Abstract
The presence of circulating tumor cells (CTCs) in a patient’s bloodstream is a hallmark of metastatic cancer. The detection and analysis of CTCs is a promising diagnostic and prognostic strategy as they may carry useful genetic information from their derived primary tumor, and the enumeration of CTCs in the bloodstream has been known to scale with disease progression. However, the detection of CTCs is a highly challenging task owing to their sparse numbers in a background of billions of background blood cells. To effectively utilize CTCs, there is a need for an assay that can detect CTCs with high specificity and can locally enrich CTCs from a liquid biopsy. We demonstrate a versatile methodology that addresses these needs by utilizing a combination of nanoparticles. Enrichment is achieved using targeted magnetic nanoparticles and high specificity detection is achieved using a ratiometric detection approach utilizing multiplexed targeted and non-targeted surface-enhanced Raman Scattering Nanoparticles (SERS-NPs). We demonstrate this approach with model prostate and cervical circulating tumor cells and show the ex vivo utility of our methodology for the detection of PSMA or folate receptor over-expressing CTCs. Our approach allows for the mitigation of interference caused by the non-specific uptake of nanoparticles by other cells present in the bloodstream and our results from magnetically trapped CTCs reveal over a 2000% increase in targeted SERS-NP signal over non-specifically bound SERS-NPs.
1. Introduction
Metastatic disease is responsible for nearly 90 percent of deaths due to cancer [1]. When a primary tumor invades the vasculature and sheds cells into circulation, the resulting circulating tumor cells (CTCs) spawn secondary tumors which can quickly become extremely difficult to therapeutically manage. Furthermore, the number of CTCs present has been shown to scale with prognostic severity [2,3]. Therefore, detection and enumeration of these CTCs can provide an important prognostic measure for clinical decision making while planning treatment options and determining the level of treatment aggressiveness [4]. Additionally, genetic analysis of CTCs is a promising means of biomarker profiling for individualizing therapy selection [5,6]. However, detection of CTCs is a highly challenging task. Typically, patients only have a few CTCs per millilitre of blood necessitating the collection of large blood volumes which contain billions of blood cells in a complex bio-environment. To effectively utilize CTCs in a liquid biopsy, there is a need to develop an assay that can enrich and quantify trapped CTCs with high specificity.
One of the currently clinically approved approaches for detecting CTCs, CellSearch, utilizes epithelial cell adhesion molecule (EpCAM) targeting for immunomagnetic purification, along with validation using DAPI, CD45, and cytokeratins 8, 18 and 19 [7]. However, the reliance on EpCAM targeting can lead to a reduction in detection sensitivity as many tumor cells down-regulate EpCAM in order to enter circulation, and it has been shown that CTCs have a greater than ten-fold decrease in EpCAM expression compared to cell lines and carcinoma cells of primary and metastatic tissues [8]. The reliance of this targeting moiety limits the applicability of CellSearch to only epithelial derived CTCs that have not undergone an epithelial-to-mesenchymal transition (EMT).
Unfortunately, many tumor cells do undergo an EMT during which the expression of targets such as EpCAM is lost and hence, these CTCs are not captured using CellSearch [9,10]. Moreover, this and other approaches still struggle to detect CTCs in low abundance and non-specific binding can lead to false positive detection events. Additionally, EpCAM targeting may not be appropriate for in vivo cytometry applications as it not a tumor-specific antigen and is also present in healthy epithelial tissues [11].
Alternatively, more specific targeting moieties that are retained past the EMT exist. For example, many epithelial sourced cancers such as breast, cervical and ovarian cancers significantly overexpress the FOLR1 folate receptor compared to healthy cells [12,13] and prostate specific membrane antigen (PSMA) is overexpressed in most prostate cancers. The upregulation of these receptors makes them ideal candidates for CTC targeting and offers promising alternatives for the enumeration of CTCs as their expression is retained past the EMT unlike currently targeted markers.
In general, there could be several other useful biomarkers for detection, enumeration, and biomarker profiling, but even low levels of non-specific binding could lead to a significant source of false positives limiting the utility of such biomarkers when there is not a large abundance of CTCs present. High-speed flow cytometry applications offer the possibility of multiplexed fluorescent antibody-based detection but does not provide a means of pre-enrichment, leading to the requirement of large blood volumes and the inability to handle large throughputs, which precludes its widespread usage for screening applications. Differentiating specific from non-specific binding could have significant implications for widening the spectrum of biomarkers used for enumerating CTCs, and lead to an increased capacity for biomarker profiling.
Sensitive detection of magnetically trapped CTCs has been accomplished using various strategies including fluorescence [14–16] and photoacoustic detection [17–23]. However, fluorescence approaches require detection above the auto-fluorescence background and lack sensitivity in deep tissues for in vivo applications. Photoacoustic approaches must achieve sufficient nanoparticle targeting density to achieve signals above the background of red blood cells and multiplexing approaches are challenging due to the broad absorption spectra of nanoparticles and the requirements for multiple laser wavelengths pulsed fast enough to avoid motion artifacts.
Microfluidic approaches have also been employed, largely utilizing methodologies such as immunocapture [24–26], immunofluorescence detection [27], immunomagnetic trapping [28,29] or ligand-interactive cell rolling detection [30]. Alternatively, label-free microfluidic approaches have also been developed which leverage physical size discrepancy between CTCs and normal blood cells. These methods include mechanical filtering [31,32], hydrodynamic isolation [33], dielectrophoresis [34] or acoustophoresis [35]. Although these methods are resistant to epitope loss, they typically suffer from a high rate of false positives due physical heterogeneities present in both CTCs and background blood cells. Microfluidic approaches are also currently not easily translated to in vivo usage and some methods such as dielectrophoresis are limited to ex vivo processing as it requires alteration of the bulk media with a conductive solution. Additionally, microfluidic approaches are fundamentally limited by low blood throughput and would be difficult to implement for sampling the systemic blood volumes required for detecting rare CTCs.
Recently, an in vivo enrichment methodology, MagWIRE [36], has been demonstrated to effectively utilize functionalized magnetic nanoparticles to enrich CTCs and a surgically implanted wire for retrieval. However, this process is still hindered by masking of CTCs by other blood borne particulates which may non-specifically sequester magnetic nanoparticles.
In vivo flow cytometry offers the promise of magnetic enrichment of CTCs using the whole blood volume by using magnetic nanoparticles targeted to CTCs to facilitate specific magnetic trapping. Galanzha et al. demonstrated in vivo flow cytometry using magnetic nanoparticles and gold-plated carbon-nanotubes for photoacoustic detection [20]. Our previous work introduced both magnetic nanoparticles (MNPs) and surface-enhanced Raman-scattering nanoparticles (SERS-NPs) for in vivo magnetic enrichment and Raman-based detection of trapping events [37]. Owing to the ultra-low background of the SERS signature, effective single-cell trapping sensitivity could be achieved. Furthermore, SERS signatures are highly unique and can be spectrally unmixed enabling their use in multiplexed applications. Previous work, however, did not investigate targeting specificity and non-specific binding of nanoparticles to non-tumor cells which could lead to false positive detection. False positives may also be due to phagocytic uptake by immune cells.
To accomplish the aims of CTC enrichment with improved differentiation of specific and non-specific detection, this paper introduces a novel method of nanoparticle-based magnetic enrichment and ratiometric detection of multiplexed SERS nanoparticles. To perform this task, we decorate magnetic nanoparticles, for trapping, and a single variant of SERS-NP, for detection, with small molecule targeting compounds to specifically target overexpressed moieties found on CTCs. In this study we will focus on targeted using folate and a peptidomimetic PSMA inhibitor respectively. The folate receptor is upregulated in many forms of cancer such as cervical and breast cancer and PSMA is upregulated in prostate cancer. When all nanoparticles are introduced into blood containing CTCs, only cells binding to both the targeted magnetic and the SERS nanoparticles will produce an increasing SERS signal over time [37]. To discriminate against non-specific uptake of these nanoparticles by phagocytic cells, the uptake of targeted SERS-NPs is compared to a second untargeted variant which exhibits receptor-independent uptake. The highly unique SERS spectra exhibit ultra-low crosstalk, ultra-high signal-to-noise, and are not affected by tissue auto-fluorescence. As such, we can determine the contribution of each nanoparticle variant to the detected SERS signal. CTCs which preferentially bind to both targeted MNPs and targeted SERS-NPs will be detectable and quantifiable against the non-specific trapping of phagocytic and other background cells. The long-term goal of this work is to demonstrate in vivo detection and trapping of sparse numbers of CTCs with multiplex biomarker profiling and ultra-specific detection. As an initial demonstration of the potential of this approach we demonstrate CTC enrichment and specific detection in an ex vivo flow phantom. As such, we have developed a powerful and highly specific means of CTC enrichment which retains its effectiveness past epithelial-to-mesenchymal transitions.
