Abstract
Plant development programs are constantly updated by information about environmental conditions, currently available resources, and sites of active organogenesis. Much of this information is encoded in modifications of transcription factors that lead to changes in their relative abundance, activity and localization. Recent work on the Auxin Response Factor family of transcription factors has highlighted the large diversity of such modifications, as well as how they may work synergistically or antagonistically to regulate downstream responses. ARFs can be regulated by alternative splicing, post-translational modification, and subcellular localization, among many other mechanisms. Beyond the many ways ARFs themselves can be regulated, they can also act cooperatively with other transcription factors to enable highly complex genetic networks with distinct developmental outcomes. Multi-level regulation like what has been documented for ARFs has the capacity to generate flexibility in transcriptional outputs, as well as resilience to short-term perturbations.
Keywords: transcription, multi-level regulation, developmental flexibility, developmental, robustness, gene regulatory networks, biomechanical regulation, post-translational, modifications, alternative splicing
Introduction
Small molecule signaling is an essential mechanism by which plants develop in response to internal cues and external environmental stimuli. At first pass, the expansive range of regulation of signaling pathways can seem overwhelming, as plants must integrate an enormous diversity of input signals into a cohesive whole. However, this complexity may be inextricably linked to survival. In systems biology theory, multi-level regulatory modules can generate arbitrarily complex computations, providing systems with the increased flexibility to discriminate between distinct stimuli (Alon, 2006). At the same time, multi-mode regulation increases robustness, as redundant encoding allows for the optimal response even with incomplete information.
Most plant hormone signaling relies of the activity of specific transcription factor families. The function of these transcription factors in turn is regulated by hormone perception pathways. In the case of auxin signaling, the activity of Auxin Response Factors (ARFs) is promoted by the addition of auxin, which targets the Auxin/Indole-3-acetic acid (Aux/IAA) repressors for proteasomal-mediated degradation. This relatively simple relief of repression module is complicated by the large gene families of ARF and Aux/IAA proteins, and allows for subfunctionalization and specificity in interaction strengths between family members, as reviewed in Li et al., 2016 and Roosjen et al., 2018. Further complicating auxin response is the large variety of regulation impinging on ARF activity independently of auxin perception. Consequently, auxin response serves as an ideal case study for integration of multiple inputs by transcriptional response. In this review, we will focus on how the multi-level regulation of ARF activity allows for flexibility and robustness in plant development.
ARFs are regulated at the transcriptional level
ARFs are grouped into three main clades, the result of retained duplications from an ancient evolutionary divergence (Mutte et al., 2018). These three clades (termed A, B, and C) are traditionally categorized as activator (A) or repressor (B and C) proteins. Recently developed transcriptional reporters of the class A ARFs showed that their spatial expression in Arabidopsis roots is strongly dependent on both a long 5’ promoter region (approximately three to five kilobases) and, in the case of ARF7, their first intron sequences (Truskina et al., 2018*). While most class A ARFs are broadly expressed in the root meristem, ARF8 expression is restricted to outer cell files and ARF19 expression is restricted to root cap, epidermal, and pericycle cells (Figure 1A). These expression patterns differ from those of previous reporters that only included a two kilobase 5’ upstream promoter sequence and no intergenic DNA (Rademacher et al., 2011). A recent study in Marchantia polymorpha found MpARF1 localizes to the entirety of the gemmae, while MpARF2 is only found on the apical edge (Kato et al., 2019). Consequently which ARFs are present in any cell is dependent on cellular development and positioning. Temporal and spatial regulation of ARF expression causes different ARFs to be present in certain tissues. Auxin response then depends on what other factors are present in these tissues and interact with the ARFs, causing distinct developmental outcomes.
Figure 1: Regulation of ARF function at the transcriptional and protein level.
