Abstract
Liquid–liquid phase separation is emerging as the universal mechanism by which membraneless cellular granules form. Despite many previous studies on condensation of intrinsically disordered proteins and low complexity domains, we lack understanding about the role of RNA, which is the essential component of all ribonucleoprotein (RNP) granules. RNA, as an anionic polymer, is inherently an excellent platform for achieving multivalency and can accommodate many RNA binding proteins. Recent findings have highlighted the diverse function of RNA in tuning phase-separation propensity up or down, altering viscoelastic properties and thereby driving immiscibility between different condensates. In addition to contributing to the biophysical properties of droplets, RNA is a functionally critical constituent that defines the identity of cellular condensates and controls the temporal and spatial distribution of specific RNP granules. In this review, we summarize what we have learned so far about such roles of RNA in the context of in vitro and in vivo studies.
Keywords: RNA, liquid–liquid phase separation, polymer, multivalency, RNA binding protein, aberrant aggregation
1. INTRODUCTION
Membrane-bound organelles are the hallmark of eukaryotic organisms. The ability to compartmentalize different cellular components and functions and to regulate the flow of molecular traffic between organelles is essential for survival. Although these organelles have long been appreciated as the primary means of cellular organization, the discovery of membraneless organelles has highlighted a new and ubiquitous mechanism through which cells can further compartmentalize critical functions, many of which require transient assembly and timely dissolution (13, 26). Membraneless organelles form by a process called liquid–liquid phase separation (LLPS) in which proteins and associated RNA molecules condense from the surrounding dilute phase, which in cells would be the cytoplasm or nucleoplasm (7, 73, 82).
LLPS occurs when multivalent biopolymers interact transiently to coalesce into a dense condensate (Figure 1a). LLPS is often promoted by high protein concentration and low salt concentration where the electrostatic, hydrophobic, π–cation, cation–cation, and π–π interactions among neighboring molecules are favorable (23). Numerous studies have demonstrated that proteins harboring intrinsically disordered regions (IDRs), also known as low-complexity domains (LCDs), are necessary and sufficient to drive LLPS in vitro and in cells (16, 58). In the cellular context, the IDR-containing proteins can interact with multiple binding partners—thus achieving multivalency (Figure 1b). Self-assembly of phase-separated condensates then becomes energetically favorable (82). The physical properties of these condensates, also referred to as droplets (in vitro) or granules (in vivo), are distinct from the surrounding environment. The viscoelastic properties of different condensates are tuned by the constituent molecules (1). Functionally, condensates can accelerate enzymatic activity of the phase-separated proteins and metabolites (7, 88). Accordingly, diverse functions within the cell, including splicing, transcription, micro RNA (miRNA) processing, heterochromatin formation, DNA damage repair, and the stress response, occur in the context of phase separated bodies (7, 17, 18, 20, 51, 74, 79, 85, 87, 99, 104). Importantly, many of these functions require RNA.
Figure 1.
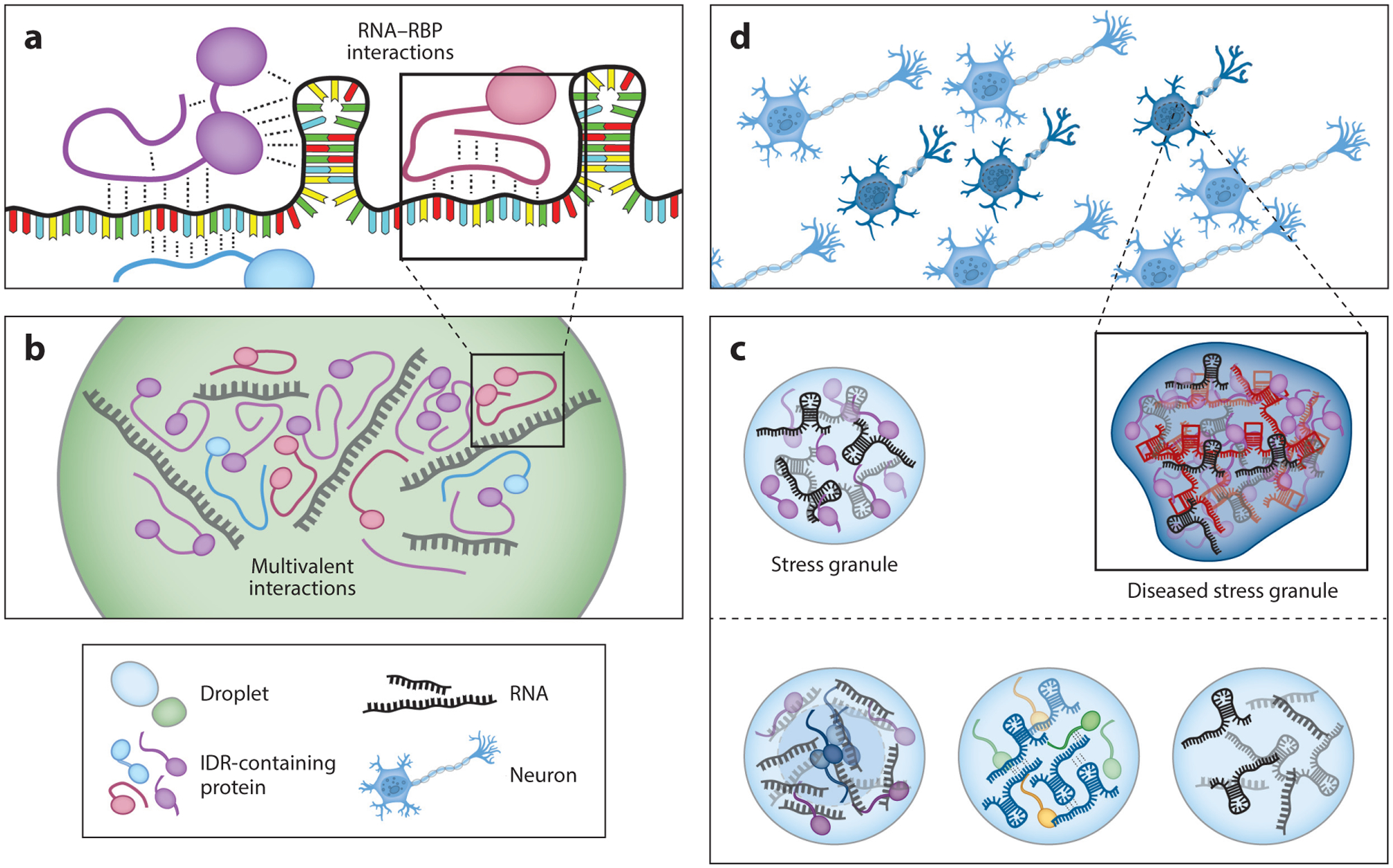
RNA droplets at every stage of formation and maturation. (a) RNA–RNA binding protein (RBP) contacts underlie the formation of ribonucleoprotein (RNP) complexes that contribute to droplet formation. Interactions are varied, and they can occur with any combination of structured and unstructured protein domains and RNA sequences. (b) Multivalent RNA–RBP interactions support the formation of phase-separated droplets. (c) Combination of different intrinsically disordered region (IDR) proteins and RNA allow for the formation of specific RNP granules with different cellular functions. (d) Accumulation of aberrant RNAs such as repeat expansion RNAs gives rise to aggregations via improper RNA–RNA and protein–RNA interactions that can lead to cellular toxicity and neurological disorders.
Many membraneless organelles are ribonucleoprotein (RNP) granules enriched with RNA and RNA binding proteins (RBPs). They are highly conserved across species and are found in the nucleus as well as the cytoplasm of cells (5). Despite the ubiquitous presence of the RNP granules, the constituents of different types of granules are vastly different, reflecting the selective recruitment of molecules according to their functional roles (64). One major focus in the phase separation field has been to determine the mechanism by which particular granules are nucleated and assembled into distinct functional condensates. Understanding how these granules form will give insight into how the cell organizes itself in a spatially, temporally, and functionally regulated manner (Figure 1c).
To date, the majority of studies have primarily focused on the role of proteins in driving LLPS; they have done this by performing deletion experiments of the IDR domain from RNP-forming proteins. When these IDR proteins are removed, reduction in granule assembly or aberrant formation has been reported (14, 27, 76, 77, 78, 80, 86). Interestingly, the majority, if not all, of these proteins are RBPs with high affinity for RNA, yet the role of RNA in RNP granule formation is far less well understood. It is well documented that RNA is required for the formation of certain RNP granules, for instance, NEAT1 in paraspeckles (21, 27). RNA plays a major role in seeding or nucleating the formation of RNP granules, such as the nucleolus, P granules, and stress granules (7, 23, 96) (Figure 1c). In this article, we review the importance of RNA in RNP granule assembly, identity, function, material property, and potential transition to a disease state (Figure 1d).
