Abstract
The cumulative of genes carrying mutations is vital for the establishment and development of cancer. However, this driver gene exploring research line has selected and used types of tools and models of analysis unsystematically and discretely. Also, the previous studies may have neglected low-frequency drivers and seldom predicted subgroup specificities of identified driver genes. In this study, we presented an improved driver gene identification and analysis pipeline that comprises the four most widely focused analyses for driver genes: enrichment analysis, clinical feature association with expression profiles of identified driver genes as well as with their functional modules, and patient stratification by existing advanced computational tools integrating multi-omics data. The improved pipeline's general usability was demonstrated straightforwardly for breast cancer, validated by some independent databases. Accordingly, 31 validated driver genes, including four novel ones, were discovered. Subsequently, we detected cancer-related significantly enriched gene ontology terms and pathways, probable drug targets, two co-expressed modules associated significantly with several clinical features, such as number of positive lymph nodes, Nottingham prognostic index, and tumor stage, and two biologically distinct groups of BRCA patients. Data and source code of the case study can be downloaded at https://github.com/hauldhut/drivergene.
Subject terms: Computational biology and bioinformatics, Data integration
Introduction
Cancer is one of the most dangerous diseases that poses as a threat to public health, second only to behind cardiovascular disease1. In recent years, we have gradually realized that genes carrying mutations are critical for the establishment and development of cancer2–6. It has rung the warning bell for cancer researchers to rapidly identify and characterize driver genes in each cancer type that will make enormous contributions to precision medicine in cancer treatment in the future.
Over the past five years, however, researchers tend to disagree on a unifying pipeline for analysis7–15. This was likely because they were used to solve specific problems regarding the cancer of interest; thus, there was no common direction that has been proposed to apply for most types of cancers. For example, iCAGES16 was used to identify driver genes and personalized treatments for clear cell renal cell carcinoma10, and then the identified driver genes were enriched with gene ontology terms and biological pathways using Gene Ontology17 and STRING18, respectively. Besides, a co-expression network was constructed with the WGCNA package19 to analyze the association between co-expressed modules and clinical features. For prostate cancer11, a set of driver identification tools such as OncodriveCLUST20, OncodriveFM21, iCAGES16 and DrGaP22 were used. Then, Gene Ontology17 and STRING18 were employed to annotate the identified driver genes. In addition, WGCNA19 was used to identify the co-expressed module-clinical feature association. For kidney cancer7, driver genes and their cancer pathways were predicted with OncodriveFM21 and OncodriveCLUST20, and then assessed for the association between high-scoring noncoding variants and their regulatory features using three computational tools such as CADD23, FunSeq224 and GWAWA25. For hepatocellular carcinoma8, the identification of driver genes and their pathways was performed with OncodriveFM21, Dendrix26, and the enrichment analysis was additionally performed with Gene Ontology17 and STRING18 data. Finally, Kaplan–Meier survival analyses were done using OncoLnc27 to observe an association between survival rates and each driver gene.
In addition, previous studies have paid close attention to recurrently mutated genes and coding driver genes in cancer patients by using high-frequency-specific tools such as MutSigCV28 and MuSiC29. However, plenty of cancer drivers was mutated at less than 1% of cancer patients14,30. Therefore, previous results may have neglected rare cancer drivers. Since cancer is heterogeneous, stratifying cancer patients with the identified driver genes is a core element in the development of precision medicine for tackling this heterogeneity. However, the previous studies have been almost rarely touched on the taxonomy of cancer patients using somatically mutated genes. This raises a tough challenge to suggest specifically clinical guidance for each cancer patient, except for some recent case studies like prostate cancer11, breast cancer (BRCA)13,14. Even ref.11 and ref.14 used a hierarchical clustering method at a basic level that can totally be improved. In addition, ref.11 also did not clarify how they identified the number of classes of patients and whether the number is optimal.
Our study was collectively developed based on previous studies to overcome the above challenges, using a case study of breast cancer from the popular database METABRIC13. The goal of this work is to help improve the available analysis pipeline, in a more systematic and efficient way, in the identification and analysis of driver genes in future studies. Moreover, in recent years, developments on multi-omics data integration related to BRCA have been useful and efficient in various aspects31–35. This is the basis for us to propose the pipeline relying on integrative multi-omics implementation. For this purpose, in the real data analysis, a total of 35 driver genes was predicted using somatic mutation data, and then 31 driver genes closely related to BRCA were validated and used for subsequent analyses. They were first significantly enriched with gene ontology terms and pathways. The associations between the identified driver genes and their two co-expressed modules with several clinical features such as survival rate, number of positive lymph nodes, Nottingham prognostic index, and cancer stage were also analyzed using gene expression data. Finally, BRCA patients were stratified into two distinct subgroups using copy number data of the identified driver genes with significant differences concerning the clinical features.
Material and methods
Overview of an improved pipeline
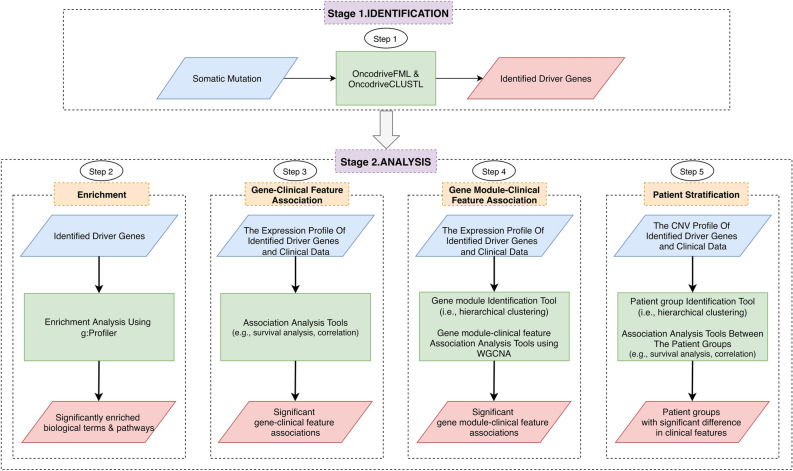
Figure 1 illustrates the improved analysis pipeline to identify and analyze cancer driver genes. The scheme is conceptually straightforward with two stages: identification and analysis. For the identification stage, somatic mutation data is inputted to identify driver genes using the OncodriveCLUSTL36 and OncodriveFML37 tools (Step 1). For the analysis stage, we provide the four most widely focused analyses in the cancer driver exploring studies7–15. Firstly, the identified driver genes will be annotated by the tool g:Profiler38 (Step 2). Secondly, they are further investigated for the association between expression levels of genes and clinical features of interest by statistical tools (Step 3). Thirdly, the association of their functional modules (e.g., co-expressed modules, which are identified with a hierarchical agglomerative clustering method39) with the clinical features is also performed by the tool WGCNA19 (Step 4). Fourthly, patients can be stratified into subgroups on the basis of the copy number profiles of the identified driver genes using a hierarchical agglomerative clustering method39 (Step 5).
Figure 1.
Improved analysis pipeline for identification and analysis of driver genes. The scheme comprises two stages: identification and analysis, in which the former uses the OncodriveFML and OncodriveCLUSTL to identify driver genes with somatic mutation data as input, and the latter performs the four most widely focused analyses to deal with those driver genes. Abbreviation: CNV, Copy number variations.
List of improvements proposed in the work
Selection of driver gene prediction tools
This work recommends the use of OncodriveFML and OncodriveCLUSTL packages for predicting driver genes. In fact, according to the previous studies, OncodriveFM and OncodriveCLUST tools were employed many times7,8,11. However, a potential weakness of the OncodriveFM compared to the OncodriveFML is only the detection of coding driver genes, whereas the OncodriveCLUST also has several weaknesses solved by the OncodriveCLUSTL, like disregarding drivers whose mutations are distributed across the sequence, or requiring a large quantity of observed mutations to ensure a good outcome. From that, the OncodriveFML and OncodriveCLUSTL packages are proven as state-of-the-art methods, even confirmedly outperforming the above two36,37. Moreover, they can not only help researchers find low-frequency and non-coding mutated genes but also were designed as friendly web-based applications.
