Abstract
Aim
To assess kallikrein (KLK) expression in recurrent and non-recurrent prostate tumors and adjacent healthy prostate tissues.
Methods
The expression levels of 15 KLK genes in 34 recurrent and 36 non-recurrent prostate cancer samples and 19 adjacent healthy prostate tissue samples was assessed with quantitative reverse-transcription polymerase chain reaction. The samples were obtained from Baylor College of Medicine, Houston, TX, USA between 2013 and 2016 .
Results
Compared with controls, prostate cancer samples showed a strong decrease in KLK1, KLK4, KLK9, and KLK14. Recurrent samples were negative for KLK1, KLK2, and KLK14 but demonstrated higher levels of KLK3, KLK4, and KLK9 than controls. Other KLKs were not significantly expressed.
Conclusion
This study for the first time showed a difference in the expression levels of the KLK gene family in recurrent prostate cancer. KLKs could be used as recurrence markers for prostate cancer.
Prostate cancer is the most common visceral malignancy and the second deadliest cancer in men. In most cases, the cancer grows gradually, is initially limited to the prostate gland, and does not cause significant morbidity. However, some prostate cancers are aggressive and rapidly spreading (1).
Prostate specific antigen (PSA) or kallikrein (KLK) 3 is an enzyme released into the seminal fluid. It is secreted from the cells surrounding and forming the inner part of the prostate acini and luminal epithelial cells (2). Elevated serum PSA levels are frequently used as prostate cancer markers but are also observed in ejaculation, transurethral catheterization, transrectal ultrasonography, trauma, prostate infections, and benign prostatic hyperplasia. PSA levels can also be used as prognostic markers of cancer recurrence. Such a “biochemical relapse” usually precedes other clinical signs and symptoms of recurrence (3). Biochemical recurrence after radical prostatectomy has been determined using blood PSA level in addition to clinical T stage and Gleason score (4).
KLK genes are localized on the 19q13 chromosome and encode 15 serine proteases, the largest protease family in the human genome. These proteases play a role in different physiological processes, such as semen liquefaction and skin shedding. Various amounts of KLK genes are expressed in a broad range of tissues, indicating a various degree of their functional involvement in physiological processes (5). For example, certain KLK genes (NES1, protease M, PSA) have a reduced expression in the prostate, breast, and some other cancers. NES1 has been shown to be a potent angiogenesis inhibitor of a new breast cancer tumor suppressor protein and PSA (6).
To the best of our knowledge, no study so far has eveluated KLKs expression levels in recurrent and non-recurrent prostate cancer tissues. Therefore, this research aimed to determine the effects of the KLK gene family on the recurrence of prostate cancer and evaluate whether KLKs could be used as biomarkers of prostate cancer progression.
MATERIALS AND METHODS
Samples
We analyzed 70 recurrent and non-recurrent prostate cancer samples and 19 adjacent healthy prostate tissue samples kindly provided by Baylor College of Medicine, Houston, TX, from 2013 to 2016. The tissues were harvested at radical prostatectomy for clinically diagnosed prostate cancer, snap frozen in liquid nitrogen, and stored at -80 °C as punch biopsies from fresh tissue. The percentage of cancer in the punch area was determined according to published procedures (7). Cancer specimens contained more than 70% tumor tissue, and benign tissue samples were free of cancer or high-grade prostate intraepithelial neoplasia tissue. Recurrence was defined as two successive serum PSA levels above 0.2 ng/mL and was deemed as a biochemical recurrence. All patients were clinically followed-up until PSA recurrence or for a minimum of four years for non-recurrent patients. Patients' clinical and pathological characteristics are summarized in Table 1. Some of these prostate tissues were previously used by our research team to determine the expression profile of cancer stem cell markers, specifically deregulated microRNAs in recurrent and non-recurrent prostate cancer samples (8,9). The study was approved by the Institutional Review Board of Baylor College of Medicine (2018/20-14).
Table 1.
