Abstract
Purpose
Most lung adenocarcinoma (LUAD) patients are diagnosed at the advanced stage and have poor prognosis. DNA methylation plays an important role in the prognosis prediction of cancers. The objective of this study was to identify new DNA methylation sites as biomarkers for LUAD prognosis.
Materials and Methods
We downloaded DNA methylation data from The Cancer Genome Atlas data portal. Cox proportional hazard regression model and random survival forest algorithm were applied to identify the DNA-methylation sites. Methylation of sites were validated in the Gene Expression Omnibus cohorts. Function annotation were done to explore the biological function of DNA methylated sites signature.
Results
Six DNA methylation sites were identified as prognosis signature. The signature yielded acceptable discrimination between the high-risk group and low-risk group. The discrimination effect of this DNA methylation signature for the OS was obvious, with a median OS of 21.89 months vs. 17.74 months for high-risk vs. low-risk groups. This prognostic prediction model was validated by the test group and GEO dataset. The predictive survival value was higher for the prognostic prediction model than that for the tumor node metastasis stage. Adjuvant hemotherapy could not affect the prediction of the signature. Functional analysis indicated that these signature genes were involved in protein binding and cytoplasm.
Conclusion
We identified the prognostic signature for LUAD by combining six DNA methylation sites. This could service as potential robust and specificity signature in the prognosis prediction of LUAD.
Keywords: Lung adenocarcinoma, prognosis signature, methylation, TCGA
INTRODUCTION
Cancer is a serious threat to human health. Previous study found that there were 18.1 million new cancer cases and 9.6 million cancer deaths worldwide in 2018, and it is estimated that nearly half of all new cancer cases and more than half of all cancer deaths occurred in Asia. Especially in China, the incidence of cancer is very serious. The incidence of cancer in China accounts for about 22% of the world's total, and the number of cancer cases there is the highest in the world.1 In recent years, lung cancer (LC) still remains one of the most common malignant tumors in human beings, and the incidence and mortality of LC are increasing every year, with a relative survival rate of about 25% in the 5 years following initial diagnosis.2,3 There are two main types of LC, small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC).4,5 However, lung adenocarcinoma (LUAD) is the main subtype of NSCLC, and its incidence is increasing. For decades, LUAD has been one of the most dangerous types of cancer in many countries.6 Previous studies have shown that patients with LUAD have a shorter survival period compared to patients with other NSCLC.7 As a common histological subtype of NSCLC, the clinical treatment of LUAD is mainly surgical resection; however, the prognosis of patients after treatment is poor, and the recurrence rate of patients after surgery is as high as 35–50%.8 Therefore, it is urgent to establish a biomarker model with high sensitivity and strong predictive power to elucidate the prognosis of LUAD.
Although the occurrence and development of cancer mainly depend on the changes of tumor-related genes, epigenetic changes such as DNA methylation of tumor-related genes play an important role in the development of tumors.9,10 DNA methylation is often considered a mechanism of gene silencing and plays an important role in many cellular processes, such as embryonic development, transcription, genomic imprinting, and x chromosome inactivation.11,12,13,14 DNA methylation biomarkers have been used in the early diagnosis and prognosis of cancer. For example, in breast cancer (BC), poor prognosis may be associated with CDH1 promoter methylation.15 In renal cell carcinoma, CRHBP, RCVRN, and DAB2IP have been identified as prognostic biomarkers of methylation.16,17,18 In addition, it has been found that some gene methylation changes in LUAD affect gene expression and finally affect the prognosis of LUAD patients.19,20
However, many previous studies on methylation in LUAD focused on the prognostic significance of monomethylated genes. Considering that the occurrence and development of LUAD is a complex process, which requires the co-regulation of multiple genes, it is necessary to establish a methylated biomarker model with high sensitivity and strong predictive power to predict the prognosis of LUAD. Therefore, in this study, the methylation data of LUAD in The Cancer Genome Atlas (TCGA) were mined to explore the methylation genes related to the prognosis of LUAD in the whole genome, laying a foundation for the development of new prognostic markers and therapeutic targets for LUAD.
