Abstract
Coronaviruses (CoVs), enveloped positive-sense RNA viruses, are characterized by club-like spikes that project from their surface, an unusually large RNA genome and a unique replication strategy. CoVs cause a variety of diseases in mammals and birds ranging from enteritis in cows and pigs and upper respiratory tract and kidney disease in chickens to potentially lethal human respiratory infections. Here we provide a brief introduction to CoVs discussing their replication, pathogenicity, and current prevention and treatment strategies. We will also discuss the outbreaks of the highly pathogenic Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV).
Keywords: Nidovirales, Coronavirus, positive-sense RNA viruses, SARS-CoV, MERS-CoV
Classification
CoVs, the largest group of viruses within the Nidovirales order, comprises Coronaviridae, Arteriviridae, Roniviridae and Mesoniviridae families. The Coronavirinae include one of two subfamilies in the Coronaviridae family, with the other subfamily being Torovirinae. The Coronavirinae are further subdivided into four genera: the α, β, γ and δ-CoVs. The viruses were initially sorted into these groups based on serology but now are divided by phylogenetic clustering and pair-wise evolutionary distances in seven key domains of the replicase-transcriptase polyprotein. The arteriviruses consist of five genera of mammalian pathogens. The roniviruses, which infect shrimp and the mosquito-borne mesoniviruses have invertebrate hosts.
All viruses in the Nidovirales order are enveloped, non-segmented positive-sense RNA viruses. They share a significant number of common features include: a) a highly conserved genomic organization, with a large replicase gene upstream of structural and accessory genes; b) expression of many non-structural protein genes by ribosomal frameshifting; c) several unique or unusual enzymatic activities encoded within the large replicase-transcriptase protein product; and d) expression of downstream genes by synthesis of 3′ nested sub-genomic mRNAs. In fact, the Nidovirales order name is derived from these nested 3′ mRNAs as nido is Latin for “nest”. The major differences within the four Nidovirus families are in the numbers, types, and sizes of their structural proteins and significant alterations in the structure and morphology of their virions and nucleocapsids.
Genomic Organization
Nidoviruses, which include CoVs, have the largest identified RNA genomes; CoVs contain approximately 30 kilobases (kb). The genome contains a 5′ cap structure along with a 3′ poly(A) tail, allowing it to act as a mRNA for translation of the replicase polyproteins. The replicase gene encoding the non-structural proteins (nsps) occupies two-thirds of the genome, about 20 kb, as opposed to the structural and accessory proteins, which make up about 10 kb of the viral genome. The 5′ end of the genome contains a leader sequence and untranslated region (UTR) that contains multiple stem loop structures required for RNA replication and transcription. Additionally, at the beginning of each structural or accessory gene are transcriptional regulatory sequences (TRSs) that are required for expression of each of these genes. The 3′UTR also contains RNA structures required for replication and synthesis of viral RNA. The organization of the CoVs genome is 5′-leader-UTR-replicase-S (Spike)–E (Envelope)-M (Membrane)-N (Nucleocapsid)-3′UTR-poly (A) tail with accessory genes interspersed within the structural genes at the 3′ end of the genome (see Figure 1). As shown using reverse genetics with deletion of these accessory genes, accessory proteins are almost exclusively non-essential for replication in tissue culture; however, some have been shown to have profound roles in viral pathogenesis(1–5). In some cases, accessory proteins inhibit the host defense response, especially innate immune mechanisms. For example, during MERS-CoV infection, accessory ORFs 3–5 antagonize the innate immune response (6); ORF4a binds to dsRNA, inhibiting type I interferon (IFN-I) expression and prevents the antiviral stress response (7, 8); ORF4b inhibits IFN-I expression(9) and blocks NF-kB signaling (10). ORF4b also encodes a cyclic phosphodiesterase, which blocks RNaseL activation (11).
Figure 1. Genome Organization of Representative α, β, γ and δ-CoVs.
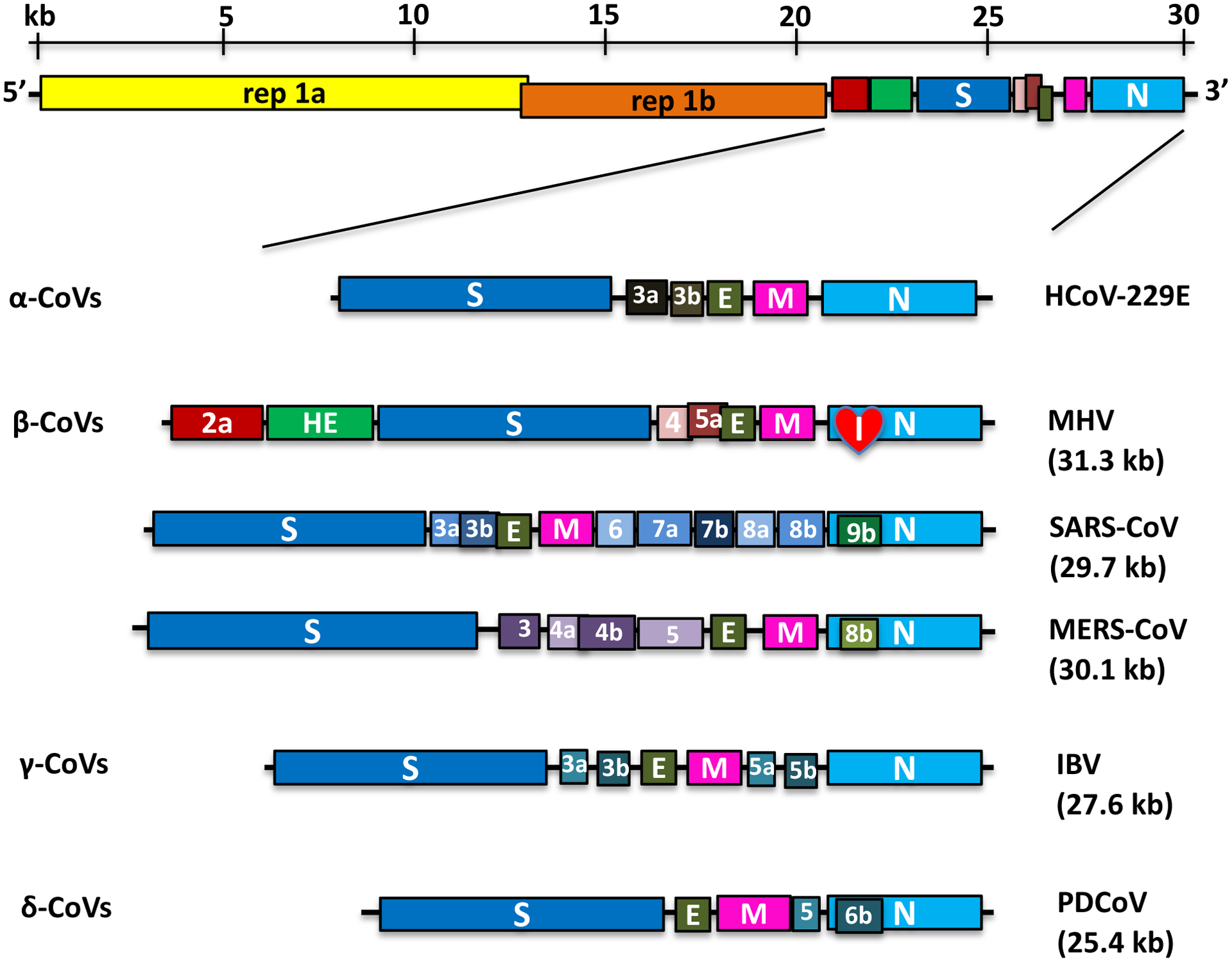
An illustration of the MHV genome is shown on top. The replicase gene constitutes two ORFs, rep 1a and rep 1b, which are expressed by a ribosomal frameshifting mechanism. The expanded regions below show the structural and accessory proteins in the 3′ regions of α-CoVs (HCoV-229E), β-CoVs (MHV, SARS-CoV, and MERS-CoV), γ-CoVs (IBV) and δ-CoVs (PDCoV). The total genome size is given for each virus. The sizes and positions of accessory genes are indicated, relative to the basic genes S, E, M, and N. The size of the genome and individual genes are approximated using the legend at the top of the diagram but are not drawn to scale.
Virion Structure
CoVs virions are spherical with diameters of approximately 125 nm as depicted in studies by cryo-electron tomography and cryo-electron microscopy (12, 13). The most prominent feature of CoVs is the club-shape spike projections emanating from the surface of the virion. These spikes are a defining feature of the virion and give them the appearance of a solar corona, prompting the name, CoVs. Within the envelope of the virion is the nucleocapsid. CoVs have helically symmetrical nucleocapsids, which is uncommon among positive-sense RNA viruses but far more common for negative-sense RNA viruses.
CoVs virus particles contain four main structural proteins. These are the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, all of which are encoded within the 3′ end of the viral genome. The distinctive spike structure on the surface of CoV is comprised of trimers of S molecules (14, 15). The S protein is a class I viral fusion protein (16). It binds to host cell receptors and mediates the earliest infection steps. In some case it also induces cell-cell fusion in late infection. The S monomer is a transmembrane protein with mass ranging from 126 to 168 kDa and is heavily N-linked glycosylated, increasing the apparent molecular weight by some 40 kDa. In most, but not all CoVs S is cleaved by a host cell furin-like protease into two separate polypeptides, S1 and S2 (17–19). The S protein contains a very large ectodomain and a small endodomain. The ectodomain structures of representative viruses from each genus of CoV, MHV(20), HCoV-HKU1 (21), HCoV-NL63(22), MERS-CoV (23), PDCoV (24), and IBV (25), have been determined by high resolution cryo-electron microscopy (cryo-EM) and were found to share a common architecture.
