Significance
Form follows function is true of living organisms, including bacteria, as the shapes and morphology they adopt contribute to their biological characteristics. But whether bacteria can actively change their shape to adapt to their environment is less understood. Vibrio cholerae, the pathogenic bacterium responsible for the diarrheal disease cholera, adopts a characteristic “comma”-shaped cell morphology. Here we show that the intracellular signaling molecule, cyclic di-GMP, drives curved V. cholerae to adopt a straight cell morphology that is advantageous to a sessile biofilm lifestyle, while curved V. cholerae are better adapted for swimming. Our research provides a clear example of how bacteria can actively alter their form to impact function.
Keywords: cell shape, crvA, cyclic di-GMP, biofilm, motility
Abstract
The cell morphology of rod-shaped bacteria is determined by the rigid net of peptidoglycan forming the cell wall. Alterations to the rod shape, such as the curved rod, occur through manipulating the process of cell wall synthesis. The human pathogen Vibrio cholerae typically exists as a curved rod, but straight rods have been observed under certain conditions. While this appears to be a regulated process, the regulatory pathways controlling cell shape transitions in V. cholerae and the benefits of switching between rod and curved shape have not been determined. We demonstrate that cell shape in V. cholerae is regulated by the bacterial second messenger cyclic dimeric guanosine monophosphate (c-di-GMP) by posttranscriptionally repressing expression of crvA, a gene encoding an intermediate filament-like protein necessary for curvature formation in V. cholerae. This regulation is mediated by the transcriptional cascade that also induces production of biofilm matrix components, indicating that cell shape is coregulated with V. cholerae’s induction of sessility. During microcolony formation, wild-type V. cholerae cells tended to exist as straight rods, while genetically engineering cells to maintain high curvature reduced microcolony formation and biofilm density. Conversely, straight V. cholerae mutants have reduced swimming speed when using flagellar motility in liquid. Our results demonstrate regulation of cell shape in bacteria is a mechanism to increase fitness in planktonic and biofilm lifestyles.
Morphology is an important biological feature because it dictates how organisms interact with their physical world. Various aspects of morphology such as shape, length, and the presence of appendages are subject to selective pressures and contribute to adaptation to specific ecological niches (1). Not surprisingly, bacteria take on diverse shapes from simple rods and cocci to helices and curves; however, the fitness benefits of each shape is not always well understood (2).
Shape in bacteria is largely determined by the structure of the peptidoglycan (PG) cell wall as bacteria without PG adopt pleiomorphic, or l-form, shapes (3, 4). A predominant morphology in gram-negative bacteria is the rod shape. During growth of a rod-shaped cell, new PG subunits are added to the growing PG layer by continuous rounds of PG cleavage and subunit insertion along the sides of the rod, at the midcell, or in some cases the pole (3, 5). The curved-rod shape is common in many aquatic organisms, such as the fresh-water bacterium Caulobacter crescentus and the opportunistic human pathogen Vibrio cholerae. In his 1928 book titled “Morphologic Variation of the Cholera Vibrio” Arthur Henrici measured V. cholerae shape as a function of time during a batch culture growth curve and observed shapes ranging from spheres to straight and curved rods (6). Curvature in the bacterium Caulobacter crescentus is mediated by the protein crescentin (7). Similarly, research by Bartlett et al. (8) identified that the gene crvA encoding the periplasmic intermediate filament-like protein CrvA introduces curvature by altering PG insertion rates. Mutants lacking crvA grow as straight rods, have reduced migration in soft agar, and are less virulent in animal models of infection (8). The adjacent ORF to crvA, annotated as crvB, functions with CrvA and, together, CrvAB are sufficient to produce curvature in normally straight cells (9).
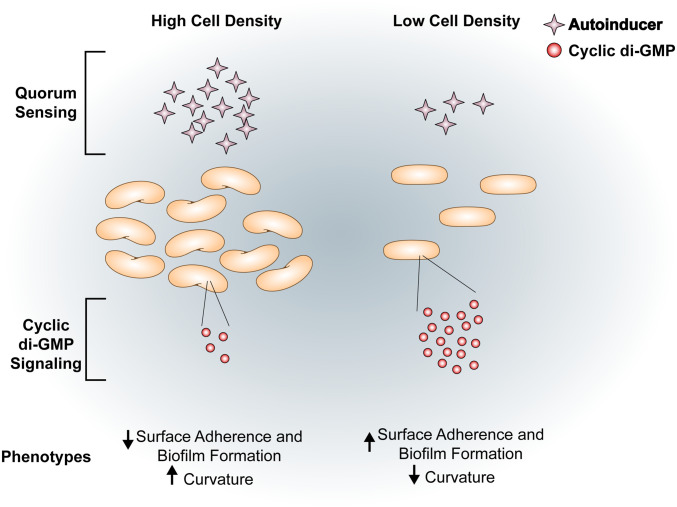
Interestingly, in V. cholerae, cell shape change is controlled by quorum sensing (QS), a phenomenon in which microorganisms communicate and coordinate behavior based on population density (8). Specifically, Henrici and Bartlett et al. both observed highly curved rods in high-cell density (HCD) populations and straighter rods in low-cell density (LCD) populations (6, 8). In addition to changing cell shape, QS regulates many population-density-dependent phenotypes such as biofilm formation through complex regulatory pathways, one of which is the bacterial second messenger cyclic dimeric guanosine monophosphate (c-di-GMP) (Fig. 1) (10, 11).
Fig. 1.
QS and cyclic di-GMP signaling and linked phenotypes in V. cholerae. V. cholerae exhibits an inverse relationship between cell density and intracellular c-di-GMP. Under HCD conditions where there is a high concentration of autoinducers, the intracellular concentration of c-di-GMP is low (11, 12). In HCD conditions, cells have increased curvature and a decreased propensity to adhere to surfaces and form biofilms. Conversely, cells in LCD conditions have low concentrations of autoinducers and high c-di-GMP (reviewed in ref. 13). Under LCD conditions, cells are more likely to form biofilms (14). Additionally, V. cholerae loses its canonical curved-rod shape and appears as straight rods (6).
Flagellar motility and biofilm formation are regulated by c-di-GMP signaling in many bacteria (15). C-di-GMP is produced by enzymes called diguanylate cyclases (DGCs) and degraded by enzymes called phosphodiesterases (PDEs). In V. cholerae, c-di-GMP binds to and activates the transcription factors VpsR and VpsT (16, 17). These transcription factors then induce expression of the operons encoding the enzymes necessary for synthesizing Vibrio polysaccharide (VPS), the major exopolysaccharide component of the biofilm matrix, and matrix-associated proteins resulting in mature biofilm formation (18–21). In addition, c-di-GMP represses motility by inhibiting transcription of the flagellar biosynthesis genes, repressing the expression of the transcription factor TfoY by binding to a c-di-GMP-dependent riboswitch, and inducing MshA pilus extension mediating surface attachment (12, 22). Intracellular c-di-GMP concentrations are controlled by a variety of signals including QS, where cells in the LCD state have higher c-di-GMP concentrations than cells in the HCD (11).
Although cell shape in V. cholerae has been demonstrated to shift between curved and straight morphologies, the regulatory pathways that control this shift and the potential ecological benefits of such changes are not fully understood (6, 8). In this study, we report that elevated intracellular c-di-GMP concentrations straighten V. cholerae by decreasing CrvA expression. Locking V. cholerae in a curved morphology reduced the formation of microcolonies and biofilm formation. Conversely, V. cholerae cells locked in a straight morphology reduced swimming speed when compared to the curved wild-type (WT) cells. Therefore, curved and straight cell morphologies are optimal for motility and biofilm formation, respectively, and our results demonstrate how bacteria can actively control cell shape to adapt to different lifestyles.
Results
High Intracellular C-di-GMP Reduces Cell Curvature.
