Abstract
Citrus bacterial canker (CBC) results from Xanthomonas citri subsp. citri (Xcc) infection and poses a grave threat to citrus production. Class III peroxidases (CIII Prxs) are key proteins to the environmental adaptation of citrus plants to a range of exogenous pathogens, but the role of CIII Prxs during plant resistance to CBC is poorly defined. Herein, we explored the role of CsPrx25 and its contribution to plant defenses in molecular detail. Based on the expression analysis, CsPrx25 was identified as an apoplast-localized protein that is differentially regulated by Xcc infection, salicylic acid, and methyl jasmone acid in the CBC-susceptible variety Wanjincheng (C. sinensis) and the CBC-resistant variety Calamondin (C. madurensis). Transgenic Wanjincheng plants overexpressing CsPrx25 were generated, and these transgenic plants exhibited significantly increased CBC resistance compared with the WT plants. In addition, the CsPrx25-overexpressing plants displayed altered reactive oxygen species (ROS) homeostasis accompanied by enhanced H2O2 levels, which led to stronger hypersensitivity responses during Xcc infection. Moreover, the overexpression of CsPrx25 enhanced lignification as an apoplastic barrier for Xcc infection. Taken together, the results highlight how CsPrx25-mediated ROS homeostasis reconstruction and cell wall lignification can enhance the resistance of sweet orange to CBC.
Subject terms: Metabolism, Biotic
Introduction
Plants possess an intricate repertoire of cell-based defense systems to maintain their resistance to potentially harmful pathogens1,2. As an immediate pathogen recognition response, oxidative bursts produced in apoplasts induce reactive oxygen species (ROS), including superoxide (O2.−) and H2O2, as a first line of defense3. The current models of plant responses include ROS and other radicals as catalysts of covalent cell-wall modifications4, as signals for cell-death reactions5,6 and as regulators of resistance-associated genes7,8. In plants, high concentrations of ROS act to strengthen the cell wall and inhibit pathogen growth, which results in the enhancement of host resistance to pathogens via hypersensitive responses (HRs) and the modulation of gene expression via signaling molecules9,10. However, high accumulation of ROS can be toxic to plant cells by inhibiting plant growth and development2. Thus, ROS homeostasis needs to be maintained by antioxidant compounds and enzymes11. In plant cells, ROS are produced by NADPH oxidase resident at the cell surface, class III peroxidases (CIII Prxs, or POD) and their associated pathways, including photosynthesis, photorespiration, and respiration12,13. In addition, the ROS scavengers superoxide dismutase (SOD), catalase (CAT), and glutathione s-transferase (GST) cooperate with ROS producers to maintain ROS homeostasis14. Moreover, antioxidant enzyme activities and ROS homeostasis are regulated by important plant hormones, including jasmonic acid (JA) and salicylic acid (SA)15–17.
CIII Prxs are heme-binding proteins that are ubiquitously expressed in all plants and comprise large multigene families18–21. For example, a total of 73 CIII Prxs are present in Arabidopsis thaliana22,23, and 138, 374, 93, 94 and 72 have been found in Oryza sativa24, Triticum aestivum25, Populus trichocarpa26, Pyrus bretschneideri27 and Citrus sinensis28, respectively. CIII Prxs regulate the loosening of cell walls, lignification and suberization29–32 and participate in ROS and RNS metabolism during abiotic and biotic stress responses33–35. CIII Prxs are key to the innate resistance of many plants to both fungal and bacterial pathogens and mediate both passive and active defense mechanisms6,36,37, and the efficiency of this mediation determines their susceptibility to pathogenic infections38. Rapid ROS production is one such exemplar defense strategy that leads to O2.− generation and H2O2 production in apoplasts. H2O2 is tightly regulated by CIII Prxs as both producers and scavengers depending on whether the enzyme participates in peroxidative cycles and hydroxylic cycles, respectively12,13. In French bean and tobacco plants, apoplastic CIII Prxs produce ROS and act as catalysts for covalent cell-wall modifications4 and cell death regulators6. Based on these functions of CIII Prxs, an increasing number of studies have identified links between this enzyme and pathogen attack and have improved host resistance due to CIII Prxs. Radwan and colleagues reported that bean yellow mosaic virus infection leads to increased levels of monodihydroascorbate (MDA) and H2O2 in Vicia faba leaves39. Enhanced CIII Prx and SOD activities have also been observed in leaves infected by yellow mosaic virus, which suggests that enzymatic antioxidants regulate ROS generation in response to pathogen infection39. Increasing the expression of a peroxidase in plants can effectively increase the resistance of the plants to disease. For example, the overexpression of HvPrx4040 and TaPrx1039,41 leads to higher levels of resistance to Blumeria graminis (wheat powdery mildew) in wheat (T. aestivum).
Xanthomonas citri subsp. citri (Xcc) pathogen is the causative agent of citrus bacterial canker (CBC), a known cause of citrus yield losses in an array of citrus-producing regions42,43. In our previous studies of the citrus transcriptomes induced by Xcc, we found that CIII Prxs were differentially expressed and explored the relationship between CBC and CIII Prxs, and our results revealed CsPrx25 as a potential gene for improving CBC resistance28. Here, we performed both a structural and functional characterization of CsPrx25. We also developed transgenic sweet orange overexpressing CsPrx25 that displayed enhanced tolerance to CBC due to ROS homeostasis accompanied by high levels of H2O2 and high lignification of the apoplastic barrier. We herein describe the utility of transgenic plants overexpressing CsPrx25 for enhancing CBC resistance.
Results
CsPrx25 encodes a CIII Prx in citrus
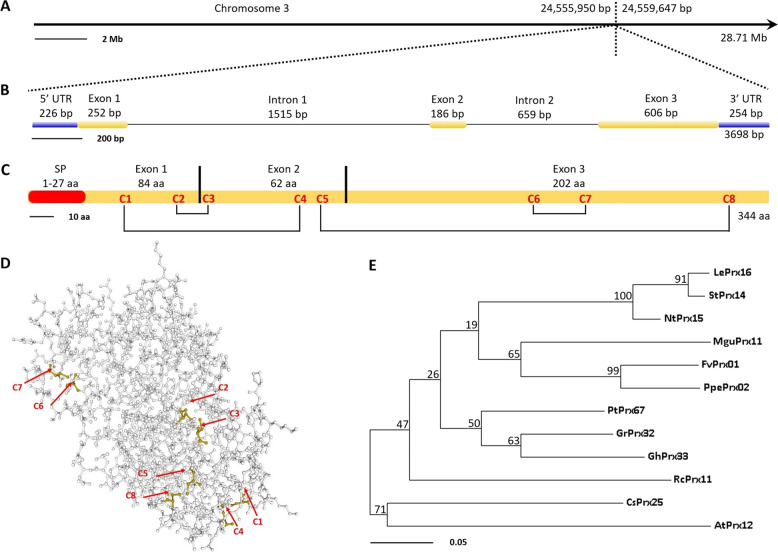
We amplified and sequenced the complete transcript of CsPrx25 using cDNA from Wanjincheng leaves as the PCR template. The primary sequences were searched in PeroxiScan, which is built in RedoxiBase44,45. The findings revealed that CsPrx25 belonged to the CIII Prx family (PeroxiScan accession: PS52045), a subgroup of nonanimal peroxidases (PeroxiScan accession: PS50873). The CsPrx25 sequence was further analysed by the Blast tool built in RedoxiBase and CAP46, and the results revealed that CsPrx25 was clustered with the CIII Prxs sequence ID 8898 in RedoxiBase and Cs3g21730 in CAP due to 100 and 98% sequence similarities, respectively. CsPrx25 is a 344-residue CIII Prx (molecular weight: 38.06 kD; isoelectric point: 8.55) present on chromosome 3 of C. sinensis (Fig. 1a) that possesses two introns (1515 bp and 659 bp, respectively) (Fig. 1b). The N-terminus of CsPrx25 contains a signal peptide of 27 residues that is required for correct trafficking to the apoplast. Throughout the sequence, eight cysteine residues were detected (C1–C8) (Fig. 1c), and these form a total of four disulfide bonds (DB) that maintain thermal stability. These 4-DB structures are common to almost all plant CIII Prxs and impart distinction from ascorbate and other plant peroxidases47. The three-dimensional (3D) structures also showed that the cysteines that form disulfide bonds are close to each other (Fig. 1d). To study the evolutive scenario of CIII Prxs between organisms, the phylogeny of CIII Prxs orthologs was assessed, and close relationships between CsPrx25 and AtPrx12 were found (Fig. 1e).
