Summary
Background
Variation in liability to cannabis use disorder has a strong genetic component (estimated twin and family heritability about 50–70%) and is associated with negative outcomes, including increased risk of psychopathology. The aim of the study was to conduct a large genome-wide association study (GWAS) to identify novel genetic variants associated with cannabis use disorder.
Methods
To conduct this GWAS meta-analysis of cannabis use disorder and identify associations with genetic loci, we used samples from the Psychiatric Genomics Consortium Substance Use Disorders working group, iPSYCH, and deCODE (20 916 case samples, 363 116 control samples in total), contrasting cannabis use disorder cases with controls. To examine the genetic overlap between cannabis use disorder and 22 traits of interest (chosen because of previously published phenotypic correlations [eg, psychiatric disorders] or hypothesised associations [eg, chronotype] with cannabis use disorder), we used linkage disequilibrium score regression to calculate genetic correlations.
Findings
We identified two genome-wide significant loci: a novel chromosome 7 locus (FOXP2, lead single-nucleotide polymorphism [SNP] rs7783012; odds ratio [OR] 1·11, 95% CI 1·07–1·15, p=1·84 × 10−9) and the previously identified chromosome 8 locus (near CHRNA2 and EPHX2, lead SNP rs4732724; OR 0·89, 95% CI 0·86–0·93, p=6·46 × 10−9). Cannabis use disorder and cannabis use were genetically correlated (rg 0·50, p=1·50 × 10−21), but they showed significantly different genetic correlations with 12 of the 22 traits we tested, suggesting at least partially different genetic underpinnings of cannabis use and cannabis use disorder. Cannabis use disorder was positively genetically correlated with other psychopathology, including ADHD, major depression, and schizophrenia.
Interpretation
These findings support the theory that cannabis use disorder has shared genetic liability with other psychopathology, and there is a distinction between genetic liability to cannabis use and cannabis use disorder.
Funding
National Institute of Mental Health; National Institute on Alcohol Abuse and Alcoholism; National Institute on Drug Abuse; Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing; The European Commission, Horizon 2020; National Institute of Child Health and Human Development; Health Research Council of New Zealand; National Institute on Aging; Wellcome Trust Case Control Consortium; UK Research and Innovation Medical Research Council (UKRI MRC); The Brain & Behavior Research Foundation; National Institute on Deafness and Other Communication Disorders; Substance Abuse and Mental Health Services Administration (SAMHSA); National Institute of Biomedical Imaging and Bioengineering; National Health and Medical Research Council (NHMRC) Australia; Tobacco-Related Disease Research Program of the University of California; Families for Borderline Personality Disorder Research (Beth and Rob Elliott) 2018 NARSAD Young Investigator Grant; The National Child Health Research Foundation (Cure Kids); The Canterbury Medical Research Foundation; The New Zealand Lottery Grants Board; The University of Otago; The Carney Centre for Pharmacogenomics; The James Hume Bequest Fund; National Institutes of Health: Genes, Environment and Health Initiative; National Institutes of Health; National Cancer Institute; The William T Grant Foundation; Australian Research Council; The Virginia Tobacco Settlement Foundation; The VISN 1 and VISN 4 Mental Illness Research, Education, and Clinical Centers of the US Department of Veterans Affairs; The 5th Framework Programme (FP-5) GenomEUtwin Project; The Lundbeck Foundation; NIH-funded Shared Instrumentation Grant S10RR025141; Clinical Translational Sciences Award grants; National Institute of Neurological Disorders and Stroke; National Heart, Lung, and Blood Institute; National Institute of General Medical Sciences.
Introduction
Cannabis use is common, but most users do not progress to cannabis use disorders. About 50–70% of liability to cannabis use disorders is due to genetic factors.1 Three genome-wide association studies (GWASs) of cannabis use disorders2, 3, 4 have identified variants reaching genome-wide significance, but inadequate sample sizes (sample size from largest study to date: 51 372, with 2387 cases) and heterogeneity among samples have contributed to a paucity of replicable findings: only one locus, tagged by a cis-eQTL for CHRNA2 (encoding a nicotinic acetylcholine receptor), has been robustly identified.3
A GWAS of lifetime cannabis use (184 765 total sample size, 43 380 cases) identified eight genome-wide significant loci and 35 significant genes.5 Twin studies suggest high genetic correlations between early stages of cannabis experimentation and later cannabis use disorder.6 However, casual cannabis use is affected by a variety of socioenvironmental influences and age-period-cohort effects, whereas progression to cannabis use disorder is related to other psychopathologies. Findings have suggested partially distinct genetic causes underlying alcohol consumption and alcohol use disorder, including different genetic associations with other psychiatric disorders and traits.7, 8 Thus, in addition to examining the genomic liability for cannabis use disorder, we tested whether the genetic influences underlying cannabis use and cannabis use disorder diverge with respect to behavioural and brain measures.
Research in context.
Evidence before this study
Cannabis use disorder is heritable (50–70% according to twin and family studies), yet identification of genomic variants associated with cannabis use disorder from genome-wide association studies (GWASs) remains sparse. We surveyed all peer-reviewed journal publications in English on GWASs of cannabis use disorder or cannabis dependence using Google Scholar and PubMed, published between Jan 1, 1990, and April 1, 2020. Search terms included “cannabis dependence”, “cannabis abuse”, “cannabis use disorder”, “marijuana dependence”, “marijuana abuse”, “marijuana use disorder”, and “GWAS”. The most promising finding to date is a variant that is a cis-eQTL for CHRNA2 (Demontis and colleagues), which was replicated in an independent dataset for cannabis use disorder. Independently, GWAS of cannabis use have identified multiple genetic risk loci; however, the extent to which the genetics of cannabis use correlates with liability to cannabis use disorder has not been determined. Although GWASs of cannabis use have been studied in the context of a variety of psychiatric and psychosocial correlates, it is expected that some divergent associations will be seen when looking at cannabis use disorder. Previous studies have drawn causal links between cannabis exposure and brain volume, but the relationship between genetic liability to cannabis use disorder and brain volume in individuals naive to cannabis has not yet been studied.
Added value of this study
Our study is the current largest GWAS of cannabis use disorder and the first to include a transancestral component. We found a novel risk locus on chromosome 7. The lead risk variant at this locus is an eQTL for FOXP2—a gene previously implicated in risk-taking behaviours. Contrasting cannabis use and cannabis use disorder, we found that increased liability for cannabis use disorder is genetically correlated with low educational attainment, early age at first birth, and high body-mass index, traits that show opposite directions of association with lifetime cannabis ever-use. We also found that genetic liability for cannabis use disorder is associated with increased risk of mental health problems, infectious diseases, and respiratory illnesses in a large independent sample. Finally, we found a significant association between increased polygenic liability for cannabis use disorder and low white matter volume in cannabis-naive children, suggesting a potential role of cannabis-related genetic predisposition in early brain development.
Implications of all the available evidence
Cannabis use disorder is a psychiatric illness that is genetically associated with many negative outcomes (including increased risk of psychiatric disorders and respiratory illnesses). Lifetime cannabis use and cannabis use disorder show at least partially divergent genetic influences and associations with relevant traits. Given increasingly permissive cannabis laws and positive perceptions of its safety, the recognition that cannabis use disorder is a serious psychiatric illness should spur prevention and treatment efforts.
Methods
Samples
We performed a GWAS of 20 samples in total: 18 from the Psychiatric Genomics Consortium Substance Use Disorders working group (European ancestry 8277 cases, 23 497 controls; African ancestry 3848 cases, 5897 controls), one iPSYCH9 sample (European ancestry 2758 cases, 53 326 controls), and one deCODE sample (European ancestry 6033 cases, 280 396 controls; table 1; appendix pp 2–8).
Table 1.
Numbers of cases and controls in meta-analysis
|
European ancestry |
African ancestry |
European ancestry—case-control individuals |
|||||
|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | ||
| Case-control studies | |||||||
| CATS | 958 | 453 | .. | .. | 958 | 453 | |
| CADD | 397 | 699 | 59 | 55 | 397 | 699 | |
| CHDS | 201 | 420 | .. | .. | 201 | 420 | |
| FSCD | 226 | 314 | 199 | 401 | 226 | 314 | |
| COGEND Nico | 306 | 607 | 154 | 313 | 306 | 607 | |
| COGEND SAGE | 228 | 830 | 79 | 187 | 228 | 830 | |
| GEDI-GSMS | 81 | 491 | .. | .. | 81 | 491 | |
| ADAA | .. | .. | 1000 | 811 | .. | .. | |
| Total | 2397 | 3814 | 1491 | 1767 | 2397 | 3814 | |
| Family-based studies | |||||||
| BLTS | 170 | 1216 | .. | .. | 147 | 662 | |
| MCTFR | 449 | 1625 | .. | .. | 389 | 1185 | |
| Yale Penn 1 | 916 | 833 | 1189 | 1857 | 839 | 657 | |
| Yale Penn 2 | 557 | 497 | 355 | 548 | 557 | 495 | |
| bigCOGA | 2206 | 5053 | 813 | 1725 | 248 | 795 | |
| CEDAR | 64 | 148 | .. | .. | 64 | 148 | |
| OZ-ALC | 593 | 4893 | .. | .. | 470 | 1534 | |
| VTSABD | 99 | 734 | .. | .. | 94 | 361 | |
| IASPSAD | 104 | 613 | .. | .. | 84 | 353 | |
| Total | 5158 | 15 612 | 2357 | 4130 | 2892 | 6190 | |
| Summary statistics | |||||||
| Add health | 722 | 4071 | .. | .. | .. | .. | |
| PGC studies total | 8277 | 23497 | 3848 | 5897 | 5289 | 10 004 | |
| iPSYCH | 2758 | 53326 | 0 | 0 | 2758 | 53 326 | |
| deCODE | 6033 | 280 396 | 0 | 0 | 6033 | 280 396 | |
| Total (European ancestry) | 17 068 | 357 219 | .. | .. | 14 080 | 343 726 | |
| Total (Transancestral) | 20 916 cases;36 3116 controls | .. | .. | .. | .. | .. | |
ADAA=Alcohol Dependence in African Americans. BLTS= Brisbane Longitudinal Twin Study. CADD=Center on Antisocial Drug Dependence. CATS=Comorbidity and Trauma Study. CEDAR= Center for Education and Drug Abuse Research. CHDS=Christchurch Health and Development Study. COGA=Collaborative Study on the Genetics of Alcoholism. COGEND=Collaborative Genetic Study of Nicotine Dependence. FSCD=Family Study of Cocaine Dependence. GEDI=Gene-Environment-Development Initiative. GSMS=Great Smoky Mountains Study. IASPSAD=Irish Affected Sib-Pair Study of Alcohol Dependence. MCTFR= Minnesota Center for Twin and Family Research. OZ-ALC=Australian Alcohol and Nicotine Studies. SAGE=Study of Addiction: Genetics and Environment. VTSABD=Virginia Twin Studies of Adolescent Behavioral Development.
