Abstract
Long noncoding RNAs have key roles in glioma progression. However, the function and mechanisms of action of the long noncoding RNA, LINC00346, in glioma remain unclear. In our study, we observed that LINC00346 levels were increased in glioma tissue samples, and according to Gene Expression Profiling Interactive Analysis, its levels were related to disease‐free survival and overall survival rates, suggesting that a high level of LINC00346 expression corresponds to a poor prognosis. We next confirmed the high levels of LINC00346 expression in glioma tissues and cell lines and showed that LINC00346 knockdown suppressed glioma cell proliferation, migration and invasion; promoted apoptosis; and delayed tumour growth. Moreover, the oncogenic function of LINC00346 may be explained, in part, by the down‐regulation of miR‐340‐5p and the de‐repression of ROCK1. We showed that LINC00346 may function as a competing endogenous RNA of miR‐340‐5p, thereby de‐repressing ROCK1. This study revealed a new regulatory network in glioma and identified potential therapeutic targets for this cancer.
Keywords: apoptosis, cell migration, cell proliferation, glioma, LINC00346, miR‐340‐5p, ROCK1
1. INTRODUCTION
Glioma is the most common, invasive and destructive primary brain tumour, with a poor prognosis and few treatment options. 1 In the past few decades, progress has been made in surgery, chemotherapy, radiotherapy and combination therapy for the treatment of gliomas, but the median survival time for patients newly diagnosed with glioblastoma remains <15 months. 2 , 3 Therefore, it is urgently necessary to identify novel therapeutic targets and gains new insights into the molecular events underlying the pathogenesis of glioma, to enable the development of more effective strategies for its treatment.
Studies have shown that the human genome encodes long noncoding RNAs (lncRNAs, ≥200 nucleotides) that have a limited or no ability to be translated into proteins. 4 , 5 Currently, several lncRNAs are known to have vital functions in cellular development, differentiation and various other biological processes. 6 , 7 , 8 Recently, lncRNAs have been shown to perform vital functions in cancer initiation and progression. Many lncRNAs, such as HAS2‐AS1, FOXD2‐AS1, LSINCT5 and SNHG1, have been shown to promote glioma tumorigenesis. 9 , 10 , 11 , 12 LncRNA LINC00346, which is an intergenic lncRNA, has been found to be involved in many cancers. For example, Shi et al showed that LINC00346 enhances pancreatic cancer cell proliferation, colony‐forming ability and tumorigenesis. 13 Xu et al showed that LINC00346 gene expression levels are increased in gastric cancer, and its expression level positively correlates with a more advanced pathologic stage and poor prognosis. 14 Yin et al observed that LINC00346 expression levels are elevated in hepatocellular carcinoma cells, thereby promoting hepatocellular carcinoma progression. 15 Nonetheless, the function of LINC00346 in glioma progression remains unclear.
Emerging studies have revealed that lncRNAs and miRNAs are closely related in the regulation of biological processes in cancers, through the competing endogenous RNA (ceRNA) hypothesis. 16 , 17 Thus, we speculated that LINC00346 may function as a ceRNA in glioma. miR‐340‐5p has important roles in the progression of many types of human cancers, by functioning as an oncogenic RNA or a tumour suppressor. For example, miR‐340‐5p acts as tumour suppressors in non‐small‐cell lung cancer, colon cancer and glioblastoma multiforme (GBM), 18 , 19 , 20 but it acts as an oncogenic RNA in ovarian cancer. 21 miR‐340‐5p expression is down‐regulated in serum exosomes from GBM patients. 22 Moreover, the miR‐340‐5p‐macrophage feedback loop has been shown to modulate tumour progression and the tumour microenvironment in GBM patients and it may represent a prognostic biomarker and a therapeutic target for GBM. 23 These findings suggest that miR‐340‐5p may play a role in glioma progression. Some studies have shown that several lncRNAs, such as LINC00662, LINC01354 and MCM3AP‐AS1, may function as miR‐340‐5p sponges and thus, affect cancer progression. 24 , 25 , 26 However, the function of these lncRNAs and their mechanisms of action in glioma require further exploration.
In our study, we showed that LINC00346 levels were increased in glioma tissues and cell lines, and the knockdown of LINC00346 expression suppressed glioma cell proliferation, migration, and invasion and induced apoptosis in vitro. Moreover, we showed that LINC00346 may act as a ceRNA of miR‐340‐5p, thereby de‐repressing ROCK1. These findings will facilitate the development of a novel strategy for the treatment of glioma.
2. MATERIALS AND METHODS
2.1. Human tissue samples
Twenty glioma tissue samples and 20 normal brain tissue samples were obtained from patients undergoing surgical resection at Fudan University Shanghai Cancer Center. Informed consent was given by all participating patients, and the study protocol was approved by the Research Ethics Committee at Fudan University Shanghai Cancer Center.
2.2. Cell culture
Normal human astrocytes (NHAs) and glioma cell lines (U87, U251, LN229 and H4) were obtained from the Shanghai Institutes for Biological Sciences Cell Resource Center. Glioma cell lines were cultured in DMEM (containing 10% foetal bovine serum; Life Technologies, Carlsbad, CA, USA). NHAs were cultured in RPMI 1640 medium (Life Technologies).
2.3. Bioinformatics analysis
LINC00346 expression levels in GBM and normal tissue were analysed using Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/). 27 The prognosis based on LINC00346 expression levels was also analysed using GEPIA. Potential LINC00346 and miR‐340‐5p binding sites were predicted using miRanda (http://www.miranda.org/) and MiRDB (http://mirdb.org/), and miR‐340‐5p and ROCK1 binding sites were predicted using Targetscan (http://www.targetscan.org/vert_72/).
