Summary
The liver is an immune‐privileged organ with a tolerogenic environment for maintaining liver homeostasis. This hepatic tolerance limits the intrahepatic CD8+ T‐cell response for eliminating infections. The tolerant microenvironment in the liver is orchestrated by liver‐specific immunoregulatory cells that can be functionally regulated by pathogen‐associated molecular patterns (PAMPs). Here, we report that flagellin, a key PAMP of gut bacteria, modulates the intrahepatic CD8+ T‐cell response by activating the TLR5 signalling pathway of hepatocytes. We found that mice treated with Salmonella‐derived recombinant flagellin (SF) by hydrodynamic injection had a significantly elevated IFN‐γ production by the intrahepatic lymphocytes in 7 days after injection. This was correlated with a reduced immune suppressive effect of primary mouse hepatocytes (PMHs) in comparison with that of PMHs from mock‐injected control mice. In vitro co‐culture of SF‐treated PMHs with splenocytes revealed that hepatocyte‐induced immune suppression is alleviated through activation of the TLR5 but not the NLRC4 signalling pathway, leading to improved activation and function of CD8+ T cells during anti‐CD3 stimulation or antigen‐specific activation. In an acute HBV replication mouse model established by co‐administration of SF together with an HBV‐replicating plasmid by hydrodynamic injection, SF significantly enhanced the intrahepatic HBV‐specific CD8+ T‐cell response against HBV surface antigen. Our results clearly showed that flagellin plays a role in modulating the intrahepatic CD8+ T‐cell response by activating the TLR5 pathway in PMHs, which suggests a potential role for gut bacteria in regulating liver immunity.
Keywords: CD8+ T‐cell response, flagellin, hepatocytes, liver tolerance, Toll‐like receptor 5
Flagellin alleviates the immune suppression effect of hepatocytes by activating the TLR5 signalling pathway, resulting in improved activation and function of CD8+ T cells. Administration of flagellin in vivo by hydrodynamic injection enhances the intrahepatic rather than the systemic viral‐specific CD8+ T‐cell response. Our work suggests a potential role of gut bacteria in regulating liver immunity by targeting the parenchymal cells.
Abbreviations
- APCs
antigen‐presenting cells
- DCs
dendritic cells
- FV
Friend virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HSCs
hepatic stellate cells
- HV
hypervariable region
- IHLs
intrahepatic lymphocytes
- KCs
Kupffer cells
- LFn
the PA recognition domain of anthrax lethal factor
- LSECs
liver endothelial cells
- NLR
NOD‐like receptor
- NPCs
non‐parenchymal liver cells
- PA
anthrax protection antigen
- PAMPs
pathogen‐associated molecular patterns
- PMHs
primary mouse hepatocytes
- SF
Salmonella flagellin
- Tg
transgenic
- TLR
Toll‐like receptor
- WT
wild type
Introduction
The liver, with its close anatomical and physiological relationships to the gastrointestinal tract, is continuously exposed to food and microbial components that are transported from the gut via the portal circulation. Because of this special microenvironment, the liver represents a unique organ that is hyporesponsive to inducing the adaptive immune responses against food‐derived antigens, liver transplants and some viruses. 1 , 2 , 3 Intensive studies have shown that the liver immune hyporesponsiveness is orchestrated by parenchymal hepatocytes and non‐parenchymal liver cells (NPCs), including liver endothelial cells (LSECs), Kupffer cells (KCs) and dendritic cells (DCs). 4 Priming of naïve T cells by these cells usually leads to distinct activation properties from the DC‐primed T cells, including low cytokine production and reduced cytotoxicity. 5 , 6 , 7 Moreover, the proliferation and function of effector T cells are suppressed due to the constitutive expression of immune suppressive molecules such as IL‐10 and PD‐L1 on the liver‐resident cells. 7 , 8 Antigen‐specific CD8+ T cells, which are the important cell type for controlling infections in the liver, are functionally suppressed at the priming or effector stage. By utilizing the hyporesponsive adaptive immune responses in the liver, pathogens are able to establish persistent infections resulting in various chronic liver infections with subsequent diseases. 9 , 10
The function of liver‐resident cells is not simply restricted to immune suppression and tolerance induction. Some bacterial products, which can often be detected in the systemic circulation of diseased people, may functionally regulate or even alter the immune activities of liver cells by activating the Toll‐like receptor (TLR) and Nod‐like receptor (NLR) signalling pathways. 11 , 12 For example, stimulation of TLR4 by bacterial LPS induces activation of KCs and leads to liver inflammation. 13 , 14 TLR9 activation by unmethylated CpG sequences in bacterial DNA molecules induces the aggregation of intrahepatic myeloid cells, which may enable the expansion of CD8+ T cells in the liver. 15 TLR1/2 ligand derived from bacterial peptidoglycan may trigger CD8+ T‐cell activation in vitro by stimulating LSECs and is also involved in mouse intrahepatic CD8+ T‐cell tolerance by stimulating KCs. 8 , 16 These examples suggest that PAMPs derived from gut microbiota or bacterial pathogens are potent immune regulators that may perform a complicated crosstalk with hepatic CD8+ T‐cell responses by targeting the liver cells. 17 , 18
Flagellin is a unique protein‐based PAMP of either pathogenic bacteria or the gut microbiota that can be recognized by the cell surface TLR5 and the cytosolic NAIP5/6 receptors. Flagellin triggers the TLR5 signalling cascade and activates proinflammatory responses via the TLR5/MyD88/NF‐κB axis. 19 Once taken up into the cell, flagellin can be detected by cytosolic NAIP5/6 receptors and trigger the NAIP/NLRC4 inflammasome pathway. 20 , 21 A recent study reported that the liver was the major organ primarily responding to flagellin by activating the TLR5‐MyD88 signalling pathway. 22 Therefore, it is interesting to investigate the role of flagellin in regulating liver immune responses while it is still poorly understood. Several in vitro or in vivo mouse investigations showed that flagellin may activate hepatocytes or NK cells in the liver, leading to resistance against Fas‐mediated apoptosis in hepatocytes or enhancement of NK cell‐related antitumor CD8+ T‐cell responses, respectively. 22 , 23 Our previous studies demonstrated that systemic administration of recombinant flagellin induces a rapid but transient influx of immune cells, mainly granulocytes and macrophages, into the liver. 24 It is intriguing but remains unclear whether TLR5 pathway activation in the liver participates in regulating the immunological function of intrahepatic CD8+ T cells, especially during infectious diseases in the liver.
As mentioned above, flagellin can also activate the intracellular NAIP/NLRC4 inflammasome pathway, which usually induces caspase‐1 cleavage and secretion of proinflammatory cytokines such as IL‐1β and IL‐18. 21 , 25 Although there is no direct evidence that the liver cells could respond to the intracellular flagellin, several studies have already pointed out that the inflammasome machinery can be activated in liver cells such as KCs, hepatic stellate cells (HSCs) and hepatocytes. 21 , 26 , 27 These studies suggested another potential pathway for flagellin to regulate liver immune responses. However, the effect of the NAIP/NLRC4 inflammasome pathway on hepatic CD8+ T‐cell responses remains elusive.
In this study, we investigated whether and how flagellin regulates intrahepatic CD8+ T‐cell responses. Our results revealed that Salmonella flagellin (SF) modulates the activity and function of intrahepatic CD8+ T cells through TLR5 activation of hepatocytes. We further confirmed that administration of SF to the liver enhances the intrahepatic CD8+ T‐cell response rather than the systemic viral‐specific CD8+ T‐cell response in an HBV replication mouse model.
Materials and methods
Mice
C57BL/6J wild‐type (WT) female mice at 6–8 weeks of age were purchased from Hunan SJA Laboratory Animal Company. C57BL/6 background TLR5−/− mice of the Jackson Laboratory origin were maintained under specific pathogen‐free conditions in the Animal Center of Wuhan Institute of Virology (WIV), Chinese Academy of Sciences (CAS). Animal studies were performed according to the Regulations for the Administration of Affairs Concerning Experimental Animals in China (1988) and the Guidelines for Animal Care and Use, WIV, CAS. Animal experiments were reviewed and approved by Ethics Committees and Biosafety Committees, Institutional Review Board (IRB), WIV, CAS (permission number: WIVA09201211). For antigen‐specific CD8+ T‐cell activation assay, inbred female DbGagL TCR transgenic (Tg) mice were used. The DbGagL TCR Tg mice were on a C57BL/6 or B6.SJL (CD45.1 congenic) background, and >90% of the CD8+ T cells contained a TCR specific for the DbGagL Friend virus (FV) epitope (FV‐TCR Tg CD8+ T cells). 28 DbGagL TCR Tg mice were kept in the Animal Care Center, University of Duisburg‐Essen (Essen, Germany). Experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and were reviewed and approved by the local Animal Care and Use Committee (Animal Care Center, University of Duisburg‐Essen, Essen, Germany, and the District Government of Dusseldorf, Germany).
Proteins and peptides
Recombinant Salmonella flagellin (SF), anthrax protection antigen (PA), LFn‐SF fusion protein (LFn: the PA recognition domain of anthrax lethal factor, LFn‐SF) and the truncated SF recombinants, including hypervariable (HV) region (amino acids 172–415, without TLR5‐ and NLRC4‐activating domain), FliCΔ90–97 (without TLR5 signalling activity) and FliC‐L3A (without NLRC4 signalling activity) were cloned, expressed and purified as described previously. 29 , 30 The endotoxin values of all purified recombinant flagellin preparations were <0.01 EU/mg.
For stimulation of murine lymphocytes, the following peptides were used: HBsAg aa 19–33 (FFLLTRILTIPQSLD), HBsAg aa 190–197 (VWLSVIWM), HBsAg aa 208–215 (ILSPFLPL), HBcAg aa 93–100 (MGLKFRQL) and HBcAg aa 129–140 (PPAYRPPNAPIL). 31 A cytomegalovirus‐derived peptide (YILEETSVM) was used as control. The peptides were purchased from Chinese Peptide Company. A FV GagL CTL epitope aa 85–93 (CCLCLTVFL) was used for antigen‐specific activation assay. 28
Hydrodynamic injection
Mice were injected by hydrodynamic injection by using a replication‐competent HBV clone harbouring a head‐to‐tail tandem dimeric HBV genome, pSM2 (provided by Dr. Hans Will, Heinrich Pette Institute, Hamburg, Germany), as described previously. 32 In brief, 10 μg pSM2 was mixed with or without 25 μg SF or HV protein in a volume of normal saline solution equivalent to 10% of the mouse body weight, and injected through the tail vein of mice within 8 s. Mice were killed at 30 min, 1 day, and 3, 7, 10, 14 and 21–28 days post‐injection for indicated detections.
Detection of SF in liver tissues
In order to detect the distribution of SF in the liver, mice were killed at 30 min after hydrodynamic injection with SF. Liver tissue samples were fixed in 10% formalin and embedded in paraffin. For immunohistochemistry staining of SF, liver sections were stained with rabbit anti‐SF polyclonal antibody or isotype control antibody, followed by secondary HRP‐conjugated goat anti‐rabbit IgG and visualized by the Envision System (Dako). 33 At 6 h after injection, the liver was perfused with 1.5 ml PBS containing 5% BSA, and the intermediate 0.5 ml of perfusate liquid was harvested. The liver lavages were prepared by centrifugation at 3000 rpm for 10 min at 4°C to remove the cells and debris. TNF‐α, IL‐10 and IL‐6 in the liver lavage samples were measured by ELISA (BioLegend).
Cell isolation
Splenocyte suspensions were prepared by homogenization as described previously. 31 CD8+ T cells were purified by MACS separation using mouse CD8a (Ly‐2) MicroBeads (Miltenyi Biotec). Peritoneal macrophages were collected from naïve animals by RPMI‐1640 lavage as described previously. 34 Bone marrow cells from tibias and femurs of mice were differentiated into dendritic cells (DCs) with GM‐CSF and IL‐4 as described previously. 35
Isolation of PMHs, LSECs, KCs and intrahepatic lymphocytes (IHLs) was performed by liver perfusion through the portal vein with 70 μg/ml Liberase Blendzymes in Grey's balanced saline solution at 37°C. 36 Parenchymal and non‐parenchymal liver cells were separated by centrifuging at 50 g for 5 min. PMHs were washed twice with complete DMEM by centrifuging at 50 g and then plated at a density of 1 × 105 cells per well on collagen I‐coated 24‐well plate in Williams' medium supplemented with 10% fetal bovine serum (Gibco). LSECs were isolated by MACS separation using anti‐CD146 MicroBeads (Miltenyi Biotec), and then, 5 × 105 cells per well were plated on collagen I‐coated 48‐well plate in DMEM supplemented with 10% fetal bovine serum. KCs were isolated by MACS separation using anti‐F4/80 MicroBeads (Miltenyi Biotec), and then, 2 × 105 cells per well were plated on collagen I‐coated 96‐well plate in DMEM supplemented with 10% fetal bovine serum.