Fig. 1. Bioinformatics features of CsPrx25.
a CsPrx25 chromosomal locus obtained via CAP. b Exons and introns of CsPrx25 obtained through GSDS V2.0. The blue rectangles represent untranslated regions (UTRs) at the 5′ and 3′ ends; the yellow rectangles represent exons; and the blank lines represent introns. c Schematic of the signal peptide and cysteines (C1–C8) of CsPrx25. The disulfide bonds formed by the cysteine residues are joined by lines. The signal peptide was predicted using SignalP V4.0. d 3D model of CsPrx25 predicted by Phyre V2.0. The cysteine residues are labeled with red arrows and C1–C8. e Maximum-likelihood (ML) phylogeny of CsPrx25 and its orthologs in several organisms. The ML tree was constructed using the CIII Prx sequences of Lycopersicon esculentum (LePrx16), Solanum tuberosum (StPrx14), Nicotiana tabacum (NtPrx15), Mimulus guttatus (MguPrx11), Fragaria vesca (FvPrx01), Prunus persica (PpePrx02), Populus trichocarpa (PtPrx67), Gossypium raimondii (GrPrx32), Gossypium hirsutum (GhPrx33), Ricinus communis (RcPrx11), A. thaliana (AtPrx12) and C. sinensis (CsPrx25) with MEGA V7.0 (bootstrap: 500, Poisson model). The clustering of the taxa is shown through the percentages of trees displaying clusters. The branches are drawn to scale, and each length is representative of the number of substitutions at each site
CsPrx25 is an apoplast-localized protein that is induced by Xcc and phytohormones
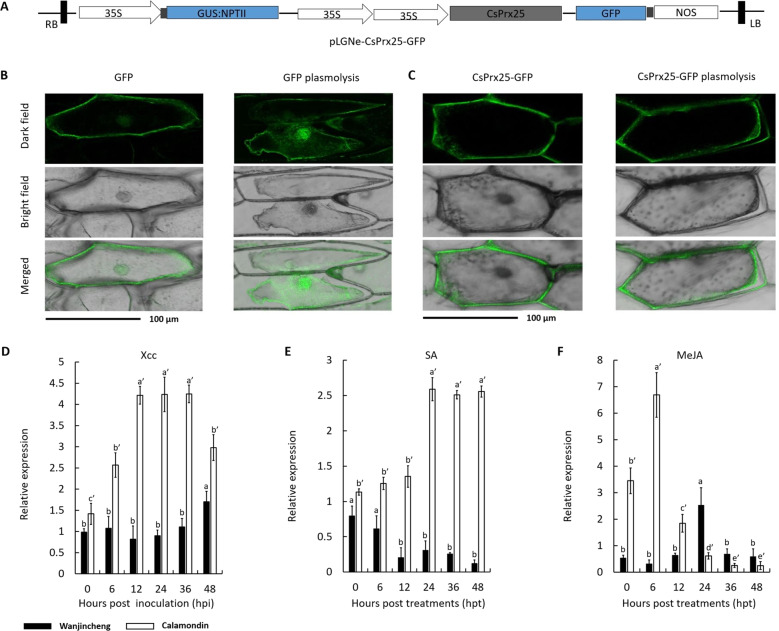
To elucidate the localization of CsPrx25, software predictions and transient expression systems were investigated. CELLO V2.5 displayed extracellular loci values of 2.46, which were larger than other loci (Supplementary Table S1). The signal peptide detected by SignalP V4.0 suggests that CsPrx25, as most of the CIII Prxs, is extracellular. To validate these predictions, the transient expression of CsPrx25 was assessed with 35S::CsPrx25-GFP (Fig. 2a). Relative to the controls, both cytoplasmic and nuclear fluorescence were observed before and after plasmolysis (Fig. 2b). In epidermal onion cells, CsPrx25-GFP showed robust cell surface expression (Fig. 2c), confirming that CsPrx25 localizes to apoplasts.
Fig. 2. Expression profiles of CsPrx25.
a Structure of the pLGNe-CsPrx25-GFP plasmid. LB: left border; RB: right border. The CsPrx25 expression plasmids included the 35 S cauliflower mosaic virus promoter (35 S), the terminator of nos (NOS), and NPTII. b Transient expression of GFP in onion epidermal cells. c Transient expression of CsPrx25-GFP. In b, c, scale bar: 100 μm. d CsPrx25 expression in response to Xcc in Wanjincheng (filled bars) and Calamondin (open bars). CsPrx25 induction in response to SA (e) and MeJA (f) in Wanjincheng (filled bars) and Calamondin (open bars). The relative expression levels were measured by qRT-PCR and normalized to that of CsActin. In d–f, the data were analyzed using Tukey’s HSD test (P = 0.05; n = 3)
Pathogens and phytohormones can mediate gene expression changes that occur in response to plant disease48,49. In Calamondin, CsPrx25 was upregulated, and maximal expression (~5-fold) was observed at 36 hpi. In contrast, Wanjincheng CsPrx25 showed little-to-no expressional changes in response to Xcc infection (Fig. 2d). To detect the effect of drought during in vitro inoculation, we tested the inducibility of CsPrx25 under drought stress. The results indicated that CsPrx25 was hardly induced by drought in both varieties, which indicated that it was specifically induced by Xcc (Supplementary Fig. S1). CsPrx25 is therefore likely to represent an Xcc resistance gene. To reveal the molecular mechanisms through which CsPrx25 mediates disease resistance, CsPrx25 transcripts were assessed in SA- and MeJA-treated leaves. The expression of CsPrx25 rapidly increased in Calamondin in response to SA. In contrast, CsPrx25 expression was downregulated in Wanjincheng (Fig. 2e). The expression of CsPrx25 induced by MeJA increased and then decreased over time in both Wanjincheng and Calamondin, and the times to maximal expression in these varieties was different (Wangjincheng: 24 hpt vs Calamondin: 6 hpt) (Fig. 2f). The different expression patterns of CsPrx25 induced by phytohormones indicate the different roles of CsPrx25 in disease resistance signaling in Calamondin and Wanjincheng.