This study was approved by the institutional review board at Washington University School of Medicine and was done in accordance with all relevant ethical regulations. Investigators for each contributing study obtained informed consent from their participants and received ethics approvals from their respective review boards in accordance with applicable regulations. Personal identifiers associated with phenotypic information and samples from deCODE were encrypted using a third-party encryption system.10 The iPSYCH group used pseudonymised unique identifications.9
Measures
Psychiatric Genomics Consortium cases met criteria for a lifetime diagnosis of DSM-IV (or DSM-III-R) cannabis abuse or dependence11 derived from clinician ratings or semi-structured interviews.7 Cases from the iPSYCH sample had ICD-10 codes of F12.1 (cannabis abuse) or F12.2 (cannabis dependence), or both in the Danish Psychiatric Central Research Register;12 the remaining individuals in the sample were used as controls. Cases in the deCODE sample met criteria for lifetime DSM-III-R or DSM-IV cannabis abuse or dependence or DSM-5 cannabis use disorder according to diagnoses made at the National Center of Addiction Medicine in Iceland, whereas controls were derived from the general population of Iceland (appendix pp 2–3). Exposure data were not available for some large groups (eg, iPSYCH and deCODE); therefore, controls were defined regardless of lifetime cannabis exposure across all datasets.
Genotyping: quality control and imputation
For the Psychiatric Genomics Consortium, standard procedures for GWAS quality control and imputation were applied using the Ricopili13 pipeline for case-control groups and the Picopili pipeline for family-based samples. Briefly, variants in each group were filtered for call rate (<5% missingness), followed by individual-level filtering for call rate (<2% missingness) and heterozygosity (|Fhet| <0·20). If available, chromosome X variants were checked to ensure concordance between genotype sex and reported sex. Variants were then filtered more stringently: variants with more than 2% missingness, differential missingness between cases and controls greater than 2%, invariant markers, and those departing from Hardy-Weinberg equilibrium in cases (p<1·00 × 10−10) or controls (p<1·00 × 10−6) were removed (appendix pp 8–10). Principal components analysis was done on a stringently quality-control set of variants using EIGENSOFT14, 15 to exclude population outliers, infer ancestry among the retained individuals (using the 1000 Genomes Phase 316 cosmopolitan reference panel), and derive ancestry-specific principal components for inclusion in analyses (appendix p 10). Sample and variant quality-controlled procedures, including filters for call rate, heterozygosity, and departure from Hardy-Weinberg equilibrium, were done within each ancestry group in each sample. Each group was phased using SHAPEIT17 and imputed using IMPUTE218 to the 1000 Genomes Phase 316 cosmopolitan reference panel (appendix pp 10–11). Duplicate individuals were removed and individuals who were cryptically related across groups were excluded from all but one group (appendix p 12). Single-nucleotide polymorphisms (SNPs) were filtered for INFO score of more than 0·8 and minor allele frequency of at least 0·01 before meta-analysis (appendix pp 13–14).
Quality control of iPSYCH data mirrored the process implemented in the Psychiatric Genomics Consortium, with minor deviations in thresholds for exclusion (appendix p 9).3 As for deCODE, samples were assayed with several Illumina arrays at deCODE genetics. SNPs with low call rate (<95%), significant deviation from Hardy-Weinberg equilibrium (p<0·001), and excessive inheritance error rates (>0·001) were excluded. We did variant imputation on the basis of the IMPUTE HMM model and long-range phasing.19 Variants were further filtered for imputation INFO score more than 0·8 and minor allele frequency at least 1% before inclusion in meta-analysis.
Statistical analysis
We did separate association analyses for each sample (ie, 18 individual samples from Psychiatric Genomics Consortium, iPSYCH, and deCODE) by ancestry. For the eight case-control studies from the Psychiatric Genomics Consortium, imputed dosages were analysed using logistic regression models, implemented in the Ricopili pipeline.13 For family-based samples of the Psychiatric Genomics Consortium, we did association analyses with imputed best-guess genotypes using generalised estimating equations for samples that included only first-degree relatives (eg, sibships), and logistic mixed models for complex pedigrees, in the Picopili pipeline.7 For calculation of SNP heritability and genetic correlations, subsets of genetically unrelated individuals were selected from each family-based sample from the Psychiatric Genomics Consortium and analysed using logistic regression through Picopili (5289 cases, 10 004 controls). These results were then meta-analysed along with the case-control groups. Psychiatric Genomics Consortium covariates included sex and five to ten within-ancestry principal components to account for population stratification (appendix pp 12–13). Because age was not available in all samples, it was not included as a covariate in the Psychiatric Genomics Consortium analyses. Sensitivity analyses in one representative sample showed this to have no impact on study-specific findings.
In the iPSYCH cohort, logistic regression was done with imputed dosages, covarying for five ancestral principal components, data processing waves, and the presence of another psychiatric disorder (because iPSYCH was established to study major psychiatric disorders, cases of cannabis use disorder and controls include comorbidity).3 Adding sex as a covariate to iPSYCH analyses has been shown not to alter findings.20
deCODE data were analysed using logistic regression of imputed dosage data with sex, age, and county of origin as covariates.21 To account for inflation due to population stratification and relatedness, test statistics were divided by an inflation factor estimated from linkage disequilibrium score regression (LDSR; appendix p 13).22
Effective sample size-weighted meta-analyses across case-control and family-based samples within ancestry were done using METAL (appendix pp 13–14).23 First, summary statistics of case-control and family-based samples from the Psychiatric Genomics Consortium were combined and weighted by the effective sample size, because effect sizes from case-control logistic regression analyses and family-based analyses using generalised estimating equations and logistic mixed models are not directly comparable. Then the Psychiatric Genomics Consortium results were meta-analysed with those from the iPSYCH and deCODE samples (between-sample genetic correlations [rg] 0·66–0·70). The summary statistics were filtered such that an SNP had to be present in at least two of the three contributing GWASs (deCODE, iPSYCH, and the Psychiatric Genomics Consortium).
We also did a meta-analysis that excluded related individuals from the family-based samples of the Psychiatric Genomics Consortium, using an inverse variance-weighted scheme, to generate summary statistics that produced effect sizes for use in follow-up analyses (14 080 cases, 343 726 controls). A transancestral meta-analysis using METAL23 combined results across the European and African ancestry cohorts, comprising 20 916 individuals with cannabis use disorder (17 068 from European ancestry, 3848 from African ancestry) and 363 116 controls (357 219 from European ancestry, 5897 African ancestry; appendix pp 13–14, 17). Conditional analyses were done in GCTA-COJO24 by conditioning the meta-analysis summary statistics on the lead variants of genome-wide significance.
The FUMA web-based platform25 version 1.3.5e was used for visualisation and annotation, and MAGMA26 was used within the FUMA framework to do gene-based association analyses, with SNPs assigned to genes on the basis of physical position (appendix pp 14–15). We also used Hi-C coupled MAGMA to assign non-coding SNPs (intergenic and intronic) to genes on the basis of their chromatin interactions (exonic and promoter SNPs are still assigned to genes on the basis of their genomic location; appendix p 15).27 Pathway analyses were done using PASCAL to test canonical pathways in the European ancestry sample.28 All variants within all genes were tested, using default settings, with the structure of linkage disequilibrium estimated using the 1000 Genomes European sample as a reference. We used S-PrediXcan29 to examine gene expression differences associated with case-control status, using our summary statistics of cannabis use disorder and transcriptome data from the PredictDB Data Repository for 11 brain regions, liver tissue, whole blood, and two types of adipose tissue. We included these tissues because cannabis use disorder is a psychiatric disorder and tetrahydrocannabinol, a key psychoactive cannabis component, accumulates in adipose tissue.30 Analyses were restricted to the European ancestry meta-analysis because the prediction models were trained on reference transcriptome data from GTEx version 831 using only individuals of European ancestry. The significance threshold was corrected for the total number of gene-tissue pairs tested (75 684 tested, α=6·69 × 10−7).
