Abstract
Background
Mitochondria support critical cellular functions, such as energy production through oxidative phosphorylation, regulation of reactive oxygen species, apoptosis, and calcium homeostasis.
Objective
Given the heightened level of cellular activity in patients with asthma, we sought to determine whether mitochondrial DNA (mtDNA) copy number measured in peripheral blood differed between individuals with and without asthma.
Methods
Whole genome sequence data was generated as part of the Trans-Omics for Precision Medicine (TOPMed) Program on participants from the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) and the Study of African Americans, Asthma, Genes, & Environment II (SAGE II). We restricted our analysis to individuals who self-identified as African American (3,651 asthma cases and 1,344 controls). Mitochondrial copy number was estimated using the sequencing read depth ratio for the mitochondrial and nuclear genomes. Respiratory complex expression was assessed using RNA-sequencing.
Results
Average mitochondrial copy number was significantly higher among individuals with asthma when compared with controls (SAPPHIRE: 218.60 vs. 200.47, P<0.001; SAGE II: 235.99 vs. 223.07, P<0.001). Asthma status was significantly associated with mitochondrial copy number after accounting for potential explanatory variables, such as participant age, sex, leukocyte counts, and mitochondrial haplogroup. Despite the consistent relationship between asthma status and mitochondrial copy number, the latter was not associated with time-to-exacerbation or patient-reported asthma control. Mitochondrial respiratory complex gene expression was disproportionately lower in individuals with asthma when compared with individuals without asthma and other protein-encoding genes.
Conclusions
We observed a robust association between asthma and higher mitochondrial copy number. Asthma having an effect on mitochondria function was also supported by lower respiratory complex gene expression in this group.
Introduction
Asthma is a chronic disease of the respiratory system often characterized by airway inflammation, excess mucus production, and bronchial constriction [1]. In the United States, it is currently estimated that 11 million adults and 11 million children suffer from asthma [2,3].
Mitochondria are subcellular organelles that produce up to 90% of cellular energy through the citric acid cycle and oxidative phosphorylation [4]. In addition to producing adenosine triphosphate, mitochondria are involved in other critical intracellular processes such maintaining calcium homeostasis, triggering apoptotic cell death, and regulating potentially toxic levels of reactive oxygen species (ROS) [5–8].
Since mitochondria possess their own genome [9], it is possible to estimate the number of mitochondrial genomes per cell (i.e., mitochondrial copy number) by obtaining the mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) ratio. Previous studies have shown that average mitochondrial copy numbers in white blood cells (WBCs) differ for individuals with respiratory diseases, such as chronic obstructive pulmonary disease (COPD) when compared to healthy individuals [10,11], but to our knowledge such studies have not been performed among individuals with asthma alone. In vitro and animal models suggest that various environmental exposures, including aeroallergens, can damage electron transport chain (ETC) respiratory complex proteins, promote ROS production, and increase oxidative stress [12,13]. Decreased expression of ETC genes in bronchial epithelial cells may predispose to allergic inflammation and increase airway responsiveness following allergen challenge [12]. Oxidative stress has also been associated with mitochondrial copy number [14,15]. Therefore, differences in mitochondrial copy number and ETC gene expression among individuals with asthma may identify subtle forms of mitochondrial dysfunction contributing to disease. Such findings and insights may ultimately improve asthma phenotyping and treatment.
In this study, we evaluated whether mitochondrial copy numbers in blood differed among individuals with and without asthma in participants from both the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity (SAPPHIRE) cohort and the Study of African Americans, Asthma, Genes & Environment (SAGE II). We also consider potential causes mtDNA copy number differences by measuring expression of genes involved in oxidative phosphorylation and by assessing mitochondrial volume and membrane potential.
Methods
Patients and settings
Both the SAPPHIRE and SAGE II cohorts and the research presented here were approved by the Institutional Review Boards at Henry Ford Health System and the University of California San Francisco. Both cohorts are part of the Asthma Translational Genomics Collaborative (ATGC) in National Heart Lung and Blood Institute’s (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program. Written informed consent was obtained from all participants or their guardians, as well as written assent from participants <18 years of age, prior to study enrollment and the collection of participant data.
All individuals in the SAPPHIRE cohort were patients from a single health system serving metropolitan Detroit and southeast Michigan. This study has been described in detail elsewhere [16]. Briefly, eligible asthma case individuals had to be 12–56 years of age at the time of recruitment, have a previously documented clinical diagnosis of asthma from a physician, and have no prior history of chronic obstructive pulmonary disease or congestive heart failure. Control individuals were recruited from the same patient population and met the same entry requirements with the exception of not having a prior diagnosis of asthma. Since whole genome sequencing (WGS) was primarily performed in African American participants in SAPPHIRE as part of the TOPMed program, we restricted our analysis to African American participants and to individuals who were ≥18 years of age at the time of enrollment.
At the time of enrollment, SAPPHIRE participants underwent a detailed clinical evaluation which included a detailed questionnaire, anthropomorphic measurements, lung function testing, and specimen collection. Level of asthma control was evaluated using the Asthma Control Test (ACT), a self-reported instrument consisting of 5 questions, each scored on a 5-point Likert scale [17]. Composite ACT scores <20 and ≥20 are considered uncontrolled asthma and controlled asthma, respectively. Lung function testing was performed in accordance with 2005 ATS/ERS guidelines [18]. Complete blood counts with 5-part white blood cell (WBC) differential were measured in blood collected in ethylenediaminetetraacetic acid tubes using a UniCel DxH 800 Coulter Cellular Analysis System (Beckman Coulter, Inc., Brea, CA).
The SAGE II cohort consisted of African American individuals with and without asthma recruited from clinics throughout the San Francisco Bay Area. As a criterion for enrollment, subjects had to self-identify as African American and report that both biological parents and all biological grandparents identified themselves as African American. Participants were between the ages of 8 and 21 years at the time of enrollment. Cases had a history of physician-diagnosed asthma and two or more asthma symptoms (wheezing, coughing, or shortness of breath) in the preceding two years. Participants underwent a detailed evaluation which included obtaining anthropomorphic measurements, testing lung function in accordance with 2005 ATS/ERS guidelines [18], completing a detailed survey with questions on medication use and smoking status, and collecting blood for various laboratory tests (e.g., blood counts). Growth charts (available at http://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm) were used to calculate the BMI percentile of SAGE II participants [19]. Only SAGE II participants <20 years of age were included in the current analysis.
Whole genome sequencing, mitochondrial copy number, mitochondrial haplogroup, and genetic ancestry
DNA was isolated from whole blood, and samples of sufficient quantity and quality were sent to the University of Washington’s Northwest Genomic Center or the New York Genome Center for sequencing. Pre-sequencing quality checks included an assessment of DNA quantity and sex discordance. Libraries for WGS were created using Illumina’s TruSeq DNA PCR-Free Library Preparation Kit. Sequencing was performed on the Illumina HiSeq X platform with paired-end 150bp reads. The Illumina package, bcl2fastq v2 15.0, was used to generate individual FASTQ files. The alignment pipeline can be found at https://github.com/CCDG/Pipeline-Standardization/blob/master/PipelineStandard.md. In brief, the TOPMed Data Coordinating Center at the Northwest Genomics Center aligned the raw sequence reads in FASTQ format to the human reference genome to create binary sequence alignment (BAM) files using the Burrows-Wheeler Aligner algorithm, BWA-MEM [20]. The alignment files were then sent to the Informatics Research Center at the University of Michigan where joint variant calls were performed. We first used existing array data to identify and remove discordant samples based on genotype mismatches. Samples with >10% genotype missingness or sex mismatched X chromosome heterozygosity were also excluded. The R package GENESIS was used to assess for relatedness and population structure [21]. In the case of monozygotic twins, one from each pair was randomly selected and removed from the analysis. Individuals with African ancestry >5 standard deviations [SD] below the mean (~20% global African ancestry) and individuals with closer than 3rd degree relationship with other study participants were excluded as a post hoc sensitivity analysis. We used the BAM files to estimate mitochondrial copy number based on the ratio of read depth using the fastMitoCalc program included in the mitoAnalyzer software package [22]. As a check of the mitochondrial copy number estimates derived using WGS data, we agnostically selected a set of 50 blood samples to quantitate copy number using both the real-time polymerase chain reaction (RT-PCR)-based method described by Ajaz et al. [23] and sequence read depth. The primers selected for this RT-PCR assay identified unique and singular portions of the mitochondrial and nuclear genomes (i.e., to avoid mitochondrial pseudogene amplification and dilutional effects from copy number differences in the nuclear genome) [24]. Mitochondrial copy number estimates were significantly lower using PCR-based method when compared with the read depth method (67.6 ± 24.4 standard deviation [SD] vs. 232.3 ± 77.3, respectively; P = 2.2x10-16 [paired T-test]). However, the measurements obtained by both methods were strongly correlated (Pearson’s r = 0.74, P = 5.9 x 10−10) as shown in Supplementary S1 Fig, suggesting that relative copy number estimates between individuals were similar using both approaches.
BAM files containing mitochondrial read alignments were separated using SAMtools v1.8 software. The pipeline for mitochondrial haplogroup assignments involved the use of Cloudgene (locally installed) with mtDNA-Server v1.1.3 and the program HaploGrep 2 [25,26]. Global ancestry (i.e., the overall proportion of continental group ancestry) was estimated using the programs RFMix and Admixture [27,28].
Medication use and asthma severity
A subset of the SAPPHIRE participants with asthma had detailed records on medication fills as a result of their health plan membership; these data included pharmacy claims from health plan and retail pharmacies. We estimated inhaled corticosteroid (ICS) use in the 6 months prior to the visit date using algorithms that we had developed and reported on previously [29,30]. Average daily exposure to short acting beta agonist (SABA) medication was estimated using records of SABA fills in the 3-month period preceding study enrollment. Separate metrics of SABA use were calculated for nebulizer use and metered dose inhaler (MDI) use, as we have previously shown that both the form and the frequency of SABA medication use may be related to asthma severity [31,32]. Average daily SABA use for each participant was estimated by summing the total number of doses (e.g., 50 puffs per canister) dispensed for that individual in the 91 days preceding enrollment and dividing by 91.
We also estimated asthma severity using a previously described metric which uses both SABA and oral corticosteroid prescription fills in the preceding year [33]. The lowest severity group, Group 1, had no oral corticosteroid (OCS) fills and ≤1 SABA fill in the 12 months prior to the time of evaluation. Group 3 had one of the following: 2 OCS fills, >6 SABA fills, or the combination of 1 OCS fill and ≥4 SABA fills. Group 4 had either ≥3 OCS fills or the combination of 2 OCS fills and >6 SABA fills. Group 2 encompassed all other combinations of SABA and OCS fills in the year prior to the study visit.