The M protein is the most abundant structural protein in the virion (26) and is thought to give the virion its shape. The M monomer, which ranges from 25 to 30 kDa, is a polytopic protein with three transmembrane domains(27). It has a small N-terminal ectodomain, and a C-terminal endodomain that accounts for the major part of the molecule and is situated in the interior of the virion or on the cytoplasmic face of intracellular membranes (28). M is usually modified by N-linked glycosylation(29, 30), although a subset of β-CoVs and δ-CoVs M proteins exhibit O-linked glycosylation(31). M protein glycosylation has been shown to affect both organ tropism and the IFN inducing capacity of certain CoVs (32, 33). Despite being co-translationally inserted in the ER membrane, most M proteins do not contain a signal sequence. Rather, the first or the third transmembrane domain of the MHV and IBV M proteins suffices as a signal for insertion and anchoring of the protein in its native membrane orientation (34, 35). M proteins of the α-CoVs species do contain cleavable amino-terminal signal peptides, but it is still not clear if these are necessary for membrane insertion (36). One study suggested that the M protein exists as a dimer in the virion and adopts two different conformations, allowing it to promote membrane curvature as well as bind to the nucleocapsid (37). The M protein of MHV binds to the packaging signal in nsp15 and in conjunction with the N protein is likely the primary determinant of selective packaging(38).
The E protein is a small protein with a size 8 to 12 kDa and is found in small quantities within the virion (39). E proteins from different CoVs are highly divergent but share a common architecture: a short hydrophilic N-terminal, followed by a large hydrophobic region, and, lastly, a large hydrophilic C-terminal tail. The membrane topology of E protein is not completely resolved(40–42), but most data suggest that it is a transmembrane protein. The E protein has ion channel activity and were observed to assemble into homo-oligomers, ranging from dimers through hexamers(43), and a pentameric α-helical bundle structure has been solved for the hydrophobic region of SARS-CoV E protein(44). An oligomeric form is consistent with the ion channel activity of the E protein, but the monomeric form of E may also play a separate role. As opposed to other structural proteins, recombinant viruses lacking the E protein are not always lethal, although this is virus type dependent (45, 46). The E protein facilitates assembly and release of the virus but also has other functions. For instance, SARS-CoV E protein is not required for viral replication but is required for pathogenesis (47, 48).
The N protein constitutes the only protein present in the helical nucleocapsid. It is composed of two independently folding domains, an N-terminal domain (NTD) and a C-terminal domain (CTD), both capable of binding RNA in vitro, but each domain uses different mechanisms to bind RNA. It has been suggested that optimal RNA binding requires contributions from both domains (49, 50). N protein is heavily phosphorylated (51), which may be important for triggering a structural change enhancing the affinity for viral versus non-viral RNA, and is ADP ribosylated (52). N protein binds the viral genome in a beads-on-a-string type conformation. Two specific RNA substrates have been identified for N protein: the transcription-regulating sequences (TRSs) (53) and the genomic packaging signal. The genomic packaging signal has been found to bind specifically to the second, or C-terminal RNA binding domain (38). N protein also binds to nsp3 (50, 54), a key component of the replicase-transcriptase complex (RTC), and to the M protein (26). These protein interactions serve to tether the viral genome to the RTC and subsequently package the encapsidated genome into viral particles.
Hemagglutinin-esterase (HE), a fifth structural protein, is present only in a subset of β-CoVs, which include MHV, BCoV, HCoV-OC43, and HCoV-HKU1. The protein acts as a hemagglutinin, binds sialic acids on surface glycoproteins, and contains acetyl-esterase activity (55). These activities are thought to enhance S protein-mediated cell entry and virus spread through the mucosa (56). Interestingly, HE enhances murine hepatitis virus (MHV) neurovirulence (1); however, it is selected against in tissue culture for unknown reasons (57).
Coronavirus Life Cycle
Attachment and Entry
The initial attachment of the virion to the host cell is initiated by interactions between the S protein and its receptor. This interaction is the primary determinant controlling CoVs host species range and tissue tropism. Individual CoVs usually infects one or a few closely-related hosts. S protein includes two subunits, the comparatively variable S1 subunit mediates the binding to receptor and the more conserved S2 subunit undergoes large conformational changes that results in fusion of virion and cell membranes. The sites of receptor binding domains (RBD) within the S1 region of a CoVs S protein vary depending on the virus: the RBD located at the N-terminal of S1 (MHV) in some cases (58) while it is present in the C-terminal of S1 in the case of SARS-CoV(59), MERS-CoV (60, 61),HCoV-229E (62), HCoV-HKU1(63), HCoV-NL63 (64) and TGEV (65).
MHV enters cells by binding to its receptor (carcinoembryonic antigen-related adhesion molecule 1, CEACAM1), the CoVs receptor that was first discovered (66–68). CEACAM1 has different isoforms which contains two and four Ig-like domains. The diversity of the receptor isoforms expressed in different genetic backgrounds results in a wide range of pathogenicity of MHV in mice (69). Many α-CoVs and δ-CoVs utilize aminopeptidase N (APN) as their cellular receptor (70–75). APN (also called CD13), a heavily glycosylated homodimer, is a cell-surface, zinc-binding protease that is resident in respiratory and enteric epithelia and in neural tissue. The α-CoVs receptor activities of APN homologs are not interchangeable among species (76, 77) while the δ-CoV PDCoV can use APN homologs from multiple mammalian and avian species as a receptor(73).
SARS-CoV uses angiotensin-converting enzyme 2 (ACE2) as its receptor(78). ACE2 is mainly expressed in epithelial cells of the lung and the small intestine, the primary targets of SARS-CoV, and also in heart, kidney and other tissues(79). ACE2 is a cell-surface, zinc-binding carboxypeptidase and plays a role in regulation of cardiac function and blood pressure. ACE2 also serves as the receptor for the α-CoV HCoV-NL63(80).
MERS-CoV uses dipeptidyl-peptidase 4 (DPP4) as its cellular receptor(81). DPP4, also called CD26, is a membrane-bound exoprotease with a wide tissue distribution; it cleaves dipeptides from hormones, chemokines, and cytokines and plays multiple other physiological functions(81). DPP4 includes an N-terminal eight-blade β-propeller domain and a C-terminal catalytic domain. Structure analysis of the MERS-CoV RBD-DPP4 complex indicates that the receptor binding surface of the RBD is a four-stranded β-sheet that contacts blades 4 and 5 of the DPP4 propeller domain (61, 82). Further, key residues of camel and human DPP4 critical for binding to the RBD are highly-conserved facilitating zoonotic transmission of MERS-CoV (82). (See Table 1 for a list of known CoVs receptors).
Table 1.
The Known Receptors for Coronaviruses
| Virus | Receptor | References |
|---|---|---|
| α-CoVs | ||
| CCoV | Canine APN (cAPN) | (76) |
| FCoV I | Unknown, but not fAPN | (224) |
| FCoV II, FIPV | Feline APN (fAPN) | (72) |
| HCoV-229E | Human APN (hAPN) | (71) |
| HCoV-NL63 | ACE2 | (80) |
| PEDV | Unknown, but not pAPN | (225, 226) |
| PRCoV | Porcine APN (pAPN) | (75) |
| TGEV | Porcine APN (pAPN) | (70) |
| β-CoVs | ||
| MHV | Murine CEACAM1 | (67) |
| BCoV | Neu5,9Ac2 | (227) |
| HCoV-OC43 | Neu5,9Ac2 | (228) |
| SARS-CoV | ACE2 | (78) |
| MERS-CoV | DPP4 | (81) |
| γ-CoVs | ||
| IBV | alpha-2,3-linked sialic acid | (229) |
| δ-CoVs | ||
| PDCoV | Porcine APN | (73, 74) |
APN, aminopeptidase N; ACE2, angiotensin-converting enzyme 2; BCoV, bovine coronavirus; CCoV, canine coronavirus; CEACAM, carcinoembryonic antigen-related adhesion molecule 1; DPP4, dipeptidyl peptidase 4; HCoV, human coronavirus; PRCoV, porcine respiratory coronavirus; TGEV, transmissible gastroenteritis virus; PEDV, porcine epidemic diarrhea virus; FIPV, feline infectious peritonitis virus; MHV, murine hepatitis virus; SARS-CoV, severe acute respiratory syndrome coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; Neu5,9Ac2, N-acetyl-9-O-acetylneuraminic acid; PDCoV,Porcine Delta-Coronavirus.
Many CoVs S proteins are cleaved during exit from the producer cells, often by a furin-like protein (17). This cleavage separates the RBD and fusion domains of the S protein (83). Following receptor binding, the virus must next gain access to the host cell cytosol. This is generally accomplished by a second proteolytic cleavage of the S protein by TMPRRS2, a cathepsin or another protease (84, 85). Following cleavage at S2′, a fusion peptide is exposed, which is followed by joining of two heptad repeats in S2 forming an antiparallel six-helix bundle (16). The formation of this bundle allows for the mixing of viral and cellular membranes, resulting in fusion and ultimately release of the viral genome into the cytoplasm. Fusion generally occurs at the plasma membrane or in some cases, within acidified endosomes (86).
