Abstract
Chromosomal instability (CIN) is one of the major forms of genomic instability in various human cancers and is recognized as a common hallmark of tumorigenesis and heterogeneity. However, some malignant tumors show a paucity of chromosomal alterations, suggesting that tumor progression and evolution can occur in the absence of CIN. It is unclear whether CIN is stable between precursor lesions, primary tumor, and metastases or if it evolves during these steps. In this review, we describe the influence of CIN on the various steps in tumor initiation and development. Given the recognized significant effects of CIN in cancer, CIN-targeted therapeutics could have a major impact on improving clinical outcomes.
Introduction
Most cancer types have the presence of a population of cells with chromosomal instability (CIN; ref. 1). This hallmark of cancer is suggested as a major modulator of tumor adaptation and evolution in response to challenges arising from the tumor microenvironment such as metastasis or therapeutic resistance (2). CIN, one of the major forms of genomic instability in various human cancers, is typically associated with structural and numerical chromosomal changes over time in tumor tissues (3-5). CIN and aneuploidy are distinct, but closely related, and have been shown to affect carcinogenesis and therapeutic responses. Although there is increasing understanding of the role of CIN in biological systems, there are currently no drugs in the clinic that can be used specifically to inhibit chromosome segregation errors (4). Hence, increasing our understanding on the dual role of CIN as a modulator of tumor development and of resistance to therapy is critical for our ability to target it for therapeutic benefit (4).
In this review, we highlight the role of CIN in tumor initiation and development, especially in precursor lesions and metastases. We also discuss the impact of CIN on clinical outcome and highlight the challenges related to specifically targeting CIN.
Interplay of CIN, Aneuploidy, and Chromothripsis in Cancer
Although CIN, aneuploidy, and chromothripsis are intimately linked, there are fundamental differences between them, especially with regard to their function in various stages of tumor progression.
CIN versus aneuploidy in cancer
Aneuploidy, one of the most common chromosomal alterations, is characterized by an unbalanced chromosome number (i.e., having missing or extra chromosomes). Aneuploidy can be a consequence of CIN, and the level of CIN is typically associated with karyotypic complexity (6). Aneuploidy has been suggested to be a logical aberration in cells with CIN; however, CIN is not an obligatory outcome of aneuploidy (6). For instance, aneuploidy rearrangements that occur early in breast cancer cells are stably maintained because there is little karyotypic variance between cells; such breast cancer cells might not have CIN (6-8). In a CIN mouse model, nonregenerating tissues can have major aneuploidy, whereas regenerating tissues do not (6, 9). Alternatively, in silico modeling of somatic genome evolution has shown that CIN was significantly involved in generating higher-grade aneuploidies, and aneuploidy tolerance in the absence of CIN was sufficient to explain lower-grade aneuploidies (10).
CIN versus chromothripsis in cancer
As with CIN and aneuploidy, CIN and chromothripsis are not necessarily directly related. Chromothripsis ["chromosome" (chromo) and "shattering into pieces" (thripsis)] is a process by which dozens to up to thousands of chromosomal rearrangements occur in localized regions of one or a few chromosomes (11). Multistep carcinogenesis requires genomic instability (12), and defects in chromosome segregation and/or the DNA damage response process can also affect carcinogenesis by stimulating chromothripsis (11). It also should be noted that de novo rearrangements caused by chromothripsis can trigger CIN in subsequent cell divisions through other possible mechanisms, such as the absence of proper templates for homologous repair (13). The chromosomal missegregation detected in cleavage-stage embryos might cause mosaic chromothripsis, similar to somatic events present in cancer, by possibly giving rise to complex genomic rearrangements after chromosome pulverization in the micronuclei (13, 14).
Chromosome segregation in mitosis
Mitosis is a process that typically entails perfect duplication and segregation of chromosomes (15). In various cancers, chromosome missegregations, as well as changes in structural chromosomes (deletions or translocations), are known to occur. In addition, translocation may be the most effective way of producing structural CIN, which can result in the overexpression or formation of oncogenes by gene fusion (16, 17). For instance, recurrent translocations that generate chimeric fusions have important functions in modulating tumor progression in blood malignancies such as acute lymphoblastic leukemia, follicular lymphoma, acute myeloid leukemia, or myelodysplastic syndrome (17, 18); recurrent gene fusions can also contribute to solid tumors such as bone or breast malignancies (17, 19). For example, the ETV6-NTRK3 fusion oncogene is thought to initiate breast cancer from committed mammary progenitors via activation of the AP1 complex (20). Extrachromosomal oncogene amplification can also enable adaptation to variable environmental conditions by enhancing the likelihood that a subpopulation of cells will express that oncogene at a level that maximizes tumor development (21). Extrachromosomal DNA is observed in nearly half of human cancers; however, it was mostly absent in normal cells (21). This notably high frequency of extrachromosomalDNA in cancer is relative to chromosomal inheritance; driver oncogenes are amplified most commonly in extrachromosomal DNA. Thereby, oncogene amplification on extrachromosomal DNA can shape genetic heterogeneity in human cancer (21). Alternatively, several potential gross defects can occur during mitosis, such as defective sister chromatid cohesion and segregation and lagging chromosomes; these defects result in aberrant karyotypes and can involve CIN in cancer cells (Fig. 1A; refs. 3, 5, 12, 22-25).