Heritability explained by common variants (h2SNP) and genetic correlations with 23 other traits chosen because of previous findings or hypothesised relationships (including cannabis use; appendix pp 15–17) were estimated using LDSR22, 32 on the results of the meta-analysis of case-control individuals of European ancestry (the number of unrelated cases of African ancestry was less than the acceptable sample size threshold for LDSR). Conversion of h2SNP estimates from observed scale to liability scale was done using a range of estimated population prevalences from 1%3 to 8·5% (because in some samples we used DSM-IV cannabis abuse or dependence).33 Significance of genetic correlations with other traits was determined using a Bonferroni correction for 23 tests (including with cannabis use; α=0·002). Finally, we examined whether the genetic correlations for cannabis use disorder were significantly different than those for cannabis use5 using the jackknife procedure implemented through LDSR.32
To investigate potential causal relationships, we did latent causal variable analyses on cannabis use disorder and the top genetically correlated traits: educational attainment, age at first birth, Townsend Deprivation Index, smoking initiation, and ADHD (appendix p 16).34
We used mtCOJO35 to condition the summary statistics of cannabis use disorder on loci associated with cannabis use at p<0·0015 to adjust for as many SNPs as possible while retaining computational efficiency. Adjusted summary statistics were used to recompute genetic correlations. Because of the high co-occurrence of cannabis use and tobacco smoking, we also did mtCOJO analyses to condition the summary statistics of cannabis use disorder for loci significantly associated with smoking initiation and cigarettes smoked per day36 (p<5·00 × 10−8; excluding 23andMe data, because of restricted access). Moreover, given long-standing interest in the comorbidity of schizophrenia and cannabis misuse, we used mtCOJO to condition the summary statistics of cannabis use disorder on significant schizophrenia loci.37
LDSR was used to estimate the genetic correlation between cannabis use disorder and a broad measure of maximum cannabis use frequency. Linear regression was then used to examine the extent to which polygenic risk scores (PRS) for cannabis use disorder predicted a pseudocontinuous measure of self-reported cannabis use frequency, while covarying for age, sex, and 20 ancestral principal components (appendix p 16). PRSice-238 was also used to do gene-set enrichment using gene sets and pathways from the Molecular Signatures Database.39
PRS for cannabis use disorder were computed using PRS-CS40 for each of the 66 915 genotyped individuals of European descent in BioVU (appendix pp 16–17). Genotyping and quality control of this sample have been described elsewhere.41 A logistic regression model was fitted to each of 1335 case or control phenotypes that had at least 100 cases to estimate the odds of each diagnosis given the PRS for cannabis use disorder, after adjustment for sex, median age of the longitudinal electronic health record measurements, and the top ten ancestral principal controls. To explore whether pleiotropic effects of the PRS for cannabis use disorder were mediated by smoking behaviours, we did two phenotype-wide association study (PheWAS) sensitivity analyses: a PheWAS on summary statistics of cannabis use disorder that had been conditioned on the top smoking initiation loci using mtCOJO,35 and a PheWAS using a diagnosis of tobacco use disorders as an additional covariate in the regression model, which is a conservative over-correction given the extremely high comorbidity expected between cannabis use disorder and tobacco use disorder. We used a Bonferroni-corrected phenome-wide significance threshold of 3·74 × 10−5; this is overly conservative because it incorrectly assumes independence between phenotypes. PheWAS analyses were run using the PheWAS R package, version 0.12.42
Data from the Adolescent Brain Cognitive Development Study (Registered; ABCD study)43 (data release 2.0.1) were used to test the association of PRS for cannabis use disorder with brain structure among 4539 cannabis-naive children (through self-reporting or hair toxicology) of European ancestry (mean age 9·93 years [SD 0·63], 2125 [47%] were girls; appendix p 17). Total bilateral white matter volume, grey matter volume, and intracranial volume were estimated using FreeSurfer 5.3.44 PRS from the cannabis use disorder GWAS were generated at nine p value thresholds (ie, p<0·0001, p<0·001, p<0·01, p<0·10, p<0·20, p<0·30, p<0·40, p<0·50, and p<1·00), as were PRS for cannabis use.5 Linear mixed models were used to include scanner (for imaging analyses) and family as nested random effects, done using the lme4 package in R, version 3.6.0. All analyses included as fixed effect covariates the first 20 ancestral principal components, age, sex, age by sex, parents combined income, caregiver education, genotyping batch, caregiver's marital status, prenatal cannabis exposure before and after knowledge of pregnancy, and twin status. Multiple testing within each brain structure phenotype was accounted for by applying random field theory correction across p value thresholds, as this method directly models the overlap across the different PRS thresholds and corrects for the statistical dependence among them.45
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all of the data and the final responsibility to submit for publication.
Results
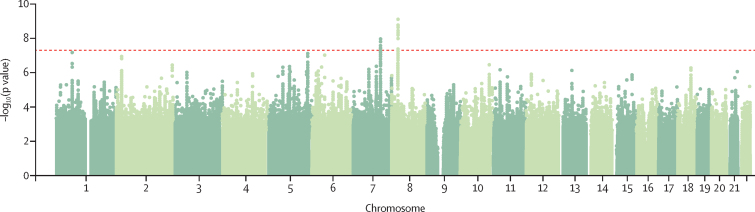
We identified two genome-wide significant loci in the transancestral meta-analysis of cannabis use disorder (African and European ancestries, 20 916 cases, 363 116 controls; appendix pp 17, 20). These loci were significant in the European ancestry meta-analysis but did not reach significance in the much smaller African ancestry analysis (17 068 cases, 357 219 controls vs 3848 cases, 5897 controls; table 2). The lead SNPs were rs4732724 on chromosome 8 (ptransancestral=2·64 × 10−9, pEuropean=6·46 × 10−9, pAfrican=0·70) and rs7783012 on chromosome 7 (ptransancestral=2·43 × 10−9, pEuropean=1·84 × 10−9, pAfrican=0·09), with the same direction of effect observed for both ancestries. No additional ancestry-specific loci were observed.
Table 2.
Association statistics for the lead genome-wide significant SNPs across each of the three primary samples (deCODE, iPSYCH, PGC) in the European ancestry and transancestral meta-analyses
| Position | SNP | Effect allele | deCODE OR (SE) | deCODE p value | iPSYCH OR (SE) | iPYSCH p value | PGC EUR unrel OR (SE) | PGC EUR unrel p value | PGC EUR comp Z score | PGC EUR comp p value | EUR meta-analysis OR (SE)* | EUR meta-analysis p value* | Transancestral Z score† | Transancestral p value† | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome 7 | 114 116 881 | rs7783012 | A | 1·10 (0·03) | 5·32 × 10−4 | 1·09 (0·03) | 2·58 × 10−3 | 1·11 (0·03) | 9·56 × 10−5 | 3·47 | 5·22 × 10−4 | 1·11 (0·02) | 1·84 × 10−9 | 5·97 | 2·43 × 10−9 |
| Chromosome 8 | 27 432 062 | rs4732724‡ | C | 0·90 (0·03) | 3·03 × 10−4 | 0·84 (0·03) | 5·73 × 10−8 | 0·98 (0·04) | 0·616 | −1·91 | 0·056 | 0·89 (0·02) | 6·46 × 10−9 | −5·95 | 2·64 × 10−9 |
comp=complete meta-analysis (including related individuals and summary statistic cohorts). EUR=European ancestry. OR=odds ratio. PGC=Psychiatric Genomics Consortium. SNP=single nucleotide polymorphism. unrel=unrelated genotyped meta-analysis.
Complete deCODE, iPSYCH, and PGC EUR meta-analysis (excluding related individuals and summary statistic cohorts in the PGC).
Transancestral meta-analysis with deCODE, iPSYCH, and PGC samples (including related individuals and summary statistic cohorts).
SNP was only present in half of the PGC samples.
Based on effect sizes and linkage disequilibrium from the case-control European ancestry meta-analysis (cases 14 080, controls 343 726), the genome-wide significant locus on chromosome 8 contains a single association (independent at R2<0·1) with lead SNP rs4732724 (odds ratio [OR] 0·89, 95% CI 0·86–0·93, SE 0·02; p=6·46 × 10−9; figure 1, appendix pp 21–23). This locus was previously associated with cannabis use disorder in the iPSYCH sample3 and includes eQTLs for CHRNA2 (cholinergic receptor nicotinic α2 subunit) in the cerebellum and cerebellar hemisphere and EPHX2 (epoxide hydrolase 2) in the cerebellum and adipose tissue. One genome-wide significant variant in the chromosome 8 locus (rs1565735) had a CADD score of 13·28, indicating high probability of deleteriousness (appendix p 17). There were additional eQTL signals at this chromosome 8 locus, for CCDC25 (coiled-coil domain containing 25, in nucleus accumbens, multiple SNPs), CLU (clusterin, in adipose, rs2640724), and STMN4 (stathmin 4, in prefrontal cortex, rs78875955 and rs72477506; appendix p 25).
Figure 1.
Manhattan plot of the European ancestry-only genome-wide meta-analysis
The chromosome 7 locus is located in an intron of FOXP2 (Forkhead box protein P2, index SNP, rs7783012; OR 1·11, 95% CI 1·07–1·15, SE 0·02; p=1·84 × 10−9; figure 1, appendix pp 21–22, 24). The index variant was an eQTL for FOXP2 in brain (prefrontal cortex, anterior cingulate cortex) and adipose tissue, and demonstrated chromatin interactions with FOXP2, MDFIC, and MIR3666 (appendix p 26).
Inflation in the test statistics (λ=1·10) probably reflects the polygenic architecture of cannabis use disorder, a conclusion supported by LDSR (LDSR intercept 0·99). Conditioning the summary statistics of cannabis use disorder on the lead SNP in each genome-wide significant locus, rs7783012 and rs4732724, did not reveal additional independent significant findings.
