Abstract
Background
The intestinal microbiota play a key role in the onset, progression, and recurrence of Crohn disease (CD). Most microbiome studies assay fecal material, which does not provide region-specific information on mucosally adherent bacteria that directly interact with host systems. Changes in luminal oxygen have been proposed as a contributor to CD dybiosis.
Methods
The authors generated 16S rRNA data using colonic and ileal mucosal bacteria from patients with CD and without inflammatory bowel disease. We developed profiles reflecting bacterial abundance within defined aerotolerance categories. Bacterial diversity, composition, and aerotolerance profiles were compared across intestinal regions and disease phenotypes.
Results
Bacterial diversity decreased in CD in both the ileum and the colon. Aerotolerance profiles significantly differed between intestinal segments in patients without inflammatory bowel disease, although both were dominated by obligate anaerobes, as expected. In CD, high relative levels of obligate anaerobes were maintained in the colon and increased in the ileum. Relative abundances of similar and distinct taxa were altered in colon and ileum. Notably, several obligate anaerobes, such as Bacteroides fragilis, dramatically increased in CD in one or both intestinal segments, although specific increasing taxa varied across patients. Increased abundance of taxa from the Proteobacteria phylum was found only in the ileum. Bacterial diversity was significantly reduced in resected tissues of patients who developed postoperative disease recurrence across 2 independent cohorts, with common lower abundance of bacteria from the Bacteroides, Streptococcus, and Blautia genera.
Conclusions
Mucosally adherent bacteria in the colon and ileum show distinct alterations in CD that provide additional insights not revealed in fecal material.
Keywords: Crohn disease, mucosally adherent microbiota, microbiome, IBD, postoperative CD
INTRODUCTION
Loss of tolerance to microbiota by the intestinal immune system is central to the onset and progression of Crohn disease (CD). In CD patients, disease-associated alterations in microbial composition, referred to as dysbiosis,1-3 involve the loss of microbial diversity; reduced abundance of commensal bacteria including those within the Firmicutes phylum, such as Faecalibacterium prausnitzii and the Clostridium groups IV and XIVa, and those within the Bacteroides genus; and concomitant increased abundance of species within the Enterobacteriaceae family, in particular Escherichia coli.4, 5 High interindividual variation in intestinal microbiota composition6 has limited the identification of a consistent microbial signal associated with CD and its heterogenous range of clinical phenotypes.7-12
Bacterial taxa highlighted as having decreased relative abundance in CD tend to be obligate anaerobes, especially Firmicutes, and taxa having increased abundance tend to be facultative anaerobes and aerobes, such as Enterbactericeae.5, 13 Thus, researchers have proposed that changes in intestinal luminal oxygen levels contribute to dysbiosis in CD, referred to as the “oxygen hypothesis.” 14 Healthy colonic and distal ileal lumen are nearly depleted of oxygen with slightly decreasing oxygen gradients present from the small to the large intestine and from intestinal tissue to the lumen.15 Accordingly, both the colon and ileum are dominated by the presence of obligate anaerobes.16 Within colonic epithelial cells, luminal oxygen is normally consumed through aerobic respiration. Increased oxygen levels in mucosal inflammation may promote or result from altered energy metabolism with a shift toward anerobic glycolysis.17-19 Recent studies have explored the oxygen hypothesis by computationally modeling multispecies biofilms, which have shown that even micromolar oxygen concentrations within the gut could induce the dysbiosis commonly seen in IBD.20 These data suggest that CD may result in an overall shift in the relative proportion of anaerobes and aerobes.
Few microbial studies in CD have been performed using samples from the colonic mucosa7, 21-23 and even fewer using samples from the ileal mucosa.7, 23 The majority have focused on fecal samples,7, 24-26 which can be limited because of (1) distinct microbial composition in feces compared with the mucosa,27 where microbes are more likely to contribute to the immune response that leads to inflammation; 28, 29 and (2) an inability to characterize microbial variation along segments of the gastrointestinal tract, which vary in microbial composition, load, and metabolic function.6, 30, 31
In this study, we used 16S rRNA sequencing to investigate differences in mucosally associated bacteria in the ileum and colon of CD patients and non-inflammatory bowel disease (non-IBD) control patients. Our results confirmed the reduction of diversity in both the colon and the ileum in CD, but we found that disease affected each intestinal segment differently. For instance, previously identified disease-linked changes in the relative abundance of Klebsiella and Escherichia/Shigella were seen in the ileum but not in the colon. Overall, bacterial alterations associated with CD focused on shifts in relative abundances of obligate and facultative anaerobes, potentially reflecting altered luminal oxygen levels. Surprisingly, overall proportions of obligate anaerobes did not change in the colon and actually increased in the ileum in CD, which did not support what may be predicted by the oxygen hypothesis. Notably, in both the colon and ileum, there was a significant increase in certain obligate anaerobes, including Bacteroides fragilis. Traditional analysis of differential bacterial abundance is challenged by interindividual variability. Thus, we identified functional bacterial groups that seemed uniquely present in high abundance in CD patients or non-IBD control patients, again mostly obligate anaerobes, providing an alternative accounting of CD-associated bacteria. Finally, using 2 independent cohorts, we found similar genera within the resected ileum in both sets of patients that were significantly associated with recurrence of disease at the anastomosis within 1 year after surgical resection.
METHODS
Patient Population and Clinical Phenotyping
Mucosal biopsies from either the ascending colon or the distal ileum were obtained at the time of surgery from 125 adult patients with well-characterized CD (Supplemental Tables 1, 2) from 2 institutions: the University of North Carolina Multidisciplinary IBD Center (UNC; ileum and/or colon; 46 patients; institutional review board (IRB) approvals 10-0355, 14-2445, and 11-0359) and Washington University in St. Louis (WashU; ileum; 79 patients; IRB approval 201204075). Noninflamed samples from CD patients were from regions with no active inflammation based on review of Haemotoxylin and Eosin stained slides. Active disease with inflammation was defined by the presence of histologic features of acute inflammation, including neutrophilic infiltration of the crypt epithelium and crypt abscess formation. Biopsies from a site distant from any pathology and confirmed by histology to be unaffected at the time of surgery were also obtained from the ascending colon and distal ileum from patients without IBD (non-IBD; UNC; ileum and/or colon; 23 patients; Supplemental Table 1).
Clinical phenotype data for all patients were recorded at the time of tissue collection. No patients had been treated with antibiotic therapy for management of their CD within the 3 months before sample collection. However, all patients received prophylactic intravenous antibiotics within 30 minutes of incision and a standard bowel preparation the day before surgery. Disease recurrence was determined via Rutgeerts scoring32 with scores of i0 and i1 categorized as “no recurrence” and scores of i2, i3, and i4 scored as “recurrence” (Supplemental Tables 1, 2).
16S rRNA Gene Amplicon Library Construction
A total of 177 biopsies were submitted for 16S rRNA gene sequencing following methods previously described in Markle et al.33 Amplicons of ~300 base pairs were generated that spanned the V1 and V2 variable regions of the 16S rRNA gene using the 27F-YM (5′-AGAGTTTGATYMTGGCTCAG) and 338R (5′-TGCTGCCTCCCGTAGGAGT) primers.34 Polymerase chain reaction products were normalized using a SequalPrepTM kit (Invitrogen, Carlsbad, CA) and pooled, lyophilized, purified, and concentrated using a DNA Clean and Concentrator Kit (Zymo, Irvine, CA). Pooled amplicons were quantified using the Qubit Fluorometer 2.0 (Invitrogen). The pool was diluted to 4 nM and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 15 pM and spiked with 25% of the Illumina PhiX control DNA before loading the sequencer. Illumina paired-end sequencing was performed on the MiSeq platform with versions v2.4 of the MiSeq Control Software and the MiSeq Reporter, using a 600-cycle version 3 reagent kit.
16S rRNA Gene Paired-End Read Processing and Contaminant Filtering
A total of 23,692,571 nonhuman paired-end sequences were generated for 177 samples (average sample size: 133,856 sequences/sample; minimum sample size: 8126; maximum sample size: 591,504; NCBI GEO accession GSE147600). Bacterial composition for each sample was estimated using DADA2, recorded as amplicon sequence variants (ASVs).35 Taxonomic labels were assigned to ASVs by aligning to the Ribosomal Database Project bacterial genome database.36 Five hundred forty ASVs, constituting only 0.067% of all sequences, that could not be assigned at the phylum level were removed. To eliminate potential batch contaminants, ASVs specific to a single sequencing batch and in ≥15% of samples in that batch were removed. In addition, ASVs specific to a single collection center and in ≥15% of those samples were also removed. Relative abundance measures for all taxa in each sample were calculated (Supplemental Table 3).
