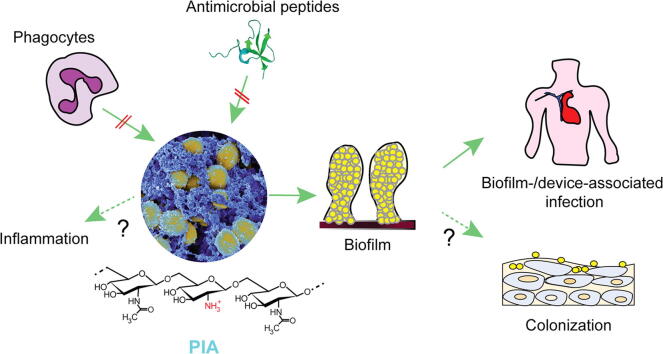
Graphical abstract
Abbreviations: CoNS, coagulase-negative staphylococci; PIA, polysaccharide intercellular adhesin; PNAG, poly-N-acetylglucosamine; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus
Keywords: Poly-N-acetylglucosamine, PNAG, PIA, Biofilm, Colonization, Polysaccharide intercellular adhesin, Staphylococcus epidermidis, Staphylococcus aureus, Device-related infection
Highlights
-
•
PIA is a key extracellular matrix component in staphylococci and other bacteria.
-
•
PIA is a cationic, partially deacetylated N-acetylglucosamine polymer.
-
•
PIA has a major role in bacterial biofilms and biofilm-associated infection.
Abstract
Exopolysaccharide is a key part of the extracellular matrix that contributes to important mechanisms of bacterial pathogenicity, most notably biofilm formation and immune evasion. In the human pathogens Staphylococcus aureus and S. epidermidis, as well as in many other staphylococcal species, the only exopolysaccharide is polysaccharide intercellular adhesin (PIA), a cationic, partially deacetylated homopolymer of N-acetylglucosamine, whose biosynthetic machinery is encoded in the ica locus. PIA production is strongly dependent on environmental conditions and controlled by many regulatory systems. PIA contributes significantly to staphylococcal biofilm formation and immune evasion mechanisms, such as resistance to antimicrobial peptides and ingestion and killing by phagocytes, and presence of the ica genes is associated with infectivity. Due to its role in pathogenesis, PIA has raised considerable interest as a potential vaccine component or target.
1. Introduction
The genus Staphylococcus comprises more than 40 species [1], of which at least 10 are found in the human skin microbiome [2]. Several species are among the most frequently isolated bacteria from the human skin and mucous membranes [3]. While many coagulase-negative staphylococci (CoNS), such as S. epidermidis, are skin colonizers in virtually all humans [3], the coagulase-positive species S. aureus persistently colonizes only ~20%, and intermittently about another 30% of the human population [4]. Many staphylococci are opportunistic pathogens with the ability to cause numerous infections. CoNS and S. aureus are both involved in subacute and chronic infections, particularly device-associated infections [5], while S. aureus can also cause severe lung, blood, and bone infections [6]. The success of the staphylococci in human colonization and infection is due to a plethora of factors. Similar to many other pathogenic bacteria [7], staphylococci produce extracellular polysaccharide (EPS), which has multiple functions in pathogenesis, including biofilm formation and immune evasion. The term EPS is used to differentiate from other bacterial polysaccharides, such as internal polysaccharides that have storage function, and capsular polysaccharides, which are also external but more closely related to the surface and usually covalently surface-linked [8].
Unlike some other bacteria, such as Pseudomonas aeruginosa, which have several types of EPS [9], staphylococci only produce one dominant EPS molecule [7]. This EPS has been named polysaccharide intercellular adhesin (PIA) based on function [10], or poly-β-1-6-N-acetylglucosamine (PNAG) based on its chemical nature [11]. We will use the original term PIA in this review. The genes necessary for PIA biosynthesis are encoded in the ica (intercellular adhesion) locus [12]. Like all Gram-positive bacteria, staphylococci also produce teichoic acids, which are polymers of sugars and alcohol phosphates and which – similar to EPS - have been implicated in colonization and biofilm formation [13]. However, due to their ubiquitous presence and covalent surface linkage, teichoic acids are generally not considered EPS.
PIA is also found in many other, phylogenetically diverse bacteria. In Escherichia coli, PIA is called PGA and the ica locus, pgaABCD [14]. In Yersinia pestis, Pseudomonas fluorescens, Bordetella bronchiseptica, B. pertussis, and B. parapertussis, homologues of pga were discovered, named hmsHFRS for Y. pestis and bpsABCD for Bordetella [15], [16], [17], [18], [19]. PIA homologues were shown to be directedly related to biofilm formation in these species [15], [16], [18], [19]. Similar findings were obtained in Acinetobacter baumannii [20], Actinobacillus actinomycetemcomitans and A. pleuropneumoniae [21], [22], Burkholderia ambifaria, B. cenocepacia, B. cepacia, B. multivorans and B. vietnamiensis [23], K. pneumoniae [24], [25] and Bacillus subtilis [26].
The wide distribution of PIA and its frequently established importance in infection has resulted in considerable interest in this molecule in recent years. While there are many reviews on biofilm formation, there is no comprehensive review on this specific key biofilm molecule. Here, we present a review of PIA in staphylococci, including its structure, biosynthesis and regulation, role in biofilm formation, colonization, and infection, and finish with a discussion of the potential of PIA-targeting therapeutics.
2. Distribution and genetic encoding of PIA in staphylococci
The production of PIA is mediated by the ica locus, which consists of a regulatory gene, icaR, and the biosynthetic operon icaADBC [12]. PIA and the ica locus were first described in S. epidermidis [12], [27] but then also found in S. aureus and other staphylococcal species with significant conservation [28], [29], [30]. Presence and expression of the icaADBC operon can vary significantly among the many staphylococcal species in which ica genes have been detected. For example, while most S. aureus strains have the ica genes [31], only some appear to rely on PIA expression for biofilm formation in vitro and in vivo [32], [33], [34]. In S. epidermidis, which has been in the focus of PIA research, recent findings indicate that presence of the ica genes is linked to a specific genetic cluster. Namely, ica genes are present in the S. epidermidis A/B cluster at ~37%, as opposed to only 4% in cluster B [35], [36]. Furthermore, the ica genes are virtually the only genes of S. epidermidis whose presence has been found to be significantly higher in isolates from device infection, or device-associated blood infection, versus colonization isolates [37], [38], [39], [40], [41], [42]. However, this association has been doubted [43], [44]. So far, other staphylococcal species have rarely been investigated for a correlation of ica gene presence, PIA production, and source from infection. In the species where this was analyzed, presence of the ica genes generally was highly strain-specific and associations with infection were similar to those found for S. epidermidis [45], [46], [47], [48], [49]. Altogether, it has been difficult to attribute roles in infection and colonization to the ica genes and their biosynthetic product solely based on epidemiological data, which is why functional research on this EPS molecule has focused on investigation of deletion strains and in some cases, purified PIA.
3. Structure of PIA
PIA was discovered when what was previously called slime underwent in-depth chemical analysis. In 1996, PIA from S. epidermidis was identified to be a linear, positively charged, partially (~15–20%) deacetylated polymer of β-1–6-N-acetylglucosamine [27], whose expression was highly correlated with biofilm formation [50]. Beside N-acetylation, around 10% of N-acetylglucosamine residues of PIA have been reported to be O-succinylated in S. epidermidis and S. aureus [27], [51], [52]. Before the chemical description of PIA by Mack et al. [27], preliminary studies had identified slime-associated staphylococcal polysaccharides that were given different names (SAA or PS/A) [53], [54]. Mack et al. described PIA to contain about 130 residues of N-acetylglucosamine (NAG) with some degree of deacetylation corresponding to an estimated molecular weight of ~30 kDa [27]. Subsequently, McKenney et al. identified PS/A from S. epidermidis and S. aureus as a >250 kDa molecule with considerable N-succinylation (65–100%) that is synthesized from the same locus as PIA (ica), ultimately calling it poly-N-succinyl-β-(1–6)-glucosamine (PNSG) [55], [56]. Later, the same group reported PS/A to have a size of 21 kDa, 100 kDa and 460 kDa but no degree of N-succinylation, henceforth calling the molecule PNAG [poly-N-acetyl-β-(1–6)-glucosamine] [11]. It was confirmed by detailed NMR analyses that N-succinylation was indeed an analytical artifact in a study that referred to S. aureus exopolysaccharide as SAE, a PIA-related molecule of high molecular weight (>300 kDa) having about 45–60% N-acetylation and 10% O-succinylation [51]. Notably, when using the same strain and growth condition as well as a similar purification strategy as used by Maira-Litran et al. [11], Sadovskaya et al. showed that PIA, PS/A, SAA and SAE are all of the same chemical entity [52]. Furthermore, all these molecules were shown to be synthesized by the ica locus [55]. Therefore, variable reports on the size and slightly different characteristics of PIA are likely due to differences in the degree of polymerization as well as variation in experimental approaches used in different studies (Fig. 1).
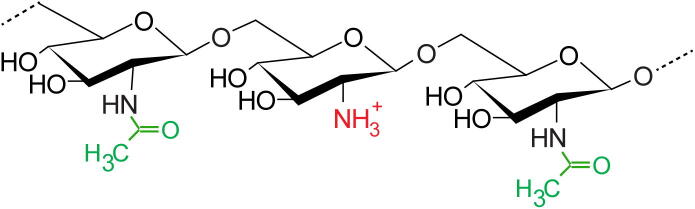
Fig. 1.
