Abstract
Advances in genome editing technologies have enabled manipulation of genomes at the single base level. These technologies are based on programmable nucleases (PNs) that include meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated 9 (Cas9) nucleases and have given researchers the ability to delete, insert or replace genomic DNA in cells, tissues and whole organisms. The great flexibility in re-designing the genomic target specificity of PNs has vastly expanded the scope of gene editing applications in life science, and shows great promise for development of the next generation gene therapies. PN technologies share the principle of inducing a DNA double-strand break (DSB) at a user-specified site in the genome, followed by cellular repair of the induced DSB. PN-elicited DSBs are mainly repaired by the non-homologous end joining (NHEJ) and the microhomology-mediated end joining (MMEJ) pathways, which can elicit a variety of small insertion or deletion (indel) mutations. If indels are elicited in a protein coding sequence and shift the reading frame, targeted gene knock out (KO) can readily be achieved using either of the available PNs. Despite the ease by which gene inactivation in principle can be achieved, in practice, successful KO is not only determined by the efficiency of NHEJ and MMEJ repair; it also depends on the design and properties of the PN utilized, delivery format chosen, the preferred indel repair outcomes at the targeted site, the chromatin state of the target site and the relative activities of the repair pathways in the edited cells. These variables preclude accurate prediction of the nature and frequency of PN induced indels. A key step of any gene KO experiment therefore becomes the detection, characterization and quantification of the indel(s) induced at the targeted genomic site in cells, tissues or whole organisms. In this survey, we briefly review naturally occurring indels and their detection. Next, we review the methods that have been developed for detection of PN-induced indels. We briefly outline the experimental steps and describe the pros and cons of the various methods to help users decide a suitable method for their editing application. We highlight recent advances that enable accurate and sensitive quantification of indel events in cells regardless of their genome complexity, turning a complex pool of different indel events into informative indel profiles. Finally, we review what has been learned about PN-elicited indel formation through the use of the new methods and how this insight is helping to further advance the genome editing field.
INTRODUCTION
Naturally occurring indels
In the study of the size distribution of nucleotide insertions and deletions in genomic DNA from human and rodents by Gu and Li in 1995 (1), the term indel (for insertion and/or deletion) was used for one of the first times. At the time, naturally occurring indels were believed to have arisen through complex combined insertion and deletion events (2,3) and in 2001, the nomenclature for sequence variations described as indel events was defined as; ‘a deletion followed by an insertion after the nucleotides affected’ (4). In more recent general terms, indels are collectively referred to as an insertion, a deletion, or an insertion and a deletion of nucleotides in genomic DNA (5). Most commonly, naturally occurring indels are less than 1 kb in length. Indels larger than 1 kb are referred to as copy number variations that typically have arisen through amplification or duplication events (6) or through deletion events resulting from two distant DSBs followed by fusion of the DNA ends (7). Naturally occurring indels are considered as polymorphisms—a nucleotide sequence that has been added or deleted in individuals, creating a polymorphism at that site. When indels occur in the coding sequence of genes and the inserted and/or deleted base pairs (bp) are divisible by 3, they are described as being ‘in-frame’ and may either retain or disrupt the function of the encoded protein depending on the importance of the deleted amino acid residues for protein structure or function. If the triplet reading code is altered, the indels are termed ‘frameshift’ polymorphisms, which result in a premature stop codon that may abrogate gene function by truncating the protein and/or eliciting degradation of the mRNA via the nonsense-mediated mRNA decay pathway (8–10). A computational report based on DNA re-sequencing traces originally generated for single-nucleotide polymorphism (SNP) discovery, demonstrated that indels are distributed throughout the human genome with an average density of one indel per 7.2 kb of DNA (5). Computational analysis using DNA re-sequencing traces has also determined that indel variations are the second most common form of genetic variation in humans after SNPs, totaling 15–20% of all variations and of these, single-base indels represent approximately one third (5,11). Of note, it is estimated that individuals possess 102–280 frameshifting single-base indels (5,12).
However, accurate identification of indels in genomic studies is not straightforward and is affected by both structural genomic features such as the presence of repeats, short interspersed elements, homopolymers/dimers and the quality of the indel detection methods used. Initial indel identification efforts were based on Sanger re-sequenced data aimed at identifying genetic variation on chromosome 22 (11,13). These and other studies were primarily based on resources from the human genome project. In 2006 and 2007, computational software packages (PolyPhred, PolyScan) were developed for indel detection based on automated Sanger sequencing (14,15). More recent naturally occurring indel detection approaches have been based on next generation sequencing (NGS) platforms for which software packages such as SOAP (16) and MAQ (17) have been developed for variant base discovery. However, the various NGS platforms have different dominant error types with respect to detection of nucleotide substitutions and indels, and comparative analyses have shown limited concordance between the indels that were identified (18). Due to the false negative rates in many NGS-based studies, it is estimated that one third of the small indels in human genomes are left undetected (17). Supporting this notion, recent studies have suggested that indels are often severely under-reported due to difficulties in accurate indel detection and consequently it is estimated that only 55% of insertions present in European and Yoruban genomes have been detected (19).
In light of the difficulties in discriminating true indels from errors in NGS analysis (20), algorithms such as KAUST, assembly read error correction tool (Karect) and other solutions have been developed to correct nucleotide substitution, insertion and deletion errors from NGS data (18). In spite of this, a common denominator for NGS methodologies is that they are all based on multi-step processes, including the generation of a large set of DNA sequences, data-reads, software-driven mapping of the generated reads to a reference genome, followed by identification of indels by analysis of the mapping results using an indel-calling software. The various steps require the use of a growing number of software programs that for mapping include; BFAST (21), Bowtie2 (22), BWA (23) and SHRIMP (24), and for indel calling; Dindel (25), GATK (26), FreeBayes (27) and SNVer (28). While the softwares are continuously being improved, the technically challenging bioinformatic alignment analysis of the massive amount of NGS data can have a profound effect on indel detection accuracy and in a recent study, indel concordance between three indel-calling pipelines (SOAPindel, SAMtools and GATK) was only 26.8% (29).
Similarly, a low concordance between GATK-UnifiedGenotyper, GATKHaplotypeCaller and Pindel was found, when re-analysing three sets of human NGS data (targeted exome sequencing (TES), whole exome sequencing (WES), and whole genome sequencing (WGS)), showing variable concordance of indel calls of the three algorithms for the three data sets, being as low as 5.70% for the TES data (30). An often overlooked, but important general concern of NGS, relates to the effects that DNA extraction and other library preparation steps have on downstream sequence integrity (31). In this regard it has been shown that technical mutagenic damage can account for a significant number of erroneous identified variants with low to moderate (1–5%) frequency (32). Taken together, improvements in benchmarking of NGS-based variant discovery methodologies remains an unmet need in the field (33).
Cellular pathways for DNA double-strand break repair
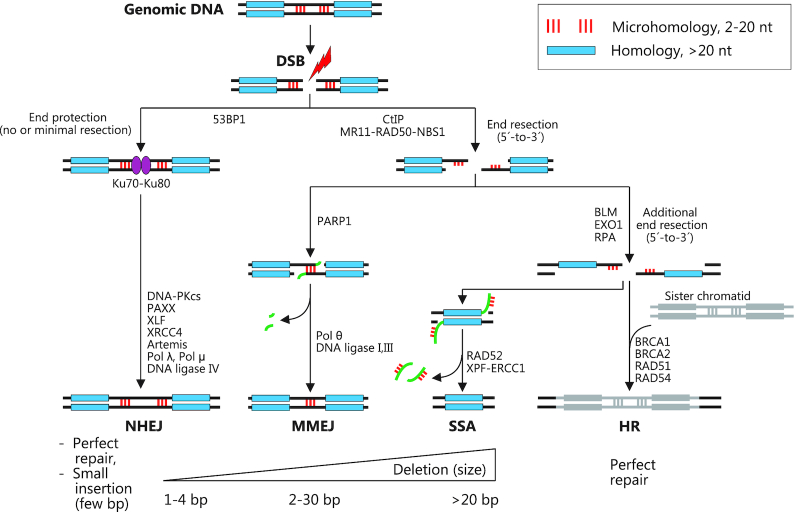
Naturally occurring indels arise through cellular repair of DNA double-strand breaks (DSBs) that may be produced by DNA damaging agents such as UV and ionizing irradiation or metabolic byproducts. Two competing repair pathways underlie the majority of indels: the classical non-homologous end joining (NHEJ) pathway and the alternative non-homologous end joining (alt-NHEJ) pathway, also known as microhomology-mediated end joining (MMEJ).
NHEJ is generally dominant, because it is active in all cell cycle phases, except for mitosis, and once a DSB has occurred, its highly abundant initiating components Ku70-Ku80 rapidly bind the DNA ends and shield them from the actions of the MMEJ pathway (34–37) (Figure 1). Ku70–Ku80 next recruits the essential NHEJ proteins DNA-PKcs and XLF-XRCC4, which in turn recruits DNA ligase IV to ligate the DNA ends. If the DNA ends are not directly ligatable due to incompatible single-stranded overhangs, the ends are processed by the nuclease Artemis or DNA polymerases to enable ligation. As indicated by its name, NHEJ can repair a DSB without the need for homologies at the DNA ends. Often, however, NHEJ exploits small homologies in single-stranded overhangs at the DSB to facilitate repair, but these can be minimal (1–2 nt). NHEJ results in either perfect repair or in small indels of typically a few bp in size.
Figure 1.
Cellular pathways for DNA double-strand break repair. Schematic representation of the four major pathways for repair of DNA double-strand breaks: NHEJ (non-homologous end joining), MMEJ (microhomology-mediated end joining), SSA (single-strand annealing) and HR (homologous recombination). Major repair proteins are shown with a focus on mammals. The figure illustrates that the repair pathways are competing. Repair pathway choice is governed by factors that include (i) the relative activities of the various repair proteins (modulated by expression level, the cell cycle etc), (ii) the absence or presence of microhomologies (red sticks) or longer homologies (blue bar) in the vicinity of the DSB and the availability of sister chromatid for homologous repair and (iii) the nature of the DSB (blunt, staggered, 5′ or 3′ overhang). DNA flaps produced and excised during MMEJ and SSA are shown in green. The major repair outcomes of the four pathways with approximate sizes of indels are indicated.
MMEJ only occurs in S and G2 phases of the cell cycle, because it is initiated by limited end resection at the DSB by the MR11-RAD50-NBS1 complex after its activation by CtIP, which takes place in these cell cycle phases only (35–37) (Figure 1). This may eliminate Ku70–Ku80-bound ends and thereby prevent NHEJ and it will generate 3′ single-stranded overhangs that may expose microhomologies of 2–20 nt on either side of the DSB, which can anneal to one another. Subsequently, the flaps will be excised by the ERCC1-XPF endonuclease, DNA polymerase Θ will fill in the gaps and finally the strands will be joined by DNA ligases I and III. The MMEJ pathway is thereby inherently mutagenic, yielding deletions that eliminate one copy of the two microhomology stretches and the intervening sequence. MMEJ elicited indels are typically larger than NHEJ indels, yet still relatively small (<30 bp).
If MMEJ does not happen, 5′-to-3′ end resection will proceed, which may expose longer homologies of 20–200 nt that can anneal and lead to larger deletions via the single-strand annealing (SSA) pathway in a fashion similar to MMEJ, except that different proteins mediate the repair (35–37) (Figure 1). SSA is a minor source of indels, one reason being that the chance of a long homology stretch in the vicinity of the DSB is much smaller than that of a microhomology. If yet further resection takes place, the homologous repair (HR) pathway may be harnessed to elicit perfect mending of the DSB, using the sister chromatid as repair template (38) (Figure 1).
The factors that govern repair pathway choice are complex and inter-dependent (35–37,39) and include: (i) the relative activities of the various repair proteins that may be modulated at the expression level, by the cell cycle or by other parameters, (ii) the absence or presence of microhomologies, longer homologies or a sister chromatid for homologous recombination and (iii) the nature of the DSB, i.e. if it is blunt, staggered and with 5′ or 3′ overhangs.
Programmable nucleases—meganucleases, ZFNs, TALENs and CRISPR/Cas9
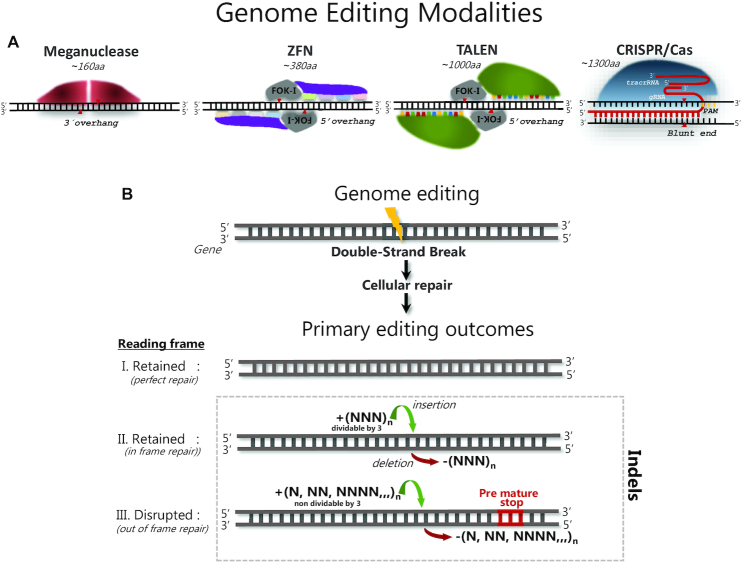
Currently, the most commonly used PN modalities include meganucleases (40,41), ZFNs (42–46), TALENs (47–49) and CRISPR/Cas9 (50–53) (Figure 2, Panel A). Although any of these PNs allow for specific targeting of genomic loci, the underlying principle for locus binding and induction of double-stranded breaks differs considerably among the modalities. Meganucleases, the first nucleases shown capable of increasing homology-directed repair integration of a double stranded DNA donor (54), are naturally occurring endonucleases, found in a large number of organisms—archaea or archaebacteria. Meganucleases are represented by two main enzyme families collectively known as homing endonucleases: intron endonucleases and intein endonucleases (55). In nature, meganucleases are encoded and expressed from mobile genetic elements, introns or inteins, and their expression produces a DSB in the complementary intron- or intein-free allele (55). Because the residues for DNA binding and cleavage show great overlap, they are difficult to redirect to user-specified target sites.
