Abstract
Multiple myeloma (MM) is a hematological malignancy characterized by abnormal plasma cell proliferation within the bone marrow which leads to progressive bone marrow failure, skeletal osteolytic lesions, and renal insufficiency, thus severely affecting the quality of life. MM is always preceded by monoclonal gammopathy of uncertain significance (MGUS), which progresses to asymptomatic-MM (aMM) or symptomatic-MM (sMM) at a rate of 1% per year. Despite impressive progress in the therapy of the disease, MM remains incurable. Based on these premises, the identification of biomarkers of MGUS progression to MM is a crucial issue in disease management. In this regard, exosomes (EXs) and their precious biomolecular cargo could play a pivotal role in MM detection, stratification, and follow-up. Raman spectroscopy, a label- and manipulation-free technique, and its enhanced version, surface-enhanced Raman spectroscopy (SERS), have been used for characterizing MGUS, aMM, and sMM patient-derived EXs. Here, we have demonstrated the capability of Raman spectroscopy for discriminating EXs along the progression from MGUS to aMM and sMM, thus providing useful clinical indications for patient care. The used SERS devices, based on random nanostructures, have shown good potential in terms of sensitivity, but further developments are needed for achieving reproducible and quantitative SERS results.
Introduction
Multiple myeloma (MM) accounts for more than 30,000 new blood cancer cases in the US alone each year, and it is responsible for about 2% of all cancer deaths.1,2 It is a hematological malignancy characterized by abnormal monoclonal plasma cell proliferation and typical clinical features such as hypercalcemia, osteolytic bone lesions, anemia, and renal failure.3 MM is always preceded by monoclonal gammopathy of uncertain significance (MGUS), which progresses to asymptomatic-MM (aMM, which requires close clinical and laboratory follow-up) or symptomatic-MM (sMM, which requires treatment) at the rate of 1% per year, while the aMM progression to sMM is about 10% per year.4
Even if MM outcome has extremely improved since the advent of biological agents (proteasome inhibitors and immunomodulatory drugs) and the introduction of monoclonal antibodies in clinical practice,5 prognostic stratification for the identification of those patient categories at higher risk to progress to sMM and/or to experience a poor outcome still plays a pivotal role for better patient care.6
MM onset from MGUS is the result of a complex interplay between aberrant plasma cell clone and a permissive bone marrow microenvironment (BMM) that sustains MM cell expansion.7 Among BMM-derived factors involved in MM pathogenesis, exosomes (EXs) have been recently considered as central players in the cross-talk between malignant plasma and BMM cells.8 Indeed, they facilitate material and, by consequence, information transfer between the cells releasing EXs and the target cells.9 These microvesicles, represented in all biofluids, such as blood, plasma, serum, urine, saliva, amnion, and cerebrospinal fluid, are characterized by a dimension ranging from 30 to 150 nm. To date, EXs have been shown to be involved in many aspects of MM pathogenesis and progression, such as escape mechanism regulation, drug resistance, and survival promotion as well as angiogenesis and osteolysis induction.8
In light of this evidence, this “vesicular dialogue” in MM patients deserves a specific consideration and requires careful evaluation and monitoring along all different phases of MM development and progression. “Omics” techniques have allowed identifying specific and detailed molecular EX components, even if these analytical methods rely on complicated and time-consuming protocols10,11 and, notably, are unfeasible for assays that require a large amount of EXs or in which their amount is low.12 Therefore, new techniques able to identify and to characterize exosomes during pathological routes could be of paramount importance in cancer monitoring.
Vibrational spectroscopies hold great potential to overcome these protocol limits. Raman spectroscopy (RS) is a non-invasive, label-free technique based on the inelastic scattering of laser light due to the interaction of photons with molecular vibrations. RS can get biochemical information from the whole biological sample. In the last decade, this spectroscopic technique has been largely proposed for cancer detection, characterization, and prognosis.13−15 In addition, RS is becoming increasingly interesting for liquid biopsy of circulating biomolecules, notably for cancer diagnostics.16
The relatively low Raman spectroscopy signal, when needed, can be enhanced with the use of metal nanoparticles, such as gold or silver, giving rise to surface-enhanced Raman scattering (SERS). The combination of geometry and materials can produce enhancements as high as 1014, which makes this technique sensitive enough to reveal single molecules lying on a surface.17,18 Because of its huge Raman signal amplification, SERS has been used for EX detection and characterization.14,19
In this work, exosomes extracted from MM patient’s serum, as well as their asymptomatic forms MGUS and aMM, have been collected and characterized first by RS and later on by SERS. Principal component analysis (PCA) was used to analyze Raman and SERS spectra. RS was able to discriminate EXs derived from the three plasma cell dyscrasia conditions (MGUS, aMM, and sMM), emphasizing the importance of this technique as a new tool for MM patient stratification.
We also compared Raman results with SERS micro-spectroscopy, although limited to cheap SERS substrates based on random nanostructures. This choice is dictated by two concerns: (i) the comparison is made between two techniques which are cost-similar and (ii) the literature widely deals with non-uniform, random SERS substrates and/or nanoparticles because of their availability. We achieved satisfactory enhancements by SERS, which constitute a good result toward few/single-molecule detection in bioclinical assays, but we also found that SERS spectra have great variations even for the EXs derived from one single individual. This irreproducibility of SERS spectra, due to the randomness of the substrate, does not lead to a fully quantitative analysis, limiting our SERS devices to qualitative dissertations only.
On the other hand, it is worthy to notice that the Raman results presented herein show, for the first time to our knowledge, the capability to discriminate between the different phases of MM disease by optical spectroscopy. Above all, the discrimination between the two asymptomatic forms, MGUS and aMM, can potentially lead to the development of a very helpful tool for clinical decisions.