Assignment of Aerotolerance Categories
Taxa were classified into one of 6 aerotolerance categories: obligate anaerobe, anaerobe, facultative anaerobe/microaerophilic, aerobe, obligate aerobe, and unknown (Supplemental Table 4). Category assignments were based on previously reported aerotolerance of taxonomic labels assigned to each ASV (Supplemental Table 4).
Identification of Highly Abundant Bacteria in Specific Cohorts
Bacteria with greater than 5% relative abundance in a tissue in a particular subset of patients were identified. To determine whether this high relative abundance was unique to that cohort, we compared these bacteria to the relative abundance in a related cohort, for example, in CD patients vs non-IBD patients. Those that were found to occur at less than 5% relative abundance in the second cohort and whose greatest relative abundance in the second cohort was 3-fold less than the greatest relative abundance in the initial cohort.
Statistical Analysis
Alpha diversity was estimated based on the Shannon diversity index using the DivNet R package.37 DivNet estimates the diversity of the ecological communities within a niche, such as the colon mucosa, using information from all samples in a given set of samples. The calculated diversity of the community of interest accounts for missing taxa, undersampled taxa, and oversampled taxa, accounting for differences in sequencing depth. Associations between therapy usage and disease behavior and location were calculated using the χ 2 test. Differential levels of individual aerotolerance categories were tested using the Wilcoxon rank-sum test. Overall aerotolerance profiles were tested using a 5-dimensional nonparametric 2-sample Cramer test. Differential taxa abundance meta-analysis was conducted using DESeq2 and LEfSe38, 39 on all phylogenetic levels. Global P values from combined DESeq2 and LEfSe analyses were constructed using the Fisher method, and a false discovery rate of 0.05 was applied. Predictive functional analysis was performed using PICRUSt2.40 Differential testing of functional pathways was conducted using LEfSe. Additional data analysis and visualization were performed using the phyloseq,41 ggplot2,42 and EnhancedVolcano R packages.
ETHICAL CONSIDERATIONS
The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice. The study protocol was approved by the IRB at UNC at Chapel Hill (approval numbers 10-0355, 14-2445, and 11-0359) and separately at Washington University in St. Louis (approval number 201204075). All patients provided written informed consent.
RESULTS
Location, Disease Activity and Behavior, and Therapy-Specific Bacterial Diversity in Non-IBD and CD Patients
Loss of bacterial diversity in the colonic mucosa observed across CD clinical phenotypes
Patient sample and clinical characteristics are summarized in Table 1. Colonic bacteria in noninflamed samples from 15 CD patients showed significant loss of diversity compared with samples from 16 non-IBD patients (P < 0.0001; Table 2). Unexpectedly, diversity increased in inflamed regions of CD colonic samples compared with noninflamed regions in unmatched samples (P < 0.00001). However, when we restricted samples to matched biopsies from 3 CD patients, there was no significant difference (P = 0.774). Noninflamed CD patient samples stratified by disease behavior [Montreal classifications: inflammation only (B1), presence of colonic strictures (B2) and presence of internal penetrating disease including fistulas (B3)] exhibited significantly lower diversity than non-IBD samples when individually tested for each behavior (P < 0.0001). Across disease behaviors, patients with penetrating disease had reduced diversity compared with those with colonic stricturing (P < 0.016), who in turn had reduced diversity compared with patients with inflammation only (P < 0.001). Patients with disease affecting both the ileum and the colon (ileocolitis) had significantly lower diversity than both non-IBD patients and patients with only colon involvement (colitis; P < 0.00001). Interestingly, diversity in colitis patients was significantly higher than in non-IBD patients (P = 0.011). Finally, we evaluated the effect of therapies administered within the 8 weeks before sample collection. Patients who received anti-tumor necrosis factor (anti-TNF) therapy or steroids had significantly higher diversity (P = 0.0001 and P = 0.002, respectively), and patients who used immunomodulators had significantly lower diversity (P < 0.0001) than those who did not undergo each therapy. We verified that therapy use was not associated with disease location or behavior and thus therapy associations were independent of these disease characteristics.
TABLE 1.
Summary of Patient Phenotypes
| UNC Non-IBD Patients (n = 23) | UNC CD Patients (n = 46) | WashU CD Patients (n = 79) | ||||
|---|---|---|---|---|---|---|
| Non-IBD Colon (n = 16) | Non-IBD Ileum (n = 9) | CD Colon (n = 21) | CD Ileum (n = 33) | CD Ileum (n = 79) | ||
| Pathology | Noninflamed | 16 | 9 | 15 | 25 | 79 |
| Inflamed | 0 | 0 | 9 | 24 | 0 | |
| Matched | 0 | 0 | 3 | 16 | 0 | |
| Race | Caucasian | 12 | 9 | 11 | 20 | 73 |
| African American | 4 | 0 | 4 | 4 | 5 | |
| Other | 0 | 0 | 0 | 1 | 1 | |
| Ethnicity | Not Hispanic | 13 | 8 | 9 | 16 | 18 |
| Hispanic | 1 | 1 | 2 | 0 | 1 | |
| Not reported | 2 | 0 | 4 | 9 | 60 | |
| Sex | Female | 11 | 7 | 13 | 13 | 48 |
| Male | 5 | 2 | 2 | 12 | 31 | |
| Agea, y | 56.7 (15.0) | 59.1 (19.0) | 34.6 (12.8) | 37.3 (14.7) | 38.0 (13.5) | |
| Disease behavior | Inflammation (B1) | N/A | N/A | 6 | 0 | 4 |
| Stricturing (B2) | N/A | N/A | 6 | 16 | 42 | |
| Penetrating (B3) | N/A | N/A | 3 | 9 | 33 | |
| Disease location | Colon-only (L1) | N/A | N/A | 1 | 4 | 39 |
| Ileum-only (L2) | N/A | N/A | 4 | 0 | 0 | |
| Colon and ileum (L3) | N/A | N/A | 10 | 21 | 40 | |
| Smoking status | Never | 9 | 5 | 8 | 12 | 36 |
| Current smoker | 3 | 1 | 3 | 5 | 17 | |
| Former smoker | 4 | 3 | 4 | 8 | 25 | |
| Recurrence | Unknown | 16 | 9 | 6 | 5 | 15 |
| No recurrence | N/A | N/A | 6 | 11 | 43 | |
| Recurrence | N/A | N/A | 3 | 9 | 21 | |
| 5-ASAs | No current use | 9 | 16 | 13 | 19 | 54 |
| Current use | N/A | N/A | 2 | 6 | 25 | |
| Immunomodulators | No current use | 9 | 16 | 8 | 16 | 46 |
| Current use | N/A | N/A | 7 | 9 | 33 | |
| Steroids | No current use | 15 | 8 | 5 | 17 | 50 |
| Current use | 1 | 1 | 10 | 8 | 29 | |
| anti-TNFs | No current use | 16 | 9 | 5 | 13 | 42 |
| Current use | N/A | N/A | 10 | 12 | 37 | |
aThe values in parenthesis are standard deviations.
5-ASAs indicate 5-aminosalicylates. B1, inflammation; B2, stricturing; B3, penetrating; L1, colon-only; L2, ileum-only; L3, colon and ileum.
TABLE 2.