MATERIALS AND METHODS
DNA methylation profiling of LUAD patients
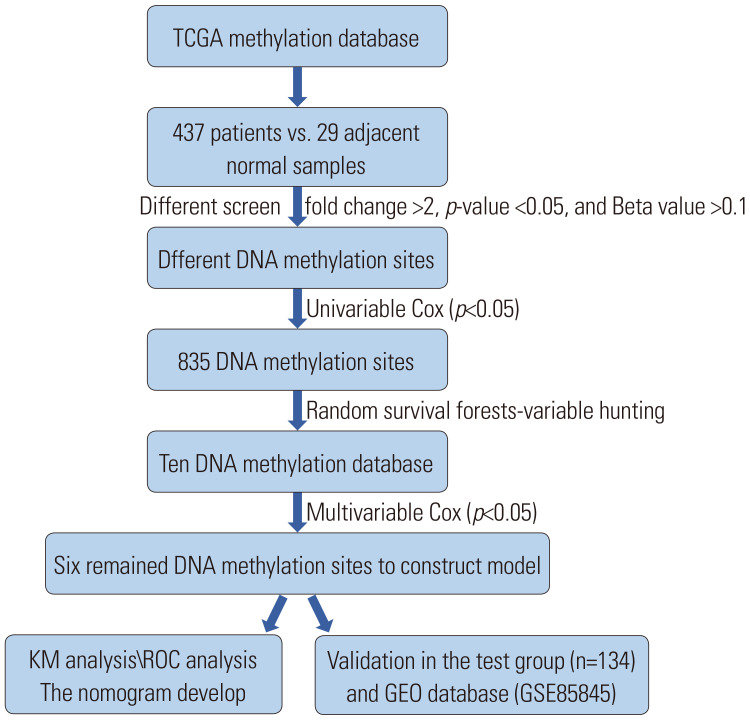
In the current study, DNA methylation profiling (Illumina Human Methylation 450K Bead Chip Array) and the following clinical data were downloaded from TCGA database. A total of 437 LUAD and 29 control specimens were enrolled in the methylation dataset, and clinical data included the survival time, status, gender, age, and clinical stage. To improve data accuracy, we preprocessed both of the two datasets. If the methylation level of a gene site is NA, which is more than 70% of the total samples, the locus is removed.21 The clinical information of methylation data are shown in Table 1. The technical route to select the DNA methylated sites signature is shown in Fig. 1. We download the GSE56044 (n=97) dataset of GEO database for signature independent validation.
Table 1. Summary of Patient Demographics and Characteristics.
| Characteristics | Training (n=267) | Test (n=134) |
|---|---|---|
| Gender | ||
| Female | 149 (56.2) | 71 (53.0) |
| Male | 118 (54.6) | 63 (47.0) |
| Age (yr) | ||
| <65 | 125 (47.2) | 63 (47.0) |
| ≥65 | 142 (52.9) | 71 (53.0) |
| Stage | ||
| I | 135 (50.9) | 86 (64.2) |
| II | 66 (24.9) | 29 (21.6) |
| III | 50 (18.9) | 14 (10.4) |
| IV | 14 (5.3) | 5 (3.8) |
| Not reported | 2 (0.7) | 0 (0) |
| Adjuvant chemotherapy | ||
| Adjuvant chemotherapy | 103 (38.5) | 68 (50.7) |
| None | 164 (61.5) | 66 (49.3) |
| Vital status | ||
| Living | 177 (66.4) | 90 (67.2) |
| Dead | 90 (33.6) | 44 (32.8) |
Data are presented as n (%).
Fig. 1. Flow chart of the study showing the analysis to develop the risk score model in training group and validate the efficiency of the model in test group. TCGA, The Cancer Genome Atlas; KM, Kaplan-Meier; ROC, receiver operating characteristic; GEO, Gene Expression Omnibus.
Identification of different methylated genes (DMGs) associated with LUAD
To identify the differentially methylation sites, we adopted the Benjamini-Hochberg false-discovery rate (FDR) method to adjusted the p value for each gene. The DMGs were identified by a fold change >2, p-value <0.05, and Beta value >0.1, respectively.
Constructing a prognostic DNA methylation signature in the training dataset
Some statistics were employed to build the model, which turned out to be a better method to construct biomarker module than the one we adopted.22,23 Thereafter, the LUAD samples were randomly divided into two groups, namely, the test group (n=134) and the training group (n=267). The two groups were unpaired and uncrossed. Univariate Cox proportional regression analysis was performed to determine the correlation between the OS and DNA methylation in the training group.24 As the model was more practical when the number of methylated genes was small, we used the random survival forests-variable hunting (RSFVH) algorithm to screen the methylation sites and identified 10 sites.25 To screen the predictive prognostic biomarker model, we used multivariate Cox regression analysis and created a model to assess the prognostic risk as follows:
In the formula above, N represent the number of methylation sites, Methi is the methylation value of the sites, and Coefi is a single factor Cox regression coefficient. Coefi < 0 was identified as a favorable prognosis site, and Coefi >0 was considered a poor prognosis site. RS is the multi-node weighted sum of the risk scores.
Statistical analysis
We adopted the median risk score as the cutoff value in the training group, and used it to divide the LUAD patients into low-risk and high-risk groups. Kaplan-Meier survival analysis was used to estimate the survival time and compare the survival curves of the two groups. The two-sided log-rank test was used to determine the statistical significance. For the test group, the prognostic value of the DNA methylation biomarker was validated using receiver operating characteristic (ROC) values and Kaplan-Meier survival analysis. To determine whether the DNA methylation biomarker was an independent factor, we assessed the biomarker using data stratification analysis and multivariate Cox regression analysis. p-values <0.05 were considered statistically significant. All analyses were performed with the R statistical program (version 3.5.1) using the survival, random Forest SRC, and pROC packages (downloaded from Bioconductor).
