Abstract
Kidney transplantation is the preferred renal replacement therapy available. Yet, long‐term transplant survival is unsatisfactory, partially due to insufficient possibilities of longitudinal monitoring and understanding of the biological processes after transplantation. Small urinary extracellular vesicles (suEVs) – as a non‐invasive source of information – were collected from 22 living donors and recipients. Unbiased proteomic analysis revealed temporal patterns of suEV protein signature and cellular processes involved in both early response and longer‐term graft adaptation. Complement activation was among the most dynamically regulated components. This unique atlas of the suEV proteome is provided through an online repository allowing dynamic interrogation by the user. Additionally, a correlative analysis identified putative prognostic markers of future allograft function. One of these markers – phosphoenol pyruvate carboxykinase (PCK2) – could be confirmed using targeted MS in an independent validation cohort of 22 additional patients. This study sheds light on the impact of kidney transplantation on urinary extracellular vesicle content and allows the first deduction of early molecular processes in transplant biology. Beyond that our data highlight the potential of suEVs as a source of biomarkers in this setting.
Keywords: biomarker, chronic kidney disease, complement, kidney transplantation, proteomics, transplant biology, urinary extracellular vesicles
1. INTRODUCTION
Chronic kidney disease has emerged as one of the major health problems and cost factors for health care systems worldwide affecting all age groups (Chen, Knicely, & Grams, 2019). While renal replacement therapy by dialysis can reinstate kidney function regarding acid‐base homeostasis, water and toxin elimination to a certain extent, it is accompanied by a dramatic increase in both cardiovascular and overall mortality (Matsushita et al., 2015; Van Der Velde et al., 2011). The treatment strategy that results in optimal quality of life and patient survival is kidney transplantation (Mange, Joffe, & Feldman, 2001; Terasaki, Cecka, Gjertson, & Takemoto, 1995). However, the number of potential recipients outweighs the number of donors considerably. This unmet need has prompted a rise in living donor transplantation circumventing the need for dialysis (Currie & Henderson, 2019).
Yet, transplant rejection and chronic allograft nephropathy limit organ survival making repetitive transplantation necessary in many cases (Sellarés et al., 2012). Diagnosis of the underlying disorder relies primarily on renal biopsy via hollow needle tissue sampling involving both a bleeding risk with possible organ damage and the potential of misdiagnosis due to sampling errors (Schinstock et al., 2019; Solez et al., 1993). With primary urine passing all cells of the nephrons, the identification of urinary markers could be of great potential to complement histology. However, to date, urinary sediment and proteinuria are still the only tools available in clinical routine (Klein, Bascands, Mischak, & Schanstra, 2016).
Recent years have shifted different extracellular vesicles (EVs) into the research focus for both their biological function as well as their potential as a source of biomarkers for various diseases (Roy, Hochberg, & Jones, 2018). In contrast to analyses from full urine, EVs – being actively secreted by nephron‐lining epithelial cells – hold the promise to reflect cellular biology more directly. Most studies have discriminated between three types of EVs (exosomes, microvesicles and apoptotic bodies) depending on their biogenesis (Karpman, Ståhl, & Arvidsson, 2017; Merchant, Rood, Deegens, & Klein, 2017).
Current investigations, however, have depicted both the shortcomings of past separation methods and vesicle characterizations as well as a considerable overlap in both size, density as well as protein and nucleic acid content of all three groups (Choi, Kim, Kim, & Gho, 2015; Jeppesen et al., 2019; Musante, Saraswat, Ravidà, Byrne, & Holthofer, 2013; Simons & Raposo, 2009; Théry et al., 2018). As a consequence, the International Society of Extracellular Vesicles (ISEV) proposes an operational terminology for separated extracellular vesicles, indicating the biochemical and physical properties (Théry et al., 2018). This terminology entails the origin indicated by biofluid, organ or cells the vesicles are separated from, specific markers (including nucleic acids, proteins or lipids) together with vesicle size and density. Our study characterizes small extracellular vesicles separated from the urine, hence termed small urinary extracellular vesicles (suEVs).
To quantitatively assess the proteome of suEVs and its changes throughout living donor transplantation, we employed a simple separation protocol for urinary EVs through differential (ultra‐)centrifugation. The separated suEVs were analysed for their protein content by mass spectrometry using label‐free quantification. The resulting data set represents a novel and unique resource giving insight into biological processes involved throughout renal transplantation. Additionally, correlation with kidney function after transplantation in combination with targeted mass spectrometry for specific markers in a validation cohort provides a first insight regarding the power of this approach for outcome prediction after kidney transplantation.
2. MATERIALS AND METHODS
2.1. Patient recruitment
Patients were recruited through the Departments of General, Visceral and Cancer Surgery, Division of Transplantation Surgery, and the Department II of Internal Medicine, Transplant Center Cologne according to the approved ethics statements 11–310 and 15–032, Ethics committee, University of Cologne, Cologne, Germany. The performed studies are registered at the German Clinical Trials Registry under the ID: DRKS00010534 and DRKS00007704.
2.2. Urine collection and pre‐processing
Urine samples (≥50 ml) were either collected as second morning midstream sample (timepoint A, D and E) or as catheter urine from a fresh collection bag (timepoint B and C) into standard urine sample cups (Sarstedt, Germany). Preservatives and protease inhibitors were added to the final concentrations 6.6 mM NaN3, 2.0 mM EGTA, 1.0 mM PMSF within 90 min after sample collection. Afterwards, cells, debris and large to medium‐sized EVs were separated via high speed centrifugation using a Beckman Avanti Centrifuge with a TLA 16,250 fixed angle rotor in 50 ml centrifugation Tubes (Greiner Bio‐One, Germany) at 17,000 × g, 4°C for 20 min. Supernatant was transferred to fresh 50 ml centrifugation tubes and samples stored at −80°C until further processing.
2.3. Small urinary extracellular vesicle separation
For suEV separation, frozen samples were thawed with subsequent repetition of centrifugation at 17,000 × g, 4°C for 20 min to pellet protein precipitates due to the freeze thaw cycle. Supernatant was afterwards transferred to polycarbonate ultracentrifugation bottles (Beckman Coulter, USA) and suEVs were separated at 200,000 × g, 4°C, 60 min using a Beckman Coulter Optima Ultracentrifuge with a 70Ti fixed angle rotor. Supernatant was discarded and suEV pellets resuspended and pooled in 4°C 20 mM Tris Buffer pH 8.6 with subsequent repetition of the ultracentrifugation step. After discarding the supernatant, the suEV pellet of originally ≥25 ml whole urine was resuspended in 50 μl of Paraformaldehyde and stored at 4°C for electron microscopy or 40 μl 8M Urea Buffer + Protease Inhibitor Mix (Sigma‐Aldrich, USA) and stored at −80°C for protein analysis.
For size exclusion chromatography 10ml of supernatant after 17,000 × g centrifugation were processed using the IZON qEV10/35nm column according to the manufacturers protocol (IZON Science LTD, USA). Resulting fractions 1–4 were pooled and processed using ultracentrifugation at 200,000 × g, 4°C, 60 min for EV separation following the aforementioned protocol.
2.4. Electron microscopy and assessment of size distribution
Samples were processed according to a modified version of previously published protocols (Harford & Bonifacino, 2011). In short: suEV pellet was resuspended in 50 μl 2% PFA (weight/vol) in PBS and incubated at 4°C over night. 5 μl was added to formwar‐coated grids and dried for 20 min at room temperature. Grids were washed seven times in PBS for 2 min at room temperature before incubation in 1% glutharaldehyde (vol/vol) for 5 min. After rinsing in distilled water, grids were contrasted in 1.5% uranylacetate in distilled water (weight/vol) in the dark for 4 min and dried afterwards. Grids were analysed at the TEM109 from Zeiss with a CCD camera by Tröndle. Size distribution was measured in 5 randomly picked images from two individual healthy subjects using the measuring tool in FIJI (Rueden et al., 2017; Schindelin et al., 2012).
2.5. Electron microscopy and immunogold labelling of PCK2
For immunogold staining, 5 μl of resuspended paraformaldehyde fixed suEVs were transferred on Formvar‐carbon coated nickel grids (20 min incubation). After washing with PBS the samples on the grids were permeabilized with 0.05% Tween (in the blocking solution) and blocked with 0.5% BSA for 30 min. suEVs were incubated with the primary rabbit‐anti‐PCK2 antibody (Abcam – ab187145) diluted 1:200 for 2 h at room temperature, washed and labelled with a goat‐anti‐rabbit secondary antibody conjugated to 1.4 nm gold particles diluted 1:200 (Jackson ImmunoResearch ‐ 711‐205‐152) for 1 h at room temperature. After washing nanogold particles were enhanced for 2 min using HQ silver kit (Nanoprobes). Grids were contrasted as described above. Imaging was performed using a Philipps CM 100 TEM and Olympus imaging software ITEM.
2.6. Western blot analysis
suEV pellets resuspened in 8M urea buffer were diluted using 1% Triton X‐100 buffer and 4% SDS sample buffer was added accordingly. Size separation was done using SDS‐PAGE. Samples were blotted onto polyvinylidene difluoride membranes and visualized with enhanced chemiluminescence after incubation of blots with corresponding antibodies (anti‐ALIX antibody BD Bioscience ‐ 611620, anti‐TSG101 antibody Abcam – ab30871).
2.7. Unbiased proteomic analysis
Samples were measured by the CECAD/CMMC Proteomics Core Facility. Peptides were analysed using a Q Exactive Plus tandem mass spectrometer (Thermo Scientific, (Michalski et al., 2011)). A 50 cm column was packed with 1.9 μm C18 beads (Dr Maisch GmbH). The column was used in a column oven at 50°C. Peptides were separated by nano‐liquid chromatography prior to MS measurement. For this purpose, a binary buffer system was used consisting of 0.1% formic acid (buffer A) and 80% acetonitrile with 0.1% formic acid (buffer B). Linear gradients of 150 min duration were used according to the following scheme: initial 4 % B, up to 6 % B over 5 min, up to 23 % B over 120 min, up to 54% B over 7 min. Washing occurred by increasing B to 85% over 6 min, keeping at 85% B for 5 min. Equilibration was performed by decreasing B to 5% over 5 min followed by 2 min equilibration at initial conditions. Peptides were acquired in Top10 DDA with the following parameters for MS1 scans: injection time 20 ms, resolution 70,000, mass range 300–1750 m/z, 3E6 as to AGC target. Peptides selected for higher‐energy collisional dissociation (HCD) fragmentation were analysed with the following parameters: 35,000 resolution, mass range 200–2000 m/z, AGC target of 5E5, max injection time 60 ms. Raw data (.raw) was analysed using MaxQuant V 1.5.0.1 (Cox & Mann, 2008) The search (target decoy strategy) was performed against a 2017 reference human Uniprot database. The default settings were used and the ‘MQLFQ” and ‘Match between runs’ options were both used (Cox et al., 2014). The FDR cut‐offs were FDR 0.01 for all peptide, PSM, and protein identifications. Perseus (v. 1.4.0.1 and v. 1.5.2.4) (Cox & Mann, 2012) was used for further analysis. To correct for multiple testing, a SAM approach was chosen (cut‐off s0 = 1, FDR < 0.001) (Tusher, Tibshirani, & Chu, 2001). Significantly enriched GeneOntology terms were determined by Fisher's Exact Test (FDR = 0.05).
2.8. Correlative analysis
Detected proteins were correlated by linear regression models to the (estimated glomerular filtration rate) eGFR. The eGFR was estimated using the CKD‐EPI Creatinine Equation (Levey et al., 2009). Models were generated using a leave‐one‐out crossvalidation procedure (Stone, 1974). Here, all values but one are used as the training set (n‐1) to calculate a linear model using the R function lm. The left out measurement (the ‘test set’) is calculated using method predict.lm and the difference between predicted and true value is used to calculate the prediction error of the model. The process is repeated according to the number of measured proteins (n) resulting in n linear models. Using these models a mean linear model is generated for each detected protein. From these models candidate proteins were determined for targeted mass spectrometry with criteria described in the main text. Additionally, multiple linear regression was performed using R function lm to fit a linear model including all clinical characteristics, i.e. donor age, donor sex, donor hypertension (yes/no), donor eGFR, recipient age and recipient sex, in addition to PCK2.
