Abstract
The human gut is important for food digestion and absorption, as well as a venue for a large number of microorganisms that coexist with the host. Although numerous in vitro models have been proposed to study intestinal pathology or interactions between intestinal microbes and host, they are far from recapitulating the real intestinal microenvironment in vivo. To assist researchers in further understanding gut physiology, the intestinal microbiome, and disease processes, a novel technology primarily based on microfluidics and cell biology, called “gut-on-chip,” was developed to simulate the structure, function, and microenvironment of the human gut. In this review, we first introduce various types of gut-on-chip systems, then highlight their applications in drug pharmacokinetics, host–gut microbiota crosstalk, and nutrition metabolism. Finally, we discuss challenges in this field and prospects for better understanding interactions between intestinal flora and human hosts, and then provide guidance for clinical treatment of related diseases.
Keywords: Microfluidics, gut-on-chip, host–microbiome interactions, drug pharmacokinetics, artificial gut
Introduction
Because of its significant roles in digestion, absorption, disease regulation, and immune function, the human intestine has attracted great attention in recent years, particularly as research involving the human microbiome progresses. Most nutrients and oral drugs need to be absorbed in the intestinal tract. More than 70% of immune cells are located in the intestinal mucosa, and the number of lymphocytes is much higher than that of other lymphoid tissues.1 Thus, intestine is a natural protective barrier to pathogens and other harmful microorganisms.2 In addition, microbes in the human intestine are reportedly associated with a series of diseases, such as indigestion, obesity, and immune damage.3–5 Therefore, in vitro simulation of the intestinal microenvironment is of great significance for drug development and disease treatment.6 At present, mammalian animal models and static Transwell models are most commonly utilized to mimic the human gut system for investigations of human gut physiology and the microbiome.7 Mammals have been used extensively to test drug metabolism, toxicity, and efficacy in preclinical periods,8,9 and two-dimensional (2D) Transwell culture systems offer a simple in vitro approach to realize short-term observations and detection of microbiome–host interactions through the growth of a monolayer intestinal epithelium with tight junctions and cellular polarity.10,11 However, deficiencies of these two models limit exploration within this field. On the one hand, animal models require a long experimental cycle, involve extensive labor, and have high costs. Furthermore, as there are large differences between animal and human physiology, the physiological response of the human intestinal tract may not be well predicted.12–14 On the other hand, the static model cannot simulate features of the intestinal microenvironment such as fluid flow, villi structures, and peristalsis. Thus, it is difficult to mimic the comprehensive functions of human intestinal tract and realize real-time observations of interactive dynamics.15 Up to date, organoid culture using induced pluripotent stem cells or primary intestinal cells is an emerging method to regenerate three-dimensional (3D) intestine architecture with fully differentiated intestinal epithelial cells.16 Nevertheless, organoid culture stays static that does not experience fluid flow and peristalsis existing in human intestine. Although intestine organoid can simulate gut physiology, it is hard to inject microbial cells or drug to the apical side of intestinal epithelium because the lumen of intestine organoid is enclosed.17 So this approach is still challenged by host-microbiome interactions and intestine physiology including absorption, drug pharmacokinetics and nutrition metabolism. As a suitable platform for in vitro cell culture, microfluidics has attracted a lot of attention for recreating diverse human organs, namely organ-on-chip technology.18,19 In particular, gut-on-chip is becoming a powerful tool to construct physiological models of the human gut, perform drug testing and development, and investigate host–microbiome interactions.17,20,21 Compared with conventional model systems, gut-on-chip offers the capabilities of real-time observation, simulation of the intestinal microenvironment, adjustable fluid flow shear stress and host–microbiome interface.22,23
In this review, we first provide comprehensive information about the human intestinal microenvironment, including the structure, function, immune components, and microbiome, and demonstrate why gut-on-chip technology has been developed. Next, we classify the design of gut-on-chip platforms into 2D models with an epithelial layer and 3D models with intestinal villi, and introduce their establishment. Finally, we summarize emerging applications of gut-on-chip in drug pharmacokinetics and development, interactions between host and intestinal microorganisms, and nutrition metabolism, and further depict what gut-on-chip systems can explore. Furthermore, we provide viewpoints on the importance and current limitations of this technology, and envision future applications in biomedical fields.
Characteristics of the human gut microenvironment
The human intestine is not only the biggest digestion and absorption organ, but also home to a wide variety of microorganisms that inhabit the human intestinal tract, estimated at 100 trillion bacteria representing 500 to 1000 species.24,25 The gut microbiome is closely related to human health and essential for facilitating gut homeostasis, contributing to nutrition absorption, drug metabolism, maturation of the immune system, and protection against pathogens.26–28 Once the intestinal flora is disturbed, changes in its structure and function (even breaking the immune balance) likely lead to a series of intestinal diseases, such as inflammatory bowel disease (IBD), and ileus or even colorectal cancer.29,30
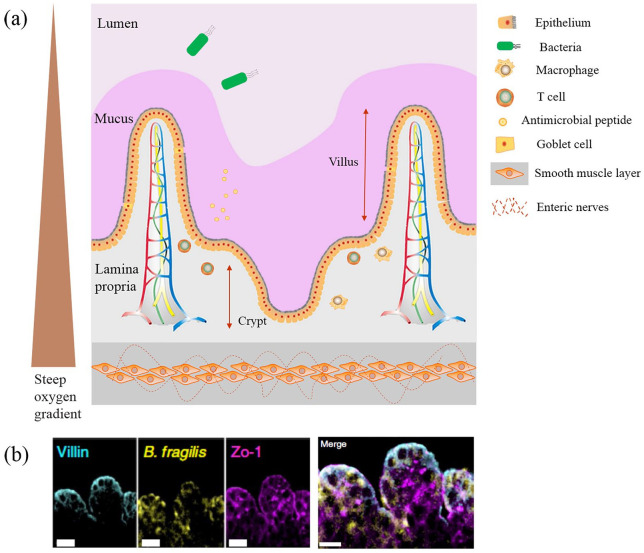
To support physiological function, intestine has a unique microenvironment, as shown in Figure 1(a). Normally, intestine consists of a closed inner cavity that folds into small finger-like projections, called villi, that tremendously increase the absorptive surface area compared with a flat surface. At the base of the intestinal glands, called crypts, proliferative stem cells produce new epithelial cells capable of differentiating into four cell types: absorptive enterocytes, goblet cells, enteroendocrine cells, and Paneth cells. The differentiated cells then migrate upwards to form the epithelial layer.15 Integrity of the intestinal epithelium is mostly characterized by tight junctions and polarity, which ensure that digested nutrients can only enter the body from the intestinal cavity to capillaries surrounding each villous.32 As one of the largest bacterial habitats, balanced host–microbe interactions within the human intestine are necessary for maintaining homeostasis. The gut microbiota has multiple functions in human health. For instance, intestinal flora is helpful for the fermentation of non-digestible carbohydrates to produce short-chain fatty acids.33 The gut microbiome provides protection against pathogens via competition for uptake of trace elements and ecological niches, and the strengthening of host defense mechanisms. When the composition of the microbiome changes, whether by dietary changes, antibiotic treatment, or incursive pathogen, disruptions in the balance of the microbiota can induce the occurrence of disease in the organism. The intestine has unique structures and functions for maintaining its homeostasis. The mucus layer covering the intestinal epithelium creates a boundary between the gut lumen and host tissue, which limits direct contact between intestinal or invasive microorganisms and human cells.34 In the presence of mucus, antimicrobial molecules produced by the epithelium can accumulate at a high local concentration, which efficiently inhibits the detrimental effects of bacterial colonization and growth.35 Even if some bacterial colonies can penetrate the mucus layer, enterocytes can further secrete responsive chemokines and cytokines to regulate the functionality of immune cells in the lamina propria.36 These local immune responses are considered to be key for solving intestinal barrier infections and promoting mucosal repair after intestinal injury.5 The epithelium is encircled by smooth muscle layers, with the enteric neural system embedded within muscle to control intestinal peristalsis and move materials within the intestinal cavity in a constant forward direction.37–39 Variations in oxygen distribution are another feature of the intestinal microenvironment. From the mucosal layer to the cavity, the oxygen gradient gradually decreases to an anaerobic environment that determines the distribution of various microbes.35,40
Figure 1.