The S protein is the major target for anti-virus neutralizing antibodies and its binding to host cell receptor is critical for a productive infection and for cross-species transmission. This was illustrated during the SARS epidemic, when the S protein showed extensive adaptation to the human ACE2 receptor(87). In contrast, the MERS S protein has changed little during the course of the MERS outbreak(88), except during the Korean outbreak when there was a single point introduction of virus(89). Remarkably, virus mutated so that the S protein exhibited reduced affinity of the DPP4 receptor (90). This may have been driven by the anti-virus neutralizing antibody response, but this is not certain. One interpretation is that, in the case of MERS-CoV, receptor binding is less important than other parts of the entry process, such as S protein cleavage by host cell proteases, particularly TMPRSS2(91).
Replicase Protein Expression
The next step in the CoVs lifecycle is the translation of the replicase gene from the virion genomic RNA. The replicase gene encodes two large ORFS, rep1a and rep1b, which express two co-terminal polyproteins, pp1a and pp1ab (Figure 1). In order to express both polyproteins, the virus utilizes a slippery sequence (5′-UUUAAAC-3′) and an RNA pseudoknot that cause ribosomal frameshifting from the rep1a reading frame into the rep1b ORF. In most cases, the ribosome unwinds the pseudoknot structure and continues translation until it encounters the rep1a stop codon. Occasionally the pseudoknot blocks the ribosome from continuing elongation, causing it to pause on the slippery sequence, changing the reading frame by moving back one nucleotide (−1 frameshift) before the ribosome is able to melt the pseudoknot structure and extend translation into rep1b, resulting in the translation of pp1ab (92, 93). In vitro studies predict the incidence of ribosomal frameshifting to be as high as 25%, but this has not been determined in the context of virus infection. Viruses probably utilize frameshifting to control the precise ratio of rep1b:rep1a proteins or delay the production of rep1b products until the products of rep1a have created a suitable environment for RNA replication (94).
Polyproteins pp1a and pp1ab contain the nsps 1–11 and 1–16 respectively. In pp1ab, nsp11 from pp1a becomes nsp12 following extension of pp1a into pp1b. However, γ-CoVs do not contain a comparable nsp1. These polyproteins are subsequently cleaved into individual nsps (95). There are two types of polyprotein cleavage activity. One or two papain-like proteases (PLpro), which are situated within nsp3, carry out the relatively specialized separations of nsp1, nsp2, and nsp3. Many PLpro also have deubiquitinase activity, which counters some host antiviral defenses(96). nsp5, the main protease (Mpro), performs the remaining 11 cleavage events(97, 98). Mpro is often designated the 3C-like protease (3CLpro) to denote its distant relationship to the 3C proteins of picornaviruses. Since PLpro and Mpro have pivotal roles early in infection, they present attractive targets for antiviral drug design(96, 99).
Next, many of the nsps assemble into the replicase-transcriptase complex (RTC) to create an environment suitable for RNA synthesis including replication and transcription of sub-genomic RNAs(100). Notably, products of rep 1a, nsp3,nsp4 and nsp6 each contain multiple transmembrane helices which anchor the RTC to intracellular membranes (101, 102). They are responsible for remodeling membranes to form organelles which are dedicated to viral RNA synthesis(103). Among them, nsp3 is the largest RTC proteins by far(104). It contains a hypervariable acidic N-terminal region that is a ubiquitin-like domain (Ubl1) and a highly conserved C-terminal region which is designated the Y domain and contains three metal-binding clusters of cysteine and histidine residues(105). Ubl1 interacts with the serine and arginine-rich region (SR region) of the N protein; this interaction may tether the genome to the RTC, facilitating formation of the RNA synthesis initiation complex (54, 106, 107). Also located within nsp3 is a conserved macrodomain (Mac1), that exhibits ADP-ribose-protein hydrolase activity(108). The macrodomain nonessential for viral replication but critical for viral pathogenesis(109). The nsps also have other functions important for RNA replication. For example, nsp10, a small non-enzymatic viral protein contributes to CoV replication fidelity by regulating nsp14 and nsp16 activity during virus replication(110); nsp12 encodes the RNA-dependent RNA polymerase (RdRp); nsp13 encodes the RNA helicase and RNA 5′-triphosphatase; nsp14 encodes the exoribonuclease (ExoN) involved in replication fidelity (111–113) and N7-methyltransferase activity(114); and nsp16 harbors 2′-O-methyltransferase activity(115). In addition to roles in replication, nsp1 blocks innate immune responses by direct inhibition of translation or by promoting degradation of host IFN mRNA by nsp1(116); nsp15 contains an endoribonuclease domain that mediates evasion of host dsRNA sensors(117, 118). For a list of non-structural proteins and their putative functions, see Table 2. Ribonucleases nsp15-NendoU and nsp14-ExoN activities are unique to the Nidovirales order and are considered genetic markers for these viruses (119).
Table 2.
Functions of Coronaviruses Non-Structural Proteins
| Protein | Function | References |
|---|---|---|
| nsp1 |
|
(116, 230, 231) |
| nsp2 |
|
(232, 233) |
| nsp3 |
|
(105, 234–240) |
| nsp4 |
|
(241, 242) |
| nsp5 |
|
(243, 244) |
| nsp6 |
|
(101) |
| nsp7 |
|
(245, 246) |
| nsp8 |
|
(245, 246) |
| nsp9 |
|
(247, 248) |
| nsp10 |
|
(110, 249, 250) |
| nsp12 |
|
(246, 251) |
| nsp13 |
|
(252, 253) |
| nsp14 |
|
(111–114, 254, 255) |
| nsp15 |
|
(117, 118, 256–258) |
| nsp16 |
|
(115, 259) |
NAB, nucleic acid binding; DMVs, double-membrane vesicles; MDA5, Melanoma differentiation associated protein 5.
Replication and Transcription
Viral RNA synthesis follows the translation and assembly of the viral replicase complexes. Viral RNA synthesis produces both genomic and sub-genomic RNAs. Sub-genomic RNAs serve as mRNAs for the structural and accessory genes which reside downstream of the replicase genes in Orf1. All positive-sense sub-genomic RNAs are 3′ co-terminal with the full-length viral genome and thus form a set of nested RNAs, a distinctive property of the order Nidovirales. Both genomic and sub-genomic RNAs are produced through negative-strand intermediates. These negative-strand intermediates are only about 1% as abundant as their positive-sense counterparts and contain both poly-uridylate and anti-leader sequences (120).
Many cis-acting sequences are important for the replication of viral RNAs. Within the 5′ UTR of the genome are seven stem-loop structures that may extend into the replicase 1a gene (121–124). The 3′ UTR contains a bulged stem-loop, a pseudoknot, and a hypervariable region (125–128). The stem-loop and the pseudoknot at the 3′ end overlap, and thus cannot form simultaneously (126, 129). Therefore, these different structures are proposed to regulate alternate stages of RNA synthesis, although exactly which stages are regulated and their precise mechanism of action are still unknown.
Perhaps the most novel aspect of CoV replication is how the leader and body TRS segments fuse during production of sub-genomic RNAs. Leader-TRS joining occurs during the discontinuous extension of negative-strand RNA (130). The current model proposes that the RdRp pauses at body TRS sequences (TRS-B); following this pause, the RdRp either continues elongation to the next TRS or switches to amplifying the leader sequence at the 5′ end of the genome guided by complementarity of the TRS-B to the leader TRS (TRS-L). Furthermore, nucleocapsid phosphorylation and RNA helicase DDX1 recruitment was shown to facilitate the transition from discontinuous to continuous transcription (131). However, many questions remain. For instance, how does the RdRp bypass all of the TRS-B sequences to produce full-length negative-strand genomic RNA? Also, how are the TRS-B sequences directed to the TRS-L and how much complementarity is necessary? Answers to these questions and others will be necessary to gain a full perspective of how RNA replication occurs in CoVs. Eventual development of an in vitro replication system will be required to fully understand these processes.
Finally, CoVs are also known for their ability to recombine by both homologous and non-homologous recombination (132, 133). The ability of these viruses to recombine is tied to the strand switching ability of the RdRp. Recombination likely plays a prominent role in viral evolution and is the basis for targeted RNA recombination(134), a reverse genetics tool used to engineer viral recombinants at the 3′ end of the genome.
Assembly and Release
Following replication and sub-genomic RNA synthesis, the viral structural proteins, S, E, and M are translated and inserted into the endoplasmic reticulum (ER). These proteins move along the secretory pathway into the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) (135, 136). There, viral genomes encapsidated by N protein bud into membranes of the ERGIC containing viral structural proteins, forming mature virions (137).
The M protein directs most protein-protein interactions required for assembly of CoVs. M protein is not sufficient for virion formation as virus-like particles (VLPs) cannot be formed by M protein expression alone. However, when M protein is expressed along with E protein, VLPs are formed, suggesting these two proteins function together to produce CoV envelopes (138). N protein enhances VLP formation, suggesting that fusion of encapsidated genomes into the ERGIC enhances viral envelopment (139). The S protein is incorporated into virions at this step but is not required for assembly. The ability of the S protein to traffic to the ERGIC and interact with the M protein is critical for its incorporation into virions.
While the M protein is relatively abundant, the E protein is only present in small quantities in the virion. Thus, it is likely that M protein interactions provide the impetus for envelope maturation. E protein may assist the M protein in virion assembly either by inducing membrane curvature (46, 140, 141), preventing the aggregation of M protein (142), or by an uncharacterized mechanism. The E protein may also have a separate role in promoting viral release by altering the host secretory pathway (143).
