Summary
Previous research has shown that polygenic risk scores (PRSs) can be used to stratify women according to their risk of developing primary invasive breast cancer. This study aimed to evaluate the association between a recently validated PRS of 313 germline variants (PRS313) and contralateral breast cancer (CBC) risk. We included 56,068 women of European ancestry diagnosed with first invasive breast cancer from 1990 onward with follow-up from the Breast Cancer Association Consortium. Metachronous CBC risk (N = 1,027) according to the distribution of PRS313 was quantified using Cox regression analyses. We assessed PRS313 interaction with age at first diagnosis, family history, morphology, ER status, PR status, and HER2 status, and (neo)adjuvant therapy. In studies of Asian women, with limited follow-up, CBC risk associated with PRS313 was assessed using logistic regression for 340 women with CBC compared with 12,133 women with unilateral breast cancer. Higher PRS313 was associated with increased CBC risk: hazard ratio per standard deviation (SD) = 1.25 (95%CI = 1.18–1.33) for Europeans, and an OR per SD = 1.15 (95%CI = 1.02–1.29) for Asians. The absolute lifetime risks of CBC, accounting for death as competing risk, were 12.4% for European women at the 10th percentile and 20.5% at the 90th percentile of PRS313. We found no evidence of confounding by or interaction with individual characteristics, characteristics of the primary tumor, or treatment. The C-index for the PRS313 alone was 0.563 (95%CI = 0.547–0.586). In conclusion, PRS313 is an independent factor associated with CBC risk and can be incorporated into CBC risk prediction models to help improve stratification and optimize surveillance and treatment strategies.
Keywords: polygenic risk score, contralateral breast cancer, epidemiology, genetic
Introduction
Due to the high incidence of breast cancer and improving survival, an increasing number of breast cancer survivors are at risk of developing contralateral breast cancer (CBC). The 10-year cumulative incidence of CBC is ∼4%,1,2 but estimates vary widely depending on factors such as germline genetics, family history, and (neo)adjuvant systemic therapy for the first breast cancer.3 The risk of developing CBC is particularly high in women with rare mutations in certain genes including BRCA1, BRCA2, and CHEK2, with approximately 2- to 4-fold higher risks reported compared with women without these mutations.3
Recently, genome-wide association studies (GWASs) have identified multiple common germline variants that are associated with first primary breast cancer risk.4,5 These are associated with small differences in risk individually, but their combined effects can be summarized in a polygenic risk score (PRS), which has been shown to stratify women according to their risk of developing breast cancer.6, 7, 8, 9 Using a large GWAS dataset from the Breast Cancer Association Consortium (BCAC), we previously developed and validated a 313-variant PRS (PRS313) among women of European descent. In independent prospective studies, this PRS313 predicted the risk of primary invasive breast cancer with an odds ratio (OR) per standard deviation (SD) of 1.61 (95% confidence interval (95%CI) = 1.57–1.65).7 The PRS313 has also been externally validated using the UK Biobank cohort.
The aim of the current study was to evaluate the association between PRS313 and CBC risk, using data from BCAC. Other studies have shown associations between risk of CBC and both a 67-variant PRS10 and individual variants,11 but not yet with PRS313, the most extensively validated PRS. Further, the dataset currently evaluated is larger than those previously tested. We carried out two types of analyses. We conducted a cohort study among studies of European ancestry women with follow-up data available and performed Cox regression analyses to estimate hazard ratios (HRs) for CBC. Potential confounding and interaction with characteristics of the individual, characteristics of the primary tumor, or treatment were tested. In addition, to directly compare with the OR reported for PRS313 and first breast cancer, we selected case-case series and performed logistic regression analyses comparing the PRS313 distribution in women with CBC versus those with unilateral breast cancer. These analyses were conducted separately in European and Asian women (follow-up was too limited to perform a cohort study for the Asian population). Use of PRS313 may lead to more accurate CBC risk prediction to support decision making for women who may or may not benefit from additional surveillance and risk-reducing treatment strategies.
Material and Methods
Study Subjects
Case-Case Series
We selected women who were diagnosed with breast cancer and women without any diagnosis of breast cancer from the BCAC including all women of European ancestry, based on genotyping data, and selecting only those studies which reported on CBC (62 studies) (Figure S1A, Table S1 and S2). BCAC database version freeze 12 was used. All women diagnosed with invasive breast cancer as a first cancer were included in the analysis; the small number of tumors with unknown invasiveness were considered invasive (Table S2). In the case-case series, a CBC was defined as a breast cancer (in situ or invasive) in the contralateral breast irrespective of the time since the first breast cancer. The case-case series comprised 81,000 women with unilateral breast cancer, 3,607 women with CBC, and 62,830 women without any diagnosis of breast cancer (Figure S1A). We also compared women with unilateral breast cancer to women without any diagnosis of breast cancer to reproduce the estimate that was previously reported for first breast cancer risk7 in our study selection.
We selected for a separate analysis women of Asian ancestry from the BCAC data, comprising 12,133 women with unilateral breast cancer, 340 women with CBC, and 13,398 women without any diagnosis of breast cancer from eight studies (Figure S1B, Table S2).
European Cohort
In the European cohort, we used metachronous CBC as the outcome, defined as a breast cancer in the contralateral breast (in situ or invasive) diagnosed at least 3 months after the first breast cancer. We used a cut-off of 3 months to reduce the likelihood that these CBCs represent metastases rather than true second primary tumors. We selected all women diagnosed with breast cancer from the European case-case series and excluded four studies that did not provide follow-up information on vital status (Figure S1A). We did not include Asian women since follow-up was too limited in these studies. We additionally excluded 6,207 women with no follow-up and 2,208 women who developed synchronous CBC, distant metastasis, or who died or were last known to be alive within 3 months after the first breast cancer diagnosis. Since BCAC also included prevalent cases, we excluded 3,796 women who developed CBC or were censored before study entry. The case-case series included women diagnosed between 1947 and 2018. In the European cohort, we excluded 2,235 women who were diagnosed with their first breast cancer before 1990 or who had missing year of first diagnosis. We restricted to women diagnosed from 1990 onward so that diagnostic procedures and treatment would be more representative of current practice. Moreover, clinico-pathological, treatment, and follow-up data were more complete after 1990. In addition, we excluded 16 studies (9,783 women) without information about metachronous CBC events (Figure S1A). After these exclusions, the cohort for this analysis comprised data from 42 studies, including 56,068 women with invasive breast cancer among whom 1,027 metachronous CBC occurred (Table S2).
All individuals provided written informed consent, and all studies were approved by the relevant institutional review boards. BCAC data were centrally harmonized and cleaned in communication with the study data managers and principal investigators. Data collection for individual studies is described in Table S1.