CsPrx25 overexpression in sweet orange induces resistance to CBC
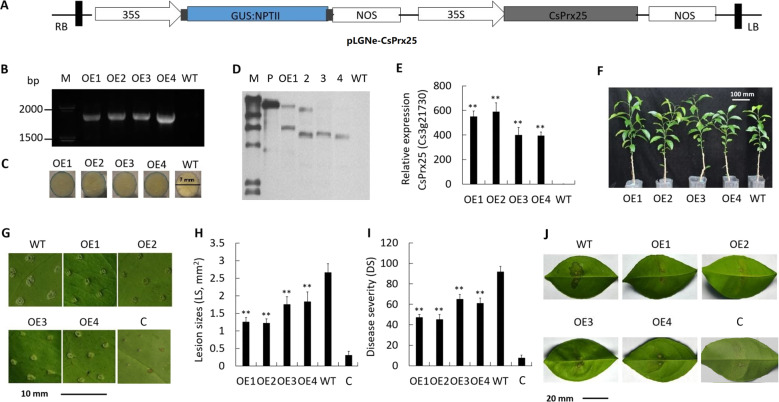
Transgenic citrus constructs overexpressing CsPrx25 were used to fully dissect the role of CsPrx25 during Xcc resistance. CsPrx25 was overexpressed using exogenous expression plasmids driven by the 35S promoter (Fig. 3a). The generation of four CsPrx25-overexpressing plants 1–4 (OE1–OE4) that successfully integrated CsPrx25 was confirmed by qRT-PCR, GUS assay and Southern blot. Through PCR, we detected an 1874-bp fragment that was not present in the wild-type (WT) lines (Fig. 3b), and the GUS assay revealed blue color on the periphery of the leaf discs (Fig. 3c). As determined by Southern blot, OE1 and OE2 contain two copies of CsPrx25, and OE3 and OE4 harbor only one copy (Fig. 3d). We confirmed that all lines expressed high levels of CsPrx25 (550-fold, 589-fold, 401-fold and 395-fold of the WT levels, respectively) by qRT-PCR analysis (Fig. 3e). According to the Southern blot assay, a certain positive correlation exists between copy number and expression (Fig. 3d). With respect to phenotypes, the four transgenic lines showed normal growth rates compared with the WT lines (Fig. 3f). Acupuncture is an effective method for quantitatively assessing the resistance to CBC and can be used to accurately quantify CBC resistance, which would allow the assessment and comparison of resistance between varieties50,51. To assess the CBC resistance of CsPrx25-OE plants, in vitro assays were performed with acupuncture inoculation at 10 dpi. Smaller lesion sizes, which are indicative of less-severe symptoms, were observed in the OE leaves compared with the WT leaves (Fig. 3g). This finding suggested that Xcc pustules are reduced by CsPrx25 overexpression, and OE2 showed the highest levels of resistance. Compared with the WT plants, OE2 showed smaller lesions (45.8% of the WT levels), OE1 exhibited comparable lesions (47.0% of the WT levels), and OE3 and OE4 displayed larger lesions (65.8% and 68.8% of the WT levels) (Fig. 3h). The disease severity decreased by 29.2% (OE3) to 50.7% (OE2) in the OE plants compared with WT plants (Fig. 3i). Using infiltration assays after 10 dpi, symptoms of canker (including pustules) were observed in then WT lines, but these symptoms were markedly reduced in the OE plants (Fig. 3j). We therefore conclude that CsPrx25 overexpression enhances Xcc resistance in the OE transgenic citrus lines.
Fig. 3. The overexpression of CsPrx25 in sweet orange confers CBC resistance.
a Structure of the pLGNe-CsPrx25 plasmid used for the overexpression assays. b Validation of the transgenic plants by PCR. c Confirmation of the transgenic lines by the GUS assay (diameter of leaf discs: 7 mm). d Southern blot of transgenic and WT lines. P: pLGNe-CsPrx25 plasmid. Exposure period: 1 h. e CsPrx25 expression assessed by qRT-PCR with CsActin as an internal control. f Transgenic lines and their phenotypes. Scale bar: 100 mm. g The symptoms of OE and WT plants inoculated with Xcc were assessed and imaged at 10 dpi. Scale bar: 10 mm. Lesion size (LS) h and disease severity (DS) i of the OE and WT plants at 10 dpi. In e, h, i, the data were normalized to the data from the WT plants and compared using Tukey’s HSD test (P = 0.05, n = 3). j Infiltration assays for measuring CBC resistance. Xcc was found to induce disease-related symptoms after 10 dpi. Scale bar: 20 mm. In g–j, OE1–4 represent the four transgenic Wangjincheng plants; WT and C represent the wild-type Wanjincheng and Calamondin plants, respectively
CsPrx25 overexpression modulates the enzymatic antioxidant system
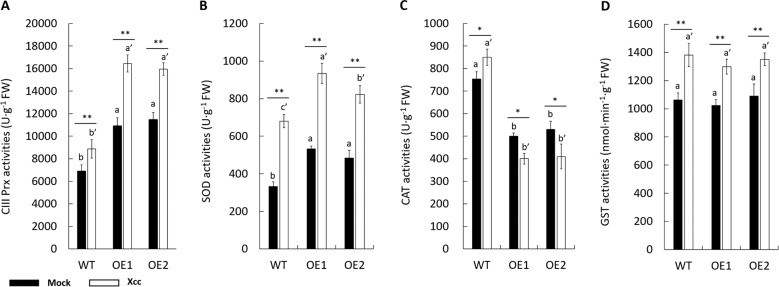
Plants possess a well-developed ROS homeostasis enzymatic system that efficiently regulates the ROS levels, and this system includes CIII Prx, SOD, CAT and GST38,52. To assess the changes in the antioxidant system following the induction of CsPrx25-mediated resistance to Xcc, the antioxidant activity in transgenic lines in these lines was compared with that in the WT plants at 12 hpi. OE plants with higher resistance (OE1 and OE2) to CBC were selected for analysis. The activities of both CIII Prx and SOD were upregulated by CsPrx25 overexpression (Fig. 4a, b). The overexpression of CsPrx25 conferred antioxidant defenses and led to the induction by Xcc infection. In contrast to CIII Prx and SOD, the activities of CAT in OE plants were downregulated, and the Xcc-induced profiles were altered compared with those observed in the WT plants (Fig. 4c). In contrast to SOD, CIII Prx and CAT, the overexpression of CsPrx25 did not affect the activity of GST compared with that found in the WT plants (Fig. 4d).
Fig. 4. CsPrx25 overexpression modulates the enzymatic antioxidant system.