Generating the nomogram
We generated a nomogram by using the “RMS” package of R software. The nomogram concordance index (C-index) of all patients were obtained by multivariate Cox regression analysis. The higher the C-index, the more accurate the prediction. The nomogram was used to calculate the total score of each patient. Overall scores were then used to predict the 1-year, 3-year, and 5-year survival rates.26
Functional annotation of the selected DNA methylation biomarker genes
In order to further study the function of survival-related DNA methylation signature genes, we used gene ontology (GO) analysis (http://www.geneontology.org) to investigate the roles of all of the selected genes, and performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.genome.jp/kegg/) to determine the significant pathways. Fisher's exact test and chi-square tests were used to select significant GO and pathway categories, with the threshold of significance of p<0.05.
RESULTS
Patient characteristics and DNA methylation profiles
All 401 patients, who had been preprocessed, were pathologically diagnosed with LUAD. The patients were randomly divided into the test group (n=134) and the training group (n=267). There were 221, 95, 64, and 19 patients with stage I, II, III, and IV LUAD, respectively. Two patients did not report their stage. The median patient age was 65.0 years (range, 33.0–88.0 years). Additional patient characteristics are shown in Table 1. Patients with missing data were excluded from this study. The selection process of the prognostic DNA methylation signature is shown in Fig. 1.
Identification of six DNA methylation sites in the training group
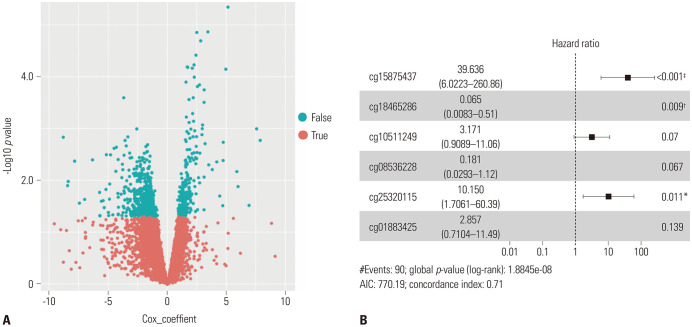
The training group (n=267) was used to investigate the association between DNA methylation sites and survival. The profiling data were analyzed using univariate Cox proportional hazards regression, with survival time and survival status as dependent variables. We identified 835 DNA methylation sites that were significantly related to the survival time (p<0.05) (Fig. 2A). The random Forest SRC algorithm was applied to identify the DNA methylation sites. Ten gene sites related to the overall survival were selected according to the permutation importance score using the RSFVH algorithm (Supplementary Fig. 1, only online). We conducted the multivariate Cox regression analysis and built a model to assess the prognosis risk until six gene sites (cg15875437, cg18465286, cg10511249, cg08536228, cg25320115, and cg01883425) were chosen (Fig. 2B). The RS of each patient was determined as follows:
Fig. 2. Screening optimal model of all methylation gene sites. Univariable (A) and multivariable Cox (B) regression analysis of the association between the methylated sites signature and the survival of lung adenocarcinoma patients in training group. *p<0.05, †p<0.01, ‡p<0.001; all the three symbols means there is a significant correlation.
| RS=(1.54×methcg15875437)+(−2.13×methcg18465286)+(1.44×methcg01883425)+(3.44×methcg25320115)+(−2.03×methcg08536228)+(2.12×methcg10511249) |
RS and meth are the risk score and methylation value, respectively.
Identification of the survival power of DNA methylation signature
The selected DNA methylation signature returned a risk score for each patient. We used the median risk score to divide the patients in the training group into either the low-risk group (n=133) or the high-risk group (n=134). We found that the patients in the low-risk group had a significantly longer OS compared to those in the high-risk group (median OS, 23.76 months vs. 20.76 months; [hazard ratio (HR)=1.37; 95% confidence interval (CI)=1.25–1.50; p<0.001] (Fig. 3A). To validate the predictive power of the DNA methylation signature, we used the same prognostic risk score model to calculate the DNA methylation signature-based risk score for the test group (Fig. 3B). The results were similar to those of the training group. For the test group, the patients of the low-risk group had a significantly higher OS than those of the high-risk group (median survival, 29.28 months vs. 18.96 month (HR=1.14; 95% CI=1.02–1.27; p<0.004).
Fig. 3. Methylated sites signature predicts prognosis of LUAD patients. Kaplan-Meier survival curves classified LUAD patients into high-risk and low-risk groups using the sites signature in the training and test datasets. p-values were calculated by log-rank test (A and B). Results of receiver operating characteristic analysis in the training and test datasets (C and D). LUAD, lung adenocarcinoma; AUC, area under curve.