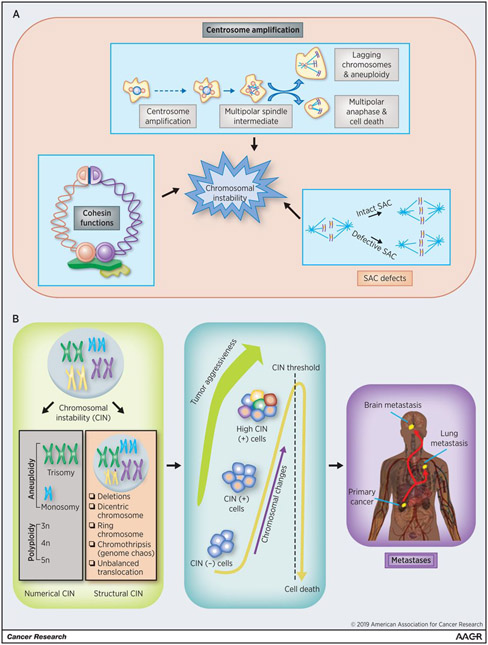
Figure 1.
CIN in cancer. A, Paths to CIN. Mechanisms leading to defects in chromosome instability (see text for details). B, The critical role of CIN in the development of cancer. CIN is included in whole-chromosomal losses and gains (numerical CIN) and subchromosomal gains, losses, and translocations (structural CIN). Increasing CIN has been correlated with key tumor features through chromosomal alterations that can contribute to metastases (see text for details).
It has been suggested that these chromosomal aberrations are highly associated with human cancers because the aberrations allow cells to rapidly acquire genetic changes (26) with diverse mechanisms; the causative functions of these aberrations in cancer progression have been reviewed in detail in previous reports (27, 28). For example, given the critical role of the spindle assembly checkpoint (SAC) for chromosome segregation fidelity, anaphase-promoting complex/cyclosome suppression was found to potentially enhance the frequency of bipolar spindle formation and reduce CIN after genome doubling (29). Alternatively, impairments of the SAC may also lead to separation of premature sister chromatids, which can result in chromosome missegregation (30, 31). In addition to the defects that arise during mitosis and immediately lead to chromosome missegregation, there are other plausible mechanisms that originate during interphase that could (indirectly) contribute to chromosome missegregation, such as centrosome amplification (32, 33), alterations in gene transcription affecting mitosis (34, 35), and replication stress (36, 37).
CIN in Tumor Initiation and Development
CIN in precursor lesions
Tumor initiation is known to be associated with multiple processes, including epigenetic and genetic alterations. Several studies have examined the role of mitotic errors in shaping cancer genomes through CIN, which can provide the evolutionary fuel for cancer progression (38). For example, a mathematical framework was used to determine the effect of CIN on the somatic evolution of cancer and showed that CIN mutations seem to initiate colorectal cancer (39); in addition, a remarkably high degree of allelic imbalance (which reflects CIN with changes in DNA copy number) has been observed in small tumors, strongly suggesting that CIN occurs early during colon cancer progression (40).
Since the 1800s, researchers have known that tumor cells missegregate their chromosomes during mitosis and that such missegregation events are more prevalent in advanced stages of cancer. At least 24 independent genetic lesions known to cause aneuploidy also enhance or initiate tumorigenesis in mice (38). In addition, X chromosome aneuploidy has been associated with breast cancer initiation and development (41). Aneuploidy can drive tumor formation in cases in which mutations at tumor suppressor or tumor promoter loci have already enhanced the potential for cellular transformation (42). For instance, overexpression of the mitotic checkpoint gene Mad2 is sufficient to lead to aneuploidy, nondisjunction, and tumor initiation in mice (43). However, whether aneuploidy alone is sufficient to initiate tumor progression is still unknown (44).
The centrosome, coordinator of most microtubule-associated processes, can play a critical role in organizing the bipolar spindle that partitions chromosomes during cell division (45). Supernumerary centrosomes have been observed early in the development of various tumors and are commonly associated with poor clinical outcome (45). In cultured cells, centrosome amplification leads to mitotic errors that may cause chromosome missegregation (45). A previous study has suggested that centrosome amplification may initiate tumorigenesis in flies (46); another study suggests that centrosome amplification stimulates aneuploidy in vivo and that extra centrosomes can trigger early tumorigenesis in a model of intestinal neoplasia (45). Furthermore, transplantation of extra centrosomes in larval brain cells stimulated the formation of metastases (47).
