Abstract
FOXP3+CD4+ regulatory T cells (Tregs) are critical for immune homeostasis and respond to local tissue cues, which control their stability and function. We explored here whether developmental endothelial locus-1 (DEL-1), which, like Tregs, increases during resolution of inflammation, promotes Treg responses. DEL-1 enhanced Treg numbers and function at barrier sites (oral and lung mucosa). The underlying mechanism was dissected using mice lacking DEL-1 or expressing a point mutant thereof, or mice with T cell–specific deletion of the transcription factor RUNX1, identified by RNA sequencing analysis of the DEL-1–induced Treg transcriptome. Specifically, through interaction with αvβ3 integrin, DEL-1 promoted induction of RUNX1-dependent FOXP3 expression and conferred stability of FOXP3 expression upon Treg restimulation in the absence of exogenous TGF-β1. Consistently, DEL-1 enhanced the demethylation of the Treg-specific demethylated region (TSDR) in the mouse Foxp3 gene and the suppressive function of sorted induced Tregs. Similarly, DEL-1 increased RUNX1 and FOXP3 expression in human conventional T cells, promoting their conversion into induced Tregs with increased TSDR demethylation, enhanced stability, and suppressive activity. We thus uncovered a DEL-1/αvβ3/RUNX1 axis that promotes Treg responses at barrier sites and offers therapeutic options for modulating inflammatory/autoimmune disorders.
Keywords: Autoimmunity, Inflammation
Keywords: Adaptive immunity, T cells
Introduction
T cell–mediated immunity entails 2 aspects, proinflammatory and regulatory, which need to be balanced for immune homeostasis (1, 2). For instance, effector CD4+ T helper cells expressing IL-17 (Th17 cells) play a crucial role in host immunity, particularly in neutrophil-mediated host defenses at mucosal barrier sites (3). However, Th17 responses need to be tightly controlled to prevent pathologic inflammation at the mucosa and other sites. In this regard, FOXP3-expressing regulatory CD4+ T cells (Tregs) can suppress unwarranted IL-17–driven inflammation by downregulating Th17 expansion and effector functions (4–6). FOXP3+ Tregs are generated either in the thymus or in peripheral tissues by induction from naive CD4+ T cells and have versatile and plastic regulatory functions. Tregs maintain immunological tolerance to self-antigens, suppress excessive immune responses against pathogens and other insults, prevent or mitigate autoimmune and inflammatory diseases, and contribute to inflammation resolution (6–9). FOXP3 is not only a lineage-specification transcription factor required for the differentiation of Tregs but is also crucial for their function by regulating specific transcriptional programs (10, 11). FOXP3 deficiency or dysfunction causes severe autoimmune diseases, known as the IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome in humans and as the scurfy phenotype in mice (12). Forced expression of FOXP3 in CD4+ T cells confers a regulatory phenotype and function and, conversely, experimental deletion of the FOXP3 gene in differentiated Tregs abrogates their regulatory activity (13).
Similarly to effector T cells, the numbers and activity of Tregs must be finely regulated to allow the development of protective immunity with no or minimal immunopathology, or to effectively block unwarranted inflammatory responses at steady state or during inflammation resolution. In this regard, Tregs respond to environmental cues (e.g., cytokines, lipid mediators, vitamin metabolites, hypoxia), which positively or negatively regulate their differentiation, stability, and function, hence modulating Treg responses at effector sites (14–17). We reasoned that local tissue-derived factors that are upregulated during inflammation resolution, such as developmental endothelial locus-1 (DEL-1), might promote Treg responses and thereby facilitate the restoration of tissue homeostasis.
DEL-1 is a locally secreted 52-kDa protein that interacts with distinct integrins and homeostatically regulates the initiation and resolution of inflammation (18–25). DEL-1 consists of 3 N-terminal EGF-like repeats and 2 C-terminal discoidin-I–like domains, hence it is also known as EDIL3 (EGF-like repeats and discoidin-I–like domains-3). The second EGF-like repeat (E2) contains an RGD (Arg-Gly-Asp) motif, which enables DEL-1 to bind αvβ3 integrin (19, 26), whereas its discoidin-I–like domains can bind the “eat-me” signal phosphatidylserine on apoptotic cells (22, 27). These interactions allow DEL-1 to serve as a molecular bridge that facilitates the phagocytosis of apoptotic neutrophils (efferocytosis) by αvβ3 integrin–bearing macrophages, thereby triggering liver-X-receptor–dependent macrophage reprogramming to a proresolving phenotype (22). We have shown that DEL-1 also interacts with β2 integrins, such as αLβ2 (LFA-1, lymphocyte function-associated antigen 1); the binding of DEL-1 to the LFA-1 integrin on neutrophils prevents the interaction of the integrin with ICAM-1 on vascular endothelial cells, resulting in suppressed neutrophil adhesion and recruitment to sites of inflammation (20, 21). Consistent with these functions, DEL-1 protects against inflammatory pathologies in preclinical murine or nonhuman primate models, such as inflammatory bone loss in periodontitis (19, 20), experimental autoimmune encephalitis (EAE)/multiple sclerosis (28), lung inflammation and fibrosis (21, 29), chemical peritonitis (22), allergic asthma (30), and inflammatory reactions associated with islet transplantation (31). Many of these pathologies are driven by IL-17–mediated inflammation; for instance, the development of periodontitis or EAE associated with DEL-1 deficiency is reversed in mice lacking both DEL-1 and IL-17 receptor (20, 28).
During active inflammation, IL-17 inhibits endothelial DEL-1 expression and thereby promotes leukocyte infiltration and tissue inflammation (20, 32). However, in both humans and mice, the expression of DEL-1 mRNA and protein resurges remarkably during the resolution of inflammation, largely driven by the increase of proresolving lipid mediators, whereas the resolution of experimental periodontitis or peritoneal inflammation fails in DEL-1 deficiency (22, 32). As DEL-1 is an integrin-binding protein and integrins are involved in the regulation of Tregs (33, 34), which are elevated during resolution of inflammation (35, 36), we investigated whether the resurgence of DEL-1 during resolution may act as a tissue-derived signal that can contribute to Treg responses.
In the present study, we showed that DEL-1 promotes Treg responses by upregulating FOXP3 expression in a manner dependent on the αvβ3 integrin and the Runt-related transcription factor-1 (RUNX1). This activity required an intact RGD motif but not the entire molecule, as truncated DEL-1 containing only the EGF-like repeats was sufficient to upregulate FOXP3 expression. DEL-1, moreover, conferred stability of FOXP3 expression upon restimulation of Tregs in the absence of exogenous TGF-β1. Mice deficient in DEL-1 or expressing a point mutant thereof incapable of interacting with αvβ3 integrin displayed defective Treg responses in 2 mucosal inflammatory disease models, periodontitis and acute lung injury. Human relevance for the mouse experimental studies was established by findings that DEL-1 increased RUNX1 and FOXP3 expression in human CD4+CD25– conventional T (Tconv) cells, promoting their conversion into induced Tregs with increased stability, accompanied by increased demethylation of the Treg-specific demethylated region (TSDR) and enhanced suppressive activity. These data show that DEL-1 promotes Treg responses, in both humans and mice, with important therapeutic implications for inflammatory or autoimmune disorders.
Results
DEL-1 deficiency is associated with decreased Treg/Th17 cell ratio: reversal by the EGF-like repeat region of DEL-1.
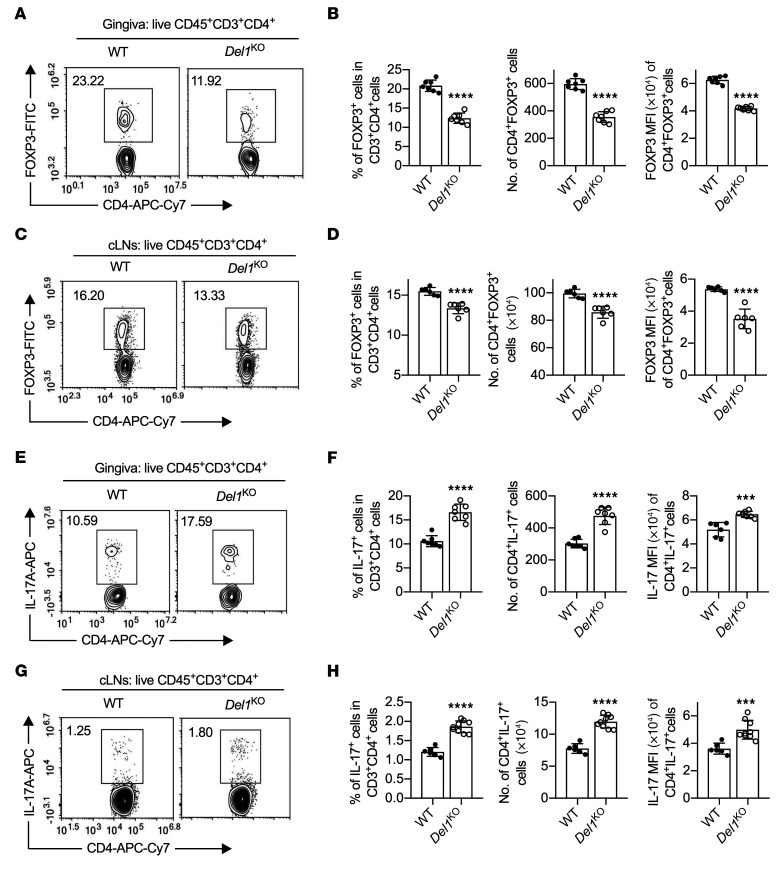
The mouse ligature-induced periodontitis (LIP) model simulates human periodontitis and its resolution; it generates a biofilm-retentive milieu leading to dysbiotic inflammation and bone loss, whereas ligature removal abrogates the dysbiotic microbial challenge, resulting in inflammation resolution (22, 37). Therefore, LIP is an ideal model to explore mechanisms operating during inflammation resolution, where Tregs play important roles (7, 9, 35). Following significant downregulation in the inductive phase of periodontitis, DEL-1 expression resurges during the resolution phase (22). However, whether DEL-1 regulates or contributes to Treg responses has not been hitherto addressed. To this end, DEL-1–deficient (Del1KO) mice and WT littermate controls were subjected to LIP and the ligatures were removed on day 10 for 5 days. The mice were euthanized on day 15 for immunological analysis of the gingival tissue and the draining cervical lymph nodes (cLNs) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI137530DS1). In the absence of DEL-1, the frequencies and absolute numbers of Tregs and the FOXP3 expression level (mean fluorescence intensity; MFI) were significantly decreased in both the gingival tissue (Figure 1, A and B) and the cLNs (Figure 1, C and D), accompanied, as expected, by defective inflammation resolution (Supplemental Figure 2). Conversely, Th17 cell frequencies/absolute numbers and IL-17A expression level (MFI) were increased in the gingival tissue and cLNs of Del1KO mice as compared with WT controls (Figure 1, E–H). Thus, DEL-1 deficiency is associated with defective Treg responses and a shift in the Treg/Th17 balance in favor of the latter during inflammation resolution.
Figure 1. DEL-1 deficiency decreases Treg numbers while it increases Th17 cells during resolution of inflammation.
Groups of littermate WT and Del1KO mice were subjected to ligature-induced periodontitis (LIP) for 10 days and ligatures were removed on day 10 (to facilitate inflammation resolution) for 5 days. (A–D) FACS plots of Tregs in gingival tissues (A) and cLNs (C) on day 15 and bar graphs showing percentage of Tregs in CD4+ T cells (left), absolute numbers (middle), and FOXP3 MFI (right) of Tregs from gingival tissues (B) and cLNs (D) of littermate WT and Del1KO mice on day 15 (n = 6–7 mice/group). (E) FACS plots of Th17 cells in gingival tissue on day 15 and (F) bar graphs showing percentage of Th17 cells in CD4+ T cells (left), absolute numbers (middle), and IL-17A MFI of Th17 cells (right) from gingival tissues of littermate WT and Del1KO mice on day 15 (n = 7 mice/group). (G) FACS plot of Th17 cells in cLNs and (H) bar graphs showing percentage of Th17 cells in CD4+ T cells (left), absolute numbers (middle), and IL-17A MFI of Th17 cells (right) from cLNs of littermate WT and Del1KO mice on day 15 (n = 6 mice for WT group and n = 9 for Del1KO group). Data are means ± SD and are pooled from 2 independent experiments. ***P < 0.001, ****P < 0.0001 vs. WT mice by 2-tailed Student’s t test.