Figure 2.
Schematic illustration of the four main programmable nuclease modalities. (A) Meganucleases are naturally occurring and represented by two main enzyme families with different molecular structures, but in both of which, the residues for DNA binding and cleavage show great overlap. ZFNs are artificial modular molecules consisting of a non-specific FOK-I endonuclease domain fused to several, specific triplet-nucleotide binding domains (color coded) and function as dimers. TALENs are also artificial modular molecules consisting of a non-specific FOK-I domain fused to several, specific single-nucleotide binding domains (color coded). CRISPR/Cas9 is represented by a diverse family of endonucleases derived from bacteria and/or archaea. Depicted is the most commonly used SpCas9 nuclease that upon complex formation with gRNA and tracrRNA enables targeting of a PAM possessing target DNA locus by Watson-Crick base pairing between gRNA and genomic DNA. (B) Common to all modalities shown in panel A, the primary outcome after binding of the PN to its target sequence is formation of a DNA double-strand break, followed by cellular repair of the break, which when targeting the coding region of a gene can result in three major outcomes; (i) perfect repair of the break, which retains the protein coding reading frame, (ii) in frame repair, where the resultant deletions or insertions retain the protein coding reading frame or (iii) out of frame repair, where the resultant deletions or insertions disrupt the protein coding reading frame, generating a knock-out.
The modular programmable ZFNs and TALENs are composed of naturally occurring, but distinct DNA binding modules that in both cases are artificially fused to a bacterial type IIS FOK-I restriction endonuclease domain that, when homodimerized, induces non-specific DNA cleavage. ZFN targeting specificity is mediated through binding of specific amino acids within the individual zinc finger DNA binding domains that contact three to four nucleotides in a sequence specific manner (56). Fusion of 3–5 ZF DNA binding domains generates the ZFN monomer that enables specific targeting of a genomic locus. Complementary binding of 2 ZFN monomers to the sense and antisense strands enables FOK-I dimerization to occur at the target site and elicit a DSB (Figure 2, panel A). For TALENs, DNA binding is mediated through specific amino acids within individual TAL-domains that each contact a single nucleotide within the target sequence (57). Fusion of 12–16 TAL-domains enable specific targeting of a genomic locus and the complementary binding of two TALEN monomers to the sense and antisense strands, similar to ZFNs, induce DSB formation to occur as a consequence of FOK-I dimerization at the target site (Figure 2, panel A). The DSBs formed by meganucleases, ZFNs and TALENs all possess single-stranded overhangs or ‘sticky ends’ (58,59) (Figure 2, panel A). Of note, meganuclease, ZFN and TALEN targeting is mediated through protein–DNA binding and therefore, user-specification of the targeting specificity requires protein engineering. This makes the engineering of meganuclease, ZFN and TALEN specificity time- and resource-intensive and requires great insight into the rules that determine DNA binding of these nucleases (60–63). These limitations have been largely overcome with the development of the CRISPR/Cas9 system (50,52–53,64). This PN is derived from an adaptive immune system of bacteria and archaea (65,66). The targeting specificity and nuclease activity of Cas9 nuclease is determined by the CRISPR RNA (crRNA), also called guide RNA (gRNA) and the presence of transactivating crRNA (tracrRNA) that are transcribed from the CRISPR locus (67): the annealed crRNA and tracrRNA is complexed with Cas9, which allosterically activates the nuclease, when the ∼20 nt gRNA binds to its genomic target site via Watson-Crick base-pairing. In addition, Cas9 must bind a small (few nt), generic so-called protospacer-adjacent motif (PAM) on the opposite strand, 3′ to the gRNA target site (68,69). Thus, Cas9 nuclease targeting is only dependent on a ∼20 nt gRNA sequence and the presence of a PAM, which greatly simplifies the redirection of CRISPR-Cas9 to any given user-selected target sequence. For gene editing purposes, Cas9 derived from Streptococcus pyogenes has been most widely used, which primarily induces a blunt-ended DSB and less frequently a DSB with a 1-nt overhang, possibly dictated by sequence features near the cut site.
Programmable nuclease-induced indels
PN-elicited indel formation is a complex process, where both the initial DSB and the subsequent repair events are difficult to predict. With respect to the latter, PN-induced DSBs are repaired by the same four major cellular repair pathways used by naturally occurring DSBs. Recent studies, including large scale analyses of indels elicited by thousands of SpCas9:gRNAs have provided a wealth of new insight into PN-elicited DSB repair, which will be reviewed in detail in the Discussion. Very briefly, PN-elicited indel formation is guided by several factors that include; (i) the PNs used and their different abilities to induce blunt-ended or staggered DSBs (70–73). The sequence flanking the PN cut site that may have microhomologies or other features, which can promote discrete indel size and frequency outcomes (73–79), (iii) the chromatin structure at the target site (80–82) and (iv) the activities of the individual repair pathways in the cells edited, which vary with cell type, may be perturbed by mutation in cancer cells and are affected by the proliferation state of the cells (cycling versus quiescent). The latter may itself be modulated by editing, since PN-elicited DSBs can induce growth arrest depending on the p53 status of the cells (83). Furthermore, if a PN-induced DSB becomes perfectly repaired by HR or error-free NHEJ and the PN is still present in the cell, the PN may cut again because its target site is preserved, and one or more of such cycles may happen until indel forming repair has occurred and disrupted the target site. This will bias PN-elicited repair towards error-prone NHEJ and MMEJ repair, in agreement with the findings that indel formation via NHEJ and MMEJ seems to be the predominant outcome after PN-induced DSBs, as reviewed in the Discussion section. Approximately 90–95% of indels elicited by most classes of PNs are deletions <30 bp or small insertions of 1 to a few bp in size (73–76,79,84). While some progress has been made with respect to predicting indel editing outcomes, the spectrum and frequency of indels elicited by a given PN can still not be predicted in the majority of editing applications. Furthermore, it has been shown that PN-elicited DSBs can induce targeted rearrangements and chromosome elimination (85–87) and recently it has been shown they also result in unintended very large deletions, insertions or chromosomal rearrangements, sometimes at frequencies up to 10% depending on editing context, by mechanisms that are largely unclear (88–92).
The factors that affect the cutting efficiency of PNs are even less clear. Specific sequence features at the target site can either promote or disfavor this process and nucleosomes may impede access of the PN to DNA. However, despite advances in incorporating this knowledge into design algorithms, the Cas9:gRNA cutting efficiency, for instance, still varies considerably between algorithm-based gRNA designs and in many cases, designs are inactive (77–78,93).
Since most KO applications require that both the efficiency and nature of indel editing is accurately determined, the need for cost-efficient, sensitive and accurate indel profiling methods remains. Furthermore, the prominent 1-bp insertion feature of some gRNA designs possesses great potential in gene re-framing applications, as recently demonstrated for restoration of dystrophin expression in a Duchenne muscular dystrophy preclinical model (94) and therefore, indel profiling methods become relevant for both KO and gene re-framing applications. Finally, for knockin applications, indel detection and quantification methods are essential for the initial steps of identifying a highly active PN for the genomic site to be edited.
This review intends to survey and guide the reader through the available methods for accurate and sensitive detection of PN-induced indels. We briefly outline the procedure and discuss the practical advantages and problems related to the use of each of these methods. Examples will be given for indel detection methodologies applicable to low as well as high-throughput gene editing workflows and identification of ex vivo and in vivo gene editing in cells, whole organs and organisms with high genome complexity.
Classic indel detection methodologies
Several mutation-screening methodologies have in the past decades been developed in the field of human genetics and hereditary disease. These include denaturing high-performance liquid chromatography (DHPLC) (95,96), single stranded conformational polymorphism (SSCP) (97), denaturing gradient gel electrophoresis (DGGE) (98) and High Resolution Melt Analysis (HRMA) of fluorescently stained PCR products . The methods are all cost-effective and robust. However, they are not well suited for evaluation of genome editing outcomes, as they fall short regarding one or more important parameters, such as not providing information on the nature of indels, poor performance for low-frequency indels or laborious assay optimization.
Indel detection methods for genome editing
During recent years, several methods have been developed or adapted to serve the specific needs for indel detection in the genome editing field. Generally, PNs induce small indels and, in the case of CRISPR/Cas9, often single-base insertions as the predominant indel that must be reliably detected (73,75,77). Furthermore, single-base resolution is essential to determine if the indels cause frameshifts or re-framing and finally the methods should be simple, cost-efficient and adaptable to the many diverse applications of genome editing. These needs impose demanding requirements to the ‘ideal’ indel detection methodology. A comprehensive overview of the available indel detection methods is provided in Table 1 and the most commonly used methods for genome editing indel detection are shown in Figure 3.
Table 1.
Indel detection methods
| Method | Principle for detection | Indel resolution/Sensitivity | Cost pr. sample | Labor intensity | High throughput amenabilityb | Sequence of indel provided | Indel quantification/indel sizes detected | Can analyse Dual gRNA/knockinc | References |
|---|---|---|---|---|---|---|---|---|---|
| DHPLC | Denaturing high-performance liquid chromatography | 1 bp/10–20% | Low | Low | Yes | No | Limited/not revealed | Yes/Yes | (95) |
| SSCP | Single stranded conformational polymorphism | 2 bp/not determined | Low | Low | Yes | No | Limited/not revealed | Yes/Yes | (97) |
| DGGE | Denaturing gradient gel electrophoresis | 1 bp/not determined | Low | Low | No | No | Limited/not revealed | Yes/Yes | (98) |
| HRMA | High resolution melt analysis | 1 bp/1.4% | Low | Low | No | No | Limited/not revealed | Yes/Yes | (165) |
| ddPCR | Mechanically emulsified droplet probe-based quantitative PCR | Probe dependent/0.2% | Medium | Low | Yes | No | Higha/not revealed | Yes/No | (119) |
| qEva-CRISPR | Multiplex ligation-based probe amplification of ligated, hybridized, half-probes | Probe dependent/5% | Medium | Medium | No | No | Mediuma/not revealed | Yes/No | (117,118) |
| RFLP | Restriction fragment length polymorphism of a diagnostic restriction site | Not revealed/2–5% | Low | Low | No | No | Limited/not revealed | Yes/Yes | (99) |
| EMC | Enzyme mismatch cleavage of heteroduplex amplicons | >2–5 bp/2–5% | Low | Medium | No | No | Limited/not revealed | Yes/No | (106,107) |
| Sanger Topo | Sanger sequencing of single colony Topo cloned amplicons | 1 bp/1% | High | High | No | Yes | High/1–1000 bp | Yes/Yes | (115) |
| TIDE | Sanger sequence trace decomposition | 1 bp/1–5% | Low | Low | No | No | High/1–50 | No/Yes | (120) |
| ICE | Sanger sequence trace decomposition | 1 bp/1–5% | Low | Low | Yes | Yes | High/1–30 bp | Yes/Yes | (122) |
| NGS | Massive parallel sequencing of target specific amplicons | 1 bp/0.1% | High | High | Yes | Yes | High/1–200 bp | Yes/Yes | (73,124) |
| IDAA | Capillary electrophoretic fragment analysis of tri-primer PCR labelled amplicons | 1 bp/0.1% | Low | Low | Yes | No | High/1–1000 bp | Yes/Yes | (115,140) |
| PacBio | SMRTbell replication | Large indels/variable | High | High | Yes | Yes | High/large indels >100 bp | Yes/Yes | (129) |
| Nano-pore | Nanopore ssDNA sequencing | Large indels/variable | Low | High | Yes | Yes | High/large indels >100 bp | Yes/Yes | (130) |
aDetection/quantification require that indel-detecting probe is affected by the indel.
bDefined as batch upload of 96 samples or more.
cKnockin is here defined as ssODN donor-specified nucleotide insertions.
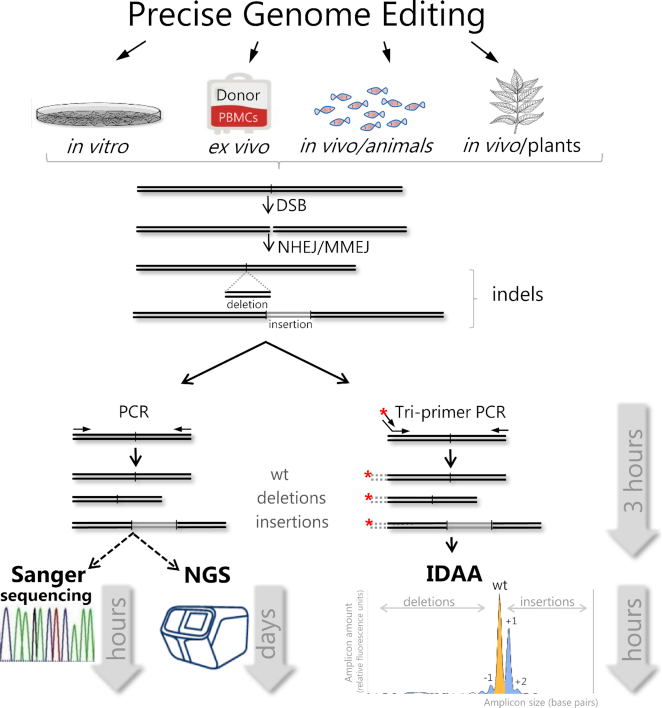
Figure 3.
Comparative presentation of the most commonly used indel detection methodologies for genome editing. Methods shown are; next-generation sequencing (NGS), Sanger sequencing based methods via cloning (Topo TA) or sequence trace decomposition (TIDE/ICE), enzyme mismatch cleavage assay (EMC), restriction fragment length polymorphism assay (RFLP) and indel detection by amplicon analysis (IDAA). Approximate effort, time and cost (from low to high) required for completing the steps for each respective method from genome edited sample (light grey hexagon) to full insight is indicated at the bottom. QC indicates some of the necessary quality control steps required for NGS.