Results and Discussion
Exosomes Characterization
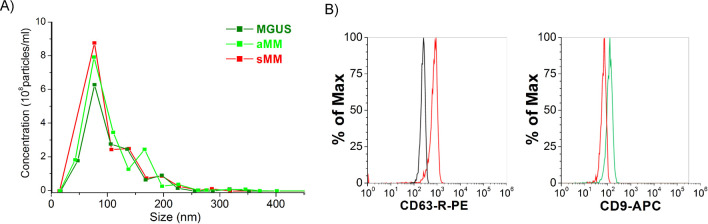
Exosomes were extracted from the patient’s sera as reported in the Materials and Method section and, later on, characterized for both their size and markers expression. The average size measured for all EX samples was around 98 ± 46 nm, as reported in Figure 1A, ranging in the typical EX range. Further characterization of the EXs has been conducted by flow cytometry in order to investigate the expression of two out of three of the classical markers, such as CD9 and CD63.20 83% of the whole EX population extracted by the miRCURY Exosome Isolation Kit was positive for CD63, and 33% of the CD63+ one was characterized by further positivity for CD9 (Figure 1B). This double vesicular characterization “certified” us to state, as far as we know, that the extracted vesicles can be identified as EXs since they respect two important parameters (size and marker expressions) set up by the International Society for Extracellular Vesicles.21
Figure 1.
Characterization of purified MM EXs by miRCURY Exosome Isolation Kit (Qiagen). MM EXs have been characterized by their size (A) and their marker expression (B). (A) EX size measurements performed by NanoSight showed as 98.9 ± 46.3 nm for all measured nano-vesicles. (B) Typical tetraspanin protein markers, CD63 and CD9, have been also used as proof of the presence of EXs within the sample fractions collected. 83% of the extracted EXs were positive for CD63. 33% of the CD63+ has shown positivity for CD9s.
Exosome Screening by Raman Spectroscopy
Samples of MGUS-, aMM-, and sMM-derived exosomes have been measured by Raman spectroscopy in the 750–1700 cm–1 range with an 830 nm laser source, recording at least 10 spectra for each sample. Subsequently, all Raman spectra have been averaged by group, and MGUS, aMM, and sMM average curves are reported in Figure 2A with grey shaded areas as standard deviations. The three curves are very similar to each other, and all of them exhibit the major Raman peaks/bands typical of cellular biochemistry: a sharp, intense peak at 1003 cm–1 corresponding to the breathing vibration of phenylalanine; a broadband at 1240–1340 cm–1 with vibrations from Amide III, nucleic acids, and several fatty acids; a broad peak centered at about 1445 cm–1 coming from CH deformations in lipids (closer to 1440 cm–1) or proteins (closer to 1450 cm–1); and the Raman band at 1640–1700 cm–1 comprising the Amide I vibration of proteins and the C=C stretches of lipids.
Figure 2.
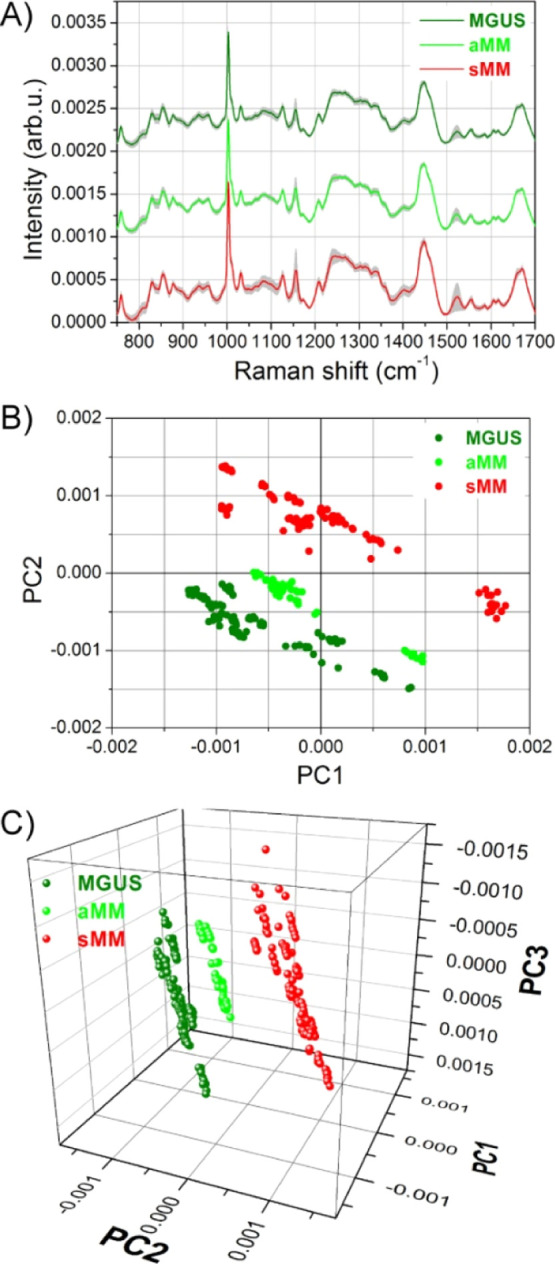
Raman spectra and PCA analysis of different exosomes groups. (A) Average curves of Raman spectra collected on exosomes derived from MGUS, aMM, and sMM patients. (B) PC2 vs PC1 scores plots for the three exosome groups. (C) 3D scatter plots of PC1, PC2, and PC3 scores produced by PCA analysis.