CIII Prx (a), SOD (b), CAT (c) and GST (d) in both OE and WT plants inoculated with mock (ddH2O) (filled bars) and Xcc (open bars) for 12 h. FW: fresh weight. In a–d, the values were compared to those found for the WT lines. The differences between the mock- and Xcc-infected samples were analyzed using Fisher’s LSD test, *P < 0.05; **P < 0.01. Tukey’s HSD test was used to compare the WT and OE plants (P = 0.05; n = 3)
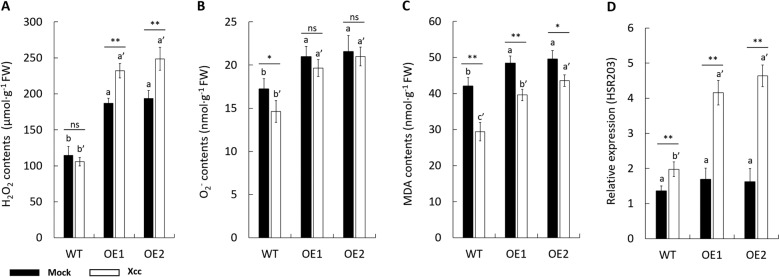
CsPrx25 overexpression establishes ROS homeostasis to confer a more sensitive HR to Xcc infection
In response to pathogen infection, ROS production intricately controls many responses, including apoptotic cell death and oxidative damage53,54. To confirm the involvement of ROS homeostasis in CsPrx25-mediated Xcc resistance, the levels of H2O2 and O2.− in WT vs. CsPrx25-OE lines were assessed. We observed higher levels of H2O2 in the OE lines. Of interest, Xcc infection did not significant change the levels of H2O2 in WT plants but increased these levels in the OE lines (Fig. 5a). This finding suggested that CsPrx25 overexpression not only increased the levels of H2O2 but also reversed the inducible patterns of H2O2 during Xcc infection. The levels of O2.− also increased in response to CsPrx25 overexpression (Fig. 5b). The cell membrane is first affected by lipid peroxidation, and MDA is the final product55. A spectroscopic analysis of the transgenic and WT plants revealed elevated levels of MDA, and these levels were modestly reduced in response to Xcc infection (Fig. 5c), which indicate lower levels of damage following Xcc infection in both the transgenic and WT plants. These data indicate that Wanjincheng has the ability to suppress the oxidative damage caused by Xcc infection, and this suppression is strengthened by CsPrx25 overexpression. H2O2 is a key mediator of an early HR. Because CsPrx25 overexpression regulates H2O2 modulation, the immediate question was whether the HR is also altered in the transgenic plants. To investigate the relationship between the increased CBC resistance induced by CsPrx25 and HR, we assessed the HR of the transgenic plants before and after Xcc infection. The expression of the HR marker gene, CsHSR20356–58 was significantly upregulated in the transgenic plants infected with Xcc but only modestly increased in the Xcc-infected WT plants. No obvious changes in the expression of CsHSR203 were observed between the transgenic and WT plants in the absence of Xcc infection (Fig. 5d). We therefore conclude that the transgenic plants are more sensitive to a HR following Xcc infection, which increases the early resistance of transgenic plants to CBC.
Fig. 5. CsPrx25 reconstructs ROS homeostasis and imparts HR sensitivity during Xcc infection.
The levels of H2O2 (a), O2.− (b) and MDA (c) in OE and WT plants were assayed at 12 h after mock inoculation (ddH2O; filled bars) and Xcc infection (open bars). In a–c, FW: fresh weight. d CsHSR203 transcript levels in WT vs. OE plants at 12 h after mock (ddH2O) (filled bars) or Xcc inoculation (open bars). The data were normalized to the CsActin levels. In a–d, the differences between the mock- and Xcc-infected samples were analyzed by Fisher’s LSD test, *P < 0.05; **P < 0.01. The differences between the OE and WT plants were analyzed using Tukey’s HSD test (P = 0.05; n = 3)
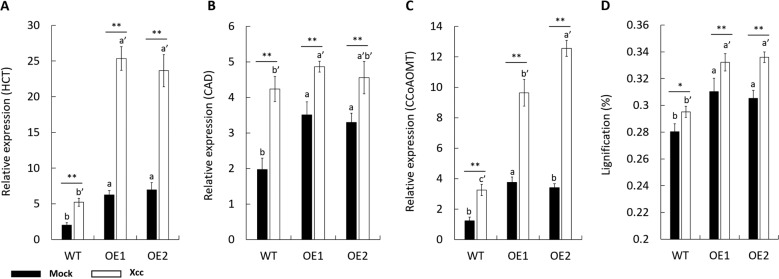
CsPrx25 overexpression enhances lignification as an apoplastic barrier for Xcc infection
CIII Prxs regulate cell wall lignification, which suggests a direct role of CIII Prxs on the cell walls of plants6,59–61. To investigate the effects of lignification on Xcc resistance in transgenic plants, the role of CsPrx25 in lignification was assessed. The transcript levels of lignin biosynthetic genes, namely, hydroxycinnamoyl transferase (CsHCT, CAP ID: Cs1g14450), cinnamyl alcohol dehydrogenase (CsCAD, CAP ID: Cs1g20590) and caffeoyl-CoA O-methyltransferase (CsCCoAOMT, CAP ID: Cs4g13430), were elevated in the leaves of the transgenic lines after mock inoculation and Xcc infection (Fig. 6a–c). These findings highlight the role of CsPrx25 in lignin biosynthesis. All the data were confirmed through lignin assays, which showed higher values in the transgenic compared with the WT plants (Fig. 6d). These data therefore reflect the role of CsPrx25 in the polymerization of lignin during its biosynthesis and highlight its importance in CBC resistance through enhanced lignification.
Fig. 6. CsPrx25 positively regulates cell wall lignin biosynthesis.
Transcripts of the genes involved int he phenylpropanoid pathway, including HCT (a), CAD6 (b) and CCoAOMT (c), in WT vs. OE plants at 12 h after mock (ddH2O) inoculation (filled bars) or Xcc (open bars) infection. d Spectroscopic analysis of lignin at 12 hpi. Leaves of OE1, OE2 and WT plants were sampled for lignification analysis. In a–d, the values were statistically compared with those found for the WT lines. The differences between the mock- and Xcc-infected samples were analyzed by Fisher’s LSD test, *P < 0.05; **P < 0.01. The differences between the OE and WT plants were analyzed using Tukey’s HSD test (P = 0.05; n = 3)
CsPrx25 enhances CBC resistance, and this effect is associated with ROS homeostasis reconstruction and lignification
CsPrx25 overexpression confers ROS homeostasis to the transgenic lines through modulation of the enzymatic antioxidant system (Figs. 4–5). The levels of lignin were also higher in the transgenic lines than in the WT plants, and some lignin biosynthetic genes were more highly expressed in the transgenic lines (Fig. 6). Based on these results, we proposed a model to explain how Calamondin and CsPrx25-OE transgenic Wanjincheng acquired CBC resistance (Fig. 7). In Calamondin, Xcc infection improves the levels of CsPrx25, and this effect enhances the H2O2 levels and HR sensitivity and induces lignification, resulting in CBC resistance. The overexpression of CsPrx25 in CBC-susceptible Wanjincheng establishes ROS homeostasis, and higher H2O2 levels confer HR sensitivity in response to Xcc infection. In the transgenic plants, CsPrx25 overexpression also enhanced lignin biosynthesis, reinforcing the apoplastic barrier for Xcc infection. Through these two mechanisms, CsPrx25 promotes CBC resistance.