Cohesin, best known for its function in chromosome segregation, has been found to play a role in cancer progression (48). Sequencing of cancer genomes has also revealed the presence of somatic mutations in cohesin in various cancers. For example, 11 mutations in the SMC1A core cohesin subunit were identified in screening a large series of early colorectal adenomas, a precocious step during progression of colon cancer, suggesting that mutant cohesin can drive CIN in early colorectal adenomas (48). Another consequence of CIN is an enhanced rate of LOH. LOH at 7q31 has been observed in early stages of prostate cancer (49); LOH preferentially occurs in early replicating regions in cancer genomes (50). Several studies have revealed the function of CIN as a result of mitotic checkpoint hyperactivation in the initiation of tumors (51).
Dysfunctional telomeres can also initiate events that cause CIN. Telomeres, when functioning correctly, are composed of repetitive G-rich sequences and can form protective caps at the ends of linear chromosomes that prevent CIN (52). However, telomeres lose their capping role in response to telomere shortening, which may stimulate CIN and facilitate initiation of tumors (53). This role of telomeres has also been detected in cancer precursor lesions, for instance in colonic adenomas with high-grade dysplasia (54) and in ductal carcinoma in situ (55). Furthermore, telomere shortening, as an early change in preneoplastic cells, has been observed in various epithelial cancers; hence, telomere dysfunction is likely one of the various driving events in early carcinogenesis in these entities (56, 57). Whereas abnormalities in telomere length occur at an early stage in the development of epithelial carcinogenesis (58), CIN and tetraploidy are early events in cervical carcinogenesis (59). In addition, tetraploidy may be a predictive marker for progression of Barrett's esophagus into esophageal adenocarcinoma (60). Researchers have also proposed that tetraploidy, much like aneuploidy (61), may stimulate tumor formation (62).
CIN, as an early event in the development of high-grade serous ovarian or fallopian tube cancer, is similar to events observed in ductal carcinoma in situ in breast tumorigenesis (55, 63-65). Oncogenic H-RasV12, which is commonly related with cancer progression, can cause telomere attrition and telomeric replication stress, and subsequently dysfunctional telomeres in cells that lack hTERT activity (66). These studies further suggested that cells in early cancer precursor lesions commonly show no or low activation of telomerase, whereas cells in more than 90% of all human cancers show reactivation of this enzyme to maintain telomere function and length (66, 67).
CIN in metastases
Comparative reports of metastatic and primary tumors suggest that CIN may contribute to the development of cancer metastases via diverse mechanisms (68), as summarized below.
Breast cancer
Chromosome segregation errors can promote the number of micronuclei that when ruptured, cause enhanced cytosolic DNA, activate downstream noncanonical NF-κB signaling, and activate the cytosolic DNA-sensing cGAS-STING pathway in CIN-high breast cancer cells, hence stimulating cancer metastases (69). The amplification and overexpression of MASTL, an essential kinase for correct progression through mitosis, correlates with enhanced CIN in breast cancer and poor patient survival, whereas knock-down of MASTL may suppress breast cancer metastasis in vivo (70). CIN has also been suggested to initiate the formation of somatic copy-number alterations (SCNA; ref. 71); SCNAs have been detected both subclonally and clonally across all histologic subtypes of breast cancer (68). In a multiregional profiling study, metastatic subclones were identifiable in a primary basal breast tumor that resulted in lung metastases (72). In that study, SCNAs were identified both subclonally and clonally across all histologic subtypes of primary breast cancer (68, 72). Topographic single-cell sequencing analysis to evaluate genomic copy-number profiles of single tumor cells suggested that most mutations and CNAs evolved within the ducts before invasion (73).
Prostate cancer
Several studies have shown the presence of CIN in prostate cancer, especially in those presenting with metastatic disease. Reactivation of telomerase after telomere dysfunction was determined to yield murine prostate tumors with bone metastases (74). High levels of aneuploidy and tetraploidy have also been generally associated with the generation of metastases (75, 76). The intratumoral evolutionary landscape of high-risk prostate cancer suggests that primary tumors of patients with metastatic disease had a higher burden of SCNAs (77). This is consistent with previous studies correlating biochemical recurrence after prostatectomy with high SCNA in localized disease (78). Thus, it is plausible that changes in SCNAs can contribute to aggressive metastatic prostate cancer.
Colorectal cancer
The progression from invasive cancer to metastatic disease in colorectal cancer is accompanied by increased CNAs (79) on the basis of a meta-analysis of chromosome CGH (80) and array CGH (81). For instance, the number of CNAs in metastases is higher than in primary tumors; small regions of gain at chromosomes 10p and 6p21 and of loss at chromosome 8p12 also occur more commonly in metastases than in primary tumors (79). In addition, no changes in known drivers of CIN were identified exclusively in chromothripsis-containing metastasis; TP53 clonal mutations in particular were detected in all of the cases studied, reinforcing the possibility that CIN has a critical role in metastases (68).
Microtubule systems are known to be important for mitosis, for CIN, and for cell migration and morphologic plasticity required by metastasis (69, 82-84). During mitosis, microtubules can form the spindle to enable correct chromosomal segregation, whereas overexpression of the nonmotile microtubule-depolymerizing kinesin-13 family proteins KIF2B and KIF2C (also known as MCAK) may contribute to destabilization of microtubule attachments to chromosomes at the kinetochores, hence directly inhibiting CIN in otherwise chromosomally unstable cells (69). In colorectal tumors, CIN has been correlated with defects in microtubule plus-end attachments led by a dominant mutation in the adenomatous polyposis coli gene (85).
