Abstract
While cancer stem cells are well established in certain hematologic and solid malignancies, their existence in T cell lymphoma is unclear and the origin of disease is not fully understood. To examine the existence of lymphoma stem cells, we utilized a mouse model of anaplastic large cell lymphoma. Established NPM-ALK+ lymphomas contained heterogeneous cell populations ranging from mature T cells to undifferentiated hematopoietic stem cells. Interestingly, CD4−/CD8− double negative (DN) lymphoma cells aberrantly expressed the T cell receptor α/β chain. Serial transplantation of sorted CD4/CD8 and DN lymphoma subpopulations identified lymphoma stem cells within the DN3/DN4 T cell population, whereas all other subpopulations failed to establish serial lymphomas. Moreover, transplanted lymphoma DN3/DN4 T cells were able to differentiate and gave rise to mature lymphoma T cells. Gene expression analyses unmasked stem-cell-like transcriptional regulation of the identified lymphoma stem cell population. Furthermore, these lymphoma stem cells are characterized by low CD30 expression levels, which might contribute to limited long-term therapeutic success in patients treated with anti-CD30-targeted therapies. In summary, our results highlight the existence of a lymphoma stem cell population in a NPM-ALK-driven CD30+ mouse model, thereby giving the opportunity to test innovative treatment strategies developed to eradicate the origin of disease.
Subject terms: Cancer stem cells, Oncogenes, Cancer models, T-cell lymphoma
Introduction
The identity of so-called cancer stem cells in malignant diseases is subject to debate [1–3]. As the clinical response of solid and hematological tumors to traditional treatments has been attributed to this small subpopulation of cells, the importance of characterizing them is crucial [4, 5]. The concept of a limited pool of tumor stem cells with capacity for self-renewal and plasticity is well established in certain malignancies such as breast cancer, colorectal cancer, brain cancer, and leukemia [6–10]. However, identification of the cell-of-origin of B cell lymphomas is limited and data on the understanding of initiation and maintaining of T cell lymphoma, especially for anaplastic large cell lymphoma (ALCL) are limited [11, 12]. ALCL belongs to a group of aggressive Non-Hodgkin lymphomas and was first described in 1985 by Stein et al., who characterized it as a cohesive proliferation of large pleomorphic cells expressing CD30 [13]. In 60% of patients a translocation t(2;5)(p23;q35) is found, which generates a fusion gene between nucleophosmin (NPM) and the receptor tyrosine kinase gene anaplastic lymphoma kinase (ALK), thereby leading to constitutive activation of the ALK tyrosine kinase [14, 15].
Molecular characterization of lymphoma stem cells and their activated pathways in ALCL may help in the design of targeted treatment strategies for the ALCL patient population [16–19]. It is a well-established principle, that therapies which eradicate the cancer stem cell population, assure higher long-term response with a lower recurrence rate than treatment regimens that focus on tumor mass reduction and may not affect cancer stem cells.
In this work, we aimed to identify and characterize the lymphoma stem cell population in a murine ALCL-like model [20]. Within this model, NPM-ALK is expressed in a lineage-specific manner in early T cells leading to characteristic CD30 expression and large cell cytology in a heterogeneous malignant T-cell population [20].
In this study, we identify and characterize a distinct, malignant T-cell subpopulation of CD30+ NPM-ALK driven lymphoma as well as their stem cell potency measured by their ability to establish lymphoma in serially transplanted mice.
Methods
Constructs, cell culture and virus generation
For virus production, Phoenix E ecotropic packaging cells (kind gift from G. Nolan, Stanford, CA) were transiently transfected with the retroviral construct MSCV-STOP-NPM-ALK-IRES-EGFP (MSNAIE) [20] and viral supernatants were collected as described previously [21, 22]. Retroviral titers were determined by transduction of NIH/3T3 cells (DSMZ) as described previously (Supplementary Fig. 1) [23].
Reagents
Recombinant murine Interleukin-3 (IL-3), IL-6, and SCF were purchased from R&D Systems (Minneapolis, MN, USA). Fetal calf serum, 5-Flouorouracil, and Polybrene was from Sigma-Aldrich (St. Louis, MO, USA). DMEM (Dulbecco’s Modified Eagle Medium) and ES cell FBS (fetal bovine serum) was from Thermo Scientific (Waltham, USA). Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA, USA).
Mice
The Lck-Cre mouse line (B6.Cg-Tg(Lck-cre)548Jxm/J) was obtained from Jackson Laboratory (Bar Harbor, Maine, USA). All mice were backcrossed to a C57BL/6 background for more than 10 generations. The animals were housed in a special caging system with autoclaved food and acidified water at the University of Freiburg. All procedures were performed in accordance with national and institutional guidelines for animal care and experiments.
Transplantation assays
Murine bone marrow (BM) was collected from Lck-Cre-transgenic and control mice and infected as described previously [21, 24]. Briefly, 12-week-old male donor mice were treated once with 5-Fluorouracil (150 mg/kg) at day −4 and BM cells were harvested from tibia and femur. After preincubation overnight in BM media (DMEM, 30% FBS, 10 ng/ml mIL-3, 10 ng/ml mIL-6, 50 ng/ml mSCF), BM cells were infected with retroviral supernatant as described previously [23]. Infection efficiency was determined by flow cytometric analysis of EGFP expression. Female C57BL/6 wildtype recipient mice were irradiated with 850 rad and transplanted with 5 × 104 infected cells/2 × 105 total cells via tail vein injection (Supplementary Fig. 2A). For serial transplantations, mice were irradiated sublethally with 450 rad and 2.5 or 1.5 × 103 EGFP positive cells were injected. Peripheral blood was taken at the indicated time points and WBCs were measured using an automated counter (ABC scil vet). Transplanted mice were monitored for signs of disease and sacrificed and analysed on observing clinical signs.
Flow cytometry analysis and cell sorting
Flow cytometric analysis was performed as described [25]. The BD LSR Fortessa and FACS Aria III (BD Biosciences, Heidelberg, Germany) were used for analyzing and sorting. Antibodies used to stain cell surface proteins were: antimouse CD4 (GK1.5), CD8a (53-6.7), CD11b (Mac-1, M1/70), CD25 (PC61.5), CD30 (mCD30.1), CD44 (IM7), CD45R/B220 (RA3-6B2), CD45 (30-F11), CD90.2 (THY1.2, 53-2.1), CD117 (c-KIT, 2B8), CD127 (IL-7Ra, A7R34), GR1 (Ly-6G, RB6-8C5), SCA1 (D7), TCR-α/β (IP26) and TER119 (TER119) and the corresponding isotypes, were obtained from BD Biosciences or eBiosciences (Frankfurt am Main, Germany). For intracellular staining cells were treated with Cytofix Buffer and Perm Buffer III (both from BD Biosciences) according to the manufacturer’s protocol. Antibodies used to perform intracellular staining were Ki67 (SolA15) (eBiosciences) and ALK (D5F3) (Cell signaling, Frankfurt am Main, Germany).
Gene expression analysis
RNA extraction was performed as described previously [25] as was generation of Microarray data [26]. Regulated genes were identified using a linear-based model, limma R package. The threshold for significance was set to an adjusted p value < 0.01 (Benjamini Hochberg).
