Summary
The apparent antagonism between salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signalling resulting in trade‐offs between defence against (hemi)biotrophic and necrotrophic pathogens has been widely described across multiple plant species. However, the underlying mechanism remains to be fully established.
The molecular and cellular functions of ANGUSTIFOLIA (AN) were characterised, and its role in regulating the pathogenic response was studied in Arabidopsis.
We demonstrated that AN, a plant homologue of mammalian C‐TERMINAL BINDING PROTEIN (CtBP), antagonistically regulates plant resistance to the hemibiotrophic pathogen Pseudomonas syringae and the necrotrophic pathogen Botrytis cinerea. Consistent with phenotypic observations, transcription of genes involved in SA and JA/ET pathways was antagonistically regulated by AN. By interacting with another nuclear protein TYROSYL‐DNA PHOSPHODIESTERASE1 (TDP1), AN imposes transcriptional repression on MYB46, encoding a transcriptional activator of PHENYLALANINE AMMONIA‐LYASE (PAL) genes which are required for SA biosynthesis, while releasing TDP1‐imposed transcriptional repression on WRKY33, a master regulator of the JA/ET signalling pathway.
These findings demonstrate that transcriptional co‐regulation of MYB46 and WRKY33 by AN mediates the coordination of SA and JA/ET pathways to optimise defences against (hemi)biotrophic and necrotrophic pathogens.
Keywords: ANGUSTIFOLIA, Arabidopsis, CtBP, (hemi)biotrophic and necrotrophic pathogens, SA and JA/ET antagonism, transcriptional reprogramming
Introduction
Rapid and selective transcriptional reprogramming is central for launching effective host immune responses to microbial infection (Tsuda & Somssich, 2015). Plants have sophisticated regulatory mechanisms to fine‐tune transcriptional responses, depending on the nature and virulence mechanism of the pathogen. The coordinated actions of transcription factors and interactions among hormone signalling pathways contribute to the selective activation of defence‐related genes (Tsuda & Somssich, 2015). Despite great progress, molecular mechanisms modulating pathogen‐specific immune responses remain to be fully elucidated.
In general, the salicylic acid (SA) pathway triggers defences against biotrophic and hemibiotrophic pathogens, which acquire nutrients from living host tissues, whereas the jasmonic acid (JA) and ethylene (ET) pathways cooperate to activate defences against necrotrophic pathogens, which feed on the remains of dead host cells. In addition to the isochorismate pathway, studies in Arabidopsis have demonstrated that SA is also synthesised via the phenylpropanoid pathway (Mauch‐Mani & Slusarenko, 1996). PHENYLALANINE AMMONIA‐LYASE (PAL) catalyses the first step of the phenylpropanoid pathway. Arabidopsis knockout mutants of PAL genes have reduced SA accumulation and exhibit the enhancement of disease symptoms induced by the hemibiotrophic bacteria Pseudomonas syringae (P. syringae) (Huang et al., 2010). The accumulation of SA promotes the translocation of the NONEXPRESSER OF PR GENES1 (NPR1) monomer into the nucleus and acts as a transcriptional co‐activator to trigger the expression of certain stress‐responsive genes (e.g. PATHOGENESIS‐RELATED PROTEIN1 (PR‐1)) to limit the growth of biotrophs (Pieterse & Van Loon, 2004). In resistance to necrotrophs, two transcription factors, ETHYLENE INSENSITIVE3 (EIN3) and ETHYLENE INSENSITIVE3‐LIKE1 (EIL1), form a regulatory hub of the JA/ET signalling pathway to activate APETALA2/ETHYLENE‐RESPONSIVE FACTOR (AP2/ERF) transcription factors, such as OCTADECANOIDRESPONSIVE ARABIDOPSIS APETALA2/ETHYLENERESPONSIVE FACTOR59 (ORA59) and ERF1 (Solano et al., 1998; Zhu et al., 2011). ORA59 and ERF1 then activate the expression of stress‐responsive genes (e.g. PLANT DEFENSIN1.2 (PDF1.2)) against necrotrophs (Lorenzo et al., 2003; Zarei et al., 2011). WRKY transcription factors are major players in plant immunity. WRKY33 is required for defence against necrotrophs (e.g. Botrytis cinerea (B. cinerea)) (Zheng et al., 2006). In Arabidopsis, WRKY33 targets ORA59, PDF1.2, as well as ethylene biosynthetic genes ACC SYNTHASE (ACS2) and ACS6 (Birkenbihl et al., 2012; Datta et al., 2015; Liu et al., 2015; Wang et al., 2015).
Trade‐offs exist in plant defences against (hemi)biotrophic and necrotrophic pathogens. Elevated resistance to (hemi)biotrophs is often correlated with increased susceptibility to necrotrophs and vice versa (Robert‐Seilaniantz et al., 2011). The antagonism between SA and JA/ET pathways has been identified to be involved in trade‐offs between (hemi)biotroph and necrotroph defences (Robert‐Seilaniantz et al., 2011). At the molecular level, transcription factors involved in SA and JA/ET pathways play crucial roles in this antagonism. For example, NPR1, the master activator of SA signalling, was shown to suppress the JA signalling pathway (Spoel et al., 2003). By contrast, overexpression of ERF1 is capable of promoting JA/ET‐dependent defences and reducing the defence against biotrophs (Berrocal‐Lobo et al., 2002). In addition to activating genes involved in JA/ET signalling, WRKY33 was found to repress some genes involved in SA signalling and host cell death pathway (Birkenbihl et al., 2012; Liu et al., 2015).
C‐TERMINAL BINDING PROTEIN (CtBP) is an ancient protein that exists widely in mammals, flies, worms and plants. Animal CtBPs play a central role in development and disease via transcriptional co‐repression (Chinnadurai, 2009). By physically interacting with transcription factors and subsequently recruiting histone modification enzymes in the nucleus, CtBPs efficiently silence the expression of numerous tumour suppressor genes and proapoptotic genes to promote cellular survival and tumorigenesis (Chinnadurai, 2009). Phylogenetic analysis has illustrated that plant and animal CtBPs are in different subfamilies, suggesting that plant CtBPs may have unique functions not shared by animal CtBPs (Kim et al., 2002). ANGUSTIFOLIA (AN) is the singular homologue of CtBP in Arabidopsis and the first plant CtBP that has been studied. AN is involved in the morphogenetic development of leaves and floral organs by controlling the arrangement of cortical microtubules. The Arabidopsis an mutant has a narrow‐leaf phenotype with fewer and smaller cells in the leaf‐width direction than those of wild‐type plant (Tsuge et al., 1996; Kim et al., 2002). In addition, AN has been shown to affect plant responses towards biotic and abiotic stresses. The an mutant has enhanced tolerance to drought and P. syringae that is accompanied by induced expression of stress‐responsive genes (Gachomo et al., 2013). Collectively, AN is involved in development and disease tolerance as found for animal CtBPs. However, the nuclear function of AN remains unclarified. To date, only physical and genetic associations with cytosolic proteins, including a receptor‐like kinase STRUBBELIG (SUB), RNA‐binding proteins in stress granules, SPIKE1 (SPK1) and DUAL‐SPECIFICITY TYROSINE PHOSPHORYLATION‐REGULATED KINASEs (DYRKPs), have been validated experimentally for AN (Bai et al., 2013; Bhasin & Hulskamp, 2017; Iwabuchi et al., 2019). Despite the lack of direct evidence, a potential transcriptional function of AN cannot be ruled out, as the dysregulation of several genes has been detected in the Arabidopsis an knockout mutants (Kim et al., 2002; Gachomo et al., 2013; Bryan et al., 2018). Moreover, dimerisation of Arabidopsis AN has been reported (Kim et al., 2002), which is an essential feature for the transcriptional role of animal CtBPs.
In the present study, we demonstrated that AN is involved in the trade‐offs between (hemi)biotroph and necrotroph defences. Alteration of AN expression in transgenic plants oppositely affected resistance to P. syringae DC3000 (hemibiotroph) and B. cinerea (necrotroph). Gene expression analysis revealed that alternation of AN expression also antagonistically altered gene expression in the SA and JA/ET pathways during pathogen infection. Molecular studies illustrated that AN has nuclear accumulation and transcriptional repressor activity that are associated with the physical interaction with TYROSYL‐DNA PHOSPHODIESTERASE 1 (TDP1), an enzyme involved in DNA damage repair in plants and animals (Lee et al., 2010; Stingele et al., 2017). AN directly targets and represses the transcription of MYB46, which encodes a master regulator of the phenylpropanoid pathway (Xie et al., 2018b). Moreover, AN displayed the capability to release the TDP1‐imposed transcriptional repression on WRKY33. The antagonistic transcriptional regulation of MYB46 and WRKY33 by AN suggested a transcriptional co‐regulatory mechanism of genes involved in the SA (e.g. PR‐1, PALs) and JA/ET pathways (e.g. PDF1.2, ACS, ERF1 and ORA59), demonstrating a transcriptional node regulating the trade‐offs between (hemi)biotrophic and necrotrophic defences.