2.4. Cell transfection
Short hairpin RNA (shRNA)‐encoding sequences targeting LINC00346 were inserted into the pLKO.1 vector at the AgeI and EcoRI sites, to generate LINC00346 shRNA expression constructs. LINC00346‐targeting shRNAs were obtained from Sangon Biotech (Shanghai, China), with the following sequences: 5′‐CCGGGCATGAGAACTCTCTGCATCTCGAGATGCAGAGAGTTCTCATGCTTTTTG‐3′ and 5′‐AATTCAAAAAGCATGAGAACTCTCTGCATCTCGAGATGCAGAGAGTTCTCATGC‐3′. 293T cells were co‐transfected with validated vector and lentivirus packaging vectors (pMD and psPAX2) using Lipofectamine 2000 (Life Technologies). Virus particles were harvested and purified after 48 hours. An miR‐340‐5p mimic and inhibitor (anti‐miR‐340‐5p) and their relevant negative controls were obtained from RiboBio (miR10004692‐1‐5 and miR20004692‐1‐5; Guangzhou, China).
2.5. RNA extraction and quantitative reverse transcription PCR
Total RNA was extracted from tissue samples and cells using TRIzol (Life Technologies). For lncRNA and mRNA analysis, total RNA was reverse‐transcribed into cDNA, and then qPCR was performed on 4 µL of cDNA using a SYBR Green PCR kit (Takara, Dalian, China). lncRNA and mRNA expression levels were normalized to GAPDH expression levels. miRNA expression levels were evaluated using Stem‐Loop Primer SYBR Green Quantitative Real‐Time PCR (RiboBio), and the levels were normalized to U6 expression levels. PCR primer sequences were as follows: LINC00346 forward, 5′‐CACCATGTTGGCCAGGCTGGT‐3′, reverse, 5′‐GGCCAAAGAGTGACCATCATC‐3′; miR‐340‐5p forward, 5′‐CCGTTAGTTACGATTCGAAG‐3′, reverse, 5′‐ AGGCCGCGCGTAGTGATGCAACA‐3′; GAPDH forward, 5′‐AGCAAGAGCACAAGAGGAAG‐3′, reverse, 5′‐GGTTGAGCACAGGGTACTTT‐3′; and U6 forward, 5′‐ AACCTTATATCGGGCGGGA‐3′, reverse, 5′‐TTACGGCGATGCATAAT‐3′.
2.6. Cell proliferation
Cell proliferation was assessed using a Cell Counting Kit‐8 (CCK‐8, Beyotime Institute of Biotechnology, Jiangsu, China) assay. Transfected cells were plated in 96‐well plates, and cell viability was evaluated at 24, 48 and 72 hours after seeding. After 2 hours of incubation with the CCK‐8 solution (10 μL), optical density was measured at 450 nm. 28
2.7. Apoptosis quantitation
Flow‐cytometric methods were utilized to analyse apoptosis. After 48 hours of infection, cells were collected, washed and stained with annexin V‐FITC for 15 minutes in the dark at room temperature, according to the manufacturer's instructions. Cells were assessed using flow cytometry (FACScan, BD Biosciences, Franklin Lakes, NJ, USA), and the percentage of stained cells was determined. 29
2.8. Cell migration and invasion assays
The migration and invasiveness of transfected U87 and U251 cells were determined using a QCM Laminin Migration Assay and a Cell Invasion Assay Kit, respectively. After incubation for 48 hours with the migration and invasion assay reagents, absorbance was quantified at 560 nm.
2.9. Dual‐luciferase reporter assay
A fragment of the miR‐340‐5p gene or the ROCK1 gene, including the putative LINC00346‐ or miR‐340‐5p‐binding sites, was subcloned into the pmirGLO dual‐luciferase vector, to construct the reporter plasmids, miR‐340‐5p‐wild‐type (miR‐340‐5p‐wt) or ROCK1 wild‐type 3′‐untranslated region (UTR, ROCK1‐wt). Mutant putative LINC00346‐ or ROCK1‐binding sites were generated to construct the reporter plasmids, miR‐340‐5p‐mutated (miR‐340‐5p‐mt) or ROCK1‐3′‐UTR‐mutated, (ROCK1‐mt). Glioma cells were co‐transfected with the construct(s) and LINC00346 or miR‐340‐5p mimic or negative controls, using Lipofectamine 2000. Luciferase activity was detected using a Dual‐Glo Luciferase Assay System (Promega, Madison, WI, USA), 48 hours after transfection.
2.10. RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assays were performed using the Magna RIP RNA‐Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA). Briefly, cells were washed and resuspended in ice‐cold RIPA lysis buffer. The resulting cell lysates were centrifuged, and the supernatant was transferred to RIP immunoprecipitation buffer containing AGO2‐ or IgG‐conjugated magnetic beads. After incubation, the magnetic beads were washed and then incubated. RNA was then isolated for quantitative reverse transcription PCR (qRT‐PCR) analysis. 30
2.10.1. Western blotting
Cells were homogenized and lysed in RIPA buffer including a cocktail of protease inhibitors. Protein extracts were loaded onto 12% polyacrylamide gels, subjected to electrophoresis, and electrotransferred onto nitrocellulose membranes. Protein bands were visualized using an ECL chemiluminescence kit. Antibodies against ROCK1 and β‐actin were obtained from Proteintech (Wuhan, China).
