Summary
Systemic lupus erythematosus (SLE) is a multi-organ autoimmune disorder with a prominent genetic component. Individuals of African ancestry (AA) experience the disease more severely and with an increased co-morbidity burden compared to European ancestry (EA) populations. We hypothesize that the disparities in disease prevalence, activity, and response to standard medications between AA and EA populations is partially conferred by genomic influences on biological pathways. To address this, we applied a comprehensive approach to identify all genes predicted from SNP-associated risk loci detected with the Immunochip. By combining genes predicted via eQTL analysis, as well as those predicted from base-pair changes in intergenic enhancer sites, coding-region variants, and SNP-gene proximity, we were able to identify 1,731 potential ancestry-specific and trans-ancestry genetic drivers of SLE. Gene associations were linked to upstream and downstream regulators using connectivity mapping, and predicted biological pathways were mined for candidate drug targets. Examination of trans-ancestral pathways reflect the well-defined role for interferons in SLE and revealed pathways associated with tissue repair and remodeling. EA-dominant genetic drivers were more often associated with innate immune and myeloid cell function pathways, whereas AA-dominant pathways mirror clinical findings in AA subjects, suggesting disease progression is driven by aberrant B cell activity accompanied by ER stress and metabolic dysfunction. Finally, potential ancestry-specific and non-specific drug candidates were identified. The integration of all SLE SNP-predicted genes into functional pathways revealed critical molecular pathways representative of each population, underscoring the influence of ancestry on disease mechanism and also providing key insight for therapeutic selection.
Keywords: SLE, ancesty, Immunochip, drug repurposing, lupus, genetics, GWAS, pathway analysis
Introduction
Systemic lupus erythematosus (SLE) (MIM: 152700) is a multi-organ autoimmune disorder associated with significant morbidity and mortality. SLE is strongly influenced by genetic factors and recent candidate gene, Immunochip, and genome-wide association studies (GWASs) have identified more than 100 SLE susceptibility loci.1, 2, 3, 4, 5, 6 However, disease development is complex and unpredictable, with considerable clinical heterogeneity among ancestral groups. Specifically, individuals of African ancestry (AA) experience more severe disease and more co-morbidities compared to European ancestry (EA) populations.7, 8, 9 Furthermore, there seems to be variability in the response of individuals within different ancestral groups to standard medications, including cyclophosphamide, mycophenolate, rituximab, and belimumab. For example, belimumab, a monoclonal antibody directed to TNFSF13B exhibiting clinical benefit in moderately active SLE, was reported to be less effective in treating AA populations.10, 11, 12, 13
Although GWASs have achieved great success in mapping disease loci in polygenic autoimmune diseases, the majority of GWAS findings have failed to impact clinical practice.14 Moreover, for many single-nucleotide polymorphisms (SNPs), the biologic implications have usually not been identified. Here we hypothesize that using the more global approach of identifying all of the genes implicated by GWASs and modeling them into biologic pathways might provide a broader view of the impact of genetics on SLE, and, also, an indication of the disparate genetic influences manifest in affected individuals of different ancestries. Utilization of expression quantitative trait loci (eQTL) mapping15, 16, 17 as well as identification of variants disrupting transcription factor binding site (TFBS) occupancy in active regulatory regions resulted in the identification of genes implicated by ancestry-specific and trans-ancestral SNP associations. We then applied a comprehensive systems biology approach to predict SLE-associated biological pathways. Putative pathways were validated by connectivity mapping to differentially expressed genes (DEGs) in SLE and candidate treatments for each ancestral group were identified. Together, these genetic and gene expression analyses have identified biological pathways common to both EA and AA, as well as those implicated by the differential strength of the ancestry-specific association as more dominant in either ancestry, and have helped identify novel drug candidates that might uniquely impact EA and AA SLE.
Material and Methods
Identification of SLE-Associated SNPs and Predicted Genes
The SLE Immunochip study18 identified single-nucleotide polymorphisms (SNPs) significantly associated with SLE in AA (2,970 case subjects, 2,452 control subjects) and EA (6,748 case subjects, 11,516 control subjects) cohorts. SNP proxies, identified via rAggr (see Web Resources) in linkage disequilibrium (LD) (r2 > 0.5) with these SLE-associated SNPs, were then determined, using the Central European Utah (CEU) population as background for EA SNPs and the Yoruban (YRI) population for AA SNPs. Expression quantitative trait loci (eQTLs) were then identified as previously described18 using GTEx v.619 and the Blood eQTL browser database20 and mapped to their associated eQTL expression genes (E-Genes). In parallel, random E-Gene datasets were generated from randomly selected SLE Immunochip SNPs using the same methodology. SNP proxies were then queried by GTEx to generate eQTLs and matched to ENSEMBL gene IDs. To find SNPs in enhancers and promoters and their associated transcription factors and downstream target genes (T-Genes), we queried the atlas of Human Active Enhancers to interpret Regulatory variants (HACER)21 and the GeneHancer database.22 To find structural SNPs in protein-coding genes (C-Genes), we queried the human Ensembl genome browser (GRCh38.p12) and dbSNP. Several additional databases were used to generate loss-of-function prediction scores, including SIFT4G,23,24 PolyPhen-2,25 PROVEAN,26 and PANTHER.27 All other SNPs were linked to the most proximal gene (P-Gene) or gene region as previously detailed.18 For overlap studies, Venn diagrams were computed and visualized using InteractiVenn.28 All predicted genes were divided into an AA, EA, or shared group depending on the ancestral designation of the original SLE-associated SNP.
Statistical Analysis
The single-locus and multi-locus ancestry-specific tests of association within each ancestral group have been previously reported.18 Specifically, to test for an association between a SNP and case/control status separately for the AA and EA ancestries, logistic regression models were computed adjusting for population substructure using admixture factors as covariates. The Benjamini-Hochberg false discovery rate adjusted p values (pFDR) were computed, and SNPs were considered for subsequent analyses in this manuscript if they met a pPDR < 0.05 threshold. The two ancestry-specific analyses (i.e., AA and EA) were meta-analyzed using the weighted inverse normal (weighted by sample size) method and tested for heterogeneity also as previously described (pHET)18). The following algorithm was used to classify significant associations in either ancestral group (pFDR < 0.05) as shared or ancestry specific (i.e., primarily driven by the EA or AA ancestry subpopulations). First, if the pHET > 0.01 then the association was considered common (shared) across the EA and AA ancestries. If the pHET < 0.01, then we considered the direction (odds ratio: OR > 1, OR < 1) and the ancestry-specific p values. If pHET < 0.01 and the OR was in the same direction with suggestive evidence of association (p < 0.05; not FDR adjusted), then the association was considered shared. If pHET < 0.01 and the OR was in the same or opposite directions without at least suggestive evidence of association in both populations (p < 0.05), then the association was considered ancestry specific and driven by the ancestry with the significant association (pFDR < 0.05). Finally, if pHET < 0.01 but the associations were significant and in opposite directions, the association was considered shared (noting the ancestry-specific direction of the associations). Graphpad PRISM 8.0 was used to perform mean, 95% confidence intervals, and unpaired t test with Welch’s correction.
Genomic Functional Categories
The Variant Effect Predictor (VEP) tool available on the Ensembl genome browser 93 was used for annotation information to specify SNPs located within non-coding regions, including micro (mi)RNAs, long non-coding (lnc)RNAs, introns, and intergenic regions. Regulatory regions include transcription factor binding sites (TFBS), promoters, enhancers, repressors, promoter flanking regions, and open chromatin. Coding regions were broken down further and include 5′ UTRs, 3′ UTRs, and synonymous and nonsynonymous (missense and nonsense) mutations. The online resource tool HaploReg (v.4.1)29 was also used to identify DNA features and regulatory elements and to assess regulatory potential.
Differential Expression Analysis of E-Genes
Predicted genes were compared to multiple differential expression datasets, as summarized in Table S1. These datasets include the log-fold changes of all genes with significant (FDR < 0.2) differential expression in whole blood (WB), peripheral blood mononuclear cells (PBMC), B cells, T cells, myeloid cells, synovium, skin, kidney glomerulus (G), and kidney tubulointerstitium (TI). The FDR was selected a priori to avoid excluding false negatives from the analysis. Cohorts are SLE versus control (CTL) unless noted otherwise. Additional cohorts include SLE synovium versus oseteoarthritis (OA) synovium, discoid lupus erythematosus (DLE) skin versus CTL skin, and subacute cutaneous lupus erythematosus (CLE) skin versus CTL skin. Datasets include GEO: GSE88884 (Illuminate 1 and 2), GSE49454, GSE22908, GSE61635, GSE29536, GSE39088, GSE50772, FDABMC3, EMTAB2713, GSE10325, GSE4588, GSE38351, GSE36700, GSE52471, GSE72535, GSE81071, and GSE32591.
Functional Gene Set Analysis and Identification of Upstream Regulators (UPRs)
For both ancestral groups, predicted gene lists were examined using Biologically Informed Gene Clustering (BIG-C; v.4.4.). BIG-C is a custom functional clustering tool developed to annotate the biological meaning of large lists of genes. Genes are sorted into 54 categories based on their most likely biological function and/or cellular localization based on information from multiple online tools and databases including UniProtKB/Swiss-Prot, gene ontology (GO) Terms, MGI database, KEGG pathways, NCBI, PubMed, and the Interactome and has been previously described.30,31
I-Scope is a custom clustering tool used to identify immune infiltrates in large gene datasets and has been described previously.32 Briefly, I-Scope was created through an iterative search of more than 17,000 genes identified in more than 50 microarray datasets. These genes were researched for immune cell-specific expression in 30 hematopoietic sub-categories: T cells, regulatory T cells, activated T cells, anergic cells, CD4 T cells, CD8 T cells, gamma-delta T cells, NK/NKT cells, T and B cells, B cells, activated B cells, T and B and monocytes, monocytes and B cells, MHC class II expressing cells, monocyte dendritic cells, dendritic cells, plasmacytoid dendritic cells, Langerhans cells, myeloid cells, plasma cells, erythrocytes, neutrophils, low-density granulocytes, granulocytes, platelets, and all hematopoietic stem cells.