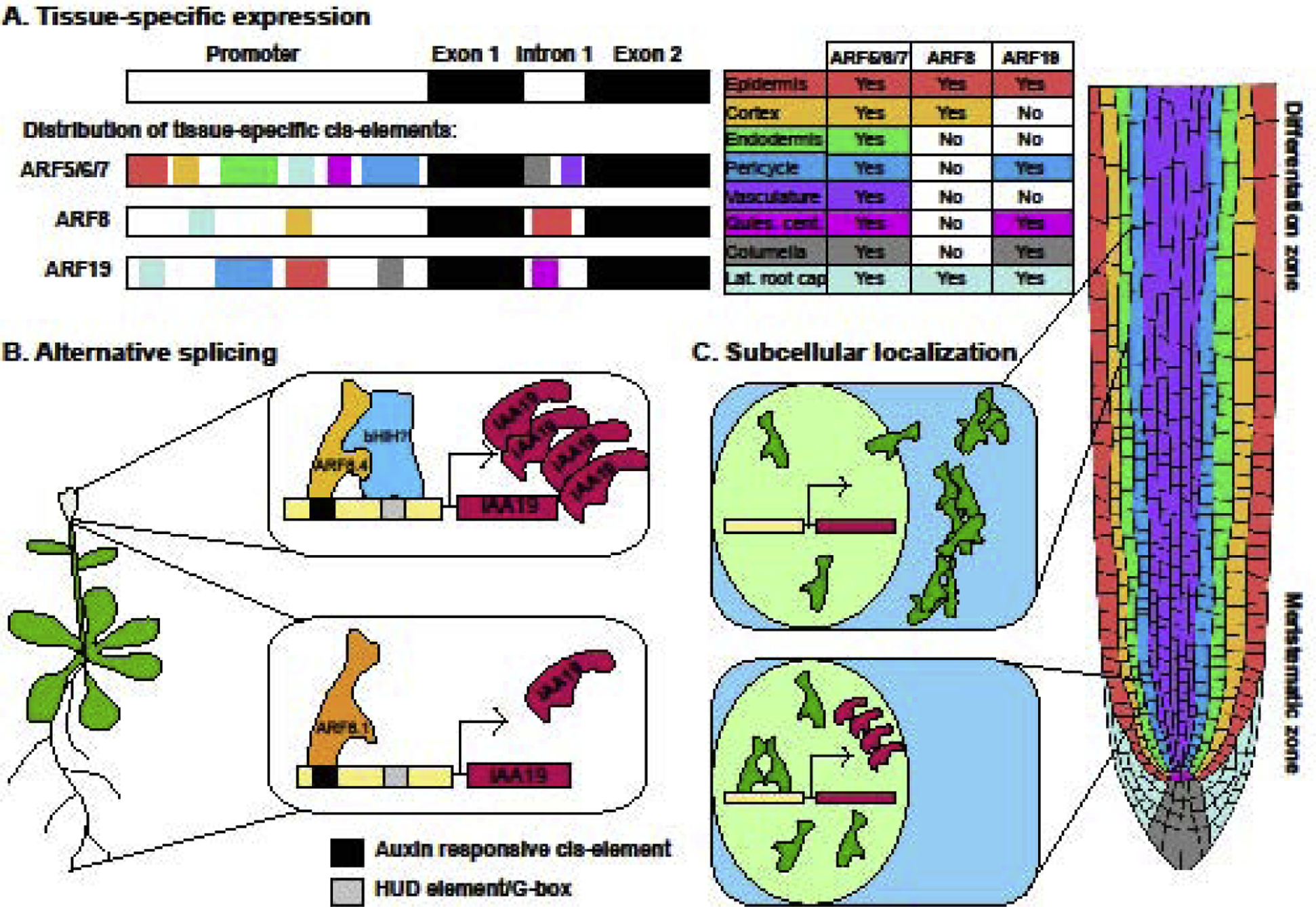
A. Different ARFs show spatially specific expression patterns, as exemplified by the pattern of class A ARF expression in the root. Likely cis-elements within the unique ARF promoters and the first intron regulate tissue-specific expression. B. Alternative splicing can regulate ARF expression profiles and function—a splice variant of ARF8, ARF8.4, is only expressed in developing flowers. This variant binds more strongly to regions of the IAA19 promoter that include G-boxes and HUD-box elements, potentially by interacting with bHLH or other transcription factors that bind to these elements. The more globally expressed variant ARF8.1 binds less strongly to this promoter, potentially because its protein structure disallows binding to interaction partners. C. ARF protein localization is regulated on the subcellular level—ARF7 and ARF19 localize to the nucleus in the meristematic region of the primary root, where they promote transcription of auxin target genes, but aggregate in the cytoplasm in the differentiation zone, where they are no longer active.
