Abstract
Streptococcus sanguinis is a primary colonizer of teeth and is typically considered beneficial due to its antagonistic relationship with the cariogenic pathogen Streptococcus mutans. However, S. sanguinis can also act as an opportunistic pathogen should it enter the bloodstream and colonize a damaged heart valve, leading to infective endocarditis. Studies have implicated manganese acquisition as an important virulence determinant in streptococcal endocarditis. A knockout mutant lacking the primary manganese import system in S. sanguinis, SsaACB, is severely attenuated for virulence in an in vivo rabbit model. Manganese is a known cofactor for several important enzymes in S. sanguinis, including superoxide dismutase, SodA, and the aerobic ribonucleotide reductase, NrdEF. To determine the effect of manganese depletion on S. sanguinis, we performed transcriptomic analysis on a ΔssaACB mutant grown in aerobic fermentor conditions after the addition of the metal chelator EDTA. Despite the broad specificity of EDTA, analysis of cellular metal content revealed a decrease in manganese, but not in other metals, that coincided with a drop in growth rate. Subsequent supplementation with manganese, but not iron, zinc, or magnesium, restored growth in the fermentor post-EDTA. Reduced activity of Mn-dependent SodA and NrdEF likely contributed to the decreased growth rate post-EDTA, but did not appear entirely responsible. With the exception of the Dps-like peroxide resistance gene, dpr, manganese depletion did not induce stress response systems. By comparing the transcriptome of ΔssaACB cells pre- and post-EDTA, we determined that manganese deprivation led to altered expression of diverse systems. Manganese depletion also led to an apparent induction of carbon catabolite repression in a glucose-independent manner. The combined results suggest that manganese limitation produces effects in S. sanguinis that are diverse and complex, with no single protein or system appearing entirely responsible for the observed growth rate decrease. This study provides further evidence for the importance of this trace element in streptococcal biology. Future studies will focus on determining mechanisms for regulation, as the multitude of changes observed in this study indicate that multiple regulators may respond to manganese levels.
Keywords: Streptococcus sanguinis, infective endocarditis, manganese, transcriptome, fermentor, carbon catabolite repression, CcpA
Introduction
Streptococcus sanguinis is a facultative anaerobe that is typically found in much greater abundance at healthy oral sites than in carious lesions or diseased gingiva (Stingu et al., 2008; Belda-Ferre et al., 2012; Griffen et al., 2012; Gross et al., 2012; Giacaman et al., 2015). The S. sanguinis genome encodes a variety of adhesins (Xu et al., 2007; Bensing et al., 2018) that allow it to act as one of the primary colonizers of the salivary pellicle (Socransky et al., 1977; Kolenbrander et al., 2010). It has the capacity to produce (Garcia-Mendoza et al., 1993; Kreth et al., 2008) and survive in (Xu et al., 2014) high concentrations of hydrogen peroxide (H2O2), which allows it to compete against the dental caries pathogen Streptococcus mutans (Kreth et al., 2005). These traits, which have evolved to ensure survival in the highly diverse oral cavity, also make S. sanguinis an opportunistic pathogen (Das et al., 2009; Turner et al., 2009; Bensing et al., 2019). Dental procedures (Kinane et al., 2005; Forner et al., 2006; Lockhart et al., 2008), routine oral hygiene practices (Silver et al., 1977, 1979; Moreillon and Que, 2004), mastication (Sreenivasan et al., 2017), and poor oral hygiene (Kholy et al., 2015) can all damage the oral mucosa, allowing bacteria to enter the bloodstream. S. sanguinis and certain other bacterial species can bind to cardiac vegetations composed of platelets and fibrin that form on damaged heart valves and endocardium (Lee et al., 2001), leading to the disease infective endocarditis (IE) (Moreillon et al., 2002; Widmer et al., 2006). Recent studies estimate that IE affects more than 40,000 people each year in the United States and kills 12–40% (Bor et al., 2013; Cahill et al., 2017; Jamil et al., 2019) due to complications such as congestive heart failure and stroke (Bashore et al., 2006). In the United States, prevention depends upon antibiotic prophylaxis prior to dental procedures for at-risk patients (Wilson et al., 2007). The economic burden, potential for side effects, and questionable efficacy (Dayer and Thornhill, 2018; Thornhill et al., 2018; Quan et al., 2020) of this practice, as well as the increasing prevalence of antibiotic resistance (Dodds, 2017) are all pressing concerns.
Manganese (Mn) is an essential human micronutrient and has been linked to virulence in many human pathogens, including streptococci (Kehres and Maguire, 2003; Papp-Wallace and Maguire, 2006; Eijkelkamp et al., 2015). Mn has been shown to play an important role in oxidative stress tolerance through several mechanisms: (i) serving as a cofactor for enzymes that break down reactive oxygen species (ROS), (ii) substituting for iron (Fe) in enzymes when cells experience oxidative stress in order to prevent Fenton reaction-mediated damage, and (iii) reacting with superoxide radicals when complexed with small molecules such as bicarbonate (Waters, 2020).
Previous work from our lab established that the ABC transporter SsaACB is important for Mn transport and essential for virulence in a rabbit model of IE (Crump et al., 2014; Baker et al., 2019; Murgas et al., 2020). In S. sanguinis, Mn acts as a cofactor for superoxide dismutase (SodA) (Parker and Blake, 1988; Poyart et al., 1988) and the aerobic class 1b ribonucleotide reductase (NrdEF) (Makhlynets et al., 2014; Rhodes et al., 2014). Loss of SodA activity alone cannot account for the reduction in virulence (Crump et al., 2014). NrdEF activity is essential for virulence (Rhodes et al., 2014), but it is likely that these are not the only two Mn-cofactored enzymes or Mn-dependent pathways in S. sanguinis. In a previous microarray analysis of Mn depletion in the closely related species Streptococcus pneumoniae (Ogunniyi et al., 2010), it was found that only a few genes were differentially expressed in response to either deletion of the pneumococcal SsaB ortholog PsaA, or growth in media without supplemental Mn. However, these data alone are insufficient to explain the decreased growth of these mutants in low-Mn media. In this study, we sought to determine the overall effect of Mn depletion on the transcriptome of S. sanguinis in an attempt to identify other Mn-dependent pathways. Here we report that while there were some similarities with this previous study, we found a larger number of differentially expressed genes, providing new insights into the role of Mn in streptococci.
Results
Selection of Fermentor Growth Conditions for Mn Depletion
For this study, we were interested in measuring transcriptional changes resulting from Mn depletion in metabolically active cells. We also wanted to examine the cells as they transitioned from Mn replete conditions to Mn insufficiency, a task that would most easily have been accomplished by addition of a strong and selective Mn chelator to growing cells. However, we were aware of no such chelator. We therefore explored the use of the non-specific chelator EDTA in conjunction with an ΔssaACB mutant. This mutant was previously found to be deficient in Mn and Fe transport and aerobic growth in low-Mn media (Murgas et al., 2020).
We achieved reproducible, large-scale growth in a fermentor using Brain Heart Infusion (BHI) broth. Typical chemostat conditions (Burne and Chen, 1998) could not be identified that supported growth of the SK36 wild-type (WT) strain but not the ΔssaACB mutant, even when aeration was increased (data not shown). However, we found that when the dilution rate was increased to 0.875 vessel volumes per h, addition of 100 μM EDTA to both the fermentor vessel and media carboy dramatically reduced the optical density (OD840–910) of the ΔssaACB mutant cultures (Supplementary Figure S1A), while not affecting the WT strain (Supplementary Figure S1B). The effect of EDTA addition on the OD of the ΔssaACB cultures typically became apparent after 38 min (Supplementary Figure S1 inset). The addition of EDTA slowed the growth of ΔssaACB but did not kill the cells entirely because when the media pumps were shut off ∼80 min post-EDTA addition, the OD began to increase immediately (data not shown).
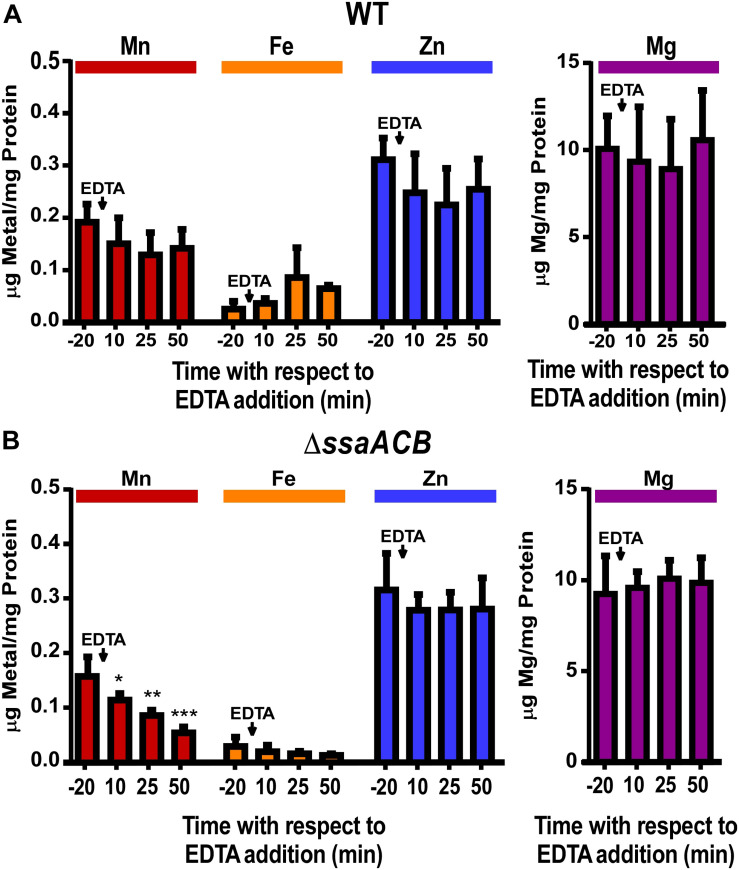
To determine if a lack of available Mn caused the EDTA-dependent reduction in the ΔssaACB growth rate, samples of both WT and ΔssaACB were collected at T–20, T10, T25, and T50, where 100 μM EDTA was added to the vessel at T0 (Supplementary Figure S1). Washed cells were analyzed using inductively coupled plasma optical emission spectroscopy (ICP-OES) (Figure 1). EDTA addition to WT did not significantly alter cellular levels of any of the four metals measured—Mn, Fe, zinc (Zn), or magnesium (Mg) (Figure 1A). Mn was the only metal significantly reduced in the post-EDTA samples as compared to pre-EDTA for ΔssaACB (Figure 1B). Fe levels were low in the ΔssaACB mutant (Figure 1B), consistent with previous results that this mutant transports less Fe than WT (Murgas et al., 2020). Fe, Zn, and Mg levels were not significantly affected by EDTA addition (Figure 1). Cobalt and copper levels were at or below the limit of detection in both strains (data not shown).
FIGURE 1.
Effect of EDTA on metal content of fermentor-grown WT and ΔssaACB cells. Aerobic fermentor-grown WT (A) and ΔssaACB (B) cells were collected at each time point and analyzed for cellular metal content using ICP-OES. Means and standard deviations of three replicates are shown. Significance was determined either by repeated measures ANOVA or by one-way ANOVA if matching was not effective, with a Tukey-Kramer multiple comparisons test to T–20. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Time points not labeled were not significantly different from T–20. For Fe, two T–20 replicates in (A) and at least two replicates for each time point in (B) were below the limit of detection.