2.9. Targeted proteomic analysis
A parallel reaction monitoring (PRM) assay was set up. Analysis was performed using the LC‐MS system as described for untargeted proteomics. Acquisition parameters for targeted proteomics data were as follows: 70 peptides were selected for scheduled PRM analysis (Table S4). MS2 resolution was set at 140k. The AGC target was set at 1,000,000 at a maximum injection time of 100 ms. Peptides were isolated using an isolation window of 1.0 Th and fragmented with a normalized collision energy of 27. The two PCK2 peptides with the amino acid sequence HGVFVGSAMR (charge state 2, m/z = 530.771468, retention time = 31.92 min and EGALDLSGLR, charge state 2, m/z = 515.780013, retention time = 46.77 min were used. A scheduled method was composed using Skyline software environment (Maclean et al., 2010; Pino et al., 2020). A 4 min retention time window was used for acquisition on a 1h gradient that was previously described. Analysis and quantification of targeted proteomics data were performed using skyline. A library of six representative suEV rawfiles from the untargeted analysis was used as to construct the spectral library. Peaks were integrated and quantified using skyline. Normalized intensities were exported and processed. Only peptides within a 5 ppm mass window were considered.
Afterwards, a focused PRM method was set up to allow for fast screening of PCK2. In addition to the peptides HGVFVGSAMR and EGALDLSGLR, the peptide TMYVLPFSMGPVGSPLSR (charge state 2, m/z = 969.99440) was considered to increase assay stability. Furthermore, the peptides from PODXL, PROM1, LG3BP and FN3K listed in Table S4 were monitored to allow for stable normalizations between samples. Samples were analysed on an Orbitrap Exploris 480 coupled to an EasyLC 1200 (both Thermo Scientific). Separation was performed on a 30 cm pico tip column (New Objective) heated to 50°C and packed with 2.7 μm PoroShell 120 (Agilent). The Orbitrap was run in PRM mode with a resolution of 60,000, an AGC target of 100%, a maximum injection time of 110 ms, a quadropol isolation window of 0.7 m/z, and HCD fragmentation set to 30%. All samples were measured by the CECAD/CMMC Proteomics Core Facility. Samples were analysed in a predefined Skyline document for rapid assessment of PCK2 presence and signal normalization.
2.10. Immunohistochemistry
The tissue was fixed in 4% paraformaldehyde in phosphate‐buffered saline (PBS) followed by paraffin embedding for histology. All steps were performed at room temperature (RT) using an autostainer 480S and protocol by Thermo Scientific. In brief, after deparaffinization with xylene and rehydration by graded series of ethanol the slides were washed with TRIS‐NaCl with Tween (Medac‐diagnostica, Germany). Peroxidase was blocked (Ultra Vision Hydrogen Peroxidase Block, Medac‐diagnostica, Germany) and a pretreatment with citrate buffer at pH6 followed (PT module Buffer 1, Thermo Scientific). The primary antibody rabbit‐anti‐PCK2 antibody (Abcam – ab187145) was diluted with antibody diluent (Medac‐diagnostica, Germany) 1:1000 and developed using poly‐HRP‐anti mouse/rabbit IgG (Bright Vision, Medac‐diagnostica, Germany) and DAB away kit (Biocare Medical, Germany). All reagents were used according to manufacturer's protocols. Samples were scanned and analysed using the Aperio Slidescanner, (Leica, Germany) with a 20× Objective, numerical aperture: 0.75 at 20°C without imaging medium and Aperio ImageScope software for the acquisition and QuPath (Bankhead et al., 2017) for analysis.
2.11. Nanoparticle tracking analysis
EVs were analysed by the ZetaView PMX‐120 device (Particle Metrix, Germany). All samples were either diluted 1:1000 or 1:5000 in PBS to a final volume of 1 ml to achieve an ideal particle per frame value of 140 to 200. Every measurement, three times 11 cell positions have been analysed after video capturing 30 frames, each position. Autofocus was set and all samples were measured with a camera sensitivity of 75, using a shutter value of 100 and a constant temperature of 25°C. Videos were analysed by the ZetaView software 8.05.10 SP1 with a minimal area of 5, a maximal area of 1000 and a minimal brightness of 25. The number of completed tracks in NTA measurements was always greater than 1000 (Bachurski et al., 2019).
3. RESULTS
3.1. A fast and widely applicable protocol of differential centrifugation yields a robust separation of suEVs
We established a fast, cost‐efficient and easy protocol for the separation of small urinary extracellular vesicles using differential ultracentrifugation to allow for a potential transfer to clinical routine in the future (Figure 1a). After the addition of preservatives and protease inhibitors, a centrifugation step of 17,000 × g performed at 4°C for 20 min pellets all cells, cellular debris and large to medium EVs. Analysis of the final suEV pellet via transmission electron microscopy (TEM) and Nanoparticle Tracking analysis via ZetaView depicted a wide variety of small EVs with a typical cup‐shaped morphology ranging from suEVs of 30–60 nm to a small fraction of large urinary EVs with a diameter up to 500 nm in diameter (Figure 1b/c and S1A). To validate the stability of suEV proteins in long‐term storage after dissolving the final pellet in 8 M urea buffer, we performed western blot analyses after 1 and 6 months at −80°C for small EV marker proteins and detected no storage‐dependent differences in immunoblot signal (Figure 1d). Additionally, our separation protocol achieved a decrease in the ratio of the ubiquitous urinary protein uromodulin to the suEV protein TSG101 (Figure S1B) and we were able to robustly detect the tetraspanins CD9, CD63 and CD81 – typically present on the surface of EVs – in a flow cytometric bead assay (Figure S1C) of the final suEV pellet. A pilot mass spectrometry analysis confirmed that our protocol separated particles containing marker proteins for all segments of the nephron (Clark et al., 2019) as well as Alix and TSG101 – two proteins typically found in EVs (Figure 1e).
FIGURE 1.
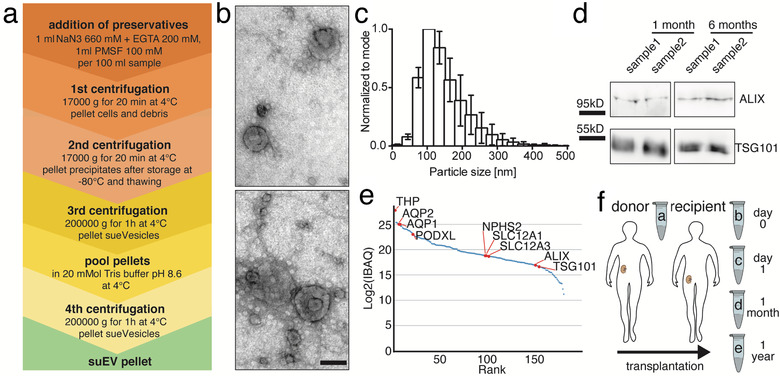
Differential centrifugation separates small urinary extracellular vesicles (suEVs) originating from all segments of the nephron. Schematic overview of the employed protocol of differential centrifugation (a). Representative scanning electron microscopy of suEV pellets depicts EVs of typical exosomal cup shape and size and as well as smaller and bigger particles. Scalebar: 100nm (b). Size distribution of suEVs measured in 4 independent suEV samples of healthy volunteers measured by Nanoparticle Tracking, bin: 30nm, errorbars: SD, (c). Representative western blot analysis for exosomal markers ALIX and TSG101 in two separate suEV pellets after 1 and 6 months of storage in 8M Urea Buffer at −80°C (d). Mass spectrometry analysis suEV pellet is able todetectmarker proteins of all nephron segments. IBAQ: Intensity based absolute quantification (a.u.) (e) Schematic overview of the employed sampling protocol throughout living donor transplantation, a: donor sample; b: recipient sample day 0; c: recipient sample day 1; d: recipient sample 1 month; e: recipient sample 1 year (f)
3.2. Clinical characteristics of donors and recipients of 22 living donor kidney transplantations (screening cohort)
We gathered suEV pellets over the course of 22 living donor transplantations. Figure 1f depicts a brief overview of the study. Urine was collected the day before transplantation from allograft donors (sample A) and their corresponding recipients directly after transplantation (sample B), 1 day after transplantation (sample C), 4 weeks (sample D) and 1 year after transplantation (sample E). Table 1 gives an overview of the clinical characteristics of the donors (Table 1A) and corresponding recipients (Table 1B). Medium age of donors was 51.5 years with roughly two thirds of the first cohort being women. Most donors showed a measured creatinine clearance of more than 100 ml/min/1.73m2 and only three donors suffered from arterial hypertension (Table 1A). Recipients were younger in age, mostly male and showed a lower body weight than the donors on average. More than two thirds underwent preemptive transplantation with the rest having been on dialysis between 2 and 39 months. Underlying renal diseases were almost tripartite between genetic, autoimmune and other causes with only one patient suffering from diabetic nephropathy. 72% of patients received basiliximab as an induction therapy and prednisolone, mycophenolic acid and tacrolimus for maintenance after transplantation (Table 1B). Three of 22 patients did not show a decrease in serum creatinine values of >1 mg/dl within the first 48 h and did not drop below a value of 3 mg/dl within the first week. For this study, these patients were classified as exhibiting ‘initial graft dysfunction’ for the following correlations.
TABLE 1.
Patient characteristics (Cohort 1). Donors (a), Recipients (b)
| Category | Subcategory | Value | ± SD |
|---|---|---|---|
| A (Donors) | |||
| Age (in years) | 51,50 | ±11,09 | |
| Sex | |||
| female (%) | 63,64 | ||
| male (%) | 36,36 | ||
| Weight (kg) | 76,95 | ±13,52 | |
| measured clearance (ml/min/1,73m2) | 126,10 | ±32,21 | |
| estimated GFR (CKD‐EPI 2009) | 92,91 | ±12,64 | |
| Hypertension (%) | 13,64 | ||
| ACE Inhibitors / AT1R Blockers (%) | 13,64 | ||
| B (Recipients) | |||
| Age (in years) | 45,68 | ±15,86 | |
| Sex | |||
| Female (%) | 45,45 | ||
| Male (%) | 54,55 | ||
| Weight (kg) | 69,89 | ±17,03 | |
| Diuresis prior to transplant (%) | 68,18 | ||
| Dialysis prior to transplant (%) | 36,36 | ||
| Haemodyalisis (%) | 100 | ||
| Peritoneal Dialysis (%) | 0 | ||
| time on dialysis in months | 15,29 | ±12,37 | |
| Underlying renal disease | |||
| genetic (%) | 31,82 | ||
| diabetic (%) | 4,55 | ||
| glomerulonephritis (%) | 31,82 | ||
| other (%) | 31,82 | ||
| Retransplant (yes (number) / no) | |||
| yes (2nd transplant) (%) | 9,09 | ||
| no (%) | 90,91 | ||
| CMV status (rec/don) | |||
| neg/neg (%) | 18,18 | ||
| neg/pos (%) | 18,18 | ||
| pos/neg (%) | 13,64 | ||
| pos/pos (%) | 50,00 | ||
| Induction | |||
| ATG (%) | 9,09 | ||
| Basiliximab (%) | 90,91 | ||
| Initial Immunosuppression | |||
| Prednisolone / Cyclosporine A / MMF (%) | 27,27 | ||
| Prednisolone / Tacrolimus / MMF (%) | 72,73 | ||
| Cold ischemia time (in min) | 174,00 | ±26,80 | |
| GFR after 6 months | 49,82 | ±15,58 | |
| GFR after 12 months | 53,23 | ±16,88 | |
| Dialysis within 1 week (Delayed Graft Function) | 0 | ||
| Rejections within 1 year | 3 | ||
| Humoral (%) | 100,00 | ||
| Borderline (%) | 66,66 | ||
| Cellular (%) | 0 |
3.3. Unbiased proteomic analysis of first 22 transplant sets reveals temporal changes of the suEV proteome
Using mass spectrometry, we detected >1700 individual proteins present in ≥50% of samples. The obtained data set is freely available at PRIDE/ProteomExchange (http://www.ebi.ac.uk/pride (Vizcaíno et al., 2016) – accession number: PXD005219). As a technical validation, we performed size exclusion chromatography (SEC) in comparison to our ultracentrifugation (UC) protocol in eight independent control samples of six healthy donors. Successful vesicle separation was validated using nanoparticle tracking analysis and flow cytometric bead assay (Figure S2A & B). The obtained data sets are freely available at PRIDE/ProteomExchange (http://www.ebi.ac.uk/pride (Vizcaíno et al., 2016) – accession number: PXD005219 & PXD021344). The proteomic profile of suEVs separated through SEC shared an overlap of 97% with the proteome of vesicles separated through UC (Figure S3A). Both separation techniques showed a minimal overlap to a proposed list of putative contaminants of separated suEVs (Dhondt et al., 2020) (Figure S3A). Likewise, only 11% of the proteins identified in suEVs for timepoint a (donor sample) were represented by this proposed list of contaminants (Figure S3B). Furthermore, the suEV proteome at timepoint a overlapped considerably with published proteomic analyses of urinary extracellular vesicles separated through different protocols (Dhondt et al., 2020; Oeyen et al., 2018) (Figure S3C). 77% of proteins detected in vesicles obtained through ultrafiltration and size exclusion chromatography were also found in our analysis and 79% of the proteins we detected were present in the vesicle enriched fraction of density‐based fractionated urine. Additionally, we were able to detect putative markers for all segments of the nephron in both suEVs separated by SEC as well as UC (Figure S3 D‐I).