Microenvironment in human intestine and gut-on-chip. (a) Illustration of the human gut microenvironment. The main components of the intestinal microenvironment include the intestinal epithelial villi, mucus layer, lamina propria, and symbiotic microbial community. Capillaries and the blood vasculature, and immune cells within the lamina propria control transport of nutrients and immune responses, respectively. Three muscle layers encircle the epithelium to regulate intestinal movements in combination with neural plexus embedded within the muscle. (b) Vertical cross-sectional confocal views through the intestinal epithelial villi–microbiome interface stained for villin (cyan) and ZO-1 (magenta) in gut-on-chip co-cultured with HADA-labelled B. fragilis (yellow) under anaerobic condition, scale bar, 50 μm. Reprint permission was obtained from Jalili-Firoozinezhad et al.31
Recent studies have found that intestinal microorganisms play a very important role in maintaining intestinal health and immune regulation. To decipher molecular mechanisms, most research thus far has employed traditional 2D models, such as Transwell technology.41 These 2D models cannot reproduce intestinal cell physiology or tissue morphology, and are unable to reconstruct key intestinal differentiation functions. Furthermore, such static models such as organoids are far removed from real physiological fluid environment, and are thus insufficient to support co-cultures of bacteria and intestinal cells. Therefore, a more realistic 3D model is imperative to simulate the core structures and key functions of intestine (Figure 1(b)).
Design of gut-on-chip
The rapid development of microfluidic technology in recent years has made it an effective way to emulate the human gastrointestinal tract. Typically called “gut-on-chip,” intrinsic features of this platform include low cost and consumption, flexible control of multiple system parameters (including fluid flow and oxygen concentration), and compatibility with cell graphic culture and real-time microscopic imaging.42,43 Over the past 10 years, gut-on-chip platforms have evolved from simple 2D structures to include more comprehensive functionality, such as villi structures, intestinal peristalsis, oxygen gradients, and even immune systems. The development of gut-on-chip system is summarized in Table 1 and Figure 2 shows a brief diagram of these models.
Table 1.
Characteristics and applications of gut-on-chip models.
| Model | Cell type | Supporting materials for cell | Static or fluid flow | Oxygen modulation | Application |
|---|---|---|---|---|---|
| 2D model | Caco-2 | Polyethylene terephthalate (PET) porous membrane | Fluid flow (1 μL/min) | No | Intestinal absorptive functionality44 |
| Caco-2 | Polycarbonate (PC) porous membrane | Fluid flow (0.1 μL/min) | No | Drug permeability45 | |
| Caco-2 | PC porous membrane | Fluid flow (25 μL/min) | Yes | Host–microbe molecular interactions46 | |
| Caco-2, HepG2 | Polyester porous membrane | Fluid flow (96 μL/h) | No | First pass metabolism of drugs47 | |
| Caco-2, U937 | PET porous membrane | Fluid flow (10–20 nL/s) | No | Nutrition metabolism and immunomodulatory function48 | |
| Scaffold-based 3D model | Caco-2 | Poly (lactic-co-glycolic acid) scaffolds | Static | No | Interaction between pathogen and probiotic49 |
| Caco-2 | Hydrogel scaffold | Fluid flow (100 μL/min) | No | Intestinal absorptive functionality50 | |
| Caco-2 | Hydrogel scaffold | Static | No | Intestinal barrier function51 | |
| Stretching-based 3D model | Caco-2 | PDMS porous membrane | Fluid flow (30 μL/h) | No | Host-microbe co-culture,52,53 enteric virus infection54 |
| Caco-2, PBMCs, capillary HMVECs, lymphatic HMVECs | PDMS porous membrane | Fluid flow (30 μL/h) | No | Gut inflammation model55 | |
| Caco-2, PBMCs | PDMS porous membrane | Fluid flow (50 μL/h) | No | Epithelial–immune interactions and host–microbiome cross-talk56 | |
| Intestinal biopsy-derived organoids | PDMS porous membrane | Fluid flow (60 μL/h) | No | Modeling normal intestinal physiology57 | |
| Caco-2, HIMECs, intestinal organoids | PDMS porous membrane | Fluid flow (60 μL/h) | Yes | Co-culture of anaerobic and aerobic commensal31 | |
| Caco-2 | PDMS porous membrane | Fluid flow (50 μL/h) | Yes | Co-culture of obligate anaerobic gut microbiome58 | |
| Caco-2, human umbilical vein endothelial cells | PDMS porous membrane | Fluid flow (60 μL/h) | No | Recapitulating disease model and countermeasure drugs screening59 |
PDMS: polydimethylsiloxane; PBMCs: peripheral blood mononuclear cells; capillary HMVECs: human capillary microvascular endothelial cells; lymphatic HMVECs: human lymphatic microvascular endothelial cells; HIMECs: human intestinal microvascular endothelial cells.