The M protein also binds to the nucleocapsid, and this interaction promotes the completion of virion assembly. These interactions have been mapped to the C-terminus of the endodomain of M with CTD 3 of the N-protein (144). However, it is unclear exactly how the nucleocapsid complexed with virion RNA traffics from the RTC to the ERGIC to interact with M protein and become incorporated into the viral envelope. Another outstanding question is how the N protein selectively packages only positive-sense full-length genomes among the many different RNA species produced during infection. A packaging signal for MHV is present in the nsp15 coding sequence (38). Mutation of this signal drastically increases sgRNA incorporation into virions, and while virus production in cultured cells is negligibly affected, the mutant virus elicits a stronger IFN response in mice (145). Furthermore, most CoVs do not contain similar sequences at this locus, indicating that packaging may be virus specific.
Following assembly, virions are transported to the cell surface in vesicles and released by exocytosis. It is not known if the virions use a traditional pathway for transport of large cargo from the Golgi or if the virus has diverted a separate, unique pathway for its own exit. Genome-wide screening has identified a host protein, valosin-containing protein (VCP/p97) that is required for release of CoV from endosomes(146). In several CoVs, S protein that does not assemble into virions transits to the cell surface where it mediates cell-cell fusion between infected cells and adjacent, uninfected cells. This leads to the formation of multinucleated cells, which allows the virus to spread within an infected organism without being detected or neutralized by virus-specific antibodies.
Pathogenesis
Animal Coronaviruses
CoVs cause a large variety of diseases in animals, and their ability to cause severe disease in livestock and companion animals such as pigs, cows, chickens, dogs, and cats led to significant research on these viruses in the last half of the 20th century. For instance, Transmissible Gastroenteritis Virus (TGEV) and Porcine Epidemic Diarrhea Virus (PEDV) cause severe gastroenteritis in young piglets, leading to significant morbidity, mortality, and ultimately economic losses. Recently an novel HKU2-related bat CoV, swine acute diarrhoea syndrome coronavirus (SADS-CoV), was shown to cause an outbreak of fetal swine disease (147). Porcine hemagglutinating encephalomyelitis virus (PHEV) causes enteric infection but has the ability to infect the nervous system, causing encephalitis, vomiting and wasting in pigs. Feline enteric coronavirus (FCoV) causes a mild or asymptomatic infection in domestic cats, but during persistent infection, mutation transforms the virus into a highly virulent strain of FCoV (Feline Infectious Peritonitis Virus, FIPV) that leads to development of a lethal disease called feline infectious peritonitis (FIP). FIPV is macrophage tropic and is believed to cause aberrant cytokine and/or chemokine expression and lymphocyte depletion, resulting in lethal disease(148). Bovine CoV, Rat CoV, and Infectious Bronchitis Virus (IBV) cause mild to severe respiratory tract infections in cattle, rats, and chickens respectively. Bovine CoV causes significant losses in the cattle industry and also has spread to infect a variety of ruminants, including elk, deer and camels. In addition to severe respiratory disease, the virus causes diarrhea (‘winter dysentery’ and ‘shipping fever’), all leading to weight loss, dehydration and decreased milk production(149). Some strains of IBV, a γ-CoV, also infect the urogenital tract of chickens causing renal disease. IBV significantly diminishes egg production and weight gain, causing substantial losses in the chicken industry each year(150). Interestingly, a novel CoV (SW1) was identified in a deceased Beluga whale(151) and shown to be a γ-CoV based on phylogenetic analysis. This is the first example of a non-avian γ-CoV, but it is not known whether this virus actually causes disease in whales.
In addition, there has been intense interest in identifying novel bat CoVs, since these are the likely ultimate source for most CoV, including SARS-CoV and MERS-CoV(152, 153). Hundreds of novel bat CoV have been identified over the past decade(154), including the agent of SADS, described above. Another novel, non-CoVs group of nidoviruses, Mesoniviridae, were recently identified as the first ones to exclusively infect insect hosts (155, 156). These viruses are highly divergent from other nidoviruses but are most closely related to the roniviruses. In size, they are ~20 kb, falling in between large and small nidoviruses. Consistent with this relatively small size, these viruses do not encode for an endoribonuclease, which is present in large nidoviruses. Recently a novel nidovirus, planarian secretory cell nidovirus (PSCNV), was identified and shown to have a 41.1 kb genome, making it the largest RNA viral genome yet discovered. The genome contains the canonical nidoviral genome organization and key replicative domains. It encodes a predicted 13556 aa polyprotein in an unconventional single ORF(157).
The most heavily studied animal CoV is murine hepatitis virus (MHV), which causes multiple diseases in mice, including respiratory, enteric, hepatic, and neurologic infections. For instance, MHV-1 causes severe respiratory disease in susceptible A/J and C3H/HeJ mice, A59 and MHV-3 induce hepatitis, and JHMV causes encephalitis and acute and chronic demyelinating diseases. MHV-3 induces cellular injury through the activation of the coagulation cascade via a fgl2/fibroleukin dependent way(158). A59 and attenuated versions of JHMV cause chronic demyelinating diseases that bears similarities to multiple sclerosis (MS), making MHV infection one of the best models for this debilitating human disease. Early studies suggested that demyelination was dependent on viral replication in oligodendrocytes in the brain and spinal cord (159, 160); however, more recent reports clearly demonstrate that the disease is immune-mediated. Irradiated mice or immunodeficient (lacking T and B cells) mice do not develop demyelination, but addition of virus-specific T cells restores the development of demyelination(161–163). Additionally, demyelination is accompanied by a large influx of macrophages and microglia that can phagocytose infected myelin (164). Microglia are especially important in the initial host defense to MHV since mice succumb to the infection if these cells are depleted(165).
Human Coronaviruses
Prior to the SARS-CoV outbreak, CoVs were only thought to cause mild, self-limiting respiratory infections in humans. Two of these human CoVs are α-CoVs (HCoV-229E and HCoV-NL63) while the other two are β-CoVs (HCoV-OC43 and HCoV-HKU1). HCoV-229E and HCoV-OC43 were isolated nearly 50 years ago (166–168), while HCoV-NL63 and HCoV-HKU1 were only identified following the SARS-CoV outbreak (169, 170). HCoV-229E and HCoV-NL63 arose from a common ancestor and diverged 200 years ago(171). HCoV-OC43 is closely related to BCoV and may have crossed species from bovids, or alternatively, may have been transmitted from humans to cows. These viruses are endemic in the human populations, causing 15–30% of upper respiratory tract infections each year. They cause more severe disease in neonates, the elderly, and in individuals with underlying illnesses, with a greater incidence of lower respiratory tract infection in these populations(172). HCoV-NL63 is also associated with acute laryngotracheitis (croup) (173). One interesting aspect of these viruses is their differences in tolerance to genetic variability. HCoV-229E isolates from around the world have only minimal sequence divergence (174), while HCoV-OC43 isolates from the same location but isolated in different years show significant genetic variability (175). Based on the ability of MHV to cause demyelinating disease, it has been suggested that human CoVs may be involved in the development of multiple sclerosis (MS)(176). However, no evidence to date suggests that human CoVs play significant roles in MS.
SARS-CoV, a group 2b β-CoV, was identified as the causative agent of the Severe Acute Respiratory Syndrome (SARS) epidemic that originated in 2002–2003 in the Guangdong Province of China. During the 2002–2003 outbreak, approximately 8098 cases occurred with 774 deaths, resulting in a mortality rate of 9%. This rate was much higher in aged individuals, with mortality rates approaching 50% in individuals over 60 years of age while no patients under 24 years died from the infection(177). Furthermore, the outbreak resulted in the loss of nearly $40 billion dollars in economic activity as the virus nearly completely shut down many activities in Southeast Asia and Toronto, Canada for several months. The epidemic began in wet markets in Guangzhou, likely originating in bats. It is widely accepted that SARS-CoV is a bat virus as a large number of Chinese horseshoe bats contain sequences of SARS-related CoVs and contain serologic evidence for a prior infection with a related CoV (178, 179). Further, two novel bat SARS-related CoVs were later identified that are more similar to SARS-CoV than any other virus identified to date, further supporting a bat origin for SARS-CoV (180).
SARS-CoV then spread from infected bats to intermediate animals such as Himalayan civet cats and raccoon dogs present in the markets and then to humans (181). An individual was infected in Guangzhou and then stayed at a hotel in Hong Kong, spreading the infection to others staying at the hotel, and ultimately, throughout the world. Although some human individuals within wet animal markets had serologic evidence of SARS-CoV infection prior to the outbreak, these individuals had no apparent symptoms (181). Thus, it is likely that SARS-like CoV circulated in the wet animal markets for some time before a series of factors facilitated its spread into larger human populations.
Transmission of SARS-CoV was relatively inefficient, as it largely spread through large droplets and direct contact with infected individuals and transmission only occurred after the onset of clinical illness. Thus, the outbreak mostly occurred within households and healthcare settings (182), except in a few cases of superspreading events where one individual was able to infect multiple contacts due to high viral burdens or an ability to aerosolize virus. As a result of the relatively inefficient transmission of SARS-CoV, the outbreak was controllable through the use of quarantining. Only a small number of SARS cases occurred after the outbreak was controlled in June 2003(183).
SARS-CoV primarily infects epithelial cells within the lung(184). The virus is capable of entering macrophages and dendritic cells but only causes an abortive infection (185, 186). Despite this, infection of these cell types may be important in inducing pro-inflammatory cytokines that may contribute to disease (187). In fact, many cytokines and chemokines are produced by these cell types and are elevated in the serum of SARS-CoV infected patients (188). Viral titers decrease when severe disease develops in both humans and in several animal models of the disease, suggesting that the host response is responsible for much of the clinical signs and symptoms. Furthermore, animals infected with rodent-adapted SARS-CoV strains show similar clinical features to the human disease, including an age-dependent increase in disease severity (189). These animals also show increased levels of proinflammatory cytokines and reduced T-cell responses, consistent with a possible immunopathological mechanism of disease (190, 191).