To highlight differences between MGUS, aMM, and sMM spectra, a PCA has been performed on the whole dataset. The first three principal components (PCs) account for nearly 90% of the total variance, with 60.4% for PC1, 20.5% for PC2, and 8.6% for PC3. The subsequent PCs mainly account for noisy variations and do not contain significant spectral features. Figure 2B shows the scatter plot of PC2 versus PC1 scores. A clear separation between sMM exosomes on one side and MGUS-aMM exosomes on the other side is evident. We can observe that the main separation occurs along the PC2 axis, while the PC1 axis is spanned by all three groups of exosomes. A 3D scatter plot of PC1, PC2, and PC3 scores is shown in Figure 2C, where the score distribution along the PC3 axis is put in evidence. As it occurs for PC1, PC3 axis also does not discriminate between the different exosome populations. Indeed, all MGUS, aMM, and sMM scores are well distributed among positive and negative values of PC3.
In Figure 2B, the scores from MGUS, aMM, and sMM exosomes roughly lay along diagonal directions parallel to each other. Hence, the direction perpendicular to these diagonal lines is the best suited for discriminating between the different groups of exosomes. To this purpose, a rotation matrix is applied to the PC axes computed by PCA. The applied rotation lays in the PC2-PC1 plane and maps the old PCs to a new set of rot.PCs (rot.PCs is a short notation used in the following for rotated-PCs). The angle of rotation, 32° counterclockwise, has been chosen so that the new rot.PC2 is the axis best dividing the exosome scores (Figure 3A). This axes rotation has the advantage that, among the new rot.PCs, the rot.PC2 alone is solely responsible for the discrimination between the exosome groups. A detailed analysis of the loading curve of rot.PC2 (Figure S2) allows for identifying which spectral regions positively contribute to MM scores and which ones are instead more expressed in the aMM and MGUS exosomes. Figure 3B shows the average spectra where reddish shaded areas indicate the spectral frequencies characteristic of sMM samples, while the greenish areas are typical of MGUS and aMM samples. Among all biomolecules contributing to the MM EX discrimination (nucleic acids, polysaccharide, carbohydrate, and β-carotene), the lipid content showed a major contribution. Fatty acid signal and the saturation degree of their chains (spectral regions at 1150–1166 and 1200–1350 cm–1, respectively),22,23 as well as the ceramide presence (1650–1685 cm–1 spectral region),24 are mainly overexpressed in the cancer EX population. Moreover, the Raman band at 1440–1450 cm–1, usually used for discriminating between lipid and protein (as reported above), is leaning here to the lipid region (1440 cm–1).23
Figure 3.
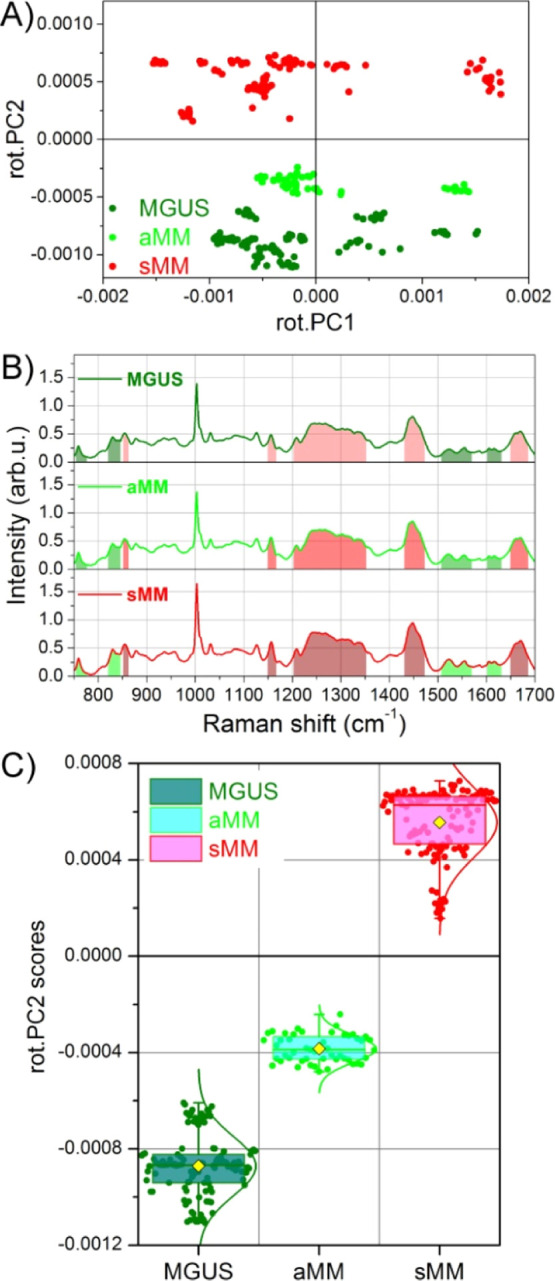
Fine tuning of PCA results and exosome screening. (A) Rotation matrix is applied to PC axes directly computed by PCA. Consequently a novel set of axes is found, named rot.PCs (after rotated PCs), and the rot.PC2 vs rot.PC1 scores plot is shown. (B) After analyzing the rot.PC2 loading curve, the spectral regions mostly contributing to sMM scores (positive values) and those contributing to MGUS scores (negative values) are, respectively, evidenced as reddish and greenish shaded regions. (C) Rot.PC2 scores of panel (A) are reported as a box-chart for MGUS, aMM, and sMM exosomes, where the yellow symbols indicate the average values and the solid curves are the Gaussian distributions. The boxes instead indicate the 25th, 50th, and 75th percentiles of each data group.