Materials and Methods
Plant materials and growth conditions
Arabidopsis plants used in this study were grown in a growth chamber under 12 h : 12 h, light : dark, 23°C : 20°C conditions with 60% relative humidity. The T‐DNA insertional mutants an‐t1 (Gachomo et al., 2013) and tdp1‐2 (Enderle et al., 2019) were obtained from the ABRC (https://abrc.osu.edu/). To generate an‐1 35S:AN and 35S:TDP1 transgenic plants, full‐length AN and TDP1 were cloned into the pENTR vector and then subcloned into binary vectors pGWB520 and pGWB515, respectively, for transformation.
Pathogen infection and analysis
To analyse disease symptoms, detached 4‐wk‐old Arabidopsis leaves were inoculated with 5 µl B. cinerea conidiospore suspension (5 × 105 spores ml−1 in potato dextrose broth) as previously described (Ingle & Roden, 2014). Leaf lesions were pictured and measured using a Zeiss dissecting microscope. For each genotype, 20 leaves (40 spots) from independent plants were used for statistical analysis. Inoculation with P. syringae DC3000 was performed as previously described (Katagiri et al., 2002). Here, c. 10 μl of P. syringae DC3000 bacteria suspension (1 × 106 CFU ml−1 in 10 mM MgCl2) was used to infiltrate the leaves from 3‐wk Arabidopsis plants. For each genotype, 15 leaves from five different plants were infiltrated. After 3 d, infiltrated leaves were pictured and measured for total weight. Leaves were then washed three times with sterile water and homogenised in 500 μl sterile water. Diluted lysis samples were plated onto LB medium containing 50 μg ml−1 rifampicin. After growing at 28°C for 2 d, the bacterial colony forming units (CFU) were counted and normalised with total weight to obtain the bacterial titer value (log10(CFU mg−1)).
To measure gene expression, 4‐wk‐old Arabidopsis plants were sprayed with B. cinerea conidiospore suspension (2.5 × 105 spores ml−1) or infiltrated with P. syringae DC3000 bacteria suspension (1 × 107 CFU ml−1). Leaves were collected at 0, 24, 48 and 72 h post infection (hpi) for B. cinerea treatment. Leaves were collected at 0 and 6 hpi for P. syringae treatment. Then, total RNAs were extracted using a Plant RNA extraction kit (Sigma, St Louis, MO, USA). Next, 2 µg of total RNA was used for cDNA synthesis. qRT‐PCR was then performed using cDNA as the template and following the Maxima SYBR Green qPCR Master Mixes manual (Thermo Scientific, Waltham, MA, USA). Gene expression was calculated using the ΔΔCT method with expression of a housekeeping gene (EF1α) for template normalisation. Three biological replicates were included in each gene expression analysis. Primers used for qPCR are listed in Supporting Information Table S1.
To analyse the nuclear accumulation changes of AN, protoplasts co‐transformed with HA‐TDP1 of AN‐Myc plasmids were incubated overnight then treated with P. syringae (1 × 106 CFU ml−1) or B. cinerea (5 × 105 spores ml−1) for 6 h. After treatment, protoplasts were collected for cell fractionation assay.
Protoplast isolation and transfection
Arabidopsis mesophyll protoplasts were isolated and transfected as described previously (Yoo et al., 2007). Protoplasts were isolated from fully expanded leaves from 3–4‐wk‐old Col‐0 plants. For subcellular localisation analysis, 8 µg of AN‐YFP plasmid DNA and 2 µg of CFP‐SHY2 or Golgi‐mCherry plasmid DNA were co‐transfected into 100 µl of protoplasts. For co‐localisation analysis, 5 µg of AN‐YFP plasmid DNA was co‐transfected with 5 µg of either mCherry‐TDP1 or mCherry‐TDP1Δ1‐122 plasmid DNA into 100 µl of protoplasts. For cell fraction analysis, 20 µg of AN‐Myc plasmid DNA were transfected into 200 µl of protoplasts. Five reactions were combined to obtain 1 ml of protoplasts for subsequent cell fraction and western blotting analyses. To determine the effect of TDP1 on AN nuclear accumulation, 15 µg of HA‐TDP1 and 5 µg of AN‐Myc plasmid DNA were co‐transfected into 200 µl of protoplasts. To balance the protein level of AN‐Myc, an AN‐Myc only reaction was performed by co‐transfecting 15 µg of blank vector and 5 µg of AN‐Myc plasmid DNA into 200 µl of protoplasts. For transcriptional activity assays, in total 10 µg plasmid DNA including reporter, effector, and/or transactivator plasmid DNA were co‐transfected into 100 µl of protoplasts. To compare the effects of different effector constructs, the same amount of effect constructs was used. For single protein chromatin immunoprecipitation (ChIP), 20 µg of plasmid DNA was transfected into 200 µl of protoplasts. For AN and TDP1 co‐expression, 10 µg of plasmid expressing AN‐Myc and 10 µg of plasmid expressing HA‐TDP1 were co‐transfected into 200 µl of protoplasts. To balance the protein level of AN‐Myc, an AN‐Myc only reaction was performed by co‐transfecting 10 µg of blank vector and 10 µg of AN‐Myc construct into 200 µl of protoplasts.
Transactivation assay
A protoplast transfection‐based transcriptional activity assay was performed in accordance with the method described by Tiwari et al. (2004). Reporter and effector constructs were co‐transfected into protoplasts and incubated in the dark for 18–20 h at room temperature. GUS activity assay was performed as described previously (Yoo et al., 2007). GUS activity was measured using a Fluoroskan microplate reader. To normalise GUS activity, 100 ng of 35S:luciferase plasmid was co‐transfected for each reaction. Luciferase activity was measured using the Promega Luciferase Assay System in accordance with the manufacturer’s manual. GUS activity in individual samples was normalised against luciferase activity (GUS/LUC). Three replicates were performed for statistical calculation.
Subcellular localisation analysis
AN‐YFP construct was co‐transfected with the nuclear marker into protoplasts to determine the subcellular localisation of AN. For co‐localisation analysis, paired constructs were co‐transfected into protoplasts. After 14 h incubation under weak light at room temperature, protoplasts were collected and resuspended in cold W5 solution (2 mM MES pH 5.7, 154 mM NaCl, 125 mM CaCl2, and 5 mM KCl) and subjected to microscopy. Images were collected using a Zeiss LSM 710 confocal microscope, equipped with 458, 514 and 561 nm laser lines for excitation of CFP, YFP and mCherry fluorescent markers, respectively. Images were processed using Zen software (Zeiss).
Cell fractionation and protein gel blots
One ml of transfected protoplasts was incubated at room temperature for 14 h to express protein and then collected by centrifugation. Cytosolic and nuclear fractions were separated as previously described (Xie et al., 2018a). Cytosolic and nuclear proteins were separated by SDS/PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Bio‐Rad). Anti‐Myc (C3956; Sigma), anti‐HA (H3663; Sigma), anti‐histone H3 (ab1791; Abcam), and anti‐UGPase (AS05 086; Agrisera) were used as primary antibodies. Anti‐rabbit IgG peroxidase antibody (A9169; Sigma) was used as the secondary antibody for anti‐Myc, anti‐histone H3 and anti‐UGPase. Anti‐mouse IgG peroxidase antibody (A9044; Sigma) was used as the secondary antibody for anti‐HA. Chemiluminescence signals were generated using ECL Western Blotting Detection Reagents (GE Health) and detected using the ChemiDoc XRS + system (Bio‐Rad). Western blotting signals were quantified using Image Lab software (Bio‐Rad).
ChIP–qPCR and µChIP–qPCR
ChIP using transgenic plants was performed as previously described (Zhang et al., 2013). Anti‐Myc (C3956; Sigma) and anti‐HA (H6908; Sigma) were used to immunoprecipitate AN‐Myc and HA‐TDP1. qPCR was performed to quantify DNA enrichment. Three biological replicates were included in each analysis. Primers used for PCR are listed in the Table S1.
µChIP was performed as described previously (Xie et al., 2018a). AN‐Myc and HA‐TDP1 plasmid DNAs were expressed transiently in protoplasts. After 14 h incubation at room temperature, c. 40 000 transfected protoplasts were used for µChIP. ChIPed DNA and the input DNA were then cleaned and concentrated using a Qiagen MinElute PCR Purification Kit (Qiagen). qPCR was performed to quantify DNA enrichment. Three biological replicates were included in each analysis. Primers used for PCR are listed in Table S1.
Electrophoretic mobility shift assay
AN was cloned into the pGEX‐6P‐1 vector (GE Healthcare, Chicago, IL, USA) using BamHI restriction enzyme for GST fusion constructs. The constructs were transformed into E. coli strain BL21(DE3) pLysS (Invitrogen) for protein expression. GST fusion proteins were extracted and purified as previously described using Glutathione Sepharose 4B beads (GE Healthcare) (Xie et al., 2012). For DNA probes, the −447‐bp to −300‐bp promoter region of AtMYB46 was amplified by PCR from Col‐0 genomic DNA, gel purified, and end labelled with biotin using a DNA 3′ End Biotinylation Kit (Thermo Scientific) in accordance with the manufacturer’s manual. The DNA binding reaction was incubated at room temperature for 20 min as previously described (Xie et al., 2018a). The reaction mixtures were then resolved in 6% DNA retardation gel (Novex) by electrophoresis at 100 V for 1–2 h and transferred to Nylon membrane. Biotin signals were detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) as suggested by the manufacturer.