2.10.2. Tumour xenografts
Six‐week‐old male BALB/c nude mice were obtained from Shanghai SIPPR‐BK Laboratory Animal Co., Ltd. (Shanghai, China). Animal experiments were performed in accordance with the guidelines for the care and use of laboratory animals at Fudan University and were approved by the Animal Research Ethics Committee of Fudan University. Transfected shLINC00346 or shNC U251 cells (3 × 106 cells) were subcutaneously injected into nude mice. Tumour volume was evaluated every 5 days via the following formula: volume (mm3) = length × width2/2. Thirty days after injection, the mice were euthanized and the tumours were excised.
2.10.3. Statistical analysis
The experimental results are presented as mean ± SD. Statistical analysis was performed using SPSS 23.0 statistical software (SPSS, Chicago, IL, USA). Student's t test and one‐way analysis of variance were used for data analysis. Results with P < .05 were considered significant.
3. RESULTS
3.1. LINC00346 was up‐regulated in glioma tissues and cell lines
According to the GEPIA database, LINC00346 expression levels are higher in GBM samples than in their corresponding normal brain tissues (Figure 1A). GEPIA database analysis revealed that LINC00346 expression was associated with disease‐free and overall survival rates, suggesting that high expression levels of LINC00346 indicate a poor prognosis (Figure 1B,C). These findings suggested that LINC00346 may play a role in promoting glioma progression. Our qRT‐PCR results showed that LINC00346 expression levels were higher in glioma tissues compared with their corresponding normal brain tissues (Figure 1D). Interestingly, LINC00346 expression was positively correlated with pathological grade (Figure 1E). We also examined LINC00346 expression in NHAs and a panel of glioma cell lines (LN229, U251, H4 and U87). qRT‐PCR analysis revealed that LINC00346 levels were clearly increased in glioma cell lines compared with NHAs (Figure 1F). These data suggested that LINC00346 is up‐regulated in glioma.
FIGURE 1.
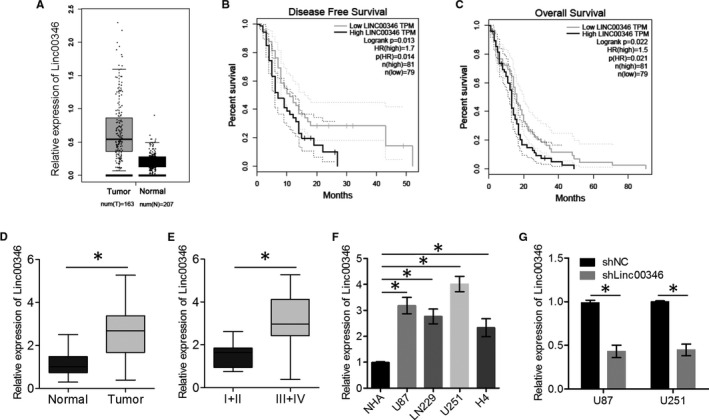
LINC00346 expression in glioma tissues and cell lines. A, Expression levels of LINC00346 in GEPIA clinical samples (normal brain tissue and GBM) are presented as a box and whisker plot. B,C, Disease‐free (DFS) and overall (OS) survival rates were computed from TCGA and GTEx data on the GEPIA server. D, The expression levels of LINC00346 in 20 glioma tissue samples and 20 normal brain tissue samples were measured by qRT‐PCR. E, The expression levels of LINC00346 in gliomas of different clinical grades. WHO grade I + II, n = 7; II + IV, n = 13. F, The expression levels of LINC00346 in glioma cell lines and NHAs were determined by qRT‐PCR. U87 and U251 cells were transfected with either shLINC00346 or shNC, and LINC00346 expression was determined by qRT‐PCR. * P < .05
3.2. LINC00346 knockdown inhibited the malignant characteristics of glioma in vitro and in vivo
To explore the biological activity of LINC00346 in glioma, LINC00346 expression was stably silenced in U87 and U251 cells (Figure 1F), and the cell proliferation was measured using the CCK‐8 assay, after confirming transfection efficiency. Glioma cells in the shLINC00346 group had significantly lower proliferative ability than those in the control group (Figure 2A). The results of flow cytometry showed that the apoptosis rate was higher in the shLINC00346 group than in the shNC group (Figure 2B). The migration and invasion abilities of U87 and U251 cells were evaluated using transwell assays. shLINC00346‐transfected cells were found to have significantly lower migration and invasion abilities than shNC‐transfected cells (Figure 2C). Furthermore, the impact of LINC00346 silencing on tumour growth was determined in vivo. These findings suggested that LINC00346 knockdown reduced tumour growth, tumour volume and tumour weight (Figure 2D‐G). These data indicated that LINC00346 knockdown suppressed the malignant characteristics of glioma.
FIGURE 2.