Structure of PIA. PIA is a homopolymer of N-acetylglucosamine (GlcNAc) residues with β-1–6 linkage. About 15 to 20% of the GlcNAc residues are de-acetylated. In the figure, acetyl groups are in green and the free amino group that results from IcaB-catalyzed deacetylation, which is positively charged at neutral or basic pH, is in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
There are reports on a similar glucosamine-containing EPS molecule in S. epidermidis of only 20 kDa, whose biosynthesis is not mediated by the ica locus [57]. This 20-kDa partially sulfated acidic polysaccharide was claimed to be both a major slime component and a distinct antigen with potential to induce specific and protective antibody in S. epidermidis [58], [59]. Later this molecule was reported to be a partially sulfated polymer of N-acetylglucosamine and glucose, expressed exclusively in S. epidermidis but not in other CoNS species [60]. It was stressed not to be synthesized from the ica locus [60], but in the absence of a defined biosynthetic locus, these reports on a second EPS molecule in staphylococci that is different from PIA have to be regarded as preliminary and in need of verification.
4. PIA biosynthesis
The icaADBC locus contains four different genes, icaA, icaD, icaB and icaC, which are arranged in an operon. The ica operon was first described in S. epidermidis in 1996 and reported to comprise three genes that are co-transcribed from one promoter [12]. Later, it was found that the locus also contains a small fourth gene, icaD, which is located between icaA and icaB [61]. Expression of all four genes is required for the synthesis of fully functional PIA [61] (Fig. 2).
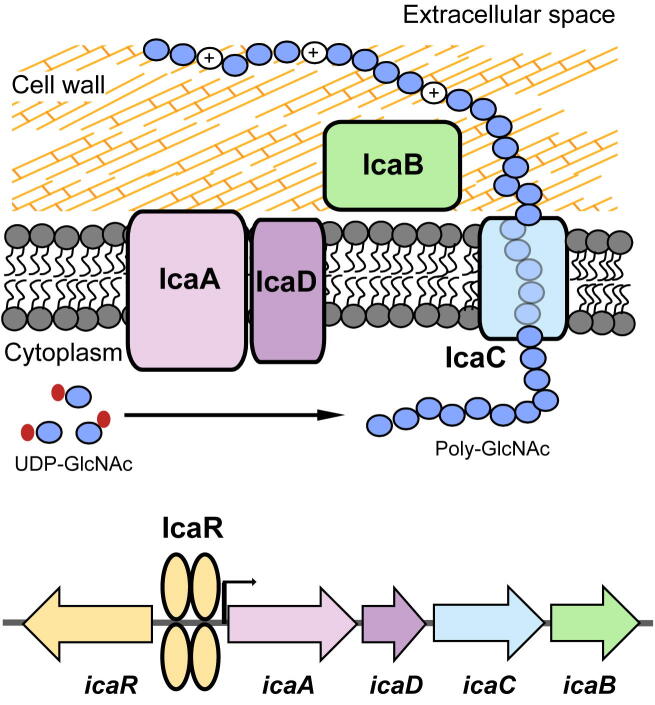
Fig. 2.
Genetic encoding and biosynthesis of PIA. PIA is synthesized by the products of the icaADBC operon. The icaADBC operon is under control of the product of the icaR gene, which is encoded upstream. IcaR, which is itself subject to control by manifold regulators and environmental conditions, binds in two dimers to the icaADBC promoter region, repressing icaADBC transcription. IcaA and IcaD, two membrane proteins, synthesize a growing poly-GlcNAc chain from activated precursor GlcNAc units. This chain is likely exported by the membrane protein IcaC, although IcaC has also, alternatively, been speculated to be involved in PIA O-succinylation. IcaB is an enzyme that is attached to the bacterial outer surface and introduces positive charges in the otherwise neutral PIA molecule by de-acetylation of some GlcNAc residues. The cationic character is vital for surface attachment and functionality of PIA.
The major PIA-synthesizing enzyme is encoded by icaA [61]. IcaA is an N-acetylglucosaminyltransferase that synthesizes PIA oligomers from UDP-N-acetylglucosamine. However, the transferase activity of IcaA is low and only reaches high efficacy in the presence of IcaD. IcaA and IcaD are located in the plasma membrane [61]. IcaA is a 412 amino-acid polypeptide having four predicted transmembrane domains, while IcaD is much smaller, having only 101 amino acids with two potential transmembrane domains [61]. IcaAD was shown to produce PIA with a maximal length of only 20 residues, while further elongation of PIA required assistance of IcaC [61]. Together with the predicted transmembrane structure of IcaC, these findings led to the assumption that IcaC exports the growing PIA chain and possibly forms a complex with IcaA and IcaD [61]. However, it has been proposed - based on comparison of ica homologues in different bacteria but without experimental evidence - that IcaAD may also export PIA, while IcaC may be responsible for modifications of PIA, such as O-succinylation, that appear to be limited to staphylococci [62]. IcaB, a 259 amino-acid polypeptide with a potential signal sequence, is a cell surface-attached enzyme that has PIA deacetylase activity [63]. Via deacetylation, IcaB introduces a positive net charge into PIA, which makes the polymer attach stably to the bacterial surface and which is crucial for PIA-mediated phenotypes [63]. IcaB enzymatic activity is metal-dependent and preferentially targets the second or third sugar residues from the reducing terminal of pentamer or hexamer PIA [64] (Fig. 2).
5. Regulation of PIA biosynthesis
PIA can be produced in large amounts in a presumably highly energy-consuming process [27]. This requires tight regulation of ica expression. PIA production and ica expression have been found to be dependent on environmental conditions, such as anaerobiosis, salt, glucose and alcohol concentration, and antibiotics [65], [66], [67], [68]. Over the years, a large number of regulatory genes and proteins have been found to regulate ica expression, which likely underlies the strongly differential expression of PIA in different staphylococcal strains [69]. It is believed that ica expression is more variant and dependent on environmental conditions in S. aureus than in S. epidermidis [70].
The ica locus contains a dedicated regulator, IcaR, which is encoded upstream of the icaADBC operon [66]. IcaR, whose crystal structure has been obtained, is a member of the TetR family of transcriptional regulators [71]. It binds to a specific DNA region upstream of icaA resulting in strong suppression of icaADBC transcription [66], [72] (Fig. 2). Deletion of icaR leads to PIA over-production [66]. Aminoglycoside antibiotics can interfere with the binding of IcaR to DNA, thus resulting in the induction of biofilm formation [71]. Some but not all of the environmental influences on PIA production as well as the impact of global regulators discussed below are mediated by IcaR. Interestingly, the 3′ untranslated region (UTR) interferes with the Shine-Dalgarno sequence of the icaR transcript, producing a substrate for RNAse III, thereby reducing icaR translation [73].
TcaR, a MarR-type transcriptional of the icaR transcript regulator, provides IcaR-independent regulation of icaADBC [74]. TcaR negatively regulates icaADBC, however to a much smaller extent than IcaR [72]. In S. epidermidis, TcaR can become the primary ica repressor in the absence of IcaR [74]. Interestingly, while IcaR binds to only one specific site upstream of icaA, TcaR can bind to multiple sites, including the binding site of IcaR as a competitor of IcaR as well as to the icaR promoter region as a repressor [74].
SarA is the most extensively studied regulator among the staphylococcal accessory regulator (Sar) family. This protein is relatively small, containing 124 amino acids with a winged-helix DNA binding domain [75]. SarA strongly activates the icaA promoter via high binding affinity [76], [77], [78]. In S. aureus, mutations in sarA decrease but do not stop the production of PIA [76], while in S. epidermidis, deletion of sarA can result in complete abortion of PIA production [77], [79]. Interestingly, in S. aureus, SarA induces not only the transcription of icaADBC but also its suppressor icaR, suggesting binary control to prevent the overproduction of PIA [78]. On the other hand, in S. epidermidis, SarA regulation of PIA production is IcaR-independent [77]. SarA represents a global regulator with manifold influences on staphylococcal physiology, many of which are mediated via its impact on the Agr quorum-sensing system [80], another major regulator of staphylococcal gene expression [81]. Agr itself does not impact ica transcription but similar to SarA impacts many unrelated biofilm factors such as proteases and phenol-soluble modulins (PSMs) [81], [82]. Overall, the impact of sarA and agr deletion on staphylococcal biofilm formation is negative and mostly PIA-independent, because both regulators strongly upregulate protease and PSMs, which are biofilm detachment factors [83]. Other members of the Sar protein family that regulate ica in S. epidermidis comprise SarX and SarZ. SarX binds to the icaADBC promoter, upregulating transcription, while SarZ also upregulates ica transcription in an unknown fashion [84], [85].
Sigma B is an alternative sigma factor that regulates a number of virulence and virulence-associated genes in response to environmental stimuli. It has been reported to be important for S. aureus and S. epidermidis biofilm formation. While initial studies reported that sigmaB increases ica transcription in a potentially IcaR-dependent way [78], [86], this has been controversial at least for S. aureus [76], and recent research suggests that the impact of sigma B on PIA production in S. aureus is due to altered proteolytic turnover of PIA biosynthesis proteins [87].
In addition to the regulators discussed in detail above, a number of other regulator factors/ systems have been shown to affect PIA synthesis, including Rbf [88], LuxS [89], Spx [90], SrrAB [91], Ygs [92], GdpS [93], and CcpA [94]. SrrAB, for example, appears to be important for the increase of PIA production under anerobic conditions [91]. Moreover, recent findings also add non-protein factors to the list of PIA regulators. IcaZ, a non-coding 400-nucleotide RNA, which is encoded downstream of icaR, was found to inhibit icaR mRNA translation, leading to increased PIA production [95]. IcaZ is found inclusively in ica-positive S. epidermidis but no other staphylococcal species [95]. Additionally, a regulatory RNA named RsaE binds in its processed form to the 5′UTR of the icaR mRNA, also increasing PIA production [96].
