Abstract
Originated in China, coronavirus disease 2019 (COVID-19)- the highly contagious and fatal respiratory disease caused by SARS-CoV-2 has already infected more than 29 million people worldwide with a mortality rate of 3.15% (according to World Health Organization’s (WHO’s) report, September 2020) and the number is exponentially increasing with no remedy whatsoever discovered till date. But it is not the first time this infectious viral disease has appeared, in 2002 SARS-CoV infected more than 8000 individuals of which 9.6% patients died and in 2012 approximately 35% of MERS-CoV infected patients have died. Literature reports indicate that a chymotripsin-like cystein protease (3CLpro) is responsible for the replication of the virus inside the host cell. Therefore, design and synthesis of 3CLpro inhibitor molecules play a great impact in drug development against this COVID-19 pandemic. In this review, we are discussing the anti-SARS effect of some small molecule 3CLpro inhibitors with their various binding modes of interactions to the target protein.
Keywords: 3CLpro, Inhibitors, SARS-CoV, SARS-CoV-2
Graphical abstract
Highlights
-
•
Comparative study of SARS-CoV and SARS-CoV-2.
-
•
Overview of various protease inhibitors.
-
•
Importance of 3CLpro enzyme for virus replication.
-
•
3CLpro as important target for drug discovery.
1. Introduction
After the official declaration of the public health emergency of international concern by World Health Organization (WHO) on 30 January 2020; there has been a proliferation of research activities on COVID-19 disease in search for an immediate remedy [1,2]. This novel coronavirus SARS-CoV-2, emerged in the Wuhan city of China in December 2019, is rapidly spreading into various countries/regions of the world within few days [3,4]. The virus was provisionally named as the 2019 novel coronavirus (2019-nCoV) which was later renamed by the International Committee on Taxonomy of Viruses as Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) on 11 February 2020 [5,6]. On the same day WHO also announced the name of the contagion disease caused by this novel coronavirus as Corona Virus Disease (COVID-19) [7]. It was the sixth public health emergency of international concern as declared by WHO after novel influenza A or H1N1 (2009), polio (2014), Ebola in West Africa (2014), Zika (2016) and Ebola in Congo (2019) [8].
1.1. Why SARS-CoV-2?
The emergence of SARS-CoV-2 is not totally new to the world as Severe Acute Respiratory Syndrome coronavirus (SARS-CoV) and Middle East Respiratory Syndrome coronavirus (MERS-CoV) already hit the various parts of the world in 2002 and 2012 respectively [9,10]. Moreover replication mechanism and pathogenesis of various coronaviruses were studied in early 1970s, but it gained much more attention after the SARS outbreak in 2002 [11]. In the history of 21st century, SARS-CoV-2 has been recognized as the most infectious, fatal and pathogenic coronavirus after SARS-CoV and MERS-CoV as until now more than 0.9 million people died out of more than 29 million infected people [the infected fatality rate (IFR) around 3.15% as compared to the SARS-CoV (800 deaths) and MERS-CoV (858 deaths)] [12,13]. From the above data it is clear that SARS-CoV-2 is much more dangerous and virulent as compared to SARS-CoV and MERS-CoV and the number of infected people/deaths are increasing at a very high rate day by day. The similar name of SARS-CoV and SARS-CoV-2 was introduced by scientific community due to the 86% genome sequence similarity of SARS-CoV-2 with SARS-CoV while MERS-CoV has almost 50% similarity to that of the SARS-CoV-2.14a In addition, bats are the primary origin of these viruses as genome sequence of SARS-CoV-2 has significant homology (96.2%) to that of the bat coronavirus (Scheme 1 ) [[14], (b)].
Scheme 1.
Pictorial representation showing the difference of SARS-CoV from SARS-CoV-2.
1.2. What is coronavirus?
Structurally, SARS-CoV-2 is encircled with positive-sense, mono-stranded RNA virus with basic reproductive number around 2–4 [15]. Coronavirus is an enveloped RNA virus which belongs to the family of coronaviridae with the classification of four genera including α, β, γ, δ-CoV [13]. As reported, α- and β-CoV are responsible for the infection in mammals while γ- and δ-CoV are responsible for the infection in birds. But out of α- and β-coronaviruses, α-CoVs have less fatal effects causing only common cold with mild respiratory problems while β-CoVs are mainly conscientious for the harsh and potentially incurable SARS-CoV and MERS-CoV [[16], [17], [18]]. As mentioned earlier, SARS-CoV-2 has 86% genome sequence similarity with that of SARS-CoV, it is clear that SARS-CoV-2 also belongs to the β-coronavirus. The primary symptoms of the SARS are the fever above 38 °C with the pneumonia and breathe related problems [19].
2. Way of transmission and outbreak of SARS-CoV-2
Literature report suggested that the advancement of SARS-CoV and SARS-CoV-2 was emerged from the non-human sources (bat) while there is probably an unidentified intermediary reservoir for SARS-CoV-2 (intermediary reservoir for SARS-CoV was Civet cat while camel was the intermediary reservoir for MERS-CoV) [20]. The key feature for the human-to-human transmission involves the transfer of respiratory droplets, direct contact, fecal-oral route, suspected mother to the new born baby, through aerosols etc. [21] Moreover, reports also suggested that SARS-CoV-2 was also independently introduced from the animals to human with or without the involvement of carrier. As reported, bat coronavirus and SARS-CoV-2 might assign the identical precursor as bats are recognized to be host reservoir for more than 30 coronaviruses with more than 92% nucleotide resemblance [22].
| Similarity between SARS-CoV and SARS-CoV-2 |
|---|
| 1. Both of them have 86% structural genome similarity. |
| 2. Bat is the primary reservoir of the both of them. |
| 3. Pneumonia is the most common symptoms in both of them with the fever above 38 °C. |
| 4. Both of them have the structurally similar to the bat coronavirus. |
| 5. Both of them are β–coronavirus. |
| 6. Pandemic potential is very high in both the cases. |
| 7. Highly efficient human-to-human transmission. |
| 8. Incubation period is 1–14 days in both the cases while the initial 3–8 days are most potent to infection. |
| 9. Fatality rate is more than 3% in both the cases (as per WHO report).∗ |
∗ For SARS-CoV-2 it is increasing day by day.
3. Structure and key features of SARS-CoV-2
SARS-CoV-2 is a mono-stranded RNA virus surrounded with positive-sense containing club-shaped spikes pointing upwards from its surface. These spikes are actually a type of glycoprotein and named as spikes proteins (S) [23]. Along with spikes proteins, the membrane of coronavirus is composed of other two proteins viz. membrane proteins (M) and a highly hydrophobic small structured envelope protein (E). Membrane protein is responsible for the extension of virus membrane while envelope protein mainly wraps the integral part of the virus [24]. Integral part of the virus mainly contains nucleocapsid protein, replicase polyproteins and other accessory proteins which hamper the host’s intrinsic resistant system. Nucleocapsid proteins are mainly responsible for the formation of helical capsids when they come in contact with genomic RNA inside the cell membranes. Replicase polyproteins are of two types’ viz. pp1a and pp1ab which are found in open reading frames of the virus and from these polyproteins, the replication of the virus is proceeded by prevalent proteolytic process [25].
4. Replication and its pathogenicity
The angiotensin-converting enzyme 2 (ACE2) is the key receptor for the cell access for both SARS-CoV and SARS-CoV-2 which infect humans while dipeptidyl peptidase 4 was the receptor for the MERS-CoV [26]. Reports have also established that SARS-CoV-2 has an analogous binding receptor (S protein of coronavirus) to SARS-CoV, which attaches the receptor protein of the infected cell, enabling the virus to raid and infect the host cells [27,28]. ACE2 which is known as cell receptor for both SARS-CoV-2 and SARS-CoV is found in the lower respiratory area of human body in which S-glycoprotein of coronavirus attaches to replicate the virus and regulates human-to-human transmission [29]. The two subunits, S1 and S2 of S-glycoprotein of coronavirus are mainly responsible for the various activities of the SARS-CoV-2. The S1 subunit is mainly responsible for the determination of range of the virus-host ability and cellular tropism whereas S2 subunit intercedes the fusion ability of the virus-cell membrane by two simultaneous domains, heptad repeats 1 and 2 (HR 1 and 2) [30,31]. Following the virus-cell membrane fusion, the viral RNA is liberated into the cytoplasm and the two replicase polyproteins, pp1a and pp1ab which convert the non-structural virus proteins to form replication complex (RTC) to infect the host cell and thus the replication goes on [32]. For proteolytic and replication processing of SARS-CoV and SARS-CoV-2, they have to encode a protease, known as chymotripsin-like cystein protease (3CLpro) or more commonly known as Main protease (Mpro) [33]. 3CLpro is a part of the replicase polyproteins (similar to the 3C protease of main picoronavirus) which plays the major role in the viral imitation through proteolysis releasing replicative proteins from their polyproteins precursor to the infected host cell at 11 different cleavage sites [34]. Therefore, for viral replication and infection process of the coronavirus inside the host cell, 3CLpro is very much requisite [35]. The practical congregation of 3CLpro is believed to be in the form of dimer where each monomer contains one independent active site making 3CLpro dimer more active than the monomer [36]. Moreover, in the active site of the enzyme 3CLpro, cysteine acts as a nucleophile and histidine residue acts as a base [37]. The key steps during the viral entry of the SARS-CoV-2 include the binding interaction of the host cells receptor, ACE2 with the spike protein of virus S protein, subsequent changes in the conformation of the S protein followed by proteolysis for the activation of the COVID-19 in the human cells [38]. In addition to that, the two replicase polyproteins, pp1a and pp1ab which are also known as the promoters for viral replication are arranged perpendicularly to one another and both of them contain three domains I, II and III. Domains I and II have similar structural β–patterns as present in picoronavirus 3C proteinases [39]. These domains are mainly responsible for the substrate binding and the autocatalytic ability containing Cys145 and His41 residues. In addition, domain III attaches to the domain II via five different α-helices with an exterior circle which primarily preserves the accurate conformation of the dimer after proteolytic activity of SARS-CoV 3CLpro.
5. Symptoms and clinical approaches
As mentioned earlier, SARS is mainly characterized by high fever above 38 °C or higher accompanied with breathing problems due to infection in lower respiratory tract, fatigue, dry cough, runny nose, atypical pneumonia and sore throat which are very common in nature [40]. Moreover, several other patients are found to have different gastrointestinal symptoms such as diarrhea, vomiting, nausea, abdominal discomfort etc. [41] The incubation period of SARS-CoV-2 infection is 1–14 days, but generally 3–7 days are the most significant to show the above mentioned symptoms. These clinical symptoms are further analyzed and classified in four different stages: a) Asymptomatic or recessive infection which occur in the initial 3–5 days of the infection; b) Respiratory tract infection which involves mild fever, sore throat, fatigue, headache and abdominal discomfort; c) Mild pneumonia which involves dry cough accompanied with or without serene fever; d) Severe pneumonia involving severe respiratory problems with high respiratory rate (more than 72 times per min), lower saturated oxygen percentage in blood (below 92%), unconsciousness etc. [42] By analysing various data related to the COVID-19, it was well reported that around 50% of the adult patients suffering from SARS-CoV-2 have diverse abnormal liver functions which may be due to dysfunction of bile duct caused by the viral infection.