Selection of enrichment tools
To understand in-depth the underlying biological phenomenon, the enrichment analysis is performed to discover involved biological processes and pathways of identified driver genes. This study recommends the use of g:Profiler package38 for enrichment analysis since it is one of the rich-annotated, friendly web-based, and up-to-date gene enrichment tools. Also, it was recommended to use for enrichment analysis step in a protocol proposed in40. Otherwise, as most previous cancer-driver-exploring studies7–11, Gene Ontology and STRING are both popular annotation resources and tools; thus, users may consider selecting them as alternative tools. Besides, users can also select other alternative options, such as GSEA41, DAVID42,43, IPA44, etc. Although they are advanced, but have their own weaknesses. For instance, the biggest weakness of the GSEA and IPA tools is that they are bulky software which require the user to run as a desktop application. Moreover, the IPA software is a commercial one. In contrast, DAVID is a web-based, but rarely updated tool.
Association analysis of individual genes with clinical features
There are several available correction methods for multiple testing (e.g., Hochberg’s method45, Bonferroni correction46, Holm’s method47, etc.) and correlation methods (i.e., Pearson, Spearman’s rank, Kendall, etc.); consequently, the previous studies selected and computed adjusted P-values (i.e., Q-values) and correlation coefficients unclearly, resulting in making the results ambiguous and running into difficulty in reproducing them. For example, ref.11 did not indicate specifically which P-value adjustment method was chosen. To produce consistent results and be convenient for users, we now built the R package ‘computeC’ (https://github.com/huynguyen250896/computeC), which computes correlation coefficients between each detected driver versus each clinical feature of interest. Then, the obtained P-values are automatically adjusted by the Benjamini–Hochberg procedure48. The problem with Bonferroni, Hochberg and Holm is that they are correction methods for a small number of tests (n). If the sample size n is large, they will reject too many null hypotheses. From that, Benjamini and Hochberg's false discovery rate (FDR) may be a better choice. Likewise, many previous works49–53 related the expression levels of each identified driver gene to prognostic value (e.g., the overall survival of patients), and the genes when P-value ≤ 0.05 (Log-rank test) were considered to define significant association. Again, FDR control is crucial, so we developed the tool ‘geneSA’ (https://github.com/huynguyen250896/geneSA) to automatically do the above task and only preserve the genes if Q-value ≤ 0.05 (Benjamini–Hochberg FDR).
Selection of tools for unsupervised identification of co-expressed gene modules and patient groups
Most co-expression network construction tools are based on unsupervised methods. Besides, as mentioned above, previous studies have not paid enough focus on driver gene-based patient stratification, which may make critical contributions to the design of therapeutic strategies13. Indeed, there is lots of evidence reporting that individual driver genes are mutated predominantly in the samples within one single subtype than in the others, suggesting that those driver genes are recognized as subtype-specific driver genes54. Those driver genes then serve as the important clues to monitor the difference among the explored subtypes54,55 as well as help to develop personalized treatments56. To this end, this study recommends the use of a hierarchical agglomerative clustering method for co-expressed module identification (Fig. 1, Stage 2—Step 4) and patient stratification (Fig. 1, Stage 2—Step 5), which is a common selection from previous studies10,11,14,49,51,52. Furthermore, these previous works performed the hierarchical clustering method using complete linkage and Euclidean distance for the clustering task in co-expression network construction and patient stratification. The work recommends adding a sub-step in this process: selecting the best agglomeration method, which designates how a hierarchical clustering method clusters objects (see the ‘Supplementary File 1′ section for the implementation). When it comes to the agglomerative coefficient, it measures the number of clustering structures found (values closer to 1 suggest strong clustering structure) and specifies the agglomeration method to be used (i.e., one of ‘complete’, ‘average’, ‘single’, or ‘ward’). More specifically, the agglomeration methods Complete/Average/Single-linkage first compute pairwise dissimilarities of the objects in group 1 and group 2. Then, those methods treat the maximum/mean/minimum value of the calculated dissimilarities as the distance between the involved groups, respectively. In contrast, Ward’s minimum variance method first minimizes the total within-cluster error sum of squares, and then, at each stage, iteratively identifies pairs of groups with minimum between-group distance and do the fusion of those two.
Selection of cluster validation
This process aims to assess the quality of the clustering results57 and determine the success or failure of the clustering works58. This work recommends the user to select the Dunn’s index46,59. This is an efficient cluster validation method, and Curtis et al.60 used it to validate the detected subgroups of patients with BRCA from METABRIC. Furthermore, to increase the reliability and robustness of results, the user should combine two or three methods at once. This study additionally recommends the user to select the average Silhouette index61 for two main reasons. First, it is among the best clustering indices62. Second, it was suggested to use in the clustering task of biomedical data63. For instance, it was selected as the index for comparing the quality of clustering results between clustering methods for breast cancer64.
Case study: breast cancer
Datasets and data preprocessing
The BRCA data were downloaded from the cBioPortal for Cancer Genomics (https://www.cbioportal.org)65. It contained the METABRIC BRCA cohort assembled from 2509 primary breast cancer patients with 548 matched normals in the United Kingdom and Canada13. The gene expression microarray data were generated using the Illumina Human v3 microarray for 1904 samples, while the CNVs data were measured on the Affymetrix SNP 6.0 platform for 2173 samples. In addition, 17,272 somatic mutations of 173 genes for 2369 samples were detected on the Illumina HiSeq 2000 platform. R scripts responsible for the work's implementation are provided in the Github repositories (https://github.com/hauldhut/drivergene) (See more detail in Supplementary File 1).
All omics data we pre-processed in the same way as in the reference paper13. Specifically, we only matched the sample labels shared between the gene expression data and clinical data, and the CNVs data and clinical data, and obtained 1904 and 2173 matched patients, respectively.
Identification of driver genes using OncodriveFML and OncodriveCLUSTL
In the first stage, we used two driver identification tools: OncodriveFML and OncodriveCLUSTL (Fig. 1, stage 1; Step 1) to detect potential cancer-related genes of BRCA. OncodriveCLUSTL 1.136 is a sequence-based clustering algorithm to identify significant clustering signals of mutations across genomic regions; meanwhile, OncodriveFML 1.037 is a method designed to analyze somatic mutations across cancer samples to positively select driver genes (i.e., Detection of positive selection in cancer genomes is to identifying genes essential for cancer growth66). Both of them are able to detect cancer driver genes in both coding regions and non-coding regions as well as non-human data. The parameters were set to default values with the exception of the sequencing parameter (targeted sequencing) and the scoring system CADD v1.323 (the latest version at the time of this writing) in the OncodriveFML tool and the selection of ‘concatenate’ option in the OncodriveCLUSTL. A gene is considered a potential driver gene identified by the OncodriveFML and OncodriveCLUSTL when Q-value < 0.25 (Figure S2, Supplementary File 1) and Q-value < 0.01 (Figure S3 Supplementary File 1) (Benjamini–Hochberg FDR), respectively. Then, according to the Cancer Gene Census database (CGC; https://cancer.sanger.ac.uk/census)67, Pereira et al. reference paper13 and Nik Zainal et al. paper14, we verified those identified driver genes. The CGC database provides a list of genes, which have been common in cancer development. Finally, driver genes were said to be bona fide ones if they met the validation process and were served for downstream analysis.
Enrichment analysis using g:Profiler
To deepen our understanding of the potential biological functions of our BRCA-associated genes, the enrichment analysis was performed using g:Profiler to discover involved biological processes and pathways. More specifically, among provided annotation resources, biological process (BP) terms, under the sub-tab ‘Gene Ontology’ (GO), and KEGG pathways, under the sub-tab ‘biological pathways’, were chosen to characterize the identified driver genes functionally. GO terms and pathways were considered to be significantly enriched if a cut-off of Q-value ≤ 0.05 (g:SCS multiple testing correction method).