Accession numbers
The accession number for the microarray data reported in this paper is GEO ID: GSE132267. Microarray data for comparison of ALCL, EL4 and Tx17 cells were submitted to Gene Omnibus database (GEO accession number pending).
Statistical analysis
A two-sided Student’s t test was used for statistical analyses. Mean ± standard deviation were analysed as indicated. The survival curves were produced using a log-rank (Mantel-Cox) test. P values were defined as indicated in figure legends: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
ALCL stem cells derive from CD4−/CD8− double negative lymphoma T cells
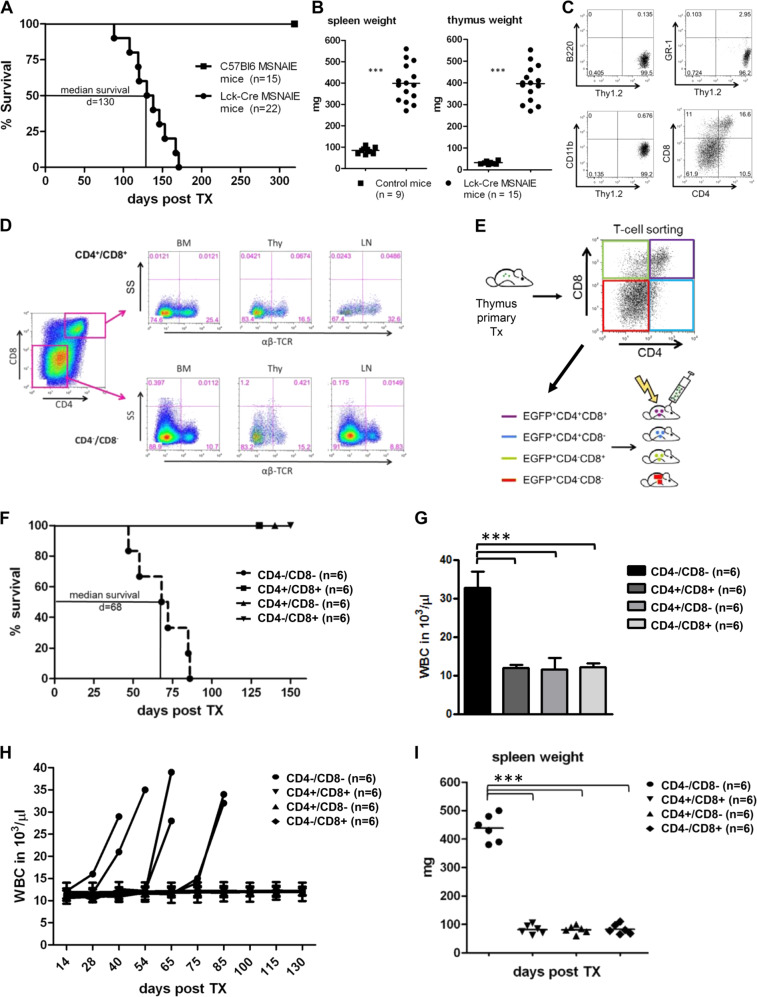
MSNAIE-infected, Lck-Cre BM transplanted, recipient mice develop ALCL-like malignancies with enlarged lymphatic organs (thymus, spleen, lymph nodes), BM infiltration, a distinct T cell phenotype and undifferentiated T cells within the EGFP positive lymphoma population (Fig 1a–c, Supplementary Figs. 1 and 2) [20]. Staining of the T cell subpopulations demonstrated high NPM-ALK expression in immature CD4−/CD8− double negative (DN) T cells and undifferentiated CD4+/CD8+ double positive T cells (Supplementary Fig. 3A). Immunophenotyping for the proliferation marker Ki-67 revealed high expression in the NPM-ALK+ T cells (Supplementary Fig. 3A). Comparison of transcriptional analyses demonstrated an upregulation of Bcl6 and Perforin in the ALCL-like lymphoma compared with other murine T cell lymphomas/leukemias (EL4 cell line and Notch-driven ALL), whereas this was not the case for Cebpb, Ptpn12, and Serpina1 (Supplementary Fig. 3B). Interestingly, the CD4−/CD8− DN lymphoma population aberrantly expressed the T cell receptor (TCR) α/β chain, which may allow these early T cells to establish a systemic lymphoma (Fig. 1d). Therefore, we hypothesized that the lymphoma stem cell population is contained within this early T cell population. To prove our hypothesis, we performed secondary transplantations with different lymphoma subpopulations depending on their CD4/CD8 T cell status. Therefore, we sorted primary ALCL-like lymphomas from the thymus for CD4 and CD8 expression (Fig. 1e) and transplanted 2500 cells of the isolated subpopulations into sublethally irradiated recipient animals. None of the CD4+/CD8+, CD4+/CD8− nor CD4−/CD8+ cell populations were able to induce lymphoma, whereas the CD4−/CD8− DN lymphoma population exclusively established T cell lymphoma in the secondary recipient mice with a median survival of 68 days (Fig. 1f). Similar to the primary transplanted animals, the serial transplanted animals developed significant splenomegaly, enlarged thymus, BM infiltration and increased white blood cell counts compared with animals transplanted with the other CD4/CD8 subpopulations (Fig. 1g–i). Flow cytometric analyses of lymphomas from CD4/CD8 DN lymphoma cell-transplanted mice showed high EGFP expression in all lymphatic organs and the BM (Supplementary Fig. 4), which indicates lymphoma induction via NPM-ALK expression.
Fig. 1. ALCL stem cells derive from CD4−/CD8− double negative lymphoma T cells.
a A Kaplan–Meier survival curve of primary transplanted mice. 50,000 EGFP positive MSNAIE Lck-Cre transgenic or wildtype BM cells were injected i.v. into lethally irradiated (8500 rad) C57Bl6 recipient mice. Median survival: 130 days. n (wildtype) = 15, n (Lck-Cre) = 22. b Comparison of spleen and thymi weights of control (n = 9) and MSNAIE Lck-Cre transplanted mice (n = 15). c Flow cytometric dot plots of EGFP positive cells in the thymus of one representative MSNAIE Lck-Cre BM transplanted mouse stained with anti-B220, anti-GR1, anti-CD11b, anti-Thy1.2, anti-CD4, and anti-CD8. d Flow cytometric dot plots of lymphatic organs and BM of primary MSNAIE Lck-Cre BM transplanted mice stained for anti-CD4, anti-CD8, and TCR α/β. e Sorting strategy for secondary transplantation of EGFP positive T cell subpopulations. Single cell suspensions of the thymus were stained with anti-CD4 and anti-CD8 and sorted for the four T cell subpopulations. f Kaplan–Meier survival curve of secondary transplanted mice. 2500 EGFP positive lymphoma cells (CD4/CD8 subpopulation) were injected i.v. into sublethally irradiated (4500 rad) C57Bl6 recipient mice. Median survival of CD4−/CD8− transplanted mice: 68 days n = 6. g White blood counts (WBC) at death of peripheral blood isolated from CD4−/CD8− transplanted recipients or at day 120 in case of the other CD4/8 subgroup. Six mice in each subgroup were analysed. h WBC counts over time of Tx of serial CD4/CD8 lymphoma subpopulation transplanted mice. Six mice in each subgroup were analysed. i Comparison of spleen weights of serial CD4/CD8 lymphoma subpopulation transplanted mice at death (CD4−/CD8−) or at the end of Tx (other subgroups). Six mice in each subgroup were analysed. ***p < 0.001. Data are represented as means ± SD.
