Abstract
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has precipitated an unprecedented and yet-unresolved health crisis worldwide. Different mammals are susceptible to SARS-CoV-2; however, few species examined so far develop robust clinical disease that mirrors severe human cases or allows testing of vaccines and drugs under conditions of severe disease. Here, we compare the susceptibilities of three dwarf hamster species (Phodopus spp.) to SARS-CoV-2 and introduce the Roborovski dwarf hamster (P. roborovskii) as a highly susceptible COVID-19 model with consistent and fulminant clinical signs. Particularly, only this species shows SARS-CoV-2-induced severe acute diffuse alveolar damage and hyaline microthrombi in the lungs, changes described in patients who succumbed to the infection but not reproduced in any experimentally infected animal. Based on our findings, we propose the Roborovski dwarf hamster as a valuable model to examine the efficacy and safety of vaccine candidates and therapeutics, particularly for use in highly susceptible individuals.
Keywords: coronavirus, animal model, COVID-19, pneumonia, histopathology, thrombosis, Campbell’s dwarf hamster (Phodopus campbelli), Djungarian hamster (Phodopus sungorus), Roborovski dwarf hamster (Phodopus roborovskii)
Graphical Abstract
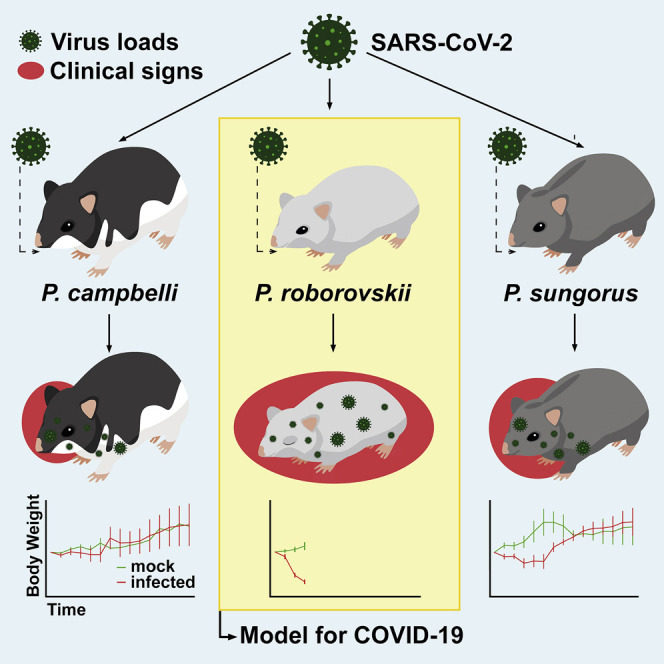
Trimpert et al. describe a highly susceptible non-transgenic rodent model for SARS-CoV-2 research. Roborovski hamsters develop fulminant clinical signs 2 to 4 days after infection. Comparison to closely related species reveals striking differences in disease susceptibility. This model mirrors severe human COVID-19 cases with great sensitivity for testing of vaccines and therapies.
Introduction
The coronavirus disease 2019 (COVID-19) epidemic is caused by the human severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Andersen et al., 2020; Bedford et al., 2020; Paules et al., 2020). The COVID-19 situation is still evolving rapidly, with alarming levels of spread in some geographic regions and disease severity, especially in the elderly and patients with pre-existing conditions (Davies et al., 2020; Dong et al., 2020). In the search for countermeasures against the pandemic, there is an unprecedented quest for the development of suitable animal models to study all presentations of COVID-19 (Lakdawala and Menachery, 2020; Yuan et al., 2020). Development of such a model is of utmost importance to unravel virus-induced pathogenesis and to evaluate vaccine candidates and potential therapeutic strategies. However, all of the currently available animal models are imperfect, and especially transgenic animals are in short supply (Callaway, 2020). Specifically, SARS-CoV-2 infection of all animal models reported so far resulted in rather mild illness, commonly characterized by no or moderate weight loss and signs of respiratory tract infection. Systemic disease has, although frequently reported in humans (Gupta et al., 2020; Tay et al., 2020), so far not consistently been observed in experimental infections of animals. Notably, virtually all experimental infections of animal models eventually lead to full recovery, which evidently is not the case in humans (Lakdawala and Menachery, 2020; Yuan et al., 2020). Currently used species were primarily selected to fit in silico predictions of susceptibility based on similarity to the human receptor of SARS-CoV-2, angiotensin converting enzyme-2 (ACE-2) (Devaux et al., 2020; Li et al., 2020; Pach et al., 2020). The animal models tested range from non-human primates (Deng et al., 2020; Lu et al., 2020b; Yu et al., 2020) to ferrets (Kim et al., 2020; Richard et al., 2020), mice (Bao et al., 2020; Dinnon et al., 2020; Gu et al., 2020; Hassan et al., 2020; Sun et al., 2020), cats (Shi et al., 2020), and hamsters (Bertzbach et al., 2020). In particular, recent SARS-CoV-2 hamster studies made use of the Syrian hamster (Mesocricetus auratus), which develops only moderate signs of clinical disease and lung pathology (Chan et al., 2020; Imai et al., 2020; Kreye et al., 2020; Sia et al., 2020), although age-related differences were reported (Osterrieder et al., 2020).
Dwarf hamster species have been used as animal models in biomedical research, mainly in endocrinology, theriogenology, and circadian biology research (Munley et al., 2018; Scherbarth and Steinlechner, 2010). To date, however, dwarf hamsters were not employed in COVID-19 research. Here, we set to investigate the outcome of SARS-CoV-2 infection in three closely related yet genetically distinct dwarf hamster species of the genus Phodopus. We observed striking differences in disease susceptibility among these species, a phenomenon apparently independent of their ACE-2 sequence. All residues shown to mediate interaction with the SARS-CoV-2 spike glycoprotein, which is deemed the decisive factor for susceptibility to the infection (Ou et al., 2020), are conserved among the hamsters compared in this study. Here, we demonstrate that all three Phodopus species tested are susceptible to the infection. However, only the Roborovski dwarf hamsters (P. roborovskii) developed a rapid onset of fulminant clinical disease. Therefore, this infection model mimics the clinical and pathological outcome observed in severe cases of COVID-19 better than any other available model. It can thus serve as a valuable alternative and a highly sensitive model in the development and testing of antiviral treatments, as well as other pro- and metaphylactic therapies.