The gene-wise association analysis of European ancestry summary statistics identified three significant genes (α=2·664 × 10−6): FOXP2 (p=7·31 × 10−8), PDE4B (p=6·66 × 10−7), and ENO4 (p=3·51 × 10−8; appendix p 17, 27). No pathways were significant (appendix p 18). Three genes, NAT6 (amygdala, cortex, frontal cortex), HYAL3 (both adipose tissues, whole blood, cerebellum, frontal cortex, hippocampus, nucleus accumbens, and spinal cord), and IFRD2 (cerebellum) were significantly related to cannabis use disorder through genetically regulated gene expression (appendix pp 18, 28). Connecting SNPs to genes via chromatin interaction data revealed significant associations in adult brain tissue (ten genes), fetal brain tissues (12 genes), iPSC-derived astrocytes (11 genes), and iPSC-derived neurons (eight genes); these genes included HYAL3, ENO4, CHRNA2, and FOXP2 (appendix pp 18, 29).
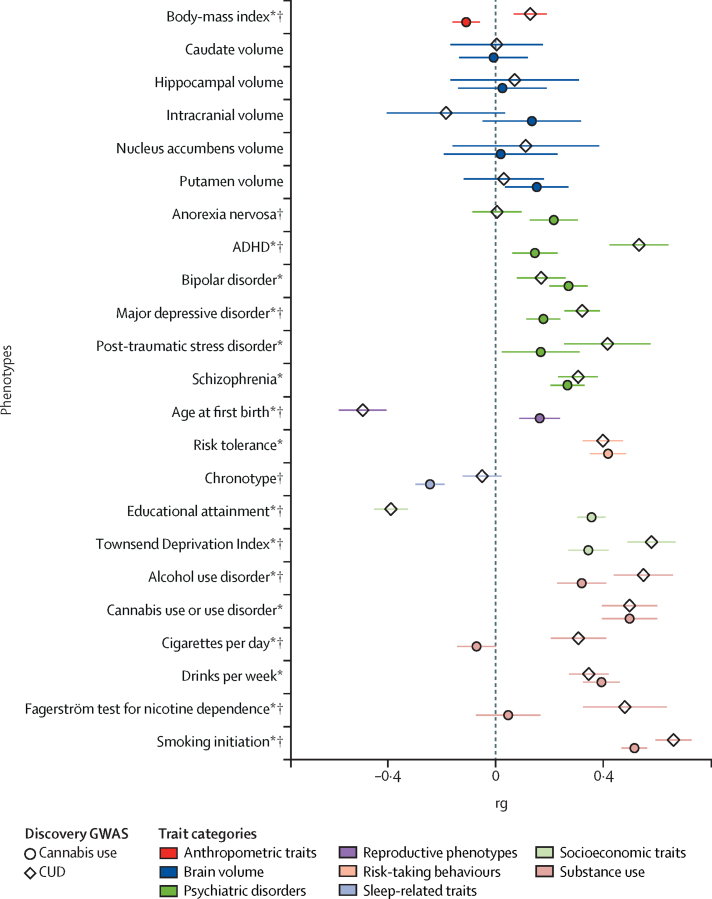
The SNP-heritability (h2SNP) for cannabis use disorder was 0·067–0·121 (SE 0·006–0·011) on the liability scale, depending on the estimated population prevalence and h2SNP0·02 (SE 0·002) on the raw scale. Cannabis use disorder showed significant rg with 16 of the 23 studied phenotypes, for which the strongest relationships were observed with smoking initiation36 (rg 0·66, p=3·20 × 10−83), Townsend Deprivation Index (a measure of regional poverty46; rg0·58, p=3·30 × 10−37), educational attainment47 (rg–0·39, p=6·70 × 10−34), and age at which first child is born (rg–0·49; p=5·40 × 10−28; figure 2, appendix p 18). Thus, increased risk of cannabis use disorder is genetically correlated with increased liability for smoking initiation, living in an area of high material poverty, having children at an early age, and low levels of educational attainment. Liability to cannabis use disorder was also positively genetically correlated with alcohol use,36 nicotine dependence,48 psychiatric disorders (eg, ADHD,20 schizophrenia,37 major depression),49 and body-mass index (BMI).50
Figure 2.
Genetic correlations between CUD, cannabis use, and other traits of interest
CUD=cannabis use disorder. GWAS=genome-wide association studies. rg=genetic correlation. *Significantly genetically correlated with CUD. †Significantly different correlations between CUD and cannabis use (α=0·002).
The rg between cannabis use and cannabis use disorder was 0·50 (SE 0·05, p=1·50 × 10−21). Of the eight genome-wide significant SNPs associated with cannabis use, only four had p<0·05 in the meta-analysis of cannabis use disorder (modest sample overlap between the two studies: genetic covariance intercept 0·014 [SE 0·005]).5 Conditioning the summary statistics of cannabis use disorder for loci associated with cannabis use neither substantially modified the effect sizes of the genome-wide significant loci (rs4732724, β=–0·11, SE 0·02, p=8·25 × 10−9; rs7783012, β=0·10, SE 0·02, p=2·62 × 10−9) nor identified additional novel loci (appendix p 18). The heritability of cannabis use disorder adjusted for cannabis use loci (using mtCOJO35) was 0·095 (SE 0·01) on the liability scale (estimated population prevalence 8·5%).
The rgs with cannabis use disorder and cannabis use were significantly different for 12 of the 22 traits compared (figure 2, appendix p 18). Cannabis use5 and cannabis use disorder were positively genetically correlated with liability to smoking initiation, schizophrenia, major depressive disorder, risk tolerance, and the Townsend Deprivation Index. Cannabis use5 was positively genetically correlated with educational achievement and later age at birth of first child, and negatively with BMI. In contrast, cannabis use disorder was genetically correlated with low education attainment, early age at birth of first child, and high BMI. Liability to cannabis use disorder was genetically correlated with nicotine dependence (rg0·48, p=1·35 × 10−9), whereas the genetic correlation of this trait with cannabis use was not significant (p=0·44). In contrast, cannabis use was significantly genetically correlated with chronotype (rg −0·24, p=6·40 × 10−19), whereas cannabis use disorder showed no significant correlation with this trait (p=0·18). Conditioning the rg of cannabis use disorder on cannabis use loci (with p<0·001) made little difference in the magnitude of the rgs (appendix p 18).
We found no evidence of genetically causal relationships between liability to cannabis use disorder and to any of the most highly correlated traits (ie, educational attainment, age at first birth, Townsend Deprivation Index, smoking initiation, or ADHD; genetic causality proportion 0·05–0·27, p=0·128–0·856; appendix p 18).
Liability to cannabis use disorder and maximum cannabis use frequency in the UK Biobank were genetically correlated (rg 0·75, p=1·80 × 10−6). PRS for cannabis use disorder were significantly associated with our pseudocontinuous measure of cannabis use frequency in the UK Biobank (maximum R2 0·04%, Z 7·42, p=1·15 × 10−13, threshold p<0·3; appendix p 18, 30). 65 of 12 461 gene sets and pathways were significantly enriched, highlighting involvement of CNS morphogenesis (transcription factor Nkx–2·2 target genes, R2 0·02%, Z 4·46, p=8·22 × 10−6) and immune responses to exogenous compounds (ZFP91 target genes R2 0·01%, Z 4·41, p=1·01 × 10−5; CD4+ T-cell R2 0·02%, Z 4·41, p=3·79 × 10−6; and macrophage gene sets R2 0·01%, Z 4·62, p=1·04 × 10−5; appendix p 18).