Alternative splicing is another method of regulating ARF expression and interaction partners. A recent paper showed ARF8 splice variants differ in their expression profiles, with the splice variant 8.4 expressed specifically in the developing Arabidopsis inflorescence (Figure 1B) (Ghelli et al., 2018**). This splice variant has a 28 amino acid insertion compared to the splice variants ARF8.1 and ARF8.2, which are more globally expressed. ARF8.2 and ARF8.4 also lack the last 38 amino acids of ARF8.1 due to an early stop codon. Overexpression of ARF8.4 in an arf8–7 mutant fully rescues transcription of the auxin-responsive gene IAA19, unlike ARF8.1 and ARF8.2. Interestingly, ARF8.4 binds regions of the IAA19 promoter that contain G-boxes and HUD elements more strongly than the other variants. As these elements are binding sites for other transcription factors, ARF8.4 may exhibit cooperative activity on this promoter that the other variants do not. Splice variants have also been observed for ARFs in tomato, and these variants also show tissue-specific expression in reproductive tissue, though in this case expression is regulated temporally during the transition from flower to fruit (Zouine et al., 2014). How splicing can impact transcription factor expression and protein-protein interactions is understudied, and may be a more universal mechanism to promote cooperativity in hormone responses.
ARFs are regulated at the level of protein structure and localization
Protein localization is another key mechanism by which plants regulate transcriptional factor activity, as transcription factors specifically act in the nucleus. A recent paper showed that subcellular localization of ARF7 and ARF19 proteins varies along the Arabidopsis root axis—ARF7 and ARF19 localize in the nucleus in the dividing region of the primary root but relocalize to large cytosolic aggregates in the differentiated region of the root (Powers et al., 2019**) (Figure 1C). This localization profile directly parallels ARF activity profiles. ARF7 and ARF19 regulate lateral root initiation, a process that only occurs in the dividing region of the root, where the ARFs localize to the nucleus and are presumably active. Aggregation and nuclear export may be an important mechanism by which transcription factors can quickly be rendered nonfunctional. Such mechanisms keep a reserve of fully folded protein present that could quickly be reactivated, allowing development to be environmentally responsive and dynamic.
