkat cells. J Biol Chem. 2004;279:18342–18352. doi: 10.1074/jbc.M400205200. [DOI] [PubMed] [Google Scholar]
- 31.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 32.Presente A, Shaw S, Nye JS, Andres AJ. Transgene-mediated RNA interference defines a novel role for notch in chemosensory startle behavior. Genesis. 2002;34:165–169. doi: 10.1002/gene.10149. [DOI] [PubMed] [Google Scholar]
- 33.Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 34.Fayyazuddin A, Zaheer MA, Hiesinger PR, Bellen HJ. The nicotinic acetylcholine receptor Dalpha7 is required for an escape behavior in Drosophila. PLoS Biol. 2006;4:e63. doi: 10.1371/journal.pbio.0040063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen MJ, Drummond JA, Moffat KG. Development of the giant fiber neuron of Drosophila melanogaster. J Comp Neurol. 1998;397:519–531. doi: 10.1002/(sici)1096-9861(19980810)397:4<519::aid-cne5>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 36.Thornhill WB, Wu MB, Jiang X, Wu X, Morgan PT, Margiotta JF. Expression of Kv1.1 delayed rectifier potassium channels in Lec mutant Chinese hamster ovary cell lines reveals a role for sialidation in channel function. J Biol Chem. 1996;271:19093–19098. doi: 10.1074/jbc.271.32.19093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


