Here, Infarinato et al. investigated the signaling factors regulating activation of quiescent melanocyte stem cells (McSCs) to generate proliferative progeny that differentiate into pigment-producing melanocytes. Using conditional lineage-specific genetic ablation studies in mice, they found that loss of BMP signaling in the lineage leads to hair graying due to a block in melanocyte maturation, and furthermore show that following WNT-mediated activation of McSCs, BMP, and WNT pathways collaborate to trigger the commitment of proliferative progeny by fueling LEF1- and MITF-dependent differentiation.
Keywords: BMP signaling, LEF1, MITF, melanocyte stem cells, pigment production, proliferative progeny, quiescence, stem cell activation
Abstract
Through recurrent bouts synchronous with the hair cycle, quiescent melanocyte stem cells (McSCs) become activated to generate proliferative progeny that differentiate into pigment-producing melanocytes. The signaling factors orchestrating these events remain incompletely understood. Here, we use single-cell RNA sequencing with comparative gene expression analysis to elucidate the transcriptional dynamics of McSCs through quiescence, activation, and melanocyte maturation. Unearthing converging signs of increased WNT and BMP signaling along this progression, we endeavored to understand how these pathways are integrated. Employing conditional lineage-specific genetic ablation studies in mice, we found that loss of BMP signaling in the lineage leads to hair graying due to a block in melanocyte maturation. We show that interestingly, BMP signaling functions downstream from activated McSCs and maintains WNT effector, transcription factor LEF1. Employing pseudotime analysis, genetics, and chromatin landscaping, we show that following WNT-mediated activation of McSCs, BMP and WNT pathways collaborate to trigger the commitment of proliferative progeny by fueling LEF1- and MITF-dependent differentiation. Our findings shed light upon the signaling interplay and timing of cues that orchestrate melanocyte lineage progression in the hair follicle and underscore a key role for BMP signaling in driving complete differentiation.
To maintain their long-term potential over the course of an organism's lifetime, adult tissue stem cells (SCs) must exert tight control over quiescence, activation, and differentiation. Melanocyte stem cells (McSCs) are nonpigmented, neural crest-derived SCs that coreside with epithelial hair follicle stem cells (HFSCs) in an anatomical structure called the bulge of the hair follicle (HF) (Nishimura et al. 2002). McSCs generate and replenish the HF's pool of pigment-producing melanocytes that provide color for mammalian hair. These specialized cells create a diverse array of pigmentation patterns in other species and protect the body surface against ultraviolet radiation (Lin and Fisher 2007).
The bulge is a dynamic SC niche where HFSCs fuel episodic bouts of active HF down-growth and hair production (anagen), followed by a destructive phase (catagen) in which most of the regenerated portion of the HF degenerates, and finally, a resting period (telogen) in which no tissue regeneration or destruction occurs. To generate melanocytes only when appropriate, quiescent McSCs (qMcSCs) must coordinate their activity with the hair cycle (Fig. 1A; Nishimura 2011). By synchronizing their brief window of activation with HFSCs at the start of each hair cycle, McSCs are able to generate proliferative progeny in early anagen that go on to mature and produce and transfer melanin to the differentiating hair cells in the hair bulb, thereby imparting color to the hair. While fueling this process, the bulge must sustain sufficient numbers of both McSCs and HFSCs to embark upon subsequent cycles of hair growth.
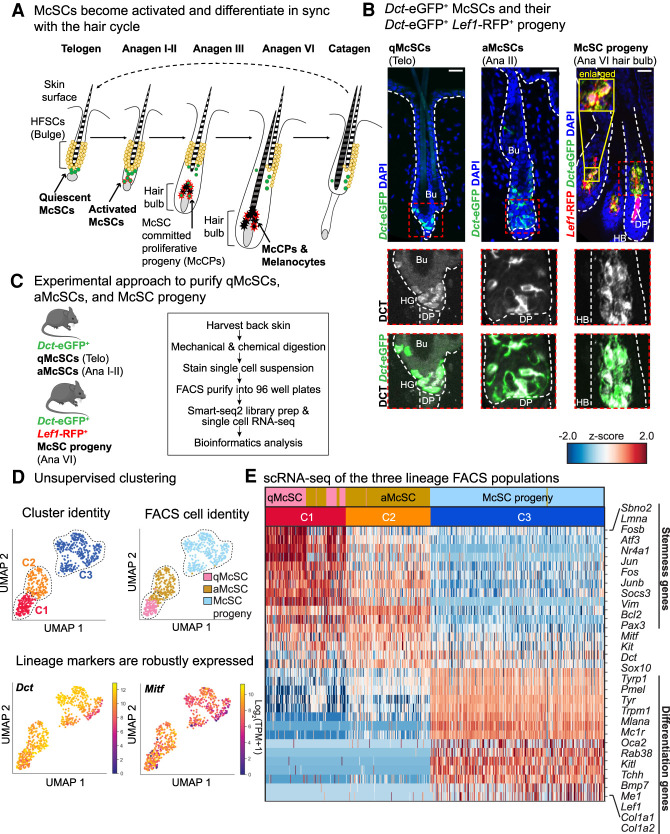
Figure 1.
McSCs and their progeny exhibit distinct transcriptional signatures during lineage progression. (A) Schematic of melanocyte lineage populations during bouts of quiescence, activation, and differentiation throughout the hair cycle. (B) IMF images of qMcSCs (left, second telogen [telo] approximately P60), aMcSCs (middle, anagen II [ana II] at P21), and hair bulb McSC progeny (right, anagen VI at P10). Scale bars, 25 μm. The white dashed line outlines HFs at the epithelial/mesenchymal junction, and the red dashed box indicates enlarged areas below. (Bu) Bulge. (C) Experimental approach used to isolate McSC lineage and perform transcriptional profiling. (D) UMAP representation and unsupervised k-NN-based clustering of single-cell data. Each dot represents a cell colored by its cluster (“C”) and known cell identity. For cell identity, pink dots are qMcSCs (n = 104 cells, n = 3 mice), gold dots are aMcSCs (n = 193 cells, n = 3 mice), and light-blue dots are hair bulb McSC progeny (n = 308 cells, n = 2 mice). (Below) Expression plots for the lineage markers, Dct and Mitf, across all three melanocyte populations. (E) Heat map illustrating differentially expressed genes between McSCs (C1 and C2) and McSC progeny (C3).
Insights into the signals involved in this process come from studying the behavior of SCs during the hair cycle. In telogen, BMPs from a layer of differentiated “inner bulge” cells regulate HFSC quiescence (Hsu et al. 2011), while a related family member, TGF-β, controls McSC quiescence (Nishimura et al. 2010). At anagen onset, WNT/β-catenin signaling stimulates the activation of both HFSCs and McSCs at the bulge base (hair germ [HG]) (Rabbani et al. 2011). An additional twist comes from cross-communication, where WNT-activated HFSCs produce endothelins that further stimulate McSC expansion (Rabbani et al. 2011; Chang et al. 2013; Takeo et al. 2016).
Once activated, HFSCs within the HG generate short-lived proliferative progeny that envelope the dermal papilla (DP) to form a hair bulb from which the hair emerges. In contrast, HFSCs within the bulge generate the downward growing outer root sheath (ORS), which then distances the bulge from the proliferative hair bulb and its DP-signaling center, thereby restoring quiescence to the bulge and upper ORS (Hsu et al. 2014; Yang et al. 2017). The McSC lineage seems to follow this trend (Fig. 1A). As qMcSCs near the HG become activated (aMcSCs), they give rise to committed, proliferative progeny (McCPs) that coexist with the proliferative HFSC progeny in the hair bulb. There, they mature into melanin-producing melanocytes concomitantly with the differentiating hair cells at the heart of the hair bulb. Melanocyte activity continues until catagen ensues, when the differentiated McSC progeny undergo apoptosis alongside HFSC progeny.
Functional studies in mice have provided valuable insights into the final maturation steps in melanocyte differentiation, which require both WNT/β-catenin (Rabbani et al. 2011) and SCF/c-Kit signaling (Botchkareva et al. 2001; Liao et al. 2017). However, our temporal knowledge of the signals governing the intermediate steps between McSCs and mature melanocytes remains limited, as does an understanding of the global changes in gene expression that occur throughout McSC lineage progression. Thus far, our transcriptional insights rest on single-cell qPCR or bulk RNA sequencing of McSCs (Osawa et al. 2005; Moon et al. 2017; Joshi et al. 2019; Zhang et al. 2020) or single-cell comparisons between McSCs and melanoma (Sun et al. 2019).
Understanding how the melanocyte lineage is orchestrated to yield normal hair pigmentation is a fundamental prerequisite to elucidating how these mechanisms are disrupted in age-related and premature hair graying, as well as in melanocyte-related diseases such as vitiligo (Mort et al. 2015). It is also at the crux of understanding how melanoma cells can exhibit features of both stemness and differentiation as they progress through malignancy.
Here, we endeavored to bridge this gap by purifying McSCs as they transition through their discrete stages of the lineage and elucidating their transcriptional features. We then harnessed bioinformatics, genetics, and molecular approaches to dissect the signals that orchestrate these transitional steps. In doing so, we unearth and elucidate a surprising role of BMP signaling downstream from McSCs and at the intersection between early McCPs and mature melanocytes. We further uncover evidence that BMP collaborates with WNT signaling to promote full differentiation.
Results
Identifying and isolating melanocyte lineage populations throughout the hair cycle
To label all melanocyte lineage populations throughout the hair cycle, we used dopachrome tautomerase-eGFP (Dct-eGFP) mice (Gong et al. 2003). We confirmed that eGFP+ patterns overlapped with endogenous DCT, expressed in both the McSCs in the bulge and differentiating progeny in the hair bulb (Fig. 1B). To distinguish the committed melanocyte populations in the hair bulb from the McSCs in the bulge region, we mated Dct-eGFP mice to Lef1-RFP transgenic mice (Rendl et al. 2005). Within the hair bulb, Lef1-RFP was active in both melanocytes and the DP, but only melanocytes coexpressed Dct-eGFP and Lef1-RFP (Fig. 1B). To determine the timing of McSC activation (proliferation), we administered 24 h pulses of 5-ethynyl-2-deoxyuridine (EdU) from late telogen through anagen (Supplemental Fig. S1A). As assessed by immunofluorescence (IMF) microscopy and quantification, McSC proliferative activity peaked at P21 (anagen I-II) (Supplemental Fig. S1B,C). At this stage, HG expansion and DP engulfment was minimal (Supplemental Fig. S1D). By P23 (anagen III), HGs had developed into hair bulbs, which became distanced from the bulge, allowing us to evaluate the McCP compartment. Of total DCT+ P23 hair bulb cells, ∼30% were proliferating (Supplemental Fig. S1D,E), suggestive of a considerable pool of McCPs.
We next devised fluorescence-activated cell sorting (FACS) strategies to isolate qMcSCs, aMcSCs, and differentiating McSC progeny (Fig. 1C; Supplemental Fig. S2A). McSCs are quiescent throughout second telogen, and because hair bulbs are not present at this stage, it was possible to enrich for qMcSCs by first excluding endothelial (CD31+), immune (CD45+), fibroblast, platelet, and adipocyte (CD140a+), epidermal/HF infundibulum (Sca-1+) and bulge HFSC (CD34+) populations, and then selecting for eGFP+ integrin α6medium-low cells. By shifting to anagen I-II, we could enrich for aMcSCs with a similar strategy. Finally, by harvesting cells at (full) anagen VI and using additional markers RFP and c-Kit (CD117), we could enrich for McSC progeny in the hair bulb, namely McCPs and melanocytes.
Single-cell transcriptional profiling dissects the melanocyte lineage into six stages
For initial validation, we used bulk RNA sequencing to profile our qMcSC and differentiating hair bulb McSC progeny populations and found robust expression of lineage markers Dct, Sox10, and c-Kit, relative to HFSCs (Supplemental Fig. S2B; Supplemental Table S1). Differential gene expression analysis with DESeq2 (Love et al. 2014) revealed major transcriptional differences between qMcSCs and differentiating McSC progeny (2385 genes up in qMcSCs; 2819 up in progeny, Padj ≤ 0.001, log2fold ≥ 1) (Supplemental Fig. S2C; Supplemental Table S1). While our independent replicates showed strong correlations and reproducibility (Supplemental Fig. S2D), we anticipated heterogeneity from the presence of intermediate melanocyte lineage steps. We therefore performed single-cell RNA sequencing (scRNA-seq) on each population.
Single-cell mRNAs were subjected to SmartSeq2, which provides higher depth of coverage than Drop-seq or 10X Genomics (Picelli et al. 2013). Dctlow and Krt15+ cells were determined to be nonmelanocyte lineage cells and were eliminated from further consideration, as were cells displaying transcript reads from <1250 genes (see the Materials and Methods). In all cases the retained cells had an average of 1 × 105 reads/cell, with few reads from the full data sets mapping to the mitochondrial genome, reflecting the high quality of our data (Supplemental Fig. S3A,B). Telogen-phase qMcSCs from independent replicates exhibited strongly overlapping profiles (Supplemental Fig. S3C, middle), reflecting minimal batch effects. Even for anagen, where slight variations in hair cycle stage can impact lineage profiles, hair bulb McSC progeny from independent mice were still more closely related to each other than to any other population. qMcSCs displayed the least complex transcriptome (Supplemental Fig. S3C, right), consistent with the findings of Osawa et al. (2005). Moreover, this transcriptome was maintained in a quiescent fraction of early anagen McSCs (Supplemental Fig. S1B,C), underscoring this low complexity molecular signature as a bona fide feature of qMcSCs.