As another test of metal specificity, 100 μM of either Mn2+ or Fe2+ was added to the vessel 70 mins post-EDTA addition. The addition of Mn2+ eliminated, and then reversed, the post-EDTA decline in OD, while Fe2+ had no discernible effect (Supplementary Figure S2). The metal content of samples collected 10 mins after addition of Mn2+ or Fe2+ (T80) revealed that both Mn and Fe were taken up by cells, resulting in significantly higher levels than at T–20 (Supplementary Figure S3). Although neither Zn nor Mg levels were significantly affected by addition of EDTA (Figure 1), 100 μM of either Zn2+ or Mg2+ was added at T70 for at least two fermentor runs each and, like Fe2+, neither produced any apparent effect (data not shown).
Overview of Transcriptional Response of S. sanguinis to Mn Depletion
In order to assess the impact of Mn depletion on the S. sanguinis transcriptome, RNA sequencing (RNA-seq) analysis was performed on ΔssaACB fermentor samples collected at the same time points as above (Supplementary Figure S1A). Principal component analysis (PCA) revealed that the samples from each time point clustered together, indicating minimal variation between independent replicates (Figure 2A). The T10 samples overlapped slightly with T–20, indicating few early changes in gene expression. The dissimilarities of the RNA-seq profiles were greater at T25 and T50, suggesting that EDTA treatment increasingly affected the gene expression of ΔssaACB during the period tested.
FIGURE 2.
Analysis of gene expression in fermentor-grown S. sanguinis ΔssaACB mutant cells. (A) Principal component analysis of the RNA-seq samples as determined by the pcaExplorer package for R. Replicates are labeled by fermentor run number. Ellipses are drawn around the 95% confidence interval for each time point. (B) Volcano plots comparing each post-EDTA time point to T–20 were generated using log2 fold changes from Geneious in the EnhancedVolcano package for R. Genes that are upregulated in the post-EDTA time point are positive on the x-axis (right) and those that are downregulated are negative (left). Genes exhibiting |log2 fold change| >1 are depicted by either red (>1) or blue (<1) spheres. (C) Pathway enrichment analysis of differentially expressed genes at T50 using DAVID classification of genes and KEGG annotations.
Volcano plot analysis of differentially expressed genes (DEGs; defined as | log2| ≥ 1, adjusted P-value ≤ 0.05) comparing post-EDTA time points to the pre-EDTA time point revealed that there were only 48 (2.1%) and 139 (6.1%) DEGs at T10 and T25, respectively (Figure 2B). In contrast, at 50 mins post-EDTA, 407 genes (17.9%) were differentially expressed, with a number of genes more severely downregulated (Figure 2B). Consistent with these results, the growth rate of ΔssaACB decreased dramatically between T25 and T50 (Supplementary Figure S1A).
Gene classification analysis of DEGs at T50 using KEGG annotations revealed that genes involved in sugar metabolism were highly enriched, with phosphotransferase systems (PTS), fructose/mannose transport, and pyruvate metabolism all in the top 12 most highly enriched pathways (Figure 2C). Other highly enriched pathways included amino acid metabolism, ribosomes, oxidative phosphorylation (ATP synthases), and biosynthesis of coenzymes and various secondary metabolites (Figure 2C).
RNA-seq trends for several genes of interest with moderate to high expression level changes were validated by measuring mRNA levels of fermentor samples via quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) (Supplementary Figure S4). The relative expression levels observed in the qRT-PCR experiments largely replicated the trends observed in the RNA-seq analysis.
In the following sections, we highlight results we believe to be most important.
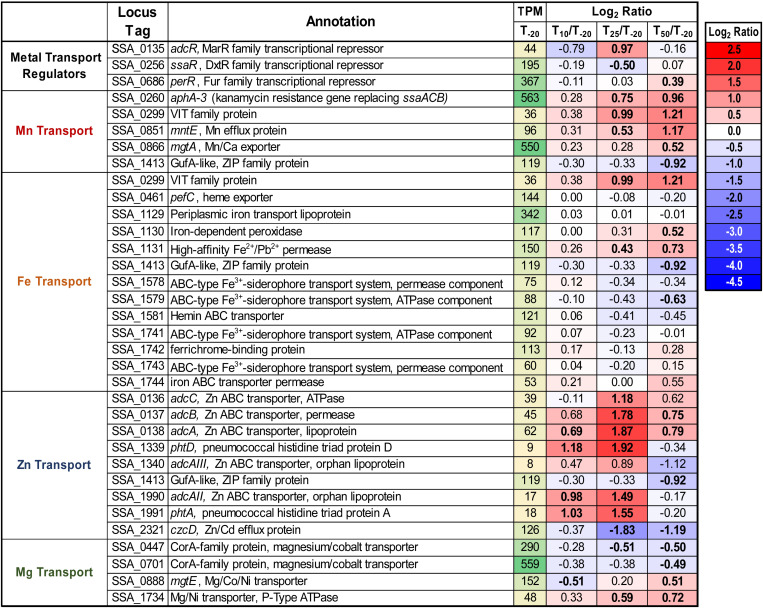
Regulation of Metal Transport Genes
As seen in Figure 1, Mn was the only tested metal whose cellular concentration was decreased upon addition of EDTA to ΔssaACB cells. To further investigate the impact of EDTA on metal transport, we examined the expression of metal transport genes (Figure 3). The kanamycin (Kan) resistance gene aphA-3 that replaced the Mn transporter operon, ssaACB, in this mutant strain was upregulated in all three post-EDTA time points (Figure 3). This is consistent with previous results from our lab showing Mn-dependent repression of SsaB expression as measured by western blot (Crump et al., 2014).
FIGURE 3.
Expression of metal transport genes post-Mn depletion. Putative metal transport genes are depicted with their average transcripts per million reads (TPM) at T–20 and log2 fold change values for each post-EDTA time point. TPM values greater than 1,000 are full saturation (green). Positive log2 fold change values (red) indicate genes upregulated after Mn depletion as compared to T–20, while negative values (blue) indicate downregulated genes. Values in bold indicate significant changes in expression by adjusted P-value (≤0.05).
Given that the cells were Mn-depleted after EDTA addition (Figure 1), it was surprising to see the upregulation at T50 of genes encoding putative orthologs of Mn-export proteins MntE (Rosch et al., 2009; Martin and Giedroc, 2016) and MgtA (Martin et al., 2019) of the closely related species S. pneumoniae (Figure 3). Expression of mntE was found to be constitutive (Martin and Giedroc, 2016) and mgtA expression was found to be positively regulated by Mn through a metal-dependent riboswitch (Martin et al., 2019). We therefore sought to test whether ΔmntE and ΔmgtA mutants generated previously in S. sanguinis (Xu et al., 2011) exhibited increased Mn sensitivity relative to WT as expected based on previous findings in S. pneumoniae (Rosch et al., 2009; Martin et al., 2019). The ΔmgtA mutant grew as expected, with a lower final density than WT in BHI with 2 mM added Mn (data not shown). The ΔmntE mutant, however, grew just like WT, which was unlike the findings in S. pneumoniae (data not shown). We tested up to 10 mM Mn in BHI and Todd Hewitt broth with 1% yeast extract and could not find a Mn concentration that prevented growth of either the ΔmntE mutant or WT (data not shown). Initial metal analysis revealed that the ΔmntE mutant accumulated slightly more Mn than WT (data not shown). These results indicate that S. sanguinis may primarily use MgtA to export excess Mn, and MntE may function differently in S. sanguinis than in S. pneumoniae. Future studies will elucidate the function of these putative exporters and their transcriptional regulation in S. sanguinis.
As seen in Figure 1, cellular Zn levels in ΔssaACB were not significantly altered by EDTA addition, despite the high affinity of this chelator for Zn (Perrin and Dempsey, 1974). Zn level maintenance may be due to the higher levels of Zn than Mn in BHI (1.7 ± 0.02 vs. 0.02 ± 0.003 μg mL–1, respectively) (Murgas et al., 2020) or the regulation of Zn transporter genes. S. sanguinis possesses orthologs of the Zn ABC transporter AdcCBA of S. pneumoniae (Dintilhac and Claverys, 1997), and all three genes were upregulated post-EDTA (Figure 3). Expression of the gene encoding the Zn2+ and Cd2+ efflux protein, CzcD (Nies, 1992), decreased after EDTA addition (Figure 3). Thus, cellular Zn levels appear to have been maintained during EDTA treatment by decreasing export of intracellular Zn and increasing import of any remaining bioavailable Zn through regulation of Zn transporters.
We also examined the regulation of other putative Zn-transport proteins. In S. pneumoniae, AdcAII and several histidine triad proteins also contribute to Zn transport. AdcAII is an orphan lipoprotein of the AdcCBA system (Bayle et al., 2011; Plumptre et al., 2014) and PhtD is a histidine triad protein encoded adjacent to AdcAII (Bersch et al., 2013; Kallio et al., 2014). S. sanguinis has two genes, SSA_1340 and SSA_1990, that encode proteins similar to AdcAII, and each is also adjacent to putative histidine triad protein genes, SSA_1339 or SSA_1991. Because AdcAII is more similar to SSA_1990, we have named this protein AdcAII, whereas we have designated SSA_1340 as AdcAIII. Consistent with a potential role in Zn uptake, all four of these genes were upregulated at T25 (Figure 3). The relative contribution of each of these proteins to Zn import remains to be determined, although we hypothesize that the upregulation of these genes contributes to the tight maintenance of Zn levels in cells post-EDTA.
Expression data for additional putative metal transporters may be found in Figure 3.
Examination of Known Mn-Cofactored Enzymes
Superoxide Dismutase
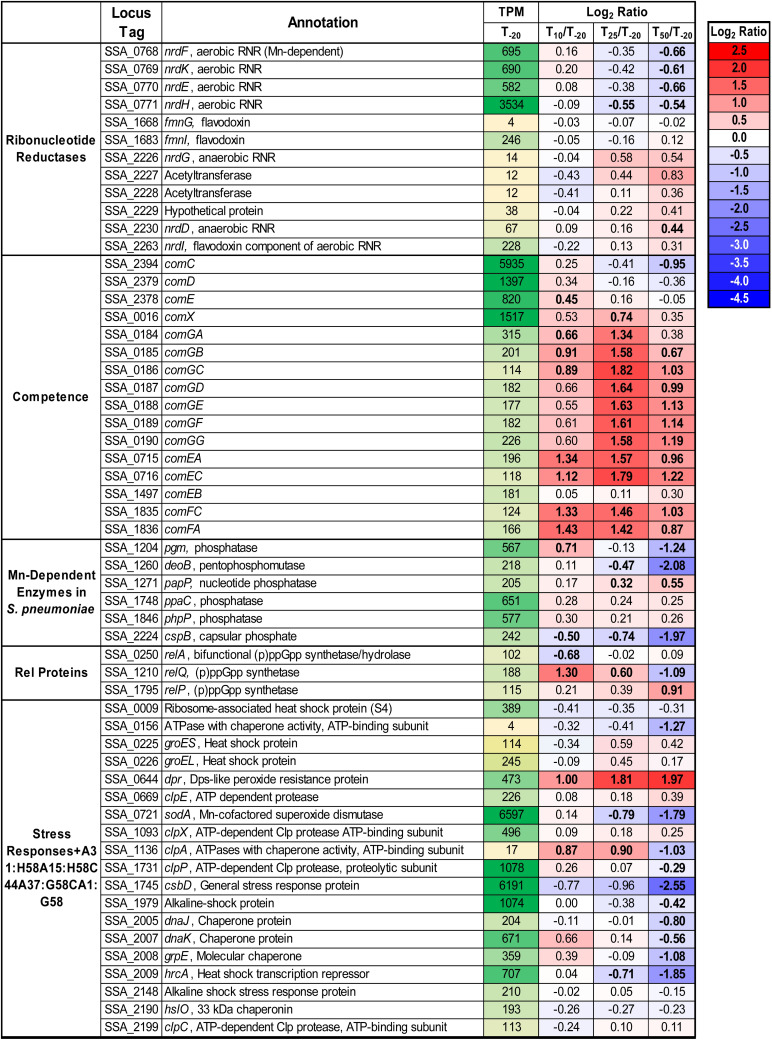
S. sanguinis possesses a single superoxide dismutase, SodA (Xu et al., 2007), and it is Mn-cofactored (Crump et al., 2014). Our previous study indicated that reduced SodA activity could account for only a portion of the reduced virulence and aerobic serum growth of the ΔssaB mutant (Crump et al., 2014). Expression of sodA decreased significantly at both T25 and T50 despite constant air input (Figure 4), which may be due to Mn-dependent positive regulation of transcription (Jakubovics et al., 2002; Eijkelkamp et al., 2014). Given that the fermentor growth conditions do not exactly replicate our previous in vitro or in vivo assays, we wondered whether SodA would be important here. To answer this question, we grew our ΔsodA knockout mutant in the fermentor under the same conditions without EDTA. The ΔsodA mutant grew similarly to WT (Supplementary Figure S5), indicating that Mn-dependent SodA activity is not essential for aerobic growth under these conditions. While this does not rule out the possibility that reduced SodA activity post-Mn depletion contributed to the reduced growth rate of ΔssaACB, it established that it was not the sole cause, thus encouraging us to investigate other possibilities.