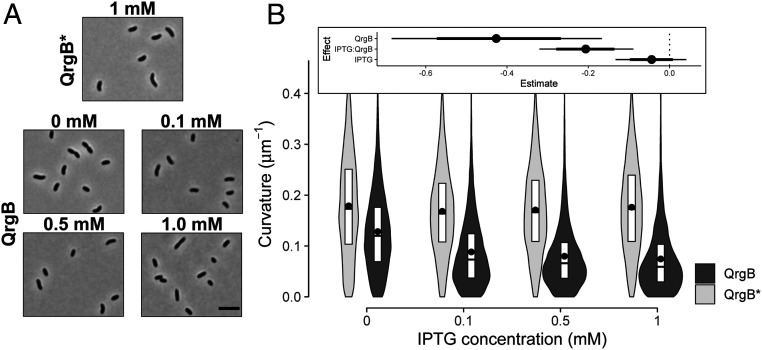
When exploring the impact of c-di-GMP on stress response pathways in V. cholerae (23, 24), we noted that strains with high intracellular concentrations of c-di-GMP were more frequently straight rods. We explored this finding by quantifying cell shape parameters under different intracellular c-di-GMP concentrations utilizing strains that ectopically express an isopropyl-β-D-thiogalactoside (IPTG)-inducible DGC (QrgB) that synthesizes c-di-GMP or a catalytically inactive DGC (QrgB*). We have previously shown such expression of QrgB in V. cholerae produces physiologically relevant concentrations of c-di-GMP (12). We observed that the curvature of the medial axis of cells decreased in a strain harboring the active DGC when no IPTG was added due to leaky expression of the inducible promoter (Fig. 2, P value <1e-3). An even further reduction of curvature was observed when IPTG was added to increase QrgB expression (P value <6e-4), supporting our hypothesis that high c-di-GMP concentrations cause cells to straighten (Fig. 2). No such change in curvature was observed when the QrgB* mutant protein was induced, indicating the change in morphology was specific to c-di-GMP synthesis (Fig. 2). Increasing IPTG in cells expressing QrgB* caused cells to increase in length by 1.16-fold and expressing QrgB, regardless of IPTG, caused width to decrease by 1.02-fold (SI Appendix 1, Fig. S1). While these subtle changes are statistically significant given our data, they likely reflect experimental noise and are not biologically relevant. Thus, c-di-GMP primarily alters V. cholerae cell morphology by decreasing curvature.
Fig. 2.
C-di-GMP decreases cell curvature in a dose-dependent manner. (A) Representative phase-contrast micrographs of early stationary phase cells. Cells harboring a plasmid encoding an IPTG-inducible inactive DGC (QrgB*) with 1 mM IPTG and an IPTG-inducible active DGC (QrgB) with 0, 0.1, 0.5, and 1 mM IPTG (OD600 = 1.3). (Scale bar on the last image of the panel, 5 µm.) (B) Distributions of cell curvature as a function of IPTG concentration in populations expressing inactive DGC (QrgB*, light) or active DGC (QrgB, dark). The box represents the first, second, and third quartiles. The dot represents the mean. Each distribution represents between 1,000 and 1,200 cells analyzed and pooled from four to five separate experiments. (Inset) Estimate of the contribution of each experimental factor to changes in the medians (dot = mean, thick line = 90% credible interval (CI), thin line = 98% CI). Effects with 90% CI that do not contain zero have a P value ≤0.05. Effects with 98% CI that do not contain zero have a P value ≤0.01.
The C-di-GMP-Dependent Transcription Factors, VpsR and VpsT, Control C-di-GMP-Mediated Changes to Cell Curvature.
In V. cholerae, three c-di-GMP-dependent transcription factors (VpsR, VpsT, and FlrA) and two c-di-GMP-dependent riboswitches (Vc1 and Vc2), are known to regulate genes that elicit diverse phenotypes such as biofilm formation, motility, DNA repair, and catalase production (16, 19, 22–28). We hypothesized that VpsT or VpsR were involved in the c-di-GMP-dependent regulation of cell curvature because of their roles in regulating multiple c-di-GMP-dependent phenotypes (23, 24, 29). To test this hypothesis, we measured cell shape parameters in ∆vpsR and ∆vpsT mutants at high (QrgB) and unaltered (QrgB*) concentrations of c-di-GMP. While width varied very little across all experimental conditions, in the absence of vpsT or vpsR, QrgB expression reduced cell length (P value <6e-3) (SI Appendix 1, Fig. S2). Additionally, overexpressing VpsT, regardless of c-di-GMP and genetic background, caused a small increase in length (P value <6e-3) (SI Appendix 1, Fig. S2). These data suggest c-di-GMP and VpsT could play a role in other aspects of cell morphology.
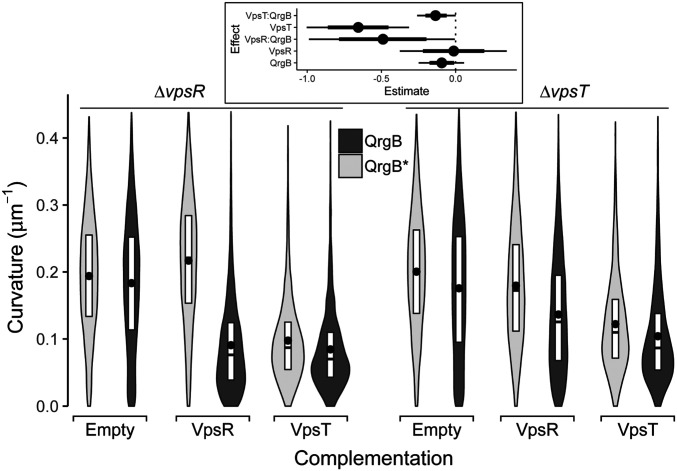
The c-di-GMP-mediated decrease in curvature observed in the parent background was lost in the ∆vpsR mutant (Fig. 3). Expression of VpsR from a multicopy plasmid in the ∆vpsR mutant restored c-di-GMP-dependent reduction of cell curvature (P value <1e-2). In the ∆vpsT background, QrgB expression caused a reduction in cell curvature (P value <4e-2), but not to the same extent as the parent background (Fig. 3). Complementation of VpsT from a multicopy plasmid resulted in decreased cell curvature regardless of c-di-GMP concentrations (P value <5e-4) although greater curvature reduction was observed at increased c-di-GMP concentrations (P value <1e-2). VpsT is a c-di-GMP-dependent transcription factor, but this result suggests that high levels of VpsT expression are sufficient to decrease curvature at the unaltered concentrations of c-di-GMP (16). To determine if VpsT required c-di-GMP to reduce curvature, we overexpressed VpsT in a V. cholerae mutant lacking 12 DGC encoding genes (Δ12 DGC) that was previously shown to have undetectable levels of c-di-GMP (30). In this background, VpsT overexpression did not decrease curvature showing this regulation required c-di-GMP (SI Appendix 1, Fig. S3).
Fig. 3.
The transcription factors VpsR and VpsT are necessary for C-di-GMP-dependent reduction of curvature. Distributions of cell curvature from different mutant strains in early stationary phase populations supplemented with 100 µM IPTG expressing inactive DGC (QrgB*, light) or active DGC (QrgB, dark) (OD600 = 1.3). The ∆vpsR strain was complemented with either an empty plasmid or a plasmid expressing VpsR or VpsT. The ∆vpsT strain was complemented with either an empty plasmid or a plasmid expressing VpsR or VpsT. The box represents the first, second, and third quartiles. The dot represents the mean. Each distribution represents between 1,000 and 1,200 cells analyzed and pooled from four to five separate experiments. (Inset) Estimated effects of QrgB, VpsT, and VpsR expression on the median population curvature. (dot = mean, thick line = 90% CI, thin line = 98% CI).
As VpsR directly activates vpsT transcription, we hypothesized the role of VpsR in controlling cell shape is to increase VpsT expression, which reduces curvature (17). We tested this hypothesis by overexpressing VpsT in the ∆vpsR mutant and found that cells appeared straight regardless of intracellular c-di-GMP concentrations (Fig. 3). Overexpression of VpsR in the vpsT mutant only partially restored c-di-GMP inhibition of curvature, indicating vpsT is necessary for complete c-di-GMP-dependent reduction in cell curvature. Collectively, this analysis supports the idea that at high c-di-GMP conditions, VpsR primarily inhibits curvature indirectly by inducing transcription of vpsT while VpsT and c-di-GMP are sufficient for reducing curvature.
C-di-GMP Decreases CrvA Expression to Inhibit Curvature.
In V. cholerae, cell curvature is generated by the intermediate filament-like protein CrvA by decreasing net growth on the minor axis relative to the major axis (8). We hypothesized that c-di-GMP-dependent inhibition of cell curvature was due to negative regulation of crvA transcription. To test this hypothesis, we generated a transcriptional reporter of the crvA promoter (PcrvA, 358 bp upstream of the crvA transcriptional start site) fused to luciferase. This promoter region was sufficient to complement a ∆crvA mutant when integrated with the crvA ORF at a heterologous site on the chromosome (SI Appendix 1, Fig. S4D).
