In this study, Petrenko et al. examined the role of pancreatic islets in β-cell regeneration after the massive ablation of β cells by doxycycline-induced expression of diphtheria toxin A (DTA) in Insulin-rtTA/TET-DTA mice. Using reporter genes expressing α- and β-cell-specific fluorescent proteins in these mice they could follow the fate of α and β cells separately, and they found that DTA induction resulted in an acute hyperglycemia, which was accompanied by dramatic changes in gene expression in residual β cells, and in arrhythmic Bmal1-deficient mice, which lack circadian clocks, no compensatory β-cell proliferation was observed, and the β-cell ablation led to aggravated hyperglycemia, hyperglucagonemia, and fatal diabetes.
Keywords: circadian clockwork, pancreatic α and β cells, Insulin-rtTA/TET-DTA mouse model, diabetes, glucose metabolism, β-cell proliferation, β-cell regeneration
Abstract
Circadian clocks in pancreatic islets participate in the regulation of glucose homeostasis. Here we examined the role of these timekeepers in β-cell regeneration after the massive ablation of β cells by doxycycline-induced expression of diphtheria toxin A (DTA) in Insulin-rtTA/TET-DTA mice. Since we crossed reporter genes expressing α- and β-cell-specific fluorescent proteins into these mice, we could follow the fate of α- and β cells separately. As expected, DTA induction resulted in an acute hyperglycemia, which was accompanied by dramatic changes in gene expression in residual β cells. In contrast, only temporal alterations of gene expression were observed in α cells. Interestingly, β cells entered S phase preferentially during the nocturnal activity phase, indicating that the diurnal rhythm also plays a role in the orchestration of β-cell regeneration. Indeed, in arrhythmic Bmal1-deficient mice, which lack circadian clocks, no compensatory β-cell proliferation was observed, and the β-cell ablation led to aggravated hyperglycemia, hyperglucagonemia, and fatal diabetes.
The circadian system allows organisms to synchronize physiology and behavior to the changes of geophysical time repeatable every 24 h. In mammals, the clock network is organized in a hierarchical manner. It comprises a master pacemaker, located in the paired suprachiasmatic nuclei (SCN) of the hypothalamus, that orchestrates the timing of peripheral oscillators situated in all organs on a daily basis (Dibner 2020). The molecular clocks rely on two coupled auto-regulatory transcriptional–translational negative feedback loops (TTFLs). In the primary TTFL, the transcriptional activators BMAL1 and CLOCK form a heterodimer that activates transcription of the genes encoding the negative limb members PER1–3 and CRY1–2 (Cox and Takahashi 2019; Sinturel et al. 2020). CRY and PER proteins form complexes that translocate to the nucleus and inhibit the action of BMAL1 and CLOCK on their own transcription (Aryal et al. 2017). In the secondary TTFL, members of the nuclear orphan receptor families ROR and REV-ERB drive the circadian transcription of the positive limb members BMAL1 and CLOCK (Dibner et al. 2010).
A coupling between the circadian oscillator and the cell division cycle has been observed in cultured fibroblasts (Nagoshi et al. 2004) and in tissues with high proliferative activity, such as oral mucosa, skin (Bjarnason et al. 2001), and intestinal epithelium (Karpowicz et al. 2013; Stokes et al. 2017). Indeed, core clock components were demonstrated to directly drive the expression of key cell cycle genes comprising Wee1, Cyclins, and Myc1 (Matsuo et al. 2003), and to gate cytokinesis to a specific time within the circadian cycle (Nagoshi et al. 2004; Bieler et al. 2014). Both the cell division cycle and the circadian clock undergo significant changes upon malignant transformation, although no clear causal links between the two processes are known (Mannic et al. 2013; Gaucher et al. 2018).
The importance of temporal coordination of cell proliferation has emerged in the context of reparative regeneration in tissues that bear intrinsic regenerative capacity, such as liver, intestine, and skeletal muscle (Matsuo et al. 2003; Karpowicz et al. 2013; Chatterjee et al. 2015; Bellet et al. 2016; Stokes et al. 2017). Rodent studies suggest that β cells have a limited potential for regeneration via replication of existing β cells (Dor et al. 2004; Nir et al. 2007; Stolovich-Rain et al. 2012; Klochendler et al. 2016) or trans-differentiation from adjacent ductal epithelial cells (Xu et al. 2008), α cells (Thorel et al. 2010; Chakravarthy et al. 2017), and δ cells (Chera et al. 2014). Unlike fast regenerating tissues including liver, the regeneration of β cells is a long-lasting process that takes a few weeks and even months in mice (Nir et al. 2007; Cheng et al. 2017). Core clock components may play a role in regulating β-cell division. Indeed, mice lacking at least one allele of Bmal1 in β cells fail to increase β-cell mass in response to diet-induced obesity (Rakshit et al. 2016), and siRNA-mediated down-regulation of RORγ attenuated glucose-driven induction of cell cycle-related genes and the proliferation rate in INS-1E cell line (Schmidt et al. 2016). Unraveling mechanisms of regulation of β-cell regeneration is of a fundamental clinical importance in the search of new therapeutic approaches for management of diabetes mellitus.
Molecular clocks operative in pancreatic islets (Pulimeno et al. 2013) play a critical role in islet cell physiology and in regulating glucose homeostasis (Marcheva et al. 2010; Petrenko et al. 2017b; Sinturel et al. 2020). Indeed, mice harboring whole-body or tissue-specific null alleles for essential core clock components develop hyperglycemia, hypoinsulinemia and glucose intolerance (Marcheva et al. 2010; Dyar et al. 2014; Perelis et al. 2015; Rakshit et al. 2016). Although perturbation of the islet clocks emerges as a part of type 2 diabetes (T2D) pathogenesis in humans (Petrenko et al. 2020), it remains unclear whether alterations of the molecular clockwork precede development of T2D or stem from the disease progression. Thus, understanding the functional link between changes of the molecular clockwork, islet dysfunction and β-cell regeneration potential is an important challenge. In this work we studied the diurnal regulation of β-cell proliferation and profiled the circadian transcriptomes of α and β cells following massive β-cell ablation in the Insulin-rtTA/TET-DTA mouse model in the presence or absence of functional clocks.
Results
β-Cell regeneration follows a diurnal pattern
In order to study the impact of the circadian system on β-cell dysfunction and regeneration, we used Insulin-rtTA/TET-DTA transgenic mice, in which the ablation of ∼80% of β cells can be induced in a controlled manner (Nir et al. 2007). This mouse strain was crossed with triple-reporter mice (Petrenko et al. 2017b) that expressed fluorescent protein reporters specific for α cells (ProGlucagon [Gcg]-Venus) and β cells (rat Insulin2 promoter [RIP]-Cherry), along with the ubiquitously expressed circadian bioluminescence reporter Period2::Luciferase (Per2::Luc) (Supplemental Fig. S1A). The resulting mouse model encompassing five transgenes permitted to conduct parallel analyses of the transcriptional landscape separately in residual β cells and neighboring α cells following massive β-cell ablation. Administration of doxycycline (DOX) to the drinking water during 7–10 d induced a massive β-cell loss in Insulin-rtTA/TET-DTA mice (Supplemental Fig. S1B), resulting in acute hyperglycaemia sustained for at least 14 d (Supplemental Fig. S1C), consistent with previous publications (Nir et al. 2007).
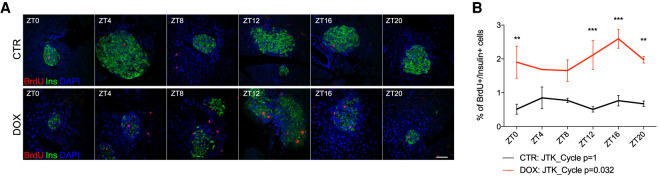
We first assessed whether entry into the S phase of the cell cycle of remnant β cells exhibited a diurnal profile. This was done by injecting 5-bromo-2′-deoxyuridine (BrdU), a marker of DNA replication during S phase, 2 h prior to sacrificing the animals at 6 time points with 4-h intervals across 24 h. In accordance to previously published data (Nir et al. 2007), we observed overall increased BrdU incorporation in the nuclei of insulin-labeled cells following massive β-cell loss when averaging the results across all time points (Supplemental Fig. S1D). Most importantly, the incorporation of BrdU by residual β cells was not only augmented overall, but it also exhibited a circadian rhythmicity (adjusted P-value = 0.032; BH.Q = 0.065) when examined using the JTK_Cycle paradigm (Fig. 1A,B; Supplemental Fig. S2; Hughes et al. 2010). These data suggest that the proliferative response in residual β cells after ablation gains a circadian pattern, with a peak observed in the middle of activity phase.
Figure 1.
Circadian regulation of β-cell regeneration following massive ablation. (A,B) Around-the-clock assessment of β-cell proliferation conducted 12 d after beginning of DOX administration. BrdU was injected intraperitoneally 2 h prior to sacrificing the animals every 4 h across 24 h. (A) Representative confocal images of pancreatic islets show localization of proliferation marker BrdU (red staining) in the nucleus of insulin-producing β cells (green) at the indicated time points (see also Supplemental Fig. S2). Scale bar, 50 µm. (B) Daily profile of β-cell proliferation. Data are expressed as mean ± SEM for n = 3 mice per time point. Two-way ANOVA test with Bonferroni post-test was used to assess the difference between control and DOX groups. (**) P < 0.01, (***) P < 0.001. JTK_Cycle was used to assess circadian rhythmicity of the averaged profiles.
