Abstract
2′-O-Methyl (Nm) is a highly abundant post-transcriptional RNA modification that plays important biological roles through mechanisms that are not entirely understood. There is evidence that Nm can alter the biological activities of RNAs by biasing the ribose sugar pucker equilibrium toward the C3′-endo conformation formed in canonical duplexes. However, little is known about how Nm might more broadly alter the dynamic ensembles of flexible RNAs containing bulges and internal loops. Here, using NMR and the HIV-1 transactivation response (TAR) element as a model system, we show that Nm preferentially stabilizes alternative secondary structures in which the Nm-modified nucleotides are paired, increasing both the abundance and lifetime of low-populated short-lived excited states by up to 10-fold. The extent of stabilization increased with number of Nm modifications and was also dependent on Mg2+. Through phi-value analysis, the Nm modification also provided rare insights into the structure of the transition state for conformational exchange. Our results suggest that Nm could alter the biological activities of Nm-modified RNAs by modulating their secondary structural ensembles as well as establish the utility of Nm as a tool for the discovery and characterization of RNA excited state conformations.
INTRODUCTION
Nm is a highly abundant post-transcriptional modification (Figure 1A) found in non-coding (1–3) and coding RNAs (4,5). It can be added to the ribose sugar moiety of all four nucleotides (Nm = Am, Um, Cm and Gm) (Figure 1A) either via stand-alone methyltransferases (6) or by the enzyme fibrillarin when guided by box C/D small nucleolar RNAs (snoRNAs) (7,8). Nm modifications are important for the cellular activity of diverse RNAs (9,10) and have been linked to diseases (11–14). For example, ribosomal RNAs (rRNA) contain more than 100 Nm sites (15), many in the decoding and peptidyl-transferase centers (16). Downregulation of fibrillarin and loss of rRNA Nm sites results in impaired ribosomes that are incapable of translating mRNA (17). Conversely, overexpression of fibrillarin and the concomitant increase in Nm modifications is associated with an increase in protein translation in rapidly dividing breast cancer cells (12). In the spliceosome, all small nuclear RNAs (snRNAs) harbor Nm modifications, some of which are required for proper spliceosome assembly and function (18,19). The loss of Nm modifications on snRNA U6 due to knockout of the La-related protein, which helps load U6 snRNA onto the box C/D snoRNA protein complex, changes splicing fidelity, impairs spermatogenesis in mice (20), and contributes to the Alazami syndrome (21). Loss of snoRNA-guided Nm modifications on snRNAs also leads to profound defects in cardiac mRNA splicing and development (13,22,23).
Figure 1.
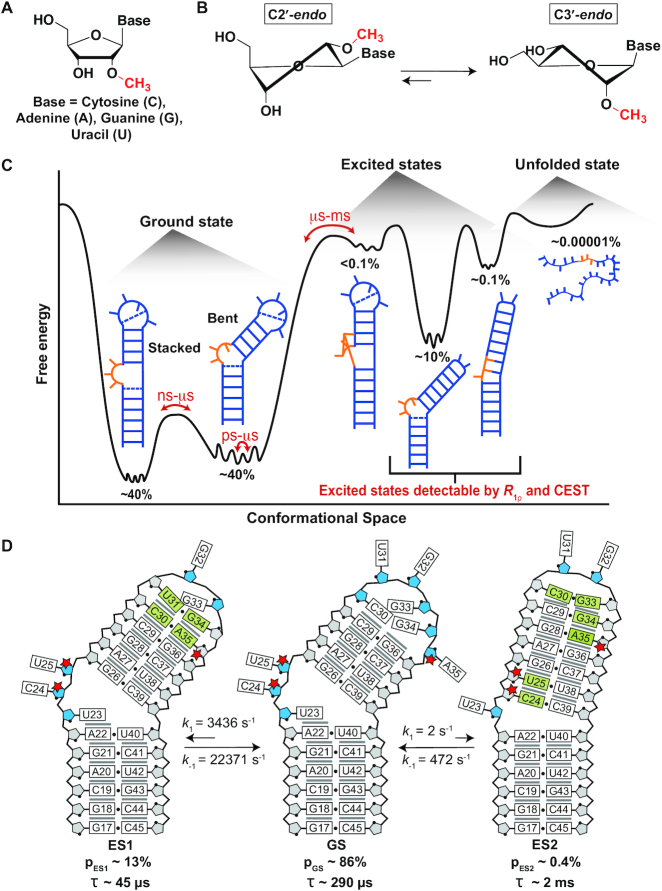
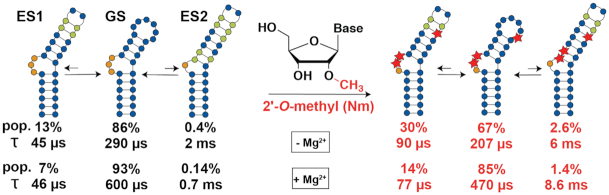
(A) Chemical structure of a 2′-O-methylated nucleoside (Nm). (B) Nm biases the sugar pucker towards C3′-endo (34). (C) The RNA free energy landscape of HIV-1 TAR includes (i) the dominant GS secondary structure that interconverts between the bent and coaxially stacked conformations, (ii) ESs that have non-native secondary structures and (iii) the unfolded/melted conformations. ESs with populations >0.1% can be studied using R1ρ and CEST NMR experiments. (D) Secondary structure of the TAR GS and ESs. The exchange parameters shown are for TAR in the absence of Mg2+ as reported previously (69,70). Nucleotides that undergo sugar repuckering from C2′-endo (blue) in the GS to C3′-endo (grey) in the ES are indicated using green bases in the ESs. Red stars indicate the Nm-modified residues. Black dots denote base pairing and grey lines denote stacking. The lifetime (τ) of each ES is equal to 1/k-1, and τ of the GS = 1/(k1,ES1 + k1,ES2).
In contrast to the well-studied post-transcriptional modification N6-methyladenosine (m6A) (24–28), which mainly exerts its biological activity by recruiting reader proteins (28–31), no reader proteins have been discovered for Nm to date. Rather, it appears that in some cases, the Nm modification serves to alter the conformational preferences of the Nm-modified RNAs. In isolated mononucleotides and unpaired pyrimidine nucleotides, the ribose moiety exists in a 60:40 dynamic equilibrium between C3′-endo and C2′-endo sugar pucker conformations (32), whereas most paired nucleotides or nucleotides that stack intra-helically predominantly adopt the C3′-endo conformation (33). In pyrimidine nucleosides (Um and Cm) and dinucleotides (UmpU and CmpC), 2′-O-methylation biases the sugar pucker equilibrium in favor of C3′-endo by 0.1–0.6 kcal/mol (Figure 1B) (34–37). This bias arises due to intra-residue steric repulsion between the 2′-O-methyl, the 3′-phosphate, and the 2-carbonyl groups when in the C2′-endo conformation (34). The Nm modification has also been shown to stabilize RNA helices by an average of ∼0.2 kcal/mol per Nm (38). This is due to a reduction in entropy loss upon duplex formation, as a result of pre-organizing the C3′-endo sugar pucker in the single-strand (34,39) and/or favorable enthalpic interactions (40). Since Nm affords increased thermal stability of duplexes and greater stability toward nuclease digestion, it has important applications in FDA-approved oligonucleotide therapeutics (41–44), siRNAs (45,46) and anti-miRNAs (47).
These biases in RNA conformation induced by Nm can serve important biological roles. For example, the bias in sugar pucker introduced by Um at the first anticodon position in tRNA is proposed to reduce misrecognition of noncognate codons, which require a C2′-endo conformation (34). In the A loop of the 23S rRNA, the universally conserved Um2552 intercalates between two bases to help maintain the active conformation for the G2553 base, which is directly involved in accommodation of the aminoacyl-tRNA (48). These and other modifications appear to play the role of maintaining distorted RNA conformations at the functional interfaces of the ribosome (48). U2–U6 and U4–U6 snRNAs are critical structural elements of the spliceosome that are rich in Nm modifications (3). These snRNAs undergo changes in secondary structure and base-pairing which are essential for the assembly and disassembly of the spliceosome as well as in cycling between different conformational states required for catalysis (49,50). Nm modifications in the active core of the spliceosome U2–U6 snRNA complex have been shown to promote a conformational transition between a three-way and four-way junction proposed to aid correct positioning of pre-mRNA for exon ligation (51). The modification can also sterically hinder stabilizing interactions between Nm modified mRNAs and ribosomal-monitoring bases, thus disrupting tRNA selection and proofreading and inhibiting translation (5,52).
For the vast majority of the cases however, the role of Nm modifications has remained elusive. While it is known that Nm modifications can stabilize canonical duplexes, little is known about how the modification might more broadly impact the propensities of RNAs to form alternative secondary structures in more complex motifs (such as U2–U6) in which they are frequently found (10,53,54). By biasing sugar pucker and favoring helical conformations, we hypothesized that Nm could redistribute the RNA secondary structural ensemble.
Studies over the past decades have emphasized the dynamic nature of RNA structures showing that they do not adopt a single conformation but rather an ensemble of conformations that have different populations and that interconvert on timescales ranging between picoseconds and seconds (Figure 1C). In addition to the dominant ground state (GS) secondary structure, included in the dynamic ensembles of RNAs are low-abundance short-lived conformations referred to as ‘excited states’ (ESs) that form on the micro- to-millisecond timescale and that have alternative secondary structures (55–62). Despite being low-populated and short-lived, ESs have been detected with the use of NMR R1ρ relaxation dispersion (RD) and chemical exchange saturation transfer (CEST) experiments (59–65) and have been shown to play critical roles in the folding (66) and regulatory functions of RNAs (56,59), and are increasingly considered to be attractive drug targets (67). Relative to the GS, RNA ESs form by reshuffling base pairs in and around bulges and loops and they tend to be enriched in helical residues, as unpaired nucleotides in bulges and loops from the GS form mismatches in the ES (Figure 1C and D).
Given the enrichment in paired residues in the ES relative to the GS, Nm could potentially increase the population and lifetime of ESs. This would potentially provide a means to redistribute RNA ensembles without changing sequence, providing an additional chemical layer with which to optimize RNA structural dynamics for folding and function. In addition to potentially explaining some of the biological roles of Nm, having an ability to control RNA conformational ensembles through Nm modifications may provide a valuable chemical tool for elucidating the characteristics of these low-abundance and short-lived conformations that are difficult to study (62). Here, we have tested our hypothesis using the well-characterized HIV-1 transactivation response element (TAR) RNA as a model system for non-canonical bulge motifs (68). Although TAR is not naturally modified with Nm, its secondary structural ensemble in its unmodified form has been extensively characterized previously using NMR relaxation dispersion (RD) (69,70). It therefore provides an excellent model system for testing our Nm-directed preferential stabilization of paired ESs hypothesis.