The six methylation biomarker sites had great survival predictive power
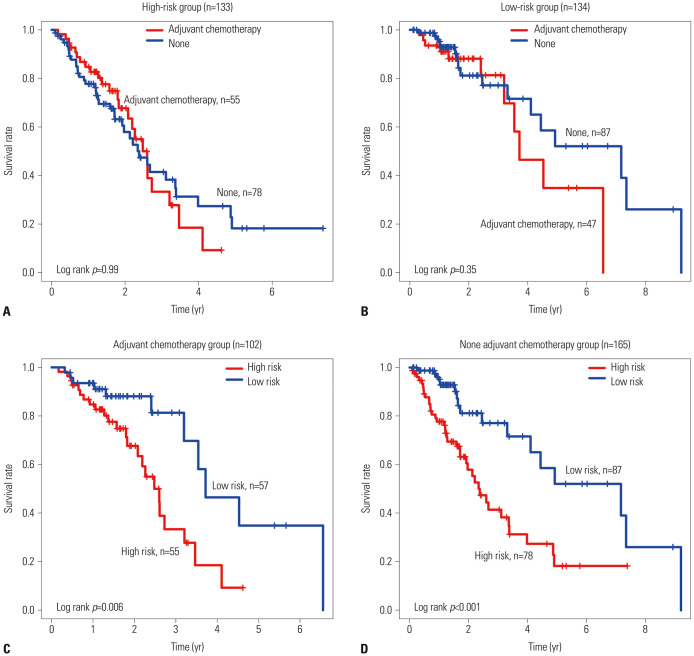
To test the predictive power of the methylation biomarker, we performed ROC analysis. In the training group, the predictive power of the six DNA methylated biomarker sites was high (AUCsignature=0.74) (Fig. 3C), which further demonstrated that the signature genes were reliable prognostic markers. Similar results were obtained for the test group (AUCsignature=0.72) (Fig. 3D). Then, we validated the predictive power of the markers through the Kaplan-Meier survival analysis and ROC analysis in the individual dataset GSE56044, and found it had a good predictive power (Fig. 4). We also compared the predictive survival power of the DNA methylation biomarker using the adjuvant chemotherapy. We found that our DNA methylation biomarker could separate the patients into the high-risk group or the low-risk group not only in the patients who received the adjuvant hemotherapy but also in the patients who did not received the adjuvant chemotherapy (Fig. 5A and B). While adjuvant chemotherapy could not divide the patients into high-risk group and the low-risk group significantly, so adjuvant chemotherapy could not as a biomarker (Fig. 5C and D). The relationship between the methylated sites signature and metastasis was also examined, and we found no significant difference in methylation level between the metastasis group and the none metastasis group (Supplementary Fig. 2, only online).
Fig. 4. Validated the predictive power of the markers through the Kaplan-Meier survival analysis (A) and receiver operating characteristic analysis (B) in the individual dataset GSE56044. AUC, area under curve.
Fig. 5. Adjuvant chemotherapy could not divided the patients into high-risk group and the low-risk group (A). DNA methylation biomarker could separate the patients into high-risk group or low-risk group either the patients were used adjuvant hemotherapy nor none adjuvant chemotherapy (B). Adjuvant chemotherapy could not divided the patients into high-risk group and the low-risk group (C and D).
Using nomogram of combined methylated sites
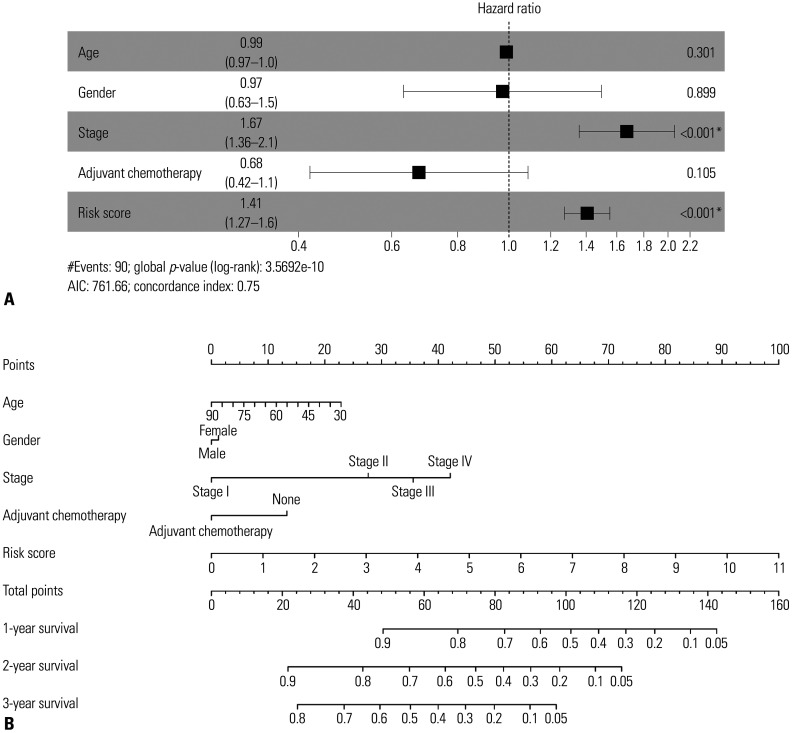
signature and clinical variables to predict patients' OS Our multivariate Cox regression model demonstrated that the predictive power of the signature risk score was independent of clinical characters (high-risk group vs. low-risk group, HR=1.36; 95% CI=1.25–1.49; p<0.05) (Fig. 6A). Based on the analysis results above, we developed a methylated gene sites nomogram. The nomogram combined the clinical-related factors (stage, age, sex and Adjuvant Chemotherapy) and methylated genes signature. In the training group, the calibration chart of the 5-year operating system was well-predicted (Fig. 6B).
Fig. 6. Multivariable Cox (A) regression analysis and nomogram of combined methylated sites signature and clinical variables predict patients' overall survival (B). *p<0.05, there is a significant correlation.