In contrast, under steady-state conditions, we did not observe significant abnormalities in Treg frequencies or absolute numbers in the cLNs, spleen, or thymus in Del1KO mice as compared with WT littermate controls (Supplemental Figure 3). However, in comparison with steady-state gingival tissue of WT mice, the frequency/absolute numbers of gingival Tregs were modestly decreased in Del1KO mice (Supplemental Figure 3), which spontaneously develop gingival inflammation featuring cytokines such as IL-6 and IL-23 (20) that may destabilize Tregs (38).
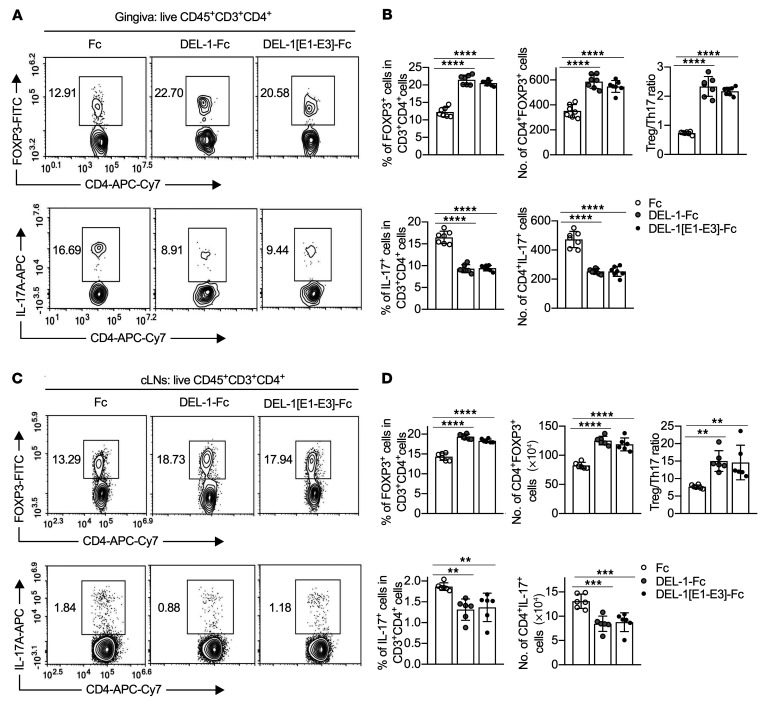
To validate the role of DEL-1 in regulating Tregs during inflammation resolution, we next assessed whether exogenous DEL-1 could rescue the attenuated Treg responses in Del1KO mice. To this end, intact DEL-1–Fc was microinjected into the gingiva of Del1KO mice after ligature removal on day 10 and daily thereafter until day 15, when mice were euthanized for analysis. DEL-1–Fc, but not Fc control, significantly increased Treg frequencies and absolute numbers as well as FOXP3 expression, while suppressing Th17 cell frequencies/absolute numbers and IL-17A expression, thus leading to an increased Treg/Th17 cell ratio in both the gingival tissue (Figure 2, A and B) and the cLNs (Figure 2, C and D), in the setting of inflammation resolution (Supplemental Figure 4). A side-by-side experiment using a version of DEL-1 containing only the EGF-like repeats (DEL-1[E1–E3]–Fc) yielded similar results to those obtained with the full-length molecule (Figure 2). Hence, the EGF-like repeat region of DEL-1 is sufficient to upregulate the Treg/Th17 cell ratio, although it fails to promote efferocytosis, as it lacks the phosphatidylserine-binding domains (22). Therefore, the capacity of DEL-1 to regulate the Treg/Th17 cell balance includes mechanism(s) that are beyond and independent of its effects on efferocytosis-associated inflammation resolution.
Figure 2. The DEL-1 EGF-like repeats (DEL-1[E1–E3]) are sufficient to upregulate the Treg/Th17 cell ratio.
(A–D) Groups of Del1KO mice were subjected to ligature-induced periodontitis (LIP) for 10 days and ligatures were removed on day 10 to facilitate inflammation resolution. The mice were locally microinjected daily with DEL-1–Fc, DEL-1-[E1–E3]–Fc, or Fc control from day 10 to day 14 for a total of 5 doses. FACS plots of Tregs (top) and Th17 cells (bottom) in gingival tissues (A) and cLNs (C) of microinjected Del1KO mice on day 15 and bar graphs showing the percentages and absolute numbers of Tregs (top left and middle) and Th17 cells (bottom left and middle) in CD4+ T cells, Treg/Th17 cell ratio (top right) from gingival tissues (B) and cLNs (D) (n = 6–7 mice/group). Data are means ± SD and are pooled from 2 independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. Fc treatment group by 1-way ANOVA with Dunnett’s multiple-comparisons test for comparison with Fc control treatment.
Given that macrophages and Tregs engage in bidirectional crosstalk (39) and that proresolving macrophages may express DEL-1 (22), we investigated whether macrophages may contribute to the increased abundance of Tregs during resolution. To examine this possibility that would further strengthen the role of DEL-1 in promoting Treg abundance during resolution, we compared mice with macrophage-specific overexpression of DEL-1 (CD68-Del1 mice; ref. 22) and WT littermates in the periodontitis resolution model. We found that CD68-Del1 mice showed significantly higher Treg and lower Th17 cell frequencies in the gingival tissue and draining cLNs as compared with their WT littermates (Supplemental Figure 5). Therefore, as a cellular source of DEL-1, macrophages can promote Treg responses during inflammation resolution.
DEL-1 promotes mouse Treg differentiation via its RGD motif in the oral and lung mucosa.
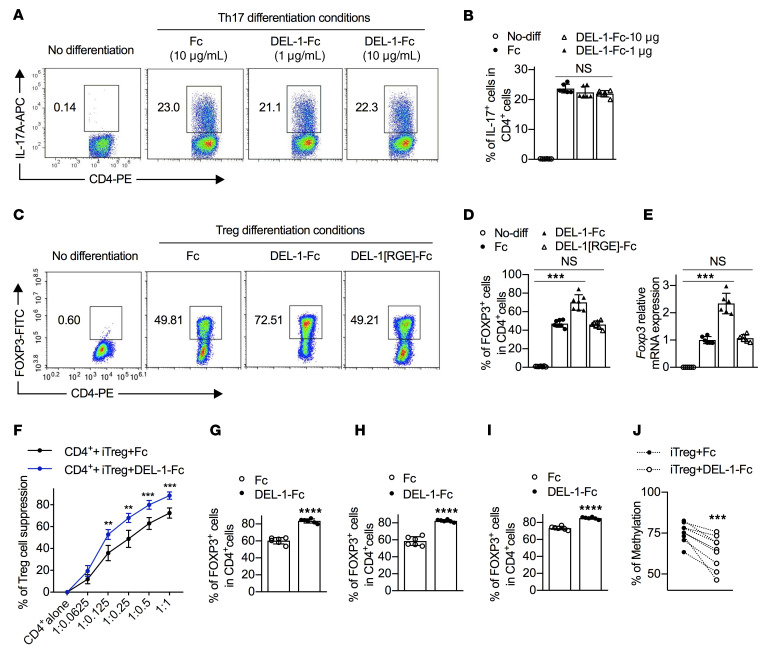
Our finding that DEL-1 deficiency is associated with increased gingival Th17 cell frequency (Figure 1, E and F) is consistent with our earlier observations that DEL-1 inhibits IL-17 production in periodontitis and multiple sclerosis models (20, 28). A conceivable explanation might be that DEL-1 directly affects the differentiation of Th17 cells. To address this possibility, we performed a standard naive CD4+ T cell differentiation assay, based on polyclonal stimulation with anti-CD3/anti-CD28 and appropriate polarizing cytokines, in the presence or absence of DEL-1. Exogenously added DEL-1–Fc failed to influence Th17 cell differentiation of mouse splenic naive CD4+ T cells under the influence of IL-6 and TGF-β1 (nonpathogenic conditions; ref. 40) (Supplemental Figure 6) or under the influence of IL-6, TGF-β1, IL-1β, and IL-23 (pathogenic conditions; ref. 40) (Figure 3, A and B), as compared with Fc control treatment. These data suggest that DEL-1 is unlikely to directly influence Th17 differentiation, prompting us to alternatively consider that DEL-1 might act on Tregs.
Figure 3. DEL-1 directly promotes de novo Treg differentiation via its RGD motif.
(A and B) Naive splenic CD4+ cells were differentiated, or not, into Th17 cells under pathogenic conditions (see Methods) in the presence of DEL-1–Fc or Fc control. (A) FACS plots and (B) percentage of IL-17A+ cells in CD4+ T cells (n = 6 replicates). (C–E) Naive splenic CD4+ cells were differentiated, or not, into Tregs in the presence of DEL-1–Fc, DEL-1[RGE]–Fc, or Fc control (10 μg/mL). (C) FACS plots and (D) percentage of FOXP3+ cells in CD4+ T cells (n = 7 replicates). (E) Relative mRNA expression of Foxp3 in Tregs (n = 6 replicates). (F) Suppression of CFSE-labeled CD4+CD25– T cell (Tconv) division by purified DEL-1–Fc−iTregs or Fc-iTregs. Numbers on x axis indicate CD4+CFSE+cell/iTreg ratio (n = 6 replicates). (G) Naive splenic CD4+ T cells were differentiated into Tregs in the presence of DEL-1–Fc or Fc control (10 μg/mL). CD4+CD25+ cells were sorted and restimulated for 4 days in medium containing IL-2 (40 ng/mL) and FOXP3 expression was assessed. (H and I) Naive splenic CD4+ cells were differentiated into Tregs. CD4+CD25+ cells were sorted and restimulated for 4 days with DEL-1–Fc or Fc control in medium containing IL-2 (40 ng/mL) without (H) or with TGF-β1 (5 ng/mL) (I), and FOXP3 expression was assessed. (J) Naive splenic CD4+ T cells were differentiated for 4 days to Tregs in the presence of DEL-1–Fc or Fc control (10 μg/mL). iTregs (CD4+CD25+) were sorted and evaluated for their methylation status of the Foxp3 CNS2 (n = 9 mice). All CD4+ T cells were isolated from WT mice. Data are means ± SD and are pooled from 2 (B and D–I) or 5 (J) independent experiments. **P < 0.01, ***P < 0.001, ****P < 0.0001 vs. Fc control (B and D–J) by 1-way ANOVA with Dunnett’s post-test for comparison with Fc control (B, D, and E), 2-way ANOVA with Holm-Šidák post hoc test for comparison with Fc control (F), or 2-tailed Student’s t test for comparison with Fc control (G–J). NS, not significant.