Various chemical and analytical techniques such as next-generation sequencing, cell-culture, real time PCR, electron microscopy etc. have been used to detect SARS-CoV-2 [43,44]. From medicinal chemistry perspectives, an imperative and promising advance to fight with COVID-19 is to develop molecules that can inhibit the essential major polyprotein processing, 3CLpro. As 3CLpro is very much crucial for the viral replication, it is widely regarded as a major drug target for the development of SARS treatment [45]. Several synthetic inhibitors for the 3CLpro have been reported but they are relatively weak in nature [46]. The inhibitors initially bind with the 3CLpro through covalent bonding with the active site of the cysteine 145 residue leading to the deactivation of 3CLpro’s enzymatic activity [47]. The binding of S-glycoprotein of the virus and ACE2 receptor is a decisive step for virus entry and the virus-receptor binding affinity is under intensive study through different approaches. Additionally this approach also suggests that the S pockets of 3CLpro have been the principal targeting sites for anti-SARS molecules which can block the cysteine site and do not make them free to attach with the ACE2 receptor molecule. The reasons behind the choice of 3CLpro inhibition as the target are a) 3CLpro has the predetermined crystal structure which makes them more approachable in the study, b) 3CLpro is the most studied enzyme of coronavirus which is also common in various bacterial systems and its functional examination such as proteolytic activity is very much easy in presence of inhibitor molecules, c) practical importance of 3CLpro in the life cycle of coronavirus make them more practically superlative as a target for antiviral drug design [48,49].
In this review, we are trying to analyse the various small molecules/drugs developed as cysteine protease inhibitors or anti-SARS therapy. As COVID-19 is virus infected disease and till now there is not a single commercially available protective drug/vaccine present in the market, therapies currently undergoing include the application of existing drugs used to treat malaria and autoimmune diseases [50]. Antiviral drugs developed for other viral diseases and antibodies from recovered COVID-19 patients have also been used as potential remedy for the disease. The SARS infected person is kept in isolation for 14 days in a low pressure zone with mechanical ventilation [51]. Moreover in case of severe breathing problems of patient a high dose of steroid is also needed. In addition to that patients with SARS may also have severe damage of their immune systems due to which various other diseases may also occur. Therefore, people are advised to keep away from the SARS infected patients or the probable SARS infected people to diminish the infection of SARS.
Most of the human diseases are directly or indirectly linked to the cysteine protease; the nucleophilic sulphur atom of Cys145 forms covalent bond with the receptor of the human body causing the disease [52]. Now if we block this nucleophilic centre with some other small molecule drugs or inhibitors, this will lead to the deactivation of the enzymatic activity of 3CLpro. After this covalent binding with the small molecule inhibitors, 3CLpro is not free to bind with the ACE2 receptor of the human body causing no infection of SARS CoV (Scheme 2 ). Literature reports clearly state that the principal targeting site for the anti-SARS molecules/drugs is the S pockets of the SARS-CoV 3CLpro [53,54].
Scheme 2.
Schematic representation showing viral binding of SARS-CoV with the receptor, ACE2.
6. Treatment
6.1. Repurpose drugs and their stage of study for SARS 3CLpro inhibitors
The conventional method of finding the biologically pronounced small molecule inhibitors is primarily based on the rigorous screening of the chemical libraries which is actually very time consuming as well as involves laborious work. In this lengthy process the success of finding the drug molecules also greatly relies on the superiority of the existing libraries. Consequently, at the initial stage most of the researchers tried different protease inhibitors which were previously used to cure and prevent various infectious diseases to inhibit the COVID-19 infection. Out of all well-known drugs, chloroquine (antimalarial drug) is the foremost drug that comes into the mind which has the ability to hold back the SARS-CoV replication through various pH-dependent steps [55]. Moreover, chloroquine has the ability to modify the various functions related to the immune system such as antibody formation and hence it is known as immunomodulatory drug (Fig. 1 ). Various literature reports reveal that functioning cellular receptors of SARS-CoV-2 was impeded by chloroquine (through glycosylation) for the entrance of the virus into the Vero E6 cells of human to keep away the COVID-19 infection from human body [56]. Similarly, teicoplanin [57], an antibiotic used for prophylaxis had also proven efficacy (less than that of the chloroquine) through in vitro manner to hold back the SARS-CoV replication [58]. But the results obtained are not so much effective towards the inhibition of the SARS-CoV, only lowers the fever of the patient as a secondary treatment.
Fig. 1.
Structure of chloroquine and hydroxychloroquine.
Several other drug molecules which have the ability to inhibit/reduce the SARS-CoV are cinanserin [59], ribavirin [60], corticosteroids [61], lopinavir/ritonavir [62], neuraminidase inhibitors, umifenovir [63] etc. (Fig. 2 )
Fig. 2.
Structure of various commercially available antiviral drugs.
Use of cinanserin significantly reduces the replication of SARS-CoV in cell culture with the evidence of negligibly lower cytotoxicity at the inhibitory concentrations. Cinanserin is also available in the form of water soluble hydrochloride salt form but its hydrochloride salt form is less active than the neutral molecule. This is due to the fact that cinanserin has less hydrophilic nature making it more efficient to penetrate the cell membrane and inhibit the viral replication [59].
Furthermore, the inhibition of SARS-CoV in tissue culture can also be shown by individual use of ribavirin (a nucleoside analogue), corticosteroids, lopinavir and type I interferon (INF α) [64,65] which were proved by the clinical trial methods and in vitro studies but their applications in human body is still not beneficial [[66], [67], [68]]. But the combined use of ribavirin and corticosteroids proved to be a beneficial antiviral agent against SARS [69].
A well known naturally occurring alkaloid reserpine which was originally used as sedative and antihypertensive drug also has a potential ability against SARS-CoV [70]. (Fig. 3 ).
Fig. 3.
Structure of reserpine.
A significant inhibitory efficiency against the replication of SARS-CoV was analyzed by He et al. [71] by the use of aurintricarboxylic acid (ATA) in cell culture. The drastic efficiency of the ATA compound as anti-SARS-CoV agent was attributed due to its polymerization ability in aqueous solution forming a stable free radical which prevents the binding of the protease enzyme to the human cell. In their study, they found that aurintricarboxylic acid has the EC50 value of 0.2 mg/ml which implies that the use of 0.2 mg per ml (concentration) of ATA gave 50% of its maximal effect (Fig. 4 ). While comparing aurintricarboxylic acid with the Interferon α and β (INF α and β), it was observed that aurintricarboxylic acid is 10 and 100 times more effective than IFN α and interferon β respectively against SARS-CoV replication.
Fig. 4.

Structure of aurintricarboxylic acid.
Most recently, SARS-CoV-2 has been found to inhibit by the use of two well known drugs viz. remdesivir and chloroquine with high efficacy [7,72]. (Fig. 5 ).
Fig. 5.

Structure of remdesivir.
6.2. Small organic compounds as SARS 3CLpro inhibitors
Because of the less effectiveness of the commercially available drugs against SARS-CoV-2, the current interest of research in the various fields is to develop more effectual and potent anti-SARS-CoV-2 inhibitors. Various techniques such as structure based discovery, virtual and experimental screening have been utilized by the researchers in the search for small molecules cysteine protease inhibitors and their implication in the mammalian cells [73].
In search of the cysteine protease inhibitors, we reported a research article describing the inhibitory effect of peptidic compounds bearing a several heteroatom ketone groups such as benzothaizoyl or thaizolyl and trifluoromethyl ketone groups as SARS-CoV 3CL protease inhibitors [74]. In our study, we found that in the series of trifluoromethyl ketone containing peptidic compounds, the compound with N-morpholine groups as substituent, 1 was found to be most potent with the inhibition constant (K i) value of 21.0 μM. In the second series of compounds bearing benzothaizoyl or thaizolyl ketone group, the compound with P1-pyrrolidone and thaizolyl warhead, 2 was found to be the most potent against 3CL protease with the inhibition constant (K i) value of 2.20 μM while the compound containing benzothaizoyl warhead, 3 was observed to be little less effective with 49.3 μM. From this report we can assure that the presence/introduction of such groups (fluorine atom, heterocyclic aromatic ring with two or more than two heteroatoms, naturally occurring compounds with such type of rings etc.) in different small molecules may be effective as anti-SARS-CoV agent or cysteine protease inhibitors [53]. (Fig. 6 ) As peptide chemistry is very complex and broad in nature, researchers usually utilized various techniques such as structure based discovery, virtual and experimental screening in the search for small molecules cysteine protease inhibitors bearing such kind of warhead groups, effectiveness and its implication in the mammalian cells.
Fig. 6.
Structure of trifluoromethyl ketone and benzothaizoyl or thaizolyl warhed containing peptidic compounds.
Ikejiri et al. [75] developed and evaluated the biological activity against SARS-CoV using nucleoside analogues based on 6-chloropurine derivatives. This kind of nucleoside based purine derivatives have already been reported as antiviral compounds and also show potent as anti-SARS-CoV activity [76]. Out of several synthesized nucleoside analogues, two compounds (4 and 5) exhibited major significant activity as anti-SARS-CoV agent with the IC50 values 48.7 and 14.5 μM respectively (Fig. 7 ). The structure-activity relationship of these compounds revealed that the presence of 6-chloropurine motif in both the compounds, unprotected 5/-hydroxyl group in 4 and protected (benzoylated) 5/-hydroxyl group in 5 mainly enhanced the antiviral activities in these compounds.
Fig. 7.
Structure of nucleoside analogues bearing 6-chloropurine derivatives.
Blanchard et al. [77] synthesized novel drug-like small molecule, 6 (named as MAC-5576) which acts as one of the potent inhibitors of the SARS coronavirus main proteinase through vigorous screening (Fig. 8 ). Actually, MAC-5576 is 2-(5-chloropyridin-3-yl)-1-(thiophen-2-yl)ethanone which showed IC50 value of 0.5 ± 0.3 μM, makes it a better one than the previous compounds.
Fig. 8.

Structure of drug-like small molecule MAC-5576.
Similarly, Niu et al. [39] also synthesized non-peptidic 3-chloropyridine derivatives which are structurally similar to that of MAC-5576 and evaluated the biological activity of these compounds as chymotripsin-like proteinase inhibitors to diminish the SARS infection.
