Abstract
Chronic alcoholism disrupts mitochondrial function and often results in alcoholic cardiomyopathy (ACM). Fas-activated serine/threonine kinase (FASTK) is newly recognized as a key post-transcriptional regulator of mitochondrial gene expression. However, the modulatory role of FASTK in cardiovascular pathophysiology remains totally unknown. In experimental ACM models, cardiac FASTK expression markedly declined. Ethanol directly suppressed FASTK expression at post-transcriptional level through NADPH oxidase-derived reactive oxygen species (ROS). Ethanol destabilized FASTK mRNA 3′-untranslated region (3′-UTR) and accelerated its decay, which was blocked by the clearance of ROS. Regnase-1 (Reg1), a ribonuclease regulating mRNA stability, was induced by ROS in ethanol-stimulated cardiomyocytes. Reg1 directly bound to FASTK mRNA 3′-UTR and promoted its degradation, whereas silencing of Reg1 reversed ethanol-induced FASTK downregulation. Compared to wild type control, alcohol-related myocardial morphological (hypertrophy, fibrosis and cardiomyocyte apoptosis) and functional (reduced ejection fraction and compromised cardiomyocyte contraction) anomalies were worsened in FASTK deficient mice. Mechanistically, FASTK ablation repressed NADH dehydrogenase subunit 6 (MTND6, a mitochondrial gene encoding a subunit of complex I) mRNA production and reduced complex I-supported respiration. Importantly, cardiomyocyte-specific upregulation of FASTK through intra-cardiac AAV9-cTNT injection mitigated myocardial mitochondrial dysfunction and restrained ACM progression. In vitro study showed that overexpression of FASTK ameliorated ethanol-induced MTND6 mRNA downregulation, complex I inactivation, and cardiomyocyte death, whereas these beneficial effects were counteracted by rotenone, a complex I inhibitor. Collectively, ROS-accelerated FASTK mRNA degradation via Reg1 underlies chronic ethanol ingestion-associated mitochondrial dysfunction and cardiomyopathy. Restoration of FASTK expression through genetic approaches might be a promising therapeutic strategy for ACM.
Keywords: Alcoholic cardiomyopathy, Fas-activated serine/threonine kinase, mRNA stability, Mitochondrial gene expression, Regnase-1
Graphical abstract

Highlights
-
•
Ethanol accelerates cardiac FASTK mRNA degradation via NOX/ROS/Regnase-1 pathway.
-
•
Genetic FASTK deletion exacerbates alcoholic cardiomyopathy (ACM) via repressing mitochondiral function.
-
•
Cardiac overexpression of FASTK protects against ACM.
1. Introduction
The harmful use of alcohol is one of the leading threats for population health worldwide and resulted in nearly 3 million deaths in 2016 [1]. Chronic alcoholism often results in devastating organ damage, such as alcoholic steatohepatitis and alcoholic cardiomyopathy (ACM). ACM accounts for nearly one third of dilated cardiomyopathy and 50% individuals with chronic alcoholism die from cardiovascular diseases [2]. Several theories have been postulated for the pathogenesis of ACM, such as direct and indirect cardiotoxicity of ethanol. However, neither precise etiology nor effective therapy is available [3].
Mitochondria play irreplaceable role in the generation of adenosine triphosphate (ATP) through oxidative phosphorylation, a process that requires five large protein complexes (complex I–V) located at the inner mitochondrial membrane [4,5]. The assembly of the electron transport chain complex I-IV requires nearly 100 proteins and most of them are encoded in the nucleus and imported into mitochondria from the cytoplasm [6]. 13 proteins are encoded within the mitochondrial genome and their proper expression is critical for the maintenance of mitochondrial respiratory function [7]. Mitochondria are the primary target for ethanol toxicity in cardiac myocytes and chronic ethanol ingestion results in gross structural and functional anomalies of the heart mitochondria in both laboratory animals and human subjects [8]. In ethanol-exposed hepatocytes and cardiomyocytes, proteins encoded by mitochondrial genome markedly decreased, suggesting that ethanol disrupts mitochondrial gene expression [9,10]. However, the molecular association between ethanol and mitochondrial genome expression remains largely obscure.
The mitochondrial genome relies heavily on post-transcriptional regulation for its proper expression, and deregulation of this process can result in mitochondrial dysfunction and diseases [11]. Fas-activated serine/threonine kinase (FASTK) is a founding member of a protein family consisting of FASTK and its five homologs FASTK domain-1 (FASTKD1) to FASTKD5 [12]. FASTK has initially been described as a protein kinase activated upon Fas signaling, but the arguments in favor of this kinase activity have been recently doubted and this molecule should no longer be considered as a kinase [13]. Emerging evidence has revealed that the FASTK family members are a group of RNA binding protein and act as key post-transcriptional regulators of mitochondrial gene expression [12]. Among the FASTK family members, FASTK, FASTKD1, FASTKD2, FASTKD3, and FASTKD4 co-localize with mitochondrial RNA granules and participate in mitochondrial RNA processing [[14], [15], [16]]. FASTK specifically interacts with NADH dehydrogenase subunit 6 (MTND6, a mitochondrial gene encoding a subunit of complex I) mRNA at multiple sites and promotes its maturation and biogenesis in mammalian cells [17]. Genetic ablation of FASTK results in aberrant MTND6 mRNA production and impairs cellular respiration with the reduced complex I activity [17].
In human beings and rodents, FASTK is highly expressed in the heart tissues [18]; however, the role of FASTK in cardiovascular diseases has never been explored. Considering the critical role of FASTK in the regulation of mitochondrial gene expression and function, we speculate that FASTK may participate in the pathogenesis of ACM. Utilizing in vivo and in vitro experiments, the present study aims: 1) to define whether FASTK involves in the progression of ACM; 2) if so, to explore the underlying mechanisms.
2. Materials and Methods
An expanded and detailed “Materials and Methods” section is available in the online supplementary materials.
2.1. Experimental animals and ACM models
Animal procedures were approved by the Animal Care and Use Committee of Air Force Medical University and were performed according to the National Institutes of Health Guidelines on Laboratory Animals. Transgenic mice with FASTK knockout (KO) were obtained from the Jackson Laboratory (Bar Harbor, Maine) and were maintained and genotyped as we and others described [19,20]. Mice were housed in a temperature-controlled environment (23 ± 2 °C) with a 12/12 h light/dark cycle. To establish cardiomyopathic models induced by alcohol, 12-week old male mice were pair-fed a modified Lieber-DeCarli alcohol or isocaloric maltose dextrin control liquid diet for 8 weeks as previously described [21]. Dietary ethanol content (%, w/v) was 4.8 (34% of total calories) at initiation, and gradually increased up to 5.4 (38% of total calories). The amount of food given to the pair-fed mice was matched to that of the ethanol-fed mice measured at the previous day. At the end of experimental period, cardiac function and blood pressure were measured. Mice were anesthetized by inhalation of 2% isoflurane and sacrificed. Heart and blood samples were harvested for the following examinations.
2.2. Myocardial gene delivery
Recombinant adeno-associated virus serotype-9 (AAV9) harboring full length mouse FASTK gene with the cardiac troponin T (cTNT) promoter (AAV9-cTNT-FASTK) and control vectors (AAV9-cTNT-GFP) were prepared and delivered as we previously described [22,23]. AAV9-cTNT-FASTK and AAV9-cTNT-GFP vectors were constructed by Hanbio Co, Ltd (Shanghai, China). pHBAAV-cTNT-FASTK-GFP was constructed by cloning the target FASTK into the pHBAAV-cTNT-GFP vector. After confirming the sequence, pHBAAV-cTNT-FASTK-GFP was cloned into the recombinant AAV9 frame vector. The AAV9 vector was amplified in HEK293 cells and the viral titer was determined. AAV9-cTNT-GFP vectors were concomitantly constructed and produced.