These results highlight the existence of a lymphoma stem cell population in our murine ALCL-like, which has the ability to induce serial malignancies in mice and is CD4−/CD8− DN supportive of a primitive origin.
Serially transplanted CD4−/CD8− lymphoma cells differentiate into mature T cells
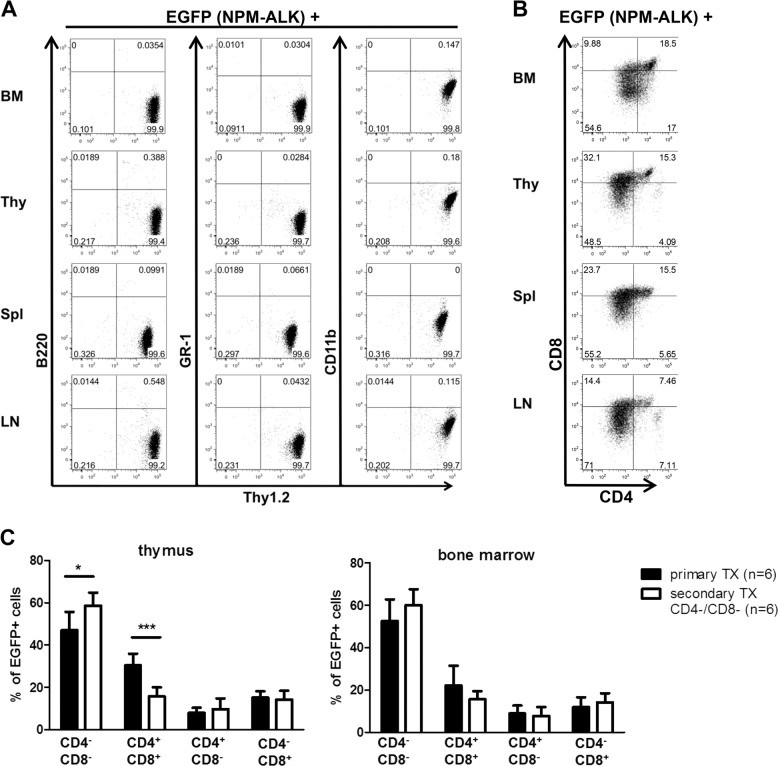
Further immunophenotypic characterization of lymphomas induced by serially transplantation of CD4−/CD8− T cells of primary murine ALCL-like lymphomas demonstrates a pure T cell phenotype (Fig. 2a). Interestingly, all of the CD4/CD8 T cell subpopulations: single positive CD4+/CD8−, CD4−/CD8+ as well as double positive CD4+/CD8+ cells, are detectable by flow cytometry in all organs analysed, although the main lymphoma population consists of DN T cells (Fig. 2b). The ability of the initially transplanted CD4−/CD8− T cells to differentiate in distinct CD4/CD8 T cell subpopulations supports the hypothesis of lymphoma stem cell presence within the CD4−/CD8− T cell subpopulation that maintains the ability to differentiate into mature T cells.
Fig. 2. Serially transplanted CD4−/CD8− lymphoma cells differentiate into mature T cells.
Representative flow cytometry profiles of single cell suspensions of lymphatic organs: thymus (Thy), spleen (Spl), lymph node (LN) and bone marrow (BM) of CD4−/CD8− lymphoma subpopulation transplanted mice. a EGFP+ lymphoma cells stained for anti-B220, anti-GR1, anti-CD11b, and anti-Thy1.2 show a clear T cell phenotype. b EGFP+ lymphoma cells stained for anti-CD4 and anti-CD8 demonstrate the ability to differentiate into mature T cell subpopulations. c Quantitative analysis of CD4/CD8 lymphoma T cell subpopulation distribution within the thymus and bone marrow of primary (black bars) and secondary (white bars) transplanted mice shows the differentiation capability of CD4−/CD8− DN subpopulations. Six mice in each subgroup were analysed. *p < 0.05; ***p < 0.001. Data are represented as mean + SD.
Quantitative analysis between the EGFP positive T cell lymphoma subpopulations of primary and CD4−/CD8− transplanted secondary mice exhibited slight differences in T cell distribution: thymic lymphomas developing in secondary CD4−/CD8− lymphoma T cell-transplanted mice contained a larger amount of CD4−/CD8− DN lymphoma cells (58.7% vs. 48.0%; p = 0.03) than the primary thymic lymphomas developing in the donors, whereas CD4+/CD8+ double positive lymphoma cells were represented to a greater extent in primary thymic lymphomas compared with those developing in secondary CD4−/CD8− transplanted mice (29.4% vs. 15.7%; p = 0.0007) (Fig. 2c, left panel). Analyses of BM from primary and secondary CD4−/CD8− lymphoma cell-transplanted animals revealed no differences in the CD4/CD8 distribution of lymphoma cells (Fig. 2c, right panel). Therefore, our data show, that serially transplanted CD4−/CD8− lymphoma stem cells have the potential to partly differentiate and give rise to mature T cell subpopulations.