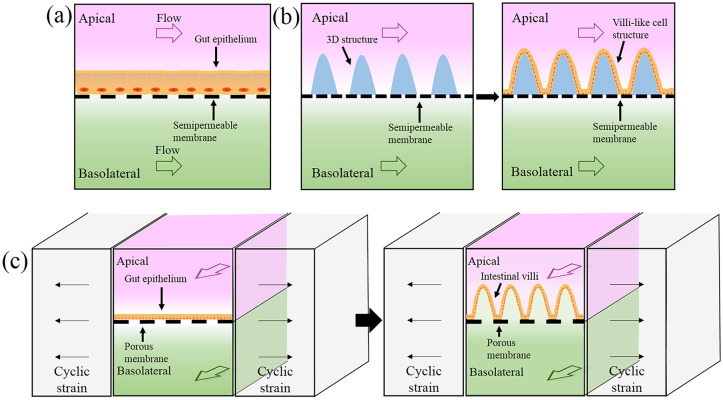
Figure 2.
Schematic illustrations of gut-on-chip models. (a) Two-dimensional gut-on-chip model comprising upper and lower channels, with a semipermeable membrane designed to serve as both a microchannel segregator and carrier for growing cells. (b) Three-dimensional villi-like structures on gut-on-chip models formed by culturing intestinal cells on designed microstructures. (c) Villi formation in gut-on-chip by mechanically stretching porous membrane.
Two-dimensional gut-on-chip model
Initially, the most common device structure contained two channels (upper and lower) separated by a semipermeable membrane, such as polycarbonate or polyester materials. Intestinal epithelial cells growing on the membrane form apical and basolateral sides of a monolayer epithelium (Figure 2(a)). This simple model has commonly been used to assess the pharmacokinetic properties of drug and nutrition absorption in vitro.22 For example, Kimura et al. developed a similar device with integrated on-chip pumping and optical detection. The two independent channels were vertically separated by a polyester semipermeable membrane, on which cells were inoculated and cultured.60 Imura et al. designed a more complex system consisting of two-tiered channels divided by a collagen-coated semipermeable membrane and the absorptive functionality was assessed as permeation of the anticancer drug cyclophosphamide.44 In addition, other gut-on-chip models attempted to integrate more advanced instruments to perform specific detection modalities.61 For instance, Gao et al. integrated an ESI-Q-TOF MS (electrospray ionization quadrupole time-of-flight mass spectrometer) to analyze molecules transported across the epithelial layer.45 The microchip consisted of two units: one simulating cell culture on a polycarbonate membrane, the other for molecule purification and concentration via miniaturized solid-phase extraction. Shah et al. designed a HuMiX (human–microbial crosstalk) system allowing co-culture of human and microbial cells at the gastrointestinal human–microbe interface; in addition, oxygen sensors were integrated to monitor gas conditions.46 Their system comprised three microchannels for medium flow, cell culture, and embedded sensors, respectively.
As described above, the 2D gut-on-chip model can simulate fluid shear stress on intestinal cells to reduce the demand on cells and medium. Continuous flow maintains the nutrition supply and washes away metabolic waste over time.62 In most cases, the viability of intestinal epithelium is better than Transwell, thus allowing long-term co-culture of microbiome and host cells.63 However, the villous microstructure, which is one of the most functional units within the intestine, fails to be simulated and investigated.
Three-dimensional gut-on-chip model with villi microstructures
The villi microstructures within the intestinal tract not only act as a physiological barrier for the epithelial layer, but more importantly increase the absorption area of the intestinal surface.64 Various types of villi-like structures have been proposed to reproduce the in vivo intestine microenvironment.31,52,65,66 For example, Sung et al. fabricated a hydrogel array based on laser ablation and soft lithography technology, and then cultured epithelial cells on the 3D microstructures to simulate the shape and distribution density of human intestinal villi.65 Similar studies revealed that these villi-shaped scaffolds could change physiological intestinal functions, such as drug absorption,67 mucus production, and resistance to bacterial invasion.49,51 When the culture time was extended to 2 weeks, the height of villi microstructures could reach 100 to 150 μm, thus improving intestinal epithelial barrier function and selective permeability.50,68 Wang et al. modified a 3D porous substrate to simulate the villi-crypt axis, and cell migration across the crypt axis could be clearly monitored.69 Costello et al. improved the scaffold using poly (lactic-co-glycolic acid) to support the co-culture of different cell lines, and to evaluate interactions among bacterial adhesion and pathogenic invasion in static culture.49,66 Furthermore, they created a bioreactor containing villus scaffolds and fluid flow.70 Besides the physical fabrication of a 3D scaffold for villi-like structure formation (Figure 2(b)), other reported protocols employed mechanical stretching against an elastic porous membrane to which intestinal epithelial cells attach.52,53 The stretching process can simulate peristaltic motions that the other microstructures above cannot perform. Artificial peristalsis can then induce the morphogenesis of 3D villi lined by the four types of intestinal epithelial cells (Figure 2(c)). Kim et al. used this strategy to induce human Caco-2 cells to undergo villus differentiation and produce intestinal villi-like structures.71 Based on this method, they found that optimized experimental conditions to transform monolayer Caco-2 cells to villous morphology were medium flow speed at 30 μL/h in both upper and lower channels, and a cyclic strain frequency of 0.15 Hz.52,53 Moreover, Caco-2 cells not only formed intestinal villi, but also developed basolateral proliferative crypts and ultimately differentiated into multiple intestinal cell types. The resulting vascular endothelial cells could then be further cultured within the lower vascular channel to recreate the intestinal tissue-tissue interface.55 This tissue-level gut-on-chip exhibited gut functions such as nutrient digestion, mucus secretion, intestinal barrier formation, and even multilineage differentiation.57,72 Thus far, all the reviewed work was performed in an aerobic environment, which is easier to fabricate and manipulate. However, human gut, especially the intestinal cavity, is a strictly anaerobic environment. Therefore, to emulate the anaerobic condition of the intestinal tract, biologists further developed anaerobic intestinal chips.31 Shin et al. developed an anoxic–oxic interface-on-a-chip (AOI Chip) to support the growth of anaerobes.58 In their work, anoxic culture medium was perfused into the upper channel for bacterial growth, whereas oxic culture medium was added to the lower channel for intestinal cells. Their results revealed that both the epithelial layer and medium flow are necessary and sufficient to create a steady-state vertical oxygen gradient in the AOI Chip. Two obligate anaerobic commensals were successfully co-cultured without compromising cell viability. Recently, a more complex anaerobic gut-on-chip system was built to maintain dynamic interactions between mucus-producing human intestinal epithelium and aerobic/anaerobic commensal microbes, and the villus structure and hollow vascular lumen were well-lined with endothelial cells.31 In this work, six micro-sensors were integrated into the device for in situ oxygen measurements, and the whole device was incubated in a custom anaerobic chamber.
In general, 3D gut-on-chip models provide a better solution to simulate the human gut in vitro, including the epithelial/endothelial layer, villous microstructures, mucus, anaerobic environment, and even peristalsis. These advanced units ensure more realistic functions of the human gut and permit increased applications, such as drug screening, host–microorganism interactions, and nutrition metabolism.