The screening potential, and consequently the promise for a diagnostic tool, is better appreciated if the rotated scores of Figure 3A are translated in a quantitative representation by means of a box plot (Figure 3C). In this plot, the box limits are, respectively, the 25th, 50th, and 75th percentiles of each group, the yellow full-symbol indicates the mean score value, and the solid curve is the Gaussian distribution of the scores data. While the difference between sMM and aMM-MGUS scores is well clear even in the scatter plot of Figure 3A, this box plot representation adequately helps to distinguish also the much closer aMM and MGUS populations. More in detail, the mean values of aMM and MGUS data are, respectively, −3.8 × 10–4 and −8.7 × 10–4 score units of rot.PC2, while the corresponding data groups are well separated both at levels of 25–75th percentiles and by 1 standard deviation (not shown for clarity sake).
Even if the biochemical characterization of EXs goes beyond the aim of this work, some of the biochemical variations reported here might be interesting for all professionals working in the field of EXs. Rot.PC2 loads revealed that the differences between EXs from the asymptomatic and symptomatic phases are mainly due to spectral features typical of lipids and their saturation degree. This result is in agreement with the recent literature reporting that lipids have a huge role in EX formation, composition, and the execution of their signaling roles.24,25 Moreover, Record et al., have shown that EXs contain two main ceramide species (C18:0 and C24:1) and that these lipid molecules are very abundant within EXs.26 This specific lipid composition might facilitate EX internalization, increase the lipid content of the recipient cell,27 and play a crucial role in several pathways of the immune response.26,27
SERS Spectroscopy Results
As several authors have already reported,19,28−30 SERS microspectroscopy has a potential added value in the characterization of exosomes and/or other microvesicles. In this work, we have used a cheap and easily reproducible SERS substrate, constituted by a regular array of micrometric circles in which Au nanoparticles are randomly self-assembled by electroless deposition (see Materials and Method section for details). SERS spectra are recorded with a 633 nm excitation laser due to surface plasmon resonance effects which typically occur close to this wavelength for Au nanostructures,31 while standard Raman spectra were collected with an 830 nm laser source (in the infrared region) to avoid both exosomes photodamage and too large fluorescence effects. In order to investigate potential wavelength-dependent effects, standard Raman spectra have been measured with both 830 and 633 nm on some exosome samples (a typical result is shown in Figure S4). The comparison shows that major Raman peaks/bands are the same for both excitation wavelengths, but some minor spectral features can be better appreciated with the 830 nm laser source (mainly because of smaller fluorescence effects).
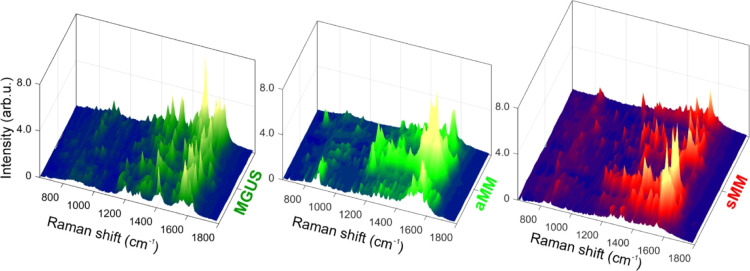
For each exosome sample, many spectra have been recorded by raster scanning over the microstructures. However, not all these spectra can be considered SERS-enhanced due to their low intensities. This behavior is well expected because of the random distribution of Au nanoparticles within each microstructure. After recording some hundreds of spectra for each exosome sample, only those with high intensities have been considered as SERS spectra in the following analysis. In Figure 4 all selected SERS spectra have been arranged one after the other and shown in a “landscape” fashion for MGUS, aMM, and sMM samples. Within each one of these three classes, we can observe a large variability of spectral features from one spectrum to the other. Indeed, Figure 4 shows three irregular landscapes, where SERS sharp peaks are not uniformly repeated across the spectra of the same sample. It is worth noticing that multivariate approaches, such as PCA and clustering analysis, did not produce satisfactory results on the SERS dataset.
Figure 4.
SERS spectra large variability. SERS efficiency and enhancement is strongly dependent upon the probed position over Au nanoparticles because of the nanoscale size of hot spots. This in turn negatively affects SERS reproducibility, even within the same experimental session. Several SERS spectra are reported close to one another, in the shape of a 3D landscape, for MGUS-, aMM-, and sMM-derived exosomes (at least 20 spectra for each group are chosen among the most intense ones). This representation shows at once the large differences occurring in SERS measurements.
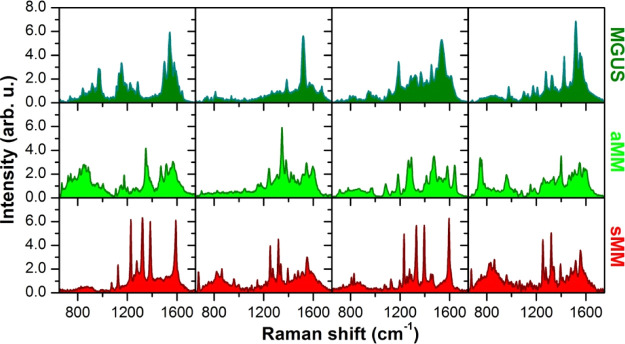
In order to deepen SERS analysis, we have restricted the problem by choosing four SERS spectra for each exosome class. Figure 5 shows the comparison between the SERS spectra with the highest intensities for MGUS, aMM, and sMM exosome samples. It is worthy to remind that SERS active modes can be quite different from the Raman ones because selection rules can be modified when the molecules are very close to the SERS nanostructures.32,33 Within each group, some major peaks are recurring in the four spectra. More in detail, the SERS curves of MGUS exosomes exhibit notable peaks at about 970, 1200, 1520, and 1535 cm–1. Also, aMM curves have peaks at about 1180, 1345, 1400, and 1580 cm–1. Finally, spectra from sMM exosomes show major peaks at about 1125, 1325, 1395, 1560, and 1590 cm–1. Except for these peaks that are recurring in more than one spectrum within each patient group, the four spectra chosen for each group do not exhibit a similar profile to each other. Also, there is a significant difference of intensity even for the peaks just mentioned above. This signal variability, which has to be expected in the case of SERS from disordered plasmonic nanoparticles, makes the SERS analysis quite difficult, and as a consequence, this SERS approach (based on random nanostructures) does not result in a robust protocol potentially suitable for clinical exploitation.