Results
Clinical Disease
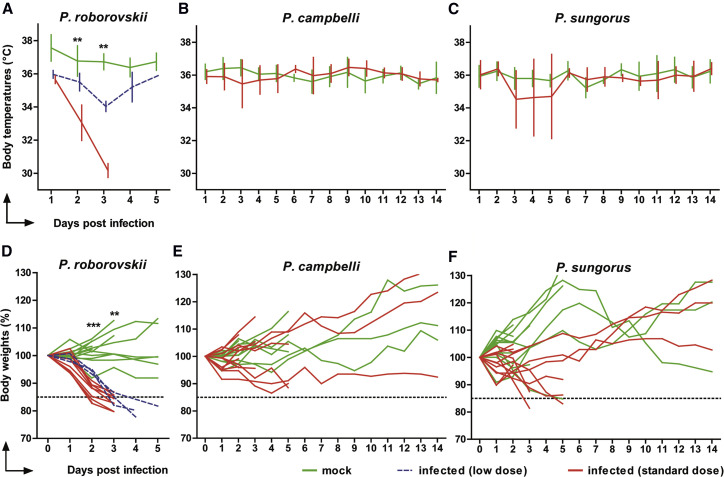
Roborovski dwarf hamsters (P. roborovskii), Campbell’s dwarf hamsters (P. campbelli), and Djungarian hamsters (P. sungorus) were infected with 1 × 105 plaque-forming units (pfu) of SARS-CoV-2 and inspected daily for up to 14 days. We observed changes in body temperatures, body weights, and clinical signs at early time points post-infection (Figure 1 ). While these parameters did not significantly differ between mock-infected and infected Campbell’s or Djungarian dwarf hamsters (Figures 1B, 1C, 1E, and 1F), infected Roborovski dwarf hamsters exhibited consistent and fulminant clinical signs within 3 days following infection. We recorded a dramatic decrease of body temperatures following infection, with a reduction in mean body temperatures from ∼36°C at 1 day post-infection (dpi) to 33.1°C ± 1.1°C standard deviation (SD) at 2 dpi and 30.2°C ± 0.4°C SD at 3 dpi (Figure 1A). Moreover, mean weight losses reached 14.35% ± 2.96% SD (2 dpi) and 19.86% ± 2.68% SD on 3 dpi when compared to the mock-infected group (Figure 1D). Roborovski dwarf hamsters also showed clear signs of clinical disease including snuffling, dyspnea, and ruffled fur. Overall, the very active behavior otherwise typical for this species was dramatically reduced. Three days after infection, multiple individuals appeared to be terminally ill. For humane reasons, all infected Roborovski dwarf hamsters were euthanized at this point.
Figure 1.
Changes in Body Temperature and Body Weight Following SARS-CoV-2 Infection in Phodopus Hamsters
(A–C) Temperature changes of (A) Roborovski dwarf hamsters, (B) Campbell’s dwarf hamsters, and (C) Djungarian hamsters over the course of infection (shown as means with SD of at least 12 [2 and 3 dpi], 7 [5 dpi], and 6 animals [after 5 dpi] per group).
(D–F) Corresponding individual relative body weights of (D) Roborovski dwarf hamsters (n = 22), (E) Campbell’s dwarf hamsters (n = 24), and (F) Djungarian hamsters (n = 24) are given. The color code represents mock-infected animals (green) or Phodopus hamsters infected with the low dose (blue) or standard dose (red) (Wichmann et al., 2020).
The dotted lines at 85% in (D)–(F) refer to 15% body weight losses (first defined humane endpoint). Mann-Whitney U tests (B, C, E, and F) and Kruskal-Wallis tests (A and D) for each time point revealed significant differences in body temperatures and body weights of infected versus non-infected Roborovski dwarf hamsters at 2 and 3 dpi (∗∗∗p ≤ 0.0001, ∗∗p ≤ 0.001).
To investigate dose dependency of the disease phenotype, we re-used three uninfected animals intended as controls for the now-obsolete later time points after infection. The three remaining animals were infected with 5,000 pfu of SARS-CoV-2, a dose reduction of 95% compared to the initial challenge. Weight loss observed in these hamsters was only slightly delayed compared to the standard dose challenge; however, the decrease of body temperatures was less pronounced, and dyspnea was the only prominent clinical sign in the low dose challenge group (Figures 1A and 1D). Regardless of the overall milder disease progression, all three animals included in the low dose challenge reached the defined humane endpoints by 4 or 5 dpi. The body weight losses observed in Djungarian hamsters (Figure 1C) were less pronounced and rather transient, mirroring the course of infection described for Syrian hamsters (Chan et al., 2020; Imai et al., 2020; Osterrieder et al., 2020; Sia et al., 2020). While decreases of mean body temperatures were observed in this species after infection, they proved to be highly inconsistent between individuals. From the three species compared here, Campbell’s dwarf hamsters were the least susceptible to clinical disease following SARS-CoV-2 infection, as evidenced by a mild or completely absent reduction of body weight and no decline of body temperature. In addition, and in particularly stark contrast to infected Roborovski dwarf hamsters, no clinical signs were observed in this species following SARS-CoV-2 infection.
Virological Assessment
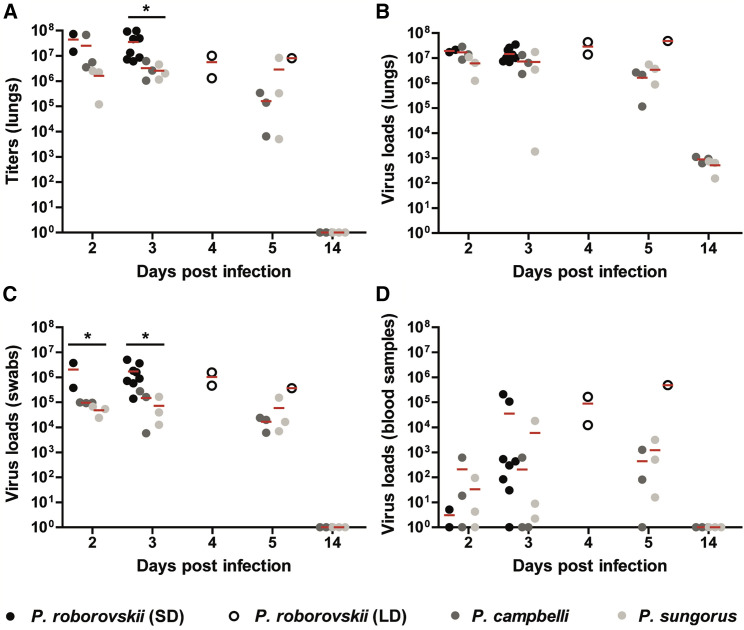
In the next series of experiments, we determined virus titers and SARS-CoV-2 RNA copy numbers in the lungs, bucco-laryngeal swabs, and whole-blood samples of all hamsters (Figure 2 ). Results confirmed readily detectable and high virus loads in respiratory samples at early time points after infection in all species tested (Figures 2A–2C). We found significantly higher virus titers in the lungs of Roborovski dwarf hamsters at 3 dpi when compared to the other two species (Figure 2A). Notably, we detected viral RNA copies of 2.06 × 106 ± 2.37 × 106 SD (2 dpi) and 1.81 × 106 ± 1.69 × 106 SD (3 dpi) in bucco-laryngeal swabs of Roborovski dwarf hamsters, while copy numbers reached a maximum of only ∼1 × 105 in Campbell’s and Djungarian hamsters at these time points (Figure 2C). Virus copy numbers detected in blood samples indicated a potential viremic spread in select individuals of all three species (Figure 1D). By virus titrations of the blood collected from infected dwarf hamsters, we detected infectious virus at titers of more than 100 pfu/mL blood in some but not all infected animals (Table S1).