Finally, another distinctly different way to regulate PIA synthesis that was found in S. epidermidis is the reversible insertion of IS256 into either icaA, icaC, rsbU or sarA, which causes a “phase variation” phenotype of abolished or decreased PIA production [97], [98], [99], [100]. Similar to ica, IS256 is associated with infection origin of S. epidermidis isolates [101], [102], suggesting that this type of PIA regulation is important for pathogenesis.
6. Role of PIA in biofilm formation
Biofilm is a consortium of microbial cells that aggregate with each other and to a surface via a self-synthesized slimy extracellular matrix (ECM). This matrix is chemically heterogenous, comprising extracellular DNA (eDNA), lipids, EPS, and proteins that frequently form amyloid fibers [103]. The types and ratio of each component depend on the bacterial species and environmental conditions. In many staphylococci, particularly S. epidermidis, the EPS PIA is the major component of the biofilm matrix [12], [104] (Fig. 3). Biofilm formation develops in at least three main stages: (i) attachment of microbial cells to a surface, followed by (ii) production of the ECM and maturation of the biofilm, and finally (iii) detachment of microbial cells or clusters [105], [106].
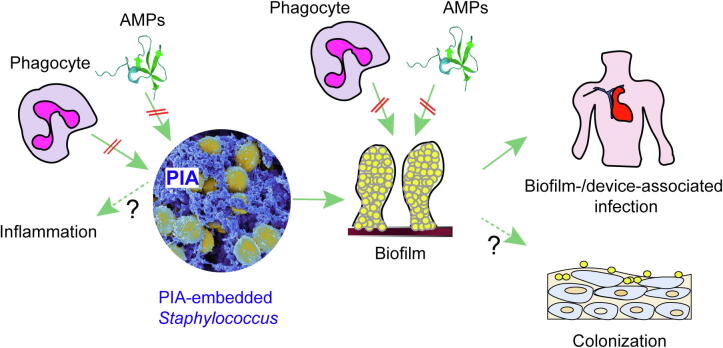
Fig. 3.
Functions of PIA. PIA embeds staphylococcal cells in a dense extracellular matrix network. This network protects the cells from attacks by mechanisms of innate host defense (AMPs, phagocytes). Furthermore, some reports have suggested direct pro-inflammatory functions of PIA. However, most of the biological functions of PIA are mediated by its contribution to biofilm formation. This includes most notably device- and other biofilm-associated infections. Biofilm formation also further contributes to the protection from AMPs and phagocytosis. Finally, PIA may contribute to epithelial colonization under specific conditions.
Attachment to an abiotic surface, such as that of an indwelling medical device, is governed by the physicochemical properties of the surface and the bacterial envelope and is reversible [107]. Studies mostly performed in S. epidermidis have attributed key roles to charge and surface hydrophobicity in staphylococcal attachment to abiotic surfaces [108], [109], [110]. However, in vivo, surface attachment is mediated predominantly via specific adhesion molecules, such as those of the MSCRAMM family, which cover the abiotic surface of an indwelling medical device soon after insertion [111].
Despite its positive charge, PIA appears to contribute to surface hydrophobicity of S. epidermidis [112] and may thus mediate initial adherence to some extent. However, the adherence properties often attributed to PIA in the literature [113] likely rather reflect its contribution to the beginning second, accumulation stage of biofilm formation. By representing a major component of the extracellular matrix, PIA fixes staphylococcal cells in the fibrous net it produces and thereby builds up biofilm mass [114], which leads to increased resistance of the biofilm to mechanical force. Accordingly, PIA is crucial for biofilm formation under high-shear flow conditions like those found inside catheters [40], [115], [116], [117], [118], [119], but it becomes less important under low-shear conditions like those in subcutaneously implanted tissue [120], ocular infections [121], or platelet concentrate [122]. Single-cell force spectroscopy data demonstrated multivalent electrostatic interaction of the cationic PIA polymer with the negatively charged wall teichoic acids on staphylococcal cells, confirming on the molecular level that the cationic character previously shown to be crucial for PIA function [63] has an important role in the attachment of PIA to the cell surface and PIA-mediated intercellular adhesion [123]. There is no evidence for a covalent linkage of PIA to the cell surface.
In the final stage of biofilm formation, staphylococcal cells are detached from the biofilm. Detachment can happen via mechanic force in a process often called sloughing, by enzymatic digestion via proteases or nucleases, or via detergents [124]. The role of the PSM detergent-like molecules in this process has been demonstrated in vivo in S. epidermidis and S. aureus and is independent of whether the biofilm is PIA-dependent or -independent [125], [126], [127], while enzymatic digestion depends on the chemical nature of the biofilm [128], [129], [130]. Notably, PIA-degrading enzymes have not been found in staphylococci. The only known enzyme to degrade PIA is dispersin B, which is found in a periodontal disease-causing pathogen, Actinobacillus actinomycetemcomitans [131]. This enzyme hydrolyzes the 1,4-β-glycosidic linkage of PIA, causing detachment and dispersion of cells from biofilms [131], effectively inhibiting PIA-dependent staphylococcal biofilm formation and immune evasion capacity [114], [132], [133]. It is often used to determine PIA dependence of staphylococcal biofilm formation [134]. Whether an open reading frame that has similarity to the dispersin B gene and is found in S. lugdunensis close to the ica locus codes for a PIA-hydrolase remains to be shown [135].
For a long time, PIA has been deemed crucial for staphylococcal biofilms, but beginning in the early 2000s, there have been reports on PIA-independent staphylococcal biofilm formation, in which isolates from biofilm infection were shown to be ica-negative and form in-vitro biofilms [33], [136], [137]. However, strains using PIA-independent biofilm formation seem to form weaker and less stable biofilm than those whose biofilms are based on PIA [33], [138]. Furthermore, PIA production results in dense, rough colonies as opposed to smooth colonies formed by PIA-negative biofilm-forming S. epidermidis [114]. It has also been reported that ica-negative and -positive clinical staphylococcal isolates show enhanced biofilm production when induced by heparin [139], staphylococcal or host proteases [134], trypsin [140] or by special conditions like those found within platelet concentrates [141], [142], [143]. Furthermore, PIA-dependent biofilm is frequently found in methicillin-sensitive (MSSA), while PIA-independent biofilm is prevalent in methicillin-resistant S. aureus (MRSA) [32], [136], [144]. There have been attempts to link this difference to the mecA gene that is responsible for methicillin resistance [145]. However, how the mecA gene is mechanistically involved in the difference of PIA usage for biofilm formation in MSSA and MRSA remains largely undetermined. Lastly, S. epidermidis appears to have the ability to switch to a protein-dependent biofilm upon disruption of the ica gene locus by IS256 [98].
7. Contribution to host colonization
Research on staphylococci has traditionally been focused on infection, while their commensal lifestyle has received only minor attention. This is now changing due to the increased interest in microbial communities and the human skin microbiome. The abundant skin commensal S. epidermidis has recently been shown to occur in two major genetic clusters (A/C and B), of which virtually only the A/C cluster isolates contain ica genes (37% versus 4% in cluster B) [35], [36]. A/C cluster isolates also exhibit ica-unrelated phenotypes, such as protease production and matrix protein binding, making them potentially more pathogenic, while B cluster isolates seem to have evolved to adapt to conditions found in sebaceous glands and hair follicles [35]. Furthermore, an earlier study performed in human volunteers showed that presence of the ica genes appeared to be disadvantageous for survival on the skin due to a high fitness cost [146]. Together with the many reports that associate presence of ica with infection, these results suggest that PIA production may only be of advantage on the skin under certain conditions and that presence of ica genes together with other genetic features makes specific S. epidermidis isolates more prone to infect the host. How PIA affects skin colonization in other staphylococci has not yet been investigated.
8. PIA in infection
Animal studies that analyzed the contribution of PIA to different types of infection have yielded conflicting results. Most frequently these studies investigated S. epidermidis device-related infection. Before the discovery of PIA, some studies reported an impact of slime production on the pathogenesis of S. epidermidis device-related infection [147] and an association with origin from infection, for example from nosocomial bacteremia [148], while others did not [148], [149]. In addition to the assumption that clumping/slime production increases the success of infections on indwelling medical devices, recent research also has suggested that this phenotype increases the chances of staphylococcal dissemination through the bloodstream [150].
After the discovery of the ica genes, isogenic deletion mutants were used to directly investigate the impact of PIA on infection. Most of those studies used the ica-negative M10 transposon mutant of S. epidermidis strain 1457 [10]. The first studies were performed in the Rupp laboratory and consistently showed a significant impact of the ica genes on catheter-related infection in mice and rats [117], [151], [152]. Later, it was shown that introduction of the ica genes alone is sufficient to render a commensal S. epidermidis strain invasive [153], and several further studies showed similarly reduced infectivity of isogenic ica-negative S. epidermidis as compared to the parental strain in device-related infection [154], independently of the used biomaterial [155]. Moreover, the importance of PIA deacetylation for device-related infection in mice, as investigated using an icaB isogenic deletion mutant, further confirmed PIA’s importance for pathogenesis [63]. Additionally, ica-positive S. aureus or S. epidermidis showed better in-vivo survival than their corresponding ica mutants in wild-type/mutant mouse co-infection models [70]. Finally, a significant impact of the ica genes on S. epidermidis infection was confirmed in a C. elegans infection model, where ica genes were required for lethal infection produced by feeding challenge [156]. Together, these results add to those already mentioned above showing increased prevalence of ica genes in infective S. epidermidis isolates to substantiate a role of ica in S. epidermidis device-related infection [37], [38], [39], [40], [41], [42] (Fig. 3).