Hierarchical clustering revealed a strong tendency of samples collected at the same timepoint during transplantation to cluster together (Figure 2a). Specifically, the two samples obtained early after transplantation (B and C) share a subcluster clearly distinguishing them from A, D and E samples – i.e. with the proteomic profiles 1 month and 1 year after transplantation showing more resemblance to the profile in the donor sample.
FIGURE 2.
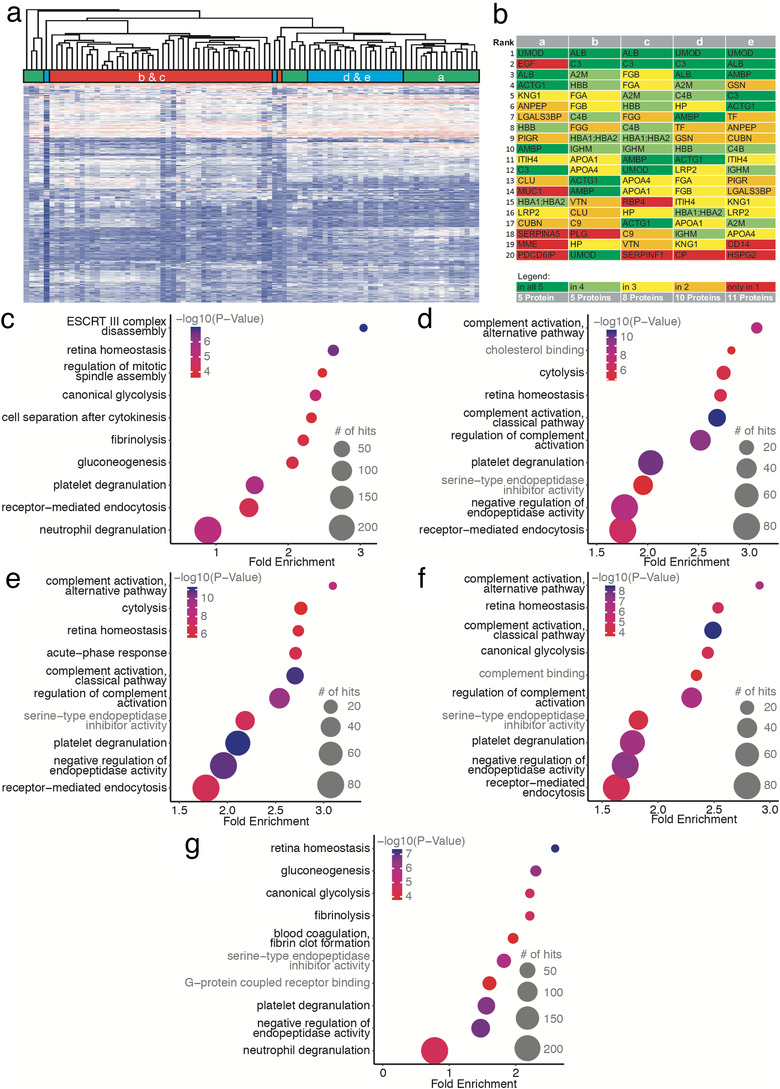
The suEV proteome clusters depending on the collection timepoint throughout living donor kidney transplantation. Hierarchical clustering of all single sample protein measurements of the first cohort (a). Top 20 annotated proteins for each timepoint of sample collection (b). Bubble plots of top 10 significant GO terms for each timepoint of sample collection. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function. (c: Timepoint A – donor sample, D: Timepoint B – day 0, E: Timepoint C – day 1, F: Timepoint D – 1 month, G: Timepoint E – 1 year)
In order to facilitate fast and uncomplicated access to a primary analysis and visualization of the data set, we have designed a browser‐based application allowing the direct search for protein LFQs, their rank within the data set as well as the temporal changes in their abundance in suEVs during living donor kidney transplantation (http://shiny.cecad.uni-koeln.de:3838/suEV/).
When comparing the top 20 proteins per timepoint ranked by label‐free quantification after excluding proteins that could not be annotated, we detected 25% of these proteins to be present among the top 20 at all timepoints ( Figure 2b). These proteins contain the ubiquitous urinary protein uromodulin (UMOD) as well as albumin (ALB), AMBP and Complement Factor 3 (C3) but, interestingly, also cytoplasmatic actin (ACTG). Despite the hierarchical clustering results, the donor samples differ most from all other timepoints regarding the top 20 suEV proteins indicating a shift induced by transplantation itself. Programmed cell death 6‐interacting protein (PDCD6IP), a bonafide marker for small extracellular vesicles, for instance, was only detected among the top 20 proteins in the donor samples. All early timepoints up to 4 weeks after transplantation (B/C/D) show an overrepresentation of proteins typically expected in serum rather than urine including fibrinogen chains alpha and beta and apolipoprotein A1 and A4. Importantly, we were able to detect most of these markers also in vesicles separated through SEC from urine samples of healthy volunteers (Figure S3J).
To further dissect the impact of renal transplantation on the suEV proteome, we performed Gene Ontology (GO) analyses of all detected proteins per timepoint (Figure 2c‐g). We observed a striking overlap among the GO terms of highest significance per timepoint. ‘Platelet degranulation’ and ‘Retina homeostasis’ was detected as highly significant at all timepoints while ‘Receptor‐mediated endocytosis’ was absent, solely, at timepoint E (Figure 2g). ‘Gluconeogenesis’ was not annotated at timepoints b and c and not among the top 20 terms of highest significance at timepoint d while ‘Cytolysis’ showed the exact opposite pattern. GO terms for complement activation and regulation were detected among the top 10 significant GO terms exclusively after transplantation (Figure 2d, e and f) but lacking in the donor sample and decreased in significance 12 months after transplantation (Figure 2c and g and http://shiny.cecad.uni-koeln.de:3838/suEV/).
We also observed a considerable overlap when ranking the top GO terms by enrichment (Figure S4). However, proteins associated with cholesterol uptake and lipoprotein binding were only enriched in the samples early after transplantation (Figure S4B and C) whilst proteins involved in Toll‐like‐receptor 4 and arachidonic acid binding were overrepresented in the donor sample and 1 year after transplantation (Figure S4A and E).
In contrast to the GO analysis performed separately for each timepoint, GO terms for the difference in the suEV proteome content of each timepoint after renal transplantation compared to the initial donor suEV proteome showed much less of an overlap both ranked by enrichment and significance (Figure S5/S6). Interestingly, we detected a significant fold change for proteins associated with the regulation of complement activation one day and 1 month after transplantation (Figure S5B, C and S6A). Four weeks after transplantation, ‘cytolysis’, ‘clearance of apoptotic cells’ and protein as well as membrane trafficking pathways were overrepresented (Figure S6C).
To further elucidate the timepoint‐dependent shift in the suEV proteome during renal transplantation, we analysed the data set for clusters of proteins showing the same temporal signatures. We were able to identify 9 clusters exhibiting specific temporal shifts in their protein abundances (Figure 3). Proteins with rising abundances after transplantation and a following decrease to levels above those detected in the initial donor suEV pellet appear to play a role in Immunity and Blood coagulation (Figure 3c) while HDL‐associated proteins decreased to donor values 1 year after transplantation. Proteins with a persistent increase after transplantation (Figure 3f) were enriched in GO terms of protease homeostasis and ‘Thrombophilia’. An S shaped temporal signature (rise in abundance at timepoints B and C, decrease at timepoint D and normalization at timepoint E) was detected for proteins involved in ribosomal and chromosomal biology as well as ‘Dynein’ activity (Figure 3g). Conversely, decreased abundances right after transplantation were observed for proteins enriched in GO terms for ‘Glycoprotein’ and Secreted proteins (Figure 3e). These terms showed a ‘recovery’ to donor values 1 year after transplantation, while the majority of proteins in the terms ‘Transmembrane’, ‘Glycosidase’ and ‘Signal‐Anchor’ remained slightly decreased in the suEV pellet after 1 year (Figure 3h). Interestingly, stress response proteins were detected within the cluster exhibiting the lowest to non‐detectable changes in the suEV proteome (Figure 3j). Two clusters, one with a pronounced and permanent decrease in abundance right after transplantation and one with a slow continuous decrease in abundance, could not be annotated using standard GO terms (Figure 3b and i).
FIGURE 3.

The suEV proteome underlies specific temporal changes indicative of specific biological processes in allograft adaptation. Hierarchical clustering of mean relative protein abundance per timepoint (a). Spaghetti plots depicting suEV protein clusters with specific temporal changes throughout renal transplantation with enriched GO keywords bargraphs below each cluster. Colour code indicates cluster branches on the heatmap depicted in 3a (b‐j). a: donor sample; b: recipient sample day 0; c: recipient sample day 1; d: recipient sample 1 month; e: recipient sample 1 year
3.4. Identification of potential biomarkers predicting kidney function 6 and 12 months after renal transplantation using a leave‐one‐out cross validation model
To assess the potential of the acquired proteomics data for identifying novel biomarkers of the outcome of renal transplantation, we performed a correlative analysis with the estimated glomerular filtration rate (eGFR) 6 and 12 months after renal transplantation using a leave‐one‐out cross validation model (for a description of the approach see Movie S1). We investigated the predictive value of the foldchange of protein abundances between timepoints A to B and A to C as well as the single protein intensities at timepoints A and C normalized to the mean protein intensity. The resulting candidate protein lists were filtered for (I) the 10 candidates exhibiting the lowest error (lowest RSS squared) and highest stability (lowest stdev RSS squared) when correlated to the estimated GFR after 12 month and, (II) the 20 candidates exhibiting the lowest error correlating both with the estimated GFR 6 and 12 months after transplantation (Table S2).
To generate a candidate list of markers for further validation we complemented this selection by 5 proteins specifically not detected and 10 proteins with significantly higher expression in patients with initial graft dysfunction at timepoints A and C (Table S2). These proteins, the 10 candidates described under (I) and the overlap of proteins found both predictive for the eGFR after 6 and 12 months described under (II) resulted in a target list of 64 proteins (Table 2).
TABLE 2.
