Abstract
As immunotherapy assumes a central role in the management of many cancers, ongoing work is directed at understanding whether immune-based treatments will be successful in patients with glioblastoma (GBM). Despite several large studies conducted in the last several years, there remain no FDA-approved immunotherapies in this patient population. Nevertheless, there are a range of exciting new approaches being applied to GBM, all of which may not only allow us to develop new treatments but also help us understand fundamental features of the immune response in the central nervous system. In this review, we summarize new developments in the application of immune checkpoint blockade, from biomarker-driven patient selection to the timing of treatment. Moreover, we summarize novel work in personalized immune-oncology by reviewing work in cancer immunogenomics–driven neoantigen vaccine studies. Finally, we discuss cell therapy efforts by reviewing the current state of chimeric antigen receptor T-cell therapy.
Keywords: cancer immunogenomics, CNS immunosurveillance, glioblastoma, neoantigen, personalized vaccine
As immunotherapy assumes an increasingly central role in cancer treatment, the search continues for effective immune-based treatments for patients with glioblastoma (GBM). Indeed, there are still no FDA-approved brain tumor immunotherapies. There are many reasons for this current lack of progress, some of which we understand and many of which we do not. Although it has become clear that the central nervous system (CNS) is not as hermetically immunoprivileged and immunologically inaccessible as previously thought,1 the CNS is nevertheless immunologically specialized such that the basic mechanisms of immune system activation as conceived in extracranial compartments may not be readily extrapolated to the brain. Specifically, there are major gaps in our understanding of fundamental components of the immune response to brain tumors, such as the identification of critical antigen presenting cells, the location of naïve T-cell priming, and the details of underlying effector T-cell trafficking into brain tumors, among others. Moreover, there appears to be a severe degree of immune dysfunction in GBM patients.2 A recent study showed that both for the case of GBM patients and mouse models, and for intracranial tumors such as brain metastasis, scant T-cell infiltration in intracranial tumors is explained by robust sequestration of these lymphocytes in the bone marrow. Yet, such sequestration phenomenon does not occur when these tumors are implanted extracranially.3 Thus, there are both biological and anatomic features specific to GBM that contribute to the difficulty of identifying and implementing effective brain tumor immunotherapies.
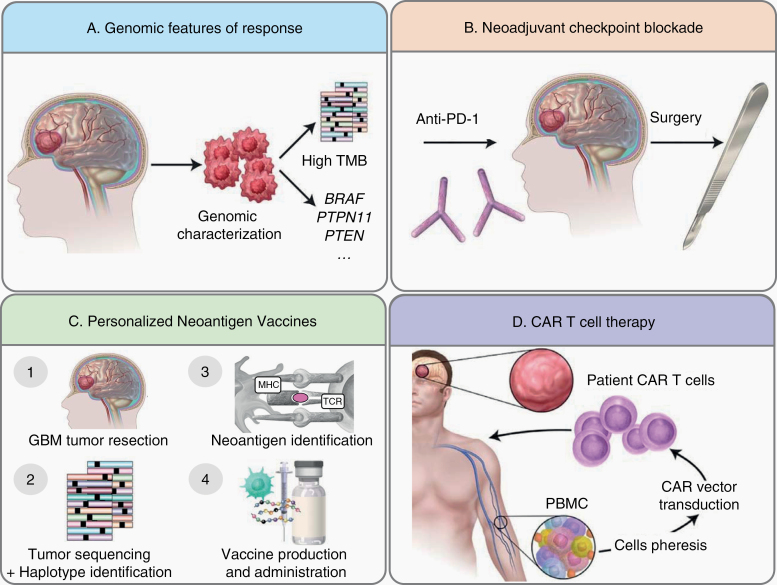
Despite the challenges to brain tumor immunotherapy development, recent work in the field points to a range of new and innovative approaches to this problem that we will review here (Fig. 1). First, we will highlight the application of cancer immunogenomics to several treatment approaches. “Cancer immunogenomics” represent an integration of genomics approaches to immunology and immunotherapy.4 Using this methodology, genomic features may be correlated with response and/or resistance to immunotherapy. Additionally, candidate tumor-specific neoantigens may be identified from expressed exome alterations predicted to bind with high affinity to a patient’s specific human leukocyte antigen (HLA) molecules.5 We will therefore review the identification of genomically defined patient groups that may be more sensitive to checkpoint blockade, as well as explore the potential for cancer immunogenomics-based personalized neoantigen vaccine trials in GBM. We will subsequently discuss new data on the presurgical, neoadjuvant administration of anti–programmed cell death 1 (PD-1) immunotherapy in GBM, which has generated promising results in several cancer types, including recent studies in GBM. Finally, we will review the opportunities in cell-based therapy for GBM by focusing on chimeric antigen receptor (CAR) T-cell efforts.
Fig. 1.
New frontiers in brain tumor immunotherapy. (A) Identifying patients with tumor genomic features that correlate with increased response to checkpoint blockade immunotherapy. (B) Administration of checkpoint blockade immunotherapy prior to surgery in the neoadjuvant setting. (C) Generating of therapeutic personalized cancer vaccines targeting GBM neoantigens. (D) Generation of CAR T cells from autologous patient peripheral blood mononuclear cells.
New Ways to Apply Checkpoint Blockade in GBM
Due to the success of checkpoint blockade immunotherapy in many cancer types,6 significant work has been directed at understanding its efficacy in GBM. To date, these efforts have been unsuccessful. Anti–PD-1 (nivolumab) immunotherapy did not improve survival in unselected recurrent GBM patients,4 and recent studies of nivolumab in newly diagnosed patients failed to reveal significant clinical responses. Several possibilities underlie these clinical outcomes. It is possible that GBM has been subjected to such significant immunoediting2 during tumor development that it is highly immunosuppressive and immunoevasive, unable to respond to checkpoint blockade and other immunotherapies. Many studies have described severe immunologic impairments observed in GBM.7 It is also possible that we have not yet determined which patients may respond to checkpoint blockade immunotherapy. Recent work showed that it be may be possible to identify specific GBM genomic features that may confer increased responsiveness to checkpoint blockade. Specific biomarkers such as PD-1 ligand (PD-L1) expression and tumor mutational burden, among others, are associated with responses to checkpoint blockade in several cancer types.8 To date, we have little understanding of the intrinsic properties that render some gliomas susceptible or resistant to immunotherapy. Due to intratumoral differences, clinical trials designed under the premise that all tumors are equally susceptible may not be able to demonstrate efficacy. Additional complementary studies have begun to explore a more fundamental question in the use of checkpoint blockade in GBM—ie, when should these agents be administered? Specifically, checkpoint blockade has traditionally been administered to GBM patients in the adjuvant setting, mirroring the use of temozolomide (TMZ). However, it is possible that neoadjuvant administration may stimulate an antitumor immune response in a more robust manner. Thus, identifying who may benefit from checkpoint blockade and when checkpoint immunotherapy should be administered represent new ways to think about the utility of these therapies in treating GBM (Fig. 1A). Ultimately, understanding what other checkpoint pathways beyond the PD-1/PD-L1 axis may be targetable will also be critical to developing these treatments for GBM patients.
Identifying Genomic Features of Response to Checkpoint Blockade: BRAF, PTPN11, and PTEN Alterations
Identification of responder patients based on clinical outcomes is critical to investigate the molecular and microenvironmental determinants of therapy response. Moreover, rational patient stratification based on known biomarkers of response is imperative for the personalized application of immunotherapy for GBM. In this context, a definition of efficacy that can be applied to individual patient outcomes is necessary. In Zhao et al,9 a subset of patients with recurrent GBM exhibiting responses to anti–PD-1 blockade was found to harbor particular shared genomic features. In order to correlate genomic alterations with clinical outcomes, response criteria were rigorously defined such that responses included (i) stable disease on MRI for >6 months from initiation of PD1 blockade or (ii) evidence of treatment effect with inflammatory infiltrate and scant tumor cells on surgical specimens obtained after treatment initiation or in the setting of recurrent GBM.9 Interestingly, whereas cases of pseudoprogression in this cohort were observed, most cases defined as responders were categorized as such based on stable appearance of MRI over 6 months.9 Notably, the subset of recurrent GBM patients who responded to PD-1 blockade (based on imaging and pathology criteria) exhibited favorable and significant overall survival (OS) compared with nonresponder patients, independent of other therapies or clinical or molecular prognostic features.9
In patients who exhibited response and prolonged survival following immunotherapy, pretreatment samples harbored an overrepresentation of B-raf murine sarcoma (BRAF) and protein tyrosine phosphatase non-receptor type 11 (PTPN11) activating mutations.9 BRAF is a serine/threonine kinase, whereas PTPN11 is a phosphatase; both are upstream activators of the mitogen-activated protein kinase (MAPK) signaling pathway with the oncogenic potential to activate extracellular signal-regulated kinase (ERK) phosphorylation within MAPK.10,11 In spite of the approximately 10-fold enrichment of BRAF/PTPN11 activating mutations in tumors of GBM patients who exhibited response and prolonged survival following PD-1 blockade immunotherapy, only 30% of responder patients—and 2–3% of all GBM—harbor such genetic alterations. Thus, although activating mutations in this pathway may be enriched in some responders, the genomic features associated with response in the majority of patients remain unclear. Presently, the mechanism of MAPK pathway involvement in response to PD1 blockade remains a subject of investigation. This signaling cascade modulates macrophage phenotypes, with some reports suggesting a pro-inflammatory and others an immunosuppressive phenotype.12–17 Regardless, the MAPK signaling observed in GBM responders was identified by activating mutations of effectors of this pathway, implying that the immune-related properties of this pathway take place in the context of its activation within tumor cells and may be mediated in a paracrine fashion.
