Summary
Circadian rhythms influence daily molecular oscillations in gene/protein expression and aspects of biology and physiology, including behaviour, body temperature and sleep–wake cycles. These circadian rhythms have been associated with a number of metabolic, immune and microbial changes that correlate with health and susceptibility to disease, including infection. While light is the main inducer of circadian rhythms, other factors, including the microbiota, can have important effects on peripheral rhythms. The microbiota have been of significant interest to many investigators over the past decade, with the development of molecular techniques to identify large numbers of species and their function. These studies have shown microbial associations with disease susceptibility, and some of these have demonstrated that alterations in microbiota cause disease. Microbial circadian oscillations impact host metabolism and immunity directly and indirectly. Interestingly, microbial oscillations also regulate host circadian rhythms, and the host circadian rhythms in turn modulate microbial composition. Thus, it is of considerable interest and importance to understand the crosstalk between circadian rhythms and microbiota and especially the microbial influences on the host. In this review, we aim to discuss the role of circadian microbial oscillations and how they influence host immunity. In addition, we discuss how host circadian rhythms can also modulate microbial rhythms. We also discuss potential connections between microbes and circadian rhythms and how these may be used therapeutically to maximize clinical success.
Keywords: circadian rhythms, immune system, Microbiota, pattern recognition receptors
Microbial oscillations provide an important feedback system to both central and peripheral circadian clocks through the oscillations in microbial metabolites and ligands, which help to fine‐tune the oscillations and ensure the host can respond more quickly to environmental changes. The oscillations in microbiota are also important for modulating their own functions including chemotaxis, motility, adherence, DNA repair and metabolism. These microbial oscillations are vital for the finetuning of immune responses, which our review discusses in detail.
Abbreviations
- BMAL1
Brain and muscle ARNT‐like 1
- CLOCK
Circadian locomotor output cycles protein kaput
- CRY
Cryptochrome
- DCs
Dendritic cells
- GF
Germ‐free
- GM‐CSF
Granulocyte–macrophage colony‐stimulating factor
- HIF
Hypoxia‐inducible factor
- IECs
Intestinal epithelial cells
- LPS
Lipopolysaccharides
- MYD88
Myeloid differentiation primary response gene 88
- NFIL3
Nuclear factor, Interleukin‐3 regulated
- Nod2
Nucleotide oligomerization domain‐containing protein 2
- PAMPs
Pathogen‐associated molecular patterns
- PER
Period circadian protein homologue
- PPARα
Peroxisome proliferator‐activated receptor alpha
- PRRs
Pattern recognition receptors
- ROR
Retinoic acid receptors
- RORE
ROR response element
- SCFA
Short‐chain fatty acids
- SCN
Suprachiasmatic nucleus
- SPF
Specific pathogen‐free
- TLRs
Toll‐like receptors
- TRIF
TIR‐domain‐containing adaptor‐inducing interferon‐β
- ZT
Zeitgeber
INTRODUCTION
The microbiota and the immune system have co‐evolved to maintain homeostasis and help protect our bodies from pathogens. 1 , 2 Approximately 1000 microbial species reside in the human intestines, encoding a metagenome of trillions of genes, which are over 100 times greater than the human genome, with millions of unique genes. 3 , 4 , 5 , 6 The microbiota influence many important aspects of host physiology, including modulating development, maturation and functions of the innate and adaptive immune systems. This has been highlighted in studies with gnotobiotic (mice with a defined microbial community) or germ‐free (GF; no microbiota) animals, which have a more naïve immune system, particularly in mucosa‐associated lymphoid tissues. 7 The microbial composition can be modulated by a number of factors, including age, diet and host health (Figure 1). Disruption of the balance between the immune system and the microbiota promotes dysbiosis and increases susceptibility to various health issues including cancers, infections, autoimmune diseases and metabolic disorders. 8 , 9 , 10 , 11 Furthermore, microbiota can also modulate responses to immunotherapy, for example anti‐PD1 treatment in cancer. 12 , 13
Figure 1.
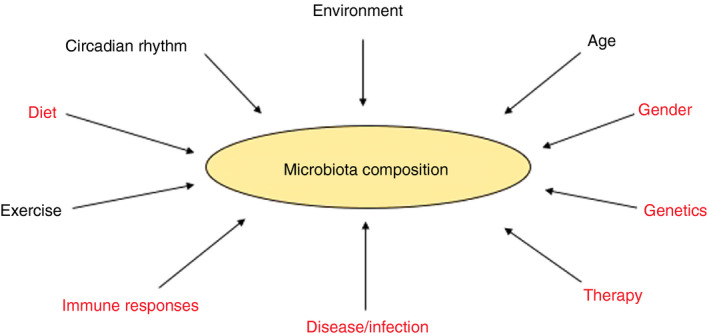
Factors influencing microbial composition. There are many factors which can alter microbial composition. Some factors can also modulate microbial composition and circadian rhythms, as discussed in this review.
Circadian rhythms, referring to daily oscillations in gene activation and repression, as well as biological and physiological processes, can be induced by light, hormones, metabolic cues and the microbiota. The circadian rhythm influences immunity, microbial dynamics and host metabolism. Circadian rhythms are controlled by highly regulated transcription/translation feedback loops (Figure 2) and are reviewed in more detail elsewhere. 14 Light is the main inducer of circadian rhythms via activation of the suprachiasmatic nucleus (SCN). 15 , 16 The SCN comprises ~20,000 specialized neurons within the hypothalamus, which control and co‐ordinate circadian rhythms in exercise, hormones, body temperature and eating. However, additional peripheral rhythms are needed for fine‐tuning the circadian clock, enhancing responses to environmental cues, for example food intake, body temperature and the microbiota. These peripheral rhythms differ from central circadian rhythms in that individual circadian clock components differentially modulate both types of rhythms. Furthermore, peripheral rhythms are subject to different influences which reset individual rhythms and control their outputs. Genes controlling these peripheral rhythms in different tissues also control individual cellular physiology. 17 , 18 Thus, peripheral rhythms temporally control many aspects of metabolism, including glucose homeostasis, lipogenesis and xenobiotic detoxification. 19 , 20 , 21 , 22 , 23 These evolutionarily conserved, cell‐autonomous, biological clocks enable organisms to both anticipate, and adapt to, important environmental changes, aiding in their survival. In circadian studies, zeitgeber (ZT) measurements identify the time from the start of the rhythm to the end of the daily oscillation. In most cases, ZT times coincide with the number of hours after light exposure. For example, ZT12 refers to 12 hours after light exposure.
Figure 2.
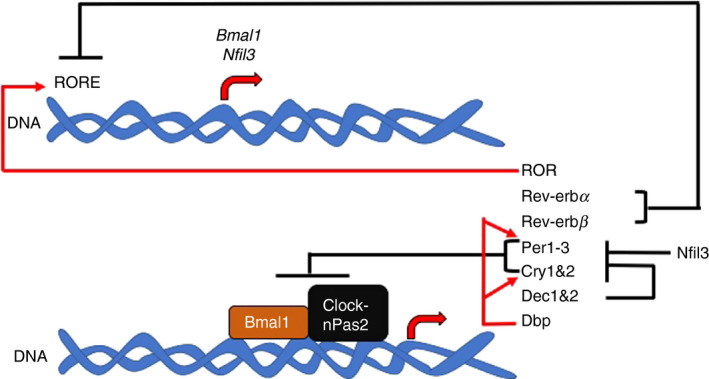
Circadian rhythm gene regulations. Initiation of the circadian rhythm requires activation of the circadian locomotor output cycles protein kaput (Clock), brain and muscle ARNT‐like 1 (Bmal1) and neuronal PAS domain‐containing protein 2 (nPas2) genes. These proteins regulate the expression of multiple genes – period circadian protein homologue 1, 2 and 3 (Per1‐3), cryptochrome 1 and 2 (Cry1 and Cry2), Rev‐erbα (Nr1d1), Rev‐erbβ (Nr1d2) and differentially expressed in chondrocytes protein 1 and 2 (Dec1 and Dec2). These proteins then repress the transcription of the Clock, Bmal1 and Npas2 genes, as well as their own transcription. Furthermore, D‐site of albumin promoter (Dbp) enhances transcription of clock genes, while nuclear factor, interleukin 3 regulated (Nfil3, also known as E4 bp4), suppresses clock gene transcription. Retinoic acid receptors (ROR) can promote Bmal1 and Nfil3 transcription from the ROR response elements (RORE). Red lines = gene activation; black lines = gene repression.