The molecular docking study of these compounds proved that the chloropyridine moieties of these inhibitors initially bind inside the S1 pocket of the inhibitor to the enzyme through in vitro manner whereas ester part specially binds to the S1 substrate binding site of 3CLpro. In their report they summarized all the molecules in the different groups of chloropyridine molecules. Group I compounds contained chloropyridine ester moiety as well as heterocyclic furan moiety (7 and 8) which is found to be more potent for 3CLpro inhibitor than the group II compounds containing simply chloropyridine ester moieties (9) along with fused rings such as naphthalene and coumarin (10 and 11 respectively). The best potent compounds of both the groups are shown in the figure with the IC50 value (Fig. 9 ).
Fig. 9.
Structure of 3-chloropyridine ester derivatives.
To evaluate the potent biological activity of 3-chloropyridine ring containing compounds as SARS-CoV 3CLpro inhibitor, Ghosh et al. [78] designed and synthesize a series of chloropyridyl ester-derived compounds which have prominent activity as 3CLpro inhibitors. The synthetic methodology involves the treatment of 5-chloro-3-pyridinol with various carboxylic acids through esterification using carbodiimide based coupling agents and racemization suppressor (mainly DMAP, N,N-dimethylaminopyridine). Out of the synthesized compounds, indole-4-carboxylic acid containing 3-chloropyridyl ester 12 shows most potent activity against SARS-CoV 3CLpro with the IC50 value of 0.03 μM (30 nM). (Fig. 10 ).
Fig. 10.

The structure of indole moiety bearing chloropyridyl ester analogue.
Docking study also reveals the molecular binding of the above mentioned most potent compound, 12 where the sulphur atom of Cys-145 is in close proximity to the carbonyl carbon atom of the compound, the imidazole nitrogen of His-163 residue to the 3-chloropyridinyl group and indole group to the His-41 residue.
Zhang et al. [79] synthesized a series of two or three aromatic rings containing aryl methylene ketones and their mono and di-fluoro derivatives. Out of various compounds synthesized; two compounds viz. 2-(5-bromopyridin-3-yl)-1-(5-(4-chlorophenyl) furan-2-yl)ethanone, 13 and its monofluorinated derivative, 14 were found to be most potent SARS 3CLpro inhibitors with the IC50 values of 13 μM and 28 μM, respectively (Fig. 11 ). They found that introduction of fluorine substituent to the aryl methylene ketones containing 4-chlorophenyl ring (most potent compound) lowers the inhibitor activity while the inhibitors with the three aromatic rings are considered to have more potent inhibitor efficiency than the inhibitors with the two aromatic rings. Herein, in 13, the oxygen atom of the furan ring binds with the -NH of Glu166 residue through hydrogen bonding interaction. Similarly, the pyridinyl moiety also binds with the Cys145 reside inside the S1 pocket and in case of the fluoro substituent, 14 the oxygen atom of carbonyl group binds with the His164 residue.
Fig. 11.
Structure of aryl methylene ketone and fluorinated methylene ketone with pyridinyl ring moiety.
Similar to the previous report, Zhang et al. [80] also reported different heteroaromatic ester compounds as inhibitors (15–18) for 3CL protease. In their study they found four most potent inhibitors bearing 3-chloropyridine ring and other heteroatoms with the ester linkage with IC50 value ranging from 50 to 65 nM (Fig. 12 ).
Fig. 12.
Structure of heteroaromatic esters with 3-halopyridinyl ring motif.
Development of new pyrazole and pyrimidine derivatives was carried out by Mohamed et al. [81] starting from 3-indolaldehyde moiety with 3-aminoacetophenone giving a chalcone. This chalcone then reacts with hydrazine hydrate or phenylhydrazine to give pyrazole derivaties, 19 while pyrimidine derivatives were obtained by the reaction of chalcone with hydroxylamine hydrochloride, thiosemicarbazide (20–23). Both pyrazole and pyrimidine moieties are well known to exhibit various biological activities [82] and this newly developed series of compounds have the activity as SARS-CoV 3CLpro protease inhibitors (Fig. 13 ). Out of these two different series of compounds the five compounds are found to be the best SARS-CoV 3CLpro protease inhibitors having prominent IC50 values ranging from 0.12 to 0.38 μM.
Fig. 13.
Preparation scheme and the structure of substituted pyrazoles and substituted pyrimidines derivatives.
Chen et al. [83] initially synthesized N-Substituted isatin derivatives by the simple reaction of isatin molecule (2,3-dioxindole) and various bromide sources. The SARS-CoV 3CLpro inhibition assays were conducted via fluorescence resonance energy transfer (FRET) followed by HPLC analysis of the fragment peaks cleaved by 3CLpro. The two thiophene containing isatin derivatives such as thiophenecarboxylic 7-bromo-3-pyridinol ester, 24 and thiophenecarboxylic 5-iodo-3-pyridinol ester, 25 are reported as the most potent SARS-CoV 3CLpro inhibitors with the IC50 values 0.98 and 0.95 μM respectively (Fig. 14 ). Molecular docking of these molecules showed that the isatin scaffold was located in the S1 site whereas S2 site was blocked by the N-substituted side chain. Moreover it also showed that both the oxygen atoms of the carbonyl group of the synthesized isatin derivative was bonded through hydrogen bond to the- NH groups of the different protein chain residues such as Cys145, His41, Ser144, and Gly143.
Fig. 14.
Preparation scheme and the structure of N-Substituted isatin derivatives with thiophenecarboxylic ester analogues.
Similarly, isatin derivatives as SARS-CoV 3C-like protease were also designed by Liu et al. [84] Out of their synthesized isatin derivatives, three compound viz. 5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 26 1-methyl-5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 27 and 1-benzyl-5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 28 showed the most potent activity as SARS-CoV 3C-like protease inhibitors with the IC50 values ranging from 1.04 to 1.68 μM (Fig. 15 ).
Fig. 15.
Preparation scheme and the structure of 5-sulfonyl isatin derivatives.
The potent inhibitory activity of the compound 5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 26 was enhanced by the presence of sulfonyl group with the simple six-membered ring in the 5-position of the isatin molecule which was well fixed at the S2 site of SARS CoV 3CLpro. Furthermore, two different N-substituted derivatives of the previous compound, 1-methyl-5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 27 was bonded through hydrogen bonding with Gly143 and Cys145 of protease and the carbonyl group of 2-position and the nitrogen atom at the 1-postion of isatin molecule respectively. Similarly, in 1-benzyl-5-[(4-methylpiperidin-1-yl)sulfonyl]-1H-indole-2,3-dione, 28 the carbonyl group of C2-position also formed hydrogen bonds with Gly143 and Ser144 residue of the protease while Asn142 residue binds with the nitrogen atom at the 1-postion of isatin molecule. Moreover, molecular docking revealed that the S1/site of SARS CoV 3CLpro was blocked by the isatin scaffold while the S2 and S1 sites are booked by the side chain of R2 and R3 of the newly synthesized 5-sulfonyl isatin derivatives respectively.
Zhou et al. [85] reported various isatin derivatives substituted at the N-1 positions with different alkyl/aryl groups and C-5 positions with the iodo and amide groups. They examined these derivatives to elucidate the various binding sites of the 3CLpro. In their analyses, they got the best potent isatin derivative with the N-substituted 2-methylnaphthalene group and C-5 substituted carboxyamide group, 29 with an IC50 value of 0.37 μM against SARS CoV 3C-like protease (Fig. 16 ). The active site residues His41 and Cys145 were attached to the C-2 and C-3 position carbonyl oxygen atoms through hydrogen bonding respectively. Similarly, the carboxamide at C-5 also formed the H-bonds with the backbone carbonyl of Phe140 and the imidazole ring of His163 residue. Moreover, the hydrophobic S2 site originally generated by the Met49 and Met165 residues of the protease was well occupied by the alkyl groups of N-1 position.
Fig. 16.

Structure of N-substituted isatin derivatives with C-5 substituted carboxyamide group.
Lee et al. [86] reported benzimidazole based SARS-CoV 3-Chymotrypsin-like protease inhibitors with IC50 value 13.9–18.2 μM (30 and 31) (Fig. 17 ). These inhibitors bind the S1 site of the His163 and Glu166 residues through hydrogen bonding interactions while hydrophobic S2 site was booked with Met, Asp, His, Pro and Tyr residues. Moreover two additional aromatic/hydrophobic interactions were also found in the S1 and S2 sites close to the catalytic cysteine residue where a donor site interacts with residues His163 and His164 in the S1 site.
Fig. 17.
Structure of benzimidazole based 3CLpro inhibitors.
Kao et al. [87] developed a 3-quinolinecarboxylic acid derivative that has the potent inhibitor efficacy against the SARS-CoV Mpro. They named the compound as MP576, 32 which has the IC50 value 2.5 μM and EC50 value 7 μM (Fig. 18 ).
Fig. 18.

Structure of 3-quinolinecarboxylic acid derivatives.
The ether linked oxygen atom and the oxygen atom of nitro group of the synthesized derivative 32 is proved to bind with the nitrogen of His41 and Cys145 residue respectively through hydrogen bonding interaction.
Shimamoto et al. [88] designed and developed novel decahydroisoquinoline motif as an inhibitor for SARS. In their study, they found three most potent derivatives, 33–35 for the inhibition of the SARS 3CLpro protease with the IC50 values 57–68 μM (Fig. 19 ).
Fig. 19.
Structure of decahydroisoquinoline motif containing 3CLpro inhibitors.
Herein, the S2 pocket originated by the residues such as His41, Met49, Met165 and Asp187 side chains of the protease was occupied by the novel decahydroisoquinoline scaffold of 33. Similarly, the S1 site which was originated by the residues Phe-140, Leu-141 and Glu-166 was occupied by the P1 site nitrogen atom of the imidazole of 33 and it forms hydrogen bonding interaction with the His-163 residue of the protease.
Ramajayam et al. [89] developed a series of 1,6-dihydropyrimidine derivatives as SARS-CoV 3CL protease inhibitors and their effectiveness was appraised by in vitro protease inhibitor analyses. These 1,6-dihydropyrimidine derivatives were synthesized by the two step reaction of substituted benzaldehyde, ethyl cyanoacetate and thiourea followed by the slow addition of different halide derivatives in DMF as solvent and K2CO3 as base at 0–5 °C. In this study, they found two compounds, 36 and 37 showing most potent activity against SARS-CoV 3CL protease with the IC50 values of 6.1 and 10.6 μM respectively (Fig. 20 ).
Fig. 20.
Preparation scheme and the structure of 1,6-dihydropyrimidine derivatives.