Intra-myocardial injection was used to deliver the gene as we previously described with minor modifications [23,24]. Briefly, mice were anesthetized using 2% isoflurane and a skin cut (about 1.5 cm) was made on the left chest. The pectoral muscle was dissected and the ribs were exposed. The heart was smoothly and gently “popped out” through a small hole at the 4th intercostal space. AAV9 vectors were diluted to 2.5 × 1011 particles/ml in saline and 25 μL was directly injected into the left ventricle free wall using a 30.5 G Hamilton syringe (Hamilton Co, Reno, NV, USA). The injection sites were as follows: (i) starting from the apex and moving toward the base in the left ventricle (LV) anterior wall; (ii) at the upper part of the LV anterior wall; and (iii) starting at the apex and moving toward the base in the LV posterior wall. After the injection, the heart was immediately placed back into the chest, followed by manual evacuation of pneumothoraxes, closure of muscle, and the skin suture. Two weeks after the injection, mice were randomized to receive control or ethanol-containing diet for the following eight weeks.
2.3. Statistical analysis
Data were shown as mean ± SD and analyzed by the GraphPad Prism 8 software (GraphPad Software). For the analysis of two groups, an unpaired two-tailed student's t-test was conducted. One-way ANOVA with a post-hoc analysis was performed when more than two groups were compared. For multiple groups following the same animals over-time, two-way ANOVA with repeated measures, followed by the Bonferroni post-hoc test was performed. A P < 0.05 was considered statistically significant.
3. Results
3.1. Ethanol suppresses myocardial FASTK expression through NADPH oxidase (NOX)-dependent reactive oxygen species (ROS) generation
Experimental ACM was induced in mice fed by the ethanol liquid diet for eight weeks. Age-matched mice pair-fed with a regular diet were set as the control group. Echocardiographic parameters and morphological analysis showed that ethanol-fed mice developed cardiomyopathic phenotypes characterized by contraction dysfunction, chamber dilation, myocyte hypertrophy, interstitial fibrosis, and lung edema due to cardiac insufficiency, mimicking the ACM clinical manifestations (figure-1a to 1f). To test the participation of FASTK family members in ACM progression, we first observed the mRNA levels of FASTK family members in myocardial tissues isolated from the control or ethanol-fed mice. The mRNA expression levels of FASTK obviously declined in the heart tissues of mice receiving the ethanol-containing diet (figure-1g). Western blot confirmed that chronic ethanol ingestion suppressed FASTK protein expression in the heart (figure-1h). These data identify myocardial FASTK decline as a molecular characteristic of ACM.
Figure-1.
Effects of chronic ethanol ingestion on cardiac function, structure, and FASTK expression. (a) Mice received ethanol-containing diet or were pair-fed with a regulator diet for eight weeks. Cardiac function was determined by m-mode echocardiography and ejection fractions, LVIDs as well as LVIDd were measured. (b) Representative images of gross structure, Masson trichrome staining and WGA staining were shown. (c) Quantitative analysis of average cardiomyocyte cross-sectional area. (d) Quantitative analysis of interstitial fibrotic area. (e) Heart weights and body weights were measured and HW/BW ratios were calculated. (f) Wet lung weights were measured and LW/BW ratios were calculated. (g) The mRNA levels of FASTK and FASTKD1 to FASTKD5 were measured by RT-PCR and normalized to β-actin levels. (h) The protein levels of FASTK were determined by Western blot and normalized to β-actin protein levels. Data are shown as mean ± SD and analyzed by unpaired student's t-test. LVIDs, left ventricular internal diameter at the end of systole; LVIDd, left ventricular internal diameter at the end of diastole; WGA, wheat-germ agglutinin; HW/BW, heart to body weight ratios; LW/BW, lung to body weight ratios.
To define whether ethanol directly suppresses FASTK expression, we exposed H9C2 cardiomyocytes to increasing concentrations of ethanol for 12 h. Consistent with in vivo observation, ethanol decreased FASTK mRNA and protein expression in a dose-dependent manner (figure-2a and 2b). We next defined the participation of NOX and its product cellular ROS in ethanol-induced FASTK decline. As expected, the expression of p47phox (a protein component of NOX2) and NOX activity were gradually upregulated when ethanol concentrations increased (figure-2c and 2d). Cellular ROS levels were increased in response to ethanol exposure (figure-2e). Meanwhile, ethanol dose-dependently upregulated mitochondrial ROS generation as evidenced by increasing fluorescence intensity of MitoSOX (figure-2f) [25,26]. Co-treatment with diphenyleneiodonium chloride (DPI, a NOX inhibitor) or N-acetyl-l-cysteine (NAC, a ROS scavenger) eliminated ROS accumulation in ethanol-stimulated cells (figure-2g). Notably, the decline of FASTK expression was reversed by either DPI or NAC co-treatment (figure-2h and 2i). These results confirm that ethanol represses FASTK expression through NOX-derived ROS production.
Figure-2.
Ethanol suppresses FASTK expression through NOX-dependent ROS generation. (a) H9C2 cardiomyocytes were exposed to different concentrations of ethanol for 12 h. FASTK mRNA was determined by RT-PCR and normalized to β-actin mRNA. (b-c) FASTK and p47phox protein expression levels were measured by Western blot and normalized to β-actin. (d) NOX activity was measured as methods described. (e) Intracellular ROS content was stained by DCFH-DA and fluorescence emission was measured. (f) H9C2 cells were exposed to 200 mM Ethanol for 12 h, co-treated with vehicle, DPI (a NOX inhibitor, 10 μM), or NAC (a ROS scavenger, 10 mM). ROS content was determined by DCFH-DA staining. (g-h) FASTK protein expression was determined and normalized to β-actin in indicated groups. Data are shown as mean ± SD and analyzed by one-way ANOVA analysis, followed by a Bonferroni post-hoc test. NOX, NADPH oxidase; ROS, reactive oxygen species; DCFH-DA, dichloro-dihydro-fluorescein diacetate; DPI, diphenyleneiodonium chloride; NAC, N-acetyl-l-cysteine.
3.2. Ethanol destabilizes FASTK mRNA 3′-UTR through the ribonuclease regnase-1
Considering that ethanol decreases FASTK expression at both the mRNA and protein level, we assumed that ethanol disrupts FASTK gene transcription. Thus, we detected the nascent synthesis of FASTK mRNA using a Click-iT nascent RNA capture system. To our surprise, compared with the control group, the nascent synthesis of FASTK mRNA remained unchanged upon ethanol exposure, no matter with or without NAC co-treatment (figure-3a). Next, the transcription inhibitor actinomycin D (ActD) was added into H9C2 cells to block de novo RNA synthesis, and the persistence of the existing FASTK mRNA levels were measured at different time points. Ethanol induced FASTK mRNA instability and promoted its degradation, which was rescued by NAC-mediated ROS clearance (figure-3b). To test which region is responsible for ethanol-induced instability, the 5′-UTR (untranslated region), coding region (CR), or 3′-UTR of mouse FASTK mRNA was separately inserted into luciferase reporter constructs. In H9C2 cells, ethanol decreased the luciferase activity of 3′-UTR containing construct and the phenomenon was counteracted by ROS elimination (figure-3c). In contrast, the luciferase activity of the construct carrying 5′-UTR or CR sequence was unchanged upon ethanol treatment (supplementary figure-1). These data collectively reveal that the sequence of 3′-UTR is responsible for ethanol-induced FASTK mRNA instability.
Figure-3.