Principle component analysis (PCA) with uniform manifold approximation and projection (UMAP) (Becht et al. 2019) coupled with k-nearest neighbors (k-NN) based community detection (Blondel et al. 2008) showed that our three FACS populations segregated into three distinct clusters, within which cells exhibited robust expression of Dct and Mitf, encoding the master melanocyte transcription factor (Fig. 1D; Goding and Arnheiter 2019). Telogen and anagen I-II McSC populations (C1 and C2) shared significantly more similarities than to the differentiating McSC progeny hair bulb population (C3). To understand what major transcriptional features define “stemness” versus “differentiation,” we combined clusters C1 and C2 and performed differential gene expression analysis against C3. Selected differentially expressed genes are shown in the heat map in Figure 1E (Materials and Methods; Supplemental Table S2). Based on global transcriptional differences, we then assigned each cell a “stemness” and “differentiation” score as a measure of their stage in lineage progression (Materials and Methods; Supplemental Fig. S4A). Cells ubiquitously expressed certain pan-lineage genes, including Sox10 (Supplemental Fig. S4B). However, C1 and C2 showed selective up-regulation of genes including Pax3, Sbno2, and Bcl2 (Nishimura et al. 2005), while C3 cells exhibited elevated transcripts involved in pigment production, including melanin production enzymes (Tyrp1 and Tyr) and melanosome-related proteins (i.e., Mlana and Oca2). C3 was also enriched for transcripts involved in well-known melanocyte differentiation pathways, including c-Kit/SCF (Kitl) (Botchkareva et al. 2001; Liao et al. 2017) and WNT signaling (Lef1) (Fig. 1E; Rabbani et al. 2011).
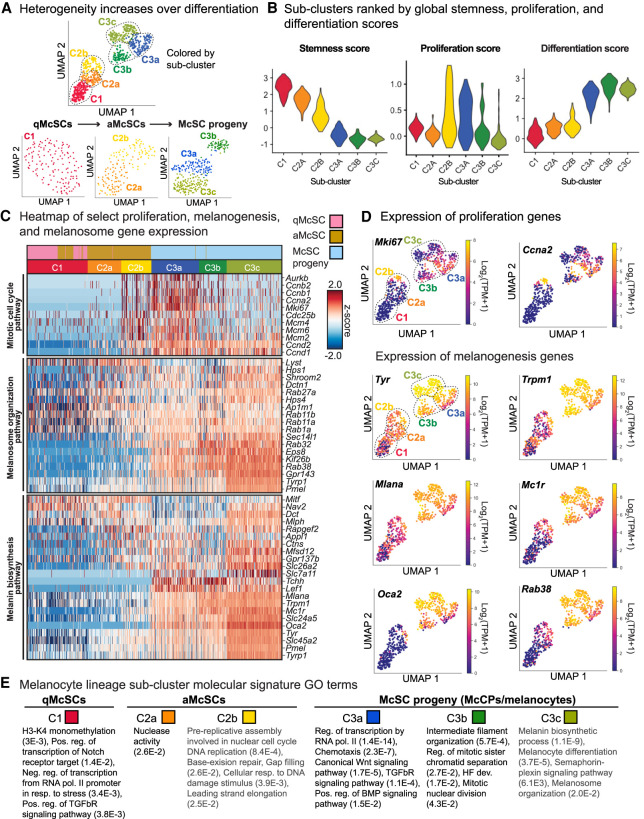
We also detected transcriptional heterogeneity within each major cluster, which increased as cells passed from quiescence through activation to differentiation (Fig. 2A). Six distinct subpopulations were identified, three of which (C1, C2a, and C2b) showed positive scores for “stemness” (Fig. 2A,B). Cells within subclusters were further distinguished by their differential expression of proliferation genes, which we used to similarly generate a global score (Fig. 2B,C; Supplemental Fig. S4C,D). From these comparisons, cluster C2a was less quiescent than C1, while clusters C2b and C3a were particularly enriched for cell cycling transcripts (i.e., Mki67 and Ccna2). Based upon their stemness, proliferation, and differentiation scores, as well as the hair cycle stage at which they were isolated, we assigned these four clusters as qMcSCs (C1), aMcSCs (C2a), a transitional state from aMcSCs to early McCPs (C2b), hair bulb McCPs (C3a), and differentiated melanocytes (C3b,c).
Figure 2.
Single-cell transcriptomes of melanocyte lineage cells suggests that heterogeneity increases during lineage progression. (A) Subclustering of each original cluster from Figure 1D. (B) Violin plots indicating stemness, proliferation, and differentiation scores throughout the six subclusters. (C) Heat map of select proliferation and melanogenesis mRNAs. (D) Expression plots of example proliferation genes and melanogenesis genes. (E) Selected GO terms for each subcluster with false discovery rate (FDR). See also Supplemental Tables S3–S6.
Although some cell cycle transcripts remained elevated in the more differentiated C3b and C3c subclusters, overall transcripts for key cell cycle genes were substantially lower than in the C3a subcluster. In contrast, C3c expressed the highest level of melanogenesis transcripts, marking this subcluster as having the most mature melanocytes (Fig. 2C,D). Overall, the six discrete melanocyte lineage cell subclusters could be demarcated into proliferative and nonproliferative populations, and then further delineated according to melanogenesis and melanosome gene expression patterns. Differential gene expression analysis and gene ontology (GO) analysis (PANTHER) (Thomas et al. 2006; Mi et al. 2019) underscored these distinctive features (Fig. 2E; Supplemental Tables S3–S6). Notably, many expected terms were enriched for each population; for example, Notch (Moriyama et al. 2006) and TGF-βR signaling (Nishimura et al. 2010) for qMcSCs in the C1 subcluster.
Uncovering signaling pathway integration during melanocyte lineage progression
We next turned our attention to signaling pathways and genes likely to be involved in lineage commitment and progression, focusing our attention on cohorts of genes that robustly change expression during transitional stages where less is understood. Expression of genes annotated under select subcluster GO terms (Fig. 2E) are illustrated in the heat map in Figure 3A (see also Supplemental Table S7).
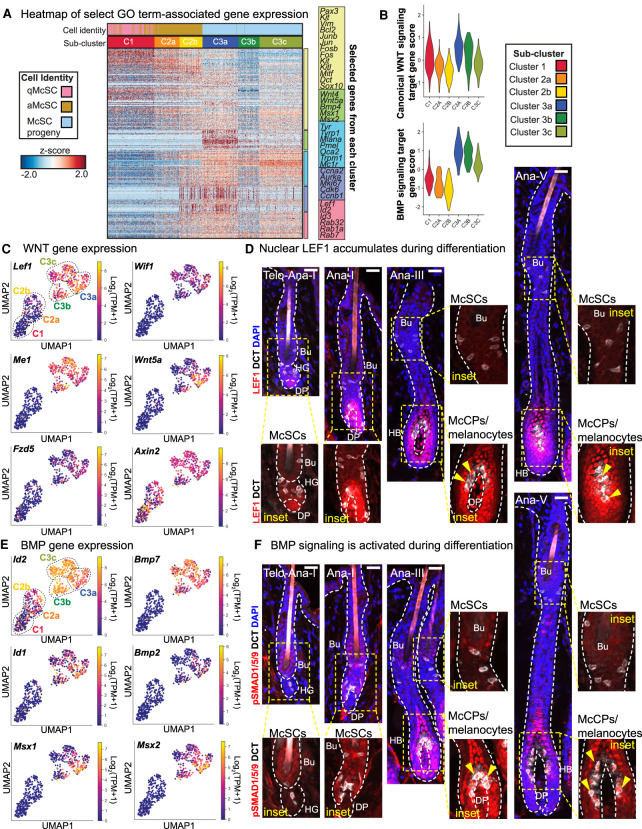
Figure 3.
WNT and BMP signaling increase during melanocyte differentiation. (A) Heat map of gene expression for select genes from GO terms in Figure 2E, including WNT and BMP signaling pathways. See also Supplemental Table S7. (B) Violin plots of canonical WNT and BMP target gene pathway scores in each subcluster. See also Supplemental Table S8. (C) Expression plots of WNT pathway genes. (D) IMF images showing LEF1 protein expression in the melanocyte lineage throughout hair cycling. Nuclear LEF1 (red) indicated by yellow arrows. (E) Expression plots of BMP pathway genes. (F) IMF images of melanocyte lineage populations in HFs throughout the hair cycle. pSMAD1/5/9 is absent in McSCs but becomes nuclear in McCPs/melanocytes (Mcs), indicated by yellow arrows. All scale bars, 25μm.
WNT-signaling is known to promote McSC proliferation and drive their differentiation into pigment-producing melanocytes (Rabbani et al. 2011). Our ability to subdivide the McSC lineage into six hierarchical subclusters now further revealed that genes implicated in WNT signaling and response were particularly elevated at the juncture between the proliferative anagen I-II C2b cells and the proliferative McCP C3a cells in the anagen VI hair bulb (Fig. 3A,B; Supplemental Fig. S5A; Supplemental Table S8). This marked transition was further reflected by the significantly enriched (FDR = 1.7 × 10−5) “canonical WNT signaling pathway” in subcluster C3a, as compared with C3c (Fig. 2E).
Of further intrigue was a dramatic elevation of bone morphogenetic protein (BMP) pathway response genes occurring at the same juncture (Fig. 3A,B; Supplemental Fig. S5B; Supplemental Table S8). Analogously, the “positive regulation of BMP signaling pathway” GO term was enriched in the C3a McCPs (FDR = 1.5 × 10−2) (Fig. 2E). BMPs have been reported to elicit ligand and context-specific effects on melanin production in cultured cells (Bilodeau et al. 2001; Jin et al. 2001; Yaar et al. 2006; Kawakami et al. 2008; Park et al. 2009; Singh et al. 2012; Yang et al. 2014). However, their role in the native tissue microenvironment, as well as their cooperation with other niche signaling components throughout differentiation, is not well understood. We therefore sought deeper understanding of the role of BMP signaling in melanocyte lineage progression, while also exploring possible interplay with WNT.
Probing deeper into WNT pathway activation throughout the lineage, we found that in addition to the elevated levels of genes such as Lef1, Wif1, Me1, and Wnt5a (Wnt5a expression was consistent with Laron et al. 2018) within the McCP/melanocyte populations (Fig. 3C), nuclear LEF1, an established proxy for active WNT signaling, correlated with these patterns, showing strongest labeling within differentiating progeny in the hair bulb (Fig. 3D). Similarly, we found robust up-regulation of BMP target genes Id1/2 and Msx1/2 and ligands BMP2/7 in McCPs/melanocytes (Fig. 3E), consistent with their BMP receptor expression (Supplemental Fig. S5C).
BMP receptor signaling triggers the phosphorylation and activation of SMAD1/5/9 proteins, which form a complex with SMAD4 that translocates to the nucleus to regulate gene expression (Wang et al. 2014). While the bulge niche is rich in BMPs that signal to pSMAD1/5/9+ HFSCs (Kobielak et al. 2007; Genander et al. 2014), surprisingly, neither quiescent nor activated McSCs displayed nuclear pSMAD1/5/9. In contrast, McCPs in anagen III HFs showed nuclear pSMAD1/5/9+, and this was maintained as McCPs/melanocytes continued to differentiate (Fig. 3F). Taken together, our comparative transcriptional analyses and IMF data were consistent with the hypothesis that BMP signaling might be acting downstream to promote differentiation, and that it might act in concert with WNT signaling to do so.
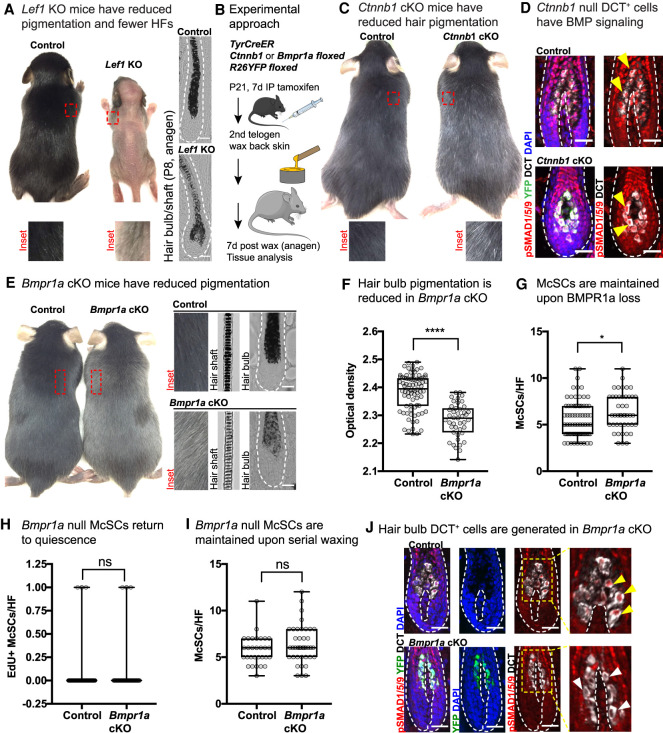
To probe deeper into how WNT and BMP signaling might intersect, we employed loss of function mouse models for each pathway. We first demonstrated LEF1's importance in melanocyte differentiation in the HF by showing that the remaining HFs in Lef1-null mice (C57/BL6J) (van Genderen et al. 1994; Kratochwil et al. 1996) had DCT+ cells in the hair bulb, but that hairs were less pigmented (Fig. 4A; Supplemental Fig. S6A). Using TyrCreER Ctnnb1fl/fl R26-fl-stop-fl-YFP mice (Brault et al. 2001; Srinivas et al. 2001; Bosenberg et al. 2006) and the strategy illustrated in Figure 4B, we then conditionally ablated LEF1's WNT signaling cofactor, β-catenin (Ctnnb1) in the melanocyte lineage to recapitulate a model similar to that used by Rabbani et al. (2011) to demonstrate a role for WNT in McSC proliferation and differentiation. As expected, after one hair cycle, second telogen Ctnnb1 cKO mice had reduced pigmentation relative to control (Fig. 4C). Having substantiated and extended the findings of prior studies (van Genderen et al. 1994; Kratochwil et al. 1996; Rabbani et al. 2011), we then turned to assessing the status of BMP signaling in the absence of WNT signaling. For this purpose, we waxed Ctnnb1 cKO mice to induce hair growth and then analyzed tissue 1 wk later when HFs were in anagen (Fig. 4B). Interestingly, Ctnnb1-null hair bulb McSC progeny still showed nuclear pSMAD1/5/9, indicating that BMP signaling is still active in the absence of functional WNT signaling (Fig. 4D).
Figure 4.
