Summary
Winter, spring and biennial varieties of Brassica napus that vary in vernalization requirement are grown for vegetable and oil production. Here, we show that the obligate or facultative nature of the vernalization requirement in European winter oilseed rape is determined by allelic variation at a 10 Mbp region on chromosome A02. This region includes orthologues of the key floral regulators FLOWERING LOCUS C (BnaFLC.A02) and FLOWERING LOCUS T (BnaFT.A02). Polymorphism at BnaFLC.A02 and BnaFT.A02, mostly in cis‐regulatory regions, results in distinct gene expression dynamics in response to vernalization treatment. Our data suggest allelic variation at BnaFT.A02 is associated with flowering time in the absence of vernalization, while variation at BnaFLC.A02 is associated with flowering time under vernalizing conditions. We hypothesize selection for BnaFLC.A02 and BnaFT.A02 gene expression variation has facilitated the generation of European winter oilseed rape varieties that are adapted to different winter climates. This knowledge will allow for the selection of alleles of flowering time regulators that alter the vernalization requirement of oilseed rape, informing the generation of new varieties with adapted flowering times and improved yields.
Keywords: flowering time, vernalization, Brassica napus, oilseed rape, FLC, FT
Introduction
Vernalization leads to an acceleration of flowering in response to cold temperatures. Manipulation of the requirement for, and responsiveness to, vernalization has facilitated the generation of novel crop varieties that are adapted to local environments (Jung and Muller, 2009). In seed crops, such as oilseed rape (Brassica napus, 2n = 4x = 38, AACC), predictable and synchronized flowering is essential for a harvestable product.
The close evolutionary relationship between B. napus and the reference plant Arabidopsis thaliana makes it possible to identify orthologues of flowering time genes (Bancroft et al., 2011; Chalhoub et al., 2014; Schiessl et al., 2017a; Schiessl et al., 2014). In A. thaliana, allelic variation at two loci, FRIGIDA (AtFRI) and FLOWERING LOCUS C (AtFLC), often underlies variation in reproductive strategy (Shindo et al., 2005; Stinchcombe et al., 2004). Mutations at AtFRI that disrupt protein function result in a loss of vernalization requirement (Johanson et al., 2000; Michaels et al., 2004; Shindo et al., 2005), while allelic variation at AtFLC is important for fine‐tuning the vernalization response (Ågren et al., 2013; Coustham et al., 2012; Duncan et al., 2015; Grillo et al., 2013; Li et al., 2014; Li et al., 2015; Shindo et al., 2006; Strange et al., 2011).
Analysis of variation for flowering time in B. napus has identified quantitative trait loci (QTL) containing orthologues of AtFRI, AtFLC and FLOWERING LOCUS T (AtFT) the floral integrator gene (Chen et al., 2018; Ferreira et al., 1995; Hou et al., 2012; Long et al., 2007; Mei et al., 2009; Murphy and Scarth, 1994; Nelson et al., 2014; Raman et al., 2016; Raman et al., 2013; Schiessl et al., 2017a; Schiessl et al., 2015; Schiessl et al., 2019; Schiessl et al., 2014; Tadege et al., 2001; Wang et al., 2009; Wang et al., 2011; Wu et al., 2019; Xu et al., 2016; Yi et al., 2018). In particular, the AtFLC homologue on chromosomes A10 and A02 and the AtFRI homologue on chromosome A03 have been identified as major candidates for variation in flowering time in both QTL and genome‐wide association studies.
Despite high sequence conservation between A. thaliana and B. napus, the presence of multiple orthologous genes complicates translation of the floral regulatory network from reference diploid to polyploid crop species (Jones et al., 2018). Multiple orthologues of flowering time genes have been preferentially retained and are expressed in B. napus (Jones et al., 2018; Schiessl et al., 2017a; Schiessl et al., 2014); however, many studies have predominately focused on Chinese semi‐winter and Australian or Canadian spring varieties of oilseed rape (Hou et al., 2012; Long et al., 2007; Raman et al., 2016; Raman et al., 2013; Wang et al., 2009; Wang et al., 2011; Xu et al., 2016). Here, we investigate the genetic variation underlying flowering time differences between European winter oilseed rape varieties. An F2 population was generated from a biparental cross between an early flowering European winter oilseed rape variety Cabriolet and a late flowering European winter oilseed rape variety Darmor. A QTL‐seq approach (Takagi et al., 2013), which combines bulked segregant analysis (Giovannoni et al., 1991; Michelmore et al., 1991) with whole‐genome resequencing (Das et al., 2015; Illa‐Berenguer et al., 2015; Lu et al., 2014; Singh et al., 2016; Takagi et al., 2013; Wang et al., 2016), was used to identify QTL for flowering time with and without vernalization. A major QTL was identified on chromosome A02 which includes orthologues of AtFLC (BnaFLC.A02) and AtFT (BnaFT.A02). Allelic variation at BnaFLC.A02 accounted for a higher proportion of variation in flowering time following a six‐week vernalization treatment, while polymorphism at BnaFT.A02 co‐segregated with flowering time in the absence of vernalization. Cis‐polymorphism and altered gene expression dynamics were detected at both genes, revealing parallels with natural accessions of A. thaliana.
Results
European winter oilseed rape segregates for an obligate or facultative vernalization requirement
To investigate variation in flowering time within European winter oilseed rape with and without vernalization, the winter oilseed rape varieties Cabriolet and Darmor (Figure 1a) were used as parents to construct a segregating F2 population. Four‐week‐old plants from this population were either given a six‐week vernalization treatment at 5 °C (VERN) before being transferred to an unheated poly‐tunnel or grown on under ambient temperature conditions (no vernalization, NVERN) in the same poly‐tunnel during the spring and summer of 2017 when the daylength always exceeded 12 h. Temperature and relative humidity were recorded for the duration of the experiment (Figure S1), and plants experienced an average temperature and relative humidity of 18.96 °C and 68.24%, respectively. Flowering time was recorded from the date of transfer to the poly‐tunnel and excluded the number of days growth before transfer.
Figure 1.
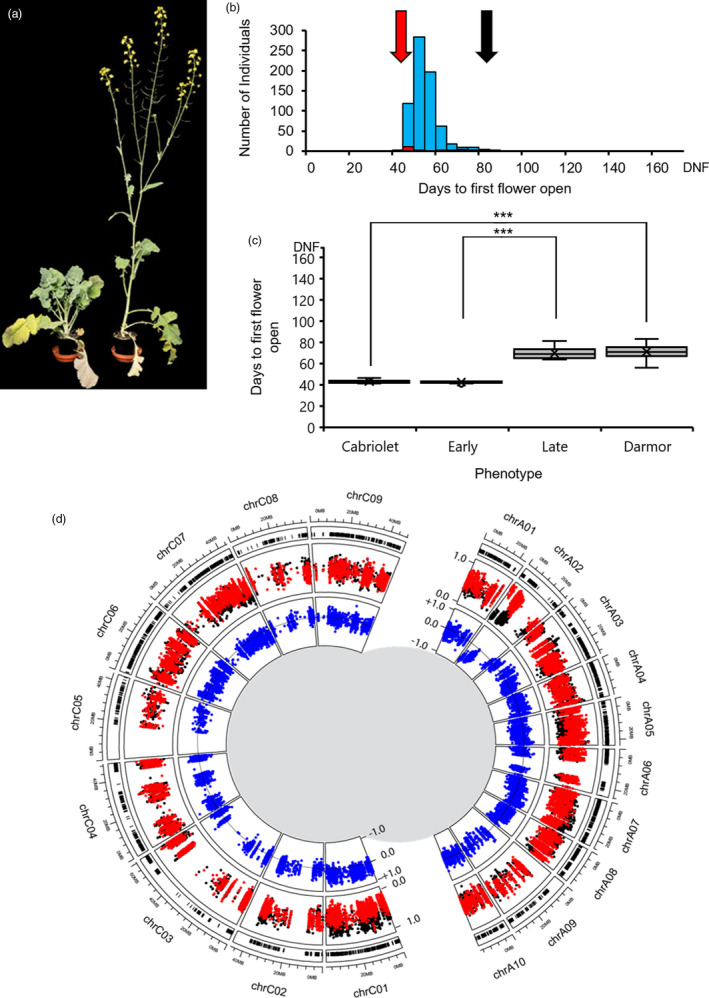
Variation for vernalization response is present in European winter oilseed rape. (a) Flowering phenotype of the late flowering variety Darmor (left) and early flowering variety Cabriolet (right) at 50 days’ growth after a 6‐week vernalization treatment. (b) Frequency distribution of flowering time of Cabriolet, Darmor and 704 F2 lines under VERN treatment. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel. (c) Flowering time distribution of Cabriolet, the early flowering bulk, the late flowering bulk and Darmor under VERN treatment; box and whisker plots represent the mean and quartile values. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel. (d) Results of a QTL‐seq approach for mapping flowering time in European winter oilseed rape under VERN treatment. Outer circle: the distribution of Cabriolet variants detected in the bulks plotted against the chromosomal position according to Darmor‐bzh. Middle circle: the SNP index values calculated for each variant in the early bulk (red) and the late bulk (black) plotted in genome order according to Darmor‐bzh. Inner circle: the difference between SNP index values between the bulks plotted as the ΔSNP index (blue) against the chromosomal position according to Darmor‐bzh, a ΔSNP index equal to zero representing no deviation in allele segregation between the bulks is plotted as a horizontal line. Each genome is plotted as separate half circles.
Under both treatments, Darmor flowered significantly later than Cabriolet (P < 0.001, Mann–Whitney U‐test, Figure 1a,c and Figure 2a,b). Under VERN, Cabriolet flowered within an average of 42.92 days after transfer to the poly‐tunnel (which equated to 114.92 days after sowing), while Darmor flowered within an average of 71.25 days (141.25 days after sowing) and exhibited a broader range of flowering times, indicative of incomplete vernalization (Table S1). Under NVERN, Cabriolet flowered within an average of 78 days (106 days after sowing) and therefore exhibited a facultative vernalization requirement, while Darmor did not flower within the timeframe of the experiment and exhibited an obligate requirement.
Figure 2.