In vitro stimulation of PMHs with recombinant flagellin
PMHs from naïve mice were stimulated with SF or HV protein at a final concentration of 1–5 μg/ml by direct administration of SF or HV protein in the medium. To monitor the activation of TLR signalling pathway, the downstream cytokines including IL‐6 and TNF‐α were detected at mRNA and protein levels. Total RNA was isolated from 1 × 106 hepatocytes using TRIzol reagent (Life Technologies) after 3–6 h. The cDNA was prepared from total RNA with a Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) using oligo‐(dT) primers. Real‐time PCR was carried out with the QuantiTect SYBR Green RT‐PCR Kit (Qiagen) on the CFX Connect Real‐Time PCR Detection System (Bio‐Rad). The following primers were used for real‐time PCR: IL‐6 (QT00098875, Qiagen); TNF‐α (QT00104006, Qiagen); CD80 sense, 5′‐CAGAAGACCCTCCTGATAG‐3′; CD80 anti‐sense, 5′‐AGGTAAGGCTGTTGTTTGT‐3′; ICAM‐1 sense, 5′‐CATCACCGTGTATTCGTTTC‐3′; ICAM‐1 anti‐sense, 5′‐GCTGGCGGCTCAGTATCT‐3′; IL‐7 sense, 5′‐TGTGATGATACAAAGGAAGC‐3′; IL‐7 anti‐sense, 5′‐TGTGCCTTGTGATACTGTTA‐3′; PD‐L1 sense, 5′‐TTTACTATCACGGCTCCA‐3′; PD‐L1 anti‐sense, 5′‐TCCTGCCACAAACTGAAT‐3′; TGF‐b2 sense, 5′‐TCGTCCGCTTTGATGTCT‐3′; TGF‐b2 anti‐sense, 5′‐CAGTTCAATCCGCTGCTC‐3′; b‐actin sense, 5′‐CAGCCTTCCTTCTTGGGTAT‐3′; and b‐actin anti‐sense, 5′‐TGGCATAGAGGTCTTTACGG‐3′. For PCR detection of IL‐12, IL‐12 sense primer 5′‐GGAAGCACGGCAGCAGAATA‐3′ and anti‐sense primer 5′‐AACTTGAGGGAGAAGTAGGAATGG‐3′ were used. GM‐CSF (detection limit: >16 pg/ml), IL‐6 (detection limit: >2 pg/ml) and TNF‐α (detection limit: >4 pg/ml) in the supernatant were measured by ELISA (BioLegend) after 24 h. CD40 (clone 3/23; BioLegend) and ICAM‐1 (clone YN1/1.7.4; BioLegend) in PMHs were measured by FACS after 24 h.
To activate the intracellular NLRC4 pathway, PMHs, KCs or peritoneal macrophages were pretreated with LPS (50 ng/ml) and stimulated with LFn‐SF (1 μg/ml) in the presence of PA protein (1 µg/ml) as previously described in the SF‐induced inflammasome activation assays. 21 , 30 By using the LFn‐SF fusion protein, SF can be efficiently transported into mammalian cytosol through endocytosis‐mediated entry after engagement of LFn with PA. Cleavage of caspase‐1 and secretion of IL‐1β were used as indicators for the activation of inflammasome pathway. For Western blot analysis of caspase‐1 cleavage, cell lysates and supernatant were collected at 1 h after stimulation as described previously. 30 IL‐1β (detection limit: >16 pg/ml) in the supernatant was measured by ELISA (BioLegend) after 6 h. Cells stimulated with LFn‐SF or PA alone were used as negative control.
In vitro activation of splenocytes, IHLs and purified CD8+ T cells
Splenocytes or IHLs isolated from the hydrodynamically injected mice were activated by stimulation with anti‐CD3 (1 μg/ml) and anti‐CD28 (1 μg/ml) for 24–48 h. Intrahepatic CD8+ T cells were activated with plate‐coated anti‐CD3 (clone 145‐2C11, 5 mg/ml; eBioscience) and anti‐CD28 (1 μg/ml).
For in vitro co‐culture experiment, WT or TLR5−/− PMHs pooled from 1 to 3 naïve mice were pre‐stimulated with SF, FliCΔ90–97, FliC‐L3A or HV for 24 h in triplicates and washed with PBS for five times. Splenocytes or CD8+ T cells were pooled from 3 to 8 naïve mice. Up to 2 × 106 splenocytes or 5 × 105 purified CD8+ T cells derived from naïve WT or TLR5−/− mice were co‐cultured with PMHs for 24–48 h in the presence of anti‐CD3 (1 μg/ml) and anti‐CD28 (1 μg/ml) in a total volume of 1 ml. Splenocytes cultured alone were used as negative control (NC; without anti‐CD3 and anti‐CD28) or reactive control (RC; with anti‐CD3 and anti‐CD28). CD8+ T cells cultured alone were used as NC (without anti‐CD3 and anti‐CD28) or RC (with plate‐coated anti‐CD3 and anti‐CD28). For hydrodynamically injected mice, PMHs, LSECs or KCs were isolated from 3 mice of each group at indicated time‐points after injection with pSM2, pSM2 + SF or pSM2 + HV. Cells from each mouse were seeded individually in triplicates and co‐cultured with splenocytes or purified CD8+ T cells with the same procedure. Mean value of the triplicates was calculated for each mouse and was used for statistical analysis between groups.
For antigen‐specific activation, CellTrace Violet (Invitrogen)‐labelled FV‐TCR Tg CD8+ T cells were pre‐activated with peptide‐loaded bone marrow‐derived dendritic cells at 5:1 ratio. 28 After 24‐h activation, 2 × 105 CD8+ T cells were co‐cultured with the SF‐ or HV‐pre‐stimulated PMHs for an extended 48 h.
For ex vivo analysis of HBV‐specific T‐cell responses, splenocytes or IHLs were stimulated with HBs‐ or HBc‐derived peptide pools at a final concentration of 2 μg/ml in the presence of anti‐CD28 (1 μg/ml) and Brefeldin A (4 μg/ml) for 4–6 h. Unstimulated cells or cells stimulated with CMV‐derived peptide were used as negative controls.
Cells were harvested for further analysis by flow cytometry. Cytokines in the supernatants were assessed by ELISA.
Cell surface and intracellular cytokine staining of lymphocytes
Cell surface staining was performed using eBioscience or BioLegend reagents including anti‐CD4 (L3T4), anti‐CD8 (53‐6.7), anti‐CD69 (H1.2F3), anti‐CD44 (eBioR2/60), anti‐CD62L (MEL‐14), anti‐PD‐1 (J43), anti‐CD25 (PC61.5), anti‐NK1.1 (PK136), CD11b (M1/70), anti‐F4/80 (BM8), anti‐Gr‐1 (RB6‐8C5), anti‐CD3e (145‐2C11) and anti‐B220 (RA3‐6B2). Dead cells were excluded from the analysis by 7‐AAD or Fixable Viability Dye (eBioscience) staining. The proliferation of CD8+ T cells was detected by the CellTrace Violet (eBioscience) dilution assay. For intracellular cytokine staining, cells were fixed and permeabilized using the Cytofix/Cytoperm intracellular staining kit (eBioscience), and stained with cytokine‐specific mAbs including IFN‐γ (XMG1.2), IL‐2 (JES6‐5H4) and TNF‐α (MP6‐XT22). For transcription factor staining, cells were fixed and permeabilized using Foxp3/Transcription Factor Staining Buffer Set (eBioscience), and stained with anti‐T‐bet (eBio4B10) and anti‐EOMES (Dan11mag).
Stained cells were analysed on FACSCanto II or LSRFortessa (BD). The data were analysed by using FlowJo software (Tree Star). The frequency of HBs‐ or HBc‐specific CD8+ T cells was calculated by subtracting the background value using the CMV peptide control.
In vitro blockage experiment
WT PMHs were pre‐stimulated by SF or HV for 24 h and washed with PBS for five times. Up to 2 × 106 TLR5−/− splenocytes were cultured with PMHs in the presence of anti‐CD3 (1 μg/ml) and anti‐CD28 (1 μg/ml) and neutralizing antibodies or isotype controls for 24 h in a total volume of 800 μl. The neutralizing antibodies or isotype controls included ICAM‐1 (clone YN1/1.7.4; Bio X Cell), IL‐6 (clone MP5‐20F3; BioLegend), GM‐CSF (clone MP1‐22E9; BioLegend), rat IgG2b, κ (clone LTF‐2; Bio X Cell), rat IgG2a, κ (clone RTK2758; BioLegend), or rat IgG1 (clone RTK2071; BioLegend). IFN‐γ (detection limit: >15 pg/ml) in the supernatant was detected by ELISA (eBioscience) after 24 h.
Serology and detection of HBV DNA
HBsAg, HBeAg and anti‐HBs in mouse serum were detected at a 1:10 dilution using the commercial HBsAg, HBeAg and anti‐HBs ELISA kits (Kehua Biotechnology) according to the manufacturer's instructions. 37 Serum HBV DNA was extracted using the QiAamp DNA Blood Mini Kit (Qiagen) and quantified by real‐time PCR using a SYBR Green Real‐Time PCR Master Mix (Toyobo) as described previously. 37
CD8+ T‐cell transfer
Recipient mice were hydrodynamically injected with pAAV‐HBV1·2 plasmid as described previously to induce persistent HBV‐replicating mice that were tolerant to HBV antigens. 38 Ten days later, 25 μg SF or HV protein was hydrodynamically injected into mice as described previously. Donor mice were immunized with a eukaryotic HBsAg‐expressing plasmid (pHBsAg‐WT) for three times at 2‐week intervals as described previously. 39 Ten days after the third immunization, the mice were killed and CD8+ T cells were pooled and purified by MACS isolation. For each recipient mouse, 2 × 106 purified CD8+ T cells suspended in 200 μl PBS were adoptive‐transferred to the recipient mice by intravenous injection at 6 days after the hydrodynamic injection of SF or HV protein. Serum HBsAg and HBeAg were monitored until 21 days after the transfer. Splenic and intrahepatic HBsAg‐specific CD8+ T cells were analysed by staining with HBs208 peptide‐loaded Mouse DimerX (Recombinant Soluble Dimeric H‐2Db:Ig Fusion Protein, BD Biosciences) and intracellular IFN‐γ.
Statistical analyses
Statistical analyses were performed using GraphPad Prism software version 5 (GraphPad Software Inc.). Statistical differences were analysed by one‐way ANOVA followed by the Bonferroni post‐test. For the dose‐dependent analysis, statistical differences were performed by two‐way ANOVA followed by the Bonferroni post‐test. NC and RC were used as the systemic controls and were not included in the statistical analysis. The p‐values < 0.05 were considered significant. All data are presented as mean ± SD. All experiments are representative of at least three independent experiments.
Results
Stimulation of PMHs by SF improves the activation and function of CD8+ T cells in vitro
Priming, activation and function of intrahepatic CD8+ T cells are regulated by the liver‐resident cells. In the co‐culture system using anti‐CD3/CD28‐activated splenocytes, we confirmed that the PMHs, LSECs and KCs represented an immune suppressive activity under basal conditions, leading to significantly reduced production of IFN‐γ by activated splenocytes (Figure S1A–C). To determine whether SF modulates the immune activities of these liver cells, PMHs, LSECs and KCs were pretreated with SF in vitro and their immunoregulatory properties were analysed by co‐culture with TLR5−/− splenocytes. A fragment of flagellin containing only the hypervariable region (HV) of SF, which has neither TLR5 nor NLRC4 activity, was used as a control protein in the subsequent experiments. Compared with the anti‐CD3/CD28‐activated CD8+ T cells in the absence of PMHs (reactive control, RC), the presence of PMHs resulted in altered expression of CD8+ T‐cell activation markers, for example upregulated CD25 but downregulated CD44 (Figure 1A,B, unstimulated PMHs). The absolute cell count of living CD8+ T cells is almost the same after co‐culture with untreated, SF‐treated and HV‐treated PMHs, indicating that the survival or proliferation of CD8+ T cells was not significantly affected by SF‐treated PMHs (Figure S1D). However, the CD8+ T cells in the presence of the SF‐treated PMHs expressed significantly more CD25 and CD44 than those in the presence of the untreated or HV‐treated PMHs (Figure 1A,B). Moreover, the population of CD8+ T cells co‐cultured with SF‐treated PMHs contained a higher percentage of CD44+CD62L− effector cells and CD44+CD62L+ memory cells but fewer CD44−CD62L+ naïve cells than that co‐cultured with untreated or HV‐treated PMHs (Figure 1C, S1E). This suggests that SF‐treated PMHs changed the expansion of effector and memory CD8+ T‐cell subsets (Figure 1C). Compared with the untreated or HV‐treated PMHs, SF‐treated PMHs significantly upregulated the expression of transcription factors T‐bet and Eomes in CD8+ T cells (Figure 1D,E) and resulted in elevated percentages of both IFN‐γ‐ and TNF‐α‐producing CD8+ T cells (Figure 1F). In contrast, the IFN‐γ production was not significantly changed by co‐culture using SF‐ or HV‐treated LSECs or KCs, indicating that these two cell types could not be affected by SF (Figure S1F,G). These results suggest that in vitro treatment of PMHs with SF not only promoted the activation but also increased the cytokine production of CD8+ T cells.
FIGURE 1.