Figure 2.
Virus Loads in the Respiratory Tract and Whole-Blood Samples
(A and B) Virus titers in 50-mg lung tissues were determined by plaque assay in Vero E6 cells (A) and virus genome copy numbers per 2.5-mg lung tissue as determined by qRT-PCR at different time points after infection (B).
(C and D) Virus loads were also determined by qRT-PCR in (C) bucco-laryngeal swabs and (D) 2.5 μl of whole-blood samples. Kruskal-Wallis tests were employed to determine significant differences between the Phodopus hamsters. Shown are results for Roborovski dwarf hamsters infected with the standard dose (black circles, n = 2 at 2 dpi, n = 8 at 3 dpi) and low dose infection (white circles, n = 2 at 4 dpi, n = 1 at 5 dpi), Campbell’s dwarf hamsters (dark gray circles, n = 3 at all time points), and Djungarian hamsters (light gray circles, n = 3 at all time points). Significant differences are marked with asterisks (∗) and indicate a p value of ≤ 0.05.
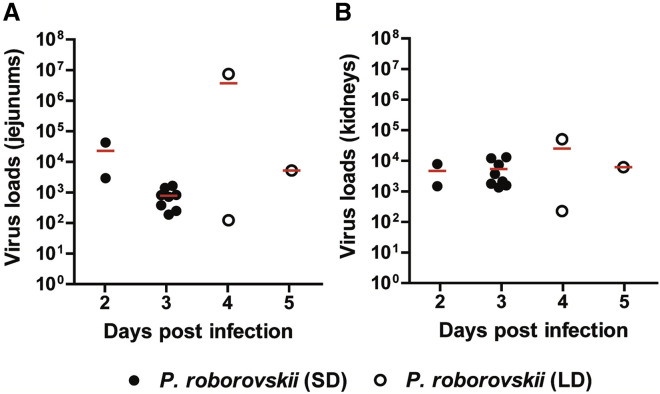
We followed up on the question of systemic virus spread by determining virus loads in jejunal and kidney samples of all infected Roborovski dwarf hamsters at different time points after infection (Figure 3A and 3B). As determined by qRT-PCR, we detected relatively high viral RNA copy numbers in both organs at all time points, confirming systemic, extrapulmonary spread of the virus in all individuals of this species.
Figure 3.
Virus Loads in Intestinal and Kidney Samples of Infected Roborovski Dwarf Hamsters
Virus loads in 2.5 mg of (A) jejunal and (B) kidney tissues of animals infected with the standard dose (black circles) or low dose (white circles). Viral RNA copies were determined by qRT-PCR at different time points after infection and revealed systemic virus spread.
Finally, using qRT-PCR, we found fewer than 1,000 copies of virus RNA per 2.5 mg of brain tissue, which suggests a lack of productive infection of the brain (Table S1).
Histopathology
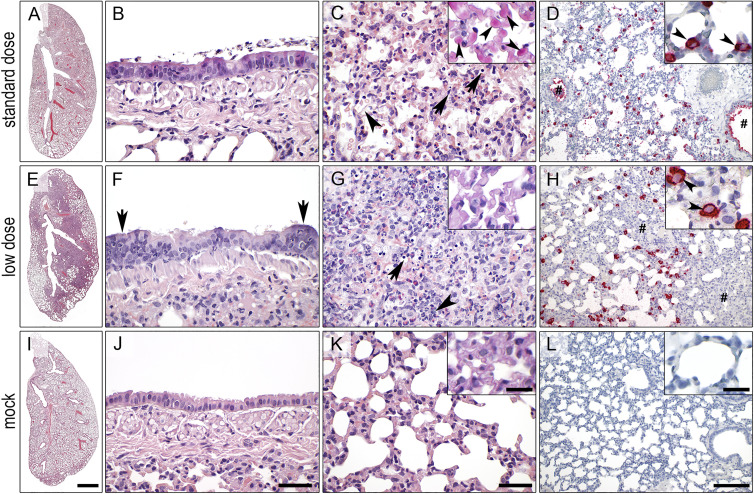
Similar to the marked clinical differences observed among the three hamster species following SARS-CoV-2 infection, a systematic histopathological examination of formalin-fixed, paraffin-embedded lung sections following recommended guidelines (Gruber et al., 2020) revealed striking differences, both in terms of lesion characters as well as their distribution and severities (Table S2). Most obviously, only the Roborovski dwarf hamsters infected with 1 × 105 pfu (standard dose) developed highly destructive and diffuse alveolar damage throughout the entire lungs with massive alveolar epithelial cell (AEC) necrosis, hyaline membranes, proteinaceous material, hemorrhages, edema, and cellular debris within alveoli as early as 2 dpi (Figures 4A and 4C). Of note, marked hyaline thrombi were present in high numbers of interalveolar wall capillaries with strong purple signals in a periodic acid Schiff (PAS) reaction (Figure 4C, inset). However, contrary to previous reports on Syrian hamsters (Gruber et al., 2020; Imai et al., 2020; Osterrieder et al., 2020), the bronchitis component was mild (Figure 4B) or absent, and vascular endothelialitis was not observed. Attempts of regeneration in terms of hyperplasia of AEC or bronchial epithelium (BE) were not observed until 3 dpi, probably due to the overwhelming and early parenchymal destruction. Importantly, these patterns were uniformly present in all hamsters of this group.
Figure 4.
