Abstract
An increasing number of commercial skincare products are being manufactured with engineered nanomaterials (ENMs), prompting a need to fully understand how ENMs interact with the dermal barrier as a major biodistribution entry route. Although animal studies show that certain nanomaterials can cross the skin barrier, physiological differences between human and animal skin, such as the lack of sweat glands, limit the translational validity of these results. Current optical microscopy methods have limited capabilities to visualize ENMs within human skin tissues due to the high amount of background light scattering caused by the dense, ubiquitous extracellular matrix (ECM) of the skin. Here, we hypothesized that organic solvent-based tissue clearing (“immunolabeling-enabled three-dimensional imaging of solvent-cleared organs”, or “iDISCO”) would reduce background light scattering from the extracellular matrix of the skin to sufficiently improve imaging contrast for both 2D mapping of unlabeled metal oxide ENMs and 3D mapping of fluorescent nanoparticles. We successfully mapped the 2D distribution of label-free TiO2 and ZnO nanoparticles in cleared skin sections using correlated signals from darkfield, brightfield, and confocal microscopy, as well as micro-spectroscopy. Specifically, hyperspectral microscopy and Raman spectroscopy confirmed the identity of label-free ENMs which we mapped within human skin sections. We also measured the 3D distribution of fluorescently labeled Ag nanoparticles in cleared skin biopsies with wounded epidermal layers using light sheet fluorescence microscopy. Overall, this study explores a novel strategy for quantitatively mapping ENM distributions in cleared ex vivo human skin tissue models using multiple imaging modalities. By improving the imaging contrast, we present label-free 2D ENM tracking and 3D ENM mapping as promising capabilities for nanotoxicology investigations.
Keywords: engineered nanomaterials, skin imaging, tissue clearing, hyperspectral microscopy, 3D imaging
1. Introduction
Metal and metal oxide engineered nanomaterials (ENMs) are becoming increasingly prevalent in pharmaceutical and cosmetic products designed for dermal application. For the past decades, chemical compounds such as avobenzone and oxybenzone have been the dominant UV blocking agents in sunscreens and other cosmetics. More recently, mineral-based compounds, such as TiO2, have been formulated as nanoparticles and are commonly used as both colorants and UV blockers in cosmetics and other personal care products (Delouise, 2012; Lin et al., 2015; Singh and Nanda, 2012; Smijs and Pavel, 2011). Similarly, ZnO nanoparticles are widely used in sunscreens because they reflect a broader spectrum of UV rays, especially in the UVA range (Lin et al., 2015; Morganti, 2010; Smijs and Pavel, 2011). The use of these and other nanomaterial additives in skincare products has attracted the attention of researchers from a variety of disciplines in an effort to understand whether the eventual permeation of ENMs across the skin can lead to toxic effects (Wang et al., 2018). As a result, acquiring comprehensive biodistribution profiles of these ENMs becomes increasingly necessary. There is therefore a growing need to devise methods to quantitatively map both two- and three-dimensional ENM interactions across different layers of the skin, the dermal microvasculature, and skin appendages such as hair follicles, sebaceous glands and sweat glands.
Recent studies have begun to explore the possible routes of ENM penetration through the skin, specifically through the hair follicles (“follicular pathway”). Some reports suggest that ENMs can penetrate hair follicles to depths of 400 μm –500 μm, although there remains no evidence that they permeate through the follicles and deposit in the epidermis or the dermis (Lekki et al., 2007; Mathes et al., 2016). The uptake of nanoparticles can be higher in depilated skin (Senzui et al., 2010), suggesting that wounded or altered skin is more susceptible to ENM permeation as previously reported in skin with damages or lesions (Larese Filon et al., 2013; Lin et al., 2011). While the skin permeation dynamics for some metal and metal oxides have been studied, newer generations of ENMs continue to be developed and used in different forms and sizes in multiple commercial products. The influence of such modifications in nanoparticle-skin interactions and the dynamics of ENM biodistribution remains elusive. To date, the most common method for mapping metal/metal oxide nanoparticle permeation or distribution in biological tissues relies on electron microscopy (e.g., energy dispersive X-ray mapping) (Lin et al., 2016; Pena Mdel et al., 2016; Zheng et al., 2011). However, this method can be destructive to biological samples, requires intricate and time-intensive sample preparation, and is not practical as an imaging tool for reconstructing large tissue areas. Multiphoton confocal imaging has been employed as an alternative method for mapping ENMs in reconstructed and patient-derived human skin, but the problem of inherent light-scattering from skin tissues still results in limited imaging depth and accuracy in determining nanoparticle permeation (Labouta et al., 2012). Hyperspectral imaging is another relatively newer approach which relies on the optical scattering of nanomaterials and does not require additional labor-intensive sample preparation steps or nanoparticle labeling (Idelchik et al., 2016; SoRelle et al., 2016). Label-free imaging techniques study nanoparticles in their native state, minimize surface modifications that can alter ENM biodistribution kinetics. However, 3D volumetric imaging is still a limitation for hyperspectral microscopy and has also slow acquisition times. Confocal and high-resolution fluorescence-based microscopy remain the most widely used tools for 3D volumetric imaging nanoparticles in tissues, but are only applicable to fluorescently labeled ENMs. Therefore, existing methods remain limited in their ability to provide a high-fidelity understanding of the toxicity, biointeractions, and potential for translocation across the dermal barrier of unmodified ENMs.
Even with advanced microscopy tools, the skin is a challenging organ to image because it contains multiple light scattering layers that limit imaging depth and contrast. The extracellular matrix (ECM) of the dermis is composed primarily of a heterogeneous and dense network of fibrillar collagen, which is positioned above a substantial layer of subcutaneous fat (Watt and Fujiwara, 2011). One promising option for overcoming these obstacles and successfully mapping ENM distribution in 2D and 3D is to use tissue clearing—which can eliminate lipid content and standardize the refractive index (RI) of the tissue while still preserving its protein and nucleic acid landscape (Richardson and Lichtman, 2015). Prior research has produced fully transparent human skin via organic solvent-based tissue clearing (Abadie et al., 2018), which entails dehydrating the fixed and stained tissue, dissolving the lipids responsible for much of the light scattering, and immersing the tissue in a solvent that homogenizes its refractive index (Renier et al., 2014; Richardson and Lichtman, 2015). However, this tissue clearing approach has not been exploited yet for understanding ENM permeation and distribution in human skin.
In this study, we present an organic solvent-based approach to tissue clearing (“iDISCO”: immunolabeling-enabled three-Dimensional Imaging of Solvent-Cleared Organs) to render intact ex vivo human skin biopsies sufficiently translucent for 2D/3D mapping of both label-free and fluorescently tagged nanoparticles. The tissue clearing technique explored herein was demonstrated to drastically reduce background signal on ex vivo human skin biopsies. This approach allowed for an improved signal-to-noise ratio when quantitatively analyzing the permeation and localization of label-free ENMs in thick human skin tissue sections or of fluorescently labeled nanoparticles in whole skin biopsies. Through these studies, we show that “iDISCO+”-cleared ex vivo human skin samples offer a versatile and promising model for studying the interaction of ENMs with dense skin tissues under non-perfused conditions. These techniques represent an important advance towards enabling future research to assess biodistribution patterns of nanomaterials with human tissues and understanding their nanotoxicological implications.
2. Materials and Methods
2.1. ENM characterization and preparation of dispersions
The ENMs used in our label-free imaging experiments were procured and characterized by the Engineered Nanomaterials Coordination Core (ERCC) - part of the Nanotechnology Health Implications Research (NHIR) Consortium at Harvard School of Public Health. Detailed characterization and biological assessment of the ENMs in this study have been described elsewhere: Lee et al. and Zhang et al. presented the characterization for TiO2-E171 (100 nm) (Lee et al., 2018; Zhang et al., 2019), while Eweje et al. presented the characterization for ZnO (50 nm) (Eweje et al., 2019). Both TiO2 and ZnO suspensions in tissue culture medium were prepared using a sonic dismembrator (acoustic power = 2.51 J/s; FB-505, Fisher Scientific, USA) according to literature protocols (Cohen et al., 2018; DeLoid et al., 2014; DeLoid et al., 2017; DeLoid et al., 2015; Taurozzi et al., 2011). The minimum amount of sonication time was calculated from the previously reported critical delivered sonication energy (DSEcr) for each particle (Eweje et al., 2019; Lee et al., 2018; Zhang et al., 2019): DSEcr, TiO2 = 0 J/mL; DSEcr, ZnO = 420 J/mL. An ENM stock solution of 500 μg/mL was first prepared with DNase/RNase free distilled water (Invitrogen, MA, USA). The 100 μg/mL working solutions for these ENMs were then prepared by diluting the sonicated 500 μg/mL stock solutions with the tissue culture medium (Genoskin, MA, USA). Dynamic light scattering (DLS, Malvern Nanosizer, Worcestershire, UK) was used to characterize the hydrodynamic diameter profiles for these solutions (Fig. S1). To produce the labeled Ag nanoparticles (NPs) for light sheet fluorescent microscopy, ~12 μM biotinylated AgNPs (20 nm, Cytodiagnostics, Ontario, Canada) were incubated with ~1.5 μM Alexa Fluor 647-conjugated streptavidin (Invitrogen, MA) for 30 min at 37°C prior to the exposure experiments.
