Abstract
Alzheimer’s disease (AD) is a progressive, mental illness without cure. Several years of intense research on postmortem AD brains using cell and mouse models of AD have revealed that multiple cellular changes are involved in the disease process, including mitochondrial abnormalities, synaptic damage, and glial/astrocytic activation, in addition to age-dependent accumulation of amyloid beta (Aβ) and hyperphosphorylated tau (p-tau). Synaptic damage and mitochondrial dysfunction are early cellular changes in the disease process. Healthy and functionally active mitochondria are essential for cellular functioning. Dysfunctional mitochondria play a central role in aging and AD. Mitophagy is a cellular process whereby damaged mitochondria are selectively removed from cell and mitochondrial quality and biogenesis. Mitophagy impairments cause the progressive accumulation of defective organelle and damaged mitochondria in cells. In AD, increased levels of Aβ and p-tau can induce reactive oxygen species (ROS) production, causing excessive fragmentation of mitochondria and promoting defective mitophagy. The current article discusses the latest developments of mitochondrial research and also highlights multiple types of mitophagy, including Aβ and p-tau-induced mitophagy, stress-induced mitophagy, receptor-mediated mitophagy, ubiquitin mediated mitophagy and basal mitophagy. This article also discusses the physiological states of mitochondria, including fission-fusion balance, Ca2+ transport, and mitochondrial transport in normal and diseased conditions. Our article summarizes current therapeutic interventions, like chemical or natural mitophagy enhancers, that influence mitophagy in AD. Our article discusses whether a partial reduction of Drp1 can be a mitophagy enhancer and a therapeutic target for mitophagy in AD and other neurological diseases.
Keywords: Alzheimer’s disease, amyloid beta, phosphorylated tau, mitochondrial dysfunction, mitophagy
1. Introduction
Alzheimer’s disease (AD), a progressive and multifactorial neurological disease, is characterized by multiple cognitive impairments and memory loss (Selkoe, 2001; Swerdlow, 2009; Reddy et al 2010, Mao and Reddy 2011). AD occurs in 2 forms – early-onset familial and late-onset sporadic (Selkoe 2001). In early-onset AD, genetic mutations in the amyloid precursor protein (APP) and in presenilin 1 and presenilin 2 loci induce a cascade of cellular changes, including synaptic damage and loss, mitochondrial failure, oxidative stress, glial and astrocytic activation, and microRNA deregulation, primarily caused by amyloid beta (Aβ) and hyperphosphorylated tau (p-tau) (Selkoe, 2001; Swerdlow, 2009; Reddy et al., 2010; Mao and Reddy, 2011; Reddy et al., 2012; Reddy et al., 2017; Amakiri et al., 2019). Although causal factors are largely unknown for late-set AD, multiple factors are involved in the disease process of AD, including the apolipoprotein allele 4 genotype accumulation of reactive oxygen species (ROS), traumatic brain injury, lifestyle factors such as unhealthy diet, lack of physical activity, and chronic conditions like diabetes/obesity and hypertension (Reddy, 2019; George and Reddy, 2019; Mao and Reddy, 2011; Amakiri et al., 2019). Above all, aging is the major risk factor for developing both familial and sporadic AD. As mentioned above, pathological and morphological features are similar in the brains of both early-onset and late-onset AD patients, but cellular and pathological changes occur late in sporadic AD.
Synaptic failure and mitochondrial abnormalities are widely accepted as early cellular changes in the AD disease process (Reddy et al., 2012). In the brains of both early and late-onset AD patients, the following mitochondrial abnormalities occur: an age-dependent accumulation of mitochondrial DNA (mtDNA) changes (Santoro et al., 2006; Salvioli et al., 2006; Rose et al., 2017), a reduced mitochondrial membrane potential, increased mitochondrial ROS production, reduced axonal transport of mitochondria, reduced mitochondrial adenosine triphosphate (ATP), altered mitochondrial enzymatic activities, and, most importantly, increased mitochondrial fragmentation. These mitochondrial changes lead to defective mitophagy in AD patients.
Mitophagy is a cellular process by which damaged mitochondria are removed from the cell via autophagy (Reddy and Oliver 2019). This cellular event is used to confirm the elimination of damaged mitochondria, critically challenged in protein levels by proteasomal degradation pathways and alterations in biogenesis (Bragoszewski et al., 2017; Ross et al., 2005; Lou et al., 2020; Wu et al., 2019; Oliver and Reddy, 2019). Measurement of mitochondrial content determines the markers of mitochondrial quality (Evans and Holzbaur, 2020; Calkins et al., 2011; Wang et al., 2009). Despite the significant recent advances in our knowledge regarding mitophagy, reliable quantitative assays still need to be developed. Interest on mitophagy has recently increased, mostly by our growing awareness that mutations in mitophagy proteins, like PTEN-induced kinase 1 (PINK1) or Parkin, occur in neurodegenerative diseases, such as Parkinson’s and Alzheimer’s, as well as Aβ and p-tau induced toxicities in mitochondria (Cai and Jeong, 2020; Liu et al., 2020; Kerr et al., 2017). Although there are several other relevant players in mitophagy other than PINK1 and Parkin, advances have been made in the mitophagy field (Chakravorty et al., 2019; Reddy et al., 2018; Koyano et al., 2014, but a better understanding of the pathological process still needs investigation.
Mitophagy is the selective degradation of mitochondria by autophagy. Mitophagy is a key process in keeping the cell healthy. It promotes turnover of mitochondria and prevents accumulation of dysfunctional mitochondria which can lead to cellular degeneration. Mitophagy is regulated by PINK1 and Parkin proteins. In addition to the selective removal of damaged mitochondria, mitophagy is also required to adjust mitochondrial numbers to changing cellular metabolic needs, for steady-state mitochondrial turnover (Fritsch et al., 2010; Fritsch et al., 2020; Wang et al., 2019; Wang et al., 2013; Zhang et al., 2009; Harper et al., 2018; Harper et al., 2006; Schulman et al., 2009; Sarraf et al., 2013; Ordureau et al., 2015; Cai and Jean, 2020).
Mitophagy is initially mediated by the several autophagy pathways, beginning with the formation of phagophores. It then expands through the acquisition of lipids, resulting in the autophagosomes (Roca-Agujetas et al., 2019; Seo et al., 2010; Weber and Reichert, 2010). In the cell, autophagosomes first engulf mitochondria via autophagosomes and their subsequent catabolism by lysosomes, which protein first binds with, what are the adaptor proteins, and who recruit the other autophagy proteins in terms of a phagophore formation around mitochondria are in debate.
The purpose of our article is to examine the status of mitophagy in healthy and disease states. We discussed 1) the tools and methods to study mitophagy; 2) the cellular events that occur in mitophagy pathways; 3) the abnormal interactions between Aβ and Drp1 and p-tau and Drp1, leading to defective mitophagy in AD and 4) the latest developments in mitophagy enhancers, using pharmacological and genetic approaches.
2. Tools and Methods to Study Mitophagy
Mitochondria are highly dynamic organelles. They undergo fusion and fission, and remove dead mitochondria by mitophagy. Mitophagy contributes to balancing mitochondrial morphology, the quantity of mitochondria, and their quality. Some of the important measures of mitophagy are: mitochondrial permeability transition, mitochondrial respiration, mitochondrial mass, mitochondrial axonal transport, mitochondrial fragmentation, mitochondrial functional parameters (free radical production, ATP levels, enzymatic activities), mitochondrial and nuclear DNA (mtDNA/nDNA) ratios, citrate synthase activity, mitochondria-autophagosome or mitochondria-lysosome colocalization and cellular and mitochondrial calcium levels (Ding and Yin, 2012; Williams et al., 2017). Table 1 summarizes tools and methods to study mitophagy in AD and other human diseases. Details of tools and methods are given below:
Table 1.
Summary of tools and methods to study mitophagy in Alzheimer’s disease and other human diseases
| Methods | Comments | |
|---|---|---|
| 1. | qRT-PCR | Measuring mRNA expressions in mitochondrial dynamics and mitochondrial biogenesis, synaptic, autophagy and mitophagy genes. |
| 2. | Western Blot | Measuring the protein levels in mitochondrial dynamics and mitochondrial biogenesis, synaptic, autophagy and mitophagy genes. |
| 3. | Localization | Immunofluorescence analysis |
| 4. | Transmission electron microscopy | Transmission electron microscopy of cellular organelles - Mitochondria Neurons Nucleus Cytoplasm Golgi body Lysosome Autophagosomes Chromatin structure Number of mitochondria Length of mitochondria Structure of the neuronal soma Synapse density |
| 5. | Mitochondrial functional assays | H2O2 production Lipid peroxidation Cytochrome c oxidase activity ATP levels |
| 6. | Mitochondrial membrane potential | Generated by proton pumps in electron transport chain (ETC) (Complexes I, III and IV). |
2.1. qRT-PCR and immunoblotting techniques
Mitochondrial gene expressions and protein levels have been assessed by qRT-PCR and immunoblotting techniques (Reddy et al., 2018; Manczak et al., 2010; Manczak et al., 2018; Kandimalla et al., 2016; Kandimalla et al., 2018). Mitochondrial fission and fusion, matrix and biogenesis genes are important genes to assess mitophagy. It is also important to study neuronal, glial, and astrocytic markers of cell, because mitophagy occurs in all cell populations. Further, it is important to study genes responsible for subcellular compartments of a cell, such as synapses, endoplasmic reticulum (ER) and lysosomes.
2.2. Immunofluorescence analysis of mitochondrial genes
Advanced fluorescence techniques, such as immunofluorescence, and confocal microscopies can be used to assess the localization of proteins related to mitophagy and mitochondrial quality control (Ding and Yin, 2012; Williams, et al., 2017; Cossarizza et al., 2001). Mito-tracker, labels mitochondria within live cells utilizing the mitochondrial membrane potential. A mito-timer is a very useful tool for monitoring both mitophagy and mitochondrial biogenesis. Detection of mitophagy is keima-red is mito-keima and can be used in in vitro and in vivo studies. Mito-QC (in vivo - mouse model that can be used to study mitochondrial network) are some of the important reagents and markers that are extensively used to study mitophagy. These are pH-sensitive mitochondrial fluorescent probes and extensively used to assess mitophagy and mitochondrial quality control.
2.3. Transmission electron microscopy (TEM) of neurons
TEM has been extensively used to assess the length, number, and structure of intact and damaged mitochondria. TEM is extremely useful in determining mitochondrial morphology and in providing a more accurate assessment of mitochondrial quality. We (Manczak et al., 2006; Manczak et al., 2010; Calkins et al., 2011; Manczak et al.; 2011; Manczak et al.; 2016; Manczak et al.; 2018; Kandimalla et al., 2016; Kandimalla et al., 2018; Manczak and Reddy, 2015) and others (Wang et al., 2008; Wang et al., 2009, Han et al., 2020) have studied postmortem AD brains, brain tissues from AD mouse models, primary neurons and mammalian cells expressing Aβ and p-tau. These studies found altered mitochondrial structure and damaged mitochondria in AD specimens.
2.4. Mitochondrial membrane potential
The mitochondrial membrane potential is generated by proton pumps (in electron transport chain complexes I, III, and IV) and is an essential component in the process of energy storage during oxidative phosphorylation. Together with the proton gradient, membrane potential forms the transmembrane potential of hydrogen ions which is harnessed to make ATP (Zorova et al., 2018). The levels of mitochondrial membrane potential and ATP in the cell are kept relatively stable although there are limited fluctuations of both these factors that can occur reflecting normal physiological activity. We (Manczak et al., 2010) and others (Cardosa et al., 2004) assessed mitochondrial membrane potential, in order to determine quality of mitochondria.
2.5. Mitochondrial function
To assess mitochondrial function, it is important to measure the levels of free radical production, ATP, and enzymatic activities. The outcome of these measures will determine quality of mitochondria in both healthy and disease states. We (Reddy et al., 2004; Manczak et al., 2006; Manczak et al., 2010; Manczak et al., 2011; Manczak and Reddy, 2012a; Manczak, et al., 2012b; Reddy et al., 2016; Reddy et al 2017; Reddy et al., 2018a; Reddy et al., 2018b; Manczak et al., 2016; Manczak et al., 2018; Manczak et al., 2019; Kandimalla et al., 2016; Kandimalla et al., 2018; Pradeepkiran et al., 2020) and others (Wang et al., 2008; Wang et al., 2009; Du et al., 2008; Du et al., 2010) extensively studied in AD cells, AD-affected and AD-unaffected tissues from AD mouse models and postmortem AD brains. The outcome of these studies is that mitochondrial function is defective and depends on the severity of disease progression.
Overall, the above-mentioned tools and procedures are important to assess quality of mitochondria and mitophagy in AD and other human diseases, but they have their own limitations in terms of accuracy. We need more research develop better and more reliable assays for assessing mitophagy.