Confirming methylation pattern of the six sites of signature in normal vs. tumor in GSE85845
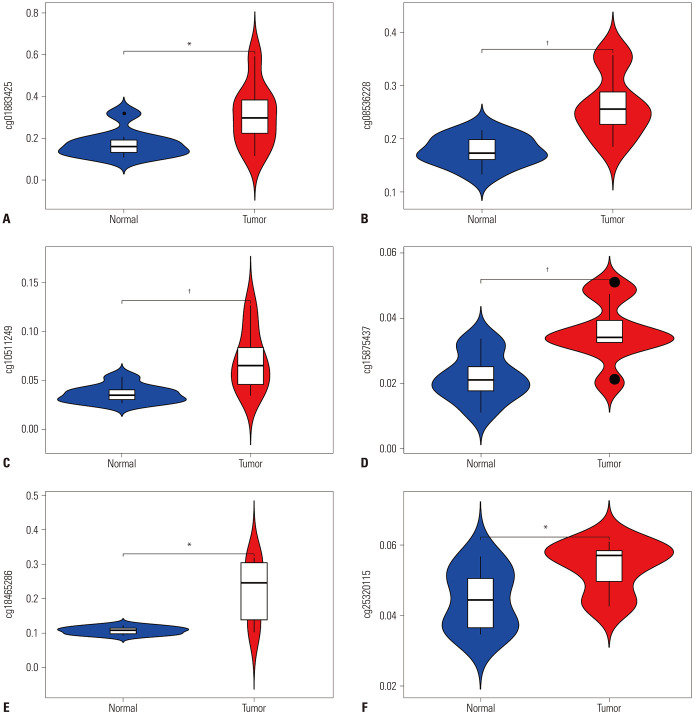
The DNA methylation profiles in GSE85845 was used to confirm the methylation pattern of the six sites of signature in normal vs. tumor. Samples included eight LC and adjacent non-tumor tissues excised from a cohort of eightg patients with LUAD. The results were the same as the ones we identified from TCGA (Fig. 7), which showed that all of the gene sites in the tumor samples were hypermethylated (Supplementary Table 1, only online).
Fig. 7. Validated methylation sites (cg01883425, cg08536228, cg10511249, cg15875437, cg18465286, cg25320115) of signature in independent GEO cohorts (A-F). It showed the results were the same as the ones we identified from TCGA data, which was all the gene sites in the tumor samples were hypermethylated. *p<0.05, †p<0.01; means there is a significant difference between the normal and tumor. GEO, Gene Expression Omnibus; TCGA, The Cancer Genome Atlas.
Functional annotation of the DNA methylated genes of signature
In order to further study the biological function of DNA methylation signature genes related to survival, we made functional annotation for these six biomarker sites. The six methylation sites were located in NAT8L, ZEB1, SYNPR, KLHDC9, and MDFI, respectively. There are two sites located in MDF1. GO term analyses showed that these the genes of signature were only enriched in two GO terms. Protein binding was the most significantly enriched in molecular function (FDR=0.0098) involving MDFI, SYNPR, and ZEB1; MDFI, ZEB1, NAT8L were significantly enriched in the function cytoplasm (FDR=0.0437). Enriched GO terms are shown in Table 2. KEGG analysis was used to analyze the signaling pathways in which these signature genes are primarily involved. We found that these genes are mainly involved in the pathways of alanine, aspartate, and glutamate metabolism and transcriptional misregulation in cancer (Supplementary Fig. 3, only online), which implies that these genes may regulate the LUAD by amino acid metabolism and transcriptional misregulation.
Table 2. Enriched GO Terms of Survival-Related DNA Methylation Signature Genes.
| Items | Items-details | p value | FDR | Genes |
|---|---|---|---|---|
| GO:0005737 | Cytoplasm (CC) | 0.0199 | 0.0437 | MDFI, ZEB1, NAT8L |
| GO:0005515 | Protein binding (MF) | 0.0053 | 0.0098 | MDFI, SYNPR, ZEB1 |
FDR, false discovery rate; MF, molecular function; CC, cellular component.
DISCUSSION
In NSCLC, LUAD is a common histological subtype with high mortality and poor prognosis in both men and women, as well as smokers and non-smokers.27 Currently, surgery remains the main treatment method for LUAD patients. However, nearly half of LUAD patients experienced recurrence or even death after surgical treatment, resulting in a low 5-year survival rate.28,29 Therefore, an effective prognostic marker to assess the survival outcomes or predict the risk of postoperative recurrence is necessary.