Complementing the association of MAPK activation with response to immunotherapy, MAPK signaling was also modulated by the presence of cluster of differentiation (CD)8+ T cells during glioma development in mouse models.18 Specifically, murine gliomas that formed in the absence of CD8 T cells not only were more immunogenic but also showed elevated MAPK signaling.18 Moreover, phosphorylation of ERK and p38 correlated with the expression of macrophage/microglial markers such as CD11b and ionized calcium binding adaptor molecule 1 (Iba1) in gliomas generated in a platelet derived growth factor B (PDGF-B+) phosphatase and tensin homolog (PTEN−/−) genetic context,19 yet this correlation was suppressed in gliomas developed in the presence of CD8+ T cells.18 In addition, analysis of gene expression data from The Cancer Genome Atlas (TCGA) revealed a strong correlation between macrophage/microglial markers, CD11b and Iba1, with several MAPK effector genes in GBMs.18 These data are consistent with findings in lung cancer in which activating mutations of KRAS (Kirsten rat sarcoma viral oncogene homolog) are associated with robust macrophage infiltration.20 Thus, given these associations, it appears that activation of the MAPK signaling cascade by tumor cells influences the microenvironment and is associated with recruitment, and possibly polarization, of tumor-associated macrophages/microglia.
Zhao et al9 also explored genomic features associated with lack of response to anti–PD-1 blockade. In this cohort of patients, PTEN mutations were associated with a decreased response to checkpoint blockade and a significantly lower T-cell infiltration following treatment compared with tumors with PTEN wild-type status.9 Similarly, PTEN mutations have also been associated with lack of response to anti–PD-1 checkpoint blockade in melanoma and metastatic uterine leiomyosarcoma.21,22 Interestingly, whereas PTEN mutant GBMs exhibited an immunosuppressive gene expression pattern, immunohistochemical analysis failed to identify differences in the abundance of T-regulatory lymphocytes based on PTEN mutation status.9 Single-cell sequencing analysis performed on cells isolated from PTEN mutant GBMs revealed that an immunosuppressive gene signature is expressed by CD44+ tumor cells.9 Thus, it is possible that the lack of immune response associated with PTEN loss relates to Akt signaling activation, which has been shown to modulate immune suppression and expression of PD-L1 in cancer.9,23,24 Together, these findings suggest that PTEN loss may represent a mechanism of glioma immunoediting during immunotherapy treatment.25,26
These data underscore the importance of tissue sampling in trying to understand mechanisms of response and resistance as well as personalized treatments for GBM patients. Although MAPK pathway activation as well as PTEN alterations point to biomarkers that may help to define patient cohorts capable of responding or not, respectively, to checkpoint blockade immunotherapy, further work is needed in larger patient cohorts to define additional features correlated with clinical outcomes.
Treating Patients with High Tumor Mutational Burden with Checkpoint Blockade
Analysis of the tumor genomic features of patients treated with checkpoint blockade immunotherapy has demonstrated that elevated tumor mutational burden (TMB) represents an important biomarker of treatment response across some tumor types. One potential hypothesis underlying this correlation is that an increased repository of mutations within a tumor is the engine for an increased neoantigen repertoire that could be recognized by licensed immune cells. In the landmark study by Le and colleagues,27 patients with hypermutated colorectal cancers due to germline mutations in DNA mismatch repair (MMR) genes demonstrated significant clinical responses to pembrolizumab. Even in this small study, the objective response rate was 40% with a disease control rate of 90% compared with 0% and 11%, respectively, in non-hypermutated, MMR-proficient patients. High objective response rates (71%) were also observed in non-colorectal MMR-deficient tumors, suggesting that elevated mutational burden may be associated with immunotherapy responses across cancer types. Additional studies have also demonstrated a strong association between increased mutational burden and response to checkpoint blockade therapy in non-small-cell lung cancer,28,29 melanoma,30–32 bladder cancer,33 and others.34–36 These clinical observations have been corroborated by preclinical studies in which well-studied murine tumors engineered to harbor higher TMB through clustered regularly interspaced short palindromic repeat (CRISPR) targeting of MMR pathway components were more sensitive to checkpoint blockade.37,38 Further analysis suggests that the clonal architecture as well as types of mutations may also represent key parameters. Swanton and colleagues showed that anti–PD-1 immunotherapy was more effective in tumors with a higher proportion of neoantigens that were clonal in nature.39 Additionally, further work underscored that the amount of particular genomic variants, such as inversion deletion and frameshift mutations, may also be correlated with responses to checkpoint blockade immunotherapy.38
There are several practical methods with which to identify the high TMB genotype reviewed comprehensively by Chan et al.36 TMB can be assessed as part of currently employed cancer gene panel sequencing platforms available either internally in many academic institutions or through external companies. Largely, this approach is based on the premise that a hypermutated genotype is distributed well enough across the exome that select cancer gene panel sequencing approaches, rather than whole exome or genome characterization, can capture this genomically disrupted state. To this end, Stadler et al clearly demonstrated that hypermutated tumors correlated with ≥20 mutations identified by a 341-gene panel, while non-hypermutated tumors harbored <20 alterations.40 Johnson et al extended these findings to show that melanoma patients with a higher mutation per megabase rate as captured on the commercial FoundationOne cancer gene panel exhibited improved responses to anti–PD-1/PD-L1 immunotherapy.32 This study was complemented by Chalmers et al in work showing that the total number of mutations across the exome was highly correlated with the number of mutations in the FoundationOne gene panel comprising 315 genes.41 Presently, both the Foundation Medicine CDx and Memorial Sloan Kettering MSK-IMPACT study are FDA approved for the purpose of identifying TMB. It will be important to determine the threshold of TMB that is needed to stratify patients for immunotherapy, which may also vary depending on the tumor type.
High TMB and Immunotherapy in GBM
The most common clinical setting in which GBM patients harbor a high mutational burden is when tumors progress or recur following TMZ therapy. Recent independent studies showed that up to 20–30% of recurrent GBMs exhibit the hypermutated genotype.42–46 Moreover, low-grade gliomas treated with temozolomide can also recur at a higher grade with the hypermutated genotype.44,47 These tumors frequently display mutations in the MMR pathway—such as mutator S homolog (MSH)6, MSH2, and PMS1 Homolog 2, Mismatch Repair System Component (PMS2)—either through acquisition during treatment or by selection of preexisting MMR-deficient subclones.47,48 The association of somatic MMR deficiency and the hypermutated genotype is consistent with the current understanding of how TMZ, as well as other alkylating agents, can induce genotoxic damage through covalent modification of nitrogen and oxygen atoms of DNA bases.49 When TMZ is metabolized in vivo to 5-amino-imidazole-4 carboxamide and a methyldiazonium cation, methyl groups are added at N7 positions of guanine, N3 positions of adenine, and the O6 positions of guanine.49,50 The latter, O6-methylguanine (O6-MeG), is the major driver of TMZ genotoxicity. If not repaired by the enzyme O6-methylguanine DNA methyltransferase (MGMT), O6-MeG can pair with thymine instead of cytosine. If MMR machinery is intact, then these base-pairing errors can either be repaired or lead to apoptosis. However, in the setting of MMR deficiency, base mispairing is not detected and can lead to the accumulation of a large number of mutations. Thus, the vast majority of post-TMZ hypermutated GBMs exhibit loss or mutation in MMR machinery.47
One hypothesis underlying the development of post-TMZ hypermutated GBMs is that pretreatment MMR-deficient populations exist de novo and become the dominant posttreatment populations when TMZ treatment is initiated. This possibility is supported by recent work from Mahlokozera et al51 in which a cohort of primary GBMs underwent multisector genomic profiling. In 2 patients, 1 of 2 sectors harbored significantly higher mutations, illustrating one scenario in which preexisting high mutational burden subclones may be selected by TMZ exposure. Further spatial and temporal work is necessary to understand the natural history of the TMZ-associated hypermutated state.