While the SARS-CoV epidemic was controlled in 2003, and the virus has not since returned, a novel human CoV emerged in the Middle East in 2012. This virus, named Middle East Respiratory Syndrome-CoV (MERS-CoV) is a group 2b β-CoV and was found to be the causative agent of a highly lethal respiratory tract infection in Saudi Arabia and other countries in the Middle East (153, 192). Since its emergence, the virus has spread to over 27 countries, including to South Korea in 2015, where it caused 186 cases and 38 deaths(193). Among those cases, 83% were transmitted from five super spreading events and 44% were due to nosocomial transmission at 16 hospitals(194). As of September 2019, there have been a total of 2468 laboratory-confirmed cases of MERS-CoV, with 851 associated deaths and a case fatality rate of approximately 35%, as reported to the World Health Organization (https://www.who.int/emergencies/mers-cov/en/). The majority of cases early in the outbreak resulted from nosocomial transmission. As better infection control measures were instituted, approximately 50% of cases are considered primary, with infection resulting from direct or indirect contact with camels, the zoonotic source of the infection(195). Serological studies have identified MERS-CoV antibodies in dromedary camels in the Middle East and Africa from samples obtained as early as 1983(196). Supporting evidence for camel to human transmission comes from studies identifying nearly identical MERS-CoVs in camels and humans in nearby proximities in Saudi Arabia (5, 23, 197). In one of these studies the human case had direct contact with an infected camel and the virus isolated from this patient was nearly identical to the virus isolated from the camel (5). MERS-CoV likely originated in bats because it is related to two previously identified bat CoV, HKU4 and HKU5 (198). Furthermore, new evidence has emerged to support the hypothesis that bats are the evolutionary source of MERS-CoV since a MERS-like CoV was identified from a Pipistrellus cf. hesperidus bat sampled in Uganda(199).
MERS-CoV utilizes Dipeptidyl peptidase 4 (DPP4) as its receptor (81). The virus is only able to use the receptor from certain species such as bats, humans, camels, rabbits, and horses to establish infection. While the virus is unable to naturally infect mouse cells due to differences in the structure of DPP4, several mouse models expressing human DPP4 have been developed which can successfully be infected with MERS-CoV (200–203).
Diagnosis, Treatment, and Prevention
In most cases of self-limited infection, diagnosis of CoVs is unnecessary. However, it is important in certain clinical and veterinary settings or in epidemiological studies to identify an etiological agent. Diagnosis is also important in locations where a severe CoV outbreak is occurring, such as, at present, in the Middle East, where MERS-CoV continues to circulate. The identification of cases will guide the development of public health measures to control outbreaks. It is also important to diagnose cases of severe veterinary CoV-induced disease, such as PEDV and IBV, to control these pathogens and protect food supplies. The primary methods to diagnose CoV infection use molecular techniques such as RT-PCR. RT-PCR has become the method of choice for diagnosis of human CoV, as multiplex real-time RT-PCR assays, such as RT-RTPA, and RT-LAMP, have been developed. They are able to detect all four respiratory human CoVs and could be further adapted to detect novel CoVs (204, 205). Serologic assays are important in cases where RNA is difficult to isolate, virus is no longer present, and for epidemiological studies. Because rapid and accurate diagnosis of MERS is important, several diagnostic tests including one in which RT-LAMP is combined with vertical flow visualization (RT-LAMP-VF) (206) have been developed.
To date, there are no antiviral therapeutics that specifically target human CoVs, so treatments are only supportive. IFNs have been used in some patients, without evidence of therapeutic benefit(207). Studies in mice indicate that the relative timing of IFN administration and virus replication are critical to either protective or pathogenic effects after infection with SARS-CoV(208) or MERS-CoV(209), which may explain the variable results that are observed in patients. The SARS and MERS outbreaks have stimulated research on these viruses, and this research has identified a large number of suitable antiviral targets, such as viral proteases, polymerases, and entry proteins but so far no specific treatment has been licensed. Multi-target treatment should be a priority as for antiviral treatment(210, 211). Significant work remains to develop drugs that target these processes and are useful in infected patients.
Many CoV-specific vaccines have been developed and some targeting veterinary CoV pathogens have been licensed. Vaccines have been approved for IBV(212), TGEV(213), and canine CoV, but these vaccines are not always used because they are either not very effective, or in some cases, have resulted in the selection of novel pathogenic CoVs via recombination of circulating strains(214). In general, it is thought that live attenuated vaccines are most efficacious in targeting CoVs. This was illustrated in the case of TGEV, where an attenuated naturally appearing variant, porcine respiratory coronavirus (PRCoV), appeared in Europe in the 1980s. This variant only caused mild disease and protected swine from lethal TGEV. This attenuated virus has prevented the recurrence of severe TGEV in Europe and the U.S. over the past 30 years (215).
In the case of SARS-CoV, several potential vaccines have been developed. The spike protein, which elicits a neutralizing antibody response, has been a major target of vaccine development (216, 217). Therapeutic SARS-CoV neutralizing human monoclonal antibodies have been generated and stockpiled (218). These antibodies would be useful for passive immunization of healthcare workers and other high-risk individuals in the event of another SARS outbreak (219). Similarly, efforts have been made to develop vaccines against MERS-CoV. Several vaccine approaches have been tried, including subunits vaccines, DNA vaccines, viral vector vaccines and live attenuated and inactivated vaccines(220, 221). Some of these have shown efficacy in animal testing and several are in clinical trials. For example, a MERS-CoV DNA vaccine recently underwent phase I Clinical Trials (222, 223), While it induced MERS-CoV-specific neutralizing antibody titers, these tended to decline substantially by 60 weeks after immunization.
Owing to the lack of effective therapeutics or vaccines, the best measures to contain human CoVs outbreaks remain a strong public health surveillance system coupled with rapid diagnostic testing and quarantine when necessary. For international outbreaks, cooperation of governmental entities, public health authorities and health care providers is critical. During outbreaks of veterinary CoV that are readily transmitted, such as PEDV, more drastic measures such as culling of entire herds of pigs may be necessary to prevent transmission of these deadly viruses.
Conclusions
Over the last half century, several varieties of CoVs have emerged to cause human and veterinary diseases. It is likely that these viruses will continue to emerge and evolve, and cause both human and veterinary outbreaks owing to their ability to recombine, mutate, and infect various animal species and cell types.
Critical problems remain to be resolved in future research. One focus should be to understand viral replication and pathogenesis in greater detail. Another is to explore the propensity of these viruses to cross species and the features that facilitate or inhibit cross-species transmission and to identify CoVs reservoirs, which will enhance our ability to predict potential future epidemics. So far, bats seem to be a primary reservoir for these viruses, but they do not develop clinically evident disease, for reasons that require further investigation. Additionally, many of the non-structural and accessory proteins encoded by CoVs are only partly characterized, and it will be important to identify their mechanisms of action and their role in viral replication and pathogenesis. These studies will help identify more suitable therapeutic targets. Finally, additional studies should probe CoV-induced immunopathological disease and delineate the relationship between CoVs and the host immune response. These will guide efforts to design vaccines and drugs that prevent and treat CoV infections.