2. Methods
2.1. Overview of system setup
In short, a whole blood sample containing varying concentrations and phenotypes of model CTCs is mixed with a combination of targeted and untargeted SERS nanoparticles and targeted magnetic nanoparticles. This mixture is then pumped through a segment of tubing at varying flow rates. A trapping region is established by placing the tubing in close proximity of a cone magnet and the center of this trapping region is probed with our custom Raman spectroscopy system. This system provides spectrographic data of the back-scatter from a single interrogation point where it is expected that concentration of trapped CTCs would increase. This spectrum is collected over time and, using a linear-unmixing algorithm, is decomposed into the signal contribution of its constituent SERS nanoparticle signatures, which in turn allows for the calculation of the concentration of each SERS nanoparticle variant. Assuming all nanoparticles exhibit comparable levels of non-specific uptake by phagocytic cells such as CTCs and WBCs and comparable levels of random aggregation to RBCs and platelets, we expect the signal from the targeted SERS nanoparticle variant to increase at a differing rate from the accumulation of untargeted SERS nanoparticle signal if appropriately displaying CTCs are present. As such, the signal increase from untargeted nanoparticles acts as a baseline for non-specific binding/uptake allowing us to more accurately detect the presence of CTCs.
2.2. Cell lines
The HeLa cell line (CCL-2, ATCC) was used for folate receptor targeting experiments as they have previously been shown to overexpress the FOLR1 folate receptor [12]. PC3 (CRL-1435, ATCC) and LNCaP (CRL-1740,ATCC) which were previously transduced using lentiviral particles to express palmitoylated green fluorescent protein (PALM-GFP) were used for prostate-specific membrane antigen (PSMA) experiments as they are both prostate derived cell lines. Transduction for PALM-GFP expression was required for validation experiments with fluorescence microscopy and these specific cell lines were chosen as LNCaP cells are known to overexpress PSMA whereas PC3 cells do not [38]. HeLa and PC3 cells were initially grown to 80% confluency in high glucose DMEM and LNCaP cells in RPMI 1640. For folate targeting experiments, the growth media was aspirated, and the cells were washed once with phosphate buffered saline (PBS). The cells were then incubated in a folate-free RPMI 1640 medium (27016021, ThermoFisher) for 6 hours prior to all experiments to eliminate the effect of free folate in the growth media as a confounding variable. All growth media used was supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin. All cell lines were within 3 – 10 passages for all experiments.
2.3. Synthesis of the thiol reactive PSMA inhibitor
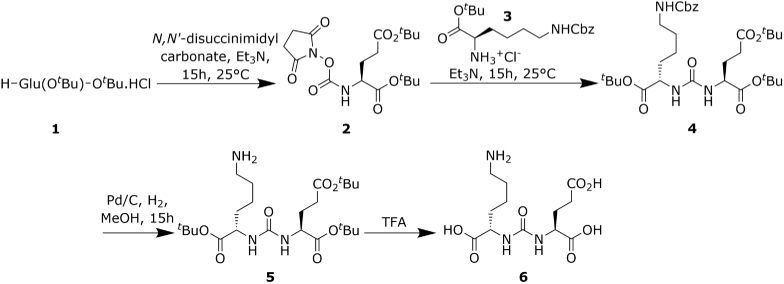
PSMA targeting was achieved using an analogue of the lysine-urea-glutamate peptidomimetic small-molecule PSMA inhibitor, DCFPyL. In this study we will refer to this amine reactive analogue as iPSMA to differentiate it from its fluorinated counterpart. This compound was synthesized as outlined in our previous work [39] and the reaction scheme is summarized in Fig. 1. In summary, first a N-succinimidyl ester of Compound 1, Glu(OtBu)-OtBu HCl, solubilized in acetonitrile was synthesized by the addition of N,-disuccinimidyl carbonate in the presence of triethylamine to produce Compound 2, (S)-Di-tert.-butyl-(((2,5-dioxopyrrolidin-1-yl)oxy)carbonyl)-l-glutamate. Compound 2 was further reacted with Compound 3, µs-benzyloxycarbonyl-l-lysine, in the presence of triethylamine. Deprotection of the benzyl chloroformate capped amine on the resultant Compound 4, (S)-2-[3-((S)-1-tert.-Butylcarboxylate-(5-benzyloxycarbonylpentyl)) ureido]-di-tert.-butyl pentanedioate, was then performed in the presence of a palladium-on-carbon catalyst in a atmosphere. Finally, Compound 6, the reactive amine PSMA inhibitor, was produced by removal of the tert-butyloxycarbonyl protecting groups using trifluoroacetic acid. TLC and 1H-NMR was performed for each intermediate and the final product.iPSMA-PEG3400-maleimide was then synthesized by mixing maliemide-PEG3400-NHS (PG2-MLNS-3k, Nanocs Inc.) with the reactive amine presenting iPSMA in a 2:11 ratio in a 4:1 DMSO:PBS buffer maintained at a pH of 7.4. The reaction proceeded to completion for 1.5 hours at room temperature. The reaction mixture was then buffer exchanged by ultracentrifugation in spin filtration tubes (14-558-443, Fisher Scientific) three times to remove DMSO and any non-coupled iPSMA, and to buffer exchange the final compound into PBS at a pH of 6.8. Identity of the maleimide-PEG linker-iPSMA compound was confirmed by NMR prior to functionalization of nanoparticles.
Fig. 1.
PSMA inhibitor synthesis reaction scheme.
2.4. Preparation of functionalized magnetic and SERS nanoparticles
Thiolated magnetic nanoparticles with a mean diameter of 0.5µm (2108-1, chemicell) were utilized for magnetic trapping. The particles consisted of a superparamagnetic maghemite core, coated with an inner layer of silicon dioxide and an outer layer of silicon dioxide doped with the fluorophore Nile Blue A (Ex/Em: 633nm/672nm). For detection of trapping events, two variants of thiolated 120-nm Surface-Enhanced Raman scattering nanoparticles (S440 and S421, Oxonica Materials) were utilized. The particles consisted of a 60-nm gold sphere with a Raman active molecular monolayer adsorbed on its surface and further coated with a 30-nm thick silica shell. The two variants of SERS-NPs used in this study were S440 (trans-1,2-Bis(4-pyridyl)-ethylene reporter) and S421 (d8-4,-dipyridyl reporter), the Raman spectra of which are shown in Fig. 2(D). All purchased nanoparticles were thiolated by the respective manufacturers. To compensate for variable number of thiols on each batch of purchased nanoparticle, available thiol groups present per particle type was assessed via microflow cytometry and a BODIPY depletion assay as described in Section 2.5.
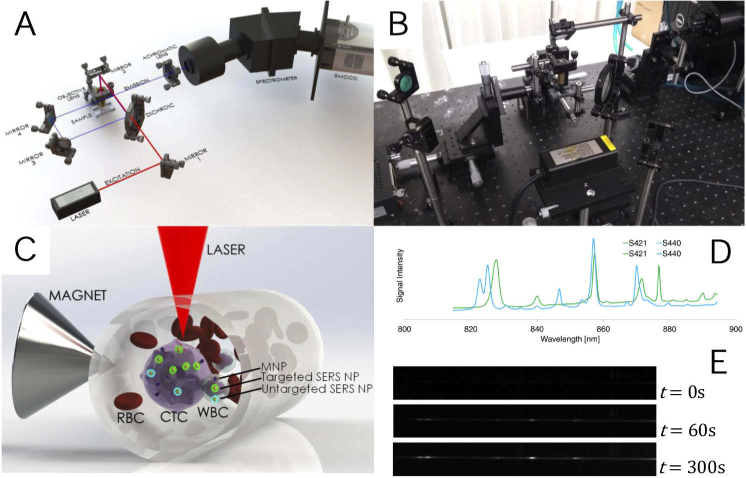
Fig. 2.
Schematic (A) and photograph (B) of Raman spectrometry system, diagram of trapping and detection scheme (C), SERS signatures of the nanoparticle variants used (D) and example output of spectrometer during trapping (E).