Genotyping and PRSs
DNA samples from participants were genotyped using the iCOGS array12,13 or the OncoArray,4,14 with genotypes for variants not on the arrays estimated by imputation.4,13 The PRS313 was calculated as a weighted sum of the minor allele dosages; the variant selection and weights are as given by Mavaddat et al.7 We also calculated estimates for a previously published PRS776 and for estrogen receptor (ER)-specific PRSs (ER-positive PRS313 and ER-negative PRS313).7 The ER-specific PRSs were constructed by defining subtype-specific weights for the 313 variants using a hybrid approach.7 Variants and corresponding coefficients used to construct the PRS are shown in Table S3. We standardized the PRS in our analyses by dividing it by the SD of the PRS of the control subjects (PRS77 SD = 0.45; PRS313 SD = 0.61; ER-positive PRS313 SD = 0.65; ER-negative PRS313 SD = 0.59) exactly as was done in the analyses of the PRS and first breast cancer risk.6,7 This allows a direct comparison of the magnitude of the CBC relative risk estimation to that of the first breast cancer.
For samples genotyped with both OncoArray and iCOGS array (9,071 samples), OncoArray data were used in preference as the imputation quality was generally higher. The intraclass correlation coefficient (ICC) between the PRSs derived from the two platforms was 0.99 (95%CI = 0.99–0.99) for the PRS77 and 0.96 (95%CI = 0.95–0.96) for PRS313 (Figure S2). Given the high correlation between the two platforms, PRS measures from both platforms were used in the analyses without adjustment.
Statistical Analysis
European Cohort
The primary outcome in the European cohort was the development of metachronous CBC. Cox proportional hazards models were used to estimate HRs for metachronous CBC risk by PRS, stratified by country. Since previous studies have shown that age at first breast cancer diagnosis is an important predictor of CBC,3 the analyses were performed with attained age as the timescale. Time at risk started 3 months after the first breast cancer diagnosis and ended at the age of CBC diagnosis, distant metastasis (where available), death, or end of follow-up, whichever came first. For women who had a study entry more than 3 months after first breast cancer diagnosis, follow-up started at the age of study entry. We also performed a fixed-effect meta-analysis of country-specific effects using the STATA command metan. We performed a fixed-effect meta-analysis over a random-effect meta-analysis since there was no evidence for heterogeneity in effect sizes between countries (I-squared = 0%, Figure S3). For some analyses, only invasive CBC was used as the outcome; in these analyses we censored on in situ CBC. Separate analyses were conducted for ER-positive CBC (censored on ER-negative and ER-unknown CBC) and ER-negative CBC (censored on ER-positive and ER-unknown CBC).
We evaluated the linearity of the association between PRS313 per unit SD and CBC risk using restricted cubic splines with three knots. There was no evidence for violation of the linearity assumption. Therefore, in the main analysis, the PRS313 was treated as a continuous covariate, and estimated the HR per unit SD of the PRS313. Violation of the proportional hazard assumption was assessed by inspection of the Schoenfeld residuals.15 As a second analysis, we used the per SD log HR of the PRS313 to calculate the predicted HR at different percentiles of the PRS313, compared to the 50th percentile. Third, the PRS313 was categorized into percentile groups (0th to 10th, 10th to 20th, 20th to 40th, 40th to 60th, 60th to 80th, 80th to 90th, 90th to 100th) to illustrate the differences between PRS313 subgroups, with the middle quintile (40th to 60th) as the reference.
We also performed multivariable Cox regression analyses to determine whether the log HR of CBC risk by PRS changed when adjusting for year of first breast cancer diagnosis, family history of breast cancer in a first degree relative, and several clinical characteristics of the first breast cancer such as nodal status, tumor size, morphology, ER status, progesterone receptor (PR) status, human epidermal growth factor receptor 2 (HER2) status, (neo)adjuvant chemotherapy, adjuvant endocrine therapy, and radiotherapy. These analyses were performed in all women, a complete case set (excluding those with unknown values for the covariates), and in a set excluding studies oversampling case subjects with family history. Potential effect modification of the PRS313 effect by the same variables was evaluated by fitting interaction terms in different models using complete case sets, including the standardized PRS313, modifier, and interaction.
The discriminative ability of different models (model 1: PRS313 alone; model 2: other risk factors [the adjustment variables from the multivariable Cox regression analyses]; model 3: PRS313 + other risk factors) was calculated using Harrell’s C-index.16 Since no standard performance measures are currently available to account for left-truncated follow-up time (i.e., to start analyses at age at study entry), we used time since first breast cancer as the timescale to calculate the C-index.
Absolute Risks
Absolute risks of developing CBC at PRS313 percentiles were calculated using the estimated log HRs per SD from the breast cancer cohort (BCAC) under the log-linear model, assuming the PRS is normally distributed. The PRS313- and age-specific incidences were constrained to the age-specific CBC incidences from women diagnosed with a first invasive breast cancer in the period 2003–2010 from the Netherlands Cancer Registry (NCR).1 The procedure for constraining the incidences has been previously described.17 The age-specific CBC incidences were calculated overall and for age-specific groups, censoring on death and distant metastasis. We used data from the NCR since this registry has complete coverage of all newly diagnosed cancers in the Netherlands. The NCR cohort included all females aged ≥18 years and follow-up for second cancers was complete until February 1, 2016.1 We then applied the competing risk of dying on the absolute CBC risks. The absolute CBC risk (ARg) by age t in PRS313 category g, taking into account the competing risk of dying was calculated by:
where μg (t) is the CBC incidence associated with PRS313 category g, Sg (t) the probability of being free of CBC to age t, and Sm (t) the probability of surviving to age t.
Case-Case Series
For the case-case series (European and Asian), logistic regression models were used to estimate the ORs for CBC risk (comparing with unilateral breast cancer) and for unilateral breast cancer risk (comparing with women without any diagnosis of breast cancer) associated with PRS313. All analyses were adjusted for age and country (Table S1). For all unilateral- and contralateral breast cancer patients, we used age at first breast cancer diagnosis, and for women without any diagnosis of breast cancer we used age at baseline questionnaire.
For direct comparison with the estimate reported for PRS313 and first breast cancer, we also performed logistic regression analyses in the same BCAC study participants included in the validation of the association between PRS313 and first breast cancer risk.7 This validation set comprised a subsample from 24 studies and included 3,781 women with unilateral breast cancer, 94 women with CBC, and 3,753 women without any diagnosis of breast cancer (Table S2). For this analysis, we adjusted for 10 principal components, in line with Mavaddat et al.7
For European women who had follow-up time available more than 3 months after the first breast cancer diagnosis, a sensitivity analysis was performed for metachronous CBC (1,702 CBCs). We also did a separate analysis for invasive CBC (N = 3,246), by excluding CBC in situ.