Figure 5.
Selected SERS spectra. Because of large variability in SERS results, four spectra of each group are selected and displayed for comparison. The chosen spectra are among those with the highest intensities so that SERS enhancement is ensured.
Conclusions
Despite remarkable progress in MM treatment, this malignancy still remains incurable.8 When considering the natural history of the disease, the development of a tool to monitor MM progression since its early phases is an attractive and fascinating challenge for the modern hemato-oncological community. The discovery of the EX role in tumor progression and their use as potential biomarkers has aroused huge interest in the field. Indeed, EX detection and interpretation of their complex content from the blood stream might be revolutionary for the patient’s stratification, therapy, and outcome. However, the lack of sturdy and standardized methods for characterizing them has limited their clinical use so far.
In this work, we have shown the potential use of EX screening, by means of Raman spectroscopy, to discriminate the patients across the three different clinical conditions: MGUS, aMM, and sMM. The combination of Raman spectroscopy with the adopted multivariate analysis (PCA) has successfully stratified patients belonging to these three groups. Interestingly, while sMM patients are clearly separated from aMM and MGUS, the latter groups present more similar although still distinct patterns. These data, once validated on larger cohorts, open to the design of an EX screening-based follow-up protocol for MGUS individuals to identify patients at risk of progression to overt MM. To our knowledge, this is the first time that Raman analysis is used to provide a stratification of MM patients using EXs from peripheral blood collection. Further improvements of the presented approach could potentially lead to a novel assay for the stratification of MGUS patients according to their risk to progress to MM. In turn, this achievement would reduce the need for multiple hospital access and allow to focus on high-risk patients that require closer follow-ups.
Finally, SERS devices based on random Au nanostructures have been used for EX analysis. SERS results have shown a much higher sensitivity, as revealed by the larger spectral intensities and sharpness of SERS peaks, but a too poor reproducibility in spectra collection. This limit is surely due to the intrinsic randomness of Au nanoparticle distribution over the SERS substrate: different spatial arrangements as well as different sizes of nanoparticles produce disparate enhancing factors, and in turn unequal SERS spectra. In order to avoid this disadvantage and to move toward a truly quantitative SERS analytical tool, the plasmonic nanostructures used for SERS substrates must fulfill very strict requirements in terms of sizes, uniformity, and spatial arrangement. Much more can be done and achieved with very regular and controlled SERS substrates, such as SERS devices produced by nano-lithographic techniques. Typically, lithographic protocols like the ones used for nano-electronics (e.g., electron beam lithography) are very well suited for this purpose, but conversely they are quite expensive and not easily scalable for mass production. These latter two drawbacks make SERS substrates fabricated by nano-lithography unaffordable nowadays for the development of widespread bioclinical assays. One compromise could be the investigation and development of unusual lithographic techniques, such as nanosphere lithography34 and template stripping replication,35 which lead to uniform SERS substrates with both low cost and great potential for mass production.
Materials and Methods
Patients and Sample Collection
We obtained whole blood samples from 14 newly diagnosed sMM, 13 MGUS, and 4 aMM patients collected between September 2016 and June 2018 at the Medical Oncology Unit of University Magna Graecia of Catanzaro (Table S1). None of the patients received therapy before the collection of blood samples. Criteria for diagnosis, clinical staging, and risk stratification were assessed according to the International Myeloma Working Group guidelines. Patients provided written informed consent in accordance with the Declaration of Helsinki. Whole blood samples were collected in red-top Vacutainer tubes.
Serum and Exosome Isolation
Blood was kept at room temperature for 30 min, followed by centrifugation at 2000g for 10 min to ensure serum separation. The supernatant was centrifuged at 3000g for 5 min to pellet cells, debris, and platelets, and samples were stored at −80 °C in aliquots of 1.5 mL before use. EXs were isolated from frozen serum samples (1.5 mL) using the miRCURY Exosome Isolation Kit (Qiagen) according to the manufacturer’s instructions.
Nanoparticle Tracking Analysis
Nanoparticle tracking analysis (NTA) was performed using a NanoSight LM10-HS microscope equipped with NTA software v3.1 (NanoSight Ltd., UK). Each sample was diluted at 1:10.000 in sterile-filtered PBS (Sigma, USA). The particle movement was analyzed by NTA software. Only measurements with >1000 completed tracks were analyzed.
Flow Cytometry
Total exosome isolation preparations were characterized by flow cytometry using human CD63-coated beads (Thermo Fisher Scientific, Inc.) in order to isolate CD63-positive subpopulations of exosomes from total exosome isolations derived from serum samples. The isolated exosomes were stained for typical exosome markers such as CD63 and CD9 analyzed with the Attune NxT Flow Cytometer (Thermo Fisher Scientific, Inc.).