Evidence that BMP signaling acts downstream from McSCs in lineage progression. (A) Representative phenotype of Lef1 KO mice at P13, when hair on the skin surface has emerged in KO (control littermate: female; Lef1 KO: male). Bright-field images of P8 anagen HFs shown at the left. (B) Experimental schematic for interrogating WNT and BMP pathways in the melanocyte lineage in vivo. (C) Representative phenotype of tamoxifen-treated TyrCreER Ctnnb1floxed R26YFP mice at second telogen. (D) IMF of Ctnnb1 cKO and control skin 1 wk after waxing. BMP signaling through nuclear pSMAD1/5/9 is indicated by yellow arrows. (E) Representative phenotype of TyrCreER Bmpr1afloxed R26YFP mice tamoxifen-treated mice in second telogen. (F) Quantification of pigment content in hair bulbs measured by optical density. (****) P < 0.0001, Mann–Whitney test; control n = 80 HFs (n = 4 mice); Bmpr1a cKO n = 43 HFs, (n = 3 mice). (G) Quantification of McSCs numbers per anagen HF 1 wk after waxing. (*) P = 0.0184, Mann–Whitney test; control n = 76 HFs (n = 5 mice); Bmpr1a cKO n = 45 HFs (n = 4 mice). (H) Quantification of EdU+ McSCs per HF 1 wk after waxing. (ns) Not significant, Mann–Whitney test; control n = 61 HFs (n = 3 mice); Bmpr1a cKO n = 45 HFs (n = 4 mice). (I) Quantification of McSCs per HF after five rounds of depilation, quantified in anagen 1 wk after the fifth wax. Mann–Whitney test; control n = 30 HFs (n = 3 mice); Bmpr1a cKO n = 40 HFs (n = 4 mice). (J) IMF shows presence of DCT+ hair bulb McSC progeny, but loss of nuclear pSMAD1/5/9 in YFP+ cells. Scale bars, 25μm.
Given this result, we then proceeded to functionally interrogate the role of BMP signaling in melanocyte lineage progression by performing conditional melanocyte lineage-specific ablation of Bmpr1a (Alk3) to extinguish downstream signaling through this pathway (Mishina et al. 2002). At P21, TyrCreER Bmpr1afl/fl R26fl-stop-fl-YFP mice were treated with tamoxifen, and anagen skins were analyzed by IMF 1 wk after waxing (Fig. 4B). Strikingly, by second telogen, Bmpr1a cKO mice exhibited gray hair, whereas mice expressing at least one Bmpr1a allele (TyrCreER−, TyrCreER+ Bmpr1a+/+or TyrCreER+ Bmpr1afl/+) showed no difference in coat color (Fig. 4E). By optical density, we found significantly reduced pigmentation in Bmpr1a cKO hair bulbs (Fig. 4F).
Adult hair graying phenotypes can be caused by defects in the McSCs themselves or at any point during differentiation (Nishimura 2011). To test whether McSC integrity was compromised by loss of Bmpr1a, we performed waxing at second telogen to induce hair cycle entry and McSC activation, and then measured the number of McSCs 1 wk later. We found that McSC numbers were comparable when BMP signaling was abrogated (Fig. 4G), and following activation, Bmpr1a-null McSCs returned to quiescence normally, as measured by proliferation during a 4-h EdU pulse (Fig. 4H).
To further challenge the stem cell compartment upon loss of BMPR1a, we performed serial waxing, depilating second telogen mice a total of five times every 3 wk. Even after this heightened demand on McSC activity, waxing did not exacerbate the hair graying phenotype (Supplemental Fig. S6B). Indeed, upon analyzing anagen skin 1 wk after the fifth round of waxing, we found no significant difference in number of McSCs between control and Bmpr1a cKO mice (Fig. 4I).
Despite efficient Bmpr1a targeting and loss of both canonical (pSMAD1/5/9) and noncanonical (pP38) BMP signaling, DCT+ McSC progeny were still present in hair bulbs of Bmpr1a cKO mice (Fig. 4J; Supplemental Fig. S6C,D). However, hair bulbs remained poorly pigmented 12 d into anagen, indicating that the defect was not merely due to delayed maturation (Supplemental Fig. S6E). Taken together, these findings suggested that McSCs were intact and could trigger melanocyte lineage progression in the absence of BMP signaling, but that melanin production in the lineage was impaired. This is in striking contrast to WNT signaling, which impacted both McSC and hair bulb compartments during anagen (Rabbani et al. 2011).
Given these results, and the fact that most prior studies into the roles of BMP signaling had been performed with cultured cells, we initially turned to CRISPR/Cas9 targeting of Bmpr1a in the mouse melanocyte cell line, Melan-a2 (Sviderskaya et al. 1995). Following sequence confirmation of successful targeting, however, we realized that upon BMP stimulation, these cells were still able to signal through pSMAD1/5/9 (Supplemental Fig. S6F). While Bmpr1a targeting in vivo resulted in loss of nuclear pSMAD1/5/9, there are other BMP/TGF-β receptor superfamily members that if coexpressed can heterodimerize in multiple ways and yield responses that differ depending upon ligand, receptor combinations, and intra- and extracellular regulator molecules (for review, see Wang et al. 2014). While this complexity precluded our ability to interrogate the functional relevance of BMP signaling in vitro, it may begin to explain the context-dependent behaviors previously described of the BMP pathway in the melanocyte lineage (Bilodeau et al. 2001; Jin et al. 2001; Sharov et al. 2005; Yaar et al. 2006; Kawakami et al. 2008; Park et al. 2009; Han et al. 2012; Singh et al. 2012; Yang et al. 2014).
Returning to studying the importance of pSMAD1/5/9 signaling in vivo, we administered tamoxifen to Bmpr1a cKO and stage-matched control mice at P21, allowed them to undergo a full round of hair cycling, and then waxed to ensure synchronous induction of anagen (Fig. 4B). To capture hair bulb McSC progeny at the point when BMP signaling had become activated and differentiation had begun, we harvested skin 1 wk later when HFs were in anagen IV (Supplemental Fig. S6G). YFP+ cells were then isolated by FACS (gated against CD31, CD45, CD140a, Sca-1 and CD34) (Supplemental Fig. S7A) and subjected to scRNA-seq.
Similar to our temporal data sets, cells from Bmpr1a cKO and control mice showed a similar depth of sequencing, and independent replicates exhibited similar clustering (Supplemental Fig. S7B). Approximately 5000 genes per cell were detected, with few reads from mitochondrial transcripts (Supplemental Fig. S7C). As before, Dctlow and Krt15+ cells were excluded from analysis. Unsupervised clustering revealed that YFP+ Bmpr1a heterozygous McSC progeny showed transcriptomes comparable with YFP+ wild-type, but Bmpr1a-null cells formed a unique cohort (Fig. 5A; Supplemental Fig. S7B).
Figure 5.
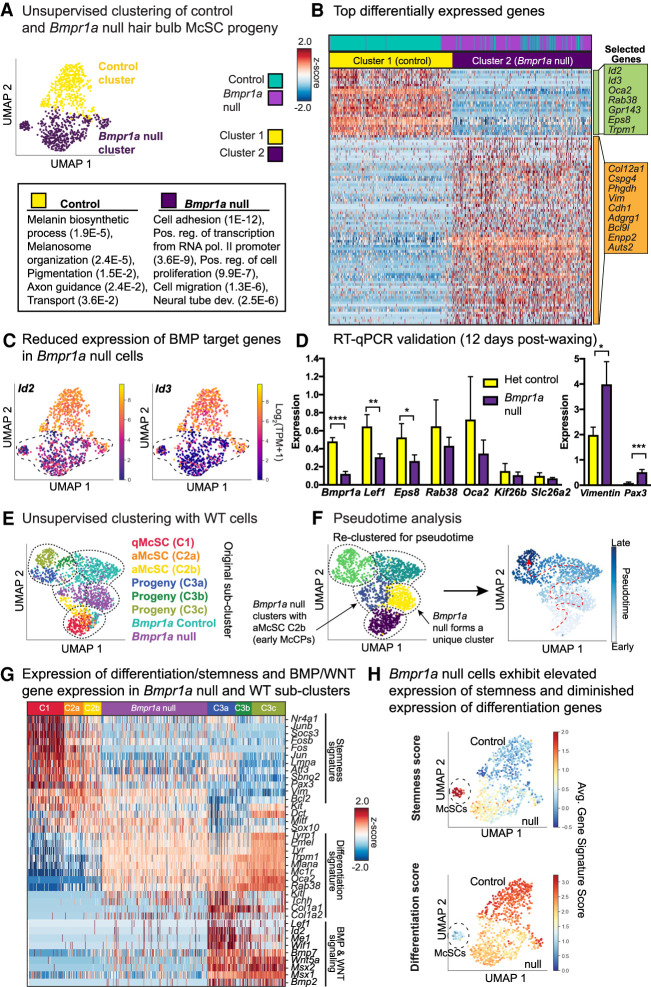
Pseudotime analysis of the Bmpr1a-null hair bulb McSC progeny suggests a block at the juncture between early activated melanocyte stem cell progeny and melanocyte maturation. (A) UMAP representation and unsupervised k-NN-based clustering of control and Bmpr1a-null cells colored by cluster identity. (B) Heat map of top differentially expressed genes for Bmpr1a-null versus stage-matched controls and selected GO terms (DAVID 6.8) to bottom left with (P-values). See also Supplemental Table S9. (C) Expression plots of Id2 and Id3 in YFP+ populations. (D) RT-qPCR expression of genes from single-cell RNA-seq list isolated 12 d postwaxing. (****) P < 0.0001 Bmpr1a, (**) P = 0.0035 Lef1, (*) P = 0.0269 Eps8, (*) P = 0.0148 Vimentin, (***) P = 0.0010 Pax3, unpaired t-test; heterozygous control n = 3 mice, Bmpr1a cKO n = 4 mice. (E) Unsupervised k-NN based clustering of original melanocyte lineage populations with Bmpr1a-null and control cells, colored by subcluster identity. (F) Pseudotime analysis of combined data sets colored by pseudotime clusters (left) and pseudotime order (right). Cells are colored by progression through pseudotime with dashed red arrow to indicate direction of progression. (G) Heat map showing expression of key signature genes in Bmpr1a-null cells relative to WT subclusters. (H) UMAP plots expressing stemness and differentiation scores for Bmpr1a-null and control cells.
Differential gene expression analysis revealed marked changes caused by Bmpr1a ablation in the melanocyte lineage (Fig. 5A,B). As expected, mRNAs encoded by direct BMP target genes, such as Id2 and Id3, were dramatically down-regulated (Fig. 5B,C). Melanogenesis transcripts such as Oca2, Rab38, and Gpr143 were also markedly diminished. Conversely, transcripts up-regulated in Bmpr1a-null cells included genes that have been implicated in stemness and/or migration; e.g., Vim, Cspg4, Adgrg1, and Enpp2 (Fig. 5A,B; Supplemental Fig. S7D). To test whether these trends persisted longer into anagen, we FACS-purified YFP+ cells 12 d postwaxing and performed RT-qPCR, with which we obtained consistent results (Fig. 5D). Altogether, 80 genes were significantly (q ≥ 0.05) down-regulated and 346 genes were up-regulated upon Bmpr1a ablation (Supplemental Table S9).
To identify the juncture along the lineage at which loss of BMP signaling exerts its effects, we integrated our Bmpr1a-null and Bmpr1a control data sets into our six subclusters of wild-type melanocyte lineage cells and again performed UMAP-based dimensionality reduction with community-based clustering (Fig. 5E). This analysis suggested that Bmpr1a-null cells were most similar to the early McCP cluster C2b, with approximately one-third of null cells found in the same community cluster (Fig. 5E,F). We then implemented pseudotime analysis using Monocle3 (Cao et al. 2019), an unbiased method to map differentiation in which all biologically significant genes are considered. This algorithm ordered each cell along the McSC lineage trajectory according to its individual transcriptome. In concordance with our collection strategies and differential gene expression analyses between subclusters, C1 qMcSCs were placed at the start of the inferred trajectory, followed by aMcSCs, then McCPs/melanocytes, with the highly melanogenic cluster C3c forming the termination point. Bmpr1a-null cells fell between the anagen I-II C2b early McCP cells and the stage-matched control cells and the anagen VI hair bulb cells, including C3a McCPs (Fig. 5E,F; Supplemental S7E).
Considering how the Bmpr1a-null cells compared with the six wild-type subclusters, we found that in contrast to McCP subcluster C3a, which highlighted melanin biosynthesis as well as WNT and BMP signaling, the Bmpr1a-null cluster displayed an enhanced stemness and a diminished differentiation (pigmentation) signature, with reduced expression of genes involved in WNT and BMP signaling (Fig. 5G). These differences were not merely attributable to differences in anagen stage, as considerably more stemness transcripts were sustained while fewer differentiation genes were elevated in the Bmpr1a-null versus stage-matched control samples. This uniqueness of signature was further reinforced by implementing the “stemness” and “differentiation” scoring parameters established for our wild-type McSC and differentiating McSC progeny populations (Fig. 5H). Overall, the Bmpr1a-null hair bulb McSC progeny most closely associated with subcluster C2b (Fig. 5E,F; Supplemental Table S10). Thus, although the level of pigmentation genes was greater in Bmpr1a-null versus wild-type C2b cells, these data indicated that without BMP signaling, the melanocyte lineage was arrested just downstream from the activated McSCs and early McCPs.