Candidate Biomarker Protein List
| List origin | Protein names |
|---|---|
| Correlating with 12 Month GFR with lowest RSSsquare mean and lowest SD | Galactokinase |
| BH3‐interacting domain death agonist;BH3‐interacting domain death agonist p15;BH3‐interacting domain death agonist p13;BH3‐interacting domain death agonist p11 | |
| Pancreatic secretory granule membrane major glycoprotein GP2 | |
| Prominin‐1 | |
| V‐type proton ATPase subunit S1 | |
| Myocilin | |
| Eukaryotic translation initiation factor 3 subunit C;Eukaryotic translation initiation factor 3 subunit C‐like protein | |
| 40S ribosomal protein SA | |
| Complement C1q subcomponent subunit B | |
| Tetranectin | |
| 2‐deoxynucleoside 5‐phosphate N‐hydrolase 1 | |
| Charged multivesicular body protein 2a | |
| DnaJ homolog subfamily A member 2 | |
| Cadherin‐16 | |
| Monocyte differentiation antigen CD14;Monocyte differentiation antigen CD14, urinary form;Monocyte differentiation antigen CD14, membrane‐bound form;Monocyte differentiation antigen CD14 | |
| Collagen alpha‐2(IV) chain;Canstatin | |
| Glutathione S‐transferase A2;Glutathione S‐transferase | |
| Villin‐1 | |
| Excitatory amino acid transporter 3 | |
| 26S protease regulatory subunit 8 | |
| Spectrin beta chain, non‐erythrocytic 1 | |
| Galectin‐3‐binding protein | |
| Receptor‐type tyrosine‐protein phosphatase eta | |
| Importin subunit beta‐1 | |
| Protein phosphatase 1 regulatory subunit 7 | |
| Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | |
| Decorin | |
| Ragulator complex protein LAMTOR1 | |
| Annexin;Annexin A4 | |
| Alpha‐(1,3)‐fucosyltransferase | |
| Protocadherin Fat 4 | |
| Sortilin‐related receptor | |
| SLIT and NTRK‐like protein 1 | |
| Protein amnionless | |
| Complement C1r subcomponent‐like protein | |
| Peflin | |
| Correlating with both 6 Month and 12 Month GFR with lowest RSSsquare | IgGFc‐binding protein |
| ATP‐dependent RNA helicase DDX3X;ATP‐dependent RNA helicase DDX3Y | |
| Cartilage oligomeric matrix protein | |
| BH3‐interacting domain death agonist;BH3‐interacting domain death agonist p15;BH3‐interacting domain death agonist p13;BH3‐interacting domain death agonist p11 | |
| DnaJ homolog subfamily A member 2 | |
| Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | |
| Haptoglobin‐related protein | |
| Eukaryotic translation initiation factor 3 subunit C;Eukaryotic translation initiation factor 3 subunit C‐like protein | |
| 40S ribosomal protein SA | |
| Syntenin‐1 | |
| Fructosamine‐3‐kinase | |
| Top5‐not detectable in early graft dysfunction | Transcription elongation factor B polypeptide 1 |
| Ras‐related protein Rab‐27B | |
| Sodium‐dependent phosphate transport protein 2A | |
| Calbindin | |
| Copine‐8 | |
| Top10‐higher abundance in non early graft dysfunction | Glutamate carboxypeptidase 2 |
| CD177 antigen | |
| Protein‐glutamine gamma‐glutamyltransferase 4 | |
| Arylsulfatase F | |
| Semenogelin‐1;Alpha‐inhibin‐92;Alpha‐inhibin‐31;Seminal basic protein | |
| 40S ribosomal protein S17‐like;40S ribosomal protein S17 | |
| 60S ribosomal protein L23a | |
| Histone H2B type 1‐D;Histone H2B | |
| Complement component C6 | |
| Podocalyxin | |
| Extracellular vesicle markers | TSG101 |
| ALIX | |
| CD81 | |
| CD40, Tumor necrosis factor receptor superfamily member 5 | |
| Annexin A5 |
3.5. Targeted mass spectrometry in suEVs of an independent validation cohort identifies PCK2 as a predictor of long‐term transplant function
To check validity of these 64 suEV candidate markers we established targeted mass spectrometry assays for these proteins. These assays were then used to measure suEV pellets collected in an independent validation cohort throughout 22 additional living donor transplantations. Table 3 gives an overview of the patient characteristics of this validation cohort.
TABLE 3.
Patient characteristics (Cohort 2). Donors (a), Recipients (b)
| Category | Subcategory | Value | ± SD |
|---|---|---|---|
| A (Donors) | |||
| Age (in years) | 53,77 | ±10,57 | |
| Sex | |||
| female (%) | 50,00 | ||
| male (%) | 50,00 | ||
| Weight (kg) | 79,53 | ±14,27 | |
| measured clearance (ml/min/1,73m2) | 116,00 | ±22,90 | |
| estimated GFR (CKD‐EPI 2009) | 94,73 | ±16,82 | |
| Hypertension (%) | 31,82 | ||
| ACE Inhibitors / AT1R Blockers (%) | 31,82 | ||
| B (Recipients) | |||
| Age (in years) | 46,05 | ±15,06 | |
| Sex | |||
| female (%) | 45,45 | ||
| male (%) | 54,55 | ||
| Weight (kg) | 81,21 | ±15,02 | |
| Diuresis prior to transplant (%) | 77,27 | ||
| Dialysis prior to transplant (%) | 50,00 | ||
| Haemodyalisis (%) | 90,90 | ||
| Peritoneal Dialysis (%) | 9,09 | ||
| time on dialysis in months | 16,24 | ±43,90 | |
| Underlying renal disease | |||
| genetic (%) | 31,82 | ||
| diabetic (%) | 0,00 | ||
| glomerulonephritis (%) | 18,18 | ||
| other (%) | 50,00 | ||
| Retransplant (yes (number) / no) | |||
| yes (2nd transplant) (%) | 9,09 | ||
| no (%) | 90,91 | ||
| CMV status (rec/don) | |||
| neg/neg (%) | 18,18 | ||
| neg/pos (%) | 18,18 | ||
| pos/neg (%) | 13,64 | ||
| pos/pos (%) | 50,00 | ||
| Induction | |||
| ATG (%) | 0,00 | ||
| Basiliximab (%) | 100,00 | ||
| Initial Immunosuppression | |||
| Prednisolone / Cyclosporine A / MMF (%) | 22,73 | ||
| Prednisolone / Tacrolimus / MMF (%) | 72,73 | ||
| Cold ischemia time (in min) | 172,70 | ±23,62 | |
| GFR after 6 months | 53,33 | ±16,18 | |
| GFR after 12 months | 59,16 | ±20,50 | |
| Dialysis within 1 week (Delayed Graft Function) | 0 | ||
| Rejections within 1 year | 8 | ||
| Humoral (%) | 75,00 | ||
| Borderline (%) | 0 | ||
| Cellular (%) | 37,50 |
Renal function as indicated by both measured creatinine clearance (116.00 ml/min/1.73m2) and estimated GFR (94.73 ml/min/1,73m2) was equal to the first cohort with 7 patients being hypertensive and receiving corresponding medication (Table 3A). Recipients were heavier than those of the initial cohort but otherwise closely resembled the latter, also when examining the renal function 6 and 12 months after transplantation (Table 3B).
We again performed both our correlative analysis and filter algorithm to identify proteins with a robust correlation to renal function 6 and 12 months after transplantation (Table S3). This validation experiment lead to the confirmation of phosphoenolpyruvate carboxykinase 2 (PCK2/PEPCK) as an early marker predicting transplant outcome after 1 year (Figure 4). PCK2 abundance in suEVs 1 day after transplantation (timepoint c) showed a robust positive correlation with the estimated GFR 6 and 12 months after renal transplantation in our first cohort (Figure 4a and S6). To examine the impact of clinical variables on the correlation of PCK2 with outcome we performed a multiple linear regression analysis including both donor and recipient characteristics. The models with the lowest P‐value (Figure 4b, S7 A/B) confirm PCK2 abundance to be an independent predictor of eGFR at 6 and 12 months. Additionally, two clinical parameters showed a potential predictive value as donor eGFR correlated significantly with the 6 month GFR and showed borderline significance regarding eGFR after 12 months. Recipient sex showed a similar tendency (without reaching a P‐value ≤ 0.05). Separate correlation of PCK2 by recipient sex with the eGFR at 12 months showed no sex‐specific differences (Figure S8A and B). In line with this finding, the abundance of PCK2 in suEVs 1 day after transplantation did not show a significant difference between male and female recipients (Figure 4c). Besides, donor eGFR (Figure 4d) and the recorded cold ischemia time (Figure 4e) did not correlate with PCK2 levels showing its potential as an independent indicator. Importantly, PCK2 abundance at timepoint c was confirmed to correlate with the estimated GFR 12 months after transplantation in the validation cohort using targeted mass spectrometry (Figure 4f). We detected no predictive value for PCK2 abundances in suEVs of the donor sample or the foldchanges directly after and 1 day after transplantation (Figure S8C‐G).
FIGURE 4.
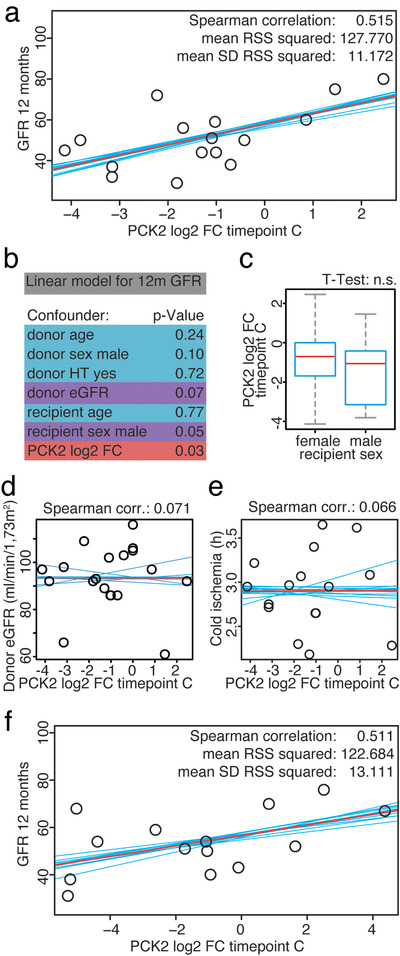
PCK2 abundance in suEVs on day one after transplantation correlates with estimated GFR 12 months after transplantation. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint C to GFR 12 months after transplantation measured in the initial transplant cohort (a). Significance levels of donor and recipient characteristics together with PCK2 intensity foldchange to mean at timepoint c correlated to GFR 12 months after transplantation through linear regression (b). Box plot indicating PCK2 intensity foldchange to mean at timepoint c compared between recipient sex (c). Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint C to donor eGFR (d) and cold ischemia (e) measured in the initial cohort. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint C to GFR 12 months after transplantation measured in the validation cohort (f). Blue: individual linear regression models; Red: merged linear regression model for all samples
3.6. suEV abundance of PCK2 rises initially during renal transplantation but does not reflect renal tissue PCK2 levels
In the initial unbiased proteomic measurement, PCK2 was detected at rank 1515 of 1728 with a mean label‐free quantification (LFQ) intensity of 21.88 at timepoint A (see (http://shiny.cecad.uni-koeln.de:3838/suEV/). Consequently, it appeared important to prove its localization within suEVs. Using the aforementioned eight independent control samples from six healthy donors separated by size exclusion chromatography in comparison to our ultracentrifugation protocol we performed targeted mass spectrometry for PCK2. We were able to robustly detect PCK2 by at least two different peptide sequences in all samples, both after ultracentrifugation and size exclusion chromatography (Figure 5a and b). Additionally, using immunogold labelling and transmission electron microscopy, we detected PCK2 within a subset of suEVs of 100—400 nm diameter (Figure 5c) separated through our ultracentrifugation protocol. Vesicles were permeabilized for 30 min (see Materials and methods) allowing antibody penetration into the vesicular lumen for successful staining. No staining was detected within non‐permeabilized vesicles (Figure S9A).
FIGURE 5.