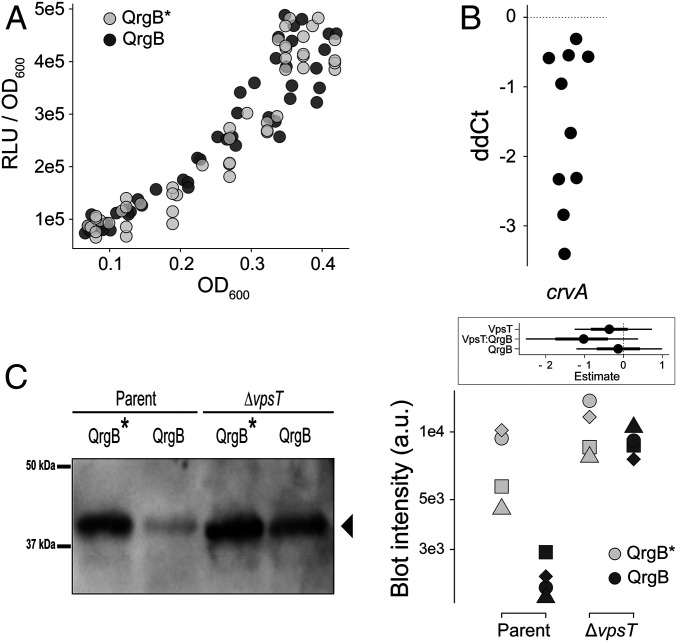
Contrary to our hypothesis, PcrvA transcriptional activity was not affected when c-di-GMP concentration was increased through QrgB expression at any of the culture densities examined (Fig. 4A) (difference in the slopes, 95% credible interval [CI] [−0.11, 0.10]). In agreement with previous studies that observed increased curvature at high cell density (6, 8), PcrvA transcriptional activity increased with cell density (Fig. 4A) (P value <2.5e-4).
Fig. 4.
Regulation of CrvA expression occurs at the posttranscriptional level. (A) Normalized luciferase reporter activity (relative light units [RLUs]) of PcrvA–luciferase transcriptional fusion as a function of cell density (OD600) under high (QrgB, dark) and unaltered (QrgB*, light) c-di-GMP conditions in populations grown to early stationary phase supplemented with 100 µm IPTG. Dots represent pooled data from four biological replicates. (B) qRT-PCR analysis of relative crvA transcript levels between high (QrgB) and unaltered (QrgB*) c-di-GMP conditions. Each dot represents one independent replicate. (C) Western blot (Left, representative plot) and quantitative intensity analysis (Right, all blots) of CrvA-HIS under high (QrgB, dark) or unaltered (QrgB*, light) c-di-GMP conditions in the parent (Left) and ∆vpsT (Right) strains grown to early stationary phase supplemented with 100 µM IPTG. Shapes represent independent replicates and y axis is log10. (Inset) Estimated effects of QrgB and VpsT expression on CrvA accumulation. (dot = mean, thick line = 90% CI, thin line = 98% CI).
Next, we tested if c-di-GMP impacted CrvA accumulation via a posttranscriptional mechanism by quantifying the effects of c-di-GMP on crvA mRNA abundance. crvA mRNA was ∼1.5-fold less abundant at high c-di-GMP conditions (Fig. 4B) (95% CI = [−2.4, −0.7]). To determine if CrvA protein levels were altered, we generated a C-terminal 6-histidine-tagged (CrvA-HIS) fusion. Cell curvature in the CrvA-HIS background was ∼82% to that of the WT (95% CI = [78 to 84%]), indicating CrvA-HIS is functional (SI Appendix 1, Fig. S5). A Western blot analysis showed that reduction of CrvA protein at high c-di-GMP concentrations was greater in the wild-type background (95% CI = [−10, −1.2] than the ∆vpsT background (95% CI = [−2.4 to 0.51]) (Fig. 4C). These data demonstrate high c-di-GMP concentrations decreases crvA mRNA and CrvA protein levels in a VpsT-dependent manner without altering crvA promoter activity, indicating that c-di-GMP negatively affects cellular curvature by decreasing CrvA abundance via a posttranscriptional mechanism.
Cell Shape Influences Biofilm Formation at the Single-Cell and Population Levels.
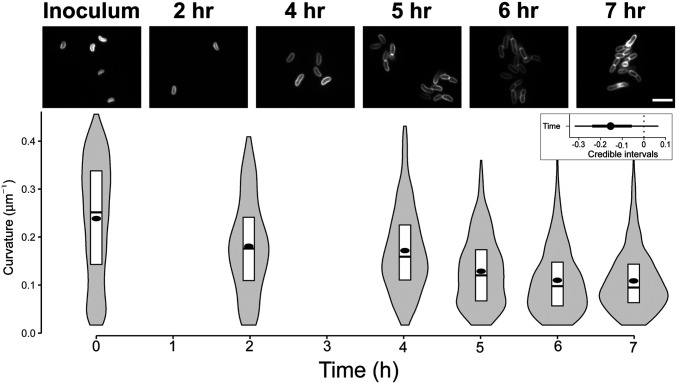
The ability of V. cholerae to attach to surfaces and initiate biofilm formation is dependent on intracellular c-di-GMP and the transcription factors VpsR and VpsT (11, 19, 25). Recent reports indicate c-di-GMP and VPS matrix production increases within hours of surface attachment (31, 32). This suggests that c-di-GMP-dependent changes to cell shape could occur as V. cholerae is initiating biofilm formation concomitantly with VpsT-inducing production of VPS and associated biofilm matrix proteins (16, 32, 33). Since form fits function, we hypothesized that straight cells of V. cholerae would be more adept at forming biofilms. To test this hypothesis, we first grew wild-type V. cholerae under static conditions and measured the morphology of attached cells over time starting from initial contact to the early stages of microcolony formation. We chose these time points because at later stages of biofilm development V. cholerae grows in dense clusters with some cells in the biofilm becoming vertical relative to the surface, making it difficult to segment and quantify curvature from two-dimensional (2D) images (34, 35). We observed that over the course of the experiment (7 h) cells attached to the glass surface became straighter than cells from the initial inoculum (Fig. 5) (P value <1.2e-2). To assess if only cells associated with the surface became straight, we harvested cells from the planktonic phase at the 8-h time point and measured curvature. Importantly, at 8 h postinoculation, both adhered and nonadhered cells were still at low-cell density due to the seeding conditions (see Materials and Methods for experimental details). Planktonic cells also exhibited a straight cell morphology similar to those attached to the surface, suggesting that the change in cell shape over the course of the experiment likely reflects QS control of cell shape (SI Appendix 1, Fig. S6). These data indicate the relationship between cell shape, quorum state, and c-di-GMP is similar in adhered and nonadhered V. cholerae cells (6, 8). Thus, in conditions that promote biofilm formation, V. cholerae cells exhibit straight morphology.
Fig. 5.
V. cholerae becomes straighter when forming microcolonies. (Top) Representative images of WT V. cholerae from the inoculum and 2, 4, 5, 6, and 7 h postattachment to glass coverslips. Cells were stained with FM4-64 prior to imaging. (Scale bar, 5 µm.) (Bottom) Distributions of cell curvature at different time points after attachment. The box represents the first, second, and third quartiles. The dot represents the mean. Each distribution represents between 500 and 1,000 cells analyzed and pooled from two separate experiments. (Inset) Estimated effect of time after surface attachment on the median population curvature. (dot = mean, thick line = 90% CI, thin line = 98% CI).
Single V. cholerae cells seeded on a surface can produce large three-dimensional biofilms through surface adhesion along the horizontal axis of the cell, replication on the surface, and a switch from horizontal to vertical cell positioning (34, 35). This process is largely dependent on VPS matrix and associated matrix proteins as well as cell–cell and cell–surface contacts (34–36). Additionally, images from single-cell studies of V. cholerae biofilm formation show relatively straight cells, including in strains that have elevated c-di-GMP concentrations, although curvature was never directly measured (32, 34, 35). Our finding that adhered V. cholerae grew as straight rods under biofilm-forming conditions suggested this morphology could be adaptive for biofilm formation. If so, we hypothesized that locking cells in a curved morphology, regardless of intracellular c-di-GMP concentrations, would negatively influence biofilm development. We tested this hypothesis by expressing CrvA from a multicopy plasmid using the PBAD promoter (pCrvA) in a ∆crvA background. We found that basal expression of this construct (i.e., no arabinose addition) was sufficient to restore curvature to the same level as wild-type planktonic cells (SI Appendix 1, Fig. S4A). Cells retain their curvature while coexpressing CrvA along with QrgB compared to QrgB*, indicating c-di-GMP does not negatively regulate ectopically expressed CrvA expression (SI Appendix 1, Fig. S4A). A slight increase in curvature was observed in cells ectopically expressing CrvA at high c-di-GMP concentrations although it is unclear if this difference is biologically meaningful (SI Appendix 1, Fig. S4A). Thus, we generated three distinct populations: 1) cells that were able to transition normally between curved and straight rods (WT harboring the control vector pHERD20C), 2) cells that were constitutively straight (∆crvA-pHERD20C), and 3) cells that were constitutively curved (∆crvA-pCrvA) regardless of c-di-GMP concentrations.