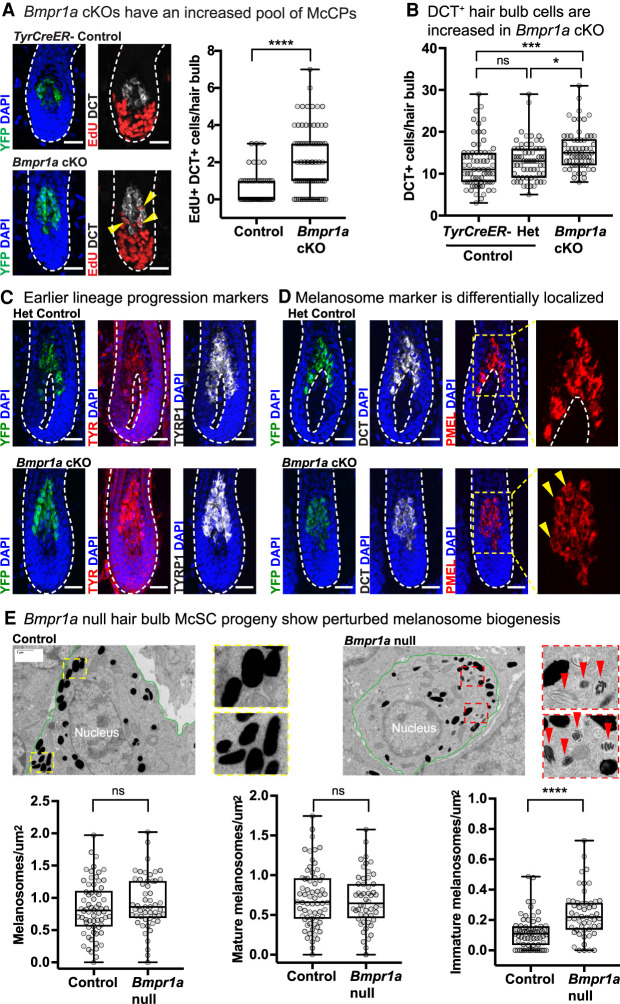
In agreement with their similarities to activated McSCs, DCT+ Bmpr1a-null McCPs in the hair bulb displayed higher EdU incorporation relative to their control counterparts (Fig. 6A). Moreover, overall numbers of DCT+ cells were increased in Bmpr1a cKO hair bulbs, indicating that the reduced pigmentation seen in these cells was not attributable to McCP depletion (Fig. 6B). In this regard, loss of BMP signaling stood in striking contrast to loss of WNT/β-catenin signaling in the melanocyte lineage, where reduced proliferation in the hair bulb was reported (Rabbani et al. 2011).
Figure 6.
Differentiating Bmpr1a-null McSC progeny exhibit signs of immaturity. (A) Four-hour EdU pulse experiment with representative IMF images and quantification. (****) P < 0.0001, Mann–Whitney test; control n = 50 HFs (n = 2 mice); Bmpr1a cKO n = 75 HFs (n = 3 mice). Yellow arrowheads indicate proliferating (EdU+) DCT+ cells. (B) Quantification of DCT+ cells per hair bulb 1 wk after waxing. (****) Padj < 0.0001, (*) Padj = 0.0103, Kruskal–Wallis test, Dunn's multiple comparisons test; TyrCreER− control n = 70 HFs (n = 3 mice); Het control n = 63 HFs (n = 3 mice); Bmpr1a cKO n = 62 HFs (n = 4 mice). (C) IMF images showing protein expression of early melanogenic enzymes in control and Bmpr1a-null cells in the hair bulb. (D) IMF image showing expression pattern of PMEL. Area within yellow dashed box is enlarged at the right, with yellow arrows to indicate perinuclear localization of signal. All IMF scale bars, 25 μm. (E) EM images of silver sections with melanocyte lineage cells outlined in green. Dashed box insets are enlarged. Red arrows indicate immature melanosomes. Scale bar, 1 μm. The bottom graphs show quantification of number of melanosomes divided by melanocyte lineage cell area. (Left) Total melanosomes per square micron. (Middle) Mature melanosomes per square micron. (Right) Immature melanosomes per square micron. (****) P < 0.0001, Mann–Whitney test; control n = 66 areas (three mice); Bmpr1a cKO n = 52 areas (three mice).
Given the increased number of McSC progeny within Bmpr1a cKO hair bulbs, we probed deeper into the underlying defect in hair pigmentation. Confirming our transcriptomic data, early melanogenic enzymes in the lineage, including DCT, tyrosinase (TYR), and tyrosinase-related protein 1 (TYRP1, reduced at the transcript level), were still present in Bmpr1a-null cells (Fig. 6A,C). Interestingly, however, even though PMEL, a structural component of melanosomes, was expressed, it exhibited an abnormal perinuclear localization in Bmpr1a-null cells, suggestive of a defect in melanosome maturation (Fig. 6D). This notion was further strengthened by the reduced expression of melanosome organization genes such as Oca2, Rab38, and Gpr143, and with diminished expression of Mc1r, which encodes a cell surface receptor that instructs melanocytes to make black/brown or yellow pigment, depending on ligand binding (Fig. 5B; Supplemental Table S9).
Melanosomes undergo four distinct maturation phases that are well-defined at the ultrastructure level (Marks and Seabra 2001). Therefore, we analyzed melanosomes by electron microscopy (EM) in Bmpr1a-null and control cells. Area densities of total and mature melanosomes were similar between conditions. However, Bmpr1a-null cells in the hair bulb displayed a significantly higher density of immature melanosomes (Fig. 6E).
Molecular underpinnings of the block in differentiation caused by loss of BMP signaling
Our collective findings pointed to a newfound physiological role of BMPR1a signaling in melanosome biogenesis and maturation and traced its loss to a block downstream from McSC activation/early McCPs and upstream of melanocyte differentiation (Fig. 7A). Seeking insights into how loss of BMP signaling arrests lineage progression, we were intrigued to see a trending down-regulation of Lef1 transcripts in Bmpr1a-null cells, which we confirmed by IMF (Fig. 7B). In cultured melanocytes, stimulation with BMP2 and BMP4, ligands in anagen skin, induced up-regulation of Lef1 mRNA and LEF1; this response was abrogated upon treatment with BMP receptor (ALK) inhibitor (Supplemental Fig. S8A,B).
Figure 7.
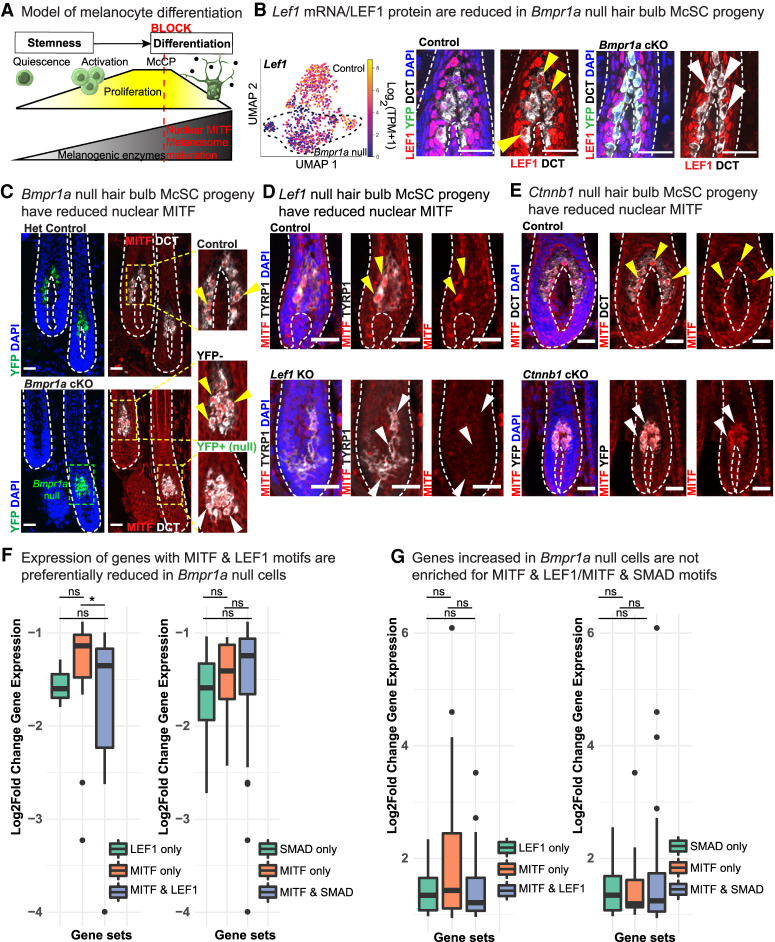
BMP-regulated LEF1 may cooperate with MITF to achieve complete differentiation. (A) Model of McSC lineage progression with the proposed timing of the differentiation block without BMP signaling. (B) Lef1 mRNA expression (left) with IMF (right) in control and Bmpr1a-null cells. Yellow arrows indicate presence and white arrows indicate reduction/absence of nuclear LEF1. (C) IMF images depicting MITF in control and Bmpr1a-null cells in anagen 1 wk after waxing. Enlarged areas show reduction (white arrows) of nuclear MITF in transduced (YFP+) versus untransduced (YFP−) as an internal control. (D,E) IMF images illustrating MITF levels in hair bulb McSC progeny of control and Lef1 KO (P4) mice (D) and Ctnnb1 cKO and control mice (E) whose HFs were analyzed 1 wk postwaxing. Yellow arrows indicate presence and white arrows indicate reduction/absence of nuclear MITF. IMF scale bars, 25μm. (F,G) Motif analysis of promotors within genes whose expression is either reduced (F) or increased (G) upon Bmpr1a loss. Box and whisker plots showing gene sets and the log2 fold change gene expression in Bmpr1a-null cells. (*) P = 0.036 by Wilcoxon test. See also Supplemental Table S11.
Pursuing further insights into the molecular underpinnings of BMP's action on the melanocyte lineage, we turned toward the master regulator of the melanocyte lineage, microphthalmia-associated transcription factor (MITF). MITF drives a myriad of lineage processes including survival, proliferation, and differentiation (Goding and Arnheiter 2019). According to the MITF rheostat model in melanoma, low MITF promotes stemness, intermediate MITF induces proliferation, and high MITF drives differentiation/melanogenesis (Carreira et al. 2006; Goding and Arnheiter 2019). Interestingly, nuclear MITF was markedly diminished in the absence of BMP signaling (Fig. 7C), although not at the mRNA level (Supplemental Fig. S8C). Moreover, we also found diminished nuclear MITF in Lef1- and Ctnnb1-null hair bulb McSC progeny (Fig. 7D,E). In cultured melanocytes, we did not observe changes in MITF upon BMP stimulation, even though LEF1 was up-regulated (Supplemental Fig. S8B–D), further underscoring the importance of studying the role of BMP signaling in a physiological context.
Given the numerous differentiation and pigmentation defects that converge on perturbations of MITF (Goding and Arnheiter 2019), our finding that nuclear MITF was diminished upon loss of BMP signaling in vivo could largely explain the block in melanocyte differentiation and the gray hair phenotype in our in Bmpr1a cKO mice. Moreover, LEF1 is known to interact with and regulate MITF (Dorsky et al. 2000; Takeda et al. 2000; Saito et al. 2002, Yasumoto et al. 2002), a feature intriguing in that LEF1 was also diminished upon loss of BMP signaling.
Evidence for cooperation between BMP-regulated LEF1 and MITF in melanocyte maturation
Our collective in vivo findings thus far led us to posit that BMP signaling might function to achieve the levels of nuclear MITF and LEF1 required to promote transcription of melanocyte maturation genes. Searching for indications of such interplay, we scanned for transcription factor motifs within the putative promoter regions of genes that were differentially expressed between control and Bmpr1a-null cells. We binned genes as either containing a MITF motif only, a LEF1 motif only, or both MITF and LEF1 motifs. In a second analyses, we binned these genes as containing either a MITF motif only, a SMAD motif only, or both SMAD and MITF motifs.
Of these comparisons, one had clear statistical significance: Putative promoters harboring both MITF and LEF1 sites exhibited a greater reduction in gene expression than MITF sites alone when BMP signaling was abrogated (Fig. 7F). These effects were not seen with genes harboring MITF and SMAD sites nor with genes whose expression increased in Bmpr1a-null cells (Fig 7F,G). Notably, the down-regulated genes whose promoters contained both LEF1 and MITF motifs encoded proteins specifically involved in transport, transmembrane transport, and melanosome organization, including Mc1r, Oca2, Trpm1, Rab38, and Shroom2 (Supplemental Table S11).
Given these results, we wondered whether further evidence for the cooperation of these factors could be found at the chromatin level. We hypothesized that chromatin openness would change as McSCs committed to melanocyte differentiation, and that sites that gain accessibility in differentiation would show transcription factor binding motifs critical for this process. To test this possibility, we performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) (Buenrostro et al. 2013) on FACS-purified qMcSCs in the bulge and on differentiating McCP/melanocyte progeny in the anagen hair bulb. In this way, we could evaluate and compare chromatin openness associated with stem and differentiating states.
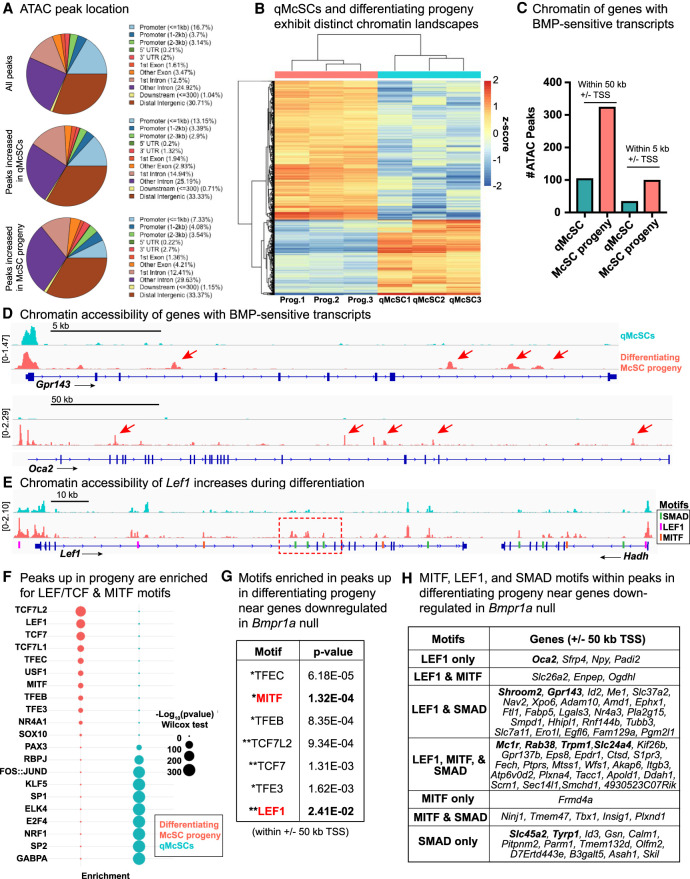
Analyses of our ATAC-seq showed high reproducibility across biological replicates (Supplemental Fig. S8E). Similar percentages of ATAC peaks were within 50 kb of canonical transcriptional start sites (TSSs) (28.2% for peaks increased by log2 fold change ≥ 1 in qMcSCs; 26.4% for McSC progeny), and peaks were similarly distributed throughout promoters, gene bodies, and intergenic regions (Fig. 8A). Differential peak analysis indicated that qMcSCs and their differentiating progeny exhibit strikingly distinct chromatin accessibility profiles (log2 fold change ≥ 1, P ≤ 0.05) (Fig. 8B; Supplemental Table S12).