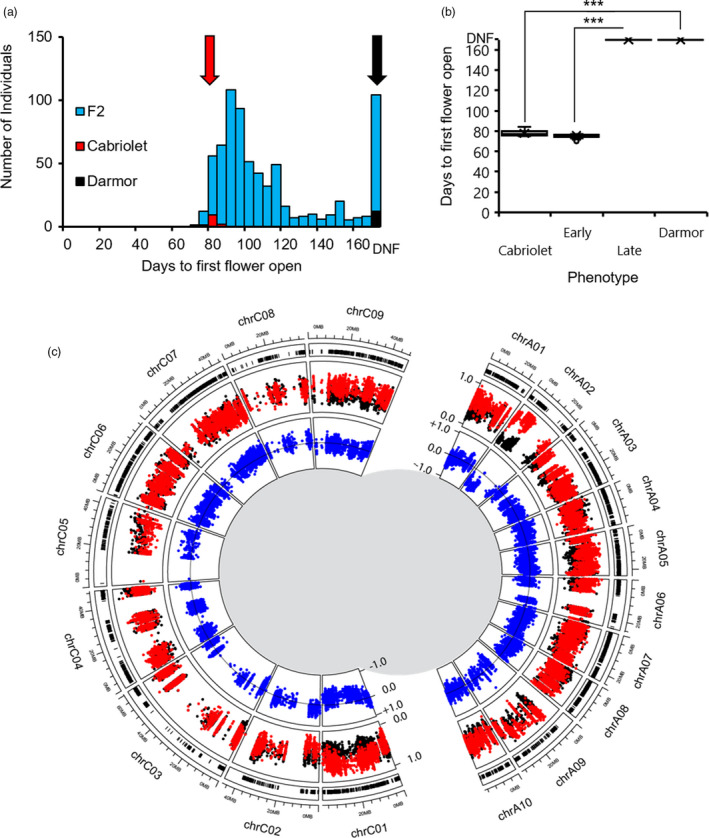
Variation for vernalization requirement is present in European winter oilseed rape. (a) Frequency distribution of flowering time of Cabriolet, Darmor and 708 F2 lines under NVERN treatment. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel. (b) Flowering time distribution of Cabriolet, the early flowering bulk, the late flowering bulk and Darmor under NVERN treatment; box and whisker plots represent the mean and quartile values. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel. (c) Results of a QTL‐seq approach for mapping flowering time in European winter oilseed rape under NVERN treatment. Outer circle: the distribution of Cabriolet variants detected in the bulks plotted against the chromosomal position according to Darmor‐bzh. Middle circle: the SNP index values calculated for each variant in the early bulk (red) and the late bulk (black) plotted in genome order according to Darmor‐bzh. Inner circle: the difference between SNP index values between the bulks plotted as the ΔSNP index (blue) against the chromosomal position according to Darmor‐bzh, a ΔSNP index equal to zero representing no deviation in allele segregation between the bulks is plotted as a horizontal line. Each genome is plotted as separate half circles.
Seven hundred and four F2 lines were assessed for flowering time following the VERN treatment. Flowering time under these conditions exhibited a continuous distribution, with a right‐hand skew suggesting flowering time in B. napus is quantitatively inherited (Figure 1b). In parallel, 708 F2 lines were assessed for flowering time under the NVERN treatment. Flowering time NVERN exhibited a bimodal distribution including a subset of 86 lines that did not flower (Figure 2a). The earliest lines flowered 70 days after transfer to the poly‐tunnel. Under both treatments, no significant difference was detected between F2 lines from each reciprocal cross (NVERN P = 0.426 ANOVA, VERN P = 0.219 ANOVA) and therefore no maternal effect on flowering time was detected. Very little transgressive segregation was observed, suggesting the two parent varieties were representative of the extremes of flowering time variation in this genetic background.
A single genomic region on chromosome A02 is associated with flowering time with and without vernalization
To identify the major genomic regions important for flowering time in the F2 population, lines were selected for bulking based on flowering time phenotype. Four DNA bulks were generated, two bulks for each treatment, and included approximately 5% of the population representing the earliest and latest flowering lines (Figures 1c and 2b). Genomic DNA from the parent plants (Cabriolet and Darmor) and the four bulks were sequenced, generating 705 million clean sequencing reads with a Q30 score of at least 90.2% (Table S1).
To identify high confidence (read depth> 20, >95% base call) variants (SNPs and small InDels (<9bp)) between Cabriolet and Darmor, we compared the sequencing reads from Darmor to the Darmor‐bzh reference genome (Chalhoub et al., 2014). Darmor‐bzh was generated by introgressing the dwarf BREIZH (Bzh) locus from a line called B192 into Darmor and is therefore predicted to carry genetic differences (Foisset et al., 1995). Twelve thousand five hundred and twenty two high confidence variants were identified in Darmor compared with Darmor‐bzh, and 9561 of these could be anchored to a chromosomal position (Figure S2). The density and location of SNPs suggest large genomic blocks vary between Darmor and Darmor‐bzh and this variation was accounted for when comparing Cabriolet and Darmor.
Thirty nine thousand seven hundred and seventeen high confidence variants were identified in Cabriolet compared with Darmor, and 32 773 could be anchored to a chromosomal position (Figure S3). SNP and ΔSNP indices were calculated for each variant under both VERN and NVERN treatments and plotted against their genomic position according to the Darmor‐bzh reference sequence (Chalhoub et al., 2014) in Figures 1d and 2c, respectively.
To identify genomic regions responsible for the difference in flowering time between the early and late flowering bulks, we chose absolute ΔSNP index threshold values of 0.686 and 0.828 for VERN and NVERN treatments, respectively. These threshold values represented the top 1% of absolute ΔSNP index values across the whole genome for each treatment. For both VERN and NVERN treatments, a deviation in SNP index values was detected across a single 10Mbp region on chromosome A02 and contained numerous absolute ΔSNP index values above these thresholds (Figures 1d, 2c, 3a and 4a). The direction of ΔSNP index values towards −1.0 in both VERN and NVERN treatments suggests the genetic contribution for early flowering was determined by Cabriolet and late flowering by Darmor. By calculating an average ΔSNP index value for every 100‐variant window across the chromosome, we could determine a peak of association within the QTL region. For VERN, a peak in average ΔSNP index values was detected 0–2.5 Mbp from the upper arm of chromosome A02 (Figure 3a). However, for NVERN this peak was detected at a region 5–7.5 Mbp from the upper arm of chromosome A02 (Figure 4a). This suggested that different genes within the same 10 Mbp region on chromosome A02 were responsible for the differences in flowering time between the bulks under VERN and NVERN treatments.
Figure 3.
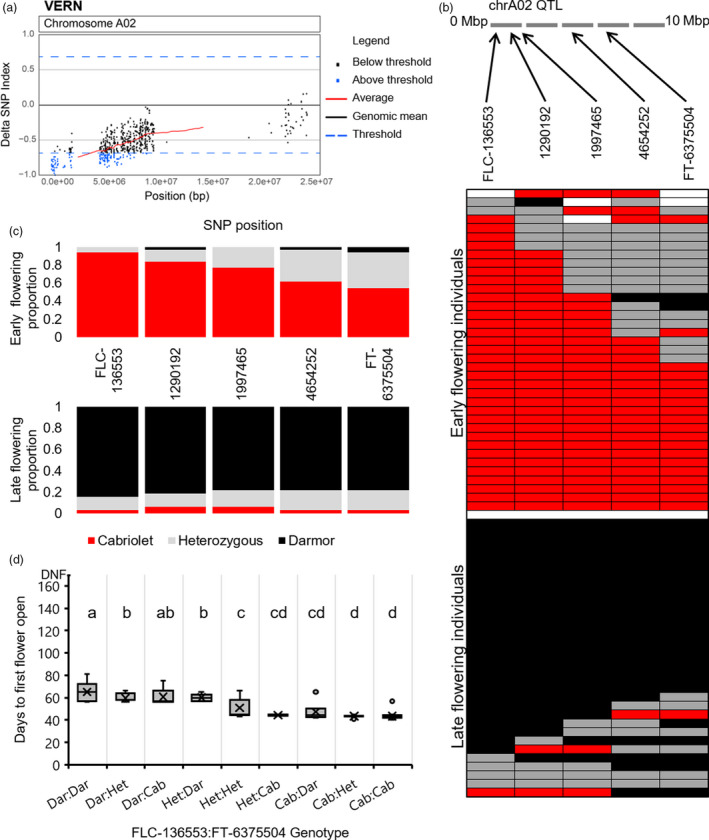
A VERN QTL for flowering time is located on chromosome A02. (a) ΔSNP index plot of chromosome A02 under VERN treatment. ΔSNP index values are plotted in Brassica chromosome A02 order, and values found within the top 1% of ΔSNP index values are coloured blue. (b) Validation of the VERN QTL region on chromosome A02 by KASP assay. Upper panel: schematic of the QTL region on chromosome A02 with the relative locations of SNPs targeted by KASP assay are highlighted. Lower panel: the genotype of all F2 lines within the VERN DNA bulks screened at five SNP positions within the QTL; SNPs homozygous for the Cabriolet allele are coloured red, SNPs homozygous for the Darmor allele are coloured black, SNPs that are heterozygous are coloured grey, and SNP genotypes that could be not determined are left white. The grid is divided into early flowering bulk and the late flowering bulk. (c) The proportion of F2 lines that were homozygous for Cabriolet alleles (red), homozygous for Darmor alleles (black) or heterozygous (grey) at five SNP positions on chromosome A02 in the DNA bulks under VERN treatment. (d) The flowering time phenotype under VERN treatment of F2 lines genotyped for SNP markers FLC‐136553 and FT‐6375504 (Cab = Cabriolet, Dar = Darmor, Het = heterozygous). Letters above the columns indicate significant differences determined by multiple pairwise comparisons using Mann–Whitney U‐test‐with an α‐value of 0.05. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel.
Figure 4.
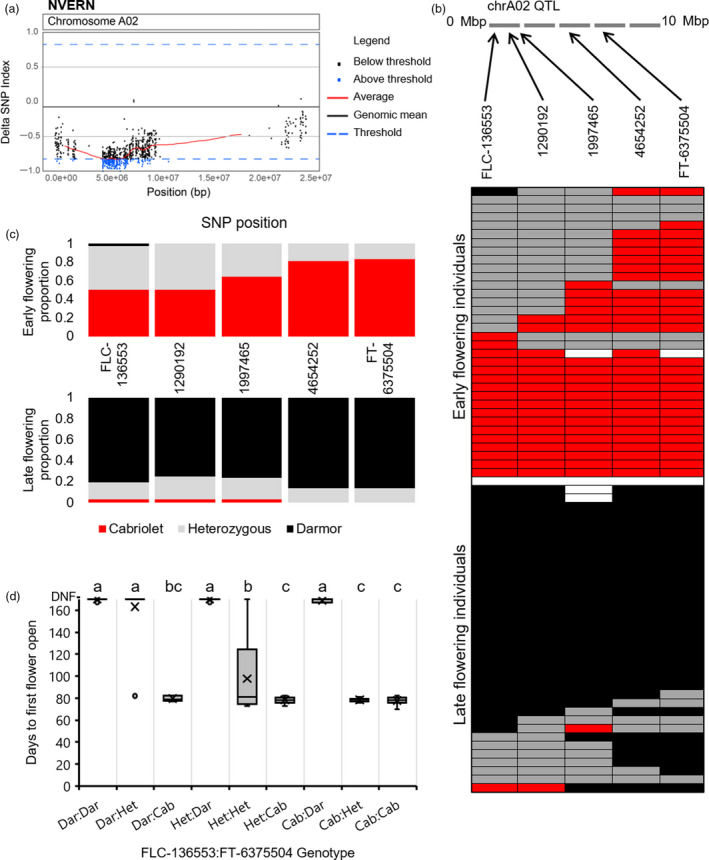
A NVERN QTL for flowering time is located on chromosome A02. (a) ΔSNP index plot of chromosome A02 under NVERN treatment. ΔSNP index values are plotted in Brassica chromosome A02 order, and values found within the top 1% of ΔSNP index values are coloured blue. (b) Validation of the NVERN QTL region on chromosome A02 by KASP assay. Upper panel: schematic of the QTL region on chromosome A02 with the relative locations of SNPs targeted by KASP assay are highlighted. Lower panel: the genotype of all F2 lines within the NVERN DNA bulks screened at five SNP positions within the QTL; SNPs homozygous for the Cabriolet allele are coloured red, SNPs homozygous for the Darmor allele are coloured black, SNPs that are heterozygous are coloured grey, and SNP genotypes that could be not determined are left white. The grid is divided into the early flowering bulk and the late flowering bulk. (c) The proportion of F2 lines that were homozygous for Cabriolet alleles (red), homozygous for Darmor alleles (black) or heterozygous (grey) at five SNP positions on chromosome A02 in the DNA bulks under NVERN treatment. (d) The flowering time phenotype under NVERN treatment of F2 lines genotyped for SNP markers FLC‐136553 and FT‐6375504 (Cab = Cabriolet, Dar = Darmor, Het = heterozygous). Letters above the columns indicate significant differences determined by multiple pairwise comparisons using Mann–Whitney U‐test‐with an α‐value of 0.05. Flowering time was recorded as days to first flower open from the first day plants were transferred to the poly‐tunnel.