2.2. Skin model and ENM exposure
Human skin tissue samples (diameter = 11 mm) collected from a donor abdomen were purchased from Genoskin (Salem, MA, USA). All samples were collected immediately after surgery and stored at 4°C until shipment. Epidermal excisions (wound diameter = 2 mm) were performed after surgical collection to produce wounded tissues. Samples were incubated under standard culture conditions (37°C, 20% O2, 5% CO2) in well plates filled with Genoskin tissue culture medium up to the bottom of the epidermis. Tissue samples in this study were obtained from a cohort of 4 female donor patients that are 30, 33, 38, and 65 years old.
Solutions of 100 μg/mL TiO2 or ZnO ENMs in Genoskin media were dispersed in the media surrounding the dermal side of the biopsies for 24 h prior to analysis. For the topical exposure with Ag-647 ENMs, 50 μL of the pre-incubated Streptavidin 647 and biotinylated Ag solution was pipetted onto the wounded site of the epidermis. After ENM exposure, whole tissue samples were immersion fixed in 4% paraformaldehyde overnight at 4°C, then washed several times in PBS over 24 h. In all experiments, control samples were incubated and fixed without exposure to ENMs.
2.3. Tissue clearing and immunostaining
2.3.1. Tissue sections
Fixed skin samples were embedded in optimal cutting temperature (OCT) compound (Tissue Tek), quick-frozen in isopentane (pre-chilled to −80°C on dry ice), and stored at −80°C until sectioning. 50 μm-thick sections were sliced at −14°C on a Leica Cryostat CM1950, collected on gelatin-coated glass slides, and stored at −20°C overnight. Tissue sections were stained and cleared following a modified version of the “iDISCO+” protocol (Renier et al., 2016), which is an updated version of the original iDISCO method (Renier et al., 2014), designed to reduce sample shrinkage. Briefly, slides were rinsed in ultrapure DI H2O to remove OCT, permeabilized in “iDISCO+” permeabilization solution for 30 min at 37°C, and blocked in “iDISCO+” blocking solution for 1 h at 37°C. To block non-specific secondary antibody binding, normal goat serum was substituted for normal donkey serum in all solutions. Following blocking, tissue sections on slides were circled with a hydrophobic PAP histology pen and incubated in “iDISCO+” primary antibody solution with primary antibodies to cytokeratin 14 (mouse, Abcam ab7800, 1:200) overnight at 4°C. After washing in “iDISCO+” PTwH (PBS with 2% v/v Tween 20 and 1% v/v heparin) solution, slides were incubated in “iDISCO+” secondary antibody solution with Alexa Fluor 488-conjugated mouse IgG (Invitrogen, RRID AB_2536161, 1:200) and 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI, Invitrogen, 1:5000) for 2 h at room temperature. All slides were washed again in PTwH solution, and any uncleared slides were mounted in PBS on coverslips and sealed with nail polish.
For clearing, slides were dehydrated in a 20-40-60-80-100-100% methanol (MeOH) series in H2O for 5 mins each. Next, the slides were gently shaken in a solution of 66% MeOH-33% dichloromethane (DCM) for 30 mins to remove lipids, then washed twice in pure DCM to remove MeOH traces. Finally, the slides were refractive index-matched in dibenzyl ether (DBE, RI = 1.55, Sigma-Aldrich, USA) for at least 1 h, then mounted in DBE on coverslips, sealed with nail polish, and cleaned of DBE residue with pure ethanol. For Raman micro-spectroscopy, slides were not mounted in DBE due to significant Raman background signals, but instead rehydrated in the H2O-MeOH series after delipidation. Final refractive index matching and mounting for Raman samples was performed with EasyIndex medium (RI = 1.46, LifeCanvas Technologies, Cambridge, MA, USA). A summary of clearing protocol is presented in Supplementary Table 1.
2.3.2. Whole tissue biopsies
Whole tissue samples for 3D imaging were immunostained and cleared following the same “iDISCO+” protocol as the tissue sections (Renier et al., 2016). We modified the published “iDISCO+” protocol by altering the durations of several steps, as described here. In pretreatment before immunostaining, samples were first dehydrated in a 20-40-60-80-100-100% MeOH series in H2O for 1 h each, then left overnight in 66% DCM - 33%. They were then washed twice for 1 h in chilled MeOH and bleached overnight in 5% H2O2 in MeOH to reduce pigmentation. Samples were then rehydrated in the MeOH series and washed 2× 1 h in PTx.2 buffer. For immunolabeling, whole samples were permeabilized overnight at 37°C, blocked overnight at 37°C (again with normal goat serum substituted for normal donkey serum), and incubated in the previously discussed primary antibodies for 48 h at 37°C with shaking. Samples were washed in PTwH solution for 24 h, then incubated in with secondary antibodies and DAPI for 48 h at 37°C with shaking. After final washing in PTwH for 24 h, samples were ready for clearing. Whole tissue samples that were not immunostained were cleared by dehydration in the MeOH series in 1 h steps, followed by delipidation in 66% MeOH-33% DCM for 3 h and in pure DCM for 2× 15 mins before final refractive index matching in DBE for at least 24 h. A summary of the iDISCO clearing protocol employed for the tissue samples studied here is presented in Supplementary Table 1.
2.4. Imaging
2.4.1. Phase contrast and darkfield microscopy for large tissue areas
For phase contrast and darkfield imaging, an Olympus ix83 inverted microscope with LUCPlanFLNPh 20× objective and a phase contrast plate were used. The condenser of the microscope was retrofitted with an oblique angle LED ring illuminator (Amscope LED-144-YK ring lamp) to allow for darkfield imaging. A Hamamatsu Orca Flash 4.0 C11440 at a 16-bit depth with a 0.16–0.33 μm pixel resolution were used to record images. Tile scans and image stitching were performed using the MetaMorph Premier (64-bit) 7.8.12.0 software. Details on image processing and data analysis can be found in the Supplementary Information.
2.4.2. Confocal microscopy
Tile scan fluorescent imaging of skin sections mounted on glass slides was performed on a Zeiss Axio Scan.Z1 automated widefield microscope (Carl Zeiss Microscopy GmbH, Germany) with a 10× Plan-Apochromat objective (NA = 0.45) and appropriate filter cubes. Individual tiles were comprised of Z stacks acquired with 5 μm intervals, after which composite images were processed in Zen Blue Lite (2.3) software by merging maximum intensity projections of each tile with 10% overlap on neighboring tiles.
2.4.3. Darkfield hyperspectral imaging and Raman spectroscopy
Cleared and uncleared skin sections were imaged on a darkfield hyperspectral microscope (Cytoviva) integrated with a confocal Raman microscope (Horiba XploRA PLUS) in transmission mode. Hyperspectral maps were processed using ENVI data analysis software (ENVI Classic 5.5) to reconstruct the spectral information for regions of interest containing ENM clusters. The corresponding darkfield images were obtained using a 10× objective under a halogen lamp (International Light Technologies Part L1090, USA). For ENM mapping, the spectral angle mapper (SAM) capability in ENVI was conducted with respect to reference spectral libraries taken from 100 μg/mL TiO2 and ZnO solutions mounted on glass slides. A threshold of 0.2 radians was applied for all conditions. The final hyperspectral darkfield images reported were treated with either “Linear 2%” or “Square Root” image enhancement options. Raman spectral measurements were conducted using a 785-nm laser excitation wavelength (25% laser filter setting) with a grating of 600 mm−1. Longer excitation wavelength was used to avoid spectral interference from tissue autofluorescence. The system was calibrated to the spectral line of a silicon reference sample at ~521 cm−1 prior to any sample measurements. All reported spectra were recorded from 75 to 2500 cm−1 for at least 10 s and 2 accumulation scans. The TiO2 and ZnO reference spectra for both imaging modalities were taken from a 100 μg/mL ENM solution prepared in ultrapure water mounted onto a glass slide. Details on image processing and data analysis can be found in the Supplementary Information.
2.4.4. Light sheet microscopy
After clearing, whole skin biopsies were mounted on an aluminum disk using a silicone adhesive (Kwik-Sil, World Precision Instruments, Sarasota, FL, USA) and imaged in DBE on an UltraMicroscope II light sheet microscope (LaVision Biotech GmbH, Bielefeld, Germany) with a combined 2× objective lens (0.5 NA) and a 10× zoom optic lens (working magnification variable from 0.63× − 6.3×). Z stack images were acquired using ImSpector software (Abberior Instruments GmbH, Gottingen, Germany) with the following parameters: dual-sided light sheet illumination, 70% sheet width, 60% laser power, 50 ms frame exposure time, optical section thickness of 5 μm. 3D image reconstructions were performed using Imaris 9.3.1 software (Imaris Bitplane, Zurich, Switzerland). Details on image processing and data analysis can be found in the Supplementary Information.