The acute loss of β-cell mass affects gene expression in residual β cells, but not in adjacent α cells
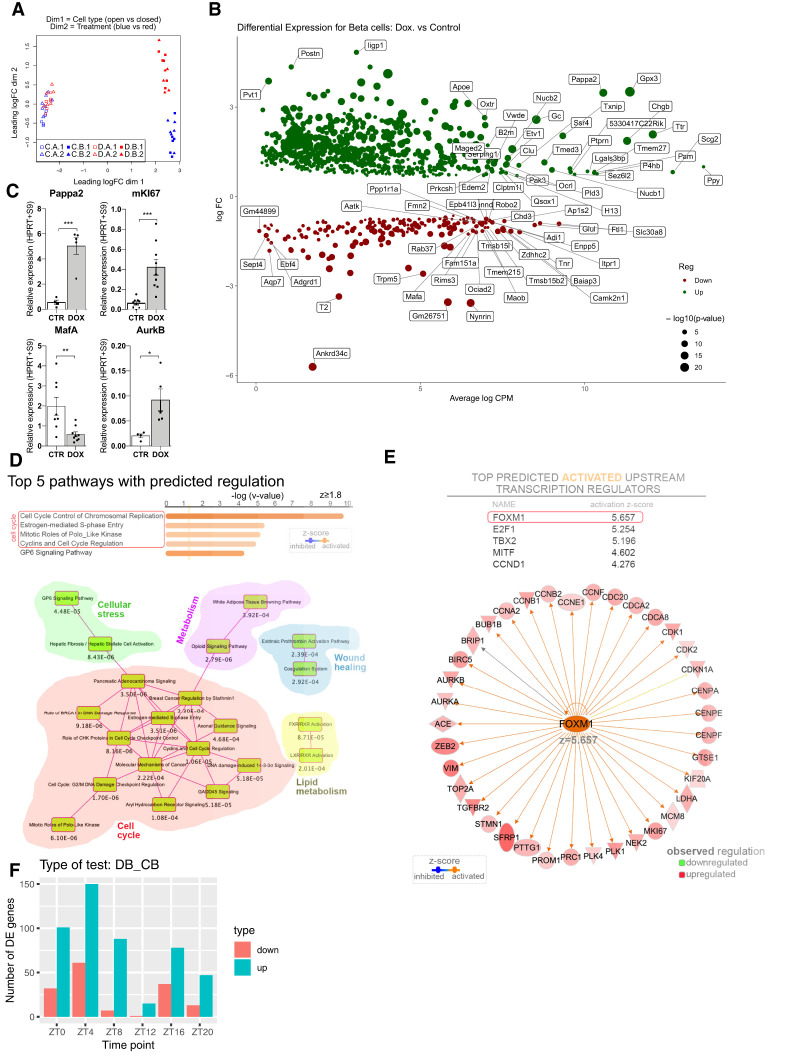
In an attempt to explore the mechanism of β-cell proliferation following massive ablation, we profiled the temporal transcriptome of FACS sorted α- and β-cell populations. Temporal analyses were conducted 7–10 d following DOX administration in Insulin-rtTA/TET-DTA-triple-reporter transgenic mice. Remnant islets were isolated every 4 h at six time points across 24 h (Supplemental Fig. S1A,B; Petrenko et al. 2017a; Petrenko and Dibner 2018). α-Cell and β-cell fractions from DOX group and untreated control counterparts were subjected to genome-wide transcriptome analysis by RNA sequencing (RNA-seq). We identified 12,912 transcripts after filtering for genes with at least 1 count per million (CPM) in at least half of all measurements (cell type, condition, and time points). A multidimensional scaling with a distance measure for microarrays (plotMDS from limma [Ritchie et al. 2015]) run on all samples showed clear separation between α and β cells (Fig. 2A, dimension 1) and major differences between β cells in intact islets and residual β cells following ablation (Fig. 2A, dimension 2). In contrast, almost no differences among α-cell samples were detected between control and DOX groups (Fig. 2A, dimension 2).
Figure 2.
Differential analysis of gene expression in α and β cells following massive β-cell ablation. A total of 12,912 transcripts were identified by RNA-seq analyses, following the experimental settings presented in Supplemental Figure S1A. (A) Multidimensional scaling across all the samples (α and β cells from mice treated or not with DOX in six time points, two experimental repetition each, total of 24 samples). (Dimension 1) Cell type, (dimension 2) treatment (DOX vs CTR). “C.A” and “C. B” (experimental repetition 1 and 2) correspond to α and β cells, respectively, from the CTR group; D.A and D. B (repetition 1 and 2) correspond to the DOX group counterparts. Note the clear separation between the transcripts specific for α and β cells, and between CTR and DOX β cells. (B) Volcano plot presents transcripts that were differentially expressed between CTR β cells and residual β cells in DOX group. For differential analysis, the average across six time points per each of four conditions (α and β cells, treated or not with DOX) was considered. Up-regulated genes (924 genes) are labeled in green; down-regulated (190 genes) are labeled in red (log2FC > 0.5 and log2FC < −0.5, respectively). Sizes of dots correspond to the log10 P-value. Volcano plots that compare α and β cells in control and DOX groups are presented in Supplemental Figure S3, A and C, respectively. (C) Examples of differentially expressed genes in β cells following ablation identified by RNA-seq (shown in B) and assessed by qRT-PCR analysis in an independent set of mice. Data are expressed as mean ± SEM for n = 4–9 mice. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. (D,E) Results of the Ingenuity Pathway Analysis (IPA) of differentially expressed transcripts (shown in B), indicating the top canonical pathways predicted regulated (z ≥ 1.8) as well as the overlapping canonical pathways map with at least five common molecules (D) and the top activated predicted upstream transcription regulators (z ≥ 2) (E). (F) Histograms presenting differential expression (number of up- and down-regulated genes) in residual β cells (DB) compared with control condition (CB) at each of six time points. See Supplemental Figure S1 for the experimental design and related Supplemental Figure S3.
Differential transcript expression analysis that compared average values across all six time points per cell type and per condition identified 924 differentially expressed genes between residual β cells from the DOX group and the untreated control counterparts (733 up-regulated and 268 down-regulated) (Fig. 2B; Supplemental Data Set S1). In contrast, only one gene (B2m) was differentially expressed in neighboring α cells after ablation as compared with controls (Supplemental Data Set S1). We next validated the changes of selected transcripts differentially expressed in β cells from the DOX group in an independent set of experimental animals by qRT-PCR analyses. Concordant with the RNA-seq results, miKi67, AurkB, and Pappa2 were up-regulated, whereas Mafa was down-regulated (Fig. 2C). Ingenuity pathway analysis (IPA) identified in the top canonical pathways characterizing the transcriptional landscape of the residual β cells the signaling pathways involved in cell cycle regulation. Based on the observed regulation of their molecular components, these were predicted as activated (Fig. 2D). Moreover, Foxm1 was predicted as a top upstream transcriptional regulator for the observed regulatory landscape (Fig. 2E).
When the analysis was conducted separately per time point, the number of differentially expressed transcripts (Supplemental Fig. S3A) was similarly distributed in β cells compared with α cells in the control condition (Supplemental Fig. S3B). Small differences across the time points were observed between α and residual β cells in the DOX group (Supplemental Fig. S3C–E). In contrast, the number of differentially regulated transcripts in β cells between DOX-treated and control mice varied greatly among the time points (Fig. 2F). There was a higher number of differentially expressed transcripts observed during the inactivity phase (between ZT0 and ZT8), and a minimal number of differentially expressed transcripts at the time point of light to dark transition (ZT12) (Fig. 2F). All in all, our findings suggest that transcripts whose expression levels change after β-cell ablation exhibit daily variability.
The temporal expression of transcripts is differentially affected in residual β- and neighboring α cells following massive β-cell loss
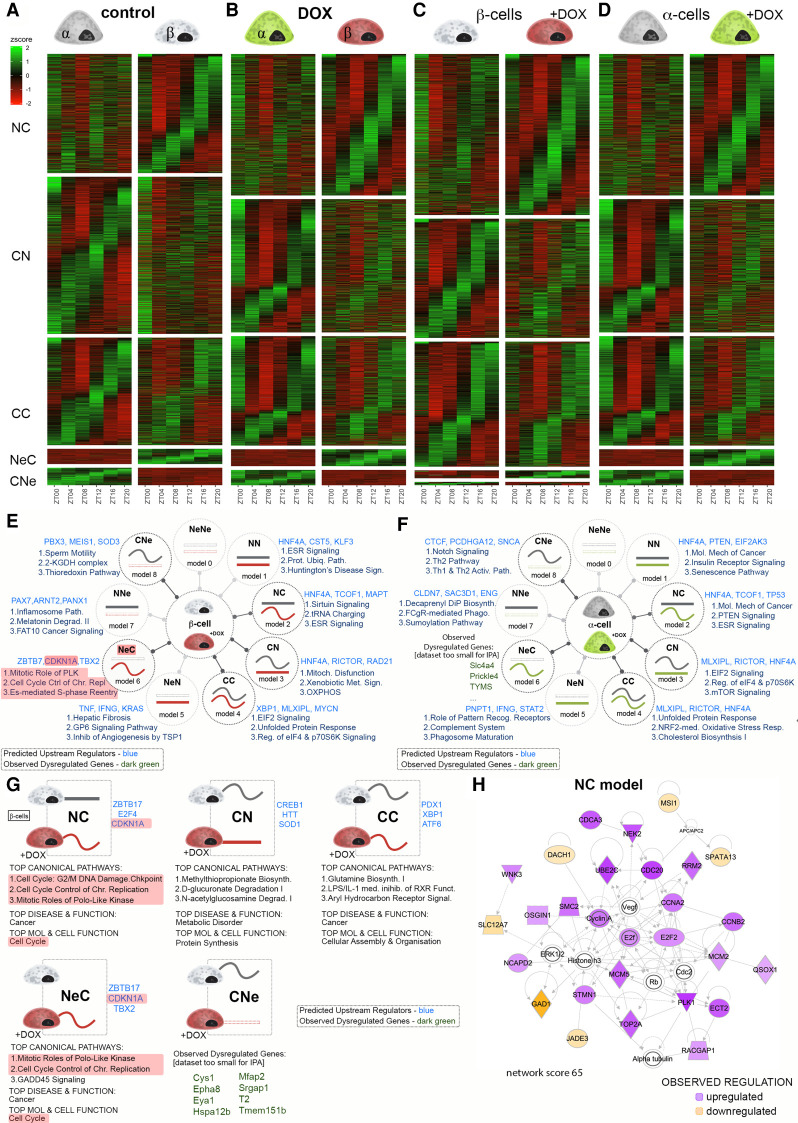
To study temporal regulation of α- and β-cell transcriptional landscapes we analyzed RNA-seq data sets collected at six time points every 4 h across 24 h from DOX Insulin-rtTA/TET-DTA-triple-reporter mice following β-cell ablation and the untreated CTR counterparts (Supplemental Fig. S1A; Supplemental Data Sets S2–S5). The rhythmicity of each gene was determined based on the best circadian profile with a 20- to 28-h period and a classification by logistic regression (Materials and Methods). Temporal gene expression profiles were grouped into a number of models, which are summarized in Figure 3, E and F. Heat maps for five selected models for the alteration of temporal gene expression between α and β cells in control and DOX groups are presented in Figure 3, A and B, respectively. The five groups of profile changes encompassed the genes expressed in both cell types that exhibited a flat (nonrhythmic) profile across 24 h in one cell type, but a circadian profile in the other cell type (NC and CN, where N stands for “nonrhythmic” and C stands for “circadian”). The CC group comprised genes that were rhythmically expressed in both cell types, with similar or distinct circadian properties. The last two categories referred to the genes that were not expressed (Ne) in one cell type but expressed in a circadian fashion in the other (CNe and NeC). A gene was considered to be expressed if it had an average RPKM across all time points of >1. Consistent with our previous study (Petrenko et al. 2017b), close to half of the detected transcripts (45.15%) exhibited a rhythmic expression in at least one cell type (Fig. 3A; Supplemental Data Set S2). Following massive β-cell loss, the overall proportion of genes defined as rhythmic in at least one cell type slightly increased to 48.5%. The number of rhythmic genes common to both cell types increased from 1505 to 1645, and the number of transcripts exclusively rhythmic in residual β cells increased from 1668 to 2128 in the DOX group (Fig. 3B; Supplemental Data Set S3). We next conducted a similar comparison of temporal gene expression in each cell type between control and DOX conditions (Fig. 3C,D). In the β-cell fraction, 1635 genes that were circadianly expressed in the control lost their rhythmic expression in the DOX group (Fig. 3C; Supplemental Data Set S4, model CN). Remarkably, 2208 transcripts that were nonrhythmic in the control group acquired rhythmicity after ablation (Fig. 3C; Supplemental Data Set S4, model NC). Noteworthy, despite the lack of differential changes in the average values of the transcript expression across all the time points in α cells following DOX treatment, the ablation of neighboring β cells strongly affected the temporal landscape of the α-cell transcriptome. Indeed, 2266 transcripts that were rhythmic in the control group became flat in the DOX group (Fig. 3D, model CN), and as many as 2225 nonrhythmic transcripts in the control α cells acquired a rhythmicity in DOX-treated mice (Fig. 3D, model NC; Supplemental Data Set S5).