In the intestine, circadian rhythms regulate digestion, including gastric acid production, gut motility and nutrient absorption. Circadian rhythms also affect intestinal stem cell regeneration and mucosal immunity. 24 , 25 , 26 , 27 , 28 Moreover, the microbiota composition and functions depend on the time of day (Figure 3), giving rise to changes in susceptibility to disease. In this review, we discuss the changes in microbial composition and functions associated with circadian rhythms. We also discuss the mechanistic interactions between the microbiota and the immune system, and how they cross‐modulate daily oscillations. We highlight various associations that require further investigation, which will greatly enhance our knowledge and understanding of the cross‐regulation between microbiota, the immune system and circadian rhythms.
Figure 3.
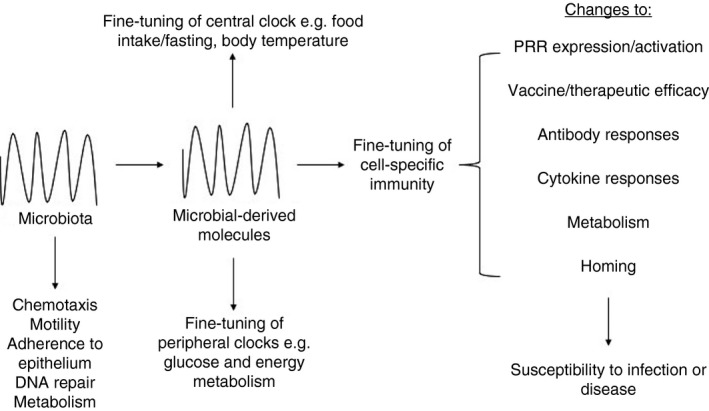
Importance of microbial oscillations on microbial functions. Microbial oscillations provide an important feedback system to both central and peripheral clocks through the oscillations in microbial metabolites, which help to fine‐tune the oscillations and ensure the host can respond more quickly to environmental changes. The oscillations in microbiota are also important for modulating their own functions including chemotaxis, motility, adherence, DNA repair and metabolism. The oscillations in microbiota and their metabolites are also vital for the fine‐tuning of immune responses. Oscillations in microbial ligands recognized by pattern recognition receptors (PRRs) are likely to result in differences in the strength of stimulation. Changes in microbial oscillations also strongly influence vaccine and therapeutic efficacy through modulating the immune functions at different times of day including antibody responses, cytokine responses, metabolic responses and homing of immune cells. Synchronization of therapy to the microbial rhythms is likely to provide the greatest efficacy, limiting susceptibility to infections and disease development.
MICROBIAL RHYTHMS ARE INFLUENCED BY NUTRIENTS AND THEIR AVAILABILITY
Similar to immune cells (discussed in the other review articles within this commissioned review series), the microbiota exhibit circadian rhythms, which modulate many important functions in the host (Figure 3). The microbial circadian dynamic is strongly associated with food intake and thus nutrient availability. In response to dietary glycans, Firmicutes thrive; however, once these glycans have been metabolized, Firmicutes decline in abundance, allowing Bacteroidetes and Verrucomicrobia to expand in response to accessible host glycans. 29 , 30 , 31 These microbial rhythms promote metabolic homeostasis and can be modulated by food availability, which influences susceptibility to metabolic diseases, as discussed next.
Time of food consumption drives microbial rhythmicity
In most laboratory animal facilities, mice are housed in 12‐hour light/dark cycles with food ad libitum. In these conditions, mice consume most of their food in the dark cycle, when they are most active. 32 The composition of the caecal microbiota in mice, maintained under these conditions, changes dynamically in a cyclical manner, as shown by the microbial 16S rRNA sequence. 33 Zarrinpar and colleagues found that the relative abundance of species belonging to the phylum Firmicutes peaked during the feeding period and was reduced during fasting, while species belonging to the phyla Bacteroidetes and Verrucomicrobia showed opposite trends (peaking during fasting and reducing when feeding). 33 Similar changes have been reported by others. 30 , 31 Variations in microbial abundance are also associated with alterations in microbial functions. In the light phase, microbial pathways associated with chemotaxis and motility, which are important for microbiota to adhere to the intestinal wall, are increased. 30 , 34 In contrast, in the dark phase, pathways related to growth, DNA repair and energy metabolism are increased, which benefit the host by the production of growth factors including vitamins. These studies in mice indicate that microbially produced molecules can modulate immunity, as discussed later. Food restriction studies in mice, limiting food availability to either the light or dark cycle, have demonstrated that these microbial oscillations are similarly induced by food intake, regardless of whether in the light or dark cycle. 30 Therefore, food availability and time of consumption drive microbial rhythmicity.
High‐fat diet
Diet can modulate gut microbiota composition in humans. 35 , 36 , 37 One of the most studied diets is a high‐fat diet, which alters the gut microbiota composition, promoting obesity and metabolic dysfunction in mice and humans. 38 , 39 , 40 , 41 , 42 In mice, a high‐fat diet induces blunted microbial diurnal rhythmicity, compared with standard chow‐fed mice. 33 , 42 Interestingly, time‐restricted feeding of mice on high‐fat diet restored cyclical microbial rhythmicity and was able to protect from diet‐induced obesity and metabolic disease. 32 , 33 Importantly, in humans with metabolic disease or obesity, restricting food intake to 8‐12 hours per day resulted in weight loss and improved metabolic parameters. 43 , 44 , 45 , 46 It is unknown whether, as observed in the mice, the gut microbiota in these individuals was similarly altered; however, a recent study showed that the microbiota in individuals with obesity and type 2 diabetes had altered rhythmicity. 47 Subsequent metagenomic analysis identified key altered microbial pathways, which associated with the clinical metabolic features of type 2 diabetes (e.g. fasting blood glucose, insulin resistance, HbA1c). Thus, the altered microbial functions could be used to predict the risk of disease development. Together, these studies highlight the importance of microbial circadian rhythms, and how they modulate disease in humans.
HOST FACTORS ALTER MICROBIAL RHYTHMICITY
Microbial rhythmicity is strongly influenced by nutrient availability. However, when nutrients were administered only intravenously to mice (no oral intake), microbial rhythmicity was still observed, 42 suggesting that host metabolic factors/rhythms, in turn, can also impact microbial rhythmicity. Many host functions are modulated by the circadian rhythm, including the secretion of hormones such as glucocorticoids and neurotransmitters, as well as immune regulation. 48 , 49 Importantly, immune cells express cell‐autonomous circadian rhythms. 50 , 51 , 52 , 53 Circadian rhythms alter immune cell differentiation, such as Th17 cell differentiation, which is regulated by Rev‐erbα‐driven repression of the Rorγt promoter via Nfil3. 54 Homing receptors for immune cells, including CCR7, sphingosine‐1‐phosphate receptor 1, IL7‐R and CXCR4, are also modulated by circadian rhythms driven by the host glucocorticoids. 55 , 56 , 57 , 58 Thus, the circadian rhythm influences both innate and adaptive immune cell migration at different times of day. Together, these circadian rhythm‐modulated immune influences protect the host from disease development. As mentioned earlier, microbiota oscillate and thus may impact immune functions. For example, Th17 cells can be induced by segmented filamentous bacteria (SFB), 59 which are members of the daily oscillating Firmicutes phylum. 30 , 31 , 33 Furthermore, microbiota can modulate intestinal epithelial cell (IEC)‐intrinsic Nfil3 circadian rhythms via type 3 innate lymphoid cells (ILC3). 23 These observations suggest that microbial oscillations not only influence immune development, but also alter the amplitude of the type of immune responses. Thus, further understanding of circadian influences on host:microbial interactions is important.
Host circadian factors modulate microbial rhythmicity
Mice deficient in key circadian rhythm genes, including Bmal1 or Per1/2, have disrupted microbial rhythmicity and composition. 30 , 31 In addition, Clock gene mutant mice, which encode a dominant negative allele (Δ19) that alters the period, precision and persistence of circadian rhythms, also exhibit changes in microbial richness and diversity in the stool microbiota. 60 These studies have demonstrated that the host circadian machinery influences microbial oscillations and composition; however, reciprocally, microbiota modulate host circadian rhythms. A comparison between GF (no microbiota present) and specific pathogen‐free (SPF; microbiota present but free of specific pathogens) mice confirmed that microbiota induce significant diurnal host circadian rhythms in liver hepatocytes. 42 Interestingly, time‐restricted feeding can restore cellular circadian rhythms in Cry1/2‐deficient and liver‐specific Bmal1 and Rev‐erbα/β‐deficient mice, preventing the development of metabolic syndrome and obesity. 61 While microbial rhythms were not investigated in this study, it is likely that they were also altered. Thus, the crosstalk between microbiota and the host clock is important and together influences a number of systemic effects in the host, including fine‐tuning peripheral circadian rhythms.