There are 1937 genes annotated within the 10Mbp QTL region on chromosome A02 according to the Darmor‐bzh reference genome. Based on homology with genes in A. thaliana, 21 are hypothesized to contribute to the control of flowering time and only six of these genes are polymorphic between Cabriolet and Darmor (Table 1). Likely candidates that associate with variation in flowering time between Cabriolet and Darmor include BnaA02g00370d (hereafter referred to as BnaFLC.A02), a homologue of AtFLC and a MADS‐box transcription factor responsible for the inhibition of flowering until after vernalization, and BnaA02g12130D (hereafter referred to as BnaFT.A02) a homologue of AtFT, a gene considered an integrator of photoperiod and vernalization signals, and a promoter of flowering. The other four potential candidates were homologues of Gibberellin 20 oxidase 2, EMBRYONIC FLOWER 2, CSTF64 and TBP2. Interestingly, no deviation in either SNP or ΔSNP indices was found on chromosomes A03, A10, C02, C03 and C09 where other orthologues of AtFLC and orthologues of AtFRI have been mapped. No high confidence variants were detected at the major candidate genes for flowering time, BnaFLC.A10 and BnaFRI.A03 (Hou et al., 2012; Wang et al., 2011) (Figure S4). Both Cabriolet and Darmor carry the previously described winter‐type allele of both BnaFLC.A10 (Hou et al., 2012; Song et al., 2020) and BnaFRI.A03 (Wang et al., 2011) which suggests neither gene contributes to flowering time variation within this population.
Table 1.
Candidate flowering time genes found within the QTL region for vernalisation requirement and response on chromosome A02
| Position (bp) | Gene name | Genetic status in Cabriolet | Homologue in A. thaliana | Function in A. thaliana |
|---|---|---|---|---|
| 134,159 – 138,121 | BnaA02g00370d | Polymorphic | AT5G10140, FLOWERING LOCUS C, FLC | MADS‐box transcription factor, repressor of flowering, responsive to vernalization |
| 408,979 – 413,943 | BnaA02g01030D | Conserved | AT5G11530, EMBRYONIC FLOWER 1, EMF1 | Involved in reproductive development |
| 565,722 – 567,762 | BnaA02g01270D | Conserved | AT5G12840, Nuclear transcription factor Y subunit A‐1, NFYA1 | Expressed in reproductive tissue |
| 773,658 – 778,149 | BnaA02g01670D | Conserved | AT5G13480, FY | Involved in regulation of flowering time, affects FCA mRNA processing |
| 893,325 – 894,144 | BnaA02g01960D | Conserved | AT5G14010, KNUCKLES, KNU | Transcription factor, mediates repression of WUS in floral meristem determinacy control |
| 2,115,552 – 2,118,359 | BnaA02g04770D | Conserved | AT5G20240, PISTILLATA, PI | Floral homeotic gene, MADS domain transcription factor, required for specification of petal and stamen identities |
| 3,037,543 – 3,043,336 | BnaA02g06350D | Conserved | AT5G60410, ATSIZ1 | |
| 3,106,207 – 3,108,826 | BnaA02g06490D | Conserved | AT5G60120, Target of early activation tagged (EAT)2, TOE2 | |
| 3,111,280 – 3,113,426 | N/A | Conserved | AT5G60100, Pseudo‐response regulator 3, PRR3 | PRR3 transcript levels vary in circadian pattern |
| 3,320,312 – 3,321,741 | BnaA02g07010D | Conserved | AT5G59560, SENSITIVITY TO RED LIGHT REDUCED 1, SRR1 | Required for normal oscillator function during circadian rhythm |
| 3,685,147 – 3,687,730 | BnaA02g07770D | Conserved | AT5G58230, MSI1 | Required for the transition to flowering |
| 3,861,836 – 3,864,833 | BnaA02g08140D | Conserved |
AT5G57380, VERNALIZATION INSENSITIVE 1, VIN3 |
Plant homeodomain protein, part of polycomb group complex of proteins, has a role in establishing FLC repression during vernalization |
| 5,851,901 – 5,853,606 | BnaA02g11210D | Polymorphic | AT5G51810, Gibberellin 20 oxidase 2, GA20OX2 | |
| 5,948,452 – 5,953,119 | BnaA02g11340D | Polymorphic | AT5G51230, EMBRYONIC FLOWER 2, EMF2 | Polycomb group protein, a negative regulator of reproductive development |
| 6,375,937 – 6,378,901 | BnaA02g12130D | Polymorphic | AT1G65480, FLOWERING LOCUS T, FT | A promoter of flowering, expressed in leaves and is induced by long day treatment |
| 7,870,038 – 7,871,424 | BnaA02g14040D | Conserved | AT1G68840, RAV2 | |
| 8,998,899 – 9,001,958 | BnaA02g15530D | Polymorphic | AT1G71800, CSTF64 | RNA 3’‐end‐processing factor of antisense FLC transcript, mediates silencing of FLC gene |
| 9,423,231 – 9,429,930 | BnaA02g15970D | Polymorphic | AT1G72390, TBP2 | |
| 9,980,484 – 9,983,268 | BnaA02g16710D | Conserved | AT2G18915, Adagio protein 2, ADO2, LKP2 | |
| 10,268,028 – 10,270,134 | BnaA02g17110D | Conserved | AT1G75060, BLH3 | |
| 10,804,098 – 10,805,192 | N/A | Conserved | AT4G20370, TWIN SISTER OF FT, TSF | A promoter of flowering and a homologue of FT, FT and TSF play overlapping roles in the transition to flowering |
Flowering time gene position on chromosome A02, the gene name according to the Darmor‐bzh genome, and genetic status in Cabriolet compared with Darmor are listed. The homologous gene name and function in A. thaliana is included for reference.
The QTL is not the result of a homeologous exchange between the A02 and C02 chromosomes
The comparatively recent origin of amphidiploid B. napus has been shown to result in frequent exchanges between homeologous regions of the A and C diploid genomes. One region where such exchanges are reported to occur is at the top of chromosomes A02 and C02 (Chalhoub et al., 2014, He et al., 2017). To determine whether a homeologous exchange (HE) contributed to the QTL on A02 in this population, we conducted HE analysis on the parental Darmor and Cabriolet genome sequences as per He et al. (2017). Some differences were observed (Figure S5), including a HE in Darmor compared with Cabriolet on chromosome C02. Although located within the QTL region, this HE did not overlap with the most strongly associated variants identified by QTL‐seq and is therefore not likely to be the cause of the QTL (Figure S5B).
Validation of the QTL region by KASP markers reveals different genomic regions of chromosome A02 are associated with flowering time with and without vernalization
To validate the region on chromosome A02 identified by QTL‐seq, KASP (Kompetitive allele‐specific PCR) primers were designed to target a 5Mbp region between the two most promising candidate genes, BnaFLC.A02 and BnaFT.A02, within the QTL. For both NVERN and VERN treatments, five SNPs including a SNP within BnaFLC.A02 (SNP 136553 hereafter referred to as FLC‐136553) and within the promoter of BnaFT.A02 (SNP 6375504 hereafter referred to as FT‐6375504) (Table S2) were targeted to determine the segregation of alleles in the earliest and latest flowering F2 lines. We first screened the lines that were included in the DNA bulks of the VERN and NVERN QTL‐seq analyses. Each line was scored for segregation of alleles at the five SNP markers, and all five SNPs were assigned a Cabriolet (Cab), Darmor (Dar) or heterozygous (Het) allele. The genotypes of each line included in the DNA bulks are summarized in Figures 3b and 4b.
Using these genotyping results, we were able to determine the frequency at which the alleles appeared within the DNA bulks. Within the VERN DNA bulks, all lines in the early flowering bulk were either homozygous (94.4%) or heterozygous (5.6%) for the Cabriolet allele at SNP marker FLC‐136553 within BnaFLC.A02 (Figure 3c). In contrast, a majority of lines within the late flowering bulk were either homozygous (84.4%) or heterozygous (12.5%) for the Darmor allele at the same SNP marker (Figure 3c). The proportion of lines in both bulks that were homozygous for each marker decreased across the 5Mbp region downstream towards SNP marker FT‐6375504 at BnaFT.A02. At this position, 54.3% of lines within the early flowering bulk were homozygous for the Cabriolet allele, and 78.1% of lines within the late flowering bulk were homozygous for the Darmor allele. We conclude that homozygosity for Darmor alleles at a region that includes BnaFLC.A02 and BnaFT.A02 is important for determining lateness of flowering, but homozygosity for Cabriolet alleles at a region that includes only BnaFLC.A02 is required for determining earliness of flowering. In agreement with the QTL‐seq analysis, a significantly higher proportion of lines were homozygous at BnaFLC.A02 compared with BnaFT.A02 (P < 0.001, Wilcoxon matched‐pairs test), confirming allelic variation at a region that includes BnaFLC.A02 is most closely associated with variation in flowering time within the F2 population under VERN conditions.
The same genotyping analysis was performed for the lines included in the DNA bulks of the NVERN treatment. Within the DNA bulks, all lines in the early flowering bulk were either homozygous (83.3%) or heterozygous (16.7%) for the Cabriolet allele at SNP marker FT‐6375504, found within the promoter of BnaFT.A02 (Figure 4c). In contrast, all lines in the late flowering bulk were either homozygous (86.1%) or heterozygous (13.9%) for the Darmor allele at the same SNP marker (Figure 4c). The proportion of lines in both bulks that were homozygous for each marker decreased across the 5Mbp region upstream towards SNP marker FLC‐136553 at BnaFLC.A02. At this position, 50% of lines within the early flowering bulk were homozygous for the Cabriolet allele, and 80.6% of lines within the late flowering bulk were homozygous for the Darmor allele. We therefore conclude that homozygosity at a region that includes BnaFLC.A02 and BnaFT.A02 is important for determining late flowering due to the high incidence of Darmor alleles within the late flowering bulk. However, homozygosity of the Cabriolet allele of BnaFLC.A02 is not required to determine early flowering under these conditions. In agreement with the QTL‐seq analysis, a significantly higher proportion of lines were homozygous at BnaFT.A02 compared with BnaFLC.A02 (P = 0.035, Wilcoxon matched‐pairs test), confirming allelic variation at a region that includes BnaFT.A02 is most closely associated with variation in flowering time within the F2 population under NVERN conditions.