References
- 1.Kazi L, Lissenberg A, Watson R, de Groot RJ, & Weiss SR (2005) Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J Virol 79(24):15064–15073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Haan CA, Masters PS, Shen X, Weiss S, & Rottier PJ (2002) The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296(1):177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz JL, et al. (2011) Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7(6):e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tangudu C, Olivares H, Netland J, Perlman S, & Gallagher T (2007) Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J Virol 81(3):1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azhar EI, et al. (2014) Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370(26):2499–2505. [DOI] [PubMed] [Google Scholar]
- 6.Menachery VD, et al. (2017) MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis. MBio 8(4). pii: e00665–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakagawa K, Narayanan K, Wada M, & Makino S (2018) Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J Virol 92(20) pii: e00902–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabouw HH, et al. (2016) Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses. PLoS Pathog 12(10):e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, et al. (2015) Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep 5:17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canton J, et al. (2018) MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog 14(1):e1006838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao L, et al. (2012) Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11(6):607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barcena M, et al. (2009) Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc Natl Acad Sci 106(2):582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman BW, et al. (2006) Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol 80(16):7918–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beniac DR, Andonov A, Grudeski E, & Booth TF (2006) Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol 13(8):751–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmas B & Laude H (1990) Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol 64(11):5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosch BJ, van der Zee R, de Haan CA, & Rottier PJ (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77(16):8801–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millet JK & Whittaker GR (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A 111(42):15214–15219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham S, Kienzle TE, Lapps W, & Brian DA (1990) Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology 176(1):296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luytjes W, et al. (1987) Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161(2):479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls AC, et al. (2016) Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531(7592):114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchdoerfer RN, et al. (2016) Pre-fusion structure of a human coronavirus spike protein. Nature 531(7592):118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walls AC, et al. (2016) Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23(10):899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Y, et al. (2017) Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong X, et al. (2018) Glycan Shield and Fusion Activation of a Deltacoronavirus Spike Glycoprotein Fine-Tuned for Enteric Infections. J Virol 92(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shang J, et al. (2018) Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog 14(4):e1007009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sturman LS, Holmes KV, & Behnke J (1980) Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol 33(1):449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rottier P, Brandenburg D, Armstrong J, van der Zeijst B, & Warren G (1984) Assembly in vitro of a spanning membrane protein of the endoplasmic reticulum: the E1 glycoprotein of coronavirus mouse hepatitis virus A59. Proc Natl Acad Sci U S A 81(5):1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo L, Hurst-Hess KR, Koetzner CA, & Masters PS (2016) Analyses of Coronavirus Assembly Interactions with Interspecies Membrane and Nucleocapsid Protein Chimeras. J Virol 90(9):4357–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobs L, van der Zeijst BA, & Horzinek MC (1986) Characterization and translation of transmissible gastroenteritis virus mRNAs. J Virol 57(3):1010–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nal B, et al. (2005) Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol 86(Pt 5):1423–1434. [DOI] [PubMed] [Google Scholar]
- 31.de Haan CA, et al. (1998) Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem 273(45):29905–29914. [DOI] [PubMed] [Google Scholar]
- 32.Laude H, Gelfi J, Lavenant L, & Charley B (1992) Single amino acid changes in the viral glycoprotein M affect induction of alpha interferon by the coronavirus transmissible gastroenteritis virus. J Virol 66(2):743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Haan CA, et al. (2003) The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology 312(2):395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Locker JK, Rose JK, Horzinek MC, & Rottier PJ (1992) Membrane assembly of the triple-spanning coronavirus M protein. Individual transmembrane domains show preferred orientation. J Biol Chem 267(30):21911–21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Machamer CE & Rose JK (1987) A specific transmembrane domain of a coronavirus E1 glycoprotein is required for its retention in the Golgi region. J Cell Biol 105(3):1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapke PA, et al. (1988) The amino-terminal signal peptide on the porcine transmissible gastroenteritis coronavirus matrix protein is not an absolute requirement for membrane translocation and glycosylation. Virology 165(2):367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neuman BW, et al. (2011) A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol 174(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo L & Masters PS (2013) Functional analysis of the murine coronavirus genomic RNA packaging signal. J Virol 87(9):5182–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masters PS (2006) The molecular biology of coronaviruses. Adv Virus Res 66:193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vennema H, et al. (1996) Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 15(8):2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corse E & Machamer CE (2002) The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J Virol 76(3):1273–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruch TR & Machamer CE (2012) A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog 8(5):e1002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson L, McKinlay C, Gage P, & Ewart G (2004) SARS coronavirus E protein forms cation-selective ion channels. Virology 330(1):322–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pervushin K, et al. (2009) Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog 5(7):e1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeDiego ML, et al. (2007) A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81(4):1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer F, Stegen CF, Masters PS, & Samsonoff WA (1998) Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol 72(10):7885–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieto-Torres JL, et al. (2014) Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog 10(5):e1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castano-Rodriguez C, et al. (2018) Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. MBio 9(3) pii: e02325–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang CK, et al. (2006) Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci 13(1):59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurst KR, Koetzner CA, & Masters PS (2009) Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J Virol 83(14):7221–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stohlman SA & Lai MM (1979) Phosphoproteins of murine hepatitis viruses. J Virol 32(2):672–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grunewald ME, Fehr AR, Athmer J, & Perlman S (2018) The coronavirus nucleocapsid protein is ADP-ribosylated. Virology 517:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stohlman SA, et al. (1988) Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol 62(11):4288–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hurst KR, Koetzner CA, & Masters PS (2013) Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol 87(16):9159–9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klausegger A, et al. (1999) Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol 73(5):3737–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelissen LA, et al. (1997) Hemagglutinin-esterase, a novel structural protein of torovirus. J Virol 71(7):5277–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lissenberg A, et al. (2005) Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol 79(24):15054–15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubo H, Yamada YK, & Taguchi F (1994) Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol 68(9):5403–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong SK, Li W, Moore MJ, Choe H, & Farzan M (2004) A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem 279(5):3197–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang N, et al. (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23(8):986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu G, et al. (2013) Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500(7461):227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, & Holmes KV (2003) Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J Virol 77(4):2530–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Qian Z, et al. (2015) Identification of the Receptor-Binding Domain of the Spike Glycoprotein of Human Betacoronavirus HKU1. J Virol 89(17):8816–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin HX, et al. (2008) Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J Gen Virol 89(Pt 4):1015–1024. [DOI] [PubMed] [Google Scholar]
- 65.Godet M, Grosclaude J, Delmas B, & Laude H (1994) Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol 68(12):8008–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams RK, Jiang GS, Snyder SW, Frana MF, & Holmes KV (1990) Purification of the 110-kilodalton glycoprotein receptor for mouse hepatitis virus (MHV)-A59 from mouse liver and identification of a nonfunctional, homologous protein in MHV-resistant SJL/J mice. J Virol 64(8):3817–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams RK, Jiang GS, & Holmes KV (1991) Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sci U S A 88(13):5533–5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dveksler GS, et al. (1991) Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J Virol 65(12):6881–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Taguchi F & Hirai-Yuki A (2012) Mouse Hepatitis Virus Receptor as a Determinant of the Mouse Susceptibility to MHV Infection. Front Microbiol 3:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Delmas B, et al. (1992) Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 357(6377):417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeager CL, et al. (1992) Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357(6377):420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tresnan DB, Levis R, & Holmes KV (1996) Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol 70(12):8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li W, et al. (2018) Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci U S A 115(22):E5135–E5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang B, et al. (2018) Porcine Deltacoronavirus Engages the Transmissible Gastroenteritis Virus Functional Receptor Porcine Aminopeptidase N for Infectious Cellular Entry. J Virol 92(12) pii:e00318–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delmas B, Gelfi J, Sjostrom H, Noren O, & Laude H (1993) Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv Exp Med Biol 342:293–298. [DOI] [PubMed] [Google Scholar]
- 76.Benbacer L, Kut E, Besnardeau L, Laude H, & Delmas B (1997) Interspecies aminopeptidase-N chimeras reveal species-specific receptor recognition by canine coronavirus, feline infectious peritonitis virus, and transmissible gastroenteritis virus. J Virol 71(1):734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Delmas B, et al. (1994) Determinants essential for the transmissible gastroenteritis virus-receptor interaction reside within a domain of aminopeptidase-N that is distinct from the enzymatic site. J Virol 68(8):5216–5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li W, et al. (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426(6965):450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hamming I, et al. (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol 203(2):631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hofmann H, et al. (2005) Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 102(22):7988–7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raj VS, et al. (2013) Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495(7440):251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li F (2015) Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89(4):1954–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belouzard S, Chu VC, & Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America 106(14):5871–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kleine-Weber H, Elzayat MT, Hoffmann M, & Pohlmann S (2018) Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep 8(1):16597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Park JE, et al. (2016) Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113(43):12262–12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.White JM & Whittaker GR (2016) Fusion of Enveloped Viruses in Endosomes. Traffic 17(6):593–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chinese SMEC (2004) Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303(5664):1666–1669. [DOI] [PubMed] [Google Scholar]