Androgens modulate microbial composition and rhythmicity
Some diseases have sex biases, 62 suggesting androgens modulate disease development. In both mice and humans, androgens modulate the gut microbiota composition. 31 , 63 , 64 , 65 Interestingly, mouse studies showed that the gut microbiota modulate androgen metabolism, 66 suggesting two‐way crosstalk. Androgens also modulate microbial oscillations. Liang and colleagues 31 found more significant diurnal oscillations in female mice compared with the male mice. Furthermore, there were sex‐dependent differences in microbiota composition in Bmal1‐deficient mice. 31 Sex differences also influence the circadian period and behaviour, including entrainment to light and food. 67 Importantly, sex modulates circadian influences on many host physiological functions including blood pressure, body temperature and adrenal functions. 68 , 69 , 70 Thus, it is imperative that any general conclusions, related to circadian influences, should be drawn from studies of both sexes in animals and humans, especially if a sex bias exists for the disease.
CIRCADIAN MODULATION OF HOST PATTERN RECOGNITION RECEPTORS
Pattern recognition receptors (PRRs) are expressed on a wide variety of cells. These recognize conserved molecular structures that are shared by both pathogens and commensal microbes alike, and are known as pathogen‐associated molecular patterns (PAMPs). 71 Through PRR signalling, the immune system is able to respond to microbial cues to regulate immunity. However, as discussed below, PRRs are also influenced by circadian rhythms (Figure 4), which can have important therapeutic effects, for example in the induction of vaccine‐induced immunity, as discussed later.
Figure 4.
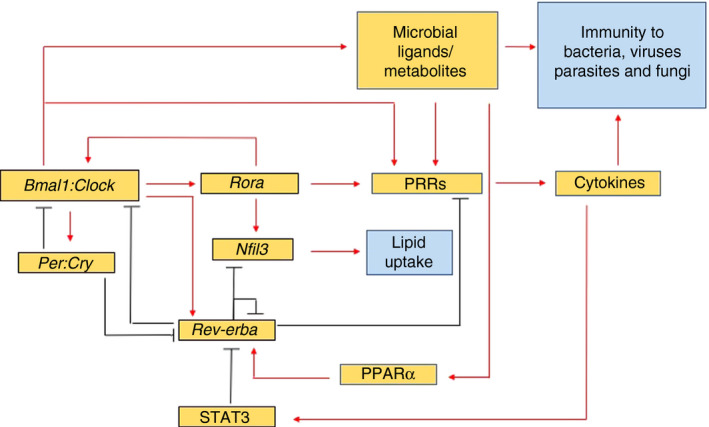
Microbial modulation of circadian rhythms. PRR oscillation (referring to published work in TLRs and Nlrp3) is driven by competition between the Rev‐erbα repressor and the Rorα activator, both of which directly bind and compete for the ROR response element, a DNA binding sequence, present in Bmal1. This competition promotes the oscillation of antimicrobial responses, for example cytokines, which are mediated by downstream NFκB and AP‐1 binding to target genes. Bmal1 binds to an Ebox element (a DNA sequence) in Rev‐erbα activating its transcription, while a DR2 element (another DNA sequence) in Rev‐erbα mediates its autorepression and activation by PPARα. DNA elements (RORE, Ebox, Dbox – not shown) are present in TLR genes, Nod2, circadian genes and many other genes, including many which influence immune processes. Circadian regulation of Nfil3 by the Rev‐erbα repressor and the Rorα activator can also modulate intestinal lipid absorption and transport 23 . Another mechanism of circadian regulation of TLRs has also been identified in peritoneal macrophages, whereby Tlr9 expression is controlled by direct binding of Bmal1 to the Ebox sequence present in the Tlr9 gene 80 . Red lines indicate activation; black lines indicate inhibition. Blue boxes indicate the known effects of microbiota in modulating circadian rhythms.
Circadian modulation of Toll‐like receptor expression
Toll‐like receptors (TLRs) are the most commonly studied family of PRRs, found both at the cell surface and within intracellular endosomes. Each TLR recognizes different PAMPs, including bacterial/viral DNA (unmethylated cytosine‐phosphate‐guanine (CpG) dinucleotides 72 ), lipopolysaccharides (LPS 73 ) and viral RNA. 74 While innate immune receptors prevent pathogen infection, they also have profound beneficial and detrimental impacts on the adaptive immune system, in both health and disease. This dichotomy is context‐dependent and has been observed in cancer, autoimmune and metabolic diseases, as reviewed elsewhere. 11 , 75 , 76 , 77 , 78 While the effects of TLR signalling can be variable, depending on the disease and model studied, it is clear that TLRs are important immune targets for therapy.
TLR expression and activation occur in response to microbial cues and can be modulated by circadian rhythms, resulting in important functional differences in responses to microbes, at different times of day. TLR expression oscillates in macrophages, dendritic cells, B cells, T cells and non‐hematopoetic cells such as IECs. 23 , 28 , 50 , 79 , 80 In macrophages, Silver and colleagues showed that different TLRs oscillate and peak at varying times; for example, the expression of Tlr2 and Tlr6 peaked at ZT19, while Tlr4 peaked earlier at ZT15. 50 As TLR2 can dimerize with TLR6, 81 it is not surprising that these are co‐regulated to peak at the same time, whereas TLR4 is a homodimer and independently regulated and peaks at a different time. TLR4, however, binds to CD14, 82 a co‐receptor. It is currently unclear whether coreceptors of PRRs, like CD14, are also regulated in a circadian manner. Furthermore, to our knowledge, there have been no studies correlating the rhythms of the microbiota with the abundance of microbial products that could activate PRRs. However, disruption to the gut microbiota by antibiotic treatment can dysregulate microbial rhythms, altering host circadian rhythms and TLR expression. 23 , 28 , 34
Mukherji and colleagues found that the depletion of gut microbiota in mice resulted in reduced microbial recognition by TLRs, which dysregulated the IEC circadian clock. 28 This increased ileal corticosterone production, altered glucose homeostasis and induced the development of prediabetes. Subsequently, Wang and colleagues found that microbial regulation of the IEC circadian clock was not directly mediated through the IEC. 23 Previous studies had identified a subepithelial intestinal signalling relay, whereby bacterially mediated TLR activation of DCs induced the secretion of IL‐23, which in turn activated ILC3 cells to secrete IL‐22 and modulate IEC gene expression. 83 , 84 Wang and colleagues further confirmed that this DC‐ILC3‐IEC circuit was required for modulating IEC circadian rhythms (Rev‐erbα and Nfil3) and that the microbiota influenced the amplitude of the circadian rhythms in IECs through STAT3. 23 Together, these data suggested that symbiosis between the microbiota and IEC requires intact circadian oscillations and a complex interplay between many cell types, which we are only beginning to decipher.
Circadian modulation of other PRRs
In addition to TLRs, other PRRs, including the nucleotide oligomerization domain‐containing protein 2 (Nod2) 28 , 85 and the nod‐like receptor pyrin domain‐containing 3 (Nlrp3) protein, a component of the inflammasome, also oscillate. Both Nod2 and Nlrp3 can be activated by bacterial products, regulating the secretion of IL‐1β and IL‐18. 86 , 87 , 88 Interestingly, in mice and humans, circadian oscillation of Nlrp3 is dependent on Rev‐erbα. 89 , 90 In mice, Rev‐erbα repressed Nlrp3 transcription by specifically binding to the promoter region. 90 Inflammasome components modulate responses to infections, as well as alter susceptibility to autoimmunity, neurological and metabolic diseases and cancer. 91 , 92 , 93 , 94 , 95 Thus, further investigating the mechanisms of circadian modulation of PRRs may prove beneficial for therapy in a broad range of diseases.
MICROBE‐DERIVED METABOLITES AND CIRCADIAN RHYTHMS
As microbiota composition and functions oscillate in a circadian manner, metabolites from gut microbiota 34 in both mice and humans also exhibit circadian variations. 96 , 97 , 98 , 99 , 100 Thaiss and co‐authors showed that antibiotic depletion of the microbiota led to the loss of microbial diurnal rhythmicity in mice. 34 In addition, they also found that the microbiota‐induced oscillations in serum ornithine and polyamines could be induced by time‐restricted feeding, in a host that had deficient circadian rhythm (Per1 and Per2 knockout mouse). Together, these data provided pivotal evidence that microbiota modulate the host circadian metabolome. While this study did not investigate the impact of microbiota and their metabolites on host immunity, there are many potential implications for the immune system, especially mucosal immunity.