Raman Measurements, Spectra Pre-processing, and Multivariate Analysis
Raman measurements were performed with an InVia Raman microscope from Renishaw in a backscattered configuration. The light from an 830 nm laser source was focused through a long working-distance objective 50×/0.5 NA. The total power at the sample level was 150 mW/cm2, and an integration time of 10 s was used for all measurements in the spectral region between 750 and 1700 cm–1. Prior to each Raman sequence, the frequency shift was calibrated by using the strong 520 cm–1 Si peak as a reference signal.
The exosome samples obtained by isolation were resuspended in PBS 1×, and the solution was deposited on a CaF2 slide. The CaF2 substrate was chosen for its negligible Raman signal. A drop coating deposition Raman (DCDR) protocol was employed.36,37 More in detail, for each exosome solution, one drop (4.0 μL) was deposited on the CaF2 slide, and Raman measurements were performed after partial evaporation of the solvent, in order to increase the exosome concentration, but before the dry-state was reached. If needed, the drop coating and DCDR protocol were repeated several times until a satisfactory number of measurements were acquired for each solution. In our experiments, at least 10 Raman spectra were collected for each exosome sample.
All measured spectra underwent the same pre-processing steps. First of all, an extended multiplicative signal correction (EMSC)38 was applied to all spectra at once for removing undesired physical variations, such as differences between spectra due to reiteration of the DCDR protocol and/or to different samples concentrations. Subsequently, all spectra were normalized to the total area under the spectrum, and a 3rd order polynomial subtraction was performed to ensure the same baseline intensity for the whole dataset. After pre-processing, PCA was performed on the spectral collection to highlight small differences between the exosome groups. The first six PCs account for nearly 90% of the total spectral variation, while the first two PCs alone account for 60% of the data variance.
All pre-processing steps, as well as the PCA, were carried out using the free software package Raman Tool Set (freely available at http://ramantoolset.sourceforge.net).39
SERS Substrate Fabrication and SERS Measurements
Microstructured arrays containing gold nanoparticles were fabricated through a standard photolithography process in combination with Au electroless deposition. Si substrates were coated with a 1.0 μm thick S1813 photoresist by spinning and then exposed to UV light using a Karl-Suss mask aligner. A hexagonal array of 8.0 μm diameter circular holes with a pitch of 20 μm was produced in the resist layer. Subsequently, the circular holes were filled with gold nanoparticles by immersing the samples in a 5 mM aqueous solution of AuCl for 2 min at 50 °C. After rinsing, the resist mask was gently removed by acetone, thus releasing an array of microcircles filled with Au nanoparticles (Figure S3). Most of the Au nanoparticles have lateral dimensions well below 30 nm, and the electroless process ensures very small gaps between the particles. Both these conditions are ideal for achieving SERS hot spots due to localized surface plasmon resonances, and this fabrication process is very cost effective and well suited for mass production of SERS devices.
SERS measurements were performed by depositing 2.0 μL drops of exosome solutions over the microstructured Au arrays. In SERS experiments, a 633 nm laser light was focused on the sample through a 100×/0.9 NA objective. The laser power was kept constant to 300 μW/cm2, while an integration time of 3.0 s was used for all samples in order to make the spectra comparable to each other as much as possible. Because of the diffraction limit of the light, the exact position of SERS hot spots (at the nanometer scale) cannot be inferred by imaging through the optical objective (spatial resolution of roughly half a micron). For this reason, SERS maps of several microstructured circles were recorded with a 100 nm stepsize in a xy raster way for each exosome sample. This approach produced a few thousands of potential SERS spectra for each exosome sample to be analyzed, but not all acquired spectra can be considered as SERS effective due to spatial variations of the enhancement factors within each microcircle.
Acknowledgments
L.T. has received funding from AIRC and from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No 800924. V.M. gratefully acknowledges financial support by Interregional Research Centre for Food Safety & Health (IRC_FSC) (PON a3-00359) by PON03PE_00078_1 and PON03PE_00078_2.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.0c03813.
Raman spectrum of exosome resuspension buffer; patients list; loading curve for rotated-PC2; peak assignment for rotated-PC2; electron microscopy pictures of SERS substrates; and comparison between standard Raman spectra recorded with 633 and 830 nm excitation sources (PDF)
Author Contributions
# M.R. and L.T. equally contributed to the present work. Conceptualization: M.R. and P.C.; Data curation: M.R. and P.C.; Formal analysis: M.R., L.T., C.R. and P.C.; Funding acquisition: G.P., V.M. and G.V.; Investigation: M.R., F.S., G.P. and N.M.; Methodology: M.R., F.S., M.L.C., C.R., N.M. and M.T.D.M.; Resources: G.P., V.M., S.G., P.T. and P.T.; Software: L.T., C.R. and P.C.; Supervision: M.R. and P.C.; Validation: M.R., L.T., M.R. and P.C.; Writing—original draft: M.R., L.T. and P.C.; Writing—review and editing: L.T., G.V. and M.R.
The authors declare no competing financial interest.