FIGURE 4.
Expression of select genes post-Mn depletion. Selected genes of interest are depicted with their average TPM at T–20 and log2 fold change values for each post-EDTA time point. TPM values greater than 1,000 are full saturation (green). Positive log2 fold change values (red) indicate genes upregulated after Mn depletion as compared to T–20, while negative values (blue) indicate downregulated genes. Values in bold indicate significant changes in expression by adjusted P-value (≤0.05).
Class Ib Aerobic Ribonucleotide Reductase
The other known Mn-cofactored enzyme in S. sanguinis is the aerobic class Ib ribonucleotide reductase (RNR), NrdEF (Makhlynets et al., 2014; Rhodes et al., 2014). RNR enzymes catalyze the production of deoxynucleotides from the corresponding ribonucleotides. It was previously found that mutant strains lacking this enzyme were unable to grow in aerobic conditions, whether in serum or BHI. These studies also suggested that Fe could not substitute for Mn as an RNR cofactor in vivo, despite its ability to do so in vitro. In addition to a likely decrease in activity, expression of nrdHEKF was downregulated after Mn depletion (Figure 4). We recently analyzed the metabolome of S. sanguinis cells under the same conditions as this study (Puccio et al., 2020). Levels of detected deoxynucleosides and deoxynucleotides in cells increased or remained constant after Mn depletion. Thus, while NrdEF requires Mn for activity, our data suggest that deoxynucleotides may not be a limiting factor for growth in our study.
Mn-Dependent Phosphatases
In the related species S. pneumoniae, there are six additional enzymes (Figure 4) that have been found to have Mn cofactors (Kuipers et al., 2016; Martin et al., 2017). Orthologs of all six enzymes are encoded in the S. sanguinis genome. Although each is a reciprocal best hit with its S. pneumoniae counterpart by NCBI BlastP, their functions have not been confirmed. Pgm and CspB are phosphatases that have been implicated in capsule biosynthesis in S. pneumoniae, although S. sanguinis lacks a true capsule. DeoB is a phosphopentomutase that connects the pentose phosphate pathway to purine biosynthesis and was also significantly downregulated at T50. Expression of papP, encoding a nucleotide phosphatase, was significantly increased at the later time points and has been shown to affect membrane lipid homeostasis (Kuipers et al., 2016). A significant morphological difference was observed in ΔpapP mutants in S. pneumoniae, but ΔssaACB cells from the T50 sample did not appear morphologically different from cells at T–20 (data not shown). Of note, we observed changes in fatty acid synthesis under these same fermentor growth conditions (Puccio et al., 2020), suggesting that activity of PapP may be reduced but not to the extent that it substantially affects morphology.
Genes encoding the other two phosphatases, PhpP and PpaC, were not differentially expressed at any time point (Figure 4). While this indicates the lack of a Mn-dependent regulation mechanism, it does not rule out the possibility that their activity was decreased due to reduction of Mn. PhpP is a serine/threonine protein phosphatase that is a key regulator of cell division and has been shown to be regulated by the bioavailable Zn:Mn ratio in S. pneumoniae (Martin et al., 2017). While the Zn:Mn ratio did increase over time in our study (Table 1), morphology did not differ from T–20 (data not shown), indicating that PhpP may not be affected by Mn limitation under these conditions. In our recent study, loss of PhpP did not significantly affect the growth of S. sanguinis in serum, a Mn-limited medium (Zhu et al., in press), which indicates that it is likely not responsible for the growth rate decrease observed here. The last phosphatase, PpaC, is essential for S. sanguinis (Xu et al., 2011), so if PpaC activity was decreased due to Mn depletion, this could have contributed to the decreased growth rate phenotype observed post-EDTA. Further studies utilizing the knockout mutants of each non-essential phosphatase (Xu et al., 2011) or an approach such as CRISPR interference (Shields et al., 2020) for PpaC would enhance our understanding of the relative contributions of each phosphatase to the growth and morphology of S. sanguinis.
TABLE 1.
Zn:Mn ratios in fermentor grown cells.
| Strain | T–20 | T10 | T25 | T50 |
| WT | 1.64 | 1.66 | 1.75 | 1.81 |
| ΔssaACB | 2.02 | 2.47* | 3.30* | 5.36* |
Values indicate ratios of Zn concentrations as compared to those of Mn. Concentrations were determined by ICP-OES and significance was determined by one-way ANOVA with a Tukey-Kramer multiple comparisons test vs. T–20 where * indicates P ≤ 0.05.
(p)ppGpp Hydrolase Domain
In streptococci and enterococci, Mn acts as a cofactor for the hydrolase domain of the bifunctional (p)ppGpp synthetase/hydrolase, RelA (also called RSH for RelA/SpoT Homologs) (Mechold et al., 1996). As an alarmone, (p)ppGpp serves as an effector of the stringent response in bacteria (Irving and Corrigan, 2018). Expression of relA was unchanged after EDTA addition, and expression of the other two small alarmone synthetase genes, relP and relQ (Lemos et al., 2007), were significantly increased and decreased, respectively (Figure 4). Both RelP and RelQ were found to produce less (p)ppGpp than RelA in S. mutans (Lemos et al., 2007) and appear to be important during different environmental conditions or growth stages in gram-positive bacteria (Yang et al., 2019).
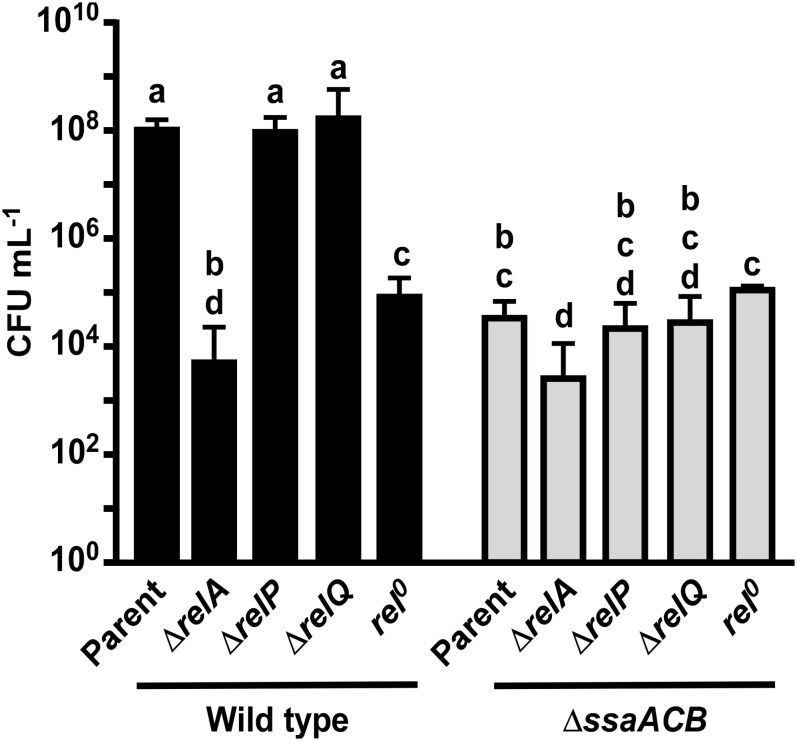
In an attempt to determine whether loss of hydrolase activity in RelA could account for the phenotypes we observed in Mn-depleted cells, we attempted to construct a hydrolase-deficient mutant by altering specific residues (R44, H62, T151) shown by Hogg et al. (2004) to be important for (p)ppGpp hydrolase, but not synthetase activity. Similar to Kaspar et al. (2016), we were unable to generate any of the three point mutants without unintended mutations arising in other regions of the gene (data not shown). This indicates that hydrolase activity may be essential for growth of S. sanguinis. We then obtained strains from the comprehensive S. sanguinis mutant knockout library created by Xu et al. (2011) that were deleted for each of the rel genes. We also generated a rel0 strain by knocking out all three rel genes utilizing a markerless mutagenesis system (Xie et al., 2011; Cheng et al., 2018). We also made these rel knockout mutants in the ΔssaACB background. We then assessed the growth of these mutants in aerobic serum-our in vitro model for infective endocarditis (Crump et al., 2014). As shown in Figure 5, neither ΔrelP nor ΔrelQ grew to a density that differed significantly from its parent strain, whether in the WT or ΔssaACB background. Likewise, in both backgrounds ΔrelA was more severely attenuated than rel0, suggesting that is it more detrimental to lose activity of RelA than to lack all (p)ppGpp.
FIGURE 5.
Aerobic serum growth of rel mutants. Various rel mutants were grown for 24 h in 6% O2 in pooled rabbit serum. The means and standard deviations of at least three replicates are displayed. Significance was determined by one-way ANOVA with a Tukey-Kramer multiple comparisons test. T24 bars with the same letter are not significantly different from each other. Initial culture concentrations were measured (not shown) and found to be not significantly different by one-way ANOVA.
Assessment of Stress and Stress Responses in Mn-Depleted Cells Through Gene Expression
We next sought to determine whether the RNA-seq data suggested anything concerning stresses experienced by the cells. Expression of various stress response genes were assessed, and most were either downregulated or unchanged at T50 (Figure 4), indicating that the reduced growth rate is likely not due to an overwhelming stress response. The only stress response-related gene to show a significant increase in expression at T50 was that encoding the Dps-like peroxide resistance protein, Dpr (Yamamoto et al., 2000). Dpr is a ferritin-like protein that has been shown to be imperative for oxidative stress tolerance in several streptococci (Yamamoto et al., 2000, 2002; Pulliainen et al., 2003; Brenot et al., 2005), including S. sanguinis SK36 (Xu et al., 2014).