The gut microbe‐derived short‐chain fatty acids (SCFAs), butyrate, acetate and propionate, modulate immune responses, including the induction of Tregs. 101 , 102 , 103 Interestingly butyrate, which is absent in germ‐free mice, modulates Per2 and Bmal1 rhythms, suggesting that the microbiota indirectly regulate circadian rhythms via their metabolites. 42 These SCFAs typically bind to the G protein‐coupled receptors (Gpr) 41/43, 104 , 105 although other receptors also bind to SCFAs. 106 Many types of host cells utilize SCFAs, and these promote deletion of autoreactive T cells and anti‐inflammatory responses in models of colitis, arthritis, type 1 diabetes and asthma. 104 , 107 , 108 SCFA utilization can also restrict tumour growth and survival. 109 , 110 , 111 Importantly, the effect of SCFAs on some cells, such as monocytes, may not always elicit the same responses from mice and humans. 112 For example, activation of human monocytes with acetate induced attenuated proinflammatory responses; however, activation of mouse monocytes (on the 129/SvEv background) with acetate promoted elevated proinflammatory GM‐CSF, IL‐1α and IL‐1β cytokine secretion. 112 Given that mouse and human monocytes respond differently to acetate, it is possible that additional regulators of SCFA utilization, and the rhythms that modulate them may vary between species. SCFAs such as butyrate can also regulate the integrity of the intestinal barrier by stabilizing hypoxia‐inducible factor (HIF). 113 , 114 Interestingly, HIFα regulates several core clock genes, allowing the cells to respond to changes in oxygen. 115 This has implications, for example, for induction of tumour hypoxia in cancer therapy. Given that microbial metabolites oscillate, there may be similar oscillations in their respective SCFA receptors, or in proteins regulating the downstream responses.
Bile metabolism is also regulated in a time‐of‐day‐dependent manner due to the need to co‐ordinate metabolic responses to food intake, enabling the esterification and absorption of dietary fats and lipids. 116 , 117 In mice, the serum bile acids peak at the beginning and end of their active (dark) phase. The majority of bile acids are reabsorbed by the liver; however, some can enter the colon and be further metabolized, producing secondary metabolites that peak in the serum at the beginning of the dark phase. 118 , 119 Many species belonging to the phyla Firmicutes and Bacteroidetes encode enzymes required to metabolize bile acids, for example bile salt hydrolases. These enzymes regulate hepatic and ileal clock genes and lipid and cholesterol metabolism. 116 , 117 , 120 Interestingly, the intestinal epithelium receptor, CD300lf, has been identified to be the receptor for mouse norovirus. 121 This CD300lf receptor undergoes structural changes upon binding to the bile acid, glycochenodeoxycholic acid, which consequently enhanced the ability of norovirus to bind to the receptor. 122 Recent studies have shown that microbiota can modulate the ability of mouse norovirus, a highly contagious viral pathogen, to infect the host. 123 Furthermore, crosstalk between mouse norovirus and the microbiota can alter susceptibility to autoimmunity, 124 , 125 , 126 , parasitic infection 127 and allergies. 128 As bile acids can be modulated in a circadian manner, it is also possible that invasion of norovirus may be enhanced at different times of the day.
In addition, microbiota synthesize a range of other molecules including xenobiotics, vitamins, polyamines and hydrogen sulphide. Little is known about how circadian rhythms modulate their production and impact on the immune system. Investment in this area would aid our understanding of their important role in health and disease.
IMPACT OF CIRCADIAN MICROBIAL OSCILLATIONS ON IMMUNITY TO INFECTIOUS ORGANISMS
Circadian rhythms, and their regulation, have an important influence on susceptibility to invading pathogens. Bellet and colleagues showed that the immune responses to Salmonella Typhimurium (a common foodborne pathogen) in infected mice were dependent on the time of day. 129 The mice infected in the active phase (ZT16) had reduced pathogen colonization and reduced inflammation, compared with mice infected in the rest phase (ZT4). 129 Host circadian rhythms are required for this response, as Clock mutant mice exhibited arrhythmic colonization and their macrophages had reduced inflammatory responses (IL‐6 and IL‐1β) to LPS (recognized by TLR4, which is known to oscillate 23 , 28 , 50 , 79 , 80 ). Similarly, in wild‐type mice, S.pneumoniae‐induced infection at ZT12 resulted in earlier neutrophilia in the bronchoalveolar lavage fluid, with reduced local (lung) and systemic (blood) bacterial counts at 48 hours post‐infection, compared with infection at ZT0. 130 In addition, intraperitoneal infection with Listeria monocytogenes confirmed that a later rest phase infection at ZT8 was important for enhanced recruitment of Ly6Chi monocytes and stronger antimicrobial immunity, compared with infection at the start of the rest phase ZT0. 53 Similar time‐of‐day‐dependent sensitivities to viral infections have also been reported. 131 , 132 , 133 Interestingly, parasitic infection of mice by oral gavage of Trichuris muris eggs at ZT0 led to enhanced anti‐parasitic IgE titres, Th2 responses and worm expulsion, compared with the mice infected at ZT12, the end of the rest phase. 134 Thus, the optimal immune responsive time to pathogens is not the same for all micro‐organisms. The different time‐dependent oscillations in pattern recognition receptors, or microbial metabolites, influence immune responsiveness. Therefore, priming the immune system at the time of greatest immune responsiveness would be vital for preventing and limiting the spread of specific infectious organisms. To date, it remains unknown as to whether the immune responses to these infectious organisms can be modulated by the commensal microbiota and whether the commensal microbial circadian rhythms are altered in pathogenic infections, which in turn subsequently change the host response. It is clear that further studies are required, especially when multiple factors are analysed in the same study.
APPLICATIONS OF KNOWLEDGE OF MICROBIAL CIRCADIAN RHYTHMS TO THERAPY
The microbiome is an important metabolic ‘organ’, aiding in many unique host functions, which include the efficacious response to a number of therapies. It is well established that the microbiome influences susceptibility to obesity. Microbiota from obese individuals can transfer metabolic dysfunction to another host, while the microbiota from lean individuals, following transfer, protect the recipient from metabolic dysbiosis. 38 , 135 , 136 Metformin, a widely used drug for the treatment of type 2 diabetes, improves insulin sensitivity. Metformin alters both the composition and the function of microbiota, enhancing the therapeutic effects. 137 However, components of the gut microbiota can also be harmful, depending on the context. In colorectal cancer, the intestinal microbiota can influence disease progression 138 , 139 , 140 or aid the therapeutic responses to both chemotherapeutics 141 , 142 and immune checkpoint inhibitors, for example CTLA‐4, PD‐1, PD‐L1. 13 , 143 , 144 Therefore, understanding the mechanisms by which specific microbiota exert their effects (e.g. circadian modulation) could improve therapeutic success. In an animal model of colorectal cancer, induced by azoxymethane and dextran sulphate sodium, Mager and colleagues identified 3 species of tumour‐associated bacteria (Bifidobacterium pseudolongum, Lactobacillus johnsonii and Olsenella sp.) which enhanced immune checkpoint blockade therapy. 145 This enhancement of therapy was orchestrated predominantly by the microbial production of inosine, which binds to the adenosine A2A receptor, promoting Th1 differentiation in the presence of IFN‐γ and consequently anti‐tumour effects. Of note, Lactobacillus johnsonii belongs to the phylum Firmicutes, the abundance of which is known to oscillate daily. 30 , 31 , 33 Lactobacillus johnsonii is also associated with modulating immunity and preventing autoimmune diabetes. 146 , 147 , 148 In a recent study assessing metabolism of 271 orally administered drugs by 76 different human gut bacteria, the authors reported that the microbiota can metabolize many more drugs than previously known. 149 In addition, the drug‐metabolizing function of the microbiota has both local intestinal effects and important systemic effects, especially on the liver. Hepatic drug metabolism is also influenced by circadian rhythm. 150 , 151 , 152 Thus, circadian microbial oscillations (whether through direct or indirect mechanisms) are likely to modulate the ability to metabolize drugs and therefore may have important therapeutic impacts. It should also be noted that peripheral circadian rhythms are often tissue‐specific, and therefore, microbial rhythms could be out of synchrony with the rhythms in different tissues. 17 , 18 This may potentially arise due to the presence of dysbiotic microbiota or stronger peripheral circadian inducers. Thus, it is critically important to identify novel pathways regulating the peripheral rhythms, which are influenced by the microbiota, and synchronize these rhythms, for maximal clinical benefits.