Enrichment of GO Biological Processes (BP) using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and the Ingenuity Pathway Analysis (IPA) platform provided additional genetic pathway identification. IPA upstream regulator (UPR) analysis was also used to identify potential transcription factors, cytokines, chemokines, etc. that can contribute to the observed gene expression pattern in the input dataset.
Network Analysis and Visualization
Visualization of protein-protein interaction and relationships between genes within datasets was done using Cytoscape (v.3.6.1) software. Briefly, STRING (v.1.3.2)-generated networks were imported into Cytoscape (v.3.8.1) and partitioned with MCODE via the clusterMaker2 (v.1.2.1) plugin. The resulting networks were further simplified into metastructures defined by the number of genes in each cluster, the number of significant intra-cluster connections predicted by MCODE, and the strength of associations connecting members of different clusters to each other.
Gene Set Variation Analysis (GSVA)
The GSVA33 (v.1.25.0) software package for R/Bioconductor and has been described previously.30,34 Briefly, GSVA is a non-parametric, unsupervised method for estimating the variation of pre-defined gene sets in case and control samples of microarray expression datasets. The input for the GSVA algorithm was a gene expression matrix of log2 microarray of expression values and a collection of pre-defined gene signatures. Enrichment scores (GSVA scores) were calculated non-parametrically using a Kolmogorov-Smirnoff (KS)-like random walk statistic and a negative value for each gene set. EA and AA SNP-predicted genes were used to create GSVA gene signatures (official gene symbols for each signature are listed in Table S2). In the case of leukotriene biosynthesis, cell cycle, ubiquitylation and sumoylation, apoptosis signaling, nuclear receptor signaling, and PKA signaling, genes were initially identified following protein-protein interaction network construction and MCODE clustering. Cluster identity was determined by BIG-C and/or IPA canonical pathway analysis, where each cluster was used as a GSVA probe. Gene signatures for diapedesis, TH1 activation pathway, unfolded protein and stress, T cell exhaustion, and SLE in B cell signaling were all informed by established IPA canonical pathways. The signature for lysosome was derived from the Lysosome BIG-C category. All interferon and cytokine signatures (core IFN, IFNB1, IFNA2, IFNW, IFNG, IL12, and TNF) have been described previously.30 Metabolic signatures for oxidative phosphorylation and glycolysis were based on literature mining and established IPA canonical pathways. Enrichment of each signature was examined in EA and AA SLE subjects and healthy control subject whole blood from GEO: GSE88884. Differences between control subjects and SLE-affected subject GSVA enrichment scores were determined using the Welch’s t test for unequal variances in Graphpad PRISM 8.0.
Drug Candidate Identification and CoLT Scoring
Drug candidates were identified using LINCS, STITCH (v.5.0), and IPA. Each of these tools includes either a programmatic method of matching existing therapeutics to their targets or else is a list of drugs and targets for achieving the same end. In addition to identifying drugs targeting predicted genes directly, these tools were also used to identify drugs targeting select upstream regulators. Where information was available, drugs were assessed by CoLT scoring to rank potential drug candidates for repositioning in SLE as previously described.35
Results
SLE-Associated Variants Predict Downstream Target Genes
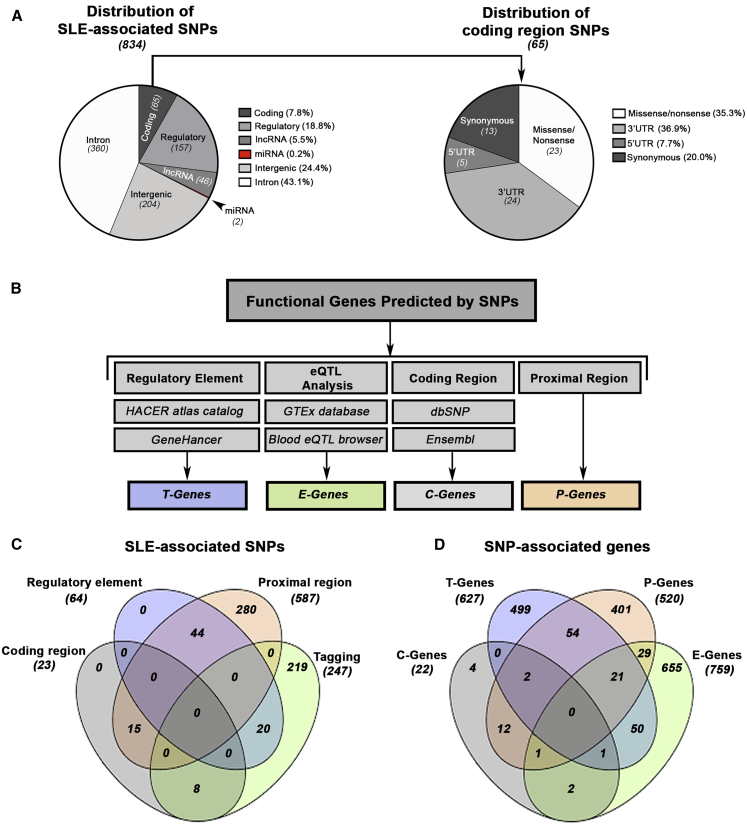
We examined the distribution (e.g., coding, non-coding) of 834 non-HLA single-nucleotide polymorphisms (SNP) reported as significantly associated with SLE in a large trans-ancestral genetic association study18 (Figure 1A; Table S3). The majority of SNPs were found in intronic (43.1%) or intergenic (24.4%) regions. Approximately 26% of SLE-associated SNPs mapped to regions encoding transcripts (7.8% exons, 5′ UTRs, 3′ UTRs) or known regulatory regions (18.8%; TFBS, promoters, enhancers, etc.). 5.7% of SNPs mapped to regions containing long non-coding (lnc)RNAs or micro (mi)RNAs.
Figure 1.
Mapping the Functional Genes Predicted by SLE-Associated SNPs
(A) Distribution of genomic functional categories for all ancestry-specific non-HLA-associated SLE SNPs (tiers 1–3). Non-coding regions include micro (mi)RNAs, long non-coding (lnc)RNAs, introns, and intergenic regions. Regulatory regions include transcription factor binding sites (TFBS), promoters, enhancers, repressors, promoter flanking regions, and open chromatin. Coding regions were broken down further and include 5′ UTRs, 3′ UTRs, and synonymous and nonsynonymous (missense and nonsense) mutations.
(B) Functional genes predicted by SNPs are derived from four sources including regulatory elements (T-Genes), eQTL analysis (E-Genes), coding regions (C-Genes), and proximal gene-SNP annotation (P-Genes).
(C and D) Venn diagram depicting the overlap of all SLE-associated SNPs (C) and all predicted E-, T-, P-, and C- Genes (D).
The reported test of SNP-by-ancestry (i.e., heterogeneity) from the meta-analysis and ancestry-specific effect size allowed determination of whether the SNP was more associated with only one ancestry (e.g., AA specific) or was a trans-ancestry association (i.e., comparable effect size across AA and EA)18 (Table S4). Next, we attempted to identify the most plausible gene(s) affected by the SLE-SNP association (Figure 1B). Using the GTEx and Blood eQTL browser databases,19,20 we identified 78 EA and 21 AA-related eQTLs linked to 207 and 29 expression genes (E-Genes) primarily associated with EA and AA, respectively. A total of 148 eQTL were common to both ancestries and were associated with 523 shared E-Genes (Table S5). Interestingly, we observed that E-Genes predicted by a single SNP tended to be enriched for a common molecular function (Figure S1; Table S6). The remaining 587 SNPs that were not associated with eQTLs were assigned to the closest proximal gene (P-Gene), revealing 520 P-Genes (465 EA, 34 AA, and 21 shared) (Figures 1C and 1D; Table S5)
Since variants that alter or disrupt transcription factor binding are also known to dysregulate gene expression,36 we sought to identify SNPs within regulatory elements (e.g., enhancers and promoters). Computational tools, including GeneHancer and HACER that connect putative regulatory SNPs with transcription factors and downstream target genes (T-Genes),21,22 were used to identify 64 SNPs overlapping regulatory elements or promoters predicted to impact the expression of 627 T-Genes (475 EA, 9 AA, and 143 shared) and the action of 95 transcription factors (Tables S5 and S7). For variants located in coding regions, 23 SNPs (14 EA, 2 AA, 7 shared) were associated with either non-synonymous or nonsense changes, affecting 22 genes (C-Genes; 14 EA, 2 AA, and 6 shared) (Table S8).
Figure 1C depicts the overlap between SNPs based on discovery method, whereas Figure 1D shows the overlap between the corresponding SNP-predicted E-, T-, C-, and P-Genes. No genes were shared within all four groups, and we observed limited commonality between T-, P-, and E-Genes, with only 21 genes shared among the three groups. Despite the overall diversity of genes observed in each list, significant overlap was observed in the number of genes shared between ancestries (Figure S2, Table S5).