Prior NMR RD studies (69,70) showed that TAR exists as a dynamic ensemble involving a GS and at least two ESs (ES1 and ES2) (Figure 1C) (71). ES1 forms rapidly (kex = k1 + k-1 ∼ 25 800 s−1) by zipping up the apical loop to form two mismatches, and has a relatively high population (pES1) of ∼13% (69). On the other hand, ES2 forms more slowly (kex = 474 s−1) through more extensive remodeling of the trinucleotide bulge, upper stem, and apical loop, and has a substantially lower population (pES2 ∼0.4%) (70). Based on a recent NMR study (33), the unpaired bulge and apical loop residues C24, U25 and A35 are highly enriched in the C2′-endo conformation in the GS whereas they are predominantly C3′-endo and helical in ES1 (A35) and ES2 (C24, U25 and A35). We reasoned that introducing Nm at these three nucleotides would stabilize both ESs relative to the GS. Additionally, we hypothesized that the extent of stabilization would be greater for ES2 than ES1, given that all three modifications favor ES2 whereas only Am35 favors ES1.
Indeed, NMR RD measurements showed that Nm increased both the abundance and the lifetime of TAR ES1 and ES2. Through a phi-value analysis (72–76), the impact of Nm on the exchange kinetics also allowed us to obtain rare conformational insights into the transition state (TS) for the GS-ES conformational exchange. Our results reveal a new mechanism by which Nm could alter the biological activity of Nm-modified RNAs as well as establish the utility of Nm as a chemical tool for the in-depth characterization of alternative ES RNA conformations.
MATERIALS AND METHODS
Preparation of RNA samples
Unlabeled TAR, TAR-A35, TAR-C24U25 and TAR-C24U25A35 and 15N and 13C site-labeled TAR and TAR-C24U25A35 oligonucleotides were synthesized using a MerMade 6 Oligo Synthesizer (BioAutomation) for solid-phase synthesis using standard phosphoramidite chemistry and deprotection protocols (77,78). Unlabeled phosphoramidites were purchased from ChemGenes and site-labeled phosphoramidites were synthesized as described previously (55,79). All RNA samples were prepared using 2′-TBDMS protected phosphoramidites using 1 μmol standard synthesis columns (1000 Å). The final 5′-DMT (4,4′-dimethoxytrityl) was removed during the synthesis for DMT-off deprotection and PAGE purification. Removal of nucleobase protecting groups and cleavage from the 1 μmol columns was achieved using 1 ml of 30% ammonium hydroxide and 30% methylamine (1:1) followed by 2 h incubation at room temperature. The solution was then air-dried. In order to remove the 2′-TBDMS protecting groups, 100 μl DMSO was added and the samples were heated at 65°C for 5 min to ensure the samples were fully dissolved. Then 125 μl TEA-3HF was added and the samples were heated at 65°C for 2.5 h. Samples were then precipitated overnight using 3 M sodium acetate and 100% ethanol, air dried, then dissolved in water for gel purification. All RNA samples were purified using a 20% (w/v) polyacrylamide gel with 8 M urea and 1× Tris/borate/EDTA. RNA was removed from the excised gel by electro-elution (Whatman, GE) in 1× Tris/acetic acid/EDTA followed by ethanol precipitation. The purified RNA was then annealed in water at a concentration of 50 μM by heating at 95°C for 5 min followed by cooling on ice for 60 min. The samples were then buffer exchanged five times using an Amicon Ultra-15 centrifugal filter (EMD Milipore) with a 3 kDa cutoff into NMR buffer (15mM sodium phosphate, 25 mM NaCl, 0.1 mM EDTA, 10% D2O) at pH 6.4 with or without 1/3 mM Mg2+ for the site-labeled/unlabeled samples. The final concentration of unlabeled and site-labeled RNA NMR samples ranged between 0.7–2.0 and 0.3–0.7 mM, respectively.
UV optical melting experiments
UV optical melting experiments were conducted on a PerkinElmer Lambda 25 UV/VIS spectrometer with RTP 6 Peltier Temperature Programmer and a PCB 1500 Water Peltier System. Samples with ∼3 μM RNA were prepared by buffer exchanging the RNA into desired buffers (15 mM sodium phosphate, 25 mM NaCl, 0.1 mM EDTA, pH 6.4 with or without 3 mM Mg2+ five times with a centrifugal concentrator (EMD Millipore). Optical melting experiments were performed in triplicates using a sample volume of 400 μl in a Teflon-stoppered 1 cm path length quartz cell. The absorbance at 260 nm (A260) was measured as a function of varying the temperature between 15 and 95°C at a ramp of 1°C/min.
The UV melting data was fit assuming two-state (in the presence and absence of Mg2+) and three-state models (in the absence of Mg2+) to obtain the thermodynamic parameters of interest using an in-house Python script available at https://github.com/alhashimilab/UV_melting. For the two-state model, the absorbance at 260 nm was fit using the standard equations:
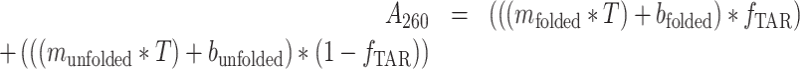 |
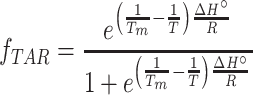 |
mfolded, bfolded, munfolded and bunfolded are coefficients representing the temperature dependence of the extinction coefficients of the folded and unfolded/single-stranded species, T is the temperature (K), fTAR is the fraction of folded TAR at a given temperature, R is the gas constant (kcal/mol/K), Tm is the melting temperature (K) and ΔHo is the enthalpy of annealing (kcal/mol). The free energy and entropy of annealing, ΔGo and ΔSo were calculated using the following equation:
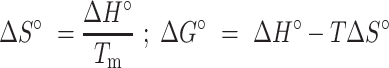 |
In the three-state model, the measured absorbance at 260 nm (A260) was fit assuming contributions from GS, ES and unfolded conformations:
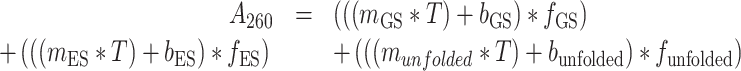 |
The relative fraction of each species is given by:
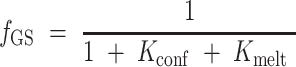 |
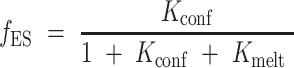 |
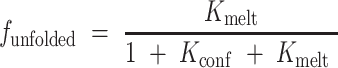 |
In which the relevant equilibrium constants Kconf and Kmelt are given by:
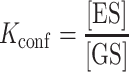 |
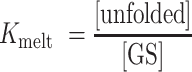 |
The free energies (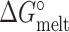 and
and 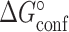 ), enthalpies (
), enthalpies (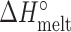 and
and 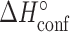 ), and entropies (
), and entropies (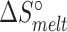 and
and 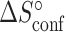 ) of the melting and conformational change can be expressed in terms of Kconf and Kmelt:
) of the melting and conformational change can be expressed in terms of Kconf and Kmelt:
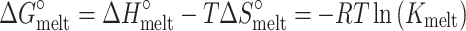 |
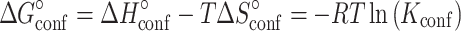 |
where T is the temperature in units Kelvin.
The value of 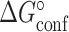 was constrained to the NMR RD derived value during the constrained three-state fit (
was constrained to the NMR RD derived value during the constrained three-state fit (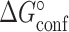 = 13% and 30% for TAR and TAR-C24U25A35 respectively in the absence of Mg2+) but was allowed to float during the unconstrained three-state fit.
= 13% and 30% for TAR and TAR-C24U25A35 respectively in the absence of Mg2+) but was allowed to float during the unconstrained three-state fit.
With the exception of the triply methylated TAR sample in the absence of Mg2+, all of the UV melts could be adequately fit to the two-state model (Supplementary Figure S1). For the triply methylated TAR sample in the absence of Mg2+, small deviations could be observed from the two-state fit particularly early on for the lower baseline (Supplementary Figure S1). However, comparison of the constrained and unconstrained three-state fits reveals that the thermodynamic parameters are poorly determined when carrying out a three-state fit, and different thermodynamic parameters can yield equally good fits. This strongly suggests that an unconstrained three-state fit would be overfitting the data. Nevertheless, the constrained three-state fits yielded thermodynamic parameters for the GS that are minimally affected relative to that obtained from a two-state fit (differences in 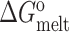 are < 0.4 kcal/mol 37°C). To avoid overfitting of the data, only the two-state fits are presented in the main paper and the constrained and unconstrained three-state fits are provided Supplementary Figure S1 and Table S1.
are < 0.4 kcal/mol 37°C). To avoid overfitting of the data, only the two-state fits are presented in the main paper and the constrained and unconstrained three-state fits are provided Supplementary Figure S1 and Table S1.
Note that we were unable to independently measure NMR RD data for TAR at 3 mM Mg2+ which are the conditions used to measure the UV melts as this results in severe line broadening. Since the ES1 population is even lower in the presence of Mg2+, we avoid an unconstrained three-state fit of these data as the danger of overfitting is even greater.
The quality of the fit (Supplementary Figure S1) was quantified using the reduced 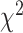 (r
(r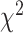 ) defined as:
) defined as:
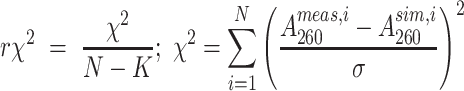 |
where N is the total number of data points, K is the number of unknowns (6 for two-state, 10 for three-state unconstrained and 9 for a three-state constrained fit), 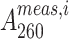 and
and 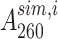 are the ith measured and simulated A260 values and σ is the error in the absorbance measurement, arbitrarily defined as the standard deviation of the first twenty A260 values. Statistical comparisons of the two- and three-state fits for the UV melting curves (Supplementary Table S2) were performed using the Akaike Information Criteria (AIC) (80) and Bayesian Information Criteria (BIC) (81) weights as described previously. Python scripts for performing the two- and three-state analyses and the statistical tests, along with the raw data and the fits can be found at https://github.com/alhashimilab/UV_melting.
are the ith measured and simulated A260 values and σ is the error in the absorbance measurement, arbitrarily defined as the standard deviation of the first twenty A260 values. Statistical comparisons of the two- and three-state fits for the UV melting curves (Supplementary Table S2) were performed using the Akaike Information Criteria (AIC) (80) and Bayesian Information Criteria (BIC) (81) weights as described previously. Python scripts for performing the two- and three-state analyses and the statistical tests, along with the raw data and the fits can be found at https://github.com/alhashimilab/UV_melting.