A large number of studies have reported that many tumor suppressor genes play a key role in tumor prevention through regulation, such as DNA repair, cell adhesion, cell cycle control, and apoptosis regulation, which can regulate the occurrence and development of tumors.30 In many cases, tumorigenesis is caused by gene silencing due to methylation of gene promoters. Therefore, it is reasonable to assume the DNA methylation status of certain genes as a useful biomarker for predicting tumor behavior. In addition, DNA methylation biomarkers have many advantages over gene and serum markers, such as DNA methylation profiles with molecular diagnostic information sources, that are more chemically and biologically stable than RNA or most proteins.31 Abnormal DNA methylation leads to dysregulation of cellular processes, such as proliferation, transformation, and antiapoptotic effects, all of which promote cancer progression. DNA methylation provides valuable information about the malignant behavior of LUAD. Most previous studies of LUAD concentrated on some of the known cancer-related genes, and most studies that searched for the biomarkers of LUAD focused on the mRNA, lncRNA, and miRNA transcription level. For example, TCGA database analysis identified hsa_circ_0000792 as a potential biomarker of LUAD,32 which could screen and diagnose LUAD in the early state. The study of mir-30a-3p also provided a new potential biomarker for the diagnosis and prognosis of LUAD.33 In addition, LncRNA-related ceRNA network studies on LUAD revealed potential RNA-related biomarkers for the diagnosis and prognosis.34 However, LUAD still lacks epigenetic biomarkers. In the present study, based on the methylation data of LUAD patients in TCGA database, a new DNA methylation biomarker model consisting of six DNA methylation gene sites was established by using bioinformatics. This methylation biomarker divided LUAD patients into high-risk and low-risk groups, with significant differences in survival. The independence of the identified DNA methylation biomarkers in the prediction of prognosis was determined by multivariate Cox regression analysis. The ROC curve further proved that the DNA methylation gene biomarker identified in this study is a new high-precision prognostic marker that offers important clinical value.
In addition, we analyzed the function of DNA methylation genes as biomarkers. GO enrichment mainly focused on the transcriptional activation activity and protein heterodimer activity, indicating that these methylated genes may affect the tumor prognosis by affecting gene transcription expression. To get a better understanding of the six genes that are involved in the prognostic signature, we performed literature review to explore their function or association in cancer. Among these six genes, several were identified to be related to cancer. In particular, cg18465286, cg15875437, and cg01883425 were located in ZEB1, NAT8L, and MDFI, respectively.
ZEB1 orchestrates EMT via the repression of epithelial genes, such as E-cadherin, a central component in adherens junctions, and the microRNA-200 family. ZEB1 represses the transcription of target genes through the epigenetic regulation of promoter chromatin architecture.35 ZEB1 enhances heterochromatinization at target gene promoters by increasing H3K27 deacetylation and tri-methylation.36 ZEB1 expression has been shown to be associated with poor clinical outcomes in different types of tumors. In colorectal cancer (CRC), the metastasis of CRC was promoted by regulating the expression of ZEB1.37 It was also found to that regulatory pathways, including ZEB1 and PKCα, could promote the activation of EMT and the occurrence of BC.38 In liver cancer, derepressing the expression of ZEB1 through DNMT3A-mediated hypermethylation suppressed miR-639 expression, and finally promoted tumorigenesis of liver cancer and provided new biomarkers for liver cancer.39 ZEB1 is also involved in regulating tumor resistance. In pancreatic cancer, the sensitivity of ZEB1 knockout cells to gemcitabine increased. In mice and pancreatic cancer cell lines, the expression of ZEB1, which activated the expression of ITGA3 and ITGB1, also increased, affecting the resistance of pancreatic cancer to chemotherapy drugs.40 ZEB1 uniquely promotes disease progression in NSCLC. In many epithelial tumors, dysregulation of ZEB1 expression is associated with poor clinical prognosis and significantly drives EMT in the pathogenesis of LC.41
Another cancer-related gene discovered in our prognostic signature is MDFI. MDFI is a known inhibitor of myogenic differentiation.42 MDFI regulates the Wnt signaling pathway and plays an important role in the development of tumors.43 A recent study showed that MDFI was methylated significantly in CRC tissues. MDFI is significantly methylated in CRC tissues when compared to adjacent normal colon tissues, and is a potential epigenetic biomarker for early detection of CRC, with a sensitivity of 81% and specificity of 91%.44 In pancreatic cancer, the aberrant DNA methylation of MDFI is a promising methylated gene marker.45 NAT8L is also a tumor-related gene, and a previous study showed that NAT8L expression was found in approximately 40% of adenocarcinoma and squamous cell carcinoma cases in NSCLC.46 Further studies focusing on the specific functions of these genes in LUAD are still required to completely understand the mechanism of LUAD recurrence at gene level. The identification of these genes in LUAD might provide a reference for further research.
This study had some limitations. First, we downloaded a small DNA methylation dataset of LUAD. Therefore, other correlations between the OS and DNA-methylated genes may have been lost. Second, we did not investigate the mechanism of DNA methylation in LUAD. Finally, although the selected DNA methylation signature had a strong prediction value, further studies are needed to determine whether this signature would be valuable to patients in different clinical settings. Despite these shortcomings, the significant correlation of the six DNA methylated genes with the OS in two independent datasets showed that they were powerful prognostic markers for LUAD. Further studies are needed to validate the prognostic value of these methylated genes in LUAD.
ACKNOWLEDGEMENTS
The present study was funded by the health industry scientific research project of Gansu province.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Min Dong.
- Data curation: Min Dong and Zengli Yang.
- Formal analysis: Min Dong and Zengli Yang.
- Funding acquisition: Min Dong.
- Investigation: Xingfang Li, Min Dong, and Zengli Yang.
- Methodology: Zhenxiang Zhang, Xingfang Li, Min Dong, and Zengli Yang.