Characterization of Gene Signatures
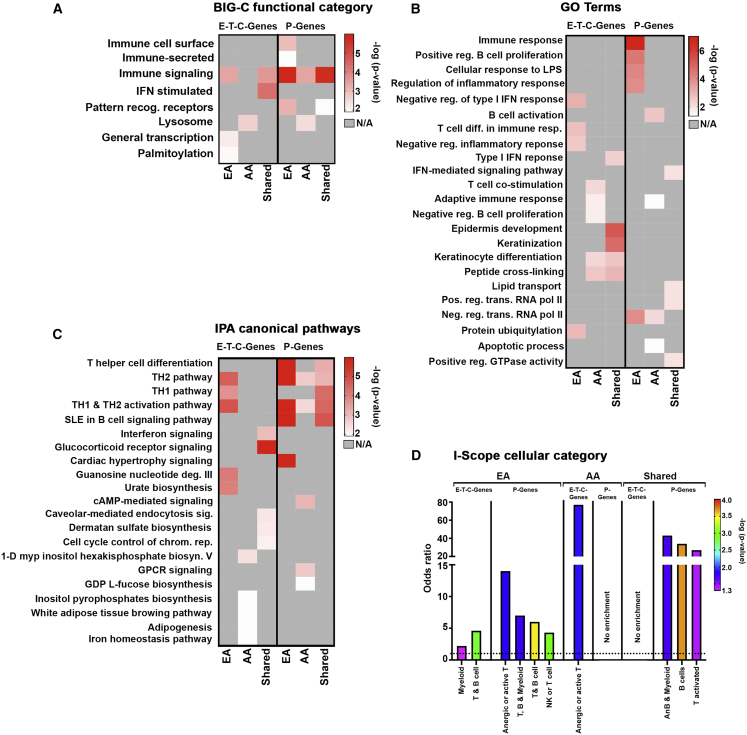
We next carried out a series of bioinformatics analyses to determine the biological function of the full array of EA (1,676) and AA (725) SNP-predicted genes (Figure S2). P-Genes were analyzed separately from E-T-C-Genes to avoid overrepresentation of immune-based processes because of the Immunochip design bias.37 Both the EA and AA gene sets exhibited similarity in biological function reflected in the number of matching functional pathways (immune signaling), IPA canonical pathways (TH1 pathway, IFN signaling pathway, glucocorticoid receptor signaling, etc.), and GO terms (epidermis development [GO: 0008544], immune response [GO: 0006955]), but also a number of clear differences. In addition, EA and AA gene sets were examined using a clustering program (I-Scope, see Material and Methods) that detects immune and inflammatory cell type signatures within large gene lists to identify dominant immune cell populations contributing to disease pathology within each ancestry.32 Both AA and EA exhibited enrichment of B cell genes, with AA exhibiting a significantly greater B cell signal. As a control, genes derived from non-associated Immunochip SNPs (“random”) were examined and found to be modestly enriched in some biosynthesis and metabolic pathways but did not overlap with any EA or AA pathways, nor did they exhibit enrichment in cellular categories.
Parallel analyses were also conducted examining associations that were more strongly linked to one ancestry over the other (because of higher allelic frequency), independent of the shared gene cohort. Analysis of representative EA genes revealed enrichment in processes related to innate immune function, including the functional category for pattern recognition receptors, GO terms for the cellular response to LPS (GO:007122), canonical pathways for JAK/STAT signaling and agranulocyte adherence and diapedesis, and cellular categories for myeloid cells and T and B cells (Figures 2A–2D and S3). AA-associated genes were differentially enriched in the adaptive immune response reflected by the cellular categories for anergic or activated T cells and GO terms for B cell activation (GO:0042113) and T cell co-stimulation (GO: 0031295), with additional enrichment in biological processes for degradation, including the functional category lysosome, and IPA pathways for autophagy and phagosome maturation (Figures 2A–2D and S3). To establish that ancestry-driven differences were not related to inequality in the number of SNPs used to predict genes for analysis (604 EA SNPs versus 77 AA SNPs), we directly compared all genes associated with AA SNPs to those predicted by the 77 most significant EA SNPs to generate an approximately equal cohort of genes. Overall, limited overlap was observed in the number of genes (3 total; c7orf72, FIGNL1, and IKZF1) and enriched pathways shared between ancestries (Figure S4).
Figure 2.
Functional Characterization of Predicted Genes
(A) Ancestry-dependent and -independent SNP-predicted genes were analyzed to determine enrichment using functional definitions from the BIG-C (Biologically Informed Gene Clustering) annotation library. E-, T-, and C-Genes were analyzed together; P-Genes were examined separately. Enrichment was defined as any category with an odds ratio (OR) > 1 and –log10(p value) > 1.33.
(B and C) Heatmap visualization of the top five significant IPA canonical pathways and gene ontogeny (GO) terms for each gene list (E-T-C-Genes and P-Genes) organized by ancestry. Top pathways with –log10(p value) > 1.33 are listed.
(D) I-Scope hematopoietic cell enrichment defined as any category with an OR > 1, indicated by the dotted line, and –log10(p value) > 1.33.
Protein Interaction-Based Clustering of SNP-Associated Genes
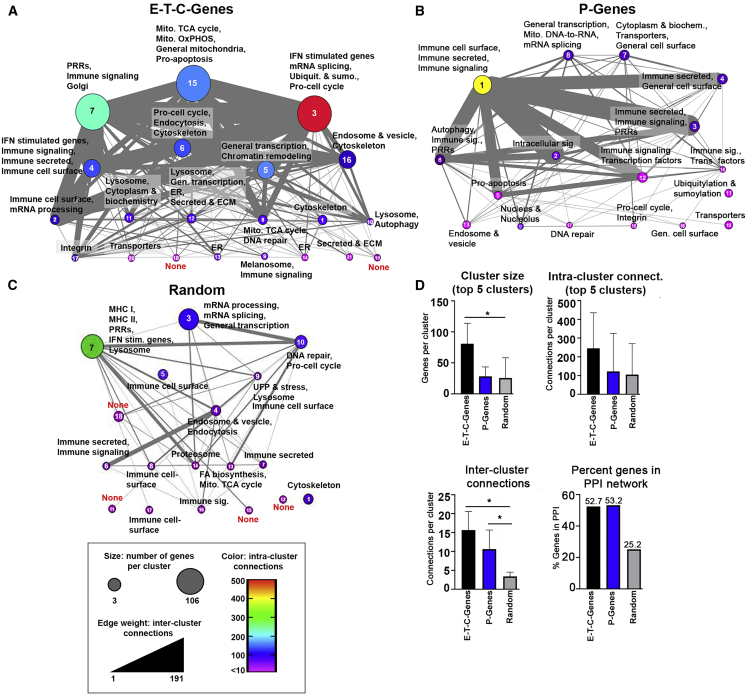
We next sought to assess the relationship between all SNP-associated genes regardless of ancestral origin to obtain information about the likely nature of the molecular pathways driving SLE. Overall, 52.7% of E-T-C-Genes (701/1,330) and 53.2% of P-Genes (272/520) were incorporated into protein-protein interaction (PPI) networks with the majority of E-T-C-Genes coalescing into several large, multi-functional clusters. The integration of all EA and AA genes into the same network highlights the overall commonality in gene function and connectivity observed among the ancestries, including cluster 7 enriched in molecules associated with immune signaling and Golgi function, cluster 15 dominated by metabolic processes, and cluster 3 containing a robust interferon signature (Figures 3A and S5). Networks constructed of all P-Genes revealed the predominance of immune function with seven out of ten of the largest, intra-connected clusters enriched in immune activity (Figure 3B).
Figure 3.
Cluster Metastructures for SLE-Predicted and Randomly Generated Genes
(A–C) Cluster metastructures were generated based on PPI networks, clustered using MCODE, and visualized in CytoScape. Size indicates the number of genes per cluster, edge weight indicates the number of inter-cluster connections, and color indicates the number of intra-cluster connections. Cluster number is indicated within each metacluster. A random gene network (1,033 genes) was clustered along-side networks for E-T-C-Genes and P-Genes. Functional enrichment for each cluster was determined using BIG-C.
(D) Quantitation of cluster size, intra-cluster connections, inter-cluster connections, and the percent of genes incorporated into each network is displayed. Error bars represent the 95% confidence interval; asterisks (∗) indicate a p value < 0.05 using Welch’s t test.
To confirm that the predicted molecular pathways derived from SNP-predicted genes were specific for SLE and not related by chance, we carried out a parallel analysis examining PPI networks composed of genes derived from non-associated (random) Immunochip SNPs (Figure 3C; Table S9). Examination of meta-structures reveals that random gene networks incorporated fewer genes overall (258/1,033; 24.9%) and exhibited significantly fewer inter-cluster connections and fewer intra-cluster connections, appearing as independent entities lacking robust functional relationships with neighboring clusters (Figure 3D).
