Abstract
Over the last 2 decades, the zebrafish (Danio rerio) has emerged as a stellar model for unraveling molecular signaling events mediated by the aryl hydrocarbon receptor (AHR), an important ligand-activated receptor found in all eumetazoan animals. Zebrafish have 3 AHRs—AHR1a, AHR1b, and AHR2, and studies have demonstrated the diversity of both the endogenous and toxicological functions of the zebrafish AHRs. In this contemporary review, we first highlight the evolution of the zebrafish ahr genes, and the characteristics of the receptors including developmental and adult expression, their endogenous and inducible roles, and the predicted ligands from homology modeling studies. We then review the toxicity of a broad spectrum of AHR ligands across multiple life stages (early stage, and adult), discuss their transcriptomic and epigenetic mechanisms of action, and report on any known interactions between the AHRs and other signaling pathways. Through this article, we summarize the promising research that furthers our understanding of the complex AHR pathway through the extensive use of zebrafish as a model, coupled with a large array of molecular techniques. As much of the research has focused on the functions of AHR2 during development and the mechanism of TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) toxicity, we illustrate the need to address the considerable knowledge gap in our understanding of both the mechanistic roles of AHR1a and AHR1b, and the diverse modes of toxicity of the various AHR ligands.
Keywords: zebrafish, aryl hydrocarbon receptor (AHR), polycyclic aromatic hydrocarbons, TCDD, cytochrome P450
Aryl Hydrocarbon Receptor
The aryl hydrocarbon receptors (AHRs) are ligand-dependent transcription factors that mediate a wide range of biological and toxicological effects in animals (Abel and Haarmann-Stemmann, 2010; Barouki et al., 2007; Esser et al., 2009; Hankinson, 1995; Nguyen et al., 2018; Safe et al., 2013). Although several endogenous ligands have been identified since the discovery of the AHR in 1976 (Poland et al., 1976), the focus has been on characterizing the toxicity of numerous environmental chemicals including the halogenated aromatic hydrocarbons (HAHs) and polycyclic aromatic hydrocarbons (PAHs), many of which cause toxicity via the AHR signaling pathway (Denison and Nagy, 2003; Nguyen and Bradfield, 2008). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD), an HAH, is the most potent and thoroughly investigated of the known AHR ligands and it elicits many species- and tissue-specific toxicological effects (Couture et al., 1990; Denison and Nagy, 2003; Mandal, 2005). By virtue of its limited metabolism (Vinopal and Casida, 1973), TCDD is typically utilized as the prototypical molecular probe to study the signaling events downstream of AHR activation (Poland and Kende, 1976) and forms the basis of investigation of many of the AHR-dependent mechanisms reviewed in this study.
Canonical signaling for the AHRs, which are part of the basic Helix-Loop-Helix Per-Arnt-Sim (bHLH/PAS) family of proteins, involves the conversion into an active form that can dimerize with another bHLH/PAS protein, the AHR nuclear translocator (ARNT) (Hoffman et al., 1991; Kewley et al., 2004). In their latent and unbound state, the AHRs are found in the cytoplasm and are stably associated with 2 molecules of the 90-kDa molecular chaperone heat shock protein 90 (Hsp90), p23, and AHR-interacting protein (AIP/XAP2/Ara9) (Carver and Bradfield, 1997; Kazlauskas et al., 1999; Ma and Whitlock, 1997; Perdew, 1988). Upon ligand binding, the AHR is activated and the AHR/Hsp90 complex translocates to the nucleus where Hsp90 is exchanged for the partner protein, ARNT (Hoffman et al., 1991; Reyes et al., 1992; Swanson, 2002). The AHR/ARNT heterodimer recognizes and regulates transcription of downstream genes such as the cytochrome P450 family of genes (CYPs) and the aryl hydrocarbon receptor repressor (AHRR) via aryl hydrocarbon response elements (AHREs; also known as DREs or XREs) in their promoter regions (Mimura et al., 1999; Watson and Hankinson, 1992). The CYP1s are among the most well-studied AHR gene targets and are involved in both the metabolic activation and detoxification of the various AHR ligands (Nebert et al., 2004). In addition to the CYPs and AHRR, the AHR can also directly or indirectly regulate expression of a large battery of genes, the identities and functions of which are still being discovered (Abel and Haarmann-Stemmann, 2010; Beischlag et al., 2008). Although our review predominantly focuses on what we know about canonical AHR signaling in zebrafish, we acknowledge that the AHRs have several noncanonical functions as well (Jackson et al., 2015). Some examples include AHR as an E3 ubiquitin ligase in cytosol (Ohtake et al., 2007), its interaction with p300, pRb, and E2F (Marlowe et al., 2004; Puga et al., 2000), and as a partner for KLF6 (Wright et al., 2017) and RelB (Vogel et al., 2007).
Evolution of the AHR in Different Species
The AHR is an ancient protein found in all eumetazoan animals, indicating that it originated more than 600 Ma (Hahn et al., 2017). A fundamental difference between AHRs in invertebrates and vertebrates is that most of the vertebrate AHR proteins exhibit high-affinity binding of halogenated and nonhalogenated aromatic hydrocarbons, whereas all invertebrate AHRs examined to date lack that ability and appear to have roles primarily in developmental processes (Butler et al., 2001; Hahn, 2002). During animal evolution, AHR genes have been duplicated, including in early vertebrate evolution (a tandem duplication and an expansion associated with 2 early vertebrate, whole-genome duplication events) and in specific vertebrate lineages, especially fish (Hahn et al., 2017). These duplications, coupled with some lineage-specific gene losses, result in the presence of between 1 and 5 AHR genes per species.
An important difference between AHR signaling in mammals and fishes is that most mammals—including most of those used as models in toxicology research—possess a single AHR gene, whereas most fishes have multiple AHRs. Fish typically possess 4 AHR genes—2 pairs of tandem AHR1-AHR2—although additional gene duplications and losses have led to some variation, including in zebrafish (Hahn et al., 2006). It is not clear why fish have retained multiple AHR genes, including the AHR2 paralogs that have been lost from most mammals, as well as the additional duplicates of AHR1 and AHR2 resulting from a fish-specific whole-genome duplication event that occurred approximately 350 Ma (Amores et al., 1998; Glasauer and Neuhauss, 2014). The maintenance of multiple AHRs in modern fish is notable considering that more than 80% of the gene duplicates formed during the fish-specific whole-genome duplication were subsequently lost. The prevailing hypothesis for retention of gene duplicates is that they have become more specialized by partitioning the multiple functions of their common ancestor (subfunctionalization; Amores et al., 1998; Force et al., 1999; Lynch and Force, 2000). However, they may also evolve new functions (neofunctionalization). Therefore, fish models can serve as an ideal platform to study the role of AHR in both physiology and toxicology.
Zebrafish as a Toxicological Model Organism
Zebrafish is a well-established vertebrate model for studying embryonic development and developmental toxicology and has been used extensively to unravel AHR pathway complexity (Garcia et al., 2016; Sipes et al., 2011; Teraoka et al., 2003a). Zebrafish embryos are transparent, and they develop externally and rapidly, with primary organogenesis complete around 48 h postfertilization (hpf), and the heart, liver, and brain well developed by 120 hpf (Kimmel et al., 1995). To this end, most early stage toxicity studies are conducted with morphological, behavioral, and molecular evaluations occurring within the first 120 h of development (Nishimura et al., 2016). Zebrafish also possess high genetic relatedness to humans; 76% of human genes have a zebrafish ortholog, and 82% of human genes that cause disease are present in zebrafish, increasing the translational value of the zebrafish model (Howe et al., 2013). Furthermore, zebrafish share similar morphology, physiology, and xenobiotic metabolic pathways with mammals (Diekmann and Hill, 2013), possessing direct orthologs of the human CYP1 enzymes like cyp1a and cyp1b1, in addition to cyp1c1 and cyp1c2 that lack human orthologs (Goldstone et al., 2010).
The Zebrafish AHRs
Zebrafish possess 3 AHR genes (ahr1a, ahr1b, and ahr2) that were named at the time of discovery according to their hypothesized evolutionary relationships to the AHR genes in other fish. Thus, the ahr1 genes were thought to be most closely related to the ahr1 genes of other fish and to the mammalian AHR (Andreasen et al., 2002a; Karchner et al., 2005); the designation “a” and “b” reflected the initial conclusion that ahr1a and ahr1b were paralogs formed during the fish-specific whole-genome duplication, and was consistent with the standard zebrafish nomenclature for such paralogs (Karchner et al., 2005). More recent analysis taking into account new AHR sequences from a variety of species, combined with analysis of shared synteny between zebrafish and human chromosomes, suggested that zebrafish ahr1a is orthologous to the mammalian AHR, rather than a paralog of ahr1b resulting from the fish-specific whole-genome duplication (Hahn et al., 2017). (Orthology refers only to evolutionary relationships, and does not necessarily imply identical functions. Two genes are orthologous if they have descended from the same gene in the most recent common ancestor of the species in which they are found [Fitch, 1970].) Zebrafish ahr2 is orthologous to ahr2b genes of other fishes (Karchner et al., 2005; Tanguay et al., 1999).
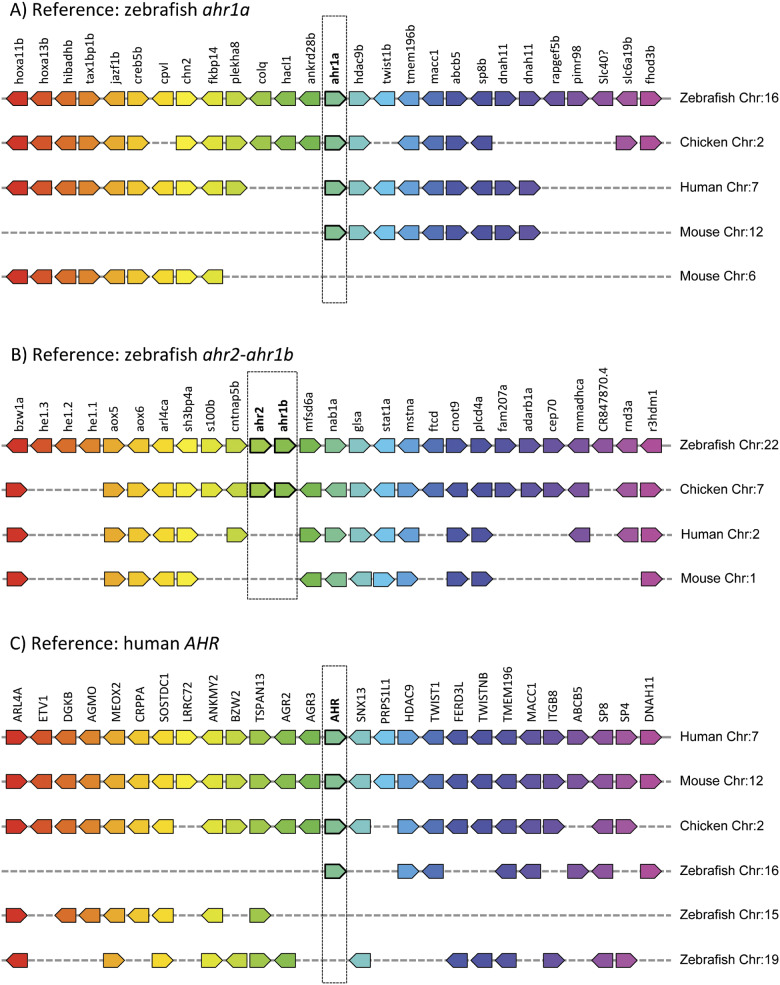
Further insight into the relationships of zebrafish ahr genes to AHR genes in other vertebrates can be obtained by additional analyses of shared synteny, which can complement gene phylogenies to help reveal evolutionary relationships (Postlethwait, 2007). A comparison of the shared synteny among AHR-containing chromosomes in zebrafish, human, mouse, and chicken using Genomicus (Muffato et al., 2010; Nguyen et al., 2018) is illustrative (Figure 1). The zebrafish ahr1a gene is located on chromosome 16 (Andreasen et al., 2002a; Barbazuk et al., 2000; Hahn et al., 2017; Le Beau et al., 1994). Zebrafish chromosome 16 exhibits extensive shared synteny with human chromosome 7, mouse chromosome 12, and chicken chromosome 2—the locations of the canonical AHR genes in each of these species (Figure 1A). This supports the earlier suggestion that ahr1a is the ortholog of human AHR (Hahn et al., 2017). A reciprocal analysis of shared synteny using the AHR on human chromosome 7 as the reference gene (Figure 1C) confirms the relationship between AHR-containing human chromosome 7, mouse chromosome 12, chicken chromosome 2, and zebrafish chromosome 16, and also reveals the loss of the predicted paralog of ahr1a that would have been expected from the fish-specific whole-genome duplication (see chromosome 19). Zebrafish ahr2 and ahr1b are located on chromosome 22 (Karchner et al., 2005; Tanguay et al., 1999; Wang et al., 1998). This chromosome exhibits extensive shared synteny with chicken chromosome 7, the location of 2 additional chicken AHR genes, designated AHR2 and AHR1B (Lee et al., 2013; Yasui et al., 2007) (Figure 1B). Both zebrafish chromosome 22 and chicken chromosome 7 exhibit shared synteny with human chromosome 2 and mouse chromosome 1, which lack the AHR2-AHR1 pair, confirming the loss of these genes in the mammalian lineages leading to human and mouse (Figure 1B). (See Table 1 for more details on the 3 zebrafish ahr genes and their respective translation products.)
Figure 1.
Shared synteny between zebrafish and other vertebrate AHR genes. Shared synteny was analyzed using Genomicus (versions 93.0 and 100.01) (Muffato et al., 2010; Nguyen et al., 2018) with manual curation using Ensembl. The AlignView tool in Genomicus was used to visualize the syntenic relationships. Genomes for human (Homo sapiens), mouse (Mus musculus), and chicken (Gallus gallus) were used to illustrate syntenic relationships with the chromosomes containing 3 zebrafish ahr s. The panels show the shared synteny obtained when using (A) zebrafish ahr1a (chromosome 16), (B) zebrafish ahr2-ahr1b (chromosome 22), and (C) human AHR (chromosome 7) as reference genes. The genes on either side of the reference gene are shown in the correct order and orientation. Orthologs (or in some cases paralogs) of the reference gene and its flanking genes are shown in the same color, below the reference chromosome, organized by species and chromosome. The position and order of genes below the reference chromosome do not necessarily reflect their position and order on the indicated chromosome.
Table 1.
Zebrafish ahr Genes and Their Respective Translation Products
| Characteristic | AHR2 | AHR1a | AHR1b |
|---|---|---|---|
| Zebrafish chromosome/linkage group | 22 | 16 | 22 |
| Mammalian orthologs | — | AHR | — |
| Amino acid length (aa) | 1027 | 805 | 954 |
| Predicted molecular mass of protein (kDa) | 113 | 90.4 | 104.8 |
| Overall amino acid identity comparison with human AHR (%) | 51 | 52 | 67 |
| Amino acid identity comparison with human AHR ligand-binding domain (%) | 71 | 68 | 71 |
| Overall amino acid identity comparison with AHR1b (%) | 45 | 44 | 100 |
| Conserved N-terminal halves identity comparison with AHR1b protein (%) | 66 | 63 | 100 |
The presence of 3 AHRs in zebrafish is intriguing. Despite the plethora of research that has been conducted, we are only beginning to understand their functional roles in development and in adult tissues, and we do not yet have a clear picture of the extent to which each paralog is involved in endogenous versus toxicological roles. In this article, we survey current AHR zebrafish toxicology research and identify specific knowledge gaps and opportunities for future research. We begin by reviewing the receptor characteristics, followed by early stage toxicity and interaction of AHRs with other signaling pathways, and conclude with adult toxicity and potential AHR-associated epigenetic effects.
RECEPTOR CHARACTERISTICS
General characteristics of the zebrafish AHRs, beginning with their baseline and chemically induced expression and endogenous roles in both developing and adult zebrafish are summarized in Table 2. We later elucidate the inducible roles of the 3 AHRs along with their known binding partners and conclude this section by reviewing homology modeling of the 3 receptors.
Table 2.