Lung Histopathology and In Situ Hybridization of SARS-CoV-2-Infected Roborovski Dwarf Hamsters
(A–D) Following infection with 1 × 105 plaque-forming units (pfu, standard dose; A–D) P. roborovskii developed devastating diffuse lung damage at 3 dpi (A) with only mild necrosis of bronchial epithelium (BE) and bronchitis (B) but severe diffuse alveolar damage, including necrosis of alveolar epithelial cells (AECs; C, arrows), hyaline membranes and fibrin extravasation (C, arrowhead), hemorrhage and alveolar edema, as well as multiple hyaline thrombi in alveolar capillaries (C, inset, arrowheads; PAS reaction). Consistent with the homogeneous distribution of lesions, in situ hybridization localized viral RNA throughout the entire lungs predominantly within AEC-II (D, inset, arrowheads) but also in bronchi (D, hash) and macrophages, while AEC-I were completely spared.
(E–H) Infection with 20-times-less virus (5 × 103 pfu, low dose; E–H) still resulted in extensive pneumonia at 4 dpi (E) with inflammation and regeneration of BE (F, arrows), necrosis of AEC and inflammatory cells (G, arrow), as well as mainly neutrophilic and heterophilic infiltration (G, arrowhead). Viral RNA was detected with a distribution similar to the standard dose group; however, areas of dense inflammation had obviously cleared the virus (H, hash). Red, signals for viral RNA; blue, hemalaun counterstain.
(I–L) Mock-infected animals developed none of the pathologic findings of animals infected with SARS-CoV-2.
Bars: 1 mm (A, E, and I), 50 μm (B, C, F, G, J, and K), 200 μm (D, H, and L), and 20 μm (insets of C, H, and K).
In contrast, all Roborovski dwarf hamsters infected with 1/20 of the standard dose (5,000 pfu, low dose) still developed pathological patterns typical of experimental SARS-CoV-2 infection, as previously described in Syrian hamsters (Chan et al., 2020; Gruber et al., 2020; Imai et al., 2020; Kreye et al., 2020; Osterrieder et al., 2020; Sia et al., 2020). These included necrosuppurative pneumonia distributed multifocally throughout the lungs as well as ongoing bronchitis with hyperplasia of BE and AEC at 4 and 5 dpi (Figures 4E–4G). Hyaline thrombi were not observed (Figure 4G), but vascular endothelialitis was sporadically present.
Infection of the Djungarian and Campbell’s dwarf hamsters resulted in overall similar tissue damage and inflammatory reactions at 2 to 5 dpi that resembled the Roborovski dwarf hamsters infected with the low dose (Figure S1). In strong contrast to the latter infected with the same (standard) dose, both species showed early and prominent onset of AEC hyperplasia from 3 dpi on, indicating active regeneration. Hyaline thrombi or vascular endothelialitis were not observed in any animal of both species (Table S2). In addition to lungs, systematic histopathological examinations of all hamster brains failed to yield any evidence of cellular or tissue damage, inflammatory responses, circulatory disturbances such as thrombosis and hemorrhage, or any other evidence of viral infection. Moreover, PAS reaction on histological sections of all brains did not reveal any evidence of hyaline thrombosis, as seen in disseminated intravascular coagulation.
In Situ Hybridization of SARS-CoV-2 RNA
In the Roborovski dwarf hamsters infected with the standard dose, viral RNA encoding for the N protein of SARS-CoV-2 was localized throughout the entire lungs, consistent with the homogeneous distribution of lesions. Specifically, the virus was found mainly in AEC-II but also in bronchi and macrophages, while AEC-I were uniformly negative (Figure 4D; Table S2). Following low dose infection, a distribution of virus nucleic acid similar to the standard dose group was observed (Figure 4H). However, under low dose conditions, regions with dense immune cell infiltration were consistently devoid of viral RNA.
In the Djungarian and Campbell’s dwarf hamsters, viral RNA was strictly localized to the vicinities of inflammatory foci. In contrast to the Roborovski dwarf hamsters, however, both AEC-I and AEC-II were equally infected (Figures S1D and S1H). Bronchial epithelial cells, detritus lining the bronchial mucosa, and macrophages also contained viral RNA, as observed in the Roborovski dwarf hamsters (Table S2).
Detailed examination of the brains of infected Roborovski dwarf hamsters by in situ hybridization for SARS-CoV-2 RNA failed to identify any viral RNA signals in the entire brains, including the rhinencephalon. Only rare signals were detected in single blood leukocytes within blood vessels, consistent with viremia.
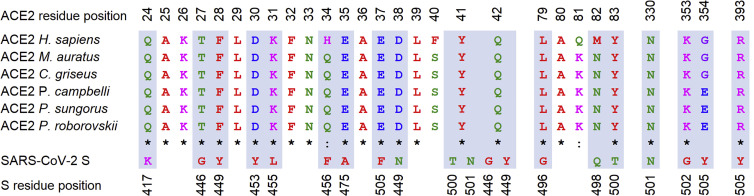
Comparison of ACE-2 Sequences between Different Hamster Species
One of the major determinants of susceptibility to SARS-CoV-2 is the ability of its spike glycoprotein (S) to bind to its cognate receptor, ACE-2, which is deemed necessary for virus entry into the host cell (Hoffmann et al., 2020). To investigate the potential role of ACE-2 sequence variation in the diverging course of SARS-CoV-2 infection in Phodopus species, we determined the full-length coding sequence of the Roborovski dwarf hamster ACE-2 by PCR amplification and Sanger sequencing and assembled the coding sequence of ACE-2 of the Djungarian hamster from publicly available shotgun sequencing data (GenBank: GCA_001707965.1). Comparison of the newly obtained and the published ACE-2 sequence of the Campbell’s dwarf hamster and the Syrian hamster revealed several single-nucleotide polymorphisms (SNPs) (Figure 5 ). However, we failed to identify any difference in any of the amino acids thought to be critically important for virus binding (Figure 5) (Lan et al., 2020; Shang et al., 2020; Wan et al., 2020). We concluded from our data that the minor difference in the primary structure of ACE-2 is likely insufficient to explain the remarkable susceptibility of the Roborovski dwarf hamster to SARS-CoV-2 infection and the development of severe disease when compared to other hamster species examined here or in previous studies (Chan et al., 2020; Imai et al., 2020; Osterrieder et al., 2020; Sia et al., 2020).
Figure 5.