Ethanol destabilizes FASTK mRNA 3′-UTR through Reg1. (a) Nascent RNA was labeled by 5-ethymyl uridine and isolated from NRVMs. Nascent FASTK mRNA levels were determined by RT-PCR. (b) ActD (2 μg/ml) was added to block RNA synthesis and FASTK mRNA levels were assessed by RT-PCR at indicated time points. (c) H9C2 cardiomyocytes were transfected with the indicated luciferase reporter vector and a Renilla reporter vector. Two days after transfection, cells were exposed to ethanol (200 mM) for 12 h, with or without NAC (10 mM) co-treatment. Luciferase activity was measured and normalized to Renilla activity. (d) Predicted stem-loop structure of mouse FASTK 3′-UTR (1753–1768). (e) Alignment of the conserved sequence in FASTK mRNA 3′-UTR among mammals. (f) H9C2 cells were transfected with WT Reg1 expression plasmid (Reg1-WT) or the empty control (Mock). The half-time of FASTK mRNA was measured after ActD treatment. (g) H9C2 cells were co-transfected with a series of luciferase reporter plasmids and the Reg1-WT or Mock plasmid. Two days after transfection, the luciferase activity was assessed. (h) H9C2 cells were transfected with WT or D141 N mutant Reg1 (D141N-Reg1) expression vector. RIP was performed using Reg1 antibody and enrichment of FASTK mRNA was measured by RT-PCR. (i) H9C2 cells were transfected with the luciferase reporter plasmid carrying FASTK 3′-UTR, co-transfected with mock, Reg1-WT, or Reg1-D141 N expression plasmid. Two days after co-transfection, the luciferase activity was determined. (j) H9C2 cells were exposed to ethanol (200 mM) for 12 h, co-treated with or without NAC (10 mM). Reg1 expression was determined and normalized to β-actin. (k) RIP assay determined Reg1 protein and FASTK mRNA combination. (l) H9C2 cells were transfected with scramble or Reg1 shRNA for two days, followed by ethanol (200 mM) treatment for 12 h. Reg1 and FASTK protein was normalized to β-actin. (m) H9C2 cells were transfected with scramble or Reg1 shRNA, co-transfected with the luciferase reporter vector carrying FASTK 3′-UTR. After exposure to ethanol (200 mM) for 12 h, the luciferase activity was measured. Data are shown as mean ± SD. Fig. 3a, b, 3c, 3h, 3i, 3j, 3k were analyzed by one-way ANOVA analysis, followed by a Bonferroni post-hoc test. Figure- 3f and 3 g were analyzed by unpaired student's t-test. Figure- 3l and 3 m were analyzed by two way ANOVA followed by a Bonferroni post-hoc test. ActD, actinomycin D; Reg1, regnase-1; WT, wild type; shRNA, short hairpin RNA; RIP, RNA immunoprecipitation.
Mouse FASTK 3′-UTR between 1755 and 1767 was predicted to form a stem-loop (SL) structure with a pyrimidine-purine-pyrimidine loop (UAU, figure-3d). This SL structure is consistent with the consensus recognition motif of regnase-1 (Reg1, encoding by Zc3h12a), a ribonuclease to regulate mRNA 3′-UTR stability and degradation [27]. This SL structure was also evolutionarily conserved among mammals, including human beings and chimpanzees (figure-3e). Biotin pulldown assay confirmed that Reg1 directly combined with the 3′-UTR but not the 5′-UTR or CR of mouse FASTK mRNA (supplementary figure-2). To determine whether Reg1 regulates FASTK mRNA stability, H9C2 cells were transfected with the expression plasmid of wild-type Reg1 (Reg1-WT). Intriguingly, Reg1 overexpression obviously accelerated FASTK mRNA degradation and shortened its half-life time upon ActD treatment (figure-3f).
To identify the region of 3′-UTR conferring Reg1 responsiveness, we built up a series of luciferase constructs containing a part of FASTK mRNA 3′-UTR sequence. The luciferase activity of the construct (1723–1841, full length) and the construct (1723–1768), which contains the SL sequence, was markedly decreased upon Reg1 overexpression (figure-3g). By contrast, the luciferase activity of the construct (1723–1754) was unchanged by Reg1 co-expression (figure-3g). These data identify that the SL structure (from 1755 to 1767) is indispensable for Reg1-mediated FASTK 3′-UTR decay. We also expressed WT or D141 N mutant Reg1 in H9C2 cells. D141 N mutant Reg1 lacks ribonuclease activity but retains RNA binding capacity [25]. RNA immunoprecipitation (RIP) assay showed that WT and D141 N Reg1 both combined with FASTK mRNA (figure-3h). However, the luciferase activity of FASTK 3′-UTR was markedly declined in the presence of WT but not D141 N mutant Reg1 (figure-3i), suggesting that Reg1 destabilizes FASTK mRNA 3′-UTR by its ribonuclease activity.
Another question is whether Reg1 participates in ethanol-induced FASTK mRNA instability. Ethanol induced Reg1 expression through promoting ROS generation because the ROS scavenger NAC reversed ethanol-induced Reg1 upregulation (figure-3j). Meanwhile, the combination between Reg1 and FASTK mRNA was also augmented by ethanol exposure, which was blocked by NAC co-treatment (figure-3k). Notably, Reg1 knockdown counteracted ethanol-induced FASTK decline and 3′-UTR instability (figure-3l and 3m). Reg1 silencing increased FASTK protein expression and 3′-UTR luciferase activity even under basal condition, supporting the notion that Reg1 is an endogenous regulator of FASTK mRNA 3′-UTR stability. Consistent with these in vitro observations, the expression of Reg1 was markedly increased in the heart tissues of ACM mice (supplementary figure-3). These results collectively demonstrate that ethanol accelerates FASTK mRNA 3′-UTR degradation via ROS-mediated upregulation of Reg1.
3.3. Genetic ablation of FASTK exacerbates cardiomyopathic phenotypes induced by chronic ethanol ingestion
An unsolved but very important question is whether FASTK involves in the progression of ACM. To test this, we established ACM models in WT and FASTK knockout (KO) mice. In the heart tissues isolated from KO animals, the mRNA and protein of FASKT were undetectable (supplementary figure-4), confirming the efficacy of FASTK KO. During the experimental period, WT and KO mice consumed equal ethanol diet per day and plasma ethanol and acetaldehyde levels were comparable between WT and KO mice receiving ethanol (supplementary table-1). Compared with the ethanol-WT group, heart to body weight ratios (HW/BW) and lung to body weight ratios (LW/BW) were markedly increased in ethanol-fed KO mice (supplementary table-1). Echocardiography showed that ethanol-induced cardiac dysfunction was worsened in KO mice as indicated by lower ejection fractions and higher left ventricular internal dimension systole (LVIDs) and diastole (LVIDd) values (figure-4a). Heart rhythm was simultaneously monitored by electrocardiogram when animals received echocardiographic examination. During the examination, premature ventricular contraction (PVC) was the only type of arrhythmia observed. Chronic ethanol ingestion induced PVC in both WT and KO mice; however, there was no significant difference in PVC incidence between WT and KO mice receiving chronic ethanol ingestion (supplementary table-1). Morphological analysis showed that FASTK deletion aggravated cardiac fibrosis, myocyte hypertrophy, and cardiomyocyte apoptosis induced by chronic ethanol ingestion (figure-4b to 4e). Molecular analysis showed that hearts isolated from KO mice displayed higher expression levels of fetal (Nppa, Nppb and Myh7) and pro-fibrotic genes (Col1a1 and Col3a1) upon chronic ethanol stress (figure-4f). Western blot results showed that cleaved caspase-3 levels in the heart tissues of KO mice were much higher than those in WT mice upon ethanol-containing diet (figure-4g). We also evaluated chronic cardiac inflammation in ethanol-fed WT and KO mice. Ethanol markedly induced the generation of inflammatory cytokines including tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and IL-1β (supplementary figure-5a to 5c). Ly6G positive neutrophils were also recruited into the heart by chronic ethanol intake (supplementary figure-5d). Unexpectedly, FASTK KO hearts were resistant to ethanol-induced chronic cardiac inflammation as evidenced by less cytokine production and lower monocyte infiltration than those of the WT group (supplementary figure-5a to 5d). Taken together, these results demonstrate that genetic ablation of FASTK worsens chronic ethanol ingestion-induced cardiomyopathy, even though the heart is less inflamed by ethanol.
Figure-4.