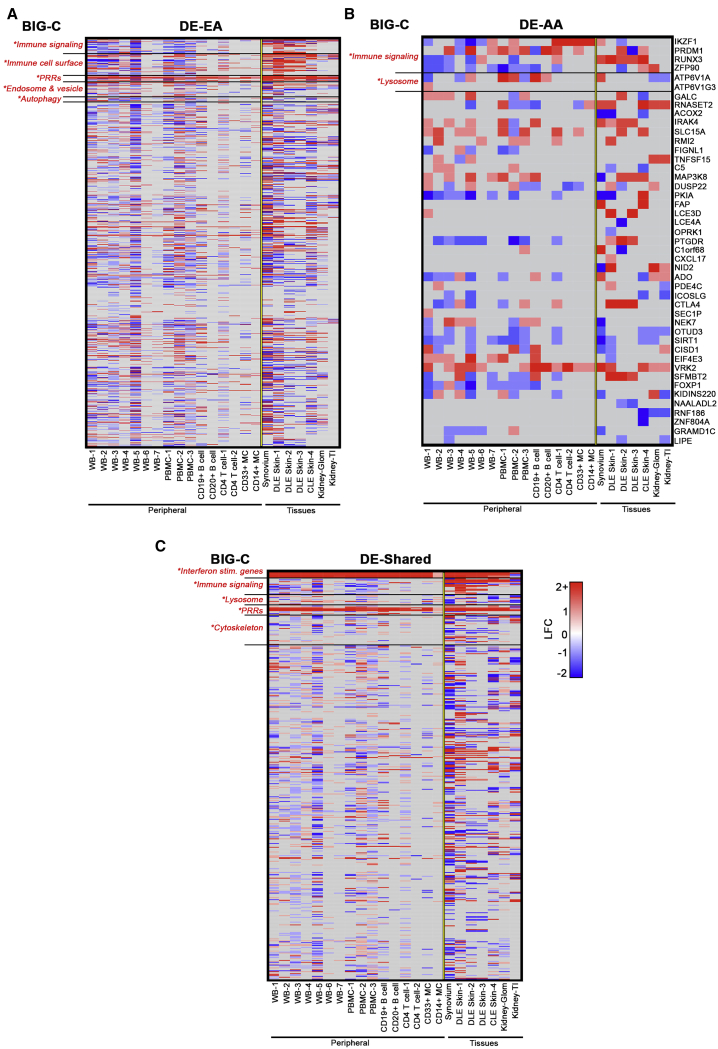
Predicted Functional Genes Are Associated with Altered Expression in SLE and Are Enriched in Differential Expression Datasets
To determine whether genes more frequently linked to specific ancestries exhibited altered expression in SLE, ancestry-driven SNP-predicted genes were matched to differentially expressed gene (DEGs) in independent SLE datasets from various tissues, including whole blood, PBMCs, B cells, T cells, synovium, skin, and kidney (Table S10). We observed that 63% of EA and 70% of AA SNP-predicted genes were identified as DEGs across all datasets (Figures 4A and 4B; Tables S10 and S11). Of the 661 genes shared between ancestries, 441 genes (66.7%) were identified as DEGs, with the interferon-stimulated genes (HERC5, IFI35, IFI44L, IFI6, IFIT1, MX1, and SPATSL2L), interferon-regulatory factors (IRF4, IRF5, and IRF7), and PRRs (OAS1, OAS2, OAS3, and SLC15A4) differentially expressed across all datasets (Figure 4C; Table S12).
Figure 4.
Comparison of EA, AA, and Shared SNP-Predicted Genes with SLE Differential Expression Datasets
SNP-predicted genes were matched with SLE differential expression (DE) data and organized by ancestry. The fold-change variation of EA, AA, and shared genes is shown. Heatmaps are organized by BIG-C category. Enriched categories indicated with an asterisk. Enrichment was defined as any category with an OR > 1 and –log10(p value) > 1.33.
Relationship of SNP-Predicted Genes and Upstream Regulators of Gene Expression Profiles
Using IPA, DEGs were used to identify potential biologic upstream regulators (UPRs) with the goal of determining whether UPRs of the altered gene expression profile in SLE were SNP-predicted genes. Overall, 141 UPRs predicted from the altered SLE gene expression profiles were also SNP-associated genes, including surface receptors, signaling molecules, cytokines, and transcription factors, many with known roles in SLE such as IRF7, ITGAM, IFNG, IKZF1, and CD40 (Table S13). In addition, 41 UPRs were transcription factors predicted to interact with transcription factor binding sites altered by an SLE-associated SNP, including MYC, EZH2, NFATC1, STAT3, STAT5a, Fos, JunB, and RelA. Thus, of the 1,238 UPRs predicted to drive SLE gene expression, 181 were SNP-predicted genes.
Delineation of Signaling Pathways Identified by Ancestry Specific SNP-Associated Genes and UPRs
Connectivity mapping of SNP-associated genes and all UPRs predicted from SLE gene expression profiles were used as input to build more complete PPI networks, and individual gene clusters were analyzed by BIG-C and IPA to identify those molecules and pathways highly associated with disease. A total of 45 pathways were representative of EA genes and UPRs, with the largest clusters (1 and 3) heavily involved in pattern recognition receptor signaling (Figures 5A and 5B; Table S14). Clusters 4 and 5 revealed enrichment in lymphocyte activation and differentiation pathways that were also common to both the AA and shared gene networks. Twenty pathways were detected only in EA, including several involved in cellular communication, cytokine signaling, and migration. The AA network was smaller (Figure 6; Table S14), with fewer SNP-predicted genes and associated UPRs, yet contained genes critical for T cell responses such as PRDM1 and IKZF1. Pathways differentially associated with AA were overwhelmingly represented by processes related to degradation and cellular stress, found in clusters 5, 3, and 11, as well as metabolic processes in cluster 6 (Figure 6B). In general, we observed good agreement between enrichment tools, with BIG-C functional categories similar or complementary to the listed canonical pathways. Discrepancies were usually the result of the wider range of pathways described by IPA utilizing a subset of genes captured by BIG-C. This was apparent in cluster 2 of the EA network (Figure 5B), where top-ranked pathways for Glucocorticoid signaling and Hepatic fibrosis include numerous genes related to the immune-secreted and immune cell surface BIG-C categories (i.e., CXCL8, CCL5, IL6, etc.). This was also the case for cluster 11 in the AA network (Figure 6B) where functional enrichment in autophagy is driven by GABARAP, a multifunctional ATG-8 subfamily member involved in both autophagosome formation as well as functioning upstream of OPRK1 (Opioid Receptor Kappa 1) within the Opioid signaling pathway.
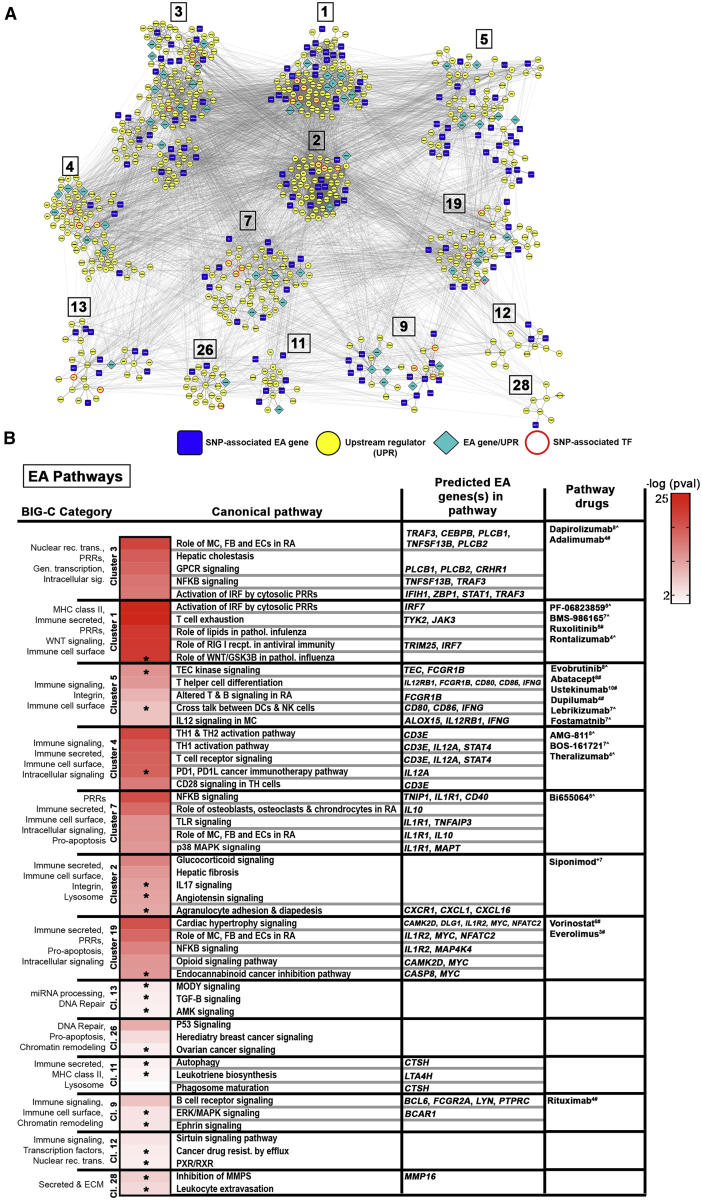
Figure 5.
Key Pathways Determined by EA Genes and Upstream Regulators
(A) Differentially expressed EA genes and their upstream regulators (UPRs) were used to create STRING-based PPI networks. DE EA genes identified as UPRs and SNP-predicted TFs are indicated. Clusters were generated via CytoScape using the MCODE plugin.
(B) Top IPA canonical pathways representing individual clusters and enriched (OR > 1, p value < 0.05) BIG-C categories are listed; heatmap depicts the –log(p value) for significant IPA pathways. Unique pathways are indicated by asterisks. Predicted EA genes and select drugs acting on direct gene targets or on any of the pathways are listed. CoLT scores (−16-+11) are in superscript; # denotes FDA-approved drugs, ˆ denotes drugs in development. SOC, standard of care.