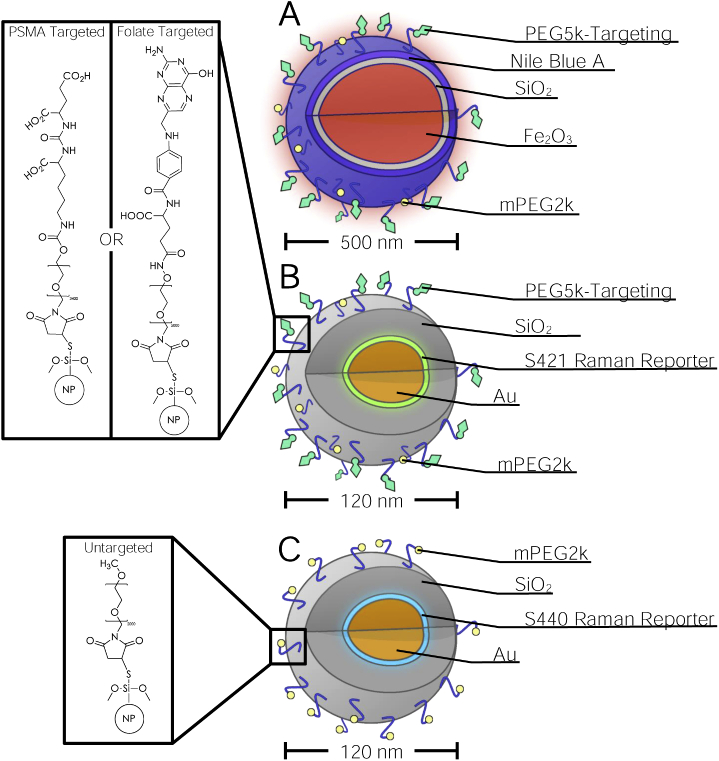
To functionalize the nanoparticles to target either folate receptors or PSMA, either folate or our peptidomimetic PSMA inhibitor was conjugated via a strong thiol-maleimide mediated linkage. First, folate-PEG5000-maleimide (PG2-FAML-5k, Nanocs) or iPSMA-PEG3400-maleimide and mPEG2000-maleimide (JKA3124, Sigma-Aldrich) were solubilized in PBS at a 1:5 molar ratio. The solution of PEG-linkers were then mixed with thiolated MNPs at 5:1 maleimide:thiol molar excess. The suspension was then thoroughly vortexed for 5 minutes and sonicated in a 37 C water bath for 15 minutes to remove aggregation of nanoparticles. Although the maleimide-thiol reaction occurs rapidly, the suspension was left overnight agitating on a rocker at room temperature to allow the reaction to reach completion and ensure maximum conjugation. The nanoparticles were then washed by pelleting the nanoparticles using centrifugation for 7 minutes at g followed by aspiration of the supernatant and resuspension in an equivalent amount of degassed PBS. This washing procedure was performed 3 times to remove all unreacted PEG linkers. The entire procedure outlined above was performed a second time to ensure maximal functionalization and lastly a final run was performed with only mPEG2000-maleimide to ensure the capping of any remaining thiols. The nanoparticles were finally pelleted down by centrifugation at and resuspended in PBS. Functionalized S421 SERS-NPs were prepared in the same manner as MNPs. Capped, non-targeted S440 SERS-NPs were also produced using the above procedure solely using mPEG2000-maleimide in all reaction steps. Schematic depictions of the nanoparticles are shown in Fig. 3.
Fig. 3.
Diagrams of targeted magnetic nanoparticles (A), targeted SERS nanoparticles (B) and untargeted SERS nanoparticles (C). Chemical structures of PSMA targeted-, folate targeted- and untargeted-nanoparticles are presented in the insets.
2.5. Nanoparticle characterization
2.5.1. SERS-NP characterization by microflow cytometry
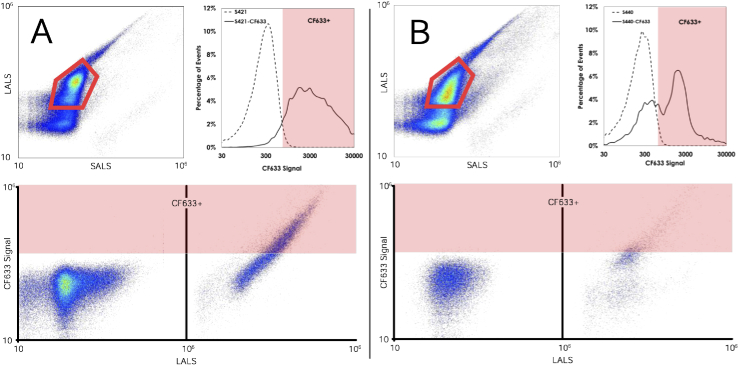
As the SERS nanoparticles purchased were subject to batch dependant variations, we employed micro-flow cytometry as a means of semi-quantitatively assessing available thiol groups for conjugation to a PEGylated folate. CF 633-maleimide dye (SCJ4600038, Sigma-Aldrich) was combined with each of the SERS-NP variants in a 1000 fluorophore per nanoparticle concentration. To ensure maximal binding of CF 633 to the nanoparticles, the suspension was vortexed for 5 minutes, sonicated for 15 minutes and left to agitate on a rocker overnight at room temperature. The nanoparticles were then separated from unbound fluorophore by 6 rounds of wash-centrifugation (, 7 min each). The final pellet was resuspended in PBS to contain a final concentration of particles/mL. This suspension was analyzed on an Apogee A50 micro-flow cytometer. Large and small angle light scatter data at 405nm along with fluorescence intensity under a 638nm excitation source were collected as summarized in Figs. 4. Gating for fluorescence intensity above an observed auto-fluorescence threshold for the undyed nanoparticles was performed to quantify the number of CF 633 positive events. All flow experiments were repeated on 5 separate aliquots of each nanoparticle. This analysis allowed us to identify that the S421 variant of SERS-NP exhibited greater amounts of available thiol groups compared to the S440 variant as on average, 88% of S421 SERS-NPs conjugated to the CF 633 above the detection threshold compared to only 35% for S440 (). As such, the S421 variant was functionalized with folate and the remaining thiol groups on the S440 were capped with DPSE-PEG5000-malemide and were used to assess non-specific uptake.
Fig. 4.
Representative Flow Cytometry Analysis of CF 633-maleimide loading on S421 (A) and S440 (B) SERS NPs.
2.5.2. BODIPY depletion assay for reactive thiol quantification
A spectrometric assay was employed to better quantify the number of thiols present on the nanoparticles which requires a much smaller number of nanoparticles than more traditional methods of thiol quantification such as Ellman’s reagent assays. Thiol-loading of the nanoparticles was determined measuring the depletion of BODIPY FL tagged maleimide present in solution due to the sequestration of the fluorophore. Firstly, BODIPY FL-maleimide (B10250, ThermoFisher) was dissolved in 20% dimethylformamide in ddH2O, and a standard curve was produced for concentrations between 0µM and 400µM. In three separate vials, 300µL of S421 (400µg dry mass), 300µL of S440 (300µg dry mass) and 200µL of screenMAG-Thiol/R nanoparticles (1000µg dry mass) were pelleted and washed with ddH2O. Next, 200µL of 100µM BODIPY FL-Maleimide solution was added to each nanoparticle suspension. The nanoparticles were allowed to thoroughly mix with BODIPY FL maleimide by vortexing and sonication for a minimum of 15 minutes each and allowed to react over agitation on a rocker for 2 hours. The nanoparticles were then pelleted by centrifugation at for 7 minutes and 100µL of nanoparticle-free supernatant was measured spectrophotometrically at 490nm. The interpolated concentrations of BODIPY FL in the supernatant were then subtracted from the 100µM value to measure the loss due to maleimide-thiol binding onto the nanoparticles. From this assay, we ascertained that S440 contained 29.0µmol thiol per gram of nanoparticles, S421 contained 40.0µmol/g, and the magnetic nanoparticles contained 3.0µmol/g. These results further support the use of the S421 variant for folate receptor targeting as they express greater number of thiols available for functionalization.
2.5.3. Nanoparticle morphology characterization by electron microscopy and energy dispersive X-ray spectroscopy
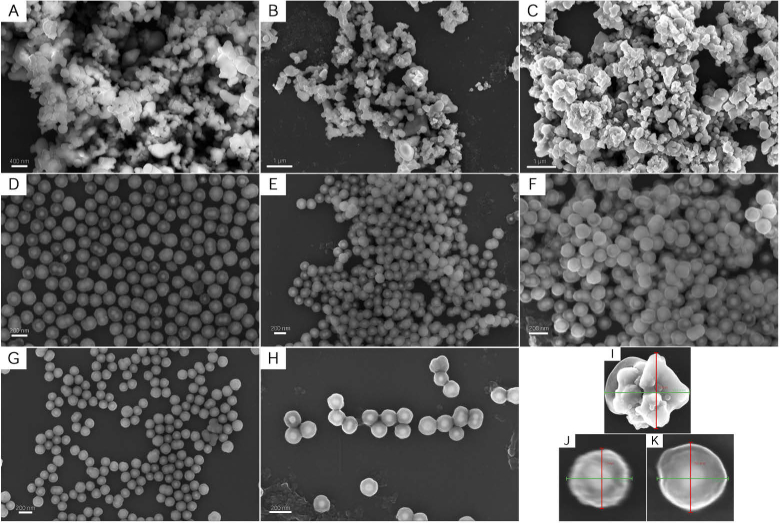
All nanoparticle suspensions used were imaged using field-emission scanning electron microscopy (FESEM) as shown in Fig. 5. Samples were prepared for imaging by spin-coating a 1-cm1-cm silicon wafer substrate with each of the nanoparticle suspensions. After the coated wafers were allowed to dry, the samples were subsequently sputter-coated with a 4nm carbon layer to improve conductivity. Samples were imaged in a Zeiss Sigma FESEM utilizing 5 kV of electron high tension voltage. As seen in Figs. 5(A)-C, the addition of varying functionalized groups on the surface of the magnetic nanoparticles had limited effect on the morphology nor size of the particle. This is also observed with the S421 SERS nanoparticles in Figs. 5(D)-F and the S440 SERS nanoparticles in Figs. 5(G) and (H). The lack of change in morphology of the SERS nanoparticles with varying functionalizations is preferable as this consistency ensures optical properties remain constant despite varying surface functionalization. Furthermore, representative small feild of view images of individual reactive thiol functionalized nanoparticles with sizing information is presented in Figs. 5(I)-K.
Fig. 5.
FESEM images of all nanoparticle formulations: magnetic nanoparticles with (A) reactive thiols, (B) folate, and (C) PSMA inhibitor functionalizations; S421 SERS nanoparticles with (D) reactive thiols, (E) folate, and (F) PSMA inhibitor functionalizations; S440 SERS nanoparticles with (G) reactive thiols, and (H) methoxy group capped functionalizations. Representative close-up images with sizing information of a reactive thiol functionalized (I) magnetic, (J) S421 SERS, and (K) S440 SERS nanoparticle.