Receptor Characteristics (Developmental Baseline and TCDD-Induced mRNA Expression, Endogenous Ligands and Roles, and Binding Partners) of AHR2, AHR1a, and AHR1b (See Text for Citations)
| Characteristic | AHR2 | AHR1a | AHR1b |
|---|---|---|---|
| Earliest detected expression |
5 hpf |
24 hpf |
24 hpf |
| mRNA localization during development |
Several regions including the head and trunk |
Liver at 52 hpf (Sugden et al., 2017), regenerating fin (Sugden et al., 2017) |
Developing eye |
| mRNA localization in adults |
Brain, heart, muscle, swim bladder, liver, gill, skin, eye, kidney, fin |
Brain (Webb et al., 2009), liver, heart, swim bladder, and kidney (Andreasen et al., 2002a) | Unknown |
| Effect of TCDD exposure on mRNA expression |
Increase in expression (Andreasen et al., 2002b; Garcia et al., 2018a; Karchner et al., 2005; Tanguay et al., 1999) |
Increase in expression |
No change in expression |
| Endogenous roles |
Several at both embryonic/larval and adult life stages (see Endogenous AHR Roles in Zebrafish) |
Possible roles in hypocretin/orexin signaling |
Crosstalk detected with NRF signaling |
| Known endogenous ligands | FICZ (Jonsson et al., 2009; Wincent et al., 2016), 3α,5α-tetrahydrocorticosterone and 3α,5β-tetrahydrocorticosterone (5α- and 5β-THB) (Wu et al., 2019) | None identified |
FICZ |
| Endogenous Cyp1a expression regulation |
Expression in the developing zebrafish eye but not in the trunk or brain |
None | None |
| In vitro binding with ARNTs |
ARNT1b, 1c, 2 b, 2c |
ARNT2b |
ARNT2b |
| In vitro binding with TCDD |
Yes |
No |
Yes |
| In vitro transactivation activity with TCDD |
Yes |
Not applicable |
Yes but less sensitive than AHR2 |
| ARNT required for in vivo activation |
ARNT1 |
Not applicable | Unknown |
Expression of AHRs in Zebrafish
Transcriptomic, in situ hybridization, and immunohistochemical techniques have been used to understand the developmental, tissue-specific, and chemically induced expression of the AHRs. Ahr2 mRNA is expressed during normal zebrafish development in several regions including the head and the trunk (Andreasen et al., 2002b; Sugden et al., 2017); expression is detected as early as 5 hpf and does not change through 120 hpf (Andreasen et al., 2002b; Tanguay et al., 1999). Upon zebrafish embryonic exposure to TCDD, ahr2 expression increases and is detected in several locations across zebrafish development from 24 to 120 hpf (Andreasen et al., 2002b; Garcia et al., 2018a; Karchner et al., 2005; Tanguay et al., 1999). Other chemicals such as beta-naphthoflavone (BNF), a synthetic flavonoid commonly used as a surrogate model PAH (Poland and Kende, 1976; Sugden et al., 2017), cardiosulfa, a sulfonamide drug (Ko et al., 2009), and the polychlorinated biphenyl (PCB), PCB-126 (Kubota et al., 2015) induce ahr2 expression in developing zebrafish. On the other hand, exposure to benzo[a]pyrene (BaP) and some other oxy-PAHs significantly reduce ahr2 expression suggesting the complexity of AHR regulation by different PAHs (Cunha et al., 2020). In adults, ahr2 mRNA is detected in the brain, heart, muscle, swim bladder, liver, gill, skin, eye, kidney, fin both in unexposed and TCDD-exposed animals (Andreasen et al., 2002a). An antibody to AHR2 has been used to investigate AHR2 function in zebrafish cell culture (Wentworth et al., 2004) and in a heterologous cell system (Evans et al., 2008), but has not been used successfully in vivo.
Ahr1a mRNA is expressed during normal zebrafish development; expression is detected from 24 hpf, increases by 72 hpf, and stays relatively constant through 120 hpf (Andreasen et al., 2002a; Karchner et al., 2005). Ahr1a has more restricted expression patterns compared with ahr2 and is detected weakly in the liver at 52 hpf (Sugden et al., 2017), in a regenerating fin 3 days postamputation (Mathew et al., 2006), and in the adult brain (Webb et al., 2009), liver, heart, swim bladder, and kidney (Andreasen et al., 2002a). Upon embryonic exposure to TCDD, ahr1a expression significantly increases at 72 and 120 hpf (Andreasen et al., 2002a; Karchner et al., 2005), whereas BNF slightly induces expression of ahr1a in 48 hpf zebrafish (Sugden et al., 2017). There are no published antibodies experimentally shown to detect AHR1a protein expression in zebrafish.
Like ahr2 and ahr1a, ahr1b mRNA is expressed during normal development; expression is detected from 24 hpf and is increased at 48 and 72 hpf (Karchner et al., 2005). Ahr1b mRNA is highly expressed in the developing eye (Karchner et al., 2017; Sugden et al., 2017). Unlike ahr2 and ahr1a, ahr1b expression does not change after exposure to TCDD or BNF (Karchner et al., 2005; Sugden et al., 2017; Ulin et al., 2019). However, BaP exposure increased expression of ahr1b in 72 hpf zebrafish (Huang et al., 2012), whereas low-level pyrene exposure did not induce expression of any of the 3 ahr genes (Zhang et al., 2012). A rabbit polyclonal antibody targeting the AHR1b protein (Ulin et al., 2019) detected protein expression by Western blot in 24-hpf zebrafish. Using this same antibody, Karchner et al., (2017) performed immunohistochemical staining of 96-hpf larvae and showed that, like its mRNA, the AHR1b protein is also expressed in the developing eye, including the retinal inner and outer plexiform layers. Overall, these studies show that the spatiotemporal expression of the AHRs is dependent on the specific AHR receptor.
Endogenous AHR Roles in Zebrafish
To effectively study the endogenous and toxicological roles of the zebrafish AHRs, reverse genetics tools including transient knockdown of translation using morpholino oligonucleotides (Heasman, 2002; Timme-Laragy et al., 2012a), and stable and heritable genetic knockout lines have been generated. Although both of these tools can greatly enable the understanding of the functions of the zebrafish AHRs, we acknowledge their limitations here. In addition to their specific targets, morpholinos can nonspecifically affect expression of other targets so without rigorous controls it can be challenging to conclude whether an observed morpholino phenotype is due to its specific or off-target effects (Kok et al.,, 2015; Stainier et al.,, 2017). On the other hand, heritable mutations—especially those generating premature termination codons and nonsense-mediated decay of the resulting mRNA—can be subject to genetic compensation, where in response to the mutation, cells upregulate related genes that rescue the mutant phenotype (El-Brolosy et al., 2019; Ma et al., 2019; Rossi et al., 2015). Additionally, a mutation presumed to be loss-of-function might be rescued by altered mRNA processing, such as exon-skipping or alternative splicing, that produces a functional or partly functional protein (Anderson et al., 2017). This means that a heritable mutation that produces no phenotype could be a false-negative result. It is important to take these drawbacks of both morpholinos and knockout lines into consideration while interpreting the results of the studies presented below.
Both splice-blocking and translation-blocking morpholinos have been designed for ahr1a (Incardona et al., 2005; Seifinejad et al., 2019), ahr1b (Goodale et al., 2012; Ulin et al., 2019), and ahr2 (Bugel et al., 2013; Prasch et al., 2003; Teraoka et al., 2003b). Morpholino knockdown of ahr2 does not produce visible phenotypes (Dong et al., 2004; Prasch et al., 2003) likely due to the incomplete and transient receptor knockdown, prompting several groups to generate stable AHR mutant lines. The first functional AHR2 knockout line was established using Targeted Induced Local Lesions in Genomes (TILLING) (Goodale et al., 2012). Later, transcription activator-like effector nucleotide-mediated mutagenesis was used to generate AHR1a, AHR1b, and AHR2 mutants (Sugden et al., 2017). More recently, 2 CRISPR-Cas9 AHR2 homozygous mutant lines (Garcia et al., 2018a; Souder and Gorelick, 2019) and CRISPR-Cas9 AHR1a and AHR1b homozygous mutant lines (Karchner et al., 2017; Souder and Gorelick, 2019) have been established. These mutant lines are being intensively used to study both the endogenous and toxicological roles of the AHRs; the endogenous roles are reviewed here, whereas the toxicological roles are examined in later sections.
Some studies have suggested that all 3 AHRs are dispensable specifically for embryonic vascular patterning, and normal larval fin development and jaw growth (Souder and Gorelick, 2019; Sugden et al., 2017). However, AHR2-null background zebrafish have fin and craniofacial malformation as adults (Garcia et al., 2018a; Goodale et al., 2012; Souder and Gorelick, 2019), and both abnormal larval and adult behavior (Garcia et al., 2018a; Knecht et al., 2017b; Wu et al., 2019). AHR2-null zebrafish are also largely infertile and show decreased survival and diminished reproductive health (Garcia et al., 2018a). Loss of AHR2 does not affect basal developmental mRNA expression of cyp1a, ahr1b, ahrra, ahrrb, cyp1b1, cyp1c1, cyp3a65, slincR, and sox9b (sry box containing 9b), all known AHR-regulated genes (Garcia et al., 2018a; Goodale et al., 2012; Prasch et al., 2003). On the other hand, AHR2 is important for endogenous cyp1a expression specifically in the developing zebrafish eye but not in the trunk or brain (Sugden et al., 2017). Only the lack of all 3 AHRs caused a complete loss of cyp1a mRNA expression (but not cyp1b1 expression) throughout the developing zebrafish (Sugden et al., 2017). These studies demonstrate the various possible roles of AHR2 in maintaining normal morphology and development. AHR2 also binds endogenous AHR ligands identified in other systems. Formylindolo[3,2-b]carbazole (FICZ), a tryptophan oxidation product formed upon exposure to UV or visible radiation, binds both AHR2 and AHR1b in vitro and induces expression of cyp1a and cyp1b1 in an AHR2-dependent manner (Jonsson et al., 2009). Morpholino knockdown experiments illustrate that FICZ has increased and decreased toxicity in the absence of cyp1a and ahr2, respectively, suggesting that the biological effects of FICZ are AHR2-dependent and regulated by its Cyp1a-mediated metabolism (Wincent et al., 2016). Zebrafish have also been used to define the endogenous roles of 3α,5α-tetrahydrocorticosterone and 3α,5β-tetrahydrocorticosterone (5α- and 5β-THB). These neuroactive steroids induce AHR2-dependent cyp1a, mbp, and sox10, the latter 2 of which are markers for myelinating cells (Wu et al., 2019). 5α-THB exposure also alters zebrafish larval behavior in an AHR2-dependent manner suggesting the importance of THB-AHR2 signaling in normal nervous system development (Wu et al., 2019).
There are no identified endogenous roles for AHR1a in zebrafish. AHR1a does not seem to play a role in normal development, larval feeding, or endogenous Cyp1a protein expression evidenced from AHR1a mutant fish that appear normal (Sugden et al., 2017). Further, neither AHR1a nor AHR1b morphants or mutants display overt phenotypes (Garner et al., 2013; Goodale et al., 2012; Sugden et al., 2017). However, a recent study showed that morpholino knockdown of ahr1a led to loss of hypocretin/orexin expression and developmental deformities (Seifinejad et al., 2019). Additionally, another study identified crosstalk between AHR1b and Nrf signaling during zebrafish development (Ulin et al., 2019). Many have suggested that partial overlapping functional redundancy of AHR2 and AHR1b allows for compensatory activity when AHR2 is lost (Prasch et al., 2003; Sugden et al., 2017); however, additional research is needed to clarify this. It is possible that the level of investigation to date has been insufficient to identify and confirm subtle development roles for these orthologs.
Inducible Roles of the Zebrafish AHRs
To understand the inducible roles of the zebrafish AHRs, a combination of in vitro binding studies, transactivation assays in COS-7 mammalian cells, and in vivo zebrafish developmental studies has been utilized.
AHR2 is a functional receptor whose signaling is modulated not only by its various ligands but also by its binding partners and downstream genes. Zebrafish have 2 ARNT genes, arnt1 and arnt2, each present as 3 splice forms (ARNT1a, 1b, 1c, and ARNT2a, 2b, 2c) (Prasch et al., 2006; Tanguay et al., 2000; Wang et al., 1998, 2000). AHR2 is capable of binding ARNT1b, ARNT1c, ARNT2b, and ARNT2c in vitro but only the complexes of AHR2 with ARNT1b, ARNT1c, or ARNT2b are able to promote transactivation by inducing AHRE-driven transcription with TCDD (Prasch et al., 2006; Tanguay et al., 2000). It was later shown using morpholino studies that both AHR2 and some form of the ARNT1 protein, but not the ARNT2 protein, are required for generating toxic responses to TCDD in developing zebrafish (Antkiewicz et al., 2006; Prasch et al., 2004, 2006). It is not yet known how the AHR2-ARNT complexes mediate responses induced by other ligands. Furthermore, the functions of the various splice variants of the ARNTs are yet to be elucidated. AHR signaling can also be subjected to downregulation by proteasomal degradation of AHR2 (Wentworth et al., 2004) as well as by transcriptional repression of its target genes by the AHRR. Zebrafish have 2 distinct AHRRs, AHRRa (originally designated AHRR1) and AHRRb (AHRR2), which are co-orthologs of the mammalian AHRR (Evans et al., 2005). Both AHRRa and AHRRb are induced in an AHR2-dependent manner similar to cyp1a, only by compounds that activate the AHR signaling pathway (Evans et al., 2005; Garcia et al., 2018a; Jenny et al., 2009; Timme-Laragy et al., 2007). AHRRa blocks AHR2 function by competing for binding to AHREs as well as by a transrepression mechanism that is independent of DNA binding (Evans et al., 2008). Knockdown of AHRRa, but not AHRRb, using a morpholino in the absence of TCDD exposure, phenocopied TCDD developmental toxicity and caused a large number of gene expression changes compared with wild-type fish, whereas knockdown of either AHRRa or AHRRb enhanced TCDD-induced pericardial edema (Aluru et al., 2014; Jenny et al., 2009). These results suggest that although AHRRa is involved in regulating constitutive AHR signaling, both AHRRa and AHRRb play a role in modulating TCDD developmental toxicity. The ability of AHRRa knockdown to both phenocopy TCDD toxicity (in the absence of TCDD exposure) and enhance TCDD toxicity is consistent with a role for AHRRa in controlling constitutive AHR activity (in unexposed embryos), and a role for the AHR2-dependent induction of AHRRa after TCDD exposure to limit the AHR-dependent TCDD effects in a negative feedback loop. Further, zebrafish embryos in which AHRRb or both AHRRs (but not AHRRa alone) were knocked down had increased TCDD-induced expression of cyp1a, cyp1b1, and cyp1c1 at 72 hpf, suggesting that AHRRb may have a role in controlling TCDD-activated AHR signaling (Jenny et al., 2009). To date, we do not know how AHRRa or AHRRb interact with AHR1a or AHR1b. Future work in single and double mutant lines for the AHRRs and AHRs will enhance our understanding of their interactions and functions in zebrafish.
The zebrafish AHR1a is a functional receptor in vivo (Goodale et al., 2012) but does not bind the canonical exogenous ligand, TCDD, in an in vitro heterologous cell system (Andreasen et al., 2002a; Karchner et al., 2005); the receptor is able to bind ARNT2b, can recognize AHREs more weakly compared with AHR2, and lacks transactivation activity with all ARNT2 proteins in vitro (Andreasen et al., 2002a). AHR1b was identified as a fully functional zebrafish receptor when assessed in vitro and in a heterologous cell system (Karchner et al., 2005). TCDD can bind AHR1b which interacts with ARNT2b, and promotes transactivation with efficacy comparable with that of AHR2 but with an 8-fold lower sensitivity (Karchner et al., 2005). It remains unknown to what extent AHR1a and AHR1b are able to interact with the ARNT1 proteins.
Homology Modeling of Zebrafish AHRs
Several in silico-based modeling studies have investigated the structure and ligand-binding properties of the 3 zebrafish AHRs (Bisson et al., 2009; Fraccalvieri et al., 2013; Zhang et al., 2018a,b). Using molecular dynamics simulations, it was determined that TCDD and many dioxin-like compounds interact with 6 amino acid residues in the AHR2 ligand-binding domain (Zhang et al., 2018b). The results supported, for the first time, the finding that polychlorinated diphenylsulfides can bind and activate AHR2 (Zhang et al., 2018b). Similarly, 2,2′,4,4′,5-penta-BDE (BDE-99) is able to bind to both the zebrafish AHR2 as well as the pregnane X receptor (PXR) (Zhang et al., 2018a). Although the ligand-binding pocket was more compact in the Bisson model (Bisson et al., 2009) compared with Fraccalvieri (Fraccalvieri et al., 2013), both models predicted that TCDD binds to AHR2 and AHR1b, but not AHR1a. Using site-directed mutagenesis coupled with functional analyses, it was determined that AHR1a was not able to bind TCDD because of differences in 3 amino acid residues in the ligand-binding domain of AHR1a compared with that of AHR2 (Fraccalvieri et al., 2013). The differences make the AHR1a binding cavity much shorter than that of AHR2 with too limited space for TCDD binding. The Bisson model (Bisson et al., 2009) has also been utilized to predict binding with molecular docking of structurally different AHR ligands to the 3 zebrafish AHRs, summarized in Table 3. The table reveals that in general, xenobiotic ligands bind to more than 1 zebrafish AHR, making it likely that their overall toxicity is mediated by a combination of the receptors.
Table 3.
Predicted Binding of Different Ligands to the Zebrafish AHRs
| Ligand | AHR2 | AHR1a | AHR1b | References |
|---|---|---|---|---|
| Anthracene | Yes | Not tested | Not tested | Goodale et al. (2015) |
| Anthrone derivative SP600125 | Yes | Not tested | Not tested | Goodale et al. (2015) |
| BAA | Yes | Not tested | Not tested | Goodale et al. (2015) |
| BaP | Yes | Not tested | Not tested | Goodale et al. (2015) |
| BEZO | Yes | Not tested | Not tested | Goodale et al. (2015) |
| CH223191 | Yes | Yes | Weak | Gerlach et al. (2014) |
| Leflunomide | Yes | Yes | Yes | Bisson et al. (2009); Goodale et al. (2012); O’Donnell et al. (2010) |
| ortho-mITP | Yes | No | No | Gerlach et al. (2014) |
| meta-mITP | Yes | Yes | Weak | Gerlach et al. (2014) |
| para-mITP | Weak | Yes | No | Gerlach et al. (2014) |
| TCDD | Yes | No | Yes | Bisson et al. (2009) |
| NPAHs, HAHs, amino PAHs | Yes | Yes | Yes | Chlebowski et al. (2017) |
Overall, the studies reviewed in this section demonstrate that the 3 zebrafish AHRs are diverse in their local expression patterns, with only partial overlap in developmental and adult expression indicating cell-type-specific regulation. AHR2 and AHR1a are more widely expressed compared with AHR1b, and although AHR2 has been associated with normal developmental and physiological functions, such roles are not yet apparent for AHR1a and AHR1b. Importantly, all 3 receptors bind a variety of ligands evidenced by empirical and homology modeling studies. Clearly, the changing levels of expression of all 3 receptors across development testify to their dynamic nature and allude to the complexity of accurately understanding the AHRs’ functional roles at different life stages.
EARLY STAGE TOXICITY
In this section, we discuss ligands that produce adverse developmental effects dependent on the presence of each of the zebrafish AHRs. The majority of the research has focused on AHR2 and environmental contaminants including PAHs, TCDD, polychlorinated biphenyls, and pharmaceuticals. It is noteworthy that several PAHs also activate AHR1a and AHR1b, and below we specifically review the Cyp1a expression patterns dependent on the 3 AHRs.
