Significance
Skeletal muscle is composed of multinucleated myofibers that are essential for movement and metabolism. Duchenne muscular dystrophy (DMD) is a devastating disease that is caused by the lack of the dystrophin protein, which maintains the integrity of muscle membranes. The absence of dystrophin results in myofiber degeneration followed by regeneration until muscle stem cells are depleted. We generated a new DMD mouse model lacking exon 51 and then used single-nucleus RNA sequencing to reveal the transcriptional diversity of individual myofiber nuclei in dystrophic muscle compared to normal muscle. Our findings uncover disease-associated pathways responsible for muscle degeneration and regeneration that might ultimately be manipulated therapeutically and reveal an unrecognized regenerative myonuclear population associated with dystrophic muscle.
Keywords: myonuclei, myofibers, DMD mouse model, dystrophin, skeletal muscle
Abstract
Duchenne muscular dystrophy (DMD) is a fatal muscle disorder characterized by cycles of degeneration and regeneration of multinucleated myofibers and pathological activation of a variety of other muscle-associated cell types. The extent to which different nuclei within the shared cytoplasm of a myofiber may display transcriptional diversity and whether individual nuclei within a multinucleated myofiber might respond differentially to DMD pathogenesis is unknown. Similarly, the potential transcriptional diversity among nonmuscle cell types within dystrophic muscle has not been explored. Here, we describe the creation of a mouse model of DMD caused by deletion of exon 51 of the dystrophin gene, which represents a prevalent disease-causing mutation in humans. To understand the transcriptional abnormalities and heterogeneity associated with myofiber nuclei, as well as other mononucleated cell types that contribute to the muscle pathology associated with DMD, we performed single-nucleus transcriptomics of skeletal muscle of mice with dystrophin exon 51 deletion. Our results reveal distinctive and previously unrecognized myonuclear subtypes within dystrophic myofibers and uncover degenerative and regenerative transcriptional pathways underlying DMD pathogenesis. Our findings provide insights into the molecular underpinnings of DMD, controlled by the transcriptional activity of different types of muscle and nonmuscle nuclei.
As the largest tissue in the body, responsible for mobility and metabolism, skeletal muscle possesses many unique and fascinating features. Skeletal muscle is a multinucleated tissue, containing vast numbers of nuclei sharing a common cytoplasm as a result of fusion of mononucleated myoblasts, and it is among the most regenerative of all tissues (1). In response to injury and disease, resident myogenic stem cells, known as satellite cells, become activated and fuse with each other and with existing myofibers to regenerate the functional tissue (2, 3). While the process of skeletal muscle regeneration has been extensively studied, little is known of possible functional differences between nuclei within the common cytoplasm of the myofiber, nor is it understood how newly recruited nuclei from satellite cells might differ transcriptionally from more mature myonuclei. Finally, the transcriptional dynamics of the many nonmuscle cells embedded in muscle tissue and their responses to injury or disease remain to be understood. Elucidation of gene expression changes at the single nucleus level offers the possibility of uncovering key regulatory pathways and possible therapeutic targets for amelioration of the many disorders that lead to muscle degeneration and the ensuing consequences on human health.
Duchenne muscular dystrophy (DMD) is among the most devasting monogenic disorders of muscle. Caused by mutations in the dystrophin gene (DMD), this disease results in myofiber degeneration followed by cycles of regeneration until the endogenous satellite cell population is depleted, at which time muscles irreversibly degenerate with consequent loss of mobility and eventual death of affected individuals (4). The DMD gene encodes dystrophin, a protein of the dystrophin–glycoprotein complex (5). DMD is the largest gene in the genome, spanning ∼2.5 megabases of DNA and containing 79 exons that are conserved in vertebrate species. Among these exons, there are several mutational “hot spots” in which exon deletions disrupt the continuity of the open reading frame (ORF) between surrounding exons, resulting in the loss of functional dystrophin protein (6). Mouse models of DMD with deletions of the exons most commonly deleted in DMD patients have been invaluable for understanding DMD pathogenesis and providing faithful animal models for therapeutic testing (7, 8).
In the present study, we created a mouse model of DMD lacking dystrophin exon 51, representing a common mutational hot spot in humans. Using these mice, we performed single-nucleus RNA sequencing (snRNA-seq) on diseased myofibers and delineated the transcriptional changes associated with DMD progression in individual myonuclei and nonmuscle nuclei. Our findings reveal fascinating and previously unrecognized heterogeneity of myonuclei within normal myofibers, as well as regenerative myonucleus subtypes, and uncover DMD-associated gene regulatory networks with therapeutic relevance.
Results
Generation of a Mouse Model with DMD Exon 51 Deletion.
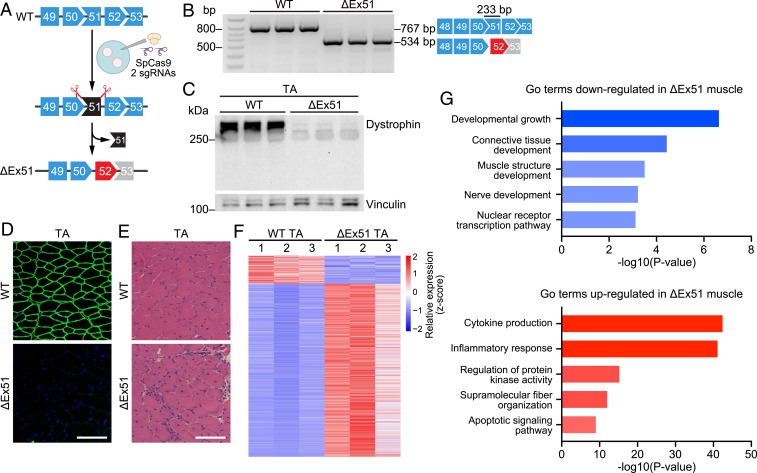
Exon 51 of the DMD gene is one of the most frequently deleted exons in DMD patients (9). To recapitulate this human mutation in a mouse model, we used CRISPR-Cas9 technology to generate mice with a deletion of exon 51 (ΔEx51) in the Dmd gene (Fig. 1A). Zygotes of C57BL/6 mice were injected with two single-guide RNAs (sgRNAs) that targeted the introns flanking exon 51 and were implanted into surrogate female mice (SI Appendix, Fig. S1A). We selected an F0 founder with a 1,516-base pair (bp) deletion that eliminated exon 51 and formed a new junction between intron 50 and intron 51, thereby disrupting the dystrophin ORF (SI Appendix, Fig. S1 B and C). We confirmed deletion of exon 51 in the resulting Dmd transcript by RT-PCR analysis and by sequencing the RT-PCR products using primers in exons 48 and 53 (Fig. 1B and SI Appendix, Fig. S1D). Western blot analysis of tibialis anterior (TA) muscle, diaphragm, and heart isolated from 4-wk-old ΔEx51 mice showed that deletion of exon 51 led to a complete loss of dystrophin protein (Fig. 1C and SI Appendix, Fig. S1E). Immunohistochemistry confirmed the absence of dystrophin protein expression in all ΔEx51 myofibers and cardiomyocytes (Fig. 1D and SI Appendix, Fig. S1F).
Fig. 1.
Creation and analysis of ΔEx51 Dmd mice. (A) CRISPR-Cas9 editing strategy used for generation of mice with dystrophin exon 51 deletion (ΔEx51). Mouse oocytes were injected with SpCas9 and 2 sgRNAs flanking exon 51. Upon deletion of exon 51, exon 52 (red) becomes out of frame with exon 50. (B) RT-PCR analysis of TA muscle to validate deletion of exon 51 (233-bp length). RT-PCR primers were in exons 48 and 53, and the amplicon size was 767 bp for WT mice and 534 bp for ΔEx51 mice. RT-PCR products are schematized on the right (n = 3). (C) Western blot analysis showing loss of dystrophin expression in the TA of ΔEx51 mice. Vinculin is the loading control (n = 3). (D) Dystrophin staining of the TA of WT and ΔEx51 mice. Dystrophin is shown in green. Nuclei are marked by DAPI stain in blue. (Scale bar, 100 μm.) (E) H&E staining of the TA of WT and ΔEx51 mice. Note extensive inflammatory infiltrate and centralized myonuclei in ΔEx51 muscle. (Scale bar, 100 μm.) (F) Heat map showing z-score–transformed expression of the differentially expressed genes between WT and ΔEx51 TA muscle (n = 3). (G) Selected top GO terms enriched in down- and up-regulated genes in ΔEx51 TA muscle.