However, other researchers found no impact of the ica genes on virulence in device-related infection models. Chokr et al. reported a lack of impact of ica on infection in a guinea pig tissue cage model for S. epidermidis and Francois et al. for both S. epidermidis and S. aureus in the same model [120], [157]. Kristian et al. reported a similar outcome when using a mouse tissue cage model and S. aureus strain SA113 [158]. Furthermore, in a C. elegans infection model no correlation of PIA-production and virulence was found comparing 30 S. epidermidis isolates from infective endocarditis [159]. The most likely reason for the conflicting reports as for the impact of ica on virulence is a differential relative effect of PIA as compared to other staphylococcal virulence factors and dependence on strains and models used. Interestingly, at least two of the three strains that were used by Francois et al. and Kristian et al. [120], [158] are Agr-dysfunctional (S. epidermidis O47, S. aureus SA113) [82], [160], suggesting that the resulting complete absence of PSM production [161] and concomitant increased compact biofilm [125], [126], [127] abrogates a measurable impact of PIA on biofilm expansion.
In-vivo investigations on the impact of ica on infection in staphylococcal species other than S. epidermidis are generally rather scarce. PIA is produced by S. aureus in vivo and significantly impacts S. aureus systemic infection in mice [162] despite variability and strain-dependence of in-vitro production [70], [163]. As for device-related infection, the abovementioned studies that did not find a role for S. aureus ica are the only studies that have been performed [120], [158]. Interestingly, loss of PIA in an S. aureus strain that overproduces PIA due to a mutation in the icaADBC promoter [164] produces a fitness gain by a compensatory mutation that was also detected in clinical isolates; however, this was only determined in vitro [165]. Later, the same mutation that leads to PIA overproduction and an associated immunoprotective “mucoid” phenotype was detected in S. aureus isolates from cystic fibrosis patients, in which also similar compensatory mutations with a non-mucoid phenotype occurred over time [166]. Altogether, these findings suggest that PIA production, while likely important for device-associated infection and associated with a high fitness cost in S. epidermidis, is subject to dynamic alterations in production, especially in vivo and in S. aureus.
During co-infection with other organisms, PIA may play a role to increase overall virulence. This was shown for coinfection with Candida albicans, a pathogenic fungus that often occurs together with staphylococci in catheter-related infection, wound infection, cystic fibrosis, periodontitis and denture stomatitis [167]. In mixed in-vitro biofilms of S. epidermidis strain RP62A and C. albicans, slime, which is mainly composed of PIA in that strain [52], protected C. albicans from fluconazole penetration [168]. EPS produced by S. epidermidis also increased the overall virulence of a mixed S. epidermidis and C. albicans challenge in C. elegans, resulting in reduced survival of the infected worms [169]. In S. aureus, MSSA and MRSA grew synergistically with C. albicans within biofilms [170], and C. albicans increased S. aureus resistance to vancomycin [171], suggesting mutual benefit.
9. PIA and the host immune system
There are multiple studies that have investigated the role of PIA in the interaction with the immune system (Fig. 3). In cell culture assays, the ica-negative mutant M10 was more susceptible to antimicrobial peptides (human beta-defensin 3, LL-37 and dermcidin) and to non-opsonic phagocytosis and killing by human polymorphonuclear leucocytes (PMNs) than the parental strain S. epidermidis 1457 [104]. PIA-mediated resistance to opsonic PMN killing was shown in another study by Kristian et al., which also demonstrated diminished immunoglobulin and complement (C3b) deposition on the surface of ica-positive S. epidermidis biofilms in a device-related infection model in addition to increased local infection, bacterial burden, and larger edema [154]. Furthermore, PIA-producing S. epidermidis biofilm was shown to lead to less pronounced granulocyte activation and cytokine release than the reduced biofilm produced by its isogenic ica mutant [172]. Finally, PIA was shown to decrease susceptibility to phagocytosis by macrophages [114] and restoration of PIA production in the M10 mutant resulted in reduced NF-kB activation and diminished IL-1β production in macrophages [114]. In S. aureus, depletion of PIA resulted in increased IL-12 production in murine dendritic cells [173], decreased blood CFU in intravenously challenged mice and increased complement-mediated phagocytic killing [162]. Together, these results indicate that most of the effects of PIA production on immune evasion are mediated by its impact on biofilm formation, which shelters the cells from recognition by phagocytes and from killing by antimicrobial peptides. Vuong et al. [104] investigated single cells after biofilm disruption and found similar immune evasion effects, suggesting that cellular “coating” with PIA provides immune evasion properties also in the absence of a biofilm, which thus may play a role also in non-biofilm-related, acute infection.
On the other hand, several ex-vivo and in-vivo studies reported increased inflammatory reactions to PIA-positive versus isogenic PIA-negative bacteria. Fredheim et al. showed increased complement activation ex vivo [172], and Ferreirinha et al. increased neutrophil recruitment in vivo by PIA-positive strains. Al-Ishaq et al. reported association of C5a concentration with PIA mode of biofilm formation in clinical samples [174]. These effects are likely due to higher bacterial survival and more pronounced infection that PIA producers cause via their above-mentioned immune evasion effects. Additionally, when assessing altered pro-inflammatory effects of PIA-negative mutants, one should keep in mind that such effects may be due to differential release of the strongly pro-inflammatory PSMs, or lipopeptides, whose release is PSM-dependent [175], in a biofilm setting [125], [126], rather than direct effects of PIA itself.
Finally, based on investigation using purified PIA, it has been suggested that the PIA molecule is itself pro-inflammatory [172], [174], [176]. For example, studies using incubation of purified PIA with human astrocytes implicated that PIA can induce IL-6, IL-8, and MCP-1 expression via TLR-2 [176]. However, the purification of PIA is difficult, making it hard to rule out effects by contaminating strongly pro-inflammatory molecules, which is why further verification of the pro-inflammatory capacities of the PIA molecule is certainly warranted.
10. PIA and antimicrobial resistance
Biofilm formation is widely known to decrease susceptibility to antibiotics and other antimicrobial agents [177]. The underlying mechanisms comprise a reduced metabolic state, persister formation, and decreased penetration through the biofilm extracellular matrix, among others [178]. As for staphylococci, oxacillin, cefotaxime and vancomycin reportedly penetrate poorly through S. aureus and S. epidermidis biofilms [179], while some other antibiotics, such as amikacin and ciprofloxacin, were unaffected by staphylococcal biofilm formation [179]. While it has also been reported that rifampin and vancomycin have at least some capacity to penetrate through the biofilm matrix [180], [181], [182], their antibacterial efficacy was shown to depend on biofilm age or infection duration [183], or concentration and conditions [184], [185], respectively.
As PIA is part of the extracellular matrix, it is reasonable to assume that it mostly affects antibiotics whose penetration through the biofilm matrix is impaired. However, due to its essentiality for biofilm formation in many isolates, PIA may also theoretically impact the activity of antibiotics that easily penetrate through the matrix. In correlative studies, ica-positive S. epidermidis and S. aureus strains showed increased resistance as compared to ica-negative strains to a variety of antibiotics, such as oxacillin, gentamicin, ciprofloxacin, levofloxacin, co-trimoxazole, erythromycin, vancomycin, and the cell-wall degrading enzyme lysostaphin [186], [187], [188], [189]. It is also noteworthy that subinhibitory concentrations of some antibiotics can increase transcription of the ica locus; yet the underlying mechanisms are not understood. This was found for tetracycline and quinupristin-dalfopristin, and to some extent erythromycin, while most antibiotics tested did not show such an effect [65]. Finally, PIA may enhance horizontal gene transfer via its impact on biofilm formation, inasmuch as plasmid transfer by conjugation in S. aureus was observed to be 10,000 times higher in biofilm than in planktonic states, which can be explained by increased cell-to-cell contact in biofilms [190].
11. PIA as an immunotherapeutic target
With surface location representing a key prerequisite of a vaccine target, PIA, as an important surface-located biofilm component, was early considered as a potential vaccine candidate. However, the immunogenicity of polysaccharides is generally low [191]; and presence of capsule or EPS generally represents an immune evasion mechanism by which the bacteria minimize opsonization [192]. Nevertheless, anti-PIA antisera may overcome such limitation if they are highly reactive. In the 1990s, immunization of rabbits with PIA [capsular polysaccharide/adhesin (PS/A)] was shown to reduce disease severity in rabbit models of catheter-related S. epidermidis bacteremia and endocarditis [193], [194]. ELISA and immunoelectron microscopy data also clearly indicated adsorption of anti-PIA antibodies by various PIA-positive staphylococcal strains [55]. A PIA-based vaccine was then developed and showed protective effects in mice against kidney infection and death caused by S. aureus strains Reynolds and MN8 [56], which interestingly produced undetectable levels of PIA in vitro [56]. Furthermore, when PIA was conjugated with diphtheria toxoid (DT), the vaccinated mice or rabbits produced significant anti-PIA antibody titers [195]. The obtained anti-PIA antibodies opsonized and induced killing of various staphylococcal strains and their transfusion cleared S. aureus from mouse blood [195]. Importantly, the conjugated deacetylated PIA (85% deacetylation)/DT was markedly more effective as a vaccine than native PIA (15% deacetylation)/DT [195]. The stronger potential of deacetylated PIA (>75% deacetylation, dPIA) compared to native PIA in inducing protective antibodies was again shown in a later study, in which dPIA was conjugated with tetanus toxoid (TT) for immunization [196], eliciting anti-PIA antibodies in mice and rabbits, mediating opsonic killing of various S. aureus strains and E. coli, and protecting the animals from skin abscess caused by S. aureus and peritonitis caused by E. coli [196]. The stronger potential of dPIA compared to native PIA in protecting challenged animals is likely due to the increase of surface attachment of PIA following deacetylation [63], [197] and may explain why natural antibody against native PIA is unable to trigger protective effects despite being common in human and animals [198], [199], [200]. In another study, a PIA vaccine in form of a bacterin preparation resulted in high production of anti-PIA antibodies and significant protection against S. aureus infection and mastitis in sheep [201]. Furthermore, PIA was expressed in E. coli in outer membrane vesicles (OMVs) together with staphylococcal IcaB and the produced PIA-decorated OMVs were highly immunogenic and protected mice from infection not only by S. aureus but also the PIA-positive Francisella tularensis subsp. holarctica [202].