In the following, we review features of the most widely used indel detection methods. Nearly all of the methods are based on genomic amplification of the PN target site by PCR, using primers located on either side of the PN cut site. Thereafter, the PCR product (hereafter designated as amplicon) is subjected to further analysis, which differs between the various methods surveyed. The shared principle of PCR amplicon analysis confers a number of common features to the methods. They are all very sensitive with respect to the genomic input required for analysis: in principle, 10 cells are sufficient input to characterize indel mutagenesis in an edited diploid, clonal cell line, where only two alleles are analysed. However, for comprehensive indel profiling with accurate quantification of several lower-to-medium frequency (5–25%) indels in a population of edited cells after ∼50% PN delivery, the number of cells typically needed as input will be at least 500 (or 3 ng genomic DNA, when assuming a DNA content of 6 pg per diploid cell). Furthermore, a 0.1% frequency indel, the lower limit of the most sensitive indel detection methods, will maximally be represented once in a pool of 500 diploid cells (=1000 template chromosomes) and therefore requires several thousand cells as input for reliable detection. As another convenient feature of the PCR-based indel detection methods, the input can be crude extracts of cells lysed in appropriate buffers for extraction of genomic DNA; i.e. there is no need for purified genomic DNA as template for the PCR in most of the methods. When applied to complex genomes, the performance of the methods can vary across target sites, and depending on genome and locus complexity, this impacts on the specificity and fidelity of the amplification reaction and on downstream amplicon analysis. All of the PCR based methods, except qEva-CRISPR, will fail to detect deletions that extend to the primer binding sites, as for instance, the recently reported large deletions and complex rearrangements sometimes elicited by CRISPR/Cas9. Often, however, this may not pose a problem, given that 90–95% of PN-elicited indels are <30 bp in size (73–76,79).
Restriction fragment length polymorphism (RFLP) analysis
RFLP assay (99), also known as Cleaved Amplified Polymorphic Sequences (CAPS) (100), was one of the first methods to be used to monitor the efficacy of PNs (46,101–102). The approach is based on the fact that the position of the PN-induced DSB is known and if placed in close proximity or ‘on top’ of a restriction endonuclease cut site, allows for identification of restriction resistant amplicons due to indel-induced destruction of the restriction site. In the first step, two PCRs amplify the PN target site of the edited sample and an unedited control sample, respectively. Next, amplicons are incubated with the appropriate restriction endonuclease and the digested amplicons are analyzed by simple agarose gel electrophoresis followed by quantification of digested amplicons (representing the wild-type allele) versus non-digested amplicons (representing the mutant allele) by free image software such as ImageJ (https://imagej.nih.gov/ij/?). The decrease in restriction endonuclease digested amplicons in the edited sample provides an estimate of the indel frequency. Complete cutting of the amplicons from the unedited control sample serves as a control for proper activity of the restriction endonuclease.
These assays are straightforward and easy to perform. If the assay design allows for a PN and a restriction endonuclease site overlap, this assay is suitable for estimating indel formation in cell pools with a detection sensitivity in the range of ≈2% (103). It is also well suited for screening clonal cell lines to identify indel mutagenesis on one or both alleles. RFLP has shown its major usefulness in monitoring of HDR-mediated knockin of donor constructs possessing a diagnostic restriction endonuclease site (104), which however, is beyond the scope of this review. As the major drawback, the assay does not provide information on the nature of the indel editing events, e.g. if the indels cause frameshifting and functional KO has been achieved. Thus, the RFLP assay may be used as an initial screening method to identify edited clonal cell lines that have been edited, followed by Sanger sequencing analysis to determine, which of the clones have frameshifting indels. Furthermore, the assay depends on the presence of a diagnostic restriction endonuclease site at or near the cut site that is destroyed by the indel(s), which is not always the case. To circumvent this problem, a recombinant version of the relevant PN may be used as the restriction endonuclease, as elegantly demonstrated with Cas9:gRNA (105). Due to the simplicity of the assay, cost effectiveness and low-tech instrumentation requirements, RFLP assays are still commonly used for determining indel and knock-in outcomes in genome editing experiments.
Enzyme mismatch cleavage (EMC) assay
The EMC assay is another first-generation but still widely-used genome editing indel detection method (106). EMC assays are based on selective endonuclease recognition and cleavage of heteroduplex DNA, but not homoduplex DNA (107), formed after reannealing of heterogenous amplicons possessing different nucleotide variations, such as indels. The method is comprised of four steps: (i) PCR amplification of the PN target site of the edited sample, (ii) denaturation and reannealing of the PCR products to allow formation of heteroduplex amplicons generating single-stranded DNA ‘bubbles’ at the mismatch position, (iii) selective cleavage of heteroduplexes by a mismatch-sensitive, single-stranded DNA specific endonuclease and (iv) detection of the cleavage event. Detection can be achieved by low-cost, size discriminatory electrophoresis in agarose gels or by capillary electrophoresis (108). Quantification of digested amplicons (representing the mutant allele) versus non-digested amplicons (representing the wild-type allele) by free image software such as ImageJ (https://imagej.nih.gov/ij/?) can provide an estimate of the indel mutation frequency. As part of the assay set up, it must first be tested on an unedited sample that the primers used do not amplify a naturally occurring SNP, which would also lead to the formation of heteroduplexes and a false-positive signal. For the digestion any of the commercially available single-stranded DNA mismatch-sensitive endonucleases can be used, including plant derived CEL-I and CEL-II (Surveyor®) endonuclease (109–111), bacteriophage derived T7 endonuclease-I (107), T4 endonuclease VII (T4E7) (112) or bacterial endonuclease V (EndoV) (113). EMC assays based on agarose gel electrophoresis are easy to perform, are low cost and depending on the endonuclease used, have a detection sensitivity of 2–3% (111) that can be increased to 0.5% using PAGE-analysis (114). With regard to CRISPR/Cas9-based genome editing, which often generates 1-bp indels, a common issue relates to the inferior ability of some of the endonucleases used for EMC to elicit cleavage at single-base loops and thus, incapable of detecting the presence of single-base indel events (111,115–116). Furthermore, EMC assays are inherently unsuitable for quantitation of high-level editing of low complexity (e.g. a specific indel of 80% frequency), since a high fraction of the mutant amplicons will reanneal to form a high fraction of endonuclease-resistant homoduplexes (111,116). The latter two scenarios often result in severe underestimation of EMC-based editing efficiencies (116). Similar to RFLP, EMC assays do not provide information of the nature of indel editing events. Despite these limitations, the simplicity, low cost and simple instrumentation requirements based on agarose gel electrophoresis have made EMC a commonly used assay for determining indel outcomes in PN targeting experiments.
qEva-CRISPR
Quantitative Evaluation of CRISPR/Cas9-mediated editing (qEva-CRISPR) (117) is a modified form of the multiplex ligation-based probe amplification (MLPA) assay developed to detect and quantitate total PN-induced indel events (118). In this method, two oligonucleotide half-probes are designed to anneal head-to-tail with the adjoining ends on top of the PN cut site. Next, the probes are incubated with genomic DNA from an edited sample as well as an unedited control sample and the samples are subjected to a ligation reaction followed by PCR using fluorescent primers specific for either half-probe. Half-probes annealed to wild-type alleles can be ligated together, and therefore PCR amplified, whereas half-probes annealed to indel-containing alleles cannot be ligated, and consequently not PCR amplified. Finally, the amount of amplicons, which is proportional to the amount of wild-type alleles present in the samples, is quantitated by capillary electrophoresis that may be performed by service providers. The extent of indel mutagenesis in the edited sample can thereby be determined by quantifying the loss of amplicon signal in the edited sample relative to the amplicon signal in the unedited control sample.
qEva-CRISPR is simple and easy to perform and only requires standard laboratory equipment, if capillary electrophoretic analysis is outsourced. The method allows for detection of indel mutagenesis in edited clones. Furthermore, it allows for detection of indel mutagenesis in edited pools of cells, with a sensitivity down to 5% frequencies. Single-nucleotide indels can be detected and, unlike the rest of the PCR-based methods, there is no upper size limit for indels that can be detected. Simultaneous (multiplex) analysis of several PN target sites is possible by using several, different probes for each of the sites in the reaction. As the major limitation, qEva-CRISPR does not provide any information on the nature of the PN-elicited indels. Furthermore, the method is relatively hands-on demanding and starting costs for probe generation are relatively high.
Digital PCR
Digital PCR, such as digital droplet PCR (ddPCR), provides an accurate and highly sensitive solution for the assessment of total gene editing frequencies (119). The principle of ddPCR is based on mechanically emulsifying (dividing) a PCR solution into thousands of single-droplet reactions. Adaptation of ddPCR for genome editing analysis requires the design of two differently fluorophore-labeled TaqMan oligonucleotide probes detecting the amplicon derived from the PN target site: one probe specific for sequence not affected by indel mutagenesis and a second probe specific for sequence spanning the PN cut site, whereby the binding of the probe will be eliminated by an indel. The fluorescence signal of the TaqMan probes bound to the amplicon is measured upon completion of the PCR reaction (end-point analysis) using a device similar to a flow cytometer, such as Bio-Rad Laboratories QX200 or similar instruments. Hereby, double-positive versus single-positive fluorescence signal in each individual PCR droplet is determined, which is counted as wild-type allele and mutant allele, respectively, enabling accurate and very sensitive quantification of indel events down to 0.2% frequencies (119). However, the method will only detect indels that eliminate the binding site of the indel-sensitive probe and thus, is best used, when the editing outcome(s) has already been defined and the probe designed accordingly. Furthermore, the method provides no information of the nature of the indel outcomes detected.
Amplicon cloning and Sanger sequencing
Specific indel detection methods have been developed based on Sanger sequencing of amplicons derived from the genomic target site (Figure 4). Depending on the application, three different Sanger sequencing-based approaches can be undertaken; (i) amplicon cloning and sequencing, (ii) direct amplicon sequencing or (iii) direct amplicon sequencing followed by sequence trace decomposition, described in the following section. The first approach may be used for indel profiling in cell pools or in samples with high indel complexity. It involves cloning of locus-derived amplicons into plasmids that are transformed into bacteria followed by agar plate spreading, clone picking, plasmid preparation, sequencing of individual clones and sequence alignment with wild-type amplicon for indel identification (115). For analysis of samples with high indel complexity or low indel representation, the sensitivity and accuracy of this procedure will directly depend on the number of clones sequenced, which may need to be rather high. For instance, to detect and quantify indels with frequencies of 10% and 1%, ≥30 and ≥300 clones would have to be analyzed, respectively. The second approach can be used for low-complexity indel analysis such as analysis of edited diploid clonal cell lines, where indels may be present on one or two alleles, giving rise to no more than two Sanger sequence traces. The locus derived amplicons are purified and sequenced directly and indels are identified by manual inspection of the composite sequence trace that is derived from the individual traces of the two different alleles present in the sample. Both Sanger sequencing procedures are simple and straightforward but require special instrumentation for Sanger sequencing. This task nowadays is outsourced to vendors specialized in Sanger sequencing services. The simplicity of both approaches makes them an accessible way of indel identification, and the methods provide the nature (sequence) of the indels. However, the methods are laborious, time-consuming and not suitable for high throughput analysis.
Figure 4.
Schematic outline of indel profiling workflows of Sanger sequence decomposition (TIDE/ICE), NGS and IDAA and sample-to-data time required. At the top, examples of genome editing applications, where these indel detection methods have been used. The primary outcome is a DNA double-strand break (DSB) followed by NHEJ/MMEJ-mediated indel formation induced by PNs. The targeted region is amplified by standard PCR or fluorophore tri-primer PCR. At bottom panels, amplicons can be gel purified, followed by analysis by Sanger sequence deconvolution using TIDE/ICE software or by NGS. Alternatively, tri-primer, fluorophore-labelled amplicons can be directly subjected to IDAA by capillary electrophoretic fragment analysis, followed by indel analysis. Cas9 targeting of human ST6GALNAC1 promoter illustrates an IDAA profile generated by ProfileIT software with typical size distribution and frequencies of WT, out-of-frame and in-frame amplicons (indels) represented by peaks colour-coded in yellow, blue and white, respectively.
Sanger sequencing and TIDE or ICE
An alternative to Sanger sequencing of individually cloned amplicons is direct Sanger sequencing of amplicons derived from the edited cells followed by deconvolution of the composite sequence traces by appropriate software to determine the size and frequency of indels. Specifically, the analysis requires two PCRs that amplify the PN target site of the edited sample and an unedited control sample (wild type). The amplicons are thereafter purified, quantitated and subjected to standard capillary electrophoresis Sanger sequencing using one of the PCR primers, which typically is outsourced to service providers. Finally, the sequencing data file and the gRNA sequence are uploaded to the software, which compares the wild-type control trace to the mixture of traces that are derived from any mutant and wild-type sequences present in the edited sample and computes indel sizes and frequencies.
Tracking of Indels by DEcomposition (TIDE) was the first such software, developed to analyze indels induced by various CRISPR/Cas9 orthologs (120). TIDE is provided as a free web service for academic institutions (https://tide.deskgen.com/) and has now been widely adopted in the field. Uploading of the required sequencing data files is easy and the output of TIDE is a comprehensive and easy-to-interpret profile of indels in the edited sample. Thus, indels are represented in a bar graph showing the size and frequency of the individual indels. A list of frequency and P-value for the individual indels and the total percentage of indel alleles in the edited sample are provided. The default range for indels analyzed is −10 bp to +10 bp, but the range can be manually increased from −50 bp to +50 bp. The TIDE variant TIDER was developed to also analyse knockin editing (121), which, however, is beyond the scope of this review. Recently, a very similar software, Inference of CRISPR Edits (ICE) was reported that is also freely available and user friendly (https://ice.synthego.com) (122). ICE builds on the method of Sanger sequence trace decomposition developed for TIDE but includes several additional features: in addition to a bar graph representation of the indel profile, the output also displays the sequence traces of the edited and the control sample to aid quality checking or interpretation of the edits. The output furthermore includes a list of the nucleotide sequences of the indels, although nucleotide insertions are represented by ‘N’. Manual inspection of the sequence traces, however, may reveal the identity of the insertion(s). The range for indels analyzed is −30 bp to +14 bp. Furthermore, ICE can determine the complex outcome of editing using up to three gRNAs that target distinct genomic sites contained within the amplicon sequence. In this application, indels of 100–150 bp or more can be analyzed, depending on sequence quality. ICE calculates the total percentage of indel alleles, as well the total percentage of knockout alleles (with indels that are frameshifting or >21 bp in size) in the edited sample. The latter calculation should be used with caution, as it does not take into consideration if the indels extend into an intron, thereby deleting a splice site, which may affect expression of the targeted gene in more complex ways. Finally, batch upload of multiple sequencing files is supported and ICE can also analyse knockin editing events.