The types of gut microbiota that adhere to the intestinal wall may also oscillate and can play an important role in intestinal immunity. Akkermansia muciniphilia, a mucin‐degrading commensal bacteria belonging to the Verrucomicrobia phylum, 153 is protective against ulcerative colitis, 154 , 155 , 156 type 1 diabetes 157 , 158 , 159 and obesity. 160 Furthermore, Akkermansia muciniphilia improves therapeutic efficacy in cancer patients who are treated with immune checkpoint blockers. 13 Given the oscillation in abundance of Verrucomicrobia, 30 , 31 , 33 appropriately timing therapeutic administration of these bacteria may enhance efficacy. As discussed earlier, it is important to note that bacteria can be beneficial or harmful, depending on the context. Akkermansia muciniphilia, while beneficial in the aforementioned studies, can also promote T‐cell‐mediated inflammation in multiple sclerosis 161 and thus microbial therapies need to be tailored appropriately.
Microbial circadian rhythms modulate long‐term immunity following immunization and vaccination. Microbiota composition can modulate vaccine efficacy to bacteria and viruses in both animal models 162 , 163 , 164 and humans. 165 , 166 , 167 , 168 Evidence for this has been obtained from vaccine studies conducted in GF, as well as antibiotic‐treated mice, resulting in impaired antibody responses. 162 , 163 Similarly, in humans, administration of broad‐spectrum antibiotics also reduced the immunogenicity of the rotavirus vaccine. 167 Rhythmic microbial and PRR rhythms are also likely to influence vaccine efficacy, although this remains to be studied. Host circadian genes can also alter the vaccine efficacy. Per2, one of the major circadian gene family members, 169 controls TLR9‐mediated innate and adaptive immune responses to infection and sepsis. 80 Moreover, Per2 also controls vaccine immune responses to TLR9 ligand‐adjuvanted immunization using CpG (a bacterial DNA, which is a TLR9 ligand and a common vaccine adjuvant 72 ). 80 These studies have provided evidence for a vital link between circadian rhythms and TLR signalling, which promotes enhanced vaccine efficacy. Furthermore, in Cry1/Cry2 double knockout mice, there are more circulating mature B cells that secrete higher titres of antibodies to T‐cell‐independent antigenic stimulation (4‐hydroxy‐3‐nitrophenyl‐acetyl (NP)‐conjugated Ficoll). 170 This again highlights host rhythms influencing immunity. Oscillations in antibody responses have also been seen in other studies, including vaccination studies. 56 , 171 , 172 , 173 , 174 A randomized trial investigating the efficacy of the trivalent inactivated influenza virus demonstrated that vaccine administration in the morning induced higher antibody titres than in the afternoon 173 ; however, the oscillating antibodies post‐vaccination are likely to be viral strain‐specific 171 and influenced by sex. 172 As TLR9 is highly expressed on B cells 175 and requires downstream MyD88 signalling to mediate functional changes, 176 , 177 it is likely that oscillating TLR9 levels may influence B‐cell antibody responses; however, this has yet to be fully investigated. This also applies to autoantibody production, which can be regulated by TLR expression. 178 , 179 , 180 Murine studies, in which peptide immunization was performed at different times of day, showed that this influenced both T‐cell immunity and susceptibility to disease. 56 , 181 , 182 Furthermore, immune‐intrinsic circadian rhythms were vital for this effect. 56 , 182 Thus, both Bcell and Tcell responses to antigen can be modulated by circadian rhythms. Further understanding of how the microbial rhythms may influence antigen‐specific immunity may prove even more beneficial.
While the role of microbiota‐mediated modulation of host immunity using antibiotic treatment or GF mice has been extensively studied, the impact of pro‐ or pre‐biotics or bacteriophages (bacteria‐targeting viruses) on either host or microbial circadian rhythms is not currently known. Studying bacteriophages and their role in modulating microbial circadian rhythms may therefore also prove to be an important research field, particularly as bacteriophages can adhere to the mucus layer and prevent infection from pathogenic bacteria. 183 , 184
CHALLENGES IN STUDYING MICROBIAL RHYTHMICITY
Reporting microbial rhythmicity
A common challenge facing researchers studying the microbial rhythmicity lies in the interpretation and presentation of the data. Relative microbial abundance is often reported in microbial circadian oscillation studies, referring to the proportion of specific bacteria within the total bacteria sequenced. However, this can potentially under‐ or over‐estimate changes in the microbiota composition and thus the inferred absolute microbial abundance is sometimes used. The inferred absolute microbial abundance is determined by multiplying the 16S rRNA copy number by the relative abundance within a given sample. Studies using different methods could lead to discrepancies between different reports of the abundance of bacteria which oscillate at different times of day. For clarity, we suggest that the reporting of these microbial rhythms should be presented both ways. An alternative method would be to conduct both microbial sequencing and a culture‐based approach (on selective media) to identify numbers of specific bacteria; however, this approach is not infallible as most gut bacteria cannot be cultured. Further developments are needed in this area before true microbial counts for all bacteria can be determined.
Limited studies in both sexes
As previously mentioned, sex influences the microbial composition and rhythmicity. While some studies do investigate sex biases, many more do not. Thus, studies should be conducted in both males and females to identify the role of androgens/oestrogens in modulating circadian rhythms.
Evaluating all variables
There are many factors, already discussed, that can influence microbial composition and circadian rhythms, including light, diet, therapies (e.g. antibiotics), genetics, hormones, sleep patterns, behaviour and disease development. In many studies, it is difficult to investigate all variables; however, in mice, many of these can be controlled but not all are studied. This is the same in human studies, where food intake is restricted 44 , 45 , 46 but exercise, genetics, light exposure, among other factors, are not considered. While these studies highlight the importance of circadian rhythms in health and disease, future studies need to consider additional factors and their contribution, if any, to the results obtained. Larger study cohorts and controlled facilities, where all participants reside and are maintained in the same light cycles and on the same diet, are likely to be required.
SUMMARY
In this review, we have discussed time‐of‐day‐dependent differences in the microbiota and how they may impact host metabolism and immunity. While it is known the microbiota are associated with the development and progression of many diseases, little is known of the influence that circadian oscillations have on the microbiota, which may also modulate disease susceptibility. We hope to have provided insight into some potential future directions in relation to microbial oscillations and how they can directly, or indirectly, regulate host immune responses in different disease settings. Given the significant influence that circadian rhythms have on host immunity, it is important that more studies consider the timing of experiments and administration of therapies. By better understanding circadian influences, we may maximize clinical success by targeting the cells/microbiota of interest, at the time they are most vulnerable. This could potentially enable reduced drug concentrations to be used, which would also limit toxic effects. Understanding these rhythms in both males and females is vital, given the importance of androgens in immunity and in shaping the host microbial communities. While the vast majority of work outlined in this review has focused on microbiota in the gut, it will also be important to study the circadian effects on microbiota in different locations that include the skin, lungs, as well as other mucosal surfaces. It is likely that most commensal microbiota, in these different locations, modulate immunity in a time‐of‐day‐dependent manner. This is a continually expanding new field and we look forward to gaining further insight, which may improve efficacy of current, as well as new therapies, for disease prevention and treatment.
FUNDING INFORMATION
This work was supported by a Medical Research Council Career Development Award (MR/T010525/1) to JAP; a NIH Research Project Grant (NIH RO1 HD097808); and a Molecular Genetic & Diabetes Mouse Core of Yale Diabetes Center grant to LW (NIH P30 DK 045735).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
OTHER ARTICLES PUBLISHED IN THIS REVIEW SERIES
It’s Time to Think about Circadian Rhythms. Immunology 2020, 161: 259‐260.
Circadian rhythms in innate immunity and stress responses. Immunology 2020, 161: 261‐267.
Circadian rhythms in adaptive immunity. Immunology 2020, 161: 268‐277.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.