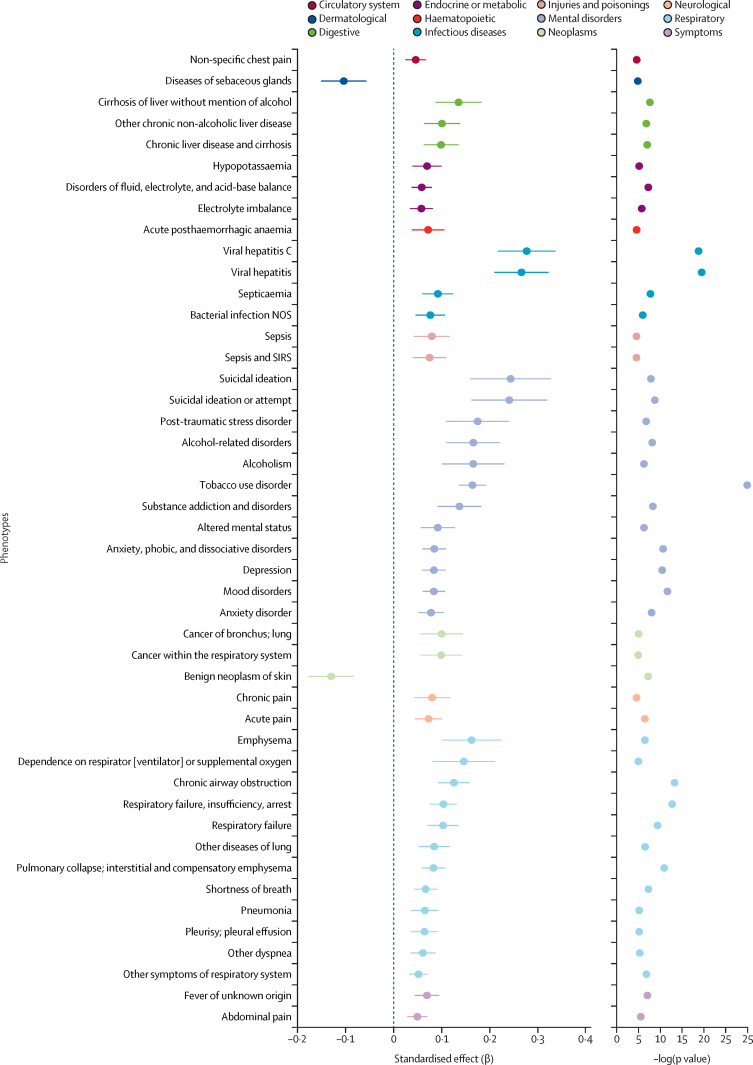
Of 1335 phenotypes in the BioVU biobank, 46 were significantly associated with the PRS for cannabis use disorder (p<3·74 × 10−5; figure 3, appendix p 18). The phenotype groups with the most abundant associations were mental disorders (n=12), the strongest associations being with tobacco use disorder (cases 5280, OR 1·18, 95% CI 1·13–1·23, SE 0·02; p=2·66 × 10−27) and substance use disorders (cases 6155, OR 1·18, SE 0·01, 95% CI 1·16–1·20; p=1·24 × 10−30), mood disorders (cases 9588, OR 1·09, SE 0·01, 95% CI 1·07–1·11; p=2·38 × 10−12) and suicidal ideation or attempt (cases 689, OR 1·27, SE 0·04, 95% CI 1·17–1·37; p=1·81 × 10−9); respiratory diseases (n=12), such as respiratory failure (cases 4485, OR 1·11, SE 0·02, 95% CI 1·07–1·15; p=4·45 × 10−10) or chronic airway obstruction (cases 4436, OR 1·13, SE 0·02, 95% CI 1·09–1·18; p=5·64 × 10−14); endocrine or metabolic conditions (n=3), such as disorders of fluid (cases 12 562, OR 1·06, SE 0·01, 95% CI 1·04–1·08; p=5·77 × 10−8); infectious diseases (n=4), such as viral hepatitis (cases 135, OR 1·3, SE 0·03, 95% CI 1·23–1·38; p=3·34 × 10−20); and digestive diseases (n=3), including cirrhosis of liver (cases 1928, OR 1·14, SE 0·02, 95% CI 1·10–1·19; p=2·49 × 10−8).
Figure 3.
PheWAS associations between polygenic risk for CUD and phenotypes in the BioVU biobank
The 46 phenotypes shown are significantly associated with CUD (p<3·74 × 10−5, corrected for 1335 phenotypes tested). CUD=cannabis use disorder. PheWAS=phenotype-wide association study. NOS=not otherwise specified. SIRS=systemic inflammatory response syndrome.
A secondary pheWAS analysis in BioVU using summary statistics of cannabis use disorder conditioned on smoking initiation revealed attenuated findings, with only ten codes now passing Bonferroni corrections; anxiety disorder, viral hepatitis, and several respiratory codes were still significant. When we conditioned the pheWAS on tobacco use disorder, some associations remained significant (respiratory conditions, viral hepatitis), whereas other associations (eg, anxiety disorder) were no longer associated with PRS for cannabis use disorder (appendix p 18).
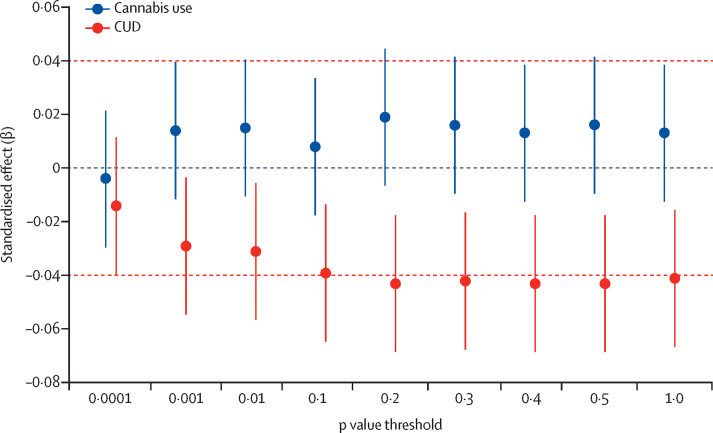
The PRS for cannabis use disorder were significantly associated with reduced total white matter volume in cannabis-naive children from the ABCD Study (standardised β=–0·04; p=0·001; figure 4), explaining up to 0·18% of the variance in white matter volume at the most predictive threshold of p<0·5 (appendix p 18). Children in the highest quartile of PRS, on average, had a white matter volume that was 1% lower than those in the lowest quartile. Results remained significant when including intracranial volume as a covariate (standardised β=–0·08, p=0·01) and when excluding 1246 (27%) of 4539 children who used any substance (standardised β=–0·05, p=0·001), or when excluding 2482 (54%) of 4539 who used any substance or were prenatally exposed to any substance (standardised β=–0·05, p=0·03). The PRS for cannabis use were not significantly correlated with white matter volume (figure 3). After correction for multiple testing, there was no association between PRS for cannabis use disorder or cannabis use and grey matter volume (all p>0·01; appendix p 18, 31).
Figure 4.
Polygenic risk score associations with white matter volume in drug-naive children
Total white matter volume was regressed on polygenic risk scores for CUD and cannabis use (in separate models). CUD=cannabis use disorder.
Discussion
This GWAS meta-analysis confirmed one previously identified locus on chromosome 8 as associated with cannabis use disorder and identified a new locus on chromosome 7. The lead variant at the chromosome 7 locus (rs7783012) is a cis-eQTL for FOXP2 expression in brain and adipose tissue. FOXP2 was also significantly implicated in gene-based tests that incorporated information about chromatin interactions in iPSC-derived astrocytes (appendix p 29). rs7783012 has also been associated with measures related to externalising behaviours (eg, ADHD,20 age at first sexual intercourse,51 generalised risk tolerance)52 and with educational attainment.47 FOXP2 is essential to synaptic plasticity and has been implicated in the normal development of speech and language acquisition53 However, because of the prominence of the protein product of FOXP2 as a regulator of numerous genes, indirect pathways of vulnerability beyond risk-taking are also possible.
Individual SNPs on chromosome 8 are eQTLs for CHRNA2 and EPHX2, extending previous work by Demontis and colleagues3 in iPSYCH, with replication in the deCODE data. Note that iPSYCH and deCODE are the main contributors to this finding in the meta-analysis (piPSYCH=5·73 × 10−8, pdeCODE=0·0003, pPGC=0·06; appendix p 23). A large GWAS of schizophrenia54 has also implicated this variant (p=3·68 × 10−6), but conditioning for top schizophrenia loci did not modify the association with cannabis use disorder (p=4·33 × 10−8; appendix p 18). Given the role of CHRNA2 variants in tobacco smoking,36 it is plausible that the findings for cannabis use disorder and schizophrenia are partially driven by the high rates of tobacco use in those populations.55 However, conditioning cannabis use disorder on the GWAS of cigarettes per day increased the significance of the lead variant rs4732724 (p=4·16 × 10−9; appendix p 18), although a different SNP was identified as the lead SNP (rs11783093). When rs11783093 was conditioned for the GWAS of smoking initiation, the signal was attenuated (p=1·55 × 10−6; appendix p 18). These findings suggest that the chromosome 8 signal might be partly driven by smoking initiation, or indicative of a pleiotropic effect with a stronger impact on cannabis use disorder than on smoking initiation.36 Despite the plausibility of CHRNA2 in the cause of cannabis use disorder, it is worth noting that EPHX2, which is involved in the metabolism of cannabinoids,56, 57, 58 was also identified in eQTL analyses but not supported by other post-hoc analyses (appendix p 29).
Cannabis use and cannabis use disorder were modestly genetically correlated (rg 0·50) but conditioning for cannabis use loci did not substantially reduce the heritability of cannabis use disorder, and although it reduced the significance of the top loci, the effect sizes remained consistent. Although this does not fully account for possible index-event bias,59 it suggests that the findings are not due to cannabis exposure alone. Cannabis use and cannabis use disorder also show divergent genetic relationships with educational attainment,47 BMI,50 and age at birth of first child, with cannabis use disorder indexing greater impairment in these psychosocial and anthropometric indices than cannabis use. This divergence is similar to that found between alcohol intake and alcohol use disorder.7, 8
We found genetic overlap between cannabis use disorder and several mental health phenotypes, respiratory illnesses, and infectious diseases in the BioVU biobank. The strongest association was with tobacco use disorder, but conditioning for loci associated with smoking initiation retained many of the pheWAS associations at significant levels, including anxiety, phobic and dissociative disorders, respiratory failure, and viral hepatitis. An even more stringent analysis that covaried for tobacco use disorder revealed independent associations with viral hepatitis, type 1 diabetes, respiratory measures, and pain, but not mental health. These associations could reflect genuine pleiotropy (eg, with risk-taking behaviours and injection drug use) or index putatively causal peripheral effects of cannabis. Cannabis use frequency in the UK Biobank was genetically correlated with cannabis use disorder as well, but, similarly to other psychiatric and behavioural traits,60 the PRS for cannabis use disorder explained only a small proportion of variance in cannabis use frequency (R2 0·04%).