AHR2
Despite much research focused on TCDD, several studies have explored a diversity of xenobiotics and suggest that in vivo toxicity may be mediated by more than 1 zebrafish AHR (Goodale et al., 2012). Majority of the research conducted so far utilizes morpholino knockdown (with regulation of cyp1a induction as confirmation for knockdown) to reveal receptor-dependent toxicity effects; however, as groups are beginning to generate stable genetic knockout lines, more AHR2 mutant studies are being conducted. Although morpholino knockdown can inform on which of the 3 receptors are important for mediating toxicity, only mutant studies with complete knockouts can definitively demonstrate toxicologically functional roles for the AHR paralogs. In this section, we focus on the xenobiotics whose toxicity is mediated primarily by AHR2 to collate what we know about AHR2’s functionality. We will begin by reviewing the early stage toxicity of PAHs and other xenobiotics, then we will summarize what we know about TCDD early stage toxicity in zebrafish. The functional role of AHR2 upon exposure to diverse ligands is summarized in Table 4.
Table 4.
Developmental Toxicity Endpoints and Cyp1a Expression Patterns Mediated by AHR2 From Morpholino Knockdown Studies
| Xenobiotic Ligand | Endpoints Mediated by Ligand x AHR2 | Results of AHR2 Knockdown | References |
|---|---|---|---|
| Polycyclic Aromatic Hydrocarbons (PAHs) | |||
| Pyrene |
|
|
Incardona et al. (2005) |
| Chrysene |
|
|
Incardona et al. (2005) |
| Dibenzothiophene |
|
|
Incardona et al. (2005) |
| Phenanthrene |
|
|
Incardona et al. (2005) |
| Benz[a]anthracene (BAA) |
|
|
Incardona et al. (2006, 2011) |
| Benzo[a]pyrene (BaP) |
|
|
Cunha et al. (2020); Incardona et al. (2011); Knecht et al. (2017b) |
|
Benzo[k]fluoranthene (BkF) |
|
|
Incardona et al. (2011) |
| Retene |
|
|
Scott et al. (2011) |
| Benz(a)anthracene-7,12-dione (7,12-B[a]AQ) |
|
|
Goodale et al. (2015); Knecht et al. (2013) |
| 1,9-Benz-10-anthrone (BEZO) |
|
|
Goodale et al. (2015) |
| 1,6-Dinitropyrene, |
|
|
Chlebowski et al. (2017) |
| benzo[j]fluoranthene, dibenzo[a.h]pyrene, dibenzo[a,i]pyrene, and benzo[b]fluoranthene |
|
|
Shankar et al. (2019) |
| 6H-benzo[cd]pyren-6-one |
|
|
Cunha et al. (2020) |
| Other chemicals: mixtures, pharmaceuticals, indoles, and halogenated aromatic hydrocarbons | |||
| PAH-containing soil extracts from a gasworks, a former wood preservation site, and a coke oven |
|
|
Wincent et al. (2015) |
| Total particulate matter (cigarette smoke) |
|
|
Massarsky et al. (2016) |
| Environmentally relevant PAH mixtures (“Supermix 3” and “Supermix 10”) |
|
|
Geier et al. (2018b) |
| BaP and 6H-benzo[cd]pyren-6-one |
|
|
Cunha et al. (2020) |
| Cardiosulfa (sulfanomide drug) |
|
|
Ko and Shin (2012); Ko et al. (2009) |
| Leflunomide |
|
|
Goodale et al. (2012); O’Donnell et al. (2010) |
| 3,3’,4,4’,5-Pentachlorobiphenyl (PCB-126) |
|
|
Garner et al. (2013); Jonsson et al. (2007) |
| Monosubstituted isopropyl triaryl phosphate (mITP) (a major component of Firemaster 550, a flame retardant mixture) |
|
|
Gerlach et al. (2014) |
| Paclobutrazol (fungicide) |
|
|
Wang et al. (2015) |
| Formylindolo[3,2-b]carbazole (FICZ) |
|
|
Wincent et al. (2016) |
| Phenanthroline |
|
|
Ellis and Crawford (2016) |
| 2,7-Dibromocarbazole and 2,3,6,7-tetrachlorocarbazole |
|
|
Fang et al. (2016) |
| 3α,5α-tetrahydrocorticosterone (5α-THB) |
|
|
Wu et al. (2019) |
| 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) |
|
|
See text |
Measured endpoints that were not altered upon AHR2 knockdown.
Polycyclic Aromatic Hydrocarbons
Many PAHs cause dioxin-like AHR2-dependent phenotypic endpoints including pericardial and yolk sac edemas, bent axes, cardiotoxicity, and eye and jaw malformations. Morpholino knockdown studies suggest that the following PAHs cause developmental toxicity primarily via AHR2: BaP (Cunha et al., 2020; Incardona et al., 2011), retene (Scott et al., 2011), benz[a]anthracene (BAA) (Incardona et al., 2006), pyrene (Incardona et al., 2005), and 1,9-benz-10-anthrone (BEZO) and benz(a)anthracene-7,12-dione (7,12-B[a]AQ) (Goodale et al., 2015). Dozens of different PAH exposures have been associated with altered embryonic and larval behavior (Geier et al., 2018a); however, only little is known about AHR2’s role in behavioral outcomes. Specifically, BaP-exposed wild-type zebrafish exhibit a hyperactive swimming response in the 120-hpf larval photomotor response (LPR) assay whereas BaP-exposed AHR2 mutants do not display a significantly altered LPR (Knecht et al., 2017b). This suggests that disruption of the AHR2 signaling pathway can lead to detrimental consequences to nervous system development and functioning. Some AHR2 knockdown studies revealed that many PAHs, including phenanthrene, dibenzothiophene, and benzo[k]fluoranthene (BkF), produce adverse developmental outcomes independent of AHR2 (Incardona et al., 2005, 2011), despite inducing AHR2-dependent Cyp1a protein expression (Incardona et al., 2005, 2011; Shankar et al., 2019). It is possible that the developmental toxicity produced by these PAHs is mediated by other zebrafish AHRs, or that incomplete morpholino knockdown confounded these studies. One recent morpholino study, however, showed that exposure to BkF and 3 other fluoranthenes produced caudal fin duplication that is AHR2 dependent (Garland et al., 2020). This suggests that AHR2 may mediate specific malformations such as the fin duplication, whereas chemical interaction with other receptors such as AHR1b may mediate other developmental toxicity endpoints. Future studies testing the toxicity of these PAHs in both AHR1b and AHR2-null backgrounds are crucial to verify these results.
Both cyp1a mRNA and protein expression are frequently used as indicators of AHR activation by PAHs. In general, PAHs that elicit AHR2-dependent toxicity also induce cyp1a mRNA expression (Goodale et al., 2015; Knecht et al., 2013). Studies have also demonstrated that the obligate AHR isoforms for PAH toxicity can be inferred by determining the larval Cyp1a protein expression pattern. For example, developmental exposure to oxy-PAHs 7,12-B[a]AQ, BEZO, and BaP produces Cyp1a protein expression in the vasculature that is partially or fully dependent on AHR2 (Goodale et al., 2015; Incardona et al., 2011; Knecht et al., 2013). PAHs such as chrysene, retene, BAA, BkF, 1,6-dinitropyrene, benzo[j]fluoranthene, dibenzo[a.h]pyrene, dibenzo[a,i]pyrene, and benzo[b]fluoranthene (Table 4) induce Cyp1a protein in both the skin and the vasculature, among other regions. Vascular Cyp1a expression in response to these latter PAHs is only partially reduced upon AHR2 knockdown; however, Cyp1a expression in the skin is lost, demonstrating its complete AHR2-dependence. Similarly, although the surrogate model PAH, BNF, induces cyp1a mRNA expression in the skin and vasculature, only the expression in the skin is completely lost in AHR2 mutants (Sugden et al., 2017). Because of this differential, we recently reported that induction of Cyp1a in the skin is a more robust and reliable biomarker for AHR2 activation in developing zebrafish (Shankar et al., 2019). Studies have also identified nitro-PAHs such as 7-nitrobenz[a]anthracene and 3,7-dinitrobenzo[k]fluoranthene that do not cause visible developmental malformations at the tested concentrations but produce AHR2-dependent Cyp1a expression in a variety of organs (Chlebowski et al., 2017). Thus, these chemicals activate AHR2 without causing visible developmental toxicity, indicative of a potential adaptive response by Cyp1a. It is also possible that the absence of developmental toxicity is due to the lack of sustained activation of AHR2 by these chemicals, which has been hypothesized for other AHR agonists such as retene (Billiard et al., 1999). Future work assessing toxicity with Cyp1a inhibition and with daily renewal of the chemical exposure solution will help clarify this hypothesis.
Several studies have investigated the specific functional role of Cyp1a induction in PAH toxicity, and the apparent direct role for Cyp1a in PAH toxicity is chemical substrate dependent. Typically, studies utilize cyp1a morphants, or known Cyp1a competitive inhibitors such as alpha-naphthoflavone (ANF) or fluoranthene. Retene (Scott et al., 2011) and BAA (Incardona et al., 2006) cause AHR2-dependent, but Cyp1a-independent cardiovascular developmental toxicity in zebrafish. On the other hand, cyp1a knockdown delays the toxic effects of pyrene but fails to entirely protect the developing zebrafish from toxicity (Incardona et al., 2005). Cyp1a morphants also display enhanced toxic responses to the strong AHR ligand BkF, suggesting a protective role for Cyp1a in BkF toxicity (Incardona et al., 2011). Strong AHR agonists BkF (Garner et al., 2013; Van Tiem and Di Giulio, 2011) and BaP (Garner et al., 2013; Jayasundara et al., 2015), and the weak AHR ligand phenanthrene (Brown et al., 2015) were more developmentally toxic when combined with fluoranthene, suggesting that inhibition of Cyp1a-mediated metabolism can enhance toxicity of these PAHs. Although the phenanthrene + fluoranthene toxicity was not AHR2 dependent (Brown et al., 2015), AHR2 knockdown offered a protective role against the cardiotoxicity induced by the BkF + fluoranthene and BaP + fluoranthene mixtures (Garner et al., 2013). Similarly, the AHR2-dependent toxicity of BNF synergistically increased in combination with either ANF or a cyp1a morpholino, further demonstrating that Cyp1a can play an important protective role against PAH toxicity (Billiard et al., 2006). It was later shown that ANF did not act as an AHR antagonist, but rather a Cyp1a enzyme inhibitor, potentially prolonging the time the AHR was being activated, enhancing developmental toxicity (Timme-Laragy et al., 2007). Other than one study demonstrating that cyp1b1 did not seem to play a role in PAH toxicity (Timme-Laragy et al., 2008), the roles of the other zebrafish cyp genes in PAH toxicity are not well understood.
For PAHs whose toxicity is AHR2 dependent based on morpholino studies, there have been a number of corresponding whole-embryonic, genome-wide transcriptomic studies (Fang et al., 2015; Goodale et al., 2015; Hawliczek et al., 2012; Shankar et al., 2019) albeit without comparing gene expression profiles in the presence and absence of AHR2. One study, however, identified several novel genes and potential mechanisms specifically in the developing zebrafish heart that could mediate cardiotoxicity via AHR2 upon exposure to BaP, fluoranthene, and BaP + fluoranthene (Jayasundara et al., 2015). It was concluded that AHR2-dependent cardiotoxicity of BaP + fluoranthene was mediated, at least in part, by perturbations to Ca2+ homoeostasis. Genome-wide transcriptomic studies for PAHs known to be primarily AHR2 agonists show that despite activating the same receptor, the transcriptomic changes downstream of AHR2 are ligand dependent (Goodale et al., 2015; Shankar et al., 2019). We hypothesize that either slight differences in how chemicals bind to the receptor, or the formation of metabolites that can activate their own receptors, or a combination of the 2, are contributing to the ligand-dependent gene-expression profiles. Future work investigating these hypotheses is needed to illuminate more specific interactions between different ligands and the zebrafish AHRs. Some studies have utilized quantitative PCR to measure expression of specific genes including the cyps (mentioned above) and have identified genes that are regulated via AHR2 upon exposures to BAAQ (cyp1b1, wfikkn1, gstp2, igfbp1a) (Goodale et al., 2015), BEZO (gstp2, igfbp1a, arg2), and BkF (cyp1a, cyp1b, cyp1c, gstp2, gpx1, gclc) (Van Tiem and Di Giulio, 2011). Although several of these genes have been identified and well studied (eg, gpx1, gstp2, gclc are involved in antioxidant responses), elucidation of functions of some genes such as wfikkn1 is ongoing.
Other Xenobiotics: Mixtures, Pharmaceuticals, and Halogenated Hydrocarbons
AHR2 can at least partially mediate toxicity of “cigarette smoke” (Massarsky et al., 2016) and “PAH-containing soil extracts” from a gasworks, a former wood preservation site, and a coke oven site (Wincent et al., 2015). A recent study found that the developmental cardiotoxicity effects of both the individual chemicals and the “mixture of BaP and the oxy-PAH, 6H-benzo[cd]pyren-6-one” were significantly reduced upon ahr2 knockdown (Cunha et al., 2020). Unlike these mixtures, “weathered crude oil” consisting of lower molecular weight PAHs elicits morphological deficits and cardiotoxicity in an AHR2-independent manner, highlighting the potential influence of the structure and size of PAHs (Incardona et al., 2005). One study investigated Cyp1a protein expression induced in 120-hpf zebrafish after exposure to an “environmentally relevant PAH mixture.” Upon ahr2 knockdown, Cyp1a vascular expression was eliminated, but there was production of Cyp1a protein in the liver attributed to the loss of AHR2 leading to the production of metabolites that had a higher affinity for AHR1a. Independently knocking down AHR1a or AHR1b did not alter Cyp1a protein expression; however, a triple morpholino knockdown of all 3 AHRs reduced Cyp1a protein expression (Geier et al., 2018b). These results reiterate the need for considering the functional roles of all 3 zebrafish AHRs, especially when studying the mechanisms of toxicity of complex mixtures.
The zebrafish AHR2 mediates developmental toxicity of other small molecules and pharmaceuticals. Although all 3 AHRs can bind leflunomide, an anti-inflammatory drug (Bisson et al., 2009; Goodale et al., 2012), AHR2 mediates the bulk of its Cyp1a vascular expression at 120 hpf (Goodale et al., 2012; O’Donnell et al., 2010). Its metabolite A771726 is not an AHR2 agonist (O’Donnell et al., 2010). The small molecule sulfonamide, cardiosulfa, also produces AHR2-dependent cardiotoxicity in developing zebrafish as seen in ahr2 morpholino knockdown studies (Ko et al., 2009; Ko and Shin, 2012). Similar to TCDD (reviewed below), Cyp1a neither reduces nor exacerbates cardiosulfa toxicity (Ko and Shin, 2012). As noted above, the indole FICZ can cause developmental toxicity in an AHR2-dependent manner, especially when Cyp1a activity is inhibited or reduced by genetic knockdown (Jonsson et al., 2009; Wincent et al., 2016).
“Other organic compounds” such as phenanthroline (Ellis and Crawford, 2016) and 2 halogenated carbazoles (Fang et al., 2016) are associated with PAH and TCDD-like developmental toxicity, respectively, and the fungicide, paclobutrazol (Wang et al., 2015) causes digestive tract toxicity, all of which are AHR2 mediated. Some flame-retardant chemicals appear to buck the trend. For instance, ahr2 knockdown does not reduce the cardiotoxicity associated with exposure to monosubstituted isopropyl triaryl phosphate (mITP), a major component of Firemaster 550 commercial mixture (Gerlach et al., 2014; McGee et al., 2013). However, ahr2 knockdown prevents vascular Cyp1a protein expression in response to mITP, suggesting that the mixture does activate AHR2 (Gerlach et al., 2014). An AHR antagonist (CH223191) was able to block heart malformations induced by mITP but it was suggested that CH223191 antagonizes another target in addition to AHR (Gerlach et al., 2014; McGee et al., 2013).
“PCB-126” (3,3′,4,4′,5-pentachlorobiphenyl) is one of the most potent AHR agonists (Kafafi et al., 1993) and is associated with developmental toxicity in zebrafish (Grimes et al., 2008). Ahr2 knockdown greatly reduces PCB-126-induced cardiac effects and mortality but only provides minimal protection against the abnormal inflation of the swim bladder (Garner et al., 2013; Jonsson et al., 2007). A follow-up study showed that, at a lower PCB-126 exposure concentration of 5 nM, ahr2 gene knockdown prevented the swim bladder phenotype. This suggests that, again, incomplete ahr2 morpholino knockdown was probably operant and thus insufficient to block toxicity at the higher concentration (Jonsson et al., 2012). This pattern was similar to ahr2 knockdown that partly mitigated cardiotoxicity caused by a lower TCDD exposure concentration of 0.3 ppb, but not at higher concentrations of 0.5 and 1 ppb (Dong et al., 2004). Ahr2 knockdown also significantly reduces cyp1a, cyp1b1, cyp1c1, and cyp1c2 mRNA expression, and Cyp1a protein activity produced by PCB-126 exposure (Garner et al., 2013; Jonsson et al., 2007, 2012). One recent study determined that PCB-126 exposure not only caused increased expression of few AHR target genes (ahrra, tiparp, and nfe2l2b) but also led to their mRNA being hypermethylated (Aluru and Karchner, 2020); future work to understand the specific role of this posttranscriptional modification is needed. Similar to TCDD, PCB-126 is not metabolized and accumulates in zebrafish, which leads to persistent expression of target genes (Garner and Di Giulio, 2012; Meyer-Alert et al., 2018). Waits and Nebert used a quantitative trait locus (QTL) approach to investigate the genetic basis for zebrafish embryo susceptibility to PCB-126-induced developmental cardiotoxicity. Among the top-ranked QTLs was a region on chromosome 22 that includes ahr2 and ahr1b, implicating 1 or both of these receptors in having a role in PCB-126 toxicity (Waits and Nebert, 2011).