Effects of FASTK KO on ethanol-induced cardiomyopathic phenotypes. (a) WT or FASTK KO mice were fed with control or ethanol-containing diet for 8 weeks. Cardiac function was analyzed by m-mode echocardiography. Ejection fractions, LVIDs and LVIDd were calculated. (b) Representative images of masson trichrome, WGA and TUNEL staining in WT and KO mice. (c) Quantitative analysis of interstitial fibrosis. (d) Quantitative analysis of average cardiomyocyte cross-sectional area. (e) Quantitative analysis of TUNEL positive apoptotic cardiomyocytes. (f) Cardiac mRNA levels of fetal remodeling genes (Nppa, Nppb and Myh7) and fibrotic genes (Col1a1 and Col3a1) were measured by RT-PCR and normalized to 18S rRNA. (g) Cleaved caspase-3 expression levels were determined by Western blotting and normalized to β-actin expression. Data are shown as mean ± SD and analyzed by two way ANOVA followed by a Bonferroni post-hoc test. WT, wild type; KO, knockout; TUNEL, TdT-mediated dUTP nick-end labeling; Nppa, natriuretic peptide A; Nppb, natriuretic peptide B; Myh7, myosin, heavy polypeptide 7; Col1a1, collagen type I alpha 1; Col3a1, collagen type III alpha 1.
We isolated adult ventricular myocytes from WT and FASTK KO mice and exposed them to ethanol. Ethanol robustly suppressed cardiac myocyte contractile function as manifested by decreased peak shortening (PS) and maximal velocity of shortening/relengthening (±dL/dt) as well as prolonged time to 90% relengthening (TR90), without any changes in resting cell length and time to peak shortening (TPS). Ethanol-induced contractile anomalies were further exacerbated in KO myocytes, which was evidenced by lower PS, lower ± dL/dt, and longer TR90, although KO itself had no obvious effect on cardiomyocyte contractile properties (figure-5a to 5g). Intracellular Ca2+ handling was evaluated using the Fura-2 fluorescence microscopy. Upon ethanol exposure, cardiac myocytes exhibited overtly decreased Ca2+ release in response to electrical stimuli (ΔFFI) and prolonged intracellular Ca2+ decay with unchanged resting intracellular Ca2+ (resting FFI). Compared with WT group, KO myocytes displayed more severe Ca2+ handling dysfunction as evidenced by much lower ΔFFI values and slower Ca2+ decay in the presence of ethanol (Figure-5h to 5j). These in vitro data provide direct evidence demonstrating that genetic FASTK deficiency in cardiomyocytes exacerbates ethanol-induced contractile dysfunction and Ca2+ handling anomalies.
Figure-5.
Effects of FASTK KO on cardiomyocyte contractile properties and Ca2+homeostasis upon ethanol stimulation. (a) adult cardiomyocytes were isolated from WT or FASTK KO mice and were exposed to ethanol (200 mM) for 6 h and contractile properties were evaluated. Representative images of cardiomyocyte shortening trace were shown. (b) Baseline cell length. (c) Peak shortening, which was normalized to baseline cell length. (d) Maximal velocity of shortening (+dL/dt). (e) Maximal velocity of relengthening (−dL/dt). (f) Time-to–peak shortening (TPS). (g) Time-to-90% relengthening (TR90). (h) Resting fura-2 fluorescence intensity (FFI). (i) Electrically stimulated rise in FFI (ΔFFI). (j) Intracellular Ca2+ decay rate. Data are obtained from eighty cells from six mice (~10–15 cells per mouse) of each group and present as mean ± SD. Data are analyzed by one-way ANOVA, followed by a Bonferroni post-hoc test.
3.4. Genetic ablation of FASTK disrupts myocardial MTND6 gene expression and worsens mitochondrial complex I dysfunction upon chronic ethanol ingestion
FASTK is a key modulator of mitochondrial RNA processing, which is essential for proper mitochondrial gene expression and function. Thus, we evaluated the structure and function of the heart mitochondria isolated from WT and KO mice under basal condition. Electric microscopy showed that no significant change was observed in the number and ultrastructure of the heart mitochondrion between WT and KO mice (supplementary figure-6a). Consistent with the pre-existing report, the mRNA abundance of MTND6 (a mitochondrial gene encoding a subunit of complex I) was robustly decreased in the KO heart tissues, whereas the mitochondrial genes encoding the other complex I subunits were normally expressed (supplementary figure-6b). Western blot showed that the steady-state levels of some subunits of mitochondrial complex I, such as NDUF88, NDUFS1, and NDUFS2, were reduced in the KO heart (supplementary figure-6c), which confirmed a disassembly of complex I in the KO hearts.
Consistent with the decline of FASTK expression, ethanol also suppressed the mRNA levels of MTND6 in cardiomyocytes (figure-6a). Correspondingly, in response to chronic ethanol ingestion, the activity of mitochondrial complex I was repressed in the heart tissues (figure-6b), whereas the activity of complex II to IV remained unchanged (supplementary figure-7). These data support that mitochondrial complex I might be a primary target for ethanol-induced mitochondrial dysfunction. To direct evaluate the effect of ethanol on mitochondrial complex I-mediated respiratory function, heart mitochondria were freshly isolated from pair-fed or ethanol-fed animals. Oxygen consumption rate was measured in the presence of complex I substrate pyruvate and malate, with or without ADP stimulation. Chronic ethanol ingestion markedly interrupted basal (state 2) and ADP-stimulated maximal (state 3) complex I respiration (figure-6c). Intriguingly, KO further exacerbated ethanol-induced complex I inactivation, MTND6 repression, and complex I respiratory dysfunction (figure-6a to 6c). These results confirmed that FASTK participates in the modulation of mitochondrial MTND6 expression and complex I function during ACM progression.
Figure-6.
Effects of FASTK KO on mitochondrial respiratory function and MTND6 mRNA expression. (a) MTND6 mRNA levels in heart tissues were determined by RT-PCR. (b) Heart tissues were collected and mitochondrial complex I activity was measured. (c) Fresh heart mitochondria were collected and treated with complex I substrate (pyruvate and malate). Basal (State 2) and ADP-stimulated maximal (State 3) mitochondrial oxygen consumption rates were measured. (d) NMVMs were isolated from WT or FASTK KO mice and were treated by ethanol (200 mM) for 12 h. MTND6 mRNA levels were determined by RT-PCR. (e) WT and KO NMVMs were exposed to ethanol and real-time oxygen consumption rate was determined by Seahorse metabolic analyzer. (f) Basal and maximal respiration as well as spare respiratory capacity were calculated based on Seahorse results. Data are presented as mean ± SD and analyzed by two-way ANOVA, followed by a Bonferroni post-hoc test. NMVMs, neonatal mouse ventricular myocytes; MTND6, mitochondrial NADH dehydrogenase subunit-6.
Next, neonatal mouse ventricular myocytes (NMVMs) were isolated from WT or KO mice and were exposed to ethanol treatment. Consistent with in vivo observations, ethanol disrupted MTND6 mRNA expression (figure-6d). In KO myocytes, the suppressive effect of ethanol on MTND6 mRNA expression was further aggravated (figure-6d). The mitochondrial oxygen consumption was evaluated in the control or ethanol-treated WT or KO myocytes in real-time by the seahorse metabolic analyzer (figure-6e). Ethanol suppressed basal and maximal respiration as well as spare respiratory capacity in comparison to the control group (figure-6f). Under basal condition, basal and maximal respiration as well as spare respiratory capacity was moderately decreased in the KO myocytes when compared to WT. Notably, the mitochondrial respiration was sharply repressed upon ethanol exposure and were much lower than those of ethanol-treated WT myocytes (figure-6f). As above in vivo and in vitro results shown, genetic loss of FASTK expression worsens ethanol-induced deregulation of MTND6 expression, complex I function, and mitochondrial respiration.