NMR experiments
All NMR experiments were performed on a 600 MHz Bruker NMR spectrometer equipped with an HCPN cryogenic probe. Data were processed using NMRpipe (82) and analyzed using SPARKY (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco). Resonances in Nm-modified TAR were assigned based on prior assignments of unmodified TAR (33) and confirmed using 2D HSQC, HMQC, and 1H-1H NOESY experiments (150 ms mixing time).
Determining ES1 population using chemical shift perturbation (CSP) analysis
Because the GS-ES1 exchange is fast on the NMR chemical shift timescale (69), the observed resonance (δobs) in the 2D [13C, 1H] HSQC spectra of unmodified TAR corresponds to a population-weighted average between the chemical shifts of the GS (δGS) and ES1 (δES1):
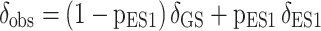 |
in which 1 − pES1 and pES1 are the populations of the GS and ES1, respectively. For unmodified TAR, pES1 ∼13% and Δδ (G34-C8) = δES1 − δGS = 2.6 ppm, as determined previously using 13C R1ρ RD (69). This allowed determination of δGS and δES1 for the various spins that experience RD (C30-C1′, U31-C1′, U31-C6, G34-C1′ and G34-C8). Note that A35-C1′ and A35-C8 are expected to exhibit CSPs towards ES1 chemical shifts; however, these resonances were excluded from analysis since they exhibit sizeable CSPs due to the chemical modification itself as verified using density functional theory (DFT) calculations (Supplementary Table S3). Based on the δobs measured for Nm-modified TAR, and the δGS and δES1 values deduced for unmodified TAR, pES1 was computed for Nm-modified TAR for each of C30-C1′, U31-C1′, U31-C6, G34-C1′ and G34-C8 (Supplementary Table S4). Similar pES1 values (average pES1 = 28.7 ± 1.5% in TAR-A35 and 28.8 ± 3.1% in TAR-C24U25A35) were obtained for these spins in the Nm-modified TAR consistent with a concerted shift in population toward ES1. Here the uncertainty represents the standard deviation across the different spins. The same approach was used to estimate pES1 for site-labeled G34-C8 TAR (pES1 = 7.3%) and TAR-C24U25A35 (pES1 = 14.5%) in 1 mM Mg2+ using the δGS and δES1 values calculated for TAR in the absence of Mg2+ (Supplementary Table S4).
13C/15N R1ρ experiments
13C/15N R1ρ RD measurements were performed on site-labeled TAR and TAR-C24U25A35 RNA in the presence or absence of 1 mM Mg2+ as described previously (62). RD measurements on the unmodified chemically synthesized site-labeled TAR were used to confirm reproducibility with previously published data on uniformly 13C/15N labeled TAR prepared using in vitro transcription (69,70). The spin lock power and offset combinations used in R1ρ experiments are listed in Supplementary Table S9.
Analysis of 13C/15N R1ρ data
The peak intensity in 13C/15N R1ρ measurements at each relaxation delay was extracted using NMRPipe (82). The R1ρ value was obtained by fitting the peak intensities at all delay points to a mono-exponential decay and the error was estimated by Monte-Carlo simulation with 500 iterations as described previously (62). The off-resonance R1ρ data was fit to a two-state exchange model using the Bloch-McConnel equations and an in-house Python script with the fitting errors estimated by a Monte-Carlo scheme (62). The fitted exchange parameters included the population (pB), exchange rate (kex), forward rate constant (k1), backward rate constant (k-1), and the difference in chemical shift between the ES and GS (Δω = ωES – ωGS). In all cases, the fitting assumed R1,GS = R1,ES = R1 and R2,GS = R2,ES = R2. In the R1ρ plots in Figures 3 and 4, ΩOBS = ωOBS − ωRF, where ωOBS is the Larmor frequency of the observed resonance and ωRF is angular frequency of the applied spin-lock power.
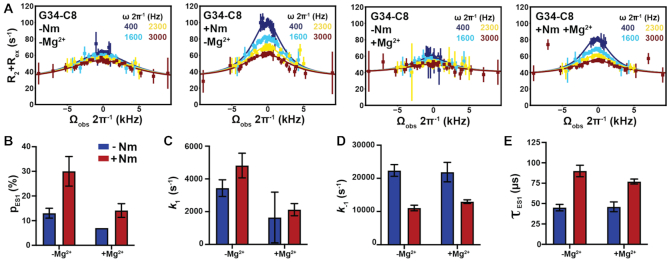
Figure 3.
Impact of Nm on GS–ES1 exchange. (A) Off-resonance R1ρ13C RD profiles for G34-C8 in TAR and TAR-C24U25A35 at 25°C, in the absence and presence of 1 mM Mg2+. R1ρ data was fit with a two-state model using Bloch-McConnell equations. Spin-lock powers are color-coded. Errors in the R1ρ data were calculated as described previously (110). (B–E) Comparison of ES1 population (pES1), forward (k1) and backward (k–1) rate constants, and lifetime (τ) between TAR and TAR-C24U25A35, in the absence and presence of 1 mM Mg2+, as obtained from fitting R1ρ data. Error bars were calculated using a Monte-Carlo scheme (110).
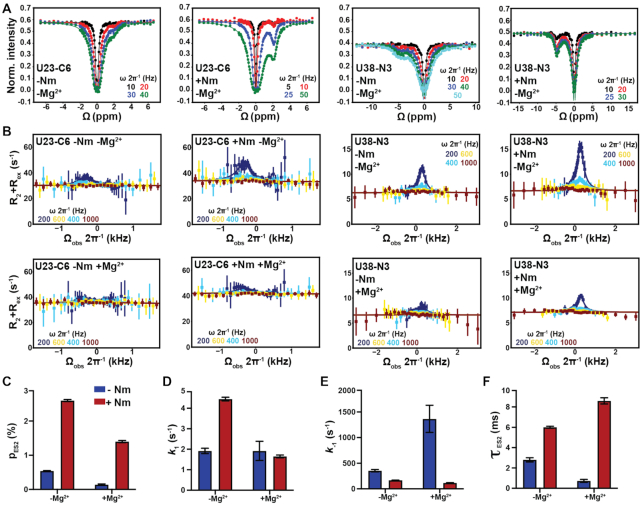
Figure 4.
Impact of Nm on GS–ES2 exchange in TAR. (A) 13C and 15N CEST profiles in TAR and TAR-C24U25A35 in the absence of Mg2+. (B) Off-resonance 13C and 15N R1ρ RD profiles for U23-C6 and U38-N3 in TAR and TAR-C24U25A35 in the absence and presence of 1 mM of Mg2+. R1ρ data was fit with a two-state model using Bloch–McConnell equations. Fits of R1ρ profiles were preformed fixing the population to the value measured using CEST. Errors in the R1ρ data were calculated as described previously (110). Spin-lock powers in CEST and R1ρ are color-coded. (C–F) Comparison of pES2, k1 and k-1 rate constants, and τ ES2 between TAR and TAR-C24U25A35 in the absence and presence of 1 mM Mg2+ obtained from fitting the CEST and R1ρ data. Error bars were calculated using a Monte-Carlo scheme (62).
GS-ES1 exchange
For the fast exchanging ES1, both in absence and presence of Mg2+, the initial alignment of the magnetization was assumed to be along the average effective field of the GS and ES (ωeff,AVG) when fitting data to the Bloch-McConnel equations (62). Because the GS-ES1 exchange is fast on the NMR chemical shift timescale (69), and only a single RD probe (G34-C8) was utilized, the exchange parameters, particularly pES1, have high uncertainty (Supplementary Table S5). Nevertheless, fitting the R1ρ data measured for unmodified TAR in the absence of Mg2+ assuming the previously reported pES1 (∼13%) (69) yielded exchange parameters (kex, k1, k−1 and Δω) that are within error to those reported previously when using many RD probes and fully labeled TAR (Supplementary Table S5). When fitting R1ρ data for TAR in the presence of 1 mM Mg2+, the population was fixed to the value determined by CSP analysis (i.e. 7%, Supplementary Table S4). Because Nm slows down the GS-ES1 exchange, the exchange parameters for TAR-C24U25A35 have lower uncertainty (Supplementary Table S5). Based on the two-state fit of the R1ρ data, ΔωG34-C8 = 2.4 ± 0.2 ppm for TAR-C24U25A35 in excellent agreement with values measured in unmodified TAR (2.6 ± 0.2 ppm). The similar Δω values indicates that the Nm does not lead to a new ES but rather changes the properties of the pre-existing ES1 (Supplementary Table S5). The fit revealed that Nm increases the ES1 population ∼2-fold, from 13% to 30% in the absence of Mg2+ and from 7% to 14% in the presence of Mg2+, in excellent agreement with the CSPs (Figure 3B and Supplementary Table S5).
GS-ES2 exchange
Because GS-ES2 is in slow exchange, the initial magnetization was assumed to be aligned along the GS when fitting R1ρ data to the Bloch-McConnel equations. Global fitting was carried by sharing pES2 and kex for U23-C6 and U38-N3. Due to degeneracy arising from slow exchange (83), R1ρ data measured for TAR in the absence of Mg2+ and for TAR-C24U25A35 in the absence and presence of Mg2+ was fit by fixing the population to the values determined using CEST (Supplementary Table S8). Based on a two-state fit of the R1ρ data, the Δω values obtained for U23-C6 and U38-N3 were in very good agreement with values measured for unmodified TAR, indicating that the Nm does not lead to a new ES but rather changes the properties of the pre-existing ES2 (Supplementary Table S8). The addition of 1 mM Mg2+ does not significantly affect the chemical shifts of the ES2 probes (U23-C6 and U38-N3) used in this study (Supplementary Figure S9). Therefore, the weak profiles measured for U23-C6 in the presence of Mg2+ (Figure 4B) can be attributed to a reduced Δω due to a Mg2+ induced downfield shift in U23-C6 GS chemical shift (Supplementary Figure S4D). A two-state fit of the U38-N3 data yielded Δω values that are in very good agreement to those measured for TAR in the absence of Mg2+.
13C/15N CEST experiment
All the 13C/15N CEST measurements for probing GS-ES2 exchange were performed on site-specific labeled TAR (without Mg2+) and TAR-C24U25A35 RNA (with and without 1 mM Mg2+) as described previously (83–85). The spin lock power and offset combinations used in CEST measurements are listed in Supplementary Table S10. The relaxation delay was 0.2 sec for U23-C6 (TAR, without Mg2+), U23-C6 (TAR-C24U25A35, without Mg2+), U38-N3 (TAR-C24U25A35, without Mg2+) and 0.3 sec for U38-N3 (TAR, without Mg2+), U23-C6 (TAR-C24U25A35, with Mg2+) and U38-N3 (TAR-C24U25A35, with Mg2+).