ΔEx51 mice exhibit histological parameters similar to mdx mice, a dystrophin-deficient mouse line most widely used for DMD research (10). Due to the absence of functional dystrophin, ΔEx51 muscle degenerates and is replaced by fibrotic tissue and inflammatory infiltration (Fig. 1E and SI Appendix, Fig. S1 G–I) as also observed in mdx muscle (11). Additionally, at 4 wk of age ΔEx51 muscle exhibits a high percentage of myofibers with centralized nuclei, indicative of muscle regeneration (Fig. 1E and SI Appendix, Figs. S1 G, H, and J). As a consequence of the muscle undergoing repeated cycles of degeneration and regeneration, ΔEx51 skeletal muscle shows an abnormal proportion of small and large myofibers, also observed in muscles of mdx mice (11) (SI Appendix, Fig. S1K). Muscle function showed a 40% decline in forelimb grip strength in ΔEx51 mice compared to wild-type (WT) mice at 4 wk of age (SI Appendix, Fig. S1L). The muscle phenotype seen in ΔEx51 mice is similar to other dystrophin-deficient DMD mouse models (12–14). Furthermore, serum creatine kinase (CK), which serves as a marker of muscle damage and an indicator of DMD in patients and mouse models (15, 16), was elevated about 20-fold in ΔEx51 mice compared to WT littermates at 4 wk of age (SI Appendix, Fig. S1M). Similar to mdx mice, ΔEx51 mice at 6 mo of age showed no impairment in cardiac function (SI Appendix, Fig. S1 N and O). Although we cannot exclude cardiomyopathy manifestations at later ages (17), we did not detect a difference in survival of ΔEx51 mice compared to WT mice, as measured up to 1 y of age. In addition, ΔEx51 male mice and ΔEx51 homozygous female mice are fertile.
To investigate the molecular basis of DMD pathology of skeletal muscle, we profiled gene expression changes in TA muscle of 4-wk-old ΔEx51 and WT mice by RNA sequencing (RNA-seq). Transcriptomic profiling showed pronounced differences in dystrophic ΔEx51 and WT muscles, confirming misregulation of gene expression due to DMD pathology (SI Appendix, Fig. S2A). In total, we identified 1,434 genes that were up-regulated and 231 genes that were down-regulated in dystrophic muscle compared to WT muscle (Fig. 1F, SI Appendix, Fig. S2B, and Dataset S1). Gene Ontology (GO) analysis identified cytokine production, inflammatory response, and apoptotic signaling as the most prominently altered pathways in ΔEx51 muscle (Fig. 1 G, Lower and Dataset S2). These data suggest high activation of the different cell types of the immune system, confirming the inflammatory infiltration observed in dystrophic muscle (Fig. 1E). Genes down-regulated in ΔEx51 muscle were related to growth stimulus and development of healthy skeletal muscle (Fig. 1 G, Upper and Dataset S2).
We compared the transcriptomic analyses of ΔEx51 muscle with the mdx/mTR muscle published previously (18) and observed overlap between differentially expressed genes between these two models (SI Appendix, Fig. S2 C and D). The different range of up- or down-regulation of the genes most likely reflects the different stages of DMD pathogenesis, as our analysis was performed at 4 wk of age, whereas the mdx/mTR model was characterized at 4 mo. Taken together, these data indicate that lack of dystrophin in ΔEx51 muscle is associated with a change in cellular composition and myonuclear activity compared to WT muscle, ultimately resulting in compromised muscle function.
snRNA-seq Reveals Heterogeneity of Myonuclei within Normal and Dystrophic Skeletal Muscle.
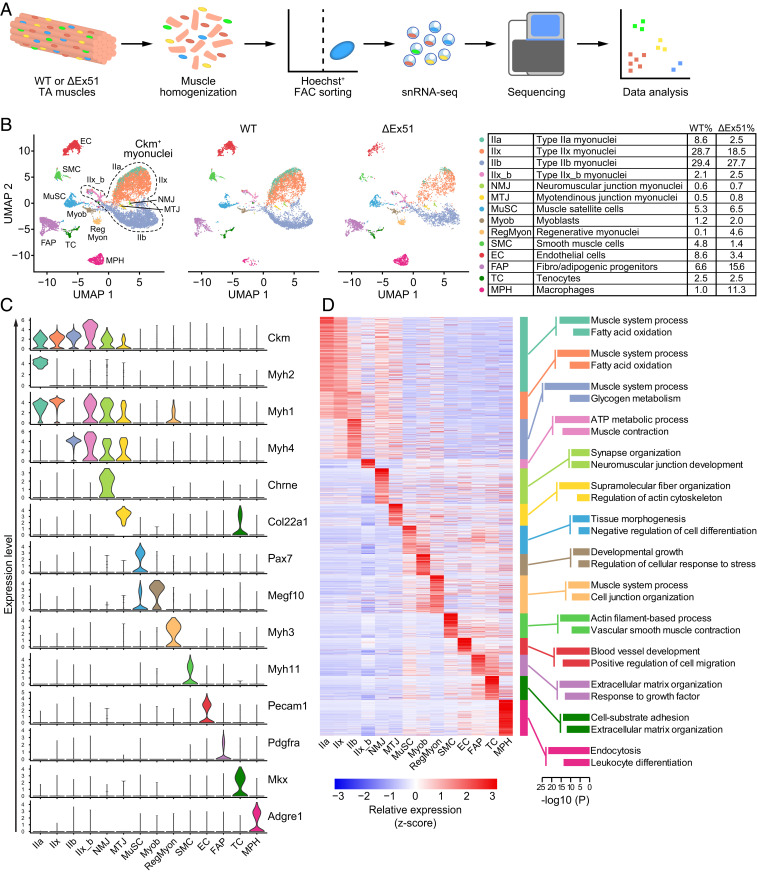
To compare different cell populations in dystrophic and WT skeletal muscles, we performed snRNA-seq on nuclei isolated from TA muscle of ΔEx51 and WT mice at 4 wk of age (Fig. 2A). We selected TA muscle for snRNA-seq analysis, as it contains both oxidative and glycolytic myofibers (19). The integrity of nuclei was preserved during the procedure by gentle homogenization of TA muscle. Nuclei of myofibers (myonuclei) and of other mononucleated cells of muscle were purified from myofibrillar debris by fluorescence-activated cell sorting using Hoechst staining that binds nuclear DNA (SI Appendix, Fig. S3A) followed by snRNA-seq using the 10× Genomics Chromium platform. From WT TA muscle we captured 7,013 single nuclei, and from ΔEx51 TA muscle we captured 4,209 single nuclei with high integrity (SI Appendix, Fig. S3B). A median of 1,067 genes per nucleus was sequenced from WT muscle and from ΔEx51 muscle there was a median of 903 genes per nucleus, for a total of 23,588 and 23,599 total genes detected, respectively (SI Appendix, Fig. S3 B and C). Our analysis showed an average of 36% of unique molecular identifiers (UMIs) confidently mapped to introns and an average of 64% of UMIs mapped to a pre-mRNA reference transcriptome (SI Appendix, Fig. S3B). Since snRNA-seq profiles nuclear RNA, our gene expression profile data reflect nascent transcription, as well as the cellular transcriptome.
Fig. 2.
snRNA-seq of TA muscle from WT and ΔEx51 mice. (A) Schematic of the experimental design for snRNA-seq on skeletal muscle nuclei. (B) UMAP visualization of all of the nuclei (11,222 nuclei, Left) from WT and ΔEx51 TA muscle colored by cluster identity. UMAPs depicting nuclei of WT TA muscle (7,013 nuclei, Center) and nuclei of ΔEx51 TA muscle (4,209 nuclei, Right). Percentages of nuclei in each cluster are indicated in the table. (C) Violin plots showing the expression of selected marker genes for each cluster of nuclei. (D) Heat map showing z-score–transformed average expression of the marker genes for each cluster of nuclei (Left). Selected top enriched GO terms of the marker genes for each cluster of nuclei (Right).