Clinicopathological characteristics of patients with recurrent and non-recurrent prostate cancer *
| No. of patients |
|||
|---|---|---|---|
| with recurrence (N = 34) | without recurrence (N = 36) | P | |
| Age, mean ± standard deviation |
61.24 ± 1.2 |
61.42 ± 1.1 |
0.28 |
| DMOS, mean ± standard deviation |
22.6 ± 4.23 |
<0.001 |
|
| DMOS mean ± standard deviation |
75.25 ± 1.56 |
||
| Gleason score |
|||
| ≤6 |
4 |
17 |
0.004 |
| 7 (3 + 4) |
14 |
11 |
0.216 |
| 7 (4 + 3) |
10 |
4 |
1.000 |
| 8-10 |
6 |
4 |
0.453 |
| Race |
|||
| African-American |
2 |
1 |
0.457 |
| Caucasian |
29 |
30 |
|
| Hispanic |
1 |
2 |
|
| NA |
2 |
3 |
|
| Pre-operative PSA | 21.73 ± 3.26 | 9.65 ± 2.84 | 0.019 |
*DMOS (R) – months from operation to first recurrence; DMOS (NR) – months from operation to last normal evaluation; NA – not available.
RNA isolation
For RNA isolation, 1 mL of Trizol solution was added to tissues pulverized in liquid nitrogen. Total RNAs were extracted as per manufacturer’s instructions. RNA purity and concentration were measured spectrophotometrically with optical density measurement using NanoDrop ND-2000c (Thermo Fisher Scientific, Dreieich, Germany) spectrophotometer device with absorbance values of 260 nm and 280 nm wavelengths.
Complementary DNA synthesis and quantitative RT-PCR
Complementary DNAs (cDNAs) were synthesized with Transcriptor High Fidelity Reverse Transcription kit (Roche, Basel, Switzerland) using the same number of samples from the isolated RNAs. Reverse transcriptase PCR protocol with Oligo (dt) primers for single chain cDNA synthesis was used as per the manufacturer’s protocol. qPCR analysis was performed with SYBR Green Master Mix (Roche, Basel, Switzerland). All primer sequences are shown in Table 2. The mix is optimized for SYBR Green reactions, and the experiments were performed in Light Cycler 480-II (Roche) qRT-PCR. β-actin was used as an internal control. The relative quantification analysis was performed by delta-delta-Ct method as reported previously (10).
Table 2.
Primer sequences of the kallikrein family
| Gene | Sequence |
|---|---|
|
KLK1-F |
CAGACTTCATGCTGTGTGTCG |
|
KLK1-R |
TTCTCCGCTATGGTGTCCTC |
|
KLK2-F |
AGCCTGCCAAGATCACAGAT |
|
KLK2-R |
CCTTCTCAGAGTAAGCTCTAGCACA |
|
KLK3-F |
CGTGACGTGGATTGGTGC |
|
KLK3-R |
GCCGCAGACTGCCCTG |
|
KLK4-F |
CTCGCTAACGACCTCATGCT |
|
KLK4-R |
TGCAGACCTCCTCAGACACC |
|
KLK5-F |
TCCTCTCATTGTCCCTCTGC |
|
KLK5-R |
CGCAGAACATGGTGTCATCT |
|
KLK6-F |
GATGGTGGTGCTGAGTCTGA |
|
KLK6-R |
CCCACAGTGGATGGATAAGG |
|
KLK7-F |
CTGTCATCCATGGTGAAGAAAGT |
|
KLK7-R |
TTGACATCCACGCACATGA |
|
KLK8-F |
GTGGCAACTGGGTCCTTACA |
|
KLK8-R |
TGCTCTGGGCCATCTTTATT |
|
KLK9-F |
TCCACCTTACTCGGCTCTTC |
|
KLK9-R |
GCTGAGGTCCTTGTTGAAGC |
|
KLK10-F |
TCTCGCTCTTCAACGGCCT |
|
KLK10-R |
CCCTACTCGAGCCCACAGT |
|
KLK11-F |
GGCAACATCACAGACACCA |
|
KLK11-R |
CCCAGGAGATAATGCCTTGA |
|
KLK12-F |
TGTGTGTTCTTGGGCTCAGC |