2.5. Inductively coupled plasma-mass spectrometry (ICP-MS)
ENM-exposed whole skin samples were paused at various phases of the tissue clearing process (before clearing: in PBS, pretreatment/immunolabeling: in MeOH delipidation: in PBS, and after clearing: in DBE). The tissues were placed into a tightly closed vial in their “wet state” (i.e., without completely drying from the last solvent in which they were submerged) and subsequently processed for Ti and Zn quantification by sector field ICP-MS (SF-ICP-MS). Tissue samples were digested, and the target metals were solubilized using a nitric acid/peroxide hot block digestion protocol in acid-washed polypropylene digestion tubes. Tared tissue samples were placed (using Teflon tweezers) into the digestion tubes along with 5.0 mL of 16 M nitric acid. The tubes/samples were pre-heated to 50ºC and 1.0 mL of hydrogen peroxide was then slowly added in 0.20 mL increments. The digestion was completed by heating the tubes for 2 hours at 80ºC. After digestion, 3 mL of the digestate was removed and diluted to 10.0 mL with high-purity water. Digestion QC samples in each analytical batch of 20 samples included 5 method blanks, 2 blank spikes, and an external reference material. The target metals (Ti and Zn) in the digests were quantified by magnetic sector ICP-MS (Thermo-Finnigan Element XR) at dilutions of 1:4 and 1:10 (two repeat experiments; n = 3 for TiO2-exposed tissues and n ≥ 5 for ZnO-exposed and unexposed (control) tissues; triplicate measurement per tissue). The isotopes (m/z) used for quantification were (primary, confirmation): Ti (49, 47) and Zn (66, 68). Ti and Zn quantification were acquired in medium resolution (R = 3950). Mean batch specific analytical blanks (with outlier detection) were applied to all sample data. Propagated uncertainty estimates for each element in all samples, as reported in Supplementary Information, include the following components: (1) sector-field ICP-MS analysis (standard deviation of triplicate analyses on each); and (2) blank subtraction (standard deviation of 5 method blanks from each batch).
2.6. Lorenz-Mie theory calculations
Scattering coefficients (Qsca) for TiO2 and ZnO (size range: 20–250 nm) were calculated in Python using the pymiecoated distribution library, following the literature protocol (Zimmerman et al., 2019). All particles were assumed to be spherical and calculations were based on the wavelength-dependent refractive index, n, and the extinction coefficient, k. Literature reference measurements were used to estimate these properties for TiO2 (Siefke et al., 2016) and ZnO (Stelling et al., 2017).
3. Results and Discussion
3.1. Organic solvent-based clearing method maximally clears human skin tissue
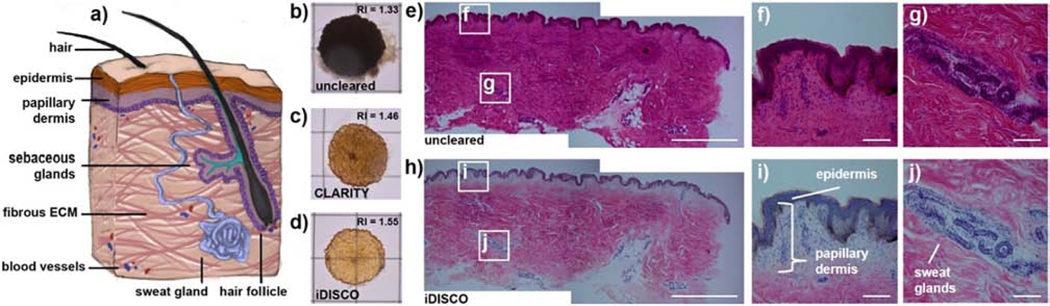
Our principal objective was to identify a human skin model that can be rendered “optically clear” (i.e., delipidated and having an RI matching its surrounding medium) to achieve an imaging contrast that will allow for 2D/3D ENM mapping with minimal background scattering from the tissues. To do so, fresh, full-thickness human skin biopsies were used to first compare the effectiveness of hydrogel-based and organic solvent-based clearing methods on rendering dense skin tissues transparent (Fig. 1a and S2). Although these methodologies differ in their clearing mechanism, both can be generally defined as ways to allow for reduced light-scattering and greater imaging depth in thick tissue sections. CLARITY (Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/ Immunostaining / in situ-hybridization-compatible Tissue hYdrogel), an aqueous clearing method, first crosslinks biomolecules in tissues within a hydrogel polymer network (Epp et al., 2015; Tomer et al., 2014). Tissue transparency is then achieved through sodium dodecyl sulfate (SDS) delipidation, followed by final aqueous refractive index (RI) homogenization. Conversely, iDISCO, an organic solvent-based clearing method, removes lipids with dichloromethane (DCM) and uses dibenzyl ether (DBE) to match the RI of the tissue (Renier et al., 2014). When used on full-thickness skin biopsies, we found that CLARITY made the skin tissues partially transparent while iDISCO yielded more transparent samples (Fig. 1b-d). As both methods remove lipids, it is likely that iDISCO resulted in better tissue transparency than CLARITY because the RI of dibenzyl ether more closely matched the RI of dermal extracellular matrix (Johnsen and Widder, 1999; Richardson and Lichtman, 2015). Consistent with the complete tissue clearing reported in a prior work (Abadie et al., 2018), our results confirm that RI matching is critical to achieving maximal optical transparency in the skin due to its abundance of extracellular matrix proteins. We then investigated the tissue histology of cleared and uncleared skin sections to assess any observable changes in skin features and architecture after the clearing protocol. Prior to clearing, H&E staining of 50 μm thick skin sections (Fig. 1e-g) revealed that skin tissues stain deeply pink, indicating a predominance of eosinophilic (acidophilic) proteins in all areas (Fischer et al., 2008). After iDISCO clearing, delipidation of cellular membranes resulted in loss of pink coloration in the lipid- and cell-dense sweat gland regions and epidermis, where blue basophilic nuclei and brown melanin-rich cells become visible with clear contrast (Fig. 1h-j). Conversely, the dermal regions remained deeply pink after clearing, indicating that elastin and collagen extracellular matrix proteins were retained during the clearing process.
Figure 1. Clearing ex vivo human skin tissues.
(a) Human skin tissue contains many light-scattering elements including hair follicles, sweat glands, and blood vessels, all within a dense, fibrous network of ECM proteins such as collagens and elastins. (b) Prior to clearing, full-thickness human skin biopsies (in water with refractive index (RI) = 1.33) are completely opaque and do not transmit light. (c) Skin tissues processed with the CLARITY method (in aqueous media with RI = 1.46) become semi-transparent after lipid removal. Skin cleared using the iDISCO method (in dibenzyl ether with RI = 1.55) yields a significantly more transparent sample (d) due to the combined effects of lipid removal and RI matching to the ECM protein RI. (e-g) Uncleared vs. (h-j) cleared skin slices (50 μm) stained with H&E demonstrate that nuclei (blue), melanocytes (brown), and ECM fibers (pink) still remain after clearing, while deeper pink lipids from cells in the epidermis and sweat glands are removed. Grid = 1 cm. Scale bars = (e, h) 2 mm; (f-g, i-j) 100 μm.
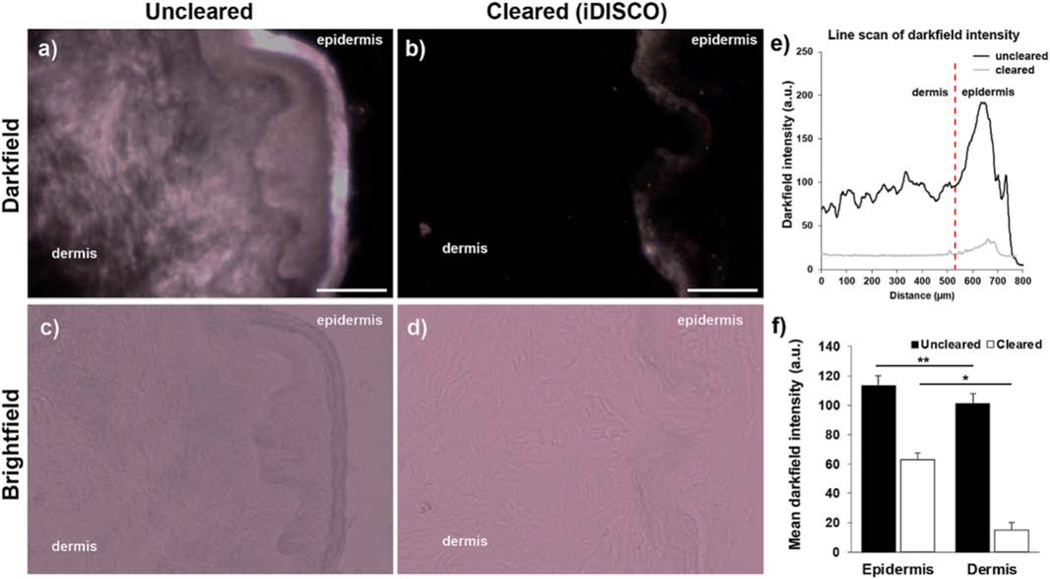
To assess the feasibility of improving the darkfield contrast for locating label-free metal oxide nanoparticles in the context of skin tissues, we quantified the reduction in background darkfield scattering in thick skin sections after iDISCO clearing (Fig. 2). Because even moderately thick skin tissues inherently scatter light strongly, we needed to first significantly reduce native tissue scattering via tissue clearing that can later assist in visually differentiating the scattering signal from ENMs and those from the background noise under darkfield. After employing iDISCO treatment on the skin biopsies not exposed to ENMs, darkfield intensity in the dermis decreased by an order of magnitude while epidermal scattering intensity was reduced by a factor of 3 (Fig. 2e-f).