3. Mitochondria
The mitochondrion is a self-autonomous double membrane organelle found in most eukaryotic organisms. The most promising role of mitochondria is the production of energy via oxidative phosphorylation in cells, ADP to ATP (Osellame et al., 2012; Bader and Winklhofer, 2020). The half-life period of mitochondria is about 2–3 weeks. Mitochondria are in a dynamic state in the cells (Reddy and Oliver, 2019). Mitochondria are synthesized in the cell body and travel along axons and dendrites to supply energy (ATP) to nerve terminals ‘synapses’ for normal neural communication. The transportation of mitochondria to synapses from axons is referred to as ‘axonal transport of mitochondria’ (Reddy et al., 2011; Kandimalla and Reddy, 2016; Oliver and Reddy, 2020; Zhu et al., 2013). In this process, mitochondria alter their shape and size, which facilitates their movement from the cell body to axons, dendrites, and synapses, and back to the cell body (Reddy, et al., 2011; Calkins, et al., 2012; Kandimalla and Reddy, 2016; Chen and Chan, 2009; Li, et al., 2004; Chan, 2006; Chen and Chan, 2009). Earlier transmission microscopy observations revealed the plasticity of mitochondria, which appear as slender filaments in which under normal conditions look like spaghetti (Nemani et al., 2012). In stressed or diseased conditions, mitochondria break up into shorter segments shaped like a torus (Ahmad et al., 2013). Mitochondria in a continual state of fission and fusion is a key flagging identity of mitochondrion is in a transient condition (Bui and Shaw, 2013; Scott and Youle, 2010). At any point in the cell cycle, a mitochondrion may undergo fission to give rise to two separate mitochondria (Lee and Yoon, 2016). Simultaneously mitochondria are undergoing fusion in which both the inner and outer membranes of the mitochondria break and rejoin to form a single intact mitochondrion. Fusion and fission events are governed by the mitochondrial shape transition by unknown mechanisms during cellular activation (Sharma et al., 2019; Liu et al., 2020). Known proteins that are associated with mitochondrial dynamics (fission-fusion balance) are - Drp1, Fis1, Miro, Opa1, Mfn1, Mfn2, Mid49, Mid51 and Mff (Reddy et al., 2011; Kandimalla and Reddy, 2016; Oliver and Reddy, 2019).
3.1. Impaired Mitochondrial Dynamics in Alzheimer’s Disease
Mitochondrial dynamics is a delicate balance between division and fusion. The shape and structure of mitochondria are maintained by fission and fusion balance (Reddy et al., 2011; Zhu et al., 2013). In healthy cells, fission and fusion events balance equally, which maintains mitochondria function. Mitochondrial fission is controlled by evolutionary conserved, dynamin-related large GTPases.
Fission is regulated by Drp1 and Fis1, the latter of which is localized in the outer membrane of the mitochondria (Reddy et al., 2011; Calkins et al., 2012; Kandimalla and Reddy, 2016; Chen and Chan, 2009; Li et al., 2004; Chan, 2006; Chen and Chan, 2009). When a mitochondrion signals to divide, Drp1 translocates from the cytosol to the outer membrane of the mitochondrion where it initiates the process of fragmentation.
In contrast, fusion is controlled by 3 GTPase proteins: Mfn1 and Mfn2, which are located in the outer mitochondrial membrane, and Opa1, which is located in the inner mitochondria membrane. The C-terminal portion of Mfn1 mediates oligomerization between Mfn molecules of adjacent mitochondria, facilitating mt fusion (Chan, 2006, Reddy et al 2011).
Recent studies from our lab (Manczak et al., 2010; Manczak et al., 2011; Manczak et al., 2011), and others (Wang et al., 2008; Wang et al., 2009: Silva et al., 2013) found increased mitochondria fission and reduced fusion, suggesting that impaired mitochondrial dynamics is present in AD-affected neurons. Recent studies of AD neurons from the lab revealed that Aβ interacts with fission protein Drp1, with a subsequent increase in free radical production, which in turn activates Drp1 and Fis1, causing excessive mitochondria fragmentation, defective transport of mitochondria to synapses, provides low synaptic ATP, and ultimately leads to synaptic dysfunction (Manczak et al., 2011). Further studies from our lab also revealed that p-tau interacts with Drp1 and enhances GTPase Drp1 enzymatic activity, leading to excessive fragmentation of mitochondria and mitochondrial dysfunction in AD (Manczak and Reddy, 2012).
Overall, abnormal mitochondrial dynamics (increased fission and reduced fusion) are present in AD and is an important biological phenomenon for defective mitophagy in AD.
3.2. Defective Mitochondrial Biogenesis and Alzheimer’s Disease
Mitochondrial biogenesis is the process by which new mitochondria are synthesized in the cell and is activated by numerous different signals during times of cellular stress (Valero, 2014). It is affected by toxins and an accumulation of mtDNA mutations. There are four genes that are involved in mitochondrial biogenesis: PPAR - peroxisome proliferator-activated receptor)-γ coactivator-1α) (PGC1α, nuclear respiratory factor 1 (NRF1), nuclear respiratory factor 2 (NRF2) and transcription factor A, mitochondrial (TFAM). In a diseased state, such as AD, mitochondrial biogenesis is reduced or defective, mainly because of the association of mutant proteins such as Aβ and p-tau with mitochondria and increased free radical production (Sheng et al., 2012; Manczak et al., 2011; Manczak et al., 2018; Manczak and Reddy, 2012; Kandimalla et al., 2018), Therefore, mitochondrial biogenesis is an important parameter to assess mitochondrial function and mitophagy, particularly in a diseased state.
Several groups found reduced mitochondrial biogenesis in AD. In studies conducted to determine mitochondrial biogenesis in postmortem AD brains and AD cells, Sheng et al. (2012) found reduced mRNA and protein levels in the mitochondrial biogenesis genes PGC1α, Nrf-1, Nrf-2, and TFAM in postmortem AD brains and AD cells. We also studied mitochondrial biogenesis in mouse models and AD cells. We found reduced levels of PGC1α, Nrf1, Nrf2, and TFAM in 6- and 12-month-old APP transgenic mice (Manczak et al., 2016; Manczak et al., 2018) and tau transgenic mice (Kandimalla et al., 2016; Kandimalla et al., 2018), suggesting that mitochondrial biogenesis is defective in AD. More recently, using mutant APP mouse primary hippocampal neurons, Reddy and colleagues (2018) studied mRNA and protein levels of mitochondrial biogenesis proteins. They found reduced levels of mRNA and proteins of mitochondrial biogenesis genes PGC1α, NRF1, NRF2 & TFAM. These observations strongly suggest that accumulation of mAPP and Aβ causes defective mitochondrial biogenesis in AD (Reddy et al., 2018).
Overall, defective mitochondrial biogenesis plays an important role in altered mitophagy in AD and other neurodegenerative diseases.
4. Mitophagy
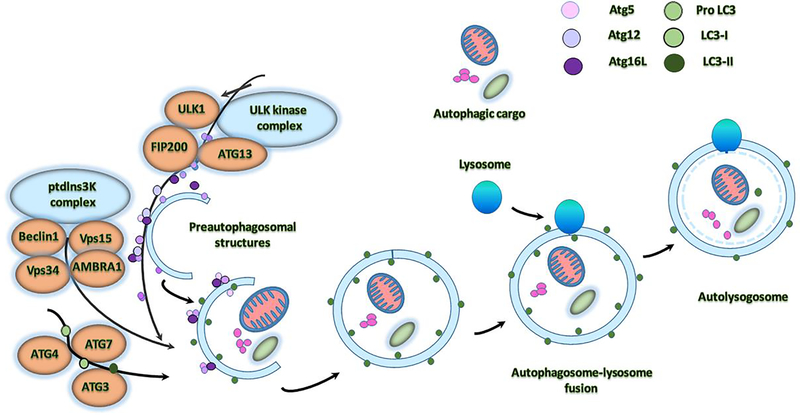
Mitophagy is nothing but removal of dead mitochondria by autophagy. Mitophagy is initiated by the formation of spherical structure double membrane ‘autophagosome’ (Figure 1). An autophagosome is a spherical structure with double layer membranes. It is the key structure in macroautophagy, the intracellular degradation system for cytoplasmic contents. After formation, autophagosomes deliver cytoplasmic components to the lysosomes (Glick et al., 2010; Tan et al., 2014). The outer membrane of an autophagosome fuses with a lysosome to form an autolysosome. The lysosome’s hydrolases degrade the autophagosome-delivered contents and its inner membrane. The formation of autophagosomes is controlled by Atg genes and LC3 complexes. The conjugate of Atg12-Atg5 also interacts with Atg16 to form larger complexes. Modification of Atg5 by Atg12 is essential for the elongation of the initial membrane. After the formation of the spherical structure, the complex of Atg12-Atg5:Atg16L1 dissociates from the autophagosome. LC3 is cleaved by ATG4 to generate LC3. LC3 cleavage is required for the terminal fusion of an autophagosome with its target membrane (Levine and Kroemer, 2008; Rajawat et al., 2009).
Figure 1. Molecular mechanism underlying autophagosome formation in normal cells.
The ULK complex consisting of ULK1/2–Atg13–FIP200–Atg101 is responsible for first initiation of autophagy, in response to certain signals. Formation of double-layered, membrane within the cytosol requires the PI3KC3 complex with contains Vps15–Vps34, Beclin-1, AMBRA1, and NBRF2. Next conjugation system is Atg, which add to the Atg5–Atg12–Atg16L complex, and LC3 conjugate system, which contains the Atg4, Atg7, Atg 3 to the elongating membrane along with the Pro LC3, LC3–1 and LC3–2. The membrane grows to enwrap a portion of the cytosol, forming an autophagosome. In the final step of the process, lysosomes fuse with the autophagosome, releasing lysosomal hydrolases into the interior, resulting in degradation of the vesicle contents.
In recent years, much progress has been made on this issue, and studies have suggested that several different organelles and potential membrane sources are the key to this process initiation (Stamatakou et al., 2020). These include the plasma membrane, the Golgi apparatus, the ER and mitochondria. Many studies reported that autophagosomes are formed by the ER-mitochondria in mammalian cells. Nearly 30–41 autophagy-related genes (Atg) are involved in autophagosome formation.
Autophagy process involves recruitment of the protein complexes to regulate the autophagy which include (1) ULK kinase complex-ULK1-ATG13-FIP200; (2) the phosphatidylinositol 3-kinase (Ptdlns3K) complex-Beclin1-ATG14-Vsp15-Vsp34-AMBRA1; (3) the Atg9-Atg2-Atg18 complex, and (4) Atg5-Atg12-Atg16 and Atg7/LC3 conjugation systems (Figure 1). This phagosome is ready to engulf the defective or damaged mitochondria with selective degradation through mitophagy (Reddy and Oliver, 2019).
5. Proteins Involved in Mitophagy
Recently, several groups extensively studied and discussed mitophagy types and proteins that are involved in mitophagy (Ding and Yin, 2012; Hirota, et al., 2012; Asrafi and Schwarz, 2013; Saita, et al., 2013; Pelligrino and Haynes, 2015; Cai and Thammineni, 2016; Burman et al., 2017; Lin et al., 2017; Pickles, et al., 2018; Safiulina, et al., 2018; Reddy, et al., 2018, Manczak, et al., 2018; Reddy and Oliver, 2019; Oliver Reddy, 2019; Cai and Jeong, 2020). These studies suggest that multiple pathways and several proteins are involved in mitophagy. The following proteins are important for mitophagy –Drp1, Fis1, Miro, Mfn1, Mfn2, Opa1, Ubiquitin, PINK1, Parkin, BNIP3, LC3, NIX, OPTN, FUNDC1, TBK1, SIAH1, and GP78. Among these, we describe important proteins for our article (Reddy and Oliver, 2019).
5.1. Drp1
Dynamin-related protein 1 (Drp1) is a fundamental component of mitochondrial fission and the key regulator of mitochondrial division in eukaryotic organisms. Drp1 is a dynamin-like GTPase, which mediates mitochondrial and peroxisomal division in a process dependent on self-assembly and coupled to GTP hydrolysis (Reddy et al., 2011; Ugarte-Uribe et al., 2014). In mitochondria fission, another GTPase called Drp1 is a mediator which facilitates mitochondrial fission, in apoptosis. Axonal transport of mitochondria allows the turnover of mitochondria in axons and axon terminals in neuron cells. Small GTPase proteins have been proposed recently as key regulators of mitochondrial motility (Kay et al., 2018). Mitochondrial Drp1 protein primarily, serves as a mitochondrial fission factor, inducing mitochondrial division, but when downregulated, indirectly promotes fusion (Osellame et al., 2012). Drp1 regulated fission may be used to excise depart damaged mitochondria for mitophagy, and reduction in Drp1 recruits parkin for excessive fission or fusion. Reports evidenced that Drp1 regulation will affect the mitochondrial axonal transport with fission or fusion exercise by promoting mitophagy mitochondrial dynamics.
