Abstract
Background
Diffuse intrinsic pontine glioma (DIPG) is an aggressive pediatric brainstem tumor. Most DIPGs harbor a histone H3 mutation, which alters histone post-translational modification (PTM) states and transcription. Here, we employed quantitative proteomic analysis to elucidate the impact of the H3.3K27M mutation, as well as radiation and bromodomain inhibition (BRDi) with JQ1, on DIPG PTM profiles.
Methods
We performed targeted mass spectrometry on H3.3K27M mutant and wild-type tissues (n = 12) and cell lines (n = 7).
Results
We found 29.2 and 26.4% of total H3.3K27 peptides were H3.3K27M in mutant DIPG tumor cell lines and tissue specimens, respectively. Significant differences in modification states were observed in H3.3K27M specimens, including at H3K27, H3K36, and H4K16. In addition, H3.3K27me1 and H4K16ac were the most significantly distinct modifications in H3.3K27M mutant tumors, relative to wild-type. Further, H3.3K36me2 was the most abundant co-occurring modification on the H3.3K27M mutant peptide in DIPG tissue, while H4K16ac was the most acetylated residue. Radiation treatment caused changes in PTM abundance in vitro, including increased H3K9me3. JQ1 treatment resulted in increased mono- and di-methylation of H3.1K27, H3.3K27, H3.3K36 and H4K20 in vitro.
Conclusion
Taken together, our findings provide insight into the effects of the H3K27M mutation on histone modification states and response to treatment, and suggest that H3K36me2 and H4K16ac may represent unique tumor epigenetic signatures for targeted DIPG therapy.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13046-020-01773-x.
Keywords: DIPG, Histone, H3K27M, H3K36me2, H4K16ac
Background
Pediatric high-grade glioma is the leading cause of cancer death in children [1]. Diffuse intrinsic pontine glioma (DIPG) is a high-grade glioma that arises in the brainstem, with an overall survival of only 11 months [2]. DIPG cannot be removed surgically, and there is no known effective treatment. Standard management is with radiotherapy, which temporarily relieves symptoms but only confers a survival benefit of three months [3]. Since DIPGs ultimately exhibit treatment resistance and disease progression, better understanding of tumor biology is desperately needed to identify effective therapeutic therapy [4].
Recent studies suggest that changes in epigenetic regulation of gene transcription play a key role in pediatric gliomagenesis and response to treatment [5–7]. Dynamic post-translational modifications (PTMs) on the histone N-terminal tail, rendered by histone modifying enzymes, alter chromatin structure to facilitate open reading (euchromatin) or silencing (heterochromatin) throughout the genome [5, 8–10]. Specific patterns of histone PTMs represent an epigenetic signature, or histone code, reflecting a relative transcriptional state. In up to 80% of DIPGs, heterozygous somatic mutations in genes encoding histone H3 isoforms H3.1 and H3.3 (HIST1H3B and H3F3A, respectively) substitute methionine for lysine at amino acid residue 27 on the H3 N-terminal tail (H3K27M) [11]. This H3K27M mutant protein alters histone H3 PTMs in DIPG, impacting chromatin structure and hence gene expression [5, 7, 12].
For example, histone H3K27 di- and tri-methylation (H3K27me2/3), transcriptionally silencing marks, are reduced in H3K27M DIPG, de-repressing transcription [13–15]. H3K27me2/3 loss is accompanied by increased H3K27ac and the formation of heterotypic H3K27M/H3K27ac nucleosomes that colocalize with bromodomain proteins 2 and 4 (BRD2/4) and RNA polymerase II, suggesting a potential role of BRD proteins in DIPG pathogenesis [7, 16]. We demonstrated pre-clinical efficacy of bromodomain inhibition (BRDi) with JQ1, a bromodomain and extra terminal domain (BET) inhibitor, resulting in reduction of H3K27ac and restoration of H3K27me3 [7]. These findings suggest that changes in epigenetic regulation of gene expression in DIPG may be reflected by histone codes, and detection of these signatures could shed light on mechanisms of tumorigenesis. Here, we employ targeted mass spectrometry via multiple reaction monitoring (MRM) to characterize 93 distinct PTM states in H3.3K27M mutant DIPG specimens, and investigate the effects of radiation and bromodomain inhibition on tumor epigenetic signatures.
Materials and methods
Tissue specimens
Pediatric DIPG tumor tissue (n = 9, H3.3K27M mutant) was collected postmortem (IRB #Pro00001339). H3.3K27 wild-type (WT) adult glioma tumor tissue specimens (n = 2), and one non-tumor frontal lobe tissue specimen were collected during the course of treatment (IRB# STU00095863). All diagnoses were made radiographically by a neuroradiologist, then confirmed by neuropathological evaluation upon tissue collection. Patient identifiers were removed and a single sequential numerical identifier assigned to each specimen (Table 1). Tissues were stored at − 80 °C prior to proteomic analysis (IRB #STU00202063).
Table 1.
Summary of specimens analyzed with histone tail mass spectrometry
| ID | Specimen type | Histone Status | Gender | Age (Years) |
|---|---|---|---|---|
| 1 | DIPG tissue | H3.3K27M | F | 5 |
| 2 | DIPG tissue | H3.3K27M | M | 8 |
| 3 | DIPG tissue | H3.3K27M | M | 7 |
| 4 | DIPG tissue | H3.3K27M | M | 19.6 |
| 5 | DIPG tissue | H3.3K27M | F | 7 |
| 6 | DIPG tissue | H3.3K27M | M | 7.6 |
| 7 | DIPG tissue | H3.3K27M | M | 6 |
| 8 | DIPG tissue | H3.3K27M | M | 6 |
| 9 | DIPG tissue | H3.3K27M | M | 6.25 |
| 10 | GBM tissue | H3.3G34R | N/A | 45 |
| 11 | GBM tissue | Wild type | N/A | 50 |
| 12 | Focal cortical dysplasia, frontal lobe | Wild type | N/A | 62 |
| SF7761 | DIPG cell line | H3.3K27M | N/A | N/A |
| SF8628 | DIPG cell line | H3.3K27M | N/A | N/A |
| DIPG 007 | DIPG cell line | H3.3K27M | N/A | N/A |
| KNS 42 | GBM cell line | H3.3G34V | N/A | N/A |
| SF9427 | Pediatric GBM cell line | Wild type | N/A | N/A |
| U87 | Adult GBM cell line | Wild type | N/A | N/A |
| NHA | Normal human astrocytes | Wild type | N/A | N/A |
Cell lines
Patient-derived pediatric DIPG cell lines SF8628, SF7761, DIPG007 (H3.3K27M mutant) and high-grade glioma cells KNS42 (H3.3G34V mutant) and SF9427 (H3.3G34 wild-type) were provided by Dr. Rintaro Hashizume (Northwestern University) as previously described [17–19]. H3 wild-type adult glioma cells (U87) and normal human astrocytes (NHA) were obtained from American Type Culture Collection and ScienCell (#1800), respectively. Cells were treated with 300 nM JQ1 (Selleckchem S7110) or DMSO (Sigma D2650) for 24 and 48 h. Cell irradiation (9 Gy) was performed with the RadSOURCE-2000 X-Ray Irradiator, and cells analyzed at 24 and 48 h. Cell pellets were generated by harvesting with trypsin and washing twice with PBS, then stored at − 80 °C prior to proteomic analysis.
Western blotting
Histones were extracted from cell pellets using Histone Extraction Kit (ab113476, Abcam) according to the manufacturer’s protocol. 0.5 μg Histones were loaded and separated by SDS-PAGE using a 12% Mini-PROTEAN® TGX™ Precast Protein Gels and transferred by using Trans-Blot Turbo Mini 0.2 μm PVDF Transfer Packs at 2.5A, 25 V for 3 min in a Trans-Blot Turbo™ Transfer Systems (Bio-Rad). The membrane was blocked prior to the addition of the primary antibody with 5% milk in Tris buffered saline with 0.1% of Tween 20. The membrane was incubated overnight with primary antibody in TBS buffer with 0.1% Tween and 5% milk. The dilution of primary antibody was as follows: Rabbit monoclonal anti-Histone H3 (mutated K27M) antibody (ab190631) at a dilution of 1:1000, Rabbit mAb Acetyl-Histone H3 (Lys27) (D5E4) XP ((#8173, Cell Signaling Technology) at a dilution of 1:1000, Mouse mAb Histone H3 (1B1B2) (#14269, Cell Signaling Technology) at a dilution of 1:1000. The membrane was washed 3 times with TBS/0.1% Tween and incubated with an anti-rabbit or anti-mouse IgG conjugated to horse radish peroxidase (7074 and 7076, Cell Signaling Technology) at a 1:2000 dilution in TBS/0.1% Tween and 5% milk. Pierce ECL Plus Western Blotting Substrate (CN: 32132, Thermo Scientific) was used according to the manufacturer’s protocol to visualize proteins and quantify band intensity. Experiments were performed in triplicate for statistical analysis. The intensity of protein bands were analyzed by ImageJ, with the relative protein value expressed via the ratio of specific protein/H3.
Gene expression profiling
Total cell RNA was extracted for gene expression profiling based on manufacturer protocol (Qiagen). All samples were adjusted to a concentration of 20 ng/μl RNA prior to submission for gene expression analysis using the NanoString nCounter Plexset Analysis System DNA damage repair 96 gene codeset (NanoString Technologies, Seattle, WA). Gene expression values were determined and converted to fold change in expression level for each treatment condition compared to DMSO controls in order to determine statistically significant differences in gene expression levels between groups (NanoString). Functional pathways and gene ontology analysis was then performed on fold change expression values (Inginuity Pathways Analysis, Qiagen).