To further validate this, we screened the next 78 lines, 94 lines in total, from each tail of the distribution in flowering time (Figures 1b and 2a) within the F2 population under both VERN and NVERN treatments using the same five SNP markers. All nine possible genotypic combinations of SNP markers were detected in the F2 population (Figures 3d and 4d). Significant differences in flowering times were observed between the nine genotypes (P < 0.001, Mann–Whitney U‐test; Figures 3d and 4d) and under both treatments, lines homozygous for FLC‐136553‐Cab and FT‐6375504‐Cab flowered significantly earlier than lines homozygous for FLC‐136553‐Dar and FT‐6375504‐Dar. Lines heterozygous for both markers exhibited an intermediate flowering time, while the flowering time of recombinant lines with one homozygous and one heterozygous allele was dependent on the genotype of the homozygous allele.
Within the DNA bulks, the instance of homozygous recombinant lines carrying BnaFLC.A02 from one parent and BnaFT.A02 from the other parent was rare (Figures 3b and 4b). This suggests both genes are important for the extreme early and late flowering phenotypes within the population. By expanding our screen to 94 lines from each tail of the distribution, we identified 11 and 14 recombinant lines under VERN and NVERN, respectively. The flowering time of recombinant lines homozygous for SNP marker FLC‐136553 from one parent and homozygous for SNP marker FT‐6375504 from the other parent (FLC‐136553‐Cab and FT‐6375504‐Dar, or FLC‐136553‐Dar and FT‐6375504‐Cab) was dependent on vernalization treatment, although not always flowering as early or late as their non‐recombinant counterparts. Under VERN treatment, lines with genotypic combination FLC‐136553‐Cab and FT‐6375504‐Dar flowered significantly earlier than lines with genotypic combination FLC‐136553‐Dar and FT‐63755040‐Cab (Figure 3d). In contrast, under NVERN treatment, lines with a genotypic combination FLC‐136553‐Cab and FT‐6375504‐Dar flowered significantly later than lines with combination FLC‐136553‐Dar and FT‐6375504‐Cab (Figure 4d). This result again supports the hypothesis that allelic variation at BnaFLC.A02 controls flowering time under VERN treatment, while allelic variation at BnaFT.A02 is responsible for differences in flowering time under NVERN treatment. A single outlier line with genotypic combination FLC‐136553‐Cab and FT‐6375504‐Dar that flowered late under VERN treatment reveals that other minor effect genes may also contribute to variation in flowering time within the population.
Cis‐polymorphism at BnaFLC.A02 affects the stability of FLC silencing
BnaFLC.A02 was previously cloned by Tadege et al. (2001) and Zou et al. (2012) and, like AtFLC, it is organized into seven exons and six introns. BnaFLC.A02 was amplified and sequenced from the two parent lines. Compared with Darmor BnaFLC.A02 (hereafter referred to as BnaFLC.A02‐Dar), the Cabriolet allele (hereafter referred to as BnaFLC.A02‐Cab) had 50 SNPs and 10 InDels; all were located within introns (Figure 5a).
Figure 5.
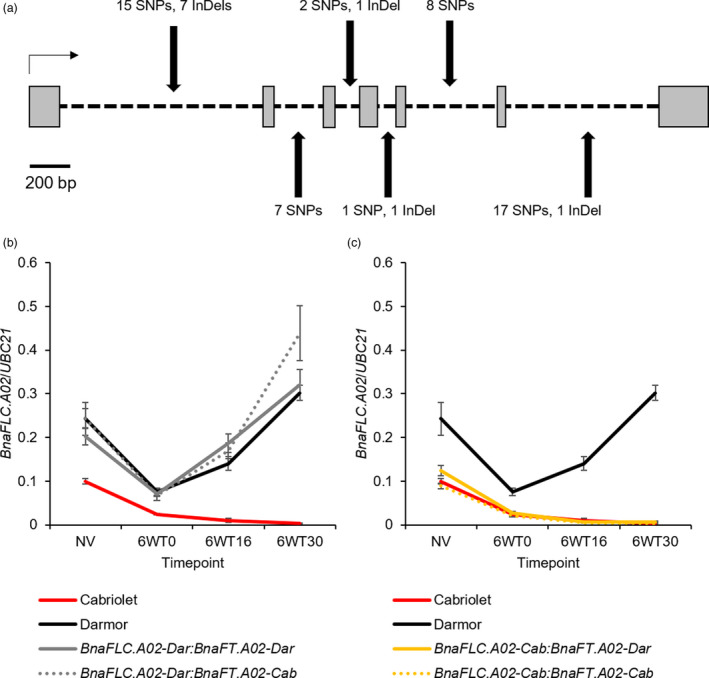
DNA sequence and gene expression variation of BnaFLC.A02 in Cabriolet and Darmor. (a) The polymorphisms identified at BnaFLC.A02 in Cabriolet compared with Darmor are highlighted with black arrows, grey boxes represent exons, black dashed lines represent introns, and the black horizontal arrow represents the direction of transcription. (b) Normalized expression of BnaFLC.A02 in Cabriolet, Darmor and genotyped F2 lines for BnaFLC.A02 and BnaFT.A02, as measured by quantitative RT‐PCR before (NV) and after a six‐week vernalization treatment (6WT0, 6WT16, 6WT32), error bars denote one standard error around the mean calculated from at least three biological replicates. (c) Normalized expression of BnaFLC.A02 in Cabriolet, Darmor and genotyped F2 lines for BnaFLC.A02 and BnaFT.A02, as measured by quantitative RT‐PCR before (NV) and after a six‐week vernalization treatment (6WT0, 6WT16, 6WT32), error bars denote one standard error around the mean calculated from at least three biological replicates.
The presence of non‐coding sequence variation at BnaFLC.A02 prompted us to investigate the expression of the two BnaFLC.A02 alleles by quantitative RT‐PCR before (NV) and after six‐week vernalization treatment in Cabriolet, Darmor and in a panel of F2 lines that had been genotyped for their alleles at BnaFLC.A02 and BnaFT.A02. BnaFLC.A02 expression was consistently lower in leaves of Cabriolet than Darmor plants at 28 days after sowing (Figure 5b,c and Figure S6). The two BnaFLC.A02 alleles also exhibited differences in expression dynamics following a six‐week vernalization treatment at 5 °C. Both alleles were repressed by six‐week vernalization, but BnaFLC.A02‐Cab was repressed to lower levels compared to BnaFLC.A02‐Dar. On return to warm conditions, BnaFLC.A02‐Cab expression remained low and therefore its expression was stably repressed, while BnaFLC.A02‐Dar reactivated to expression levels not dissimilar to those levels detected before vernalization. The F2 lines could be grouped into four genotypes dependent on their homozygous alleles at BnaFLC.A02 and BnaFT.A02 (Cab:Cab, Dar:Dar, Cab:Dar, Dar:Cab). F2 lines that were homozygous for BnaFLC.A02‐Cab exhibited similar expression patterns to those detected in Cabriolet, and lines carrying BnaFLC.A02‐Dar showed expression dynamics similar to Darmor plants at 28 days. BnaFLC.A02 expression in recombinant F2 lines (BnaFLC.A02‐Cab and BnaFT.A02‐Dar, or BnaFLC.A02‐Dar and BnaFLC.A02‐Cab) was independent of BnaFT.A02 (Figure 5b,c and Figure S6).
BnaFT.A02 is highly expressed in Cabriolet but not in Darmor
BnaFT.A02 was previously cloned by Wang et al. (2009) and, like AtFT, the gene is organized into four exons and three introns. Due to the presence of an AT rich insertion in intron 2 of BnaFT.A02, exons 1–2 of the gene were amplified and sequenced from the parental varieties. Alignment and analysis of this partial BnaFT.A02 fragment from both varieties identified polymorphisms in both coding and non‐coding regions of the gene (Figure 6a). Compared with BnaFT.A02 in Darmor (hereafter referred to as BnaFT.A02‐Dar), 4 SNPs and 1 InDel were identified in the Cabriolet BnaFT.A02 sequence (hereafter referred to as BnaFT.A02‐Cab; Figure 6a). One non‐synonymous SNP in exon 1 is predicted to cause an amino acid change from isoleucine to leucine (I48L; Figure 6b). The remaining SNPs and deletions were detected within intron 1, including sequence variation in Cabriolet that overlapped with the predicted CArG box motif sequence of BnaFT.A02 where, in A. thaliana, the AtFLC protein binds and represses AtFT (Helliwell et al., 2006) (Figure 6a).
Figure 6.
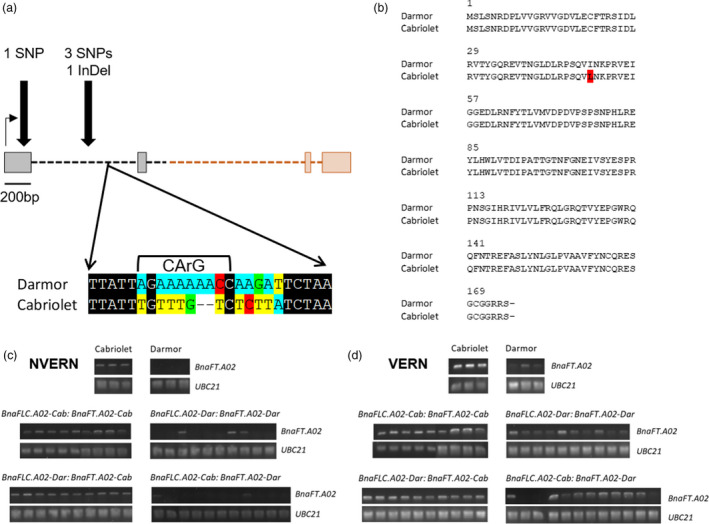
DNA sequence and gene expression variation of BnaFT.A02 in Cabriolet and Darmor. (a) The polymorphisms identified at BnaFT.A02 in Cabriolet compared with Darmor are highlighted with black arrows, grey boxes represent exons, black dashed lines represent introns, the black horizontal arrow represents the direction of transcription, and sequences not confirmed by capillary sequencing are highlighted in orange. Included is a zoom of the predicted CArG box motif sequence in Darmor and Cabriolet, conserved DNA sequences are coloured black, while polymorphisms are highlighted. (b) Comparison of the predicted amino acid sequence of BnaFT.A02 in Cabriolet and Darmor, amino acid substitution I48L is highlighted in red. (c) Qualitative expression analysis of BnaFT.A02 in Cabriolet, Darmor and F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 at 28 days after sowing and without vernalization treatment. Expression of BnaFT.A02 is detectable when a band is present on the gel. (d) Qualitative expression analysis of BnaFT.A02 in Cabriolet, Darmor and F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 at 30 days under glasshouse conditions after a six‐week vernalization treatment. Expression of BnaFT.A02 is detectable when a band is present on the gel.