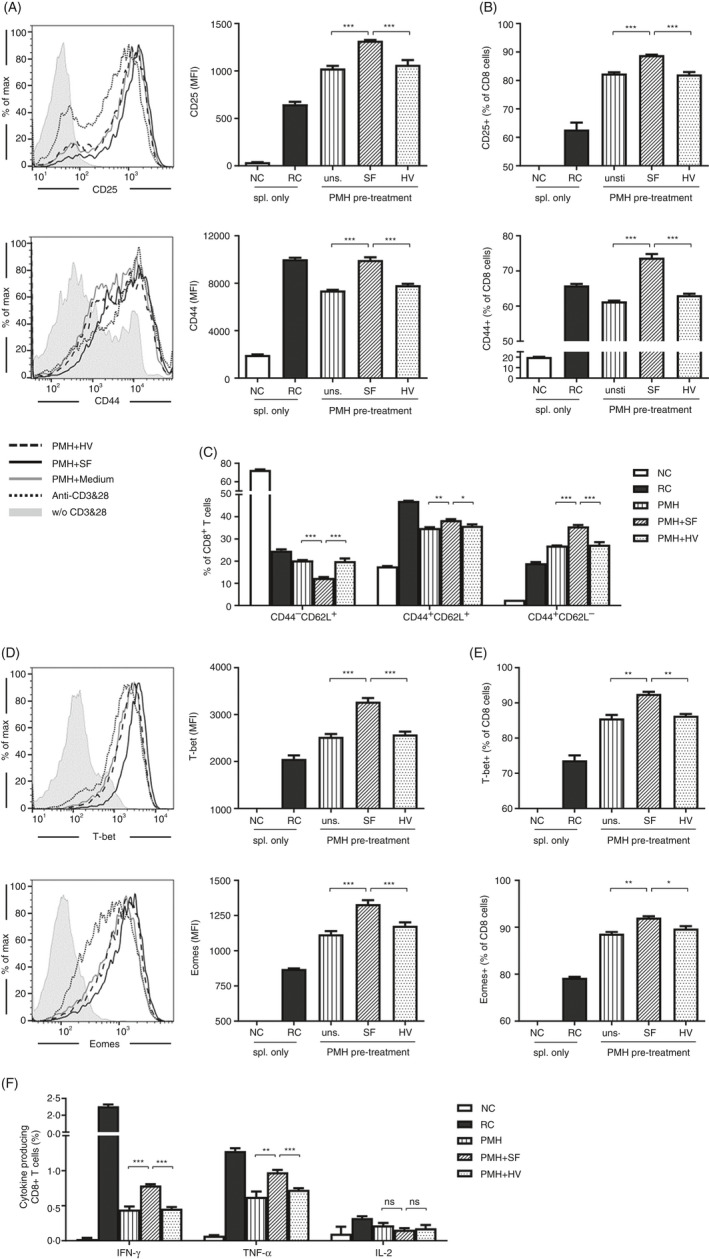
Stimulation of PMHs by SF improved the activation and function of CD8+ T cells. PMHs from WT C57BL/6 mice were pretreated with SF or HV protein (2.5 µg/ml) for 24 h and then co‐cultured with TLR5−/− splenocytes in the presence of anti‐CD3 and anti‐CD28 for an additional 24 h. (A, B) Expression of the activation markers CD25 and CD44 on CD8+ T cells. Grey shadow, without (w/o) CD3 and CD28; dark dotted line, anti‐CD3 and anti‐CD28; grey solid line, PMH + medium; dark solid line, PMH + SF; dark dashed line, PMH + HV. (C) Differentiation of CD8+ T cells was characterized as naïve cells (CD62L+CD44−), effector cells (CD62L−CD44+) and memory cells (CD62L+CD44+). (D, E) Expression of the transcription factors T‐bet and Eomes and (F) production of IFN‐γ, TNF‐α and IL‐2 in CD8+ T cells were analysed by flow cytometry. Data are representative of at least three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by one‐way ANOVA.
Activation of the TLR5 signalling pathway ameliorates the immune suppressive activity of PMHs
SF is a PAMP that could be recognized by dual signalling pathways, including the TLR5‐MyD88 and the NAIP/NLRC4 inflammasome pathway. It has been reported that both signalling pathways of hepatocytes can be activated by in vitro stimulation with TLR/NLR agonists, infection or liver damage. 40 , 41 , 42 Based on this notion, we further investigated whether the PMHs could respond to SF stimulation by activating the extracellular TLR5 or the intracellular NLRC4 inflammasome pathway and thus subsequently regulate the function of T cells. Thus, PMHs separated from naïve C57BL/6 mice were stimulated with SF or HV, and the expression of IL‐6/TNF‐α or caspase‐1 cleavage was assayed to monitor the activation of MyD88 or the NLRC4 pathway, respectively. In vitro stimulation of PMHs with SF induced significantly upregulated IL‐6 and TNF‐α mRNA expression at 6 h post‐stimulation (Figure 2A) and upregulated secretion of IL‐6 and TNF‐α at 24 h post‐stimulation (Figure 2B), indicating efficient activation of the TLR5 signalling pathway in PMHs. However, neither caspase‐1 cleavage nor IL‐1β secretion could be detected in PMHs by Western blot (Figure 2C) or ELISA (Figure 2D), in contrast to that of the positive control of macrophages (Figure 2C) or Kupffer cells (Figure 2D). These results indicate that SF can predominantly stimulate PMHs by the extracellular TLR5 pathway but hardly activate the intracellular NLRC4 inflammasome pathway.
FIGURE 2.
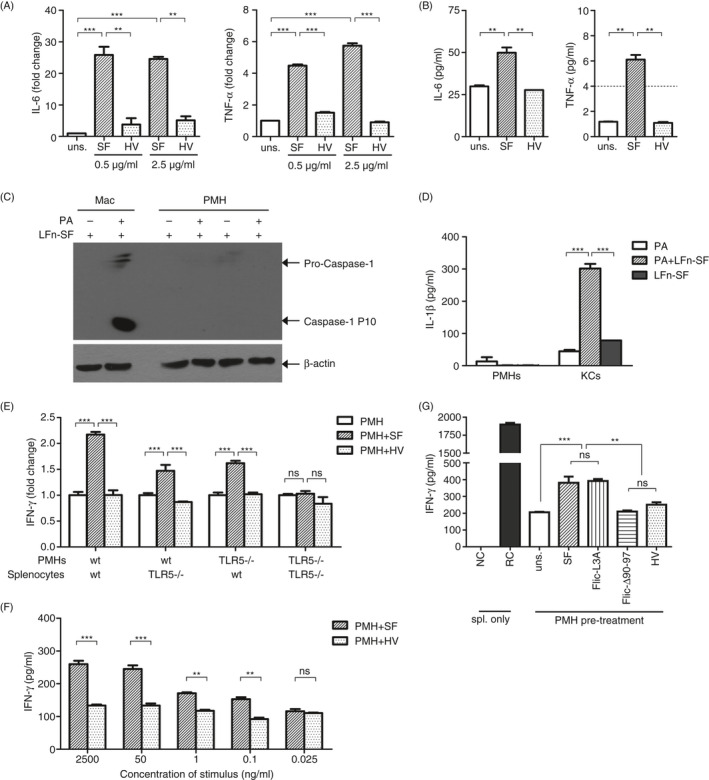
Activation of the TLR5 signalling pathway reduced the immunosuppressive properties of PMHs. PMHs were isolated from naïve C57BL/6 mice and stimulated with SF or HV protein. (A) The expression of TNF‐α and IL‐6 at the mRNA level was quantified by real‐time RT‐PCR after 6 h, and (B) TNF‐α and IL‐6 in the supernatant were measured by ELISA after 24 h. Dashed line indicates the sensitivity threshold of the ELISA kit. (C, D) PMHs were pretreated with 50 ng/ml LPS for 3 h and then transfected with the LFn‐SF + PA system. (C) Caspase‐1 cleavage and (D) IL‐1β secretion were detected by Western blot or ELISA after 1 or 6 h, respectively. (E) WT or TLR5−/− hepatocytes pretreated with SF or HV were co‐cultured with WT or TLR5−/− splenocytes in the presence of anti‐CD3 and anti‐CD28. (F) WT PMHs were pretreated with SF or HV at the indicated concentrations and co‐cultured with TLR5−/− splenocytes. (G) WT hepatocytes were pretreated with SF, FliC‐L3A, FliCΔ90‐97 or HV and co‐cultured with TLR5−/− splenocytes. IFN‐γ in the supernatants was detected by ELISA after 24 h of co‐culture. Unstimulated splenocytes were used as a negative control (NC). Splenocytes stimulated with anti‐CD3/anti‐CD28 alone were used as reactive controls (RC). Data are representative of at least three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by one‐way ANOVA or two‐way ANOVA (f).
To further confirm whether TLR5 pathway activation of PMHs affects the function of T cells, PMHs separated from WT or TLR5−/− mice were stimulated with SF or HV for 24 h and then co‐cultured with splenocytes derived from WT or TLR5−/− mice. IFN‐γ production was upregulated in both WT and TLR5−/− splenocytes that were co‐cultured with SF‐treated WT PMHs (Figure 2E, WT PMH). In contrast, no upregulated IFN‐γ production could be detected in the TLR5−/− splenocytes in the presence of SF‐treated TLR5−/− PMHs compared with that of untreated or HV‐treated PMH control groups (Figure 2E, TLR5−/− PMH; TLR5−/− splenocytes). These data indicate that SF‐induced upregulation of IFN‐γ production in TLR5−/− splenocytes is dependent on the TLR5 expression of PMHs. Moreover, SF modulated PMHs and upregulated the IFN‐γ production in splenocytes in a dose‐dependent manner over a broad range of concentrations (Figure 2F). The lowest threshold concentration of SF that modulated the PMHs was as low as 0.1 ng/ml, further indicating the susceptivity of PMHs on TLR5 for regulating T‐cell activity in response to SF. However, when TLR5−/− PMHs were tested by co‐culture with WT splenocytes, slightly elevated IFN‐γ production could also be detected, suggesting that residual SF in the co‐culture system might activate the T cells by stimulating the splenic antigen‐presenting cells (APCs) (Figure 2E, TLR5−/− PMH; WT splenocytes). To avoid this interference, splenocytes from TLR5−/− mice were used for all of the in vitro experiments.
Furthermore, the dual signalling pathway activities of SF protein were tested with WT PMHs stimulated by using truncated SF proteins that retain a single signalling pathway (either activating TLR5 or NLRC4) and co‐cultured with TLR5−/− splenocytes. As shown in Figure 2G, TLR5‐deficient FliCΔ90‐97 protein‐stimulated PMHs could not upregulate IFN‐γ production in splenocytes. In contrast, the NLRC4‐deficient but TLR5‐retaining FliC‐L3A protein‐stimulated PMHs upregulated the IFN‐γ production of splenocytes, similar to the wild‐type SF‐stimulated PMHs (Figure 2G). These results demonstrate that SF stimulated the TLR5 signalling pathway of PMHs and thus upregulated the function of lymphocytes.
Soluble molecules produced by SF‐treated PMHs play role in regulating the function of T cells
Further studies were performed to study how TLR5‐activated PMHs upregulate the function of lymphocytes, especially T cells. PMHs isolated from naïve mice were stimulated by SF for 6–24 h, and the expression of immune molecules was measured. The molecules, including the costimulators CD40 and CD80, the cytokines IL‐12 and IL‐7 and the inhibitory molecules PD‐L1 and TGF‐β2, were undetectable or unchanged in hepatocytes after stimulation with SF or HV (Figure S2A–C). To analyse whether the soluble molecules or membrane molecules of PMHs play role in regulating the T cells, the trans‐well co‐culture system with activated TLR5−/− splenocytes in the top chamber and PMHs in the 12‐well plates was used. After 24–48 h of culturing, the IFN‐γ in the trans‐well chamber was increased by co‐culture with SF‐treated PMHs than by HV‐treated PMHs (Figure S2D). Moreover, when activated TLR5−/− splenocytes were cultured with the conditioned medium from PMHs, medium from SF‐treated PMHs induced a significant higher amount of IFN‐γ than the medium from HV‐treated PMHs did (Figure S2E). These results indicate that the elevated cytokine production of splenocytes is not dependent on the cell–cell attachment with the SF‐treated PMHs. In the supernatant of SF‐treated PMHs, several proinflammatory cytokines such as IL‐6, TNF‐α and GM‐CSF in the supernatant were secreted at low levels (Figure 2B, S2F). Notably, ICAM‐1, an adhesion molecule that is found in both membrane‐bound form and soluble form, was significantly upregulated on PMHs at both mRNA and protein levels (Figure S2G,H). Therefore, we analysed whether these molecules produced by PMHs participate in modulating T‐cell function by measuring the inhibition effects using neutralizing antibodies acting against each cytokine. Neutralizing antibodies against ICAM‐1 inhibited IFN‐γ production by T cells. However, the IFN‐γ of the SF‐treated PMH group was still higher than that of the HV‐treated PMH group in the co‐culture experiment (Figure S2I). Similarly, blocking GM‐CSF or IL‐6 did not abolish the IFN‐γ enhancement in co‐culture of SF‐treated PMHs (Figure S2J,K). These results suggest that the SF‐induced expression of ICAM‐1, GM‐CSF or IL‐6 might not be indispensable regulators for PMHs to regulate T‐cell function in the in vitro co‐culture system. The mechanism by which SF‐treated PMHs regulate T‐cell function requires further investigation.
SF‐treated PMHs improve the non‐specific and antigen‐specific activation and function of CD8+ T cells
As SF‐treated PMHs improved the anti‐CD3‐induced multiclonal activation of CD8+ T cells, we further investigate whether SF‐treated PMHs could regulate the activation and function of antigen‐specific CD8+ T cells. Firstly, we wonder whether the SF‐treated PMHs regulated the CD8+ T cells directly or via other cells in the co‐culture system. By MACS separation, purified CD8+ T cells (>98%) were obtained (Figure S3A). Stimulation of MACS‐purified WT CD8+ T cells with SF did not change the IFN‐γ production, indicating that the purified CD8+ T cells were not directly responding to SF although a small portion (<0.5%) of CD11c+MHC‐II+ dendritic cells were included (Figure S3A,B). By co‐culture of MACS‐purified TLR5−/− CD8+ T cells with PMHs, the cell count of living CD8+ T cells is almost the same after co‐culture with the untreated, SF‐treated and HV‐treated PMHs (Figure S3C). Upregulated T‐bet expression and IFN‐γ secretion were similarly detected in the purified CD8+ T cells by co‐culture with SF‐treated PMHs for 24 h (Figure 3A–C). These results demonstrated that SF‐treated PMHs can directly modulate and improve the function of CD8+ T cells independently of other cell types such as splenic DCs or CD4+ helper T cells.
FIGURE 3.