PIA has also been combined with other molecules in vaccines. For example, covalent conjugation of dPIA to clumping factor A (ClfA), but not a mixture of the two unconjugated molecules, was highly immunogenic in mice, rabbits, goats and rhesus monkeys [203]. Transfusion of goat antisera to dPIA-ClfA vaccine to mice significantly reduced blood CFU of different S. aureus strains [203]. When glycerol teichoic acid (Gly-TA) and PIA were used to immunize mice, both anti-Gly-TA and anti-PIA antibodies were obtained and the anti-Gly-TA/-PIA sera were able to inhibit biofilm formation of S. epidermidis and S. aureus in vitro significantly better than anti-Gly-TA or anti-PIA sera alone [204]. Combination of PIA and recombinant SesC protein as a conjugated vaccine induced the production of opsonic antibodies, suppressed biofilm production and protected mice from intravenous challenge with S. epidermidis [205].
Despite multiple promising results in animal models, the clinical potential of a PIA vaccine remains somewhat questionable because of the limited prevalence of ica in several clinically important staphylococci, such as S. epidermidis, and the varying expression of PIA. However, due to the fact that there is no effective S. aureus vaccine despite numerous attempts, it may be worth to further invest in PIA-based vaccine development [206], particularly as PIA immunization may be valuable for infections also by Streptococcus pneumoniae [207], Rhodococcus equi [208], and multiple other species [209]. Finally, immunization against PIA only affects pathogens but leaves microbial diversity virtually unaffected [210].
12. Summary and outlook
Despite increasing reports in the last 20 years on PIA-independent biofilm formation, PIA is still recognized as a major biofilm component particularly in S. epidermidis, many other CoNS, and in MSSA. It contributes to immune evasion via its biofilm-forming ability and possibly even independently of it, and affects several directly and indirectly biofilm-related infection types.
Important open questions comprise PIA’s role in colonization and how this is related to the association of ica gene presence with specific clades. As for PIA’s role in infection, a thorough investigation of its contribution especially to the many different types of S. aureus infection, relative to the contribution of other virulence factors, is warranted. Furthermore, it should be analyzed what the precise function of IcaC is and whether and how the PIA molecule has direct pro-inflammatory effects. Finally, given the problems with obtaining a working S. aureus vaccine, PIA should not be given up on as a vaccine component or target.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), U.S. National Institutes of Health (NIH).
References
- 1.Schleifer K.H., Bell J. Staphylococcus. In: Whitman W.B., editor. Bergey's Manual of Systematics of Archaea and Bacteria. Wiley; 2015. [Google Scholar]
- 2.Grice E.A., Segre J.A. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloos W.E., Musselwhite M.S. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30(3):381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kluytmans J.A., Wertheim H.F. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005;33(1):3–8. doi: 10.1007/s15010-005-4012-9. [DOI] [PubMed] [Google Scholar]
- 5.Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004;9:841–863. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- 6.Lowy F.D. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 7.Joo H.S., Otto M. Molecular basis of in vivo biofilm formation by bacterial pathogens. Chem Biol. 2012;19(12):1503–1513. doi: 10.1016/j.chembiol.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid J., Sieber V., Rehm B. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol. 2015;6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryder C., Byrd M., Wozniak D.J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10(6):644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack D. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62(8):3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maira-Litran T. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70(8):4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heilmann C. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 13.Weidenmaier C., Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6(4):276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 14.Wang X., Preston J.F., 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186(9):2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobrov A.G. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ Microbiol. 2008;10(6):1419–1432. doi: 10.1111/j.1462-2920.2007.01554.x. [DOI] [PubMed] [Google Scholar]
- 16.Itoh Y. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187(1):382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parise G. Role of a putative polysaccharide locus in Bordetella biofilm development. J Bacteriol. 2007;189(3):750–760. doi: 10.1128/JB.00953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganguly T. The Bordetella pertussis Bps polysaccharide enhances lung colonization by conferring protection from complement-mediated killing. Cell Microbiol. 2014;16(7):1105–1118. doi: 10.1111/cmi.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson T.L. The bordetella bps polysaccharide is required for biofilm formation and enhances survival in the lower respiratory tract of swine. Infect Immun. 2017;85(8) doi: 10.1128/IAI.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi A.H. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191(19):5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan J.B. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186(24):8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izano E.A. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog. 2007;43(1):1–9. doi: 10.1016/j.micpath.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yakandawala N. Characterization of the poly-beta-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl Environ Microbiol. 2011;77(23):8303–8309. doi: 10.1128/AEM.05814-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K.M. The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb Pathog. 2014;77:89–99. doi: 10.1016/j.micpath.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Lery L.M. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. doi: 10.1186/1741-7007-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roux D. Identification of Poly-N-acetylglucosamine as a major polysaccharide component of the bacillus subtilis biofilm matrix. J Biol Chem. 2015;290(31):19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mack D. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammendolia M.G. Slime production and expression of the slime-associated antigen by staphylococcal clinical isolates. J Clin Microbiol. 1999;37(10):3235–3238. doi: 10.1128/jcm.37.10.3235-3238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cramton S.E. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moretro T. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl Environ Microbiol. 2003;69(9):5648–5655. doi: 10.1128/AEM.69.9.5648-5655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohde H. Correlation of Staphylococcus aureus icaADBC genotype and biofilm expression phenotype. J Clin Microbiol. 2001;39(12):4595–4596. doi: 10.1128/JCM.39.12.4595-4596.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Neill E. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45(5):1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohde H. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28(9):1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 34.Toledo-Arana A. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187(15):5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espadinha D. Distinct phenotypic and genomic signatures underlie contrasting pathogenic potential of staphylococcus epidermidis clonal lineages. Front Microbiol. 2019;10:1971. doi: 10.3389/fmicb.2019.01971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meric G. Disease-associated genotypes of the commensal skin bacterium Staphylococcus epidermidis. Nat Commun. 2018;9(1):5034. doi: 10.1038/s41467-018-07368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galdbart J.O. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J Infect Dis. 2000;182(1):351–355. doi: 10.1086/315660. [DOI] [PubMed] [Google Scholar]
- 38.Yao Y. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect Immun. 2005;73(3):1856–1860. doi: 10.1128/IAI.73.3.1856-1860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebuhr W. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 1997;65(3):890–896. doi: 10.1128/iai.65.3.890-896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arciola C.R., Baldassarri L., Montanaro L. In catheter infections by Staphylococcus epidermidis the intercellular adhesion (ica) locus is a molecular marker of the virulent slime-producing strains. J Biomed Mater Res. 2002;59(3):557–562. doi: 10.1002/jbm.10006. [DOI] [PubMed] [Google Scholar]
- 41.Frebourg N.B. PCR-Based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J Clin Microbiol. 2000;38(2):877–880. doi: 10.1128/jcm.38.2.877-880.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherifi S. Comparative epidemiology of Staphylococcus epidermidis isolates from patients with catheter-related bacteremia and from healthy volunteers. J Clin Microbiol. 2013;51(5):1541–1547. doi: 10.1128/JCM.03378-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohde H. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J Clin Microbiol. 2004;42(12):5614–5619. doi: 10.1128/JCM.42.12.5614-5619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris L.G. Biofilm morphotypes and population structure among staphylococcus epidermidis from commensal and clinical samples. PLoS One. 2016;11(3) doi: 10.1371/journal.pone.0151240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barros E.M. Phenotypic and genotypic characterization of biofilm formation in Staphylococcus haemolyticus. Curr Microbiol. 2015;70(6):829–834. doi: 10.1007/s00284-015-0794-x. [DOI] [PubMed] [Google Scholar]
- 46.Szczuka E., Telega K., Kaznowski A. Biofilm formation by Staphylococcus hominis strains isolated from human clinical specimens. Folia Microbiol (Praha) 2015;60(1):1–5. doi: 10.1007/s12223-014-0332-4. [DOI] [PubMed] [Google Scholar]
- 47.Giormezis N. Virulence factors among Staphylococcus lugdunensis are associated with infection sites and clonal spread. Eur J Clin Microbiol Infect Dis. 2015;34(4):773–778. doi: 10.1007/s10096-014-2291-8. [DOI] [PubMed] [Google Scholar]
- 48.Tseng S.P. Genotypes and phenotypes of Staphylococcus lugdunensis isolates recovered from bacteremia. J Microbiol Immunol Infect. 2015;48(4):397–405. doi: 10.1016/j.jmii.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Dimitriou G. Clinical and microbiological profile of persistent coagulase-negative staphylococcal bacteraemia in neonates. Clin Microbiol Infect. 2011;17(11):1684–1690. doi: 10.1111/j.1469-0691.2011.03489.x. [DOI] [PubMed] [Google Scholar]