3.5. Overexpression of FASTK ameliorates ethanol-induced deregulation of MTND6 expression and complex I function
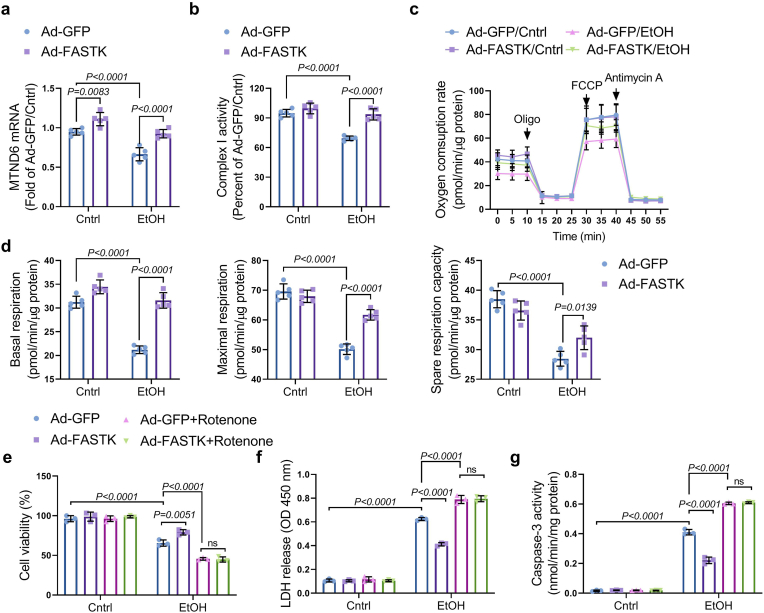
Above-mentioned results have demonstrated that myocardial FASTK deficiency contributes to mitochondrial dysfunction and ACM progression. We next defined the therapeutic potential of FASTK overexpression strategy for the management of ACM. First, we overexpressed FASTK in NMVMs via adenovirus (Ad) transfection. The efficacy of FASTK overexpression was confirmed by Western blot (supplementary figure-8). Compared with Ad vector carrying green fluorescent protein (Ad-GFP), Ad-FASTK significantly maintained MTND6 mRNA expression and restored mitochondrial complex I activity upon ethanol exposure (figure-7a and 7b). Seahorse metabolic analysis showed that FASTK overexpression ameliorated ethanol-mediated mitochondrial respiratory dysfunction (figure-7c and 7d). Collectively, these results further support the conclusion that FASTK deficiency underlies ethanol-induced mitochondrial respiratory dysfunction, and indicate that genetic overexpression of FASTK is an effective approach to ameliorate ethanol-related mitochondrial dysfunction. Furthermore, we observed that ethanol-induced cell death, lactate dehydrogenase (LDH) release, and caspase-3 activation were ameliorated in FASTK overexpressed myocytes (figure-7e to 7g). It is notable that protective effects of FASTK overexpression were totally blocked by co-treatment with rotenone, a potent complex I inhibitor (figure-7e to 7g). These results suggest that mitochondrial complex I activity is required for FASTK-mediated protective effects against ethanol toxicity. Although rotenone itself was non-toxic to NMVMs, ethanol-induced cell death was markedly aggravated by co-treatment with rotenone (figure-7e to 7g). Above-mentioned results emphasize a critical role of mitochondrial complex I activity for cardiomyocytes to resist ethanol toxicity.
Figure-7.
Effects of FASTK overexpression on mitochondrial respiration and MTND6 expression. NMVMs were isolated and cultured from WT mice and were transfected with adenovirus carrying GFP (Ad-GFP) or FASTK (Ad-FASTK), followed by ethanol stimulation for 12 h (a) MTND6 mRNA levels were determined by RT-PCR. (b) Complex I activity was measured. (c) Mitochondrial respiratory function was assessed by Seahorse. (d) Basal and maximal respiration as well as spare respiration capacity were calculated based on Seahorse results. (e) Ad-GFP or Ad-FASTK transfected NMVMs were exposed to ethanol for 12 h, co-treated with or without rotenone (0.1 μM). Cell viability was determined by MTT assay. (f) LDH concentrations in the supernatant were measured. (g) Caspase-3 activity was measured and normalized to intracellular protein content. Data are shown as mean ± SD and analyzed by two-way ANOVA, followed by a Bonferroni post-hoc test. GFP, green fluorescent protein; Ad, adenovirus; MTT, methylthiazolyldiphenyl-tetrazolium bromide; LDH, lactate dehydrogenase. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Cardiac specific overexpression of FASTK restrains ACM progression
We have explored the potential of FASTK overexpression strategy to resist ethanol toxicity in vitro. The concomitant question is how to overexpress FASTK in vivo to protect the heart against ACM. We used an adeno-associated virus of serotype 9 (AAV9) carrying FASKT gene under the control of the cardiomyocyte-specific cardiac troponin T (cTNT) promoter (AAV9-cTNT-FASTK). In vivo gene transfer of AAV9-cTNT-FASKT or negative control (AAV9-cTNT-GFP) was performed via intra-myocardial injection as we previously described [23,24]. Four weeks post intramyocardial AAV9-cTNT-GFP injection, the expression of GFP was specifically detected in the heart tissues, but not in the liver, lung, white adipose tissue, and skeletal muscle (figure-8a), suggesting that this method ensures in vivo cardiac specific gene delivery. Fourteen days after AAV9-cTNT-GFP or AAV9-cTNT-FASTK injection, mice were randomized to receive control or ethanol diet for the following eight weeks (figure-8b). Western blot confirmed that AAV9-cTNT-FASKT significantly elevated FASTK expression in the heart and reversed ethanol-induced FASTK downregulation (figure-8c). Daily ethanol consumption and plasma ethanol/acetaldehyde levels were comparable between AAV9-cTNT-GFP and AAV9-cTNT-FASTK group (supplementary table-3). Notably, upon chronic ethanol ingestion, the HW/BW and LW/BW were significantly reduced in AAV9-cTNT-FASKT group when compared to AAV9-cTNT-GFP group, suggesting that AAV9-cTNT-FASTK improved cardiac hypertrophy and lung edema (supplementary table-3). However, the incidence of PVC induced by chronic ethanol ingestion was not changed by AAV9-cTNT-FASTK injection (supplementary table-3). M-mode echocardiographic parameters showed that AAV9-cTNT-FASTK ameliorated ethanol-induced cardiac dysfunction as indicated by higher ejection fractions and lower LVIDs and LVIDd values (figure-8d). Structural analysis showed that cardiac specific overexpression of FASTK ameliorated ethanol-induced cardiac fibrosis, myocyte hypertrophy, and cardiomyocyte apoptosis (figure-8e). Molecular analysis showed that FASTK overexpression suppressed ethanol-induced remodeling and pro-fibrotic gene expression (figure-8f). Western blot revealed that FASTK overexpression repressed the activation/cleavage of caspase-3 induced by ethanol (Figure-8g). Consistent with in vitro observations, overexpression of FASTK rescued ethanol-induced MTND6 mRNA expression suppression (figure-8f) and ameliorated mitochondrial complex I inactivation (Figure-8h). Taken together, these data demonstrate that cardiac-specific upregulation of FASTK through gene transfer with AAV9 vectors bearing the cTNT promoter protects against ACM progression.
Figure-8.
Effects of cardiac-specific FASTK overexpression on ACM progression. (a) Saline or AAV9 vectors carrying GFP were delivered into WT mice by intra-myocardial injection. Twenty eight days post-injection, GFP expression in heart, liver, lung, kidney, WAT and skeletal muscle was detected by Western blot. (b) Schematic diagram of animal experiment: AAV9-cTNT-GFP or AAV9-cTNT-FASTK vectors were delivered into WT mice by intra-cardiac injection. Two weeks after injection, mice were randomized to receive control (pair-fed) or ethanol-containing diet for the following eight weeks. (c) Myocardial FASTK protein expression was determined by Western blot and normalized to β-actin. (d) Cardiac function was assessed by m-mode echocardiography and ejection fraction, LVIDs and LVIDd were calculated. (e) Representative images of Masson trichrome, WGA staining and TUNEL staining were shown. Interstitial fibrosis, myocyte cross-sectional area, and TUNEL positive apoptotic cardiomyocytes were quantified. (f) Cardiac mRNA levels of fetal genes (Nppb and Myh7), fibrotic genes (Col1a1 and Col3a1) and MTND6 were determined by RT-PCR. (g) Cleaved caspase-3 expression in heart tissues was determined by Western blot and normalized to β-actin expression. (h) The activity of complex I in fresh mitochondria isolated from heart tissues was measured and normalized to protein content. Data are present as mean ± SD and analyzed by two-way ANOVA, followed by a Bonferroni post-hoc test. AAV9, adeno-associated virus serotype-9; cTNT, cardiac troponin T.