Recent comparative studies showed that TIDE/ICE and targeted NGS assays provide very similar editing profiles for pools of cells with respect to size and frequency of indels (116,122). Thus, TIDE and ICE can provide quantitative determination of indels in complex editing spectra with single-base discrimination. The methods are very robust, yielding near-identical editing profiles in replicate experiments and the indel detection sensitivity is 2–4% (120,122). TIDE and ICE can only analyse indels elicited by Cas9. The sensitivity and accuracy of TIDE and ICE depend on high-quality Sanger sequences for the control and edited samples. For this reason, amplicons must be column purified to remove PCR reagents or agarose gel purified if unspecific PCR products are present. Furthermore, as the quality of Sanger sequencing traces deteriorates with length, determination of large indels can be less accurate. The ease, accessibility, reproducibility, accuracy and low cost make TIDE and ICE preferred methods for indel profiling of edited pools and clones of cells.
Next-generation sequencing (NGS)
Targeted NGS (amplicon deep sequencing) has recently been widely adopted as one of the preferred methods for indel profiling in the genome editing field (Figures 3 and 4), as it represents the gold standard with respect to the amount of information, accuracy and sensitivity provided in a single analysis. All NGS strategies are based on massive parallel sequencing of hundreds of thousands of amplicons derived from the PN target site, followed by bioinformatics analyses to determine the distribution of indel sizes and frequencies. Whereas the other indel detection methods reviewed here are relatively mature, NGS methods constantly evolve and the exact procedures also vary among manufacturers. However, the most commonly used procedure for amplicon NGS involves an initial amplification of the PN target site with primers containing common overhangs that form binding sites for the primers of a re-amplification step. The latter primer pairs contain overhangs with specific index sequence, which allows barcoding of amplicons derived from a given PN target site. In addition, these primers contain adaptors for the sequencing reaction. After the second PCR, amplicons derived from individual PN target sites are purified, quantified, pooled at equal ratios and finally, the amplicon pool is prepared for the sequencing. The indexing typically allows sequencing of up to 96 samples in one run. While the procedure is thus quite high-throughput, this number of samples requires an entire day of ‘hands-on’ work. Amplicon NGS is often performed as sequencing-by-synthesis on a MiSeq instrument (Illumina) that may run overnight. Other chemistries and sequencing platforms include Ion semiconductor sequencing (Ion Torrent/ThermoFisher), Combinatorial probe anchor synthesis (cPAS- BGI/MGI), Sequencing by Oligonucleotide Ligation and Detection (SOLiD/ABI-ThermoFisher). Often, amplicon NGS is outsourced to vendors operating in the field such as Beijing Genomics Institute, Genewiz or Eurofins that only require a PCR sample of the PN target site amplified using standard primers (see Table 2 for listing and information).
Table 2.
Providers for standard PN indel analytical services by NGS or IDAA
| Service provider | Servicea | Service/platform | Link |
|---|---|---|---|
| Genewiz | NGS FA PacBio | Amplicon Sequencing /Ion Proton/ABI Instrument Sequel/PacBio | https://www.genewiz.com/ |
| Eurofins Genomics | NGS FLA | Amplicon sequencing/Illumina/Ion Proton/ABI Instrument | https://www.eurofinsgenomics.eu/ |
| Applied Biological Materials | NGS | Amplicon Sequencing/Illumina | https://www.abmgood.com/ |
| CeGat | NGS/Sanger | Targeted Sequencing/Illumina | https://www.cegat.de/en/ |
| Lucigen | NGS | Amplicon Sequencing/ Illumina | https://www.lucigen.com/ |
| BGI | NGS Nanopore | Amplicon Sequencing/Illumina PacBio and Nanoporeb | https://www.bgi.com/ |
| CD Genomics | NGS | Amplicon Sequencing//Illumina PacBiob | https://www.cd-genomics.com/ |
| SeqMatic | NGS | Amplicon Sequencing/Illumina | https://www.seqmatic.com/ |
| CD Genomics | PacBio | PacBiob | https://www.cd-genomics.com/ |
| BaseClear | PacBio Nanopore | PacBiob and Nanoporeb | https://www.baseclear.com/ |
| Cobo Technologies | CIPP | Indel Detection by Amplicon Analysis/ABI Instrument | https://cobotechnologies.com/ |
aDepending on provider, IDAA service is covered by FA (Fragment Analysis), FLA (Fragment Length Analysis) or CIPP (CRISPR InDel Profiling Platform).
bPlatform used on request.
Amplicon NGS data can be analysed by the free and user-friendly software CRISPResso2 (http://crispresso.pinellolab.partners.org/) that displays the indel spectrum as bar graphs and other outputs, which collectively provide complete information on indel sizes, frequencies and the actual sequence of the indels in the sample (123). In addition, several alternative online software resources have been developed for genome editing induced indel detection by NGS (124), including CRISPR-DAV (https://github.com/pinetree1/crispr-dav), CRISPR Genome Analyzer (http://54.80.152.219/), CRISP-R (https://bioconductor.org/packages/release/bioc/html/CrispRVariants.html) and AmpliCan (https://bioconductor.org/packages/release/bioc/html/amplican.html). The recently developed Rational InDel Meta-Analysis (RIMA) software is a particularly useful tool for the analysis of PN-elicited indels with respect to the role of microhomologies in determining indel outcomes, the impact of small molecule compounds on repair outcomes and for elucidation of the role of specific genes in repair mechanisms (73).
The output of NGS is the most comprehensive indel profiling currently achievable. Due to the large number of sequences (called reads) obtained from each PN target site (typically 10 000–100 000), indel frequencies can be quantitated with high accuracy and sensitivity that can be down to 0.1%. To take advantage of the high accuracy and sensitivity, however, the number of PN manipulated cells used as input/template for target amplification in the PCR must be high, otherwise the data will represent sequencing of the PN target site at futile, high coverage. Frequently, 100 ng genomic DNA is used as PCR template, which corresponds to ∼17,000 diploid cells.
While the workflow for standard indel profiling by amplicon NGS is simple, demanding applications such as for example whole-genome off-target indel analysis requires much more complicated NGS procedures and trained bioinformatics support for processing of the raw sequencing data. The previously mentioned issues relating to the dominant NGS error types in detection of naturally occurring indels and the effects that DNA extraction and other library preparation steps have on downstream sequence integrity also apply to genome editing related indel detection by NGS. What the genome editing field in this respect is awaiting, are ways of standardization and ‘best practices’ of the various NGS platforms, their differing chemistries and downstream data processing procedures. Amplicon NGS approaches are often based on amplicons up to 500 bp and the maximum sizes of deletions and insertions that can be detected are ∼450 and 50 bp, respectively. The preparation and sequencing costs for an individual sample is relatively high, unless some 50 or more samples are analysed by multiplex NGS.
Amplicon NGS is primarily used in applications, where indel sensitivity, accuracy or sequence is of special importance. Furthermore, direct evaluation of the full spectra of repair outcomes in millions of amplicons from hundreds of PN target sites through NGS can provide important insight into indel repair mechanisms, as exemplified in the Discussion section. However, the time, labor and cost constraints associated with NGS analysis limit widespread adoption of this method for most editing applications. Thus, for standard editing tasks such as gRNA testing, indel profiling of cell pools or clonal analysis of a few or even hundreds of cell clones, analyses like TIDE/ICE or IDAA can provide the needed information, conveniently and at low cost.
Emerging single-molecule sequencing technologies (third-generation sequencing)
Recent reports have demonstrated relatively high incidences of very large insertions/deletions and chromosomal rearrangements after PN on-target editing that cannot be detected by amplicon NGS (88–90). These NGS short comings have largely been overcome by single-molecule sequencing technologies (so called ‘third-generation sequencing’) that are not based on breakdown of DNA into short fragments or amplification of DNA, but on direct sequencing of single DNA molecules (125). Current third-generation sequencing platforms enable generation of >100 kb sequence read-lengths (126–128). The longer read lengths enable assessment of large DNA insertions and deletions and structural rearrangements induced by gene editing. These emerging third-generation single-molecule sequencing technologies are primarily based on two differing methodologies, single-molecule real-time (SMRT) sequencing by PacBio (Pacific Biosciences) (129) and nanopore sequencing (Oxford Nanopore Technologies (ONT)) (130).
The PacBio SMRT principle is based on single-stranded circular DNA sequencing of a doublestranded DNA molecule ligated with hairpin adaptors. This template is designated a SMRTbell. SMRTbell sequencing takes place in a zero-mode waveguide (ZMW) detection well loaded with a single immobilized DNA polymerase and is initiated when the SMRTbell adaptor hairpin starts replication (129). The ZMW well, wherein the replication process takes place, enables detection of light emitted from the single fluorescently labelled bases that are continuously being incorporated during DNA strand synthesis, so called continuous long read (CLR). The circular nature of the SMRTbell allows for sequencing of the template many times, which increases polymerase processivity and strongly improves overall accuracy. With the recent PacBio RS II system average read lengths over 20 kb and up to 60 kb can be achieved (https://www.pacb.com/applications/targeted-sequencing/). However, the PacBio hardware and running costs have prevented it from being applied more broadly in the scientific community.
The concept of using membrane attached nanopore single-stranded DNA (ssDNA) sequencing originated in the 1980’s (131,132), but it was not until 2014 before nanopore sequencing became commercially available through ONT. The Nanopore flow cell is made of an electrical resistant membrane with a tiny pore with a diameter of one nanometer (hence the name). The pore enables measurement of the ionic current fluctuations, when single-stranded DNA passes through a biological nanopore. Since nucleotides differ in size, the size of the pore opening will be different for each base and therefore, each nucleotide will result in a unique electrical signature that is detected. Thus, Oxford Nanopore sequencers measure the ionic current fluctuations, when single-stranded DNA is electrophoretically fed and passes through into a matrix-embedded biological nanopore. Nanopore sequencing is not limited in read length, but merely the length of the ssDNA molecule to be sequenced, and extremely long reads of 1 Mb (127) and more than 2 Mb (133) have been reported. ONT offers a cost-effective iPod size MinION miniature size sequencer, which makes it very portable and independent from established sequencing infrastructure.
Although superior read lengths can be achieved with either platform, a current limitation of both methodologies relate to read accuracy that in both cases tend to be in the range of 85–99% (126,134). However, with continuous improvements of both technologies and enhanced development of software tools for base calling and error correction (128,135–136) the beginning of the third revolution in sequencing technology shows promise in shedding light on the frequencies and mechanisms by which the recently reported large deletions, insertions and chromosomal rearrangements occur after gene editing.
Indel detection by amplicon analysis (IDAA)
Fast, accurate and cost-efficient indel detection with down to single-base discrimination power can also be provided by Indel Detection by Amplicon Analysis (IDAA) (115) (Figure 4). In contrast to amplicon labelling strategies based on target-specific fluorophore-labeled primers (137–139), the IDAA principle is based on the use of a universal fluorophore-labelled primer that by tri-primer PCR enables homogenous labelling of amplicons derived from a given PN genomic target site. Following capillary electrophoresis, the amplicon fragments are detected and quantified as peaks that are called based on size and fluorescence intensity (Figure 4). Specifically, the analysis requires two tri-primer PCRs that amplify the PN target site of the edited sample and an unedited control sample (wild type). Thereafter, amplicons are directly subjected to standard capillary electrophoresis, which typically is outsourced to service providers (Table 2). Finally, the electrophoretic data files can be analysed using GeneMapper™, the free but less sophisticated Peak Scanner™ (https://www.thermofisher.com) (see (140) for additional links and instructions) or by the user-friendly software ProfileItTM (https://viking.sdu.dk/pages/software/profileit/). In the latter case, the output is a comprehensive and easy-to-interpret profile of indel alleles in the edited sample from which total and out-of-frame indel efficiencies can be quantified (141). Specifically, each indel is represented by a fragment peak for which size and fluorescence signal reveals indel size and frequency, respectively. The range of indels that can be analysed by IDAA is large as every indel located between the primer target sites will be detected, that can range from 1 to 400 bp and 1 bp to 1 kb for deletions and insertions, respectively. IDAA can determine the complex outcome of editing using two gRNAs that target distinct genomic sites covered by the amplicon sequence. Since IDAA is based solely on fragment analysis, it can analyse editing outcomes in very complex polyploid/multi-allelic genomes, where the complexity would preclude sequencing-based indel analysis. Recent comparative analyses showed that IDAA, targeted NGS and digital droplet PCR assays provide very similar editing profiles for pools of cells with respect to size and frequency of indels (116,140,142). Thus, IDAA can provide quantitative determination of indels in complex editing profiles with single-base discrimination. Because the background signal levels are low in fragment analysis, IDAA is very sensitive, showing indel detection sensitivity down to 0.1%, i.e. similar to NGS (140). Furthermore, IDAA is very robust, generating near-identical profiles in replicate experiments (108,116,140). The method benefits from a fast turn-around time that from sample to full insight takes <6 h. Preparation of the amplicon sample is very simple; no purification is required, and the crude tri-primer PCR needs only dilution prior to capillary electrophoretic analysis. In case unspecific PCR products are present, these will be identified in the wild-type control sample and can be subtracted from the edited sample (141). The sequence of the indels detected by IDAA, however, is not provided, but for KO editing applications, the ease, accessibility, reproducibility, accuracy and low cost make IDAA a preferred method for indel profiling of edited pools and clones of cells.
Examples of indel analysis in CRISPR/Cas9 editing applications
Below, we present some examples of indel detection using Sanger sequence deconvolution and IDAA to highlight various features of the two methods. Amplicon NGS could alternatively have been used in some of the examples, if it were important to also know the exact sequence of the indels.
