Abstract
Premise
Significant paleobotanical discoveries in recent decades have considerably improved our understanding of the early evolution of angiosperms and their flowers. However, our ability to test the systematic placement of fossil flowers on the basis of phylogenetic analyses has remained limited, mainly due to the lack of an adequate, angiosperm‐wide morphological data set for extant taxa. Earlier attempts to place fossil flowers phylogenetically were, therefore, forced to make prior qualitative assessments of the potential systematic position of fossils and to restrict phylogenetic analyses to selected angiosperm subgroups.
Methods
We conduct angiosperm‐wide molecular backbone analyses of 10 fossil flower taxa selected from the Cretaceous record. Our analyses make use of a floral trait data set built within the framework of the eFLOWER initiative. We provide an updated version of this data set containing data for 28 floral and two pollen traits for 792 extant species representing 372 angiosperm families.
Results
We find that some fossils are placed congruently with earlier hypotheses while others are found in positions that had not been suggested previously. A few take up equivocal positions, including the stem branches of large clades.
Conclusions
Our study provides an objective approach to test for the phylogenetic position of fossil flowers across angiosperms. Such analyses may provide a complementary tool for paleobotanical studies, allowing for a more comprehensive understanding of fossil phylogenetic relationships in angiosperms. Ongoing work focused on extending the sampling of extant taxa and the number of floral traits will further improve the applicability and accuracy of our approach.
Keywords: angiosperms, eFLOWER, floral evolution, floral structure, fossil flowers, mesofossils, molecular backbone, phylogenetic analysis
The fossil record is critical to our understanding of the evolutionary history of angiosperms (Crane et al., 1995; Crepet et al., 2004; Friis et al., 2006, 2011; Doyle and Endress, 2010; Sauquet and Magallón, 2018; Coiro et al., 2019). During the past few decades, paleobotanists have recovered and analyzed a rich fossil record dating back to the early stages of angiosperm evolution. In particular, the recovery of countless fossil flowers and dispersed floral organs from Cretaceous mesofossil localities in North America, Europe, and Japan has provided novel and unexpected details about the early structural and phylogenetic diversity of angiosperms (e.g., Crepet, 1996; Herendeen et al., 1999; Takahashi et al., 1999; Friis et al., 2011, 2019). Many of these localities typically yield small, three‐dimensionally preserved, charcoalified or lignitized fossils, including various angiosperm reproductive structures (Friis et al., 2005; Schönenberger, 2005). These exceptionally well‐preserved fossils, in combination with novel technical applications such as high‐resolution x‐ray computed tomography (Friis et al., 2014), have revolutionized the study of the early angiosperm fossil record. However, our ability to test the systematic placement of Cretaceous flowers on the basis of explicit phylogenetic analyses across angiosperms has not kept pace with these recent developments, mainly because of a lack of adequate morphological data sets for extant taxa (Crepet and Nixon, 1998b; Friis et al., 2011; Sauquet and Magallón, 2018).
The first and most crucial step in any attempt to systematically place a newly discovered fossil specimen entails a careful and extensive morphological analysis (e.g., Schönenberger, 2005; Smith et al., 2009; Friis et al., 2014). Subsequently, paleobotanical studies usually follow one of the following two trajectories, with the goal of establishing the systematic placement of a given fossil: either they take the direct approach, without any phylogenetic analysis, and assign the fossil to the “most similar” lineage, based on a list of characters shared with a number of extant taxa (and potentially also with other fossil taxa; e.g., Friis et al., 2003; Takahashi et al., 2008; Schönenberger et al., 2012); or they first assign the fossil to a clade to which it is deemed to belong on the basis of its general morphological characters, then conduct a restricted, clade‐specific phylogenetic analysis based on morphological characters to identify the placement of the fossil among the extant branches of the clade, and finally assign the fossil to the lineage that was identified as containing the “most parsimonious” positions in the analysis (e.g., Crepet and Nixon, 1998b; von Balthazar et al., 2008; Doyle and Endress, 2010, 2018; Herendeen et al., 2016). From a purely scientific point of view, neither of these two approaches is entirely satisfactory, because both make prior qualitative assessments as to which extant taxa the new fossil is compared to.
Overall, earlier studies that included phylogenetic analyses fall largely into three categories. (1) The first category comprises original paleobotanical studies that describe a new fossil flower and then conduct an unconstrained phylogenetic analysis, based solely on a newly compiled morphological data set or relying on already existing morphological matrices (no molecular data involved, treating fossils as terminals). Such analyses have been conducted at different taxonomic levels, including, for instance, the monocots (Gandolfo et al., 2002) and different orders (e.g., Piperales: Smith and Stockey, 2007; Magnoliales and Laurales: Mohr et al., 2013; Cornales: Atkinson, 2018). (2) A second category comprises original paleobotanical studies that have conducted phylogenetic analyses largely relying on previously published molecular or combined molecular/morphological data sets. These studies have usually focused on particular sets of angiosperm taxa, such as basal angiosperms (e.g., von Balthazar et al., 2008; Friis et al., 2009; Friis and Pedersen, 2011), or various smaller clades, such as Saxifragales (Hermsen et al., 2003), Nymphaeales (Gandolfo et al., 2004), and Ericales (Martínez‐Millán et al., 2009), selected on the basis of a prior qualitative assessment of the phylogenetic position of a given fossil. These studies most often constrain the tree topology to the results from previous analyses of the molecular or combined molecular/morphological data and only subsequently add the fossil to let it find its place in the constrained tree based on a parsimony analysis (called “backbone analysis” or “DNA scaffold analysis”; Manos et al., 2007). Alternatively, and more rarely, studies combine molecular/morphological data of extant taxa and fossils from the start and conduct “total‐evidence” analyses (e.g., Manos et al., 2007; Larson‐Johnson, 2016; Martínez et al., 2016; Matsunaga et al., 2019; for discussion, see also Doyle and Endress, 2010, and citations therein). (3) The third and final category of studies does not describe any new fossils themselves but rather conducts independent phylogenetic analyses with the aim of testing earlier hypotheses on the systematic placement of selected fossil flowers. Particularly important in this category are the studies by Endress and Doyle (2010) and Doyle and Endress (2014, 2018), who, on the basis of their earlier morphological phylogenetic analyses in basal angiosperms (Doyle and Endress, 2000; Endress and Doyle, 2009), tested the phylogenetic position of a series of fossil flowers related to this group of taxa.
Here, we take an approach similar to that of Doyle and Endress (2010) by conducting molecular backbone analyses using maximum parsimony with the goal of phylogenetically placing selected fossil flowers. The important difference in comparison to these earlier studies is that we attempt to test phylogenetic hypotheses for these fossil flowers on the basis of angiosperm‐wide phylogenetic analyses. Our main motivation for conducting such broad‐scale analyses is that it allows testing for the phylogenetic position of a given fossil flower without having to rely on qualitative assessments of its potential phylogenetic position within angiosperms. Clearly, many scientific studies, particularly in evolutionary biology, are forced to work with qualitative approaches—and there is nothing wrong with that. In paleontology, for instance, careful comparative morphological analyses with the goal of assessing the systematic position of fossil taxa have certainly proven their validity in many instances. Nevertheless, we consider it important to strive toward explicit and reproducible types of analyses whenever possible, as they can help to avoid some of the potential biases that qualitative assessments may entail.
Our analyses make use of the updated version of a recently published angiosperm‐wide floral data set for 792 extant species representing all but one order and 372 families (86%) of angiosperms, which we originally compiled to infer ancestral floral structure across the angiosperm tree (Sauquet et al., 2017). For the present study, we have supplemented the original data set with additional floral and pollen traits, now adding up to a total of 30 characters.
To demonstrate the potential of our approach, we scored and analyzed a selection of 10 published Cretaceous flowers, and we discuss the resulting phylogenetic hypotheses with respect to those presented in earlier studies. In addition, we discuss the advantages and challenges of our approach and point out possible further developments and improvements.
MATERIALS AND METHODS
Morphological data set
For the phylogenetic analyses in this study, we use the largest data set of floral traits currently available, compiled from the PROTEUS database (Sauquet, 2019). The original data set was assembled within the eFLOWER initiative, an international collaborative project aimed at answering key questions on flower evolution (Sauquet et al., 2017). The data set contains floral trait data for 792 species (exemplar approach), which were sampled to match the taxa of a recent dated phylogeny of angiosperms (Magallón et al., 2015), representing 63 orders (all but one of the orders recognized in APG IV, 2016) and 372 families (86%). As in Sauquet et al. (2017), we transformed all characters, as they are scored in PROTEUS (primary characters), into secondary characters for analysis by converting continuous characters into discrete characters and by reducing the number of character states of discrete characters (see Appendix S1 for the full data set analyzed in this study, and Appendix S2 for details on scoring philosophy). The original data set of Sauquet et al. (2017) comprised data for 21 primary floral characters (13,444 data points in total). For the present study, we have updated and expanded this data set in three ways. First, we have corrected some data and filled in a number of gaps for previously included characters by consulting additional sources, as part of our continued efforts to improve the quality and completeness of this data set (e.g., Reyes et al., 2018; Sauquet et al., 2018). In total, we have updated 190 records, deleted 70, and added 417 new records for the traits and species included in our original data set (Sauquet et al., 2017). Second, we have added six androecial and gynoecial characters (previously unpublished) and two pollen characters (most of the data originally compiled for Prieu et al., 2017). In total, these eight traits add 3926 new records to our data set. Third, we have scored the same 29 primary characters in 10 selected fossil species (see below), adding another 256 data records to our data set. Thus, our new data set (hereafter “Paleo‐eFLOWER data set”) comprises a total of 17,973 data records, each linked to an explicit source (1148 sources in total), all of which are provided along with the final matrix in Appendix S1.
Contrary to our study focused on ancestral state reconstruction (Sauquet et al., 2017), where we analyzed a total of 27 secondary characters (some of them multiple versions of the same primary character), we here limited ourselves to a single secondary character for each primary character because phylogenetic reconstruction requires all characters to be presumed independent. However, we maintained two secondary characters derived from the “Number of perianth parts” primary character because they are essentially capturing different information (absence/presence of a perianth, and number of perianth parts when present). As a result, the morphological matrix used in this paper has a total of 30 characters.
In addition to the updates and expansion of the data set outlined above, we have modified two characters compared to our original study (Sauquet et al., 2017). First, we have simplified (and accordingly rescored) the “floral sex” primary character, but retained the same secondary character (for details, see Appendix S2). Hence, this change affects only the structure of our updated raw data set, not the analyses. Second, we have here opted for a different “perianth differentiation” secondary character, treating all forms of differentiation, including weak or continuous, as a differentiated perianth (Appendix S2). Analyses of the same data set with the original secondary character yielded results remarkably similar and congruent to those presented here, but we believe the new secondary character is more suited for future analyses of this dataset.