2,3,7,8-Tetrachlorodibenzo-p-Dioxin
Developmental toxicity
TCDD is the most studied AHR2 ligand. In experiments using in vitro-translated AHR proteins or expression in heterologous cells, TCDD does not bind or activate AHR1a, but it binds AHR2 and AHR1b and activates them with comparable efficacies (Andreasen et al., 2002a; Karchner et al., 2005). When zebrafish are developmentally exposed to TCDD, they display reduced survival, and several phenotypes such as (but not limited to) cardiotoxicity, pericardial and yolk sac edemas, and craniofacial malformations (Henry et al., 1997). The various adverse developmental outcomes are reviewed in Carney et al. (2006b). Knockdown and knockout studies have demonstrated the role of AHR2 in mediating TCDD-induced pericardial and yolk sac edemas, cardiovascular and craniofacial malformations, decrease in body length, and increased apoptosis, in addition to a significant increase in the mRNA levels of cyp1a, cyp1b1, cyp1c1, and cyp1c2 in zebrafish (Carney et al., 2004; Dong et al., 2004; Garcia et al., 2018a; Goodale et al., 2012; Jonsson et al., 2007; Prasch et al., 2003; Souder and Gorelick, 2019; Teraoka et al., 2003b; Yin et al., 2008). One study also demonstrated that the constitutive activation of the AHR2 in zebrafish cardiac myocytes led to not only TCDD-like cardiotoxicity but also other defects in craniofacial development and failure to form swim bladders, suggesting the importance of the heart as a target organ (Lanham et al., 2014). We note that ahr2 knockdown, however, was unable to protect against TCDD-induced inhibition of swim bladder inflation and mortality; this was attributed to the short half-life of morpholinos after injection or a potential role of the other zebrafish AHRs (Prasch et al., 2003). Future work clarifying these results in an AHR2-null background zebrafish is necessary.
Mechanisms of TCDD developmental toxicity
The mechanisms of TCDD toxicity in humans and several vertebrate model organisms, including zebrafish, have been reviewed in Carney et al. (2006b), King-Heiden et al. (2012), Yoshioka et al. (2011), and Yoshioka and Tohyama (2019). TCDD-induced toxicity in zebrafish is associated with an array of transcriptomic changes, including modest changes to microRNA expression, in both developing zebrafish and specific adult organs such as the heart (Alexeyenko et al., 2010; Carney et al., 2006a; Chen et al., 2008; Garcia et al., 2018b; Handley-Goldstone et al., 2005; Jenny et al., 2012). The gene expression changes may be a function of (1) large-scale toxicological phenotypes associated with TCDD, (2) downstream effects of the AHR2/ARNT1 complex binding AHREs of various genes, or (3) the interaction of the AHR with other pathways or transcription factors (Carney et al., 2006b). Here, we review what is known about the role of AHR2-regulated genes in TCDD-induced developmental malformations in zebrafish.
Binding of TCDD to AHR2 induces expression of the cyp1 gene family. Upon exposure to TCDD, cyp1a mRNA and protein are expressed early in development in a variety of organs at the different development stages (Andreasen et al., 2002b; Kim et al., 2013; Yamazaki et al., 2002; Zodrow et al., 2004). One study noted that the Cyp1a protein is first localized to the skin and the vasculature after which it transitions to the vasculature, kidney, and liver by 120 hpf (Andreasen et al., 2002b). When AHR2-null zebrafish are exposed to TCDD, Cyp1a protein expression at 120 hpf is almost completely prevented (Goodale et al., 2012). It was initially thought that the induction of cyp1a is required for TCDD developmental toxicity (Teraoka et al., 2003b); however, a later study demonstrated that, consistent with mammalian literature, TCDD produces developmental toxicity endpoints independent of cyp1a (Carney et al., 2004). TCDD also induces expression of cyp1b1; however, this does not appear to have a direct role in TCDD-induced pericardial edema and craniofacial malformations (Yin et al., 2008). cyp1c1 and cyp1c2 likely play roles in TCDD-induced circulation failure in the midbrain but the exact mechanism is unknown (Kubota et al., 2011). Unlike the other zebrafish cyp1s, cyp1d1 does not seem to be transcriptionally activated by TCDD or PCB-126 (Goldstone et al., 2009).
Cyclooxygenase-2 (COX-2) enzymes, a family of heme-containing enzymes thought to be involved in acute inflammatory responses, have been studied in the context of the AHR signaling pathway in zebrafish and other model organisms. Zebrafish have 2 cox-2 genes, cox2a and cox2b (Ishikawa et al., 2007); both are induced in an AHR2-dependent manner upon exposure to PCB-126, but future work is needed to identify whether these genes are direct targets of AHR2 (Jonsson et al., 2012). To the best of our knowledge, induction of these genes has not been demonstrated upon TCDD exposure. However, it is suggested that cox2a, in combination with the thromboxane receptor and thomboxane A synthase 1 (also known as cyp5a), is involved in local circulation failure in the dorsal midbrain of developing zebrafish (Teraoka et al., 2009). Another study discovered the role of the cox2b-thromboxane pathway in TCDD-induced AHR2-dependent “precadiac” edema, the increased area of the small cavity between the heart and the body wall (Teraoka et al., 2014). The study demonstrated that knockdown of cox2b, but not cox2a, prevented formation of the precardiac edema in the TCDD-exposed zebrafish, and also showed the involvement of the thromboxane pathway, concluding that thromboxane release by TCDD probably led to the edema in the developing zebrafish. Additionally, other factors such as oxidative stress are involved, as an antioxidant was able to inhibit both precardiac edema and circulation failure caused by TCDD exposure (Dong et al., 2004).
The sox9b gene, a critical chondrogenic transcription factor, has been linked to cardiotoxicity caused by TCDD (Hofsteen et al., 2013b). Upon exposure to TCDD, sox9b expression is significantly reduced in an AHR2-dependent manner (Garcia et al., 2018a; Xiong et al., 2008), and 1 study found that sox9b knockdown resulted in TCDD-like heart malformations (Hofsteen et al., 2013b). Morpholino knockdown of sox9b also caused a phenotype similar to TCDD-induced jaw malformation, and restoration of sox9b with mRNA injection prevented craniofacial malformations suggesting sox9b’s role in TCDD-induced craniofacial defects (Xiong et al., 2008). A sox9b promoter-eGFP transgenic reporter fish (uw101Tg) (Plavicki et al., 2014) was produced and used to identify the TCDD-induced repression of sox9b in the developing zebrafish heart and brain (Garcia et al., 2017; Hofsteen et al., 2013b). A mechanism for linking activation of AHR2 and sox9b repression was recently suggested (Garcia et al., 2017, 2018b). The sox9b long intergenic noncoding RNA (slincR) is significantly induced by TCDD in an AHR2-dependent manner and appears to interact with the 5′ untranslated region of the sox9b gene (Garcia et al., 2017, 2018b). Exposure of slincR morphants to TCDD resulted in altered jaw cartilage structure and reduced incidence of hemorrhaging, suggesting a possible functional role of slincR in both TCDD-induced craniofacial malformations and cardiotoxicity (Garcia et al., 2018b). The study also highlighted several PAHs that induce slincR expression at high levels without causing sox9b repression, indicating that slincR could not only have tissue-specific effects but could also regulate other genes beyond sox9b (Garcia et al., 2018b).
A member of the forkhead box family of transcription factors, originally designated foxq1b but now known as foxq1a, is highly induced by TCDD exposure at a rate faster than cyp1a in developing zebrafish, in an AHR2-dependent manner (Planchart and Mattingly, 2010). In situ hybridization experiments showed that the transcript is expressed in the jaw primordium and is hypothesized to play a role in craniofacial abnormalities (Planchart and Mattingly, 2010). TCDD also induces the paralogous gene foxq1b in zebrafish (Hahn et al., 2014). More work is needed to identify the functional role of the foxq1 paralogs in the TCDD toxicity pathway.
AHR1a
Initial in vitro studies concluded that AHR1a was nonfunctional because it did not bind TCDD and was transcriptionally inactive when expressed in cells together with ARNT2b (Andreasen et al., 2002a). These results are supported by in vivo studies in AHR1a mutant fish, from which it was concluded that AHR1a was not required for TCDD-induced toxicity and Cyp1a activity in zebrafish (Souder and Gorelick, 2019). BNF also does not activate AHR1a, and it was suggested that the zebrafish ahr1a is a possible pseudogene (Karchner et al., 2005). However, more recent in vivo studies demonstrate that AHR1a is functional and can be activated by chemicals including leflunomide (Goodale et al., 2012), the oxy-PAH xanthone (Knecht et al., 2013), several nitro-PAHs like 5-nitroacenaphthalene, 9-nitrophenanthrene, and 7-nitrobenzo[k]fluoranthene (Chlebowski et al., 2017), and the parent PAHs pyrene (Incardona et al., 2006) and chrysene (Incardona et al., 2005). Upon ahr1a knockdown and developmental exposure to each of these chemicals, either a reduction of toxicity (Chlebowski et al., 2017; Incardona et al., 2006) or a reduction of induced Cyp1a protein expression (Chlebowski et al., 2017; Goodale et al., 2012; Incardona et al., 2005; Knecht et al., 2013) was confirmed. Furthermore, AHR1a is the dominant receptor involved in regulating induction of larval hepatic Cyp1a; ahr1a knockdown reduces Cyp1a liver expression induced by pyrene (Incardona et al., 2006); leflunomide (Goodale et al., 2012), xanthone (Knecht et al., 2013), and the nitro-PAHs, 5-nitroacenaphthalene, 9-nitrophenanthrene, and 7-nitrobenzo[k]fluoranthene (Chlebowski et al., 2017).
In contrast to the above-mentioned studies, Garner et al. (2013) found that morpholino knockdown of ahr1a exacerbated the developmental toxicity caused by both PCB-126 and a mixture of PAHs, BkF and fluoranthene. Although ahr1a knockdown did not affect cyp1a, cyp1b1, and cyp1c1 gene expression, Cyp1a protein activity, measured using the ethoxyresorufin-O-deethylase (EROD) assay, increased. From this study, the authors hypothesized that AHR1a likely mimics AHRR and consequently the absence of AHR1a results in excessive AHR2 signaling and enhances the cardiotoxicity measured by pericardial edema (Garner et al., 2013). The study also highlights that AHR1a seems to inhibit Cyp1a protein activity, as ahr1a knockdown led to increased activity. This was in contrast to AHR2, which mediated an increase in Cyp1a activity (Garner et al., 2013). Another study found that the prevalence of mITP-induced cardiotoxicity, but not its severity, increased when all 3 ahrs were knocked down compared with just ahr1b/ahr2 knockdown, suggesting that AHR1a may play a role in mITP-induced cardiotoxicity (Gerlach et al., 2014). However, it is noteworthy that cyp1a transcript expression was not altered by ahr1a knockdown like it was by ahr1b/ahr2 knockdown, indicating that mITP likely does not activate AHR1a (Gerlach et al., 2014). Overall, AHR1a appears to have relevance and ligand-specific functions that are currently enigmatic. Table 5 summarizes the effects mediated by AHR1a in developing zebrafish.
Table 5.
Developmental Toxicity Endpoints and Cyp1a Expression Patterns Mediated by AHR1a From Morpholino Knockdown Studies
| Xenobiotic Ligand |
|
Results of AHR1a Knockdown | References |
|---|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Measured endpoints that were not altered upon AHR1a knockdown.
AHR1b
Zebrafish morpholino studies reveal that AHR1b does not play a role in early life toxicity caused by PCB-126, a PAH mixture of BkF and fluoranthene (Garner et al., 2013), or TCDD (Souder and Gorelick, 2019). Although ahr1b knockdown did not prevent mITP-induced cardiotoxicity in zebrafish, there was a significant decrease in the prevalence of mITP-induced pericardial edema in an AHR2 mutant line injected with the ahr1b morpholino compared with control morpholino-injected AHR2 mutants. This suggests AHR1b’s possible role in mediating mITP-induced cardiotoxicity (Gerlach et al., 2014). Additionally, studies suggest that AHR1b may be involved in not only developmental toxicity but also adult toxicity effects of chemicals like TCDD, some PAHs, and PCB-126 (Garner et al., 2013; Goodale et al., 2012); however, a closer look with histopathology or immunohistochemistry may be necessary to reveal possible subtle effects missed in gross morphology studies. It was also suggested that AHR1b could be functionally redundant with AHR2, but so far, it seems evident that AHR2 has a greater role in regulating the expression of xenobiotic-metabolizing enzymes and mediating toxicity compared with AHR1b (Garner et al., 2013).
Although knockdown studies have not definitively shown a role for AHR1b in the developmental toxicity of xenobiotics, AHR1b appears to be important for leflunomide-induced Cyp1a protein expression in the vasculature, but not in the liver (Goodale et al., 2012). AHR1b may also play a role in Cyp1a protein expression induced by mITP in the vasculature, heart, and liver (Gerlach et al., 2014), and by nitro-PAHs like 7-nitrobenzo[k]fluoranthene in the vasculature, skin, and the neuromasts (Chlebowski et al., 2017). Ahr1b knockdown in mITP-exposed zebrafish also reduced cyp1a mRNA levels (Gerlach et al., 2014). These studies demonstrate that AHR1b can be activated by various chemicals and they concur with earlier studies that showed AHR1b is a fully functional receptor (Karchner et al., 2005) with partially overlapping functions with AHR2. Unlike AHR2 and AHR1a, AHR1b does not appear to mediate Cyp1a expression in any specific tissue. Table 6 summarizes the evidence for AHR1b’s role in developmental toxicity in zebrafish.
Table 6.
Developmental Toxicity Endpoints and Cyp1a Expression Patterns Mediated by AHR1b From Morpholino Knockdown Studies
| Xenobiotic Ligand | Endpoints Mediated by Ligand x AHR1b | Results of AHR1b Knockdown | References |
|---|---|---|---|
| Leflunomide | — Vasculature Cyp1a expression | — Prevention | Goodale et al. (2012) |
| mITP | — Pericardial edema | — Reduction in prevalence of pericardial edema | Gerlach et al. (2014) |
| 7-Nitrobenzo[k]fluoranthene | — Vasculature, skin, neuromast Cyp1a protein expression | — Slight reduction | Chlebowski et al. (2017) |
Overall, AHR2 is predominant in mediating the early stage toxicity of a large variety of ligands. AHR1a and AHR1b can also mediate developmental toxicity albeit to a lesser extent, and this supports the idea that the 3 AHRs have partitioned multiple AHR roles. The 3 AHRs have distinct ligand profiles, and even when different chemicals activate the same receptor, the downstream gene expression and developmental toxicity endpoints can be considerably different, suggesting ligand-specific activation of the AHRs.
INTERACTION BETWEEN AHR AND OTHER PATHWAYS
In addition to the direct AHR-mediated toxicity, the AHRs and AHR-responsive genes can directly or indirectly interact with genes from several different signaling pathways while modulating toxicological responses to a ligand. The developmental zebrafish model provides an ideal platform to study these interactions, as most signaling mechanisms are concurrently and dynamically at play during development. In this section, we focus on studies that have explored the crosstalk between AHR signaling and other pathways using embryonic zebrafish.
AHR and Oxidative Stress
Oxidative stress—the disruption of redox signaling and control (Jones, 2006)—is a well-studied toxicological phenomenon that occurs in response to several classes of chemicals that produce reactive oxygen species (ROS) or disrupt thiol homeostasis (Di Giulio and Hinton, 2008; Sies et al., 2017). AHR-mediated oxidative stress (Di Giulio and Hinton, 2008) occurs through a variety of mechanisms, including stimulation of inflammatory responses and induction of pro-oxidant enzymes such as xanthine oxidase and CYP-dependent monooxygenases, which can release ROS or generate redox-cycling metabolites (Dalton et al., 2002; Reichard et al., 2006). Developmental exposures to AHR ligands such as PAHs (Van Tiem and Di Giulio, 2011), oxy-PAHs (Knecht et al., 2013), heterocyclic and nitro-PAHs (Chlebowski et al., 2017), or PCB-126 (Liu et al., 2016) result in induction of redox-responsive antioxidant genes such as glutathione peroxidase (gpx1), glutamate cysteine ligase (gclc1), and superoxidase dismutase (sod1). In addition, PCB-126 also induces a significant increase in lipid peroxidation, a result of ROS-induced cellular damage (Liu et al., 2016). In fact, AHR activation and antioxidant responses act synchronously in response to toxicant exposures—this was evidenced by the mirroring of the activities of total SOD and Cyp enzymes in whole homogenates of fish exposed to the PAHs, phenanthrene and anthracene (Wang et al., 2018). Taken together, these studies suggest a robust antioxidant response as well as some levels of oxidative damage associated with AHR activation. A number of knockdown studies have also supported these outcomes. For example, ahr2 knockdown blocks the increased expression of gpx1, gclc1, and sod1 by the AHR agonist BkF (Van Tiem and Di Giulio, 2011). Ahr2 knockdown also prevents the induction of ROS and 8-OHdG (8-hydroxy-2′-deoxyguanosine, a marker of oxidative DNA damage) by the chlorinated solvent trichloroethylene (Jin et al., 2020), although wild-type trichloroethylene-exposed embryos do not show an induction of cyp1a1 or ahr2 transcripts. Nevertheless, these studies confirm the specific role of AHR2 in mediating both oxidative damage and antioxidant responses.