Some previous cross-sectional studies have linked differences in grey matter volume with cannabis use and dependence;61 however, a large mega-analysis did not find reductions in global or regional volumes in cannabis-dependent adults compared with controls.62 In our study, the association between PRS for cannabis use disorder and white matter volume persisted in the subset of children who were not exposed to any substance, including prenatally. This finding suggests that polygenic liability for cannabis use disorder might index differences in white matter volume in the developing brain, independently of the onset of substance use behaviours. Still, the association between PRS for cannabis use disorder and white matter was small (R2 0·15–0·18%), and additional studies are needed to confirm this association.
Some limitations are noteworthy. Our African ancestry sample was under-powered; more data are needed, particularly in light of potential disparities that result from a majority of genetic studies focusing on European-ancestry populations.63, 64 We had little or no information regarding comorbid psychiatric disorders for the majority of PGC samples; however, we did conditional analyses to account for these and it made little difference. Information regarding lifetime cannabis exposure and the potency of cannabis used was scarce. Our estimates of genome-wide SNP-h2 were far lower than the h2 estimated from twin and family studies (0·07–0·12 vs 0·5–0·7). This discrepancy between pedigree-estimated heritability and SNP-heritability is common across essentially all substance use disorders, and might be due to low power, some heritability residing in variants too rare to be included in our GWAS, and insufficient coverage of optimal common-variant genomic coverage in available microarray data even after imputation. An additional limitation is that we did not do formal Mendelian randomisation65 analysis. To do this analysis, we would have needed to remove sample overlap between our cannabis use disorder GWAS and the other GWASs of interest, which would have greatly decreased our statistical power. However, after doing latent causal variable analyses,34 an approach related to mendelian randomisation that can account for sample overlap among the input GWAS, there was no significant evidence of causal relationships between liability to cannabis use disorder and to any of the top genetically correlated traits: educational attainment, age at first birth, Townsend Deprivation Index, smoking initiation, or ADHD (appendix p 16). Overall, estimates of genetic overlap might also be sensitive to sample characteristics, such as older volunteers in the UK Biobank cohort66 and some younger registry-based cohorts in our GWAS. In addition, imbalance between cases and controls could have affected our findings, although we did not observe substantial genetic heterogeneity (appendix pp 23–24).
In conclusion, our findings provide further evidence that cannabis use disorder is a serious, psychiatric illness with genetic and neurobiological influences that diverge at least partially from cannabis use. From a public health perspective, the recognition that cannabis use disorder is a serious form of psychopathology should spur efforts to identify and aid at-risk individuals in the face of escalating cannabis use worldwide, especially among adolescents.
Data sharing
Summary statistics will be made available for download at https://www.med.unc.edu/pgc/download-results/.
Acknowledgments
Acknowledgments
The Psychiatric Genomics Consortium's Substance Use Disorders working group is supported by MH109532 with funding from the National Institute of Mental Health (NIMH) and the National Institute on Drug Abuse (NIDA). We gratefully acknowledge previous support from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and thank all our contributing investigators and study participants who make this research possible. We acknowledge Dr E Jane Costello for leadership in collecting and characterising the Gene-Environment-Development Initiative–Great Smoky Mountains Study sample and her valuable insights on the development of this study and interpretation of findings. This research was done using the UK Biobank Resource, application numbers 4844 and 58146. Individual funding was provided by the following grants: MH109532 (AA, HJE, JG); F32AA027435 (ECJ); K02DA032573 (AA); 1U01MH109514-01 (ADB); 5T32DA007261 (AH); T32MH018951 (DAAB); NIH F32 MH122058 (FRW); Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (ADB); the European Commission, Horizon 2020, grant number 667302 (ADB); R01-DA034076 (TET); R01HD060726 (BWD, KMH, JDB, MBM); Health Research Council of New Zealand 16/600 (JB, JHo, MAK, JFP); Health Research Council of New Zealand 11/792 (JB, JHo); AA027827, AG061162 (RB); U24DA041147 (SAB); R01HD093651, R01MH117559, P30DA023026 (WEC); DA011015, DA038504 (RPC); K02 AA018755 (DMD); P01-HD031921, R01-HD073342 (KMH);R01 DA018673, DA025109, DA024413, DA016977 (HHM); Wellcome Trust (104036/Z/14/Z, 216767/Z/19/Z), UKRI MRC (MC_PC_17209, MR/S035818/1) (AMM); K01MH113848, The Brain & Behavior Research Foundation NARSAD grant 28632 (REP); R21 DA047527, R21 DC018098 (RP); SAMHSA Grant # 1H79TI081668 (MDR); R01DA026911, R21DA046791 (NLS); R01HD050735, U54EB020403 (subaward), NHMRC Australia (496682; 1009064) (MJW); Tobacco-Related Disease Research Program of the University of California Grant Number T29KT0526, Families for Borderline Personality Disorder Research (Beth and Rob Elliott) 2018 NARSAD Young Investigator Grant #27676 (SS-R). The iPSYCH team was supported by grants from the Lundbeck Foundation (R165–2013–15320, R102-A9118, R155–2014–1724, and R248–2017–2003), the European Commission (Horizon 2020, grant number 667302), and the universities and university hospitals of Aarhus and Copenhagen, Denmark. The Danish National Biobank resource was supported by the Novo Nordisk Foundation. Data handling and analysis on the GenomeDK HPC facility was supported by NIMH (1U01MH109514–01 to ADB). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Center for Genomics and Personalized Medicine and the Centre for Integrative Sequencing, iSEQ, Aarhus University, Denmark (grant to ADB). Work at deCODE genetics was partly funded by the NIDA (R01−DA034076). The BioVU dataset(s) used for the PheWAS analyses described were obtained from Vanderbilt University Medical Center's BioVU, which is supported by numerous sources: institutional funding, private agencies, and federal grants. These grants include the NIH funded Shared Instrumentation Grant S10RR025141, and Clinical Translational Sciences Award grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, and R01HD074711, and additional funding sources https://victr.vumc.org/biovu-funding/]. Dr Degenhardt is funded by the NIH (R01 DA144740) and the Australian National Health and Medical Research Council Research (1135991). Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development (ABCD) Study, held in the NIMH Data Archive. The ABCD Study is supported by the NIH and additional federal partners under award numbers U01DA041022, U01DA041028, U01DA041048, U01DA041089, U01DA041106, U01DA041117, U01DA041120, U01DA041134, U01DA041148, U01DA041156, U01DA041174, U24DA041123, U24DA041147, U01DA041093, and U01DA041025. A listing of participating sites and a complete listing of the study investigators can be found online. ABCD consortium investigators designed and implemented the study or provided data, or both, but did not necessarily participate in analysis or writing of this study. This manuscript reflects the views of the authors and might not reflect the opinions or views of the NIH or ABCD consortium investigators. The ABCD data repository grows and changes over time. The ABCD data used in this study came from the NIMH Data Archive 2.0.1. Funding support for the Comorbidity and Trauma Study (dbGAP accession number, phs000277.v1.p1) was provided by NIDA (R01 DA17305); GWAS genotyping services at the Center for Inherited Disease Research at The Johns Hopkins University were supported by NIH (contract N01-HG-65403). Funding support for the Center for Education and Drug Abuse Research (dbGAP accession number, phs001649.v1.p1) was provided by NIDA (P50 DA005605). The Christchurch Health and Development Study has been supported by funding from the Health Research Council of New Zealand, the National Child Health Research Foundation (Cure Kids), the Canterbury Medical Research Foundation, the New Zealand Lottery Grants Board, the University of Otago, the Carney Centre for Pharmacogenomics, the James Hume Bequest Fund, US NIH grant MH077874, and NIDA grant, a developmental model of gene-environment interplay in substance use disorders (R01DA024413) 2007–12. The Collaborative Study on the Genetics of Alcoholism (COGA; Principal Investigators B Porjesz, V Hesselbrock, and T Foroud; Scientific Director, A Agrawal; Translational Director, D Dick) includes 11 different centres: University of Connecticut (V Hesselbrock); Indiana University (H J Edenberg, T Foroud, J Nurnberger Jr, Y Liu); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (B Porjesz, J Meyers, C Kamarajan, A Pandey); Washington University in St