2. CHARGED POLYMERS ARE THE BUILDING BLOCKS OF LIQUID–LIQUID PHASE SEPARATION
Two-phase systems arise when a polymer-rich phase segregates from the polymer-deficient phase (15). This process, termed coacervation, results in colloidal droplets that have unique viscoelastic properties compared to the dilute, polymer-deficient phase (15). Phase separation can occur in one-component systems, such as with RNA (10, 38, 95), DNA (55), peptides (101), and full-length proteins alone (73, 99) (Figure 2). By contrast, oppositely charged polymers have long been shown to promote condensation (complex coacervation), as in the interaction between DNA and the polycation spermine, which is abundant in the nucleus (3, 9, 63, 90, 95).
Figure 2.
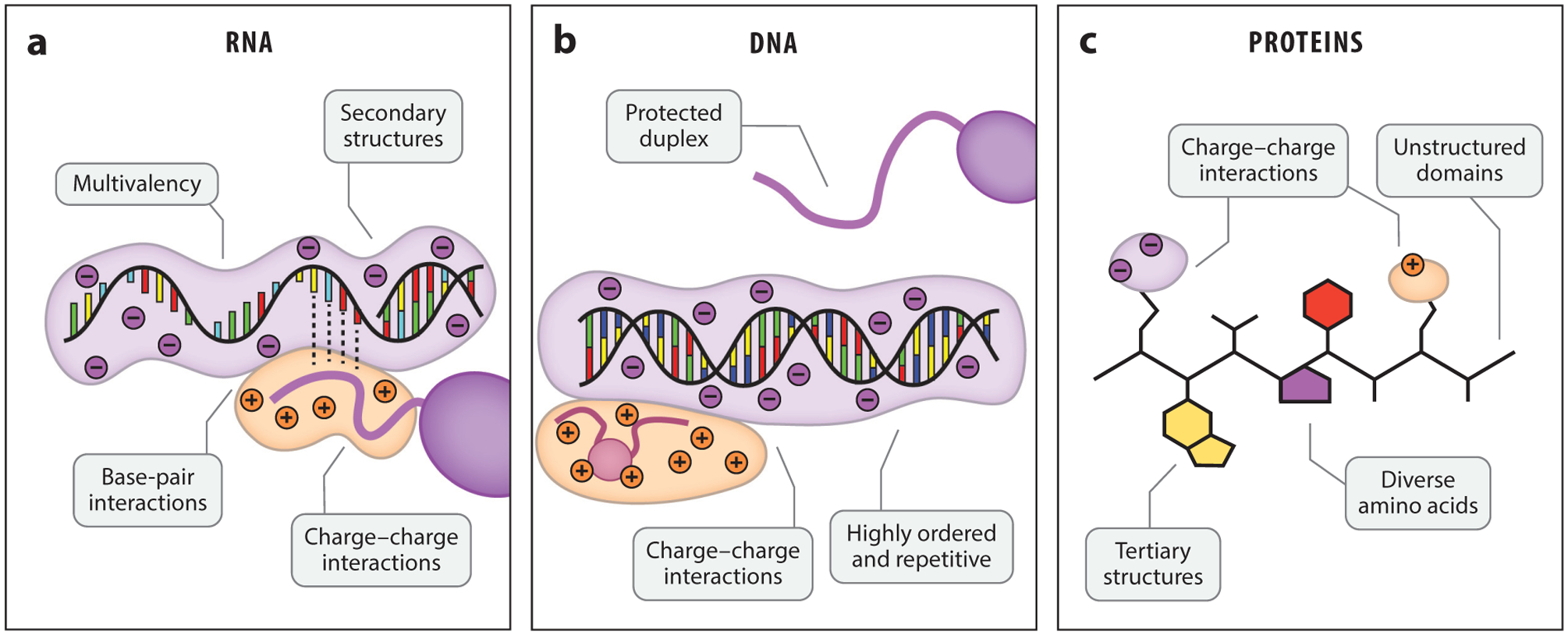
RNA is uniquely poised for liquid–liquid phase separation (LLPS). (a) Single-stranded RNA has a mixture of structured and unstructured regions, along with varied facets for charge–charge and other molecular interactions. (b) DNA has a protected duplex that limits interactions to the grooved or charged surface. (c) Proteins, like RNA, can be structured or unstructured and have varied opportunities to bind RNA.
With recent discoveries of LLPS in the cytosol and nucleus of cells (13, 26), the biopolymeric interactions that drive cellular phase-separation are now coming into focus. Proteins with LCDs and IDRs are well known to undergo phase separations through homotypic self-association (58, 73). LCDs are naturally LLPS prone, with π–π interactions between aromatic residues and non-covalent charged interactions driving phase separation (105). However, binding between polycationic protein domains (e.g., arginine- and lysine-rich motifs) and polyanionic nucleic acids can dramatically increase phase separation propensity through increased multivalency and favorable charge interactions (3). Proteins can engage in phase separation with both DNA (87) and RNA (58, 106), as well as unconventional polymers like poly(ADP-ribose) (50, 85). RNA is a uniquely potent polymer to control phase separation (Figure 2); RNA structure, sequence, and length are all parameters that can tune LLPS propensity and condensate properties, as discussed below.
2.1. RNA Innately Mimics Low-Complexity Domains
Unlike genomic DNA, the single-stranded nature of cellular RNA allows it to adopt many unique conformational structures that can promiscuously interact with many proteins (93) (Figure 2a). RNA can fold to form well-defined hairpins that are recognized by specific proteins such as MS2, tertiary structures within transfer RNAs that confer specificity to ribosome, and highly folded and enzymatically active ribozymes including Twister (8, 70, 72, 93). For example, the protein Whi3 (from the multinucleated fungus Ashbya gossypii) has a specific binding sequence found in the RNAs CLN3 and BNI1. These RNAs form secondary hairpin structures that also interact with Whi3 (47). In addition, the R motifs of the protein NPM1 specifically interact with ribosomal RNA (rRNA) to achieve multivalency and form the nucleolus (66). Small noncoding RNAs, including cis-acting riboswitches, can remain structured until base pairing with complementary DNA and RNA targets to control gene expressions in multiple cellular pathways (65, 67). Some messenger RNAs (mRNAs) are built with structured domains that reduce degradation and unstructured domains that engage in productive interactions (61).
In most unstructured cellular RNAs, both the phosphate backbone and RNA bases are exposed, enabling RNA binding partners to engage in many different specific and nonspecific interactions (57). Physiological salt conditions can stabilize charge–charge interactions between RNA and positively charged peptide sequences (6, 23, 52, 106). The physiological salt concentration is simultaneously low enough to permit protein–RNA binding but high enough to confer some selectivity to dedicated RNA binding domains (46, 56, 75). Certain RNA motifs can form additional structures, such as G-quadruplexes, which may help support LLPS (10, 28). Importantly, long RNA chains can bind multiple partners, and this multivalency is critical for LLPS (42). The ability to have numerous interactions of varying molecular specificity—and the flexibility to adopt many conformations—suggests that RNA mimics the properties of a protein’s LCD.
2.2. RNA–Protein Interaction Promotes Liquid–Liquid Phase Separation
Many RBPs harbor IDRs enriched with arginine, lysine, glycine, tyrosine, and proline (16, 71). These residues drive LLPS and control the fluidity of the resulting condensate. Accordingly, deletion of IDRs greatly dampens LLPS propensity (52, 58). Although the IDRs of RBPs are sufficient for LLPS, association with RNA can significantly promote LLPS, likely by enabling multimerization of RBPs (52).
RBP interactions with RNA can be specific and nonspecific (40, 76, 81). Many RBPs have distinct and structured RNA binding domains, such as RNA recognition motifs (RRMs) and zinc fingers (ZnFs) (16, 58), which can associate with RNA in the context of RNP granules (39, 106). Meanwhile, nonspecific contacts between positively charged arginine-glycine-glycine (RGG) domains, such as those found in FUS or PGL-3, and negatively charged RNA can strengthen the binding affinity of existing RNA binding domains and provide alternative interaction modes (23, 52, 76, 81, 105). Many classical RBPs are good candidates for phase separation given that RNA binding domains tend to be intrinsically disordered. This group of RBPs also includes the RNA helicases LAF1 and DDX-3, which have RecA-like domains that can interact with single-stranded RNA, as well as IDRs rich in RGG motifs (36, 44, 80). Above, we discuss Whi3, which also contains an RNA recognition motif, as well as a poly-Q IDR (106). The combination of protein subdomains and RNA parameters—such as structure, length, and sequence composition—can modulate phase separation.