Our philosophy for scoring morphological data remains the same as in our earlier paper using the original version of the dataset (Sauquet et al., 2017). In connection with this, detailed information on how we dealt with issues such as polymorphic data, missing data, and continuous (quantitative) vs. discrete (qualitative) characters is given in Appendix S2, which also contains adapted/complemented definitions and explanations for all characters used in this study.
Selection of fossil flowers
Here, we infer the phylogenetic positions of 10 published fossil flowers (listed in Table 1). We selected these fossils to represent (based on the hypotheses presented in original studies) different parts of the angiosperm tree (including early‐diverging angiosperms, magnoliids, early‐diverging eudicots, rosids, and asterids) and different levels of information (the number of characters scored per fossil ranges from a minimum of 12 in Chloranthistemon endressii to a maximum of 29 (all but one character in our data set) in Dakotanthus cordiformis and Paradinandra suecica; see Table 2). In a few cases (e.g., some characters of Microvictoria svitkoana; Gandolfo et al., 2004), we have interpreted and scored characters differently than in the original paper in which the fossil was described. We did this because of alternative and better‐supported interpretations of floral morphology published in later studies (for details, see Results and Discussion). We provide all fossil data, each record linked to an explicit source, along with the final morphological matrix for extant species, in Appendix S1.
Table 1.
Information on fossil species included in phylogenetic backbone analyses.
| Species | References | Type locality | Age | Type of preservation |
|---|---|---|---|---|
| Chloranthistemon endressii Crane, Friis and Pedersen | Crane et al. (1989); Eklund et al. (1997) | Åsen, Höganäs AB, Scania, Sweden | Late Cretaceous (Late Santonian/Early Campanian) | Charcoalified/lignitized |
| Dakotanthus cordiformis (Lesq.) Manchester, Dilcher, Judd and Basinger | Basinger and Dilcher (1984); Manchester et al. (2018) | Ottawa County, Kansas, USA | Mid‐Cretaceous (Late Albian/Early Cenomanian) | Compression/impression/casts and molds |
| Kajanthus lusitanicus Mendes, Grimm, Pais and Friis | Mendes et al. (2014) | Chicalhão (opencast clay pit), Juncal, Portugal | Early Cretaceous (Late Aptian/Early Albian) | Charcoalified |
| Mauldinia mirabilis Drinnan, Crane, Friis and Pedersen | Drinnan et al. (1990) | West of Mauldin Mountain, Elk Neck Peninsula, Maryland, USA | Mid‐Cretaceous (Cenomanian) | Charcoalified/lignitized |
| Microvictoria svitkoana Nixon, Gandolfo and Crepet | Gandolfo et al. (2004) | Old Crossman Clay Pit, Sayreville, New Jersey, USA | Late Cretaceous (Turonian) | Charcoalified |
| Paleoclusia chevalieri Crepet and Nixon | Crepet and Nixon (1998a) | Old Crossman Clay Pit, Sayreville, New Jersey, USA | Late Cretaceous (Turonian) | Charcoalified |
| Paradinandra suecica Schönenberger and Friis | Schönenberger and Friis (2001) | Åsen, Höganäs AB, Scania, Sweden | Late Cretaceous (Late Santonian/Early Campanian) | Charcoalified |
| Spanomera mauldiniensis Drinnan, Crane, Friis and Pedersen | Drinnan et al. (1991) | West of Mauldin Mountain, Elk Neck Peninsula, Maryland, USA | Late Cretaceous (Cenomanian) | Charcoalified/lignitized |
| Tylerianthus crossmanensis Gandolfo, Nixon and Crepet | Gandolfo et al. (1998) | Old Crossman Clay Pit, Sayreville, New Jersey, USA | Late Cretaceous (Turonian) | Charcoalified |
| Virginianthus calycanthoides Friis, Eklund, Pedersen and Crane | Friis et al. (1994) | Puddledock, Hopewell, Virginia, USA | Early Cretaceous (Early or Middle Albian) | Charcoalified |
Table 2.
Overview of results from phylogenetic backbone analyses (MP = most parsimonious).
| Species | Number of analyzed characters (of 30) | Number of most parsimonious positions (MP/MP+1) | Systematic (MP) placement according to this study a | Systematic placement according to earlier studies b |
|---|---|---|---|---|
| Chloranthistemon endressii | 12 | 1/3 | CG Chloranthaceae (Chloranthales) | Chloranthaceae (Chloranthales); Eklund et al. (2004, PA) |
| Dakotanthus cordiformis | 29 | 23/118 | CG Rosidae (including many MP positions in Fabidae and Malvidae) | “Basal lineage within Fabales” (Fabidae); Manchester et al. (2018) |
| Kajanthus lusitanicus | 27 | 11/10 | CG Ranunculales (Eudicotyledoneae), with several MP positions in Lardizabalaceae and Berberidaceae | Lardizabalaceae (Ranunculales, Eudicotyledoneae); Mendes et al. (2014) |
| Mauldinia mirabilis | 24 | 9/13 | CG Laurales (Magnoliidae), with 9 MP positions in the clade with Lauraceae, Hernandiaceae, and Monimiaceae |
Lauraceae (Laurales, Magnoliidae); Drinnan et al. (1990) Sister to a clade with Lauraceae and Hernandiaceae; Doyle and Endress (2010, PA) |
| Microvictoria svitkoana | 20 | 13/7 | 12 MP positions in CGs and SGs of the ANA grade (Amborellales, Nymphaeales, and Austrobaileyales); 1 MP position in CG Calycanthaceae (Laurales, Magnoliidae) | Nymphaeaceae (Nymphaeales); Gandolfo et al. (2004, PA) |
| Paleoclusia chevalieri | 27 | 14/16 | CG and SG of the clusioid clade (Malpighiales, Fabidae); 14 MP positions, most in CG Clusiaceae and CG Hypericaceae | Clusiaceae (Malpighiales, Fabidae); Crepet and Nixon (1998a, PA); Ruhfel et al. (2013, PA) |
| Paradinandra suecica | 29 | 21/82 | Numerous MP positions in CG Ericales (Asteridae); additional MP positions in different clades of CG Pentapetalae (e.g., Caryophyllales, Malpighiales, and Dilleniales) | Ericales (Asteridae); Schönenberger and Friis (2001) |
| Spanomera mauldinensis | 25 | 4/8 | 3 MP positions in CG and SG Buxales and 1 MP position in CG Gunnerales (Eudicotyledoneae) |
“Closely related to Buxaceae” (Buxales, Eudicotyledoneae); Drinnan et al. (1991) Sister to Buxaceae; Doyle and Endress (2010, PA) |
| Tylerianthus crossmanensis | 29 | 1/12 | 1 MP position in CG Saxifragaceae (Saxifragales, Superrosidae) | Hydrangeaceae (Cornales, Asteridae) or Saxifragaceae (Saxifragales, Superrosidae) ; Gandolfo et al. (1998) |
| Virginianthus calycanthoides | 23 | 3/18 | 3 MP positions in CG and SG Calycanthaceae (Laurales, Magnoliidae) |
“Stem group Calycanthaceae (Laurales, Magnoliidae)”; Friis et al. (1994) “Stem relative of either Calycanthaceae (including Idiospermum) or the remaining Laurales”; Doyle and Endress (2010, PA) |
Only families mentioned in APG IV; CG = crown group, SG = stem group (i.e., the stem node and all of its descendants exclusive of the crown group).
PA = studies including a restricted phylogenetic analysis.
The 10 selected fossils cover a range of Cretaceous stages, from the Albian to the Campanian (Table 2). We focus on the Cretaceous because flowers are abundant in this period and are very important for our understanding of the evolutionary history of angiosperms (Friis et al., 2011). At the same time, a systematic placement of such early flowers is often particularly challenging because of their often mosaic‐like character combinations compared to extant taxa (Crepet et al., 2004), and our angiosperm‐wide approach promises to be particularly useful for this type of fossil. It is clear, however, that the methods proposed here can be applied to any angiosperm flower irrespective of its age.
Phylogenetic analyses
We conducted molecular backbone analyses with parsimony using the phylogenetic tree of 792 taxa from Magallón et al. (2015) and the expanded morphological data set of 802 taxa (792 extant + 10 fossils) described above. Specifically, we used the same transformed version of the maximum clade credibility tree of Magallón et al. (2015) that we prepared for our previous study (Sauquet et al., 2017; therein referred to as the “A tree series”), whereby original chimeric tip names were converted into explicit exemplar species. In addition, we conducted sensitivity analyses using four alternative backbones to test the impact of phylogenetic uncertainty on our results. For this, we used the corresponding maximum clade credibility trees from the B, C, D, and E tree series reconstructed in our previous study (Sauquet et al., 2017). These trees differ mainly in the relationships among the main lineages of angiosperms, which remain in flux according to data and analytical methods (e.g., One Thousand Plant Transcriptomes Initiative, 2019), but also in some finer details of relationships among families and orders that remain weakly supported. Hence, they provide a reasonable initial assessment of the sensitivity of our approach to future improvements in angiosperm phylogenetics. Although all of our backbone trees are dated and drawn as such (chronogram), branch lengths were not used in the analyses presented here.
All analyses were performed in R (R Core Team, 2019) using a custom script relying on the package “phangorn” version 2.0.2 (Schliep, 2011) to compute unordered (Fitch) parsimony scores. Briefly, for each fossil, we performed the following steps. The total number of possible branching positions for any fossil on the extant phylogeny is 1582 (792 terminal branches + 790 internal branches). Each fossil taxon was successively grafted onto each of the 1582 possible branches to obtain a parsimony score for each of these locations (based on the 30 characters in the dataset). To visualize the most parsimonious (MP) positions, the entire tree was then colored according to a gradient based on the parsimony score, with a high contrast for the lowest scores: black for MP positions, green for positions within one parsimony step of the MP position. This approach is very similar to that of previous studies (e.g., Doyle et al., 2008; Endress and Doyle, 2009; Sauquet et al., 2009), with the notable exception that we exhaustively searched for all possible positions instead of conducting a heuristic search in PAUP (Swofford, 2002), allowing a more straightforward examination of the results (the outcome in terms of MP positions and scores, however, is expected to be identical).