REFERENCES
- 1. Simon AK, Hollander GA, McMichael A. Evolution of the immune system in humans from infancy to old age. Proc Biol Sci 2015;282:20143085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cheng J, Ringel‐Kulka T, Heikamp‐de Jong I, Ringel Y, Carroll I, de Vos WM, et al Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016;10:1002–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 2014;32:834–41. [DOI] [PubMed] [Google Scholar]
- 6. Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 2016;14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012;336:1268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015;26:26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol 2019;4:35–45. [DOI] [PubMed] [Google Scholar]
- 10. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med 2019;25:377–88. [DOI] [PubMed] [Google Scholar]
- 11. Pearson JA, Agriantonis A, Wong FS, Wen L. Modulation of the immune system by the gut microbiota in the development of type 1 diabetes. Hum Vaccin Immunother 2018;14:2580–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 14. Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 2017;18:164–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meijer JH, Groos GA, Rusak B. Luminance coding in a circadian pacemaker: the suprachiasmatic nucleus of the rat and the hamster. Brain Res 1986;382:109–18. [DOI] [PubMed] [Google Scholar]
- 16. Meijer JH, Rusak B, Gänshirt G. The relation between light‐induced discharge in the suprachiasmatic nucleus and phase shifts of hamster circadian rhythms. Brain Res 1992;598:257–63. [DOI] [PubMed] [Google Scholar]
- 17. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002;109:307–20. [DOI] [PubMed] [Google Scholar]
- 18. Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al Extensive and divergent circadian gene expression in liver and heart. Nature 2002;417:78–83. [DOI] [PubMed] [Google Scholar]
- 19. Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR‐domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 2006;4:25–36. [DOI] [PubMed] [Google Scholar]
- 20. Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci USA 2008;105:15172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 2010;466:627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gachon F, Leuenberger N, Claudel T, Gos P, Jouffe C, Fleury Olela F, et al Proline‐ and acidic amino acid‐rich basic leucine zipper proteins modulate peroxisome proliferator‐activated receptor alpha (PPARalpha) activity. Proc Natl Acad Sci USA 2011;108:4794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL. Science 2017;357:912–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Larsen KR, Moore JG, Dayton MT. Circadian rhythms of acid and bicarbonate efflux in fasting rat stomach. Am J Physiol 1991;260:G610–G614. [DOI] [PubMed] [Google Scholar]
- 25. Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res 2009;50:1800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoogerwerf WA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, et al Rhythmic changes in colonic motility are regulated by period genes. Am J Physiol Gastrointest Liver Physiol 2010;298:G143–G150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep 2013;3:996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013;153:812–27. [DOI] [PubMed] [Google Scholar]
- 29. Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 2013;23:1038–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thaiss CA, Zeevi D, Levy M, Zilberman‐Schapira G, Suez J, Tengeler AC, et al Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014;159:514–29. [DOI] [PubMed] [Google Scholar]
- 31. Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci USA 2015;112:10479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, et al Time‐restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high‐fat diet. Cell Metab 2012;15:848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 2014;20:1006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, et al Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 2016;167(6):1495–1510. [DOI] [PubMed] [Google Scholar]
- 35. De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet‐induced extinctions in the gut microbiota compound over generations. Nature 2016;529:212–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013;341:1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sonnenburg JL, Bäckhed F. Diet‐microbiota interactions as moderators of human metabolism. Nature 2016;535:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, et al Effects of diurnal variation of gut microbes and high‐fat feeding on host circadian clock function and metabolism. Cell Host Microbe 2015;17:681–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gill S, Panda S. A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab 2015;22:789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gabel K, Hoddy KK, Haggerty N, Song J, Kroeger CM, Trepanowski JF, et al Effects of 8‐hour time restricted feeding on body weight and metabolic disease risk factors in obese adults: A pilot study. Nutr Healthy Aging 2018;4:345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hutchison AT, Regmi P, Manoogian ENC, Fleischer JG, Wittert GA, Panda S, et al Time‐restricted feeding improves glucose tolerance in men at risk for type 2 diabetes: a randomized crossover trial. Obesity 2019;27:724–32. [DOI] [PubMed] [Google Scholar]
- 46. Wilkinson MJ, Manoogian ENC, Zadourian A, Lo H, Fakhouri S, Shoghi A, et al Ten‐hour time‐restricted eating reduces weight, blood pressure, and atherogenic lipids in patients with metabolic syndrome. Cell Metab 2020;31(1):92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, et al Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 2020;28(2):258–72. [DOI] [PubMed] [Google Scholar]
- 48. Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000;289:2344–7. [DOI] [PubMed] [Google Scholar]
- 49. Logan RW, Sarkar DK. Circadian nature of immune function. Mol Cell Endocrinol 2012;349:82–90. [DOI] [PubMed] [Google Scholar]
- 50. Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun 2012;26:407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hemmers S, Rudensky AY. The Cell‐Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell Rep 2015;11:1339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bollinger T, Leutz A, Leliavski A, Skrum L, Kovac J, Bonacina L, et al Circadian clocks in mouse and human CD4+ T cells. PLoS One 2011;6:e29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 2013;341:1483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, et al TH17 cell differentiation is regulated by the circadian clock. Science 2013;342:727–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang JE, Zhang D, et al Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 2012;37:290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Druzd D, Matveeva O, Ince L, Harrison U, He W, Schmal C, et al Lymphocyte circadian clocks control lymph node trafficking and adaptive immune responses. Immunity 2017;46:120–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, et al Circadian expression of migratory factors establishes lineage‐specific signatures that guide the homing of leukocyte subsets to tissues. Immunity 2018;49(6):1175–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimba A, Cui G, Tani‐Ichi S, Ogawa M, Abe S, Okazaki F, et al Glucocorticoids drive diurnal oscillations in T cell distribution and responses by inducing interleukin‐7 receptor and CXCR4. Immunity 2018;48(2):286–98. [DOI] [PubMed] [Google Scholar]
- 59. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009;139:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Voigt RM, Summa KC, Forsyth CB, Green SJ, Engen P, Naqib A, et al The circadian clock mutation promotes intestinal dysbiosis. Alcohol Clin Exp Res 2016;40:335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaix A, Lin T, Le HD, Chang MW, Panda S. Time‐restricted feeding prevents obesity and metabolic syndrome in mice lacking a circadian clock. Cell Metab 2018;29:303–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Regitz‐Zagrosek V. Sex and gender differences in health. Science & Society Series on Sex and Science. EMBO Rep 2012;13:596–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al Gender bias in autoimmunity is influenced by microbiota. Immunity 2013;39:400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Markle JG, Frank DN, Mortin‐Toth S, Robertson CE, Feazel LM, Rolle‐Kampczyk U, et al Sex differences in the gut microbiome drive hormone‐dependent regulation of autoimmunity. Science 2013;339:1084–8. [DOI] [PubMed] [Google Scholar]
- 65. Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, et al The mouse microbiome is required for sex‐specific diurnal rhythms of gene expression and metabolism. Cell Metab 2019;29(2):362–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Colldén H, Landin A, Wallenius V, Elebring E, Fändriks L, Nilsson ME, et al The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am J Physiol Endocrinol Metab 2019;317:E1182–E1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Krizo JA, Mintz EM. Sex differences in behavioral circadian rhythms in laboratory rodents. Front Endocrinol (Lausanne) 2014;5:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ochi M, Sono S, Sei H, Oishi K, Kobayashi H, Morita Y, et al Sex difference in circadian period of body temperature in Clock mutant mice with Jcl/ICR background. Neurosci Lett 2003;347:163–6. [DOI] [PubMed] [Google Scholar]
- 69. Kloehn I, Pillai SB, Officer L, Klement C, Gasser PJ, Evans JA. Sexual differentiation of circadian clock function in the adrenal gland. Endocrinology 2016;157:1895–904. [DOI] [PubMed] [Google Scholar]
- 70. Douma LG, Solocinski K, Holzworth MR, Crislip GR, Masten SH, Miller AH, et al Female C57BL/6J mice lacking the circadian clock protein PER1 are protected from nondipping hypertension. Am J Physiol Regul Integr Comp Physiol 2019;316:R50–R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Akira S, Takeda K, Kaisho T. Toll‐like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001;2:675–80. [DOI] [PubMed] [Google Scholar]