Primary MSNAIE Lck-Cre induced lymphomas contain a heterogeneous cell population ranging from undifferentiated hematopoietic stem cells to mature T cells
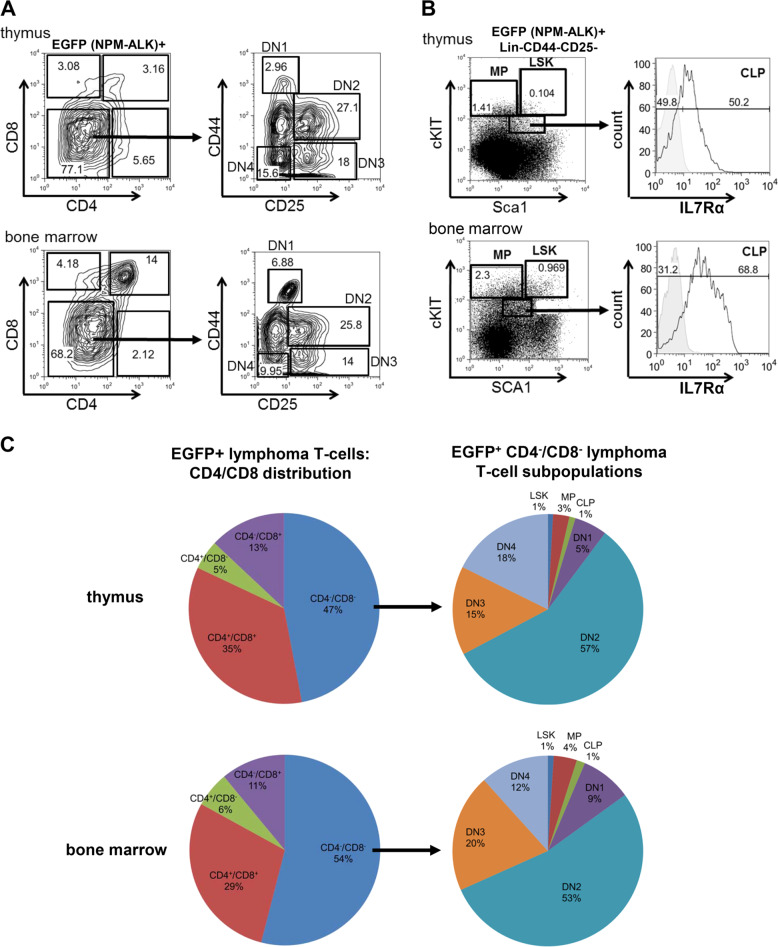
Having identified lymphoma stem cells in the CD4−/CD8− cell population of lymphomas developing in primary MSNAIE Lck-Cre mice, we set out to further characterize this distinct cell population. Hence, the CD4−/CD8− DN T cell lymphoma population of primary MSNAIE-infected Lck-Cre transgenic BM transplanted mice was comprehensively immunophenotyped. To differentiate in the EGFP (NPM-ALK) positive population between T cell DN stages (DN1–DN4) and immature LSK (lineage− SCA1+cKIT+), MP (multipotent progenitors, lineage− SCA1−cKIT+) and CLP (common lymphoid progenitor, lineage− SCA1low cKITlow Il7Rα+) populations, cells were stained for T cell markers (CD25 and CD44) as well as stem and progenitor cell markers (cKIT, SCA1, and Il7Rα) (Fig. 3a, b, Supplementary Fig. 5). As shown in Fig. 3c, the composition of primary EGFP+ thymic and BM lymphoma cells from MSNAIE-infected Lck-Cre transgenic BM transplanted mice was heterogeneous: regarding the CD4/CD8 distribution (Fig. 3c, left pie charts), the majority of cells belong to the CD4−/CD8− DN population, whereas all other CD4/CD8 subgroups were detected with CD4+/CD8− single positive cells being the minor population. Interestingly, in-depth immunophenotypic analyses of the CD4−/CD8− DN population (Fig. 3c, right pie charts) identified T cell DN stages 1–4 as well as lymphatic precursors (CLP), multipotent progenitors (MP), and hematopoietic stem cells (LSK). These results showed that the DN T cell pool is considerably heterogeneous and consists of different immature cell populations. It thereby points to a necessity to further define and specify the lymphoma stem cells within these immunophenotypically different cell populations.
Fig. 3. Primary MSNAIE Lck-Cre induced lymphomas contain a heterogeneous cell population ranging from undifferentiated hematopoietic stem cells to mature T cells.
a Representative flow cytometry profiles of EGFP+ thymus and BM cells of MSNAIE Lck-Cre transgenic BM transplanted mice. Cells were stained for anti-CD4, anti-CD8, anti-CD25, and anti-CD44. b Representative flow cytometry profiles of EGFP+ lineage negative (lin-) thymus and BM cells of MSNAIE Lck-Cre transgenic BM transplanted mice. Cells were stained for hematopoetic stem cell and lymphatic precursor proteins anti-SCA1, anti-cKIT, and anti-IL7Rα. LSK: lineage−SCA1+cKIT+; MP multipotent progenitors, lineage−SCA1−cKIT+; CLP common lymphoid progenitors, lineage−SCA1lowcKITlowIl7Rα+. c Pie chart of EGFP+ T cell subpopulations differentiated for CD4 and CD8 (left charts) of thymus and BM of five independent MSNAIE Lck-Cre transgenic BM transplanted mice. Distribution over the different cell types within the EGFP+ CD4−/CD8− DN lymphoma subpopulation are shown from five independent diseased animals on the right. CLP common lymphoid progenitors, lineage−SCA1lowcKITlowIl7Rα+, LSK lineage−SCA1+cKIT+, MP multipotent progenitors, lineage−SCA1−160cKIT+.
Lymphoma stem cells reside within the DN3/DN4 T cell population, but not the DN1/DN2 population
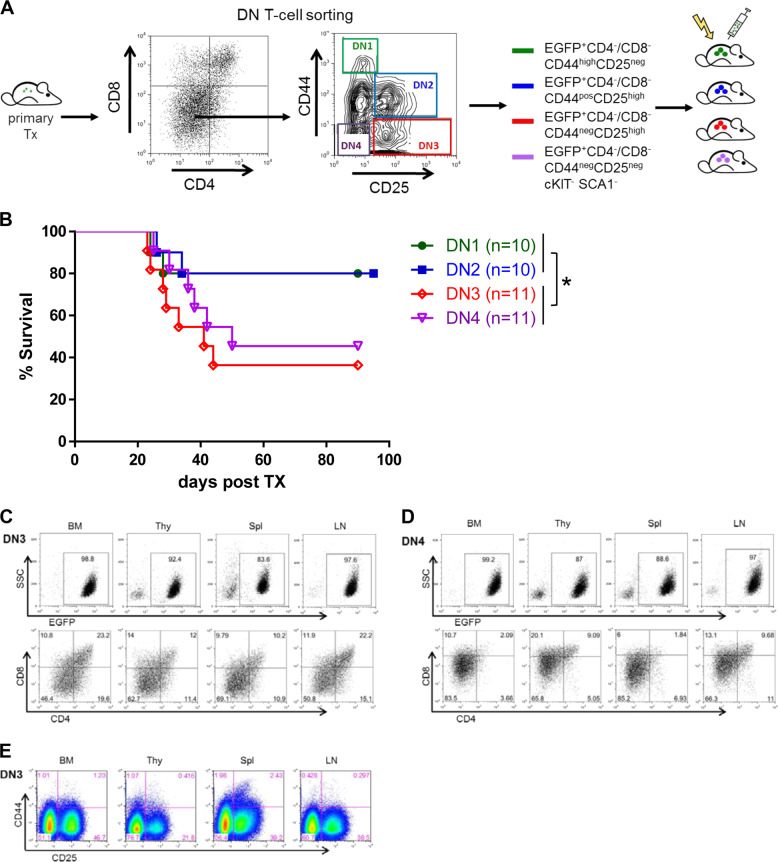
To further investigate and specify the origin of the lymphoma stem cell population we performed multiple serial transplantation experiments, where we isolated and sorted DN1, DN2, DN3, and DN4 T cell subpopulations from established tumors (Fig. 4a) and transplanted 1500 cells into sublethally irradiated recipient mice. Interestingly, development of serial lymphomas was only detected in mice, which were transplanted with DN3 or DN4 lymphoma T cells, whereas sorted DN1 or DN2 lymphoma T cells failed to establish serial disease (p = 0.018; Fig. 4b). Immunophenotypic analyses of serial lymphomas induced following transplantation of DN3 or DN4 lymphoma cells showed a range of CD4/8 subpopulations (CD4−/CD8− DN T cells as well as CD4+ and CD8+ single positive cells and CD4+/CD8+ double positive cells) as observed previously (Fig. 4c, d). These results imply that the DN3/DN4 population contains lymphoma stem cells, which are responsible for induction of serially transplantable disease and give rise to differentiated cells. Staining for early T cell markers (CD25 and CD44) demonstrated the co-occurrence of DN3 and DN4 cells. Dedifferentiation of transplanted DN3 cells to a DN1/2 status was not observed (Fig. 4e).