Figure 3.
Temporal analysis of gene expression in α and residual β cells following partial β-cell ablation. (A–D) Heat maps comparing temporal profiles of transcripts between α and β cells in control condition (A) or following β-cell ablation (DOX condition; B), between α cells in control and DOX groups (C), and between β cells (D) in control and DOX groups for the temporal transcript profile changes from nonrhythmic to circadian (NC), circadian to nonrhythmic (CN), circadian rhythmic in both conditions (CC), nonexpressed in one condition and circadian rhythmic in the other (NeC and CNe), nonexpressed in one condition and nonrhythmic in the other (NeN and NNe), and nonexpressed in both conditions (NeNe). (E,F) IPA of different models of rhythmic transcripts expressed in β cells (E) and in α cells (F), in control versus DOX-treated conditions. Three predicted upstream regulators (blue) and three most enriched functional pathways are listed next to the model drawings. For data sets too small to allow pathway analysis, a selection of genes belonging to the respective model is indicated (green). (G) IPA of sub-groups of rhythmic transcripts (Fig. 3C, models CC, NC, CN, NeC, and CNe) that were differentially expressed in β cells between control and DOX-treated conditions. Predicted upstream regulators (blue) are listed next to the schematic drawing of the temporal profile change. For data sets too small to allow pathway analysis, a selection of DEGs (green) is indicated. Top canonical pathways, disease, and molecular/cellular function are listed below the drawing. (H) Leading IPA-generated organic network of up-regulated and down-regulated transcripts that acquired rhythmicity following DOX treatment in the model NC in presented in G displaying the regulation of cell cycle related genes.
We next performed pathway analysis by applying IPA to each of the five groups of transcripts that presented temporal profile changes in β and α cells between the control and DOX groups (Fig. 3E,F, respectively, referring to the temporal heat maps in Fig. 3C,D). Interestingly, nonrhythmic transcripts that acquired rhythmicity in residual β cells were involved into Sirtuin signaling, tRNA charging, and estrogen receptor (ESR) signaling (Fig. 3E). Those in neighboring α cells were related to ESR and PTEN signaling, and to molecular mechanisms related to cancer (Fig. 3F). In turn, rhythmic transcripts that lost rhythmicity in residual β cells were related to mitochondrial dysfunctions, oxidative phosphorylation, and xenobiotic metabolism. Those in neighboring α- cells were related to mTOR signaling, to EIF2 signaling, and to the regulation of eIF4 and the ribosomal protein p70S6k, which are involved in the tuning of protein synthesis.
Two additional groups of genes were identified in each cell type: (1) genes that were defined as nonexpressed in control condition, and that were up-regulated and acquired rhythmic profiles following DOX treatment (model NeC in Fig. 3E,F); and (2) rhythmically expressed genes in the control condition that became silent following DOX treatment (model CNe in Fig. 3E,F). Interestingly, newly expressed rhythmic genes in residual β cells were related to mitosis, with the mitotic role of Polo-like kinase, cell cycle control of chromosome replication, and estrogen-mediated S-phase reentry signaling being identified as the top three canonical pathways (Fig. 3E).
Finally, we applied IPA to the genes that were both differentially expressed according to the analyses of mean values (Fig. 2B) and exhibited distinct temporal rhythmic profiles in β cells following the β-cell ablation compared with the control group (Fig. 3C). Strikingly, a high number of genes coding for the key regulators of the cell cycle were comprised in this group. The transcripts of cell cycle regulators exhibited higher expression in residual β cells (Fig. 2B) and became rhythmic after ablation (models NC and NeC in Fig. 3G,H). This further supports the possibility that compensatory β-cell proliferation is controlled in a circadian fashion. Accordingly, the top-rated network of differentially expressed genes within the NC model consisted of genes involved in cell cycle regulation (Fig. 3H), further substantiating the role of circadian regulation in the proliferation of β cells during regeneration.
The temporal expression of core clock and cell cycle-controlling genes in residual β cells and adjacent α cells following β-cell ablation
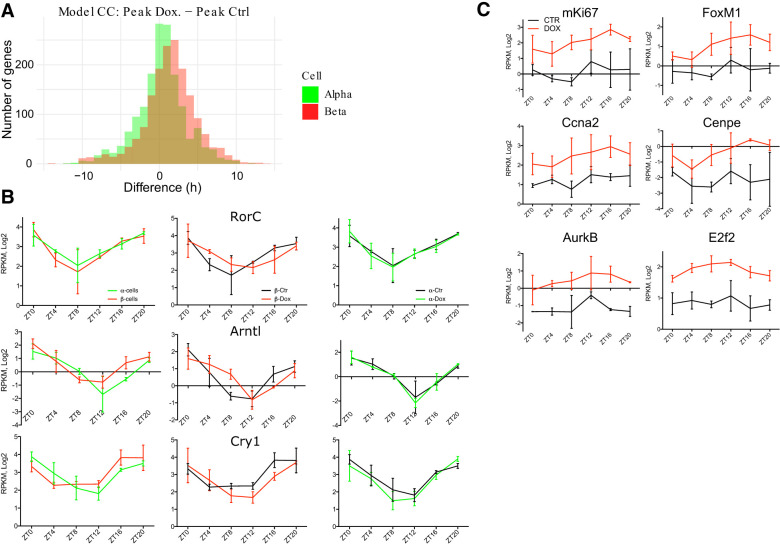
Remarkably, a comparison of the circadian phases between the transcripts that exhibited rhythmic expression profiles in control and DOX groups in β cells (Fig. 3C) revealed that most of the transcripts rhythmic in the DOX group had a phase delay of ∼2.5 h when compared with the corresponding transcripts of the control group (Fig. 4A, red bars; Supplemental Data Set S6). In contrast, no significant phase shifts were found for the transcripts that were rhythmically expressed in α cells between control and DOX conditions (Fig. 4A, green bars; Supplemental Data Set S6).
Figure 4.
Temporal profiles of core clock and proliferation regulating genes in residual β cells and adjacent α cells following massive ablation. (A) Phase shift between rhythmically expressed transcripts in CTR and DOX conditions (model CC) identified in α cells (in green) and β cells (in red) according to the temporal analysis of RNA-seq presented in Figure 3. Phase differences were calculated for each rhythmic transcript and plotted on the graph. 0 corresponds to identical phase between the transcript expression in CTR and DOX conditions. (B) Temporal profiles of selected core clock transcripts (RorC, Arntl [Bmal1], and Cry1) in control α and β cells (in green and in red, respectively; left panels), in control β cells versus residual counterparts (middle panels), and in control and DOX-treated α cells (right panels) assessed by RNA-seq. (C) Temporal profiles of selected transcripts encoding for the proliferation regulating proteins (see also Fig. 3G) in sorted β cells. Data are expressed as mean ± SD for two experimental repetitions per each time point with at least three mice in each plicate.
To address the question of whether the core clock machinery operative in the islet cells had been altered by massive β-cell ablation, we compared profiles of the core clock components in DOX and control groups in α and β cells. Rorc, Bmal1 (Arntl), and Cry1 oscillatory profiles were phase-advanced in β cells compared with α cells in untreated control mice, concordant with our previous findings (Petrenko et al. 2017b). Noteworthy, these transcript profiles were phase-delayed in β cells following DOX treatment when compared with β cells of untreated mice. However, these phase differences could not be observed in the neighboring α cells (Fig. 4B). This finding was further supported by a KEGG pathway enrichment analysis of RNA-seq database (Supplemental Data Set S6), with circadian clock pathway appearing among the most altered ones (Supplemental Fig. S4A). We next assessed the molecular clockwork in isolated remnant islets after massive β-cell ablation and in separated α and β cells synchronized by a forskolin pulse in vitro (Supplemental Fig. S4B,C). These experiments suggest that cell-autonomous molecular clocks are fully operative in isolated α and β cells of remnant pancreatic islets following massive β-cell ablation.
An additional prominent group of transcripts whose accumulation appeared altered following DOX treatment by IPA analysis was related to the regulation of the cell cycle. Noteworthy, most of the up-regulated cell cycle-related transcripts exhibited a circadian expression profile (see Fig. 3G). These transcripts include mRNAs encoding the proliferation regulators mKi67, Foxm1, Pbk, E2f2, cyclins (Ccna2 and Ccnb2), aurora kinases (Aurkb and Aurka), Cdk1, Cdc20, and other regulators of cellular cycle progression (Fig. 4C; Supplemental Data Set S3).