- 50.Mack D. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis. 1996;174(4):881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- 51.Joyce J.G. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr Res. 2003;338(9):903–922. doi: 10.1016/s0008-6215(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 52.Sadovskaya I. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect Immun. 2005;73(5):3007–3017. doi: 10.1128/IAI.73.5.3007-3017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tojo M. Isolation and characterization of a capsular polysaccharide adhesin from Staphylococcus epidermidis. J Infect Dis. 1988;157(4):713–722. doi: 10.1093/infdis/157.4.713. [DOI] [PubMed] [Google Scholar]
- 54.Baldassarri L. Purification and characterization of the staphylococcal slime-associated antigen and its occurrence among Staphylococcus epidermis clinical isolates. Infect Immun. 1996;64(8):3410–3415. doi: 10.1128/iai.64.8.3410-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenney D. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect Immun. 1998;66(10):4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKenney D. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284(5419):1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
- 57.Arvaniti A. Isolation and characterization of a novel 20-kDa sulfated polysaccharide from the extracellular slime layer of Staphylococcus epidermidis. Arch Biochem Biophys. 1994;308(2):432–438. doi: 10.1006/abbi.1994.1061. [DOI] [PubMed] [Google Scholar]
- 58.Karamanos N.K. The major 20-kDa polysaccharide of Staphylococcus epidermidis extracellular slime and its antibodies as powerful agents for detecting antibodies in blood serum and differentiating among slime-positive and -negative S. epidermidis and other staphylococci species. Arch Biochem Biophys. 1997;342(2):389–395. doi: 10.1006/abbi.1997.0107. [DOI] [PubMed] [Google Scholar]
- 59.Georgakopoulos C.D. Immunization with specific polysaccharide antigen reduces alterations in corneal proteoglycans during experimental slime-producing Staphylococcus epidermidis keratitis. Curr Eye Res. 2006;31(2):137–146. doi: 10.1080/02713680500516540. [DOI] [PubMed] [Google Scholar]
- 60.Spiliopoulou A.I. An extracellular Staphylococcus epidermidis polysaccharide: relation to Polysaccharide Intercellular Adhesin and its implication in phagocytosis. BMC Microbiol. 2012;12:76. doi: 10.1186/1471-2180-12-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gerke C. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273(29):18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 62.Atkin K.E. A different path: revealing the function of staphylococcal proteins in biofilm formation. FEBS Lett. 2014;588(10):1869–1872. doi: 10.1016/j.febslet.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Vuong C. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 64.Pokrovskaya V. Functional characterization of Staphylococcus epidermidis IcaB, a de-N-acetylase important for biofilm formation. Biochemistry. 2013;52(32):5463–5471. doi: 10.1021/bi400836g. [DOI] [PubMed] [Google Scholar]
- 65.Rachid S. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob Agents Chemother. 2000;44(12):3357–3363. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conlon K.M., Humphreys H., O'Gara J.P. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J Bacteriol. 2002;184(16):4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cramton S.E. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(6):4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dobinsky S. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J Bacteriol. 2003;185(9):2879–2886. doi: 10.1128/JB.185.9.2879-2886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cue D., Lei M.G., Lee C.Y. Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect Microbiol. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fluckiger U. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect Immun. 2005;73(3):1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeng W.Y. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucl Acids Res. 2008;36(5):1567–1577. doi: 10.1093/nar/gkm1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jefferson K.K. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz de los Mozos I. Base pairing interaction between 5'- and 3'-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 2013;9(12) doi: 10.1371/journal.pgen.1004001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoang T.M. Transcriptional Regulation of icaADBC by both IcaR and TcaR in Staphylococcus epidermidis. J Bacteriol. 2019;201(6) doi: 10.1128/JB.00524-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheung A.L. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40(3):355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valle J. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48(4):1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 77.Tormo M.A. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J Bacteriol. 2005;187(7):2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cerca N., Brooks J.L., Jefferson K.K. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J Bacteriol. 2008;190(19):6530–6533. doi: 10.1128/JB.00482-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Handke L.D. SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can J Microbiol. 2007;53(1):82–91. doi: 10.1139/w06-108. [DOI] [PubMed] [Google Scholar]
- 80.Heinrichs J.H., Bayer M.G., Cheung A.L. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178(2):418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le K.Y., Otto M. Quorum-sensing regulation in staphylococci-an overview. Front Microbiol. 2015;6:1174. doi: 10.3389/fmicb.2015.01174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vuong C. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J Infect Dis. 2003;188(5):706–718. doi: 10.1086/377239. [DOI] [PubMed] [Google Scholar]
- 83.Le K.Y. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front Cell Infect Microbiol. 2014;4:167. doi: 10.3389/fcimb.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L. SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J Infect Dis. 2008;197(9):1254–1262. doi: 10.1086/586714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rowe S.E. A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology. 2011;157(Pt 4):1042–1049. doi: 10.1099/mic.0.046581-0. [DOI] [PubMed] [Google Scholar]
- 86.Knobloch J.K. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect Immun. 2004;72(7):3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Valle J., Echeverz M., Lasa I. sigma(B), inhibits poly-N-acetylglucosamine exopolysaccharide synthesis and biofilm formation in staphylococcus aureus. J Bacteriol. 2019;201(11) doi: 10.1128/JB.00098-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cue D. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191(20):6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu L. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pamp S.J. Spx is a global effector impacting stress tolerance and biofilm formation in Staphylococcus aureus. J Bacteriol. 2006;188(13):4861–4870. doi: 10.1128/JB.00194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ulrich M. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol. 2007;65(5):1276–1287. doi: 10.1111/j.1365-2958.2007.05863.x. [DOI] [PubMed] [Google Scholar]
- 92.Wang X. ygs is a novel gene that influences biofilm formation and the general stress response of Staphylococcus epidermidis. Infect Immun. 2011;79(3):1007–1015. doi: 10.1128/IAI.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holland L.M. A staphylococcal GGDEF domain protein regulates biofilm formation independently of cyclic dimeric GMP. J Bacteriol. 2008;190(15):5178–5189. doi: 10.1128/JB.00375-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Seidl K. Staphylococcus aureus CcpA affects biofilm formation. Infect Immun. 2008;76(5):2044–2050. doi: 10.1128/IAI.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lerch M.F. A non-coding RNA from the intercellular adhesion (ica) locus of Staphylococcus epidermidis controls polysaccharide intercellular adhesion (PIA)-mediated biofilm formation. Mol Microbiol. 2019;111(6):1571–1591. doi: 10.1111/mmi.14238. [DOI] [PubMed] [Google Scholar]
- 96.Schoenfelder S.M.K. The small non-coding RNA RsaE influences extracellular matrix composition in Staphylococcus epidermidis biofilm communities. PLoS Pathog. 2019;15(3) doi: 10.1371/journal.ppat.1007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conlon K.M., Humphreys H., O'Gara J.P. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J Bacteriol. 2004;186(18):6208–6219. doi: 10.1128/JB.186.18.6208-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hennig S., Nyunt Wai S., Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive Staphylococcus epidermidis isolate. Int J Med Microbiol. 2007;297(2):117–122. doi: 10.1016/j.ijmm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 99.Valle J. sigmaB regulates IS256-mediated Staphylococcus aureus biofilm phenotypic variation. J Bacteriol. 2007;189(7):2886–2896. doi: 10.1128/JB.01767-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ziebuhr W. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32(2):345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 101.Kozitskaya S. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004;72(2):1210–1215. doi: 10.1128/IAI.72.2.1210-1215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gu J. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J Hosp Infect. 2005;61(4):342–348. doi: 10.1016/j.jhin.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 103.Hobley L. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39(5):649–669. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vuong C. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6(3):269–275. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 105.Tolker-Nielsen T. Biofilm development. Microbiol Spectr. 2015;3(2) doi: 10.1128/microbiolspec.MB-0001-2014. p. MB-0001-2014. [DOI] [PubMed] [Google Scholar]
- 106.Otto M. Staphylococcal Biofilms. Microbiol Spectr. 2018;6:4. doi: 10.1128/microbiolspec.gpp3-0023-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Carniello V. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv Colloid Interface Sci. 2018;261:1–14. doi: 10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 108.Hogt A.H. Cell surface characteristics of coagulase-negative staphylococci and their adherence to fluorinated poly(ethylenepropylene) Infect Immun. 1986;51(1):294–301. doi: 10.1128/iai.51.1.294-301.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu Y., Strauss J., Camesano T.A. Adhesion forces between Staphylococcus epidermidis and surfaces bearing self-assembled monolayers in the presence of model proteins. Biomaterials. 2008;29(33):4374–4382. doi: 10.1016/j.biomaterials.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 110.Patel J.D. S. epidermidis biofilm formation: effects of biomaterial surface chemistry and serum proteins. J Biomed Mater Res A. 2007;80(3):742–751. doi: 10.1002/jbm.a.31103. [DOI] [PubMed] [Google Scholar]