A major driver for AHR-induced antioxidant responses is the crosstalk between AHR and their prime regulator, Nrf2 (Baird and Yamamoto, 2020). This mechanism is particularly important for AHR2 ligands such as TCDD, which do not undergo substantial metabolism and hence, redox cycling (Dietrich, 2016). Nrf2 (also called Nfe2l2) is a transcription factor that regulates the expression of a number of antioxidant enzymes such as NAD(P)H: quinone oxidoreductase (NQO1) as well as xenobiotic-metabolizing enzymes such as glutathione-S-transferases (Dietrich, 2016). In mammals, AHR regulates Nrf2 expression and Nrf2 mediates the AHR-dependent induction of several xenobiotic-metabolizing enzymes by TCDD (Miao et al., 2005; Yeager et al., 2009). Zebrafish have 2 Nrf2 genes, nrf2a and nrf2b, both of which contain AHREs within their promoter regions (Timme-Laragy et al., 2012b). A number of chemicals such as TCDD (Hahn et al., 2014; Timme-Laragy et al., 2012b), PAHs (Knecht et al., 2013), and PCBs (Timme-Laragy et al., 2012b, 2015) induce nrf2a or nrf2b mRNA expression at different life stages, in an AHR2-dependent manner (Timme-Laragy et al., 2012b). For example, embryonic exposures to the oxy-PAH 7,12-B[a]AQ result in increased expression of nrf2 and nqo1 in addition to genes associated with the glutathione redox cycle (gst, gpx, and sod families) (Knecht et al., 2013). One study showed that although exposures to PCB-126 in wild-type embryos did not elicit any antioxidant responses, an nrf2a mutant displayed altered both basal expression and PCB-inducibility of certain ahr, nrf2, and gst family genes (Rousseau et al., 2015). Other nrf2-family genes and AHR forms may also be involved in this crosstalk; for example, AHR1b regulates the constitutive and TCDD-inducible expression of nrf2a as well as other members of the nrf gene family, nrf1a and nrf1b (Ulin et al., 2019). These studies suggest that antioxidant responses to TCDD, PCBs, and PAHs may be driven by a combination of oxidative stress and AHR-Nrf2 crosstalk. Indeed, it is well known that the glutathione and Nrf2 pathways are interdependent, and nrf2 knockdown can perturb the glutathione redox state (Sant et al., 2017). In contrast, TCDD, a strong inducer of nrf2 in both zebrafish (Hahn et al., 2014; Ulin et al., 2019), and in mammals (Miao et al., 2005; Yeager et al., 2009), does not induce expression of antioxidant genes such as sod, gst, or nqo1 (Alexeyenko et al., 2010; Hahn et al., 2014). Overall, these studies provide evidence of the complexity of the role of AHR, the glutathione redox state, and the Nrf2 pathway in trying to maintain oxidative homeostasis in response to xenobiotics.
AHR-Wnt Crosstalk and Tissue Regeneration
Although mammals, including humans, have a limited regenerative capacity restricted to some organs such as liver and skin, other vertebrates possess high regenerative capacity of the heart, liver, limbs, etc. (Marques et al., 2019). The process of regeneration involves cellular migration, blastema formation, differentiation, and proliferation that are all regulated by multiple signaling pathways (Akimenko et al., 2003; Santamaria and Becerra, 1991), and external stressors can potentially inhibit regeneration (Mathew et al., 2009). Zebrafish, in particular, has been widely used as a model for studying tissue regeneration following surgical amputation of organs (Akimenko et al., 2003). Exposure to AHR ligands such as TCDD and leflunomide (an anti-inflammatory drug) following fin amputation results in a failure of adult and larval fin regeneration (Andreasen et al., 2007; Mathew et al., 2006; O’Donnell et al., 2010; Zodrow and Tanguay, 2003). Morpholino knockdown of ahr2 and arnt1 results in both unexposed and TCDD-exposed morphants showing normal fin regeneration, indicating that the inhibition of regeneration by TCDD is AHR2 and ARNT1-dependent (Mathew et al., 2006). Following TCDD exposures, adult regenerative tissues also show widespread changes in the transcripts that regulate cellular differentiation, cartilage, collagen, cell growth, tissue regeneration, and extracellular matrix—all important factors involved in tissue regeneration (Andreasen et al., 2007). Specifically, TCDD exposure is associated with both transcriptional activation of R-spondin 1, and a repression of sox9b (a transcription factor regulated by AHR2, as discussed previously) in embryos (Mathew et al., 2007). R-spondin 1 is a Wnt/β catenin signaling gene that contains an AHRE in its promoter region. Morphants resulting from partial suppression of both R-spondin 1 and LRP6, a Wnt coreceptor, show normal caudal fin regeneration following TCDD exposure, demonstrating that activation of these Wnt signaling genes is required for TCDD to inhibit regeneration (Mathew et al., 2007). This result is also supported by the induction of a number of other Wnt/β-catenin signaling genes by TCDD in regenerating tissues. In conjunction with transcriptional activation of R-spondin 1, the expression of sox9b is repressed within regenerating fin tissues after TCDD exposure (Mathew et al., 2007). Interestingly, although sox9b morphants show some levels of regeneration of caudal fins, the regenerative tissue still possesses defective structures, indicating that this process is not completely dependent on sox9b (Mathew et al., 2007). In humans, SOX9 is also a Wnt target gene and is directly regulated by R-spondin 1 (Yano et al., 2005). Furthermore, SOX9 also inhibits expression of β catenin-associated genes and promotes degradation of β catenin (Yano et al., 2005). Therefore, it is likely that the inverse expression patterns between R-spondin 1 and sox9b observed within regenerative fin tissues result from a crosstalk between the AHR and Wnt signaling mechanisms to regulate tissue regeneration. In addition to the proposed AHR-Wnt crosstalk, other AHR-mediated mechanisms can govern tissue regeneration, depending on the tissue type. For example, 1 study showed that TCDD exposures of adult zebrafish with partially amputated hearts led to an inhibition of regeneration of myocardial tissues, but there was no impact on sox9b, R-spondin 1, or other Wnt signaling genes although, as seen with caudal fin amputation, expression of genes associated with tissue regeneration and extracellular matrix was altered (Hofsteen et al., 2013a). The lack of change in transcript levels of sox9b and fin tissue regeneration, while largely governed by similar molecular factors, have some differences; for example, although fin regeneration is coregulated by Wnt signaling, myocardial regeneration is coregulated by TGF-β and NF-κβ pathways (Sehring et al., 2016). Despite these differences, the studies show that only the chemical activation of AHR inhibits tissue regeneration.
Estrogen Receptor
Both mammalian and zebrafish studies show clear evidence of crosstalk between AHR and estrogen receptor (ER). In mammals, AHR interactions with estrogen signaling pathways have long been known to occur through a variety of mechanisms (Safe and Wormke, 2003; Swedenborg and Pongratz, 2010), including the role of AHR as an E3 ubiquitin ligase controlling proteasomal degradation of ER (Ohtake et al.,, 2003, 2007; Wormke et al., 2003). It is not known whether fish AHRs can act in this way, but there is other evidence for AHR-ER crosstalk. In zebrafish, Cyp3c can be induced by both AHR and ER ligands, suggesting that there may be a crosstalk between these 2 receptor mechanisms in regulation of CYP3 (Shaya et al., 2019). The direct interaction between AHR and ER pathways has been shown in other studies where the transcriptional induction of ER-target cyp19b or vitellogenin by ER ligands 17α-ethynylestradiol and 17ß-estradiol was reversed by TCDD (Bugel et al., 2013; Cheshenko et al., 2007). In addition, this effect was partially blocked by ANF, an AHR antagonist (Cheshenko et al., 2007). Interestingly, a chemically induced pan-ER inhibition does not block TCDD-induced, AHR2-mediated cardiotoxicity, suggesting that AHR2 is not a constitutive partner of ER (Souder and Gorelick, 2019). However, these authors also conclude that an AHR-ER crosstalk may be tissue dependent. This was supported by another study, where 17β-estradiol increased ahr1a mRNA expression only in the 4-day-old zebrafish brain, but not in other organs (Hao et al., 2013). Likewise, an adult zebrafish study showed that TCDD inhibited levels of the genes regulating the ER as well as estrogen synthesis and follicular development in the zebrafish ovary (King-Heiden et al., 2008), highlighting the role of the AHR-ER crosstalk in the reproductive system (discussed later). Therefore, interactions between AHR and ER in zebrafish is likely complicated and highly dependent on the specific target tissues.
Pregnane X Receptor
AHR displays some levels of crosstalk with the transcription factor PXR, which regulates a number of CYP2 and CYP3 enzymes and is involved in detoxification of an array of xenobiotics, primarily steroids. Embryonic zebrafish exposures to either pregnenolone (a PXR agonist) or PCB-126 (an AHR agonist) result in the increased transcript levels of pxr, ahr2, cyp1a as well as a number of cyp2 and cyp3 genes (Kubota et al., 2015). Furthermore, knockdown of ahr2 reverses the PCB-126-induced transcriptional activation of cyp1a and pxr, as well as cyp2 and cyp3 genes. Taken together, these studies suggest an inevitable crosstalk between AHR and PXR in regulation of cyp2 and cyp3 genes.
Fibroblast Growth Factor
Studies have explored the interaction between AHR and the fibroblast growth factor (FGF) pathway in developmental processes. In mammals, the FGF pathway gene, fgf21, is a known AHR target gene (Cheng et al., 2014). In zebrafish, similar to AHR, the FGF pathway is known to independently regulate tissue regeneration. However, a study comparing fin regeneration after exposure to TCDD as well as an FGF pathway inhibitor, SU5042, showed that, although both AHR activation and FGF inhibition lead to inhibition of fin regeneration, the phenotypes were morphometrically different and there was no evidence of interaction between the 2 pathways in the regenerative process (Mathew et al., 2006).
In summary, we note that much of the work concerning crosstalk with other signaling pathways has focused on AHR2. Although it is evident that the AHR2 signaling pathway interacts with several other transcription factors and signaling pathways, the interactions are highly complex, and we have only begun to understand them in zebrafish.
ADULT TOXICITY, EPIGENETICS, AND MULTIGENERATIONAL EFFECTS
Compared with the many studies assessing the role of AHRs in during development, only a limited number have investigated the role of the zebrafish AHRs in both post-developmental physiology in juveniles and adults, and across generations. To the best of our knowledge, AHR1a- or AHR1b-specific adult and multigenerational toxicity after exposure to xenobiotic chemicals has not been investigated. In this section, we first review the functional roles of AHR2 in TCDD-induced adult toxicity of the reproductive and musculoskeletal systems, and then describe what is known about the epigenetic effects of the ligands TCDD, PCB-126, and BaP, whose toxicity endpoints are mediated primarily by AHR2.
Adult Toxicity
Reproductive System
AHR plays a modest constitutive role in the reproductive system; a study with AHR mutants showed altered follicular development in ovaries of the AHR2-null zebrafish compared with wild-type zebrafish (Garcia et al., 2018a). However, more profound effects on reproductive organs have been shown to be triggered by xenobiotic activation of AHR2 by TCDD (King-Heiden et al., 2012) and these impacts are expected due to the crosstalk between AHR and ER as described in the previous section. Indeed, dietary TCDD exposure reduced mRNA levels of genes regulating the ER and follicular development in adult zebrafish ovaries, while inducing expression of cyp1a (King-Heiden et al., 2008). Additionally, zebrafish exposed to TCDD at 3- and 7-week postfertilization displayed reduced fecundity and reduced percentage of fertilized eggs (Baker et al., 2013). Paired spawning also showed that these impacts were independent of the sex of the fish. Although female fish displayed abnormalities in ovarian structures, the testes of TCDD-exposed male fish displayed decreased spermatozoa with increase in spermatogonia, decreased germinal epithelial thickness, and altered responses in genes regulating testis development and steroidogenesis (Baker et al., 2016). Interestingly, TCDD-exposed fish also experienced a prevalent shift toward feminization, but a significant percentage of female fish possessed male gonads (Baker et al., 2013). From these studies, it is evident that both AHR knockout (Garcia et al., 2018a) and its activation (other studies) result in reproductive deficiencies, suggesting that any disruption to the normal AHR signaling mechanism can have deleterious effects on reproductive physiology. These effects on the reproductive system may also be mediated through AHR-associated epigenetic mechanisms, discussed in Epigenetics and Multigenerational Effects. Taken together, these studies unequivocally highlight the significant role of AHR in reproductive development, sex determination, and reproductive functions.
Musculoskeletal System
TCDD exposures have also been shown to affect musculoskeletal development in adults, with zebrafish exposed to TCDD at 3- and 7-week postfertilization displaying skeletal deficits during adulthood, including skeletal kinks, shortened jaw structures, and abnormal operculum and bone structures (Baker et al., 2013). Interestingly, AHR2-null fish also showed similar deficits in skeleton and fins, including defective fins, abnormal dentary, operculum and frontal structures, smaller orbital and supraorbital bones (Baker et al., 2014b; Garcia et al., 2018a; Goodale et al., 2012; Souder and Gorelick, 2019). These identical responses to both AHR2 deficiency and AHR2 activation mimic the equivocality of responses on reproductive development and highlight the need for more detailed studies on the role of AHR2 in musculoskeletal development. They also reiterate the sensitive nature of the AHR2 signaling pathway, where any alteration—either its deficiency or activation—can have profound impacts on the musculoskeletal system.
Epigenetics and Multigenerational Effects
“TCDD” exposure results in several transgenerational effects (Baker et al., 2014a, 2014c; King-Heiden et al., 2005; Meyer et al., 2018) which have been reviewed recently (Viluksela and Pohjanvirta, 2019). It has been hypothesized that epigenetic mechanisms mediate these effects of TCDD, although specific epigenetic modifications have not been identified as having a causal role in the responses observed in F1 and F2 generations (Baker et al., 2014a). There is increasing evidence in other organisms showing how epigenetic modifications are related to AHR signaling and TCDD toxicity (Patrizi and Siciliani de Cumis, 2018; Viluksela and Pohjanvirta, 2019). The expression of cyp1a1 and cyp1b1 in the F1 generation of a TCDD-exposed zebrafish lineage was significantly higher than in control-lineage F1 animals, suggesting an AHR role (Olsvik et al., 2014). Within the genome region queried, there was no effect of TCDD on the methylation pattern of the ahr2 promoter in developing zebrafish (Aluru et al., 2015). However, the study found altered promoter methylation of AHR target genes, ahrra and c-fos. TCDD exposure also altered dnmt expression in the F0 generation, and in vitro transactivation studies identified that 3 of the 5 tested dnmt promoters caused transactivation of luciferase reporter by AHR2/ARNT2 in the presence of TCDD (Aluru et al., 2015). A recent study that examined genome-wide changes in DNA methylation in adult testes of zebrafish exposed to TCDD found differential methylation of genes involved in reproductive and epigenetic processes (Akemann et al., 2018) and some of these histological and transcriptomic effects persisted in subsequent F1 and F2 generations (Meyer et al., 2018), suggesting a methylation-dependent transfer of biomarkers across generations. “PCB-126” exposure of adult zebrafish caused extensive alteration in genome-wide DNA methylation patterns in liver and brain, which were not strongly correlated with altered gene expression, suggesting a complex relationship between DNA methylation and gene regulation (Aluru et al., 2018). Taken together, these studies suggest the potential role of methylation-dependent epigenetics in driving TCDD- and PCB-126-induced reproductive outcomes.
“BaP” exposure leads to transgenerational effects including alterations to locomotor activity, decreased heartbeat and mitochondrial function, reduced hatch rate, reduced egg production and offspring survival, increased mortality and incidence of malformations up to the F2 generation (Corrales et al., 2014a,b; Fang et al., 2013; Knecht et al., 2017a). These effects were likely a result of global hypomethylation in conjunction with alterations of expression of developmental and cancer-related genes in BaP-exposed F0 zebrafish (Corrales et al., 2014a; Fang et al., 2013; Knecht et al., 2017a). Dnmt expression was generally reduced with BaP exposure, an effect that was further strengthened by AHR2 knockdown (Knecht et al., 2017a). This was unexpected and may have been due to a mechanism different from AHR2/ARNT acting via AHREs in the dnmt promoters. Another study found that, following BaP exposure, dnmt1 and dnmt3a had increased mRNA expression in 96-hpf larvae and in adult brains; however, the role of the AHRs in mediating the altered expression was not investigated (Gao et al., 2017).
In summary, the functional role of AHR2 in mediating adult toxicity and epigenetic perturbation is in the early stages of investigation and more studies are being conducted across several labs to better understand these mechanisms.
CONCLUSIONS AND FUTURE DIRECTIONS
The studies conducted so far have demonstrated the vast diversity of both the endogenous and toxicological functional roles of the zebrafish AHRs. Research in zebrafish (and other fish models) has enhanced our understanding of AHR biology in all its richness, including endogenous and toxicological AHR roles. This research has complemented studies in other animal models, and in particular has contributed to knowledge about AHR functions during vertebrate development. Additionally, research on the zebrafish AHRs has explored and revealed the heterogeneity of ligands that are able to bind to each of the 3 receptors. The majority of the research so far on all aspects of functionality has focused on AHR2; future work concentrating on how AHR1a and AHR1b contribute to normal physiology and toxicity of xenobiotics will not only inform us of their roles, but could also reveal unknown functions of the AHR, many of which are conserved across vertebrates. Further, understanding the downstream signaling events upon AHR activation has centered on TCDD as a ligand. Exploration of the functions of AHR-regulated genes upon exposure to other chemicals will facilitate a gene biomarker approach to further characterize and classify xenobiotics that act via AHR. With the remarkable diversity of AHR ligands, the wide-ranging downstream AHR-regulated genes, and the crosstalk interactions with other signaling pathways, it is clear we must discern how the activation of the AHRs by its various ligands can differentially modulate signaling pathways that dictate biological outcomes.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ACKNOWLEDGMENTS
We would like to thank Dr Michael Simonich (Tanguay Sinnhuber Aquatic Research Laboratory) for help editing the review and Dr Sibel Karchner (Woods Hole Oceanographic Institution) for advice on the synteny analysis.