Our goal in this study was to evaluate the potential of the molecular backbone approach to perform a first “scan” of the possible phylogenetic positions of a fossil across the entire phylogeny of angiosperms, and to test the behavior of our floral trait data set (with its current sampling) for this purpose. For these reasons, we refer to this approach informally as a “phyloscan.” Although this approach is attractive in being simple and computationally very fast, we certainly acknowledge that it has limitations. First, uncertainty in phylogenetic relationships among extant taxa is not taken into account, because we use a single fixed tree (but see sensitivity analyses outlined above). Second, parsimony does not take branch lengths (or time) into account; comparing parsimony inference with model‐based approaches would be desirable. Lastly, by analyzing each fossil individually, we do not allow for the potential of reciprocal improvements in phylogenetic placement accuracy through the combined interactions of multiple fossil taxa with extant taxa (e.g., Manos et al., 2007; Ronquist et al., 2012). However, none of these issues are simple, and current total‐evidence approaches entail other limitations, including difficulty in extracting results and disentangling fossil placement uncertainty from other sources of uncertainty, as well as the risk of using overly simplistic models for morphological evolution (e.g., assuming that all characters are linked and evolve according to the same symmetrical rate regime; Wright et al., 2016).
In addition to the data set and matrix (Appendix S1), we provide the R script and electronic data files (including the backbone trees) in an online public repository (https://github.com/eflowerproject/paleoeflower). This will allow anyone to replicate the analyses we did for this study or to test the phylogenetic position of any other fossil flower by simply adding its floral characters to the data set and running the backbone analysis.
RESULTS AND DISCUSSION
The fossil record plays an important role in reconstructing and understanding the evolutionary history of most organismal lineages—including, of course, the angiosperms. However, the degree of utility of any fossil in evolutionary research is directly related to the level of accuracy and support for its hypothesized phylogenetic placement among other taxa. The often highly fragmentary nature of the plant fossil record and the mosaic character combinations of many fossil taxa compared to extant species (Crepet and Nixon, 1998b) make the establishment of phylogenetic relationships a daunting task. In the past, at least part of this process often relied solely on taxonomic expertise and qualitative morphological comparisons. Our angiosperm‐wide floral data set opens the way for direct, explicit, and efficient phylogenetic analyses to test the phylogenetic position of any crown group angiosperm flower. With this study, we show that our approach is applicable to a variety of fossil flowers, with different degrees of floral character preservation, and across different parts of the angiosperm tree. Note that we originally built our data set for extant taxa independently from the specific goals of this paper and it is, therefore, free from any potential biases derived from the aim of resolving the phylogenetic position of specific fossils.
Despite the limited number of characters used in our data set, our angiosperm‐wide phylogenetic analyses yielded mostly unequivocal results, with the total number of most parsimonious positions per fossil ranging from 1 (C. endressii and Tylerianthus crossmanensis) to 23 (D. cordiformis) (Table 2 and Figs. 1, 2, 3, 4; Appendix S3). However, the outcome in terms of comparison with previously proposed placements differed markedly among fossils. Seven of the 10 selected fossil flowers were found in positions that have already been proposed in earlier studies (Chloranthistemon endressii, Kajanthus lusitanicus, Mauldinia mirabilis, Paleoclusia chevalieri, Spanomera mauldinensis, Tylerianthus crossmanensis, and Virginianthus calycanthoides; Table 2). For one fossil, our results clearly deviate from earlier hypotheses by suggesting alternative positions (Microvictoria svitkoana). The remaining two fossils (Dakotanthus cordiformis and Paradinandra suecica) were found in various positions distant from each other, including the stem branches of very large clades. In the following sections, we detail our findings for four of the 10 fossil species that we have included in our analyses. The remaining six species are dealt with in the Supplemental Data (see Appendix S3).
FIGURE 1.
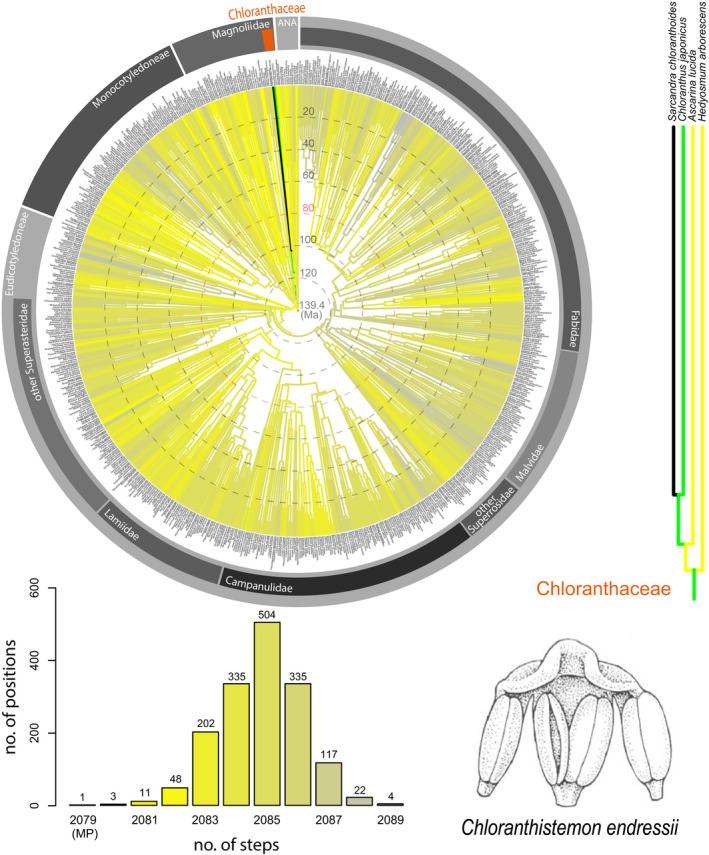
Results for Chloranthistemon endressii from angiosperm‐wide molecular backbone analyses using maximum parsimony, based on 12 floral characters that could be scored for this fossil (of 30 secondary characters in the data set). The backbone tree (circular chronogram) is a transformed version of the maximum clade credibility tree from Magallón et al. (2015), and the floral morphological data set for the 792 extant taxa is an updated version of the data set from Sauquet et al. (2017). Black branches in trees (circular tree and scaled‐up partial tree on the right) indicate most parsimonious (MP) positions; green branches indicate positions that are one step less parsimonious (MP+1). A gradient from yellow to gray branches indicates positions three or more steps less parsimonious. The same color coding applies to the legend in the lower left, indicating the number of most and less parsimonious positions. The drawing in the lower right shows the three‐lobed androecium of C. endressii (reproduced from Friis et al., 2011, with permission of Cambridge University Press). The position of Chloranthaceae is indicated by an orange bar in the circular chronogram. The concentric circles superimposed on the chronogram indicate a time scale with intervals of 20 million years, with the age of the most recent common ancestor of all angiosperms in the center of the figure set to 139.4 million years according to Magallón et al. (2015). The pink circle indicates the approximate age of C. endressii according to Crane et al. (1989).
FIGURE 2.
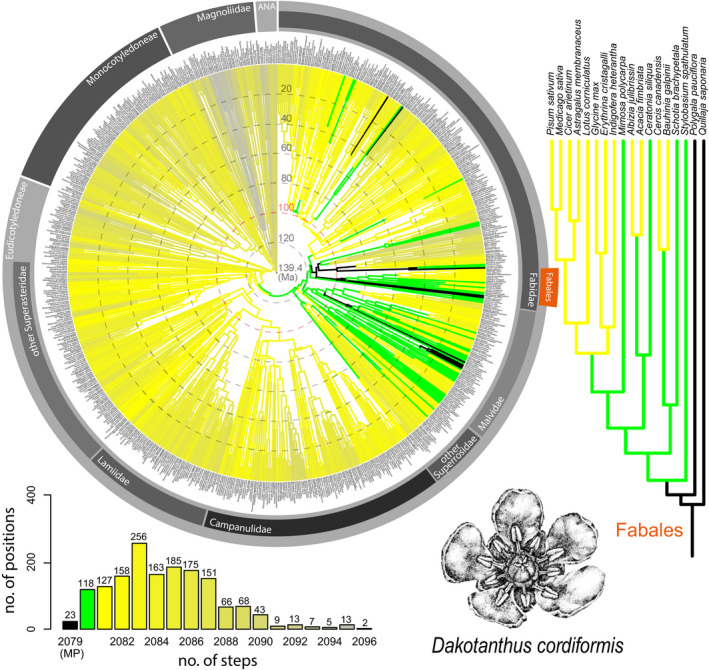
Results for Dakotanthus cordiformis from angiosperm‐wide molecular backbone analyses using maximum parsimony, based on 29 floral characters that could be scored for this fossil. The color coding, the time scale superimposed on the circular chronogram, and the sources of the backbone tree and morphological data set of extant taxa are as in Figure 1. The drawing in the lower right shows the pentamerous flower of D. cordiformis (reproduced from Manchester et al., 2018, with permission of De Gruyter). The position of the order Fabales is indicated by an orange bar in the circular chronogram. The pink circle indicates the approximate age of D. cordiformis according to Manchester et al. (2018).
FIGURE 3.
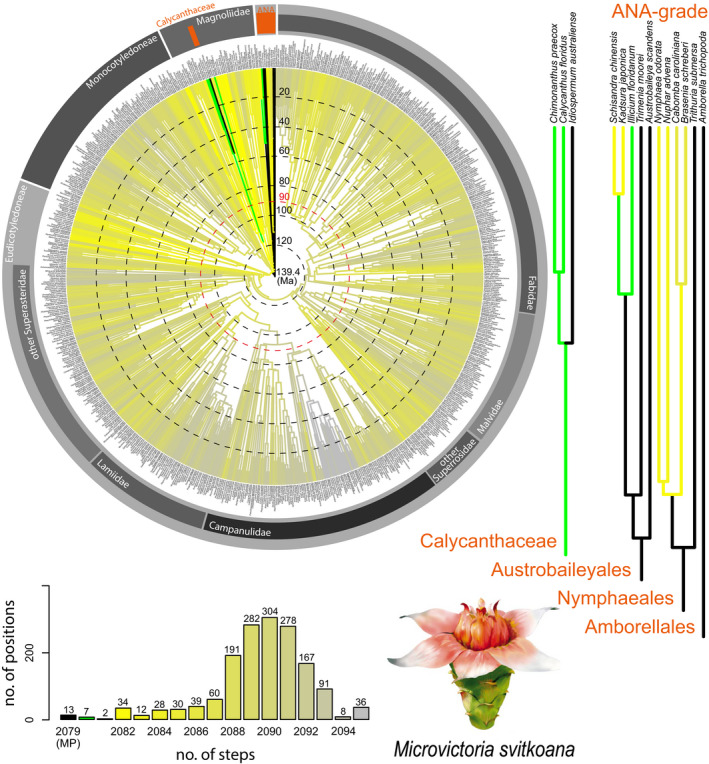
Results for Microvictoria svitkoana from angiosperm‐wide molecular backbone analyses using maximum parsimony, based on 20 floral characters that could be scored for this fossil. The color coding, the time scale superimposed on the circular chronogram, and the sources of the backbone tree and morphological data set of extant taxa are as in Figure 1. The drawing in the lower right shows a reconstruction of the complex flowers of M. svitkoana (reproduced from Crepet et al., 2004, with permission of Wiley). The positions of the ANA grade (Amborellales, Nymphaeales, and Austrobaileyales) and of Calycanthaceae (Laurales), respectively, are indicated by orange bars in the circular chronogram. The pink circle indicates the approximate age of M. svitkoana according to Gandolfo et al. (2004).