- 111.Foster T.J. The MSCRAMM family of cell-wall-anchored surface proteins of gram-positive cocci. Trends Microbiol. 2019;27(11):927–941. doi: 10.1016/j.tim.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 112.Nuryastuti T., Krom B.P. Ica-status of clinical Staphylococcus epidermidis strains affects adhesion and aggregation: a thermodynamic analysis. Antonie Van Leeuwenhoek. 2017;110(11):1467–1474. doi: 10.1007/s10482-017-0899-2. [DOI] [PubMed] [Google Scholar]
- 113.Olson M.E. Adherence of Staphylococcus epidermidis to biomaterials is augmented by PIA. Clin Orthop Relat Res. 2006;451:21–24. doi: 10.1097/01.blo.0000229320.45416.0c. [DOI] [PubMed] [Google Scholar]
- 114.Schommer N.N. Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect Immun. 2011;79(6):2267–2276. doi: 10.1128/IAI.01142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schaeffer C.R. Versatility of biofilm matrix molecules in staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere. 2016;1(5) doi: 10.1128/mSphere.00165-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chaieb K., Mahdouani K., Bakhrouf A. Detection of icaA and icaD loci by polymerase chain reaction and biofilm formation by Staphylococcus epidermidis isolated from dialysate and needles in a dialysis unit. J Hosp Infect. 2005;61(3):225–230. doi: 10.1016/j.jhin.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 117.Rupp M.E. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect Immun. 1999;67(5):2656–2659. doi: 10.1128/iai.67.5.2656-2659.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Foka A. The combined effect of surface chemistry and flow conditions on Staphylococcus epidermidis adhesion and ica operon expression. Eur Cell Mater. 2012;24:386–402. doi: 10.22203/ecm.v024a28. [DOI] [PubMed] [Google Scholar]
- 119.Weaver W.M. Fluid flow induces biofilm formation in Staphylococcus epidermidis polysaccharide intracellular adhesin-positive clinical isolates. Appl Environ Microbiol. 2012;78(16):5890–5896. doi: 10.1128/AEM.01139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Francois P. Lack of biofilm contribution to bacterial colonisation in an experimental model of foreign body infection by Staphylococcus aureus and Staphylococcus epidermidis. FEMS Immunol Med Microbiol. 2003;35(2):135–140. doi: 10.1016/S0928-8244(02)00463-7. [DOI] [PubMed] [Google Scholar]
- 121.Juarez-Verdayes M.A. Staphylococcus epidermidis with the icaA(-)/icaD(-)/IS256(-) genotype and protein or protein/extracellular-DNA biofilm is frequent in ocular infections. J Med Microbiol. 2013;62(Pt 10):1579–1587. doi: 10.1099/jmm.0.055210-0. [DOI] [PubMed] [Google Scholar]
- 122.Loza-Correa M. The peptidoglycan and biofilm matrix of Staphylococcus epidermidis undergo structural changes when exposed to human platelets. PLoS One. 2019;14(1) doi: 10.1371/journal.pone.0211132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Formosa-Dague C. Sticky matrix: adhesion mechanism of the staphylococcal polysaccharide intercellular adhesin. ACS Nano. 2016;10(3):3443–3452. doi: 10.1021/acsnano.5b07515. [DOI] [PubMed] [Google Scholar]
- 124.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 125.Periasamy S. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci U S A. 2012;109(4):1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang R. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J Clin Invest. 2011;121(1):238–248. doi: 10.1172/JCI42520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Le K.Y. Role of phenol-soluble modulins in staphylococcus epidermidis biofilm formation and infection of indwelling medical devices. J Mol Biol. 2019;431(16):3015–3027. doi: 10.1016/j.jmb.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chaignon P. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75(1):125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 129.Kiedrowski M.R. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mootz J.M. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun. 2013;81(9):3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaplan J.B. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185(16):4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kaplan J.B. Extracellular polymeric substance (EPS)-degrading enzymes reduce staphylococcal surface attachment and biocide resistance on pig skin in vivo. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0205526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Izano E.A. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74(2):470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rohde H. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 135.Frank K.L., Patel R. Poly-N-acetylglucosamine is not a major component of the extracellular matrix in biofilms formed by icaADBC-positive Staphylococcus lugdunensis isolates. Infect Immun. 2007;75(10):4728–4742. doi: 10.1128/IAI.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fitzpatrick F., Humphreys H., O'Gara J.P. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol. 2005;43(4):1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kogan G. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255(1):11–16. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]
- 138.Dice B. Biofilm formation by ica-positive and ica-negative strains of Staphylococcus epidermidis in vitro. Biofouling. 2009;25(4):367–375. doi: 10.1080/08927010902803297. [DOI] [PubMed] [Google Scholar]
- 139.Shanks R.M. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73(8):4596–4606. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinez-Garcia S. Non-biofilm-forming commensal Staphylococcus epidermidis isolates produce biofilm in the presence of trypsin. Microbiologyopen. 2019;8(10) doi: 10.1002/mbo3.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ali H. Characterization of the growth dynamics and biofilm formation of Staphylococcus epidermidis strains isolated from contaminated platelet units. J Med Microbiol. 2014;63(Pt 6):884–891. doi: 10.1099/jmm.0.071449-0. [DOI] [PubMed] [Google Scholar]
- 142.Greco-Stewart V.S. Biofilm formation by Staphylococcus capitis strains isolated from contaminated platelet concentrates. J Med Microbiol. 2013;62(Pt 7):1051–1059. doi: 10.1099/jmm.0.050500-0. [DOI] [PubMed] [Google Scholar]
- 143.Hodgson S.D. Enhanced pathogenicity of biofilm-negative Staphylococcus epidermidis isolated from platelet preparations. Transfusion. 2014;54(2):461–470. doi: 10.1111/trf.12308. [DOI] [PubMed] [Google Scholar]
- 144.Sugimoto S. Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus. Sci Rep. 2018;8(1):2254. doi: 10.1038/s41598-018-20485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pozzi C. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8(4) doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rogers K.L., Rupp M.E., Fey P.D. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl Environ Microbiol. 2008;74(19):6155–6157. doi: 10.1128/AEM.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Christensen G.D. Experimental foreign body infections in mice challenged with slime-producing Staphylococcus epidermidis. Infect Immun. 1983;40(1):407–410. doi: 10.1128/iai.40.1.407-410.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ishak M.A. Association of slime with pathogenicity of coagulase-negative staphylococci causing nosocomial septicemia. J Clin Microbiol. 1985;22(6):1025–1029. doi: 10.1128/jcm.22.6.1025-1029.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Patrick C.C. Role of the Staphylococcus epidermidis slime layer in experimental tunnel tract infections. Infect Immun. 1992;60(4):1363–1367. doi: 10.1128/iai.60.4.1363-1367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gronnemose R.B. A novel in vitro model for haematogenous spreading of S. aureus device biofilms demonstrating clumping dispersal as an advantageous dissemination mechanism. Cell Microbiol. 2017;19(12) doi: 10.1111/cmi.12785. [DOI] [PubMed] [Google Scholar]
- 151.Rupp M.E. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J Infect Dis. 2001;183(7):1038–1042. doi: 10.1086/319279. [DOI] [PubMed] [Google Scholar]
- 152.Rupp M.E. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect Immun. 1999;67(5):2627–2632. doi: 10.1128/iai.67.5.2627-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li H. Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect Immun. 2005;73(5):3188–3191. doi: 10.1128/IAI.73.5.3188-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kristian S.A. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197(7):1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 155.Hudetz D. Weak effect of metal type and ica genes on staphylococcal infection of titanium and stainless steel implants. Clin Microbiol Infect. 2008;14(12):1135–1145. doi: 10.1111/j.1469-0691.2008.02096.x. [DOI] [PubMed] [Google Scholar]
- 156.Begun J. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4) doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chokr A. Neither the presence of ica locus, nor in vitro-biofilm formation ability is a crucial parameter for some Staphylococcus epidermidis strains to maintain an infection in a guinea pig tissue cage model. Microb Pathog. 2007;42(2–3):94–97. doi: 10.1016/j.micpath.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 158.Kristian S.A. The ability of biofilm formation does not influence virulence of Staphylococcus aureus and host response in a mouse tissue cage infection model. Microb Pathog. 2004;36(5):237–245. doi: 10.1016/j.micpath.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 159.Monk A.B. Analysis of the genotype and virulence of Staphylococcus epidermidis isolates from patients with infective endocarditis. Infect Immun. 2008;76(11):5127–5132. doi: 10.1128/IAI.00606-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Herbert S. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78(6):2877–2889. doi: 10.1128/IAI.00088-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Queck S.Y. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32(1):150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kropec A. Poly-N-acetylglucosamine production in Staphylococcus aureus is essential for virulence in murine models of systemic infection. Infect Immun. 2005;73(10):6868–6876. doi: 10.1128/IAI.73.10.6868-6876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.McKenney D. Vaccine potential of poly-1-6 beta-D-N-succinylglucosamine, an immunoprotective surface polysaccharide of Staphylococcus aureus and Staphylococcus epidermidis. J Biotechnol. 2000;83(1–2):37–44. doi: 10.1016/s0168-1656(00)00296-0. [DOI] [PubMed] [Google Scholar]