|
KLK12-R |
CCCACCTGTGGTCAATAAGGAC |
|
KLK13-F |
GCACAAAAGAGGGTGGCAA |
|
KLK13-R |
CGGATCCACAGGACGTATCT |
|
KLK14-F |
GCCTATCCTAGAACCATCACG |
|
KLK14-R |
CTGGAGCTGTCCTCTGCA |
|
KLK15-F |
GGAAGGTGACGAGTGTGC |
|
KLK15-R |
TTGCGCAGGTTGTGCTCT |
|
β-actin-F |
GCCTCGCCTTTGCCGATC |
| β-actin-R | CCCACGATGGAGGGGAAG |
Statistical analysis
Normality testing was performed with the Kolmogorov-Smirnov test. Data are expressed as mean ± standard deviation. Significance of differences in KLKs expression between the groups was assessed with the t test. Spearman’s rho coefficient was used to assess the correlation between KLK genes, serum PSA levels, and Gleason scores. For normally distributed variables, the significance of differences among groups was assessed with one way ANOVA with post-hoc Tamhane T2 for the groups in which the variances were not equal. If the variances were homogeneous, the Fisher LSD test was performed. For not normally distributed variables (KLK9), the Kruskal-Wallis H test was used. All significance values were adjusted by the Bonferroni correction. Recurrent and non-recurrent patients were compared according to race with the χ2 test. The level of statistical significance was set at P ≤ 0.05. Statistical analyses were performed with SPSS v. 16.0 (SPSS Inc., Chicago, IL, USA), and graphs were created with the GraphPad Prism Trial Version (GraphPad, San Diego, CA, USA).
RESULTS
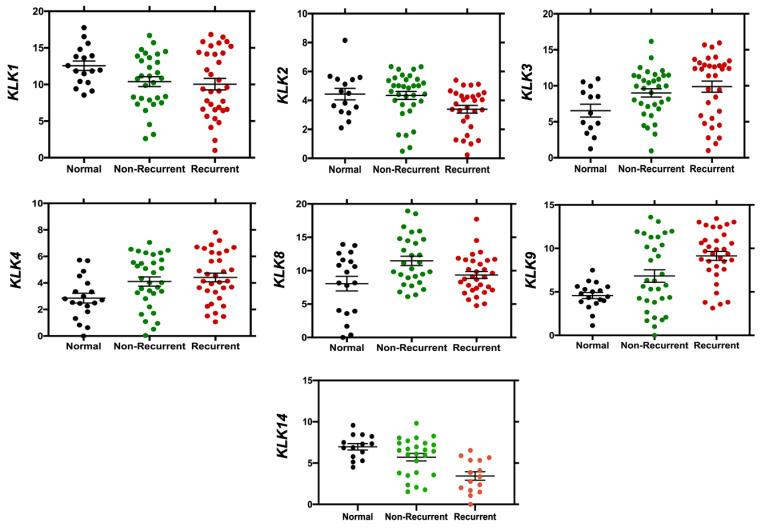
KLK1 expression was significantly decreased in recurrent (P = 0.048), non-recurrent (P = 0.03), and both tumor tissues combined (R+NR) compared with healthy tissues (P = 0.007). KLK2 expression was significantly decreased in recurrent tumor tissues compared with non-recurrent tissues (P = 0.021), and in non-recurrent tissues compared with healthy tissues (P = 0.026). There was no significant difference between healthy tissues and R + NR tissues. The expression of both KLK3 and KLK4 was increased in recurrent tissues (P = 0.049, P = 0.007, respectively) and R + NR tissues compared with healthy tissues (P = 0.025, P = 0.005, respectively). KLK8 expression did not significantly differ between recurrent tissues and healthy tissues and between R + NR tissues and healthy tissues, and it was increased in non-recurrent tissues compared with recurrent tissues (P = 0.045). KLK9 was significantly increased in recurrent tissues (P < 0.001) and R + NR tissues compared with healthy tissues (P < 0.001). KLK14 was the only KLK significantly decreased in R + NR tissues compared with healthy tissues (P = 0.004). KLK14 was significantly decreased in recurrent tissues compared with healthy tissues (P < 0.001) and significantly increased in recurrent tissues compared with non-recurrent tissues (P = 0.001) (Figure 1). We did not observe a significant expression differences in KLK5, KLK6, KLK7, KLK10, KLK11, KLK12, KLK13, and KLK15.