Comparisons of Bacterial Diversity
| Increased Diversity | Decreased Diversity | |||
|---|---|---|---|---|
| Group (sample size) | Mean ± SD (95% CI) | Group (sample size) | Mean ± SD (95% CI) | P |
| Tissue and disease | ||||
| Colon non-IBD (16) | 2.52 ± 0.96 (2.04-2.99) | Colon CD (15) | 1.54 ± 2.56 (0.22-2.85) | <0.00001 |
| Ileum non-IBD (9) | 2.30 ± 1.29 (1.44-3.17) | Ileum CD (25) | 1.45 ± 1.2 (0.96-1.94) | 0.001 |
| Colon non-IBD (16) | 2.51 ± 0.44 (2.3-2.73) | Ileum non-IBD (9) | 2.34 ± 1.92 (1.07-3.62) | 0.128 |
| Colon CD (15) | 1.52 ± 1.16 (0.91-2.13) | Ileum CD (25) | 1.45 ± 0.95 (1.08-1.83) | 0.144 |
| Ileum CD—B2, B3 (25) | 1.44 ± 0.85 (1.1-1.77) | Colon CD—B2, B3 (9) | 1.33 ± 0.93 (0.71-1.96) | 0.084 |
| Inflammation | ||||
| Colon CD, inflamed (9) | 1.93 ± 0.48 (1.61-2.24) | Colon CD, noninflamed (15) | 1.51 ± 1.43 (0.76-2.26) | <0.00001 |
| Colon CD, inflamed, matched (3) | 2.74 ± 0.71 (1.93-3.55) | Colon CD, noninflamed, matched (3) | 2.7 ± 0.4 (2.25-3.15) | 0.774 |
| Ileum CD noninflamed (25) | 1.45 ± 1.45 (0.88-2.02) | Ileum CD, inflamed (24) | 1.4 ± 1.22 (0.89-1.91) | 0.029 |
| Ileum CD, –inflamed, matched (16) | 1.74 ± 0.24 (1.62-1.85) | Ileum CD, non–inflamed, matched (16) | 1.26 ± 1.24 (0.65-1.87) | <0.00001 |
| Disease behavior | ||||
| Colon non-IBD (16) | 2.51 ± 0.8 (2.12-2.91) | Colon CD—B1 (6) | 2.12 ± 2.3 (0.25-4.0) | <0.00001 |
| Colon non-IBD (16) | 2.51 ± 1.56 (1.73-3.28) | Colon CD—B2 (6) | 1.65 ± 2.3 (-0.23-3.53) | <0.00001 |
| Colon non-IBD (16) | 2.51 ± 0.52 (2.25-2.76) | Colon CD—B3 (3) | 1.36 ± 1.54 (-0.43-3.15) | <0.00001 |
| Colon CD—B1 (6) | 2.11 ± 0.76 (1.49-2.73) | Colon CD—B2 (6) | 1.63 ± 1.1 (0.73-2.53) | 0.001 |
| Colon CD—B1 (6) | 2.11 ± 0.61 (1.62-2.60) | Colon CD—B3 (3) | 1.35 ± 0.95 (0.25-2.44) | <0.00001 |
| Colon CD—B2 (6) | 1.62 ± 1.27 (0.59-2.65) | Colon CD—B3 (3) | 1.33 ± 0.62 (0.61-2.05) | 0.016 |
| Ileum non-IBD (9) | 2.29 ± 1.14 (1.54-3.05) | Ileum CD—B2 (16) | 1.68 ± 0.8 (1.27-2.09) | <0.00001 |
| Ileum non-IBD (9) | 2.28 ± 2.01 (0.93-3.63) | Ileum CD—B3 (9) | 1.22 ± 1.29 (0.35-2.08) | <0.00001 |
| Ileum CD—B2 (16) | 1.67 ± 1.2 (1.06-2.27) | Ileum CD—B3 (9) | 1.2 ± 1.05 (0.5-1.91) | <0.00001 |
| Colon CD—B2, colonic strictures only (4) | 1.82 ± 0.84 (0.98-2.67) | Ileum CD—B2, ileal strictures only (15) | 1.65 ± 1.2 (1.04-2.27) | 0.002 |
| Colon CD—B3 (3) | 1.33 ± 0.42 (0.85-1.81) | Ileum CD—B3 (9) | 1.18 ± 0.93 (0.55-1.81) | 0.037 |
| Disease location | ||||
| Colon CD colon-only (4) | 2.69 ± 0.72 (1.98-3.41) | Colon non-IBD (16) | 2.51 ± 0.76 (2.13-2.9) | 0.011 |
| Colon non-IBD (16) | 2.51 ± 0.52 (2.25-2.77) | Colon CD, ileum-colon (10) | 1.3 ± 2.47 (-0.27-2.86) | <0.00001 |
| Colon CD, colon-only (4) | 2.67 ± 0.26 (2.41-2.93) | Colon CD, ileum-colon (10) | 1.28 ± 1.52 (0.32-2.23) | <0.00001 |
| Ileum non-IBD (9) | 2.27 ± 0.72 (1.79-2.75) | Ileum CD, ileitis (4) | 1.35 ± 1.06 (0.29-2.42) | <0.00001 |
| Ileum non-IBD (9) | 2.31 ± 1.74 (1.14-3.47) | Ileum CD, ileum-colon (21) | 1.58 ± 1.83 (0.78-2.37) | <0.00001 |
| Ileum CD, ileum-colon (21) | 1.55 ± 1.19 (1.03-2.08) | Ileum CD, ileitis (4) | 1.35 ± 0.78 (0.58-2.12) | <0.00001 |
| Ileum CD, ileum-colon (21) | 1.57 ± 0.64 (1.3-1.84) | Colon CD, ileum-colon (10) | 1.29 ± 0.89 (0.73-1.84) | <0.00001 |
| Colon CD, ileum-colon, no B1 (7) | 1.57 ± 0.56 (1.15-1.98) | Ileum CD, ileum-colon, no B1 (21) | 1.56 ± 1.33 (0.99-2.13) | 0.93 |
| Medication usage—colon CD | ||||
| No immunomodulators (8) | 1.84 ± 0.91 (1.19-2.49) | Immunomodulators (7) | 1.37 ± 1.38 (0.32-2.41) | <0.00001 |
| Anti-TNFs (10) | 1.79 ± 1.49 (0.85-2.73) | No anti-TNFs (5) | 1.35 ± 0.42 (0.98-1.73) | <0.00001 |
| Steroids (10) | 1.66 ± 0.25 (1.49-1.82) | No steroids (5) | 1.39 ± 1.36 (0.16-2.61) | 0.002 |
| Medication usage—ileum CD | ||||
| No steroids (17) | 1.73 ± 1.32 (1.09-2.37) | Steroids (8) | 1.14 ± 0.49 (0.89-1.39) | <0.00001 |
| No anti-TNFs (13) | 1.65 ± 1.01 (1.1-2.2) | Anti-TNFs (12) | 1.46 ± 0.9 (0.94-1.98) | <0.00001 |
| No immunomodulators (16) | 1.54 ± 0.92 (1.08-2.0) | Immunomodulators (9) | 1.41 ± 0.87 (0.84-1.99) | 0.28 |
| Recurrence | ||||
| Colon CD, no recurrence (6) | 2.09 ± 0.73 (1.49-2.69) | Colon CD, recurrence (3) | 1.18 ± 0.55 (0.54-1.82) | 0.0007 |
| Ileum CD, no recurrence (11) | 1.68 ± 0.8 (1.21-2.15) | Ileum CD, recurrence (9) | 1.2 ± 0.6 (0.8-1.59) | <0.00001 |
Numbers of samples for each cohort are reported. Means, SD, 95% CI, and P values were calculated using DivNet (see Methods section).
CI indicates confidence interval. B1, inflammation; B2, stricturing; B3, penetrating.
Bacterial diversity also reduced in the ileal mucosa in CD
Noninflamed ileal mucosa samples from 25 CD patients also showed significant loss in bacterial diversity compared with samples from 9 non-IBD patients (P = 0.001; Table 2). In contrast to samples from the colon, samples from the inflamed mucosa exhibited significantly lower diversity than unmatched inflamed mucosa (P = 0.029). Interestingly, when we restricted our comparison to 16 matched samples, inflamed regions had higher diversity (P < 0.0001). Together with the results from the colon, the effect of inflammation on diversity is not clear, although our data provide some evidence for increased diversity because of inflammation.
Noninflamed samples from patients with ileal stricturing or with penetrating phenotypes had lower diversity compared with samples from non-IBD patients (P < 0.0001), with the penetrating phenotype again having lower diversity than the stricturing phenotype (P < 0.0001). No patient had inflammation-only disease. As with the colon, non-IBD patients had higher diversity than those with solely ileal involvement (ileitis; P < 0.0001) and patients with ileocolitis (P < 0.0001), but patients with ileitis had lower diversity than those with ileocolitis (P < 0.0001). In contrast to the colon, patients receiving anti-TNF therapy or steroids exhibited significantly lower, not higher, diversity in the ileum than patients not using those therapies (P < 0.0001 and P < 0.0001, respectively). Again, we verified the lack of an association between disease phenotypes and therapies administered.