Applications of in vitro gut-on-chip models
Oral delivery is considered to be the most convenient drug administration method. Before reaching the systemic circulation, orally administrated drugs are absorbed and metabolized first by the small intestine epithelium.64 Thus, understanding oral drug absorption, intestinal transportation, and toxicity is indispensable to the drug development process.73 Many foods can cause allergic reactions in different people. Similar to drug pharmacokinetics, nutrients of food are absorbed first in the small intestine and then delivered to the rest of the body through blood vessels.74 Thus, it is important to understand absorption, metabolism, and distribution of digested food to identify treatments for food allergies as well. Traditional in vitro models based on an intestinal epithelial monolayer lack gut physiological characteristics such as intestinal structures, tissue-tissue interactions, and host–microbiome crosstalk, which is strongly related to human health. Although animal models are physiologically relevant to human, there are huge discrepancies between different animal models and humans in terms of absorption, metabolism, microbiome, and gastrointestinal tract. Gut-on-chip allows engineering of an artificial gut containing various human cell types (intestinal epithelial, endothelial, and immune) with a controlled biochemical microenvironment. Importantly, the creation of gut-on-chip models supports drug pharmacokinetic research, the testing of food allergenicity, and prediction of drug efficacy, thus accelerating both the drug development process and analysis of relationships between food and inflammation.75,76 Furthermore, the balance between the gut microbiome or gut flora and host is of great significance to maintaining intestinal homeostasis. Human gut-on-chip can be exploited for co-culture with the microbiome, to recreate intestinal disease models, test targeted therapeutics, and dissect complex disease mechanisms. In this section, we review applications of gut-on-chip systems for evaluating drug pharmacokinetics, host–gut microbiota crosstalk, and nutrition metabolism (Figure 3).
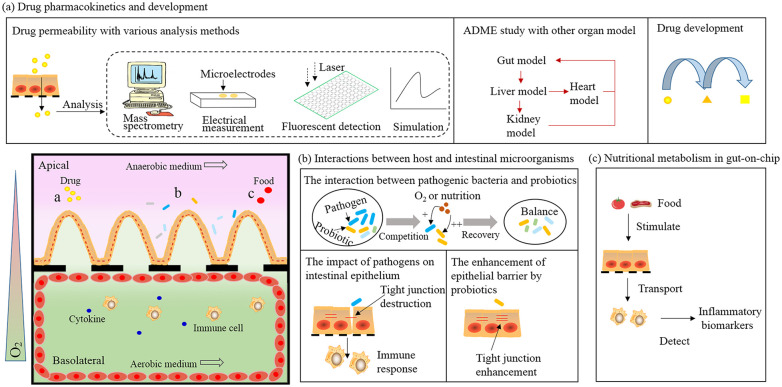
Figure 3.
Applications of gut-on-chip system (bottom left). (a) Drug permeability and pharmacokinetics. By combining with multiple analysis tools, gut-on-chip is easy to realize drug absorption and metabolism with the other organ model to accelerate drug development. (b) Gut-on-chip can be used to understand pathogen-induced pathogenesis, analyze host responses, and identify probiotic therapies. (c) Nutritional metabolism in gut-on-chip. Multiple stimuli are applied to the intestinal villi layer, together with digested nutrients, then transported into the basolateral layer. Ultimately, the inflammatory biomarkers are detected to evaluate the ability of nutrient modulation on inflammatory activities.
Drug pharmacokinetics and development
Drug absorption mainly occurs in the intestinal tract, but studying intestinal permeability in animal models in vivo is very challenging. Although drugs absorbed by passive transport show a good correlation between animals and humans, there are significant distinctions between animal models and humans. Thus, many alternative in vitro assays have been developed to investigate intestinal absorption mechanisms for diverse types of drugs.76 With regard to these investigations, gut-on-chip provides an efficient method because of the aforementioned advantages (Figure 3(a)).47,64 For instance, Park et al. employed a gut-on-chip platform with a microhole-trapping array to measure the cellular permeability of ten well-known drugs commonly used as antihypolipidemic agents or diuretics.77 Gao et al. integrated a microdevice with mass spectrometry to measure the permeability of curcumin (a polyphenolic antioxidant), that could further improve detection capability.45 More importantly, gut-on-chip can serve as one part of a body-on-chip system to evaluate continuous pharmacokinetic processes such as absorption, distribution, metabolism, and excretion (ADME) of various drug administration routes, and predict drug efficacy.78–80 An et al. created a laminated microfluidic device to mimic the drug ADME response in vivo. They measured three drugs (propranolol, thiopentone and pentobarbital) pharmacokinetics on chip and found that propranolol was better absorbed than thiopentone and pentobarbital.78 Kimura et al. developed a body-on-chip model including an intestinal barrier, liver metabolism, and lung model for in vitro evaluation of three anticancer drugs (epirubicin, irinotecan, and cyclophosphamide).81 Imura et al. integrated an intestine–liver model to evaluate correlations among intestinal drug absorption, hepatic metabolism, and bioactivity of breast cancer cells.82 Li et al. designed an intestine–kidney chip to investigate the absorption of a drug (digoxin) from the small intestine, as well as toxicity to the downstream kidney, thus providing a powerful platform to test drug absorption and nephrotoxicity. Meanwhile, gut-on-chip platforms have also been applied for drug screening and development. Sasan et al. engineered an intestinal epithelium-endothelium model to identify new radioprotective drugs by physiologically mimicking radiation damage.59 They exposed the above model to dimethyloxaloylglycine, a compound that prevents radiation injury in rats, and noticed that intestinal permeability and microvillus injury were efficiently mitigated. Moreover, apoptosis, reactive oxygen species generation, and lipid degradation were significantly reduced in both epithelial and endothelial cells. From another perspective, the gut-on-chip could also be used to create more human-relevant pathological models for screening and development of appropriate countermeasures.83 By utilizing patient-derived stem cells, this platform also provides a novel way to potentially develop personalized therapies in the future.84
Interactions between host and intestinal microorganisms
Through long-term co-evolution, the gut microbiome has become inseparable from the host and closely related to human health.85,86 In recent years, gut microbes have been implicated in many diseases, such as obesity, inflammatory bowel disease, and neurobehavioral disorders.87–89 To decipher the detailed mechanism of such correlations, gut-on-chip technology can provide an appropriate platform by simulating the gut microenvironment, simplifying the microbiome, and permitting monitoring on interactions in vitro (Figure 3(b)).63
In the typical two-channel design separated by a porous membrane, the upper channel can be used to mimic the host intestinal cavity, where the interaction between epithelium and microbes occurs, while the lower channel can represent blood vessels that provide nutrients and oxygen to epithelial cells, and even be supplemented with immune cells to understand the critical role of the immune system.71 Kim et al. engineered such a chip and found that the co-culture of non-pathogenic E. coli and Lactobacillus rhamnosus GG significantly increased epithelial integrity. The bacterial strains and mammalian cells could be labeled with different fluorescence markers, thus allowing interactive dynamics to be detected.52 To expand the capabilities of this technology, the authors further recreated a gut inflammation model by combining immune cells, commensal strains, pathogens, probiotics, and antibiotics.55 They noticed that the colonization of pathogenic E. coli (EIEC) destroyed villi structure and caused gut injury even in the presence of immune cells. They proposed that the pre-colonization with the probiotic VSL#3 could inhibit the growth of EIEC and protect against gut injuries. Moreover, the simulated peristalsis implicated that ileus could be exacerbated by peristaltic interruption and bacterial overgrowth within the small intestine. This work provided a good idea to study the repair effect of probiotics on intestinal inflammation on chip. Shin et al. reconstructed a similar intestinal inflammation model by simulating physiological flow and movement, and found that a complete epithelial barrier was necessary to maintain gut homeostasis in host–intestinal microbiota crosstalk.56 Moreover, pathogen infection model is a significant application area of gut-on-chip. Grassart group, who utilized gut-on-chip to interrogate the impact of intestinal mechanical forces on Shigella infection, found that a 3D gut-on-chip with intestinal flow and peristaltic motion enhanced Shigella invasion and infectivity compared with a conventional 2D system with no mechanical forces.90 Other than bacteria, gut-on-chip has also been explored as an in vitro model for interactions between the host and infective viruses, and to depict mechanisms of enterovirus pathogenesis.54 Pathogen infection model as descripted above can offer a new insight in pathogenesis study and even provide guidance for the clinical treatment of infectious diseases.