These findings suggest a critical association between CIN and metastases, especially because CIN is enhanced in metastases compared with primary tumors in cancer patients with multiple tumor types (Fig. 1B; refs. 4, 86). In addition, tumor metastasis may be characterized as clonal progression modulated by CIN (87). Table 1 lists a number of important associations with clinical implications. First, if the primary tumor can be stopped from repeatedly seeding metastases, then its removal might halt further metastatic development. Second, the ability to estimate the timing of metastatic spread from the primary tumor could be important for cancer treatment; this is because once the primary tumor is clinically detectable after metastatic dissemination occurs, early surgical resection may fail to reduce metastatic disease (68).
Table 1.
Associations between CIN and tumor initiation and metastases
| Cancer type |
Event | Methodology | Phenomenon and potential mechanism | Refs. |
|---|---|---|---|---|
| Breast cancer | SCNAs | Profiling of 52 single cells from a primary basal breast tumor and 48 cells from the tumor's associated liver metastasis | • A high level of concordance was observed at the level of SCNAs | (109) |
| • Tumors grew by punctuated clonal expansions with few persistent intermediates | ||||
| Comparing somatic mutations and gene CNAs of primary breast cancers and their matched metastases from patients with estrogen receptor (ER)-negative breast cancer | • There was a large subset of gene CNAs (55%) and nonsynonymous somatic mutation sharing between primary tumors and paired metastases | (110) | ||
| • Synchronous metastases displayed higher concordance with the paired primary tumor than did metachronous metastases | ||||
| • The repertoires of somatic genetic alterations in ER-negative breast cancer metastases may differ from those of their primary tumors | ||||
| Investigating the genomic evolution between primary and matched metastatic ER+ breast cancers after failure of adjuvant treatment | • ESR1 mutations were in the metastases, but none were in the primary tumor | (111) | ||
| • Although there was a high level of concordance between primary tumor and matched metastases for the investigated molecular alterations, ESR1 mutations as potential actionable targets were identified only in metastases | ||||
| Performing DNA exome and RNA-sequencing of matched primary tumors and multiple metastases from 83 distinct specimens of 16 patients | • Genetic drivers unique to metastasis were identified as somatic mutations in the androgen and ER genes | (112) | ||
| • Most metastatic drivers might be established in the primary tumor despite the substantial heterogeneity observed in the metastases | ||||
| High-depth whole-exome sequencing of distinct core biopsies of primary breast cancers and synchronous distant metastases | • Synchronous primary breast cancers and metastases differed in their repertoire of somatic genetic alterations | (113) | ||
| • Mutational signature shifts could affect spatial intratumoral genetic heterogeneity | ||||
| Genome-wide sequencing of ctDNA from plasma of 162 patients with biopsy-proven metastatic triple-negative breast cancer (TNBC) | • Percent genome altered and copy-number profiles were similar between primary tumor and metastases in TNBCs | (114) | ||
| • SCNAs were enriched in TNBC metastases | ||||
| Whole-exome sequencing (WES) of a base-like breast cancer primary tumor and a metachronous brain metastasis | • More than 90% of the SCNAs in the primary tumor were propagated in metastases, whereas ~80% of SCNAs in metastases were not shared by the primary tumor | (115) | ||
| • Enhanced CNAs were in metastases | ||||
| Prostate cancer | SCNAs | Comparing analysis of 333 primary prostate cancers (represented by single biopsies) and an unrelated cohort of 150 soft tissue and bone metastases from castration-resistant prostate cancers | • There was a remarkably higher SCNA and mutational burden in the metastases than in primary tumors | (68) |
| • Patients who had a high SCNA burden had an elevated risk of relapse | ||||
| TP53 mutations | Whole-genome and ultradeep targeted sequencing of longitudinally collected metastatic and primary tumors | • Both primary tumor and metastatic clones were detected | (116) | |
| • Enrichment of TP53 mutations was present in metastases | ||||
| WES of multiple metastases arising from prostate tumors in 10 patients | • Metastasis-to-metastasis seeding may occur either by a branching or a linear pattern of spread | (117) | ||
| Colorectal cancer | Combing WES CNAs for 15 triplets | • The primary colorectal carcinomas and about half the metastatic colorectal carcinomas had the same clonal origin | (118) | |
| Whole-genome sequencing of two primary colorectal cancer tumors and their metastases | • Most of the somatic alterations existed in both sites, and distinct clonal evolution patterns were identified in the two cases | (119) | ||
| Performing targeted next-generation sequencing on liver metastases and primary tumors from 18 patients with liver-limited metastatic colorectal cancer | • There was high genomic concordance between metastases and primary tumors, in support of the linear progression model in liver-limited metastatic colorectal cancer | (120) |
Future Perspectives of CIN
CIN as a prognostic tool
CIN is commonly associated with tumorigenesis and clinical outcomes (Supplementary Table S1); various reports have shown that aneuploidy that arises as a consequence of CIN in malignant tumors favors the evolution of tumors (88-90). Investigation of CIN signatures from specific genes whose expression was consistently associated with total functional aneuploidy in many different cancer types and the net overexpression of this signature were predictive of poor clinical outcome in 12 cancer data sets representing 6 cancer types (91). In addition, intratumoral heterogeneity mediated through CIN was correlated with increased risk of death or recurrence. This investigation subsequently confirmed the potential prognostic value of CIN (92). Another study demonstrated the prognostic and predictive value of the centromere and of the kinetochore gene expression score, which indicated a role for centromere misregulation in the development of cancer and supported the notion that tumors with extremely high CIN are less tolerant to specific genotoxic therapies (93). In addition, a correlation has also been observed between improved prognosis and CIN, which was detected in non–small cell lung, gastric, and ovarian cancers; this correlation was not observed in estrogen receptor–positive breast cancers (90, 94).