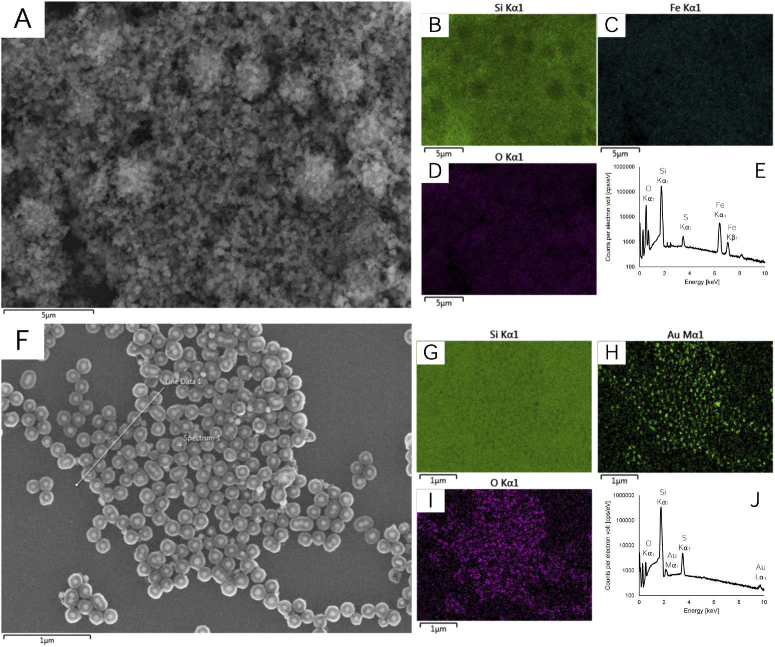
Furthermore, electron disperive X-ray spectroscopy (EDX) was perfromed to provide compositional information as shown in Fig. 6. Figures 6(A)-D show a representative region of reactive thiol functionalized magnetic nanoparticles and Fig. 6(E) presents the elemental mapping information; notable peaks are observed correspondig to the presence of an iron core, silica coating and thiol surface functionalization. Figures 6(F)-I show a representative region of reactive thiol functionalized S421 SERS nanoparticles and Fig. 6(J) presents the elemental mapping information; notable peaks are observed corresponding to the presence of a gold core, silica coating and thiol surface functionalization.
Fig. 6.
EDX analysis of reactive thiol functionalized (A-E) magnetic nanoparticles and (F-J) S421 SERS nanoparticles.
2.5.4. Hydrodynamic radius measurement by nanoparticle tracking analysis
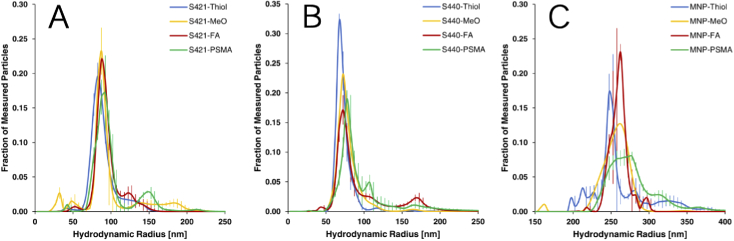
Nanoparticle Tracking Analysis was employed to measure the hydrodynamic radii of all populations of produced nanoparticles. These measurements were performed on a NanoSight LM10 (Malvern) utilizing a 45-mW, 405-nm light source and a sCMOS camera which was calibrated using a 100-nm polystyrene size standards prior to experimental measurements. To avoid senor over-saturation, the nanoparticle formulations were diluted in PBS to attain 20-80 particles per frame. For each nanoparticle formulation, three 60-second videos were captured at a camera level of 8. Temperature was maintained between 25 C and 27 C and the temperature value used in analysis was adjusted accordingly. After capture, the videos were analyzed using the NanoSight Software NTA 3.3 with a detection threshold of 4. The results of hydrodynamic radii measurement is presented in Fig. 7 and statistically summarized in Table 1. Notably, particle species containing reactive thiols (ie. prior to fuctionalization with PEG linkers) consistently had lower hydrodynamic radii to their fuctionalized counterparts. This is to be expected as the addition of long chain PEG linkers is expected to increase the hydrodynamic radius and gives further credence to the effective functionalization of nanoparticle formulations. All nanoparticle formulations were also fairly monodisperse with only a single primary population peak.
Fig. 7.
Nanoparticle Tracking Analysis measurements of (A) S421 SERS, (B) S440 SERS and (C) magnetic nanoparticle species with either reactive thiol, methoxy group capped, folate or PSMA inhibitor functionalizations.
Table 1. Statistical summary of hydrodynamic radii measurements of all nanoparticle formulations.
| Functionalization | S421 | S440 | MNP | |||
|---|---|---|---|---|---|---|
| mean [nm] | SD [nm] | mean [nm] | SD [nm] | mean [nm] | SD [nm] | |
| Reactive thiol (-Thiol) | ||||||
| Methoxy capped (-MeO) | ||||||
| Folate (-FA) | ||||||
| PSMA inhibitor (-iPSMA) | ||||||
2.5.5. Zeta potential
Zeta potential measurements of the nanoparticle formulations dispersed in phosphate buffered saline were measured using a phase analysis light scattering-based analyzer (ZetaPALS, Brookhaven) and summarized in Table 2. Utilizing 30mV as a cut-off for stability in accordance with the literature, the original thiolated nanoparticle formulations exist within the stable regime and would exhibit limited flocculation. The addition of a surface coating for methoxy capped, folate functionalized and PSMA inhibitor functionalized formulations is verified by the significant reduction in the magnitude of zeta potentials. It is also noteable that the change in zeta potential is far greater in the formulations functionalized with folate or PSMA inhibitor as those are bound using longer PEG-chain linkers. These changes in zeta potential are expected however and do not reduce formulation stability as the addition of a PEG layer induces the formation of a hydration shell that imbues sterical stability. Furthermore, the reduction of zeta potential magnitude is preferable for biocompatibility as recognition by the immune and complement system is suppressed [40].
Table 2. Statistical summary of hydrodynamic radii measurements of all nanoparticle formulations.
| Functionalization | S421 | S440 | NP | |||
|---|---|---|---|---|---|---|
| mean [mV] | SD [mV] | mean [mV] | SD [mV] | mean [mV] | SD [mV] | |
| Reactive thiol (-Thiol) | −37.5 | 3.7 | −32.6 | 5.9 | −45.3 | 7.2 |
| Methoxy capped (-MeO) | −7.3 | 5.1 | −10.4 | 5.8 | −22.1 | 11.7 |
| Folate (-FA) | −1.2 | 0.5 | −2.4 | 0.8 | −9.8 | 5.3 |
| PSMA inhibitor (-iPSMA) | −3.2 | 0.4 | −4.5 | 0.9 | −9.7 | 1.7 |
2.5.6. UV-Vis absorption spectroscopy
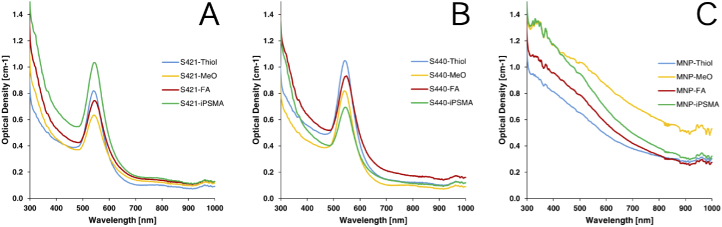
UV-Vis absorption spectroscopy measurements of all nanoparticle formulations is presented in Fig. 8. These measurements were performed using a plate spectrometer (FLUOstar Omega, BMG Labtech). As shown in Fig. 8(A) and 8(B), in all S421 and S440 SERS nanoparticle formulations, an absorption peak is noted at approximately 545 nm which corresponds to the characteristic localized surface plasmon resonance peak of gold nanoparticles in the 100-nm diameter size range. However, to prevent dampening of the SERS effect due to the interband transition of gold [41], we utilized the supplier suggested exciation wavelength at 785-nm for Raman spectroscopy experiments. No such absorption peak was noted for the magnetic nanoparticle formulations as shown in Fig. 8(C) suggesting these particles would not interfere with Raman Spectroscopy. Furthermore, it is also worth noting the lack of appreciable change in the absorption peaks for varying functionalizations which suggests the surface functionalizations used for targeting would not have an appreciable effect on the nanoparticle optical properties.
Fig. 8.
UV-Vis absoprtion spectroscopy measurements of (A) S421 SERS, (B) S440 SERS and (C) magnetic nanoparticle species with either reactive thiol, methoxy group capped, folate or PSMA inhibitor functionalizations.
2.6. Fluorescence microscopy for targeting validation
Cells were grown on coverslips with the same growth parameters as earlier stated and incubated with MNPs/mL for 30 minutes. The cells were then washed 3 times in PBS and fixed in 4% paraformaldehyde. Hoechst 33342 (14533, Sigma-Aldrich) was used to stain the nucleus and HeLa cell membranes were stained with Alexa Fluor 488, wheat germ agglutin (W11261, FisherScientific). Imaging was performed on a Nikon A1r MP microscope equipped with a 63x/1.40 Oil objective.