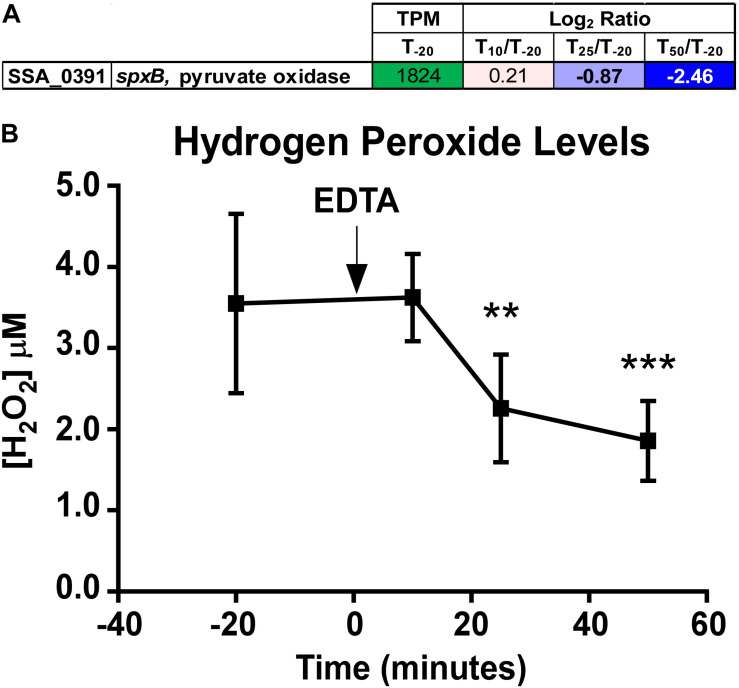
S. sanguinis is known to generate copious amounts of H2O2, presumably to more effectively compete against other oral species, such as the caries-forming pathogen S. mutans (Kreth et al., 2005; Chen et al., 2011). Simple Mn compounds have been reported to prevent oxidative stress by catalyzing the decomposition of H2O2 (Liochev and Fridovich, 2004) and superoxide (Barnese et al., 2008, 2012). We observed a significant decrease in expression of the gene encoding the H2O2-generating enzyme pyruvate oxidase, spxB (Spellerberg et al., 1996; Chen et al., 2011), at T25 and T50 (Figure 6A). To determine whether the decreased growth rate of the ΔssaACB strain during aerobic fermentor growth after Mn depletion was due to excess H2O2 generation or the inability of cells to cope with H2O2 without Mn, H2O2 levels were measured in spent supernatant. Concentrations ranged between 1 and 5 μM (Figure 6B), far lower than has been observed in previous studies employing SK36 (Kreth et al., 2008) despite the constant influx of air into the vessel. H2O2 levels also decreased significantly at T25 and T50 as compared to T–20 (Figure 6B), which correlates with the decreased expression of spxB (Figure 6A). These results indicate that oxidative stress related to excess H2O2 levels is unlikely to be the cause of the growth rate decrease observed upon Mn depletion.
FIGURE 6.
Quantitation of spxB expression and hydrogen peroxide in ΔssaACB fermentor cultures. (A) Expression of the spxB gene in fermentor-grown ΔssaACB cells as determined by RNA-seq analysis. Average TPM at T–20 and log2 fold change values for each after Mn depletion time point are displayed. TPM values greater than 1,000 are full saturation (green). Positive log2 fold change values (red) are genes upregulated in after Mn depletion samples as compared to T–20, while negative values (blue) indicate downregulated genes. Values in bold indicate significant changes in expression by adjusted P-value (≤0.05). (B) H2O2 levels of the BHI culture supernatant were measured at each time point. Means and standard deviations of at least four replicates are shown. Significance was determined by one-way ANOVA with a Tukey-Kramer multiple comparisons test, comparing each post-EDTA time point to T–20. **P ≤ 0.01, ***P ≤ 0.001.
Analysis of Carbon Catabolite Repression and Sugar Transport
Examination of DEG clusters revealed that the majority of those thought to transport sugars were downregulated (Supplementary Table S2), and of these, the majority belonged to the PTS family, which is regulated by carbon catabolite repression (CCR). CCR is a regulatory mechanism that gives bacteria the ability to utilize carbon sources in order of preference (Gorke and Stulke, 2008). In gram-positive bacteria, a carbon catabolite protein such as CcpA binds to catabolite responsive elements (cre) and represses transcription of genes encoding non-preferred carbon source transport and utilization systems (Warner and Lolkema, 2003). To determine the extent to which CcpA binding could be responsible for the observed downregulation, cre sites identified previously by RegPrecise (Novichkov et al., 2013) and by our custom searches were collected and compared. Using these methods, 393 putative binding sites were identified (Supplementary Table S2). Several PTS and sugar ABC transport genes were predicted to have 5’ cre sites, the majority of which were downregulated at T50. Other genes known to be CcpA-regulated in SK36, such as spxB (Zheng et al., 2011; Redanz et al., 2018), were downregulated as well. This is surprising given that the glucose-containing media was replenished at a constant and rapid rate throughout the experiment, indicating that there could be a Mn-related mechanism for CcpA repression. Cells also did not appear to be starved for glucose; when excess glucose (1.8%) was added to the media, the post-EDTA growth rate was similar to that of normal BHI, which contains 0.2% glucose (data not shown). This is not entirely unexpected, as it has been established by Redanz et al. (2018) that CcpA repression of spxB in S. sanguinis is glucose-independent.
Several CcpA-regulated genes were annotated as amino acid transporters and synthetases, which led us to examine the expression of genes with these putative functions. Many of these genes were differentially expressed (Supplementary Figure S6), which may be due to the intertwined nature of amino acid and carbohydrate metabolism (Radin et al., 2016). We also observed decreased expression of large, contiguous loci encoding ethanolamine utilization (Fox et al., 2009; Kaval and Garsin, 2018; Kaval et al., 2019), a type IV pilus system (Chen et al., 2019), and CRISPR-associated proteins (Makarova et al., 2015; Gong et al., 2020; Supplementary Figure S7), which are all also tied to CcpA-regulation (Bai et al., 2019).
Discussion
Only two enzymes have been confirmed to be Mn-dependent in S. sanguinis, and few others have been identified in other streptococci. Despite this, we observed changes in a wide variety of systems after Mn depletion of the ΔssaACB mutant using EDTA. One possible explanation for this discrepancy is that Mn binds with low affinity to most proteins, resulting in Mn loss or replacement during purification. In fact, initial studies of the aerobic class Ib RNR identified Fe as the exclusive cofactor based on RNR activity in vitro and the fact that Fe was present in many different bacterial RNRs heterologously expressed in E. coli. Only later was it discovered that these enzymes were Mn-cofactored when natively expressed (Cotruvo and Stubbe, 2012), and despite the in vitro activity of both forms of the S. sanguinis RNR, only the Mn-cofactored version was active in vivo (Makhlynets et al., 2014; Rhodes et al., 2014). Additional Mn-dependent enzymes may have similarly escaped detection. Another possible explanation is that Mn depletion impacts several key regulatory systems, such as CCR and (p)ppGpp, which leads to changes in the expression of many different genes. Mn levels have been found to be related to each of these systems in other gram-positive bacteria (Kehres and Maguire, 2003; Colomer-Winter et al., 2017). Mn depletion does not appear to induce a traditional stress response, although expression of an oxidative stress tolerance protein, Dpr, significantly increased, which is consistent with a recent study on Mn depletion in S. mutans (Kajfasz et al., 2020). Here we highlight Mn-related systems we identified in this study of S. sanguinis for future investigation.
Mn Depletion Leads to Glucose-Independent Changes in the Regulon of CcpA
Given that BHI contains glucose, it was expected that S. sanguinis would preferentially transport and utilize it as a preferred carbon source under standard fermentor conditions. This is supported by the fact that glucose levels decreased in the media after cell growth (Puccio et al., 2020) as well as by the high expression of putative glucose transporters SSA_1752, SSA_1918-1920, and SSA_1298-1300 (Ajdic and Pham, 2007) at T–20 (Supplementary Table S2). Surprisingly, expression of nearly all sugar transport systems decreased after Mn depletion (Supplementary Table S2), despite constant levels of glucose in the cells (Puccio et al., 2020). CcpA is known to repress its own expression in a glucose-dependent manner (Bai et al., 2019), and yet much like the glucose transporters, ccpA expression was high at T–20 and significantly decreased by T50 (Supplementary Table S2). Potential explanations could include: (i) 2 g/L glucose in BHI is not sufficient to induce CcpA repression; (ii) other regulatory mechanisms are preventing proper CCR under these conditions; or (iii) much like spxB, many other systems in S. sanguinis are subject to glucose-independent CcpA repression. Redanz et al. (2018) used 0.3% as the low-glucose condition in their study of CcpA repression of spxB in S. sanguinis, whereas Bai et al. (2019) used BHI alone (0.2% glucose) to observe differences in the transcriptome between the WT and ΔccpA S. sanguinis strains. Thus, the glucose concentration in BHI may indeed be low, yet sufficient to induce some repression of its regulon.
It is interesting that Mn depletion leads to an apparent increase in CcpA repression because we are not aware of this having ever been reported. The strongest evidence for CcpA-dependent regulation is spxB. In S. sanguinis, spxB expression has been shown to be positively regulated by SpxA1 (Chen et al., 2012) and VicK (Moraes et al., 2014) and negatively regulated by CcpA (Zheng et al., 2011). The spxA1 gene was in the top 10% of all genes based on expression at T–20 and remained unchanged after EDTA addition (Supplementary Table S1), indicating that repression by CcpA is likely responsible for the decrease in spxB expression as opposed to changes in induction by SpxA1. The mechanism by which CcpA represses spxB expression in S. sanguinis is unique from other streptococci in that it is independent of glucose (Redanz et al., 2018). It was previously determined that Mn may play a role in spxB expression in S. pneumoniae, as a ΔmntE mutant in S. pneumoniae accumulated Mn and produced more H2O2 than WT under excess Mn conditions (Rosch et al., 2009).
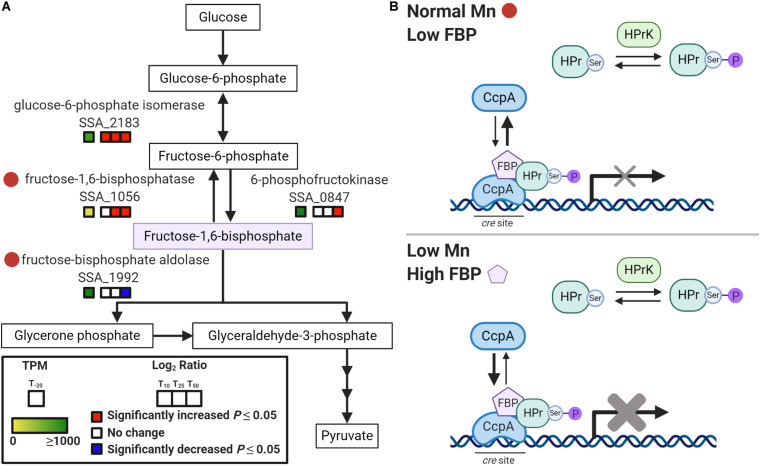
The connection between Mn and sugar catabolism is not unprecedented, as previous studies have implicated Mn as important for the activity of carbon catabolism enzymes in other bacteria (Kehres and Maguire, 2003). Additionally, recent studies in S. pneumoniae and S. mutans have shown that fluctuations in metal homeostasis influence the regulation of carbohydrate metabolism (Burcham et al., 2020; Kajfasz et al., 2020). One possible explanation for the Mn-dependent, glucose-independent CcpA repression observed in our study is the accumulation of the glycolytic intermediate, fructose-1,6-bisphosphate (FBP) during Mn depletion (Figure 7). In Firmicutes, phosphorylation of histidine phosphocarrier protein (HPr) to HPr(Ser-P) occurs when FBP and ATP levels are high (Gorke and Stulke, 2008). HPr(Ser-P) then binds to CcpA, which in turn induces the binding of the repressor to cre sites on the DNA. Additionally, FBP enhances the binding interaction of HPr(Ser-P) and CcpA, increasing repression. In our concurrent metabolomics study, we found that ΔssaACB cells accumulated FBP at T50 after Mn depletion (Puccio et al., 2020), which may explain the strong evidence for CcpA repression. As expected if CcpA were responsible for the changes in expression, we found that 48 out of the 169 DEGs found by Bai et al. (2019) when comparing a ΔccpA mutant to the SK36 WT were changed in the opposite direction as our T50 sample. However, 15 significant DEGs were in the same direction, and the remainder were unchanged in our study. Additionally, most of the DEGs we observed in our study did not overlap with those of Bai et al. (2019). This comparison indicates that CcpA-dependent repression could be responsible for some of the changes in expression after Mn depletion, but it does not explain all of the observed results.
FIGURE 7.