In an in vitro Treg differentiation assay using mouse splenic naive CD4+ T cells stimulated with anti-CD3/anti-CD28 and IL-2 plus TGF-β1 in the presence of DEL-1–Fc or Fc control, DEL-1–Fc–treated cells contained a significantly higher (~50% increase) percentage of FOXP3+ cells on day 4 (Figure 3, C and D) and exhibited significantly higher Foxp3 mRNA expression on day 3 (Figure 3E) as compared with Fc control–treated cells. Thus, DEL-1 induces Treg differentiation in vitro, over and beyond Treg induction driven by strong polarizing conditions (TGF-β1 and IL-2). Importantly, moreover, DEL-1–Fc–treated inducible Tregs (iTregs) exhibited significantly stronger suppressive activity against CD4+ T cell proliferation than Fc-treated iTregs (Figure 3F). Consistently, WT mice showed a significantly higher frequency of Tregs expressing markers associated with their suppressive function (IL-10, CTLA-4, ICOS) than their counterparts from Del1KO mice during inflammation resolution (Supplemental Figure 7). In vitro–induced mouse FOXP3+ Tregs may have an unstable phenotype and may thus lose FOXP3 expression upon subsequent restimulation in the absence of exogenous TGF-β (41, 42). To evaluate the stability of DEL-1–Fc–induced Tregs, we generated Tregs as described above (culture of splenic naive CD4+ T cells stimulated with anti-CD3/anti-CD28 and IL-2 plus TGF-β1 in the presence of DEL-1–Fc or Fc control) and, after 4 days, CD4+CD25+ Tregs were sorted to high purity and restimulated for another 4 days without TGF-β1 or DEL-1–Fc. The restimulated cultures of sorted Tregs that were originally induced in the presence of DEL-1–Fc displayed a significantly higher frequency of FOXP3-expressing cells (>80% of CD4+ cells) than those originally induced in the presence of Fc control (Figure 3G and Supplemental Figure 8A), suggesting that DEL-1–Fc can induce stable FOXP3 expression (maintained even in the absence of TGF-β1). Next, sorted CD4+CD25+ Tregs (induced as above but in the absence of DEL-1–Fc or Fc control) were restimulated for another 4 days without TGF-β1 but in the presence of DEL-1–Fc or Fc control. The DEL-1–Fc–containing restimulated cultures had a higher percentage of FOXP3-expressing cells than Fc control–containing restimulated cultures (Figure 3H and Supplemental Figure 8B), suggesting that DEL-1–Fc stabilizes TGF-β1–induced FOXP3 expression. Interestingly, even if the restimulation (anti-CD3/anti-CD28 and IL-2) was performed in the presence of TGF-β1 in the cultures, DEL-1 still significantly increased the numbers of FOXP3-expressing cells as compared with Fc control (Figure 3I and Supplemental Figure 8C). Therefore, although TGF-β1 promotes the stability of FOXP3 expression in the restimulated cultures (75% vs. 58%; see Fc groups in Figure 3, H and G), DEL-1–Fc promotes the stability of FOXP3 expression even further than what TGF-β1 can achieve on its own.
The expression of FOXP3 in Tregs is stabilized by demethylation of CpG motifs in a conserved noncoding sequence 2 (CNS2) region that serves as an enhancer for FOXP3 transcription (42, 43). This major TSDR of CNS2 can distinguish Tregs from naive CD4+CD25– Tconv cells, in which this region is heavily methylated (42, 43). To determine whether the ability of DEL-1 to stabilize FOXP3 expression is associated with increased demethylation of the CNS2 TSDR, we determined the methylation status of this region in mouse iTregs induced in the presence of DEL-1–Fc or Fc control. We found that DEL-1–Fc significantly promoted the demethylation of the FOXP3 CNS2 region (Figure 3J), which may thus represent a mechanism whereby DEL-1 contributes to stabilization of FOXP3 expression in iTregs.
Integrins are involved in the regulation of Tregs (33, 34), which are elevated upon administration of the DEL-1 EGF-like repeat region containing an integrin-binding RGD motif in Del1KO mice (Figure 2). To determine whether the ability of DEL-1 to promote FOXP3 expression requires integrin function, we tested in parallel DEL-1[RGE]–Fc, wherein the integrin-binding RGD motif of the second EGF-like repeat was mutated to render the molecule incapable of interacting with αvβ3 integrin (22, 44). In stark contrast to DEL-1–Fc, DEL-1[RGE]–Fc failed to influence Treg differentiation and FOXP3 expression (Figure 3, C–E), indicating that DEL-1 promotes Treg differentiation via its integrin-binding RGD motif in vitro. Interestingly, moreover, DEL-1–Fc, but not DEL-1[RGE]–Fc, could promote the expression of FOXP3 even in T cells cultured under Th17 differentiation conditions (i.e., 1 ng/mL TGF-β1 and 50 ng/mL IL-6), as compared with Fc control (Supplemental Figure 9).
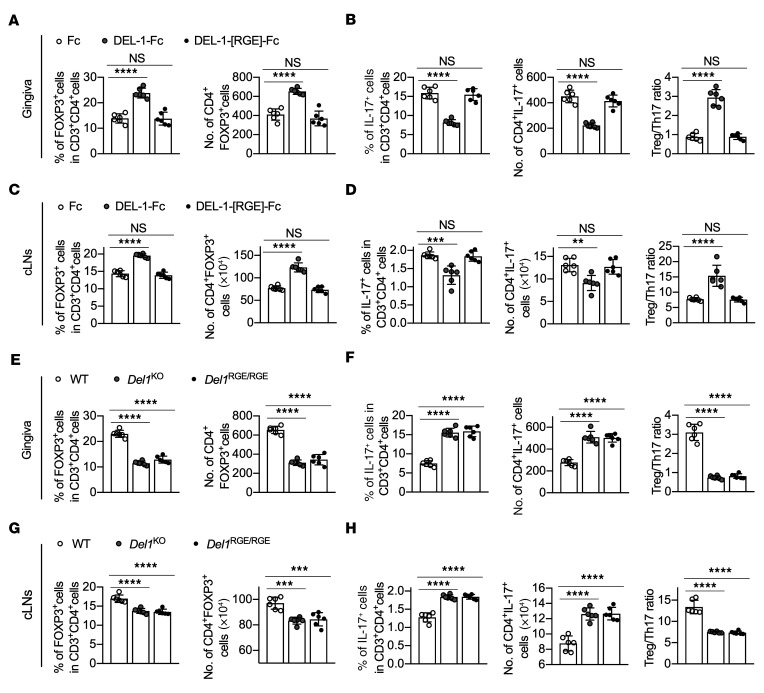
To validate the role of the RGD motif under in vivo conditions, DEL-1[RGE]–Fc was locally microinjected into the gingiva of Del1KO mice after ligature removal on day 10 and daily thereafter until day 15. In contrast to the WT molecule, DEL-1[RGE]–Fc failed to enhance Treg or suppress Th17 cell frequencies and absolute numbers both in the gingival tissue and the cLNs (Figure 4, A–D). To corroborate the involvement of the RGE motif of DEL-1 in Treg regulation, we used mice that express the RGE point mutant of DEL-1 (Del1RGE/RGE mice; ref. 45). Similarly to Del1KO mice, Del1RGE/RGE mice displayed significantly lower Treg and higher Th17 cell frequencies and absolute numbers (and hence decreased Treg/Th17 cell ratio), as compared with WT mice, in both the gingival tissue and cLNs (Figure 4, E–H) on day 15. Together, these data suggest that, during inflammation resolution, DEL-1 increases the abundance of FOXP3+ Tregs via a mechanism that depends on its integrin-binding RGD motif. On the other hand, similarly to Del1KO mice, Del1RGE/RGE mice did not exhibit significant alterations in Treg frequencies and absolute numbers in the cLNs, spleen, or thymus under steady-state conditions, as compared with WT mice (Supplemental Figure 3).
Figure 4. The DEL-1 RGD motif is critical for Treg responses in vivo.
(A–D) Groups of Del1KO mice were subjected to ligature-induced periodontitis (LIP) for 10 days, at which time the ligatures were removed. The mice were then locally microinjected daily with DEL-1–Fc, DEL-1[RGE]–Fc, or Fc control from day 10 to day 14 for a total of 5 doses. Percentages and absolute numbers of Tregs (A and C) and Th17 cells and Treg/Th17 cell ratio (B and D) from gingival tissues (A and B) and cLNs (C and D) on day 15 (n = 6 mice/group). (E–H) Groups of WT, Del1KO, and Del1RGE/RGE mice were subjected to LIP for 10 days and ligatures were removed on day 10 for 5 days. Percentages and absolute numbers of Tregs (E and G) and Th17 cells and Treg/Th17 cell ratio (F and H) from gingival tissues (E and F) and cLNs (G and H) of WT, Del1KO, and Del1RGE/RGE mice on day 15 (n = 6 mice/group). Data are means ± SD and are pooled from 2 independent experiments. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Dunnett’s post hoc test vs. Fc control group (A–D) or vs. WT mice (E–H).
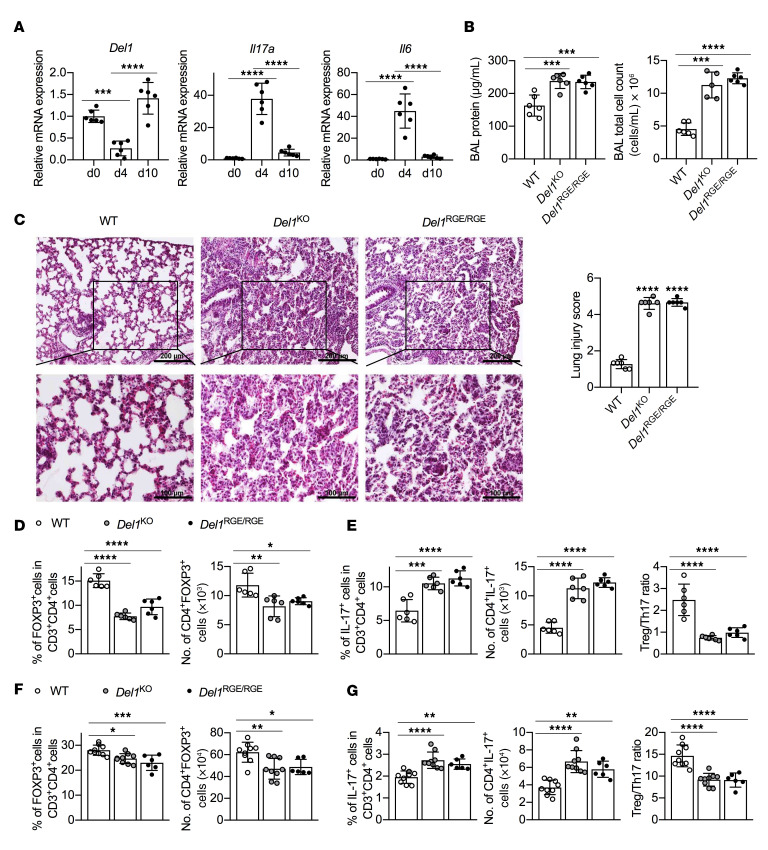
DEL-1 is also expressed in the lungs (21), where Tregs have been reported to mediate resolution of experimental lung injury in mice (7). To investigate whether the observed RGD motif–dependent effect of DEL-1 on Treg differentiation in experimental periodontitis represents a more general principle applicable to other inflammatory settings, the acute lung injury model (7) was engaged. Specifically, WT, Del1KO, and Del1RGE/RGE mice were challenged intratracheally with LPS and were monitored for 10 days thereafter. Consistent with findings in mouse and human periodontitis (22), Del1 expression in the lungs of WT mice was significantly downregulated at the peak of inflammation (day 4) but rebounded on day 10 (resolution) to an even higher level than the baseline on day 0 (Figure 5A). The Del1 expression pattern exhibited a converse relationship with Il6 and Il17a expression levels (Figure 5A). Compared with WT controls, Del1KO and Del1RGE/RGE mice had significantly higher total protein and total number of cells in the BAL (Figure 5B) and persistent interstitial thickening and cellular infiltration (Figure 5C) on day 10, indicating impaired resolution of pulmonary inflammation. In line with these data and the importance of Tregs in the resolution of pulmonary inflammation (7), Del1KO and Del1RGE/RGE mice exhibited significantly lower frequencies and absolute numbers of FOXP3+ Tregs and, conversely, higher frequencies of Th17 cells than seen in WT mice, resulting in significantly reduced Treg/Th17 cell ratios both in the BAL (Figure 5, D and E) and the draining mediastinal LNs (mLNs) (Figure 5, F and G). In comparison with WT mice under steady-state conditions, Del1KO and Del1RGE/RGE mice displayed only modestly reduced Treg frequencies and numbers in the BAL but similar Treg frequencies and numbers in mLNs (Supplemental Figure 3). Taken together, these data show that, during inflammation resolution, DEL-1 promotes Treg responses in the lungs in a manner dependent on its RGD motif, as seen in the gingival tissue.