The essential core clock transcription factor BMAL1 is required for β-cell proliferation following massive β-cell ablation
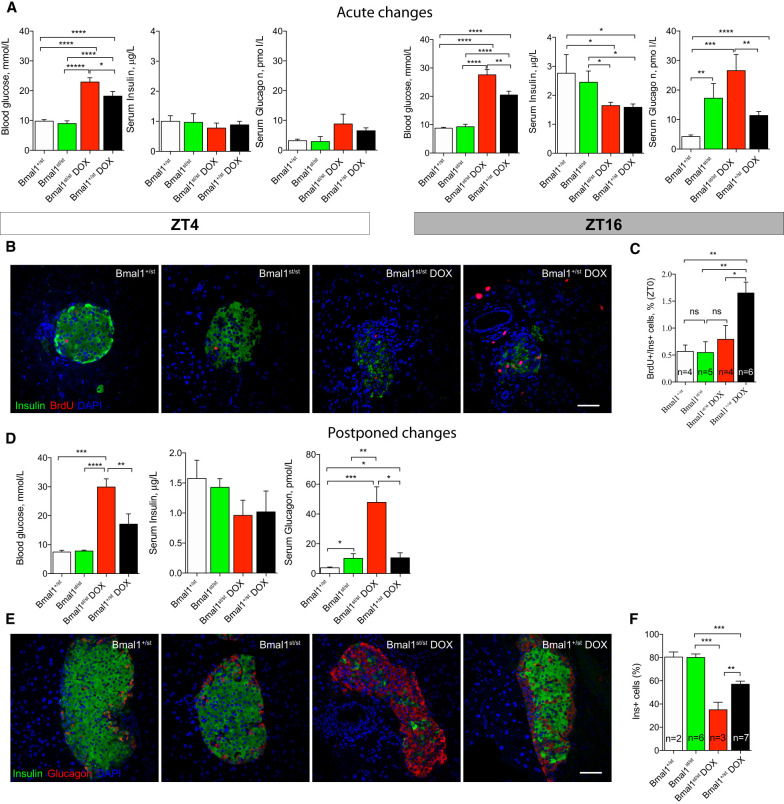
To scrutinize the role of the molecular clocks in the development of diabetic phenotypes and in the regulation of compensatory β-cell regeneration, β-cell ablation was triggered in arrhythmic Bmal1 deficient mice. To this end, we crossed mice in which BMAL1 expression was blocked by a transcriptional-translational stop cassette knock-in into the Bmal1 locus (Bmal1st/st mice) (Supplemental Fig. S5A) with Insulin-rtTA/TET-DTA-triple-reporter mice. As expected, the absence of functional clocks in isolated islets from these transgenic animals resulted in a flat Per2-luc bioluminescence profile following forskolin synchronization (Supplemental Fig. S5B). Clock-compromised Bmal1st/stDTA mice and their heterozygous Bmal1+/stDTA counterparts were treated with DOX according to the design depicted in Supplemental Figure S1A. Of note, nontreated Bmal1+/st mice and their Bmal1+/+ counterparts exhibited comparable levels of glucose, insulin, and glucagon (Supplemental Fig. S6A–C). Induction of β-cell loss in Bmal1st/st mice led to a significantly more severe hyperglycemia as compared with DOX-treated heterozygous mice (Fig. 5A, cf. Bmal1+/stDOX and Bmal1st/stDOX). Thus, in DOX-treated Bmal1st/stDTA animals, peripheral blood glycemia was ∼10 mmol/L higher than in heterozygotes control mice in the middle of the activity phase (ZT16). This difference was slightly smaller, but still significant, at ZT4 (Fig. 5A). Bmal1st/stDTA and heterozygotes mice exhibited similar levels of hypoinsulinemia after DOX treatment at ZT16 as compared with nontreated Bmal1st/stDTA and control mice, with no significant changes in blood insulin observed at ZT4 (Fig. 5A). Bmal1st/stDTA mice not treated with DOX exhibited higher levels of blood glucagon as compared with the clock proficient controls. Strikingly, β-cell ablation resulted in a significantly more dramatic hyperglucagonemia in Bmal1+/stDTA mice at ZT16. This phenotype was even more pronounced in Bmal1st/stDTA mice. A trend for hyperglucagonemia was also observed for Bmal1+/stDTA and Bmal1st/stDTA mice treated with DOX at ZT4; however this tendency did not reach statistical significance (Fig. 5A, Bmal1+/st DOX, Bmal1st/stDOX).
Figure 5.
Bmal1 is required for functional recovery and compensatory β-cell proliferation following massive β-cell loss. (A) Assessment of blood glucose, insulin and glucagon levels 12 d after beginning of DOX administration at ZT16 and ZT4. Data are expressed as mean ± SEM for n = 8–16 mice per group. One-way ANOVA was applied to test the difference between groups, followed by t-test comparisons. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. (B,C) β-Cell proliferation 12 d following DOX administration, assessed by 2-h BrdU pulse prior to sacrificing at ZT0. Representative confocal images of pancreatic islets (B) showing the colocalization of proliferation marker BrdU (red staining) with nuclei of insulin-producing β cells (green) (see also Supplemental Fig. S7); and corresponding histogram (C) showing quantification of double-positive cells within insulin-labeled populations. Data are expressed as mean ± SEM for n = 4–6 mice per phenotype. One-way ANOVA was applied to test the difference between groups, followed by t-test comparisons. (*) P < 0.05, (**) P < 0.01. (D) Assessment of blood glucose, insulin and glucagon at ZT4, one month after withdrawing DOX. Data are expressed as mean ± SEM for n = 4–9 mice per phenotype. One-way ANOVA was applied to test the difference between groups, followed by t-test comparisons. (*) P<0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. (E,F) Representative confocal images of islets on pancreatic sections (E) showing α- and β-cell composition (stained for glucagon in red and for insulin in green, respectively) of the islets from Bmal1st/st and Bmal1+/st mice one month after DOX withdrawal (Bmal1st/stDOX and Bmal1+/stDOX, respectively). Control (Bmal1+/st) and Bmal1st/st mice did not receive DOX treatment. Percentage of insulin-labeled β-cell out of total number of α and β cells is presented in adjacent histogram (F). Data are expressed as mean ± SEM for n = 2–7 mice per phenotype. A one-way ANOVA was applied to test the difference between groups, followed by t-test comparisons (except for Bmal1+/st). (**) P < 0.01, (***) P < 0.001. Scale bars in B and E, 50 µm.
Next, we assessed the proliferation rate of remnant β cells by injecting the thymidine analog BrdU intraperitoneally 2 h before sacrificing the animals at ZT0. In agreement with our previous data (Fig. 1A,B), the number of BrdU/Insulin double-positive β cells was significantly increased in DOX-treated Bmal1+/stDTA mice following β-cell ablation as compared with both Bmal1+/stDTA and Bmal1st/stDTA control mice without DOX treatment (Fig. 5B,C). In striking contrast, β-cell loss failed to enhance the number of BrdU/Insulin double-positive cells in DOX-treated Bmal1st/stDTA (Bmal1st/stDOX) (Fig. 5B,C; Supplemental Fig. S7).
Finally, we followed the regeneration and functional recovery of β cells during a longer period after massive ablation. Four weeks after DOX withdrawal the blood glucose levels measured at ZT4 in Bmal1st/stDTA animals stayed consistently high when compared with Bmal1+/stDTA counterparts (Fig. 5D, left panel, cf. Bmal1st/stDOX and Bmal1+/stDOX). The level of serum insulin was comparable between DOX-treated Bmal1+/stDTA and Bmal1st/st DTA animals, with a tendency to be lower in Bmal1st/st group that did not reach statistical significance (Fig. 5D, middle panel). Furthermore, untreated Bmal1st/stDTA mice exhibited a significantly more pronounced hyperglucagonemia as compared with heterozygous controls. Observed hyperglucagonemia was further elevated in DOX treated Bmal1st/stDTA animals compared with their Bmal1st/st counterparts without such a treatment (Fig. 5D, right panel).
Strikingly, 6 wk following DOX withdrawal the survival rate for Bmal1st/stDTA animals was ∼33% only (three out of nine mice), whereas all of the Bmal1+/stDTA animals (n = 8) survived at this point. Consistent with the blood glycemia and hormone status, histological examination of the pancreas from the animals following 6 wk of recovery after DOX administration revealed a significantly lower proportion of insulin-positive β cells in Bmal1st/stDTA mice among overall insulin- and glucagon-labeled cells as compared with their Bmal1+/st counterparts (Fig. 5E,F, cf. Bmal1st/stDOX and Bmal1+/stDOX). Hence, in contrast to the β-cell regeneration and recovery from diabetes observed in rhythmic mice, β-cell regeneration did not occur in Bmal1st/st arrhythmic mice. Consequently, this led to a life-threatening diabetes, with pronounced hyperglycemia and hyperglucagonemia.
The expression of cell cycle-regulating genes is strongly inhibited in residual β cells of Bmal1st/stDTA mice following β-cell ablation
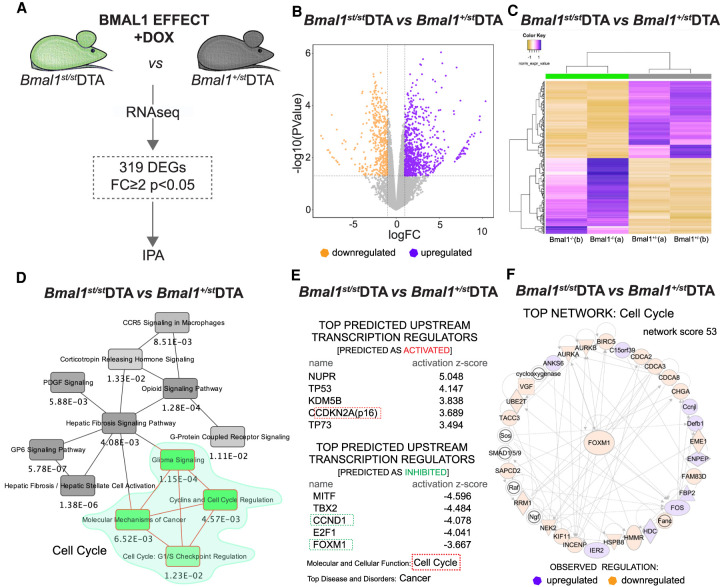
In an attempt to dissect the molecular mechanism underlying the failure of β-cell regeneration in the absence of the core clock transcription factor BMAL1, we performed RNA-seq on FACS-purified residual β cells from Bmal1st/stDTA mice 10 d following DOX administration (Fig. 6A; Supplemental Data Set S7). First, we compared DOX-treated Bmal1st/stDTA with untreated controls (Supplemental Data Set S8). We identified only three differentially expressed genes between these two conditions: transcripts related to fructose metabolism Aldob and Pfkfb3 were up-regulated, while H2-Ea-ps was down-regulated. We failed to observe activation of cell cycle related pathways. When compared DOX-treated Bmal1st/stDTA and Bmal1+/stDTA mice, an impaired expression of functional Bmal1 pre-mRNA in Bmal1st/stDTA mice was confirmed by a decreased RNA sequence read coverage of the Bmal1 gene region downstream from the stop cassette (between exon 5 and 6) as compared with heterozygotes mice (Supplemental Fig. S5A,C). An up-regulation of incomplete Bmal1 transcript was observed in Bmal1st/st mice (Supplemental Fig. S5E,F; Supplemental Data Set S7), supposedly because the expression levels of REV-ERBα/β, that act as repressors of Bmal1 transcription (Preitner et al. 2002), were diminished in these animals (Supplemental Data Set S7).