Fig. 4. Lymphoma stem cells reside within the DN3/DN4 T cell population, but not the DN1/DN2 population.
a Strategy for secondary transplantation of lymphoma subpopulations: 1500 EGFP positive CD4−/CD8− double negative lymphoma T cells of stages DN1 to DN4 were injected i.v. into subl-ethally irradiated (4500 rad) C57Bl6 recipient mice. b Kaplan–Meier survival curve of serially transplanted mice. Median survival: DN3 transplanted mice: 41 days, DN4 transplanted mice: 50 days. Representative flow cytometry profiles of lymphatic organs, thymus (Thy), spleen (Spl), lymph node (LN) and bone marrow (BM) of DN3 (c, e) and DN4 (d) transplanted mice stained for anti-CD4, anti-CD8, anti-CD44, and anti-CD25. *p < 0.05; ***p < 0.001.
Taken together, serial transplantation with sorted DN1-4 lymphoma T cell subpopulations identified the existence of lymphoma stem cells within the DN3/DN4 T cell population. In addition, these lymphoma stem cells are able to differentiate and give rise to mature lymphoma T cells.
The gene expression signature of the lymphoma stem cell population reveals a stem-cell-like transcriptional profile
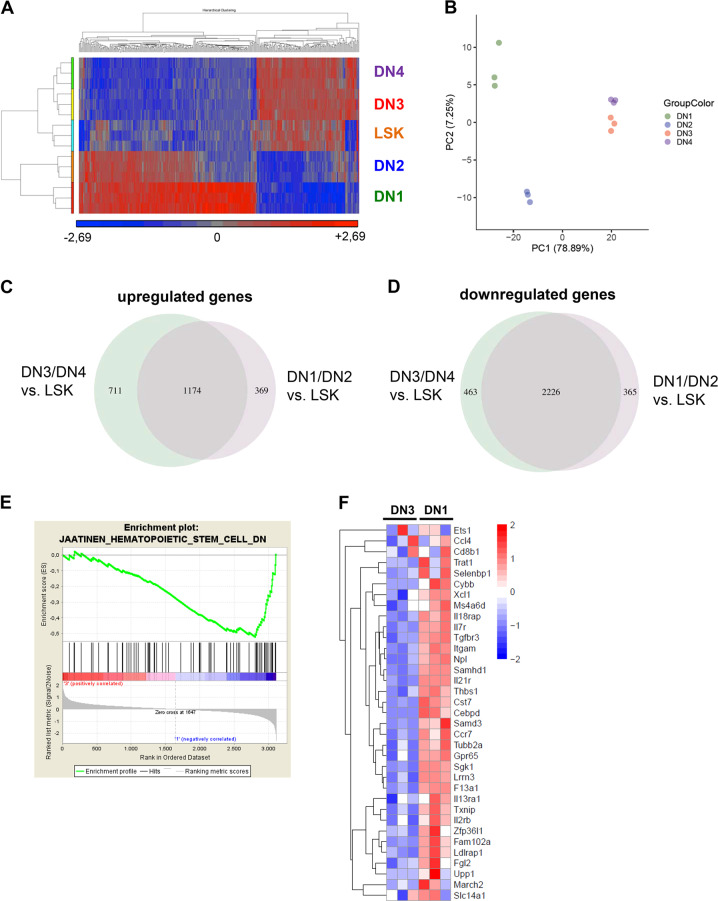
To further characterize the lymphoma stem cell population, we performed Affymetrix-based global gene expression analysis of the sorted lymphoma cell subsets. We hypothesized that lymphoma stem cells might have a gene expression pattern, which resembles hemopoietic stem cells. Indeed, microarray analyses showed not only a high similarity of gene expression patterns between DN3 and DN4 lymphoma T cell subpopulations but importantly also resembled stem cell (LSK cells) gene expression profiles as seen in the heatmap (Fig. 5a, Supplementary Fig. 6A). Principle component analysis separated lymphoma T cell subpopulations DN1–DN4 according to their variance not only of the most 1000 variable genes (Fig. 5b) but also all analysed genes (Supplementary Fig. 6B), thereby substantiating our hypothesis as DN3 and DN4 cluster together and are significantly separate from DN1 and DN2 populations. Using a Venn diagram, we were able to show significantly more up- and downregulated genes in DN3/DN4 lymphoma T cell subpopulations in comparison to the DN1/DN2 lymphoma T cell subpopulation vs. LSK cells with a cut-off < 0.01 (Fig. 5c, d), which might suggest a higher similarity of expression profiles of DN3/DN4 lymphoma T cells and LSK cells. Gene set enrichment analyses for genes, which are downregulated in hematopoietic stem cells revealed a stem cell signature in DN3 lymphoma T cells, whereas for example, the DN1 lymphoma T cell population did not show similarity to transcriptional profiles of hematopoeitic stem cells (Fig. 5e). The most deregulated genes of the gene set enrichment analysis in 5E and their expression can be seen in Fig. 5f.
Fig. 5. The gene expression signature of the lymphoma stem cell population reveals a stem-cell-like transcriptional profile.
a Heatmap comparing gene expression levels of DN1-DN4 lymphoma subpopulations with the primitive LSK population. Color scale represents the raw Z-score mRNA intensity value. b Principle component analysis (PCA) separating lymphoma subpopulations DN1–DN4 according to their variance of all genes analysed by microarray. PCA was performed on the top 1000 variable genes based on the Median Absolute Deviation (MAD). c Venn diagram showing significantly upregulated genes comparing DN1/DN2 and DN3/DN4 lymphoma subpopulations vs. LSK cells analysed by microarray with a q value cut-off < 0.01. d Venn diagram showing significantly downregulated genes comparing DN1/DN2 and DN3/DN4 lymphoma subpopulations vs. LSK cells analysed by microarray with a q value cut-off < 0.01. e Signature enrichment plot comparing DN3 vs. DN1 lymphoma subpopulations for genes downregulated in CD133+ hematopoietic stem cells compared with CD133- cells (M6905 gene set) analysed by microarray. FDR q value = 0.027. f Heatmap comparing DN3 vs. DN1 lymphoma subpopulations for expression levels of genes downregulated in hematopoietic stem cells analysed by microarray. Color scale represents raw Z-score mRNA intensity values (red = high expression, blue = low expression).
To summarize, bioinformatic analyses of Affymetrix-based global gene expression data clearly separated DN3 and DN4 lymphoma T cell subpopulations from DN1 and DN2 lymphoma T-cell subpopulations. Interestingly, heatmap analyses as well as Venn diagram and gene set enrichment analyses suggest that the DN3 and DN4 lymphoma T cells exhibit a gene expression signature resembling that seen in LSK cells which show more stem-like features than the DN1 lymphoma T cells, although being immunophenotypically more differentiated.
The lymphoma stem cell population is characterized by relatively low CD30 expression levels
CD30 expression on human ALCL cells is not only a diagnostic tool but is also exploited for targeted therapeutic approaches whereby the antibody-drug conjugate Brentuximab vedotin [27] binds and eliminates CD30-positive cells [28, 29]. However, resistance can develop due to down-regulation of CD30 expression on tumor cells [30, 31] and hence durable remissions are limited in ALCL patients treated with single-agent brentuximab vedotin. Therefore, one may hypothesize, that CD30 expression on lymphoma stem cells might facilitate long-term treatment response.