To compare ΔEx51 and WT samples, we integrated datasets from both preparations (total of 11,222 nuclei) for downstream bioinformatic analysis. Nuclear transcriptomes were visualized using a uniform manifold approximation and projection (UMAP) plot (Fig. 2B). UMAP is a nonlinear dimensionality reduction algorithm for the analysis of high-dimensionality data which shows the greatest power to resolve different cell or nuclear populations in comparison with other methods (20). Using UMAP, we identified 14 different clusters of nuclei, according to their transcriptional signatures (Fig. 2B). Overall, we identified a total of 4,748 marker genes for the different types of skeletal muscle nuclei (Dataset S3). The identity of each cluster was assigned based on the expression of known cellular marker genes and the GO analysis of the marker genes of each cluster of nuclei (Fig. 2 C and D and SI Appendix, Fig. S4 A and B).
As seen by UMAP, several clusters of nuclei were detected that express Ckm, a marker of mature myonuclei (Fig. 2 B and C and SI Appendix, Fig. S4B). These Ckm+ nuclei express different isoforms of myosin heavy chain (Myh), the classical marker for myofiber classification (19). We identified two large clusters of myonuclei with high levels of expression of Myh isoforms: Cluster IIx expressed high levels of Myh1, which encodes myosin heavy chain protein IIx (MyHC-IIx), and Cluster IIb had high levels of expression of Myh4, encoding MyHC-IIb. Both MyHC-IIx and MyHC-IIb are markers of fast-glycolytic myofibers, consistent with the known fiber-type composition of TA muscle. A smaller fraction of myonuclei (as seen in Cluster IIa) expressed Myh2, which encodes MyHC-IIa, indicative of fast-oxidative myofibers. Additionally, Cluster IIx_b, which highly expresses both Myh1 and Myh4 transcripts, displayed high levels of the transcripts of fast isoforms of troponins (Tnnc2 and Tnni2), myoglobin (Mb), enzymes of cytochrome C oxidase (Cox6a2 and Cox6c), and adenosine 5′-triphosphate synthase complexes (Atp5e and Atp5g1) (SI Appendix, Fig. S4C and Dataset S3). Expression of these marker genes and the position in the UMAP suggest that Cluster IIx_b contains myonuclei with high metabolic activity, instead of originating from a population that is in a transitional state between Cluster IIx and Cluster IIb.
We labeled another cluster as Cluster NMJ, since the myonuclei in this cluster express transcripts associated with the neuromuscular junction (NMJ), such as Chrne (coding for the acetylcholine receptor subunit epsilon), Colq, Prkar1a, and Col25a1 (21, 22) (Fig. 2C, SI Appendix, Fig. S4B, and Dataset S3). Another myonuclear cluster was labeled Cluster MTJ, as the myonuclei in this cluster selectively express Col22a1, a transcript that encodes a member of the collagen family that contributes to the stabilization of myotendinous junctions (MTJ) and strengthens skeletal muscle attachments during contraction (23) (Fig. 2C and SI Appendix, Fig. S4B). On the UMAP plot, the NMJ and MTJ clusters appeared to localize to the center of the Ckm+ myonuclei. However, the three-dimensional (3D) UMAP showed distinct separation from other myonuclei (SI Appendix, Fig. S4D). Additionally, a subset of genes expressed in the NMJ and MTJ clusters is also highly expressed in the nuclei of other mononucleated cell types (e.g., Lama2 in fibro/adipogenic progenitor [FAP] cells and tenocytes), but these genes are not expressed in the myonuclei of Clusters IIa, IIx, or IIb, supporting the uniqueness of Clusters NMJ and MTJ.
Especially interesting was the identification of three clusters of nuclei related to skeletal muscle regeneration. One cluster, Cluster MuSC, is characterized by expression of Pax7, a marker of satellite cells. The second cluster, Cluster Myob, expresses high levels of Megf10, a marker of skeletal myoblasts (24). The third cluster, RegMyon, expresses Myh3 and Myh8 transcripts that encode embryonic and perinatal MyHC isoforms, respectively, which have been shown to be distinctive to regenerative myofibers (Fig. 2C, SI Appendix, Fig. S4B, and Dataset S3).
Analysis of Nonmuscle Nuclei by snRNA-seq.
As expected, snRNA-seq of skeletal muscle also detected clusters of nuclei from other mononucleated cell types, such as Cluster SMC, containing nuclei from smooth muscle cells, as identified by expression of Myh11; Cluster EC, containing nuclei from endothelial cells, as characterized by expression of the adhesive stress-response protein Pecam-1 (25); Cluster FAP, containing nuclei from FAP cells, as characterized by expression of Pdgfra (26); Cluster TC, containing nuclei from tenocytes, as characterized by expression of the tenogenesis-promoting homeoprotein Mkx (27); and Cluster MPH, containing nuclei from macrophages, as characterized by expression of Adgre1 (28) (Fig. 2C and SI Appendix, Fig. S4B). In addition to identifying known markers, we identified previously uncharacterized genes enriched in specific clusters, enabling further characterization of the molecular features of each cluster (Dataset S3).
Changes in Nuclear Abundance in ΔEx51 Muscle.
snRNA-seq analysis identifies the relative abundance of nuclei for each cluster in ΔEx51 and WT muscles. Compared to WT muscle, we observed an overall reduction of the percentage of mature myonuclei in ΔEx51 TA muscle, specifically with a decrease from 69.9 to 52.7%. In particular, Cluster IIa showed a decrease from 8.6 to 2.5% and Cluster IIx showed a decrease from 28.7 to 18.5% (Fig. 2B). In contrast, Cluster RegMyon showed an increase of nuclei from 0.1 to 4.6% and Cluster MPH showed an increase of nuclei from 1.0 to 11.3% in ΔEx51 dystrophic muscle. The percentage of nuclei in Cluster FAP dramatically increased in ΔEx51 muscle, from 6.6 to 15.6% (Fig. 2B). These data are consistent with the histological analysis of ΔEx51 muscle showing a different composition of cell types compared to WT muscle, mainly due to an increase in fibrosis, inflammatory infiltration, and muscle regeneration (Fig. 1E).
snRNA-seq Analysis Reveals Degenerative Pathways in DMD Muscle.
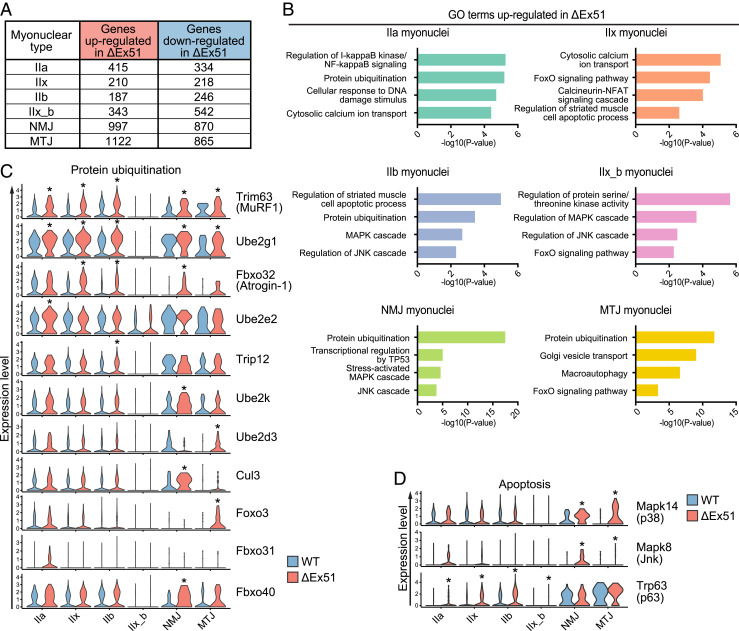
To uncover aberrant transcriptional activity in nuclei isolated from different cell types in ΔEx51 muscle, we analyzed the differentially expressed genes of each cluster. Myonuclei in Clusters NMJ and MTJ showed a greater number of up-regulated and down-regulated genes compared to those in Clusters IIa, IIx, IIb, and IIx_b (Fig. 3A and Dataset S4). This indicates that these two small clusters of specialized myonuclei are the most affected in dystrophic muscle. Interestingly, the myonuclei in Clusters IIa, IIx, and IIb isolated from ΔEx51 muscle exhibited up-regulation of an aberrant dystrophin transcript (ΔEx51), most likely as a compensatory response to the absence of functional dystrophin protein in the myofibers (SI Appendix, Fig. S5A). Furthermore, in the ΔEx51 myonuclei in Clusters IIa, IIx, and IIb, we observed a significant down-regulation of transcripts encoding titin (Ttn), nebulin (Neb), and specific Myh isoforms, suggesting a reduction in the transcriptional activity of sarcomeric genes in dystrophic myonuclei (SI Appendix, Fig. S5A).