2.7. Magnetic trapping and raman imaging system
Magnetic trapping was achieved using a 1″ cone magnet (Cone0100N, SuperMagnetMan) with the tip pointed towards a 0.015″ ID polyurethane tube (PU-033, SAI Infusion Technologies) positioned at the focal point of the optical system. Nanoparticles were added to 1mL of heparinized whole rat blood and mixed for 5 minutes on a rocker at room temperature. The blood suspensions were then loaded into 1mL syringes (309659, Becton-Dickinson) connected to 27G × 1/2″ hypodermic needles (305109, Becton-Dickinson) and were pumped through the tube by a syringe pump (NE-300, New Era Pump Systems). An illustration of the optical system is shown in Fig. 2(A). The excitation source used for imaging was a 785nm Raman laser (FB-785-350-FM-FS-1-1, RGBLase) which provides a maximum output of 350mW with a 20-dB spectral linewidth of 0.4 nm and side lobe suppression ratio >40dB. An aspheric lens (A240TM-B, Thorlabs) with a 0.5 numerical aperture and 8-mm focal length was used to focus both the excitation beam and resultant backscatter. Collimated backscattered light from the sample was redirected towards the input aperture of the imaging spectrometer using an 805nm cut-off dichroic mirror (DMSP805L, Thorlabs) and focused via a 0.3 numerical aperture lens (AC127-019-B-ML, Thorlabs). Optical alignment of both the excitation beam and backscatter was controlled by adjusting four 750 – 1100nm broadband dielectric mirrors (BB1-E03, Thorlabs). Raman spectra were captured using a custom Raman imaging spectrometer (RASPPEC-785-HR, P&P Optica) which offers a spectral resolution of 10 with an f-number of 3.5. Backscattered light entered the spectrometer through an input aperture slit size of 15µm 26mm and was collimated through a gel grating (900 L/cm, = 815 to 895nm), passed through a 21 bend and refocused onto a 1600-pixel 400-pixel front-illuminated EMCCD array (DU971NUV, Andor Technology). Pixel elements were 16 μm × 16 μm and only 5-pixel vertical spatial channels were used for point illumination and collection in this study. In future studies, multiple points could be spectrally imaged simultaneously on unused channels. Algorithms to correct for spherical aberrations on the camera sensor were provided by the spectrometer manufacturer. All imaging was performed with an EM gain of 10 over a 3 second integration time. To minimize electronic noise, the system was cooled to −55 C prior to imaging. Spectral unmixing was preformed in MATLAB using a linear least-squares method with a positivity constraint as per the following equation: , in which contains individual component spectra, is the measured spectra at time , and is the unknown component concentrations. The spectral signatures from the following components were spectrally unmixed: S421 SERS-NPs, S440 SERS-NPs, MNPs, whole rat blood, HeLa cells in PBS, polyurethane tubing and noise expected from ambient light. The spectral signature of each component was re-acquired immediately prior to each experimental run and compared to maintain consistency.
2.8. Trapping experiments
Trapping experiments were performed at an internal tube flow rate of 10mm/s (1.14µL/s) inside the tubing unless otherwise stated. Cells were counted using a hemocytometer and diluted into whole rat blood at HeLa cells/mL blood. MNPs and both variants of SERS-NPs were added such that the blood suspension had a final concentration of MNPs/mL and SERS nanoparticles/mL. For blocking studies, folate was added to the blood suspensions achieve a 50µM concentration. All samples were subsequently agitated on a rocker for 20 minutes at room temperature prior to flow-trapping experiments. All flow-trapping experiments were performed on 1 mL total of blood-model CTC-NP mixtures flowed over approximately 15 minutes at room temperature and the resultant magnetically trapped bolus was imaged and unmixed to calculate the contribution of each SERS-NP variant to the detected signal. All imaging was performed under an excitation of 60mW at the focal point. Trapping was performed for each experimental condition in a minimum of 5 trials and the results were compared using two-tailed -tests assuming unequal variance (, ).
3. Results
3.1. Assessment of non-specific uptake of thiolated nanoparticles
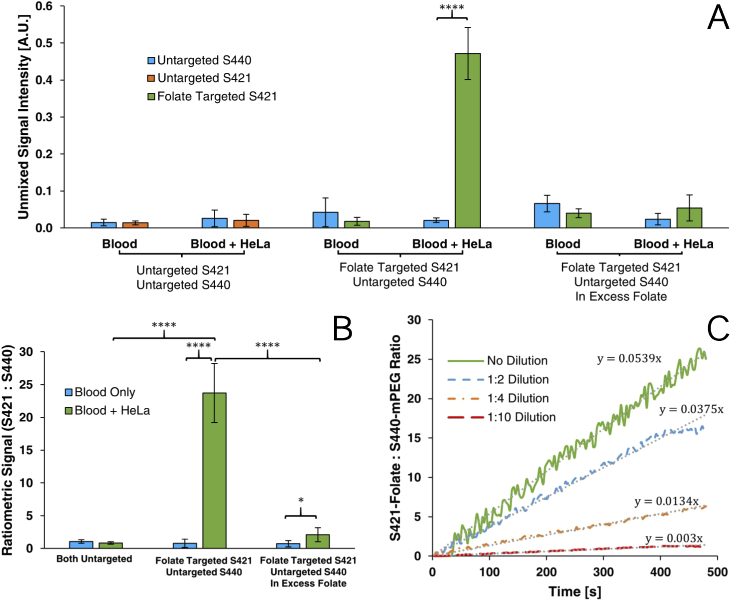
We first established that both variants of SERS-NPs exhibited equivalent non-specific uptake. This was performed by incubating HeLa cells with folate-MNPs, thiol-S440-NPs and thiol-S421-NPs. Nanoparticles were also added to whole blood lacking tumor cells to assess uptake by phagocytic cells. The non-specific uptake of both variants of thiolated SERS nanoparticles by both HeLa and blood cells was assessed and the results are shown in the first four datasets in Fig. 9(A). As expected, the uptake of both untargeted SERS-NP variants was within error of each other in both, blood alone and blood containing HeLa cells. Ratiometrically comparing the uptake of untargeted S421-NPs versus untargeted S440-NPs, HeLa cells did not show a significant increase in preferential uptake compared to blood () and was within error of unity as shown in Fig. 9(B). The observation of a signal due to non-specific uptake validates the need for a method that discriminates between specific and non-specific trapping events.
Fig. 9.
Unmixed SERS-NP signal intensities (A), ratiometric analysis (B) of folate targeting experiments and time resolved trapping curves (C) of ratiometric SERS-NP signal at various model CTC dilutions.
3.2. Assessment of specific uptake of folate targeted nanoparticles
Flow trapping experiments were performed using the folate receptor targeted S421 and untargeted S440 nanoparticles to demonstrate the enhanced specificity afforded by using spectrally distinct variants of SERS-NPs. As shown in Fig. 9(A), suspensions containing HeLa cells had 22.7 times greater uptake of folate conjugated S421 than untargeted S440 () whereas the comparative uptake in only blood was not significantly different ().
Comparing the ratiometric analysis in Fig. 9(B), the disparity in uptake by HeLa cells was significantly higher than in only blood (). Furthermore, there is a significant increase in ratiometric uptake by HeLa containing samples with one targeted variant compared to the uptake seen when both variants of SERS-NPs were left untargeted (). These results confidently demonstrate the efficacy of targeting as the increase in folate-S421-NP signal is due to the binding of both folate receptor targeting MNPs and S421s bound to HeLa cells. Ratiometric detection here is shown to provide an effective discriminatory measure to ensure specificity of detection.
3.3. Blocking study and microscopy for validation of folate targeting mechanism
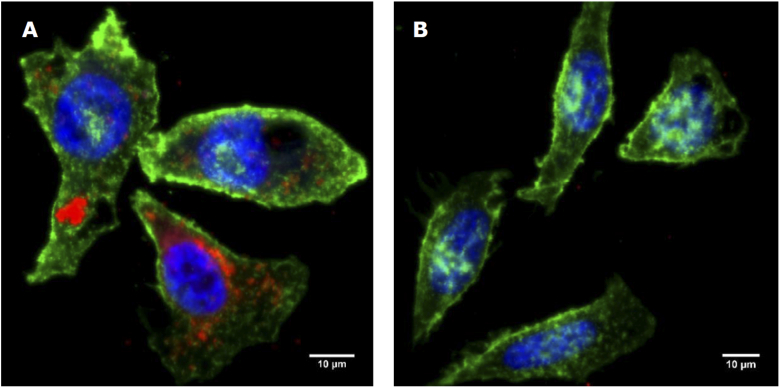
To further validate the folate-mediated mechanism of targeting, a blocking study was performed by introducing an excess of free folate to saturate folate receptors prior to nanoparticle loading. Blocking was performed by adding 50µM of free folate to the blood/HeLa suspension prior to incubation with nanoparticles. As shown in Fig. 9(A), in either blood suspensions with or without HeLa cells, there was no significant difference in the calculated uptake of S421-folate versus S440-mPEG ( and respectively) after blocking. When compared ratiometrically, there was a significant difference in the ratiometric uptake of particles between blood suspensions with or without HeLa cells (). This implies that 50µM of free folate may not have been enough to fully block targeting. However, as shown in Fig. 9(B), there is a marked decrease in ratiometric uptake in HeLa cell suspensions after blocking () which does not occur in blood alone verifying that folate receptor targeted nanoparticles indeed experience preferential uptake by HeLa cells. Furthermore, fluorescence microscopy was used to validate folate mediated targeting. As apparent in Fig. 10, blocking in excess folate resulted in lower uptake of fluorescent folate conjugated magnetic nanoparticles.