Combination with chemotherapy to overcome resistance
Resistance to anticancer drugs is a complex process (5, 95-102), and CIN-related phenotypic and genetic diversity in tumors has implications for chemotherapy-resistance including innate and acquired resistance (89). CIN-positive tumors seem to be more sensitive to DNA-damaging agents or radiotherapy (6). The SAC mainly contributes to CIN through the cell division and mono-polar spindle 1 kinase [MPS-1; also known as Thr/Tyr kinase (TTK); ref. 103]. Various studies using xenograft models have demonstrated that TTK/MPS-1 suppression may improve the effect of paclitaxel in the treatment of melanoma, glioblastoma, and triple-negative breast cancer (6). Improving microtubule stability by using inhibitors of the microtubule-destabilizing kinase, Aurora B (104), may enhance chromosome missegregation; Aurora B inhibitors have shown efficacy in primary as well as taxane-resistant tumors (105, 106).
Identifying a group or kinetochore and centromere protein genes that are related to sensitivity to therapy and cancer patient outcome could be attractive targets (107). These chromosomal roles are distinct from many existing drug targets involved in modulation of tumor suppression or oncogenic pathways. Hence, these proteins (i.e., centromere and kinetochore protein genes; ref. 107) might provide novel drug targets that could overcome the drug resistance associated with CIN, especially when combined with therapies that target known tumor suppression or oncogenic pathways or signal transduction in cancer patients. Some studies indicate that CIN modulates tumor heterogeneity in patients with myeloma and contributes to acquired drug resistance in multiple myeloma (108). As a result, targeting CIN upfront can be a potential approach to prevent genetic heterogeneity from occurring; new experimental model systems of CIN-dependent malignancy are needed for developing new therapies (108).
Conclusions
Whole-genome sequencing efforts have demonstrated that no two malignant tumors are the same; thus, careful matching of individual cancer patients with specific drugs, an approach referred to as personalized medicine, will be important to improving cancer therapy. We suggest that CIN can significantly affect tumor evolution and therapy response and could be an option for selecting therapy and targeting chemotherapy-resistant cancers.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by NIH grants (P50CA217685, P50 CA098258, UH3TR000943, R35 CA209904, and CA016672), the Frank McGraw Memorial Chair in Cancer Research, and the American Cancer Society Research Professor Award, the Blanton-Davis Ovarian Cancer Research Program to A.K. Sood, a fellowship from the National Foundation for Cancer Research, a Hanes and Wills Family endowed professorship in cancer at the Wake Forest Baptist Comprehensive Cancer Center, the Cancer Center Support Grant from the NCI to the Comprehensive Cancer Center of Wake Forest Baptist Medical Center (P30 CA012197) to W. Zhang. The authors thank Tamara K. Locke from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for editorial assistance.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
A.K. Sood reports receiving commercial research grant from M-Trap; has an ownership interest (including stock, patents, etc.) in BioPath; and is a consultant/advisory board member for Kiyatec and Merck. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Nicholson JM, Cimini D. Cancer karyotypes: survival of the fittest. Front Oncol 2013;3:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gronroos E, Lopez-Garcia C.Tolerance of chromosomal instability in cancer: mechanisms and therapeutic opportunities. Cancer Res 2018;78:6529–35. [DOI] [PubMed] [Google Scholar]
- 3.Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol 2018;15:139–50. [DOI] [PubMed] [Google Scholar]
- 4.Bakhoum SF, Cantley LC.The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018;174:1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach D-H, Lee SK.Long noncoding RNAs in cancer cells. Cancer Lett 2018;419:152–66. [DOI] [PubMed] [Google Scholar]
- 6.van Jaarsveld RH, Kops G. Difference makers: chromosomal instability versus aneuploidy in cancer. Trends Cancer 2016;2:561–71. [DOI] [PubMed] [Google Scholar]
- 7.Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 2016;48:1119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Waters J, Leung ML, Unruh A, Roh W, Shi X, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 2014;512:155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pfau SJ, Silberman RE, Knouse KA, Amon A. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev 2016;30:1395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valind A, Jin Y, Gisselsson D. Elevated tolerance to aneuploidy in cancer cells: estimating the fitness effects of chromosome number alterations by in silico modelling of somatic genome evolution. PLoS ONE 2013;8:e70445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer 2012;12:663–70. [DOI] [PubMed] [Google Scholar]
- 12.Jones MJ, Jallepalli PV. Chromothripsis: chromosomes in crisis. Dev Cell 2012;23:908–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloosterman WP, Cuppen E.Chromothripsis in congenital disorders and cancer: similarities and differences. Curr Opin Cell Biol 2013;25:341–8. [DOI] [PubMed] [Google Scholar]