In the most potent molecule, 36 the -NH group of the pyrimidine ring was constrained in the distance of 2.0 Å with the oxygen atom of Glu-166 residue. Herein, the nitro group is the main functional group for showing the potent inhibitor efficacy and its one of the oxygen atoms is in secure vicinity to the Gly-143 residue while the other oxygen atom binds with the Cys-145 residue of the protease through hydrogen bond interaction. Furthermore, the S2 pocket was blocked by the chlorophenyl ring with Met-49 and Gln-189 through hydrophobic interactions.
Ramajayam et al. [90] designed and synthesized a series of pyrazolone derivatives and evaluated the in vitro SARS-coronavirus 3C-like protease inhibitor activity. These compounds were synthesized by the two step reaction of different aromatic hydrazine hydrate derivatives with the ethyl benzoylacetate in presence of triethylamine as base and acetic acid as solvent to give pyrazolone product followed by the reaction with aromatic aldehydes in presence of piperidine affording substituted pyrazolone compounds. From their study, three pyrazolone compounds 38, 39 and 40 were found to be most potent against SARS 3CLpro with the IC50 values 5.5, 6.8 and 8.4 μM respectively (Fig. 21 ). In 40, S1/pocket of the 3CLpro is blocked by the N1-phenyl group while one of the oxygen atoms of the nitro group forms H-bonding interaction with the Gly-143 residue of 3CLpro. Similarly, the carbonyl group of the pyrazolone ring also forms H-bonding interaction with the Glu-166 residue. Moreover, the C-3 phenyl ring is situated in the S2 pocket and forms hydrophobic interactions with Met-49, Arg-188, and Gln-189 residue meanwhile the carboxyl benzylidene group fits into the S3 pocket of the 3CLpro and forms H-bonding interaction with the Gln-192 residue.
Fig. 21.
Preparation scheme and the structure of novel pyrazolone derivatives.
Jacobs et al. [91] designed a series of furyl amide derivatives with 3-pyridyl ring and applied for the inhibition of 3CLpro. These compounds are derivatives of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides. The pyridyl ring of the best inhibitor compound interacts with the His163 residue through hydrogen bond while furan ring oxygen and the amide carbonyl oxygen have a diverse interaction with Gly143 residue. After that, the enantiomers of the compound was separated and isolated through supercritical fluid chromatography and they found that only R-stereoisomer of the compound, 41 (named as MT188) was inherently active for the inhibition of 3CLpro with IC50 value of 1.5 μM while S-stereoisomer of the compound was found inactive (Fig. 22 ).
Fig. 22.
Structural separation of pyridine containing acetamide analogues.
Jacobs et al. [92] also extended their research for the further development of 3CL protease inhibitors by the simple modification of the previous compound ML188. The new compounds have the same structural moiety containing diamide bonds and the newly introduced benzo-1,2,3-triazolyl ring (42–45) with the IC50 values 0.051–6.2 μM (Fig. 23 ). In the molecular docking study of the compound 42, it was observed that N3 atom of the benzotriazolyl ring forms hydrogen bonding interaction with the His-163 while the carbonyl oxygen attached to the benzotriazolyl ring also forms hydrogen bonding interaction with the Glu166 residue of the protease.
Fig. 23.
Structure of benzo-1,2,3-triazolyl ring containing diamaide derivatives.
Li et al. [93] synthesized various derivatives of stilbenes by Michaelis–Arbuzov reaction followed by Wittig–Horner reaction and then finally demethylated with BBr3 in dichloromethane. In their study, they found that only two compounds viz. [(E)-2′,3,5′,5-tetrthydroxystilben], 46 and [(E)-2′,5,5′,6-tetrahyroxystilbene-2-nitrogen], 47 have the potential ability to inhibit the replication of SARS virus in concentration ≤ 0.5 mg ml−1 with no cytotoxicity (Fig. 24 ).
Fig. 24.
Structure of stilbene derivatives.
Wu et al. [94] reported two different series of stable benzotriazole esters as inactivators of 3CLpro. One series was synthesized from the well known peptide coupling agent HBTU (1H-Benzotriazolyl Tetramethyl Uronium hexafluorophosphate) along with carboxylic acid derivatives in presence of N,N-diisopropylethylamine and N,N-diethylformamide while the other series was synthesized by the reaction of benzotriazole with the aromatic α–halo ketone derivatives in presence of TBAF (1M in THF) as catalyst. Herein, they found two benzotriazole esters as the post potent inactivators of 3CLpro viz. 1H-indole-5-carboxylic acid benzotriazol-1-yl ester, 48 and 2-(1H-benzo[d][1,2,3]triazol-1-yl)-1-(4-(diethylamino)phenyl)ethanone, 49 with the inhibition constant value 7.5 nM and 1.0 μM respectively (Fig. 25 ).
Fig. 25.
Preparation scheme and the structure of benzotriazole structure conataining 3CLpro inhibitors.
The molecular docking of compound 48 reveals that the benzotriazole moiety fits into the pocket formed by Cys145, Ser144, and Gly14 residues of 3CLpro. The carbonyl group of the benzotriazole ester binds the S-atom of Cys145 residue in the course of a nucleophilic attack. Moreover, the indole moiety is situated in the pocket created by Thr25, Thr26, His41, Thr45, Ala46 and Met49 residue while its -NH group was bonded through hydrogen bonding interaction with the side chain -OH group of Thr25 residue of 3CLpro.
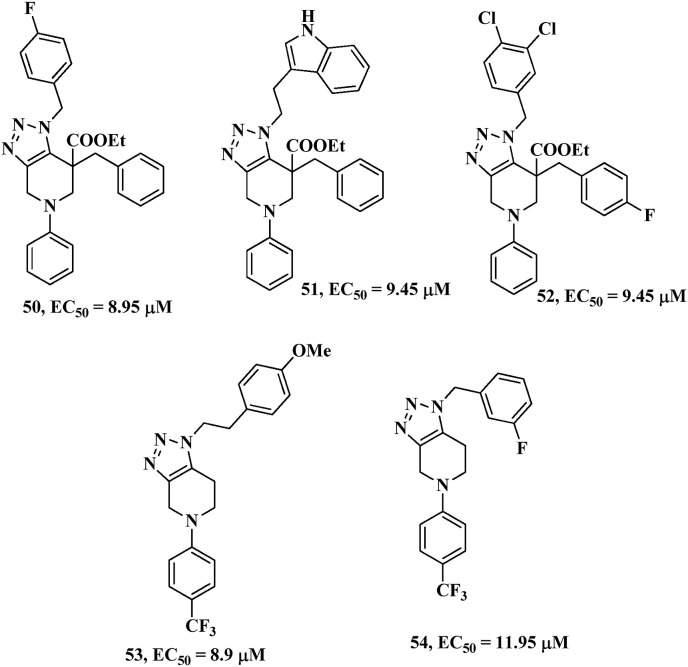
Karypidou et al. [95] synthesized a new library of fused 1,2,3-triazoles and evaluated the biological activity as potential anti-coronavirus agents (against human coronavirus 229E). In their research, they found five most potent compounds with the half maximal effective concentration (EC50) ranging from 8.90 to 11.95 μM (compounds 50–54) (Fig. 26 ). Furthermore, in silico study also showed various molecular binding interactions of the synthesized novel fused 1,2,3-triazoles with the 3-chymotrypsin-like protease.
Fig. 26.
Structure of fused 1,2,3-triazole derivatives as potential anti-coronavirus agents.
In the molecular docking of these bioactive fused 1,2,3-triazoles, they observed that compounds 50 and 51 have two hydrogen bond interactions with His163 and Glu166 residues with the fused triazole ring positioning in the close proximities of the catalytic dyad of 3CLpro. Moreover, in case of other derivatives, 53 and 54 similar behaviors were also observed in the molecular docking study enclosing two established hydrogen bond interactions within the catalytic site of 3CLpro. In brief, the compound 53 is attached through a stable binding with His163 residue and two additional interactions with Glu166 and Thr25 residues within the catalytic site. Compound 54 established stable hydrogen bonding interaction with both Glu166 and His163 residues and additionally anchored to the Gln189 residue.
Shie et al. [96] developed a series of peptide based anilide derivatives and out of all these, Niclosamide type anilides were found to be potent inhibitors of 3CLpro. Literature reveals that Niclosamide, 55 itself acts as a 3CLpro inhibitor with IC50 value > 50 μM [97]. Therefore, the authors designed and synthesized a series of Niclosamide type anilides and the best compound was found by the reaction of 2-chloro-4-nitroaniline, l-phenylalanine and 4-(dimethylamino)benzoic acid, 56 with IC50 value of 0.06 μM (Fig. 27 ).
Fig. 27.
Structure of Niclosamide and Niclosamide type anilides derivatives.
In the molecular docking study, the pocket created by Thr25, His41, Cys44, Thr45, and Ala46 residues was occupied by the 2-chloro-4-nitroanilide moiety of 56. In addition to that, the nitro group in 56 is envisaged to be bonded with the -NH of Ala46 residue of 3CLpro through hydrogen bonding, while the S-atom of Cys145 and N2 atom of His41 residues was fitted with the chlorine atom of the compound 56. Similarly, the site formed by Gln189-Gln192 and Met165-Pro68 residues was filled by the dimethylaminophenyl group of the compound.
A series of benzoquinoline compounds were designed, synthesized and evaluated against 3C-like protease (3CLpro) inhibitors by Ahn et al. [98] They found that alkylated benzoquinolines with N-phenoltetrazole moieties linked through sulphur atom, 57 was found to be more potent than the non-alkylated derivative, 58 with the IC50 values 2.0 and 10.0 μM respectively (Fig. 28 ). Computer modelling of the compound 57 shows that the benzyl group attached to the phenyl ring is fitted into the extended S3 subsite and makes interaction with the Glu166 residue through hydrophobic interaction. The other side chain (methoxy group) of 57 is situated between the region of S2 and S3 sites while the benzoquinoline nitrogen and N3 nitrogen of tetrazole ring are located in the S1/and S1 subsite of the 3CLpro. Furthermore the N3 nitrogen of tetrazole ring also binds to the Gly145 residue through hydrogen bonding.
Fig. 28.
Structure of benzoquinoline derivatives as SARS-CoV 3CL protease inhibitors.
Kuo et al. [99] recognized numerous new inhibitors of SARS-CoV 3CLpro by high throughput screening of over 6000 molecules. They found that an oxazole motif bearing acetate group, 59 and N-aryl imino group and a pyrazole moiety with four different substituents, 60 are found to be most potent inhibitor of SARS-CoV 3CLpro with IC50 values of 3.3 and 2.5 μM respectively (Fig. 29 ).
Fig. 29.
Structure of oxazole and pyrazole moiety bearing SARS-CoV 3CL protease inhibitors.