Because hypermutated GBMs harbor a significantly higher predicted neoantigen load than non-hypermutated tumors,52 it is important to determine if this patient population exhibits an improved response to checkpoint blockade immunotherapy. Several clinical trial efforts are currently under way to address this question. Two currently open trials include “Nivolumab in People with IDH-Mutant Glioma With and Without Hypermutator Phenotype/NCT03718767” and “Pembrolizumab (MK-3475) in Patients With Recurrent Malignant Glioma With a Hypermutator Phenotype/NCT02658279.” In addition, there is a larger phase II study sponsored by the Neuro-Oncology Committee of the Alliance for Clinical Trials in Oncology, “A Phase II Study of Checkpoint Blockade Immunotherapy in Patients with Somatically Hypermutated Recurrent Glioblastoma,” which has been approved by the National Cancer Institute (NCI) and will open shortly. Thus, several studies will directly test the hypothesis that increased TMB will lead to enhanced response to checkpoint blockade.
Immunotherapy in Germline Hypermutation Settings
Hypermutated GBMs also occur in patients with newly diagnosed disease. There are hereditary cancer syndromes in which patients harboring germline variants in genes involved either in MMR or DNA replication predispose patients to the development of a range of cancers, including GBM, that exhibit a hypermutated genotype. Bouffet et al reported the genomic landscape of pediatric GBM associated with germline bialleic MMR deficiency.53 Tumors arising in this setting harbor a hypermutated genotype and, in turn, extremely high neoantigen loads. Two siblings carrying biallelic germline PMS2 deficiency developed tumors carrying over 20 000 mutations. Strikingly, both patients in this study were treated with nivolumab at recurrence and demonstrated durable clinical responses. A clinical trial studying the response of pediatric patients with hypermutated tumors to nivolumab is ongoing (Pilot Study of Nivolumab in Pediatric Patients with With Hypermutant Cancers/NCT02992964). In the adult setting, a 31-year-old man with a GBM harboring over 10 000 DNA exome mutations in the setting of a polymerase epsilon gene (POLE) L212V germline mutation was treated with pembrolizumab.54 Using a cancer immunogenomics pipeline,55,56 1245 expressed predicted neoantigens were identified within the founder clone alone. Following tumor recurrence as a spinal metastasis, he was treated with pembrolizumab. The patient underwent surgery for another spinal lesion which proved to represent pseudoprogression. Histologic and genomic analyses of pre- and post-pembrolizumab tissue revealed that checkpoint blockade induced a significant T-cell infiltrate associated with a robust pro-inflammatory gene expression signature. These studies underscore the importance of tumor genomic annotation in all malignant glioma settings.
Thus, those patients with resulting hypermutated recurrent tumors may represent cohorts suitable for checkpoint blockade immunotherapy (Fig. 1A). The pursuit of this possibility will change management for the majority of patients with recurrent GBM because most patients do not undergo tissue sampling of their recurrence unless a redo-craniotomy or biopsy at recurrence is performed. It may be time to consider placing more importance on sampling recurrent disease to search for those patients who are hypermutated and may benefit from checkpoint blockade instead of exposing them to potentially futile second-line chemotherapy agents, as MMR-deficient tumors may be completely resistant.37 Importantly, whether the clonal architecture of this abundant neoantigen landscape in the hypermutated setting will influence responses to checkpoint blockade in brain tumors remains an open question.39
It is important to keep in mind that in the case of TMZ-induced hypermutator GBM phenotype, the number of mutations is considerably lower than that exhibited by hypermutator tumors derived from germline alterations in DNA replication and repair pathways. In the former cases, the number of somatic mutations is approximately 1000–2500,44,46 whereas in tumors derived from germline DNA repair deficiencies, and in particular, mutations in polymerases, the mutations tend to be more frequent. Indeed, an analysis of cancers from patients with germline mismatch repair deficiency revealed that resulting tumors harbor ~1500 somatic mutations, with up to ~17 000 mutations in tumors that also contained secondary polymerase mutations.53 GBM from patients with germline POLE mutations harbor 9000 to 11 000 non-synonymous mutations,54 and gliomas from patients with germline PMS2 loss harbor 20 000 mutations. On the other hand, in our recent analysis of molecular features associated with recurrent GBM response to PD-1 blockade, we did not find significant differences in the number of mutations between tumors that responded and those that did not respond to this form of immunotherapy.9 Similarly, mutational burden did not correlate with OS in another recent retrospective series of glioma patients receiving anti–PD-1 therapy.57 In this analysis, hypermutation was detected in 5.4% of 10 294 gliomas, and did not correlate with survival among GBM patients, but actually predicted poorer OS of oligodendroglioma and astrocytoma patients regardless of isocitrate dehydrogenase 1 and 2 mutational status. Thus, whereas a definitive answer on whether TMB can serve as a predictive biomarker for response on recurrent GBM can only be determined on clinical trials, it is possible that the hypermutator phenotype induced by TMZ treatment might not be informative of response to immunotherapy for patients who do not have germline alterations in DNA repair and replication pathways. This is important to consider, as gliomas are regarded as immunologically cold tumors in part because of a characteristic low mutational burden. Therefore, for most gliomas, neoantigen-based immunotherapy will not be an option and strategies to circumvent this poor antigenicity might be required.
Neoadjuvant Immunotherapy for GBM
As we begin to understand more about identifiable biomarkers that correlate with response and resistance to checkpoint blockade, recent studies have started to examine whether these drugs are “on target” and truly stimulate observable immune responses in the GBM microenvironment. Conceptually, in order to undergo a clinically relevant response to agents blocking the PD-1/PD-L1 molecules, the host must have first developed an antitumor response, likely antigen specific, which was prevented from effectively attacking tumor cells due to the engagement of the PD-1/PD-L1 pathway. However, unless the unique features of the tissue-specific GBM microenvironment are characterized in patients following checkpoint blockade, it is difficult to determine the effects of these treatments on the tumor-immune system dynamic and to understand the molecular basis for intrinsic resistance. To this end, “window of opportunity” clinical studies are well suited to enable comprehensive interrogation of the immune response in GBM following treatment.58 In this method, patients are treated neoadjuvantly prior to surgery or biopsy. Importantly, recent preclinical and clinical studies have both provided compelling evidence that the timing of immunotherapy and surgery can improve antitumor immune responses59 and lead to objective clinical response rates and survival in other solid tumors.59–64 For example, in a recent comparison study in stage III melanoma, 2 doses of concurrent ipilimumab (3 mg/kg) and nivolumab (1 mg/kg) prior to surgery resulted in a complete pathologic response at surgery in 7 of 9 patients.61 In another recent study, the combination of combined PD-1 and blockade of cytotoxic T-lymphocyte antigen 4 in the neoadjuvant setting resulted in higher overall response rates and antitumor T-cell responses.60 In patients with non-small-cell lung cancer treated neoadjuvantly with nivolumab, pathologic responses were observed in 45% of patients, and T-cell clones identified within tumors and blood in a subset of patients were shown to expand.
This approach has recently been extended to recurrent GBM with provocative data from 2 independent studies underscoring the need for further study (Fig. 1B). In Cloughesy et al, patients were enrolled in a randomized clinical trial designed to compare neoadjuvant plus adjuvant PD-1 blockade with adjuvant PD-1 blockade only.65 One dose of pembrolizumab (200 mg) was administered prior to surgical resection in the neoadjuvant group, and both groups were treated with the same dosing every 3 weeks following resection until progression. In responding patients, a transcriptional signature highlighted by high expression of T-cell and interferon genes, but downregulation of cell cycle–related genes, was differentially expressed in tumor tissue of patients who received neoadjuvant pembrolizumab. Moreover, an expansion of T-cell clones in peripheral blood and induction of PD-L1 in the tumor tissue was observed. These data were concordant with the study by Schalper and colleagues,66 who also observed the induction of an interferon signature in the tumor tissue and changes in T-cell receptor clonality. Schalper et al confirmed PD-1 receptor occupancy by nivolumab in brain tumor tissue, addressing a long-standing question that the blood–brain barrier may not limit the reinvigoration of antitumor immune responses in situ. Together with the study by Zhao et al,9 these studies showed similar immunologic findings that, in patients with recurrent GBM, exhausted T cells exist in the tumor microenvironment and can be activated with PD-1 blockade. However, when these data are considered along with the negative large randomized trials in GBM, it is likely that reinvigoration of exhausted T cells alone is necessary but not sufficient to provide a therapeutic effect.