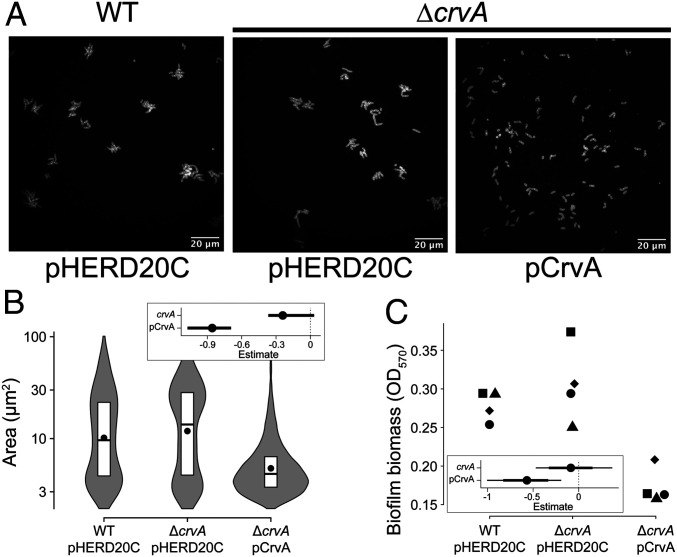
Through microcolony analysis at 8 h postinoculation, we found that the area of individual microcolonies of the WT and the straight populations were 10 µm2 and 12 µm2 on average, indicating the straight mutant is not impaired in microcolony formation (Fig. 6 A and B). Microcolonies formed by the curved mutant were smaller than the size of WT (5 µm2, P value <2.5e-4) and contained only two to three cells (area of one cell = 2.6 ± 1.4 µm2) (Fig. 6 A and B). By analyzing the total surface fluorescence from the images in Fig. 6, we found that WT and straight populations had similar levels of fluorescence. The curved population had a higher level of fluorescence; however, that difference was not statistically significant (SI Appendix 1, Fig. S7). Regardless, the data indicate that reduced microcolony area of the locked curved cells was not caused by an inability to adhere to the surface under the conditions tested.
Fig. 6.
Curvature influences microcolony development and population level biofilm formation. (A) Representative images of V. cholerae microcolonies of WT harboring the control vector pHERD20C (Left), ∆crvA harboring the control vector pHERD20C (Center), and ∆crvA harboring the CrvA expression vector pCrvA (Right). Microcolonies were stained with FM4-64 prior to imaging. (Scale bars at Bottom Right of each image, 20 µm.) (B) Distributions of microcolony areas pooled from two independent experiments totaling between 700 and 2,000 microcolonies. The box represents the first, second, and third quartiles. The dot represents the mean. Each distribution represents between 500 and 1,000 cells analyzed and pooled from two separate experiments. (C) Biofilm biomass formed by different strains. Dot shapes represent independent replicates. (Insets) Estimated effects of genomic crvA or ectopic CrvA expression on the median microcolony area and biofilm biomass (dot = mean, thick line = 90% CI, thin line = 98% CI).
Based on the altered microcolony formation, we hypothesized that locking cells into curved rods would lead to a decrease in the biomass of mature biofilms. Using crystal violet to quantify the total biomass of attached cells from static culture in glass tubes, we found that WT and straight populations had indistinguishable biofilm biomass (Fig. 6C) (difference in biomass, 95% CI = [−0.40, 0.22]). The curved mutant had reduced biofilm biomass compared to the other strains, supporting the idea that reduced microcolony formation led to reduced overall biofilm biomass (Fig. 6C) (difference in biomass, 95% CI = [−0.91, −0.28], P value = 1.8e-3). Given that surface adhesion and growth rates are indistinguishable for these strains (SI Appendix 1, Fig. S8), we conclude that the observed differences in microcolony and biofilm formation are due to cell shape. Our analysis of cell curvature during biofilm formation indicates that cell shape is a regulated process and that decoupling of curvature from changes in c-di-GMP negatively affects biofilm formation in V. cholerae.
Cell Curvature Increases the Swimming Speed of Flagellated Cells.
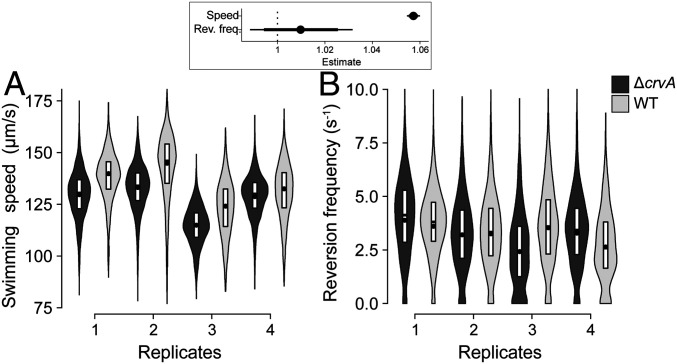
Because wild-type V. cholerae is curved in growth conditions that promote a planktonic lifestyle such as low intracellular concentrations of c-di-GMP, we hypothesized that curvature provides an advantage for cells swimming in liquid using flagellar motility. To test this hypothesis, we grew wild-type and the ∆crvA mutant in minimal medium supplemented with pyruvate to late exponential phase (most cells are highly motile in these conditions) and tracked single cells to quantify their swimming speed. We determined that curved rods swim on average 5.5% faster than straight rods (Fig. 7A) (relative speed increase, 95% CI = [5.5%, 5.9%], P value <1e-5). The difference in the reversal frequency between the trajectories from each strain was not significant (relative difference in reversion frequency, 95% CI = [−0.87%, 2.8%]) (Fig. 7B), indicating that cell curvature does not significantly affect cell behavior. In addition, while ∆crvA cells had a 1.08-fold increase in cell length compared to WT cells, the major characteristic was the 4-fold difference in curvature (SI Appendix, Fig. S9). Therefore, curvature is advantageous for motile cells by providing a significant boost in speed for bacteria that are already swimming relatively fast. Overall, our results support that V. cholerae modulates its cell shape by regulating CrvA expression through c-di-GMP signaling to increase its fitness both during biofilm formation and when swimming freely.
Fig. 7.
Curvature increases the swimming speed of flagellated cells. Distributions of (A) swimming speed and (B) reversion frequency of the WT (light) and ∆crvA mutant (dark) determined by single-cell tracking on four independent replicates grown to early stationary phase. The dot represents the mean. Each distribution represents more than 1,000 cell trajectories. (Inset) Estimated effect of crvA deletion on swimming speed and reversion frequency (dot = mean, thick line = 90% CI, thin line = 98% CI).
Discussion
Derivatives of the rod shape, such as helical or vibrioid, are adaptations produced by genes with specific shape-altering functions (2). While significant progress has been made understanding how bacteria build and manipulate their cell wall to alter cell shape, if and how these processes are influenced by environmental conditions is poorly understood. With multiple c-di-GMP synthesis and degradation enzymes that sense and respond to specific environmental cues, c-di-GMP signaling networks are one of the major mechanisms by which bacteria integrate environmental information to regulate their lifestyles, such as motility and biofilm formation. In this study, we show that c-di-GMP is a regulator of cell shape in the aquatic organism V. cholerae, where high c-di-GMP concentrations decrease cell curvature to generate straight rods. This response is dependent on the gene crvA and biofilm-promoting transcription factors, VpsR and VpsT, prompting us to assess the implications of shape in biofilm formation. We found that adhered cells in developing microcolonies adopt straight morphologies and that curvature retention caused defects in microcolony formation and mature biofilm production. Conversely, straight rods swim slower than curved cells when propelled by a single flagellum in liquid environments. This work highlights the importance of controlling cell shape when switching from a single-cell planktonic lifestyle to the development of multicellular communities such as biofilms. In addition, the regulation of crvA provides an example of cell-shape control by second messengers in bacteria.
In 1928, the microbiology pioneer Arthur Henrici discovered that V. cholerae cell shape changes with culture density, stating: “The embryonic cells [those cells early in the growth curve] are therefore large, plump, and straight […] Therefore, the mature cells are slender and curved, the typical Vibrio form” (6). Additionally, Bartlett et al. discovered cells were more curved when sampled from stationary phase populations and determined this result was caused by QS (8). Studies comparing V. cholerae transcripts from cells in LCD to cells in HCD identified crvA as a QS-regulated gene, with higher expression in the HCD state (37). V. cholerae utilizes QS as one mechanism to change intracellular c-di-GMP where cells in LCD cultures have higher c-di-GMP concentrations than cells in HCD cultures (Fig. 1) (11, 38). Additionally, cells in the HCD can form biofilms if c-di-GMP production is stimulated through another mechanism (e.g., ectopically express a DGC), indicating c-di-GMP is epistatic to QS (11). In our study, we also observe a similar phenomenon where HCD cells in early stationary phase have decreased curvature with the expression of QrgB (Fig. 2). Thus, the finding that cells at LCD, which have high c-di-GMP concentrations, appear straight support our conclusion that c-di-GMP controls cell shape in V. cholerae (6, 8, 11).