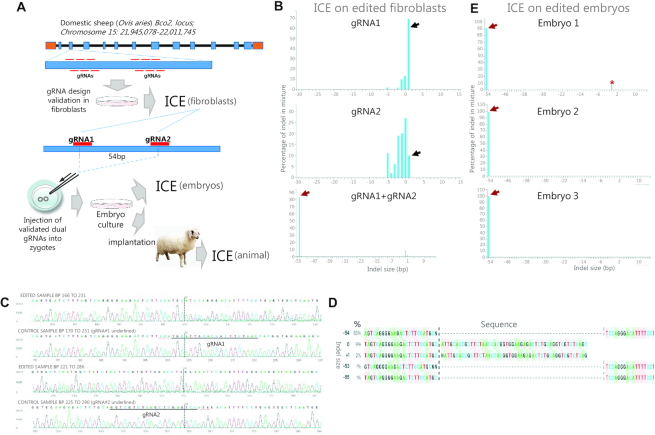
Genome editing in sheep zygotes
Genome editing in zygotes is a powerful approach for improving economically important traits in livestock, as exemplified by knockout of the beta-carotene oxygenase (BCO2) gene associated with yellow fat disease in Tan sheep (143) (Figure 5A). Figure 5B shows the use of ICE to validate 2 gRNA designs used individually or combined in sheep fibroblasts, illustrating how ICE portrays the various detected indels as bars on the x-axis and their relative frequencies on the y-axis. Note that the gRNAs used individually elicited a mixture of small indels, whereas their dual application elicited a predominant deletion of 54 bp between the two gRNA target sites, but hardly any indels at the individual target sites. The sequence chromatograms of the dual gRNA-edited fibroblasts and the control sample are shown in Figure 5C and the list of detected indels in Figure 5D, as displayed by the ICE software. In the next step, the validated dual gRNAs were injected into sheep zygotes and indel analysis was performed on derived embryos. As shown in Figure 5E, ICE determined that the 54 bp deletion was the sole (i.e. biallelic) editing outcome in two embryos, whereas a third embryo also had a low-frequency 4-bp deletion, thereby revealing a low level of mosaicism in this embryo. Thus, the example illustrates the ability of ICE to profile single as well as dual gRNA editing, providing a comprehensive identification of indels of high as well as low frequencies.
Figure 5.
Genome editing in sheep zygotes—use of ICE for screening and validation of gRNA designs and genotyping of edited embryos. (A) Work flow for BCO2 gene editing in Tan sheep, indicating the use of ICE to validate gRNA designs in fibroblasts and genotype embryos prior to generation of lambs. (B) The predominant 1-bp insertion often observed after single gRNA editing is indicated by black arrow. The major 54-bp deletion resulting from dual gRNA editing is indicated by red arrow. (C) Sequence chromatograms from the dual gRNA-edited and control fibroblasts. Note the composite sequence trace from the edited sample as opposed to the single trace from the control sample. (D) List of sequences from the dual gRNA-edited fibroblasts, as displayed by ICE. Note that ICE does not provide the full sequence of indels, which must be deduced by inspecting the boundaries. Note that the list provides indel size, frequency and sequence of the various alleles, although some nucleotide identities are not defined (indicated by ‘N’). (E) ICE indel profiles for three embryos edited with dual gRNAs. The major 54-bp deletion is indicated by red arrow and the low frequency 4-bp deletion by an asterisk.
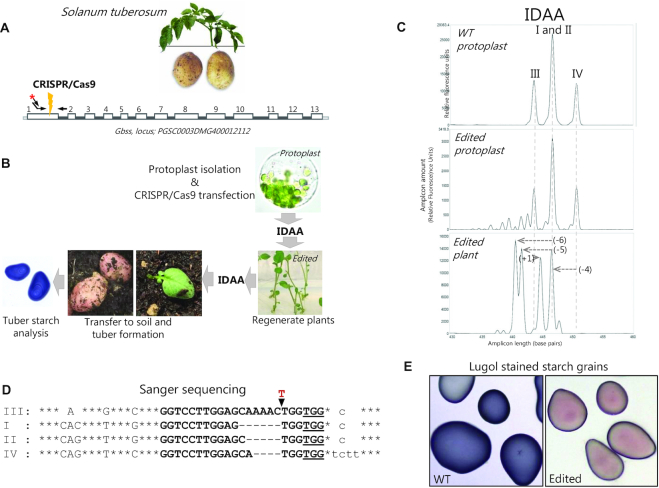
Genome editing of tetraploid Solanum tuberosum
In many plant species, profiling and quantitation of CRISPR/Cas9-induced indels is a demanding task due to the presence of complex and high-ploidy genomes (144). This is illustrated by editing of the granule bound starch synthase gene GBSS in Solanum tuberosum (potato) (Figure 6A, B), a tetraploid organism, where GBSS furthermore is represented by three allelic variants. IDAA on wild-type protoplasts (144,145) can reveal the presence of the four alleles, as they can be distinguished by size in the GBSS gRNA target region (Figure 6C, upper panel). Editing of potato was achieved through delivery of CRISPR/Cas9 to protoplasts that were subsequently analysed by IDAA, which revealed that indels had been induced (Figure 6C, middle panel). Consequently, plants were regenerated from the pool of edited protoplasts and IDAA was used to identify individual plants with major indel mutation of all four GBSS alleles (Figure 6C, lower panel). The sequences of the major indels were determined by amplicon cloning and Sanger sequencing (Figure 6D) and functional knockout validated by starch staining on potatoes from the regenerated ex-plant (Figure 6E). Thus, the example illustrates the ability of IDAA to indel profile a complex locus, where sequence heterogeneity in the wild-type cells would preclude Sanger sequence decomposition approaches. It also illustrates the ability of IDAA to provide a comprehensive identification of indels of high as well as low frequencies.
Figure 6.
Genome editing of tetraploid Solanum tuberosum (potato) - use of IDAA to assess indel mutation in edited protoplasts and derived ex-plants. (A) Schematic of the targeted GBSS gene. (B) Work flow for GBSS gene editing in potato, indicating the use of IDAA to assess indel mutagenesis in edited protoplasts and derived ex-plants. (C) Upper panel: IDAA on wild-type protoplasts displaying the polymorphic GBSS alleles (I–IV) that can be discriminated by size, 426 bp (III), 429 bp (I/II), 433 bp (IV), in the amplified target region. As the two non-polymorphic alleles I and II have same size, they generate a single IDAA peak with double height relative to the alleles III and IV. Middle panel: IDAA on protoplast pool 24 h post CRISPR/Cas9 transfection shows that extensive indel mutagenesis has occurred. Lower panel: IDAA on an individual plant derived from a single edited protoplast shows indel mutation in all 4 alleles, as indicated by a shift in all positions of the WT peak positions in the profile. (D) Sanger sequencing results from the ex-plant IDAA profiled in C, lower panel, displaying the indel sequences of the four alleles. Capital letters denote exon/coding sequence, small letters denote intron sequence, bold letters denote the gRNA target sequence and PAM sequence (underlined), common to all four alleles. Asterisks denote identical nts outside of the gRNA target region and SNPs listed for the individual alleles. Blank spaces indicate naturally occurring deletions in individual alleles. Hyphens indicate gene editing induced deletions in individual alleles. The red (T) denotes an insertion (E) Loss of Lugol′s staining (blue) of starch from potatoes of the edited plant demonstrates functional knockout of GBSS.
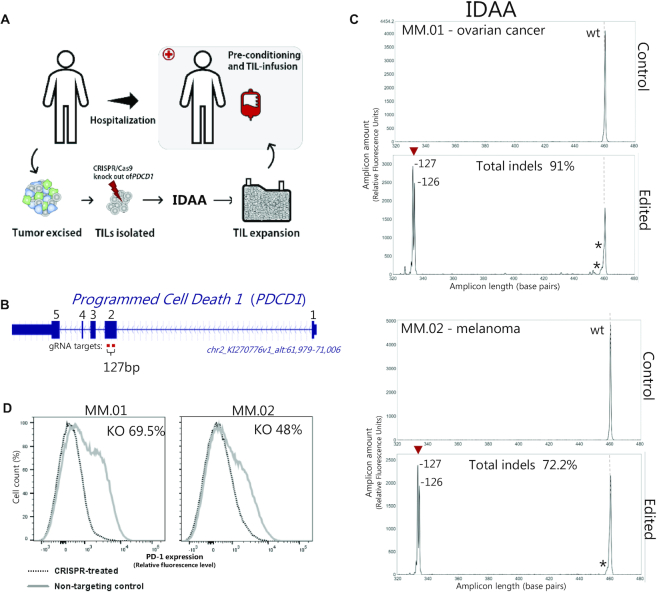
Genome editing for T-cell cancer therapeutics
Genome editing holds great promise as one of the next-generation therapies for the correction of genetic disorders or treatment of non-genetic diseases that remain refractory to traditional treatments (146,147). One therapeutic application being explored in the adaptive cancer immunotherapy space, involves the use of CRISPR/Cas9 to knockout the immunoregulatory gene PDCD1 in patient-derived tumor infiltrating lymphocytes (TILs) in order to enhance the anti-tumor activity of the T-cell population, which is subsequently infused back into the patient (Figure 7A, B) (148). Prior to infusion, PDCD1 knockout must be validated by a fast and robust method, such as IDAA (Figure 7C). This example also illustrates the ability of IDAA to detect and characterize large indels 126 or 127 bp generated by the use of two gRNAs.
Figure 7.
Generation of T-cell cancer therapeutics—use of IDAA for fast and accurate ex vivo validation of PDCD1-edited T-cells. (A) Potential clinical regimen for generation and use of PDCD1-edited tumour-infiltrating lymphocytes (TILs), indicating the use of IDAA for validating PDCD1 knockout in TILs prior to infusion in patients. (B) Schematic of the targeted PDCD1 gene with indication of the two gRNAs used that are interspaced by 127 bp between the cut sites. (C) IDAA PDCD1 profiles of TILs from patients with ovarian cancer or melanoma. Upper panels represent TILs prior to editing. Lower panels represent edited TILs, showing efficient deletion of the genomic sequence between the two gRNAs with two major repair outcomes of 1 bp size difference. A low level of small indels (*) at the individual gRNA target sites was also detected. Note the similarity of the two profiles, illustrating the nonrandom nature of indel repair as well as the high reproducibility of IDAA. (D) Loss of PD-1 immunofluorescence signal in FACS analysis of edited T-cells (stippled curve) versus control T-cells (grey curve) demonstrates functional knockout of PDCD1.
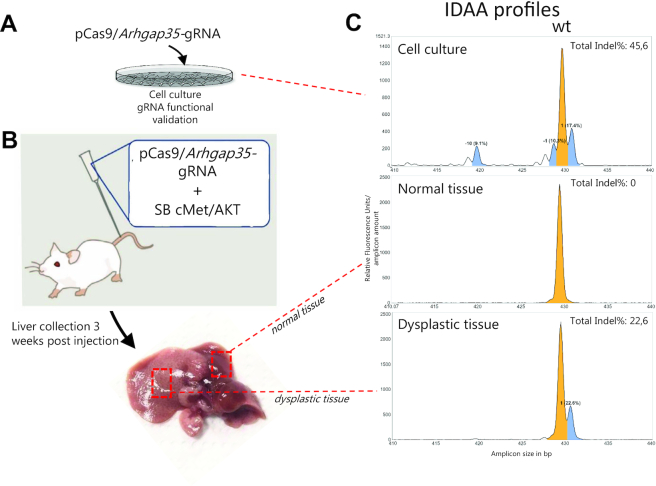
Generation of mouse models of liver cancer through in vivo liver editing
Somatic genome editing in mice is a powerful approach to study functionally the large number of putative cancer genes emanating from tumor genome sequencing. As one example, candidate tumor suppressor genes can be knocked out in adult mouse liver via hydrodynamic tail vein injection of CRISPR/Cas9 to generate new models of liver cancer within weeks, as illustrated with Arhgap35 (Figure 8). After initial in vitro screening for gRNA designs with high indel-inducing activity (Figure 8A,C), the ability of the chosen gRNA to achieve knockout in vivo must be validated at an early time point after injection, because tumor modeling are long-term (6–12 months) and costly experiments (Figure 8B). To this end, IDAA is a robust and sensitive assay to test and quantify if frame-shifting indels have been elicited (Figure 8C).
Figure 8.
Generation of mouse models of liver cancer through in vivo editing of tetraploid hepatocytes–use of IDAA to assess indel mutation of Arhgap35 candidate tumor suppressor gene. (A) gRNA designs for knockout of Arhgap35 were first tested in the mouse Neuro2a cancer cell line (B) Plasmids expressing pCas9 and the most active Arhgap35 gRNA identified in (A) were delivered to the adult mouse liver via hydrodynamic tail vein injection along with Sleeping Beauty (SB) transposon vectors for genomic insertion of cMet and AKT oncogenes. Three weeks post-injection, liver tissue showing normal or dysplastic morphology (indicative of transfection) was isolated. (C) IDAA profiles for the Arhgap35 gRNA target site, as determined in Neuro2a cells in vitro (upper panel), in normally appearing liver tissue (middle panel) or in the dysplastic liver tissue (lower panel). The ProfileIT software indicates the wild-type allele in yellow and out-of-frame indel alleles of >5% frequency in blue. Note the presence of the predominant 1-bp insertion in the cell line as well as in the liver cells, whereas the deletions are absent from the liver cells.
DISCUSSION
The new indel detection methods have not only greatly facilitated the practical procedures needed to perform a genome editing experiment; their application in a broad range of settings is currently providing essential new insight into the mechanisms underlying PN-elicited indel formation, which in turn, is greatly instrumental in further improving the genome editing technology.
The bulk of insight has been obtained studying Streptococcus pyogenes (Sp)Cas9 in mammalian cells. An early major discovery was that a given SpCas9:gRNA elicits a highly discrete and reproducible indel spectrum (‘finger print’), as revealed through profiling of hundreds of different gRNA designs by amplicon NGS (76–77,79,149–153), IDAA (93) or TIDE (120). Thus, the predominant indels and the relative frequencies elicited by a given SpCas9:gRNA are nearly identical between replicate experiments in a given cell type and often relatively similar across different cell types from the same species. The reason is that the DNA sequence flanking the cut site (73–79) and the nature of the cut (72–73,79,151) are the main determinants for which types of indels are formed. This realization is of great practical importance, since once the indel spectrum of a SpCas9:gRNA has been determined, its performance in subsequent experiments carried out under similar conditions can be predicted with high accuracy.