Individual gene-clinical feature association analysis
In this section, we analyzed associations between clinical features of interest and the identified driver genes. More specifically, gene expression data were used to examine the associations between individual drivers and several familiar clinical features, including survival rates11,13,15,49, numbers of positive lymph nodes11,13, Nottingham prognostic index, and pathologic stages11,15 of 1,904 patients.
For survival rate, we performed a survival analysis for the expression profiles of each driver gene, like in refs.49–53. In brief, given a driver gene, the median expression of that gene was calculated across the patients, then the patients were classified into two groups based on the expression of the gene. The first group ‘up-regulation’ includes patients having the expression of the genes was greater than the median; meanwhile, the second group ‘down-regulation’ includes patients having the expression of the genes was less than the median. Then, a log-rank test in univariate Cox regression analysis with a proportional hazards model68 (i.e., implemented by the ‘geneSA’ function) was used to compare the survival rates between the two groups. Next, hazard ratios (HR) with their 95% confidence intervals (CI), P-values, and Q-values were reported. Driver genes were considered to be significantly associated with survival rate if Q-value ≤ 0.05. Finally, we validated those prognostic driver genes by using KMplot website (https://kmplot.com/analysis/index.php?p=service&start=1)69 with the ‘Survival’ option of ‘OS (n = 1402)’ and the ‘Split patients by’ option of ‘median’, the remaining options were left at default.
Additionally, for the three clinical features (i.e., the number of positive lymph nodes, the Nottingham prognostic index and the pathologic stages), we correlated them with the expression of each driver gene using Spearman's rank correlation method (i.e., implemented by the ‘computeC’ function in R). Driver genes were considered to be significantly associated with positive lymph nodes, Nottingham prognostic index, or pathologic stages if Q-value ≤ 0.05.
Co-expressed module-clinical feature association analysis using WGCNA
We first found the optimal soft threshold β to make the co-expression matrix of the identified driver genes fit a scale-free topology model, then the Topological Overlap Matrix (TOM)-based dissimilarity matrix of the identified driver genes was computed using Pearson’s correlation (see Supplementary File 1: User manual). Next, the co-expressed modules were detected by two steps: (1) an agglomerative clustering algorithm, i.e., Ward’s method70, implemented by R function ‘hclust’ in the package ‘flashClust’71 was used to hierarchically cluster the TOM-based dissimilarity matrix into a gene dendrogram; (2) Then, those genes were distributed to each resulting module with the minimum number of genes was set as ten by the function ‘cutreeDynamic’ implemented in the package ‘dynamicTreeCut’72. Notably, to make the gene network consistent, according to prior studies10,11,53, we suggest that users should choose the ten number of genes existing in each module minimally. Genes with high intra-modular connectivity were considered as hub genes. The association between resulting co-expressed modules and the clinical features was then analyzed using the correlation between the modular eigengene and the clinical features.
Patient stratification
Similar to step 4 (Fig. 1, stage 2), hierarchical agglomerative clustering of all the patients using Ward’s method and Euclidean distance creates a patient dendrogram. Then, the function ‘clValid’73 reports how many patient groups were optimal by connectivity, Dunn’s index, and Average Silhouette algorithm, and the BRCA patients were distributed to each group using the ‘cutree’ with ‘agnes’ functions. To observe the differences, we further performed the analyses between groups in terms of the clinical features. For survival rate, the log-rank test in univariate Cox regression analysis with a proportional hazards model was employed to compare the survival rates of the patients between the involved groups, and Kaplan–Meier curves were then plotted by the R package ‘survminer’74. In addition, the number of positive lymph nodes, the Nottingham prognostic index, and the tumor stages were also compared between the discovered groups. Subsequently, we tested the significance between the given results in terms of these clinical features using the function ‘compareGroups’75. P-value ≤ 0.05 was predefined as statistically significant.
Results
Identification of driver genes using OncodriveFML and OncodriveCLUSTL
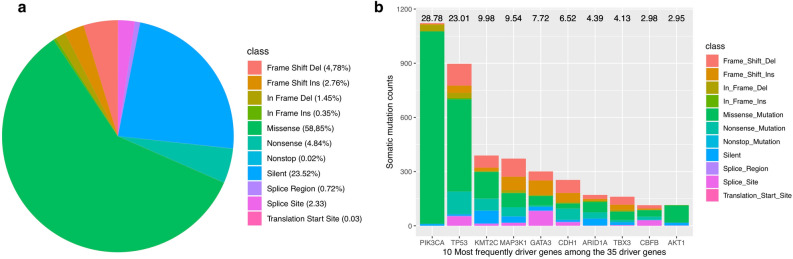
Pathological clinical features (e.g., survival time, tumor stage, number of lymph nodes, and Nottingham prognostic index) were collected (Table 1). All of the 17,272 somatic mutations were used as input to both of the tools. These mutations include 10,165 missense (non-synonymous mutations), 4,063 silent, 402 splice-site, 836 nonsense, 124 splice-region, 5 translation start site, 4 nonstop mutations, and 1,613 insertions or deletions (indels) (Fig. 2a). Of the 1,613 indels, 1,302 is frameshift, 311 is in-frame. A total of 35 unique driver genes were detected by the two tools, in which 30 and 10 driver genes were predicted by OncodriveFML and OncodriveCLUSTL, respectively (Supplementary File 2, Table S1). Both of the tools detected five of them simultaneously, including AKT1, CDH1, ERBB2, ERBB3, and TP53. Among the 35 driver genes, PIK3CA, TP53, KMT2C, MAP3K1, GATA3, CDH1, ARID1A, TBX3, CBFB and AKT1 were the 10 most frequently mutated genes in BRCA, with mutation rates of 28.78%, 23.01%, 9.96%, 9.54%, 7.72%, 6.52%, 4.39%, 4.13%, 2.98% and 2.95%, respectively (Fig. 2b). Interestingly, some of the involved genes were lowly mutated ones in BRCA, including CDKN2A, 0.6% and KRAS, 0.6% (we checked mutation frequencies through cBioPortal website65).
Table 1.
Description of the clinical features of the patients included in the study.
| Clinical features | Overall study cohort (n = 2509) | Data type | |
|---|---|---|---|
| n | % | ||
| Tumor stage | Nominal variable | ||
| 0 | 24 | 1.0 | |
| 1 | 630 | 25.1 | |
| 2 | 979 | 39.0 | |
| 3 | 144 | 5.8 | |
| 4 | 11 | 0.4 | |
| Unknown | 721 | 28.7 | |
| Number of positive lymph nodes | Continuous variable | ||
| 0—≤ 3 | 695 | 27.7 | |
| > 3—≤ 9 | 233 | 9.3 | |
| > 10 | 119 | 4.7 | |
| Unknown | 1462 | 58.3 | |
| Nottingham prognostic index | |||
| 1—≤ 2.4 | 219 | 8.7 | |
| > 1—≤ 3.4 | 555 | 22.1 | |
| > 3.4—≤ 5.4 | 1256 | 50.1 | |
| > 5.4 | 252 | 10.0 | |
| Unknown | 227 | 9.1 | |
| Follow-up months, median* | 116.47 ± 76.11 | Survival | |
| Survival status | |||
| Alive (= 0) | 837 | 33.4 | |
| Death (= 1) | 1144 | 45.6 | |
| Unknown | 528 | 21.0 | |
*Missing information is excluded.
Figure 2.
Characterization of somatic mutations and driver genes in BRCA. (a) Summarization of mutation classes in BRCA. (b) Number and rate of the ten most frequently mutated driver genes.