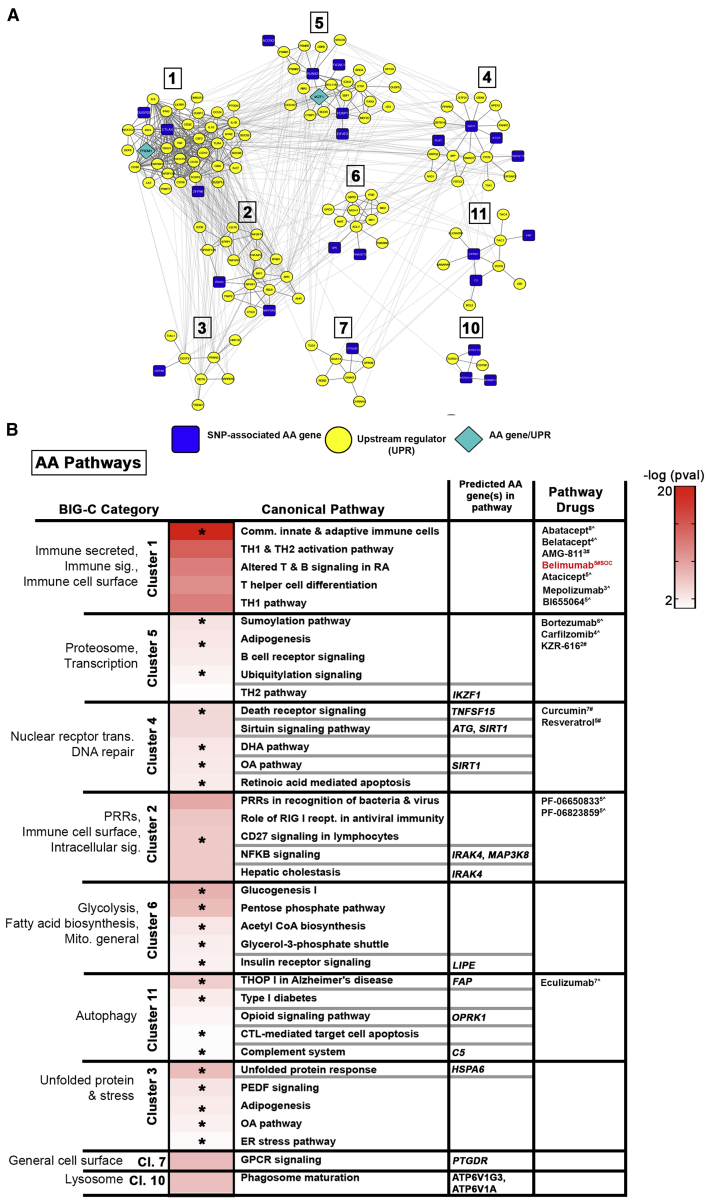
Figure 6.
Key Pathways Determined by AA Genes and Upstream Regulators
(A) Differentially expressed AA genes and their upstream regulators (UPRs) were used to create STRING-based PPI networks. DE AA genes identified as UPRs are indicated. Clusters were generated via CytoScape using the MCODE plugin.
(B) Top IPA canonical pathways representing individual clusters and enriched (OR > 1, p value < 0.05) BIG-C categories are listed; heatmap depicts the –log(p value) for significant IPA pathways. Unique pathways are indicated by asterisks. Predicted AA genes and select drugs acting on direct gene targets or on any of the pathways are listed. CoLT scores (−16 to +11) are in superscript; # denotes FDA-approved drugs; ˆ denotes drugs in development. SOC, standard of care.
Pathways that were enriched in both EA and AA (shared) included IL12 signaling and production by macrophages, TLR signaling, and activation of IRFs by cytosolic PRRs, as well as PRRs in the recognition of bacteria and virus (Figure 7). Figure 8A depicts both the unique and overlapping canonical pathways predicted by the EA and AA gene sets, whereas Figure 8B shows the broad overall pathway categories shared between EA and AA and those that are differentially enriched within each ancestral group.
Figure 7.
Key Pathways Determined by Shared Genes and Upstream Regulators
(A) Differentially expressed shared genes and their upstream regulators (UPRs) were used to create STRING-based PPI networks. DE shared genes identified as UPRs and SNP-predicted TFs are indicated. Clusters were generated via CytoScape using the MCODE plugin.
(B) Top IPA canonical pathways representing individual clusters and enriched (OR > 1, p value < 0.05) BIG-C categories are listed; heatmap depicts the –log(p value) for significant IPA pathways. Unique pathways are indicated by asterisks. Predicted shared genes and select drugs acting on direct gene targets or on any of the pathways are listed. CoLT scores (−16 to +11) are in superscript; # denotes FDA-approved drugs; ˆ denotes drugs in development. SOC, standard of care.
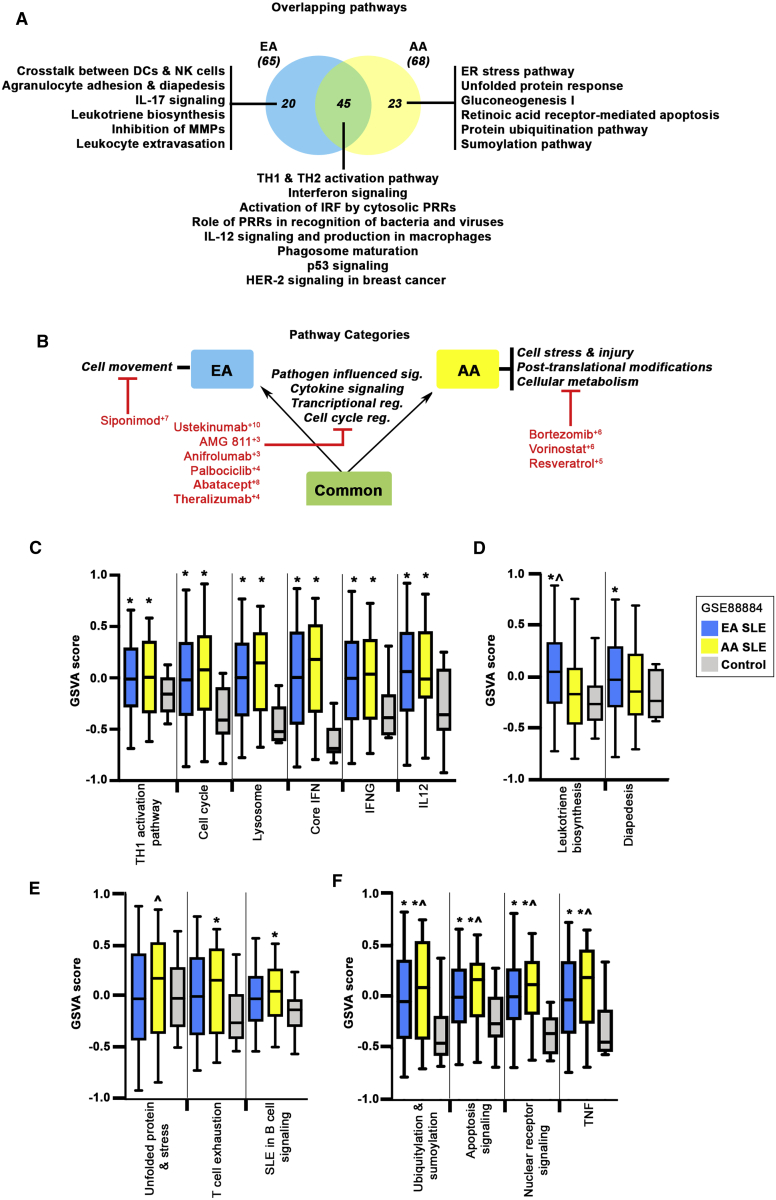
Figure 8.
Overlapping Pathways and Categories Defining the EA and AA Predicted Gene Sets
(A) Venn diagram showing the number of overlapping pathways between EA and AA predicted genes and their UPRs. Representative IPA canonical pathways are indicated.
(B) Overall pathway categories are defined; shared categories are between the arrows, EA-specific (left) and AA-specific categories (right) are indicated. Select drugs at points of intervention are noted. Superscript denotes CoLT score.
(C–F) GSVA enrichment scores were calculated for ancestry-specific and independent gene signatures in patient WB (GEO: GSE88885). GSVA signature scores (C) separating SLE-affected individuals (EA and AA) from control subjects, signature scores (D) distinguishing EA SLE-affected individuals from AA subjects and/or healthy control subjects, signature scores (E) distinguishing AA SLE-affected individuals from EA subjects or control subjects, and signature scores (F) separating SLE-affected individuals (EA and AA) from control subjects and that are additionally elevated in AA subjects compared to EA subjects. Error bars indicate 95% confidence interval. Asterisks (∗) indicate a p value < 0.05 using Welch’s t test comparing SLE to control; ˆ indicates a p value < 0.05 using Welch’s t test comparing EA to AA.
To support our pathway predictions, Gene Set Variation Analysis (GSVA)33 was applied to identify differentially enriched gene signatures in whole blood (WB) samples from SLE-affected subjects (EA and AA) and control subjects (Table S15). SNP-associated genes were used to create a collection of signatures informed by protein-protein interaction networks and IPA canonical pathways, or were previously defined as SLE associated (Table S2).30 As shown in Figure 8C (and Figure S6), enrichment of a number of pathways in both EA and AA SLE was noted, including TH1 activation pathway, cell cycle, and lysosome, as well as cytokine-based signatures for core interferon (IFN), IFNG, IL12, and the IFN subtypes IFNA2, IFNB1, and IFNW. GSVA enrichment scores using signatures for leukotriene biosynthesis and diapedesis were able to separate EA SLE-affected individuals, but not AA subjects, from healthy control subjects (Figure 8D), whereas signatures for unfolded protein response, T cell exhaustion, and B cell signaling were specifically enriched in AA SLE (Figure 8E). A number of pathways including ubiquitylation and sumoylation, apoptosis signaling, nuclear receptor signaling, and TNF separated SLE-affected individuals from control subjects with additional enrichment observed in AA (versus EA) SLE (Figure 8F).
Gene signatures for metabolic pathways, including mitochondrial oxidative phosphorylation and glycolysis, were also investigated but did not demonstrate any significant change between SLE-affected individuals and control subjects or between ancestries, although there was a trend toward enrichment in SLE-affected individuals (Figure S6). It is possible that the relatively few control samples available in this dataset may have masked subtle differences between disease and control. In contrast, GSVA scores for the PKA signaling gene signature were significantly lower in SLE-affected individuals compared to control subjects.