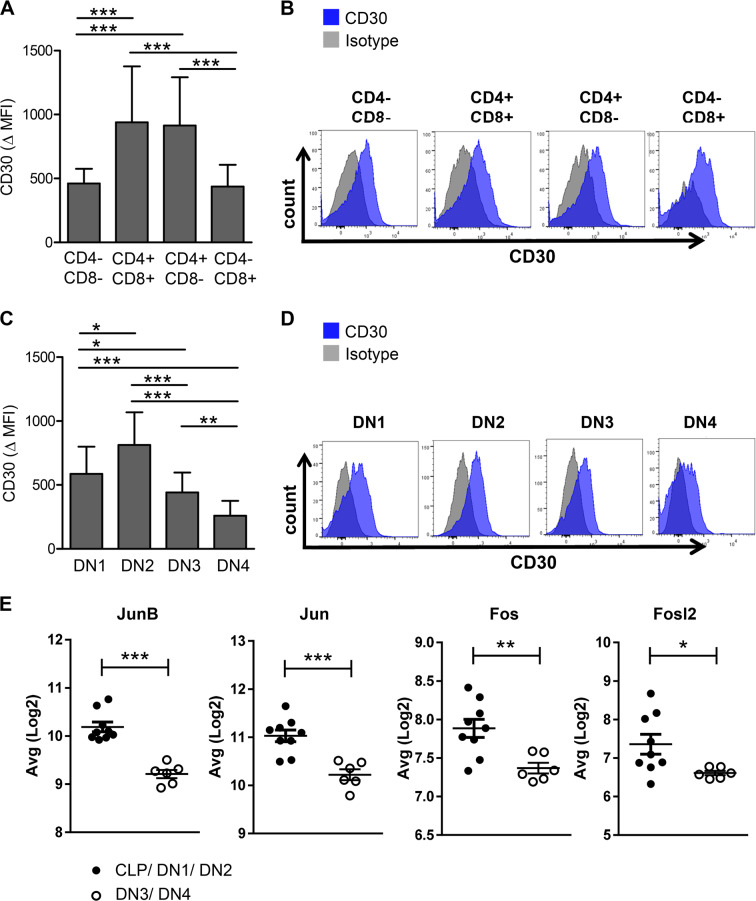
To test this hypothesis, we analysed the CD30 expression level on lymphoma cells in the ALCL-like mouse model. Interestingly, although the T cell lymphoma showed overall high CD30 expression [20], the level differed between the distinct lymphoma T cell subpopulations: focusing on T cell lymphoma subpopulations according to CD4 and/or CD8 expression, the DN CD4−/CD8− as well as the single positive CD8+ lymphoma subpopulation showed significantly lower CD30 expression compared with the double positive CD4+/CD8+ and single positive CD4+ lymphoma population (Fig. 6a, b). As our previous data indicate that the lymphoma stem cell population is immunophenotypically included in the DN3/DN4 lymphoma T cell subpopulation, we analysed CD30 expression levels on different DN stages of lymphoma cells. As shown in Fig. 6c, DN3 and DN4 T cell lymphoma subpopulations in particular showed significantly lower CD30 expression compared with DN1 and DN2 cells (Fig. 6c, d). It is noteworthy, that DN4 lymphoma cells show even lower CD30 expression than the single positive CD8+ lymphoma subpopulation and this phenomenon was consistently seen on lymphoma cells extracted from thymus, lymph node or spleen (∆MFI (DN4) = 259 vs. ∆MFI (CD8+) = 437; p = 0.003; Supplementary Fig. 7). We hypothesized that the alteration in CD30 expression is caused by transcriptional regulation. To verify our hypothesis, we analyzed well known CD30 transcriptional regulators of the activating protein-1 for their expression levels in the different lymphoma subpopulations and were indeed able to show, that they are downregulated in the DN3/4 lymphoma stem cell population compared with CLP, DN1, and DN2 lymphoma subpopulation (Fig. 6e) [32, 33].
Fig. 6. The lymphoma stem cell population is characterized by low CD30 expression levels.
a CD30 expression levels of the indicated EGFP+ T cell subpopulations differentiated for CD4 and CD8 of lymphoma cells isolated from MSNAIE Lck-Cre transgenic BM transplanted mice. ∆ Mean fluorescence intensity (MFI) = MFI of anti-CD30 stained sample minus MFI isotype n = 14. b Representative flow cytometry histograms of EGFP+ lymphoma cells differentiated for CD4 and CD8 and stained for anti-CD30 isolated from MSNAIE Lck-Cre transgenic BM transplanted mice. c CD30 expression levels on the indicated EGFP+ T cell lymphoma subpopulations differentiated by DN stages isolated from MSNAIE Lck-Cre transgenic BM transplanted mice. ∆ Mean fluorescence intensity (MFI) = MFI of anti-CD30 stained sample minus MFI isotype n = 14. d Representative flow cytometry histograms of EGFP+ lymphoma cells of MSNAIE Lck-Cre transgenic BM transplanted mice differentiated by DN stages and stained for anti-CD30. e Gene expression levels for the indicated genes comparing CLP/DN1/DN2 and DN3/DN4 lymphoma subpopulations. n(CLP/DN1/DN2) = 9, n(DN3/DN4) = 6. *p < 0.05; **p < 0.01; ***p < 0.001. Data are represented as mean + SD.
These results imply, that the lymphoma stem cell DN3/DN4 population is characterized by relatively low CD30 expression levels due to transcriptional regulation, which might contribute to limited long-term response in patients treated with anti-CD30-targeted therapies.
Our results therefore highlight the existence of a lymphoma stem cell population originating from the DN3/DN4 lymphoma T cell population in a relevant NPM-ALK positive CD30-expressing ALCL-like mouse model, which might contribute to treatment evasion due to low CD30 expression levels.
Discussion
In this study, we have identified and specified the existence of a lymphoma stem cell population using a retroviral murine BM transplantation model resembling human ALCL in terms of CD30 expression and large cell cytology whereas other morphologic characteristics differ from the human pathology. Although the murine tumor cells do not show the characteristic hallmark cells and variants (like doughnut cells and horse shoe or kidney shaped nuclei) [34], we believe that the large cell morphology with open chromatin and prominent nucleoli reproduced the human counterpart well. However, mice with advanced disease show diffuse pattern of infiltration like seen in advanced human disease. Histophathologic analyses of mice with diffuse lymph node involvement and splenic disease did not show sinusoidal pattern of infiltrated lymph nodes as observed in human ALCL. As the pattern of lymph node involvement changes due the course of disease, different lymph node infiltration patterns may be observed in earlier stage diseased animals. The observed cytological differences may also be influenced by microenvironmental distinctions. It is noteworthy that most murine mouse models mimic hardly all features of the human counterpart disease, but serve as important and useful platforms for understanding molecular and cellular mechanisms of the investigated human original.