By comparing the predicted driver genes to TSGene (i.e., a database of tumor suppressor genes; Supplementary File 2, Table S2)76 and ONGene (i.e., a database of oncogenes; Supplementary File 2, Table S3)77, this study detected 13 tumor suppressor genes including MAP2K4, ARID1A, TP53, PTEN, CDH1, NF1, RB1, CDKN2A, FOXO3, SMAD4, BRCA2, BAP1, and MEN1 as well as 11 known oncogenes including PIK3CA, TBX3, CBFB, AKT1, RUNX1 CDH1, PIK3R1, CDKN1B, ERBB2, ERBB3, and KRAS. According to the CGC database and two in vitro experiments13,14, we realized that 31 out of 35 driver genes were genuine (Supplementary File 2, Table S1). On top of that, to the best of our knowledge, several genes, including AHNAK, DNAH2, PDE4DIP, and SYNE1 were detected as driver genes in BRCA for the first time (Supplementary File 2, Table S1).
Enrichment analysis using g:Profiler
As a result, 483 biological processes (Supplementary File 2, Table S4), and 71 pathways (Supplementary File 2, Table S5) were significantly overrepresented for the gene set. Tables 2 and 3 show ten most enriched biological processes and ten most enriched KEGG pathways, respectively. The majority of biological process terms and pathways were widely known as cancer-related, such as “negative regulation of developmental process”34,78, “positive regulation of gene expression”, “regulation of gene expression”34,… for biological processes, whereas for KEGG pathways, cancer-specific ones were ‘Chronic myeloid leukemia’, “Endometrial cancer”, “Non-small cell lung cancer”, … These results further confirmed that cancer driving-genes detected by the two tools OncodriveFML and OncodriveCLUSTL have key functions in cancer in general and breast cancer in particular.
Table 2.
Ten most enriched gene ontology terms. Q-value is computed by using g:SCS multiple testing correction method. Abbreviation: Num, Number of genes involving in the term.
| GO ID | Term name | Num | Q-value |
|---|---|---|---|
| GO:0010604 | Positive regulation of macromolecule metabolic process | 29 | 3.44 × 10–15 |
| GO:0009893 | Positive regulation of metabolic process | 29 | 3.14 × 10–14 |
| GO:0051173 | Positive regulation of nitrogen compound metabolic process | 27 | 2.38 × 10–13 |
| GO:0051093 | Negative regulation of developmental process | 19 | 7.33 × 10–13 |
| GO:0010628 | Positive regulation of gene expression | 24 | 8.55 × 10–13 |
| GO:0031325 | Positive regulation of cellular metabolic process | 27 | 9.02 × 10–13 |
| GO:0045596 | Negative regulation of cell differentiation | 17 | 1.22 × 10–12 |
| GO:0048513 | Animal organ development | 27 | 5.77 × 10–12 |
| GO:0048518 | Positive regulation of biological process | 31 | 1.97 × 10–11 |
| GO:0010468 | Regulation of gene expression | 29 | 4.16 × 10–11 |
Table 3.
Ten most enriched KEGG pathways. Q-value is computed by using g:SCS multiple testing correction method. Abbreviation: Num, Number of genes involving in the pathway.
| Pathway ID | Pathway name | Num | Q-value |
|---|---|---|---|
| KEGG:05213 | Chronic myeloid leukemia | 10 | 5.02 × 10–12 |
| KEGG:01522 | Endometrial cancer | 9 | 2.24 × 10–11 |
| KEGG:05223 | Endocrine resistance | 10 | 5.67 × 10–11 |
| KEGG:05218 | Non-small cell lung cancer | 9 | 1.01 × 10–10 |
| KEGG:05166 | Melanoma | 9 | 1.73 × 10–10 |
| KEGG:01521 | Human T-cell leukemia virus 1 infection | 12 | 3.62 × 10–10 |
| KEGG:05215 | EGFR tyrosine kinase inhibitor resistance | 9 | 4.12 × 10–10 |
| KEGG:05226 | Prostate cancer | 9 | 2.74 × 10–09 |
| KEGG:04218 | Gastric cancer | 10 | 4.55 × 10–09 |
| KEGG:05213 | Cellular senescence | 10 | 7.69 × 10–09 |
Individual gene-clinical feature association analysis
As a result, we found nine genes, including AKT1, KMT2C, KRAS, PIK3R1, PTEN, SMAD4, MAP3K1, MAP2K4 and TBX3, significantly correlated with the survival rate (Supplementary File 2, Table S6). Through the validation process using the KMplot database (Supplementary File 1, Figure S4), we realized that five out of nine driver genes were prognostic ones that we may be interested in best (Table 4). Among them, KRAS with up-regulation levels, and four other genes, including MAP2K4, MAP3K1, PIK3R1, and TBX3 with down-regulation levels significantly associated with shortened lifespan (down-regulation is the reference) (Table 4). This suggested that those driver genes were related to the pathophysiology of breast cancer in varying degrees. They could also help observe the rigor of breast cancer or anticipate the survival rate of patients.
Table 4.
Validated association between the expression of driver genes and the overall survival of BRCA patients.
| Gene | HR (95% CI) | P-value | Q-value |
|---|---|---|---|
| KRAS | 1.20 (1.07–1.35) | 2.30 × 10–03 | 1.19 × 10–02 |
| MAP2K4 | 0.76 (0.67–0.85) | 4.57 × 10–06 | 1.42 × 10–05 |
| MAP3K1 | 0.82 (0.73–0.93) | 1.23 × 10–03 | 7.61 × 10–03 |
| PIK3R1 | 0.84 (0.75–0.95) | 4.37 × 10–03 | 1.93 × 10–02 |
| TBX3 | 0.84 (0.75–0.95) | 4.91 × 10–03 | 1.90 × 10–02 |
One gene, including KRAS with above-median expression levels and four genes, including MAP2K4, MAP3K1, PIK3R1, and TBX3 with below-median expression levels significantly associated with a shortened lifespan. HR is a measure that helps determine whether either of two expression levels of each driver gene will result in an increased (i.e., HR > 1) or decreased (i.e., HR < 1) probability of experiencing the defined event (i.e., death), at any time (below-median expression level is the reference). P-value is computed by the Cox proportional hazard method to test the statistical difference of the given results. Q-value is computed following the Benjamini–Hochberg procedure. HR: hazard ratio. 95% CI: 95% confidence interval.
For the three clinical features (i.e., the number of positive lymph nodes, the Nottingham prognostic index, and the pathologic stages), there are a large number of driver genes negatively correlated with the number of positive lymph nodes (12 genes; A, Supplementary File 2, Table S7), Nottingham prognostic index (16 genes; Supplementary File 2, Table S8), and the pathologic stages (10 genes, Supplementary File 2, Table S9). Similarly, several genes are positively correlated with the number of positive lymph nodes (four genes; Supplementary File 2, Table S10), the Nottingham prognostic index (seven genes; Supplementary File 2, Table S11), and the pathologic stages (three genes; Supplementary File 2, Table S12). A total of 10 genes, including ARID1A, RUNX1, GATA3, TBX3, NF1, MAP2K4, PTEN, SMAD4, MAP3K1, and SF3B1 showed significant associations with all of the three clinical features (Table 5).
Table 5.