Figure 1.
Relative expression levels of KLK1, KLK2, KLK3, KLK4, KLK8, KLK9, and KLK14 in recurrent and non-recurrent tumor tissues.
There was no significant correlation between age and Gleason score, except for Gleason score ≤6 and race, whereas pre-operative PSA levels were significantly different between the recurrent and non-recurrent groups (P = 0.019, Table 1). There was no correlation between PSA levels and KLKs expression, except for KLK1 and KLK14, in the recurrent group. We only found a significant negative correlation between Gleason score and KLK14 in the non-recurrent group (Table 3).
Table 3.
Differences in kallikreins expression between tumor and control samples, and correlations between gene expression of kallikreins, serum prostate specific antigen (PSA), and Gleason score*
| Relative expression |
PSA |
Gleason score |
||||||
|---|---|---|---|---|---|---|---|---|
| Genes | T/H P Value | R/NR P Value | R/H P Value | R
P Value |
NR P Value | R P Value (correlation coefficients) | NR P Value (correlation coefficients) | |
|
KLK1 |
0.007 |
0.978 |
0.048 |
0.026 |
0.544 |
0.295 (0.188) |
0.103 (-0.303) |
|
|
KLK2 |
0.906 |
0.021 |
0.026 |
0.842 |
0.986 |
0.418 (-0.156) |
0.495 (0.123) |
|
|
KLK3 |
0.025 |
0.081 |
0.049 |
0.665 |
0.056 |
0.541 (-0.110) |
0.364 (-0.163) |
|
|
KLK4 |
0.005 |
0.106 |
0.007 |
0.427 |
0.849 |
0.666 (-0.780) |
0.649 (-0.084) |
|
|
KLK5 |
0.451 |
0.249 |
0.233 |
0.512 |
0.246 |
0.564 (-0.253) |
0.526 (0.274) |
|
|
KLK6 |
0.231 |
0.315 |
0.345 |
0.816 |
0.267 |
0.209 (-0.279) |
0.895 (0.430) |
|
|
KLK7 |
0.082 |
0.245 |
0.203 |
0.305 |
0.283 |
0.246 (-0.369) |
0.269 (0.342) |
|
|
KLK8 |
0.120 |
0.045 |
0.335 |
0.752 |
0.842 |
0.655 (-0.376) |
0.093 (0.324) |
|
|
KLK9 |
<0.001 |
0.086 |
<0.001 |
0.545 |
0.617 |
0.745 (-0.584) |
0.905 (0.322) |
|
|
KLK10 |
0.303 |
0.272 |
0.023 |
0.813 |
0.605 |
0.767 (-0.465) |
0.465 (0.352) |
|
|
KLK11 |
0.165 |
0.312 |
0.392 |
0.127 |
0.624 |
0.115 (-0.478) |
0.541 (0.432) |
|
|
KLK12 |
0.209 |
0.203 |
0.065 |
0.642 |
0.403 |
0.234 (-0.336) |
0.237 (0.159) |
|
|
KLK13 |
0.096 |
0.305 |
0.245 |
0.265 |
0.534 |
0.426 (-0.257) |
0.267 (0.279) |
|
|
KLK14 |
0.004 |
0.001 |
<0.001 |
0.047 |
0.586 |
0.353 (-0.258) |
0.049 (-0.398) |
|
| KLK15 | 0.712 | 0.613 | 0.349 | 0.635 |
0.451 | 0.798 (-0.158) | 0.458 (0.536) | |
*T – tumor; H – healthy; R – recurrent; NR – non-recurrent.
DISCUSSION
In this study, KLK1, KLK4, KLK9, and KLK14 were strongly decreased in prostate cancer samples compared with controls. Recurrent samples were negative for KLK1, KLK2, and KLK14 but demonstrated higher levels of KLK3, KLK4, and KLK9 than controls. Other KLKs were not significantly expressed. To the best of our knowledge, this is the first study that evaluated KLKs expression levels in R+NR prostate cancer tissues.