The existence of lymphoma stem cells is supported by several case stories, in which both -recipient and donor- develop the same lymphoid disease after allogeneic transplant. Genetic sequence homology proved the same origin of the recipient and donor tumors [35]. Our data identify a distinct lymphoma stem cell population, which displays ambiguous features: while these lymphoma stem cells show immunophenotypical features of DN3/DN4 cells, gene expression profiles resemble primitive stem-cell-like features. This might be caused by reprogramming of the immunophenotypically “more differentiated” DN3/DN4 population -although still being CD4− and CD8− negative- to a stem-cell-like origin, thereby giving this population the capacity to robustly induce and maintain lymphoma in the ALCL-like mouse model. In support of our data, Moti et al., showed the existence of a tumor-propagating cell population in ALCL cell lines and primary ALCL patient samples, which could not be defined solely by immunophenotyping but shows a “stemness” gene expression profile [36]. Furthermore, the authors showed in this lymphoma initiating population a gene expression pattern that is reminiscent of early thymic progenitors [36]. Consistent with this, Hassler et al. demonstrated that DNA methylation patterns in ALCL cells closely resemble that of undifferentiated thymic progenitor cells [37]. These data strengthen our hypothesis that the lymphoma stem cell population originated from the DN3/DN4 lymphoma population, which fits to the published concept of a thymic origin for ALCL [38, 39].
In murine thymocyte development, DN3 cells are fully committed to the T cell lineage after transition from the DN2 stage due to successful rearrangement of their TCR β chain, which is associated with an invariant pre-TCR α chain and CD3 signaling molecules forming the pre-TCR complex. Due to β-selection, DN3 cells develop to the DN4 stage with further maturation capacity [40]. Using a CD4/NPM-ALK thymic T cell lymphoma model [41], Malcolm et al. showed the importance of TCR expression for thymic egress of tumor cells [38]. These data further support the hypothesis that aberrant expression of TCR α/β chains on DN lymphoma cells is important for the establishment of systemic lymphoma. In addition, analyses of clonal TCR rearrangements in human ALCL demonstrate major in-frame TCRα clonal rearrangements in two-thirds of patient cases [38]. Recent analyses furthermore support the existence of a lymphoma stem cell population, which is able to differentiate into mature lymphoma T cell subpopulations, as they presented similar findings in human ALCL cell lines and primary patient samples, demonstrating the existence of a side population with serial tumor-propagating capacity, characterized by self-renewal-ability and potency to differentiate to the bulk tumor population [36].
CD30 expression is a hallmark of ALCL and is exploited for targeted therapeutic approaches such as brentuximab vedotin or C2NP-functionalized nanoparticles encapsulated with doxorubicin [27, 42]. Our data demonstrate variable CD30 expression levels within different lymphoma subpopulations in this ALCL-like mouse model, suggesting that the responses to anti-CD30 treatment may differ between the lymphoma subpopulations. In particular, as the lymphoma stem cell population demonstrates relatively low CD30 expression, limited long-term responses of patients treated with anti-CD30-targeted therapies may reflect the insufficient lymphoma stem cell eradication of this treatment.
In summary, our results highlight the existence of a lymphoma stem cell population that originates from the DN3/DN4 cell population in a relevant NPM-ALK positive CD30-expressing ALCL-like mouse model, thereby providing the opportunity to test innovative treatment strategies (e.g., NPM-ALK-/JAK-/TYK2- inhibitors) [16], which aim to eradicate the lymphoma stem cell population and may overcome common treatment unresponsiveness due to low CD30 expression levels.
Supplementary information
Acknowledgements
We thank Sabina Schaberg for excellent technical assistance. This work was supported by a Grant from the DJCLS (R14/22) and MSCA-ITN-2015-ETN Alkatras to ALI, TP, JD, IG-M, FF, LQ-M and SDT and grants from the DFG to JD (FOR 2033 B1). GA and MB are supported by the DFG within the CRC850. MB is funded by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (DeCaRe, FKZ 01ZX1409B). ALI was supported by a Grant from the Government of Baden-Württemberg (BSL).
Author contributions
SK and CK performed and designed experiments, analysed data and wrote the manuscript. AK, GA, DP, and MB performed and analysed the array data. MF supported flow cytometry and sorting analyses. TP, CM, CL, RZ, SDT, KS, IG-M, FF, and LQ-M provided critical material. JD developed the concept and analysed the data. ALI developed the overall concept, supervised the experiments, analysed data and wrote the manuscript. All Authors edited and reviewed the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Stefanie Kreutmair, Cathrin Klingeberg
Supplementary information
The online version of this article (10.1038/s41375-020-0789-x) contains supplementary material, which is available to authorized users.
References
- 1.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 2.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 3.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23:1124–34. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 8.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 9.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 10.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 11.Mathur R, Sehgal L, Braun FK, Berkova Z, Romaguerra J, Wang M, et al. Targeting Wnt pathway in mantle cell lymphoma-initiating cells. J Hematol Oncol. 2015;8:63. doi: 10.1186/s13045-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laurent C, Do C, Gascoyne RD, Lamant L, Ysebaert L, Laurent G, et al. Anaplastic lymphoma kinase-positive diffuse large B-cell lymphoma: a rare clinicopathologic entity with poor prognosis. J Clin Oncol. 2009;27:4211–6. doi: 10.1200/JCO.2008.21.5020. [DOI] [PubMed] [Google Scholar]
- 13.Stein H, Mason DY, Gerdes J, O’Connor N, Wainscoat J, Pallesen G, et al. The expression of the Hodgkin’s disease associated antigen Ki-1 in reactive and neoplastic lymphoid tissue: evidence that Reed-Sternberg cells and histiocytic malignancies are derived from activated lymphoid cells. Blood. 1985;66:848–58. doi: 10.1182/blood.V66.4.848.848. [DOI] [PubMed] [Google Scholar]
- 14.Ambrogio C, Martinengo C, Voena C, Tondat F, Riera L, di Celle PF, et al. NPM-ALK oncogenic tyrosine kinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009;69:8611–9. doi: 10.1158/0008-5472.CAN-09-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner MT, Zhao C, Zhang Q, Wasik MA. Nucleophosmin-anaplastic lymphoma kinase: the ultimate oncogene and therapeutic target. Blood. 2017;129:823–31. doi: 10.1182/blood-2016-05-717793. [DOI] [PubMed] [Google Scholar]