Figure 6.
RNA-seq analysis of residual β cells in Bmal1st/st mice following massive β-cell loss. (A) Schematic drawing of RNA-seq differential analysis of FACS-separated β cells from Bmal1st/st and Bmal1+/st mice treated with DOX. Data represent two biological replicates collected at ZT0 (each replicate is a mix of cells from three mice). (B) Volcano plot presenting 319 differentially expressed transcripts (showing significance [−log10 {P-value with FDR}] versus fold change [logFC]). Vertical lines highlight log2 fold changes of −2 and +2, while a horizontal line represents a P-value with FDR of 0.05. (C) Dendrogram with heat maps highlighting clusters of up-regulated and down-regulated genes between Bmal1st/st and Bmal1+/st mice treated with DOX in two biological replicates (a and b). (D–F) The IPA of differentially expressed genes, revealing the overlapping canonical pathways map (minimum number of common molecules: four; D); top predicted upstream transcription regulators (activated or inhibited), top molecular and cellular function, and top disease and disorders (E); and the top 1 network (F) of the analyzed differential transcriptional landscape, all suggesting the modulation of cell cycle regulation in Bmal1st/st mice.
Within all of the detected 14,649 expressed genes, a subset of 319 differentially expressed genes (DEGs) defined by FC ≥ 2 with P < 0.05 was identified between homozygous Bmal1st/stDTA and heterozygous Bmal1+/stDTA counterparts (Fig. 6B,C). The IPA of this DEG subset revealed regulators involved in cell cycle regulation and circadian rhythm in the top 5 canonical pathways (Fig. 6D, green island; Supplemenal Fig. S5D). KEGG pathway enrichment analysis revealed a differential expression of modulators of the neuroactive ligand-receptor interaction pathway, comprising key hormonal regulators of β-cell function Gcgr and Ptger3 up-regulation and Glp1r, Oxtr, Ucn3, and Lpar6 down-regulation (Supplemental Fig. S5G; Supplemental Data Set S7). Functional networks directly linked to impaired Bmal1 expression are presented in Supplemental Figure S5, E andF.
Furthermore, based on the observed changes, the pathways involved in cell cycle regulation were predicted as inhibited (Supplemental Fig. S5D), suggesting a decreased β-cell proliferation in the Bmal1st/st mice. Supporting this observation, key cell cycle regulators such as senescence markers Cdkn2a (p16) and Cdkn1a (p21, P-value of overlap 6.82E-28, activation z-score 2.701) and tumor suppressor genes p53 and p73, were inferred in the top predicted upstream transcription regulators with expected activation (Fig. 6E). Furthermore, CcnD1 (cyclin D1) (Fig. 6E) and Foxm1 (Fig. 6E,F) were included in the top predicted upstream regulators inferred as inhibited. Accordingly, cell cycle progression was identified as a top molecular and cellular function of the analyzed DEG set, while cancer was suggested to be the leading predicted associated pathology (Fig. 6E). Strikingly, all three top hits revealed by the network analysis were connected to cell cycle regulation (Fig. 6F). Taken together, these findings may explain the low proliferative potential of Bmal1 deficient β cells after β-cell ablation (Fig. 5B,C, E,F).
Discussion
In this study, we induced massive β-cell ablation to identify mechanisms participating in the regulation of islet cell regeneration and to characterize the transcriptional landscape in separated α cells and residual β cells. Our results provide evidence that proliferation of residual β cells triggered by massive ablation follows a circadian pattern (Fig. 1). Virtually no compensatory β-cell regeneration occurred in arrhythmic Bmal1st/st mice that may suggest a role of the functional oscillators in coordinating β-cell division. Importantly, massive β-cell ablation led to a fatal noncompensated diabetes in the absence of the core clock transcription factor BMAL1 (Fig. 5).
Coupling between the circadian clock and β-cell proliferation
Replication of terminally differentiated β cells, rather than differentiation of stem cells, represents a major source of newly generated β cells after birth. This holds true not only under physiological conditions, but also during regeneration following lesions (Dor et al. 2004; Brennand et al. 2007; Nir et al. 2007; Teta et al. 2007; Klochendler et al. 2016). Noteworthy, in the absence of residual β-cell upon near-total ablation the conversion of α or δ cells into insulin-producing cells was also described (Thorel et al. 2010; Chera et al. 2014). Our data indicate that β-cell reparative proliferation follows a daily rhythm and that it requires functional core clock protein Bmal1 (Figs. 1, 5). We now show that the highest proliferation rate occurs during the activity phase in the middle of night (Fig. 1), and that it coincides with the peaks of rhythmic expression of the genes encoding key regulators of the cell cycle (Fig. 4B). Importantly, whole-body knockout of the core clock component BMAL1 strongly disrupts β cells regeneration following massive ablation in transgenic mice (Fig. 5). A β-cell-specific disruption of the Bmal1 gene was previously reported to prevent a high-fat diet-induced expansion of β-cell mass (Rakshit et al. 2016). These previously published data suggest that the lack of β-cell mass expansion following diet-induced obesity takes place in the absence of at least one allele of Bmal1 in β cells, and it is likely attributed to the increased cell death, rather than to decreased proliferation per se. Moreover, it is important to highlight that expansion of β-cell mass upon obesity and β-cell regeneration have a different nature. In contrast to the expansion of β cells upon obesity, we show that regeneration requires functional clock and it is still present in rhythmic mice heterozygotes for Bmal1.
We found that a number of the up-regulated cell cycle-related transcripts exhibited a circadian rhythmicity in residual β cells following ablation (Fig. 4C) and were likely under the positive control of the transcription factor FOXM1 (Fig. 2E). The transcription of Foxm1 itself was circadian and peaked in the middle of the active phase, coinciding with the peak of proliferation (Fig. 4C–E). Strikingly, unlike heterozygous animals, no up-regulation of Foxm1 mRNA and transcripts specifying other cell cycle regulators was observed in Bmal1st/st mice following massive β-cell ablation, as compared with their nontreated counterparts (used as controls) (Fig. 6F; Supplemental Data Set S8). This provides a mechanistic basis for the lack of β-cell regeneration (Fig. 5E,F) and further suggests a key role of circadian clocks in the expression of cell cycle genes. FOXM1 is one of the key transcription factors required for postnatal β-cell proliferation, but not for neogenesis of β cells from progenitors (Zhang et al. 2006; Ackermann Misfeldt et al. 2008). Its expression is enhanced in different models of β-cell expansion associated with obesity (Davis et al. 2010; Yamamoto et al. 2017), pregnancy (Zhang et al. 2010) and regeneration following partial pancreatectomy (Ackermann Misfeldt et al. 2008). Moreover, induction of the activated form of FOXM1 restored the replicative potential of β cells in aged animals (Golson et al. 2015). Finally, stimulation of cell proliferation in juvenile human islets by activation of GLP-1r signaling triggered the expression of Foxm1. This makes FOXM1 a molecular candidate for coupling the cell cycle to molecular clocks. The precise mechanistic details of these interactions need to be elucidated in future work.
Proliferating β cells bear functional clocks
In adulthood, both α and β cells possess cell-autonomous and self-sustained circadian oscillators in mice and humans (Pulimeno et al. 2013; Petrenko et al. 2017b; Petrenko and Dibner 2018). Here we report that in adult mice the residual β cells maintain a circadian rhythmicity both in vivo and in vitro following ablation (Fig. 4B; Supplemental Fig. S4B,C). The characteristics of rhythmic expression patterns of selected core clock genes (Arntl, RorC, and Cry1) in residual β cells of DOX-treated mice were, however, altered when compared with nontreated mice (Fig. 4B; Supplemental Fig. S4A). These differences may stem from β-cell proliferation, similar to what has previously been reported for dividing and nondividing NIH3T3 fibroblasts (Nagoshi et al. 2004). Alternatively, altered paracrine or systemic signals along with the observed changes in the molecular landscape of residual β cells may lead to differences in the resetting of the molecular oscillator in residual β cells. For example, the loss of β cells leads to an enhanced proportion of neighboring α cells and an augmented secretion of glucagon (Fig. 5A), a potential synchronizer of molecular clocks in β cells (Petrenko and Dibner 2018).
Interestingly, residual β cells exhibited decreased levels of the mRNA encoding the important differentiation regulator MafA (Fig. 2B,C). This may be the consequence of dedifferentiation and dysfunction of these cells (Wang et al. 2007; Aguayo-Mazzucato et al. 2011; Swisa et al. 2017). Moreover, the circadian amplitude and magnitude of MafA mRNA accumulation were attenuated (Supplemental Fig. S4D) in DOX-treated mice. Besides core clock transcripts and MafA mRNA, numerous groups of transcripts changed their temporal expression profiles in residual β cells and neighboring α cells (Fig. 3), suggesting a modified temporal transcription landscape within regenerating islets upon diabetes.