Post-translational modifications of ARFs theoretically produce autonomous, stackable layers of transcriptional control, but how these modifications interact with each other or contribute to cooperativity is unknown. Phosphorylation of the ARFs has been shown to be critical for ARF function for several family members. Phosphorylation of the B class ARF2 by the brassinosteroid-regulated kinase BIN2 decreases its binding to DNA (Vert et al., 2008), and, as ARF2 is a transcriptional repressor, presumably increases auxin response (Figure 2A). ARF2 phosphorylation is upregulated by low potassium conditions (Zhao et al., 2016), decreasing ARF2 binding to the promoter of a potassium transporter HAK5 and relieving repression on this gene. Phosphorylation of ARF2 can consequently integrate at least three hormonal and environmental signaling pathways: 1) auxin response, 2) brassinosteroid response, and 3) potassium deficiency response. BIN2 can also phosphorylate class A ARF7 and ARF19, though this phosphorylation event promotes ARF-DNA binding and auxin response by disturbing ARF interactions with their IAA repressors (Figure 2B) (Cho et al., 2014). The identified phosphorylation sites on ARF2 and ARF7 are not shared, suggesting that these post-translational modifications may be ARF-specific.
Figure 2: Post-translational regulation of ARF activity.
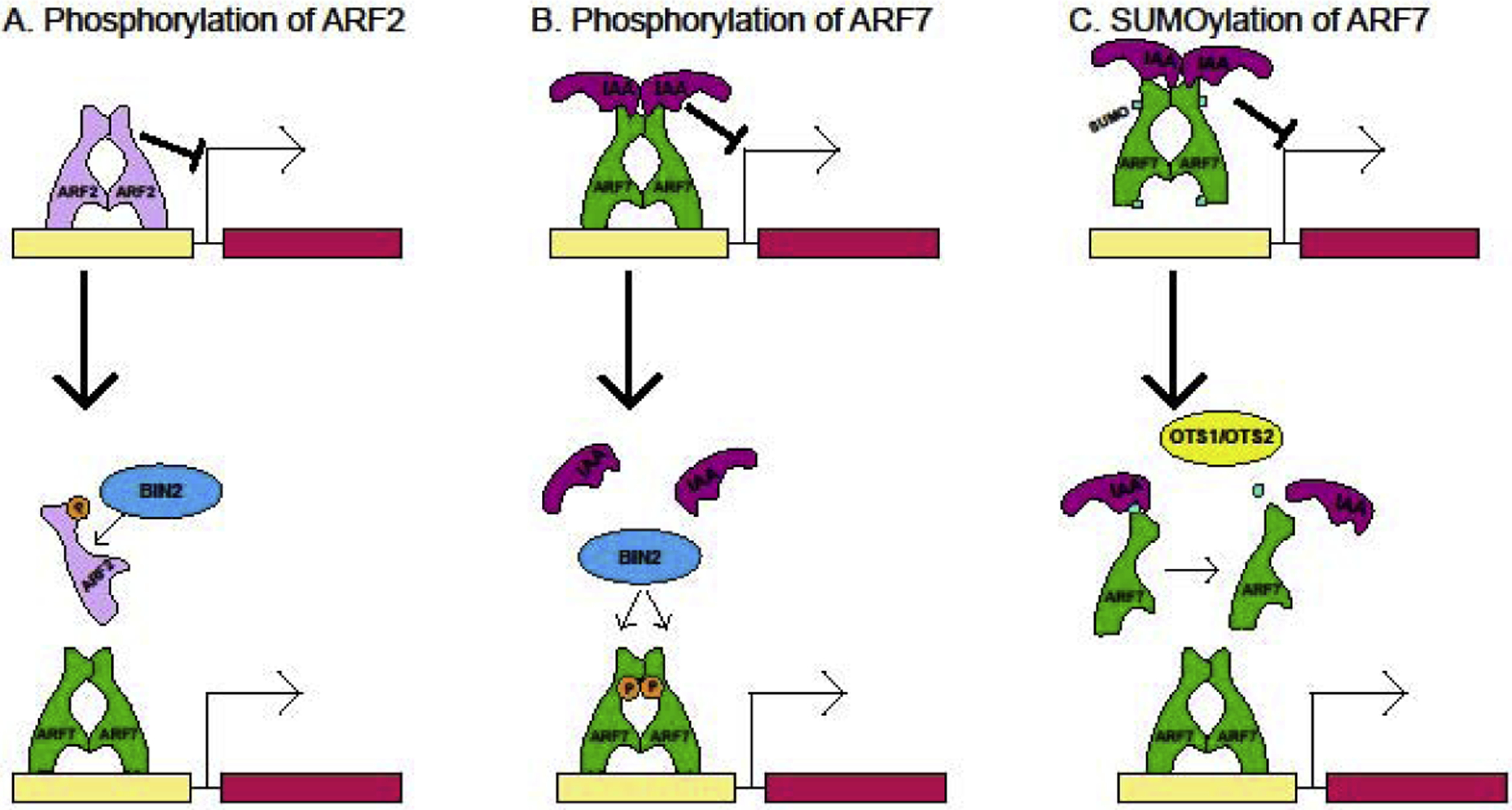
ARF activity is regulated by phosphorylation and sumoylation. A. Phosphorylation of the B class ARF2 by GSK3 kinase BIN2 decreases its affinity for DNA, potentially decreasing competition for binding sites and allowing A class ARFs to bind promoters and activate transcription. B. Phosphorylation of ARF7 and ARF19 by BIN2 promotes transcription by interfering with the interacting between ARF7 and its repressor IAA proteins. C. Sumoylation of ARF7 promotes the interaction between ARF7 and the IAA3 repressor and decreases ARF DNA-binding affinity. Removal of the SUMO group by proteases OTS1/OTS2 decreases the strength of IAA3-ARF7 interactions and increases ARF7 binding to and activity on the promoters of target genes.
Other post-translational modifications further modulate auxin responsiveness. Sumoylation of ARF7 interferes with its DNA-binding capacity and promotes orientation of Arabidopsis lateral root emergence towards a water source (Figure 2C) (Orosa-Puente et al., 2018*). Directly contrary to phosphorylation of ARF7, sumoylation promotes ARF-IAA recruitment. Consequently removal of the SUMO group by proteases OTS1/OTS2 promotes auxin response on the water-rich side of the root and causes lateral root emergence. Sumoylation promotion of ARF-IAA interaction is specific to IAA3, which acts during lateral root emergence, exhibiting how post-translational modifications can regulate protein-protein interactions in a highly specific manner. Furthermore only ARF7 activity was shown to affect hydropatterning, though it is possible SUMOylation of other ARFs occurs and may have distinct developmental outcomes. Interestingly, none of the identified phosphorylation sites for ARF2 (Zhao et al., 2016) and ARF7/ARF19 (Cho et al., 2014) or sumoylation sites for ARF7 (Orosa-Puente et al., 2018*) are near their proposed site of action (the DNA-binding domain for ARF2 phosphorylation and the ARF-IAA interaction face for ARF7/ARF19 phosphorylation and ARF7 sumoylation).