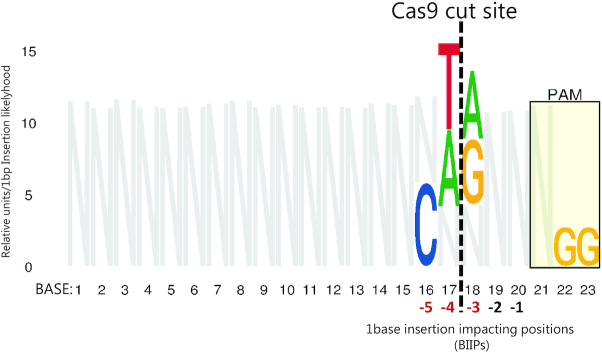
The overall spectrum of indels induced by SpCas9:gRNAs and how they may arise have been revealed by bioinformatics analysis of large indel data sets from amplicon NGS. The most common indel is a 1-bp insertion, accounting for 10–25% of all events, varying with cells/cell conditions studied (73–75,77,79). It is thought to result from the ability of SpCas9 to generate not only blunt-ended DSBs, but also staggered cuts with a one-nucleotide 5′-overhang, which is filled by DNA polymerase, followed by ligation of the DNA ends via the NHEJ pathway (73,79,151,154). The second most frequent indels are 1- and 2-bp deletions (together accounting for 20–25% of all events) (73,75,77). These are often caused by deletion of one copy of a repeating pair of one and two nucleotides, respectively, on either side of the cut, probably via microhomology-mediated annealing, processing and ligation of the DNA ends by the NHEJ pathway (75). Then follows a tail of increasingly larger deletions of steadily declining frequency up til ∼30 bp (73,75,79). MMEJ repair based on short stretches (typically 2–3 nt) of homology accounts for a majority of the deletions >2 bp, amounting to 30–40% of all indels (73,75,77). One study estimated that >75% of all deletions can be ascribed to microhomology-mediated repair (with microhomologies down to 1 bp) via either the NHEJ or the MMEJ pathway (79). Of note, the above-described indels created by NHEJ and MMEJ repair show highly reproducible frequencies in replicate experiments. By contrast, the remainder of deletions >2 bp that show no associated microhomologies can vary significantly in frequency between replicate experiments and may account for 20–30% of all events. These deletions may arise in a more stochastic manner by mechanisms that are not clear (75), possibly involving Cas9 exonuclease activity (155). Deletions, whether homology-mediated or not, are overwhelmingly unidirectional, meaning that they extend either upstream or downstream from the DSB, rather than spanning it (79). Altogether, 90–95% of all SpCas9:gRNA elicited indels are deletions <30 bp or 1-bp insertions that arise through above-outlined mechanisms (73–75,152).
However, the indel spectrum elicited by an individual SpCas9:gRNA does not conform to the average trend described above. Instead, it is typically composed of one or up to a small hand-full of predominant indels as well as several low-frequency indels, as revealed by amplicon NGS (73,75–77,79,149,151,153), IDAA (115) or TIDE (120). Furthermore, the indel spectra elicited by individual gRNAs show great variability relative to each other, as expected, given the major role of the target site sequence in dictating indel repair (for examples of different spectra, see Figures 5, 6 and 8). A majority of SpCas9:gRNAs also elicit low-frequency (<1%), relatively large insertions of up to 85 bp, often representing copies of sequence of adjacent or distal chromosomal regions (78). Finally, long-read sequencing has recently revealed that SpCas9:gRNAs may elicit several-kb deletions and complex rearrangements at significant frequencies depending on context, as discussed above (89,92).
The finding that NHEJ and MMEJ pathways both contribute substantially to indel mutagenesis induced by the typical SpCas9:gRNA (73–76) has important practical implications: specifically, factors that affect the relative activities of these two pathways, such as cell cycle status (83) or mutation of genes in the pathways or in genes affecting the pathways, as may occur in cancer cells (76), will impact indel outcomes in the cells being edited. Such factors help account for the differences in indel profiles often observed for a given SpCas9:gRNA across cell types (see Figure 8 for an example). As another practical implication, chemical inhibition or knockdown of either NHEJ or MMEJ pathway components can be used as a means to bias indel mutagenesis towards desired outcomes (73,76,151,156). In addition to above factors, the chromatin state has been found to modestly influence SpCas9:gRNA indel spectra by mechanisms that are not clear (77).
The indel detection methods have also provided important knowledge on the dynamics of PN-elicited indel formation. Strikingly, repair rates at SpCas9:gRNA cuts appear to be slow (many hours-one day) compared to the faster repair of naturally occurring DSBs and repair rates vary greatly between target sites, as demonstrated by amplicon NGS and mathematical modelling (156). The slow repair may be a consequence of the binding of SpCas9:gRNA to DNA ends after cutting (156,157), although studies using single-particle tracking analysis have suggested that the SpCas9:gRNA-target site interaction is very transient (158). As another key insight, NHEJ repair is predominant in the early phase after DSB induction, whereas MMEJ repair contributes mainly after 1–3 days (73,76,156). This has the important practical implication that indel characterization should be performed ∼72 h after DSB induction in order to determine the full spectrum of indel mutagenesis, as for instance, when characterizing a new gRNA design. In addition to the intrinsic repair kinetics, indel dynamics are also a function of PN delivery format. Thus, IDAA measurements from hours to days after delivery of SpCas9:gRNA by RNP/electroporation, plasmid liposomal transfection, transposon integration or lentiviral transduction showed great temporal differences in indel induction (159).
The non-random, reproducible and target sequence-specified nature of indel mutagenesis has motivated the development of algorithms to predict the indel spectrum elicited by a given SpCas9:gRNA, based on the large indel data sets derived from amplicon NGS. These studies have, for example, revealed specific nucleotides around the cut site that strongly promote the common 1-bp insertion, possibly by promoting SpCas9:gRNA to make the 1-nt 5′-overhang staggered cut that can be filled-in and ligated to produce a 1-bp insertion (73–75,78–79). Data generated from analysis of >800 gRNA designs using IDAA have confirmed the motif (Figure 9). Other rules that promote specific 1-bp and 2-bp deletions as well as microhomology-mediated deletions have also been delineated (73–75,78–79). The studies have resulted in web tools to assist the design of gRNAs for SpCas9, which include: Lindel (https://lindel.gs.washington.edu) (79), inDelphi (https://indelphi.giffordlab.mit.edu) (74), FORECasT (https://partslab.sanger.ac.uk/FORECasT) (75) and SPROUT (https://zou-group.github.io/SPROUT) (78). While these CRISPR/Cas9 prediction tools are all based on indel data from thousands of gRNA design, they used very different modeling approaches, experimental designs and cell systems to build the algorithms. In brief, the Lindel model is based on 4790 gRNA designs and SpCas9 stably expressed in human embryonic kidney (HEK) 293T cells. inDelphi is based mainly on 1872 gRNA designs and SpCas9 stably expressed in mouse embryonic stem cells (mESCs) and human osteosarcoma U2OS cells. FORECasT is based on 5000 gRNA designs and SpCas9 stably expressed in human leukemic K562 cells. These three models were based on exogenously integrated target sites for the gRNAs. By contrast, SPROUT is based on endogenous target sites for 1656 gRNA designs delivered as RNPs to human primary T cells. Not surprisingly, the predictions of the various models showed significant differences, when compared side-by-side (78,79). However, these tools do present significant advances on certain aspects, in a particular with respect to predicting gRNAs with increased probability of eliciting a frameshifting indel spectrum and 1-bp insertions (78,79) and can therefore reduce the number of gRNA designs that need be tested in pursuit of an efficient KO tool.
Figure 9.
IDAA based identification of 1-bp insertion impacting positions (BIIPs) after SpCas9:gRNA editing. Starting with the indel summary data generated from >800 individually validated gRNA design IDAA profiles (GlycoCRISPRcollection Addgene https://www.addgene.org/browse/article/28192658/), a BBAM model (BennettBrodyAnalysis Model) was trained for each type of indel that occurred more than 30 times as a sign of a significant repair outcome. From these models, the 1-bp insertion prediction BBAM model performed the best and was further investigated. The 1-bp insertion rate was calculated for each base in each position across the protospacer (number of 1-bp insertions/number of non-1-bp insertions). From this, outliers were defined as those bases where the +1 insertion rate had a modified Z-score greater than 3.0. Following that, we let the value of N at each base (except for the last two bases) be the sum of the value of the bases not determined to be an outlier. Only significant BIIPs are shown in color and all other sequence positions with N (in gray).
The factors that affect the cutting efficiency of SpCas9:gRNAs are much less clear, since direct measurements have not been performed at scale. Many large-scale gRNA screens have shown that overall GC content and specific nucleotides in or close to the target site as well as chromatin state influence gRNA efficiency (160,161). These studies have produced web tools for design of efficient gRNAs, such as The CRISPR Guide RNA design tool (https://www.benchling.com/crispr/) (161) or CRISPOR (http://crispor.tefor.net/) (162). However, the screens underlying these studies nearly all used gene KO as readout, which is a combined measure for cutting efficiency and frameshift indel repair. A few screens have linked gRNA design to indel mutagenesis, as assessed by amplicon NGS. One such study found that G at position 20 in the target site (next to the PAM) and DNA accessibility, as in open chromatin of actively transcribed genes, are factors that promote gRNA efficiency (152). The latter agrees with studies showing that active gRNAs cluster to regions of low nucleosome occupancy and that nucleosomes directly impede SpCas9:gRNA binding and cleavage of DNA (81,163). Another study showed that SpCas9:gRNAs that elicit one/few predominant indels are on average twice as active as those eliciting multiple indels (77). Despite the progress, up to 20–30% of designs in large-scale SpCas9:gRNA studies were found to be inactive or have very low activity, as determined by IDAA (93) or amplicon NGS (78), even though all were based on prediction algorithms. In another large scale study, many gRNAs with high predicted activity scores were found to be inactive (77).
Several of the principles outlined above for SpCas9 are general for PNs and some of the concepts were, in fact, discovered through early studies on meganucleases, although the small number of analysed target sites limited the depth of the investigations (reviewed in (164)). However, there are important variations on the common theme. For instance, while ZFNs and TALENs obey the common rules of highly reproducible indel finger prints (140,153), deletions as preferred editing outcomes (84,140,153) and substantial contributions of homology-based repair (153), these PNs elicit indel spectra that are overall distinct relative to each other and to SpCas9 and, for TALENs in particular, are typically more complex than those elicited by SpCas9 (84,140,153). The different nature of DSBs generated by the various classes of PNs is one major determinant for the distinct indel spectra. For instance, ZFNs and Francisella novicida Cas12a (Cpf1) generate staggered cuts with 4-nucleotide 5′-overhangs. These were shown to be duplicated to produce predominant 4-bp insertions, probably via the fill-in and ligation mechanism described above for the predominant 1-bp insertion of SpCas9 (Taheri-Ghahfarokhi et al., 2018). As another example, SpCas9 nickase pairs are typically designed to generate staggered cuts with large overhangs (40–70 nucleotides), as a result of 2 independent SpCas9 nickase induced nicks on opposing DNA strands. These generate very complex spectra with numerous, large indels up to 200 bp in size, varying with distance between the individual nickase cut sites and the polarity of the overhangs (5′ or 3′) (72). Editing using dual SpCas9:gRNAs may elicit perfect excision of the sequence between the cuts or imperfect excision with additional insertions and/or deletions at either cut (151,156).
In summary, significant progress has been made in understanding the mechanism of indel formation induced by SpCas9:gRNA. Yet, in the majority of editing applications, we still cannot accurately predict the complete spectrum of indels elicited by a particular gRNA or its absolute activity. This may not be surprising, since the prediction algorithms are most accurate, when the new SpCas9:gRNAs are used under conditions similar to those used for developing the tool, which is rarely the case. When a gRNA is used under other conditions, factors like cell cycle/p53 status, repair pathway status, chromatin status, stochastic microhomology-less deletions, delivery method (transient versus stable) and gRNA secondary structure formation will introduce various degrees of unpredictability regarding the editing outcome. Furthermore, when using modified versions of SpCas9, somewhat different indel spectra and efficiencies may result (73). Finally, the other classes of PNs are largely unexplored with respect to editing forecasting and SpCas9 nickase pairs and TALENs produce indel spectra so complex that with current knowledge, it is hard to see how they would become predictable.
For these reasons, experimental characterization of indel spectra and efficiencies remains an essential task in any genome editing application. It is therefore fortunate that a plethora of indel detection methods have now been developed. The various methods are very diverse in terms of accuracy, ease, cost, throughput, instrument requirement and information output. Naturally, it is still possible to further optimize some of the methods, in particular amplicon NGS, where simpler workflows and lower costs would be welcomed. Nevertheless, collectively the methods cover nearly all of the requirements of the genome editing field: for any given genome editing experiment, it is now possible to find one or several methods that will suit the particular need for proper indel detection and characterization.
ACKNOWLEDGEMENTS
We thank Camilla Andersen from Copenhagen Center for Glycomics, Department of Odontology, University of Copenhagen for excellent technical assistance and Leigh Brody and Mark Dunne, Desktop Genetics, 28 Hanbury St, London E1 6QR for generating the algorithm underlying the BBAM model.
Contributor Information
Eric Paul Bennett, Copenhagen Center for Glycomics, Department of Odontology and Molecular and Cellular Medicine, Faculty of Health Sciences, University of Copenhagen, DK-2200 Copenhagen N, Denmark.
Bent Larsen Petersen, Department of Plant and Environmental Sciences, University of Copenhagen, DK-1871 Frederiksberg C, Denmark.
Ida Elisabeth Johansen, Department of Plant and Environmental Sciences, University of Copenhagen, DK-1871 Frederiksberg C, Denmark.
Yiyuan Niu, Biotech Research and Innovation Centre (BRIC), Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark; College of Animal Science and Technology, Northwest A&F University, Yangling Shaanxi, China.
Zhang Yang, Copenhagen Center for Glycomics, Department of Odontology and Molecular and Cellular Medicine, Faculty of Health Sciences, University of Copenhagen, DK-2200 Copenhagen N, Denmark.