FUNDING
National Institute of Environmental Health Sciences (NIEHS) through the Oregon State University (P42 ES016465) and Boston University (P42 ES007381) Superfund Research Programs; the Woods Hole Center for Oceans and Human Health (NIEHS Grant P01 ES028938 and National Science Foundation Grant OCE-1840381). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- Abel J., Haarmann-Stemmann T. (2010). An introduction to the molecular basics of aryl hydrocarbon receptor biology. Biol. Chem. 391, 1235–1248. [DOI] [PubMed] [Google Scholar]
- Akemann C., Meyer D. N., Gurdziel K., Baker T. R. (2018). Developmental dioxin exposure alters the methylome of adult male zebrafish gonads. Front. Genet. 9, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimenko M. A., Mari-Beffa M., Becerra J., Geraudie J. (2003). Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 226, 190–201. [DOI] [PubMed] [Google Scholar]
- Alexeyenko A., Wassenberg D. M., Lobenhofer E. K., Yen J., Linney E., Sonnhammer E. L., Meyer J. N. (2010). Dynamic zebrafish interactome reveals transcriptional mechanisms of dioxin toxicity. PLoS One 5, e10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru N., Karchner S. I. 2020. PCB126 exposure revealed alterations in m6A RNA modifications in transcripts associated with AHR activation. Toxicol. Sci. [DOI] [PMC free article] [PubMed]
- Aluru N., Jenny M. J., Hahn M. E. (2014). Knockdown of a zebrafish aryl hydrocarbon receptor repressor (AHRRA) affects expression of genes related to photoreceptor development and hematopoiesis. Toxicol. Sci. 139, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru N., Karchner S. I., Krick K. S., Zhu W., Liu J. (2018). Role of DNA methylation in altered gene expression patterns in adult zebrafish (Danio rerio) exposed to 3, 3', 4, 4', 5-pentachlorobiphenyl (PCB 126). Environ. Epigenet. 4, dvy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru N., Kuo E., Helfrich L. W., Karchner S. I., Linney E. A., Pais J. E., Franks D. G. (2015). Developmental exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase (dnmt) expression in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 284, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A. (1998). Zebrafish hox Clusters and Vertebrate Genome Evolution. Science 282, 1711–1714. 10.1126/science.282.5394.1711 [DOI] [PubMed] [Google Scholar]
- Anderson J. L., Mulligan T. S., Shen M. C., Wang H., Scahill C. M., Tan F. J., Du S. J., Busch-Nentwich E. M., Farber S. A. (2017). mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 13, e1007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen E. A., Hahn M. E., Heideman W., Peterson R. E., Tanguay R. L. (2002. a). The zebrafish (Danio rerio) aryl hydrocarbon receptor type 1 is a novel vertebrate receptor. Mol. Pharmacol. 62, 234–249. [DOI] [PubMed] [Google Scholar]
- Andreasen E. A., Mathew L. K., Lohr C. V., Hasson R., Tanguay R. L. (2007). Aryl hydrocarbon receptor activation impairs extracellular matrix remodeling during zebra fish fin regeneration. Toxicol. Sci. 95, 215–226. [DOI] [PubMed] [Google Scholar]
- Andreasen E. A., Spitsbergen J. M., Tanguay R. L., Stegeman J. J., Heideman W., Peterson R. E. (2002. b). Tissue-specific expression of AHR2, ARNT2, and CYP1A in zebrafish embryos and larvae: effects of developmental stage and 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure. Toxicol. Sci. 68, 403–419. [DOI] [PubMed] [Google Scholar]
- Antkiewicz D. S., Peterson R. E., Heideman W. (2006). Blocking expression of ahr2 and arnt1 in zebrafish larvae protects against cardiac toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 94, 175–182. [DOI] [PubMed] [Google Scholar]
- Baird L. Yamamoto M. (2020). The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell Biol. 40, e00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B. B., Yee J. S., Meyer D. N., Yang D., Baker T. R. (2016). Histological and transcriptomic changes in male zebrafish testes due to early life exposure to low level 2,3,7,8-tetrachlorodibenzo-p-dioxin. Zebrafish 13, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. R., King-Heiden T. C., Peterson R. E., Heideman W. (2014. a). Dioxin induction of transgenerational inheritance of disease in zebrafish. Mol. Cell. Endocrinol. 398, 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. R., Peterson R. E., Heideman W. (2013). Early dioxin exposure causes toxic effects in adult zebrafish. Toxicol. Sci. 135, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. R., Peterson R. E., Heideman W. (2014. b). Adverse effects in adulthood resulting from low-level dioxin exposure in juvenile zebrafish. Endocr. Disruptors (Austin) 2, e28309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. R., Peterson R. E., Heideman W. (2014. c). Using zebrafish as a model system for studying the transgenerational effects of dioxin. Toxicol. Sci. 138, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W. B., Korf I., Kadavi C., Heyen J., Tate S., Wun E., Bedell J. A., McPherson J. D., Johnson S. L. (2000). The syntenic relationship of the zebrafish and human genomes. Genome Res. 10, 1351–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R., Coumoul X., Fernandez-Salguero P. M. (2007). The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 581, 3608–3615. [DOI] [PubMed] [Google Scholar]
- Beischlag T. V., Luis Morales J., Hollingshead B. D., Perdew G. H. (2008). The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 18, 207–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard S., Timme-Laragy A. R., Wassenberg D. M., Cockman C., Di Giulio R. T. (2006). The role of the aryl hydrocarbon receptor pathway in mediating synergistic developmental toxicity of polycyclic aromatic hydrocarbons to zebrafish. Toxicol. Sci. 92, 526–536. [DOI] [PubMed] [Google Scholar]
- Billiard S. M., Querbach K., Hodson P. V. (1999). Toxicity of retene to early life stages of two freshwater fish species. Environ. Toxicol. Chem. 18, 2070–2077. [Google Scholar]
- Bisson W. H., Koch D. C., O’Donnell E. F., Khalil S. M., Kerkvliet N. I., Tanguay R. L., Abagyan R., Kolluri S. K. (2009). Modeling of the aryl hydrocarbon receptor (AhR) ligand binding domain and its utility in virtual ligand screening to predict new AhR ligands. J. Med. Chem. 52, 5635–5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. R., Clark B. W., Garner L. V., Di Giulio R. T. (2015). Zebrafish cardiotoxicity: the effects of CYP1A inhibition and AHR2 knockdown following exposure to weak aryl hydrocarbon receptor agonists. Environ. Sci. Pollut. Res. Int. 22, 8329–8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugel S. M., White L. A., Cooper K. R. (2013). Inhibition of vitellogenin gene induction by 2,3,7,8-tetrachlorodibenzo-p-dioxin is mediated by aryl hydrocarbon receptor 2 (AHR2) in zebrafish (Danio rerio). Aquat. Toxicol. 126, 1–8. [DOI] [PubMed] [Google Scholar]
- Butler R. A., Kelley M. L., Powell W. H., Hahn M. E., Van Beneden R. J. (2001). An aryl hydrocarbon receptor (AHR) homologue from the soft-shell clam, Mya arenaria: Evidence that invertebrate AHR homologues lack 2,3,7,8-tetrachlorodibenzo-p-dioxin and beta-naphthoflavone binding. Gene 278, 223–234. [DOI] [PubMed] [Google Scholar]
- Carney S. A., Chen J., Burns C. G., Xiong K. M., Peterson R. E., Heideman W. (2006. a). Aryl hydrocarbon receptor activation produces heart-specific transcriptional and toxic responses in developing zebrafish. Mol. Pharmacol. 70, 549–561. [DOI] [PubMed] [Google Scholar]
- Carney S. A., Peterson R. E., Heideman W. (2004). 2,3,7,8-tetrachlorodibenzo-p-dioxin activation of the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator pathway causes developmental toxicity through a CYP1A-independent mechanism in zebrafish. Mol. Pharmacol. 66, 512–521. [DOI] [PubMed] [Google Scholar]
- Carney S. A., Prasch A. L., Heideman W., Peterson R. E. (2006. b). Understanding dioxin developmental toxicity using the zebrafish model. Birth Defects Res. A Clin. Mol. Teratol. 76, 7–18. [DOI] [PubMed] [Google Scholar]
- Carver L. A., Bradfield C. A. (1997). Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 272, 11452–11456. [DOI] [PubMed] [Google Scholar]
- Chen J., Carney S. A., Peterson R. E., Heideman W. (2008). Comparative genomics identifies genes mediating cardiotoxicity in the embryonic zebrafish heart. Physiol. Genomics 33, 148–158. [DOI] [PubMed] [Google Scholar]
- Cheng X., Vispute S. G., Liu J., Cheng C., Kharitonenkov A., Klaassen C. D. (2014). Fibroblast growth factor (Fgf) 21 is a novel target gene of the aryl hydrocarbon receptor (AhR). Toxicol. Appl. Pharmacol. 278, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshenko K., Brion F., Le Page Y., Hinfray N., Pakdel F., Kah O., Segner H., Eggen R. I. (2007). Expression of zebra fish aromatase CYP19A and CYP19B genes in response to the ligands of estrogen receptor and aryl hydrocarbon receptor. Toxicol. Sci. 96, 255–267. [DOI] [PubMed] [Google Scholar]
- Chlebowski A. C., Garcia G. R., La Du J. K., Bisson W. H., Truong L., Massey Simonich S. L., Tanguay R. L. (2017). Mechanistic investigations into the developmental toxicity of nitrated and heterocyclic PAHs. Toxicol. Sci. 157, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales J., Fang X., Thornton C., Mei W., Barbazuk W. B., Duke M., Scheffler B. E., Willett K. L. (2014. a). Effects on specific promoter DNA methylation in zebrafish embryos and larvae following benzo[a]pyrene exposure. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 163, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales J., Thornton C., White M., Willett K. L. (2014. b). Multigenerational effects of benzo[a]pyrene exposure on survival and developmental deformities in zebrafish larvae. Aquat. Toxicol. 148, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture L. A., Abbott B. D., Birnbaum L. S. (1990). A critical review of the developmental toxicity and teratogenicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin: recent advances toward understanding the mechanism. Teratology 42, 619–627. [DOI] [PubMed] [Google Scholar]
- Cunha V., Vogs C., Le Bihanic F., Dreij K. (2020). Mixture effects of oxygenated PAHs and benzo [a] pyrene on cardiovascular development and function in zebrafish embryos. Environ. Int. 143, 105913. [DOI] [PubMed] [Google Scholar]
- Dalton T. P., Puga A., Shertzer H. G. (2002). Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chem. Biol. Interact. 141, 77–95. [DOI] [PubMed] [Google Scholar]
- Denison M. S., Nagy S. R. (2003). Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43, 309–334. [DOI] [PubMed] [Google Scholar]
- Di Giulio R. T., Hinton D. E. (2008). The Toxicology of Fishes. CRC Press, Boca Raton, FL. [Google Scholar]
- Diekmann H., Hill A. (2013). Admetox in zebrafish. Drug Discov. Today Dis. Models 10, e31–e35. [Google Scholar]
- Dietrich C. (2016). Antioxidant functions of the aryl hydrocarbon receptor. Stem Cells Int. 2016, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W., Teraoka H., Tsujimoto Y., Stegeman J. J., Hiraga T. (2004). Role of aryl hydrocarbon receptor in mesencephalic circulation failure and apoptosis in zebrafish embryos exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 77, 109–116. [DOI] [PubMed] [Google Scholar]
- El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Günther S., Fukuda N., Kikhi K., Boezio G. L. M., Takacs C. M., Lai S.-L., et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T. R., Crawford B. D. (2016). Experimental dissection of metalloproteinase inhibition-mediated and toxic effects of phenanthroline on zebrafish development. Int. J. Mol. Sci. 17, 1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C., Rannug A., Stockinger B. (2009). The aryl hydrocarbon receptor in immunity. Trends Immunol. 30, 447–454. [DOI] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Allan L. L., Pollenz R. S., Tanguay R. L., Jenny M. J., Sherr D. H., Hahn M. E. (2008). Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol. Pharmacol. 73, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans B. R., Karchner S. I., Franks D. G., Hahn M. E. (2005). Duplicate aryl hydrocarbon receptor repressor genes (ahrr1 and ahrr2) in the zebrafish Danio rerio: structure, function, evolution, and AHR-dependent regulation in vivo. Arch. Biochem. Biophys. 441, 151–167. [DOI] [PubMed] [Google Scholar]
- Fang M., Guo J., Chen D., Li A., Hinton D. E., Dong W. (2016). Halogenated carbazoles induce cardiotoxicity in developing zebrafish (Danio rerio) embryos. Environ. Toxicol. Chem. 35, 2523–2529. [DOI] [PubMed] [Google Scholar]
- Fang X., Corrales J., Thornton C., Clerk T., Scheffler B. E., Willett K. L. (2015). Transcriptomic changes in zebrafish embryos and larvae following benzo[a]pyrene exposure. Toxicol. Sci. 146, 395–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Thornton C., Scheffler B. E., Willett K. L. (2013). Benzo[a]pyrene decreases global and gene specific DNA methylation during zebrafish development. Environ. Toxicol. Pharmacol. 36, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W. M. (1970). Distinguishing homologous from analogous proteins. Syst. Zool. 19, 99–113. [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F. B., Amores A., Yan Y. L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraccalvieri D., Soshilov A. A., Karchner S. I., Franks D. G., Pandini A., Bonati L., Hahn M. E., Denison M. S. (2013). Comparative analysis of homology models of the Ah receptor ligand binding domain: verification of structure-function predictions by site-directed mutagenesis of a nonfunctional receptor. Biochemistry 52, 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wang C., Xi Z., Zhou Y., Wang Y., Zuo Z. (2017). Early-life benzo[a]pyrene exposure causes neurodegenerative syndromes in adult zebrafish (Danio rerio) and the mechanism involved. Toxicol. Sci. 157, 74–84. [DOI] [PubMed] [Google Scholar]
- Garcia G. R., Bugel S. M., Truong L., Spagnoli S., Tanguay R. L. (2018. a). Ahr2 required for normal behavioral responses and proper development of the skeletal and reproductive systems in zebrafish. PLoS One 13, e0193484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. R., Goodale B. C., Wiley M. W., La Du J. K., Hendrix D. A., Tanguay R. L. (2017). In vivo characterization of an AHR-dependent long noncoding RNA required for proper sox9b expression. Mol. Pharmacol. 91, 609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. R., Noyes P. D., Tanguay R. L. (2016). Advancements in zebrafish applications for 21st century toxicology. Pharmacol. Ther. 161, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia G. R., Shankar P., Dunham C. L., Garcia A., La Du J. K., Truong L., Tilton S. C., Tanguay R. L. (2018. b). Signaling events downstream of AHR activation that contribute to toxic responses: the functional role of an AHR-dependent long noncoding RNA (slincR) using the zebrafish model. Environ. Health Perspect. 126, 117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland M. A., Geier M. C., Bugel S. M., Shankar P., Dunham C. L., Brown J. M., Tilton S. C., Tanguay R. L. (2020). Aryl hydrocarbon receptor mediates larval zebrafish fin duplication following exposure to benzofluoranthenes. Toxicol. Sci. 176, 46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner L. V., Di Giulio R. T. (2012). Glutathione transferase pi class 2 (GSTp2) protects against the cardiac deformities caused by exposure to PAHs but not PCB-126 in zebrafish embryos. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 155, 573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner L. V., Brown D. R., Di Giulio R. T. (2013). Knockdown of AHR1A but not AHR1B exacerbates PAH and PCB-126 toxicity in zebrafish (Danio rerio) embryos. Aquat. Toxicol. 142–143, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M. C., Chlebowski A. C., Truong L., Massey Simonich S. L., Anderson K. A., Tanguay R. L. (2018. a). Comparative developmental toxicity of a comprehensive suite of polycyclic aromatic hydrocarbons. Arch. Toxicol. 92, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geier M. C., James Minick D., Truong L., Tilton S., Pande P., Anderson K. A., Teeguardan J., Tanguay R. L. (2018. b). Systematic developmental neurotoxicity assessment of a representative PAH superfund mixture using zebrafish. Toxicol. Appl. Pharmacol. 354, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C. V., Das S. R., Volz D. C., Bisson W. H., Kolluri S. K., Tanguay R. L. (2014). Mono-substituted isopropylated triaryl phosphate, a major component of Firemaster 550, is an AHR agonist that exhibits AHR-independent cardiotoxicity in zebrafish. Aquat. Toxicol. 154, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasauer S. M., Neuhauss S. C. (2014). Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289, 1045–1060. [DOI] [PubMed] [Google Scholar]