Pathway Analysis Facilitates Drug Prediction
Identified pathways were employed to facilitate identification of possible new therapeutic interventions and numerous drug candidates were predicted. Canonical pathways related to T cell function are shared among ancestries, as are many predicted drugs targeting T cell activity including abatacept, theralizumab, and AMG-811 (Figure 8B). Broader analysis of common pathway categories also suggested the utility of targeting T cell signaling, as well as cytokine pathways such as IL12/23 signaling with ustekinumab and/or interferon signaling with anifrolimab (Figure 8B). Drugs targeting pathways associated with EA include BMS-986165, a small molecular inhibitor of TYK2 (Figure 5B). Therapeutic candidates targeting pathways representative of AA include the proteasome inhibitor bortezomib, as well as PF-06650833, an IRAK4-specific inhibitor (Figure 6B). Unique pathway categories identified for EA and AA suggest additional ancestry-driven interventions, such as the small molecule inhibitor of sphingosine-1-phosphate receptor 1 (S1PR1) siponimod for EA (prevents leukocyte egress) and the HDAC inhibitor vorinostat for AA SLE (Figure 8B).
Discussion
SLE is a chronic autoimmune disease with a strong genetic component.38 Although socioeconomic disparities are also known to impact disease development,39,40 genetic heterogeneity between ancestral populations is widely acknowledged to be important in SLE risk, since affected individuals of African descent have a higher prevalence of lupus and experience the disease more severely than those of European ancestry.7,8 Despite improved understanding of how inherited genetic variation impacts disease risk, genetic analyses to date have failed to provide a clear path toward novel therapeutic development. This is of particular concern with respect to AA populations where the control of disease activity remains suboptimal.10, 11, 12 Furthermore, most GWASs only consider the most significant variants (lead SNP, marker with the lowest p value), typically following-up on those that are novel.2 Here, we propose a novel strategy employing integration of all SLE SNP association-predicted genes into functional pathways to identify the genetic contributions to SLE disease pathogenesis and possible differences contributed by ancestry. To accomplish this, we employed statistical and computational analyses along with data acquired from functional genomic assays and differential gene expression studies to map the global gene expression landscape of SLE and further define the disease-associated pathways responsible for the inherent disparities influencing SLE progression.
It is important to reiterate that the SNPs examined here are generally present in both ancestral populations and are associated with a significant cohort of common genes. However, differences in allele frequencies suggest that some SNP associations and pathways may be more representative of one ancestry over another. Here, eQTL analysis identified 247 tagging SNPs associated with 759 E-Genes (77 EA, 21 AA, 523 shared). Given that the majority of eQTLs identified here map to multiple E-Genes (many within the same functional network), eQTL-based gene prediction may be particularly valuable for network modeling and disease analysis. Recent studies have also shown that disease-susceptibility variants frequently lie in regulatory enhancer elements.41 However, only 20% (157) of SNPs analyzed here were located in known regulatory regions. While other methodologies, such as promoter capture Hi-C, link regulatory SNPs to putative target genes,42 we were particularly interested in predicting the effect of variants in non-coding regions, especially since the majority of SNPs examined here were intergenic. Using computational gene prediction algorithms that incorporate chromatin interaction data and intergenic enhancer annotation from several hundred cell lines,21 additional regulatory SNPs were identified that were predicted to change transcription factor binding and were associated with 627 downstream targets (T-Genes; 472 EA, 9 AA, 143 shared). Although some regulatory SNPs also exhibit eQTL effects, we nonetheless uncovered 496 unique T-Genes enriched in a diverse array of functional categories. Finally, we identified 23 variants resulting in nonsense or non-synonymous amino acid changes affecting 22 genes (C-Genes), 12 of which were predicted to negatively impact protein function. The remaining 587 risk SNPs were mapped to the nearest, most proximal gene, resulting in 520 P-Genes (465 EA, 34 AA, 21 shared).
One major limitation to the current study is that all computational and experimental approaches outlined here are inferential; by attempting to provide a more comprehensive translation of GWAS findings, a major challenge remains in determining those genes that are causative. To address this, PPI networks and clustering based on interaction strength helped exclude those genes lacking strong connections to molecules within or between similarly functioning clusters. Compared to SNP-predicted E-, T-, C-, and P-Genes where we observed large, highly connected clusters, randomly generated genes generally formed smaller clusters, exhibited fewer intra- and inter-cluster connections, and ultimately appeared as independent entities. In addition, SNP-predicted genes were compared to SLE datasets (SLE versus control) to determine those genes that were differentially expressed in active disease. This was important to amplify the GTEx predictions that were based on data from a collection of cell lines and not primary SLE cells.19 We observed a high percentage of SNP-predicted genes differentially expressed across all datasets, although expression was not always in the same direction (i.e., all upregulated or all downregulated). In EA, for example, functionally enriched categories included immune signaling and immune cell surface whereas DE genes falling into these categories were more upregulated in the tissues (synovium, skin, and kidney) and predominantly downregulated in the periphery (WB, PBMC, T cells, B cells, and myeloid cells). This could reflect a cellular response to different inflammatory stimuli, such as the enhanced recruitment of myeloid cells to tissue in response to immune complex deposition. All DE genes were then used as input into IPA to generate upstream and downstream regulators and combined for further network and clustering analysis. This allowed us to identify biologically relevant pathways unique to each ancestry, a strategy that revealed essential differences between EA and AA SLE, as well as many pathways that were shared.
A second caveat to the current study is the use of the Immunochip which was constructed to cover multiple major autoimmune diseases and enable the identification of top-ranked SNPs associated with disease.37 This platform was designed for use in EA populations and is therefore less informative for other ancestral groups, especially in non-HLA-associated regions. Furthermore, as chip coverage was confined to autoimmune and inflammatory diseases, SNPs affecting non-immune-related processes could be under-represented.
Despite these drawbacks, the pathway-based analysis of predicted genes and their upstream regulators presented here helps clarify the complex polygenic risk associated with SLE in multiple ancestries. We observed key dysregulated EA pathways centered around innate immune function and the response to inflammation, including cell movement, cytokine signaling, and cell-cell communication. Remarkably, GSVA gene signatures for leukotriene biosynthesis and diapedesis were sufficiently selective to separate EA SLE-affected individuals from control subjects, providing additional evidence for these pathways in SLE pathogenesis. Furthermore, SNP-predicted EA genes were enriched in myeloid and NK cell signatures, along with T and B cell signatures, findings that are consistent with previous reports showing increased myeloid lineage cell modules in EA subjects.43,44
Pathways identified by AA-associated SNPs included those linked to protein degradation, such as the sumoylation pathway, ubiquitylation signaling, the ER stress pathway, unfolded protein response, osteoarthritis pathway (cell stress), and the neuroprotective role of THOP1 in Alzheimer disease, a pathway involved in the presentation of antigen generated by the proteasome. To ensure that the differences in ancestrally related genes were not related to imbalances in the number of SNPs assessed, we separately looked only at the most highly associated genes in AA and EA populations and still were able to detect ancestry-related pathway differences. The importance of these pathways was confirmed by GSVA which demonstrated the unique enrichment of cellular stress mechanisms and B cell signaling in AA SLE-affected indviduals. These observations are in line with reports showing increased B cell activation and plasma cells in AA case subjects.43, 44, 45 In fact, a recent comprehensive analysis of gene expression profiling in whole blood44 showed that a major AA ancestral difference is related to the propensity to form autoantibodies to RNP and Sm and to have more types of autoantibodies in general. With these results together with the data presented in the current study demonstrating an underlying genetic predisposition toward B cell-driven disease, dysregulated B cell responses in AA case subjects likely contribute to the overall higher prevalence of tissue damage in this ancestral population. The increase in B cell-driven responses in AA subjects manifested by an increase in circulating B lineage cells was determined by the use of I-Scope, a validated analytic tool that converts gene expression profiles from bulk mRNA into the most likely cellular constituents.44 Finally, it is increasingly recognized that ER stress and the UPR signaling pathway in dysregulated immune responses is closely tied to aberrant B cell activity in SLE.46,47
Gene signatures representing cellular processes shared between ancestries provide further validation for our comprehensive pathway analysis. GSVA enrichment scores for the interferon response (IFN core, IFNA2, IFNB1, IFNW1, and IFNG) and inflammatory cytokines (IL-12 and TNF) exhibited the greatest difference between SLE and control, independent of ancestry. In line with this, work by Catalina et al.30 showed that multiple IFN signatures are operative in an array of SLE subject samples from whole blood and tissues. Despite the report that metabolic abnormalities, including heightened glycolysis and mitochondrial glucose oxidation,48 are associated with SLE, metabolic gene signatures for OXPHOS and glycolysis did not distinguish SLE-affected individuals from control subjects. However, examination of protein kinase A (PKA) signaling, a pathway that participates in the regulation of immune effector functions in T cells,49 demonstrated significantly lower GSVA scores for the PKA signaling signature in both EA and AA SLE-affected individuals compared to control subjects. Consistent with these findings, previous reports have shown that T cells from SLE-affected individuals have a metabolic disorder of the PKA pathway characterized by markedly diminished PKA activity and dysfunctional T cell activity.50,51
By focusing on pathways instead of individual genes, the current approach identified “actionable” points of therapeutic intervention with the potential to impact EA and AA SLE specifically. Thus, EA subjects may derive particular benefit from treatments that prevent leukocyte or lymphocyte infiltration into tissues, highlighting drugs that modulate this process. For example, sphingosine-1 phosphate receptor (S1PR) is a pleiotropic lipid mediator involved in the regulation of many cellular functions, including proliferation, survival, and cell motility.52,53 Siponimod, an FDA-approved treatment for multiple sclerosis, promotes the internalization of S1PR expressed on lymphocytes preventing cell migration to sites of inflammation.54, 55, 56 Given its high combined lupus treatment score (CoLTS),34,35,57 siponimod represents a high-priority small-molecule drug with potential for repurposing in SLE.