Fig. 3.
Analysis of gene expression changes in ΔEx51 myonuclei compared to WT myonuclei. (A) Number of up- and down-regulated genes in different clusters of ΔEx51 myonuclei. (B) Selected top GO terms enriched in up-regulated genes from the different clusters of ΔEx51 myonuclei. (C) Violin plots showing the differential expression of selected marker genes of protein ubiquitination between WT and ΔEx51 myonuclei. *Genes significantly differentially expressed. (D) Violin plots showing the differential expression of selected marker genes of apoptosis in WT and ΔEx51 myonuclei. *Genes significantly differentially expressed.
To define the myofiber degenerative pathways that lead to the decline of the number of myonuclei in ΔEx51 muscle, we performed GO analysis of the ΔEx51 up-regulated genes of each myonuclear cluster (Fig. 3B and Dataset S5). We identified the protein ubiquitination pathway as enriched in almost all of the myonuclear types isolated from ΔEx51 muscle, reflecting the breakdown of myofibrillar proteins by the ubiquitin–proteasome system (29). In the ubiquitin–proteasome system, proteins are marked for degradation by three classes of ubiquitinating enzymes (E1, E2, and E3), after which the proteasome degrades the ubiquitinated proteins. Muscle RING finger 1 (MuRF1) and Atrogin-1 are muscle-specific E3 ubiquitin ligases that are markers of muscle atrophy. However, it is unknown if their up-regulation occurs in only specific myonuclear subtypes. snRNA-seq allows the detection of expression of different genes of the ubiquitination pathway within each cluster of myonuclei (Fig. 3C). With the exception of the specialized Cluster IIx_b, all ΔEx51 myonuclei have high expression of MuRF1 (Trim63) transcript. Up-regulation of Atrogin-1 (Fbxo32) was not detected in Cluster IIa; however, we identified up-regulation of Fbxo31 in this cluster (30). For Cluster NMJ, Fbxo40 was up-regulated (31), indicating a redundant and complementary role of these ubiquitin-ligases in different types of dystrophic myonuclei. The transcript encoding the skeletal muscle-specific ubiquitin-conjugating enzyme UBE2G1, was among the ubiquitination enzymes up-regulated in almost all ΔEx51 myonuclei (32). Interestingly, the dystrophic myonuclei in different clusters up-regulate different E3 ligase enzymes. For example, the ΔEx51 myonuclei in Cluster IIa showed up-regulation of Ube2e2, in Cluster IIb showed up-regulation of the E3 ligase Trip12 (33), in Cluster NMJ showed up-regulation of Ube2k and Cul3, and in Cluster MTJ showed up-regulation of Ube2d3 (Fig. 3C and Dataset S4). In atrophying myofibers, the ubiquitin–proteasome pathway is regulated by the activation of FOXO (forkhead box O) transcription factors (34). We observed a substantial up-regulation of Foxo3, especially in the ΔEx51 myonuclei in Cluster MTJ. It was shown that Foxo3 acts on the Atrogin-1 promoter to induce atrophy of myotubes in vitro and to activate the ubiquitin–proteasomal system of protein breakdown in skeletal muscle in vivo (30, 35, 36). Interestingly, MTJ myonuclei are the nuclei of myofibers most susceptible to mechanical stress (37). This revealed a previously unrecognized expression pattern of Foxo3 that is restricted to a specific cluster of myonuclei in dystrophic myofibers.
In addition to muscle atrophic markers, we also identified apoptotic markers that were expressed in specific ΔEx51 myonuclei (Fig. 3D). ΔEx51 myonuclei of Clusters IIa, IIx, IIb, and IIx_b showed up-regulation of the apoptotic marker p63 (Trp63), whereas the proapoptotic factors Jnk (Mapk8) and p38 (Mapk14) were up-regulated in ΔEx51 myonuclei of Clusters NMJ and MTJ. Alteration of the p38 pathway has previously been demonstrated in satellite cells in association with loss of muscle mass (38). p53, which is known to mediate myonuclear apoptosis, is a downstream target of the p38 pathway (39). Thus, up-regulation of the p38 transcript in ΔEx51 myonuclei in Cluster NMJ is consistent with enhanced transcriptional regulation by the p53 pathway in Cluster NMJ (Fig. 3B).
Among the down-regulated GO terms in ΔEx51 myonuclei, we identified pathways of negative regulation of apoptosis, such as Bcl6. Consistent with this, we observed that messenger RNA (mRNA) metabolism pathways, which are important for myofiber growth and whose down-regulation is considered a marker of muscle atrophy, were down-regulated in ΔEx51 myonuclei (SI Appendix, Fig. S5B and Dataset S5).
Genetic Determinants of the Regenerative Myonuclear Population of DMD Muscle.
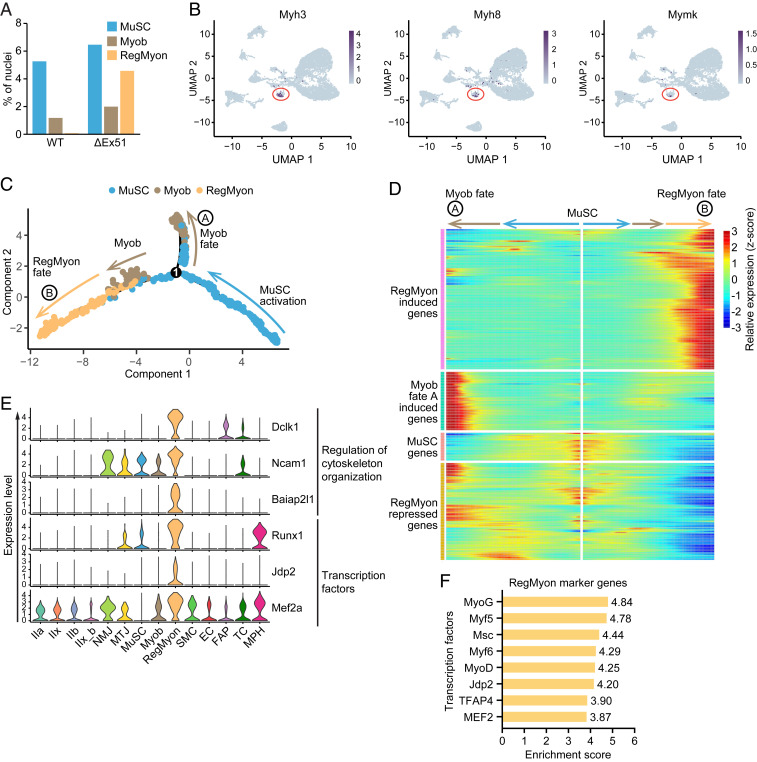
In TA muscles of 4-wk-old ΔEx51 mice, myonuclear degeneration is accompanied by high regenerative activity, as indicated by the presence of centralized nuclei (Fig. 1E). Following myofiber damage, satellite cells are activated and can differentiate into myoblasts that repair injured myofibers by fusing at the sites of injury (4). In both WT and ΔEx51 muscles, MuSC and Myob nuclei were detected, whereas we discovered a cluster, labeled RegMyon, that exists exclusively in dystrophic muscles (Fig. 4A). To explore the specific transcriptional changes in the MuSC and Myob clusters in WT and ΔEx51 muscles, we identified marker genes for each of these clusters. We found 56 marker genes that were up-regulated and 54 that were down-regulated in the ΔEx51 MuSC cluster (Dataset S4). Additionally, we found 63 genes that were up-regulated and 56 that were down-regulated in the ΔEx51 Myob cluster (Dataset S4). Interestingly, the long noncoding RNA Meg3, which is necessary for differentiation of satellite cells (40), was the most down-regulated marker in both ΔEx51 MuSC and Myob clusters (SI Appendix, Fig. S6 A and B). These data reveal distinct alterations in the transcriptional programs of muscle precursors in ΔEx51 muscles.