Diversity across colonic and ileal mucosa not significantly different
Diversity between ileal and colonic samples was not significantly different in either non-IBD (P = 0.128; Table 2) or CD patients (P = 0.144). Patients with ileal stricturing or penetrating disease had significantly lower diversity in the ileum compared with the colon of patients with the same disease complication (P = 0.002 and P = 0.037, respectively). When we compared all patients with ileocolitis, we found that diversity in the ileal mucosa seemed to be significantly lower than in the colonic mucosa, but this was driven by colon samples from inflammation-only patients. When these samples were removed, the difference was no longer significant.
Ileal and Colonic Bacteria Aerotolerance Profiles in CD
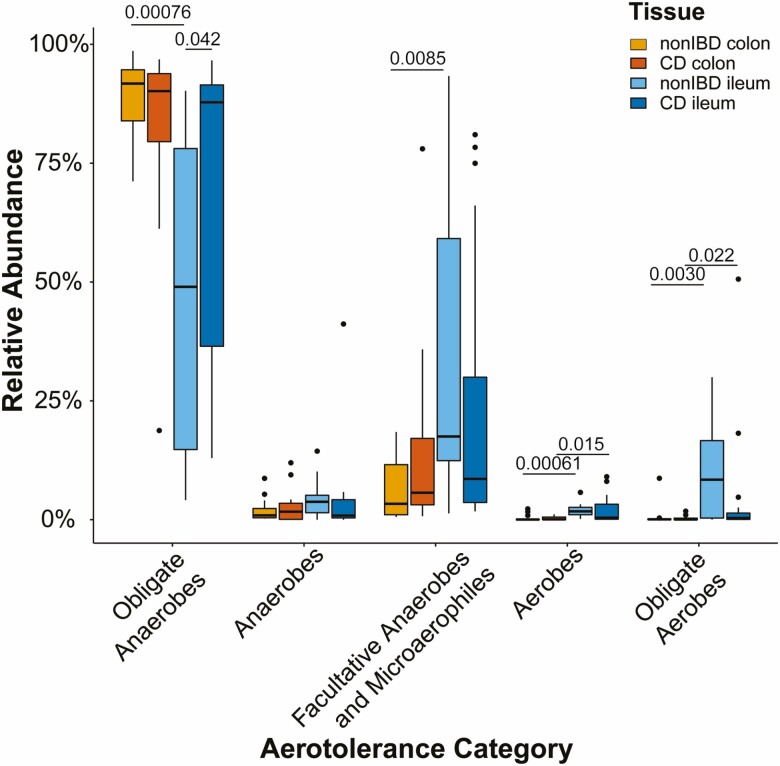
The oxygen hypothesis proposes that increased oxygen tension in the gut promotes dysbiosis in IBD patients.14 We hypothesized that higher levels of oxygen would result in CD patients having a decreased overall proportion of anaerobic species and an increased proportion of facultative anaerobic and aerobic species. To investigate whether our data supported this hypothesis, we generated aerotolerance profiles for each sample that reflected the relative abundance of bacteria in each category. As expected, obligate anaerobes were the dominant species in nearly all samples from the colon and most samples from the ileum in non-IBD patients (Fig. 1). We used these profiles to determine significant changes in proportions of bacteria from aerotolerance categories across cohorts.
FIGURE 1.
Ileal mucosa adopts a more colon-like aerotolerance profile in CD. Boxplots of the relative abundance of taxa assigned to 1 of 5 aerotolerance categories (obligate anaerobes, anaerobes, facultative anaerobes and microaerophiles, aerobes, obligate aerobes) in non-IBD colon (yellow), CD colon (orange), non-IBD ileum (light blue), and CD ileum (dark blue) mucosa. P values calculated based on Wilcoxon rank-sum test.
Negligible effect of CD on aerotolerance profiles in the colonic mucosa
We found no significant differences in the relative abundance of any individual aerotolerance category within colon samples from non-IBD and CD patients, including obligate anaerobes and facultative anaerobes (Fig. 1), or in the overall aerotolerance profile (P = 0.26). Upon investigating individual CD patient profiles, we did observe one patient with a significantly lower proportion of obligate anaerobes and a corresponding higher proportion of facultative anaerobes, consistent with the oxygen hypothesis, but all others maintained similar category levels as non-IBD patients (Supplemental Fig. 1). No differences in aerotolerance profiles associated with therapy usage or disease phenotype were found.
Relative abundance of obligate anaerobes increased in ileal mucosa in CD
Significantly higher relative levels of obligate anaerobes were found in the ileum of CD patients compared with non-IBD patients (P = 0.04), but the distribution across categories in the overall aerotolerance profile remained the same (P = 0.12; Fig. 1). Similar to the colon, there was no significant alteration in aerotolerance associated with disease behavior. Considering therapy usage, patients who recently received 5-aminosalicylate therapy showed a significantly higher relative abundance of obligate anaerobes compared with patients not undergoing that therapy (P = 0.01).
Aerotolerance profile in ileal mucosa becomes similar to colon in the presence of CD
In non-IBD patients, the ileum showed a significantly lower relative abundance of obligate anaerobes (P = 0.00076) and a significantly higher relative abundance of obligate aerobes (P = 0.003), aerobes (P = 0.00061), and facultative anaerobes (P = 0.0085) compared with the colon (Fig. 1). Overall aerotolerance profiles were significantly different as well (P = 0.001). We noted greater consistency in relative abundances across categories within colon samples compared with ileum samples (Supplemental Fig. 2). In the presence of CD, the ileal mucosa had a significantly higher relative abundance of aerobes (P = 0.015) and obligate aerobes (P = 0.022). However, the relative abundance of obligate and facultative anaerobes no longer differed between intestinal segments, and the composite aerotolerance profiles were not statistically different (P = 0.19; Fig. 1).
Compositional Shifts in Bacteria Corresponded to Differences in Diversity and Aerotolerance Profiles
Obligate anaerobes both increased and decreased in colonic mucosa in the presence of CD
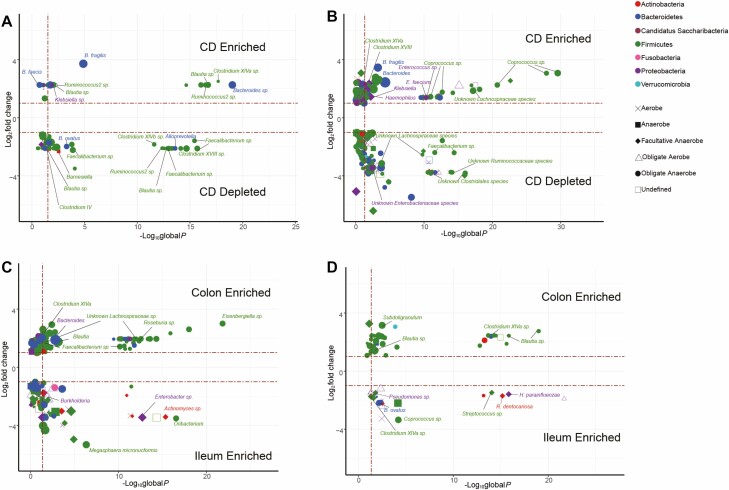
We first determined the differential relative abundance of individual bacterial taxa in the colon of the 15 CD patients and 16 nonIBD patients to better understand differences across intestinal location and disease phenotypes (Supplemental Tables 5–25). We also calculated the mean relative abundance (MRA) of the individual or collection of differential bacteria to provide a measure of the magnitude of the change and the potential impact on the total community. In the colon, the presence of CD was associated with a decreased abundance of 46 taxa (CD MRA = 0.012; non-IBD MRA = 0.065) of which 39, all obligate anerobes, were from the Clostridia class within the Firmicutes phylum, including 5 Faecalibacterium species (Fig. 2A, Supplemental Table 5). We found 15 taxa increased (CD MRA = 0.071; non-IBD MRA = 0.00015), again dominated by Firmicutes (11 taxa), of which 8 taxa were from the Lachnospiraceae family including 2 species each from the Blautia and Ruminococcus genera. The remaining 3 increased Firmicutes taxa were Klebsiella, Bacteroides faecis, and Bacteroides fragilis. As reflected in the mean relative abundance measures, even though there were a number of significant differences, together these bacteria encompassed less than 8% of the total relative abundance on average in these individuals.