Figure 8.
Chromatin accessibility is dynamic during lineage progression and reveals cooperation between MITF, LEF1, and SMAD. (A) Location of ATAC peaks expressed as percentages. (B) Heat map of differential peaks in qMcSCs versus their differentiating hair bulb progeny (prog.) (log2 fold change ≥ 1, P ≤ 0.05). See also Supplemental Table S12. (C) Number of peaks increased in either qMcSCs or their differentiating progeny within ±5 or 50 kb of the TSS of genes (80) whose expression is diminished upon loss of BMPR1a signaling by scRNA-seq. (D) ATAC tracks showing differential peaks of select differentiation genes. (E) ATAC track of Lef1 gene showing chromatin opening in differentiating progeny in regions with SMAD motifs, indicated by dashed red boxes. Merged (mean averaged) of three biological replicates are shown for D and E. (F) Plot expressing enrichment of select motifs in differential ATAC peaks between each cell type. See also Supplemental Table S13. Motifs with −log10(P-value) >300 are expressed as a maximum of 300 here. (G) Motif analysis within peaks subset by the following parameters: peaks enriched in differentiating progeny relative to McSCs and 50 kb ± TSS of genes down-regulated in Bmpr1a-null cells. (*) MITF family, (**) LEF/TCF family. See Supplemental Table S14. (H) Motif analysis within differentiating McSC progeny peaks within 50 kb ± TSS of genes whose expression is diminished in Bmpr1a-null cells. Table highlights LEF1, MITF, and SMAD(2, 3, and/or 4) motifs. Genes in bold specify those encoding proteins involved in melanogenesis. See also Supplemental Table S15.
Interestingly, a number of these lineage sensitive peaks appeared to be responsive to the status of BMP signaling, as peaks associated with differentiation resided within the promoter/enhancer regions of genes down-regulated upon Bmpr1a ablation (Fig. 8C,D). Notably, Lef1 showed chromatin remodeling as McSCs became activated and progressed along the lineage. Moreover, many of the ATAC peaks within the Lef1 locus that were more open in differentiating progeny contained SMAD motifs (Fig. 8E; Supplemental Fig. S8F). These analyses offered new insights into why Lef1 transcripts were down-regulated upon loss of BMP signaling and provided support for direct transcriptional regulation of Lef1 by BMP signaling.
To ask what transcription factors may bind these differentially accessible regions, we performed an unbiased motif analysis. Strikingly, LEF/TCF and MITF family factors were the top most enriched factors in peaks that increased in differentiation (Fig. 8F; full list in Supplemental Table S13). Conversely, peaks increased in qMcSCs showed enrichment of SP/KLF and AP1 family motifs, as well as those expected to play a role in SC biology, such as PAX3 and RBPJ (Notch) (Moriyama et al. 2006). Motifs such as SOX10 were present in both qMcSC and McCP peaks (Fig. 8F).
Focusing on the entire cohort of genes whose expression was diminished upon loss of BMPR1a, we analyzed differentiation-enriched ATAC peaks that were ±50 kb (putative promoter and enhancer regions) of the TSS of these genes (Fig. 8C). Again, we found significant enrichment of motifs for the binding of LEF/TCF (TCF7L2, TCF7, and LEF1) and MITF (Fig. 8G; Supplemental Table S14). Consistent with our single-cell analysis of stemness factors (Fig. 1E), AP1 family motifs dominated the motifs in peaks increased in qMcSCs (Supplemental Table S14). Finally, to assess how SMAD-mediated regulation might impact chromatin dynamics, we analyzed differentiating progeny for their promoter/enhancer chromatin peaks that were associated with genes whose expression was markedly changed upon BMPR1a loss. In agreement with our predictive motif scanning (Fig. 7F,G), our analysis suggested that many of the key genes involved in melanin biosynthesis, transport, and trafficking (i.e., Mc1r, Rab38, and Trpm1) may require cooperation of SMAD as well as MITF and LEF1 in controlling their lineage-specific activation (Fig. 8H; Supplemental Table S15).
Discussion
BMP signaling at the helm of melanocyte maturation
By devising methods to purify and transcriptionally profile individual cells from their native tissue context and at discrete steps along the melanocyte lineage, we made major inroads into illuminating the signals involved in this progression. Taking an unbiased approach, we harnessed the power of scRNA-seq to map the landscape of global gene expression changes that occur as McSCs transition from quiescence to activation to differentiation. In all, we discovered six discrete transcriptional profiles and uncovered signaling pathways and a plethora of transcriptional changes that occur as McSC progress through differentiation.
McSCs and HFSCs must coordinate their behaviors. A priori, because BMP signaling functions to maintain HFSCs in a quiescent state (Kobielak et al. 2007; Genander et al. 2014; Adam et al. 2018), this pathway might have been predicted to also govern McSC quiescence. However, qMcSCs appear to be largely refractory to BMP signaling status, as reflected by the paucity of pSMAD1/5/9 IMF in these cells. Even when BMP signaling was completely abrogated in qMcSCs, they still became activated at early anagen and then returned to quiescence later in anagen. Reflecting on the distinct BMP responses of McSCs and HFSCs, it is notable that these two SCs occupy distinct locales within their shared niche. While quiescent HFSCs exist as a single layer flanking the BMP-rich inner bulge, qMcSCs reside near the base of the bulge, where BMP signaling is lower (Hsu et al. 2011; Yang et al. 2017). Intriguingly, TGF-β signaling is elevated at the bulge base (Oshimori and Fuchs 2012), which could explain the paradoxical preferential sensitivity of McSCs to this related signaling pathway (Nishimura et al. 2010).
In contrast to the proliferative effects of WNT signaling on McSCs (Rabbani et al. 2011), BMP signaling appeared to function after McCP specification. At this stage, cells were proliferative, but showed considerable signs of differentiation. Interestingly upon TyrCreER-mediated ablation of Ctnnb1 in the lineage, nuclear pSMAD1/5/9 was still seen McSC hair bulb progeny, suggesting that BMP signaling is not dependent on WNT signaling. In contrast, LEF1 was diminished in Bmpr1a hair bulb McSC progeny, indicative of a reliance of BMP signaling in WNT-mediated melanocyte maturation.
The reduction in MITF in Bmpr1a-null cells further positioned BMP signaling at the helm of melanocyte maturation. Indeed, loss of BMP signaling skewed the hair bulb population toward proliferative and less differentiated McCPs. In this regard, it is notable that in normal skin, BMPs are elevated in the apical region of the dermal papilla; i.e., the region of the hair bulb where proliferative precursors of both the hair shaft and the melanocytes reside (Yang et al. 2017). BMP signaling also directly regulates a cohort of hair-specific genes in the proliferative progenitors of the hair shaft (Genander et al. 2014; Adam et al. 2018).
Our new findings here suggest that BMP signaling plays a similar role in the melanocyte lineage by governing the expression of later stage pigmentation genes involved in melanosome maturation and transport. For melanocyte maturation, however, at least some of BMP's effects may be indirect, operating through BMP-mediated activation of Lef1, followed by LEF1-mediated activation of Mitf and/or stabilization of MITF, and finally LEF1 and MITF collaborative activation of melanosome machinery genes. While hammering out the precise details is beyond the scope of this study, our findings begin to illuminate how different SCs and their lineage progenitors can utilize similar signals from the microenvironment through distinct avenues. Our findings place BMP signaling as a gatekeeper in coordinating maturation of these two SC lineages so that pigment can be transferred to differentiated hair cells on cue. Our findings further raise the intriguing possibility that differential levels of BMP signaling may be a means of accomplishing different human hair colors and coat and feather color patterning throughout the animal kingdom.
BMP signaling at the crossroads of MITF and LEF1
In placing BMP signaling upstream of Lef1 expression as well as both nuclear LEF1 and MITF, we could also begin to understand the profound effects of BMP signaling on pigment production, as well as a failure to progress appreciably past the immature McCP stage in the hair bulb. While the “rheostat model” alone might explain why some MITF targets were still expressed in Bmpr1a-null cells (i.e., low levels induce expression of genes involved in survival and proliferation) and others are diminished (i.e., high levels induce genes involved in melanosome biogenesis and transport), our data also hinted at the importance of cooperation with the transcription factor LEF1 to completely achieve differentiation. Previous studies have provided some insights into how this interplay might occur, including a physical interaction of the two factors (Yasumoto et al. 2002) or stabilization of MITF protein through enhanced WNT signaling (Ploper et al. 2015).
The temporal nature of our gene expression data set made it a powerful tool with which to elucidate the precise coordination of niche pathways and transcription factor responses that interact to achieve quiescence, activation, proliferation, and differentiation in the melanocyte lineage. Most interesting was the marked sensitivity to BMP signaling that occurred in genes whose putative promoters harbor both LEF1- and MITF-binding motifs. When coupled with the specific genes implicated, these findings support a model whereby BMP signaling acts to activate Lef1, and LEF1 then promotes nuclear accumulation of and cooperates with MITF to govern a cohort of melanocyte maturation genes. Importantly, our chromatin landscaping analyses of McSCs in the bulge and their differentiating progeny in the hair bulb provided compelling evidence in support of this model. First, they revealed that a number of regions within the Lef1 gene locus that show enhanced chromatin-accessibility upon McSC lineage progression harbor SMAD motifs, corroborating our finding that Lef1 mRNAs are down-regulated upon Bmpr1a ablation, and that Lef1 is likely a direct BMP target gene. Second, they revealed that among the cohort of genes whose expression diminishes upon Bmpr1a ablation, many show ATAC peaks whose accessibility increases in differentiating wild-type McCPs/melanocytes and that have binding motifs for SMAD, LEF1 and/or MITF. Together, these data provide new insights into how BMP signals in this lineage.
BMP signaling and melanoma: Are there lessons to be learned?
BMP signaling has been correlated with melanoma invasiveness and migration (Hsu et al. 2005; Rothhammer et al. 2005; Braig and Bosserhoff 2013; Sinnberg et al. 2018; Venkatesan et al. 2018; Gramann et al. 2019). Indeed, in zebrafish models of melanoma, signaling through BMP family member GDF6 has been shown to suppress differentiation through down-regulation of MITF and promote malignant progression (Venkatesan et al. 2018; Gramann et al. 2019). Our findings provide compelling evidence that at least in the mammalian context, blocking BMP signaling suspends the melanocyte lineage in a committed progenitor-like state, where cells are proliferative but unable to fully differentiate. In this way, BMP signaling might have both oncogenic and tumor suppressive roles in melanoma, reminiscent of studies in the lineage where both promelanogenic/differentiation and anti-melanogenic/differentiation effects have been observed (Bilodeau et al. 2001; Jin et al. 2001; Sharov et al. 2005; Yaar et al. 2006; Kawakami et al. 2008; Park et al. 2009; Han et al. 2012; Singh et al. 2012; Yang et al. 2014).
Indeed, related family member TGF-β can enhance proliferation of tumor-initiating cells when signaling is silenced, while simultaneously playing a powerful counter role in promoting invasion and metastasis (Guasch et al. 2007; Oshimori et al. 2015; David et al. 2016; David and Massagué 2018; Su et al. 2020). It is tempting to speculate that in melanoma, the juncture between the proliferative progeny of McSCs and differentiating melanocytes might become plastic, allowing cells to flip in their reliance upon BMP signaling, depending on the stage of disease progression. Future studies in mammalian models of melanoma may illuminate whether such mechanisms might be at work in disease.
Materials and methods
Contact for reagent and resource sharing
Additional requests for resources or information should be directed to and will be fulfilled by Elaine Fuchs (fuchslb@rockefeller.edu).
Experimental models and subject details
Mice and procedures
Mice were maintained in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility (Comparative Bioscience Center) of The Rockefeller University. All procedures were performed under Institutional Animal Care and Use Committee (IACUC)-approved protocols. Genotyping was performed using the primers listed in Supplemental Table S16.
Dct-eGFP mice [Tg(Dct-EGFP)NY2Gsat/Mmucd, RRID:MMRRC_032849-UCD] were obtained from Mutant Mouse Resource and Research Center (MMRRC) at University of California at Davis, an National Institutes of Health-funded strain repository, and was donated to the MMRRC by Nathaniel Heintz, Ph.D., of The Rockefeller University (Gene Expression Nervous System Atlas [GENSAT]) (Gong et al. 2003). Lef1-RFP mice were generated by Rendl et al. (2005). Dct-eGFP (mixed C57BL6/J (Jax 000664) males were crossed to CD1 females (CRL, strain 022) (to increase litter size and improve maternal care relative to C57BL6/J females) to generate mice used for qMcSC and aMcSC studies (Dct-eGFP+). Background strain was kept as consistent as possible to generate Dct-eGFP+ Lef1-RFP+ mice, where Dct-eGFP (mixed C57BL6/J) males were mated to Lef1-RFP (CD1 background) females. For all RNA and ATAC-seq experiments, male mice were used to minimize hair cycle variation and to acquire more cells and tissue per experiment.
To assess McSC proliferation (activation), two 100-µL doses of 5 mg/mL 5-ethynyl-2′-deoxyuridine (EdU) were administered by intraperitoneal injection. Mice were euthanized and skin was collected for IMF analysis 24 h after the first injection. To assess McSC and hair bulb progeny proliferation 1 wk after waxing, mice were given one 100- to 150-µL intraperitoneal injection of EdU 4 h prior to sacrifice and skin collection.