- 88.Cotten M, et al. (2013) Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet 382(9909):1993–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim DW, et al. (2016) Variations in Spike Glycoprotein Gene of MERS-CoV, South Korea, 2015. Emerg Infect Dis 22(1):100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim Y, et al. (2016) Spread of Mutant Middle East Respiratory Syndrome Coronavirus with Reduced Affinity to Human CD26 during the South Korean Outbreak. MBio 7(2):e00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Earnest JT, et al. (2017) The tetraspanin CD9 facilitates MERS-coronavirus entry by scaffolding host cell receptors and proteases. PLoS Pathog 13(7):e1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baranov PV, et al. (2005) Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology 332(2):498–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brierley I, Digard P, & Inglis SC (1989) Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57(4):537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Araki K, et al. (2010) Pathogenic virus-specific T cells cause disease during treatment with the calcineurin inhibitor FK506: implications for transplantation. J Exp Med 207(11):2355–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ziebuhr J, Snijder EJ, & Gorbalenya AE (2000) Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81(Pt 4):853–879. [DOI] [PubMed] [Google Scholar]
- 96.Mielech AM, Chen Y, Mesecar AD, & Baker SC (2014) Nidovirus papain-like proteases: multifunctional enzymes with protease, deubiquitinating and deISGylating activities. Virus Res 194:184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Anand K, et al. (2002) Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J 21(13):3213–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stobart CC, Lee AS, Lu X, & Denison MR (2012) Temperature-sensitive mutants and revertants in the coronavirus nonstructural protein 5 protease (3CLpro) define residues involved in long-distance communication and regulation of protease activity. J Virol 86(9):4801–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang H, et al. (2005) Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol 3(10):e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Neuman BW, Chamberlain P, Bowden F, & Joseph J (2014) Atlas of coronavirus replicase structure. Virus Res 194:49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oostra M, et al. (2008) Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J Virol 82(24):12392–12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanjanahaluethai A, Chen Z, Jukneliene D, & Baker SC (2007) Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361(2):391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neuman BW (2016) Bioinformatics and functional analyses of coronavirus nonstructural proteins involved in the formation of replicative organelles. Antiviral Res 135:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lei J, Kusov Y, & Hilgenfeld R (2018) Nsp3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res 149:58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Neuman BW, et al. (2008) Proteomics analysis unravels the functional repertoire of coronavirus nonstructural protein 3. J Virol 82(11):5279–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hurst KR, Ye R, Goebel SJ, Jayaraman P, & Masters PS (2010) An interaction between the nucleocapsid protein and a component of the replicase-transcriptase complex is crucial for the infectivity of coronavirus genomic RNA. J Virol 84(19):10276–10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keane SC & Giedroc DP (2013) Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein. J Virol 87(6):3502–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li C, et al. (2016) Viral Macro Domains Reverse Protein ADP-Ribosylation. J Virol 90(19):8478–8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fehr AR, Jankevicius G, Ahel I, & Perlman S (2018) Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol 26(7):598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Smith EC, et al. (2015) Mutations in coronavirus nonstructural protein 10 decrease virus replication fidelity. J Virol 89(12):6418–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eckerle LD, et al. (2010) Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6(5):e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eckerle LD, Lu X, Sperry SM, Choi L, & Denison MR (2007) High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol 81(22):12135–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Minskaia E, et al. (2006) Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 103(13):5108–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Y, et al. (2009) Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106(9):3484–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Decroly E, et al. (2008) Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2’O)-methyltransferase activity. J Virol 82(16):8071–8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kindler E, Thiel V, & Weber F (2016) Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv Virus Res 96:219–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deng X, et al. (2017) Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114(21):E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kindler E, et al. (2017) Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13(2):e1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Snijder EJ, et al. (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sethna PB, Hofmann MA, & Brian DA (1991) Minus-strand copies of replicating coronavirus mRNAs contain antileaders. Journal of virology 65(1):320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brown CG, Nixon KS, Senanayake SD, & Brian DA (2007) An RNA stem-loop within the bovine coronavirus nsp1 coding region is a cis-acting element in defective interfering RNA replication. J Virol 81(14):7716–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guan BJ, Wu HY, & Brian DA (2011) An optimal cis-replication stem-loop IV in the 5’ untranslated region of the mouse coronavirus genome extends 16 nucleotides into open reading frame 1. J Virol 85(11):5593–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu P, et al. (2009) Mouse hepatitis virus stem-loop 2 adopts a uYNMG(U)a-like tetraloop structure that is highly functionally tolerant of base substitutions. J Virol 83(23):12084–12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Raman S, Bouma P, Williams GD, & Brian DA (2003) Stem-loop III in the 5’ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J Virol 77(12):6720–6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Q, Johnson RF, & Leibowitz JL (2001) Secondary structural elements within the 3’ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J Virol 75(24):12105–12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Goebel SJ, Miller TB, Bennett CJ, Bernard KA, & Masters PS (2007) A hypervariable region within the 3’ cis-acting element of the murine coronavirus genome is nonessential for RNA synthesis but affects pathogenesis. J Virol 81(3):1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Williams GD, Chang RY, & Brian DA (1999) A phylogenetically conserved hairpin-type 3’ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol 73(10):8349–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hsue B & Masters PS (1997) A bulged stem-loop structure in the 3’ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol 71(10):7567–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hsue B, Hartshorne T, & Masters PS (2000) Characterization of an essential RNA secondary structure in the 3’ untranslated region of the murine coronavirus genome. J Virol 74(15):6911–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sawicki SG, Sawicki DL, & Siddell SG (2007) A contemporary view of coronavirus transcription. J Virol 81(1):20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu CH, Chen PJ, & Yeh SH (2014) Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe 16(4):462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Keck JG, et al. (1987) RNA recombination of coronavirus. Adv Expt Med Biol 218:99–107. [DOI] [PubMed] [Google Scholar]
- 133.Lai MM, et al. (1985) Recombination between nonsegmented RNA genomes of murine coronaviruses. J Virol 56(2):449–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kuo L, Godeke GJ, Raamsman MJ, Masters PS, & Rottier PJ (2000) Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol 74(3):1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krijnse-Locker J, Ericsson M, Rottier PJM, & Griffiths G (1994) Characterization of the budding compartment of mouse hepatitis virus: Evidence that transport from the RER to the golgi complex requires only one vesicular transport step. J. Cell Biol 124:55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tooze J, Tooze S, & Warren G (1984) Replication of coronavirus MHV-A59 in sac- cells: determination of the first site of budding of progeny virions. Euro J Cell Biol 33(2):281–293. [PubMed] [Google Scholar]
- 137.de Haan CA & Rottier PJ (2005) Molecular interactions in the assembly of coronaviruses. Adv Virus Res 64:165–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bos EC, Luytjes W, van der Meulen HV, Koerten HK, & Spaan WJM (1996) The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Siu YL, et al. (2008) The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 82(22):11318–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Raamsman MJ, et al. (2000) Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol 74(5):2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Corse E & Machamer CE (2000) Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J Virol 74(9):4319–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Boscarino JA, Logan HL, Lacny JJ, & Gallagher TM (2008) Envelope protein palmitoylations are crucial for murine coronavirus assembly. J Virol 82(6):2989–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ye Y & Hogue BG (2007) Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol 81(7):3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hurst KR, et al. (2005) A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J Virol 79(21):13285–13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Athmer J, et al. (2018) Selective Packaging in Murine Coronavirus Promotes Virulence by Limiting Type I Interferon Responses. MBio 9(3) pii:e00272–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wong HH, et al. (2015) Genome-Wide Screen Reveals Valosin-Containing Protein Requirement for Coronavirus Exit from Endosomes. J Virol 89(21):11116–11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhou P, et al. (2018) Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556(7700):255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.de Groot-Mijnes JD, van Dun JM, van der Most RG, & de Groot RJ (2005) Natural history of a recurrent feline coronavirus infection and the role of cellular immunity in survival and disease. J Virol 79(2):1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Saif LJ (2010) Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 26(2):349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ignjatovic J & Sapats S (2000) Avian infectious bronchitis virus. Rev Sci Tech 19(2):493–508. [DOI] [PubMed] [Google Scholar]
- 151.Mihindukulasuriya KA, Wu G, St Leger J, Nordhausen RW, & Wang D (2008) Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J Virol 82(10):5084–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.de Groot RJ, et al. (2013) Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 87(14):7790–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, & Fouchier RA (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- 154.He B, et al. (2014) Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in china. J Virol 88(12):7070–7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nga PT, et al. (2011) Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog 7(9):e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lauber C, et al. (2012) Mesoniviridae: a proposed new family in the order Nidovirales formed by a single species of mosquito-borne viruses. Arch. Virol 157(8):1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, & Gorbalenya AE (2018) A planarian nidovirus expands the limits of RNA genome size. PLoS Pathog 14(11):e1007314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Levy GA, et al. (2000) Molecular and functional analysis of the human prothrombinase gene (HFGL2) and its role in viral hepatitis. Am J Pathol 156:1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lampert PW, Sims JK, & Kniazeff AJ (1973) Mechanism of demyelination in JHM virus encephalomyelitis. Acta Neuropathol. 24:76–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weiner LP (1973) Pathogenesis of demyelination induced by a mouse hepatitis virus (JHM virus). Arch. Neurol 28:298–303. [DOI] [PubMed] [Google Scholar]
- 161.Houtman JJ & Fleming JO (1996) Pathogenesis of mouse hepatitis virus-induced demyelination. J Neurovirol 2(6):361–376. [DOI] [PubMed] [Google Scholar]
- 162.Wang F, Stohlman SA, & Fleming JO (1990) Demyelination induced by murine hepatitis virus JHM strain (MHV-4) is immunologically mediated. J. Neuroimmunol 30:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu GF, Dandekar AA, Pewe L, & Perlman S (2000) CD4 and CD8 T cells have redundant but not identical roles in virus-induced demyelination. J. Immunol 165:2278–2286. [DOI] [PubMed] [Google Scholar]
- 164.Wu GF & Perlman S (1999) Macrophage infiltration, but not apoptosis, is correlated with immune-mediated demyelination following murine infection with a neurotropic coronavirus. J. Virol 73:8771–8780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wheeler DL, Sariol A, Meyerholz DK, & Perlman S (2018) Microglia are required for protection against lethal coronavirus encephalitis in mice. J Clin Invest 128(3):931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McIntosh K, Becker WB, & Chanock RM (1967) Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci 58:2268–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bradburne AF, Bynoe ML, & Tyrell DAJ (1967) Effects of a “new” human respiratory virus in volunteers. Br. Med. J 3:767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hamre D & Procknow JJ (1966) A new virus isolated from the human respiratory tract. Proc Sco Exp Biol Med 121(1):190–193. [DOI] [PubMed] [Google Scholar]
- 169.Woo PC, et al. (2005) Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79(2):884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.van der Hoek L, et al. (2004) Identification of a new human coronavirus. Nat Med 10(4):368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huynh J, et al. (2012) Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86(23):12816–12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jean A, Quach C, Yung A, & Semret M (2013) Severity and outcome associated with human coronavirus OC43 infections among children. Pediatr Infect Dis J 32(4):325–329. [DOI] [PubMed] [Google Scholar]
- 173.van der Hoek L, et al. (2005) Croup is associated with the novel coronavirus NL63. PLoS Med 2(8):e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chibo D & Birch C (2006) Analysis of human coronavirus 229E spike and nucleoprotein genes demonstrates genetic drift between chronologically distinct strains. J Gen Virol 87(Pt 5):1203–1208. [DOI] [PubMed] [Google Scholar]