KLK1 was downregulated in recurrent and non-recurrent prostate cancer tissues compared with healthy samples. This is in contrast to the findings of Mingxin (11), who observed KLK1 expression in gallbladder cancer, especially in female patients. Other studies reported KLK2, KLK3, and KLK4 expression in the breast and prostate (12). The KLK2 protein product is emerging as a new prostate tumor marker. It activates PSA by converting its pre-form to an active mature form. Lower KLK2 is also correlated with prostate cancer risk and higher percentage of free PSA, both of which are related to lower total PSA (13). Similar to a previous study, in our study KLK2 was under-expressed in recurrent prostate cancer tissues compared with healthy tissues (14).
PSA (KLK3) plays a vital role in both normal biology and tumor growth and progression (15). In our study, it was overexpressed in recurrent and non-recurrent cancer tissues, as was the case with KLK4. Another study (15) also found KLK4 to be significantly expressed in 55% of ovarian cancer tissue samples. KLK4 expression was reported to be strongly positively correlated with clinical stage and tumor grade (16).
KLK5 was reported to be overexpressed in ovarian cancer (17), while in prostate cancer its expression levels were notably decreased compared with control tissues (18). Similar to KLK5, KLK6 is a biomarker for ovarian cancer, but is also overexpressed in colorectal cancers (19). When it comes to KLK7, its mRNA was significantly lower in either stage I or stage II breast cancer patients, and its high mRNA expression was related to a favorable prognosis. It was also expressed at unexpectedly elevated levels in ovarian cancer (19). Although in our study KLK8 was upregulated in both recurrent and non-recurrent prostate cancer tissues, its expression was lower in recurrent than in non-recurrent cancer. Patients with recurrent prostate cancer might have a loss of KLK8 gene sequences or are affected by other factors. In ovarian tumors, the expression of the KLK8 and its spliced variants indicate the frequent expression of novel variants. This full-length KLK8 expression is an independent and favorable marker for ovarian cancer (20).
KLK9 in our study was upregulated in recurrent and non-recurrent prostate cancer tissues. Other studies observed elevated KLK9 expression in the early stages of breast cancer compared with advanced stages and in patients with tumor size <2 cm compared with bigger tumors (21). In addition, higher KLK9 expression is a favorable prognostic marker of ovarian cancer (22). Elevated levels of KLK11 are found in the prostate, gastric tissue, trachea, colon, and skin samples (23). When it comes to KLK13, a recently identified family member of kallikreins, it is significantly upregulated in metastatic lung adenocarcinoma. KLK13 overexpression is reported to result in an increased tumor malignant potential, knockdown of its internal gene expression, and decreased cell migration and invasive characteristics. Functional studies further showed that KLK13 was activated via de-methylation of its upstream site (24).
Another study showed that seven genes (KLK5-8, KLK10, KLK11, and KLK14) were elevated in ovarian cancer tissue samples and cell lines compared with the healthy ovary (25). KLK12, KLK13, and KLK14 genes were downregulated in breast cancer (17), while KLK15 was overexpressed in more aggressive forms of prostate cancer (26). Another study has shown trypsin-like activity of KLK15 in prostate cell line (27), but we did not observe a significant change related to this gene.
Our results point to the fact that KLK1, KLK2, and KLK14 (only in recurrent prostate cancer tissues) are underexpressed in prostate cancer tissues, while KLK3, KLK4, KLK8, and KLK9 are overexpressed in both recurrent and non-recurrent tissues. The limitations of this study include a relatively small size, retrospective study design, and a single-center experience.
In conclusion, we propose that KLK1, KLK2, KLK3 KLK4, KLK8, KLK9, and KLK14, which are differentially expressed in prostate cancer, could be used as promising biomarkers of prostate cancer progression. In our opinion, KLKs play a role in promoting or inhibiting tumor growth and metastasis by regulated gene expression. However, our findings need to be validated in studies with a much larger number of participants. In addition, more novel markers should be assessed.
Acknowledgments
Funding This research was supported by Scientific and Technological Research Council of Turkey (115Z417).
Ethical approval given by the Institutional Review Board of Baylor College of Medicine (2018/20-14).