Louis (L Bierut, J Rice, K Bucholz, A Agrawal); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield, A Brooks, R Hart); The Children's Hospital of Philadelphia, University of Pennsylvania (L Almasy); Virginia Commonwealth University (D Dick, J Salvatore); Icahn School of Medicine at Mount Sinai (A Goate, M Kapoor, P Slesinger); and Howard University (D Scott). Other COGA collaborators include L Bauer (University of Connecticut); L Wetherill, X Xuei, D Lai, S O'Connor, M Plawecki, S Lourens (Indiana University); L Acion (University of Iowa); G Chan (University of Iowa; University of Connecticut); D B Chorlian, J Zhang, S Kinreich, G Pandey (SUNY Downstate); M Chao (Icahn School of Medicine at Mount Sinai); A Anokhin, V McCutcheon, S Saccone (Washington University); F Aliev, P Barr (Virginia Commonwealth University); and H Chin, and A Parsian (NIAAA Staff Collaborators). We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding Principal Investigator and Co-Principal Investigator of COGA, and also owe a debt of gratitude to other past organisers of COGA, including Ting-Kai Li, P Michael Conneally, Raymond Crowe, and Wendy Reich for their crucial contributions. This national collaborative study is supported by NIH Grant U10AA008401 from NIAAA and NIDA. We thank Kim Doheny and Elizabeth Pugh from the Center for Inherited Disease Research and Justin Paschall from the NCBI dbGaP staff for valuable assistance with genotyping and quality control in developing the dataset available at dbGaP (phs000125.v1.p1, phs000763.v1.p1, phs000976.v1.p1). Support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative (GEI; U01 HG004422; dbGaP study accession phs000092.v1.p1). SAGE is one of the GWAS funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonisation and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by COGA (U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (P01 CA089392; see also phs000404.v1.p1), and the Family Study of Cocaine Dependence (R01 DA013423, R01 DA019963). Funding support for genotyping, which was done at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), NIAAA, NIDA, and the NIH contract High throughput genotyping for studying the genetic contributions to human disease (HHSN268200782096C). The Great Smoky Mountains Study project (phs000852.v1.p1) was supported by NIDA (U01DA024413, R01DA11301), NIMH (R01MH063970, R01MH063671, R01MH048085, K01MH093731, and K23MH080230), NARSAD, and the William T Grant Foundation. We are grateful to all the Great Smoky Mountains Study and Caring for Children in the Community study participants who contributed to this work. The following grants supported data collection and analysis of Center on Antisocial Drug Dependence (dbGAP in progress): DA011015, DA012845, DA021913, DA021905, DA032555, and DA035804. Alcohol Dependence in African Americans was funded by NIH grant R01 AA017444. Brisbane Longitudinal Twin Study was supported by the US NIDA (R00DA023549), and by the Australian Research Council (DP0343921, DP0664638, 464914, 619667, FT110100548). Gene-Environment-Development Initiative Virginia Commonwealth University (VTSABD; dbGAP in progress) was supported by NIDA (U01DA024413, R01DA025109), NIMH (R01MH045268, R01MH055557, R01MH068521), and the Virginia Tobacco Settlement Foundation, grant 8520012. We are grateful to all the Virginia Twin Studies of Adolescent Behavioral Development, Young Adult Follow-Up, Transitions to Substance Use study participants who contributed to this work. Minnesota Center for Twin and Family Research (phs000620.v1.p1) contributing to this publication was supported by the NIH under award numbers DA005147, DA013240, DA024417, DA036216, AA009367, MH066140, DA042755, DA037904, HG008983, and DA044283. Funding for Substance Use Disorder Liability: Candidate System Genes study was supported by R01 DA019157 and P50 DA005605. Yale-Penn (phs000425.v1.p1; phs000952.v1.p1) was supported by NIH grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 and the Veterans Affairs Connecticut and Philadelphia Veterans Affairs Mental Illness Research, Educational, and Clinical Centers. Australian Alcohol and Nicotine studies (OZ-ALC-NAG; phs000181.v1.p1) were supported by NIH grants AA07535,AA07728, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, and DA019951, by grants from the Australian National Health and Medical Research Council (241944, 339462, 389927,389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, and 552498), by grants from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, and DP0343921); and by the 5th Framework Programme (FP-5) GenomEUtwin Project (QLG2-CT-2002–01254). GWAS genotyping at Center for Inherited Disease Research was supported by a grant to the late Richard Todd, MD, PhD, former Principal Investigator of grant AA13320. Irish Affected Sib-Pair Study of Alcohol Dependence GWAS data collection and analysis was supported by NIAAA grants P20-AA-017828 and P50-AA-022537. Sample collection was supported by R01-AA-011408. Control genotyping was supported by NIMH grant R01-MH-083094 and Wellcome Trust Case Control Consortium 2 grant WTCCC-084710. This research uses data from Add Health, a programme project directed by Kathleen Mullan Harris and designed by J Richard Udry, Peter S Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill, and funded by grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 23 other federal agencies and foundations. No direct support was received from grant P01-HD31921 for this analysis. We additionally thank the groups who directly shared GWAS results. We would like to acknowledge all participating groups of the International Cannabis Consortium, and in particular the members of the working group including Sven Stringer, Camelia Minica, Karin Verweij, Hamdi Mbarek, Eske Derks, Nathan Gillespie, and Jacqueline Vink. We also wish to thank the ENIGMA consortium for providing GWAS results on subcortical brain volumes. Finally, we acknowledge the valuable contribution of groups who have publicly released summary statistics from their respective GWAS. Specifically, thanks to researchers from Schumann and colleagues (2016) including the CHARGE+ and AlcGen consortia and to all members of Psychiatric Genomics Consortium (PGC). Individual-level data from the genotyped cohorts and cohort-level summary statistics will be made available to researchers following an approved analysis proposal through the PGC Substance Use Disorder group with agreement of the cohort Principal Investigators; contact the corresponding authors for details. Some cohort data are also available from dbGaP except when prohibited by Institutional Review Board, funding requirements or European Union data restrictions (accession numbers: phs000277.v1.p1, phs000092.v1.p1, phs000852.v1.p1, phs000404.v1.p1, phs000092.v1.p1, phs000620.v1.p1, phs001649.v1.p1, phs000425.v1.p1, phs000181.v1.p1, phs000763.v1.p1, phs000125.v1.p1).
Contributors
SAB, DD, ADB, JHo, GWM, MN, AA, RB, HJE, IBH, JG, ECJ, TET, and RKW designed the study. MJA, SAB, JDB, AMG, AMM, HRK, DL, BTW, RET, S-AB, VH, BSM, AEA, KSK, WEC, ADB, FRW, SEM, JB-G, LD, NGM, DBH, JHo, RPC, LW, MAK, DMD, JPR, MDR, MCS, MMV, BPR, LJB, LF, JB, KKB, JG, NAG, RAG, DFG, KMH, SMH, ACH, JKH, IBH, DMH, WGI, EOJ, KK, PAFM, HHM, MM, MBM, GWM, OM, PBM, ECN, JFP, BP, VR, NLS, RS, JLS, TET, TT, SV, TLW, TW, MJW, and RW collected and interpreted the data. DAAB, AMM, RP, ASH, IBH, DD, RET, BSM, FRW, SEM, TBB, SEP, LMH, DBH, LW, MAK, DEA, SZ, JA, GWR, AA, RB, SS-R, T-KC, BWD, NAG, SDG, DFG, SMH, JHe, ECJ, EOJ, REP, PAL, GWM, TET, RKW, and RW analysed the data. MJA, DAAB, DL, RP, ASH, DD, S-AB, DBH, DEA, GWR, AA, RB, SS-R, JDB, LKD, HJE, JG, DFG, JHo, JHe, ECJ, NLS, TET, RKW, and HZ gave the statistical input. ASH, ADB, FRW, SEP, JLM, AA, RB, T-KC, HJE, JG, ECJ, TET, and RKW wrote the manuscript. SAB, AMG, HRK, DL, LAF, RP, ASH, DD, RET, WEC, ADB, SEP, DBH, JHo, RPC, LW, MAK, DMD, MCS, MMV, GWR, AA, RB, JNM, SS-R, JDB, KKB, LKD, HJE, JG, ECJ, ECN, NLS, TET, RKW, and HZ edited the manuscript. AMM, KSK, SEM, LD, DMD, JLM, BPR, AA, RB, HJE, JG, NAG, ACH, EOJ, HHM, ECN, and TET provided phenotype expertise. CJH, AMM, LAF, ADB, KSK, NGM, JHo, AA, RB, LKD, HJE, TMF, JG, KMH, JKH, WGI, HHM, ECN, BP, KS, TET, SV, RKW, TW, and MJW supervised the study.
Declaration of interests
TW has acted as an advisor and a lecturer to H Lundbeck A/S. LJB and the spouse of NLS are listed as inventors on Issued US Patent 8 080 371, Markers for Addiction, covering the use of particular SNPs in determining the diagnosis, prognosis, and treatment of addiction. AMM has received research support from Eli Lilly, Janssen, Pfizer, and the Sackler Foundation. HRK is a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which was supported in the last 3 years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences. HRK and JG are named as inventors on PCT patent application number 15/878 640, entitled Genotype-guided dosing of opioid agonists, filed Jan 24, 2018. LD reports untied educational grant funding to research studies of new opioid medications in Australia from Indivior, Mundipharma, Seqirus, and Reckitt Benckiser.
Contributor Information
Emma C Johnson, Email: emma.c.johnson@wustl.edu.