- 72. Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, et al A Toll‐like receptor recognizes bacterial DNA. Nature 2000;408:740–5. [DOI] [PubMed] [Google Scholar]
- 73. Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, et al Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 1998;282:2085–8. [DOI] [PubMed] [Google Scholar]
- 74. Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double‐stranded RNA and activation of NF‐kappaB by Toll‐like receptor 3. Nature 2001;413:732–8. [DOI] [PubMed] [Google Scholar]
- 75. Könner AC, Brüning JC. Toll‐like receptors: linking inflammation to metabolism. Trends Endocrinol Metab 2011;22:16–23. [DOI] [PubMed] [Google Scholar]
- 76. Wu YW, Tang W, Zuo JP. Toll‐like receptors: potential targets for lupus treatment. Acta Pharmacol Sin 2015;36:1395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mohammad Hosseini A, Majidi J, Baradaran B, Yousefi M. Toll‐like receptors in the pathogenesis of autoimmune diseases. Adv Pharm Bull 2015;5:605–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Urban‐Wojciuk Z, Khan MM, Oyler BL, Fåhraeus R, Marek‐Trzonkowska N, Nita‐Lazar A, et al The role of TLRs in anti‐cancer immunity and tumor rejection. Front Immunol 2019;10:2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Silver AC, Buckley SM, Hughes ME, Hastings AK, Nitabach MN, Fikrig E. Daily oscillations in expression and responsiveness of Toll‐like receptors in splenic immune cells. Heliyon 2018;4:e00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll‐like receptor 9‐mediated innate and adaptive immunity. Immunity 2012;36:251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, et al The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll‐like receptors. Proc Natl Acad Sci USA 2000;97:13766–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 1990;249:1431–3. [DOI] [PubMed] [Google Scholar]
- 83. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22‐producing NKp46+ cells. Nat Immunol 2009;10:83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al An IL‐23R/IL‐22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell 2015;163:381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mukherji A, Kobiita A, Damara M, Misra N, Meziane H, Champy MF, et al Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc Natl Acad Sci USA 2015;112:E6691–E6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, et al Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 2003;278:8869–72. [DOI] [PubMed] [Google Scholar]
- 87. Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL‐1beta‐processing inflammasome with increased activity in Muckle‐Wells autoinflammatory disorder. Immunity 2004;20:319–25. [DOI] [PubMed] [Google Scholar]
- 88. Yang Y, Wang H, Kouadir M, Song H, Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis 2019;10:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pourcet B, Zecchin M, Ferri L, Beauchamp J, Sitaula S, Billon C, et al Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology 2018;154(5):1449–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang S, Lin Y, Yuan X, Li F, Guo L, Wu B. REV‐ERBα integrates colon clock with experimental colitis through regulation of NF‐κB/NLRP3 axis. Nat Commun 2018;9:4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Elinav E, Strowig T, Kau AL, Henao‐Mejia J, Thaiss CA, Booth CJ, et al NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011;145:745–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell 2014;157:1013–22. [DOI] [PubMed] [Google Scholar]
- 93. Hu C, Ding H, Li Y, Pearson JA, Zhang X, Flavell RA, et al NLRP3 deficiency protects from type 1 diabetes through the regulation of chemotaxis into the pancreatic islets. Proc Natl Acad Sci USA 2015;112:11318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med 2015;21:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Moossavi M, Parsamanesh N, Bahrami A, Atkin SL, Sahebkar A. Role of the NLRP3 inflammasome in cancer. Mol Cancer 2018;17:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, et al Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA 2009;106:9890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kasukawa T, Sugimoto M, Hida A, Minami Y, Mori M, Honma S, et al Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA 2012;109:15036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA 2012;109:2625–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ang JE, Revell V, Mann A, Mäntele S, Otway DT, Johnston JD, et al Identification of human plasma metabolites exhibiting time‐of‐day variation using an untargeted liquid chromatography‐mass spectrometry metabolomic approach. Chronobiol Int 2012;29:868–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Eckel‐Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone‐Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA 2012;109:5541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly‐Y M, et al The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013;341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013;504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013;504:446–50. [DOI] [PubMed] [Google Scholar]
- 104. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009;461:1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al Effects of the gut microbiota on host adiposity are modulated by the short‐chain fatty‐acid binding G protein‐coupled receptor, Gpr41. Proc Natl Acad Sci USA 2008;105:16767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ahmed K, Tunaru S, Offermanns S. GPR109A, GPR109B and GPR81, a family of hydroxy‐carboxylic acid receptors. Trends Pharmacol Sci 2009;30:557–62. [DOI] [PubMed] [Google Scholar]
- 107. Mariño E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J, et al Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017;18:552–62. [DOI] [PubMed] [Google Scholar]
- 108. Huang J, Pearson JA, Peng J, Hu Y, Sha S, Xing Y, et al Gut microbial metabolites alter IgA immunity in type 1 diabetes. JCI Insight 2020;5:e135718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short‐chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res 1994;54:3288–93. [PubMed] [Google Scholar]
- 110. Ruemmele FM, Dionne S, Qureshi I, Sarma DS, Levy E, Seidman EG. Butyrate mediates Caco‐2 cell apoptosis via up‐regulation of pro‐apoptotic BAK and inducing caspase‐3 mediated cleavage of poly‐(ADP‐ribose) polymerase (PARP). Cell Death Differ 1999;6:729–35. [DOI] [PubMed] [Google Scholar]
- 111. Tang Y, Chen Y, Jiang H, Robbins GT, Nie D. G‐protein‐coupled receptor for short‐chain fatty acids suppresses colon cancer. Int J Cancer 2011;128:847–56. [DOI] [PubMed] [Google Scholar]
- 112. Ang Z, Er JZ, Tan NS, Lu J, Liou YC, Grosse J, et al Human and mouse monocytes display distinct signalling and cytokine profiles upon stimulation with FFAR2/FFAR3 short‐chain fatty acid receptor agonists. Sci Rep 2016;6:34145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP‐activated protein kinase in Caco‐2 cell monolayers. J Nutr 2009;139:1619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al Crosstalk between microbiota‐derived short‐chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 2015;17:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Adamovich Y, Ladeuix B, Golik M, Koeners MP, Asher G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab 2017;25:93–101. [DOI] [PubMed] [Google Scholar]
- 116. Labbé A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn's, ulcerative colitis and type 2 diabetes metagenomes. PLoS One 2014;9:e115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Fiorucci S, Distrutti E. Bile acid‐activated receptors, intestinal microbiota, and the treatment of metabolic disorders. Trends Mol Med 2015;21:702–14. [DOI] [PubMed] [Google Scholar]
- 118. Zhang YK, Guo GL, Klaassen CD. Diurnal variations of mouse plasma and hepatic bile acid concentrations as well as expression of biosynthetic enzymes and transporters. PLoS One 2011;6:e16683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Eggink HM, Oosterman JE, de Goede P, de Vries EM, Foppen E, Koehorst M, et al Complex interaction between circadian rhythm and diet on bile acid homeostasis in male rats. Chronobiol Int 2017;34:1339–53. [DOI] [PubMed] [Google Scholar]
- 120. Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, et al Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci USA 2014;111:7421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, Hykes BL, et al Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 2018;360:204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Nelson CA, Wilen CB, Dai YN, Orchard RC, Kim AS, Stegeman RA, et al Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc Natl Acad Sci USA 2018;115:E9201–E9210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, et al Commensal microbes and interferon‐λ determine persistence of enteric murine norovirus infection. Science 2015;347:266–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, et al Virus‐plus‐susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 2010;141:1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schröder B, et al Norovirus triggered microbiota‐driven mucosal inflammation in interleukin 10‐deficient mice. Inflamm Bowel Dis 2014;20:431–43. [DOI] [PubMed] [Google Scholar]