- Goldstone J., McArthur A., Kubota A., Zanette J., Parente T., Jonsson M., Nelson D., Stegeman J. (2010). Identification and developmental expression of the full complement of cytochrome p450 genes in zebrafish. BMC Genomics 11, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone J. V., Jonsson M. E., Behrendt L., Woodin B. R., Jenny M. J., Nelson D. R., Stegeman J. J. (2009). Cytochrome P450 1D1: A novel CYP1A-related gene that is not transcriptionally activated by PCB126 or TCDD. Arch. Biochem. Biophys. 482, 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale B. C., La Du J., Tilton S. C., Sullivan C. M., Bisson W. H., Waters K. M., Tanguay R. L. (2015). Ligand-specific transcriptional mechanisms underlie aryl hydrocarbon receptor-mediated developmental toxicity of oxygenated PAHs. Toxicol. Sci. 147, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodale B. C., La Du J. K., Bisson W. H., Janszen D. B., Waters K. M., Tanguay R. L. (2012). AHR2 mutant reveals functional diversity of aryl hydrocarbon receptors in zebrafish. PLoS One 7, e29346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes A. C., Erwin K. N., Stadt H. A., Hunter G. L., Gefroh H. A., Tsai H. J., Kirby M. L. (2008). PCB126 exposure disrupts zebrafish ventricular and branchial but not early neural crest development. Toxicol. Sci. 106, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. E. (2002). Aryl hydrocarbon receptors: diversity and evolution. Chem. Biol. Interact. 141, 131–160. [DOI] [PubMed] [Google Scholar]
- Hahn M. E., Karchner S. I., Merson R. R. (2017). Diversity as opportunity: Insights from 600 million years of AHR evolution. Curr. Opin. Toxicol. 2, 58–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. E., Karchner S. I., Evans B. R., Franks D. G., Merson R. R., Lapseritis J. M. (2006). Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: Insights from comparative genomics. J. Exp. Zool. A Comp. Exp. Biol. 305, 693–706. [DOI] [PubMed] [Google Scholar]
- Hahn M. E., McArthur A. G., Karchner S. I., Franks D. G., Jenny M. J., Timme-Laragy A. R., Stegeman J. J., Woodin B. R., Cipriano M. J., Linney E. (2014). The transcriptional response to oxidative stress during vertebrate development: Effects of tert-butylhydroquinone and 2,3,7,8-tetrachlorodibenzo-p-dioxin. PLoS One 9, e113158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley-Goldstone H. M., Grow M. W., Stegeman J. J. (2005). Cardiovascular gene expression profiles of dioxin exposure in zebrafish embryos. Toxicol. Sci. 85, 683–693. [DOI] [PubMed] [Google Scholar]
- Hankinson O. (1995). The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35, 307–340. [DOI] [PubMed] [Google Scholar]
- Hao R., Bondesson M., Singh A. V., Riu A., McCollum C. W., Knudsen T. B., Gorelick D. A., Gustafsson J. A. (2013). Identification of estrogen target genes during zebrafish embryonic development through transcriptomic analysis. PLoS One 8, e79020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawliczek A., Nota B., Cenijn P., Kamstra J., Pieterse B., Winter R., Winkens K., Hollert H., Segner H., Legler J. (2012). Developmental toxicity and endocrine disrupting potency of 4-azapyrene, benzo[b]fluorene and retene in the zebrafish Danio rerio. Reprod. Toxicol. 33, 213–223. [DOI] [PubMed] [Google Scholar]
- Heasman J. (2002). Morpholino oligos: Making sense of antisense? Dev. Biol. 243, 209–214. [DOI] [PubMed] [Google Scholar]
- Henry T. R., Spitsbergen J. M., Hornung M. W., Abnet C. C., Peterson R. E. (1997). Early life stage toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 142, 56–68. [DOI] [PubMed] [Google Scholar]
- Hoffman E. C., Reyes H., Chu F. F., Sander F., Conley L. H., Brooks B. A., Hankinson O. (1991). Cloning of a factor required for activity of the Ah (dioxin) receptor. Science 252, 954–958. [DOI] [PubMed] [Google Scholar]
- Hofsteen P., Mehta V., Kim M.-S., Peterson R. E., Heideman W. (2013. a). TCDD inhibits heart regeneration in adult zebrafish. Toxicol. Sci. 132, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofsteen P., Plavicki J., Johnson S. D., Peterson R. E., Heideman W. (2013. b). Sox9b is required for epicardium formation and plays a role in TCDD-induced heart malformation in zebrafish. Mol. Pharmacol. 84, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Wang C., Zhang Y., Li J., Zhong Y., Zhou Y., Chen Y., Zuo Z. (2012). Benzo[a]pyrene exposure influences the cardiac development and the expression of cardiovascular relative genes in zebrafish (Danio rerio) embryos. Chemosphere 87, 369–375. [DOI] [PubMed] [Google Scholar]
- Incardona J. P., Carls M. G., Teraoka H., Sloan C. A., Collier T. K., Scholz N. L. (2005). Aryl hydrocarbon receptor-independent toxicity of weathered crude oil during fish development. Environ. Health Perspect. 113, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J. P., Day H. L., Collier T. K., Scholz N. L. (2006). Developmental toxicity of 4-ring polycyclic aromatic hydrocarbons in zebrafish is differentially dependent on Ah receptor isoforms and hepatic cytochrome P4501A metabolism. Toxicol. Appl. Pharmacol. 217, 308–321. [DOI] [PubMed] [Google Scholar]
- Incardona J. P., Linbo T. L., Scholz N. L. (2011). Cardiac toxicity of 5-ring polycyclic aromatic hydrocarbons is differentially dependent on the aryl hydrocarbon receptor 2 isoform during zebrafish development. Toxicol. Appl. Pharmacol. 257, 242–249. [DOI] [PubMed] [Google Scholar]
- Ishikawa T.-O., Griffin K. J., Banerjee U., Herschman H. R. (2007). The zebrafish genome contains two inducible, functional cyclooxygenase-2 genes. Biochem Bioph Res Co 352, 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D. P., Joshi A. D., Elferink C. J. (2015). Ah receptor pathway intricacies; signaling through diverse protein partners and DNA-motifs. Toxicol. Res. (Camb.) 4, 1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasundara N., Van Tiem Garner L., Meyer J. N., Erwin K. N., Di Giulio R. T. (2015). AHR2-mediated transcriptomic responses underlying the synergistic cardiac developmental toxicity of PAHs. Toxicol. Sci. 143, 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M. J., Aluru N., Hahn M. E. (2012). Effects of short-term exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin on microRNA expression in zebrafish embryos. Toxicol. Appl. Pharmacol. 264, 262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M. J., Karchner S. I., Franks D. G., Woodin B. R., Stegeman J. J., Hahn M. E. (2009). Distinct roles of two zebrafish AHR repressors (AHRRa and AHRRb) in embryonic development and regulating the response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 110, 426–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Ji C., Ren F., Aniagu S., Tong J., Jiang Y., Chen T. (2020). AHR-mediated oxidative stress contributes to the cardiac developmental toxicity of trichloroethylene in zebrafish embryos. J. Hazard. Mater. 385, 121521. [DOI] [PubMed] [Google Scholar]
- Jones D. P. (2006). Redefining oxidative stress. Antioxid. Redox Signal. 8, 1865–1879. [DOI] [PubMed] [Google Scholar]
- Jonsson M. E., Franks D. G., Woodin B. R., Jenny M. J., Garrick R. A., Behrendt L., Hahn M. E., Stegeman J. J. (2009). The tryptophan photoproduct 6-formylindolo[3,2-b]carbazole (FICZ) binds multiple AHRs and induces multiple CYP1 genes via AHR2 in zebrafish. Chem. Biol. Interact. 181, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson M. E., Jenny M. J., Woodin B. R., Hahn M. E., Stegeman J. J. (2007). Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3',4,4',5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Sci. 100, 180–193. [DOI] [PubMed] [Google Scholar]
- Jonsson M. E., Kubota A., Timme-Laragy A. R., Woodin B., Stegeman J. J. (2012). Ahr2-dependence of PCB126 effects on the swim bladder in relation to expression of CYP1 and cox-2 genes in developing zebrafish. Toxicol. Appl. Pharmacol. 265, 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafafi S. A., Afeefy H. Y., Ali A. H., Said H. K., Kafafi A. G. (1993). Binding of polychlorinated biphenyls to the aryl hydrocarbon receptor. Environ. Health Perspect. 101, 422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchner S., Jenny M., Aluru N., Franks D., Laub L., Linney E., Williams L., Teraoka H., Hahn M. (2017). Evidence for developmental versus toxicological roles for zebrafish Ahr1b. Toxicol. Sci. 156, S39 (Abstract #1165). [Google Scholar]
- Karchner S. I., Franks D. G., Hahn M. E. (2005). AHR1B, a new functional aryl hydrocarbon receptor in zebrafish: Tandem arrangement of ahr1b and ahr2 genes. Biochem. J. 392, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas A., Poellinger L., Pongratz I. (1999). Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (aryl hydrocarbon) receptor. J. Biol. Chem. 274, 13519–13524. [DOI] [PubMed] [Google Scholar]
- Kewley R. J., Whitelaw M. L., Chapman-Smith A. (2004). The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 36, 189–204. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Park H. J., Kim J. H., Kim S., Williams D. R., Kim M. K., Jung Y. D., Teraoka H., Park H. C., Choy H. E., et al. (2013). Cyp1a reporter zebrafish reveals target tissues for dioxin. Aquat. Toxicol. 134–135, 57–65. [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. [DOI] [PubMed] [Google Scholar]
- King-Heiden T. C., Mehta V., Xiong K. M., Lanham K. A., Antkiewicz D. S., Ganser A., Heideman W., Peterson R. E. (2012). Reproductive and developmental toxicity of dioxin in fish. Mol. Cell. Endocrinol. 354, 121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Heiden T. C., Struble C. A., Rise M. L., Hessner M. J., Hutz R. J., Carvan M. J. III (2008). Molecular targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) within the zebrafish ovary: Insights into TCDD-induced endocrine disruption and reproductive toxicity. Reprod. Toxicol. 25, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Heiden T. K., Hutz R. J., Carvan M. J. III (2005). Accumulation, tissue distribution, and maternal transfer of dietary 2,3,7,8,-tetrachlorodibenzo-p-dioxin: Impacts on reproductive success of zebrafish. Toxicol. Sci. 87, 497–507. [DOI] [PubMed] [Google Scholar]
- Knecht A. L., Goodale B. C., Truong L., Simonich M. T., Swanson A. J., Matzke M. M., Anderson K. A., Waters K. M., Tanguay R. L. (2013). Comparative developmental toxicity of environmentally relevant oxygenated PAHs. Toxicol. Appl. Pharmacol. 271, 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht A. L., Truong L., Marvel S. W., Reif D. M., Garcia A., Lu C., Simonich M. T., Teeguarden J. G., Tanguay R. L. (2017. a). Transgenerational inheritance of neurobehavioral and physiological deficits from developmental exposure to benzo[a]pyrene in zebrafish. Toxicol. Appl. Pharmacol. 329, 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht A. L., Truong L., Simonich M. T., Tanguay R. L. (2017. b). Developmental benzo[a]pyrene (B[a]P) exposure impacts larval behavior and impairs adult learning in zebrafish. Neurotoxicol. Teratol. 59, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S. K., Shin I. (2012). Cardiosulfa induces heart deformation in zebrafish through the Ahr-mediated, CYP1A-independent pathway. Chembiochem 13, 1483–1489. [DOI] [PubMed] [Google Scholar]
- Ko S. K., Jin H. J., Jung D. W., Tian X., Shin I. (2009). Cardiosulfa, a small molecule that induces abnormal heart development in zebrafish, and its biological implications. Angew. Chem. Int. Ed. Engl. 48, 7809–7812. [DOI] [PubMed] [Google Scholar]
- Kok F. O., Shin M., Ni C. W., Gupta A., Grosse A. S., van Impel A., Kirchmaier B. C., Peterson-Maduro J., Kourkoulis G., Male I., et al. (2015). Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 32, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A., Goldstone J. V., Lemaire B., Takata M., Woodin B. R., Stegeman J. J. (2015). Role of pregnane X receptor and aryl hydrocarbon receptor in transcriptional regulation of pxr, CYP2, and CYP3 genes in developing zebrafish. Toxicol. Sci. 143, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota A., Stegeman J. J., Woodin B. R., Iwanaga T., Harano R., Peterson R. E., Hiraga T., Teraoka H. (2011). Role of zebrafish cytochrome P450 CYP1C genes in the reduced mesencephalic vein blood flow caused by activation of AHR2. Toxicol. Appl. Pharmacol. 253, 244–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanham K. A., Plavicki J., Peterson R. E., Heideman W. (2014). Cardiac myocyte-specific AHR activation phenocopies TCDD-induced toxicity in zebrafish. Toxicol. Sci. 141, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Beau M., Carver L., Espinosa I. I. R., Schmidt J., Bradfield C. (1994). Chromosomal localization of the human AHR locus encoding the structural gene for the Ah receptor to 7p21→ p15. Cytogenet. Genome Res. 66, 172–176. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Iwabuchi K., Nomaru K., Nagahama N., Kim E. Y., Iwata H. (2013). Molecular and functional characterization of a novel aryl hydrocarbon receptor isoform, AHR1beta, in the chicken (Gallus gallus). Toxicol. Sci. 136, 450–466. [DOI] [PubMed] [Google Scholar]
- Liu H., Nie F. H., Lin H. Y., Ma Y., Ju X. H., Chen J. J., Gooneratne R. (2016). Developmental toxicity, oxidative stress, and related gene expression induced by dioxin-like PCB 126 in zebrafish (Danio rerio). Environ. Toxicol. 31, 295–303. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A. (2000). The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q., Whitlock J. P. Jr (1997). A novel cytoplasmic protein that interacts with the ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 272, 8878–8884. [PubMed] [Google Scholar]
- Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J. (2019). PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. [DOI] [PubMed] [Google Scholar]
- Mandal P. K. (2005). Dioxin: A review of its environmental effects and its aryl hydrocarbon receptor biology. J. Comp. Physiol. B 175, 221–230. [DOI] [PubMed] [Google Scholar]
- Marlowe J. L., Knudsen E. S., Schwemberger S., Puga A. (2004). The aryl hydrocarbon receptor displaces p300 from E2F-dependent promoters and represses S phase-specific gene expression. J. Biol. Chem. 279, 29013–29022. [DOI] [PubMed] [Google Scholar]
- Marques I. J., Lupi E., Mercader N. (2019). Model systems for regeneration: Zebrafish. Development 146, dev167692. [DOI] [PubMed] [Google Scholar]
- Massarsky A., Bone A. J., Dong W., Hinton D. E., Prasad G. L., Di Giulio R. T. (2016). Ahr2 morpholino knockdown reduces the toxicity of total particulate matter to zebrafish embryos. Toxicol. Appl. Pharmacol. 309, 63–76. [DOI] [PubMed] [Google Scholar]
- Mathew L. K., Andreasen E. A., Tanguay R. L. (2006). Aryl hydrocarbon receptor activation inhibits regenerative growth. Mol. Pharmacol. 69, 257–265. [DOI] [PubMed] [Google Scholar]
- Mathew L. K., Andreasen E. A., Tanguay R. L. (2007). Misexpression of R-Spondin1 impairs tissue regeneration. FASEB J. 21, A620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew L. K., Simonich M. T., Tanguay R. L. (2009). AHR-dependent misregulation of Wnt signaling disrupts tissue regeneration. Biochem. Pharmacol. 77, 498–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee S. P., Konstantinov A., Stapleton H. M., Volz D. C. (2013). Aryl phosphate esters within a major pentabde replacement product induce cardiotoxicity in developing zebrafish embryos: Potential role of the aryl hydrocarbon receptor. Toxicol. Sci. 133, 144–156. [DOI] [PubMed] [Google Scholar]
- Meyer-Alert H., Ladermann K., Larsson M., Schiwy S., Hollert H., Keiter S. H. (2018). A temporal high-resolution investigation of the Ah-receptor pathway during early development of zebrafish (Danio rerio). Aquat. Toxicol. 204, 117–129. [DOI] [PubMed] [Google Scholar]
- Meyer D. N., Baker B. B., Baker T. R. (2018). Ancestral TCDD exposure induces multigenerational histologic and transcriptomic alterations in gonads of male zebrafish. Toxicol. Sci. 164, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W., Hu L., Scrivens P. J., Batist G. (2005). Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: Direct cross-talk between phase I and II drug-metabolizing enzymes. J. Biol. Chem. 280, 20340–20348. [DOI] [PubMed] [Google Scholar]
- Mimura J., Ema M., Sogawa K., Fujii-Kuriyama Y. (1999). Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev. 13, 20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffato M., Louis A., Poisnel C. E., Crollius H. R. (2010). Genomicus: A database and a browser to study gene synteny in modern and ancestral genomes. Bioinformatics 26, 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert D. W., Dalton T. P., Okey A. B., Gonzalez F. J. (2004). Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J. Biol. Chem. 279, 23847–23850. [DOI] [PubMed] [Google Scholar]
- Nguyen L. P., Bradfield C. A. (2008). The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 21, 102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N. T. T., Vincens P., Roest Crollius H., Louis A. (2018). Genomicus 2018: Karyotype evolutionary trees and on-the-fly synteny computing. Nucleic Acids Res. 46, D816–D822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Inoue A., Sasagawa S., Koiwa J., Kawaguchi K., Kawase R., Maruyama T., Kim S., Tanaka T. (2016). Using zebrafish in systems Toxicology for developmental toxicity testing. Congenit. Anom. 56, 18–27. [DOI] [PubMed] [Google Scholar]
- O’Donnell E. F., Saili K. S., Koch D. C., Kopparapu P. R., Farrer D., Bisson W. H., Mathew L. K., Sengupta S., Kerkvliet N. I., Tanguay R. L., et al. (2010). The anti-inflammatory drug leflunomide is an agonist of the aryl hydrocarbon receptor. PLoS One 5, e13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F., Baba A., Takada I., Okada M., Iwasaki K., Miki H., Takahashi S., Kouzmenko A., Nohara K., Chiba T., et al. (2007). Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446, 562–566. [DOI] [PubMed] [Google Scholar]
- Ohtake F., Takeyama K., Matsumoto T., Kitagawa H., Yamamoto Y., Nohara K., Tohyama C., Krust A., Mimura J., Chambon P., et al. (2003). Modulation of oestrogen receptor signalling by association with the activated dioxin receptor. Nature 423, 545–550. [DOI] [PubMed] [Google Scholar]