Long RNAs can engage in multivalent protein interactions, which efficiently promotes LLPS. Most RBPs have RNA binding footprints of approximately 10–20 nucleotides (33). Therefore, the minimum size of RNA needed to enable multivalency is likely 20–40 nucleotides; smaller RNAs such as miRNAs and piwi-interacting RNAs (piRNAs) may not be able to seed phase separation independently (42, 95). Indeed, RNAs found in stress granules tend to skew longer in length (42, 81, 95). In vitro, longer RNA also more effectively phase separates into functional condensates (88). RNA concentration is also important for controlling LLPS. The high RNA-to-protein concentration ratio in the nucleus buffers phase separation of RBPs such as FUS and TAR DNA-binding protein 43 (TDP-43). By contrast, the low RNA-to-protein ratio leads to condensation of the same proteins in the cytoplasm (58). This indicates that both RNA concentration and length, which together define the total number of potential RNA binding sites, are key regulators of LLPS in cells.
2.3. RNA Can Tune the Fluidity of Ribonucleoprotein Liquid–Liquid Phase Separation
RNA properties can be altered depending on which ribonucleotides are enriched. Certain nucleotide sequences promote promiscuous RBP binding. For instance, FUS favors binding to uracil-rich sequences, though it does not have a strong sequence preference (82). By contrast, TDP-43 prefers structured RNAs (37). The poly-pyrimidine tracts (poly-uracil and poly-cytosine) tend to be more flexible due to the availability of only one aromatic ring for base stacking (3, 10, 95). These sequences may promote more dynamic RNA–protein interactions and more fluidic condensates. By contrast, poly-adenine sequences may promote stiffer interactions due to the bulky purine base, which presents two rings for stronger base stacking interactions with neighboring ribonucleotides (3, 10, 95). Poly-guanine tracts can fold into G-quadruplex structures, which are not recognized by most RBPs (10, 95). G-quadruplexes, however, may promote RNA–RNA contacts, contributing to RNA seeding (25). However, if G-quadruplexes do form within droplets, then they will likely dramatically reduce the fluidity of the resulting droplet (10).
Although many RBPs can bind and condense with unstructured or poorly structured RNAs, some RBPs require structured RNAs for binding. Secondary folds, such as hairpins, may sequester certain proteins into phase-separated droplets (47, 60, 107). For example, Whi3 binds differently shaped mRNAs to form biophysically (viscosity) and functionally distinct condensates, which become immiscible in vitro and in vivo (47, 48, 106). Disruption of the distinct RNA structure led to loss of specific Whi3–RNA interactions, resulting in miscibility between the two condensates (47). Therefore, long mRNAs encompassing both structured and unstructured regions may accommodate different types of RBPs in a multivalent manner. Tight binding with structured RNA elements may help form stable, solid-like droplet cores (see Section 3.2) (96). The multivalent nature of RNA makes it an ideal candidate for scaffolding and organizing LLPS. RNA in the form of homopolymers or highly repeated sequences, which are also pathogenic, can undergo LLPS or aggregation depending on the sequence composition and length (10, 38). Taken together, RNA is important for controlling phase separation and altering the properties of phase-separated condensates.
3. RNA CONTRIBUTES TO PHASE SEPARATION OF PROTEINS INTO CONDENSATES
Many cellular granules contain RNA. Indeed, RNA can play a pivotal role in condensing proteins into distinct phases, acting as a scaffold to increase the frequency of protein–protein contacts. When droplets form, they exhibit different properties from the dilute phase, including altered viscosity, fluidity, size, circularity, and elasticity. Concentrating RNA and RBPs in close proximity can trigger functional cellular responses including the DNA damage response, splicing, and transcription. In this section, we discuss how RNA seeds RBP-enriched droplets, and how RNA efficiently promotes the formation of substructures within biphasic droplets.
3.1. RNA Seeds and Accelerates Condensation of RNA-Binding Proteins
Many proteins can form phase-separated droplets at a high concentration threshold, referred to as the saturation concentration or Csat. Therefore, a higher Csat means that a higher protein concentration is needed to achieve phase separation (100). Addition of RNA can decrease Csat, likely through its multivalent scaffolding of RBPs (7, 23, 52, 58, 82, 106) (Figure 3a). Multiple RBPs can bind to a single RNA molecule, promoting oligomeric interactions (82). For instance, the DEAD-box ATPase Dhh1 multimerizes with RNA in the presence of Pat1, and FUS is known to oligomerize with long RNAs (80, 82). In agreement with these results, disabling RNA contacts can effectively prevent LLPS of RBPs, suggesting that RNA–protein interactions are critical for RNP condensation (69, 86). The multivalency enabled by long RNAs can permit phase separation of proteins that would not ordinarily phase separate at physiological concentrations. In repeat-expansion disorders, the longer length of the encoded RNA can increase binding multivalency and lead to disease (38). Another example of the importance of multivalency is FUS, which does not usually phase separate at its concentration in the cytoplasm, 2 μM (73, 100). However, addition of RNA can drive droplet formation at this low concentration. This may be because RNA–protein contacts can stimulate subsequent protein–protein, protein–RNP, or RNP–RNP contacts, thereby concentrating RBPs in close proximity (82). RNA is also critical for the maintenance of droplet structure; RNase treatment leads to loss of the two-phase system (58). The formation and dissolution of RNA-containing droplets can occur on the order of seconds to minutes, depending on the kinetics and binding strength of the protein–RNA interaction, and RNA can readily transit between phase-separated granules in cells (68, 102). RNA-containing granules often persist in cells during all stages of the cell cycle, except during M phase, in which these granules often disassemble and reassemble during the next G1 phase (21, 24). Although RBPs can often undergo phase separations independently, RNA is important for seeding and maintaining RNP droplet structure at physiological conditions.
Figure 3.
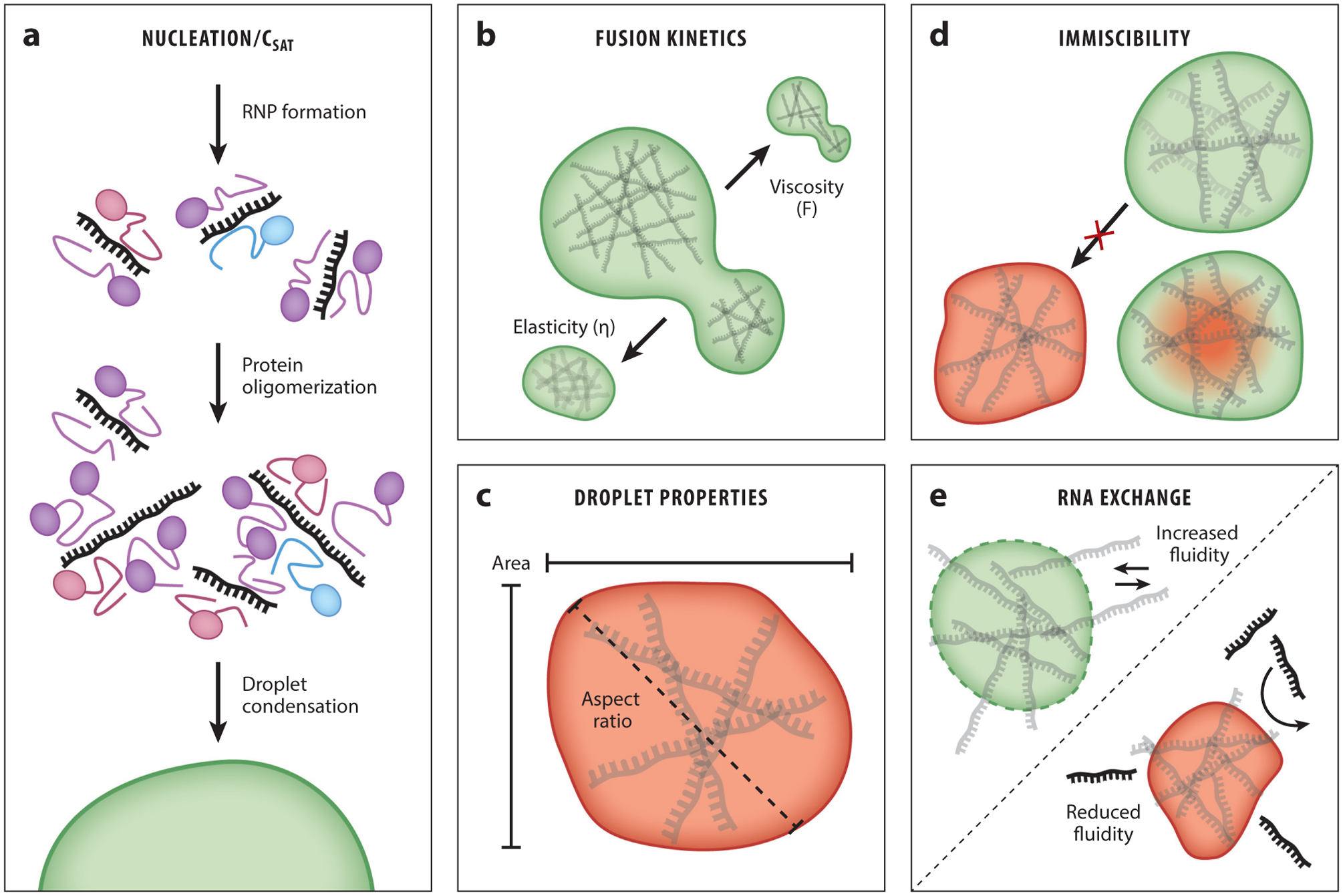
RNA controls the physical properties of phase-separated droplets. (a) RNA seeds droplet formation through multivalent contacts with RNA binding protein (RBP) partners. After additional RNA–RBP interactions, droplet condensation can occur. (b) The viscoelastic properties of droplets can be tuned by the RNAs incorporated into the droplet. These RNAs can affect the kinetics of the droplet fusion event (viscosity) or the return to a normal circular morphology (elasticity). (c) Droplet size and circularity (also referred to as the aspect ratio) can be controlled by RNA content. (d) Heterotypic RNA droplets can have widely divergent viscoelastic properties that prevent mixing (miscibility) or lead to formation of biphasic droplets. (e) RNA exchange rates with the surrounding environment can vary depending on the RNA contacts within the droplet and the viscoelastic properties of the droplet.