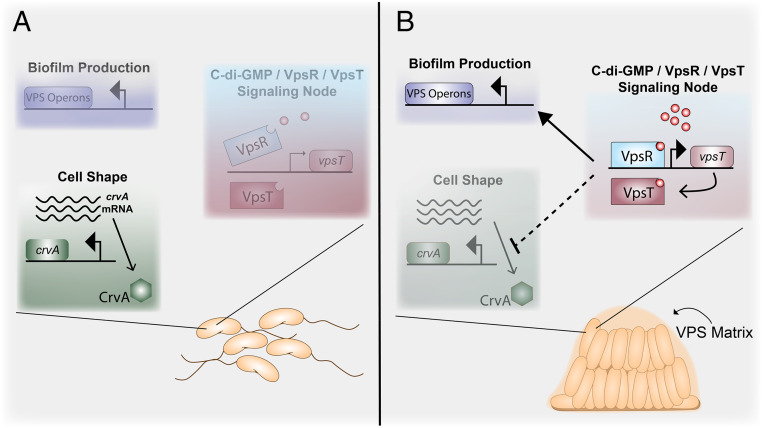
The c-di-GMP-dependent transcription factor VpsT was sufficient to inhibit curvature in planktonic cells. Additionally, our data implicate a potential direct role for VpsR in regulating curvature, since VpsR and c-di-GMP partially decreased curvature in a ∆vpsT background (Fig. 3). These data suggest that the VpsR/VpsT regulatory node is responsible for the straightening of cells during microcolony formation. While we could not directly test this, because vpsR and vpsT are required microcolony formation, several studies strongly support this model (16, 25, 26, 33). Specifically, adhered cells produce VPS within 15 min and structural matrix proteins within an hour of surface attachment, both of which are positively regulated by the VpsR/VpsT regulatory node and high c-di-GMP concentrations (32). Additionally, c-di-GMP production increases over time in surface-attached V. cholerae, peaking around 6 h after attachment (31). Similar increases in c-di-GMP have been observed for other bacteria such as Pseudomonas aeruginosa after surface attachment (39, 40). These studies support a model whereby upon attaching to a surface, c-di-GMP concentrations increase and activate the VpsR/VspT regulatory node, resulting in surface adhesion. As cells divide on the surface, the negative regulation of crvA causes the resulting changes in progeny cell curvature during biofilm development (Fig. 8).
Fig. 8.
Model of c-di-GMP control of cell shape and biofilm formation in V. cholerae. (A) Under low c-di-GMP conditions (e.g., HCD, presence of bicarbonate) (11, 41), VpsR is unable to induce VpsT and the VPS operons. Under these conditions, V. cholerae is flagellated and exists as a curved rod due to CrvA production (8). (B) Under high c-di-GMP conditions (e.g., low temperature, LCD) (11, 42), c-di-GMP-bound VpsR is active and induces expression of VpsT, which in turn binds to c-di-GMP. Together, VpsR and VpsT form a node in the c-di-GMP signaling network that induces expression of the VPS operons resulting in mature biofilm formation (16, 28). Additionally, the c-di-GMP-activated VpsR/VpsT node negatively inhibits crvA expression at the posttranscriptional level via an unknown mechanism (dashed arrow indicating repression), resulting in decreased CrvA protein levels and production of straight rods as cells divide.
Pandemic isolates of V. cholerae often retain their vibrioid morphology when examined microscopically (43, 44). However, Wucher et al. found that nutrient deprivation in pandemic isolates induced cell filamentation, which aided in chitin colonization and VPS-independent biofilm formation under flow conditions (44). While the cells were longer than the reference strain, they appeared to retain their curvature, suggesting this mechanism is independent of crvA and c-di-GMP (8, 44). Single V. cholerae cells in VPS-dependent microcolonies, however, appear straight rather than curved in submerged flow cell conditions in multiple reports; yet the degree of change in cell shape was never quantified (32, 34, 35). Thus, this research quantifies curvature in 2D developing microcolonies (Fig. 6).
While V. cholerae curvature has been shown to promote motility in plate-based motility assays, how shape influences specific parameters of motility such as chemotaxis or swimming speed at the single-cell level was not determined (8). Our findings indicate the influence of cell shape on motility extends beyond promoting movement in restricted spaces such as the matrices formed in low-agar motility assays. By tracking single cells swimming in liquid media, we found that constitutively straight cells swim slower than curved cells (Fig. 7). Our study provides experimental evidence supporting theoretical results that predicted intermediate curvature improves cellular swimming efficiency of slender rods in solution by increasing rotational resistance more than translational resistance, resulting in more power transferred to the flagellum and faster swimming speed (45, 46).
Examples of vibriod or helical morphologies include C. crescentus, Helicobacter pylori, and V. cholerae as well as many bacteria found in aquatic reservoirs (47). Curvature is likely to influence how each organism adapts to their respective niche. For example, curvature in C. crescentus is important for positioning the cell pole near the surface during biofilm growth in flow conditions. Straight mutants are unable to form biofilms under these conditions, but there was no defect under static conditions, which is opposite of the conclusions for V. cholerae drawn here (48). The helical shape of the human pathogen H. pylori is thought to be an adaptive trait that promotes fitness in a human host; however, straight H. pylori clinical isolates have been identified that provide distinct advantages and disadvantages in certain physiological niches during infection (49). These results emphasize that cell shape can differentially impact bacterial species based on their own specific physiologies.
Rod-shaped cells are observed in all three domains of life, suggesting this shape provides evolutionary advantages compared to others (5). For example, mutations that alter the morphology of the rod-shaped bacteria Escherichia coli or Rhodobacter sphaeroides also negatively impact biofilm formation, which could impact fitness in certain environments (50). Further, analysis of cell packaging in dense clusters highlighted the importance of the rod shape in cell positioning and self-organization in developing microbial communities (51, 52). While helical or curved bacteria like H. pylori and C. crescentus can access the adaptive benefits of curved or straight cells through mutagenesis and loss of function, V. cholerae has evolved to accomplish this by utilizing environmental sensing and c-di-GMP signaling (48, 49). Our work suggests cell shape in bacteria is a malleable feature that is regulated by environmental signaling to maximize swimming speed in the planktonic state and cell-surface interactions during biofilm development.
Materials and Methods
DNA Manipulations and Growth Conditions.
Strains derived from V. cholerae C6706 Str2 were used in all experiments except for SI Appendix, Fig. S3 (SI Appendix S1, which used V. cholerae E7946 and the ∆12DGC mutant). (See SI Appendix 2, Table S1 for the complete list of strains used in this study.) In Figs. 2–4, the biofilm mutant derivative (∆vpsL) was used as the parental strain to prevent aggregate formation at high c-di-GMP so that individual bacteria could be imaged in solution. The ∆vpsT and ∆vpsR mutants as well as expression vectors for VpsT and VpsR were previously generated (17). pKAS32 was used to construct deletion and knockin strains (53). Unless otherwise stated, all cloning was done by Gibson Assembly (NEB). For the deletion construct pKAS32_∆crvA, pKAS32 was digested with XbaI and SacI and purified by gel extraction (Promega). Primers (Integrated DNA Technologies) were designed with NEBuilder (www.nebuilder.com, NEB) to incorporate the appropriate 5′ ends necessary for Gibson Assembly and 3′ gene specific ends. Specifically, 700 bp upstream and downstream of crvA (VCA1075) were amplified by PCR (Q5 Polymerase, NEB) using V. cholerae gDNA as a template. For the knockin construct pKAS32_crvA-HIS, the replacement allele harboring a 6-histidine tag before the stop codon was amplified by PCR with the HIS tag incorporated into the 5′ of the oligonucleotide primer. For the knockin construct pKAS32_pcrvA-CrvA, the native promoter and open reading frame (ORF) of crvA (−358 bp relative to the ATG start codon until the translational stop codon) were amplified using PCR and inserted into the locus VC1807, a pseudogene encoding an authentic frameshift that provides a neutral site for insertion into the genome (54). For pBBRLux_pcrvA, the upstream sequence of crvA from −358 to −1 bp relative to the ATG start codon was amplified and Gibson cloned into the BamHI and SpeI restriction site of pBBRlux. For pHERD20C_CrvA, the ORF of crvA was amplified and cloned into the KpnI and SacI sites of pHERD20C. Successful clones were screened by colony PCR using GoTaq polymerase (Promega) and sequenced by Sanger sequencing (Genewiz Inc.) to ensure no mutations were incorporated during the cloning process. Construction of knockout and knockin strains was carried out by the protocol from Skorupski et al. (53). Plasmids were moved from S17-λpir E. coli into V. cholerae by conjugation using Polymixin B for counterselection (10 U/mL). All plasmids and oligonucleotides can be found in SI Appendix, Tables S2 and S3 (SI Appendix S2) (55, 56). Unless otherwise stated, both E. coli and V. cholerae were propagated in LB with ampicillin (100 µg/mL), kanamycin (100 µg/mL), and/or chloramphenicol (10 µg/mL) when needed. The inducer IPTG was routinely used at 100 µM unless stated otherwise.