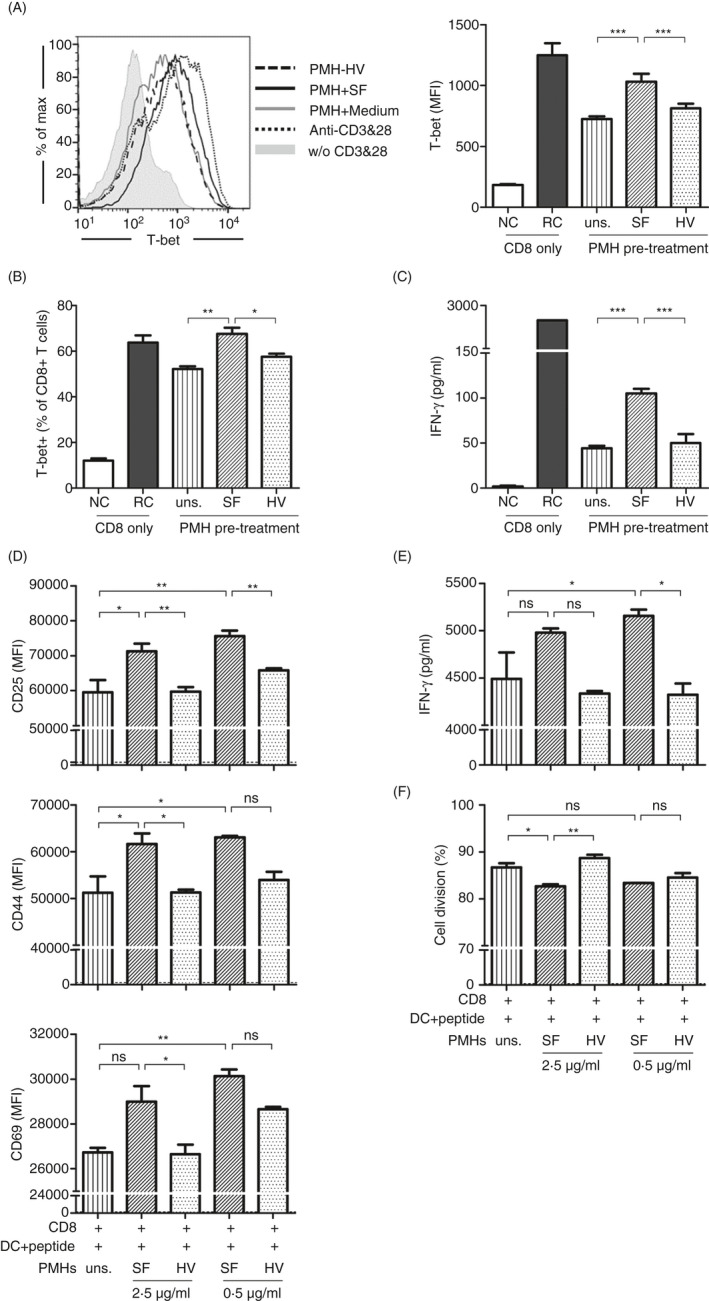
Antigen‐specific CD8+ T‐cell activation and function were improved by SF‐treated PMHs. PMHs from WT C57BL/6 mice were pretreated with SF or HV protein (0.5 or 2.5 µg/ml) for 24 h. MACS‐purified TLR5−/− CD8+ T cells were co‐cultured with pretreated PMHs in the presence of anti‐CD3 and anti‐CD28 for 24 h. (A, B) The expression of the transcription factors T‐bet and (C) the production of IFN‐γ were analysed by flow cytometry and ELISA, respectively. Grey shadow, without (w/o) CD3 and CD28; dark dotted line, anti‐CD3 and anti‐CD28; grey solid line, PMH + medium; dark solid line, PMH + SF; dark dashed line, PMH + HV. (d–f) FV‐TCR Tg CD8+ T cells were pre‐activated by DbGagL FV epitope‐loaded dendritic cells for 24 h and then co‐cultured with the pretreated PMHs. (D) Activation of FV‐TCR Tg CD8+ T cells was evaluated by staining for CD25, CD44 and CD69. (E) IFN‐γ in the supernatant was detected by ELISA. (F) Proliferation of CD8+ T cells was detected by CellTrace Violet division assay. Dash line, results from the FV‐TCR Tg CD8+ T cells pre‐activated with the non‐loaded dendritic cells. Data are representative of at least three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by one‐way ANOVA.
To analyse the antigen‐specific activation of CD8+ T cells, TCR transgenic CD8+ T cells specific to the DbGagL Friend virus epitope (FV‐TCR Tg CD8+ T cells) were primarily activated by peptide‐loaded DCs for 24 h. The CD8+ T cells together with the DCs were subjected to an additional 48‐h co‐culture with PMHs that were untreated or treated with SF. In accordance with our previous results, priming with peptide‐loaded DCs strongly activated the FV‐TCR Tg CD8+ T cells. 16 By co‐culturing with SF‐treated PMHs, the expression of CD25, CD44 and CD69 (Figure 3D) and the secretion of IFN‐γ (Figure 3E) were significantly upregulated in the FV‐TCR Tg CD8+ T cells compared with those of FV‐TCR Tg CD8+ T cells co‐cultured with untreated or HV‐treated PMHs. Stimulation of PMHs with either 2.5 or 0.5 μg/ml SF induced comparable levels of CD25, CD44 and CD69 expression and IFN‐γ production in CD8+ T cells. Over 80% of DC‐activated FV‐TCR Tg CD8+ T cells were proliferating in the present system. The CD8+ T cells show similarly high division rates in all experimental groups (Figure 3F). Taken together, SF‐treated PMHs upregulate both activation markers and cytokine production of antigen‐specific CD8+ T cells.
Hydrodynamic injection of SF regulates the activation of CD8+ T cells in the mouse liver
Therefore, we further asked whether the intrahepatic flagellin could regulate the activation and function of intrahepatic CD8+ T cells in vivo. In our previous study, we found that intraperitoneal injection of recombinant SF into mice resulted in significant recruitment of macrophages and neutrophils into the liver in a short period. 24 However, when mice received a single intraperitoneal injection of SF or HV (10 μg per mouse), PMHs derived from the SF‐ or HV‐injected mice show similar activities to regulate the IFN‐γ production of splenocytes (Figure S4A). The immunosuppressive activity of PMHs was alleviated after continuous administration of SF for 3–6 times at 24 h post‐injection (Figure S4B), while the activation of intrahepatic CD8+ T cells was not significantly changed after the injection (Figure S4C). These results confirm that SF activates the TLR5 signalling pathway of PMHs and regulates their function in vivo. However, delivery of SF by intraperitoneal injection is not sufficient to regulate the intrahepatic CD8+ T cells probably because of the mild and retarded response in the liver.
Alternatively, SF was designed to be administrated by the hydrodynamic injection route to enhance the local response in the liver. SF could be detected by immunohistochemistry staining with anti‐SF antibodies in hepatocytes as early as 30 min post‐injection, indicating successful transfer of flagellin protein into the liver and hepatocytes by hydrodynamic injection (Figure S5A). Significant secretion of IL‐6 was detected in liver lavage at 6 h after injection of SF, which then decreased to baseline at 24 h post‐injection. In contrast, IL‐1β, as the downstream molecule of the inflammasome pathway, was undetectable in liver lavage either with or without injection of SF (Figure 4A). These results suggest that although hydrodynamic injection successfully delivered SF into hepatocytes, SF predominantly activated the TLR5‐ but rarely activated the NLRC4‐related innate immune response in the liver.
FIGURE 4.
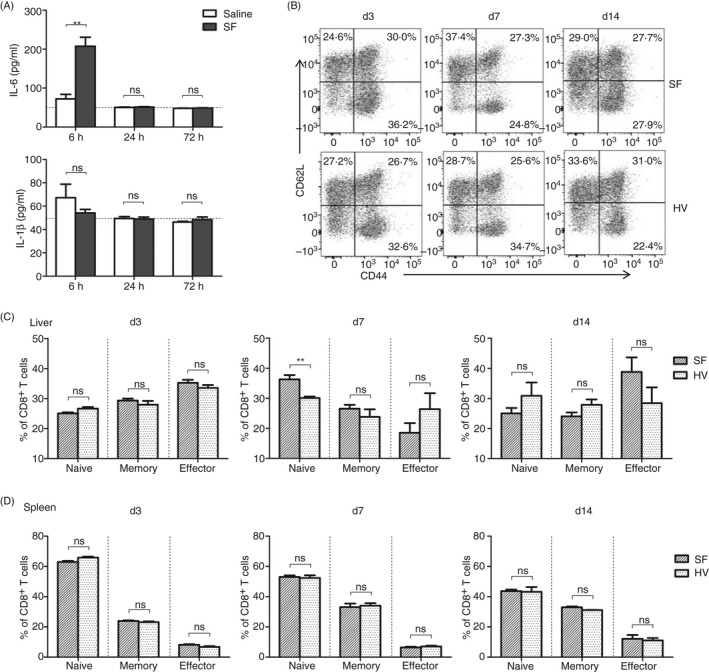
Hydrodynamic injection of SF regulates the activation of lymphocyte subsets in the liver. C57BL/6 mice were hydrodynamically injected with 25 µg SF or HV protein via the tail vein. (A) IL‐6 and IL‐1β in the liver lavage samples at 6, 48 and 72 h post‐injection were measured by ELISA. (B) Differentiation of intrahepatic CD8+ T cells was indicated by CD62L and CD44 expression at 3, 7 and 14 days post‐injection. (C, D) Frequency of naïve (CD62L+CD44−), memory (CD62L+CD44+) and effector (CD62L−CD44+) CD8+ T cells in the liver (C) and spleen (D). n = 6–8 mice per group. Data are representative of at least three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by unpaired t‐test (two‐tailed).
To investigate the role of intrahepatic SF in modulating liver immune cells, component of intrahepatic lymphocytes (IHLs) was monitored by FACS analysis of NK cells, NKT cells, T cells, B cells, macrophages and neutrophils for surface markers at 1 day (24 h), and 3, 7 and 14 days post‐injection (Figure S5B). The total number of IHLs was rapidly increased within 24 h and returned to the baseline after 3 days in all hydrodynamically injected mice with no significant differences between SF‐ or HV‐injected mice (Figure S5C). A transiently increased frequency of NK cells was observed at 24 h and returned to a similar level in SF‐ and HV‐injected mice at 72 h (Figure S5D). The expression of NKG2D is related to the activation of T cells maintained at the similar level on NK cells in all groups after 3 days (Figure S5E). These results suggest that injection of SF activates the innate immune response in the liver, leading to a quick but transient intrahepatic immune cell influx.
In contrast, the absolute number of total CD8+ T cells in the liver was not significantly changed during the monitoring period except for a slight decrease on day 7 in all groups (Figure S5F). The intrahepatic CD8+ T‐cell subsets, including CD62L+CD44+ memory cells and CD62L−CD44+ effector cells, were not changed by SF at the early time‐points until 3 days. However, the frequency of CD62L+CD44− population of intrahepatic CD8+ T cells was significantly increased at 7 days after injection of SF (Figure 4B,C), suggesting that SF regulated the differentiation or accumulation of CD8+ T‐cell subsets in the liver. This was followed by a slightly and not significantly increased CD62L−CD44+ effector CD8+ T‐cell population in the liver of SF‐injected mice at 14 days. In contrast, no significant changes were observed in the splenic CD8+ T cells of mice injected with either SF or HV protein at all time‐points (Figure 4D). Furthermore, the expression of the activation markers, such as CD25 and CD44, and the transcription factors, such as T‐bet and Eomes, was not significantly changed in intrahepatic CD8+ T cell of SF‐ or HV‐injected mice at 7 days post‐injection, along with a slightly increased frequency of CD62L+ CD8+ cells in SF‐injected mice (Figure S5G). These results indicate that hydrodynamically injected SF may regulate the intrahepatic CD8+ T‐cell activation in a liver‐specific manner after several days.
Hepatocytes participate in modulating the functional activation of IHLs induced by hydrodynamic injection of SF
As intrahepatic SF was efficient to regulate the intrahepatic CD8+ T cells alone, we further analysed whether SF could regulate the function of IHLs induced by viral infection. To this aim, SF was administrated together with an HBV‐replicating plasmid (pSM2), which results in an HBV replication in the liver in a 2‐week period and induces HBV‐specific CD8+ T‐cell responses in the spleen and liver. The splenic and intrahepatic lymphocytes were isolated at 3, 5, 7 and 14 days post‐injection and analysed by in vitro stimulation with anti‐CD3 and anti‐CD28 for 48 h (Figure 5A). As a result, slightly increased IFN‐γ secretion from splenocytes was observed in the pSM2 + SF‐injected animals compared with the pSM2‐only or the pSM2 + HV‐injected animals at 3 days post‐injection. No differences in IFN‐γ production were observed among the splenocytes from the mice injected with pSM2 only, pSM2 + SF or pSM2 + HV from 5 to 14 days after injection. The results suggest that hydrodynamic injection of SF into the liver only slightly impacts T cells in splenocytes (Figure 5B).
FIGURE 5.