Association between the expression of driver genes and the other clinical features.
| Gene | Number of lymph nodes | Nottingham prognostic index | Cancer Stage | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CC | P-value | Q-value | CC | P-value | Q-value | CC | P-value | Q-value | |
| ARID1A | − 0.06 | 0.01 | 0.02 | − 0.13 | 1.31 × 10–8 | 3.69 × 10–8 | − 0.10 | 1.13 × 10–4 | 5.82 × 10–4 |
| RUNX1 | − 0.14 | 1.65 × 10–9 | 5.12 × 10–8 | − 0.25 | 6.20 × 10–29 | 9.61 × 10–28 | − 0.11 | 2.97 × 10–5 | 4.61 × 10–4 |
| GATA3 | − 0.01 | 1.27 × 10–5 | 1.31 × 10–4 | − 0.28 | 1.28 × 10–35 | 3.98 × 10–34 | − 0.12 | 3.83 × 10–5 | 3.95 × 10–4 |
| TBX3 | − 0.10 | 9.10 × 10–6 | 1.41 × 10–4 | − 0.18 | 1.44 × 10–15 | 7.41 × 10–15 | − 0.12 | 6.12 × 10–5 | 4.74 × 10–4 |
| NF1 | − 0.09 | 5.77 × 10–5 | 4.47 × 10–4 | − 0.08 | 2.23 × 10–4 | 3.64 × 10–4 | − 0.07 | 6.36 × 10–3 | 0.02 |
| MAP2K4 | − 0.08 | 4.61 × 10–4 | 1.79 × 10–3 | − 0.22 | 2.04 × 10–21 | 1.58 × 10–20 | − 0.07 | 6.99 × 10–3 | 0.02 |
| PTEN | − 0.08 | 6.02 × 10–4 | 2.07 × 10–3 | − 0.23 | 1.22 × 10–23 | 1.26 × 10–22 | − 0.11 | 2.93 × 10–5 | 9.08 × 10–4 |
| SMAD4 | − 0.06 | 0.01 | 0.03 | − 0.10 | 1.79 × 10–5 | 3.47 × 10–5 | − 0.08 | 1.48 × 10–3 | 5.11 × 10–3 |
| MAP3K1 | − 0.06 | 0.01 | 0.03 | − 0.16 | 6.35 × 10–13 | 2.46 × 10–12 | − 0.09 | 7.16 × 10–4 | 2.77 × 10–3 |
| SF3B1 | 0.09 | 7.83 × 10–5 | 4.84 × 10–4 | 0.07 | 1.78 × 10–3 | 3.76 × 10–3 | 0.10 | 8.48 × 10–5 | 5.26 × 10–4 |
ARID1A, RUNX1, GATA3, TBX3, NF1, MAP2K4, PTEN, SMAD4, MAP3K1 and SF3B1 significantly associated with all of the three clinical features. The column ‘CC’ (i.e., correlation coefficient denoted by r) measures the degree of association between the two variables: each driver gene versus each clinical feature. It takes on values ranging between − 1 and + 1. When r = 0, there is no relationship between the two variables. When r closer to 1, there is an increasingly strong positive (uphill) relationship between the two variables; otherwise, there is an increasingly strong negative (downhill) relationship between the two variables. CC: correlation coefficient.
Co-expressed module-clinical feature association analysis using WGCNA
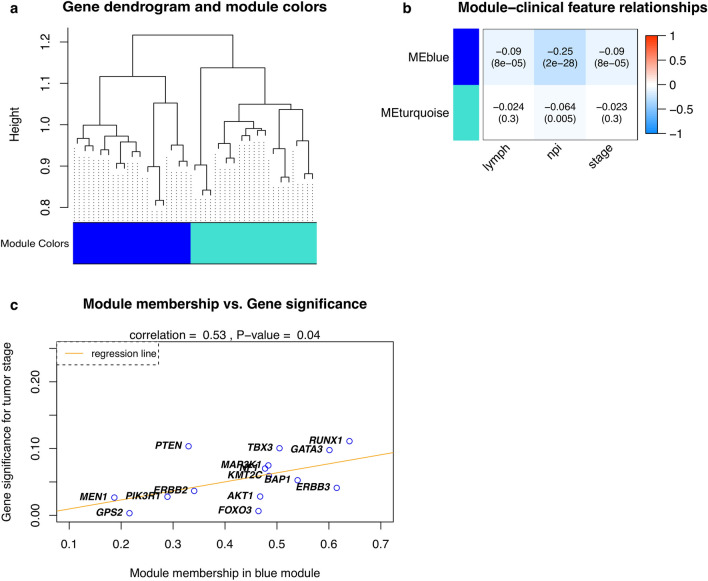
Accordingly, Fig. 3A illustrates the dendrogram of the identified driver genes on their TOM-based dissimilarity (On top of Fig. 3a and Supplementary File 1, Figure S6). The height of the dendrogram indicates dissimilarity of two driver genes, in which low dissimilarities indicate that two driver genes are close (similar), whereas the high dissimilarities imply two driver genes are far apart (dissimilar). In addition, a total of two distinct gene co-expressed modules were found and represented in different colors, and they were arranged from large to small by the number of genes they included (i.e., 15 and 16 genes in the blue and turquoise modules, respectively).
Figure 3.
Co-expression network analysis for module-clinical feature associations. (a) Dendrogram of the identified driver genes on Topology Overlap Matrix-based dissimilarity. The dendrogram height corresponds to the coefficient of dissimilarity matrix that is for every pair of 31 driver genes, in which the low dissimilarities indicate two driver genes are close, whereas the high dissimilarities imply two driver genes are distant apart. Two co-expressed modules were detected and are shown in different colors. (b) Module–feature associations. Each row corresponds to a module eigengene (ME), column to a feature. Each cell contains the corresponding correlation coefficient and P-value. (c) A correlation between gene significance for tumor stage and module membership in the blue module. There is a significantly strong positive correlation between Gene significance and Module membership in this module (r = 0.58, P-value = 0.04). Significant genes (i.e., high Gene significance and high Module membership) in a single module are the ones having a significant association with a clinical feature considered. Abbreviation: lymph: number of positive lymph nodes, and npi: the Nottingham prognostic index, respectively.
Results of the module-clinical feature association analysis (Fig. 3b) indicated that the blue module was significantly negatively correlated (i.e., r < 0 and corresponding P-values ≤ 0.05) with all three clinical features (i.e., numbers of positive lymph nodes, the Nottingham prognostic index, and the tumor stages), whereas the turquoise module showed a significant negative correlation with the Nottingham prognostic index (i.e., r = 0.064 and corresponding P-value = 0.005). In addition, Fig. 3c shows that module membership and gene significance in the blue module for tumor stages are moderately correlated (i.e., r = 0.53 with P-value = 0.04). Also, GATA3, ERRB3, RUNX1, BAP1, and TBX3 were the top five hub genes in the blue module, whereas in the turquoise module was RB1, ZFP36L1, SMAD4, SF3B1, and CDKN1B. RUNX1, GATA3, and TBX3 were the top three significant and module memberships for tumor stages in the blue module. However, we realized that Table 5 and Fig. 3b show extremely modest effects with correlation coefficients close to zero. These results demonstrated that our identified driver genes are weakly correlated with selected clinical features.
Patient stratification
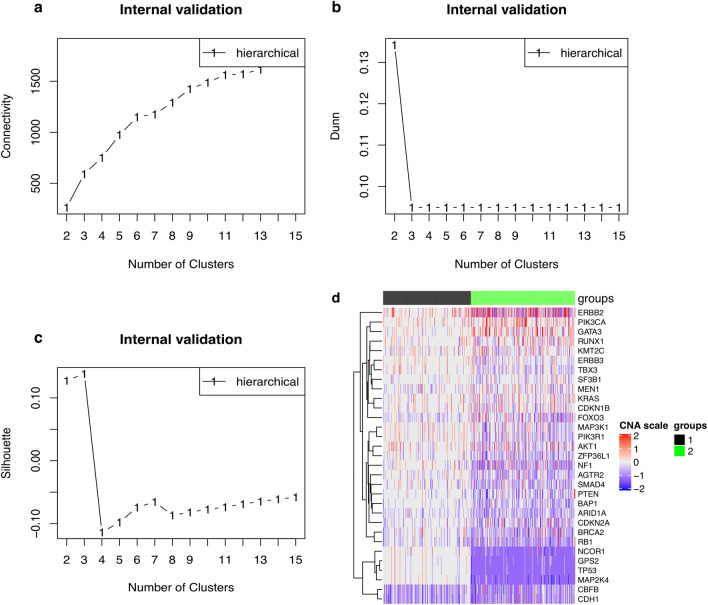
Similar to identifying the above co-expressed modules, here we classified BRCA patients into different groups based on the CNV data of the identified driver genes. Firstly, the agglomerative clustering algorithm (i.e., the Ward algorithm) was used to cluster into patient dendrogram hierarchically. Then, an optimal number of groups was determined by the connectivity (Fig. 4a), the Dunn’s index (Fig. 4b) and the average Silhouette width (Fig. 4c) and the BRCA patients were distributed to each group. Collectively, Fig. 4A–C show that two optimal number of groups for the 2,173 BRCA patients were identified, which implied that clustering the patients into two subgroups was the best solution. Finally, we visualized the given result using a heatmap plot implemented in the function ‘Heatmap’ of the R package, ComplexHeatmap79. The heatmap reports the differences in CNV events between the two patient groups (group 1: 993 patients and group 2: 1180 patients; Fig. 4d).