The human gut microbiome contains thousands of different bacterial strains, many of which are actually obligate anaerobes.91 Thus, another pivotal role for gut-on-chip is the investigation of crosstalk between the gut microbiome and host under anaerobic conditions. One recent example is the HuMix model, which was proposed to investigate the interface across host and microbiota.46 For HuMix, the authors co-cultured human intestinal epithelial cells with the obligate anaerobe Bacteroides caccae and facultative anaerobe Lactobacillus rhamnosus GG (LGG), and analyzed interactions by transcriptomics, metabolomics, and immunological assays. They found that the addition of B. caccae resulted in significant changes in transcriptional and metabolic responses of intestinal cells compared with intestinal cells co-cultured with only LGG. Shin et al. co-cultured obligate anaerobic symbiotic Bifidobacterium adolescentis and Eubacterium hallii on a similar gut chip. Epithelial cell viability was unaffected, even after 1 week, and the two bacterial strains gradually colonized. It is noteworthy that the integrity of the epithelial barrier was significantly higher in the presence of B. adolescentis in anoxic medium compared with control culture in oxic medium. However, co-culture with E. hallii did not cause any significant difference in barrier integrity, possibly because of complicated anaerobic interactions among the microenvironment, bacterial growth, and epithelial regulation.58 Using an anaerobic gut-on-chip, Jalili-Firoozinezhad et al. also illustrated that anaerobic conditions could physiologically maintain higher microbial diversity than aerobic conditions.31 In this work, the authors co-cultured microbial cells and epithelial cells in anaerobic and aerobic conditions, respectively, by modulating the oxygen concentration. This method provided a new opportunity to simulate the real interface between anaerobe and host as the human intestinal environment. With the gut-on-chip technology developing from initial two-dimensional models to current more comprehensive systems, studies on host-microbial interactions range from a monolayer to the multi-tissue interface, from a single strain to the complex microbiome community, even from aerobic to anaerobic environment that highly restores the intestinal microenvironment and reveals the role of intestinal microorganisms in maintaining human health. Thus, gut-on-chip as a precisely controllable system for biochemical microenvironment can effectively maintain the cell viability and monitor real-time responses from the cells, providing excellent technical support for the mechanisms between host-microorganisms interactions.
Nutritional metabolism
Food is not only a source of nutrients, but also a modulator of physiological functions that can induce inflammatory responses in the human GI tract. Different types of diets cause various body responses. High-fat meals elicit a systemic rise in pro-inflammatory cytokines that contributes to the development of chronic inflammation. In contrast, several food products, such as orange juice, tomato, and dairy products, have been demonstrated to reduce the postprandial inflammatory response to high-fat meals.92 However, clear interactions between nutrition and organismal inflammation have not been expounded. Thus, understanding the molecular fate of nutrients in ingested food, including ADME, is crucial to human health. During the process of screening food products using in vitro models, digested nutrients are transported through an intestinal cell monolayer, and the ability of transported nutrients to ultimately modulate the inflammatory activity of immune cells is measured.93 Microfluidic-based gut-on-chip models provide a fast and more efficient approach to study the immunomodulatory potential of food and model food allergies compared with traditional Transwell systems.92 Most gut-on-chip systems for nutritional analysis contain an intestinal cell culture module on the apical side and an immune cell co-culture module on the basolateral side.48,75 Specific food stimulates the gut epithelium in the upper channel to simulate nutrition digestion, and detecting the secretion of pro-inflammatory cytokines by immune cells in the lower channel helps to identify the stimulating food (Figure 3(c)). For example, researchers used lipopolysaccharide (LPS) to stimulate immune cells and then quantified different responses. Their results indicated that LPS induced interleukins 1 and 6 secretion by immune cells.75 By combining microfluidic technology-based cell culture and manipulation with time-resolved fluorescent imaging, gut-on-chip platforms have the potential to optimize nutrient-kinetic measurements and alleviate some restrictions of traditional systems in vitro.75 By providing a physiologically relevant gut microenvironment and sensitive real-time analysis, gut-on-chip offers a simpler, faster platform for screening products before they enter the increasingly rich and varied food market. Despite the potential of microfluidic devices, such as a controlled fluid environment that more closely mimics human physiology, use of gut-on-chip systems for nutrition analysis has been challenged. Food is a very complex sample, thus, to analyze food at a micro-scale, complex sample processing and analysis methods need to be developed. Some foods stimulate the host by interacting with gut cells and gut bacteria, leading to an immune response.94,95 Therefore, co-culture of different types of cells, even interacting with bacteria in the microfluidic network, is indispensable. We believe gut-on-chip technology and nutraceutical science can be integrated to establish platforms for investigating food allergies on-chip, as well as the influence of food quality on health, thus supporting the identification of treatments for nutrition-related disease.