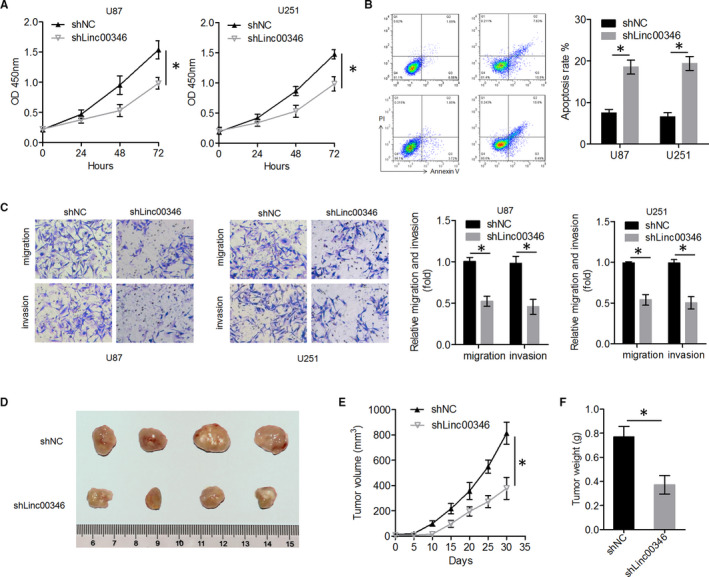
LINC00346 knockdown suppressed glioma cell proliferation, migration, invasion and tumour growth in vitro and in vivo. A, Cell proliferation was examined using the CCK‐8 assay in U87 and U251 cells after LINC00346 knockdown. B, The percentage of apoptotic cells was determined by flow cytometry with an annexin V‐FITC antibody and propidium iodide staining. C, Effects of LINC00346 knockdown on the migration and invasiveness of U87 and U251 cells. D, Xenograft tumours from nude mice. E, The expression levels of LINC00346 in tumour tissue samples from two groups were measured by qRT‐PCR. F, Tumour growth curves were constructed by measuring tumour volume every 5 d for 30 d after glioma cell injection. G, The weight of tumours excised from the mice in each treatment group was determined on day 30 after injection (n = 4, each group). * P < .05
3.3. miR‐340‐5p knockdown reversed the LINC00346 knockdown–induced suppression of glioma cell proliferation and invasion
Previous studies have shown that lncRNAs function as molecular sponges of miRNAs; thus, we hypothesized that LINC00346 may have sponging activity. The databases, miRDB and miRanda, revealed that LINC00346 has five putative binding sites for miR‐340‐5p, which serves as a tumour suppressor in several cancers (Figure 3A). Reporter assays showed that the activity of a luciferase gene linked to LINC00346 was repressed in a dose‐dependent manner in miR‐340‐5p mimic‐transfected glioma cells, compared with control cells (Figure 3B). Of note, mutations introduced into the seed sequence of miR‐340‐5p (Figure 3A) abrogated its suppressive effects (Figure 3B). Collectively, these data suggested that LINC00346 directly binds to miR‐340‐5p in glioma cell lines.
FIGURE 3.
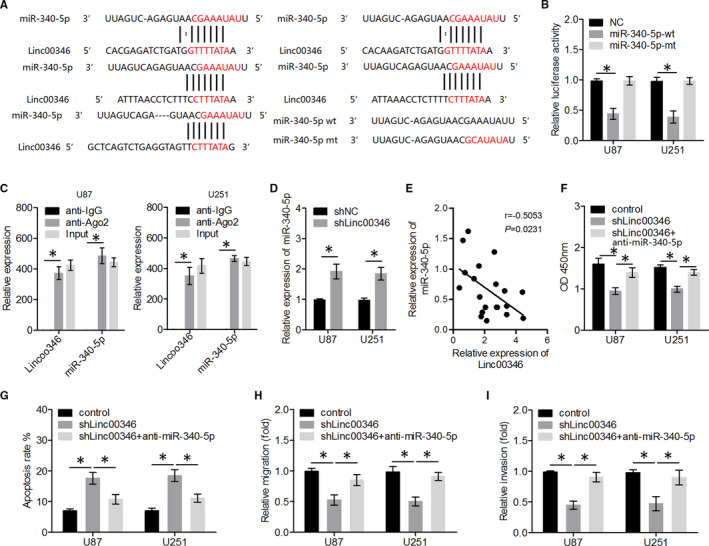
LINC00346 knockdown significantly inhibited glioma cell proliferation, migration, and invasion and promoted apoptosis by reducing the targeting (sponging) of miR‐340‐5p. A, Schematic representation of putative miR‐340‐5p–binding sites in LINC00346, and the sequence of miR‐340‐5p‐mt. B, Dual‐luciferase reporter and (C) RIP assays were performed to determine whether LINC00346 directly binds to miR‐340‐5p. D, The expression levels of miR‐340‐5p were assessed in U87 and U251 cells transfected with either shLINC00346 or shNC. E, The expression levels of miR‐340‐5p were negatively correlated with LINC00346 expression levels in glioma tissue samples. F‐I, Glioma cell proliferation, apoptosis, and migration and invasion were quantified by CCK‐8, flow cytometry and transwell assays, respectively. * P < .05
The RNA‐induced silencing complex is a factor vital for the biological effects of miRNAs, and AGO2 is a key catalytic constituent involved in RNA cleavage. 31 To determine the possible interactions between LINC00346 and miR‐340‐5p, RIP assays were performed on U87 and U251 cells. As depicted in Figure 3C, the enrichment levels of LINC00346 and miR‐340‐5p were higher in the anti‐AGO2 group than in the anti‐IgG group, suggesting that as a ceRNA, LINC00346 is capable of sponging miR‐340‐5p. We also showed that LINC00346 knockdown increased miR‐340‐5p expression levels in glioma cells (Figure 3D). In addition, we observed that the level of miR‐340‐5p expression in glioma tissues was negatively correlated with LINC00346 levels (Figure 3E). Next, to identify the mechanism by which LINC00346 regulates the glioma cell activity, U87 and U251 cells were either transfected with shLINC00346 or co‐transfected with shLINC00346 and an miR‐340‐5p inhibitor. As presented in Figure 3F‐I, LINC00346 knockdown suppressed glioma cell proliferation, migration, and invasion and increased apoptosis, whereas miR‐340‐5p inhibitor transfection significantly ameliorated the suppressive effects of LINC00346 knockdown on glioma cells.