FIGURE 4.
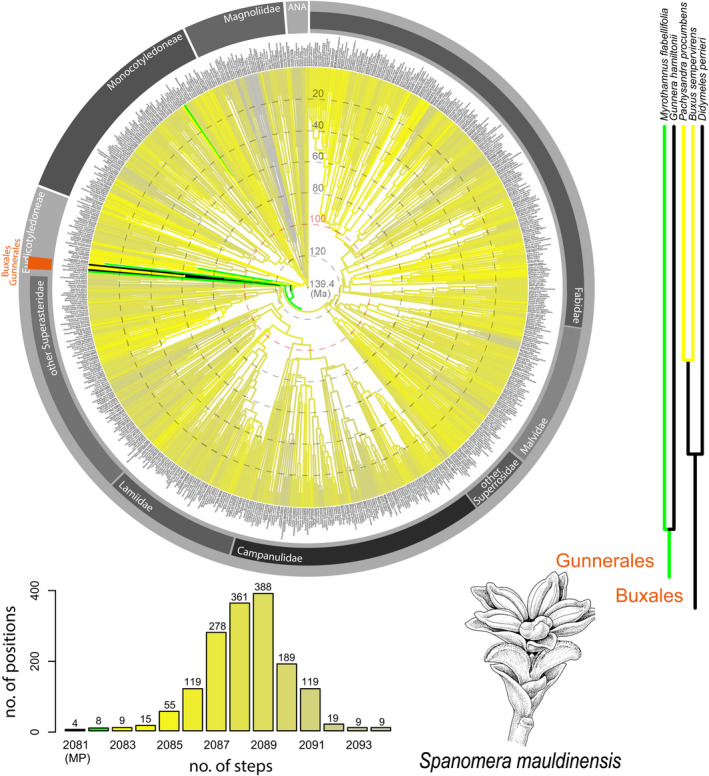
Results for Spanomera mauldinensis from angiosperm‐wide molecular backbone analyses using maximum parsimony, based on 25 floral characters that could be scored for this fossil. The color coding, the time scale superimposed on the circular chronogram, and the sources of the backbone tree and morphological data set of extant taxa are as in Figure 1. The drawing in the lower right shows a female and a male flower of S. mauldinensis (reproduced from Friis et al., 2006, with permission of Elsevier). The position of the Gunnerales and Buxales is indicated by an orange bar in the circular chronogram. The pink circle indicates the approximate age of S. mauldinensis according to Drinnan et al. (1991).
Chloranthistemon endressii
Chloranthistemon endressii was originally established on the basis of a few charcoalified or lignitized three‐lobed androecia and androecial fragments from Late Santonian/Early Campanian sediments in southern Sweden (Crane et al., 1989). Later, more complete specimens including gynoecial organs where recovered from the same locality (Eklund et al., 1997). Because the flowers of C. endressii have no perianth, our data set for this species comprises only 12 characters scored for the androecium and gynoecium characters (Appendix S1). In spite of the relatively low number of characters, our analysis places C. endressii with a single MP position unequivocally within Chloranthaceae (Fig. 1). Chloranthistemon endressii shares with Chloranthaceae a unique combination of floral traits, including the absence of a perianth, extensive connective tissue, and valvate anther dehiscence (Friis et al., 2011). The peculiar three‐lobed androecium attached to the upper part of the ovary (a feature not included in our data set) of C. endressii closely resembles that of an extant representative of the genus Chloranthus, and a previous phylogenetic analysis focused on extant and fossil Chloranthaceae (Eklund, 1999) placed C. endressii in a nested position among extant Chloranthus species. Similarly, the phylogenetic analyses by Doyle and Endress (2018), which are based on a taxon sample spanning early‐diverging angiosperms and early‐diverging eudicots, place Chloranthistemon most parsimoniously on the stem of Chloranthus. In our analysis, the most parsimonious position of C. endressii corresponds with the terminal branch of Sarcandra chloranthoides rather than with the representative of the genus Chloranthus, C. japonicus, in our data set. This may be explained by the low sampling density of Chloranthaceae in our data set (one species from each of the four extant genera) and the absence of Chloranthaceae‐specific traits such as “persistent cup‐shaped floral bract” or “androecium‐lobation” (Eklund, 1999; Eklund et al., 2004). In spite of the lack of any such traits, our angiosperm‐wide approach largely agrees with earlier phylogenetic analyses of C. endressii.
Dakotanthus cordiformis
Dakotanthus cordiformis was first described only informally (Basinger and Dilcher, 1984) and was long referred to as the “Rose Creek flower” (e.g., Schönenberger and von Balthazar, 2006), based on the Mid‐Cretaceous “Rose Creek” locality in Nebraska, from which numerous specimens were collected over the years. A formal description was published only recently (Manchester et al., 2018), combining the original material with fruit specimens that had been published much earlier under now synonymized names such as Carpites cordiformis and Nordenskioldia borealis (Lesquereux, 1892). The holotype of D. cordiformis is an isolated fruit valve from a locality in Ottawa County, Kansas, USA (Manchester et al., 2018). However, the species description is based on numerous flowers, flower fragments, and fruits preserved as compressions, impressions, and sandstone casts recovered at multiple localities from the mid‐Cretaceous (late Albian to early Cenomanian) Dakota Formation, which represents deposition along the eastern margin of the North American Cretaceous Epicontinental Seaway (Manchester et al., 2018). Although the quality of preservation of individual specimens is relatively low (compared to charcoalified fossils), the high number of specimens has allowed for a detailed reconstruction of floral morphology, and we were able to run our analysis with all but one of the 30 floral characters in our data set (i.e., all except number of ovules; Appendix S1). Our analysis identifies relatively many (23) MP positions for D. cordiformis (Fig. 2). However, MP positions are clearly restricted to the rosid clade. The flowers of D. cordiformis display a combination of traits that we have inferred as ancestral for Pentapetalae (Sauquet et al., 2017) and that have remained common in Rosidae (pentamery, a differentiated perianth, free petals, a diplostemonous androecium with tricolporate pollen grains, a syncarpous gynoecium with five carpels). However, given that all of these characters are widespread across the Rosidae and that most major rosid clades lack clear structural synapomorphies (Endress and Matthews, 2006; Soltis et al., 2018), it is not surprising that earlier hypotheses have associated D. cordiformis with different rosid lineages (Basinger and Dilcher, 1984; Richardson et al., 2000; Calvillo‐Canadell and Cevallos‐Ferriz, 2007). The latest and most complete interpretation of the floral morphology and phylogenetic position (without explicit phylogenetic analysis) of D. cordiformis points out similarities with flowers of the extant family Quillajaceae in the order Fabales (Manchester et al., 2018). In particular, the authors suggested two potential synapomorphies (crescent‐shaped nectary lobes and stamen positioning at nectary pads and sinuses) shared between the single extant genus Quillaja and D. cordiformis. It is noteworthy that while our data set does not comprise these two characters, our analysis still identifies Quillajaceae and other fabalean branches as possible MP positions of D. cordiformis (Fig. 2). Our analysis, therefore, agrees with Manchester et al.’s (2018) statement that D. cordiformis may represent a “basal lineage within the Fabales.” However, our analysis identifies a few more MP positions in Rosales (also part of fabids) and in Sapindales (malvids) and highlights the need for future comparative work on this fossil taxon.
Microvictoria svitkoana
Microvictoria svitkoana is based on a few charcoalified flowers from Turonian sediments in New Jersey, USA (Gandolfo et al., 2004). The specimens are very well preserved and we were able to score 20 characters in our data set (several characters, including the number of organ whorls and merism of perianth and androecium, are inapplicable because the flowers have spiral phyllotaxis). Microvictoria svitkoana has numerous floral organs, and the organization and construction of the flowers are highly complex. Because of this and the lack of cross sections or tomographic analyses in the original study describing the flowers (Gandolfo et al., 2004), some of M. svitkoana’s floral traits are difficult to establish and the original interpretation of its floral structure has partly been questioned (Endress, 2008; Friis et al., 2009, 2011; Doyle and Endress, 2014). On the basis of Endress (2008), and after carefully reviewing additional images of the fossil that are provided on the website of the Herbarium of the L.H. Bailey Hortorium at Cornell University (http://tcf.bh.cornell.edu/), we have interpreted and scored (see Appendix S1) some of the floral traits of M. svitkoana differently from the original description by Gandolfo et al. (2004). In particular, we have scored ovary position as superior (interpreted as inferior by Gandolfo et al., 2004; but see Doyle and Endress, 2014), anther attachment as basifixed (not described in Gandolfo et al., 2004), anther connective with long extension (not described in Gandolfo et al., 2004), and the gynoecium as apocarpous (no clear interpretation in Gandolfo et al., 2004).