3.2. RNAs Alter the Physical Properties of Droplets
Phase-separated droplets exhibit physical properties that are distinct from the surrounding dilute phase, and these properties are determined by the constituent polymers of the droplet. Visual characteristics, such as droplet size and circularity, are easily influenced by protein concentration (1) (Figure 3c). Moreover, time can also influence the droplet properties. Aged droplets are more likely to be more solid like, especially as disordered proteins adopt conformations at the energetic minima of their folding potential energies (73). Increased droplet size and decreased circularity may result as phase separation reaches equilibrium with the dilute phase and ages. Importantly, such in vitro aging of droplets leads to changed properties that recapitulate the aberrant state found in age-dependent neurodegenerations such as amyotrophic lateral sclerosis (ALS) and frontotemporal lobar dementia (FTD), in which liquid-like RNP droplets are converted to solid-like fibrils in motor neurons (see Section 6) (73).
Because RNA alters the Csat of RBPs, RNA can also increase the observable droplet size—when more proteins can engage in LLPS, the area of the droplet increases (7, 106). Some RNAs have been observed to readily exchange between neighboring RBP granules in vivo (Figure 3e), implying that these condensates are dynamically interacting with the resident RNA (68). Similarly, addition of RNA in vitro increases droplet fluidity (6, 23, 58, 91, 106) and viscosity (58) (Figure 3b). In the case of the P granule protein LAF-1, the effect of RNA on droplet fluidity was measured by microrheology and fluorescence recovery after photobleaching (23). With both experimental methods, the addition of RNA increased the observed fluidity of LAF-1 condensates. RNA granules also may have biphasic partitioning between a solid-like core and a fluid-like shell, and highly structured RNAs may be important for scaffolding the least fluidic subcompartments of the droplet (52). Despite these findings, how RNA affects the liquid properties of phase-separated droplets is poorly understood.
3.3. RNAs Can Help Subcompartmentalize Phase-Separated Droplets
Although most droplets appear symmetric and devoid of internal substructures, phase separations can occur within droplets, driven by differing viscosities and surface tensions (26, 84) (Figure 3d). Biphasic droplets have been observed in vivo and in vitro, and they can be modeled in simulations (26, 31, 47). The functional roles of biphasic droplets remain unknown, but they may play a role in spatial organization of nuclear compartments, as shown in Drosophila melanogaster embryos (45). RNAs have been shown to direct biphasic partitioning of granules. As stated above, distinct and immiscible droplets can form using different RNAs that bind to specific RBPs; Whi3 can phase separate independently with differing RNAs (48, 106). However, the functional role of biphasic RNA droplets remains unknown.
3.4. RNAs May Disaggregate Droplets by Preventing Additional Protein–Protein Contacts
Based on the aberrant LLPS observed in pathological inclusions in neurodegeneration, recent efforts have focused on how to disaggregate solid-like droplets. Potent disaggregases that disentangle specific droplets do so through strong interactions with target RBPs that eject protein molecules from aggregated droplets. For example, FUS is disaggregated by the nuclear import receptor Karyopherin-β2 (Kapβ2), which sequesters the PY-NLS located in FUS’s C terminus and thereby dissolves FUS condensates (32, 35, 77, 105). Likewise, the potentiated Hsp104 disaggregates TDP-43 and other phase separation–prone proteins (89). Although ejection of the RBP from the condensed phase was a hallmark of these and other disaggregases, high-affinity RNAs or highly concentrated RNAs may also serve as disaggregases by sequestering RBPs and thereby buffering phase separation (58). For example, if a particular structure of RNA is able to trap the RBP and preclude additional protein–protein or protein–RNA interactions, then it would limit multivalency and thus lower the phase separation potential of the RNP complex (29). This may be an effective mechanism for reducing phase separation through treatment with otherwise inert RNAs.
3.5. RNAs Are a Fundamental Component of RNA-Binding Protein Droplets
As discussed above, many in vitro studies have demonstrated that RNAs have the powerful ability to concentrate RBPs and promote phase separation. They can lower Csat, control intradroplet substructures, and tune the physical properties of droplets. All of these roles constitute means through which the cell can compartmentalize biological functions without the permanence of membrane-bound organelles. In cells, RNA-containing condensates can be reversibly created and destroyed, and—as with protein expression—RNA acts as another layer of droplet regulation.
4. RNA NUCLEATES RIBONUCLEOPROTEIN GRANULE FORMATION IN VIVO
Cellular RNP granules are comprised of RNA and RBPs that together form a liquid-like complex to carry out a variety of processes in vivo. Both RNA and RBPs are necessary for the formation of RNP granules. The role of RNA in nucleating these bodies can vary. In many RNP granules, RNA serves as a scaffold for RNA–RNA and RNA–protein interactions to form these structures (Figure 4). These RNAs are often long, and some RNAs have expanded 3′ untranslated regions (UTRs) that allow for multivalent RNA–RNA and protein–RNA interactions (34). RNAs nucleate granules through RNA–RNA interactions and recruit RBPs to join these structures (52). Cellular granule formation can be promoted by RNA, as demonstrated in the nucleation of the nucleolus (7, 24, 66), P granules (23, 81, 86), bacterial RNP (BR) bodies of α-proteobacteria (2), processing bodies (PBs) (14, 22, 30, 54, 74, 78, 80, 83, 92), and stress granules (41–43, 96).
Figure 4.