We assessed BnaFT.A02 expression in Cabriolet, Darmor and in F2 lines (the same lines that had been genotyped for homozygosity at BnaFLC.A02 and BnaFT.A02 as described previously) with and without vernalization treatment using both qualitative and quantitative RT‐PCR (Figure 6c,d and Figure S7). Although lowly expressed, BnaFT.A02‐Cab expression was detectable in leaves at 28 days after sowing, while BnaFT.A02‐Dar expression was not detected (Figure 6c and Figure S7A). Under ambient temperature conditions, and without vernalization, BnaFT.A02‐Cab was consistently detectable up to 100 days after sowing, while BnaFT.A02‐Dar was not (Figure S7A). After a six‐week vernalization treatment and upon return to warm glasshouse conditions, the expression of BnaFT.A02‐Cab increased, while the expression of BnaFT.A02‐Dar remained low but detectable (Figures 6d and Figure S7B). To confirm that the absence of BnaFT.A02 expression in Darmor was not due to a homeologous exchange, we quantified the expression of BnaFT.C02 in both parent lines. Both BnaFT.A02 and BnaFT.C02 expression were activated after vernalization in Cabriolet, but the expression of both genes was not detectable in Darmor (Figure S7C). BnaFT.A02 expression was assessed in a panel of F2 lines that could be grouped into four genotypes dependent on their homozygous alleles at BnaFLC.A02 and BnaFTA02 (Cab:Cab, Dar:Dar, Cab:Dar, Dar:Cab). BnaFT.A02 expression was detectable at 28 days after sowing in F2 lines that were homozygous for BnaFT.A02‐Cab (Figure 6c). For most cases with lines carrying BnaFT.A02‐Dar (18 out of 21 lines), its expression was not detectable before vernalization (Figure 6c). In recombinant F2 lines, BnaFT.A02‐Cab expression was detectable regardless of the BnaFLC.A02 allele present (Figure 6c). This suggests that, before vernalization, BnaFT.A02‐Cab expression is independent of the allele present at the BnaFLC.A02 locus. After vernalization, expression of both alleles of BnaFT.A02 was detectable in all F2 lines (Figure 6d). However, lines that carried the BnaFLC.A02‐Cab allele expressed BnaFT.A02 at quantitatively higher levels compared with lines that carried the BnaFLC.A02‐Dar allele (Figure S7D,E).
Discussion
Varieties of European winter oilseed rape are traditionally considered to have an obligate vernalization requirement, exhibiting an extended vegetative growth during the winter and flowering at the onset of spring. Here, we provide evidence that European winter oilseed rape varieties can exhibit an obligate or facultative vernalization requirement and we identify the same 10 Mbp genomic region on chromosome A02, which includes BnaFT.A02 and BnaFLC.A02, as a candidate for controlling this requirement.
Previous reports (Chen et al., 2018; Nelson et al., 2014; Raman et al., 2016; Schiessl et al., 2015; Wu et al., 2019; Xu et al., 2016) have identified associations between regions on chromosome A02 and flowering time in B. napus. Variation at BnaFLC.A02 and BnaFT.A02 has been reported to contribute a significant proportion of variation in flowering time between crop types of oilseed rape (Chen et al., 2018; Wu et al., 2019), but here we report their role in flowering time within winter oilseed rape. Although our QTL encompassed a large genomic region and included several flowering time genes, genotypic data suggest that allelic variation at a region including BnaFT.A02 contributed more strongly to flowering time without vernalization, while allelic variation at a region including BnaFLC.A02 contributed to flowering time after vernalization.
We detected differences in the pre‐vernalization expression level of BnaFLC.A02 between the early and late flowering varieties Cabriolet and Darmor, respectively. Striking differences in the expression profiles of both alleles of BnaFLC.A02 were also detected in response to vernalization. The expression of the BnaFLC.A02 in Cabriolet was stably silenced after six‐week vernalization, while the BnaFLC.A02 allele in Darmor reactivated upon return to ambient temperatures. Variation in the epigenetic silencing of both BnaFLC.A02 alleles highlight parallels with reports in A. thaliana and Brassica oleracea (Coustham et al., 2012; Irwin et al., 2016; Li et al., 2014). Variation in FLC silencing in both species (AtFLC and BolFLC.C02) is associated with sequence variation within non‐coding regions of the gene (Irwin et al., 2016; Li et al., 2014). Here, we report the presence of sequence polymorphisms at BnaFLC.A02 within non‐coding intronic regions, while the BnaFLC.A02 protein of both alleles is predicted to be identical. The cis‐polymorphisms responsible for the variation in BnaFLC.A02 expression are yet to be determined; however, based on reports in A. thaliana (Coustham et al., 2012; Li et al., 2014; Li et al., 2015; Questa et al., 2016) and due to conservation of AtFLC sequence between A. thaliana and Brassica sp. (Irwin et al., 2016; Schranz et al., 2002; Tadege et al., 2001; Wu et al., 2012; Xiao et al., 2013; Yuan et al., 2009; Zhao et al., 2010), it is reasonable to hypothesize that cis‐regulatory variation within intron 1 of BnaFLC.A02 is likely to underpin the differential expression dynamics detected. As has been reported for winter accessions of A. thaliana (Coustham et al., 2012; Duncan et al., 2015; Li et al., 2014), variation in the length of cold required to induce epigenetically stable silencing of BnaFLC.A02 is likely a major determinant of flowering time in European winter oilseed rape.
BnaFT.A02 has previously been identified as a candidate gene for flowering time in B. napus (Long et al., 2007; Raman et al., 2016; Wang et al., 2009). Here, we have shown allelic variation at BnaFT.A02 was associated with variation for flowering time in the absence of vernalization and is therefore similar to reports in Lupin (Nelson et al., 2017) and wheat (Yan et al., 2006). Although we were not able to amplify and sequence the whole BnaFT.A02 gene from both varieties, we identified a non‐synonymous SNP in exon 1 of BnaFT.A02 which distinguished Cabriolet from Darmor. This is predicted to encode an amino acid substitution (I48L) in the early flowering variety Cabriolet. The same amino acid substitution has been reported for a homologous gene BraFT.A07 in B. rapa (Schiessl et al., 2017b; Zhang et al., 2015) but detected in a late flowering cultivar. As the I48L mutation involves substitution between two very similar amino acids and given it is associated with contrasting flowering time phenotypes in B. rapa and B. napus, we consider it unlikely that it confers the flowering time variation detected. However, this remains to be tested.
Vernalization length had a quantitative effect on FT expression; however, we detected differences between our early and late flowering varieties. BnaFT.A02 was detectable in the early flowering variety Cabriolet before vernalization, and its expression increased after vernalization. In contrast, in the late flowering variety Darmor, BnaFT.A02 expression was not detectable by RT‐PCR before vernalization, and although detectable after vernalization, the gene was expressed at low levels as determined by qRT‐PCR. A previous report in B. rapa identified an insertion within intron 2 of BraFT.A07 which was associated with late flowering due to failed transcription of BraFT.A07 (Zhang et al., 2015). We detected the presence of polymorphisms at the CArG box motif sequence within intron 1 of BnaFT.A02 in Cabriolet compared with Darmor. In A. thaliana, the AtFLC protein binds directly with the CArG box motif sequence within intron 1 of AtFT to inhibit its expression (Helliwell et al., 2006). It is therefore plausible that variation at cis‐regulatory regions of BnaFT.A02, such as the polymorphisms detected at the CArG box motif, has resulted in a lack of FLC protein binding in intron 1 of BnaFT.A02 leading to low but detectable expression of BnaFT.A02 in Cabriolet prior to vernalization.
Genotypic data confirmed that the allelic combination BnaFLC.A02‐Cab and BnaFT.A02‐Cab conferred the earliest flowering, while BnaFLC.A02‐Dar and BnaFT.A02‐Dar conferred the latest flowering, with and without vernalization. Analysis of recombinant F2 lines segregating for BnaFLC.A02 and BnaFT.A02 indicated the phenotypic effect of both genes was dependent on genotype and vernalization treatment. BnaFLC.A02‐Cab when in combination with BnaFT.A02‐Dar conferred late flowering in the absence of vernalization, but early flowering after vernalization. In contrast, BnaFLC.A02‐Dar when in combination with BnaFT.A02‐Cab conferred early flowering in the absence of vernalization, but late flowering after vernalization. This suggests that BnaFT.A02 was more strongly associated with earliness of flowering in the absence of vernalization and that BnaFLC.A02 played a more dominant role in flowering after vernalization. Analysis of expression of both genes in recombinant F2 lines demonstrated that BnaFT.A02 and BnaFLC.A02 acted independently from one another before vernalization. Expression of the Cabriolet allele of BnaFT.A02 was detectable before vernalization treatment, regardless of BnaFLC.A02 allele, and the Darmor allele of BnaFLC.A02 exhibited reactivation of expression after vernalization independent of BnaFT.A02 allele. We hypothesize polymorphisms that affect the expression dynamics of both genes underpin the association with flowering time variation under vernalized and non‐vernalized growth conditions.
Cis‐variation at orthologues of AtFLC and AtFT in B. napus is likely to have a major influence on gene expression and ultimately the flowering time phenotype of the crop. Like in A. thaliana, cis‐variation resulting in changes to transcription factor binding sites may have played an important role in flowering time evolution in B. napus. We hypothesize plant breeding for varieties adapted to varying winter climates has selected for variation at BnaFLC.A02 and BnaFT.A02 in European winter oilseed rape adapted to different environmental conditions. This knowledge will allow for the selection of alleles of flowering time regulators that alter the vernalization requirement of oilseed rape, informing the generation of new varieties with adapted flowering times and improved yields.
Methods
Plant materials and growth conditions
An F2 population generated from a reciprocal cross between two inbred lines derived from the winter oilseed rape varieties Cabriolet and Darmor (both sourced from the oilseed rape genetic improvement network (OREGIN) B. napus diversity fixed foundation set population; http://www.herts.ac.uk/oregin) were used in the present study. Five heterozygous F1 siblings from each reciprocal cross were selected and carried forward to the F2 generation by self‐fertilization.
Seeds were sown directly onto soil (Levington F2 compost, 600 L peat, 100 L 4 mm grit, 196 g ‘Exemptor’ chloronicotinyl insecticide) and grown under glasshouse conditions (16‐h light/8‐h dark, 600 W HPS lamps provided supplementary lighting when required, 18 °C day temperature, 15 °C night temperature and 70% humidity) for 28 days before receiving one of two vernalization treatments; six‐week vernalization (VERN) or no vernalization (NVERN). For the NVERN treatment, plants were grown for 28 days under glasshouse conditions. For the VERN treatment, after 28 days’ growth under glasshouse conditions, plants were transferred to a vernalization chamber (5 °C, 8‐h light/16‐h dark, 70% humidity) for six weeks. For both vernalization treatments, a total F2 population of 720 lines (72 randomly selected seed from each of the ten F1 plants) in addition to 12 plants each of Cabriolet and Darmor were grown and assessed for flowering time variation. Sowing was staggered so that all plants from the NVERN and VERN treatments were transferred to a concrete floored poly‐tunnel (Keder house; https://www.kedergreenhouse.co.uk/) at the John Innes Centre, Norwich on the 5th and 6th April 2017, respectively. Plants received no supplementary heating or lighting, were transplanted to 1 L pots and arranged in a randomized complete block design containing four blocks at a density of 36 plants/m2. Plants were watered twice daily by automatic irrigation and chemically sprayed when required. Temperature and humidity within the poly‐tunnel were recorded by Tinytag (Gemini Data Loggers, Chichester, UK) at 30‐min intervals for the duration of the experiment.