RNA sequence (RNA-seq) data
Blood was collected from SAPPHIRE participants at the time of enrollment for later RNA analysis. Blood was collected and stored in PAXgene Blood RNA tubes (BD Biosciences, San Jose, CA) at -80°C until the time of analysis. The TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero Globin (Illumina, San Diego, CA) was used to create sequencing libraries on a random sample of African American SAPPHIRE participants with and without asthma. Libraries were sequenced on an Illumina HiSeq. FastQC v0.11.5 (available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and MultiQC v0.8 were used to assess RNA-seq quality, including the read length distribution and depth [34]. The program BBDuk (BBMap v36.49) was used to trim reads and remove Illumina adapter sequence (available at https://sourceforge.net/projects/bbmap/). The software program HISAT2 2.1.0 was used to map reads to human genome build GRCh38.p5 [35], and mapped reads were quantified at the transcript level using StringTie v1.3.3 [36,37]. RSeQC (v2.6.4) was used to provide post-alignment QC, including the distribution of mapped reads, coverage uniformity, strand specificity after alignment [38]. Samples that did not pass any of the above QC measures were excluded from the analysis, we also excluded samples from batches that didn’t include both cases and control participants. We restricted our assessment to mitochondrial and nuclear genes encoding proteins making up ETC complexes. These complexes compose the final critical pathway for oxidative metabolism of foodstuffs; complexes I-IV participate in the transfer of electrons and the creation of a proton potential across the inner mitochondrial membrane, and this gradient drives the formation of cellular energy as ATP by complex V [39]. Gene expression was quantified as transcripts per kilobase per million reads (TPM); this measure normalizes read counts for a gene by accounting for its length (in kilobases) and the total number of reads for a sample (in millions). In addition, the R-package, DESeq2, was used to normalize read counts across individuals and all protein-encoding genes in order to assess for differences in expression by asthma status [40].
Mitochondrial assessment in peripheral blood leukocytes by flow cytometry
We invited a subset of SAPPHIRE participants to provide a new blood sample for mitochondrial assessment by flow cytometry. This group included individuals with asthma and a high mitochondrial copy number (>300) (eligible n = 61), and individuals with (eligible n = 119) and without (eligible n = 107) asthma who had a mitochondrial copy number near the average for their group (± 10 from the mean). We evaluated the first to respond from these eligible groups—4 (average copy number 371.3 ± 25 SD), 4 (average copy number 217.9 ± 1.3 SD), and 5 (average copy number 198.1 ± 5.7 SD), respectively. Approximately 12 milliliters (ml) of blood were collected from each participant into heparinized tubes; blood was centrifuged with 14 ml of Ficoll-Paque Plus (GE Healthcare Life Sciences) to isolate WBCs. After centrifugation, both the buffy coat layer and the granulocyte layer were removed. The latter was combined with 20 ml of FACS lysing solution (BD Biosciences); the sample was then mixed and incubated for 30 minutes. The peripheral blood mononuclear cells (PBMCs) from the buffy coat were centrifuged, and the resulting cell pellet was washed multiple times and resuspended in sterile 15 ml of 1x phosphate-buffered saline solution. A 10 microliter (μl) sample from each cell suspension was stained with trypan blue, and the cells were counted manually using a hemacytometer. We used MitoTracker Red CMXRos (Molecular Probes, Inc., Eugene, OR) to quantify mitochondria–dye accumulation was dependent on both mitochondrial number and membrane potential. Following the manufacture’s guidelines, cells were incubated for 30 minutes with prewarmed staining solution containing a 100 nanomolar concentration of MitoTracker Red CMXRos. Cells were then fixed with buffer containing 4% formaldehyde and stored at -80°C until the time of analysis. For the flow cytometry analysis, granulocytes were stained with 2.5 μl of anti-CD203c antibody conjugated with allophycocyanin (APC) dye (BioLegend, San Diego, CA), 3 μl of anti-CD117 antibody conjugated with APC-Cy7TM dye (BioLegend), 3 μl of anti-Siglec 8 antibody conjugated with phycoerythrin (PE) dye (BioLegend), and 5 μl of anti-CD45 antibody conjugated with Brilliant Violet 421TM dye (BioLegend); PBMCs were stained with 5 μl of anti-CD45 antibody conjugated with Brilliant Violet 421TM dye (BioLegend), 2 μl of anti-CD3 antibody conjugated with APC-Cy7TM dye (BioLegend), 5 μl of anti-CD4 antibody conjugated with peridinin-chlorophyll-protein-Cy5.5 dye (BD Biosciences), and 3 μl of anti-CD56 antibody conjugated with PE-CF594 dye (BioLegend). Eosinophils (CD45+, Siglec 8+, and CD203c-), T-helper cells (CD45+, CD3+, and CD4+), and natural killer (NK) cells (CD45+, CD3-, and CD56+) were assessed for MitoTracker staining intensity using a LSRFortessa flow cytometer (BD Biosciences). Compensation beads (BD CompBead, BD Biosciences) were used to correct for spectral overlap between fluorochrome-labeled antibodies, and the gates for positively stained cells were positioned using “fluorescence minus one” controls. Leukocyte populations were identified using the light scatter characteristics of live cells in the forward scatter and side scatter channels to collect 100,000 gated events. Flow cytometry data were analyzed using FacsDiva v8.0.1 software (BD Biosciences).
Statistical analysis
The primary study aim was to assess the relationship between mitochondrial copy number (outcome) and asthma status (predictor variable). Secondary analyses included assessing factors associated with mitochondrial copy number (outcome) among individuals with asthma and, conversely, assessing whether mitochondrial copy number (predictor variable) was separately associated with the outcomes of asthma status, exacerbations, and control.
Differences in baseline (i.e., at the time of the initial study visit) characteristics between individuals with and without asthma were assessed using chi-squared tests and t-tests for categorical and continuous variables, respectively. We then used Welch’s two-sample t-test to compare copy number differences between mitochondrial haplogroups categorized by location of origin. We compared copy numbers between individuals with either African mitochondrial haplogroups (L0, L1, L2, L3, and L4) or east Eurasian haplogroups combined (B, F, A, D, E, and C/Z) with west Eurasian haplogroups combined (U/K [U2, U3, U4, U5, U7, and K], J/T, HV/H/V, W, and I); individuals with west Eurasian haplogroups were the reference group for these comparisons. The above population distribution of haplogroup assignments has been described elsewhere [4,41,42]. We also compared copy numbers between individuals with African mitochondrial haplogroups (L0, L1, L2, L3, and L4), M haplogroups (D, E, C/Z, M7, and other M), and N + R haplogroups (N macrohaplogroups [X, A, W, and I] and the R sub-macrohaplogroups [U/K, B, F, HV/H/V, and J/T]); in these comparisons, individuals with N + R haplogroups were considered the reference group. Haplogroup categories were also stratified by asthma status in order to assess within haplogroup differences in mitochondrial copy numbers by asthma status. The R package, metafor, was used to meta-analyze the standardized mean difference in mitochondrial copy number between asthma case and control participants from the SAPPHIRE and SAGE II cohorts [43].
To understand the factors contributing to mitochondrial copy number, we used linear regression to model copy number as a function of patient asthma status, patient age, sex (male = 0, female = 1), proportion of African ancestry, body mass index (BMI) in kilograms per meter squared, smoking status (past or never smoker = 0, active smoker = 1), percent of predicted forced expiratory volume at 1 second (FEV1), absolute WBC counts for neutrophils, monocytes, lymphocytes, and eosinophils (basophils were not included given their scant number), platelet count, and mitochondrial haplogroup (included using dummy variables for African haplogroups [L0, L1, L2, and L3] and combined east Eurasian haplogroups as compared with the combined west Eurasian haplogroups). Given the small number of individuals with the L4 haplogroup, we did not include them in these analyses. Since most SAGE II participants were children, BMI percentile was used in lieu of BMI [19]. Only a small subset of SAGE II participants had WBC counts, so we performed analyses with and without this variable.
In order to identify potential factors within individuals with asthma contributing to higher mitochondrial copy number, we restricted the model to SAPPHIRE participants with asthma. In this analysis, we included variables related to asthma treatment and severity, such as the composite ACT score, asthma severity score, SABA exposure, and ICS exposure. These particular variables were only available for SAPPHIRE participants.
Logistic regression was used to model asthma status (dependent variable) as a function of mitochondrial copy number with other potential explanatory and confounding variables, such as patient age, sex, proportion of African ancestry, BMI, smoking status, percent of predicted FEV1, absolute WBC counts for neutrophils, monocytes, lymphocytes, and eosinophils (basophils were not included given their scant number), and mitochondrial haplogroups.
As a sensitivity analysis, we reassessed the relationship between asthma status and mitochondrial copy number after removing individuals with a low proportion of African ancestry or those who were closely related to other study participants. Specifically, we removed 23 individuals from SAPPHIRE and 1 individual from SAGE II whose African ancestry was >5 SD below the mean (i.e., a global African ancestry proportion of approximately 20% or less), and we excluded 203 individuals from SAPPIRE and 0 individuals from SAGE II whose kinship suggested closer than a 3rd degree relationship (coefficient of relationship >0.125) with another study participant.
To evaluate whether mitochondrial copy number was associated with asthma complications and control, we used a Cox proportional hazards regression to prospectively model the time to severe asthma exacerbations and we used logistic regression to model baseline asthma control (ACT score <20 [uncontrolled] vs. ≥20 [controlled]). Prospective data were only available in SAPPHIRE participants. Severe asthma exacerbations were defined as those requiring oral corticosteroid burst treatment, an emergency department visit, or hospitalization. The survival analysis modeled the time between study enrollment and the first severe exacerbation. Individuals were censored on their last recorded clinical visit or at the time of disenrollment from the health plan.
The non-parametric Mann–Whitney U test was used for the univariate comparison of gene expression TPM values between asthma cases and controls. DESeq2 was used to assess for differential gene expression by asthma status while also adjusting for patient age, sex, global proportion of African ancestry, and absolute cells counts for neutrophils, monocytes, lymphocytes, eosinophils, and platelets. We also used surrogate variable analysis (SVA) implemented in the package svaseq to account for hidden batch effects; 10 surrogate variables were used to adjust the models [44]. The Fisher’s exact test was used to compare the expression distribution (i.e., number of higher, lower, and not-significantly different expressed genes for individuals with asthma as compared with individuals without asthma) between ETC genes and other protein-encoding genes (restricted to genes with >10 total read counts across all individuals).