TyrCreER (Bosenberg et al. 2006) were purchased from The Jackson Laboratories (JAX 012328) and crossed to our in house colonies of Bmpr1a floxed (Mishina et al. 2002) R26YFP floxed (JAX 006148; Srinivas et al. 2001) (mixed C57BL/6 background). Fifty microliters of 2% tamoxifen in corn oil (Sigma) was administered by intraperitoneal injection for seven consecutive days starting at P21 (weaning age). In second telogen (P60-75), the back skin was waxed under isoflurane anesthesia, and buprenorphine was given for pain management. Male mice were singly housed for 1 wk after the procedure until the time of euthanasia. Male mice were used for sequencing studies and microscopy quantifications; male and female mice were used for IMF analysis (sex-matched for comparison), and female mice were used for RT-qPCR. The Lef1 KO strain (C57BL/6J background) was obtained from Dr. Rudolf Grosschedl (Kratochwil et al. 1996). Ctnnb1 floxed (C57BL/6J) were obtained from the Jackson Laboratories (JAX 004152) (Brault et al. 2001). Photographs of mice were acquired with iPhone 6, 7 Plus, and 11 Pro Max cameras and brightened in Fijj/ImageJ as necessary.
Method details
Fluorescence-activated cell sorting
To obtain single-cell suspensions for FACS, male back skin was dissected and subjected to chemical and mechanical digestion. Telogen (P58–P80 for qMcSCs) and anagen I-II (P21 for aMcSCs) skin was dissected and scraped with a dull scalpel to remove excess fat prior to incubation in trypsin EDTA with the dermis side down for 45–60 min at 37°C. The hairy side of the skin was scraped with a dull scalpel against the direction of hair growth to release cells in the HFs. Anagen skin (P9–P10 or 1 wk or 12 d postwaxing in second telogen) was digested dermis side down in 0.25% collagenase (Sigma) in HBSS (Gibco) for 25–30 min at 37°C; DNase (Roche) and 5 mM MgCl2 were sometimes added to minimize clumping. Cells were then scraped from the dermal side to release cells in the hair bulb. The resulting cell suspensions were quenched with FACS buffer (5% FBS in PBS), filtered, spun down, and washed before incubating with primary antibodies for 25 min on ice. Cells were washed with FACS buffer again before incubating in secondary antibody ∼10 min on ice. For bulk RNA-seq, cells were sorted with a 70-µm nozzle directly into TRIzol (Thermo Fisher). For single-cell RNA-seq, cells were sorted with a 100-µm nozzle into 96-well plates, gating more conservatively to minimize the capture of contaminating nonmelanocyte lineage cells.
The following antibodies were used for labeling the resulting single-cell suspensions: PerCP-Cy5.5- or APC-Cy7-Sca-1 (1:1000), e-Fluor660-CD34, BV421-CD49f or PE-Cy-7-CD49f, PE-Cy7-CD117 (1:100), Biotin-CD140a, Biotin-CD31, and Biotin-CD45 (1:150). For biotin-conjugated antibodies, cells were washed and incubated for 10 min in PE-Streptavidin (1:1500) or APC-Cy7-Streptavidin (1:500). DAPI was used for live/dead cell exclusion. Flow cytometry plots were generated using FlowJo to illustrate the strategies used to isolate each cell population. Manual compensation was performed for presentation.
Immunofluorescence and microscopy
For IMF analysis of tissue, mouse back skin was prefixed in 4% paraformaldehyde immediately after dissection for 30 min at room temperature or 1–2 h at 4°C. After fixing, tissue was washed with PBS and incubated in 30% sucrose in PBS at 4°C overnight. Tissue was embedded in OCT Compound, and frozen tissue blocks were sectioned (10–18 µm) with a cryostat (Leica) and mounted on SuperFrost Plus slides (Thermo Fisher). Slides were blocked in staining buffer (5% normal donkey serum, 1% bovine serum albumin, 2% fish gelatin, 0.3% Triton X-100 in PBS) for at least 1 h at room temperature before incubation in primary antibodies. For tyrosinase and MITF IMF, slides were further fixed in cold 100% methanol for 20 min at 20°C before blocking. Sections were washed three times with PBS prior to incubation with secondary antibodies for 1 h at room temperature. For EdU IMF, Click-iT EdU Alexa Fluor 647 imaging kit (C10340) was used according to the manufacturer's instructions before proceeding to secondary antibody labeling. Sections were washed an additional three times before mounting slides with ProLong Antifade Gold with DAPI Mountant (Thermo Fisher). Counterstaining with DAPI in PBS prior to mounting was sometimes performed to enhance DAPI signal.
Microscope images were acquired with an Axio Observer.Z1 epifluorescence microscope with a Hamamatsu ORCA-ER camera (Hamamatsu Photonics), and with an ApoTome.2 (Carl Zeiss) slider using 20×, 40×, or 63× objectives and Zen software (Carl Zeiss). Differential interference contrast (DIC) bright-field images were taken on the Axio Observer.Z1 using the Axiocam 305 camera (Zeiss). Images were processed by channel using Fiji/ImageJ using brightness/contrast where appropriate. Images with Z-stacks were displayed as max intensity projections. Optical density (OD) was determined with MetaMorph software (Molecular Devices), where OD = log10 (1/transmittance), and transmittance = transmitted light/incident light. Measurements were made of a single plane of DIC bright-field image with integrated morphometry analysis within a freehand drawn region outlining the DCT-stained region (based on maximum intensity projection).
The following primary antibodies and dilutions were used: DCT/TRP2 (D-18; 1:200–250; Santa Cruz Biotechnology sc-10451), TYRP1 (1:100; laboratory made), PEP7/tyrosinase (1:750; a kind gift from V.J. Hearing), CD104 (1:100; BD Pharmingen 553745), PMEL (anti-melanoma gp100 [EP4863{2}]; 1:750; Abcam ab137078), GFP (1:2000; Abcam ab13970), RFP (laboratory made), phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad9 (Ser465/467) (D5B10; 1:100; Cell Signaling), phospho-p38 MAPK (Thr180/Tyr182) (D3F9; 1:100; XP Cell Signaling), MITF (1:100; Abcam ab122982), and LEF1 (1:250; laboratory made). Alexa Fluor-RRX-, 488-, or 647-conjugated secondary antibodies (Life Technologies) were used at 1:500.
Electron microscopy
Skin samples were fixed in 2% glutaraldehyde, 4% PFA, and 2 mM CaCl2 in 0.1 M sodium cacodylate buffer (pH 7.2) for >1 h at room temperature, postfixed in 1% osmium tetroxide, and processed for Epon embedding; ultrathin sections (60–65 nm) were counterstained with uranyl acetate and lead citrate. Images were taken with a transmission electron microscope (Tecnai G2-12; FEI) equipped with a digital camera (AMT BioSprint29). For quantification, melanosomes were counted within hair bulb McSC progeny areas ≥10 µm2. Melanosomes were scored as immature by the presence of striations/translucency and as mature if they were opaque. Measurements were expressed as melanosome number/cell area captured.
Cell culture
The Melan-a2 mouse melanocyte cell line was generated by Sviderskaya, et al. (1995) and purchased from the Wellcome Trust Functional Genomics Cell Bank (http://anatomy.sgul.ac.uk/pages). Cells were cultured in RPMI 1640 with L-glutamine supplemented with 10% FBS, 5% penicillin–streptomycin, 200 pM cholera toxin, and 200 nM phorbol 12-myristate 13-acetate and grown at 7.5% CO2. Cells were plated 24 h before treatment in regular growth media, and serum-starved (0.5% FBS) during the course of treatment (10 ng/mL recombinant human/murine/rat BMP2, human BMP6 [Peprotech] and or 250 nM ALK inhibitor K02288 [Tocris]).
To generate a Bmpr1a KO Melan-a2 cell line with CRISPR, we first FACS-sorted Melan-a2 cells into 96-well plates to generate a clonal parental cell line to minimize experimental variability. We then purchased predesigned gRNA from Integrated DNA Technologies (IDT) (control nontargeting and DesignID:Mm.Cas9.BMPR1A.1.AB Bmpr1a, exon 9, 5′-CAUGACGCAUUAACACCGUCGUUUUAGAGCUAUGCU-3′), and cells were transfected with Lipofectamine RNAiMAX (Thermo Fisher) to deliver ribonucleoproteins (RNPs) consisting of duplexed gRNA and tracrRNA-Atto550 with Alt-R S.p. Cas9 Nuclease V3 (IDT). Media was changed 12–24 h posttransfection, and then genomic DNA was extracted with QuickExtract (Lucigen). The cut site region was PCR amplified (primers in Supplemental Table S16), and cutting efficiency was tested by T7 endonuclease assay (New England Biolabs). To generate the Bmpr1a knockout cell line, the parental clonal cell line was transfected and again sorted as single cells into 96-well plates. Gene knockout was confirmed by MiSeq (sequencing primers in Supplemental Table S16) and the rgenome.net Cas analyzer tool.
RT-qPCR
Cultured cells were washed with PBS and harvested in TRI Reagent LS (Sigma); FACS-purified cells were sorted directly into TRI Reagent LS. Total RNA was purified using the Direct-zol RNA MicroPrep kit (Zymo Research) according to the kit protocol. cDNA was made with the SuperScript VILO cDNA synthesis kit (Thermo Fisher) and diluted before RT-qPCR using SYBR Green PCR master mix (Thermo Fisher). Ppib2 was used as a housekeeping gene. Primer sequences are listed in Supplemental Table S16.
Immunoblotting
To harvest, cultured cells were washed on the plate in cold PBS, lysed in RIPA buffer (Millipore) supplemented with protease and phosphatase inhibitors (Roche), and collected by scraping. Cells were lysed for 30 min on ice and then centrifuged to collect the supernatant. Protein concentration was determined by BCA assay (Pierce) against a bovine serum albumin standard curve. Up to 50 µg of protein was run on NuPAGE 4%–12% Bis-Tris gels (Invitrogen) for 1 h at 200 V in NuPAGE MOPS SDS running buffer (Invitrogen). Protein was transferred overnight onto PVDF membrane at in NuPage transfer buffer (Invitrogen) with methanol at 30 V at 4°C. Membranes were blocked in Odyssey PBS blocking buffer for at least 1 h at room temperature before incubating with primary antibodies overnight at 4°C in Odyssey buffer with Tween-20. Membranes were washed several times in 0.1% Tween-20 in PBS before incubating with fluorescent secondary antibody.
The following primary antibodies and dilutions were used: phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad9 (Ser465/467) D5B10 (1:1000; Cell Signaling), SMAD 1/5/9 (1:1000; Invitrogen), LEF1 (1:1000; Cell Signaling), MITF (1:1000; Cell Signaling), and α-tubulin (1:10,000; Invitrogen). Fluorescent secondary antibodies were used at 1:10,000 (donkey anti-rabbit 800 and donkey anti-mouse 680 [LI-COR]). Membranes were imaged with an Odyssey CLx machine (LI-COR).
Bulk RNA-seq
Cells were FACS purified into TRI Reagent LS, and total RNA was purified using the Direct-zol RNA MicroPrep kit (Zymo Research). For McSCs, cells from 4 male mice at P60 were pooled for replicate 1, and cells from four male mice at P62 were pooled for replicate 2. For differentiating McSC progeny in the hair bulb, cells from male P10 mice were also pooled: three mice for replicate 1, two mice for replicate 2, and four mice for replicate 3. The Weill Cornell Medical College Genomic Core facility performed quality control analysis and sequencing. Briefly, RNA quality was determined by Agilent 2100 Bioanalyzer, and cDNA libraries were prepared using the Illumina TrueSeq mRNA sample preparation kit (nonstranded, poly-A selection) and sequenced on an Illumina HiSeq 4000 instrument.
Single-cell cDNA synthesis and library preparation
Single-cell RNA-seq libraries were prepared using a slightly modified version of the Smart-seq2 protocol described by Picelli et al. (2014). Cells were sorted into 96-well plates containing lysis buffer and flash frozen in liquid nitrogen and stored at -80°C until further processing. External RNA (ERCC) spike-ins were implemented to assess technical noise associated with library preparation and sequencing and to facilitate comparative analyses (Brennecke et al. 2013). Plates were thawed on ice, then lysed at 72°C for 3 min. The RNA was subject to reverse transcription (4U/μL Maxima H transcriptase), template switching reaction, and whole transcriptome amplification, and cDNA was purified with AMPure XP beads. To test for low quality libraries or empty wells, RT-qPCR for GAPDH was performed before proceeding. Illumina sequencing libraries were then prepared using the Nextera XT DNA library preparation kit (Illumina). After barcoding, samples were pooled and cleaned up with 0.9X by volume AMPure XP beads (Beckman Coulter). Pooled libraries were sequenced on an Illumina NextSeq 500 instrument using a 75-bp paired-end read mid-output setting.
ATAC library preparation
Cells were FACS purified using an 85-µm nozzle for gentler sorting to improve viability. qMcSCs were isolated as described in Supplemental Figure S2A from Dct-eGFP+× CD1 second telogen skin at P60 (replicate 1, cells from three male mice pooled) and P80 (replicates 2 and 3, cells from four male mice pooled for each); differentiating progeny were isolated from Dct-eGFP+ (C57BL6/J background with some CD1) anagen skin at P9 (replicate 1, cells from one male mouse; replicates 2 and 3, cells from two male mice pooled for each) using the following FACS strategy: dump (CD31, CD45, CD140a)−, Sca-1−, CD34−, eGFPhigh, and CD117+ cells. Library preparation was performed as described (Buenrostro et al. 2013) with some modifications. Briefly, after sorting, cells were washed with PBS, pelleted, and resuspended in cold lysis buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Igepal CA-630). Lysis buffer was removed by centrifugation, and samples were incubated in the transposition reaction for 30 min at 37°C (Illumina Nextera DNA preparation kit; TDE1 enzyme, 10 µL). The reaction was terminated by adding Tagmentation Clean Up buffer (300 mM EDTA, 900 mM NaCl) and purified with the Qiagen MiniElute PCR purification kit. The DNA was then PCR amplified with barcode identifiers for 12–18 cycles, and the products taken at three cycling times were tested by D1000 tape station (Agilent). Optimal samples were then pooled and bead purified before sequencing on NextSeq High Output 75 single read (40 × 40-bp paired-end).
Schematics
Some graphics from SMART Servier Medical Art (https://smart.servier.com) were used to make to make schematics. Other illustrations were made in Adobe Illustrator and Microsoft PowerPoint.