- 16.Prutsch N, Gurnhofer E, Suske T, Liang HC, Schlederer M, Roos S, et al. Dependency on the TYK2/STAT1/MCL1 axis in anaplastic large cell lymphoma. Leukemia. 2019;33:696–709. doi: 10.1038/s41375-018-0239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merkel O, Hamacher F, Griessl R, Grabner L, Schiefer AI, Prutsch N, et al. Oncogenic role of miR-155 in anaplastic large cell lymphoma lacking the t(2;5) translocation. J Pathol. 2015;236:445–56. doi: 10.1002/path.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garces de Los Fayos Alonso I, Liang HC, Turner SD, Lagger S, Merkel O, Kenner L. The role of activator protein-1 (AP-1) family members in CD30-positive lymphomas. Cancers. 2018;10. [DOI] [PMC free article] [PubMed]
- 19.Hwang SR, Murga-Zamalloa C, Brown N, Basappa J, McDonnell SR, Mendoza-Reinoso V, et al. Pyrimidine tract-binding protein 1 mediates pyruvate kinase M2-dependent phosphorylation of signal transducer and activator of transcription 3 and oncogenesis in anaplastic large cell lymphoma. Lab Investig; a J Tech methods Pathol. 2017;97:962–70. doi: 10.1038/labinvest.2017.39. [DOI] [PubMed] [Google Scholar]
- 20.Shoumariyeh K, Schneider N, Poggio T, Veratti P, Ehrenfeld S, Redhaber DM, et al. A novel conditional NPM-ALK-driven model of CD30+ T-cell lymphoma mediated by a translational stop cassette. Oncogene. 2019;39:1904–13. doi: 10.1038/s41388-019-1058-1. [DOI] [PubMed] [Google Scholar]
- 21.Illert AL, Albers C, Kreutmair S, Leischner H, Peschel C, Miething C, et al. Grb10 is involved in BCR-ABL-positive leukemia in mice. Leukemia. 2015;29:858–68. doi: 10.1038/leu.2014.283. [DOI] [PubMed] [Google Scholar]
- 22.Ewerth D, Kreutmair S, Schmidts A, Ihorst G, Follo M, Wider D, et al. APC/C(Cdh1) regulates the balance between maintenance and differentiation of hematopoietic stem and progenitor cells. Cell Mol life Sci. 2019;76:369–80. doi: 10.1007/s00018-018-2952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerboth S, Frittoli E, Palamidessi A, Baltanas FC, Salek M, Rappsilber J, et al. Phosphorylation of SOS1 on tyrosine 1196 promotes its RAC GEF activity and contributes to BCR-ABL leukemogenesis. Leukemia. 2018;32:820–7. doi: 10.1038/leu.2017.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudorf A, Muller TA, Klingeberg C, Kreutmair S, Poggio T, Gorantla SP, et al. NPM1c alters FLT3-D835Y localization and signaling in acute myeloid leukemia. Blood. 2019;134:383–8. doi: 10.1182/blood.2018883140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller TA, Grundler R, Istvanffy R, Rudelius M, Hennighausen L, Illert AL, et al. Lineage-specific STAT5 target gene activation in hematopoietic progenitor cells predicts the FLT3(+)-mediated leukemic phenotype. Leukemia. 2016;30:1725–33. doi: 10.1038/leu.2016.72. [DOI] [PubMed] [Google Scholar]
- 26.Klein C, Zwick A, Kissel S, Forster CU, Pfeifer D, Follo M, et al. Ptch2 loss drives myeloproliferation and myeloproliferative neoplasm progression. J Exp Med. 2016;213:273–90. doi: 10.1084/jem.20150556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pro B, Advani R, Brice P, Bartlett NL, Rosenblatt JD, Illidge T, et al. Five-year results of brentuximab vedotin in patients with relapsed or refractory systemic anaplastic large cell lymphoma. Blood. 2017;130:2709–17. doi: 10.1182/blood-2017-05-780049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkel O, Hamacher F, Sifft E, Kenner L, Greil R. Novel therapeutic options in anaplastic large cell lymphoma: molecular targets and immunological tools. Mol cancer therapeutics. 2011;10:1127–36. doi: 10.1158/1535-7163.MCT-11-0042. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz S, O’Connor OA, Pro B, Illidge T, Fanale M, Advani R, et al. Brentuximab vedotin with chemotherapy for CD30-positive peripheral T-cell lymphoma (ECHELON-2): a global, double-blind, randomised, phase 3 trial. Lancet. 2019;393:229–40. doi: 10.1016/S0140-6736(18)32984-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskowitz CH, Nademanee A, Masszi T, Agura E, Holowiecki J, Abidi MH, et al. Brentuximab vedotin as consolidation therapy after autologous stem-cell transplantation in patients with Hodgkin’s lymphoma at risk of relapse or progression (AETHERA): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2015;385:1853–62. doi: 10.1016/S0140-6736(15)60165-9. [DOI] [PubMed] [Google Scholar]
- 31.Moskowitz CH, Walewski J, Nademanee A, Masszi T, Agura E, Holowiecki J, et al. Five-year PFS from the AETHERA trial of brentuximab vedotin for Hodgkin lymphoma at high risk of progression or relapse. Blood. 2018;132:2639–42. doi: 10.1182/blood-2018-07-861641. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe M, Sasaki M, Itoh K, Higashihara M, Umezawa K, Kadin ME, et al. JunB induced by constitutive CD30-extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase signaling activates the CD30 promoter in anaplastic large cell lymphoma and reed-sternberg cells of Hodgkin lymphoma. Cancer Res. 2005;65:7628–34. doi: 10.1158/0008-5472.CAN-05-0925. [DOI] [PubMed] [Google Scholar]
- 33.van der Weyden CA, Pileri SA, Feldman AL, Whisstock J, Prince HM. Understanding CD30 biology and therapeutic targeting: a historical perspective providing insight into future directions. Blood Cancer J. 2017;7:e603. doi: 10.1038/bcj.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montes-Mojarro IA, Steinhilber J, Bonzheim I, Quintanilla-Martinez L, Fend F The Pathological Spectrum of Systemic Anaplastic Large Cell Lymphoma (ALCL). Cancers 2018;10. [DOI] [PMC free article] [PubMed]
- 35.Husby S, Gronbaek K. Mature lymphoid malignancies: origin, stem cells, and chronicity. Blood Adv. 2017;1:2444–55. doi: 10.1182/bloodadvances.2017008854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moti N, Malcolm T, Hamoudi R, Mian S, Garland G, Hook CE, et al. Anaplastic large cell lymphoma-propagating cells are detectable by side population analysis and possess an expression profile reflective of a primitive origin. Oncogene. 2015;34:1843–52. doi: 10.1038/onc.2014.112. [DOI] [PubMed] [Google Scholar]
- 37.Hassler MR, Pulverer W, Lakshminarasimhan R, Redl E, Hacker J, Garland GD, et al. Insights into the pathogenesis of anaplastic large-cell lymphoma through genome-wide DNA methylation profiling. Cell Rep. 2016;17:596–608. doi: 10.1016/j.celrep.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malcolm TI, Villarese P, Fairbairn CJ, Lamant L, Trinquand A, Hook CE, et al. Anaplastic large cell lymphoma arises in thymocytes and requires transient TCR expression for thymic egress. Nat Commun. 2016;7:10087. doi: 10.1038/ncomms10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malcolm TI, Hodson DJ, Macintyre EA, Turner SD. Challenging perspectives on the cellular origins of lymphoma. Open Biology 2016;6. [DOI] [PMC free article] [PubMed]
- 40.Shah DK, Zuniga-Pflucker JC. An overview of the intrathymic intricacies of T cell development. J Immunol. 2014;192:4017–23. doi: 10.4049/jimmunol.1302259. [DOI] [PubMed] [Google Scholar]
- 41.Chiarle R, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–27. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 42.Luo X, Yang Y, Kong F, Zhang L, Wei K. CD30 aptamer-functionalized PEG-PLGA nanoparticles for the superior delivery of doxorubicin to anaplastic large cell lymphoma cells. Int J Pharm. 2019;564:340–9. doi: 10.1016/j.ijpharm.2019.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.