Fig. 7. Hypothetical model of the molecular mechanisms mediating improved CBC resistance through CsPrx25.
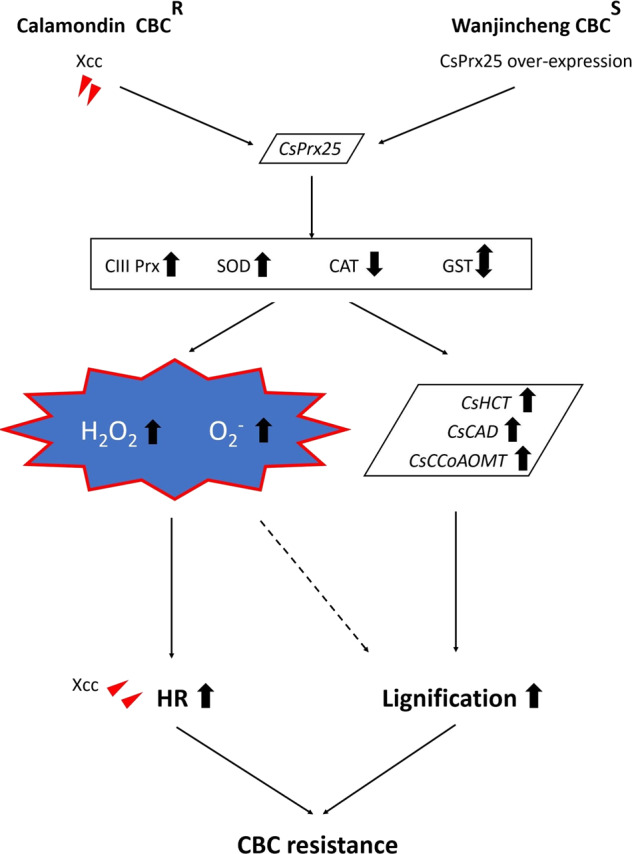
Increases, decreases and no changes are represented by up arrows, down arrows, and double arrows, respectively. The genes involved in this model are written in the trapezoids and in italics. Antioxidants are shown in rectangles. The ROS content is written in starburst
Materials and methods
Plants, bacteria and growth conditions
All the plants were obtained from the National Citrus Germplasm Repository. Wanjincheng (C. sinensis) was used for gene transformations. All the plants were grown at 28 °C in a greenhouse. The Xcc variants were derived from citrus leaves that are susceptible to natural infections. The Xcc cultures were grown at 28 °C in peptone-yeast extract-malt extract containing 1.5% (w/v) d-glucose.
In silico characterization of CsPrx25
The complete transcript sequence of CsPrx25 was amplified from Wanjincheng leaves using the primers Fclone (ATGGCAACTGCTTCAGCTTCT) and Rclone (TTAGATAATCCCAGACCAAGC). PeroxiScan was used for the family classification of CsPrx2520. Blast tools built in RedoxiBase20,44, CAP46, CitGVD62 and SMART63 were used to reconfirm the sequence of CsPrx25 retrieved by PCR. The chromosomal loci and the locations of exons and introns were defined using GSDS V2.064 based on the genome assembly of C. sinensis in CAP. SignalP V4.065 was used for signal peptide predictions, and CELLO V2.566 was used for cellular localization prediction. Phyre V2.067 was used for the 3D assessments of CsPrx25. The gene, protein and coding sequences (CDSs) of CsPrx25 are shown in Table S2.
Transient expression of GFP-tagged CsPrx25
The coding sequence (CDS) of CsPrx25 lacking a stop codon was amplified with flanking restriction sites using the primers FSC (CGGGGTACCATGGCTGTTCATCAACATTATCTGG) (KpnI) and RSC (TCCCCCGGGTCACTGGTTTGAAATTAAAGGATCT) (SmaI), digested, recovered and cloned into pLGNe-GFP driven by the 35S promoter to construct the recombinant plasmid pLGNe-CsPrx25-GFP. The pLGNe-CsPrx25-GFP plasmid encodes a fusion protein composed of CsPrx25 and GFP. The plasmids were heat-shocked into Agrobacterium EHA105. The transformed EHA105 was infiltrated into onion epidermal cells, and the GFP fluorescence signals were observed at 48 hpi by laser-scanning confocal microscopy (LSM 510 Meta, Zeiss).
Treatments with Xcc and phytohormones
The expression of CsPrx25 in excised leaves maintained in culture plates for 16 h of light and 8 h of darkness was assessed. Diluted Xcc (OD600: 0.8) was inoculated onto the leaves at 28 °C, and after defined durations, the expression of CsPrx25 was assessed by qRT-PCR. For phytohormone assessments, leaf discs were soaked in 10 μmol L-1 SA or 100 μmol L-1 MeJA and collected for qRT-PCR assays of exogenous phytohormones. The primers used for CsPrx25 detection were FRT (CCCCACTTCGGATTCCAACA) and RRT (CAACCCCTGTCGGTTCATCA).
Overexpression vector construction and plant transformation
For the generation of overexpression lines, full-length CsPrx25 was PCR amplified using FOEC (GGGGTACCATGGCAACTGCTTCAGCTTC) and ROEC (CGGGATCCTTAGATAATCCCAGACCAAGCC) and cloned into pLGNe to yield the recombinant plasmid pLGNe-CsPrx25. Wanjincheng shoot transformations were performed using Agrobacterium tumefaciens as previously described by Li and He48,50.
Validation of the transgenic lines by PCR and GUS assays
PCR assays were used to confirm the presence of the transgenic gene with the primers FOED (CGACACGCTTGTCTACTCCA) and ROED (CGGGATCCTTAGATAATCCCAGACCAAGCC). GUS activity was assessed through histochemical analysis48,51.
Southern blot assay
Total genomic DNA (gDNA) was extracted from the leaves of the transgenic plants and WT plants using a CTAB kit (Zoonbio, China). The gDNA was fragmented using the restriction enzyme EcoRI, and the DNA fragments were separated on a 0.7% agarose gel and transferred to a Hybond-N+ membrane (Amersham, UK). The NPTII coding gene labeled by digoxin (DIG) was used to hybridize the membrane-bound DNA (Roche, Switzerland). The nylon membrane was then exposed using nonradioactive probe detection. In the Southern blot assay, the pLGNe-CsPrx25 plasmid was used as the positive control.
Assessment of CBC resistance
CBC resistance analyses were performed as previously described48,68,69. Briefly, six punctures were made in six healthy mature leaves of each transgenic line via 0.5-mm pins, and 1 µL of each Xcc suspension (1 × 105 cfu mL-1) was subsequently inoculated. CBC development was assessed at 10 dpi, and both the disease severity (DS) and lesion size (LS) of the diseased spots were used for the assessment of CBC resistance. The DS was calculated as previously described51,70. CBC resistance was further evaluated through Xcc infiltration assays (1 × 105 cfu mL-1), and canker symptoms were imaged at 10 dpi.