Quantification and statistical analysis
Statistical analyses
Statistical tests for microscopy quantifications and RT-qPCR were performed with GraphPad Prism 7. Column data was first subjected to D'Agostino and Pearson normality testing. For data without normal distribution, two-tailed Mann–Whitney test was used to compare two groups, and Kruskal–Wallis test with Dunn's with multiple comparisons was used to compare between more than two groups. For RT-qPCR, where n number was too low for normality testing, unpaired t-test or ordinary one-way ANOVA with Dunnett's multiple comparison tests were performed.
Bulk RNA-seq analysis
Sequence and transcript coordinates for mouse mm10 UCSC genome and gene models were retrieved from the Bioconductor Bsgenome.Mmusculus.UCSC.mm10 (version 1.4.0) and TxDb.Mmusculus.UCSC.mm10.knownGene (version 3.4.0) Bioconductor libraries, respectively. Read length was 51 bp. Transcript expressions were calculated using the Salmon quantification software (version 0.8.2) (Patro et al. 2017) and gene expression levels as TPMs and counts were obtained using Tximport (version 1.8.0) (Soneson et al. 2015). Normalization and rlog transformation of raw read counts in genes were performed using DESeq2 (version 1.20.0) (Love et al. 2014). Variability between samples was assessed with hierarchical clustering and heat maps of between sample distances implemented in the Pheatmap R package (1.0.10), as described by Love et al. (2014), and sample similarity is expressed as Euclidean distance as well as Pearson correlation. Published HFSC RNA-seq data from Ge et al. (2017) was obtained from Gene Expression Omnibus (GEO) accession number GSE89928 (GSM2656733 CL_BuA and GSM2656734 CL_BuB).
Single-cell RNA-seq analysis
Sequence and transcript coordinates for mouse release M23 (GRCm38.p6) genome and gene models were downloaded from GENCODE (https://www.gencodegenes.org/mouse/release_M23.html). Adaptors were first trimmed from reads using Skewer (version 0.2.2). Sequencing reads for scRNA-seq libraries were aligned to the mouse reference genome, combined with sequences for ERCC spike-ins as artificial chromosomes, using STAR (version 2.5.2a) (Dobin et al. 2013) with default parameters for paired-end reads. Transcript expressions were calculated using the Salmon quantification software (version 0.14.1) (Patro et al. 2017) and gene expression levels as TPMs and counts were obtained using Tximport (version 1.12.3) (Soneson et al. 2015). TPMs were transformed to log2(TPM + 1). For downstream analyses, cells with <1250 genes detected per cell and genes expressed in <5% of the cell population were removed. We also excluded cells expressing lower levels of the lineage marker Dct (log2(TPM+1) <6) and those expressing the HF lineage gene Krt15 [log2(TPM + 1) > 6]. After filtering, the number of cells in our data set were as follows: qMcSCs n = 104 cells (n = 3 mice), aMcSCs n = 193 cells (n = 3 mice), hair bulb McSC progeny n = 308 cells (n = 2 mice), Bmpr1a wild-type control n = 205 cells (n = 1 mouse), Bmpr1a heterozygous n = 304 cells (n = 2 mice), and Bmpr1a-null n = 421 cells (n = 2 mice).
Analyses and visualization of data were conducted in a Python environment built on the Numpy, SciPy, matplotlib, scikit-learn package, and Pandas libraries (Pedregosa et al. 2011). Batch effect variation between sequencing runs and biological replicates was assessed by examining the Euclidean distances in PCA space between replicates within each specific time point data set (corresponding to one of qMcSCs, aMcSCs, or various stages of melanocyte differentiation) versus across time points, which showed closer relationships between replicates compared with variation between biologically distinct time points. Furthermore, PCA analysis of the only data set that showed some evidence of replicate-specific clustering patterns (P10 anagen VI) captured biologically relevant differences in gene expression separating replicates, including variation in M/G1 cell cycle phase, melanosome organization, and BMP signaling pathway genes. This strongly suggests the differences between P10 replicates is likely to represent true biological variation between animals in this highly dynamic cell population, rather than technical batch effects.
To distinguish true biological variability in gene expression from technical noise, we used a statistical model for identifying highly variable genes compared with ERCC spike-ins as described by Brennecke et al. (2013). Briefly, we used a custom script based on the methodology described by Brennecke et al. 2013, running in R version 3.6.1, to identify those genes with higher level of variation (at least 10% above the technical variation) and false discovery rate (FDR) value less than 0.1.
To identify cell clusters and visualize the data, we first centered and scaled the highly variable gene data set and performed PCA on the list of highly variable genes. A Jack-straw approach for the first 100 components (based on Seurat v3.1.1) (Stuart et al. 2019) was implemented in R to calculate a statistical significance for each principal component (PC), and components with P < 0.05 were retained for future analysis. Significant PCs were used as input for nonlinear dimensionality reduction, performed using UMAP implemented in scikit-learn. To identify clusters we used a graph-based clustering approach based on building a k-nearest neighbors (kNN) graph and clustering with the Louvain algorithm (with k set to one-fifth of the data set size, and resolution parameter of 1 × 10−4). Euclidean distance in PCA space served as input for both UMAP generation and Louvain clustering. During second level analysis, we used the same strategy for dimensionality reduction, clustering and visualization, with variable genes and PCs determined separately for each of the main cell clusters.
To construct pseudotime trajectories we used the same strategy for dimensionality reduction and clustering of each data set as before, constructed a principal graph based on the community analysis, and computed pseudotime values for each cell using Monocle 3 (version 0.2.0) (Trapnell et al. 2014; Cao et al. 2019). To identify genes that change as a function of pseudotime we used the “graph_test()” function of Monocle 3, to implement Moran's I test. Genes were considered to be significantly changing with pseudotime with a q-value<1.0 × 10−4.
Differential gene expression was used to identify genes specific to each cluster. Briefly, we used raw count matrices for the biologically variable genes within each data set and applied them to the DESeq2 package (version 1.24.0) (Love et al. 2014) using R. We used a negative binomial fit to model differential gene expression, factored the data set based on the Louvain cluster assignments, and used a threshold of 0.75 to construct Wald tests of significance. Genes were considered to be differentially expressed if their log2(fold change) ≥|1| and P-adjusted value (q) ≤0.05. Lowly expressed differential genes (baseMean expression ≤5) were discarded from visualization and further analysis. The expression levels of specific genes of interest were visualized as log2(TPM + 1) values on the corresponding UMAP representation of the data. The “stemness” and “differentiation” signature gene sets used to generate cumulative “stemness” or “differentiation” scores are the top 100 up-regulated or down-regulated differentially expressed genes between McSCs and differentiating hair bulb McSC progeny, and largely encompassed genes previously implicated in their biology. The “AddModuleScore” function from Seurat v3.1.1 was used to calculate the average expression levels of each gene set on a single-cell level, subtracted by the aggregated expression of control feature sets, as was originally described in Tirosh et al. (2016). Resulting stemness or differentiation score values for each cell are color-coded on corresponding UMAP visualizations of the data, or on violin plots binned by subcluster cell identity.
Gene ontology (GO) analyses of differentially expressed genes were performed using PANTHER tools overrepresentation test (version 14.0), and annotated using the GO biological process complete list (Thomas et al. 2006; Mi et al. 2019). Significant associations were calculated using the Fisher's exact test, and multiple comparisons were corrected for using the false discovery rate (FDR). GO terms were arranged hierarchically, and parent terms with greater than twofold enrichment and FDR < 0.05 were selected for presentation. GO terms explicitly linked to tissues other than skin/HFs were excluded from presentation in the main text, but are included in Supplemental Tables. Where indicated, GO analyses were also annotated using DAVID Bioinformatics Resources 6.8 “Functional Annotation,” “BP Direct” lists (Huang et al. 2009a,b).
To visualize changes in gene expression across data sets, heat maps were constructed in a Python environment built on the Numpy, SciPy, matplotlib, scikit-learn package and pandas libraries (Pedregosa et al. 2011). For specific GO annotations, the corresponding Mus musculus gene lists were obtained from AmiGO 2 through the Gene Ontology Consortium. Each GO gene list was used to filter the biologically variable genes associated with the data set to generate normalized expression matrices, and genes characteristic for clusters of cells (expressed in >50% of the cluster cells) were retained for visualization. As pseudotime analysis was based solely on biologically variable genes, the pseudotime heat maps by-passed this filtering step. Each expression matrix was scaled and centered, and hierarchical linkages between genes calculated based on pairwise correlation distances using the farthest point algorithm. The matplotlib extension Seaborn visualization library (version 0.9.1) was used to create heat maps using the clustermap() function with the previously generated gene-based hierarchical linkages fed in.
To generate comprehensive gene sets based on GO term analyses, for example for BMP or canonical WNT signaling pathways, the corresponding Mus musculus gene lists were obtained from AmiGO 2 through the Gene Ontology Consortium. BMP or canonical WNT target gene lists were culled from mouse literature. To be included, target genes had to be validated as direct targets of the signaling pathway in at least two mesenchymal or epithelial tissues, and be expressed in our data sets. Complete target gene lists are available in Supplemental Table S8. Cell cycle stage specific gene lists were obtained from Macosko et al. (2015). To generate a global proliferation gene set score across all stages of the cell cycle an amalgamated list of cell cycle stage specific gene expression was used. The “AddModuleScore” function from Seurat v3.1.1 was used to calculate the average expression levels of each gene set on a single-cell level, subtracted by the aggregated expression of control feature sets, as was originally described in Tirosh et al. (2016). Resulting gene set scores for each cell are color-coded on corresponding UMAP visualizations of the data, or on violin plots binned by subcluster cell identity.
Promoter analysis
HOMER genome-wide motif predictions were used to scan for LEF1, MITF, or SMAD motifs within 2.5 kb upstream of the transcription start site (TSS) of the genes that were significantly (Padj ≤ 0.05, baseMean ≥ 5) changed in the Bmpr1a-null cluster relative to the control cluster. Motifs had to be found in at least one TSS to be considered positive for that group. Genes were then grouped based on the presence or absence of motifs in the TSS. The difference in fold change between two groups were tested by Wilcoxon test with the null hypothesis that there is no difference in the distribution between the test groups. The results were plotted with R (3.6.1) and ggplot2.
ATAC-seq analysis
ATAC-seq reads are aligned to the mm10 genome from the Bsgenome.Mmusculus.UCSC.mm10 Bioconductor package (version 1.4.0) using Rsubread'’s align method in paired-end mode with fragments between 1 and 5000 base-pairs considered properly paired (Liao et al. 2013). Normalized, fragment signal bigWigs were created using the rtracklayer package. Peak calls were made with MACS2 software in BAMPE mode (Zhang et al. 2008; Feng et al. 2012) and sequences under these peaks used with the motifMatcher Bioconductor package and JASPAR motif database (Fornes et al. 2020) to identify known motifs within ATAC-seq peaks. Differential ATAC-seq signal was identified using the DESeq2 package (Ross-Innes et al. 2012; Love et al. 2014) and enrichment for motifs identified using the Limma packages geneSetTest function (Ritchie et al. 2015). Integrative Genomics Viewer (IGV) was used for visualization.
Data and software availability
The data sets and code generated during this study are available at GEO (GSE147299; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147299).
Competing interest statement
E.F. is a scientific advisory board member of L'Oreal and of Arsenal Biosciences.
Supplementary Material
Acknowledgments
We thank all current and former Fuchs laboratory members for their feedback on this project, especially B. Hurwitz (for help generating the Bmpr1a KO cell line), L. Yuan, S. Baksh, P. Miao, H. Yang, M. Laurin, S. Ellis, S. Naik, R.C. Adam, and K. Gonzales. We thank N.R.I.’s Thesis Committee (S. Tavazoie and M.E. Hatten) and the Rockefeller University Dean's Office, especially A. Morris. We thank M. Sribour (for help with Lef1 KO mice), E. Wong, J. Racelis, M. Nikolova, J. Levorse, P. Nasseir, S. Mereby, and S. Chai for technical assistance. We thank the Rockefeller University Flow Cytometry Resource Center for FACS (S. Mazel, director), the Rockefeller University Genomics Resource Center for scRNA-seq (C. Zhao, director), N. Saplop for EM training, the Rockefeller University Comparative Bioscience Center for mouse care, the Rockefeller University Bio-Imaging Resource Center for assistance with microscopy and image quantification (A. North, director), and the Genomics Core at Weill Cornell Medical College for RNA-seq. E.F. is an Investigator of the Howard Hughes Medical Institute. N.R.I. is the recipient of a National Institute of Arthritis and Musculoskeletal and Skin Diseases National Research Service Award (NRSA; F31AR073110). K.S.S. is a recipient of a Canadian Institute of Health Research Postdoctoral Fellowship. N.C.G. is a recipient of a National Cancer Institute NRSA F32 Postdoctoral Award (F32CA221353) and a Burroughs Wellcome Fund Postdoctoral Enrichment Program. The work was supported by grants from the National Institutes of Health to E.F. (AR050452) and the Melanoma Research Alliance.
Author contributions: N.R.I. and E.F. designed the experiments and wrote the manuscript. K.S.S. performed single-cell RNA-seq analysis. Y.Y. performed motif analysis and provided bioinformatics/experimental support. N.C.G. performed preprocessing of single-cell data and provided bioinformatics/experimental support. H.A.P. performed EM. L.H. and L.P. maintained animal colonies and helped perform mouse work. T.S.C. performed bulk RNA and ATAC-seq analyses. N.R.I. performed most experiments. E.F. supervised this project. All authors provided valuable intellectual input and reviewed, edited, and approved the final manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340281.120.