Histone preparation for targeted mass spectrometry
Nuclei were isolated in buffer (15 mM Tris-HCl pH 7.5, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 250 mM sucrose; 1 mM DTT, 1:100 HALT protease inhibitor (Thermo Scientific)), and 10 mM sodium butyrate added immediately prior to use containing 0.3% NP-40. Nuclei were centrifuged at 600 x g for five minutes at 4 °C and washed twice with nuclear isolation buffer. Histones were extracted by via five volumes of H2SO4 and incubated at room temperature one hour. Debris was removed by centrifugation at 4000 x g for five minutes. Trichloroacetic acid was added to the supernatant at a final concentration of 20% (v/v) and incubated overnight at 4 °C. Precipitated histones were pelleted at 10,000 x g for five minutes, washed once with ice-cold acetone with 0.1% HCl and twice with acetone at 15,000 x g for five minutes. Residual acetone was dried in a fume hood, and samples stored at − 20 °C until further processing. Histones were derivatized with a single round of propionylation, digested with 1 μg trypsin (Promega, Madison, WI), and derivatized again with a single round of propionylation according to Garcia et al. [20].
Targeted LC-MS/MS of histone peptides
Histone peptides were resuspended in Loading Buffer (0.1% TFA in water) and analyzed on a Thermo TSQ Quantiva (Thermo Scientific) in-line with a Dionex nano-LC (Sunnyvale, CA). Peptides were loaded onto a trapping column (3 cm × 150 μm, packed with ProntoSIL C18-AQ, 3 μm, 200 Å resin (New Objective, Woburn, MA)) for 10 min at 2.5 μL/min with 100% Loading Buffer. Elution from the trapping column and separation on a PicoChip analytical capillary column (10 cm × 75 μm packed with ProntoSIL C18-AQ, 3 μm, 200 Å resin (New Objective)) was achieved by decreasing the percentage of Solvent A (0.1% formic acid in water) and increasing the percentage of Solvent B (0.1% formic acid in 95% acetonitrile) from 1 to 35% at a flow rate of 0.30 μL/min over 45 min. The peptides were introduced into the triple quadrupole mass spectrometer by electrospray from an emitter with a 10 μm tip. The instrument settings were as follows: collision gas pressure of 1.5 mTorr; Q1 peak width of 0.7 (FWHM); cycle time of 3 s; skimmer offset of 10 V; electrospray voltage of 2.5 kV. All injections were performed in technical triplicate. Targeted methods were performed as previously published [21]. A list of monitored peptide transitions is provided (Supplementary material 1).
Quantitation of histone modification states
Raw data were imported into Skyline with Savitzky-Golay smoothing for analysis. Peak area assignments were manually confirmed, and total peak areas used to determine the percent relative abundance of histone modifications. The percent relative peptide abundance was determined by dividing the area under the curve for a given peptide of interest by the sum of the areas under the curve for all peptides with the same sequence, then multiplying by 100. Heatmaps were generated with R using gplots package, and show modifications that were above the limit of detection in all samples.
Statistical analysis
Differences in peptide abundance were analyzed using independent-sample t test for between-group comparisons, or one-way ANOVA and Tukey’s post hoc test for multi-group analyses using SPSS (IBM, Version 25), and expressed as mean ± standard deviation (SD). Pearson Correlation was performed to analyze the relationship between H3.3K27M mutation and detected histone codes. Multivariable analysis was performed using linear regression with Stepwise entry (p = 0.05) and removal (p = 0.10). All p values were two-sided, with p < 0.05 considered statistically significant.
Results
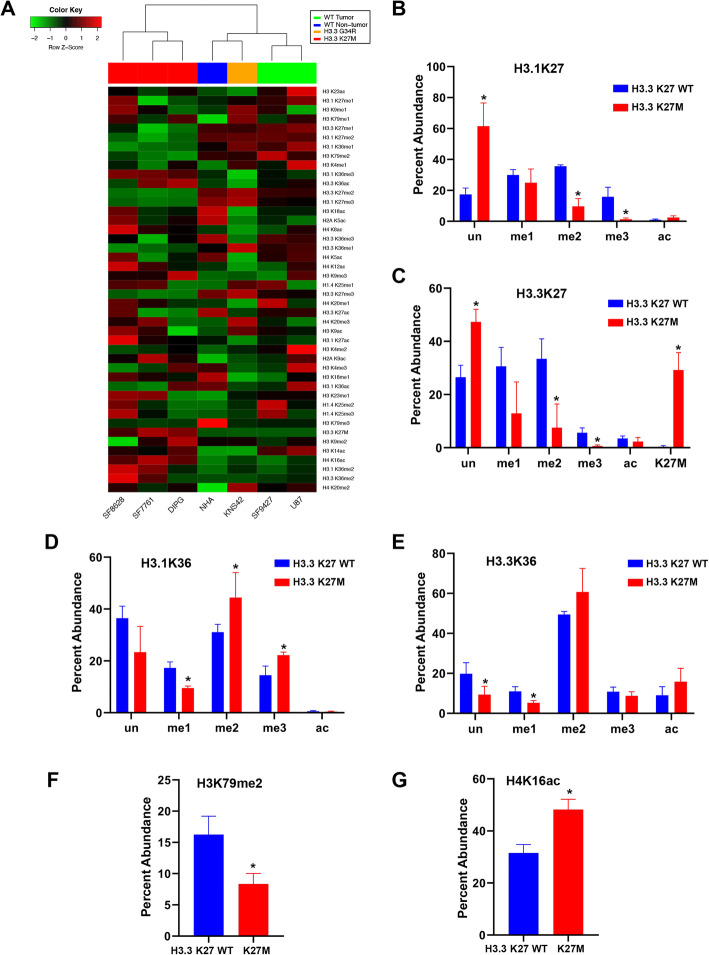
Glioma cell histone codes are distinct by H3.3K27M mutation status
Using MRM, we determined histone N-terminal tail signatures (histone codes) in H3.3K27M mutant and wild type (WT) glioma cell lines and astrocytes (Supplementary material 2, Table 1). Histone codes were distinct by H3 mutation status (Fig. 1a). K27 di- and tri-methylation (K27me2/3) were significantly decreased in H3.3K27M mutant cells on H3.1 and H.3.3 isoforms (Fig. 1a, Supplementary material 3, 4). A significant difference in H3.1K27me3 was also observed, with a mean relative abundance of 15.9% versus 1.4% in mutant versus WT lines (Fig. 1b). A similar trend was observed on H3.3, with H3.3K27me3 relative abundance of 5.6% in H3.3K27 WT cells and only 0.6% in mutants (Fig. 1c). We observed greater K36 di- and tri-methylation in mutants, coupled with less abundant H3.1K36me (Fig. 1d, e). H3K79me2 was less abundant in H3.3K27M mutant cells compared to WT (mean = 8.3 and 16.3%, respectively, Fig. 1f).
Fig. 1.
Histone peptide modification states in pediatric glioma cell lines. a Unsupervised hierarchical clustering of pediatric glioma cell line histone tail modification profiles reveals clustering by H3K27M mutation status. b-e Bar graph showing that relative abundance of modifications at H3.1K27/K36 and H3.3K27/K36 in glioma cell lines are distinct by H3.3K27M mutation status. f-g Statistically significant differences in H3K79me2 and H4K16ac abundance are observed in H3.3K27M mutant glioma cell lines (n = 3) compared to H3.3K27 WT (n = 4). * p < 0.05 compared to H3.3K27 WT group, independent-sample t test, two-tailed. un = unmodified, me1 = mono-methylation, me2 = di-methylation, me3 = tri-methylation, ac = acetylation
Histone acetylation also differed based on mutation status. Acetylation at H4K16 was lower in H3.3K27 WT cells compared to mutant (31.5 and 48.2%, respectively Fig. 1g). H3.1K27me2 and H3.1K36me2 were the most common PTMs associated with H3.3K27M mutant peptide on multivariate analysis (F = 428.7, p < 0.01). Importantly, because the H3F3A mutation yielding the H3.3K27M mutant protein is heterozygous, we quantified relative abundance of H3.3K27 WT and H3.3K27M peptides in a given cell sample: 29.2% of H3.3K27 peptides in our mutant cell lines were H3.3K27M (SD = 6.6, range 22.2–35.4%, n = 3. Figure 1c). As expected, the H3.3K27M peptide was not detected in H3.3K27 WT cells.