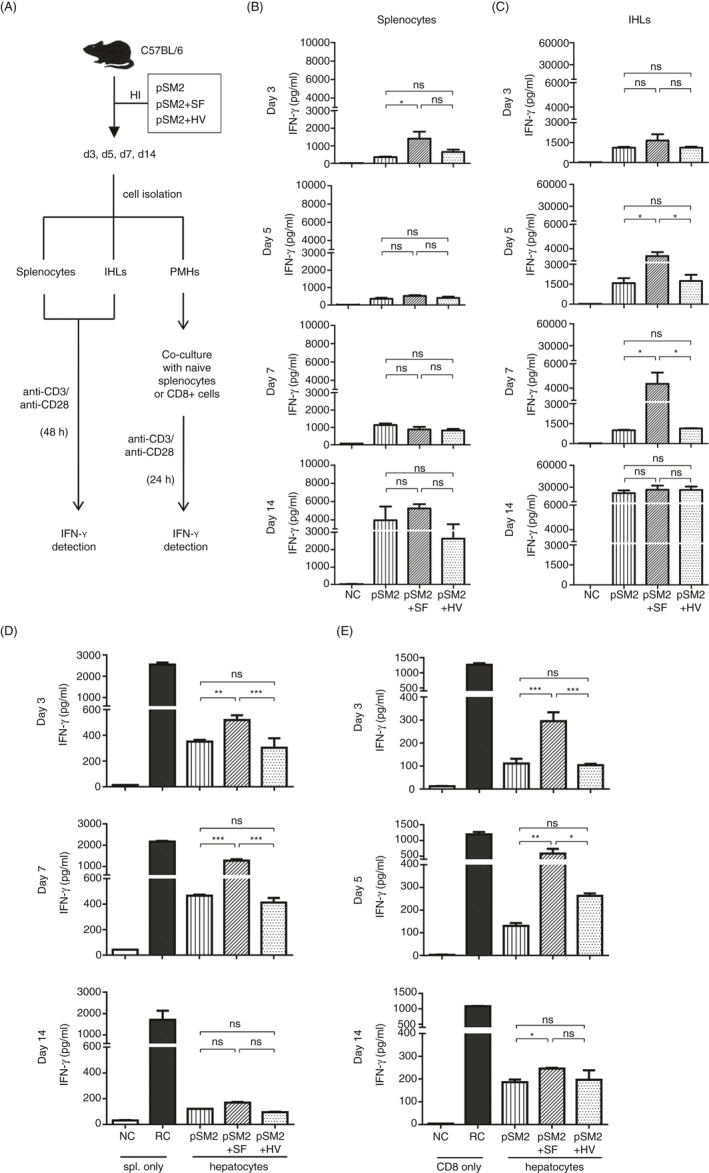
Hydrodynamic injection of SF regulated the function of intrahepatic T cells and hepatic cells. (A) Experimental design of SF treatment, cell isolation and analysis. C57BL/6 mice were hydrodynamically injected with pSM2, pSM2 + SF or pSM2 + HV protein. Splenocytes, intrahepatic lymphocytes (IHLs) and PMHs were isolated at 3, 5, 7 and 14 days post‐injection. (B) Splenocytes and (C) IHLs were stimulated with anti‐CD3 and anti‐CD28 for 48 h. PMHs were co‐cultured with TLR5−/− splenocytes (D) or MACS‐purified TLR5−/− CD8+ T cells (E) that were freshly isolated from naïve C57BL/6 mice in the presence of anti‐CD3 and anti‐CD28 for 24 h. IFN‐γ in the supernatant was detected by ELISA. Unstimulated splenocytes or CD8+ T cells were used as a negative control (NC). Splenocytes or CD8+ T cells stimulated with anti‐CD3/anti‐CD28 alone were used as reactive controls (RC). n = 3–4 mice per group. Data are representative of at least three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by one‐way ANOVA.
Upon anti‐CD3 and anti‐CD28 stimulation, IFN‐γ production by IHLs from the mice injected with pSM2 + SF was also slightly increased compared with that by IHLs from the mice injected with pSM2 only or pSM2 + HV at 3 days after injection (Figure 5C, day 3). However, IFN‐γ production by IHLs from pSM2 + SF group was significantly increased at 5 and 7 days post‐injection (Figure 5C, day 5 and day 7). The IFN‐γ production significantly raised and reached a similar level in all groups at 14 days (Figure 5C, day 14). Intracellular IFN‐γ staining of the anti‐CD3‐stimulated IHLs showed significantly increased IFN‐γ production by the CD8+ T cells isolated from pSM2 + SF‐injected mice at 5 days post‐injection but dropped to the similar level as the control groups in some animals at 7 days post‐injection (Figure S6A). These results suggest that hydrodynamic injection of SF might improve the functional activation of CD8+ T cells in the liver at an early stage.
To determine whether PMHs participated in regulating the function of IHLs by injection of SF, PMHs, LSECs and KCs were isolated at the same time‐points post‐injection, and their immunoregulatory properties were analysed by co‐culture with TLR5−/− splenocytes or purified CD8+ T cells (Figure 5A). Consistent with the LSECs or KCs stimulated by SF in vitro, no significant changes in IFN‐γ secretion were observed between the co‐cultures using LSECs or KCs derived from pSM2 + SF‐ or pSM2 + HV‐injected mice, indicating that these two cell types could not be affected by either TLR5 or inflammasome activities of SF (Figure S6B,C). However, when co‐cultured with hepatocytes isolated from the pSM2 + SF group at 3 or 7 days post‐injection, splenocytes secreted significantly higher levels of IFN‐γ than in the presence of those from either the pSM2 only or the pSM2 + HV group (Figure 5D, day 3, day 7). This difference lasted until 14 d post‐injection (Figure 5D, day 14). Similarly, the IFN‐γ production of purified CD8+ T cells was significantly improved by co‐culture with PMHs separated from pSM2 + SF group at 3 and 5 days post‐injection, but only non‐significant and slight differences were observed when co‐culture with PMHs was separated at 7 days post‐injection (Figure 5E). These results indicate that SF‐experienced PMHs might not only regulate CD8+ T cells within 5 days but also affect other types of IFN‐γ‐producing cells in 7 days. In agreement with the in vitro‐stimulated PMHs, an increased expression of ICAM‐1 on the PMHs of SF‐injected mice was observed on 7 days post‐injection (Figure S6D). Similar results were obtained using the hepatocytes from the mice injected with SF alone but without pSM2, indicating that the SF regulates the immune suppressive ability of hepatocytes independent of viral infection (Figure S6E). These results confirm that hepatocytes, but not LSECs or KCs, participate in the functional modulation of intrahepatic lymphocytes induced by SF in the HBV hydrodynamic injection mouse model.
SF upregulates the intrahepatic CD8+ T‐cell response against HBV in the hydrodynamically injected mouse model
To demonstrate that SF may regulate intrahepatic antigen‐specific CD8+ T cells in vivo, the splenic and intrahepatic viral‐specific CD8+ T‐cell responses were analysed using an HBV hydrodynamically injected mouse model. The HBV‐replicating plasmid pSM2 mixed with SF or HV protein was delivered into mice by hydrodynamic injection via the tail vein. Serum HBsAg and HBV DNA could be detected and showed the typical kinetics of viral replication after hydrodynamic injection within 2 weeks. SF administration only transiently and slightly impaired viral DNA at a very early stage, while the kinetics of HBsAg and HBV DNA clearance were not changed, suggesting that SF does not directly affect viral clearance (Figure S7A,B).
At 2–3 weeks after viral clearance, the virus induced a similar level of peripheral immune response in the pSM2, pSM2 + SF or pSM2 + HV groups, including anti‐HBs antibody levels in the serum (Figure 6A) and HBs‐ and HBc‐specific CD8+ T‐cell responses in the spleen (Figure 6B, S7C). In contrast to the spleen, the frequencies of intrahepatic HBs‐specific IFN‐γ+ CD8+ T cells in pSM2 + SF‐injected mice were significantly increased compared with those of the pSM2 or pSM2 + HV mice (Figure 6C,D). The intrahepatic HBc‐specific IFN‐γ+ CD8+ T cells were also slightly elevated in pSM2 + SF‐injected mice (Figure 6C,D). However, IL‐2‐ and TNF‐α‐producing CD8+ T cells showed no significant differences among the three groups (Figure S7D,E). Moreover, in the persistent HBV‐replicating mice that were immunotolerant to HBV antigens after hydrodynamically injected with the pAAV‐HBV1.2 plasmid, transfer of purified CD8+ T cells containing around 0.7% HBs‐specific IFN‐γ+ CD8+ T cells resulted in suppression of serum HBV antigens both in SF‐ and in HV‐pretreated mice (Figure S8A–C). In comparison with HV, pretreatment with SF led to a slightly lower level of HBsAg and HBeAg at day 8 after transfer, indicating a transiently and mildly improved antiviral activity of CD8+ T cells induced by SF (Figure S8B). At day 21 after transfer, the frequency of HBs208‐specific CD8+ T cells was maintained at a higher level in the liver of SF‐pretreated mice (Figure S8D), although the IFN‐γ production of these HBs208‐specific CD8+ T cells was exhausted in both groups (Figure S8E). In contrast, no significant differences were observed in the spleen between SF‐ and HV‐pretreated mice. These results demonstrate that SF predominantly improves the intrahepatic but not systemic HBV‐specific CD8+ T‐cell responses.
FIGURE 6.
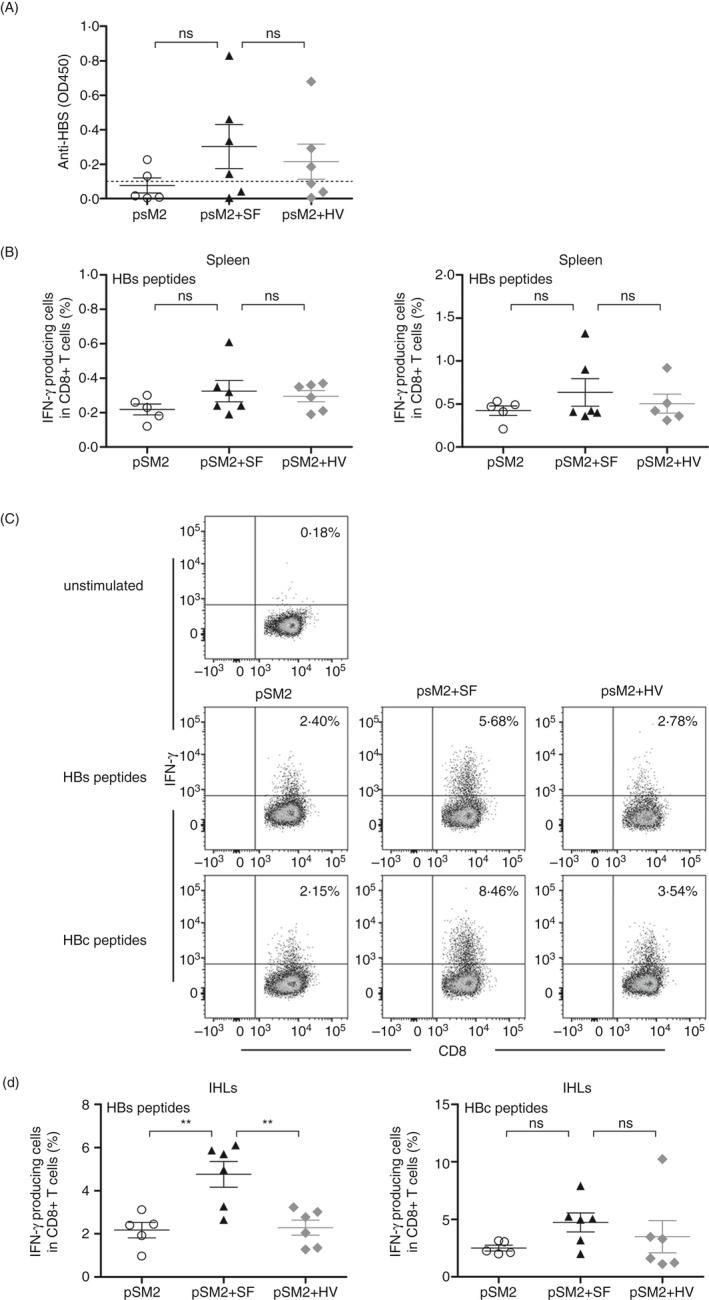
SF improved the intrahepatic viral‐specific CD8+ T‐cell response in the HBV‐replicating mouse model. C57BL/6 mice were hydrodynamically injected with pSM2, pSM2 + SF or pSM2 + HV protein. Animals were killed at day 28 post‐injection. (A) Serum anti‐HBs antibodies were detected by ELISA. Frequencies of HBs‐ or HBc‐specific CD8+ T cells in the spleen (B) or liver (C, D) were detected by intracellular staining for IFN‐γ after 4.5 h ex vivo stimulation with HBs‐ or HBc‐derived peptides. n = 5–6 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: p < 0.05; **: p < 0.01; ***: p < 0.001, statistical relevance was determined by one‐way ANOVA.
Discussion
In the present study, we demonstrated that a unique protein‐based PAMP, flagellin, can modulate liver immunity by activating hepatocytes through the TLR5 signalling pathway. We found that flagellin not only changes the components of immune cell subsets in the liver of mice but also regulates the function of IHLs in a liver‐specific manner. We observed that hydrodynamic injection of flagellin counteracts hepatocyte‐mediated immune suppression. The flagellin‐activated TLR5 signalling pathway in hepatocytes, activated by either in vitro stimulation or in vivo administration, provides a benefit for CD8+ T‐cell activation and enhanced IFN‐γ production when CD8+ T cells encounter hepatocytes. Furthermore, activation of TLR5 by administration of flagellin in an acute HBV‐replicating mouse model showed a significantly elevated intrahepatic HBV‐specific CD8+ T‐cell response without affecting the systemic antibody or T‐cell responses. Our results confirm that flagellin‐triggered TLR5 activation in hepatocytes plays a role in regulating liver immune status and in subsequent modulation of the CD8+ T‐cell responses in the liver.