The mechanisms preventing a clinical response in recurrent GBM are currently unknown but are likely multifactorial. Notably, patients who were randomized to receive neoadjuvant pembrolizumab in the Cloughesy et al study65 lived, on average, twice as long as the patients who received only adjuvant pembrolizumab,65 although no survival benefit compared with historical controls was observed among neoadjuvant nivolumab recipients in the Schalper study.66 Although this finding might be simply related to chance, another plausible hypothesis is that the neoadjuvant priming and expansion of exhausted T cells within the GBM microenvironment, followed by the removal of an immune suppressive tumor microenvironment through surgical resection, led to the observed prolonged survival. A mechanistic underpinning for these data is not yet fully understood. However, preclinical studies and clinical immune monitoring work have shown that neoadjuvant immunotherapy was associated with the rapid expansion of tumor-specific CD8+ T cells in other solid tumors.59,63 Some of these expanded CD8+ T-cell clones found in the systemic circulation could be identified in the primary tumor, while other expanded tumor-specific T-cell populations seem to be clonally replaced following PD-1 blockade.59–62,67 Crucially, the presence of the primary tumor prior to extirpation appears to be critical for the efficacy of the neoadjuvantly administered immunotherapy, suggesting that factors such as (i) the absolute amount of antigen burden during immune cell licensing as well as (ii) the presence of tumor-involved antigen presenting cells are critically involved in the reinvigoration of antigen-specific T cells. In mouse models, cross-presentation by Batf3+ or CD103+ dendritic cells is involved in effector T-cell trafficking into the tumor microenvironment.68,69 Thus, available data suggest that a complex dynamic of T-cell reinvigoration, chemokine cues for trafficking, and available antigen presentation capacity regulates the infiltration and function of effective antitumor immune responses induced by checkpoint blockade.
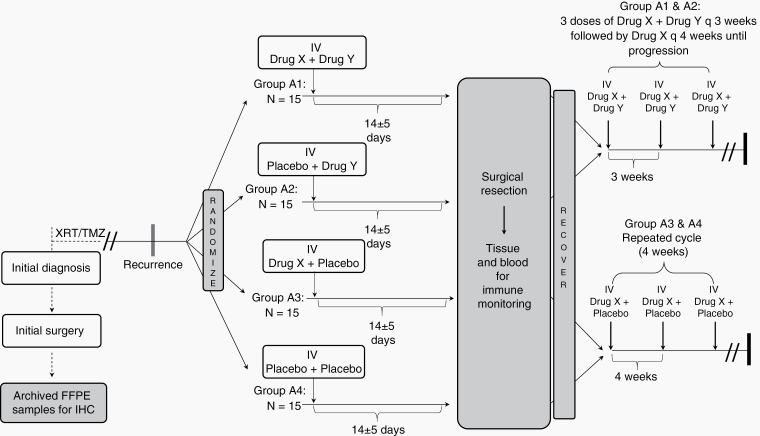
In the study by Cloughesy et al,65 a randomization strategy was employed into the neoadjuvant component of the clinical trial design that ultimately provided 2 distinct advantages. First, this approach enabled the use of a contemporary randomized control group prior to surgery, with the same inclusion and exclusion criteria from which to make comparisons of tissue and systemic immune responses against the experimental arm. Randomization is particularly important for these studies and provides additional confidence that observed immunologic and transcriptional findings were robust. Second, randomization facilitated reduced bias in evaluation when comparing arms for a clinical efficacy endpoint. Despite the underpowering of the clinical efficacy endpoints, large effect sizes can be statistically evaluated in order to provide further insight into clinical development opportunities. Randomization will likely be incorporated into the neoadjuvant element of future studies, and the appropriate trial design of such studies will ideally allow for meaningful comparisons across datasets (Fig. 2). Moving forward, additional trials should be initiated in the neoadjuvant setting to better understand how particular drug combinations alter the immune cellular composition and activation states with the tumor microenvironment. Importantly, harmonized assays needed to decode the immune responses may allow for meaningful and effective comparisons between trials. As it stands, the current data suggest that there may be opportunities to tune the immunogenicity of GBM if the timing of treatment is “moved up” to the presurgical setting. Rigorous studies of these approaches may help to deconstruct the key features of GBM immunoediting currently limiting immunotherapeutic approaches.
Fig. 2.
Schema for randomized neoadjuvant studies. In recurrent GBM, patients are randomized to receive neoadjuvant immunotherapeutics (drug X and/or drug Y) or placebo combinations in the presurgical window. Following surgery, tissue is carefully characterized for immunologic and genomic changes. In the adjuvant setting, patients may receive either a single drug X or the combination of drugs X and Y.
Personalized Cancer Immunogenomics: Neoantigen Cancer Vaccines
Whereas checkpoint blockade immunotherapy is an “off the shelf” treatment, recent work has been directed at designing truly personalized immunotherapies that target a patient’s unique tumor-associated features. To this end, the field of cancer immunogenomics has led to a reinvigoration of the idea that therapeutic—rather than prophylactic—cancer vaccines represent promising new approaches to GBM treatment. After a myriad of unsuccessful efforts at cancer vaccination targeting mostly wild-type proteins, enthusiasm for this approach was limited.70 These efforts were likely hampered by prior host tolerization to vaccine targets as well as the lack of checkpoint blockade augmentation. In contrast, discovery that tumor-specific, mutant-derived peptides—termed “neoantigens”—can be recognized by the immune system71 has created new and exciting opportunities to study antitumor immune responses and to create personalized, precision immunotherapies.
The foundation of neoantigen targeting is that some structural genomic abnormalities generate immunogenic protein antigens. Whereas a subset of founder DNA alterations confers phenotypic advantages to cancer cells and can be considered “driver” mutations, the majority of DNA mutations are functionally inconsequential and are therefore referred to as “passenger” mutations. In some cancers, driver mutations are true dependencies in that inhibiting the mutant proteins, if possible, can impair tumor growth. In GBM, no high incidence of shared dependencies have been identified to date. However, even if mutant proteins are not excellent drug targets, they may represent bona fide immunogenic antigens that can be used to direct immune responses to cancer. Regardless of biological function, some tumor mutations are transcribed and ultimately translated into novel, tumor-specific neoantigens that undergo proteasomal degradation, transport to the endoplasmic reticulum, and subsequent presentation on the surface of tumor cells via major histocompatibility complex (MHC) class I or of antigen-presenting cells on MHC class I or II molecules. In turn, T cells can react specifically against MHC-presented neoantigens.
The identification of candidate neoantigens through cancer immunogenomic approaches represents the synthesis of next-generation sequencing and immunology. Rather than distinguishing somatic mutations according to perceived biological function, cancer immunogenomics prioritizes tumor-specific mutations by predicted immunogenicity. These “neoantigens” are identified by whole exome DNA and RNA sequencing as expressed alterations predicted to bind with high affinity to specific MHC molecules.5 This approach was first conceptualized in silico by Segal et al71 and validated in preclinical sarcoma,72,73 colorectal cancer,74 melanoma,75 and GBM.76 Importantly, identification of neoantigens in patients77,78 has raised the exciting possibility of leveraging this approach to develop new treatment strategies. By definition, neoantigens are expressed exclusively by tumor cells, and therefore are capable of generating perhaps more robust immune responses because they, unlike tumor-associated antigens which share expression with normal tissues, did not exist during immunologic development.5,55,79,80 Thus, it is likely that patients are not tolerant to this class of epitopes.
Many mechanisms contribute to neoantigen formation in tumor cells.81 The most common mechanism is the generation of single nucleotide variants (SNVs), which arise from non-synonymous nucleotide substitutions to generate mutant peptides that differ from wild-type sequences by a single amino acid. In addition, the insertion or deletion of base pairs within a protein coding DNA sequence leads to “indel” mutations which generate open reading frames that are postulated to be highly immunogenic due to their unique coding sequence downstream of the indel. A pan-cancer analysis of indel mutations and their corresponding immunogenicity has recently been described from TCGA and identified renal cell carcinomas (RCCs) as the cancer subtype having the highest percentage of indel mutations.82 Indel mutation burden and immunogenicity likely contribute to the therapeutic benefit of immune checkpoint inhibition among RCCs, despite the overall low mutational burden and frequency of SNV mutations exhibited by these tumors.83 Neoantigens can also be generated by interchromosomal fusion mutations,84 insertion of endogenous retro-elements such as long-terminal repeats and retroviruses,85 posttranscriptional RNA splice variants,86 and posttranslational modifications.87 Thus, neoantigens may arise from any pre- or posttranslational process that leads to the presentation by the MHC of a peptide sequence to which the host was not tolerized during immunologic development.