Fig. 4.
Analysis of gene expression dynamics in WT and ΔEx51 myonuclei. (A) Percentage of nuclei of clusters involved in the regeneration of skeletal muscle (Clusters MuSC, Myob, and RegMyon) in WT and ΔEx51 TA muscles. (B) UMAPs depicting the expression of Myh3, Myh8, and Mymk in the RegMyon cluster. (C) Pseudotime ordering of all of the nuclei of Clusters MuSC, Myob, and RegMyon. Each dot represents one nucleus (color-coded by its identity) and each branch represents one cell state. Activation of the MuSC cluster can lead to Myob fate A or to RegMyon fate B. (D) Heatmap showing z-score–transformed average expression of the differentially expressed gene dynamics toward Myob fate A and RegMyon fate B along pseudotime. The differentially expressed genes were clustered into four gene sets according to k-means. (E) Violin plots showing the expression of selected genes (involved in the regulation of cytoskeleton organization or encoding transcriptional factors) highly expressed in the nuclei of Cluster RegMyon. (F) Enrichment score of the top transcription factor binding motifs identified by iRegulon from the promoter regions of the marker genes of the Cluster RegMyon.
The myonuclei in Cluster RegMyon express Myh3 and Myh8, which encode the embryonic and perinatal MyHC isoforms, respectively (Fig. 4B). Myomaker (Mymk), a muscle-specific membrane protein required for fusion of myoblasts during regeneration (41), is also expressed in myonuclei of Cluster RegMyon (Fig. 4B). To infer a hierarchical trajectory among the RegMyon, MuSC, and Myob clusters, we applied Monocle analysis to these three nuclear populations (42). This analysis aligned the nuclei into a one-dimensional pseudotime axis, based on the differentially expressed genes across the three different clusters. Labeling each nucleus according to its cluster, the pseudotime trajectory shows an organized branched progression from MuSC to Myob (fate B) and to RegMyon (Fig. 4C and SI Appendix, Fig. S6 C, Upper). A subset of genes in Cluster Myob diverts at the central node into a third branch composed of both WT and ΔEx51 Myob (fate A) that do not develop into myonuclei in Cluster RegMyon (Fig. 4C and SI Appendix, Fig. S6 C, Lower). GO analysis of the differentially expressed genes between Myob of fate A and fate B displayed a higher transcriptional activity and identified genes associated with myoblast proliferation for fate A Myob (SI Appendix, Fig. S6 D, Upper and Datasets S4 and S5). Distinctively, fate B Myob are enriched in genes encoding proteins of muscle contractility or involved in myotube differentiation or cell junction assembly, most likely indicating the predisposition of these cells to fuse to form myotubes (SI Appendix, Fig. S6 D, Lower and Datasets S4 and S5). Next, we identified genes that are significantly branch-dependent in the pseudotime trajectory. We plotted the 583 genes in a special type of heat map, where columns are points in pseudotime, rows are genes, and the beginning of pseudotime is in the middle of the heat map. This heat map shows gene expression changes along pseudotime progression in both lineages at the same time (Fig. 4D).
To explore the key genes that determine the RegMyon fate, we analyzed RegMyon marker genes in the top cluster of the pseudotime heatmap. In particular, we focused on two sets of genes: 1) those encoding proteins involved in regulation of cytoskeleton organization, for their putative role in cell fusion, and 2) those encoding transcription factors, for their putative roles as genetic regulators of regenerative myogenesis (Fig. 4E). Three of the top markers in the Cluster RegMyon were Dclk1, Ncam1, and Baiap2l1, which encode proteins involved in cytoskeleton organization (Fig. 4 E, Upper). Dclk1 encodes a protein that binds and regulates microtubule polymerization and dynamics and is involved in motility of muscle cell precursors during forelimb muscle development (43). Ncam1 encodes a cell-surface protein with a well-established role in adult myogenesis (44). Baiap2l1 encodes a protein that facilitates the formation of F-actin protrusions (45). This suggests a potential role of Baiap2l1 in muscle regeneration. Among the transcription factor markers expressed in the myonuclei of Cluster RegMyon, we identified Runx1, Jdp2, and Mef2a (Fig. 4 E, Lower). Mef2a is a well-documented regulator of skeletal muscle regeneration (46). Similarly, Runx1 has been reported to be necessary for myoblast proliferation during skeletal muscle regeneration (47). Although Jdp2 has been reported to promote muscle cell differentiation in vitro (48), its role in muscle regeneration remains unknown. Despite the prior implication of these transcription factors in muscle regeneration, it has not been previously possible to assign their expression to specific subsets of myonuclei.
Next, we performed enrichment analysis of the transcription factor binding sites of the marker genes of Cluster RegMyon using iRegulon (Dataset S6). Interestingly, associated with the most common myogenic regulatory factors (such as MyoG, Myf5, Myf6, Msc, MyoD, and MEF2) we found that the basic-leucine zipper protein Jdp2 is predicted to bind the promoter regions of a significant number of genes expressed in the Cluster RegMyon (Fig. 4F and SI Appendix, Fig. S6E), underlying its putative role as a regulator of regenerative myogenesis.
Discussion
By identifying and characterizing myonuclear populations of skeletal muscle, our study provides insights into normal skeletal muscle biology and DMD pathology. Since myofibers contain hundreds of nuclei, it has been technically challenging to examine potential functional distinctions and transcriptional heterogeneity between individual myonuclei within the common cytoplasm of the myofiber (49). Recently, high-resolution transcriptional analyses were performed on isolated myofibers or mononucleated cells in skeletal muscle, but the definition of nuclei populations within myofibers was not possible (50–56). In this study, we used snRNA-seq technology to unmask the transcriptional profile of individual nuclei of muscle fibers, providing invaluable information about the different cell type populations and gene expression profiles in normal and diseased skeletal muscle.
Using normal and dystrophic TA muscles, generated by deletion of exon 51 in the Dmd gene, we were able to identify 14 different populations of nuclei that we grouped into three classes: 1) myonuclei of myofibers, 2) nuclei of the regenerative pathway, and 3) nuclei of mononucleated cells of skeletal muscle. Furthermore, we quantified changes in abundance of individual nuclei in response to DMD pathogenesis and identified marker genes for each nuclear subtype. Elucidating the transcriptional responses of individual nuclei within and surrounding diseased myofibers sheds light on fundamental biological processes of skeletal muscle and offers insights into potential strategies to target nuclei with specific functions in dystrophic muscle.
Transcriptional Heterogeneity of Individual Myonuclei.
The syncytial nature of skeletal muscle in which hundreds of nuclei share a common cytoplasm raises fascinating questions about the extent to which individual nuclei may display transcriptional diversity. We identified and characterized not only the myonuclei expressing the typical MyHC isoforms that characterize the myofibers of the TA muscle (IIa, IIx, and IIb) (19) but also three specialized types of myonuclei: NMJ, MTJ, and IIx_b. NMJ and MTJ nuclei are specialized cell–cell contact nuclei with well-defined positions within the myofiber (37). The third cluster, IIx_b myonuclei, coexpresses both IIx and IIb Myh isoforms and has high levels of myoglobin (Mb) and mitochondrial enzymes. This is consistent with previous reports showing that Mb overexpression up-regulates mitochondrial activity (57). We anticipate that IIx_b myonuclei have a high metabolic activity and are likely localized near blood vessels, but further in situ localization experiments are needed to confirm this possibility.