Gandolfo et al. (2004) included M. svitkoana in a phylogenetic analysis, in which it resulted in a clade with the extant Nymphaeaceae genera Euryale, Nymphaea, and Victoria. However, as pointed out by Yoo et al. (2005) and Doyle and Endress (2014), the analysis of Gandolfo et al. (2004) could not test the hypothesis that M. svitkoana belonged to any other angiosperm clade because the taxon sampling of that analysis was restricted to Nymphaeales (not including Hydatellaceae at that time). Our angiosperm‐wide phylogenetic analysis of M. svitkoana finds 12 MP positions among members of the ANA grade (Amborellales, Nymphaeales, Austrobaileyales). Importantly, none of these 12 MP positions belongs to the clade with Cabombaceae and Nymphaeaceae. An additional MP position is present in Calycanthaceae in the magnoliid order Laurales (Fig. 3). Endress (2008) and Doyle and Endress (2014) hypothesized that the phylogenetic position of M. svitkoana is most likely not with Nymphaeaceae and not even with Nymphaeales, albeit without suggesting any alternative position. A phylogenetic placement among ANA‐grade lineages, as suggested here, seems plausible given the spiral floral organization and the laminar stamens of M. svitkoana. Both of these characters are widespread within the ANA grade. However, a closer relationship of M. svitkoana with the Nymphaeceae, as suggested by Gandolfo et al. (2004), seems unlikely, given clear differences in floral phyllotaxis (Nymphaeaceae and Cambombaceae both have whorled flowers) and other floral traits (see also Endress, 2008; Friis et al., 2011). To place M. svitkoana in a position in Nymphaeaceae would require several more steps than the MP positions suggested here. A close relationship to Calycanthaceae has, to our knowledge, not been suggested so far. Such a relationship is supported by various floral traits, including a floral cup with tepals and stamens placed on its rim (not scored in this study), relatively high numbers of spirally arranged perianth organs and stamens, inner staminodes (not scored), and the presence of a sheltered floral chamber (e.g., in extant Idiospermum [Worboys and Jackes, 2005] and Sinocalycanthus [Staedler et al., 2007], not scored).
It is not yet possible to draw a final conclusion on the phylogenetic relationships of M. svitkoana. Its flowers are highly complex and its floral organization and construction, in particular with respect to the gynoecium, are still not sufficiently well understood. We hope that our analyses will inspire further morphological investigations of these fossil flowers. Modern tomographic analyses would certainly help clarify many of the remaining issues and allow for a more detailed comparison with Calycanthaceae and other extant and fossil taxa.
Spanomera mauldinensis
Spanomera mauldinensis is based on series of charcoalified or lignitized inflorescence fragments, flowers, and dispersed floral organs extracted from mid‐Cretaceous (early Cenomanian) sediments at the Mauldin Mountain locality in North America (Drinnan et al., 1991). Floral structure is well understood and we were able to analyze 25 of the 30 floral characters in our data set (Table S1). Our analysis identifies four MP positions, three MP positions in Buxales and one in Gunnerales for S. mauldinensis (Fig. 4). The unisexual and basically dimerous flowers (seemingly pentamerous specimens such as the one shown in Figure 4 are also interpreted as dimerous, but with two organs instead of one in the adaxial position of one of the tepal whorls and one of the stamen whorls, respectively; Drinnan et al., 1991) of S. mauldinensis are closely similar to those of extant Buxaceae (von Balthazar and Endress, 2002) and, accordingly, were suggested to be closely related to Buxaceae when first described (Drinnan et al., 1991). Next to Buxaceae, the latter study also pointed out similarities between Spanomera and Myrothamnaceae, a family now included in Gunnerales. A phylogenetic analysis focused on basal angiosperms (also including basal eudicots, but not Gunnerales) also supported a close relationship of Spanomera with extant Buxaceae and, in addition, suggested possible relationships to Trochodendrales, another lineage among basal eudicots (Doyle and Endress, 2010). Trochodendrales, Buxales, and Gunnerales are successive sister groups to the remainder of the eudicots (i.e., to the Pentapetalae; Soltis et al., 2011; Ruhfel et al., 2014; APG IV, 2016). Based on the results of our analysis, the age of S. mauldinensis (~100 million years; Drinnan et al., 1991), and the estimated crown group ages of Buxales and Gunnerales (median ages 85.1 and 104.6 million years, respectively; Magallón et al., 2015), there are three plausible hypotheses for the phylogenetic position of S. mauldinensis. It may either belong to a separate but now extinct lineage among early‐diverging eudicots, it may belong to the stem lineages of either Buxales or Gunnerales, or it may even be part of either crown group. A data set with denser taxon sampling among basal eudicots and additional traits that are particularly informative in this part of the angiosperm tree (e.g., additional pollen characters, stigma shape, fruit type, and inflorescence characters; see Drinnan et al., 1990; Doyle and Endress, 2010) might help to fine tune hypotheses on the exact phylogenetic placement of S. mauldinensis.
GENERAL DISCUSSION
Benefits of the Paleo‐eFLOWER approach
In addition to introducing a useful and easy‐to‐handle tool that can be incorporated into the process of placing fossil flowers within the phylogenetic framework of the angiosperms, our phyloscan approach offers several other advantages. First, it allows for the possibility of highlighting potential assignments to deep branches in the phylogeny, which are in accordance with the Cretaceous age of the fossils tested here, rather than crown group families and orders. Hence, the capability of a method to relate fossils to internal, deeper branches (and their character combinations) in a phylogeny may allow for a more comprehensive discussion of relationships and characters going beyond a comparison with only extant (terminal) taxa. However, we caution against a direct comparison of fossil age with estimated ages of the nodes defining branches with most parsimonious positions as an acceptable measure of plausibility. This is because of (1) the risk of circularity (many of the early fossil records considered here have been used as calibration points to produce time trees), (2) the large uncertainties remaining in divergence times across the phylogeny of angiosperms, and (3) the related unsolved question of the crown‐group age of angiosperms (Magallón et al., 2015; Foster et al., 2017; Barba‐Montoya et al., 2018; Coiro et al., 2019). Only integrated approaches estimating fossil relationships simultaneously with divergence times based on combined morphological and molecular data sets (i.e., “tip‐dating” or “total‐evidence dating” analyses; e.g., Ronquist et al., 2012; Gavryushkina et al., 2017) provide a satisfying avenue to allow fossil age to contribute to the reconstruction of fossil relationships.
Second, angiosperm‐wide analyses such as ours allow not only for an explicit test of phylogenetic relationships, but also for the simultaneous identification of alternative, equally or perhaps somewhat less parsimonious positions for any given fossil (e.g., Dakotanthus cordiformis, Fig. 2; Microvictoria svitkoana, Fig. 3; and Paradinandra suecica, Appendix S3; see also Appendix S4 for a compilation of all phyloscan trees). Such potential alternative positions may easily be missed without such a broad‐scale phylogenetic approach, but may be helpful for identifying additional taxa that could be considered in comparative morphological investigations of a given fossil and potential extant relatives.
Third, due to incomplete preservation and taphonomic transformation of fossils, the interpretation of some characters is likely to be difficult and equivocal in many cases, which suggests the desirability of testing the influence of alternative character scorings (e.g., Doyle et al., 2008) or of alternative subsets of characters (e.g., von Balthazar et al., 2008) on phylogenetic analyses. Likewise, the discovery of additional specimens of a fossil taxon or alternative interpretations based on new investigations (e.g., D. cordiformis; Manchester et al., 2018) might make it desirable to adapt earlier scorings and to rerun phylogenetic analyses at a later stage. Our approach, with a flexible data set and database (PROTEUS), in which each data record is unequivocally referenced (Sauquet et al., 2017; Sauquet, 2019), makes it easy to replicate and verify any part of the analyses. This open and transparent approach to which the eFLOWER initiative is committed has already proven fruitful in allowing constructive discussion and further improvement of the data set (Sauquet et al., 2017, 2018; Sokoloff et al., 2018).
Impact of phylogenetic uncertainty
Broad‐scale phylogenomic analyses (e.g., One Thousand Plant Transcriptomes Initiative, 2019) and other molecular phylogenetic studies focusing on particular subclades of angiosperms with much denser taxon sampling (e.g., Ericales; Rose et al., 2018) have shown that some higher‐level relationships are still not well supported and might change in future studies. To assess the potential impact of how such future improvements in angiosperm phylogenetics might affect the outcome of this study, we repeated all analyses using four alternative molecular backbones derived from our previous study (Sauquet et al., 2017). Our results summarized in Appendix S5 indicate remarkable robustness across the five backbones. For instance, the number and location of MP positions remained unchanged in five fossils (albeit with slight variations in the number of MP+1 positions in three of these fossils). For the remaining fossils, we observed variations in the number of MP positions, but their location in the phylogeny of angiosperms remained entirely consistent across backbones. These results are consistent with previous work using a similar approach (e.g., Doyle & Endress, 2010) and of comparable robustness as ancestral state reconstructions of floral traits using a similar data set and the same five trees in our previous study (Sauquet et al., 2017). They suggest that the information content in the floral traits used here, and their phylogenetic pattern of variation among angiosperms, is sufficient for a reliable initial assessment of potential relationships using the phyloscan approach. While we consider these results very encouraging, we caution that these tests do not capture the full breadth of phylogenetic uncertainty that remains across angiosperms. Hence, additional work will be required, including total‐evidence analyses (e.g., Manos et al., 2007), which are well suited for this purpose.
Clade‐specific analyses based on the Paleo‐eFLOWER data set
Once an initial angiosperm‐wide phyloscan analysis has been conducted and has helped identify a particular angiosperm subclade (an order or a clade with several orders) as the most likely phylogenetic “home” for a given fossil, it might be advisable to build a clade‐specific data set (or use an existing one) with denser taxon sampling and additional, clade‐specific characters, with the goal of placing the fossil more accurately within that clade. Furthermore, while such clade‐focused data sets may be built entirely independently from the angiosperm‐wide data set presented here, we propose that using the PROTEUS and eFLOWER framework to develop such data sets would not only allow one to build on existing data (where taxa and characters overlap), but, in turn, would also contribute toward a much more densely sampled angiosperm‐wide data set that will allow improved future phyloscan analyses. Although the PROTEUS database is not yet directly accessible, it is our aim to make it open to all in the future.
Current limitations and future challenges for Paleo‐eFLOWER
The need for more focused, clade‐specific data sets and analyses mentioned above also reflects some of the limitations of our current data set. In accordance with our original goal to reconstruct character evolution across angiosperms as a whole (Sauquet et al., 2017), we have restricted character selection to characters that are broadly applicable and meaningful across angiosperms. Accordingly, most of the characters correspond to basic parameters of floral organization (sensu Endress, 1996), such as the basic structure, the number, and the arrangement of floral organs. We have not included characters at the level of floral construction and floral mode (e.g., size and shape of flowers and flower organs, indumentum). These characters tend to be highly variable even among closely related taxa and therefore provide very limited phylogenetic signal for sparsely sampled angiosperm‐wide analyses. This means that our phyloscan analyses do not make use of the full breadth of morphological information available for some of the fossils. For instance, for various Cretaceous taxa referred to the asterid order Ericales, peculiar peltate or likely glandular, multicellular trichomes have been described—for example, in Glandulocalyx (Schönenberger et al., 2012), Parasaurauia (Keller et al., 1996), Rariglanda (Martínez et al., 2016), Raritaniflora (Crepet et al., 2013), and Teuschestanthes (Crepet et al., 2018). Because there are currently no indumentum characters in our data set, our analyses cannot make use of these potentially phylogenetically informative characters. Likewise, among the many fossil flowers from the Cretaceous presumably related to magnoliid order Laurales—for example, Cohongarootonia (von Balthazar et al., 2011), Lauranthus (Takahashi et al., 2001), Mauldinia (Drinnan et al., 1990; Viehofen et al., 2008), and Neusenia (Eklund, 1999)—the presence or absence of nectariferous glandular appendages on the stamen filaments is a key character that has been shown to be parsimony informative within the clade Lauraceae (von Balthazar et al., 2011) and should be part of any phylogenetic analysis focused on Lauraceae and the order Laurales as a whole. Further room for improvement in our data set pertains also to entire character complexes, such as features of the pollen wall, the fruit, and the seed coat, which are readily observable in many fossil taxa (Friis et al., 2011) and are also potentially informative across broader taxonomic groups.