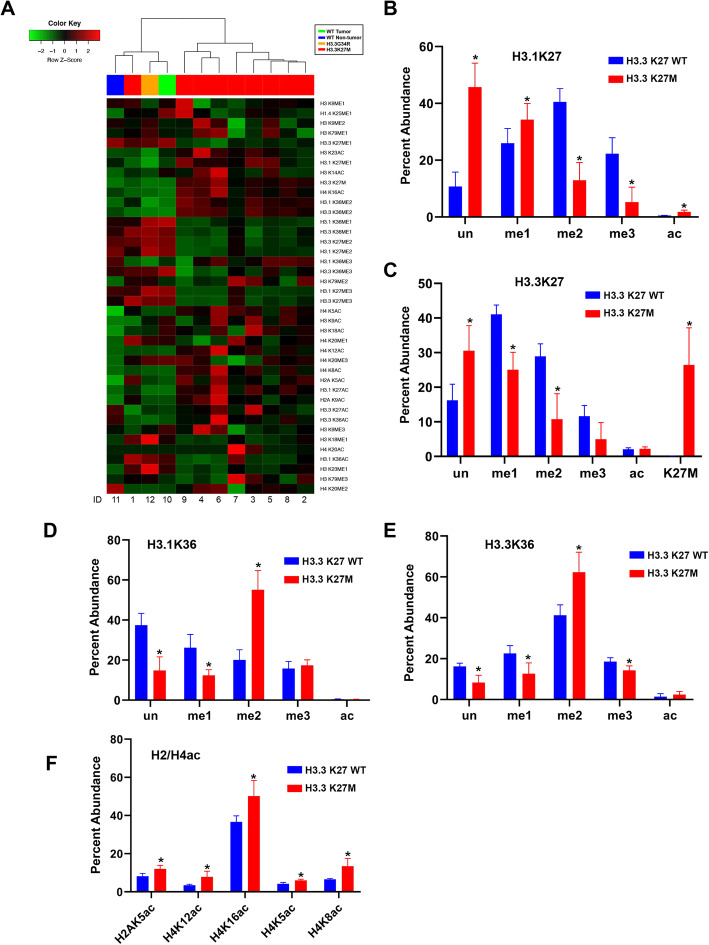
Glioma tissue histone codes are distinct by H3.3K27M mutation status
Tissue histone PTM profiles also clustered by H3.3K27M mutation status (Fig. 2a). The relative abundance of 16 PTMs were statistically significantly different between H3.3K27M mutant and WT tissues (Supplementary material 5). As with cell lines, the greatest differences in PTMs between H3.3K27M mutant and WT tissues were K27 and K36 methylation states (Fig. 2b-e, Supplementary material 5). We also detected significantly greater acetylation of H2AK5, H4K5, H4K8, H4K12, and H4K16 (Fig. 2f). H3.3K27me1 and H4K16ac were the most common PTM states associated with H3.3K27M peptide abundance on multivariate analysis (F = 214.0, p < 0.01).
Fig. 2.
Tissue histone peptide modification states differ by H3.3K27M mutation status. a Unsupervised hierarchical clustering of glioma and normal brain tissue histone tail modification profiles reveals clustering by H3.3K27M mutation status. b-e Bar graph showing relative modifications at H3.1K27/K36 and H3.3K27/K36 in tissue specimens are distinct by H3.3K27M mutation status. f Statistically significant modifications of H2 and H4 acetylation are observed in H3.3K27M mutant tissues (n = 9) compared to H3.3K27 WT (n = 3).* p < 0.05 compared to H3.3K27 WT group, independent-sample t test, two-tailed. un = unmodification, me1 = mono-methylation, me2 = di-methylation, me3 = tri-methylation, ac = acetylation
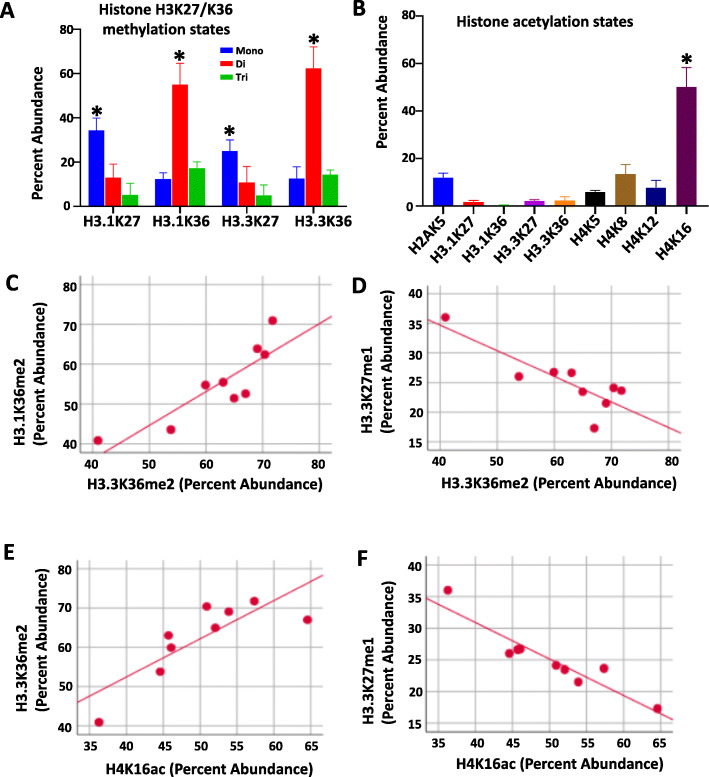
As the greatest difference in relative abundance of PTM states in H3.3K27M mutant specimens were observed at H3K27, H3K36 and multiple H4 tail amino acids, we queried for the predominant modification states at these residues to determine correlations between specific modification states at these locations (Fig. 3). Overall, K27me1 and K36me2 were the predominant modifications observed on H3.1 and H3.3 isoforms (Fig. 3a), while acetylation was greatest at H4K16 (Fig. 3b). H3.3K36me2 abundance positively correlated with H3.1K36me2 levels, and negatively correlated with H3.3K27me1 (Fig. 3c, d). Also, H4K16ac abundance correlated positively with H3.3K36me2, and negatively with H3.3K27me1 (Fig. 3e, f).
Fig. 3.
Predominant histone modification states in DIPG tumor tissue. a K27 mono-methylation and K36 di-methylation are predominant modification states on H3.1 and H3.3 tails in DIPG tumor tissue (n = 9). * p < 0.05 compared to all the other modification states (Tukey HSD, Post Hoc Tests, one way ANOVA). b H4K16ac is the predominant acetylated residue in DIPG tumor tissue (n = 9). * p < 0.05 compared to all the other modification states (Tukey HSD, Post Hoc Tests, one way ANOVA). c,d H3.3K36me2 abundance positively correlates with H3.1K36me2, and negatively correlates with H3.3K27me1 (Pearson Correlation = 0.87 and − 0.84, respectively, all p < 0.01. Pearson Correlation, two-tailed). e, f H4K16ac abundance positively correlates with H3.3K36me2, and negatively correlates with H3.3K27me1 (Pearson Correlation = 0.82 and − 0.94, respectively, all p < 0.01. Pearson Correlation, two-tailed)
We also quantified the relative abundance of H3.3K27 WT and H3.3K27M peptides in a given tissue sample. Similar to cell lines, 26.44% of H3.3K27 peptides in our mutant tumor specimens were H3.3K27M (SD = 10.7, range 5.2–44.5%, n = 9), with no mutant peptides detected in WT tissues.
Lastly, we compared DIPG cell and tumor tissue epiproteomic profiles. Overall, modifications on the H3.1/H3.3K27 N-terminal tail were similar in DIPG cell lines and tumor tissues. Specifically, there was no significant difference in abundance of the majority of H3.1/H3.3K27 peptides quantified, with differential fold change (FC) in abundance > 2 or < 0.5 for only six peptide modification states between DIPG cells and tissues (Table 2, Supplementary material 6). Significant differences in peptide modification states between groups were observed on H1.4K25, H3.3K36, H3K79, H3K9, and H4K20 peptides. Importantly, no difference in relative H3.3K27M peptide abundance was detected between DIPG tissue and cell lines (mean = 26.4 and 29.2, respectively, p = 0.686). Of note, unmodified peptides and modifications with < 5% relative abundance were excluded from this comparative analysis to ensure biological significance.
Table 2.
