We read with great interest the paper by de Vries et al 1 which provides an interesting and unbiased characterisation of the immune contexture of colorectal cancer (CRC), the third most common cancer worldwide.2 By mass cytometric analysis together with single cell RNA sequencing, the authors identified several clusters of immune cells infiltrating CRC. They show that natural killer (NK) cells (identified as CD127−CD56+CD45RO+) are the prevalent innate lymphoid cell (ILC) population. NK cells are cytotoxic cells and can be distinguished from the non-cytotoxic CD127+ ‘helper’ ILC subsets (hILCs), which are specialised in the secretion of different sets of cytokines.3 Because of the low numbers of CD127+ hILCs, the authors did not characterise further hILC subsets. However, given their capacity to rapidly respond to environmental signals and pathogenic challenges, hILCs serve as important sentinels of mucosal tissue homeostasis.4 Therefore, their possible role both in CRC pathogenesis and in antitumour response has yet to be determined. Data on the ILC subset composition in CRC may allow improvement in therapeutic strategies for the control of this tumour by either suppressing or harnessing ILC function.
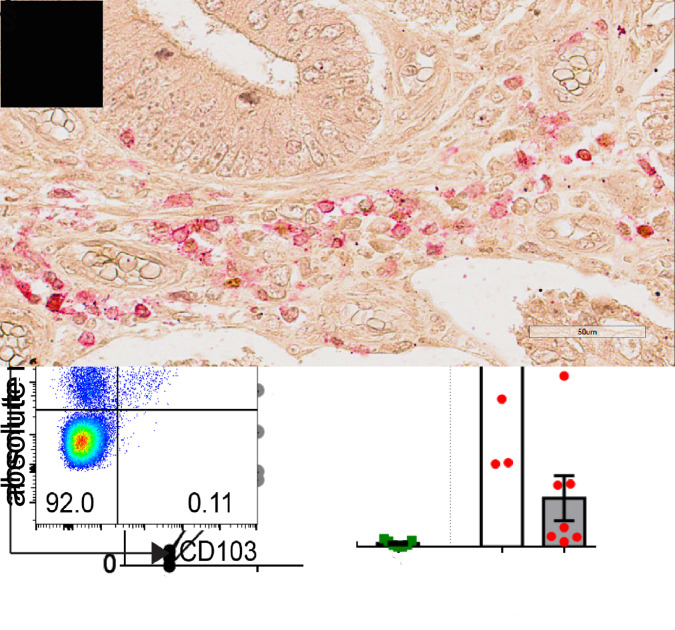
In this context, we analysed ILC subsets in samples from 33 resected primary CRC (online supplementary table 1) in comparison with matched normal mucosa (from the same patient), sampled 10 cm distant from the tumour (see online supplementary data). Single cell suspensions were analysed by multiparametric flow cytometry (online supplementary figure 1). No difference was found in the number of leukocytes (CD45+ cells) infiltrating the normal or tumour tissues (figure 1A). Among lineage (CD34, CD123, CD3, CD19, CD14) negative cells, we gated on CD56+CD127− cells (NK cells), and on both CD56+ and CD56- CD127+ cells (hILCs). While NK cell numbers were similar in the normal and tumour tissues (figure 1B), a significant decrease in the overall number and frequency of hILCs among the total innate lymphocytes was detected (figure 1B–E). In agreement with the conventional classification of ILCs,3 we further gated hILCs into CD117−CRTH2− (ILC1s), CD117bimodalCRTH2+ (ILC2s) and CD117+CRTH2− (ILC3s). As shown in figure 1D–F, the hILC subset composition was different between the normal and tumour samples. In the normal colon mucosa, ILC3s represented the most abundant subset, followed by ILC1s and ILC2s (the least represented subset among hILCs). In tumour samples, there was a marked reduction in total hILCs that was mainly reflecting a loss of interleukin (IL)-22-producing ILC3s (figure 1D–F). The ILC2 frequency was also low in tumour tissues, while the ILC1/ILC3 proportion was altered, reflecting both the decrease of ILC3s and the increase of ILC1s (figure 1D, E). Immunohistochemical analysis of epithelial crypts revealed higher numbers of infiltrating ILC3s (RORγt+CD3−) in the normal as compared with the tumour tissues (figure 1G). ILC1s are characterised by the secretion of the proinflammatory cytokines interferon (IFN)-γ and tumour necrosis factor-α,5 while ILC3s are thought to play a protective role against cancer as suggested by different studies.6 7 Thus, their sharp reduction, together with the increase of ILC1s (which may favour chronic intestinal inflammation), could play a role in malignant transformation/tumour progression. Notably, the altered ILC1/ILC3 balance may be, at least in part, dependent on the plasticity of ILC3s driven by the CRC microenvironment in which cytokines such as IL-1β, IL-15 and IL-12 may convert ILC3s into IFN-γ-producing ILC1-like cells.5 8 In addition, another subset of IFN-γ-producing ILC1 (NKp44+CD103+CD127−), called intraepithelial ILC1 and previously identified in intestinal lamina propria,9 was also enriched in the tumour as compared with the normal tissues (figure 1H, I).