Phase Contrast Microscopy and Single-Cell Analysis.
Overnight cultures of V. cholerae were diluted 1:100 into 2 mL LB supplemented with ampicillin and IPTG at appropriate concentrations. Cultures were grown until an OD600 of 1.3 to 1.5, at which point cells were diluted to an OD600 of 0.5 in microcentrifuge tubes. The 1% agarose pads in deionized water were cut into squares of ∼20 × 20 mm and placed on microscope slides (75 × 25 × 1.0 mm [length {L} × height {H} × width {W}], Alkali Scientific Inc.). A total of 2 µL of diluted cultures was spotted onto glass coverslips (22 × 22 mm, #1.0 thickness, Alkali Scientific) and the coverslip was gently placed onto the agarose pad. Phase-contrast microscopy was carried out with a Nikon Eclipse Ti-E inverted microscope equipped with a 100× phase contrast oil immersion objective (1.4 NA), a Nikon Perfect Focus System, a Prior H117 ProScan motorized stage, a Lumencor PEKA white LED diascopic light source, and an Andor Zyla 4.2 sCMOS camera. The microscope and camera were controlled by a computer workstation with MATLAB (Mathworks Inc.) and Micromanager version 1.4 (micromanager.org). Uneven illumination and image artifacts were corrected using a flat field image of a clean slide. Cells were detected and segmented using the medial axis method in the Fiji plugin MicrobeJ with the following threshold settings: area (0 to 4.5, µm2), length (1.5 max, µm) and width (0 to 3, µm) (57). Data from segmented images were analyzed using R to plot curvature (defined as the average of the reciprocal of the radius of curvature taken along the medial axis of a given cell) (µm−1), width (defined as the average of width measurements taken along the medial axis of a given cell) (µm), and length (defined as the pole-to-pole distance) (µm) (57). Representative images were cropped and the scale bar was added using Fiji software (58). The median cell length, width, and curvature were calculated for each biological replicate. The posterior probability distributions and the P values for the different factors tested (QrgB, VpsR, VpsT, and CrvA inductions) were calculated for the medians using different linear mixed-effect models with a lognormal distribution link function. Fig. 1 and ED1: (curvature, length, width) ∼ QrgB* IPTG + (QrgB* IPTG|replicate), Fig. 2: (curvature, length, width) ∼ complement + QrgB: complement + (complement + QrgB: complement|replicate), Fig. 4: (curvature, length, width) ∼ time + (time|replicate).
Analysis of Curvature of Adhered Cells in Microcolonies and Nonadhered in Solution.
Overnight cultures were diluted 1:1,000 in 1 mL 1× PBS (Sigma) by 10-fold serial dilutions. UV-sterilized #1 coverslips (22 × 22 cm) were placed in six-well plates (Costar) submerged in 1 mL LB. Cultures, in six sets of biological replicates, were seeded in the wells with slides by further diluting 5-fold, resulting in a final dilution of 1:5,000, followed by gentle swirling. Microcolonies were allowed to develop during static incubation at 21 °C. At the given time point, the media for two biological replicates were removed by aspiration, the wells washed with 1 mL 1× PBS, and the resulting adhered bacteria on the coverslip were stained with 200 µL of the membrane stain N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64) (Sigma) at a final concentration of 20 µg/mL for 5 min. The stain was removed by an additional wash with 1 mL 1× PBS. Small sections of agarose pads (∼5 × 5 mm) were arranged in a 20 × 20 cm square pattern on glass microscope slides. The glass coverslip containing the stained microcolonies was then inverted and placed on top of the agarose pads. Biofilm imaging was carried out using a Leica DM5000b epifluorescence microscope with a 100×-brightfield objective (1.4 NA) equipped with a Spot Pursuit CCD camera and an X-cite 120 Illumination system. Images were acquired using the dsRed filter set (excitation 560/40 nm, emission 620/60 nm). Each slide was imaged with at least 20 fields of view for each biological replicate at each time point. Cells within microcolonies were manually outlined in MicrobeJ using the manual interface option with at least 500 to 1,000 cells outlined per replicate. Data from MicrobeJ analysis were exported into R for analysis. The posterior probability distribution of the effect of time on the morphology of attached cells was calculated using a linear mixed-effect model with a lognormal distribution link function [(curvature, length, width) ∼ time + (time|replicate)].
For imaging of nonadhered cells, overnight cultures were diluted 1:100 in 1 mL of 1× PBS (Sigma) by 10-fold serial dilutions. The 1:100 diluted overnight cultures were additionally diluted by adding 20 μL in three individual wells in a six-well plate (Costar) containing 4 mL LB, followed by gentle swirling and static incubation at 21 °C for 7 to 8 h. For each biological replicate, 3 out of 4 mL of the cultures were collected and subjected to membrane filtration using a stainless-steel vacuum filter (Millipore 13 mm) without touching the bottom of the wells. The cells were collected on a presoaked 0.45 µm polyvinylidene difluoride membrane filter (Millipore), moved to a 1.5-mL microcentrifuge tube, and resuspended in 20 µL of the membrane stain FM4-64 (20 µg/mL) for 5 min. A total of 5 µL of stained cells was spotted onto glass coverslips and the coverslip was gently placed onto the agarose pad. Cell imagining was carried out by the same method as microcolonies imaging.
Microcolony Area Analysis.
For single time point biofilm analysis, eight-well microchamber slides (µ-Slide, eight-well glass bottom, ibidi) were used. The 1:1,000 diluted overnight cultures were additionally diluted 1:5 in an individual well of the microchamber slide in 200 µL LB supplemented with chloramphenicol at the appropriate concentration. Each slide had three strains in biological duplicate. Microcolonies were developed by incubating the microchamber slide statically at 21 °C for 8 h, resulting in WT microcolony sizes between 10 and 20 µm2. Media were removed from slides by aspiration, each well was washed with 200 µL 1× PBS, and the microcolonies were stained with FM4-64 (150 µL) at a final concentration of 20 µg/mL for 5 min. The remaining stain was washed with 200 µL 1× PBS and biofilms were imaged by inverting the microchamber (positioning the glass bottom upwards) by fluorescence microscopy using a Leica DM5000b epifluorescence microscope as previously described. At least 20 fields of vision were captured per replicate per strain and the resulting images were processed using Fiji (enhance contrast, saturated pixel %, 0.3) (58). Processed images were then analyzed using MicrobeJ with the following settings: Background type (dark), mode of detection (basic), area (4.5 to 100, µm2), and circularity (0 to 1). Data were exported into R for analysis. For all fluorescence microscopy, representative images were cropped, and the scale bar was added using Fiji software (58). The posterior probability distributions of the effects of mutation and complementation of the microcolony areas were calculated using a linear mixed-effect model with a lognormal distribution link function [area ∼ strain + complement + (strain + complement|replicate)]. For measurement of total surface fluorescence, images above were processed using Fiji (subtract background, rolling ball radius: 50 pixels) (58) and analyzed for total integrated density using the measurement function in Fiji. Data were exported into R for analysis.
Luciferase Reporter Assay.
Overnight cultures of four biological replicates of V. cholerae harboring PcrvA transcriptional fusions to luciferase in pBBRlux were diluted 1:100 in 1 mL LB supplemented with ampicillin, chloramphenicol, and IPTG in 1.5-mL microcentrifuge tubes. A total of 150 µL of cell solution was aliquoted into wells of a black 96-well plates (Costar) in technical duplicates. Plates were incubated at 35 °C with shaking at 220 RPM. Every hour, luciferase (relative light units [RLUs]) and OD595 measurements were taken using an Envision plate reader (Perkin-Elmer). The posterior probability distributions of the relationship between the light emission and optical density was calculated using a linear mixed-effect model with a normal distribution link function [RLU ∼ OD600:QrgB + (OD600:QrgB|replicate)].
RNA Isolation and qRT-PCR.
RNA isolation and qRT-PCR analysis were carried out following protocols described previously (24). In brief, overnight cultures were diluted to a starting OD600 of 0.040 in 2 mL LB supplemented with ampicillin and IPTG and grown until an OD600 of ∼1.3 at 35 °C and 220 RPM. A total of 1 mL of each replicate was pelleted, and RNA was extracted using the TRIzol reagent following the manufacturer’s directions (Thermo Fisher Scientific). Purified RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific). A total of 5 µg of purified RNA was treated with DNase (Turbo DNase, Thermo Fisher Scientific). cDNA synthesis was carried out using the GoScript Reverse Transcription kit (Promega). cDNA was diluted 1:30 into molecular biology grade water and amplification was quantified using SYBR Green (Applied Biosystems). Reactions consisted of 5 µL of 2.5 µM primer 1, 5 µL of 2.5 µM primer 2, 5 µL of diluted cDNA template, and 15 µL of 2× SYBR Green consisting of dNTPs and AmpliTaq Gold DNA polymerase. Each plate had technical duplicates and biological triplicate samples as well as no reverse transcriptase reactions to detect genomic DNA contamination. The StepOnePlus Real Time PCR system was used for qRT-PCR with the following thermocycling conditions: 95 °C for 20 s then 40 cycles of 95 °C for 2 s and 60 °C for 30 s. Melting curves were included to ensure PCR products had single amplicons. Data were analyzed by the ∆∆Ct method using gyrA as a reference target. The experiment was repeated with 10 biological replicates on separate days and the data from each experiment were pooled.