All p values are two sided; tests with p < .05 are referred to as statistically significant. Analyses were performed using STATA, v.13.1 (StataCorp) and R v.3.3.2.
Results
European (Cohort) Cox Regression Analyses
The European cohort included 56,068 women diagnosed with first invasive breast cancer with 1,027 metachronous CBC events. Median follow-up was 8.4 years. Patient, tumor, and treatment characteristics are summarized in Table S4.
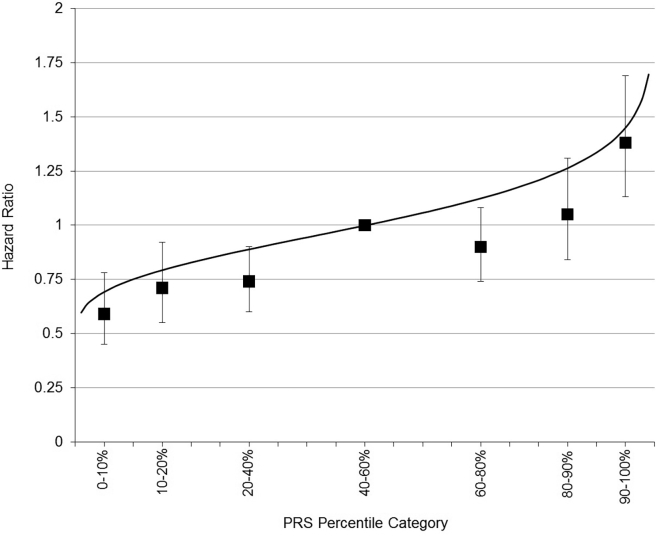
The associations between the different PRSs and CBC risk are shown in Table 1. The HR for CBC per SD of PRS313 was 1.25 (95%CI = 1.18–1.33). For comparison, the HR per SD for PRS77 was 1.21 (95%CI = 1.14–1.29). Women within the 0th to 10th and the 90th to 100th percentile of the PRS313 had 0.59-fold (95%CI = 0.45–0.78) and 1.38-fold (95%CI = 1.13–1.69) risk of CBC, respectively, compared with women within the 40th to 60th percentile (Figure 1, Table S5). The predicted HRs of CBC for women at the 10th and 90th percentile of the PRS313 were 0.75 and 1.33, respectively, compared to the 50th percentile (Figure 1). Since we observed evidence of departure from the proportional hazards assumption (p = 0.02),15 we also calculated HRs stratified for follow-up duration (<5 and ≥5 years). The HR by SD of the PRS313 was 1.21 (95%CI = 1.10–1.32) for CBC diagnosed ≤5 years after first breast cancer diagnosis (CBC N = 428) and 1.28 (95%CI = 1.18–1.38) for CBC diagnosed >5 years after first diagnosis (CBC N = 599).
Table 1.
Association between PRSs and Contralateral Breast Cancer Risk in the European Cohort (N = 56,068)
| Polygenic Risk Score (PRS) | No. of CBC | HR per Unit SDa | 95%CI | p Value |
|---|---|---|---|---|
| PRS77b | ||||
| All CBC | 1,027 | 1.21 | 1.14–1.29 | <.001 |
| Invasive CBC | 923 | 1.21 | 1.13–1.29 | <.001 |
| PRS313b | ||||
| All CBC | 1,027 | 1.25 | 1.18–1.33 | <.001 |
| Invasive CBC | 923 | 1.24 | 1.16–1.32 | <.001 |
| ER-positive invasive CBCd | 275 | 1.38 | 1.23–1.55 | <.001 |
| ER-negative invasive CBCd | 97 | 0.92 | 0.75–1.12 | .39 |
| ER-Positive PRS313b,c | ||||
| All CBC | 1,027 | 1.23 | 1.16–1.31 | <.001 |
| Invasive CBC | 923 | 1.22 | 1.15–1.30 | <.001 |
| ER-positive invasive CBCd | 275 | 1.37 | 1.22–1.54 | <.001 |
| ER-Negative PRS313b,c | ||||
| All CBC | 1,027 | 1.25 | 1.17–1.33 | <.001 |
| Invasive CBC | 923 | 1.24 | 1.16–1.33 | <.001 |
| ER-negative invasive CBCd | 97 | 1.06 | 0.86–1.30 | .58 |
Abbreviations: PRS, polygenic risk score; No., number; CBC, contralateral breast cancer; HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; SD, standard deviation.
All analyses were performed with attained age as timescale.
Coefficients to construct the PRSs are shown in Table S3. All PRSs were standardized by the same SD as was used by Mavaddat et al.7 The SD was 0.45 for overall breast cancer PRS77, 0.61 for overall breast cancer PRS313, 0.65 for ER-positive PRS313, and 0.59 for ER-negative PRS313.
ER-specific PRSs were constructed using a hybrid method, as described by Mavaddat et al.7
Women with ER-unknown CBC (N = 551) were censored in these analyses.
Figure 1.
Estimates for Contralateral Breast Cancer Risk by Percentile Categories of the 313-Variant PRS (PRS313)
The figure shows the hazard ratios per SD and 95% confidence intervals for percentiles of the PRS313 relative to the middle quintile (underlying table can be found in Table S5). The solid line denotes the estimates for contralateral breast cancer risk with the PRS313 fitted as a continuous covariate. Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by SD = 0.61, in line with Mavaddat et al.7 The analyses were performed with attained age as timescale. PRS, polygenic risk score; SD, standard deviation.
The HR per SD of PRS313 for ER-positive invasive CBC was 1.38 (95%CI = 1.23–1.55) compared to a HR per SD of the ER-positive PRS313 of 1.37 (95%CI = 1.22–1.54) (Table 1). For ER-negative invasive CBC, the HR per SD was 0.92 (95%CI = 0.75–1.12) for PRS313 and 1.06 (95%CI = 0.86–1.30) for the ER-negative PRS313.
Sensitivity analysis using the overall PRS313 showed a HR per SD of 1.24 (95%CI = 1.16–1.32) for invasive CBC risk. When we used time since first breast cancer as the timescale, we found similar results (HR per SD = 1.25, 95%CI = 1.18–1.33). Meta-analysis of country-specific effects showed a HR per SD of 1.25 (95%CI = 1.18–1.33) for CBC risk by PRS313 (Figure S3).