gutjnl-2020-320908supp001.pdf (20.3KB, pdf)
gutjnl-2020-320908supp002.pdf (32.4KB, pdf)
gutjnl-2020-320908supp003.pdf (460.1KB, pdf)
Figure 1.
hILCs infiltrate CRC. After Percoll, single cell suspensions of normal and tumour colon biopsies from patients with CRC were stained and analysed by flow cytometry. (A, B) Absolute number of CD45+ cells (n=10), NK cells and hILC (CD127+) (n=8) per gram. (C, D) Proportion of NK cells (purple) and hILC (black) among live CD45+ cells and proportion of ILC1 (blue), ILC2 (green) and ILC3 (red) among hILCs (n=13). (E) Frequency of ILC1, ILC2 and ILC3 among live/CD45+/lin− (CD3, CD19, CD14, CD34, CD123) CD127+ cells (n=13). (F) Absolute number of ILC1, ILC2 and ILC3 (n=7) per gram. (G) Immunohistochemical analysis of normal and tumorous specimens from a patient with CRC using RORγt (brown) and CD3 (red). The arrows indicate RORγt+CD3− ILC3. Scale bars: 50 mm. (H) Absolute number of ieILC1 per gram (n=5) and (I) representative dot plots showing ieILC1s gated as live/CD45+/lin− (CD3, CD19, CD14) NKp44+CD103+ cells. CRC, colorectal cancer; hILC, helper ILC; ieILC1, intraepithelial ILC1; ILC, innate lymphoid cell; NK, natural killer; RORγt, RAR-related orphan receptor gamma. * p<0.05, ** p<0.01, *** p<0.001.
In conclusion, our data complement the study by de Vries et al,1 providing information on the hILC populations present in CRC tissues. Our data may offer a clue for therapeutic strategies, for example, harnessing ILC3s and/or targeting ILC1s. It will also be important to define whether altered ILC1/ILC3 proportions in CRC samples may have a prognostic value.
Footnotes
Contributors: PC designed the research, performed the experiments and revised the manuscript. PO performed the immunohistochemical experiments. LQ and NT analysed the data and wrote the manuscript. RV, RB and AP selected and processed the samples and revised the manuscript. SS provided samples from the patients. MCM critically revised the manuscript. LM wrote the manuscript. PV conceived and supervised the research and wrote the paper.
Funding: This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC)-Special Program Metastatic disease: the key unmet need in oncology 5 per mille 2018 Id 21147 (LM) and AIRC IG 2017 Id 19920 (LM); and Ministero della Salute RF-2013, GR-2013-02356568 (PV), RC-2020 OPBG (LM, PV) and RC 5x1000 (MCM). LQ has received funding from AIRC and from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no 800924. NT is recipient of a fellowship awarded by AIRC.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; internally peer reviewed.
References
- 1. de Vries NL, van Unen V, Ijsselsteijn ME, et al. High-Dimensional cytometric analysis of colorectal cancer reveals novel mediators of antitumour immunity. Gut 2020;69:691–703. 10.1136/gutjnl-2019-318672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brenner H, Chen C. The colorectal cancer epidemic: challenges and opportunities for primary, secondary and tertiary prevention. Br J Cancer 2018;119:785–92. 10.1038/s41416-018-0264-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vivier E, Artis D, Colonna M, et al. Innate lymphoid cells: 10 years on. Cell 2018;174:1054–66. 10.1016/j.cell.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 4. Atreya I, Kindermann M, Wirtz S. Innate lymphoid cells in intestinal cancer development. Semin Immunol 2019;41:101267 10.1016/j.smim.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 5. Bernink JH, Krabbendam L, Germar K, et al. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity 2015;43:146–60. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- 6. Carrega P, Loiacono F, Di Carlo E, et al. NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat Commun 2015;6:8280 10.1038/ncomms9280 [DOI] [PubMed] [Google Scholar]
- 7. Tumino N, Martini S, Munari E, et al. Presence of innate lymphoid cells in pleural effusions of primary and metastatic tumors: functional analysis and expression of PD-1 receptor. Int J Cancer 2019;145:1660–8. 10.1002/ijc.32262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cella M, Gamini R, Sécca C, et al. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol 2019;20:980–91. 10.1038/s41590-019-0425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuchs A, Vermi W, Lee JS, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 2013;38:769–81. 10.1016/j.immuni.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2020-320908supp001.pdf (20.3KB, pdf)
gutjnl-2020-320908supp002.pdf (32.4KB, pdf)
gutjnl-2020-320908supp003.pdf (460.1KB, pdf)