- Olsvik P. A., Williams T. D., Tung H. S., Mirbahai L., Sanden M., Skjaerven K. H., Ellingsen S. (2014). Impacts of TCDD and MeHG on DNA methylation in zebrafish (Danio rerio) across two generations. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 165, 17–27. [DOI] [PubMed] [Google Scholar]
- Patrizi B., Siciliani de Cumis M. (2018). TCDD toxicity mediated by epigenetic mechanisms. Int. J. Mol. Sci. 19, 4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdew G. H. (1988). Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. 263, 13802–13805. [PubMed] [Google Scholar]
- Planchart A., Mattingly C. J. (2010). 2,3,7,8-Tetrachlorodibenzo-p-dioxin upregulates Foxq1b in zebrafish jaw primordium. Chem. Res. Toxicol. 23, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plavicki J. S., Baker T. R., Burns F. R., Xiong K. M., Gooding A. J., Hofsteen P., Peterson R. E., Heideman W. (2014). Construction and characterization of a sox9b transgenic reporter line. Int. J. Dev. Biol. 58, 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland A., Kende A. (1976). 2,3,7,8-Tetrachlorodibenzo-p-dioxin: Environmental contaminant and molecular probe. Fed. Proc. 35, 2404–2411. [PubMed] [Google Scholar]
- Poland A., Glover E., Kende A. S. (1976). Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J. Biol. Chem. 251, 4936–4946. [PubMed] [Google Scholar]
- Postlethwait J. H. (2007). The zebrafish genome in context: Ohnologs gone missing. J. Exp. Zool. B Mol. Dev. Evol. 308, 563–577. [DOI] [PubMed] [Google Scholar]
- Prasch A. L., Heideman W., Peterson R. E. (2004). Arnt2 is not required for TCDD developmental toxicity in zebrafish. Toxicol. Sci. 82, 250–258. [DOI] [PubMed] [Google Scholar]
- Prasch A. L., Tanguay R. L., Mehta V., Heideman W., Peterson R. E. (2006). Identification of zebrafish arnt1 homologs: 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in the developing zebrafish requires ARNT1. Mol. Pharmacol. 69, 776–787. [DOI] [PubMed] [Google Scholar]
- Prasch A. L., Teraoka H., Carney S. A., Dong W., Hiraga T., Stegeman J. J., Heideman W., Peterson R. E. (2003). Aryl hydrocarbon receptor 2 mediates 2,3,7,8-tetrachlorodibenzo-p-dioxin developmental toxicity in zebrafish. Toxicol. Sci. 76, 138–150. [DOI] [PubMed] [Google Scholar]
- Puga A., Barnes S. J., Dalton T. P., Chang C. Y., Knudsen E. S., Maier M. A. (2000). Aromatic hydrocarbon receptor interaction with the retinoblastoma protein potentiates repression of E2F-dependent transcription and cell cycle arrest. J. Biol. Chem. 275, 2943–2950. [DOI] [PubMed] [Google Scholar]
- Reichard J. F., Dalton T. P., Shertzer H. G., Puga A. (2006). Induction of oxidative stress responses by dioxin and other ligands of the aryl hydrocarbon receptor. Dose Response 3, 306–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes H., Reisz-Porszasz S., Hankinson O. (1992). Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science 256, 1193–1195. [DOI] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Holper S., Kruger M., Stainier D. Y. (2015). Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524, 230–233. [DOI] [PubMed] [Google Scholar]
- Rousseau M. E., Sant K. E., Borden L. R., Franks D. G., Hahn M. E., Timme-Laragy A. R. (2015). Regulation of Ahr signaling by Nrf2 during development: Effects of Nrf2a deficiency on PCB126 embryotoxicity in zebrafish (Danio rerio). Aquat. Toxicol. 167, 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S., Wormke M. (2003). Inhibitory aryl hydrocarbon receptor-estrogen receptor alpha cross-talk and mechanisms of action. Chem. Res. Toxicol. 16, 807–816. [DOI] [PubMed] [Google Scholar]
- Safe S., Lee S. O., Jin U. H. (2013). Role of the aryl hydrocarbon receptor in carcinogenesis and potential as a drug target. Toxicol. Sci. 135, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant K. E., Hansen J. M., Williams L. M., Tran N. L., Goldstone J. V., Stegeman J. J., Hahn M. E., Timme-Laragy A. (2017). The role of Nrf1 and Nrf2 in the regulation of glutathione and redox dynamics in the developing zebrafish embryo. Redox Biol. 13, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria J. A., Becerra J. (1991). Tail fin regeneration in teleosts: Cell-extracellular matrix interaction in blastemal differentiation. J. Anat. 176, 9–21. [PMC free article] [PubMed] [Google Scholar]
- Scott J. A., Incardona J. P., Pelkki K., Shepardson S., Hodson P. V. (2011). AHR2-mediated, CYP1A-independent cardiovascular toxicity in zebrafish (Danio rerio) embryos exposed to retene. Aquat. Toxicol. 101, 165–174. [DOI] [PubMed] [Google Scholar]
- Sehring I. M., Jahn C., Weidinger G. (2016). Zebrafish fin and heart: What's special about regeneration? Curr. Opin. Genet. Dev. 40, 48–56. [DOI] [PubMed] [Google Scholar]
- Seifinejad A., Li S., Mikhail C., Vassalli A., Pradervand S., Arribat Y., Pezeshgi Modarres H., Allen B., John R. M., Amati F., et al. (2019). Molecular codes and in vitro generation of hypocretin and melanin concentrating hormone neurons. Proc. Natl Acad. Sci. U.S.A. 116, 17061–17070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar P., Geier M. C., Truong L., McClure R. S., Pande P., Waters K. M., Tanguay R. L. (2019). Coupling genome-wide transcriptomics and developmental toxicity profiles in zebrafish to characterize polycyclic aromatic hydrocarbon (PAH) hazard. Int. J. Mol. Sci. 20, 2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaya L., Jones D. E., Wilson J. Y. (2019). CYP3C gene regulation by the aryl hydrocarbon and estrogen receptors in zebrafish. Toxicol. Appl. Pharmacol. 362, 77–85. [DOI] [PubMed] [Google Scholar]
- Sies H., Berndt C., Jones D. P. (2017). Oxidative stress. Annu. Rev. Biochem. 86, 715–748. [DOI] [PubMed] [Google Scholar]
- Sipes N. S., Padilla S., Knudsen T. B. (2011). Zebrafish: As an integrative model for twenty-first century toxicity testing. Birth Defects Res. C Embryo Today 93, 256–267. [DOI] [PubMed] [Google Scholar]
- Souder J. P., Gorelick D. A. (2019). ahr2, but not ahr1a or ahr1b, is required for craniofacial and fin development and TCDD-dependent cardiotoxicity in zebrafish. Toxicol. Sci. 170, 25–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D. Y. R., Raz E., Lawson N. D., Ekker S. C., Burdine R. D., Eisen J. S., Ingham P. W., Schulte-Merker S., Yelon D., Weinstein B. M., et al. (2017). Guidelines for morpholino use in zebrafish. PLoS Genet. 13, e1007000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden W. W., Leonardo-Mendonca R. C., Acuna-Castroviejo D., Siekmann A. F. (2017). Genetic dissection of endothelial transcriptional activity of zebrafish aryl hydrocarbon receptors (AHRs). PLoS One 12, e0183433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson H. I. (2002). DNA binding and protein interactions of the AHR/ARNT heterodimer that facilitate gene activation. Chem. Biol. Interact. 141, 63–76. [DOI] [PubMed] [Google Scholar]
- Swedenborg E., Pongratz I. (2010). AhR and ARNT modulate ER signaling. Toxicology 268, 132–138. [DOI] [PubMed] [Google Scholar]
- Tanguay R. L., Abnet C. C., Heideman W., Peterson R. E. (1999). Cloning and characterization of the zebrafish (Danio rerio) aryl hydrocarbon receptor. Biochim. Biophys. Acta 1444, 35–48. [DOI] [PubMed] [Google Scholar]
- Tanguay R. L., Andreasen E., Heideman W., Peterson R. E. (2000). Identification and expression of alternatively spliced aryl hydrocarbon nuclear translocator 2 (ARNT2) cdnas from zebrafish with distinct functions. Biochim. Biophys. Acta 1494, 117–128. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Dong W., Hiraga T. (2003. a). Zebrafish as a novel experimental model for developmental toxicology. Congenit. Anom. (Kyoto) 43, 123–132. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Dong W., Tsujimoto Y., Iwasa H., Endoh D., Ueno N., Stegeman J. J., Peterson R. E., Hiraga T. (2003. b). Induction of cytochrome P450 1A is required for circulation failure and edema by 2,3,7,8-tetrachlorodibenzo-p-dioxin in zebrafish. Biochem. Biophys. Res. Commun. 304, 223–228. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Kubota A., Dong W., Kawai Y., Yamazaki K., Mori C., Harada Y., Peterson R. E., Hiraga T. (2009). Role of the cyclooxygenase 2-thromboxane pathway in 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced decrease in mesencephalic vein blood flow in the zebrafish embryo. Toxicol. Appl. Pharmacol. 234, 33–40. [DOI] [PubMed] [Google Scholar]
- Teraoka H., Okuno Y., Nijoukubo D., Yamakoshi A., Peterson R. E., Stegeman J. J., Kitazawa T., Hiraga T., Kubota A. (2014). Involvement of cox2-thromboxane pathway in TCDD-induced precardiac edema in developing zebrafish. Aquat. Toxicol. 154, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Cockman C. J., Matson C. W., Di Giulio R. T. (2007). Synergistic induction of AHR regulated genes in developmental toxicity from co-exposure to two model PAHs in zebrafish. Aquat. Toxicol. 85, 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Karchner S. I., Hahn M. E. (2012. a). Gene knockdown by morpholino-modified oligonucleotides in the zebrafish (Danio rerio) model: Applications for developmental toxicology. Methods Mol. Biol. 889, 51–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Karchner S. I., Franks D. G., Jenny M. J., Harbeitner R. C., Goldstone J. V., McArthur A. G., Hahn M. E. (2012. b). Nrf2b, novel zebrafish paralog of oxidant-responsive transcription factor NF-E2-related factor 2 (NRF2). J. Biol. Chem. 287, 4609–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Noyes P. D., Buhler D. R., Di Giulio R. T. (2008). CYP1B1 knockdown does not alter synergistic developmental toxicity of polycyclic aromatic hydrocarbons in zebrafish (Danio rerio). Mar. Environ. Res. 66, 85–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy A. R., Sant K. E., Rousseau M. E., diIorio P. J. (2015). Deviant development of pancreatic beta cells from embryonic exposure to pcb-126 in zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 178, 25–32. [DOI] [PubMed] [Google Scholar]
- Ulin A., Henderson J., Pham M. T., Meyo J., Chen Y., Karchner S. I., Goldstone J. V., Hahn M. E., Williams L. M. (2019). Developmental regulation of nuclear factor erythroid-2 related factors (NRFs) by AHR1B in zebrafish (Danio rerio). Toxicol. Sci. 167, 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tiem L. A., Di Giulio R. T. (2011). AHR2 knockdown prevents PAH-mediated cardiac toxicity and XRE- and ARE-associated gene induction in zebrafish (Danio rerio). Toxicol. Appl. Pharmacol. 254, 280–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viluksela M., Pohjanvirta R. (2019). Multigenerational and transgenerational effects of dioxins. Int. J. Mol. Sci. 20, 2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinopal J. H., Casida J. E. (1973). Metabolic stability of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in mammalian liver microsomal systems and in living mice. Arch. Environ. Contam. Toxicol. 1, 122–132. [DOI] [PubMed] [Google Scholar]
- Vogel C. F., Sciullo E., Matsumura F. (2007). Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem. Biophys. Res. Commun. 363, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waits E. R., Nebert D. W. (2011). Genetic architecture of susceptibility to PCB126-induced developmental cardiotoxicity in zebrafish. Toxicol. Sci. 122, 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Li Y., Xia X., Xiong X. (2018). Relationship between metabolic enzyme activities and bioaccumulation kinetics of PAHs in zebrafish (Danio rerio). J. Environ. Sci. (China) 65, 43–52. [DOI] [PubMed] [Google Scholar]
- Wang W.-D., Chen Y.-M., Hu C.-H. (1998). Detection of Ah receptor and Ah receptor nuclear translocator mRNAs in the oocytes and developing embryos of zebrafish (Danio rerio). Fish Physiol. Biochem. 18, 49–57. [Google Scholar]
- Wang W. D., Chen G. T., Hsu H. J., Wu C. Y. (2015). Aryl hydrocarbon receptor 2 mediates the toxicity of paclobutrazol on the digestive system of zebrafish embryos. Aquat. Toxicol. 159, 13–22. [DOI] [PubMed] [Google Scholar]
- Wang W. D., Wu J. C., Hsu H. J., Kong Z. L., Hu C. H. (2000). Overexpression of a zebrafish ARNT2-like factor represses CYP1A transcription in ZLE cells. Mar. Biotechnol. (NY) 2, 376–386. [DOI] [PubMed] [Google Scholar]
- Watson A. J., Hankinson O. (1992). Dioxin- and Ah receptor-dependent protein binding to xenobiotic responsive elements and G-rich DNA studied by in vivo footprinting. J. Biol. Chem. 267, 6874–6878. [PubMed] [Google Scholar]
- Webb K. J., Norton W. H., Trumbach D., Meijer A. H., Ninkovic J., Topp S., Heck D., Marr C., Wurst W., Theis F. J., et al. (2009). Zebrafish reward mutants reveal novel transcripts mediating the behavioral effects of amphetamine. Genome Biol. 10, R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentworth J. N., Buzzeo R., Pollenz R. S. (2004). Functional characterization of aryl hydrocarbon receptor (zfahr2) localization and degradation in zebrafish (Danio rerio). Biochem. Pharmacol. 67, 1363–1372. [DOI] [PubMed] [Google Scholar]
- Wincent E., Jonsson M. E., Bottai M., Lundstedt S., Dreij K. (2015). Aryl hydrocarbon receptor activation and developmental toxicity in zebrafish in response to soil extracts containing unsubstituted and oxygenated PAHs. Environ. Sci. Technol. 49, 3869–3877. [DOI] [PubMed] [Google Scholar]
- Wincent E., Kubota A., Timme-Laragy A., Jonsson M. E., Hahn M. E., Stegeman J. J. (2016). Biological effects of 6-formylindolo[3,2-b]carbazole (FICZ) in vivo are enhanced by loss of CYP1A function in an Ahr2-dependent manner. Biochem. Pharmacol. 110–111, 117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormke M., Stoner M., Saville B., Walker K., Abdelrahim M., Burghardt R., Safe S. (2003). The aryl hydrocarbon receptor mediates degradation of estrogen receptor alpha through activation of proteasomes. Mol. Cell Biol. 23, 1843–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. J., De Castro K. P., Joshi A. D., Elferink C. J. (2017). Canonical and non-canonical aryl hydrocarbon receptor signaling pathways. Curr. Opin. Toxicol. 2, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. Y., Chuang P. Y., Chang G. D., Chan Y. Y., Tsai T. C., Wang B. J., Lin K. H., Hsu W. M., Liao Y. F., Lee H. (2019). Novel endogenous ligands of aryl hydrocarbon receptor mediate neural development and differentiation of neuroblastoma. ACS Chem. Neurosci. 10, 4031–4042. [DOI] [PubMed] [Google Scholar]
- Xiong K. M., Peterson R. E., Heideman W. (2008). Aryl hydrocarbon receptor-mediated down-regulation of sox9b causes jaw malformation in zebrafish embryos. Mol. Pharmacol. 74, 1544–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki K., Teraoka H., Dong W., Stegeman J. J., Hiraga T. (2002). Cdna cloning and expressions of cytochrome P450 1A in zebrafish embryos. J. Vet. Med. Sci. 64, 829–833. [DOI] [PubMed] [Google Scholar]
- Yano F., Kugimiya F., Ohba S., Ikeda T., Chikuda H., Ogasawara T., Ogata N., Takato T., Nakamura K., Kawaguchi H., et al. (2005). The canonical Wnt signaling pathway promotes chondrocyte differentiation in a Sox9-dependent manner. Biochem. Biophys. Res. Commun. 333, 1300–1308. [DOI] [PubMed] [Google Scholar]
- Yasui T., Kim E. Y., Iwata H., Franks D. G., Karchner S. I., Hahn M. E., Tanabe S. (2007). Functional characterization and evolutionary history of two aryl hydrocarbon receptor isoforms (AhR1 and AhR2) from avian species. Toxicol. Sci. 99, 101–117. [DOI] [PubMed] [Google Scholar]
- Yeager R. L., Reisman S. A., Aleksunes L. M., Klaassen C. D. (2009). Introducing the “TCDD-inducible AhR-Nrf2 gene battery”. Toxicol. Sci. 111, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H. C., Tseng H. P., Chung H. Y., Ko C. Y., Tzou W. S., Buhler D. R., Hu C. H. (2008). Influence of TCDD on zebrafish CYP1B1 transcription during development. Toxicol. Sci. 103, 158–168. [DOI] [PubMed] [Google Scholar]
- Yoshioka W., Tohyama C. (2019). Mechanisms of developmental toxicity of dioxins and related compounds. Int. J. Mol. Sci. 20, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka W., Peterson R. E., Tohyama C. (2011). Molecular targets that link dioxin exposure to toxicity phenotypes. J. Steroid Biochem. Mol. Biol. 127, 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jin Y., Han Z., Liu H., Shi L., Hua X., Doering J. A., Tang S., Giesy J. P., Yu H. (2018. a). Integrated in silico and in vivo approaches to investigate effects of BDE-99 mediated by the nuclear receptors on developing zebrafish. Environ. Toxicol. Chem. 37, 780–787. [DOI] [PubMed] [Google Scholar]
- Zhang R., Wang X., Zhang X., Song C., Letcher R. J., Liu C. (2018. b). Polychlorinated diphenylsulfides activate aryl hydrocarbon receptor 2 in zebrafish embryos: Potential mechanism of developmental toxicity. Environ. Sci. Technol. 52, 4402–4412. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wang C., Huang L., Chen R., Chen Y., Zuo Z. (2012). Low-level pyrene exposure causes cardiac toxicity in zebrafish (Danio rerio) embryos. Aquat. Toxicol. 114–115, 119–124. [DOI] [PubMed] [Google Scholar]
- Zodrow J. M., Tanguay R. L. (2003). 2,3,7,8-Tetrachlorodibenzo-p-dioxin inhibits zebrafish caudal fin regeneration. Toxicol. Sci. 76, 151–161. [DOI] [PubMed] [Google Scholar]
- Zodrow J. M., Stegeman J. J., Tanguay R. L. (2004). Histological analysis of acute toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in zebrafish. Aquat. Toxicol. 66, 25–38. [DOI] [PubMed] [Google Scholar]