- 14.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012;482:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jefford CE, Irminger-Finger I.Mechanisms of chromosome instability in cancers. Crit Rev Oncol Hematol 2006;59:1–14. [DOI] [PubMed] [Google Scholar]
- 16.Vargas-Rondón N, Villegas VE, Rondón-Lagos M. The role of chromosomal instability in cancer and therapeutic responses. Cancers 2017;10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007;7:233–45. [DOI] [PubMed] [Google Scholar]
- 18.Nambiar M, Kari V, Raghavan SC. Chromosomal translocations in cancer. Biochim Biophys Acta 2008;1786:139–52. [DOI] [PubMed] [Google Scholar]
- 19.Thompson SL, Compton DA. Chromosomes and cancer cells. Chromosome Res 2011;19:433–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Tognon CE, Godinho FJ, Yasaitis L, Hock H, Herschkowitz JI, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell 2007;12:542–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 2017;543:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 2001;1:109–17. [DOI] [PubMed] [Google Scholar]
- 23.Targa A, Rancati G. Cancer: a CINful evolution. Curr Opin Cell Biol 2018;52:136–44. [DOI] [PubMed] [Google Scholar]
- 24.Milunović-Jevtić A, Mooney P, Sulerud T, Bisht J, Gatlin JC. Centrosomal clustering contributes to chromosomal instability and cancer. Curr Opin Biotechnol 2016;40:113–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci 2008;13:2075–90. [DOI] [PubMed] [Google Scholar]
- 26.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998;396:643–9. [DOI] [PubMed] [Google Scholar]
- 27.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 2010;10:102–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 2012;13:189–203. [DOI] [PubMed] [Google Scholar]
- 29.Sansregret L, Patterson JO, Dewhurst S, Lopez-Garcia C, Koch A, McGranahan N, et al. APC/C dysfunction limits excessive cancer chromosomal instability. Cancer Discov 2017;7:218–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012;22:R966–80. [DOI] [PubMed] [Google Scholar]
- 31.Musacchio A Spindle assembly checkpoint: the third decade. Philos Transa R Soc Lond B Biol Sci 2011;366:3595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderhub SJ, Kramer A, Maier B. Centrosome amplification in tumorigenesis. Cancer Lett 2012;322:8–17. [DOI] [PubMed] [Google Scholar]
- 33.Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature 2004;428:77–81. [DOI] [PubMed] [Google Scholar]
- 34.Kabeche L, Compton DA.Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol 2012;22:638–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning AL, Longworth MS, Dyson NJ. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev 2010;24:1364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakhoum SF, Silkworth WT, Nardi IK, Nicholson JM, Compton DA, Cimini D. The mitotic origin of chromosomal instability. Curr Biol 2014;24:R148–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013;494:492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schvartzman J-M, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 2010;10:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak MA, Komarova NL, Sengupta A, Jallepalli PV, Shih Ie M,Vogelstein B, et al. The role of chromosomal instability in tumor initiation. Proc Natl Acad Sci U S A 2002;99:16226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih I-M, Zhou W, Goodman SN, Lengauer C, Kinzler KW, Vogelstein B. Evidence that genetic instability occurs at an early stage of colorectal tumorigenesis. Cancer Res 2001;61:818–22. [PubMed] [Google Scholar]
- 41.Zhang H, Yang X, Feng X, Xu H, Yang Q, Zou L, et al. Chromosome-wide gene dosage rebalance may benefit tumor progression. Mol Genet Genomics 2018;293:895–906. [DOI] [PubMed] [Google Scholar]
- 42.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 2009;10:478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sotillo R, Hernando E, Díaz-Rodríguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 2007;11:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giam M, Rancati G.Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div 2015;10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell 2017;40:313–22.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell 2008;133:1032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pino MS, Chung DC.The chromosomal instability pathway in colon cancer. Gastroenterology 2010;138:2059–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cucco F, Servadio A, Gatti V, Bianchi P, Mannini L, Prodosmo A, et al. Mutant cohesin drives chromosomal instability in early colorectal adenomas. Hum Mol Genet 2014;23:6773–8. [DOI] [PubMed] [Google Scholar]
- 49.Latil A, Cussenot O, Fournier G, Baron JC, Lidereau R. Loss of heterozygosity at 7q31 is a frequent and early event in prostate cancer. Clinical Cancer Res 1995;1:1385–9. [PubMed] [Google Scholar]
- 50.Pedersen BS, De S.Loss of heterozygosity preferentially occurs in early replicating regions in cancer genomes. Nucleic Acids Res 2013;41:7615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A 2008;105:16719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailey SM, Murnane JP. Telomeres, chromosome instability and cancer. Nucleic Acids Res 2006;34:2408–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meena J, Rudolph KL, Günes C. Telomere dysfunction, chromosomal instability and cancer In:Ghadimi BM, Ried T, editors. Chromosomal instability in cancer cells. Cham: Springer Intl Publishing; 2015. P. 61–79. [DOI] [PubMed] [Google Scholar]