Interactions between ARFs and other transcription factors impact development
Combinatorial control of transcription, where transcription factors from multiple pathways interact on a promoter to cause a response qualitatively or quantitatively different than either of the factors alone, is a signature of cooperativity between signaling pathways. Despite a plethora of research on how auxin response is affected by other signaling pathways, data on the physical interactions between ARFs and other transcription factors on promoters are relatively sparse (reviewed in Table 1). An analysis of natural variation in auxin response proteins among Arabidopsis ecotypes found that ARF natural variation is enriched in the variable middle and C-terminal domains of several family members (Hamm et al., 2019). These regions are protein interaction domains, suggesting that ARF-protein interactions may occur more broadly than characterized and act as targets for evolutionary diversification.
Table 1:
Cooperative interactions characterized between ARFs and other transcription factors
| ARF | Other TF | Signaling pathway/class of other TF | Interaction characterized | Citation |
|---|---|---|---|---|
| ARF3 | IND, RPL, BP | Gynoecium development/bH LH (IND)/homeobox signaling (RPL, BP) (Liljegren et al., 2004; Roeder et al., 2003; Venglat et al., 2002) | Direct (BiFC, Y2H) | Simonini et al., 2018** |
| ARF3/ARF4 | FIL | Floral initiation/YABBY signaling (Kumaran et al., 2002) | Direct (BiFC, Y2H, co-IP) | Chung et al., 2019* |
| ARF5 | BES1 | Brassinosteroid signaling (Zhao et al., 2002) | Cooperative activity on promoter | Walcher and Nemhauser, 2012; Galstyan and Nemhauser, 2019 |
| ARF6/ARF8 | FUL | Fruit development/MA Ds-box signaling (Gu et al., 1998) | Direct (BiFC, Y2H) | Ripoll et al., 2015 |
| ARF7 | MYB77 | Potassium signaling (Shin and Schachtman, 2004) | Direct (BiFC, in vitro pull-down) | Shin et al., 2007 |
| ARF8 | BIGPETALp | Petal growth/bHLH signaling (Szécsi et al., 2006) | Direct (BiFC, Y2H) | Varaud et al., 2011 |
A recent study on ARF3 (ETTIN) is an excellent example of how protein-protein interactions can generate cooperative responses. In Arabidopsis ARF3 directly interacts with multiple transcription factors of varying classes, specifically INDEHISCENT (IND), BREVIPEDICELLUS (BP), and REPLUMLESS (RPL), to shape the development of the gynoecium (Simonini et al., 2018**). All of these factors bind to the promoter of cell-wall modification enzyme XYLOGLUCAN ENDOTRANSGLYCOSYLASE/HYDROLASE 7 (XTH7) and regulate its expression. ARF3 upregulates XTH7 expression while IND, BP, and RPL downregulate its expression, suggesting that the composition of the protein complex of these factors may change the effect the complex has on transcription (Simonini et al., 2018**). Consequently which proteins are active in a specific tissue and at a specific developmental stage can generate unique responses. The observed variation in style morphologies across different plants might be explained by species-specific affinities between interacting factors, generating diversity in plant form on the evolutionary scale. Earlier in reproductive development, ARF3 and its close homologue ARF4 directly interact with another transcription factor, FILAMENTOUS FLOWER (FIL) (Chung et al., 2019*), to promote flower formation, further supporting the hypothesis that ARF-protein interactions are dynamic, and that their function depends on which interacting factors are present.
A high-throughput study of Arabidopsis ARF interactions using a novel yeast two hybrid method called CrY2H-seq identified many candidates for cooperative action (Trigg et al., 2017). CrY2H-seq uses a Cre recombinase downstream of a yeast two hybrid protein-protein interaction reporter, which then is able to link interacting bait and prey plasmids via Cre-mediated recombination. The study queried interactions among more than a thousand transcription factors, including eight ARFs, and revealed more than eight thousand interactions, the vast majority of them novel. Some of these interactions are quite ARF-specific. For instance, ARF18 interacts with abscisic acid response factors, while other ARFs in the study do not. Intriguingly ARF19 interacts with 10-fold more proteins than any of the other ARFs tested, suggesting it may serve as an integration hub for diverse signals.