Figure 5. Del1KO and Del1RGE/RGE mice have impaired Treg induction and resolution of inflammation in a model of acute lung injury.
(A–G) Groups of WT, Del1KO, or Del1RGE/RGE mice were intratracheally instilled with Escherichia coli LPS at 3.75 μg/g bodyweight or PBS control. (A) Relative mRNA expression of Del1, Il17a, and Il6 in WT mice at the indicated time points (n = 6 mice/group). (B) BAL from WT, Del1KO, or Del1RGE/RGE mice was analyzed for total protein concentration (left) and total cell numbers (right) on day 10 after LPS instillation (n = 6 mice/group). (C) H&E staining of lung sections (left) and lung injury scoring (right) from WT, Del1KO, or Del1RGE/RGE mice on day 10 after LPS instillation (n = 6 mice/group). Scale bars: 200 μm (top panels) and 100 μm (bottom panels). (D and E) BAL from WT, Del1KO, or Del1RGE/RGE mice was analyzed for (D) the percentage and absolute numbers of Tregs and (E) Th17 cells in CD4+ T cells and Treg/Th17 cell ratio (n = 6 mice/group). (F) The percentage and absolute numbers of Tregs and (G) Th17 cells in CD4+ T cells and Treg/Th17 cell ratio in mediastinal LNs from WT, Del1KO, or Del1RGE/RGE mice on day 10 (n = 6–9 mice/group as indicated). Data are means ± SD and are pooled from 2 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Dunnett’s post hoc test for comparison between indicated groups.
αvβ3 Integrin mediates DEL-1–induced FOXP3 expression in Tregs.
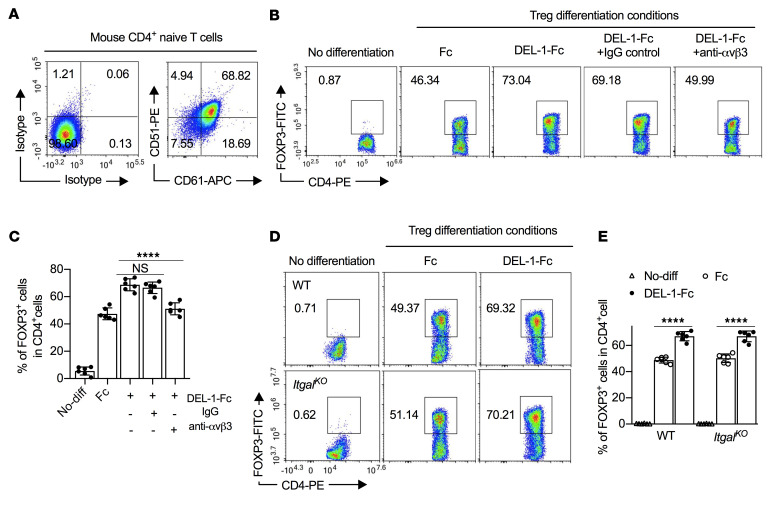
Among several RGD-binding integrins, αvβ3 is a well-established receptor for DEL-1 (22, 44, 46). To assess possible involvement of αvβ3 in DEL-1–induced FOXP3 expression, we first showed that αvβ3 integrin (CD51/CD61) is expressed by splenic CD4+ naive T cells (~70% positive cells; Figure 6A), and by thymus-derived natural Treg (nTreg) cells and in vitro–induced iTregs (Supplemental Figure 10). We next pretreated WT CD4+ naive T cells with a neutralizing antibody against αvβ3 integrin. The use of the anti-αvβ3 antibody (but not of IgG control) significantly inhibited the ability of DEL-1–Fc to induce FOXP3+ Tregs (to levels comparable to those seen in the Fc control group; Figure 6, B and C). As DEL-1 also interacts with β2 integrins (21, 47), we examined whether LFA-1, a T cell–expressed β2 integrin, also mediates DEL-1–dependent Treg differentiation. To this end, we performed in vitro differentiation of Tregs from splenic naive CD4+ T cells isolated from WT or LFA-1–deficient (ItgalKO) mice. In this system, DEL-1–Fc promoted the induction of FOXP3+ Tregs regardless of the presence or absence of the LFA-1 integrin (Figure 6, D and E). Taken together, these data show that DEL-1 promotes the induction of FOXP3+ Tregs in a manner dependent on the αvβ3 integrin.
Figure 6. DEL-1 enhances FOXP3 expression in Tregs in a β3 integrin–dependent manner.
(A) FACS plots of CD51 (αv) and CD61 (β3) expression on mouse naive splenic CD4+ T cells. (B and C) Naive splenic CD4+ cells isolated from WT mice were differentiated, or not, into Tregs in medium containing anti-CD3/anti-CD28, TGF-β1 (5 ng/mL), IL-2 (40 ng/mL), and Fc control or DEL-1–Fc (10 μg/mL) in the presence or not of IgG control or anti-αvβ3 antibody (10 μg/mL; added 15 minutes before DEL-1–Fc treatment). Shown are (B) FACS plots and (C) data analysis of the percentage of FOXP3+ cells in CD4+ T cells from the in vitro culture system on day 4 (n = 6 replicates from 2 separate cell isolations). (D and E) Naive splenic CD4+ cells isolated from WT and ItgalKO mice were differentiated, or not, into Tregs in medium containing anti-CD3/anti-CD28, TGF-β1 (5 ng/mL), IL-2 (40 ng/mL), and Fc control or DEL-1–Fc (10 μg/mL). Shown are (D) FACS plots and (E) data analysis of the percentage of FOXP3+ cells in CD4+ T cells from the in vitro culture system on day 4 (n = 6 replicates from 2 separate cell isolations). Data are means ± SD and are pooled from 2 independent experiments. ****P < 0.0001 between indicated groups by 1-way ANOVA with Dunnett’s post hoc test for comparisons with DEL-1–Fc treatment (C) or by 2-tailed Student’s t test (E). NS, not significant.
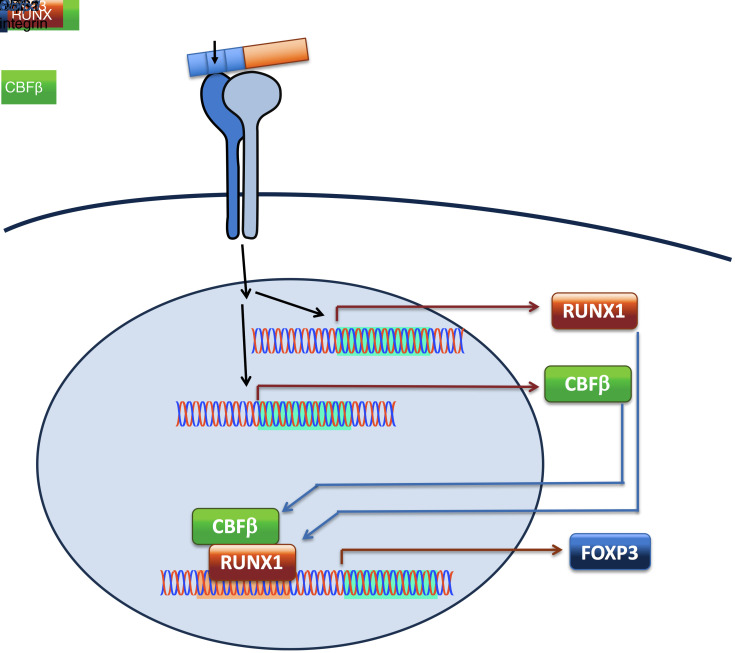
DEL-1 upregulates RUNX1 and is required for its ability to induce Treg differentiation.
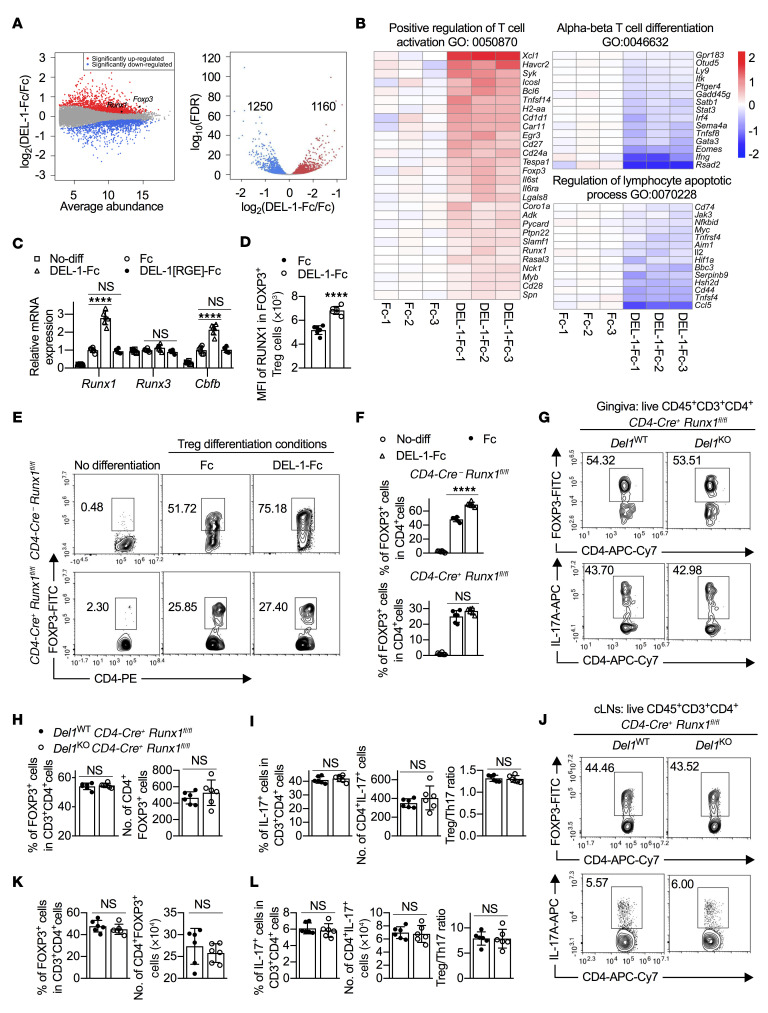
To obtain insights into how DEL-1 may promote FOXP3 expression, we applied RNA sequencing (RNA-seq) to Tregs that were differentiated for 3 days in vitro in the presence of DEL-1–Fc or Fc control. Among the differentially regulated genes (false discovery rate [FDR] < 0.05) by DEL-1–Fc versus Fc control, 1250 genes were significantly downregulated and 1160 genes were significantly upregulated, respectively (Figure 7A). Further, we applied PANTHER Gene Ontology (GO) enrichment analysis of those genes that were significantly regulated. Notably, the GO term “Positive regulation of T cell activation (GO: 0050870)” was highly enriched (fold enrichment 3.23; FDR = 6.53 × 10–5) among the significantly enriched GO terms of significantly upregulated genes. Consistent with our earlier observation (Figure 3, C–E), Foxp3 was among those 28 genes of this term that were significantly upregulated by DEL-1–Fc (Figure 7B). Other significantly upregulated genes, Xcl1, Havcr, Icosl, Bcl6, Tnfsf14, Cd1d1, Car11, Egr3, Cd27, Cd24a, Lgals8, Coro1a, Adk, Slamf1, Runx1, Myb, Cd28, and Spn (Figure 7B), are either highly expressed in Tregs or are closely related to their stability or suppressive function (1, 2, 6). Importantly, RUNX1 is pivotal for the suppressive function of nTregs and iTregs by modulating the expression and stability of FOXP3 or by physically interacting with FOXP3 for regulating Treg-associated target genes (48–51). The DEL-1–Fc–induced increase in Runx1 mRNA expression was validated by qPCR (Figure 7C), while FACS analysis showed that DEL-1–Fc also significantly promoted RUNX1 protein abundance within FOXP3+ iTregs (Figure 7D). Importantly, DEL-1–induced Runx1 upregulation absolutely required an intact RGD motif on DEL-1 (Figure 7C), as seen with Foxp3 upregulation (Figure 3E), thus suggesting a common, αvβ3 integrin–dependent pathway for the regulation of both Runx1 and Foxp3 by DEL-1. In contrast, DEL-1–Fc did not affect the expression of Runx3 (Figure 7C), another transcription factor involved in the establishment of lineage specification of T cells (51–53). The ability of RUNX1 to bind specific DNA consensus sequences is stabilized by associating with a non–DNA-binding cofactor, the core-binding factor β (CBFβ) (54). Interestingly, DEL-1–Fc also significantly promoted Cbfb expression in Tregs in a manner dependent on an intact RGD motif on DEL-1 (Figure 7C).