Analysis of 13C/15N CEST data
For each spin lock power and offset, the peak intensities in the 1D 13C/15N CEST were extracted using NMRPipe (82). The uncertainty in peak intensity was obtained from the intensity variations in the regions of CEST profiles that do not contain any intensity dips (85). The exchange parameters were obtained by fitting the data to the two-state Bloch-McConnel equations using an in-house Python script with the fitting errors estimated by a Monte-Carlo scheme with 100 iterations (83). In all cases, the observed peak was assumed to be the GS peak, as the system is in slow exchange regime (62). ES2 global fitting was carried out by sharing pB and kex between U23-C6 and U38-N3 and assuming R1,GS = R1,ES = R1 and R2,GS = R2,ES = R2. In the CEST profiles shown in Figure 4 and Supplementary Figure S7, Ω = ωRF-ωOBS where ωOBS is the Larmor frequency of the observed resonance and ωRF is angular frequency of the applied spin-lock power.
Density functional theory calculations
To understand the contributions to the chemical shift induced by introduction of Nm, due to introduction of the methyl group and/or sugar re-puckering, sugar chemical shifts were computed with and without the 2′-O-methyl group for both C2′-endo and C3′-endo sugar pucker conformations. Density functional theory (DFT) calculations using Gaussian 09c (Gaussian Inc.) (86) were used to compute chemical shifts for methylated and unmethylated RNA mono-nucleosides as described previously (87). Starting structures for the mononucleotides rA and rU with C3′-endo sugar pucker were generated using the RNA fiber module of the 3DNA suite with the χ torsion angle fixed around -160° (88). Starting structures for rA and rU with C2′-endo sugar were derived from the corresponding dA and dT model by adding a 2′-hydroxl group on the deoxyribose and removing the 7-methyl group from dT in GaussView (https://gaussian.com/gaussview6). The χ torsion angle was fixed around the most common glycosidic torsion angle (−100°) for C2′-endo RNA (89). The nucleoside motif was generated by removing the nucleotide phosphate group in GaussView. The γ torsion angle was fixed at 60° (89). As a benchmark, the calculated chemical shift of both C2′-endo and C3′-endo for unmodified nucleosides were in good agreement with prior DFT calculation on the RNA with different sugar pucker (89,90) and with experimental measurements (33). The structure of 2′-O-methyl modified rA and rU were generated by adding a 2′-O-methyl group on the ribose. In order to examine the effect of the chemical shifts with and without 2′-O-methyl group, geometry optimizations were only applied on the added 2′-O-methyl functional group using the B3LYP functional (91) with a single run of 6-311+G (2d, p) basis sets (92). 13C chemical shifts were then calculated using the GIAO method (93) with the B3LYP/6-311+G (2d, p) basis sets. The calculated chemical shifts were then referenced to isotropic magnetic shielding of TMS.
Phi-value analysis
Phi-value analysis (73) was carried out by computing phi (Supplementary Table S7) using the following equation:
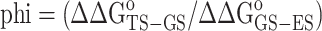 |
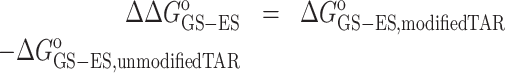 |
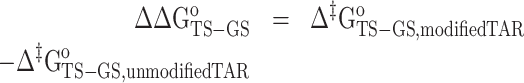 |
where ΔΔGoGS-ES is the change in the free-energy difference between GS and ES induced by the Nm modification obtained from ES population (pES) obtained using R1ρ and CEST experiments (Supplementary Tables S5 and S8):
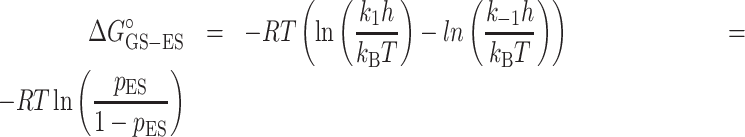 |
where R is the gas constant, T is temperature, h is Planck's constant, kB is Boltzmann's constant, k1 is the forward rate constant, and k−1 backward rate constant.
ΔΔGoTS-GS is the change in the forward free-energy barrier (Δ‡Go) induced by Nm and calculated using the forward rate constant k1 obtained from R1ρ and CEST experiments (Supplementary Tables S5 and S8):
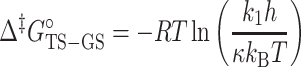 |
where 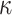 is the transmission coefficient and is assumed to be unity. Δ‡Go values are reported in Supplementary Table S6.
is the transmission coefficient and is assumed to be unity. Δ‡Go values are reported in Supplementary Table S6.
A phi-value = 0 indicates an early TS, where the site of the modification in the TS exhibits a similar environment as in the GS (i.e. paired/unpaired or helical/bulged out) (73). A phi-value = 1 indicates that the site of the modification in the TS is as paired/unpaired as in the ES. Phi-value analysis is not feasible to compare TAR in the absence of Mg2+ with TAR in the presence of Mg2+ since adding Mg2+ shifts the GS conformation to the coaxially stacked state that is ∼3 kcal/mol more stable.
RESULTS
Nm modifications have a small impact on the thermal stability of the TAR GS
We first used UV melting experiments to assess the impact of the modification on the overall thermal stability of TAR, which is dominated by the GS (population ∼ 86%) and to a lesser extent ES1 (population ∼ 13%) (see Materials and Methods). Given that other post-transcriptional modifications such as m6A have been shown to affect RNA thermal stability and structural dynamics in a Mg2+-dependent manner (83,94,95), and that the TAR bulge binds Mg2+ in a unique conformation (96,97), experiments were performed in the absence and presence of Mg2+.
Using solid-phase oligonucleotide synthesis, TAR samples were prepared in which A35 alone (TAR-A35) or in conjunction with C24 and U25 (TAR-C24U25A35) were Nm modified (Figure 1D). A third reference sample was also prepared lacking any modifications (TAR). The single Am35 modification had a minimal effect on the free energy of melting (Table 1, Supplementary Table S1, and Figure 2A) both in the absence (ΔΔGo ∼ -0.1 kcal/mol) and presence (ΔΔGo ∼ +0.4 kcal/mol) of 3mM Mg2+. The triple modification slightly stabilized TAR by ΔΔGo ∼ −0.3 kcal/mol in the absence of Mg2+ and resulted in a of destabilization (ΔΔGo ∼ +0.6 kcal/mol) in the presence of 3 mM Mg2+ (Table 1 and Figure 2A). The addition of 3 mM Mg2+ stabilized both Nm-modified and unmodified TAR samples by a more significant amount of ∼3 kcal/mol (Table 1) consistent with prior studies (96,97) showing that TAR interacts favorably with divalent metals. Given the high population of ES1, particularly in the absence of Mg2+, we also considered unconstrained and constrained three-state fits of the UV data (see methods) and this analysis yielded similar thermodynamic parameters for the dominant GS relative to those obtained with a two-state fit (Supplementary Tables S1, S2, and Figure S1).
Table 1.
Melting temperature and thermodynamic parameters derived from melting curves in the absence and presence of Mg2+
| No Mg2+ | + 3 mM Mg2+ | |||||||
|---|---|---|---|---|---|---|---|---|
| Sample | T m (°C) |
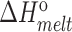 (kcal/mol) (kcal/mol) |
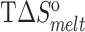 (kcal/mol) (kcal/mol) |
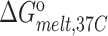 (kcal/mol) (kcal/mol) |
T m (°C) |
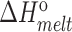 (kcal/mol) (kcal/mol) |
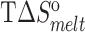 (kcal/mol) (kcal/mol) |
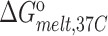 (kcal/mol) (kcal/mol) |
| TAR | 65.2 ± 0.2 | −62.8 ± 1.1 | −57.5 ± 1.0 | −5.2 ± 0.1 | 77.8 ± 0.4 | −75.1 ± 2.8 | −66.4 ± 2.4 | −8.7 ± 0.4 |
| TAR-A35 | 65.4 ± 0.2 | −63.0 ± 0.9 | −57.7 ± 0.8 | −5.3 ± 0.1 | 77.6 ± 0.2 | −71.7 ± 2.2 | −63.4 ± 1.8 | −8.3 ± 0.2 |
| TAR-C24U25A35 | 65.5 ± 0.1 | −65.0 ± 2.4 | −59.5 ± 2.2 | −5.5 ± 0.2 | 77.5 ± 0.3 | −70.0 ± 1.6 | −61.9 ± 1.4 | −8.1 ± 0.1 |
Experiments were run in triplicates at a ramp of 1 °C/min and the values with standard deviation are reported. 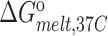 =
= 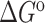 (folded TAR) –
(folded TAR) – 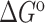 (single-stranded TAR). Experiments were performed Samples were ∼3.0–3.5 μM in NMR buffer (15 mM NaPO4, 25 mM NaCl and 0.1 mM EDTA) at pH 6.4.
(single-stranded TAR). Experiments were performed Samples were ∼3.0–3.5 μM in NMR buffer (15 mM NaPO4, 25 mM NaCl and 0.1 mM EDTA) at pH 6.4.
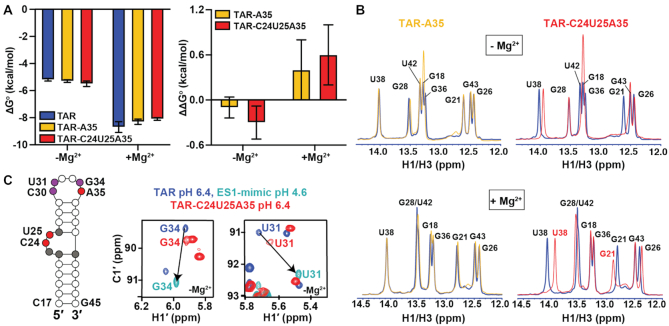
Figure 2.
Impact of Nm on the thermal stability of TAR GS. (A) ΔG°and ΔΔG° values derived from melting curves in the absence and presence of 3 mM Mg2+. Error bars on ΔG° denote standard deviation of triplicate measurements as described in the methods. 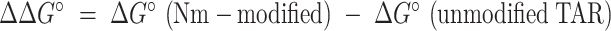 . Error bars on ΔΔG° were obtained by propagating the errors from triplicate measurements (see Methods). (B) Overlays comparing 1D 1H imino spectra for unmodified TAR (blue), TAR-A35 (yellow) and TAR-C24U25A35 (red) in the absence and presence of 3 mM Mg2+. All samples were unlabeled. (C) Secondary structure of TAR with Nm-modified residues shown in red, residues showing CSPs towards ES1 are in purple, and residues exhibiting CSPs due to bulge modifications are in grey. Overlay of natural abundance 2D [13C, 1H] HSQC spectra for the C1′-H1′ region in TAR (pH 6.4), TAR ES1-mimic (pH 4.6) (69), and TAR-C24U25A35 (pH 6.4), in the absence of Mg2+. Lowering the pH to 4.6 shifts the population of TAR (structure shown in Supplementary Figure S3) to stabilize ES1 as the dominant conformation due to protonation of the A35-C30 mismatch unique to ES1 (69). Spectra with complete assignments are shown in Supplementary Figures S2 and S3.