Figure 2. Tissue clearing reduces darkfield light scattering in skin sections.
(a-b) Darkfield images of 50 μm skin sections before and after clearing. (c-d) Brightfield images of (a) and (b). (e) Representative intensity profile of skin sections before and after clearing (6 line scans from 1 skin tissue section). (f) Darkfield signal intensity before and after clearing in the dermal and epidermal regions (total of 14 fields of view (FOV) from n = 2 sections from 1 sample). Scale bars = 200 μm. *p < 0.0005; **p < 0.0001. Error bars represent SEM.
3.2. ENM retention in ex vivo skin biopsies after tissue clearing
Despite the known advantages of tissue clearing, one of the major concerns for its application in nanoparticle mapping is the ENM retention throughout the process (Sindhwani et al., 2017). In this regard, iDISCO presents an advantage over other clearing techniques of not having to process the tissues with electrophoresis (i.e., needed in some cases for CLARITY treatment), which may cause more ENM loss. Here, sector-field-inductively coupled plasma-mass spectrometry (ICP-MS) was used to quantify and report the total amount of unlabeled ENMs (TiO2 and ZnO) retained in skin tissues at different stages of the clearing process. We selected TiO2 and ZnO as the test label-free ENMs throughout our studies because these are commonly used in commercial skin products (e.g., sunscreens, lotions, cosmetic products) and are more effective blockers of UVA/UVB rays than most chemical sunscreens (Smijs and Pavel, 2011). To test the compatibility of our proposed sample processing technique with pre-clinical nanotoxicological skin models, we exposed the dermis of ex vivo human skin biopsies to TiO2 and ZnO nanoparticle dispersions at 100 μg/mL. As an initial proof of principle and to maximize the signal per tissue area, our first exposure model only involved the dermal region of human skin biopsies fully exposed to ENM suspensions in tissue culture media for 24 h as a closer simulation of a systemic ENM exposure (Fig. S2).
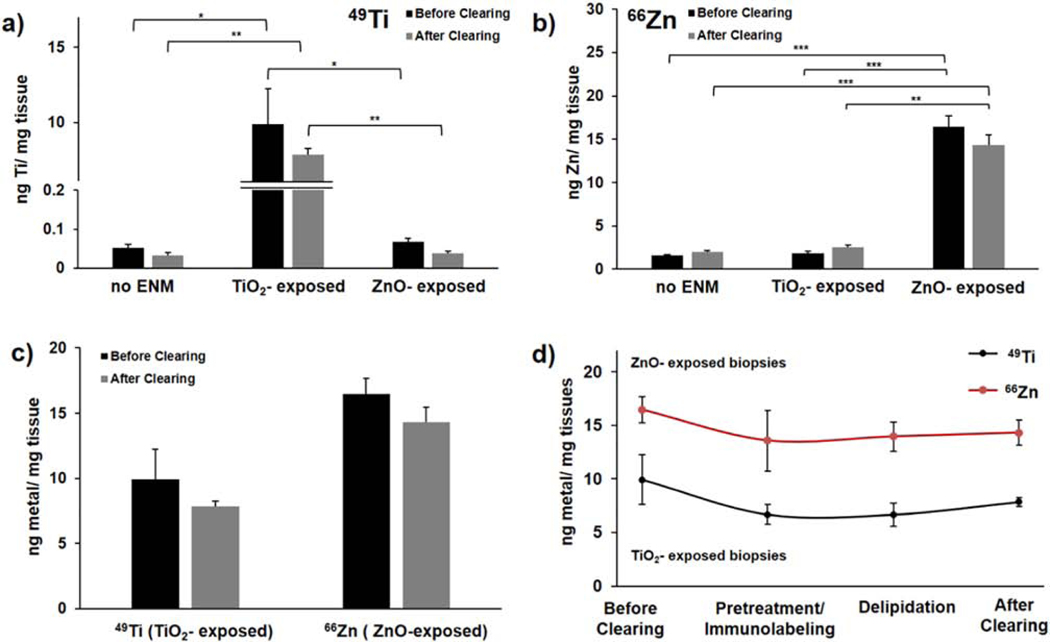
TiO2- and ZnO-exposed tissues were then fixed and immediately collected for digestion to analyze the levels of trace metals remaining after each major phase of the iDISCO method: (i) before clearing; (ii) pretreatment/immunolabeling; (iii) delipidation; and (iv) RI matching (Fig. 3). Fully cleared nanoparticle-exposed tissues retained 79% and 87% of their starting TiO2 and ZnO content, respectively. By examining the trace metal levels at each stage of clearing (Fig. 3d), we found that the pretreatment/ immunolabeling stage, which is the longest part of the treatment process and had the greatest number of washes, removed the majority of the initially embedded ENMs from the tissue. In general, the later delipidation stage typically affects the tightly packed layers of the epidermis. Thus, any ENMs unable to permeate past the epidermis during cutaneous exposure would be most likely to be released during delipidation compared to other phases. In our case, because only the dermal region was exposed to the ENM suspension, ENM content of samples did not significantly vary between the pretreatment/ immunolabeling and delipidation stages.
Figure 3. ICP-MS analysis of ENM retention after tissue clearing.
ICP-MS analysis of (a) 49Ti and (b) 66Zn content per tissue condition before and after clearing (no ENM, 24 h TiO2-exposed and 24 h ZnO-exposed); reported as ng metal/ mg tissue. (c) 49Ti content analysis for TiO2-exposed tissues plotted together with the 66Zn content analysis for ZnO-exposed tissues (TiO2, ~79% retention; ZnO, ~87% retention). (d) Monitoring the metal content at different phases of iDISCO tissue clearing process. Error bars represent SEM between biological replicates (n = 3 for TiO2-exposed tissues; n ≥ 5 for ZnO-exposed and unexposed/control tissues). Complete ICP-MS results for solvent blanks are summarized in Supplementary Table 2 with standard deviation per data point (from triplicate analyses each sample; technical replicates); *p ≥ 0.05, *p ≥ 0.005, *p ≥ 0.0005. Solvents: before clearing, delipidation = in PBS; pretreatment/immunolabeling = in methanol; after clearing = dibenzyl ether.
3.3. Confocal and darkfield imaging for visualizing ENMs in cleared human skin tissues
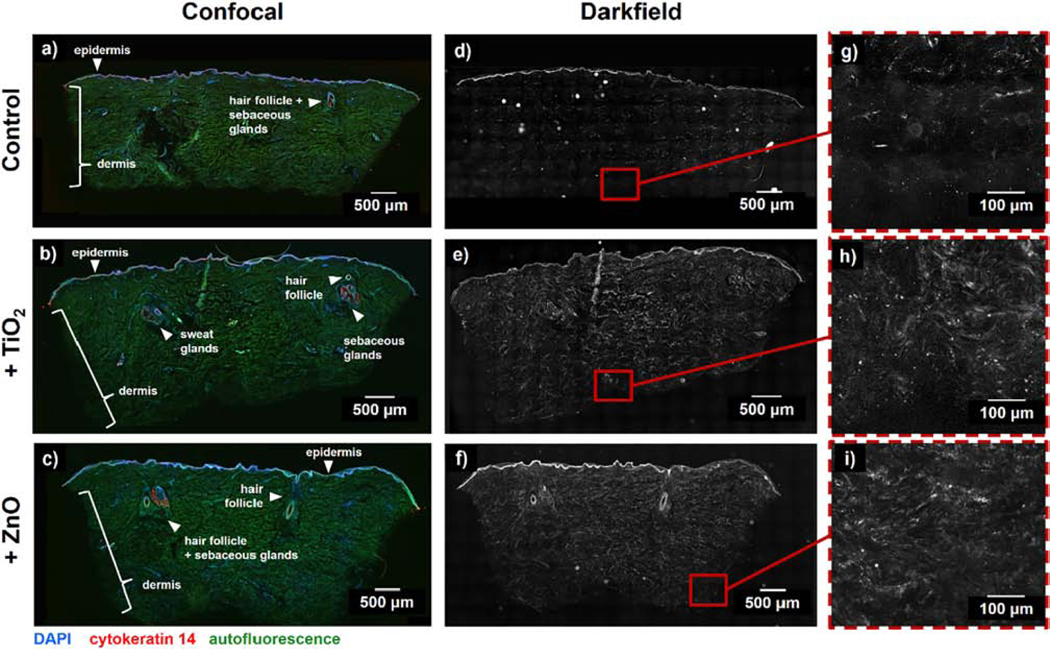
Once we confirmed that iDISCO effectively reduces background darkfield scattering caused by skin tissue without completely losing the permeated ENMs, we then used a nanoparticle scattering approach to image the retained ENMs in exposed skin tissues. Similar to the exposure protocol employed for the ENM retention analysis, after 24 h exposure of dermis to the ENM dispersions, the human skin samples were then fixed, sectioned, immunostained, and cleared. Interestingly, the remaining intact tissues and skin appendages exhibited autofluorescence at different excitation wavelengths (Fig. S3 and Video S1-S2). These signals, along with labeling the nuclei (DAPI) and epidermal regions (cytokeratin 14), allowed us to visualize major features of interest within the skin, including blood vessels, sweat glands, and hair follicles (Fig. 4a-c). These skin features and appendages therefore remained intact throughout the complete tissue clearing process, as presented in the representative confocal images of full tissue sections. On the other hand, darkfield imaging of the cleared tissue sections show signals that predominantly come from parts of the skin that effectively scatter light (i.e., epidermis) and the nanoparticles retained by the tissues after the clearing process (Fig. 4d-i). These representative images further support how background signal reduction from tissue autofluorescence and scattering can be achieved by an organic solvent-based clearing, therefore, improving the imaging contrast to differentiate the scattering signal coming from nanoparticles.