Figure 7. RUNX1 is required for the ability of DEL-1 to induce Treg differentiation.
(A and B) Naive splenic CD4+ cells from WT mice were differentiated into Tregs in the presence of DEL-1–Fc or Fc control for 3 days. RNA-seq analysis of the Treg transcriptome is presented as expression in the DEL-1–Fc group relative to the Fc control (log2 values) (n = 3 biological repeats/group). (A) MA (left) and volcano (right) plots show the distribution of gene expression. (B) Heatmaps show the significantly regulated genes (FDR < 0.05) annotated with ontology terms (Gene Ontology [GO] and PANTHER protein class). (C) Relative mRNA expression of Runx1, Runx3, and Cbfb in Tregs in in vitro culture on day 3 by qPCR (n = 6 replicates; 2 separate cell isolations). (D) MFI of RUNX1 in RUNX1+FOXP3+ iTregs (n = 6 replicates; 2 separate cell isolations). (E and F) Naive splenic CD4+ cells from CD4-Cre+ Runx1fl/fl mice or CD4-Cre– Runx1fl/fl littermate controls were differentiated, or not, into Tregs in the presence of Fc control or DEL-1–Fc for 4 days. (E) FACS plots and (F) percentage of FOXP3+ cells in CD4+ T cells from the in vitro culture system on day 4 (n = 6 replicates; 2 separate cell isolations). (G–L) Groups of littermate Del1WT CD4-Cre+ Runx1fl/fl and Del1KO CD4-Cre+ Runx1fl/fl mice were subjected to ligature-induced periodontitis for 10 days and ligatures were removed on day 10 for 5 days. FACS plots of Tregs (top) and Th17 cells (bottom) in gingival tissues (G) and cLNs (J) and percentages and absolute numbers of Tregs (H and K) and Th17 cells as well as Treg/Th17 cell ratio (I and L) in gingival tissues (H and I) and cLNs (K and L) of littermate Del1WT CD4-Cre+ Runx1fl/fl and Del1KO CD4-Cre+ Runx1fl/fl mice on day 15 (n = 6 mice/group). Data are means ± SD and are pooled from 2 independent experiments. ****P < 0.0001 by 1-way ANOVA with Dunnett’s post hoc test for comparison with Fc control (C) or 2-tailed Student’s t test (D, F, H, I, K, and L). NS, not significant.
PANTHER GO enrichment analysis based on significantly downregulated genes also revealed high enrichment of “alpha-beta T cell differentiation GO: 0046632” (fold enrichment 4.86, FDR = 0.000136) (Figure 7B) and “Regulation of lymphocyte apoptotic process GO: 0070228” (fold enrichment 3.99, FDR = 0.00143) (Figure 7B). Interestingly, in “alpha-beta T cell differentiation GO:0046632,” the genes downregulated by DEL-1, Rsad2, Gata3, Sema4a, and Irf4, are required for Th2 cell differentiation; Ifng encodes a Th1 signature cytokine (IFN-γ) and Gadd45 g is important for Th1 induction and function; Tnfsf8, Stat3, Ptger4, and Ly9 control Th17 cell differentiation. Among 14 downregulated genes of “Regulation of lymphocyte apoptotic process GO: 0070228,” Ccl5, Cd44, Bbc3, Hif1a, Tnfrsf4, and Myc have been reported to promote T cell apoptosis, suggesting that DEL-1–Fc might suppress apoptosis of Tregs.
To test the hypothesis that RUNX1 may link the DEL-1–αvβ3 integrin interaction to FOXP3 upregulation and Treg induction, we generated mice with T cell–specific conditional deletion of RUNX1 (CD4-Cre+ Runx1fl/fl mice). In vitro Treg differentiation assays showed that deletion of RUNX1 from T cells resulted in lower yield of Tregs (by ~50%) as compared with RUNX1-expressing controls, i.e., naive CD4+ T cells from CD4-Cre– Runx1fl/fl mice (Figure 7, E and F), thus confirming the importance of RUNX1 in Treg differentiation. Importantly, compared with Fc control, DEL-1–Fc failed to induce FOXP3+ Tregs in the cultures of CD4-Cre+ Runx1fl/fl CD4+ cells, whereas it readily increased the numbers of FOXP3+ cells in RUNX1-expressing CD4+ cell cultures (Figure 7, E and F). Thus, RUNX1 is required for the ability of DEL-1 to promote Treg induction in vitro.
To determine whether RUNX1 is required in vivo for the capacity of DEL-1 to increase the abundance of Tregs in the LIP resolution model, we generated Del1WT and Del1KO littermates in the CD4-Cre+ Runx1fl/fl background. Consistent with the role of RUNX1 in Treg induction (48–51), we noticed that the absolute numbers of FOXP3+ Tregs in the gingival tissue of mice with T cell–specific deletion of RUNX1 were reduced compared with WT mice expressing RUNX1 in T cells (Figure 7H [Del1WT CD4-Cre+ Runx1fl/fl mice] vs. Figure 1B [WT mice]); such reduction was even more obvious in the cLNs (Figure 7K [Del1WT CD4-Cre+ Runx1fl/fl mice] vs. Figure 1D [WT mice]). In contrast, the percentage of FOXP3+ Tregs was remarkably increased in the absence of RUNX1 from T cells (compare Figure 7, H and K [Del1WT CD4-Cre+ Runx1fl/fl] and Figure 1, B and D [WT]). This seeming paradox is explained by the established function of RUNX1 in T cell development and homeostasis (55, 56); indeed, T cell–specific deletion of RUNX1 predominantly affects the development of FOXP3−CD4+ Tconv cells (55). In our earlier experiments using mice with RUNX1-expressing CD4+ T cells, endogenous DEL-1 increased the absolute numbers of FOXP3+ cells in the gingival tissue and the cLNs at the resolution phase (Figure 1, B and D). In stark contrast, endogenous DEL-1 in mice with T cell–specific deletion of RUNX1 failed to increase the absolute numbers of Tregs in the gingival tissue (Figure 7H) and the draining cLNs (Figure 7K). Specifically, no difference was observed in Treg numbers in the gingival tissue and the cLNs between DEL-1–sufficient and –deficient mice in the absence of T cell RUNX1 (Figure 7, G, H, J, and K). Consistent with this finding and data showing that DEL-1 does not directly influence Th17 differentiation (Figure 3, A and B, and Supplemental Figure 6), no differences were observed between Del1WT CD4-Cre+ Runx1fl/fl and Del1KO CD4-Cre+ Runx1fl/fl mice regarding Th17 cell frequencies, absolute numbers, or the Treg/Th17 cell balance, in either the gingival tissue (Figure 7, G and I) or the cLNs (Figure 7, J and L). Overall, our data show that, during inflammation resolution, DEL-1 increases the abundance of Tregs via an αvβ3/RUNX1/FOXP3 axis.
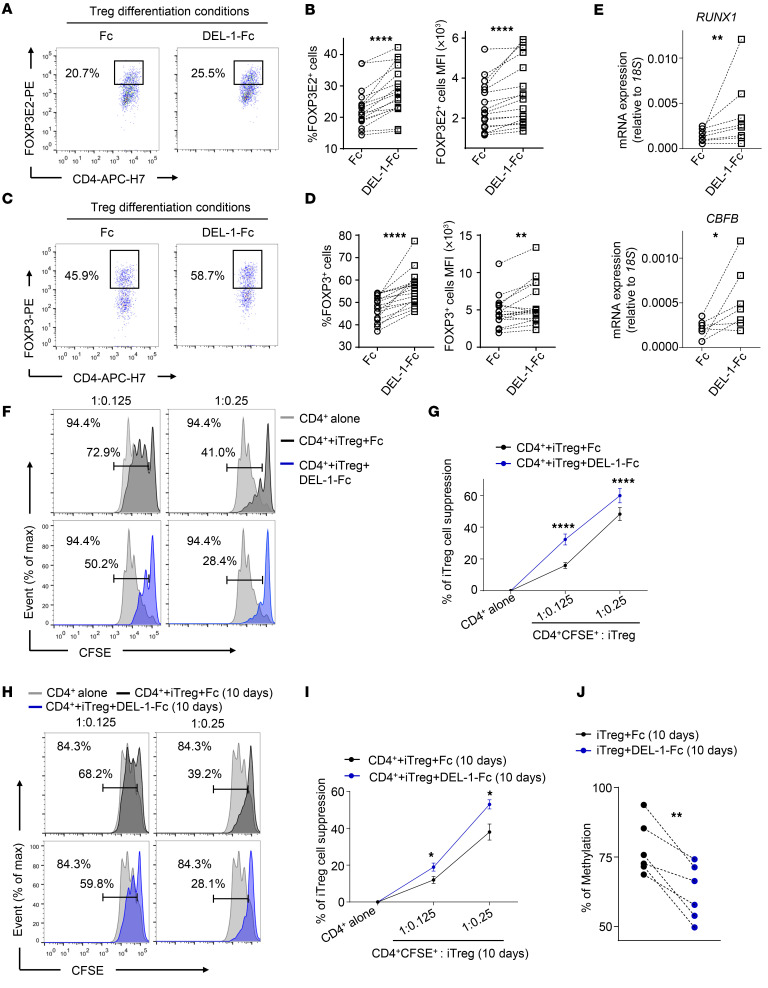
DEL-1 promotes human Treg differentiation and function.
Next, we evaluated whether DEL-1 could directly influence the differentiation of human CD4+CD25– Tconv cells into iTregs upon suboptimal stimulation via the T cell antigen receptor (TCR) (57, 58). In contrast to mice, multiple FOXP3 splicing variants have been described in humans and their role is now becoming increasingly clear (59, 60). Among them, we evaluated the induction of the splice variants containing exon 2 (FOXP3E2), which is required for proper Treg function (58–62). We observed that DEL-1–Fc treatment increased the expression levels of FOXP3E2 and of all the other FOXP3 splice variants during iTreg induction, both as percentage of positive cells and MFI (Figure 8, A–D). Consistently, DEL-1–Fc treatment also upregulated the mRNA expression of FOXP3E2 and FOXP3 transcripts during iTreg induction (Supplemental Figure 11). In agreement with the data obtained in the mouse system (Figure 7C), DEL-1–Fc significantly increased the mRNA expression levels of RUNX1 and CBFB in human Tconv cells stimulated with anti-CD3/anti-CD28 for 24 hours, as compared with treatment with Fc control (Figure 8E). Importantly, flow-sorted iTregs generated in a 36-hour culture in the presence of DEL-1–Fc exhibited significantly higher suppressive function than in the presence of Fc control, as evidenced by increased inhibition of the proliferation of CSFE+CD4+ T cells after 72 hours of coculture (Figure 8, F and G). We further asked whether DEL-1–Fc could also influence the function of nTregs freshly isolated from peripheral blood. DEL-1–Fc treatment did not affect the proliferation of TCR-stimulated nTregs, or of Tconv cells, as compared with Fc control (Supplemental Figure 12, A and B). Moreover, DEL-1–Fc treatment did not affect the suppressive function of nTregs, evaluated as their ability to suppress Tconv cell proliferation in coculture experiments (Supplemental Figure 12C).
Figure 8. DEL-1 increases FOXP3E2 expression and the suppressive capacity of human iTregs.