FIGURE 2.
Obligate anaerobic taxa are predominantly differentially abundant between mucosal region and disease. Volcano plots of differential abundance meta-analysis data exhibiting that the presence of CD leads to the alterations in Bacteroidetes and Firmicutes taxa that are classified as obligate anaerobes in the colonic mucosa (A), with the relative of abundance of obligate anaerobic taxa increased in the presence of CD reflecting the aerotolerance analysis seen in the ileum (B). Within-disease non-IBD mucosa shows that the colonic mucosa is a signficantly more anaerobic environment compared to the ileal mucosa (C); this difference is greatly reduced in the presence of disease. Differential taxa abundance meta-analysis conducted using DESeq2 and LEfSe on all phylogenetic levels. Global P values from combined DESeq2 and LEfSe analyses constructed using Fisher method (FDR ≤ 0.05). Taxa with sp labels indicate that an exact species could not be ascertained.
Of note, B. fragilis was the most abundant obligate anaerobe on average within the CD patient group. Upon investigating abundance levels of B. fragilis in individual patients, we found this average statistic misleading because it was driven by extremely (8–18%) or nominally (0.8–5%) high abundance in 8 patients, with a complete absence of the species in the remaining 7 patients, highlighting the interindividual variability seen in these data. We note that in non-IBD patients, we detected B. fragilis in only 3 of the 16 patients, with a maximum abundance of 0.2%, thus explaining the significance of this comparison.
Because traditional differential analyses consider abundance across all samples, extreme intraclass variation often renders insignificant species differences characterized by large increased abundance in just 1 or a few samples.43 To provide an alternative view, we determined species with high relative abundance (>5%) in at least 1 CD sample that were not found in similar high abundance in any of the non-IBD samples (<5% and at least 3-fold less abundant). Based on this criteria, we found 20 species, 16 being obligate anaerobes, with high relative abundance in 1 or more CD patients (CD MRA = 0.21; non-IBD MRA = 0.018; Supplemental Table 26). These species accounted for >9% of the total abundance in 12 of the 15 CD patients, with >25% in 7 of these patients, whereas they accounted for <6% of the abundance in all 16 non-IBD patients, with <1% in 8 of these patients. Of these 20 species, 17 were not identified by traditional differential analyses including Bacteroides vulgatus and E. coli, previously associated with IBD.44B. vulgatus was found in high abundance in 2 CD samples (5.4% and 7.9%) and was at most 3.1% in non-IBD samples, and E. coli was found in nontrivial amounts in 4 CD samples (8.2%, 7.2%, 3.5%, and 1.1%) and <0.5% in all non-IBD samples. We likewise determined the species that were more highly abundant in non-IBD patients (Supplemental Table 27). Only 11 taxa, including species from the Bifidobacterium, Ruminococcus2, and Parabacteroides genera, were found, none of which were considered significantly different using the traditional analyses. These 11 taxa accounted for a more modest overall increased relative abundance in non-IBD patients (range = 1.6%-32%; 4 of 16 > 9%; mean = 0.09) compared with CD patients (range = 0%-6%; 7 of 15 < 1%; mean = 0.02).
We performed a predictive metagenomic analysis and found that overall, bacteria involved in biosynthetic pathways were enriched in CD and those involved in degradation pathways were depleted (Supplemental Table 28). Among these pathways, nucleic acid biosynthetic pathways were enriched and degradation pathways were depleted. Branched-chain and aromatic amino acid biosynthetic pathways were also enriched in CD patients, with other amino acid–related degradation pathways being depleted. Interestingly, the aerobic tricarboxylic acid (TCA) cycle was enriched, indicating a potential shift in energy metabolism.
Patients with colonic strictures or penetrating disease showed a decreased relative abundance of Alistipes putredinis and 4 Bacteroides species including Bacteroides ovatus, all from the Bacteroidetes phylum, compared with non-IBD patients (Supplemental Tables 9, 10). Patients with colonic strictures also had a decreased abundance of 15 Faecalibacterium species (strictures MRA = 0; non-IBD MRA = 0.027). Medication usage primarily altered species from the Firmicutes phylum, the majority from the Lachnospiraceae family. Anti-TNF therapy influenced the frequency of 76 taxa, with 13 taxa decreased (no anti-TNF MRA = 0.062; anti-TNF MRA = 0.00015) and 63 increased (no anti-TNF MRA = 0.0017; anti-TNF MRA = 0.083). Steroid usage influenced 69 taxa, with 20 taxa decreased (no steroids MRA = 0.074; steroids MRA = 0.0035) and 49 increased (no steroids MRA = 0.0000022; steroids MRA = 0.070). Immunomodulator usage influenced 68 taxa, with 22 taxa decreased (no immunomodulator MRA = 0.076; immunomodulator MRA = 0.00083) and 48 increased (no immunomodulator MRA = 0.0032; immunomodulator MRA = 0.14). These results show that bacteria associated with medication usage were not in high relative abundance, but those that increased were rarely found or were found in very small amounts on average when that medication was not being used (<0.5% in all cases).
Greater diversity of taxa altered in the ileal mucosa of CD patients
We compared 25 ileum samples from CD patients with 9 ileum samples from nonIBD patients. CD was associated with many more altered taxa than in the colon (159 vs 61) with a significantly increased abundance of 61 taxa from 4 phyla (CD MRA = 0.46; non-IBD MRA = 0.059; 1 Actinobacteria, 11 Bacteroidetes, 37 Firmicutes, 12 Proteobacteria) and a decreased abundance of 98 taxa from 6 phyla (CD MRA = 0.02; non-IBD MRA = 0.3; 4 Actinobacteria, 21 Bacteroidetes, 1 Candidatus Saccharibacteria, 50 Firmicutes, 21 Proteobacteria, 1 Verrucamicrobia; Fig. 2B). As in the colon, the number of decreased taxa in the ileum was much greater than the number of increased taxa, in line with the reduced diversity shown above. Unlike in the colon, note that in the ileum the increased taxa, including 27 from the highly abundant obligate anaerobic Lachnospiraceae family (CD MRA = 0.138; non-IBD MRA = 0.018) and the obligate anaerobic genera Bacteroides including B. fragilis (CD MRA = 0.027; non-IBD MRA = 0.00074), constituted a large proportion of the altered bacteria on average (46%) in the ileum in CD. We also saw an increased abundance of facultative anaerobes from the Proteobacteria phylum, including the known pathobiont Klebsiella and the pathogen Haemophilus, whereas no taxa from this phylum were differentially abundant in the colon. Taxa that decreased spanned 30 different families (CD MRA = 0.0014; non-IBD MRA = 0.025), with 12 Lachnospiraceae and 10 Ruminococcaceae taxa contributing the most.
Performing similar analyses as in the colon, we found 62 species in the ileum highly abundant in 1 or more CD patients, 17 of which were found using traditional analyses, that together constituted between 12.7% and 84% of the total abundance in each patient (mean abundance = 0.46; Supplemental Table 29). In 8 of the 9 non-IBD patients, these species represented <4% of the total abundance, with 1 outlier at 12.2% (mean = 0.036). These species were split fairly even across the Firmicutes, Bacteroidetes, and Proteobacteria phyla but were again dominated by obligate anaerobes (44 of 62). Interestingly, 10 of these highly abundant species in the ileum were also among the 20 highly abundant species in the colon of CD patients, all but 1 being obligate anaerobes. The 45 species not identified in the traditional differential analysis included E. coli and species from the Haemophilus and Bacteroides genera. In non-IBD patients, only 12 highly abundant species relative to CD patients were found, but these accounted for >18% of the abundance in 6 of 9 non-IBD patients (overall range = 0.9%-85%; mean abundance = 0.27; Supplemental Table 30) and accounted for <1% in 20 of 25 CD patients (range = 0%-3.5%; mean abundance = 0.0054). Of the 12 highly abundant species, only 5 were obligate anaerobes and 6 were facultative anaerobes. The imbalance in the number of high-abundance taxa found in CD and non-IBD ileum samples may partially reflect the number of samples in each group (25 CD, 9 non-IBD). But together, these results again help explain the overall increase in obligate anaerobes in the ileum of CD patients and emphasize the dramatic differences seen specifically in the ileum compared with the colon.