Figure 4.
Optimal group number detection and difference in CNVs events between the identified groups. (a) Two optimal groups were determined by the connectivity. The connectivity computes the degree of connectedness of a given group partitioning. The connectivity shows the connectedness of a given cluster partitioning and has a value between 0 and infinity. The user should choose a point reaching the most minimized value (y-axis). (b) Two optimal groups were also determined by the Dunn’s index. The Dunn’s index (y-axis) has a value between zero (poorly clustered observations) and infinity (well clustered observations), and the place where the black line of Dunn’s index plot peaks at, which implies that that group number is optimal. (c) Three optimal groups were determined by the Silhouette width. The average Silhouette has a value between -1 (poorly clustered observations) and 1 (well clustered observations), and the place where the black line of the Silhouette plot peaks at, which implies that that group number is optimal. (d) The heatmap indicates the differences in CNV event distribution of two subgroups. The dark red, red, grey, blue and dark blue represent high-level amplification, amplification, copy‐neutral, deletion and high-level deletion, respectively.
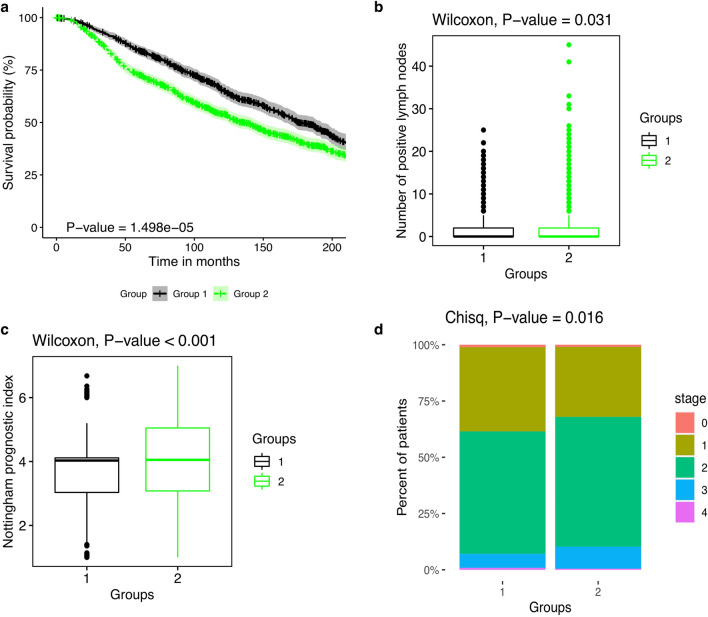
Particularly, the tumors in the second group exhibit significantly worse outcomes (HR is 1.29 with 95% CI (1.15–1.45), P-value < 0.01; group 1 is the reference) (Fig. 5a), higher numbers of positive lymph nodes (P-value = 0.02, Wilcoxon rank-sum test) (Fig. 5b; Supplementary File 2, Table S13), higher Nottingham prognostic index (P-value < 0.01, Wilcoxon rank-sum test) (Fig. 5c; Supplementary File 2, Table S13) and more advanced tumor stages than those in the first group (P-value < 0.01, Pearson’s χ2 test) (Fig. 5d; Supplementary File 2, Table S13).
Figure 5.
Differences between groups of BRCA patients in terms of clinical features. (a) survival rates, (b) the number of positive lymph nodes, (c) the Nottingham prognostic index and (d) cancer stage. Abbreviation: Chisq, Pearson’s χ2 test.
Discussions and conclusion
In this study, the two driver gene identification tools were used to detect the 35 driver genes in 2,369 BRCA samples, in which it showed that the 31 genes overlap with previously published common BRCA driver genes, whereas, to the best of our knowledge, the four leftover genes are yet-to-be-discovered BRCA drivers. The two OncodriveFML and OncodriveCLUSTL tools detected BRCA-related driver genes that are well established in other cancer types, such as KRAS, ARID1A, CDKN2A, MEN1, BAP1, SMAD4. This implies that therapies used in other clinical settings could be appropriate for BRCA with mutations in these genes13. Then, collectively, the five genes show a significant association with survival rate, whereas the ten genes are significantly but weakly correlated with all the three remaining clinical features through analyses between the expression levels of individual driver genes and clinical features. Nevertheless, this result is understandable since it is believed that genes do not function separately but work in concert to affect human health jointly. Indeed, recent studies have shown that individual genes averagely interact with at least four other genes80 and are relevant to 10 biological functions81. Cancer is a complex human disease caused by multiple molecular mechanisms, so gene co-expression networks are a potential approach to detecting a set of cancer-related genes that may be targeted for therapeutic interventions82,83 as well as the identification of hub genes that serve as fundamental roles in cancer. From such, we continue to implement the WGCNA tool to construct weighted driver gene co-expression networks. As a result, two co-expressed modules are detected; among them, one module is significantly negatively associated with the numbers of positive lymph nodes, the Nottingham prognostic index, and the tumor stage, whereas the other is significantly negatively associated with the Nottingham prognostic index. The top five hub genes are correspondingly identified in the two modules, indicating possession of a vast range of interactions with other genes and playing crucial roles in the co-expression network of those genes. Finally, hierarchical clustering analysis of all of the identified driver genes reveals two subgroups of BRCA patients. Further mining the heatmap, we realize that the mutation frequencies of all the driver genes are disparate across the groups: minimal frequencies in the first group and substantial frequencies in the second group. In addition, the second group is significantly poorer than the first group with regard to the three clinical features and patient survival. Therefore, more intensive treatment and frequent follow-up may be necessary for those patients assigned to the second group.
For our work, Fig. 1 describes an improved pipeline to perform the four most widely used analyses developed collectively based on prior works7–15, each of which could be highly adjustable. We envision this pipeline as a general but unlimited solution for cancer researchers wishing to deal with driver genes integrating multi-omics data for which parts of the proposed protocol can permit users to explore any other implementation suitable for their research. At Steps 2 and 4, the workflow implements the g:Profiler and WGCNA tools with basic settings, consistent with the implementation of previous studies. Users totally may refer to ref.40 and ref.84 to perform intensive analyses with them, respectively; however, it may lead to a considerable increase in computation time. For WGCNA, users must face a question: should one choose a ‘signed’ or ‘unsigned’ network? This selection affects how WGCNA treats the correlation of driver genes, in which ‘signed’ considers negative correlation coefficients of pairs of nodes (i.e., pairs of driver genes) to be unconnected, whereas ‘unsigned’ treats positive and negative correlations equally. There is no right answer existing, and previous works could make an inconsistent decision with one another. To avoid any confusion and follow the suggestion of WGCNA19, this study currently recommends a ‘signed’ network. At Step 3, for survival analysis, there are five survival types: overall survival, disease-specific survival, disease-free survival, recurrence-free survival, and distant metastasis-free survival that users can perform with single (i.e., univariate analysis) or combined (i.e., multivariate analysis) clinical features of interest10–13,85 to assess the association between individual driver genes and survival rates. At Steps 3 and 4, the work issues the main focus on the associations between clinical features and changed expression levels of identified driver genes. Based on the belief that gene expression is considered the first level of phenotype affected by the mutation/change on the gene, it is reasonable to investigate how the mutation affects its phenotype7,8,10,12,15,49. For example, a large number of non-coding drivers can regulate the expression of genes and driver genes86,87. However, it is optional, and you can use other omics data types for this step. At Step 5, in the patient stratification process, to limit computational burden, one prior study only selects 20 driver genes with the most frequent changes as input, including the ten most frequently amplified and the ten most frequently deleted driver genes11. Nevertheless, for methylation data, there is alternative to choose driver genes for this step9,12. In this study, we even use all the detected driver genes as input as already mentioned in the reference paper13. Finally, at Steps 3 and 5, in most cases, the distribution of omics data is skewed, so it is recommended that users should priorly select non-parametric methods for testing, such as preferring the Spearman’s rank correlation to the Pearson’s correlation. However, there may be applications in which parametric statistical methods are preferable.