Comparison of DIPG Tumor tissue and cell line epiproteomic profiles. Differential fold change (FC) in abundance (> 2 or < 0.5) between DIPG tumor tissue and cell lines was detected only in the six H3.1/H3.3K27 peptide modification states listed
| Histone code | Mean | Fold_Change | p value | |
|---|---|---|---|---|
| Tissue | Cells | Tissue/Cells | ||
| H3.3K36ac | 2.4 | 15.8 | 0.2 | < 0.01 |
| H3K79me2 | 18.0 | 8.3 | 2.2 | < 0.05 |
| H3.3K36me1 | 12.7 | 5.3 | 2.4 | < 0.05 |
| H3K9ac | 5.6 | 2.0 | 2.8 | < 0.01 |
| H4K20me3 | 8.5 | 2.2 | 3.9 | < 0.01 |
| H1.4K25me1 | 29.7 | 5.1 | 5.8 | < 0.01 |
Co-occurring histone modification states in tissue specimens
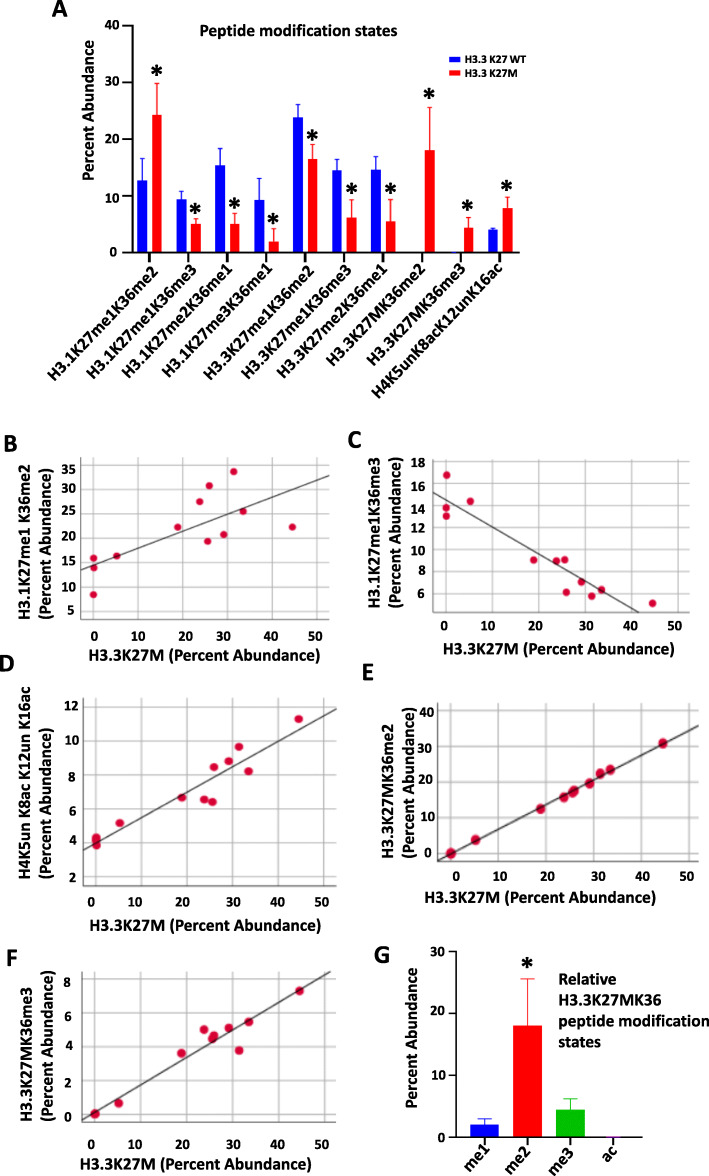
Because our method requires protein digestion for subsequent peptide mass spectrometry, determination of co-occurring PTMs at distant amino acid residues along the N-terminal tail is not possible. However, PTMs at proximal amino acids on the same peptide can be assessed. Therefore, to further explore the relationship between the H3.3K27M mutant peptide and histone PTM states, we investigated the PTMs of H3K27/K36 and H4K5/K8/K12/K16 peptides. The relative abundance of ten combinations of PTM states along these peptides were significantly different by tissue H3.3K27M mutation status (Fig. 4a, Supplementary material 7). Of these, H3.1K27me1K36me2, H3.3K27MK36me2, and H4K5unK8acK12unK16ac were increased in mutants compared to WT. These findings are consistent with the single amino acid analysis presented above. Further, we observed a positive correlation between H3.1K27me1K36me2 and H3.3K27M peptide abundance, and a negative correlation between H3.1K27me1K36me3 and H3.3K27M peptide abundance (Fig. 4b, c, Supplementary material 7). The abundance of the H4K5unK8acK12unK16ac peptide also positively correlated with H3.3K27M peptide abundance (Fig. 4d, Supplementary material 7). In contrast, H3.1K27me2K36me1, H3.1K27me3K36me1, H3.3K27me1K36me2/3 and H3.3K27me2K36me1 peptide abundance negatively correlated with H3.3K27M abundance (Supplementary material 7). On multivariate analysis, K36me2/3 were the most common modification states on the mutant H3.3K27MK36 peptide (F = 18,444.8, p < 0.01). As such, H3.3K27MK36me2 and H3.3K27MK36me3 peptide abundance also positively correlated with H3.3K27M abundance (Fig. 4e, f, Supplementary material 7). Importantly, di-methylation was the predominant modification state of the K36 residue on the H3.3K27MK36 peptide, while K36Ac was least abundant (Fig. 4g).
Fig. 4.
Distinct peptide modification states detected in DIPG tumor tissue. a Distinct combinations of modification states are observed on ten peptides in H3.3K27M mutant tumor tissue (n = 9) relative to H3.3K27 wild-type (WT) tissue (n = 3).* p < 0.05 compared to H3.3K27 WT group, Independent-sample t test, two-tailed. b-f Scatter plots showing the correlation between relative abundance of H3.1K27me1K36me2/me3, H4K5un K8acK12unK16ac, and H3.3K27MK36me2/3 peptides with the H3.3K27M peptide in DIPG tumor tissue (Pearson Correlation = 0.72, −0.79, 0.95, 0.99, and 0.97, respectively, all p < 0.01. Pearson Correlation, two-tailed, n = 12). g Relative abundance of H3.3K27MK36 peptide modification states in DIPG tumor tissue reveals K36me2 as the most abundant co-occurring peptide with H3.3K27M mutation peptide. * p < 0.05 compared to all the other groups (Tukey HSD, Post Hoc Tests, one way ANOVA, n = 9)
Radiation treatment alters DIPG histone codes in vitro
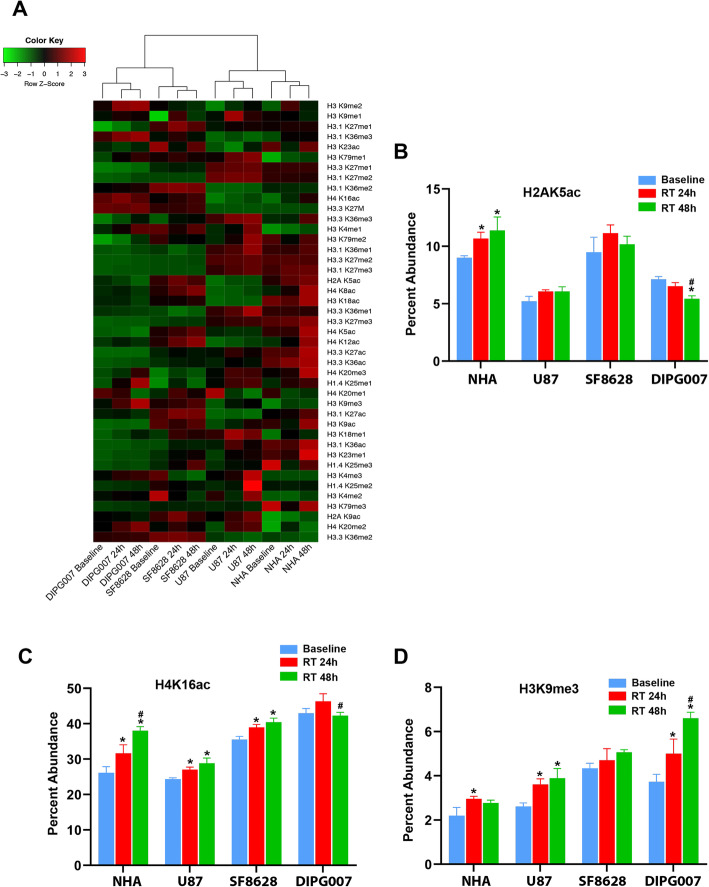
As radiation treatment (RT) is the standard therapy for DIPG, we examined the effects of RT on the glioma histone code in vitro. We treated H3.3K27M mutant DIPG cells (SF8628, DIPG007), H3.3K27 WT glioma cells (U87) and normal astrocytes (NHA) with 9 Gy radiation, and performed targeted histone mass spectrometry at 24 and 48 h post-treatment. Cell histone codes clustered first by origin, then by treatment condition (Fig. 5a). Comparative analysis revealed distinct differences in relative peptide abundance after RT, most notably in acetylated peptides (Supplementary material 8). In DIPG007 cells, the relative abundance of H2AK5ac decreased with RT at 48 h (Fig. 5b). In contrast, H2AK5ac abundance increased in NHA at 24 and 48 h, with no significant change in SF8628 or U87 (Fig. 5b). At 24 and 48 h post-RT, all lines except DIPG007 showed a significant increase in H4K16ac (Fig. 5c, Supplementary material 7). H3K9me3 also significantly increased in NHA, U87, and DIPG007, and trended towards a significant increase in SF8628, after RT (Fig. 5d, Supplementary material 8).
Fig. 5.