The association between the PRS313 and CBC risk did not change when adjusting for characteristics of the individual, tumor, or treatment, nor when excluding studies oversampling case subjects with a family history (Table S6). When considering potential modifiers of the effect of the PRS313 on CBC risk (Table 2), we found that the HR was the lowest in women aged <40 years at first breast cancer diagnosis (HR per SD = 1.13; 95%CI = 0.98–1.31) and tended to increase with age, although these effects were not statistically significant (Pheterogeneity = 0.26; Ptrend = 0.05). We found no indication for effect modification by family history (Pheterogeneity = 0.63), morphology (Pheterogeneity = 0.14), ER status (Pheterogeneity = 0.13), PR status (p = 0.26), HER2 status (Pheterogeneity = 0.42), chemotherapy (Pheterogeneity = 0.60), endocrine therapy (Pheterogeneity = 0.79), or radiotherapy (Pheterogeneity = 0.40) (Table 2).
Table 2.
Association between the 313-Variant PRS (PRS313) and Contralateral Breast Cancer Risk for Subgroups
| Subgroups | No. of Patients | No. of CBC | HR per Unit SDa,b | 95%CI | p Value | Phetero-geneityc,d | Ptrendc,e |
|---|---|---|---|---|---|---|---|
| All patients | 56,068 | 1,027 | 1.25 | 1.18–1.33 | <.001 | - | - |
| Age at First Breast Cancer Diagnosis (Years) | .26 | .05 | |||||
| <40 | 5,877 | 171 | 1.13 | 0.98–1.31 | .09 | ||
| 40–49 | 11,928 | 265 | 1.25 | 1.11–1.41 | <.001 | ||
| 50–59 | 16,882 | 320 | 1.22 | 1.09–1.36 | <.001 | ||
| 60+ | 21,381 | 271 | 1.36 | 1.21–1.52 | <.001 | ||
| Family History (First Degree Relative) | .63 | - | |||||
| No | 33,623 | 618 | 1.26 | 1.16–1.36 | <.001 | ||
| Yes | 10,369 | 302 | 1.22 | 1.09–1.36 | <.001 | ||
| Morphology | .14 | - | |||||
| Ductal | 37,324 | 621 | 1.21 | 1.12–1.31 | <.001 | ||
| Lobular | 5,878 | 118 | 1.32 | 1.10–1.59 | .002 | ||
| Mixed (ductal and lobular) | 2,174 | 46 | 1.52 | 1.15–2.02 | .004 | ||
| Other | 3,344 | 70 | 1.20 | 0.96–1.50 | .11 | ||
| ER Status | .13 | - | |||||
| Negative | 9,527 | 194 | 1.13 | 0.98–1.30 | .08 | ||
| Positive | 38,090 | 670 | 1.28 | 1.19–1.38 | <.001 | ||
| PR Status | .26 | - | |||||
| Negative | 13,098 | 244 | 1.16 | 1.03–1.32 | .02 | ||
| Positive | 27,044 | 554 | 1.27 | 1.17–1.38 | <.001 | ||
| HER2 Status | .42 | - | |||||
| Negative | 23,787 | 352 | 1.29 | 1.17–1.44 | <.001 | ||
| Positive | 4,969 | 60 | 1.45 | 1.13–1.85 | .004 | ||
| (Neo)adjuvant Chemotherapy | .60 | - | |||||
| No | 18,110 | 361 | 1.28 | 1.16–1.42 | <.001 | ||
| Yes | 18,559 | 363 | 1.24 | 1.12–1.37 | <.001 | ||
| (Neo)adjuvant Endocrine Therapy | .79 | - | |||||
| No | 10,781 | 242 | 1.28 | 1.13–1.44 | <.001 | ||
| Yes | 27,322 | 460 | 1.30 | 1.19–1.43 | <.001 | ||
| Radiotherapy | .40 | - | |||||
| No | 11,023 | 188 | 1.33 | 1.15–1.53 | <.001 | ||
| Yes | 29,142 | 617 | 1.24 | 1.15–1.34 | <.001 | ||
Abbreviations: PRS, polygenic risk score; No., number; CBC, contralateral breast cancer; HR, hazard ratio; CI, confidence interval; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
HR for CBC risk by unit SD of PRS313. All analyses were performed with attained age as timescale.
Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by standard deviation = 0.61, in line with Mavaddat et al.7
The interaction between the PRS313 and each subgroup was tested in different models including the standardized PRS313, modifier, and interaction. Patients with unknown values were excluded from these analyses. Since attained age was used as timescale in all models, the model with age at first breast cancer only included the PRS313 and interaction.
P for interaction based on test for heterogeneity across categories.
P for interaction based on a trend test with age as continuous variable.
The C-index was 0.563 (95%CI = 0.547–0.586) for the model only including PRS313, 0.605 (95%CI = 0.591–0.629) for the model only including other risk factors, and 0.623 (95%CI = 0.608–0.645) for the complete model (Table 3).
Table 3.
Discriminatory Ability (C-Index) of the 313-Variant PRS (PRS313) and Other Risk Factors for Contralateral Breast Cancer Risk in the European Cohort
| C-Index (95%CI)a,b | |
|---|---|
| Model 1: PRS313c alone | 0.563 (0.547–0.586) |
| Model 2: Other risk factorsd | 0.605 (0.591–0.629) |
| Model 3: PRS313c + other risk factorsd | 0.623 (0.608–0.645) |
Abbreviations: PRS, polygenic risk score; CI, confidence interval.
The Harrell’s C-index was obtained by the STATA stcox postestimation command “estat concordance,” using time since first breast cancer on the timescale without taking delayed entry (prevalent cases) into account. We did not consider delayed entry since no standard performance measures are currently available in the statistical literature to account for left-truncated follow-up time. The median of delayed entry was 0.4 years (standard deviation = 2.7) in our study.
The 95% CIs were obtained by use of the somersd package in STATA.
Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by SD = 0.61, in line with Mavaddat et al.7
Including age at first diagnosis, year of first diagnosis, family history for breast cancer in a first degree relative, and clinical characteristics of the first breast cancer (nodal status, tumor size, differentiation grade, morphology, estrogen receptor status, human epidermal growth factor receptor 2 status, chemotherapy, endocrine therapy, radiotherapy).
Absolute Risks
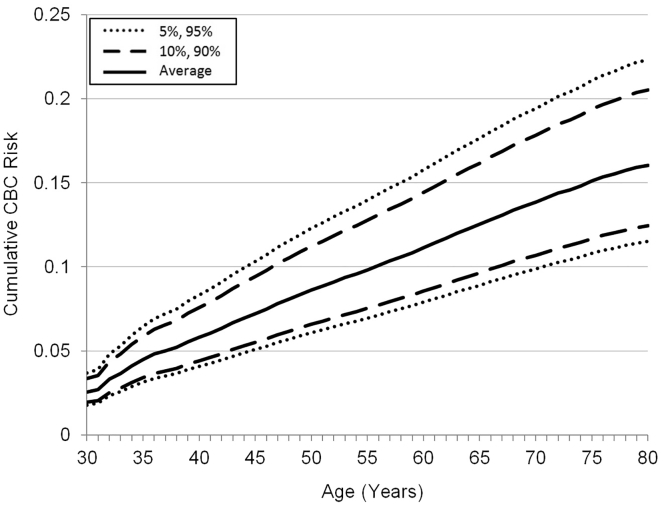
Based on the HR estimates for PRS313, the predicted CBC risk by age 80 years was 12.4% at the 10th percentile of the PRS313, compared with 20.5% at the 90th percentile of the PRS313 (Figure 2), accounting for death as competing risk. When death was not taken into account as competing risk, the corresponding predicted risks by age 80 were 17.0% at the 10th percentile and 27.9% at the 90th percentile of the PRS313 (Figure S4). Table 4 shows the 5- and 10-year cumulative CBC risks by PRS313 for different age groups, accounting for death as competing risk (Table S7 shows results without competing risks).