Comparison of normal and ΔEx51 myonuclei showed unique and common degenerative pathways associated with DMD. We observed that, compared to other myonuclei, the NMJ and MTJ myonuclei in dystrophic muscle express a higher number of genes encoding proteins involved in the ubiquitin–proteasome system, which is responsible for the breakdown of the majority of the myofibrillar proteins during muscular atrophy. In addition, we found genes encoding enzymes (e.g., Ube2k in NMJ myonuclei and Ube2d3 in MTJ myonuclei) involved in muscle degeneration to be exclusively up-regulated in specific types of dystrophic myonuclei, offering candidates for therapeutic targets.
Since the TA muscle is a frequently used model system in muscular dystrophy research, in this study we focused our attention on the myonuclei of TA skeletal muscle that are composed of both fast-glycolytic and fast-oxidative myofiber types. It will be of interest to analyze the myonuclear transcriptional activity of other muscle groups in order to gain information about myonuclei of slow-twitch myofibers and compare the specialized nuclei in slow and fast myofibers. From a clinical point of view, analyzing the transcriptional activity of the myonuclei of the respiratory muscle fibers of the diaphragm (58) could provide insights into respiratory insufficiency in DMD patients. In addition, future snRNA-seq analysis on DMD hearts could identify the physiological changes leading to dystrophic heart failure (59).
Transcriptional Heterogeneity of Nuclei Involved in Muscle Regeneration.
Our findings uncover and characterize populations of regenerative nuclei that express known and previously unknown regulators of muscle regeneration. Interestingly, among the three regenerative populations of myonuclei, classified as MuSC, Myob, and RegMyon clusters, we recognized two different trajectories for MuSCs that define the cell state transitions: one leading to definitive Myob (fate A) and another leading to Myob (fate B) that can advance to RegMyon. Fate A Myob express markers of proliferation of muscle cells, such as Ppard (60) or Sirt1 (61), whereas the fate B Myob express characteristic markers of myotube differentiation, such as Rbm24 (62) and Stac3 (63). The cluster of RegMyon nuclei is particularly interesting as it is specific to dystrophic muscle. Among the known markers of this cluster of nuclei, we detected the embryonic and perinatal MyHC isoforms Myh3 and Myh8 and the fusogenic factor Myomaker, which is required for regenerative fusion (64). In addition, we identified putative regulators of muscle regeneration acting as transcription factors or involved in myoblast fusion. Particularly interesting, the transcription factor Jdp2 is expressed only in RegMyon and appears to regulate the expression of many genes contained in the RegMyon cluster.
The DMD mouse model ΔEx51 will serve as another animal model to study DMD pathogenesis. As mouse DMD models show different phases of the disease (65), snRNA-seq can be used to investigate the populations of regenerative myonuclei during disease progression.
Transcriptional Diversity of Nonmuscle Nuclei during DMD Pathogenesis.
As expected from the histological analysis, compared to normal samples ΔEx51 skeletal muscle contained fewer myonuclei, and consequently a higher percentage of nuclei are from mononucleated cells. In particular, we detected an increase in macrophage nuclei that determine the high inflammatory response of dystrophic muscle and an increase in FAPs, which are responsible for fibrotic and fat deposition of dystrophic muscle (66). In contrast, in ΔEx51 muscle we detected fewer endothelial cells and smooth muscle cells, both components of blood vessels, indicating a decrease in vascularity of DMD muscles (67).
Concluding Remarks
Our work represents a step toward the decoding of distinct transcriptional programs characteristic of different myonuclei populations in normal and dystrophic muscles. This atlas of transcriptome landscapes of normal and DMD muscle at single-nucleus resolution provides an in-depth view of the different types of nuclei within and surrounding myofibers, identifying specific markers that define each population. How genes respond to positional information within different myonuclei of the multinucleated muscle tissue represents an interesting question for the future. In addition, snATAC-seq will allow the analysis of chromatin accessibility of the single myonucleus, providing a deeper understanding of gene regulatory mechanisms (68).
A major limitation of snRNA-seq technology is that it does not provide information about the spatial location of the myonuclei in the muscle. New technologies for spatial transcriptomics are emerging to capture in situ polyadenylated RNA transcripts, but these techniques are still under optimization and have low resolution and sensitivity (69–71). It will be interesting to apply this approach to skeletal muscle tissue to reveal the spatial positions of each type of myonuclei within the myofiber to better define the volume of cytoplasm or “myonuclear domain” governed by a given myonucleus (72, 73).
Due to the massive amount of new information generated by snRNA-seq analysis, it is reasonable to assume that this technology will be increasingly applied in the field of muscle biology to better characterize skeletal muscle myonuclei in response to exercise, development, aging, and in different myopathies. Intriguingly, snRNA-seq of skeletal muscle can provide a detailed understanding of the transcriptional changes of diseased myonuclei after pharmacological treatments or gene editing therapies.
Materials and Methods
Mice.
Animal work described in this manuscript has been approved and conducted under the oversight of the University of Texas Southwestern Institutional Animal Care and Use Committee. Mice were housed in a barrier facility and maintained on standard chow. ΔEx51 mice were generated using the CRISPR-Cas9 system in C57/BL6N mice. The two sgRNAs specific to the intronic regions surrounding exon 51 of the mouse Dmd locus were synthetized by Synthego. Injection of Cas9 protein and sgRNAs into zygotes was performed as described (74). ΔEx51 mice and WT littermates were genotyped using primers encompassing the targeted region as indicated in SI Appendix, Fig. S1A as described (13).
RT-PCR.
RNA was purified using TRIzol (Thermo Fisher Scientific) and complementary DNA was synthesized using the iScript Reverse Transcription Supermix (Bio-Rad Laboratories) according to the manufacturer’s protocols. RT-PCR was performed using primers in Dmd exons 48 and 53: (forward, 5′-AGGTTCACTTAAAAGATTTTAGGCAG -3′; reverse, 5′-TAACATTTCATTCAACTGTTGTCTCC -3′).
Dystrophin Western Blot Analysis.
Preparation of protein lysates and Western blot analysis were performed as described (14). Dystrophin was detected using mouse anti-dystrophin antibody (MANDYS8, D8168; Sigma-Aldrich). The loading control was determined by blotting with mouse anti-vinculin antibody (V9131; Sigma-Aldrich).
Histological Analysis of Muscles.
Muscles from WT and ΔEx51 mice were cryoembedded for hematoxylin/eosin (H&E) staining according to established staining protocols (75). Dystrophin immunohistochemistry was performed using MANDYS8 monoclonal antibody (D8168; Sigma-Aldrich) with modifications to the manufacturer’s instructions as previously described (14). Image analyses were performed using ImageJ software (76) on at least three muscles for each condition. Myofiber diameter was calculated as minimal Feret’s diameter, a geometrical parameter that allows reliable measurement of cross-sectional size (11).
Grip Strength Test.
Muscle strength was assessed by a grip strength meter (Columbus Instruments). Each measurement was repeated five or six times.
Serum CK Measurement.
Blood was collected and serum CK levels were measured by the Metabolic Phenotyping Core at the University of Texas Southwestern Medical Center using VITROS MicroSlide technology.
Cardiac Function.
Cardiac function was determined by two-dimensional echocardiography using a Visual Sonics Vevo 2100 Ultrasound on conscious 6-mo-old mice as previously described (77).
RNA-seq.
RNA-seq was performed as previously described (78), with minor modifications. TA muscles were mechanically homogenized in TRIzol (Thermo Fisher Scientific) before RNA purification. Stranded mRNA-seq libraries were generated using KAPA mRNA HyperPrep Kit (KK8580; Roche) following the manufacturer’s protocol. Sequencing was performed on an Illumina NextSeq 500 system, using the 75-bp high-output sequencing kit (Illumina) for single-end sequencing.
snRNA-seq.
snRNA-seq was performed as previously described (79), with modifications. snRNA-seq libraries were generated using Chromium Next GEM Single Cell 3′ Gene Expression v3.1 kit (10× Genomics) according to the manufacturer’s protocol. Sequencing was performed on an Illumina NextSeq 500 system with the pair-end sequencing settings Read1 – 28 bp, i7 index – 8 bp, and Read2 – 122 bp.
snRNA-seq Data Preprocessing.