Biochemical analysis
The activities of CIII Prx, SOD, CAT and GST and the concentrations of H2O2, O2.−, MDA, and lignin were measured via SinoBestBio assays (Shanghai, China). The experiments were repeated three times, and the results are shown as the means ± SEs.
RNA isolation, cDNA synthesis and qRT-PCR assay
Miniprep kits (AidLab) were used for RNA isolation, and cDNA was synthesized using TaKaRa kits. qRT-PCR was performed using QuantStudio 7. The values were normalized to the CsActin levels (GenBank accession: GU911361.1, CAP ID: Cs1g05000) obtained using FActin (CATCCCTCAGCACCTTCC) and RActin (CCAACCTTAGCACTTCTCC). The qRT-PCR parameters were as follows: 95 °C for 5 min followed by 40 cycles of 95 °C for 10 s and 56 °C for 30 s. The reaction mixtures (total volume of 12 μL) contained 50 ng of cDNA, 0.5 μM primers and 6 μL of the PCR mix. The relative gene expression levels were assessed using the 2-∆∆CT method71. NCBI was used for qRT-PCR primer design (Supplementary Table S3). The data are presented as the means from three independent biological repeats.
Statistics
The data were analyzed using SPSS V22. Gene expression was compared by analysis of variance (ANOVA). The statistical significance was analyzed by Fisher’s LSD test. *P < 0.05 and **P < 0.01 indicate significant and extremely significant differences, respectively. The plant lines were compared using Tukey’s HSD test (P = 0.05).
Discussion
CIII Prxs belong to a plant-specific multigene family that promotes disease resistance18,33,34, lignification, the flexibility of cell walls and suberization29,30. In sweet orange, 72 CIII Prxs have been identified28. The expression of each isoform varies across tissues and can be influenced by environmental factors, which suggests that different peroxidase isoenzymes regulate distinct processes72. The distribution of enzymes to either the cell walls or vacuoles and their destinations reflect their specific functions31. In CBC-resistant and CBC-susceptible varieties, CsPrx25 exhibits altered expression patterns (Fig. 2d–f), which suggests its role during CBC development. The importance of CIII Prxs for the resistance of plants to pathogenic diseases was identified through reverse genetics. CIII Prxs mediate innate resistance both passively and actively6. HvPrx4040 and TaPrx10 39,41 enhance the resistance of wheat against wheat powdery mildew. Here, CsPrx25 was found to mediate protection against Xcc pathogenesis, which confirmed its role as a CIII Prx and further highlighted the importance of this family in pathogen immunity in sweet orange. We explored its functional role using overexpression strategies and found that CsPrx25 strongly conferred CBC resistance to the transgenic plants (Fig. 3g–j).
Oxidative bursts, particularly the production of H2O2 and O2.−, are common innate responses in plant cells in response to pathogen infection38. As key enzymes for ROS homeostasis in plants, CIII Prxs have multiple functions and are proposed to serve as key regulators of the extracellular H2O2 and O2− levels depending on peroxidative cycles (ROS scavenging) or hydroxylic cycles (ROS production)73. Plant defense responses are governed by the ROS levels and peroxidase-generated radicals, which mediate cell wall reinforcement, damage repair4,6 and apoptotic responses to induce plant resistance5,6. In this study, the molecular mechanisms of CsPrx25 were explored. Based on our analysis of ROS homeostasis and enzymatic antioxidant activities in the transgenic plants, we concluded that CsPrx25 overexpression enhances CIII Prx activities and leads to a simultaneous improvement in the H2O2 and O2.− content (Fig. 5a, b). In plants, the HR is directly related to plant disease resistance and represents the classic response to pathogen infection6. These reactions lead to both rapid and localized necrosis of the infected tissues and thus prevent the spread of infection56,57. H2O2 is key to the HR and is related to programmed cell death (PCD) in infected plants58. To investigate the relationship between the CBC resistance induced by CsPrx25 and the HR, we assessed the HR of the transgenic plants before and after Xcc infection (Fig. 5d). HSR203 is upregulated by plant HRs and is used as a marker for the HR levels56,58. In this study, the links among CsPrx25 activity, ROS content and HR level were established. Cell wall lignification was further shown to mediate CBC resistance, which was also demonstrated in rice due to the enhancement in Xanthomonas oryzae resistance conferred by CIII Prx-mediated lignification73.
Due to the evolutionary diversity and functional diversity of CIII Prxs, different studies have drawn different links between CIII Prx and disease resistance. Increased LePrx06 makes tomato more susceptible to Pseudomonas syringae infection. In contrast to CsPrx25, the suppression of LePrx06 can enhance resistance to this pathogen74. Long-term studies of the relationship between the ROS levels and the development of CBC have revealed increased peroxidase activity and thus a reduced ROS content. Furthermore, the reduction in the ROS levels was associated with CBC resistance. These effects parallel the overexpression of MdATG18a and can enhance resistance to Diplocarpon mali infection via H2O2 scavenging75. Cybrids of grapefruit with a kumquat plastid genome exhibit increased CBC resistance through an early upregulation of ROS-controlling genes upon Xcc infection76. These findings illustrate potential links between ROS homeostasis mediated by plastid ROS-controlling genes and Xcc resistance. However, this study revealed that CsPrx25 is an apoplast-localized enzyme rather than a plastid enzyme (Fig. 2b, c), and this knowledge expands the list of ROS-controlling enzymes that can upregulate CBC resistance.
In this study of CsPrx25, regulation of the ROS levels by CsPrx25 and improvements in HR sensitivity were the major mechanisms through which transgenic citrus developed resistance to CBC. Although CsPrx25 overexpression greatly improved the resistance of Wanjincheng to CBC, CsPrx25-overexpressing Wanjincheng cells were still not as resistant as Calamondin cells, which might be due to the fact that Calamondin also has other mechanisms to maintain an even higher level of CBC resistance. Anyway, this study explores new insights into the mechanisms of CIII Prxs in CBC resistance and provides potential clues for breeding CBC-resistant citrus.
Supplementary information
Acknowledgements
The National Key Research and Development Program of China (2018YFD1000306), the Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX1064), the Fundamental Research Funds for the Central Universities (SWU115025), the Earmarked Funds for the China Agriculture Research System (CARS-26), and the Key Project of Guangxi Science and Technology (GuiKeAA18118046-6) funded this study.
Author contributions
Q.L. and S.C. conceived the experiments; Q.L., J.Q., X.Q. and W.D. conduced the experiments; Q.L. and C.D. conducted the bioinformatics analysis; Q.L. and W.D. were responsible for the data analyses; and Q.L. and Y.H. wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Shanchun Chen, Email: chenshanchun@cric.cn.
Yongrui He, Email: heyongrui@cric.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-020-00415-9).