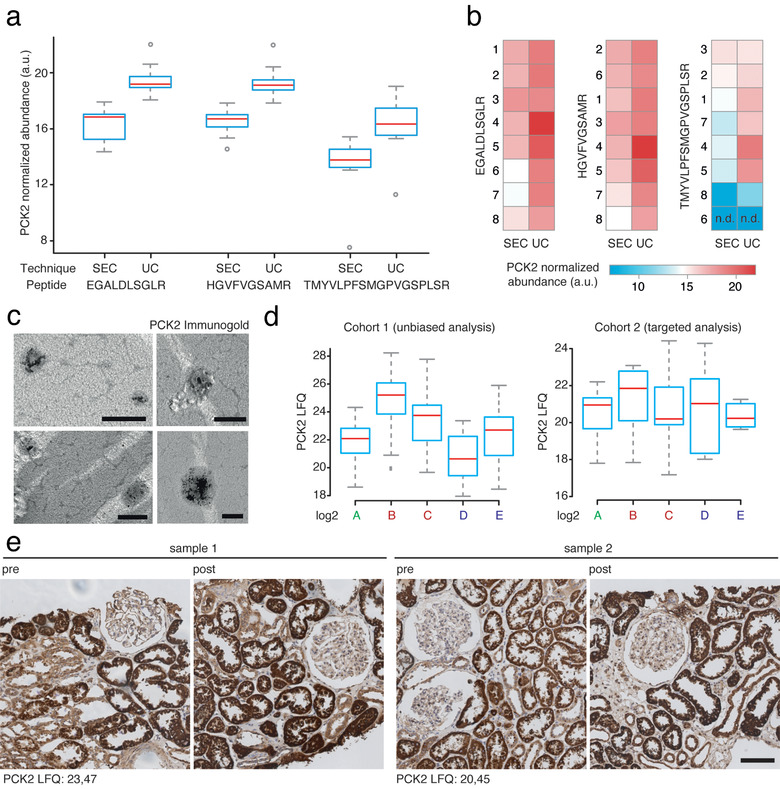
PCK2 localizes to suEVs, increases during the initial stages of transplantation and does not correlate to tissue PCK2 levels after reperfusion. PCK2 abundance (shown for three different PRM assays / peptides) in suEVs separated through size exclusion chromatography (SEC) in comparison to ultracentrifugation (UC) depicted as box plot diagrams with mean (red line), upper and lower quartile (blue box), 95% confidence interval (grey whiskers) and outliers (grey circles) (a) or heatmap exemplifying single sample values, numbers represent individual samples; n.d.: not detected (b). Electron microscopy depicting permeabilized small urinary extracellular vesicles harbouring immunogold labelled PCK2, scalebar 200nm (c). Box plot diagrams depicting suEV PCK2 abundance at different timepoints throughout living donor kidney transplantation in the initial and validation cohort. Mean (red line), upper and lower quartile (blue box) 95% confidence interval (grey whiskers) (d). Immunohistochemistry staining for PCK2 in renal biopsies taken from two donors / patients (sample 1 / 2) included in the suEV analysis before explantation (pre) and after reperfusion (post). suEV PCK2 LFQs at timepoint C indicated in the lower left corner, scalebar 100μm (e). a: donor sample; b: recipient sample day 0; c: recipient sample day 1; d: recipient sample 1 month; e: recipient sample 1 year
PCK2 shows a rising abundance in suEVs until 4 weeks after transplantation, with a decrease towards values measured in the donor sample 1 year after transplantation (Figure 5d). To examine, whether the changes in suEV levels of PCK2 were due to differences in renal expression, we performed immunohistochemistry for PCK2 in renal biopsies taken during living donor kidney transplantation before explantation (pre) and right after reperfusion (post) from a subset of patients also included in the suEV analysis. We detected a strong signal predominantly in proximal tubules, as described before, but no overt changes after reperfusion or evidence for a correlation to the measured LFQs in the suEV proteome of the donor samples (Figure 5e).
4. DISCUSSION
Despite the acceptance of kidney transplantation as the preferred renal replacement therapy, the precise effects cold ischemia time and reperfusion exert on renal cells are incompletely understood. Mouse models can only partially shed light on these mechanisms, as their organism differs greatly concerning their immune systems, one of the key players involved in transplant biology (Mestas & Hughes, 2004). Nevertheless, a better understanding could impact future therapeutic approaches to renal transplantation and lead to a better overall outcome both of organ function in the early days and long‐term organ survival. This study represents the first in‐human analysis of these temporal proteomic changes deduced from suEVs and allows novel insights into the molecular processes taking place in the early days and weeks after transplantation as well as those underlying the long‐term recovery.
Former studies in donors of renal allografts investigated the correlation between EVs carrying specific markers to the tissue characteristics obtained through renal biopsy during transplantation (Turco et al., 2016). While an impressive number of patients was investigated and valuable information was obtained, a deeper understanding was limited due to the use of a specific antibody panel. Whole urine proteomics have been employed in the setting of renal transplantation in the past (Jahnukainen et al., 2006; Kim, Page, & Knechtle, 2014; Suhail, 2014). Yet, it is important to note, that, apart from a ‘core urinary proteome’, free urinary proteins showed a fairly high inter‐ and intra‐individual variability (Nagaraj & Mann, 2011). Employing an unbiased proteomic analysis, we were able to detect >1,700 proteins enriched through our separation protocol.
The suEV proteome showed a strong tendency to form timepoint‐dependent subclusters. This finding confirms technical reproducibility in a clinical setting, as the individual vesicle separation was performed by various trained members of our group, and the storage time of the initially collected urine samples varied from sample to sample. Ancillary, we were able to reproduce the finding, that channel proteins AQP1 and AQP2 were shown to be decreased in urinary EVs after ischemia reperfusion injury in a rat model (Sonoda et al., 2009) and in human patients undergoing renal transplantation (Oshikawa‐Hori, Yokota‐Ikeda, Sonoda, & Ikeda, 2019) in our unbiased proteomic approach.
A deeper understanding of the response to transplantation was delineated through the investigation of both single protein abundances and overall processes identified through gene ontology terms. When considering the top 20 proteins of highest abundance, five proteins were detectable to a high degree at all timepoints. While UMOD, ALB, C3 could be interpreted as a sign for co‐precipitation of free proteins, it is worth noting that all three proteins were also present in proteomic analyses of urinary extracellular vesicles separated through protocols involving a sucrose cushion or immune affinity (Goetzl, Schwartz, Abner, Jicha, & Kapogiannis, 2018; Musante et al., 2013; Raj, Fiume, Capasso, & Pocsfalvi, 2012). Interestingly, PDCD6IP or ALIX, which, as a bona fide marker for the late endosome and the vesicles derived from it, is an integral interactor with the ESCRT machinery in vesicle formation, was only present among the proteins of highest abundance in suEVs before transplantation. This also holds true when broadening the filter to the top 50 proteins, indicating a decrease in ALIX‐positive vesicles with potential implications for vesicle formation and late endosome maturation throughout renal transplantation. Further indication for an alteration in the release of vesicles is given by the reduction in TSG101 right after transplantation as TSG101 was proposed as a surrogate marker for urinary vesicle number in a recent study (Koritzinsky et al., 2019).
The increase of serum‐associated proteins at timepoints B‐D can reflect differences in EV packing as well as a decreased intercellular barrier upon reperfusion resulting in the crossing of plasma extracellular vesicles into the urinary space (Gonzales et al., 2009, Pienimaeki‐Roemer et al., 2015). Yet, the fact that these proteins are still detected as late as 4 weeks after transplantation (timepoint d), speaks for them rather to be derived from cells of the nephron. This is particularly interesting for the detected complement factors as our findings point towards a source within the kidney and complement has shown to be involved in many renal pathologies and transplant dysfunction (Biglarnia, Huber‐Lang, Mohlin, Ekdahl, & Nilsson, 2018; Ricklin, Mastellos, Reis, & Lambris, 2017).
Regarding the GO term analyses, the term ‘platelet degranulation’ was detected as highly significant at all timepoints (Figure 2c‐g). As the common denominator of most proteins associated to the term is their incorporation in platelet‐derived extracellular vesicles (Pienimaeki‐Roemer et al., 2015), we believe that there is constant passage of these vesicles into the urinary space either through the glomerular filtration barrier, leaky intercellular space or active transcellular transport. A potential mechanism for this was proposed by Lenzini et al., who reported that fluctuating stiffness and relaxation of polymeric matrixes allows EVs larger than the actual mesh size to migrate through without the necessity of matrix degradation (Lenzini, Bargi, Chung, & Shin, 2020). This process seems particularly likely at the glomerular filtration barrier, as the capillaries exhibit a constant pulsatile distension and contraction with the blood flow. Future investigations should address the effects of platelet derived EVs on renal cell populations as well as whether the depletion of such vesicles has direct effects on the kidney.
The surge in proteins associated with both complement activation, its regulation and cytolysis most likely reflects the initial immune response triggered by both ischemia‐reperfusion and exposure to the allograft. This allows for the hypothesis that an early blockade of these responses – probably focusing on the complement system – may be beneficial (Nauser, Farrar, & Sacks, 2017; Zuber et al., 2019). Furthermore, the analysis of suEVs could become a diagnostic strategy to monitor complement activation in the setting of renal transplantation.
Toll‐like receptor 4 (TLR4) signalling has long been investigated as a mediator for renal fibrosis after transplantation and over the course of various nephropathies (Bergler et al., 2012; Pulskens et al., 2010; Pushpakumar et al., 2017; Souza et al., 2015; Zhao, Perez, Lu, George, & Ma, 2014; Zmonarski, Banasik, Madziarska, Mazanowska, & Krajewska, 2019). In contrast to that, derivatives of arachidonic acids and their mimics were shown to exhibit anti‐fibrotic effects and polymorphisms in their CYP‐dependent metabolism to influence susceptibility to renal injury (Gervasini et al., 2015; Hye Khan, Stavniichuk, Sattar, Falck, & Imig, 2019; Yeboah et al., 2016). The overrepresentation of these processes in the donor sample and the 1‐year sample does not necessarily reflect two counter mechanisms. In fact, EVs activating TLR4 baring cells mitigated the expression of genes leading to the resolve of inflammatory processes, in comparison to activation through LPS (Manček‐Keber et al., 2015). As these processes are highly enriched in suEVs in the donor sample and 1 year after transplantation, this could indicate an organ specific anti‐inflammatory environment, moderated through extracellular vesicles.
GO term analysis of the differences in the suEV proteome of each timepoint to the healthy (donor) state revealed the vast and quickly adapting changes in EV assembly and cargo. Again, the significant enrichment of complement pathway‐activating proteins is not surprising and confirms published data based on tissue analyses (Nauser et al., 2017). Conversely, the enrichment of proteins associated to ‘multivesicular body assembly’ and ‘viral (vesicle) budding’ point to a heightened activity of EV production, potentially triggered through the ischemia reperfusion injury, which is also reflected by a continuous rise in TSG101 protein abundance after an initial dip following transplantation (http://shiny.cecad.uni-koeln.de:3838/suEV/). Induction of vesicle production has been described both in cell culture damage systems as well as cardiac ischemia in humans and mice and has been associated with the recruitment of monocytes to the damaged tissue (Akbar et al., 2017, Collino et al., 2019). Given the fact that EV administration has been shown to facilitate renal recovery after ischemia‐reperfusion damage (Dominguez et al., 2017), our finding could point towards an intrinsic counter mechanism.
Besides being a resource for the biological temporal changes after transplantation, we hypothesized that the suEV proteome could be a future source for the identification of biomarkers. A set of proteins detected in EVs early after transplantation correlated clearly with eGFR 6 and 12 months later. Since the robust identification of marker proteins cannot be achieved using untargeted proteomics in a limited set of patients, we aimed for the validation in a separate cohort employing targeted mass spectrometry. This validation strongly narrowed down the list of potential markers. Most recognizable, the markers derived from the patients exhibiting initial graft dysfunction proved of little stability which is probably due to the low number of patients in this group. PCK2 abundance in the suEV pellet one day after transplantation, however, was confirmed to correlate with eGFR 1 year after transplantation (Figure 4a and b). This correlation was not detected when measured at any other timepoint or investigating the difference in abundance between timepoints (Figure S6). Strikingly, PCK2 abundance did not correlate with the initial donor clearance or cold ischemia time during transplantation (Figure 4c and d).
This finding points towards unknown mechanisms in transplant adaptation or resistance to ischemia‐reperfusion, revealed by the investigation of the suEV proteome. Mechanistically, PCK2 has been shown to react to states of metabolic starvation, particularly in cancer. In glucose deprivation, PCK2 upregulation with ensuing gluconeogenesis stabilizes nutrient supply and tumour growth with growth impairment upon PCK2 inhibition (Leithner et al., 2015, Montal et al., 2019). This process may well take place also in the renal allograft, as PCK2 is highly expressed in the proximal tubule and the tubular compartment is the prime target of ischemic and acute damage in the kidney (Späth et al., 2019).
An increase in PCK2 levels could potentially counteract the observed decrease in gluconeogenesis (Figure 2c‐g) at timepoints b, c and d as indicated by our GO analysis. It is possible that this PCK2 induction is activated after reperfusion, but takes time to reach a measurable peak in tissue analyses. This would explain why we did not detect evidence for a correlation of tissue PCK2 levels right after reperfusion in immunohistochemistry with suEV PCK2 levels one day after transplantation. Albeit, there is ongoing debate as to whether suEV protein content does correlate with tissue protein content at all (Blijdorp & Hoorn, 2019, Sabaratnam et al., 2019). PCK2 is a mitochondrial protein. An alternative explanation of the increased PCK2 content in suEVs without a change in the cellular content may be an activation of mitochondrial activity in individuals protected from damage and a consecutive rise in the release of this protein. However, this remains speculation at this point and will require further cell biological studies.