Model of Mn-dependent CcpA repression. (A) Depiction of a segment of the glycolysis and gluconeogenesis pathway in S. sanguinis from KEGG (Kanehisa and Goto, 2000). Red circles indicate potentially Mn-cofactored enzymes. Gene expression for each post-EDTA time point compared to T–20 is indicated by the colored boxes. Significant changes (P < 0.05) are indicated by red (increased) or blue (decreased). TPM values at T–20 are colored as indicated, with values ≥ 1,000 fully saturated (green). (B) Model of Mn-dependent CcpA repression based on CCR in Firmicutes as described by Gorke and Stulke (2008). In the top panel, normal Mn levels in BHI result in low FBP, which leads to less phosphorylation of HPr to HPr(Ser-P) by HPr kinase/phosphorylase (HPrK). With low FBP and HPr(Ser-P), CcpA exists mainly in its free state, unbound to cre sites in the DNA. This results in little to no repression of the CcpA regulon. The bottom panel depicts Mn depletion, where reduced activity of fructose-1,6-bisphosphatase and fructose-bisphosphate aldolase leads to an accumulation of FBP. This induces the phosphorylation of HPr so that it is primarily in the HPr(Ser-P) state. Increased FBP levels also enhance binding of CcpA to HPr(Ser-P) and to DNA. This results in augmented repression of the CcpA regulon.
To determine potential causes for the accumulation, we examined the enzymes required for synthesizing and catabolizing FBP (Figure 7A). Both enzymes responsible for metabolizing FBP, fructose-1,6-bisphosphatase (Fbp; SSA_1056) and fructose-bisphosphate aldolase (Fba; SSA_1992), may require a Mn cofactor according to BRENDA (RRID:SCR_002997)1 (Jeske et al., 2019). Thus, it is possible that Mn depletion led to reduced activity of both Fba and Fbp and this resulted in accumulation of FBP (Puccio et al., 2020), which in turn induced CcpA repression after Mn depletion (Figure 7B). Enzymatic activity assays will be required to determine the true cofactor for these enzymes in S. sanguinis, but accumulation of FBP is strong evidence that the activity of at least one enzyme in this pathway is affected by Mn depletion.
Reduced (p)ppGpp Hydrolase Activity May Contribute to the Post-Mn Depletion Phenotype
The relationship between (p)ppGpp and carbon source utilization has been previously established; S. mutans strains lacking RelA or all Rel proteins showed delayed growth rates when transitioned from media containing glucose to lactose (Zeng et al., 2018). When we examined the genome for cre sites, relQ was predicted to have a 5’ cre site and was downregulated at T50 (Supplementary Table S2), indicating that it could be under CcpA control. Little is known about the transcriptional regulation of relA in streptococci (Nascimento et al., 2008; Irving and Corrigan, 2018), although regulation of activity has been established in other species (Gratani et al., 2018). In S. mutans, expression of relP is activated by a two-component system, RelRS, which is thought to sense oxidative stressors (Seaton et al., 2011). It was hypothesized by Kim et al. (2012) that (p)ppGpp production by RelP in S. mutans may be an attempt by the cell to slow growth to minimize damage from oxygen radicals produced during metabolism. While it was observed in this study that H2O2 levels decreased in response to Mn depletion, it is possible that other ROS were present due to a decrease in SodA activity. Thus, increased expression and activity of RelP, in addition to lack of hydrolase activity by RelA, could be at least partly responsible for the reduced growth rate.
In analyzing previous transcriptome studies using ΔrelA or rel0 mutants in the related species S. mutans, S. pneumoniae, and Enterococcus faecalis (Nascimento et al., 2008; Kazmierczak et al., 2009; Colomer-Winter et al., 2019), we noted many expression patterns similar to our study. While these studies utilized different species and growth conditions and thus are not a direct comparison, it is remarkable that reduction in (p)ppGpp levels would lead to similar changes in gene expression as Mn depletion. Of interest to us, several PTS genes were downregulated in all three previous studies. Similar to the results we observed in this study, the S. pneumoniae ΔrelA mutant showed decreased expression of spxB and sodA (Kazmierczak et al., 2009). These comparisons indicate either that dysregulation of (p)ppGpp levels leads to changes in expression of these genes in response to stress or that decreased activity of the Rel hydrolase domain due to Mn depletion is not responsible for the observed changes in expression of these genes in S. sanguinis.
Based on these results, we hypothesize that reduced activity of the RelA hydrolase domain may contribute to the observed reduction in growth rate in the fermentor studies but is not entirely responsible. Specifically, our inability to eliminate the cell’s only known (p)ppGpp hydrolase, combined with our finding that the ΔrelA strains, which also have no hydrolase, exhibited worse growth than rel0 mutants having no synthetase and therefore no (p)ppGpp, suggests that (p)ppGpp accumulation is highly detrimental to growth. A definitive test of this hypothesis will require measurement of (p)ppGpp levels in fermentor-grown cells. We are currently assessing various approaches for feasibility. In addition, the significant decrease in growth of the ΔssaACB ΔrelA strain as compared to the ΔssaACB parent shows that there is an additive effect, indicating that the impact of the loss of RelA is not entirely Mn-dependent.
Conclusion
The effect of Mn depletion on a multitude of diverse systems indicates that the impact of Mn is not relegated to only a few enzymes. Depletion of Mn does not induce a traditional stress response, instead inducing what appears to be dysregulation of many different genes that leads to rapid reduction in the growth rate, despite plentiful nutrients and other metals. While decreased function of the known Mn-cofactored enzymes, such as NrdF, SodA, and the hydrolase domain of RelA, likely contributed to the decreased growth rate we observed upon Mn depletion, it is probably a combination of multiple systems leading to the observed phenotype. Additionally, many of the affected systems appear to be regulated by CCR through CcpA-dependent repression in a glucose-independent manner. Future research will focus on determining the respective contribution of each putative Mn-dependent enzyme as well as whether there is a direct relationship between Mn and CCR.
Materials and Methods
Bacterial Strains and Growth Conditions
The S. sanguinis strain SK36 is a human oral isolate from Mogens Killian, Aarhus University, Denmark. All mutant strains were generated in the SK36 background (Table 2) using the primers listed in Supplementary Table S3. The ΔssaACB strains were generated previously (Baker et al., 2019) with all three genes replaced with either a kanamycin (Kan) resistance gene, aphA-3, or tetracycline (Tet) resistance gene, tetM, using gene splicing by overlap extension (SOEing) PCR (Ho et al., 1989). The ΔssaACB::aphA-3 mutation is non-polar, as confirmed by complementation (Murgas et al., 2020). With the exception of Figure 5, all ΔssaACB experiments were completed with the KanR strain, JFP169. The ΔrelA, ΔrelP, and ΔrelQ mutants were re-created for this study by amplifying the aphA-3 gene and flanking DNA from the corresponding, previously created mutants (Xu et al., 2011) using primers from the same study. All PCR products were purified using a Qiagen MinElute PCR kit prior to transformation. Transformations employing antibiotic selection were performed using the protocol described previously (Paik et al., 2005). Briefly, an overnight culture of the parent strain was grown in BD BactoTM Todd Hewitt broth with horse serum (Invitrogen), then diluted 200-fold and incubated at 37°C. Optical density (OD600) of tube cultures was determined using a Thermo Scientific BioMate 3S UV-VIS spectrophotometer. Knockout construct DNA (100 ng) and S. sanguinis competence stimulating peptide (70 ng) were added to the culture (OD600 ∼0.07) and incubated at 37°C for 1.5 h prior to selective plating on BHI (BD) agar plates with antibiotics. Kan (Sigma-Aldrich) was added to a final concentration of 500 μg mL–1. All plates were incubated for 24 h at 37°C under anaerobic conditions, where atmospheric composition was adjusted using a programmable Anoxomat Mark II jar-filling system (AIG, Inc.) and a palladium catalyst was included in the jars. All mutants were confirmed to have the expected composition by sequence analysis of the DNA flanking the insertion sites.
TABLE 2.
Strains used in this study.
| Strain | Description | Source or reference |
| SK36 | Human oral plaque isolate | M. Killian, Aarhaus University; Xu et al., 2007 |
| JFP132 | ΔsodA::aphA-3 | Crump et al., 2014 |
| JFP169† | ΔssaACB::aphA-3 | Baker et al., 2019 |
| JFP173‡ | ΔssaACB::tetM | Baker et al., 2019 |
| SSX_0250 | ΔrelA::aphA-3 | Xu et al., 2011 |
| JFP259 | ΔrelA::aphA-3 | This study |
| JFP260 | ΔssaACB::tetM ΔrelA::aphA-3 | This study |
| SSX_1210 | ΔrelQ::aphA-3 | Xu et al., 2011 |
| JFP279 | ΔrelQ::aphA-3 | This study |
| JFP281 | ΔssaACB::tetM ΔrelQ::aphA-3 | This study |
| SSX_1795 | ΔrelP::aphA-3 | Xu et al., 2011 |
| JFP275 | ΔrelP::aphA-3 | This study |
| JFP277 | ΔssaACB::tetM ΔrelP::aphA-3 | This study |
| JFP276 | ΔrelA::aphA-3ΔrelP | This study |
| JFP278 | ΔssaACB::tetM ΔrelA::aphA-3ΔrelP | This study |
| JFP280 | ΔrelA::aphA-3ΔrelPΔrelQ (rel0) | This study |
| JFP282 | ΔssaACB::tetM ΔrelA::aphA-3ΔrelPΔrelQ (rel0) | This study |
| SK36 IFDC | pheS* ermAM | Cheng et al., 2018 |
†Designated ΔssaACB throughout the manuscript. ‡Only used for ΔssaACB strains in Figure 5.
To generate the rel0 strain, ΔrelP and ΔrelQ mutations were made using a markerless mutation system described previously (Xie et al., 2011; Cheng et al., 2018). Briefly, the IFDC2 cassette was amplified from the S. sanguinis IFDC2 strain and combined with flanking region from relP using gene SOEing. The two parent ΔrelA strains (WT and ΔssaACB::tetM backgrounds) were then transformed as described above, plating on BHI agar plates containing 10 μg mL–1 erythromycin (Erm; Fisher Scientific). A gene SOEing product merging the two flanking regions of relP was then generated. This SOEing product was then used to transform the ErmR colonies from the first transformation. Immediately prior to plating on agar plates containing 20 mM 4-chloro-phenylalanine (Sigma-Aldrich), the cells were washed twice with phosphate-buffered saline (PBS) to remove excess media. Resulting colonies were then screened for Erm sensitivity and sequenced to confirm removal of the desired gene and IFDC2 cassette. The process was then repeated for relQ, converting the two ΔrelA ΔrelP parent strains into rel0 strains.
Overnight BHI cultures (pre-cultures) were inoculated from single-use aliquots of cryopreserved cells by 1,000-fold dilution. Antibiotics were included in mutant pre-cultures at the aforementioned concentrations or 5 μg mL−1 for Tet (Sigma Aldrich). Cultures were then incubated for approximately 18 h at 6% O2 (Anoxomat jar set to 6% O2, 7% H2, 7% CO2, and 80% N2) at 37°C. To determine CFUs, samples were sonicated for 90 s using an ultrasonic homogenizer (Biologics, Inc) to disrupt chains prior to dilution in PBS and plated using an Eddy Jet 2 spiral plater (Neutec Group, Inc.). For static growth studies, tubes containing 100% pooled rabbit serum (Gibco) were pre-incubated at 37°C in an atmosphere of 6% O2. Each tube was then inoculated with a 10–6-fold dilution of the overnight pre-culture, as described above. The inoculated tubes were returned to incubate at the same oxygen concentration. Cultures were removed after 24 h, sonicated, and diluted in PBS prior to plating on BHI agar. Plates were incubated for 24 h in 0% O2 prior to colony enumeration.