Neoantigens can be identified several ways. The most common approach is to use any one of a suite of prediction algorithms that input (i) expressed variants with (ii) HLA haplotype information to identify mutant peptides that may bind with high affinity to MHC molecules.88–90 Importantly, there are a number of computational algorithms designed to predict neoantigens that take into account binding affinity, peptide processing, and transcript abundance, among other features (reviewed by Richters et al88). There is no consensus on the best method of prediction. Because these approaches attempt to infer the presence of the actual peptides bound to MHC, the biochemical characterization of peptides physically bound to MHC molecules using mass spectrometry represents another approach to neoantigen identification.74 Although proteomic approaches are appealing because they may provide information on the actual immunopeptidome, they are highly technical and may not be available at every center. Moreover, because T cells may be activated by as few as 1–2 MHC:peptide complexes, sensitivity of mass spectrometry may miss low abundance neoantigens. Ideally, neoantigen identification would integrate both in silico and proteomic approaches.
Accumulating data implicate neoantigens as therapeutic targets for cancer therapy. First, higher neoantigen load has been associated with improved outcome to checkpoint blockade in some cancers as described above.28–36 Second, neoantigen reactive T cells have been demonstrated to expand in the setting of an effective antitumor immune response.28,91 Third, neoantigen reactive T cells can kill target expressing tumor cells in vivo.75,92 Finally, loss of tumor neoantigens has been implicated as a mechanism of acquired resistance to immune checkpoint blockade.93 Although neoantigens represent a potentially potent source of antitumor recognition, not all neoantigens are created equally, as some elicit more robust immune responses than others. As described above, indel neoantigens are predicated to be highly immunogenic and are estimated to generate 3–9 times more neoantigens per mutation than SNVs.82 Enhanced benefit following immune checkpoint blockade has also been associated with neoantigens that are clonally distributed within tumors,39 as well as those with a higher “fitness” score, which reflects an enhanced likelihood of MHC presentation as well as subsequent T-cell recognition.94 A variant neoantigen fitness model based on similarity of neoantigens to infectious disease–derived immune epitopes combined with CD8+ T-cell tumor density has recently been demonstrated to predict improved survival of GBM patients.95 Combining whole-exome and transcriptome sequencing with mass spectrometric analysis of MHC-presented peptides represents another approach to better define immunogenic neoepitope peptides.74,96 Nonetheless, given that <1% of neoantigen mutations are capable of generating spontaneous immune responses,97 strategies to sensitize immune responses to tumor neoantigens have evolved.
A more recent, exciting therapeutic application of neoantigen discovery is the development of clinical trials evaluating the potential of personalized vaccines against tumor-specific neoepitopes for cancer patients98,99 (Fig. 1C). The first personalized vaccines were developed to treat advanced melanoma, a tumor with known high mutational burden and inherent immunogenicity. The first treatment was developed to treat 3 patients with stage III melanoma with a personalized, neoantigen peptide-loaded dendritic cell vaccine in which immune responses to a subset of vaccine peptides were detected in all treated patients.100 Subsequent studies by 2 additional groups showed that CD4+ and CD8+ neoantigen-specific T-cell responses could be generated in patients with stage III–IV melanoma vaccinated with either polyvalent peptide101 or RNA vaccines.102 These studies demonstrated the potential of generating immune responses to neoantigens and reflect the various vaccine platforms employed to this end.
A priori, key challenges to incorporating neoantigen vaccination approaches in GBM include: (i) incorporation of vaccination into an established cytotoxic therapy regimen; (ii) immune specialization of the CNS1,103; (iii) frequent administration of immunosuppressive corticosteroids to treat symptomatic cerebral edema; (iv) inherent low immunogenicity and mutational burden of GBM104; and (v) known intratumoral molecular heterogeneity.51,105,106 Nonetheless, neoantigen vaccination efforts have demonstrated safety, feasibility, and encouraging immunogenicity among GBM patients. The first neoantigen vaccine targeted the junctional neoantigen created by epidermal growth factor receptor variant III (EGFRvIII), although predicted HLA binding was not a part of the trial design.107 Recent studies have applied cancer immunogenomics methods to vaccinating GBM patients.4,108 Johanns et al reported the outcome of a single patient treated with a personalized vaccine consisting of autologous tumor lysate pulsed dendritic cells followed by administration of 8 synthetic long peptides corresponding to tumor-specific neoepitopes with polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose (poly-ICLC) adjuvant.109 HLA classes I and II reactivity to tumor neoantigens was detected in peripheral blood with a similar pattern of reactivity observed among tumor infiltrating T cells isolated post-vaccination following redo surgery.
Two larger studies demonstrated the potential of personalized GBM vaccination. In the “Glioma Actively Personalized Vaccine Consortium” (GAPVAC) study, 15 newly diagnosed GBM patients who underwent a gross total resection not on steroids were treated with personalized, dual-product vaccines along with radiation and TMZ.110 Patients were first vaccinated with up to 7, best-ranked—via criteria including expression, physical HLA binding, and immunogenicity—“off-the-shelf” tumor-associated antigen peptides (GAPVAC1). Patients were then vaccinated with 2 neoantigen peptides identified by sequencing and HLA binding affinity (GAPVAC2). CD4+ and CD8+ T-cell responses were observed to both GAPVAC 1 and GAPVAC 2 vaccines in 11 patients. Vaccines were well tolerated, and efficacy was encouraging, including median progression-free survival and OS of 14.5 and 29.0 months, respectively. Independently, Keskin and colleagues vaccinated 8 patients with newly diagnosed and MGMT unmethylated GBM with a median of 13 neoepitope peptides (range, 7–20) identified by sequencing and HLA prediction using a prime/boost schedule with adjuvant poly-ICLC.111 All patients received standard radiotherapy, but TMZ was withheld due to its nominal benefit among MGMT unmethylated patients.112 Most patients had residual measurable tumor, and 6 were on dexamethasone at the time of vaccine priming. No patients developed vaccine-induced, serious adverse events. Notably, none of the patients who were on dexamethasone at vaccine priming demonstrated vaccine-specific immune responses. In contrast, both patients not on dexamethasone generated tumor-specific CD4+ and CD8+ T-cell responses to multiple vaccinated neoepitope peptides. Analysis of tumor resected after vaccination revealed a robust infiltration of immune effector T cells among responding patients who were confirmed by T-cell receptor clonotypic analyses to be specific for vaccinated neoepitope peptides.111 Together, these studies confirm that neoantigen vaccination is safe, feasible, and capable of generating neoantigen-specific T-cell responses that can traffic successfully to intracranial tumors. Importantly, these data suggest that dexamethasone should be used with caution as it may attenuate vaccine-induced immune responses. Further studies evaluating neoantigen vaccination for GBM patients are under way, including combination with PD-1 checkpoint blockade (NCT02287428) and a novel, DNA-based plasmid approach (NCT04015700). Ongoing studies will be necessary to determine which vaccine platforms are most effective, what combination therapies are necessary, and how best to target heterogeneity within tumors.
Chimeric Antigen Receptor T-Cell Therapy
Adoptive immunotherapy with engineered T cells is part of the broad explosion in immunotherapeutic approaches to cancer. Transferring T cells in GBM may be particularly advantageous because it bypasses endogenous antigen presentation and T-cell clonal expansion (Fig. 1D). After nearly three decades of research in antigen receptors, ex vivo T-cell culture, and gene transfer, it has become possible to redirect autologous human T cells on a scale suitable for therapeutic use. The combination of these technologies is used to collect peripheral blood mononuclear cells and confer a large fraction of that population of T cells with specificity to a defined antigen using gene transfer of a well-characterized antigen receptor. Typically, transferred receptors are high-affinity, multifunctional structures in which (i) antibody-based binding domains dictate recognition and specificity and (ii) T-cell signal transduction molecules to which the recognition domain is fused trigger T-cell activation, cytokine production, and proliferation. The chimerism of B cell–based target recognition with T cell–based functionality within the same receptor led to the term “chimeric antigen receptors,” or CARs. Importantly, CARs recognize targets in their native confirmation on the surfaces of tumor cells, independent of the HLA:peptide complex. Autologous T cells redirected with CARs targeting the B-cell marker CD19 have shown remarkable and durable efficacy in B-cell malignancies, including acute lymphoblastic leukemia and most lymphomas,113–119 and are approved by the FDA for subsets of these indications. Similarly, autologous CAR T cells targeting B-cell maturation antigen expressed by plasma cells have shown significant efficacy in patients with relapsed or refractory multiple myeloma.120 Thus, CAR T cells are a promising therapeutic modality in advanced, chemotherapy-resistant hematologic malignancies. Importantly, CAR T cells can also traffic to the CNS121 and have induced remissions of secondary CNS lymphoma.122,123 The ability of CAR T cells to penetrate the blood–brain barrier opens the possibility for their deployment in malignant brain tumors, where it can be difficult to achieve therapeutic concentrations of small molecule–based chemotherapies or larger antibody-based therapeutics.