Radiation treatment alters relative histone peptide modification states in DIPG, in vitro. a Unsupervised hierarchical clustering of pediatric glioma cell line histone tail modification profiles reveals clustering by cell line and treatment condition. b-d Relative abundance of H2AK5ac, H4K16ac and H3K9me3 peptides are distinct after radiation treatment. * p < 0.05 compared to Baseline; # p < 0.05 compared to 24 h post-RT treatment (Tukey HSD, Post Hoc Tests, one way ANOVA)
To determine the potential biological impact of observed differences in histone modifications after RT relative to controls, we compared cell gene expression profiles in these same treatment groups. On functional pathways analysis of genes identified as statistically significantly differentially expressed by RT treated cells relative to controls (fold change in expression > 2 or < − 2, adjusted p-value < 0.05), we identified DNA double-strand break repair as a top canonical pathway activated in RT treated DIPG cell lines (p-value 1.10 × 10− 21, Supplementary material 9). The top implicated molecular and cellular function in RT treated cells, as well as the top network of molecular interaction, was DNA replication, recombination and repair (p-value 2.06 × 10− 6),
Bromodomain inhibition alters DIPG histone codes in vitro
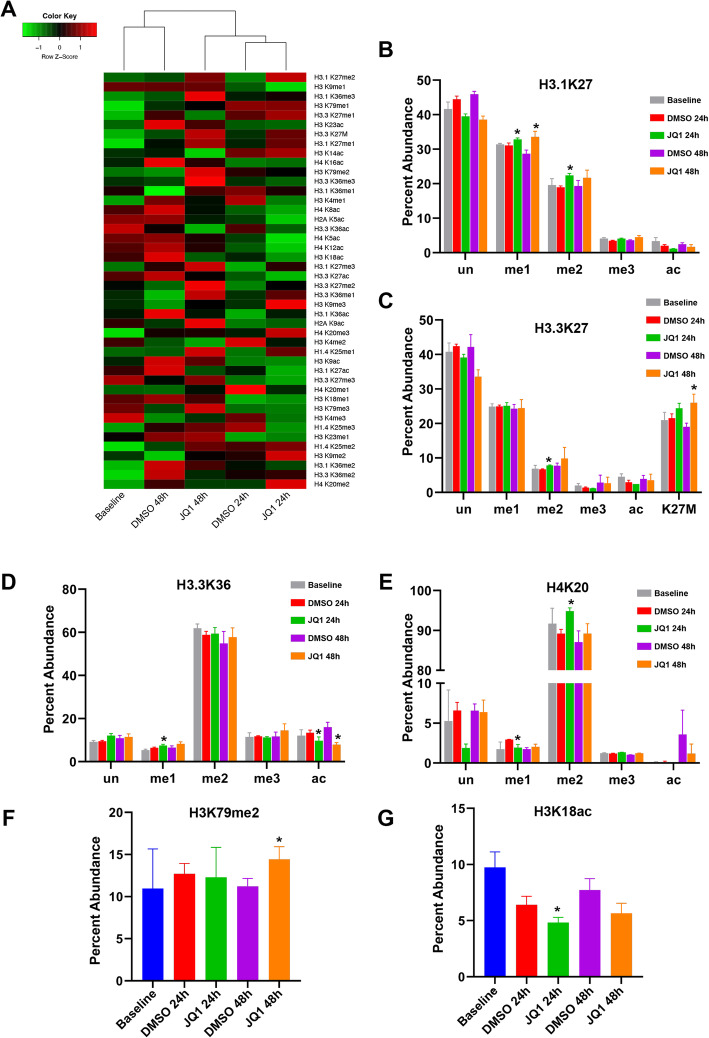
We previously reported pre-clinical efficacy of JQ1, a BET/Bromodomain inhibitor (BRDi), in DIPG [7]. We observed that BRD proteins are enriched at acetylated residues on histone H3 in DIPG cells and animal models, and that JQ1 treatment results in decreased global H3K27ac, with restoration of global H3K27 methylation [7]. To further elucidate the epigenetic effects of JQ1 in DIPG, we examined its effects on histone codes in SF8628, DIPG007, NHA, and U87 cells. As observed with RT, PTM profiles clustered by cell line, then treatment condition (Supplementary material 10). When analyzed individually, cell profiles cluster by treatment condition (Fig. 6a, Supplementary material 11). In DIPG007 cells, H3.3K27me1 decreased by 1.5% at 48 h after JQ1 treatment compared to vehicle control, representing the only significant change in modification state observed at that time point (Supplementary material 11). A small increase in H3.3K27M abundance was also detected in DIPG007 at 48 h (31.4% vs 26.9%, p = 0.067, Supplementary material 11). Surprisingly, in DIPG007, H3.3K27ac abundance was not significantly different from control at 48 h post-treatment, though a marginal decrease was observed at 24 h (1.65 and 1.95%, respectively, p = 0.065, Supplementary material 11). These results were validated in DIPG007 and SF8628 cells via western blot for H3K27M and H3K27ac (Supplementary material 12).
Fig. 6.
Bromodomain inhibition with JQ1 alters relative histone peptide modification states in DIPG, in vitro. a Unsupervised hierarchical clustering of DIPG cell line SF8628 histone tail modification profiles reveals clustering by treatment condition. b-e Relative abundance of H3.1/3.3 K27, H3.3K36, and H4K20 peptide modification states in SF8628 cells are distinct after JQ1 treatment. f-g Relative abundance of H3K79me2 and H3K18ac peptides in SF8628 cell lines are distinct after JQ1 treatment. * p < 0.05 compared to DMSO group (independent-sample t test, two-tailed)
In contrast, we observed statistically significant differences in abundance of multiple PTMs in SF8628 cells in response to JQ1 (Supplementary material 11). Increased H3.1K27me1 and decreased H3.3K36ac were seen at both 24 h and 48 h (Fig. 6b, Table 3, Supplementary material 11). Additional changes in relative modification states were observed after either 24 or 48 h post-treatment. For example, increased abundance of H3.1/H3.3K27me2, H3.3K36me1, and H4K20me2 were seen 24 h (Fig. 6b-e, Table 3) and H3K79me2 at 48 h (Fig. 6f, Table 3, Supplementary material 11). In addition, acetylation of H3K18 significantly decreased at 24 h (Fig. 6g, Table 3, Supplementary material 11). Surprisingly, we also observed a significant increase in H3.3K27M peptides at 24 and 48 h after JQ1 treatment in SF8628 cells (Fig. 6b, Table 3, Supplementary material 11).
Table 3.
Effects of bromodomain inhibition with JQ1 on relative histone peptide abundance in SF8628 cells
| Histone code | Mean | Fold (JQ1/DMSO) | p_SF8628 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | DMSO 24 h | DMSO 48 h | JQ1 24 h | JQ1 48 h | 24 h | 48 h | 24 h | 48 h | |
| H3.1K27me1 | 31.4 | 31.1 | 28.7 | 32.8 | 33.5 | 1.1 | 1.2 | < 0.05 | < 0.05 |
| H3.1K27me2 | 19.6 | 19.0 | 19.3 | 22.4 | 21.7 | 1.2 | 1.1 | < 0.01 | 0.21 |
| H3.3K27me2 | 6.9 | 6.7 | 7.7 | 7.8 | 9.8 | 1.2 | 1.3 | < 0.01 | 0.33 |
| H3.3K27M | 21.0 | 21.5 | 19.0 | 24.4 | 26.0 | 1.1 | 1.4 | < 0.05 | < 0.05 |
| H3.3K36me1 | 5.4 | 6.4 | 6.5 | 7.5 | 8.3 | 1.2 | 1.3 | < 0.05 | 0.07 |
| H3.3K36ac | 12.0 | 13.5 | 16.0 | 9.7 | 7.9 | 0.7 | 0.5 | < 0.05 | < 0.01 |
| H3K18ac | 9.7 | 6.4 | 7.7 | 4.8 | 5.7 | 0.8 | 0.7 | < 0.05 | 0.05 |
| H3K79me2 | 11.0 | 12.7 | 11.2 | 12.3 | 14.4 | 1.0 | 1.3 | 0.86 | < 0.05 |
| H4K20me1 | 1.7 | 2.9 | 1.7 | 1.9 | 2.0 | 0.7 | 1.2 | < 0.05 | 0.29 |
| H4K20me2 | 91.7 | 89.2 | 87.1 | 94.8 | 89.2 | 1.1 | 1.0 | < 0.01 | 0.38 |
Changes in histone PTM profiles were also observed in H3.3K27 WT cell lines after exposure to JQ1. The relative abundance of multiple PTMs were observed in U87 cells, including H3.1K27me2/me3, H3.1K36me1/me3, H3.3K36me1/ac, H3K14ac, H3K79me1, H3K4me1, and H4K5ac (Table 4, Supplementary material 11). Lastly, distinct changes were observed in NHAs after JQ1 treatment, including methylation of H3.1K27, H3.1/3.3 K36, and H3K79me1, and acetylation of H2AK5, H3.3K36, H3K18, and H4K16ac (Table 5, Supplementary material 11).
Table 4.
Effects of bromodomain inhibition with JQ1 on relative histone peptide abundance in U87 cells
| Histone code | Mean | Fold (JQ1/DMSO) | p_U87 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | DMSO 24 h | DMSO 48 h | JQ1 24 h | JQ1 48 h | 24 h | 48 h | 24 h | 48 h | |
| H3.1K27me2 | 34.7 | 34.2 | 35.2 | 37.0 | 38.6 | 1.1 | 1.1 | < 0.05 | < 0.05 |
| H3.1K27me3 | 10.0 | 11.0 | 12.2 | 13.1 | 14.4 | 1.2 | 1.2 | < 0.05 | < 0.05 |
| H3.1K36me1 | 19.9 | 19.6 | 18.3 | 17.9 | 19.0 | 0.9 | 1.0 | < 0.05 | 0.39 |
| H3.1K36me3 | 15.4 | 16.4 | 15.3 | 14.9 | 14.1 | 0.9 | 0.9 | < 0.05 | < 0.05 |
| H3.3K36me1 | 10.7 | 11.1 | 10.4 | 11.4 | 12.5 | 1.0 | 1.2 | 0.51 | < 0.05 |
| H3.3K36ac | 13.3 | 11.9 | 14.1 | 10.9 | 9.6 | 0.9 | 0.7 | 0.30 | < 0.01 |
| H3K14ac | 45.0 | 37.3 | 28.5 | 30.2 | 26.8 | 0.8 | 0.9 | < 0.05 | 0.38 |
| H3K79me1 | 24.8 | 27.7 | 26.6 | 23.9 | 23.7 | 0.9 | 0.9 | < 0.01 | 0.14 |
| H3K4me1 | 21.1 | 17.6 | 16.7 | 12.8 | 15.5 | 0.7 | 0.9 | < 0.01 | 0.69 |
| H4K5ac | 4.9 | 5.6 | 5.4 | 4.6 | 4.6 | 0.8 | 0.8 | < 0.01 | < 0.05 |
Table 5.