Fermentor Growth Conditions and Sample Collection
A BIOSTAT B bioreactor (Sartorius Stedim) with a 1.5-L capacity UniVessel glass vessel® was used for growth of 800-mL cultures at 37°C, as described in Puccio and Kitten (2020). Briefly, cultures were stirred at 250 rpm and pH was maintained by the automated addition of 2 N KOH (Fisher Chemical). A 40-mL overnight pre-culture of S. sanguinis was grown as described above and centrifuged for 10 min at 3,740 × g in an Allegra X-142 centrifuge at 4°C (Beckman-Coulter). The supernatant was discarded and the cells were resuspended in BHI prior to inoculation. The air flow was increased stepwise, based on the OD of the fermentor culture. At the peak OD, air flow was increased to 0.50 liters per min, input flow of fresh BHI was set to 17% (∼700 mL h–1) and output flow of waste was set to 34%. Cells were allowed to acclimate to the new conditions for 1 h. The T–20 sample was aseptically removed for total RNA isolation or metal analysis (described below). EDTA (Invitrogen) was introduced to the carboy 16 mins later (T–4) to achieve a final concentration of 100 μM. EDTA was then introduced directly to the vessel 4 min later (T0), corresponding to the time at which EDTA from the carboy would reach the vessel, to achieve a final concentration of 100 μM. Samples were taken for each post-EDTA time point (T10, T25, T50). In some experiments, MnSO4 or FeSO4 (PuratronicTM; Alfa Aesar) was added to the carboy (T66) and vessel (T70) at a final concentration of 100 μM and samples were taken for metal analysis at T80.
Metal Analysis
Additional 40-mL cell culture samples were collected from WT and ΔssaACB cells at the same fermentor growth time points as described above. The cells were immediately centrifuged at 3,740 × g for 10 min at 4°C. The supernatant was decanted and the cell pellet was washed twice with cold cPBS (PBS treated with Chelex-100 resin (Bio-Rad) for 2 h, then filter sterilized and supplemented with EDTA to 1 mM). The pellet was then divided for subsequent acid digestion or protein concentration determination. Trace metal grade (TMG) nitric acid (15%) (Fisher Chemical) was added to one portion of the pellet. The pellet was digested using an Anton Paar microwave digestion system using a modified Organic B protocol: 120°C for 10 min, 180°C for 20 min, with the maximum temperature set to 180°C. The digested samples were then diluted 3-fold with Chelex-treated dH2O. Metal concentrations were determined using an Agilent 5110 inductively coupled plasma-optical emission spectrometer (ICP-OES). Concentrations were determined by comparison with a standard curve created with a 10 μg mL–1 multi-element standard (CMS-5; Inorganic Ventures) diluted in 5% TMG nitric acid. Pb (Inorganic Ventures) was used as an internal standard (10 μg mL–1). The other portion of the pellet was resuspended in PBS and mechanically lysed using a FastPrep-24 instrument with Lysing Matrix B tubes (MP Biomedicals) as described previously (Rhodes et al., 2014). Insoluble material was removed by centrifugation. Protein concentrations were determined using a bicinchoninic acid Protein Assay Kit (Pierce) as recommended by the manufacturer, with bovine serum albumin as the standard. Absorbance was measured in a black, flat-bottom 96-well plate (Greiner) using a microplate reader (BioTek).
Total RNA Isolation
For each RNA sample, 2 mL of fermentor culture was added to 4 mL RNAprotect Bacteria Reagent (Qiagen) and immediately vortexed for 10 s. The sample was then incubated at room temperature for 5–90 min. The samples were then centrifuged for 10 min at 3,740 × g at 4°C. The supernatant was discarded and the samples stored at −80°C. RNA isolation and on-column DNase treatment were completed using the RNeasy Mini Kit and RNase-Free DNase Kit, respectively (Qiagen). RNA was eluted in 50 μL RNase-Free water (Qiagen). A second DNase treatment was then performed on the samples (Invitrogen). Total RNA was quantified and purity was assessed using a Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific).
RNA-Seq Library Preparation and Sequencing
Total RNA quantity and integrity were determined using an Agilent Bioanalyzer RNA Pico assay. All samples passed quality control assessment with RNA Integrity Numbers (RIN) above 8. Two sequential rounds of ribosomal reduction were then performed on all samples using Illumina’s Ribo-Zero rRNA Removal Kit. The resulting depleted RNA was assessed using an Agilent Bioanalyzer RNA Pico assay to confirm efficient rRNA removal. Stranded RNA-seq library construction was then performed on the rRNA-depleted RNA using the Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs), following manufacturer’s specifications for library construction and multiplexing. Final Illumina libraries were assessed for quality using an Agilent Bioanalyzer DNA High Sensitivity Assay and qPCR quantification was performed using NEBNext Library Quant kit for Illumina (New England Biolabs). Individual libraries were pooled equimolarly, and the final pool was sequenced on an Illumina MiSeq, with 2 × 150-bp paired-end reads. Demultiplexing was performed on the Illumina MiSeq’s on-board computer and resulting demultiplexed files uploaded to Illumina BaseSpace for data delivery. The University of Virginia Department of Biology Genomics Core Facility (Charlottesville, Virginia) completed all RNA-seq library preparation and sequencing.
RNA-Seq Analysis Pipeline
Using Geneious 11.1 (RRID:SCR_010519)2, sequence reads were paired and then trimmed using the BBDuk Trimmer prior to mapping to a modified SK36 genome, in which the ssaACB operon was replaced with the aphA-3 sequence. The locus tags are from the Genbank annotation (Benson et al., 2013) available at the time; the annotations were updated shortly before publication and the new locus tags are included in Supplementary Table S1 for reference. PATRIC annotations (RRID:SCR_004154)3 (Wattam et al., 2017) are also included for reference. Reads for each post-EDTA sample were compared to the corresponding pre-EDTA (T–20) sample using DESeq2 (Love et al., 2014) (RRID: SCR_015687) in Geneious to determine log2 fold changes and adjusted P-values. Principal component analysis was completed using R (version 3.6.1) and RStudio (version 1.2.5033-1) with Bioconductor (Bioconductor, RRID:SCR_006442) package pcaExplorer version 2.13.0 (Marini and Binder, 2019). Volcano plots were generated using R and RStudio with Bioconductor package EnhancedVolcano (Blighe et al., 2018). All DEGs were input into the DAVID database (RRID:SCR_001881)4 (Dennis et al., 2003). The KEGG_pathway option was chosen for functional annotation clustering. The P-value shows the significance of pathway enrichment. Figure 2C was generated using an R script5.
Hydrogen Peroxide Quantification
Culture supernatants without RNAprotect were collected at each time point and stored at −20°C. Hydrogen peroxide concentration was measured using a Fluorometric Hydrogen Peroxide Assay Kit (Sigma). Standards were prepared from 3% hydrogen peroxide provided with the kit as recommended by the manufacturer. Fluorescence was measured in a black-walled, flat-bottom 96-well plate (Greiner) using a microplate reader (BioTek).
Data Analysis and Presentation
All statistical tests, excluding RNA-seq DESeq2 calculations, were performed in GraphPad InStat6. Significance was determined by analysis of variance (ANOVA) as indicated in the figure legends. For ANOVA, a Tukey-Kramer test for multiple comparisons was used when P ≤ 0.05. DESeq2 calculations were completed in Geneious 11.1 or in the pcaExplorer R package, as indicated above. Confidence intervals (95%) of replicate samples were determined by the pcaExplorer R package. P ≤ 0.05 were considered significant. All graphs and the heat map were constructed using GraphPad Prism6; Figure 7 was made using Biorender.com.
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE150593 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE150593).
Author Contributions
TP and TK designed the experiments and wrote all drafts of the manuscript. TP and KK performed the experiments. TP, KK, and BZ generated the figures. TP, TK, KK, BZ, and PX analyzed the data. All authors reviewed and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Shannon Green, Seon-Sook An, Brittany Spivey, and Rachel Korba for meaningful discussions and assistance with experiments. We appreciate Jody Turner (VCU Department of Chemistry) for advice and technical support regarding ICP-OES. We thank Jens Kreth and Nyssa Cullin (Oregon Health & Science University) for providing the IFDC2 S. sanguinis strain and corresponding protocol and Stephanie Neal for construction of JFP169.
Funding. This work was supported by the National Institutes of Health: award no. R01 AI114926 to TK from the National Institute of Allergy and Infectious Diseases; awards no. R01 DE023078 to PX and no. F31 DE028468 to TP from the National Institute of Dental and Craniofacial Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.592615/full#supplementary-material
RNA-seq Analysis.
Analysis of putative cre sites and CcpA regulon.
Primers used in this study.
Figures, Methods, and References.