3.4. miR‐340‐5p inhibited glioma cell proliferation, migration and invasion by reducing ROCK1 expression levels
We then aimed to identify the target mRNA of miR‐340‐5p via bioinformatic analysis. Bioinformatic software predicted that ROCK1 was the target gene of miR‐340‐5p (Figure 4A). To determine whether ROCK1 was a functional target gene of miR‐340‐5p, we constructed the reporter plasmids, ROCK1‐wt and ROCK1‐mt, which carried the 3′UTR of ROCK1, containing either a wild‐type or mutant miR‐340‐5p–binding site, respectively. Co‐transfection with an miR‐340‐5p‐overexpressing construct decreased the luciferase activity of the ROCK1‐wt plasmid but not of the ROCK1‐mt plasmid (Figure 4B). In addition, we showed that ROCK1 levels were higher in glioma tissues than in normal brain tissue, and ROCK1 levels were significantly negatively correlated with miR‐340‐5p levels in glioma samples (Figure 4C,D). Moreover, to clarify whether ROCK1 was involved in the tumour‐suppressive effects of miR‐340‐5p in glioma cells, transfection combinations were performed prior to the assessment of glioma cell proliferation, migration, invasion and apoptosis. The results of qRT‐PCR and Western blot assays showed that ROCK1 levels were significantly lower in the miR‐340‐5p group, but this difference was smaller in the miR‐340‐5p + ROCK1 group (Figure 4E,F).
FIGURE 4.
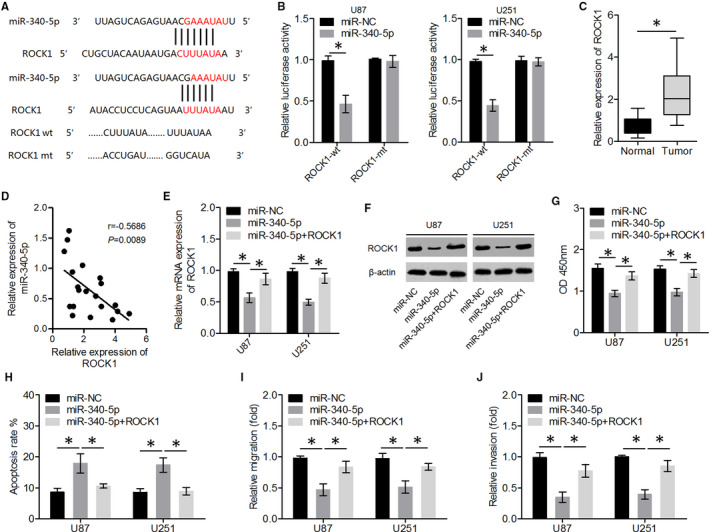
miR‐340‐5p inhibited glioma cell proliferation, migration, and invasion and promoted apoptosis by directly targeting ROCK1 mRNA. A, A schematic representation of putative miR‐340‐5p‐binding sites in ROCK1 mRNA. B, Dual‐luciferase reporter assays were performed to determine whether miR‐340‐5p directly binds ROCK1 mRNA. C, The expression levels of ROCK1 in glioma tissues and normal brain tissues were quantitated by qRT‐PCR. D, The expression levels of ROCK1 were negatively correlated with miR‐340‐5p expression levels in glioma tissue samples. E, F, The expression levels of ROCK1 were determined in U87 and U251 cells transfected with miR‐340‐5p or co‐transfected with miR‐340‐5p and a ROCK1‐overexpressing plasmid. G‐J, Glioma cell proliferation, apoptosis, migration and invasion were then measured by CCK‐8, flow cytometry and transwell assays, respectively. * P < .05
Overexpression of miR‐340‐5p suppressed the proliferation, migration, and invasiveness and promoted the apoptosis of glioma cells, whereas ectopic ROCK1 overexpression significantly attenuated the inhibitory effects of miR‐340‐5p on glioma cells (Figure 4G‐I). These results suggested that ROCK1 is a functional target gene of miR‐340‐5p and that ectopic expression of ROCK1 attenuated the tumour‐suppressive effects of miR‐340‐5p.
3.5. Overexpression of ROCK1 reversed the LINC00346 knockdown‐induced suppression of glioma cell proliferation and invasion
Transfection of an miR‐340‐5p inhibitor clearly restored ROCK1 levels in shLINC00346‐transfected cells (Figure 5A,B). In addition, we showed that ROCK1 levels positively correlated with LINC00346 levels in glioma tissues (Figure 5C). We performed rescue experiments to confirm that ROCK1 mediated the effects of LINC00346 on glioma cell growth and invasion. As expected, overexpression of ROCK1 reversed the suppressive effects of LINC00346 knockdown on glioma cell proliferation, migration, and invasion and the stimulatory effect on glioma cell apoptosis (Figure 5D‐G). Overall, these data revealed that the knockdown of LINC00346 suppressed glioma progression, because of ROCK1 down‐regulation. Therefore, LINC00346 may function as a ceRNA of miR‐340‐5p, thus up‐regulating ROCK1.
FIGURE 5.