Western Blot Analysis.
Overnight cultures of the parent or ∆vpsT backgrounds with 6-histidine tagged crvA integrated at the crvA locus (CrvA-HIS) harboring QrgB* or QrgB were diluted 1:100 in 10 mL LB supplemented with ampicillin and IPTG in 50 mL baffled flasks. Cultures were grown until the OD600 reached ∼1.3, collected by centrifugation (2.5 min at 7,000 × g), resuspended in 200 µL lysis buffer (20 mM Tris⋅HCl, 1% SDS, pH 6.8), and moved to 1.5-mL microcentrifuge tubes, and boiled for 10 min at 95 °C. Boiled lysates were centrifuged (1 min at 12,000 × g) to pellet insoluble material and the supernatants were moved to new 1.5-mL microcentrifuge tubes. Samples were diluted 1:20 in 1× PBS and protein concentration was quantified by the detergent compatible protein assay with bovine serum albumin as the standard (BioRad). Lysates were normalized to 5 µg/µL in 2× loading buffer (lysis buffer supplemented with 5 mM beta-mercaptoethanol and Coomassie blue). A total of 20 µL of normalized lysates were loaded into precast 4 to 20% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) gels (4 to 20% Mini-PROTEAN TGX Precast Protein Gels, Bio-Rad) alongside size standards (Precision Protein Plus, Bio-Rad). Gels were run at room temperature for 90 min at 90 V in 1× Tris/glycine/SDS running buffer (Bio-Rad). Blots were transferred to nitrocellulose membranes using the Mini Trans-Blot system (Bio-Rad) with TBST (Tris buffered saline with Tween 20, pH 8.0)-methanol (20% vol/vol). Transfers were carried out at 4 °C for 2 h at 250 mAmps. After the transfer, blots were removed and placed in blocking buffer (TBST supplemented with 5% skim milk) and incubated with agitation for 1.5 h at room temperature. Blocking buffer was removed and replaced with 20-mL blocking buffer supplemented with 4 µL anti-HIS primary antibody (mAb, mouse, GenScript) and the blot was incubated overnight at 4 °C with agitation. The next day, the blot was washed three times by removing spent blocking buffer, addition of 20 mL blocking buffer, and agitation for 2 to 3 min at room temperature. After the last wash, 20 mL blocking buffer supplemented with 5 µL of horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody (Bio-Rad) was added and incubated for 2 h at room temperature with agitation. After incubation with the secondary antibody, the blot was washed with five cycles of 10 mL blocking buffer with 1-min agitations. Reagents 1 and 2 from the Pierce ECL kit were mixed according to manufacturer’s instructions and chemiluminescence was detected using Amersham Imager 600 with the “chemilumescence” settings. Images were removed from the imager, uploaded to Fiji, cropped to remove the protein standards, and processed by enhancing contrast (enhance contrast, 0.3% saturated pixels). Band intensities were measured using the gel analyzer function. Two separate experiments with two biological replicates were repeated on separate days (n = 4). The posterior probability distributions of the effects of VpsT and QrgB on the accumulation of CrvA-His were calculated using a linear mixed-effect model with a lognormal distribution link function [intensity ∼ QrgB*VpsT + (QrgB*VpsT|replicate)].
Crystal Violet Biofilm Assay.
Overnight cultures of V. cholerae were diluted 1:100 into 1 mL LB supplemented with chloramphenicol into new, sterile 18 × 150 mm borosilicate test tubes and statically incubated at 21 °C for 8 h. After 8 h, media and unattached bacteria were removed, and the biofilms were washed twice with 1 mL 1× PBS (PBS washes were removed by aspiration). A 1-mL crystal violet (CV) (0.4%) solution was added to each tube and the biofilm was stained for 10 min. The stain was removed by aspiration and the stained biofilm was washed twice with 1 mL 1× PBS. CV was eluted with ethanol and the absorbance at 570 nm (OD570) was measured. The posterior probability distributions of the effects of mutation and complementation of the biofilm biomass were calculated using a linear mixed-effect model with a lognormal distribution link function [biomass ∼ strain + complement + (strain + complement |replicate)].
Tracking and Analysis of Swimming Cells.
Wild-type and crvA mutant cells were grown in M9 minimal salts (52 mM Na2HPO4, 18 mM K2HPO4, 18.69 mM NH4Cl, 2 mM MgSO4, pH 7) supplemented with 10 µM FeSO4, 20 µM C6H9Na3O9 and 36.4 mM sodium pyruvate shaking (200 RPM) in liquid cultures at 37 °C. Cultures were sampled at early stationary phase (1.9 × 109 colony forming units [c.f.u.]/mL) and diluted in fresh medium to 107 c.f.u./mL and incubated for 15 min before tracking. Swimming cells were tracked in liquid following the protocol previously described (59). Briefly, polyvinylpyrrolidone (PVP) was added at 0.05% wt/vol to the samples to prevent attachment on the glass slide. A total of 6 µL of each sample was dropped on a glass slide and trapped under a 22 × 22 mm, #1.5 coverslip sealed with wax and paraffin to create a thin water film (10 ± 2 µm) for video microscopy. The samples were kept at 37 °C during tracking. Images of swimming cells were recorded using a sCMOS camera (Andor Zyla 4.2, Oxford Instruments) at 20 frames per second using a 40× objective (Plan Fluor 40×, Nikon Instruments, Inc.) mounted on an inverted microscope (Eclipse Ti-E, Nikon Instruments, Inc.). Cell were illuminated using phase contrast. Images were analyzed to detect and localize cells using custom scripts (59) and cell trajectories were reconstructed using the µ-track package (60). The analysis of the cell trajectories was done in MATLAB (The Mathworks, Inc.) as previously described (59). The mean swimming speed of each cell was calculated by averaging instantaneous speed along the trajectory when the cell is not doing a reversal. The posterior probability distributions and the P values for the differences in mean swimming speed and reversal frequency for each strain were calculated using a linear mixed-effect model with a lognormal distribution link function [(mean speed, reversal frequency) ∼ strain + (1|replicate)].
Growth Rate Measurements.
Overnight cultures were diluted 1:100 into LB supplemented with appropriate antibiotics, grown until the cultures reach exponential phase, and further diluted to OD590 of 0.002. A total of 200 μL of the diluted cultures were transferred to a 96-well microplate (Costar). The cultures were grown at 37 °C for 24 h in a Sunrise (Tecan) plate reader, with OD590 measurements every 10 min. To calculate the growth rate, the OD measurements were trimmed to the exponential phase of growth by computing the first and second derivatives of OD590 with respect to time, then keeping only the measurements where the first and second derivatives were positive. Growth rates were extracted by fitting an exponential function to the trimmed data using a nonlinear least-squares algorithm.
Statistical Analyses.