5.1.2. Drp1 Structure
Drp1 protein is encoded by the Dnm1 gene, a member of the dynamin superfamily of GTPases, with 736 amino acids length with six different isoforms by peroxisome inhibition property. Drp1 has several splice variants: variant 1 consists of 736 amino acids with a calculated molecular mass of 81.6 kDa; in variant 2, exon 15 is spliced out and has a calculated molecular mass of 710 amino acids; in variant 3, exons 15 and 16 are spliced out and have a total of 699 amino acids; variant 4 has 725 amino acids; variant 5, 710 amino acids; and variant 6, 749 amino acids (Reddy et al., 2011). Similar to humans, mice have multiple variants, variant 1 consists of 712 amino acids, with a calculated molecular mass of 78.3 kDa; in variant 2, exon 3 is spliced out, and in variant 3, exons 15 and 16 are spliced out. It is possible that multiple splice variants are preset in mouse Drp1 (Reddy et al., 2011). The presence of a highly conserved GTPase domain in Drp1 indicates that Drp1 is important for several essential cellular functions.
5.1.3. Drp1 Function
Increasing evidence suggests that Drp1 is involved in several functions, including: 1) mitochondrial division, 2) mitochondrial distribution, 3) peroxisomal fragmentation, 4) phosphorylation, 5) ubiquitination, and 6) SUMOylation (Reddy et al 2011, Kandimalla and Reddy 2016).
Drp1 is primarily involved in mitochondrial division, a loss of Drp1 function increases mitochondrial fusion and mitochondrial connectivity. Drp1 interacts with Fis1 at the outer mitochondrial membrane, forms constriction and initiates mitochondrial division. Other than Drp1 receptors like Mff, Mid49 and Mid51 and Drp1 have been reported to be involved with fission process.
Drp1 activity is altered by its post-translational modifications like hyperphosphorylation activity. This affects the rate of mitochondrial fission and mitochondrial dynamics in cellular events. Drp1 has two major phosphorylated sites CDK S579, and PKA site is at S600 in Drp1 isoform 3. These sites are mostly responsible for hyperphosphorylation events of Drp1 (Kandimalla and Reddy, 2016; Oliver and Reddy, 2019).
As mentioned above, Drp1 is also involved in other modifications like S-nitrosylation, sumoylation, and ubiquitination, which alter the Drp1 activity. Drp1 has been shown to interact with toxic Aβ aggregates and p-tau intervening thought to play an important role in defective autophagy and mitophagy in AD. Drp1+Aβ and p-tau mediated autophagy and mitophagy exacerbating the disease and mitochondrial death promoted mitophagy in neurodegeneration with increased mitochondrial fission and reduced fusion in AD (Reddy and Oliver, 2019).
5.2. Fis1
Fission 1 (Fis1) is a mitochondrial division protein and plays a large role in mitochondrial dynamics and mitophagy. Fis1 encodes 150 amino acids with a molecular mass of 17 kDa (Reddy et al., 2011). Fis1 is a component of the mitochondrial complex that promotes mitochondrial fission. The fission protein Fis1 induced mitochondrial fragmentation and enhanced the formation of autophagosomes, which could enclose mitochondria (Figure 1). These changes correlated with mitochondrial dysfunction rather than with fragmentation, as substantiated by Fis1 mutants with different effects on organelle shape and function (Gomes and Scorrano, 2008). A recent study by Yu and colleagues revealed that human Fis1- mediated mitochondrial fragmentation occurs in the absence of Drp1 and Dyn2, suggesting that they are dispensable for human Fis1 function. Human Fis1 binds to Mfn1, Mfn2, and OPA1 and inhibits their GTPase activity, thus blocking the fusion machinery (Yu et al., 2019). They suggested a novel role for human Fis1 as an inhibitor of the fusion machinery, revealing an important functional evolutionary divergence between yeast and mammalian Fis1 proteins.
Overall, these studies indicate that Fis1 is involved in mitochondrial fission-fusion machinery and mitophagy activities.
5.3. Miro
Mitochondrial Rho GTPase protein miro is encoded by the Rhot1 gene on chromosome 17 plays a significant role in the mitochondrial transport. Miro protein is a member of the GTPase family and contains two isoforms Miro: Rhot1 (Miro1) and Rhot2 (Miro2) only expressed in the mitochondria where the mitochondrial transport is required (Tang 2015; Eberhardt, 2020; Fransson et al., 2006). Miro associates with Milton, forms a trimer complex and this complex contains multiple proteins, including trafficking kinesin protein 1/2 (TRAK1/2), motor protein kinesin and dynein to form the mitochondrial motor/adaptor complex. This complex helps to transport the mitochondrion via microtubules within cells by anterograde and retrograde transport (Cai and Thammineni, 2016).
Miro proteins are conserved one-way based transmembrane integral proteins with N-terminal regions exposed to the cytosolic side and consist of two GTPase domains separated by a pair of canonical EF hands (Klosowiak et al., 2013). These miro proteins are sensitive to Ca2+ binding with the EF hands resulting in stopping the stopping mobility of mitochondria on the microtubules structures. Ca2+ flows in and out in order to maintain a beneficial feedback cycle between Ca2+ buffering and mitochondrial dynamics that allows mitochondrial to the cellular events. Upon dysregulation of Ca2+ flow either side of the feedback mechanism causes signaling that leads both mitochondrial dysfunction and the activation of mitophagy events which can trap the cell by autophagosomes.
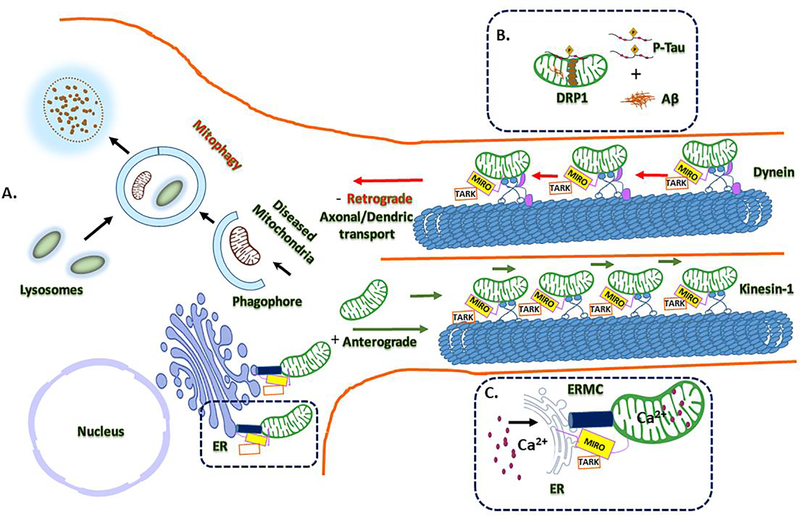
Miro, the mitochondrial outer membrane protein, involved in mitochondrial transport has recently gained great attention in mitochondrial biology. Studies reported that miro is involved in Ca2+ regulation at ER mitochondria encounter structures (ERMCS) and control of mitochondrial shape in response to Ca2+ (Csordás et al., 2018; Lee et al., 2018; Modi et al., 2019). Miro controls Ca2+ mitochondrial homeostasis at ERMCS by regulating the polo kinase-mediated phosphorylation of miro. Loss of Miro results in Ca2+ mitochondrial depletion and overexpression in mitochondrial Ca2+ burden to cell. The Polo/Miro based signaling pathway in the regulation of Ca2+ mitochondrial uptake in mitochondrial dynamics in this process PINK1 and leucine-rich repeat kinase 2 (LRRK2) are crucially involved. The increased mitochondrial Ca2+ levels, present in PINK1 and LRRK2 mutants, lead to mitochondrial swelling and eventually neuronal death observed in Parkinson diseases (Angelika and Hees, 2019). Miro-dependent mitochondrial shape transition maintains the Ca2+ homeostasis, this further validated by the mitochondrial quality control for autophagy and mitophagy by miro mediated Ca2+ sensing in mitochondrial dynamics (Nemani et al., 2018) (Figure 2).
Figure 2. A schematic illustrating of mitochondrial mitophagy events and mitochondria axonal transport.
(A) Microtubule-based transport of healthy mitochondria in neurons transfer from nucleus to cell soma (+) anterograde transportation through highlight interacting partners of kinesin-1-MIRO/TARK in three functional aspects. MIRO is the mediated by PINK-1/parkin and could be Ca2+ efflux is key for the transportation, which is mediated by the mitochondrial encounter structure (ERMES). (B) This attenuates mitochondrial mobility with Aβ and p-tau interactions foreword to the mitophagy of damaged mitochondria with abnormal mitochondrial dynamics. (C) Milton/TRAK and dynein mediates the retrograde axonal and dendritic transport. MIRO/TARK complex found to shown to be part of the ER- found at ERMES mitochondrial contact sites and showing Ca2+ exchange regulation in mitochondrial transport mechanism.
5.4. OPA1
The optic atrophy 1 (Opa1) is a mitochondrial dynamin like GTPase class, inner membrane protein which is essential for mitochondrial fusion. Opa1 protein is associated with different functions, such as maintenance of the mitochondrial respiratory chain, cristae organization, apoptosis control and balanced membrane potentials (Lee and Yoon, 2018; Wu et al., 2019; Ehses et al., 2018). Current studies support that mitochondrial inner membrane Opa1 associated with Mfn1 and Mfn2 mitochondrial outer membrane proteins is necessary for mitochondrial fusion and the process is regulated by proteolytic cleavage of Opa1 (Ali and McStay, 2018). Many different proteases are directly and indirectly involved in leading Opa1 processing associated with a number of cellular activities that includes matrix depended of inter-membrane ATPases. The proteases like presenilin-associated (PARL), high temperature requirement A2 protease (HTRA2) and overlapping activity of metalloendopeptidase 1(OMA1) (Jiang et al., 2014), are all associated with neurodegenerative diseases (Lebeau et al., 2018).
5.5. MFNs 1 and 2
Mitofusin1 and 2 proteins are encoded by the Mfn gene. These proteins are responsible for mitochondrial fusion. Mfn1 and Mfn2 have been implicated in regulating mitochondrial morphology (i.e., balancing fusion and fission events) and, when mutated, result in various diseases including AD (Filadi et al., 2018). Numerous studies suggest mitochondrial fusion is influenced by the pro-fission factors, such as Fis1 and the Mff interactions of Drp1 with Aβ and p-tau (Balog et al., 2016; Oliver and Reddy, 2019; Kandimalla et al., 2016). A number of papers state that dramatic remodeling of the mitochondrial network with a wide range of pathophysiological conditions, which include Ca2 + transfer inner and outer, apoptosis and autophagy leads to the progression of mitochondrial fission and fusion functions. Aβ and p-tau can also influence the regulation of mitochondrial dynamics through proteins such as Drp1, which are thought to alter processes of mitochondrial fission in AD and other neurological diseases (Qi et al., 2019). The activities of these GTPases proteins are highly regulated to control the mitochondrial morphology. These mechanisms include Aβ and p-tau induced defective autophagy and mitophagy in AD (Figure 3).
Figure 3.
A brief autophagy and mitophagy events in Alzheimer’s disease with key toxic players like Aβ and p-tau and Drp1 inducers that progress the activation of PINK1/parkin mediated abnormal mitochondrial dynamics.
6. Mitochondrial Autophagy
Mitochondrial autophagy (mitophagy) plays an essential role in response to a variety of pathological stimuli, which includes Aβ, and p-tau in AD. There are a number of mitophagy pathways that have been identified mainly on - 1. Aβ and p-tau-induced mitophagy, 2. Stress induced mitophagy 3. Receptor mediated PINK1-Parkin-mediated mitophagy, 4. Ubiquitin mediated mitophagy and 5. Basal mitophagy. Among all, receptor mediated PINK1-Parkin-mediated mitophagy is well studied and the best-understood mitophagy pathway (Cai and Jeong, 2020). We briefly describe these pathways below:
6.1. Aβ and p-Tau-induced Mitophagy
Major pathological hallmarks of AD include intracellular deposition of neurofibrillary tangles (NFTs), which were associated with paired helical filaments (PHF), p-tau, and extracellular accumulation of Aβ peptide in the senile plaques (Pradeepkiran et al., 2019a; Pradeepkiran et al., 2019b; Oliver and Reddy, 2019). Aβ is generated predominantly in a form containing 40–42 amino acids from APP on sequential cleavage by β- and γ- secretes. Indeed, Aβ is a neurotoxic peptide and intra- and extra-cellular accumulation of this peptide characterized by aggregation, insolubility, protease resistance and delayed turnover (Reddy and Beal, 2008).
The sequential functional modifications of the mitochondrial dysfunction, including inflammation, impaired energy metabolism, overwhelmed oxidative stress and synaptic dysfunction leads to cognitive decline (Reddy and Oliver, 2019). Accumulation of Aβ within mitochondria by interacting with mitochondrial proteins cause mitochondrial damage through ROS formation and mitochondrial dysfunction (Pagani and Eckert, 2011).
Recently, Reddy group found mitochondrial fission Drp1 interacts with Aβ monomers and oligomers in brain tissues from AD patients and APP mice, and these abnormal interactions are increased with disease progression. Their findings suggest that increased production of Aβ and the interaction of Aβ with Drp1 are crucial factors in mitochondrial fragmentation, abnormal mitochondrial dynamics, and synaptic damage in AD (Manczak et al., 2011). In another study, the Reddy lab (Manczak and Reddy, 2012) found P-tau interacts with Drp1 and p-tau brain tissues from three different lines of transgenic APP, APP/PS1, and 3XTg.AD mice and postmortem AD brains. Mitochondrial fission-linked GTPase Drp1 activity was significantly elevated. Based on these findings, they concluded that Drp1 interacts with Aβ and P-tau, likely leading to excessive mitochondrial fragmentation and mitochondrial and synaptic deficiencies.