Because CARs target cell surface proteins, identifying appropriate and specific cell surface antigens to target remains a big challenge in CAR development. Several antigens have been targeted with CAR T cells in GBM clinical trials, including human epidermal growth factor receptor 2 (Her2),124,125 interleukin (IL)-13Rα2,126,127 and EGFRvIII.128–130 Importantly, these trials have demonstrated the safety of using CAR T cells in brain tumors and have not resulted in the cytokine-mediated toxicities commonly observed in hematologic malignancies—namely, cytokine release syndrome and immune-effector cell related neurologic syndrome (ICANS). ICANS is a transient syndrome characterized by altered mental status, language difficulty, and in some cases, seizures and cerebral edema. Unfortunately, with the exception of a single case,127 no complete responses have been reported in these trials, though occasional partial responses have been observed. The reasons behind lack of efficacy remain unclear, but it is likely that target antigen expression, the fitness of the starting T-cell population, and insufficient proliferation in the brain tumor parenchyma, due to trafficking or the inhibitory tumor microenvironment, all contribute.
The ideal GBM CAR target should be expressed on the tumor initiating/stem cell population and harbor a biological role in maintaining the tumor phenotype—ie, is a true cancer dependency. EGFRvIII, for example, is a constitutively active oncogenic mutation, but it is expressed in only ~30% of GBM patients, and its expression is heterogeneous within each tumor. Her2 is expressed more frequently and homogeneously, but its expression in other nontumor tissues narrows the therapeutic window. IL-13Rα2 is frequently expressed in GBM (~58%)131 but is not essential for tumor maintenance, and escape has been noted with both fusion ligands and T cells directed to it.127,132 It is likely that targeting multiple antigens will be necessary to achieve clinical efficacy. Studies of combined dual-specificity CARs have been proposed to minimize antigen escape,133,134 and are just entering clinical trials. One recently reported strategy targeted 2 antigens, one of which is tumor specific (EGFRvIII) and another which would be considered “undruggable” due to systemic expression and lack of effective penetration of targeted therapies (EGFR). In this strategy, CAR T cells serve as blood-brain barrier–penetrating carriers and can lyse tumor cells expressing EGFRvIII; concomitantly, CAR T cells also produce bispecific T-cell engager antibodies, which serve as a bridge to trigger lysis by both CAR T cells and bystander T cells, including immunosuppressive regulatory T cells. Thus, EGFR is targeted locally in the brain tumor while systemic toxicity is minimized due to low systemic concentrations and rapid clearance of the engager antibodies.135 Overall the combination of CAR T-cell therapy with bispecific engagers to repurpose resident T cells against tumor cells is an exciting new possibility that might provide advantages of each of these 2 complementary approaches for T cell–based immunotherapy for GBM. Another recent technological development is the rapid advancement in T-cell receptor discovery and engineering, which might ultimately allow elegant and efficient design of CAR T cells against prevalent tumor-specific antigens such as H3.3 mutations.136
In addition to multiple targets, there are other variables to consider in designing and administering cellular therapy with engineered T cells. CARs may contain constructs with different signaling domains influencing their long-term cellular proliferation and survival, may be cultured under different conditions during manufacturing conditions, and may be affected biologically by their transduced vectors.137 It is also important to consider route of administration,25 as has been demonstrated in preclinical and clinical studies of other solid tumors that are confined to a tissue compartment.132 In the reported case of a complete response in a patient with multifocal GBM, the response occurred after multiple intraventricular infusions of IL-13Rα2 CAR T cells.127 Thus, maximizing trafficking into the CNS may be critical to optimizing efficacy.
The GBM microenvironment is known to be highly immunosuppressive,2 and infiltrating T cells often show exhaustion characteristics.26 Even with successful initial GBM CAR T-cell antigen targeting, sustained responses may require strategies that make CAR T cells resistant to exhaustion or immunosuppression. One way to fortify transferred immune cells from immunosuppression is to combine them with checkpoint blockade, which is currently being tested. Alternatively, gene-editing technologies can knock out genes that mediate T-cell exhaustion or dampen T-cell responses, such as PD-1.135 In a case report of a chronic lymphocytic leukemia patient, a near-knockout mutation of TET2 (ten-eleven translocation methylcytosine dioxygenase 2), an epigenetic modifier, resulted in “forever young” CAR T cells,138 indicating that strategic gene editing may be used to enhance T-cell persistence and modify key biological programs. The observed T-cell dysfunction in GBM patients7 may thus be another area where novel therapeutics or technologies could be implemented to improve T-cell therapy efficacy.
Conclusions
We have summarized exciting recent progress in several areas germane to GBM immunotherapy—new ways to apply checkpoint blockade immunotherapy, highly personalized approaches to therapeutic GBM vaccination, and methods for “off the shelf” CAR T-cell technologies. Additional compelling areas of investigation, such as oncoviral treatments, also converge in GBM immunobiology but are beyond the scope of this review. The ongoing work in these areas suggests that, although challenging, many investigators in the field are not yet prepared to “write off” the potential promise of immune-based treatments for patients with GBM. Indeed, despite all of the work in GBM immunotherapy to date, there is a general sense that the field is still at the beginning, albeit unsure of where the end might be. Rational clinical trial design and prudent patient stratification remain critical to these efforts in order to not only identify those subsets of patients most likely to benefit from new treatments but also avoid ignoring a clinical signal within the noise of a heterogeneous treated population. To this end, it is likely that cancer immunogenomics approaches will remain particularly important in order to understand the genomic basis of response and resistance as well as to enable the identification and targeting of tumor-specific neoantigens. Combining these strategies rationally will likely prove pivotal in bringing more effective treatments to patients. Ultimately, as we study these exciting approaches in larger studies, we will not just learn about their therapeutic efficacy but also gain new insights into the fundamental immunobiology of the CNS itself.
Funding
This work received support to the following authors: G.P.D: R01NS112712–01 (National Institute of Neurological Disorders and Stroke), K08NS092912-05 (NINDS), Damon Runyon Cancer Research Foundation. T.F.M.: P50CA211015 (NCI), U54CA199090 (National Cancer Institute [NCI]), Ben and Catherine Ivy Foundation. R.M.P.: P50CA211015 (NCI), U54CA199090 (NCI), R01CA222695-01 (NCI), Ben and Catherine Ivy Foundation, Parker Institute for Cancer Immunotherapy, Brain Tumor Funders’ Collaborative. A.M.S.: 1R01NS110703-01A1 (NINDS), 5DP5OD021356-05 (NIH Office of the Director), P50CA221747 (NCI), P30CA060553 (NCI), philanthropic support from Dan and Sharon Moceri.
Conflict of interest statement.
G.P.D.: co-founder of Immunovalent Therapeutics.
T.F.M.: Paid consulting: Roche, Trizel, Medscape, Bayer, Amgen, Odonate Therapeutics, Pascal Biosciences, Bayer, Del Mar Pharmaceuticals, Tocagen, Karyopharm, GW Pharma, Kiyatec, Abbvie, Boehinger Ingelheim, VBI, Deciphera, VBL, Agios, Merck, Roche, Genocea, Celgene, Puma, Lilly, BMS, Cortice, Wellcome Trust, Novocure, Novogen, Boston Biomedical, Sunovion, Human Longevity, Insys, ProNai, Pfizer, Notable labs, Medqia. Stock options: Notable Labs. Other: Member of the board for the 501c3 Global Coalition for Adaptive Research. Patents: U.S. Provisional Application No.: 62/819,322 Compositions and Methods for Treating Cancer. A.M.S.: Paid Consulting: Abbive. Patents: Systems and Methods for Predicting Clinical Responses to Immunotherapies. D.A.R.: Paid Consulting: Abbvie; Advantagene; Agenus; Amgen; Bayer; Bristol-Myers Squibb; Celldex; Delmar; Emd Serono; Genentech/Roche; Inovio; Merck; Merck KGaA; Monteris; Novocure; Oncorus; Oxigene; Regeneron; Stemline; Taiho Oncology, Inc. Research support: Acerta Phamaceuticals; Agenus; Celldex; EMD Serono; Incyte; Inovio; Midatech; Omniox; Tragara.