Another, perhaps less obvious limitation of our data set pertains to the fact that it only includes characters states of extant taxa and, therefore, cannot consider any extinct character states—that is, states found only in a now extinct lineage, such as the unique, cladode‐like inflorescence units of Mauldinia (Drinnan et al., 1990), which are not present in any extant representative of the Lauraceae (Friis et al., 2011). Accordingly, fossils that are part of completely or largely extinct lineages (and therefore also with many potentially extinct features) may be problematic to place with our approach, which could produce incorrect, potentially misleading results in these circumstances. At the same time, there is thus far no widely accepted, formally named extinct family or order of angiosperms that is clearly distinct from all extant families and that includes more than one or two fossils. This is in stark contrast with the fossil record of non‐angiospermous seed plants, where many extinct higher taxa (e.g., Bennettitales, Caytoniales, Glossopteridales) that are clearly distinct from any extant seed plant lineage have been described (Doyle, 2006, 2018; Rothwell et al., 2009; Friis et al., 2011). The apparent lack of similarly distinct cases within the crown group of angiosperms might be a descriptive bias (see Sauquet and Magallón, 2018) but also suggests that there is not yet any strong evidence that such a large, entirely extinct, morphologically distinct clade of angiosperms ever existed. Among the 10 fossils that we have included in this study, there is no clear‐cut case of a fossil that might be part of such an extinct lineage. Even if this type of limitation in our approach is currently rather speculative, we believe that this needs to be kept in mind during future analyses.
A final limitation of our current analyses has to do with our sampling of extant taxa and is linked with the issue of missing characters or character states raised just above. Our data set currently comprises data for 792 extant species representing all but one of the 64 currently recognized angiosperm orders (according to APG IV, 2016), but only 372 (~86%) of the 435 families (according to Stevens, 2001, onward; see also Sauquet et al., 2017). Even if our data set is focused on characters of floral organization, which tend to be relatively stable across larger lineages, it is still possible that our data set misses crucial aspects of floral diversity in parts of the angiosperm tree that are currently underrepresented (but see below).
CONCLUSIONS AND PERSPECTIVES
In summary, the present study, which makes use of a carefully compiled and curated angiosperm‐wide data set of floral characters, allows testing hypotheses on the relationships of fossils through comprehensive phylogenetic analyses. In our view, this is a crucial step forward in our attempts to integrate knowledge of extant and extinct angiosperms and their flowers (see also Friis et al., 2011; Sauquet and Magallón, 2018). This approach not only allows for reproducible tests on the phylogenetic position of fossil flowers, but will also ultimately contribute to a better understanding of extant floral morphology and the evolutionary history of angiosperms in general. The next step in the development of Paleo‐eFLOWER, on which we are now working, is to alleviate the current potential limitation of our taxon sampling.
We are well aware of the possibility that our proposed phyloscanning approach may seduce people to draw premature systematic conclusions. Therefore, we would like to stress here that any phylogenetic analysis intended to place a fossil taxon needs to be preceded by a careful morphological investigation and description of the fossil specimens at hand and be followed by an equally careful comparison of the fossils with their hypothesized closest extant and extinct relatives, taking into account all the characters that are available. If such a course of action is followed, we are convinced that our phyloscan approach will provide a most useful, objective tool that allows the formulation of novel hypotheses on the phylogenetic position of fossil flowers. A particularly fruitful use of our approach may be to apply it to fossils that have no obvious position on first sight. This seems to be especially the case for fossils with likely affinities to the eudicots (e.g., Dakotanthus cordiformis and Paradinandra suecica). The sheer size and the overwhelming morphological diversity of the eudicot clade (e.g., Endress, 2010) make it very difficult to systematically place fossils solely on the basis of qualitative character comparisons among a few selected taxa or even the use of restricted phylogenetic analyses. Broad‐scale analyses such as the ones we propose here might help to develop sound and well‐supported hypotheses even for difficult fossils.
AUTHOR CONTRIBUTIONS
J.S., M.v.B., S.M., and H.S. designed the study. B.A. and C.P. contributed new pollen data to the data set of extant taxa. J.S., M.v.B., A.L.M, S.M., and H.S. coordinated and curated the extant data set and scored the fossil taxa. S.M. provided the backbone phylogeny. H.S. conducted all analyses. J.S. wrote the first draft of the paper, with subsequent contributions from all co‐authors.
Supporting information
APPENDIX S1. Appendix S1 contains the full list of referenced data records scored in PROTEUS and the final matrix used for all analyses in this study.
APPENDIX S2. Appendix S2 comprises information on scoring philosophy as well as definitions and explanations of all floral characters scored for this study. It is a complemented and updated version of the Supplementary Methods published online as part of Sauquet et al. (2017).
APPENDIX S3. Appendix S3 contains Results and Discussion (including figures) for the six fossils not detailed in the main text.
APPENDIX S4. Appendix S4 contains a compilation of the molecular backbone trees with color‐coded results of phyloscan parsimony analyses for the 10 fossils analyzed in this study.
APPENDIX S5. Appendix S5 contains an overview and comparison of results of phyloscan analyses using five distinct phylogenetic backbones.
Acknowledgments
We thank P. Crane, president of the Oak Spring Garden Foundation (OSGF, Virginia, USA) and the participants of the second eFLOWER Summer School, held at OSGF during September 2018, for valuable discussions on our Paleo‐eFLOWER initiative. We thank U. Schachner and M. Lachmayer for help with figures, tables, and references during manuscript preparation; R. Buchner for help with administering the eFLOWER server; B. Crepet, J. Doyle, P. Endress, and E. M. Friis for critical input and helpful discussions during the early stages of this project; and two anonymous reviewers for their constructive and valuable comments on our study.
Schönenberger, J. , von Balthazar M., López Martínez A., Albert B., Prieu C., Magallón S., and Sauquet H.. 2020. Phylogenetic analysis of fossil flowers using an angiosperm‐wide data set: proof‐of‐concept and challenges ahead. American Journal of Botany 107(10): 1433–1448.
Contributor Information
Jürg Schönenberger, Email: juerg.schoenenberger@univie.ac.at.
Hervé Sauquet, Email: herve.sauquet@gmail.com.
Data Availability
All data records from PROTEUS used in this paper are provided as Appendix S1. R code and input files (trees and data matrix) used in the analyses are provided separately via the following public link: https://github.com/eflowerproject/paleoeflower.
LITERATURE CITED
- APG IV . 2016. An update of the Angiosperms Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181: 1–20. [Google Scholar]
- Atkinson, B. A. 2018. The critical role of fossils in inferring deep‐node phylogenetic relationships and macroevolutionary patterns in Cornales. American Journal of Botany 105: 1401–1411. [DOI] [PubMed] [Google Scholar]
- Barba‐Montoya, J. , dos Reis M., Schneider H., Donoghue P. C., and Yang Z.. 2018. Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous Terrestrial Revolution. New Phytologist 218: 819–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basinger, J. F. , and Dilcher D. L.. 1984. Ancient bisexual flowers. Science 224: 511–513. [DOI] [PubMed] [Google Scholar]
- Calvillo‐Canadell, L. , and Cevallos‐Ferriz S. R.. 2007. Reproductive structures of Rhamnaceae from the Cerro del Pueblo (Late Cretaceous, Coahuila) and Coatzingo (Oligocene, Puebla) Formations, Mexico. American Journal of Botany 94: 1658–1669. [DOI] [PubMed] [Google Scholar]
- Coiro, M. , Doyle J. A., and Hilton J.. 2019. How deep is the conflict between molecular and fossil evidence on the age of angiosperms? New Phytologist 223: 83–99. [DOI] [PubMed] [Google Scholar]
- Crane, P. R. , Friis E. M., and Pedersen K. R.. 1989. Reproductive structure and function in Cretaceous Chloranthaceae. Plant Systematics and Evolution 165: 211–226. [Google Scholar]
- Crane, P. R. , Friis E. M., and Pedersen K. R.. 1995. The origin and early diversification of angiosperms. Nature 374: 27. [Google Scholar]
- Crepet, W. L. 1996. Timing in the evolution of derived floral characters: Upper Cretaceous (Turonian) taxa with tricolpate and tricolpate‐derived pollen. Review of Palaeobotany and Palynology 90: 339–359. [Google Scholar]
- Crepet, W. L. , and Nixon K. C.. 1998a. Fossil Clusiaceae from the Late Cretaceous (Turonian) of New Jersey and implications regarding the history of bee pollination. American Journal of Botany 85: 1122–1133. [PubMed] [Google Scholar]
- Crepet, W. L. , and Nixon K. C.. 1998b. Two new fossil flowers of magnoliid affinity from the Late Cretaceous of New Jersey. American Journal of Botany 85: 1273–1288. [PubMed] [Google Scholar]
- Crepet, W. L. , Nixon K. C., and Gandolfo M. A.. 2004. Fossil evidence and phylogeny: the age of major angiosperm clades based on mesofossil and macrofossil evidence from Cretaceous deposits. American Journal of Botany 91: 1666–1682. [DOI] [PubMed] [Google Scholar]
- Crepet, W. L. , Nixon K. C., and Daghlian C. P.. 2013. Fossil Ericales from the Upper Cretaceous of New Jersey. International Journal of Plant Sciences 174: 572–584. [Google Scholar]
- Crepet, W. L. , Nixon K. C., and Weeks A.. 2018. Mid‐Cretaceous angiosperm radiation and an asterid origin of bilaterality: diverse and extinct “Ericales” from New Jersey. American journal of Botany 105: 1412–1423. [DOI] [PubMed] [Google Scholar]
- Doyle, J. A. 2006. Seed ferns and the origin of angiosperms. The Journal of the Torrey Botanical Society 133: 169–210. [Google Scholar]
- Doyle, J. A. 2018. Phylogenetic analyses and morphological innovations in land plants. Annual Plant Reviews book series, Volume 45: The Evolution of Plant Form: 1–50. [Google Scholar]
- Doyle, J. A. , and Endress P. K.. 2000. Morphological phylogenetic analysis of basal angiosperms: comparison and combination with molecular data. International Journal of Plant Sciences 161: S121–S153. [DOI] [PubMed] [Google Scholar]
- Doyle, J. A. , and Endress P. K.. 2010. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: Magnoliidae and eudicots. Journal of Systematics and Evolution 48: 1–35. [Google Scholar]
- Doyle, J. A. , and Endress P. K.. 2014. Integrating Early Cretaceous fossils into the phylogeny of living angiosperms: ANITA lines and relatives of Chloranthaceae. International Journal of Plant Sciences 175: 555–600. [Google Scholar]