- 164.Jefferson K.K. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol Microbiol. 2003;48(4):889–899. doi: 10.1046/j.1365-2958.2003.03482.x. [DOI] [PubMed] [Google Scholar]
- 165.Brooks J.L., Jefferson K.K. Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog. 2014;10(7) doi: 10.1371/journal.ppat.1004292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Schwartbeck B. Dynamic in vivo mutations within the ica operon during persistence of Staphylococcus aureus in the airways of cystic fibrosis patients. PLoS Pathog. 2016;12(11) doi: 10.1371/journal.ppat.1006024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Carolus H., Van Dyck K., Van Dijck P. Candida albicans and Staphylococcus Species: a threatening twosome. Front Microbiol. 2019;10:2162. doi: 10.3389/fmicb.2019.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Adam B., Baillie G.S., Douglas L.J. Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J Med Microbiol. 2002;51(4):344–349. doi: 10.1099/0022-1317-51-4-344. [DOI] [PubMed] [Google Scholar]
- 169.Holt J.E. Role of extracellular polymeric substances in polymicrobial biofilm infections of Staphylococcus epidermidis and Candida albicans modelled in the nematode Caenorhabditis elegans. Pathog Dis. 2017;75(5) doi: 10.1093/femspd/ftx052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zago C.E. Dynamics of biofilm formation and the interaction between Candida albicans and methicillin-susceptible (MSSA) and -resistant Staphylococcus aureus (MRSA) PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Harriott M.M., Noverr M.C. Ability of Candida albicans mutants to induce Staphylococcus aureus vancomycin resistance during polymicrobial biofilm formation. Antimicrob Agents Chemother. 2010;54(9):3746–3755. doi: 10.1128/AAC.00573-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fredheim E.G. Staphylococcus epidermidis polysaccharide intercellular adhesin activates complement. FEMS Immunol Med Microbiol. 2011;63(2):269–280. doi: 10.1111/j.1574-695X.2011.00854.x. [DOI] [PubMed] [Google Scholar]
- 173.Lund L.D., Ingmer H., Frokiaer H. D-alanylation of teichoic acids and loss of poly-N-acetyl glucosamine in staphylococcus aureus during exponential growth phase enhance IL-12 production in murine dendritic cells. PLoS ONE. 2016;11(2) doi: 10.1371/journal.pone.0149092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Al-Ishaq R. Effects of polysaccharide intercellular adhesin (PIA) in an ex vivo model of whole blood killing and in prosthetic joint infection (PJI): a role for C5a. Int J Med Microbiol. 2015;305(8):948–956. doi: 10.1016/j.ijmm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 175.Hanzelmann D. Toll-like receptor 2 activation depends on lipopeptide shedding by bacterial surfactants. Nat Commun. 2016;7:12304. doi: 10.1038/ncomms12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stevens N.T. Staphylococcus epidermidis polysaccharide intercellular adhesin induces IL-8 expression in human astrocytes via a mechanism involving TLR2. Cell Microbiol. 2009;11(3):421–432. doi: 10.1111/j.1462-5822.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 177.Mah T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012;7(9):1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 178.Hall C.W., Mah T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 179.Singh R. Penetration of antibiotics through Staphylococcus aureus and Staphylococcus epidermidis biofilms. J Antimicrob Chemother. 2010;65(9):1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 180.Dunne W.M., Jr., Mason E.O., Jr., Kaplan S.L. Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother. 1993;37(12):2522–2526. doi: 10.1128/aac.37.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zheng Z., Stewart P.S. Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2002;46(3):900–903. doi: 10.1128/AAC.46.3.900-903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Darouiche R.O. Vancomycin penetration into biofilm covering infected prostheses and effect on bacteria. J Infect Dis. 1994;170(3):720–723. doi: 10.1093/infdis/170.3.720. [DOI] [PubMed] [Google Scholar]
- 183.Zimmerli W., Sendi P. Role of Rifampin against Staphylococcal biofilm infections in vitro, in animal models, and in orthopedic-device-related infections. Antimicrob Agents Chemother. 2019;63(2) doi: 10.1128/AAC.01746-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Post V. Vancomycin displays time-dependent eradication of mature Staphylococcus aureus biofilms. J Orthop Res. 2017;35(2):381–388. doi: 10.1002/jor.23291. [DOI] [PubMed] [Google Scholar]
- 185.Rose W.E., Poppens P.T. Impact of biofilm on the in vitro activity of vancomycin alone and in combination with tigecycline and rifampicin against Staphylococcus aureus. J Antimicrob Chemother. 2009;63(3):485–488. doi: 10.1093/jac/dkn513. [DOI] [PubMed] [Google Scholar]
- 186.Qin Z. Formation and properties of in vitro biofilms of ica-negative Staphylococcus epidermidis clinical isolates. J Med Microbiol. 2007;56(Pt 1):83–93. doi: 10.1099/jmm.0.46799-0. [DOI] [PubMed] [Google Scholar]
- 187.Mahmoudi H. Biofilm formation and antibiotic resistance in meticillin-resistant and meticillin-sensitive Staphylococcus aureus isolated from burns. J Wound Care. 2019;28(2):66–73. doi: 10.12968/jowc.2019.28.2.66. [DOI] [PubMed] [Google Scholar]
- 188.Kivanc S.A. Biofilm forming capacity and antibiotic susceptibility of Staphylococcus spp. with the icaA/icaD/bap genotype isolated from ocular surface of patients with diabetes. Malawi Med J. 2018;30(4):243–249. doi: 10.4314/mmj.v30i4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cafiso V. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10(12):1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- 190.Savage V.J., Chopra I., O'Neill A.J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob Agents Chemother. 2013;57(4):1968–1970. doi: 10.1128/AAC.02008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Weintraub A. Immunology of bacterial polysaccharide antigens. Carbohydr Res. 2003;338(23):2539–2547. doi: 10.1016/j.carres.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 192.Rajagopal M., Walker S. Envelope structures of gram-positive bacteria. Curr Top Microbiol Immunol. 2017;404:1–44. doi: 10.1007/82_2015_5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kojima Y. Antibody to the capsular polysaccharide/adhesin protects rabbits against catheter-related bacteremia due to coagulase-negative staphylococci. J Infect Dis. 1990;162(2):435–441. doi: 10.1093/infdis/162.2.435. [DOI] [PubMed] [Google Scholar]
- 194.Takeda S. Protection against endocarditis due to Staphylococcus epidermidis by immunization with capsular polysaccharide/adhesin. Circulation. 1991;84(6):2539–2546. doi: 10.1161/01.cir.84.6.2539. [DOI] [PubMed] [Google Scholar]
- 195.Maira-Litran T. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal Poly-N-acetyl-beta-(1–6)-glucosamine. Infect Immun. 2005;73(10):6752–6762. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Gening M.L. Synthetic {beta}-(1->6)-linked N-acetylated and nonacetylated oligoglucosamines used to produce conjugate vaccines for bacterial pathogens. Infect Immun. 2010;78(2):764–772. doi: 10.1128/IAI.01093-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cerca N. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-beta-(1–6)-glucosamine. Infect Immun. 2007;75(7):3406–3413. doi: 10.1128/IAI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Skurnik D. Animal and human antibodies to distinct Staphylococcus aureus antigens mutually neutralize opsonic killing and protection in mice. J Clin Invest. 2010;120(9):3220–3233. doi: 10.1172/JCI42748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Skurnik D. Natural antibodies in normal human serum inhibit Staphylococcus aureus capsular polysaccharide vaccine efficacy. Clin Infect Dis. 2012;55(9):1188–1197. doi: 10.1093/cid/cis624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kelly-Quintos C. The role of epitope specificity in the human opsonic antibody response to the staphylococcal surface polysaccharide poly N-acetyl glucosamine. J Infect Dis. 2005;192(11):2012–2019. doi: 10.1086/497604. [DOI] [PubMed] [Google Scholar]
- 201.Perez M.M. Protection from Staphylococcus aureus mastitis associated with poly-N-acetyl beta-1,6 glucosamine specific antibody production using biofilm-embedded bacteria. Vaccine. 2009;27(17):2379–2386. doi: 10.1016/j.vaccine.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Stevenson T.C. Immunization with outer membrane vesicles displaying conserved surface polysaccharide antigen elicits broadly antimicrobial antibodies. Proc Natl Acad Sci U S A. 2018;115(14):E3106–E3115. doi: 10.1073/pnas.1718341115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maira-Litran T. Synthesis and evaluation of a conjugate vaccine composed of Staphylococcus aureus poly-N-acetyl-glucosamine and clumping factor A. PLoS One. 2012;7(9) doi: 10.1371/journal.pone.0043813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Gholami S.A. Evaluation of polysaccharide intercellular adhesion (PIA) and glycerol teichoic acid (Gly-TA) arisen antibodies to prevention of biofilm formation in Staphylococcus aureus and Staphylococcus epidermidis strains. BMC Res Notes. 2019;12(1):691. doi: 10.1186/s13104-019-4736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mirzaei B. Synthesis of conjugated PIA-rSesC and immunological evaluation against biofilm-forming Staphylococcus epidermidis. J Med Microbiol. 2019;68(5):791–802. doi: 10.1099/jmm.0.000910. [DOI] [PubMed] [Google Scholar]
- 206.Skurnik D., Cywes-Bentley C., Pier G.B. The exceptionally broad-based potential of active and passive vaccination targeting the conserved microbial surface polysaccharide PNAG. Expert Rev Vaccines. 2016;15(8):1041–1053. doi: 10.1586/14760584.2016.1159135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zaidi T.S., Zaidi T., Pier G.B. Antibodies to conserved surface polysaccharides protect mice against bacterial conjunctivitis. Invest Ophthalmol Vis Sci. 2018;59(6):2512–2519. doi: 10.1167/iovs.18-23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cywes-Bentley C. Antibody to Poly-N-acetyl glucosamine provides protection against intracellular pathogens: mechanism of action and validation in horse foals challenged with Rhodococcus equi. PLoS Pathog. 2018;14(7) doi: 10.1371/journal.ppat.1007160. p. e1007160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cywes-Bentley C. Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc Natl Acad Sci U S A. 2013;110(24):E2209–E2218. doi: 10.1073/pnas.1303573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hulsdunker J. Immunization against poly-N-acetylglucosamine reduces neutrophil activation and GVHD while sparing microbial diversity. Proc Natl Acad Sci U S A. 2019;116(41):20700–20706. doi: 10.1073/pnas.1908549116. [DOI] [PMC free article] [PubMed] [Google Scholar]