(A–E) Human Tconv cells were stimulated with anti-CD3/anti-CD28 (0.1 beads/cell) in the presence of DEL-1–Fc or Fc control for 36 hours. (A and C) Representative FACS plots of FOXP3E2+ cells (A) or FOXP3+ cells (C) in CD4+CD25+ cells. (B and D) Percentage (left) and MFI (right) of FOXP3E2+ cells (B) or FOXP3+ cells (D) in CD4+CD25+ cells (n = 17 from 13 [A and B] or 11 [C and D] independent experiments). (E) Relative mRNA expression of RUNX1 (up) (n = 8 from 8 independent experiments) and CBFB (bottom) (n = 7 from 7 independent experiments) measured at 24 hours of Tconv cell stimulation during iTreg generation. (F–I) CFSE-labeled CD4+ T cells were cultured for 72 hours with anti-CD3/anti-CD28 (0.2 beads/cell) alone or in the presence of FACS-isolated DEL-1–Fc–iTregs or Fc control–iTregs (F and G), or long-term-cultured (10 days) DEL-1–Fc–iTregs or Fc control–iTregs (H and I). Representative FACS plots of proliferation of CFSE-labeled CD4+ T cells. Numbers in plots indicate percentage of CFSE dilution in CD4+ T cells cultured alone (top left); numbers above bracketed lines indicate percentage of CFSE dilution in CD4+ T cells cultured with either FACS-isolated DEL-1–Fc–iTregs or Fc control–treated iTregs (F), or long-term-cultured (10 days) DEL-1–Fc–iTregs or Fc control–iTregs (H). Percentage of iTreg suppression in the above conditions (G, n = 29 from 5 independent experiments; I, n = 12 from 4 independent experiments). (J) Methylation of CpG island of FOXP3 CNS2 evaluated in DEL-1–Fc–iTregs or Fc control–iTregs cultured for 10 days in the presence anti-CD3/anti-CD28 (0.1 beads/cell) and IL-2 (50 IU/mL) (n = 6 from 4 independent experiments). (B, D, E, and J) Lines connect paired data for each individual. (G and I) Data are means ± SEM. *P < 0.05, **P < 0.01, ****P < 0.0001 vs. Fc control (B, D, E, G, I, and J) by 2-tailed, paired Wilcoxon’s test (B, D, E, G, and I) or 2-tailed, paired Student’s t test (J).
We next evaluated the functional stability of iTregs generated in the presence of DEL-1–Fc (DEL-1–Fc−iTregs) or Fc control (Fc-iTregs). To this end, iTregs were generated as described above, FACS isolated after 36 hours, and cultured with anti-CD3/anti-CD28 and IL-2 for 10 days. After this period, iTregs were FACS isolated and tested for their suppressive capacity in coculture with CSFE+CD4+ T cells for 72 hours. We found that DEL-1–Fc−iTregs exhibited significantly stronger suppressive activity against CD4+ T cell proliferation than Fc-iTregs (Figure 8, H and I). Thus, even after a prolonged culture, DEL-1–Fc−iTregs maintain significantly increased suppressive activity, consistent with the data showing that DEL-1–Fc can induce stable FOXP3 expression in mouse Tregs, thereby promoting their stability (Figure 3, G–I, and Supplemental Figure 8). Moreover, we also determined the methylation status of the FOXP3 TSDR in humans and found that 10-day iTregs that were generated in the presence of DEL-1–Fc exhibited a reduction in CNS2 methylation as compared with that seen in the Fc-iTregs (Figure 8J). In conclusion, the ability of DEL-1–Fc to increase FOXP3 expression during iTreg generation is paralleled by a significant induction of RUNX1 and CBFB and increased demethylation of the TSDR, which associate with increased stability and suppressive function of the generated iTregs.
Discussion
It is becoming increasingly appreciated that Tregs mediate activities beyond their originally characterized antigen-specific immunosuppressive function; for instance, Tregs engage in activities such as tissue repair and regeneration following inflammation resolution (63–68). In this regard, we have shown that a local tissue homeostatic factor that emerges during resolution, DEL-1, increases the stability and abundance of Tregs in mucosal tissues, thereby promoting their capacity for restoration of tissue homeostasis after infectious or inflammatory insults. Although TGF-β appears to be essential for iTreg development in peripheral tissues, iTregs seem to enhance the stability of their FOXP3 expression by responding to certain environmental stimuli, such as retinoic acid and bacterial short-chain fatty acids (69–71). Our present findings indicate that, at least in the oral mucosa and the lung, DEL-1 is an environmental cue that enhances the generation, functional stability, and suppressive activity of iTregs.
In both human and mouse models, DEL-1 induced the expression of RUNX1 and CBFβ, which are known to form a transcription complex that is indispensable for the suppressive function of Tregs. Indeed, the RUNX1-CBFβ heterodimer, which binds the CNS2 enhancer at the Foxp3 locus, regulates the level and stability of FOXP3 expression (49, 72). Moreover, the RUNX1-CBFβ complex physically interacts with FOXP3 and together regulate the expression of Treg-associated target genes (e.g., CD25, cytotoxic T lymphocyte antigen 4 [CTLA-4], and glucocorticoid-induced TNF receptor [GITR]) (48–51). Using DEL-1–proficient or –deficient mice with T cell–specific conditional deletion of RUNX1, we showed that RUNX1 is essential for the capacity of endogenous DEL-1 to increase the abundance of Tregs during inflammation resolution. This in vivo upregulatory effect is likely a direct DEL-1 effect on Tregs, since, in vitro, DEL-1 failed to induce FOXP3+ Tregs in T cell cultures that lack RUNX1.
DEL-1 might increase the abundance and function of Tregs not only via RUNX1 upregulation and FOXP3 expression but, as suggested by GO enrichment analysis, also by downregulating genes that could promote apoptosis of Tregs or restrain FOXP3 expression. In the latter regard, DEL-1 downregulated Eomes, which was shown to inhibit FOXP3 expression (73). Moreover, DEL-1 downregulated factors associated with differentiation toward Th effector cell subsets, such as Itk, a member of the Tec family of kinases that regulates Th2 and Th17 cytokine expression (74).
DEL-1 not only promoted the induction of FOXP3 expression in the mouse and human system, but also conferred stability of FOXP3 expression upon restimulation of Tregs in the absence of exogenous TGF-β1 in the mouse system. Moreover, even after prolonged culture (10 days), human iTregs generated in the presence of DEL-1 displayed significantly increased suppressive activity against CD4+ T cell proliferation (vs. Fc control−iTregs) together with a significant reduction in TSDR methylation of the CNS2 enhancer. The demethylation of CpG islands within the TSDR of the CNS2 enhancer allows CNS2 to bind critical transcription factors, such as the RUNX1-CBFβ complex and FOXP3, the binding of which is RUNX1-CBFβ dependent (41, 75, 76). The stable binding of FOXP3 and the RUNX1-CBFβ heterodimer to CNS2 upon demethylation of the TSDR further sustains TSDR demethylation, perhaps by physically blocking recruitment of DNA methyltransferases (41, 75, 76). RUNX1 may promote DNA demethylation also through its ability to recruit the DNA demethylation enzymes TET2 and TET3 (77). Besides a global effect on FOXP3 induction in human Tregs, DEL-1 also promoted the transcription of the FOXP3E2 splicing variants, key regulators of human Treg generation and function (60–64). The action of RUNX1 and CBFβ, which form a trimeric complex with FOXP3 at the CNS2 (51, 71), together with increased TSDR demethylation, could establish a feed-forward loop enabling and stabilizing FOXP3E2 expression.
It is thus conceivable that the promotional effect of DEL-1 on DNA demethylation of the FOXP3 TSDR, and hence on increasing the stability of FOXP3+ Tregs, may be mediated through its capacity to increase RUNX1 expression. This notion is consistent with the failure of endogenous DEL-1 in mice with T cell–specific deletion of RUNX1 to increase Treg abundance in the gingival tissue and the draining cLNs, whereas endogenous DEL-1 in WT mice significantly increased the abundance of Tregs (as compared with Del1KO mice) in the same tissues.
In addition to the direct effect of DEL-1 in promoting FOXP3 expression, an indirect effect might potentially be mediated through induction of TGF-β1, which is a potent inducer of FOXP3 expression (78). In this regard, we have previously shown that DEL-1 facilitates inflammation resolution by promoting the ability of macrophages to efferocytose apoptotic neutrophils, which in turn stimulates macrophage secretion of TGF-β1 (22). DEL-1–mediated efferocytosis requires the full-length molecule; the N-terminal EGF-like repeat region of DEL-1 is necessary to bind αvβ3 integrin–bearing macrophages, whereas its C-terminal discoidin-I–like domains are indispensable for binding the “eat-me” signal phosphatidylserine on the apoptotic neutrophil surface (22). Nevertheless, the N-terminal EGF-like repeat region of DEL-1 was sufficient to skew the Treg/Th17 cell balance toward Tregs during inflammation resolution. This further confirms the ability of DEL-1 to increase the abundance of Tregs through a mechanism that is not secondary to the enhancing effect of DEL-1–mediated efferocytosis on TGF-β1 production and inflammation resolution (22). DEL-1, secreted by macrophages or other resident cells, may thus promote Treg induction and function through both direct and indirect, synergistically operating mechanisms.
Infections and inflammatory or autoimmune diseases can cause instability of Tregs, mainly via induction of proinflammatory cytokines, such as IL-6, which destabilizes FOXP3 expression in Tregs in vitro (79) and reprograms FOXP3+ Tregs to produce IL-17 in vivo (80). In this context, fate mapping analysis showed that the frequency of FOXP3+ Treg–derived Th17 (exFoxp3Th17) cells increases dramatically in LIP in a manner strongly dependent on IL-6 (81). Conversely, Th17 cells can convert into FOXP3+ exTh17 cells with potent regulatory activity during resolution of inflammation in the presence of TGF-β1 (82). Therefore, DEL-1 may increase the abundance of Tregs during resolution by either contributing to their stability or by promoting the conversion of exTh17 cells into FOXP3+ Tregs. These effects could in part be mediated by the antiinflammatory effects of DEL-1 that include inhibition of IL-6 production and promotion of TGF-β1 production (19, 20, 22). However, it is clear from both in vitro and in vivo experiments of the present study that DEL-1 also exerts direct modulatory effects on Tregs. As discussed above, these direct effects of DEL-1 require only its N-terminal EGF-like repeat region and are distinct from its antiinflammatory or even its proefferocytic effects, which require the entire DEL-1 structure (22).
DEL-1 and IL-17 are reciprocally negatively regulated, which is consistent with findings that the tissue expression levels of DEL-1 are inversely correlated with those of IL-17 in both humans and animal models (19, 20, 22, 28, 30, 32, 83). However, although the signaling pathway whereby IL-17 downregulates DEL-1 is well understood at the molecular level (32), the mechanism whereby DEL-1 inhibits IL-17 expression has been uncertain. The present findings from the human studies suggest that one mechanism by which DEL-1 can inhibit IL-17 is by promoting the generation of FOXP3E2+ Tregs. These cells can strongly suppress Th17 cells, a major cellular source of IL-17 in different inflammatory pathologies, including periodontitis (37) and multiple sclerosis (84).
In conclusion, we established DEL-1 as a regulator of Tregs using human models of Treg differentiation and function and distinct mouse models of mucosal inflammation resolution. We have identified an αvβ3 integrin–dependent pathway for the regulation of both Runx1 and Foxp3 that not only provides improved mechanistic understanding of Treg regulation, but can also be targeted to regulate Treg function in inflammatory or autoimmune disorders.
Methods
Mice.
C57BL/6 Del1KO mice (21) and ItgalKO mice (The Jackson Laboratory) were crossed with WT C57BL/6 mice to generate KO and WT littermates. Mice with a conditional allele of Runx1 (Runx1tm3.1Spe; Runx1fl/fl) were provided by Nancy A. Speck (University of Pennsylvania) (85). Runx1fl/fl mice and CD4-Cre transgenic mice (The Jackson Laboratory) were crossed to generate CD4-Cre+ Runx1fl/fl and CD4-Cre– Runx1fl/fl control mice. Del1WT and Del1KO littermates in the CD4-Cre+ Runx1fl/fl background were generated through a series of breedings starting with the crossing of Del1KO mice with CD4-Cre+ Runx1fl/fl mice. Mice overexpressing DEL-1 in macrophages (CD68-Del1) and Del1RGE/RGE mice (which express a DEL-1 point mutant with an Asp-to-Glu substitution in the RGD motif) were previously described (22, 45). Intervention experiments with DEL-1−Fc, DEL-1 derivatives, or Fc control were performed in randomly assigned mice. Sex- and age-matched mice (8 to 10 weeks old) were used in experiments and were maintained in individually ventilated cages under specific pathogen–free conditions.
