Abstract
Motivation
Biological rhythmicity is fundamental to almost all organisms on Earth and plays a key role in health and disease. Identification of oscillating signals could lead to novel biological insights, yet its investigation is impeded by the extensive computational and statistical knowledge required to perform such analysis.
Results
To address this issue, we present DiscoRhythm (Discovering Rhythmicity), a user-friendly application for characterizing rhythmicity in temporal biological data. DiscoRhythm is available as a web application or an R/Bioconductor package for estimating phase, amplitude and statistical significance using four popular approaches to rhythm detection (Cosinor, JTK Cycle, ARSER and Lomb-Scargle). We optimized these algorithms for speed, improving their execution times up to 30-fold to enable rapid analysis of -omic-scale datasets in real-time. Informative visualizations, interactive modules for quality control, dimensionality reduction, periodicity profiling and incorporation of experimental replicates make DiscoRhythm a thorough toolkit for analyzing rhythmicity.
Availability and implementation
The DiscoRhythm R package is available on Bioconductor (https://bioconductor.org/packages/DiscoRhythm), with source code available on GitHub (https://github.com/matthewcarlucci/DiscoRhythm) under a GPL-3 license. The web application is securely deployed over HTTPS (https://disco.camh.ca) and is freely available for use worldwide. Local instances of the DiscoRhythm web application can be created using the R package or by deploying the publicly available Docker container (https://hub.docker.com/r/mcarlucci/discorhythm).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Biological rhythmicity, including circadian and other cycles (Schibler and Naef, 2005), are fundamentally important to life on Earth (Bell-Pedersen et al., 2005). Numerous lines of evidence have indicated that disturbance of biological rhythms is a risk factor for human morbidities, including psychiatric, metabolic and malignant diseases (Roenneberg and Merrow, 2016). Several approaches can be used to estimate the phase, amplitude and statistical significance of these rhythms in time-series data, where each methodology has strengths and weaknesses under various conditions (Deckard et al., 2013). Current implementations of these algorithms in R (e.g. JTK Cycle, ARSER and Lomb-Scargle) tend to be slow and difficult to use. Additionally, no unified toolkit exists for performing pre-processing, dimensionality reduction, period detection and visualization of oscillation statistics, all of which require specialized knowledge and expertise. To address these challenges, we developed DiscoRhythm (Discovering Rhythmicity), a web application and accompanying R/Bioconductor package for analyzing rhythmicity in temporal biological datasets. DiscoRhythm provides a unified interface to execute four methods of rhythm estimation, and heuristically selects suitable approaches for the data being analyzed. By providing interactive modules for outlier detection, analysis of replicates and periodicity profiling, DiscoRhythm offers a framework for accessible analysis of periodic datasets in a web browser or in R.
2 Results
DiscoRhythm is implemented as a package in the R programming language (ver. 3.6+) with the web interface based on the R Shiny platform (Chang et al., 2018), capable of reproducing findings in transcriptomic (Li et al., 2013), epigenomic (Oh et al., 2019), metabolomic (Krishnaiah et al., 2017), proteomic (Hurley et al., 2018) and other similar datasets. A workflow in DiscoRhythm begins with a matrix of temporal data (Fig. 1), where two metrics are computed to filter outlier samples, followed by a feature selection procedure based on the ratio of biological signal to technical noise. Dominant periods are determined using dataset-wide period evaluation procedures, and finally, multiple rhythm detection methods are executed on each feature to infer the presence of rhythms. Results of the web session may be emailed or downloaded upon completion as a zip file also containing the R data and code required for future reproducibility.
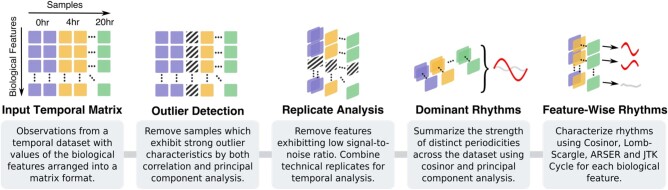
Fig. 1.
Overview of the analysis procedures performed by DiscoRhythm. The illustration shows the step-wise operations being performed on the input circadian data matrix. All procedures are performed on a matrix with the biological features represented by rows and temporal samples by columns. Columns with the same color (e.g. purple and yellow) represent technical replicates, while stripped boxes indicate samples or features removed for downstream analysis. Red and gray oscillating lines show significant and non-significant rhythms, respectively. hr, hours
2.1 Input
Input for the web interface is a single table in a CSV (comma separated values) format. Columns contain samples named according to their time of collection, and rows contain values of observed features. Experimental design specifications regarding technical replicates, units of time and the main period of interest are also required. A circadian gene expression dataset simulated using simphony (Singer et al., 2019) is provided in order to highlight the available features and demonstrate the sample naming scheme. In addition to the tabular input of the graphical interface, the DiscoRhythm R package also accepts SummarizedExperiment objects commonly returned by other R packages in Bioconductor (Gentleman et al., 2004).
2.2 Outlier detection and feature selection
Sample quality is assessed using two commonly utilized metrics for outlier detection. The first metric is the average inter-sample correlation, computed as a mean pairwise correlation between a given sample and all other samples (Oldham et al., 2008), while the second metric(s) is the sample score returned by principal component analysis (PCA). For both metrics, samples that deviate considerably from the rest (beyond a user-defined threshold) are flagged as outliers for removal from further analysis.
If present, technical replicates can be used to determine the signal-to-noise ratio for each feature (i.e. F statistic of biological versus technical variation). For further analysis, the user is able to only select the features exhibiting high signal-to-noise ratio, determined either by effect size or statistical significance. Technical replicates can then be combined by taking the mean, median or by choosing one replicate at random to prevent inflated sample size stemming from non-independent measurements.