As cells of an immune‐tolerant organ, liver structural cells, including the parenchymal hepatocytes and NPCs, such as LSECs, KCs and HSCs, usually serve as immunoregulatory cells in liver tissue to maintain liver tolerance and homeostasis. There has been evidence showing that NPCs play important roles in regulating intrahepatic T‐cell responses in the presence of pathogens or PAMPs. 8 , 16 Hepatocytes, which are the most abundant cell type in the liver, usually result in T‐cell suppression due to their lack of costimulatory factors or direct endocytosis and even killing of T cells in the liver. 43 , 44 , 45 However, it is not clear whether hepatocyte‐induced tolerance is modulated by pathogens or PAMPs. Our results in the present study show the possibility of alleviating the immune tolerance of the liver by stimulating hepatocytes with flagellin. Different from the LSECs or KCs, the hepatocytes were found to be responsive to SF in vitro and to mitigate immune suppression of T‐cell activation and function at a broad range of dosages. The hepatocytes isolated from SF‐hydrodynamically injected mice around days 3–7 post‐injection were observed to mitigate immune suppression of T‐cell function by the production of IFN‐γ. The hepatocytes isolated at day 14 were found to return to full immune suppression of T cells, indicating a recovery of hepatocytes after SF stimulation. In a previous report, flagellin induces a rapid activation of the NF‐κB signalling pathway in hepatocytes at 20 min after subcutaneous injection. 22 We did not analyse the functional regulation of hepatocytes immediately after injection because that hydrodynamic injection induced mechanical damages to hepatocytes within 3 days post‐injection. 46 Our results indicated that a single dose of SF efficiently regulates hepatocytes in a specific period.
The TLR5 pathway seems to play a key role in the modulation of hepatocytes during flagellin stimulation. By an in vitro co‐culture of PMHs together with purified CD8+ T cells, we confirmed that SF‐activated hepatocytes can modulate CD8+ T cells independent of the assistance of other APCs or CD4+ T cells in vitro. We tried to explore how TLR5‐activated hepatocytes modulate the activation and function of CD8+ T cells by further detection of the traditional regulatory molecules for CD8+ T‐cell activation. Unfortunately, most of them were either undetectable or non‐responsive to SF in the SF‐treated hepatocytes, except for a slight but constant upregulated expression of ICAM‐1 that was observed. LFA‐1/ICAM‐1 interactions in the liver tissue play a role in regulating liver‐resident T cells to patrol and remain in the hepatic sinusoids, suggesting that ICAM‐1 expression is involved in local immune surveillance and response. 47 , 48 However, our in vitro ICAM‐1 blockade experiment could not provide further evidence to support the possible role of TLR5‐induced ICAM‐1 expression on hepatocytes in regulating intrahepatic CD8+ T cells. The underlying mechanism of TLR5‐activated hepatocytes in modulating CD8+ T cells might be different from traditional dogma. Global characterization of the TLR5‐activated hepatocytes and the modulated CD8+ T cells by single‐cell sequencing may provide us with an opportunity to understand more of the interaction between the antigen‐specific CD8+ T cells and the hepatocytes.
An intracellular NLRC4 inflammasome pathway is also known to be activated by flagellin through NAIP5/6. However, this intracellular pathway is usually activated by a flagellin dose of as much as 1000 times that for cell surface TLR5, which we tested previously. 26 Though some studies have shown that the inflammasome pathway is available in hepatocytes, 40 , 41 activation of this pathway in hepatocytes by intracellular SF was usually undetectable. Alternatively, the inflammasome pathway was reported to be more activated by flagellin in macrophages in vitro. 26 However, no specific immunoregulatory changes in KCs after hydrodynamic injection of SF could be observed in our present study. It is likely that effective TLR5 activation with a normal flagellin dose even by hydrodynamic injection into the liver usually has no significant effect on the intracellular inflammasome pathway.
CD8+ T cells in the liver are regulated by many complicated local environmental factors. TLR agonists selectively activate the responding cells and play multiple roles in the direct or indirect regulation of intrahepatic CD8+ T cells. For example, TLR7 and TLR9 may predominantly activate LSECs and DCs, while TLR2 stimulates numerous cell types in the liver. 17 , 49 , 50 In contrast to TLR2, TLR7 and TLR9, TLR5 is predominantly expressed in hepatocytes, and thus, the target cell types of flagellin are relatively limited in liver tissue. By comparing the activities of different TLR agonists in stimulated PMHs, we found that TLR5‐activated PMHs exert relatively mild activity in regulating CD8+ T‐cell function (Figure S9). Our in vivo experiment showed that SF can increase the intrahepatic CD8+ T‐cell frequency in acute HBV‐replicating mice in which the viral‐specific CD8+ T cells are significantly induced. In contrast, injection of SF did not significantly alter the CD8+ T‐cell response in persistent HBV‐replicating mice in which the viral‐specific CD8+ T cells are tolerant (data not shown). Additionally, by adoptive transferring of the viral‐specific CD8+ T cells to persistent HBV‐replicating mice, injection of SF enhanced the antiviral activity of these CD8+ T cells in a short term, and provided a benefit for maintaining the frequency of HBs‐specific CD8+ T cells in the liver. These results suggest that the enhancement of intrahepatic CD8+ T cells by flagellin might be dependent on the presence of a systemic antigen‐specific CD8+ T‐cell response. It is possible that the specific CD8+ T cells tended to be trapped in the TLR5‐stimulated liver, and the TLR5‐activated hepatocytes may provide an opportunity for antigen‐specific CD8+ T cells to retain their function. Taken together, we acknowledge that flagellin induces hepatocyte‐mediated mild regulation of the function of intrahepatic CD8+ T cells.
In recent decades, intensive studies have revealed that gut microbiota contribute to shaping immune cell responses in the liver, which is continuously exposed to metabolites of intestinal commensal bacteria through the portal vein. The intestinal mucosa is usually protected by the mucus barrier, and thus, only a minority of bacteria or fragments of bacteria might occasionally breach the gut epithelium and arrive in the liver. However, the permeability of the gut increases during alcohol stimulation, infection or liver cirrhosis. 51 These pathological statuses allow increased translocation of gut bacterial products, such as LPS, flagellin and bacterial DNA. 12 Pan et al 52 found that the dynamic changes in plasma LPS were correlated with disease severity in patients with acute or chronic hepatitis B liver failure. Our results suggest that in addition to LPS, other gut bacterial derivatives, such as flagellin, should also be involved in the regulation of liver immune surveillance and responses. The results of our in vitro stimulation of primary hepatocytes showed that hepatocytes respond to flagellin at a low threshold and mediate improved CD8+ T‐cell activation and IFN‐γ production, indicating high TLR5 sensitivity of hepatocytes to flagellin. In vivo administration of SF, either by multiple intraperitoneal injection at low dose or by a single hydrodynamic injection at high dose, shows efficient effect on reducing the hepatocyte‐induced suppression of CD8+ T‐cell function. This suggests that the presence of flagellin in the portal vein may trigger the hepatocytes for immune surveillance by activating the TLR5/MyD88 signalling pathway, and prepare a reduced immune tolerance status by mitigating the immune suppression of hepatocytes for T cells in the liver. These intrahepatic immunological changes may lead to at least two subsequent outcomes. On the one hand, CD8+ T cells that are specific to intrahepatic pathogens could be more efficiently activated and thus improve immune surveillance in the liver. On the other hand, the hepatocyte‐induced immune tolerance might be reduced and result in hypersensitivity to CTL‐related immunopathogenesis. Regarding the notion of immune alerting of hepatocytes by TLR5, we envisage further study to clarify the role of flagellin in modulating liver immunity during acute and chronic viral infection.
Disclosures
The authors declare no commercial or financial conflict of interest.
Supporting information
Figure S1. Immune suppressive activity of liver‐resident cells. (A) PMHs, (B) KCs and (C) LSECs from WT C57BL/6 mice were co‐cultured with TLR5−/− splenocytes in the presence of anti‐CD3 and anti‐CD28 for 24 or 48 hr, and IFN‐γ in the supernatants was detected by ELISA. (D‐G) PMHs, KCs or LSECs from WT C57BL/6 mice were pre‐treated with SF or HV protein (2·5 µg/ml) for 24 hr and then co‐cultured with TLR5−/− splenocytes or CD8+ T cells in the presence of anti‐CD3 and anti‐CD28 for an additional 24 hr. (D) The count and (E) differentiation of living CD8+ T cells in splenocytes were analyzed by flow cytometry after co‐culture with PMHs. (F, G) IFN‐γ in the supernatants was detected by ELISA after co‐culture with LSECs or KCs. Data are representative of two independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S2. Expression of immune regulatory molecules in hepatocytes in response of SF stimulation. PMHs from WT C57BL/6 mice were stimulated with SF or HV protein (2·5 μg/ml). (A, B) Expression of CD80, IL‐7, PD‐L1 and TGFβ2 mRNA were measured by RT‐qPCR, and IL‐12 mRNA was detected by PCR after 6h. (C) The expression of CD40 was measured by flow cytometry was detected by ELISA after 24 hr stimulation. (D) PMHs were seed in the 12‐well plate and stimulated with SF or HV protein. After 24 hr, TLR5−/− splenocytes were added in the trans‐well chamber and cultured in the present of anti‐CD3 and anti‐CD28 for an additional 24 hr. (E) Conditioned medium of PMHs was harvested at 24 hr after stimulation, and the TLR5−/− splenocytes were stimulated by anti‐CD3 and anti‐CD28 for 24 hr in the presence of conditioned medium. IFN‐γ in the supernatants was detected by ELISA. (F) The expression of GM‐CSF was measured by ELISA after 24 hr stimulation. Expression of ICAM‐1 was detected by RT‐qPCR after 6 hr (G) or by flow cytometry after 24 hr stimulation (H), respectively. Anti‐CD3/CD28 activated TLR5−/− splenocytes were co‐cultured with SF‐ or HV‐pretreated PMHs in the presence of neutralizing ICAM‐1 (I), GM‐CSF (J), IL‐6 (K) or corresponding isotype controls. IFN‐γ in the supernatant was measured by ELISA after 24 hr. n = 4–6 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S3. The purity and count of living CD8+ T cells. (A) The purity of MACS‐purified CD8+ T cells. MACS‐purified CD8+ T cells were stained with anti‐CD45, CD8, CD11c and MHC‐Ⅱ. Dead cells were excluded by fixable viability dye. The frequencies of CD45+ CD8+ T cells and CD11c+ MHCII+ dendritic cells after purification were shown. (B) MACS‐purified CD8+ T cells were stimulated with 5 μg/ml of SF or HV in the presence of plate‐coated anti‐CD3 and anti‐CD28, and the IFN‐ γ in supernatant was detected by ELISA after 24 hr. (C) The count of living CD8+ T cells after co‐culture with PMHs for 24 hr. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S4. SF regulated the immune suppressive activities of PMHs after intraperitoneal injection. C57BL/6 mice were intraperitoneal injected with 5 µg SF or HV protein every other day for three times by intraperitoneal injection. PMHs were isolated at 24 hr after the first injection (A) or the final injection (B). PMHs were co‐cultured with TLR5−/− splenocytes derived from naïve C57BL/6 mice in the present of anti‐CD3 and anti‐CD28 for 24 hr. The production of IFN‐γ is detected by ELISA. (C) IHLs were isolated at 24 hr after the final injection. The frequencies of naïve (CD62L+CD44−), memory (CD62L+CD44+) and effector (CD62L−CD44+) CD8+ T cells in the IHLs were detected by Flow cytometry. n = 3–4 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01, statistical relevance was determined by two‐tailed, unpaired t‐test.
Figure S5. Hydrodynamic injection of SF modulated the component of intrahepatic lymphocytes. C57BL/6 mice were hydrodynamically injected with 25 µg SF or HV protein via the tail vein. (A) Immunohistochemistry analysis of SF in liver tissue at 30min after injection. (B) Gating strategy of T, B, NK, NKT, macrophages and neutrophils. Splenocytes or IHLs were stained with anti‐CD3, NK1.1, B220, F4/80 and Gr‐1. Dead cells were excluded by 7aad staining. (C) The number of IHLs was counted at 3 d, 7 d and 14 d post‐infection. (D) The NK cells (CD3−NK1.1+), NKT cells (CD3+NK1.1+), T cells (CD3+B220−NK1.1−), B cells (CD3−B220+NK1.1−), macrophages (F4/80hiGr‐1low) and neutrophils (F4/80−Gr‐1high) in the IHLs were analyzed at 24 and 72 hr post‐injection. (E) The expression of NKG2D on NK cells was analyzed by flow cytometry at 3, 7 and 14 days post‐injection. (F) The count and (G) expression of CD25, CD44, CD62L, T‐bet and Eomes in the intrahepatic CD8+ T cells at day 7 after SF or HV injection. n = 3–4 mice per group. Data are representative of at least three independent experiments. Bars: mean ± SD. Statistical relevance was determined by one‐way ANOVA.
Figure S6. SF regulated the function of PMHs but not LSECs or KCs in vivo. C57BL/6 mice were hydrodynamically injected with pSM2, pSM2 + SF or pSM2 + HV protein. IHLs, PMHs, LSECs and KCs were isolated at 3, 5, 7 or 14 days post‐injection. (A) IHLs were isolated at 5 or 7 days post‐injection, and the IFN‐γ producing CD8+ T cells were analyzed by intracellular staining after stimulation with anti‐CD3 and anti‐CD28 for 6h in the presence of Brefildin A. LSECs, KCs and PMHs were isolated at 3, 7 or 14 days post‐injection. (B) LSECs and (C) KCs were co‐cultured with splenocytes that were freshly isolated from naïve C57BL/6 mice in the presence of anti‐CD3 and anti‐CD28 for 24 hr. IFN‐γ in the supernatants was detected by ELISA. Unstimulated splenocytes were used as a negative control (NC). Splenocytes stimulated with anti‐CD3/anti‐CD28 alone were used as reactive controls (RC). (D) The expression of ICAM‐1 on PMHs was detected by flow cytometry at 3, 7 and 14 days post‐injection. (E) PMHs were isolated at 7 or 14 days after hydrodynamic injection of saline, SF or HV protein in the absence of pSM2. PMHs were co‐cultured with splenocytes for 24 hr. The production of IFN‐γ is detected by ELISA and described as fold change compared to the splenocytes co‐cultured with PMHs derived from saline group at the corresponding time points. n = 3–4 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by two‐tailed, unpaired t‐test.