Yeast two‐hybrid assay
pGADT7 and pGBKT7 plasmid pairs containing various genes were co‐transformed into yeast Y2H Gold (Clontech Laboratories, Mountain View, CA, USA). SD−Leu−Trp plates were used to select yeast containing both constructs. The resulting clones were diluted in 50 µl water and 5 µl was used for spot assay on SD−Leu−Trp−Ade−His plates. Interactions between AN and TDP1 were based on the growth of yeast cells in the selection medium without Ade and His. Three independent clones were tested for each plasmid pair.
SA measurement
Here, c. 85–100 mg of frozen ground plant material was twice extracted overnight with 2 ml 80% ethanol. The extracts were combined before drying in a 1 ml aliquot under a stream of nitrogen. Sorbitol (50 µl; 1 mg ml−1) was added before extraction as an internal standard. Dried extracts were silylated by addition of 500 µl of acetonitrile (TS‐20062; ThermoFisher) and 500 µl of MSTFA plus 1% TMCS (TS‐48915; ThermoFisher) and heated for 1 h at 70°C to produce trimethylsilyl (TMS) derivatives. After 2 d, 1 µl was analysed by injection into an Agilent 7890A gas chromatograph coupled to an Agilent 5975C inert XL mass spectrometer (Tschaplinski et al., 2019). SA and SA 2‐O‐glucoside metabolite peaks were quantified by extraction of the key m/z 267 and then scaled back up to the total ion current using predetermined scaling factors. Data were normalised to extracted plant mass, internal standard recovered, extraction volume, derivatisation volume and injection volume. For each genotype, at least two biological replicates were measured for statistical analysis.
Accession numbers
Sequence data from this article can be found under the following Arabidopsis Genome Initiative accession numbers: AN (AT1G01510), TDP1 (AT5G15170), MYB46 (AT5G12870), WRKY33 (AT2G38470), PR‐1 (AT2G14610), PDF1.2 (AT5G44420), PAL1 (AT2G37040), PAL2 (AT3G53260), PAL3 (AT5G04230), PAL4 (AT3G10340), ACS2 (AT1G01480), ACS6 (AT4G11280), ERF1A (AT4G17500), ORA59 (AT1G06160), ACTION (AT5G09810), EF1α (AT5G60390), ICS1 (AT1G74710), PBS3 (AT5G13320) and EDS5 (AT4G39030).
Results
AN antagonistically affects plant resistance to P. syringae and B. cinerea
In a previous RNA‐seq analysis, we found the enrichment of genes involved in hormone and defence pathways among differentially expressed genes in the Arabidopsis T‐DNA insertional mutant ofAN (an‐t1) (Bryan et al., 2018). Knockout of AN was also shown to enhance plant tolerance to the hemibiotrophic bacteria P. syringae (Gachomo et al., 2013). These results prompted us to investigate AN’s involvement in plant immunity.
Firstly, we generated transgenic Arabidopsis overexpressing AN‐Myc in an‐t1. As shown in Fig. S1, overexpression of Myc‐tagged AN (AN‐Myc) in an‐t1 (an‐t1 35S:AN) reversed its narrow‐leaf phenotype to wild‐type morphology, indicating that AN‐Myc is functional in Arabidopsis plants. By assessing the responses of an‐t1 and an‐t1 35S:AN plants to pathogen infection, we observed that AN oppositely affected the resistance to the hemibiotrophic bacteria P. syringae DC3000 and the necrotrophic fungi B. cinerea. In our analysis, an‐t1 displayed weaker disease symptoms (e.g. chlorosis, water‐soaked) and lower bacterial populations than Col‐0 after 72 h of inoculation with P. syringae (Fig. 1a), consistent with a previous study (Gachomo et al., 2013). By contrast, an‐t1 35S:AN had more severe disease symptoms than Col‐0 (Fig. 1a). Opposite to the resistance to P. syringae, an‐t1 exhibited clear lesion areas of dead cells after 72 h of inoculation with B. cinerea, indicative of its increased susceptibility towards B. cinerea infection (Fig. 1b). Compared with Col‐0, an‐t1 35S:AN displayed limited lesions, suggesting that it had enhanced resistance to B. cinerea (Fig. 1b).
Fig. 1.
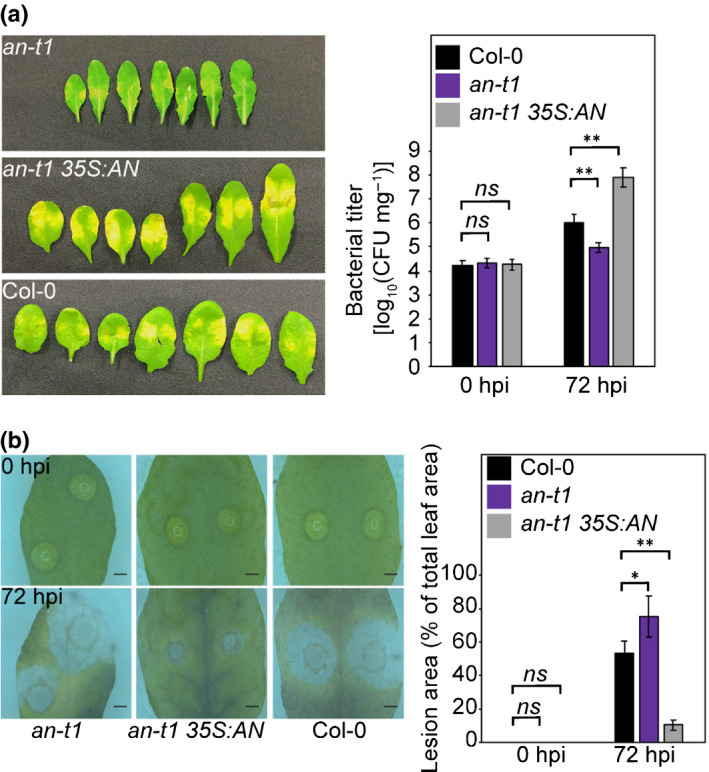
AN antagonistically regulates Arabidopsis resistance to Pseudomonas syringae and Botrytis cinerea. (a) P. syringae inoculation of Col‐0, an‐t1, and an‐t1 35S:AN. Bacteria titers in 0 h post infection (hpi) and 72 hpi are shown in the bar graph. Values represent means ± SE, n = 15. (b) B. cinerea inoculation of Col‐0, an‐t1 and an‐t1 35S:AN. Bars, 1 mm. Lesion areas (% of total leaf area) in 0 hpi and 72 hpi are shown in the bar graph. Values represent means ± SE, n = 40. Statistical significance was determined by two‐tailed Student’s t‐tests against Col‐0 (**, P < 0.01; *, P < 0.05; ns, P > 0.05).
These results demonstrated that AN antagonistically regulates resistance towards P. syringae and B. cinerea, implying that AN may be involved in the regulation of the trade‐offs between (hemi)biotroph and necrotroph defences.
AN antagonistically affects gene expression of SA and JA/ET pathways
Host cell death limits (hemi)biotrophic but benefits necrotrophic pathogens. These distinct lifestyles led to trade‐offs between defence responses against these two types of pathogens, with elevated resistance to (hemi)biotrophs being correlated with increased susceptibility to necrotrophs and vice versa. The SA pathway is primarily responsible for defence against (hemi)biotrophic pathogens, whereas the JA/ET pathway is primarily responsible for defence against necrotrophic pathogens. The antagonism between SA and JA/ET pathways has been shown to be central to the regulation of these trade‐offs (Robert‐Seilaniantz et al., 2011).
qPCR results showed that AN antagonistically affected the expression of SA‐responsive marker gene PR‐1 and JA/ET‐responsive marker gene PDF1.2. As shown in the Fig. S2, P. syringae infection activated PR‐1 expression at 6 hpi. In an‐t1, P. syringae infection induced PR‐1 expression to a higher level than that in Col‐0 whereas PR‐1 expression in an‐t1 35S:AN was lower than that in Col‐0 at 6 hpi. By contrast, PDF1.2 expression was lower in an‐t1 and higher in an‐t1 35S:AN than that in Col‐0 at 24 hpi of B. cinerea infection (Fig. S2).
In addition to defence genes (PR‐1 and PDF1.2), in our analysis AN exhibited antagonistic regulation on the expression of other genes involved in the SA and JA/ET pathways. As the enzyme catalysing the first step of the phenylpropanoid pathway, PAL was shown to be involved in SA biosynthesis in Arabidopsis (Mauch‐Mani & Slusarenko, 1996; Huang et al., 2010). Arabidopsis has four genes that encode PAL (PAL1‐4). Among these, PAL1 and PAL2 were shown to be highly induced under environmental stress (Rohde et al., 2004; Olsen et al., 2008). In our qPCR analysis, PAL1 and PAL2 showed a significant induction at 6 hpi of P. syringae infection (seven‐fold and two‐fold in Col‐0, respectively; Fig. S2a). PAL1 and PAL2 were upregulated to a higher level in an‐t1, but to a lower level in an‐t1 35S:AN compared with that in Col‐0 (Fig. S2a). Although the majority of plant defence‐related SA is synthesised via the isochorismate pathway, our qPCR analysis revealed that AN only has minor effect on this pathway. ISOCHORISMATE SYNTHASE 1 (ICS1), avrPphB SUSCEPTIBLE 3 (PBS3), and ENHANCED DISEASE SUSCEPTIBILITY 5 (EDS5) are essential components of the isochorismate pathway (Rekhter et al., 2019). During P. syringae infection, AN knockout and overexpression plants exhibited no significant change in ICS1 or PBS3 expression (Fig. S2a). In an‐t1, we observed the reduction of P. syringae‐induced EDS5 expression. Although an‐t1 35S:AN showed higher EDS5 induction than that in an‐t1, its EDS5 expression was still slightly lower than that in Col‐0 at 0 and 6 hpi of P. syringae infection (Fig. S2a). In the JA/ET pathway, ACS catalyses the rate‐limiting step of ET biosynthesis (Kende, 1993). Transcription factors ERF1 and ORA59 are involved in JA/ET signalling to induce PDF1.2 expression (Lorenzo et al., 2003; Zarei et al., 2011). We observed that ACS2, ERF1A and ORA59 exhibited a similar expression pattern to that of PDF1.2 during infection with B. cinerea, with lower expression in an‐t1 and higher expression in an‐t1 35S:AN (Fig. S2b).