We used CellRanger Software Suite (10× Genomics, v3.1.0) for data demultiplexing, transcriptome alignment, and UMI counting. Raw base call files were demultiplexed using cellranger mkfastq. A custom “pre-mRNA” mm10 reference package was generated and was used as reference for read alignments and gene counting with cellranger count, as described in ref. 79. Ambient RNA background was evaluated for each library and removed using R package SoupX (80).
Data Processing, Clustering, Dimensional Reduction, and Differential Expression Analysis.
Downstream analysis was performed using R package Seurat (v3.1.5) (68, 81). For each sample, single-nucleus transcriptomes with fewer than 200 or more than 4,000 genes or with more than 15,000 UMIs were further filtered out from the analysis. Subsequently, data were log-normalized and scaled, and principle component analysis was performed using the top 2,000 genes that showed highly variable expression in the integrated dataset. Cell clusters were called using the first 15 principle components under a clustering resolution of 0.6. Dimensional reduction was performed by UMAP using the first 15 principle components. Marker genes were unbiasedly analyzed using Seurat function FindAllMarkers. Next, cluster identities were annotated based on differentially expressed genes, as well as expression of known marker genes for different cell types as described in the manuscript. IIa, NMJ, and MTJ clusters were further subclustered based on known marker gene expression. After cluster annotation, marker genes for each nuclei cluster were analyzed using the FindAllMarkers function. Three-dimensional UMAP was generated by plot3d function from R package rgl. Differential expression analysis was performed using the FindMarkers function with absolute log (fold change) >0.25 and adjusted P value < 0.01 considered as significant.
Pseudotime Analysis.
R package Monocle2 was used to align MuSC, Myob and RegMyon clusters into pseudotime trajectories (42). The top 1,000 differentially expressed genes were used to order cells in the pseudotime trajectories, determined using the Monocle2 function differentialGeneTest. Branched expression analysis modeling was performed using BEAM to analyze genes whose expression changed along different pseudotime trajectories.
Pathway and Transcription Factor Analysis.
Pathway analysis was performed using Metascape (82). Upstream regulator analysis for RegMyon marker genes was performed using iRegulon (v1.3) (83). MyoD target genes were defined based on previous MyoD chromatin immunoprecipitation-sequencing data (Gene Expression Omnibus accession numbers: GSM915165, GSM915183, GSM915185, and GSM915186) generated by the ENCODE consortium.
RNA-seq Data Analysis.
Quality control of RNA-seq data were performed using FastQC Tool (Version 0.11.4). Sequencing reads were aligned to mouse GRCm38 (mm10) reference genome using HiSTAT2 (v2.0.4) with default settings and–rna-strandness F (84). Aligned reads were counted using feature count (v1.6.0) per gene ID (85). Differential gene expression analysis was performed with the R package edgeR (v3.30.1) using the generalized linear model approach (86). Cutoff values of absolute fold change greater than 2.0 and false discovery rate less than 0.01 were used to define differentially expressed genes. Normalized gene cpm (counts per million reads mapped) values were used to calculate RPKM (reads per kilobase per million mapped reads) values, which were then used for heat-map plotting.
Supplementary Material
Acknowledgments
We thank Jose Cabrera for graphics, Alex A. Mireault for help in tissue-processing experiments and animal studies, Dr. Efrain Sanchez-Ortiz for help in Western blot analysis and imaging, Dr. Wei Tan for echocardiography, and John M. Shelton for performing immunohistochemistry and H&E staining. We thank Drs. Jian Xu and Yoon Jung Kim from the Next Generation Sequencing Core Facility at Children’s Research Institute at the University of Texas Southwestern Medical Center for performing the Illumina sequencing. We thank Nicholas Loof and Shengsyong Shih from the Moody Foundation Flow Cytometry Facility at Children’s Research Institute at the University of Texas Southwestern Medical Center for sorting skeletal muscle nuclei. We thank Wenbin Guo and Drs. Jialei Duan, Kenian Chen, and Lin Xu for helpful advice and discussion on bioinformatic analysis. This work was supported by funds from NIH (HL130253 and AR067294), Senator Paul D. Wellstone Muscular Dystrophy Cooperative Research Center (U54 HD 087351), and the Robert A. Welch Foundation (grant 1-0025 to E.N.O.). Z.W. was supported by a predoctoral fellowship from the American Heart Association and the Harry S. Moss Heart Trust (19PRE34380436).
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018391117/-/DCSupplemental.
Statistics and Data Availability.
Statistical analyses were performed using GraphPad Prism 8 using a two-tailed unpaired t test, with P value < 0.05 considered significant. All data are displayed as mean ± SEM unless otherwise indicated. All sequencing data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE156498.
References
- 1.Chal J., Pourquie O., Making muscle: Skeletal myogenesis in vivo and in vitro. Development 144, 2104–2122 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Yin H., Price F., Rudnicki M. A., Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doles J. D., Olwin B. B., Muscle stem cells on the edge. Curr. Opin. Genet. Dev. 34, 24–28 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Wallace G. Q., McNally E. M., Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annu. Rev. Physiol. 71, 37–57 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Michele D. E., Campbell K. P., Dystrophin-glycoprotein complex: Post-translational processing and dystroglycan function. J. Biol. Chem. 278, 15457–15460 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Flanigan K. M., et al. , Mutational spectrum of DMD mutations in dystrophinopathy patients: Application of modern diagnostic techniques to a large cohort. Hum. Mutat. 30, 1657–1666 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durbeej M., Campbell K. P., Muscular dystrophies involving the dystrophin-glycoprotein complex: An overview of current mouse models. Curr. Opin. Genet. Dev. 12, 349–361 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Wasala N. B., Chen S. J., Duan D., Duchenne muscular dystrophy animal models for high-throughput drug discovery and precision medicine. Expert Opin. Drug Discov. 15, 443–456 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echigoya Y., Lim K. R. Q., Nakamura A., Yokota T., Multiple exon skipping in the Duchenne muscular dystrophy hot spots: Prospects and challenges. J. Pers. Med. 8, 41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulfield G., Siller W. G., Wight P. A., Moore K. J., X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc. Natl. Acad. Sci. U.S.A. 81, 1189–1192 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briguet A., Courdier-Fruh I., Foster M., Meier T., Magyar J. P., Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 14, 675–682 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Amoasii L., et al. , Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl Med. 9, eaan8081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Min Y. L., et al. , CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 5, eaav4324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min Y. L., et al. , Correction of three prominent mutations in mouse and human models of Duchenne muscular dystrophy by single-cut genome editing. Mol. Ther. 28, 2044–2055 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendell J. R., Lloyd-Puryear M., Report of MDA muscle disease symposium on newborn screening for Duchenne muscular dystrophy. Muscle Nerve 48, 21–26 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Hathout Y., et al. , Discovery of serum protein biomarkers in the mdx mouse model and cross-species comparison to Duchenne muscular dystrophy patients. Hum. Mol. Genet. 23, 6458–6469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan J. G., et al. , Evolution of the mdx mouse cardiomyopathy: Physiological and morphological findings. Neuromuscul. Disord. 14, 491–496 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Van Pelt D. W., et al. , Multiomics analysis of the mdx/mTR mouse model of Duchenne muscular dystrophy. Connect. Tissue Res., 1–16 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Schiaffino S., Reggiani C., Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Becht E., et al. , Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W., Mammalian nicotinic acetylcholine receptors: From structure to function. Physiol. Rev. 89, 73–120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sigoillot S. M., Bourgeois F., Lambergeon M., Strochlic L., Legay C., ColQ controls postsynaptic differentiation at the neuromuscular junction. J. Neurosci. 30, 13–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charvet B., et al. , Knockdown of col22a1 gene in zebrafish induces a muscular dystrophy by disruption of the myotendinous junction. Development 140, 4602–4613 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Holterman C. E., Le Grand F., Kuang S., Seale P., Rudnicki M. A., Megf10 regulates the progression of the satellite cell myogenic program. J. Cell Biol. 179, 911–922 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lertkiatmongkol P., Liao D. Y., Mei H., Hu Y., Newman P. J., Endothelial functions of platelet/endothelial cell adhesion molecule-1 (CD31). Curr. Opin. Hematol. 23, 253–259 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wosczyna M. N., et al. , Mesenchymal stromal cells are required for regeneration and homeostatic maintenance of skeletal muscle. Cell Rep. 27, 2029–2035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milet C., Duprez D., The Mkx homeoprotein promotes tenogenesis in stem cells and improves tendon repair. Ann. Transl. Med. 3, S33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waddell L. A., et al. , ADGRE1 (EMR1, F4/80) is a rapidly-evolving gene expressed in mammalian monocyte-macrophages. Front. Immunol. 9, 2246 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandri M., Protein breakdown in muscle wasting: Role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol. 45, 2121–2129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milan G., et al. , Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 6, 6670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J. W., Zhang Y., Xu J. L., Zhang Q., Zhu D. H., FBXO40, a gene encoding a novel muscle-specific F-box protein, is upregulated in denervation-related muscle atrophy. Gene 404, 53–60 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Watanabe T. K., et al. , Molecular cloning of UBE2G, encoding a human skeletal muscle-specific ubiquitin-conjugating enzyme homologous to UBC7 of C-elegans. Cytogenet. Cell Genet. 74, 146–148 (1996). [DOI] [PubMed] [Google Scholar]