4. Discussion
Excessive ethanol intake represses myocardial mitochondrial respiration and disrupts energy homeostasis in heart tissues, finally resulting in cardiac dysfunction, structural remodeling, and heart failure progression [28]. Several studies have documented that ethanol suppresses mitochondrial gene expression in cultured cardiomyocytes and hepatocytes, as evidenced by reduced protein expression encoded by the mitochondrial genome [9,10]. However, the relationship between ethanol and mitochondrial gene regulation is not well understood. Emerging evidence has revealed that FASTK and its family homologs are key post-transcriptional regulators of mitochondrial RNA processing and biogenesis, which is essential for the proper mitochondrial gene-encoding protein expression such as electron transporter chain (ETC) complex I to IV [12]. Utilizing in vivo and in vitro models, the present study for the first time addresses the crucial role of FASTK in the modulation of mitochondrial gene expression and function during ACM progression.
We observe that, among the six family members, FASTK expression specifically declines in the ethanol-challenged hearts and cardiomyocytes through NOX-mediated ROS generation. NOX-derived ROS has been implicated in the pathogenesis of ethanol cardiotoxicity [29]. Ethanol upregulates NOX and promotes ROS production through the protein kinase-c (PKC) β1-dependent fashion [21]. Co-treatment with the NOX inhibitor or the ROS scavenger alleviates ethanol-mediated cardiomyocyte apoptosis and restrains ACM pathologies [21]. Given that NOX-derived ROS is a direct culprit of ethanol cardiotoxicity, we determine the involvement of NOX and its product ROS in ethanol-induced FASTK decline. The decrease of FASTK expression upon ethanol exposure is totally reversed by the NOX inhibitor or the ROS scavenger, demonstrating that NOX-derived ROS is the major culprit of ethanol-induced FASTK insufficiency. NOX2 and NOX4 are induced in cardiac myocytes upon ethanol stimulation and they both contribute to ethanol-induced intracellular ROS generation [30]. DPI is a pan NOX inhibitor than can block both NOX2 and NOX4 activity. Therefore, the upregulation of FASTK induced by DPI treatment should be attributed to the inhibition of both NOX2 and NOX4 activity [31]. These results for the first time provide the evidence suggesting that the abundance of FASTK is strictly controlled by intracellular redox state, especially upon ethanol exposure.
We further identify that ethanol post-transcriptionally suppresses FASTK expression. Nascent mRNA capture analysis shows that the transcription of FASTK gene is unchanged in response to ethanol, although the expression of FASTK is declined at both mRNA and protein levels by ethanol exposure. Further analysis shows that the half-life time of FASTK mRNA is markedly shortened by ethanol, which is rescued by ROS clearance. Therefore, the insufficiency of FASTK in cardiomyocytes upon ethanol exposure is definitely due to the accelerated FASTK mRNA degradation induced by ROS. Utilizing luciferase reporter assay, we confirm that the 3′-UTR is the key element responsible for FASTK mRNA stability because ethanol reduces the luciferase activity of the construct carrying FASTK mRNA 3′-UTR, but not the CR or 5′-UTR. These data clarify a brand new post-transcriptional regulation of FASTK expression in mammalian cells, especially under the condition of oxidative stress.
Mammalian mRNA degradation is tightly regulated by RNA-binding proteins (RBP) that recognize specific RNA sequences or structures (generally named ‘elements’), which are often localized in the 3′-UTR of mRNAs [32]. By bioinformatics analysis, we find that there is a conserved SL structure with a pyrimidine-purine-pyrimidine loop, which is consistent with the consensus recognition motif of Reg1, a RBP that promotes mRNA degradation via its ribonuclease activity [33]. Reg1 is newly identified as a key regulator of proinflammatory cytokine mRNA stability in both immune and non-immune cells [34,35]. Here we find that Reg1 directly binds to FASTK mRNA 3′-UTR through the SL structure and accelerates its decay by the ribonuclease activity. Upon ethanol exposure, NOX-derived ROS upregulates Reg1 expression and promotes its binding to FASTK mRNA, thus resulting in the instability and accelerated degradation of FASTK mRNA. These data confirm that Reg1 is a previously unrecognized modulator of FASTK mRNA stability. Moreover, deregulation of Reg1 has been implicated in autoimmune, cancer, and cardiovascular diseases [36,37]. However, the genetic/environmental factors in the control of Reg1 expression remain unknown. This study finds that Reg1 expression is upregulated by ethanol-stimulated ROS generation, suggesting that the expression of Reg1 is regulated by intracellular redox homeostasis.
FASTK is recently recognized as a RBP to regulate mitochondrial RNA processing and biogenesis, which is essential for proper mitochondrial protein expression [12]. In the heart mitochondria, FASTK specifically binds to MTND6 mRNA and promotes its biogenesis [17]. Loss of FASTK results in a significant decline of MTND6 mRNA level and a subsequent repression of mitochondrial complex I activity [38]. Here we observe that ethanol disrupts MTND6 mRNA generation and represses mitochondrial complex I activity. Genetic ablation of FASTK further exacerbates ethanol-disturbed MTND6 mRNA expression and complex I activity, whereas ethanol-induced mitochondrial anomalies are rescued by FASTK overexpression. The loss-of-function and gain-of-function experiments establish a causative relationship between FASTK insufficiency and deregulated mitochondrial gene expression as well as function in the ethanol-challenged hearts and cardiomyocytes.
Mitochondrial genetic and functional homeostasis is essential to maintain normal cardiac physiology, and mitochondrial dysfunction contributes to the progression of cardiomyopathy and heart failure [[39], [40], [41]]. Complex I contains nearly 45 subunits and it is the first step of mitochondrial ETC to transfer electron from NADH to the ETC for ATP synthesis [42]. Decreased complex I activity has been observed in the cardiomyopathic and failing hearts [43]. Genetic loss of complex I-dependent respiration disrupts mitochondrial energy and redox homeostasis and renders the mice highly susceptible to the development of heart failure after chronic stress [43]. These data indicate that complex I is a critical regulator of the energy and redox balance in the heart mitochondria, which is indispensable for cardiac response to chronic stress. Compromised complex I activity has also been implicated in the pathogenesis of ACM [44]. However, the molecular relationship between ethanol ingestion and complex I inactivation is obscure. The present study confirms that FASTK insufficiency is a node point between ethanol intake, complex I inactivation, and mitochondrial respiratory dysfunction.
Chronic myocardial inflammation, such as neutrophil infiltration and cytokine production, has been implicated in the pathogenesis of ACM [45]. Emerging evidence has raised the possibility that ethanol-induced cardiac injury may occur secondary to inflammatory responses [46]. Here we show that FASTK KO mice were resistant to chronic cardiac inflammation upon chronic ethanol ingestion as evidenced by lower neutrophil recruitment and proinflammatory cytokine generation. These findings are consistent with the pre-existing report that genetic FASTK knockout eases lipopolysaccharides (LPS) or dust-induced lung inflammation [44]. Although chronic myocardial inflammation is alleviated, genetic deletion of FASTK markedly worsens ACM progression as evidenced by the functional, structural, and molecular analyses. These results confirm that FASTK KO influences ACM progression through a mechanism parallel to myocardial inflammatory responses.