- 54.Rudolph KL, Millard M, Bosenberg MW, DePinho RA. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet 2001;28:155–9. [DOI] [PubMed] [Google Scholar]
- 55.Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, et al. In situ analyses of genome instability in breast cancer. Nat Genet 2004;36:984–8. [DOI] [PubMed] [Google Scholar]
- 56.Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 2000;406:641–5. [DOI] [PubMed] [Google Scholar]
- 57.Gisselsson D, Jonson T, Petersén, Strömbeck B, Dal Cin P, Höglund M, et al. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci U S A 2001;98:12683–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meeker AK, Hicks JL, Iacobuzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, et al. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis.Clin Cancer Res 2004;10:3317–26. [DOI] [PubMed] [Google Scholar]
- 59.Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, et al. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis 2006;27:337–43. [DOI] [PubMed] [Google Scholar]
- 60.Reid BJ.Early events during neoplastic progression in Barrett's esophagus. Cancer Biomark 2010;9:307–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 2007;11:25–36. [DOI] [PubMed] [Google Scholar]
- 62.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005;437:1043–7. [DOI] [PubMed] [Google Scholar]
- 63.Salvador S, Rempel A, Soslow RA, Gilks B, Huntsman D, Miller D. Chromosomal instability in fallopian tube precursor lesions of serous carcinoma and frequent monoclonality of synchronous ovarian and fallopian tube mucosal serous carcinoma. Gynecol Oncol 2008;110:408–17. [DOI] [PubMed] [Google Scholar]
- 64.Meeker AK, Argani P. Telomere shortening occurs early during breast tumorigenesis: a cause of chromosome destabilization underlying malignant transformation? J Mammary Gland Biol Neoplasia 2004;9:285–96. [DOI] [PubMed] [Google Scholar]
- 65.Kuhn E, Meeker A, Wang T-L, Sehdev AS, Kurman RJ, Shih I-M. Shortened telomeres in serous tubal intraepithelial carcinoma: an early event in ovarian high-grade serous carcinogenesis. Am J Surg Pathol 2010;34:829–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suram A, Kaplunov J, Patel PL, Ruan H, Cerutti A, Boccardi V, et al. Oncogene-induced telomere dysfunction enforces cellular senescence in human cancer precursor lesions. EMBO J 2012;31:2839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Artandi SE, DePinho RA.Telomeres and telomerase in cancer. Carcinogenesis 2010;31:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turajlic S, Swanton C. Metastasis as an evolutionary process. Science 2016;352:169–75. [DOI] [PubMed] [Google Scholar]
- 69.Bakhoum SF, Ngo B, Laughney AM, Cavallo J-A, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers S, McCloy RA, Parker BL, Gallego-Ortega D, Law AMK, Chin VT, et al. MASTL overexpression promotes chromosome instability and metastasis in breast cancer. Oncogene 2018;37:4518–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buccitelli C, Salgueiro L, Rowald K, Sotillo R, Mardin BR, Korbel JO. Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation. Genome Res 2017;27:501–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature Med 2015;21:751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W, et al. Multi-clonal invasion in breast tumors identified by topographic single cell sequencing. Cell 2018;172:205–17.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, et al. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 2012;148:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Camps J, Morales C, Prat E, Ribas M, Capella G, Egozcue J, et al. Genetic evolution in colon cancer KM12 cells and metastatic derivates. Int J Cancer 2004;110:869–74. [DOI] [PubMed] [Google Scholar]
- 77.Linch M, Goh G, Hiley C, Shanmugabavan Y, McGranahan N, Rowan A, et al. Intratumoural evolutionary landscape of high-risk prostate cancer: the PROGENY study of genomic and immune parameters. Ann Oncol 2017;28:2472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, et al. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 2014;111:11139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Orsetti B, Selves J, Bascoul-Mollevi C, Lasorsa L, Gordien K, Bibeau F, et al. Impact of chromosomal instability on colorectal cancer progression and outcome. BMC Cancer 2014;14:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer 2006;45:31–41. [DOI] [PubMed] [Google Scholar]
- 81.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106:7131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waterman-Storer CM, Worthylake RA, Liu BP, Burridge K, Salmon ED. Microtubule growth activates Rac1 to promote lamellipodial protrusion in fibroblasts. Nature Cell Biol 1999;1:45–50. [DOI] [PubMed] [Google Scholar]
- 83.Parker AL, Kavallaris M, McCarroll JA. Microtubules and their role in cellular stress in cancer. Front Oncol 2014;4:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov 2010;9:790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Green RA, Kaplan KB.Chromosome instability in colorectal tumor cells is associated with defects in microtubule plus-end attachments caused by a dominant mutation in APC. The J Cell Biol 2003;163:949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 2012;13:528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao C, Su Y, Koeman J, Haak E, Dykema K, Essenberg C, et al. Chromosome instability drives phenotypic switching to metastasis. Proc Natl Acad Sci U S A 2016;113:14793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vargas-Rondon N, Villegas VE, Rondon-Lagos M. The role of chromosomal instability in cancer and therapeutic responses. Cancers 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McClelland SE.Role of chromosomal instability in cancer progression. Endocr Relat Cancer 2017;24:T23–t31. [DOI] [PubMed] [Google Scholar]