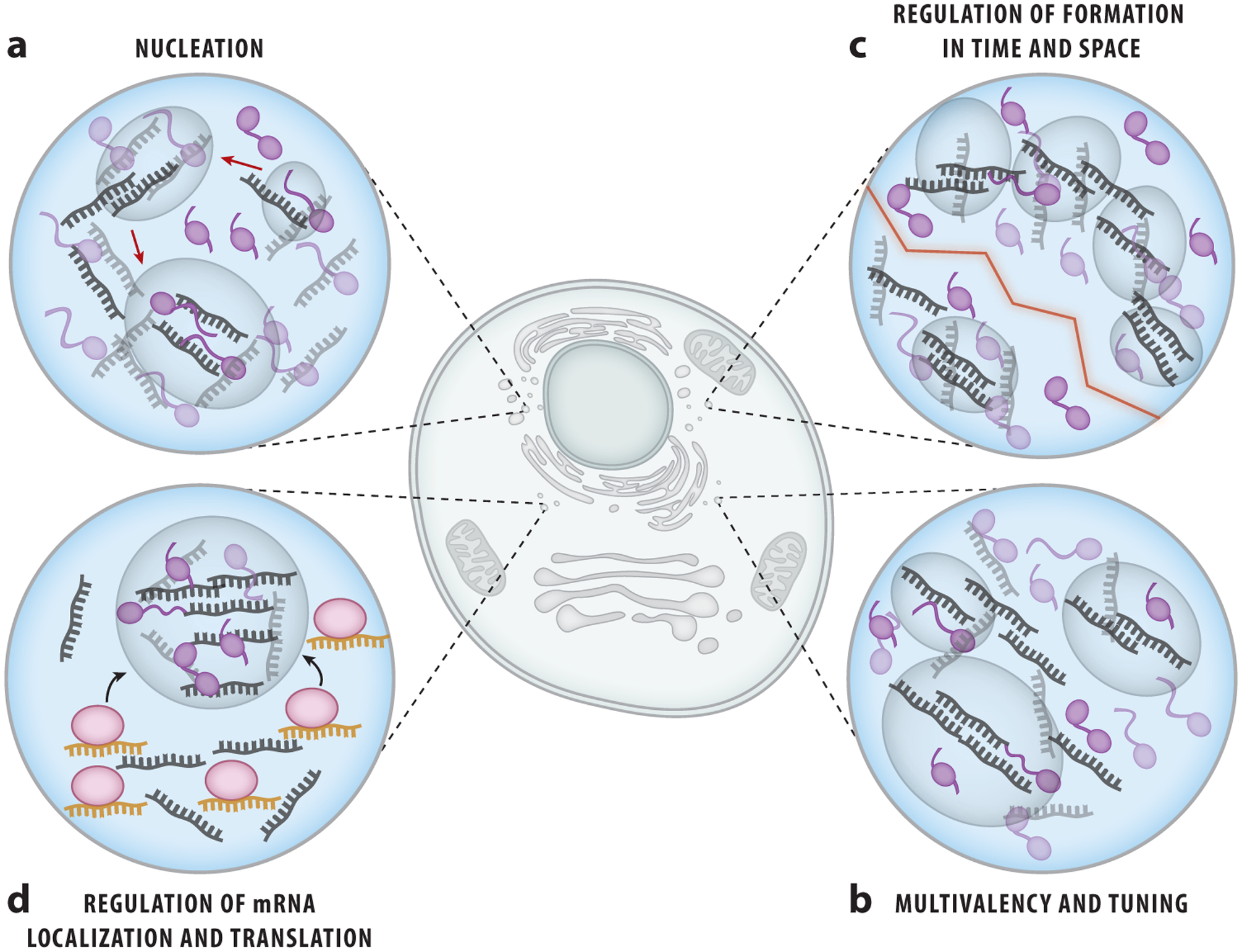
RNA contributes to ribonucleoprotein (RNP) granule formation and function. RNA can assist in the formation of RNP granules in many different ways. (a) RNA serves as a scaffold for RNA–RNA and protein–RNA interactions to seed the formation of RNP granules. (b) Specific RNAs can tune the elasticity or fluidity of RNP granules, forming distinct RNP granules. (c) The targeted localization of RNAs can control the spatial and temporal formation of RNP granules. (d) RNAs such as micro RNAs and piwi-interacting RNAs can assist in the recruitment of messenger RNAs to RNP granules for translational control and proper cell specification and function.
In the nucleolus, rRNA is necessary for nucleation (7). Pre-rRNA recruits nucleolar proteins, and reduced transcription of rRNA leads to a delay in the formation of the nucleolus with variable assembly outcomes (24). Similarly, for P granules in Caenorhabditis elegans, long mRNA molecules associate with PGL-3 protein with low sequence specificity and promote phase separation. This is seen at physiological conditions in vitro (81). Since MEX-5 has higher affinity for mRNA than does PGL-3, the addition of MEX-5 inhibits granule assembly of PGL-3 and RNA, thereby effectively preventing the RNA–RNA and RNA–PGL-3 interactions required to form P granules (81). Thus, RNA can tune up or down the RNP condensation propensity in cells and may be critical for temporal and spatial regulation of cellular processes such as embryogenesis.
BR bodies are found in bacteria and form via interactions between positively charged patches of RNase E and negatively charged RNA. Subsequently, these condensates recruit proteins that function in mRNA decay pathway (2). PBs and stress granules both rely on the presence of RNA for nucleation and recruitment of proteins for assembly. In particular, PBs form by multimerization of DHH1, PAT1, and mRNA (78, 80, 92). The removal of RNA prevents PAT1 from promoting the formation of P bodies as multimerization with DHH1 is not supported without mRNA (78, 80, 92).
In certain nuclear bodies, specific RNAs are essential for formation. Assembly of paraspeckles requires NEAT1, a 4-kilobase-pair-long noncoding RNA (11, 19, 21, 27, 53, 61, 62). Accordingly, removal of NEAT1 by RNA interference completely eradicates paraspeckles, while the overexpression of NEAT1 leads to an increased abundance of paraspeckles. NEAT1 contains a self-complementary sequence that allows for intramolecular interaction, as well as intermolecular hybrids to create an interconnected lattice. In addition to nucleating paraspeckles, NEAT1 is also required for the recruitment of paraspeckle proteins PSP1 and p54 to the condensates (21, 27). Thus, certain RNP granules require specific RNAs to nucleate and recruit RBPs to further oligomerize and carry out their function.
RNA alone may be sufficient to form condensates, as demonstrated by repeat expansion RNAs that have been implicated in neurological disorders, including Huntington’s and ALS. In a long chain of repeated segments, such RNAs can fold up into secondary or tertiary structures and engage in multivalent base pairing that promotes RNA focus formation in vivo and in vitro. Longer repeats promote more foci, and the RNA foci may exhibit liquid-like properties in vivo, as revealed by FRAP analysis (4, 25, 38). This finding demonstrates the importance of RNA–RNA interactions in RNP granule formation. Interestingly, the increased level of free mRNA in the cytoplasm induces the formation of stress granules, while an increase in protein concentration leads to their dissolution, suggesting the RNA-to-protein ratio as the key parameter in stress granule formation (12, 96). RNA is often observed to serve as the initial seeding for granule formation (Figure 4a). Therefore, RNA can impact the temporal and spatial formation of granules, regulate the recruitment of specific proteins for oligomerization, and serve as the key element that defines specific RNP granules.
4.1. RNA Provides Multivalent Interactions and Tunes Ribonucleoprotein Granule Properties In Vivo
In addition to providing a nucleation platform for RNP granule formation, RNAs also play a role in tuning the biophysical properties of RNP granules. The addition of total yeast RNA to a nucleolar protein FIB-1 lowered the Csat for droplet formation, but it also led to accelerated coarsening of the droplets, suggesting a diverse role of RNA in phase separation dynamics (Figure 4a–b) (7). In contrast, LAF-1 droplets formed with RNA exhibited diminished viscosity and enhanced fluidity within the droplets, which points to a role of RNA as a fluidizer (23). In both cases, RNA altered the liquid-like dynamics within the droplets, reminiscent of the RNP granules formed in vivo. However, specific RNAs can also determine how liquid an RNP granule may be and create granules that are distinct from those formed by other RNAs. As we describe above, Whi3 form distinct granules with particular sets of mRNAs built for different cellular roles. CLN3 and BNI1 are the two mRNAs of different sequence and shape that condense with Whi3 into droplets with differing viscosity. Once formed, the CLN3 and BNI1 droplets become immiscible and physically segregated in the organism, indicating the critical function of RNA structure, which governs assembly of distinct granules in vivo (47, 106). In summary, RNA provides a scaffold to initiate the formation of many RNP granules in vivo and tune the characteristic liquid-like properties of the cellular condensates (52, 76).