References
- 1. Dunn GP, Okada H. Principles of immunology and its nuances in the central nervous system. Neuro Oncol. 2015; 17(Suppl 7):vii3–vii8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dunn GP, Fecci PE, Curry WT. Cancer immunoediting in malignant glioma. Neurosurgery. 2012;71(2):201–222; discussion 222–223. [DOI] [PubMed] [Google Scholar]
- 3. Chongsathidkiet P, Jackson C, Koyama S, et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat Med. 2018;24(9):1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johanns TM, Dunn GP. Applied cancer immunogenomics: leveraging neoantigen discovery in glioblastoma. Cancer J. 2017;23(2):125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. [DOI] [PubMed] [Google Scholar]
- 6. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woroniecka KI, Rhodin KE, Chongsathidkiet P, Keith KA, Fecci PE. T-cell dysfunction in glioblastoma: applying a new framework. Clin Cancer Res. 2018;24(16):3792–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zappasodi R, Merghoub T, Wolchok JD. Emerging concepts for immune checkpoint blockade-based combination therapies. Cancer Cell. 2018;34(4):690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ruess DA, Heynen GJ, Ciecielski KJ, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med. 2018;24(7):954–960. [DOI] [PubMed] [Google Scholar]
- 11. Lin L, Asthana S, Chan E, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci U S A. 2014;111(7):E748–E757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Esposito G, De Filippis D, Cirillo C, Sarnelli G, Cuomo R, Iuvone T. The astroglial-derived S100beta protein stimulates the expression of nitric oxide synthase in rodent macrophages through p38 MAP kinase activation. Life Sci. 2006;78(23):2707–2715. [DOI] [PubMed] [Google Scholar]
- 13. Nadra I, Mason JC, Philippidis P, et al. Proinflammatory activation of macrophages by basic calcium phosphate crystals via protein kinase C and MAP kinase pathways: a vicious cycle of inflammation and arterial calcification? Circ Res. 2005;96(12):1248–1256. [DOI] [PubMed] [Google Scholar]
- 14. Pruett SB, Zheng Q, Schwab C, Fan R. Sodium methyldithiocarbamate inhibits MAP kinase activation through toll-like receptor 4, alters cytokine production by mouse peritoneal macrophages, and suppresses innate immunity. Toxicol Sci. 2005;87(1):75–85. [DOI] [PubMed] [Google Scholar]
- 15. Rao KM. MAP kinase activation in macrophages. J Leukoc Biol. 2001;69(1):3–10. [PubMed] [Google Scholar]
- 16. Sharma RK, Sodhi A, Batra HV, Tuteja U. Phosphorylation of p42/44 MAP kinase is required for rF1-induced activation of murine peritoneal macrophages. Mol Immunol. 2005;42(11):1385–1392. [DOI] [PubMed] [Google Scholar]
- 17. Zhao M, Liu Y, Wang X, New L, Han J, Brunk UT. Activation of the p38 MAP kinase pathway is required for foam cell formation from macrophages exposed to oxidized LDL. APMIS. 2002;110(6):458–468. [DOI] [PubMed] [Google Scholar]
- 18. Kane JR, Zhao J, Tsujiuchi T, et al. CD8+ T-cell-mediated immunoediting influences genomic evolution and immune evasion in murine gliomas. Clin Cancer Res. May 19, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lei L, Sonabend AM, Guarnieri P, et al. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6(5):e20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang SH, Mirabolfathinejad SG, Katta H, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A. 2014;111(15):5664–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. George S, Miao D, Demetri GD, et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46(2):197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lastwika KJ, Wilson W 3rd, Li QK, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. [DOI] [PubMed] [Google Scholar]
- 24. Noh KH, Kang TH, Kim JH, et al. Activation of Akt as a mechanism for tumor immune evasion. Mol Ther. 2009;17(3):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brown CE, Aguilar B, Starr R, et al. Optimization of IL13Ralpha2-targeted chimeric antigen receptor T cells for improved anti-tumor efficacy against glioblastoma. Mol Ther 2018;26(1):31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woroniecka K, Fecci PE. T-cell exhaustion in glioblastoma. Oncotarget. 2018;9(82):35287–35288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018; 33(5):843–852 e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson DB, Frampton GM, Rioth MJ, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4(11):959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387(10031):1909–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goodman AM, Sokol ES, Frampton GM, Lippman SM, Kurzrock R. Microsatellite-stable tumors with high mutational burden benefit from immunotherapy. Cancer Immunol Res. 2019;7(10):1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17(4):209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Germano G, Lamba S, Rospo G, et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552(7683):116–120. [DOI] [PubMed] [Google Scholar]
- 38. Mandal R, Samstein RM, Lee KW, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stadler ZK, Battaglin F, Middha S, et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next-generation sequencing panels. J Clin Oncol. 2016;34(18):2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. TCGA Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J, Lee IH, Cho HJ, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. [DOI] [PubMed] [Google Scholar]
- 46. Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Thuijl HF, Mazor T, Johnson BE, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7):2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi S, Yu Y, Grimmer MR, Wahl M, Chang SM, Costello JF. Temozolomide-associated hypermutation in gliomas. Neuro Oncol. 2018;20(10):1300–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12(2):104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mahlokozera T, Vellimana AK, Li T, et al. Biological and therapeutic implications of multisector sequencing in newly diagnosed glioblastoma. Neuro Oncol. 2018;20(4):472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sa JK, Choi SW, Zhao J, et al. Hypermutagenesis in untreated adult gliomas due to inherited mismatch mutations. Int J Cancer. 2019;144(12):3023–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bouffet E, Larouche V, Campbell BB, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. [DOI] [PubMed] [Google Scholar]
- 54. Johanns TM, Miller CA, Dorward IG, et al. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6(11):1230–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest. 2015;125(9):3413–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hundal J, Carreno BM, Petti AA, et al. pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 2016;8(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Touat M, Li YY, Boynton AN, et al. Mechanisms and therapeutic implications of hypermutation in gliomas. Nature. 2020;580(7804):517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Glimelius B, Lahn M. Window-of-opportunity trials to evaluate clinical activity of new molecular entities in oncology. Ann Oncol. 2011;22(8):1717–1725. [DOI] [PubMed] [Google Scholar]
- 59. Huang AC, Orlowski RJ, Xu X, et al. A single dose of neoadjuvant PD-1 blockade predicts clinical outcomes in resectable melanoma. Nat Med. 2019;25(3):454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amaria RN, Reddy SM, Tawbi HA, et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat Med. 2018;24(11):1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blank CU, Rozeman EA, Fanchi LF, et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat Med. 2018;24(11):1655–1661. [DOI] [PubMed] [Google Scholar]
- 62. Forde PM, Chaft JE, Pardoll DM. Neoadjuvant PD-1 blockade in resectable lung cancer. N Engl J Med. 2018;379(9):e14. [DOI] [PubMed] [Google Scholar]
- 63. Liu J, Blake SJ, Yong MC, et al. Improved efficacy of neoadjuvant compared to adjuvant immunotherapy to eradicate metastatic disease. Cancer Discov. 2016;6(12):1382–1399. [DOI] [PubMed] [Google Scholar]
- 64. Carthon BC, Wolchok JD, Yuan J, et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin Cancer Res. 2010;16(10):2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25(3):477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schalper KA, Rodriguez-Ruiz ME, Diez-Valle R, et al. Neoadjuvant nivolumab modifies the tumor immune microenvironment in resectable glioblastoma. Nat Med. 2019;25(3):470–476. [DOI] [PubMed] [Google Scholar]
- 67. Yost KE, Satpathy AT, Wells DK, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med. 2019;25(8):1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu J, Rozeman EA, O’Donnell JS, et al. Batf3+ DCs and type I IFN are critical for the efficacy of neoadjuvant cancer immunotherapy. Oncoimmunology. 2019;8(2):e1546068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Spranger S, Dai D, Horton B, Gajewski TF. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 2017;31(5):711–723 e714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rosenberg SA, Yang JC, Restifo NP. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Segal NH, Parsons DW, Peggs KS, et al. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68(3):889–892. [DOI] [PubMed] [Google Scholar]
- 72. Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2014;515(7528):572–576. [DOI] [PubMed] [Google Scholar]
- 75. Castle JC, Kreiter S, Diekmann J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res. 2012;72(5):1081–1091. [DOI] [PubMed] [Google Scholar]
- 76. Johanns TM, Ward JP, Miller CA, et al. Endogenous neoantigen-specific CD8 T cells identified in two glioblastoma models using a cancer immunogenomics approach. Cancer Immunol Res. 2016;4(12):1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. van Rooij N, van Buuren MM, Philips D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31(32):e439–e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med. 2013;19(6):747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Desrichard A, Snyder A, Chan TA. Cancer neoantigens and applications for immunotherapy. Clin Cancer Res. 2016;22(4):807–812. [DOI] [PubMed] [Google Scholar]