Collectively, results from gene expression analysis suggested that AN is involved in the regulation of trade‐offs between defence responses to P. syringae and B. cinerea that are associated with the antagonism between SA and JA/ET pathways. Differential gene expression induced by AN implies that AN may be involved in transcriptional regulatory mechanisms, which prompted us to examine AN’s potential nuclear function.
AN has nuclear accumulation
To determine whether AN has nuclear functions, we first examined the subcellular localisation of yellow fluorescent protein (YFP)‐tagged AN (AN‐YFP) using the Arabidopsis mesophyll protoplast transient expression system. Signals of AN‐YFP (green colour) were present in both the cytoplasm and the nucleus (Fig. 2a). In 38.75% transfected cells (N = 80), AN‐YFP signals partially overlapped with signals from the nuclear marker mCherry‐VirD2NLS (Lee et al., 2008), as indicated by the yellow colour (Fig. 2a). Consistently, using western blotting, Myc‐tagged AN (AN‐Myc) was detected in both cytosolic and nuclear fractions (Fig. 2b).
Fig. 2.
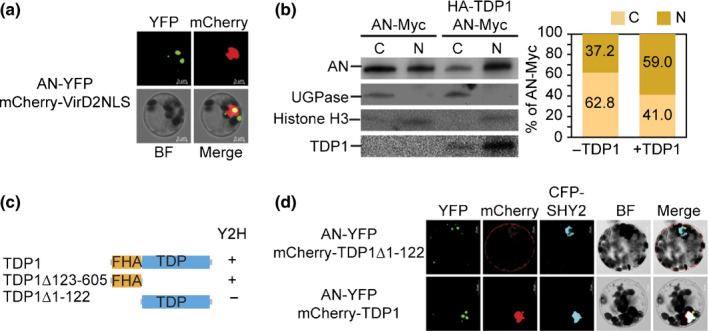
TDP1 enhances the nuclear accumulation of AN in Arabidopsis. (a) Subcellular localisation analysis showing the partial nuclear localisation of AN in the Arabidopsis protoplasts. Overlapping of AN‐YFP (green) and mCherry‐VirD2NLS (nuclear marker, red) is indicated as a yellow colour. BF, bright field. Bars, 5 µm. (b) Immunoblot analysis of the nuclear accumulation of AN with or without TDP1. AN‐Myc, HA‐TDP1, UGPase (cytosolic marker), histone H3 (nuclear marker) are examined in cytosolic (C) and nuclear (N) fractions. AN‐Myc signals were quantified using Image Lab software (Bio‐Rad) and then normalised against the corresponding nuclear or cytosolic marker to calculate the relative ratio of AN signals in cytosolic and nuclear fractions. (c) Yeast two‐hybrid analysis of the interaction between AN and TDP1. Domain structures of truncated TDP1 are displayed. ‘+’ indicates interaction; ‘−’ indicates no interaction. (d) Co‐localisation analysis of AN with full‐length TDP1 and truncated TDP1 without FHA domain (TDP1Δ1–122). CFP‐SHY2 (blue) is a nuclear marker. Bars, 10 µm.
The observed nuclear accumulation of AN is not likely to be due to its nuclear localisation signal (NLS; KKRH), which was demonstrated in a previous study to be nonfunctional (Minamisawa et al., 2011). To determine the mechanism translocating AN into the nucleus, we sought to identify proteins interacting with AN. Gachomo et al. (2013) identified nine potential interacting partners of AN using the high‐throughput integrated knowledge‐based Arabidopsis protein interaction network analysis (ANAP) (Gachomo et al., 2013). Among these nine proteins, only At5G15170 (TDP1) has been reported to localise in the nucleus (Lee et al., 2010), which prompted us to evaluate its interaction with AN. As shown in Figs 2(c), S3, we observed the interaction between AN and TDP1 using the yeast two‐hybrid (Y2H) assay. Consistent with the Y2H result, AN‐YFP showed co‐localisation with mCherry‐TDP1 in both the cytoplasm and the nucleus (Fig. 2d). TDP1 is a key enzyme catalysing DNA repair in both plants and animals. In addition to the TDP domain that is present in all TDP1 proteins, Arabidopsis TDP1 contains an FHA domain at its N‐terminal region (Kim et al., 2012). The FHA domain of TDP1 contains an NLS and is indispensable for its nuclear localisation (Kim et al., 2012). Our Y2H and co‐localisation analyses of AN and truncated TDP1 showed that AN was specifically associated with the FHA domain (TDP1Δ123–605), whereas it had no interaction with the TDP domain (TDP1Δ1–122) (Fig. 2c,d).
Given the result that AN interacts with the NLS‐containing domain of TDP1 (FHA domain), we hypothesised that TDP1 may be involved in the nuclear accumulation of AN. To test this hypothesis, AN‐Myc and HA‐tagged TDP1 (HA‐TDP1) were co‐expressed in the protoplasts and their accumulations in the cytoplasm and nucleus were detected using western blotting and then quantified using Image Lab software (Bio‐Rad). Western blotting analysis of total proteins showed that co‐expression of HA‐TDP1 did not affect AN‐Myc expression (Fig. S4). Compared with the empty vector control, co‐expression of HA‐TDP1 increased the percentage of AN nuclear signals from 37.2% to 59.0% (Fig. 2b), suggesting that TDP1 is capable of enhancing the nuclear localisation of AN. Moreover, the presence of a small amount of TDP1 protein in the cytosolic fraction (Fig. 2b) implied that TDP1 may bind to AN in the cytoplasm and translocate AN into the nucleus.
AN has transcriptional repressor activity and targets the transcription factor gene MYB46
The nuclear accumulation of AN suggested potential transcriptional functions. We then analysed whether AN could regulate transcription using the transactivation assays (Xie et al., 2018a). In this assay, the promoter region of β‐glucuronidase (GUS) reporter gene contains two DNA binding sites, Gal4 and LexA (Fig. 3a). The LexA binding domain (LD)‐fused Herpes simplex virus VP16 (LD‐VP16) was used to constitutively activate GUS expression (Fig. 3a). The finding that Gal4‐binding‐domain (GD)‐fused AN (GD‐AN), but not GD only, reduced GUS expression by c. 25% suggested that AN has moderate transcriptional repressor activity (Fig. 3a). No transcriptional activator activity of AN was detected (Fig. S5).
Fig. 3.
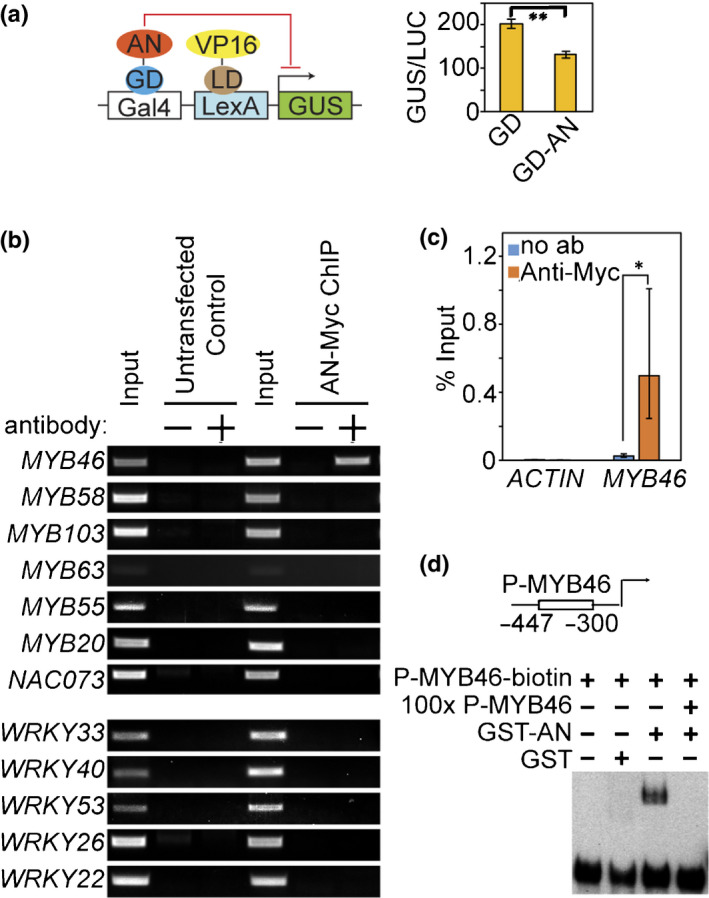
AN has transcriptional repressor activity and directly targets MYB46 in Arabidopsis. (a) Transactivation analysis of the repressor activity of AN. Top scheme displays the three vectors used in transactivation analyses: reporter construct containing Gal4 binding site and LexA binding site upstream of the GUS reporter gene; transactivation construct expressing LexA binding domain (LD) fused VP16; effector construct expressing Gal4 binding domain (GD) fused AN. GUS activity in individual samples was normalised against luciferase activity (GUS/LUC). Values represent means ± SE, n = 3. Statistical significance was determined by two‐tailed Student’s t‐tests (**, P < 0.01). (b) µChIP‐PCR analyses of AN association with promoters of genes upregulated in an‐t1 (MYB46, MYB58, MYB63, MYB55, MYB20, MYB103, NAC073) and genes downregulated in an‐t1 (WRKY33, WRKY40, WRKY53, WRKY26 or WRKY22). Two negative controls are shown: no antibody control and protoplasts without expression of AN‐Myc (untransfected control). +, indicates reactions with anti‐Myc antibody. −, indicates reactions without anti‐Myc antibody. (c) µChIP‐qPCR analyses of AN association with MYB46 promoter. No antibody assays were used as negative controls. Values represent means ± SE, n = 3. Statistical significance was determined by two‐tailed Student’s t‐tests (*, P < 0.05). (d) EMSA analysis showing the direct binding of AN to the MYB46 promoter. The 148‐bp MYB46 promoter (−447 to −300 bp from the start codon) was labelled with biotin as the probe (P‐MYB46‐biotin). The competition assay was performed using 100× unlabelled MYB46 promoter DNA (100× P‐MYB46).