The presented study entails limitations due to the separation protocol and the patient cohort chosen. Due to the protocol applied, we cannot draw conclusions of the actual vesicle type contributing to the protein signals. To this day, there is no agreed gold standard separation protocol and most studies still use the widely employed protocol of differential (ultra‐)centrifugation (Merchant et al., 2017, Théry et al., 2018). We, as well, chose a modification of these protocols, due to several reasons. Differential ultracentrifugation enables the use of higher volumes of the initial sample and, therefore, the separation of more EVs (Musante et al., 2013). Differential ultracentrifugation fails to separate a pure vesicle fraction, yet filtration and precipitation techniques were shown to unspecifically concentrate free urinary proteins as well while often excluding specific EV fractions from the separated pellet. The fact that mass spectrometry of suEVs separated by SEC nicely recapitulates the data we obtained in vesicles separated by differential ultracentrifugation confirms the validity of this approach. More laborious techniques such as sucrose cushion gradient separation, however, would make a translation to the clinical setting extremely difficult.
We, therefore, compared our data set to published proteomic analyses of urinary vesicles obtained through different separation techniques, supposedly leading to a higher purity of vesicles (Dhondt et al., 2020, Oeyen et al., 2018) (Figure S2). The striking overlap of our data set with these analyses and the low amount of proposed putative contaminants speaks for its reliability and biological relevance. The remaining discrepancies, in our view, result from several differences in these protocols. Oeyen et al. analysed only two size exclusion chromatography fractions by mass spectrometry following a very strict censoring of all other fractions, potentially losing vesicles we separate more efficiently. The urinary samples in the study of Dhondt et al. partially originated from patients following digital rectal examination with prostate stimulation. This comparison nicely illustrates the constant discussion and need for further standardization not only of separation techniques but also of the ensuing proteomic analyses. This is further underlined by the fact that PCK2, a protein we robustly detect in our samples ‐ both prepared by UC as well as SEC ‐ and locate directly to EVs using immunogold labelling in electron microscopy, was not detected by either one of the compared studies. Likewise, several groups have proposed an (unspecific) protein coating of EVs that is co‐separated with all available techniques and needs further in‐depth analyses (De Wever & Hendrix, 2019, Hadjidemetriou et al., 2015).
The fact, that patients undergoing living donor transplantation were chosen, leads to a cohort with very short cold ischemia periods and excellent outcome, that is not necessarily comparable to deceased donor transplantations. Nevertheless, this allowed us to include samples of a well‐ standardized cohort with sufficient data available for the donor's history and access to representative donor samples. Whilst it will be extremely interesting in the future to extend this approach to deceased donor transplantation, this approach was crucial to obtain a first and complete atlas of the suEV proteome from a healthy donor kidney over periprocedural samples to 1 year after transplantation. Due to the study design, we cannot completely rule out, that the proteomic differences observed between donors and recipients partially result from the change of a two kidney status (donor) to a single kidney status (recipient). However, in the hierarchical clustering analysis of the proteomic data set of the first cohort, we detect the suEV proteome after 1 month and 1 year (d and e samples) to share a subcluster with the donor samples (a). Consequently, the early timepoints (like timepoint c at which we analyse PCK2 abundance) are primarily influenced by the process of kidney transplantation itself in our view.
Whilst our study shows the potential of suEVs as a source of biomarkers, the small number of patients (22 in the screening cohort, 22 in the validation cohort) does not allow for definite statement on the predictive capacity of suEV PCK2. Therefore, a potential use of this marker will depend on future larger trials. Our data set can serve as the first step to inform the design of such studies. Since PCK2 was detected in vesicles separated through size exclusion chromatography and located to suEVs between 100–400nm in size in immuno gold stainings, future analyses can employ separation methods such as SEC to open the investigation of PCK2 in suEVs to a broad range of groups worldwide. Likewise, the timepoint of measurement provides the possibility to validate the predictive value in deceased donor transplantation.
In conclusion, using differential (ultra‐) centrifugation we assembled and analysed a concise atlas reflecting the temporal changes in the proteome of small urinary extracellular vesicles throughout the course of living donor kidney transplantation. We detected specific temporal profiles, e.g. a surge in complement‐associated proteins closely after transplantation reflective of early immunologic effects. Furthermore, our approach revealed the abundance of phosphoenol pyruvate carboxykinase (PCK2) in the suEV proteome one day after transplantation to have a predictive value for overall kidney function 1 year after transplantation. This study underlines the potential of analysing suEVs for monitoring immune response activity and for biomarker discovery using a fast and easy protocol for their separation.
CONFLICTS OF INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
The research was designed by F.B., M.R., A.B., D.S. and R.‐U.M. F.B., M.R., K.B., C.K., I.P., D.Bu., D.Ba., R.W., M.R.S., H.G., J.W.L., O.K., V.G.P. P.A. and C.E.K. conducted the experiments and acquired the data. F.B., M.R., K.B., C.K., D.B., C.E.K. and R.‐U.M. analyzed the data. F.B. and R.‐U.M. wrote the manuscript. F.B., M.R., V.G.P., O.K., D.Ba., M.H., T.B.H., B.S., T.B., D.S., C.E.K. & R.‐U.M. revised the final version. All authors approved the final version of the paper.
DATA AND MATERIALS AVAILABILITY
The obtained data sets are freely available at PRIDE/ProteomExchange (http://www.ebi.ac.uk/pride (Vizcaíno et al., 2016) ‐ Project name: Longitudinal study of the small urinary extracellular vesicle proteome during living donor kidney transplantation; Project accession: PXD005219 & PXD021344; Reviewer account details: Username: reviewer76822@ebi.ac.uk, Password: 3FR3kDp8. For direct and visual access see http://shiny.cecad.uni-koeln.de:3838/suEV/.
Supporting information
Fig.S1: Differential centrifugation leads to a decrease in Uromodulin to TSG ratio and separates small tetraspanin‐containing vesicles. Size distribution of suEVs measured in SEM of 2 independent samples of healthy volunteers (a). Western Blot Analysis and Densitometry of Uromodulin and TSG101 in 2 independent samples of healthy volunteers (b). Flow cytometric bead assay for tetraspanins CD9, CD63, CD81 and Phosphatidylserine (PS) of 4 independent samples of healthy volunteers (c)
Fig.S2: Validation of suEV separation by size exclusion chromatography compared to ultracentrifugation. Size distribution of suEVs measured in 3 independent suEV samples of healthy volunteers separated by ultracentrifugation (UC) or size exclusion chromatography (SEC) measured by Nanoparticle Tracking, bin: 30nm, errorbars: SD, (a). Flow cytometric bead assay for tetraspanins CD9, CD63 and CD81 of 3 independent samples of healthy volunteers separated by ultracentrifugation (UC) or size exclusion chromatography (SEC) (b)
Fig.S3: Comparison of suEV proteome separated through SEC and UC and overlap of donor sample (timepoint a) to published data. Venn diagram of overlap between suEV proteome of vesicles separated by size exclusion chromatography (SEC) or ultracentrifugation (UC) with proposed putative urinary vesicle contaminants (a). Venn diagram of overlap between suEV proteome at timepoint a with proposed putative urinary vesicle contaminants (Dhondt et al., 2020) (b). Venn diagram of overlap between suEV proteome at timepoint a with published data (Dhondt et al., 2020, Oeyen et al., 2018) (c). Scatter plot depicting the mean LFQ to protein rank of proteins detected in 50% of vesicle samples separated from urine samples of healthy volunteers by SEC or UC. Protein‐Rank ranging from 1–510 in SEC and 1–1248 in UC samples. Plots depicting marker proteins for extracellular vesicles (d), podocytes (e), proximal tubulus (f), distal tubulus (g), collecting duct (h), connecting tubulus (I) and the top 20 proteins detected at all timepoints in the transplant suEV cohort (J)
Fig.S4: Enriched GO term analysis of absolute protein quantification per timepoint. Bubble plots of top 10 GO terms enriched for each timepoint of sample collection. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint A, B: Timepoint B, C: Timepoint C, D: Timepoint D, E: Timepoint E)
Fig.S5: Significant GO term analysis of protein fold changes compared to initial donor sample. Top 10 significant GO terms for each timepoint after transplantation compared to the initial donor samples. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint B vs A, B: Timepoint C vs A, C: Timepoint D vs A, D: Timepoint E vs A)
Fig.S6: Enriched GO term analysis of protein fold changes compared to initial donor sample. Top 10 GO terms enriched for each timepoint after transplantation compared to the initial donor samples. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint B vs A, B: Timepoint C vs A, C: Timepoint D vs A, D: Timepoint E vs A)
Fig.S7: Linear regression analysis correlating clinical parameters and PCK2 abundance to long term transplant outcome. Regression tables for multiple linear regression with (a) 12 month eGFR, or (b) 6 month eGFR as the dependent variable. OLS = ordinary least squares, Adj. R² = adjusted R², Est. = Estimate, S.E. = Standard error, t val. = t value, and p = p‐value. Predictors with a p‐value ≤0.05 were marked in red
Fig.S8: PCK2 abundance in suEVs at other timepoints does not correlate with estimated GFR 12 months after transplantation. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint C to GFR 12 months after transplantation measured in the initial transplant cohort split by female (a) and male (b) recipeitns. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint A (c) and B (d) and foldchanges between timepoints A vs. B (e), A vs. C (f), and B vs. C (g), to GFR 12 months after transplantation measured in the validation cohort. Blue: individual linear regression models; Red: merged linear regression model for all samples
Fig.S9: PCK2 localizes to suEVs, increases during the initial stages of transplantation and does not correlate to tissue PCK2 levels after reperfusion. Electron microscopy depicting non permeabilized small urinary extracellular vesicles after immunogold staining for PCK2, scalebar 100nm (a)
Supplementary information Table S1
Supplementary information Table S2
Supplementary information Table S3
Supplementary information Table S4
Movie S1: Schematic overview of leave‐one‐out crossvalidation procedure. After plotting of all measured values per timepoint or fold change analysis, all values but one are used to create a correlative model. Subsequently, the missing value is predicted with the generated model, with the differences between predicted and true value indicating the prediction error and the correlative value of the model. The process is repeated according to the number of initial values creating correlative models of that number. All created models are summarized into an average model per protein. By using this mean model, the predictive value of protein abundance or fold change to GFR is calculated
ACKNOWLEDGEMENTS
We acknowledge the excellent technical support of Ruth Herzog, Beatrix Martini, Valerie Oberüber, Martyna Brütting, Serena Greco‐Torres, Israa Kambar, Andrea Eckardt, Maria Schiffer, Svenja Lademann, Monika Wagner, Nadine Friedelt, Frederik Sand, Constantin Rill, Charlotte Dellmann, Martin Vitus, Ugur Keser, Ismini Halmer, Dervla Reilly, Eva Schulze, Vincent Köntges, Bastian Trinsch, Philip Lützen, Nora Krasniqi, Babak Mochtarzadeh, Denise Krauß, Steffen Hinrichs, Jakob Kadereit as well as the Proteomics and Imaging Facilities at CECAD, Cologne. We thank Ingrid Becker for excellent advice regarding statistical analyses. This work was supported by the Nachwuchsgruppen.NRW program of the Ministry of Science North Rhine Westfalia (MIWF, to R.‐U.M.). T.B., B.S. and R.‐U.M. received funding from the German Research Foundation (DFG; KFO329 to T.B./B.S./R.‐U.M., MU 3629/3‐1 to R.‐U.M., SCHE1562/8‐1 to B.S., BE2212 to T.B. and MU3629/2‐1 to R.‐U.M.). The University of Cologne (Köln Fortune Program) and the Marga und Walter Boll Foundation provided additional support to R.‐U.M. V.G.P. was supported by the DFG (CRC 1192) and by the BMBF (eMed Consortia Fibromap). M.R.S. was supported by the Koeln Fortune Program and by the Cologne Clinician Scientist Program (Faculty of Medicine / University of Cologne, funded by the DFG [FI 773/15‐1]). T.B.H. was supported by the DFG (CRC 1192, HU 1016/8‐2), by the BMBF (STOP‐FSGS 01GM1901C) and by the European Research Council (ERC grant 61689, DNCure). M.M.R. is supported by a Novo Nordisk Foundation Young Investigator Grant, grant number NNF19OC0056043.
Open access funding enabled and organized by Projekt DEAL.