Effects of bromodomain inhibition with JQ1 on relative histone peptide abundance in NHA cells
| Histone code | Mean | Fold (JQ1/DMSO) | p_NHA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline | DMSO 24 h | DMSO 48 h | JQ1 24 h | JQ1 48 h | 24 h | 48 h | 24 h | 48 h | |
| H2AK5ac | 8.8 | 6.4 | 6.2 | 3.9 | 4.7 | 0.6 | 0.8 | 0.06 | < 0.05 |
| H3.1K27me3 | 19.7 | 18.3 | 18.2 | 22.0 | 24.0 | 1.2 | 1.3 | < 0.05 | < 0.05 |
| H3.1K36me1 | 14.6 | 14.5 | 13.9 | 16.4 | 15.9 | 1.1 | 1.1 | 0.21 | < 0.05 |
| H3.1K36me2 | 27.6 | 29.2 | 29.7 | 25.6 | 24.1 | 0.9 | 0.8 | 0.08 | < 0.05 |
| H3.3K36me1 | 8.7 | 9.2 | 8.5 | 10.6 | 11.0 | 1.2 | 1.3 | 0.17 | < 0.01 |
| H3.3K36ac | 8.5 | 8.8 | 10.0 | 6.3 | 7.1 | 0.7 | 0.7 | < 0.05 | < 0.01 |
| H3K18ac | 10.1 | 8.1 | 7.3 | 6.2 | 6.0 | 0.8 | 0.8 | 0.10 | < 0.05 |
| H3K79me1 | 14.3 | 15.2 | 16.1 | 17.5 | 14.2 | 1.2 | 0.9 | 0.27 | < 0.01 |
| H4K16ac | 28.5 | 27.5 | 27.3 | 26.2 | 30.8 | 1.0 | 1.1 | 0.66 | < 0.01 |
To determine the potential biological impact of observed differences in histone modifications after BRD inhibition relative to controls, we also compared cell gene expression profiles in these treatment groups. On functional pathways analysis of genes identified as statistically significantly differentially expressed by JQ1 treated cells relative to controls (fold change in expression > 2 or < − 2, adjusted p-value < 0.05), we identified nucleotide excision repair as the top canonical pathway activated in JQ1 treated DIPG cell lines (p-value 2.51 × 10− 48, Supplementary material 9). Other canonical pathways implicated by JQ1 treated cell gene expression profiles included DNA mismatch repair via non-homologous end joining (NEMJ, p-value 2.53 × 10− 32), while G2/M DNA damage check point regulation was the top toxicology list match (p-value 2.68 × 10− 6). Top upstream regulator effect networks implicated ANXA7 activation (resulting in DNA binding and conformational modification, score 9.615), and TP73 inhibition (resulting in apoptosis, score − 15.254) in JQ1 treated DIPG cells relative to controls.
Discussion
DIPG is an incurable pediatric brain tumor, with a high rate of somatic missense mutations in genes encoding histone H3.1 and H3.3 proteins. Here, we present comprehensive characterization of histone N-terminal tail modification states in pediatric glioma specimens, including H3.3K27M mutant DIPG tissue, using novel, high-throughput proteomic analysis, revealing the effects of radiotherapy and BRDi on the tumor epigenetic landscape. The greatest differences in PTMs between H3K27M mutant and WT specimens were observed at H3K27, H3K36, and H4K16ac, while H3K36me2 was the highest co-occurring modification on the H3.3K27M peptide, and H4K16 was the most acetylated residue. These results suggest that H3K36me2 and H4K16ac may serve as epigenetic signatures in DIPG, and that targeting these modification states may represent a potential therapeutic strategy.
Multiple groups have identified distinct DIPG molecular subtypes, with H3.3K27M mutant tumors associated with shorter survival [22–30]. We found that H3.3K27M significantly affects the histone PTM landscape, with cell and tissue profiles clustering by mutation status. Further, a decrease in global H3K27me2/3 has been observed in H3.3K27M tumors, and our results corroborate these data, [5, 7, 8, 10, 12, 13, 31]. Of note, a new subgroup of DIPG lacking H3K27M mutation but still exhibiting genomic H3K27me3 loss was recently reported [32]: we did not observe this phenomenon in our cohort (Supplementary material 4). Our H3K27M mutant specimens also exhibit distinct modification states at H3K36 and H4K16. Previous work showed that PRC2 loss coincides with increased H3K36me2, and that an increase in H3K36 methylation impacts methylation at H3K27 [33–35]. Our results also confirm an increase in H3K36me2 in DIPG specimens on the H3.3K27M mutant peptide, as previously detected via mass spectrometry of H3.3K27M transgenic PDGF-driven murine glioblastoma [13]. Further, we detected high H4K16ac abundance in DIPG cells and tissue specimens. A recently published report characterizing high-throughput screening with nucleosome substrate methods demonstrates small-molecule inhibitors of the human histone lysine methyltransferase NSD2 [36], the writer of mono- and di-methylation of H3K36 [37]; another report identified DC_M01_7 as a novel inhibitor of MOF, the H4K16ac-specific acetyltransferase [38, 39]. Therefore, H3K36me2 and H4K16ac may represent potential therapeutic targets in DIPG, and further investigation of this strategy is warranted.
Using our method, we also characterized H3K27 and K36 modification states in cis, confirming me2/3 as the predominant modifications, and acetylation as the least abundant, on the H3.3K27MK36 peptide. We observed positive correlation between H3.3K27M and H3.1K27K36me2, and negative correlation between H3.3K27M and H3.1K27K36me3. Taken together, these results confirm that H3.3K27M affects PTMs on both mutant and WT H3.1 and H3.3 N-terminal tails. Importantly, our study is the first, to our best knowledge, to quantify the relative amount of mutant H3.3K27M peptide in DIPG tumor specimens. It is reported that relative abundance of mutant epidermal growth factor receptor (EGFR) can predict benefit from EGFR-tyrosine kinase inhibitors for advanced non-small-cell lung cancer [40]. Longitudinal quantification of the H3.3K27M peptide in patient-derived specimens might similarly serve as a predictive biomarker for targeted therapy, and hence is worthy of consideration when designing future clinical trials.
Radiation therapy (RT) is standard treatment for DIPG, providing transient symptom relief but no change in disease outcome [41]. Unfortunately, the mechanism of radiation resistance is unknown, and radio-sensitizers provide no benefit [42]. Patients with the H3.3K27M mutation also respond more poorly to RT [24]. Our study reveals RT can induce cell-specific and peptide-specific changes in histone acetylation, underscoring the heterogeneity of DIPG and warranting consideration when using histone deacetylase / acetyltransferase inhibitors in combination with RT. Importantly, H3K9me3 abundance increased in all four cell lines after RT, which kills rapidly proliferating cancer cells by inducing DNA damage beyond cellular capacity for repair, including DNA double-strand breaks (DSBs) [43–45]. DSB repair is linked to rapid changes in epigenetic modifications, including increased H3K9 methylation, to recruit DNA repair proteins. Importantly, H3K9 methyltransferase G9a inhibition (BIX-01294) increases radio-sensitivity in glioma [46]. Indeed, on functional pathways analysis of cell gene expression profiles, we found DSB repair pathways implicated in RT treated cell lines relative to controls. These data suggest that combination therapy RT with G9a inhibition, aimed at reducing H3K9me3 levels and hence DNA DSB repair, may also be a useful therapeutic strategy in DIPG.
We found distinct DIPG histone codes after treatment with JQ1, a BET/ bromodomain inhibitor. BET domain proteins are epigenetic readers that bind acetylated histones via their bromodomain region to regulate gene transcription [46–48]. BET inhibitors dislodge acetylated histone readers from chromatin, leading to oncogene repression [49–51]. JQ1 is the best known BET inhibitor, and induces a dose-dependent reduction in DIPG cell viability and genomic H3K27ac in vitro and in vivo [7, 52, 53]. Our results showed increased mono- and/or di-methylation of multiple residues with JQ1 treatment, consistent with previous reports that JQ1 globally reduces acetylation and increases methylation [7]. Since mono-methylation of H3K27, H3K79, and H4K20 are all linked to gene activation, whereas tri-methylation of H3K27 and H3K79 are linked to repression [54], these findings may provide additional insight into the mechanism by which BRDi alters tumorigenic gene expression in DIPG. H4K20me, which is functional in DNA repair, represents a binding site for the 53BP1 protein; H3K9me3 and H4K20me3 represent epigenetic markers important to the function of 53BP1 in non-homologous end joining (NHEJ) repair [55]. In line with these known functions, pathways analysis of differential gene expression profiles implicated nucleotide excision repair and DNA mismatch repair via NHEJ as top activated pathways in JQ1 treated cells, compared to controls. As such, additional studies of the mechanism by which JQ1 may augment response to RT are necessary. Lastly, we also observed that reduction of histone acetylation in DIPG was greatest at 24 h, which may be important to consider when determining JQ1 dosing regimens.
Because fresh DIPG tissue is very rare, we chose to analyze archival post-mortem tissue specimens for this study, raising the possibility of treatment-induced effects. However, the patterns of peptide abundance observed in tumor tissues were similar to treatment-naïve cell lines. Analysis of treatment fresh tumor tissue specimens with this approach will be crucial to teasing out tumor biology from the effects of therapy. Optimization of our technical approach to minimize the amount of starting tumor tissue necessary for this purpose is currently underway.
Conclusions
In summary, targeted histone tail mass spectrometry of high grade glioma cell lines and tissues, including rare DIPG tumor tissue specimens, reveals distinct peptide modification states by H3.3K27M mutation status. Our study is the first to quantify the H3.3K27M peptide in DIPG, representing an exciting new opportunity to apply this technique on clinical specimens at diagnosis, and potentially throughout the course of treatment to guide clinical management. We also distinct epigenetic signatures of DIPG, including H3K36me2 and H4K16ac, which may represent novel therapeutic vulnerabilities for more effective treatment of this devastating disease.
Supplementary Information
Additional file 1: S1. Table of monitored peptide transitions.
Additional file 2: S2. Targeted mass spectrometry for histone tail analysis. Overview of study design and analysis of histone tail post-translational modification states in pediatric glioma specimens. Tumor tissue specimens were obtained post-mortem or during the course of treatment. Patient-derived tumor cell lines were established from tissue specimens. Extracted histones were analyzed using targeted mass spectrometry of the histone H3 and H4 N-terminal tail, with quantitation of histone post-translational modifications (me1, me2, me3, ac) and unmodified peptide on lysine (K) residues.