Despite tremendous progress has been made in mitochondrial biology in AD, the precise pathophysiology of AD neurons in relation to the mitochondria-inflammatory responses remains elusive.
Overall, these observations strongly suggest that Aβ and p-tau-induced mitophagy is a major component in AD.
6.2. Stress Induced Mitophagy
Mitophagy that is induced by mild and transient stress is referred to as ‘stress induced mitophagy’. It is caused by several ways – ROS is the major factor that induces stress related mitophagy. Several recent findings reported that advanced glycation end products (AGEs) are largely involved in many human diseases, including diabetes mellitus, inflammation, renal failure, atherosclerosis, and neurodegenerative diseases (Glass et al., 2010; Pinkas and Aschner, 2016; Min et al., 2020). AGEs produce ROS that induce stress induced mitophagy. Both AGE and its receptor, advanced glycation end product receptor (RAGE) were expressed in astrocytes, microglia and neurons, a high level of RAGE expression was reported in AD brain samples. They induce ROS production; contribute significantly to stress induced mitophagy.
AGEs can influence mitochondrial dynamics by impairing fusion-fission balance, with a significant increment of mitochondrial fission in AD. This effect of AGEs may be attributable to their role in upregulating the expression of fission proteins Drp1 and Fis1 and down-regulating fusion proteins Mfn1, Mfn2, and Opa1. AGEs are also involved in abnormal APP processing and Aβ production via ROS.
AGEs are the end products of post-translational modifications of proteins, lipids and nucleic acids that should be facilitated by non-enzymatic reaction of reducing sugars and protein amino groups (Sharma et al., 2015). AGEs formation is accelerated under the oxidative stress, hypoxia, hyperglycemia and inflammation. The pathophysiological consequences of AGEs have been mediated by ROS, especially excessive superoxide and hydrogen peroxide production within the mitochondria. These pathological actions are mediated by cascade events of inflammation (Cepas et al., 2020), which induce lowered glucose consumption, lowered ATP levels, and down regulated mitochondrial activity that leads to neuronal cell death (Sivitz and Yorek, 2010).
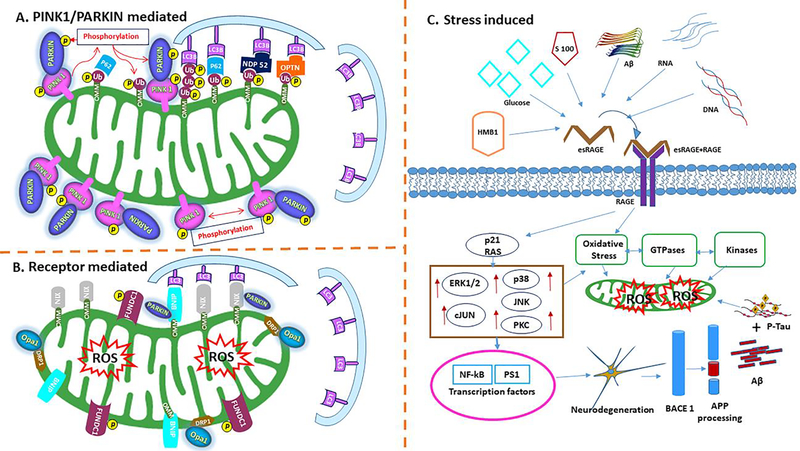
AGEs have been reported to impair the neuronal cells by cross-linking with the elevated kinases, GTPases and abnormal APP processing resulting increased Aβ deposition in AD (Li et al., 2013). Oxidative stress and neuronal inflammation caused by AGE-RAGE and/or RAGE-Aβ interactions lead to defective autophagy and mitophagy (Srikanth et al., 2011). Increased evidences of RAGE actions perturbation with Aβ interactions in neurons causes neuroinflammation and levies to increased Aβ levels in the brain by enhancing the rate of Aβ influx (Figure 4) (Stock et al., 2016). Additional aspects of AD via RAGE-Aβ interaction driven by overwhelmed Aβ and p-tau interactions and increased ROS production leads the stress-induced mitophagy in AD (Figure 5C).
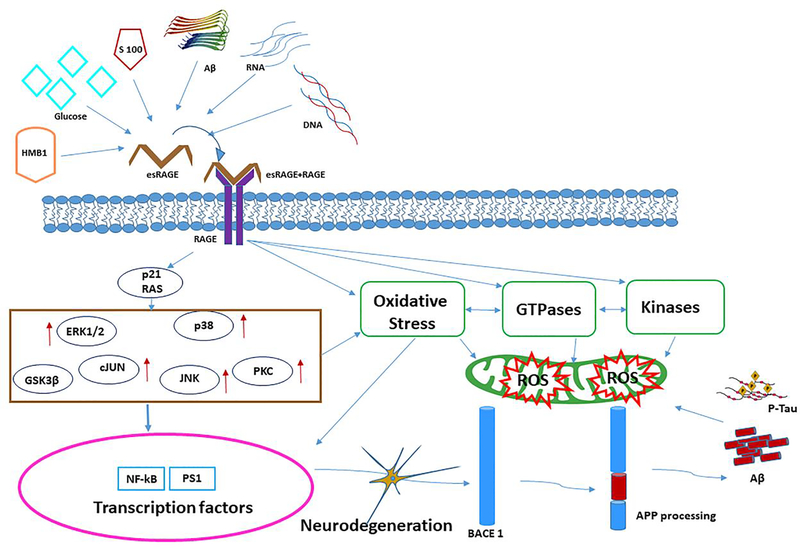
Figure 4. The extracellular and intracellular effects of AGEs induced ROS mechanism in AD.
The receptor for advanced glycation end products (RAGE) is a transmembrane, immunoglobulin-like receptor that exists and binds to ligands like HMB1, Glucose, S100, Aβ peptide, RNA, DNA. Ligand molecules binding at the extracellular domain of eRAGE initiates a complex with RAGE to form complex with esRAGE-RAGE and intracellular signaling cascade p21 RAS, stimulating NAD(P)H oxidase resulting in the production of reactive oxygen species (ROS). Up-regulation followed by ERK1/2, c-JUN, p38, JNK, PCK, GSK3β cellular events, and/or apoptosis with concomitant up regulation of and activation of NF-kB and PS1. The processing of APP has mainly focused on the correlation between RAGE activity and pathological conditions, such as BACE1 processing and Aβ accumulation and p-tau interaction in neuronal cells. The ROS formation influenced oxidative damage NADPH leads increased GTPases and kinases regulation to mitochondria mitophagy events leads neurodegeneration in RAGE mediated ROS signaling.
Figure 5.
(A) Mechanism of mitophagy with PINK1–parkin based. PINK1 recruited on outer mitochondria membrane (OMM) and promoting parkin recruitment. PINK1 phosphorylates and recruiting parkin and ubiquitinated several outer membrane components with activated phospho mechanism. Ubiquitinated chains are attached to the OMM and polymerized subsequently phosphorylated by PINK1 serving as signal for the autophagic machinery. Adaptor proteins (p62, OPTN, NDP52) recognize phosphorylated poly-Ub chains on mitochondrial proteins and initiate autophagosome formation through binding with LC3B.
(B). Receptor-mediated mitophagy. Specialized receptors, like NIX, BNIP3 and FUNDC1, expressed on the OMM in response to different stimuli. These receptors directly interact with LC3 to mediate mitochondrial elimination. NIX and BNIP3 phosphorylation enhances their association with LC3. FUNDC1 phosphorylation status, regulating mitochondrial dynamics during hypoxia. Mitophagy receptors promote fission of damaged organelles through the recruitment of DRP1, Opa1on the mitochondrial surface. Only parkin-dependent ubiquitination of NIX and BNIP3 mediated mitophagy in hypoxic condition.
(C). Stress-mediated mitophagy. Ligand molecules binding at the extracellular domain of receptor for advanced glycation end products (eRAGE) initiates a complex with RAGE to form complex with esRAGE-RAGE and intracellular signaling cascade p21 RAS, stimulating NAD(P)H oxidase resulting the production of reactive oxygen species (ROS). Upregulated ERK1/2, c-JUN, p38, JNK, PCK, GSK3β cellular events, and/or apoptosis with concomitant up regulation of and activation of NF-kB and PS1. Processing of APP, RAGE activity and BACE1 processing leads Aβ accumulation and p-tau interaction resulting increased oxidative stress on mitochondria leads abnormal mitochondrial defects in AD.
Overall, these observations indicate that stress induced mitophagy is common in aging, AD and other neurological diseases. The reduction of stress is a suggested therapeutic approach for defective mitophagy.
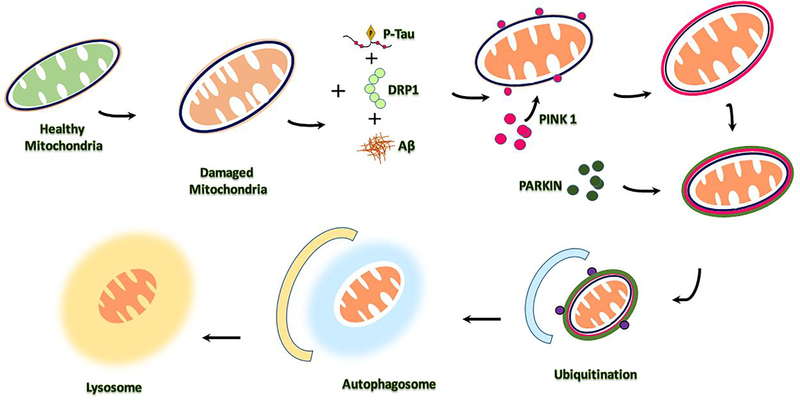
6.3. Receptor Mediated PINK1/Parkin Mitophagy
Receptor based putative kinase protein 1 (PINK1)-parkin-mediated mitophagy comes under ubiquitin-guided mitophagy. The PINK1/Parkin mitophagy is well investigated and a popular mitophagy in human diseases (Cai and Jeong 2020). In receptor mediated mitophagy, PINK1 protein stabilizes and promotes parkin recruitment to mitochondria to trigger the mitophagy actions (Vives-Bauza et al., 2010). The dysfunctional mitochondria tend to have depolarized membranes due to accumulated PINK1 on the outer membrane of mitochondria (OMM) and generates proton gradients across the inner mitochondrial membranes (IMM) (Sekine and Youle, 2018). This depolarization allows Pink1 kinase to accumulate at the OMM inability of the ETC to phosphorylate mitochondrial surface proteins, and activate parkin, an E3 ubiquitin ligase, through phosphorylation of ubiquitin. E3 ubiquitin ligase is activated by PINK1 and ubiquitinates several substrates in the outer mitochondrial membrane. Ubiquitination leads to the recruitment of several autophagy receptors to interact with ubiquitin chain, such as adaptor proteins (p62, OPTN, NDP52) and recognize the phosphorylated poly-ubiquitinated chains on mitochondria that will drive mitophagy by binding with LC3 (Figure 5A) (Chen et al., 2019). PINK1 mediated mitophagy triggered by proteins other than PINK1 and Parkin are NIX /BNIP3L, LC3 FUNDC1 to LC3 (Ye et al., 2015; Cai et a., 2012). These proteins mediate engulfment of damaged mitochondria by the autophagosome in PINK1/parkin-independent mitophagy in hypoxic condition (Yoo and Jung, 2018; Palikaras et al., 2018).
6.4. Ubiquitin Mediated Mitophagy
In ubiquitin mediated mitophagy, the protein kinase PINK1 and E3 ligase Parkin play key roles in building the ubiquitin chains on the outer surface of damaged mitochondria. The outer membrane proteins of mitochondria BNIP, NIX, initiate receptor dependent mitophagy and FUNDC1 are the receptor proteins that localize the autophagy machinery to the surface of damaged mitochondria (Hamacher-Brady and Brady, 2018; Han et al., 2020a; Han et al., 2020b). These receptor proteins bind directly to LC3 and mediate the mitochondrial elimination process. NIX protein plays an important role in mitochondrial differentiation in mitophagy by flagging that deficient NIX on outer membrane cells, leading to signaling of apoptosis process (Thomas and Ashcroft, 2019). Several studies reported that NIX associated with LC3 in stress conditions and BNIP3 involving in mitophagy through Opa1 release and Drp1 recruitment on mitochondrial surface (Zhang et al., 2016). BNIP3 and PINK1 stabilize the mitochondrial homeostasis by recruiting the parkin by interacting with PINK1-parkin mediated mitophagy. FUNDC1 receptor promotes the mitochondrial clearances during hypoxic condition; it interacts with ER and recruits the Drp1 during mitochondrial fragmentation. FUNDC1 is a good marker protein to identify the stress-mediated mitochondrial mitophagy (Figure 5B) (Wu et al., 2016).