Figure 2.
Predicted Contralateral Breast Cancer Risk by Percentile of the 313-Variant PRS (PRS313) with Death as Competing Risk
Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by SD = 0.61, in line with Mavaddat et al.7 The CBC incidences were calculated based on incidence data from the Netherlands Cancer Registry1 and relative risks estimated as described in the Material and Methods. PRS, polygenic risk score; CBC, contralateral breast cancer.
Table 4.
5- and 10-Year Cumulative Risks of Contralateral Breast Cancer by the 313-Variant PRS (PRS313) for Different Age Groups with Death as Competing Risk
|
5-Year Cumulative CBC Risks (%) Range by Age |
10-Year Cumulative CBC risks (%) Range by Age |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Age at First Breast Cancer Diagnosis (years) | 5thPercentile PRS313 | 10thPercentile PRS313 | 50thPercentile PRS313 | 90thPercentile PRS313 | 95thPercentile PRS313 | 5thPercentile PRS313 | 10thPercentile PRS313 | 50thPercentile PRS313 | 90thPercentile PRS313 | 95thPercentile PRS313 |
| 30–34 | 1.9–3.1 | 2.1–3.4 | 2.7–4.5 | 3.6–5.9 | 4.0–6.5 | 3.1–4.1 | 3.4–4.5 | 4.5–5.9 | 5.9–7.7 | 6.5–8.5 |
| 35–39 | 0.8–2.1 | 0.9–2.3 | 1.2–3.0 | 1.5–3.9 | 1.7–4.3 | 2.1–3.5 | 2.3–3.8 | 3.0–5.0 | 3.9–6.6 | 4.3–7.2 |
| 40–44 | 1.5–2.8 | 1.7–3.1 | 2.2–4.1 | 2.9–5.3 | 3.2–5.9 | 2.8–4.6 | 3.1–5.0 | 4.1–6.6 | 5.3–8.6 | 5.9–9.4 |
| 45–49 | 1.4–2.5 | 1.5–2.7 | 2.0–3.6 | 2.6–4.7 | 2.9–5.2 | 2.5–3.9 | 2.7–4.3 | 3.6–5.6 | 4.7–7.4 | 5.2–8.1 |
| 50–54 | 1.4–2.8 | 1.5–3.0 | 1.9–4.0 | 2.6–5.2 | 2.8–5.8 | 2.8–4.5 | 3.0–4.9 | 4.0–6.4 | 5.2–8.4 | 5.8–9.3 |
| 55–59 | 1.6–3.1 | 1.8–3.4 | 2.3–4.5 | 3.1–5.9 | 3.4–6.5 | 3.1–4.8 | 3.4–5.2 | 4.5–6.9 | 5.9–9.0 | 6.5–9.9 |
| 60–64 | 1.7–3.3 | 1.9–3.6 | 2.5–4.7 | 3.3–6.2 | 3.6–6.8 | 3.3–5.0 | 3.6–5.4 | 4.7–7.1 | 6.2–9.3 | 6.8–10.2 |
| 65–70 | 1.5–3.2 | 1.6–3.5 | 2.1–4.6 | 2.8–6.1 | 3.1–6.7 | 3.2–4.1 | 3.5–4.5 | 4.6–5.9 | 6.1–7.7 | 6.7–8.5 |
Abbreviations: PRS, polygenic risk score; CBC, contralateral breast cancer. Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by SD = 0.61, in line with Mavaddat et al.7 The CBC incidences for each age group were calculated based on incidence data from the Netherlands Cancer Registry1 and relative risks estimated as described in the Material and Methods. Death was taken into account as competing risk.
European and Asian (Case-Case Series) Logistic Regression Analyses
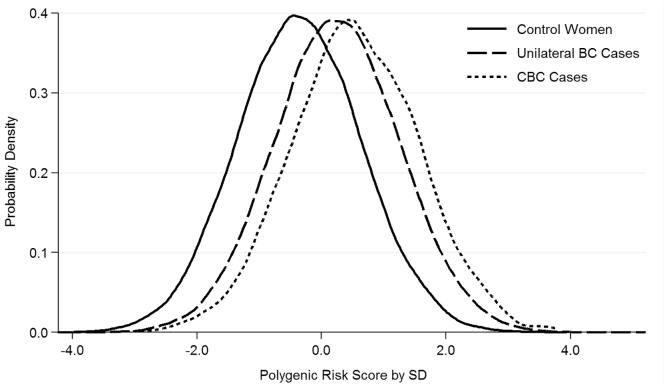
Figure 3 shows the distribution of the PRS313 per SD in the European case-case series. Median PRS313 was −0.4 (interquartile range [IQR] = 1.35) for control women without any diagnosis of breast cancer (N = 81,000), 0.2 (IQR = 1.36) for women with unilateral breast cancer (N = 62,830), and 0.5 (IQR = 1.40) for women with CBC (N = 3,607). The OR for unilateral breast cancer per SD of the PRS313, compared to control women, was 1.82 (95%CI = 1.80–1.84) (Table S8). The OR for CBC per SD of PRS313, compared to unilateral breast cancer, was 1.30 (95%CI = 1.26–1.35).
Figure 3.
Distribution of the 313-Variant PRS (PRS313) in 62,830 Control Women without Any Diagnosis of Breast Cancer, 81,000 Women with Unilateral Breast Cancer, and 3,607 Women with Contralateral Breast Cancer
Coefficients to construct the PRS313 are shown in Table S3. The PRS313 was standardized by SD = 0.61, in line with Mavaddat et al.7 PRS, polygenic risk score; BC, breast cancer; CBC, contralateral breast cancer; SD, standard deviation.