Comparison of Genomic ACE-2 Sequences
ACE-2 residues that interact with the receptor-binding domain (RBD) of SARS-CoV-2 spike glycoprotein (S) are almost completely conserved among different species of the family Cricetidae (the only exception is ACE-2 residue 354G/E). The interacting residues of the two proteins are highlighted in grey shading. Amino acids are colored according to their physico-chemical properties: red, hydrophobic; blue, negatively charged; pink, positively charged; purple, small nonpolar; green, polar residue. Asterisk, fully conserved residue; colon, residue with very similar properties.
Discussion
In this study, we compare the susceptibility of three Phodopus dwarf hamster species to SARS-CoV-2 infection and introduce the Roborovski dwarf hamster (P. roborovskii) as a highly susceptible species that develops—as the only naturally susceptible COVID-19 animal model described to date—significant body weight loss, severe reduction in body temperature, consistent systemic viral spread, and dramatic clinical signs within 48 h of infection. Dose dependency is observed in the course of disease; however, even animals challenged with a 20-fold-reduced dose develop only slightly protracted, yet still fulminant and consistent, disease progression. Notably, our findings stand in contrast to data that suggest rather low SARS-CoV-1 susceptibility of dwarf hamsters (Li et al., 2012).
Compared to the Syrian hamster and other available naturally susceptible animal models, the Roborovski dwarf hamster offers the advantage of rapid and consistent development of clinical signs following infection. This is important not only to increase the validity and significance of preclinical trials, but it may also allow a reduction of animal numbers, as statistically significant differences may be achieved more easily. Alongside the particular features described in this study, the Roborovski dwarf hamster offers the practical advantage of a much smaller size compared to Syrian hamsters: it does not, in fact, reach the size of a mouse and is thus the smallest model available. Compared to other hamster species, it also was less prone to aggressive behavior, which facilitated both handling and housing in larger groups. Notably, Roborovski dwarf hamsters are commercially available and bred in large numbers for pet trade. The species reaches sexual maturity at 4 weeks of age and can easily be bred in a laboratory environment (Kubiak, 2020).
The rapid and fatal course of experimental infection of Roborovski dwarf hamsters mirrors the situation in severe COVID-19 cases in humans in the lower respiratory tract, in that diffuse alveolar damage and hyaline microthrombi were readily detectable in infected animals within days after standard dose challenge (McGonagle et al., 2020). Of note, no other animal model described so far developed these lesions at a comparable severity observed in the Roborovski dwarf hamster (Bertzbach et al., 2020; Chan et al., 2020; Cleary et al., 2020; Gruber et al., 2020; Imai et al., 2020; Kreye et al., 2020; Rockx et al., 2020; Yu et al., 2020). It is worth mentioning that we could exclude relevant involvement of the brain in SARS-CoV-2 infection of Roborovski dwarf hamsters. Importantly, transgenic mice that show robust disease development frequently suffer from severe SARS-CoV-2-induced encephalitis (Jiang et al., 2020; Winkler et al., 2020), a cause of death not observed in humans. Thus, the severe pneumonia with thrombosis observed in the Roborovski dwarf hamster is a much closer resemblance of severe and fatal human cases of COVID-19.
Our findings allow us to conclude that comparison of disease progression is possible within a wide range of virus infection doses. This aspect is particularly important for future testing of the robustness of therapeutics and vaccines with respect to severe outcomes as well as the safety of live attenuated vaccines. Considering the unabated spread of the virus in large parts of the world, such vaccines are desperately needed to effectively combat the pandemic. A major and valid concern regarding the safety of live attenuated vaccines is the potential for residual virulence or reversion to virulence of the vaccine virus that may then cause disease in vaccines. Reducing the potential hazard of releasing an even mildly virulent vaccine virus can be minimized only by testing vaccine candidates in a highly susceptible model prior to clinical trials. The Roborovski dwarf hamster seems, so far, to be the only naturally susceptible animal that would provide the sensitivity required for such absolutely necessary studies.
Moreover, infection of the highly susceptible Roborovski dwarf hamster as demonstrated here suggests that COVID-19 disease outcome in terms of clinical symptoms might be somewhat independent of the infectious dose, provided that the minimal infectious dose is reached. However, the dynamics of disease progression in an individual may well be a function of infectious dose, and informed decisions regarding therapeutic intervention or pro- and metaphylactic measures are absolutely critical for an appropriate medical response at the different stages of infection with SARS-CoV-2. A complete understanding of disease development over time following infection with different doses, including the important question of systemic spread of the virus, in the absence or presence of any type of intervention will be absolutely necessary for an adequate individual and population response (Tay et al., 2020). This aspect may become even more important in the future, as new strains have been evolving that seem to spread extremely effectively in the human population but may also undergo attenuation (Forster et al., 2020; Korber et al., 2020; Lu et al., 2020a). In such a scenario, the availability of a highly susceptible animal model will be key to addressing pertinent questions of mitigation strategies and spread of infection.
The ability of ACE-2 to bind the SARS-CoV-2 receptor-binding domain (RBD) located in the spike protein is so far deemed to be the major determinant of species susceptibility to SARS-CoV-2 infection (Ou et al., 2020; Shang et al., 2020; Wan et al., 2020; Zhai et al., 2020). While our data do not provide any reason to question this paradigm, we discovered only minor differences in the ACE-2 protein of Phodopus species. Since none of the amino acids described to be of importance for virus RBD binding are altered among the three species, we consider it unlikely that the substantial differences in disease progression can be accounted for by differences in ACE-2 RBD binding.
Differences in other factors important for virus infection, such as the transmembrane serine protease 2 (TMPRSS2), are less understood. The same holds true for other intrinsic host factors, such as differences in innate and adaptive immune responses, different expression levels and tissue distributions of ACE-2, and the tendency to develop coagulopathy following SARS-CoV-2 infection. We believe the Roborovski dwarf hamster can serve as a model to better understand these principles underlying the highly variable pathologies of COVID-19. It seems important to emphasize that we find that all tested species are susceptible to the infection, and virological parameters—albeit slightly different between species—do not appear to be sufficient to explain the striking differences in the observed disease phenotypes. The Roborovski dwarf hamster may, thus, also offer insights into the mechanisms and modulating factors underlying the severe course of infection. Still, this model comes with the caveat of a lack of generally available tools and reagents that would allow to readily measure host factors including immune responses (e.g., chemokines or cytokines), at least on the protein level.