- 126. Pearson JA, Tai N, Ekanayake‐Alper DK, Peng J, Hu Y, Hager K, et al Norovirus changes susceptibility to type 1 diabetes by altering intestinal microbiota and immune cell functions. Front Immunol 2019;10:2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Osborne LC, Monticelli LA, Nice TJ, Sutherland TE, Siracusa MC, Hepworth MR, et al Coinfection. Virus‐helminth coinfection reveals a microbiota‐independent mechanism of immunomodulation. Science 2014;345:578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bouziat R, Biering SB, Kouame E, Sangani KA, Kang S, Ernest JD, et al Murine Norovirus Infection Induces T. Cell Host Microbe 2018;24:677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, et al Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci USA 2013;110:9897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Gibbs J, Ince L, Matthews L, Mei J, Bell T, Yang N, et al An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med 2014;20:919–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Gagnidze K, Hajdarovic KH, Moskalenko M, Karatsoreos IN, McEwen BS, Bulloch K. Nuclear receptor REV‐ERBα mediates circadian sensitivity to mortality in murine vesicular stomatitis virus‐induced encephalitis. Proc Natl Acad Sci USA 2016;113:5730–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Edgar RS, Stangherlin A, Nagy AD, Nicoll MP, Efstathiou S, O'Neill JS, et al Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc Natl Acad Sci USA 2016;113:10085–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ehlers A, Xie W, Agapov E, Brown S, Steinberg D, Tidwell R, et al BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol 2018;11:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Hopwood TW, Hall S, Begley N, Forman R, Brown S, Vonslow R, et al The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci Rep 2018;8:3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, et al Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012;143(4):913–6. [DOI] [PubMed] [Google Scholar]
- 136. Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J 2013;7:880–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås‐Holm L, et al Metformin alters the gut microbiome of individuals with treatment‐naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med 2017;23:850–8. [DOI] [PubMed] [Google Scholar]
- 138. Arthur JC, Perez‐Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al Intestinal inflammation targets cancer‐inducing activity of the microbiota. Science 2012;338:120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018;359:592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE, et al Bacteroides fragilis toxin coordinates a Pro‐carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe 2018;23(2):203–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sivan A, Corrales L, Hubert N, Williams JB, Aquino‐Michaels K, Earley ZM, et al Commensal Bifidobacterium promotes antitumor immunity and facilitates anti‐PD‐L1 efficacy. Science 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mager LF, Burkhard R, Pett N Cooke NCA, Brown K, Ramay H, Paik S, et al Microbiomederived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020. 369(6510):1481‐1489 [DOI] [PubMed] [Google Scholar]
- 146. Valladares R, Sankar D, Li N, Williams E, Lai KK, Abdelgeliel AS, et al Lactobacillus johnsonii N6.2 mitigates the development of type 1 diabetes in BB‐DP rats. PLoS One 2010;5(5):e10507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Marcial GE, Ford AL, Haller MJ, Gezan SA, Harrison NA, Cai D, et al Lactobacillus johnsonii N6.2 Modulates the Host Immune Responses: A Double‐Blind, Randomized Trial in Healthy Adults. Front Immunol 2017;8:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lau K, Benitez P, Ardissone A, Wilson TD, Collins EL, Lorca G, et al Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2‐mediated Th17 bias. J Immunol 2011;186:3538–46. [DOI] [PubMed] [Google Scholar]
- 149. Zimmermann M, Zimmermann‐Kogadeeva M, Wegmann R, Goodman AL. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 2019;570:462–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kim YC, Lee SJ. Temporal variation in hepatotoxicity and metabolism of acetaminophen in mice. Toxicology 1998;128:53–61. [DOI] [PubMed] [Google Scholar]
- 151. Wang S, Lin Y, Zhou Z, Gao L, Yang Z, Li F, et al Circadian Clock Gene Bmal1 Regulates Bilirubin Detoxification: A Potential Mechanism of Feedback Control of Hyperbilirubinemia. Theranostics 2019;9:5122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lin Y, Wang S, Zhou Z, Guo L, Yu F, Wu B. Bmal1 regulates circadian expression of cytochrome P450 3a11 and drug metabolism in mice. Commun Biol 2019;2:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin‐degrading bacterium. Int J Syst Evol Microbiol 2004;54:1469–76. [DOI] [PubMed] [Google Scholar]
- 154. Lavelle A, Lennon G, O'Sullivan O, Docherty N, Balfe A, Maguire A, et al Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015;64:1553–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Earley H, Lennon G, Balfe Á, Coffey JC, Winter DC, O'Connell PR. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci Rep 2019;9:15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium‐induced ulcerative colitis in mice. Front Microbiol 2019;10:2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, et al Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012;55:2285–94. [DOI] [PubMed] [Google Scholar]
- 158. Marietta EV, Gomez AM, Yeoman C, Tilahun AY, Clark CR, Luckey DH, et al Low incidence of spontaneous type 1 diabetes in non‐obese diabetic mice raised on gluten‐free diets is associated with changes in the intestinal microbiome. PLoS One 2013;8:e78687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Hänninen A, Toivonen R, Pöysti S, Belzer C, Plovier H, Ouwerkerk JP, et al induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut 2018;67:1445–53. [DOI] [PubMed] [Google Scholar]
- 160. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al Cross‐talk between Akkermansia muciniphila and intestinal epithelium controls diet‐induced obesity. Proc Natl Acad Sci USA 2013;110:9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Cekanaviciute E, Yoo BB, Runia TF, Debelius JW, Singh S, Nelson CA, et al Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA 2017;114:10713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al TLR5‐mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014;41:478–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al Early‐life antibiotic‐driven dysbiosis leads to dysregulated vaccine immune responses in mice. Cell Host Microbe 2018;23(5):653–60. [DOI] [PubMed] [Google Scholar]
- 164. Seekatz AM, Panda A, Rasko DA, Toapanta FR, Eloe‐Fadrosh EA, Khan AQ, et al Differential response of the cynomolgus macaque gut microbiota to Shigella infection. PLoS One 2013;8:e64212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al Significant correlation between the infant gut microbiome and rotavirus vaccine response in Rural Ghana. J Infect Dis 2017;215:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Harris V, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, et al Rotavirus vaccine response correlates with the infant gut microbiota composition in Pakistan. Gut Microbes 2018;9:93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL, et al Effect of Antibiotic‐Mediated Microbiome Modulation on Rotavirus Vaccine Immunogenicity: A Human, Randomized‐Control Proof‐of‐Concept Trial. Cell Host Microbe 2018;24(2):197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Eloe‐Fadrosh EA, McArthur MA, Seekatz AM, Drabek EF, Rasko DA, Sztein MB, et al Impact of oral typhoid vaccination on the human gut microbiota and correlations with S. Typhi‐specific immunological responses. PLoS One 2013;8:e62026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, et al The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 1999;400:169–73. [DOI] [PubMed] [Google Scholar]
- 170. Cao Q, Zhao X, Bai J, Gery S, Sun H, Lin DC, et al Circadian clock cryptochrome proteins regulate autoimmunity. Proc Natl Acad Sci USA 2017;114:12548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Langlois PH, Smolensky MH, Glezen WP, Keitel WA. Diurnal variation in responses to influenza vaccine. Chronobiol Int 1995;12:28–36. [DOI] [PubMed] [Google Scholar]
- 172. Phillips AC, Gallagher S, Carroll D, Drayson M. Preliminary evidence that morning vaccination is associated with an enhanced antibody response in men. Psychophysiology 2008;45:663–6. [DOI] [PubMed] [Google Scholar]
- 173. Long JE, Drayson MT, Taylor AE, Toellner KM, Lord JM, Phillips AC. Morning vaccination enhances antibody response over afternoon vaccination: A cluster‐randomised trial. Vaccine 2016;34:2679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Suzuki K, Hayano Y, Nakai A, Furuta F, Noda M. Adrenergic control of the adaptive immune response by diurnal lymphocyte recirculation through lymph nodes. J Exp Med 2016;213:2567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hornung V, Rothenfusser S, Britsch S, Krug A, Jahrsdörfer B, Giese T, et al Quantitative expression of toll‐like receptor 1–10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J Immunol 2002;168:4531–7. [DOI] [PubMed] [Google Scholar]
- 176. Isnardi I, Ng YS, Srdanovic I, Motaghedi R, Rudchenko S, von Bernuth H, et al IRAK‐4‐ and MyD88‐dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity 2008;29:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Kang SM, Yoo DG, Kim MC, Song JM, Park MK, Eunju O, et al MyD88 plays an essential role in inducing B cells capable of differentiating into antibody‐secreting cells after vaccination. J Virol 2011;85:11391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak‐Rothstein A. Chromatin‐IgG complexes activate B cells by dual engagement of IgM and Toll‐like receptors. Nature 2002;416:603–7. [DOI] [PubMed] [Google Scholar]
- 179. Christensen SR, Kashgarian M, Alexopoulou L, Flavell RA, Akira S, Shlomchik MJ. Toll‐like receptor 9 controls anti‐DNA autoantibody production in murine lupus. J Exp Med 2005;202:321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll‐like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity 2006;25:417–28. [DOI] [PubMed] [Google Scholar]
- 181. Fortier EE, Rooney J, Dardente H, Hardy MP, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol 2011;187:6291–300. [DOI] [PubMed] [Google Scholar]
- 182. Nobis CC, Dubeau Laramée G, Kervezee L, Maurice De Sousa D, Labrecque N, Cermakian N. The circadian clock of CD8 T cells modulates their early response to vaccination and the rhythmicity of related signaling pathways. Proc Natl Acad Sci USA 2019;116:20077–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Dubos RJ, Straus JH, Pierce C. The multiplication of bacteriophage in vivo and its protective effects against an experimental infection with Shigella dysenteriae. J Exp Med 1943;78:161–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, et al Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J Infect Dis 2003;187:613–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study.