In sensitivity analyses, the OR per SD of PRS313 was 1.27 (95%CI = 1.21–1.33) for metachronous CBC and the OR per SD was 1.29 (95%CI = 1.24–1.33) for invasive CBC, compared to unilateral breast cancer. When analyses were restricted to the validation set of Mavaddat et al.,7 the OR for unilateral breast cancer per SD of the PRS313 was 1.67 (95%CI = 1.59–1.76) compared to control women, and the OR for CBC per SD of PRS313 was 1.39 (95%CI = 1.13–1.70) compared to unilateral breast cancer (Table S8).
For women of Asian descent, the OR for unilateral breast cancer per SD of the PRS313 was 1.56 (95%CI = 1.52–1.60) compared to control women, and the OR for CBC per SD of PRS313 was 1.15 (95%CI = 1.02–1.29) compared to women with unilateral breast cancer (Table S8).
Discussion
Previous studies have shown that a PRS, summarizing the effects of common germline variants, can be used to stratify women with respect to their risk to develop a primary breast cancer.6, 7, 8, 9 In this study, we observed a clear association between the PRS313 and CBC risk in women of both European and Asian ancestry. The association was observed in both the case-case series and the European cohort. The HRs per SD of CBC for women at the 10th and 90th percentile of the continuous predicted PRS313 were 0.75 and 1.33, respectively, compared to the 50th percentile. This translates to absolute risks at the 10th and the 90th percentile of the PRS313 of 12.4% and 20.5%, respectively, by age 80 years. We estimated a C-index for the PRS313, summarizing its discriminatory ability, of 0.563 in the European cohort.
One previous study has investigated the effect of a PRS, including 67 variants, and CBC risk.10 This study found a risk ratio of 1.75 (95%CI = 1.41–2.18) for women in the upper quartile of the PRS compared with women in the lowest quartile. To facilitate comparison, we performed a similar analysis in our case-case series, showing an OR of 1.98 (95%CI = 1.79–2.18), adjusted for country and age at first diagnosis, for women in the upper quartile of the PRS313. This indicates that the PRS313 improves stratification relative to PRSs including fewer variants. Moreover, in our European cohort, the C-index for the PRS alone improved from 0.547 (95%CI = 0.536–0.575) for the previously reported PRS776 to 0.563 (95%CI = 0.547–0.586) for the PRS313.
We found no evidence that the association between the PRS313 and CBC risk was confounded by family history, adjuvant therapy, morphology, age, or tumor receptor status of the first breast cancer, nor that there was effect modification by those factors. The absence of notable effect modification is in line with the abovementioned study of a 67-variant PRS and CBC risk; no heterogeneity in association was found by age, family history, morphology, ER status, and adjuvant treatment.10
To provide an external validation of our findings, we examined data from UK Biobank, which includes many women diagnosed with breast cancer with data available on the PRS313 (Supplemental Note). Unfortunately, UK Biobank has no information available on the laterality of the tumor, and it is, therefore, not possible to distinguish between contralateral and ipsilateral breast cancers. We therefore performed analyses using any second breast cancer as the endpoint. This secondary analysis did confirm the association between the PRS313 and second breast cancer risk (HR per SD = 1.13, 95%CI = 1.01–1.27), but with a lower estimate than in our European cohort. The lower estimate may be explained by the inclusion of the ipsilateral breast cancers, which may be more likely to be recurrences than new primary breast cancers compared to CBCs. Indeed, when we used ipsilateral breast cancer as the outcome in our European cohort, we found no association with the PRS313 (HR = 1.02, 95%CI = 0.90–1.15).
The association between the PRS313 and CBC risk (OR per SD = 1.30; 95%CI = 1.26–1.35) in the BCAC database was weaker (expressed in terms of an OR) than was found for first breast cancer among independent prospective studies (OR per SD = 1.61; 95%CI = 1.57–1.65). Under a simple polygenic model, the relative risk would be expected to be similar for the second breast cancer. The attenuated estimate for CBC might however be explained by several factors. Some attenuation of the estimate might have been due to dilution in the end-point definition, i.e., if some of the CBCs were metastases. Previous studies investigating the clonal relatedness of first breast cancers and CBCs using tumor sequencing have shown that 6%–12% of CBCs represent metastases.18,19 This hypothesis would be consistent with our finding of a slightly stronger association between the PRS313 and late CBCs, diagnosed >5 years after the first breast cancer, than for early CBCs, diagnosed ≥5 years after the first cancer, since the latter are more likely to be metastases. In addition, 3%–5% of the women with breast cancer will have a mutation in BRCA1 or BRCA2,20,21 who have high CBC risks. It has been shown that the relative risk associated with PRS is lower (for the first breast cancer) for women with a BRCA1 and BRCA2 mutation than in the general population,22 diluting the overall relative risk for CBC. More generally, it is possible that the CBC association may be attenuated due to the effect of other, unmeasured, genetic or other risk factors. If the risks are high, case subjects with higher PRS313 will have, on average, lower values of other risk factors, due to elimination of the highest risk individuals, again attenuating the CBC association. Finally, given the limited information on family history in our dataset, the estimate could have been biased due to a family history effect not detected in our data.
There was some suggestion that the relative risk associated with PRS313 decreased with younger age (Ptrend = 0.05) and, specifically, was lower for women aged <40 years (HR per SD = 1.13; 95%CI = 0.98–1.31). Interestingly, Mavaddat et al.7 also found a lower relative risk below age 40 for first breast cancer. This effect may reflect the different characteristics of breast cancers at young ages, both in terms of germline susceptibility and pathology.23,24 For example, the proportion of ER-negative breast cancers is higher at young ages, and the PRS is less predictive for ER-negative disease.6,7,24
In the logistic regression analyses in Asian women, the association between the PRS313 and CBC risk was slightly weaker than in European women. This finding is consistent with a recent analysis investigating the association between a 287-variant PRS and first breast cancer risk in the Asian population,25 which showed an attenuated OR in Asian women (OR = 1.52, 95%CI = 1.49–1.56) compared to European women (OR = 1.61, 95%CI = 1.57–1.66). The lower estimate for Asian women might reflect the fact the PRS313 was developed in European populations, and the different LD structure in Asians may attenuate the association since the variants in the PRS are likely to be surrogates for the causal variants. Other explanations for the attenuated estimate may be the slightly younger age at first breast cancer diagnosis and the higher proportion ER-negative CBCs in Asian women compared to European women in our study. Finally, the imputation quality for variants was somewhat lower, on average, for the Asian than for the European dataset, with three variants on OncoArray and four variants on ICOGs with an imputation quality score < 0.3 (Table S3). Nevertheless, we included those variants in the PRS for both European and Asian women, to keep the PRS comparable between ethnicities and studies. Future studies including larger numbers of Asian women, and women of other ethnicities, are needed to generate population-specific PRSs and to validate our findings in these groups.