- 80. Castle JC, Uduman M, Pabla S, Stein RB, Buell JS. Mutation-derived neoantigens for cancer immunotherapy. Front Immunol. 2019;10:1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS. Alternative tumour-specific antigens. Nat Rev Cancer. 2019;19(8):465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Turajlic S, Litchfield K, Xu H, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol. 2017;18(8):1009–1021. [DOI] [PubMed] [Google Scholar]
- 83. Motzer RJ, Escudier B, McDermott DF, et al. ; CheckMate 025 Investigators Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373(19):1803–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yu YP, Liu P, Nelson J, et al. Identification of recurrent fusion genes across multiple cancer types. Sci Rep. 2019;9(1):1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Smith CC, Beckermann KE, Bortone DS, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128(11):4804–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jayasinghe RG, Cao S, Gao Q, et al. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 2018;23(1):270–281 e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004;427(6971):252–256. [DOI] [PubMed] [Google Scholar]
- 88. Richters MM, Xia H, Campbell KM, Gillanders WE, Griffith OL, Griffith M. Best practices for bioinformatic characterization of neoantigens for clinical utility. Genome Med. 2019;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol Res. 2013;1(1):11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liu XS, Mardis ER. Applications of immunogenomics to cancer. Cell. 2017;168(4):600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Prickett TD, Crystal JS, Cohen CJ, et al. Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol Res. 2016;4(8):669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Anagnostou V, Smith KN, Forde PM, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7(3):264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Łuksza M, Riaz N, Makarov V, et al. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature. 2017;551(7681):517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang J, Caruso FP, Sa JK, et al. The combination of neoantigen quality and T lymphocyte infiltrates identifies glioblastomas with the longest survival. Commun Biol. 2019;2:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Abelin JG, Keskin DB, Sarkizova S, et al. Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity. 2017;46(2):315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science. 2015;350(6266):1387–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sahin U, Türeci Ö. Personalized vaccines for cancer immunotherapy. Science. 2018;359(6382):1355–1360. [DOI] [PubMed] [Google Scholar]
- 99. Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol. 2018;18(3):168–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Carreno BM, Magrini V, Becker-Hapak M, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348(6236): 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547(7662):222–226. [DOI] [PubMed] [Google Scholar]
- 103. Engelhardt B, Vajkoczy P, Weller RO. The movers and shapers in immune privilege of the CNS. Nat Immunol. 2017;18(2):123–131. [DOI] [PubMed] [Google Scholar]
- 104. Hodges TR, Ott M, Xiu J, et al. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19(8):1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Weller M, Butowski N, Tran DD, et al. ; ACT IV trial investigators Rindopepimut with temozolomide for patients with newly diagnosed, EGFRvIII-expressing glioblastoma (ACT IV): a randomised, double-blind, international phase 3 trial. Lancet Oncol. 2017;18(10):1373–1385. [DOI] [PubMed] [Google Scholar]
- 108. Johanns TM, Bowman-Kirigin JA, Liu C, Dunn GP. Targeting neoantigens in glioblastoma: an overview of cancer immunogenomics and translational implications. Neurosurgery. 2017;64(CN_suppl_1):165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Johanns TM, Miller CA, Liu CJ, et al. Detection of neoantigen-specific T cells following a personalized vaccine in a patient with glioblastoma. Oncoimmunology. 2019;8(4):e1561106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hilf N, Kuttruff-Coqui S, Frenzel K, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565(7738):240–245. [DOI] [PubMed] [Google Scholar]
- 111. Keskin DB, Anandappa AJ, Sun J, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565(7738):234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Stupp R, Hegi ME, Mason WP, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumour and Radiation Oncology Groups ; National Cancer Institute of Canada Clinical Trials Group. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 113. Maude SL, Teachey DT, Porter DL, Grupp SA. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood. 2015;125(26):4017–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Porter DL, Hwang WT, Frey NV, et al. Chimeric antigen receptor T cells persist and induce sustained remissions in relapsed refractory chronic lymphocytic leukemia. Sci Transl Med. 2015;7(303):303ra139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Turtle CJ, Hanafi LA, Berger C, et al. Immunotherapy of non-Hodgkin’s lymphoma with a defined ratio of CD8+ and CD4+ CD19-specific chimeric antigen receptor-modified T cells. Sci Transl Med. 2016;8(355):355ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Turtle CJ, Hanafi LA, Berger C, et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest. 2016;126(6):2123–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Davila ML, Riviere I, Wang X, et al. Efficacy and toxicity management of 19-28z CAR T cell therapy in B cell acute lymphoblastic leukemia. Sci Transl Med. 2014;6(224):224ra225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lee DW, Kochenderfer JN, Stetler-Stevenson M, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kochenderfer JN, Dudley ME, Feldman SA, et al. B-cell depletion and remissions of malignancy along with cytokine-associated toxicity in a clinical trial of anti-CD19 chimeric-antigen-receptor-transduced T cells. Blood. 2012;119(12):2709–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Raje N, Berdeja J, Lin Y, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N Engl J Med. 2019;380(18):1726–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Grupp SA, Kalos M, Barrett D, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N Engl J Med. 2013;368(16):1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Abramson JS, McGree B, Noyes S, et al. Anti-CD19 CAR T cells in CNS diffuse large-B-cell lymphoma. N Engl J Med. 2017;377(8):783–784. [DOI] [PubMed] [Google Scholar]
- 123. Frigault MJ, Dietrich J, Martinez-Lage M, et al. Tisagenlecleucel CAR T-cell therapy in secondary CNS lymphoma. Blood. 2019;134(11):860–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ahmed N, Salsman VS, Kew Y, et al. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors. Clin Cancer Res. 2010;16(2):474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ahmed N, Brawley V, Hegde M, et al. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 2017;3(8):1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Brown CE, Badie B, Barish ME, et al. Bioactivity and safety of IL13Rα2-redirected chimeric antigen receptor CD8+ T cells in patients with recurrent glioblastoma. Clin Cancer Res. 2015;21(18):4062–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Brown CE, Alizadeh D, Starr R, et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N Engl J Med. 2016;375(26):2561–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Maus MV. Designing CAR T cells for glioblastoma. Oncoimmunology. 2015;4(12):e1048956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Johnson LA, Scholler J, Ohkuri T, et al. Rational development and characterization of humanized anti-EGFR variant III chimeric antigen receptor T cells for glioblastoma. Sci Transl Med. 2015;7(275):275ra222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. O’Rourke DM, Nasrallah MP, Desai A, et al. A single dose of peripherally infused EGFRvIII-directed CAR T cells mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma. Sci Transl Med. 2017;9(399):eaaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Brown CE, Warden CD, Starr R, et al. Glioma IL13Rα2 is associated with mesenchymal signature gene expression and poor patient prognosis. PLoS One. 2013;8(10):e77769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Adusumilli PS, Cherkassky L, Villena-Vargas J, et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci Transl Med. 2014;6(261):261ra151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hegde M, Corder A, Chow KK, et al. Combinational targeting offsets antigen escape and enhances effector functions of adoptively transferred T cells in glioblastoma. Mol Ther. 2013;21(11):2087–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Grada Z, Hegde M, Byrd T, et al. TanCAR: a novel bispecific chimeric antigen receptor for cancer immunotherapy. Mol Ther Nucleic Acids. 2013;2:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Choi BD, Yu X, Castano AP, et al. CRISPR-Cas9 disruption of PD-1 enhances activity of universal EGFRvIII CAR T cells in a preclinical model of human glioblastoma. J Immunother Cancer. 2019;7(1):304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chheda ZS, Kohanbash G, Okada K, et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J Exp Med. 2018;215(1):141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Maus MV, June CH. Making better chimeric antigen receptors for adoptive T-cell therapy. Clin Cancer Res. 2016;22(8):1875–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Fraietta JA, Nobles CL, Sammons MA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558(7709):307–312. [DOI] [PMC free article] [PubMed] [Google Scholar]