Subsequently, we attempted to identify target genes of AN using microchromatin immunoprecipitation (µChIP) following PCR (Xie et al., 2018a). Given that the expression levels of a number of transcription factor genes were altered in the an knockout mutant (an‐t1) (Bryan et al., 2018), we hypothesised that AN may function upstream of these transcription factors. We focused on transcription factors with significant gene expression changes (>2‐fold) in an‐t1 (Fig. S6). Among 12 tested transcription factors (seven upregulated genes: MYB58, MYB46, MYB63, MYB55, MYB20, MYB103 and NAC073; and five downregulated genes: WRKY33, WRKY40, WRKY53, WRKY26 and WRKY22), AN‐Myc showed association with only MYB46 promoter in the ChIP assays (Fig. 3b, c). Subsequently, an in vitro DNA binding assay (electrophoretic mobility shift assay, EMSA) was used to determine the direct binding of AN to the MYB46 promoter. Purified GST‐tagged AN protein (GST‐AN), but not the GST tag alone, was found to bind to the biotin‐labelled 148‐bp MYB46 promoter fragment (−447 bp to −300 bp) (Fig. 3d), suggesting that AN has DNA binding activity and directly targets MYB46. A competition assay using 100× unlabelled MYB46 promoter DNA abolished the shifted band, suggesting that the binding of AN to MYB46 promoter is specific (Fig. 3d).
AN–TDP1 interaction enhances transcriptional repression on MYB46
The association of AN with the MYB46 promoter was further confirmed via ChIP assays using transgenic plants expressing AN‐Myc (an‐t1 35S:AN) (Fig. 4a). As TDP1 was shown to enhance the nuclear accumulation of AN, it is possible that TDP1 may also impact the transcriptional repressor effect of AN. To test this hypothesis, a transactivation assay was performed to measure the transcriptional repression of AN on MYB46 promoter. Because AN was shown to act as a transcriptional repressor (Fig. 3a), VND6 (Zhong et al., 2007), a transcriptional activator for the MYB46 gene was used to constitutively activate MYB46 expression in the protoplast transient expression system. As shown in Fig. 4(b), AN exhibited moderate repression on MYB46 promoter activity (c. 25% reduction). Addition of full‐length TDP1 was capable of enhancing the repression of AN on MYB46 (c. 40% reduction). However, the truncated TDP1 lacking the FHA domain (TDP1Δ1–122) had little effect on the repression of MYB46 in the transactivation assay (Fig. 4b). Collectively, these results suggested that the nuclear accumulation of AN (enhanced by TDP1) is important for its transcriptional repressor function.
Fig. 4.
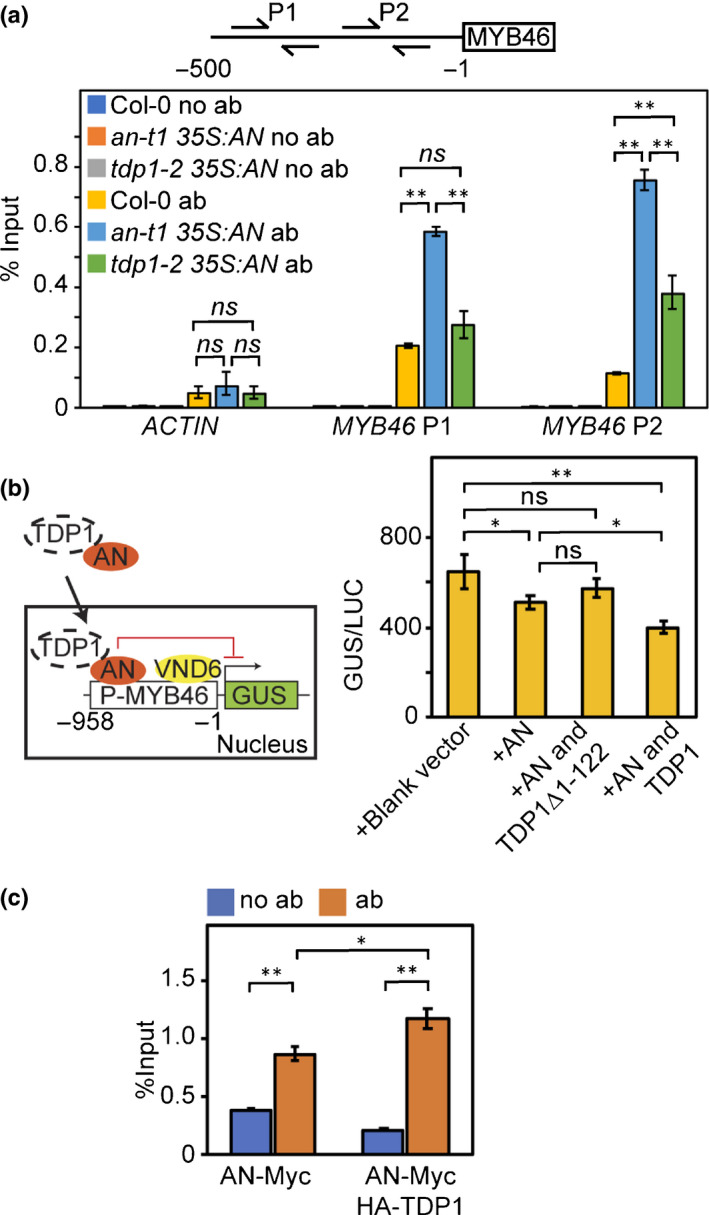
TDP1 is required for AN‐mediated transcriptional repression on MYB46 in Arabidopsis. (a) ChIP‐qPCR analysis on an‐t1 and tdp1‐2 backgrounds. Upper scheme indicates primers for the amplification of MYB46 promoters (MYB46 P1 and P2). Nontransgenic plants (Col‐0) and no antibody assays were used as negative controls. (b) Transactivation analysis showing that TDP1 enhances AN transcriptional repression on MYB46. Left scheme displays the four vectors used in transactivation analyses: reporter construct containing MYB46 promoter and GUS reporter gene; transactivation construct expressing VND6; effector construct expressing AN; effector construct expressing TDP1. GUS activity in individual assays was normalised against luciferase activity (GUS/LUC). (c) µChIP‐qPCR analysis demonstrating the enhancement of association of AN with MYB46 promoter by TDP1. Assays without antibody (no ab) were performed as negative controls. Values represent means ± SE, n = 3. Statistical significance was determined by two‐tailed Student’s t‐tests (**, P < 0.01; *, P < 0.05; ns, P > 0.05).
We then investigated whether TDP1 could increase the binding of AN to the MYB46 promoter in vivo. AN‐Myc and HA‐TDP1 were co‐expressed in the protoplasts and subjected to μChIP‐qPCR assay. Consistent with the increased nuclear accumulation, the chromatin association of AN with MYB46 promoter was significantly increased (P < 0.05) when co‐expressed with TDP1 (Fig. 4c). Meanwhile, co‐expression of AN and TDP1 also significantly increased the chromatin association of TDP1 with MYB46 promoter (Fig. S4b). Conversely, AN‐Myc expressed in the TDP1 T‐DNA insertional mutant tdp1‐2 (Enderle et al., 2019) exhibited dramatically reduced association with MYB46 promoter in the ChIP analysis (Fig. 4a).
Collectively, these molecular and genetic studies support that physical interaction with TDP1 is required for AN to fully execute its transcriptional repressor function on MYB46.