Similarly, given the enrichment of pathways linked to proteasomal and degradative processes in AA, therapeutic intervention may include proteasome inhibitors, such as bortezomib (BZ), an FDA-approved drug for mantle cell lymphoma and multiple myeloma,58 as well as B cell-targeting therapies. Despite reports of adverse events, BZ was claimed to have efficacy in treating refractory SLE in a small, uncontrolled clinical trial and was shown to deplete bone marrow and peripheral plasma cells.59 In addition, AA SLE-affected individuals showed a better response to rituximab (RTX), an anti-CD20 inhibitor,60 and a trend toward better response in AA subjects with lupus nephritis.61 However, the anti-BAFF inhibitor belimumab (BEL) failed to demonstrate a positive effect among AA subjects. The reasons for this discrepancy are unclear but may be related to the fact that anti-CD20 therapies like RTX induce a broad and deep B cell depletion, whereas BEL has a significantly more restricted and/or attenuated B cell effect.62,63 Together, these data indicate the possibility that BZ (and/or more selective immunoproteasome inhibitors), and therapies broadly targeting B cell may hold promise for AA subjects who respond poorly to conventional therapies.
In conclusion, our study demonstrates that multilevel analysis is capable of defining gene regulatory pathways which not only reflect differences in EA and AA populations but also represents candidate pathways that may be the target of ancestry-specific therapies. Indeed, the ancestral SNP-associated predicted genes and gene expression profiles outlined here illustrate fundamental differences in lupus molecular pathways between ancestries and indicate that unique sets of drugs may be particularly effective at treating lupus within each ancestral group.
Data and Code Availability
All microarray datasets listed in this publication are available on the NCBI’s database Gene Expression Omnibus (GEO).
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The work presented in this manuscript was funded by a grant awarded to P.E.L. and A.C.G. of the RILITE Research Institute by the John and Marcia Goldman Foundation. The funder provided support in the form of salaries for authors (K.A.O., A.P., B.N.A., P.B., M.D.C., J.M.D., K.M.K., A.C.L., R.D.R.). The authors gratefully acknowledge additional support from the Lupus Research Alliance (LRA) and the Department of the Army (W81XWH-20-1-0686).
Published: October 7, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.09.007.
Web Resources
CytoScape, https://cytoscape.org
DAVID, https://david.ncifcrf.gov/
Ensembl, http://www.ensembl.org
GeneHancer, https://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=geneHancer
GeneCards, https://www.genecards.org
Genotype-Tissue Expression (GTEx) project portal, http://www.gtexportal.org
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
Human Active Enhancer to Interpret Regulatory variants (HACER), http://bioinfo.vanderbilt.edu/AE/HACER
HaploReg, https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php
InteractiVenn, http://www.interactivenn.net
Ingenuity Pathway Analysis (IPA), https://www.qiagenbioinformatics.com
John and Marcia Goldman Foundation, jmgoldmanfoundation.org
Library of Integrated Network-based Cellular Signatures (LINCS), http://www.lincsproject.org/
Lupus Research Alliance, lupusresearch.org
OMIM, https://www.omim.org/
Protein Analysis Through Evolutionary Relationships (PANTHER), http://www.pantherdb.org
Polymorphisms Phenotyping v2 (PolyPhen-2), genetics.bwh.harvard.edu
PROVEAN, http://provean.jcvi.org
rAggr (site no longer live; please use SNiPA instead), http://raggr.usc.edu
SNiPA, https://snipa.helmholtz-muenchen.de/snipa3/index.php?task=proxy_search
Sorting Intolerant From Tolerant (SIFT-4G), http://sift-dna.org/sift4g
STITCH, http://stitch.embl.de
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), https://string-db.org
Variant Effect Predictor (VEP), http://www.ensembl.org
Supplemental Information
References
- 1.Rullo O.J., Tsao B.P. Recent insights into the genetic basis of systemic lupus erythematosus. Ann. Rheum. Dis. 2013;72(Suppl 2):ii56–ii61. doi: 10.1136/annrheumdis-2012-202351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris D.L., Sheng Y., Zhang Y., Wang Y.F., Zhu Z., Tombleson P., Chen L., Cunninghame Graham D.S., Bentham J., Roberts A.L. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat. Genet. 2016;48:940–946. doi: 10.1038/ng.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lessard C.J., Sajuthi S., Zhao J., Kim K., Ice J.A., Li H., Ainsworth H., Rasmussen A., Kelly J.A., Marion M. Identification of a Systemic Lupus Erythematosus Risk Locus Spanning ATG16L2, FCHSD2, and P2RY2 in Koreans. Arthritis Rheumatol. 2016;68:1197–1209. doi: 10.1002/art.39548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alarcón-Riquelme M.E., Ziegler J.T., Molineros J., Howard T.D., Moreno-Estrada A., Sánchez-Rodríguez E., Ainsworth H.C., Ortiz-Tello P., Comeau M.E., Rasmussen A. Genome-Wide Association Study in an Amerindian Ancestry Population Reveals Novel Systemic Lupus Erythematosus Risk Loci and the Role of European Admixture. Arthritis Rheumatol. 2016;68:932–943. doi: 10.1002/art.39504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanscombe K.B., Morris D.L., Noble J.A., Dilthey A.T., Tombleson P., Kaufman K.M., Comeau M., Langefeld C.D., Alarcon-Riquelme M.E., Gaffney P.M. Genetic fine mapping of systemic lupus erythematosus MHC associations in Europeans and African Americans. Hum. Mol. Genet. 2018;27:3813–3824. doi: 10.1093/hmg/ddy280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams E.M., Bruner L., Adkins A., Vrana C., Logan A., Kamen D., Oates J.C. I too, am America: a review of research on systemic lupus erythematosus in African-Americans. Lupus Sci. Med. 2016;3:e000144. doi: 10.1136/lupus-2015-000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnado A., Carroll R.J., Casey C., Wheless L., Denny J.C., Crofford L.J. Phenome-wide association study identifies marked increased in burden of comorbidities in African Americans with systemic lupus erythematosus. Arthritis Res. Ther. 2018;20:69. doi: 10.1186/s13075-018-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goulielmos G.N., Zervou M.I., Vazgiourakis V.M., Ghodke-Puranik Y., Garyfallos A., Niewold T.B. The genetics and molecular pathogenesis of systemic lupus erythematosus (SLE) in populations of different ancestry. Gene. 2018;668:59–72. doi: 10.1016/j.gene.2018.05.041. [DOI] [PubMed] [Google Scholar]
- 10.Navarra S.V., Guzmán R.M., Gallacher A.E., Hall S., Levy R.A., Jimenez R.E., Li E.K., Thomas M., Kim H.Y., León M.G., BLISS-52 Study Group Efficacy and safety of belimumab in patients with active systemic lupus erythematosus: a randomised, placebo-controlled, phase 3 trial. Lancet. 2011;377:721–731. doi: 10.1016/S0140-6736(10)61354-2. [DOI] [PubMed] [Google Scholar]
- 11.Furie R., Petri M., Zamani O., Cervera R., Wallace D.J., Tegzová D., Sanchez-Guerrero J., Schwarting A., Merrill J.T., Chatham W.W., BLISS-76 Study Group A phase III, randomized, placebo-controlled study of belimumab, a monoclonal antibody that inhibits B lymphocyte stimulator, in patients with systemic lupus erythematosus. Arthritis Rheum. 2011;63:3918–3930. doi: 10.1002/art.30613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamore R., 3rd, Parmar S., Patel K., Hilas O. Belimumab (benlysta): a breakthrough therapy for systemic lupus erythematosus. P&T. 2012;37:212–226. [PMC free article] [PubMed] [Google Scholar]
- 13.Stohl W. Inhibition of B cell activating factor (BAFF) in the management of systemic lupus erythematosus (SLE) Expert Rev. Clin. Immunol. 2017;13:623–633. doi: 10.1080/1744666X.2017.1291343. [DOI] [PubMed] [Google Scholar]
- 14.Fike A.J., Elcheva I., Rahman Z.S.M. The Post-GWAS Era: How to Validate the Contribution of Gene Variants in Lupus. Curr. Rheumatol. Rep. 2019;21:3. doi: 10.1007/s11926-019-0801-5. [DOI] [PubMed] [Google Scholar]
- 15.Schadt E.E., Monks S.A., Drake T.A., Lusis A.J., Che N., Colinayo V., Ruff T.G., Milligan S.B., Lamb J.R., Cavet G. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 16.Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 17.Stranger B.E., Montgomery S.B., Dimas A.S., Parts L., Stegle O., Ingle C.E., Sekowska M., Smith G.D., Evans D., Gutierrez-Arcelus M. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 2012;8:e1002639. doi: 10.1371/journal.pgen.1002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langefeld C.D., Ainsworth H.C., Cunninghame Graham D.S., Kelly J.A., Comeau M.E., Marion M.C., Howard T.D., Ramos P.S., Croker J.A., Morris D.L. Transancestral mapping and genetic load in systemic lupus erythematosus. Nat. Commun. 2017;8:16021. doi: 10.1038/ncomms16021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra H.J., Peters M.J., Esko T., Yaghootkar H., Schurmann C., Kettunen J., Christiansen M.W., Fairfax B.P., Schramm K., Powell J.E. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J., Dai X., Berry L.D., Cogan J.D., Liu Q., Shyr Y. HACER: an atlas of human active enhancers to interpret regulatory variants. Nucleic Acids Res. 2019;47(D1):D106–D112. doi: 10.1093/nar/gky864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017 doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 24.Sim N.L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40(Web Server issue, W1):W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;Chapter 7(SUPPL.76):20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi H., Huang X., Muruganujan A., Tang H., Mills C., Kang D., Thomas P.D. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catalina M.D., Bachali P., Geraci N.S., Grammer A.C., Lipsky P.E. Gene expression analysis delineates the potential roles of multiple interferons in systemic lupus erythematosus. Commun. Biol. 2019;2:140. doi: 10.1038/s42003-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Catalina M.D., Owen K.A., Labonte A.C., Grammer A.C., Lipsky P.E. The pathogenesis of systemic lupus erythematosus: Harnessing big data to understand the molecular basis of lupus. J. Autoimmun. 2020;110:102359. doi: 10.1016/j.jaut.2019.102359. [DOI] [PubMed] [Google Scholar]