- Doyle, J. A. , and Endress P. K.. 2018. Phylogenetic analyses of Cretaceous fossils related to Chloranthaceae and their evolutionary implications. The Botanical Review 84: 156–202. [Google Scholar]
- Doyle, J. A. , Endress P. K., and Upchurch G. R.. 2008. Early Cretaceous monocots: a phylogenetic evaluation. Acta Musei Nationalis Pragae, Series B, Historia Naturalis 64: 59–87. [Google Scholar]
- Drinnan, A. N. , Crane P. R., Friis E. M., and Pedersen K. R.. 1990. Lauraceous flowers from the Potomac Group (mid‐Cretaceous) of eastern North America. Botanical Gazette 151: 370–384. [Google Scholar]
- Drinnan, A. N. , Crane P. R., Friis E. M., and Pedersen K. R.. 1991. Angiosperm flowers and tricolpate pollen of buxaceous affinity from the Potomac Group (mid‐Cretaceous) of eastern North America. American Journal of Botany 78: 153–176. [Google Scholar]
- Eklund, H. 1999. Big survivors with small flowers: fossil history and evolution of Laurales and Chloranthaceae. Ph.D. dissertation, University Uppsala, Sweden. Acta Universitatis Upsaliensis. [Google Scholar]
- Eklund, H. , Doyle J. A., and Herendeen P. S.. 2004. Morphological phylogenetic analysis of living and fossil Chloranthaceae. International Journal of Plant Sciences 165: 107–151. [Google Scholar]
- Eklund, H. , Friis E. M., and Pedersen K. R.. 1997. Chloranthaceous floral structures from the Late Cretaceous of Sweden. Plant Systematics and Evolution 207: 13–42. [Google Scholar]
- Endress, P. K. 1996. Diversity and evolutionary biology of tropical flowers. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Endress, P. K. 2008. Perianth biology in the basal grade of extant angiosperms. International Journal of Plant Sciences 169: 844–862. [Google Scholar]
- Endress, P. K. 2010. Flower structure and trends of evolution in eudicots and their major subclades. Annals of the Missouri Botanical Garden 97: 541–583. [Google Scholar]
- Endress, P. K. , and Doyle J. A.. 2009. Reconstructing the ancestral angiosperm flower and its initial specializations. American Journal of Botany 96: 22–66. [DOI] [PubMed] [Google Scholar]
- Endress, P. K. , and Matthews M. L.. 2006. First steps towards a floral structural characterization of the major rosid subclades. Plant Systematics and Evolution 260: 223–251. [Google Scholar]
- Foster, C. S. , Sauquet H., Van der Merwe M., McPherson H., Rosetto M., and Ho S. Y.. 2017. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Systematic Biology 66: 338–351. [DOI] [PubMed] [Google Scholar]
- Friis, E. M. , and Pedersen K. R.. 2011. Canrightia resinifera gen. et sp. nov., a new extinct angiosperm with Retimonocolpites‐type pollen from the Early Cretaceous of Portugal: Missing link in the eumagnoliid tree? Grana 50: 3–29. [Google Scholar]
- Friis, E. M. , Crane P. R., and Pedersen K. R.. 2011. Early flowers and angiosperm evolution. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Friis, E. M. , Crane P. R., and Pedersen K. R.. 2019. Extinct diversity among Early Cretaceous angiosperms: mesofossil evidence of early Magnoliales from Portugal. International Journal of Plant sciences 180: 93–127. [Google Scholar]
- Friis, E. M. , Eklund H., Pedersen K. R., and Crane P. R.. 1994. Virginianthus calycanthoides gen. et sp. nov.‐ a calycanthaceous flower from the Potomac Group (Early Cretaceous) of eastern North America. International Journal of Plant Sciences 155: 772–785. [Google Scholar]
- Friis, E. M. , Marone F., Pedersen K. R., Crane P. R., and Stampanoni M.. 2014. Three‐dimensional visualization of fossil flowers, fruits, seeds, and other plant remains using synchrotron radiation X‐ray tomographic microscopy (SRXTM): new insights into Cretaceous plant diversity. Journal of Paleontology 88: 684–701. [Google Scholar]
- Friis, E. M. , Pedersen K. R., and Crane P. R.. 2005. When Earth started blooming: insights from the fossil record. Current opinion in plant biology 8: 5–12. [DOI] [PubMed] [Google Scholar]
- Friis, E. M. , Pedersen K. R., and Crane P. R.. 2006. Cretaceous angiosperm flowers: innovation and evolution in plant reproduction. Palaeogeography, palaeoclimatology, palaeoecology 232: 251–293. [Google Scholar]
- Friis, E. M. , Pedersen K. R., and Schönenberger J.. 2003. Endressianthus, a new Normapolles‐producing plant genus of fagalean affinity from the Late Cretaceous of Portugal. International Journal of Plant Sciences 164: S201–S223. [Google Scholar]
- Friis, E. M. , Pedersen K. R., von Balthazar M., Grimm G. W., and Crane P. R.. 2009. Monetianthus mirus gen. et sp. nov., a nymphaealean flower from the Early Cretaceous of Portugal. International Journal of Plant Sciences 170: 1086–1101. [Google Scholar]
- Gavryushkina, A. , Heath T. A., Ksepka D. T., Stadler T., Welch D., and Drummond A. J.. 2017. Bayesian total‐evidence dating reveals the recent crown radiation of penguins. Systematic biology 66: 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfo, M. A. , Nixon K. C., and Crepet W. L.. 1998. Tylerianthus crossmanensis gen. et sp. nov. (aff. Hydrangeaceae) from the Upper Cretaceous of New Jersey. American Journal of Botany 85: 376–386. [PubMed] [Google Scholar]
- Gandolfo, M. A. , Nixon K. C., and Crepet W. L.. 2002. Triuridaceae fossil flowers from the Upper Cretaceous of New Jersey. American Journal of Botany 89: 1940–1957. [DOI] [PubMed] [Google Scholar]
- Gandolfo, M. A. , Nixon K. C., and Crepet W. L.. 2004. Cretaceous flowers of Nymphaeaceae and implications for complex insect entrapment pollination mechanisms in early angiosperms. Proceedings of the National Academy of Sciences 101: 8056–8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herendeen, P. S. , Doyle J. A., Endress P. K., and Takahashi M.. 2016. Cecilanthus polymerus, a novel multiparted flower from the mid‐Cretaceous Rocky Point locality, Maryland. Botany 94: 787–803. [Google Scholar]
- Herendeen, P. S. , Magallón‐Puebla S., Lupia R., Crane P. R., and Kobylinska J.. 1999. A preliminary conspectus of the Allon flora from the Late Cretaceous (late Santonian) of central Georgia, USA. Annals of the Missouri Botanical Garden 86: 407–471. [Google Scholar]
- Hermsen, E. J. , Gandolfo M. A., Nixon K. C., and Crepet W. L.. 2003. Divisestylus gen. nov. (aff. Iteaceae), a fossil saxifrage from the Late Cretaceous of New Jersey, USA. American Journal of Botany 90: 1373–1388. [DOI] [PubMed] [Google Scholar]
- Keller, J. A. , Herendeen P. S., and Crane P. R.. 1996. Fossil flowers and fruits of the Actinidiaceae from the Campanian (Late Cretaceous) of Georgia. American Journal of Botany 83: 528–541. [Google Scholar]
- Larson‐Johnson, K. 2016. Phylogenetic investigation of the complex evolutionary history of dispersal mode and diversification rates across living and fossil Fagales. New Phytologist 209: 418–435. [DOI] [PubMed] [Google Scholar]
- Lesquereux, L. 1892. The flora of the Dakota Group: a posthumous work. Monographs of United States Geological Survey 17: 1–287. [Google Scholar]
- Magallón, S. , Gómez‐Acevedo S., Sánchez‐Reyes L. L., and Hernández‐Hernández T.. 2015. A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Manchester, S. R. , Dilcher D. L., Judd W. S., Corder B., and Basinger J. F.. 2018. Early Eudicot flower and fruit: Dakotanthus gen. nov. from the Cretaceous Dakota Formation of Kansas and Nebraska, USA. Acta Palaeobotanica 58: 27–40. [Google Scholar]
- Manos, P. S. , Soltis P. S., Soltis D. E., Manchester S. R., Oh S. H., Bell C. D., Dilcher D. L., and Stone D. E.. 2007. Phylogeny of extant and fossil Juglandaceae inferred from the integration of molecular and morphological data sets. Systematic Biology 56: 412–430. [DOI] [PubMed] [Google Scholar]
- Martínez, C. , Choo T. Y., Allevato D., Nixon K. C., Crepet W. L., Harbert R. S., and Daghlian C. P.. 2016. Rariglanda jerseyensis, a new ericalean fossil flower from the Late Cretaceous of New Jersey. Botany 94: 747–758. [Google Scholar]
- Martínez‐Millán, M. , Crepet W. L., and Nixon K. C.. 2009. Pentapetalum trifasciculandricus gen. et sp. nov., a thealean fossil flower from the Raritan Formation, New Jersey, USA (Turonian, Late Cretaceous). American Journal of Botany 96: 933–949. [DOI] [PubMed] [Google Scholar]
- Matsunaga, K. K. , Manchester S. R., Srivastava R., Kapgate D. K., and Smith S. Y.. 2019. Fossil palm fruits from India indicate a Cretaceous origin of Arecaceae tribe Borasseae. Botanical Journal of the Linnean Society 190: 260–280. [Google Scholar]
- Mendes, M. M. , Grimm G. W., Pais J., and Friis E. M.. 2014. Fossil Kajanthus lusitanicus gen. et sp. nov. from Portugal: floral evidence for Early Cretaceous Lardizabalaceae (Ranunculales, basal eudicot). Grana 53: 283–301. [Google Scholar]
- Mohr, B. A. , Coiffard C., and Bernardes‐de‐Oliveira M. E.. 2013. Schenkeriphyllum glanduliferum, a new magnolialean angiosperm from the Early Cretaceous of Northern Gondwana and its relationships to fossil and modern Magnoliales. Review of palaeobotany and Palynology 189: 57–72. [Google Scholar]
- One Thousand Plant Transcriptomes Initiative . 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieu, C. , Sauquet H., Gouyon P. H., and Albert B.. 2017. More than sixty origins of pantoporate pollen in angiosperms. American Journal of Botany 104: 1837–1845. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2019. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria: ). Website: http://www.R‐project.org/. [Google Scholar]
- Reyes, E. , Nadot S., von Balthazar M., Schönenberger J., and Sauquet H.. 2018. Testing the impact of morphological rate heterogeneity on ancestral state reconstruction of five floral traits in angiosperms. Scientific Reports 8: 9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, J. E. , Fay M. F., Cronk Q. C., Bowman D., and Chase M. W.. 2000. A phylogenetic analysis of Rhamnaceae using rbcL and trnL‐F plastid DNA sequences. American Journal of Botany 87: 1309–1324. [PubMed] [Google Scholar]
- Ronquist, F. , Klopfstein S., Vilhelmsen L., Schulmeister S., Murray D. L., and Rasnitsyn AP A. P.. 2012. A total‐evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Systematic Biology 61: 973–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, J. P. , Kleist T. J., Löfstrand S. D., Drew B. T., Schönenberger J., and Sytsma K. J.. 2018. Phylogeny, historical biogeography, and diversification of angiosperm order Ericales suggest ancient Neotropical and East Asian connections. Molecular Phylogenetics and Evolution 122: 59–79. [DOI] [PubMed] [Google Scholar]
- Rothwell, G. W. , Crepet W. L., and Stockey R. A.. 2009. Is the anthophyte hypothesis alive and well? New evidence from the reproductive structures of Bennettitales. American Journal of Botany 96: 296–322. [DOI] [PubMed] [Google Scholar]
- Ruhfel, B. R. , Stevens P. F., and Davis C. C.. 2013. Combined morphological and molecular phylogeny of the clusioid clade (Malpighiales) and the placement of the ancient rosid macrofossil Paleoclusia . International Journal of Plant Sciences 174: 910–936. [Google Scholar]
- Ruhfel, B. R. , Gitzendanner M. A., Soltis P. S., Soltis D. E., and Burleigh J. G.. 2014. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evolutionary Biology 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet, H. 2019. PROTEUS: A database for recording morphological data and fossil calibrations. Version 1.27. http://eflower.myspecies.info/proteus.