Psychiatric Genomics Consortium Substance Use Disorders Workgroup:
Raymond Walters, Renato Polimanti, Emma Johnson, Jeanette McClintick, Alexander Hatoum, June He, Frank Wendt, Hang Zhou, Mark Adams, Amy Adkins, Fazil Aliev, Silviu-Alin Bacanu, Anthony Batzler, Sarah Bertelsen, Joanna Biernacka, Tim Bigdeli, Li-Shiun Chen, Toni-Kim Clarke, Yi-Ling Chou, Franziska Degenhardt, Anna Docherty, Alexis Edwards, Pierre Fontanillas, Jerome Foo, Louis Fox, Josef Frank, Ina Giegling, Scott Gordon, Laura Hack, Annette Hartmann, Sarah Hartz, Stefanie Heilmann-Heimbach, Stefan Herms, Colin Hodgkinson, Per Hoffman, Jouke Hottenga, Martin Kennedy, Mervi Alanne-Kinnunen, Bettina Konte, Jari Lahti, Marius Lahti-Pulkkinen, Dongbing Lai, Lannie Ligthart, Anu Loukola, Brion Maher, Hamdi Mbarek, Andrew McIntosh, Matthew McQueen, Jacquelyn Meyers, Yuri Milaneschi, Teemu Palviainen, John Pearson, Roseann Peterson, Samuli Ripatti, Euijung Ryu, Nancy Saccone, Jessica Salvatore, Sandra Sanchez-Roige, Melanie Schwandt, Richard Sherva, Fabian Streit, Jana Strohmaier, Nathaniel Thomas, Jen-Chyong Wang, Bradley Webb, Robbee Wedow, Leah Wetherill, Amanda Wills, Jason Boardman, Danfeng Chen, Doo-Sup Choi, William Copeland, Robert Culverhouse, Norbert Dahmen, Louisa Degenhardt, Benjamin Domingue, Sarah Elson, Mark Frye, Wolfgang Gäbel, Caroline Hayward, Marcus Ising, Margaret Keyes, Falk Kiefer, John Kramer, Samuel Kuperman, Susanne Lucae, Michael Lynskey, Wolfgang Maier, Karl Mann, Satu Männistö, Bertram Müller-Myhsok, Alison Murray, John Nurnberger, Aarno Palotie, Ulrich Preuss, Katri Räikkönen, Maureen Reynolds, Monika Ridinger, Norbert Scherbaum, Marc Schuckit, Michael Soyka, Jens Treutlein, Stephanie Witt, Norbert Wodarz, Peter Zill, Daniel Adkins, Joseph Boden, Dorret Boomsma, Laura Bierut, Sandra Brown, Kathleen Bucholz, Sven Cichon, E. Jane Costello, Harriet de Wit, Nancy Diazgranados, Danielle Dick, Johan Eriksson, Lindsay Farrer, Tatiana Foroud, Nathan Gillespie, Alison Goate, David Goldman, Richard Grucza, Dana Hancock, Kathleen Mullan Harris, Andrew Heath, Victor Hesselbrock, John Hewitt, Christian Hopfer, John Horwood, William Iacono, Eric Johnson, Jaakko Kaprio, Victor Karpyak, Kenneth Kendler, Henry Kranzler, Kenneth Krauter, Paul Lichtenstein, Penelope Lind, Matt McGue, James MacKillop, Pamela Madden, Hermine Maes, Patrik Magnusson, Nicholas Martin, Sarah Medland, Grant Montgomery, Elliot Nelson, Markus Nöthen, Abraham Palmer, Nancy Pederson, Brenda Penninx, Bernice Porjesz, John Rice, Marcella Rietschel, Brien Riley, Richard Rose, Dan Rujescu, Pei-Hong Shen, Judy Silberg, Michael Stallings, Ralph Tarter, Michael Vanyukov, Scott Vrieze, Tamara Wall, John Whitfield, Hongyu Zhao, Benjamin Neale, Joel Gelernter, Howard Edenberg, and Arpana Agrawal
Supplementary Material
References
- 1.Verweij KJH, Zietsch BP, Lynskey MT. Genetic and environmental influences on cannabis use initiation and problematic use: a meta-analysis of twin studies. Addiction. 2010;105:417–430. doi: 10.1111/j.1360-0443.2009.02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal A, Lynskey MT, Hinrichs A. A genome-wide association study of DSM-IV cannabis dependence. Addict Biol. 2011;16:514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demontis D, Rajagopal VM, Thorgeirsson TE. Genome-wide association study implicates CHRNA2 in cannabis use disorder. Nat Neurosci. 2019;22:1066–1074. doi: 10.1038/s41593-019-0416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherva R, Wang Q, Kranzler H. Genome-wide association study of cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry. 2016;73:472–480. doi: 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasman JA, Verweij KJH, Gerring Z. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 2018;21:1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie NA, Neale MC, Jacobson K, Kendler KS. Modeling the genetic and environmental association between peer group deviance and cannabis use in male twins. Addiction. 2009;104:420–429. doi: 10.1111/j.1360-0443.2008.02457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters RK, Polimanti R, Johnson EC. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci. 2018;21:1656–1669. doi: 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kranzler HR, Zhou H, Kember RL. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pedersen CB, Bybjerg-Grauholm J, Pedersen MG. The iPSYCH2012 case–cohort sample: new directions for unravelling genetic and environmental architectures of severe mental disorders. Mol Psychiatry. 2018;23:6–14. doi: 10.1038/mp.2017.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulcher JR, Kristjánsson K, Gudbjartsson H, Stefánsson K. Protection of privacy by third-party encryption in genetic research in Iceland. Eur J Hum Genet. 2000;8:739–742. doi: 10.1038/sj.ejhg.5200530. [DOI] [PubMed] [Google Scholar]
- 11.American Psychiatric Association . 4th edn. American Psychiatric Publishing; Arlington, VA: 2000. Diagnostic and statistical manual of mental disorders. [Google Scholar]
- 12.Mors O, Perto GP, Mortensen PB. The Danish psychiatric central research register. Scand J Public Health. 2011;39:54–57. doi: 10.1177/1403494810395825. [DOI] [PubMed] [Google Scholar]
- 13.Lam M, Awasthi S, Watson HJ. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinsky KJ, Bhatia G, Loh P-R. Fast Principal-Component Analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98:456–472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 16.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 18.Hancock DB, Levy JL, Gaddis NC. Assessment of genotype imputation performance using 1000 Genomes in African American studies. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudbjartsson DF, Helgason H, Gudjonsson SA. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 20.Demontis D, Walters RK, Martin J. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Helgason A, Palsson S. The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulik-Sullivan BK, Loh P-H, Hilary K Finucane. LD core regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Ferreira T, Morris AP. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–3S3. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sey NYA, Hu B, Mah W. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23:583–593. doi: 10.1038/s41593-020-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbeira AN, Dickinson SP, Bonazzola R. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreuz DS, Axelrod J. Delta-9-tetrahydrocannabinol: localization in body fat. Science. 1973;179:391–393. doi: 10.1126/science.179.4071.391. [DOI] [PubMed] [Google Scholar]
- 31.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulik-Sullivan B, Finucane HK, Anttila V. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stinson FS, Ruan WJ, Pickering R, Grant BF. Cannabis use disorders in the USA: prevalence, correlates and co-morbidity. Psychol Med. 2006;36:1447–1460. doi: 10.1017/S0033291706008361. [DOI] [PubMed] [Google Scholar]
- 34.O'Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Z, Zheng Z, Zhang F. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Jiang Y, Wedow R. Association studies of up to 1·2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 2019;51:237–244. doi: 10.1038/s41588-018-0307-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi SW, O'Reilly PF. PRSice-2: polygenic risk score software for biobank-scale data. Gigascience. 2019;8 doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis J, Sealock J, Levinson RT. Genetic risk for major depressive disorder and loneliness in sex-specific associations with coronary artery disease. Mol Psychiatry. 2019 doi: 10.1038/s41380-019-0614-y. published online Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30:2375–2376. doi: 10.1093/bioinformatics/btu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lisdahl KM, Sher KJ, Conway KP. Adolescent brain cognitive development (ABCD) study: overview of substance use assessment methods. Dev Cogn Neurosci. 2018;32:80–96. doi: 10.1016/j.dcn.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischl B. FreeSurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nichols TE. Multiple testing corrections, nonparametric methods, and random field theory. Neuroimage. 2012;62:811–815. doi: 10.1016/j.neuroimage.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Morris R, Carstairs V. Which deprivation? A comparison of selected deprivation indexes. J Public Health Med. 1991;13:318–326. [PubMed] [Google Scholar]
- 47.Lee JJ, Wedow R, Aysu Okbay A. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1·1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock DB, Guo Y, Reginsson GW. Genome-wide association study across European and African American ancestries identifies a SNP in DNMT3B contributing to nicotine dependence. Mol Psychiatry. 2018;23:1911–1919. doi: 10.1038/mp.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wray NR, Ripke S, Mattheisen M. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speliotes EK, Willer CJ, Berndt SI. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe K, Stringer S, Frei O. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 52.Linnér RK, Biroli P, Kong E. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 2019;51:245–257. doi: 10.1038/s41588-018-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai CSL, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- 54.Pardiñas AF, Holmans P, Pocklington AJ. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fowler IL, Carr VJ, Carter NT, Lewin TJ. Patterns of current and lifetime substance use in schizophrenia. Schizophr Bull. 1998;24:443–455. doi: 10.1093/oxfordjournals.schbul.a033339. [DOI] [PubMed] [Google Scholar]
- 56.McDougle DR, Watson JE, Abdeen AA. Anti-inflammatory ω-3 endocannabinoid epoxides. Proc Natl Acad Sci USA. 2017;114:E6034–E6043. doi: 10.1073/pnas.1610325114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snider NT, Nast JA, Tesmer LA, Hollenberg PF. A cytochrome P450-derived epoxygenated metabolite of anandamide is a potent cannabinoid receptor 2-selective agonist. Mol Pharmacol. 2009;75:965–972. doi: 10.1124/mol.108.053439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner K, Inceoglu B, Hammock BD. Soluble epoxide hydrolase inhibition, epoxygenated fatty acids and nociception. Prostaglandins Other Lipid Mediat. 2011;96:76–83. doi: 10.1016/j.prostaglandins.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dudbridge F, Allen RJ, Sheehan NA. Adjustment for index event bias in genome-wide association studies of subsequent events. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09381-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Battistella G, Fornari E, Annoni J-M. Long-term effects of cannabis on brain structure. Neuropsychopharmacology. 2014;39:2041–2048. doi: 10.1038/npp.2014.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mackey S, Allgaier N, Chaaran B. Mega-analysis of gray matter volume in substance dependence: general and substance-specific regional effects. Am J Psychiatry. 2019;176:119–128. doi: 10.1176/appi.ajp.2018.17040415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peterson RE, Kuchenbaecker K, Walters RK. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell. 2019;179:589–603. doi: 10.1016/j.cell.2019.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 66.Fry A, Littlejohns TJ, Sudlow C. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–1034. doi: 10.1093/aje/kwx246. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics will be made available for download at https://www.med.unc.edu/pgc/download-results/.