6.5. Basal Mitophagy
Basal mitophagy is referred to as artificially induced mitophagy. It can be induced with chemicals and/or supplements. It is mostly studied in cell cultures and is characterized for altered components and proteins in cells. As reported, mitophagy is a process integral to normal mitochondrial network homeostasis maintained by the various factors (Reddy and Oliver, 2019). Rate of mitophagy varies by tissue type, suggesting a reasonable independent control of basal mitophagy. Basal mitophagy proceeds in the mammalian dopaminergic system in vivo in the absence of PINK1. Neural tissue has a high-energy demand and maintenance of mitochondrial homeostasis is essential to sustain this. Currently, the role of PINK1 in basal mitophagy is not completely understood. Therefore, it is important to study PINK1 knockout (KO) mice to understand the involvement PINK1 in basal mitophagy (McWilliams, et al., 2018).
Various human cell culture studies of heart, skeletal muscle, neuron and hepatic cells reported that PINK1-independent in chemically induced basal mitophagy. Tissue specific mitophagy activation and KO studies are required for further investigation of PINK1 and PINK1/Parkin aspects of basal mitophagy (Lee et al., 2018).
7. Reduced Drp1 as Mitophagy Therapeutic Target for Alzheimer’s disease
Impaired mitochondrial dynamics (increased fission-reduced fusion or vice versa) and defective mitophagy conditions contributes to many pathological states including the synaptic dysfunction and cognitive deficits by triggering Aβ and p-tau accumulation in disease process of AD (Wang et al., 2008; Wang et al., 2009; Manczak et al., 2010; Manczak et al., 2011; Calkins et al., 2011; Reddy et al., 2017; Reddy et al., 2018; Cai and Thammineni, 2016). Several others also extensively reported impaired mitochondrial dynamics and defective mitophagy in other neurodegenerative diseases such as Huntington’s (Song et al., 2011; Shirendeb et al., 2011; Shirendeb et al., 2012), Parkinson’s (Bueler 2009; Krebiehl et al., 2010; Wang et al., 2010) and ALS (Joshi et al., 2018; Kodavati et al., 2020). These studies strongly suggested reduced excessively fragmented mitochondria and enhanced mitophagy as therapeutic targets for these diseases. Therefore, a partial reduction of Drp1 is important and can be used to reduce excessively fragmented mitochondria and maintain and/or enhance mitophagy and autophagy events in AD and other neurodegenerative diseases. Figure 6 illustrates the Drp1 involvement in mitophagy in AD and other human diseases. In AD, we extensively studied the protective effects of reduced Drp1 as therapeutic target by 2 approaches – 1) genetic and 2) pharmacological.
Figure 6.
Factors contributing mitophagy players in response to a variety of pathological stimuli connected to centric protein, dynamin related protein 1 of neuronal cellular changes in AD.
7.1. Genetics approach
Using genetic crossings of Drp1 heterozygote knockout (Drp1+/−) mice with APP tg and Tau tg mice, we studied mitochondrial dynamics, biogenesis and synaptic alterations (Manczak et al., 2016; Kandimalla et al., 2016).
7.1.2. Drp1+/− X APP mice
We found reduced mRNA expressions and protein levels of Drp1,Fis1 (fission), CypD (matrix) genes, increased levels of Mfn1, Mfn2 and Opa1 (fusion), Nrf1, Nrf2, PGC1α, TFAM (biogenesis) and synaptophysin, PSD95, synapsin 1, synaptobrevin 1, neurogranin, GAP43 and synaptopodin (synaptic) were found in 6-month-old APP X Drp1+/− mice relative to APP mice (Manczak et al., 2016). Mitochondrial functional assays revealed that mitochondrial dysfunction is reduced in Drp1+/− X APP mice relative to APP mice, suggesting that reduced Drp1 reduces mitochondrial fragmentation, enhances mitochondrial fusion, mitochondrial function and synaptic activity in AD neurons. Sandwich ELISA assay revealed that soluble Aβ levels were significantly reduced in Drp1+/− X APP mice relative to APP mice, indicating that reduced Drp1 decreases soluble Aβ production in AD progression. These findings may have implications for the development of Drp1 based therapeutics for AD patients.
7.1.3. Drp1+/− X Tau Mice
Similar to APP mice with reduce Drp1, decreased mRNA and protein levels of fission (Drp1 and Fis1), matrix (CypD) and increased levels of fusion (Mfn1, Mfn2 and Opa1), mitochondrial biogenesis (Nrf1, Nrf2, PGC1α, TFAM), and synaptic genes (Nrf1, Nrf2, PGC1α, TFAM) were found in 6-month-old Drp1+/− X Tau mice relative to age-matched Tau mice (Kandimalla et al., 2016). Mitochondrial dysfunction was reduced in Drp1+/− X Tau mice. Phosphorylated Tau found to be reduced in Drp1+/− X Tau mice relative to Tau mice. These findings suggest that a partial reduction of Drp1 decreases the production of p-tau, reduces mitochondrial dysfunction, maintains mitochondrial dynamics, and enhances mitochondrial biogenesis and synaptic activity in Tau mice.
7.2. Pharmacological approach
Studies on AD
We sought to determine the protective effects of mitochondrial division inhibitor 1 (Mdivi-1) against Aβ- and mitochondrial fission protein, Drp1-induced excessive fragmentation of mitochondria in AD progression using cell cultures (Reddy et al., 2017). We also studied preventive (Mdivi1+Aβ42) and intervention (Aβ42+Mdivi1) effects against Aβ42 in N2a cells. Using real-time RT-PCR and immunoblotting analysis, mRNA and protein levels of mitochondrial dynamics, mitochondrial biogenesis, and synaptic genes were measured. Mitochondrial function was measured by H2O2, lipid peroxidation, cytochrome oxidase activity, and mitochondrial ATP. The cell viability was assessed using MTT assay. Aβ42 was found to impair mitochondrial dynamics, lower mitochondrial biogenesis, lower synaptic activity, and lower mitochondrial function. On the contrary, Mdivi-1 enhanced mitochondrial fusion activity, lowered fission machinery, and increased biogenesis and synaptic proteins. Mitochondrial function and cell viability were elevated in Mdivi-1-treated cells. Interestingly, Mdivi-1 pre- and post-treated cells treated with Aβ showed reduced mitochondrial dysfunction, and maintained cell viability, mitochondrial dynamics, mitochondrial biogenesis, and synaptic activity. The protective effects of Mdivi-1 were stronger in N2a+Aβ42 pre-treated with Mdivi-1, than in N2a+Aβ42 cells than Mdivi-1 post-treated cells, indicating that Mdivi-1 works better in prevention than treatment in AD like neurons.
Similar observations were found in a synergetic study of both Mdivi-1 and SS31 treated in N2a cells (Reddy et al., 2018). Significantly reduced levels of Aβ40 and Aβ42 were found in mutant AβPP cells treated with SS31, Mdivi-1, and SS31+Mdivi-1, and the reduction of Aβ42 levels were much higher in SS31+Mdivi-1 treated cells than individual treatments of SS31 and Mdivi-1. The levels of mtDNA copy number and cell survival were significantly increased in SS31, Mdivi1, and SS31+Mdivi-1 treated mutant AβPP cells; however, the increased levels of mtDNA copy number and cell survival were much higher in SS31+Mdivi-1 treated cells than individual treatments of SS31 and Mdivi-1. Mitochondrial dysfunction is significantly reduced in SS31, Mdivi-1, and SS31+Mdivi-1 treated mutant AβPP cells; however, the reduction is much higher in cells treated with both SS31+Mdvi-1.
In 2017, two groups independently studied the protective effects of Mdivi-1 in 2 different mouse models of AD. Baek and colleagues (2017) treated APP/PS1 mice with Mdivi-1 and studied cognitive behavior, mitochondrial fragmentation, mitochondrial transport and synaptic activities. They found that mice untreated with Mdivi-1 improved cognitive behavior and prevented mitochondrial fragmentation. They also noticed that the levels of lipid peroxidation, BACE1 expression, and Aβ deposition were reduced in APP/PS1 mice treated with Mdivi-1 (Baek et al., 2017). In another study, Wang and colleagues (2017) also studied the effect of Mdivi-1 on mitochondrial dynamics, particularly fission in CRND8 mice (tg APP strain). In Mdivi-1 treated 3-months-old CRND8 mice, they found rescued mitochondrial fragmentation and distribution deficits and improved mitochondrial function. The mitochondrial dynamic deficits, Aβ1–42/Aβ1–40 ratio and Aβ levels were reduced by Mdivi-1 treatment. Further, cognitive deficits were reduced in Mdivi-1 treated CRND8 mice relative to Mdivi-1-untreated CRND8 mice (Wang et al., 2017).
Overall, these studies indicate that Mdivi-1 reduced mitochondrial fragmentation, Aβ levels, improved synaptic function and ameliorated cognitive decline in disease process of AD mice.
Studies on HD and PD
Several others found reduced mitochondrial fragmentation and enhanced mitochondrial fusion, biogenesis and mitochondrial function in striatal progenitor cells (STHDhQ111/Q111) of HD knockin mice in HD and dopaminergic neuronal cells in PD treated with Mdivi-1 (Yin et al., 2016; Cui et al., 2010; Rapplod et al., 2014; Fan et al., 2019).
Overall, findings from cell culture and mouse model studies of AD, PD and HD revealed that Mdivi-1 reduces excessive fragmentation of mitochondria, enhances mitochondrial fusion, mitochondrial biogenesis, mitochondrial function and mitophagy.
8. Mitophagy Enhancers as new Therapeutic Targets for Alzheimer’s disease
Increasing evidence suggests that accumulation of defective mitochondria, primarily due to impaired mitophagy is involved in AD and other neurodegenerative diseases (Xie et al., 2019; Reddy and Oliver 2019). Recent evidence from our lab suggests that defective mitophagy is an early event in the disease process of AD (Reddy et al., 2004; Manczak et al., 2006 and Manczak et al., 2018). Age-dependent increased production and accumulation of Aβ and p-tau and their abnormal interactions with Drp1, VDAC, CypD and ABAD, cause excessive fragmentation of mitochondria, impaired mitochondrial dynamics and defective mitophagy, ultimately leading to synaptic and cognitive dysfunction in AD (Lustbader et al., 2004; Du et al., 2008; Manczak et al., 2011; Manczak and Reddy 2012). These studies also suggest that reduced levels of Drp1, VDAC, CypD and ABAD may inhibit abnormal interactions with Aβ and p-tau and maintain quality of mitochondria and mitophagy in AD.
The other important factor is that increase and/or restoration of mitophagy with pharmacological approach, in other words, treat AD cells, AD mice and AD patients mitophagy enhancers. Currently, several mitophagy enhancers, urolithin, tomatidine, NAD+ riboside and others are being investigated in cell cultures and AD mouse models. The outcome of these studies will provide new information. Several other drugs/molecules like rapamycin, torin1, perhexiline, niclosamide, rottlerin, targeting by attenuate mTORC1 (AMP-activated protein) helped to enhance mitochondrial biogenesis in disease states (Balgi et al., 2009; Fang et al., 2019; Lou et al., 2020). Natural compounds such as resveratrol and quercetin have beneficial effects by increasing mitophagy transcriptome in cardiac and hepatic cells (Yu et al., 2016). A recent study reported that resveratrol reduces defective mitophagy by activating mitochondrial biogenesis (Malek et al., 2018). Rosmarinic acid was also reported to attenuate insulin resistance in rat skeletal muscle (Naoi et al., 2019). However, further research on identification of mitophagy enhancers is still needed in order to develop potential mitophagy enhancers.
9. Conclusions and Future Directions
The molecular and cellular mechanisms of mitophagy have been extensively studied in the past. However, mitophagy deficit has only been recognized recently as a key player involved in aging, AD and other neurodegenerative diseases. Several studies revealed that in both early- and late-onset AD, defective mitophagy contributes to synaptic dysfunction and cognitive deficits by triggering Aβ and p-tau accumulations. It is therefore imperative to understand the molecular basis of Aβ and p-tau-induced defective mitophagy in AD and other neurodegenerative diseases. Our lab has been focusing on mitochondrial biology and/or pathological aspects of AD for the last 20 years, including (1) the defective mitochondrial biogenesis and impaired mitochondrial dynamics; (2) interactions between Aβ and Drp1, p-tau, Drp1 and defective mitophagy and (3) small molecule therapeutic interventions and genetic crossing studies on rescue and/or prevention of defective mitophagy in AD. In terms of therapeutics of mitophagy in AD, both genetic and pharmacological approaches are possible options: 1) genetic crossings of Drp+/− mice with Aβ and tau mice; 2) treating Aβ and tau mice with small molecules such as Mdivi-1, SS31, curcumin, DDQ and other mitophagy enhancers (Olive and Reddy, 2019; Fang et al., 2019, Kuruva et al., 2017) and validating the defective mitophagy and cellular events in both disease and normal state. We published several articles on abnormal mitochondrial interactions between Aβ and Drp1, p-tau and Drp1 leading to defective autophagy and mitophagy proteins in AD. These abnormal interactions lead to increased mitochondrial fragmentation in mitophagy (PINK1, parkin, P62, BNIP3, FUNDC1, LC3–1, OPTN, TBK1), autophagy (ATG proteins, LC3-I and II), and ubiquitination (SIAH1, Gp78, MUL1, ARIH1, SMURF1) fusion and fission process involving important genes such as Drp1, Fis1, Miro, Opa1, Mfn1, and Mfn2. Currently, we and others are focusing on identification of mitophagy modulators, lysosomal activators, mitochondrial protectors, small molecules such as SS31, Mdivi-1 and DDQ to protect mitochondria by enhancing mitochondrial quality, mitochondrial dynamics and mitophagy and autophagy in AD and other neurological diseases. However, further research is still needed for 1) better understanding the biology of mitophagy/autophagy both in normal and disease states and 2) to test the currently available mitophagy/autophagy enhancers using preclinical rodent models.
