Abstract
Rungia pectinata is an important traditional Chinese herbal medicine from the family Acanthaceae. The complete chloroplast genome (cp genome) of the genus Rungia was sequenced for the first time. The cp genome of R. pectinata was 149,627 bp in length. It was consisted of a large single copy (LSC) region (81,976 bp) and a small single copy (SSC) region (16,626 bp), which were separated by two inverted repeats (IRs, 25,511 bp). This plastome contained 114 unique genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. The overall GC content was 38.0%. Phylogenetic analysis of nine species in Acanthaceae was also conducted. This newly sequenced cp genome will be useful to further evolutionary studies, phylogenetic studies, and pharmacognostic identification in the genus Rungia.
Keywords: Rungia pectinata, chloroplast genome, phylogenetic, herbal medicine
Rungia Nees (Acanthaceae) is a genus comprising about 50 species distributed in the tropical and subtropical regions of the Old World (Hu et al. 2011; Mabberley 2017). Rungia pectinata (L.) Nees is a widespread species in Asia, the Chinese name of R. pectinata is “infant’s herb” as it is effective in treatment of infantile indigestion (Lo 1978). In this study, the complete chloroplast genome of the genus Rungia was sequenced for the first time, which will be useful to pharmacognostic identification in medicinal species of Rungia.
The complete chloroplast genome of R. pectinata was successfully assembled and characterized based on the Illumina pair-end (PE) sequencing data. The silica-gel dried leaves of R. pectinata were collected from China, Hainan Province, Baisha Xian, Nankai Xiang, Daoyin Cun (18°59′0.93″N, 109°19′45.88″E, 400 m). The voucher specimen (Z. L. Lin & Q. L. Wang 14022302) was deposited in the South China Botanical Garden Herbarium, Guangzhou, China. Total genomic DNA was isolated with a modified CTAB (Cetyl Trimethyl Ammonium Bromide) method (Doyle and Doyle 1987). Illumina Pair-end (PE) sequencing was performed on the Illumina HiSeq 2500 instruments at BGI-Wuhan. The sequenced clean PE reads were filtered using GetOrganelle pipeline (Jin et al. 2018) to get plastid-like reads. The filtered reads were assembled using SPAdes version 3.11.1 (Bankevich et al. 2012). The genome was automatically annotated using Plastid Genome Annotator (PGA) (Qu et al. 2019) coupled with manual corrections. The final complete plastome was deposited into GenBank (accession number MK946456).
The complete cp genome of R. pectinata was 149627 bp in length and presented a typical quadripartite structure including a large single copy (LSC) region (81,979 bp), a small single copy (SSC) region (16,626 bp), and two inverted repeat regions (IRs, 25,511 bp). The overall GC content of R. pectinata plastome was 38.0%. The R. pectinata plastome contained 114 different genes, including 80 protein-coding genes, 30 tRNA genes, and four rRNA genes. We also detected owing to the presence of internal stop codons, the gene ycf15 was identified as pseudogene in the plastome of R. pectinata.
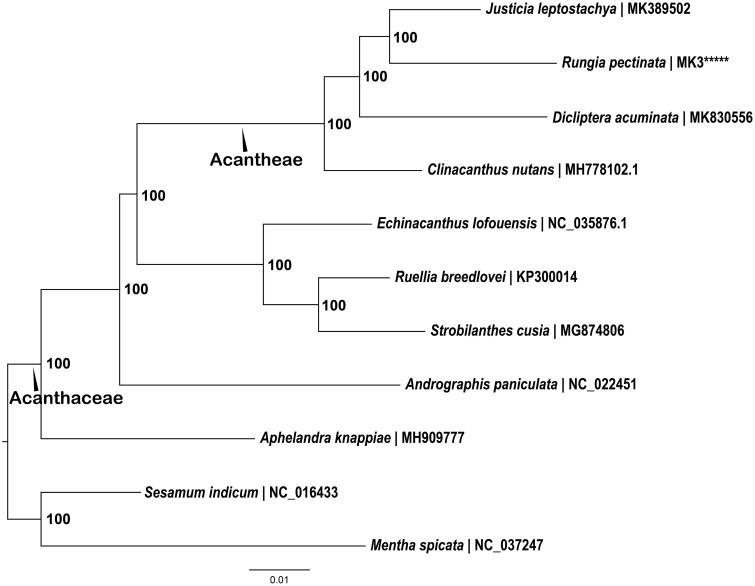
To reconstruct the phylogeny of Acanthaceae, nine Acanthaceae cp genomes were included in the phylogenetic analysis; plastomes of two relative families were used as outgroups (see Figure 1 for details). The plastomes (each excluding one IR) of all the species were aligned using MAFFT (version 1.3.7) (Katoh and Standley 2013) implemented in Geneious v. 11.0.4 and adjusted manually when necessary. The maximum-likelihood (ML) phylogeny was reconstructed using RAxML version 8.0.0 (Stamatakis 2014).
Figure 1.
Maximum-likelihood (ML) phylogeny of Acanthaceae based on complete chloroplast genome sequences. Numbers at the right of nodes are bootstrap support values. GenBank accession numbers: Andrographis paniculata (NC_022451), Ruellia breedlovei (KP300014), Strobilanthes cusia (MG874806), Echinacanthus lofouensis (NC_035876.1), Aphelandra knappiae (MH909777), Justicia leptostachya (MK389502), Clinacanthus nutans (MH778102.1), Dicliptera acuminata (MK830556), Sesamum indicum (NC_016433), Mentha spicata (NC_037247), and Rungia pectinata (MK946456).
The present phylogenetic analyses strongly confirmed the monophyly of the family as previous reported (McDade et al. 2008) (Figure 1). Furthermore, all of the four Acantheae taxa formed a monophyletic group. Among the nine Acanthaceae species in this work, R. pectinata is most close to Justicia leptostachya, which all belong to Subtribe Justiciinae. All the nodes received 100% bootstrap support. The genome data in this paper can be subsequently used for phylogenetic studies in the genus Rungia and will contribute to further understanding of the phylogeny of Subtribe Justiciinae.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Hu J, Deng Y, John RIW, Thomas FD. 2011. Rungia in Flora of China 19 In: Wu Z, Raven PH, Hong D, editors. Flora of China. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 443–477. [Google Scholar]
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 4:256479. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HS. 1978. Notes on the genus Rungia (Acanthaceae) in China. Acta Phytotaxon Sin. 16:91–95. [Google Scholar]
- Mabberley DJ. (2017) Mabberley's plant-book: a portable dictionary of plants, their classifications and uses. 4th ed Cambridge: Cambridge University Press. [Google Scholar]
- McDade LA, Daniel TF, Kiel CA. 2008. Toward a comprehensive understanding of phylogenetic relationships among lineages of Acanthaceae sl (Lamiales). Am J Bot. 95:1136–1152. [DOI] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and fexible batch annotation of plastomes. Plant Methods. 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]