In vitro mouse T cell differentiation, restimulation, and suppressive assay.
All cytokines and antibodies used in these experiments were from BioLegend. Naive splenic CD4+ T cells were isolated using an EasySep Mouse Naive CD4+ T Cell Isolation Kit (STEMCELL). The purity of the isolated CD4+ T cells was 95%. The cells were activated by plate-bound anti-CD3 (clone 145-2C11, catalog 100314, BioLegend; 2 μg/mL) and anti-CD28 (clone 37.51, catalog 102111, BioLegend; 1 μg/mL) in the presence of different sets of cytokines in complete RPMI 1640 (Gibco), supplemented with 10% FBS (Atlanta Biologicals), 1% penicillin/streptomycin (Thermo Fisher Scientific), and 50 μM 2-mercaptoethanol (Thermo Fisher Scientific). For Treg differentiation, CD4+ T cells were incubated with TGF-β1 (5 ng/mL) and IL-2 (40 ng/mL) for 3 or 4 days (for analysis of Foxp3 mRNA or protein expression, respectively). For Th17 polarization under nonpathogenic conditions (40), CD4+ T cells were incubated with IL-6 (50 ng/mL) and TGF-β1 (1 ng/mL) for 3 days. For Th17 polarization under pathogenic conditions (40), CD4+ T cells were incubated with IL-6 (50 ng/mL), TGF-β1 (1 ng/mL), IL-1β (10 ng/mL), and IL-23 (10 ng/mL) for 3 days. Using an EasySep Mouse CD4+CD25+ Regulatory T Cell Isolation Kit II (STEMCELL), CD4+CD25+ cells were sorted to high purity (>95%) from Treg induction cultures (with or without DEL-1–Fc/Fc control), and then restimulated in the absence or presence of DEL-1–Fc or Fc control. Foxp3 expression in the restimulated cultures was assessed 4 days later. For Treg suppression assay, CD4+CD25– T cells (Tconv) were isolated using CD4 positive selection and CD25 negative selection (EasySep Mouse CD4+CD25+ Regulatory T Cell Isolation Kit II). The cells (90%–95% pure) were labeled with CFSE (5 μM; Invitrogen) and plated at 2.5 × 104 cells/well. Flow cytometry analyzing CFSE dilution was performed by gating on CD4+CFSE+ cells stimulated for 72 hours with anti-CD3 (2 μg/mL) and anti-CD28 (1 μg/mL), which were cultured alone or with purified DEL-1–Fc−iTregs or Fc-iTregs.
RNA-seq.
Naive splenic CD4+ cells isolated from 6-week-old WT mice were differentiated into Tregs for 3 days as described above in medium containing TGF-β1 (5 ng/mL), IL-2 (40 ng/mL), and DEL-1–Fc or Fc control. Three independent isolations from 3 mice were performed to attain 3 biological replicates. Total RNA was extracted by TRIzol (Life Technologies) and DNA was removed by using a TURBO DNA-free Kit (Invitrogen). PolyA-selected mRNA libraries were generated following the manufacturer’s protocols (BGI). Samples were sequenced on the BGISEQ-500 platform to generate 50-bp single-end reads with an average depth of 76.46 million reads per sample. Clean reads were mapped to the mouse genome (Ensembl assembly GRCm38) using STAR (86) with default settings after filtering low-quality, adaptor-polluted, and high content of unknown base (N) reads. The average mapping ratio with reference genome is 94.92%, the average mapping ratio with gene is 84.23%. A total of 17,294 genes were detected. Most transcripts were completely covered and reads were evenly distributed throughout the transcript.
Bioinformatic analysis of RNA-seq and GO analysis.
Read counts in each gene were calculated by htseq-count (87). Subsequently, differential gene expression was analyzed using DEGseq2 (88) and significantly differentially expressed genes (DEGs) were defined by FDR-adjusted P value less than 0.05 in the DEL-1–Fc–treated group relative to their expression in the Fc control group. Significantly up- or downregulated DEGs were subjected to GO enrichment analyses using PANTHER (Protein ANalysis THrough Evolutionary Relationships) (http://www.pantherdb.org) with default background and default threshold. Significantly enriched Biological Process GO terms were defined by FDR-adjusted P value less than 0.05. Volcano plots, MA plots, and heatmaps were generated by customized R script.
RNA-seq data generated in this study (Figure 7, A and B) were deposited in the NCBI’s Gene Expression Omnibus (GEO) under accession number GSE131315 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131315).
Human subjects, cell purification, and iTreg induction.
We isolated human peripheral Treg (CD4+CD25+CD127–) and Tconv (CD4+CD25–) cells from buffy coats of healthy subjects. Briefly, after Ficoll-Hypaque gradient centrifugation (GE Healthcare), peripheral Treg (CD4+CD25+CD127–) and Tconv (CD4+CD25–) cells were purified (90%–95% pure) by magnetic cell separation with a Regulatory CD4+CD25+ T Cell Kit (Thermo Fisher Scientific). For proliferation assays, Treg and Tconv cells were cultured (1 × 104 cells/well) for 72 hours in round-bottom, 96-well plates (Corning Falcon) in RPMI 1640 medium supplemented with 5% AB human serum (EuroClone) and stimulated with anti-CD3/anti-CD28 mAb–coated beads (0.2 beads/cell; Thermo Fisher Scientific), in the presence of DEL-1–Fc or Fc control (10 μg/mL). After 60 hours, [3H]thymidine (0.5 μCi/well; Amersham Pharmacia Biotech) was added to the cell cultures, and cells were harvested 12 hours later. Radioactivity was measured with a β cell-plate scintillation counter (Wallac). For the generation of iTregs, Tconv cells were cultured (2 × 106 cells/well) in flat-bottom 6-well plates (Corning Falcon) with RPMI 1640 medium supplemented with penicillin (100 UI/mL), streptomycin (100 μg/mL), and 5% AB human serum and stimulated with anti-CD3/anti-CD28 mAb–coated beads (0.1 beads/cell), for 12, 24, or 36 hours, in the presence of DEL-1–Fc or Fc control (10 μg/mL). iTregs were obtained by flow cytometric sorting of CD4+CD25hi (cell purity >95%) with a BD FACSJazz (Becton-Dickinson). For long-term induction, iTregs were generated as described above, FACS isolated after 36 hours as CD4+CD25hi (cell purity >95%) using a BD FACSJazz, and cultured (5 × 105 cells/well) in the presence of anti-CD3/anti-CD28 mAb–coated beads (0.1 beads/cell) and human recombinant IL-2 (50 IU/mL; Roche) in flat-bottom 48-well plates (Corning Falcon) with RPMI 1640 medium supplemented with penicillin (100 UI/mL), streptomycin (100 μg/mL), and 5% AB human serum, for 10 days. After this period, we evaluated the methylation status of the CpG island within the CNS2 region of FOXP3 through methyl-sensitive PCR. Also, 10-day iTregs were FACS isolated with BD FACSJazz (Becton-Dickinson) as the population with the highest CD25 expression (CD4+CD25hi; cell purity >95%) and tested for their suppressive capacity in coculture with CSFE+CD4+ T cells for 72 hours.
For further technical information, see Supplemental Methods.
Statistics.
After testing for normality, data were analyzed by parametric or nonparametric tests. Parametric tests included 2-tailed unpaired or paired Student’s t tests (2-group comparisons), 1-way ANOVA (multiple-group comparisons) followed by a multiple comparison test (Dunnett’s or Tukey’s as appropriate), and 2-way ANOVA with Holm-Šidák’s multiple-comparisons test. Two-tailed Mann-Whitney U test was used to analyze non-normally distributed unpaired sample data. Two-tailed paired Wilcoxon’s test was used to analyze non-normally distributed paired sample data. All statistical analyses were performed using GraphPad Prism software. P values less than 0.05 were considered statistically significant.
Study approval.
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania and were performed in compliance with institutional, state, and federal policies. Human study was approved by the Institutional Review Board of the Università degli Studi di Napoli “Federico II.” Buffy coats were collected from 41 healthy subjects (18 males and 23 females) after signing written informed consent approved by the Institutional Review Board. All subjects were 20 years of age or older (20–55 years old) with no history of inflammatory, endocrine, or autoimmune disease. All in vitro and in vivo experiments were performed 2 or more times for verification. No samples (mouse or human) were excluded from analysis.
Author contributions
XL designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. AC designed and performed experiments and analyzed and interpreted data. LK and CF performed experiments and analyzed data. TK, HW, and JHL generated reagents and performed experiments. KB generated critical genetic tools and interpreted data. KJC and XY analyzed data. AP and SDS performed experiments and interpreted data. GM contributed to experimental design and interpreted data. TC designed experiments, interpreted data, and critically revised the manuscript. VDR and GH conceived and designed the study, supervised research, interpreted data, and wrote the manuscript. All authors read the manuscript and offered comments. Authorship order between co–first authors was determined by coin flipping.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (DE024153, DE024716, and DE029436 to GH; DE028561 and DE026152 to GH and TC; NS091793 to KB), the German Research Foundation (SFB1181 to TC), the European Research Council (DEMETINL to TC), Fondazione Italiana Sclerosi Multipla (FISM no. 2018/R/4 to VDR, FISM n. 2016/R/18 and FISM no. 2018/S/5 to GM), the Università degli Studi di Napoli “Federico II” (STAR Program Linea 1-2018 funded by UniNA and by Compagnia di San Paolo to VDR), Ministry of Health (Bando Ricerca Finalizzata 2016 (GR-2016-02363725 to VDR) and Ministry of Education, University and Research (MIUR) (Bando PRIN 2017 Prot. 2017K7FSYB to VDR and Bando PRIN 2017 Prot. 2017K55HLC to GM) and Telethon (no. GGP 17086 to GM).
Version 1. 08/20/2020
In-Press Preview
Version 2. 10/26/2020
Electronic publication
Version 3. 12/01/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(12):6261–6277.https://doi.org/10.1172/JCI137530.
Contributor Information
Xiaofei Li, Email: xiaofeil@upenn.edu.
Alessandra Colamatteo, Email: alessandracolamatteo@gmail.com.
Lydia Kalafati, Email: Lydia.Kalafati@uniklinikum-dresden.de.
Tetsuhiro Kajikawa, Email: kajikawa@upenn.edu.
Hui Wang, Email: hwang02@upenn.edu.
Jong-Hyung Lim, Email: limhyung@upenn.edu.
Khalil Bdeir, Email: bdeir@mail.med.upenn.edu.
Kyoung-Jin Chung, Email: Kyoung-Jin.Chung@uniklinikum-dresden.de.
Xiang Yu, Email: yuxiang8605@gmail.com.
Clorinda Fusco, Email: c.fusco@ieos.cnr.it.
Antonio Porcellini, Email: antonio.porcellini@unina.it.
Salvatore De Simone, Email: saldesimone@libero.it.
Giuseppe Matarese, Email: giuseppe.matarese@cnr.it.
Triantafyllos Chavakis, Email: triantafyllos.chavakis@uniklinikum-dresden.de.
Veronica De Rosa, Email: veronica.derosa@cnr.it.
George Hajishengallis, Email: geoh@upenn.edu.