Supplementary Material
References
- Landtblom A. R.; Bower H.; Andersson T. M.-L.; Dickman P. W.; Samuelsson J.; Björkholm M.; Kristinsson S. Y.; Hultcrantz M. Second malignancies in patients with myeloproliferative neoplasms: a population-based cohort study of 9379 patients. Leukemia 2018, 32, 2203–2210. 10.1038/s41375-018-0027-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T.; Baughn L.; Kumar S.; Van Ness B. The future of myeloma precision medicine: integrating the compendium of known drug resistance mechanisms with emerging tumor profiling technologies. Leukemia 2019, 33, 863–883. 10.1038/s41375-018-0362-z. [DOI] [PubMed] [Google Scholar]
- Dispenzieri A.; Kyle R. A. Multiple myeloma: clinical features and indications for therapy. Best Pract. Res., Clin. Haematol. 2005, 18, 553–568. 10.1016/j.beha.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Kwizera E. A.; O’Connor R.; Vinduska V.; Williams M.; Butch E. R.; Snyder S. E.; Chen X.; Huang X. Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 2018, 8, 2722–2738. 10.7150/thno.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgren O.; Iskander K. Modern Multiple Myeloma Therapy: Deep, Sustained Treatment Response and Good Clinical Outcomes. J. Intern. Med. 2017, 281, 365–382. 10.1111/joim.12590. [DOI] [PubMed] [Google Scholar]
- Rajkumar S. V. Multiple myeloma: 2016 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2016, 91, 719–734. 10.1002/ajh.24402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloudizargari M.; Abdollahi M.; Asghari M. H.; Zimta A. A.; Neagoe I. B.; Nabavi S. M. The emerging role of exosomes in multiple myeloma. Blood Rev. 2019, 38, 100595. 10.1016/j.blre.2019.100595. [DOI] [PubMed] [Google Scholar]
- Moloudizargari M.; Asghari M. H.; Abdollahi M. Modifying exosome release in cancer therapy: How can it help?. Pharmacol. Res. 2018, 134, 246–256. 10.1016/j.phrs.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Vardaki I.; Sanchez C.; Fonseca P.; Olsson M.; Chioureas D.; Rassidakis G.; Ullén A.; Zhivotovsky B.; Björkholm M.; Panaretakis T. Caspase-3-dependent cleavage of Bcl-xL in the stroma exosomes is required for their uptake by hematological malignant cells. Blood 2016, 128, 2655–2665. 10.1182/blood-2016-05-715961. [DOI] [PubMed] [Google Scholar]
- Stremersch S.; Marro M.; Pinchasik B.-E.; Baatsen P.; Hendrix A.; De Smedt S. C.; Loza-Alvarez P.; Skirtach A. G.; Raemdonck K.; Braeckmans K. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016, 12, 3292–3301. 10.1002/smll.201600393. [DOI] [PubMed] [Google Scholar]
- Iaccino E.; Mimmi S.; Dattilo V.; Marino F.; Candeloro P.; Di Loria A.; Marimpietri D.; Pisano A.; Albano F.; Vecchio E.; Ceglia S.; Golino G.; Lupia A.; Fiume G.; Quinto I.; Scala G. Monitoring multiple myeloma by idiotype-specific peptide binders of tumor-derived exosomes. Mol. Canc. 2017, 16, 159. 10.1186/s12943-017-0730-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H.; Shao H.; Park Y. I.; Peterson V. M.; Castro C. M.; Weissleder R.; Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beljebbar A.; Bouché O.; Diébold M. D.; Guillou P. J.; Palot J. P.; Eudes D.; Manfait M. Identification of Raman spectroscopic markers for the characterization of normal and adenocarcinomatous colonic tissues. Crit. Rev. Oncol. Hematol. 2009, 72, 255–264. 10.1016/j.critrevonc.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Tirinato L.; Gentile F.; Di Mascolo D.; Coluccio M. L.; Das G.; Liberale C.; Pullano S. A.; Perozziello G.; Francardi M.; Accardo A.; De Angelis F.; Candeloro P.; Di Fabrizio E. SERS analysis on exosomes using super-hydrophobic surfaces. Microelectron. Eng. 2012, 97, 337–340. 10.1016/j.mee.2012.03.022. [DOI] [Google Scholar]
- Harder S. J.; Isabelle M.; DeVorkin L.; Smazynski J.; Beckham W.; Brolo A. G.; Lum J. J.; Jirasek A. Raman spectroscopy identifies radiation response in human non-small cell lung cancer xenografts. Sci. Rep. 2016, 6, 21006. 10.1038/srep21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhan A. R.; Jackman J. A.; Park J. H.; Cho N.-J.; Kim D.-H. Nanoplasmonic sensors for detecting circulating cancer biomarkers. Adv. Drug Delivery Rev. 2018, 125, 48–77. 10.1016/j.addr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Nie S.; Emory S. R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- Kneipp K.; Wang Y.; Kneipp H.; Perelman L. T.; Itzkan I.; Dasari R. R.; Feld M. S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667. 10.1103/physrevlett.78.1667. [DOI] [Google Scholar]
- Avella-Oliver M.; Puchades R.; Wachsmann-Hogiu S.; Maquieira A. Label-free SERS analysis of proteins and exosomes with large-scale substrates from recordable compact disks. Sens. Actuators, B 2017, 252, 657–662. 10.1016/j.snb.2017.06.058. [DOI] [Google Scholar]