The t-test was used to compare mean MitoTracker intensities in blood leukocyte populations (i.e., eosinophils, T-helper cells, and NK cells) between individuals with asthma and high mitochondrial copy number vs. controls with average mitochondrial number, as well as between individuals with asthma and average mitochondrial copy number vs. controls with average mitochondrial number. All analyses were performed using R statistical software [45]. Unless otherwise specified, a p-value <0.05 was considered statistically significant.
Results
The characteristics of participants used in this analysis are shown in Table 1. All individuals were African American by self-report. The SAPPHIRE cohort analytic set consisted of 3,676 individuals (2,828 cases and 848 controls), and SAGE II set consisted of 1,319 individuals (823 cases and 496 controls). While SAPPHIRE participants were older at the time of enrollment (≥18 years) when compared with SAGE II participants (<20 years), both groups were similar in their proportion of African ancestry (~80% in both cohorts) and both had a similar mitochondrial haplogroup distribution. Approximately, 92%, 91%, 87%, 88% of SAPPHIRE cases, SAPPHIRE controls, SAGE II cases, and SAGE II controls, respectively, had mitochondrial haplogroups of African origin (i.e., haplogroups L0, L1, L2, L3, and L4). A detailed breakdown of mitochondrial haplogroup distribution for each cohort can be seen in Fig 1A and 1B.
Table 1. Characteristics of African American participants from the SAPPHIRE and SAGE II cohorts stratified by asthma status*.
| Variable | SAPPHIRE cohort | SAGE II cohort | ||||
|---|---|---|---|---|---|---|
| Participants with asthma (n = 2,828) | Participants without asthma (n = 848) | P-value† | Participants with asthma (n = 823) | Participants without asthma (n = 496) | P-value† | |
| Age (years)–mean ± SD | 36.98 ± 12.12 | 39.60 ± 12.55 | <0.001 | 14.06 ± 3.69 | 15.85 ± 3.71 | < 0.001 |
| Female–no. (%) | 1955 (69.1) | 581 (68.5) | 0.766 | 407 (49.5) | 284 (57.3) | 0.007 |
| African ancestry (%)–mean ± SD‡ | 0.80 ± 0.12 | 0.81 ± 0.12 | 0.226 | 0.79 ± 0.13 | 0.78 ± 0.12 | 0.431 |
| BMI (kg/m2)–mean ± SD | 33.17 ± 9.39 | 31.75 ± 7.89 | <0.001 | -- | -- | -- |
| BMI percentile—mean ± SD§ | -- | -- | -- | 76.39 ± 24.74 | 70.95 ± 27.21 | 0.005 |
| Smoking status–no. (%) | -- | -- | <0.001 | -- | -- | 0.001 |
| Current | 765 (27.1) | 107 (12.6) | -- | 68 (8.3) | 68 (13.7) | -- |
| Past | 322 (11.4) | 98 (11.6) | -- | 1 (0.1) | 2 (0.4) | |
| Never | 1740 (61.5) | 643 (75.8) | -- | 753 (91.6) | 426 (85.9) | -- |
| Percent of predicted FEV1 –mean ± SD|| | 85.14 ± 19.67 | 96.70 ± 15.29 | <0.001 | 99.01 ± 13.79 | 101.64 ± 9.22 | < 0.001 |
| Composite ACT score <20 –no. (%)¶ | 1,580 (55.9) | -- | -- | -- | -- | -- |
| Asthma severity score–mean ± SD** | 1.68 ± 0.85 | -- | -- | -- | -- | -- |
| ICS medication use at baseline–no. (%)†† | 237 (46.3) | -- | -- | 282 (58.51) | -- | -- |
| Mitochondrial copy number–mean ± SD‡‡ | 218.60 ± 58.80 | 200.47 ± 64.95 | <0.001 | 235.99 ± 59.22 | 223.07 ± 61.48 | <0.001 |
| WBC counts (1000/μL)–mean ± SD | 6.68 ± 2.32 | 6.10 ± 1.80 | <0.001 | 6.22 ± 1.98 | 6.20 ± 2.12 | 0.951 |
| Neutrophil counts | 3.70 ± 1.92 | 3.24 ± 1.33 | <0.001 | -- | -- | -- |
| Monocyte counts | 0.46 ± 0.17 | 0.43 ± 0.15 | 0.006 | -- | -- | -- |
| Lymphocyte counts | 2.28 ± 0.80 | 2.26 ± 0.72 | 0.633 | -- | -- | -- |
| Eosinophil counts | 0.21 ± 0.19 | 0.14 ± 0.11 | <0.001 | -- | -- | -- |
| Hemoglobin (g/dL)–mean ± SD | 13.29 ± 1.55 | 13.24 ± 1.40 | 0.492 | -- | -- | -- |
| Platelet count (1000/μL)–mean ± SD | 242.35 ± 67.00 | 242.30 ± 61.09 | 0.985 | -- | -- | -- |
| Mitochondrial haplogroup—no. (%)§§ | -- | -- | 0.429 | -- | -- | 0.735 |
| L0 | 122 (4.3) | 42 (5.0) | -- | 42 (5.1) | 22 (4.4) | -- |
| L1 | 516 (18.2) | 166 (19.6) | -- | 149 (18.1) | 94 (19.0) | -- |
| L2 | 826 (29.2) | 262 (30.9) | -- | 225 (27.3) | 145 (29.2) | -- |
| L3 | 1110 (39.2) | 298 (35.1) | -- | 291 (35.4) | 174 (35.1) | -- |
| L4 | 22 (0.8) | 7 (0.8) | -- | 5 (0.6) | 2 (0.4) | -- |
| M | 45 (1.6) | 11 (1.3) | -- | 17 (2.1) | 10 (2.0) | -- |
| N + R | 187 (6.6) | 62 (7.3) | -- | 94 (11.4) | 49 (9.9) | -- |
| East Eurasian | 48 (1.7) | 13 (1.5) | -- | 32 (3.9) | 17 (3.4) | -- |
| West Eurasian | 141 (5.0) | 40 (4.7) | -- | 61 (7.4) | 27 (5.4) | -- |
SAPPHIRE denotes Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SAGE II, Study of African Americans, Asthma, Genes, & Environment II; SD, standard deviation; BMI, body mass index; FEV1, forced expiratory volume at 1 second; ACT, asthma control test; ICS, inhaled corticosteroid; and WBC, white blood cell.
*The SAPPHIRE study sample was restricted to participants aged ≥18 years at enrollment and the SAGE II study samples was restricted to participants aged <20 years at enrollment.
†P-value for the difference between study participants with and without asthma in each cohort.
‡The average proportion of African ancestry among African Americans was estimated using a set of autosomal markers which spanned the nuclear genome.
§Growth charts specific for age and sex (available at http://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm) were used to calculate the BMI percentile of SAGE II participants.
||Based on the predictive equations from Hankinson et al. (Am J Respir Crit Care Med. 1999 Jan;159[1]:179–87).
¶Composite ACT scores <20 are considered “uncontrolled” asthma.
**This scoring algorithm, which is based on asthma rescue medication use, was developed by Allen-Ramey et al. (J Manag Care Pharm 2006; 12:310–21). This measure was calculated on the 512 individuals with asthma and available pharmacy claims information.
††This measure was calculated on the 512 individuals with asthma and available pharmacy claims information in SAPPHIRE participants and was based on cross-sectional self-report of use in SAGE II participants.
‡‡The mitochondrial copy number estimate was for whole blood. It was based on the sequencing read depth ratio between mitochondrial and nuclear DNA isolated from blood leukocytes.
§§Number of study individuals with a given mitochondrial haplogroup. As shown in Fig 1, the M haplogroups consist of D, E, C/Z, M7 and other M; the N macrohaplogroups consist of X, A, W, I, and the R sub-macrohaplogroups; and R sub-macrohaplogroups consist of U/K, B, F, HV/H/V, and J/T. Geographical mitochondrial haplogroup assignments were based on those described by Pereira et al. (Am J Hum Genet. 2009; 84:628–40). Haplogroups L0, L1, L2, L3, and L4 are considered to be African; haplogroups B, F, A, D, E, and C/Z are considered to be East Eurasian; and haplogroups U/K, J/T, HV/H/V, I, and W are considered to be West Eurasian.
Fig 1. Mitochondrial haplogroup distributions.
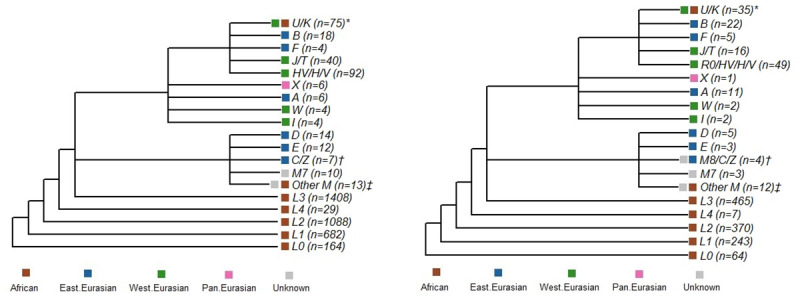
Shown are the mitochondrial haplogroup distributions of the 3,676 SAPPHIRE participants (A) and the 1,319 SAGE II participants (B). Branching shows haplogroup derivation and color coding denotes the geographic identity of the haplogroup. *In SAPPHIRE this group consisted of 34 individuals with an African haplogroup (U6) and 41 with a west Eurasian haplogroup (U2, U3, U4, U5, U7, and K), and in SAGE II this group consisted of 16 individuals with an African haplogroup (U6) and 19 with a west Eurasian haplogroup (U4, U5, U8, and K). †In SAPPHIRE this group consisted of 7 individuals with an east Eurasian haplogroup (C, Z), and in SAGE II this group consisted of 3 individuals with an east Eurasian haplogroup (C, Z) and 1 individual with a haplogroup of unknown origin (M8). ‡In SAPPHIRE this group consisted of 2 individuals with an African haplogroup (M1) and 11 individuals with a haplogroup of unknown origin (M2, M23, M30, and M32), and in SAGE II this group consisted of 6 individuals with an African haplogroup (M1) and 6 individuals with a haplogroup of unknown origin (M3, M5, M6, and M23).