- Project administration: Ankang Yin.
- Resources: Ankang Yin and Min Dong.
- Software: Min Dong and Zengli Yang.
- Supervision: Ankang Yin.
- Validation: Ankang Yin and Min Dong.
- Visualization: Zhenxiang Zhang and Xingfang Li.
- Writing—original draft: Min Dong and Zengli Yang.
- Writing—review & editing: Min Dong, Xingfang Li, and Zengli Yang.
- Approval of final manuscript: all authors.
AVAILABILITY OF DATA AND MATERIALS
Data sets used and/or analyzed in the current study are available in TCGA database.
SUPPLEMENTARY MATERIALS
Ten gene sites related to the overall survival were selected according to the permutation importance score using the random survival forests-variable hunting algorithm. The error rate was the lowest.
The relationship between the methylated sites signature and metastasis group and the non-metastasis group.
All of the Gene Sites in the Tumor Samples Were Hypermethylated
KEGG analysis of signature genes. These genes are mainly involved in the pathways of alanine, aspartate, and glutamate metabolism and transcriptional misregulation in cancer.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Eberhardt WE, Stuschke M. Multimodal treatment of non-small-cell lung cancer. Lancet. 2015;386:1018–1020. doi: 10.1016/S0140-6736(15)61083-2. [DOI] [PubMed] [Google Scholar]
- 3.Lee JM, Kim T, Kim EY, Kim A, Lee DK, Kwon NH, et al. Methionyl-tRNA synthetase is a useful diagnostic marker for lymph node metastasis in non-small cell lung cancer. Yonsei Med J. 2019;60:1005–1012. doi: 10.3349/ymj.2019.60.11.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013;11:645–653. doi: 10.6004/jnccn.2013.0084. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 6.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 7.Youlden DR, Cramb SM, Baade PD. The international epidemiology of lung cancer: geographical distribution and secular trends. J Thorac Oncol. 2008;3:819–831. doi: 10.1097/JTO.0b013e31818020eb. [DOI] [PubMed] [Google Scholar]
- 8.Chansky K, Sculier JP, Crowley JJ, Giroux D, Van Meerbeeck J, Goldstraw P, et al. The international association for the study of lung cancer staging project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol. 2009;4:792–801. doi: 10.1097/JTO.0b013e3181a7716e. [DOI] [PubMed] [Google Scholar]
- 9.Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2014;14:293–305. doi: 10.1586/14737159.2014.897610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765–780. doi: 10.1038/nrg3554. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 12.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 13.Xu X. DNA methylation and cognitive aging. Oncotarget. 2015;6:13922–13932. doi: 10.18632/oncotarget.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Sun X, Qin S, Wang H, DU N, Li Y, et al. CDH1 promoter methylation correlates with decreased gene expression and poor prognosis in patients with breast cancer. Oncol Lett. 2016;11:2635–2643. doi: 10.3892/ol.2016.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tezval H, Dubrowinskaja N, Peters I, Reese C, Serth K, Atschekzei F, et al. Tumor specific epigenetic silencing of corticotropin releasing hormone-binding protein in renal cell carcinoma: association of hypermethylation and metastasis. PLoS One. 2016;11:e0163873. doi: 10.1371/journal.pone.0163873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golovastova MO, Tsoy LV, Bocharnikova AV, Korolev DO, Gancharova OS, Alekseeva EA, et al. The cancer-retina antigen recoverin as a potential biomarker for renal tumors. Tumour Biol. 2016;37:9899–9907. doi: 10.1007/s13277-016-4885-5. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZR, Wei JH, Zhou JC, Haddad A, Zhao LY, Kapur P, et al. Validation of DAB2IP methylation and its relative significance in predicting outcome in renal cell carcinoma. Oncotarget. 2016;7:31508–31519. doi: 10.18632/oncotarget.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L, Xu G, Xu C, Liu B, Liu D. Potential prognostic biomarkers identified by DNA methylation profiling analysis for patients with lung adenocarcinoma. Oncol Lett. 2018;15:3552–3557. doi: 10.3892/ol.2018.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He W, Ju D, Jie Z, Zhang A, Xing X, Yang Q. Aberrant CpG-methylation affects genes expression predicting survival in lung adenocarcinoma. Cancer Med. 2018;7:5716–5726. doi: 10.1002/cam4.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo WM, Wang ZY, Zhang X. Identification of four differentially methylated genes as prognostic signatures for stage I lung adenocarcinoma. Cancer Cell Int. 2018;18:60. doi: 10.1186/s12935-018-0547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Li Y, Lu J, Pan T, Ding N, Wang Z, et al. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Yin X, Zhang G, Meng F. Identification of DNA methylation signature to predict prognosis in gastric adenocarcinoma. J Cell Biochem. 2019;120:11708–11715. doi: 10.1002/jcb.28450. [DOI] [PubMed] [Google Scholar]