In the compound bearing oxazole motif, 59 the free rotation of the phenyl and oxazole moiety was restricted by the 1,2-steric interaction between the N-aryl imino group and the acetate group. Based on the molecular modelling interactions, the S1, S2 and S3 sites of 3CLpro are fitted with the N-aryl imino group, phenyl ring and acetate group respectively. Moreover the carbonyl oxygen of the acetate group binds with the Glu166 residue of 3CLpro through hydrogen bonding interaction.
Bacha et al.46(b) developed different bifunctional aryl boronic acid derivatives and found that these newly synthesized compounds are predominantly effective at inhibiting the protease. Out of these compounds, one of the bifunctional aryl boronic acids, 61 (named as FL-166) has inhibition constants (K i) 40 nM (0.04 μM) and considered as the most potent inhibitor for SARS Coronavirus Main Protease 3CLpro (Fig. 30 ).
Fig. 30.

Structure of bifunctional aryl boronic acid derivatives.
The evaluation of well-known possible inhibitory potency of the compounds was identified by the reactivity of hydroxyl group in serine residues with the synthesized bifunctional aryl boronic acid compounds. The boronic acid compounds targeted the nearest active sites of the protease formed by a cluster of serine residues such as Ser139, Ser144, and Ser147.
Kaeppler et al. [100] synthesized and evaluated the potent inhibitory activity of etacrynic acid derivatives against SARS-CoV Main Protease. Out of several etacrynic acid derivatives, etacrynic acid tert-butylamide, 62 and etacrynic acid amide, 63 showed the most potent ability against SARS-CoV Main Protease with the inhibitor constants (K i) 45.8 and 35.3 μM respectively while the parent compound etacrynic acid, 64 was observed with very less amount of the inhibitor constants (K i) 375 μM (Fig. 31 ).
Fig. 31.
Structure of Etacrynic acid derivatives against SARS-CoV Main Protease.
In the molecular modeling pose for etacrynic acid tert-butylamide, 62 it was found that the hydrophobic S4 pocket was fitted with tert-butyl group, S3 pocket was situated near to the dichlorobenzyl motif while the terminal ethyl group side chain at the Michael system directing away from the active site of the enzyme residue. Moreover various ligating sites of the molecule bonded to different amino acid residues His163, Glu166, and Gln189 of the protease through hydrogen bonding interaction. Herein, the molecular modelling interactions of etacrynic acid amide, 63 were similar to that of the etacrynic acid tert-butylamide, 62 interactions. In compound 63, two hydrogen bonds are formed between the terminal amino group and the Thr190 and Gln192 residues whereas in the compound 62 the hydrogen bond interaction was found between the carbonyl group linked to the Michael system and the His163 residue of the protein which make the compound 63 more potent than compound 62.
Apart from all the synthesized compounds some of the nature derived products are also found to have the inhibitor efficiency against SARS-CoV 3CLpro. Ryu et al. [101] isolated eight diterpenoids and four biflavonoids from the ethanol extract of Torreya nucifera leaves (traditionally used as medicinal plant in Asia, mostly found in Japan and South Korea, for the treatment of several parasitic conditions including hookworm, tapeworms, pinworms and roundworms) and evaluated the SARS-CoV 3CLpro inhibition. Out of all these natural product molecules, the biflavone amentoflavone 65 was found to be the most effective inhibitor against SARS-CoV 3CLpro with IC50 8.3 μM (Fig. 32 ).
Fig. 32.
Structure of amentoflavone isolated from Torreya nucifera leaves.
Molecular docking study of amentoflavone 65 indicates the formation of two hydrogen bonds between the C5 hydroxyl group of A ring of amentoflavone with the nitrogen atom of the imidazole group of His163 and -OH group of Leu141 residue situated in the S1 site of 3CLpro. Similarly, the hydroxyl group of B ring of amentoflavone also forms hydrogen bonding interaction with Gln189 residue and situated in the S2 subsite of 3CLpro. Two more hydrogen bonding interactions were also found between the Val186 residue and the hydroxyl group of the ring C and the Gln192 residue with the carbonyl group of the ring D.
Toney et al. [102] developed a non-peptidyl inhibitor of 3CLpro known as Sabadinine, 66 which has the criteria of lowest energy conformers as similar to that of the AG7088, 67 which is a peptidyl molecule with human rhinovirus (HRV) serotype 2 3CLpro inhibitor efficacy (Fig. 33 ).
Fig. 33.
Structure of sabadinine and AG7088.
In the molecular docking study, it was found that the molecule (Sabadinine) 66 is sited next to the residues His44 and Cys144 covering the catalytic dyad of the protease. Moreover, hydrogen bonding interactions was also found between the hydroxyl oxygen at C(20) position of sabadinine and the nitrogen atom of His44 residue.
Park et al. [103] isolated seven tanshinone derivatives from ethanol extract of Salvia miltiorrhiza root and examined their biological activity against SARS-CoV 3CLpro, viral cysteine protease as well as Papain-like protease (PLpro) inhibitors. Out of seven compounds, dihydrotanshinone I, 68, Methyl tanshinonate, 69 and rosmariquinone, 70 were found to be most potent compounds as cysteine protease inhibitors with the IC50 values 14.4, 21.1 and 21.1 μM respectively (Fig. 34 ).
Fig. 34.
Structure of tanshinone derivatives isolated from Salvia miltiorrhiza root.
Lin et al. [104] isolated various polyphenolic compounds from the water extract of different tea leaves and investigated their 3CLpro inhibitory activity. Out of different tea leaves, black tea containing polyphenolic compounds showed better inhibitory activity against 3CLpro and it is mainly due to the presence of theaflavin composition in black tea. Theaflavin derivatives, 3-isotheaflavin-3-gallate (TF2B), 71 and theaflavin-3,3/-digallate (TF3), 72 have the most potent 3CLpro inhibitory activity with the IC50 values of 7.0 and 9.5 μM respectively (Fig. 35 ).
Fig. 35.
Structure of polyphenolic derivatives isolated from tea leaves with 3CLpro inhibitory activity.
Moreover various other naturally occurring derivatives such as phenanthroindolizidines, 73 and phenanthroquinolizidines, 74 [105], (Fig. 36 ) Emodin (6-methyl-1,3,8-trihydroxyanthraquinone), 75 [106], (Fig. 37 ) Cinnamomi Cortex extract and Procyanidin flavonoids, (Procyanidin A2, 76, Procyanidin B1, 77, Cinnamtannin B1, 78 [107]), (Fig. 38 ) along with several nucleoside analogues such as 4-fluorophenylsulfonyl-Val-Leu-CHO (Calpin inhibitor VI), 79 Bn-Val-Phe-CHO (Calpin inhibitor III), 80, β-D-N [4]-hydroxycytidine, 81 [108], (Fig. 39 ) are found to act as inhibitors of the SARS coronavirus.
Fig. 36.
Structure of phenanthroindolizidines and phenanthroquinolizidines derivatives.
Fig. 37.

Structure of Emodin.
Fig. 38.
Structure of Cinnamomi Cortex extract and Procyanidin flavonoids.
Fig. 39.
Structure of different nucleoside analogues.
In addition to all these, Kim et al. [109] developed 5-hydroxychromones derivative and evaluated their activity as anti-hepatitis C virus (HCV) agent as well as anti-SARS agent (NTPase and Helicase enzyme, not 3CL protease). Out of different chromone derivatives, 6-(3-chloro-benzyloxy)-5-hydroxy-2-(3-iodobenzyloxy)-chromen-4-one, 82 was observed to exhibit best result in case of both anti-HCV and anti-SARS agent (Fig. 40 ). The IC50 value of the compound 82 was found to be 4.0 μM for HCV while 4.0 and 11.0 μM for SARS NTPase and Helicase respectively.
Fig. 40.

Structure of hydroxychromones derivatives as anti-SARS agent.
A very interesting literature report by Keyaerts et al. describes organic nitric oxide donor compounds as inhibitors of SARS-CoV in Vero E6 cells [110]. Herein, S-nitroso-N-acetylpenicillamine (SNAP), 83 and is tested for anti-SARS agents with broad concentration range giving efficient IC50 values of 222 μM. Moreover the same compound gives CC50 values of 587 μM which suggests that the compound, 83 acts as an efficient anti-SARS agent (Fig. 41 ).
Fig. 41.

Structure of S-nitroso-N-acetylpenicillamine (SNAP).
Anthocyanin derivative based anti-SARS agents are reported recently by Fakhar et al. [111] which acquired noteworthy binding affinity and apparently leads to inhibition of the target. In their study, they observed that the compounds, 84 and 85, showed the best activity as anti-SARS agents with the binding energies −12.37 and −10.94 kcal/mol respectively. In compound 84, eight different hydrogen bonding interactions are found which binds with the Asn142, Gly143, Thr190, Gln189, Pro168, Glu166 and His163 active residues of the protease while in compound 85, ligating sites of the molecule bonded to different amino acid residues Thr190, Gln189, Gly143, Leu141, His163, Glu166, Pro168, Thr24, Asn142 of protease through hydrogen bonding interactions (Fig. 42 ).
Fig. 42.
Structure of anthocyanin derivatives as anti-SARS agents.
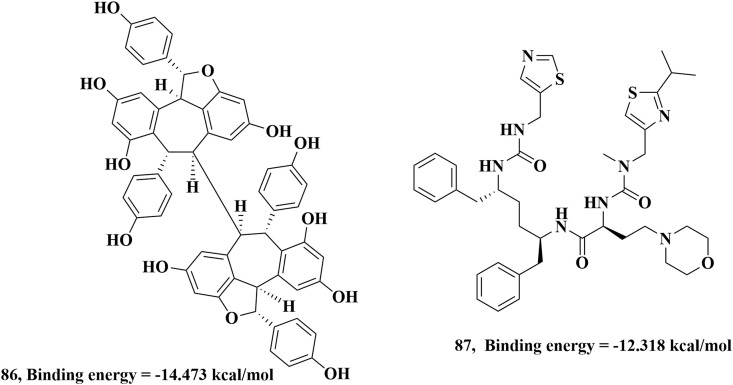
A natural compound known as Hopeaphenol (a resveratrol tetramer) can also act as SARS inhibitor in the recent study carried out by P. Sharma and co-workers through extensive computational studies [112]. Hopeaphenol, 86 binds with various amino acid residues Ser46, Glu47, Met49, Asn142, Gly143, His164, Glu166, Gln189, Thr190 and Gln192 of main protease through hydrogen bonding interactions with binding energy of −14.473 kcal/mol. In their study, they also found that Cobicistat, a common HIV drug also has SARS inhibition ability where Cobicistat, 87 molecule binds with the His41, Cys145, Glu166, Gln189 and Thr190 residues of protease through hydrogen bonding interactions with binding energy of −12.318 kcal/mol (Fig. 43 ).
Fig. 43.