Asthma cases and controls also differed in consistent ways in both cohorts, such as case participants were significantly younger, had greater a BMI (greater BMI percentile in SAGE II), and lower percent of predicted FEV1 when compared with their respective control group. Sex distribution and smoking patterns differed between cohorts (cases were more likely to have smoked in SAPPHIRE but less likely to have smoked in SAGE II). In SAPPHIRE participants, asthma cases had significantly greater cell counts when compared with controls: total WBC counts (6,680 cells/μL vs. 6,100 cells/μL), neutrophil counts (3,700 cells/μL vs. 3,240 cells/μL), monocyte counts (460 cells/μL vs. 430 cells/μL), and eosinophil counts (210 cells/μL vs. 140 cells/μL). In both SAPPHIRE and SAGE II, mitochondrial copy numbers were significantly higher among individuals with asthma when compared with those without asthma (SAPPHIRE: 218.60 vs. 200.47, P<0.001; SAGE II: 235.99 vs. 223.07, P<0.001).
Table 2 evaluates the relationship between mitochondrial haplogroup and mitochondrial copy number. Among SAPPHIRE participants, individuals with west Eurasian mitochondrial haplogroups had lower average mitochondrial counts than did individuals with L1, L3, and east Eurasian haplogroups. SAPPHIRE individuals with other African haplogroups (L0 and L2) and SAGE II individuals did not have significantly different mitochondrial copy numbers when compared with individuals with west Eurasian haplogroups. Defining haplogroup clusters based on the branching shown in Fig 1 (as opposed to geographical haplogroup assignments) demonstrated essentially the same results (Supplementary S1 Table), since the majority of individuals in the N + R haplogroup branch had haplogroups of west Eurasian origin (181 [72.7%] of 249 in SAPPHIRE and 88 [61.5%] of 143 in SAGE II).
Table 2. Relationship between mitochondria haplogroup and copy number among African American participants in the SAPPHIRE and SAGE II cohorts*.
| Mitochondrial haplogroup† | SAPPHIRE cohort | SAGE II cohort | ||||||
|---|---|---|---|---|---|---|---|---|
| Number of participants‡ | African ancestry (mean ± SD)§ | Copy number (mean ± SD)|| | P-value¶ | Number of participants‡ | African ancestry (mean ± SD)§ | Copy number (mean ± SD)|| | P-value¶ | |
| L0 | 164 | 0.81 ± 0.09 | 201.95 ± 59.25 | 0.827 | 64 | 0.82 ± 0.09 | 228.63 ± 59.02 | 0.734 |
| L1 | 682 | 0.82 ± 0.09 | 215.91 ± 60.95 | 0.010 | 243 | 0.80 ± 0.11 | 228.75 ± 52.67 | 0.618 |
| L2 | 1088 | 0.82 ± 0.09 | 211.52 ± 58.99 | 0.079 | 370 | 0.81 ± 0.10 | 228.66 ± 60.10 | 0.621 |
| L3 | 1408 | 0.82 ± 0.09 | 218.27 ± 62.84 | 0.001 | 465 | 0.81 ± 0.09 | 237.46 ± 65.52 | 0.063 |
| East Eurasian | 61 | 0.77 ± 0.16 | 222.52 ± 44.71 | 0.008 | 49 | 0.67 ± 0.18 | 222.18 ± 56.82 | 0.739 |
| West Eurasian | 181 | 0.56 ± 0.27 | 203.33 ± 57.60 | Reference | 88 | 0.58 ± 0.18 | 225.48 ± 52.70 | Reference |
SAPPHIRE denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SAGE II, Study of African Americans, Asthma, Genes, & Environment II; and SD, standard deviation.
*The SAPPHIRE study sample was restricted to participants aged ≥18 years at enrollment and the SAGE II study samples was restricted to participants aged <20 years at enrollment.
†Geographical mitochondrial haplogroup assignments were based on those described by Pereira et al. (Am J Hum Genet. 2009; 84:628–40). Haplogroups L0, L1, L2, and L3 are considered to be African. Given the small number of study individuals with non-African haplogroups, we grouped most of the remaining haplogroups into the broad categories of East Eurasian and West Eurasian.
‡Includes individuals with and without asthma in each cohort.
§African ancestry was estimated using a set of autosomal markers which spanned the nuclear genome.
||The mitochondrial copy number estimate was for whole blood. It was based on the sequencing read depth ratio between mitochondrial and nuclear DNA isolated from blood leukocytes.
¶P-values were calculated using the Welch two sample t-test to compare mitochondrial copy numbers between West Eurasian haplogroups (referent) and the other haplogroups.
Within haplogroup categories, average mitochondrial copy numbers were consistently higher among individuals with asthma when compared with those without asthma (Table 3). These differences were statistically significant for L1, L2, L3, and west Eurasian haplogroups in SAPPHIRE and the L2 haplogroup in SAGE II. Defining haplogroup clusters based on the branching shown in Fig 1 (as opposed to geographical haplogroup assignments) demonstrated essentially the same results with the exception of the small number of individuals with M haplogroups who showed a non-significant relationship in the opposite direction (Supplementary S2 Table). There were too few individuals with the L4 haplogroup for comparison in SAGE II, so this group was dropped from all subsequent analyses.
Table 3. Relationship between mitochondria haplogroup and copy number among African American participants in the SAPPHIRE and SAGE II cohorts stratified by asthma status*.
| Mitochondrial haplogroup† | SAPPHIRE cohort | SAGE II cohort | Meta-analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Participants with asthma | Participants without asthma | P-value§ | Participants with asthma | Participants without asthma | P-value§ | Standardized difference (95% CI) | Heterogeneity P-value|| | P-value¶ | |||||
| No. | Mitochondria copy number (mean ± SD)‡ | No. | Mitochondria copy number (mean ± SD) ‡ | No. | Mitochondria copy number (mean ± SD) ‡ | No. | Mitochondria copy number (mean ± SD) ‡ | ||||||
| L0 | 122 | 204.66 ± 53.87 | 42 | 194.09 ± 72.85 | 0.392 | 42 | 232.17 ± 49.65 | 22 | 221.87 ± 74.60 | 0.564 | 0.18 (-0.11–0.47) | 0.988 | 0.233 |
| L1 | 516 | 220.56 ± 60.50 | 166 | 201.44 ± 60.22 | <0.001 | 149 | 232.97 ± 48.13 | 94 | 222.06 ± 58.82 | 0.133 | 0.28 (0.14–0.43) | 0.496 | <0.001 |
| L2 | 826 | 216.40 ± 56.96 | 262 | 196.14 ± 62.65 | <0.001 | 225 | 237.64 ± 59.24 | 145 | 214.73 ± 58.95 | <0.001 | 0.36 (0.24–0.48) | 0.754 | <0.001 |
| L3 | 1110 | 221.65 ± 60.33 | 298 | 205.67 ± 70.11 | <0.001 | 291 | 240.19 ± 66.28 | 174 | 232.89 ± 64.14 | 0.242 | 0.21 (0.10–0.32) | 0.214 | <0.001 |
| East Eurasian | 48 | 222.98 ± 44.89 | 13 | 220.80 ± 45.79 | 0.880 | 32 | 227.06 ± 57.03 | 17 | 212.99 ± 56.99 | 0.417 | 0.15 (-0.27–0.58) | 0.648 | 0.485 |
| West Eurasian | 141 | 208.98 ± 56.32 | 40 | 183.41 ± 58.31 | 0.016 | 61 | 227.25 ± 55.11 | 27 | 221.47 ± 47.55 | 0.619 | 0.32 (0.04–0.60) | 0.245 | 0.024 |
SAPPHIRE denotes the Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; SAGE II, Study of African Americans, Asthma, Genes, & Environment II; SD, standard deviation; and CI, confidence interval.
*The SAPPHIRE study sample was restricted to participants aged ≥18 years at enrollment and the SAGE II study samples was restricted to participants aged <20 years at enrollment.
†Geographical mitochondrial haplogroup assignments were based on those described by Pereira et al. (Am J Hum Genet. 2009; 84:628–40). Haplogroups L0, L1, L2, and L3 are considered to be African. Given the small number of study individuals with non-African haplogroups, we grouped most of the remaining haplogroups into the broad categories of East Eurasian and West Eurasian.
‡The mitochondrial copy number estimate was for whole blood. It was based on the sequencing read depth ratio between mitochondrial and nuclear DNA isolated from blood leukocytes.
§P-values were calculated using the Welch two sample t-test to compare mitochondrial copy numbers between individuals with and without asthma.
||Assessment of the difference in P-values from the SAPPHIRE and SAGE II cohorts. A P-value<0.05 would signify a statistically significant difference in the P-values between cohorts.
¶Meta-analysis P-value for the standardized difference in mitochondrial copy number between individuals within and without asthma for the SAPPHIRE and SAGE II cohorts combined.
Effect estimates for the factors associated with mitochondrial copy number were remarkably consistent between the SAPPHIRE and SAGE II cohorts (Tables 4 and 5, respectively). Asthma was consistently and significantly associated with greater mitochondrial copy numbers in all models tested for both cohorts. Excluding individuals whose African ancestry was >5 SD below the mean or who had a coefficient of relationship >0.125 among SAPPHIRE participants did not substantively affect the significant relationship between asthma status and mitochondrial copy number (Supplementary S3 Table). Similarly, excluding the one SAGE II with African ancestry >5 SD below the mean also did not affect the results (data not shown). White blood cell counts were inversely associated with mitochondrial copy number in both groups (also shown in Supplementary S4 Table for SAGE II). In the SAPPHIRE cohort, all leukocyte counts were inversely associated with mitochondrial copy number, and all but monocytes were significantly associated with copy number in the multivariable analysis (Table 4). Platelet counts were positively associated with mitochondrial copy number. The relationship between cell counts by cell type and mitochondrial copy number stratified by asthma status is shown in Supplementary S2 Fig. Significant interactions by asthma status were seen for neutrophils (P = 2.68x10-4) and platelets (P = 0.009); however, asthma was consistently associated with higher mitochondrial counts regardless of cell type count (i.e., a cross-over interaction was not observed for any cell type).