- 23.Guo JC, Wu Y, Chen Y, Pan F, Wu ZY, Zhang JS, et al. Protein-coding genes combined with long noncoding RNA as a novel transcriptome molecular staging model to predict the survival of patients with esophageal squamous cell carcinoma. Cancer Commun (Lond) 2018;38:4. doi: 10.1186/s40880-018-0277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazzaferro V, Llovet JM, Miceli R, Bhoori S, Schiavo M, Mariani L, et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: a retrospective, exploratory analysis. Lancet Oncol. 2009;10:35–43. doi: 10.1016/S1470-2045(08)70284-5. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A. Letter to the editor: role of sorafenib in liver transplantation outcomes of patients with hepatocellular carcinoma presenting beyond milan criteria? Hepatology. 2020;72:1499. doi: 10.1002/hep.31285. [DOI] [PubMed] [Google Scholar]
- 26.Wu YS, Lin H, Chen D, Yi Z, Zeng B, Jiang Y, et al. A four-miRNA signature as a novel biomarker for predicting survival in endometrial cancer. Gene. 2019;697:86–93. doi: 10.1016/j.gene.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 27.Zeng Z, Yang F, Wang Y, Zhao H, Wei F, Zhang P, et al. Significantly different immunoscores in lung adenocarcinoma and squamous cell carcinoma and a proposal for a new immune staging system. Oncoimmunology. 2020;9:1828538. doi: 10.1080/2162402X.2020.1828538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esposito L, Conti D, Ailavajhala R, Khalil N, Giordano A. Lung cancer: are we up to the challenge? Curr Genomics. 2010;11:513–518. doi: 10.2174/138920210793175903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan X, Zhang X, Wang H, Jin B. Reevaluation of survival and prognostic factors in pathologic stage I lung adenocarcinoma by the new 2009 TNM classification. Tumour Biol. 2014;35:5905–5910. doi: 10.1007/s13277-014-1781-8. [DOI] [PubMed] [Google Scholar]
- 30.Shames DS, Minna JD, Gazdar AF. DNA methylation in health, disease, and cancer. Curr Mol Med. 2007;7:85–102. doi: 10.2174/156652407779940413. [DOI] [PubMed] [Google Scholar]
- 31.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Sun X, Miao S, Lu T, Wang Y, Liu J, et al. hsa_circ_0000729, a potential prognostic biomarker in lung adenocarcinoma. Thorac Cancer. 2018;9:924–930. doi: 10.1111/1759-7714.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, An Z, Li P, Liu H. A prognostic model for lung adenocarcinoma patient survival with a focus on four miRNAs. Oncol Lett. 2017;14:2991–2995. doi: 10.3892/ol.2017.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Li B, Ran P, Wang L. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett. 2018;16:5697–5708. doi: 10.3892/ol.2018.9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roche J, Nasarre P, Gemmill R, Baldys A, Pontis J, Korch C, et al. Global decrease of histone H3K27 acetylation in ZEB1-induced epithelial to mesenchymal transition in lung cancer cells. Cancers (Basel) 2013;5:334–356. doi: 10.3390/cancers5020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M, et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7:831–847. doi: 10.15252/emmm.201404396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Miao F, Huang R, Liu W, Zhao Y, Jiao T, et al. RHBDD1 promotes colorectal cancer metastasis through the Wnt signaling pathway and its downstream target ZEB1. J Exp Clin Cancer Res. 2018;37:22. doi: 10.1186/s13046-018-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010;296:216–224. doi: 10.1016/j.canlet.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao J, Liu Y, Wu F, Liu R, Xie Y, Yang Q, et al. miR-639 expression is silenced by DNMT3A-mediated hypermethylation and functions as a tumor suppressor in liver cancer cells. Mol Ther. 2020;28:587–598. doi: 10.1016/j.ymthe.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Liu M, Zhang Y, Yang J, Cui X, Zhou Z, Zhan H, et al. ZIP4 increases expression of transcription factor ZEB1 to promote integrin α3β1 signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology. 2020;158:679–692. doi: 10.1053/j.gastro.2019.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–3235. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Kusano S, Raab-Traub N. I-mfa domain proteins interact with Axin and affect its regulation of the Wnt and c-Jun N-terminal kinase signaling pathways. Mol Cell Biol. 2002;22:6393–6405. doi: 10.1128/MCB.22.18.6393-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Chen C, Bi X, Zhou C, Huang T, Ni C, et al. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 45.Omura N, Li CP, Li A, Hong SM, Walter K, Jimeno A, et al. Genomewide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lou TF, Sethuraman D, Dospoy P, Srivastva P, Kim HS, Kim J, et al. Cancer-specific production of N-acetylaspartate via NAT8L overexpression in non-small cell lung cancer and its potential as a circulating biomarker. Cancer Prev Res (Phila) 2016;9:43–52. doi: 10.1158/1940-6207.CAPR-14-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ten gene sites related to the overall survival were selected according to the permutation importance score using the random survival forests-variable hunting algorithm. The error rate was the lowest.
The relationship between the methylated sites signature and metastasis group and the non-metastasis group.
All of the Gene Sites in the Tumor Samples Were Hypermethylated
KEGG analysis of signature genes. These genes are mainly involved in the pathways of alanine, aspartate, and glutamate metabolism and transcriptional misregulation in cancer.
Data Availability Statement
Data sets used and/or analyzed in the current study are available in TCGA database.