AN–TDP1 interaction releases the TDP1‐imposed transcriptional repression on WRKY33
WRKY transcription factors play critical roles in plant immunity. Although multiple WRKY transcription factors were downregulated in the an‐t1 mutant, we did not detect the association of AN with promoters of these WRKY transcription factors (Fig. 3b). However, previous studies in Medicago suggested that TDP1 may regulate the expression of WRKY33 in plants. Deletion of TDP1 in Medicago was shown to activate the expression of WRKY33 (Medtr2g033820 and Medtr3g031220) (Dona et al., 2013). Given that Arabidopsis and Medicago TDP1 proteins share high amino acid identity (62.8%) and a similar structure (both of them have the FHA domain and TDP domain; Fig. S7), we hypothesised that Arabidopsis TDP1 may negatively regulate WRKY33 expression. To examine this possibility, we found that Arabidopsis TDP1 and WRKY33 genes had a negative co‐expression relationship across different developmental stages and pathogen infections (Fig. 5a). Furthermore, transient expression of TDP1 in Arabidopsis protoplasts was capable of reducing the transcriptional activity of the WRKY33 promoter by 34.9% and reducing the transcript level of endogenous WRKY33 by 46.0% (Fig. 5b,c). In addition, co‐expression of AN and TDP1 alleviated the repression of WRKY33 expression by TDP1 (Fig. 5c), suggesting that AN may negatively affect the transcriptional role of TDP1.
Fig. 5.
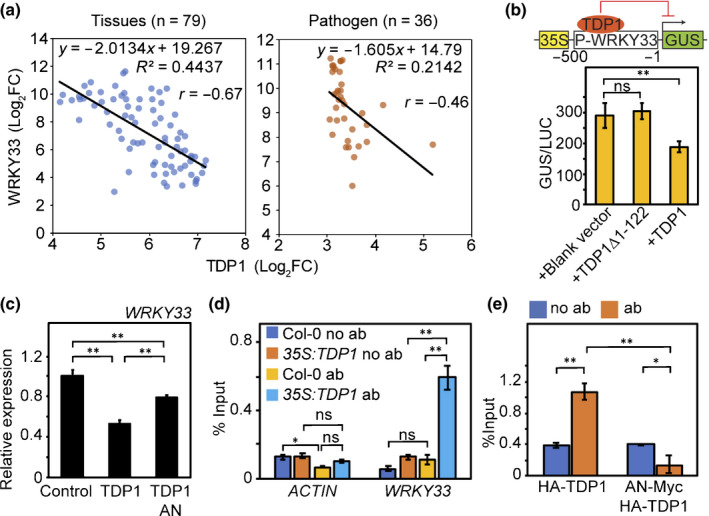
The AN–TDP1 interaction releases the TDP1‐imposed transcriptional repression on WRKY33 in Arabidopsis. (a) Negative co‐expression of TDP1 and WRKY33 across various tissues and pathogen infections. Gene expression data were obtained from the AtGenExpress Visualization Tool (AVT) (http://jsp.weigelworld.org/expviz/expviz.jsp?experiment=development&normalization=absolute&probesprobe=At5G61850&action=Run). (b) Transactivation analysis of TDP1 repression of WRKY33. Upper panel is a scheme of two constructs used in the transactivation analysis: reporter constructs containing 35S promoter, WRKY33 promoter and GUS reporter gene; effector construct expressing TDP1. (c) qRT‐PCR results showing effects of AN and TDP1 on the expression of WRKY33. Gene expression was normalised against the expression of EF1α. WRKY33 expression in Col‐0 was set as 1. Values represent means ± SE, n = 3. (d) ChIP‐qPCR analysis of HA–TDP1 in transgenic plants. Nontransgenic plants (Col‐0) and no antibody assays were used as negative controls. (e) µChIP–qPCR analysis demonstrated that AN reduces TDP1 association with WRKY33 promoter. Values represent means ± SE, n = 3. Statistical significance was determined by two‐tailed Student’s t‐tests (**, P < 0.01; *, P < 0.05; ns, P > 0.05).
To understand the molecular mechanisms underlying AN‐TDP1 regulation on WRKY33 expression, we measured the association of TDP1 with WRKY33 promoter using transgenic plants expressing HA‐TDP1 (35S:TDP1). Relative to the nontransgenic control and no antibody control, WRKY33 promoter fragments were enriched in HA‐TDP1 immunoprecipitates (Fig. 5d). A similar result was obtained via µChIP‐qPCR using protoplasts transiently expressing HA‐TDP1 (Fig. 5e). Moreover, the association between TDP1 and WRKY33 promoter was significantly reduced by the co‐expression of AN and TDP1 (Fig. 5e), suggesting that AN may release the TDP1‐imposed transcriptional repression on WRKY33 by sequestering TDP1 away from the WRKY33 promoter.
Discussion
Being continuously challenged by pathogens with different lifestyles in the natural environment, plants have evolved a sophisticated regulatory network to fine‐tune the activation of specific immune genes in response to a given pathogen. This network includes complex interconnections of hormones and transcription factors. Due to their distinct lifestyles, plants activate contrasting mechanisms in defence against (hemi)biotrophic and necrotrophic pathogens that depend on SA and JA/ET pathways, respectively. The antagonism between SA and JA/ET pathways has been demonstrated to be critical for the selective activation of defence against biotrophic and necrotrophic pathogens. However, the mechanisms coordinating SA and JA/ET pathways remain largely unknown. Using well characterised pathogens with either biotrophic or necrotrophic lifestyle (P. syringae and B. cinerea), we found that ANGUSTIFOLIA (AN) antagonistically regulated Arabidopsis resistance to biotrophic and necrotrophic pathogens. Our pathogen inoculation analyses demonstrated that AN is capable of enhancing resistance to necrotrophic pathogen and reducing the resistance to biotrophic pathogens (Fig. 1). By measuring the expression levels of marker genes for plant defence during pathogen infection, we found that AN was involved in the antagonistic transcriptional regulation of SA pathway genes (PR‐1 and PALs) and JA/ET pathway genes (PDF1.2, ACS2, ERF1A and ORA59) (Fig. S2). These results suggested that the AN‐mediated regulation of host immune responses was associated with regulation of SA–JA/ET antagonism, a well known mechanism involved in trade‐offs between (hemi)biotroph and necrotroph defences (Robert‐Seilaniantz et al., 2011). In Arabidopsis, SA is synthesised via the isochorismate pathway and the phenylpropanoid pathway. Our results that AN knockout and overexpression altered PAL expression, but not ICS1 or PBS3 expression, during P. syringae infection demonstrated that AN is likely to regulate the phenylpropanoid pathway rather than the isochorismate pathway. Although the expression of EDS5 in the isochorismate pathway was reduced in the an‐t1 mutant, AN overexpression did not increase EDS5 expression to a level higher than that in Col‐0, suggesting that the change in EDS5 expression may not be due to the direct effect of AN disruption.
Through investigating the transcriptional function of AN (Figs 2, 3, 4), we found that AN directly targeted and repressed the expression of MYB46, a transcription factor directly activating PAL genes in Arabidopsis (Zhong & Ye, 2012). Conversely, the results of our molecular analyses suggested that AN is capable of releasing transcriptional repression of WRKY33 by sequestering TDP1 away from the WRKY33 promoter (Fig. 5). WRKY33 is a transcription factor that directly targets JA/ET pathway genes (e.g. ACS2, ORA59 and PDF1.2) and is critical for plant defences against B. cinerea (Zheng et al., 2006; Birkenbihl et al., 2012; Datta et al., 2015; Liu et al., 2015; Wang et al., 2015). These results suggested that AN is involved in the antagonistic transcriptional regulation of MYB46 and WRKY33, and may explain how AN antagonistically regulates SA and JA/ET pathway genes. Collectively, our discoveries indicated that AN mediates the transcriptional co‐regulation of MYB46 and WRKY33, which play an important role in SA–JA antagonism and trade‐offs between (hemi)biotroph and necrotroph defences (Fig. S8). Consistent with our model, the an‐t1 mutant had increased P. syringae‐induced SA accumulation and an‐t1 35S:AN had reduced P. syringae‐induced SA accumulation compared with Col‐0 (Fig. S9). In this study, we have provided multiple lines of evidence supporting AN’s transcriptional regulatory role and determining its target genes as well as its interacting partner (TDP1). However, we do not rule out the possibility that AN/TDP1 may also target other genes. In a previous study, knockout of MYB46 via T‐DNA insertion did not exhibit any phenotypic effect on the Arabidopsis response to P. syringae although pathogen‐induced expression levels of bacterial disease marker gene Ep5C and SA pathway stress‐responsive gene PR‐1 were altered in the myb46 mutants (Ramírez et al., 2011). Interestingly, the transcriptomic analysis did not reveal significant change in PAL gene expression (Ramírez et al., 2011). As an essential enzyme involved in phenylpropanoid and lignin biosynthesis, the expression of PALs is controlled by an NAC–MYB transcriptional network in which MYB46 is a major player (Nakano et al., 2015). Within this NAC–MYB transcriptional network, redundant, feed‐back and feed‐forward regulations exist widely (Xie et al., 2018b). This may possibly explain why knocking‐out a single transcription factor (i.e. MYB46) within the NAC–MYB transcriptional network did not result in the alteration of PAL expression and phenotypic response to P. syringae. Together with the results in the present study, we speculated that, in addition to MYB46, AN may target and regulate other members in the NAC–MYB transcriptional network to control PAL gene expression and pathogenic responses. In future studies, genome‐wide AN‐based and TDP1‐based ChIP‐seq together with RNA‐seq analysis upon pathogen infection may help to uncover additional target genes for AN and TDP1, and reveal both overlapping and nonoverlapping target genes in response to pathogen infection. This is a fruitful area for future investigations. Conversely, AN seems to be under the regulation of plant–microbe interaction mechanisms, because we observed the alteration of AN transcription and nuclear accumulation by P. syringae and B. cinerea (Fig. S10). As shown in Fig. S10, the alteration of AN transcription occurred at the early stages of P. syringae (6 hpi) and B. cinerea (24 hpi) infections. We also found that P. syringae can induce the reduction of AN nuclear accumulation at the early stage of infection (6 hpi). Because nuclear accumulation of AN is essential for its transcriptional function, we speculated that this regulation occurred at the early stage of pathogen infection. However, answers to the questions which factors trigger the interaction between AN and TDP1, at which point the AN–TDP1 protein complex turns ON or OFF the promoter of their target genes, and which factor controls the ON or OFF, are completely unknown. Now that we have established the transcriptional regulatory role of AN, future studies focusing on addressing each of these questions may help to reveal the precise role of the AN–TDP1 complex and AN–TDP1 interaction in response to pathogen infection.