Table 4. Factors associated with mitochondrial copy number among African American SAPPHIRE participants.
| Variable | Univariable Analysis | Model 1† | Model 2‡ | Model 3§ | Model 4|| | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| R2* | Unadjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | |
| Asthma status | 0.016 | 18.13 | <0.001 | 19.42 | <0.001 | 34.19 | <0.001 | 18.19 | <0.001 | 33.25 | <0.001 |
| Age (years) | 0.003 | -0.28 | <0.001 | -0.17 | 0.039 | -- | -- | -- | -- | -0.20 | 0.096 |
| Female Sex | <0.001 | 0.17 | 0.937 | 1.68 | 0.447 | -- | -- | -- | -- | -6.54 | 0.048 |
| African Ancestry | 0.003 | 29.64 | <0.001 | 33.97 | <0.001 | -- | -- | -- | -- | 9.44 | 0.483 |
| BMI (kg/m2) | 0.002 | -0.33 | 0.003 | -0.39 | <0.001 | -- | -- | -- | -- | 0.05 | 0.790 |
| Smoking status | <0.001 | 0.36 | 0.879 | -2.93 | 0.219 | -- | -- | -- | -- | 7.00 | 0.058 |
| Percent of predicted FEV1 | <0.001 | -0.04 | 0.452 | 0.05 | 0.383 | -- | -- | -- | -- | -0.09 | 0.253 |
| Total WBC count | 0.133 | -11.70 | <0.001 | -- | -- | -- | -- | -- | -- | -- | -- |
| Neutrophils | 0.120 | -13.79 | <0.001 | -- | -- | -14.58 | <0.001 | -- | -- | -14.36 | <0.001 |
| Monocytes | 0.043 | -87.33 | <0.001 | -- | -- | -13.84 | 0.148 | -- | -- | -18.32 | 0.067 |
| Lymphocytes | 0.033 | -16.62 | <0.001 | -- | -- | -13.14 | <0.001 | -- | -- | -12.67 | <0.001 |
| Eosinophils | 0.003 | -21.91 | 0.013 | -- | -- | -20.21 | 0.014 | -- | -- | -24.34 | 0.004 |
| Platelet count | 0.008 | 0.10 | <0.001 | -- | -- | 0.217 | <0.001 | -- | -- | 0.23 | <0.001 |
| Mitochondrial haplogroup | 0.005 | -- | -- | -- | -- | -- | -- | -- | -- | -- | -- |
| L0 vs West Eurasian | -- | -1.38 | 0.833 | -- | -- | -- | -- | -0.74 | 0.909 | -1.47 | 0.875 |
| L1 vs West Eurasian | -- | 12.58 | 0.014 | -- | -- | -- | -- | 12.98 | 0.010 | 9.47 | 0.218 |
| L2 vs West Eurasian | -- | 8.19 | 0.094 | -- | -- | -- | -- | 8.55 | 0.078 | 6.38 | 0.390 |
| L3 vs West Eurasian | -- | 14.94 | 0.002 | -- | -- | -- | -- | 14.77 | 0.002 | 13.21 | 0.073 |
SAPPHIRE denotes Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity; BMI, body mass index; FEV1, forced expiratory volume at 1 second; and WBC, white blood count.
*The coefficient of determination (R2) represented the percent of the variation in the outcome variable (i.e., mitochondrial copy number) that could be accounted for by each variable in the univariable analyses.
†Model 1 assessed the relationship between mitochondrial copy number in blood leukocytes (dependent variable) and asthma status (main explanatory variable). This model adjusted for patient age in years, sex (female = 1, male = 0), proportion of African ancestry, BMI, smoking status (past or never smoker = 0, active smoker = 1), and percent of predicted FEV1. Complete data were available for 3,675 individuals in Model 1, which had an adjusted R2 = 0.024.
‡Model 2 assessed the relationship between mitochondrial copy number in blood leukocytes (dependent variable) and asthma status (main explanatory variable). This model adjusted for absolute white blood cell counts and platelet counts (in increments of 1000 cells per microliter). Complete data were available for 2,031 individuals in Model 2, which had an adjusted R2 = 0.219.
§Model 3 assessed the relationship between mitochondrial copy number in blood leukocytes (dependent variable) and asthma status (main explanatory variable). This model adjusted for mitochondrial haplogroup, and only individuals with the L0, L1, L2, L3, and West Eurasian haplogroups were included. Complete data were available for 3,523 individuals in Model 3, which had an adjusted R2 = 0.020.
||Model 4 assessed the relationship between mitochondrial copy number in blood leukocytes (dependent variable) and asthma status (main explanatory variable); this model included all of the variables from models 1–3. Complete data were available for 1,942 individuals in Model 4, which had an adjusted R2 = 0.229.
Table 5. Factors associated with mitochondrial copy number among African American SAGE II participants.
| Variable | Univariable Analysis | Model 1† | Model 2‡ | Model 3§ | |||||
|---|---|---|---|---|---|---|---|---|---|
| R2* | Unadjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | Adjusted parameter estimate | P-value | |
| Asthma status | 0.010 | 12.91 | <0.001 | 28.31 | <0.001 | 12.86 | <0.001 | 27.62 | <0.001 |
| Age (years) | 0.033 | -2.94 | <0.001 | -2.43 | <0.001 | -- | -- | -2.28 | <0.001 |
| Female sex | 0.001 | -5.52 | 0.097 | -5.12 | 0.145 | -- | -- | -6.54 | 0.072 |
| African ancestry proportion | 0.001 | 17.17 | 0.190 | 32.25 | 0.023 | -- | -- | 36.18 | 0.039 |
| BMI percentile | 0.001 | 0.07 | 0.312 | 0.01 | 0.852 | -- | -- | 0.0076 | 0.916 |
| Smoking status | <0.001 | -27.19 | 0.436 | -46.66 | 0.237 | -- | -- | -45.15 | 0.251 |
| Percent of predicted FEV1 | 0.003 | -0.30 | 0.024 | -0.14 | 0.294 | -- | -- | -0.18 | 0.201 |
| Total WBC count | 0.243 | -11.11 | <0.001 | -- | -- | -- | -- | -- | -- |
| Mitochondrial haplogroup | 0.002 | -- | -- | -- | -- | -- | -- | -- | -- |
| L0 vs West Eurasian | -- | 3.15 | 0.750 | -- | -- | 3.63 | 0.713 | -11.84 | 0.285 |
| L1 vs West Eurasian | -- | 3.28 | 0.663 | -- | -- | 4.31 | 0.565 | -7.48 | 0.389 |
| L2 vs West Eurasian | -- | 3.19 | 0.656 | -- | -- | 4.28 | 0.548 | -2.65 | 0.750 |
| L3 vs West Eurasian | -- | 11.98 | 0.088 | -- | -- | 12.85 | 0.066 | 0.013 | 0.999 |
SAGE II denotes the Study of African Americans, Asthma, Genes, & Environment II; BMI, body mass index; FEV1, forced expiratory volume at 1 second; and WBC, white blood count.
*The coefficient of determination (R2) represented the percent of the variation in the outcome variable (i.e., mitochondrial copy number) that could be accounted for by each variable in the univariable analyses.
†Multivariable linear regression model 1 (Model 1) assessed the relationship between asthma and overall mitochondrial copy number in blood. This model adjusted for patient age in years, sex (female = 1, male = 0), proportion of African ancestry per individual (continuous), BMI percentile (continuous), smoking status (past or never smoker = 0, active smoker = 1), and percent of predicted FEV1 (continuous). Complete data were available for 1017 individuals in Model 1, which had an adjusted R2 = 0.087.
‡Multivariable linear regression model 2 (Model 2) assessed the relationship between overall mitochondrial copy number in blood and asthma status as well as mitochondrial haplogroup. Haplogroup was included as categorical variable with West Eurasian as reference level. Only cases with haplogroup L0, L1, L2, L3 and West Eurasian were included in the model. Complete data were available for 1230 individuals in Model 2, which had an adjusted R2 = 0.012.
§Multivariable linear regression model 3 (Model 3) assessed the relationship between overall mitochondrial copy number in blood, and all variables in models 1 and 2. Complete data were available for 953 individuals in Model 3, which had an adjusted R2 = 0.084.
Among individuals with asthma, SABA and ICS medication exposure was inversely associated with mitochondrial copy number (i.e., use associated with lower counts–see Supplementary S5 Table); therefore, asthma medication exposure is unlikely to account for differences in copy number between asthma cases and controls. Inverting the models and predicting asthma status did not diminish the statistical significance of the relationship between asthma and mitochondrial copy number in either the SAPPHIRE or SAGE II cohorts (Supplementary S6 and S7 Tables, respectively). Among SAPPHIRE participants with asthma, mitochondrial copy number was not associated with either asthma severity, as measured by time to exacerbation (Supplementary S8 Table), or asthma control (Supplementary S9 Table).
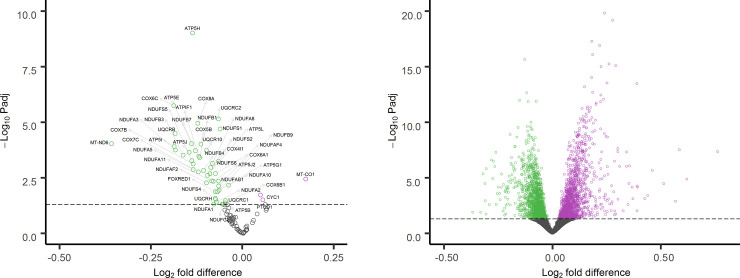
RNA-seq data from SAPPHIRE participants with (n = 197) and without (n = 419) asthma were used to evaluate differences in ETC respiratory complex gene expression (Supplementary S10 Table). In the adjusted expression analysis, 48 of 100 respiratory complex genes were significantly (FDR adjusted P-value <0.05) differentially expressed between individuals with and without asthma. Only 3 (PTCD1, CYC1 and MT-CO1) of the 48 significantly differentially expressed genes were more highly expressed in individuals with asthma; the other 45 had lower expression in individuals with asthma as compared with individuals without asthma (Fig 2A). In contrast, an evaluation of the other 18,484 protein-encoding genes with >10 total reads across all individuals showed a symmetric distribution for genes with significantly higher (n = 1,627) and significantly lower (n = 1,603) expression for individuals with asthma as compared with those without asthma (Fig 2B). The distribution observed for ETC genes differed significantly from the distribution observed for the other protein-encoding genes (P = 3.97x10-21).
Fig 2. Volcano plots of gene expression differences for SAPPHIRE participants with asthma (n = 197) as compared with participants without asthma (n = 419).
Plot A presents expression differences for 100 electron transport chain (ETC) genes derived from RNA-seq of whole blood. Plot B presents expression differences for 18,484 protein-encoding genes not involved in the ETC from the same participant samples. Genes with significantly higher expression in individuals with asthma are shown in purple; genes with significantly lower expression are shown in green; and genes whose expression does not differ significantly between individuals with and without asthma are shown in gray.