- 90.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep 2012;13:528–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet 2006;38:1043–8. [DOI] [PubMed] [Google Scholar]
- 92.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109–21. [DOI] [PubMed] [Google Scholar]
- 93.Zhang W, Mao JH, Zhu W, Jain AK, Liu K, Brown JB, et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun 2016;7:12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 2011;71:3447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huong PT, Nguyen LT, Nguyen X-B, Lee SK, Bach D-H. The role of platelets in the tumor-microenvironment and the drug resistance of cancer cells. Cancers 2019;11:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bach DH, Luu TT, Kim D, An YJ, Park S, Park HJ, et al. BMP4 upregulation is associated with acquired drug resistance and fatty acid metabolism in EGFR-mutant non-small-cell lung cancer cells. Mol Therapy Nucleic acids 2018;12:817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bach DH, Kim D, Bae SY, Kim WK, Hong JY, Lee HJ, et al. Targeting nicotinamide N-methyltransferase and miR-449a in EGFR-TKI-resistant non-small-cell lung cancer cells. Mol Therapy Nucleic Acids 2018;11:455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bach D-H, Lee SK, Sood AK. Circular RNAs in cancer. Mol Therapy Nucleic Acids 2019;16:118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bach DH, Park HJ, Lee SK. The dual role of bone morphogenetic proteins in cancer. Mol Therapy Oncolytics 2018;8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bach DH, Long NP, Luu TT, Anh NH, Kwon SW, Lee SK. The dominant role of Forkhead box proteins in cancer. Int J Mol Sci 2018;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer 2017;141:220–30. [DOI] [PubMed] [Google Scholar]
- 102.Bach DH, Lee SK. The potential impacts of tylophora alkaloids and their derivatives in modulating inflammation, viral infections, and cancer. Curr Med Chem 2018;25:1–16. [DOI] [PubMed] [Google Scholar]
- 103.Szymiczek A, Carbone M, Pastorino S, Napolitano A, Tanji M, Minaai M, et al. Inhibition of the spindle assembly checkpoint kinase Mps-1 as a novel therapeutic strategy in malignant mesothelioma. Oncogene 2017;36:6501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 2009;323:1350–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 2012;122:1138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Payton M, Bush TL, Chung G, Ziegler B, Eden P, McElroy P, et al. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res 2010;70:9846–54. [DOI] [PubMed] [Google Scholar]
- 107.Zhang W, Mao J-H, Zhu W, Jain AK, Liu K, Brown JB, et al. Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy. Nat Commun 2016;7:12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang W, Zhang Y, Chen R, Tian Z, Zhai Y, Janz S, et al. Chromosomal instability and acquired drug resistance in multiple myeloma. Oncotarget 2017;8:78234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schrijver W, Selenica P, Lee JY, Ng CKY, Burke KA, Piscuoglio S, et al. Mutation profiling of key cancer genes in primary breast cancers and their distant metastases. Cancer Res 2018;78:3112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fumagalli D, Wilson TR, Salgado R, Lu X, Yu J, O'Brien C, et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann Oncol 2016;27:1860–6. [DOI] [PubMed] [Google Scholar]
- 112.Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett AL, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 2018;128:1371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ng CKY, Bidard FC, Piscuoglio S, Geyer FC, Lim RS, de Bruijn I, et al. Genetic heterogeneity in therapy-naive synchronous primary breast cancers and their metastases. Clin Cancer Res 2017;23:4402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stover DG, Parsons HA, Ha G, Freeman SS, Barry WT, Guo H, et al. Association of cell-free DNA tumor fraction and somatic copy number alterations with survival in metastatic triple-negative breast cancer. J Clin Oncol 2018;36:543–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010;464:999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong MKH, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 2015;6:6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaem-manuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee SY, Haq F, Kim D, Jun C, Jo HJ, Ahn SM, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS ONE 2014;9:e90459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie T, Cho YB, Wang K, Huang D, Hong HK, Choi Y-L, et al. Patterns of somatic alterations between matched primary and metastatic colorectal tumors characterized by whole-genome sequencing. Genomics 2014;104:234–41. [DOI] [PubMed] [Google Scholar]
- 120.Tan IB, Malik S, Ramnarayanan K, McPherson JR, Ho DL, Suzuki Y, et al. High-depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver-limited metastatic colorectal cancer. Genome Biol 2015;16:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.