Flowering time was measured as the number of days to opening of the first flower (BBCH60 according to Meier et al. (2009)). Flowering time measurements commenced on the date of transfer to the poly‐tunnel (28 days after sowing for NVERN and 70 days after sowing for VERN) and continued for 170 days. All plants that had no open flowers at the end of the experiment were given a did not flower (DNF) score of 170 days. Statistical analysis was performed using GenStat 18th Edition (VSN International, Hemel Hempstead, UK).
Construction of bulks and Illumina sequencing
To identify sequence variants between the parents of the F2 population, DNA was extracted from leaf tissue from both parent plants of the reciprocal cross, Cabriolet and Darmor, and sequenced by whole‐genome sequencing. The DNA bulks were generated based on the flowering time measurements obtained for the F2 population. Leaf material was pooled prior to DNA extraction, and each pool contained a 1cm leaf disc taken from lines that represented the phenotypic extremes of the population (approximately 5% of lines from both tail ends of the flowering time distribution). The number of F2 lines in each bulk is listed in Table S1. A total of four DNA bulks; early flowering bulk for VERN treatment (VERN_EARLY), late flowering bulk for VERN treatment (VERN_LATE), early flowering bulk for the NVERN treatment (NVERN_EARLY) and late flowering bulk for the NVERN treatment (NVERN_EARLY), were sequenced by whole‐genome sequencing.
DNA was extracted using a CTAB‐based method and samples prepared for Illumina sequencing by Novogene Co., Ltd., Hong Kong (https://www.novogene.com). DNA libraries were prepared using TruSeq DNA Sample HT Sample Preparation Kit (Illumina, San Diego, CA) following the manufacturer's recommendations. 1µg of DNA was fragmented using Covaris cracker, end‐repaired and adapter‐ligated. After PCR enrichment, DNA libraries were purified (AMPure XP system) and analysed for size distribution by Agilent2100 Bioanalyzer. The DNA libraries were sequenced on Illumina HiSeq X platform (Illumina Inc.) to generate 300‐base paired‐end reads at an average of 30x coverage. Sample quality control, library construction and sequencing were performed by Novogene Co., Ltd., HK.
Construction of sequence assemblies
The Darmor‐bzh B. napus genome sequence (Chalhoub et al., 2014, Genome Assembly: AST_PRJEB5043_v1, plants.ensembl.org) was used as a reference. The sequencing reads from the six samples (Cabriolet, Darmor, VERN_EARLY, VERN_LATE, NVERN_EARLY, NVERN_LATE) were aligned to the Darmor‐bzh genome reference sequence using Bowtie‐2 v2.2.3 (Langmead et al., 2009) to create six separate alignment files in Sequence Alignment/Map (SAM) format which were converted to BAM (.bam) file format using SAMtools v1.5 (Li et al., 2009). For stringency, sequence data from non‐uniquely mapped reads were excluded from the alignment by filtering for the ‘‐q 42’ parameter in SAMtools. A sequence PileUp was then generated using the ‘mpileup’ command in SAMtools (Li et al., 2009).
Variants (SNPs and small Insertion/Deletions or InDels (<9bp)) were called in Cabriolet compared with Darmor, and their genomic position was assigned according to the Darmor‐bzh reference genome (Chalhoub et al., 2014). High confidence variants were called when the read depth was greater than, or equal to, 20 and the alternative sequence was found in ≥ 95% of Cabriolet sequence reads. All other genomic positions were excluded from further analysis. Genomic positions that were genetically identical, that is no alternative sequence was detected in Cabriolet compared with Darmor, were also excluded.
Calculation of SNP index
SNP index values were calculated for the bulks according to Abe et al. (2012) and Takagi et al. (2013). In this study, the SNP index was a measure of the proportion of sequencing reads at a given variant position that differed from our chosen reference variety, Darmor. At each variant position where the read depth was ≥20 in both bulks, the proportion of total reads matching the alternative variant found in Cabriolet was calculated to give a SNP index value. Although the sequence read depth filtering parameter used in this study was stringent, a relatively even distribution of genomic positions was included in the analysis (Figure S8). A SNP index value of 1 indicated all sequencing reads at that position were derived from Cabriolet, while a SNP index value of 0 indicated all sequencing reads were derived from Darmor. A SNP index value equal to 0.5, however, indicated equal contribution of alleles from both parents, Cabriolet and Darmor, in the DNA bulk. A deviation in SNP index value away from 0.5 indicated a bias in the genetic contribution of both parents between the DNA bulks. Large deviations in SNP index values from 0.5 would identify associations between a genomic region and the phenotypic differences between the DNA bulks. A delta (Δ) SNP index value was then calculated by subtracting the SNP index of the early bulk from the SNP index of the late bulk at each variant position. To reduce ambiguity introduced by sequencing error, SNP index values that were <0.3 in both bulks were excluded (as per Takagi et al., 2013). Regions of the genome representing the top 1% of absolute ΔSNP index values were considered to be strongly associated with flowering time. The code used for this analysis is available open source (https://github.com/marc-jones/brassica-napus-bulk-segregant).
Validation of alleles within the QTL region
To validate the QTL region identified on chromosome A02, allele‐specific KASP (Kompetitive allele‐specific PCR) primers (LGC Genomics, https://www.biosearchtech.com/services/genotyping-services) were designed to target five SNPs within a 6Mbp region located between base pair positions 136 553 and 6 375 504 on chromosome A02 (Table S2). The genomic positions for each SNP were according to the Darmor‐bzh reference genome and included SNPs within orthologues of the flowering time genes AtFLC (SNP FLC‐136553 within BnaFLC.A02) and AtFT (SNP FT‐6375504 within BnaFT.A02). DNA was extracted from the 94 earliest and 94 latest F2 lines under both NVERN and VERN treatments, in addition to the parental lines Cabriolet and Darmor, and assayed for SNP genotype using the KASP genotyping chemistry according to the manufacturer's instructions (LGC Genomics, Hoddesdon, UK).
Sequence analysis of BnaFLC.A02 and BnaFT.A02
To assess for DNA polymorphisms, sequences for BnaFLC.A02 and BnaFT.A02 were extracted from the Cabriolet and Darmor sequence PileUp and aligned against the Darmor‐bzh BnaFLC.A02 and BnaFT.A02 sequences (Chalhoub et al., 2014), downloaded from plants.ensembl.org. Sequence polymorphisms were identified, and amino acid sequence changes were predicted using AlignX (Vector NTI Advance®, InvitrogenTM, now Thermo Fisher Scientific, https://www.thermofisher.com). To confirm the presence of the polymorphisms in Cabriolet and Darmor, primers were designed to amplify and sequence regions of the BnaFLC.A02 and BnaFT.A02 genes from both varieties (Table S3). DNA was isolated from both varieties using the Edwards DNA extraction method (Edwards et al., 1991), and PCR was performed using AmpliTaq GoldTM DNA polymerase (Applied BiosystemsTM, now Thermo Fisher Scientific) according to the manufacturer's instructions with an annealing temperature of 58 °C. PCR products were prepared for sequencing using the Big Dye V3.1TM terminator protocol (Applied BiosystemsTM, now Thermo Fisher Scientific), and capillary sequencing was performed by Eurofins Genomics, EU. Sequences were aligned and analysed using AlignX.
Quantitative expression analysis of BnaFLC.A02 and BnaFT.A02
For quantitative expression analysis of BnaFLC.A02 and BnaFT.A02, leaf material was taken from the newest expanded leaf of Cabriolet, Darmor and F2 plants genotyped and determined homozygous by KASP assay for BnaFLC.A02 and BnaFT.A02 before (NV), at the end (T0) and after (T16, T30) a six‐week vernalization treatment. Leaf material was sampled at the same time of day at the ¾ point of the photoperiod regime to account for photoperiodic effects on gene expression. Total RNA was extracted from individual leaf samples using the E.Z.N.A.® Plant RNA Kit (Omega Bio‐tek, Georgia, USA) and contaminating DNA were removed using the on‐column RNase‐free DNase Set I (Omega Bio‐tek) according to the manufacturer's instructions. Two micrograms RNA was converted to cDNA using SuperscriptTM III Reverse Transcriptase (InvitrogenTM, now Thermo Fisher Scientific) and gene‐specific reverse primers (Table S4) according to the manufacturer's instructions. qRT‐PCR was performed using LightCycler® 480 SYBR Green I Master Mix on the LightCycler® 480 II instrument (both Roche, www.roche.com). Second‐derivative maximum values were calculated using the LightCycler® Software to give absolute expression values. Expression values of BnaFLC.A02 and BnaFT.A02 were normalized to the internal reference gene UBC21 (Orsel et al., 2014) using the ΔΔCT method (Livak and Schmittgen, 2001). For qualitative expression analysis of BnaFT.A02, the same cDNA was amplified by PCR using AmpliTaq GoldTM DNA polymerase (Thermo Fisher Scientific) and PCR products were visualized by agarose gel electrophoresis.
Accession numbers
All raw sequence reads for Cabriolet, Darmor and the four DNA bulks have been deposited in the European Nucleotide Archive under PRJEB33550. BnaFLC.A02 and BnaFT.A02 sequence data for varieties Cabriolet and Darmor can be found under GenBank accession numbers MN218571, MN218572, MN218573 and MN218574.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
E.H.T., J.A.I. and C.D. conceived the project. All in vivo experiments were conducted by E.H.T. D.M.J. performed all bioinformatics associated with the QTL‐seq analysis. Z. H. and I.B. performed the HE analysis. M.T. contributed to bulk sampling design and conducted initial bioinformatics analysis. R.W. assisted with population development and experimental design. E.H.T., J.A.I. and C.D. wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Figure S1 Temperature and humidity recorded within the Keder plastic poly‐tunnel during the 2017 flowering time phenotyping analysis of F2 lines at JIC.
Figure S2 Frequency distributions of sequence variants (SNPs and small InDels) detected in Darmor compared with Darmor‐bzh. Each chromosome is plotted separately, and the frequency of variants detected are plotted by genome order.
Figure S3 Frequency distributions of sequence variants (SNPs and small InDels) detected in Cabriolet compared with Darmor. Each chromosome is plotted separately, and the frequency of variants detected are plotted by genome order.