- 33.An C. I., Ganio E., Hagiwara N., Trip12, a HECT domain E3 ubiquitin ligase, targets Sox6 for proteasomal degradation and affects fiber type-specific gene expression in muscle cells. Skelet. Muscle 3, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stitt T. N., et al. , The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Sandri M., et al. , Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mammucari C., et al. , FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Perillo M., Folker E. S., Specialized positioning of myonuclei near cell-cell junctions. Front. Physiol. 9, 1531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernet J. D., et al. , p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 20, 265–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers S. K., Kavazis A. N., McClung J. M., Oxidative stress and disuse muscle atrophy. J. Appl. Physiol. 102, 2389–2397 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Wust S., et al. , Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the DlK1-Dio3 mega gene cluster. Cell Metab. 27, 1026–1039 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Millay D. P., Sutherland L. B., Bassel-Duby R., Olson E. N., Myomaker is essential for muscle regeneration. Genes Dev. 28, 1641–1646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X., et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell A. L., Eng D., Gross M. K., Kioussi C., Prediction of gene network models in limb muscle precursors. Gene 509, 16–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capkovic K. L., Stevenson S., Johnson M. C., Thelen J. J., Cornelison D. D., Neural cell adhesion molecule (NCAM) marks adult myogenic cells committed to differentiation. Exp. Cell Res. 314, 1553–1565 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scita G., Confalonieri S., Lappalainen P., Suetsugu S., IRSp53: Crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 18, 52–60 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Liu N., et al. , Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 111, 4109–4114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umansky K. B., et al. , Runx1 transcription factor is required for myoblasts proliferation during muscle regeneration. PLoS Genet. 11, e1005457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostrovsky O., Bengal E., Aronheim A., Induction of terminal differentiation by the c-Jun dimerization protein JDP2 in C2 myoblasts and rhabdomyosarcoma cells. J. Biol. Chem. 277, 40043–40054 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Bruusgaard J. C., Liestol K., Ekmark M., Kollstad K., Gundersen K., Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467–478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blackburn D. M., et al. , High-resolution genome-wide expression analysis of single myofibers using SMART-Seq. J. Biol. Chem. 294, 20097–20108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chemello F., et al. , Transcriptomic analysis of single isolated myofibers identifies miR-27a-3p and miR-142-3p as regulators of metabolism in skeletal muscle. Cell Rep. 26, 3784–3797.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Giordani L., et al. , High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol. Cell 74, 609–621.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 53.De Micheli A. J., et al. , Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 30, 3583–3595.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oprescu S. N., Yue F., Qiu J. M., Brito L. F., Kuang S., Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration. iScience 23, 100993 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xi H. B., et al. , A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells. Cell Stem Cell 27, 181–185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubenstein A. B., et al. , Single-cell transcriptional profiles in human skeletal muscle. Sci. Rep. 10, 229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada T., et al. , Myoglobin and the regulation of mitochondrial respiratory chain complex IV. J. Physiol. 594, 483–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polla B., D’Antona G., Bottinelli R., Reggiani C., Respiratory muscle fibres: Specialisation and plasticity. Thorax 59, 808–817 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamdar F., et al. , Stem cell-derived cardiomyocytes and beta-adrenergic receptor blockade in Duchenne muscular dystrophy cardiomyopathy. J. Am. Coll. Cardiol. 75, 1159–1174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Angione A. R., Jiang C., Pan D., Wang Y. X., Kuang S., PPARdelta regulates satellite cell proliferation and skeletal muscle regeneration. Skelet. Muscle 1, 33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rathbone C. R., Booth F. W., Lees S. J., Sirt1 increases skeletal muscle precursor cell proliferation. Eur. J. Cell Biol. 88, 35–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin D., Hidaka K., Shirai M., Morisaki T., RNA-binding motif protein 24 regulates myogenin expression and promotes myogenic differentiation. Genes Cells 15, 1158–1167 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Bower N. I., et al. , Stac3 is required for myotube formation and myogenic differentiation in vertebrate skeletal muscle. J. Biol. Chem. 287, 43936–43949 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrany M. J., Song T., Sadayappan S., Millay D. P., Myocyte-derived Myomaker expression is required for regenerative fusion but exacerbates membrane instability in dystrophic myofibers. JCI Insight 5, e136095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McGreevy J. W., Hakim C. H., McIntosh M. A., Duan D., Animal models of Duchenne muscular dystrophy: From basic mechanisms to gene therapy. Dis. Model. Mech. 8, 195–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kharraz Y., Guerra J., Pessina P., Serrano A. L., Munoz-Canoves P., Understanding the process of fibrosis in Duchenne muscular dystrophy. BioMed Res. Int. 2014, 965631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsakas A., Yadav V., Lorca S., Narkar V., Muscle ERRgamma mitigates Duchenne muscular dystrophy via metabolic and angiogenic reprogramming. FASEB J. 27, 4004–4016 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stahl P. L., et al. , Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Rodriques S. G., et al. , Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vickovic S., et al. , High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall Z. W., Ralston E., Nuclear domains in muscle cells. Cell 59, 771–772 (1989). [DOI] [PubMed] [Google Scholar]
- 73.Pavlath G. K., Rich K., Webster S. G., Blau H. M., Localization of muscle gene products in nuclear domains. Nature 337, 570–573 (1989). [DOI] [PubMed] [Google Scholar]
- 74.Long C., et al. , Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 345, 1184–1188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long C., et al. , Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351, 400–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makarewich C. A., et al. , The DWORF micropeptide enhances contractility and prevents heart failure in a mouse model of dilated cardiomyopathy. eLife 7, e38319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Z., et al. , Mechanistic basis of neonatal heart regeneration revealed by transcriptome and histone modification profiling. Proc. Natl. Acad. Sci. U.S.A. 116, 18455–18465 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cui M., et al. , Dynamic transcriptional responses to injury of regenerative and non-regenerative cardiomyocytes revealed by single-nucleus RNA sequencing. Dev. Cell 53, 102–116.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young M. D., Behjati S., SoupX removes ambient RNA contamination from droplet based single-cell RNA sequencing data. bioRxiv: 10.1101/303727 (3 February 2020). [DOI] [PMC free article] [PubMed]
- 81.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janky R. et al., iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 10, e1003731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 86.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistical analyses were performed using GraphPad Prism 8 using a two-tailed unpaired t test, with P value < 0.05 considered significant. All data are displayed as mean ± SEM unless otherwise indicated. All sequencing data have been deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE156498.