References
- Adam RC, Yang H, Ge Y, Lien WH, Wang P, Zhao Y, Polak L, Levorse J, Baksh SC, Zheng D, et al. 2018. Temporal layering of signaling effectors drives chromatin remodeling during hair follicle stem cell lineage progression. Cell Stem Cell 22: 398–413.e7. 10.1016/j.stem.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2019. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37: 38–44. 10.1038/nbt.4314 [DOI] [PubMed] [Google Scholar]
- Bilodeau ML, Greulich JD, Hullinger RL, Bertolotto C, Ballotti R, Andrisani OM. 2001. BMP-2 stimulates tyrosinase gene expression and melanogenesis in differentiated melanocytes. Pigment Cell Res 14: 328–336. 10.1034/j.1600-0749.2001.140504.x [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech 2008: P10008 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, Nogueira C, Horner JW II, Depinho R, Chin L. 2006. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis 44: 262–267. 10.1002/dvg.20205 [DOI] [PubMed] [Google Scholar]
- Botchkareva NV, Khlgatian M, Longley BJ, Botchkarev VA, Gilchrest BA. 2001. SCF/c-kit signaling is required for cyclic regeneration of the hair pigmentation unit. FASEB J 15: 645–658. 10.1096/fj.00-0368com [DOI] [PubMed] [Google Scholar]
- Braig S, Bosserhoff AK. 2013. Death inducer-obliterator 1 (Dido1) is a BMP target gene and promotes BMP-induced melanoma progression. Oncogene 32: 837–848. 10.1038/onc.2012.115 [DOI] [PubMed] [Google Scholar]
- Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. 2001. Inactivation of the β-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128: 1253–1264. [DOI] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, et al. 2013. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 10: 1093–1095. 10.1038/nmeth.2645 [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for multimodal regulatory analysis and personal epigenomics. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566: 496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Goodall J, Denat L, Rodriguez M, Nuciforo P, Hoek KS, Testori A, Larue L, Goding CR. 2006. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev 20: 3426–3439. 10.1101/gad.406406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Pasolli HA, Giannopoulou EG, Guasch G, Gronostajski RM, Elemento O, Fuchs E. 2013. NFIB is a governor of epithelial-melanocyte stem cell behaviour in a shared niche. Nature 495: 98–102. 10.1038/nature11847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Massagué J. 2018. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol 19: 419–435. 10.1038/s41580-018-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, Iacobuzio-Donahue CA, Massagué J. 2016. TGF-β Tumor suppression through a lethal EMT. Cell 164: 1015–1030. 10.1016/j.cell.2016.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Raible DW, Moon RT. 2000. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev 14: 158–162. [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 48: D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CP, Polak L, Yuan S, Elemento O, et al. 2017. Stem cell lineage infidelity drives wound repair and cancer. Cell 169: 636–650.e14. 10.1016/j.cell.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genander M, Cook PJ, Ramsköld D, Keyes BE, Mertz AF, Sandberg R, Fuchs E. 2014. BMP signaling and Its pSMAD1/5 target genes differentially regulate hair follicle stem cell lineages. Cell Stem Cell 15: 619–633. 10.1016/j.stem.2014.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding CR, Arnheiter H. 2019. MITF – the first 25 years. Genes Dev 33: 983–1007. 10.1101/gad.324657.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925. 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- Gramann AK, Venkatesan AM, Guerin M, Ceol CJ. 2019. Regulation of zebrafish melanocyte development by ligand-dependent BMP signaling. Elife 8: e50047 10.7554/eLife.50047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guasch G, Schober M, Pasolli HA, Conn EB, Polak L, Fuchs E. 2007. Loss of TGFβ signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12: 313–327. 10.1016/j.ccr.2007.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Beppu H, Lee YK, Georgopoulos K, Larue L, Li E, Weiner L, Brissette JL. 2012. A pair of transmembrane receptors essential for the retention and pigmentation of hair. Genesis 50: 783–800. 10.1002/dvg.22039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu MY, Rovinsky S, Penmatcha S, Herlyn M, Muirhead D. 2005. Bone morphogenetic proteins in melanoma: angel or devil? Cancer Metastasis Rev 24: 251–263. 10.1007/s10555-005-1575-y [DOI] [PubMed] [Google Scholar]
- Hsu Y-C, Pasolli HA, Fuchs E. 2011. Dynamics between stem cells, niche and progeny in the hair follicle. Cell 144: 92–105. 10.1016/j.cell.2010.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Li L, Fuchs E. 2014. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157: 935–949. 10.1016/j.cell.2014.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, DW, Sherman BT, Lempicki RA. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, DW, Sherman BT, Lempicki RA. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jin EJ, Erickson CA, Takada S, Burrus LW. 2001. Wnt and BMP signaling govern lineage segregation of melanocytes in the avian embryo. Dev Biol 233: 22–37. 10.1006/dbio.2001.0222 [DOI] [PubMed] [Google Scholar]
- Joshi SS, Tandukar B, Pan L, Huang JM, Livak F, Smith BJ, Hodges T, Mahurkar AA, Hornyak TJ. 2019. CD34 defines melanocyte stem cell subpopulations with distinct regenerative properties. PLoS Genet 15: e1008034 10.1371/journal.pgen.1008034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Kimura S, Kawa Y, Kato M, Mizoguchi M, Soma Y. 2008. BMP-4 upregulates Kit expression in mouse melanoblasts prior to the Kit-dependent cycle of melanogenesis. J Invest Dermatol 128: 1220–1226. 10.1038/sj.jid.5701136 [DOI] [PubMed] [Google Scholar]
- Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. 2007. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci 104: 10063–8. 10.1073/pnas.0703004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratochwil K, Dull M, Fariñas I, Galceran J, Grosschedl J. 1996. Lef 1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev 10: 1382–1394. 10.1101/gad.10.11.1382 [DOI] [PubMed] [Google Scholar]
- Laron EA, Aamar E, Enshell-Seijffers D. 2018. The mesenchymal niche of the hair follicle induces regeneration by releasing primed progenitors from inhibitory effects of quiescent stem cells. Cell Rep 24: 909–921.e3. 10.1016/j.celrep.2018.06.084 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2013. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41: e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C, Booker RC, Morrison S, Le LQ. 2017. Identification of hair shaft progenitors that create a niche for hair pigmentation. Genes Dev 31: 744–756. 10.1101/gad.298703.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Fisher DE. 2007. Melanocyte biology and skin pigmentation. Nature 445: 843–850. 10.1038/nature05660 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161: 1202–1214. 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MS, Seabra MC. 2001. The melanosome: membrane dynamics in black and white. Nat Rev Mol Cell Biol 2: 738–748. 10.1038/35096009 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res 47: D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Hanks MC, Miura S, Tallquist MD, Behringer RR. 2002. Generation of Bmpr/Alk3 conditional knockout mice. Genesis 32: 69–72. 10.1002/gene.10038 [DOI] [PubMed] [Google Scholar]
- Moon H, Donahue LR, Choi E, Scumpia PO, Lowry WE, Grenier JK, Zhu J, White AC. 2017. Melanocyte stem cell activation and translocation initiate cutaneous melanoma in response to UV exposure. Cell Stem Cell 21: 665–678.e6. 10.1016/j.stem.2017.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama M, Osawa M, Mak SS, Ohtsuka T, Yamamoto N, Han H, Delmas V, Kageyama R, Beermann F, Larue L, et al. 2006. Notch signaling via Hes1 transcription factor maintains survival of melanoblasts and melanocyte stem cells. J Cell Biol 173: 333–339. 10.1083/jcb.200509084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort RL, Jackson IJ, Patton EE. 2015. The melanocyte lineage in development and disease. Development 142:620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura EK. 2011. Melanocyte stem cells: a melanocyte reservoir in hair follicles for hair and skin pigmentation. Pigment Cell Melanoma Res 24: 401–410. 10.1111/j.1755-148X.2011.00855.x [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, Jackson IJ, Barrandon Y, Miyachi Y, Nishikawa S. 2002. Dominant role of the niche in melanocyte stem-cell fate determination. Nature 416: 854–860. 10.1038/416854a [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, Fisher DE. 2005. Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307: 720–724. 10.1126/science.1099593 [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Suzuki M, Igras V, Du J, Lonning S, Miyachi Y, Roes J, Beermann F, Fisher DE. 2010. Key roles for transforming growth factor β in melanocyte stem cell maintenance. Cell Stem Cell 6: 130–140. 10.1016/j.stem.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Egawa G, Mak SS, Moriyama M, Freter R, Yonetani S, Beermann F, Nishikawa S. 2005. Molecular characterization of melanocyte stem cells in their niche. Development 132: 5589–5599. 10.1242/dev.02161 [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. 2012. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10: 63–75. 10.1016/j.stem.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, Oristian D, Fuchs E. 2015. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160: 963–976. 10.1016/j.cell.2015.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-Y, Wu C, Yaar M, Stachur CM, Kosmadaki M, Gilchrest BA. 2009. Role of BMP-4 and its signaling pathways in cultured human melanocytes. Int J Cell Biol 2009: 750482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14: 417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, et al. 2011. Scikit-learn: machine learning in Python. J Mach Learn Res 12: 2825–2830. [Google Scholar]
- Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. 2013. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 11: 1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using smart-seq2. Nat Protoc 9: 171–181. 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- Ploper D, Taelman VF, Robert L, Perez BS, Titz B, Chen HW, Graeber TG, von Euw E, Ribas A, De Robertis EM. 2015. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc Natl Acad Sci 112: E420–E429. 10.1073/pnas.1424576112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani P, Takeo M, Chou W, Myung P, Bosenberg M, Chin L, Taketo MM, Ito M. 2011. Coordinated activation of Wnt in epithelial and melanocyte stem cells initiates pigmented hair regeneration. Cell 145: 941–955. 10.1016/j.cell.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M, Lewis L, Fuchs E. 2005. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol 3: e331 10.1371/journal.pbio.0030331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK. 2005. Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res 65: 448–456. [PubMed] [Google Scholar]
- Saito H, Yasumoto K, Takeda K, Takahashi K, Fukuzaki A, Orikasa S, Shibahara S. 2002. Melanocyte-specific microphthalmia-associated transcription factor isoform activates its own gene promoter through physical interaction with lymphoid-enhancing factor 1. J Biol Chem 277: 28787–28794. 10.1074/jbc.M203719200 [DOI] [PubMed] [Google Scholar]
- Sharov AA, Fessing M, Atoyan R, Sharova TY, Haskell-Luevano C, Weiner L, Funa K, Brissette JL, Gilchrest BA, Botchkarev VA. 2005. Bone morphogenetic protein (BMP) signaling controls hair pigmentation by means of cross-talk with the melanocortin receptor-1 pathway. Proc Natl Acad Sci 102: 93–98. 10.1073/pnas.0408455102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Abbas WA, Tobin DJ. 2012. Bone morphogenetic proteins differentially regulate pigmentation in human skin cells. J Cell Science 125: 4306–4319. 10.1242/jcs.102038 [DOI] [PubMed] [Google Scholar]
- Sinnberg T, Niessner H, Levesque MP, Dettweiler C, Garbe C, Busch C. 2018. Embryonic bone morphogenetic protein and nodal induce invasion in melanocytes and melanoma cells. Biol Open 7: bio032656 10.1242/bio.032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4: 1521 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol 1: 4 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive integration of single-cell data. Cell 177: 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Morgani SM, David CJ, Wang Q, Er EE, Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al. 2020. TGF-β orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature 577: 566–571. 10.1038/s41586-019-1897-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Lee W, Mohri Y, Takeo M, Lim CH, Xu X, Myung P, Atit RP, Taketo MM, Moubarak RS, et al. 2019. A novel mouse model demonstrates that oncogenic melanocyte stem cells engender melanoma resembling human disease. Nat Commun 10: 5023 10.1038/s41467-019-12733-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sviderskaya EV, Wakeling WF, Bennett DC. 1995. A cloned, immortal line of murine melanoblasts inducible to differentiate to melanocytes. Development 121: 1547–1557. [DOI] [PubMed] [Google Scholar]
- Takeda K, Yasumoto K, Takada R, Takada S, Watanabe K, Udono T, Saito H, Takahashi K, Shibahara S. 2000. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J Biol Chem 275: 14013–6. 10.1074/jbc.C000113200 [DOI] [PubMed] [Google Scholar]
- Takeo M, Lee W, Rabbani P, Sun Q, Hu H, Lim CH, Manga P, Ito M. 2016. Ednrb governs regenerative response of melanocyte stem cells by crosstalk with Wnt signaling. Cell Rep 15: 1291–1302. 10.1016/j.celrep.2016.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. 2006. Applications for protein sequence–function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res 34: W645–W650. 10.1093/nar/gkl229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352: 189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. Pseudo-temporal ordering of individual cells reveals dynamics and regulators of cell fate decisions. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Genderen C, Okamura RM, Fariñas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. 1994. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev 8: 2691–2703. 10.1101/gad.8.22.2691 [DOI] [PubMed] [Google Scholar]
- Venkatesan AM, Vyas R, Gramann AK, Dresser K, Gujja S, Bhatnagar S, Chhangawala S, Gomes CBF, Xi HS, Lian CG, et al. 2018. Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma. J Clin Invest 128: 294–308. 10.1172/JCI92513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RN, Green J, Wang Z, Deng Y, Qiao M, Peabody M, Zhang Q, Ye J, Yan Z, Denduluri S, et al. 2014. Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis 1: 87–105. 10.1016/j.gendis.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaar M, Wu C, Park HY, Panova I, Schutz G, Gilchrest BA. 2006. Bone morphogenetic protein-4, a novel modulator of melanogenesis. J Biol Chem 281: 25307–25314. 10.1074/jbc.M600580200 [DOI] [PubMed] [Google Scholar]
- Yang J, Wang J, Pan L, Li H, Rao C, Zhang X, Niu G, Qu J, Hou L. 2014. BMP4 is required for the initial expression of MITF in melanocyte precursor differentiation from embryonic stem cells. Exp Cell Res 320: 54–61. 10.1016/j.yexcr.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Yang H, Adam RC, Ge Y, Hua ZL, Fuchs E. 2017. Epithelial–mesenchymal micro-niches govern stem cell lineage choices. Cell 169: 483–496.e13. 10.1016/j.cell.2017.03.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumoto K, Takeda K, Saito H, Watanabe K, Takahashi K, Shibahara S. 2002. Microphthalmia-associated transcription factor interacts with LEF-1, a mediator of Wnt signaling. EMBO J 21: 2703–2714. 10.1093/emboj/21.11.2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Ma S, Rachmin I, He M, Baral P, Choi S, Gonçalves WA, Shwartz Y, Fast EM, Su Y, et al. 2020. Hyperactivation of sympathetic nerves drives depletion of melanocyte stem cells. Nature 577: 676–681. 10.1038/s41586-020-1935-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets and code generated during this study are available at GEO (GSE147299; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147299).