4.2. RNA Regulates Timing and Spatial Formation of Ribonucleoprotein Granules
RNA functions in regulating the temporal and spatial formation of granules in vivo (Figure 4c). In the Whi3 system, the G1 cyclin mRNAs, BNI1 and SPA2, form complexes with Whi3 in a polarized manner, thereby localizing these transcripts for the translation of polarity-setting proteins to induce symmetry-breaking events for the proper cell growth (48) (Figure 4c–d). Additionally, in Drosophila, approximately 200 mRNAs are enriched in the germplasm and play roles in germ cell formation, specification, survival, and migrations (94). For the development of C. elegans, MEG-3’s access to RNA allows for the regulation of the timing and orientation of P granule formation. MEG-3 phase separates with the addition of RNA in vitro, but MEX-5 disperses MEG-3–RNA condensate by sequestering RNA needed to seed P granule formation in vivo. This mechanism allows for mRNA to act as the key regulator for P granule formation and dissolution in a space- and time-dependent manner (86). In addition, miRNAs play a role in targeting mRNAs to P bodies for directed mRNA decay (54, 74). Similarly, piRNAs that are in complex with Aub have been shown to localize to germ granules and use partial base pairing to bind mRNAs and recruit them into the germplasm (98). This suggests a mechanism that allows for localization of certain mRNAs to the posterior for translational control and thus effective germ cell fate specification and function.
5. MUTATIONS AND ABNORMAL ACCUMULATION OF RNA LEAD TO TOXICITY AND NEURODEGENERATIVE DISEASE
Disease-linked RNAs, such as G4C2 repeat expansion RNAs, can lead to an aberrant phase separation stimulated by unusual RNA structures that associate and aggregate with neighboring proteins. These pathogenic RNA granules can drive cellular toxicity and diseases including ALS and FTD (Figure 5). The addition of G4C2 RNA to cells promotes the formation of stress granules in a repeat length–dependent manner (25). The connection between repeat RNA focus formation and cellular toxicity was demonstrated by nuclear retention of pathogenic 38x and 72x G4C2 repeats, which caused apoptotic cell death (49).
Figure 5.
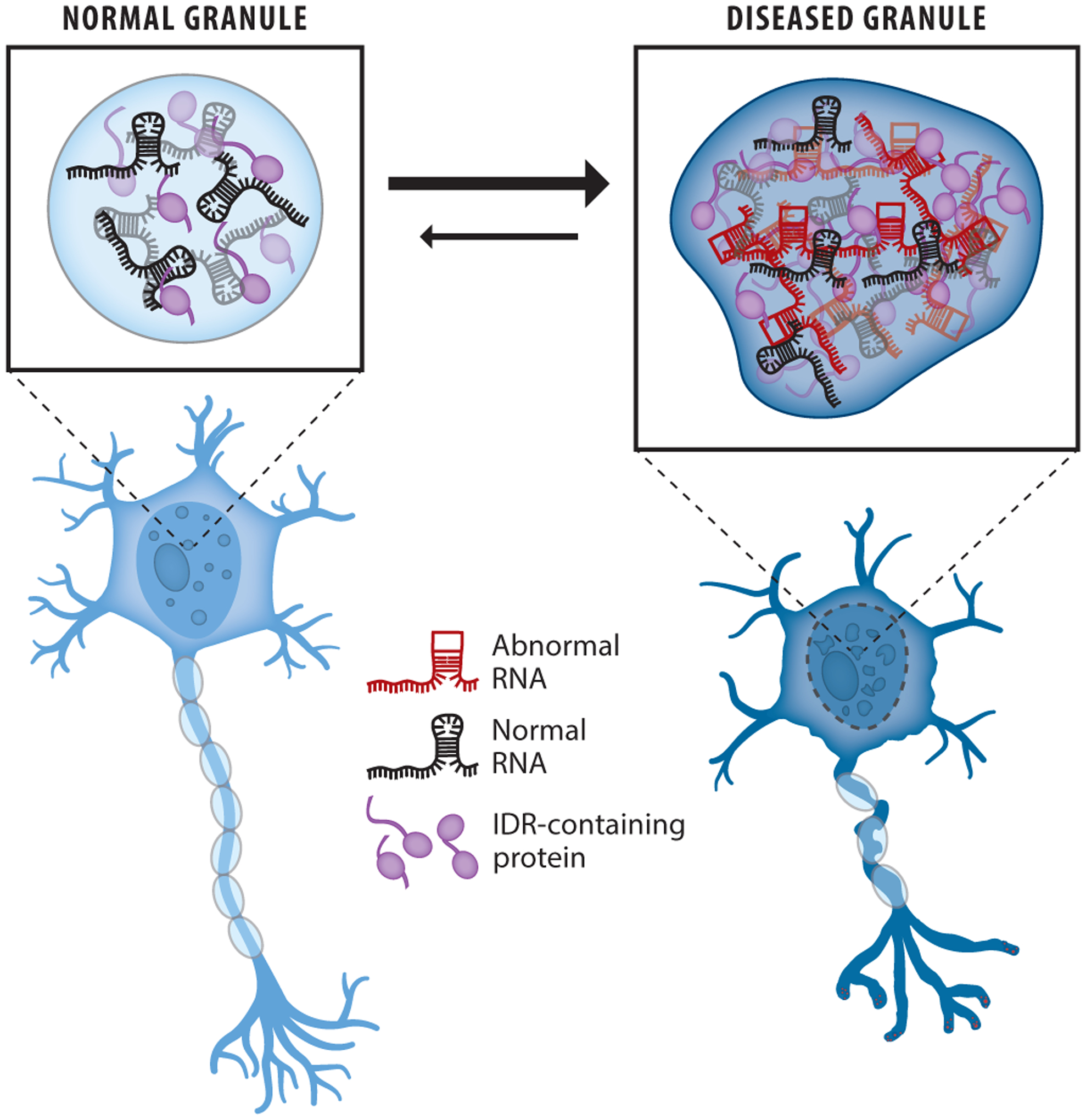
Role of RNA in aberrant phase separation. Expansion repeat RNAs with abnormal structures can cause accumulation and aggregation of RNA and intrinsically disordered proteins in RNP granules to promote cellular toxicity and neurological disease. Abbreviations: IDR, intrinsically disordered region; RNP, ribonucleoprotein.
In the case of myotonic dystrophies, the repeat expansion of CTG in the 3′ UTR of the distrophia myotonica protein kinase (DMPK) gene leads to a long repeat of CUG RNAs that are retained in the nucleus as a condensed focus (103). This nuclear accumulation of mutant RNA leads to the pathogenesis of both myotonic dystrophy types 1 and 2 (59, 97). Taken together, expansion of repeat RNA (both tri- and hexanucleotides) leads to the improper accumulation and aggregation of RNA in the cytoplasm and nucleus, which subsequently generates cellular toxicity and development of neurodegenerative diseases.
6. RNA CAN SEQUESTER RNA-BINDING PROTEINS TO PREVENT AGGREGATION FORMATION
The ratio of RNA to protein is critical for the formation of phase-separated droplets in vitro as well as in vivo. The high RNA concentration in the nucleus buffers phase separation of FUS-like RNA binding proteins (58). TDP-43 is an RNA binding protein that forms cytoplasmic inclusions and causes neurotoxicity implicated in ALS and FTD. Adding RNAs with the TDP-43 cognate sequence (Clip_34nt, rich in UG motifs) showed a loss of cytoplasmic assemblies in cells expressing optoTDP43, suggesting that RNA can rescue the formation of aberrant phase transitions and neurotoxicity. This may arise from the RNA, which buffers condensation by sequestering TDP-43 (60).
7. DISCUSSION
The role of RNA in the formation and the maintenance of RNP granules has been underappreciated. In this review, we show that RNA plays a major role in nucleating and assembling RNP droplets in vitro and in vivo via multivalent interactions (Figure 1). RNA may also serve to tune the different biophysical properties of condensates, including size, circularity, viscosity, elasticity, and surface tension. RNA can help regulate the timing and spatial formation of cellular granules by controlling the nucleation and properties of the resulting condensate. Importantly, RNAs are a functional entity of granules. RNAs can also drive liquid-like granules to an aggregated state to induce neurodegeneration. In particular, RNAs with repeat sequences can accumulate and create aberrant condensation in both the nucleus and cytoplasm that leads to plaque formation and neurodegeneration. In contrast, RNAs can also serve to fluidize RNP droplets. Physiologically, RNA-to-protein ratio is the key parameter throughout the cell, as the low and high ratios tune up and down the phase separation propensity, respectively. Therefore, RNAs are key players in cellular granule assembly and have many functions that will need to be further characterized moving forward.