Braun F, Rinschen M, Buchner D, et al. The proteomic landscape of small urinary extracellular vesicles during kidney transplantation. J Extracell Vesicles. 2020;10:e12026 10.1002/jev2.12026
Contributor Information
Fabian Braun, Email: fa.braun@uke.de.
Dirk Stippel, Email: dirk.stippel@uk-koeln.de.
Roman‐Ulrich Müller, Email: roman-ulrich.mueller@uk-koeln.de.
REFERENCES
- Akbar, N. , Digby, J. E. , Cahill, T. J. , Tavare, A. N. , Corbin, A. L. , Saluja, S. , … Choudhury, R. P. (2017). Endothelium‐derived extracellular vesicles promote splenic monocyte mobilization in myocardial infarction. JCI Insight, 2(17), e93344 10.1172/jci.insight.93344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachurski, D. , Schuldner, M. , Nguyen, P.‐H. , Malz, A. , Reiners, K. S. , Grenzi, P. C. , … Pogge Von Strandmann, E. (2019). Extracellular vesicle measurements with nanoparticle tracking analysis – An accuracy and repeatability comparison between NanoSight NS300 and ZetaView. Journal of Extracellular Vesicles, 8(1), 1596016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead, P. , Loughrey, M. B. , Fernández, J. A. , Dombrowski, Y. , Mcart, D. G. , Dunne, P. D. , … Hamilton, P. W. (2017). QuPath: Open source software for digital pathology image analysis. Scientific Reports, 7(1), 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergler, T. , Hoffmann, U. , Bergler, E. , Jung, B. , Banas, M. C. , Reinhold, S. W. , … Banas, B. (2012). Toll‐like receptor 4 in experimental kidney transplantation: Early mediator of endogenous danger signals. Nephron Experimental Nephrology [Electronic Resource], 121(3–4), e59–e70. [DOI] [PubMed] [Google Scholar]
- Biglarnia, A.‐R. , Huber‐Lang, M. , Mohlin, C. , Ekdahl, K. N. , & Nilsson, B. (2018). The multifaceted role of complement in kidney transplantation. Nature Reviews Nephrology, 14(12), 767–781. [DOI] [PubMed] [Google Scholar]
- Blijdorp, C. J. , & Hoorn, E. J. (2019). Urinary extracellular vesicles: Mothership connection. American Journal of Renal Physiology, 103, 2583. [DOI] [PubMed] [Google Scholar]
- Chen, T. K. , Knicely, D. H. , & Grams, M. E. (2019). Chronic kidney disease diagnosis and management. JAMA, Journal of the American Medical Association, 322(13), 1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, D.‐S. , Kim, D.‐K. , Kim, Y.‐K. , & Gho, Y. S. (2015). Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrometry Reviews, 34(4), 474–490. [DOI] [PubMed] [Google Scholar]
- Clark, J. Z. , Chen, L. , Chou, C.‐L. , Jung, H. J. , Lee, J. W. , & Knepper, M. A. (2019). Representation and relative abundance of cell‐type selective markers in whole‐kidney RNA‐Seq data. Kidney International, 95(4), 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collino, F. , Lopes, J. A. , Corrêa, S. , Abdelhay, E. , Takiya, C. M. , Wendt, C. H. C. , … Lindoso, R. S. (2019). Adipose‐derived mesenchymal stromal cells under hypoxia: Changes in extracellular vesicles secretion and improvement of renal recovery after ischemic injury. Cellular Physiology and Biochemistry, 52(6), 1463–1483. [DOI] [PubMed] [Google Scholar]
- Cox, J. , Hein, M. Y. , Luber, C. A. , Paron, I. , Nagaraj, N. , & Mann, M. (2014). Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular and Cellular Proteomics, 13(9), 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , & Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nature Biotechnology, 26(12), 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox, J. , & Mann, M. (2012). 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high‐throughput data. BMC Bioinformatics, 13(Suppl 16), S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie, I. S. , & Henderson, L. K. (2019). ABO‐incompatible renal transplantation. Lancet, 393(10185), 2014–2016. [DOI] [PubMed] [Google Scholar]
- De Wever, O. , & Hendrix, A. (2019). A supporting ecosystem to mature extracellular vesicles into clinical application. Embo Journal, 38(9), e101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt, B. , Geeurickx, E. , Tulkens, J. , Van Deun, J. , Vergauwen, G. , Lippens, L. , … Hendrix, An (2020). Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density‐based fractionation of urine. Journal of Extracellular Vesicles, 9(1), 1736935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, J. H. , Liu, Y. , Gao, H. , Dominguez, J. M. , Xie, D. , & Kelly, K. J. (2017). Renal tubular cell‐derived extracellular vesicles accelerate the recovery of established renal ischemia reperfusion injury. Journal of the American Society of Nephrology, 28, 3533–3544,, ASN.2016121278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasini, G. , García‐Cerrada, M. , Vergara, E. , García‐Pino, G. , Alvarado, R. , Fernández‐Cavada, M. J. , … Cubero, J. J. (2015). Polymorphisms in CYP‐mediated arachidonic acid routes affect the outcome of renal transplantation. European Journal of Clinical Investigation, 45(10), 1060–1068. [DOI] [PubMed] [Google Scholar]
- Goetzl, E. J. , Schwartz, J. B. , Abner, E. L. , Jicha, G. A. , & Kapogiannis, D. (2018). High complement levels in astrocyte‐derived exosomes of Alzheimer disease. Annals of Neurology, 83(3), 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, P. A. , Pisitkun, T. , Hoffert, J. D. , Tchapyjnikov, D. , Star, R. A. , Kleta, R. , … Knepper, M. A. (2009). Large‐scale proteomics and phosphoproteomics of urinary exosomes. Journal of the American Society of Nephrology, 20(2), 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjidemetriou, M. , Al‐Ahmady, Z. , Mazza, M. , Collins, R. F. , Dawson, K. , & Kostarelos, K. (2015). In vivo biomolecule corona around blood‐circulating, clinically used and antibody‐targeted lipid bilayer nanoscale vesicles. ACS Nano, 9(8), 8142–8156. [DOI] [PubMed] [Google Scholar]
- Harford, J. B. , & Bonifacino, J. S. Biology JBCP in C, 2011 . (2011). Subcellular fractionation and isolation of organelles. Wiley Online Library. 52(1), 3.0.1–3.0.8. 10.1002/0471143030.cb0300s52 [DOI] [Google Scholar]
- Hye Khan, Md. A. , Stavniichuk, A. , Sattar, M. A. , Falck, J. R. , & Imig, J. D. (2019). Epoxyeicosatrienoic acid analog EET—A blunts development of lupus nephritis in mice. Front Pharmacol, 10, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnukainen, T. , Malehorn, D. , Sun, M. , Lyons‐Weiler, J. , Bigbee, W. , Gupta, G. , … Vats, A. (2006). Proteomic analysis of urine in kidney transplant patients with BK virus nephropathy. Journal of the American Society of Nephrology, 17(11), 3248–3256. [DOI] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , … Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177(2), 428–445.e18.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpman, D. , Ståhl, A.‐L. , & Arvidsson, I. (2017). Extracellular vesicles in renal disease. Nature Reviews Nephrology, 13(9), 545–562. [DOI] [PubMed] [Google Scholar]
- Kim, S. C. , Page, E. K. , & Knechtle, S. J. (2014). Urine proteomics in kidney transplantation. Transplantation Reviews, 28(1), 15–20. [DOI] [PubMed] [Google Scholar]
- Klein, J. , Bascands, J.‐L. , Mischak, H. , & Schanstra, J. P. (2016). The role of urinary peptidomics in kidney disease research. Kidney International, 89(3), 539–545. [DOI] [PubMed] [Google Scholar]
- Koritzinsky, E. H. , Street, J. M. , Chari, R. R. , Glispie, D. M. , Bellomo, T. R. , Aponte, A. M. , … Yuen, P. S. T. (2019). Circadian variation in the release of small extracellular vesicles can be normalized by vesicle number or TSG101. American Journal of Renal Physiology, 317, F1098–F1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leithner, K. , Hrzenjak, A. , Trötzmüller, M. , Moustafa, T. , Köfeler, H. C. , Wohlkoenig, C. , … Olschewski, H. (2015). PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene, 34(8), 1044–1050. [DOI] [PubMed] [Google Scholar]
- Lenzini, S. , Bargi, R. , Chung, G. , & Shin, J.‐W. (2020). Matrix mechanics and water permeation regulate extracellular vesicle transport. Nature Nanotechnology, 15, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey, A. S. , Stevens, L. A. , Schmid, C. H. , Zhang, Y. L. , Castro, A. F. , Feldman, H. I. , … Coresh, J. (2009). A new equation to estimate glomerular filtration rate. Annals of Internal Medicine, 150(9), 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean, B. , Tomazela, D. M. , Shulman, N. , Chambers, M. , Finney, G. L. , Frewen, B. , … Maccoss, M. J. (2010). Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics, 26(7), 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manček‐Keber, M. , Frank‐Bertoncelj, M. , Hafner‐Bratkovič, I. , Smole, Anže , Zorko, M. , Pirher, N. , … Jerala, R. (2015). Toll‐like receptor 4 senses oxidative stress mediated by the oxidation of phospholipids in extracellular vesicles. Science Signaling, 8(381), ra60. [DOI] [PubMed] [Google Scholar]
- Mange, K. C. , Joffe, M. M. , & Feldman, H. I. (2001). Effect of the use or nonuse of long‐term dialysis on the subsequent survival of renal transplants from living donors. The New England Journal of Medicine, 344(10), 726–731. [DOI] [PubMed] [Google Scholar]
- Matsushita, K. , Coresh, J. , Sang, Y. , Chalmers, J. , Fox, C. , Guallar, E. , … Ärnlöv, J. (2015). Estimated glomerular filtration rate and albuminuria for prediction of cardiovascular outcomes: A collaborative meta‐analysis of individual participant data. Lancet Diabetes Endocrinol, 3(7), 514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, M. L. , Rood, I. M. , Deegens, J. K. J. , & Klein, J. B. (2017). Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nature Reviews Nephrology, 13, 731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas, J. , & Hughes, C. C. W. (2004). Of mice and not men: Differences between mouse and human immunology. Journal of Immunology, 172(5), 2731–2738. [DOI] [PubMed] [Google Scholar]
- Michalski, A. , Damoc, E. , Hauschild, J.‐P. , Lange, O. , Wieghaus, A. , Makarov, A. , … Horning, S. (2011). Mass spectrometry‐based proteomics using Q Exactive, a high‐performance benchtop quadrupole Orbitrap mass spectrometer. Molecular and Cellular Proteomics, 10(9), M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal, E. D. , Bhalla, K. , Dewi, R. E. , Ruiz, C. F. , Haley, J. A. , Ropell, A. E. , … Girnun, G. D. (2019). Inhibition of phosphoenolpyruvate carboxykinase blocks lactate utilization and impairs tumor growth in colorectal cancer. Cancer & Metabolism, 7, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante, L. , Saraswat, M. , Ravidà, A. , Byrne, B. , & Holthofer, H. (2013). Recovery of urinary nanovesicles from ultracentrifugation supernatants. Nephrology, Dialysis, Transplantation, 28(6), 1425–1433. [DOI] [PubMed] [Google Scholar]
- Nagaraj, N. , & Mann, M. (2011). Quantitative analysis of the intra‐ and inter‐individual variability of the normal urinary proteome. Journal of Proteome Research, 10(2), 637–645. [DOI] [PubMed] [Google Scholar]
- Nauser, C. L. , Farrar, C. A. , & Sacks, S. H. (2017). Complement recognition pathways in renal transplantation. Journal of the American Society of Nephrology, 28(9), 2571–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeyen, E. , Van Mol, K. , Baggerman, G. , Willems, H. , Boonen, K. , Rolfo, C. , …, Mertens, I. (2018). Ultrafiltration and size exclusion chromatography combined with asymmetrical‐flow field‐flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J Extracell Vesicles, 7(1), 1490143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshikawa‐Hori, S. , Yokota‐Ikeda, N. , Sonoda, H. , & Ikeda, M. (2019). Urinary extracellular vesicular release of aquaporins in patients with renal transplantation. BMC Nephrology, 20(1), 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienimaeki‐Roemer, A. , Kuhlmann, K. , Böttcher, A. , Konovalova, T. , Black, A. , Orsó, E. , … Schmitz, G. (2015). Lipidomic and proteomic characterization of platelet extracellular vesicle subfractions from senescent platelets. Transfusion, 55(3), 507–521. [DOI] [PubMed] [Google Scholar]