- Sauquet, H. , and Magallón S.. 2018. Key questions and challenges in angiosperm macroevolution. New Phytologist 219: 1170–1187. [DOI] [PubMed] [Google Scholar]
- Sauquet, H. , von Balthazar M., Doyle J. A., Endress P. K., Magallón S., Staedler Y., and Schönenberger J.. 2018. Challenges and questions in reconstructing the ancestral flower of angiosperms: A reply to Sokoloff et al. American Journal of Botany 105: 127–135. [DOI] [PubMed] [Google Scholar]
- Sauquet, H. , von Balthazar M., Magallón S., Doyle J. A., Endress P. K., Bailes E. J., Barroso de Morais E., et al. 2017. The ancestral flower of angiosperms and its early diversification. Nature Communications 8: 16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauquet, H. , Weston P. H., Anderson C. L., Barker N. P., Cantrill D. J., Mast A. R., and Savolainen V.. 2009. Contrasted patterns of hyperdiversification in Mediterranean hotspots. Proceedings of the National Academy of Sciences, USA 106: 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep, K. P. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 27: 592–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönenberger, J. 2005. Rise from the ashes–the reconstruction of charcoal fossil flowers. Trends in Plant Science 10: 436–443. [DOI] [PubMed] [Google Scholar]
- Schönenberger, J. , and Friis E. M.. 2001. Fossil flowers of ericalean affinity from the Late Cretaceous of Southern Sweden. American Journal of Botany 88: 467–480. [PubMed] [Google Scholar]
- Schönenberger, J. , and von Balthazar M.. 2006. Reproductive structures and phylogenetic framework of the rosids‐progress and prospects. Plant Systematics and Evolution 260: 87–106. [Google Scholar]
- Schönenberger, J. , von Balthazar M., Takahashi M., Xiao X., Crane P. R., and Herendeen P. S.. 2012. Glandulocalyx upatoiensis, a fossil flower of Ericales (Actinidiaceae/Clethraceae) from the Late Cretaceous (Santonian) of Georgia, USA. Annals of Botany 109: 921–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. Y. , and Stockey R. A.. 2007. Establishing a fossil record for the perianthless Piperales: Saururus tuckerae sp. nov. (Saururaceae) from the Middle Eocene Princeton Chert. American Journal of Botany 94: 1642–1657. [DOI] [PubMed] [Google Scholar]
- Smith, S. Y. , Collinson M. E., Rudall P. J., Simpson D. A., Marone F., and Stampanoni M.. 2009. Virtual taphonomy using synchrotron tomographic microscopy reveals cryptic features and internal structure of modern and fossil plants. Proceedings of the National Academy of Sciences 106: 12013–12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff, D. D. , Remizowa M. V., Bateman R. M., and Rudall P. J.. 2018. Was the ancestral angiosperm flower whorled throughout? American Journal of Botany 105: 5–15. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E. , Smith S. A., Cellinese N., Wurdack K. J., Tank D. C., Brockington S. F., Refulio‐Rodriguez N. F., et al. 2011. Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany 98: 704–730. [DOI] [PubMed] [Google Scholar]
- Soltis, D. , Soltis P., Endress P., Chase M. W., Manchester S., Judd W., Majure L., and Mavrodiev E.. 2018. Phylogeny and evolution of the angiosperms. University of Chicago Press, Chicago, IL, USA. [Google Scholar]
- Staedler, Y. M. , Weston P. H., and Endress P. K.. 2007. Floral phyllotaxis and floral architecture in Calycanthaceae (Laurales). International Journal of Plant Sciences 168: 285–306. [Google Scholar]
- Stevens, P. F. 2001. onward. Angiosperm Phylogeny Website. Version 14, July 2017 (and more or less continuously updated since).
- Swofford, D. L. 2002. PAUP*: Phylogenetic analysis using parsimony (*and other methods) version 4.0b10 for Macintosh, and version 4.0b10 for UNIX. Sinauer Associates, Sunderland. [Google Scholar]
- Takahashi, M. , Crane P. R., and Ando H.. 1999. Fossil flowers and associated plant fossils from the Kamikitaba locality (Ashizawa Formation, Futaba Group, lower Coniacian, upper Cretaceous) of Northeast Japan. Journal of Plant Research 112: 187. [Google Scholar]
- Takahashi, M. , Friis E. M., Uesugi K., Suzuki Y., and Crane P. R.. 2008. Floral evidence of Annonaceae from the Late Cretaceous of Japan. International Journal of Plant Sciences 169: 908–917. [Google Scholar]
- Takahashi, M. , Herendeen P. S., and Crane P. R.. 2001. Lauraceous fossil flower from the Kamikitaba locality (Lower Coniacian; Upper Cretaceous) in northeastern Japan. Journal of Plant Research 114: 429–434. [DOI] [PubMed] [Google Scholar]
- von Balthazar, M. , and Endress P. K.. 2002. Development of inflorescences and flowers in Buxaceae and the problem of perianth interpretation. International Journal of Plant Sciences 163: 847–876. [Google Scholar]
- von Balthazar, M. , Crane P. R., Pedersen K. R., and Friis E. M.. 2011. New flowers of Laurales from the Early Cretaceous (early to middle Albian) of eastern North America In Wanntorp L. and Ronse De Craene L. P.eds.], Flowers on the tree of life, 49–87. Cambridge University Press, Cambridge, UK. [Google Scholar]
- von Balthazar, M. , Pedersen K. R., Crane P. R., and Friis E. M.. 2008. Carpestella lacunata gen. et sp. nov., a new basal angiosperm flower from the Early Cretaceous (Early to Middle Albian) of eastern North America. International Journal of Plant Sciences 169: 890–898. [Google Scholar]
- Viehofen, A. , Hartkopf‐Fröder C., and Friis E. M.. 2008. Inflorescences and flowers of Mauldinia angustiloba sp. nov. (Lauraceae) from middle Cretaceous karst infillings in the Rhenish Massif, Germany. International Journal of Plant Sciences 169: 871–889. [Google Scholar]
- Worboys, S. J. , and Jackes B. R.. 2005. Pollination processes in Idiospermum australiense (Calycanthaceae), an arborescent basal angiosperm of Australia’s tropical rain forests. Plant Systematics and Evolution 251: 107–117. [Google Scholar]
- Wright, A. M. , Lloyd G. T., and Hillis D. M.. 2016. Modeling character change heterogeneity in phylogenetic analyses of morphology through the use of priors. Systematic Biology 65: 602–611. [DOI] [PubMed] [Google Scholar]
- Yoo, M. J. , Bell C. D., Soltis P. S., and Soltis D. E.. 2005. Divergence times and historical biogeography of Nymphaeales. Systematic Botany 30: 693–704. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Appendix S1 contains the full list of referenced data records scored in PROTEUS and the final matrix used for all analyses in this study.
APPENDIX S2. Appendix S2 comprises information on scoring philosophy as well as definitions and explanations of all floral characters scored for this study. It is a complemented and updated version of the Supplementary Methods published online as part of Sauquet et al. (2017).
APPENDIX S3. Appendix S3 contains Results and Discussion (including figures) for the six fossils not detailed in the main text.
APPENDIX S4. Appendix S4 contains a compilation of the molecular backbone trees with color‐coded results of phyloscan parsimony analyses for the 10 fossils analyzed in this study.
APPENDIX S5. Appendix S5 contains an overview and comparison of results of phyloscan analyses using five distinct phylogenetic backbones.
Data Availability Statement
All data records from PROTEUS used in this paper are provided as Appendix S1. R code and input files (trees and data matrix) used in the analyses are provided separately via the following public link: https://github.com/eflowerproject/paleoeflower.