7.1. The Molecular Grammar of RNA Structure and Phase Separation
The protein domains that contribute to phase separation have been extensively studied. Recently, certain amino acids have been implicated in controlling the physical properties of phase-separated condensates, revealing clues about the molecular grammar of disordered proteins (100, 105). How RNA sequence and structure modulate these properties is not understood. Above, we discuss several examples of proteins that display an RNA substrate specificity and different RNA shapes that drive condensates of varying viscosity and immiscibility (47, 106). Long and structured RNA may be associated with the stable core rather than the dynamic shell of stress granules. Meanwhile, unstructured RNAs that can engage in more promiscuous interactions may better support multi-valency. In addition, there is a wide spectrum of RNA structural diversity that exists in dynamic equilibrium, but little evidence exists to demonstrate which structures may promote phase separation more effectively. Whereas a molecular grammar of amino acid composition in a subset of RNA binding proteins has been linked with droplet properties, the molecular grammar that links RNA structure to phase separation remains unsolved. Each disordered RBP has its own compendium of RNA binding specificities, but we have yet to identify how RNA folding affects phase separation of even one of these RBPs. The combinatorial effect of various structured and unstructured elements on a single RNA polymer is also unknown.
Importantly, certain RNA structures may preclude multivalency, such as GU-rich RNA sequences that inhibit TDP-43 oligomerization (29). If multivalency is inhibited, phase-separation propensity is drastically reduced (42). Therefore, a better understanding of how RNA structure promotes, inhibits, and tunes phase separation is critically important for designing RNA molecules that may act as potential disaggregases, for example. Because aberrant phase separation of certain RBPs—such as FUS and TDP-43—is linked with neurodegeneration, developing disaggregases of these proteins is important for treating patients. Nuclear import receptors are known to potently disaggregate these proteins (32), but overexpression of importins may lead to adverse effects in patients. By contrast, a designer RNA that specifically binds a certain RBP and can inhibit phase separation may be a potential therapeutic approach for neurodegenerative diseases such as ALS and FTD. Therefore, clearly linking RNA structure with phase separation is an important emerging area of study in the coming years.
7.2. RNA and Granule Function In Vivo
Many recent studies report on phase-separated granules that function in central biological processes including embryogenesis, epigenetic regulation, cell differentiation, signaling, replication, transcription, and translation. Further studies are required to fully understand RNP granules in vivo; these studies should encompass not only RBPs, but also RNAs and other polymers such as poly(ADP-ribose) that are generally found in these bodies. The understanding of RNA in the nucleation of these bodies and tuning of condensate properties demands molecular probing at the level of single RNA and single protein molecules. In addition, understanding the role of RNA–RNA interactions that can potentially seed inherently different condensates is critical, especially for disease-linked aberrant aggregates. As it did when performed on stress granules, more transcriptomic analysis of the components of RNP granules will inform us about the types of RNA that constitute the RNP granules. Finally, it will be critical to understand the normal function of RNA and RBPs in the formation and cellular activity of RNP granules to determine how normal RNP assembly can become pathological RNP granule assembly. Understanding how aberrant RNAs nucleate disparate assembly of RNA and RBPs to induce neurotoxicity will be important for our fundamental understanding of neurodegenerative diseases such as ALS, FTD, and myotonic dystrophies and may assist in developing therapies for treating these incurable diseases.
SUMMARY POINTS.
Charge–charge and other molecular binding modes allow RNA to form multivalent interactions with proteins.
Phase-separated droplet properties such as viscosity, elasticity, size, area, and fluidity are tuned by constituent RNA molecules.
In vivo granule formation is accelerated with RNA via RNA–RBP contacts.
Mutations and abnormal accumulation of RNA, especially repeat-expansion RNAs, can lead to toxicity and neurodegenerative diseases such as ALS and FTD.
ACKNOWLEDGMENTS
Our research in RNP phase separation is supported by funding from the National Institute of Neurological Disorders and Stroke, grant 1RF1NS113636, to K.R., V.V., and S.M. We also acknowledge the support from grants 1F31GM134641-01 to V.V. and 5T32GM007231-45 to K.R. and V.V.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alberti S, Gladfelter A, Mittag T. 2019. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176:419–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Husini N, Tomares DT, Bitar O, Childers WS, Schrader JM. 2018. α-Proteobacterial RNA degradosomes assemble liquid-liquid phase-separated RNP bodies. Mol. Cell 71:1027–39.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aumiller WM Jr., Keating CD. 2016. Phosphorylation-mediated RNA/peptide complex coacervation as a model for intracellular liquid organelles. Nat. Chem 8:129–37 [DOI] [PubMed] [Google Scholar]
- 4.Aumiller WM Jr., Pir Cakmak F, Davis BW, Keating CD. 2016. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir 32:10042–53 [DOI] [PubMed] [Google Scholar]
- 5.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, et al. 2016. Compositional control of phase-separated cellular bodies. Cell 166:651–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee PR, Milin AN, Moosa MM, Onuchic PL, Deniz AA. 2017. Reentrant phase transition drives dynamic substructure formation in ribonucleoprotein droplets. Angew. Chem. Int. Ed. Engl 56:11354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. 2015. RNA transcription modulates phase transition-driven nuclear body assembly. PNAS 112:E5237–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437–45 [DOI] [PubMed] [Google Scholar]
- 9.Bloomfield VA. 1997. DNA condensation by multivalent cations. Biopolymers 44:269–82 [DOI] [PubMed] [Google Scholar]
- 10.Boeynaems S, Holehouse AS, Weinhardt V, Kovacs D, Van Lindt J, et al. 2019. Spontaneous driving forces give rise to protein–RNA condensates with coexisting phases and complex material properties. PNAS 116:7889–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond CS, Fox AH. 2009. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol 186:637–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, et al. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42:8678–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, et al. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324:1729–32 [DOI] [PubMed] [Google Scholar]
- 14.Buchan JR, Muhlrad D, Parker R. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol 183:441–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bungenberg de Jong HG, Kruyt HR. 1929. Coacervation (partial miscibility in colloid systems). Proc. R. Acad. Amsterdam 33:849–56 [Google Scholar]
- 16.Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149:1393–406 [DOI] [PubMed] [Google Scholar]
- 17.Cho WK, Spille JH, Hecht M, Lee C, Li C, et al. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361:412–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, et al. 2018. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361:eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chujo T, Hirose T. 2017. Nuclear bodies built on architectural long noncoding RNAs: unifying principles of their construction and function. Mol. Cells 40:889–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cid-Samper F, Gelabert-Baldrich M, Lang B, Lorenzo-Gotor N, Ponti RD, et al. 2018. An integrative study of protein-RNA condensates identifies scaffolding RNAs and reveals players in fragile X-associated tremor/ataxia syndrome. Cell Rep. 25:3422–34.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, et al. 2009. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33:717–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cougot N, Babajko S, Séraphin B. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol 165:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, et al. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. PNAS 112:7189–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. 2016. Nucleation by rRNA dictates the precision of nucleolus assembly. Curr. Biol 26:277–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fay MM, Anderson PJ, Ivanov P. 2017. ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 21:3573–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, et al. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165:1686–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fox AH, Bond CS, Lamond AI. 2005. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 16:5304–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, et al. 2012. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep 2:1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.French RL, Grese ZR, Aligireddy H, Dhavale DD, Reeb AN, et al. 2019. Detection of TAR DNA-binding protein 43 (TDP-43) oligomers as initial intermediate species during aggregate formation. J. Biol. Chem 294:6696–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fromm SA, Kamenz J, Nöldeke ER, Neu A, Zocher G, Sprangers R. 2014. In vitro reconstitution of a cellular phase-transition process that involves the mRNA decapping machinery. Angew. Chem. Int. Ed. Engl 53:7354–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasior K, Zhao J, McLaughlin G, Forest MG, Gladfelter AS, Newby J. 2019. Partial demixing of RNA-protein complexes leads to intradroplet patterning in phase-separated biological condensates. Phys. Rev. E 99:012411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, et al. 2018. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173:677–92.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, et al. 2010. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp 2:2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, et al. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149:768–79 [DOI] [PubMed] [Google Scholar]
- 35.Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, et al. 2018. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173:706–19.e13 [DOI] [PubMed] [Google Scholar]
- 36.Hondele M, Sachdev R, Heinrich S, Wang J, Vallotton P, et al. 2019. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 573:144–48 [DOI] [PubMed] [Google Scholar]
- 37.Ishiguro T, Sato N, Ueyama M, Fujikake N, Sellier C, et al. 2017. Regulatory role of RNA chaperone TDP-43 for RNA misfolding and repeat-associated translation in SCA31. Neuron 94:108–24.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546:243–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin L, Zhang K, Xu Y, Sternglanz R, Neiman AM. 2015. Sequestration of mRNAs modulates the timing of translation during meiosis in budding yeast. Mol. Cell Biol 35:3448–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin M, Fuller GG, Han T, Yao Y, Alessi AF, et al. 2017. Glycolytic enzymes coalesce in G bodies under hypoxic stress. Cell Rep. 20:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, et al. 2000. Dynamic of shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol 151:1257–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68:808–20.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khong A, Parker R. 2018. mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compation. J. Cell Biol 217:4124–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y, Myong S. 2016. RNA remodeling activity of DEAD-box proteins tuned by protein concentration, RNA length and ATP. Mol. Cell 63:865–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kistler KE, Trcek T, Hurd TR, Chen R, Liang FX, et al. 2018. Phase transitioned nuclear Oskar promotes cell division of Drosophila primordial germ cells. eLife 7:e37949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laing LG, Gluick TC, Draper DE. 1994. Stabilization of RNA structure by Mg ions: specific and nonspecific effects. J. Mol. Biol 237:577–87 [DOI] [PubMed] [Google Scholar]
- 47.Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, et al. 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360:922–27 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper finds that RNA secondary structure controls entry into and exclusion from droplets.