References
- 1.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327(5969):1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu L, Barbi J, Pan F. The regulation of immune tolerance by FOXP3. Nat Rev Immunol. 2017;17(11):703–717. doi: 10.1038/nri.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khader SA, Gaffen SL, Kolls JK. Th17 cells at the crossroads of innate and adaptive immunity against infectious diseases at the mucosa. Mucosal Immunol. 2009;2(5):403–411. doi: 10.1038/mi.2009.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 5.Weaver CT, Elson CO, Fouser LA, Kolls JK. The Th17 pathway and inflammatory diseases of the intestines, lungs, and skin. Annu Rev Pathol. 2013;8:477–512. doi: 10.1146/annurev-pathol-011110-130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133(5):775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 7.D’Alessio FR, et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J Clin Invest. 2009;119(10):2898–2913. doi: 10.1172/JCI36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Paiva R, Flavell RA. Harnessing the power of regulatory T-cells to control autoimmune diabetes: overview and perspective. Immunology. 2018;153(2):161–170. doi: 10.1111/imm.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L, et al. Active players in resolution of shock/sepsis induced indirect lung injury: immunomodulatory effects of Tregs and PD-1. J Leukoc Biol. 2014;96(5):809–820. doi: 10.1189/jlb.4MA1213-647RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 11.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 12.Bennett CL, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27(1):20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 13.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10(7):490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 14.Liu G, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10(7):769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strober W. Vitamin A rewrites the ABCs of oral tolerance. Mucosal Immunol. 2008;1(2):92–95. doi: 10.1038/mi.2007.22. [DOI] [PubMed] [Google Scholar]
- 16.Ghoreishi M, Bach P, Obst J, Komba M, Fleet JC, Dutz JP. Expansion of antigen-specific regulatory T cells with the topical vitamin D analog calcipotriol. J Immunol. 2009;182(10):6071–6078. doi: 10.4049/jimmunol.0804064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clambey ET, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci U S A. 2012;109(41):E2784–2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajishengallis G, Chavakis T. Endogenous modulators of inflammatory cell recruitment. Trends Immunol. 2013;34(1):1–6. doi: 10.1016/j.it.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin J, et al. DEL-1 restrains osteoclastogenesis and inhibits inflammatory bone loss in nonhuman primates. Sci Transl Med. 2015;7(307):307ra155. doi: 10.1126/scitranslmed.aac5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskan MA, et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol. 2012;13(5):465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi EY, et al. Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science. 2008;322(5904):1101–1104. doi: 10.1126/science.1165218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kourtzelis I, et al. DEL-1 promotes macrophage efferocytosis and clearance of inflammation. Nat Immunol. 2019;20(1):40–49. doi: 10.1038/s41590-018-0249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajishengallis G, Chavakis T. DEL-1-regulated immune plasticity and inflammatory disorders. Trends Mol Med. 2019;25(5):444–459. doi: 10.1016/j.molmed.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chavakis T, Mitroulis I, Hajishengallis G. Hematopoietic progenitor cells as integrative hubs for adaptation to and fine-tuning of inflammation. Nat Immunol. 2019;20(7):802–811. doi: 10.1038/s41590-019-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziogas A, et al. DHEA inhibits leukocyte recruitment through regulation of the integrin antagonist DEL-1. J Immunol. 2020;204(5):1214–1224. doi: 10.4049/jimmunol.1900746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hidai C, et al. Cloning and characterization of developmental endothelial locus-1: an embryonic endothelial cell protein that binds the alphavbeta3 integrin receptor. Genes Dev. 1998;12(1):21–33. doi: 10.1101/gad.12.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanayama R, Tanaka M, Miwa K, Nagata S. Expression of developmental endothelial locus-1 in a subset of macrophages for engulfment of apoptotic cells. J Immunol. 2004;172(6):3876–3882. doi: 10.4049/jimmunol.172.6.3876. [DOI] [PubMed] [Google Scholar]
- 28.Choi EY, et al. Developmental endothelial locus-1 is a homeostatic factor in the central nervous system limiting neuroinflammation and demyelination. Mol Psychiatry. 2015;20(7):880–888. doi: 10.1038/mp.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang YY, Kim DY, Lee SH, Choi EY. Deficiency of developmental endothelial locus-1 (Del-1) aggravates bleomycin-induced pulmonary fibrosis in mice. Biochem Biophys Res Commun. 2014;445(2):369–374. doi: 10.1016/j.bbrc.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Yan S, et al. Developmental endothelial locus-1 (Del-1) antagonizes interleukin-17-mediated allergic asthma. Immunol Cell Biol. 2018;96(5):526–535. doi: 10.1111/imcb.12023. [DOI] [PubMed] [Google Scholar]
- 31.Kourtzelis I, et al. Developmental endothelial locus-1 modulates platelet-monocyte interactions and instant blood-mediated inflammatory reaction in islet transplantation. Thromb Haemost. 2016;115(4):781–788. doi: 10.1160/TH15-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maekawa T, et al. Antagonistic effects of IL-17 and D-resolvins on endothelial Del-1 expression through a GSK-3β-C/EBPβ pathway. Nat Commun. 2015;6:8272. doi: 10.1038/ncomms9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klann JE, et al. Integrin activation controls regulatory T cell-mediated peripheral tolerance. J Immunol. 2018;200(12):4012–4023. doi: 10.4049/jimmunol.1800112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worthington John J, et al. Integrin αvβ8-mediated TGF-β activation by effector regulatory T cells is essential for suppression of T-cell-mediated inflammation. Immunity. 2015;42(5):903–915. doi: 10.1016/j.immuni.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega-Gomez A, Perretti M, Soehnlein O. Resolution of inflammation: an integrated view. EMBO Mol Med. 2013;5(5):661–674. doi: 10.1002/emmm.201202382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proto JD, et al. Regulatory T cells promote macrophage efferocytosis during inflammation resolution. Immunity. 2018;49(4):666–677 e666. doi: 10.1016/j.immuni.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dutzan N, et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med. 2018;10(463):eaat0797. doi: 10.1126/scitranslmed.aat0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol. 2016;16(3):149–163. doi: 10.1038/nri.2015.18. [DOI] [PubMed] [Google Scholar]
- 39.Okeke EB, Uzonna JE. The pivotal role of regulatory T cells in the regulation of innate immune cells. Front Immunol. 2019;10:680. doi: 10.3389/fimmu.2019.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13(10):991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polansky JK, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38(6):1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 42.Floess S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5(2):e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanamori M, Nakatsukasa H, Okada M, Lu Q, Yoshimura A. Induced regulatory T cells: their development, stability, and applications. Trends Immunol. 2016;37(11):803–811. doi: 10.1016/j.it.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Mitroulis I, et al. Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J Clin Invest. 2017;127(10):3624–3639. doi: 10.1172/JCI92571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuh DY, et al. The secreted protein DEL-1 activates a β3 integrin-FAK-ERK1/2-RUNX2 pathway and promotes osteogenic differentiation and bone regeneration. J Biol Chem. 2020;295(21):7261–7273. doi: 10.1074/jbc.RA120.013024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Penta K, Varner JA, Liaw L, Hidai C, Schatzman R, Quertermous T. Del1 induces integrin signaling and angiogenesis by ligation of alphaVbeta3. J Biol Chem. 1999;274(16):11101–11109. doi: 10.1074/jbc.274.16.11101. [DOI] [PubMed] [Google Scholar]
- 47.Mitroulis I, et al. Developmental endothelial locus-1 attenuates complement-dependent phagocytosis through inhibition of Mac-1-integrin. Thromb Haemost. 2014;111(5):781–1006. doi: 10.1160/TH14-03-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitoh A, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31(4):609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10(11):1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono M, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446(7136):685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 51.Klunker S, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206(12):2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–633. doi: 10.1016/S0092-8674(02)01111-X. [DOI] [PubMed] [Google Scholar]
- 53.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8(2):145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 54.Speck NA. Core binding factor and its role in normal hematopoietic development. Curr Opin Hematol. 2001;8(4):192–196. doi: 10.1097/00062752-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 55.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204(8):1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–633. doi: 10.1016/S0092-8674(02)01111-X. [DOI] [PubMed] [Google Scholar]
- 57.Walker MR, et al. Induction of FoxP3 and acquisition of T regulatory activity by stimulated human CD4+CD25– T cells. J Clin Invest. 2003;112(9):1437–1443. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Rosa V, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol. 2015;16(11):1174–1184. doi: 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith EL, Finney HM, Nesbitt AM, Ramsdell F, Robinson MK. Splice variants of human FOXP3 are functional inhibitors of human CD4+ T-cell activation. Immunology. 2006;119(2):203–211. doi: 10.1111/j.1365-2567.2006.02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mailer RKW. Alternative splicing of FOXP3-virtue and vice. Front Immunol. 2018;9:530. doi: 10.3389/fimmu.2018.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joly AL, Liu S, Dahlberg CI, Mailer RK, Westerberg LS, Andersson J. Foxp3 lacking exons 2 and 7 is unable to confer suppressive ability to regulatory T cells in vivo. J Autoimmun. 2015;63:23–30. doi: 10.1016/j.jaut.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Allan SE, et al. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115(11):3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arpaia N, et al. A distinct function of regulatory T cells in tissue protection. Cell. 2015;162(5):1078–1089. doi: 10.1016/j.cell.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, et al. Mesenchymal stem cell-based tissue regeneration is governed by recipient T lymphocytes via IFN-γ and TNF-α. Nat Med. 2011;17(12):1594–1601. doi: 10.1038/nm.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, Tan J, Martino MM, Lui KO. Regulatory T-cells: potential regulator of tissue repair and regeneration. Front Immunol. 2018;9:585. doi: 10.3389/fimmu.2018.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyagi AM, et al. The microbial metabolite butyrate stimulates bone formation via T regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018;49(6):1116–1131.e7. doi: 10.1016/j.immuni.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharma A, Rudra D. Emerging functions of regulatory T cells in tissue homeostasis. Front Immunol. 2018;9:883. doi: 10.3389/fimmu.2018.00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delacher M, et al. Genome-wide DNA-methylation landscape defines specialization of regulatory T cells in tissues. Nat Immunol. 2017;18(10):1160–1172. doi: 10.1038/ni.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33(3):313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 72.Dominguez-Villar M, Hafler DA. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19(7):665–673. doi: 10.1038/s41590-018-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lupar E, et al. Eomesodermin expression in CD4+ T cells restricts peripheral Foxp3 induction. J Immunol. 2015;195(10):4742–4752. doi: 10.4049/jimmunol.1501159. [DOI] [PubMed] [Google Scholar]
- 74.Gomez-Rodriguez J, Kraus ZJ, Schwartzberg PL. Tec family kinases Itk and Rlk/Txk in T lymphocytes: cross-regulation of cytokine production and T-cell fates. FEBS J. 2011;278(12):1980–1989. doi: 10.1111/j.1742-4658.2011.08072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ohkura N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37(5):785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki T, et al. RUNX1 regulates site specificity of DNA demethylation by recruitment of DNA demethylation machineries in hematopoietic cells. Blood Adv. 2017;1(20):1699–1711. doi: 10.1182/bloodadvances.2017005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlenner SM, Weigmann B, Ruan Q, Chen Y, von Boehmer H. Smad3 binding to the foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J Exp Med. 2012;209(9):1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang XO, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29(1):44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu L, Kitani A, Fuss I, Strober W. Cutting edge: regulatory T cells induce CD4+CD25-Foxp3- T cells or are self-induced to become Th17 cells in the absence of exogenous TGF-beta. J Immunol. 2007;178(11):6725–6729. doi: 10.4049/jimmunol.178.11.6725. [DOI] [PubMed] [Google Scholar]
- 81.Tsukasaki M, et al. Host defense against oral microbiota by bone-damaging T cells. Nat Commun. 2018;9(1):701. doi: 10.1038/s41467-018-03147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gagliani N, et al. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523(7559):221–225. doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Folwaczny M, Karnesi E, Berger T, Paschos E. Clinical association between chronic periodontitis and the leukocyte extravasation inhibitors developmental endothelial locus-1 and pentraxin-3. Eur J Oral Sci. 2017;125(4):258–264. doi: 10.1111/eos.12357. [DOI] [PubMed] [Google Scholar]
- 84.Hu D, et al. Transcriptional signature of human pro-inflammatory TH17 cells identifies reduced IL10 gene expression in multiple sclerosis. Nat Commun. 2017;8(1):1600. doi: 10.1038/s41467-017-01571-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Growney JD, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.