In contrast to the colon, metagenomic functional analysis indicated that pathways involving bacteria depleted in the ileum in CD patients were mainly biosynthetic pathways, including fatty acid biosynthesis and the biosynthesis of molecules involved in amino acid, glycogen, and carbohydrate metabolism. Similar to the colon, those involving bacteria enriched in CD patients were also biosynthetic pathways (Supplemental Table 31), including ones involved in the TCA cycle.
Patients with ileal strictures or penetrating disease shared 64 taxa that were significantly altered compared with non-IBD patients (Supplemental Tables 16, 17). These alterations included a decreased relative abundance of 53 taxa (stricture MRA = 0.010; penetrating MRA = 0.0022; non-IBD MRA = 0.21) and an increased abundance of 11 taxa (stricture MRA = 0.32; penetrating MRA = 0.28; non-IBD MRA = 0.046), reflecting the decreased diversity in these patients. When we compared patients with ileal strictures and those with penetrating disease, we found 76 taxa, primarily Firmicutes (48 taxa), significantly altered: 59 taxa were enriched in patients with ileal strictures (stricture MRA = 0.20; penetrating MRA = 0.026) and 17 taxa were enriched in penetrating disease (stricture MRA = 0.0043; penetrating MRA = 0.059). Altered taxa associated with medication usage were predominantly from the Lachnospiraceae family, similar to the colon (Supplemental Tables 19–22).
Colonic and ileal mucosa exhibited region-specific alterations in CD
In non-IBD patients, we found 178 significantly altered taxa between the ileum and the colon, emphasizing the unique communities present in each tissue (Fig. 2C; Supplemental Table 23). The 83 taxa with higher abundance in the ileum (ileum MRA = 0.50; colon MRA = 0.04) were more diverse with respect to phylum (7 distinct phyla) and aerotolerance categories, corresponding to differences in aerotolerance profiles in the ileum and colon of nonIBD patients. The 95 taxa with higher abundance in the colon (ileum MRA = 0.19; colon MRA = 0.62) came from 4 phyla. Within the Firmicutes phylum, all but 5 were from the Clostridia class, predominantly obligate anaerobes. Overall, we noted that these differences were for taxa that encompassed a large relative proportion of bacteria in each tissue. When alternatively determining highly abundant species in the colon or ileum of non-IBD patients, we found relatively few species that met our criteria (16 in colon, Supplemental Table 32; 13 in ileum, Supplemental Table 33), likely reflecting the higher diversity in both tissues with fewer individual species dominating the relative abundance. Imputed metagenomic functional analyses identified only 4 differential pathways (Supplemental Table 34), but this included the incomplete reductive TCA cycle enriched in the colon and the complete TCA cycle was enriched in the ileum.
Patients with CD showed far fewer altered taxa between ileal and colonic tissues than in non-IBD patients (178 vs 80), with 46 taxa increased in the colon (colon MRA = 0.13; ileum MRA = 0.029) and 34 taxa increased in the ileum (colon MRA = 0.0057; ileum MRA = 0.064). Those taxa enriched in the colon were dominated by obligate anaerobes from the Clostridia class, and those in the ileum again came from a wider range of phyla and aerotolerance categories (Fig. 2D; Supplemental Table 24). The mean relative abundance numbers indicate that these differences affected a smaller proportion of the total abundance in each tissue than in the non-IBD samples. We also noted that several individual taxa more abundant in the colon of non-IBD patients with a high colon MRA were no longer differential, including Bacteroides (non-IBD colon MRA = 0.20), Blautia (non-IBD colon MRA = 0.056), and Clostridium XIVa (non-IBD colon MRA = 0.033). In fact, B. ovatus switched from an increased abundance in the colon of non-IBD patients to an increased abundance in the ileum of CD patients.
Possibly reflecting this trend, our alternative analyses only identified 10 species with high abundance specifically in the colon that accounted for <10% of bacteria in patients with CD, on average (Supplemental Table 35). We did find 45 species with high abundance primarily in the ileum (Supplemental Table 36), only 3 of which were previously found in high abundance in the ileum in non-IBD samples. These may partially reflect the difference in the number of patient samples (25 ileum, 15 colon). Metagenomic functional analyses identified only a single pathway involved with 4-aminobutanoate degradation enriched in the ileum (Supplemental Table 37). In patients with ileocolitis, the ileum showed an increased abundance of taxa from the Actinobacteria and Proteobacteria phyla and of B. ovatus, and a lower relative abundance of Alistipes finegoldii and a Subdoligranulum species compared with the colon. Altogether, these analyses suggest that the large number of compositional differences between the ileum and the colon in non-IBD patients is severly reduced in the presence of CD, with each tissue having potentially unique sets of bacteria that thrive in disease.
Bacterial Alterations Seen in Postoperative Recurrence Across 2 Cohorts
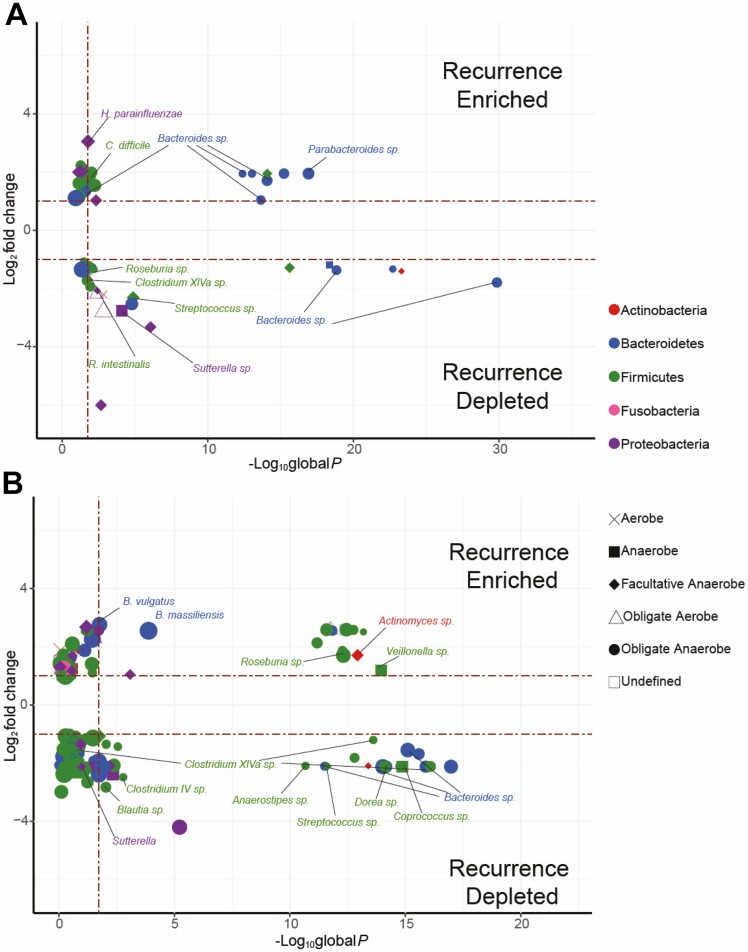
For 20 UNC CD patients, the recurrence of disease was endoscopically determined within 12 months postsurgery and scored using the Rutgeerts criteria32 (see Methods; Supplemental Table 1). Of these patients, 9 were found to have disease recurrence. Recurrent patients had significantly lower diversity than nonrecurrent patients in the resected colon (P = 0.007) and the ileum (P < 0.0001; Table 2). We did not find any significant differences in aerotolerance profiles in either tissue. Within the ileum, nonrecurrent patients had an increased relative abundance of 32 taxa (nonrecurrent MRA: 0.088; recurrent MRA: 0.0030) including 12 within the Lachnospiraceae family and 7 from the Bacteroidetes phylum (Fig. 3A; Supplemental Table 38).
FIGURE 3.