Challenge and prospect
Despite its advantages and wide applications, the technology itself still faces many challenges. For example, one of the largest concerns is reproducibility and robustness in different laboratories. Therefore, strategies to standardize different modules for gut-on-chip technology are imperative, particularly for commercialization. Secondly, although simplification and modulization speed up development, current gut-on-chip platforms do not fully comprise the real complexity of the human gut, thus, parts of gut functions have been lost. Therefore, how to add more functional modules while maintaining the current merits of this platform requires more wisdom and practice. For instance, the human intestine is surrounded by large numbers of enteric nerves that control peristaltic contractions,39 maintain gut barrier functions, tune endocrine secretions, and regulate blood flow.38,96 Indeed, the absence or lack of such enteric nerves has been implicated as the primary cause of several gastrointestinal disorders, such as Hirschsprung’s disease.97,98 There is no denying that gut-on-chip as an excellent biomimetic technology in vitro, it has the potential to be exploited to study more biological and medical problems. One of the potentials of gut-on-chip is to construct different enteropathy models, for instance, IBD, ileus and endotoxemia.55,99 Importantly, the system can create combination of different kinds of cell and independently control each parameter, so that it paves a new way to reveal the corresponding pathogenesis. In addition to intestinal diseases, we envision that gut-on-chip will be used in many other fields in the near future. For example, antibiotic misuse has led to the emergence of drug-resistant bacteria. Using gut-on-chip, we can establish a drug resistance model to investigate how resistance evolves in vivo, and potentially identify other treatments such as phage therapies.100,101 Probiotics play an important role in resisting intestinal dysfunction, which can fight against pathogenic invasion and regulate intestinal immune functions. It is noteworthy that there have already been several related probiotic research on-chip, such as LGG, VSL#3.52,55 Thus, gut-on-chip can be then explored to develop new commensal-derived therapeutics. Meanwhile, the multi-organ model established by gut-on-chip and other organ models (liver, kidney) can be effectively evaluate drug toxicity and accelerate the process of drug development. Furthermore, the next possible milestone for gut-on-chip could be the pioneering of patient-derived gut-on-chip prepared from diseased tissue or human intestinal stem cells from patients in situ. Using organoid culture derived from the primary patient intestinal cells, immune cells and local gut microbiome can build a personalized platform.102 This would allow researchers to conduct personalized diagnostics and provide precision medicine at the same time.
Conclusion
The complete function of human gut, the relevance of gut microbes to human diseases, and the corresponding treatment protocols for intestinal diseases still remain unclear in a large extent. One reason for this is the complexity of human intestine, and the numerous challenges associated with decoupling hundreds of different parameters. Gut-on-chip platforms can be employed to resolve many of these challenges by extracting key parameters to simplify the whole system, combining different modules to uncover interactions, and realizing in vitro detection to monitor real-time dynamics. In this article, we reviewed various types of gut-on-chip systems and described the characteristics of different models in structure, function and application. Gut-on-chip can be applied in drug pharmacokinetics, host-microbiome interaction research and nutrition metabolism but no limited within it. As in practical clinics, gut-on-chip can soon be served as a flexible, valuable and powerful tool to predict drug responses. In the process of continuous development, gut-on-chip with human intestine physiology will provide a personalized platform for intestinal diseases research and accelerate the personalized medicine.
Footnotes
Author contributions: Y.X. performed the literature search and wrote the manuscript. H.W. participated in the discussion. H.W., Y. Y., M.L., and S.H. edited and revised the manuscript. Y.X., H.W., X.F., and S.H. conceived this review.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Key Research and Development Program of China (2018YFA0903400), National Natural Science Foundation of China (31770111, 31971350, 31800083), Shenzhen Science Technology and Innovation Commission (JCYJ20170413153329565, JCYJ20170818160418654, JCYJ20180302145817753, KQTD2016112915000294), CAS Interdisciplinary Innovation Team (JCTD-2019-16), Instrumental Project from Chinese Academy of Science (YJKYYQ20170063), China Postdoctoral Science Foundation Grant (2017M622832, 2018M631002) and Shenzhen Biofoundry.
ORCID iD: Yunqing Xiang  https://orcid.org/0000-0003-2187-0390
https://orcid.org/0000-0003-2187-0390
References
- 1. Maynard CL, Elson CO, Hatton RD, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agace WW, McCoy KD. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 2017; 46: 532–548. [DOI] [PubMed] [Google Scholar]
- 3. Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 4. Ding S, Chi MM, Scull BP, et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE 2010; 5: e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov 2015; 14: 248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci 2018; 75: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patterson DM, Shohet JM, Kim ES. Preclinical models of pediatric solid tumors (neuroblastoma) and their use in drug discovery. Curr Protoc Pharmacol 2011; 52: 14.17.11–14.17.18. [DOI] [PubMed] [Google Scholar]
- 9. Rowland M. Influence of route of administration on drug availability. J Pharm Sci 1972; 61: 70–74. [DOI] [PubMed] [Google Scholar]
- 10. Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989; 96: 736–749. [PubMed] [Google Scholar]
- 11. Hubatsch I, Ragnarsson EGE, Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat Protoc 2007; 2: 2111–2119. [DOI] [PubMed] [Google Scholar]
- 12. Mestas J, Hughes C. Of mice and not men: differences between mouse and human immunology. J Immunol 2004; 172: 2731–2738. [DOI] [PubMed] [Google Scholar]
- 13. Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Hum Me 2009; 4: 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Seok J, Warren HS, Cuenca AG, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A 2013; 110: 3507–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brugmann SA, Wells JM. Building additional complexity to in vitro-derived intestinal tissues. Stem Cell Res Ther 2013; 4: S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 2013; 340: 1190–1194. [DOI] [PubMed] [Google Scholar]
- 17. Park GS, Park MH, Shin W, et al. Emulating host-microbiome ecosystem of human gastrointestinal tract in vitro. Stem Cell Rev 2017; 13: 321–334. [DOI] [PubMed] [Google Scholar]
- 18. Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol 2011; 21: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao D, Liu H, Jiang Y, et al. Recent developments in microfluidic devices for in vitro cell culture for cell-biology research. Trend Anal Chem 2012; 35: 150–164. [Google Scholar]