Declaration of authorship FBB, EGT, MI, and MO conceived and designed the study; FBB and EGT acquired the data; FBB, EGT, and ESA analyzed and interpreted the data; ESA and MO drafted the manuscript; FBB, EGT, and MI critically revised the manuscript for important intellectual content; all authors gave approval of the version to be submitted; all authors agree to be accountable for all aspects of the work.
Competing interests All authors have completed the Unified Competing Interest form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: no support from any organization for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work.
REFERENCES
- 1.Schrecengost R, Knudsen KE. Molecular pathogenesis and progression of prostate cancer. Semin Oncol. 2013;40:244–58. doi: 10.1053/j.seminoncol.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg Z, Cooperberg MR, Spratt DE, Feng FY. Genomic biomarkers in prostate cancer. Transl Androl Urol. 2018;7:459–71. doi: 10.21037/tau.2018.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marks LS, Bostwick DG. Prostate Cancer Specificity of PCA3 Gene Testing: Examples from Clinical Practice. Rev Urol. 2008;10:175–81. [PMC free article] [PubMed] [Google Scholar]
- 4.D’Amico AV, Cote K, Loffredo M, Renshaw AA, Chen MH. Pretreatment predictors of time to cancer specific death after prostate specific antigen failure. J Urol. 2003;169:1320–4. doi: 10.1097/01.ju.0000049200.30192.d1. [DOI] [PubMed] [Google Scholar]
- 5.Wang C, Moya L, Clements JA, Nelson CC, Batra J. Mining human cancer datasets for kallikrein expression in cancer: the ‘KLK-CANMAP’ Shiny web tool. Biol Chem. 2018;399:983–95. doi: 10.1515/hsz-2017-0322. [DOI] [PubMed] [Google Scholar]
- 6.Diamandis EP, Yousef GM, Luo LY, Magklara A, Obiezu CV. The new human kallikrein gene family: implications in carcinogenesis. Trends Endocrinol Metab. 2000;11:54–60. doi: 10.1016/S1043-2760(99)00225-8. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler TM, Lebovitz RM. Fresh tissue harvest for research from prostatectomy specimens. Prostate. 1994;25:274–9. doi: 10.1002/pros.2990250507. [DOI] [PubMed] [Google Scholar]
- 8.Guzel E, Karatas OF, Duz MB, Solak M, Ittmann M, Ozen M. Differential expression of stem cell markers and ABCG2 in recurrent prostate cancer. Prostate. 2014;74:1498–505. doi: 10.1002/pros.22867. [DOI] [PubMed] [Google Scholar]
- 9.Suer I, Guzel E, Karatas OF, Creighton CJ, Ittmann M, Ozen M. MicroRNAs as prognostic markers in prostate cancer. Prostate. 2019;79:265–71. doi: 10.1002/pros.23731. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Zuo M, Rashid A, Wang Y, Jain A, Li D, Behari A, et al. RNA sequencing-based analysis of gallbladder cancer reveals the importance of the liver X receptor and lipid metabolism in gallbladder cancer. Oncotarget. 2016;7:35302–12. doi: 10.18632/oncotarget.9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yousef GM, Diamandis EP. The new kallikrein-like gene, KLK-L2. Molecular characterization, mapping, tissue expression, and hormonal regulation. J Biol Chem. 1999;274:37511–6. doi: 10.1074/jbc.274.53.37511. [DOI] [PubMed] [Google Scholar]
- 13.Klein RJ, Halldén C, Cronin AM, Ploner A, Wiklund F, Bjartell AS, et al. Blood biomarker levels to aid discovery of cancer-related single-nucleotide polymorphisms: kallikreins and prostate cancer. Cancer Prev Res (Phila) 2010;3:611–9. doi: 10.1158/1940-6207.CAPR-09-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonk S, Kluth M, Jansen K, Hube-Magg C, Makrypidi-Fraune G, Höflmayer D, et al. Reduced KLK2 expression is a strong and independent predictor of poor prognosis in ERG-negative prostate cancer. Prostate. 2020;80:1097–107. doi: 10.1002/pros.24038. [DOI] [PubMed] [Google Scholar]