Christopher Aled Chamberlain, Center for Cancer Immune Therapy, Department of Oncology, Copenhagen University Hospital, Herlev, Denmark.
Özcan Met, Center for Cancer Immune Therapy, Department of Oncology, Copenhagen University Hospital, Herlev, Denmark; Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
Hans H Wandall, Copenhagen Center for Glycomics, Department of Odontology and Molecular and Cellular Medicine, Faculty of Health Sciences, University of Copenhagen, DK-2200 Copenhagen N, Denmark.
Morten Frödin, Biotech Research and Innovation Centre (BRIC), Faculty of Health Sciences, University of Copenhagen, Copenhagen, Denmark.
FUNDING
Lundbeck Foundation (to Z.Y.); Novo Nordisk Foundation; the Danish National Research Foundation [DNRF107]; ERC-2017-COG Type of action [ERC-COG; 772735]; GlycoSkin, Remodel EU-H2020(870133); EU H2020-MSCA-ITN (765269); The Lundbeck Foundation (R313-2019-869); Neye Foundation (to H.H.W.); Danish Councils for Strategic and Independent Research [12-125709 to B.L.P.); Kartoffelafgiftsfonden (to I.E.J.); Danish Cancer Society [R124-A7632-15-S2]; Novo Nordisk Foundation [NNF17OC0028380]; Independent Research Fund Denmark [9039-00450B to M.F.]; University of Copenhagen Excellence Programme for Interdisciplinary Research [CDO2016]; European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement [765269]. Funding for open access charge: Danish National Research Foundation [DNRF107]. Remodel EU-H2020(870133); EU H2020-MSCA-ITN (765269); The Lundbeck Foundation (R313-2019-869); Neye Foundation.
Conflict of interest statement. E.P.B. declares that a patent application covering the IDAA method is pending and acts as a scientific advisor of Cobo Technologies. IDAA and ProfileIt are commercialized by Cobo Technologies. E.P.B. currently holds a Senior Advisor position at NovoNordisk A/S.
REFERENCES
- 1. Gu X., Li W.H.. The size distribution of insertions and deletions in human and rodent pseudogenes suggests the logarithmic gap penalty for sequence alignment. J. Mol. Evol. 1995; 40:464–473. [DOI] [PubMed] [Google Scholar]
- 2. Fernie B.A., Hobart M.J.. An unusual combined insertion/deletion polymorphism in intron 10 of the human complement C6 gene. Hum. Genet. 1997; 100:104–108. [DOI] [PubMed] [Google Scholar]
- 3. Chuzhanova N.A., Anassis E.J., Ball E.V., Krawczak M., Cooper D.N.. Meta-analysis of indels causing human genetic disease: mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum. Mutat. 2003; 21:28–44. [DOI] [PubMed] [Google Scholar]
- 4. Den Dunnen J.T., Antonarakis E.. Nomenclature for the description of human sequence variations. Hum. Genet. 2001; 109:121–124. [DOI] [PubMed] [Google Scholar]
- 5. Mills R.E., Luttig C.T., Larkins C.E., Beauchamp A., Tsui C., Pittard W.S., Devine S.E.. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006; 16:1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reams A. Mechanisms of gene duplication and amplification. Cold Spring Harb. Perspect. Biol. 2015; 7:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sehn J.K. Kulkarni S., Pfeifer J.. Insertions and deletions (indels). Clinical Genomics. 2014; Elsevier; 129–150. [Google Scholar]
- 8. Brogna S., McLeod T., Petric M.. The meaning of NMD: translate or perish. Trends Genet. 2016; 32:395–407. [DOI] [PubMed] [Google Scholar]
- 9. Kurosaki T., Myers J.R., Maquat L.E.. Defining nonsense-mediated mRNA decay intermediates in human cells. Methods. 2019; 155:68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pérez-Ortín J.E., Alepuz P., Chávez S., Choder M.. Eukaryotic mRNA decay: methodologies, pathways, and links to other stages of gene expression. J. Mol. Biol. 2013; 425:3750–3775. [DOI] [PubMed] [Google Scholar]
- 11. Dawson E., Chen Y., Hunt S., Smink L.J., Hunt A., Rice K., Livingston S., Bumpstead S., Bruskiewich R., Sham P. et al.. A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res. 2001; 11:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abecasis G.R., Altshuler D.L., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., Abecasis G.R., Bentley D.R. et al.. A map of human genome variation from population-scale sequencing. Nature. 2010; 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bentley D.R., Mullikin J.C., Hunt S.E., Cole C.G., Mortimore B.J., Rice C.M., Burton J., Matthews L.H., Pavitt R., Plumb R.W. et al.. An SNP map of human chromosome 22. Nature. 2000; 407:516–520. [DOI] [PubMed] [Google Scholar]
- 14. Bhangale T.R., Stephens M., Nickerson D.A.. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat. Genet. 2006; 38:1457–1462. [DOI] [PubMed] [Google Scholar]
- 15. Chen K., McLellan M.D., Ding L., Wendl M.C., Kasai Y., Wilson R.K., Mardis E.R.. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007; 17:659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li R., Li Y., Kristiansen K., Wang J.. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008; 24:713–714. [DOI] [PubMed] [Google Scholar]
- 17. Mullaney J.M., Mills R.E., Stephen Pittard W., Devine S.E.. Small insertions and deletions (INDELs) in human genomes. Hum. Mol. Genet. 2010; 19:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allam A., Kalnis P., Solovyev V.. Karect: accurate correction of substitution, insertion and deletion errors for next-generation sequencing data. Bioinformatics. 2015; 31:3421–3428. [DOI] [PubMed] [Google Scholar]
- 19. Jiang Y., Turinsky A.L., Brudno M.. The missing indels: an estimate of indel variation in a human genome and analysis of factors that impede detection. Nucleic Acids Res. 2015; 43:7217–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim B.Y., Park J.H., Jo H.Y., Koo S.K., Park M.H.. Optimized detection of insertions/deletions (INDELs) in whole-exome sequencing data. PLoS One. 2017; 12:e0182272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Homer N., Merriman B., Nelson S.F.. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009; 4:e7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. David M., Dzamba M., Lister D., Ilie L., Brudno M.. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics. 2011; 27:1011–1012. [DOI] [PubMed] [Google Scholar]
- 25. Albers C.A., Lunter G., MacArthur D.G., McVean G., Ouwehand W.H., Durbin R.. Dindel: Accurate indel calls from short-read data. Genome Res. 2011; 21:961–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garrison E., Marth G.. Haplotype-based variant detection from short-read sequencing. 2012; arXiv doi:20 July 2012, preprint: not peer reviewedhttps://arxiv.org/abs/1207.3907.
- 28. Wei Z., Wang W., Hu P., Lyon G.J., Hakonarson H.. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011; 39:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O’Rawe J., Jiang T., Sun G., Wu Y., Wang W., Hu J., Bodily P., Tian L., Hakonarson H., Johnson W.E. et al.. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013; 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ghoneim D.H., Myers J.R., Tuttle E., Paciorkowski A.R.. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC Res. Notes. 2014; 7:864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Costello M., Pugh T.J., Fennell T.J., Stewart C., Lichtenstein L., Meldrim J.C., Fostel J.L., Friedrich D.C., Perrin D., Dionne D. et al.. Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013; 41:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen L., Liu P., Evans T.J., Ettwiller L.. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017; 355:752–756. [DOI] [PubMed] [Google Scholar]
- 33. Krusche P., Trigg L., Boutros P.C., Mason C.E., De F.M., Vega L., Moore B.L., Gonzalez-Porta M., Eberle M.A., Tezak Z. et al.. Best practices for benchmarking germlinesmall-variant calls in human genomes. Nat Biotechnol. 2019; 37:555–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis A.J., Chen D.J.. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013; 2:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sfeir A., Symington L.S.. Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heyer W.-D., Ehmsen K.T., Liu J.. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010; 44:113–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeh C.D., Richardson C.D., Corn J.E.. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019; 12:1468–1478. [DOI] [PubMed] [Google Scholar]
- 40. Rouet P., Smih F., Jasin M.. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:6064–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chevalier B.S., Stoddard B.L.. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001; 29:3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li L., Chandrasegaran S.. Alteration of the cleavage distance of Fok I restriction endonuclease by insertion mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2764–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Miller J.C., Holmes M.C., Wang J., Guschin D.Y., Lee Y.-L., Rupniewski I., Beausejour C.M., Waite A.J., Wang N.S., Kim K.A. et al.. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007; 25:778–785. [DOI] [PubMed] [Google Scholar]
- 44. Szczepek M., Brondani V., Büchel J., Serrano L., Segal D.J., Cathomen T.. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat. Biotechnol. 2007; 25:786–793. [DOI] [PubMed] [Google Scholar]
- 45. Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C.. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005; 435:646–651. [DOI] [PubMed] [Google Scholar]
- 46. Smith J., Bibikova M., Whitby F.G., Reddy A.R., Chandrasegaran S., Carroll D.. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000; 28:3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mahfouz M.M., Li L., Shamimuzzaman M., Wibowo A., Fang X., Zhu J.-K.. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:2623–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bogdanove A.J., Voytas D.F.. TAL effectors: customizable proteins for DNA targeting. Science. 2011; 333:1843–1846. [DOI] [PubMed] [Google Scholar]
- 49. Mussolino C., Morbitzer R., Lütge F., Dannemann N., Lahaye T., Cathomen T.. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011; 39:9283–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cho S.W., Kim S., Kim J.M., Kim J.-S.. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013; 3:230–232. [DOI] [PubMed] [Google Scholar]
- 53. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. Elife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rouet P., Smih F., Jasin M.. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994; 14:8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stoddard B.L. Homing endonuclease structure and function. Q. Rev. Biophys. 2005; 38:49–95. [DOI] [PubMed] [Google Scholar]
- 56. Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010; 79:213–231. [DOI] [PubMed] [Google Scholar]
- 57. Deng D., Yan C., Pan X., Mahfouz M., Wang J., Zhu J.-K., Shi Y., Yan N.. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012; 335:720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li L., Chandrasegaran S.. Alteration of the cleavage distance of Fok I restriction endonuclease by insertion mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2006; 90:2764–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bonocora R.P., Belfort M.. Mapping homing endonuclease cleavage sites using in vitro generated protein. Methods Mol. Biol. 2014; 1123:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith J., Grizot S., Arnould S., Duclert A., Epinat J.-C., Chames P., Prieto J., Redondo P., Blanco F.J., Bravo J. et al.. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006; 34:e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sander J.D., Maeder M.L., Joung J.K.. Engineering designer nucleases with customized cleavage specificities. Current Protocols in Molecular Biology. 2011; John Wiley; 12.13.1–12.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maeder M.L., Thibodeau-beganny S., Sander J.D., Voytas D.F., Joung J.K.. Oligomerized pool engineering (OPEN): an ‘open-source’ protocol for making customized zinc-finger arrays. Nat. Protoc. 2009; 4:1471–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reyon D., Tsai S.Q., Khayter C., Foden J.A., Sander J.D., Joung J.K.. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012; 30:460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mali P., Esvelt K.M., Church G.M.. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013; 10:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007; 315:1709–1712. [DOI] [PubMed] [Google Scholar]
- 66. Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V.. A putative RNA-interference-based immune system in prokaryotes: Computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E.. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011; 471:602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Anders C., Niewoehner O., Duerst A., Jinek M.. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014; 513:569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Belfort M., Bonocora R.P.. Homing endonucleases- methods and protocols. Methods Mol. Biol. 2014; 1123:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bothmer A., Phadke T., Barrera L.A., Margulies C.M., Lee C.S., Buquicchio F., Moss S., Abdulkerim H.S., Selleck W., Jayaram H. et al.. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 2017; 8:13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Taheri-Ghahfarokhi A., Taylor B.J., Nitsch R., Lundin A., Cavallo A.-L., Madeyski-Bengtson K., Karlsson F., Clausen M., Hicks R., Mayr L.M. et al.. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 2018; 46:8417–8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I.. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018; 563:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Páleníková P., Khodak A., Kiselev V., Kosicki M. et al.. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2019; 37:64–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. van Overbeek M., Capurso D., Carter M.M., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Vidal B., Zheng J., Hoffman G.R. et al.. DNA repair profiling reveals nonrandom outcomes at. Mol. Cell. 2016; 63:633–646. [DOI] [PubMed] [Google Scholar]
- 77. Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P.. Target-specific precision of CRISPR-mediated genome editing. Mol. Cell. 2019; 73:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Leenay R.T., Aghazadeh A., Hiatt J., Tse D., Roth T.L., Apathy R., Shifrut E., Hultquist J.F., Krogan N., Wu Z. et al.. Large dataset enables prediction of repair after CRISPR–Cas9 editing in primary T cells. Nat. Biotechnol. 2019; 37:1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen W., McKenna A., Schreiber J., Haeussler M., Yin Y., Agarwal V., Noble W.S., Shendure J.. Massively parallel profiling and predictive modeling of the outcomes of CRISPR/Cas9-mediated double-strand break repair. Nucleic Acids Res. 2019; 47:7989–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lazzarotto C.R., Malinin N.L., Li Y., Zhang R., Yang Y., Lee G., Cowley E., He Y., Lan X., Jividen K. et al.. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol. 2020; doi:10.1038/s41587-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S.. Nucleosomes impede cas9 access to DNA in vivo and in vitro. Elife. 2016; 5:e12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D.. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:9351–9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J.. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018; 24:927–930. [DOI] [PubMed] [Google Scholar]
- 84. Kim Y., Kweon J., Kim J.-S.S.. TALENs and ZFNs are associated with different mutation signatures. Nat. Methods. 2013; 10:185. [DOI] [PubMed] [Google Scholar]
- 85. Choi P.S., Meyerson M.. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 2014; 5:3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Blasco R.B., Karaca E., Ambrogio C., Cheong T.C., Karayol E., Minero V.G., Voena C., Chiarle R.. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014; 9:1219–1227. [DOI] [PubMed] [Google Scholar]