- 20. Paul W, Marta C, Tom VW. Resolving host-microbe interactions in the gut: the promise of in vitro models to complement in vivo research. Curr Opin Microbiol 2018; 44: 28–33. [DOI] [PubMed] [Google Scholar]
- 21. Kang TH, Kim HJ. Farewell to animal testing: innovations on human intestinal microphysiological systems. Micromachines (Basel) 2016; 7: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bein A, Shin W, Jalili-Firoozinezhad S, et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol 2018; 5: 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Calatayud M, Dezutter O, Hernandez-Sanabria E, et al. Development of a host–microbiome model of the small intestine. FASEB J 2018; 33: 3985–3996. [DOI] [PubMed] [Google Scholar]
- 24. Karkman A, Lehtimäki J, Ruokolainen L. The ecology of human microbiota: dynamics and diversity in health and disease. Ann N Y Acad Sci 2017; 1399: 78–92. [DOI] [PubMed] [Google Scholar]
- 25. Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci 2016; 371: 20150504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang MJ, Kim HG, Kim JS, et al. The effect of gut microbiota on drug metabolism. Expert Opin Drug Metab Toxicol 2013; 9: 1295–1308. [DOI] [PubMed] [Google Scholar]
- 27. Kamada N, Chen GY, Inohara N, et al. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 2013; 14: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 2011; 94: 58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Louis P, Hold GL, Flint HJJNRM. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12: 661. [DOI] [PubMed] [Google Scholar]
- 30. Kamada N, Seo S-U, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol 2013; 13: 321. [DOI] [PubMed] [Google Scholar]
- 31. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng 2019; 3: 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Santa Barbara P, van den Brink GR, Roberts DJ. Development and differentiation of the intestinal epithelium. Cell Mol Life Sci 2003; 60: 1322–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li L, Liang M, Ping F. Gut microbiota-derived short-chain fatty acids and kidney diseases. Drug Des Dev Ther 2017; 11: 3531–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Birchenough GMH, Johansson ME, Gustafsson JK, et al. New developments in goblet cell mucus secretion and function. Mucosal Immunol 2015; 8: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 2016; 14: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allaire JM, Crowley SM, Law HT, et al. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol 2018; 39: 677–696. [DOI] [PubMed] [Google Scholar]
- 37. Sinagoga KL, Wells JM. Generating human intestinal tissues from pluripotent stem cells to study development and disease. EMBO J 2015; 34: 1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mawe GM, Hoffman JM. Serotonin signalling in the gut—functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol 2013; 10: 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goldstein AM, Hofstra RM, Burns AJ. Building a brain in the gut: development of the enteric nervous system. Clin Genet 2013; 83: 307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Colgan SP, Campbell EL. Oxygen metabolism and innate immune responses in the gut. J Appl Physiol 2017; 123: 1321–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Balimane V P, Chong, et al. Cell culture-based models for intestinal permeability: a critique. Drug Discov Today 2005; 10: 335–343. [DOI] [PubMed] [Google Scholar]
- 42. Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32: 760–772. [DOI] [PubMed] [Google Scholar]
- 43. Ingber DE. Reverse engineering human pathophysiology with organs-on-chips. Cell 2016; 164: 1105–1109. [DOI] [PubMed] [Google Scholar]
- 44. Imura Y, Asano Y, Sato K, et al. A microfluidic system to evaluate intestinal absorption. Anal Sci 2009; 25: 1403–1407. [DOI] [PubMed] [Google Scholar]
- 45. Gao D, Liu H, Lin JM, et al. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip 2013; 13: 978–985. [DOI] [PubMed] [Google Scholar]
- 46. Shah P, Fritz JV, Glaab E, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun 2016; 7: 11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Choe A, Ha SK, Choi I, et al. Microfluidic gut-liver chip for reproducing the first pass metabolism. Biomed Microdevices 2017; 19: 4. [DOI] [PubMed] [Google Scholar]
- 48. Ramadan Q, Jing L. Characterization of tight junction disruption and immune response modulation in a miniaturized Caco-2/U937 coculture-based in vitro model of the human intestinal barrier. Biomed Microdevices 2016; 18: 11. [DOI] [PubMed] [Google Scholar]
- 49. Costello CM, Sorna RM, Goh YL, et al. 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol Pharm 2014; 11: 2030–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shim KY, Lee D, Han J, et al. Microfluidic gut-on-a-chip with three-dimensional villi structure. Biomed Microdevices 2017; 19: 37. [DOI] [PubMed] [Google Scholar]
- 51. Kim SH, Chi M, Yi B, et al. Three-dimensional intestinal villi epithelium enhances protection of human intestinal cells from bacterial infection by inducing mucin expression. Integr Biol (Camb) 2014; 6: 1122–1131. [DOI] [PubMed] [Google Scholar]
- 52. Kim HJ, Huh D, Hamilton G, et al. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012; 12: 2165–2174. [DOI] [PubMed] [Google Scholar]
- 53. Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 2013; 5: 1130–1140. [DOI] [PubMed] [Google Scholar]
- 54. Villenave R, Wales SQ, Hamkins-Indik T, et al. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS ONE 2017; 12: e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim HJ, Li H, Collins JJ, et al. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 2016; 113: E7–E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin W, Kim HJ. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci U S A 2018; 115: E10539–E10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kasendra M, Tovaglieri A, Sontheimer-Phelps A, et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci Rep 2018; 8: 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shin W, Wu A, Massidda MW, et al. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front Bioeng Biotechnol 2019; 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jalili-Firoozinezhad S, Prantil-Baun R, Jiang A, et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human gut-on-a-chip. Cell Death Dis 2018; 9: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kimura H, Yamamoto T, Sakai H, et al. An integrated microfluidic system for long-term perfusion culture and on-line monitoring of intestinal tissue models. Lab Chip 2008; 8: 741–746. [DOI] [PubMed] [Google Scholar]
- 61. Gao D, Li H, Wang N, et al. Evaluation of the absorption of methotrexate on cells and its cytotoxicity assay by using an integrated microfluidic device coupled to a mass spectrometer. Anal Chem 2012; 84: 9230–9237. [DOI] [PubMed] [Google Scholar]