A major strength of this study is the very large sample size in the BCAC dataset, including genotype information for ∼150,000 women and a large number of CBC events. A limitation of this study is missing data on the patient, tumor, and treatment characteristics, which reduces the power of the multivariable Cox regression analyses and interaction analyses. In addition, registration of CBC was not complete; the 10-year cumulative CBC incidence was 2.2% in the BCAC dataset, compared to 3.8% using complete data from the Netherlands Cancer Registry.1 For this reason, we estimated relative risk estimates using the BCAC data and applied these to external registry data to obtain absolute risk estimates. The underreporting of CBC should not bias our HR estimates, given that the event rate is low and reporting of CBC is unlikely to be related to the PRS313. Moreover, we reran the cohort analysis in the subset of countries with a 10-year cumulative CBC incidence ≥ 3.0% in the BCAC dataset, and the estimates were very similar to the main analyses (HR per SD = 1.23, 95%CI = 1.14–1.33) (Figure S3).
In conclusion, the PRS313 is predictive for the development of CBC. We found no evidence for confounding or effect modification by other previously established CBC risk factors. The PRS313 is therefore likely to be an independent risk factor for CBC. Since the predictive ability of the PRS on its own is modest, it should be combined with other breast cancer risk factors to provide more useful CBC risk prediction models. More accurate risk prediction will help identify women at high CBC risk who will benefit from additional surveillance and/or risk reducing mastectomy, and equally important, to identify those women at low risk in order to avoid unnecessary surgeries.
Data and Code Availability
Data used in this manuscript may be requested through the original providers. Data of the Breast Cancer Association Consortium may be requested for non-profit research through an application procedure with the Breast Cancer Association Consortium. Data of the UK Biobank needs to be requested through UK Biobank.
Declaration of Interests
M.W.B. conducts research funded by Amgen, Novartis, and Pfizer, outside the submitted work. P.A.F. conducts research funded by Amgen, Novartis, and Pfizer, outside the submitted work. He received honoraria from Roche, Novartis, and Pfizer. H.N. received honorarium from Astra Zeneca outside the submitted work.
Published: October 5, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.09.001.
Contributor Information
Marjanka K. Schmidt, Email: mk.schmidt@nki.nl.
NBCS Collaborators:
Anne-Lise Børresen-Dale, Kristine Sahlberg, Lars Ottestad, Rolf Kåresen, Ellen Schlichting, Marit Muri Holmen, Toril Sauer, Vilde Haakensen, Olav Engebråten, Bjørn Naume, Alexander Fosså, Cecile Kiserud, Kristin Reinertsen, Åslaug Helland, Margit Riis, Jürgen Geisler, and Grethe Grenaker Alnæs
ABCTB Investigators:
Christine Clarke, Deborah Marsh, Rodney Scott, Robert Baxter, Desmond Yip, Jane Carpenter, Alison Davis, Nirmala Pathmanathan, Peter Simpson, J. Dinny Graham, and Mythily Sachchithananthan
kConFab Investigators:
David Amor, Lesley Andrews, Yoland Antill, Rosemary Balleine, Jonathan Beesley, Ian Bennett, Michael Bogwitz, Leon Botes, Meagan Brennan, Melissa Brown, Michael Buckley, Jo Burke, Phyllis Butow, Liz Caldon, Ian Campbell, Deepa Chauhan, Manisha Chauhan, Georgia Chenevix-Trench, Alice Christian, Paul Cohen, Alison Colley, Ashley Crook, James Cui, Margaret Cummings, Sarah-Jane Dawson, Anna deFazio, Martin Delatycki, Rebecca Dickson, Joanne Dixon, Ted Edkins, Stacey Edwards, Gelareh Farshid, Andrew Fellows, Georgina Fenton, Michael Field, James Flanagan, Peter Fong, Laura Forrest, Stephen Fox, Juliet French, Michael Friedlander, Clara Gaff, Mike Gattas, Peter George, Sian Greening, Marion Harris, Stewart Hart, Nick Hayward, John Hopper, Cass Hoskins, Clare Hunt, Paul James, Mark Jenkins, Alexa Kidd, Judy Kirk, Jessica Koehler, James Kollias, Sunil Lakhani, Mitchell Lawrence, Geoff Lindeman, Lara Lipton, Liz Lobb, Graham Mann, Deborah Marsh, Sue Anne McLachlan, Bettina Meiser, Roger Milne, Sophie Nightingale, Shona O'Connell, Sarah O'Sullivan, David Gallego Ortega, Nick Pachter, Briony Patterson, Amy Pearn, Kelly Phillips, Ellen Pieper, Edwina Rickard, Bridget Robinson, Mona Saleh, Elizabeth Salisbury, Christobel Saunders, Jodi Saunus, Rodney Scott, Clare Scott, Adrienne Sexton, Andrew Shelling, Peter Simpson, Melissa Southey, Amanda Spurdle, Jessica Taylor, Renea Taylor, Heather Thorne, Alison Trainer, Kathy Tucker, Jane Visvader, Logan Walker, Rachael Williams, Ingrid Winship, and Mary Ann Young
Web Resources
Breast Cancer Association Consortium, http://bcac.ccge.medschl.cam.ac.uk/bcacdata/
UK Biobank, https://www.ukbiobank.ac.uk/researchers/
Supplemental Data
References
- 1.Kramer I., Schaapveld M., Oldenburg H.S.A., Sonke G.S., McCool D., van Leeuwen F.E., Van de Vijver K.K., Russell N.S., Linn S.C., Siesling S. The influence of adjuvant systemic regimens on contralateral breast cancer risk and receptor subtype. J. Natl. Cancer Inst. 2019;111:709–718. doi: 10.1093/jnci/djz010. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Z., Yang L., Deng G., Huang X., Li X., Xie X., Wang J., Shuang Z., Wang X. Patterns of Occurrence and Outcomes of Contralateral Breast Cancer: Analysis of SEER Data. J. Clin. Med. 2018;7:7. doi: 10.3390/jcm7060133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akdeniz D., Schmidt M.K., Seynaeve C.M., McCool D., Giardiello D., van den Broek A.J., Hauptmann M., Steyerberg E.W., Hooning M.J. Risk factors for metachronous contralateral breast cancer: A systematic review and meta-analysis. Breast. 2019;44:1–14. doi: 10.1016/j.breast.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., NBCS Collaborators. ABCTB Investigators. ConFab/AOCS Investigators Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milne R.L., Kuchenbaecker K.B., Michailidou K., Beesley J., Kar S., Lindström S., Hui S., Lemaçon A., Soucy P., Dennis J., ABCTB Investigators. EMBRACE. GEMO Study Collaborators. HEBON. kConFab/AOCS Investigators. NBSC Collaborators Identification of ten variants associated with risk of estrogen-receptor-negative breast cancer. Nat. Genet. 