Activation of β-cell regeneration following ablation
The massive β-cell loss induced the transcription of genes involved in cell cycle progression and replication in the residual β cells (Fig. 2D; Supplemental Data Set S3; Dor et al. 2004), but not in neighboring α cells (Fig. 2A). Likewise, the activation of GLP-1 signaling by exendine-4 exclusively induced proliferation in β-cell, and not in α or δ cells (Dai et al. 2017). This raises the question of which β-cell-specific mitogenic signals become important after β-cell ablation. The function of pancreatic islet is regulated by systemic and paracrine stimuli mediating distinct cell type-specific effects (Koh et al. 2012; Petrenko et al. 2018). Such differences in receptor repertoire and signaling molecules between α and β cells also define the cell-specific entrainment of circadian oscillators by physiologically relevant inputs that may regulate coupling between cell cycle and circadian oscillators (Petrenko et al. 2017b; Petrenko and Dibner 2018). In addition, the difference in glucose sensing and metabolism in α and β cells (Olsen et al. 2005) may account for the cell type-specific activation of β-cell proliferation. Indeed, high glucose plays a role in triggering β-cell proliferation capacity via the glucose sensor glucokinase (Porat et al. 2011). This conjunction may involve the molecular clocks machinery, as the core clock component RORγ was reported to regulate glucose-stimulated proliferation in INS-1E cells in vitro (Schmidt et al. 2016). Concordantly, our data indicate that the enhanced proliferation rate following β-cell ablation parallels a hyperglycemia. Nonetheless, no significant proliferation of residual β cells was induced following ablation in absence of BMAL1, although the levels of glycemia were even higher in Bmal1st/st animals. These data suggest that high glucose alone is insufficient to trigger proliferation of β cells, and that functional oscillators are required to induce β-cell proliferation. We wish to emphasize, however, that we do not know whether the effects on glucose metabolism and β-cell proliferation observed in Bmal1st/st mice (Figs. 5, 6) were attributed to the elimination of the transcriptional factor BMAL1 per se, or to its function as a key core clock regulator. The discrimination between circadian and noncircadian activities of core clock transcription factors has been a notoriously difficult problem in the field. So-called “resonance experiments,” in which phenotypes are compared in organisms whose oscillators do or do not resonate with environmental cycles (T cycles), are probably the only approaches revealing the importance of rhythmic functions (Ma et al. 2013). Unfortunately, such endeavors are not feasible in the system described here.
Elegant work by Okamura and colleagues (Matsuo et al. 2003) reported that the initiation of mitotic division in regenerating mouse livers follows a robust circadian rhythm. These investigators provided compelling evidence that rhythmic cell cycle progression in regenerating livers is governed by the circadian expression of Wee1, a key regulator of mitosis. Interestingly, the kinetics of S-phase hepatocytes was not controlled by molecular clock. In our study, the kinetics of S-phase regenerating β cells was circadian, and the temporal expression levels of Wee1 transcript were rhythmic in β cells in both control and DOX conditions, with absolute levels being unchanged (Supplemental Fig. S4E). Wee1 expression exhibited a tendency for down-regulation in Bmal1st/st mice as compared with Bmal1+/st controls (FC = −1.45) (Supplemental Data Set S7) that did not reach statistical significance. Moreover, liver regeneration was delayed, but not arrested, in arrhythmic Cry1/Cry2 knockout mice. In contrast, the regeneration of pancreatic β cells did not proceed in the absence of the essential core clock component BMAL1, leading to fatal diabetes in a high proportion of animals (Fig. 5).
Systemic loss of BMAL1 aggravates diabetes
Disruption of the core clock components impairs insulin secretion by mouse and human β cells (Perelis et al. 2015; Petrenko et al. 2020) and results in a diabetic phenotype in mice (Marcheva et al. 2010). We now demonstrate that in the absence of a functional clock mice develop severe fatal diabetes following massive β-cell loss, contrary to their rhythmic counterparts that partially recover from thus-induced diabetes (Fig. 5D–F; Nir et al. 2007). While we failed to demonstrate a difference in blood insulin levels between Bmal1st/st mice and heterozygotes counterparts at the examined time points, arrhythmic animals developed strong hyperglucagonemia (Fig. 5D). Since glucagon stimulates gluconeogenesis in the liver (Perry et al. 2020), this may contribute to the strongly elevated blood glucose levels following massive β-cell ablation in Bmal1st/st mice. In addition, loss of functional clocks evoked insulin resistance in skeletal muscles (Dyar et al. 2014; Harfmann et al. 2016) and impaired cell-autonomous adoptative changes to increased insulin needs upon diet-induced obesity (Rakshit et al. 2016). Taken together, these data suggest that complex interaction between different pathogenetic pathways in pancreatic islets and metabolic tissues may account for the deterioration of diabetes upon dysfunctional clocks.
In summary, our data strongly suggest that regeneration of β cells is tightly coupled to diurnal rhythm. Moreover, loss of functional clocks aggravates the development of diabetic phenotype in mice. Indeed, human pancreatic islets from T2D individuals bear dysfunctional clocks associated with perturbed temporal regulation of insulin and glucagon secretion (Petrenko et al. 2020). These findings have important translational potential and should be considered in patients with T1D and T2D, as reboosting circadian rhythms by lifestyle adaptations may help preventing the aggravation of the disease.
Materials and methods
Animals
Animal studies were performed according to the regulations of the veterinary office of the State of Geneva (authorization number GE/47/19). Experimental mouse strain proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA was established by crossing triple reporter mouse line proGcg-Venus/RIP-Cherry/PER2::Luc (Petrenko et al. 2017b) to Insulin-rtTA/TET-DTA mice (Supplemental Fig. S1; Nir et al. 2007). ProGcg-Venus and RIP-Cherry reporters exhibited very high specificity and expression levels in α and β cells, respectively (Dusaulcy et al. 2016; Petrenko et al. 2017a). Bmal1st/st mice were developed by introducing (knock-in) a transcriptional-translational stop cassette between exons 5 and 6 of the Bmal1 locus, containing a splice acceptor and a triple repeat of a polyadenylation signal sequence, flanked by LoxP sites. Bmal1st/st mice were crossed to proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA. The obtained proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA/Bmal1st/st mice allowed to genetically induce massive β-cell ablation in the absence of functional clocks by administering 300 mg of doxycycline (DOX) in the drinking water supplemented with 2% sucrose. The control group received only water supplemented with 2% sucrose. All the experiments were done in mice aged 6–8 wk, under standard animal housing conditions with ad libitum access to food and water and under 12-h light/12-h dark cycles (LD; lights on at 7:00 a.m., lights off at 7:00 p.m.). For the islet isolation and BrdU experiments, male mice were fed exclusively during the night 10 d prior to the experiment and during the experiment (Supplemental Fig. S1A), allowing to reduce the effect of individual feeding rhythm (Atger et al. 2015). For the islet isolation across the 24-h period, half of the animals were entrained by inverted LD and feeding cycles for 2 wk preceding the experiments.
Pancreatic islet isolation and separation of α and β cells
Islets of Langerhans were isolated by standard procedure based on collagenase (type XI; Sigma) digestion of pancreas followed by Ficoll purification (Petrenko et al. 2017a). Islet cells were gently dissociated by 0.05% trypsin (GIBCO) treatment, resuspended in KRB solution (pH 7.4, supplemented with 0.3% free fatty acid bovine serum albumin [BSA; Sigma], 1.4 mM glucose, 0.5 mM EDTA). α-Cell and β-cell populations were separated by flow cytometry fluorescence activated cell sorting (FACS; Astrios sorter [Beckman Coulter]) based on fluorescence wavelength and intensity, cell singlet nature, size, and viability as described previously (Petrenko et al. 2017b).
RNA sequencing (RNA-seq)
For around-the-clock RNA-seq experiments underlying Figures 2 and 3, total RNA was prepared from FACS-sorted α and β cells collected from male proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA control and DOX-treated mice every 4 h around the clock in biological duplicates, using RNeasy Plus microkit (Qiagen). A total of 48 samples, representing RNA pool of three to six mice each, was prepared. RNA-seq was performed by the iGE3 Genomics Platform (University of Geneva, Switzerland). The Illumina TruSeq RNA sample preparation kits were used for the library preparation with 25–50 ng of total RNA as input. Library molarity and quality was assessed with Qubit (Life Technologies) and Tapestation using a DNA high sensitivity chip (Agilent Technologies). Paired-end reads of 50 bases were generated using SMART-Seq v4 chemistry on an Illumina HiSeq 2000 (first replicate) and Illumina HiSeq 4000 (second replicate) sequencers.
For RNA-seq experiments in Bmal1st/st mice (Fig. 6), total RNA was prepared from FACS-sorted β cells collected from males and females of proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA/Bmal1st/st mice and their proGcg-Venus/RIP-Cherry/PER2::Luc/Insulin-rtTA/TET-DTA/Bmal1+/st littermates following 10 d of DOX administration at ZT0 in biological duplicates, using RNeasy Plus microkit (Qiagen) (a total of four samples, representing RNA pool of ∼7000 cells from three mice each). RNA-seq was performed by the iGE3 Genomics Platform (University of Geneva, Switzerland). The Clontech SMARTer ultralow input RNA was used to generate cDNA followed by Nextera XT kit for library preparation on samples with low RNA input. Library molarity and quality was assessed with Qubit (Life Technologies) and Tapestation using a DNA high sensitivity chip (Agilent Technologies). Single-end reads of 50 bases were generated using on an Illumina HiSeq 4000 sequencer.
RNA-seq data processing and analysis
For around-the-clock gene expression assessments, paired end reads were aligned to the mouse genome (assembly GRCm38/mm10) using STAR v2.5.3ausing Gencode vM16 annotation. The quantification was performed with QTLtools quan (https://qtltools.github.io/qtltools/). The mapping quality threshold was set at 255, and the number of mismatches allowed in both reads together was set at five (reads below these thresholds were not considered for the quantification) (Dobin et al. 2013). The genome index was built using Ensembl annotation to improve splice junction's accuracy. For each Ensembl gene, all of the exons of the respective annotated protein-coding transcripts were considered. Using custom Perl script, reads up to one mismatch were counted considering only reads in the right gene orientation. Both uniquely mapping and multimapping reads were reported. Normalization and differential expression analysis were performed with the R/Bioconductor packages edgeR v.3.28 (Robinson et al. 2010) and limma 3.42.2 (Ritchie et al. 2015). Log2 expression levels were calculated.
For the assessment of differential gene expression in β cells between Bmal1st/st and Bmal1st/stDOX; and between Bmal1st/stDOX and Bmal1+/stDOX, a quality control was performed with FastQC v.0.11.5. Single-end reads were mapped to mouse genome (assembly GRCm38/mm10) using STAR v.7.0f (Dobin et al. 2013). The biological quality control was done with PicardTools v.2.21.6. Raw counts were obtained using HTSeq v.0.9.1. Normalization and differential expression analysis were performed with the R/Bioconductor package edgeR v.3.26.8 (Robinson et al. 2010). The overall workflow of RNA-seq analysis is presented in Supplemental Figure S8.