Combinatorial control of transcription has signatures in promoter structure
Promoter structure is another lens by which one can look for combinatorial control of transcription, and recent studies focusing on auxin-responsive promoters suggest broad transcription factor co-occupancy on auxin-responsive promoters in Arabidopsis (Mironova et al., 2014; Omelyanchuk et al., 2017). One analysis tested for the enrichment of different transcription factor binding sites in auxin-responsive promoters (Cherenkov et al., 2018*). The authors found that the promoters of genes that are upregulated rapidly in response to auxin, and consequently may be direct ARF targets, are enriched for binding sites for TCP, bZIP, and bHLH transcription factor classes. Binding sites for these factors were also significantly enriched in ARF5- and ARF6-binding data sets. Notably, even when direct interactions are not observed, promoters that contain cis-elements for both interacting partners can still exhibit cooperativity, suggesting that binding of each factor may increase affinity or residency of the other on the promoter (Walcher and Nemhauser, 2012). For example binding of ARF5 to promoters that contain both auxin-responsive and brassinosteroid-responsive cis-elements is increased in a stabilized mutant of the brassinosteroid response transcription factor BES1 (Galstyan and Nemhauser, 2019).
Cooperative transcription factor interactions are often thought of as promoting transcriptional activity, but equally important for robust and replicable development is the repression of response. Repression of auxin response could occur through the competition between activator and repressor ARFs for binding sites, as has been shown in Physcomitrella patens (Lavy et al., 2016). Recent work in Marchantia posits a similar competition mechanism, though intriguingly B-class ARF are auxin-insensitive, providing an auxin-independent mechanism of decreasing activation (Kato et al., 2019). Similar transcription repression can be conferred by non-auxin transcription factors, as was shown in the Arabidopsis shoot apical meristem. High auxin response promotes cell differentiation in the shoot apical meristem, but in order to retain a stem cell niche in the central zone a low auxin response is necessary. This low response is maintained without leading to differentiation by the the activity of the transcription factor WUSCHEL, which directly represses ARF targets (Ma et al., 2019**). On promoters of these ARF targets WUSCHEL acts in opposition to ARFs promoting transcription. The combinatorial effect of the factors leads to a low auxin response that does not reach the threshold to cause differentiation, permitting stem cell persistence. Antagonistic pathways allow for balancing and threshold models of development and prevent noisiness within a pathway and unchecked development.
Biophysical constraints act independently of transcription to produce developmental resilience
Cooperativity in transcriptional regulation generates developmental complexity, but such complexity carries the risk of increasing noise within the system and impeding a coherent response. As plants must develop in environments that are in constant flux, excessive responsiveness to minor environmental perturbations would negatively impact plant fitness. Consequently, plants use biomechanical constraints to restrict this noisiness by acting as a method of regulating developmental response independently of transcription. This gives resilience to growth processes that must occur in all environmental conditions. On the cellular level, one of these processes is cell expansion, required for anisotropic growth in the root and shoot that determines plant form. Vacuole size and morphology regulate cell expansion, and vacuole size itself is regulated by multiple regulatory modules, including auxin. However, a recent paper showed that Arabidopsis vacuole size can also be regulated independently of auxin by the actin-binding protein NET4, which localizes to the vacuolar membrane and promotes its constriction (Kaiser et al., 2019). NET4’s effect on vacuole size does not depend on auxin treatment, suggesting that this cytoskeleton regulator provides an orthogonal mechanism for regulating organ growth and tissue expansion that can act as a failsafe against deleterious transcriptional fluctuations.
Oscillations in auxin response in the primary root meristem specify which cells are competent for lateral root development (De Smet et al., 2007). Auxin transport and programmed cell death play a role in generating these oscillations (Moreno-Risueno et al., 2010; Xuan et al., 2016). However, a recent modeling paper showed that such oscillations can be generated purely by the stochasticity of rates of cell division in the meristem, which leads to cells of differing sizes and differing capacity for auxin loading (Berg and Tusscher, 2018**). Specification of lateral root-competent cells is important for plant fitness, as a plant without the capacity to form lateral roots will have lowered ability to acquire nutrients and maintain vertical growth. Which of these competent cells is activated to form a root is the product of a complex processing of a large number of environmental, metabolic and genetic factors. Because plants require the ability to form lateral roots in all environmental conditions, a model where specification occurs independently of this complex processing and is environmentally agnostic may increase plant fitness.