References
- 1.Molina L, Kahmann R. An ustilago maydis gene involved in H2O2 detoxification is required for virulence. Plant Cell. 2007;19:2293–2309. doi: 10.1105/tpc.107.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters LP, et al. Functional analysis of oxidative burst in sugarcane smut-resistant and -susceptible genotypes. Planta. 2017;245:749–764. doi: 10.1007/s00425-016-2642-z. [DOI] [PubMed] [Google Scholar]
- 3.Pitino M, Armstrong CM, Duan Y. Rapid screening for citrus canker resistance employing pathogen-associated molecular pattern-triggered immunity responses. Hortic. Res. 2015;2:15042. doi: 10.1038/hortres.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostergaard L, et al. Expression and high-resolution structure of a plant peroxidase with implications for lignification. Plant Mol. Biol. 2000;44:231–243. doi: 10.1023/A:1006442618860. [DOI] [PubMed] [Google Scholar]
- 5.Passardi F, Penel C, Dunand C. Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci. 2004;9:534–540. doi: 10.1016/j.tplants.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Schweizer P. Tissue-specific expression of a defence-related peroxidase in transgenic wheat potentiates cell death in pathogen-attacked leaf epidermis. Mol. Plant Pathol. 2008;9:45–57. doi: 10.1111/j.1364-3703.2007.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu G, et al. Profiling of wheat class III peroxidase genes derived from powdery mildew-attacked epidermis reveals distinct sequence-associated expression patterns. Mol. Plant Microbe Interact. 2005;18:730–741. doi: 10.1094/MPMI-18-0730. [DOI] [PubMed] [Google Scholar]
- 8.Han FP, Fedak G, Ouellet T, Dan H, Somers DJ. Mapping of genes expressed in Fusarium graminearum-infected heads of wheat cultivar ‘Frontana’. Genome. 2005;48:88–96. doi: 10.1139/g04-098. [DOI] [PubMed] [Google Scholar]
- 9.Mittler R, et al. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc. Natl Acad. Sci. USA. 1999;96:14165–14170. doi: 10.1073/pnas.96.24.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barna B, Fodor J, Harrach BD, Pogány M, Király Z. The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 2012;59:37–43. doi: 10.1016/j.plaphy.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 12.Torres MA. ROS in biotic interactions. Physiol. Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 13.Aviello G, Knaus UG. NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol. 2018;11:1011–1023. doi: 10.1038/s41385-018-0021-8. [DOI] [PubMed] [Google Scholar]
- 14.Peters LP, et al. Differential responses of the antioxidant system of ametryn and clomazone tolerant bacteria. PLoS ONE. 2014;9:e112271. doi: 10.1371/journal.pone.0112271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- 16.Thomma BP, et al. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. USA. 1998;95:15107–15111. doi: 10.1073/pnas.95.25.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieterse CM, et al. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, et al. Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biol. Evol. 2015;7:1068–1081. doi: 10.1093/gbe/evv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Q, San Clemente H, He YR, Fu YY, Dunand C. Global evolutionary analysis of 11 gene families part of reactive oxygen species (ROS) gene network in four Eucalyptus species. Antioxidants. 2020;9:19. doi: 10.3390/antiox9030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savelli B, et al. RedoxiBase: a database for ROS homeostasis regulated proteins. Redox Biol. 2019;26:101247. doi: 10.1016/j.redox.2019.101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mbadinga Mbadinga D, Li Q, Ranocha P, Martinez Y, Dunand C. Global analysis of non-animal peroxidases provides insights into the evolution of this gene family in the green lineage. J. Exp. Bot. 2020;71:3350–3360. doi: 10.1093/jxb/eraa141. [DOI] [PubMed] [Google Scholar]
- 22.Tognolli M, Penel C, Greppin H, Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/S0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- 23.Cosio C, Dunand C. Transcriptome analysis of various flower and silique development stages indicates a set of class III peroxidase genes potentially involved in pod shattering in Arabidopsis thaliana. BMC Genom. 2010;11:528. doi: 10.1186/1471-2164-11-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passardi F, Longet D, Penel C, Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry. 2004;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, et al. Genome-wide and evolutionary analysis of the class III peroxidase gene family in wheat and Aegilops tauschii reveals that some members are involved in stress responses. BMC Genom. 2019;20:666. doi: 10.1186/s12864-019-6006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren LL, et al. Subcellular relocalization and positive selection play key roles in the retention of duplicate genes of populus class III peroxidase family. Plant Cell. 2014;26:2404–2419. doi: 10.1105/tpc.114.124750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y, et al. Structural, evolutionary, and functional analysis of the class III peroxidase gene family in chinese pear. Front. Plant Sci. 2016;7:1874. doi: 10.3389/fpls.2016.01874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Q, et al. Genomewide analysis of the CIII peroxidase family in sweet orange (Citrus sinensis) and expression profiles induced by Xanthomonas citri subsp. citri and hormones. J. Genet. 2020;99:13. doi: 10.1007/s12041-019-1163-5. [DOI] [PubMed] [Google Scholar]
- 29.Fernández-Pérez F, Pomar F, Pedreño MA, Novo-Uzal E. Suppression of arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci. 2015;239:192–199. doi: 10.1016/j.plantsci.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Pandey VP, Dwivedi UN. A ripening associated peroxidase from papaya having a role in defense and lignification: heterologous expression and in-silico and in-vitro experimental validation. Gene. 2015;555:438–447. doi: 10.1016/j.gene.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Shigeto J, Tsutsumi Y. Diverse functions and reactions of class III peroxidases. N. Phytol. 2016;209:1395–1402. doi: 10.1111/nph.13738. [DOI] [PubMed] [Google Scholar]
- 32.Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a swiss army knife. Plant Cell Rep. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- 33.Liszkay A, Kenk B, Schopfer P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta. 2003;217:658–667. doi: 10.1007/s00425-003-1028-1. [DOI] [PubMed] [Google Scholar]
- 34.McInnis SM, Desikan R, Hancock JT, Hiscock SJ. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? N. Phytol. 2006;172:221–228. doi: 10.1111/j.1469-8137.2006.01875.x. [DOI] [PubMed] [Google Scholar]
- 35.Mei W, Qin Y, Song W, Li J, Zhu Y. Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J. Genet. Genomics. 2009;36:141–150. doi: 10.1016/S1673-8527(08)60101-0. [DOI] [PubMed] [Google Scholar]
- 36.Cao J, Jiang M, Li P, Chu Z. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium distachyon. BMC Genom. 2016;17:175. doi: 10.1186/s12864-016-2526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Loon LC, Rep M, Pieterse CM. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006;44:135–162. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 38.Almagro L, et al. Class III peroxidases in plant defence reactions. J. Exp. Bot. 2009;60:377–390. doi: 10.1093/jxb/ern277. [DOI] [PubMed] [Google Scholar]