Figure S7. Kinetics of HBV replication and T cell response after hydrodynamic injection of HBV replicating plasmid. C57BL/6 mice were hydrodynamic injected by pSM2 plasmid together with 25 μg SF or HV protein via the tail veins. (A) HBsAg, HBeAg and (B) HBV DNA in the serum were detected on day 3, 7 and 14 post injection by ELISA or realtime‐PCR. Frequencies of HBs‐ or HBc‐specific CD8+ T cells in the spleen (C) or liver (D, E) were detected by intracellular staining for IFN‐γ, TNF‐α or IL‐2 after 4·5 hr ex vivo stimulation with HBs‐ or HBc‐derived peptides. n = 5–6 mice per group. Data are representative of three independent experiments. n = 4–6 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S8. SF improved the intrahepatic viral‐specific CD8+ T cell responses in the persistent HBV replicating mouse model. (A) Schematic of experimental design. The persistent HBV replicating mice were constructed by hydrodynamic injection of pAAV‐HBV1.2 plasmid. Saline, SF or HV were hydrodynamically injected after 10 days. CD8+ T cells were purified from pHBsAg‐WT immunized mice and transferred into SF‐ or HV‐injected recipient. HI: Hydrodynamic injection. (B) Serum HBsAg and HBeAg were detected at indicated timepoints by ELISA. *: P < 0·05; **: P < 0·01; ***: P < 0·001; ****: P < 0·0001, statistical differences between w/o transfer and SF pretreated group; #: P < 0·05, statistical differences between SF pretreated and HV pretreated group; $: P < 0·05; $$: P < 0·01, statistical differences between w/o transfer group and HV pretreated group. Statistical relevance was determined by Two‐way ANOVA. (C) Frequency of HBs‐specific IFN‐γ producing cells in the donor CD8+ T cells. (D, E) The splenocytes and IHLs were separated from mice at 21 days post‐transfer. (D) HBs‐specific CD8+ T cells were analyzed by HBs208 peptide‐loaded dimer staining. (E) IFN‐γ producing cells in the Dimer+ cells were analyzed intracellular staining after 4·5 hr ex vivo stimulation with HBs208 peptide. n = 5–7 mice per group. Bars: mean ± SD. *: P < 0·05, statistical relevance was determined by one‐way ANOVA (D, E). Data are representative of two independent experiments.
Figure S9. The CD8+ T cell activation and function was regulated by TLR‐stimulated PMHs. PMHs from WT C57BL/6 mice were pre‐stimulated with TLR1‐9 agonists (InvivoGen) including TLR2 (P3C, 4 μg/ml), TLR3 (poly I:C, 50 μg/ml), TLR4 (LPS, 30 μg/ml), TLR5 (SF, 2·5 μg/ml), TLR7 (imiquimod, 20 ng/ml), TLR7/8 (R848, 10 μg/ml), TLR9 (ODN1826, 31·8 μg/ml) for 24 hr, then washed and co‐cultured with WT splenocytes in the presence of anti‐CD3 and anti‐CD28. (A) Expression of activating markers CD25, CD44 and CD69 on CD8+ T cells. (B) Production of IFN‐γ in the CD8+ T cells were analyzed by intracellular staining. Data are representative of at least three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Acknowledgements
HY, EZ, ML and HMY conceived and designed the study and wrote the paper. HY, EZ, JG and YZ performed the experiments and analysed the data. YY, JY and BZ set up the detection methods. JYY, MZ and UD provided the materials and reagents. HY, ZM, JW and CW performed the hydrodynamic injection of animals. HY and EZ drafted the paper. UD, DY, ML and HMY revised the paper. HMY supervised this project. All authors have read and approved the final version of this paper.
We thank Xuefang An, Fan Zhang and Juan Min in the core facility for technical support of the Wuhan Institute of Virology for their kind help in animal experiments and flow cytometry analysis, respectively.
This work was supported by grants from Deutsche Forschungsgemeinschaft (TRR60) and the National Natural Science Foundation of China (Nos. 81771688, 81461130019, 81971568 and 31970878), the Natural Science Foundation of Guangzhou Province (2016A030311046) and Scientific and Technological Project of Guangdong Province (2017B020226006).
Contributor Information
Ejuan Zhang, Email: zhangejuan@wh.iov.cn.
Huimin Yan, Email: hmyan@wh.iov.cn.
Data availability statement
Data availability is not applicable to this work.
References
- 1. Crispe IN, Giannandrea M, Klein I, John B, Sampson B, Wuensch S. Cellular and molecular mechanisms of liver tolerance. Immunol Rev. 2006;213:101–18. [DOI] [PubMed] [Google Scholar]
- 2. Zheng M, Tian Z. Liver‐mediated adaptive immune tolerance. Front Immunol. 2019;10:2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Spangenberg HC, Viazov S, Kersting N, Neumann‐Haefelin C, McKinney D, Roggendorf M, et al. Intrahepatic CD8+ T‐cell failure during chronic hepatitis C virus infection. Hepatology. 2005;42:828–37. [DOI] [PubMed] [Google Scholar]
- 4. Kubes P, Jenne C. Immune responses in the liver. Annu Rev Immunol. 2018;36:247–77. [DOI] [PubMed] [Google Scholar]
- 5. Pillarisetty VG, Shah AB, Miller G, Bleier JI, DeMatteo RP. Liver dendritic cells are less immunogenic than spleen dendritic cells because of differences in subtype composition. J Immunol. 2004;172:1009–17. [DOI] [PubMed] [Google Scholar]
- 6. Bottcher JP, Schanz O, Wohlleber D, Abdullah Z, Debey‐Pascher S, Staratschek‐Jox A, et al. Liver‐primed memory T cells generated under noninflammatory conditions provide anti‐infectious immunity. Cell Rep. 2013;3:779–95. [DOI] [PubMed] [Google Scholar]
- 7. Wahl C, Bochtler P, Chen L, Schirmbeck R, Reimann J. B7–H1 on hepatocytes facilitates priming of specific CD8 T cells but limits the specific recall of primed responses. Gastroenterology. 2008;135:980–8. [DOI] [PubMed] [Google Scholar]
- 8. Liu J, Yu Q, Wu W, Huang X, Broering R, Werner M, et al. TLR2 stimulation strengthens intrahepatic myeloid‐derived cell‐mediated T cell tolerance through inducing Kupffer cell expansion and IL‐10 production. J Immunol. 2018;200:2341–51. [DOI] [PubMed] [Google Scholar]
- 9. Crispe IN. Immune tolerance in liver disease. Hepatology. 2014;60:2109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knolle PA, Thimme R. Hepatic immune regulation and its involvement in viral hepatitis infection. Gastroenterology. 2014;146:1193–207. [DOI] [PubMed] [Google Scholar]
- 11. Schierwagen R, Alvarez‐Silva C, Madsen MSA, Kolbe CC, Meyer C, Thomas D, et al. Circulating microbiome in blood of different circulatory compartments. Gut 2019;68 3:578–580. [DOI] [PubMed] [Google Scholar]
- 12. Chassaing B, Etienne‐Mesmin L, Gewirtz AT. Microbiota‐liver axis in hepatic disease. Hepatology. 2014;59:328–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Imajo K, Fujita K, Yoneda M, Nozaki Y, Ogawa Y, Shinohara Y, et al. Hyperresponsivity to low‐dose endotoxin during progression to nonalcoholic steatohepatitis is regulated by leptin‐mediated signaling. Cell Metab. 2012;16:44–54. [DOI] [PubMed] [Google Scholar]
- 14. El Kasmi KC, Anderson AL, Devereaux MW, Fillon SA, Harris JK, Lovell MA, et al. Toll‐like receptor 4‐dependent Kupffer cell activation and liver injury in a novel mouse model of parenteral nutrition and intestinal injury. Hepatology. 2012;55:1518–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang LR, Wohlleber D, Reisinger F, Jenne CN, Cheng RL, Abdullah Z, et al. Intrahepatic myeloid‐cell aggregates enable local proliferation of CD8(+) T cells and successful immunotherapy against chronic viral liver infection. Nat Immunol. 2013;14:574–83. [DOI] [PubMed] [Google Scholar]
- 16. Liu J, Jiang M, Ma Z, Dietze KK, Zelinskyy G, Yang D, et al. TLR1/2 ligand‐stimulated mouse liver endothelial cells secrete IL‐12 and trigger CD8+ T cell immunity in vitro. J Immunol. 2013;191:6178–90. [DOI] [PubMed] [Google Scholar]
- 17. Ma Z, Zhang E, Yang D, Lu M. Contribution of Toll‐like receptors to the control of hepatitis B virus infection by initiating antiviral innate responses and promoting specific adaptive immune responses. Cell Mol Immunol. 2015;12:273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gehring AJ, Protzer U. Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology. 2019;156:325–37. [DOI] [PubMed] [Google Scholar]
- 19. Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll‐like receptor 5. Nature 2001;410:1099–103. [DOI] [PubMed] [Google Scholar]
- 20. Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 2011;477:592–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011;477:596–600. [DOI] [PubMed] [Google Scholar]
- 22. Burdelya LG, Brackett CM, Kojouharov B, Gitlin II, Leonova KI, Gleiberman AS, et al. Central role of liver in anticancer and radioprotective activities of Toll‐like receptor 5 agonist. Proc Natl Acad Sci USA. 2013;110:E1857–E1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brackett CM, Kojouharov B, Veith J, Greene KF, Burdelya LG, Gollnick SO, et al. Toll‐like receptor‐5 agonist, entolimod, suppresses metastasis and induces immunity by stimulating an NK‐dendritic‐CD8+ T‐cell axis. Proc Natl Acad Sci USA. 2016;113:E874–E883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao Y, Liu F, Yang J, Zhong M, Zhang E, Li Y, et al. Over‐activation of TLR5 signaling by high‐dose flagellin induces liver injury in mice. Cell Mol Immunol. 2015;12:729–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franchi L, Kamada N, Nakamura Y, Burberry A, Kuffa P, Suzuki S, et al. NLRC4‐driven production of IL‐1beta discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat Immunol. 2012;13:449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J, Zhang E, Liu F, Zhang Y, Zhong M, Li Y, et al. Flagellins of Salmonella Typhi and nonpathogenic Escherichia coli are differentially recognized through the NLRC4 pathway in macrophages. J Innate Immun. 2014;6:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boaru SG, Borkham‐Kamphorst E, Tihaa L, Haas U, Weiskirchen R. Expression analysis of inflammasomes in experimental models of inflammatory and fibrotic liver disease. J Inflamm (Lond). 2012;9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akhmetzyanova I, Drabczyk M, Neff CP, Gibbert K, Dietze KK, Werner T, et al. PD‐L1 expression on retrovirus‐infected cells mediates immune escape from CD8+ T cell killing. PLoS Pathog. 2015;11:e1005224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li W, Yang J, Zhang E, Zhong M, Xiao Y, Yu J, et al. Activation of NLRC4 downregulates TLR5‐mediated antibody immune responses against flagellin. Cell Mol Immunol. 2016;13:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang J, Zhao Y, Li P, Yang Y, Zhang E, Zhong M, et al. Sequence determinants of specific pattern‐recognition of bacterial ligands by the NAIP‐NLRC4 inflammasome. Cell Discov. 2018;4:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Z, Liu J, Wu W, Zhang E, Zhang X, Li Q, et al. The IL‐1R/TLR signaling pathway is essential for efficient CD8(+) T‐cell responses against hepatitis B virus in the hydrodynamic injection mouse model. Cell Mol Immunol. 2017;14:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, et al. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA‐1. Hepatology 2011;53:1476–85. [DOI] [PubMed] [Google Scholar]
- 33. Cao Y, Zhang E, Yang J, Yang Y, Yu J, Xiao Y, et al. Frontline Science: Nasal epithelial GM‐CSF contributes to TLR5‐mediated modulation of airway dendritic cells and subsequent IgA response. J Leukoc Biol. 2017;102:575–87. [DOI] [PubMed] [Google Scholar]
- 34. Buzzo CL, Campopiano JC, Massis LM, Lage SL, Cassado AA, Leme‐Souza R, et al. A novel pathway for inducible nitric‐oxide synthase activation through inflammasomes. J Biol Chem. 2010;285:32087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Machen J, Harnaha J, Lakomy R, Styche A, Trucco M, Giannoukakis N. Antisense oligonucleotides down‐regulating costimulation confer diabetes‐preventive properties to nonobese diabetic mouse dendritic cells. J Immunol. 2004;173:4331–41. [DOI] [PubMed] [Google Scholar]
- 36. Wu J, Lu M, Meng Z, Trippler M, Broering R, Szczeponek A, et al. Toll‐like receptor‐mediated control of HBV replication by nonparenchymal liver cells in mice. Hepatology 2007;46:1769–78. [DOI] [PubMed] [Google Scholar]
- 37. Wu C, Deng W, Deng L, Cao L, Qin B, Li S, et al. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol. 2012;86:4658–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang LR, Wu HL, Chen PJ, Chen DS. An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci USA. 2006;103:17862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu C, Zhang X, Tian Y, Song J, Yang D, Roggendorf M, et al. Biological significance of amino acid substitutions in hepatitis B surface antigen (HBsAg) for glycosylation, secretion, antigenicity and immunogenicity of HBsAg and hepatitis B virus replication. J Gen Virol. 2010;91:483–92. [DOI] [PubMed] [Google Scholar]