- Pino, L. K. , Searle, B. C. , Bollinger, J. G. , Nunn, B. , Maclean, B. , & Maccoss, M. J. (2020). The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrometry Reviews, 39, 229–244, [published online ahead of print: 2017]; 10.1002/mas.21540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulskens, W. P. , Rampanelli, E. , Teske, G. J. , Butter, L. M. , Claessen, N. , Luirink, I. K. , … Leemans, J. C. (2010). TLR4 promotes fibrosis but attenuates tubular damage in progressive renal injury. Journal of the American Society of Nephrology, 21(8), 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakumar, S. , Ren, Lu , Kundu, S. , Gamon, A. , Tyagi, S. C. , & Sen, U. (2017). Toll‐like Receptor 4 Deficiency Reduces Oxidative Stress and Macrophage Mediated Inflammation in Hypertensive Kidney. Scientific Reports, 7(1), 6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj, D. A. A. , Fiume, I. , Capasso, G. , & Pocsfalvi, G. (2012). A multiplex quantitative proteomics strategy for protein biomarker studies in urinary exosomes. Kidney International, 81(12), 1263–1272. [DOI] [PubMed] [Google Scholar]
- Ricklin, D. , Mastellos, D. C. , Reis, E. S. , & Lambris, J. D. (2017). The renaissance of complement therapeutics. Nature Reviews Nephrology, 14(1), 26–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Hochberg, F. H. , & Jones, P. S. (2018). Extracellular vesicles: The growth as diagnostics and therapeutics; a survey. Journal of Extracellular Vesicles, 7(1), 1438720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden, C. T. , Schindelin, J. , Hiner, M. C. , Dezonia, B. E. , Walter, A. E. , Arena, E. T. , & Eliceiri, K. W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics, 18(1), 529–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabaratnam, R. , Geertsen, L. , Skjødt, K. , Højlund, K. , Dimke, H. , Lund, L. , & Svenningsen, P. (2019). In human nephrectomy specimens, the kidney level of tubular transport proteins does not correlate with their abundance in urinary extracellular vesicles. American Journal of Renal Physiology, 317(3), F560–F571. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , … Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinstock, C. A. , Sapir‐Pichhadze, R. , Naesens, M. , Batal, I. , Bagnasco, S. , Bow, L. , … Kraus, E. (2019). Banff survey on antibody‐mediated rejection clinical practices in kidney transplantation: Diagnostic misinterpretation has potential therapeutic implications. American Journal of Transplantation, 19(1), 123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellarés, J. , De Freitas, D. G. , Mengel, M. , Reeve, J. , Einecke, G. , Sis, B. , … Halloran, P. F. (2012). Understanding the causes of kidney transplant failure: The dominant role of antibody‐mediated rejection and nonadherence. American Journal of Transplantation, 12(2), 388–399. [DOI] [PubMed] [Google Scholar]
- Simons, M. , & Raposo, G. (2009). Exosomes–vesicular carriers for intercellular communication. Current Opinion in Cell Biology, 21(4), 575–581. [DOI] [PubMed] [Google Scholar]
- Solez, K. I. M. , Axelsen, R. A. , Benediktsson, H. , Burdick, J. F. , Cohen, A. H. , Colvin, R. B. , … Yamaguchi, Y. (1993). International standardization of criteria for the histologic diagnosis of renal allograft rejection: The Banff working classification of kidney transplant pathology. Kidney International, 44(2), 411–422. [DOI] [PubMed] [Google Scholar]
- Sonoda, H. , Yokota‐Ikeda, N. , Oshikawa, S. , Kanno, Y. , Yoshinaga, K. , Uchida, K. , … Ikeda, M. (2009). Decreased abundance of urinary exosomal aquaporin‐1 in renal ischemia‐reperfusion injury. American Journal of Renal Physiology, 297(4), F1006–F1016. [DOI] [PubMed] [Google Scholar]
- Souza, A. C. P. , Tsuji, T. , Baranova, I. N. , Bocharov, A. V. , Wilkins, K. J. , Street, J. M. , … Star, R. A. (2015). TLR4 mutant mice are protected from renal fibrosis and chronic kidney disease progression. Physiological Reports, 3(9), e12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Späth, M. R. , Bartram, M. P. , Palacio‐Escat, N. , Hoyer, K. J. R. , Debes, C. , Demir, F. , … Rinschen, M. M. (2019). The proteome microenvironment determines the protective effect of preconditioning in cisplatin‐induced acute kidney injury. Kidney International, 95(2), 333–349. [DOI] [PubMed] [Google Scholar]
- Stone, M. (1974). Cross‐validatory choice and assessment of statistical predictions. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 36(2), 111–133. [Google Scholar]
- Suhail, S. M. (2014). Significance of urinary proteome pattern in renal allograft recipients. Journal of Transplantation, 2014(5), 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki, P. I. , Cecka, J. M. , Gjertson, D. W. , & Takemoto, S. (1995). High survival rates of kidney transplants from spousal and living unrelated donors. The New England Journal of Medicine, 333(6), 333–336. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018), a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco, A. E. , Lam, W. , Rule, A. D. , Denic, A. , Lieske, J. C. , Miller, V. M. , … Jayachandran, M. (2016). Specific renal parenchymal‐derived urinary extracellular vesicles identify age‐associated structural changes in living donor kidneys. Journal of Extracellular Vesicles, 5, 29642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher, V. G. , Tibshirani, R. , & Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences, 98(9), 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Velde, M. , Matsushita, K. , Coresh, J. , Astor, B. C. , Woodward, M. , Levey, A. S. , … Gansevoort, R. T. (2011). Lower estimated glomerular filtration rate and higher albuminuria are associated with all‐cause and cardiovascular mortality. A collaborative meta‐analysis of high‐risk population cohorts. Kidney International, 79(12), 1341–1352. [DOI] [PubMed] [Google Scholar]
- Vizcaíno, J. A. , Csordas, A. , Del‐Toro, N. , Dianes, J. A. , Griss, J. , Lavidas, I. , … Hermjakob, H. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Research, 44(22), 11033–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeboah, M. M. , Khan, Md A. H. , Chesnik, M. A. , Sharma, A. , Paudyal, M. P. , Falck, J. R. , & Imig, J. D. (2016). The epoxyeicosatrienoic acid analog PVPA ameliorates cyclosporine‐induced hypertension and renal injury in rats. American Journal of Renal Physiology, 311(3), F576‐F585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H. , Perez, J. S. , Lu, K. , George, A. J. T. , & Ma, D. (2014). Role of Toll‐like receptor‐4 in renal graft ischemia‐reperfusion injury. American Journal of Renal Physiology, 306(8), F801‐F811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmonarski, S. C. , Banasik, M. , Madziarska, K. , Mazanowska, O. , & Krajewska, M. (2019). The role of toll‐like receptors in multifactorial mechanisms of early and late renal allotransplant injury, with a focus on the TLR4 receptor and mononuclear cells. Advances in Clinical and Experimental Medicine, 28(7), 885–891. [DOI] [PubMed] [Google Scholar]
- Zuber, Julien, Frimat, M. , Caillard, S. , Kamar, N. , Gatault, P. , Petitprez, F. , … Frémeaux‐Bacchi, V. (2019). Use of highly individualized complement blockade has revolutionized clinical outcomes after kidney transplantation and renal epidemiology of atypical hemolytic uremic syndrome. Journal of the American Society of Nephrology, 30(12), 2449–2463. ASN.2019040331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1: Differential centrifugation leads to a decrease in Uromodulin to TSG ratio and separates small tetraspanin‐containing vesicles. Size distribution of suEVs measured in SEM of 2 independent samples of healthy volunteers (a). Western Blot Analysis and Densitometry of Uromodulin and TSG101 in 2 independent samples of healthy volunteers (b). Flow cytometric bead assay for tetraspanins CD9, CD63, CD81 and Phosphatidylserine (PS) of 4 independent samples of healthy volunteers (c)
Fig.S2: Validation of suEV separation by size exclusion chromatography compared to ultracentrifugation. Size distribution of suEVs measured in 3 independent suEV samples of healthy volunteers separated by ultracentrifugation (UC) or size exclusion chromatography (SEC) measured by Nanoparticle Tracking, bin: 30nm, errorbars: SD, (a). Flow cytometric bead assay for tetraspanins CD9, CD63 and CD81 of 3 independent samples of healthy volunteers separated by ultracentrifugation (UC) or size exclusion chromatography (SEC) (b)
Fig.S3: Comparison of suEV proteome separated through SEC and UC and overlap of donor sample (timepoint a) to published data. Venn diagram of overlap between suEV proteome of vesicles separated by size exclusion chromatography (SEC) or ultracentrifugation (UC) with proposed putative urinary vesicle contaminants (a). Venn diagram of overlap between suEV proteome at timepoint a with proposed putative urinary vesicle contaminants (Dhondt et al., 2020) (b). Venn diagram of overlap between suEV proteome at timepoint a with published data (Dhondt et al., 2020, Oeyen et al., 2018) (c). Scatter plot depicting the mean LFQ to protein rank of proteins detected in 50% of vesicle samples separated from urine samples of healthy volunteers by SEC or UC. Protein‐Rank ranging from 1–510 in SEC and 1–1248 in UC samples. Plots depicting marker proteins for extracellular vesicles (d), podocytes (e), proximal tubulus (f), distal tubulus (g), collecting duct (h), connecting tubulus (I) and the top 20 proteins detected at all timepoints in the transplant suEV cohort (J)
Fig.S4: Enriched GO term analysis of absolute protein quantification per timepoint. Bubble plots of top 10 GO terms enriched for each timepoint of sample collection. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint A, B: Timepoint B, C: Timepoint C, D: Timepoint D, E: Timepoint E)
Fig.S5: Significant GO term analysis of protein fold changes compared to initial donor sample. Top 10 significant GO terms for each timepoint after transplantation compared to the initial donor samples. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint B vs A, B: Timepoint C vs A, C: Timepoint D vs A, D: Timepoint E vs A)
Fig.S6: Enriched GO term analysis of protein fold changes compared to initial donor sample. Top 10 GO terms enriched for each timepoint after transplantation compared to the initial donor samples. P value depicted as color code, number of annotated proteins corresponding to bubble size, black GO terms indicating biological processes, gray GO terms indicating molecular function (A: Timepoint B vs A, B: Timepoint C vs A, C: Timepoint D vs A, D: Timepoint E vs A)
Fig.S7: Linear regression analysis correlating clinical parameters and PCK2 abundance to long term transplant outcome. Regression tables for multiple linear regression with (a) 12 month eGFR, or (b) 6 month eGFR as the dependent variable. OLS = ordinary least squares, Adj. R² = adjusted R², Est. = Estimate, S.E. = Standard error, t val. = t value, and p = p‐value. Predictors with a p‐value ≤0.05 were marked in red
Fig.S8: PCK2 abundance in suEVs at other timepoints does not correlate with estimated GFR 12 months after transplantation. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint C to GFR 12 months after transplantation measured in the initial transplant cohort split by female (a) and male (b) recipeitns. Correlation plots of PCK2 intensity foldchange to mean intensity at timepoint A (c) and B (d) and foldchanges between timepoints A vs. B (e), A vs. C (f), and B vs. C (g), to GFR 12 months after transplantation measured in the validation cohort. Blue: individual linear regression models; Red: merged linear regression model for all samples
Fig.S9: PCK2 localizes to suEVs, increases during the initial stages of transplantation and does not correlate to tissue PCK2 levels after reperfusion. Electron microscopy depicting non permeabilized small urinary extracellular vesicles after immunogold staining for PCK2, scalebar 100nm (a)
Supplementary information Table S1
Supplementary information Table S2
Supplementary information Table S3
Supplementary information Table S4
Movie S1: Schematic overview of leave‐one‐out crossvalidation procedure. After plotting of all measured values per timepoint or fold change analysis, all values but one are used to create a correlative model. Subsequently, the missing value is predicted with the generated model, with the differences between predicted and true value indicating the prediction error and the correlative value of the model. The process is repeated according to the number of initial values creating correlative models of that number. All created models are summarized into an average model per protein. By using this mean model, the predictive value of protein abundance or fold change to GFR is calculated