Individuals with asthma and high mitochondrial copy number had lower average MitoTracker intensities in blood eosinophils (16,279.8 ± 5,414.7 SD) when compared with controls without asthma (25,693.3 ± 21,181.4 SD), but these differences were not statistically significant (P = 0.525) (Supplementary S11 Table). Individuals with both asthma and an average mitochondrial number had a similar average eosinophil MitoTracker intensity (29,350.5 ± 21,897.1 SD) when compared with controls (P = 0.833). Neutrophils showed the same pattern among individuals with asthma and high mtDNA copy number, asthma with average mitochondrial number, and controls– 13,510.2 (± 5,453.0 SD), 24,581.5 (± 18,482.5 SD), and 23,865.7 (± 21,196.3 SD), respectively. With the exception of monocytes, average MitoTracker intensity was lower in all other cell types assessed (i.e., NK-cells, B-lymphocytes, and T-helper cells) among individuals with asthma and high mitochondrial counts when compared with control individuals.
Discussion
To our knowledge this is the first study to investigate the association between mitochondrial copy number and asthma status. Specifically, individuals with asthma were observed to have on average higher mitochondrial copy numbers when compared with individuals without asthma. Both cohorts comprised individuals from a similar ancestral background, yet the two study populations were largely non-overlapping with respect to age (i.e., <20 years vs. ≥ 18 years at the time of enrollment) and U.S. locale (i.e., West Coast vs. Midwest). Therefore, the association was robust to these differences, as well as adjustment for multiple potential confounders and explanatory variables. Although leukocytes and platelets are the source of mitochondria in peripheral blood, white cell counts were actually inversely associated with intracellular mtDNA numbers and platelet counts were positively associated with mitochondrial copy numbers; these findings have been reported previously [46]. Mitochondrial copy number appeared to be consistently higher among individuals with asthma at all cell count levels, and the relationship between asthma and copy number persisted after adjustment for leukocyte and platelet numbers. Mitochondrial copy number also appeared to be unrelated to asthma severity or level of control. Together these observations suggest that higher mitochondrial copy numbers in WBCs may be intrinsic to asthma.
Higher mitochondrial copy numbers in peripheral blood leukocytes have been associated with either higher or lower risk for a number of other disease conditions, including development of chronic kidney disease (lower risk) [47], cardiovascular disease (lower risk) [48], diabetes (lower risk and later onset) [49,50], rheumatoid arthritis (lower risk) [51], and multiple malignancies, such as breast cancer (greater risk) [52], chronic lymphocytic leukemia/small lymphocytic lymphoma (greater risk) [53,54], and renal cell carcinoma (lower risk) [55]. Given these multiple associations and the substantial overlap in the distribution of counts between asthma cases and controls, it is unlikely that mitochondrial copy number alone will prove useful as a predictive or diagnostic measure for asthma. Nevertheless, understanding the mechanisms driving the higher mitochondrial counts in individuals with asthma may result in better disease phenotyping and treatment. One twin study has suggested that the heritability of mitochondrial copy number in lymphocytes is 65% [55]. Given the likely polygenic etiology of both mitochondrial copy number and asthma, it is conceivable these two traits share one or more genetic risk factors.
Oxidative stress and the overproduction of damaging ROS are considered features of asthma [56,57]. Inflammatory cells, such as neutrophils, macrophages, and eosinophils, are capable of producing significant increases in ROS when activated [58]. Without sufficient enzymatic and non-enzymatic antioxidant activity, these oxygen radicals can promote cellular apoptosis, damage tissue, and increase airway reactivity [59]. Leukocytes from individuals with asthma have been shown to generate greater levels of superoxide (O2-) and have lower antioxidant activity when compared with cells from healthy controls [60,61]. Zmijewski and colleagues found that mitochondrial membrane complexes play a central role in regulating ROS levels, and that selective inhibition of complexes I and III could reduce lipopolysaccharide induced lung injury in mice [62,63]. We found that the expression of both nuclear- and mitochondrial-encoded respiratory complex genes were more likely to be lower among individuals with asthma when compared with individuals without asthma. Hence, the lower expression of these genes among individuals with asthma could be a compensatory response to oxidative stress in order to limit inflammatory tissue damage. Oxidative stress can also stimulate an increase in mitochondrial copy number [14]. With diminished ETC function, a proliferative reaction to ROS production could be a compensatory response to maintain cellular energy levels [15].
We also used flow cytometry to identify whether rhodamine-based mitochondrial dye retention in blood leukocytes was influenced by asthma status and mitochondrial copy number. Cellular uptake of rhodamine-based dyes is influenced by mitochondrial number and membrane potential, such that both higher mitochondrial numbers and functioning electron transport with lower proton leak result in greater dye retention [64]. We obtained white blood cells from individuals with asthma and high mtDNA copy numbers, as well as from individuals with and without asthma with average mitochondrial copy numbers. Among cells capable of high ROS production (i.e., eosinophils and neutrophils), MitoTracker staining was lower in individuals with asthma and high mitochondrial copy number when compared with individuals with average mitochondrial copy numbers (with and without asthma). These differences were not statistically different; however, their consistency suggests that rhodamine dye uptake was more profoundly affected by the loss of mitochondrial membrane potential than by the overall increase in mitochondrial copy number. This would be consistent with the observed lower ETC gene expression among participants with asthma, and it suggests that copy number increases are a response to intracellular oxidative stress and reduced ETC function in individuals with asthma. Since a small number of samples were evaluated, these interpretations should be considered highly speculative; nevertheless, these results are useful for generating hypotheses and providing a direction for future investigations.
It is possible that differences in mitochondrial copy number by asthma status reflect environmental exposures. For example, exposure to volatile organic compounds, such as benzene, have been separately associated with higher blood mitochondrial copy numbers and childhood asthma [65,66]. However, the effect of environmental pollutants may differ by exposure type. Exposure to elemental carbon and particulate matter (PM10) has been associated with lower mitochondrial copy numbers in the blood of adults [67], but is associated with an increased risk of asthma diagnosis in young children [68]. In our study, we did not observe a significant relationship between smoking status and mitochondrial copy number in blood. However, other studies of individuals exposed to either tobacco smoke or high levels of polycyclic aromatic hydrocarbons suggest that the relationship to mitochondrial copy number is complex and may require better accounting of environmental exposures and individual respiratory complex function than performed here [69,70].
Other study limitations must be considered when evaluating this study. First, since we included only individuals who self-identified as African American in our analysis, our findings may not be generalizable to other populations groups, such as Europeans and Asians. However, as many African Americans have both European and African ancestry [71,72], we assessed and did not find that adjusting for genetic ancestry and mitochondrial haplogroup abrogated the significant relationship between asthma status and mitochondrial copy number. Second, PCR-based quantification is the most common and accepted method to assess mitochondrial DNA copy number [23,73], whereas we used the results of massively parallel WGS to estimate mtDNA copy number. Methods have been developed to rapidly calculate copy number based on WGS data and have been employed in large studies [22,74], but there have been few head-to-head comparisons with PCR-based methods. A recent study using exome sequence data showed moderately good agreement with real time PCR methods for quantifying mitochondrial copy number [75]. In a set of 50 samples, we also showed that PCR-based and sequencing-based assessments of mitochondrial copy number were strongly correlated (albeit different in absolute number). Third, we did not specifically evaluate rhodamine-based mitochondrial dye uptake in platelets. The observed significant interaction between platelet counts and asthma status on mitochondrial copy number suggests that platelets may be a source of some of the higher mitochondrial counts seen in individuals with asthma; this deserves further evaluation. Lastly, we did not directly measure intracellular differences in ROS production or mitochondrial respiration between individuals with and without asthma, and this could be an important next step to further support our observations and hypotheses.
In summary, we performed the first assessment of mitochondrial copy number with asthma status and identified an association that was robust to adjustment for multiple potential explanatory and confounding variables. We further evaluated this relationship in the context of mitochondrial ETC gene expression and rhodamine-based dye uptake by leukocytes. From the resulting observations, we hypothesize that mitochondrial copy number is a manifestation of underlying intracellular oxidative stress in individuals with asthma. Our findings are supported by a number of studies showing that stimulated and unstimulated inflammatory cells (i.e., eosinophils, neutrophils, and monocytes) from individuals with asthma produce more ROS when compared with individuals without asthma [76,77], and also by a study showing that exhaled air condensate from children with stable asthma had higher peroxide levels when compared with healthy controls [78]. Inhaled corticosteroid treatment has been shown to restore antioxidant superoxide dismutase enzyme activity in bronchial epithelial cells from individuals with asthma to levels similar to healthy controls [79], and we found that ICS use among individuals with asthma was significantly and negatively associated with mtDNA copy number. Therefore, mitochondrial copy number may prove to be a subtle sign of ongoing inflammation, a measure of therapeutic response, or an indicator for specific types of treatments (e.g., anti-oxidants) among individuals with asthma; however, this will require additional research.
Supporting information
Correlation between mitochondrial DNA (mtDNA) copy number measurements in whole blood using whole genome sequencing (WGS) read depth (y-axis) and real-time PCR quantification (x-axis). Mitochondrial copy number estimated by WGS is on the y-axis and copy number estimated by real-time PCR quantification is on the x-axis.
(TIF)
Subfigures show the relationship for neutrophils (A), monocytes (B), lymphocytes (C), eosinophils (D), and platelets (E). Individuals with asthma are shown in red and individuals without asthma are shown in blue. The interaction P-value is derived from the linear adjusted model for the differences in the relationships by asthma status. The shaded areas represent the 95% confidence interval.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The analysis was performed in SAPPHIRE participants with and without a diagnosis of asthma.
(DOCX)
(DOCX)
Acknowledgments
We gratefully acknowledge the studies and participants who provided biological samples and data for the ATGC as part of the TOPMed program. We are thankful to all those involved in the TOPMed Consortium, especially the members of the Lung, Asthma, and Mitochondrial working groups. A complete list of TOPMed Consortium participants can be found at https://www.nhlbiwgs.org/topmed-banner-authorship. We also thank all of the staff who worked and the SAPPHIRE study, as well as SAGE study collaborators–Harold J. Farber, Emerita Brigino-Buenaventura, Michael A. LeNoir, Kelley Meade, Luisa N. Borrell, Adam Davis, and Fred Lurmann; SAGE laboratory director—Celeste Eng; SAGE study coordinator–Sandra Salazar; and SAGE recruiters–Lisa Caine, Elizabeth Castellanos, Brenda Lopez, and Shahdad Saeedi. Lastly, we thank all of the participants, families and clinic staff who made these studies possible.