Highlights.
Healthy mitochondria are essential for functional bioenergetics, cellular functions and balanced homeostasis.
Mitophagy is a cellular process whereby damaged mitochondria are selectively removed.
In Alzheimer’s disease, toxic amyloid beta and hyperphosphorylated tau formation and accumulation, mitochondrial damage, are the early cellular events.
Abnormal interactions between Aβ and Drp1 and p-Tau and Drp1, causes defective mitophagy and autophagy in AD,
Reduced Drp1 enhances mitophagy and autophagy in AD.
Acknowledgements
Authors sincerely thank Ms. Hallie Morton for critical reading of the manuscript.
Funding: The research presented in this article was supported by NIH grants AG042178, AG047812, NS105473, AG060767, AG069333 (to PHR).
List of Abbreviations
- AD
Alzheimer’s disease
- ROS
Reactive Oxygen Species
- mtDNA
Mitochondrial DNA
- Aβ
Amyloid beta
- Drp1
Dynamin-1-like protein
- ER
Endoplasmic reticulum
- TEM
Transmission electron microscopy
- Fis1
Fission 1 protein
- Miro
Mitochondrial Rho GTPase
- Opa1
Optic atrophy 1
- Mfn1
Mitofusin-1
- Mfn2
Mitofusin-2
- Mid49
Mitochondrial dynamics protein MID49
- Mid51
Mitochondrial dynamics protein MID51
- Mff
Mitochondrial fission factor
- PGC-1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- NRF1
Nuclear respiratory factor 1
- NRF2
Nuclear respiratory factor 2
- TFAM
Transcription factor A, mitochondrial
- mAPP
Mutant amyloid beta precursor protein
- PINK1
PTEN-induced kinase 1
- BNIP3
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3
- LC3
Microtubule-associated protein 1A/1B-light chain 3
- OPTN
Optineurin
- FUNDC1
FUN14 domain containing 1
- TBK1
TANK-binding kinase 1
- SIAH1
E3 ubiquitin-protein ligase SIAH1
- GSK3β
Glycogen synthase kinase 3 beta
- CDK
Cyclin-dependent kinases
- PKA
Protein kinase A
- EF
Helix-loop-helix structural domain
- ERMCS
ER mitochondria encounter structures
- LRRK2
Leucine-rich repeat kinase 2
- PARL
Proteases like presenilin-associated
- OMA1
Metalloendopeptidase 1
- AGEs
Advanced glycation end products
- RAGE
Receptor for advanced glycation end products
- OMM
Outer membrane of mitochondria
- ETC
Electron transport chain
- SIRT1
Sirtuin 1
- SS-31
Szeto Schiller peptide 31
- Mdivi-1
Mitochondrial division inhibitor 1
Footnotes
Conflicts of Interest: The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad T, Aggarwal K, Pattnaik B, Mukherjee S, Sethi T, Tiwari BK, Kumar M, Micheal A, Mabalirajan U, Ghosh B, Sinha Roy S, Agrawal A 2013Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis 4:e461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, McStay GP 2018. Regulation of Mitochondrial Dynamics by Proteolytic Processing and Protein Turnover. Antioxidants (Basel) 7, pii: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amakiri N, Kubosumi A, Tran J, Reddy PH 2019. Amyloid Beta and MicroRNAs in Alzheimer’s Disease. Front Neurosci. 13, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelika BH, Hees JT 2019. Calcium Dysregulation and Mitochondrial Dysfunction Form A Vicious Cycle in Parkinson’s Disease. American Journal of Biomedical Science & Research 5, 246–249. [Google Scholar]
- Ashrafi G, Schwarz TL 2013. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ, 20, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader V, Winklhofer KF 2020. Mitochondria at the interface between neurodegeneration and neuroinflammation. Semin Cell Dev Biol 99, 163–171. [DOI] [PubMed] [Google Scholar]
- Baek SH, Park SJ, Jeong JI, Kim SH, Kyung JW, Baik SH, Choi Y, Choy BY, Park JS, Bahm G, Shin JH, Jo DS, Lee JY, Jang CG, Arumugam TV, Kim J, Han JW, Koh JY, Cho DH, Jo DG 2017. Inhibition of Drp1 Ameliorates Synaptic Depression, Aβ Deposition, and Cognitive Impairment in an Alzheimer’s Disease Model. J Neurosci. 37, 5099–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balgi AD, Fonseca BD/, Donohue E, Tsang TC, Lajoie P, Proud CG, Nabi IR, Roberge M 2009. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4:e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balog J, Mehta SL, Vemuganti R, 2016. Mitochondrial fission and fusion in secondary brain damage after CNS insults. J Cereb Blood Flow Metab 36, 2022–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bido S, Soria FN, Fan RZ, Bezard E, Tieu K 2017. Mitochondrial division inhibitor-1 is neuroprotective in the A53T-α-synuclein rat model of Parkinson’s disease. Sci Rep 7, 7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boengler K, Kosiol M, Mayr M, Schulz R, Rohrbach S. 2017. Mitochondria and ageing: role in heart, skeletal muscle and adipose tissue. J Cachexia Sarcopenia Muscle. 8, 349–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongarzone S, Savickas V, Luzi F, Gee AD 2017. Targeting the Receptor for Advanced Glycation Endproducts (RAGE): A Medicinal Chemistry Perspective. J Med Chem 60, 7213–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragoszewski P, Turek M, Chacinska A 2017. Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol. 7, pii: 170007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitzig MT, Alleyn MD, Lockey RF, Kolliputi N. 2018. A mitochondrial delicacy: dynamin related protein 1 and mitochondrial dynamics. Am J Physiol Cell Physiol.315, C80–C90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büeler H 2009. Impaired mitochondrial dynamics and function in the pathogenesis of Parkinson’s disease. Exp Neurol. 218, 235–246. [DOI] [PubMed] [Google Scholar]
- Buettner GR 2011. Superoxide dismutase in redox biology: the roles of superoxide and hydrogen peroxide. Anticancer Agents Med Chem. 11, 341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HT, Shaw JM 2013. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol. 23, R891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman JL, Pickles S, Wang C, et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J Cell Biol. 2017;216(10):3231–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Jeong YY 2020. Mitophagy in Alzheimer’s Disease and Other Age-Related Neurodegenerative Diseases. Cells. 9, pii: E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Tammineni P 2016. Alterations in Mitochondrial Quality Control in Alzheimer’s Disease. Front Cell Neurosci. 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Tammineni P 2017. Mitochondrial Aspects of Synaptic Dysfunction in Alzheimer’s Disease. J Alzheimers Dis.57, 1087–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A, Sheng ZH. 2012. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 22,545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins MJ, Manczak M, Mao P, Shirendeb U, Reddy PH 2011. Impaired mitochondrial biogenesis, defective axonal transport of mitochondria, abnormal mitochondrial dynamics and synaptic degeneration in a mouse model of Alzheimer’s disease. Hum Mol Genet 20, 4515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso SM, Santana I, Swerdlow RH, Oliveira CR 2004. Mitochondria dysfunction of Alzheimer’s disease cybrids enhances Abeta toxicity. J Neurochem, 89, 1417–1426. [DOI] [PubMed] [Google Scholar]
- Cenini G, Lloret A, Cascella R 2019. Oxidative Stress in Neurodegenerative Diseases: From a Mitochondrial Point of View. Oxid Med Cell Longev 2105607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepas V, Collino M, Mayo JC, Sainz RM 2020. Redox Signaling and Advanced Glycation Endproducts (AGEs) in Diet-Related Diseases. Antioxidants (Basel) 9, pii: E142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty A, Jetto CT, Manjithaya R 2019. Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front Aging Neurosci 11, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Chen YH, Huang TY 2019. Ubiquitin-mediated regulation of autophagy. J Biomed Sci. 26, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CH, Lin CC, Yang MC, et al. 2012. GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS One.7, e49112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A, Salvioli S 2001. Analysis of mitochondria during cell death. Methods Cell Biol. 63, 467–486. [DOI] [PubMed] [Google Scholar]
- Csordas G, Weaver D, Hajnoczky G 2018. Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol 28, 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Tang X, Christian WV, Yoon Y, Tieu K 2010. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J Biol Chem 285, 11740–11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalli P, Mitic T, Caporali A, Lauriola A, D’Arca, ROS D, 2016. Cell Senescence, and Novel Molecular Mechanisms in Aging and Age-Related Diseases. Oxid Med Cell Longev 3565127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane R, Singh I, Sagare AP, Bell RD, Ross NT, LaRue B, Love R, Perry S, Paquette N, Deane RJ, Thiyagarajan M, Zarcone T, Fritz G, Friedman AE, Miller BL, Zlokovic BV 2012. A multimodal RAGE-specific inhibitor reduces amyloid β mediated brain disorder in a mouse model of Alzheimer disease. J Clin Invest 122, 1377–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Yin XM 2012. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 393:547–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guo L, Fang F, et al. 2008. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer’s disease. Nat Med. 14, 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS 2010. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 107, 18670–18675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt EL, Ludlam AV, Tan Z, Cianfrocco MA 2020. Miro: A molecular switch at the center of mitochondrial regulation. Protein Sci. doi: 10.1002/pro.3839 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T 2009. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol 187, 1023–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CS, Holzbaur EL 2020. Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. Elife 9 pii: e50260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan RZ, Guo M, Luo S, Cui M, Tieu K 2019. Exosome release and neuropathology induced by α-synuclein: new insights into protective mechanisms of Drp1 inhibition. Acta Neuropathol Commun 7, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, et al. 2019. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat Neurosci 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filadi R, Pendin D, Pizzo P 2018. Mitofusin 2: from functions to disease. Cell Death Dis 9, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenström P 2006. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun 344, 500–10. [DOI] [PubMed] [Google Scholar]
- Fritsch LE, Moore ME, Sarraf SA, Pickrell AM 2020. Ubiquitin and Receptor-Dependent Mitophagy Pathways and Their Implication in Neurodegeneration. J Mol Biol 432, 2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch LE, Moore ME, Sarraf SA, Pickrell AM 2020. Ubiquitin and Receptor-Dependent Mitophagy Pathways and Their Implication in Neurodegeneration. J Mol Biol. 432(8):2510–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EK, Reddy PH 2019. Can Healthy Diets, Regular Exercise, and Better Lifestyle Delay the Progression of Dementia in Elderly Individuals?. J Alzheimers Dis, 72, S37–S58. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH 2010. Mechanisms underlying inflammation in neurodegeneration. Cell, 140, 918–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF 2010. Autophagy: cellular and molecular mechanisms. J Pathol 221, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Scorrano L 2008. High levels of Fis1, a pro-fission mitochondrial protein, trigger autophagy. Biochim Biophys Acta.1777, 860–866. [DOI] [PubMed] [Google Scholar]
- Hamacher-Brady A, Brady NR 2016. Mitophagy programs: mechanisms and physiological implications of mitochondrial targeting by autophagy. Cell Mol Life Sci, 73, 775–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Jeong YY, Sheshadri P, Su X, Cai Q 2020a. Mitophagy regulates integrity of mitochondria at synapses and is critical for synaptic maintenance [published online ahead of print, 2020 Jul 6]. EMBO Rep. 2020;e201949801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Jeong YY, Sheshadri P, Cai Q 2020bMitophagy coordination with retrograde transport ensures the integrity of synaptic mitochondria [published online ahead of print, 2020 Aug 19]. Autophagy. 10.1080/15548627.2020.1810919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Ordureau A, Heo JM 2018. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol, 19, 93–108. [DOI] [PubMed] [Google Scholar]
- Harper JW, Schulman BA 2006. Structural complexity in ubiquitin recognition. Cell. 124, 1133–1136. [DOI] [PubMed] [Google Scholar]
- Hirota Y, Kang D, Kanki T 2012. The physiological role of mitophagy: new insights into phosphorylation events. Int J Cell Biol, 2012:354914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellinger KA 2010. Basic mechanisms of neurodegeneration: a critical update. J Cell Mol Med 14, 457–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Jiang H, Shen Z, Wang X 2014. Activation of mitochondrial protease OMA1 by Bax and Bak promotes cytochrome c release during apoptosis. Proc Natl Acad Sci U S A 111, 14782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AU, Saw NL, Vogel H, Cunnigham AD, Shamloo M, Mochly-rosen D 2018. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Molecular Medicine, 10, e8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla R, Manczak M, Fry D, Suneetha Y, Sesaki H, Reddy PH, 2016. Reduced dynamin-related protein 1 protects against phosphorylated Tau-induced mitochondrial dysfunction and synaptic damage in Alzheimer’s disease. Hum Mol Genet, 25, 4881–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandimalla R, Manczak M, Yin X, Wang R, Reddy PH 2018. Hippocampal phosphorylated tau induced cognitive decline, dendritic spine loss and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum Mol Genet, 27, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay L, Pienaar IS, Cooray R, Black G, Soundararajan M 2018. Understanding Miro GTPases: Implications in the Treatment of Neurodegenerative Disorders. Mol Neurobiol 55, 7352–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, Fang EF 2017. Mitophagy and Alzheimer’s Disease: Cellular and Molecular Mechanisms. Trends Neurosci 40, 151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DK, Mook-Jung I 2019. The role of cell type-specific mitochondrial dysfunction in the pathogenesis of Alzheimer’s disease. BMB Rep. 52, 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosowiak JL, Focia PJ, Chakravarthy S, Landahl EC, Freymann DM, Rice SE 2013. Structural coupling of the EF hand and C-terminal GTPase domains in the mitochondrial protein Miro. EMBO Rep 14, 968–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodavati M, Wang H, Hegde ML 2020. Altered Mitochondrial Dynamics in Motor Neuron Disease: An Emerging Perspective. Cells. 9, 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H Yoshihara H, Hirokawa T, Endo T, Fon EA, Trempe JF, Saeki Y, Tanaka K, Matsuda N 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–6. [DOI] [PubMed] [Google Scholar]