. Error bars on ΔΔG° were obtained by propagating the errors from triplicate measurements (see Methods). (B) Overlays comparing 1D 1H imino spectra for unmodified TAR (blue), TAR-A35 (yellow) and TAR-C24U25A35 (red) in the absence and presence of 3 mM Mg2+. All samples were unlabeled. (C) Secondary structure of TAR with Nm-modified residues shown in red, residues showing CSPs towards ES1 are in purple, and residues exhibiting CSPs due to bulge modifications are in grey. Overlay of natural abundance 2D [13C, 1H] HSQC spectra for the C1′-H1′ region in TAR (pH 6.4), TAR ES1-mimic (pH 4.6) (69), and TAR-C24U25A35 (pH 6.4), in the absence of Mg2+. Lowering the pH to 4.6 shifts the population of TAR (structure shown in Supplementary Figure S3) to stabilize ES1 as the dominant conformation due to protonation of the A35-C30 mismatch unique to ES1 (69). Spectra with complete assignments are shown in Supplementary Figures S2 and S3.
These results show that even three Nm modifications have minor contributions to the overall thermal stability of RNAs when placed on unpaired flexible nucleotides. Such small energetic perturbations on the GS make the modification particularly attractive for carrying out phi-value analysis to gain insights into the transition-state (TS) (72–76). As we show below, while the modifications only weakly affected the energetic stability of the TAR folded state relative to the unfolded one, they had a bigger effect on the energetic stabilities of the two ESs relative to the GS, particularly for ES2.
Nm modifications impact the TAR conformational preferences
Comparison of the 1D 1H NMR spectra of TAR-A35 and TAR-C24U25A35 with their unmodified counterparts (Figure 2B), revealed that Nm minimally impacts the secondary structure of the dominant GS both in the presence and absence of 3 mM Mg2+. Both Nm-modified samples showed the imino resonances belonging to the TAR Watson–Crick base-pairs (bps) (Figure 2B). The modifications did however induce chemical shift perturbations (CSPs) in 1D 1H NMR (Figure 2B) and 2D [13C,1H] HSQC spectra (Figure 2C and Supplementary Figures S2-S4).
The methyl group can give rise to CSPs by two mechanisms. First, CSPs can be caused by changes in the electronic environment of a nucleus in the methylated residue purely due to the addition of the methyl group. Such changes would occur even if the structure of the residue did not change upon methylation. Based on density functional theory (DFT) calculations (Supplementary Table S3) (98), this contribution is expected to cause 2–3 ppm upfield shift in the C1′ chemical shift. This is consistent with the sizeable CSPs (2.0–2.2 ppm) observed at the C1′ sugar resonances of the Nm-modified residues C24, U25 and A35 (Supplementary Figures S2 and S3). This chemical contribution is local and will not impact other residues in the RNA structure. The second mechanism involves the methyl group changing the RNA structural ensemble. This contribution can affect both the methylated residues as well as other (methylated or unmethylated) residues as long as their conformational properties are impacted by the modification. As described below, the CSPs observed for many unmethylated apical loop residues in the 2D [13C,1H] HSQC spectra of TAR-A35 and TAR-C24U25A35 are as expected if the Nm modifications were to increase the abundance of ES1.
Additionally, small CSPs were also observed in TAR-C24U25A35 at residues in and around the bulge (grey residues in Figure 2C), indicating a subtle change in the GS conformational ensemble. New conformational states unique to Nm other than ES1 and ES2 cannot be entirely ruled out; however, this requires further investigation and is beyond the scope of the current work.
Nm increases the abundance of ES1 relative to the GS
The GS-ES1 exchange is fast on the NMR chemical shift timescale. Consequently, if the Nm modification at A35 were to increase the abundance of ES1 without substantially slowing the exchange kinetics, we would expect to observe CSPs in apical loop residues that are directed specifically towards the ES1 1H and 13C chemical shifts, which have been determined previously (69). Furthermore, we would expect similar CSPs for both TAR-A35 and TAR-C24U25A35 given that only Am35 is expected to bias the equilibrium in favor of ES1 (Figure 1D). The magnitude of the shift should be proportional to the degree to which the modification increases the ES1 population (69).
Indeed, for both TAR-A35 and TAR-C24U25A35 in the absence of Mg2+, 1H and 13C CSPs were observed at C30-C1′, U31-C1′, U31-C6, G34-C1′ and G34-C8 that are specifically directed towards the ES1 chemical shifts (Figure 2C and Supplementary Figure S3). As a negative control, no CSPs were observed at apical loop residues G32 and G33, which have similar chemical shifts in the GS and ES1 as they remain unpaired in the two cases (33) (Supplementary Figure S3). Based on the magnitude of the CSPs (see Methods), Nm increases the ES1 population ∼2-fold from 13 ± 2% to 29 ± 3% in both TAR-A35 and TAR-C24U25A35 (Supplementary Table S4). The ∼0.6 kcal/mol stabilization of ES1 relative to the GS (Supplementary Figure S5) is larger than the ∼0.2 kcal/mol stabilization reported previously from incorporating a single Am into RNA duplexes (99). This is not surprising given that the net effect of Nm on thermodynamic parameters is context-dependent (99). The Am35 modification at the Am35+-C30 mismatch could have a more stabilizing effect on ES1 and/or the modification may also destabilize the unpaired conformation of A35 in the GS.
The above results indicate that Nm at the apical loop residue A35 is responsible for the increased stabilization of ES1 and the Nm modifications at the distant bulge do not significantly affect the ES1 population. To further test this hypothesis, we prepared a new sample only containing Nm modification at the bulge residues C24 and U25 (TAR-C24U25). Unlike TAR-A35 and TAR-C24U25A35, modification of the bulge did not result in any CSPs in the apical loop residues of TAR-C24U25 (Supplementary Figure S6). However, CSPs were observed at residues in and around the bulge similar to those observed for TAR-C24U25A35 (Supplementary Figure S6). This also confirms that the CSPs observed at the bulge arise from modification of the bulge residues.
To assess the ES1 abundance in the presence of Mg2+, we prepared unmodified TAR and TAR-C24U25A35 samples site-labeled (55,79) with 13C at G34-C8 to probe ES1, and with 13C at U23-C6 and 15N at U38-N3 to probe ES2 (69) (see below). This was also necessary to boost NMR sensitivity given that Mg2+ significantly broadened the resonances in the 2D NMR spectra (100) (Supplementary Figure S4A-C). The addition of 1 mM Mg2+ to TAR resulted in CSPs at G34-C8H8 that are directed away from ES1 toward the GS, as expected if Mg2+ were to stabilize the GS relative to ES1 (Supplementary Figure S4D). Based on the magnitude of these CSPs, Mg2+ reduced the abundance of ES1 two-fold (from 13% to ∼7%, Supplementary Table S4) corresponding to 0.4 kcal/mol destabilization of ES1 relative to the GS (Supplementary Figure S5). Thus, Mg2+ preferentially binds and stabilizes the apical loop in the more open GS conformation relative to the zipped up ES1.
Although Mg2+ decreased the ES1 population, introducing Nm once again induced CPSs directed towards ES1 (Supplementary Figure S4D). Based on these CSPs, Nm increased the abundance of ES1 2-fold (from 7% to ∼14%, Supplementary Table S4). Therefore, Nm stabilizes ES1 relative to GS in a manner independent of Mg2+, whereas Mg2+ stabilizes the GS relative to ES1 in a manner independent of Nm (Supplementary Figures S4D and S5). This independence could arise because the Mg2+ ions that stabilize the GS relative to the ES1 interact with apical loop residues distant from the modified A35 site and are therefore unaffected by the Nm modification.
Nm increases the lifetime of ES1 in the absence and presence of Mg2+
To further confirm that Nm stabilizes ES1, and to gain insights into how Nm affects the GS-ES1 exchange kinetics, we preformed off-resonance spin relaxation in the rotating frame (R1ρ) RD NMR experiments on the isotopically labeled unmodified TAR and modified TAR-C24U25A35 samples (Figure 3A). The Nm modification and Mg2+ had opposite effects on the R1ρ profiles measured for G34-C8. Nm enhanced the R1ρ profile whereas the addition of Mg2+ diminished it (Figure 3A). In accord with the CSP analysis, a two-state fit of the RD data showed that Nm increased the ES1 population ∼2-fold both in the presence and absence of Mg2+, whereas Mg2+ decreased the population by a similar amount in the presence and absence of Nm (Figure 3B and Supplementary Table S5).
The RD data also revealed that Nm did not significantly change the forward rate (Figure 3C). Rather, it robustly reduced the backward rate (k-1) (Figure 3D) and increased the ES1 lifetime by ∼2-fold in the absence and presence of Mg2+ (Figure 3E). Conversely, Mg2+ robustly increased the abundance and lifetime of the GS in the absence and presence of Nm modifications. Thus, the opposing effects of Nm and Mg2+ on the GS-ES1 exchange are independent of one another at both the thermodynamic as well as the kinetic level. While this suggests that the modification similarly impacts the energetic stabilities of the GS and the transition state for ES1 (TS1) (Supplementary Table S5 and Figure S5), the nature of the TS1 was difficult to ascertain given the large uncertainty in the phi-value (73) (Supplementary Table S7).
Nm increases the abundance and lifetime of ES2 in a Mg2+-dependent manner
We next turned our attention to how Nm impacts the GS-ES2 exchange using U23-C6 and U38-N3 as RD probes. Here, we expected a greater effect because all three Nm modifications should energetically favor ES2 relative to the GS. To overcome a degeneracy arising due to slow exchange kinetics (Supplementary Figure S7), we supplemented the R1ρ data with measurements of chemical exchange saturation transfer (CEST) experiments (84,85), which are better suited for studying slower GS-ES exchange process (83).
We first benchmarked the unmodified site-labeled TAR sample in the absence of Mg2+. As expected, the exchange parameters obtained by fitting CEST (Figure 4A) and R1ρ (Figure 4B) data measured for U23-C6 and U38-N3 (Figure 4C-F and Supplementary Table S8) were in excellent agreement with the values reported previously (70). As predicted, relative to unmodified TAR, TAR-C24U25A35 showed enhanced CEST (Figure 4A) and R1ρ profiles (Figure 4B), consistent with an increase in ES2 population (Figure 4C). A two-state fit of the RD data revealed that Nm increased the ES2 population ∼5-fold (from ∼0.5% to ∼2.6%), stabilizing ES2 relative to the GS by ∼1 kcal/mol (Supplementary Table S8 and Figure S8). The ∼2.5-fold greater stabilization of ES2 relative to ES1 was as expected from application of three versus a single Nm modification. As with ES1, the Nm modifications increased ES2 lifetime by ∼2-fold from ∼2.8 to ∼6.0 ms (Supplementary Table S8). However, unlike for ES1, the Nm modifications also increased the forward rate ∼2.3-fold (Figure 4D and Supplementary Table S8). This results in an intermediate phi-value of ∼0.5 for Nm-modified TAR in the absence of Mg2+, which is difficult to interpret to obtain structural insights into the transition state for ES2 formation (TS2) (Supplementary Table S7).