- 40. Szabo G, Petrasek J. Inflammasome activation and function in liver disease. Nat Rev Gastroenterol Hepatol. 2015;12:387–400. [DOI] [PubMed] [Google Scholar]
- 41. Han CY, Rho HS, Kim A, Kim TH, Jang K, Jun DW, et al. FXR inhibits endoplasmic reticulum stress‐induced NLRP3 inflammasome in hepatocytes and ameliorates liver injury. Cell Rep. 2018;24:2985–99. [DOI] [PubMed] [Google Scholar]
- 42. Broering R, Lutterbeck M, Trippler M, Kleinehr K, Poggenpohl L, Paul A, et al. Long‐term stimulation of Toll‐like receptor 3 in primary human hepatocytes leads to sensitization for antiviral responses induced by poly I: C treatment. J Viral Hepat. 2014;21:480–90. [DOI] [PubMed] [Google Scholar]
- 43. Muhlbauer M, Fleck M, Schutz C, Weiss T, Froh M, Blank C, et al. PD‐L1 is induced in hepatocytes by viral infection and by interferon‐alpha and ‐gamma and mediates T cell apoptosis. J Hepatol. 2006;45:520–8. [DOI] [PubMed] [Google Scholar]
- 44. Bertolino P, McCaughan GW, Bowen DG. Role of primary intrahepatic T‐cell activation in the 'liver tolerance effect'. Immunol Cell Biol. 2002;80:84–92. [DOI] [PubMed] [Google Scholar]
- 45. Benseler V, Warren A, Vo M, Holz LE, Tay SS, Le Couteur DG, et al. Hepatocyte entry leads to degradation of autoreactive CD8 T cells. Proc Natl Acad Sci USA. 2011;108:16735–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu LL, Peng WH, Wu HL, Miaw SC, Yeh SH, Yang HC, et al. Lymphocyte antigen 6 complex, locus C(+) monocytes and Kupffer cells orchestrate liver immune responses against hepatitis B virus in mice. Hepatology 2019;69:2364–80. [DOI] [PubMed] [Google Scholar]
- 47. McNamara HA, Cai Y, Wagle MV, Sontani Y, Roots CM, Miosge LA, et al. Up‐regulation of LFA‐1 allows liver‐resident memory T cells to patrol and remain in the hepatic sinusoids. Sci Immunol. 2017;2:eaaj1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Un K, Kawakami S, Yoshida M, Higuchi Y, Suzuki R, Maruyama K, et al. Efficient suppression of murine intracellular adhesion molecule‐1 using ultrasound‐responsive and mannose‐modified lipoplexes inhibits acute hepatic inflammation. Hepatology 2012;56:259–69. [DOI] [PubMed] [Google Scholar]
- 49. Wu J, Meng Z, Jiang M, Zhang E, Trippler M, Broering R, et al. Toll‐like receptor‐induced innate immune responses in non‐parenchymal liver cells are cell type‐specific. Immunology 2010;129:363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang E, Yan H, Li Q, Dittmer U, Yan H, Lu M. Activation of the TLR signaling pathway in CD8+ T cells counteracts liver endothelial cell‐induced T cell tolerance. Cell Mol Immunol. 2019;16:774–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tilg H, Cani PD, Mayer EA. Gut microbiome and liver diseases. Gut 2016;65:2035–44. [DOI] [PubMed] [Google Scholar]
- 52. Pan C, Gu Y, Zhang W, Zheng Y, Peng L, Deng H, et al. Dynamic changes of lipopolysaccharide levels in different phases of acute on chronic hepatitis B liver failure. PLoS One 2012;7:e49460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immune suppressive activity of liver‐resident cells. (A) PMHs, (B) KCs and (C) LSECs from WT C57BL/6 mice were co‐cultured with TLR5−/− splenocytes in the presence of anti‐CD3 and anti‐CD28 for 24 or 48 hr, and IFN‐γ in the supernatants was detected by ELISA. (D‐G) PMHs, KCs or LSECs from WT C57BL/6 mice were pre‐treated with SF or HV protein (2·5 µg/ml) for 24 hr and then co‐cultured with TLR5−/− splenocytes or CD8+ T cells in the presence of anti‐CD3 and anti‐CD28 for an additional 24 hr. (D) The count and (E) differentiation of living CD8+ T cells in splenocytes were analyzed by flow cytometry after co‐culture with PMHs. (F, G) IFN‐γ in the supernatants was detected by ELISA after co‐culture with LSECs or KCs. Data are representative of two independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S2. Expression of immune regulatory molecules in hepatocytes in response of SF stimulation. PMHs from WT C57BL/6 mice were stimulated with SF or HV protein (2·5 μg/ml). (A, B) Expression of CD80, IL‐7, PD‐L1 and TGFβ2 mRNA were measured by RT‐qPCR, and IL‐12 mRNA was detected by PCR after 6h. (C) The expression of CD40 was measured by flow cytometry was detected by ELISA after 24 hr stimulation. (D) PMHs were seed in the 12‐well plate and stimulated with SF or HV protein. After 24 hr, TLR5−/− splenocytes were added in the trans‐well chamber and cultured in the present of anti‐CD3 and anti‐CD28 for an additional 24 hr. (E) Conditioned medium of PMHs was harvested at 24 hr after stimulation, and the TLR5−/− splenocytes were stimulated by anti‐CD3 and anti‐CD28 for 24 hr in the presence of conditioned medium. IFN‐γ in the supernatants was detected by ELISA. (F) The expression of GM‐CSF was measured by ELISA after 24 hr stimulation. Expression of ICAM‐1 was detected by RT‐qPCR after 6 hr (G) or by flow cytometry after 24 hr stimulation (H), respectively. Anti‐CD3/CD28 activated TLR5−/− splenocytes were co‐cultured with SF‐ or HV‐pretreated PMHs in the presence of neutralizing ICAM‐1 (I), GM‐CSF (J), IL‐6 (K) or corresponding isotype controls. IFN‐γ in the supernatant was measured by ELISA after 24 hr. n = 4–6 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S3. The purity and count of living CD8+ T cells. (A) The purity of MACS‐purified CD8+ T cells. MACS‐purified CD8+ T cells were stained with anti‐CD45, CD8, CD11c and MHC‐Ⅱ. Dead cells were excluded by fixable viability dye. The frequencies of CD45+ CD8+ T cells and CD11c+ MHCII+ dendritic cells after purification were shown. (B) MACS‐purified CD8+ T cells were stimulated with 5 μg/ml of SF or HV in the presence of plate‐coated anti‐CD3 and anti‐CD28, and the IFN‐ γ in supernatant was detected by ELISA after 24 hr. (C) The count of living CD8+ T cells after co‐culture with PMHs for 24 hr. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S4. SF regulated the immune suppressive activities of PMHs after intraperitoneal injection. C57BL/6 mice were intraperitoneal injected with 5 µg SF or HV protein every other day for three times by intraperitoneal injection. PMHs were isolated at 24 hr after the first injection (A) or the final injection (B). PMHs were co‐cultured with TLR5−/− splenocytes derived from naïve C57BL/6 mice in the present of anti‐CD3 and anti‐CD28 for 24 hr. The production of IFN‐γ is detected by ELISA. (C) IHLs were isolated at 24 hr after the final injection. The frequencies of naïve (CD62L+CD44−), memory (CD62L+CD44+) and effector (CD62L−CD44+) CD8+ T cells in the IHLs were detected by Flow cytometry. n = 3–4 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01, statistical relevance was determined by two‐tailed, unpaired t‐test.
Figure S5. Hydrodynamic injection of SF modulated the component of intrahepatic lymphocytes. C57BL/6 mice were hydrodynamically injected with 25 µg SF or HV protein via the tail vein. (A) Immunohistochemistry analysis of SF in liver tissue at 30min after injection. (B) Gating strategy of T, B, NK, NKT, macrophages and neutrophils. Splenocytes or IHLs were stained with anti‐CD3, NK1.1, B220, F4/80 and Gr‐1. Dead cells were excluded by 7aad staining. (C) The number of IHLs was counted at 3 d, 7 d and 14 d post‐infection. (D) The NK cells (CD3−NK1.1+), NKT cells (CD3+NK1.1+), T cells (CD3+B220−NK1.1−), B cells (CD3−B220+NK1.1−), macrophages (F4/80hiGr‐1low) and neutrophils (F4/80−Gr‐1high) in the IHLs were analyzed at 24 and 72 hr post‐injection. (E) The expression of NKG2D on NK cells was analyzed by flow cytometry at 3, 7 and 14 days post‐injection. (F) The count and (G) expression of CD25, CD44, CD62L, T‐bet and Eomes in the intrahepatic CD8+ T cells at day 7 after SF or HV injection. n = 3–4 mice per group. Data are representative of at least three independent experiments. Bars: mean ± SD. Statistical relevance was determined by one‐way ANOVA.
Figure S6. SF regulated the function of PMHs but not LSECs or KCs in vivo. C57BL/6 mice were hydrodynamically injected with pSM2, pSM2 + SF or pSM2 + HV protein. IHLs, PMHs, LSECs and KCs were isolated at 3, 5, 7 or 14 days post‐injection. (A) IHLs were isolated at 5 or 7 days post‐injection, and the IFN‐γ producing CD8+ T cells were analyzed by intracellular staining after stimulation with anti‐CD3 and anti‐CD28 for 6h in the presence of Brefildin A. LSECs, KCs and PMHs were isolated at 3, 7 or 14 days post‐injection. (B) LSECs and (C) KCs were co‐cultured with splenocytes that were freshly isolated from naïve C57BL/6 mice in the presence of anti‐CD3 and anti‐CD28 for 24 hr. IFN‐γ in the supernatants was detected by ELISA. Unstimulated splenocytes were used as a negative control (NC). Splenocytes stimulated with anti‐CD3/anti‐CD28 alone were used as reactive controls (RC). (D) The expression of ICAM‐1 on PMHs was detected by flow cytometry at 3, 7 and 14 days post‐injection. (E) PMHs were isolated at 7 or 14 days after hydrodynamic injection of saline, SF or HV protein in the absence of pSM2. PMHs were co‐cultured with splenocytes for 24 hr. The production of IFN‐γ is detected by ELISA and described as fold change compared to the splenocytes co‐cultured with PMHs derived from saline group at the corresponding time points. n = 3–4 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by two‐tailed, unpaired t‐test.
Figure S7. Kinetics of HBV replication and T cell response after hydrodynamic injection of HBV replicating plasmid. C57BL/6 mice were hydrodynamic injected by pSM2 plasmid together with 25 μg SF or HV protein via the tail veins. (A) HBsAg, HBeAg and (B) HBV DNA in the serum were detected on day 3, 7 and 14 post injection by ELISA or realtime‐PCR. Frequencies of HBs‐ or HBc‐specific CD8+ T cells in the spleen (C) or liver (D, E) were detected by intracellular staining for IFN‐γ, TNF‐α or IL‐2 after 4·5 hr ex vivo stimulation with HBs‐ or HBc‐derived peptides. n = 5–6 mice per group. Data are representative of three independent experiments. n = 4–6 mice per group. Data are representative of three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Figure S8. SF improved the intrahepatic viral‐specific CD8+ T cell responses in the persistent HBV replicating mouse model. (A) Schematic of experimental design. The persistent HBV replicating mice were constructed by hydrodynamic injection of pAAV‐HBV1.2 plasmid. Saline, SF or HV were hydrodynamically injected after 10 days. CD8+ T cells were purified from pHBsAg‐WT immunized mice and transferred into SF‐ or HV‐injected recipient. HI: Hydrodynamic injection. (B) Serum HBsAg and HBeAg were detected at indicated timepoints by ELISA. *: P < 0·05; **: P < 0·01; ***: P < 0·001; ****: P < 0·0001, statistical differences between w/o transfer and SF pretreated group; #: P < 0·05, statistical differences between SF pretreated and HV pretreated group; $: P < 0·05; $$: P < 0·01, statistical differences between w/o transfer group and HV pretreated group. Statistical relevance was determined by Two‐way ANOVA. (C) Frequency of HBs‐specific IFN‐γ producing cells in the donor CD8+ T cells. (D, E) The splenocytes and IHLs were separated from mice at 21 days post‐transfer. (D) HBs‐specific CD8+ T cells were analyzed by HBs208 peptide‐loaded dimer staining. (E) IFN‐γ producing cells in the Dimer+ cells were analyzed intracellular staining after 4·5 hr ex vivo stimulation with HBs208 peptide. n = 5–7 mice per group. Bars: mean ± SD. *: P < 0·05, statistical relevance was determined by one‐way ANOVA (D, E). Data are representative of two independent experiments.
Figure S9. The CD8+ T cell activation and function was regulated by TLR‐stimulated PMHs. PMHs from WT C57BL/6 mice were pre‐stimulated with TLR1‐9 agonists (InvivoGen) including TLR2 (P3C, 4 μg/ml), TLR3 (poly I:C, 50 μg/ml), TLR4 (LPS, 30 μg/ml), TLR5 (SF, 2·5 μg/ml), TLR7 (imiquimod, 20 ng/ml), TLR7/8 (R848, 10 μg/ml), TLR9 (ODN1826, 31·8 μg/ml) for 24 hr, then washed and co‐cultured with WT splenocytes in the presence of anti‐CD3 and anti‐CD28. (A) Expression of activating markers CD25, CD44 and CD69 on CD8+ T cells. (B) Production of IFN‐γ in the CD8+ T cells were analyzed by intracellular staining. Data are representative of at least three independent experiments. Bars: mean ± SD. *: P < 0·05; **: P < 0·01; ***: P < 0·001, statistical relevance was determined by one‐way ANOVA.
Data Availability Statement
Data availability is not applicable to this work.