Fig. 10.
Maximum intensity projection of fluorescence microscopy images of HeLa cells incubated with folate conjugated fluorescent magnetic nanoparticles(red) without(A) and with(B) blocking in excess folate. Counterstained with Alexa Fluor 488, WGA membrane stain(green) and Hoechst 33342 nuclear stain(blue).
3.4. Assessment of trapping and detection sensitivity in serial dilutions
To validate the detection of discrete trapping events and the capacity to detect increasingly sparse concentrations of circulating tumor cells, serial dilutions of HeLa cells in blood were used, the results of which are shown in Fig. 9(C). The same quantities of each SERS-NP variant and MNPs as prior experiments were incubated with blood containing , , and HeLa cells/mL. For all endpoint experiments, serial dilutions were flowed through the tubing at 15mm/s (1.71µL/s) and the Raman signal was measured over 480 seconds. After spectral unmixing, the ratiometric S421-folate/S440-mPEG signal increased over time as greater trapping events occurred with a slower rate of increase with each subsequent dilution. Furthermore, using linear regression, it is observed that rate of valid trapping events observed approximately corresponds with the serial dilution used (i.e. 1:4 dilution resulted in around rate of trapping). We have previously observed that approximately 300 SERS-NPs result in a unitary signal-to-noise ratio of 123 [42]. Given that tumor cells may have anywhere from to folate-receptor binding sites [43], this suggests that near single-cell detection capabilities are possible and our ratiometric approach would be a powerful method of identifying trapped CTCs.
3.5. Assessment of specific uptake of PSMA targeted nanoparticles
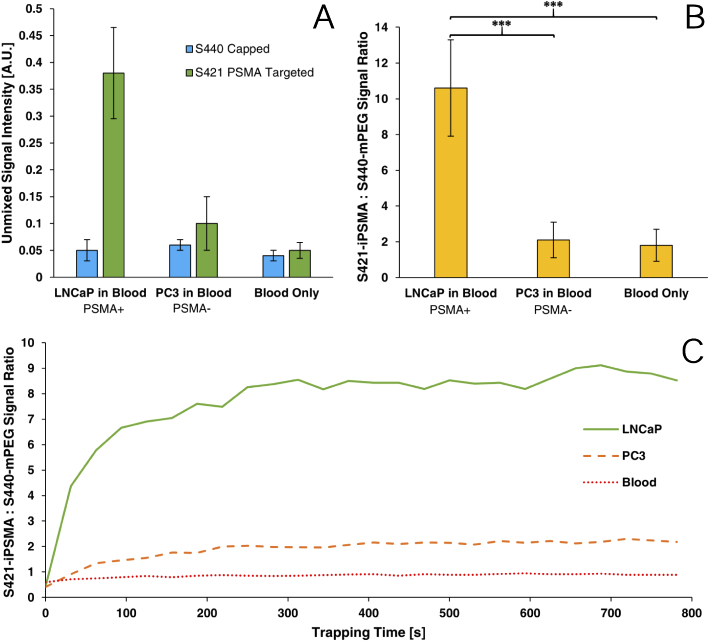
To further expand this methodology for use in prostatic cancers, flow trapping experiments were performed using the PSMA targeted S421 and untargeted S440 nanoparticles. Trapping experiments were performed with PSMA(+) LNCaP model CTCs and PSMA(-) PC3 model CTCs and as shown in Fig. 11, samples containing LNCaP derived model CTCs, on average, resulted in 5 times greater ratiometric signal compared to PC3 cells or blood alone. The ratiometric analysis shows that there is a significantly higher signal from LNCaP cells than PC3 cells () and in only blood (). These results confidently demonstrate the efficacy of PSMA targeting as the increase in iPSMA-S421-NP signal is due to the binding of both PSMA targeting MNPs and S421s bound to LNCaP cells.
Fig. 11.
Unmixed SERS-NP signal intensities (A), ratiometric analysis (B) and representative time resolved trapping curves (C) of PSMA targeting experiments..
Trapping curves over time are shown in Fig. 11(C). The ratiometric signal is observed to be near unity for PC3 and blood-born cells lines however LNCaP model CTC suspensions show a dramatic rise in signal during the first 5 minutes. Although individual SERS signals continue to increase throughout the entirety of the trapping experiment, the ratiometric signal reaches a plateau at 5 minutes as the further trapping of additional LNCaP cells does not change the detected ratio of targeted to untargeted SERS particles.
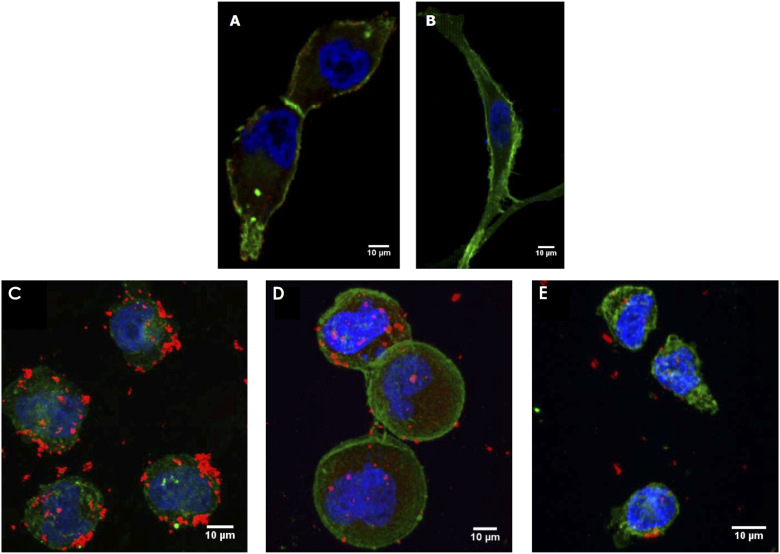
Furthermore, fluorescence microscopy was performed to validate the PSMA-mediate mechanism of targeting. First, a conventional fluorescent antibody was used to verify that the LNCaP and PC3 cell lines used in this experiment were PSMA positive and negative respectively as shown in Fig. 12(A) and (B). These images were then compared to microscopy performed with LNCaP and PC3 cells incubated in our iPSMA targeted fluorescent magnetic nanoparticles. As shown in Fig. 12(C) and (E), the attachment of MNPs coincides with the presence of PSMA expression in the cell line. Some targeted nanoparticles however still attach to the PSMA- PC3s and this can be attributed to the large size of the MNPs compared to antibodies which promotes non-specific uptake through means such as phagocytosis. This further demonstrates the need for our ratiometric approach to help mitigate the presence of non-specific uptake which will always be present for nanoparticle-based assays. A blocking experiment was also performed in Fig. 12(D) using excess free PSMA inhibitor which resulted in lower targeted MNP attachment. Lastly, to better illustrate magnetic trapping, LNCaP-PALM-GFP cells incubated with iPSMA-targeted magnetic nanoparticles were flowed through a tube phantom in contact with a cone magnet and trapping was imaged using a low magnification objective fluorescence microscope. The results of this demonstration are shown in Visualization 1 (81MB, mov) , wherein we can visually observe an accumulation of fluorescent cells within the magnetic trapping region over time.
Fig. 12.
Maximum intensity projection of fluorescence microscopy images of LNCaP-PALM-GFP (A) and PC3-PALM-GFP (B) cells stained with anti-PSMA-Cy5 (red) as well as LNaP-PALM-GFP without (C) and with (D) blocking in excess free PSMA inhibitor and PC3-PALM-GFP (E) cells incubated with PSMA inhibitor conjugated fluorescent magnetic nanoparticles(red). Counterstained and Hoechst 33342 nuclear stain(blue) and endogenously expressing PALM-GFP(green) on the membrane.
4. Discussion
In this study, we were able to use the highly unique Raman spectra of two variants of SERS nanoparticles to ratiometrically discriminate the uptake due to targeting versus non-specific mechanisms. Increased SERS signal from targeted nanoparticles was observed and the mechanism of targeting was verified by a blocking study. The ratiometric signal was also observed to increase with greater trapping times and the rate of signal increase corresponded to the concentration of model CTCs present. This work demonstrates the feasibility of targeted SERS-based multiplex detection and enrichment of model CTCs in an ex vivo model and offers several advantages: (1) the Raman scattering signature of each Raman reporter on the SERS nanoparticles is highly unique allowing for extremely effective multiplexing and can be detected with high sensitivity unlike traditional fluorophores which suffer from spectral overlap and low signal intensities. (2) Detectable Raman signals are only produced by SERS nanoparticles allowing for a higher signal to noise ratio unlike fluorescence-based approaches which suffer from auto-fluorescence background. (3) folate receptor and PSMA targeting allows us to maintain targeting efficacy past epithelial-to-mesenchymal transitions. (4) Targeting using small molecule compounds is a more consistent and readily available targeting moiety compared to antibodies. (5) Versatile thiol-maleimide mediated nanoparticle functionalization enables the use of a large variety of compounds for functionalization past folate receptor and PSMA targeting. (6) The ratiometric approach effectively normalizes the observed signal from the targeted nanoparticle against background non-specific uptake which acts as a built-in standard to account for injection-to-injection variations.