- 48.Lee C, Occhipinti P, Gladfelter AS. 2015. PolyQ-dependent RNA-protein assemblies control symmetry breaking. J. Cell Biol 208:533–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y-B, Chen H-J, Peres JN, Gomez-Deza J, Attig J, et al. 2013. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 5:1178–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leung AK. 2014. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol 205:613–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42:489–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Y, Protter DS, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60:208–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Y, Schmidt BF, Bruchez MP, McManus CJ. 2018. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 46:3742–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol 7:719–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu K, Shuai M, Chen D, Tuchband M, Gerasimov JY, et al. 2015. Solvent-free liquid crystals and liquids from DNA. Chemistry 21:4898–903 [DOI] [PubMed] [Google Scholar]
- 56.Lohman TM, DeHaseth PL, Record MT Jr. 1978. Analysis of ion concentration effects on the kinetics of protein–nucleic acid interactions: application to Lac repressor–operator interactions. Biophys. Chem 8:281–94 [DOI] [PubMed] [Google Scholar]
- 57.Lunde BM, Moore C, Varani G. 2007. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol 8:479–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, et al. 2018. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360:918–21 [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper highlights how concentrated RNA in the nucleus can buffer LLPS.
- 59.Mankodi A, Urbinati CR, Yuan Q-P, Moxley RT, Sansone V, et al. 2001. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet 10:2165–70 [DOI] [PubMed] [Google Scholar]
- 60.Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, et al. 2019. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102:321–38.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao Y, Liu H, Liu Y, Tao S. 2014. Deciphering the rules by which dynamics of mRNA secondary structure affect translation efficiency in Saccharomyces cerevisiae. Nucleic Acids Res. 42:4813–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mao YS, Sunwoo H, Zhang B, Spector DL. 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol 13:95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marianelli AM, Miller BM, Keating CD. 2018. Impact of macromolecular crowding on RNA/spermine complex coacervation and oligonucleotide compartmentalization. Soft Matter 14:368–78 [DOI] [PubMed] [Google Scholar]
- 64.Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, et al. 2018. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell 172:590–604.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mattick JS. 2001. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2:986–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mitrea DM, Cika JA, Guy CS, Ban D, Banerjee PR, et al. 2016. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 5:e13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montange RK, Batey RT. 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441:1172–75 [DOI] [PubMed] [Google Scholar]
- 68.Moon SL, Morisaki T, Khong A, Lyon K, Parker R, Stasevich TJ. 2019. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol 21:162–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niewidok B, Igaev M, Pereira da Graca A, Strassner A, Lenzen C, et al. 2018. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol 217:1303–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–30 [DOI] [PubMed] [Google Scholar]
- 71.Oldfield CJ, Dunker AK. 2014. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem 83:553–84 [DOI] [PubMed] [Google Scholar]
- 72.Panja S, Hua B, Zegarra D, Ha T, Woodson SA. 2017. Metals induce transient folding and activation of the Twister ribozyme. Nat. Chem. Biol 13:1109–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, et al. 2015. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162:1066–77 [DOI] [PubMed] [Google Scholar]
- 74.Pitchiaya S, Mourao MDA, Jalihal AP, Xiao L, Jiang X, et al. 2019. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol. Cell 74:521–33.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Printz MP, von Hippel PH. 1968. On the kinetics of hydrogen exchange in deoxyribonucleic acid: pH and salt effects. Biochemistry 7:3194–206 [DOI] [PubMed] [Google Scholar]
- 76.Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, et al. 2018. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 22:1401–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, et al. 2018. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173:720–34.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rao BS, Parker R. 2017. Numerous interactions act redundantly to assemble a tunable size of P bodies in Saccharomyces cerevisiae. PNAS 114:E9569–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, et al. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sachdev R, Hondele M, Linsenmeier M, Vallotton P, Mugler CF, et al. 2019. Pat1 promotes processing body assembly by enhancing the phase separation of the DEAD-box ATPase Dhh1 and RNA. eLife 8:e41415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, et al. 2016. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell 166:1572–84.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schwartz JC, Wang X, Podell ER, Cech TR. 2013. RNA seeds higher-order assembly of FUS protein. Cell Rep. 5:918–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shiina N 2019. Liquid- and solid-like RNA granules form through specific scaffold proteins and combine into biphasic granules. J. Biol. Chem 294:3532–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, et al. 2019. PARP-1 activation directs FUS to DNA damage sites to form PARG-reversible compartments enriched in damaged DNA. Cell Rep. 27:1809–21.e5 [DOI] [PubMed] [Google Scholar]
- 86.Smith J, Calidas D, Schmidt H, Lu T, Rasoloson D, Seydoux G. 2016. Spatial patterning of P granules by RNA-induced phase separation of the intrinsically-disordered protein MEG-3. eLife 5:e21337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. 2017. Phase separation drives heterochromatin domain formation. Nature 547:241–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. 2012. RNA catalysis through compartmentalization. Nat. Chem 4:941–46 [DOI] [PubMed] [Google Scholar]
- 89.Sweeny EA, Jackrel ME, Go MS, Sochor MA, Razzo BM, et al. 2015. The Hsp104 N-terminal domain enables disaggregase plasticity and potentiation. Mol. Cell 57:836–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tabor H 1962. The protective effect of spermine and other polyamines against heat denaturation of deoxyribonucleic acid. Biochemistry 1:496–501 [DOI] [PubMed] [Google Scholar]
- 91.Taylor N, Elbaum-Garfinkle S, Vaidya N, Zhang H, Stone HA, Brangwynne CP. 2016. Biophysical characterization of organelle-based RNA/protein liquid phases using microfluidics. Soft Matter 12:9142–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tinoco I Jr., Bustamante C. 1999. How RNA folds. J. Mol. Biol 293:271–81 [DOI] [PubMed] [Google Scholar]
- 94.Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. 2015. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun 6:7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Van Treeck B, Parker R. 2018. Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174:791–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. PNAS 115:2734–39 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors report that RNA–RNA contacts may help condense stress granules in yeast.
- 97.Vogler TO, Wheeler JR, Nguyen ED, Hughes MP, Britson KA, et al. 2018. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563:508–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vourekas A, Alexiou P, Vrettos N, Maragkakis M, Mourelatos Z. 2016. Sequence-dependent but not sequence-specific piRNA adhesion traps mRNAs to the germ plasm. Nature 531:390–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, et al. 2018. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 37:e97452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, et al. 2018. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174:688–99.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors’ mapping of FUS’s LLPS grammar may serve as a roadmap for a similar effort with RNA.
- 101.Wang Y, Lomakin A, Kanai S, Alex R, Benedek GB. 2017. Liquid-liquid phase separation in oligomeric peptide solutions. Langmuir 33:7715–21 [DOI] [PubMed] [Google Scholar]
- 102.Wilbertz JH, Voigt F, Horvathova I, Roth G, Zhan Y, Chao JA. 2019. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell 73:946–58.e7 [DOI] [PubMed] [Google Scholar]
- 103.Wojciechowska M, Krzyzosiak WJ. 2011. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet 20:3811–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang L, Gal J, Chen J, Zhu H. 2014. Self-assembled FUS binds active chromatin and regulates gene transcription. PNAS 111:17809–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, et al. 2018. Nuclear import receptor inhibits phase separation of FUS through binding to multiple sites. Cell 173:693–705.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, et al. 2015. RNA controls polyQ protein phase transitions. Mol. Cell 60:220–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, et al. 2017. RNA stores tau reversibly in complex coacervates. PLOS Biol. 15:e2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]