In summary, we document that the P. roborovskii small-animal model for SARS-CoV-2 infection is an asset to animal models described to date because infection, even with low virus doses, results in rapid and robust development of severe clinical disease due to early destructive and fatal lung pathology, while relevant central nervous system infection is not observed. Importantly, variation of virus doses of infection allows for following disease progression and spans a wide spectrum of clinical signs, from a respiratory-only to a severe and systemic outcome. Therefore, we contend that the Roborovski dwarf hamster may serve as a valuable addition to the animal models available to examine SARS-CoV-2 pathogenesis; the safety and efficacy of vaccines, particularly for modeling highly susceptible individuals; as well as the safety and efficacy of therapeutic or metaphylactic interventions.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| SARS-CoV-2 | (Wölfel et al., 2020) | N/A |
| Biological Samples | ||
| Hamster lungs, swabs, blood, intestines, kidneys | This paper | N/A |
| Critical Commercial Assays | ||
| innuPREP Virus RNA kit | Analytic Jena | Cat# 845-KS-4700250 |
| NEB Luna Universal Probe One-Step RT-qPCR kit | New England Biolabs | Cat# E3006L |
| Deposited Data | ||
| P. roborovskii ACE-2 sequence | This paper | GenBank: MW075232 |
| P. sungorus ACE-2 sequence | This paper | GenBank: MW075233 |
| P. campbelli ACE-2 | GenBank | GenBank: GQ262790 |
| Complete genomic sequence of P. sungorus | GenBank | GenBank: GCA_001707965.1 |
| Experimental Models: Cell Lines | ||
| Vero E6 cells | ATCC | CRL-1586 |
| Experimental Models: Organisms/Strains | ||
| Hamster: P. roborovskii | German pet trade | N/A |
| Hamster: P. campbelli | German pet trade | N/A |
| Hamster: P. sungorus | German pet trade | N/A |
| Oligonucleotides | ||
| RT-qPCR primers and probe | (Corman et al., 2020) | N/A |
| Sequencing primers, see Table S3 | This paper | N/A |
| Synthetic probes for the in situ-detection of the N gene RNA of SARS-CoV-2 (NCBI database NC_045512.2, nucleotides 28,274 to 29,533) | Invitrogen by Thermo Fisher Scientific, Darmstadt, Germany | assay ID: VPNKRHM |
| Software and Algorithms | ||
| GraphPad PRISM | N/A | graphpad.com |
Resource availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jakob Trimpert (trimpert.jakob@fu-berlin.de).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The datasets generated during this study are available at GenBank. The accession number for the Phodopus roborovskii ACE-2 sequence reported in this paper is Genbank: MW075232. The accession number for the Phodopus sungorus ACE-2 sequence reported in this paper is Genbank: MW075233.
Experimental model and subject details
Ethics statement
In vitro and animal work was conducted under appropriate biosafety conditions in the BSL-3 facility at the Institut für Virologie, Freie Universität Berlin, Germany. All animal experimentation was approved by the Landesamt für Gesundheit und Soziales in Berlin, Germany (permit number 0086/20) and performed in compliance with relevant national and international guidelines for care and humane use of animals.
Viruses and cells
We used the previously published SARS-CoV-2 München isolate (Wölfel et al., 2020) for virus challenge. Virus stocks were propagated on Vero E6 cells (ATCC CRL-1586) in minimal essential medium (MEM; PAN Biotech) that was supplemented with 10% fetal bovine serum (PAN Biotech), 100 IU/ml penicillin G and 100 μg/ml streptomycin (Carl Roth). The virus stocks were stored at −80°C prior to experimental infections.
Hamsters
This study involved the three members of the genus Phodopus of the hamster subfamily Cricetinae: Roborovski dwarf hamsters (P. roborovskii), Campbell’s dwarf hamsters (P. campbelli) and Djungarian hamsters (P. sungorus) obtained through the German pet trade. All animals were sourced from the same breeding facility. For each species, 22 to 24 female and male hamsters of 5 to 7 weeks of age were kept in groups of 6 animals in individually ventilated cages. Enrichment was provided bountiful according to provisions of the animal experimentation guidelines and legal requirements. Hamsters had unrestricted access to food and water, and were allowed acclimatization to the experimental conditions for seven days prior to SARS-CoV-2 infection. According to stipulations of the animal use permit, high caloric feed (oilseed) was offered to all animals once the first animal in a group had presented with a body weight loss of > 10% at day two post infection. Cage temperatures and relative humidities were recorded daily and ranged from 22 to 24°C and 45 to 60%, respectively. The defined humane endpoints included > 15% weight loss for > 48 h or > 20% weight loss.
An obvious association between disease progression or any other parameter examined in this report and the sex of the animals was not observed.
Method details
Infection Experiments
Hamsters of each species were randomly distributed into two groups: SARS-CoV-2-infected (Roborovski dwarf hamsters: n = 10, Campbell’s dwarf hamsters: n = 12 and Djungarian hamsters: n = 12; 1x105 plaque forming units (pfu) in a volume of 30 μl) and mock-infected (n = 12 per species; 30 μl of cell culture supernatant from uninfected Vero E6 cells). The infection was performed as previously described (Osterrieder et al., 2020). In extension of our initial trial, we infected three of the previously mock-infected Roborovski dwarf hamsters one week later with 5x103 pfu (in 20 μL volume) to test for dose-dependent outcomes of the infection. Body weights of all hamsters and body temperatures of at least 3 hamsters per group were recorded daily and clinical signs of all animals were monitored twice daily throughout the experiment. On days 2, 3, 5 and 14 post infection, three hamsters were euthanized either randomly assigned or because they had reached defined humane endpoints by the respective day post infection. Euthanasia was applied by exsanguination under general anesthesia as described (Nakamura et al., 2017). Whole-blood, tracheal swabs, lungs (left and right), kidneys and parts of the jejunum were collected for virus titrations, RT-qPCR and/or histopathological examinations. All organs were preserved in 4% formalin for subsequent in-depth histopathological investigations.