2.3 Period detection
Two approaches are available for identifying the dominant period of rhythmicity. First, a goodness of fit can be evaluated for each period using a cosinor model across all selected features, returning the median coefficient of determination (R2) of the fits. Alternatively, global rhythmic patterns may be investigated using PCA scores. If ‘circular time’ is used for sample collection [e.g. time of day, in hours, is recorded over multiple days as 2, 4, …, 24, 2, 4, …, where samples with the same collection times are assumed to be biological replicates (Hughes et al., 2017)], DiscoRhythm will limit the candidate periods for rhythm detection to p/k where p is the length of the cycle and k is a positive integer (Cornelissen, 2014).
2.4 Estimating rhythm characteristics
Oscillations can be detected for each feature using a user-specified period. The period should be chosen by an a priori hypothesis or detected by the procedures in Section 2.3. An interface is provided to four commonly used approaches to oscillation detection [Cosinor (Cornelissen, 2014), ARSER (Yang and Su, 2010), JTK Cycle (Hughes et al., 2010) and Lomb-Scargle (Glynn et al., 2006)]. Each is heuristically made available if the input dataset satisfies algorithm-specific criteria, such as: the presence (or absence) of missing values, biological replicates, uneven sampling frequencies or non-integer intervals. To make DiscoRhythm suitable for -omic-scale and real-time analysis, high-performance implementations of each algorithm were developed, with runtime improvements of up to 30-fold [Supplementary Table S1 and Fig. S1; parallelized ARSER, JTK Cycle and Lomb-Scargle were contributed directly to MetaCycle version 1.2 (Wu et al., 2016)]. Each method returns estimated phases, amplitudes, and P-values, both raw and adjusted for multiple testing (Benjamini and Hochberg, 1995). These feature-specific rhythm characteristics can be interactively visualized and downloaded for further exploration.
3 Discussion
Rhythmicity is a common topic of discussion for most biological researchers. Yet the quantitative analysis is difficult and, therefore, almost exclusively performed by researchers with specialization in statistics and computation. To democratize the field of chronobiology, we developed DiscoRhythm—a suite of standardized analytical procedures made approachable through interactivity, informative visualizations and key statistics for characterizing rhythmic patterns of temporal datasets. This new tool will enable even non-computational researchers to extract insights on the rhythmicity of biological data in a highly efficient manner. While our workflow is optimized for -omic-scale experiments, future versions of DiscoRhythm will also tailor to lower throughput datasets. Lastly, to maintain and extend accessibility to relevant periodic analysis approaches, we plan to adopt new methods as they become available in R/Bioconductor.
Supplementary Material
Acknowledgements
The authors thank all of those involved in the testing of DiscoRhythm at all stages of the project, especially Edward Oh and Paul Liu. We thank MetaCycle maintainer, Gang Wu, for reviewing and publishing the performance improvements to the oscillation detection algorithms. Computations were performed on the CAMH Specialized Computing Cluster funded by The Canada Foundation for Innovation, Research Hospital Fund. We also thank the three anonymous reviewers of this article whose comments resulted in improvements of the manuscript and software.
Funding
This work was supported by the Lithuanian Science Foundation (S-MIP-19-66), the Canadian Institutes of Health Research [MOP-119451, MOP-133496, PJT 148719, NTC-154084, IGH-155180, TGH-158223], the National Institute of Mental Health [1R01MH105409-01], Brain Canada and CAMH Foundation [554] and the Krembil Foundation.
Conflict of Interest: none declared.
References
- Bell-Pedersen D. et al. (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat. Rev. Genet., 6, 544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol., 57, 289–300. [Google Scholar]
- Chang W. et al. (2018) shiny: web application framework for R, 2015. R Package Version, 1, 14. [Google Scholar]
- Cornelissen G. (2014) Cosinor-based rhythmometry. Theor. Biol. Med. Model., 11, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckard A. et al. (2013) Design and analysis of large-scale biological rhythm studies: a comparison of algorithms for detecting periodic signals in biological data. Bioinformatics, 29, 3174–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R.C. et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol., 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E.F. et al. (2006) Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. Bioinformatics, 22, 310–316. [DOI] [PubMed] [Google Scholar]
- Hughes M.E. et al. (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms, 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M.E. et al. (2017) Guidelines for genome-scale analysis of biological rhythms. J. Biol. Rhythms, 32, 380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.M. et al. (2018) Circadian proteomic analysis uncovers mechanisms of post-transcriptional regulation in metabolic pathways. Cell Syst., 7, 613–626.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaiah S.Y. et al. (2017) Clock regulation of metabolites reveals coupling between transcription and metabolism. Cell Metab., 25, 961–974.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z. et al. (2013) Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. USA, 110, 9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh G. et al. (2019) Circadian oscillations of cytosine modification in humans contribute to epigenetic variability, aging, and complex disease. Genome Biol., 20, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham M.C. et al. (2008) Functional organization of the transcriptome in human brain. Nat. Neurosci., 11, 1271–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T., Merrow M. (2016) The circadian clock and human health. Curr. Biol., 26, R432–R443. [DOI] [PubMed] [Google Scholar]
- Schibler U., Naef F. (2005) Cellular oscillators: rhythmic gene expression and metabolism. Curr. Opin. Cell Biol., 17, 223–229. [DOI] [PubMed] [Google Scholar]
- Singer J.M. et al. (2019) Simphony: simulating large-scale, rhythmic data. PeerJ, 7, e6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. et al. (2016) MetaCycle: an integrated R package to evaluate periodicity in large scale data. Bioinformatics, 32, 3351–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Su Z. (2010) Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics, 26, i168–i174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.