Figure 4. Retention of key skin features and reduction of background signal after tissue clearing.
(a-c) Whole skin tissue sections (thickness ~50 μm) were stained for DAPI and cytokeratin 14, cleared, and imaged with a confocal microscope to identify keratin- and cell-rich skin layers and appendages including the epidermis, hair follicles, sebaceous glands and sweat glands. (d-i) After confocal imaging, sections were imaged with darkfield microscopy to identify regions of increased scattering where applied ENMs permeated into the tissue. Skin samples in (a, d, g) did not have any treatment applied, while other samples were exposed to 100 μg/mL of (b, e, h) TiO2 and (c, f, i) ZnO nanoparticles for 24 hours. For these exposure experiments, only the dermal side of the biopsies was exposed to ENM suspensions.
3.4. Mapping 2D ENM distributions within skin sections using micro-spectroscopy
To quantify the distribution of label-free ENMs retained within the cleared tissues, we examined both TiO2 and ZnO nanoparticles that have visible optical scattering under darkfield illumination. Lorenz-Mie (LM) theory was employed here to predict the relative optical scattering intensities of these test ENMs, which has previously been identified as a reliable method for modeling the optical scattering of nanoparticles in biological samples (Zimmerman et al., 2019; Zimmerman et al., 2016). LM calculations show that the scattering intensity of a 100 nm TiO2 particle is expected to be higher in the visible range than a 50 nm ZnO particle (Fig. 5a-b). This is consistent with what was observed in both the exposed tissue samples and reference solutions. To map the distribution of the ENMs retained after clearing, hyperspectral darkfield imaging was performed on tissue sections. When working with ENMs with indistinct morphologies, darkfield imaging alone becomes insufficient for differentiating the signal from ENMs or background. Micro-spectroscopy techniques such as hyperspectral imaging and Raman spectroscopy can integrate darkfield images with a spatially resolved spectral profile of the scanned region that can be compared with a reference spectral fingerprint of the ENM of interest. To map the ENM distribution within exposed skin sections, the Spectral Angle Mapper (SAM) feature for hyperspectral imaging was used. Here, the scattering profile of each pixel of a hyperspectral image was compared to the separately recorded scattering spectra of aqueous TiO2 and ZnO solutions (Fig. 5c-d). For each particle type, the scattering profile does not only rely on the identity of the particle, but also on the size and extent of aggregation. Aggregated particle clusters, which often form under physiological conditions, can elicit a more red-shifted scattering profile than individual particles (Fig. S4-S5). Because of this phenomenon, particles and small aggregates in the reference solutions can have slight differences in spectra. Nonetheless, we used the most predominant spectral profile from the reference solutions in the reference spectral library when generating the nanoparticle maps. These maps positively co-registered with certain regions of the darkfield images. Compared against the average spectrum from the dermal tissue background, ZnO shows a more distinct profile from the background than TiO2 (Fig. 5e-f). To confirm the particle identity of the pixels that co-register with a red-shifted scattering spectrum, we examined these particles using Raman spectroscopy. Specifically, TiO2 is a strong Raman scatterer (maximum shift, ~144 cm−1) and its 2D spatial distribution can therefore be confirmed using Raman microscopy (Fig. 5g-h). In the representative darkfield image of a dermal region in Fig. 5g, merged with the brightfield image, the darkfield signals co-registered with Raman spectra that positively matched that of the reference TiO2. Moreover, larger particle clusters exhibit stronger apparent Raman scattering, therefore providing higher intensity Raman peaks that match with the spectrum of the reference.
Figure 5. Label-free mapping of TiO2 and ZnO within skin tissue sections using micro-spectroscopy techniques.
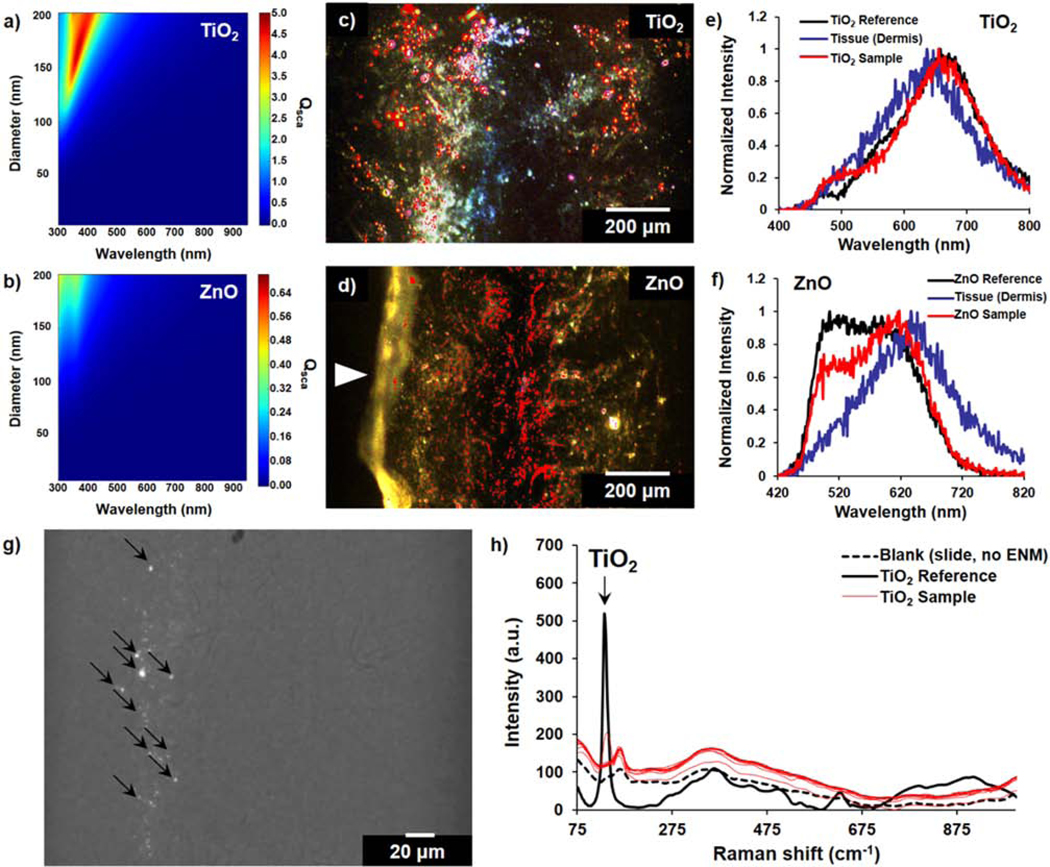
(a, b) Calculated scattering efficiency (Qsca) of TiO2 and ZnO at different diameters and multiple wavelengths, based on Lorenz-Mie theory model of particle scattering. (c, d) Enhanced darkfield images of ENM-exposed skin tissue sections overlaid with (c) TiO2 and (d) ZnO distribution map; annotated as red highlighted regions. White arrow in (d) indicates the epidermal region. (e, f) Representative scattering spectra taken from hyperspectral scans of tissue samples compared to a reference slide with TiO2 or ZnO aqueous solutions and background dermal tissue signal. (g) Darkfield-visible TiO2 ENMs in cleared skin sections; merged with brightfield image. (h) Raman scans of points in a representative cleared skin section with darkfield-visible scattering signal; compared to a reference TiO2 and blank glass slide. Black arrows in (g) represent the spots corresponding to the reported Raman spectra in (h). For these 24 h exposure experiments, only the dermal side of the biopsies was exposed to ENM suspensions.
After clearing, there are certain features of the skin tissues that still have apparent optical scattering and contribute to background noise. Blood vessels and hair follicles of the ex vivo human skin samples both show high background scattering even after the clearing process. Clusters of some nanomaterials like TiO2 that have overlapping spectra with the tissue can therefore complicate the measurements in such regions. This case was observed in the epidermal regions of untreated tissues, which can show false positive maps (Fig. S6). We then focused on ZnO-exposed samples to demonstrate how reduced background noise enables a better visualization of these ENMs under darkfield microscopy. These darkfield-visible particles co-registered well with pixels that are ZnO-positive in hyperspectral mapping (Fig. 6). No positive mapping was observed for untreated (“no ENM”) samples, indicating that this technique is robust against false positives for ENMs that have a scattering profile which do not significantly overlap with the tissue background. Additionally, a higher density of ZnO-positive distribution maps was observed in the dermal than epidermal region since only the dermis was exposed to the ENMs here. Interestingly, we observed an apparent localization of the retained ZnO nanoparticles within the microvasculature region. This observation can be attributed to the high amount of ECM surrounding these vessels and the known uptake efficiency of vascular cells (Behzadi et al., 2017; Gomez-Garcia et al., 2018; Samuel et al., 2012). While follicular permeation of nanoparticles is a common dermal translocation route, our darkfield images revealed a low density of ENMs (i.e., only around the hair follicle bulb) surrounding hair follicles. These observations are also consistent with our experimental model since only the dermal region was exposed. However, follicular localization would have been expected if the exposure was done through the epidermal layer. Together, our hyperspectral mapping findings in human skin sections demonstrate that tissue clearing improved imaging contrast, enabling us to examine the localization behavior of those ENMs which are retained throughout the clearing process.