Histopathology and in situ-hybridization
For histopathology and localization of viral RNA by in situ-hybridization (ISH), the left lung lobe was carefully removed and immersion fixed in buffered 4% formalin, pH 7.0, for 48 h. Lungs were embedded in paraffin and cut at 2 μm thickness. Sections were stained with hematoxylin and eosin (HE) and periodic acid-Schiff (PAS) reaction followed by blinded microscopic evaluation by board certified veterinary pathologists (K.D., A.D.G.) (Gruber et al., 2020). For ISH, the ViewRNA ISH Tissue Assay Kit (Invitrogen by Thermo Fisher Scientific, Darmstadt, Germany) was used following the manufacturer’s instructions with minor adjustments. Probe for the detection of the N gene RNA of SARS-CoV-2 (NCBI database NC_045512.2, nucleotides 28,274 to 29,533, assay ID: VPNKRHM) was employed. Lung sections of 2 μm thickness mounted on adhesive glass slides were dewaxed in xylol and dehydrated in graded ethanols. Tissues were incubated at 95°C for 10 min and subsequently protease digested for 20 min. Sections were fixed with 4% paraformaldehyde dissolved in phosphate buffered saline (PBS) and hybridized with the probes. Amplifier and label probe hybridizations were performed according to the manufacturer’s instructions using fast red as chromogen. Sections were counterstained with hematoxylin for 45 s, washed in tap water for 5 min, and mounted with Roti®-Mount Fluor-Care DAPI (4, 6-diaminidino-2-phenylindole; Carl Roth). An irrelevant probe for the detection of streptococcal pneumolysin was used as a control for sequence-specific binding. HE and PAS-stained and ISH slides were analyzed and photographed using an Olympus BX41 microscope with a DP80 Microscope Digital Camera and the cellSens Imaging Software, Version 1.18 (Olympus Corporation, Münster, Germany). For overviews with lower magnification, slides were automatically digitized using the Aperio CS2 slide scanner (Leica Biosystems Imaging Inc., Vista, CA, USA) and image files were generated using the Image Scope Software (Leica Biosystems Imaging Inc.). The same experimental procedures were used to evaluate brain tissues.
Virus titrations, RNA extractions and RT-qPCR
To assess virus titers from 50 mg lung tissue, tissue homogenates were prepared using a bead mill (Analytic Jena) and 10-fold serial dilutions were prepared in MEM-10% FBS, which were then added to Vero E6 cells in 12-well-plates Likewise, titrations were performed on 10-fold serial dilutions of whole blood samples. The dilutions were removed after 2 h and cells were overlaid with 1.25% carboxy-methylcellulose (viscosity 400 cP, Sigma) in MEM-10% FBS. Three days later, cells were formalin-fixed, stained with crystal violet, and plaques were counted. RNA was extracted from 25 mg lung homogenates, 25 mg jejunal and kidney tissue samples, 25 μl blood and tracheal swabs using the innuPREP Virus RNA kit (Analytic Jena). Viral RNA copies were quantified in 10% of the obtained eluate volume with a one-step RT-qPCR reaction using a standard curve and the NEB Luna Universal Probe One-Step RT-qPCR kit (New England Biolabs) and previously published TaqMan primers and probe (SARS-CoV-2 E_Sarbeco; Table S3) (Corman et al., 2020) on a StepOnePlus RealTime PCR System (Thermo Fisher Scientific).
ACE-2 PCR and sequencing
The complete coding sequence (CDS) of ACE-2 of P. campbelli was determined previously (GQ262790). The complete protein coding sequence of ACE-2 of P. roborovskii was determined from its mRNA sequence. Total RNA was isolated from the lungs of 2 different Roborovski dwarf hamsters. Harvested lung tissue samples were submersed in RNAlater solution (Thermo Fisher Scientific) to stabilize and protect cellular RNA. The tissues were homogenized and total RNA was isolated with InviTrap Spin Cell RNA Mini kit (Invitec Molecular). RNA (500 ng) was converted to complementary DNA (cDNA) by using the Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamer primers. Next, cDNA was amplified as three overlapping fragments spanning the entire protein coding region of ACE-2 using ACE-2-specific primers and the high-fidelity DNA polymerase Phusion (Thermo Fisher Scientific). The primers were designed to bind regions that are conserved across four species of the family Cricetidae (Cricetulus griseus, Mesocricetus auratus, Peromyscus maniculatus bairdii and Phodopus campbelli) with known complete cDNA (Table S3). PCR products were purified and sequenced by Sanger sequencing. The complete CDS of ACE-2 of P. sungorus was predicted from the genomic sequence of P. sungorus (GenBank GCA_001707965.1) based on sequence similarity. The sequence of ACE-2 is highly conserved across mammalian species (Zhai et al., 2020). The ACE-2 gene of Cricetulus griseus, Mesocricetus auratus, Peromyscus maniculatus bairdii and Phodopus campbelli contains 18 exons. To identify the CDS of ACE-2 of P. sungorus, the ACE-2 exon sequences of P. campbelli and the ACE-2 exon sequences of P. roborovskii that were identified in this study, were aligned individually against the available genomic reads of P. sungorus using the BLAST program and the complete CDS was derived from the obtained alignments. The complete predicted ACE-2 CDS of P. roborovskii and P. sungorus were submitted to GenBank (Accession numbers pending). Complete alignments of ACE-2 protein sequences are supplied as supplementary information (Figure S2).
Quantification and statistical analysis
Statistical analyses were performed using Graph-Pad Prism (GraphPad Software Inc., San Diego, CA, USA). Statistical details of all analyzed experiments can be found in the respective figure legends. Data were considered significant if p ≤ 0.05.
Acknowledgments
We thank Ann Reum, Annett Neubert, Angela Linke, and Michaela Scholz for excellent technical assistance. We are grateful to Christa Thöne-Reineke for expert support in animal welfare and husbandry and to Carfil Inc. for their support of our animal husbandry. This research was supported by the Deutsche Forschungsgemeinschaft (DFG) grant SFB-TR84/Z01b awarded to A.D.G. and J.T. and by COVID-19 grants from Freie Universität Berlin and Berlin University Alliance to N.O.
Author Contributions
Conceptualization, J.T. and N.O.; Investigation, J.T., D.V., K.D., A.A., D.K., S.D., A.V., A.D.G., and L.D.B.; Writing, J.T., K.D., A.D.G., D.K., N.O., and L.D.B. All authors had the opportunity to comment on the draft manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: November 19, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108488.