Notably, our pipeline is considered as an improved and refined solution to those mentioned in previous studies. We attempt to make the analysis pipeline for the identification and characterization of driver genes more consistent and reproducible than old investigations. Also, most current driver identification tools are developed to detect genes with coding mutation, whereas the number of non-coding driver identification methods is considerably limited. However, an enormous number of mutations exist in non-coding regions (due to only around 2% of the human genome comprising of protein-coding regions); meanwhile, many previous studies7,8,11 selected coding-driver-specific tools such as OncodriveFM21. In this improved pipeline, we priorly select the two tools OncodriveFML and OncodriveCLUSTL that are two minor cases can identify both non-coding driver genes and infrequently mutated genes in the hope of encouraging researchers interested in this field to take this challenge into account when building a new tool or conducting a driver-gene-related study. For future work, the only way to validate non-coding cancer drivers is to do the literature review manually86,88 and most of the available databases used to validate only coding drivers; therefore, new resources for non-coding drivers should be built as rapidly as possible.
Last, the following limitations are essential to consider before performing the pipeline. Besides, we also make them feasible with the solutions attached. Firstly, cancer is a common disease, and most driver genes are now known, such as breast cancer14. Therefore, we suggest that when applying the proposed scheme for any cancer, we may skip Stage 1 (Fig. 1) and go directly to Stage 2 (Fig. 1) with well-established genes in that cancer type. In contrast, users should maintain Stage 1 to predict new drivers. A hint for the latter case is a combining approach using many driver identification tools simultaneously, for example, as seen in ref.11. Secondly, users can encounter several other limitations with Step 2 (Fig. 1, Stage 2) using g:Profiler that can see ref.40. Thirdly, the work proposes the computeC tool at Step 3 (Fig. 1, Stage 2) to perform correlation analysis using simple methods, but not more sophisticated methods for gene expression data like the R package ‘limma’89, etc. In the future, we will take this issue into account to improve the tool. Finally, a potential restriction when performing survival analysis at Steps 3 and 5 is that the pipeline deals with the censored data in a simple way. More specifically, for missing information, the function ‘coxph’ ignores it automatically, whereas, for end-of-study and loss-to-follow-up censoring, we select the approach of analyzing dichotomized data (see detailed implementations at Supplementary File 1). Consequently, these processes may pose problems to the analysis. Our solution is making assumptions about censoring to selecting the most appropriate statistical methods. For example, if the clinical data whose missing information is limited, the user can remove it; conversely, imputation methods should be taken into consideration.
In summary, we proposed an improved pipeline integrating state-of-the-art computational tools to identify and characterize the driver genes more efficiently and refinedly. Through the successful use of the proposed pipeline, many exciting results were identified, from revealing the four new driver genes, then discovering potential druggable targets as well as the two co-expressed modules, to detecting the two prognostic groups of BRCA patients. Obviously, it is valuable to develop individualized treatments for patients with BRCA in the future. Furthermore, we believe that this success, plus accompanying public codes, demonstrate the efficacy of the work as well as persuade other researchers to use the pipeline.
Supplementary information
Acknowledgements
The authors would like to thank Hoang-Anh Tran at Colby college, US, for proofreading the work.
Author contributions
Q.H.N. conceived the idea and wrote the manuscript, which was edited by D.H.L. Q.H.N. wrote the code, ran the model, and analyzed output data. D.H.L. supervised the work. Q.H.N. and D.H.L. read and approved the final manuscript.
Funding
This research is supported by Vingroup Innovation Foundation (VINIF) in project code VINIF.2019.DA18.
Data availability
The raw data used in the study are available in the cBioportal website: (https://www.cbioportal.org/study/summary?id=brca_tcga_pub). Approval by a local ethics committee was not required, and all the data can be immediately downloaded from the cBioportal website. R packages of computeC and geneSA are available on GitHub (https://github.com/huynguyen250896/computeC and https://github.com/huynguyen250896/geneSA), respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77318-1.
References
- 1. Mortality, G. B. D. & Causes of Death, C. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet388, 1459–1544, 10.1016/S0140-6736(16)31012-1 (2016). [DOI] [PMC free article] [PubMed]
- 2.Barbieri CE, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research, N. Integrated genomic characterization of papillary thyroid carcinoma. Cell159, 676–690, 10.1016/j.cell.2014.09.050 (2014). [DOI] [PMC free article] [PubMed]
- 4.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sato Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 6.Collisson EA, et al. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long C, Jian J, Li X, Wang G, Wang J. A comprehensive analysis of cancer-driving mutations and genes in kidney cancer. Oncol Lett. 2017;13:2151–2160. doi: 10.3892/ol.2017.5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z, Xu L, Sun C. Comprehensive characterization of cancer genes in hepatocellular carcinoma genomes. Oncol. Lett. 2018;15:1503–1510. doi: 10.3892/ol.2017.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Chai JL, Zhongwei L. An integrated analysis of cancer genes in thyroid cancer. Oncol. Rep. 2016 doi: 10.3892/or.2015.4466. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Guo L, Chai L, Ai Z. Comprehensive analysis of driver genes in personal genomes of clear cell renal cell carcinoma. Technol. Cancer Res. Treat. 2019;18:1533033819830966–1533033819830966. doi: 10.1177/1533033819830966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, et al. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol. Med. Rep. 2019;19:2707–2715. doi: 10.3892/mmr.2019.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Guo L, Ai Z. An integrated analysis of cancer genes in clear cell renal cell carcinoma. Future Oncol. 2017;13:715–725. doi: 10.2217/fon-2016-0473. [DOI] [PubMed] [Google Scholar]
- 13.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kong F, et al. Integrative analysis of highly mutated genes in hepatitis B virus-related hepatic carcinoma. Cancer Med. 2020;9:2462–2479. doi: 10.1002/cam4.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong C, et al. iCAGES: integrated CAncer GEnome Score for comprehensively prioritizing driver genes in personal cancer genomes. Genome Med. 2016;8:135–135. doi: 10.1186/s13073-016-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashburner M, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013;29:2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012;40:e169–e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hua X, et al. DrGaP: a powerful tool for identifying driver genes and pathways in cancer sequencing studies. Am. J. Hum. Genet. 2013;93:439–451. doi: 10.1016/j.ajhg.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Y, et al. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15:480–480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie GRS, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat. Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandin F, Upfal E, De Raphael BJ. novo discovery of mutated driver pathways in cancer. Genome Res. 2012;22:375–385. doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016 doi: 10.7717/peerj-cs.67. [DOI] [Google Scholar]