- 62. Thuenauer R, Rodriguez-Boulan E, Romer W. Microfluidic approaches for epithelial cell layer culture and characterisation. Analyst 2014; 139: 3206–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trujillo-de Santiago G, Lobo-Zegers MJ, Montes-Fonseca SL, et al. Gut-microbiota-on-a-chip: an enabling field for physiological research. Microphysiol Syst 2018; 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yu J, Carrier RL, March JC, et al. Three dimensional human small intestine models for ADME-tox studies. Drug Discov Today 2014; 19: 1587–1594. [DOI] [PubMed] [Google Scholar]
- 65. Sung JH, Yu J, Luo D, et al. Microscale 3-D hydrogel scaffold for biomimetic gastrointestinal (GI) tract model. Lab Chip 2011; 11: 389–392. [DOI] [PubMed] [Google Scholar]
- 66. Costello CM, Hongpeng J, Shaffiey S, et al. Synthetic small intestinal scaffolds for improved studies of intestinal differentiation. Biotechnol Bioeng 2014; 111: 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu J, Peng S, Luo D, et al. In vitro 3D human small intestinal villous model for drug permeability determination. Biotechnol Bioeng 2012; 109: 2173–2178. [DOI] [PubMed] [Google Scholar]
- 68. Kim SH, Lee JW, Choi I, et al. A microfluidic device with 3-d hydrogel villi scaffold to simulate intestinal absorption. J Nanosci Nanotechnol 2013; 13: 7220–7228. [DOI] [PubMed] [Google Scholar]
- 69. Wang Y, Gunasekara DB, Reed MI, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 2017; 128: 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Costello CM, Phillipsen MB, Hartmanis LM, et al. Microscale bioreactors for in situ characterization of GI epithelial cell physiology. Sci Rep 2017; 7: 12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim HJ, Lee J, Choi JH, et al. Co-culture of living microbiome with microengineered human intestinal villi in a gut-on-a-chip microfluidic device. J Vis Exp 2016; 114: e54344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pearce SC, Coia HG, Karl JP, et al. Intestinal in vitro and ex vivo models to study host-microbiome interactions and acute stressors. Front Physiol 2018; 9: 1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lipinski CA, Lombardo, Franco, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev 2001; 46: 3–26. [DOI] [PubMed] [Google Scholar]
- 74. Pali-Schöll I, Untersmayr E, Klems M, et al. The effect of digestion and digestibility on allergenicity of food. Nutrients 2018; 10: 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ramadan Q, Jafarpoorchekab H, Huang C, et al. NutriChip: nutrition analysis meets microfluidics. Lab Chip 2013; 13: 196–203. [DOI] [PubMed] [Google Scholar]
- 76. Lee SH, Choi N, Sung JH. Pharmacokinetic and pharmacodynamic insights from microfluidic intestine-on-a-chip models. Expert Opin Drug Metab Toxicol 2019; 15: 1005–1019. [DOI] [PubMed] [Google Scholar]
- 77. Yeon JH, Park J-K. Drug permeability assay using microhole-trapped cells in a microfluidic device. Anal Chem 2009; 81: 1944–1951. [DOI] [PubMed] [Google Scholar]
- 78. An F, Qu Y, Luo Y, et al. A laminated microfluidic device for comprehensive preclinical testing in the drug ADME process. Sci Rep 2016; 6: 25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kimura H, Sakai Y, Fujii T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab Pharmacokinet 2018; 33: 43–48. [DOI] [PubMed] [Google Scholar]
- 80. Sung JH, Srinivasan B, Esch MB, et al. Using physiologically-based pharmacokinetic-guided “body-on-a-chip” systems to predict mammalian response to drug and chemical exposure. Exp Biol Med (Maywood) 2014; 239: 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kimura H, Ikeda T, Nakayama H, et al. An on-chip small intestine-liver model for pharmacokinetic studies. J Lab Autom 2015; 20: 265–273. [DOI] [PubMed] [Google Scholar]
- 82. Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem 2010; 82: 9983–9988. [DOI] [PubMed] [Google Scholar]
- 83. Astolfi M, Péant B, Lateef MA, et al. Micro-dissected tumor tissues on chip: an ex vivo method for drug testing and personalized therapy. Lab Chip 2016; 16: 312–325. [DOI] [PubMed] [Google Scholar]
- 84. Ma L, Barker J, Zhou C, et al. Towards personalized medicine with a three-dimensional micro-scale perfusion-based two-chamber tissue model system. Biomaterials 2012; 33: 4353–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev 2010; 90: 859–904. [DOI] [PubMed] [Google Scholar]
- 86. Gayer CP, Basson MDJCs. The effects of mechanical forces on intestinal physiology and pathology. Cell Signal 2009; 21: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. de La Serre CB, Ellis CL, Lee J, et al. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol 2010; 299: G440–G448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gao R, Zhu C, Li H, et al. Dysbiosis signatures of gut microbiota along the sequence from healthy, young patients to those with overweight and obesity. Obesity 2018; 26: 351–361. [DOI] [PubMed] [Google Scholar]
- 89. Clemente JC, Ursell LK, Parfrey LW, et al. The impact of the gut microbiota on human health: an integrative view. Cell 2012; 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Grassart A, Malarde V, Gobaa S, et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting Shigella infection. Cell Host Microbe 2019; 26: 435–444 [DOI] [PubMed] [Google Scholar]
- 91. Walsh CJ, Guinane CM, O’Toole PW, et al. Beneficial modulation of the gut microbiota. FEBS Lett 2014; 588: 4120–4130. [DOI] [PubMed] [Google Scholar]
- 92. Vergeres G, Bogicevic B, Buri C, et al. The NutriChip project – translating technology into nutritional knowledge. Br J Nutr 2012; 108: 762–768. [DOI] [PubMed] [Google Scholar]
- 93. Selimović Š, Dokmeci MR, Khademhosseini A. Research highlights. Lab Chip 2013; 13: 2863–2865. [Google Scholar]
- 94. Suez J, Korem T, Zeevi D, et al. Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014; 514: 181–186. [DOI] [PubMed] [Google Scholar]
- 95. Burton-Freeman B, Talbot J, Park E, et al. Protective activity of processed tomato products on postprandial oxidation and inflammation: a clinical trial in healthy weight men and women. Mol Nutr Food Res 2012; 56: 622–631. [DOI] [PubMed] [Google Scholar]
- 96. Andrews ZB. Central mechanisms involved in the orexigenic actions of ghrelin. Peptides 2011; 32: 2248–2255. [DOI] [PubMed] [Google Scholar]
- 97. McKeown SJ, Stamp L, Hao MM, et al. Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip Rev 2013; 2: 113–129. [DOI] [PubMed] [Google Scholar]
- 98. Chang DF, Zuber SM, Gilliam EA, et al. Induced pluripotent stem cell-derived enteric neural crest cells repopulate human aganglionic tissue-engineered intestine to form key components of the enteric nervous system. J Tissue Eng 2020; 11: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Maurer M, Gresnigt MS, Last A, et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials 2019; 220: 119396. [DOI] [PubMed] [Google Scholar]
- 100. Smith H.W. Successful treatment of experimental Escherichia coli infections in mice using phage: its general superiority over antibiotics. Microbiology 1982; 128: 307–318. [DOI] [PubMed] [Google Scholar]
- 101. Viertel TM, Ritter K, Horz H-P. Viruses versus bacteria—novel approaches to phage therapy as a tool against multidrug-resistant pathogens. J Antimicrob Chemother 2014; 69: 2326–2336. [DOI] [PubMed] [Google Scholar]
- 102. Richmond CA, Breault DTJC, Gastroenterology M, et al. Move over Caco-2 cells: human-induced organoids meet gut-on-a-chip. Cell Mol Gastroenterol Hepatol 2018; 4: 634–635. [DOI] [PMC free article] [PubMed] [Google Scholar]