Although this methodology shows promise, we do recognize several hurdles that would need to be dealt with to achieve in vivo utility. Most notable would be the need to gain FDA-approval for in vivo nanoparticle use [44]. Although they are made of relatively inert, biocompatible materials such as gold and silica and PEGylation greatly reduces cytotoxicity, the systemic toxicity and clearance rates of such particle constructs would need to be properly established. We have however demonstrated one approach to reduce cytotoxicity by utilizing low toxicity thiol-maleimide mediated functionalization. Compared to the commonly used biotin-streptavidin functionalization methods, thiol-maleimide exhibits far lesser toxicity while still providing a versatile functionalization platform to conjugate various biomarker targeting agents. Further experimentation could also be performed with other bio-orthogonal and click-chemistry procedures to further increase in vivo applicability.
Furthermore, clearance rates of these nanoparticle constructs in the bloodstream would also need to be assessed and larger dosages may be required to offset short clearance times. Varying levels and lengths of PEGylation again have shown to mitigate this to some extent as it masks these agents from the immune system and decreases renal clearance rates [40]
Even with ex vivo blood samples, our approach could prove useful for rapid assessment of the presence of CTCs with high specificity, which, if present, would help guide the decision to do more costly biomarker or single cell assays, and to do these with enriched cell populations.
Our previous work has determined a sensitivity of approximately 100 SERS-NPs [37] and we expect that we would be able to detect CTCs with near single cell capabilities as each CTC could bind to nearly 20,000 nanoparticles. Therefore, only a single cell would need to be trapped in order to detect an increased signal. Additionally, by utilizing more spatial channels on our spectrometer, we can also expand our optical system to simultaneously image multiple spots to allow for a larger trapping region. Deeper imaging may also be possible using higher fluence and longer optical wavelengths where tissue scattering is weaker.
We have effectively leveraged the multiplexing capability of SERS nanoparticles to discriminate between specific and non-specific uptake of targeted nanoparticles. This combined with earlier demonstrations of magnetic enrichment of CTCs allows to produce a highly sensitive and specific assay for detection of CTCs that overexpress cancer specific markers such as PSMA. By discriminating between non-specific and specific trapping events, we can better quantify the number of CTCs trapped as opposed to uptake from irrelevant phagocytic cells which can allow us to determine when an adequate number of CTCs have been trapped.
Funding
SPIE (Optics and Photonics Education Scholarship); Canadian Federation of University Women10.13039/501100000151 Margaret Brine Graduate Scholarships for Women; Ministry of Advanced Education, Government of Alberta10.13039/100014407, Small Equipment Grants Program (URSI09007SEG); Canada Foundation for Innovation10.13039/501100000196 (18472); Canadian Institutes of Health Research10.13039/501100000024 (PS 153067); Canadian Cancer Society (CCS 2011-700718); Natural Sciences and Engineering Research Council of Canada10.13039/501100000038 (RGPIN-2018-05788, STPGP 396444); Prostate Cancer Canada10.13039/501100000109 ((355544-2008, 375340-2009).
Disclosures
R.J. Zemp is a founder and shareholder of Illumisonics Inc. and CliniSonix Inc., which, however, did not support this work.
References
- 1.Chaffer C. L., Weinberg R. A., “A Perspective on Cancer Cell Metastasis,” Science 331(6024), 1559–1564 (2011). 10.1126/science.1203543 [DOI] [PubMed] [Google Scholar]
- 2.Stathopoulou A., Vlachonikolis I., Mavroudis D., Perraki M., Kouroussis C., Apostolaki S., Malamos N., Kakolyris S., Kotsakis A., Xenidis N., Reppa D., Georgoulias V., “Molecular Detection of Cytokeratin-19–Positive Cells in the Peripheral Blood of Patients With Operable Breast Cancer: Evaluation of Their Prognostic Significance,” J. Clin. Oncol. 20(16), 3404–3412 (2002). 10.1200/JCO.2002.08.135 [DOI] [PubMed] [Google Scholar]
- 3.Zach O., Kasparu H., Wagner H., Krieger O., Lutz D., “Prognostic value of tumour cell detection in peripheral blood of breast cancer patients,” Acta medica Austriaca. Supplement 59, 32–34 (2002). [PubMed] [Google Scholar]
- 4.Shiomi-Mouri Y., Kousaka J., Ando T., Tetsuka R., Nakano S., Yoshida M., Fujii K., Akizuki M., Imai T., Fukutomi T., Kobayashi K., “Clinical significance of circulating tumor cells (CTCs) with respect to optimal cut-off value and tumor markers in advanced/metastatic breast cancer,” Breast Cancer 23(1), 120–127 (2016). 10.1007/s12282-014-0539-x [DOI] [PubMed] [Google Scholar]
- 5.Bredemeier M., Edimiris P., Tewes M., Mach P., Aktas B., Schellbach D., Wagner J., Kimmig R., Kasimir-Bauer S., “Establishment of a multimarker qPCR panel for the molecular characterization of circulating tumor cells in blood samples of metastatic breast cancer patients during the course of palliative treatment,” Oncotarget 7(27), 41677–41690 (2016). 10.18632/oncotarget.9528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredemeier M., Edimiris P., Mach P., Kubista M., Sjöback R., Rohlova E., Kolostova K., Hauch S., Aktas B., Tewes M., Kimmig R., Kasimir-Bauer S., “Gene Expression Signatures in Circulating Tumor Cells Correlate with Response to Therapy in Metastatic Breast Cancer,” Clin. Chem. (Washington, DC, U. S.) 63(10), 1585–1593 (2017). 10.1373/clinchem.2016.269605 [DOI] [PubMed] [Google Scholar]
- 7.Lianidou E. S., Markou A., Strati A., “Molecular characterization of circulating tumor cells in breast cancer: Challenges and promises for individualized cancer treatment,” Cancer Metastasis Rev. 31(3-4), 663–671 (2012). 10.1007/s10555-012-9366-8 [DOI] [PubMed] [Google Scholar]
- 8.Rao C., Chianese D., Doyle G., Miller M., Russell T., Sanders R., Terstappen L., “Expression of epithelial cell adhesion molecule in carcinoma cells present in blood and primary and metastatic tumors,” Int. J. Oncol. 27, 49–57 (2005). 10.3892/ijo.27.1.49 [DOI] [PubMed] [Google Scholar]
- 9.Hyun K.-A., Goo K.-B., Han H., Sohn J., Choi W., Kim S.-I., Jung H.-I., Kim Y.-S., Hyun K.-A., Goo K.-B., Han H., Sohn J., Choi W., Kim S.-I., Jung H.-I., Kim Y.-S., “Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer,” Oncotarget 7(17), 24677–24687 (2016). 10.18632/oncotarget.8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakobsen K. R., Demuth C., Sorensen B. S., Nielsen A. L., “The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer,” Transl. Lung Cancer Res. 5(2), 172–182 (2016). 10.21037/tlcr.2016.04.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balzar M., Winter M. J., de Boer C. J., Litvinov S. V., “The biology of the 17-1A antigen (Ep-CAM),” J. Mol. Med. 77(10), 699–712 (1999). 10.1007/s001099900038 [DOI] [PubMed] [Google Scholar]
- 12.Destito G., Yeh R., Rae C. S., Finn M. G., Manchester M., “Folic Acid-Mediated Targeting of Cowpea Mosaic Virus Particles to Tumor Cells,” Chem. Biol. (Oxford, U. K.) 14(10), 1152–1162 (2007). 10.1016/j.chembiol.2007.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudimack J., Lee R. J., “Targeted drug delivery via the folate receptor,” (2000). [DOI] [PubMed]
- 14.Georgakoudi I., Solban N., Novak J., Rice W. L., Wei X., Hasan T., Lin C. P., “In Vivo Flow Cytometry,” Cancer Res. 64(15), 5044–5047 (2004). 10.1158/0008-5472.CAN-04-1058 [DOI] [PubMed] [Google Scholar]
- 15.He W., Wang H., Hartmann L. C., Cheng J.-X., Low P. S., “In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry,” Proc. Natl. Acad. Sci. 104(28), 11760–11765 (2007). 10.1073/pnas.0703875104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hristozova T., Konschak R., Budach V., Tinhofer I., “A simple multicolor flow cytometry protocol for detection and molecular characterization of circulating tumor cells in epithelial cancers,” Cytometry, Part A 81A(6), 489–495 (2012). 10.1002/cyto.a.22041 [DOI] [PubMed] [Google Scholar]