As with ES1, the addition of Mg2+ to unmodified TAR diminished the R1ρ profiles of U38-N3 and U23-C6 (Figure 4B) due to a ∼4-fold reduction in ES2 population (from ∼0.54 to ∼0.14%, Supplementary Table S8). The greater degree to which Mg2+ stabilizes the GS relative to ES2 versus ES1 is as expected given that a change in the bulge conformation is required to form ES2 and that four divalent metal ions specifically bind and stabilize the TAR bulge in the GS conformation (97).
When combined together, Nm and Mg2+ produced an unexpected result. Based on the R1ρ and CEST profiles, introducing the Nm modifications in the presence of Mg2+ increased the abundance of ES2 by 10-fold (Figure 4C, F and Supplementary Table S8). The greater stabilization of ES2 relative to the GS in the presence (ΔΔGo = 1.4 kcal/mol) versus absence of Mg2+ (ΔΔGo = 1.0 kcal/mol) might in part be explained by the UV melting data (Figure 2A and Table 1), where Nm slightly destabilizes TAR GS in the presence of Mg2+ by ∼0.6 kcal/mol. Consequently, the Nm modifications at the bulge could potentially be disrupting favorable Mg2+ interactions, thus destabilizing the GS and stabilizing the helical conformations formed in the ES2. This underscores the importance of examining how the Nm modifications may impact the conformational properties of non-canonical motifs.
Strikingly, in the presence of Mg2+, the introduction of the three Nm sites minimally impacted the forward rate (1.2-fold), whereas they decreased the backward rate by 12-fold and increased the ES2 lifetime by the same amount. Compared to the unmodified TAR ES2 in the presence of Mg2+, the addition of Nm leads to a phi-value of −0.07 ± 0.10 (Supplementary Table S7) that strongly implies an early transition state for ES2 formation. These results indicate that in the presence of Mg2+, the Nm modified bulge residues C24 and U25 remain unpaired and enriched in C2′-endo sugar pucker in the TS2 as is observed in the GS.
Direct observation of ES2 in Nm-modified TAR
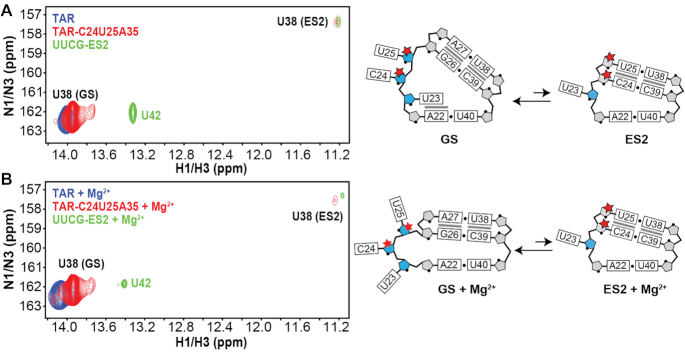
If three Nm modifications do indeed increase the population of ES2 to >1%, it should be possible to directly observe ES2 in the NMR spectra of TAR-C24U25A35, given that the GS-ES2 exchange is slow on the NMR chemical shift timescale, and is further slowed down by the Nm modifications. Indeed, a minor imino resonance was observed in 2D [15N,1H] HSQC spectra of the site-labeled TAR-C24U25A35 but not in unmodified TAR that could be assigned to U38 when forming the wobble U38-U25 mismatch in ES2 both in the absence (Figure 5A) and presence of 1 mM Mg2+ (Figure 5B). This resonance shows excellent overlap in both 1H and 15N chemical shifts with the imino resonance for U38 in a previously described unmodified ES2-mutant (UUCG-ES2) (70) which stabilizes ES2 as the major conformation (Supplementary Figure S9). Based on integrated volumes of the GS and ES2 resonances, the ES2 population is estimated to be ∼3% and ∼1% in the absence and presence of Mg2+, respectively, in excellent agreement with the ES2 populations (∼3% and ∼1%, respectively) obtained from the CEST and R1ρ data. Thus, Nm increases the abundance of ES2 to a degree that makes it directly observable in NMR 2D spectra.
Figure 5.
Direct observation of ES2 U38-N3H3 resonance in the Nm-modified TAR sample in NMR 2D spectra. Overlay of 2D [1H,15N] HSQC spectra of uniformly 15N/13C labeled UUCG-ES2 with 15N(U38) site-labeled TAR and TAR-C24U25A35 (left) and zoom in showing the bulge region in the GS and ES2 (right) in the (A) absence or (B) presence of 1 mM Mg2+. The ES2 U38-N3H3 resonance is absent in unmodified TAR and clearly visible in TAR-C24U25A35. Full [1H,15N] 2D HSQC spectra of UUCG-ES2 are shown in Supplementary Figure S9.
DISCUSSION
Our results show that beyond biasing the local sugar pucker equilibrium, Nm can have broader effects on RNA secondary structural ensembles by preferentially stabilizing and prolonging the lifetime of conformations where Nm-modified residues adopt a paired helical conformation. Interestingly, in the U2-U6 snRNA complex, Nm modifications stabilize a four-way junction where the Nm-modified residues are paired relative to a three-way junction where they are unpaired (19), suggesting a similar mechanism to the one uncovered in our studies. Given the prevalence of secondary structural transitions during the folding, assembly, and function of both the spliceosome (101) and ribosome (102), and given the high abundance of Nm at functionally important non-canonical motifs, there may be analogous roles for Nm that involve the modulation of secondary structural equilibria in these and other Nm-modified RNAs. The preferential stabilization of paired conformations uncovered in this work should provide a useful framework for investigating such roles for Nm in RNA biology.
Nm also expands the toolbox of modifications currently available for studying RNA ESs (62). Point substitution mutations and nucleobase modifications that render an ES the dominant conformation have proven critically important for visualizing the structures of RNA ESs as well as for characterizing their functional properties (62,67,69,70,103,104) and testing their validity as drug targets (105). Yet because the sequence is altered, ES-mutants may not fully recapitulate the properties of the wild-type ES and finding an ES-mutant is not always feasible (59,104). Any putative model for an RNA ES could be tested by introducing Nm at residues that are unpaired in the GS but are proposed to be paired in the ES and then testing for an increase in ES abundance by NMR or chemical probing (106–108). Nm is an attractive strategy for rationally modulating RNA ensembles because it preserves the sequence identify, can be used to target all four nucleotides, and because the modification is inexpensive and commercially available for in vitro studies. In addition, snoRNAs can be engineered to guide novel Nm modifications in vivo (109), and this could provide a route for studying RNA ESs in vivo or provide a means for altering RNA cellular activity as a therapeutic strategy.
Nm also provides a rare opportunity to gain insights into the TS of conformational exchange through phi-value analysis, and specifically, to gain insights into whether or not a given residue is paired (C3′-endo) or unpaired (enriched in C2′-endo) in the TS. The results for ES2 in the presence of Mg2+ suggest an early TS2 in which all the modified residues remain unpaired and enriched in the C2′-endo sugar pucker as in the GS. An Nm scan across the TAR sequence combined with RD measurements will permit broader characterization of the TS structure, and help to elucidate the base pairs that might partially melt in the TS to initiate pair reshuffling and produce ES2. These structural insights into the TS2 are of great interest in the development of anti-HIV therapeutics given that the stabilization of ES2 potently inhibits the cellular activity of TAR (103) and that stabilizing the TS2 in addition to ES2 by small molecules would provide a route for accelerating production of the inhibitory ES2 conformation.
As is the case for other post-transcriptional modifications such as m6A (83,94,95), the impact of Nm on the TAR dynamic ensemble cannot be fully understood without an appreciation for how Mg2+ impacts the ensemble as well (Figure 6). Whereas Nm perturbed the GS-ES1 exchange in a largely Mg2+ independent manner, both the kinetics and thermodynamics of GS-ES2 exchange were strongly dependent on Mg2+. Based on the crystal structure of TAR bound to Ca2+ (97), there are no direct contacts between bound metal ions and the 2′-OH of the Nm-modified residues (97). Rather, Nm modifications could potentially destabilize conformations within the TAR GS ensemble that preferentially bind Mg2+. Further studies are needed to understand how this inter-play between Nm and Mg2+ impacts RNA folding and function.
Figure 6.
Summary of the effect of Nm on GS-ES exchange in the presence and absence of Mg2+. Bulge nucleotides are shown in orange. Nucleotides that transition from being unpaired in GS to paired in the ES are shown in green. Red stars indicate the Nm-modified residues. Pop and τ refer to the population and lifetime of the conformational state (GS, ES1 or ES2).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Duke Magnetic Resonance Spectroscopy Center for their technical support and resources. We also thank Megan Kelly for technical assistance and support and Dr Richard Brennan for providing access to the UV–Vis spectrophotometer.
Notes
Present address: Mary C. Clay, Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.
Contributor Information
Hala Abou Assi, Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA; Department of Medicine, Duke University School of Medicine, Durham, NC 27710, USA.
Atul K Rangadurai, Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA.
Honglue Shi, Department of Chemistry, Duke University, Durham, NC 27708, USA.
Bei Liu, Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA.
Mary C Clay, Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA.
Kevin Erharter, Institute of Organic Chemistry and Center for Molecular Biosciences Innsbruck (CMBI), University of Innsbruck, 6020 Innsbruck, Austria.
Christoph Kreutz, Institute of Organic Chemistry and Center for Molecular Biosciences Innsbruck (CMBI), University of Innsbruck, 6020 Innsbruck, Austria.
Christopher L Holley, Department of Medicine, Duke University School of Medicine, Durham, NC 27710, USA.
Hashim M Al-Hashimi, Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710, USA; Department of Chemistry, Duke University, Durham, NC 27708, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health [1R01GM132899 and U54GM103297 to H.M.A., T32HL007101 to H.A.A.]; Duke Strong Start Award (to C.L.H.); Austrian Science Fund [P30370 and P32773 to C.K.]. Funding for open access charge: US National Institutes of Health [1R01GM132899 and U54GM103297].
Conflict of interest statement. H.M.A. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company. The remaining authors declare no competing interests.