Structure of natural compound Hopeaphenol and HIV drug Cobicistat.
A recent in-silico study reveals that the leaves extract of Justicia adhatoda can be used as a good inhibitor for SARS-CoV [113]. Justicia adhatoda leaves extract contains various organic compounds which are actually available as a drug in market. Among all the organic compounds present in the Justicia adhatoda leaves extract, it is found that the compounds viz. Vasicoline, 88, Anisotine, 89 and Pemirolast, 90 act as a good SARS inhibitors with binding energy ranging from −6.9 to −7.8 kcal/mol. The first two compounds, (88 and 89) are quinazoline based compounds (vasicine derivatives) while the last compound, 90 is composed of complex fused heterocycles with a tetrazole ring moiety. As vasicine and vasicinone compounds are very well known for their various biological activities therefore these compounds are also able to exhibit as anti-SARS agents (Fig. 44 ).
Fig. 44.
Structure of compounds derived from Justicia adhatoda leaves extract as SARS inhibitors.
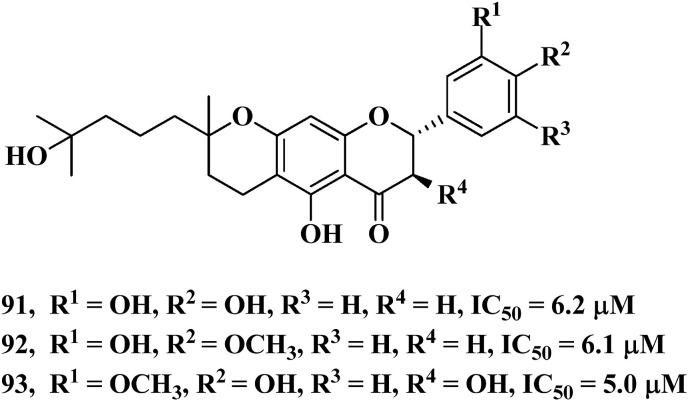
Another natural product derived SARS-CoV Papain-like protease (PLpro) inhibitor is reported by Cho et al. [114] by the use of methanol extract of Paulownia tomentosa fruit. This methanol extract mainly contains geranylated flavonoids as well as tomentin compounds which are actually 3,4-dihydro-2H-pyran derivatives as confirmed by NMR analyses. There are two geranylated flavonoids, 91 and 92 present in the methanol extract and both of them showed good efficiency as Papain-like protease inhibitor with the IC50 values of 6.2 and 6.1 μM respectively. While among the tomentin compounds, tomentin E, 93 showed the best inhibitor efficiency against Papain-like protease with IC50 values of 5.0 μM (Fig. 45 ).
Fig. 45.
Structure of geranylated flavonoids and Tomentin E.
7. Conclusion
In summary, the use of small molecules as cysteine protease inhibitor reduces the feasible replication of SARS corona virus. There is no report of corona virus protease inhibitors developed as drug candidate until now, but the approach of targeting 3CLpro to find novel anti-SARS CoV protease inhibitor with drug like properties is expected to lead the drug discovery approach against COVID-19 and other SARS related disease. Therefore, we hope that the summarization of all these small molecule cysteine protease inhibitors in this review will undeniably help medicinal chemists for further development/modification of the new/existing structures in near future to find effective therapy for corona viruses.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
D.S. is thankful to DBT, New Delhi, India for a research grant [No. BT/PR24684/NER/95/810/2017] and DST, New Delhi, India for a research grant [No. EMR/2016/002345]. The authors also acknowledge the Department of Science and Technology for financial assistance under DST-FIST program and UGC, New Delhi for Special Assistance Programme (UGC-SAP) to the Department of Chemistry, Dibrugarh University.
Biographies

Diganta Sarma did his PhD from National Chemical Laboratory, Pune, India in 2007. He then moved to Kyoto Pharmaceutical University as JSPS Postdoctoral Fellow. After completing 2 year tenure as JSPS fellow he joined University of Kansas, Lawrence, USA as Research Associate in 2009. He joined Dibrugarh University as Assistant Professor in 2012 and currently he is an Associate professor and Head of the Dept. of Chemistry, Dibrugarh University. His research areas of interest are: Green Chemistry, Peptide Chemistry, Click Chemistry and Medicinal Chemistry. He has published more than 50 articles in international journal of high repute. He has also authored a Green Chemistry Book published by Kalyani Publications. In addition to that three more book chapters are in press which are going to be published soon; two book chapters in Elsevier and one in Spinger. His is also the recipient of JSPS Bridge Fellowship Award, 2018.

Manashjyoti Konwar did his Masters degree (M.Sc.) in Organic Chemistry from Dibrugarh University in 2014. He then joined the same department for PhD program and in 2019 he got his PhD degree pursuing his research in the field of green chemistry. Dr. Konwar has already published 11 research articles in international journals from his green, medicinal chemistry work so far. He is also a co-author of one of the book chapters along with Dr. Diganta Sarma in the Elsevier publishing house which will be published soon. Currently he is an assistant professor in the Dept. of Chemistry, Dibru College, Dibrugarh, Assam.
References
- 1.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiatong S., Wenjun L. COVID-19 epidemic: disease characteristics in children. J. Med. Virol. 2020:1–8. doi: 10.1002/jmv.25807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlman S. Another decade, another coronavirus. N. Engl. J. Med. 2020;382:760–762. doi: 10.1056/NEJMe2001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su L., Ma X., Yu H., Zhang Z., Bian P., Han Y., Sun J., Liu Y., Yang C., Geng J., Zhang Z., Zhang Z. The different clinical characteristics of corona virus disease cases between children and their families in China–the character of children with COVID-19. Emerg. Microb. Infect. 2020;9:707–713. doi: 10.1080/22221751.2020.1744483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spinelli A., Pellino G. COVID-19 pandemic: perspectives on an unfolding crisis. Br. J. Surg. 2020:1–3. doi: 10.1002/bjs.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect. Genet. Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoo J.H. The fight against the 2019-nCoV outbreak: an arduous march has just begun. J. Kor. Med. Sci. 2019;35:e56. doi: 10.3346/jkms.2020.35.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramadan N., Shaib H. Middle East respiratory syndrome coronavirus (MERSCoV): a review. Germs. 2019;9:35–42. doi: 10.18683/germs.2019.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luk H.K.H., Li X., Fung J., Lau S.K.P., Woo P.C.Y. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect. Genet. Evol. 2019;71:21–30. doi: 10.1016/j.meegid.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Zhong N., Zeng G. What we have learnt from SARS epidemics in China. Br. Med. J. 2006;333:389–391. doi: 10.1136/bmj.333.7564.389. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- 12.(a) Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Estimating Mortality from COVID-19, Scientific Brief as Reported on 4 August 2020 by World Health Organization. WHO); 2020. [Google Scholar]
- 13.https://www.who.int/classifications/icd/Guidelines_Cause_of_Death_COVID-19.pdf?ua=1
- 14.(a) Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wilder-Smith A., Chiew C.J., Lee V.J. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect. Dis. 2020:1–6. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satija N., Lal S.K. The molecular biology of SARS coronavirus. Ann. Ny. Acad. Sci. 2007;1102:26–38. doi: 10.1196/annals.1408.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) de Groot R.J., Baker S.C., Baric R., Enjuanes L., Gorbalenya A.E., Holmes K.V., Perlman S., Poon L., Rottier P.J.M., Talbot P.J., Woo P.C.Y., Ziebuhr J. Family Coronaviridae. Virus Taxonomy. 2012:806–828. [Google Scholar]; (b) King A.M.Q., Lefkowitz E., Adams M.J., Carstens E.B. Elsevier Academic Press; Oxford: 2011. Virus Taxonomy: in Ninth Report of the International Committee on Taxonomy of Viruses; pp. 806–828. [Google Scholar]
- 17.Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbalenya A.E., Snijder E.J., Spaan W.J. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 2004;78:7863–7866. doi: 10.1128/JVI.78.15.7863-7866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sligl W.I., Marrie T.J. Severe community-acquired pneumonia. Crit. Care Clin. 2013;29:563–601. doi: 10.1016/j.ccc.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Y., Gao G.F. Towards our understanding of SARS-CoV, an emerging and devastating but quickly conquered virus. Comput. Immunol. Microb. 2007;30:309–327. doi: 10.1016/j.cimid.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Qing H., Li Z., Yang Z., Shi M., Huang Z., Song J., Song Z. The possibility of COVID-19 transmission from eye to nose. Acta Ophthalmol. 2020 doi: 10.1111/aos.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Xu J., Zhao S., Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet. Microbiol. 2020;244:108693. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W., Morse J.S., Lalonde T., Xu S. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020 doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.V’kovski P., Al-Mulla H., Thiel V., Neuman B.W. New insights on the role of paired membrane structures in coronavirus replication. Virus Res. 2015;202:33–40. doi: 10.1016/j.virusres.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.(a) Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A.A., Zaki A., Fouchier R.A.M., Thiel V., Drosten C., Rottier P.J.M., Osterhaus A.D.M.E., Bosch B.J., Thiel V. Dipeptidyl peptidase4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Lenane C.W., Perlman S., McCray P.B. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fehr A.R., Perlman S. Springer-Nature publication; 2015. Coronaviruses: an Overview of Their Replication and Pathogenesis. Methods in Molecular Biology, Chapter 1-Coronaviruses; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frieman M. The art of war: battles between virus and host. Curr. Opin. Virol. 2014;6:76–77. doi: 10.1016/j.coviro.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.(a) Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microb. Infect. 2020;22:74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020:1–3. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grum-Tokars V., Ratia K., Begaye A., Baker S.C., Mesecar A.D. Evaluating the 3C-like protease activity of SARS-Coronavirus: recommendations for standardized assays for drug discovery. Virus Res. 2008;133:63–73. doi: 10.1016/j.virusres.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao F., Ou H.Y., Chen L.L., Zheng W.X., Zhang C.T. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 2003;553:451–456. doi: 10.1016/S0014-5793(03)01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt M.F., Isidro-Llobet A., Lisurek M., El-Dahshan A., Tan J., Hilgenfeld R., Rademann J. Sensitized detection of inhibitory fragments and iterative development of non-peptidic protease inhibitors by dynamic ligation screening. Angew. Chem. Int. Ed. 2008;47:3275–3278. doi: 10.1002/anie.200704594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akaji K., Konno H., Mitsui H., Teruya K., Shimamoto Y., Hattori Y., Ozaki T., Kusunoki M., Sanjoh A. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J. Med. Chem. 2011;54:7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh A.K., Xi K., Grum-Tokars V., Xu X., Ratia K., Fu W., Houser K.V., Baker S.C., Johnson M.E., Mesecar A.D. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2007;17:5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhardwaj K. How prepared are we to control severe acute respiratory syndrome in future. Am. J. Virol. 2013;2:8–19. [Google Scholar]