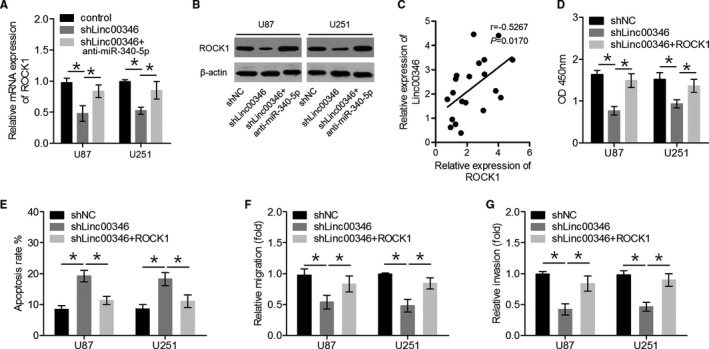
Overexpression of ROCK1 reversed the suppressive effects of LINC00346 knockdown on glioma cell proliferation and invasion. A, B, The expression levels of ROCK1 were quantified in U87 and U251 cells either transfected with shLINC00346 or co‐transfected with shLINC00346 and an miR‐340‐5p inhibitor. C, The expression levels of ROCK1 were positively correlated with LINC00346 expression levels in glioma tissue samples. D‐G, Glioma cell proliferation, apoptosis, migration and invasion were quantitated by CCK‐8, flow cytometry and transwell assays, respectively. * P < .05
4. DISCUSSION
Glioma is one of the most common malignant brain tumours; therefore, investigation of the molecular mechanisms involved in glioma progression is essential for the effective treatment of this cancer. 32 LINC00346 levels are increased in hepatocellular carcinoma, pancreatic cancer and gastric cancer, thus contributing to cancer progression. 14 , 15 , 33 In the present study, GEPIA data analysis revealed that LINC00346 was up‐regulated in GBM tissue samples and was associated with a poor prognosis, suggesting that LINC00346 may be involved in glioma progression. In addition, we confirmed higher LINC00346 expression levels in glioma cell lines and glioma samples than in normal cells and tissues. Moreover, we observed that the knockdown of LINC00346 suppressed glioma cell proliferation, migration, and invasion and promoted apoptosis in vitro, and inhibited tumour growth in vivo. These findings revealed that LINC00346 serves as an oncogenic RNA in glioma and contributes to the progression of this cancer.
Mounting evidence points to a close link between lncRNAs and miRNAs in the regulation of various biological processes. 16 We predicted miR‐340‐5p to be the target miRNA of LINC00346 using the bioinformatic tools, miRDB and miRanda. This finding was confirmed using dual‐luciferase reporter, qRT‐PCR and RIP assays. miR‐340‐5p performs an antitumour function in lung cancer, pancreatic cancer, ovarian cancer and angiosarcoma. 34 , 35 , 36 , 37 Li et al reported that miR‐340‐5p suppresses glioblastoma cell proliferation by repressing CDK6, cyclin D1 and cyclin D2. 38 The miR‐340‐5p‐macrophage feedback loop modulates the progression of GBM and may represent a prognostic biomarker and a therapeutic target for GBM [23]. In our study, we showed that miR‐340‐5p interacted with LINC00346, and that miR‐340‐5p levels negatively correlate with LINC00346 levels in glioma tissues. Furthermore, an miR‐340‐5p inhibitor attenuated the antitumour effects of LINC00346 knockdown in glioma cells. These results suggested that LINC00346 knockdown inhibited glioma progression via up‐regulation of miR‐340‐5p.
Bioinformatic software predicted that binding sites for miR‐340‐5p are present in ROCK1 mRNA. Next, we found that ROCK1 was a downstream target of miR‐340‐5p, which is consistent with data from a previous study. 39 ROCK1 is a member of the Rho‐associated protein kinase family and has a role in cell invasion in neoplasms. 40 ROCK1 is also overexpressed in several cancers, such as hepatocellular carcinoma, non‐small cell lung cancer, breast cancer and oral squamous cell carcinoma. 41 , 42 , 43 , 44 An et al showed that miR‐124 suppresses glioma cell invasion by down‐regulating ROCK1. 45 In addition, ROCK1 has been shown to be a novel target of miR‐145, thus promoting glioma cell invasion. 46 An miRNA can target several mRNAs and an mRNA can also be regulated by one or several miRNAs involved in several different processes. LINC00346 may also be able to regulate the expression of ROCK1 through miR‐145 or miR‐124, thus participating in the development of glioma. This study focused on miR‐340‐5p. The roles of miR‐145 and miR‐124 will be investigated in future studies. Here, we found that ROCK1 levels were negatively correlated with miR‐340‐5p levels, but were positively correlated with LINC00346 levels in glioma tissues. We also showed that an miR‐340‐5p mimic suppressed the proliferation, migration, and invasiveness and promoted the apoptosis of glioma cells, whereas ROCK1 overexpression abrogated these effects of miR‐340‐5p overexpression. Furthermore, we demonstrated that LINC00346 promotes glioma cell proliferation, migration, and invasion and inhibits apoptosis by increasing the output of the miR‐340‐5p‐ROCK1 axis.
In conclusion, we found that LINC00346 levels were increased in glioma tissues and cell lines and LINC00346 knockdown suppressed glioma cell proliferation, migration, and invasion and promoted apoptosis in vitro, and inhibited tumour growth in vivo. Moreover, the oncogenic role of LINC00346 in glioma may be partly explained by the inactivation of miR‐340‐5p and thus, the disinhibition of ROCK1 expression. This study uncovered a new regulatory network in glioma and may help identify potential therapeutic targets for glioma.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
AUTHOR CONTRIBUTION
Xin Chen: Investigation (lead); Writing‐original draft (lead). Deheng Li: Investigation (equal). Lei Chen: Investigation (lead); Resources (equal); Writing‐original draft (lead). Bin Hao: Data curation (equal); Software (equal). Yang Gao: Formal analysis (equal); Methodology (equal); Validation (equal). Liangdong Li: Methodology (equal); Resources (equal); Validation (equal). Changshuai Zhou: Investigation (equal); Supervision (equal); Visualization (equal). Xiayun He: Funding acquisition (supporting); Project administration (lead); Writing‐review & editing (lead). Yiqun Cao: Formal analysis (equal); Funding acquisition (supporting); Project administration (lead); Writing‐review & editing (lead).