Similar and divergent alterations seen between cohorts in postoperative recurrence of CD. Volcano plots of differential abundance meta-analysis of alterations between patients with and without postoperative recurrence at UNC (A) and WashU (B) show little overlap between the 2 locations. Differential taxa abundance meta-analysis was conducted using DESeq2 and LEfSe on all phylogenetic levels. Global P values from combined DESeq2 and LEfSe analyses constructed using Fisher method (FDR ≤ 0.05). Taxa with sp labels indicate that an exact species could not be ascertained.
To determine the generalizability of these and other results, we independently generated data from the ileal mucosa of 79 CD patients undergoing resection at WashU (Table 1). We found that CD patients from WashU had a significantly lower diversity in the ileum than did patients from UNC, and that 272 taxa (86 Bacteroidetes, 102 Firmicutes, 84 Proteobacteria) were significantly differentially abundant between the 2 sites. The majority of the altered taxa were increased in WashU patients (257; UNC MRA = 0.33; WashU MRA = 0.71; Supplemental Table 39). Aerotolerance profiles from the 2 sites were not significantly different, indicating that intestinal bacterial communities were highly divergent across these geographically distinct populations but retained similar global aerotolerance properties.
Using the same criteria, we found that 21 WashU patients had disease recurrence and 43 did not (Supplemental Table 2). Similar to the patients at UNC, there was decreased diversity in recurrent patients at WashU. We found 42 taxa with decreased relative abundance (nonrecurrent MRA = 0.045; recurrent MRA = 0.0048) and 21 taxa with increased abundance (nonrecurrent MRA = 0.0080; recurrent MRA = 0.066; Fig. 3B; Supplemental Table 40). Comparing the 2 cohorts using traditional differential analysis, we noted some overlap at the genera level, such as Bacteroides, Streptococcus, and Sutterella being found in recurrent patients in both cohorts, although similar species within those genera were not identified. We found a number of species that satisfied our highly abundant designation between recurrent and nonrecurrent patients in each of the 2 cohorts (Supplemental Tables 41–44). We only found 1 common species, Haemophilus parainfluenzae, highly abundant in 1 or more in each of the UNC and WashU recurrent samples and 2 species, Bacteroides and Clostridium sensu stricto, in the nonrecurrent samples. Together, these analyses suggest that consistent taxa that defined recurrence did not exist in the UNC and WashU cohorts.
DISCUSSION
We present a study of bacterial alterations in the colonic and ileal mucosa of non-IBD and CD patients. Prior studies in healthy patients have shown increased bacterial load within the colon compared with the ileum45 along with compositional differences.30, 31, 46, 47 Although our data do not allow for the measure of total bacterial abundance, there were substantial differences in the bacterial communties within the ileum and colon in non-IBD patients, and these differential taxa accounted for the majority of the overall abundance in each tissue. These differences did not lead to a significant change in bacterial diversity but showed that in the absence of CD, each intestinal segment harbors distinct bacteria.
We examined aerotolerance profiles of mucosa-adherent intestinal bacteria to determine whether these provided evidence for the proposed oxygen hypothesis. The oxygen hypothesis posits that dysbiotic microbial shifts in CD from obligate anaerobes to facultative anaerobes and obligate aerobes results from a change in oxygen concentrations in the intestinal tract, or a “dysanaerobiosis.” 14 To establish a baseline, we found an increased abundance of obligate anaerobes in the colon compared with the ileum in non-IBD patients, in agreement with a study in mice that showed a decreasing oxygen gradient along the intestinal tract48 and a study in humans that showed lower ratios of obligate anaerobes in the ileum than in the colon.14 Interestingly, though, the presence of CD did not change the aerotolerance profile in the colon, and in the ileum, we actually saw an increased relative abundance of obligate anaerobes. This increased relative abundance of obligate anaerobes in the ileum was no longer significantly different from that in the colon of CD patients. Our differential analyses showed a decreased relative abundance of several obligate anaerobes and an increased abundance of several facultative anaerobes, as has been previously reported. We note that these taxa were of low relative abundance, in general, having small effects on the overall levels of these aerotolerance categories. These effects were offset by other taxa that were not consistently increased or dereased across samples and thus did not obtain significance in the differential analyses. These results motivated the second differential analyses we performed that identified taxa with high relative abundance in only 1 subset but just a few samples in that subset.
We note that this analysis is highly dependent on the accuracy of the aerotolerance classsifications available, which may change as new information is obtained on specific species. If in fact some key obligate anaerobes are found to be facultative, then the results of this type of analysis may change. In addition, some taxa could only be assigned to more general aerobe and anaerobe categories and others could not be assigned because of the lack of information on these taxa. We note that on average, taxa encompassing ~95% of the relative abundance of each patient could be assigned to one of the obligate anaerobe, faculatative anaerobe, or obligate aerobe categories, so the poorly annotated taxa were unlikely to significantly affect our results. Overall, these analyses do not necessarily refute the oxygen hypothesis, but they also do not support it and provide a novel way to examine this phenomenon. Studies to objectively measure oxygen concentration in the ileum and colon in the setting of CD are necessary to provide more direct evidence.
We found similar highly abundant obligate anaerobes that increased in CD patients in both tissues compared to non-IBD patients, such as B. fragilis, which was nearly 40 times more abundant in the ileum and >200 times more abundant in the colon of CD patients. This species, considered an obligate anaerobe in our study,49 has been associated with anaerobic infections and antibiotic resistance but is also among the most aerotolerant obligate anaerobes.49 It is possible that increased B. fragilis and other highly adaptable species do in fact reflect increased oxygen levels in CD, where instead of promoting the growth of facultative anaerobic and aerobic bacteria, the most oxygen-sensitive anaerobes are eliminated. Those anaerobes that better tolerate this increased oxygen environment become more relatively abundant and may explain the shift toward increased abundance of that aerotolerance category in the ileum in CD. 16S rRNA sequence data do not allow for calculation of total bacterial load. Thus, it is unclear whether more aerotolerant obligate anaerobes increased in total number or simply in relative abundance because of the loss of the more oxygen-sensitive anaerobes.
Our results support many previously reported associations in bacterial shifts between nonIBD and CD patients, although previous studies have primarily used fecal material. Diversity in the colon and ileum were lower in CD, similar to previous fecal and mucosa-adherent bacterial studies,5, 7, 50-52 and species from Escherichia/Shigella, Klebsiella, and Clostridium increased whereas species from Faecalibacterium and Lachnospiraceae decreased in disease. With data from both the colon and the ileum, we were able to investigate intestinal segment–specific changes and found an interesting dynamic in how bacterial composition changed in each location. For instance, increased B. fragilis and Escherichia/Shigella species and the loss of Faecalibacterium species were seen in both the colon and the ileum. Most changes were region-specific, though, such as the increased abundance of Proteobacteria within the ileum and the loss of B. ovatus within the colon. B. ovatus has been shown to cause serum antibody responses in IBD patients, but its pathogenesis is still unknown.53 Overall, however, disease affected substantially more taxa in the ileum, constituting a greater proportion of the total bacterial abundance in that tissue. These changes seemed to increase the similarity in bacterial composition between the ileum and the colon in CD, with a decreased number of differentially abundant taxa and increased similarity in their aerotolerance profiles. It is unclear whether these changes were because of the adoption of a common disease environment or because of the ability of similar taxa to survive and/or thrive in the presence of CD regardless of intestinal location.
Our data reflect the previously noted large interindividual variation in the gut microbiota, both between intestinal segments and in the presence of CD. Because of this heterogeneity, disease-associated bacteria may not be identified using more traditional differential analyses because of their high abundance in only a fraction of patients. We defined criteria to identify bacteria that were highly abundant in 1 or more patients in one stratification but not highly abundant in any patients in the comparative group. With this analysis, we showed that several species previously associated with CD that were not identified using traditional analyses had a high relative abundance in a few CD patients. The criteria we defined are somewhat arbitrary, and we acknowledge that not all species identified in this way will have disease relevance. However, we propose that analyses like these are necessary to find additional bacteria that may substantially contribute to CD pathogenesis and outcome in a patient subset, just not across a majority of patients. As with any such analyses, further mechanistic studies are required to validate the roles of specific bacteria in CD.