- 39.Radwan MA, El-Gendy KS, Gad AF. Biomarkers of oxidative stress in the land snail, Theba pisana for assessing ecotoxicological effects of urban metal pollution. Chemosphere. 2010;79:40–46. doi: 10.1016/j.chemosphere.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 40.Johrde A, Schweizer P. A class III peroxidase specifically expressed in pathogen-attacked barley epidermis contributes to basal resistance. Mol. Plant Pathol. 2008;9:687–696. doi: 10.1111/j.1364-3703.2008.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altpeter F, et al. Stable expression of a defense-related gene in wheat epidermis under transcriptional control of a novel promoter confers pathogen resistance. Plant Mol. Biol. 2005;57:271–283. doi: 10.1007/s11103-004-7564-7. [DOI] [PubMed] [Google Scholar]
- 42.Omar AA, Murata MM, El-Shamy HA, Graham JH, Grosser JW. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018;27:179–191. doi: 10.1007/s11248-018-0065-2. [DOI] [PubMed] [Google Scholar]
- 43.Schaad NW, et al. Reclassification of Xanthomonas campestris pv. citri (ex Hasse 1915) Dye 1978 forms A, B/C/D, and E as X. smithii subsp. citri (ex Hasse) sp. nov. nom. rev. comb. nov., X. fuscans subsp. aurantifolii (ex Gabriel 1989) sp. nov. nom. rev. comb. nov., and X. alfalfae subsp. citrumelo (ex Riker and Jones) Gabriel et al., 1989 sp. nov. nom. rev. comb. nov.; X. campestris pv malvacearum (ex smith 1901) Dye 1978 as X. smithii subsp. smithii nov. comb. nov. nom. nov.; X. campestris pv. alfalfae (ex Riker and Jones, 1935) dye 1978 as X. alfalfae subsp. alfalfae (ex Riker et al., 1935) sp. nov. nom. rev.; and “var. fuscans” of X. campestris pv. phaseoli (ex Smith, 1987) Dye 1978 as X. fuscans subsp. fuscans sp. nov. Syst. Appl. Microbiol. 2005;28:494–518. doi: 10.1016/j.syapm.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Fawal N, et al. PeroxiBase: a database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013;41:D441–D444. doi: 10.1093/nar/gks1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Savelli B, et al. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019;26:5. doi: 10.1016/j.redox.2019.101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, et al. Citrus sinensis annotation project (CAP): a comprehensive database for sweet orange genome. PLoS ONE. 2014;9:e87723. doi: 10.1371/journal.pone.0087723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Welinder KG, et al. Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 2002;269:6063–6081. doi: 10.1046/j.1432-1033.2002.03311.x. [DOI] [PubMed] [Google Scholar]
- 48.He Y, et al. Functional analysis of citrus AP2 transcription factors identified CsAP2-09 involved in citrus canker disease response and tolerance. Gene. 2019;707:178–188. doi: 10.1016/j.gene.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 49.Zuo W, et al. A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 2015;47:151–157. doi: 10.1038/ng.3170. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, et al. CsWAKL08, a pathogen-induced wall-associated receptor-like kinase in sweet orange, confers resistance to citrus bacterial canker via ROS control and JA signaling. Hort. Res. 2020;7:15. doi: 10.1038/s41438-020-0241-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sendín LN, et al. Inducible expression of Bs2 R gene from Capsicum chacoense in sweet orange (Citrus sinensis L. Osbeck) confers enhanced resistance to citrus canker disease. Plant Mol. Biol. 2017;93:607–621. doi: 10.1007/s11103-017-0586-8. [DOI] [PubMed] [Google Scholar]
- 52.Du XM, Yin WX, Zhao YX, Zhang H. The production and scavenging of reactive oxygen species in plants. Sheng Wu Gong. Cheng Xue Bao. 2001;17:121–125. [PubMed] [Google Scholar]
- 53.Hückelhoven R, Kogel KH. Reactive oxygen intermediates in plant-microbe interactions: who is who in powdery mildew resistance? Planta. 2003;216:891–902. doi: 10.1007/s00425-003-0973-z. [DOI] [PubMed] [Google Scholar]
- 54.Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP. Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 2005;3:789–798. doi: 10.1038/nrmicro1239. [DOI] [PubMed] [Google Scholar]
- 55.Mishra MK, et al. Overexpression of WsSGTL1 gene of Withania somnifera enhances salt tolerance, heat tolerance and cold acclimation ability in transgenic Arabidopsis plants. PLoS ONE. 2013;8:e63064. doi: 10.1371/journal.pone.0063064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pontier D, Tronchet M, Rogowsky P, Lam E, Roby D. Activation of hsr203, a plant gene expressed during incompatible plant-pathogen interactions, is correlated with programmed cell death. Mol. Plant Microbe Interact. 1998;11:544–554. doi: 10.1094/MPMI.1998.11.6.544. [DOI] [PubMed] [Google Scholar]
- 57.Pontier D, Godiard L, Marco Y, Roby D. Hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 1994;5:507–521. doi: 10.1046/j.1365-313X.1994.5040507.x. [DOI] [PubMed] [Google Scholar]
- 58.Tronchet M, Ranty B, Marco Y, Roby D. Hsr203 antisense suppression in tobacco accelerates development of hypersensitive cell death. Plant J. 2001;27:115–127. doi: 10.1046/j.1365-313x.2001.01072.x. [DOI] [PubMed] [Google Scholar]
- 59.Herrero J, et al. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta. 2013;237:1599–1612. doi: 10.1007/s00425-013-1865-5. [DOI] [PubMed] [Google Scholar]
- 60.Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H. A large family of class III plant peroxidases. Plant Cell Physiol. 2001;42:462–468. doi: 10.1093/pcp/pce061. [DOI] [PubMed] [Google Scholar]
- 61.Elfstrand M, Sitbon F, Lapierre C, Bottin A, von Arnold S. Altered lignin structure and resistance to pathogens in spi 2-expressing tobacco plants. Planta. 2002;214:708–716. doi: 10.1007/s00425-001-0681-5. [DOI] [PubMed] [Google Scholar]
- 62.Li Q, et al. CitGVD: a comprehensive database of citrus genomic variations. Hort. Res. 2020;7:12. doi: 10.1038/s41438-019-0234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu B, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 66.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 67.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Q, et al. CsBZIP40, a BZIP transcription factor in sweet orange, plays a positive regulatory role in citrus bacterial canker response and tolerance. PloS ONE. 2019;14:e0223498. doi: 10.1371/journal.pone.0223498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peng A, et al. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017;15:1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Q, et al. Systematic analysis and functional validation of citrus XTH genes reveal the role of CsXTH04 in citrus bacterial canker resistance and tolerance. Front. Plant Sci. 2019;10:1109. doi: 10.3389/fpls.2019.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Caruso C, et al. A basic peroxidase from wheat kernel with antifungal activity. Phytochemistry. 2001;58:743–750. doi: 10.1016/S0031-9422(01)00226-6. [DOI] [PubMed] [Google Scholar]
- 73.Kawano T. Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep. 2003;21:829–837. doi: 10.1007/s00299-003-0591-z. [DOI] [PubMed] [Google Scholar]
- 74.Coego A, et al. An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell. 2005;17:2123–2137. doi: 10.1105/tpc.105.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun X, et al. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018;41:469–480. doi: 10.1111/pce.13110. [DOI] [PubMed] [Google Scholar]
- 76.Murata MM, et al. Novel plastid-nuclear genome combinations enhance resistance to citrus canker in cybrid grapefruit. Front. Plant Sci. 2018;9:1858. doi: 10.3389/fpls.2018.01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.