Although CtBP is an ancient protein, widely existing in mammals, flies, worms and plants, the evolution of its molecular and biological roles remains poorly studied. AN protein has only 26.6% identity with animal CtBPs, and this was thought to result from high diversity in protein structure and the complete loss of nuclear function of AN. AN was believed to only have cytosolic function, because previous localisation analyses using stable transgenic plants expressing green florescent protein‐tagged AN failed to detect AN in the nucleus (Minamisawa et al., 2011). However, using the transient expression system, we were able to detect the nuclear accumulation of AN protein (Fig. 2). Furthermore, we identified the transcriptional repressor function of AN and the physical interaction of AN with the nuclear protein TDP1 (Figs 2,3). Our results suggested that AN has a nuclear function as found in animal CtBPs. It is plausible that AN may have rapid nucleocytoplasmic shuttling as found for CtBP2 (Zhao et al., 2006), which forms occasional nuclear localisation and may lead to the lack of nuclear localisation in stable transgenic plants. Our molecular analyses of AN nuclear function further indicated that the divergence in protein sequences of AN and animal CtBPs may have given rise to their different mechanisms of action, which can be specified by two aspects:
Different from animal CtBPs, the NLS of AN is nonfunctional (Minamisawa et al., 2011). Our results showed that AN physically associates with the NLS‐containing FHA domain of TDP1, suggesting that AN may utilise the NLS of TDP1 rather than its own NLS for nuclear localisation.
AN lacks residues to bind to PXDLS‐containing transcription factors (Stern et al., 2007).
In this study we have provided results from yeast two‐hybrid assays, cell fractionation, co‐localisation and transactivation to demonstrate that TDP1 is capable of enhancing AN nuclear accumulation and function. ChIP assay of AN in the tdp1‐2 mutant further illustrated that TDP1 is required for AN binding to the MYB46 promoter, which is crucial for AN transcriptional function. Instead of depending on associated transcription factors to repress target genes, we found that AN itself was sufficient to execute transcriptional repressor function and directly bind to the MYB46 promoter. Despite these distinct molecular functions, our discoveries of the involvement of AN in the regulation of host tolerance to pathogens, which is analogous to animal CtBPs involvement in tumour progression, suggested that plant and animal CtBPs are evolutionary conserved from the point of their biological function. This point is supported by the fact that AN is involved in the morphogenetic development of plants (Kim et al., 2002) analogous to animal CtBP role in development. In conclusion, our studies on the nuclear function and the involvement in plant immune responses of AN provide new insights into the molecular and biological functions of AN, which enable a better understanding of the evolution of CtBP in animals and plants.
TDP1 is a tyrosyl‐DNA phosphodiesterase hydrolysing DNA–protein cross‐links to prevent double‐stranded DNA damage in plants and animals (Lee et al., 2010; Stingele et al., 2017). However, little information is known about its role in transcriptional regulation. The existence of cross‐talk between DNA damage mechanisms and the plant immune responses has been previously reported, such as the dual role of SUPPRESSOR OF NPR1 INDUCIBLE1 (SNI1), RAD51, and BREAST CANCER2 (BRCA2) in the DNA damage response and the transcriptional regulation of defence response genes (Durrant et al., 2007; Wang et al., 2010). However, the complex interaction between these two fundamental biological processes is still poorly studied. Our results suggested that TDP1 negatively regulates the expression of WRKY33, encoding an essential transcription factor activating plant defence mechanisms against necrotrophic pathogens. Although TDP1 has been reported to be involved in plant responses to abiotic stresses (Sabatini et al., 2017), its role in biotic stress responses has not been reported previously. Our genetic analyses using the tdp1‐2 mutant demonstrated that TDP1 is involved in plant responses towards P. syringae and B. cinerea, because the tdp1‐2 mutant exhibited enhanced tolerance towards P. syringae and B. cinerea (Fig. S11). These results are consistent with our proposed model that TDP1 is involved in the transcriptional repression of both MYB46 and WRKY33 (Fig. S8).
In summary, our study revealed that AN is involved in plant immunity against biotrophic and necrotrophic pathogens. We demonstrated that the AN–TDP1 interaction is essential for the antagonistic transcriptional co‐regulation of MYB46 and WRKY33, which in turn regulates the expression of genes in the SA and JA/ET pathways.
Author contributions
WM and J‐GC initiated the project and coordinated data analysis and manuscript preparation; MX performed all biochemical assays; JZ, ACB and GAT contributed to bioinformatics analysis; MX, NE, TY, YP, AJR and TJT contributed to physiological analysis; MX, JL and DAP contributed to the pathogenic response assay; MX, JLM contributed to imaging analysis; MX, JZ, TY, ACB and JS contributed to expression analysis; MX, FC, WM and J‐GC drafted the manuscript and all authors critically reviewed and approved the final version of the manuscript for publication.
Supporting information
Fig. S1 Overexpression of Myc‐tagged AN (AN‐Myc) in an‐t1 (an‐t1 35S:AN) reverses its narrow‐leaf phenotype to wild‐type morphology.
Fig. S2 AN antagonistically regulates gene expression in SA and JA/ET pathways.
Fig. S3 Yeast two‐hybrid analysis of AN and TDP1 interaction.
Fig. S4 µChIP‐qPCR analysis using protoplasts co‐expressing AN‐Myc and HA‐TDP1.
Fig. S5 AN has no transcriptional activator activity.
Fig. S6 Transcription factors upregulated and downregulated in the an‐t1 mutant.
Fig S7 Amino acid sequence alignment of Arabidopsis TDP1 (AT5G15170) and Medicago TDP1 (XM_003622639).
Fig. S8 Model showing AN‐mediated transcriptional regulation of plant resistance to P. syringae and B. cinerea.
Fig. S9 Total amounts of salicylic acid (SA) and salicylic acid 2‐O‐glucoside (SAG) in Col‐0, an‐t1, and an‐t1 35S:AN in 0 hpi and 24 hpi of P. syringae inoculation.
Fig. S10 Pathogen infection changes the transcription and nuclear accumulation of AN.
Fig. S11 tdp1‐2 mutant has increased resistance to P. syringae and B. cinerea.
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by the Plant–Microbe Interfaces Scientific Focus Area, the Center for Bioenergy Innovation, and the BioEnergy Science Center by the Office of Biological and Environmental Research in the US Department of Energy Office of Science. Oak Ridge National Laboratory is managed by UT‐Battelle, LLC, for the United States Department of Energy under contract DE‐AC05‐00OR22725. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract no. DE‐AC02‐05CH11231. The authors declare no competing financial interests.
Contributor Information
Wellington Muchero, Email: mucherow@ornl.gov.
Jin‐Gui Chen, Email: chenj@ornl.gov.