Figure S4 No sequence variants (SNPs and small InDels) are detected in Cabriolet compared with Darmor at the major flowering time genes BnaFRI.A03 and BnaFLC.A10. (A) The relative position of BnaFRI.A03 on chromosome A03 is plotted with the closest sequence variant found up‐ and down‐stream highlighted with a red arrow. (B) The relative position of BnaFLC.A10 on chromosome A10 is plotted with the closest sequence variant found up‐ and down‐stream highlighted with a red arrow.
Figure S5 Visualization of homeologous genome exchanges in Cabriolet and Darmor based on DNA resequencing.
(A & B): The relative redundancy of coverage of A and C genome homeologous gene pairs is represented in CMYK colour space, with cyan component representing coverage of the Brassica A genome copy and magenta component representing coverage of the Brassica C genome copy.
(A) Genome‐wide homeologous genome exchanges in Cabriolet and Darmor. The gene pairs are plotted in Brassica C genome order (chromosomes denoted C1 to C9).
(B) Homeologous exchanges present on chromosome A02/C02. The gene pairs are plotted in Brassica chromosome C02 gene order, the relative position of BnaFLC.A02/C02 and BnaFT.A02/C02 gene pairs are highlighted.
Figure S6 Expression of BnaFLC.A02 varies between Cabriolet and Darmor before and after vernalisation.
(A‐D) Normalised expression of BnaFLC.A02 in Cabriolet, Darmor, and genotyped F2 individuals as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30). The expression levels, normalised to UBC21, detected in each line plant are plotted here.
(A) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Dar/BnaFT.A02‐Dar.
(B) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Dar/BnaFT.A02‐Cab.
(C) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Cab/BnaFT.A02‐Dar.
(D) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Cab/BnaFT.A02‐Cab.
Figure S7 Expression of BnaFT is detected in Cabriolet, but not detectable in Darmor.
(A) Normalised expression of BnaFT.A02 in Cabriolet and Darmor over time and under ambient temperature conditions as measured by quantitative RT‐PCR error bars denote one standard error around the mean calculated from at least three biological replicates, T= days from sowing.
(B) Normalised expression of BnaFT.A02 in Cabriolet and Darmor as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from three biological replicates.
(C) Normalised expression of BnaFT.C02 in Cabriolet and Darmor as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from three biological replicates.
(D) Normalised expression of BnaFT.A02 in F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from at least three biological replicates.
(E) Normalised expression of BnaFT.A02 in F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from at least three biological replicates.
Figure S8 The distribution of genomic positions included in the QTL‐seq analysis. Each chromosome is plotted as separate histograms of the frequency of genomic positions with read depth coverage of more than 20 reads and included in SNP and ΔSNP indices calculation.
Table S1 Summary of flowering time and Illumina sequencing data of parent lines and bulks for treatments VERN and NVERN.
Table S2 KASP primers used for validation of QTL region.
Table S3 Primers used to amplify and sequence BnaFLC.A02 and BnaFT.A02.
Table S4 Primers used for quantitative RT‐PCR analysis of BnaFLC.A02 and BnaFT.A02.
Supplementary Material
Acknowledgements
We thank Emily Hawkes for the BnaFLC.A02 qPCR primers and Richard Goram at the JIC Genotyping Services for DNA extraction and genotyping of F2 lines by KASP. Thank you to Jo Hepworth, Richard Morris and Lars Østergaard for useful contributions and editing of this manuscript. This work was supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) iCASE Studentship (BB/L016079/1) with BASF to E.H.T., J.A.I. and C.D. acknowledge funding from BBSRC Institute Strategic Programme (BB/P013511/1), and I.B. and Z.H. acknowledge funding from BBSRC (BB/L002124/1). Additional funding was provided by BBSRC sLoLa (BB/P003095/1).
Tudor, E. H. , Jones, D. M. , He, Z. , Bancroft, I. , Trick, M. , Wells, R. , Irwin, J. A. and Dean, C. (2020) QTL‐seq identifies BnaFT.A02 and BnaFLC.A02 as candidates for variation in vernalization requirement and response in winter oilseed rape (Brassica napus). Plant Biotechnol. J., 10.1111/pbi.13421
References
- Abe, A. , Kosugi, S. , Yoshida, K. , Natsume, S. , Takagi, H. , Kanzaki, H. , Matsumura, H. et al (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 30, 174. [DOI] [PubMed] [Google Scholar]
- Ågren, J. , Oakley, C.G. , McKay, J.K. , Lovell, J.T. and Schemske, D.W. (2013) Genetic mapping of adaptation reveals fitness tradeoffs in Arabidopsis thaliana. Proc. Natl Acad. Sci. 110, 21077–21082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I. , Morgan, C. , Fraser, F. , Higgins, J. , Wells, R. , Clissold, L. , Baker, D. et al (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 29, 762–766. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A. , Tang, H. , Wang, X. , Chiquet, J. et al (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Dong, F. , Cai, J. , Xin, Q. , Fang, C. , Liu, L. , Wan, L. et al (2018) A 2.833‐kb insertion in BnFLC. A2 and its homeologous exchange with BnFLC. C2 during breeding selection generated early‐flowering rapeseed. Mol. Plant, 11, 222–225. [DOI] [PubMed] [Google Scholar]
- Coustham, V. , Li, P. , Strange, A. , Lister, C. , Song, J. and Dean, C. (2012) Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science, 337, 584–587. [DOI] [PubMed] [Google Scholar]
- Das, S. , Upadhyaya, H.D. , Bajaj, D. , Kujur, A. , Badoni, S. , Kumar, V. , Tripathi, S. et al (2015) Deploying QTL‐seq for rapid delineation of a potential candidate gene underlying major trait‐associated QTL in chickpea. DNA Res. 22, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, S. , Holm, S. , Questa, J. , Irwin, J. , Grant, A. and Dean, C. (2015) Seasonal shift in timing of vernalization as an adaptation to extreme winter. Elife, 4, e06620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, K. , Johnstone, C. and Thompson, C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M.E. , Satagopan, J. , Yandell, B.S. , Williams, P.H. and Osborn, T.C. (1995) Mapping loci controlling vernalization requirement and flowering time in Brassica napus. Theor. Appl. Genet. 90, 727–32. [DOI] [PubMed] [Google Scholar]
- Foisset, N. , Delourme, R. , Barret, P. and Renard, M. (1995) Molecular tagging of the dwarf BREIZH (Bzh) gene in Brassica napus . Theor. Appl. Genet. 91, 756–761. [DOI] [PubMed] [Google Scholar]
- Giovannoni, J.J. , Wing, R.A. , Ganal, M.W. and Tanksley, S.D. (1991) Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Res. 19, 6553–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillo, M.A. , Li, C. , Hammond, M. , Wang, L. and Schemske, D.W. (2013) Genetic architecture of flowering time differentiation between locally adapted populations of Arabidopsis thaliana. New Phytol. 197, 1321–1331. [DOI] [PubMed] [Google Scholar]
- He, Z. , Wang, L. , Harper, A.L. , Havlickova, L. , Pradhan, A.K. , Parkin, I.A. and Bancroft, I. (2017) Extensive homoeologous genome exchanges in allopolyploid crops revealed by mRNA seq‐based visualization. Plant Biotechnol. J. 15(5), 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A. , Wood, C.C. , Robertson, M. , Peacock, W.J. and Dennis, E.S. (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high‐molecular‐weight protein complex. Plant J. 46(2), 183–192. [DOI] [PubMed] [Google Scholar]
- Hou, J. , Long, Y. , Raman, H. , Zou, X. , Wang, J. , Dai, S. , Xiao, Q. et al (2012) A Tourist‐like MITE insertion in the upstream region of the BnFLC. A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 12, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illa‐Berenguer, E. , van Houten, J. , Huang, Z. and van der Knaap, E. (2015) Rapid and reliable identification of tomato fruit weight and locule number loci by QTL‐seq. Theor. Appl. Genet. 128, 1329–1342. [DOI] [PubMed] [Google Scholar]
- Irwin, J.A. , Soumpourou, E. , Lister, C. , Ligthart, J.D. , Kennedy, S. and Dean, C. (2016) Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 87, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson, U. , West, J. , Lister, C. , Michaels, S. , Amasino, R. and Dean, C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science, 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Jones, D.M. , Wells, R. , Pullen, N. , Trick, M. , Irwin, J.A. and Morris, R.J. (2018) Spatio‐temporal expression dynamics differ between homologues of flowering time genes in the allopolyploid Brassica napus. Plant J. 96, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, C. and Muller, A.E. (2009) Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–73. [DOI] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Filiault, D. , Box, M.S. , Kerdaffrec, E. , van Oosterhout, C. , Wilczek, A.M. , Schmitt, J. et al (2014) Multiple FLC haplotypes defined by independent cis‐regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 28, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Tao, Z. and Dean, C. (2015) Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29, 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2− ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Long, Y. , Shi, J. , Qiu, D. , Li, R. , Zhang, C. , Wang, J. , Hou, J. et al (2007) Flowering time quantitative trait Loci analysis of oilseed brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics, 177, 2433–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Lin, T. , Klein, J. , Wang, S. , Qi, J. , Zhou, Q. , Sun, J. et al (2014) QTL‐seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor. Appl. Genet. 127, 1491–1499. [DOI] [PubMed] [Google Scholar]
- Mei, D. , Wang, H. , Hu, Q. , Li, Y. , Xu, Y. and Li, Y. (2009) QTL analysis on plant height and flowering time in Brassica napus. Plant Breed. 128, 458–465. [Google Scholar]
- Meier, U. , Bleiholder, H. , Buhr, L. , Feller, C. , Hack, H. , Heß, M. , Lancashire, P.D. et al (2009) The BBCH system to coding the phenological growth stages of plants–history and publications. J. Kulturpflanzen, 61, 41–52. [Google Scholar]
- Michaels, S.D. , Bezerra, I.C. and Amasino, R.M. (2004) FRIGIDA‐related genes are required for the winter‐annual habit in Arabidopsis. Proc. Natl Acad. Sci. USA, 101, 3281–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W. , Paran, I. and Kesseli, R. (1991) Identification of markers linked to disease‐resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl Acad. Sci. 88, 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, L. and Scarth, R. (1994) Vernalization response in spring oilseed rape (Brassica napus L.) cultivars. Can. J. Plant Sci. 74, 275–277. [Google Scholar]
- Nelson, M.N. , Rajasekaran, R. , Smith, A. , Chen, S. , Beeck, C.P. , Siddique, K.H. and Cowling, W.A. (2014) Quantitative trait loci for thermal time to flowering and photoperiod responsiveness discovered in summer annual‐type Brassica napus L. PLoS One, 9, e102611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M.N. , Książkiewicz, M. , Rychel, S. , Besharat, N. , Taylor, C.M. , Wyrwa, K. , Jost, R. et al (2017) The loss of vernalization requirement in narrow‐leafed lupin is associated with a deletion in the promoter and de‐repressed expression of a Flowering Locus T (FT) homologue. New Phytol. 213(1), 220–232. [DOI] [PubMed] [Google Scholar]