Chen X, Li D, Chen L, et al. Long noncoding RNA LINC00346 promotes glioma cell migration, invasion and proliferation by up‐regulating ROCK1. J Cell Mol Med. 2020;24:13010–13019. 10.1111/jcmm.15899
Xin Chen, Deheng Li and Lei Chen these authors contributed equally to this work.
Funding information
This study was supported by the Chinese National Natural Science Foundation (Grant No. 81802494), the Shanghai Municipal Commission of Health and Family Planning Foundation, China (Grant No. 20184Y0107), and the Wu Jieping Medical Foundation, China (Grant No. 320.6750.17150).
Contributor Information
Xiayun He, Email: hexiayun1962@126.com.
Yiqun Cao, Email: fudancaoyiqun@163.com.
REFERENCES
- 1. Jue TR, McDonald KL. The challenges associated with molecular targeted therapies for glioblastoma. J Neurooncol. 2016;127:427‐434. [DOI] [PubMed] [Google Scholar]
- 2. Chen D, Li D, Xu XB, et al. Galangin inhibits epithelial‐mesenchymal transition and angiogenesis by downregulating CD44 in glioma. J Cancer. 2019;10:4499‐4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Preusser M, de Ribaupierre S, Wohrer A, et al. Current concepts and management of glioblastoma. Ann Neurol. 2011;70:9‐21. [DOI] [PubMed] [Google Scholar]
- 4. Lv J, Liu H, Huang Z, et al. Long non‐coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 2013;41:10044‐10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non‐small cell lung cancer and affects cell apoptosis by regulating Bcl‐2. Cell Death Dis. 2015;6:e1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298‐1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen X, Jiang C, Qin B, et al. LncRNA ZNF503‐AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017;8:e3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Lima DS, Cardozo LE, Maracaja‐Coutinho V, et al. Long noncoding RNAs are involved in multiple immunological pathways in response to vaccination. Proc Natl Acad Sci USA. 2019;116:17121‐17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L, Wang H, Xu M, et al. Long noncoding RNA HAS2‐AS1 promotes tumor progression in glioblastoma via functioning as a competing endogenous RNA. J Cell Biochem. 2020;121(1):661‐671. [DOI] [PubMed] [Google Scholar]
- 10. Wang J, Li B, Wang C, Luo Y, Zhao M, Chen P. Long noncoding RNA FOXD2‐AS1 promotes glioma cell cycle progression and proliferation through the FOXD2‐AS1/miR‐31/CDK1 pathway. J Cell Biochem. 2019;120(12):19784‐19795. [DOI] [PubMed] [Google Scholar]
- 11. Liu B, Cao W, Ma H. Knockdown of lncRNA LSINCT5 suppresses growth and metastasis of human glioma cells via up‐regulating miR‐451. Artif Cells Nanomed Biotechnol. 2019;47:2507‐2515. [DOI] [PubMed] [Google Scholar]
- 12. Liu L, Shi Y, Shi J, et al. The long non‐coding RNA SNHG1 promotes glioma progression by competitively binding to miR‐194 to regulate PHLDA1 expression. Cell Death Dis. 2019;10:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi W, Zhang C, Ning Z, et al. Long non‐coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR‐188‐3p to derepress BRD4 expression. J Exp Clin Cancer Res. 2019;38:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu TP, Ma P, Wang WY, et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26(11):2179‐2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yin YZ, Zheng WH, Zhang X, Chen YH, Tuo YH. LINC00346 promotes hepatocellular carcinoma progression via activating the JAK‐STAT3 signaling pathway. J Cell Biochem. 2020;121(1):735‐742. [DOI] [PubMed] [Google Scholar]
- 16. Wei W, Liu Y, Lu Y, Yang B, Tang L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR‐133a/EGFR. J Cell Biochem. 2017;118:3349‐3358. [DOI] [PubMed] [Google Scholar]
- 17. Yao Y, Ma J, Xue Y, et al. Knockdown of long non‐coding RNA XIST exerts tumor‐suppressive functions in human glioblastoma stem cells by up‐regulating miR‐152. Cancer Lett. 2015;359:75‐86. [DOI] [PubMed] [Google Scholar]
- 18. Chen Z, Chen X, Lu B, et al. Up‐regulated LINC01234 promotes non‐small‐cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng B, Rong A, Zhou Q, Li W. LncRNA LINC00662 promotes colon cancer tumor growth and metastasis by competitively binding with miR‐340‐5p to regulate CLDN8/IL22 co‐expression and activating ERK signaling pathway. J Exp Clin Cancer Res. 2020;39:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim S, Choi JY, Seok HJ, Park MJ, Chung HY, Bae IH. miR‐340‐5p suppresses aggressiveness in glioblastoma multiforme by targeting Bcl‐w and Sox2. Mol Ther Nucleic Acids. 2019;17:245‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J, Lin Y, Jia Y, Xu T, Wu F, Jin Y. LncRNA HAND2‐AS1 exerts anti‐oncogenic effects on ovarian cancer via restoration of BCL2L11 as a sponge of microRNA‐340‐5p. J Cell Physiol. 2019;234:23421‐23436. [DOI] [PubMed] [Google Scholar]
- 22. Ebrahimkhani S, Vafaee F, Hallal S, et al. Deep sequencing of circulating exosomal microRNA allows non‐invasive glioblastoma diagnosis. NPJ Precision Oncol. 2018;2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y, Li X, Zhang Y, et al. An miR‐340‐5p‐macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene. 2019;38:7399‐7415. [DOI] [PubMed] [Google Scholar]