Abbreviations
- ACT
Asthma Control Test
- ETC
electron transport chain
- FEV1
percent of predicted forced expiratory volume at 1 second
- MDI
metered dose inhaler
- mtDNA
mitochondrial DNA
- ROS
reactive oxygen species
- SABA
short-acting beta-agonist
- SAPPHIRE
Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity
- SAGE II
Study of African Americans, Asthma, Genes & Environment
- TOPMed
Trans-Omics for Precision Medicine
- WBC
white blood cel
Data Availability
The data used in this study are available through dbGaP but with restrictions as approved by the individual study Institutional Review Boards due to their sensitive nature which includes biogeographic ancestry information. Requesting individual(s) must have IRB approval to use the data, and obtain a letter of collaboration from each study's principal investigator. The IRBs with purview over these data are as follows: Research Administration - IRB Henry Ford Health System 1 Ford Place, 2F Detroit, MI 48202 Reseach_admin@hfhs.org University of California San Francisco - IRB 490 Illinois Street, Floor 6 San Francisco, CA 94143 IRB@ucsf.edu.
Funding Statement
Whole genome sequencing (WGS) for the Trans-Omics for Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for “NHLBI TOPMed: Study of African Americans, Asthma, Genes and Environment (SAGE) Study” (phs000921.v1.p1) was performed at the New York Genome Center (3R01HL117004-02S3). WGS for “NHLBI TOPMed: Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-ethnicity” (phs001467.v1.p1) was performed at University of Washington’s Northwest Genome Center (HHSN268201600032I). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample-identity QC, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1; contract HHSN268201800001I). This work was also supported by the Fund for Henry Ford Hospital (AML, DEL, and LKW); the American Asthma Foundation (LKW, EGB); the Sandler Family Foundation (EGB); the RWJF Amos Medical Faculty Development Program (EGB); Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II (EGB); and the following institutes of the National Institutes of Health: National Institute of Allergy and Infectious Diseases (U19AI077439 to DJE, and R01AI079139 and R01AI061774 to LKW), the National Heart Lung and Blood Institute (R01HL117004 and X01HL134589 to EGB; and R01HL079055, R01HL118267, R01HL141845, and X01HL134589 to LKW), the National Institute of Diabetes and Digestive and Kidney diseases (R01DK064695 and R01DK113003 to LKW), the National Institute of Health and Environmental (R01ES015794 and R21ES024844 to EGB), and the National Institute on Minority Health and Health Disparities (P60MD006902 and R01MD010443 to EGB). GA reported receiving grants from the National Heart Lung and Blood Institute of the National Institutes of Health during the conduct of the current study, as well as personal fees and salary support from Regeneron Pharmaceuticals which is outside of the submitted work. None of the above funding organizations played a role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fanta CH. Asthma. The New England journal of medicine. 2009;360(10):1002–14. 10.1056/NEJMra0804579 [DOI] [PubMed] [Google Scholar]
- 2.Mazurek JM, Syamlal G. Prevalence of Asthma, Asthma Attacks, and Emergency Department Visits for Asthma Among Working Adults—National Health Interview Survey, 2011–2016. MMWR Morb Mortal Wkly Rep. 2018;67(13):377–86. 10.15585/mmwr.mm6713a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zahran HS, Bailey CM, Damon SA, Garbe PL, Breysse PN. Vital Signs: Asthma in Children—United States, 2001–2016. MMWR Morb Mortal Wkly Rep. 2018;67(5):149–55. 10.15585/mmwr.mm6705e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DC. A mitochondrial bioenergetic etiology of disease. The Journal of clinical investigation. 2013;123(4):1405–12. 10.1172/JCI61398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Armada MJ, Riveiro-Naveira RR, Vaamonde-Garcia C, Valcarcel-Ares MN. Mitochondrial dysfunction and the inflammatory response. Mitochondrion. 2013;13(2):106–18. 10.1016/j.mito.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 6.Aravamudan B, Thompson MA, Pabelick CM, Prakash YS. Mitochondria in lung diseases. Expert Rev Respir Med. 2013;7(6):631–46. 10.1586/17476348.2013.834252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelova PR, Abramov AY. Functional role of mitochondrial reactive oxygen species in physiology. Free Radic Biol Med. 2016;100:81–5. 10.1016/j.freeradbiomed.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 8.Diebold L, Chandel NS. Mitochondrial ROS regulation of proliferating cells. Free Radic Biol Med. 2016;100:86–93. 10.1016/j.freeradbiomed.2016.04.198 [DOI] [PubMed] [Google Scholar]
- 9.Mengel-From J, Thinggaard M, Dalgard C, Kyvik KO, Christensen K, Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum Genet. 2014;133(9):1149–59. 10.1007/s00439-014-1458-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu SF, Kuo HC, Tseng CW, Huang HT, Chen YC, Tseng CC, et al. Leukocyte Mitochondrial DNA Copy Number Is Associated with Chronic Obstructive Pulmonary Disease. PLoS One. 2015;10(9):e0138716 10.1371/journal.pone.0138716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpagnano GE, Lacedonia D, Malerba M, Palmiotti GA, Cotugno G, Carone M, et al. Analysis of mitochondrial DNA alteration in new phenotype ACOS. BMC Pulm Med. 2016;16:31 10.1186/s12890-016-0192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. Journal of immunology. 2009;183(8):5379–87. 10.4049/jimmunol.0900228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldogh I, Bacsi A, Choudhury BK, Dharajiya N, Alam R, Hazra TK, et al. ROS generated by pollen NADPH oxidase provide a signal that augments antigen-induced allergic airway inflammation. The Journal of clinical investigation. 2005;115(8):2169–79. 10.1172/JCI24422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. The Biochemical journal. 2000;348 Pt 2:425–32. [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. The international journal of biochemistry & cell biology. 2005;37(4):822–34. 10.1016/j.biocel.2004.09.010 [DOI] [PubMed] [Google Scholar]
- 16.Levin AM, Gui H, Hernandez-Pacheco N, Yang M, Xiao S, Yang JJ, et al. Integrative approach identifies corticosteroid response variant in diverse populations with asthma. The Journal of allergy and clinical immunology. 2019;143(5):1791–802. 10.1016/j.jaci.2018.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nathan RA, Sorkness CA, Kosinski M, Schatz M, Li JT, Marcus P, et al. Development of the asthma control test: a survey for assessing asthma control. J Allergy ClinImmunol. 2004;113(1):59–65. 10.1016/j.jaci.2003.09.008 [DOI] [PubMed] [Google Scholar]
- 18.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, et al. Standardisation of spirometry. EurRespir J. 2005;26(2):319–38. 10.1183/09031936.05.00034805 [DOI] [PubMed] [Google Scholar]
- 19.Borrell LN, Nguyen EA, Roth LA, Oh SS, Tcheurekdjian H, Sen S, et al. Childhood obesity and asthma control in the GALA II and SAGE II studies. American journal of respiratory and critical care medicine. 2013;187(7):697–702. 10.1164/rccm.201211-2116OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gogarten SM, Sofer T, Chen H, Yu C, Brody JA, Thornton TA, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35(24):5346–8. 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding J, Sidore C, Butler TJ, Wing MK, Qian Y, Meirelles O, et al. Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools. PLoS genetics. 2015;11(7):e1005306 10.1371/journal.pgen.1005306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ajaz S, Czajka A, Malik A. Accurate measurement of circulating mitochondrial DNA content from human blood samples using real-time quantitative PCR. Methods in molecular biology. 2015;1264:117–31. 10.1007/978-1-4939-2257-4_12 [DOI] [PubMed] [Google Scholar]
- 24.Malik AN, Shahni R, Rodriguez-de-Ledesma A, Laftah A, Cunningham P. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochemical and biophysical research communications. 2011;412(1):1–7. 10.1016/j.bbrc.2011.06.067 [DOI] [PubMed] [Google Scholar]
- 25.Weissensteiner H, Forer L, Fuchsberger C, Schopf B, Kloss-Brandstatter A, Specht G, et al. mtDNA-Server: next-generation sequencing data analysis of human mitochondrial DNA in the cloud. Nucleic acids research. 2016;44(W1):W64–9. 10.1093/nar/gkw247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissensteiner H, Pacher D, Kloss-Brandstatter A, Forer L, Specht G, Bandelt HJ, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic acids research. 2016;44(W1):W58–63. 10.1093/nar/gkw233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maples BK, Gravel S, Kenny EE, Bustamante CD. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. American journal of human genetics. 2013;93(2):278–88. 10.1016/j.ajhg.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–64. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams LK, Peterson EL, Wells K, Ahmedani BK, Kumar R, Burchard EG, et al. Quantifying the proportion of severe asthma exacerbations attributable to inhaled corticosteroid nonadherence. J AllergyClinImmunol. 2011;128(6):1185–91. 10.1016/j.jaci.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wells KE, Peterson EL, Ahmedani BK, Severson RK, Gleason-Comstock J, Williams LK. The relationship between combination inhaled corticosteroid and long-acting beta-agonist use and severe asthma exacerbations in a diverse population. J Allergy ClinImmunol. 2012;129(5):1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paris J, Peterson EL, Wells K, Pladevall M, Burchard EG, Choudhry S, et al. Relationship between recent short-acting beta-agonist use and subsequent asthma exacerbations. Ann Allergy Asthma Immunol. 2008;101(5):482–7. 10.1016/S1081-1206(10)60286-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cajigal S, Wells KE, Peterson EL, Ahmedani BK, Yang JJ, Kumar R, et al. Predictive Properties of the Asthma Control Test and Its Component Questions for Severe Asthma Exacerbations. J Allergy Clin Immunol Pract. 2017;5(1):121–7 e2. 10.1016/j.jaip.2016.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen-Ramey FC, Bukstein D, Luskin A, Sajjan SG, Markson LE. Administrative claims analysis of asthma-related health care utilization for patients who received inhaled corticosteroids with either montelukast or salmeterol as combination therapy. J ManagCare Pharm. 2006;12(4):310–21. 10.18553/jmcp.2006.12.4.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewels P, Magnusson M, Lundin S, Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–8. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature protocols. 2016;11(9):1650–67. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–5. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- 39.Nicholls DG, Ferguson SJ. Bioenergetics. Fourth edition/ed Amsterdam: Academic Press, Elsevier; 2013. xiv, 419 pages p. [Google Scholar]
- 40.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15(12):550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pereira L, Freitas F, Fernandes V, Pereira JB, Costa MD, Costa S, et al. The diversity present in 5140 human mitochondrial genomes. American journal of human genetics. 2009;84(5):628–40. 10.1016/j.ajhg.2009.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen YS, Torroni A, Excoffier L, Santachiara-Benerecetti AS, Wallace DC. Analysis of mtDNA variation in African populations reveals the most ancient of all human continent-specific haplogroups. American journal of human genetics. 1995;57(1):133–49. [PMC free article] [PubMed] [Google Scholar]
- 43.Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36(3):1–48. [Google Scholar]
- 44.Leek JT. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic acids research. 2014;42(21). 10.1093/nar/gku864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R: A language and environment for statistical computing. [Internet]. R Foundation for Statistical Computing. 2014. Available from: http://www.R-project.org/.