- 39.Niu C., Yin J., Zhang J., Vederas J.C., James M.N. Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1 pocket of SARS-CoV Mpro. Bioorg. Med. Chem. 2008;16:293–302. doi: 10.1016/j.bmc.2007.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.(a) Hui D.S.C., Wong P.C., Wang C. SARS: clinical features and diagnosis. Respirology. 2003;8:S20–S24. doi: 10.1046/j.1440-1843.2003.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020:1–7. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.(a) Carlos W.G., Dela Cruz C.S., Cao B., Pasnick S., Jamil S. Novel wuhan (2019-nCoV) coronavirus. Am. J. Respir. Crit. Care Med. 2020;201:P7–P8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]; (b) Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Guo F., Cao Y., Li L., Guo Y. Insight into COVID-2019 for pediatricians. Pediatr. Pulmonol. 2020:1–4. doi: 10.1002/ppul.24734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.(a) Radford A.D., Chapman D., Dixon L., Chantrey J., Darby A.C., Hall N. Application of next-generation sequencing technologies in virology. J. Gen. Virol. 2012;93:1853–1868. doi: 10.1099/vir.0.043182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Colson P., Lagier J.C., Baudoin J.P., Khalil J.B., La Scola B., Raoult D. Ultrarapid diagnosis, microscope imaging, genome sequencing, and culture isolation of SARS-CoV-2. Eur. J. Clin. Microbiol. Infect. Dis. 2020:1–3. doi: 10.1007/s10096-020-03869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cetron M., Simone P. Battling 21st-century scourges with a 14th-century toolbox. Emerg. Infect. Dis. 2004;10:2053–2054. doi: 10.3201/eid1011.040797_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leyssen P., De Clercq E., Neyts J. Molecular strategies to inhibit the replication of RNA viruses. Antivir. Res. 2008;78:9–25. doi: 10.1016/j.antiviral.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.(a) Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 47.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 48.(a) Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]; (b) Kuo C.J., Chi Y.H., Hsu J.T.A., Liang P.H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318:862–867. doi: 10.1016/j.bbrc.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H., Luo H., Yu C., Sun T., Chen J., Peng S., Qin J., Shen J., Yang Y., Xie Y., Chen K., Wang Y., Shen X., Jiang H., Chen K. Molecular cloning, expression, purification, and mass spectrometric characterization of 3C-like protease of SARS coronavirus. Protein Expr. Purif. 2003;32:302–308. doi: 10.1016/j.pep.2003.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S.J., Silverman E., Bargman J.M. The role of antimalarial agents in the treatment of SLE and lupus nephritis. Nat. Rev. Nephrol. 2011;7:718–729. doi: 10.1038/nrneph.2011.150. [DOI] [PubMed] [Google Scholar]
- 51.Cheng V.C., Wong S.C., To K.K., Ho P.L., Yuen K.Y. Preparedness and proactive infection control measures against the emerging novel coronavirus in China. J. Hosp. Infect. 2020;104:254–255. doi: 10.1016/j.jhin.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee P., Shah F., Desai P., Avery M. Inhibitors of SARS-3CLpro: virtual screening, biological evaluation, and molecular dynamics simulation studies. J. Chem. Inf. Model. 2011;51:1376–1392. doi: 10.1021/ci1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.H. An Overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.(a) Kumar V., Tan K.P., Wang Y.M., Lin S.W., Liang P.H. Identification, synthesis and evaluation of SARS-CoV and MERS-CoV 3C-like protease inhibitors Bioorg. Med. Chem. 2016;24:3035–3042. doi: 10.1016/j.bmc.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Skorenski M., Sienczyk M. Viral proteases as targets for drug design. Curr. Pharmaceut. Des. 2013;19:1126–1153. doi: 10.2174/1381612811319060013. [DOI] [PubMed] [Google Scholar]
- 55.Savarino A., Di Trani L., Donatelli I., Cauda R., Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.(a) Colson P., Rolain J.M., Raoult D. Chloroquine for the 2019 novel coronavirus. Int. J. Antimicrob. Agents. 2020;55:105923. doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Colson P., Rolain J.M., Lagier J.C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020;55:105932. doi: 10.1016/j.ijantimicag.2020.105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.(a) Brogden R.N., Peters D.H. Teicoplanin. Drugs. 1994;47:823–854. doi: 10.2165/00003495-199447050-00008. [DOI] [PubMed] [Google Scholar]; (b) Balzarini J., Keyaerts E., Vijgen L., Egberink H., De Clercq E., Van Ranst M Printsevskaya S.S., Olsufyeva E.N., Solovieva S.E., Preobrazhenskaya M.N. Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antivir. Res. 2006;72:20–33. doi: 10.1016/j.antiviral.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen L., Gui C., Luo X., Yang Q., Gunther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Vonderen M.G.A., Bos J.C., Prins J.M., Wertheim-van Dillen P., Speelman P. Ribavirin in the treatment of severe acute respiratory syndrome (SARS) Neth. J. Med. 2003;61:238–241. [PubMed] [Google Scholar]
- 61.Barnes P.J., Pedersen S., Busse W.W. Efficacy and safety of inhaled corticosteroids: new developments. Am. J. Respir. Crit. Care Med. 1998;157:S1–S53. doi: 10.1164/ajrccm.157.3.157315. [DOI] [PubMed] [Google Scholar]
- 62.Cvetkovic R.S., Goa K.L. Lopinavir/ritonavir. Drugs. 2003;63:769–802. doi: 10.2165/00003495-200363080-00004. [DOI] [PubMed] [Google Scholar]
- 63.(a) Proskurnina E.V., Izmailov D.Y., Sozarukova M.M., Zhuravleva T.A., Leneva I.A., Poromov A.A. Antioxidant potential of antiviral drug umifenovir. Molecules. 2020;25:1577. doi: 10.3390/molecules25071577. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Haviernik J., Štefánik M., Fojtíková M., Kali S., Tordo N., Rudolf I., Hubálek Z., Eyer L., Ruzek D. Arbidol (Umifenovir): a broad-spectrum antiviral drug that inhibits medically important arthropod-borne flaviviruses. Viruses. 2018;10:184. doi: 10.3390/v10040184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cinatl J., Morgenstern B., Bauer G., Chandra P., Rabenau H., Doerr H.W. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/S0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K.S., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-coV inhibitors for inhibition of SARS-coV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 66.(a) Chiou H.E., Liu C.L., Buttrey M.J., Kuo H.P., Liu H.W., Kuo H.T., Lu Y.T. Adverse effects of ribavirin and outcome in severe acute respiratory syndrome: experience in two medical centers. Chest. 2005;128:263–272. doi: 10.1378/chest.128.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muller M.P., Dresser L., Raboud J., McGeer A., Rea E., Richardson S.E., Mazzulli T., Loeb M., Louie M. Adverse events associated with high-dose ribavirin: evidence from the Toronto outbreak of severe acute respiratory syndrome. Pharmacotherapy. 2007;27:494–503. doi: 10.1592/phco.27.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.(a) Al-Tawfiq J.A., Momattin H., Dib J., Memish Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: an observational study. Int. J. Infect. Dis. 2014;20:42–46. doi: 10.1016/j.ijid.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wang H., Ding Y., Li X., Yang L., Zhang W., Kang W. Fatal aspergillosis in a patient with SARS who was treated with corticosteroids. N. Engl. J. Med. 2003;349:507–508. doi: 10.1056/NEJM200307313490519. [DOI] [PubMed] [Google Scholar]
- 68.Auyeung T.W., Lee J.S.W., Lai W.K., Choi C.H., Lee H.K., Lee J.S., Li P.C., Lok K.H., Ng Y.Y., Wong W.M., Yeung Y.M. The use of corticosteroid as treatment in SARS was associated with adverse outcomes: a retrospective cohort study. J. Infect. 2005;51:98–102. doi: 10.1016/j.jinf.2004.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stockman L.J., Bellamy R., Garner P.S.A.R.S. Systematic review of treatment effects. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030343. e343- e343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S.E., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He R., Adonov A., Traykova-Adonova M., Cao J., Cutts T., Grudesky E., Deschambaul Y., Berry J., Drebot M., Li X. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 2004;320:1199–1203. doi: 10.1016/j.bbrc.2004.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.(a) Regnier T., Sarma D., Hidaka K., Bacha U., Freire E., Hayashi Y., Kiso Y. New developments for the design, synthesis and biological evaluation of potent SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2009;19:2722–2727. doi: 10.1016/j.bmcl.2009.03.118. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Silva D.G., Ribeiro J.F., De Vita D., Cianni L., Franco C.H., Freitas-Junior L.H., Moraes C.B., Rocha J.R., Burtoloso A.C.B., Kenny P.W., Leitão A., Montanaria C.A. A comparative study of warheads for design of cysteine protease inhibitors. Bioorg. Med. Chem. Lett. 2017;27:5031–5035. doi: 10.1016/j.bmcl.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 75.Ikejiri M., Saijo M., Morikawa S., Fukushi S., Mizutani T., Kurane I., Maruyama T. Synthesis and biological evaluation of nucleoside analogues having 6-chloropurine as anti-SARS-CoV agents. Bioorg. Med. Chem. Lett. 2007;17:2470–2473. doi: 10.1016/j.bmcl.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.(a) Honjo M., Maruyama T., Horikawa M., Balzarini J., De Clercq E. Synthesis and biological evaluation of phosphonopyrimidine and phosphonopurine ribonucleosides. Chem. Pharm. Bull. 1987;35:3227. doi: 10.1248/cpb.35.3227. [DOI] [PubMed] [Google Scholar]; (b) Maruyama T., Sato Y., Oto Y., Takahashi Y., Snoeck R., Andrei G., Witvrouw M., De Clercq E. Synthesis and antiviral activity of 6-chloropurine arabinoside and its 2’-deoxy-2’-fluoro derivative. Chem. Pharm. Bull. 1996;44:2331. doi: 10.1248/cpb.44.2331. [DOI] [PubMed] [Google Scholar]