- 175.Vijgen L, et al. (2005) Circulation of genetically distinct contemporary human coronavirus OC43 strains. Virology 337(1):85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Arbour N, Day R, Newcombe J, & Talbot PJ (2000) Neuroinvasion by human respiratory coronaviruses. J Virol 74(19):8913–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Peiris JS, Guan Y, & Yuen KY (2004) Severe acute respiratory syndrome. Nat Med 10(12 Suppl):S88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lau SK, et al. (2005) Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci 102(39):14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li W, et al. (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676–679. [DOI] [PubMed] [Google Scholar]
- 180.Ge XY, et al. (2013) Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503(7477):535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guan Y, et al. (2003) Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. [DOI] [PubMed] [Google Scholar]
- 182.Peiris JS, Yuen KY, Osterhaus AD, & Stohr K (2003) The severe acute respiratory syndrome. New Engl J Med 349(25):2431–2441. [DOI] [PubMed] [Google Scholar]
- 183.Christian MD, Poutanen SM, Loutfy MR, Muller MP, & Low DE (2004) Severe acute respiratory syndrome. Clin Infect Dis 38(10):1420–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nicholls JM, et al. (2003) Lung pathology of fatal severe acute respiratory syndrome. Lancet 361(9371):1773–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Peiris JS, et al. (2003) Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361(9371):1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Spiegel M, Schneider K, Weber F, Weidmann M, & Hufert FT (2006) Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J Gen Virol 87(Pt 7):1953–1960. [DOI] [PubMed] [Google Scholar]
- 187.Law HK, et al. (2005) Chemokine upregulation in SARS coronavirus infected human monocyte derived dendritic cells. Blood 106:2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lau YL & Peiris JSM (2005) Pathogenesis of severe acute respiratory syndrome. Curr Op Immunol 17:404–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Roberts A, et al. (2005) Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J Virol 79(9):5833–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhao J, Zhao J, & Perlman S (2010) T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol 84(18):9318–9325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhao J, Zhao J, Legge K, & Perlman S (2011) Age-related increases in PGD(2) expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest 121(12):4921–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Aly M, Elrobh M, Alzayer M, Aljuhani S, & Balkhy H (2017) Occurrence of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) across the Gulf Corporation Council countries: Four years update. PLoS One 12(10):e0183850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Choi WS, et al. (2016) Clinical Presentation and Outcomes of Middle East Respiratory Syndrome in the Republic of Korea. Infect Chemother 48(2):118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ki M (2015) 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health 37:e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Conzade R, et al. (2018) Reported Direct and Indirect Contact with Dromedary Camels among Laboratory-Confirmed MERS-CoV Cases. Viruses 10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mohd HA, Al-Tawfiq JA, & Memish ZA (2016) Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virol J 13:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Memish ZA, et al. (2014) Human Infection with MERS Coronavirus after Exposure to Infected Camels, Saudi Arabia, 2013. Emerg Inf Dis 20(6):1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.van Boheemen S, et al. (2012) Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6) pii: e00473–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Anthony SJ, et al. (2017) Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. MBio 8(2) pii:e00373–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li K, et al. (2017) Mouse-adapted MERS coronavirus causes lethal lung disease in human DPP4 knockin mice. Proc Natl Acad Sci U S A 114(15):E3119–E3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Agrawal AS, et al. (2015) Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol 89(7):3659–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zhao J, et al. (2014) Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci 111(13):4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cockrell AS, et al. (2016) A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2:16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Emery SL, et al. (2004) Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Inf Dis 10(2):311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Gaunt ER, Hardie A, Claas EC, Simmonds P, & Templeton KE (2010) Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol 48(8):2940–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Huang P, et al. (2018) A Rapid and Specific Assay for the Detection of MERS-CoV. Front Microbiol 9:1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Omrani AS, et al. (2014) Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: a retrospective cohort study. Lancet Infect Dis 14(11):1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Channappanavar R, et al. (2016) Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 19(2):181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Channappanavar R, et al. (2019) IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 130:3625–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Wang L, et al. (2018) Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape. J Virol 92(10) pii:e02002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Mo Y & Fisher D (2016) A review of treatment modalities for Middle East Respiratory Syndrome. J Antimicrob Chemother 71(12):3340–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Jordan B (2017) Vaccination against infectious bronchitis virus: A continuous challenge. Vet Microbiol 206:137–143. [DOI] [PubMed] [Google Scholar]
- 213.Welter MW, Horstman MP, Welter CJ, & Welter LM (1993) An overview of successful TGEV vaccination strategies and discussion on the interrelationship between TGEV and PRCV. Adv Exp Med Biol 342:463–468. [DOI] [PubMed] [Google Scholar]
- 214.Jones KE, et al. (2008) Global trends in emerging infectious diseases. Nature 451(7181):990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Laude H, Van Reeth K, & Pensaert M (1993) Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res 24(2):125–150. [PubMed] [Google Scholar]
- 216.Pogrebnyak N, et al. (2005) Severe acute respiratory syndrome (SARS) S protein production in plants: development of recombinant vaccine. Proc Natl Acad Sci U S A 102(25):9062–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.McPherson C, et al. (2016) Development of a SARS Coronavirus Vaccine from Recombinant Spike Protein Plus Delta Inulin Adjuvant. Methods Mol Biol 1403:269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tsunetsugu-Yokota Y, Ohnishi K, & Takemori T (2006) Severe acute respiratory syndrome (SARS) coronavirus: application of monoclonal antibodies and development of an effective vaccine. Rev Med Virol 16(2):117–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Coughlin MM & Prabhakar BS (2012) Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev Med Virol 22(1):2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Schindewolf C & Menachery VD (2019) Middle East Respiratory Syndrome Vaccine Candidates: Cautious Optimism. Viruses 11(1) pii:E74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zumla A, Memish ZA, Hui DS, & Perlman S (2019) Vaccine against Middle East respiratory syndrome coronavirus. Lancet Infect Dis 19(10):1054–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Modjarrad K, et al. (2019) Safety and immunogenicity of an anti-Middle East respiratory syndrome coronavirus DNA vaccine: a phase 1, open-label, single-arm, dose-escalation trial. Lancet Infect Dis 19(9):1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yoon IK & Kim JH (2019) First clinical trial of a MERS coronavirus DNA vaccine. Lancet Infect Dis 19(9):924–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Dye C, Temperton N, & Siddell SG (2007) Type I feline coronavirus spike glycoprotein fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J Gen Virol 88(Pt 6):1753–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Shirato K, et al. (2016) Porcine aminopeptidase N is not a cellular receptor of porcine epidemic diarrhea virus, but promotes its infectivity via aminopeptidase activity. J Gen Virol 97(10):2528–2539. [DOI] [PubMed] [Google Scholar]
- 226.Li W, et al. (2017) Aminopeptidase N is not required for porcine epidemic diarrhea virus cell entry. Virus Res 235:6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Schultze B & Herrler G (1992) Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J Gen Virol 73 (Pt 4):901–906. [DOI] [PubMed] [Google Scholar]
- 228.Kunkel F & Herrler G (1993) Structural and functional analysis of the surface protein of human coronavirus OC43. Virology 195(1):195–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Promkuntod N, van Eijndhoven RE, de Vrieze G, Grone A, & Verheije MH (2014) Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology 448:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zust R, et al. (2007) Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 3(8):e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Nakagawa K, et al. (2018) The Endonucleolytic RNA Cleavage Function of nsp1 of Middle East Respiratory Syndrome Coronavirus Promotes the Production of Infectious Virus Particles in Specific Human Cell Lines. J Virol 92(21) pii: e01157–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Graham RL, Sims AC, Brockway SM, Baric RS, & Denison MR (2005) The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J Virol 79(21):13399–13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Cornillez-Ty CT, Liao L, Yates JR 3rd, Kuhn P, & Buchmeier MJ (2009) Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J Virol 83(19):10314–10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Chatterjee A, et al. (2009) Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold. J Virol 83(4):1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Egloff MP, et al. (2006) Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80(17):8493–8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Eriksson KK, Cervantes-Barragan L, Ludewig B, & Thiel V (2008) Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1"-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol 82(24):12325–12334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Frieman M, Ratia K, Johnston RE, Mesecar AD, & Baric RS (2009) Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83(13):6689–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Serrano P, et al. (2007) Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus. J Virol 81(21):12049–12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Serrano P, et al. (2009) Nuclear magnetic resonance structure of the nucleic acid-binding domain of severe acute respiratory syndrome coronavirus nonstructural protein 3. J Virol 83(24):12998–13008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Ziebuhr J, Thiel V, & Gorbalenya AE (2001) The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J Biol Chem 276(35):33220–33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Clementz MA, Kanjanahaluethai A, O’Brien TE, & Baker SC (2008) Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375(1):118–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Gadlage MJ, et al. (2010) Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J Virol 84(1):280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lu Y, Lu X, & Denison MR (1995) Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol 69(6):3554–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhu X, et al. (2017) Porcine deltacoronavirus nsp5 inhibits interferon-beta production through the cleavage of NEMO. Virology 502:33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zhai Y, et al. (2005) Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol 12(11):980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kirchdoerfer RN & Ward AB (2019) Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10(1):2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Egloff MP, et al. (2004) The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci 101(11):3792–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Zeng Z, et al. (2018) Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity. J Virol 92(17). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Bouvet M, et al. (2010) In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog 6(4):e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Decroly E, et al. (2011) Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2’-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7(5):e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Xu X, et al. (2003) Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Nucl Acids Res 31(24):7117–7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ivanov KA, et al. (2004) Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 78(11):5619–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ivanov KA & Ziebuhr J (2004) Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5’-triphosphatase activities. J Virol 78(14):7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Becares M, et al. (2016) Mutagenesis of Coronavirus nsp14 Reveals Its Potential Role in Modulation of the Innate Immune Response. J Virol 90(11):5399–5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Case JB, et al. (2018) Murine Hepatitis Virus nsp14 Exoribonuclease Activity Is Required for Resistance to Innate Immunity. J Virol 92(1) pii: e01531–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bhardwaj K, Sun J, Holzenburg A, Guarino LA, & Kao CC (2006) RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol 361(2):243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ivanov KA, et al. (2004) Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci 101(34):12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Athmer J, et al. (2017) In Situ Tagged nsp15 Reveals Interactions with Coronavirus Replication/Transcription Complex-Associated Proteins. MBio 8(1) pii:e02320–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zust R, et al. (2011) Ribose 2’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12(2):137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]