Conclusions
Considering the massive body of research on hormone signaling’s regulation of transcription, it is surprisingly difficult to draw a link between any one transcription factor and a specific morphological outcome. Many nuclear auxin response proteins do not show single null mutant phenotypes, due to the partial redundancy of these large gene families and the interconnectivity of their members. On the other hand, ARFs necessary for embryonic development, such as ARF5, play key roles in the adult plant that are not apparent from their early-arresting null mutants. While redundancy, pleiotropy, and polygenism are often explanations of this disconnect between genetic perturbations and downstream development, how cooperativity generates complexity in small molecule hormone signaling is another likely culprit. The relative simplicity of auxin signaling makes the multiplicity of regulation at every step of this pathway clear—as there are only a handful of pathway components to act as loci for regulation, each component is subject to multiple modes of regulation acting at diverse temporal and spatial scales.
In this review, we focused on the ARF transcription factors as exemplar signaling integrators but multi-level regulation impacts all auxin signaling proteins, generating even further diversity in downstream responses. Auxin can even impact plant form independently of transcription, such as in the case of root gravitropism (Fendrych et al., 2018). Intriguingly, the auxin receptor chiefly responsible for mediating the gravitropic response does not regulate the auxin nuclear response (Prigge et al., 2020). Such orthogonality is possible because auxin response proteins are comprised of large gene families, each member of which is differently regulated, as observed in the case of ARFs. As reviewed here ARF expression, localization, post-translational modifications, and protein-protein interactions are all highly ARF-specific. Which ARFs are subject to which regulatory models adds yet another level of complexity, and this is equally true for other auxin response proteins. Multi-level modes of regulations are not unique to auxin signaling, as recently reviewed for the PHYTOCHROME INTERACTING FACTOR (PIF) proteins (Favero, 2020). Such diversity in modes of regulation is likely necessary to provide balance between the plasticity needed to rapidly adapt to new environments and the robustness required to maintain functional morphogenesis.
Highlights.
Multi-level regulation of small molecule signaling generates flexibility and robustness in plant development
Auxin signaling converges on transcriptional control by the auxin responsive transcription factors (ARFs), which act as signaling integrators
ARF function is regulated by temporally and spatially specific expression, protein structure and localization, and post-translational modifications
Protein-protein interactions and cooperative activity on promoters integrate signaling from multiple pathways at the locus of transcriptional control
Biomechanical regulatory pathways generate developmental resilience by promoting responses that are required for plant fitness in a wide range of environments
Acknowledgements
Thank you to members of the Nemhauser lab for guidance and feedback, especially Alex Leydon, Hardik Gala, and Sarah Guiziou for careful reading of the manuscript. This work was supported by the National Science Foundation (MCB-1411949), and National Institute of Health (R01- GM107084) and the Howard Hughes Medical Institute Faculty Scholar Award. AL was supported by an NSF Graduate Research Fellowship DGE-1256082. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
X The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alon U (2006) An Introduction to Systems Biology: Design Principles of Biological Circuits, 1 edition. Chapman and Hall/CRC, Boca Raton, FL [Google Scholar]
- Berg T van den, Tusscher KH ten (2018) Lateral Root Priming Synergystically Arises from Root Growth and Auxin Transport Dynamics. bioRxiv 361709 [Google Scholar]
- Cherenkov P, Novikova D, Omelyanchuk N, Levitsky V, Grosse I, Weijers D, Mironova V (2018) Diversity of cis-regulatory elements associated with auxin response in Arabidopsis thaliana. J Exp Bot 69: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. (2014) A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol 16: 66–76 [DOI] [PubMed] [Google Scholar]
- Chung Y, Zhu Y, Wu M-F, Simonini S, Kuhn A, Armenta-Medina A, Jin R, Østergaard L, Gillmor CS, Wagner D (2019) Auxin Response Factors promote organogenesis by chromatin-mediated repression of the pluripotency gene SHOOTMERISTEMLESS. Nat Commun 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frei dit Frey N, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. (2007) Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Dev Camb Engl 134: 681–690 [DOI] [PubMed] [Google Scholar]