- 87. Zuo E., Huo X., Yao X., Hu X., Sun Y., Yin J., He B., Wang X., Shi L., Ping J. et al.. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017; 18:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Weisheit I., Kroeger J.A., Malik R., Klimmt J., Crusius D., Dannert A., Dichgans M., Paquet D.. Detection of deleterious on-target effects after HDR-Mediated CRISPR editing. Cell Rep. 2020; 31:107689. [DOI] [PubMed] [Google Scholar]
- 89. Cullot G., Boutin J., Toutain J., Prat F., Pennamen P., Rooryck C., Teichmann M., Rousseau E., Lamrissi-Garcia I., Guyonnet-Duperat V. et al.. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019; 10:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F. et al.. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020; 367:eaba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q.. Brief Communications Arising Inter-homologue repair in fertilized human eggs. Nature. 2018; 560:E8–E9. [DOI] [PubMed] [Google Scholar]
- 92. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Narimatsu Y., Joshi H.J., Yang Z., Gomes C., Chen Y.H., Lorenzetti F.C., Furukawa S., Schjoldager K.T., Hansen L., Clausen H. et al.. A validated gRNA library for CRISPR/Cas9 targeting of the human glycosyltransferase genome. Glycobiology. 2018; 28:295–305. [DOI] [PubMed] [Google Scholar]
- 94. Amoasii L., Li H., Sanchez-Ortiz E., Caballero D., Harron R., Massey C., Shelton J., Piercy R., Olson E.. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018; 362:86–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Eshaghpour H., Crothers D.M.. Preparative separation of the complementary strands of DNA restriction fragments by alkaline RPC-5 chromatography. Nucleic Acids Res. 1978; 5:1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liu W., Smith D.I., Rechtzigel K.J., Thibodeau S.N., James C.D.. Denaturing high performance liquid chromatography (DHPLC) used in the detection of germline and somatic mutations. Nucleic Acids Res. 2002; 26:1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Inazuka M., Wenz H., Sakabe M., Tahira T., Hayashi K.. A Streamlined mutation detection system: multicolor Post-PCR fluorescence labeling and single-strand conformational polymorphism analysis by capillary electrophoresis. Genome Res. 1997; 7:1094–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Børresen A.L., Hovig E., Brøgger A.. Detection of base mutations in genomic DNA using denaturing gradient gel electrophoresis (DGGE) followed by transfer and hybridization with gene-specific probes. Mutat. Res. 1988; 202:77–83. [DOI] [PubMed] [Google Scholar]
- 99. Botstein D., White R.L., Skolnick M., Davis R.W.. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Gen. 1980; 32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 100. Barth S., Melchinger A.E., Lübberstedt T.H.. Genetic diversity in Arabidopsis thaliana L. Heynh. investigated by cleaved amplified polymorphic sequence (CAPS) and inter-simple sequence repeat (ISSR) markers. Mol. Ecol. 2002; 11:495–505. [DOI] [PubMed] [Google Scholar]
- 101. Smith J., Berg J.M., Chandrasegaran S.. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999; 27:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bibikova M., Carroll D., Segal D.J., Trautman J.K., Smith J., Kim Y.G., Chandrasegaran S.. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001; 21:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen F., Pruett-miller S.M., Huang Y., Gjoka M., Duda K., Taunton J., Collingwood T.N., Frodin M., Davis G.D.. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods. 2011; 8:753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kim J.M., Kim D., Kim S., Kim J.S.. Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat. Commun. 2014; 5:3157. [DOI] [PubMed] [Google Scholar]
- 106. Yeung A.T., Hattangadi D., Blakesley L., Nicolas E.. Enzymatic mutation detection technologies. BioTechniques. 2005; 38:749–758. [DOI] [PubMed] [Google Scholar]
- 107. Mashal R.D., Koontz J., Sklar J.. Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 1995; 2:177–183. [DOI] [PubMed] [Google Scholar]
- 108. Bennett E.P., Jacobi A.M., Garrett R.R., Behlke M.A.. Appasani K. Detection of insertion/deletion (indel) events after genome targeting: Pro's and con's of the available methods. Genome Editing and Engineering: From Talens, ZFNs and CRISPRs to Molecular Surgery. 2018; Cambridge University Press; 181–194. [Google Scholar]
- 109. Oleykowski C.A., Mullins C.R.B., Godwin A.K., Yeung A.T.. Mutation detection using a novel plant endonuclease. Nucleic Acids Res. 1998; 26:4597–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pimkin M., Caretti E., Canutescu A., Yeung J.B., Cohn H., Chen Y., Oleykowski C., Bellacosa A., Yeung A.T.. Recombinant nucleases CEL I from celery and SP I from spinach for mutation detection. BMC Biotechnol. 2007; 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Qiu P., Shandilya H., D’Alessio J.M., O’Connor K., Durocher J., Gerard G.F.. Mutation detection using Surveyor nuclease. BioTechniques. 2004; 36:702–707. [DOI] [PubMed] [Google Scholar]
- 112. Youil R., Kemper B.W., Cotton R.G.. Screening for mutations by enzyme mismatch cleavage with T4 endonuclease VII. Proc. Natl. Acad. Sci. USA. 1995; 92:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gao H., Huang J., Barany F., Cao W.. Switching base preferences of mismatch cleavage in endonuclease V: an improved method for scanning point mutations. Nucleic Acids Res. 2007; 35:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zhu X., Xu Y., Yu S., Lu L., Ding M., Cheng J., Song G., Gao X., Yao L., Fan D. et al.. An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep. 2014; 4:6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang Z., Steentoft C., Hauge C., Hansen L., Thomsen A.L., Niola F., Vester-Christensen M.B., Frodin M., Clausen H., Wandall H.H. et al.. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 2015; 43:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sentmanat M.F., Peters S.T., Florian C.P., Connelly J.P., Pruett-Miller S.M.. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 2018; 8:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Dabrowska M., Czubak K., Juzwa W., Krzyzosiak W.J., Olejniczak M., Kozlowski P.. qEva-CRISPR: a method for quantitative evaluation of CRISPR/Cas-mediated genome editing in target and off-target sites. Nucleic Acids Res. 2018; 46:e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G.. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002; 30:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mock U., Hauber I., Fehse B.. Digital PCR to assess gene-editing frequencies (GEF-dPCR) mediated by designer nucleases. Nat. Protoc. 2016; 11:598–615. [DOI] [PubMed] [Google Scholar]
- 120. Brinkman E.K., Chen T., Amendola M., van Steensel B.. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014; 42:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Brinkman E.K., Kousholt A.N., Harmsen T., Leemans C., Chen T., Jonkers J., van Steensel B.. Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res. 2018; 46:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hsiau T., Maures T., Waite K., Yang J., Kelso R., Holden K., Stoner R.. Inference of CRISPR edits from sanger trace data. 2018; bioRxiv doi:10 August 2019, preprint: not peer reviewed 10.1101/251082. [DOI] [PubMed]
- 123. Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M.A., Liu D.R., Joung J.K., Bauer D.E. et al.. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019; 37:224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Bell C.C., Magor G.W., Gillinder K.R., Perkins A.C.. A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics. 2014; 15:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Midha M.K., Wu M., Chiu K.P.. Long-read sequencing in deciphering human genetics to a greater depth. Hum. Genet. 2019; 138:1201–1215. [DOI] [PubMed] [Google Scholar]
- 126. Ardui S., Ameur A., Vermeesch J.R., Hestand M.S.. Single molecule real-time (SMRT) sequencing comes of age: applications and utilities for medical diagnostics. Nucleic Acids Res. 2018; 46:2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T. et al.. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 2018; 36:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. van Dijk E.L., Jaszczyszyn Y., Naquin D., Thermes C.. The third revolution in sequencing technology. Trends Genet. 2018; 34:666–681. [DOI] [PubMed] [Google Scholar]
- 129. Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B. et al.. Real-time DNA sequencing from single polymerase molecules. Science. 2009; 323:133–138. [DOI] [PubMed] [Google Scholar]
- 130. Clarke J., Wu H.C., Jayasinghe L., Patel A., Reid S., Bayley H.. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 2009; 4:265–270. [DOI] [PubMed] [Google Scholar]
- 131. Niedringhaus T.P., Milanova D., Kerby M.B., Snyder M.P., Barron A.E.. Landscape of next-generation sequencing technologies. Anal. Chem. 2011; 83:4327–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Deamer D., Akeson M., Branton D.. Three decades of nanopore sequencing. Nat. Biotechnol. 2016; 34:518–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Payne A., Holmes N., Rakyan V., Loose M.. Bulkvis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics. 2019; 35:2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Petersen L.M., Martin I.W., Moschetti W.E., Kershaw C.M., Tsongalis G.J.. Third-generation sequencing in the clinical laboratory: sequencing. J. Clin. Microbiol. 2020; 58:e01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Sedlazeck F.J., Lee H., Darby C.A., Schatz M.C.. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet. 2018; 19:329–346. [DOI] [PubMed] [Google Scholar]
- 136. Chu J., Mohamadi H., Warren R.L., Yang C., Birol I.. Innovations and challenges in detecting long read overlaps: an evaluation of the state-of-the-art. Bioinformatics. 2017; 33:1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Pratt J., Venkatraman N., Brinker A., Xiao Y., Blasberg J., Thompson D.C., Bourner M.. Use of zinc finger nuclease technology to knock out efflux transporters in C2BBe1 cells. Current protocols in toxicology / editorial board, Mahin D. Maines (editor-in-chief) … [et al.]. 2012; United States: John Wiley; 23.2.1–23.2.22. [DOI] [PubMed] [Google Scholar]
- 138. Foley J.E., Maeder M.L., Pearlberg J., Joung J.K., Peterson R.T., Yeh J.-R.J.. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 2009; 4:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Linke B., Bolz A.F., von Hofen M., Pott C., Bertram J., Hiddemann W., Kneba M.. Automated high resolution PCR fragment analysis for identification of clonally rearranged immunoglobulin heavy chain genes. Leukemia. 1997; 11:1055–1062. [DOI] [PubMed] [Google Scholar]
- 140. Lonowski L.A., Narimatsu Y., Riaz A., Delay C.E., Yang Z., Niola F., Duda K., Ober E.A., Clausen H., Wandall H.H. et al.. Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. Nat. Protoc. 2017; 12:581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. König S., Yang Z., Wandall H.H., Mussolino C., Bennett E.P.. Luo Y. Fast and quantitative identification of ex vivo precise genome targeting-induced indel events by IDAA. Methods in Molecular Biology. 2019; 1961:Springer Nature; 45–66. [DOI] [PubMed] [Google Scholar]
- 142. Carballar-lejarazú R., Kelsey A., Pham T.B., Bennett E.P., James A.A.. CRISPR indel edits in the malaria species Anopheles. BioTechniques. 2020; 68:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang X., Niu Y., Zhou J., Yu H., Kou Q., Lei A., Zhao X., Yan H., Cai B., Shen Q. et al.. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 2016; 6:32271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jørgensen B., Liu Y., Bennett E.P., Andreasson E., Nielsen K.L., Blennow A., Petersen B.L.. High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 2019; 9:17715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Petersen B.L., Möller S.R., Mravec J., Jørgensen B., Christensen M., Liu Y., Wandall H.H., Bennett E.P., Yang Z.. Improved CRISPR/Cas9 gene editing by fluorescence activated cell sorting of green fluorescence protein tagged protoplasts. BMC Biotechnol. 2019; 19:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Cox D.B.T., Platt R.J., Zhang F.. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015; 21:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Porteus M.H. A new class of medicines through DNA editing. N. Engl. J. Med. 2019; 380:947–959. [DOI] [PubMed] [Google Scholar]
- 148. Met Ö., Jensen K.M., Chamberlain C.A., Donia M., Svane I.M.. Principles of adoptive T cell therapy in cancer. Semin. Immunopathol. 2018; 41:49–58. [DOI] [PubMed] [Google Scholar]
- 149. Koike-Yusa H., Li Y., Tan E.-P., Velasco-Herrera M.D.C., Yusa K.. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2013; 32:267–273. [DOI] [PubMed] [Google Scholar]
- 150. Bae S., Park J., Kim J.-S.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014; 30:1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Shou J., Li J., Liu Y., Wu Q.. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-Mediated nucleotide insertion. Mol. Cell. 2018; 71:498–509. [DOI] [PubMed] [Google Scholar]
- 152. Chari R., Mali P., Moosburner M., Church G.M.. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015; 12:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Bae S., Kweon J., Kim H.S., Kim J.-S.. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods. 2014; 11:705–706. [DOI] [PubMed] [Google Scholar]
- 154. Lemos B.R., Kaplan A.C., Bae J.E., Ferrazzoli A.E., Kuo J., Anand R.P., Waterman D.P., Haber J.E.. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E2010–E2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Stephenson A.A., Raper A.T., Suo Z.. Bidirectional degradation of DNA cleavage products catalyzed by CRISPR/Cas9. J. Am. Chem. Soc. 2018; 140:3743–3750. [DOI] [PubMed] [Google Scholar]
- 156. Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B.. Kinetics and fidelity of the repair of Cas9-Induced Double-Strand DNA breaks. Mol. Cell. 2018; 70:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E.. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016; 34:339–344. [DOI] [PubMed] [Google Scholar]
- 158. Knight S.C., Xie L., Deng W., Guglielmi B., Witkowsky L.B., Bosanac L., Zhang E.T., Beheiry M.E., Masson J.B., Dahan M. et al.. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015; 350:823–826. [DOI] [PubMed] [Google Scholar]
- 159. Kosicki M., Rajan S.S., Lorenzetti F.C., Wandall H.H., Narimatsu Y., Metzakopian E., Bennett E.P.. Dynamics of indel profiles induced by various CRISPR / Cas9 delivery methods. Progress in Molecular Biology and Translational Science. 2017; 152:Elsevier Inc; 49–67. [DOI] [PubMed] [Google Scholar]
- 160. Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E.. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014; 32:1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. et al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016; 34:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Concordet J.P., Haeussler M.. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018; 46:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Isaac R.S., Jiang F., Doudna J.A., Lim W.A., Narlikar G.J., Almeida R.. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. Elife. 2016; 5:e13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Gallagher D.N., Haber J.E.. Repair of a site-specific DNA cleavage: old-school lessons for Cas9-mediated gene editing. ACS Chem. Biol. 2018; 13:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., Grunwald D.J.. Simple methods for generating and detecting locus- specific mutations induced with TALENs in the zebrafish genome. Plos Genet. 2012; 8:e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]