- Krebiehl G, Ruckerbauer S, Burbulla LF, et al. 2010. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One; 5(2):e9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebiehl G, Ruckerbauer S, Burbulla LF, et al. 2010. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One 5, e9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuruva CS, Manczak M, Yin X, Ogunmokun G, Reddy AP and Reddy PH 2017. Aqua-soluble DDQ reduces the levels of Drp1 and Aβ and inhibits abnormal interactions between Aβ and Drp1 and protects Alzheimer’s disease neurons from Aβ- and Drp1-induced mitochondrial and synaptic toxicities. Hum Mol Genet. 26, 3375–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau J, Rainbolt TK, Wiseman RL 2018. Coordinating Mitochondrial Biology Through the Stress-Responsive Regulation of Mitochondrial Proteases. Int Rev Cell Mol Biol 340, 79–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Yoon Y 2016. Mitochondrial fission and fusion. Biochem Soc Trans 44, 1725–1735. [DOI] [PubMed] [Google Scholar]
- Lee H, Yoon Y 2018. Mitochondrial Membrane Dynamics-Functional Positioning of OPA1. Antioxidants (Basel) 7, pii: E186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Sanchez-Martinez A, Zarate AM, Benincá C, Mayor U, Clague MJ, Whitworth AJ 2018. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J Cell Biol, 217, 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Huh S, Lee S, Wu Z, Kim AK, Kang HY, Lu B, 2018. Altered ER mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc Natl Acad Sci U S A 115, E8844–E8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Levine B, Debnath J 2018. Autophagy and the cell biology of age-related disease. Nat Cell Biol. 20, 1338–1348. [DOI] [PubMed] [Google Scholar]
- Levine B Klionsky DJ 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroeme r G. 2008. Autophagy in the pathogenesis of disease. Cell 132, 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2019. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XH, Du LL, Cheng XS, Jiang X, Zhang Y, Lv BL, Liu R, Wang JZ, Zhou XW 2013. Glycation exacerbates the neuronal toxicity of β-amyloid. Cell Death Dis 4:e673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M 2004. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 119(6):873–887. [DOI] [PubMed] [Google Scholar]
- Lin MY, Cheng XT, Xie Y, Cai Q, Sheng ZH 2017. Removing dysfunctional mitochondria from axons independent of mitophagy under pathophysiological conditions. Autophagy, 13, 1792–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MY, Cheng XT, Xie Y, Cai Q, Sheng ZH 2017. Removing dysfunctional mitochondria from axons independent of mitophagy under pathophysiological conditions. Autophagy 13, 1792–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chan DC. 2015. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol Biol Cell, 26, 4466–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Liao X, Wu H, Li Y, Zhu Y, Chen Q 2020. Mitophagy and Its Contribution to Metabolic and Aging-Associated Disorders. Antioxid Redox Signal, 32, 906–927. [DOI] [PubMed] [Google Scholar]
- Liu YJ, McIntyre RL, Janssens GE, Houtkooper RH 2020. Mitochondrial fission and fusion: A dynamic role in aging and potential target for age-related disease. Mech Ageing Dev, 186, 111212. [DOI] [PubMed] [Google Scholar]
- Lou G, Palikaras K, Lautrup S, Scheibye-Knudsen M, Tavernarakis N, Fang EF 2020. Mitophagy and Neuroprotection. Trends Mol Med, 26, 8–20. [DOI] [PubMed] [Google Scholar]
- Lustbader JW, Cirilli M, Lin C, et al. 2004. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 304, 448–452. [DOI] [PubMed] [Google Scholar]
- Malek MH, Hüttemann M, Lee I 2018. Mitochondrial Structure, Function, and Dynamics: The Common Thread across Organs, Disease, and Aging. Oxid Med Cell Longev, 1863414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH 2006. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet, 15, 1437–1449. [DOI] [PubMed] [Google Scholar]
- Manczak M, Calkins MJ, Reddy PH 2011. Impaired mitochondrial dynamics and abnormal interaction of amyloid beta with mitochondrial protein Drp1 in neurons from patients with Alzheimer’s disease: implications for neuronal damage. Hum Mol Genet, 20, 2495–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Kandimalla R, Fry D, Sesaki H, Reddy PH 2016. Protective effects of reduced dynamin-related protein 1 against amyloid beta-induced mitochondrial dysfunction and synaptic damage in Alzheimer’s disease. Hum Mol Genet. 25, 5148–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Kandimalla R, Yin X, Reddy PH 2018. Hippocampal mutant APP and amyloid beta induced cognitive decline, dendritic spine loss, defective autophagy, mitophagy and mitochondrial abnormalities in a mouse model of Alzheimer’s disease. Hum Mol Genet, 27:1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Kandimalla R, Yin X, Reddy PH 2019. Mitochondrial division inhibitor 1 reduces dynamin-related protein 1 and mitochondrial fission activity [published correction appears in Hum Mol Genet. 2019 Mar 1;28(5):875–876]. Hum Mol Genet. 28(2):177–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Mao P, Calkins MJ, Cornea A, Reddy AP, Murphy MP, Szeto HH, Park B, Reddy PH. 2010. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer’s disease neurons. J Alzheimers Dis. 20 Suppl 2(Suppl 2):S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Mao P, Calkins MJ, et al. 2010. Mitochondria-targeted antioxidants protect against amyloid-beta toxicity in Alzheimer’s disease neurons. J Alzheimers Dis;20 Suppl 2(Suppl 2):S609–S631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Reddy PH 2012. Abnormal interaction between the mitochondrial fission protein Drp1 and hyperphosphorylated tau in Alzheimer’s disease neurons: implications for mitochondrial dysfunction and neuronal damage. Hum Mol Genet. 21(11):2538–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Reddy PH 2015. Mitochondrial division inhibitor 1 protects against mutant huntingtin-induced abnormal mitochondrial dynamics and neuronal damage in Huntington’s disease. Hum Mol Genet, 24, 7308–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manczak M, Sesaki H, Kageyama Y, Reddy PH 2012. Dynamin-related protein heterozygote knockout mice do not have synaptic and mitochondrial deficiencies. Biochim Biophys Acta. 1822, 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao P, Reddy PH 2011. Aging and amyloid beta-induced oxidative DNA damage and mitochondrial dysfunction in Alzheimer’s disease: implications for early intervention and therapeutics. Biochim Biophys Acta; 1812, 1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Montava-Garriga L, Ball G, Singh F, Barini E, Muqit MMK, Brooks SP, Ganley IG 2018. Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab 27, 439–449.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min LJ, Iwanami J, Shudou M, Bai HY, Shan BS, Higaki A, Mogi M, Horiuchi M 2020. Deterioration of cognitive function after transient cerebral ischemia with amyloid-β infusion-possible amelioration of cognitive function by AT2 receptor activation. J. Neuroinflammation 17, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S, López-Doménech G, Halff EF, Covill-Cooke C, Ivankovic D, Melandri D, Arancibia-Cárcamo IL, Burden JJ, Lowe AR, Kittler JT 2019. Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat Commun 10, 4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP 2009. How mitochondria produce reactive oxygen species. Biochem J. 417, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoi M, Wu Y, Shamoto-Nagai M, Maruyama W 2019. Mitochondria in Neuroprotection by Phytochemicals: Bioactive Polyphenols Modulate Mitochondrial Apoptosis System, Function and Structure. Int J Mol Sci 20, pii: E2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani N, Carvalho E, Tomar D, Dong Z, Ketschek A, Breves SL, et al. 2018, MIRO-1 Determines Mitochondrial Shape Transition upon GPCR Activation and Ca2+ Stress. Cell Rep 23, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D, Reddy PH 2019. Dynamics of Dynamin-Related Protein 1 in Alzheimer’s Disease and Other Neurodegenerative Diseases. Cells 8, pii: E961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DMA, Reddy PH, 2019. Molecular Basis of Alzheimer’s Disease: Focus on Mitochondria. J Alzheimers Dis 72, S95–S116. [DOI] [PubMed] [Google Scholar]
- Oliver DMA, Reddy PH, 2019. Small Molecules as Therapeutic Drugs for Alzheimer’s Disease. Mol Cell Neurosci 96, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Heo JM, Duda DM et al. 2015. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A; 112, 6637–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Singh AP, Stroud DA, et al. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J Cell Sci. 2016; 129(11):2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Blacker TS, Duchen MR 2012. Cellular and molecular mechanisms of mitochondrial function. Best Pract Res Clin Endocrinol Metab 26, 711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani L, Eckert A 2011. Amyloid-Beta interaction with mitochondria. Int J Alzheimers Dis, 925050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N 2018. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol, 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- Pellegrino MW, Haynes CM 2015. Mitophagy and the mitochondrial unfolded protein response in neurodegeneration and bacterial infection. BMC Biol. 13:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Gao P, Shi L, Chen L, Liu J, Long J 2020. Central and Peripheral Metabolic Defects Contribute to the Pathogenesis of Alzheimer’s Disease: Targeting Mitochondria for Diagnosis and Prevention. Antioxid Redox Signal. 32, 1188–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Vigié P, Youle RJ 2018. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr Biol. 28, R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkas A, Aschner M 2016. Advanced Glycation End-Products and Their Receptors: Related Pathologies, Recent Therapeutic Strategies, and a Potential Model for Future Neurodegeneration Studies. Chem Res Toxicol 29, 707–14. [DOI] [PubMed] [Google Scholar]
- Pradeepkiran JA, Reddy AP, Reddy PH 2019. Pharmacophore-based models for therapeutic drugs against phosphorylated tau in Alzheimer’s disease. Drug Discov Today 24, 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeepkiran JA, Reddy PH 2019. Structure Based Design and Molecular Docking Studies for Phosphorylated Tau Inhibitors in Alzheimer’s Disease. Cells 8, pii: E260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Huang Z, Xie F, Chen L 2019. Dynamin-related protein 1: A critical protein in the pathogenesis of neural system dysfunctions and neurodegenerative diseases. J Cell Physiol 234, 10032–10046. [DOI] [PubMed] [Google Scholar]
- Rajawat Y, Hilioti Z, Bossis I 2010. Autophagy: a target for retinoic acids. Autophagy. 6, 1224–1226. [DOI] [PubMed] [Google Scholar]
- Rajawat YS, Bossis I 2008. Autophagy in aging and in neurodegenerative disorders. Hormones (Athens) 7 46–61. [DOI] [PubMed] [Google Scholar]
- Rajawat YS, Hilioti Z, Bossis I 2009. Aging: central role for autophagy and the lysosomal degradative system. Ageing Res Rev 8,199–213. [DOI] [PubMed] [Google Scholar]
- Rappold PM, Cui M, Grima JC, et al. 2014. Drp1 inhibition attenuates neurotoxicity and dopamine release deficits in vivo. Nat Commun. 5:5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold PM, Cui M, Grima JC, et al. 2014. Drp1 inhibition attenuates neurotoxicity and dopamine release deficits in vivo. Nat Commun. 5, 5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH 2006. Amyloid Precursor Protein mediated free radicals and oxidative damage: Implications for progression and pathogenesis of Alzheimer’s disease. J Neurochem. 96 1–13. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Beal MF 2008. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer’s disease. Trends Mol Med 14, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Manczak M, Mao P, Calkins MJ, Reddy AP, Shirendeb U 2010. Amyloid beta and mitochondria in aging and Alzheimer’s disease: implications for synaptic damage and cognitive decline. J Alzheimers Dis. 20 Suppl 2, S499–S512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Manczak M, Yin X, et al. 2016. Protective effects of a natural product, curcumin, against amyloid β induced mitochondrial and synaptic toxicities in Alzheimer’s disease. J Investig Med. 64, 1220–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Manczak M, Yin X, Reddy AP 2018. Synergistic Protective Effects of Mitochondrial Division Inhibitor 1 and Mitochondria-Targeted Small Peptide SS31 in Alzheimer’s Disease. J Alzheimers Dis. 62, 1549–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, McWeeney S, Park BS, et al. 2004. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer’s disease. Hum Mol Genet, 13, 1225–1240. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Oliver DM 2019. Amyloid Beta and Phosphorylated Tau-Induced Defective Autophagy and Mitophagy in Alzheimer’s Disease. Cells, 8, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Oliver DM 2019. Amyloid Beta and Phosphorylated Tau-Induced Defective Autophagy and Mitophagy in Alzheimer’s Disease. Cells 8, pii: E488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Reddy TP, Manczak M, Calkins MJ, Shirendeb U, Mao P 2011. Dynamin related protein 1 and mitochondrial fragmentation in neurodegenerative diseases. Brain Res Rev 67, 103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Tonk S, Kumar S, et al. 2017. A critical evaluation of neuroprotective and neurodegenerative MicroRNAs in Alzheimer’s disease. Biochem Biophys Res Commun. 483(4):1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Tripathi R, Troung Q, et al. 2012. Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer’s disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim Biophys Acta.1822, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PH, Yin X, Manczak M, Kumar S, Pradeepkiran JA, Vijayan M, Reddy AP 2018. Mutant APP and amyloid beta-induced defective autophagy, mitophagy, mitochondrial structural and functional changes and synaptic damage in hippocampal neurons from Alzheimer’s disease. Hum Mol Genet 27, 2502–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Agujetas V, de Dios C, Lestón L, Marí M, Morales A, Colell A 2019. Recent Insights into the Mitochondrial Role in Autophagy and Its Regulation by Oxidative Stress. Oxid Med Cell Longev 3809308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Santoro A, Salvioli S 2017. Mitochondria and mitochondria-induced signalling molecules as longevity determinants. Mech Ageing Dev.165, 115–128. [DOI] [PubMed] [Google Scholar]