References
- Bai Y, Vaddepalli P, Fulton L, Bhasin H, Hulskamp M, Schneitz K. 2013. ANGUSTIFOLIA is a central component of tissue morphogenesis mediated by the atypical receptor‐like kinase STRUBBELIG. BMC Plant Biology 13: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal‐Lobo M, Molina A, Solano R. 2002. Constitutive expression of ETHYLENE‐RESPONSE‐FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29: 23–32. [DOI] [PubMed] [Google Scholar]
- Bhasin H, Hulskamp M. 2017. ANGUSTIFOLIA, a plant homolog of CtBP/BARS localizes to stress granules and regulates their formation. Frontiers in Plant Science 8: 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Diezel C, Somssich IE. 2012. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiology 159: 266–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan AC, Zhang J, Guo J, Ranjan P, Singan V, Barry K, Schmutz J, Weighill D, Jacobson D, Jawdy S et al 2018. A variable polyglutamine repeat affects subcellular localization and regulatory activity of a populus ANGUSTIFOLIA protein. Genes, Genomes, Genetics 8: 2631–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. 2009. The transcriptional corepressor CtBP: a foe of multiple tumor suppressors. Cancer Research 69: 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R, Kumar D, Sultana A, Bhattacharyya D, Chattopadhyay S. 2015. Glutathione regulates 1‐aminocyclopropane‐1‐carboxylate synthase transcription via WRKY33 and 1‐aminocyclopropane‐1‐carboxylate oxidase by modulating mRNA stability to induce ethylene synthesis during stress. Plant Physiology 169: 2963–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dona M, Confalonieri M, Minio A, Biggiogera M, Buttafava A, Raimondi E, Delledonne M, Ventura L, Sabatini ME, Macovei A et al 2013. RNA‐Seq analysis discloses early senescence and nucleolar dysfunction triggered by Tdp1alpha depletion in Medicago truncatula . Journal of Experimental Botany 64: 1941–1951. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X. 2007. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proceedings of the National Academy of Sciences, USA 104: 4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle J, Dorn A, Beying N, Trapp O, Puchta H. 2019. The protease WSS1A, the endonuclease MUS81, and the phosphodiesterase TDP1 are involved in independent pathways of DNA‐protein crosslink repair in plants. The Plant Cell 31: 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachomo EW, Jimenez‐Lopez JC, Smith SR, Cooksey AB, Oghoghomeh OM, Johnson N, Baba‐Moussa L, Kotchoni SO. 2013. The cell morphogenesis ANGUSTIFOLIA (AN) gene, a plant homolog of CtBP/BARS, is involved in abiotic and biotic stress response in higher plants. BMC Plant Biology 13: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z. 2010. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiology 153: 1526–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Roden LC. 2014. Circadian regulation of plant immunity to pathogens. Methods in Molecular Biology 1158: 273–283. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Ohnishi H, Tamura K, Fukao Y, Furuya T, Hattori K, Tsukaya H, Hara‐Nishimura I. 2019. ANGUSTIFOLIA regulates actin filament alignment for nuclear positioning in leaves. Plant Physiology 179: 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY. 2002. The Arabidopsis thaliana–Pseudomonas syringae interaction. Arabidopsis Book 1: e0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H. 1993. Ethylene biosynthesis. Annual Review of Plant Physiology and Plant Molecular Biology 44: 283–307. [Google Scholar]
- Kim GT, Shoda K, Tsuge T, Cho KH, Uchimiya H, Yokoyama R, Nishitani K, Tsukaya H. 2002. The ANGUSTIFOLIA gene of Arabidopsis, a plant CtBP gene, regulates leaf‐cell expansion, the arrangement of cortical microtubules in leaf cells and expression of a gene involved in cell‐wall formation. EMBO Journal 21: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Na SH, Lee S‐Y, Jeong Y‐M, Hwang H‐J, Hur JY, Park S‐H, Woo J‐C, Kim S‐G. 2012. Structure–function studies of a plant tyrosyl‐DNA phosphodiesterase provide novel insights into DNA repair mechanisms of Arabidopsis thaliana . Biochemical Journal 443: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. 2008. Vectors for multi‐color bimolecular fluorescence complementation to investigate protein‐protein interactions in living plant cells. Plant Methods 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Kim H, Hwang HJ, Jeong YM, Na SH, Woo JC, Kim SG. 2010. Identification of tyrosyl‐DNA phosphodiesterase as a novel DNA damage repair enzyme in Arabidopsis. Plant Physiology 154: 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Kracher B, Ziegler J, Birkenbihl RP, Somssich IE. 2015. Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. elife 4: e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sanchez‐Serrano JJ, Solano R. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch‐Mani B, Slusarenko AJ. 1996. Production of salicylic acid precursors is a major function of phenylalanine ammonia‐lyase in the resistance of Arabidopsis to Peronospora parasitica . Plant Cell 8: 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamisawa N, Sato M, Cho KH, Ueno H, Takechi K, Kajikawa M, Yamato KT, Ohyama K, Toyooka K, Kim GT et al 2011. ANGUSTIFOLIA, a plant homolog of CtBP/BARS, functions outside the nucleus. The Plant Journal 68: 788–799. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M. 2015. NAC‐MYB‐based transcriptional regulation of secondary cell wall biosynthesis in land plants. Frontiers in Plant Science 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C. 2008. Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental‐triggered flavonoid synthesis. Journal of Plant Physiology 165: 1491–1499. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van Loon LC. 2004. NPR1: the spider in the web of induced resistance signaling pathways. Current Opinion in Plant Biology 7: 456–464. [DOI] [PubMed] [Google Scholar]
- Ramírez V, Agorio A, Coego A, García‐Andrade J, Hernández MJ, Balaguer B, Ouwerkerk PB, Zarra I, Vera P. 2011. MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiology 155: 1920–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I. 2019. Isochorismate‐derived biosynthesis of the plant stress hormone salicylic acid. Science 365: 498–502. [DOI] [PubMed] [Google Scholar]
- Robert‐Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylate antagonism. Annual Review of Phytopathology 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M et al 2004. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far‐reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini ME, Pagano A, Araujo S, Balestrazzi A, Macovei A. 2017. The tyrosyl‐DNA phosphodiesterase 1beta (Tdp1beta) gene discloses an early response to abiotic stresses. Genes (Basel) 8: 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. 1998. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE‐INSENSITIVE3 and ETHYLENE‐RESPONSE‐FACTOR1. Genes & Development 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Metraux JP, Brown R, Kazan K et al 2003. NPR1 modulates cross‐talk between salicylate‐ and jasmonate‐dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MD, Aihara H, Cho KH, Kim GT, Horiguchi G, Roccaro GA, Guevara E, Sun HH, Negeri D, Tsukaya H et al 2007. Structurally related Arabidopsis ANGUSTIFOLIA is functionally distinct from the transcriptional corepressor CtBP. Development Genes and Evolution 217: 759–769. [DOI] [PubMed] [Google Scholar]
- Stingele J, Bellelli R, Boulton SJ. 2017. Mechanisms of DNA‐protein crosslink repair. Nature Reviews Molecular Cell Biology 18: 563–573. [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ. 2004. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschaplinski TJ, Abraham PE, Jawdy SS, Gunter LE, Martin MZ, Engle NL, Yang X, Tuskan GA. 2019. The nature of the progression of drought stress drives differential metabolomic responses in Populus deltoides . Annals of Botany 124: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE. 2015. Transcriptional networks in plant immunity. New Phytologist 206: 932–947. [DOI] [PubMed] [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H. 1996. Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana (L.) Heynh. Development 122: 1589–1600. [DOI] [PubMed] [Google Scholar]
- Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z. 2015. The Arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum . Plant Physiology 169: 856–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Durrant WE, Song J, Spivey NW, Dong X. 2010. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proceedings of the National Academy of Sciences, USA 107: 22716–22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Muchero W, Bryan AC, Yee K, Guo HB, Zhang J, Tschaplinski TJ, Singan VR, Lindquist E, Payyavula RS et al 2018a. A 5‐enolpyruvylshikimate 3‐phosphate synthase functions as a transcriptional repressor in Populus . Plant Cell 30: 1645–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Ren G, Costa‐Nunes P, Pontes O, Yu B. 2012. A subgroup of SGS3‐like proteins act redundantly in RNA‐directed DNA methylation. Nucleic Acids Research 40: 4422–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen JG, Muchero W. 2018b. Regulation of lignin biosynthesis and its role in growth‐defense tradeoffs. Frontiers in Plant Science 9: 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zarei A, Korbes AP, Younessi P, Montiel G, Champion A, Memelink J. 2011. Two GCC boxes and AP2/ERF‐domain transcription factor ORA59 in jasmonate/ethylene‐mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Molecular Biology 75: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xie M, Ren G, Yu B. 2013. CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proceedings of the National Academy of Sciences, USA 110: 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Subramanian T, Zhou Y, Chinnadurai G. 2006. Acetylation by p300 regulates nuclear localization and function of the transcriptional corepressor CtBP2. Journal of Biological Chemistry 281: 4183–4189. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. 2006. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. The Plant Journal 48: 592–605. [DOI] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. 2007. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19: 2776–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. 2012. MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant and Cell Physiology 53: 368–380. [DOI] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, Jiang Z, Kim J‐M, To TK, Li W, Zhang X et al 2011. Derepression of ethylene‐stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis . Proceedings of the National Academy of Sciences, USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Overexpression of Myc‐tagged AN (AN‐Myc) in an‐t1 (an‐t1 35S:AN) reverses its narrow‐leaf phenotype to wild‐type morphology.
Fig. S2 AN antagonistically regulates gene expression in SA and JA/ET pathways.
Fig. S3 Yeast two‐hybrid analysis of AN and TDP1 interaction.
Fig. S4 µChIP‐qPCR analysis using protoplasts co‐expressing AN‐Myc and HA‐TDP1.
Fig. S5 AN has no transcriptional activator activity.
Fig. S6 Transcription factors upregulated and downregulated in the an‐t1 mutant.
Fig S7 Amino acid sequence alignment of Arabidopsis TDP1 (AT5G15170) and Medicago TDP1 (XM_003622639).
Fig. S8 Model showing AN‐mediated transcriptional regulation of plant resistance to P. syringae and B. cinerea.
Fig. S9 Total amounts of salicylic acid (SA) and salicylic acid 2‐O‐glucoside (SAG) in Col‐0, an‐t1, and an‐t1 35S:AN in 0 hpi and 24 hpi of P. syringae inoculation.
Fig. S10 Pathogen infection changes the transcription and nuclear accumulation of AN.
Fig. S11 tdp1‐2 mutant has increased resistance to P. syringae and B. cinerea.
Table S1 Primers used in this study.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