Supplemental Information
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.D., Sall A.A., WHO Strategic and Technical Advisory Group for Infectious Hazards COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertzbach L.D., Vladimirova D., Dietert K., Abdelgawad A., Gruber A.D., Osterrieder N., Trimpert J. SARS-CoV-2 infection of Chinese hamsters (Cricetulus griseus) reproduces COVID-19 pneumonia in a well-established small animal model. Transbound. Emerg. Dis. 2020 doi: 10.1111/tbed.13837. Published online September 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Labs rush to study coronavirus in transgenic animals - some are in short supply. Nature. 2020;579:183. doi: 10.1038/d41586-020-00698-x. [DOI] [PubMed] [Google Scholar]
- Chan J.F., Zhang A.J., Yuan S., Poon V.K., Chan C.C., Lee A.C., Chan W.M., Fan Z., Tsoi H.W., Wen L. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa325. Published online March 26, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Klepac P., Liu Y., Prem K., Jit M., Eggo R.M., CMMID COVID-19 working group Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 2020;26:1205–1211. doi: 10.1038/s41591-020-0962-9. [DOI] [PubMed] [Google Scholar]
- Deng W., Bao L., Gao H., Xiang Z., Qu Y., Song Z., Gong S., Liu J., Liu J., Yu P. Ocular conjunctival inoculation of SARS-CoV-2 can cause mild COVID-19 in rhesus macaques. Nat. Commun. 2020;11:4400. doi: 10.1038/s41467-020-18149-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux C.A., Pinault L., Osman I.O., Raoult D. 2020. Can ACE2 receptor polymorphism predicts species susceptibility to SARS-CoV-2? Research Square. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A.D., Osterrieder N., Bertzbach L.D., Vladimirova D., Greuel S., Ihlow J., Horst D., Trimpert J., Dietert K. Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2 Infected Hamsters - What Matters? Am. J. Respir. Cell Mol. Biol. 2020 doi: 10.1165/rcmb.2020-0280LE. Published September 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y. Rapid adaptation of SARS-CoV-2 in BALB/c mice: Novel mouse model for vaccine efficacy. bioRxiv. 2020 doi: 10.1101/2020.05.02.073411. [DOI] [Google Scholar]
- Gupta A., Madhavan M.V., Sehgal K., Nair N., Mahajan S., Sehrawat T.S., Bikdeli B., Ahluwalia N., Ausiello J.C., Wan E.Y. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.D., Liu M.Q., Chen Y., Shan C., Zhou Y.W., Shen X.R., Li Q., Zhang L., Zhu Y., Si H.R. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell. 2020;182:50–58.e8. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.I., Kim S.G., Kim S.M., Kim E.H., Park S.J., Yu K.M., Chang J.H., Kim E.J., Lee S., Casel M.A.B. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe. 2020;27:704–709.e702. doi: 10.1016/j.chom.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E.E., Bhattacharya T., Parker M.D. Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.29.069054. [DOI] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.D. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069.e19. doi: 10.1016/j.cell.2020.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubiak M. Hamsters. In: Kubiak M., editor. Handbook of Exotic Pet Medicine. John Wiley & Sons Ltd.; 2020. pp. 83–98. [Google Scholar]
- Lakdawala S.S., Menachery V.D. The search for a COVID-19 animal model. Science. 2020;368:942–943. doi: 10.1126/science.abc6141. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li K.K.B., Yip C.W., Hon C.C., Lam C.Y., Zeng F., Leung F.C.C. Characterisation of animal angiotensin-converting enzyme 2 receptors and use of pseudotyped virus to correlate receptor binding with susceptibility of SARS-CoV infection. Hong Kong Med. J. 2012;18:35–38. [PubMed] [Google Scholar]
- Li H., Chen L., Xu Y., Wang Q., Wang Y., Zhang P., Liu Y., Li M., Yang Y., Zheng M. 2020. In Silico Analysis of Intermediate Hosts and Susceptible Animals of SARS-CoV-2. ChemRxiv, doi: 10.26434/chemrxiv.12057996.v1. [Google Scholar]
- Lu J., Cui J., Qian Z., Wang Y., Zhang H., Duan Y., Wu X., Yao X., Song Y., Li X. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Zhao Y., Yu W., Yang Y., Gao J., Wang J., Kuang D., Yang M., Yang J., Ma C. Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv. 2020 doi: 10.1101/2020.04.08.031807. [DOI] [Google Scholar]
- McGonagle D., O’Donnell J.S., Sharif K., Emery P., Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munley K.M., Rendon N.M., Demas G.E. Neural Androgen Synthesis and Aggression: Insights From a Seasonally Breeding Rodent. Front. Endocrinol. (Lausanne) 2018;9:136. doi: 10.3389/fendo.2018.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Karakida N., Dantsuka A., Ichii O., Elewa Y.H.A., Kon Y., Nagasaki K.I., Hattori H., Yoshiyasu T. Effects of a mixture of medetomidine, midazolam and butorphanol on anesthesia and blood biochemistry and the antagonizing action of atipamezole in hamsters. J. Vet. Med. Sci. 2017;79:1230–1235. doi: 10.1292/jvms.17-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Bertzbach L.D., Dietert K., Abdelgawad A., Vladimirova D., Kunec D., Hoffmann D., Beer M., Gruber A.D., Trimpert J. Age-Dependent Progression of SARS-CoV-2 Infection in Syrian Hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pach S., Ngoc Nguyen T., Trimpert J., Kunec D., Wolber G. ACE2-Variants Indicate Potential SARS-CoV-2-Susceptibility in Animals: An Extensive Molecular Dynamics Study. bioRxiv. 2020 doi: 10.1101/2020.05.14.092767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules C.I., Marston H.D., Fauci A.S. Coronavirus Infections-More Than Just the Common Cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M.A., Fentener van Vlissingen M., Rockx B., Haagmans B.L., Koopmans M.P.G. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherbarth F., Steinlechner S. Endocrine mechanisms of seasonal adaptation in small mammals: from early results to present understanding. J. Comp. Physiol. B. 2010;180:935–952. doi: 10.1007/s00360-010-0498-2. [DOI] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., Huang J., He J., Zhu A., Zhao J. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743.e5. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann D., Sperhake J.P., Lütgehetmann M., Steurer S., Edler C., Heinemann A., Heinrich F., Mushumba H., Kniep I., Schröder A.S. Autopsy Findings and Venous Thromboembolism in Patients With COVID-19: A Prospective Cohort Study. Ann. Intern. Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yu P., Qi F., Xu Y., Li F., Liu P., Liu J., Bao L., Deng W., Gao H., Xiang Z. Age-related rhesus macaque models of COVID-19. Animal Model. Exp. Med. 2020;3:93–97. doi: 10.1002/ame2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Tang Q., Cheng T., Xia N. Animal models for emerging coronavirus: progress and new insights. Emerg. Microbes Infect. 2020;9:949–961. doi: 10.1080/22221751.2020.1764871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai X., Sun J., Yan Z., Zhang J., Zhao J., Zhao Z., Gao Q., He W.T., Veit M., Su S. Comparison of SARS-CoV-2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J. Virol. 2020;94 doi: 10.1128/JVI.00831-20. e00831-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available at GenBank. The accession number for the Phodopus roborovskii ACE-2 sequence reported in this paper is Genbank: MW075232. The accession number for the Phodopus sungorus ACE-2 sequence reported in this paper is Genbank: MW075233.