- Ross JM, Olson L, Coppotelli G 2015. Mitochondrial and Ubiquitin Proteasome System Dysfunction in Ageing and Disease: Two Sides of the Same Coin? Int J Mol Sci 16, 19458–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzepka R, Dołęgowska B, Rajewska A, Kwiatkowski S 2014. On the significance of new biochemical markers for the diagnosis of premature labour. Mediators Inflamm 251451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiulina D, Kuum M, Choubey V, et al. 2019. Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J; 38(2):e99384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saita S, Shirane M, Nakayama KI 2013. Selective escape of proteins from the mitochondria during mitophagy. Nat Commun, 4:1410. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K, Kauppinen A, et al. 2013. Impaired autophagy and APP processing in Alzheimer’s disease: The potential role of Beclin 1 interactome. Prog Neurobiol.106 107, 33–54. [DOI] [PubMed] [Google Scholar]
- Salvioli S, Olivieri F, Marchegiani F et al. 2006. Genes, ageing and longevity in humans: problems, advantages and perspectives. Free Radic Res. 40, 1303–1323. [DOI] [PubMed] [Google Scholar]
- Santoro A, Salvioli S, Raule N, et al. 2006. Mitochondrial DNA involvement in human longevity. Biochim Biophys Acta.1757,1388–1399. [DOI] [PubMed] [Google Scholar]
- Santos RX, Correia SC, Wang X, et al. 2010. A synergistic dysfunction of mitochondrial fission/fusion dynamics and mitophagy in Alzheimer’s disease. J Alzheimers Dis. 20, S401–S412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, et al. 2013. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 496, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Harper JW 2009. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol; 10, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Youle RJ 2010. Mitochondrial fission and fusion. Essays Biochem 47:85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Youle RJ 2018. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol 16, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ 2001. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 81, 741–766. [DOI] [PubMed] [Google Scholar]
- Seo AY, Joseph AM, Dutta D, Hwang JC, Aris JP, Leeuwenburgh C 2010. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci 123, 2533–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Smith HJ, Yao P, Mair WB 2019. Causal roles of mitochondrial dynamics in longevity and healthy aging. EMBO Rep 20, e48395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Kaur A, Thind SS, Singh B, Raina S 2015. Advanced glycation End-products (AGEs): an emerging concern for processed food industries. J Food Sci Technol 52, 7561–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng SP, Lei B, James ML, et al. 2012. Xenon neuroprotection in experimental stroke: interactions with hypothermia and intracerebral hemorrhage. Anesthesiology, 117(6):1262–1275. [DOI] [PubMed] [Google Scholar]
- Sheng ZH, Cai Q 2012. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci; 13, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirendeb U, Reddy AP, Manczak M, et al. 2011. Abnormal mitochondrial dynamics, mitochondrial loss and mutant huntingtin oligomers in Huntington’s disease: implications for selective neuronal damage. Hum Mol Genet. 20, 1438–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirendeb UP, Calkins MJ, Manczak M, et al. 2012. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum Mol Genet; 21, 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DF, Selfridge JE, Lu J, et al. 2013. Bioenergetic flux, mitochondrial mass and mitochondrial morphology dynamics in AD and MCI cybrid cell lines. Hum Mol Genet, 22, 3931–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz WI, Yorek MA 2010. Mitochondrial dysfunction in diabetes: from molecular mechanisms to functional significance and therapeutic opportunities. Antioxid Redox Signal 12, 537–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Chen J, Petrilli A, et al. 2011. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med; 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth V, Maczurek A, Phan T, Steele M, Westcott B, Juskiw D, Münch G 2011. Advanced glycation endproducts and their receptor RAGE in Alzheimer’s disease. Neurobiol Aging 32, 763–77. [DOI] [PubMed] [Google Scholar]
- Stamatakou E, Wróbel L, Hill SM, Puri C, Son SM, Fujimaki M, Zhu Y, Siddiqi F 2020. Fernandez-Estevez M, Manni MM, Park SJ, Villeneuve J, Rubinsztein DC. Mendelian neurodegenerative disease genes involved in autophagy. Cell Discov 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AJ, Kasus-Jacobi A, Wren JD, Sjoelund VH, Prestwich GD, Pereira HA 2016. The Role of Neutrophil Proteins on the Amyloid Beta-RAGE Axis. PLoS One 11, e0163330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe DF, Camara AK 2009. Mitochondrial reactive oxygen species production in excitable cells: modulators of mitochondrial and cell function. Antioxid Redox Signal 11, 1373–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow RH 2009. The neurodegenerative mitochondriopathies. J Alzheimers Dis.17, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammineni P, Cai Q 2017. Defective retrograde transport impairs autophagic clearance in Alzheimer disease neurons. Autophagy.13, 982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CC, Yu JT, Tan MS, Jiang T, Zhu XC, Tan L 2014. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging 35, 941–57. [DOI] [PubMed] [Google Scholar]
- Tang BL 2015. MIRO GTPases in Mitochondrial Transport, Homeostasis and Pathology. Cells 5, pii: E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezze C, Romanello V, Desbats MA, et al. 2017. Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence. Cell Metab.25, 1374–1389.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LW 2019. Ashcroft M. Exploring the molecular interface between hypoxia-inducible factor signalling and mitochondria. Cell Mol Life Sci 76, 1759–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarte-Uribe B, Müller HM, Otsuki M, Nickel W, García-Sáez AJ 2014. Dynamin related protein 1 (Drp1) promotes structural intermediates of membrane division. J Biol Chem 289, 30645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valero T 2014. Mitochondrial biogenesis: pharmacological approaches. Curr Pharm Des. 20, 5507–5509. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J et al. 2010. PINK1 dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107, 378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yin J, Ma X, et al. 2017. Inhibition of mitochondrial fragmentation protects against Alzheimer’s disease in rodent model. Hum Mol Genet, 26, 4118–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yin J, Ma X, Zhao F, Siedlak SL, Wang Z, Torres S, Fujioka H, Xu Y, Perry G, Zhu X 2017. Inhibition of mitochondrial fragmentation protects against Alzheimer’s disease in rodent model. Hum Mol Genet. 26, 4118–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhao F, Ma X, Perry G, Zhu X 2020. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener 15, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Petrie TG, Liu Y, Liu J, Fujioka H, Zhu X 2012. Parkinson’s disease-associated DJ-1 mutations impair mitochondrial dynamics and cause mitochondrial dysfunction. J Neurochem 121, 830–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Lee HG, et al. 2009. Impaired balance of mitochondrial fission and fusion in Alzheimer’s disease. J Neurosci; 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Liu W, et al. 2011. DLP1-dependent mitochondrial fragmentation mediates 1-methyl-4-phenylpyridinium toxicity in neurons: implications for Parkinson’s disease. Aging Cell, 10, 807–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Siedlak SL, et al. 2008. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A; 105, 19318–19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X 2009. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Neurochem, 109 Suppl 1(Suppl 1):153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Zheng L, Perry G, Smith MA, Zhu X 2009. The role of abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Neurochem 109, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han R, Liang ZQ, et al. 2008. An autophagic mechanism is involved in apoptotic death of rat striatal neurons induced by the non-N-methyl-D-aspartate receptor agonist kainic acid. Autophagy. 4, 214–226. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu N, Lu B 2019. Mechanisms and roles of mitophagy in neurodegenerative diseases. CNS Neurosci Ther 25, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qin ZH 2013. Coordination of autophagy with other cellular activities. Acta Pharmacol Sin.34, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Su B, Fujioka H, Zhu X 2008. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am J Pathol.173, 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TA, Reichert AS 2010. Impaired quality control of mitochondria: aging from a new perspective. Exp Gerontol 45, 503–11. [DOI] [PubMed] [Google Scholar]
- Williams JA, Zhao K, Jin S, Ding WX 2017. New methods for monitoring mitochondrial biogenesis and mitophagy in vitro and in vivo. Exp Biol Med(Maywood). 242:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lin C, Wu K, Jiang L, Wang X, Li W et al. 2016. FUNDC1 regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO J 35,1368–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Zhao D, Shah SZA, Zhang X, Lai M, Yang D, et al. , 2019. OPA1 overexpression ameliorates mitochondrial cristae remodeling, mitochondrial dysfunction, and neuronal apoptosis in prion diseases. Cell Death Dis 10, 710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Chen M, Jiang J 2019. Mitochondrial dysfunction in neurodegenerative diseases and drug targets via apoptotic signaling. Mitochondrion 49, 35–45. [DOI] [PubMed] [Google Scholar]
- Xie C, Aman Y, Adriaanse BA, et al. Culprit or Bystander: Defective Mitophagy I Alzheimer’s Disease. Front Cell Dev Biol. 2020;7:391 Published 2020 Jan 17. doi: 10.3389/fcell.2019.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Manczak M, Reddy PH 2016. Mitochondria-targeted molecules MitoQ and SS31 reduce mutant huntingtin-induced mitochondrial toxicity and synaptic damage in Huntington’s disease. Hum Mol Genet. 25, 1739–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Sun X, Starovoytov V, Cai Q 2015. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer’s disease patient brains. Hum Mol Genet. 24, 2938–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SM, Jung YK 2018. A Molecular Approach to Mitophagy and Mitochondrial Dynamics. Mol Cells 41, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younessi P, Yoonessi A 2011. Advanced glycation end-products and their receptor-mediated roles: inflammation and oxidative stress. Iran J Med Sci 36, 154–66. [PMC free article] [PubMed] [Google Scholar]
- Yu R, Jin SB, Lendahl U, Nistér M, Zhao J 2019. Human Fis1 regulates mitochondrial dynamics through inhibition of the fusion machinery. EMBO J. 38, e99748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Liu T, Jin SB, et al. 2017. MIEF1/2 function as adaptors to recruit Drp1 to mitochondria and regulate the association of Drp1 with Mff. Sci Rep.7, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Xu Y, Zhang S, Sun J, Liu P, Xiao L, Tang Y, Liu L, Yao P 2016. Quercetin Attenuates Chronic Ethanol-Induced Hepatic Mitochondrial Damage through Enhanced Mitophagy. Nutrients 8, pii: E27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Xue L, Li L, Tang C, Wan Z, Wang R et al. 2016. BNIP3 Protein Suppresses PINK1 Kinase Proteolytic Cleavage to Promote Mitophagy. J Biol Chem 291, 21616–21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XD, Wang Y, Wang Y, et al. 2009. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy 5, 339–350. [DOI] [PubMed] [Google Scholar]
- Zhu X, Perry G, Smith MA, Wang X 2013. Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J Alzheimers Dis, 33 Suppl 1(0 1):S253–S262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorova LD, Popkov VA, Plotnikov EY, et al. 2018. Mitochondrial membrane potential. Anal Biochem, 552:50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]