References
- Ajdic D., Pham V. T. (2007). Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189 5049–5059. 10.1128/JB.00338-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Shang M., Xu M., Wu A., Sun L., Zheng L. (2019). Transcriptome, phenotypic, and virulence analysis of Streptococcus sanguinis SK36 wild type and its CcpA-null derivative (ΔCcpA). Front. Cell Infect. Microbiol. 9:411. 10.3389/fcimb.2019.00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. P., Nulton T. J., Kitten T. (2019). Genomic, phenotypic, and virulence analysis of Streptococcus sanguinis oral and infective-endocarditis isolates. Infect. Immun. 87:e00703-18. 10.1128/IAI.00703-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnese K., Gralla E. B., Cabelli D. E., Valentine J. S. (2008). Manganous phosphate acts as a superoxide dismutase. JACS 130 4604–4606. 10.1021/ja710162n [DOI] [PubMed] [Google Scholar]
- Barnese K., Gralla E. B., Valentine J. S., Cabelli D. E. (2012). Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds. Proc. Natl. Acad. Sci. U.S.A. 109 6892–6897. 10.1073/pnas.1203051109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashore T. M., Cabell C., Fowler V., Jr. (2006). Update on infective endocarditis. Curr. Probl. Cardiol. 31 274–352. 10.1016/j.cpcardiol.2005.12.001 [DOI] [PubMed] [Google Scholar]
- Bayle L., Chimalapati S., Schoehn G., Brown J., Vernet T., Durmort C. (2011). Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol. Microbiol. 82 904–916. 10.1111/j.1365-2958.2011.07862.x [DOI] [PubMed] [Google Scholar]
- Belda-Ferre P., Alcaraz L. D., Cabrera-Rubio R., Romero H., Simon-Soro A., Pignatelli M., et al. (2012). The oral metagenome in health and disease. ISME J. 6 46–56. 10.1038/ismej.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B. A., Li L., Yakovenko O., Wong M., Barnard K. N., Iverson T. M., et al. (2019). Recognition of specific sialoglycan structures by oral streptococci impacts the severity of endocardial infection. PLoS Pathog. 15:e1007896. 10.1371/journal.ppat.1007896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensing B. A., Li Q., Park D., Lebrilla C. B., Sullam P. M. (2018). Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: implications for infective endocarditis. Glycobiology 28 601–611. 10.1093/glycob/cwy052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D. A., Cavanaugh M., Clark K., Karsch-Mizrachi I., Lipman D. J., Ostell J., et al. (2013). GenBank. Nucleic Acids Res. 41 D36–D42. 10.1093/nar/gks1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersch B., Bougault C., Roux L., Favier A., Vernet T., Durmort C. (2013). New insights into histidine triad proteins: solution structure of a Streptococcus pneumoniae PhtD domain and zinc transfer to AdcAII. PLoS One 8:e81168. 10.1371/journal.pone.0081168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blighe K., Rana S., Lewis M. (2018). EnhancedVolcano: Publication-ready Volcano Plots with Enhanced Colouring and Labeling [Online]. Available online at: https://github.com/kevinblighe/EnhancedVolcano (accessed on 10 January 2020). [Google Scholar]
- Bor D. H., Woolhandler S., Nardin R., Brusch J., Himmelstein D. U. (2013). Infective endocarditis in the U.S., 1998-2009: a nationwide study. PLoS One 8:e60033. 10.1371/journal.pone.0060033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenot A., King K. Y., Caparon M. G. (2005). The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55 221–234. 10.1111/j.1365-2958.2004.04370.x [DOI] [PubMed] [Google Scholar]
- Burcham L. R., Hill R. A., Caulkins R. C., Emerson J. P., Nanduri B., Rosch J. W., et al. (2020). Streptococcus pneumoniae metal homeostasis alters cellular metabolism. Metallomics 12 1416–1427. 10.1039/d0mt00118j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne R. A., Chen Y.-Y. M. (1998). The use of continuous flow bioreactors to explore gene expression and physiology of suspended and adherent populations of oral streptococci. Methods Cell Sci. 20 181–190. 10.1007/978-94-017-2258-2_20 [DOI] [Google Scholar]
- Cahill T. J., Baddour L. M., Habib G., Hoen B., Salaun E., Pettersson G. B., et al. (2017). Challenges in infective endocarditis. J. Am. Coll. Cardiol. 69 325–344. 10.1016/j.jacc.2016.10.066 [DOI] [PubMed] [Google Scholar]
- Chen L., Ge X., Dou Y., Wang X., Patel J. R., Xu P. (2011). Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology 157 13–20. 10.1099/mic.0.039669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ge X., Wang X., Patel J. R., Xu P. (2012). SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS One 7:e40034. 10.1371/journal.pone.0040034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Chiang Y. C., Tseng T. Y., Wu H. Y., Chen Y. Y., Wu C. H., et al. (2019). Molecular and functional analysis of the type IV pilus gene cluster in Streptococcus sanguinis SK36. Appl. Environ. Microbiol. 85:e02788-18. 10.1128/AEM.02788-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Redanz S., Cullin N., Zhou X., Xu X., Joshi V., et al. (2018). Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral streptococci. Appl. Environ. Microbiol. 84:e01697-17. 10.1128/AEM.01697-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Winter C., Flores-Mireles A. L., Kundra S., Hultgren S. J., Lemos J. A. (2019). (p)ppGpp and CodY promote Enterococcus faecalis virulence in a murine model of catheter-associated urinary tract infection. mSphere 4:e00392-19. 10.1128/mSphere.00392-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Winter C., Gaca A. O., Lemos J. A. (2017). Association of metal homeostasis and (p)ppGpp regulation in the pathophysiology of Enterococcus faecalis. Infect. Immun. 85:e00260-17. 10.1128/IAI.00260-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotruvo J. A., Jr., Stubbe J. (2012). Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4 1020–1036. 10.1039/c2mt20142a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump K. E., Bainbridge B., Brusko S., Turner L. S., Ge X., Stone V., et al. (2014). The relationship of the lipoprotein SsaB, manganese and superoxide dismutase in Streptococcus sanguinis virulence for endocarditis. Mol. Microbiol. 92 1243–1259. 10.1111/mmi.12625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Kanamoto T., Ge X., Xu P., Unoki T., Munro C. L., et al. (2009). Contribution of lipoproteins and lipoprotein processing to endocarditis virulence in Streptococcus sanguinis. J. Bacteriol. 191 4166–4179. 10.1128/JB.01739-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayer M., Thornhill M. (2018). Is antibiotic prophylaxis to prevent infective endocarditis worthwhile? J. Infect. Chemother. 24 18–24. 10.1016/j.jiac.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., et al. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:3. [PubMed] [Google Scholar]
- Dintilhac A., Claverys J.-P. (1997). The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res. Microbiol. 148 119–131. 10.1016/S0923-2508(97)87643-7 [DOI] [PubMed] [Google Scholar]
- Dodds D. R. (2017). Antibiotic resistance: a current epilogue. Biochem. Pharmacol. 134 139–146. 10.1016/j.bcp.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A. E. (2002). Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., McDevitt C. A., Kitten T. (2015). Manganese uptake and streptococcal virulence. Biometals 28 491–508. 10.1007/s10534-015-9826-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Morey J. R., Ween M. P., Ong C. L., McEwan A. G., Paton J. C., et al. (2014). Extracellular zinc competitively inhibits manganese uptake and compromises oxidative stress management in Streptococcus pneumoniae. PLoS One 9:e89427. 10.1371/journal.pone.0089427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner L., Larsen T., Kilian M., Holmstrup P. (2006). Incidence of bacteremia after chewing, tooth brushing and scaling in individuals with periodontal inflammation. J. Clin. Periodontol. 33 401–407. 10.1111/j.1600-051X.2006.00924.x [DOI] [PubMed] [Google Scholar]
- Fox K. A., Ramesh A., Stearns J. E., Bourgogne A., Reyes-Jara A., Winkler W. C., et al. (2009). Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 106 4435–4440. 10.1073/pnas.0812194106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mendoza A., Liebana J., Castillo A. M., Higuera A. D. L., Piedrola G. (1993). Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J. Med. Microbiol. 39 434–439. 10.1099/00222615-39-6-434 [DOI] [PubMed] [Google Scholar]
- Giacaman R. A., Torres S., Gomez Y., Munoz-Sandoval C., Kreth J. (2015). Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Arch. Oral Biol. 60 154–159. 10.1016/j.archoralbio.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Gong T., Zeng J., Tang B., Zhou X., Li Y. (2020). CRISPR-Cas systems in oral microbiome: from immune defense to physiological regulation. Mol. Oral Microbiol. 35 41–48. 10.1111/omi.12279 [DOI] [PubMed] [Google Scholar]
- Gorke B., Stulke J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6 613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- Gratani F. L., Horvatek P., Geiger T., Borisova M., Mayer C., Grin I., et al. (2018). Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 14:e1007514. 10.1371/journal.pgen.1007514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffen A. L., Beall C. J., Campbell J. H., Firestone N. D., Kumar P. S., Yang Z. K., et al. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6 1176–1185. 10.1038/ismej.2011.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross E. L., Beall C. J., Kutsch S. R., Firestone N. D., Leys E. J., Griffen A. L. (2012). Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59. 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117 57–68. 10.1016/s0092-8674(04)00260-0 [DOI] [PubMed] [Google Scholar]
- Irving S. E., Corrigan R. M. (2018). Triggering the stringent response: signals responsible for activating (p)ppGpp synthesis in bacteria. Microbiology 164 268–276. 10.1099/mic.0.000621 [DOI] [PubMed] [Google Scholar]
- Jakubovics N. S., Smith A. W., Jenkinson H. F. (2002). Oxidative stress tolerance is manganese (Mn2 +)-regulated in Streptococcus gordonii. Microbiology 148 3255–3263. 10.1099/00221287-148-10-3255 [DOI] [PubMed] [Google Scholar]
- Jamil M., Sultan I., Gleason T. G., Navid F., Fallert M. A., Suffoletto M. S., et al. (2019). Infective endocarditis: trends, surgical outcomes, and controversies. J. Thorac. Dis. 11 4875–4885. 10.21037/jtd.2019.10.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeske L., Placzek S., Schomburg I., Chang A., Schomburg D. (2019). BRENDA in 2019: a European ELIXIR core data resource. Nucleic Acids Res. 47 D542–D549. 10.1093/nar/gky1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajfasz J. K., Katrak C., Ganguly T., Vargas J., Wright L., Peters Z. T., et al. (2020). Manganese uptake, mediated by SloABC and MntH, is essential for the fitness of Streptococcus mutans. mSphere 5:e00764-19. 10.1128/mSphere.00764-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio A., Sepponen K., Hermand P., Denoel P., Godfroid F., Melin M. (2014). Role of pneumococcal histidine triad (Pht) proteins in attachment of Streptococcus pneumoniae to respiratory epithelial cells. Infect. Immun. 82 1683–1691. 10.1128/IAI.00699-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar J., Kim J. N., Ahn S. J., Burne R. A. (2016). An essential role for (p)ppGpp in the integration of stress tolerance, peptide signaling, and competence development in Streptococcus mutans. Front. Microbiol. 7:1162. 10.3389/fmicb.2016.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaval K. G., Garsin D. A. (2018). Ethanolamine utilization in bacteria. mBio 9:e00066-18. 10.1128/mBio.00066-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaval K. G., Gebbie M., Goodson J. R., Cruz M. R., Winkler W. C., Garsina D. A. (2019). Ethanolamine utilization and bacterial microcompartment formation are subject to carbon catabolite repression. J. Bacteriol. 201:e00703-18 10.1128/JB [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak K. M., Wayne K. J., Rechtsteiner A., Winkler M. E. (2009). Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72 590–611. 10.1111/j.1365-2958.2009.06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres D. G., Maguire M. E. (2003). Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27 263–290. 10.1016/s0168-6445(03)00052-4 [DOI] [PubMed] [Google Scholar]
- Kholy K. E., Genco R. J., Van Dyke T. E. (2015). Oral infections and cardiovascular disease. Trends Endocrinol. Metab. 26 315–321. 10.1016/j.tem.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Kim J. N., Ahn S. J., Seaton K., Garrett S., Burne R. A. (2012). Transcriptional organization and physiological contributions of the relQ operon of Streptococcus mutans. J. Bacteriol. 194 1968–1978. 10.1128/jb.00037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinane D. F., Riggio M. P., Walker K. F., MacKenzie D., Shearer B. (2005). Bacteraemia following periodontal procedures. J. Clin. Periodontol. 32 708–713. 10.1111/j.1600-051X.2005.00741.x [DOI] [PubMed] [Google Scholar]
- Kolenbrander P. E., Palmer R. J., Jr., Periasamy S., Jakubovics N. S. (2010). Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8 471–480. 10.1038/nrmicro2381 [DOI] [PubMed] [Google Scholar]
- Kreth J., Merritt J., Shi W., Qi F. (2005). Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187 7193–7203. 10.1128/JB.187.21.7193-7203.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J., Zhang Y., Herzberg M. C. (2008). Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190 4632–4640. 10.1128/jb.00276-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers K., Gallay C., Martinek V., Rohde M., Martinkova M., van der Beek S. L., et al. (2016). Highly conserved nucleotide phosphatase essential for membrane lipid homeostasis in Streptococcus pneumoniae. Mol. Microbiol. 101 12–26. 10.1111/mmi.13312 [DOI] [PubMed] [Google Scholar]