Figure 6. ENM localization on skin layers and appendages after 24 h exposure.
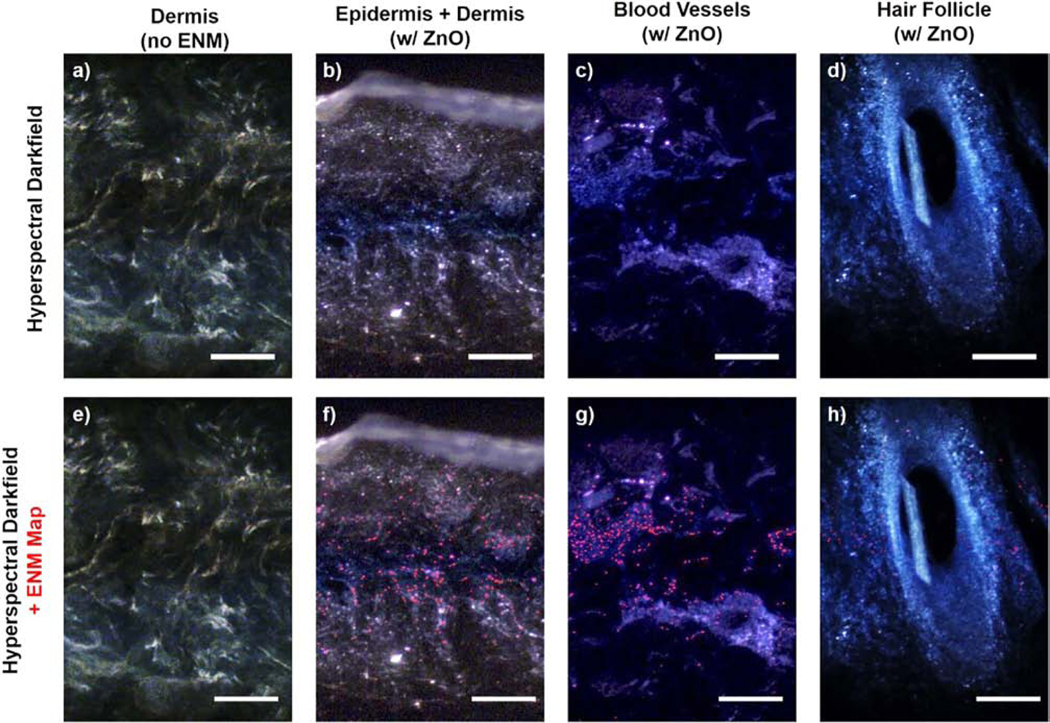
(a-d) Darkfield images with (e-h) hyperspectral maps of ZnO-positive pixels in areas with high background scattering even after iDISCO-clearing. (a, e) Dermal region of an unexposed tissue is shown for reference. Scale bars = 200 μm. For these exposure experiments, only the dermal side of the biopsies was exposed to ENM suspensions.
3.5. 3D mapping of fluorescently labeled ENMs
In addition to mapping the 2D distribution of darkfield-visible metal oxides, we sought to extend our methodology to 3D analytical techniques to better capture the localization patterns of ENMs in skin tissues. Previously, tissue clearing in rat brain, liver, and kidney showed the feasibility of quantifying the distribution of quantum dots and fluorescently labeled ENMs through 3D imaging (Syed et al., 2017). Additionally, another group pioneered the concept of multi-wavelength autofluorescence imaging in 3D cleared human skin, but without any investigation into nanoparticle mapping (Abadie et al., 2018). We reasoned that we could unify these approaches to be able to evaluate the 3D distribution of ENMs within human skin tissue.
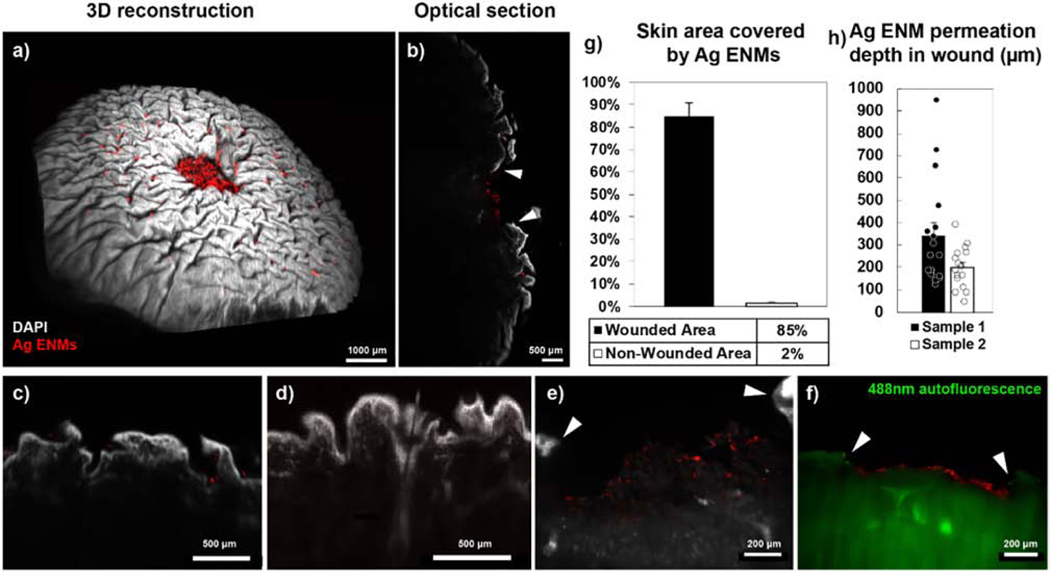
Due to the inherent limitations of hyperspectral imaging and Raman spectroscopy for 3D volumetric imaging, we focused on a fluorescence-based approach to locate ENMs in 3D human skin tissues. Here, fluorescently tagged Ag nanoparticles were used as test ENMs for investigating their permeation across compromised skin biopsies (epidermal wound diameter = 2 mm). Since longer wavelengths of light scatter less in biological tissue, we labeled biotinylated Ag ENMs with a streptavidin-conjugated Alexa Fluor 647 dye and exposed compromised human skin samples to these labeled ENMs through the wounded epidermal layer. After fixing, immunostaining, and clearing the tissues, we imaged them using light sheet fluorescence microscopy (LSFM) due to its speed advantage over confocal microscopy for imaging millimeter-sized tissues (Fig. 7a-f and Video S3-S5). Our imaging analysis clearly showed that ENMs localized in the wounded area and only in the ridges of the non-wounded epidermal layer (Fig. 7g). Our analysis also showed that the fluorescently labeled Ag ENMs were unable to penetrate deeper than the papillary dermis (i.e., < 500 μm) during the exposure period (Fig. 7h). Since previous in vitro studies have suggested that certain ENMs such as cobalt are able to penetrate human skin (Larese Filon et al., 2013), our results support the need for particle-specific imaging protocols with for testing cutaneous nanoparticle translocation in a translatable human model.
Figure 7. 3D mapping of fluorescently labeled Ag nanoparticles in cleared human skin tissues after 24 h exposure.
Biotinylated Ag nanoparticles (~20 nm) labeled with Alexa Fluor 647-conjugated streptavidin were applied to wounded human skin biopsies, then imaged in 3D on a light sheet fluorescent microscope (LSFM) after tissue immunolabeling and clearing. Large deposits of Ag ENMs remaining in and around the wound site are visible in both (a) 3D reconstruction and (b) optical sections of a wounded skin biopsy. (c-f) No apparent Ag localization was observed in regions with hair follicles and sweat glands at the wound surface (h, green = 488 nm autofluorescence). White arrowheads indicate the epidermal boundary of the wound site. Wound diameter = 2 mm. (g) Within the wound site (top view), 85% of exposed dermal surface area is covered by Ag ENMs, while only 2% of intact epidermal surface area is covered (503 light sheet sections from 2 wounded skin samples). (h) Ag ENMs remained confined to the superficial papillary dermis and did not permeate into the deeper dermal layers (mean < 500 μm; 12 ENMs from 4 light sheet sections of 2 wounded skin samples). Error bars are SEM.
3.6. Protocol strengths and limitations
In this study, we demonstrated a strategy for nanoparticle distribution mapping in non-perfused human skin biopsies that involves a tissue clearing protocol. Cleared skin samples offer improved signal-to-noise ratio over uncleared samples for both label-free 2D mapping and 3D mapping of fluorescent nanoparticles (Sindhwani et al., 2017; Sindhwani et al., 2016; Syed et al., 2017). The technique is also less intensive as compared to the sample preparation required for traditional electron microscopy approaches (Pena Mdel et al., 2016). Additionally, the imaging modalities compatible with tissue cleared samples are non-destructive and are better suited to mapping large tissue segments. However, the micro-spectroscopy techniques for label-free mapping used in this study are not currently suited for 3D volumetric analysis and are limited to relative quantitation (Idelchik et al., 2016). By using fluorescently labeled test nanoparticles, we were able to perform 3D ENM mapping on cleared human skin tissues. In terms of the skin model used, a non-perfused human ex vivo tissue allows for studying the colocalization of nanoparticles in skin features that are not present in animal models (such as sweat glands) but does not mimic the physiological blood flow involved in systemic biodistribution of ENMs. The lack of perfusion presents a limitation because it eliminates the effects that pressure differences and fluid flow might have on nanoparticle localization and clearance in vivo. Thus, the static nature of our current model must be considered when interpreting the translational validity of the ENM distribution patterns obtained using ex vivo skin tissue models to the actual biodistribution profiles in vivo. In the future, this can potentially be addressed by performing ENM exposure experiments in fluidically-assisted in vitro skin models (Abaci et al., 2015; Abaci et al., 2016; Alberti et al., 2017; Lee et al., 2017).