This study also emphasizes that restoration of FASTK expression might be a promising therapeutic strategy for ACM. In vitro experiments show that overexpression of FASTK ameliorates ethanol-induced MTND6 insufficiency, complex I inactivation, and mitochondrial dysfunction, thus alleviating ethanol-induced cell death. Notably, these protective effects mediated by FASTK overexpression are counteracted by the complex I inhibitor rotenone, suggesting that FASTK alleviates ethanol cardiotoxicity probably through preserving complex I activity. Furthermore, cardiac specific overexpression of FASTK via intra-myocardial AAV9-cTNT injection restrains cardiomyopathic phenotypes in chronic ethanol ingested mice, highlighting the therapeutic potential of AAV9-mediated FASTK gene delivery against ACM. The current study is the first pre-clinical study pointing out the critical role of FASTK in the modulation of the heart mitochondrial homeostasis and heart diseases. Considering the necessity of heart mitochondria in the regulation of cardiovascular pathophysiology, we believe that FASTK has the potential to be an attractive therapeutic target for the management of heart diseases [47]. We previously showed that FASTK deletion protects against obesity-associated fatty liver injury through directly modulating hepatic lipogenesis and gluconeogenesis, which are two major metabolic processes involved in the progression of obesity and non-alcoholic fatty liver disease [19]. We believe that it is dependent on the difference of pathogenic mechanisms whether FASTK exerts protective or detrimental role in the different diseases.
In conclusion, the present study demonstrates that, in ethanol-challenged hearts and cardiomyocytes, NOX-derived ROS upregulates the ribonuclease Reg1, which binds to and accelerates FASTK mRNA degradation. In turn, downregulation of FASTK contributes to dysregulated MTND6 expression and compromised complex I activity, finally resulting in mitochondrial dysfunction and ACM progression (figure-9). These findings delineate a novel post-transcriptional mechanism by which ethanol disturbs the heart mitochondrial homeostasis during ACM progression.
Figure-9.
Schematic illustration. Ethanol stimulates NOX activation and ROS generation, therefore upregulating Reg1 expression. Reg1 directly binds to the 3′-UTR of FASTK mRNA and accelerates its decay, resulting in insufficient FASTK expression in chronic ethanol-challenged hearts. The decline of FASTK abundance contributes to impaired mitochondrial MTND6 biogenesis, complex I inactivation, and mitochondrial respiratory dysfunction, finally leading to the progression of alcoholic cardiomyopathy. Restoration of FASTK expression via genetic approaches might be a promising strategy for ACM prevention and treatment.
Conflicts of interest
None to declare.
CRediT authorship contribution statement
Fuyang Zhang: Formal analysis, Data curation, Writing - original draft, Writing - review & editing, performed the experiments, analyzed the data, conceived the study, drafted, and revised the manuscript. Kai Wang: performed the experiments. Shumiao Zhang: performed the experiments. Juan Li: performed the experiments. Rong Fan: performed the experiments. Xiyao Chen: Formal analysis, Data curation, Writing - original draft, Writing - review & editing, designed the study, analyzed the data, conceived the study, drafted, and revised the manuscript. Jianming Pei: Formal analysis, Data curation, Writing - original draft, Writing - review & editing, designed the study, analyzed the data, conceived the study, drafted, and revised the manuscript.
Declaration of competing interest
Authors declare no competing interests.
Data availability: All the data of this study are free to obtain in the paper or in the supplementary materials. Raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgement
This work was financially supported by the Program for National Science Funds of China (No. 81800306, 81800326, 8770243, 82070051 and 81800226), the China Postdoctoral Science Foundation Grant (No. 2017M613347), the Natural Science Basic Research Plan in Shaanxi Province of China (No. 2018JQ8062), and the Shaanxi Provincial Scientific and Technological Project (No. 2013K12-01-06).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101778.
Contributor Information
Xiyao Chen, Email: cxyfmmu@163.com.
Jianming Pei, Email: jmpei8@fmmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.reportGlobal Status Report on Alcohol and Health 2018, World Health Organization.
- 2.Adam A., Nicholson C., Owens L. Alcoholic dilated cardiomyopathy. Nurs. Stand. 2008;22(38):42–47. [PubMed] [Google Scholar]
- 3.Hantson P. Mechanisms of toxic cardiomyopathy. Clin. Toxicol. 2019 Jan;57(1):1–9. doi: 10.1080/15563650.2018.1497172. [DOI] [PubMed] [Google Scholar]
- 4.Sousa J.S., D'Imprima E., Vonck J. Mitochondrial respiratory chain complexes. Subcell. Biochem. 2018;87:167–227. doi: 10.1007/978-981-10-7757-9_7. [DOI] [PubMed] [Google Scholar]
- 5.Dorn G.W., 2nd, Vega R.B., Kelly D.P. Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 2015;29(19):1981–1991. doi: 10.1101/gad.269894.115. Oct 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Souza A.R., Minczuk M. Mitochondrial transcription and translation: Overview. Essays Biochem. 2018;62(3):309–320. doi: 10.1042/EBC20170102. Jul 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott M., Amunts A., Brown A. Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. Jun 2. [DOI] [PubMed] [Google Scholar]
- 8.Piano M.R. Alcohol's Effects on the cardiovascular system. Alcohol Res. 2017;38(2):219–241. [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy J.M. Mitochondrial gene expression is impaired by ethanol exposure in cultured chick cardiac myocytes. Cardiovasc. Res. 1998 Jan;37(1):141–150. doi: 10.1016/s0008-6363(97)00198-3. [DOI] [PubMed] [Google Scholar]
- 10.Hoek J.B., Cahill A., Pastorino J.G. Alcohol and mitochondria : A dysfunctional relationship. Gastroenterology. 2002;122(7):2049–2063. doi: 10.1053/gast.2002.33613. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sirey T.M., Ponting C.P. Insights into the post-transcriptional regulation of the mitochondrial electron transport chain. Biochem. Soc. Trans. 2016;44(5):1491–1498. doi: 10.1042/BST20160100. Oct 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srivastava S., Syed S.B., Kumar V., Islam A., Ahmad F. Fas-activated serine/threonine kinase: Structure and function. Gene Reports. 2017;8:117–127. Sept. [Google Scholar]
- 13.Simarro M., Mauger D., Rhee K., Pujana M.A., Kedersha N.L. Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc. Natl. Acad. Sci. U. S. A. 2007;104(27):11370–11375. doi: 10.1073/pnas.0704964104. Jul 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boehm E., Zornoza M., Jourdain A.A., Delmiro Magdalena A., García-Consuegra I. Role of FAST kinase domains 3 (FASTKD3) in post-transcriptional regulation of mitochondrial gene expression. J. Biol. Chem. 2016;291(50):25877–25887. doi: 10.1074/jbc.M116.730291. Dec 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popow J., Alleaume A.M., Curk T., Schwarzl T., Sauer S. FASTKD2 is an RNA-binding protein required for mitochondrial RNA processing and translation. RNA. 2015;21(11):1873–1884. doi: 10.1261/rna.052365.115. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm E, Zaganelli S, Maundrell K, Jourdain AA, Thore S, et al. FASTKD1 and FASTKD4 Have Opposite Effects on Expression of Specific Mitochondrial RNAs, Depending upon Their Endonuclease-like RAP Domain. [DOI] [PMC free article] [PubMed]