This study investigated the existence of reproducible signals associated with postoperative recurrence of CD within 1 year of surgery. We found significant differences in bacterial profiles across the 2 geographic locations in resected tissues which far outnumbered the differences in recurrent and nonrecurrent patients. In fact, between the 2 cohorts there were no common taxa associated with recurrence status. We also found a difference in bacterial diversity measures between the 2 cohorts, which may indicate that certain disease phenotypes that affect diversity may vary across cohorts. We did note that altered species in both groups came from similar genera: increased levels of species from Bacteroides, Blautia, and Streptococcus in recurrence. These results extend previous observations in small studies (6–13 patients) showing that decreased F. prausnitzii mucosal concentrations54 and altered mucosal bacterial profiles55 were associated with a risk of postoperative recurrence of CD. Moving forward, the challenge for finding predictors of postoperative recurrence in multicohort studies will be to unravel associations that are dependent on that disease phenotype while conditioning on batch effects, such as geography and other patient characteristics, that affect bacterial composition.
As with most clinical studies involving the human microbiota, our study has some limitations. Most important, the number of patients for which data were obtained was modest, especially when considering the interindividual heterogeneity and the number of CD phenotypes, which limited our power to detect significant meaningful differences for certain specific clinical characteristics. We acknowledge that in some of the results we report the sample sizes were very small, which must be considered when assessing the strength for evidence of significant differences. This is especially true for medication usage, where combinations of drugs may have different effects than when a drug is taken in isolation. Despite these limitations, we believe that our overall findings provide a baseline for further studies that will be better powered for more detailed phenotypic analyses.
In summary, we conducted a comprehensive region-specific characterization of the mucosa-association intestinal microbiota. We presented a novel approach to analyzing, and potentially functionally understanding, the intestinal microbiota from the perspective of aerotolerance. Furthermore, we provided evidence that the presence of CD is associated with region-specific alterations in bacterial composition that correspond to an increasing similarity of bacteria from major aerotolerance categories across intestinal segments.
Supplementary Material
Supported by: Helmsley Charitable Trust (SHARE Project 2), NIDDK P01 DK094779, NIDDK 1R01DK104828-01A1, and NIDDK P30-DK034987. The University of North Carolina Translational Pathology Laboratory is supported in part by grants from the National Cancer Institute (3P30CA016086). Biospecimens collected by the Washington University DDRCC are supported by NIDDK P30 DK052574.
Conflicts of interest: None declared.
REFERENCES
- 1. Eckburg PB, Relman DA. The role of microbes in Crohn’s disease. Clin Infect Dis. 2007;44:256–262. [DOI] [PubMed] [Google Scholar]
- 2. Elson CO, Cong Y, McCracken VJ, et al. . Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. [DOI] [PubMed] [Google Scholar]
- 3. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology. 2017;152:327–339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frank DN, Robertson CE, Hamm CM, et al. . Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huttenhower C, Gevers D, Knight R, et al. . Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gevers D, Kugathasan S, Denson LA, et al. . The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haberman Y, Tickle TL, Dexheimer PJ, et al. . Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kugathasan S, Denson LA, Walters TD, et al. . Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389:1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis RT, Maron DJ. Efficacy and complications of surgery for Crohn’s disease. Gastroenterol Hepatol. 2010;6:587–596. [PMC free article] [PubMed] [Google Scholar]
- 11. Pascal V, Pozuelo M, Borruel N, et al. . A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wright EK, Kamm MA, Teo SM, et al. . Recent advances in characterizing the gastrointestinal microbiome in Crohnʼs disease. Inflamm Bowel Dis. 2015;21:1219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lupp C, Robertson ML, Wickham ME, et al. . Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:119–129. [DOI] [PubMed] [Google Scholar]
- 14. Rigottier-Gois L. Dysbiosis in inflammatory bowel diseases: the oxygen hypothesis. Isme J. 2013;7:1256–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albenberg L, Esipova TV, Judge CP, et al. . Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2016;14:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Litvak Y, Byndloss MX, Bäumler AJ. Colonocyte metabolism shapes the gut microbiota. Science. 2018;362:eaat9076. doi: 10.1126/science.aat9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rivera-Chávez F, Lopez CA, Bäumler AJ. Oxygen as a driver of gut dysbiosis. Free Radic Biol Med. 2017;105:93–101. [DOI] [PubMed] [Google Scholar]
- 19. Rivera-Chávez F, Zhang LF, Faber F, et al. . Depletion of butyrate-producing clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe. 2016;19:443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henson MA, Phalak P. Microbiota dysbiosis in inflammatory bowel diseases: in silico investigation of the oxygen hypothesis. BMC Syst Biol. 2017;11. Article number 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank DN, St Amand AL, Feldman RA, et al. . Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Swidsinski A, Ladhoff A, Pernthaler A, et al. . Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. [DOI] [PubMed] [Google Scholar]
- 23. Tyler AD, Kirsch R, Milgrom R, et al. . Microbiome heterogeneity characterizing intestinal tissue and inflammatory bowel disease phenotype. Inflamm Bowel Dis. 2016;22:807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halfvarson J, Brislawn CJ, Lamendella R, et al. . Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2. Article number 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang S, Denman SE, Morrison M, et al. . Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034–2042. [DOI] [PubMed] [Google Scholar]
- 26. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. . Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zoetendal EG, von Wright A, Vilpponen-Salmela T, et al. . Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol. 2002;68:3401–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cobrin GM, Abreu MT. Defects in mucosal immunity leading to Crohn’s disease. Immunol Rev. 2005;206:277–295. [DOI] [PubMed] [Google Scholar]
- 29. Whitfield-Cargile CM, Cohen ND, Chapkin RS, et al. . The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes. 2016;7:246–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Aidy S, van den Bogert B, Kleerebezem M. The small intestine microbiota, nutritional modulation and relevance for health. Curr Opin Biotechnol. 2015;32:14–20. [DOI] [PubMed] [Google Scholar]
- 31. Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. [DOI] [PubMed] [Google Scholar]
- 32. Rutgeerts P, Geboes K, Vantrappen G, et al. . Predictability of the postoperative course of Crohn’s disease. Gastroenterology. 1990;99:956–963. [DOI] [PubMed] [Google Scholar]
- 33. Markle JG, Frank DN, Mortin-Toth S, et al. . Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. [DOI] [PubMed] [Google Scholar]
- 34. Frank JA, Reich CI, Sharma S, et al. . Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74:2461–2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Callahan BJ, McMurdie PJ, Rosen MJ, et al. . DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cole JR, Wang Q, Fish JA, et al. . Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Willis AD, Martin BD. DivNet: Estimating diversity in networked communities. bioRxiv. 2018:305045. doi: 10.1101/305045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15. Article number 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Segata N, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Douglas GM, Maffei VJ, Zaneveld J, et al. . PICRUSt2: an improved and extensible approach for metagenome inference. bioRxiv. 2019:672295. doi: 10.1101/672295. [DOI] [Google Scholar]
- 41. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wickham H. Ggplot2: elegrant graphics for data analysis. Accessed February 26, 2019. https://ggplot2.tidyverse.org/authors.html
- 43. Weiss S, Xu ZZ, Peddada S, et al. . Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5. Article number 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagao-Kitamoto H, Kamada N. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 2017;17:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morgan XC, Tickle TL, Sokol H, et al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13. Article number R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X, Heazlewood SP, Krause DO, et al. . Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J Appl Microbiol. 2003;95:508–520. [DOI] [PubMed] [Google Scholar]
- 48. Friedman ES, Bittinger K, Esipova TV, et al. . Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A. 2018;115:4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meehan BM, Baughn AD, Gallegos R, et al. . Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad Sci U S A. 2012;109:12153–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. Ilar J. 2015;56:192–204. [DOI] [PubMed] [Google Scholar]
- 51. Dicksved J, Halfvarson J, Rosenquist M, et al. . Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. Isme J. 2008;2:716–727. [DOI] [PubMed] [Google Scholar]
- 52. Willing B, Halfvarson J, Dicksved J, et al. . Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. [DOI] [PubMed] [Google Scholar]
- 53. Saitoh S, Noda S, Aiba Y, et al. . Bacteroides ovatus as the predominant commensal intestinal microbe causing a systemic antibody response in inflammatory bowel disease. Clin Diagn Lab Immunol. 2002;9:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sokol H, Pigneur B, Watterlot L, et al. . Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dey N, Soergel DA, Repo S, et al. . Association of gut microbiota with post-operative clinical course in Crohn’s disease. BMC Gastroenterol. 2013;13. Article number 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.