Differential and temporal analysis
For around-the-clock gene expression assessment, genes with less than one CPM in half of the all measurements were discarded. Differential expression analysis between α and β cells was assessed using the EdgeR and limma packages, following the protocol of (Chen et al. 2016). A gene was defined as differentially expressed when the absolute value of the log2 fold change (log2FC) was >0.5 (same cell type) or 1 (otherwise), and the FDR-corrected P-value was <0.05 from the resulting differential expression analysis. Genes were considered as “nonexpressed” (Ne in Fig. 3) if their respective expression value was below an arbitrary threshold of 1 RPKM averaged over time. Rhythmicity of the transcript profiles was determined in two steps: First, 17 sin/cos models with intercept were regressed on each of two independent experiments corresponding to different periods in 20–28 h. The model with the lowest P-value from the F test was kept. Second, the gene was deemed rhythmically expressed based on a logistic regression model with the previous P-value for the two samples used as the input. The logistic regression was trained on a gold standard of 31 manually labeled genes (see Supplemental Data Set S9). All fitted coefficients were significant (P-value < 0.008). Based on a ROC curve, a cutoff of 0.6 was decided leading to a 95% accuracy on the golden standard between 0.849 and 0.988. Rhythmic parameters (i.e., phase, period length, and amplitude) can be shared between the two different cell types and between two conditions (CTR and DOX).
Differential gene expression analysis for β cells between Bmal1st/st and Bmal1st/stDOX, and between Bmal1st/stDOX and Bmal1+/stDOX was assessed using a generalized linear model with quasi-likelihood F-test of the EdgeR package (v 3.26.8). A gene was defined as differentially expressed when the absolute value of the log2 fold change (log2FC) was >1 and the FDR-corrected P-value was <0.05 (Benjamini and Hochberg correction).
Ingenuity pathway analysis (IPA) and KEGG pathway enrichment analysis
The pathway analyses were generated by Qiagen's IPA program (IPA; Qiagen, http://www.qiagen.com/ingenuity). Briefly, the analyses were performed with the following settings: expression value type (exp fold change), reference set (ingenuity knowledge base [genes only]), relationships to consider (direct and indirect relationships), interaction networks (35 molecules/network; 25 networks/analysis), node types (all), data sources (all), confidence (experimentally observed), species (human, mouse, rat), tissue and cell lines (all), and mutations (all).
The KEGG pathway enrichment analysis was generated by Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 based on the gene lists (Supplemental Data Set S6) comprising rhythmic genes with phase difference >2 h (Supplemental Fig. S4A), and on the gene list (Supplemental Data Set S7) comprising differentially expressed genes between Bmal1st/stDOX and Bmal1+/stDOX phenotypes.
Quantitative polymerase chain reaction (qPCR)
Total RNA was prepared from FACS separated islet cells using RNeasy Plus microkit (Qiagen). The RNA concentration was measured by Qubit RNA SH kit (Invitrogen). Next, 0.1 μg of total RNA was reverse-transcribed using TAKARA (Roche) and random hexamers and was PCR-amplified on a LightCycler 480 (Roche Diagnostics AG). Mean values for each sample were calculated from technical duplicates of each quantitative RT-PCR (qRT-PCR) analysis and normalized to the mean of housekeeping genes hypoxanthine–guanine phosphoribosyltransferase (Hprt) and S9. Primers used for this study are listed in Supplemental Table S1.
Assessment of blood glucose, insulin, and glucagon levels
Blood glucose (Fig. 5) was measured by the Accu-Chek glucometer (Roche). For insulin and glucagon assessment, blood was collected from the tail vein at ZT4 (11:00), or at ZT16 (23:00) with subsequent serum preparation by immediate centrifugation at 94,000 rcf for 15 min at 4°C and stored at −80°C. Protease inhibitors aprotinin (Sigma) and DPP4 (Millipore) were added to the samples to preserve the hormones from degradation. Insulin and glucagon concentrations were assessed by ultrasensitive mouse insulin and glucagon 100μL ELISA kits (Mercodia).
Assessment of cell proliferation
5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) was injected intraperitoneally (100 mg/kg) 2 h prior to sacrifice at the indicated time points.
Immunostaining
Paraffin sections (5 μm thick) were rehydrated, and antigen retrieval was performed using a Biocare pressure cooker and citrate (pH 6) buffer. The following primary antibodies were used: guinea pig anti-insulin (1:200; Dako), mouse anti-glucagon (1:800; Abcam), rabbit anti-glucagon (1:8000; Abcam), and mouse anti-BrdU (1:300; GE healthcare). For DNA counterstaining we used DAPI (Sigma). Secondary antibodies from Jackson ImmunoResearch were as follows: anti-guinea pig Alexa Fluor 488 (1:200), anti-mouse Cy3 (1:500), and anti-mouse Alexa Fluor 647 (1:500). Immunofluorescence images were captured using a Nikon confocal microscope. To assess β cell replication, at least 2000 β cells were counted per animal, and at least two slides at a distance of >200 µm were analyzed.
Image analysis
All immunofluorescence images were captured on a Nikon C1 confocal microscope at a magnification of 40× randomly from at least two different slices. The islets were identified by insulin labeling and captured throughout the entire slide. The number of labeled cells was quantified using Fiji software. The numbers of insulin-labeled cells bearing BrdU-labeled nuclei was normalized to the total number of insulin-positive cells in islet, and expressed in percentage. The numbers of insulin- and glucagon-labeled cells were quantified on slices immunostained for these two hormones. The proportion of β cells was expressed as the ratio between number of insulin-positive cells among overall insulin- and glucagon-labeled cells.
Quantification and statistical analysis
The results are expressed as means ± SEM (standard error of mean) for the indicated number of donors (as given in the figure legends) or illustrated as mean value of all experiments (for bioluminescence profiles). The statistical difference was tested by a Student's t-test to compare the two groups. A nonpaired Student's t-test was used to compare group donors. A one-way ANOVA test followed by a nonpaired Student's t-test comparisons was used to compare multiple conditions. All statistical tests were conducted with GraphPad Prism 8 software. Statistical significance was defined at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). To assess the circadian characteristics of proliferation a JTK_Cycle fitting method was used (Hughes et al. 2010). The profile was considered as circadian if adjusted P-value were <0.05.
Data availability
The accession numbers for the deposited data in GEO are GSE151338 and GSE152751.
Supplementary Material
Acknowledgments
We thank the colleagues from the University of Geneva—Ueli Schibler, Jacques Philippe, and Claes Wollheim—for constructive discussions; Marie-Claude Brulhart-Meynet for skillful technical assistance; Mylène Docquier, Céline Delucinge-Vivier, Didier Chollet, Emmanouil Dermitzakis, Luciana Romano, Deborah Bielser, Cédric Howald, and Keith Harshman for conducting RNA sequencing; Jean-Pierre Aubry-Lachainaye for help with cell sorting; and Charles Weitz from Harvard Medical School for generously sharing the Bmal1st/st mouse strain. This work was funded by Swiss National Science Foundation grants 31003A_166700/1, and 310030_184708/1; European Foundation for the Study of Diabetes/Novo Nordisk Programme for Diabetes Research; the Vontobel Foundation; the Novartis Consumer Health Foundation; the Bo and Kerstin Hjelt Foundation for diabetes type 2; the Swiss Life Foundation; the Olga Mayenfisch Foundation (to C.D.); University of Geneva-The Hebrew University of Jerusalem Joint Seed Money Funding Scheme (to Y.D. and C.D.); Research Council of Norway (NFR 251041, SC); and the Novo Nordic Foundation (NNF15OC0015054 to S.C.).
Author contributions: C.D., Y.D., and V.P. designed the study, conducted data analysis, and wrote the manuscript. V.P. performed the experiments. M.S.-R. performed histological data collection. K.-F.S. developed the Bmal1st/st mouse line. L.G. participated in the islet isolations. B.V. and S.C. conducted RNA-seq data analyses. All authors contributed to the manuscript preparation and approved the final version.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.343137.120.