- 17.Zharov V. P., Galanzha E. I., Shashkov E. V., Khlebtsov N. G., Tuchin V. V., “In vivo photoacoustic flow cytometry for monitoring of circulating single cancer cells and contrast agents,” Opt. Lett. 31(24), 3623 (2006). 10.1364/OL.31.003623 [DOI] [PubMed] [Google Scholar]
- 18.Holan S. H., Viator J. A., “Automated wavelet denoising of photoacoustic signals for circulating melanoma cell detection and burn image reconstruction,” Phys. Med. Biol. 53(12), N227 (2008). 10.1088/0031-9155/53/12/N01 [DOI] [PubMed] [Google Scholar]
- 19.Galanzha E. I., Kim J. W., Zharov V. P., “Nanotechnology-based molecular photoacoustic and photothermal flow cytometry platform for in-vivo detection and killing of circulating cancer stem cells,” J. Biophotonics 2(12), 725–735 (2009). 10.1002/jbio.200910078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galanzha E. I., Shashkov E. V., Kelly T., Kim J.-W., Yang L., Zharov V. P., “In vivo magnetic enrichment and multiplex photoacoustic detection of circulating tumour cells,” Nat. Nanotechnol. 4(12), 855–860 (2009). 10.1038/nnano.2009.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galanzha E. I., Zharov V. P., “Circulating Tumor Cell Detection and Capture by Photoacoustic Flow Cytometry in Vivo and ex Vivo,” Cancers 5(4), 1691–1738 (2013). 10.3390/cancers5041691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedosekin D. A., Juratli M. A., Sarimollaoglu M., Moore C. L., Rusch N. J., Smeltzer M. S., Zharov V. P., Galanzha E. I., “Photoacoustic and photothermal detection of circulating tumor cells, bacteria and nanoparticles in cerebrospinal fluid in vivo and ex vivo,” J. Biophotonics 6(6-7), 523–533 (2013). 10.1002/jbio.201200242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galanzha E. I., Shashkov E. V., Spring P. M., Suen J. Y., Zharov V. P., “In vivo, noninvasive, label-free detection and eradication of circulating metastatic melanoma cells using two-color photoacoustic flow cytometry with a diode laser,” Cancer Res. 69(20), 7926–7934 (2009). 10.1158/0008-5472.CAN-08-4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S., Liu K., Liu J., Yu Z. T.-F., Xu X., Zhao L., Lee T., Lee E. K., Reiss J., Lee Y.-K., Chung L. W. K., Huang J., Rettig M., Seligson D., Duraiswamy K. N., Shen C. K.-F., Tseng H.-R., “Highly Efficient Capture of Circulating Tumor Cells by Using Nanostructured Silicon Substrates with Integrated Chaotic Micromixers,” Angew. Chem., Int. Ed. 50(13), 3084–3088 (2011). 10.1002/anie.201005853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stott S. L., Hsu C.-H., Tsukrov D. I., Yu M., Miyamoto D. T., Waltman B. A., Rothenberg S. M., Shah A. M., Smas M. E., Korir G. K., Floyd F. P., Gilman A. J., Lord J. B., Winokur D., Springer S., Irimia D., Nagrath S., Sequist L. V., Lee R. J., Isselbacher K. J., Maheswaran S., Haber D. A., Toner M., “Isolation of circulating tumor cells using a microvortex-generating herringbone-chip,” Proc. Natl. Acad. Sci. 107(43), 18392–18397 (2010). 10.1073/pnas.1012539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., Toner M., “Isolation of rare circulating tumour cells in cancer patients by microchip technology,” Nature 450(7173), 1235–1239 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao M., Schiro P. G., Kuo J. S., Koehler K. M., Sabath D. E., Popov V., Feng Q., Chiu D. T., “An Automated High-Throughput Counting Method for Screening Circulating Tumor Cells in Peripheral Blood,” Anal. Chem. 85(4), 2465–2471 (2013). 10.1021/ac400193b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoshino K., Huang Y.-Y., Lane N., Huebschman M., Uhr J. W., Frenkel E. P., Zhang X., “Microchip-based immunomagnetic detection of circulating tumor cells,” Lab Chip 11(20), 3449 (2011). 10.1039/c1lc20270g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plouffe B. D., Mahalanabis M., Lewis L. H., Klapperich C. M., Murthy S. K., “Clinically Relevant Microfluidic Magnetophoretic Isolation of Rare-Cell Populations for Diagnostic and Therapeutic Monitoring Applications,” Anal. Chem. 84(3), 1336–1344 (2012). 10.1021/ac2022844 [DOI] [PubMed] [Google Scholar]
- 30.Choi S., Karp J. M., Karnik R., “Cell sorting by deterministic cell rolling,” Lab Chip 12(8), 1427 (2012). 10.1039/c2lc21225k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosokawa M., Hayata T., Fukuda Y., Arakaki A., Yoshino T., Tanaka T., Matsunaga T., “Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells,” Anal. Chem. 82(15), 6629–6635 (2010). 10.1021/ac101222x [DOI] [PubMed] [Google Scholar]
- 32.Park E. S., Jin C., Guo Q., Ang R. R., Duffy S. P., Matthews K., Azad A., Abdi H., Todenhöfer T., Bazov J., Chi K. N., Black P. C., Ma H., “Continuous Flow Deformability-Based Separation of Circulating Tumor Cells Using Microfluidic Ratchets,” Small 12(14), 1909–1919 (2016). 10.1002/smll.201503639 [DOI] [PubMed] [Google Scholar]
- 33.Loutherback K., D’Silva J., Liu L., Wu A., Austin R. H., Sturm J. C., “Deterministic separation of cancer cells from blood at 10 mL/min,” AIP Adv. 2(4), 042107 (2012). 10.1063/1.4758131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shim S., Stemke-Hale K., Tsimberidou A. M., Noshari J., Anderson T. E., Gascoyne P. R. C., “Antibody-independent isolation of circulating tumor cells by continuous-flow dielectrophoresis,” Biomicrofluidics 7(1), 011807 (2013). 10.1063/1.4774304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P., Mao Z., Peng Z., Zhou L., Chen Y., Huang P.-H., Truica C. I., Drabick J. J., El-Deiry W. S., Dao M., Suresh S., Huang T. J., “Acoustic separation of circulating tumor cells,” Proc. Natl. Acad. Sci. 112(16), 4970–4975 (2015). 10.1073/pnas.1504484112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermesh O., Aalipour A., Ge T. J., Saenz Y., Guo Y., Alam I. S., Park S.-m., Adelson C. N., Mitsutake Y., Vilches-Moure J., Godoy E., Bachmann M. H., Ooi C. C., Lyons J. K., Mueller K., Arami H., Green A., Solomon E. I., Wang S. X., Gambhir S. S., “An intravascular magnetic wire for the high-throughput retrieval of circulating tumour cells in vivo,” Nat. Biomed. Eng. 2(9), 696–705 (2018). 10.1038/s41551-018-0257-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi W., Paproski R. J., Moore R., Zemp R., “Detection of circulating tumor cells using targeted surface-enhanced Raman scattering nanoparticles and magnetic enrichment,” J. Biomed. Opt. 19(5), 056014 (2014). 10.1117/1.JBO.19.5.056014 [DOI] [PubMed] [Google Scholar]
- 38.Ghosh A., Wang X., Klein E., Heston W. D. W., “Novel role of prostate-specific membrane antigen in suppressing prostate cancer invasiveness,” Cancer Res. 65(3), 727–731 (2005). [PubMed] [Google Scholar]
- 39.Bouvet V., Wuest M., Jans H.-S., Janzen N., Genady A. R., Valliant J. F., Benard F., Wuest F., “Automated synthesis of [(18)F]DCFPyL via direct radiofluorination and validation in preclinical prostate cancer models,” EJNMMI Res. 6(1), 40 (2016). 10.1186/s13550-016-0195-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knop K., Hoogenboom R., Fischer D., Schubert U. S., “Poly(ethylene glycol) in Drug Delivery: Pros and Cons as Well as Potential Alternatives,” Angew. Chem., Int. Ed. 49(36), 6288–6308 (2010). 10.1002/anie.200902672 [DOI] [PubMed] [Google Scholar]
- 41.Rycenga M., Hou K. K., Cobley C. M., Schwartz A. G., Camargo P. H., Xia Y., “Probing the surface-enhanced raman scattering properties of au–ag nanocages at two different excitation wavelengths,” Phys. Chem. Chem. Phys. 11(28), 5903–5908 (2009). 10.1039/b903533h [DOI] [PubMed] [Google Scholar]
- 42.Shi W., Paproski R. J., Shao P., Forbrich A., Lewis J. D., Zemp R. J., “Multimodality Raman and photoacoustic imaging of surface-enhanced-Raman-scattering-targeted tumor cells,” J. Biomed. Opt. 21(2), 020503 (2016). 10.1117/1.JBO.21.2.020503 [DOI] [PubMed] [Google Scholar]
- 43.Forster M. D., Ormerod M. G., Agarwal R., Kaye S. B., Jackman A. L., “Flow cytometric method for determining folate receptor expression on ovarian carcinoma cells,” Cytometry, Part A 71A(11), 945–950 (2007). 10.1002/cyto.a.20456 [DOI] [PubMed] [Google Scholar]
- 44.Andreou C., Kishore S. A., Kircher M. F., “Surface-Enhanced Raman Spectroscopy: A New Modality for Cancer Imaging,” J. Nucl. Med. 56(9), 1295–1299 (2015). 10.2967/jnumed.115.158196 [DOI] [PMC free article] [PubMed] [Google Scholar]