- Kowal J.; Arras G.; Colombo M.; Jouve M.; Morath J. P.; Primdal-Bengtson B.; Dingli F.; Loew D.; Tkach M.; Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, E968–E977. 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.; Kaslan M.; Lee S. H.; Yao J.; Gao Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Volponi J. V.; Oliver A. E.; Parikh A. N.; Simmons B. A.; Singh S. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 3809–3814. 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czamara K.; Majzner K.; Pacia M. Z.; Kochan K.; Kaczor A.; Baranska M. Raman spectroscopy of lipids: a review. J. Raman Spectrosc. 2015, 46, 4–20. 10.1002/jrs.4607. [DOI] [Google Scholar]
- Gualerzi A.; Niada S.; Giannasi C.; Picciolini S.; Morasso C.; Vanna R.; Rossella V.; Masserini M.; Bedoni M.; Ciceri F.; Bernardo M. E.; Brini A. T.; Gramatica F. Raman spectroscopy uncovers biochemical tissue-related features of extracellular vesicles from mesenchymal stromal cells. Sci. Rep. 2017, 7, 9820. 10.1038/s41598-017-10448-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Mó M.; Siljander P. R. M.; Andreu Z.; Zavec A. B.; Borras F. E.; Buzas E. I.; Buzas K.; Casal E.; Cappello F.; Carvalho J.; Colas E.; Cordeiro-da Silva A.; Fais S.; Falcon-Perez J. M.; Ghobrial I. M.; Giebel B.; Gimona M.; Graner M.; Gursel I.; Gursel M.; Heegaard N. H. H.; Hendrix A.; Kierulf P.; Kokubun K.; Kosanovic M.; Kralj-Iglic V.; Kramer-Albers E. M.; Laitinen S.; Lasser C.; Lener T.; Ligeti E.; Line A.; Lipps G.; Llorente A.; Lotvall J.; Mancek-Keber M.; Marcilla A.; Mittelbrunn M.; Nazarenko I.; Nolte-t’ Hoen E. N. M.; Nyman T. A.; O’Driscoll L.; Olivan M.; Oliveira C.; Pallinger E.; del Portillo H. A.; Reventos J.; Rigau M.; Rohde E.; Sammar M.; Sanchez-Madrid F.; Santarem N.; Schallmoser K.; Ostenfeld M. S.; Stoorvogel W.; Stukelj R.; Van der Grein S. G.; Vasconcelos M. H.; Wauben M. H. M.; De Wever O. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M.; Carayon K.; Poirot M.; Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2014, 1841, 108–120. 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Xu F.; Yang C.-C.; Gomillion C.; Burg K. J. L. Effect of Ceramide on Mesenchymal Stem Cell Differentiation Toward Adipocytes. Appl. Biochem. Biotechnol. 2010, 160, 197–212. 10.1007/s12010-008-8505-8. [DOI] [PubMed] [Google Scholar]
- Kerr L. T.; Gubbins L.; Gorzel K. W.; Sharma S.; Kell M.; McCann A.; Hennelly B. M.. Raman spectroscopy and SERS analysis of ovarian tumour derived exosomes (TEXs): a preliminary study. Proceedings of SPIE: Photonic Solutions for Better Health Care IV; International Society for Optics and Photonics, 2014; p 9129.
- Zong S.; Wang L.; Chen C.; Lu J.; Zhu D.; Zhang Y.; Wang Z.; Cui Y. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. 10.1039/c6ay00406g. [DOI] [Google Scholar]
- Wang Z.; Zong S.; Wang Y.; Li N.; Li L.; Lu J.; Wang Z.; Chen B.; Cui Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. 10.1039/c7nr09162a. [DOI] [PubMed] [Google Scholar]
- Huang X.; El-Sayed M. A. Gold nanoparticles: Optical properties and implementations in cancer diagnosis and photothermal therapy. J. Adv. Res. 2010, 1, 13–28. 10.1016/j.jare.2010.02.002. [DOI] [Google Scholar]
- Moskovits M.; Suh J. S. Surface selection rules for surface-enhanced Raman spectroscopy: calculations and application to the surface-enhanced Raman spectrum of phthalazine on silver. J. Phys. Chem. 1984, 88, 5526–5530. 10.1021/j150667a013. [DOI] [Google Scholar]
- Smith E.; Dent G.. Modern Raman Spectroscopy: A Practical Approach, 1st ed.; John Wiley and Sons, 2005. [Google Scholar]
- Haynes C. L.; Van Duyne R. P. Nanosphere lithography: A versatile nanofabrication tool for studies of size-dependent nanoparticle optics. J. Phys. Chem. B 2001, 105, 5599–5611. 10.1021/jp010657m. [DOI] [Google Scholar]
- Candeloro P.; Iuele E.; Perozziello G.; Coluccio M. L.; Gentile F.; Malara N.; Mollace V.; Di Fabrizio E. Plasmonic nanoholes as SERS devices for biosensing applications: An easy route for nanostructures fabrication on glass substrates. Microelectron. Eng. 2017, 175, 30–33. 10.1016/j.mee.2016.12.015. [DOI] [Google Scholar]
- Zhang D.; Xie Y.; Mrozek M. F.; Ortiz C.; Davisson V. J.; Ben-Amotz D. Raman detection of proteomic analytes. Anal. Chem. 2003, 75, 5703–5709. 10.1021/ac0345087. [DOI] [PubMed] [Google Scholar]
- Zolea F.; Biamonte F.; Candeloro P.; Di Sanzo M.; Cozzi A.; Di Vito A.; Quaresima B.; Lobello N.; Trecroci F.; Di Fabrizio E.; Levi S.; Cuda G.; Costanzo F. H ferritin silencing induces protein misfolding in K562 cells: A Raman analysis. Free Radical Biol. Med. 2015, 89, 614–623. 10.1016/j.freeradbiomed.2015.07.161. [DOI] [PubMed] [Google Scholar]
- Martens H.; Nielsen J. P.; Engelsen S. B. Light scattering and light absorbance separated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 2003, 75, 394–404. 10.1021/ac020194w. [DOI] [PubMed] [Google Scholar]
- Candeloro P.; Grande E.; Raimondo R.; Di Mascolo D.; Gentile F.; Coluccio M. L.; Perozziello G.; Malara N.; Francardi M.; Di Fabrizio E. Raman database of amino acids solutions: a critical study of Extended Multiplicative Signal Correction. Analyst 2013, 138, 7331–7340. 10.1039/c3an01665j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.