References
- Ackermann Misfeldt A, Costa RH, Gannon M. 2008. β-Cell proliferation, but not neogenesis, following 60% partial pancreatectomy is impaired in the absence of FoxM1. Diabetes 57: 3069–3077. 10.2337/db08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguayo-Mazzucato C, Koh A, El Khattabi I, Li WC, Toschi E, Jermendy A, Juhl K, Mao K, Weir GC, Sharma A, et al. 2011. Mafa expression enhances glucose-responsive insulin secretion in neonatal rat β cells. Diabetologia 54: 583–593. 10.1007/s00125-010-2026-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryal RP, Kwak PB, Tamayo AG, Gebert M, Chiu PL, Walz T, Weitz CJ. 2017. Macromolecular assemblies of the mammalian circadian clock. Mol Cell 67: 770–782.e6. 10.1016/j.molcel.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atger F, Gobet C, Marquis J, Martin E, Wang J, Weger B, Lefebvre G, Descombes P, Naef F, Gachon F. 2015. Circadian and feeding rhythms differentially affect rhythmic mRNA transcription and translation in mouse liver. Proc Natl Acad Sci 112: E6579–E6588. 10.1073/pnas.1515308112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet MM, Masri S, Astarita G, Sassone-Corsi P, Della Fazia MA, Servillo G. 2016. Histone deacetylase SIRT1 controls proliferation, circadian rhythm, and lipid metabolism during liver regeneration in mice. J Biol Chem 291: 23318–23329. 10.1074/jbc.M116.737114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler J, Cannavo R, Gustafson K, Gobet C, Gatfield D, Naef F. 2014. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol 10: 739 10.15252/msb.20145218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, Hrushesky WJ, Ben-David Y. 2001. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol 158: 1793–1801. 10.1016/S0002-9440(10)64135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Huangfu D, Melton D. 2007. All β cells contribute equally to islet growth and maintenance. PLoS Biol 5: e163 10.1371/journal.pbio.0050163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy H, Gu X, Enge M, Dai X, Wang Y, Damond N, Downie C, Liu K, Wang J, Xing Y, et al. 2017. Converting adult pancreatic islet α cells into β cells by targeting both Dnmt1 and Arx. Cell Metab 25: 622–634. 10.1016/j.cmet.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Yin H, Nam D, Li Y, Ma K. 2015. Brain and muscle Arnt-like 1 promotes skeletal muscle regeneration through satellite cell expansion. Exp Cell Res 331: 200–210. 10.1016/j.yexcr.2014.08.041 [DOI] [PubMed] [Google Scholar]
- Chen Y, Lun AT, Smyth GK. 2016. From reads to genes to pathways: differential expression analysis of RNA-seq experiments using rsubread and the edgeR quasi-likelihood pipeline. F1000Res 5: 1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CW, Villani V, Buono R, Wei M, Kumar S, Yilmaz OH, Cohen P, Sneddon JB, Perin L, Longo VD. 2017. Fasting-mimicking diet promotes Ngn3-driven β-cell regeneration to reverse diabetes. Cell 168: 775–788.e12. 10.1016/j.cell.2017.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chera S, Baronnier D, Ghila L, Cigliola V, Jensen JN, Gu G, Furuyama K, Thorel F, Gribble FM, Reimann F, et al. 2014. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature 514: 503–507. 10.1038/nature13633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Takahashi JS. 2019. Circadian clock genes and the transcriptional architecture of the clock mechanism. J Mol Endocrinol 63: R93–R102. 10.1530/JME-19-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Hang Y, Shostak A, Poffenberger G, Hart N, Prasad N, Phillips N, Levy SE, Greiner DL, Shultz LD, et al. 2017. Age-dependent human β cell proliferation induced by glucagon-like peptide 1 and calcineurin signaling. J Clin Invest 127: 3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DB, Lavine JA, Suhonen JI, Krautkramer KA, Rabaglia ME, Sperger JM, Fernandez LA, Yandell BS, Keller MP, Wang IM, et al. 2010. Foxm1 is up-regulated by obesity and stimulates β-cell proliferation. Mol Endocrinol 24: 1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C. 2020. The importance of being rhythmic: living in harmony with your body clocks. Acta Physiol (Oxf) 228: e13281 10.1111/apha.13281 [DOI] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. 2010. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549. 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. 2004. Adult pancreatic β-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429: 41–46. 10.1038/nature02520 [DOI] [PubMed] [Google Scholar]
- Dusaulcy R, Handgraaf S, Heddad-Masson M, Visentin F, Vesin C, Reimann F, Gribble F, Philippe J, Gosmain Y. 2016. α-Cell dysfunctions and molecular alterations in male insulinopenic diabetic mice are not completely corrected by insulin. Endocrinology 157: 536–547. 10.1210/en.2015-1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar KA, Ciciliot S, Wright LE, Biensø RS, Tagliazucchi GM, Patel VR, Forcato M, Paz MI, Gudiksen A, Solagna F, et al. 2014. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab 3: 29–41. 10.1016/j.molmet.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher J, Montellier E, Sassone-Corsi P. 2018. Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol 28: 368–379. 10.1016/j.tcb.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Golson ML, Dunn JC, Maulis MF, Dadi PK, Osipovich AB, Magnuson MA, Jacobson DA, Gannon M. 2015. Activation of FoxM1 revitalizes the replicative potential of aged β-cells in male mice and enhances insulin secretion. Diabetes 64: 3829–3838. 10.2337/db15-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfmann BD, Schroder EA, Kachman MT, Hodge BA, Zhang X, Esser KA. 2016. Muscle-specific loss of Bmal1 leads to disrupted tissue glucose metabolism and systemic glucose homeostasis. Skelet Muscle 6: 12 10.1186/s13395-016-0082-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK_Cycle: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25: 372–380. 10.1177/0748730410379711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. 2013. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep 3: 996–1004. 10.1016/j.celrep.2013.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klochendler A, Caspi I, Corem N, Moran M, Friedlich O, Elgavish S, Nevo Y, Helman A, Glaser B, Eden A, et al. 2016. The genetic program of pancreatic β-cell replication in vivo. Diabetes 65: 2081–22093. 10.2337/db16-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DS, Cho JH, Chen L. 2012. Paracrine interactions within islets of Langerhans. J Mol Neurosci 48: 429–440. 10.1007/s12031-012-9752-2 [DOI] [PubMed] [Google Scholar]
- Ma P, Woelfle MA, Johnson CH. 2013. An evolutionary fitness enhancement conferred by the circadian system in cyanobacteria. Chaos Solitons Fractals 50: 65–74. 10.1016/j.chaos.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannic T, Meyer P, Triponez F, Pusztaszeri M, Le Martelot G, Mariani O, Schmitter D, Sage D, Philippe J, Dibner C. 2013. Circadian clock characteristics are altered in human thyroid malignant nodules. J Clin Endocrinol Metab 98: 4446–4456. 10.1210/jc.2013-2568 [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. 2010. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466: 627–631. 10.1038/nature09253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. 2003. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259. 10.1126/science.1086271 [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. 10.1016/j.cell.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Nir T, Melton DA, Dor Y. 2007. Recovery from diabetes in mice by β cell regeneration. J Clin Invest 117: 2553–2561. 10.1172/JCI32959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen HL, Theander S, Bokvist K, Buschard K, Wollheim CB, Gromada J. 2005. Glucose stimulates glucagon release in single rat α-cells by mechanisms that mirror the stimulus-secretion coupling in β-cells. Endocrinology 146: 4861–4870. 10.1210/en.2005-0800 [DOI] [PubMed] [Google Scholar]
- Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. 2015. Pancreatic β cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350: aac4250 10.1126/science.aac4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RJ, Zhang D, Guerra MT, Brill AL, Goedeke L, Nasiri AR, Rabin-Court A, Wang Y, Peng L, Dufour S, et al. 2020. Glucagon stimulates gluconeogenesis by INSP3R1-mediated hepatic lipolysis. Nature 579: 279–283. 10.1038/s41586-020-2074-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko V, Dibner C. 2018. Cell-specific resetting of mouse islet cellular clocks by glucagon, glucagon-like peptide 1 and somatostatin. Acta Physiol (Oxf) 222: e13021 10.1111/apha.13021 [DOI] [PubMed] [Google Scholar]
- Petrenko V, Gosmain Y, Dibner C. 2017a. High-resolution recording of the circadian oscillator in primary mouse α- and β-cell culture. Front Endocrinol (Lausanne) 8: 68 10.3389/fendo.2017.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko V, Saini C, Giovannoni L, Gobet C, Sage D, Unser M, Heddad Masson M, Gu G, Bosco D, Gachon F, et al. 2017b. Pancreatic α- and β-cellular clocks have distinct molecular properties and impact on islet hormone secretion and gene expression. Genes Dev 31: 383–398. 10.1101/gad.290379.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrenko V, Philippe J, Dibner C. 2018. Time zones of pancreatic islet metabolism. Diabetes Obes Metab 20: 116–126. 10.1111/dom.13383 [DOI] [PubMed] [Google Scholar]
- Petrenko V, Gandasi NR, Sage D, Tengholm A, Barg S, Dibner C. 2020. In pancreatic islets from type 2 diabetes patients, the dampened circadian oscillators lead to reduced insulin and glucagon exocytosis. Proc Natl Acad Sci 117: 2484–2495. 10.1073/pnas.1916539117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat S, Weinberg-Corem N, Tornovsky-Babaey S, Schyr-Ben-Haroush R, Hija A, Stolovich-Rain M, Dadon D, Granot Z, Ben-Hur V, White P, et al. 2011. Control of pancreatic β cell regeneration by glucose metabolism. Cell Metab 13: 440–449. 10.1016/j.cmet.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110: 251–260. 10.1016/S0092-8674(02)00825-5 [DOI] [PubMed] [Google Scholar]
- Pulimeno P, Mannic T, Sage D, Giovannoni L, Salmon P, Lemeille S, Giry-Laterriere M, Unser M, Bosco D, Bauer C, et al. 2013. Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia 56: 497–507. 10.1007/s00125-012-2779-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K, Hsu TW, Matveyenko AV. 2016. Bmal1 is required for β cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice. Diabetologia 59: 734–743. 10.1007/s00125-015-3859-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SF, Madsen JG, Frafjord KO, Poulsen L, Salö S, Boergesen M, Loft A, Larsen BD, Madsen MS, Holst JJ, et al. 2016. Integrative genomics outlines a biphasic glucose response and a ChREBP–RORγ axis regulating proliferation in β cells. Cell Rep 16: 2359–2372. 10.1016/j.celrep.2016.07.063 [DOI] [PubMed] [Google Scholar]
- Sinturel F, Petrenko V, Dibner C. 2020. Circadian clocks make metabolism run. J Mol Biol 432: 3680–3699. 10.1016/j.jmb.2020.01.018 [DOI] [PubMed] [Google Scholar]
- Stokes K, Cooke A, Chang H, Weaver DR, Breault DT, Karpowicz P. 2017. The circadian clock gene BMAL1 coordinates intestinal regeneration. Cell Mol Gastroenterol Hepatol 4: 95–114. 10.1016/j.jcmgh.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovich-Rain M, Hija A, Grimsby J, Glaser B, Dor Y. 2012. Pancreatic β cells in very old mice retain capacity for compensatory proliferation. J Biol Chem 287: 27407–27414. 10.1074/jbc.M112.350736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisa A, Glaser B, Dor Y. 2017. Metabolic stress and compromised identity of pancreatic β cells. Front Genet 8: 21 10.3389/fgene.2017.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. 2007. Growth and regeneration of adult β cells does not involve specialized progenitors. Dev Cell 12: 817–826. 10.1016/j.devcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Thorel F, Népote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. 2010. Conversion of adult pancreatic α-cells to β-cells after extreme β-cell loss. Nature 464: 1149–1154. 10.1038/nature08894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. 2007. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia 50: 348–358. 10.1007/s00125-006-0490-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, D'Hoker J, Stangé G, Bonné S, De Leu N, Xiao X, Van de Casteele M, Mellitzer G, Ling Z, Pipeleers D, et al. 2008. β Cells can be generated from endogenous progenitors in injured adult mouse pancreas. Cell 132: 197–207. 10.1016/j.cell.2007.12.015 [DOI] [PubMed] [Google Scholar]
- Yamamoto J, Imai J, Izumi T, Takahashi H, Kawana Y, Takahashi K, Kodama S, Kaneko K, Gao J, Uno K, et al. 2017. Neuronal signals regulate obesity induced β-cell proliferation by FoxM1 dependent mechanism. Nat Commun 8: 1930 10.1038/s41467-017-01869-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, Costa RH, Gannon M. 2006. The FoxM1 transcription factor is required to maintain pancreatic β-cell mass. Mol Endocrinol 20: 1853–1866. 10.1210/me.2006-0056 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Pope CF, Crawford LA, Vasavada RC, Jagasia SM, Gannon M. 2010. Gestational diabetes mellitus resulting from impaired β-cell compensation in the absence of FoxM1, a novel downstream effector of placental lactogen. Diabetes 59: 143–152. 10.2337/db09-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the deposited data in GEO are GSE151338 and GSE152751.