- Lee S., Kim K.-K., Choe S.-J. (2001). Binding of oral streptococci to human fibrinogen. Oral Microbiol. Immunol. 16 88–93. 10.1034/j.1399-302x.2001.016002088.x [DOI] [PubMed] [Google Scholar]
- Lemos J. A., Lin V. K., Nascimento M. M., Abranches J., Burne R. A. (2007). Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65 1568–1581. 10.1111/j.1365-2958.2007.05897.x [DOI] [PubMed] [Google Scholar]
- Liochev S. I., Fridovich I. (2004). Carbon dioxide mediates Mn(II)-catalyzed decomposition of hydrogen peroxide and peroxidation reactions. Proc. Natl. Acad. Sci. U.S.A. 101 12485–12490. 10.1073/pnas.0404911101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P. B., Brennan M. T., Sasser H. C., Fox P. C., Paster B. J., Bahrani-Mougeot F. K. (2008). Bacteremia associated with toothbrushing and dental extraction. Circulation 117 3118–3125. 10.1161/CIRCULATIONAHA.107.758524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13 722–736. 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhlynets O., Boal A. K., Rhodes D. V., Kitten T., Rosenzweig A. C., Stubbe J. (2014). Streptococcus sanguinis class Ib ribonucleotide reductase: high activity with both iron and manganese cofactors and structural insights. J. Biol. Chem. 289 6259–6272. 10.1074/jbc.M113.533554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F., Binder H. (2019). pcaExplorer: an R/Bioconductor package for interacting with RNA-seq principal components. BMC Bioinform. 20:331. 10.1186/s12859-019-2879-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. E., Giedroc D. P. (2016). Functional determinants of metal ion transport and selectivity in paralogous cation diffusion facilitator transporters CzcD and MntE in Streptococcus pneumoniae. J. Bacteriol. 198 1066–1076. 10.1128/JB.00975-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. E., Le M. T., Bhattarai N., Capdevila D. A., Shen J., Winkler M. E., et al. (2019). A Mn-sensing riboswitch activates expression of a Mn2 + /Ca2 + ATPase transporter in Streptococcus. Nucleic Acids Res. 47 6885–6899. 10.1093/nar/gkz494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. E., Lisher J. P., Winkler M. E., Giedroc D. P. (2017). Perturbation of manganese metabolism disrupts cell division in Streptococcus pneumoniae. Mol. Microbiol. 104 334–348. 10.1111/mmi.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U., Gentry D., Cashel M., Steiner K., Malke H. (1996). Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J. Bacteriol. 178 1401–1411. 10.1128/jb.178.5.1401-1411.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes J. J., Stipp R. N., Harth-Chu E. N., Camargo T. M., Hofling J. F., Mattos-Graner R. O. (2014). Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect. Immun. 82 4941–4951. 10.1128/IAI.01850-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreillon P., Que Y.-A. (2004). Infective endocarditis. Lancet 363 139–149. 10.1016/s0140-6736(03)15266-x [DOI] [PubMed] [Google Scholar]
- Moreillon P., Que Y. A., Bayer A. S. (2002). Pathogenesis of streptococcal and staphylococcal endocarditis. Infect. Dis. Clin. North Am. 16 297–318. 10.1016/s0891-5520(01)00009-5 [DOI] [PubMed] [Google Scholar]
- Murgas C. J., Green S. P., Forney A. K., Korba R. M., An S. S., Kitten T., et al. (2020). Intracellular metal speciation in Streptococcus sanguinis establishes SsaACB as critical for redox maintenance. ACS Infect. Dis. 6 1906–1921. 10.1021/acsinfecdis.0c00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento M. M., Lemos J. A., Abranches J., Lin V. K., Burne R. A. (2008). Role of RelA of Streptococcus mutans in global control of gene expression. J. Bacteriol. 190 28–36. 10.1128/JB.01395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies D. H. (1992). CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174 8102–8110. 10.1128/jb.174.24.8102-8110.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov P. S., Kazakov A. E., Ravcheev D. A., Leyn S. A., Kovaleva G. Y., Sutormin R. A., et al. (2013). RegPrecise 3.0 - A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. 10.1186/1471-2164-14-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunniyi A. D., Mahdi L. K., Jennings M. P., McEwan A. G., McDevitt C. A., Van der Hoek M. B., et al. (2010). Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. J. Bacteriol. 192 4489–4497. 10.1128/JB.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S., Senty L., Das S., Noe J. C., Munro C. L., Kitten T. (2005). Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 73 6064–6074. 10.1128/IAI.73.9.6064-6074.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Maguire M. E. (2006). Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60 187–209. 10.1146/annurev.micro.60.080805.142149 [DOI] [PubMed] [Google Scholar]
- Parker M. W., Blake C. C. F. (1988). Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 229 377–382. 10.1016/0014-5793(88)81160-8 [DOI] [PubMed] [Google Scholar]
- Perrin D. D., Dempsey B. (1974). Buffers for pH and Metal Ion Control. Bristol: J. W. Arrowsmith Ltd. [Google Scholar]
- Plumptre C. D., Eijkelkamp B. A., Morey J. R., Behr F., Counago R. M., Ogunniyi A. D., et al. (2014). AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Mol. Microbiol. 91 834–851. 10.1111/mmi.12504 [DOI] [PubMed] [Google Scholar]
- Poyart C., Quesne G., Coulon S., Berche P., Trieu-Cuot P. (1988). Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36 41–47. 10.1128/jcm.36.1.41-47.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puccio T., Kitten T. (2020). Fermentor growth of Streptococcus sanguinis. Protocols.io. 10.17504/protocols.io.bkayksfw [DOI] [Google Scholar]
- Puccio T., Misra B. B., Kitten T. (2020). Time-course analysis of Streptococcus sanguinis after manganese depletion reveals changes in glycolytic, nucleotide, and redox metabolites. bioRxiv 10.1101/2020.08.30.274233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliainen A. T., Haataja S., Kahkonen S., Finne J. (2003). Molecular basis of H2O2 resistance mediated by streptococcal Dpr: demonstration of the functional involvement of the putative ferroxidase center by site-directed mutagenesis in Streptococcus suis. J. Biol. Chem. 278 7996–8005. 10.1074/jbc.M210174200 [DOI] [PubMed] [Google Scholar]
- Quan T. P., Muller-Pebody B., Fawcett N., Young B. C., Minaji M., Sandoe J., et al. (2020). Investigation of the impact of the NICE guidelines regarding antibiotic prophylaxis during invasive dental procedures on the incidence of infective endocarditis in England: an electronic health records study. BMC Med. 18:84. 10.1186/s12916-020-01531-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radin J. N., J. L., Kelliher P. K., Parraga Solorzano, Kehl-Fie T. E. (2016). The two-component system ArlRS and alterations in metabolism enable Staphylococcus aureus to resist calprotectin-induced manganese starvation. PLoS Pathog 12:e1006040. 10.1371/journal.ppat.1006040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redanz S., Masilamani R., Cullin N., Giacaman R. A., Merritt J., Kreth J. (2018). Distinct regulatory role of carbon catabolite protein A (CcpA) in oral streptococcal spxB expression. J. Bacteriol. 200:e00619-17. 10.1128/JB.00619-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. V., Crump K. E., Makhlynets O., Snyder M., Ge X., Xu P., et al. (2014). Genetic characterization and role in virulence of the ribonucleotide reductases of Streptococcus sanguinis. J. Biol. Chem. 289 6273–6287. 10.1074/jbc.M113.533620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J. W., Gao G., Ridout G., Wang Y. D., Tuomanen E. I. (2009). Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 72 12–25. 10.1111/j.1365-2958.2009.06638.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaton K., Ahn S. J., Sagstetter A. M., Burne R. A. (2011). A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacteriol. 193 862–874. 10.1128/JB.01257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields R. C., Walker A. R., Maricic N., Chakraborty B., Underhill S. A. M., Burne R. A. (2020). Repurposing the Streptococcus mutans CRISPR-Cas9 system to understand essential gene function. PLoS Pathog. 16:e1008344. 10.1371/journal.ppat.1008344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. G., Martin A. W., McBride B. C. (1977). Experimental transient bacteraemias in human subjects with varying degrees of plaque accumulation and gingival inflammation. J. Clin. Periodontol. 4 92–99. 10.1111/j.1600-051x.1977.tb01888.x [DOI] [PubMed] [Google Scholar]
- Silver J. G., Martin A. W., McBride B. C. (1979). Experimental transient bacteraemias in human subjects with clinically healthy gingivae. J. Clin. Periodontol. 6 33–36. 10.1111/j.1600-051x.1979.tb02288.x [DOI] [PubMed] [Google Scholar]
- Socransky S. S., Manganiello A. D., Propas D., Oram V., Houte J. V. (1977). Bacteriological studies of developing supragingival dental plaque. J. Periodontal Res. 12 90–106. 10.1111/j.1600-0765.1977.tb00112.x [DOI] [PubMed] [Google Scholar]
- Spellerberg B., Cundell D. R., Sandros J., Pearce B. J., Idanpaan-Heikkila I., Rosenow C., et al. (1996). Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19 803–813. 10.1046/j.1365-2958.1996.425954.x [DOI] [PubMed] [Google Scholar]
- Sreenivasan P. K., Tischio-Bereski D., Fine D. H. (2017). Reduction in bacteremia after brushing with a triclosan/copolymer dentifrice-A randomized clinical study. J. Clin. Periodontol. 44 1020–1028. 10.1111/jcpe.12798 [DOI] [PubMed] [Google Scholar]
- Stingu C. S., Eschrich K., Rodloff A. C., Schaumann R., Jentsch H. (2008). Periodontitis is associated with a loss of colonization by Streptococcus sanguinis. J. Med. Microbiol. 57(Pt 4), 495–499. 10.1099/jmm.0.47649-0 [DOI] [PubMed] [Google Scholar]
- Thornhill M. H., Gibson T. B., Cutler E., Dayer M. J., Chu V. H., Lockhart P. B., et al. (2018). Antibiotic prophylaxis and incidence of endocarditis before and after the 2007 AHA recommendations. J. Am. Coll. Cardiol. 72 2443–2454. 10.1016/j.jacc.2018.08.2178 [DOI] [PubMed] [Google Scholar]
- Turner L. S., Kanamoto T., Unoki T., Munro C. L., Wu H., Kitten T. (2009). Comprehensive evaluation of Streptococcus sanguinis cell wall-anchored proteins in early infective endocarditis. Infect. Immun. 77 4966–4975. 10.1128/IAI.00760-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner J. B., Lolkema J. S. (2003). CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67 475–490. 10.1128/mmbr.67.4.475-490.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. S. (2020). Bacterial manganese sensing and homeostasis. Curr. Opin. Chem. Biol 55 96–102. 10.1016/j.cbpa.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A. R., Davis J. J., Assaf R., Boisvert S., Brettin T., Bun C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45 D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer E., Que Y.-A., Entenza J. M., Moreillon P. (2006). New concepts in the pathophysiology of infective endocarditis. Curr. Infect. Dis. Rep. 8 271–279. 10.1007/s11908-006-0071-z [DOI] [PubMed] [Google Scholar]
- Wilson W., Taubert K. A., Gewitz M., Lockhart P. B., Baddour L. M., Levison M., et al. (2007). Prevention of infective endocarditis: guidelines from the american heart association. Circulation 116 1736–1754. 10.1161/CIRCULATIONAHA.106.183095 [DOI] [PubMed] [Google Scholar]
- Xie Z., Okinaga T., Qi F., Zhang Z., Merritt J. (2011). Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl. Environ. Microbiol. 77 8025–8033. 10.1128/AEM.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Alves J. M., Kitten T., Brown A., Chen Z., Ozaki L. S., et al. (2007). Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189 3166–3175. 10.1128/JB.01808-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Ge X., Chen L., Wang X., Dou Y., Xu J. Z., et al. (2011). Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 1:125. 10.1038/srep00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Itzek A., Kreth J. (2014). Comparison of genes required for H2O2 resistance in Streptococcus gordonii and Streptococcus sanguinis. Microbiology 160 2627–2638. 10.1099/mic.0.082156-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Higuchi M., Poole L. B., Kamio Y. (2000). Role of the dpr product in oxygen tolerance in Streptococcus mutans. J. Bacteriol. 182 3740–3747. 10.1128/jb.182.13.3740-3747.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Poole L. B., Hantgan R. R., Kamio Y. (2002). An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol. 184 2931–2939. 10.1128/jb.184.11.2931-2939.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Xie S., Tang N. Y., Choi M. Y., Wang Y., Watt R. M. (2019). The Ps and Qs of alarmone synthesis in Staphylococcus aureus. PLoS One 14:e0213630. 10.1371/journal.pone.0213630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Chen L., Burne R. A. (2018). Preferred hexoses influence long-term memory in and induction of lactose catabolism by Streptococcus mutans. Appl. Environ. Microbiol. 84:e00864-18. 10.1128/AEM.00864-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Chen Z., Itzek A., Ashby M., Kreth J. (2011). Catabolite control protein A controls hydrogen peroxide production and cell death in Streptococcus sanguinis. J. Bacteriol. 193 516–526. 10.1128/JB.01131-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNA-seq Analysis.
Analysis of putative cre sites and CcpA regulon.
Primers used in this study.
Figures, Methods, and References.
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE150593 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE150593).