- 17.Jourdain A.A., Koppen M., Rodley C.D., Maundrell K., Gueguen N. A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 2015;10(7):1110–1121. doi: 10.1016/j.celrep.2015.01.063. Feb 24. [DOI] [PubMed] [Google Scholar]
- 18.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36(1):40–45. doi: 10.1038/ng1285. Jan. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F., Pei J., Chen X. Genetic ablation of Fas-activated serine/threonine kinase ameliorates obesity-related hepatic glucose and lipid metabolic disorders via sirtuin-1 signaling. Biochem. Biophys. Res. Commun. 2020;529(4):1066–1072. doi: 10.1016/j.bbrc.2020.06.049. Sept 3. [DOI] [PubMed] [Google Scholar]
- 20.Simarro M., Giannattasio G., De la Fuente M.A., Benarafa C., Subramanian K.K. Fas-activated serine/threonine phosphoprotein promotes immune-mediated pulmonary inflammation. J. Immunol. 2010;184(9):5325–5332. doi: 10.4049/jimmunol.1000104. May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y., Li X., Prabhu S.D., Brittian K.R., Chen Q. Angiotensin II plays a critical role in alcohol-induced cardiac nitrative damage, cell death, remodeling, and cardiomyopathy in a protein kinase C/nicotinamide adenine dinucleotide phosphate oxidase-dependent manner. J. Am. Coll. Cardiol. 2012;59(16):1477–1486. doi: 10.1016/j.jacc.2011.12.034. Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C., Sun W., Gu C., Yang Z., Quan N. Targeting ALDH2 for therapeutic interventions in chronic pain-related myocardial ischemic susceptibility. Theranostics. 2018;8(4):1027–1041. doi: 10.7150/thno.22414. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Y., Zhang F., Zhao S., Li Y., Chen X. Adiponectin determines farnesoid X receptor agonism-mediated cardioprotection against post-infarction remodelling and dysfunction. Cardiovasc. Res. 2018;114(10):1335–1349. doi: 10.1093/cvr/cvy093. Aug 1. [DOI] [PubMed] [Google Scholar]
- 24.Zhao S., Xia Y., Zhang F., Xiong Z., Li Y. Nucleostemin dysregulation contributes to ischemic vulnerability of diabetic hearts: Role of ribosomal biogenesis. J. Mol. Cell. Cardiol. 2017;108:106–113. doi: 10.1016/j.yjmcc.2017.05.010. Jul. [DOI] [PubMed] [Google Scholar]
- 25.Xie W., Santulli G., Reiken S.R., Yuan Q., Osborne B.W. Mitochondrial oxidative stress promotes atrial fibrillation. Sci. Rep. 2015;5:11427. doi: 10.1038/srep11427. Jul 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santulli G., Xie W., Reiken S.R., Marks A.R. Mitochondrial calcium overload is a key determinant in heart failure. Proc. Natl. Acad. Sci. U. S. A. 2015;112(36):11389–11394. doi: 10.1073/pnas.1513047112. Sep. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H. Regnase-1 and roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 2015;161(5):1058–1073. doi: 10.1016/j.cell.2015.04.029. May 21. [DOI] [PubMed] [Google Scholar]
- 28.Steiner J.L., Lang C.H. Etiology of alcoholic cardiomyopathy: Mitochondria, oxidative stress and apoptosis. Int. J. Biochem. Cell Biol. 2017;89:125–135. doi: 10.1016/j.biocel.2017.06.009. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moritz B., Venkata G., Matthias O., Efthymios S., Maike K. NOX2 amplifies acetaldehyde-mediated cardiomyocyte mitochondrial dysfunction in alcoholic cardiomyopathy. Sci. Rep. 2016;6:32554. doi: 10.1038/srep32554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moe K.T., Yin N.O., Naylynn T.M., Khairunnisa K., Wutyi M.A. Nox2 and Nox4 mediate tumour necrosis factor-α-induced ventricular remodelling in mice. J. Cell Mol. Med. 2011;15(12):2601–2613. doi: 10.1111/j.1582-4934.2011.01261.x. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Augsburger F., Filippova A., Rasti D., Seredenina T., Lam M. Pharmacological characterization of the seven human NOX isoforms and their inhibitors. Redox Biol. 2019;26:101272. doi: 10.1016/j.redox.2019.101272. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ayesha H., Cristina C., Caia D.S.D., Juan M. Systematic analysis of the role of RNA-binding proteins in the regulation of RNA stability. PLoS Genet. 2014;10(11) doi: 10.1371/journal.pgen.1004684. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao R., Yang R., Chen X., Harhaj E.W., Wang X. Regnase-1, a rapid response ribonuclease regulating inflammation and stress responses. Cell. Mol. Immunol. 2017;14(5):412–422. doi: 10.1038/cmi.2016.70. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilamowski M., Gorecki A., Wasylewska M.D. J Jura. Substrate specificity of human MCPIP1 endoribonuclease. Sci. Rep. 2018;8(1):7381. doi: 10.1038/s41598-018-25765-2. May 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Omiya S., Omori Y., Taneike M., Murakawa T., Ito J. Cytokine mRNA degradation in cardiomyocytes restrains sterile inflammation in pressure-overloaded hearts. Circulation. 2020;141(8):667–677. doi: 10.1161/CIRCULATIONAHA.119.044582. Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei J., Long L., Zheng W., Dhungana Y., Lim S.A. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature. 2019;576(7787):471–476. doi: 10.1038/s41586-019-1821-z. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153(5):1036–1049. doi: 10.1016/j.cell.2013.04.034. May 23. [DOI] [PubMed] [Google Scholar]
- 38.Gomez-Niño A., Docio I., Prieto-Lloret J., Simarro M., de la Fuente M.A. Mitochondrial complex I dysfunction and peripheral chemoreflex sensitivity in a FASTK-deficient mice model. Adv. Exp. Med. Biol. 2018;1071:51–59. doi: 10.1007/978-3-319-91137-3_6. [DOI] [PubMed] [Google Scholar]
- 39.Meyers D.E., Basha H.I., Koenig M.K. Mitochondrial cardiomyopathy: pathophysiology, diagnosis, and management. Tex. Heart Inst. J. 2013;40(4):385–394. [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou H., Toan S., Zhu P., Wang J., Ren J. DNA-PKcs promotes cardiac ischemia reperfusion injury through mitigating BI-1-governed mitochondrial homeostasis. Basic Res. Cardiol. 2020;115(2):11. doi: 10.1007/s00395-019-0773-7. Jan 9. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H., Zhu P., Wang J., Zhu H., Ren J. Pathogenesis of cardiac ischemia reperfusion injury is associated with CK2α-disturbed mitochondrial homeostasis via suppression of FUNDC1-related mitophagy. Cell Death Differ. 2018;25(6):1080–1093. doi: 10.1038/s41418-018-0086-7. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang J.Y., Blum A., Liu J., Finkel T. The role of mitochondria in aging. J. Clin. Invest. 2018;128(9):3662–3670. doi: 10.1172/JCI120842. Aug 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karamanlidis G., Lee C.F., Garcia-Menendez L., Kolwicz S.C., Jr., Suthammarak W. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell Metabol. 2013;18(2):239–250. doi: 10.1016/j.cmet.2013.07.002. Aug 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matyas C., Varga Z.V., Mukhopadhyay P., Paloczi J., Lajtos T. Chronic plus binge ethanol feeding induces myocardial oxidative stress, mitochondrial and cardiovascular dysfunction, and steatosis. Am. J. Physiol. Heart Circ. Physiol. 2016;310(11):H1658–H1670. doi: 10.1152/ajpheart.00214.2016. Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lucas D.L., Brown R.A., Wassef M., Giles T.D. Alcohol and the cardiovascular system: research challenges and opportunities. J. Am. Coll. Cardiol. 2005;45(12):1916–1924. doi: 10.1016/j.jacc.2005.02.075. Jun 21. [DOI] [PubMed] [Google Scholar]
- 46.Kesteloot H. Alcohol intake and markers of inflammation. Eur. Heart J. 2004;25(23):2075–2076. doi: 10.1016/j.ehj.2004.09.029. Dec. [DOI] [PubMed] [Google Scholar]
- 47.Murphy Elizabeth, Ardehali Hossein, Balaban Robert S., DiLisa Fabio, Dorn Gerald W. AHA position paper on Mitochondrial function, biology and Role in disease. Circ. Res. 2016;118(12):1960–1991. doi: 10.1161/RES.0000000000000104. Jun 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.