- 77.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghosh A.K., Gong G., Grum-Tokars V., Mulhearn D.C., Baker S.C., Coughlin M., Prabhakar B.S., Sleeman K., Johnson M.E., Mesecar A.D. Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors. Bioorg. Med. Chem. Lett. 2008;18:5684–5688. doi: 10.1016/j.bmcl.2008.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J., Huitema C., Niu C., Yin J., James M.N., Eltis L.D., Vederas J.C. Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase. Bioorg. Chem. 2008;36:229–240. doi: 10.1016/j.bioorg.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., Pettersson H.I., Huitema C., Niu C., Yin J., James M.N.G., Eltis L.D., Vederas J.C. Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters. J. Med. Chem. 2007;50:1850–1864. doi: 10.1021/jm061425k. [DOI] [PubMed] [Google Scholar]
- 81.Mohamed S.F., Ibrahiem A.A., Amr A.E., Abdalla M.M. SARS-CoV 3C-like protease inhibitors of some newly synthesized substituted pyrazoles and substituted pyrimidines based on 1-(3-aminophenyl)-3-(1H-indol-3-yl) prop-2-en-1-one. Int. J. Pharmacol. 2015;11:749–756. [Google Scholar]
- 82.(a) Konwar M., Phukan P., Chaliha A.K., Buragohain A.K., Damarla K., Gogoi D., Kumar A., Sarma D. Lewis acidic catalytic system for synthesis of pyrazole and its biaryls derivatives with antimicrobial activities through cycloaddition-Iodination-Suzuki reaction. ChemistrySelect. 2019;4:10236–10245. [Google Scholar]; (b) Akbas E., Berber I., Sener A., Hasanov B. Synthesis and antibacterial activity of 4-benzoyl-1-methyl-5-phenyl-1H-pyrazole-3-carboxylic acid and derivatives. Farmaco. 2005;60:23–26. doi: 10.1016/j.farmac.2004.09.003. [DOI] [PubMed] [Google Scholar]; (c) Ding L., Grehn L., De Clercq E., Andrei G., Snoeck R., Balzarini J., Fransson B., Ragnarsson U. Synthesis and antiviral activity of three pyrazole analogues of distamycin A. Acta Chem. 1994;48:498–505. doi: 10.3891/acta.chem.scand.48-0498. Scandinavica (Copenhagen, Denmark: 1989) [DOI] [PubMed] [Google Scholar]
- 83.Chen L.R., Wang Y.C., Lin Y.W., Chou S.Y., Chen S.F., Liu L.T., Wu Y.T., Kuo C.J., Chen T.S.S., Juang S.H. Synthesis and evaluation of isatin derivatives as effective SARS coronavirus 3CL protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3058–3062. doi: 10.1016/j.bmcl.2005.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu W., Zhu H.M., Niu G.J., Shi E.Z., Chen J., Sun B., Chen W.Q., Zhou H.G., Yang C. Synthesis, modification and docking studies of 5-sulfonyl isatin derivatives as SARS-CoV 3C-like protease inhibitors. Bioorg. Med. Chem. 2014;22:292–302. doi: 10.1016/j.bmc.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou L., Liu Y., Zhang W., Wei P., Huang C., Pei J., Yuan Y., Lai L. Isatin compounds as noncovalent SARS coronavirus 3C-like protease inhibitors. J. Med. Chem. 2006;49:3440–3443. doi: 10.1021/jm0602357. [DOI] [PubMed] [Google Scholar]
- 86.Lee H., Mittal A., Patel K., Gatuz J.L., Truong L., Torres J., Mulhearn D.C., Johnson M.E. Identification of novel drug scaffolds for inhibition of SARS-CoV 3-Chymotrypsin-like protease using virtual and high-throughput screenings. Bioorg. Med. Chem. 2014;22:167–177. doi: 10.1016/j.bmc.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kao R.Y., Tsui W.H.W., Lee T.S.W., Tanner J.A., Watt R.M., Huang J.D., Hu L., Chen G., Chen Z., Zhang L., He T., Chan K.H., Tse H., To A.P.C., Ng L.W.Y., Wong B.C.W., Tsoi H.W., Yang D., Ho D.D., Yuen K.Y. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shimamoto Y., Hattori Y., Kobayashi K., Teruya K., Sanjoh A., Nakagawa A., Yamashita E., Akaji K. Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors. Bioorg. Med. Chem. 2015;23:876–890. doi: 10.1016/j.bmc.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010;20:3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramajayam R., Tan K.P., Liu H.G., Liang P.H. Synthesis and evaluation of pyrazolone compounds as SARS-coronavirus 3C-like protease inhibitors. Bioorg. Med. Chem. 2010;18:7849–7854. doi: 10.1016/j.bmc.2010.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M., Baker S.C., Lindsley C.W., Hodder P., Mesecar A., Stauffer S.R. Discovery, synthesis, and structure-based optimization of a series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turlington M., Chun A., Tomar S., Eggler A., Grum-Tokars V., Jacobs J., Daniels J.S., Dawson E., Saldanha A., Chase P., Baez-Santos Y.M., Lindsley C.W., Hodder P., Mesecar A.D., Stauffer S.R. Discovery of N-(benzo [1,2,3] triazol-1-yl)-N-(benzyl) acetamido) phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding. Bioorg. Med. Chem. Lett. 2013;23:6172–6177. doi: 10.1016/j.bmcl.2013.08.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li Y.Q., Li Z.L., Zhao W.J., Wen R.X., Meng Q.W., Zeng Y. Synthesis of stilbene derivatives with inhibition of SARS coronavirus replication. Eur. J. Med. Chem. 2006;41:1084–1089. doi: 10.1016/j.ejmech.2006.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu C.Y., King K.Y., Kuo C.J., Fang J.M., Wu Y.T., Ho M.Y., Liao C.L., Shie J.J., Liang P.H., Wong C.H. Stable benzotriazole esters as mechanism-based inactivators of the severe acute respiratory syndrome 3CL protease. Chem. Biol. 2006;13:261–268. doi: 10.1016/j.chembiol.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Karypidou K., Ribone S.R., Quevedo M.A., Persoons L., Pannecouque C., Helsen C., Claessens F., Dehaen W. Synthesis, biological evaluation and molecular modeling of a novel series of fused 1,2,3-triazoles as potential anti-coronavirus agents. Bioorg. Med. Chem. Lett. 2018;28:3472–3476. doi: 10.1016/j.bmcl.2018.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shie J.J., Fang J.M., Kuo C.J., Kuo T.H., Liang P.H., Huang H.J., Yang W.B., Lin C.H., Chen J.L., Wu Y.T., Wong C.H. Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease. J. Med. Chem. 2005;48:4469–4473. doi: 10.1021/jm050184y. [DOI] [PubMed] [Google Scholar]
- 97.Wu C.J., Jan J.T., Chen C.M., Hsieh H.P., Hwang D.R., Liu H.W., Liu C.Y., Huang H.W., Chen S.C., Hong C.F., Lin R.K., Chao Y.S., Hsu J.T.A. Inhibition of severe acute respiratory syndrome coronavirus replication by niclosamide. Antimicrob. Agents Chemother. 2004;48:2693–2696. doi: 10.1128/AAC.48.7.2693-2696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahn T.Y., Kuo C.J., Liu H.G., Ha D.C., Liang P.H., Jung Y.S. Synthesis and evaluation of benzoquinolinone derivatives as sars-cov 3CL protease inhibitors. Bull. Kor. Chem. Soc. 2010;31:87–91. [Google Scholar]
- 99.Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S., Liang P.H. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583:549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kaeppler U., Stiefl N., Schiller M., Vicik R., Breuning A., Schmitz W., Rupprecht R., Schmuck C., Baumann K., Ziebuhr J., Schirmeister T. A new lead for nonpeptidic active-site-directed inhibitors of the severe acute respiratory syndrome coronavirus main protease discovered by a combination of screening and docking methods. J. Med. Chem. 2005;48:6832–6842. doi: 10.1021/jm0501782. [DOI] [PubMed] [Google Scholar]
- 101.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Naguyen T.T.H., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toney J.H., Navas-Martín S., Weiss S.R., Koeller A. Sabadinine. A potential non-peptide anti-severe acute-respiratory-syndrome agent identified using structure-aided design. J. Med. Chem. 2004;47:1079–1080. doi: 10.1021/jm034137m. [DOI] [PubMed] [Google Scholar]
- 103.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen C.N., Lin C.P., Huang K.K., Chen W.C., Hsieh H.P., Liang P.H., Hsu J.T.A. Inhibition of SARS-CoV 3C-like protease activity by theaflavin-3,3’-digallate (TF3) Evid.-Based Complementary Altern. Med. 2005;2:209–215. doi: 10.1093/ecam/neh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang C.W., Lee Y.Z., Kang I.J., Barnard D.L., Jan J.T., Lin D., Huang C.W., Yeh T.K., Chao Y.S., Lee S.J. Identification of phenanthroindolizines and phenanthroquinolizidines as novel potent anti-coronaviral agents for porcine enteropathogenic coronavirus transmissible gastroenteritis virus and human severe acute respiratory syndrome coronavirus. Antivir. Res. 2010;88:160–168. doi: 10.1016/j.antiviral.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwarz S., Wang K., Yu W., Sun B., Schwarz W. Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 2011;90:64–69. doi: 10.1016/j.antiviral.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhuang M., Jiang H., Suzuki Y., Li X., Xiao P., Tanaka T., Ling H., Yang B., Saitoh H., Zhang L., Qin C., Sugamura K., Hattori T. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antivir. Res. 2009;82:73–81. doi: 10.1016/j.antiviral.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barnard D.L., Hubbard V.D., Burton J., Smee D.F., Morrey J.D., Otto M.J., Sidwell R.W. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and β-D-N4-hydroxycytidine. Antivir. Chem. Chemother. 2004;15:15–22. doi: 10.1177/095632020401500102. [DOI] [PubMed] [Google Scholar]
- 109.Kim M.K., Yu M.S., Park H.R., Kim K.B., Lee C., Cho S.Y., Kang J., Yoon H., Kim D.E., Choo H., Jeongb Y.J., Chong Y. 2, 6-Bis-arylmethyloxy-5-hydroxychromones with antiviral activity against both hepatitis C virus (HCV) and SARS-associated coronavirus (SCV) Eur. J. Med. Chem. 2011;46:5698–5704. doi: 10.1016/j.ejmech.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Keyaerts E., Vijgen L., Chen L., Maes P., Hedenstierna G., Van Ranst M. Inhibition of SARS-coronavirus infection in vitro by S-nitroso-N-acetylpenicillamine, a nitric oxide donor compound. Int. J. Infect. Dis. 2004;8:223–226. doi: 10.1016/j.ijid.2004.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fakhar Z., Faramarzi B., Pacifico S., Faramarzi S. Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: an in-silico perspective of therapeutic targets against COVID-19 pandemic. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1801510. [DOI] [PubMed] [Google Scholar]
- 112.Sharma P., Vijayan V., Pant P., Sharma M., Vikram N., Kaur P., Singh T.P., Sharma S. Identification of potential drug candidates to combat COVID-19: a structural study using the main protease (mpro) of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1798286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bag A., Bag A. Justicia adhatoda leaves extract is a strong remedy for COVID-19–Case report analysis and docking based study. ChemRxiv. 2020:1–7. doi: 10.26434/chemrxiv.12038604.v1. [DOI] [Google Scholar]
- 114.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21:3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]