REFERENCES
- 1. Marchand V., Pichot F., Thuring K., Ayadi L., Freund I., Dalpke A., Helm M., Motorin Y.. Next-generation sequencing-based RiboMethSeq protocol for analysis of tRNA 2′-O-methylation. Biomolecules. 2017; 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N. et al.. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018; 46:9289–9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krogh N., Kongsbak-Wismann M., Geisler C., Nielsen H.. Substoichiometric ribose methylations in spliceosomal snRNAs. Org. Biomol. Chem. 2017; 15:8872–8876. [DOI] [PubMed] [Google Scholar]
- 4. Dai Q., Moshitch-Moshkovitz S., Han D., Kol N., Amariglio N., Rechavi G., Dominissini D., He C.. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods. 2017; 14:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elliott B.A., Ho H.T., Ranganathan S.V., Vangaveti S., Ilkayeva O., Abou Assi H., Choi A.K., Agris P.F., Holley C.L.. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat. Commun. 2019; 10:3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Somme J., Van Laer B., Roovers M., Steyaert J., Versees W., Droogmans L.. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA. 2014; 20:1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavaille J., Nicoloso M., Bachellerie J.P.. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996; 383:732–735. [DOI] [PubMed] [Google Scholar]
- 8. Kiss-Laszlo Z., Henry Y., Bachellerie J.P., Caizergues-Ferrer M., Kiss T.. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996; 85:1077–1088. [DOI] [PubMed] [Google Scholar]
- 9. Dimitrova D.G., Teysset L., Carre C.. RNA 2′-O-methylation (Nm) modification in human diseases. Genes (Basel). 2019; 10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ayadi L., Galvanin A., Pichot F., Marchand V., Motorin Y.. RNA ribose methylation (2′-O-methylation): occurrence, biosynthesis and biological functions. Biochim Biophys. Acta Gene Regul. Mech. 2019; 1862:253–269. [DOI] [PubMed] [Google Scholar]
- 11. Koh C.M., Gurel B., Sutcliffe S., Aryee M.J., Schultz D., Iwata T., Uemura M., Zeller K.I., Anele U., Zheng Q. et al.. Alterations in nucleolar structure and gene expression programs in prostatic neoplasia are driven by the MYC oncogene. Am. J. Pathol. 2011; 178:1824–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marcel V., Ghayad S.E., Belin S., Therizols G., Morel A.P., Solano-Gonzalez E., Vendrell J.A., Hacot S., Mertani H.C., Albaret M.A. et al.. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell. 2013; 24:318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagasawa C., Ogren A., Kibiryeva N., Marshall J., O’Brien J.E., Kenmochi N., Bittel D.C.. The role of scaRNAs in adjusting alternative mRNA splicing in heart development. J. Cardiovasc. Dev. Dis. 2018; 5:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guy M.P., Shaw M., Weiner C.L., Hobson L., Stark Z., Rose K., Kalscheuer V.M., Gecz J., Phizicky E.M.. Defects in tRNA anticodon Loop 2′-O-Methylation are implicated in Nonsyndromic X-Linked intellectual disability due to mutations in FTSJ1. Hum. Mutat. 2015; 36:1176–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Piekna-Przybylska D., Decatur W.A., Fournier M.J.. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic Acids Res. 2008; 36:D178–D183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monaco P.L., Marcel V., Diaz J.J., Catez F.. 2′-O-Methylation of ribosomal RNA: towards an epitranscriptomic control of translation. Biomolecules. 2018; 8:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Erales J., Marchand V., Panthu B., Gillot S., Belin S., Ghayad S.E., Garcia M., Laforets F., Marcel V., Baudin-Baillieu A. et al.. Evidence for rRNA 2′-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl Acad. Sci. U.S.A. 2017; 114:12934–12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donmez G., Hartmuth K., Luhrmann R.. Modified nucleotides at the 5′ end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004; 10:1925–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Karunatilaka K.S., Rueda D.. Post-transcriptional modifications modulate conformational dynamics in human U2-U6 snRNA complex. RNA. 2014; 20:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang X., Li Z.T., Yan Y., Lin P., Tang W., Hasler D., Meduri R., Li Y., Hua M.M., Qi H.T. et al.. LARP7-Mediated U6 snRNA modification ensures splicing fidelity and spermatogenesis in mice. Mol. Cell. 2020; 77:999–1013. [DOI] [PubMed] [Google Scholar]
- 21. Hasler D., Meduri R., Bak M., Lehmann G., Heizinger L., Wang X., Li Z.T., Sement F.M., Bruckmann A., Dock-Bregeon A.C. et al.. The Alazami Syndrome-Associated protein LARP7 guides U6 small nuclear RNA modification and contributes to splicing robustness. Mol. Cell. 2020; 77:1014–1031. [DOI] [PubMed] [Google Scholar]
- 22. O’Brien J.E. Jr, Kibiryeva N., Zhou X.G., Marshall J.A., Lofland G.K., Artman M., Chen J., Bittel D.C.. Noncoding RNA expression in myocardium from infants with tetralogy of Fallot. Circ. Cardiovasc. Genet. 2012; 5:279–286. [DOI] [PubMed] [Google Scholar]
- 23. Patil P., Kibiryeva N., Uechi T., Marshall J., O’Brien J.E. Jr, Artman M., Kenmochi N., Bittel D.C.. scaRNAs regulate splicing and vertebrate heart development. Biochim. Biophys. Acta. 2015; 1852:1619–1629. [DOI] [PubMed] [Google Scholar]
- 24. Roost C., Lynch S.R., Batista P.J., Qu K., Chang H.Y., Kool E.T.. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J. Am. Chem. Soc. 2015; 137:2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desrosiers R., Friderici K., Rottman F.. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl Acad. Sci. U.S.A. 1974; 71:3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rottman F., Shatkin A.J., Perry R.P.. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: possible implications for processing. Cell. 1974; 3:197–199. [DOI] [PubMed] [Google Scholar]
- 27. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meyer K.D., Jaffrey S.R.. Rethinking m(6)A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017; 33:319–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 30. Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. et al.. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017; 6:e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C.. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017; 27:315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Davies D.B. Conformations of nucleosides and nucleotides. Prog. Nucl. Magn. Reson. Spectrosc. 1978; 12:135–225. [Google Scholar]
- 33. Clay M.C., Ganser L.R., Merriman D.K., Al-Hashimi H.M.. Resolving sugar puckers in RNA excited states exposes slow modes of repuckering dynamics. Nucleic Acids Res. 2017; 45:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S.. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992; 31:1040–1046. [DOI] [PubMed] [Google Scholar]
- 35. Malek-Adamian E., Patrascu M.B., Jana S.K., Martinez-Montero S., Moitessier N., Damha M.J.. Adjusting the structure of 2′-modified nucleosides and oligonucleotides via C4′-alpha-F or C4′-alpha-OMe substitution: synthesis and conformational analysis. J. Org. Chem. 2018; 83:9839–9849. [DOI] [PubMed] [Google Scholar]
- 36. Cheng D.M., Sarma R.H.. Nuclear magnetic resonance study of the impact of ribose 2′-O-methylation on the aqueous solution conformation of cytidylyl-(3′ leads to 5′)-cytidine. Biopolymers. 1977; 16:1687–1711. [DOI] [PubMed] [Google Scholar]
- 37. Mahto S.K., Chow C.S.. Synthesis and solution conformation studies of the modified nucleoside N(4),2′-O-dimethylcytidine (m(4)Cm) and its analogues. Bioorg. Med. Chem. 2008; 16:8795–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yildirim I., Kierzek E., Kierzek R., Schatz G.C.. Interplay of LNA and 2′-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: a molecular dynamics study using the revised AMBER force field and comparison with experimental results. J. Phys. Chem. B. 2014; 118:14177–14187. [DOI] [PubMed] [Google Scholar]
- 39. Kool E.T. Preorganization of DNA: design principles for improving nucleic acid recognition by synthetic oligonucleotides. Chem. Rev. 1997; 97:1473–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Auffinger P., Westhof E.. Hydrophobic groups stabilize the hydration shell of 2′-O-Methylated RNA duplexes. Angew. Chem. 2001; 40:4648–4650. [DOI] [PubMed] [Google Scholar]
- 41. Khvorova A., Watts J.K.. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017; 35:238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng E.W., Shima D.T., Calias P., Cunningham E.T. Jr, Guyer D.R., Adamis A.P.. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006; 5:123–132. [DOI] [PubMed] [Google Scholar]
- 43. Shen X., Corey D.R.. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018; 46:1584–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khvorova A. Oligonucleotide therapeutics - a new class of cholesterol-lowering drugs. N. Engl. J. Med. 2017; 376:4–7. [DOI] [PubMed] [Google Scholar]
- 45. Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B.. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005; 48:901–904. [DOI] [PubMed] [Google Scholar]
- 46. Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K. et al.. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006; 12:1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lennox K.A., Behlke M.A.. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011; 18:1111–1120. [DOI] [PubMed] [Google Scholar]
- 48. Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A.. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015; 22:342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Didychuk A.L., Butcher S.E., Brow D.A.. The life of U6 small nuclear RNA, from cradle to grave. RNA. 2018; 24:437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Montemayor E.J., Curran E.C., Liao H.H., Andrews K.L., Treba C.N., Butcher S.E., Brow D.A.. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-A resolution. Nat. Struct. Mol. Biol. 2014; 21:544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mefford M.A., Staley J.P.. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009; 15:1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi J., Indrisiunaite G., DeMirci H., Ieong K.W., Wang J., Petrov A., Prabhakar A., Rechavi G., Dominissini D., He C. et al.. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 2018; 25:208–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bohnsack M.T., Sloan K.E.. Modifications in small nuclear RNAs and their roles in spliceosome assembly and function. Biol. Chem. 2018; 399:1265–1276. [DOI] [PubMed] [Google Scholar]
- 54. Incarnato D., Anselmi F., Morandi E., Neri F., Maldotti M., Rapelli S., Parlato C., Basile G., Oliviero S.. High-throughput single-base resolution mapping of RNA 2-O-methylated residues. Nucleic Acids Res. 2017; 45:1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Juen M.A., Wunderlich C.H., Nussbaumer F., Tollinger M., Kontaxis G., Konrat R., Hansen D.F., Kreutz C.. Excited states of nucleic acids probed by proton relaxation dispersion NMR spectroscopy. Angew. Chem. 2016; 55:12008–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen B., LeBlanc R., Dayie T.K.. SAM-II riboswitch samples at least two conformations in solution in the absence of ligand: implications for recognition. Angew. Chem. 2016; 55:2724–2727. [DOI] [PubMed] [Google Scholar]