- 24. Liu Y, Gao X, Tian X. High expression of long intergenic non‐coding RNA LINC00662 contributes to malignant growth of acute myeloid leukemia cells by upregulating ROCK1 via sponging microRNA‐340‐5p. Eur J Pharmacol. 2019;859:172535. [DOI] [PubMed] [Google Scholar]
- 25. Yang G, Yang C, She Y, Shen Z, Gao P. LINC01354 enhances the proliferation and invasion of lung cancer cells by regulating miR‐340‐5p/ATF1 signaling pathway. Artif Cells Nanomed Biotechnol. 2019;47:3737‐3744. [DOI] [PubMed] [Google Scholar]
- 26. Li X, Yu M, Yang C. YY1‐mediated overexpression of long noncoding RNA MCM3AP‐AS1 accelerates angiogenesis and progression in lung cancer by targeting miR‐340‐5p/KPNA4 axis. J Cell Biochem. 2020;121:2258‐2267. [DOI] [PubMed] [Google Scholar]
- 27. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98‐W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR‐185/ANXA2 loop contributes to glioblastoma growth and progression. J Expe Clin Cancer Res. 2019;38:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi J, Zhang Y, Qin B, Wang Y, Zhu X. Long non‐coding RNA LINC00174 promotes glycolysis and tumor progression by regulating miR‐152‐3p/SLC2A1 axis in glioma. J Exp Clin Cancer Res. 2019;38:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H, Xue Y, Ma J, et al. SNHG1 promotes malignant biological behaviors of glioma cells via microRNA‐154‐5p/miR‐376b‐3p‐ FOXP2‐ KDM5B participating positive feedback loop. J Exp Clin Cancer Res. 2019;38:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nie W, Ge HJ, Yang XQ, et al. LncRNA‐UCA1 exerts oncogenic functions in non‐small cell lung cancer by targeting miR‐193a‐3p. Cancer Lett. 2016;371:99‐106. [DOI] [PubMed] [Google Scholar]
- 32. Chen Y, Bao C, Zhang X, Lin X, Huang H, Wang Z. Long non‐coding RNA HCG11 modulates glioma progression through cooperating with miR‐496/CPEB3 axis. Cell Prolif. 2019;52:e12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng WX, He RZ, Zhang Z, Yang L, Mo YY. LINC00346 promotes pancreatic cancer progression through the CTCF‐mediated Myc transcription. Oncogene. 2019;38(41):6770‐6780. [DOI] [PubMed] [Google Scholar]
- 34. Lu G, Zhang Y. MicroRNA‐340‐5p suppresses non‐small cell lung cancer cell growth and metastasis by targeting ZNF503. Cell Mol Biol Lett. 2019;24:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou Y, Li Z, Ding Y, Zhang P, Wang J. MicroRNA‐340 suppresses pancreatic cancer growth by targeting BICD2. Pancreatology. 2019;S1424‐3903:30556–30563. [DOI] [PubMed] [Google Scholar]
- 36. Huang Z, Li Q, Luo K, et al. miR‐340‐FHL2 axis inhibits cell growth and metastasis in ovarian cancer. Cell Death Dis. 2019;10:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang X, Song Y. MicroRNA‐340 inhibits the growth and invasion of angiosarcoma cells by targeting SIRT7. Biomed Pharmacother. 2018;103:1061‐1068. [DOI] [PubMed] [Google Scholar]
- 38. Li X, Gong X, Chen J, Zhang J, Sun J, Guo M. miR‐340 inhibits glioblastoma cell proliferation by suppressing CDK6, cyclin‐D1 and cyclin‐D2. Biochem Biophys Res Comm. 2015;460:670‐677. [DOI] [PubMed] [Google Scholar]
- 39. Cai H, Lin L, Cai H, Tang M, Wang Z. Combined microRNA‐340 and ROCK1 mRNA profiling predicts tumor progression and prognosis in pediatric osteosarcoma. Int J Mol Sci. 2014;15:560‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hu C, Zhou H, Liu Y, et al. ROCK1 promotes migration and invasion of nonsmallcell lung cancer cells through the PTEN/PI3K/FAK pathway. Int J Oncol. 2019;55:833‐844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen H, Li M, Huang P. LncRNA SNHG16 Promotes Hepatocellular Carcinoma Proliferation, Migration and Invasion by Regulating miR‐186 Expression. J Cancer. 2019;10:3571‐3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tang H, Du W, Jiang Y, Li H, Bo H, Song S. Upregulated expression of ROCK1 promotes cell proliferation by functioning as a target of miR‐335‐5p in non‐small cell lung cancer. J Cell Physiol. 2019. [DOI] [PubMed] [Google Scholar]
- 43. Xu F, Li H, Hu C. MiR‐202 inhibits cell proliferation, invasion, and migration in breast cancer by targeting ROCK1 gene. J Cell Biochem. 2019;120:16008‐16018. [DOI] [PubMed] [Google Scholar]
- 44. Jin Z, Jiang S, Jian S, Shang Z. Long noncoding RNA MORT overexpression inhibits cancer cell proliferation in oral squamous cell carcinoma by downregulating ROCK1. J Cell Biochem. 2019;120(7):11702‐11707. [DOI] [PubMed] [Google Scholar]
- 45. An L, Liu Y, Wu A, Guan Y. microRNA‐124 inhibits migration and invasion by down‐regulating ROCK1 in glioma. PLoS One. 2013;8:e69478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan X, Cheng Q, Peng R, et al. ROCK1, a novel target of miR‐145, promotes glioma cell invasion. Molecular medicine reports. 2014;9:1877‐1882. [DOI] [PubMed] [Google Scholar]