Additional file 3: S3. Table of results from targeted histone tail mass spectroscopy (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in glioma cell lines by H3.3K27M mutation status (Independent-sample t test, two-tailed).
Additional file 4: S4. H3.3K27M, H3.1K27me3, and H3.3K27me3 peptide abundance in cell lines. Targeted mass spectroscopy reveals differences in H3.3K27M, H3.1K27me3, and H3.3K27me3 peptide abundance in pediatric glioma and adult glioma cells, and normal human astrocytes. Of note, loss of H3.1K27 and H3.3K27 trimethylation is observed in H3.3K27M cell lines.
Additional file 5: S5. H3.3K27M mutation is associated with distinct histone modification states (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in glioma tissues by H3.3K27M mutant status (Independent-sample t test, two-tailed).
Additional file 6: S6. Modification states along short peptides (Microsoft Excel format). Distinct combinations of histone peptide modification states with H3.3K27M mutation status in tissues (Independent-sample t test, two-tailed).
Additional file 7: S7. Radiation treatment is associated with changes in histone modification states in vitro (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in NHA, U87, SF8628, and DIPG007 cells treated with radiation (one-way ANOVA).
Additional file 8: S8. Bromodomain inhibition with JQ1 is associated with changes in histone modification states in vitro. Unsupervised analysis of histone tail modification profiles of glioma cell lines (SF8628, DIPG007 and U87) and normal astrocytes (NHA) reveals clustering by cell line and treatment condition.
Additional file 9: S9. Peptide modification states observed after bromodomain inhibition with in vitro. (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in NHA, U87, SF8628, and DIPG007 cells after bromodomain inhibition with JQ1 (Independent-sample t test, two-tailed).
Additional file 10: S10. Functional pathways analysis of differentially expressed genes after RT and Bromodomain inhibition. A Differentially expressed genes by DIPG cells after 48 h RT compared to DMSO control enrich for DNA double-strand break repair as a top canonical pathway on functional analysis (p-value 1.10 × 10− 21). B Differentially expressed genes by DIPG cells after 48 h JQ1 treatment compared to DMSO control enrich for nucleotide excision repair as the top canonical pathway (p-value 2.51 × 10− 48).
Additional file 11: S11. Validation of changes in H3K27M and H3K27Ac in DIPG cells after Bromodomain inhibition. A Western blot of extracted histones for H3K27M and H3K27Ac from DIPG cell lines SF8628 and DIPG007 after 24 h of Bromodomain inhibition was performed in triplicate and compared to DMSO treated controls. B, C Bar graphs depicting mean OD value ± SEM across replicates for each treatment group, with p-values for between group comparisons (Independent-sample t test, two-tailed).
Acknowledgements
The authors would like to thank Rintaro Hashizume, MD, PhD for generous donation of cell lines, and all patients and their families for generous donation of tissue specimens for study.
Abbreviations
- DIPG
Diffuse intrinsic pontine glioma
- BRDi
Bromodomain inhibition
- PTMs
Post-translational modifications
- BRD2/4
Bromodomain proteins 2 and 4
- BET
Bromodomain and extra terminal domain
- WT
wild-type
- K27me2/3
K27 di- and tri-methylation
- RT
Radiation treatment
- DSBs
DNA double-strand breaks
Authors’ contributions
SA and JMC performed the experiments, analyzed the data, and wrote the manuscript, TH, DL, JAM MAZ, JQ, and MB performed the experiments, MK, JN, NLK, and PMT provided conceptualization and resources, AMS provided conceptualization and resources, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Institute of Neurological Disorders and Stroke (K08NS097624), the National Resource for Translational and Developmental Proteomics (P41 GM108569), National Cancer Institute (CCSG P30 CA060553), the Sherman Fairchild Foundation, the Northwestern Proteomics Core Facility, the Pediatric Cancer Research Foundation, the Frankel Foundation, the Lurie Children’s Hospital Faculty Practice Plan, and Northwestern University Clinical and Translational Sciences Institute. Adult tissue was provided by the Northwestern Nervous System Tumor Bank, which is supported by the P50CA221747 SPORE for Translational Approaches to Brain Cancer.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Ethics approval and consent to participate
Pediatric glioma tumor tissue specimens were collected postmortem in accordance to Children’s National Medical Center Institutional Protocol (IRB #Pro00001339). H3.3K27 wild-type (WT) adult glioma tumor tissue specimens (n = 2), and one non-tumor frontal lobe tissue specimen from a patient with focal cortical dysplasia, were collected during the course of treatment for the Nervous System Tumor Bank at Northwestern University, in accordance to Northwestern University IRB (STU00095863). Proteomic analysis was conducted with approval from the Northwestern University IRB (STU00202063).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shejuan An and Jeannie M. Camarillo contributed equally to this work.
References
- 1.Ostrom QT, de Blank PM, Kruchko C, Petersen CM, Liao P, Finlay JL, Stearns DS, Wolff JE, Wolinsky Y, Letterio JJ, et al. Alex's lemonade Stand Foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro-Oncology. 2015;16(Suppl 10):x1–x36. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kasper LH, Baker SJ. Invited review: emerging functions of histone H3 mutations in paediatric diffuse high-grade gliomas. Neuropathol Appl Neurobiol. 2020;46(1):73–85. 10.1111/nan.12591. [DOI] [PMC free article] [PubMed]
- 3.Langmoen IA, Lundar T, Storm-Mathisen I, Lie SO, Hovind KH. Management of pediatric pontine gliomas. Childs Nerv Syst. 1991;7(1):13–15. doi: 10.1007/BF00263825. [DOI] [PubMed] [Google Scholar]
- 4.Dasgupta T, Haas-Kogan DA. The combination of novel targeted molecular agents and radiation in the treatment of pediatric gliomas. Front Oncol. 2013;3:110. doi: 10.3389/fonc.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev. 2013;27(9):985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krug B, De Jay N, Harutyunyan AS, Deshmukh S, Marchione DM, Guilhamon P, Bertrand KC, Mikael LG, McConechy MK, Chen CCL, et al. Pervasive H3K27 acetylation leads to ERV expression and a therapeutic vulnerability in H3K27M Gliomas. Cancer Cell. 2019;35(5):782–797. doi: 10.1016/j.ccell.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi YH, Woodfin AR, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med. 2017;23(4):493–500. doi: 10.1038/nm.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24(5):660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Igolkina AA, Zinkevich A, Karandasheva KO, Popov AA, Selifanova MV, Nikolaeva D, Tkachev V, Penzar D, Nikitin DM, Buzdin A. H3K4me3, H3K9ac, H3K27ac, H3K27me3 and H3K9me3 Histone Tags Suggest Distinct Regulatory Evolution of Open and Condensed Chromatin Landmarks. Cells. 2019;8(9):1034. 10.3390/cells8091034. [DOI] [PMC free article] [PubMed]
- 10.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol. 2013;23(5):558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan YCE, Liu J, Chan KM. Histone H3 mutations in Cancer. Curr Pharmacol Rep. 2018;4(4):292–300. doi: 10.1007/s40495-018-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang T, Garcia R, Qi J, Lulla R, Horbinski C, Behdad A, Wadhwani N, Shilatifard A, James C, Saratsis AM. Detection of histone H3 K27M mutation and post-translational modifications in pediatric diffuse midline glioma via tissue immunohistochemistry informs diagnosis and clinical outcomes. Oncotarget. 2018;9(98):37112–37124. doi: 10.18632/oncotarget.26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647(1–2):21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345(6200):1065–1070. doi: 10.1126/science.1255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi J, Esfahani DR, Huang T, Ozark P, Bartom E, Hashizume R, Bonner ER, An S, Horbinski CM, James CD, et al. Tenascin-C expression contributes to pediatric brainstem glioma tumor phenotype and represents a novel biomarker of disease. Acta Neuropathol Commun. 2019;7(1):75. doi: 10.1186/s40478-019-0727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med. 2014;20(12):1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashizume R, Smirnov I, Liu S, Phillips JJ, Hyer J, McKnight TR, Wendland M, Prados M, Banerjee A, Nicolaides T, et al. Characterization of a diffuse intrinsic pontine glioma cell line: implications for future investigations and treatment. J Neuro-Oncol. 2012;110(3):305–313. doi: 10.1007/s11060-012-0973-6. [DOI] [PubMed] [Google Scholar]