- 57. Rinnenthal J., Buck J., Ferner J., Wacker A., Furtig B., Schwalbe H.. Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc. Chem. Res. 2011; 44:1292–1301. [DOI] [PubMed] [Google Scholar]
- 58. Tian S., Cordero P., Kladwang W., Das R.. High-throughput mutate-map-rescue evaluates SHAPE-directed RNA structure and uncovers excited states. RNA. 2014; 20:1815–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao B., Guffy S.L., Williams B., Zhang Q.. An excited state underlies gene regulation of a transcriptional riboswitch. Nat. Chem. Biol. 2017; 13:968–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao B., Zhang Q.. Characterizing excited conformational states of RNA by NMR spectroscopy. Curr. Opin. Struct. Biol. 2015; 30:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sekhar A., Kay L.E.. NMR paves the way for atomic level descriptions of sparsely populated, transiently formed biomolecular conformers. Proc. Natl Acad. Sci. U.S.A. 2013; 110:12867–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rangadurai A., Szymaski E.S., Kimsey I.J., Shi H., Al-Hashimi H.M.. Characterizing micro-to-millisecond chemical exchange in nucleic acids using off-resonance R1ρ relaxation dispersion. Prog. Nucl. Magn. Reson. Spectrosc. 2019; 112–113:55–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Palmer A.G. 3rd, Kroenke C.D., Loria J.P.. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001; 339:204–238. [DOI] [PubMed] [Google Scholar]
- 64. Palmer A.G. 3rd, Massi F.. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 2006; 106:1700–1719. [DOI] [PubMed] [Google Scholar]
- 65. Palmer A.G., 3rd Chemical exchange in biomacromolecules: past, present, and future. J. Magn. Reson. 2014; 241:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xue Y., Gracia B., Herschlag D., Russell R., Al-Hashimi H.M.. Visualizing the formation of an RNA folding intermediate through a fast highly modular secondary structure switch. Nat. Commun. 2016; 7:doi:10.1038/ncomms11768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ganser L.R., Kelly M.L., Patwardhan N.N., Hargrove A.E., Al-Hashimi H.M.. Demonstration that small molecules can bind and stabilize low-abundance short-lived RNA excited conformational states. J. Mol. Biol. 2019; 432:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Puglisi J.D., Tan R., Calnan B.J., Frankel A.D., Williamson J.R.. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992; 257:76–80. [DOI] [PubMed] [Google Scholar]
- 69. Dethoff E.A., Petzold K., Chugh J., Casiano-Negroni A., Al-Hashimi H.M.. Visualizing transient low-populated structures of RNA. Nature. 2012; 491:724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lee J., Dethoff E.A., Al-Hashimi H.M.. Invisible RNA state dynamically couples distant motifs. Proc. Natl Acad. Sci. U.S.A. 2014; 111:9485–9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mulder F.A., Mittermaier A., Hon B., Dahlquist F.W., Kay L.E.. Studying excited states of proteins by NMR spectroscopy. Nat. Struct. Biol. 2001; 8:932–935. [DOI] [PubMed] [Google Scholar]
- 72. Alvey H.S., Gottardo F.L., Nikolova E.N., Al-Hashimi H.M.. Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat. Commun. 2014; 5:4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fersht A.R., Matouschek A., Serrano L.. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992; 224:771–782. [DOI] [PubMed] [Google Scholar]
- 74. Neudecker P., Zarrine-Afsar A., Davidson A.R., Kay L.E.. Phi-value analysis of a three-state protein folding pathway by NMR relaxation dispersion spectroscopy. Proc. Natl Acad. Sci. U.S.A. 2007; 104:15717–15722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bartley L.E., Zhuang X., Das R., Chu S., Herschlag D.. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J. Mol. Biol. 2003; 328:1011–1026. [DOI] [PubMed] [Google Scholar]
- 76. Cho J.H., O’Connell N., Raleigh D.P., Palmer A.G. 3rd. Phi-value analysis for ultrafast folding proteins by NMR relaxation dispersion. J. Am. Chem. Soc. 2010; 132:450–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Damha M.J., Ogilvie K.K.. Oligoribonucleotide synthesis. The silyl-phosphoramidite method. Methods Mol. Biol. 1993; 20:81–114. [DOI] [PubMed] [Google Scholar]
- 78. Ogilvie K.K., Beaucage S.L., Schifman A.L., Theriault N.Y., Sadana K.L.. Synthesis of oligoribonucleotides. 2. Use of silyl protecting groups in nucleoside and nucleotide chemistry 7. Can. J. Chem. 1978; 56:2768–2780. [Google Scholar]
- 79. Wunderlich C.H., Spitzer R., Santner T., Fauster K., Tollinger M., Kreutz C.. Synthesis of (6-(13)C)pyrimidine nucleotides as spin-labels for RNA dynamics. J. Am. Chem. Soc. 2012; 134:7558–7569. [DOI] [PubMed] [Google Scholar]
- 80. Wagenmakers E.J., Farrell S.. AIC model selection using Akaike weights. Psychon. Bull. Rev. 2004; 11:192–196. [DOI] [PubMed] [Google Scholar]
- 81. Burnham K.P., Anderson D.R.. Multimodel inference: understanding AIC and BIC in model selection. Sociological Methods & Research. 2004; 33:261–304. [Google Scholar]
- 82. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. Nmrpipe - a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 83. Shi H., Liu B., Nussbaumer F., Rangadurai A., Kreutz C., Al-Hashimi H.M.. NMR chemical exchange measurements reveal that N(6)-Methyladenosine slows RNA annealing. J. Am. Chem. Soc. 2019; 141:19988–19993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhao B., Hansen A.L., Zhang Q.. Characterizing slow chemical exchange in nucleic acids by carbon CEST and low spin-lock field R(1rho) NMR spectroscopy. J. Am. Chem. Soc. 2014; 136:20–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vallurupalli P., Bouvignies G., Kay L.E.. Studying “invisible” excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 2012; 134:8148–8161. [DOI] [PubMed] [Google Scholar]
- 86. Frisch M. J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Petersson G.A., Nakatsuji H. et al.. Gaussian 09, Revision D.01. 2016; Wallingford CT: Gaussian Inc. [Google Scholar]
- 87. Rangadurai A., Kremser J., Shi H., Kreutz C., Al-Hashimi H.M.. Direct evidence for (G)O6···H2-N4(C)+ hydrogen bonding in transient G(syn)-C+ and G(syn)-m5C+ Hoogsteen base pairs in duplex DNA from cytosine amino nitrogen off-resonance R1ρ relaxation dispersion measurements. J. Magn. Reson. 2019; 308:106589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lu X.J., Olson W.K.. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003; 31:5108–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Fonville J.M., Swart M., Vokacova Z., Sychrovsky V., Sponer J.E., Sponer J., Hilbers C.W., Bickelhaupt F.M., Wijmenga S.S.. Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment. Chem.-Eur. J. 2012; 18:12372–12387. [DOI] [PubMed] [Google Scholar]
- 90. Dejaegere A.P., Case D.A.. Density functional study of ribose and deoxyribose chemical shifts. J. Phys. Chem. A. 1998; 102:5280–5289. [Google Scholar]
- 91. Lee C.T., Yang W.T., Parr R.G.. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron-density. Phys Rev B. 1988; 37:785–789. [DOI] [PubMed] [Google Scholar]
- 92. Krishnan R., Binkley J.S., Seeger R., Pople J.A.. Self‐consistent molecular orbital methods. XX. A basis set for correlated wave functions. J. Chem. Phys. 1980; 72:650–654. [Google Scholar]
- 93. Wolinski K., Hinton J.F., Pulay P.. Efficient implementation of the gauge-independent atomic orbital method for NMR chemical-shift calculations. J. Am. Chem. Soc. 1990; 112:8251–8260. [Google Scholar]
- 94. Chu C.C., Liu B., Plangger R., Kreutz C., Al-Hashimi H.M.. m6A minimally impacts the structure, dynamics, and Rev ARM binding properties of HIV-1 RRE stem IIB. PLoS One. 2019; 14:e0224850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Liu B., Merriman D.K., Choi S.H., Schumacher M.A., Plangger R., Kreutz C., Horner S.M., Meyer K.D., Al-Hashimi H.M.. A potentially abundant junctional RNA motif stabilized by m(6)A and Mg(2). Nat. Commun. 2018; 9:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Merriman D.K., Yuan J., Shi H., Majumdar A., Herschlag D., Al-Hashimi H.M.. Increasing the length of poly-pyrimidine bulges broadens RNA conformational ensembles with minimal impact on stacking energetics. RNA. 2018; 24:1363–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ippolito J.A., Steitz T.A.. A 1.3-A resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl Acad. Sci. U.S.A. 1998; 95:9819–9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fonville J.M., Swart M., Vokacova Z., Sychrovsky V., Sponer J.E., Sponer J., Hilbers C.W., Bickelhaupt F.M., Wijmenga S.S.. Chemical shifts in nucleic acids studied by density functional theory calculations and comparison with experiment. Chemistry. 2012; 18:12372–12387. [DOI] [PubMed] [Google Scholar]
- 99. Kumar S., Mapa K., Maiti S.. Understanding the effect of locked nucleic acid and 2′-O-methyl modification on the hybridization thermodynamics of a miRNA-mRNA pair in the presence and absence of AfPiwi protein. Biochemistry. 2014; 53:1607–1615. [DOI] [PubMed] [Google Scholar]
- 100. Merriman D.K., Xue Y., Yang S., Kimsey I.J., Shakya A., Clay M., Al-Hashimi H.M.. Shortening the HIV-1 TAR RNA Bulge by a single nucleotide preserves motional modes over a broad range of time scales. Biochemistry. 2016; 55:4445–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Matera A.G., Wang Z.. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014; 15:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Klinge S., Woolford J.L. Jr. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2019; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ganser L.R., Chu C.C., Bogerd H.P., Kelly M.L., Cullen B.R., Al-Hashimi H.M.. Probing RNA conformational equilibria within the functional cellular context. Cell Rep. 2020; 30:2472–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chu C.C., Plangger R., Kreutz C., Al-Hashimi H.M.. Dynamic ensemble of HIV-1 RRE stem IIB reveals non-native conformations that disrupt the Rev-binding site. Nucleic Acids Res. 2019; 47:7105–7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ganser L.R., Kelly M.L., Herschlag D., Al-Hashimi H.M.. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 2019; 20:474–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Leamy K.A., Assmann S.M., Mathews D.H., Bevilacqua P.C.. Bridging the gap between in vitro and in vivo RNA folding. Q. Rev. Biophys. 2016; 49:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Weeks K.M. Advances in RNA structure analysis by chemical probing. Curr. Opin. Struct. Biol. 2010; 20:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mustoe A.M., Lama N.N., Irving P.S., Olson S.W., Weeks K.M.. RNA base-pairing complexity in living cells visualized by correlated chemical probing. Proc. Natl Acad. Sci. U.S.A. 2019; 116:24574–24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ge J., Liu H., Yu Y.T.. Regulation of pre-mRNA splicing in Xenopus oocytes by targeted 2′-O-methylation. RNA. 2010; 16:1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kimsey I.J., Petzold K., Sathyamoorthy B., Stein Z.W., Al-Hashimi H.M.. Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature. 2015; 519:315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.