- Favero DS Mechanisms regulating PIF transcription factor activity at the protein level. Physiol Plant. doi: 10.1111/ppl.13075 [DOI] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J (2018) Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A, Nemhauser JL (2019) Auxin promotion of seedling growth via ARF5 is dependent on the brassinosteroid-regulated transcription factors BES1 and BEH4. Plant Direct 3: e00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelli R, Brunetti P, Napoli N, De Paolis A, Cecchetti V, Tsuge T, Serino G, Matsui M, Mele G, Rinaldi G, et al. (2018) A Newly Identified Flower-Specific Splice Variant of AUXIN RESPONSE FACTOR8 Regulates Stamen Elongation and Endothecium Lignification in Arabidopsis. Plant Cell 30: 620–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm MO, Moss BL, Leydon AR, Gala HP, Lanctot A, Ramos R, Klaeser H, Lemmex AC, Zahler ML, Nemhauser JL, et al. (2019) Accelerating structure-function mapping using the ViVa webtool to mine natural variation. Plant Direct. doi: 10.1002/pld3.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S, Eisa A, Kleine-Vehn J, Scheuring D (2019) NET4 Modulates the Compactness of Vacuoles in Arabidopsis thaliana. Int J Mol Sci 20: 4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Mutte SK, Suzuki H, Crespo I, Das S, Radoeva T, Fontana M, Yoshitake Y, Hainiwa E, van den Berg W, et al. (2019) Design principles of a minimal auxin response system. bioRxiv 760876. [DOI] [PubMed] [Google Scholar]
- Lavy M, Prigge MJ, Tao S, Shain S, Kuo A, Kirchsteiger K, Estelle M (2016) Constitutive auxin response in Physcomitrella reveals complex interactions between Aux/IAA and ARF proteins. eLife 5: e13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-B, Xie Z-Z, Hu C-G, Zhang J-Z (2016) A Review of Auxin Response Factors (ARFs) in Plants. Front Plant Sci. doi: 10.3389/fpls.2016.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Miotk A, Šutiković Z, Ermakova O, Wenzl C, Medzihradszky A, Gaillochet C, Forner J, Utan G, Brackmann K, et al. (2019) WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat Commun 10: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova VV, Omelyanchuk NA, Wiebe DS, Levitsky VG (2014) Computational analysis of auxin responsive elements in the Arabidopsis thaliana L. genome. BMC Genomics 15: S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329: 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GK-S, Weijers D (2018) Origin and evolution of the nuclear auxin response system. eLife. doi: 10.7554/eLife.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelyanchuk NA, Wiebe DS, Novikova DD, Levitsky VG, Klimova N, Gorelova V, Weinholdt C, Vasiliev GV, Zemlyanskaya EV, Kolchanov NA, et al. (2017) Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci Rep 7: 2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. (2018) Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410 [DOI] [PubMed] [Google Scholar]
- Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, et al. (2019) Nucleocytoplasmic Partitioning of ARF Proteins Controls Auxin Responses in Arabidopsis thaliana. Mol Cell 76: 177–190.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. (2020) Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher EH, Möller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D (2011) A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J Cell Mol Biol 68: 597–606 [DOI] [PubMed] [Google Scholar]
- Roosjen M, Paque S, Weijers D (2018) Auxin Response Factors: output control in auxin biology. J Exp Bot 69: 179–188 [DOI] [PubMed] [Google Scholar]
- Simonini S, Stephenson P, Østergaard L (2018) A molecular framework controlling style morphology in Brassicaceae. Dev Camb Engl. doi: 10.1242/dev.158105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigg SA, Garza RM, MacWilliams A, Nery JR, Bartlett A, Castanon R, Goubil A, Feeney J, O’Malley R, Huang SC, et al. (2017) CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat Methods 14: 819–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truskina J, Han J, Galvan-Ampudia CS, Lainé S, Brunoud G, Porco S, Bågman A-M, Smit ME, Bennett M, Roudier F, et al. (2018) A network of transcriptional repressors mediates auxin response specificity. bioRxiv 448860 [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci. doi: 10.1073/pnas.0803996105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher CL, Nemhauser JL (2012) Bipartite Promoter Element Required for Auxin Response. Plant Physiol 158: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan W, Band LR, Kumpf RP, Van Damme D, Parizot B, De Rop G, Opdenacker D, Möller BK, Skorzinski N, Njo MF, et al. (2016) Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 351: 384–387 [DOI] [PubMed] [Google Scholar]
- Zhao S, Zhang M-L, Ma T-L, Wang Y (2016) Phosphorylation of ARF2 Relieves Its Repression of Transcription of the K+ Transporter Gene HAK5 in Response to Low Potassium Stress[OPEN]. Plant Cell 28: 3005–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouine M, Fu Y, Chateigner-Boutin A-L, Mila I, Frasse P, Wang H, Audran C, Roustan J-P, Bouzayen M (2014) Characterization of the Tomato ARF Gene Family Uncovers a Multi-Levels Post-Transcriptional Regulation Including Alternative Splicing. PLoS ONE. doi: 10.1371/journal.pone.0084203 [DOI] [PMC free article] [PubMed] [Google Scholar]