- 20.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2(4):933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camarillo JM, Swaminathan S, Abshiru NA, Sikora JW, Thomas PM, Kelleher NL. Coupling fluorescence-activated cell sorting and targeted analysis of histone modification profiles in primary human leukocytes. J Am Soc Mass Spectrom. 2019;30(12):2526–2534. doi: 10.1007/s13361-019-02255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silveira AB, Kasper LH, Fan Y, Jin H, Wu G, Shaw TI, Zhu X, Larson JD, Easton J, Shao Y, et al. H3.3 K27M depletion increases differentiation and extends latency of diffuse intrinsic pontine glioma growth in vivo. Acta Neuropathol. 2019;137(4):637–655. doi: 10.1007/s00401-019-01975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buczkowicz P, Bartels U, Bouffet E, Becher O, Hawkins C. Histopathological spectrum of paediatric diffuse intrinsic pontine glioma: diagnostic and therapeutic implications. Acta Neuropathol. 2014;128(4):573–581. doi: 10.1007/s00401-014-1319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pages M, Taylor KR, Saulnier P, Lacroix L, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130(6):815–827. doi: 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124(3):439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, et al. Integrated molecular meta-analysis of 1,000 pediatric high-grade and diffuse intrinsic Pontine Glioma. Cancer Cell. 2017;32(4):520–537. doi: 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagaraja S, Quezada MA, Gillespie SM, Arzt M, Lennon JJ, Woo PJ, Hovestadt V, Kambhampati M, Filbin MG, Suva ML, et al. Histone variant and cell context determine H3K27M reprogramming of the enhancer landscape and oncogenic state. Mol Cell. 2019;76(6):965–980. doi: 10.1016/j.molcel.2019.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saratsis AM, Kambhampati M, Snyder K, Yadavilli S, Devaney JM, Harmon B, Hall J, Raabe EH, An P, Weingart M, et al. Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol. 2014;127(6):881–895. doi: 10.1007/s00401-013-1218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D, Sill M, Buchhalter I, Northcott PA, Leis I, et al. New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell. 2016;164(5):1060–1072. doi: 10.1016/j.cell.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ, Sullivan LM, Martinez D, Perry A, Lewis PW, Thompson CB, et al. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol. 2014;128(5):743–753. doi: 10.1007/s00401-014-1338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anastas JN, Zee BM, Kalin JH, Kim M, Guo R, Alexandrescu S, Blanco MA, Giera S, Gillespie SM, Das J, et al. Re-programing chromatin with a Bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36(5):528–544. doi: 10.1016/j.ccell.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Castel D, Kergrohen T, Tauziede-Espariat A, Mackay A, Ghermaoui S, Lechapt E, Pfister SM, Kramm CM, Boddaert N, Blauwblomme T, et al. Histone H3 wild-type DIPG/DMG overexpressing EZHIP extend the spectrum diffuse midline gliomas with PRC2 inhibition beyond H3-K27M mutation. Acta Neuropathol. 2020;139(6):1109–1113. doi: 10.1007/s00401-020-02142-w. [DOI] [PubMed] [Google Scholar]
- 33.Stafford JM, Lee CH, Voigt P, Descostes N, Saldana-Meyer R, Yu JR, Leroy G, Oksuz O, Chapman JR, Suarez F, et al. Multiple modes of PRC2 inhibition elicit global chromatin alterations in H3K27M pediatric glioma. Sci Adv. 2018;4(10):eaau5935. doi: 10.1126/sciadv.aau5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42(3):330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coussens NP, Kales SC, Henderson MJ, Lee OW, Horiuchi KY, Wang Y, Chen Q, Kuznetsova E, Wu J, Chakka S, et al. High-throughput screening with nucleosome substrate identifies small-molecule inhibitors of the human histone lysine methyltransferase NSD2. J Biol Chem. 2018;293(35):13750–13765. doi: 10.1074/jbc.RA118.004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Ahn JH, Wang GG. Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci. 2019;76(15):2899–2916. doi: 10.1007/s00018-019-03144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R, Wang J, Zhao L, Liu S, Du D, Ding H, Chen S, Yue L, Liu YC, Zhang C, et al. Identification of novel inhibitors of histone acetyltransferase hMOF through high throughput screening. Eur J Med Chem. 2018;157:867–876. doi: 10.1016/j.ejmech.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Jang Y, Lee JE, Ahn J, Xu L, Holden MR, Cornett EM, Krajewski K, Klein BJ, Wang SP, et al. Selective binding of the PHD6 finger of MLL4 to histone H4K16ac links MLL4 and MOF. Nat Commun. 2019;10(1):2314. doi: 10.1038/s41467-019-10324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Zhang XC, Chen ZH, Yin XL, Yang JJ, Xu CR, Yan HH, Chen HJ, Su J, Zhong WZ, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2011;29(24):3316–3321. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 41.Gwak HS, Park HJ. Developing chemotherapy for diffuse pontine intrinsic gliomas (DIPG) Crit Rev Oncol Hematol. 2017;120:111–119. doi: 10.1016/j.critrevonc.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 42.Vanan MI, Eisenstat DD. DIPG in children - what can we learn from the past? Front Oncol. 2015;5:237. doi: 10.3389/fonc.2015.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulston M, Fulford J, Jenner T, de Lara C, O'Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and nth sensitive sites. Nucleic Acids Res. 2002;30(15):3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma J, Benitez JA, Li J, Miki S, Ponte de Albuquerque C, Galatro T, Orellana L, Zanca C, Reed R, Boyer A, et al. Inhibition of nuclear PTEN tyrosine phosphorylation enhances Glioma radiation sensitivity through attenuated DNA repair. Cancer Cell. 2019;35(3):504–518. doi: 10.1016/j.ccell.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S, Kozono D, Yang X, Fendler W, Fitts W, Ni J, Alberta JA, Zhao J, Liu KX, Bian J, et al. Dual HDAC and PI3K inhibition abrogates NFkappaB- and FOXM1-mediated DNA damage response to Radiosensitize pediatric high-grade Gliomas. Cancer Res. 2018;78(14):4007–4021. doi: 10.1158/0008-5472.CAN-17-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bennett RL, Licht JD. Targeting epigenetics in Cancer. Annu Rev Pharmacol Toxicol. 2018;58:187–207. doi: 10.1146/annurev-pharmtox-010716-105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faivre EJ, McDaniel KF, Albert DH, Mantena SR, Plotnik JP, Wilcox D, Zhang L, Bui MH, Sheppard GS, Wang L, et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature. 2020;578(7794):306–310. doi: 10.1038/s41586-020-1930-8. [DOI] [PubMed] [Google Scholar]
- 48.Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, Carroll TS, Li Y, Feng L, Chen G, et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020;577(7788):121–126. doi: 10.1038/s41586-019-1842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502(7472):480–488. doi: 10.1038/nature12751. [DOI] [PubMed] [Google Scholar]
- 50.Jung M, Philpott M, Muller S, Schulze J, Badock V, Eberspacher U, Moosmayer D, Bader B, Schmees N, Fernandez-Montalvan A, et al. Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1. J Biol Chem. 2014;289(13):9304–9319. doi: 10.1074/jbc.M113.523019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi J. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol. 2014;6(12):a018663. doi: 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, Li M, Meng W, Ponnuswami A, Sun W, et al. Transcriptional dependencies in diffuse intrinsic Pontine Glioma. Cancer Cell. 2017;31(5):635–652. doi: 10.1016/j.ccell.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 55.Svobodova Kovarikova A, Legartova S, Krejci J, Bartova E. H3K9me3 and H4K20me3 represent the epigenetic landscape for 53BP1 binding to DNA lesions. Aging (Albany NY) 2018;10(10):2585–2605. doi: 10.18632/aging.101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: S1. Table of monitored peptide transitions.
Additional file 2: S2. Targeted mass spectrometry for histone tail analysis. Overview of study design and analysis of histone tail post-translational modification states in pediatric glioma specimens. Tumor tissue specimens were obtained post-mortem or during the course of treatment. Patient-derived tumor cell lines were established from tissue specimens. Extracted histones were analyzed using targeted mass spectrometry of the histone H3 and H4 N-terminal tail, with quantitation of histone post-translational modifications (me1, me2, me3, ac) and unmodified peptide on lysine (K) residues.
Additional file 3: S3. Table of results from targeted histone tail mass spectroscopy (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in glioma cell lines by H3.3K27M mutation status (Independent-sample t test, two-tailed).
Additional file 4: S4. H3.3K27M, H3.1K27me3, and H3.3K27me3 peptide abundance in cell lines. Targeted mass spectroscopy reveals differences in H3.3K27M, H3.1K27me3, and H3.3K27me3 peptide abundance in pediatric glioma and adult glioma cells, and normal human astrocytes. Of note, loss of H3.1K27 and H3.3K27 trimethylation is observed in H3.3K27M cell lines.
Additional file 5: S5. H3.3K27M mutation is associated with distinct histone modification states (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in glioma tissues by H3.3K27M mutant status (Independent-sample t test, two-tailed).
Additional file 6: S6. Modification states along short peptides (Microsoft Excel format). Distinct combinations of histone peptide modification states with H3.3K27M mutation status in tissues (Independent-sample t test, two-tailed).
Additional file 7: S7. Radiation treatment is associated with changes in histone modification states in vitro (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in NHA, U87, SF8628, and DIPG007 cells treated with radiation (one-way ANOVA).
Additional file 8: S8. Bromodomain inhibition with JQ1 is associated with changes in histone modification states in vitro. Unsupervised analysis of histone tail modification profiles of glioma cell lines (SF8628, DIPG007 and U87) and normal astrocytes (NHA) reveals clustering by cell line and treatment condition.
Additional file 9: S9. Peptide modification states observed after bromodomain inhibition with in vitro. (Microsoft Excel format). Statistically significant differences in histone acetylation and methylation are observed in NHA, U87, SF8628, and DIPG007 cells after bromodomain inhibition with JQ1 (Independent-sample t test, two-tailed).
Additional file 10: S10. Functional pathways analysis of differentially expressed genes after RT and Bromodomain inhibition. A Differentially expressed genes by DIPG cells after 48 h RT compared to DMSO control enrich for DNA double-strand break repair as a top canonical pathway on functional analysis (p-value 1.10 × 10− 21). B Differentially expressed genes by DIPG cells after 48 h JQ1 treatment compared to DMSO control enrich for nucleotide excision repair as the top canonical pathway (p-value 2.51 × 10− 48).
Additional file 11: S11. Validation of changes in H3K27M and H3K27Ac in DIPG cells after Bromodomain inhibition. A Western blot of extracted histones for H3K27M and H3K27Ac from DIPG cell lines SF8628 and DIPG007 after 24 h of Bromodomain inhibition was performed in triplicate and compared to DMSO treated controls. B, C Bar graphs depicting mean OD value ± SEM across replicates for each treatment group, with p-values for between group comparisons (Independent-sample t test, two-tailed).
Data Availability Statement
All data generated or analyzed during this study are included in this article.