- 15.Yousef GM, Diamandis EP. The new human tissue kallikrein gene family: structure, function, and association to disease. Endocr Rev. 2001;22:184–204. doi: 10.1210/edrv.22.2.0424. [DOI] [PubMed] [Google Scholar]
- 16.Obiezu CV, Scorilas A, Katsaros D, Massobrio M, Yousef GM, Fracchioli S, et al. Higher human kallikrein gene 4 (KLK4) expression indicates poor prognosis of ovarian cancer patients. Clin Cancer Res. 2001;7:2380–6. [PubMed] [Google Scholar]
- 17.Diamandis EP, Yousef GM. Human tissue kallikrein gene family: a rich source of novel disease biomarkers. Expert Rev Mol Diagn. 2001;1:182–90. doi: 10.1586/14737159.1.2.182. [DOI] [PubMed] [Google Scholar]
- 18.Yousef GM, Scorilas A, Chang A, Rendl L, Diamandis M, Jung K, et al. Down-regulation of the human kallikrein gene 5 (KLK5) in prostate cancer tissues. Prostate. 2002;51:126–32. doi: 10.1002/pros.10067. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa K, Utsunomiya T, Mimori K, Tanaka F, Inoue H, Nagahara H, et al. Clinical significance of human kallikrein gene 6 messenger RNA expression in colorectal cancer. Clin Cancer Res. 2005;11:2889–93. doi: 10.1158/1078-0432.CCR-04-2281. [DOI] [PubMed] [Google Scholar]
- 20.Magklara A, Scorilas A, Katsaros D, Massobrio M, Yousef GM, Fracchioli S, et al. The human KLK8 (neuropsin/ovasin) gene: identification of two novel splice variants and its prognostic value in ovarian cancer. Clin Cancer Res. 2001;7:806–11. [PubMed] [Google Scholar]
- 21.Yousef GM, Scorilas A, Nakamura T, Ellatif MA, Ponzone R, Biglia N, et al. The prognostic value of the human kallikrein gene 9 (KLK9) in breast cancer. Breast Cancer Res Treat. 2003;78:149–58. doi: 10.1023/A:1022931403825. [DOI] [PubMed] [Google Scholar]
- 22.Yousef GM, Kyriakopoulou LG, Scorilas A, Fracchioli S, Ghiringhello B, Zarghooni M, et al. Quantitative expression of the human kallikrein gene 9 (KLK9) in ovarian cancer: a new independent and favorable prognostic marker. Cancer Res. 2001;61:7811–8. [PubMed] [Google Scholar]
- 23.Diamandis EP, Okui A, Mitsui S, Luo LY, Soosaipillai A, Grass L, et al. Human kallikrein 11: a new biomarker of prostate and ovarian carcinoma. Cancer Res. 2002;62:295–300. [PubMed] [Google Scholar]
- 24.Chou RH, Lin SC, Wen HC, Wu CW, Chang WS. Epigenetic activation of human kallikrein 13 enhances malignancy of lung adenocarcinoma by promoting N-cadherin expression and laminin degradation. Biochem Biophys Res Commun. 2011;409:442–7. doi: 10.1016/j.bbrc.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Yousef GM, Polymeris ME, Yacoub GM, Scorilas A, Soosaipillai A, Popalis C, et al. Parallel overexpression of seven kallikrein genes in ovarian cancer. Cancer Res. 2003;63:2223–7. [PubMed] [Google Scholar]
- 26.Stephan C, Yousef GM, Scorilas A, Jung K, Jung M, Kristiansen G, et al. Quantitative analysis of kallikrein 15 gene expression in prostate tissue. J Urol. 2003;169:361–4. doi: 10.1016/S0022-5347(05)64127-4. [DOI] [PubMed] [Google Scholar]
- 27.Filippou PS, Ren AH, Bala S, Papaioannou MD, Brinc D, Prassas I, et al. Biochemical characterization of human tissue kallikrein 15 and examination of its potential role in cancer. Clin Biochem. 2018;58:108–15. doi: 10.1016/j.clinbiochem.2018.06.007. [DOI] [PubMed] [Google Scholar]