- 46.Knez J, Winckelmans E, Plusquin M, Thijs L, Cauwenberghs N, Gu Y, et al. Correlates of Peripheral Blood Mitochondrial DNA Content in a General Population. American journal of epidemiology. 2016;183(2):138–46. 10.1093/aje/kwv175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tin A, Grams ME, Ashar FN, Lane JA, Rosenberg AZ, Grove ML, et al. Association between Mitochondrial DNA Copy Number in Peripheral Blood and Incident CKD in the Atherosclerosis Risk in Communities Study. J Am Soc Nephrol. 2016;27(8):2467–73. 10.1681/ASN.2015060661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yue P, Jing S, Liu L, Ma F, Zhang Y, Wang C, et al. Association between mitochondrial DNA copy number and cardiovascular disease: Current evidence based on a systematic review and meta-analysis. PLoS One. 2018;13(11):e0206003 10.1371/journal.pone.0206003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong J, McLennan SV, Molyneaux L, Min D, Twigg SM, Yue DK. Mitochondrial DNA content in peripheral blood monocytes: relationship with age of diabetes onsetand diabetic complications. Diabetologia. 2009;52(9):1953–61. 10.1007/s00125-009-1424-6 [DOI] [PubMed] [Google Scholar]
- 50.Lee HK, Song JH, Shin CS, Park DJ, Park KS, Lee KU, et al. Decreased mitochondrial DNA content in peripheral blood precedes the development of non-insulin-dependent diabetes mellitus. Diabetes research and clinical practice. 1998;42(3):161–7. 10.1016/s0168-8227(98)00110-7 [DOI] [PubMed] [Google Scholar]
- 51.Svendsen AJ, Tan Q, Jakobsen MA, Thyagarajan B, Nygaard M, Christiansen L, et al. White blood cell mitochondrial DNA copy number is decreased in rheumatoid arthritis and linked with risk factors. A twin study. J Autoimmun. 2019;96:142–6. 10.1016/j.jaut.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 52.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10(1):62–8. 10.1016/j.mito.2009.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, et al. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112(10):4247–9. 10.1182/blood-2008-05-157974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim C, Bassig BA, Seow WJ, Hu W, Purdue MP, Huang WY, et al. Mitochondrial DNA copy number and chronic lymphocytic leukemia/small lymphocytic lymphoma risk in two prospective studies. Cancer Epidemiol Biomarkers Prev. 2015;24(1):148–53. 10.1158/1055-9965.EPI-14-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100(15):1104–12. 10.1093/jnci/djn213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henricks PA, Nijkamp FP. Reactive oxygen species as mediators in asthma. Pulm Pharmacol Ther. 2001;14(6):409–20. 10.1006/pupt.2001.0319 [DOI] [PubMed] [Google Scholar]
- 57.Sahiner UM, Birben E, Erzurum S, Sackesen C, Kalayci O. Oxidative stress in asthma. World Allergy Organ J. 2011;4(10):151–8. 10.1097/WOX.0b013e318232389e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dworski R. Oxidant stress in asthma. Thorax. 2000;55 Suppl 2:S51–3. 10.1136/thorax.55.suppl_2.s51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comhair SA, Erzurum SC. Redox control of asthma: molecular mechanisms and therapeutic opportunities. Antioxid Redox Signal. 2010;12(1):93–124. 10.1089/ars.2008.2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nadeem A, Chhabra SK, Masood A, Raj HG. Increased oxidative stress and altered levels of antioxidants in asthma. The Journal of allergy and clinical immunology. 2003;111(1):72–8. 10.1067/mai.2003.17 [DOI] [PubMed] [Google Scholar]
- 61.Sackesen C, Ercan H, Dizdar E, Soyer O, Gumus P, Tosun BN, et al. A comprehensive evaluation of the enzymatic and nonenzymatic antioxidant systems in childhood asthma. The Journal of allergy and clinical immunology. 2008;122(1):78–85. 10.1016/j.jaci.2008.03.035 [DOI] [PubMed] [Google Scholar]
- 62.Zmijewski JW, Lorne E, Zhao X, Tsuruta Y, Sha Y, Liu G, et al. Mitochondrial respiratory complex I regulates neutrophil activation and severity of lung injury. American journal of respiratory and critical care medicine. 2008;178(2):168–79. 10.1164/rccm.200710-1602OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zmijewski JW, Lorne E, Banerjee S, Abraham E. Participation of mitochondrial respiratory complex III in neutrophil activation and lung injury. Am J Physiol Lung Cell Mol Physiol. 2009;296(4):L624–34. 10.1152/ajplung.90522.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ronot X, Benel L, Adolphe M, Mounolou JC. Mitochondrial analysis in living cells: the use of rhodamine 123 and flow cytometry. Biol Cell. 1986;57(1):1–7. 10.1111/j.1768-322x.1986.tb00458.x [DOI] [PubMed] [Google Scholar]
- 65.Carugno M, Pesatori AC, Dioni L, Hoxha M, Bollati V, Albetti B, et al. Increased mitochondrial DNA copy number in occupations associated with low-dose benzene exposure. Environ Health Perspect. 2012;120(2):210–5. 10.1289/ehp.1103979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rumchev K, Spickett J, Bulsara M, Phillips M, Stick S. Association of domestic exposure to volatile organic compounds with asthma in young children. Thorax. 2004;59(9):746–51. 10.1136/thx.2003.013680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou L, Zhang X, Dioni L, Barretta F, Dou C, Zheng Y, et al. Inhalable particulate matter and mitochondrial DNA copy number in highly exposed individuals in Beijing, China: a repeated-measure study. Part Fibre Toxicol. 2013;10:17 10.1186/1743-8977-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clark NA, Demers PA, Karr CJ, Koehoorn M, Lencar C, Tamburic L, et al. Effect of early life exposure to air pollution on development of childhood asthma. Environ Health Perspect. 2010;118(2):284–90. 10.1289/ehp.0900916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giordano L, Deceglie S, d'Adamo P, Valentino ML, La Morgia C, Fracasso F, et al. Cigarette toxicity triggers Leber's hereditary optic neuropathy by affecting mtDNA copy number, oxidative phosphorylation and ROS detoxification pathways. Cell Death Dis. 2015;6:e2021 10.1038/cddis.2015.364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pavanello S, Dioni L, Hoxha M, Fedeli U, Mielzynska-Svach D, Baccarelli AA. Mitochondrial DNA copy number and exposure to polycyclic aromatic hydrocarbons. Cancer Epidemiol Biomarkers Prev. 2013;22(10):1722–9. 10.1158/1055-9965.EPI-13-0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rumpel JA, Ahmedani BK, Peterson EL, Yang M, Levin AM, Yang JJ, et al. Genetic ancestry and its association with asthma exacerbations among African American patients with asthma. Journal of Allergy and Clinical Immunology. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–44. 10.1126/science.1172257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rooney JP, Ryde IT, Sanders LH, Howlett EH, Colton MD, Germ KE, et al. PCR based determination of mitochondrial DNA copy number in multiple species. Methods in molecular biology. 2015;1241:23–38. 10.1007/978-1-4939-1875-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qian Y, Butler TJ, Opsahl-Ong K, Giroux NS, Sidore C, Nagaraja R, et al. fastMitoCalc: an ultra-fast program to estimate mitochondrial DNA copy number from whole-genome sequences. Bioinformatics. 2017;33(9):1399–401. 10.1093/bioinformatics/btw835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang P, Lehmann BD, Samuels DC, Zhao S, Zhao YY, Shyr Y, et al. Estimating relative mitochondrial DNA copy number using high throughput sequencing data. Genomics. 2017;109(5–6):457–62. 10.1016/j.ygeno.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 76.Chanez P, Dent G, Yukawa T, Barnes PJ, Chung KF. Generation of oxygen free radicals from blood eosinophils from asthma patients after stimulation with PAF or phorbol ester. The European respiratory journal. 1990;3(9):1002–7. [PubMed] [Google Scholar]
- 77.Vachier I, Chanez P, Le Doucen C, Damon M, Descomps B, Godard P. Enhancement of reactive oxygen species formation in stable and unstable asthmatic patients. The European respiratory journal. 1994;7(9):1585–92. 10.1183/09031936.94.07091585 [DOI] [PubMed] [Google Scholar]
- 78.Jobsis Q, Raatgeep HC, Hermans PW, de Jongste JC. Hydrogen peroxide in exhaled air is increased in stable asthmatic children. The European respiratory journal. 1997;10(3):519–21. [PubMed] [Google Scholar]
- 79.De Raeve HR, Thunnissen FB, Kaneko FT, Guo FH, Lewis M, Kavuru MS, et al. Decreased Cu,Zn-SOD activity in asthmatic airway epithelium: correction by inhaled corticosteroid in vivo. The American journal of physiology. 1997;272(1 Pt 1):L148–54. 10.1152/ajplung.1997.272.1.L148 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation between mitochondrial DNA (mtDNA) copy number measurements in whole blood using whole genome sequencing (WGS) read depth (y-axis) and real-time PCR quantification (x-axis). Mitochondrial copy number estimated by WGS is on the y-axis and copy number estimated by real-time PCR quantification is on the x-axis.
(TIF)
Subfigures show the relationship for neutrophils (A), monocytes (B), lymphocytes (C), eosinophils (D), and platelets (E). Individuals with asthma are shown in red and individuals without asthma are shown in blue. The interaction P-value is derived from the linear adjusted model for the differences in the relationships by asthma status. The shaded areas represent the 95% confidence interval.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
The analysis was performed in SAPPHIRE participants with and without a diagnosis of asthma.
(DOCX)
(DOCX)
Data Availability Statement
The data used in this study are available through dbGaP but with restrictions as approved by the individual study Institutional Review Boards due to their sensitive nature which includes biogeographic ancestry information. Requesting individual(s) must have IRB approval to use the data, and obtain a letter of collaboration from each study's principal investigator. The IRBs with purview over these data are as follows: Research Administration - IRB Henry Ford Health System 1 Ford Place, 2F Detroit, MI 48202 Reseach_admin@hfhs.org University of California San Francisco - IRB 490 Illinois Street, Floor 6 San Francisco, CA 94143 IRB@ucsf.edu.