All of the statistical analyses were done using Bayesian sampling of the respective mixed-effect generalized linear models (see each section) using the RSTAN (61) and BRMS packages (62) in R (63) with four chains, each with 1,000 warmup iterations and at least 2,500 sampling iterations. Uninformative priors were set to the defaults generated by BRMS. The plots were generated using the ggplot2 and tidybayes packages (64, 65). The posterior probability distribution for the magnitude of the effect for each experimental treatment was used to calculate the 90% and 98% credible intervals reported in the figure Insets. Posterior probabilities are calculated given the model and the data using the Bayesian rule to update the prior distribution of each parameter. The credible interval is the Bayesian analog of the confidence interval. It is defined as the interval within which a parameter value falls with a particular probability. There is 90% probability that the population parameter is within the 90% credible interval given the model and the sample data. The 90% confidence interval is defined as an interval that if constructed multiple times with different samples from the same population will contain the population parameter 90% of the time. Credible intervals provide a more intuitive and direct representation of the likely values for the population parameters. Detailed explanations can be found in refs. 66, 67.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants GM109259, GM110444, and AI143098 to C.M.W.; NSF Grant 1714612 to Y.S.D. and C.M.W.; and funds from Michigan State University to Y.S.D.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010199117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Young K. D., Bacterial morphology: Why have different shapes? Curr. Opin. Microbiol. 10, 596–600 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang D. C., Blair K. M., Salama N. R., Staying in shape: The impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80, 187–203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Teeseling M. C. F., de Pedro M. A., Cava F., Determinants of bacterial morphology: From fundamentals to possibilities for antimicrobial targeting. Front. Microbiol. 8, 1264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allan E. J., Hoischen C., Gumpert J., “Bacterial L-Forms” in Advances in Applied Microbiology, A. Laskin, G. Gadd, S. Sariaslani, Eds., (Elsevier, 2009), pp. 1–39. [DOI] [PubMed] [Google Scholar]
- 5.Chang F., Huang K. C., How and why cells grow as rods. BMC Biol. 12, 54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrici A., “Morphologic variations of the Cholera vibrio” in Morphologic Variation and the Rate of Growth of Bacteria, Thomas C. C., Ed. (C. C. Thomas, 1928), pp. 117–124. [Google Scholar]
- 7.Ausmees N., Kuhn J. R., Jacobs-Wagner C., The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 115, 705–713 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Bartlett T. M., et al. , A periplasmic polymer curves Vibrio cholerae and promotes pathogenesis. Cell 168, 172–185.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin N. R., Blackman E., Bratton B. P., Bartlett T. M., Gitai Z., The evolution of bacterial shape complexity by a curvature-inducing module. bioRxiv:2020.02.20.954503 (20 February 2020). [Google Scholar]
- 10.Waters C. M., Bassler B. L., The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev. 20, 2754–2767 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L., Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pursley B. R., et al. , Cyclic di-GMP regulates TfoY in Vibrio cholerae To control motility by both transcriptional and posttranscriptional mechanisms. J. Bacteriol. 200, 200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srivastava D., Waters C. M., A tangled web: Regulatory connections between quorum sensing and cyclic Di-GMP. J. Bacteriol. 194, 4485–4493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim B., Beyhan S., Meir J., Yildiz F. H., Cyclic-diGMP signal transduction systems in Vibrio cholerae: Modulation of rugosity and biofilm formation. Mol. Microbiol. 60, 331–348 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Conner J. G., Zamorano-Sánchez D., Park J. H., Sondermann H., Yildiz F. H., The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol. 36, 20–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasteva P. V., et al. , Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327, 866–868 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava D., Harris R. C., Waters C. M., Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 193, 6331–6341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yildiz F. H., Schoolnik G. K., Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 96, 4028–4033 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid M. H., et al. , Role of exopolysaccharide, the rugose phenotype and VpsR in the pathogenesis of epidemic Vibrio cholerae. FEMS Microbiol. Lett. 230, 105–113 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Fong J. C. N., Yildiz F. H., The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189, 2319–2330 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong J. C. N., Syed K. A., Klose K. E., Yildiz F. H., Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156, 2757–2769 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava D., Hsieh M.-L., Khataokar A., Neiditch M. B., Waters C. M., Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 90, 1262–1276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandez N. L., Srivastava D., Ngouajio A. L., Waters C. M., Cyclic di-GMP positively regulates DNA repair in Vibrio cholerae. J. Bacteriol. 200, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez N. L., Waters C. M., Cyclic di-GMP increases catalase production and hydrogen peroxide tolerance in Vibrio cholerae. Appl. Environ. Microbiol. 85, e01043–e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casper-Lindley C., Yildiz F. H., VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186, 1574–1578 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyhan S., Bilecen K., Salama S. R., Casper-Lindley C., Yildiz F. H., Regulation of rugosity and biofilm formation in Vibrio cholerae: Comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR. J. Bacteriol. 189, 388–402 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kariisa A. T., Grube A., Tamayo R., Two nucleotide second messengers regulate the production of the Vibrio cholerae colonization factor GbpA. BMC Microbiol. 15, 166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsieh M.-L., Hinton D. M., Waters C. M., VpsR and cyclic di-GMP together drive transcription initiation to activate biofilm formation in Vibrio cholerae. Nucleic Acids Res. 46, 8876–8887 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloup R. E., et al. , Cyclic di-GMP and VpsR induce the expression of Type II secretion in Vibrio cholerae. J. Bacteriol. 199, 10–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalia T. N., et al. , Enhancing multiplex genome editing by natural transformation (MuGENT) via inactivation of ssDNA exonucleases. Nucleic Acids Res. 45, 7527–7537 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamorano-Sánchez D., et al. , Functional specialization in Vibrio cholerae diguanylate cyclases: Distinct modes of motility suppression and c-di-GMP production. MBio 10, e00670-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berk V., et al. , Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337, 236–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zamorano-Sánchez D., Fong J. C. N., Kilic S., Erill I., Yildiz F. H., Identification and characterization of VpsR and VpsT binding sites in Vibrio cholerae. J. Bacteriol. 197, 1221–1235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drescher K., et al. , Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 113, E2066–E2072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan J., Sharo A. G., Stone H. A., Wingreen N. S., Bassler B. L., Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc. Natl. Acad. Sci. U.S.A. 113, E5337–E5343 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beroz F., et al. , Verticalization of bacterial biofilms. Nat. Phys. 14, 954–960 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papenfort K., Förstner K. U., Cong J.-P., Sharma C. M., Bassler B. L., Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 112, E766–E775 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee S., Bassler B. L., Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laventie B. J., et al. , A surface-induced asymmetric program promotes tissue colonization by Pseudomonas aeruginosa. Cell Host Microbe 25, 140–152.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Lee C. K., et al. , Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 115, 4471–4476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koestler B. J., Waters C. M., Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect. Immun. 82, 3002–3014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Townsley L., Yildiz F. H., Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ. Microbiol. 17, 4290–4305 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albert M. J., Vibrio cholerae O139 Bengal. J. Clin. Microbiol. 32, 2345–2349 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wucher B. R., et al. , Vibrio cholerae filamentation promotes chitin surface attachment at the expense of competition in biofilms. Proc. Natl. Acad. Sci. U.S.A. 116, 14216–14221 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuech R., Hoehfurtner T., Smith D. J., Humphries S., Motile curved bacteria are Pareto-optimal. Proc. Natl. Acad. Sci. U.S.A. 116, 14440–14447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lauga E., Powers T. R., The hydrodynamics of swimming microorganisms. Rep. Prog. Phys. 72, 096601 (2009). [Google Scholar]
- 47.Young K. D., The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70, 660–703 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Persat A., Stone H. A., Gitai Z., The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat. Commun. 5, 3824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez L. E., O’Brien V. P., Leverich C. K., Knoblaugh S. E., Salama N. R., Nonhelical Helicobacter pylori mutants show altered gland colonization and elicit less gastric pathology than helical bacteria during chronic infection. Infect. Immun. 87, e00904-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin T.-Y., Santos T. M. A., Kontur W. S., Donohue T. J., Weibel D. B., A cardiolipin-deficient mutant of Rhodobacter sphaeroides has an altered cell shape and is impaired in biofilm formation. J. Bacteriol. 197, 3446–3455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho H., et al. , Self-organization in high-density bacterial colonies: Efficient crowd control. PLoS Biol. 5, e302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith W. P. J., et al. , Cell morphology drives spatial patterning in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 114, E280–E286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skorupski K., Taylor R. K., Positive selection vectors for allelic exchange. Gene 169, 47–52 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Dalia A. B., McDonough E., Camilli A., Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U.S.A. 111, 8937–8942 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn A. K., Millikan D. S., Adin D. M., Bose J. L., Stabb E. V., New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72, 802–810 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu D., Damron F. H., Mima T., Schweizer H. P., Yu H. D., PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74, 7422–7426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ducret A., Quardokus E. M., Brun Y. V., MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dufour Y. S., Gillet S., Frankel N. W., Weibel D. B., Emonet T., Direct correlation between motile behavior and protein abundance in single cells. PLOS Comput. Biol. 12, e1005041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaqaman K., et al. , Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods 5, 695–702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stan Development Team , RStan: the R interface to Stan. R package version 2.21.2. http://mc-stan.org/ (2019). Accessed 28 October 2020.
- 62.Bürkner P.-C., Advanced Bayesian multilevel modeling with the R package brms. R J. 10, 395 (2018). [Google Scholar]
- 63.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria).
- 64.Gómez-Rubio V., ggplot2—Elegant Graphics for Data Analysis (Journal of Statistical Software, ed. 2, 2017), vol. 77. [Google Scholar]
- 65.Kay M., tidybayes: Tidy Data and Geoms for Bayesian Models. R package version 2.1.1. http://mjskay.github.io/tidybayes/ (2020).
- 66.McElreath R., Statistical Rethinking: A Bayesian Course with Examples in R and Stan (CRC Press, 2020). [Google Scholar]
- 67.Kruschke J., Doing Bayesian Data Analysis: A Tutorial with R, JAGS, and Stan (Academic Press, 2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.