2017;49:1767–1778. doi: 10.1038/ng.3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mavaddat N., Pharoah P.D., Michailidou K., Tyrer J., Brook M.N., Bolla M.K., Wang Q., Dennis J., Dunning A.M., Shah M. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 2015;107:107. doi: 10.1093/jnci/djv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavaddat N., Michailidou K., Dennis J., Lush M., Fachal L., Lee A., Tyrer J.P., Chen T.H., Wang Q., Bolla M.K., ABCTB Investigators. kConFab/AOCS Investigators. NBCS Collaborators Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019;104:21–34. doi: 10.1016/j.ajhg.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brentnall A.R., van Veen E.M., Harkness E.F., Rafiq S., Byers H., Astley S.M., Sampson S., Howell A., Newman W.G., Cuzick J., Evans D.G.R. A case-control evaluation of 143 single nucleotide polymorphisms for breast cancer risk stratification with classical factors and mammographic density. Int. J. Cancer. 2020;146:2122–2129. doi: 10.1002/ijc.32541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shieh Y., Hu D., Ma L., Huntsman S., Gard C.C., Leung J.W., Tice J.A., Vachon C.M., Cummings S.R., Kerlikowske K., Ziv E. Breast cancer risk prediction using a clinical risk model and polygenic risk score. Breast Cancer Res. Treat. 2016;159:513–525. doi: 10.1007/s10549-016-3953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robson M.E., Reiner A.S., Brooks J.D., Concannon P.J., John E.M., Mellemkjaer L., Bernstein L., Malone K.E., Knight J.A., Lynch C.F. Association of Common Genetic Variants With Contralateral Breast Cancer Risk in the WECARE Study. J. Natl. Cancer Inst. J. Natl. Cancer Inst. 2017;109:djx051. doi: 10.1093/jnci/djx051. djx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teraoka S.N., Bernstein J.L., Reiner A.S., Haile R.W., Bernstein L., Lynch C.F., Malone K.E., Stovall M., Capanu M., Liang X., WECARE Study Collaborative Group Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women’s Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Res. 2011;13:R114. doi: 10.1186/bcr3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M., BOCS. kConFab Investigators. AOCS Group. NBCS. GENICA Network Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., Milne R.L., Schmidt M.K., Chang-Claude J., Bojesen S.E., Bolla M.K., Breast and Ovarian Cancer Susceptibility Collaboration. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) kConFab Investigators. Australian Ovarian Cancer Study Group. GENICA (Gene Environment Interaction and Breast Cancer in Germany) Network Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013;45:353–361, e1–e2. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amos C.I., Dennis J., Wang Z., Byun J., Schumacher F.R., Gayther S.A., Casey G., Hunter D.J., Sellers T.A., Gruber S.B. The OncoArray Consortium: A Network for Understanding the Genetic Architecture of Common Cancers. Cancer Epidemiol. Biomarkers Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfeld D.A. Sample-size formula for the proportional-hazards regression model. Biometrics. 1983;39:499–503. [PubMed] [Google Scholar]
- 16.Harrell F.E., Jr., Califf R.M., Pryor D.B., Lee K.L., Rosati R.A. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 17.Antoniou A.C., Beesley J., McGuffog L., Sinilnikova O.M., Healey S., Neuhausen S.L., Ding Y.C., Rebbeck T.R., Weitzel J.N., Lynch H.T., Ontario Cancer Genetics Network. SWE-BRCA. HEBON. EMBRACE. GEMO. GEMO. Breast Cancer Family Registry. kConFab. CIMBA Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klevebring D., Lindberg J., Rockberg J., Hilliges C., Hall P., Sandberg M., Czene K. Exome sequencing of contralateral breast cancer identifies metastatic disease. Breast Cancer Res. Treat. 2015;151:319–324. doi: 10.1007/s10549-015-3403-6. [DOI] [PubMed] [Google Scholar]
- 19.Begg C.B., Ostrovnaya I., Geyer F.C., Papanastasiou A.D., Ng C.K.Y., Sakr R.A., Bernstein J.L., Burke K.A., King T.A., Piscuoglio S. Contralateral breast cancers: Independent cancers or metastases? Int. J. Cancer. 2018;142:347–356. doi: 10.1002/ijc.31051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson D., Easton D. The genetic epidemiology of breast cancer genes. J. Mammary Gland Biol. Neoplasia. 2004;9:221–236. doi: 10.1023/B:JOMG.0000048770.90334.3b. [DOI] [PubMed] [Google Scholar]
- 21.van den Broek A.J., van ’t Veer L.J., Hooning M.J., Cornelissen S., Broeks A., Rutgers E.J., Smit V.T., Cornelisse C.J., van Beek M., Janssen-Heijnen M.L. Impact of Age at Primary Breast Cancer on Contralateral Breast Cancer Risk in BRCA1/2 Mutation Carriers. J. Clin. Oncol. 2016;34:409–418. doi: 10.1200/JCO.2015.62.3942. [DOI] [PubMed] [Google Scholar]
- 22.Kuchenbaecker K.B., McGuffog L., Barrowdale D., Lee A., Soucy P., Dennis J., Domchek S.M., Robson M., Spurdle A.B., Ramus S.J. Evaluation of Polygenic Risk Scores for Breast and Ovarian Cancer Risk Prediction in BRCA1 and BRCA2 Mutation Carriers. J. Natl. Cancer Inst. 2017;109:109. doi: 10.1093/jnci/djw302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azim H.A., Jr., Michiels S., Bedard P.L., Singhal S.K., Criscitiello C., Ignatiadis M., Haibe-Kains B., Piccart M.J., Sotiriou C., Loi S. Elucidating prognosis and biology of breast cancer arising in young women using gene expression profiling. Clin. Cancer Res. 2012;18:1341–1351. doi: 10.1158/1078-0432.CCR-11-2599. [DOI] [PubMed] [Google Scholar]
- 24.Anders C.K., Hsu D.S., Broadwater G., Acharya C.R., Foekens J.A., Zhang Y., Wang Y., Marcom P.K., Marks J.R., Febbo P.G. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J. Clin. Oncol. 2008;26:3324–3330. doi: 10.1200/JCO.2007.14.2471. [DOI] [PubMed] [Google Scholar]
- 25.Ho W.K., Tan M.M., Mavaddat N., Tai M.C., Mariapun S., Li J., Ho P.J., Dennis J., Tyrer J.P., Bolla M.K. European polygenic risk score for prediction of breast cancer shows similar performance in Asian women. Nat. Commun. 2020;11:3833. doi: 10.1038/s41467-020-17680-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this manuscript may be requested through the original providers. Data of the Breast Cancer Association Consortium may be requested for non-profit research through an application procedure with the Breast Cancer Association Consortium. Data of the UK Biobank needs to be requested through UK Biobank.