- Orsel, M. , Moison, M. , Clouet, V. , Thomas, J. , Leprince, F. , Canoy, A.‐S. , Just, J. et al (2014) Sixteen cytosolic glutamine synthetase genes identified in the Brassica napus L. genome are differentially regulated depending on nitrogen regimes and leaf senescence. J. Exp. Bot. 65, 3927–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Questa, J.I. , Song, J. , Geraldo, N. , An, H. and Dean, C. (2016) Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science, 353, 485–8. [DOI] [PubMed] [Google Scholar]
- Raman, H. , Raman, R. , Eckermann, P. , Coombes, N. , Manoli, S. , Zou, X. , Edwards, D. et al (2013) Genetic and physical mapping of flowering time loci in canola (Brassica napus L.). Theor. Appl. Genet. 126, 119–132. [DOI] [PubMed] [Google Scholar]
- Raman, H. , Raman, R. , Coombes, N. , Song, J. , Prangnell, R. , Bandaranayake, C. , Tahira, R. et al (2016) Genome‐wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant, Cell Environ. 39, 1228–1239. [DOI] [PubMed] [Google Scholar]
- Schiessl, S. , Samans, B. , Hüttel, B. , Reinhard, R. and Snowdon, R.J. (2014). Capturing sequence variation among flowering‐time regulatory gene homologs in the allopolyploid crop species Brassica napus . Front. Plant Sci. 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl, S. , Iniguez‐Luy, F. , Qian, W. and Snowdon, R.J. (2015) Diverse regulatory factors associate with flowering time and yield responses in winter‐type Brassica napus. BMC Genom. 16, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl, S. , Huettel, B. , Kuehn, D. , Reinhardt, R. and Snowdon, R.J. (2017a) Targeted deep sequencing of flowering regulators in Brassica napus reveals extensive copy number variation. Sci Data, 4, 170013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl, S.V. , Huettel, B. , Kuehn, D. , Reinhardt, R. and Snowdon, R.J. (2017b) Flowering time gene variation in brassica species shows evolutionary principles. Front. Plant Sci. 8, 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl, S.V. , Quezada‐Martinez, D. , Tebartz, E. , Snowdon, R.J. and Qian, L. (2019) The vernalisation regulator FLOWERING LOCUS C is differentially expressed in biennial and annual Brassica napus. Sci. Rep. 9(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz, M.E. , Quijada, P. , Sung, S.‐B. , Lukens, L. , Amasino, R. and Osborn, T.C. (2002) Characterization and effects of the replicated flowering time gene FLC in Brassica rapa . Genetics 162, 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C. , Aranzana, M.J. , Lister, C. , Baxter, C. , Nicholls, C. , Nordborg, M. and Dean, C. (2005) Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138, 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C. , Lister, C. , Crevillen, P. , Nordborg, M. and Dean, C. (2006) Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev. 20, 3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, V.K. , Khan, A.W. , Jaganathan, D. , Thudi, M. , Roorkiwal, M. , Takagi, H. , Garg, V. et al (2016) QTL‐seq for rapid identification of candidate genes for 100‐seed weight and root/total plant dry weight ratio under rainfed conditions in chickpea. Plant Biotechnol. J. 14, 2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.M. , Guan, Z. , Hu, J. , Guo, C. , Yang, Z. , Wang, S. , Liu, D. et al (2020) Eight high‐quality genomes reveal pan‐genome architecture and ecotype differentiation of Brassica napus. Nat. Plants, 6, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe, J.R. , Weinig, C. , Ungerer, M. , Olsen, K.M. , Mays, C. , Halldorsdottir, S.S. , Purugganan, M.D. et al (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc. Natl Acad. Sci. USA, 101, 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange, A. , Li, P. , Lister, C. , Anderson, J. , Warthmann, N. , Shindo, C. , Irwin, J. et al (2011) Major‐effect alleles at relatively few loci underlie distinct vernalization and flowering variation in Arabidopsis accessions. PLoS One, 6, e19949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege, M. , Sheldon, C.C. , Helliwell, C.A. , Stoutjesdijk, P. , Dennis, E.S. and Peacock, W.J. (2001) Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28, 545–553. [DOI] [PubMed] [Google Scholar]
- Takagi, H. , Abe, A. , Yoshida, K. , Kosugi, S. , Natsume, S. , Mitsuoka, C. , Uemura, A. et al (2013) QTL‐seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Long, Y. , Wu, B. , Liu, J. , Jiang, C. , Shi, L. , Zhao, J. et al (2009) The evolution of Brassica napus FLOWERING LOCUS T paralogues in the context of inverted chromosomal duplication blocks. BMC Evol. Biol. 9, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. , Qian, W. , Suppanz, I. , Wei, L. , Mao, B. , Long, Y. , Meng, J. et al (2011) Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA. FRI. a. J. Exp. Bot. 62, 5641–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H. , Cheng, H. , Wang, W. , Liu, J. , Hao, M. , Mei, D. , Zhou, R. et al (2016) Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL‐seq. Sci. Rep. 6, 38493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Wei, K. , Cheng, F. , Li, S. , Wang, Q. , Zhao, J. , Bonnema, G. et al (2012) A naturally occurring InDel variation in BraA. FLC. b (BrFLC2) associated with flowering time variation in Brassica rapa . BMC Plant Biol. 12, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, D. , Liang, Z. , Yan, T. , Xu, Y. , Xuan, L. , Tang, J. , Zhou, G. et al (2019) Whole‐genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant 12(1), 30–43. [DOI] [PubMed] [Google Scholar]
- Xiao, D. , Zhao, J.J. , Hou, X.L. , Basnet, R.K. , Carpio, D.P. , Zhang, N.W. , Bucher, J. et al (2013) The Brassica rapa FLC homologue FLC2 is a key regulator of flowering time, identified through transcriptional co‐expression networks. J. Exp. Bot. 64, 4503–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Hu, K. , Zhang, Z. , Guan, C. , Chen, S. , Hua, W. , Li, J. et al (2016) Genome‐wide association study reveals the genetic architecture of flowering time in rapeseed (Brassica napus L.). DNA Res. 23, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, L. , Fu, D. , Li, C. , Blechl, A. , Tranquilli, G. , Bonafede, M. , Sanchez, A. et al (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl Acad. Sci. 103, 19581–19586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, L. , Chen, C. , Yin, S. , Li, H. , Li, Z. , Wang, B. , King, G.J. et al (2018) Sequence variation and functional analysis of a FRIGIDA orthologue (BnaA3. FRI) in Brassica napus. BMC Plant Biol. 18, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.X. , Wu, J. , Sun, R.F. , Zhang, X.W. , Xu, D.H. , Bonnema, G. and Wang, X.W. (2009) A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J. Exp. Bot. 60, 1299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Meng, L. , Liu, B. , Hu, Y. , Cheng, F. , Liang, J. , Aarts, M.G. et al (2015) A transposon insertion in FLOWERING LOCUS T is associated with delayed flowering in Brassica rapa. Plant Sci. 241, 211–220. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Kulkarni, V. , Liu, N. , del Carpio, D.P. , Bucher, J. and Bonnema, G. (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa . J. Exp. Bot. 61, 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, X. , Suppanz, I. , Raman, H. , Hou, J. , Wang, J. , Long, Y. , Jung, C. et al (2012) Comparative analysis of FLC homologues in Brassicaceae provides insight into their role in the evolution of oilseed rape. PLoS One, 7, e45751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Temperature and humidity recorded within the Keder plastic poly‐tunnel during the 2017 flowering time phenotyping analysis of F2 lines at JIC.
Figure S2 Frequency distributions of sequence variants (SNPs and small InDels) detected in Darmor compared with Darmor‐bzh. Each chromosome is plotted separately, and the frequency of variants detected are plotted by genome order.
Figure S3 Frequency distributions of sequence variants (SNPs and small InDels) detected in Cabriolet compared with Darmor. Each chromosome is plotted separately, and the frequency of variants detected are plotted by genome order.
Figure S4 No sequence variants (SNPs and small InDels) are detected in Cabriolet compared with Darmor at the major flowering time genes BnaFRI.A03 and BnaFLC.A10. (A) The relative position of BnaFRI.A03 on chromosome A03 is plotted with the closest sequence variant found up‐ and down‐stream highlighted with a red arrow. (B) The relative position of BnaFLC.A10 on chromosome A10 is plotted with the closest sequence variant found up‐ and down‐stream highlighted with a red arrow.
Figure S5 Visualization of homeologous genome exchanges in Cabriolet and Darmor based on DNA resequencing.
(A & B): The relative redundancy of coverage of A and C genome homeologous gene pairs is represented in CMYK colour space, with cyan component representing coverage of the Brassica A genome copy and magenta component representing coverage of the Brassica C genome copy.
(A) Genome‐wide homeologous genome exchanges in Cabriolet and Darmor. The gene pairs are plotted in Brassica C genome order (chromosomes denoted C1 to C9).
(B) Homeologous exchanges present on chromosome A02/C02. The gene pairs are plotted in Brassica chromosome C02 gene order, the relative position of BnaFLC.A02/C02 and BnaFT.A02/C02 gene pairs are highlighted.
Figure S6 Expression of BnaFLC.A02 varies between Cabriolet and Darmor before and after vernalisation.
(A‐D) Normalised expression of BnaFLC.A02 in Cabriolet, Darmor, and genotyped F2 individuals as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30). The expression levels, normalised to UBC21, detected in each line plant are plotted here.
(A) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Dar/BnaFT.A02‐Dar.
(B) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Dar/BnaFT.A02‐Cab.
(C) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Cab/BnaFT.A02‐Dar.
(D) Normalised expression of BnaFLC.A02 detected in 3 Cabriolet, 3 Darmor and 10 F2 individuals with genotypic combination BnaFLC.A02‐Cab/BnaFT.A02‐Cab.
Figure S7 Expression of BnaFT is detected in Cabriolet, but not detectable in Darmor.
(A) Normalised expression of BnaFT.A02 in Cabriolet and Darmor over time and under ambient temperature conditions as measured by quantitative RT‐PCR error bars denote one standard error around the mean calculated from at least three biological replicates, T= days from sowing.
(B) Normalised expression of BnaFT.A02 in Cabriolet and Darmor as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from three biological replicates.
(C) Normalised expression of BnaFT.C02 in Cabriolet and Darmor as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from three biological replicates.
(D) Normalised expression of BnaFT.A02 in F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from at least three biological replicates.
(E) Normalised expression of BnaFT.A02 in F2 lines genotyped for BnaFLC.A02 and BnaFT.A02 as measured by quantitative RT‐PCR before (NV) and after vernalisation (6WT0, 6WT16, 6WT30), error bars denote one standard error around the mean calculated from at least three biological replicates.
Figure S8 The distribution of genomic positions included in the QTL‐seq analysis. Each chromosome is plotted as separate histograms of the frequency of genomic positions with read depth coverage of more than 20 reads and included in SNP and ΔSNP indices calculation.
Table S1 Summary of flowering time and Illumina sequencing data of parent lines and bulks for treatments VERN and NVERN.
Table S2 KASP primers used for validation of QTL region.
Table S3 Primers used to amplify and sequence BnaFLC.A02 and BnaFT.A02.
Table S4 Primers used for quantitative RT‐PCR analysis of BnaFLC.A02 and BnaFT.A02.
Supplementary Material