Lastly, we have reported the degree of ENM retention for label-free TiO2 and ZnO after tissue clearing as exemplary cases. Although there are previous studies in non-skin animal organs which sought to prevent nanoparticle loss during tissue clearing, particle modification was not employed for TiO2 and ZnO for the studies presented herein as we focused on investigating label-free ENMs that preserve their native size and surface chemistry. Among the reported approaches include coating nanoparticles with serum proteins and subsequently crosslinking them to the tissue during the polyacrylamide gelation step of the CLARITY method (Sindhwani et al., 2016). The analysis consistently showed between 60%–80% quantum dot retention in the mouse liver, spleen, and kidney after CLARITY clearing. Protocols for particle modification are therefore available for improving particle retention rate during tissue clearing and can be performed prior to exposure experiments. Such strategies can be employed as long as any influence of particle surface modification on the biodistribution kinetics of the ENM being studied is properly considered.
4. Conclusions
In this study, a strategy for mapping nanoparticle distributions in ENM-exposed ex vivo human skin tissues using multiple imaging modalities is presented. An organic solvent-based tissue clearing approach was shown to improve the imaging contrast for label-free ENM mapping in 2D skin tissues and to enable 3D nanoparticle mapping for larger skin sections. This clearing approach successfully achieved the optical clearing of thick tissue sections and whole skin biopsies while retaining the key features of the skin and without completely washing out the tissue embedded ENMs. Here, micro-spectroscopy techniques such as hyperspectral and Raman darkfield microscopy were both presented as imaging techniques compatible with mapping the 2D distribution of TiO2 and ZnO nanoparticles in exposed dermal regions of cleared skin tissue sections. The strong optical and Raman scattering of TiO2 nanoparticles allowed for the use of correlated hyperspectral and Raman microscopy to quantify its 2D distribution within the context of a skin tissue section. Minimal penetration of fluorescently tagged Ag nanoparticles (i.e., < 500 μm) was also measured by imaging cleared human skin biopsies with cutaneous wounds using light sheet fluorescence microscopy. Overall, the approach presented herein expands the available imaging and sample preparation techniques for exploring metal/metal oxide nanoparticle-tissue interactions in skin with significantly improved imaging contrast. The insights learned from these studies could therefore be translated towards developing a better imaging approach for evaluating the biodistribution and toxicological implications of various commercial products that contain ENMs as active ingredients.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number (under award numbers U01ES027272 and U24ES026946) as part of the Nanotechnology Health Implications Research (NHIR) Consortium. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The TiO2-E171 and ZnO reference nanomaterials used in the research presented in this publication have been procured and characterized by the Engineered Nanomaterials Resource and Coordination Core (ERCC) at the Center for Nanotechnology and Nanotoxicology at Harvard School of Public Health, part of the NIEHS/NHIR consortium. This work was partially supported by the Wyss Institute for Biologically Inspired Engineering at Harvard University. For the imaging of nanoparticles using hyperspectral darkfield Raman microscope, we thank the Harvard Center for Nanoscale Systems (CNS), which is a member of the National Nanotechnology Infrastructure Network (NNIN) under NSF Award No. 1541959. This study was also partly supported by the Harvard University Materials Research Science and Engineering Center (MRSEC), NSF Award No. DMR-1420570. We thank the Harvard Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported the 3D imaging of cleared samples in this work. This facility is supported in part by the Neural Imaging Center as part of an NINDS P30 Core Center grant #NS072030. We acknowledge the Harvard Center for Biological Imaging for infrastructure and support of the confocal microscopy presented in this work. We also thank the Harvard Stem Cell and Regenerative Biology Histology Core for assisting with histology services. Our group would also like to acknowledge Dr. Martin Shafer and his Trace Element Research team at the University of Wisconsin State Laboratory of Hygiene for the ICP-MS analysis of our cleared tissue samples. G. J. T. is supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1745303. H. A. M. A. thanks the American Chemical Society for generous support through the Irving S. Sigal Postdoctoral Fellowship. We also thank Grace Matthews for graphical assistance and Gabrielle Nekrasas for providing guidance on the topical exposure experiments.
Abbreviations:
- AgNPs
silver nanoparticles
- CLARITY
Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/ Immunostaining / in situ-hybridization-compatible Tissue hYdrogel
- DAPI
4’,6-diamidino-2-phenylindole dihydrochloride
- DBE
dibenzyl ether
- DCM
dichloromethane
- DLS
dynamic light scattering
- DSEcr
critical delivered sonication energy
- ECM
extracellular matrix
- ENM
engineered nanomaterial
- ICP-MS
inductively coupled plasma-mass spectrometry
- iDISCO/iDISCO+
immunolabeling-enabled three-Dimensional Imaging of Solvent-Cleared Organs
- LM
Lorenz-Mie
- LSFM
light sheet fluorescent microscopy
- MeOH
methanol
- OCT
optimal cutting temperature
- PAP
peroxidase-antiperoxidase
- PBS
phosphate-buffered saline
- PTwH
PBS with Tween-20 and Heparin
- PTx.2
PBS with 0.2% Triton X-100
- Qsca
optical scattering coefficient
- RI
refractive index
- SAM
spectral angle mapping
- SDS
sodium dodecyl sulfate
Footnotes
Appendix A. Supplementary data
Supplementary information to this article containing a more comprehensive description of experimental methods, quantification protocols, nanoparticle characterization, 3D renderings of light sheet images and other supporting images can be found online at: http://dx.doi.org/10.1016/j.impact.2019.xx.xxx.
Declaration of Competing Interest
The authors declare no conflict of interest regarding the publication of this article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abaci HE, Gledhill K, Guo Z, Christiano AM, and Shuler ML (2015). Pumpless microfluidic platform for drug testing on human skin equivalents. Lab Chip 15, 882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaci HE, Guo Z, Coffman A, Gillette B, Lee WH, Sia SK, and Christiano AM (2016). Human Skin Constructs with Spatially Controlled Vasculature Using Primary and iPSC-Derived Endothelial Cells. Adv Healthc Mater 5, 1800–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abadie S, Jardet C, Colombelli J, Chaput B, David A, Grolleau JL, Bedos P, Lobjois V, Descargues P, and Rouquette J. (2018). 3D imaging of cleared human skin biopsies using light-sheet microscopy: A new way to visualize in-depth skin structure. Skin Research and Technology 24, 294–303. [DOI] [PubMed] [Google Scholar]
- Alberti M, Dancik Y, Sriram G, Wu B, Teo YL, Feng Z, Bigliardi-Qi M, Wu RG, Wang ZP, and Bigliardi PL (2017). Multi-chamber microfluidic platform for high-precision skin permeation testing. Lab Chip 17, 1625–1634. [DOI] [PubMed] [Google Scholar]
- Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC, and Mahmoudi M. (2017). Cellular uptake of nanoparticles: journey inside the cell. Chem Soc Rev 46, 4218–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JM, Beltran-Huarac J, Pyrgiotakis G, and Demokritou P. (2018). Effective delivery of sonication energy to fast settling and agglomerating nanomaterial suspensions for cellular studies: Implications for stability, particle kinetics, dosimetry and toxicity. NanoImpact 10, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoid G, Cohen JM, Darrah T, Derk R, Rojanasakul L, Pyrgiotakis G, Wohlleben W, and Demokritou P. (2014). Estimating the effective density of engineered nanomaterials for in vitro dosimetry. Nature Communications 5, 3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoid GM, Cohen JM, Pyrgiotakis G, and Demokritou P. (2017). Preparation, characterization, and in vitro dosimetry of dispersed, engineered nanomaterials. Nat Protoc 12, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoid GM, Cohen JM, Pyrgiotakis G, Pirela SV, Pal A, Liu J, Srebric J, and Demokritou P. (2015). Advanced computational modeling for in vitro nanomaterial dosimetry. Part Fibre Toxicol 12, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delouise LA (2012). Applications of nanotechnology in dermatology. Journal of Investigative Dermatology 132, 964–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp JR, Niibori Y, Hsiang HL, Mercaldo V, Deisseroth K, Josselyn SA, and Frankland PW (2015). Optimization of CLARITY for Clearing Whole-Brain and Other Intact Organs. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eweje F, Ardoña HAM, Zimmerman JF, O’Connor BB, Ahn S, Grevesse T, Rivera K, Bitounis D, Demokritou P, and Parker KK (2019). Quantifying the effects of engineered nanomaterials on endothelial cell architecture and vascular barrier integrity quantified using a cell pair model. Nanoscale 11, 17878–17893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, and Zeller R. (2008). Hematoxylin and Eosin Staining of Tissue and Cell Sections. Cold Spring Harbor Protocols 2008, pdb.prot4986-pdb.prot4986. [DOI] [PubMed] [Google Scholar]