- 28.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dees ND, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Maity AK, Lee SC, Mallick BK, Sarkar TR. Bayesian structural equation modeling in multiple omics data with application to circadian genes. Bioinformatics. 2020;36:3951–3958. doi: 10.1093/bioinformatics/btaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, et al. SALMON: survival analysis learning with multi-omics neural networks on breast cancer. Front. Genet. 2019;10:166–166. doi: 10.3389/fgene.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy Sarkar T, Maity AK, Niu Y, Mallick BK. Multiple omics data integration to identify long noncoding RNA responsible for breast cancer-related mortality. Cancer Inform. 2019;18:1176935119871933–1176935119871933. doi: 10.1177/1176935119871933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen Q-H, Nguyen H, Nguyen T, Le D-H. Multi-omics analysis detects novel prognostic subgroups of breast cancer. Front. Genet. 2020 doi: 10.3389/fgene.2020.574661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham T, et al. Meta-analysis of computational methods for breast cancer classification. Int. J. Intell. Inf. Database Syst. 2020;13:89. doi: 10.1504/IJIIDS.2020.108226. [DOI] [Google Scholar]
- 36.Arnedo-Pac C, Mularoni L, Muiños F, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUSTL: a sequence-based clustering method to identify cancer drivers. Bioinformatics. 2019;35:4788–4790. doi: 10.1093/bioinformatics/btz501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mularoni L, Sabarinathan R, Deu-Pons J, Gonzalez-Perez A, López-Bigas N. OncodriveFML: a general framework to identify coding and non-coding regions with cancer driver mutations. Genome Biol. 2016;17:128. doi: 10.1186/s13059-016-0994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lance GN, Williams WT. A general theory of classificatory sorting strategies: 1. hierarchical systems. Comput. J. 1967;9:373–380. doi: 10.1093/comjnl/9.4.373. [DOI] [Google Scholar]
- 40.Reimand J, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA Cytoscape and EnrichmentMap. Nat. Prot. 2019;14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Protoc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–802. doi: 10.1093/biomet/75.4.800. [DOI] [Google Scholar]
- 46.Bezdek JC, Pal NR. Some new indexes of cluster validity. IEEE Trans. Syst. Man Cybern. Part B (Cybernetics) 1998;28:301–315. doi: 10.1109/3477.678624. [DOI] [PubMed] [Google Scholar]
- 47.Holm S. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 1979;6:65–70. [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodological) 1995;57:289–300. [Google Scholar]
- 49.Jin H, et al. Integrated bioinformatics analysis to identify 15 hub genes in breast cancer. Oncol. Lett. 2019;18:1023–1034. doi: 10.3892/ol.2019.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei J, et al. Integrative analysis of MicroRNA and gene interactions for revealing candidate signatures in prostate cancer. Front. Genet. 2020;11:176. doi: 10.3389/fgene.2020.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou B, Guo R. Integrative analysis of genomic and clinical data reveals intrinsic characteristics of bladder urothelial carcinoma progression. Genes (Basel) 2019;10:464. doi: 10.3390/genes10060464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z, et al. Co-expression network analysis identifies four hub genes associated with prognosis in soft tissue sarcoma. Front. Genet. 2019;10:37. doi: 10.3389/fgene.2019.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, et al. Co-expression network analysis identified gene signatures in osteosarcoma as a predictive tool for lung metastasis and survival. J. Cancer. 2019;10:3706–3716. doi: 10.7150/jca.32092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cyll K, et al. Tumour heterogeneity poses a significant challenge to cancer biomarker research. Br. J. Cancer. 2017;117:367–375. doi: 10.1038/bjc.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alizadeh AA, et al. Toward understanding and exploiting tumor heterogeneity. Nat. Med. 2015;21:846–853. doi: 10.1038/nm.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 57.Maulik U, Bandyopadhyay S. Performance evaluation of some clustering algorithms and validity indices. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24:1650–1654. doi: 10.1109/TPAMI.2002.1114856. [DOI] [Google Scholar]
- 58.Dubes RC, Jain AK. Algorithms for Clustering Data. Upper Saddle River: Prentice-Hall; 1988. [Google Scholar]
- 59.Dunn JC. Well-separated clusters and optimal fuzzy partitions. J. Cybern. 1974;4:95–104. doi: 10.1080/01969727408546059. [DOI] [Google Scholar]
- 60.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rousseeuw P, Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 62.Chaimontree S, Atkinson K, Coenen F. Best clustering configuration metrics: towards multiagent based clustering. In: Cao L, Feng Y, Zhong J, editors. Proceedings of the 6th International Conference on Advanced Data Mining and Applications. Berlin, Heidelberg: Springer; 2010. [Google Scholar]
- 63.Wiwie C, Baumbach J, Röttger R. Comparing the performance of biomedical clustering methods. Nat. Methods. 2015;12:1033–1038. doi: 10.1038/nmeth.3583. [DOI] [PubMed] [Google Scholar]
- 64.Mathews JC, et al. Robust and interpretable PAM50 reclassification exhibits survival advantage for myoepithelial and immune phenotypes. NPJ Breast Cancer. 2019;5:30. doi: 10.1038/s41523-019-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.cd-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martincorena I, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171:1029–1041.e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andersen P, Gill R. Cox's regression model for counting processes: a large sample study. Ann. Stat. 1982 doi: 10.1214/aos/1176345976. [DOI] [Google Scholar]
- 69.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 70.Ward JH. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 71.Langfelder P, Horvath S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012;46:i11. doi: 10.18637/jss.v046.i11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the dynamic tree cut package for R. Bioinformatics. 2007;24:719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]
- 73.Brock G, Pihur V, Datta S, Datta S. clValid : an R package for cluster validation. J. Stat. Softw. 2008 doi: 10.18637/jss.v025.i04. [DOI] [Google Scholar]
- 74.Alboukadel, K. M. K., Przemyslaw, B. Survminer: Drawing Survival Curves Using 'ggplot2', https://cran.r-project.org/package=survminer (2019).
- 75.Subirana I, Sanz H, Vila J. Building bivariate tables: the comparegroups package for R. J. Stat. Softw. 2014;1(12):1–16. [Google Scholar]
- 76.Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Sun J, Zhao M. ONGene: a literature-based database for human oncogenes. J. Genet. Genom. 2016 doi: 10.1016/j.jgg.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 78.Tripathi MK, Misra S, Chaudhuri G. Negative regulation of the expressions of cytokeratins 8 and 19 by SLUG repressor protein in human breast cells. Biochem. Biophys. Res. Commun. 2005;329:508–515. doi: 10.1016/j.bbrc.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 80.Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development. 1997;124:1851–1864. doi: 10.1242/dev.124.10.1851. [DOI] [PubMed] [Google Scholar]
- 81.Miklos GL, Rubin GM. The role of the genome project in determining gene function: insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 82.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J, et al. Application of weighted gene co-expression network analysis for data from paired design. Sci. Rep. 2018;8:622. doi: 10.1038/s41598-017-18705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu J, et al. BCIP: a gene-centered platform for identifying potential regulatory genes in breast cancer. Sci. Rep. 2017;7:45235. doi: 10.1038/srep45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puente XS, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015;526:519–524. doi: 10.1038/nature14666. [DOI] [PubMed] [Google Scholar]
- 87.Weinhold N, Jacobsen A, Schultz N, Sander C, Lee W. Genome-wide analysis of noncoding regulatory mutations in cancer. Nat. Genet. 2014;46:1160–1165. doi: 10.1038/ng.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poulos RC, Sloane MA, Hesson LB, Wong JW. The search for cis-regulatory driver mutations in cancer genomes. Oncotarget. 2015;6:32509–32525. doi: 10.18632/oncotarget.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data used in the study are available in the cBioportal website: (https://www.cbioportal.org/study/summary?id=brca_tcga_pub). Approval by a local ethics committee was not required, and all the data can be immediately downloaded from the cBioportal website. R packages of computeC and geneSA are available on GitHub (https://github.com/huynguyen250896/computeC and https://github.com/huynguyen250896/geneSA), respectively.