- 32.Ren J., Catalina M.D., Eden K., Liao X., Read K.A., Luo X., McMillan R.P., Hulver M.W., Jarpe M., Bachali P. Selective Histone Deacetylase 6 Inhibition Normalizes B Cell Activation and Germinal Center Formation in a Model of Systemic Lupus Erythematosus. Front. Immunol. 2019;10:2512. doi: 10.3389/fimmu.2019.02512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Labonte A.C., Kegerreis B., Geraci N.S., Bachali P., Madamanchi S., Robl R., Catalina M.D., Lipsky P.E., Grammer A.C. Identification of alterations in macrophage activation associated with disease activity in systemic lupus erythematosus. PLoS ONE. 2018;13:e0208132. doi: 10.1371/journal.pone.0208132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grammer A.C., Ryals M.M., Heuer S.E., Robl R.D., Madamanchi S., Davis L.S., Lauwerys B., Catalina M.D., Lipsky P.E. Drug repositioning in SLE: crowd-sourcing, literature-mining and Big Data analysis. Lupus. 2016;25:1150–1170. doi: 10.1177/0961203316657437. [DOI] [PubMed] [Google Scholar]
- 36.Chorley B.N., Wang X., Campbell M.R., Pittman G.S., Noureddine M.A., Bell D.A. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutat. Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes A., Brown M.A. Promise and pitfalls of the Immunochip. Arthritis Res. Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alarcón-Segovia D., Alarcón-Riquelme M.E., Cardiel M.H., Caeiro F., Massardo L., Villa A.R., Pons-Estel B.A., Grupo Latinoamericano de Estudio del Lupus Eritematoso (GLADEL) Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 39.Barber M.R.W., Clarke A.E. Socioeconomic consequences of systemic lupus erythematosus. Curr. Opin. Rheumatol. 2017;29:480–485. doi: 10.1097/BOR.0000000000000416. [DOI] [PubMed] [Google Scholar]
- 40.Carter E.E., Barr S.G., Clarke A.E. The global burden of SLE: prevalence, health disparities and socioeconomic impact. Nat. Rev. Rheumatol. 2016;12:605–620. doi: 10.1038/nrrheum.2016.137. [DOI] [PubMed] [Google Scholar]
- 41.van Arensbergen J., Pagie L., FitzPatrick V.D., de Haas M., Baltissen M.P., Comoglio F., van der Weide R.H., Teunissen H., Võsa U., Franke L. High-throughput identification of human SNPs affecting regulatory element activity. Nat. Genet. 2019;51:1160–1169. doi: 10.1038/s41588-019-0455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Javierre B.M., Burren O.S., Wilder S.P., Kreuzhuber R., Hill S.M., Sewitz S., Cairns J., Wingett S.W., Várnai C., Thiecke M.J., BLUEPRINT Consortium Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016;167:1369–1384.e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banchereau R., Hong S., Cantarel B., Baldwin N., Baisch J., Edens M., Cepika A.M., Acs P., Turner J., Anguiano E. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 2016;165:1548–1550. doi: 10.1016/j.cell.2016.05.057. [DOI] [PubMed] [Google Scholar]
- 44.Catalina M., Bachali P., Yeo A., Geraci N., Petri M., Grammer A., Lipsky P. The Influence of Ancestry, Serology and Treatment on the Transcriptomic Fingerprint of Systemic Lupus Erythematosus. JCI Insight. 2020;5:e140380. doi: 10.1172/jci.insight.140380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menard L.C., Habte S., Gonsiorek W., Lee D., Banas D., Holloway D.A., Manjarrez-Orduno N., Cunningham M., Stetsko D., Casano F. B cells from African American lupus patients exhibit an activated phenotype. JCI Insight. 2016;1:e87310. doi: 10.1172/jci.insight.87310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navid F., Colbert R.A. Causes and consequences of endoplasmic reticulum stress in rheumatic disease. Nat. Rev. Rheumatol. 2017;13:25–40. doi: 10.1038/nrrheum.2016.192. [DOI] [PubMed] [Google Scholar]
- 47.Lam W.Y., Bhattacharya D. Metabolic Links between Plasma Cell Survival, Secretion, and Stress. Trends Immunol. 2018;39:19–27. doi: 10.1016/j.it.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lightfoot Y.L., Blanco L.P., Kaplan M.J. Metabolic abnormalities and oxidative stress in lupus. Curr. Opin. Rheumatol. 2017;29:442–449. doi: 10.1097/BOR.0000000000000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wehbi V.L., Taskén K. Molecular mechanisms for cAMP-mediated immunoregulation in T cells - role of anchored protein kinase a signaling units. Front. Immunol. 2016;7:222. doi: 10.3389/fimmu.2016.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kammer G.M., Khan I.U., Malemud C.J. Deficient type I protein kinase A isozyme activity in systemic lupus erythematosus T lymphocytes. J. Clin. Invest. 1994;94:422–430. doi: 10.1172/JCI117340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kammer G.M. Deficient protein kinase a in systemic lupus erythematosus: a disorder of T lymphocyte signal transduction. Ann. N Y Acad. Sci. 2002;968:96–105. doi: 10.1111/j.1749-6632.2002.tb04329.x. [DOI] [PubMed] [Google Scholar]
- 52.Cartier A., Hla T. Sphingosine 1-phosphate: Lipid signaling in pathology and therapy. Science. 2019;366:eaar5551. doi: 10.1126/science.aar5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aoki M., Aoki H., Ramanathan R., Hait N.C., Takabe K. Sphingosine-1-Phosphate Signaling in Immune Cells and Inflammation: Roles and Therapeutic Potential. Mediators Inflamm. 2016;2016:8606878. doi: 10.1155/2016/8606878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kappos L., Bar-Or A., Cree B.A.C., Fox R.J., Giovannoni G., Gold R., Vermersch P., Arnold D.L., Arnould S., Scherz T., EXPAND Clinical Investigators Siponimod versus placebo in secondary progressive multiple sclerosis (EXPAND): a double-blind, randomised, phase 3 study. Lancet. 2018;391:1263–1273. doi: 10.1016/S0140-6736(18)30475-6. [DOI] [PubMed] [Google Scholar]
- 55.Faissner S., Gold R. Progressive multiple sclerosis: latest therapeutic developments and future directions. Ther. Adv. Neurol. Disorder. 2019;12 doi: 10.1177/1756286419878323. 1756286419878323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gajofatto A. Spotlight on siponimod and its potential in the treatment of secondary progressive multiple sclerosis: the evidence to date. Drug Des. Devel. Ther. 2017;11:3153–3157. doi: 10.2147/DDDT.S122249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kingsmore K., Grammer A., Lipsky P. Drug repurposing to improve treatment of rheumatic autoimmune inflammatory diseases. Nature Review Rheumatology. 2020;16:32–52. doi: 10.1038/s41584-019-0337-0. [DOI] [PubMed] [Google Scholar]
- 58.Robak P., Robak T. Bortezomib for the Treatment of Hematologic Malignancies: 15 Years Later. Drugs R D. 2019;19:73–92. doi: 10.1007/s40268-019-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander T., Sarfert R., Klotsche J., Kühl A.A., Rubbert-Roth A., Lorenz H.M., Rech J., Hoyer B.F., Cheng Q., Waka A. The proteasome inhibitior bortezomib depletes plasma cells and ameliorates clinical manifestations of refractory systemic lupus erythematosus. Ann. Rheum. Dis. 2015;74:1474–1478. doi: 10.1136/annrheumdis-2014-206016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrill J.T., Neuwelt C.M., Wallace D.J., Shanahan J.C., Latinis K.M., Oates J.C., Utset T.O., Gordon C., Isenberg D.A., Hsieh H.J. Efficacy and safety of rituximab in moderately-to-severely active systemic lupus erythematosus: the randomized, double-blind, phase II/III systemic lupus erythematosus evaluation of rituximab trial. Arthritis Rheum. 2010;62:222–233. doi: 10.1002/art.27233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rovin B.H., Furie R., Latinis K., Looney R.J., Fervenza F.C., Sanchez-Guerrero J., Maciuca R., Zhang D., Garg J.P., Brunetta P., Appel G., LUNAR Investigator Group Efficacy and safety of rituximab in patients with active proliferative lupus nephritis: the Lupus Nephritis Assessment with Rituximab study. Arthritis Rheum. 2012;64:1215–1226. doi: 10.1002/art.34359. [DOI] [PubMed] [Google Scholar]
- 62.Wallace D.J., Stohl W., Furie R.A., Lisse J.R., McKay J.D., Merrill J.T., Petri M.A., Ginzler E.M., Chatham W.W., McCune W.J. A phase II, randomized, double-blind, placebo-controlled, dose-ranging study of belimumab in patients with active systemic lupus erythematosus. Arthritis Rheum. 2009;61:1168–1178. doi: 10.1002/art.24699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobi A.M., Huang W., Wang T., Freimuth W., Sanz I., Furie R., Mackay M., Aranow C., Diamond B., Davidson A. Effect of long-term belimumab treatment on B cells in systemic lupus erythematosus: extension of a phase II, double-blind, placebo-controlled, dose-ranging study. Arthritis Rheum. 2010;62:201–210. doi: 10.1002/art.27189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microarray datasets listed in this publication are available on the NCBI’s database Gene Expression Omnibus (GEO).