- Gomez-Garcia MJ, Doiron AL, Steele RRM, Labouta HI, Vafadar B, Shepherd RD, Gates ID, Cramb DT, Childs SJ, and Rinker KD (2018). Nanoparticle localization in blood vessels: dependence on fluid shear stress, flow disturbances, and flow-induced changes in endothelial physiology. Nanoscale 10, 15249–15261. [DOI] [PubMed] [Google Scholar]
- Idelchik M.d.P.S., Neu-Baker NM, Chandrasekaran A, Friedman AJ, Frame MD, and Brenner SA (2016). Relative quantitation of metal oxide nanoparticles in a cutaneous exposure model using enhanced darkfield microscopy and hyperspectral mapping. NanoImpact 3-4, 12–21. [Google Scholar]
- Johnsen S, and Widder EA (1999). The Physical Basis of Transparency in Biological Tissue: Ultrastructure and the Minimization of Light Scattering. Journal of Theoretical Biology 199, 181–198. [DOI] [PubMed] [Google Scholar]
- Labouta HI, Hampel M, Thude S, Reutlinger K, Kostka K-H, and Schneider M. (2012). Depth profiling of gold nanoparticles and characterization of point spread functions in reconstructed and human skin using multiphoton microscopy. Journal of Biophotonics 5, 85–96. [DOI] [PubMed] [Google Scholar]
- Larese Filon F, Crosera M, Timeus E, Adami G, Bovenzi M, Ponti J, and Maina G. (2013). Human skin penetration of cobalt nanoparticles through intact and damaged skin. Toxicology in Vitro 27, 121–127. [DOI] [PubMed] [Google Scholar]
- Lee JY, Wang H, Pyrgiotakis G, DeLoid GM, Zhang Z, Beltran-Huarac J, Demokritou P, and Zhong W. (2018). Analysis of lipid adsorption on nanoparticles by nanoflow liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem 410, 6155–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Jin S-P, Kim YK, Sung GY, Chung JH, and Sung JH (2017). Construction of 3D multicellular microfluidic chip for an in vitro skin model. Biomedical Microdevices 19, 22. [DOI] [PubMed] [Google Scholar]
- Lekki J, Stachura Z, Dąbroś W, Stachura J, Menzel F, Reinert T, Butz T, Pallon J, Gontier E, Ynsa MD, et al. (2007). On the follicular pathway of percutaneous uptake of nanoparticles: Ion microscopy and autoradiography studies. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 260, 174–177. [Google Scholar]
- Lin L, Nufer K, Tomihara S, and Prow T. (2015). Non-Invasive Nanoparticle Imaging Technologies for Cosmetic and Skin Care Products. Cosmetics 2, 196–210. [Google Scholar]
- Lin LL, Grice JE, Butler MK, Zvyagin AV, Becker W, Robertson TA, Soyer HP, Roberts MS, and Prow TW (2011). Time-correlated single photon counting for simultaneous monitoring of zinc oxide nanoparticles and NAD(P)H in intact and barrier-disrupted volunteer skin. Pharmaceutical Research 28, 2920–2930. [DOI] [PubMed] [Google Scholar]
- Lin LL, Yamada M, and Prow TW (2016). Imaging Nanoparticle Skin Penetration in Humans. In Nanoscience in Dermatology, pp. 353–366. [Google Scholar]
- Mathes C, Brandner JM, Laue M, Raesch SS, Hansen S, Failla AV, Vidal S, Moll I, Schaefer UF, and Lehr C-M (2016). Tight junctions form a barrier in porcine hair follicles. European Journal of Cell Biology 95, 89–99. [DOI] [PubMed] [Google Scholar]
- Morganti P. (2010). Use and potential of nanotechnology in cosmetic dermatology. Clinical, cosmetic and investigational dermatology 3, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena Mdel P, Gottipati A, Tahiliani S, Neu-Baker NM, Frame MD, Friedman AJ, and Brenner SA (2016). Hyperspectral imaging of nanoparticles in biological samples: Simultaneous visualization and elemental identification. Microsc Res Tech 79, 349–358. [DOI] [PubMed] [Google Scholar]
- Renier N, Adams EL, Kirst C, Wu Z, Azevedo R, Kohl J, Autry AE, Kadiri L, Umadevi Venkataraju K, Zhou Y, et al. (2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier N, Wu Z, Simon DJ, Yang J, Ariel P, and Tessier-Lavigne M. (2014). iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910. [DOI] [PubMed] [Google Scholar]
- Richardson DS, and Lichtman JW (2015). Clarifying Tissue Clearing . Cell 162, 246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel SP, Jain N, O’Dowd F, Paul T, Kashanin D, Gerard VA, Gun’ko YK, Prina-Mello A, and Volkov Y. (2012). Multifactorial determinants that govern nanoparticle uptake by human endothelial cells under flow. Int J Nanomedicine 7, 2943–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzui M, Tamura T, Miura K, Ikarashi Y, Watanabe Y, and Fujii M. (2010). Study on penetration of titanium dioxide (TiO(2)) nanoparticles into intact and damaged skin in vitro. The Journal of toxicological sciences 35, 107–113. [DOI] [PubMed] [Google Scholar]
- Siefke T, Kroker S, Pfeiffer K, Puffky O, Dietrich K, Franta D, Ohlídal I, Szeghalmi A, Kley E-B, and Tünnermann A. (2016). Materials Pushing the Application Limits of Wire Grid Polarizers further into the Deep Ultraviolet Spectral Range. Advanced Optical Materials 4, 1780–1786. [Google Scholar]
- Sindhwani S, Syed AM, Wilhelm S, and Chan WC (2017). Exploring Passive Clearing for 3D Optical Imaging of Nanoparticles in Intact Tissues. Bioconjug Chem 28, 253–259. [DOI] [PubMed] [Google Scholar]
- Sindhwani S, Syed AM, Wilhelm S, Glancy DR, Chen YY, Dobosz M, and Chan WCW (2016). Three-Dimensional Optical Mapping of Nanoparticle Distribution in Intact Tissues. ACS Nano 10, 5468–5478. [DOI] [PubMed] [Google Scholar]
- Singh P, and Nanda A. (2012). Nanotechnology in cosmetics: a boon or bane? Toxicological & Environmental Chemistry 94, 1467–1479. [Google Scholar]
- Smijs TG, and Pavel S. (2011). Titanium dioxide and zinc oxide nanoparticles in sunscreens: focus on their safety and effectiveness. Nanotechnol Sci Appl 4, 95–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SoRelle ED, Liba O, Campbell JL, Dalal R, Zavaleta CL, and de la Zerda A. (2016). A hyperspectral method to assay the microphysiological fates of nanomaterials in histological samples. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling C, Singh CR, Karg M, Konig TA, Thelakkat M, and Retsch M. (2017). Plasmonic nanomeshes: their ambivalent role as transparent electrodes in organic solar cells. Sci Rep 7, 42530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed AM, Sindhwani S, Wilhelm S, Kingston BR, Lee DSW, Gommerman JL, and Chan WCW (2017). Three-Dimensional Imaging of Transparent Tissues via Metal Nanoparticle Labeling. Journal of the American Chemical Society 139, 9961–9971. [DOI] [PubMed] [Google Scholar]
- Taurozzi JS, Hackley VA, and Wiesner MR (2011). Ultrasonic dispersion of nanoparticles for environmental, health and safety assessment – issues and recommendations. Nanotoxicology 5, 711–729. [DOI] [PubMed] [Google Scholar]
- Tomer R, Ye L, Hsueh B, and Deisseroth K. (2014). Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nature Protocols 9, 1682–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lai X, Shao L, and Li L. (2018). Evaluation of immunoresponses and cytotoxicity from skin exposure to metallic nanoparticles. International Journal of Nanomedicine Volume 13, 4445–4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, and Fujiwara H. (2011). Cell-extracellular matrix interactions in normal and diseased skin. Cold Spring Harbor perspectives in biology 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang R, Xiao H, Bhattacharya K, Bitounis D, Demokritou P, and McClements DJ (2019). Development of a standardized food model for studying the impact of food matrix effects on the gastrointestinal fate and toxicity of ingested nanomaterials. NanoImpact 13, 13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Nagashima K, Parmiter D, de la Cruz J, and Patri AK (2011). SEM X-Ray Microanalysis of Nanoparticles Present in Tissue or Cultured Cell Thin Sections In Characterization of Nanoparticles Intended for Drug Delivery, McNeil SE, ed. (Totowa, NJ: Humana Press; ), pp. 93–99. [DOI] [PubMed] [Google Scholar]
- Zimmerman J, Ardoña HAM, Pyrgiotakis G, Dong J, Moudgil BM, Demokritou P, and Parker KK (2019). Scatter Enhanced Phase Contrast Microscopy for Discriminating Mechanisms of Active Nanoparticle Transport in Living Cells. Nano Letters. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JF, Parameswaran R, Murray G, Wang Y, Burke M, and Tian B. (2016). Cellular uptake and dynamics of unlabeled freestanding silicon nanowires. Science Advances 2, e1601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.